Abstract
Although Escherichia coli has long been recognized as the best-understood living organism, little was known about its abilities to use aromatic compounds as sole carbon and energy sources. This review gives an extensive overview of the current knowledge of the catabolism of aromatic compounds by E. coli. After giving a general overview of the aromatic compounds that E. coli strains encounter and mineralize in the different habitats that they colonize, we provide an up-to-date status report on the genes and proteins involved in the catabolism of such compounds, namely, several aromatic acids (phenylacetic acid, 3- and 4-hydroxyphenylacetic acid, phenylpropionic acid, 3-hydroxyphenylpropionic acid, and 3-hydroxycinnamic acid) and amines (phenylethylamine, tyramine, and dopamine). Other enzymatic activities acting on aromatic compounds in E. coli are also reviewed and evaluated. The review also reflects the present impact of genomic research and how the analysis of the whole E. coli genome reveals novel aromatic catabolic functions. Moreover, evolutionary considerations derived from sequence comparisons between the aromatic catabolic clusters of E. coli and homologous clusters from an increasing number of bacteria are also discussed. The recent progress in the understanding of the fundamentals that govern the degradation of aromatic compounds in E. coli makes this bacterium a very useful model system to decipher biochemical, genetic, evolutionary, and ecological aspects of the catabolism of such compounds. In the last part of the review, we discuss strategies and concepts to metabolically engineer E. coli to suit specific needs for biodegradation and biotransformation of aromatics and we provide several examples based on selected studies. Finally, conclusions derived from this review may serve as a lead for future research and applications.
Next to glucosyl residues, the benzene ring is the most widely distributed unit of chemical structure in nature (59, 132). The complex aromatic polymer lignin comprises about 25% of the land-based biomass on Earth, and the recycling of this and other plant-derived aromatic compounds is vital for maintaining the Earth's carbon cycle. The degradation of such chemicals is accomplished mainly by microorganisms (129, 321), and in recent years there has been considerable interest in exploring their ability to degrade and detoxify the increasing amounts of aromatic compounds which enter the environment as by-products of many industrial processes (239). Although many genera of microorganisms degrade aromatic compounds other than aromatic amino acids, with Pseudomonas being the most extensively analyzed (132, 335), this ability has been only occasionally studied in enteric bacteria. The early literature contains reports on the formation of phenol and p-cresol by Escherichia coli bacterial cultures growing in natural media, peptone and casein media, and in chemically defined media containing l-tyrosine or p-hydroxybenzoic acid (316). The decarboxylation of substituted cinnamic acids with the production of volatile phenolic compounds that provide phenolic flavors in fermented beverages has been reported for many enterobacteria of the genera Klebsiella, Enterobacter, and Hafnia, as well as for Escherichia intermedia (179). The use of benzoic acid, p-hydroxybenzoic acid, and phenylacetic acid (PA) by Enterobacter aerogenes as the sole carbon and energy source was one of the first reports of the catabolism of aromatic acids by an enterobacterium (119). In the mid-1970s, Chapman and coworkers found that most enterics could use aromatic compounds, with the ability to utilize hydroxyphenylacetic acid (HPA) being the most widespread (38). It was the work of Cooper and Skinner in 1980 on the ability of E. coli to mineralize 3- and 4-hydroxyphenylacetic acids (3HPA and 4HPA) that delineated for the first time a complete catabolic pathway in enteric bacteria (50). Later, Burlingame and Chapman (36) reported that many laboratory strains and clinical isolates of E. coli can catabolize various aromatic acids. A historical perspective on the mineralization of aromatic compounds by E. coli is summarized in Table 1.
TABLE 1.
Historical perspective on the mineralization of aromatic compounds by E. coli
| Yr | Author(s) and location | Eventa |
|---|---|---|
| 1980 | R. A. Cooper and M. A. Skinner (Leicester) | First report on a pathway for the catabolism of aromatic acids in E. coli (mineralization of 3HPA and 4HPA) |
| 1983 | R. P. Burlingame and P. J. Chapman (Minnesota) | E. coli mineralizes a variety of aromatic acids (PA, HPA, PP, 3HPP, 3HCI); biochemical characterization of the PP and 3HPP catabolic pathway. |
| 1985 | R. A. Cooper, D. C. Jones, and S. Parrot (Leicester) | Isolation of the first E. coli mutants defective in catabolism of aromatic acids (PA− mutants) |
| 1986 | R. P. Burlingame, L. Wyman, and P. J. Chapman (Minnesota) | Isolation and characterization of E. coli mutants defective in PP and 3HPP degradation |
| 1987 | S. Parrot, S. Jones, and R. A. Cooper (Leicester) | First report on the catabolism of aromatic amines in E. coli (PEA can be used as nitrogen and carbon source) |
| 1987 | N. Abdulrashid and D. P. Clark (Illinois) | Degradation of furans and thiophenes by E. coli mutants |
| 1988 | J. R. Jenkins and R. A. Cooper (Leicester) | Molecular cloning of the meta cleavage route (hpc genes) involved in HPC degradation |
| 1990 | D. I. Roper and R. A. Cooper (Leicester) | First nucleotide sequence of a gene (hpcD) involved in mineralization of aromatic compounds (HPA) in E. coli |
| 1993 | D. I. Roper, T. Fawcett, and R. A. Cooper (Leicester) | First nucleotide sequence of a regulatory gene (hpcR) that controls the expression of an aromatic catabolic operon (hpc) in E. coli |
| 1994 | M. A. Prieto and J. L. García (Madrid) | Characterization of the HpaBC monooxygenase from E. coli; first primary structure of a TC-FDM |
| 1996 | M. A. Prieto, E. Díaz, and J. L. García (Madrid) | Molecular characterization of a complete aromatic catabolic pathway from E. coli (hpa cluster) and its use to construct a mobile degradative cassette; expansion of the degradative abilities of P. putida by expressing some hpa genes |
| 1997 | M. A. Prieto and J. L. García (Madrid) | Characterization of the first gene (hpaX) responsible of the transport of an aromatic acid (HPA) in E. coli |
| 1997 | A. Ferrández, J. L. García, and E. Díaz (Madrid) | Heterologous expression of a complete aromatic catabolic pathway (mhp) from E. coli to improve the catabolic abilities of different environmentally relevant bacteria |
| 1998 | E. Díaz, A. Ferrández, and J. L. García (Madrid) | Molecular characterization of the hca cluster encoding a multicomponent aromatic initial dioxygenase in E. coli |
| 1998 | A. Ferrández, B. Miñambres, B. García, E. R. Olivera, J. M. Luengo, J. L. García, and E. Díaz (Madrid) | Molecular characterization of an aerobic hybrid pathway for the catabolism of PA in E. coli |
Abbreviations: HPA, hydroxyphenylacetic acid; HPC, homoprotocatechuate; 3HPP, 3-hydroxyphenylpropionic acid; 3HCI, 3-hydroxycinnamic acid; PA, phenylacetic acid; PEA, 2-phenylethylamine; PP, phenylpropionic acid; TC-FDM, two-component nonheme flavin-diffusible monooxygenase.
At first view, the ability of E. coli to degrade aromatic compounds appears to be an unexpected finding since this feature has always been associated with typical soil bacteria and E. coli is mainly regarded as an inhabitant of the animal gut. However, when one analyzes the ecology of E. coli, it becomes clear that this bacterium may easily encounter aromatic compounds in both the intestinal and extraintestinal habitats that it colonizes, which explains its catabolic potential to use such compounds as carbon and energy source.
The Two Habitats of E. coli
The intestine of warm-blooded animals, the primary habitat of E. coli, contains some 400 to 500 different bacterial species, with E. coli being the most abundant (making up about 1% of the total fecal bacterial flora) (233). The success of E. coli in the gut ecosystem is thought to reflect its abilities to occupy different ecological niches. Thus, since E. coli grows both anaerobically and aerobically, it is able to colonize intestinal habitats in which oxygen offers some ecological advantage. Such habitats could be ones in close proximity to epithelial cells, where oxygen molecules might pass from the blood through the epithelium to the microbes attached to it. By assimilating such molecules, E. coli may be important in developing and maintaining the oxygen-free conditions and low oxidation-reduction potential favoring strict anaerobes in the large intestine (280). While embedded within the mucus layer overlying intestinal epithelial cells, E. coli grows with a generation time of 40 to 80 min (245, 246). In contrast, the population of E. coli cells in the cecal luminal contents are essentially static with respect to growth and are excreted in the feces (246).
Although E. coli is a highly successful commensal of the intestines of warm-blooded animals as well as a pathogen of the enteric, urinary, pulmonary, and nervous systems, this facultative anaerobe must survive and grow outside the animal host to effect successful interhost spread. E. coli is not famous for extracorporeal existence, but it nonetheless shares with its soil-inhabiting relatives such as Klebsiella species the ability to thrive under a wide range of various physical and chemical conditions, including adaptation for survival over long periods of nongrowth (210). Soil, water, sediment, and perhaps food are other habitats of E. coli, and the bacterium might spend comparable times in each of its two main habitats (88, 281). While pollution from human sources may be the most important source of E. coli in the environment (29, 260), the fact that this bacterium was found in pristine tropical waters, where it remained physiologically active and grew at rates dependent on nutrient levels, suggests that it can be a natural inhabitant in these environments and that it may be part of a previously established community (22). E. coli can also replicate and survive in soil protozoa. Since protozoa are widely distributed in soils and effluents, they may also constitute an environmental reservoir for transmission of this enterobacterium (17). Thus, unlike host-specific or obligate parasites, E. coli is a highly adaptable microorganism with an extensive repertoire of metabolic and regulatory genes that facilitate the colonization of widely different environments (78).
Sources of Aromatic Compounds in the E. coli Life Cycle
As stated above, aromatic compounds are highly abundant in soil and water, and therefore it is obvious that they can constitute a normal carbon source for E. coli when this bacterium reaches its extraintestinal habitat. Although it is still not known which substrates E. coli grows on in the large intestine and which pathways provide it with the metabolic advantage necessary for it to compete with the hundreds of other bacteria with which it shares this habitat, it is likely that aromatic compounds can also be a frequent carbon source for E. coli in the animal gut. Aromatic amino acids and plant constituents are the major sources of aromatic compounds in the gastrointestinal tract. Minor sources of aromatic compounds in the human gut include certain steroids and some drugs and food constituents (additives, colorants, and contaminants) (117). Estimates suggest that between 3 and 25 g of protein and peptides enters the large bowel every day from the diet, as well as from endogenous sources such as host tissues, bacterial debris, pancreatic enzymes, and other secretions (296). In the large gut, these substances are depolymerized by a mixture of residual pancreatic endopeptidases and bacterial proteases and peptidases. The resulting short peptides and amino acids then become available for fermentation by many intestinal anaerobes such as Bacteroides, Lactobacillus, Bifidobacterium, Clostridium, and Streptococcus species, generating a wide range of phenolic and indolic compounds in a series of deamination, transamination, decarboxylation and dehydrogenation reactions. Phenol, p-cresol, HPA, hydroxyphenylpropionic acid (HPP), and hydroxybenzoic acid are the principal products of tyrosine fermentation in the human large intestine, while PA, phenylpropionic acid (PP), and benzoic acid are produced from phenylalanine. PP may, however, also be formed from tyrosine (296, 320). Phenylethylamine (PEA) and tyramine are produced from decarboxylation of phenyalanine and tyrosine, respectively (320). Since the production of these aromatic compounds was inhibited in the presence of a readily fermentable source of carbohydrate, carbohydrate availability may be a critical factor affecting aromatic amino acid fermentation in the large intestine (296). The aromatic compounds generated by the intestinal anaerobes meet a variety of fates in the body. Thus, they may be detoxified by glucuronide or sulfate conjugation or may remain unabsorbed and be voided in the feces and urine. They can also completely break down under local aerobic conditions in the large intestine as a result of the action of some facultative anaerobes such as E. coli, such that mono- and dioxygenases are able to incorporate molecular oxygen into the aromatic ring (296, 320). A second major source of aromatic compounds in the animal gut involves the dietary plant constituents such as ferulic and caffeic acid, which result in HPP acids (235), as well as different flavonoid glycosides that are ingested in daily quantities of 1 to 2 g by humans. A collaborative bacterial catabolism, i.e., syntrophic interactions among microorganisms, of flavonoids in the human gut has been suggested. Thus, a number of obligately anaerobic bacteria from the human intestinal flora, e.g., Bacteroides species, are capable of cleaving the glycosidic bond of flavonoids, generating the corresponding aglycones such as quercetin, kaempferol, naringenin, and catechin (338). These aglycones are then subject to ring cleavage by different bacteria, e.g., Clostridium and Eubacterium species, giving rise to a variety of aromatic acids, such as 4HPA (from kaempferol), 3,4-dihydroxyphenylacetic acid (from quercetin), PA (from naringenin), and HPP (from tricetin and tricin) (121, 285, 338), that can be utilized by other intestinal bacteria such as E. coli.
Intraspecies Variation in E. coli and the Catabolism of Aromatic Compounds
Phylogenetic analyses have shown that E. coli strains fall into four main phylogenetic groups (A, B1, B2, and D), where virulent extraintestinal strains belong mainly to groups B2 and D, whereas most commensal strains belong to group A (46). E. coli K-12 (group A) is by far the most extensively studied E. coli strain, and it represents the best-understood living organism at the biochemical and genetic levels. The complete sequences of the 4.6-Mb genome of two E. coli K-12 derivatives, MG1655 (27; EcoGene database accessible using the Colibri website, http://www.genolist.pasteur.fr/Colibri/) and W3110 (202; GenoBase database accessible at the website http://ecoli.aist-nara.ac.jp/), have been reported, and functional genomic analyses are being performed (291). The wild-type strain of E. coli K-12 was isolated from the feces of a convalescent diphtheria patient in 1922 at Stanford University, and subcultures and derivatives of this strain were first reported in 1944 (120). Since the K-12 strains are unable to colonize the human gut (297), the K-12 lineage is considered to be the prototype of a biologically safe vehicle for the propagation of many efficient gene-cloning and expression systems. However, the presence of K-12 strains among E. coli isolates is extremly low, since no K-12 strains were detected among 226 environmental and human stool isolated samples (29). Other E. coli laboratory strains not derived from K-12 are, for instance, E. coli C, B, and W. E. coli C (no. 122 of the National Collection of Type Cultures, London, United Kingdom) is a prototroph F− strain (24) and was one of the first strains shown to be able to recombine (acting as a recipient) with K-12 (177). E. coli B is a wild-type E. coli strain (67). A mutant of strain B that is resistant to radiations, E. coli B/r, has been frequently used in laboratory studies (339). The W (or Waksman) wild-type strain of E. coli (ATCC 9637) was apparently isolated from the soil of a graveyard by S. A. Waksman (E. Ron, personal communication). A vitamin B12 auxotroph derivative of the W wild-type strain, E. coli ATCC 11105 (hereafter referred as E. coli W) (63), is a well-known penicillin G acylase producer. During the writing of this review, the 5.4-Mb genome of the enterohemorrhagic E. coli O157:H7 strain (group D) was reported (236). The genomes of E. coli K-12 and O157:H7, two strains that last had a common ancestor about 4.5 million years ago, revealed an unexpectedly complex segmented relationship (236). This diversity within the E. coli species is reflected in significant differences in gene content among different strains whose chromosome size ranges from 4.5 to 5.5 Mb (214, 262). Genes found in most individuals, that is, the core set of genes for that species (core gene pool), are the genes that determine those properties characteristic of all members of the species. Additionally, each strain has auxiliary genes distributed throughout the genome (flexible gene pool), e.g., pathogenicity islands and metabolic pathway genes, which determine properties found in some but not all members of the species and that contribute to maintaining the dynamic gene pool in E. coli (78, 125, 171, 236, 262). In this sense, the E. coli intraspecies variation in the ability to use different aromatic acids as sole carbon and energy sources (36) (Table 2) is a clear example that E. coli strains possess different capacities for utilizing growth-limiting nutrients and that they are likely to be used to increase the fitness and to expand the ecological niches of individual E. coli cells. The work of Burlingame and Chapman (36) revealed that all seven laboratory strains tested grew using PP, 3HPP, or 3-hydroxycinnamic acid (3HCI) as the sole carbon and energy source. However, while E. coli W is also able to grow on PA and 4HPA or 3HPA, E. coli K-12 and E. coli NCTC 5928 grew on PA but not on 4HPA or 3HPA and E. coli B and E. coli C grew on 4HPA or 3HPA but not on PA (Table 2). None of the strains tested was able to grow on 2HPA, cinnamic acid (CI) or its 2- or 4-hydroxy derivatives, or with the 2- or 4-hydroxy derivatives of PP (36). Among the clinical isolates analyzed, 48 (72%) of 67 could grow on at least one of the six aromatic acids tested and 27 (40%) could use all six (Table 2). Strains that could grow on 4HPA could also grow on 3HPA, and those that grew on 3HPP also grew on 3HCI, consistent with a common catabolic pathway for each of these two pair of compounds (see below).
TABLE 2.
Aromatic acids degraded by different E. coli strainsa
| Strain | No. examined | Aromatic acid utilized as sole carbon and energy sourceb:
|
||||
|---|---|---|---|---|---|---|
| PA | HPA | PP | 3HPP | 3HCI | ||
| B | 1 | − | + | + | + | + |
| C | 1 | − | + | + | + | + |
| K-12 | 2 | +c | − | + | + | + |
| W | 2 | + | + | + | + | + |
| NCTC 5928 | 1 | + | − | + | + | + |
| Clinical isolates | 19 | − | − | − | − | − |
| 5 | + | − | − | − | − | |
| 2 | − | − | − | + | + | |
| 8 | − | − | + | + | + | |
| 1 | − | + | − | − | − | |
| 2 | + | + | − | − | − | |
| 3 | + | − | + | + | + | |
| 27 | + | + | + | + | + | |
Modified from reference 36.
Abbreviations: HPA, 3- and 4-hydroxyphenylacetic acid; 3HCI, 3-hydroxycinnamic acid; 3HPP, 3-hydroxyphenylpropionic acid; PA, phenylacetic acid; PP, phenylpropionic acid.
Some E. coli K-12 derivatives such as DH5α, HB101, JM109, CC118, and DH1 do not grow on PA.
The aim of this article is to review the genetics and biochemistry of the catabolism of aromatic compounds in E. coli. Our understanding of the utilization of these compounds by E. coli has leapt forward in recent years with the genetic characterization of the cognate catabolic pathways. Additional information has become available through the sequencing of the E. coli genome. Homologues of many of the E. coli genes involved in the catabolism of aromatic compounds can be identified on other bacterial genomes, some of which have been completely sequenced at the time of writing. Where possible, data derived from these genomes have been also included in this article and some evolutionary considerations have been pointed out. Finally, the use of E. coli as a biocatalyst for biotransformation or biodegradation of aromatic compounds will be addressed. Conclusions derived from this review may provide useful starting points for future research.
GENERAL ORGANIZATION OF THE GENE CLUSTERS FOR THE CATABOLISM OF AROMATIC ACIDS AND AMINES IN E. COLI
As indicated in the Introduction, E. coli is able to mineralize several aromatic compounds (Table 2). In this section, the general organization of the gene clusters for the degradation of 4HPA, 3HPA, 3HPP, 3HCI, PP, PA, and some aromatic amines is revised.
4HPA/3HPA Catabolic Pathway
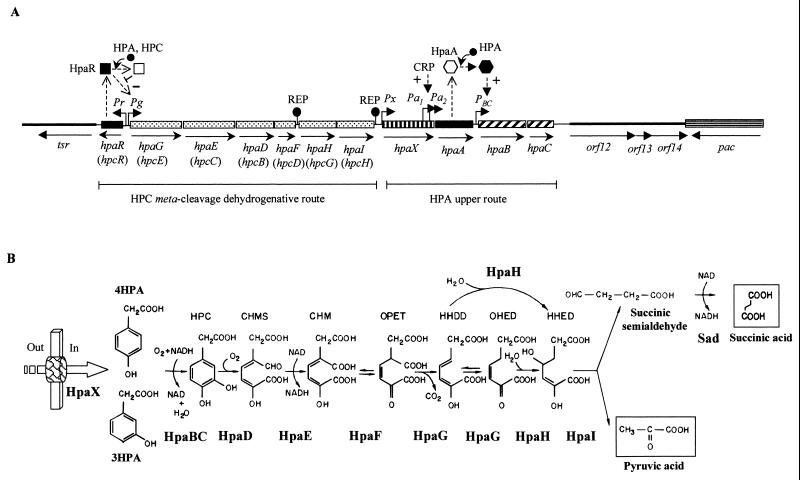
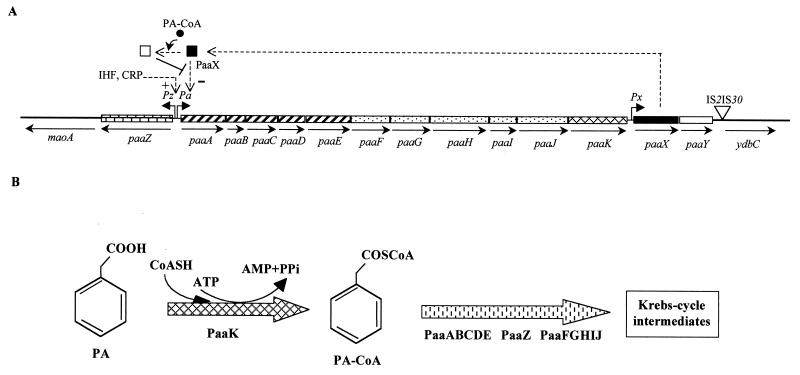
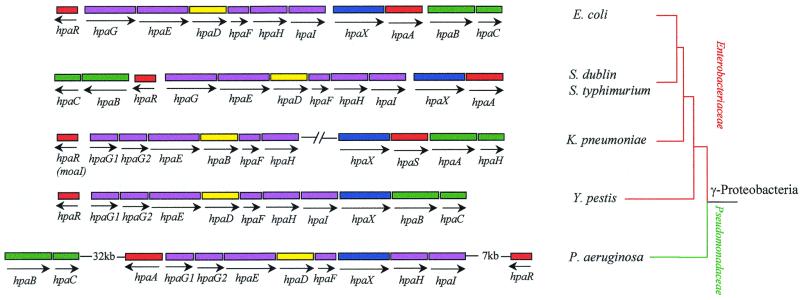
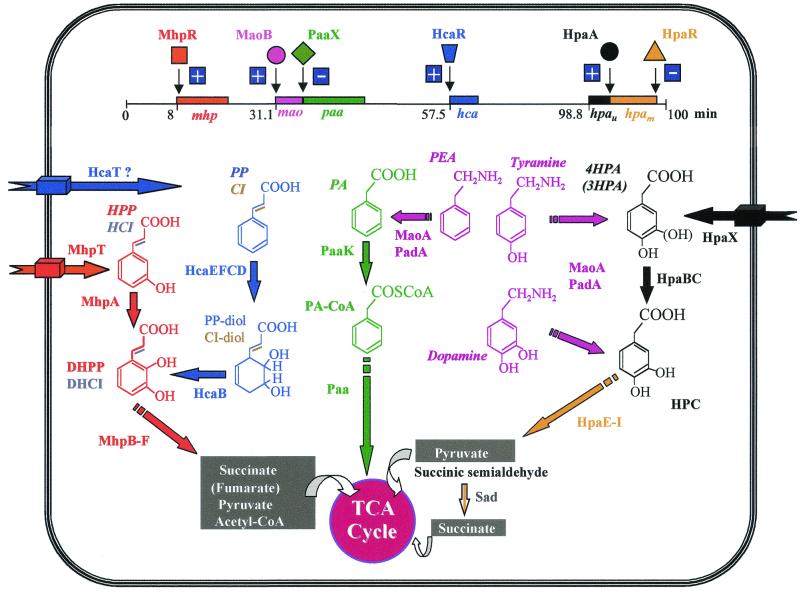
Some E. coli laboratory strains such as E. coli B, C, and W, but not E. coli K-12, are able to degrade 4HPA, 3HPA, and homoprotocatechuate (3,4-dihydroxyphenylacetate [HPC]) via an inducible chromosomally encoded meta-cleavage pathway (36, 50). The genes encoding the HPC meta-cleavage degradative route from E. coli C (hpc cluster) and E. coli W (hpa cluster) have been cloned and sequenced (143, 250, 270). The upper hpa gene cluster encoding the enzymes responsible for the hydroxylation of 4HPA and 3HPA to the catecholic intermediate HPC in E. coli W has also been cloned and sequenced, and it is located immediately adjacent to the HPC meta-cleavage genes (Fig. 1A). The homologous upper hpa cluster of E. coli C has also been cloned and partially sequenced (251, 254). In addition, near the hpa cluster E. coli W contains the pac gene (Fig. 1A), which encodes penicillin G acylase, an enzyme able to hydrolyze a wide range of amides and esters of HPA and PA (251, 254) (see below). The biochemical pathway for the catabolism of HPC in E. coli W (250) is similar to that previously delineated in E. coli C (50, 143, 270) (Fig. 1B). The HPC dehydrogenative route in E. coli W involves meta-cleavage of HPC by HPC 2,3-dioxygenase (HpaD), to give 5-carboxymethyl-2-hydroxymuconic semialdehyde (CHMS), which undergoes dehydrogenation to 5-carboxymethyl-2-hydroxymuconic acid (CHM) by the action of the CHMS dehydrogenase (HpaE). CHM isomerizes to 5-oxo-pent-3-ene-1,2,5-tricarboxylic acid (OPET) through a CHM isomerase (HpaF), and the OPET undergoes decarboxylation to 2-hydroxy-hept-2,4-diene-1,7-dioic acid (HHDD) by the HpaG decarboxylase. The formation of 2,4-dihydroxy-hept-2-ene-1,7-dioic acid (HHED) through the action of the HpaH hydratase on the product of the HpaG-catalyzed reaction is followed by its HpaI-mediated cleavage to give pyruvate and succinic semialdehyde as final products (Fig. 1B). Although pyruvate is a central metabolite, succinic semialdehyde requires previous dehydrogenation to succinic acid to enter the Krebs cycle, and this enzymatic step is not encoded within the hpa cluster (see below).
FIG. 1.
Pathway for the catabolism of HPA (4HPA and 3HPA) in E. coli. (A) Genetic map of the chromosomal hpa (in E. coli W) and hpc (in E. coli C) regions. Relevant genes are indicated by blocks: genes with similar shading participate in the same enzymatic step or in the same functional unit (route) of the pathway. The hpc genes are indicated in brackets. Regulatory and transport genes are shown by solid and vertically striped blocks, respectively. The genes flanking the hpa cluster (tsr, orf12, orf13, and orf14) are contiguous in the genome of E. coli K-12 and are represented by thick lines. orf12 and orf13 correspond to the yjiY gene from E. coli K-12. orf14 corresponds to the yjiA gene from E. coli K-12. The arrows show the directions of gene transcription. Bent arrows represent the Pr, Pg, Px, Pa1, Pa2 and PBC promoters. REP sequences are shown. The HpaR repressor and HpaA activator are represented by a square and hexagon, respectively; empty and solid symbols indicate inactive and active regulators, respectively; − and + indicate transcriptional repression and activation, respectively. The inducer molecule (HPA and HPC) is represented by a solid circle. (B) Biochemistry of the HPA catabolic pathway. The metabolites are 4HPA and 3HPA, HPC (homoprotocatechuate), CHMS (5-carboxymethyl-2-hydroxy-muconic semialdehyde), CHM (5-carboxymethyl-2-hydroxy-muconic acid), OPET (5-oxo-pent-3-ene-1,2,5-tricarboxylic acid), HHDD (2-hydroxy-hept-2,4-diene-1,7-dioic acid), OHED (2-oxo-hept-3-ene-1,7-dioic acid), and HHED (2,4-dihydroxy-hept-2-ene-1,7-dioic acid). The enzymes are HpaBC (HPA monooxygenase), HpaD (HPC 2,3-dioxygenase), HpaE (CHMS dehydrogenase), HpaF (CHM isomerase), HpaG (OPET decarboxylase), HpaH (hydratase), HpaI (HHED aldolase), and Sad (succinic semialdehyde dehydrogenase). The HPA transport protein (HpaX) is represented by a thick arrow. Out and In indicate outside and inside the cell, respectively.
The hpa catabolic cluster of E. coli W (Fig. 1A) is composed of 11 genes arranged as follows: (i) eight enzyme-encoding genes organized in two putative operons, the 4HPA/3HPA hydroxylase operon (hpaBC) and the HPC meta-cleavage operon (hpaGEDFHI) homologous to the hpcECBDGH operon from E. coli C; (ii) two regulatory genes, hpaR (hpcR in E. coli C) and hpaA, that control the expression of the meta-cleavage and HPA hydroxylase operons, respectively; and (iii) the hpaX gene, encoding the 4HPA/3HPA transport protein. All the genes are transcribed in the same direction with the sole exception of hpaR (250) (Fig. 1A). Analysis of the intergenic regions revealed the presence of five REP (repetitive extragenic palindromic) or PU (palindromic unit) sequences (16). While two REP sequences are located in the largest intercistronic region of the meta-cleavage operon between the hpaF and hpaH genes, the other three REP sequences are found at the 3′ end of such operon and might act as transcription termination signals (Fig. 1A). Two putative hairpin loops that might act as transcriptional terminators are also located downstream of the hpaA gene and hpaBC operon (250).
Analysis of the regions flanking the hpa cluster from E. coli W showed the presence of several open reading frames (orf genes) (250) (Fig. 1A). Downstream of hpaR is located an incomplete orf that corresponds to the tsr gene mapped in the chromosome at 98.9 min (hereafter, chromosomal mapping in E. coli refers to that of E. coli K-12 strain MG1655 [EcoGene database at the Colibri website]) and that encodes the serine chemoreceptor (6). At the other end of the hpa cluster, that is, downstream of the hpaC gene, there are several orf genes that correspond to the yjiY (orf12 and orf13) and yjiA (orf14) genes of E. coli K-12 (EcoGene database at the Colibri website) (Fig. 1A).
3HPP/3HCI Catabolic Pathway
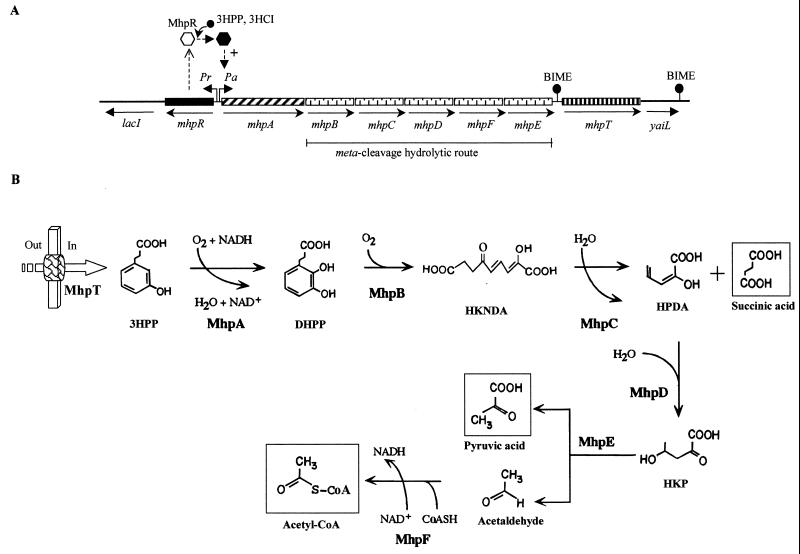
Most E. coli strains are able to degrade 3HPP via a meta-fission pathway (36) (Table 2), and several mutants defective in this catabolism have been isolated and characterized (39). The biochemical pathway for the catabolism of 3HPP in E. coli K-12 (36, 39) is shown in Fig. 2B. The first step is catalyzed by the MhpA hydroxylase, which inserts one atom of molecular oxygen into the 2 position of the phenyl ring of 3HPP to give 2,3-dihydroxyphenylpropionate (DHPP), which, when acted on by 2,3-dihydroxyphenylpropionate 1,2 dioxygenase (MhpB), undergoes an extradiol cleavage, yielding the meta ring fission product, 2-hydroxy-6-keto-nona-2,4-diene 1,9 dioic acid (HKNDA). HKNDA is cleaved by the MhpC hydrolase to give succinate and 2-hydroxy-penta-2,4-dienoic acid (HPDA), which is hydrated to 4-hydroxy-2-ketopentanoic acid (HKP) by the MhpD hydratase. Then the MhpE aldolase catalyzes the fission of HKP to give pyruvate and acetaldehyde, with the latter being converted to acetyl coenzyme A (acetyl-CoA) through the action of the MhpF acetaldehyde dehydrogenase (Fig. 2B).
FIG. 2.
Pathway for the catabolism of 3HPP in E. coli. (A) Genetic map of the chromosomal mhp cluster. Relevant genes are indicated by blocks: genes with similar shading participate in the same enzymatic step or in the same functional unit (route) of the pathway. Regulatory and transport genes are shown by solid and vertically striped blocks, respectively. Genes flanking the mhp cluster (lacI and yaiL) are represented by thick lines. The arrows show the directions of gene transcription. Bent arrows represent the Pr and Pa promoters. The location of the BIME is shown. The inactive and active forms of the MhpR activator are represented by empty and solid hexagons, respectively. + indicates transcriptional activation. The inducer molecule (3HPP and 3HCI) is represented by a solid circle. (B) Biochemistry of the 3HPP catabolic pathway. The metabolites are 3HPP, DHPP (2,3-dihydroxyphenlypropionic acid), HKNDA (2-hydroxy-6-keto-nona-2,4-diene 1,9-dioic acid), HPDA (2-hydroxy-penta-2,4-dienoic acid), and HKP (4-hydroxy-2-ketopentanoic acid). The enzymes are MhpA (3HPP monooxygenase), MhpB, (DHPP 1,2 dioxygenase), MhpC (HKNDA hydrolase), MhpD (HPDA hydratase), MhpE (HKP aldolase), and MhpF (acetaldehyde dehydrogenase [acylating]). The 3HPP transport protein (MhpT) is represented by a thick arrow. Out and In indicate outside and inside the cell, respectively.
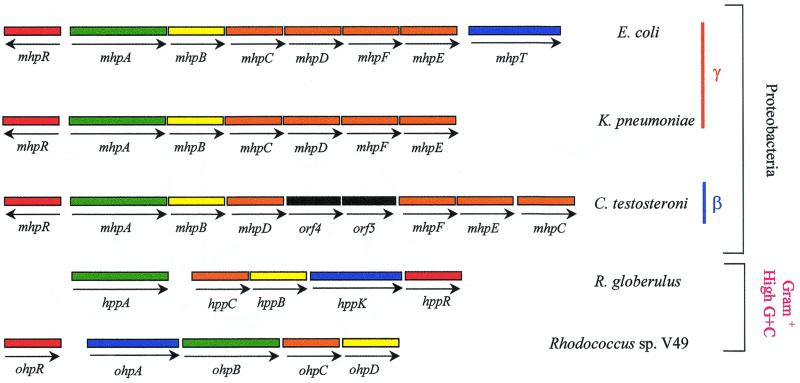
Analysis of the 9.8-kb mhp cluster (92) (Fig. 2A) reveals the existence of eight genes arranged as follows: (i) six genes encoding the initial monocomponent monooxygenase (mhpA), the extradiol dioxygenase (mhpB), and the hydrolytic meta-cleavage enzymes (mhpCDFE); (ii) one regulatory gene (mhpR); and (iii) one gene (mhpT) that encodes a transporter and is flanked by two bacterial interspersed mosaic elements (BIMEs) that belong to the BIME-2 subfamily (16). All the genes appear to be transcribed in the same direction, with the sole exception of mhpR (Fig. 2A). Although the Shine-Dalgarno sequences of the mhpR, mhpA, and mhpC genes are located in intergenic regions, those of mhpB, mhpD, mhpF, and mhpE overlap the preceding genes, suggesting that the most common mechanism of translational coupling may occur (92).
The mhp cluster maps at min 8.0 of the chromosome, between the lacI gene, encoding the transcriptional repressor of the lac operon, and the adhC gene, encoding an alcohol-acetaldehyde dehydrogenase, that is in the vecinity of the tau operon for taurine metabolism (EcoGene database at the Colibri website). The order of the catabolic genes in the meta-cleavage operon, with the single exception of mhpF, parallels that of the enzymatic steps in the 3HPP catabolic pathway (92) (Fig. 2).
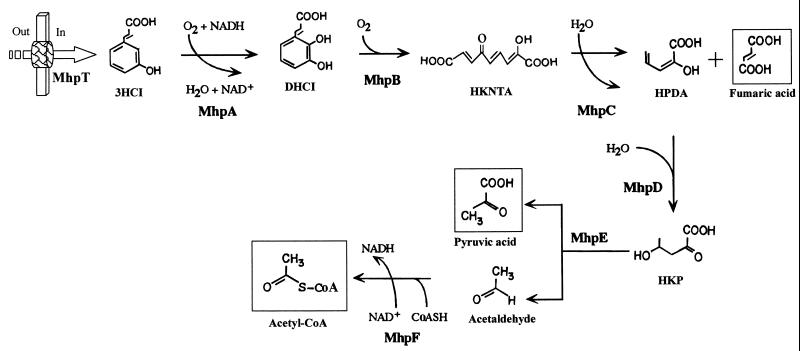
It is known that E. coli is also able to grow with 3HCI as the sole carbon and energy source (36) (Table 2). Growth with 3HCI induces the synthesis of the MhpA and MhpB enzymes, which are responsible for the initial attack on 3HPP (36), and it has been shown that the mhp cluster from E. coli confers to Salmonella enterica serovar Typhimurium LT2, a strain unable to grow on 3HPP and 3HCI, the ability to use these two aromatics as the sole carbon and energy source (92). Hence, the mhp genes are also responsible for the catabolism of 3HCI via 2,3-dihydroxycinnamic acid (DHCI) with the formation of pyruvate, acetyl-CoA, and fumarate (Fig. 3).
FIG. 3.
Biochemistry of the 3HCI catabolic pathway. The metabolites are 3HCI, DHCI (2,3-dihydroxycinnamic acid), HKNTA (2-hydroxy-6-keto-nona-2,4,7-triene 1,9-dioic acid), HPDA (2-hydroxy-penta-2,4-dienoic acid) and HKP (4-hydroxy-2-ketopentanoic acid). The enzymes are MhpA (3HCI monooxygenase), MhpB, (DHCI 1,2-dioxygenase), MhpC (HKNTA hydrolase), MhpD (HPDA hydratase), MhpE (HKP aldolase), and MhpF (acetaldehyde dehydrogenase [acylating]). The 3HCI transport protein (MhpT) is represented by a thick arrow. Out and In indicate outside and inside the cell, respectively.
