Abstract
The concepts that we generally associate with the field of data science are strikingly descriptive of the way that life, in general, processes information about its environment. The ‘information life cycle’, which enumerates the stages of information treatment in data science endeavours, also captures the steps of data collection and handling in biological systems. Similarly, the ‘data–information–knowledge ecosystem’, developed to illuminate the role of informatics in translating raw data into knowledge, can be a framework for understanding how information is constantly being transferred between life and the environment. By placing the principles of data science in a broader biological context, we see the activities of data scientists as the latest development in life's ongoing journey to better understand and predict its environment. Finally, we propose that informatics frameworks can be used to understand the similarities and differences between abiotic complex evolving systems and life.
Keywords: information, informatics, data science, evolution, definitions of life
1. Introduction
One of the most enigmatic questions in science continues to be what is life? (e.g. [1–4]). Despite numerous attempts to define life, there is no single agreed upon characterization of the living state—or even a consensus on whether one is needed [5–7]. This lack of agreement reveals a major gap in scientific understanding with implications for the search for life elsewhere and the creation of de novo life [8]. Few would argue with the idea that information processing is one of the central pillars of life, but a universal definition of information and how exactly information creates a distinction between life and non-life is far from settled.
Today, information is so prevalent in our lives that we have created new domains of science—e.g. data science and informatics—that are centred upon exploring information's multifaceted nature and how it can be used to reveal trends, patterns and truths about our world. The development of informatics has resulted in heuristic methods that elucidate the role of information in data science endeavours. In this contribution, we illustrate how two of these concepts—namely, the ‘information life cycle’ and the ‘data–information–knowledge ecosystem’—can also be used to describe the ways in which information flows through living systems in general. We propose that an informatics perspective may be a particularly illuminating lens through which to understand the differences between living and non-living systems. In our framework, living systems constitute a subset of complex evolving systems that perform the full information life cycle and are characterized by a rich data–information–knowledge ecosystem.
2. The role of information in data science