PP Catabolic Pathway
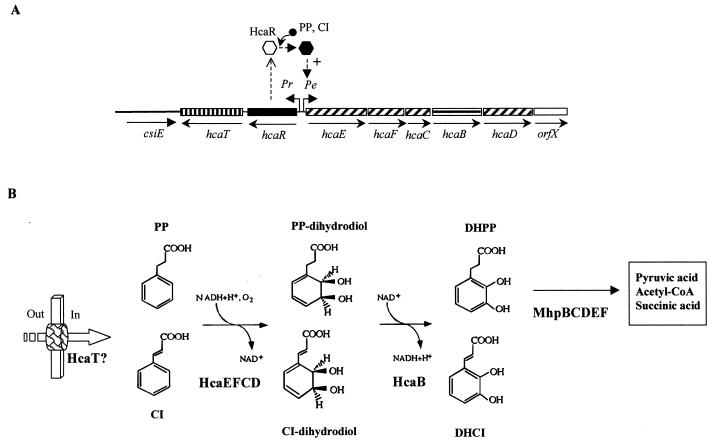
The catabolism of PP in E. coli is initiated by a dioxygenolytic pathway (36, 39) (Fig. 4B). The first step is catalyzed by a PP dioxygenase, which inserts an atoms of molecular oxygen into each of positions 2 and 3 of the phenyl ring of PP, yielding cis-3-(3-carboxyethyl)-3,5-cyclohexadiene-1,2-diol (PP-dihydrodiol), which is subsequently oxidized by the PP-dihydrodiol dehydrogenase to give DHPP (36, 39) (Fig. 4B). DHPP is the substrate of the meta-cleavage pathway described above for 3HPP degradation, and therefore it links the catabolism of PP and 3HPP in E. coli. The nucleotide sequence of a 7.2-kb DNA fragment that carries the hca cluster for PP catabolism revealed (i) five genes encoding the PP-dioxygenase (hcaEFCD; formerly named as hcaA1A2CD) and PP-dihydrodiol dehydrogenase (hcaB), (ii) a regulatory gene (hcaR), and (iii) a gene (hcaT) that might encode a transporter (71). Downstream of hcaD, the closely linked orfX (Fig. 4A) could also belong to the hca cluster, and it codes for a 155-amino-acid (aa) product of unknown function. The Shine-Dalgarno sequences of hcaE, hcaF, hcaC, hcaB, and hcaD overlap the preceding genes, suggesting that translational coupling occurs (71). Immediately downstream of orfX there is an inverted repeat sequence that could act as a transcriptional terminator of a potential operon. hcaR and hcaT are located upstream of hcaEFCBDorfX, but they are transcribed in the opposite direction from the other genes (Fig. 4A). Although the intergenic spacing between hcaR and hcaT was 159 bp, no typical transcriptional terminator and promoter sequences were detected in this DNA fragment (71).
FIG. 4.
Pathway for the catabolism of PP in E. coli. (A) Genetic map of the chromosomal hca cluster. Relevant genes are indicated by blocks: genes with similar shading encode the subunits of the PP dioxygenase. Regulatory (solid block) and putative transport (vertically striped block) genes are also shown. The horizontally striped block indicates the gene encoding the PP dihydrodiol dehydrogenase. The empty block represents a gene of unknown function. The csiE gene flanking the hca cluster is represented by a thick line. The arrows show the directions of gene transcription. Bent arrows represent the Pr and Pe promoters. The inactive and active forms of the HcaR activator are represented by empty and solid hexagons, respectively. + indicates transcriptional activation. The inducer molecule (PP and CI) is represented by a solid circle. (B) Biochemistry of the PP catabolic pathway. The metabolites are PP, CI, PP dihydrodiol, CI-dihydrodiol, DHPP, and DHCI (see the legends to Fig. 2 and 3). The enzymes are HcaEFCD (PP dioxygenase), HcaB (PP-dihydrodiol dehydrogenase), and MhpBCDEF (see the legend to Fig. 2). The putative PP/CI transport protein (HcaT) is represented by a thick arrow. Out and In indicate outside and inside the cell, respectively.
The hca cluster maps immediately downstream of csiE, which encodes the stationary-phase-inducible protein CsiE, at 57.5 min of the E. coli chromosome (EcoGene database at the Colibri website) and therefore far from min 8, where the mhp cluster responsible for DHPP degradation is located (see above).
The HcaEFCD dioxygenase and HcaB dihydrodiol dehydrogenase are able to transform CI into DHCI (Fig. 4B) (36, 71). Thus, when the hca and mhp clusters from E. coli are expressed in S. enterica serovar Typhimurium LT2, the resulting strain can grow on minimal medium containing CI as the sole carbon and energy source (71). Therefore, while in some soil Pseudomonas species and in Lactobacillus pastorianus the catabolism of CI could be accomplished by an initial reduction of the double bond of the side chain with formation of PP, the catabolism of CI by the Hca enzymes in serovar Typhimurium produces DHCI, which, following the mhp-encoded pathway, will be finally mineralized to pyruvate, acetyl-CoA, and fumarate (71) (Fig. 3). Since CI can induce the expression of the hca genes and is converted to DHCI by E. coli cells in a resting-cell process, it is difficult to explain the unexpected observation that this bacterium does not grow when cultivated on CI as the sole carbon and energy source (71). A possible explanation of this lack of growth could be that the DHCI generated in the reactions catalyzed by the Hca enzymes (and not the DHCI generated from 3HCI by the MhpA hydroxylase) or other intermediates farther down the mhp-encoded pathway accumulate to a toxic level that prevents the normal metabolic flux of the cell (71). Why this toxicity is not observed in serovar Typhimurium is also an open question.
PA Catabolic Pathway
E. coli W and K-12, but not E. coli B and C, mineralize PA (Table 2) through a novel catabolic pathway which does not follow the conventional routes for the aerobic catabolism of aromatic compounds (such as those of E. coli for HPA, 3HPP, and PP degradation) and whose first step is the activation of PA to phenylacetyl-coenzyme A (PA-CoA) by the action of a PA-CoA ligase (94, 328, 182) (Fig. 5B). The second step of the PA catabolic pathway will probably involve the hydroxylation of PA-CoA followed by cleavage of the aromatic ring and further degradation of the resulting aliphatic compound through a β-oxidation-like pathway (Fig. 5B). Since the participation of CoA ligases in the initial step of the catabolism of aromatic compounds is a typical feature of anaerobic catabolism (131), the aerobic PA degradation in E. coli constitutes one of the few examples of aerobic hybrid pathways (94, 182).
FIG. 5.
Pathway for the catabolism of PA in E. coli. (A) Genetic map of the chromosomal paa cluster. Relevant genes are indicated by blocks: genes with similar shading participate in the same enzymatic step or in the same functional unit of the pathway. Genes flanking the paa cluster (maoA and ydbC) are represented by thick lines. The arrows show the directions of gene transcription. Bent arrows represent the Pz, Pa, and Px promoters. The locations of the IS2 and IS30 insertion sequences within ydbA in E. coli K-12 are shown. The regulatory gene (paaX) is represented by a black block. The inactive and active forms of the PaaX repressor are indicated by empty and solid squares, respectively. − and + indicate transcriptional repression and activation, respectively. The inducer molecule (PA-CoA) is represented by a solid circle. (B) Biochemistry of the PA catabolic pathway. The first intermediate of the pathway is PA-CoA (phenylacetyl CoA). The enzymes are PaaK (PA-CoA ligase), PaaABCDE (putative multicomponent oxygenase), PaaZ (putative ring-cleavage enzyme), and PaaFGHIJ (putative β-oxidation-like enzymatic system).
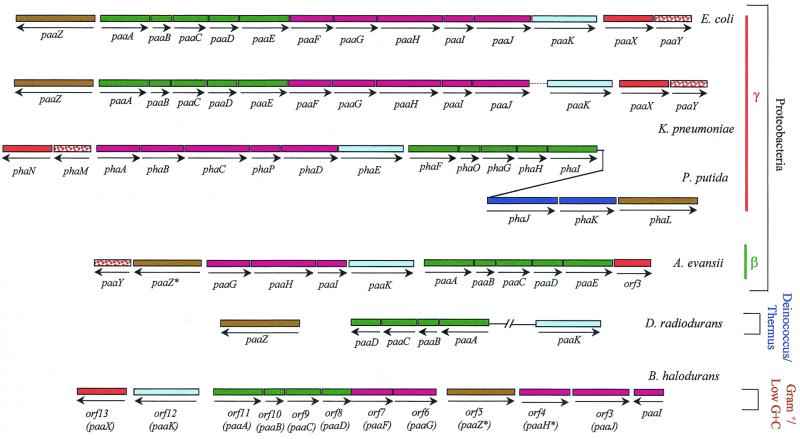
The paa genes responsible for PA degradation in E. coli K-12 and E. coli W strains are organized as a chromosomal cluster of about 14 kb (Fig. 5A), which is absent in the PA-deficient E. coli C strain. However, the ability of E. coli K-12 to grow on PA is strain dependent, with point mutations or small gene rearrangements in the paa cluster being the most probable reason for the PA-deficient phenotype of some K-12 laboratory strains (94) (Table 2).
The E. coli paa cluster contains 14 genes organized in three transcriptional units; two of them, paaZ and paaABCDEFGHIJK, encode the catabolic genes, and the third, paaXY, contains the paaX regulatory gene. All the genes are transcribed in the same direction with the sole exception of paaZ (Fig. 5A). Located downstream of paaZ, paaK, and paaY, three inverted-repeat sequences may act as transcriptional terminators (94). Although the paaK gene product catalyzes the first enzymatic step of the PA catabolic pathway, paaK is located at the 3′ end of the paa operon (Fig. 5A), a gene arrangement that explains why Tn1000 insertions within the paa catabolic operon caused polar effects leading to E. coli mutants that did not attack PA (94). The paa genes map at the right end of the mao cluster (Fig. 5A), which is involved in the transformation of PEA into PA (see below), at 31.1 min near the replication terminus of the chromosome (94). This location confirms previous observations that mapped the mutations in two PA-deficient mutants of E. coli K-12 in this chromosomal region (48). Although the left end of the paa cluster is adjacent to the maoA gene both in E. coli W and K-12, the right end of this cluster differs in these two strains. Thus, while the paaY stop codon was found 231 bp upstream of the ATG start codon of the ydbC gene in E. coli W (94), a 9.2-kb sequence containing a long open reading frame (ydbA) disrupted by two insertion sequences (IS2D and IS30C) was found between paaY and ydbC in E. coli K-12 (EcoGene database at the Colibri website) (Fig. 5A). The presence of insertion sequences near the paa cluster and the location of this cluster in a nonessential region of the chromosome (135) provide some clues to the possible mechanisms of gene mobilization of a catabolic cassette and could explain the heterogeneity observed among different E. coli strains to mineralize PA (see below).
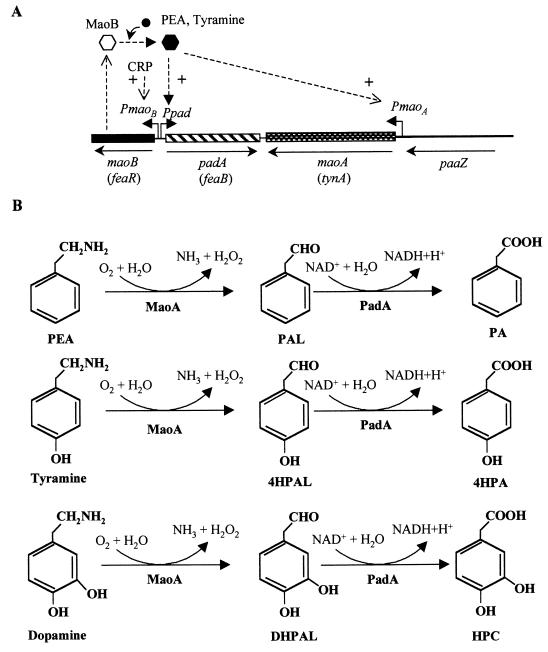
Upper Pathway for the Catabolism of Aromatic Amines
Growth of E. coli on PEA as the sole carbon and energy source induces two enzymatic activities that constitute the upper pathway for the catabolism of aromatic amines: an amine oxidase (MaoA) that converts PEA into phenylacetaldehyde (PAL) and a phenylacetaldehyde dehydrogenase (PadA or FeaB) that oxidizes the latter to PA (228) (Fig. 6B). E. coli mutants defective in PadA cannot grow on PEA as a carbon source but can still use it as a nitrogen source (228). Tyramine and dopamine are also substrates of MaoA and PadA, leading to formation of the corresponding aromatic acids, i.e., 4HPA and HPC, respectively (228) (Fig. 6B). Therefore, while E. coli K-12, which lacks the hpa cluster (see above), is able to use tyramine and dopamine only as nitrogen sources, E. coli W, which catabolizes 4HPA and HPC, can use these two aromatic amines as both carbon and nitrogen sources. The reaction catalyzed by MaoA leads to the formation of H2O2; therefore, to avoid the toxic effects of the latter, catalase is also induced within E. coli cells growing in the presence of aromatic amines (228).
FIG. 6.
Upper pathway for the catabolism of aromatic amines (PEA, tyramine, and dopamine) in E. coli. (A) Genetic map of the chromosomal cluster for the initial catabolism of aromatic amines. Relevant genes are indicated by blocks. Alternative gene names are in parentheses. The paaZ gene from the paa cluster (see Fig. 5) is indicated by a thick line. The arrows show the directions of gene transcription. Bent arrows represent the PmaoB PmaoA and Ppad promoters. The regulatory gene maoB (feaR) is shown by a solid block. The inactive and active forms of the MaoB activator are represented by empty and solid hexagons, respectively. + indicates transcriptional activation. The inducer molecule (PEA, tyramine) is represented by a solid circle. (B) Biochemistry of the initial catabolism of aromatic amines. The metabolites are PAL (phenylacetaldehyde), 4HPAL (4-hydroxyphenylacetaldehyde), DHPAL (3,4-dihydroxyphenylacetaldehyde), PA, 4HPA, and HPC (homoprotocatechuate). The enzymes are MaoA (monoamine oxidase) and PadA (phenylacetaldehyde dehydrogenase).
Expression of the maoA and padA genes is controlled by the MaoB (FeaR) regulator. The maoA, padA, and maoB genes (alternative gene names, tynA, feaB, and feaR, respectively) have been sequenced in E. coli K-12 and W strains and are located at 31.1 min on the chromosome (94, 127, 307). The padA gene is transcribed in the opposite direction to that of maoA and maoB (Fig. 6A), indicating that these three genes, although involved in the same metabolic pathway and physically associated, do not constitute an operon (95). Since these genes map immediately adjacent to the left end of the paa cluster (Figs. 5A and 6A), this supraoperonic clustering of two related pathways, i.e., the upper pathway for PEA catabolism and the PA catabolic pathway, represents an example of physical linkage within the PA-CoA catabolon (182, 217).
ENZYMES FOR THE CATABOLISM OF AROMATIC ACIDS AND AMINES IN E. COLI
The success of a particular catabolic pathway depends on two major elements, i.e., the catabolic enzymes, which must be assembled in an optimal chain of sequential transformations leading to the mineralization of the compound, and the regulatory elements. In this section we review the different catabolic genes and the corresponding encoded proteins that allow the activation and cleavage of the aromatic ring, as well as the further degradation of the nonaromatic intermediates for entry into the Krebs cycle. Although the catabolic genes of the aromatic biodegradative pathways in E. coli have been characterized, most of the catabolic steps for PA degradation still require an experimental demonstration.
Aromatic Ring-Hydroxylating Oxygenases
The initial step in the aerobic catabolism of 4HPA/3HPA, 3HPP/3HCI, and PP in E. coli is carried out by oxygenases that introduce one (monooxygenases) or two (dioxygenases) atoms of molecular oxygen into the phenyl ring. These oxygenases belong to three different families of ring-hydroxylating oxygenases (Table 3).
TABLE 3.
The ring-hydroxylating oxygenases for the catabolism of aromatic acids in E. coli
| Gene | Location (min)a | Gene product | Size in aa (kDa) | Oxygenase protein family | Natural substrate(s) | % Identity to other described proteinsc |
|---|---|---|---|---|---|---|
| hpaB | 98.8 | Oxygenase component of HPA MOxb | 520 (58.7) | TC-FDMb | 4HPA, 3HPA | 93 HpaA_Kp; 54 PheA_Btl; 48 PheA1_Btg |
| hpaC | 98.8 | Reductase component of HPA MOx | 170 (18.6) | TC-FDM | 4HPA, 3HPA | 80 HpaH_Kp; 26 SnaC_Sp; 25 ActVB_Sc; 22 PheA2_Btg; 21 StyB_P |
| mhpA | 8.0 | 3HPP MOx | 554 (62.0) | Monocomponent flavin MOx | 3HPP, 3HCI | 53 MhpA_Ct; 39 HppA_Rg; 27 TcmG_Sg; 22 OhpB_Rs; 22 PheA_Ps |
| hcaE | 57.4 | Large terminal subunit of PP DOxb | 453 (51.1) | Class IIB DOx | PP, CI | 48 TcbAa_Ps; 48 IpbA1_Re; 47 BphA1_Rs; 47 TodC1_Pp; 47 TecA1_Bs |
| hcaF | 57.4 | Small terminal subunit of PP DOx | 172 (20.5) | Class IIB DOx | PP, CI | 38 BphE_Ct; 34 CumA2_Pf; 34 IpbA2_Ps |
| hcaC | 57.4 | Ferredoxin subunit of PP DOx | 106 (11.3) | Class IIB DOx | PP, CI | 48 BphA3_Rs; 47 IpbA3_Re; 44 CumA3_Pf |
| hcaD | 57.4 | Ferredoxin reductase subunit of PP DOx | 400 (43.9) | Class IIB DOx | PP, CI | 35 CmtAa_Pp; 31 IpbA4_Ps; 31 CumA4_Pf; 31 BphG_Ct; 30 TecA4_Bs |
| paaA | 31.2 | Putative oxygenase of a diiron multicomponent oxygenased | 309 (35.4) | Diiron multicomponent oxygenased | PA-CoA?d | 64 PhaF_Pp |
| paaB | 31.2 | 95 (10.9) | 63 PhaO_Pp | |||
| paaC | 31.2 | 248 (27.8) | 53 PhaG_Pp | |||
| paaD | 31.2 | 167 (18.6) | 33 PhaH_Pp | |||
| paaE | 31.2 | Putative reductase of a diiron multicomponent oxygenase | 356 (39.3) | Class IA-like reductase | PA-CoA? | 41 PhaI_Pp; 30 TdnB_Pp; 28 TsaB_Ct; 27 Hcr_Ec; 26 Pdr_Bc |
The location of the genes is with respect to the E. coli K-12 genome. The location of the hpa genes (lacking in E. coli K-12) was inferred from the position of their flanking regions that are present in the chromosome of the K-12 strain.
Abbreviations: MOx monooxygenase: DOx, dioxygenase; TC-FDM, two-component nonheme flavin-diffusible monooxygenase.
The names of the proteins, with their GenBank accession numbers in parentheses, are as follows. HpaA_Kp and HpaH_Kp, oxygenase and reductase components of HPA MOx from Klebsiella pneumoniae (L41068); PheA_Btl, phenol MOx from Bacillus thermoleovorans (AF031325); PheA1_Btg and PheA2_Btg, oxygenase and reductase components of phenol MOx from B. thermoglucosidasius (AF140605); SnaC_Sp, flavin reductase in pristinamycin IIA biosynthesis from Streptomyces pristinaespiralis (P54994); ActVB_Sc, flavin reductase in biosynthesis of actinorhodin in S. coelicolor (X58833); StyB_P, reductase component of styrene MOx from Pseudomonas strains (AJ000330, AF031161, Z92524); MhpA_Ct and HppA_Rg, 3HPP MOx from Comamonas testosteroni (AB024335) and Rhodococcus globerulus (U89712), respectively; TcmG_Sg, hydroxylase from S. glaucescens (P39888); PheA_Ps, phenol MOx from Pseudomonas sp. strain EST1001 (P31020); OhpB_Rs, 2HPP MOx from Rhodococcus sp. strain V49 (AF274045); TcbAa_Ps, IpbA1_Re, BphA1_Rs, TodC1_Pp, and TecA1_Bs, large terminal subunits of the multicomponent DOx for degradation of chlorobenzene in Pseudomonas sp. strain P51 (U15298), isopropylbenzene in R. erythropolis BD2 (U24277), biphenyl/polychlorobiphenyl (PCB) in Rhodococcus sp. strain RHA1 (D32142), toluene in P. putida F1 (J04996), and tetrachlorobenzene in Burkholderia sp. strain PS12 (U78099), respectivelyl; BphE_Ct, CumA2_Pf, and IpbA2_Ps, small terminal subunits of DOx for degradation of PCB in C. testosteroni B-356 (U47637, U47638), cumene in P. fluorescens IP01 (D37828), and isopropylbenzene in Pseudomonas sp. strain JR1 (U53507), respectively; BphA3_Rs, IpbA3_Re, and CumA3_Pf, ferredoxin subunits of multicomponent DOx for degradation of PCB in Rhodococcus sp. strain RHA1 (D32142), isopropylbenzene in R. erythropolis BD2 (U24277), and cumene in P. fluorescens IP01 (D37828), respectively; CmtAa_Pp, IpbA4_Ps, CumA4_Pf, BphG_Ct, and TecA4_Bs, ferredoxin reductase components of multicomponent DOx for degradation of p-cumate in P. putida F1 (U24215), isopropylbenzene in Pseudomonas sp. strain JR1 (U53507), cumene in P. fluorescens IP01 (D37828), PCB in C. testosteroni B-356 (U47637, U47638), and tetrachlorobenzene in Burkholderia sp. strain PS12 (U78099), respectively; PhaF, PhaO, PhaG, and PhaH, components of the putative PA-CoA oxygenase from P. putida U (AF029714); Phal, putative reductase component of a PA-CoA oxygenase from P. putida U (AF029714); TdnB_Pp, TsaB_Ct, and Pdr_Bc, reductase components of aniline DOx from P. putida UCC22 (D85415), toluenesulfonate MOx from C. testosteroni T-2 (U32622), and phthalate Dox from Burkholderia cepacia DBO1 (P33164), respectively; Hcr_Ec, reductase component of the hybrid cluster protein (prismane protein) from E. coli (P75824).
Also for paaB, paaC, and paaD.
4HPA/3HPA monooxygenase.
The hpaBC genes located at the 3′ end of the hpa cluster in E. coli W encode the two-component monooxygenase that initiates the catabolism of 4HPA and 3HPA in this strain (251) (Fig. 1). This enzyme is a member of the two-component nonheme flavin diffusible monooxygenases (TC-FDM) family, which can be defined according to the following properties: (i) the reductase and the oxygenase components are encoded by two different genes; (ii) the reductase component uses NAD(P)H to catalyze the reduction of a flavin that diffuses to the oxygenase component for oxidation of the substrate by molecular oxygen; and (iii) the two components are not flavoproteins and lack typical ferredoxin and/or flavin/NAD(P)H binding motifs (104).
The HpaBC monooxygenase is the best-studied ring-hydroxylating oxygenase in the catabolism of aromatic compounds in E. coli. E. coli K-12 strains expressing the hpaBC genes from a plasmid produced black pigments when they were growing in minimal glucose medium supplemented with l-Tyr, N-acetyl-l-Tyr, l-Tyr-methyl ester, 4HPA, 3HPA, or phenol. Since catechol derivatives form spontaneously black or brown oxidation products, the appearance of a black pigment in the culture medium reflected the HpaBC-mediated monooxygenation of the aromatic compounds added to the growth medium (254). The substrate range of the HpaBC monooxygenase was checked by measuring NADH oxidation. Although the best substrate was 4HPA followed by 3HPA, the enzyme was capable to oxidize NADH in the presence of chloro- and methylphenols such as 3-chloro-4HPA, 4-chloro-PA, 4-chlorophenol, 3-chlorophenol, and p-cresol. The HpaBC monooxygenase can also oxidize some dihydroxylated aromatic compounds such as HPC, 2,5-dihydroxyphenylacetic acid, and, with lower efficiency, catechol, resorcinol, hydroquinone, and 3,4-dihydroxyphenylalanine (l-Dopa) (254). The equivalent 4HPA monooxygenase from Klebsiella pneumoniae also exhibits a broad substrate specificity range and transforms the dihydroxylated compounds into quinones which subsequently polymerize to produce a melanin-like pigment (114). The HPA monooxygenase from E. coli does not consume NADH in the presence of PA, 2HPA, l-Phe, o-cresol, m-cresol, or 2-chlorophenol. It seems, therefore, that a substituent at the para position in the aromatic ring is very important in substrate recognition by the HpaBC monooxygenase (254).
The purified HpaC protein (Table 3) is a dimer that shows flavin reductase activity. The HpaC oxidoreductase is colorless, and the UV-visible spectrum shows no evidence for any chromogenic cofactor (251). Although the most effective substrates are NADH and flavin mononucleotide FMN, flavin adenine dinucleotide FAD and riboflavin can be also turned over by the enzyme with similar Km values (104). This low specificity could be ascribed to the fact that in these reductases the flavin behaves as a real substrate and not as a tightly bound prosthetic group, as is the case in the majority of flavin enzymes (112). Although HpaC can also use NADPH as substrate, its specific activities on different flavins are about 2 orders of magnitude lower than those obtained with NADH (104). The HpaC oxidoreductase can also reduce, in a FMN- and NADH-dependent way, cytochrome c and iron (III) (104).
The HpaB protein (Table 3) is a dimer that constitutes the oxygenase component of the HPA monooxygenase (251). The formation of HPC by purified HpaB protein is absolutely dependent on the presence of HpaC, which confirms that both components are required for HPA hydroxylation (104) (Fig. 1B). This hydroxylating activity was NADH and FAD dependent, and neither FMN nor riboflavine could replace FAD in the reaction (104, 340). However, the HpaB oxygenase component does not require a direct interaction with the HpaC oxidoreductase to hydroxylate 4HPA/3HPA, and therefore any flavin reductase present in the host cell able to release FADH2 into the cytoplasm would replace the role of HpaC (104). In this sense, when the E. coli Fre flavin reductase was used to generate FADH2 in vitro, HpaB was able to use FADH2 and O2 for the oxidation of 4HPA. HpaB also used chemically produced FADH2 for 4HPA oxidation, further confirming that HpaB is a FADH2-utilizing monooxygenase (340).
In general, the oxygenase components of members of the TC-FDM family show marked differences in their primary structures, which might reflect the fact that the substrate specificity of these enzymes resides in such components (104, 149). Nevertheless, the HpaB oxygenase shows high similarity to the equivalent HpaA oxygenase component of the HPA monooxygenase from K. pneumoniae (115) and to the phenol hydroxylases PheA and PheA1 from Bacillus thermoleovorans and B. thermoglucosidasius, respectively (79) (Table 3). This high amino acid sequence identity agrees with the observation that phenol is also a satisfactory substrate for HpaB (250, 251), suggesting that these enzymes might have evolved from a common ancestor able to hydroxylate phenol derivatives.
In contrast to the oxygenase components, an extended similarity among the reductase components of the TC-FDM proteins has been observed (104). The similarities among the reductases correlates with the fact that all of them use the same substrates, i.e., FAD/FMN and NAD(P)H. Amino acid sequence comparison analyses revealed that the HpaC-like reductases are not similar to other nonflavoprotein flavin reductases described so far; therefore, they appear to constitute a novel subfamily sharing several conserved motifs (104). Some HpaC-like flavin reductases have been found in different microorganisms (Table 3). In the genomes of several enteric bacteria (Salmonella species, Yersinia pestis, and Photorabdus luminescens [GenBank accession no. AF021838-9]) and in that of Pseudomonas aeruginosa (see below), there are two contiguous genes, hpaB and hpaC, whose products are homologous to HpaB and HpaC from E. coli, thus also suggesting the existence of a HPA monooxygenase of the TC-FDM family in these bacteria.
The existence in E. coli of several reductases such as Fre reductase (139), the sulfite reductase (53), the SsuE reductase of the alkanesulfonatase (86), and the HpaC reductase, capable of producing free reduced flavins, and the apparent functional interchangeability between them (53, 229), poses some questions. A potential adaptative significance of this redundancy is to provide a readily available backup if an enzyme is lost by a mutational event (53). Similarly, the existence of Fre or other flavin reductases could explain the puzzling result observed in an E. coli K-12 strain that expressed the oxygenase HpaB component alone but showed a significant 4HPA hydroxylating activity (251). Nevertheless, since the amount of FADH2 provided by the host reductases is likely to be insufficient to achieve an optimal HPA monooxygenase activity, the acquisition of the hpaC reductase gene cotranscribed (coregulated) with the hpaB oxygenase gene might represent an evolutionary advantage to develop a highly efficient HPA catabolic pathway.
3HPP monooxygenase.
The first catabolic gene of the mhp cluster, that is, mhpA, encodes the MhpA monooxygenase, which hydroxylates 3HPP and 3HCI to DHPP and DHCI, respectively (36, 39) (Figs. 2 and 3). MhpA shows similarity to typical monocomponent bacterial flavin-type aromatic hydroxylases (Table 3). In these proteins, there are three regions of special relevance involved in binding to the FAD molecule: (i) the NH2 terminus contains the putative βαβ fold consensus fingerprint sequence, which is involved in binding of the ADP moiety of FAD; (ii) middle of the protein contains the A(C)DG motif; and (iii) the COOH terminus contains a stretch of amino acids that are likely to participate in binding to the flavin moiety of FAD (75, 92). Although MhpA requires NADH for its activity (36), no NADH binding site could be inferred from the analysis of its amino acid sequence, a characteristic also observed with other flavin monooxygenases (75). Two 3HPP monooxygenases that show similarity to MhpA have been described so far, i.e., HppA from Rhodococcus globerulus PWD1 (18) and MhpA from Comamonas testosteroni TA441 (10) (Table 3). The HppA enzyme appears to have a relatively broad substrate specificity, although PP, 2HPP, 2HCI, and CI were not oxygenated (18). 2HPP (melilotate) is also not oxidized by MhpA, which explains why this aromatic acid cannot be used as carbon source by E. coli (36). Melilotate is the substrate recognized by the flavoprotein melilotate hydroxylase isolated from Pseudomonas spp., an enzyme that does not recognize 3HPP (302). A second melilotate hydroxylase, OhpB, has been described in Rhodococcus sp. strain V49. Although the OhpB enzyme also oxidizes 2HCI, it has low activity on 3HPP, 3HCI, and CI (247). The different substrate specificities of the 3HPP monooxygenases (MhpA and HppA) and the 2HPP monooxygenase (OhpB) reflect their low amino acid sequence similarity (Table 3).
PP dioxygenase.
Sequence comparison analyses of the hcaE (formerly hcaA1), hcaF (formerly hcaA2), hcaC, and hcaD gene products revealed significant similarities to the corresponding four protein subunits of the three-component class IIB ring-activating dioxygenases (41) (Table 3). The hcaEFCD genes are suggested, therefore, to encode the HcaEFCD initial dioxygenase of the PP catabolic pathway. This enzyme, together with the HcaB dihydrodiol dehydrogenase, is able to oxidize PP and CI to DHPP and DHCI, respectively (71) (Fig. 4B).
Although the hcaE gene product shows similarity to the corresponding large (α) subunit of the terminal oxygenase component of other class IIB ring-hydroxylating dioxygenases (41, 71, 152), mainly to the analogous ipb, tod, and bph gene products of the toluene/biphenyl family (Table 3), it does not fall within any of the clusters on the phylogenetic tree of the α subunits of Rieske nonheme iron oxygenases (116). The hcaF and hcaC genes encode proteins whose amino acid sequences show significant identity to those of the small (β) subunit of the terminal oxygenase and the ferredoxin component of multicomponent dioxygenases, respectively (12) (Table 3). The hcaD gene product is homologous to the ferredoxin reductase subunit of other dioxygenases (41) (Table 3).
The hcaB gene (Fig. 4) encodes a protein that shows a significant identity to cis-dihydrodiol dehydrogenases that participate in pathways involving class IIB dioxygenases and convert the cis-dihydrodiols formed by the initial dioxygenases into the corresponding dihydroxy derivatives with regeneration of NADH (41). Therefore, HcaB is postulated to be the PP-dihydrodiol dehydrogenase (71) (Fig. 4). The length (270 aa; 28.4 kDa) and amino acid sequence of HcaB fall within the average range for members of the short-chain alcohol dehydrogenase/reductase family (138, 323).
The accumulation of DHPP by E. coli cells lacking the mhp cluster but expressing the hcaEFBCD genes in PP-containing rich medium generates a reddish-brown color due to autooxidation of this dihydroxylated compound to the corresponding quinones and semiquinones (39, 71). The formation of this red pigment has been used to monitor hca gene expression (39, 71) and allows the potential use of the hcaEFCBD cluster as a reporter system for gene expression studies.
Aromatic Ring Cleavage Dioxygenases
The reaction catalyzed by the ring-opening enzymes metabolizing catecholic compounds is a critical step in the bacterial assimilation of aromatic compounds (129). Thus far, only two ring cleavage dioxygenases have been identified in the catabolism of aromatic compounds in E. coli, i.e., MhpB for DHPP and DHCI cleavage in the PP/3HPP and 3HCI pathways, respectively (Fig. 2 and 3), and HpaD (HpcB) for HPC cleavage in the HPA pathway (Fig. 1). Both enzymes cluster within the type II (87) or class III (298) extradiol dioxygenases, although they belong to two different subfamilies. The HpaD subfamily shows lower similarity among its members, which, in addition, have a deletion of about 40 residues in the central region of their primary structures (175). The two histidines and one glutamate (or aspartate) that form the iron coordination sphere in the class III extradiol dioxygenase whose three-dimensional structure is known, i.e., the LigAB protocatechuate 4,5-dioxygenase from Sphingomonas paucimobilis SYK-6 (306), are conserved in MhpB (His-10, His-53, and Glu-271) and HpaD (His-12, His-57, and Asp-257). Moreover, the histidine residue that functions as the catalytic base (306) is also conserved in MhpB (His-179) and HpaD (His-185).
The hpaD gene from E. coli W encodes a protein of 32 kDa (283 aa) homologous to the HPC 2,3-dioxygenase (276 aa) of E. coli C encoded by the hpcB gene (268). The most relevant difference between the two enzymes was observed in their C-terminal ends, where a 2-bp insertion produces a premature termination in HpcB. Another HPC 2,3-dioxygenase (HpaB) has been described in K. pneumoniae M5a1 (GenBank accession no. AJ000054) and shows 87% amino acid sequence identity to its orthologous enzymes HpaD and HpcB of E. coli. Putative HPC 2,3-dioxygenases that show 93, 85, and 65% identity to those of E. coli are also encoded in the putative hpa clusters of Salmonella species, Y. pestis, and P. aeruginosa, respectively (see below). Both HpcB from E. coli C (268) and the equivalent HpaB protein from K. pneumoniae M5a1 (113) are highly unstable enzymes, and the addition of reagents such as glycerol, acetone, dithiothreitol, or iron salts, which are known to stabilize some ring cleavage dioxygenases, had no effect on these HPC dioxygenases. Unexpectedly, the HPC dioxygenase from K. pneumoniae appears to contain Mg2+ instead of Fe2+ in its tetrameric structure (113). Whether the HPC 2,3-dioxygenase from E. coli also requires Mg2+ for its activity or requires Fe2+ (like most extradiol dioxygenases) or Mn2+ (like the HPC 2,3-dioxygenase from Arthrobacter globiformis [333]) remains to be determined.
The MhpB dioxygenase responsible for the meta cleavage of DHPP was unstable in cellular crude extracts but could be substantially stabilized by addition of ethanol, glycerol, and iron(II) to the purification buffer (34). The purified protein could be fully reactivated by treatment with iron(II) and a reducing agent such as ascorbate. Therefore, as was found for most of the extradiol dioxygenases, MhpB requires a nonheme Fe2+ cofactor for activity and exists as a tetramer (34). The extradiol oxidative cleavage is a complex multistep reaction that highlights the multifaceted catalytic properties of nonheme iron(II): activation of dioxygen, activation of the aromatic ring, carbon-carbon bond cleavage, and insertion of both atoms of molecular oxygen. Semiquinone and lactone intermediates have been identified in the MhpB-mediated extradiol cleavage of DHPP (35). Chemical modification with group-specific reagents revealed that the MhpB enzyme was inactivated by treatment with the histidine-directed reagent diethyl pyrocarbonate, which is in agreement with the identification of conserved histidine residues that may function as iron(II) ligands in the primary structure of MhpB (see above) (298). p-Hydroxymercuribenzoate and dithionitrobenzoic acid, two bulky cysteine-directed reagents, also inactivated MhpB, suggesting that a cysteine residue is present at or near the active site of the enzyme (298).
The MhpB dioxygenase shows a broad specificity toward 3-substituted catechols, with the optimum side chain being that of propionate. Catechol and 4-methylcatechol were also cleaved by MhpB. The methyl esther of the natural substrate, DHPP, was processed at almost identical rates, implying that the carboxylate group is not bound through an electrostatic interaction at the respective active sites of MhpB. Similarly, both 3-ethylcatechol and 3-propylcatechol were processed efficiently by MhpB. The importance of the catecholic hydroxyl groups was confirmed by the observation that neither 2,3-dimethoxyphenylpropionic acid nor 2-aminophenol was accepted as a substrate. Although MhpB processed 3-phenethylcatechol, 2,3-dihydroxybenzoic acid was not used as a substrate (298). DHCI was accepted as a good substrate (Fig. 3), indicating that the enzyme is able to bind the alkyl side chain in a transoid conformation. At high substrate concentrations, substrate inhibition was observed; however, at low substrate concentrations, a sigmoidal kinetics was detected, suggesting cooperativity between protein subunits (298). A similar substrate specificity pattern was observed for the MpcI dioxygenase of Ralstonia eutropha (298), which suggests that this enzyme is also a DHPP dioxygenase. Two other DHPP dioxygenases, MhpB from C. testosteroni TA441 (10) and OhpD from Rhodococcus sp. strain V49 (247), also accept catechol and 3-methylcatechol as substrates. In contrast, the DHPP dioxygenase (HppB) from R. globerulus PWD1 appears to be highly specific for catechols with a relatively large acid group (18). Interestingly, the two DHPP dioxygenases from gram-positive bacteria, i.e., HppB and OhpD, cluster as different subfamilies within the phylogenetic tree of class III extradiol dioxygenases.
HPC meta Cleavage Dehydrogenative Route
As indicated above, following meta cleavage of HPC during the catabolism of 4HPA and 3HPA in E. coli W and C, the resulting CHMS gives rise to succinic semialdehyde and pyruvate through a dehydrogenative route (Fig. 1B).
hpaE and hpcC encode the CHMS dehydrogenases of E. coli W (250) and E. coli C (271), respectively. These two proteins show 98% amino acid sequence identity and catalyze the dehydrogenation of CHMS to CHM (Fig. 1B). The HpcC enzyme has been purified and appears to be a NAD-dependent homodimer (50, 90). Comparisons of HpaE and HpcC with proteins of the aldehyde dehydrogenase superfamily revealed all the motifs that characterize these enzymes (237). Moreover, about 40% identity was observed to other members of the hydroxymuconic semialdehyde dehydrogenase family such as the DmpC and XylG proteins from the phenol pathway of Pseudomonas sp. strain CF600 (249) and the toluene pathway of P. putida (129), respectively (250).
The protein encoded by the hpaF and hpcD genes catalyzes the isomerization of CHM to OPET in E. coli W and C, respectively (267) (Fig. 1B). The HpcD isomerase is a trimeric enzyme with a subunit size of 14 kDa (126 aa). The crystal structure of HpcD has been determined, and a large pocket that is proposed to be the active site was identified. The catalytic mechanism involves a single catalytic base that is the N-terminal proline residue. Two arginine residues interact with the substrate in the active-site region (304, 312). Although the HpcD isomerase catalyzes a reaction analogous to that catalyzed by the 4-oxalocrotonate tautomerase of the meta-cleavage pathway of phenol and toluene on a chemically similar substrate, the two enzymes do not have any apparent sequence similarity. However, comparison of the overall three-dimensional structures of the two enzymes revealed similarities in the catalytic pocket, which confirms that the same catalytic mechanism is operative in both systems. Nevertheless, the two isomerases are specific, and each shows a preference for its own substrate. This substrate specificity in the isomerases seems to be determined by steric constraints in the catalytic cleft (304, 312).
The first catabolic gene of the HPC operon, hpaG in E. coli W (Fig. 1), encodes a protein of 46.9 kDa (429 aa), almost identical to the hpcE gene product from E. coli C (269). However, HpaG is 24 aa longer than HpcE due to a 7-bp deletion at the 3′ end of hpcE that produces a premature termination (250). HpcE was reported to be a bifunctional decarboxylase/isomerase monomeric enzyme that catalyzes the magnesium-dependent decarboxylation of OPET to 2-oxo-hept-3-ene-1,7-dioic acid (OHED) through HHDD as an intermediate (Fig. 1B) (108, 145, 269). This type of bifunctional decarboxylase/isomerase activity has not been detected in other aromatic catabolic pathways where the enol-keto isomerization appears to be a dispensable enzymatic step and the actual substrate for the next enzyme in the pathway, a hydratase, is the enol form instead of the keto form (128). Since there are no conclusive data to assume that OHED is the substrate of the HpaH hydratase, it is possible that transformation of HHDD into HHED might occur without a previous enzymatic isomerization step (250) (Fig. 1B). The HpaG and HpcE enzymes may have arisen from a gene duplication event since their N-terminal halves are very similar to their C-terminal ones (269). Reinforcing this assumption, in the genomes of Y. pestis, K. pneumoniae, and P. aeruginosa, the putative HPC meta-cleavage operon contains two hpaG-like genes in tandem (hpaG1 and hpaG2), which through gene fusion might have evolved as the two halves of hpaG in E. coli and Salmonella species (see below). It is worth mentioning that while HpaG (HpcE) shows low identity (below 20%) to other aromatic decarboxylases, such as DmpH and XylI for phenol and toluene catabolism, respectively (250), it shows a significant similarity to fumarylacetoacetate hydrolase that catalyzes the last step in the homogentisate pathway for the catabolism of PA and phenylalanine in Aspergillus nidulans (91) and to fumarylpyruvate hydrolases that catalyze the last step in gentisate degradation (329, 351).
The hpaH gene from E. coli W codes for a protein of 267 aa, whose primary structure shows 94% amino acid sequence identity to that of HpcG (264 aa), the hydratase from E. coli C (97, 271) (Fig. 1). The HpcG enzyme requires metal ions for its activity, and the native protein seems to be an hexamer (97). Although the substrate of HpaH (HpcG) is a dioic acid with two carbon atoms more than the 2-hydroxy-penta-2,4-dienoic acid (HPDA), i.e., the substrate of hydratases of the catechol or catechol-like meta-cleavage pathways such as that of 3HPP (Fig. 2B), there is a significant amino acid sequence identity between HpaH (HpcG) and the hydratases of the latter pathways. For instance, the MhpD hydratase from the 3HPP degradation pathway of E. coli (see below) shows 34% amino acid sequence identity to HpaH, which suggests that they have the same ancestral source. Moreover, as previously reported for the hydratases of catechol meta-cleavage pathways, the HpaH (HpcG) protein presents a relevant similarity to aromatic decarboxylases (250), reinforcing the previous suggestion about the common origin for the hydratases/decarboxylases involved in catabolism of aromatic compounds (269, 271).
The next enzyme of the HPC meta-cleavage dehydrogenative route, the HHED aldolase, is encoded by the last gene in the HPC operon, i.e., hpaI in E. coli W and hpcH in E. coli C (Fig. 1) (250, 303). Unlike the situation mentioned above for the aromatic hydratases, there is no striking identity between HHED aldolase (262 aa) and most of the aldolases of catechol meta-fission pathways. However, HpaI (HpcH) aldolase shows significant amino acid sequence identity (57%) to the BphF aldolase of the catechol meta-cleavage pathway for biphenyl/polychlorinated biphenyl degradation in Rhodococcus sp. strain RHA1. Moreover, since the HpaH (HpcG) hydratase (see above) also reveals a significant similarity to the corresponding BphE hydratase from strain RHA1, the RHA1 meta-cleavage pathway genes may have evolved from the same ancestor as the HPC meta-cleavage pathway genes (189). Two other putative aldolases that show significant similarity to HpaI were found encoded in the genome of Deinococcus radiodurans R1 (331) and in the 184-kb catabolic plasmid of Sphingomonas aromaticivorans F199 (266). Interestingly, two E. coli proteins, YfaU (267 aa) and YhaF (256 aa), show 56 and 48% amino acid sequence identity to HpaI, respectively, suggesting that their genes might encode aldolases that act on substrates chemically similar to HHED. The genes encoding such putative aldolases are located at 50.7 min (yfaU) and 70.5 min (yhaF) on the E. coli K-12 map, and in both cases they are followed by a gene, yfaV and yhaU, respectively, encoding a putative transporter of the anion:cation (H+ or Na+) symporter (ACS) family of transport proteins (223). A similar gene arrangement is present in the hpa cluster from E. coli W; i.e., the hpaI gene encoding HHED aldolase is followed by hpaX encoding the HPA transporter, also a member of the ACS family of transport proteins (see below) (Fig. 1). An aldol cleavage reaction that resembles that on HHED is found in the catabolism of glucarate and galactarate, and therefore the YhaF protein has been suggested to be the α-dehydro-β-deoxy-d-glucarate aldolase that produces pyruvate and tartronic semialdehyde (151, 303), with the YhaU protein being a putative glucarate transporter (223).
Of the two products generated by the action of HpaI (HpcH) on HHED, i.e., pyruvate and succinic semialdehyde (Fig. 1B), the latter needs to be oxidized to succinic acid before it can enter the Krebs cycle. E. coli strains have two succinic semialdehyde dehydrogenases, a NADP-dependent enzyme encoded by the gabD gene and a NAD-dependent enzyme encoded by a gene (sad) that was mapped at 34 min and has been characterized yet (184, 262, 295). While the GabD enzyme is induced by growth on γ-aminobutyrate, the NAD-dependent succinic semialdehyde dehydrogenase is induced by growth of the cells in 4HPA or succinic semialdehyde. A NAD-linked succinic semialdehyde dehydrogenase participating in the catabolism of 4HPA in K. pneumoniae has also been described (277). The NAD-dependent enzyme from E. coli is a dimer with a subunit size of 55 kDa (77). E. coli C sad mutants were unable to grow on 4HPA, and this defect was repaired by replacing the affected gene with its orthologous gene from E. coli K-12 (295). Since the sad gene is not within the hpa cluster and is present in E. coli K-12 strains, which lack the 4HPA pathway, it is likely that the NAD-dependent succinic semialdehyde dehydrogenase may fulfill different physiological roles in E. coli, such as preventing the toxic effect of the succinic semialdehyde generated when the cells grow on γ-aminobutyrate as the sole carbon and nitrogen source (77).
DHPP meta Cleavage Hydrolytic Route
During the catabolism of 3HPP and PP in E. coli, ring cleavage of DHPP by the MhpB dioxygenase (see above) give rise to 2-hydroxy-6-keto-nona-2,4-diene 1,9 dioic acid (HKNDA) (Fig. 2B). Further catabolism of HKNDA through a hydrolytic route ends with the formation of succinic acid, pyruvic acid, and acetyl-CoA, which enter the Krebs cycle (36, 39) (Fig. 2).
The mhpC gene encodes the HKNDA hydrolase (MhpC), which catalyzes the hydrolytic cleavage of the extradiol ring fission product of DHPP, giving rise to succinic acid (which enters the Krebs cycle) and HPDA (Fig. 2B). The purified MhpC protein (287 aa) lacks the initial methionine residue and is a homodimer that does not require cofactors for its catalytic activity (170). In contrast to the ring cleavage dioxygenase MhpB (see above), the MhpC hydrolase shows some substrate selectivity for the carboxylate of the side chain. Slow turnover was observed for the ring fission products of 3-methylcatechol or catechol. However, the ring fission product of DHCI was a fairly efficient substrate, generating fumaric acid and HPDA (170) (Fig. 3). A single active site is responsible for the enzymatic reaction, which involves a fast initial ketonization of the dienol substrate followed by a stereospecific C-C fragmentation step. The dissociation of the keto intermediate from the enzyme prior to hydrolysis explains the uncoupling of substrate utilization and product formation (134, 169, 170). This leakiness has been also found in other C-C bond hydrolases for the catabolism of aromatic compounds such as BphD from Burkholderia cepacia LB400 (288). Extensive sequence similarity has been detected between MhpC and other C-C bond hydrolases cleaving vinylogous 1,5-diketones from, for example, the meta-cleavage biphenyl (BphD and PcbD), xylene (XylF), toluene (TodF), and phenol (DmpD) biodegradative pathways (92). A dendrogram resulting from the comparison of amino acid sequences of hydrolases involved in meta-cleavage pathways of aromatic compounds revealed four different groups, with the MhpC protein being the only exception within group I of a hydrolase not involved in biodegradation of bicyclic molecules (136). All C-C bond hydrolases cleaving 1,5-diketones are members of the α/β-hydrolase family (136), with residues Ser-114, Asp-239, and His-267 forming the putative catalytic triad of MhpC. Although there is a large body of biochemical and crystallographic evidence to suggest that the active-site serine group acts as a nucleophile in members of the α/β-hydrolase family, no acyl enzyme intermediate has been found in the catalytic mechanism of MhpC. In contrast, a gem-diol intermediate was observed during the MhpC-mediated hydrolytic reaction, indicating a catalytic mechanism involving base-catalyzed attack of water rather than nucleophilic attack of an active-site serine. Therefore, the previously identified active-site serine residue in the C-C bond hydrolases appears to act as a base and not as a nucleophile (98). Both MhpC and BphD from B. cepacia LB400 have been crystallized, and it is hoped that the knowledge of their three-dimensional structure will provide an insight into the role of the active-site serine residue and will allow us to identify the structural determinants of substrate specificity (98). Three other hydrolases acting on the ring cleavage product of DHPP and DHCI, i.e., MhpC from C. testosteroni, HppC from R. globerulus, and OhpC from Rhodococcus sp. strain V49, have been described, and all of them appear to be highly specific for the cleavage products of acid catechols (10, 18, 247). While a 68% amino acid sequence identity was observed between MhpC from E. coli and from C. testosteroni, less than 30% identity was observed between MhpC and the HKNDA hydrolases HppC and OhpC from the gram-positive Rhodococcus strains.
The mhpD gene codes for a 269-aa protein that converts HPDA to HKP (Fig. 2B). The MhpD hydratase is a highly efficient catalyst that requires a divalent metal ion for activity, with Mn2+ being the one that showed that highest activation. Inactivation by group-specific reagents revealed the presence of essential cysteine and tryptophan residues at or near the enzyme active site (242). The E. coli MhpD hydratase shows 61% amino acid sequence identity to the isofunctional MhpD enzyme from C. testosteroni (10), and they are also homologous to hydratases of other meta-cleavage pathways such as the HpaH (HpcG) protein from the HPC dehydrogenative route from E. coli (see above). Amino acid sequence alignments revealed several regions of high sequence conservation, and some or all of the conserved aspartate and glutamate residues in these regions may function as manganese(II) ligands. The potent inhibition of the MhpD-mediated reaction by oxalate suggests that it proceeds via tautomerization to cis-2-ketopent-3-enoic acid, followed by conjugate addition of water and formation of HKP (37, 242). Although in P. putida the XylJ hydratase for toluene degradation forms a complex with the preceding enzyme in the TOL pathway, the XylI decarboxylase, to avoid the release of the chemically labile HPDA intermediate (128), there is no evidence for a physical association between MhpD and MhpC in E. coli. In this case, the high catalytic efficiency of MhpD will ensure that HPDA is processed rapidly in vivo before decomposition (242).
The last two genes of the meta-cleavage route, mhpF and mhpE, are the most closely related to analogous genes of other aromatic meta-cleavage pathways, and their gene products have been ascribed to the acetaldehyde dehydrogenase (acylating) and HKP aldolase, respectively (Fig. 2). Equivalent mhpF and mhpE genes have been found in the 3HPP catabolic pathway of C. testosteroni (10) (see below). The MhpE enzyme (337 aa) from E. coli has been purified, and it cleaves HKP to acetaldehyde and pyruvic acid (Fig. 2B). Despite the high similarity between MhpE and class II aldolases such as DmpG and XylK from Pseudomonas sp. strain CF600 and P. putida mt-2, respectively (92), the E. coli enzyme is a class I aldolase that shows no dependence on divalent metal ions and utilizes an imine linkage between the C-2 carbonyl carbon of HKP and the ɛ-amino group of a lysine residue at the active site (243). Like the DmpG and XylK aldolases (249), the MhpE enzyme is selective for one enantiomer of the substrate, presumably the 4S enantiomer produced by the preceding enzyme on the pathway (243). MhpE also catalyzes the reverse reaction at 13% of the rate of the forward reaction, being highly selective for the acetaldehyde acceptor but showing a relaxed specificity for the α-keto acid carbonyl donor, which offers the possibility of using such an enzyme for stereospecific C-C bond formation reactions with nonnatural substrates (243). The generation of chiral hydroxy-2-keto-pentanoic acids and analogues thereof is under study and would be of great interest for the further synthesis of hydroxy alpha-amino acids (T. D. H. Bugg, personal communication).
The acetaldehyde dehydrogenase (acylating) is the enzyme that catalyzes the terminal reaction in catechol or catechol-like meta-cleavage pathways, i.e., the transformation of acetaldehyde into acetyl-CoA (92) (Fig. 2B). The MhpF acetaldehyde dehydrogenase (316 aa) conserves the catalytic thiol (Cys-131), which is present in all members of the aldehyde dehydrogenases superfamily (237). Although two other acetaldehyde dehydrogenases (acylating) have been reported in E. coli, that is, the AdhE aldehyde-alcohol dehydrogenase (154) and the putative EutE acetaldehyde dehydrogenase (GenBank accession no. P77445) for ethanolamine utilization, they do not show significant similarity to MhpF. Moreover, MhpF is shorter and does not show similarity to HpaE and PadA, two aldehyde dehydrogenases involved in catabolism of aromatic compounds in E. coli. Whether MhpF and MhpE need to be physically associated in vivo to form an active complex, as has been shown for DmpGF (237) and suggested for the XylKQ and NahMO aldolase-acetaldehyde dehydrogenase pairs (241), is still unknown.
Upper Pathway for the Catabolism of Aromatic Amines
Growth of E. coli K-12 and W in the presence of tyramine, dopamine, and PEA induces the maoA and padA genes, which are responsible for the transformation of such aromatic amines into ammonia and the corresponding aromatic acids (228, 282) (Fig. 6B).
The maoA (tynA) gene encoding the monoamine oxidase of E. coli K-12 (49) and W (95) has been cloned and overexpressed (13, 95, 299). The periplasmic MaoA protein (757 aa) shows 84% amino acid sequence identity to the equivalent soluble monoamine oxidase from Klebsiella aerogenes, although the similarity to amino oxidases of gram-positive bacteria and eukaryotic organisms is much lower (124). The MaoA enzyme is a member of the copper-topaquinone (2,4,5-trihydroxyphenylalanine quinone [TPQ]) family of amine oxidases that catalyzes the two-electron oxidative deamination of amines. The formation of the TPQ cofactor by the posttranslational modification of a tyrosine residue within the active site involves self-processing events requiring both copper and molecular oxygen (126, 190). The MaoA enzyme oxidizes PEA, tyramine, and dopamine to the corresponding PAL, 4-hydroxyphenylacetaldehyde, and 3,4-dihydroxyphenylacetaldehyde, respectively, with the concomitant release of ammonia (Fig. 6B). 3-Phenylpropylamine was also oxidized, although the catalytic efficiency was about 10-fold lower than that for PEA (228).
The MaoA protein is a homodimer, with each subunit containing a single copper ion and a TPQ cofactor at the active site. The structure of MaoA has been determined crystallographically, and each subunit consists of four distinct domains with the active site being located in the C-terminal one (206, 230). The central water-filled cavity of the dimer interface is proposed to be the entry site for molecular oxygen. The copper is coordinated by three conserved histidine residues and two water molecules. The TPQ is close to the copper and appears to have high rotational mobility (206, 265). The catalytic reaction of the enzyme divides into a reductive half-reaction and an oxidative half-reaction. Crystallographic and solution studies have shown that the enzyme is reduced through dissociation of a Schiff base from position 5 of the quinone, leading to the release of the product aldehyde. The oxidative half-reaction involves the copper ion and molecular oxygen and recycles the reduced enzyme back to its oxidized resting state, accompanied by the release of ammonia and hydrogen peroxide. The Asp-383 residue performs multiple roles in the catalytic mechanism, acting not only as the active-site base at different stages of the catalytic cycle but also in regulating the mobility and regenerating the TPQ cofactor that is essential to catalysis (206, 336).
The second step in the upper pathway for the catabolism of aromatic amines, that is, the oxidation of the aromatic aldehyde generated by MaoA to the corresponding aromatic acid, is catalyzed by the protein encoded by the padA gene in E. coli W (95) or the homologous feaB gene in E. coli K-12 (127) (Fig. 6). The PadA (FeaB) protein (499 aa) shows significant amino acid sequence identity to other aldehyde dehydrogenases such as the equivalent PAL dehydrogenase SytD (45.3% identity) from the styrene upper pathway of different Pseudomonas strains (19, 221, 324), the putative aldehyde dehydrogenases DhaS (GenBank accession no. AF027868) from B. subtilis (44.5% identity) and FldD (GenBank accession no. AJ277295) from Sphingomonas sp. strain LB126 (43.6% identity), the aldehyde dehydrogenase AldH (GenBank accession no. P23883) from E. coli (40.5% identity), and the p-cumic aldehyde dehydrogenase CymC (GenBank accession no. U24215) from P. putida F1 (40.3% identity). PadA also has 35% amino acid sequence identity to the HpaE (HpcC) semialdehyde dehydrogenase from the HPC dehydrogenative route of E. coli (see above). All the conserved motifs that characterize the superfamily of aldehyde dehydrogenases (237) are present in the primary structure of PadA. The PadA protein was partially purified and was shown to be an homodimer (95). Although many of the aldehyde dehydrogenases so far characterized are homotetramers, the quaternary structure in this superfamily of enzymes is evolutionarily variable, and several homodimers, such as the HpaE (HpcC) and Sad aldehyde dehydrogenases from the HPC dehydrogenative route of E. coli (see above), have also been described. As already reported for the PAL dehydrogenase from Achromobacter eurydice (101), a high concentration of PAL inhibited the PadA (FeaB) enzyme (95, 127). Substrate inhibition has also been observed in other aldehyde dehydrogenases (140). The PadA enzyme prefers NAD+ over NADP+ as coenzyme, and the stoichiometry of the reaction between NAD+ and PAL was shown to be 1:1. This result is in agreement with the observation that most aldehyde dehydrogenases from both prokaryotic and eukaryotic organisms are NAD specific (101, 237), even though NADP-dependent PAL dehydrogenases have also been reported (51, 215). Although the affinity of PadA for NAD+ was 2 orders of magnitude lower than that for PAL (Km, 3 μM), this may be compensated for the high concentration (806 μM) of the NAD+/NADH pool in E. coli. On the other hand, since poor expression of the padA gene can be predicted based on its codon usage, the high affinity of PadA for PAL might constitute an effective safety mechanism to eliminate free aldehyde that could be toxic to the cell (95).
The substrate specificity of PadA (FeaB) was examined, and the enzyme was shown to act almost equally well on PAL, 4-hydroxyphenylacetaldehyde, and 3,4-dihydroxyphenylacetaldehyde (95, 127, 228). While benzaldehyde and trans-cinnamaldehyde were not substrates, phenylpropionaldehyde was oxidized, although with a catalytic efficiency about 15-fold lower than that for PAL (95). The PadA enzyme was also able to catalyze the hydrolysis of p-nitrophenyl acetate, but this esterase activity was less than 0.3% of the dehydrogenase activity and its physiological role, if any, is still unknown (95).
PA Aerobic Hybrid Pathway
The initial reaction in aerobic catabolism of PA in E. coli is the activation of the aromatic ring through an unusual strategy based on CoA thioesterification of the carboxy group by the PA-CoA ligase (PaaK) (Fig. 5). The participation of CoA ligases in the initial step of the aerobic catabolism of PA in P. putida U (182, 187), Pseudomonas sp. strain Y2 (324) and Azoarcus evansii (formerly Pseudomonas sp. strain KB740) (200), 2-aminobenzoate and benzoate in A. evansii (5, 344), benzoate in Bacillus stearothermophilus (344), ferulate in P. putida (325, 345), Pseudomonas sp. strain HR199 (219), and P. fluorescens (109), and 2-furoic acid in P. putida Ful (160) has also been reported, and the existence of a CoA ligase has been suggested for the aerobic catabolism of salicylate (122) and thiophen-2-carboxylate (56). Moreover, some dehalogenation mechanisms of aromatic compounds also involve CoA thioester formation in aerobiosis (81). Although the rationale for utilizing such hybrid pathways, i.e., aerobic catabolic pathways endowed with typical features of an anaerobic catabolism, is not known, it has been suggested that they could represent a strategy of the microorganisms to cope with fluctuations of oxygen supply (212). In this sense, the existence of a hybrid pathway for the catabolism of PA in E. coli could reflect the facultative anaerobic character of this bacterium.
The PA-CoA ligase activity from E. coli is dependent on the presence of ATP, Mg2+, CoA, and PA (94). The PaaK protein (49 kDa, 437 aa) shows significant amino acid sequence identity to equivalent PA-CoA ligases involved in PA catabolism in other organisms, such as PhaE (65.6% identity) from P. putida U (182, 199) and PaaK from Pseudomonas sp. strain Y2 (324) (67.3% identity) and A. evansii (200) (64.6% identity). Putative PA-CoA ligases are Orf12 from a potential PA degradation pathway from Bacillus halodurans (310, 311) and Orf03286 (PaaK) from the D. radiodurans genome (GenBank accession no. AE001863) (331), which show about 50% amino acid sequence identity to PaaK from E. coli (182). Moreover, FtsAII and FtsAIII from Methanobacterium thermoautotrophicum (GenBank accession no. AE000938 and AE000804) and Archaeoglobus fulgidus (GenBank accession no. AE000988 and AE000964), two homologues of the FtsAI enzyme, which catalyzes coenzyme F390 formation (326), also show about 50% amino acid sequence identity to PaaK. Analysis of the primary structure of PaaK revealed the conserved motifs for AMP and substrate binding in acyl-adenylate-forming enzymes (43, 94).
The paaABCDE genes appear to be responsible for the second enzymatic step in the catabolism of PA in E. coli, i.e., hydroxylation of the aromatic ring of PA-CoA (94). Thus, the expression of the paaK and paaABCDE genes in E. coli W14, an E. coli W mutant strain lacking the paa and mao genes and therefore unable to grow on PA and PEA, caused the consumption of PA and the accumulation of 2HPA in the culture medium. However, 2HPA appears not to be a true intermediate in the PA catabolic pathway since it does not support the growth of E. coli W and is not consumed even when E. coli W cells are growing also in the presence of PA (94). A similar lack of growth on 2HPA and accumulation of this compound after addition of PA to some cultures of PA-deficient mutant strains from E. coli K-12 (48) and P. putida U (217), has been also observed. Although the possibility that exogenous 2HPA does not enter the cells cannot be ruled out, the fact that 2HPA formation requires the simultaneous expression of the paaK and paaABCDE genes strongly suggests that 2HPA is not a true intermediate in PA degradation but is derived from the accumulation of a hydroxylated PA-CoA intermediate that cannot be further degraded. The excretion to the culture medium of a hydroxylated aromatic compound derived from the intracellular accumulation of a hydroxylated CoA derivative has also been reported in the hybrid pathway for the catabolism of 2-aminobenzoate (5). A similar phenomenon was observed in E. coli during a blockade at the MenB-catalyzed step in menaquinone biosynthesis, when o-succinylbenzoyl-CoA (Fig. 7D) accumulates and becomes transformed nonenzymatically to the corresponding spirodilactone with elimination of CoA and excretion of the lactone to the growth medium (195). The excretion to the culture medium of an aromatic compound as a dead-end product derived from the intracellular accumulation of a CoA derivative could be a general strategy of the cells to prevent the possible metabolic risk of depletion of the intracellular pool of CoA (94, 217).
FIG. 7.
Relevant enzymatic activities of E. coli for the metabolism of chorismate-derived compounds. The enzymes catalyzing the different reactions are those described in Table 4. SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; R, octaprenyl side chain.
Sequence comparison analyses of the paaABCDE gene products revealed that the PaaE protein shows significant similarity to class IA-like reductases (Table 3), which are members of the ferredoxin-NADP reductase (FNR) family (52, 209, 272). Other proteins from E. coli with a modular organization similar to that of PaaE are YeaX (GenBank accession no. P76254), a protein of unknown function, and Hcr (Table 3), a protein that catalyzes the reduction of Hcp (prismane protein) with NADH as electron donor and FAD and [2Fe-2S] as cofactors and that might be involved in nitrate and/or nitrite respiration (319). The primary structure of the PaaA protein shows the two repeats of residues (EX2H) that characterize the dinuclear iron binding site of the large (α) subunit of the heteromultimeric (αβγ) oxygenase component in aromatic diiron monooxygenases such as the phenol and toluene monooxygenases (94, 248). Moreover, the amino acid sequence of PaaB shows the strictly conserved residues found in the low-molecular-weight dissociable activator protein that is required for optimal turnover of the oxygenase component in multicomponent diiron monooxygenases (255). Therefore, it is tempting to speculate that paaABCDE may encode the five subunits of a diiron multicomponent oxygenase acting on a CoA-activated aromatic acid (Table 3), with PaaB being the effector protein and PaaE being the reductase that mediates electron transfer between NAD(P)H and the PaaACD oxygenase component. Another putative multicomponent oxygenase (Box protein) acting on benzoyl-CoA has been identified in A. evansii (201). A similar set of genes encoding the oxygenase of the PA catabolic pathway has been reported in P. putida U (217) (Table 3) and is also present in other gram-negative bacteria such as K. pneumoniae and A. evansii (182). paaABCD orthologs have also been found in a similar gene arrangement in the putative PA catabolic clusters of two gram-positive bacteria, D. radiodurans (331) and the extremophile B. halodurans (310, 311) (see below), strongly suggesting both a functional linkage between the encoded proteins and their participation in PA catabolism (182). It is worth mentioning that the gene cluster encoding the putative multicomponent oxygenase from the two gram-positive bacteria is lacking a paaE homologue (see below), suggesting that electron transfer might be carried out by a different enzyme system in these bacteria.
The PaaZ protein (681 aa) has an N-terminal region (residues 1 to 527) containing all the conserved motifs that characterize the aldehyde dehydrogenase superfamily of proteins (94, 237). The amino acid sequence of the C-terminal region of PaaZ shows high similarity (97%) to the last 157 aa of MaoC from K. aerogenes, a protein that was predicted to be a regulatory element (307). Nevertheless, translation of the maoC gene can be extended toward the 5′ end of the reported sequence, revealing a single protein that is homologous to PaaZ. Therefore, it is likely that the extended MaoC protein is the PaaZ ortholog in K. aerogenes. Thus, it appears that PaaZ and its orthologs in K. aerogenes (MaoC), P. putida U (PhaL), and D. radiodurans (OrfDR2381) (see below) are bifunctional proteins consisting of an aldehyde dehydrogenase domain fused to a C-terminal domain of unknown function. Interestingly, Orf5 from the putative PA-degradative pathway from B. halodurans and PaaZ∗ from A. evansii (see below) show significant similarity to the aldehyde dehydrogenase domain of PaaZ but lack the C-terminal domain present in the latter. This observation reinforces the assumption that in the course of evolution, two independent domains became fused in PaaZ-like proteins of some organisms but are still maintained as separate proteins in other organisms. As has been suggested for the analogous PhaL protein of P. putida U (182, 217), the paaZ gene product in E. coli might catalyze the aromatic-ring cleavage of the hydroxylated CoA derivative formed during PA degradation. In this sense, it is worth mentioning that the C-terminal domain of PaaZ (and other PaaZ-like proteins) has some similarity enoyl-CoA hydratases, such as that from P. aeruginosa (315), which, in turn, appear to be related to the ring cleavage enzyme in anaerobic benzoate degradation pathways (131).
The paaF, paaG, paaH, and paaJ gene products can tentatively constitute a β-oxidation-like pathway (another typical feature of anaerobic catabolism of aromatic compounds), involved in the successive oxidations of the nonaromatic CoA intermediate to Krebs cycle compounds (94, 182). While the primary structures of the PaaF (255 aa) and PaaG (262 aa) proteins show similarity to the structures of members of the enoyl-CoA hydratase/isomerase superfamily (81, 204), the PaaH protein (475 aa) shares the signature sequence motifs of 3-hydroxyacyl-CoA dehydrogenases (133), suggesting that it could attack the product of the reaction catalyzed by the PaaF and/or PaaG enzymes. A C-terminal region of unknown function (133), which is not found in other 3-hydroxyacyl-CoA dehydrogenases, including the equivalent Orf4 (283 aa) from B. halodurans, is present in PaaH and in its orthologs from P. putida, K. pneumoniae, and A. evansii (see below). The paaJ gene product (401 aa) reveals a significant sequence similarity to the PcaF and CatF β-ketoadipyl-CoA thiolases. Since β-ketoadipyl-CoA is formed during the aerobic hybrid pathway for benzoate degradation in A. evansii (344) and since PcaF and CatF catalyze the last step in the β-ketoadipate pathway for the aerobic degradation of protocatechuate and catechol, respectively (132), it is tempting to speculate that PaaJ, and its analogous proteins from other bacteria, such as PhaD from P. putida U (see below), are also responsible for the last enzymatic step in PA degradation (94).
Since the paaI gene product (140 aa) shows 20% amino acid sequence identity to the Fbc thioesterase from Arthrobacter spp, a putative role for the former in cleaving a thioester bond during PA degradation can be anticipated (94).
REGULATORY ELEMENTS THAT CONTROL EXPRESSION OF THE GENE CLUSTERS FOR THE CATABOLISM OF AROMATIC ACIDS AND AMINES IN E. COLI
Regulatory proteins and regulated promoters are the key elements that allow catabolic operons to be transcribed only when required and at levels sufficient to guarantee an adequate metabolic return when the particular substrate is abundant and can serve as a nutrient source. Nevertheless, promoter activity in vivo is not just dependent on the performance of the specific regulator-promoter couple that responds to a given signal but also relies on superimposed mechanisms that connect the activity of individual promoters to the metabolic and energetic status of the cell (72).
All the E. coli aromatic catabolic clusters described above contain their cognate regulatory protein. Four transcriptional activators (HpaA, MaoB, MhpR, and HcaR) and two repressors (PaaX and HpaR) have been identified. Although all of them bind to their cognate promoters through a helix-turn-helix (HTH) motif and recognize structurally similar inducer molecules, they cluster (with the exception of only HpaA and MaoB) within different families of regulators. This observation suggests a divergent evolution in the regulation of aromatic catabolic pathways in E. coli and reinforces the previous observation that there are no rules for anticipating the type of regulator that responds to a particular chemical structure (68).
Transcriptional Activators
HpaA protein.
The hpaA gene is located upstream of the hpaBC operon of E. coli W (Fig. 1A) and encodes the HpaA regulatory protein (295 aa). Using monocopy and multicopy lacZ reporter systems, it was shown that HpaA is a transcriptional activator of the PBC promoter, which drives the expression of the hpaBC genes encoding the two-component HPA monooxygenase (253) (Fig. 1A). 4HPA was the best inducer of HpaA when tested on a PBC::lacZ reporter fusion, with the induction produced by 3HPA and PA being 70 and 35%, respectively, of that produced by 4HPA. It is surprising that PA, which is not a substrate of the HPA hydroxylase, is capable of a low but detectable induction. Although the PA effect can be ascribed to its structural similarity to 4HPA, it is worth noting that 2HPA, HPC, and other structurally related aromatic compounds are not able to increase the expression of the PBC::lacZ fusion (253). Whether some metabolic intermediates of the PA pathway might activate HpaA or whether a different regulator activated by PA could cross-interact with the PBC promoter is still unknown.
The C terminus of the HpaA protein shows a significant amino acid sequence identity to that of members of the AraC family of regulators (105, 251). This region comprises two potential HTH DNA binding motifs, of which only the second is present in all proteins of the family (105, 183). This is also the case in HpaA, where the potential second HTH motif (Val-257 to Gly-281) fits the signature pattern of AraC proteins much better than the potential first HTH motif (Trp-206 to Pro-234). The small region of high sequence conservation found outside the second HTH motif and toward the C-terminal end of the AraC-like proteins (183) is also present in HpaA. While the variation in the first HTH motif in AraC-like proteins may represent the recognition of different target sequences at the cognate promoters by the different regulators, conservation at the second HTH motif may reflect structural requirements rather than functionality in specific DNA interactions (183). The modular structure of the XylS protein, a prototype of the AraC family that controls benzoate and toluate degradation in P. putida, revealed that the N-terminal and central regions of the protein are involved in binding to the effector molecule (150). In agreement with this finding, the highest amino acid sequence identities between HpaA and other members of the AraC family are those to regulators that control the catabolism of structural analogs of 4HPA, such as the PobR proteins that activate gene expression for 4-hydroxybenzoate degradation in P. aeruginosa, Rhizobium leguminosarum, and Agrobacterium tumefaciens (72).
The transcription initiation site of the PBC promoter has been identified 56 bp upstream of the hpaB start codon (253). The −35 box of PBC clearly deviates from the consensus sequence of ς70-dependent promoters of E. coli, and two direct repeats have been found upstream of this box (253). The presence of direct repeats in this position appears to be a common feature of the binding sites for AraC-like regulators such as the XylS protein, which recognizes direct repeats leading to the formation of a dimer that interacts with the RNA polymerase and hence activates transcription from the cognate promoter (118).
The expression of hpaA is controlled by at least two promoters, Pa1 and Pa2, that are located within the hpaX coding region at 107 and 60 bp, respectively, from the translational start codon of hpaA (253) (Fig. 1A). The −35 and −10 boxes that can be ascribed to both promoters match only one or two nucleotides of the corresponding consensus sequences of ς70-dependent promoters. On the other hand, a putative CRP (catabolite repression protein) binding sequence was located upstream of the Pa1 start site, which agrees with the observation that glucose decreases the expression of hpaA through a CRP-dependent mechanism (253). Since hpaX and hpaA appear to form an operon, the putative promoter of hpaX, Px (Fig. 1A), may also contribute to the expression of hpaA. The physiological reasons that favor the existence of several promoters for controlling the expression of the hpaA regulatory gene remain to be elucidated, but the existence of promoters embedded within the coding sequence of an operon is not uncommon in E. coli (253).
MaoB protein.
The MaoB (FeaR) protein (301 aa) is a transcriptional regulator of the maoA and padA genes, which encode the monoamine oxidase and PAL dehydrogenase, respectively, in the catabolism of aromatic amines in E. coli (Fig. 6A). Amino acid sequence comparisons reveal that MaoB is also a member of the AraC family of regulators, having 22% sequence identity to the other AraC family member involved in catabolism of aromatics in E. coli, i.e., the HpaA protein (see above). The significant similarity between the C-terminal domain of MaoB and that of other members of the AraC family suggests that this domain binds to specific DNA in the maoA and padA promoters (PmaoA and Ppad), with the nonhomologous N-terminal and central region of MaoB being involved in the recognition of the inducer molecules, that is, tyramine and PEA (127, 342) (Fig. 6A).
By using lacZ gene fusions with the maoA and maoB genes, it was shown that maoA is activated by tyramine and the MaoB regulator, with the expression of maoB being subjected to catabolite repression by glucose. Thus, the catabolite repression of the PmaoA promoter, rather than being a direct effect on such promoter, may reflect the catabolite repression of the maoB gene (342). By using crp cya E. coli mutant strains that contained the maoB::lacZ fusion, it was shown that the catabolite repression by glucose was mediated by the cyclic AMP (cAMP)-CRP complex on the PmaoB promoter (Fig. 6A) and a potential CRP-binding site was identified in such promoter region (342).
MhpR protein.