The advent of data science in recent decades has reshaped science and society alike [9]. In the physical sciences (e.g. astronomy, geochemistry and mineralogy), life sciences (e.g. agricultural science, genomics and public health) and social sciences (e.g. economics, linguistics and political science), nearly every sector of modern life has been affected by the so-called ‘big data revolution’. Concomitantly, we have witnessed the rise of informatics—a field that focuses on understanding the structure, properties and activities of scientific information, rather than just its content [10]. In the digital age, it is easy to see how the human world is critically dependent upon flows of information. But could it be that life has been practicing the principles of data science and informatics ever since its inception?
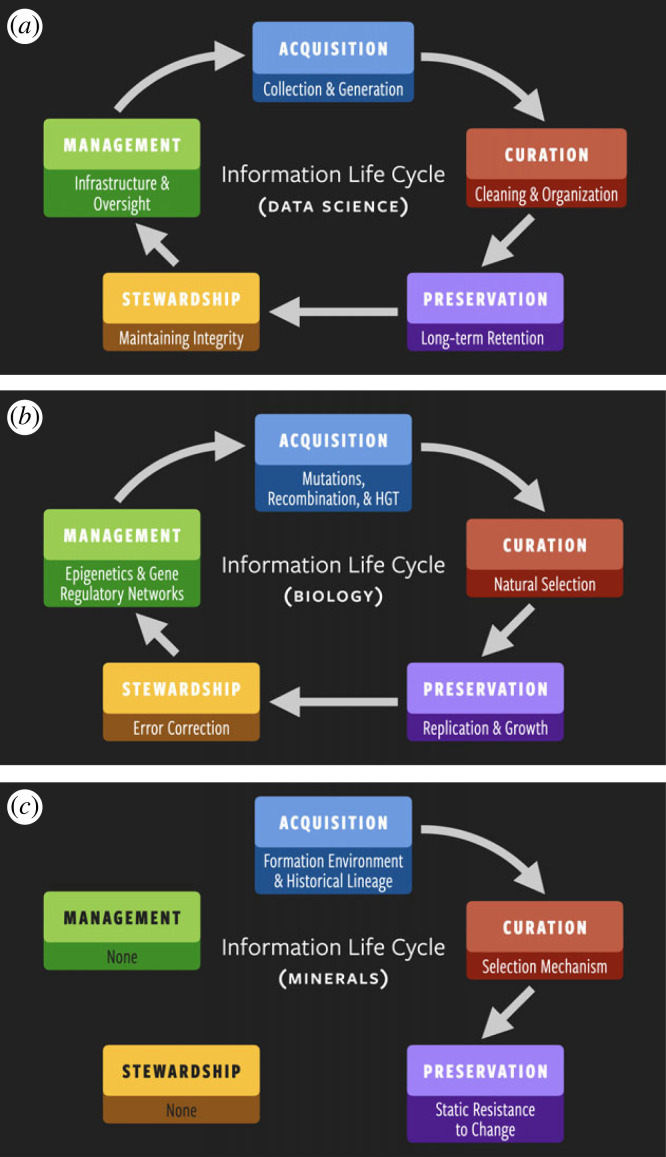
Central to data science endeavours is the so-called ‘information life cycle’, which describes the steps that stored information goes through, from its creation to its deletion or archiving (figure 1a) [10,11]. The first step is acquisition: data must be gathered, perhaps by direct observation or experiment, generated by theory, or rescued from existing but scattered resources. The second step is curation: after its collection, data must be cleaned and processed into a state where it is useful; e.g. disparate pieces of information may be standardized and ‘datafied’. The third step is preservation: the data must be retained in usable form so that they can be accessed in the future, both for their original purpose and for purposes not yet imagined at the time of collection. The fourth step is stewardship: the process of maintaining integrity across acquisition, curation and preservation. The fifth and final step is management: creating the infrastructure that oversees all of the other processes, ensuring that previously acquired data are always available to access and use and facilitating further data collection. The functional use of information drives the information life cycle: each step advances the goal of making information more usable, reliable and acquirable.
Figure 1.
The information life cycle in (a) informatics, (b) biology and (c) minerals. Inspired by [10].
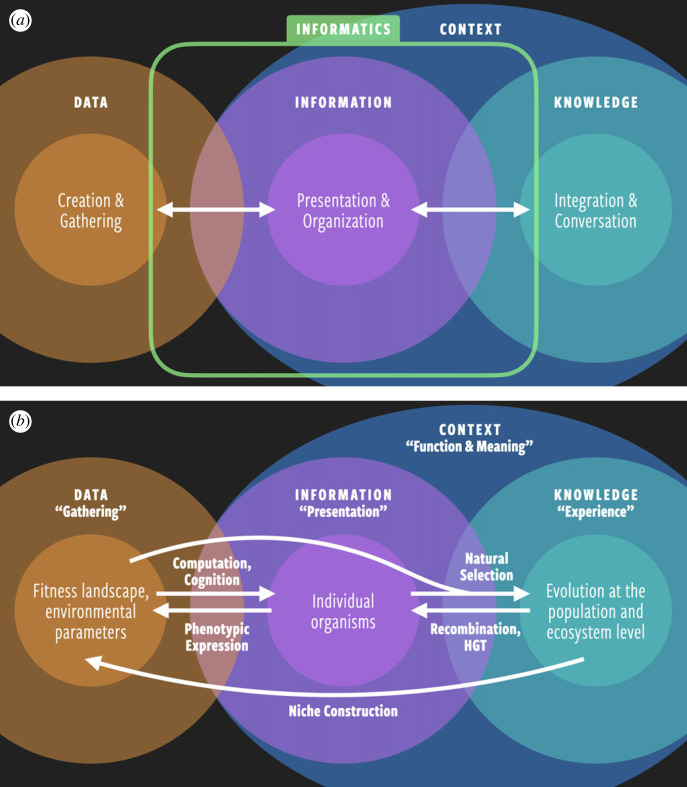
Another key concept in informatics is the ‘data–information–knowledge ecosystem’, which describes how data are processed, represented and communicated in a meaningful way (figure 2a) [10,11]. Raw data are generally useless to the majority of people, so informatics methods must be used to transform data into information, which is a representation of the raw data that can be understood by the public. For example, raw data for natural events like hurricanes, earthquakes or volcanoes may include readings from sensors, satellite data and even data from social media posts documenting these natural events. When sensor data or satellite data are plotted into a visualization like a map or a graph, we can finally see the geographical boundaries for the affected areas of a particular hurricane or earthquake.
Figure 2.
(a) The data–information–knowledge ecosystem in data science, highlighting the role of informatics in converting raw data into human knowledge. (b) The data–information–knowledge ecosystem in biological systems, cataloguing various transformations and feedbacks between the three domains. In both data science and living systems, information and knowledge are reliant on context. Inspired by [10].