The mhpR gene is oriented in the opposite direction to that of the other mhp genes (Fig. 2A) and exhibits common features of regulatory genes such as a codon usage that predicts a low level of expression (92). Enzymatic assays carried out using crude extracts of E. coli cells cultured in the absence (uninduced) or presence (induced) of 3HPP demonstrated that MhpR fostered inducible expression of the mhp catabolic genes, hence acting as a transcriptional activator (92) (Fig. 2A). The amino acid sequence of MhpR (277 aa) revealed significant identities to those of aromatic regulators that belong to the IclR family, such as PobR (23% identity) and PcaU (23% identity) from the 4-hydroxybenzoate and protocatechuate degradation pathways of Acinetobacter sp. strain ADP1, PcaR (21% identity) from the protocatechuate degradation pathways of P. putida PRS2000, and CatR (23% identity) and PcaR (27% identity) from the catechol and protocatechuate degradation pathways of Rhodococcus opacus 1CP (72). As expected, the highest identity was observed to the MhpR regulator of the equivalent 3HPP degradation pathway from C. testosteroni (10) and the putative mhp cluster from K. pneumoniae (see below). Although the HppR regulator of the 3HPP catabolic pathway from R. globerulus PWD1 has been classified as an IclR-type regulator (18), it does not show a significant amino acid sequence identity to MhpR, which might reflect differences not only in the regulatory mechanism but also in the chemical structure of the inducer molecule. In this sense, it has been observed that while some aromatic IclR-type regulators respond to aromatic compounds, other regulators of this family respond to nonaromatic compounds (89). Whereas near the N terminus of MhpR a stretch of residues matches the potential HTH structural motif involved in binding to the target DNA, the amino acid sequence pattern that characterizes the members of the IclR family is located at the C terminus of the MhpR protein (92). Mutations that do not affect DNA binding but that are involved in the recognition of the inducer molecule are found toward the middle of the protein in some aromatic IclR-type regulators (161).
Usually, IclR-type regulators are divergently transcribed with respect to the genes that they control, and the corresponding promoters are superimposed in a short DNA segment (72). A similar genetic arrangement has been observed within the mhp cluster from E. coli (Fig. 2A), K. pneumoniae, and C. testosteroni (see below). Thus, a 178-bp intergenic region with a higher A+T content (58%) than that of the mhp coding regions (44.2%) is located between the translational start sites of the divergently transcribed mhpR and mphA genes in E. coli, suggesting that the mhpR promoter is located near or overlapping the regulated promoter of the mhpA gene (92). A different genetic arrangement was observed with the hppR regulatory gene from the 3HPP catabolic pathway from R. globerulus (see below), which could reflect a different regulatory mechanism in this gram-positive bacterium, as pointed out above. Analyses of the mhpR-mhpA intergenic region in E. coli and C. testosteroni showed the presence of a 7-bp inverted repeat sequence equivalent to palindromic repeats that have been shown to be the binding site (operator) for other aromatic IclR-type regulators (92, 111). Moreover, putative IHF (integration host factor) and CRP binding sites have been mapped within the mhp intergenic region (92), revealing that expression of the mhp cluster in E. coli is the subject of a complex regulatory mechanism that requires further study.
HcaR protein.
The hcaR gene encodes a protein with a size (296 aa) and an amino acid sequence similar to those of LysR-type transcriptional regulators (LTTRs) (72). Within the LTTR family, HcaR shows the highest degrees of identity to the AlsR regulator of acetoin synthesis and to a select group of regulators, such as CatR, CatM, TfdR, TcbR, ClcR, and TfdT, that activate the genes encoding muconate- or chloromuconate-lactonizing enzymes and/or those encoding oxygenases that act on catechol or chlorinated aromatic compounds (71). Like the majority of the genes encoding LTTRs, hcaR is transcribed divergently from the catabolic genes (hcaEFCBD) that it regulates (Fig. 4A). Near the N terminus of HcaR, an HTH motif that characterizes LTTRs (283) can be found (71). The C-terminal domain of LTTRs seems to be involved in multimerization, and its consensus motif (283) fits with the amino acid sequence between residues 232 and 242 of HcaR (71).
To study the regulation of the hca genes, these genes were cloned and expressed in S. enterica serovar Typhimurium LT2, a strain lacking such genes. By monitoring the formation of the DHPP pathway intermediate (Fig. 4B), it was shown that HcaR was an activator of the hcaEFCBD catabolic genes in the presence of PP and CI (71). Therefore, as already described for other aromatic LTTRs (72), HcaR behaves as a transcriptional activator. A 135-bp intergenic region is located between the potential translational start sites of the divergently transcribed hcaR and hcaE genes (Fig. 4A), suggesting that the hcaR promoter is located near or overlaps the regulated promoter of the hca catabolic operon. As has been noted with other LysR-type regulatory targets (226), the A+T content (66%) of the hcaR-hcaE intergenic region is higher than that of the hca genes (47%) (71). Within the hca intergenic region and located 85 nucleotides upstream of the putative hcaE translation start site, a sequence (TAG-N7-CTA) matches the consensus binding motif of LTTRs (71, 283).
It has been reported that when E. coli is exposed to mixtures of glucose (an easily degradable substrate) and PP in batch culture, cells utilized the two compounds sequentially, i.e., the utilization of PP was inmediately repressed by glucose, regardless of whether glucose was present in the initial substrate mixture or was pulsed to cells growing with PP alone (164). However, PP and glucose were consumed simultaneously in carbon-limited continuous culture (165). The latter situation probably resembles that under environmental conditions, where growth is mostly carbon limited and microorganisms utilize a variety of carbon and energy substrates simultaneously (163). The molecular mechanisms underlying such carbon-dependent hca gene expression remain to be elucidated.
Interestingly, the two convergent branches of the DHPP meta-cleavage pathway in E. coli, that is, the 3HPP and the PP degradation branches, are controlled by members of different families of regulatory proteins, i.e., the MhpR (IclR family) and HcaR (LysR family) activators, respectively. Although the physiological meaning of such branch specificity of regulatory proteins is not known, it might facilitate the interactions between branches and could reflect a kind of hierarchy of pathway utilization in E. coli when this bacterium is provided with 3HPP and PP simultaneously. A similar finding was reported for the protocatechuate and catechol degradation branches of the β-ketoadipate pathway in P. putida PRS2000 and Acinetobacter sp. strain ADP1 (161).
Transcriptional Repressors
HpaR protein.
Upstream of the first catabolic gene in the HPC dehydrogenative cluster of E. coli W and in the equivalent hpc cluster of E. coli C, there is a divergently oriented gene, hpaR and hpcR, respectively, that encodes the cognate transcriptional regulator (143, 250, 270) (Fig. 1A). The 148-aa HpaR (HpcR) protein has significant identity to members of the MarR family of bacterial regulatory proteins involved in the metabolism of aromatic compounds, such as the CinR repressor (30% identity) from Butyrivibrio fibrisolvens, which responds to cinnamoyl esters (61), and the BadR activator (27% identity) from Rhodopseudomonas palustris, which most probably responds to benzoyl-CoA (84). The HTH motif associated with the MarR family (3) is also present in the central region of the HpaR (HpcR) regulator. HpaR orthologs within the predicted hpa clusters of other enteric bacteria, such as K. pneumoniae, Salmonella species, and Y. pestis, and from P. aeruginosa have been also identified (see below).
HpcR is a negative regulator of the hpc cluster when expressed in trans from a plasmid in E. coli K-12 harboring the hpc catabolic genes. Moreover, when the hpcR gene is inactivated in E. coli C, there is a constitutive expression of the hpc catabolic genes. HPC and 4HPA relieve the repressor effect of HpcR on the cognate catabolic promoter in vivo (143, 270) (Fig. 1A). A transcriptional start point (+1 site) was found 40 bp upstream from the initiation codon of the first catabolic gene in the hpc cluster, i.e., the hpcE gene (Fig. 1A), and two sequences that match the typical −10 and −35 boxes of ς70-dependent promoters are found at the right distance from such start point. At position −62.5 from the +1 site there is a potential CRP-binding site, which is in agreement with the reported repression by glucose of the HPC catabolism (270). A similar repressor effect on the expression of the hpa meta-cleavage operon, as well as a peculiar CRP-mediated control by glucose, has been characterized for the analogous HpaR regulator from E. coli W (B. Galán, A. Kolb, J. L. García, and M. A. Prieto, submitted for publication).
PaaX protein.
The paa-encoded pathway for PA degradation in E. coli K-12 and W is inducible after addition of PA to the cells. Tn1000 insertions within the paaX gene cause a constitutive expression of the paa-encoded pathway, thus suggesting that the paaX gene product (316 aa) behaves as a negative regulator of the paa catabolic genes (94). Three promoters, Pz, Pa, and Px, which drive the expression of genes paaZ, paaABCDEFGHIJK, and paaXY, respectively (Fig. 5A), have been identified in the paa cluster (94). By using genetic (lacZ translational fusions) and biochemical (gel retardation assays) approaches, the PaaX protein was shown to act as a transcriptional repressor of the catabolic Pa and Pz promoters. The region within the Pa and Pz promoters that is protected by the PaaX repressor in DNase I footprinting assays is about 50 bp and contains a conserved 15-bp imperfect palindromic sequence motif (WWTRTGATTCGYGWT) that was shown, through mutational analyses, to be indispensable for PaaX binding and repression (93). The region protected by PaaX is located inmediately downstream of the transcription start sites within the Pa promoter, and it spans the +1 and the −10 region in the Pz promoter, probably revealing a different mechanism of transcriptional repression in each of these two promoters. PA-CoA but not PA specifically inhibited the binding of PaaX to the target sequences, thus confirming the first intermediate of the pathway (PA-CoA) as the true inducer (93) (Fig. 5A).
The PaaX protein contains a stretch of 25 residues at aa 39 to 64 that has similarity to the HTH motif of transcriptional regulators of the GntR family such as GntR (102) and FadR (76, 318). PaaX is highly similar (41% identity) to the PhaN repressor that controls the pha cluster for PA degradation in P. putida U (see below) and that also recognizes PA-CoA as the inducer molecule (107, 182). A significant similarity (31.5% identity) was also observed to Orf13 of the putative PA catabolic cluster from B. halodurans (see below) and the putative repressor (24.1% identity) of the 4-chlorobenzoate dehalogenation pathway involving CoA derivatives in Arthrobacter sp. strain TM1 (284). Thus, the PaaX and PhaN repressors, as well as the putative regulators mentioned above, may constitute a subfamily of GntR-like regulators that respond to aryl-CoA compounds.
When E. coli cells are grown in PA-containing minimal medium in the presence of their preferred carbon source, glucose, gene expression from the Pa and Pz promoters is subject to carbon catabolite repression. Thus, the operon-specific regulation mediated by the PaaX repressor is subordinated to a superimposed regulation mediated by global regulators such as CRP and IHF, which connect the expression of the paa catabolic genes to the metabolic and energetic status of the cell (93). E. coli CRP-deficient strains failed to express the Pa-lacZ and Pz-lacZ translational fusions, indicating that CRP acts as an activator of the gene expression driven by the Pa and Pz promoters. Gel retardation assays confirmed the binding of the cAMP-CRP complex to the Pa promoter region (93). Since a potential CRP binding site was identified at position −61.5 with respect to the major transcription start site of Pa, this promoter might follow a CRP-dependent activation mechanism similar to that described for class I promoters (40). Although CRP is also necessary for activity of the Pz promoter, no binding of CRP to Pz was observed in gel retardation assays, suggesting that CRP bound to Pa is able to activate the divergent Pz promoter. IHF also binds to the paaZ-paaA intergenic region and stimulates transcription from the Pa and Pz promoters (93). Whether IHF-induced bending of the paaZ-paaA intergenic region can bring together RNA polymerase bound to Pz and CRP bound to Pa is an interesting model that remains to be confirmed.
Overlapping the 3′ end of the paaX gene is the putative translation initiation codon of the paaY gene (Fig. 5A). A palindromic sequence followed by a T7 tract is located 42 bp downstream of the TAA stop codon of paaY and may act as a ρ-independent transcription terminator of the putative paaXY operon (94). Although the primary structures of the paaY gene product (196 aa) and its analogous PhaM protein from P. putida U (182, 217) show several repeats of the hexapeptide motif that characterizes the members of the bacterial transferases family, e.g., the CaiE protein from the carnitine operon of E. coli and the Fbp ferripyochelin binding protein of P. aeruginosa (recently classified as a member of family 3 of carbonic anhydrases) (94), the physiological role of these proteins in PA catabolism is still unknown. Although the paaX and paaY genes (or their homologues) cluster within the PA catabolic operons from proteobacteria of the γ subgroup, only the paaX-like or paaY-like genes are present in the putative PA operons from B. halodurans (gram-positive bacterium) and A. evansii (β proteobacteria), respectively (see below).
TRANSPORT PROTEINS OF AROMATIC COMPOUNDS IN E. COLI
Although aromatic compounds, and particularly the lipophilic weak aromatic acids, can enter the cells by passive diffusion when present at high (millimolar) concentrations, active transport increases the efficiency and rate of substrate acquisition and thus may impart a growth advantage in natural environments, where these compounds are present at low (micromolar) concentrations (211).
Besides the tryptophan permease (TnaB), which is involved in the catabolism of tryptophan (see below), two permeases for the uptake of 4HPA/3HPA and 3HPP/3HCI have been characterized in E. coli. Two potential permeases for the uptake of PP (HcaT) and PA can be also identified through sequence analysis of the E. coli genome. The transporters involved in catabolism of aromatic compounds in E. coli usually belong to the large and diverse major facilitator superfamily (MFS) of transport proteins, whose members generally possess 12 α-helical transmembrane spanners (TMS) (223). These TMSs form a channel for transport through the cytoplasmic membrane, and most of the MFS transport proteins contain a consensus motif [(R/K)XXX(R/K)] between TMS2 and TMS3 and a similar but less well conserved motif between TMS8 and TMS9, which may be important in promoting the global conformational changes that accompany transport (223).
HpaX Permease
When the transport of radioactively labelled 4HPA was analyzed during the growth of E. coli W, the maximum uptake was observed during the early exponential phase. The time course of this transport showed that after 3 min the rate of 4HPA uptake plateaued, and the saturation kinetics revealed a Kt of 25 μM and a Vmax of 3 nmol/min per 109 cells (252). Similar values were observed for the transport of 4HPA in K. pneumoniae M5a1 (4) and 4-hydroxybenzoate in P. putida (211). Since l-tyrosine did not inhibit 4HPA uptake, the transporter involved in the uptake of this aromatic amino acid (see below) seems to be not involved in 4HPA transport (252). Transport of 4HPA was abolished by cyanide and the transport uncoupler 4-nitrophenol; whereas azide and arsenate resulted in a decrease of the transport rate of up to 62 and 21%, respectively, the ATPase inhibitor N,N′′-dicyclohexylcarbodiimide did not affect the transport. These data suggest that 4HPA transport is controlled by an active transport system and not by a passive or facilitated mechanism (252).
The insertion of the hpa cluster from E. coli W into the chromosome of E. coli K-12 conferred on the latter the capacity to transport and metabolize 4HPA (250), indicating that the hpa catabolic cluster also encoded the 4HPA transport protein (252). Analysis of the hpa cluster revealed the existence of a gene, hpaX, that appears to be cotranscribed with the hpaA regulatory gene (Fig. 1A) and whose expression from the Px promoter seems to be constitutive in E. coli K-12 (252). The hpaX gene product (458 aa) shows significant similarity to aromatic transport proteins of the ACS family of MFS transporters (223, 275), such as the phthalate permeases from B. cepacia 17616 (GenBank accession no. AF152094) and P. putida NMH102-2 (GenBank accession no. Q05181), and homologous permeases can be found encoded within the hpa clusters from other bacteria (see below).
While the uptake of 4HPA in E. coli W is inducible by 4HPA, 3HPA, and, to a lesser extent, PA, the structural analogues 2HPA and HPC did not induce 4HPA transport (252). This pattern of induction is similar to that reported for the HpaA regulator, which activates expression of the hpaBC catabolic genes (see above), suggesting that 4HPA transport is tightly linked to its catabolism. Although the uptake activity can be a direct consequence of the expression of a transport machinery, it might be also ascribed to a simultaneous or independent expression of the catabolic pathway that rapidly metabolizes the internalized substrate, maintaining a downhill concentration gradient and allowing the continued diffusion of substrate into the cell by mass action (47). Nevertheless, the role of the HpaX protein as a 4HPA transporter was confirmed by observing a significant uptake and accumulation of 4HPA in E. coli K-12 cells that expressed the hpaX gene but that were unable to metabolize 4HPA since they lacked the hpa catabolic genes (252). By coupling the 4HPA uptake to the expression of a PBC::lacZ reporter fusion regulated by the 4HPA-responsive HpaA activator (see above), it was possible to detect the highest β-galactosidase production in E. coli K-12 cells at a 4HPA concentration as low as 1 μM. In contrast, E. coli cells lacking the hpaX gene reached the maximum β-galactosidase activity at a 1,000-fold-higher concentration of 4HPA inducer (252). Interestingly, while 3HPA was also able to induce the production of β-galactosidase at low concentrations, PA induced only at high concentrations (1 mM) (252). These results are in agreement with the observation that the 4HPA pathway can also metabolize 3HPA but not PA (see above), and they strongly suggest that hpaX encodes a transporter specific for 4HPA and 3HPA. The physiological role of HpaX will be important for optimal growth of E. coli at the low 4HPA/3HPA concentrations that are likely to occur in natural environments. The effects observed at high substrate concentrations (in the millimolar range) most probably should be ascribed to passive diffusion.
MhpT Permease
Located 0.35 kb downstream of the 3′ end of the mhp catabolic operon and flanked by two BIME sequences there is a gene, mhpT (Fig. 2A), which encodes the 3HPP transporter (B. Torres, J. L. García, and E. Díaz, unpublished data). The MhpT protein (418 aa) shows similarity to members of the aromatic acid:H+ symporter (AAHS) family of MFS proteins, a family of transporters that includes most of the permeases so far described for the uptake of aromatic compounds (223). The hydrophilicity profile of MhpT revealed that it could be divided by a central hydrophilic region into two halves, each containing six TMSs (92). A multiple-sequence alignment of members of the AAHS family revealed a family-specific signature sequence between the predicted second half of TMS1 and the end of TMS2 and a common domain at the C terminus of the molecule (223). Remarkably, MhpT is smaller than the other members of the AAHS family, and this is mainly due to its short central hydrophilic region between TMS6 and TMS7. Although MhpT and the putative 3HPP transporter from R. globerulus (HppK) (18) may have a similar substrate specificity, they do not cluster within the AAHS family (223). This divergence is even higher with the putative permease OhpA of the 2HPP pathway from Rhodococcus sp. strain V49 (GenBank accession no. AF274045), a protein that does not show significant amino acid sequence similarity to members of the AAHS family of MFS proteins. This heterogeneity among the putative HPP transporters, together with the fact that some mhp clusters are lacking the gene encoding such a permease (see below), suggests that uptake of HPP in different bacteria may be accomplished through different mechanisms.
HcaT Protein
At the 5′ end of the hca cluster is located a gene, hcaT (Fig. 4A), which encodes a protein with similarity to MFS proteins (71). Interestingly, the predicted HcaT protein (379 aa) is smaller than the majority of MFS proteins (about 400 aa), and some common amino acid sequences that characterize the members of this superfamily are not found in the primary structure of HcaT (71). The HcaT protein shows the highest similarity to nucleoside transport proteins such as NupG and XapB of E. coli (289). Although the HcaT protein has been classified as the only representative of the phenylpropionic permease family of MFS proteins (information found at the Transport Commission website [http://www-biology.ucsd.edu/∼msaier/transport/]), an experimental demonstration that this protein is involved in the uptake of PP in E. coli is still required.
A Putative PA Permease
Although the genes responsible for the catabolism of PA in E. coli and P. putida are homologous, it is remarkable that the phaJ and phaK genes of P. putida U, encoding a permease and a porin for the uptake of PA, respectively (217), are absent in the paa cluster from E. coli (94, 182). However, the PhaJ protein from P. putida U shows significant amino acid sequence identity to the product of the yjcG gene, which is located at 92.2 min in the E. coli K-12 linkage map (EcoGene database at the Colibri website). The PhaJ permease (520 aa) and the putative YjcG transporter (549 aa) cluster within the solute:sodium symporter (SSS) family of carrier-type facilitators. Members of the SSS family catalyze solute uptake via Na+ symport and possess 12 to 15 TMSs with a periplasmic N terminus and a cytoplasmic C terminus (275). Whether a permease, such as the putative YjcG protein, and a channel-forming protein are required for the catabolism of PA in E. coli is still an open question. Alternatively, it is possible that, like for the catabolism of long-chain fatty acids in E. coli (14), the activation of PA to PA-CoA could be linked to the transport of this aromatic acid through the cytoplasmic membrane, dispensing with the need for a permease.
Uptake of Other Aromatic Compounds
An E. coli mutant strain blocked in the common pathway for the biosynthesis of aromatic compounds (multiple aromatic auxotroph) is able to grow well on a medium with glucose as the carbon source in the presence of aromatic amino acids (l-phenylalanine, l-tyrosine, and l-tryptophan, each at 0.1 mM) and some aromatic acids such as p-aminobenzoate and 2,3-dihydroxybenzoate (each at 0.01 mM) and p-hydroxybenzoate (0.1 mM) (54). Thus, E. coli is able to take up aromatic amino acids (see below) and aromatic acids other than those used as carbon sources. However, the transport systems of such aromatic acids are still unknown (240).
Early studies established that E. coli possesses transport systems for uptake of all three of the aromatic amino acids. Thus, five different aromatic amino acid permeases that have the general characteristics of carrier-type facilitators (275) have been reported so far: a general aromatic amino acid permease (AroP), two tryptophan-specific proteins (Mtr and TnaB), a tyrosine-specific protein (TyrP), and a phenylalanine-specific permease (PheP) (240). The genes encoding the aromatic amino acid permeases are spread along the E. coli chromosome at 2.59 min (aroP), 12.96 min (pheP), 42.85 min (tyrP), 71.18 min (mtr), and 83.80 min (tnaB) (EcoGene database at the Colibri website). While expression of pheP appears to be constitutive, the aroP, tyrP, and mtr genes are under the control of the TyrR regulatory protein and therefore form part of the TyrR regulon (240, 350). The AroP, PheP, Mtr, and TyrP permeases are designed primarily to provide aromatic amino acids for protein synthesis and perhaps to prevent the leakage of endogenous pools outside the cell (240). The Mtr permease is also able to transport inside the cell another aromatic compound (indole) when tryptophan synthetase mutants use tryptophanase to convert the exogenously provided indole into tryptophan (343).
Of the five genes encoding aromatic amino acid transporters, only tnaB is organized, together with tnaA and tnaL, in an operon (tryptophan catabolic operon). The role of the specific tryptophan permease TnaB is clearly different from the roles of the other four permeases. TnaB is formed when tryptophan is used as a source of carbon, and it was referred to as a high-capacity, low-affinity system with a Km for tryptophan of 10−5 M (83). The TnaB protein (415 aa) is a tryptophan:H+ symport permease that belongs to the aromatic amino acid permease family of carrier-type facilitators, and it shows a significant similarity to the Mtr and TyrP transporters (275). The hydrophaty profiles, distribution of charged residues, and incidence of proposed β turns in Mtr, TnaB, and TyrP permeases led to a model of polytopic proteins with 11 spans across the membrane, with a cytoplasmically located amino terminus and a periplasmically located carboxyl terminus (240, 279). TnaB synthesis is induced by tryptophan when cells are growing under conditions of no catabolite repression. The TnaA tryptophanase is induced at the same time, and this enzyme degrades the tryptophan that is transported into the cell (see below). The regulation of the tna operon by substrate induction coupled to catabolite repression ensures that this catabolic pathway is not expressed unless it can be useful, hence avoiding the depletion of the expensive intracellular pool of tryptophan (194). The capacity of the TnaB permease to transport tryptophan is approximately 40 times greater than that of the Mtr transporter. This high capacity is clearly required to maintain internal pools of tryptophan in the presence of high levels of tryptophanase when tryptophan is used as a carbon source (240, 343).
OTHER ENZYMATIC ACTIVITIES ACTING ON AROMATIC COMPOUNDS IN E. COLI
Some of the proteins involved in the metabolism of different aromatic compounds in E. coli resemble those described above for the mineralization of aromatic acids, or they catalyze reactions similar to those found during the catabolism of certain aromatic compounds in other bacteria. These enzymatic activities are reviewed here.
Catabolism of Heterocyclic Aromatic Compounds in E. coli
Although wild-type E. coli K-12 does not degrade heterocyclic aromatic compounds, successive mutations yielded some strains that were able to use as the sole carbon and energy source some thiophene and furan derivatives such as furfuryl alcohol, furan-2-carboxylate, ascorbic acid, thiophene-2-carboxylate, thiophene-2-acetate, and thiophene-2-methylamine. However, none of these strains releases inorganic sulfur from thiophene derivatives, and hence these compounds cannot be used as sulfur sources (1). The pathway for the catabolism of thiophene and furan derivatives in such E. coli mutant strains is still unknown (1). Thus, although a pathway for the degradation of furan-2-carboxylate involving initial activation of the carboxyl group with CoA has been proposed (31), formation of thiophene-2-carboxyl CoA thioester was not observed in extracts from the E. coli mutant strains (1). A genetic analysis of the thiophene and furan degrader strains revealed mutations at three different loci, thdA, thdC, and thdD, that have been mapped at 10.5, 92, and 97.5 min, respectively, but uncharacterized. In addition, constitutivity at both the fadR and atoC genes (which encode the regulatory proteins of the long-chain and short-chain fatty acid degradation pathways, respectively) was required for efficient thiophene breakdown, which may reflect the need for the Ato and Fad systems to degrade the putative four- and five-carbon ring fission products (1). The thdC and thdD loci improve the phenotypic response to thiophenes, but probably as a result of increased resistance to toxicity rather than of improved metabolism. The thdA locus may be responsible for controlling several genes, one of which codes for an uncharacterized methylene blue-linked sulfone oxidase activity that can use both heterocyclic and simple aliphatic sulfones as substrate (148). Another gene, thdF, which was mapped at 83 min, confers a thiophene oxidation-positive phenotype when present in a high-copy-number plasmid (2).
Tryptophan Catabolism
Of the three aromatic amino acids, only tryptophan can be catabolized by E. coli and used as carbon source (194). Nevertheless, the early literature contains some reports of the production of phenol from l-tyrosine by certain E. coli strains (316). In this context, a tyrosine-phenol lyase (which hydrolyzes l-tyrosine to phenol, pyruvate and ammonia) has been isolated from Escherichia intermedia and is responsible for the growth of this species on a medium with l-tyrosine as the sole carbon source (166). Therefore, whether the reported tyrosine degraders were authentic E. coli strains or were other Escherichia species remains to be probed.
Most E. coli strains utilize l-tryptophan as the carbon and nitrogen source via an inducible l-tryptophan indole-lyase (l-tryptophanase) (TnaA) and l-tryptophan permease (TnaB, see above) system (194). The TnaA enzyme (Table 4) is a homotetramer with a pyridoxal 5′-phosphate cofactor linked to a lysine residue in each subunit (65, 66, 194). TnaA is a cytoplasmic protein that catalyzes the hydrolytic β-elimination of l-tryptophan to indole (an aromatic product which does not suffer further ring cleavage), pyruvate (used as a carbon source), and ammonia (used as a nitrogen source) (Fig. 7A). Since this reaction is reversible at high concentrations of pyruvate and ammonia, the enzyme can substitute for tryptophan synthetase in an E. coli trpAB mutant strain (194). Amino acid biosynthesis is expensive for the cell, and it is therefore essential that individual amino acids cannot be catabolized unless they are present in excess of the expected cellular requirements. Thus, the relatively high Km (0.33 mM) of TnaA for l-tryptophan could be explained as a mechanism to avoid depletion of the cellular pool of this amino acid (194). l-Tryptophan is transported by E. coli via three permease systems, i.e., AroP, Mtr, and TnaB transporters (see above). While AroP and Mtr are concerned with the provision of amino acids for protein synthesis, the primary function of TnaB is the scavenging of tryptophan as a carbon source (343).
TABLE 4.
Enzymatic reactions on aromatic compounds other than those involved in their mineralization
| Genea | Location (min)b | Gene product | Size (aa) | Substrate | Pathway |
|---|---|---|---|---|---|
| tnaA | 83.8 | l-Tryptophanase | 471 | l-Tryptophan | Tryptophan catabolism |
| ubiD (yigC) | 86.7 | Hydroxybenzoate decarboxylase | 497 | 3-Octaprenyl-4-hydroxybenzoate | Ubiquinone biosynthesis |
| ubiX (dedF) | 52.3 | Decarboxylase | 189 | 3-Octaprenyl-4-hydroxybenzoate, p- and o-aminobenzoate? | Ubiquinone biosynthesis? |
| ubiE | 86.7 | Methyltransferase | 251 | 2-Octaprenyl-6-methoxy-1,4-benzoquinol | Ubiquinone biosynthesis |
| ubiG | 50.3 | Methyltransferase | 240 | 2-Octaprenyl-3-methyl-5-hydroxy-6-methoxy-1,4-benzoquinol | Ubiquinone biosynthesis |
| ubiB (yigR) | 86.7 | Putative kinase involved in hydroxylation reaction | 546 | 2-Octaprenylphenol? | Ubiquinone biosynthesis |
| ubiH (visB) | 65.7 | Monocomponent flavin-type monooxygenase | 392 | 2-Octaprenyl-6-methoxyphenol | Ubiquinone biosynthesis |
| ubiF (yleB) | 14.9 | Monocomponent flavin-type monooxygenase | 391 | 2-Octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol | Ubiquinone biosynthesis |
| entA | 13.4 | Dihydrodiol dehydrogenase | 248 | trans-2,3-Dihydro-2,3-dihydroxybenzoate | Enterobactin biosynthesis |
| entE | 13.4 | AMP ligase | 536 | 2,3-Dihydroxybenzoate | Enterobactin biosynthesis |
| menE | 51.1 | CoA Ligase | 451 | o-Succinylbenzoic acid | Menaquinone biosynthesis |
| nfsA (nfnA) | 19.2 | Nitroreductase IA | 240 | Nitroaromatics (nitrofurazone, nitrobenzoate, nitrophenol, nitrotoluene, etc.) and azo dyes. | Defense against oxidative stress? |
| nfsB (nfnB) | 13.0 | Nitroreductase IB1 (pteridine reductase) | 217 | Recycling pterin cofactors | |
| nhoA | 33.0 | Putative arylamine N-acetyltransferase | 281 | 2-Aminofluorene, p-aminobenzoate | Detoxification? |
| aslA | 85.8 | 551 | |||
| ydeN | 34.0 | Putative arylsulfatases | 571 | Aryl sulfatesd | Catabolism of arylsulfates? |
| yidJ | 83.0 | 497 | |||
| pacc | 98.8 | Penicillin G acylase | 846 | Esters and amides of PA and HPA | Catabolism of natural esters and amides of PA and HPA ? |
Additional gene designations are indicated in parentheses.
The location of the genes is with respect to the E. coli K-12 genome as in Table 3.
The pac gene is present in E. coli W.
The substrates are thought to be arylsulfates because of the amino acid sequence similarities of the three gene products with known arylsulfatases.
The tnaAB genes make up an operon that is located at 83.8 min of the E. coli K-12 linkage map. The tna promoter is highly sensitive to catabolite repression, and tryptophanase expression is reduced 100-fold when the cells grow in glucose. A consensus CRP binding site is located about 60 bp upstream of the transcription start site, and the cAMP-CRP complex is necessary to RNA polymerase binding at that site and to tna transcription initiation in vitro (66). Transcription initiation is not subject to tryptophan control. Rather, the presence of tryptophan induces a transcriptional antitermination mechanism that prevents ρ-dependent termination in the 319-bp leader region of the operon between the transcription start site and the tnaA translation start codon. This leader region contains a short coding region, tnaL or tnaC, encoding a 24-aa leader peptide whose synthesis in the presence of tryptophan blocks the access of ρ to the boxA and rut sites of the transcript and thereby avoids transcription termination (162, 172). Recently, it has been shown that the tnaAB operon is also activated by self-produced extracellular signals (quorum sensing) that are secreted to the medium when the bacterial culture reaches the stationary phase, a growth stage where uptake and catabolism of amino acids may be important for energy production (15).
Enzymatic Reactions in Ubiquinone Biosynthesis
During the biosynthesis of ubiquinone (coenzyme Q) a number of decarboxylation, methylation, and hydroxylation reactions (Fig. 7B) occur that resemble similar reactions carried out during the bacterial catabolism of aromatic compounds.
The third step in the biosynthesis of ubiquinone is the decarboxylation of 3-octaprenyl-4-hydroxybenzoate to 2-octaprenylphenol (Fig. 7B). This decarboxylase activity is absent in ubiD mutants (195). The ubiD (yigC) gene product (Table 4) seems to be an hexameric and membrane-associated protein with similarity to decarboxylases of other organisms such as the 4-hydroxybenzoate decarboxylase from Clostridium hydroxybenzoicum (349). However, a number of ubiD mutants formed about 20% of the wild-type levels of ubiquinone, suggesting that there is an isoenzyme of the UbiD decarboxylase in E. coli. In fact, the ubiX (dedF) gene within the purF operon encodes such isoenzyme (Table 4), which is 48.6% identical to the PAD1 protein (242 aa) from Saccharomyces cerevisiae, an enzyme that decarboxylates CI and other phenylacrylic acids (coumaric and ferulic acids) to the corresponding styrenes (45, 213). A comparison between the amino acid sequences of the ubiX and ubiD gene products revealed no significant similarities (349). Enzymes similar to UbiD and UbiX are responsible for the carboxylation of phenylphosphate during the anaerobic catabolism of phenol in Thauera aromatica (30). Interestingly, it has been reported that E. coli O111:B4 is able to decarboxylate p-aminobenzoic and o-aminobenzoic (anthranilic) acids, giving rise to aniline under aerobic conditions (193). Since the substrate of UbiX (DedF) is also a substituted benzoate, a role for this enzyme in the decarboxylation of p- and o-aminobenzoate could be envisaged (Fig. 8B). A decarboxylase activity acting on p-coumaric and ferulic acids, with formation of the corresponding volatile phenolic compounds, was reported for E. intermedia (179). Whether the UbiX (DedF) protein could be also involved in the partial catabolism of ferulic acid by some E. coli isolates (218) (Fig. 8C) remains to be checked.
FIG. 8.
Biotransformation activities of E. coli on some aromatic compounds. The known or putative proteins catalyzing the different enzymatic reactions are indicated in boldface type and are described in Table 4.
In the biosynthesis of ubiquinone, three hydroxylation reactions alternate with three methylation reactions of the aromatic ring (Fig. 7B). The ubiE and ubiG gene products (Table 4) are two methyltransferases that transfer the methyl group from S-adenosylmethionine to the aromatic ring (195). The three hydroxyl groups introduced at positions 4, 5, and 6 of the benzene nucleus via flavin-linked monooxygenases are derived from oxygen (157) (Fig. 7B). Mutants blocked in each hydroxylation reaction were isolated and designated the ubiB, ubiH, and ubiF mutants. Interestingly, when grown anaerobically, these mutants were able to synthesize ubiquinone, indicating that specific hydroxylases which use oxygen atoms derived from water are involved in the anaerobic pathway for the synthesis of ubiquinone, but these hydroxylases are still unknown (195). The first hydroxylation reaction in the biosynthesis of ubiquinone is the monooxygenation of 2-octaprenylphenol to the corresponding catechol (2-octaprenyl-6-hydroxyphenol) (Fig. 7B), a compound that has never been isolated and characterized and may not exit as a free product but rather as an enzyme-bound intermediate. The ubiB (yigR) gene product seems to be involved in this hydroxylation reaction (195, 244) (Table 4). The ubiB gene product does not show sequence identity to other monooxygenases, but it has similarity to the ABC1 protein of S. cerevisiae, revealing motifs found in eukaryotic-type protein kinases. Although it is not known whether UbiB has kinase activity and the substrates on which it may act, this protein may play a role in ubiquinone biosynthesis by activating, via phosphorylation, the unknown protein(s) necessary for the monooxygenase step (244). The second and third monooxygenation reactions in ubiquinone biosynthesis are those catalyzed by the ubiH (visB) and ubiF (yleB) gene products (Fig. 7B; Table 4), which show in their primary structure the three regions of special relevance in monocomponent flavin-type aromatic hydroxylases (see above) (168, 207). Immediately downstream of ubiH (visB) is another gene, visC, that encodes a protein whose primary structure shows 30.5% identity to that of UbiH. Although VisC presents monooxygenase activity on aromatic compounds, it seems to be unnecessary for the production of ubiquinone (207). Therefore, although an extensive biochemical characterization of the ubiH, visC, and yleB gene products needs to be done, it appears that E. coli possesses at least four different monocomponent flavin-type aromatic monooxygenases, i.e., VisC, MhpA for 3HPP/3HCI catabolism (see above), and UbiH and UbiF for ubiquinone biosynthesis.
Enzymatic Reactions in Enterobactin and Menaquinone Biosynthesis
The entCEBA genes responsible for enterobactin (a catecholate siderophore) biosynthesis are clustered on the E. coli chromosome at 13.4 min (181). Interestingly, the entA and entE genes code for enzymes that show similarity to proteins involved in the catabolism of aromatic compounds. The entA gene product converts 2,3-dihydro-2,3-dihydroxybenzoate into the aromatic 2,3-dihydroxybenzoate (181) (Table 4; Fig. 7C). The EntA dihydrodiol dehydrogenase has similarity to dihydrodiol dehydrogenases involved in catabolism of aromatic compounds, although its C terminus does not show the consensus pattern present in such dehydrogenases. Although most dehydrogenases are tetramers, the native EntA protein is an octamer (181). The EntA protein, unlike other aromatic diol dehydrogenases from bacteria, uses a trans-diol substrate (trans-2,3-dihydro-2,3-dihydroxybenzoate) and proceeds by oxidation of the C-3 alcohol group to the corresponding ketone (2-hydroxy-3-oxo-4,6-cyclohexadiene-1-carboxylate), which undergoes rapid aromatization to give 2,3-dihydroxybenzoate. The stereospecificity of the C-3 allylic alcohol group oxidation was confirmed to be 3R in a 1R,3R-dihydro substrate, and hydride transfer occurs to the si face of enzyme-bound NAD+ (276). It is worth noting that EntA and HcaB (see above) have 23% amino acid sequence identity and that they are the only dihydrodiol dehydrogenases so far reported in E. coli that catalyze the formation of an aromatic compound.
The entE gene product is a homodimer that participates in the second phase of enterobactin biosynthesis by activating 2,3-dihydroxybenzoate to (2,3-dihydroxybenzoyl)adenylate, consuming ATP and releasing PPi (274) (Fig. 7C; Table 4). The resulting acyladenylate remains bound to the enzyme for further reactions in the overall biosynthesis of enterobactin (85). Although the natural substrate, 2,3-dihydroxybenzoate, is by far the best substrate, additional aromatic substrates have a requirement for a hydroxyl group ortho to the carboxyl group, and it appears that the enzyme can tolerate substituents at all the remaining positions of the aromatic ring except the sixth position (274). The EntE protein belongs to the thiol-template synthetases, a family of enzymes that cluster within the acyl-adenylate-forming enzyme superfamily (43). Two additional acyl-adenylate-forming enzymes that use aromatic compounds as substrates in E. coli are the PA-CoA ligase (PaaK) (see above) and the MenE protein, which acts on o-succinylbenzoic acid during the biosynthesis of menaquinone (vitamin K2) (Fig. 7D; Table 4). The three proteins share the three conserved motifs that characterize the members of the acyl-adenylate-forming enzyme superfamily (43), although their global amino acid sequence identity is below 20%.
Penicillin G Acylase
Penicillin G acylase (Pac) is a member of a large enzyme family known as β-lactam acylases because they are industrially used for the semisynthesis of β-lactam antibiotics (317). Although several Pac proteins have been identified in different microorganisms, the best-studied enzyme is that from E. coli W (317) (Table 4). Although Pac is used mainly because of its capacity to hydrolyze penicillin G into PA and 6-aminopenicillanic acid (Fig. 8A), it does not seem to play a role in bacterial antibiotic resistance (182, 196). The production of Pac in E. coli is subjected to different regulatory controls at the translational and transcriptional levels (196, 216). Pac has a very unusual structure within the prokaryotic enzymes since it is produced as an inactive precursor that is secreted to the periplasmic space after the hydrolysis of the signal peptide. Then the enzyme becomes active by an autoproteolytic process that releases an internal peptide and divides the protein into two dissimilar subunits (α and β) (137). Pac activity is consistent with an acyl enzyme mechanism involving the N-terminal serine of the β chain as the active site (327). The three-dimensional structure of the Pac protein is known (80), and the enzyme has been classified within a new Ntn-hydrolase family (308). The pac gene of E. coli W is located in the vicinity of the hpa cluster (250) (Fig. 1A) and is transcribed as a monocistronic unit using different alternative promoters (190, 263). The expression of the pac gene is induced by PA and phenoxyacetic acid and repressed by metabolic carbohydrates and polyalcohols (glucose, fructose, and glycerol) (309). Although the repression by glucose appears to be a typical CRP-dependent process that is partially overcome by the addition of cAMP (106, 196, 263, 300), the regulatory proteins involved in PA induction remain unknown. In this context, a pacR regulatory gene located inside the pac structural gene and transcribed in the opposite direction to pac has been identified (144). The PacR repressor mediates the PA-dependent induction of the pac gene (60). However, induction of pac expression by PA has also been observed using Ppac::lacZ fusions in the absence of the pac gene (196, 263). Involvement of IHF as an additional regulatory protein in pac transcription was also reported (300).
The Pac enzyme has a broad substrate range, being able to hydrolyze different esters and amides of PA and of many derivatives, e. g., 4HPA (Table 4). Moreover, it hydrolyzes esters and amides of other aromatic and aliphatic acids such as thyenylacetic acid and hexanoic acid (327). Therefore, it appears to play a role as a scavenger enzyme for many different natural esters and amides which after their hydrolysis can be transported and mineralized by central catabolic pathways (182). In this sense, Pac activity was shown to confer on E. coli the ability to use the PA group of penicillin G as a carbon source when this bacterium also carries a β-lactamase gene (196). The hypothesis that Pac is an enzyme involved in degradation of phenylacetylated compounds is further supported by the thermoregulation of its autocatalytic activation. This autoproteolytic process is more efficient at 28°C, which explains why the production of Pac decreases when E. coli is cultured above this temperature and is practically negligible at 37°C (60, 216). Therefore, when E. coli abandons its intestinal habitat, the lower temperatures might act as a signal for the detection of the new environment and, in conjunction with the phenylacetylated compounds present in such ecological niche, activation of the Pac enzyme would take place and the generated aromatic acids could then be used as carbon source for the cells (317).
Reduction of Nitroaromatic Compounds
E. coli is able to reduce nitroaromatic compounds (many of which are toxic or mutagenic) to the corresponding amines via two types of enzymes, the oxygen-insensitive (type I) and oxygen-sensitive (type II) nitroreductases (188). The oxygen-sensitive nitroreductases catalyze nitroreduction through single-electron increments, yielding an additional product, the nitroanion free radical, that in the presence of oxygen accounts for the oxygen sensitivity of these enzymes (238). E. coli cells grown anaerobically are able to reduce two of the nitro groups of 2,4,6-trinitrotoluene (TNT), generating 2,4-diamino-6-nitrotoluene (Fig. 8D), and a cell extract of anaerobically grown E. coli in the presence of added hydrogen donor is able to completely reduce TNT to 2,4,6-triaminotoluene (187). However, the genes encoding type II nitroreductases have not yet been identified in E. coli. Metabolism of nitroaromatic compounds by the oxygen-insensitive nitroreductases proceeds through a two-electron reduction of the nitro group and yields nitroso and hydroxylamine intermediates and an amino-substituted product. Three type I nitroreductases have been found in E. coli (332). The nfsA gene product, the major E. coli nitroreductase (nitroreductase IA) (Table 4), is a homodimeric FMN-containing protein that uses NADPH as an electron donor (347). The nfsB gene encodes the minor nitroreductase, previously referred to as IB1, an FMN-containing flavoprotein that can use either NADH or NADPH as a source of reducing equivalents and whose three-dimensional structure has been determined (33, 346). The NfsA and NfsB flavoenzymes catalyze the divalent reduction of a wide variety of nitroaromatics: nitrofurazone, nitrofurantoin, methyl 4-nitrobenzoate, 4-nitrobenzoate, 4-nitroacetophenone, 4-nitrotoluene, 4-nitrophenol, 4-nitroaniline, and some azo dyes (347) (Fig. 8D; Table 4). Aerobically growing E. coli cells are also able to reduce TNT to 2,4-diamino-6-nitrotoluene (192) (Fig. 8D).
The NfsA and NfsB nitroreductases from E. coli show a close evolutionary relationship to the Frp and Frase I reductases, respectively, from luminescent Vibrio strains. In this sense, the crystal structure of NfsA is similar to that of Frp (159). However, in contrast to the two luminescent bacterial enzymes, the E. coli enzymes exhibit little or no FMN reductase activity, suggesting that progenitors of the NfsA-Frp and NfsB-Frase I pairs lost FMN reductase activity during evolution in E. coli cells or acquired FMN reductase activity during evolution in luminescent bacteria (347). The physiological role of nitroreductases in E. coli is unknown. Recent work shows that the nfsA gene is a member of the soxRS regulon, and therefore it is likely that nitroreductase A1 contributes to the defenses against oxidative stress. The paradoxical findings that mutants lacking nitroreductase NfsA or NfsB were more resistant to some nitroaromatics may be due to the greater toxicity of the products of reduction of those nitro compounds (198, 332). However, the nitroaromatic compounds used in the previous studies are not the ones that elicited the evolution of nitroreductases. It might be supposed that nitro compounds produced by nitration of endogenous targets, such as tyrosyl residues and quinones, impose a toxicity by autoxidation that can be avoided by the action of the oxygen-insensitives nitroreductases (180, 332). An additional function of nitroreductase NfsB in E. coli could be that of recycling pterin cofactors (322) (Table 4).
Arylamine N-Acetyltransferase Activity
Arylamine N-acetyltransferases (NATs) are cytosolic enzymes which acetylate arylamines and hydrazines by transfer of the acetyl group from acetyl-CoA to the free amino group, forming an acetylamide, or to the oxygen of an arylhydroxylamine, generating unstable N-acetoxy species. E. coli shows NAT activity toward isoniazid, 2-aminofluorene, and p-aminobenzoate (Fig. 8E), and this activity is inhibited by idoacetamide and divalent cations (42, 232). During a search of the E. coli genome for putative NATs, it was found that the nhoA gene product showed the common features of NAT proteins (231, 232) (Table 4). The existence of highly conserved NAT sequences in bacteria suggests that the protein is conserved in evolution. Although the role of this enzyme in endogenous metabolism is unclear, the activity profile of NATs in prokaryotes is more likely to be the detoxification of carcinogens and drugs (231, 232).
Arylsulfatase-Like Genes
Arylsulfatases hydrolyze arylsulfate esters to the corresponding phenols and inorganic sulfate (153). In E. coli an arylsulfatase-like protein (AslA) has been detected by cross-reaction with antibodies raised against the well-known AtsA arylsulfatase from K. aerogenes; however, no activity of the AslA protein has been reported (341). The presence in the E. coli genome of three genes encoding two putative Ser-type arylsulfatases (AslA and YdeN) (62) and a Cys-type arylsulfatase (YidJ) (Table 4) is particularly interesting because this organism does not grow with sulfate esters as sulfur sources. It will be interesting to learn whether these are all pseudogenes or whether the encoded proteins are expressed under conditions that have not yet been discovered and catalyze the desulfation of substrates that have not yet been tested (153).
Dehalogenation Reactions
The toxic γ-hexachlorocyclohexane, known as the insecticide lindane, gives rise to aromatic compounds during its catabolism by various microorganisms (294). The aerobic conversion of lindane to γ-pentachlorocyclohexene, a compound that is 1,000 times less toxic than lindane, was observed for the first time with a pure culture of an E. coli strain isolated from rat feces (100). A similar conversion is catalyzed by the LinA dehydrochlorinase in Sphingomonas paucimobilis UT26 (294), but no LinA equivalent can be identified in E. coli by amino acid sequence comparison, and hence the mechanism and genes responsible for such a reaction remain to be characterized in E. coli. Anaerobic degradation of lindane has been found in both strictly and facultatively anerobic bacteria. Several enteric bacteria such as E. coli DSM 30083 (serotype O1:K1:H7; pathogenic to chickens) are able to attack lindane under anaerobic conditions. All species formed γ-tetrachlorocyclohexene as the main intermediate metabolite, and no γ-pentachlorocyclohexene was observed. The mechanism of dechlorination in these enteric bacteria has not yet been characterized (142).
Dechlorination of the 1,1,1-trichlor-2,2-bis(p-chlorophenyl)ethane (DDT) peticide to 1,1,-dichloro-2,2-bis(p-chlorophenyl)ethane (DDD) has been reported to occur in pure E. coli culture systems, (147), but the genes and proteins involved in this process are still unknown.
EVOLUTIONARY CONSIDERATIONS ABOUT THE AROMATIC CATABOLIC CLUSTERS OF E. COLI
The HPA cluster of E. coli W is a 11.4-kb DNA catabolic cassette (250) (see above). Putative HPA catabolic clusters can be also found in other enteric bacteria such as Salmonella species, K. pneumoniae, and Y. pestis (Fig. 9). The order of the genes within the hpa clusters from S. enterica Dublin and serovar Typhimurium (hpaS) and from Y. pestis (hpaYp) is similar to that found in E. coli, with the only exceptions being that the hpaA gene is absent in hpaYp and that the hpaBC genes, encoding a putative HPA monooxygenase, were subject of an inversion and are located upstream of hpaR in hpaS (Fig. 9). In K. pneumoniae (185) the hpa genes are arranged in two clusters, the hpaXSAH cluster encoding the HPA upper route and the hpaRG1G2EBFH cluster encoding the HPC dehydrogenative route, separated by an unknown distance. Although the order of the genes is similar to that found in the hpa cluster of E. coli, the hpaI gene is absent in the hpaKp clusters (Fig. 9). In a nonenteric bacterium such as P. aeruginosa (58, 301), the putative hpa genes are organized in three clusters, hpaBC, hpaAG1G2EDFXHI, and hpaR, and they show an arrangement different to that observed in enteric bacteria (Fig. 9). Thus, the hpaX gene (which encodes a putative HPA transporter) from P. aeruginosa is located within the HPC dehydrogenative cluster and the two regulatory genes, hpaA and hpaR, are located far from the genes that they are supposed to regulate. Interestingly, the hpaG gene in E. coli and Salmonella species seems to be a fusion of the tandem hpaG1 and hpaG2 genes present in the hpa clusters of Y. pestis, K. pneumoniae, and P. aeruginosa. The genes flanking the hpa clusters differ from one species to another, revealing the mobile character of this catabolic DNA cassette. In E. coli, the hpa catabolic cassette does not contain a recombinase, indicating that it is not a typical transposable element and opening new insights on the evolutionary mechanisms that enhance bacterial adaptability (250). Since most enterics utilize 4HPA/3HPA (38), it is likely that the strains of E. coli that do not grow on these aromatic compounds as the sole carbon source, e.g., E. coli K-12, have lost the hpa cluster present in the original ancestor cell.
FIG. 9.
Comparison of hpa clusters involved in HPA catabolism in different proteobacteria from the γ subgroup. Arrows show the directions of gene transcription. Genes are indicated by blocks: red (regulatory genes), blue (transport genes), green (genes encoding the initial HPA monooxygenase), yellow (ring cleavage dioxygenase gene), and purple (genes encoding the meta-cleavage dehydrogenative route). A broken line indicates an unknown distance. The hpaR gene from K. pneumoniae corresponds to the previously characterized moaI gene (in parentheses). References of the sequences are as follows: E. coli (strain W) (accession no. Z37980), S. enterica serovar Dublin (accession no. AF144422), S. enterica serovar Typhimurium and Y. pestis (obtained from the ERGO database website) K. pneumoniae (accession no. L41068 and AJ000054 and ERGO database), and P. aeruginosa PAO1 (Pseudomonas Genome Project, at http://www.pseudomonas.com/).
A gene cluster similar to that for 3HPP degradation in E. coli can be also found in the genome of K. pneumoniae (Fig. 10). Outside the γ subgroup of proteobacteria, a mhp-like cluster has been identified and characterized in C. testosteroni TA441, a 3HPP degrader that belongs to the β subgroup of proteobacteria (10) (Fig. 10). Although the mhp genes from strain TA441 are homologous to those from E. coli, a different gene arrangement can be observed between the two mhp clusters. Thus, in C. testosteroni the mhpC gene is located at the 3′ end of the cluster and two orf genes (orf4 and orf5) encoding proteins of unknown function are inserted between the mhpD and mhpF genes. The mhpT gene encoding the 3HPP transporter of E. coli was not found in the mhpCt and mhpKp clusters, although a mhpT homolog can be found by sequence comparison in the genome of K. pneumoniae (ERGO database at website http: //wit.integratedgenomics.com/IGwit/). The mhpCDFE genes from E. coli have the same organization and appear to be homologous to other hydrolytic meta-cleavage pathway genes such as xylFJQK (toluene/xylene degradation), dmpDEFG (phenol degradation), and nahNLOM (naphthalene degradation) from different Pseudomonas strains (129, 321, 335). It is possible that the proteobacterial meta-cleavage pathways for catechol and catechol-like derivatives have coevolved with the different proteobacterial subgroups that are all derived from a common ancestor (123). This would agree with the observation that the gene order in meta-cleavage pathway clusters is similar in E. coli, Klebsiella, and Pseudomonas (which are all from the γ subgroup), but differs from that in C. testosteroni (which is from the β subgroup). Nevertheless, this segregation is not the only factor contributing to the evolution for these operons since horizontal gene transfer between proteobacterial subgroups has been extensively reported (123). The hpp and ohp clusters (Fig. 10) containing genes responsible for the catabolism of 3HPP and 2HPP have been characterized in the gram-positive bacteria Rhodococcus globerulus PWD1 and Rhodococcus sp. strain V49, respectively (18, 247). In both cases, the clusters do not account for the complete mineralization of the aromatic compound since they lack the genes encoding the last three enzymatic steps, i.e., the hydratase, aldolase, and acylating aldehyde dehydrogenase enzymes (Fig. 2 and 10). The order of the regulatory, transport, and catabolic genes is different in the hpp and ohp clusters, and it also differs from that observed in the mhp clusters from gram-negative bacteria (Fig. 10). Although the HPC (dehydrogenative) and DHPP (hydrolytic) meta-fission routes coexist is some E. coli strains such as W and C (Table 2), sequence comparison and gene arrangement analyses revealed that these two routes have arisen independently, with only the hydratase function being recruited from the same ancestral source.
FIG. 10.
Comparison of gene clusters involved in HPP catabolism in different bacteria. Arrows show the directions of gene transcription. Genes are indicated by blocks: red (regulatory genes), blue (transport genes), green (genes encoding the HPP monooxygenase), yellow (ring cleavage dioxygenase gene), orange (genes encoding the meta-cleavage hydrolytic route), and black (genes of unknown function). The ohpR gene from Rhodococcus sp. strain V49 encodes a regulator that, in contrast to the regulators of the other four gene clusters, does not belong to the IclR protein family. β and γ indicate the β and γ subgroups of proteobacteria. The references of the sequences are as follows: E. coli strain K-12 (EcoGene database), K. pneumoniae (ERGO database), C. testosteroni TA441 (accession no. AB024335), R. globerulus PWD1 (accession no. U89712), and Rhodococcus sp. strain V49 (accession no. AF274045).
The hca catabolic cluster encoding the PP dioxygenase from E. coli is similar to analogous clusters encoding class IIB dioxygenases from different microorganisms (11, 259). However, it is remarkable that the hcaD gene encoding the reductase component is separated from the hcaEFC genes, encoding the other two components of the PP dioxygenase, by the gene (hcaB) that codes for the next enzyme of the pathway (Fig. 4). On the other hand, while a close association between the csiE (a gene flanking the hca cluster in E. coli) (Fig. 4A) and hcaT genes has been also found in the genomes of other enteric bacteria such as K. pneumoniae, Y. pestis, and E. enterica serovar Typhimurium (ERGO database), the remaining hca genes have not been so far identified in the latter organisms. The linkage between hcaT and the hcaR regulatory gene reinforces the putative role of HcaT as a PP transporter in E. coli (see above). In this sense, a close association between the genes encoding the regulator and the transporter in clusters responsible for the catabolism of aromatic compounds has been also found for the permease HpaX and the transcriptional activator HpaA in E. coli W (Fig. 1A). A similar gene arrangement was reported for the permeases PcaK (4-hydroxybenzoate transporter) and HppK (putative 3HPP transporter) and the corresponding transcriptional regulators PcaR and HppR in P. putida and R. globerulus, respectively (18, 132, 211). In Acinetobacter sp. strain ADP1, the benK and benM genes, which encode a benzoate transporter and a transcriptional regulator that responds to this aromatic acid, respectively, are also contiguous in the genome but transcribed in opposite directions (47). It appears reasonable to assume that the observed association of genes encoding regulators and transporters in some catabolic clusters may reflect the fact that permeases for aromatic compounds can be indirectly involved in the regulation of the catabolic pathways by bringing these substrates (inducers) inside the cell and hence leading to the induction of their respective regulatory proteins.
Two genes, yjiY (orf12 and orf13 in E. coli W) and csiE, encoding putative carbon starvation proteins, are flanking the hpa and hca clusters, respectively (Fig. 1A and 4A). Moreover, the cstA gene encoding the CstA carbon starvation protein is also located downstream from a gene cluster (entCEBAybdB) which encodes enzymatic activities responsible for the metabolism of an aromatic compound (2,3-dihydroxybenzoic acid) during the formation of enterobactin in E. coli (181) (EcoGene database). Whether carbon starvation proteins could play a role in the metabolism of aromatic compounds is unknown, but it has been demonstrated that these proteins are induced by different aromatic pollutants (28), which could reflect, in some cases, an adaptive response of the bacterial cell to the lack of an easily fermentable carbon source.
A paa cluster similar to that of E. coli can be found through sequence comparisons in the genomes of other enteric bacteria such as K. pneumoniae (Fig. 11). However, the gene arrangement within the paa cluster in enteric bacteria differs significantly from that found in P. putida strains (GenBank accession no. AF029714, and database referenced at website http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html). Thus, while the pha genes from P. putida U and P. putida KT2440 appear to be cotranscribed in four discrete DNA segments or modules encoding the six different functional units for the catabolism of PA, i.e., the β-oxidation and activation (phaABCPDE), hydroxylation (phaFOGHI), transport and dearomatization (phaJKL), and regulation (phaMN) units (182, 217), the paa cluster from E. coli shows the transcriptional coupling of the hydroxylation, β-oxidation, and activation functional units into the single paaABCDEFGHIJK operon (94, 182) (Fig. 5 and 11). Since there is good evidence that operons coding for the catabolism of aromatic compounds are assembled in a stepwise manner from existing catabolic genes (321), it is tempting to speculate that the paa cluster from E. coli arose by the fusion of some gene blocks that are contiguous but separately regulated in the pha cluster of P. putida and that it could therefore be considered a further step in the evolution toward a single regulon of a common ancestral gene cluster involved in PA catabolism. Moreover, the differences in gene order within some of the DNA modules and the relative locations of these modules in the clusters of P. putida and E. coli suggest that various DNA rearrangements have occurred during their evolution within each particular host (94, 182). Gene context conservation of a higher order than operons has been called uber-operon (173), and the genes responsible for PA degradation therefore, constitute a clear example of such a conserved context, since although the exact neighborhood of each particular gene is not necessarily conserved, the gene is invariably maintained in a transcriptional neighborhood of associated genes from a discrete set. Especially remarkable is the observation that the phaJ and phaK genes of P. putida U, encoding a permease and a specific channel-forming protein for the uptake of PA, respectively (217), are absent in the paa cluster from E. coli (182) (Fig. 11). The paa cluster from A. evansii resembles the pha cluster from P. putida, but it lacks the transport and some catabolic (paaFJ) genes and contains a truncated paaZ gene (paaZ∗) in the vicinity of paaY (Fig. 11). This gene arrangement might indicate that during evolution the paa cluster was broken up in A. evansii and some of the genes are now present in separate genomic locations. In this sense, two putative regulatory genes, paaY and orf3 (which encodes a putative transcriptional regulator of the TetR family), are flanking the paa cluster in A. evansii (Fig. 11). The breakup of paa clusters is even greater in gram-positive bacteria. Thus, in the genome of B. halodurans the putative paa cluster contains truncated paaH (paaH∗) and paaZ (paaZ∗) genes and lacks paaE and paaY orthologs (310, 311) (Fig. 11). In D. radiodurans (331), while a putative PA hydroxylation unit lacking the paaE gene is located close to a paaZ ortholog within chromosome 1, a paaK homolog is located in chromosome 2, and the putative regulatory, transport, and β-oxidation-like genes are absent from these clusters (182) (Fig. 11).
FIG. 11.
Comparison of gene clusters involved in PA catabolism in different bacteria. Arrows show the directions of gene transcription. Genes are indicated by blocks: red (regulatory genes), dark blue (transport genes), light blue (genes encoding the PA-CoA ligase), green (genes encoding a putative multicomponent oxygenase), brown (genes encoding a putative ring cleavage enzyme), purple (genes encoding a putative β-oxidation-like pathway), and dotted (genes of unknown function). The paaI ortholog in P. putida U has not been described previously (182, 217), and it was named phaP here. orf3 is an incomplete orf gene that encodes a putative transcriptional regulator of the TetR family. An asterisk indicates a truncated gene. A discontinuous line means that the sequence has not been yet reported. The broken line shows that the genes are not adjacent. γ and β represent the γ and β subgroups of proteobacteria. The references of the sequences are as follows: E. coli (strain W) (accession no. X97452), K. pneumoniae (ERGO database), P. putida U and KT2440 (accession no. AF029714 and database referenced at website http: //www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html, respectively), A. evansii KB740 (paa or pac genes under accession no. AF176259 or AJ278756, respectively), D. radiodurans R1 (accession no. AE002069), and B. halodurans C-125 (accession no. AB011837).
Althoug aromatic catabolic pathways are frequently plasmid encoded (129), all the aromatic catabolic clusters described in E. coli have a chromosomal location. The G+C content of the hpa, mhp, hca, and paa clusters averaged 54.4, 55.7, 53.8, and 52.5%, respectively. These values are close to the mean G+C content of the genomic E. coli DNA (51.5%) (208) and reflect the fact that these sets of catabolic genes have been imprisoned within the chromosome of this enterobacterium over a long period of evolution. In contrast to some bacterial strains, such as Acinetobacter sp. strain ADP1, where the aromatic catabolic genes are clustered in a limited region of the chromosome (227), this clustering is not observed in E. coli. In this enteric bacterium the aromatic catabolic clusters are dispersed, with cluster mhp at min 8, paa at min 31.1, hca at min 57.5, and hpa (absent in E. coli K-12 but present in other E. coli strains) at min 98.8 of the E. coli K-12 genome (see Fig. 13). However, the physical association between genes belonging to the same catabolon may constitute an important evolutionary and adaptive advantage (182, 217), and some examples can be found in E. coli. For instance, E. coli W contains near the hpa cluster the pac gene encoding the penicillin G acylase enzyme, which hydrolyzes a wide range of amides and esters of 4HPA (see above), thus releasing this aromatic acid and feeding it into the hpa-encoded 4HPA catabolic pathway. A comparison of the pac flanking sequences with the tsr-cst-yijA-mrr region of E. coli K-12 revealed that the pac gene has been inserted just downstream of the stop codon of the yjiA (orf14) gene in E. coli W (250) (Fig. 1A). The fact that pac is not present in E. coli strains other than W (254) suggests that this gene is a recent acquisition to improve the ability of this strain to metabolize a wider range of substrates, and it draws attention to the selective forces that may favor supraoperonic clustering of genes that channel new substrates into a common catabolic pathway (catabolon). The clustering of genes responsible for the hydrolysis of aromatic esters with those encoding the catabolism of the aromatic moiety has been also described in other bacteria (146). Another example of such clustering in E. coli are the mao genes, responsible for the catabolism of PEA to PA, which lie adjacent to the paa cluster involved in PA degradation (94, 182) (Fig. 5A and 6A).
FIG. 13.
Schematic representation of the gene clusters and the encoded catabolic pathways for the aerobic degradation of aromatic compounds in E. coli. The gene clusters mhp, mao, paa, hca, hpau (upper route), and hpam (meta-cleavage route) are indicated by blocks and correspond to those described in Fig. 1 to 6. The locations of the clusters refer to the E. coli K-12 linkage map (the hpa cluster is absent in E. coli K-12). The regulatory proteins are indicated by different symbols that reflect the different regulatory protein families. + and − indicate transcriptional activation and repression, respectively. The transporters are represented by thick arrows spanning the cellular envelope. Abbreviations of the metabolites and enzymes are the same than those used in Fig. 1 to 6. The different colors correspond to the different catabolic pathways (or to different routes within the same pathway). A discontinuous arrow indicates more than one enzymatic step. TCA, tricarboxylic acids.
Despite their chromosomal location, most of the aromatic catabolic clusters from E. coli are closely linked to mobile genetic elements that could facilitate their mobilization. Thus, the mhp and mao-paa clusters are located at chromosomal regions that are rich in IS2 and IS30 insertion elements (26) (EcoGene database) (Fig. 5A). Moreover, the presence of BIME and REP sequences (16) within the mhp and hpa clusters, respectively (Fig. 1A and 2A), might also contribute to the spread and genetic rearrangements of these clusters. Horizontal transfer of such catabolic cassettes will enhance bacterial adaptability and could explain the heterogeneity observed among different E. coli strains respect to their ability to mineralize aromatic compounds (Table 2). In this sense, gene comparison among E. coli strains suggests that the mao-paa cluster, which lies between an acknowledged horizontally transferred gene and an IS element in E. coli K-12 (214) and which is missing from other E. coli strains such as O157:H7, ECOR37, and ECOR40 (214) and E. coli C (94), represents a horizontally transferred sequence (despite having a base composition similar to that of the E. coli chromosome) that was not present in the ancestral E. coli (214). Interestingly, a gene cluster likely to be involved in 3-hydroxybenzoate and gentisate degradation is located in the genome of strain O157:H7 as a catabolic cassette between two genes, yohG and yohI, that map at min 48 of the E. coli K-12 chromosome. Since a similar catabolic cassette is also present in the genome of S. enterica serovar Typhimurium, one can argue that this gene cluster was present in the common ancestor of all present-day E. coli strains but then it was lost in a number of lineages such as, in E. coli K-12 (M. J. Hernáez, J. L. García, and E. Díaz, unpublished data). In contrast, the hca cluster present in E. coli K-12 is absent in three Salmonella species and in the more distant relative K. pneumoniae (191), suggesting an unique acquisition by the former.
It has been suggested that pathways for the catabolism of aromatic compounds widely available in nature, such as those used by E. coli as carbon source, are among the most ubiquitous aromatic catabolic systems and that they are closer to central metabolism than are those involved in the degradation of xenobiotic compounds (18). These pathways that occupy central positions within the secondary metabolism may have been one of the commonest sources for the initial recruitment of genes for many of the routes involved in the degradation of anthropogenic compounds, which are more peripheral to the natural carbon cycle (18). In this sense, since the whole structure and organization of the hca cluster resembles that of clusters responsible for initial dioxygenation of the highly recalcitrant polychlorobiphenyls (PCBs) and chlorinated benzenes (259), it seems likely that these peripheral pathways have evolved from a central one, such as hca, through mutation, recombination, and gene transfer events. Since lignin is one of the major natural sources of phenylpropanoid compounds, it is worth mentioning that PCB degraders have been found associated with plant lignin degraders (155). Moreover, it has been postulated that in the PCB degrader Rhodococcus sp. strain RHA1, the meta-cleavage pathway genes could have evolved from the same ancestor as the HPC meta-cleavage genes of E. coli (189). Therefore, it is tempting to speculate that the catabolic pathways for mineralization of PCBs (and other xenobiotic compounds) might be derived from aromatic central pathways through assembling of hca-like clusters with mhp- or hpa-like clusters.
Several genome sequencing programs for specific E. coli isolates are under way, and they will allow us to define more clearly the E. coli flexible gene pool (see above) (125). Since evolution is highly active on bacteria, more novel genes are expected to enter the E. coli aromatic catabolic gene pool in the future. Natural selection events will finally determine whether such genes will remain in the catabolic pool of this enteric bacterium (78).
BIOTECHNOLOGICAL APPLICATIONS OF THE CATABOLISM OF AROMATIC COMPOUNDS IN E. COLI
Relevant Properties of E. coli To Tackle Environmental Pollution by Aromatic Compounds
Several properties of E. coli facilitate its use as a biocatalyst for biodegradation and biotransformation of aromatic compounds. Some of these properties are discussed here.
Increased solvent tolerance.
Usually aromatic compounds are highly hydrophobic and toxic for microorganisms, mainly because they accumulate in and disrupt cell membranes. However, bacteria resistant to solvents have been isolated and characterized, and they constitute interesting recipient cells to engineer suitable biocatalysts that can be used in environmental biotechnology for removal of aromatic pollutants as well as in biotechnological production processes in two-liquid water-solvent systems (64). The level of aromatic solvent tolerance is variable among E. coli strains, and some mutants able to grow in the presence of toxic solvents such as xylenes have been isolated (7). The increased solvent resistance of the mutants can be due to a less hydrophobic cell surface (8) and/or to the overexpression of efflux pumps (158), such as the AcrA AcrB TolC proton motive force-dependent efflux pump that extrudes organic solvents and multiple hydrophobic antibiotics (9, 64). Consequently, there is a relationship between aromatic-solvent tolerance and antibiotic resistance in E. coli. Thus, organic-solvent tolerance levels increase by overexpression of marA, robA, and soxS, three stress response genes that control the expression of the mar-sox regulon involved in the multiple antibiotic resistance response of E. coli (9). Some aromatic solvents such as tetralin (1,2,3,4-tetrahydronaphthalene) enter the cell and are oxidized, leading to formation of highly toxic hydroperoxides. Nevertheless, E. coli mutant strains that overexpress the AhpCF alkylhydroperoxide reductase, an enzyme involved in detoxification of organic hydroperoxides, are resistant to tetralin and other organic solvents (96), and they could be also used as suitable host cells to engineer efficient biocatalysts.
Heavy-metal resistance.
Frequently, environments containing aromatic compounds are contaminated with heavy metals and radionuclides. Therefore, it may be desirable to express aromatic catabolic pathways in bacterial biocatalysts resistant to heavy metals. There are several reports on heavy-metal resistance in E. coli (292). Some E. coli strains contain heavy-metal resistance plasmids such as pDU1358, R773, pRJ1004, and pMG101, which confer organomercury, arsenic, copper, and silver resistance, respectively (292). Thus, the mer operon from plasmid pBD7 of E. coli K-12 strain BL308 (derived from the R831 plasmid originally isolated from Serratia marcescens [286]) has been successfully used to engineer the radiation-resistant D. radiodurans strain to detoxify mercury in mixed radioactive wastes (32).
Aerobic/anaerobic life-style.
Since E. coli is a facultative anaerobe, it can grow in both the presence and absence of oxygen. This feature can be very useful for some biotransformation processes that require a two-step anaerobic-aerobic treatment (156). 1,2-Dihydroxynaphthalene, an industrially and pharmaceutically interesting catechol, can be formed from naphthalene by biocatalysts expressing naphthalene dioxygenase and the corresponding dihydrodiol dehydrogenase. The main difficulty with this biotransformation is represented by the oxidation and polymerization of the catechol in the presence of the trace amounts of oxygen needed for the dioxygenation reaction. To overcome this problem, a strategy based on the aerobic/anaerobic life-style of E. coli was approached. Thus, an E. coli recombinant strain carrying both naphthalene dioxygenase and dihydrodiol dehydrogenase genes from P. fluorescens N3 was engineered in such a way that selectively induces the expression of the dioxygenase and dehydrogenase under aerobic and anaerobic conditions, respectively. Bioconversion experiments performed under aerobic conditions showed dihydrodiol production and dehydrogenase repression; as soon as the cultures were switched to a nitrogen atmosphere, dihydrodiol dehydrogenation allowed efficient production of 1,2-dihydroxyderivatives (74). Similar strategies for the catabolism of other aromatic compounds such as the azo dyes can be forseen.
Surface display.
A number of vehicles, including subunits of cellular appendages or outer membrane proteins, have been used as carriers in vivo for the display and action of enzymes, peptides, antigenic determinants, or single-chain antibodies on the surface of E. coli host cells (110). Some of these surface display vehicles have been used for the removal of aromatic pollutants. Thus, surface display of antipollutant antibodies provides a virtually unlimited range of specific, whole-cell adsorbents capable of separating and removing substances from contaminated sources (130). As an example of this emerging technology, the peptidoglycan-associated lipoprotein has been fused to an antibody fragment (scFv) specific to the herbicide and environmental pollutant atrazine, and the fusion was successfully targeted to the cell surface of E. coli (69).
Surface display of enzymes that attack aromatic compounds has been also carried out with recombinant E. coli strains. For example, an organophosphorus hydrolase (OPH) from a Flavobacterium sp. was successfully anchored and displayed onto the cell surface of E. coli using a Lpp-OmpA fusion system. E. coli cultures with surface-expressed OPH degraded organophosphate pesticides, such as parathion and paraoxon, very efficiently without the diffusional limitation observed in cells expressing the enzyme intracellularly and avoiding the toxic effects of the pollutants inside the cells (261). The recombinant cultures had a very long shelf-life, retaining full activity for a month in a resting state (44). The immobilization of this novel biocatalyst facilitates its recycle and reuse and paves the way for an efficient, simple, and cost-effective method for detoxification of organophosphate nerve agents (203).
Expression of E. coli Aromatic Catabolic Genes in Heterologous Hosts
Some of the E. coli gene clusters involved in catabolism of aromatic compounds have been expressed in different E. coli strains and in other bacterial species to expand and/or increase their endogenous catabolic abilities.
hpa gene cluster.
The complete hpa cluster from E. coli W was expressed either in multicopy or single copy in E. coli K-12, and all the resulting recombinant strains were able to grow in 4HPA as the sole carbon source. Moreover, since the recombinant strains cultured in glycerol as the carbon source transformed phenol to catechol after induction by 4HPA, they represent interesting biocatalysts for the production of catechols (250).
A mobile catabolic segment carrying the HPA monooxygenase operon (hpaBC) under the control of the Ptrc promoter was engineered and introduced into the chromosome of P. putida KT2442, a strain unable to metabolize phenol since it does not contain the gene(s) required to hydroxylate this compound. The new recombinant strain, P. putida KTH2, was able to grow on phenol as the sole carbon and energy source because this aromatic compound was converted by the HPA hydroxylase to catechol (250), which was further mineralized through the chromosomally encoded ortho-cleavage pathway of P. putida (132). This result illustrates the utility of a broad-substrate-range catabolic enzyme, the two-component HPA monooxygenase of E. coli, to increase the ability of heterologous hosts to degrade new aromatic compounds, and it constitutes an example of in vitro pathway evolution by vertical expansion of a catabolic route naturally present in the recipient cell.
The recombinant P. putida KTH2 strain (see above) efficiently expresses the hpaC gene from E. coli and hence shows high levels of flavin:NAD(P)H activity. This feature was exploited to clone and express in strain KTH2 the dsz genes from Rhodococcus sp. strain IGTS8 that are involved in desulfurization of dibenzothiophene, an energy-intensive process that requires large amounts of reducing equivalents in form of FMNH2. The resulting strain was shown to be a desulfurizer that was significantly more efficient than the initial P. putida KT2442 dsz+ strain (103). Moreover, hpaC was combined with the dsz genes in a broad-host-range DNA cassette that conferred the desulfurization phenotype to a wide variety of bacteria regardless of the expression of putative housekeeping flavin reductases within the host cells (103).
The hpaBC genes from E. coli W have also been expressed from a plasmid in E. coli K-12 and the recombinant strain was used as a biocatalyst for the production of l-Dopa (Fig. 12A), the most widely used drug for Parkinson's disease. This process exploits the ability of the two-component HpaBC monooxygenase to hydroxylate the cheap substrate l-tyrosine to l-Dopa (176).
FIG. 12.
Selected biotransformations of aromatic compounds in recombinant E. coli strains. The enzymes catalyzing the different reactions are indicated in boldface type. (A) Production of 3,4-dihydroxyphenylalanine (l-Dopa) from tyrosine through the 4HPA monooxygenase (HpaBC) from E. coli W. (B) Biotransformation of styrene into (S)-styrene oxide (epoxystyrene), phenylacetaldehyde (PAL), and PA by the StyAB, StyC, and StyD enzymes from different Pseudomonas strains. Formation of (S)-styrene oxide from styrene was also reported using the xylene monooxygenase (XylAM) from the TOL plasmid of P. putida. (C) Conversion of benzene into l-Dopa using the toluene dioxygenase and toluene cis-dihydrodiol dehydrogenase (DH) from P. putida F1 and the tyrosine phenol-lyase from C. freundii. (D) Biotransformation of glucose into the dye indigo in a recombinant E. coli strain that converts glucose into indole and then oxidizes the latter through naphthalene dioxygenase (DOx) or xylene monooxygenase (MOx) from P. putida. (E) Conversion of l-phenylalanine to cinnamic acid (CI) and ammonia through the phenylalanine ammonia-lyase of R. toruloides.