Once consumers are armed with the appropriate information, they use that information, in combination with other experiences, to gain knowledge. For example, meteorologists are able to track the movement of a hurricane and predict the path and intensity of that hurricane based on their expertise and insights they obtained from looking at the information presented to them from a combination of data sources. Knowledge, therefore, is not something acquired in solitude; it is a collective phenomenon reliant on social experiences and context.
The modus operandi of data science is to amass and process large quantities of data in order to draw statistically robust correlations, find previously undiscovered associations or make predictions and/or recommendations that rival or even supersede those of analytical theory. The characterization of complex systems is one arena where data science shines. It is often challenging to derive analytical laws for phenomena with many dynamical components that are influenced by forces at a wide range of spatial and temporal scales. Instead, data science methods can be used to glean accurate and predictive statistical laws. Although big data analytics alone may not be able to divine causality, the discoveries that data science makes can be used to motivate new physical explanations and novel scientific narratives. For example, mineral occurrence data can be used to form mineral association rules that give us the ability to predict previously unknown mineral occurrences and also provide insights into formational environments of minerals and their characteristics [12]. For myriad complex phenomena, data science has helped us reach beyond the limitations of traditional reductionist theory. Will data science approaches eventually lead us to an ultimate description of reality that is superior, in predictive and explanatory power, to compact, ‘physicsy’ descriptions of nature? We leave this question to the future and a discussion of its implications to a more philosophical text.
3. Seeing information in life through a data science lens
Information gathering, processing and transference are fundamental aspects of life. Biological systems are learning systems: they record information about their environment and process that information to enhance their ability to survive [13]. Life has evolved manifold ways to perceive its environment, from chemical receptors to vision to magnetoreception. Our mammalian neurological architecture allows us to integrate huge amounts of data to compute our surroundings and make useful predictions about the world—e.g. if I climb a tree, is that lion less likely to eat me?—but even the simplest unicellular life forms (including their ‘dormant’ spores) can perform complex cognitive tasks, like memory and decision-making [14,15]. Indeed, populations of replicating entities can be understood to perform Bayesian inference, and Darwinian evolution can be equated to a search algorithm built upon the basic principles of replication, variation and selection [16–18]. We propose that life does (and has always done) data science–analogue activities without necessarily being ‘conscious’ of it.
The information life cycle of data science is also a valid summary of major informational processes in biological evolution (figure 1b). The acquisition stage describes the generation of novel genetic sequences, primarily driven by mutations to the germline, recombination and horizontal gene transfer. The curation stage is performed by natural selection, pruning a wide range of possible information-bearing states to a smaller number of viable ones. The preservation stage is achieved through replication, reproduction and growth, ensuring that genomic information persists and proliferates through time. (We note that in biological systems, curation and preservation are intimately linked: natural selection operates upon reproducing systems. However, it is useful to keep these stages distinct because non-living systems can exhibit selection without replication (e.g. mineral paragenesis) and growth without curation (e.g. wildfire).) The stewardship stage is performed by error-correcting mechanisms, such as enzymes that perform kinetic proofreading during DNA replication (e.g. [19–23]). The management stage involves a host of mechanisms that control gene expression, such as epigenetic markers (e.g. [24]), gene regulatory networks (e.g. [25]) and factors that tune mutation rate and the uptake of new genes from the environment (e.g. [26,27]).
The modern biological mechanisms responsible for stewardship and management are the result of billions of years of evolution, and it is likely that near the origin of life, steps four and five emerged from the first three steps of the information life cycle. Once evolved, stewardship and management cemented themselves in the information life cycle by benefiting preservation and evolvability—a feedback akin to how stewardship and management in data science facilitate data sharing and collaboration, which in turn help with the acquisition and processing of yet more data.
There is also an analogue to the data–information–knowledge ecosystem present in living systems (figure 2b). Here, data represent the physical features of the environment—temperature, salinity, chemical gradients, seasonal cycles, random fluctuations, etc. By sensing and computing their environment, biological systems transform data into information that is relevant to the survival of an organism or a group of organisms. Examples of the transduction of environmental data into biological information include: how the retina converts photons into nerve impulses [28], how cells transform physical forces into chemical signals [29] and how any number of stimuli can result in modifications to the biochemical circuitry of the proteome [30,31].