The regulatory (HpaA/PBC) and transport (HpaX) systems of the HPA catabolic pathway from E. coli W have been used together with the lacZ reporter gene to engineer an E. coli K-12 highly sensitive HPA biosensor. Concentrations of 4HPA as low as 10 nM trigger the production of β-galactosidase, an enzyme that can be easily monitored in the recombinant strains (252).
As discussed above, the pac gene encoding the penicillin G acylase (Pac) is physically and functionally linked to the hpa cluster in E. coli W. Since Pac produces 6-aminopenicillanic acid from penicillin G (see above), this enzyme has been extensively used in industry for the synthesis of many semisynthetic penicillin derivatives (182, 317).
mhp and hca gene clusters.
A DNA cassette containing the mhpRABCDFET cluster involved in 3HPP catabolism in E. coli has been engineered and expressed in both enteric and nonenteric bacteria that are unable to degrade such aromatic acids. Thus, when the mhp cassette was introduced into proteobacteria of the γ subgroup, such as S. enterica serovar Typhimurium LT-2 and P. putida KT2442, or into proteobacteria of the α subgroup, such as Rhizobium meliloti Rm1021, all recombinant strains were able to grow efficiently on minimal medium containing 3HPP as the sole carbon and energy source (92). The combination of the mhp cassette with that containing the hca cluster responsible for the initial degradation of PP in E. coli was carried out using serovar Typhimurium LT2 as host cells. The recombinant serovar Typhimurium mhp+ hca+ strain was able to grow in different phenylpropanoid compounds (3HPP, 3HCI, PP, and CI) as the sole carbon and energy source (71). Since HPP, PP, and other phenyl carboxylates are intermediates produced during the catabolism of linear alkylbenzenes present in commercial detergents (293), the use of the mhp and hca cassettes to improve the catabolic abilities of bacteria to degrade such pollutants becomes of biotechnological interest.
The regulatory and transport systems of the 3HPP degradation pathway have also been used to engineer cellular biosensors. Recombinant E. coli strains expressing the MhpR/Pa::lacZ reporter system and the transport (mhpT) gene of the 3HPP catabolic pathway behave as efficient 3HPP biosensors (B. Torres, J. L. Garcia, and E. Diaz, unpublished data).
paa gene cluster.
The complete paa cluster was cloned and efficiently expressed in different E. coli PA-deficient strains, conferring on the latter the ability to use PA as the sole carbon and energy source (94). The paaK gene (encodes the PA-CoA ligase) and the paaABCDE functional unit (involved in the hydroxylation of the PA-CoA intermediate) (Fig. 5B) have been used to engineer efficient biocatalysts for the bioconversion of PA into 2HPA, a compound of interest in the pharmaceutical industry for the synthesis of different biotechnological products (182; A. Ferrandez, B. Miñambres, B. García, E. R. Olivera, J. M. Luengo, J. L. García, and E. Díaz, March 2001, Spanish patent 2148084).
Expression of Heterologous Genes in E. coli for Biodegradation and Biotransformation of Aromatic Compounds
Since E. coli is the best-known microorganism both at the biochemical and genetic levels, it has been extensively used for expression of single genes or gene clusters involved in catabolism of aromatic compounds. The properties of some E. coli strains (see above) make them suitable for designing efficient biocatalysts. E. coli strains containing catabolic operons equipped with efficient foreign expression signals and which can be grown to very high cell densities on simple carbon sources may provide efficient performance (313). Moreover, the use of a well-characterized host such as E. coli might also minimize the regulatory concerns about releasing genetically modified organisms into the environment (70, 256, 337). The purpose of this section is not to review the huge amount of work on the use of recombinant E. coli strains in biodegradation and biotransformation of aromatic compounds but, rather, to highlight some examples on this subject.
Expansion of the abilities of E. coli to grow on aromatic compounds.
Catabolic plasmids contribute to the metabolic versatility of microorganisms by specifying enzymes responsible for the catabolism of a wide variety of aromatic compounds. One of the most extensively studied family of catabolic plasmids is the family of TOL plasmids, which determine the catabolism of toluene, m-xylene, p-xylene, m-ethyltoluene, and 1,2,4-trimethylbenzene through their corresponding benzoate derivatives via a meta-cleavage pathway (xyl genes) in P. putida strains (129). When the xyl genes were transposed to the broad-host-range plasmid RP4 and this derivative was transferred to E. coli J53, the resulting strain was able to grow on toluene, m-xylene, and p-xylene, although the level of expression of the xyl genes was very low in comparison with that observed in Pseudomonas strains (141). Transfer to and expression of the TOL derivative pWWO-EB62 in E. coli EEZ8000 allowed growth of the recombinant strain on p-ethylbenzoate as the sole source of carbon, but no expression of the catabolic xyl genes was observed in other E. coli K-12 strains (257). Similarly, E. coli C600 derivatives bearing the TOL plasmid did not express the TOL catabolic pathways (20). The reason for the strain-dependent expression of TOL catabolic pathways in E. coli is still unknown, and this phenomenon requires further research.
While some enteric bacteria such as K. pneumoniae and S. enterica serovar Typhimurium are able to grow on m-hydroxybenzoate, E. coli K-12 cannot use this aromatic compound as the sole carbon and energy source. The chromosomal mhb genes from K. pneumoniae encode the enzymes responsible for the catabolism of m-hydroxybenzoate via 2,5-dihydroxybenzoate (gentisate), and they were cloned and expressed in E. coli K-12 (264). The recombinant E. coli strain acquired the ability to use m-hydroxybenzoate as the carbon source, which indicates that the gentisate route is also functional in E. coli K-12; this expands the catabolic potential of this bacterium toward aromatic compounds that are degraded via the gentisate pathway (264).
The cloning and expression of heterologous catabolic pathways whose final products are incorporated into the housekeeping catabolic routes of the host cell constitutes a common method of rational expansion of catabolic pathways (313). An example of this strategy of vertical expansion of catabolic abilities was the cloning and expression in E. coli of the upper pathway for styrene degradation. Thus, when the complete set of sty genes from Pseudomonas sp. strain Y2 that are responsible for the oxidation of styrene to PA (Fig. 12B) were transferred to the PA-degrading E. coli W strain, the resulting bacterium was able to grow on styrene as the sole carbon and energy source, demonstrating that E. coli can be engineered to degrade highly toxic aromatic hydrocarbons such as styrene (324).
Engineering of E. coli strains as biocatalysts for selected biotransformations.
The use of enzymes in biocatalytic processes that involve aromatic compounds is not evenly distributed over the different classes of enzymes; the hydrolases and oxidoreductases are the most heavily utilized (287). Oxidoreductases are complex enzymes, usually multicomponent proteins, which require cofactors and are therefore mostly used as whole-cell biocatalysts. Despite their difficulty of handling, these biocatalysts are of great biotechnological value because of their regio- and stereoselectivity. A number of E. coli biocatalysts have been designed to express aromatic ring mono- and/or dioxygenases for the biotransformation of particular aromatic compounds to cis-dihydrodiols, epoxides, and catechols that have diverse environmental and pharmaceutical applications (224, 258, 348). The biosynthesis of epoxides, versatile chemical building blocks in the manufacture of optically active compounds, illustrates this point. One attractive route to efficiently access enantiomerically pure epoxides in a one-step reaction is the epoxidation of double bonds by monooxygenases. In this context, recombinant E. coli cells have been equipped with the genes encoding the xylene monooxygenase from the TOL plasmid of P. putida (220) or the styrene monooxygenase from P. fluorescens ST (73) or Pseudomonas sp. strain VLB120 (222) to oxidize styrene to chiral (S)-styrene oxide (Fig. 12B). The broad substrate preference of the styrene monooxygenase allowed the engineered E. coli cells to oxidize different aryl-vinyl and aryl-ethenyl compounds to their corresponding optically pure epoxides (73). The combination in recombinant E. coli cells of the alkane-responsive AlkS/Palk regulatory system from P. oleovorans GPo1, driving the expression of the genes encoding the StyAB styrene monooxygenase, with an efficient two-liquid phase fed-batch process led to an optimized system for the production of enantiopure styrene oxide (222).
The rational design of hybrid pathways based on the combination of enzymes from different sources has also been performed using E. coli as host cells. For instance, recombinant biocatalysts have been developed that are able to use benzene, one of the most toxic components in petroleum, as a cheap raw material for the synthesis of the high value product l-Dopa. Thus, a hybrid pathway consisting of toluene dioxygenase and toluene cis-glycol dehydrogenase from P. putida F1 and tyrosine phenol-lyase from Citrobacter freundii was engineered in E. coli cells (Fig. 12C). In this pathway, catechol is formed from benzene through the sequential action of toluene dioxygenase and toluene cis-glycol (dihydrodiol) dehydrogenase, and l-Dopa is synthesized from the resulting catechol in the presence of pyruvate and ammonia by tyrosine phenol-lyase (225). Although a toxic effect of benzene on the recombinant E. coli cells was observed (225), the use of solvent-resistant E. coli cells (see above) may increase the efficacy of the process.
Most of the approaches to the development of microbial strains for overproduction of aromatic compounds have relied on improving the enzymatic steps within the specific pahtway of the desired product. However, further improvements can be achieved only by redirecting the carbon flux from the central metabolism to the product-forming pathway (23). A nice example of biocatalysis by means of an engineered whole cell is the biosynthesis of indigo using recombinant E. coli cells that can directly synthesize this dye from glucose via the formation of indole (Fig. 12D). To accomplish this task, a number of comprehensive series of genetically engineered metabolic transformations were optimized: (i) more glucose-derived carbon was redirected to the production of chorismic acid by amplifying genes of the shikimate pathway and by increasing the levels of available phosphoenolpyruvate (99); (ii) diverting the excess chorismate to tryptophan synthesis was solved by amplifying a mutant tryptophan synthetase enzyme complex that released free indole (99); and (iii) the E. coli biocatalysts were endowed with a naphthalene dioxygenase of P. putida that was genetically modified to increase its activity and enhance its half-life, such that the produced indole was efficiently oxidized to indoxyl, a reactive compound that undergoes oxidative dimerization to form indigo (205). The xylene monooxygenase from the TOL plasmid has also been used to oxidize indole to indigo in recombinant E. coli strains (197) (Fig. 12D). A major disadvantage of the biologically produced indigo was the presence of a red dye (indirubin) formed during the fermentation process by reaction of indoxyl with an oxidative derivative (isatin). This problem was solved by the cloning and expression of an isatin hydrolase in the engineered indigo-producing cells. At present, the indigo production strain contains over 18 kb of stable and coordinately regulated additional genetic information (25). Another important biotransformation process using a recombinant E. coli biocatalyst and involving the conversion of glucose into an aromatic compound is that of vanillin synthesis (178).
Recombinant E. coli cells endowed with additional abilities to transform aromatic compounds have also been used as a novel approach to treat some genetic diseases such as phenylketonuria. This prototypical human genetic disease consists of a hyperphenylalaninemia due to impaired activity of phenylalanine hydroxylase, the enzyme responsible for disposal, by oxidative catabolism, of the majority of nutrient phenylalanine intake. The cloning and expression in E. coli of the gene encoding the phenylalanine ammonia-lyase from Rhodosporidium toruloides, allows the recombinant cells to convert phenylalanine to metabolically insignificant amounts of ammonia and CI (Fig. 12E), a harmless metabolite that is converted to benzoic acid and rapidly excreted in urine as hippurate (278). When given by oral gavage, recombinant E. coli cells producing phenylalanine ammonia-lyase lowered phenylalanine levels in the mouse intestinal lumen and therefore in body fluids in the whole animal (278). These findings are just the beginning of an exciting and potentially applicable strategy based on the use of transgenic commensal bacteria for preventing disease (330).
Construction of E. coli whole-cell biosensors and contained biocatalysts.
Tightly regulated expression systems based on a regulatory protein and its cognate promoter from aromatic catabolic pathways have been extensively studied in a wide variety of organisms. The combination of these regulatory systems with suitable reporters or with broad-host-range lethal genes has allowed the construction of E. coli biosensors and contained biocatalysts, respectively (72).
Whole-cell biosensors are particularly valuable for examining pollutant bioavailability since contaminants must be accessible to the cells for induction of the intracellular reporter fusion. E. coli luminescent biosensors have been engineered to detect different aromatic pollutants. Thus, the IpbR/PA transcriptional couple, which is involved in the regulation of the isopropylbenzene catabolism from plasmid pRE4 of P. putida RE204, was fused to the luxCDABE genes from Vibrio fischeri to create a reporter fusion that conferred to E. coli cells the ability to produce light in the presence of inducers of the ipb operon. Bacteria carrying the ipb-lux reporter fusion were used as bioindicators of monoalkylbenzenes, substituted benzenes and toluenes, chlorinated solvents, naphthalenes, and complex hydrocarbon mixtures such as gasoline, diesel fuel, jet fuels, and creosote (290). A similar bacterial biosensor was constructed by expressing in E. coli cells the luc gene, encoding firefly luciferase, under the control of the XylR/Pu regulatory system from the TOL plasmid of P. putida mt-2. Luminescence from the cells was induced in the presence of benzene, toluene, and xylene contamination in water and soil samples (334). Transport genes can increase the sensitivity of cellular biosensors by allowing the detection of very low external concentrations of the aromatic compounds, and this strategy has been successfully used in E. coli to design a 4HPA biosensor (see above).
Although recombinant microorganisms offer obvious benefits in environmental applications, their behavior in open environments and, in some cases, even in physically contained fermentation tanks is difficult to predict and raises serious public and scientific concerns (256, 337). One of the major concerns is how recombinant DNA can spread among indigenous bacterial populations and what might be the impact of such transfer on the microbial ecosystem. Horizontal DNA transfer from transgenic organisms may also be undesirable for process protection and process optimization reasons. An appropriate approach to reduce the transfer of recombinant DNA is to provide to the engineered organisms with efficient gene containment systems based on the controlled expression of some lethal functions. E. coli has been used as a source of different lethal functions, such as colicin E3 RNase, EcoRI restriction endonuclease, and the RelF membrane protein, and as a model system to check the efficiency of different gene containment systems (70, 314). To control the life cycle of the recombinant biocatalysts such that their survival in the target habitat will be limited in time and space, different biological containment systems have been designed. To this end, the regulatory elements of some aromatic catabolic pathways, e.g., the XylS/Pm regulatory couple, have been engineered to control the expression of different lethal functions in E. coli, restricting the survival of the biocatalyst to the presence of the target aromatic compound (70, 256). Since these active biological containment systems use competitive wild-type microorganisms, they represent an advantage with respect to the more classical biosafety approaches of employing disabled E. coli mutant strains, e.g., E. coli X1776 (57), that cannot perform and compete successfully in a heterogeneous microbial community. In addition to diminishing the potential risks associated with the deliberate or unintentional release of recombinant bacteria into the open environment, the conditional suicide systems might be of interest as a biomass control method in bioreactors (55).
CONCLUSIONS AND OUTLOOK
Although E. coli is the model organism for cellular and molecular biology and it is better characterized than any other cell, only 63% of the reading frames present in its genome have identified functions, and many of these correspond to phenotypes rather than to specific reactions (273). In particular, the rapid progress made in the catabolism of natural aromatic compounds by E. coli over the past 10 years illustrates that the narrowing of this gap is a fertile research area at the interface of chemistry and biology. The current state of knowledge of the biodegradative abilities of E. coli reveals that this bacterium is not an “empty box” for the catabolism of aromatic compounds but instead is endowed with typical aerobic degradation routes highly similar to those described in environmental relevant bacteria such as those of the genus Pseudomonas. Moreover, just as different Pseudomonas strains have specific aromatic substrates for growth, intraspecies variation concerning the ability to mineralize different aromatic compounds (PA, HPA, PP, 3HPP, 3HCI, PEA, tyramine, and dopamine) has also been observed in E. coli. Therefore, E. coli turns out to be a very useful model system to decipher biochemical, genetic, evolutionary, and ecological aspects of the catabolism of natural aromatic compounds, an unnoticed finding for most researchers dealing with this laboratory “workhorse” (174, 234).
The fact that E. coli is endowed with its own set of genes and enzymes for the catabolism of aromatic compounds (Fig. 13) should be taken into consideration when cloning and expressing heterologous aromatic catabolic clusters in this enterobacterium. In this sense, several reports on the cloning and expression of aromatic ring-hydroxylating dioxygenases in E. coli claim that equivalent enzymes from the host could explain the unexpected activities observed when some of the subunits of the cloned dioxygenases were lacking in the recombinant bacteria (82, 167, 186, 305). The described PP-dioxygenase activity of E. coli (see above) could explain some of these unexpected findings.
Despite our current extensive knowledge about the aerobic catabolism of aromatic compounds in E. coli, there remains so much more to learn. For instance, the large knowledge gap between sequence information and function for enzymes responsible for PA catabolism and involving unprecedented reactions that have not yet been reconstituted in vitro is a major challenge to field of functional genomics. PA catabolism through a hybrid aerobic pathway like the one described in E. coli is likely to be a widespread route for the metabolism of this aromatic compound, and therefore the cloned paa genes should be useful as probes to identify homologous genes from distinct groups of bacteria.
Regulatory proteins and regulated promoters driving the expression of catabolic genes have been identified in most of the aromatic catabolic pathways described in E. coli, and they reveal a great structural and functional diversity. However, further research is needed to unravel the molecular mechanisms that allow regulators to sense the inducer metabolite and to connect the induced conformational changes with promoter output. On the other hand, since the pathway-specific regulation of some catabolic pathways, e.g., the paa-, mhp-, and hpa-encoded pathways, has been shown to be subordinated to a more general control that adjusts the particular transcriptional output to the physiological status of the cell, E. coli becomes a useful organism to gain a complete picture of the regulatory mechanisms that control the catabolism of aromatic compounds within a particular bacterial cell. Cross talk among different aromatic catabolic pathways and preferential utilization of some metabolites present in a mixture of biodegradable aromatic compounds (a scenario that, by the way, is the normal situation in natural environments) are also important issues that can be addressed by using E. coli.
The acquisition of aromatic compounds from the environment is governed by transport processes (132). Thus far, only three transporters, the HpaX, MhpT, and TnaB proteins, have been shown to be involved in catabolism of aromatic compounds in E. coli. However, a repertoire of other aromatic transport proteins can be predicted when analyzing the genome of this enteric bacterium. Since E. coli has a motile life-style, the ability of this bacterium to sense and swim toward aromatic compounds (chemotaxis) is an important issue that deserves study and that may be of great interest to fully understand the behavioral responses of this organism to the presence of aromatic compounds.
Our current knowledge about the overall catabolic versatility of E. coli toward aromatic compounds may still be far from complete. Thus, analysis of the whole E. coli genome has shown the presence of several unknown genes that are likely to be involved in the aerobic degradation and transformation of aromatic compounds. Moreover, the facultatively anaerobic life-style of E. coli allows us to presume the existence of anaerobic catabolic pathways that might facilitate the growth of this bacterium on aromatic compounds in the absence of oxygen. Anaerobic catabolism of aromatic compounds in E. coli is therefore a research field that needs to be explored. All the efforts toward a better understanding of the complete set of aromatic catabolic abilities of E. coli will pave the way for the rational design of efficient and predictable biocatalysts for many biotechnological applications.
ACKNOWLEDGMENTS
Work in our laboratory was supported by grants from the Comisión Interministerial de Ciencia y Tecnología (AMB97-063-C02-02, 2FD97-1326-C02-01, BMC2000-0125-C04-02, and BIO2000-1076), Comunidad Autónoma de Madrid (07M/0050/99 and 07M/0127/2000), and European Union (BIO4-CT97-2313 and QLK3-2000-00170).
REFERENCES
- 1.Abdulrashid N, Clark D P. Isolation and genetic analysis of mutations allowing the degradation of furans and thiophenes by Escherichia coli. J Bacteriol. 1987;169:1267–1271. doi: 10.1128/jb.169.3.1267-1271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alam K Y, Clark D P. Molecular cloning and sequence of the thdF gene, which is involved in thiophene and furan oxidation by Escherichia coli. J Bacteriol. 1991;173:6018–6024. doi: 10.1128/jb.173.19.6018-6024.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alekshun M N, Kim Y S, Levy S B. Mutational analysis of MarR, the negative regulator of marRAB expression in Escherichia coli, suggests the presence of two regions required for DNA binding. Mol Microbiol. 2000;35:1394–1404. doi: 10.1046/j.1365-2958.2000.01802.x. [DOI] [PubMed] [Google Scholar]
- 4.Allende J L, Gibello A, Martin M, Garrido-Pertierra A. Transport of 4-hydroxyphenylacetic acid in Klebsiella pneumoniae. Arch Biochem Biophys. 1992;292:583–588. doi: 10.1016/0003-9861(92)90034-t. [DOI] [PubMed] [Google Scholar]
- 5.Altenschmidt U, Oswald B, Steiner E, Herrmann H, Fuchs G. New aerobic benzoate oxidation pathway via benzoyl-coenzyme A and 3-hydroxybenzoyl-coenzyme A in a denitrifying Pseudomonas sp. J Bacteriol. 1993;175:4851–4858. doi: 10.1128/jb.175.15.4851-4858.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ames P, Parkinson J. Constitutively signaling fragments of Tsr, the Escherichia coli serine chemoreceptor. J Bacteriol. 1994;176:6340–6348. doi: 10.1128/jb.176.20.6340-6348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aono R, Aibe K, Inoue A, Horikoshi K. Preparation of organic solvent tolerant mutants from Escherichia coli K-12. Agric Biol Chem. 1991;55:1935–1938. [Google Scholar]
- 8.Aono R, Kobayashi H. Cell surface properties of organic solvent-tolerant mutants of Escherichia coli K-12. Appl Environ Microbiol. 1997;63:3637–3642. doi: 10.1128/aem.63.9.3637-3642.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aono R, Tsukagoshi N, Yamamoto M. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance level of Escherichia coli K-12. J Bacteriol. 1998;180:938–944. doi: 10.1128/jb.180.4.938-944.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arai H, Yamamoto T, Ohishi T, Shimizu T, Nakata T, Kudo T. Genetic organization and characteristics of the 3-(3-hydroxyphenyl)propionic acid degradation pathway of Comamonas testosteroni TA441. Microbiology. 1999;145:2813–2820. doi: 10.1099/00221287-145-10-2813. [DOI] [PubMed] [Google Scholar]
- 11.Armengaud J. Molecular genetics of the degradation of dioxins by bacteria. In: Wittich R-M, editor. Biodegradation of dioxins and furans. Austin, Tex: Springer-Verlag and R. G. Landes; 1998. pp. 75–123. [Google Scholar]
- 12.Asturias J A, Díaz E, Timmis K N. The evolutionary relationships of biphenyl dioxygenase from Gram-positive Rhodococcus globerulus P6 to multicomponent dioxygenases from Gram-negative bacteria. Gene. 1995;156:11–18. doi: 10.1016/0378-1119(94)00530-6. [DOI] [PubMed] [Google Scholar]
- 13.Azakami H, Yamashita M, Roh J-H, Suzuki H, Kumagai H, Murooka Y. Nucleotide sequence of the gene for monoamine oxidase (maoA) from Escherichia coli. J Ferment Bioeng. 1994;77:315–319. [Google Scholar]
- 14.Azizan A, Sherin D, DiRusso C C, Black P N. Energetics underlying the process of long-chain fatty acid transport. Arch Biochem Biophys. 1999;365:299–306. doi: 10.1006/abbi.1999.1171. [DOI] [PubMed] [Google Scholar]
- 15.Baca-DeLancey R R, South M M T, Ding X, Rather P N. Escherichia coli gene regulated by cell-to-cell signaling. Proc Natl Acad Sci USA. 1999;96:4610–4614. doi: 10.1073/pnas.96.8.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bachellier S, Clément J-M, Hofnung M. Short palindromic repetitive DNA elements in enterobacteria: a survey. Res Microbiol. 1999;150:627–639. doi: 10.1016/s0923-2508(99)00128-x. [DOI] [PubMed] [Google Scholar]
- 17.Barker J, Humphrey T J, Brown M W R. Survival of Escherichia coli 0157 in a soil protozoan: implications for disease. FEMS Microbiol Lett. 1999;173:291–295. doi: 10.1111/j.1574-6968.1999.tb13516.x. [DOI] [PubMed] [Google Scholar]
- 18.Barnes M R, Duetz W A, Williams P A. A 3-(3-hydroxyphenyl)propionic acid catabolic pathway in Rhodococcus globerulus PWD1: cloning and characterization of the hpp operon. J Bacteriol. 1997;179:6145–6153. doi: 10.1128/jb.179.19.6145-6153.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beltrametti F, Marconi A M, Bestetti G, Colombo C, Galli E, Ruzzi M, Zennaro E. Sequencing and functional analysis of styrene catabolism genes from Pseudomonas fluorescens ST. Appl Environ Microbiol. 1997;63:2232–2239. doi: 10.1128/aem.63.6.2232-2239.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benson S, Shapiro J. TOL is a broad-host-range plasmid. J Bacteriol. 1978;135:278–280. doi: 10.1128/jb.135.1.278-280.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergthorsson U, Ochman H. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol Biol Evol. 1998;15:6–16. doi: 10.1093/oxfordjournals.molbev.a025847. [DOI] [PubMed] [Google Scholar]
- 22.Bermudez M, Hazen T C. Phenotypic and genotypic comparison of Escherichia coli from pristine tropical waters. Appl Environ Microbiol. 1988;54:979–983. doi: 10.1128/aem.54.4.979-983.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berry A. Improving production of aromatic compounds in Escherichia coli by metabolic engineering. Trends Biotechnol. 1996;14:250–256. doi: 10.1016/0167-7799(96)10033-0. [DOI] [PubMed] [Google Scholar]
- 24.Bertani G, Weigle J J. Host controlled variation in bacterial viruses. J Bacteriol. 1953;65:113–121. doi: 10.1128/jb.65.2.113-121.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bialy H. Biotechnology, bioremediation, and blue genes. Nat Biotechnol. 1997;15:110. doi: 10.1038/nbt0297-110. [DOI] [PubMed] [Google Scholar]
- 26.Birkenbihl R P, Vielmetter W. Complete maps of IS1, IS2, IS3, IS4, IS5, IS30 and IS150 locations in Escherichia coli K12. Mol Gen Genet. 1989;220:147–153. doi: 10.1007/BF00260869. [DOI] [PubMed] [Google Scholar]
- 27.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 28.Blom A, Harder W, Matin A. Unique and overlapping pollutant stress proteins of Escherichia coli. Appl Environ Microbiol. 1992;58:331–334. doi: 10.1128/aem.58.1.331-334.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blum G, Mühldorfer I, Kuhnert P, Frey J, Hacker J. Comparative methodology to investigate the presence of Eschrichia coli K-12 strains in environmental and human stool samples. FEMS Microbiol Lett. 1996;143:77–82. doi: 10.1111/j.1574-6968.1996.tb08464.x. [DOI] [PubMed] [Google Scholar]
- 30.Breinig S, Schiltz E, Fuchs G. Genes involved in anaerobic metabolism of phenol in the bacterium Thauera aromatica. J Bacteriol. 2000;182:5849–5863. doi: 10.1128/jb.182.20.5849-5863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bressler D C, Norman J A, Fedorak P M. Ring cleavage of sulfur heterocycles: how does it happen? Biodegradation. 1998;8:297–311. doi: 10.1023/a:1008283207090. [DOI] [PubMed] [Google Scholar]
- 32.Brim H, McFarlan S C, Fredrickson J K, Minton K W, Zhai M, Wackett L P, Daly M J. Engineering Deinococcus radiodurans for metal remediation in radioactive mixed waste environments. Nat Biotechnol. 2000;18:85–90. doi: 10.1038/71986. [DOI] [PubMed] [Google Scholar]
- 33.Bryant D W, McCalla D R, Leeksma M, Laneuville P. Type I nitroreductases of Escherichia coli. Can J Microbiol. 1981;27:81–86. doi: 10.1139/m81-013. [DOI] [PubMed] [Google Scholar]
- 34.Bugg T D H. Overproduction, purification and properties of 2,3-dihydroxyphenylpropionate 1,2-dioxygenase from Escherichia coli. Biochim Biophys Acta. 1993;1202:258–264. doi: 10.1016/0167-4838(93)90013-h. [DOI] [PubMed] [Google Scholar]
- 35.Bugg T D H, Sanvoisin J, Spence E L. Exploring the catalytic mechanism of the extradiol catechol dioxygenases. Biochem Soc Trans. 1997;25:81–85. doi: 10.1042/bst0250081. [DOI] [PubMed] [Google Scholar]
- 36.Burlingame R, Chapman P J. Catabolism of phenylpropionic acid and its 3-hydroxy derivative by Escherichia coli. J Bacteriol. 1983;155:113–121. doi: 10.1128/jb.155.1.113-121.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burlingame R, Chapman P J. Stereospecificity in meta-fission catabolic pathways. J Bacteriol. 1983;155:424–426. doi: 10.1128/jb.155.1.424-426.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burlingame R P. Ph.D. thesis. Minneapolis: University of Minnesota; 1983. [Google Scholar]
- 39.Burlingame R P, Wyman L, Chapman P J. Isolation and characterization of Escherichia coli mutants defective for phenylpropionate degradation. J Bacteriol. 1986;168:55–64. doi: 10.1128/jb.168.1.55-64.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Busby S, Ebright R H. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 41.Butler C S, Mason J R. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases. Adv Microb Physiol. 1997;38:47–84. doi: 10.1016/s0065-2911(08)60155-1. [DOI] [PubMed] [Google Scholar]
- 42.Chang F C, Chung J G. Evidence for arylamine N-acetyltransferase activity in the Escherichia coli. Curr Microbiol. 1998;36:125–130. doi: 10.1007/pl00006755. [DOI] [PubMed] [Google Scholar]
- 43.Chang K-H, Xiang H, Dunaway-Mariano D. Acyl-adenylate motif of the acyl-adenylate/thioester-forming enzyme superfamily: a site-directed mutagenesis study with the Pseudomonas sp. strain CBS3 4-chlorobenzoate:coenzyme A ligase. Biochemistry. 1997;36:15650–15659. doi: 10.1021/bi971262p. [DOI] [PubMed] [Google Scholar]
- 44.Chen W, Mulchandani A. The use of live biocatalysts for pesticide detoxification. Trends Biotechnol. 1998;16:71–76. doi: 10.1016/s0167-7799(97)01160-8. [DOI] [PubMed] [Google Scholar]
- 45.Clausen M, Lamb C J, Megnet R, Doerner P W. PAD1 encodes phenylacrylic acid decarboxylase which confers resistance to cinnamic acid in Saccharomyces cerevisiae. Gene. 1994;142:107–112. doi: 10.1016/0378-1119(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 46.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–4558. doi: 10.1128/aem.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collier L S, Nichols N N, Neidle E L. benK encodes a hydrophobic permease-like protein involved in benzoate degradation by Acinetobacter sp. strain ADP1. J Bacteriol. 1997;179:5943–5946. doi: 10.1128/jb.179.18.5943-5946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cooper R A, Jones D C N, Parrot S. Isolation and mapping of Escherichia coli K12 mutants defective in phenylacetate degradation. J Gen Microbiol. 1985;131:2753–2757. doi: 10.1099/00221287-131-10-2753. [DOI] [PubMed] [Google Scholar]
- 49.Cooper R A, Knowles P F, Brown D E, McGuirl M A, Dooley D M. Evidence for copper and 3,4,6-trihydroxyphenylalanine quinone cofactors in an amine oxidase from the Gram-negative bacterium Escherichia coli K-12. Biochem J. 1992;288:337–340. doi: 10.1042/bj2880337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooper R A, Skinner M A. Catabolism of 3- and 4-hydroxyphenylacetate by the 3,4-dihydroxyphenylacetate pathway in Escherichia coli. J Bacteriol. 1980;143:302–306. doi: 10.1128/jb.143.1.302-306.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corkery D M, O'Connor K E, Buckley C M, Dobson A D W. Ethylbenzene degradation by Pseudomonas fluorescens strain CA-4. FEMS Microbiol Lett. 1994;124:23–28. doi: 10.1111/j.1574-6968.1994.tb07256.x. [DOI] [PubMed] [Google Scholar]
- 52.Correll C C, Batie C J, Ballou D P, Ludwig M L. Phthallate dioxygenase reductase: a modular structure for electron transfer from pyridine nucleotides to [2Fe-2S] Science. 1992;258:1604–1610. doi: 10.1126/science.1280857. [DOI] [PubMed] [Google Scholar]
- 53.Covès J, Nivière V, Eschenbrenner M, Fontecave M. NADPH-sulfite reductase from Escherichia coli. A flavin reductase participating in the generation of the free radical of the ribonucleotide reductase. J Biol Chem. 1993;268:18604–18609. [PubMed] [Google Scholar]
- 54.Cox G B, Gibson F, Pittard J. Mutant strains of Escherichia coli K-12 unable to form ubiquinone. J Bacteriol. 1968;95:1591–1598. doi: 10.1128/jb.95.5.1591-1598.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cox H H J, Deshusses M A. Biological waste air treatment in biotrickling filters. Curr Opin Biotechnol. 1998;9:256–262. doi: 10.1016/s0958-1669(98)80056-6. [DOI] [PubMed] [Google Scholar]
- 56.Cripps R E. The microbial metabolism of thiophen-2-carboxylate. Biochem J. 1973;134:353–366. doi: 10.1042/bj1340353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Curtiss R, III, Inoue R M, Pereira D, Hsu J C, Alexander L, Rock L. Construction and use of safer bacterial host strains for recombinant DNA research. In: Scott WA, Werner R, editors. Molecular cloning of recombinant DNA. New York, N.Y: Academic Press, Inc.; 1977. pp. 99–111. [Google Scholar]
- 58.Cuskey S M, Olsen R H. Catabolism of aromatic biogenic amines by Pseudomonas aeruginosa PAO1 via meta cleavage of homoprotocatechuic acid. J Bacteriol. 1988;170:393–399. doi: 10.1128/jb.170.1.393-399.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dagley S. New perspectives in aromatic catabolism. In: Leisinger T, Cook A M, Hütter R, Nüesch J, editors. Degradation of xenobiotics and recalcitrant compounds. New York, N.Y: Academic Press, Inc.; 1981. pp. 181–186. [Google Scholar]