In our framework, information is distinct from data in that information contains ‘meaning’ in a given context. Here we draw inspiration from the field of informatics, in which the term ‘data’ refers to raw bits in nature, whereas ‘information’ refers to data that have gained meaning through context or function. In other words, information is a product of data going through the information life cycle; it is data in use. For example, a sequence of nucleobases in a DNA polymer means nothing without the enzymatic machinery required to transcribe and translate it into a polypeptide. In the context of life on Earth, DNA exists within the biological context of a living cell, so it serves a specific function and derives its meaning from its role in the so-called ‘central dogma of molecular biology’ [32]. Should a molecule of DNA exist in some extraterrestrial biosphere whose exobiochemistry has no use for it, that DNA strand would not contain information in our sense of the word; rather, it would merely be a piece of raw data in the environment.
An individual biological unit of selection is essentially a proposal from the biosphere to the environment—a prediction put to the test against new data. Via natural selection over multi-generational time, certain predictions will be strengthened while others are discarded, and populations of individual agents will gain lasting knowledge of their environment—recipes of success written in genetic code. Hence, knowledge in the biological sense is like knowledge in the data science sense: it can only be gained through ‘experience’ over time at the communal level.
Biological systems create information from data and knowledge from information (rightward arrows in figure 2b), but so too do they create new information from knowledge and new data from information (leftward arrows in figure 2b). By existing within the environment with respect to other organisms, and by influencing the environment through phenotypic expression and niche construction, life creates more environmental data and alters the fitness landscape for other life [33–35]. Through recombination (e.g. exercising the ‘knowledge’ of sexual reproduction) and horizontal gene transfer, populations steadily produce new individual variations. Thus, the data–information–knowledge ecosystem is made complete by the ways in which individual organisms and ecosystems impact their environment, changing the data that must be gathered by future generations and compelling the coevolution of life and planet.
Information processing and preservation is an energetically costly undertaking; life's emergence and continual evolution must be driven by the non-negligible complexity—i.e. the high data content—of its environment [36]. Although the emergence of life is still shrouded in mystery, information processing was probably a key feature responsible for the transition between non-living and living systems [37,38]. Numerous proposals have been made for how abiotic systems could have begun transforming environmental data into meaningful information, including but not limited to RNA template-driven replication (e.g. [39]), amyloidal self-propagation (e.g. [40]), mineral templating (e.g. [41,42]) and associative learning in chemical systems [43]. Despite the great diversity of environments and materials proposed to be responsible for the origin of life, in each of these scenarios, information in a proto-living system promotes that system's persistence over time, reflecting the general principle that information processing is a hypothesis-agnostic pillar of life [13].
Life is a data collection and information-processing system built from organic chemistry, powered by free energy gradients and streamlined via natural selection. In our view, life has been honing the core activities of data science for over 3.5 billion years. Over deep time, biology has created genetic knowledge of nearly every kind of environment on Earth (and is now experimenting with living beyond it). As life spread and gained influence over its surroundings, it created new environments and learned how to survive in those too. But life doesn't merely learn; it learns to learn better (e.g. [44–49]). Through a series of evolutionary innovations and major transitions, biology has enhanced its data-gathering and information-processing abilities [50], invented minds that can infer causality [51], produced the dataome [52], expanded its cognitive horizon by orders of magnitude from the microscopic to the planetary [53] and may potentially begin to consciously influence its own evolutionary trajectory [54–56].
Strikingly, many artificial intelligence techniques and computer algorithms are inspired by natural systems [57,58] including: artificial neural networks [59,60], evolutionary algorithms (e.g. genetic algorithms) [61,62], swarm intelligence [63,64], artificial immune systems [65,66], communication networks [67] and even a slime mould-inspired algorithm for mapping the distribution of dark matter across the universe [68]. Just as new discoveries in biology will be enabled by innovations in data science, data science will continue to benefit from a deeper understanding of how biological systems organize and process information and learn from their surroundings.
In this light, the difference between the informatics activities of data scientists and the rest of the living world can be viewed as one not of kind but of degree. Combining the evolutionary gifts of neurological cognition with the technological powers and mathematical techniques of today, human informaticians are engaging in data acquisition, curation, preservation, stewardship and management at unprecedented scale and velocity to wield predictions about nearly every aspect of the known universe with extraordinary precision. But this is hardly a unique endeavour—we humans are just the first to notice the unity of informatics principles at play across the tree of life. The principles of data science are expressed across the expanse of biology, from nanoscopic virions to the largest clonal tree to your friendly AI engineer. To think, it all started with a cell!
4. Leveraging an informatics perspective in the quest for a theory of life
Through the lens presented here, the essence of informatics is fundamental to what life is (or, perhaps better put, what life does). We propose that atoms, molecules, stars, planets, minerals, hurricanes and other prebiotic or non-living systems do not display the same kind of information life cycle or data–information–knowledge ecosystem as biological systems. Thus, the way that life assimilates data and uses it to enhance its own persistence could be a defining distinction between abiotic and biotic systems. Furthermore, although we take Darwinian evolution as an exemplar of biological information processing over deep time on Earth, the abstract principles of informatics and learning may be more universal frameworks for understanding extraterrestrial life, which may not necessarily use Darwinian evolution to update its knowledge about its surroundings [13,69].
The act of transforming data into a state that increases a system's survivability generates functional [70] or semantic [71] information. Take, for example, a bacterial gene that produces an enzyme that metabolizes a certain nutrient in the environment. This gene is the result of mutations and/or recombinations that explored the vast combinatorial space of nucleobase strings (acquisition) and was selected for by differential reproductive success (curation and preservation). If this gene can be turned on and off depending on the concentration of the nutrient in the environment, the cell will be spared the expense of producing a needless enzyme when the nutrient is scarce, further enhancing survivability (management). Via the information life cycle, raw data, in the form of fluctuating concentrations of nutrients, have been turned into various layers of functional information, in the form of the genetic, epigenetic and enzymatic apparati that help the cell persist and proliferate.
Life represents a subset of all known complex evolving systems, which are broadly defined as systems where (i) a large number of interacting components results in a potentially large combinatorial space, (ii) one or more mechanisms exist to generate numerous configurations within that combinatorial space, and (iii) a selection mechanism favours certain configurations over others (e.g. [72–78]). While non-biological complex evolving systems contain information, the information life cycle—and hence the degree of functional information within them—is stunted compared with biological systems.
Let us take mineral evolution as a characteristic example. In general, the minerals that form are those that minimize the free energy of the system at the pressure–temperature–composition conditions during crystallization. Hence, data about the paragenetic environment are recorded in the mineral's chemical and isotopic composition, crystal structure, habit and its context within a mineral assemblage (e.g. [79–90]). Furthermore, the increasing diversity of mineral species and natural kinds through time reflects the increasing chemical complexification of the cosmos [91].
However, the data that minerals contain do not participate fully in the information life cycle (figure 1c). First, in minerals, preservation is static (resistance to chemical change) rather than a dynamic process (like cellular replication); the identity of a crystal is tied to its physical substrate, whereas the information in an organism will be replicated many times from new material. Second, the steps of stewardship and management are non-existent for minerals. Third, while the information content of mineralogical systems can be updated by subsequent alteration in changing environments, there is no feedback between the information that has already been generated and the acquisition of new data.
In other words, minerals are essentially geological flash drives: they record data imparted on them by external forces—information that can be erased, updated or overwritten with time (e.g. the conversion of graphite to diamond)—but they do not correct defects in acquired data or otherwise use their stored data to ensure the fidelity of the other steps in the information life cycle. These kinds of differences limit the amount of functional information in abiotic systems to a narrow range of modalities (in minerals, to dissipation and static persistence) and prevent them from evolving open-endedly. Perhaps one axis for measuring ‘lifelikeness’ is the degree to which a system performs informatics processes and exhibits a data–information–knowledge ecosystem.
Finally, we wish to emphasize that on a living planet, information flows between biotic and abiotic systems (figure 2), blurring the distinction between life and its environment. Roughly half of the mineral species on Earth are biologically mediated [92], and the nature of Earth's atmosphere has been reshaped over evolutionary time through the exchange of metabolic gases (e.g. [93–95]). At the macroscopic scale, the information life cycle that the biosphere engages in will certainly include traditionally non-biological factors. A fuller exploration of this idea is saved for future work.
We contend that understanding how data are acquired and processed into functional information will be instrumental in developing a richer understanding of complex evolving systems, and to building a general theory of life. A full examination of the role that information plays in various biotic and abiotic systems requires a more granular level of exploration than we can cover here. The details of information content and the degree of information processing differ greatly across non-living systems; for instance, how would one weigh the functional information content of a star versus that of a river channel? Future work in information theory is required before we can truly characterize and compare these disparate physical systems with one another—and with life—on an equal footing. With the tools of data science and informatics at our disposal, perhaps the answers to these fundamental questions are finally within reach.