- 60.Dai M, Yingmin Z, Yang Y, Wang E, Xie Y, Zhao G, Jiang W. Expression of penicillin G acylase from the cloned pac gene of Escherichia coli ATCC11105. Eur J Biochem. 2001;268:1298–1303. doi: 10.1046/j.1432-1327.2001.01994.x. [DOI] [PubMed] [Google Scholar]
- 61.Dalrymple B P, Swadling Y. Expression of a Butyrivibrio fibrisolvens E14 gene (cinB) encoding an enzyme with cinnamoyl ester hydrolase activity is negatively regulated by the product of an adjacent gene (cinR) Microbiology. 1997;143:1203–1210. doi: 10.1099/00221287-143-4-1203. [DOI] [PubMed] [Google Scholar]
- 62.Daniels D L, Plunkett G, Burland V D, Blattner F R. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science. 1992;257:771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- 63.Davis B D, Mingioli E S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950;60:17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Bont J A M. Solvent-tolerant bacteria in biocatalysis. Trends Biotechnol. 1998;16:493–499. [Google Scholar]
- 65.Deeley M C, Yanofsky C. Nucleotide sequence of the structural gene for tryptophanase of Escherichia coli K-12. J Bacteriol. 1981;147:787–796. doi: 10.1128/jb.147.3.787-796.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deeley M C, Yanofsky C. Transcription initiation at the tryptophanase promoter of Escherichia coli K-12. J Bacteriol. 1982;151:942–951. doi: 10.1128/jb.151.2.942-951.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Delbrück M, Luria S E. Interference between bacterial viruses. I Arch Biochem. 1942;1:111–141. [Google Scholar]
- 68.de Lorenzo V, Pérez-Martín J. Regulatory noise in prokaryotic promoters: how bacteria learn to respond to novel environmental signals. Mol Microbiol. 1996;19:1177–1184. doi: 10.1111/j.1365-2958.1996.tb02463.x. [DOI] [PubMed] [Google Scholar]
- 69.Dhillon J K, Drew P D, Porter A J R. Bacterial surface display of an anti-pollutant antibody fragment. Lett Appl Microbiol. 1999;28:350–354. doi: 10.1046/j.1365-2672.1999.00552.x. [DOI] [PubMed] [Google Scholar]
- 70.Díaz E, Chithila-Munthali M T, Jaenecke S, de Lorenzo V, Timmis K N. Design of genetic circuits for restricting gene and biocatalyst dispersal. 1999. pp. 1–18. (unit 6.1.14). In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbiology ecology manual.Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 71.Díaz E, Ferrández A, García J L. Characterization of the hca cluster encoding the dioxygenolytic pathway for initial catabolism of 3-phenylpropionic acid in Escherichia coli K-12. J Bacteriol. 1998;180:2915–2923. doi: 10.1128/jb.180.11.2915-2923.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Díaz E, Prieto M A. Bacterial promoters triggering biodegradation of aromatic pollutants. Curr Opin Biotechnol. 2000;11:467–475. doi: 10.1016/s0958-1669(00)00126-9. [DOI] [PubMed] [Google Scholar]
- 73.Di Gennaro P, Colmegna A, Galli E, Sello G, Pelizzoni F, Bestetti G. A new biocatalyst for production of optically pure aryl epoxides by styrene monooxygenase from Pseudomonas fluorescens ST. Appl Environ Microbiol. 1999;65:2794–2797. doi: 10.1128/aem.65.6.2794-2797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Di Gennaro P, Galli E, Orsini F, Pelizzoni F, Sello G, Bestetti G. Development of biocatalysts carrying naphthalene dioxygenase and dihydrodiol dehydrogenase genes inducible in aerobic and anaerobic conditions. Res Microbiol. 2000;151:383–391. doi: 10.1016/s0923-2508(00)00161-3. [DOI] [PubMed] [Google Scholar]
- 75.DiMarco A A, Averhoff B, Kim E E, Ornston L N. Evolutionary divergence of pobA, the structural gene encoding p-hydroxybenzoate hydroxylase, in an Acinetobacter calcoaceticus strain well-suited for genetic analysis. Gene. 1993;125:25–33. doi: 10.1016/0378-1119(93)90741-k. [DOI] [PubMed] [Google Scholar]
- 76.DiRusso C C, Heimert T L, Metzger A K. Characterization of FadR, a global transcriptional regulator of fatty acid metabolism in Escherichia coli. Interaction with the fadB promoter is prevented by long chain fatty acyl coenzyme As. J Biol Chem. 1992;267:8685–8691. [PubMed] [Google Scholar]
- 77.Donnelly M I, Cooper R A. Succinic semialdehyde dehydrogenase of Escherichia coli. Their role in the degradation of p-hydroxyphenylacetate and γ-aminobutyrate. Eur J Biochem. 1981;113:555–561. doi: 10.1111/j.1432-1033.1981.tb05098.x. [DOI] [PubMed] [Google Scholar]
- 78.Dougan G, Haque A, Pickard D, Frankel G, O'Goara P, Wain J. The Escherichia coli gene pool. Curr Opin Microbiol. 2001;4:90–94. doi: 10.1016/s1369-5274(00)00170-3. [DOI] [PubMed] [Google Scholar]
- 79.Duffner F M, Kirchner U, Bauer M P, Müller R. Phenol/cresol degradation by the thermophilic Bacillus thermoglucosidasius A7: cloning and sequence analysis of five genes involved in the pathway. Gene. 2000;256:215–221. doi: 10.1016/s0378-1119(00)00352-8. [DOI] [PubMed] [Google Scholar]
- 80.Duggleby H J, Tolley S P, Hill C P, Dodson E J, Dodson G, Moody P C. Penicillin acylase has a single-amino-acid catalytic centre. Nature. 1995;373:264–268. doi: 10.1038/373264a0. [DOI] [PubMed] [Google Scholar]
- 81.Dunaway-Mariano D, Babbitt P C. On the origins and functions of the enzymes of the 4-chlorobenzoate to 4-hydroxybenzoate converting pathway. Biodegradation. 1994;5:259–276. doi: 10.1007/BF00696464. [DOI] [PubMed] [Google Scholar]
- 82.Eaton R W, Timmis K N. Characterization of a plasmid-specified pathway for catabolism of isopropylbenzene in Pseudomonas putida RE204. J Bacteriol. 1986;168:123–131. doi: 10.1128/jb.168.1.123-131.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Edwards R M, Yudkin M D. Location of the gene for the low-affinity tryptophan-specific permease of Escherichia coli. Biochem J. 1982;204:617–619. doi: 10.1042/bj2040617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Egland P G, Harwood C S. BadR, a new MarR family member, regulates anaerobic benzoate degradation by Rhodopseudomonas palustris in concert with AadR, a Fnr family member. J Bacteriol. 1999;181:2102–2109. doi: 10.1128/jb.181.7.2102-2109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ehmann D E, Shaw-Reid C A, Losey H C, Walsh C T. The EntF and EntE adenylation domains of Escherichia coli enterobactin synthetase: sequestration and selectivity in acyl-AMP transfers to thiolation domain cosubstrates. Proc Natl Acad Sci USA. 2000;97:2509–2514. doi: 10.1073/pnas.040572897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Eichhorn E, van der Ploeg J R, Leisinger T. Characterization of a two component alkanesulfonate monooxygenase from Escherichia coli. J Biol Chem. 1999;274:26639–26646. doi: 10.1074/jbc.274.38.26639. [DOI] [PubMed] [Google Scholar]
- 87.Eltis L D, Bolin J T. Evolutionary relationships among extradiol dioxygenases. J Bacteriol. 1996;178:5930–5937. doi: 10.1128/jb.178.20.5930-5937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Espinosa-Urgel M, Kolter R. Escherichia coli genes expressed preferentially in an aquatic environment. Mol Microbiol. 1998;28:325–332. doi: 10.1046/j.1365-2958.1998.00796.x. [DOI] [PubMed] [Google Scholar]
- 89.Eulberg D, Schlömann M. The putative regulator of catechol catabolism in Rhodococcus opacus 1CP—an IclR-type, not a LysR-type transcriptional regulator. Antonie Leeuwenhoek. 1998;74:71–82. doi: 10.1023/a:1001755928637. [DOI] [PubMed] [Google Scholar]
- 90.Fawcett T, Garrido-Pertierra A, Cooper R A. 5-Carboxymethyl-2-hydroxymuconic semialdehyde dehydrogenases of Escherichia coli C and Klebsiella pneumoniae M5a1 show very high N-terminal sequence homology. FEMS Microbiol Lett. 1989;57:307–312. doi: 10.1016/0378-1097(89)90319-4. [DOI] [PubMed] [Google Scholar]
- 91.Fernández-Cañón J M, Peñalva M A. Fungal metabolic model for human type I hereditary tyrosinaemia. Proc Natl Acad Sci USA. 1995;92:9132–9136. doi: 10.1073/pnas.92.20.9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ferrández A, García J L, Díaz E. Genetic characterization and expression in heterologous hosts of the 3 (3-hydroxyphenyl)propionate catabolic pathway of Escherichia coli K-12. J Bacteriol. 1997;179:2573–2581. doi: 10.1128/jb.179.8.2573-2581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ferrández A, García J L, Díaz E. Transcriptional regulation of the divergent paa catabolic operons for phenylacetic acid degradation in Escherichia coli. J Biol Chem. 2000;275:12214–12222. doi: 10.1074/jbc.275.16.12214. [DOI] [PubMed] [Google Scholar]
- 94.Ferrández A, Miñambres B, García B, Olivera E R, Luengo J M, García J L, Díaz E. Catabolism of phenylacetic acid in Escherichia coli. Characterization of a new aerobic hybrid pathway. J Biol Chem. 1998;273:25974–25986. doi: 10.1074/jbc.273.40.25974. [DOI] [PubMed] [Google Scholar]
- 95.Ferrández A, Prieto M A, García J L, Díaz E. Molecular characterization of PadA, a phenylacetaldehyde dehydrogenase from Escherichia coli. FEBS Lett. 1997;406:23–27. doi: 10.1016/s0014-5793(97)00228-7. [DOI] [PubMed] [Google Scholar]
- 96.Ferrante A A, Augliera J, Lewis K, Klibanov A M. Cloning of an organic solvent-resistance gene in Escherichia coli: the unexpected role of alkylhydroperoxide reductase. Proc Natl Acad Sci USA. 1995;92:7617–7621. doi: 10.1073/pnas.92.17.7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ferrer E, Cooper R A. Studies with a cloned Escherichia coli C 2-oxo-hept-3-ene-1,7-dioate hydratase gene. FEMS Microbiol Lett. 1988;52:155–160. [Google Scholar]
- 98.Fleming S M, Robertson T A, Langley G J, Bugg T D H. Catalytic mechanism of a C-C hydrolase enzyme: evidence for a Gem-diol intermediate, not an acyl enzyme. Biochemistry. 2000;39:1522–1531. doi: 10.1021/bi9923095. [DOI] [PubMed] [Google Scholar]
- 99.Flores N, Xiao J, Berry A, Bolivar F, Valle F. Pathway engineering for the production of aromatic compounds in Escherichia coli. Nat Biotechnol. 1996;14:620–623. doi: 10.1038/nbt0596-620. [DOI] [PubMed] [Google Scholar]
- 100.Francis A J, Spanggord R J, Ouchi G I. Degradation of lindane by Escherichia coli. Appl Microbiol. 1975;29:567–568. doi: 10.1128/am.29.4.567-568.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fujioka M, Morino Y, Wada H. Phenylacetaldehyde dehydrogenase from Achromobacter eurydice. Methods Enzymol. 1970;17A:593–596. [Google Scholar]
- 102.Fujita Y, Miwa Y. Identification of an operator sequence for the Bacillus subtilis gnt operon. J Biol Chem. 1989;264:4201–4206. [PubMed] [Google Scholar]
- 103.Galán B, Díaz E, García J L. Enhancing desulphurization by engineering a flavin reductase-encoding gene cassette in recombinant biocatalysts. Environ Microbiol. 2000;2:687–694. doi: 10.1046/j.1462-2920.2000.00151.x. [DOI] [PubMed] [Google Scholar]
- 104.Galán B, Díaz E, Prieto M A, García J L. Functional analysis of the small component of the 4-hydroxyphenylacetate 3-monooxygenase of Escherichia coli W: a prototype of a new flavin:NAD(P)H reductase subfamily. J Bacteriol. 2000;182:627–636. doi: 10.1128/jb.182.3.627-636.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gallegos M T, Schleif R, Bairoch A, Hofmann K, Ramos J L. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gang D M, Shaikh K. Regulation of penicillin acylase in Escherichia coli by cyclic AMP. Biochim Biophys Acta. 1976;425:110–114. doi: 10.1016/0005-2787(76)90220-3. [DOI] [PubMed] [Google Scholar]
- 107.García B, Olivera E R, Miñambres B, Carnicero D, Muñiz C, Naharro G, Luengo J M. Phenylacetyl-coenzyme A is the true inducer of the phenylacetic acid catabolism pathway in Pseudomonas putida U. Appl Environ Microbiol. 2000;66:4575–4578. doi: 10.1128/aem.66.10.4575-4578.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Garrido-Pertierra A, Cooper R A. Identification and purification of distinct isomerase and decarboxylase enzymes involved in the 4-hydroxyphenylacetate catabolic pathway of Escherichia coli. Eur J Biochem. 1981;117:581–584. doi: 10.1111/j.1432-1033.1981.tb06377.x. [DOI] [PubMed] [Google Scholar]
- 109.Gasson M J, Kitamura Y, McLauchlan W R, Narbad A, Parr A J, Parsons E L H, Payne J, Rhodes M J C, Walton N J. Metabolism of ferulic acid to vanillin. A bacterial gene of the enoyl-SCoA hydratase/isomerase superfamily encodes an enzyme for the hydratation and cleavage of a hydroxycinnamic acid SCoA thioester. J Biol Chem. 1998;273:4163–4170. doi: 10.1074/jbc.273.7.4163. [DOI] [PubMed] [Google Scholar]
- 110.Georgiou G, Poetschke H L, Stathopoulos C, Francisco J A. Practical applications of engineering Gram-negative bacterial cell surfaces. Trends Biotechnol. 1993;11:6–10. doi: 10.1016/0167-7799(93)90068-K. [DOI] [PubMed] [Google Scholar]
- 111.Gerischer U, Segura A, Ornston L N. PcaU, a transcriptional activator of genes for protocatechuate utilization in Acinetobacter. J Bacteriol. 1998;180:1512–1524. doi: 10.1128/jb.180.6.1512-1524.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ghisla S, Massey V. Mechanism of flavoprotein-catalyzed reactions. Eur J Biochem. 1989;181:1–17. doi: 10.1111/j.1432-1033.1989.tb14688.x. [DOI] [PubMed] [Google Scholar]
- 113.Gibello A, Ferrer E, Martín M, Garrido-Pertierra A. 3,4-Dihydroxyphenylacetate 2,3-dioxygenase from Klebsiella pneumoniae, a Mg2+-containing dioxygenase involved in aromatic catabolism. Biochem J. 1994;301:145–150. doi: 10.1042/bj3010145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gibello A, Ferrer E, Sanz J, Martin M. Polymer production by Klebsiella pneumoniae 4-hydroxyphenylacetic acid hydroxylase genes cloned in Escherichia coli. Appl Environ Microbiol. 1995;61:4167–4171. doi: 10.1128/aem.61.12.4167-4171.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gibello A, Suárez M, Allende J L, Martín M. Molecular cloning and analysis of the genes encoding the 4-hydroxyphenylacetate hydroxylase from Klebsiella pneumoniae. Arch Microbiol. 1997;167:160–166. [PubMed] [Google Scholar]
- 116.Gibson D T, Parales R E. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr Opin Biotechnol. 2000;11:236–243. doi: 10.1016/s0958-1669(00)00090-2. [DOI] [PubMed] [Google Scholar]
- 117.Goldman P. Biochemical pharmacology and toxicology involving the intestinal flora. In: Hentges D J, editor. Human intestinal microflora in health and disease. New York, N.Y: Academic Press, Inc.; 1983. pp. 241–263. [Google Scholar]
- 118.González-Pérez M M, Ramos J L, Gallegos M-T, Marqués S. Critical nucleotides in the upstream region of the XylS-dependent TOL meta-cleavage pathway operon promoter as deduced from analysis of mutants. J Biol Chem. 1999;274:2286–2290. doi: 10.1074/jbc.274.4.2286. [DOI] [PubMed] [Google Scholar]
- 119.Grant D J W. Kinetic aspects of the growth of Klebsiella aerogenes with some benzenoid carbon sources. J Gen Microbiol. 1967;46:213–224. doi: 10.1099/00221287-46-2-213. [DOI] [PubMed] [Google Scholar]
- 120.Gray C H, Tatum E I. X-ray induced growth factor requierements in bacteria. Proc Natl Acad Sci USA. 1944;30:404–411. doi: 10.1073/pnas.30.12.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Griffiths L A, Smith G E. Metabolism of myricetin and related compounds in the rat metabolite formation in vivo and by the intestinal microflora in vitro. Biochem J. 1972;130:141–151. doi: 10.1042/bj1300141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Grund E, Denecke B, Eichenlaub R. Naphthalene degradation via salicylate and gentisate by Rhodococcus sp. strain B4. Appl Environ Microbiol. 1992;58:1874–1877. doi: 10.1128/aem.58.6.1874-1877.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gupta R S. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol Mol Biol Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hacisalihoglu A, Jongejan J A, Duine J A. Distribution of amine oxidases and amine dehydrogenases in bacteria grown on primary amines and characterization of the amine oxidase from Klebsiella oxytoca. Microbiology. 1997;143:505–512. doi: 10.1099/00221287-143-2-505. [DOI] [PubMed] [Google Scholar]
- 125.Hacker J, Carniel E. Ecological fitness, genomic islands and bacterial pathogenicity. EMBO Rep. 2001;2:376–381. doi: 10.1093/embo-reports/kve097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hanlon S P, Carpenter K, Hassan A, Cooper R A. Formation in vitro of the 3,4,6-trihydroxyphenylalanine quinone cofactor. Biochem J. 1995;306:627–630. doi: 10.1042/bj3060627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hanlon S P, Hill T K, Flavell M A, Stringfellow J M, Cooper R A. 2-Phenylethylamine catabolism by Escherichia coli K-12: gene organization and expression. Microbiology. 1997;143:513–518. doi: 10.1099/00221287-143-2-513. [DOI] [PubMed] [Google Scholar]
- 128.Harayama S, Rekik M, Ngai K-L, Ornston L N. Physically associated enzymes produce and metabolize 2-hydroxy-2,4-dienoate, a chemically unstable intermediate formed in catechol metabolism via meta cleavage in Pseudomonas putida. J Bacteriol. 1989;171:6251–6258. doi: 10.1128/jb.171.11.6251-6258.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Harayama S, Timmis K N. Aerobic biodegradation of aromatic hydrocarbons by bacteria. In: Sigel H, Sigel A, editors. Metal ions in biological systems. New York, N.Y: Marcel Dekker, Inc; 1992. pp. 99–156. [Google Scholar]
- 130.Harris B. Exploiting antibody-based technologies to manage environmental pollution. Trends Biotechnol. 1999;17:290–296. doi: 10.1016/s0167-7799(99)01308-6. [DOI] [PubMed] [Google Scholar]
- 131.Harwood C S, Burchhardt G, Herrmann H, Fuchs G. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol Rev. 1999;22:439–458. [Google Scholar]
- 132.Harwood C S, Parales R E. The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 133.He X-Y, Deng H, Yang S-Y. Importance of the γ-carboxyl group of glutamate-462 of the large α-subunit for the catalytic function and the stability of the multienzyme complex of fatty acid oxidation from Escherichia coli. Biochemistry. 1997;36:261–268. doi: 10.1021/bi961841e. [DOI] [PubMed] [Google Scholar]
- 134.Henderson I M J, Bugg T D H. Pre-steady-state kinetic analysis of 2-hydroxy-6-keto-nona-2,4-diene-1,9-dioic acid 5,6-hydrolase: kinetic evidence for enol/keto tautomerization. Biochemistry. 1997;36:12252–12258. doi: 10.1021/bi971116j. [DOI] [PubMed] [Google Scholar]
- 135.Henson J M, Kopp B, Kuempel P L. Deletion of 60 kilobase pairs of DNA from the terC region of the chromosome of Escherichia coli. Mol Gen Genet. 1984;193:263–268. doi: 10.1007/BF00330678. [DOI] [PubMed] [Google Scholar]
- 136.Hernáez M J, Andújar E, Ríos J L, Kaschabek S R, Reineke W, Santero E. Identification of a serine hydrolase which cleaves the alicyclic ring of tetralin. J Bacteriol. 2000;182:5448–5453. doi: 10.1128/jb.182.19.5448-5453.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hewitt L, Kasche V, Lummer K, Lewis R J, Murshudov G N, Verma C S, Dodson G G, Wilson K S. Structure of a slow processing precursor penicillin acylase from Escherichia coli reveals the linker peptide blocking the active-site cleft. J Mol Biol. 2000;302:887–898. doi: 10.1006/jmbi.2000.4105. [DOI] [PubMed] [Google Scholar]
- 138.Hülsmeyer M, Hecht H-J, Niefend K, Hofer B, Eltis L D, Timmis K N, Schomburg D. Crystal structure of cis-biphenyl-2,3-dihydrodiol-2,3-dehydrogenase from a PCB degrader at 2.0 Å resolution. Protein Sci. 1998;7:1286–1293. doi: 10.1002/pro.5560070603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ingelman M, Ramaswamy S, Nivière V, Fontecave M, Eklund H. Crystal structure of NAD(P)H:flavin oxidoreductase from Escherichia coli. Biochemistry. 1999;38:7040–7049. doi: 10.1021/bi982849m. [DOI] [PubMed] [Google Scholar]
- 140.Inoue J, Shaw J P, Rekik M, Harayama S. Overlapping substrate specificities of benzaldehyde dehydrogenase (the xylC gene product) and 2-hydroxymuconic semialdehyde dehydrogenase (the xylG gene product) encoded by TOL plasmid pWW0 of Pseudomonas putida. J Bacteriol. 1995;177:1196–1201. doi: 10.1128/jb.177.5.1196-1201.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jacoby G A, Rogers J E, Jacob A E, Hedges R W. Transposition of Pseudomonas toluene-degrading genes and expression in Escherichia coli. Nature. 1978;274:179–180. doi: 10.1038/274179a0. [DOI] [PubMed] [Google Scholar]
- 142.Jagnow G, Haider K, Ellwardt P-C H R. Anaerobic dechlorination and degradation of hexachlorocyclohexane isomers by anaerobic and facultative anaerobic bacteria. Arch Microbiol. 1977;115:285–292. doi: 10.1007/BF00446454. [DOI] [PubMed] [Google Scholar]
- 143.Jenkins J R, Cooper R A. Molecular cloning, expression, and analysis of the genes of the homoprotocatechuate catabolic pathway of Escherichia coli C. J Bacteriol. 1988;170:5317–5324. doi: 10.1128/jb.170.11.5317-5324.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jiang L, Wang Y, Yang S. Location of regulatory gene in penicillin acylase operon (pacR) of E. coli D816. Genet Anal Biomol Eng. 1997;14:51–54. doi: 10.1016/s1050-3862(97)00011-9. [DOI] [PubMed] [Google Scholar]
- 145.Johnson W H, Jr, Hajipour G, Whitman C P. Characterization of a dienol intermediate in the 5-(carboxymethyl)-2-oxo-3-hexene-1,6-dioate decarboxylase reaction. J Am Chem Soc. 1992;114:11001–11003. [Google Scholar]
- 146.Jones R M, Pagmantidis V, Williams P A. sal genes determining the catabolism of salicylate esters are part of a supraoperonic cluster of catabolic genes in Acinetobacter sp. strain ADP1. J Bacteriol. 2000;182:2018–2025. doi: 10.1128/jb.182.7.2018-2025.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Juhasz A L, Naidu R. Enrichment and isolation of non-specific aromatic degraders from unique uncontaminated (plant and faecal material) sources and contaminated soils. J Appl Microbiol. 2000;89:642–650. doi: 10.1046/j.1365-2672.2000.01161.x. [DOI] [PubMed] [Google Scholar]
- 148.Juhl M J, Clark D P. Thiophene-degrading Escherichia coli mutants possess sulfone oxidase activity and show altered resistance to sulfur-containing antibiotics. Appl Environ Microbiol. 1990;56:3179–3185. doi: 10.1128/aem.56.10.3179-3185.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kadiyala V, Spain J C. A two-component monooxygenase catalyzes both the hydroxylation of p-nitrophenol and the oxidative release of nitrite from 4-nitrocatechol in Bacillus sphaericus JS905. Appl Environ Microbiol. 1998;64:2479–2484. doi: 10.1128/aem.64.7.2479-2484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kaldalu N, Toots U, de Lorenzo V, Ustav M. Functional domains of the TOL plasmid transcription factor XylS. J Bacteriol. 2000;182:1118–1126. doi: 10.1128/jb.182.4.1118-1126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Karp P, Riley M, Paley S, Pellegrini-Toole A, Krummenacker M. EcoCyc: electronic encyclopedia of E. coli genes and metabolism. Nucleic Acids Res. 1999;27:55. doi: 10.1093/nar/27.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kauppi B, Lee K, Carredano E, Parales R E, Gibson D T, Eklund H, Ramaswamy S. Structure of an aromatic ring-hydroxylating dioxygenase—naphthalene 1,2-dioxygenase. Structure. 1998;6:571–586. doi: 10.1016/s0969-2126(98)00059-8. [DOI] [PubMed] [Google Scholar]
- 153.Kertesz M A. Riding the sulfur cycle- metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS Microbiol Rev. 1999;24:135–175. doi: 10.1016/S0168-6445(99)00033-9. [DOI] [PubMed] [Google Scholar]
- 154.Kessler D, Leibrecht I, Knappe J. Pyruvate-formate-lyase-deactivase and acetyl-CoA reductase activities of Escherichia coli reside on a polymeric protein particle encoded by adhE. FEBS Lett. 1991;281:59–63. doi: 10.1016/0014-5793(91)80358-a. [DOI] [PubMed] [Google Scholar]
- 155.Kimura N, Nishi A, Goto M, Furukawa K. Functional analyses of a variety of chimeric dioxygenases constructed from two biphenyl dioxygenases that are similar structurally but different functionally. J Bacteriol. 1997;179:3936–3943. doi: 10.1128/jb.179.12.3936-3943.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Knackmuss H-J. Basic knowledge and perspectives of bioelimination of xenobiotic compounds. J Biotechnol. 1996;51:287–295. [Google Scholar]
- 157.Knoell H-E. On the nature fo the monooxygenase system involved in ubiquinone-8 synthesis. FEMS Microbiol Lett. 1981;10:63–65. [Google Scholar]
- 158.Kobayashi K, Tsukagoshi N, Aono R. Suppression of hypersensitivity of Escherichia coli acrB mutant to organic solvents by integrational activation of the acrEF operon with the IS1 or IS2 element. J Bacteriol. 2001;183:2646–2653. doi: 10.1128/JB.183.8.2646-2653.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kobori T, Sasaki H, Cheol Lee W, Zenno S, Saigo K, Murphy M E P, Tanokura M. Structure and site-directed mutagenesis of a flavoprotein from Escherichia coli that reduces nitrocompounds. J Biol Chem. 2001;276:2816–2823. doi: 10.1074/jbc.M002617200. [DOI] [PubMed] [Google Scholar]
- 160.Koenig K, Andreesen J R. Molybdenum involvement in aerobic degradation of 2-furoic acid by Pseudomonas putida Ful. Appl Environ Microbiol. 1989;55:1829–1834. doi: 10.1128/aem.55.7.1829-1834.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kok R G, D'Argenio D A, Ornston L N. Mutation analysis of PobR and PcaU, closely related transcriptional activators in Acinetobacter. J Bacteriol. 1998;180:5058–5069. doi: 10.1128/jb.180.19.5058-5069.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Konan K V, Yanofsky C. Rho-dependent transcription termination in the tna operon of Escherichia coli: roles of the boxA sequence and the rut site. J Bacteriol. 2000;182:3981–3988. doi: 10.1128/jb.182.14.3981-3988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kovárová-Kovar K, Egli T. Growth kinetics of suspended microbial cells: from single-substrate-controlled growth to mixed-substrate kinetics. Microbiol Mol Biol Rev. 1998;62:646–666. doi: 10.1128/mmbr.62.3.646-666.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kovárová K, Käch A, Chaloupka V, Egli T. Cultivation of Escherichia coli with mixtures of 3-phenylpropionic acid and glucose: dynamics of growth and substrate consumption. Biodegradation. 1996;7:445–453. doi: 10.1007/BF00115291. [DOI] [PubMed] [Google Scholar]
- 165.Kovárová K, Käch A, Zehnder A J B, Egli T. Cultivation of Escherichia coli with mixtures of 3-phenylpropionic acid and glucose: steady-state growth kinetics. Appl Environ Microbiol. 1997;63:2619–2624. doi: 10.1128/aem.63.7.2619-2624.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kumagai H, Yamada H, Matsui H, Ohkishi H, Ogata K. Tyrosine phenol lyase. I. Purification, crystallization, and properties. J Biol Chem. 1970;245:1767–1772. [PubMed] [Google Scholar]
- 167.Kurkela S, Lehväslaiho H, Palva E T, Teeri T H. Cloning, nucleotide sequence and characterization of genes encoding naphthalene dioxygenase of Pseudomonas putida strain NCIB 9816. Gene. 1988;73:355–362. doi: 10.1016/0378-1119(88)90500-8. [DOI] [PubMed] [Google Scholar]
- 168.Kwon O, Kotsakis A, Meganathan R. Ubiquinone (coenzyme Q) biosynthesis in Escherichia coli: identification of the ubiF gene. FEMS Microbiol Lett. 2000;186:157–161. doi: 10.1111/j.1574-6968.2000.tb09097.x. [DOI] [PubMed] [Google Scholar]
- 169.Lam W W Y, Bugg T D H. Chemistry of extradiol aromatic ring cleavage: isolation of a stable dienol ring fission intermediate and stereochemistry of its enzymatic hydrolytic cleavage. J Chem Soc Chem Commun. 1994;1994:1163–1164. [Google Scholar]
- 170.Lam W W Y, Bugg T D H. Purification, characterization, and stereochemical analysis of a C-C hydrolase: 2-hydroxy-6-keto-nona-2,4-diene-1,9-dioic acid 5,6-hydrolase. Biochemistry. 1997;36:12242–12251. doi: 10.1021/bi971115r. [DOI] [PubMed] [Google Scholar]
- 171.Lan R, Reeves P R. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 2000;8:396–401. doi: 10.1016/s0966-842x(00)01791-1. [DOI] [PubMed] [Google Scholar]
- 172.Landick R, Turnbough J R, Yanofsky C. Transcription attenuation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1263–1286. [Google Scholar]
- 173.Lathe W C, III, Snel B, Bork P. Gene context conservation of a higher order than operons. Trends Biochem Sci. 2000;25:474–479. doi: 10.1016/s0968-0004(00)01663-7. [DOI] [PubMed] [Google Scholar]
- 174.Lau P. Dirt and degradation and E. coli. ASM News. 1998;64:1. [Google Scholar]
- 175.Laurie A D, Lloyd-Jones G. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J Bacteriol. 1999;181:531–540. doi: 10.1128/jb.181.2.531-540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lee J-Y, Xun L. Novel biological process for l-DOPA production from l-tyrosine by p-hydroxyphenylacetate 3-hydroxylase. Biotechnol Lett. 1998;20:479–482. [Google Scholar]
- 177.Lieb M, Weigle J J, Kellenberger E. A study of hybrids between two strains of Escherichia coli. J Bacteriol. 1955;69:468–471. doi: 10.1128/jb.69.4.468-471.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Liese A, Filho M V. Production of fine chemicals using biocatalysis. Curr Opin Biotechnol. 1999;10:595–603. doi: 10.1016/s0958-1669(99)00040-3. [DOI] [PubMed] [Google Scholar]
- 179.Lindsay R F, Priest F G. Decarboxylation of substituted cinnamic acids by Enterobacteria: the influence on beer flavour. J Appl Bacteriol. 1975;39:181–187. doi: 10.1111/j.1365-2672.1975.tb00560.x. [DOI] [PubMed] [Google Scholar]
- 180.Liochev S I, Hausladen A, Fridovich I. Nitroreductase A is regulated as a member of the soxRS regulon of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:3537–3539. doi: 10.1073/pnas.96.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liu J L, Duncan K, Walsh C T. Nucleotide sequence of a cluster of Escherichia coli enterobactin biosynthesis genes: identification of entA and purification of its product 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase. J Bacteriol. 1989;171:791–798. doi: 10.1128/jb.171.2.791-798.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Luengo J M, García J L, Olivera E R. The phenylacetyl-CoA catabolon: a complex catabolic unit with broad biotechnological applications. Mol Microbiol. 2001;39:1434–1442. doi: 10.1046/j.1365-2958.2001.02344.x. [DOI] [PubMed] [Google Scholar]
- 183.Manzanera M, Marqués S, Ramos J L. Mutational analysis of the highly conserved C-terminal residues of the XylS protein, a member of the AraC family of transcriptional regulators. FEBS Lett. 2000;476:312–317. doi: 10.1016/s0014-5793(00)01749-x. [DOI] [PubMed] [Google Scholar]
- 184.Marek L E, Henson J M. Cloning and expression of the Escherichia coli K-12 sad gene. J Bacteriol. 1988;170:991–994. doi: 10.1128/jb.170.2.991-994.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Martín M, Gibello A, Fernández J, Ferrer E, Garrido-Pertierra A. Catabolism of 3- and 4-hydroxyphenylacetic acid by Klebsiella pneumoniae. J Gen Microbiol. 1991;132:621–628. doi: 10.1099/00221287-137-3-621. [DOI] [PubMed] [Google Scholar]
- 186.Martin V J J, Mohn W W. A novel aromatic-ring-hydroxylating dioxygenase from the diterpenoid-degrading bacterium Pseudomonas abietaniphila BKME-9. J Bacteriol. 1999;181:2675–2682. doi: 10.1128/jb.181.9.2675-2682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Martínez-Blanco H, Reglero A, Rodríguez-Aparicio L B, Luengo J M. Purification and biochemical characterization of phenylacetyl-CoA ligase from Pseudomonas putida. J Biol Chem. 1990;265:7084–7090. [PubMed] [Google Scholar]
- 188.Marvin-Sikkema F D, de Bont J A M. Degradation of nitroaromatic compounds by microorganisms. Appl Microbiol Biotechnol. 1994;42:499–507. doi: 10.1007/BF00173912. [DOI] [PubMed] [Google Scholar]
- 189.Masai E, Sugiyama K, Iwashita N, Shimizu S, Hauschild J E, Hatta T, Kimbara K, Yano K, Fukuda M. The bphDEF meta-cleavage pathway genes involved in biphenyl/polychlorinated biphenyl degradation are located on a linear plasmid and separated from the initial bphACB genes in Rhodococcus sp. strain RHA1. Gene. 1997;187:141–149. doi: 10.1016/s0378-1119(96)00748-2. [DOI] [PubMed] [Google Scholar]
- 190.Matsuzaki R, Fukui T, Sato H, Ozaki Y, Tanizawa K. Generation of the topaquinone cofactor in bacterial monoamine oxidase by cupric ion-dependent autoxidation of a specific tyrosyl residue. FEBS Lett. 1994;351:360–364. doi: 10.1016/0014-5793(94)00884-1. [DOI] [PubMed] [Google Scholar]
- 191.McClelland M, Florea L, Sanderson K, Clifton S W, Parkhill J, Churcher C, Dougan G, Wilson R K, Miller W. Comparison of the Escherichia coli K-12 genome with sampled genomes of a Klebsiella pneumoniae and three Salmonella enterica serovars, Typhimurium, Typhi and Paratyphi. Nucleic Acids Res. 2000;28:4974–4986. doi: 10.1093/nar/28.24.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.McCormick N G, Feeherry F E, Levinson H S. Microbial transformation of 2,4,6-trinitrotoluene and other nitroaromatic compounds. Appl Environ Microbiol. 1976;31:949–958. doi: 10.1128/aem.31.6.949-958.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.McCullough W G, Piligian J T, Daniel I J. Enzymatic decarboxylation of the aminobenzoates. J Am Chem Soc. 1957;79:628–630. [Google Scholar]
- 194.McFall E, Newman E B. Amino acids as carbon sources. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 358–379. [Google Scholar]
- 195.Meganathan R. Biosynthesis of the isoprenoid quinones menaquinone (vitamin K2) and ubiquinone (coenzyme Q) In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 642–656. [Google Scholar]
- 196.Merino E, Balbas P, Recillas F, Becerril B, Valle F, Bolivar F. Carbon regulation and the role in nature of the Escherichia coli penicillin acylase (pac) gene. Mol Microbiol. 1992;6:2175–2182. doi: 10.1111/j.1365-2958.1992.tb01391.x. [DOI] [PubMed] [Google Scholar]
- 197.Mermod N, Harayama S, Timmis K N. New route to bacterial production of indigo. Bio/Technology. 1986;4:321–324. [Google Scholar]
- 198.Michael N P, Brehm J K, Anlezark G M, Minton N P. Physical characterisation of the Escherichia coli B gene encoding nitroreductase and its over-expression in Escherichia coli K12. FEMS Microbiol Lett. 1994;124:195–202. doi: 10.1111/j.1574-6968.1994.tb07284.x. [DOI] [PubMed] [Google Scholar]
- 199.Miñambres B, Martínez-Blanco H, Olivera E R, García B, Díez B, Barredo J L, Moreno M A, Schleissner C, Salto F, Luengo J M. Molecular cloning and expression in different microbes of the DNA encoding Pseudomonas putida U phenylacetyl-CoA ligase. J Biol Chem. 1996;271:33531–33538. doi: 10.1074/jbc.271.52.33531. [DOI] [PubMed] [Google Scholar]
- 200.Mohamed M E-S. Biochemical and molecular characterization of phenylacetate-coenzyme A ligase, an enzyme catalyzing the first step in aerobic metabolism of phenylacetic acid in Azoarcus evansii. J Bacteriol. 2000;182:286–294. doi: 10.1128/jb.182.2.286-294.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Mohamed M E-S, Zaar A, Ebenau-Jehle C, Fuchs G. Reinvestigation of a new type of aerobic benzoate metabolism in the proteobacterium Azoarcus evansii. J Bacteriol. 2001;183:1899–1908. doi: 10.1128/JB.183.6.1899-1908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mori H, Isono K, Horiuchi T, Miki T. Functional genomics of Escherichia coli in Japan. Res Microbiol. 2000;151:121–128. doi: 10.1016/s0923-2508(00)00119-4. [DOI] [PubMed] [Google Scholar]
- 203.Mulchandani A, Kaneva I, Chen W. Detoxification of organophosphate nerve agents by immobilized Escherichia coli with surface-expressed organophosphorus hydrolase. Biotechnol Bioeng. 1999;63:216–223. doi: 10.1002/(sici)1097-0290(19990420)63:2<216::aid-bit10>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 204.Müller-Newen G, Janssen U, Stoffel W. Enoyl-CoA hydratase and isomerase form a superfamily with a common active-site glutamate residue. Eur J Biochem. 1995;228:68–73. doi: 10.1111/j.1432-1033.1995.tb20230.x. [DOI] [PubMed] [Google Scholar]
- 205.Murdock D, Ensley B D, Serdar C, Thalen M. Construction of metabolic operons catalyzing the de novo biosynthesis of indigo in Escherichia coli. Bio/Technology. 1993;11:381–386. doi: 10.1038/nbt0393-381. [DOI] [PubMed] [Google Scholar]
- 206.Murray J M, Saysell C G, Wilmot C M, Tambyrajah W S, Jaeger J, Knowles P F, Phillips S E V, McPherson M J. The active site base controls cofactors reactivity in Escherichia coli amine oxidase: X-ray crystallographic studies with mutational variants. Biochemistry. 1999;38:8217–8227. doi: 10.1021/bi9900469. [DOI] [PubMed] [Google Scholar]
- 207.Nakahigashi K, Miyamoto K, Nishimura K, Inokuchi H. Isolation and characterization of a light-sensitive mutant of Escherichia coli K-12 with a mutation in a gene that is required for the biosynthesis of ubiquinone. J Bacteriol. 1992;174:7352–7359. doi: 10.1128/jb.174.22.7352-7359.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from the international DNA sequence databases. Nucleic Acids Res. 1997;25:244–245. doi: 10.1093/nar/25.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Nakatsu C H, Straus N A, Wyndham R C. The nucleotide sequence of the Tn5271 3-chlorobenzoate 3,4-dioxygenase genes (cbaAB) unites the class IA oxygenases in a single lineage. Microbiology. 1995;141:485–495. doi: 10.1099/13500872-141-2-485. [DOI] [PubMed] [Google Scholar]
- 210.Neidhardt F C. The enteric bacterial cell and the age of bacteria. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1–3. [Google Scholar]
- 211.Nichols N N, Harwood C S. PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J Bacteriol. 1997;179:5056–5061. doi: 10.1128/jb.179.16.5056-5061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Niemetz R, Altenschmidt U, Brucker S, Fuchs G. Benzoyl-coenzyme-A 3-monooxygenase, a flavin-dependent hydroxylase. Purification, some properties and its role in aerobic benzoate oxidation via gentisate in a denitrifying bacterium. Eur J Biochem. 1995;227:161–168. doi: 10.1111/j.1432-1033.1995.tb20372.x. [DOI] [PubMed] [Google Scholar]
- 213.Nonet M L, Marvel C C, Tolan D R. The hisT-purF region of the Escherichia coli K-12 chromosome. Identification of additional genes of the hisT and purF operons. J Biol Chem. 1987;262:12209–12217. [PubMed] [Google Scholar]
- 214.Ochman H, Jones I B. Evolutionary dynamics of full genome content in Escherichia coli. EMBO J. 2000;19:6637–6643. doi: 10.1093/emboj/19.24.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.O'Connor K, Buckley C M, Hartmans S, Dobson A D W. Possible regulatory role for nonaromatic carbon sources in styrene degradation by Pseudomonas putida CA-3. Appl Environ Microbiol. 1995;61:544–548. doi: 10.1128/aem.61.2.544-548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Oh S-J, Kim Y-C, Park Y-W, Min S-Y, Kim I-S, Kang H-S. Complete nucleotide sequence of the penicillin acylase gene and the flanking regions, and its expression in Escherichia coli. Gene. 1987;56:87–97. doi: 10.1016/0378-1119(87)90161-2. [DOI] [PubMed] [Google Scholar]
- 217.Olivera E R, Miñambres B, García B, Muñiz C, Moreno M A, Ferrández A, Díaz E, García J L, Luengo J M. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc Natl Acad Sci USA. 1998;95:6419–6424. doi: 10.1073/pnas.95.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ötük G. Degradation of ferulic acid by Escherichia coli. J Ferment Technol. 1985;63:501–506. [Google Scholar]
- 219.Overhage J, Priefert H, Steinbüchel A. Biochemical and genetic analyses of ferulic acid catabolism in Pseudomonas sp. strain HR199. Appl Environ Microbiol. 1999;65:4837–4847. doi: 10.1128/aem.65.11.4837-4847.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Panke S, Meyer A, Huber C M, Witholt B, Wubbolts M G. An alkane-responsive expression system for the production of fine chemicals. Appl Environ Microbiol. 1999;65:2324–2332. doi: 10.1128/aem.65.6.2324-2332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Panke S, Witholt B, Schmid A, Wubbolts M G. Towards a biocatalyst for (S)-styrene oxide production: characterization of the styrene degradation pathway of Pseudomonas sp. strain VLB120. Appl Environ Microbiol. 1998;64:2032–2043. doi: 10.1128/aem.64.6.2032-2043.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Panke S, Wubbolts M G, Schmid A, Witholt B. Production of enantiopure styrene oxide by recombinant Escherichia coli synthesizing a two-component styrene monooxygenase. Biotechnol Bioeng. 2000;69:91–100. doi: 10.1002/(sici)1097-0290(20000705)69:1<91::aid-bit11>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 223.Pao S S, Paulsen I T, Saier M H., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Parikh A, Gillam E M J, Guengerich F P. Drug metabolism by Escherichia coli expressing human cytochromes P450. Nat Biotechnol. 1997;15:784–788. doi: 10.1038/nbt0897-784. [DOI] [PubMed] [Google Scholar]
- 225.Park H-S, Lee J-Y, Kim H-S. Production of l-DOPA (3,4-dihydroxyphenyl-l-alanine) from benzene by using a hybrid pathway. Biotechnol Bioeng. 1998;58:339–343. doi: 10.1002/(sici)1097-0290(19980420)58:2/3<339::aid-bit36>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 226.Parke D. Characterization of PcaQ, a LysR-type transcriptional activator required for catabolism of phenolic compounds, from Agrobacterium tumefaciens. J Bacteriol. 1996;178:266–272. doi: 10.1128/jb.178.1.266-272.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Parke D, D'Argenio D A, Ornston L N. Bacteria are not what they eat: that is why they are so diverse. J Bacteriol. 2000;182:257–263. doi: 10.1128/jb.182.2.257-263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Parrot S, Jones S, Cooper R A. 2-Phenylethylamine catabolism by Escherichia coli K12. J Gen Microbiol. 1987;133:347–351. doi: 10.1099/00221287-133-2-347. [DOI] [PubMed] [Google Scholar]
- 229.Parry R J, Wenying L. An NADPH:FAD oxidoreductase from the valanimycin producer Streptomyces viridifaciens. Arch Biochem Biophys. 1997;339:47–54. doi: 10.1006/abbi.1996.9857. [DOI] [PubMed] [Google Scholar]
- 230.Parsons M R, Convery M A, Wilmot C M, Yadav K D S, Blakeley V, Corner A S, Philips S E V, McPherson M J, Knowles P F. Crystal structure of a quinoenzyme:copper amine oxidase of Escherichia coli at 2Å resolution. Structure. 1995;3:1171–1184. doi: 10.1016/s0969-2126(01)00253-2. [DOI] [PubMed] [Google Scholar]
- 231.Payton M, Auty R, Delgoda R, Everett M, Sim E. Cloning and characterization of arylamine N-acetyltransferase genes from Mycobacterium smegmatis and Mycobacterium tuberculosis: increased expression results in isoniazid resistance. J Bacteriol. 1999;181:1343–1347. doi: 10.1128/jb.181.4.1343-1347.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Payton M, Mushtaq A, Yu T-W, Wu L-J, Sinclair J, Sim E. Eubacterial arylamine N-acetyltransferases—identification and comparison of 18 members of the protein family with conserved active site cysteine, histidine and aspartate residues. Microbiology. 2001;147:1137–1147. doi: 10.1099/00221287-147-5-1137. [DOI] [PubMed] [Google Scholar]
- 233.Peekhaus N, Conway T. What's for dinner? Entner-Doudoroff metabolism in Escherichia coli. J Bacteriol. 1998;180:3495–3502. doi: 10.1128/jb.180.14.3495-3502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Pennisi E. Laboratory workhorse decoded. Science. 1997;277:1432–1434. doi: 10.1126/science.277.5331.1432. [DOI] [PubMed] [Google Scholar]
- 235.Peppercorn M A, Goldman P. Caffeic acid metabolism by bacteria of the human gastrointestinal tract. J Bacteriol. 1971;108:996–1000. doi: 10.1128/jb.108.3.996-1000.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Perna N T, Plunkett III G, Burland V, Mau B, Glasner J D, Rose D J, Mayhew G F, Evans P S, Gregor J, Kirkpatrick H A, Pósfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck E J, Davis N W, Lim A, Dimalanta E T, Potamousis K D, Apodaca J, Anantharaman T S, Lin J, Yen G, Schwartz D C, Welch R A, Blattner F R. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 237.Perozich J, Nicholas H, Wang B-C, Lindahl R, Hempel J. Relationships within the aldehyde dehydrogenase extended family. Protein Sci. 1999;8:137–146. doi: 10.1110/ps.8.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Peterson F J, Mason R P, Hovsepian J, Holtzman J L. Oxygen-sensitive and -insensitive nitroreduction by Escherichia coli and rat hepatic microsomes. J Biol Chem. 1979;254:4009–4014. [PubMed] [Google Scholar]
- 239.Pieper D H, Reineke W. Engineering bacteria for bioremediation. Curr Opin Biotechnol. 2000;11:262–270. doi: 10.1016/s0958-1669(00)00094-x. [DOI] [PubMed] [Google Scholar]
- 240.Pittard A J. Biosynthesis of the aromatic amino acids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 458–484. [Google Scholar]
- 241.Platt A, Shingler V, Taylor S C, Williams P A. The 4-hydroxy-2-oxovalerate aldolase and acetaldehyde dehydrogenase (acylating) encoded by the nahM and nahO genes of the naphthalene catabolic plasmid pWW60–22 provide further evidence of conservation of meta-cleavage pathway gene sequences. Microbiology. 1995;141:2223–2233. doi: 10.1099/13500872-141-9-2223. [DOI] [PubMed] [Google Scholar]
- 242.Pollard J R, Bugg T D H. Purification, characterisation and reaction mechanism of monofunctional 2-hydroxypentadienoic acid hydratase from Escherichia coli. Eur J Biochem. 1998;251:98–106. doi: 10.1046/j.1432-1327.1998.2510098.x. [DOI] [PubMed] [Google Scholar]
- 243.Pollard J R, Rialland D, Bugg T D H. Substrate selectivity and biochemical properties of 4-hydroxy-2-keto-pentanoic acid aldolase from Escherichia coli. Appl Environ Microbiol. 1998;64:4093–4094. doi: 10.1128/aem.64.10.4093-4094.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Poon, W. W., D. E. Davis, H, T. Ha, T. Jonassen, P. N. Rather, and C. F. Clarke. 2000. Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. J. Bacteriol. 182:5139–5146. [DOI] [PMC free article] [PubMed]
- 245.Poulsen L K, Lan F, Kristensen C S, Hobolth P, Molin S, Krogfelt K A. Spatial distribution of Escherichia coli in the mouse large intestine inferred from rRNA in situ hybridization. Infect Immun. 1994;62:5191–5194. doi: 10.1128/iai.62.11.5191-5194.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Poulsen L K, Licht T R, Rang C, Krogfelt K A, Molin S. Physiological state of Escherichia coli BJ4 growing in the large intestines of streptomycin-treated mice. J Bacteriol. 1995;177:5840–5845. doi: 10.1128/jb.177.20.5840-5845.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Powell J A C, Archer J A C. Molecular characterisation of a Rhodococcus ohp operon. Antonie Leeuwenhoek. 1998;74:175–188. doi: 10.1023/a:1001784702230. [DOI] [PubMed] [Google Scholar]
- 248.Powlowski J, Sealy J, Shingler V, Cadieux E. On the role of DmpK, an auxiliary protein associated with multicomponent phenol hydroxylase from Pseudomonas sp. strain CF600. J Biol Chem. 1997;272:945–951. doi: 10.1074/jbc.272.2.945. [DOI] [PubMed] [Google Scholar]
- 249.Powlowski J, Shingler V. Genetics and biochemistry of phenol degradation by Pseudomonas sp. CF600. Biodegradation. 1994;5:219–236. doi: 10.1007/BF00696461. [DOI] [PubMed] [Google Scholar]
- 250.Prieto M A, Díaz E, García J L. Molecular characterization of the 4-hydroxyphenylacetate catabolic pathway of Escherichia coli W: engineering a mobile aromatic degradative cluster. J Bacteriol. 1996;178:111–120. doi: 10.1128/jb.178.1.111-120.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Prieto M A, García J L. Molecular characterization of 4-hydroxyphenylacetate 3-hydroxylase of Escherichia coli. J Biol Chem. 1994;269:22823–22829. [PubMed] [Google Scholar]
- 252.Prieto M A, García J L. Identification of the 4-hydroxyphenylacetate transport gene of Escherichia coli W: construction of a highly sensitive cellular biosensor. FEBS Lett. 1997;414:293–297. doi: 10.1016/s0014-5793(97)01012-0. [DOI] [PubMed] [Google Scholar]
- 253.Prieto M A, García J L. Identification of a novel positive regulator of the 4-hydroxyphenylacetate catabolic pathway of Escherichia coli. Biochem Biophys Res Commun. 1997;232:759–765. doi: 10.1006/bbrc.1997.6368. [DOI] [PubMed] [Google Scholar]
- 254.Prieto M A, Pérez-Aranda A, García J L. Characterization of an Escherichia coli aromatic hydroxylase with a broad substrate range. J Bacteriol. 1993;175:2162–2167. doi: 10.1128/jb.175.7.2162-2167.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Qian H, Edlund U, Powlowski J, Shingler V, Sethson I. Solution structure of phenol hydroxylase protein component P2 determined by NMR spectroscopy. Biochemistry. 1997;36:495–504. doi: 10.1021/bi9619233. [DOI] [PubMed] [Google Scholar]
- 256.Ramos J L, Andersson P, Jensen L B, Ramos C, Ronchel M C, Díaz E, Timmis K N, Molin S. Suicide microbes on the loose. Bio/Technology. 1995;13:35–37. doi: 10.1038/nbt0195-35. [DOI] [PubMed] [Google Scholar]
- 257.Ramos-González M-I, Duque E, Ramos J L. Conjugational transfer of recombinant DNA in cultures and in soils: host range of Pseudomonas putida TOL plasmids. Appl Environ Microbiol. 1991;57:3020–3027. doi: 10.1128/aem.57.10.3020-3027.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Reddy J, Lee C, Neeper M, Greasham R, Zhang J. Development of a bioconversion process for production of cis-1S,2R-indandiol from indene by recombinant Escherichia coli constructs. Appl Microbiol Biotechnol. 1999;51:614–620. doi: 10.1007/s002530051440. [DOI] [PubMed] [Google Scholar]
- 259.Reineke W. Development of hybrid strains for the mineralization of chloroaromatics by patchwork assembly. Annu Rev Microbiol. 1998;52:287–331. doi: 10.1146/annurev.micro.52.1.287. [DOI] [PubMed] [Google Scholar]
- 260.Rhodes G, Saunders J R, Pickup R W. Detection and distribution of insertion sequence 1 (IS1)-containing bacteria in the freshwater environment. FEMS Microbiol Ecol. 2000;34:81–90. doi: 10.1111/j.1574-6941.2000.tb00757.x. [DOI] [PubMed] [Google Scholar]
- 261.Richins R D, Kaneva I, Mulchandani A, Chen W. Biodegradation of organophosphorous pesticides by surface-expressed organophosphorus hydrolase. Nat Biotechnol. 1997;15:984–987. doi: 10.1038/nbt1097-984. [DOI] [PubMed] [Google Scholar]
- 262.Riley M, Serres M H. Interim report on genomics of Escherichia coli. Annu Rev Microbiol. 2000;54:341–411. doi: 10.1146/annurev.micro.54.1.341. [DOI] [PubMed] [Google Scholar]
- 263.Roa A, García J L. New insights into the regulation of the pac gene from Escherichia coli W ATCC 11105. FEMS Microbiol Lett. 1999;177:7–14. doi: 10.1111/j.1574-6968.1999.tb13706.x. [DOI] [PubMed] [Google Scholar]
- 264.Robson N D, Parrot S, Cooper R A. In vitro formation of a catabolic plasmid carrying Klebsiella pneumoniae DNA that allows growth of Escherichia coli K-12 on 3-hydroxybenzoate. Microbiology. 1996;142:2115–2120. doi: 10.1099/13500872-142-8-2115. [DOI] [PubMed] [Google Scholar]
- 265.Roh J H, Takenaka Y, Suzuki H, Yamamoto K, Kumagai H. Escherichia coli K-12 copper-containing monoamine oxidase: investigation of the copper binding ligands by site-directed mutagenesis, elemental analysis and TOPA quinone formation. Biochem Biophys Res Commun. 1995;212:1107–1114. doi: 10.1006/bbrc.1995.2083. [DOI] [PubMed] [Google Scholar]
- 266.Romine M F, Stillwell L C, Wong K-K, Thurston S J, Sisk E C, Sensen C, Gaasterland T, Fredrickson J K, Saffer J D. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J Bacteriol. 1999;181:1585–1602. doi: 10.1128/jb.181.5.1585-1602.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Roper D I, Cooper R A. Purification, some properties and nucleotide sequence of 5-carboxymethyl-2-hydroxymuconate isomerase of Escherichia coli C. FEBS Lett. 1990;266:63–66. doi: 10.1016/0014-5793(90)81507-k. [DOI] [PubMed] [Google Scholar]
- 268.Roper D I, Cooper R A. Subcloning and nucleotide sequence of the 3,4-dihydroxyphenylacetate (homoprotocatechuate) 2,3-dioxygenase gene from Escherichia coli C. FEBS Lett. 1990;275:53–57. doi: 10.1016/0014-5793(90)81437-s. [DOI] [PubMed] [Google Scholar]
- 269.Roper D I, Cooper R A. Purification, nucleotide sequence and some properties of a bifunctional isomerase/decarboxylase from the homoprotocatechuate degradative pathway of Escherichia coli C. Eur J Biochem. 1993;217:575–580. doi: 10.1111/j.1432-1033.1993.tb18279.x. [DOI] [PubMed] [Google Scholar]
- 270.Roper D I, Fawcett T, Cooper R A. The Escherichia coli C homoprotocatechuate degradative operon: hpc gene order, direction of transcription and control of expression. Mol Gen Genet. 1993;237:241–250. doi: 10.1007/BF00282806. [DOI] [PubMed] [Google Scholar]
- 271.Roper D I, Stringfellow J M, Cooper R A. Sequence of the hpcC and hpcG genes of the meta-fission homoprotocatechuic acid pathway of Escherichia coli C: nearly 40% amino-acid identity with the analogous enzymes of the catechol pathway. Gene. 1995;156:47–51. doi: 10.1016/0378-1119(95)00082-h. [DOI] [PubMed] [Google Scholar]
- 272.Rosche B, Tshisuaka B, Hauer B, Lingens F, Fetzner S. 2-Oxo-1,2-dihydroquinoline 8-monooxygenase: phylogenetic relationship to other multicomponent nonheme iron oxygenases. J Bacteriol. 1997;179:3549–3554. doi: 10.1128/jb.179.11.3549-3554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Rudd K E. New tools for an old workhorse. Nat Biotechnol. 2000;18:1241–1242. doi: 10.1038/82339. [DOI] [PubMed] [Google Scholar]
- 274.Rusnak F, Faraci W S, Walsh C T. Subcloning, expression, and purification of the enterobactin biosynthetic enzyme 2,3-dihydroxybenzoate-AMP-ligase: demonstration of enzyme-bound (2,3-dihydroxybenzoyl)adenylate product. Biochemistry. 1989;28:6827–6835. doi: 10.1021/bi00443a008. [DOI] [PubMed] [Google Scholar]
- 275.Saier M H., Jr Molecular phylogeny as a basis for the classification of transport proteins from bacteria, archaea and eukarya. Adv Microb Physiol. 1998;40:81–136. doi: 10.1016/s0065-2911(08)60130-7. [DOI] [PubMed] [Google Scholar]
- 276.Sakaitani M, Rusnak F, Quinn N R, Tu C, Frigo T B, Berchtold G A, Walsh C T. Mechanistic studies on trans-2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase (EntA) in the biosynthesis of the iron chelator enterobactin. Biochemistry. 1990;29:6789–6798. doi: 10.1021/bi00481a006. [DOI] [PubMed] [Google Scholar]
- 277.Sánchez M, Fernández J, Martin M, Gibello A, Garrido-Pertierra A. Purification and properties of two succinic semialdehyde dehydrogenases from Klebsiella pneumoniae. Biochim Biophys Acta. 1989;990:225–231. doi: 10.1016/s0304-4165(89)80038-8. [DOI] [PubMed] [Google Scholar]
- 278.Sarkissian C N, Shao Z, Blain F, Peevers R, Su H, Heft R, Chang T M S, Scriver C R. A different approach to treatment of phenylketonuria: phenylalanine degradation with recombinant phenylalanine ammonia lyase. Proc Natl Acad Sci USA. 1999;96:2339–2344. doi: 10.1073/pnas.96.5.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Sarsero J P, Pittard A J. Membrane topology analysis of Escherichia coli K-12 Mtr permease by alkaline phosphatase and β-galactosidase fusions. J Bacteriol. 1995;177:297–306. doi: 10.1128/jb.177.2.297-306.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Savage D C. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 281.Savageau M A. Regulation of differentiated cell-specific functions. Proc Natl Acad Sci USA. 1983;80:1411–1415. doi: 10.1073/pnas.80.5.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Scarlett F A, Turner J M. Microbial metabolism of amino alcohols. Ethanolamine catabolism mediated by coenzyme B12-dependent ethanolamine ammonia-lyase in Escherichia coli and Klebsiella aerogenes. J Gen Microbiol. 1976;95:173–176. doi: 10.1099/00221287-95-1-173. [DOI] [PubMed] [Google Scholar]
- 283.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 284.Schmitz A, Gatermann K-H, Fiedler J, Grund E, Eichenlaub R. Cloning and sequence analysis of genes for dehalogenation of 4-chlorobenzoate from Arthrobacter sp. strain SU. Appl Environ Microbiol. 1992;58:4068–4071. doi: 10.1128/aem.58.12.4068-4071.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Schneider H, Schwiertz A, Collins M D, Blaut M. Anaerobic transformation of quercetin-3-glucoside by bacteria from human intestinal tract. Arch Microbiol. 1999;171:81–91. doi: 10.1007/s002030050682. [DOI] [PubMed] [Google Scholar]
- 286.Schottel J L. The mercuric and organomercurial detoxifying enzymes from a plasmid-bearing strain of Escherichia coli. J Biol Chem. 1978;253:4341–4349. [PubMed] [Google Scholar]
- 287.Schulze B, Wubbolts M G. Biocatalysis for industrial production of fine chemicals. Curr Opin Biotechnol. 1999;10:609–615. doi: 10.1016/s0958-1669(99)00042-7. [DOI] [PubMed] [Google Scholar]
- 288.Seah S Y K, Terracina G, Bolin J T, Riebel P, Snieckus V, Eltis L D. Purification and preliminary characterization of a serine hydrolase involved in the microbial degradation of polychlorinated biphenyls. J Biol Chem. 1998;273:22943–22949. doi: 10.1074/jbc.273.36.22943. [DOI] [PubMed] [Google Scholar]
- 289.Seeger C, Poulsen C, Dandanell G. Identification and characterization of genes (xapA, xapB, and xapR) involved in xanthosine catabolism in Escherichia coli. J Bacteriol. 1995;177:5506–5516. doi: 10.1128/jb.177.19.5506-5516.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Selifonova O V, Eaton R W. Use of ipb-lux fusion to study regulation of the isopropylbenzene catabolism operon of Pseudomonas putida RE204 and to detect hydrophobic pollutants in the environment. Appl Environ Microbiol. 1996;62:778–783. doi: 10.1128/aem.62.3.778-783.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Selinger D W, Cheung K J, Mei R, Johansson E M, Richmond C S, Blattner F R, Lockhart D J, Church G M. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat Biotechnol. 2000;18:1262–1268. doi: 10.1038/82367. [DOI] [PubMed] [Google Scholar]
- 292.Silver S. Bacterial resistances to toxic metal ions—a review. Gene. 1996;179:9–19. doi: 10.1016/s0378-1119(96)00323-x. [DOI] [PubMed] [Google Scholar]
- 293.Simoni S, Klinke S, Zipper C, Angst W, Kohler H-P E. Enantioselective metabolism of chiral 3-phenylbutyric acid, an intermediate of linear alkylbenzene degradation by Rhodococcus rhodochrous PB1. Appl Environ Microbiol. 1996;62:749–755. doi: 10.1128/aem.62.3.749-755.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Singh B K, Kuhad R C, Singh A, Tripathi K K, Ghosh P K. Microbial degradation of the pesticide lindane (γ-hexachlorocyclohexane) Adv Appl Microbiol. 2000;47:269–298. doi: 10.1016/s0065-2164(00)47007-3. [DOI] [PubMed] [Google Scholar]
- 295.Skinner M A, Cooper R A. An Escherichia coli mutant defective in the NAD-dependent succinate semialdehyde dehydrogenase. Arch Microbiol. 1982;132:270–275. doi: 10.1007/BF00407964. [DOI] [PubMed] [Google Scholar]
- 296.Smith E A, Macfarlane G T. Enumeration of human colonic bacteria producing phenolic and indolic compounds: effects of pH, carbohydrate avalability and retention time on dissimilatory aromatic amino acid metabolims. J Appl Bacteriol. 1996;81:288–302. doi: 10.1111/j.1365-2672.1996.tb04331.x. [DOI] [PubMed] [Google Scholar]
- 297.Smith H W. Survival of orally administered E. coli K12 in alimentary tract of human. Nature. 1975;55:500–502. doi: 10.1038/255500a0. [DOI] [PubMed] [Google Scholar]
- 298.Spence E L, Kawamukai M, Sanvoisin J, Braven H, Bugg T D H. Catechol dioxygenases from Escherichia coli (MhpB) and Alcaligenes eutrophus (MpcI): sequence analysis and biochemical properties of a third family of extradiol dioxygenases. J Bacteriol. 1996;178:5249–5256. doi: 10.1128/jb.178.17.5249-5256.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Steinebach V, Benen J A E, Bader R, Postma P W, de Vries S, Duine J A. Cloning the maoA gene that encodes aromatic amino oxidase of Escherichia coli W3350 and characterization of the overexpressed enzyme. Eur J Biochem. 1996;237:584–591. doi: 10.1111/j.1432-1033.1996.0584p.x. [DOI] [PubMed] [Google Scholar]
- 300.Stojcevic N, Moric I, Begovic J, Radoja S, Konstantinovic M. DNA architecture and transcriptional regulation of the Escherichia coli penicillin amidase (pac) gene. Biomol Eng. 2001;17:113–117. doi: 10.1016/s1389-0344(00)00074-5. [DOI] [PubMed] [Google Scholar]
- 301.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S, Hufnagle W O, Kowalik D J, Lagrou M, Garber R L, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody L L, Coulter S N, Folger K R, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong G K, Wu Z, Paulsen I T. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 302.Strickland S, Massey V. The purification and properties of the flavoprotein melilotate hydroxylase. J Biol Chem. 1973;248:2944–2952. [PubMed] [Google Scholar]
- 303.Stringfellow J M, Turpin B, Cooper R A. Sequence of the Escherichia coli C homoprotocatechuic acid degradative operon completed with that of the 2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase-encoding gene (hpcH) Gene. 1995;166:73–76. doi: 10.1016/0378-1119(95)00596-8. [DOI] [PubMed] [Google Scholar]
- 304.Subramanya H S, Roper D I, Dauter Z, Dodson E J, Davies G J, Wilson K S, Wigley D B. Enzymatic ketonization of 2-hydroxymuconate: specificity and mechanism investigated by the crystal structures of two isomerases. Biochemistry. 1996;35:792–802. doi: 10.1021/bi951732k. [DOI] [PubMed] [Google Scholar]
- 305.Suen W-C, Haigler B E, Spain J C. 2,4-Dinitrotoluene dioxygenase from Burkholderia sp. strain DNT: similarity to naphthalene dioxygenase. J Bacteriol. 1996;178:4926–4934. doi: 10.1128/jb.178.16.4926-4934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Sugimoto K, Senda T, Aoshima H, Masai E, Fukuda M, Mitsui Y. Crystal structure of an aromatic ring opening dioxygenase LigAB, a protocatechuate 4,5-dioxygenase, under aerobic conditions. Structure. 1999;7:953–965. doi: 10.1016/s0969-2126(99)80122-1. [DOI] [PubMed] [Google Scholar]
- 307.Sugino H, Sasaki M, Azakami H, Yamashita M, Murooka Y. A monoamine-regulated Klebsiella aerogenes operon containing the monoamine oxidase structural gene (maoA) and the maoC gene. J Bacteriol. 1992;174:2485–2492. doi: 10.1128/jb.174.8.2485-2492.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Suresh C G, Pundle A V, SivaRaman H, Rao K N, Brannigan J A, McVey C E, Verma C S, Dauter Z, Dodson E J, Dodson G G. Penicillin V acylase crystal structure reveals new Ntn-hydrolase family members. Nat Struct Biol. 1999;6:414–416. doi: 10.1038/8213. [DOI] [PubMed] [Google Scholar]
- 309.Szentimai A. Production of penicillin acylase. Appl Microbiol. 1964;12:185–187. doi: 10.1128/am.12.3.185-187.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Takami H, Nakasone K, Ogasawara N, Hirama C, Nakamura Y, Masui N, Fuji F, Takaki Y, Inoue A, Horikoshi K. Sequencing of three lambda clones from the genome of alkaliphilic Bacillus sp. strain C-125. Extremophiles. 1999;3:29–34. doi: 10.1007/s007920050096. [DOI] [PubMed] [Google Scholar]
- 311.Takami H, Nakasone K, Takaki Y, Maeno G, Sasaki R, Masui N, Fuji F, Hirama C, Nakamura Y, Ogasawara N, Kuhara S, Horikoshi K. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 2000;28:4317–4331. doi: 10.1093/nar/28.21.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Taylor A B, Czerwinski R M, Johnson W H, Jr, Whitman C P, Hackert M L. Crystal structure of 4-oxalocrotonate tautomerase inactivated by 2-oxo-3-pentynoate at 2.4 Å resolution: analysis and implications for the mechanism of inactivation and catalysis. Biochemistry. 1998;37:14692–14700. doi: 10.1021/bi981607j. [DOI] [PubMed] [Google Scholar]
- 313.Timmis K N, Steffan R J, Unterman R. Designing microorganisms for the treatment of toxic wastes. Annu Rev Microbiol. 1994;48:525–557. doi: 10.1146/annurev.mi.48.100194.002521. [DOI] [PubMed] [Google Scholar]
- 314.Torres B, Jaenecke S, Timmis K N, García J L, Díaz E. A gene containment strategy based on a restriction-modification system. Environ Microbiol. 2000;2:555–563. doi: 10.1046/j.1462-2920.2000.00138.x. [DOI] [PubMed] [Google Scholar]
- 315.Tsuge T, Fukui T, Matsusaki H, Taguchi S, Kobayashi G, Ishizaki A, Doi Y. Molecular cloning of two (R)-specific enoyl-CoA hydratase genes from Pseudomonas aeruginosa and their use for polyhydroxyalkanoate synthesis. FEMS Microbiol Lett. 2000;184:193–198. doi: 10.1111/j.1574-6968.2000.tb09013.x. [DOI] [PubMed] [Google Scholar]
- 316.Updegraff D M. The production of phenol and para-cresol by marine bacteria J. Bacteriol. 1949;57:555–564. doi: 10.1128/jb.57.5.555-564.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Valle F, Balbás P, Merino E, Bolivar F. The role of penicllin amidase in nature and in industry. Trends Biochem Sci. 1991;16:36–40. doi: 10.1016/0968-0004(91)90014-m. [DOI] [PubMed] [Google Scholar]
- 318.van Aalten D M F, DiRusso C C, Knudsen J, Wierenga R K. Crystal structure of FadR, a fatty acid-responsive transcription factor with a novel acyl coenzyme A-binding fold. EMBO J. 2000;19:5167–5177. doi: 10.1093/emboj/19.19.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.van den Berg W A M, Hagen W R, van Dongen W M A M. The hybrid-cluster protein (‘prismane protein’) from Escherichia coli. Characterization of the hybrid-cluster protein, redox properties of the [2Fe-2S] and [4Fe-2S–2O] clusters and identification of an associated NADH oxidoreductase containing FAD and [2Fe-2S] Eur J Biochem. 2000;267:666–676. doi: 10.1046/j.1432-1327.2000.01032.x. [DOI] [PubMed] [Google Scholar]
- 320.van der Heiden C, Wadmn S K, Ketting D, de Bree P K. Urinary and faecal excretion of metabolites of tyrosine and phenylalanine in patient with cystic fibrosis and severely impaired amino acid absortion. Clin Chim Acta. 1971;31:133–141. doi: 10.1016/0009-8981(71)90370-6. [DOI] [PubMed] [Google Scholar]
- 321.van der Meer J R, de Vos W M, Harayama S, Zehnder A J B. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev. 1992;56:677–694. doi: 10.1128/mr.56.4.677-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Vasudevan S G, Shaw D C, Armarego W L F. Dihydropteridine reductase from Escherichia coli. Biochem J. 1988;255:581–588. [PMC free article] [PubMed] [Google Scholar]
- 323.Vedadi M, Barriault D, Sylvestre M, Powlowski J. Active site residues of cis-2,3-dihydro-2,3-dihydroxybiphenyl dehydrogenase from Comamonas testosteroni strain B-356. Biochemistry. 2000;39:5028–5034. doi: 10.1021/bi992232k. [DOI] [PubMed] [Google Scholar]
- 324.Velasco A, Alonso S, García J L, Perera J, Díaz E. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J Bacteriol. 1998;180:1063–1071. doi: 10.1128/jb.180.5.1063-1071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Venturi V, Zennaro F, Degrassi G, Okeke B C, Bruschi C V. Genetics of ferulic acid bioconversion to protocatechuic acid in plant-growth-promoting Pseudomonas putida WCS358. Microbiology. 1998;144:965–973. doi: 10.1099/00221287-144-4-965. [DOI] [PubMed] [Google Scholar]
- 326.Vermeij P, Vinke E, Keltjens J T, van der Drift C. Purification and properties of coenzyme F390 hydrolase from Methanobacterium thermoautotrophicum (strain Marburg) Eur J Biochem. 1995;234:592–597. doi: 10.1111/j.1432-1033.1995.592_b.x. [DOI] [PubMed] [Google Scholar]
- 327.Virden R. Structure, processing and catalytic action of penicillin acylase. Biotechnol Genet Eng Rev. 1990;8:189–218. [Google Scholar]
- 328.Vitovski S. Phenylacetate-coenzyme A ligase is induced during growth on phenylacetic acid in different bacteria of several genera. FEMS Microbiol Lett. 1993;108:1–6. doi: 10.1016/0378-1097(93)90477-j. [DOI] [PubMed] [Google Scholar]
- 329.Werwath J, Arfmann H A, Pieper D H, Timmis K N, Wittich R M. Biochemical and genetic characterization of a gentisate 1,2-dioxygenase from Sphingomonas sp. strain RW5. J Bacteriol. 1998;180:4171–4176. doi: 10.1128/jb.180.16.4171-4176.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Whaley K J, Zeitlin L. Transgenic commensals as mucosal protectants. Nat Biotechnol. 2000;18:1038–1039. doi: 10.1038/80220. [DOI] [PubMed] [Google Scholar]
- 331.White O, Eisen J A, Heidelberg J F, Hickey E K, Peterson J D, Dodson R J, Haft D H, Gwinn M L, Nelson W C, Richardson D L, Moffat K S, Qin H, Jiang L, Pamphile W, Crosby M, Shen M, Vamathevan J J, Lam P, McDonald L, Utterback T, Zalewski C, Makarova K S, Aravind L, Daly M J, Minton K W, Fleischmann R D, Ketchum K A, Nelson K E, Salzberg S, Smith H O, Venter J C, Fraser C M. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Whiteway J, Koziarz P, Veall J, Sandhu N, Kumar P, Hoecher B, Lambert I B. Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nsfB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J Bacteriol. 1998;180:5529–5539. doi: 10.1128/jb.180.21.5529-5539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Whiting A K, Boldt Y R, Hendrich M P, Wackett L P, Que L Jr. Manganese(II)-dependent extradiol-cleaving catechol dioxygenase from Arthrobacter globiformis CM-2. Biochemistry. 1996;35:160–170. doi: 10.1021/bi951979h. [DOI] [PubMed] [Google Scholar]
- 334.Willardson B M, Wilkins J F, Rand T A, Schupp J M, Hill K K, Keim P, Jackson P J. Development and testing of a bacterial biosensor for toluene-based environmental contaminants. Appl Environ Microbiol. 1998;64:1006–1012. doi: 10.1128/aem.64.3.1006-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Williams P A, Sayers J R. The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation. 1994;5:195–217. doi: 10.1007/BF00696460. [DOI] [PubMed] [Google Scholar]
- 336.Wilmot C M, Hajdu J, McPherson M J, Knowles P F, Phillips S E V. Visualization of dioxygen bound to copper during enzyme catalysis. Science. 1999;286:1724–1728. doi: 10.1126/science.286.5445.1724. [DOI] [PubMed] [Google Scholar]
- 337.Wilson M, Lindow S E. Release of recombinant microorganisms. Annu Rev Microbiol. 1993;47:913–944. doi: 10.1146/annurev.mi.47.100193.004405. [DOI] [PubMed] [Google Scholar]
- 338.Winter J, Moore L H, Dowell V R, Jr, Bokkenheuser V D. C-ring cleavage of flavonoids by human intestinal bacteria. Appl Environ Microbiol. 1989;55:1203–1208. doi: 10.1128/aem.55.5.1203-1208.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Witkin E M. Inherited differences in sensitivity to radiation in Escherichia coli. Proc Natl Acad Sci USA. 1946;32:59–68. doi: 10.1073/pnas.32.3.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Xun L, Sandvik E R. Characterization of 4-hydroxyphenylacetate 3-hydroxylase (HpaB) of Escherichia coli as a reduced flavin adenine dinucleotide-utilizing monooxygenase. Appl Environ Microbiol. 2000;66:481–486. doi: 10.1128/aem.66.2.481-486.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Yamada T, Murooka Y, Harada T. Comparative immunological studies on arylsulfatase in bacteria of the family Enterobacteriaceae: occurrence of latent arylsulfatase protein regulated by sulfur compounds and tyramine. J Bacteriol. 1978;133:536–541. doi: 10.1128/jb.133.2.536-541.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Yamashita M, Azakami H, Yokoro N, Roh J-H, Suzuki H, Kumagai H, Murooka Y. maoB, a gene that encodes a positive regulator of the monoamine oxidase gene (maoA) in Escherichia coli. J Bacteriol. 1996;178:2941–2947. doi: 10.1128/jb.178.10.2941-2947.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Yanofsky C, Horn V, Gollnick P. Physiological studies of tryptophan transport and tryptophanase operon induction in Escherichia coli. J Bacteriol. 1991;173:6009–6017. doi: 10.1128/jb.173.19.6009-6017.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Zaar A, Eisenreich W, Bacher A, Fuchs G. A novel pathway of aerobic benzoate catabolism in the bacteria Azoarcus evansii and Bacillus stearothermophilus. J Biol Chem. 2001;276:24997–25004. doi: 10.1074/jbc.M100291200. [DOI] [PubMed] [Google Scholar]
- 345.Zenk M H, Ulbrich B, Busse J, Stöckigt J. Procedure for the enzymatic synthesis and isolation of cinnamoyl-CoA thioesters using a bacterial system. Anal Biochem. 1980;101:182–187. doi: 10.1016/0003-2697(80)90058-5. [DOI] [PubMed] [Google Scholar]
- 346.Zenno S, Kobori T, Tanokura M, Saigo K. Conversion of NfsA, the major Escherichia coli nitroreductase, to a flavin reductase with an activity similar to that of Frp, a flavin reductase in Vibrio harveyi, by a single amino acid substitution. J Bacteriol. 1998;180:422–425. doi: 10.1128/jb.180.2.422-425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Zenno S, Koike H, Kumar A N, Jayaraman R, Tanokura M, Saigo K. Biochemical characterization of NfsA, the Escherichia coli major nitroreductase exhibiting a high amino acid sequence homology to Frp, a Vibrio harveyi flavin oxidoreductase. J Bacteriol. 1996;178:4508–4514. doi: 10.1128/jb.178.15.4508-4514.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Zeyer J, Lehrbach P R, Timmis K N. Use of cloned genes of Pseudomonas TOL plasmid to effect biotransformation of benzoates to cis-dihydrodiols and catechols by Escherichia coli cells. Appl Environ Microbiol. 1985;50:1409–1413. doi: 10.1128/aem.50.6.1409-1413.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Zhang H, Javor G T. Identification of the ubiD gene on the Escherichia coli chromosome. J Bacteriol. 2000;182:6243–6246. doi: 10.1128/jb.182.21.6243-6246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Zhao S, Zhu Q, Somerville R L. The ς70 transcription factor TyrR has zinc-stimulated phosphatase activity that is inhibited by ATP and tyrosine. J Bacteriol. 2000;182:1053–1061. doi: 10.1128/jb.182.4.1053-1061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Zhou N-Y, Fuenmayor S L, Williams P A. nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J Bacteriol. 2001;183:700–708. doi: 10.1128/JB.183.2.700-708.2001. . R. P. [DOI] [PMC free article] [PubMed] [Google Scholar]