Acknowledgements
We are immensely grateful to our two anonymous peer reviewers for insightful comments that strengthened our paper. We are indebted to Robert M. Hazen, Stuart Bartlett, Marshall Ma, Sue Rhee and Elena Litchman for their responses to early drafts of this manuscript. We also thank the Carnegie Science ‘Missing Law’ Study Group for many stimulating conversations.
Contributor Information
Michael L. Wong, Email: mwong@carnegiescience.edu.
Anirudh Prabhu, Email: aprabhu@carnegiescience.edu.
Data accessibility
This article has no additional data.
Authors' contributions
M.L.W.: conceptualization, investigation, project administration, visualization, writing—original draft and writing—review and editing; A.P.: investigation, visualization, writing—original draft and writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
The authors declare no competing interests.
Funding
Support for this work was provided by (i) the Carnegie Postdoctoral Fellowship; (ii) NASA through the NASA Hubble Fellowship Program grant no. HST-HF2-51521.001-A awarded by the Space Telescope Science Institute, which is operated by the Association of Universities for Research in Astronomy, Inc., for NASA, under contract NAS5-26555; (iii) the John Templeton Foundation and (iv) a private foundation.
References
- 1.Margulis L, Sagan D. 1995. What is life? Berkeley, CA: University of California Press. [Google Scholar]
- 2.Nurse P. 2021. What is life? Five great ideas in biology. New York, NY: W. W. Norton & Company. [Google Scholar]
- 3.Schrödinger E. 1944. What is life? Cambridge, UK: Cambridge University Press. [Google Scholar]
- 4.Zimmer C. 2021. Life's edge: the search for what it means to be alive. New York, NY: Penguin Random House LLC. [Google Scholar]
- 5.Cleland CE. 2019. Moving beyond definitions in the search for extraterrestrial life. Astrobiology 19, 722-729. ( 10.1089/ast.2018.1980) [DOI] [PubMed] [Google Scholar]
- 6.Machery E. 2012. Why I stopped worrying about the definition of life… and why you should as well. Synthese 185, 145-164. ( 10.1007/s11229-011-9880-1) [DOI] [Google Scholar]
- 7.Parke EC. 2020. Dimensions of life definitions. In Social and conceptual issues in astrobiology (eds Smith KC, Mariscal C), pp. 79-91. Oxford, UK: Oxford University Press. [Google Scholar]
- 8.Cleland CE. 2019. The quest for a universal theory of life. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 9.Mayer-Schönberger V, Cukier K. 2013. Big data: a revolution that will transform how we live, work, and think. New York, NY: Houghton Mifflin Harcourt Publishing Company. [Google Scholar]
- 10.Prabhu A. 2019. Informatics. In Encyclopedia of big data (eds Schintler L, McNeely C). Cham, Switzerland: Springer. [Google Scholar]
- 11.Fox P. 2016. Xinformatics 2016. See https://tw.rpi.edu/classes/xinformatics-2016 (accessed 7 November 2022).
- 12.Prabhu A, et al. 2022. What Is mineral informatics? Authorea. ( 10.2138/am-2022-8613) [DOI] [Google Scholar]
- 13.Bartlett S, Wong ML. 2020. Defining lyfe in the universe: from three privileged functions to four pillars. Life 10, 42. ( 10.3390/life10040042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baluška F, Levin M. 2016. On having no head: cognition throughout biological systems. Front. Psychol. 7, 902. ( 10.3389/fpsyg.2016.00902) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kikuchi K, Galera-Laporta L, Weatherwax C, Lam JY, Moon EC, Theodorakis EA, Garcia-Ojalvo J, Süel GM. 2022. Electrochemical potential enables dormant spores to integrate environmental signals. Science (1979) 378, 43-48. ( 10.1126/science.abl7484) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Czégel D, Giaffar H, Tenenbaum JB, Szathmáry E. 2022. Bayes and Darwin: how replicator populations implement Bayesian computations. Bioessays 44, 2100255. ( 10.1002/bies.202100255) [DOI] [PubMed] [Google Scholar]
- 17.Watson RA, et al. 2016. Evolutionary connectionism: algorithmic principles underlying the evolution of biological organisation in evo-devo, evo-eco and evolutionary transitions. Evol. Biol. 43, 553-581. ( 10.1007/s11692-015-9358-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson RA, Szathmáry E. 2016. How can evolution learn? Trends Ecol. Evol. 31, 147-157. ( 10.1016/j.tree.2015.11.009) [DOI] [PubMed] [Google Scholar]
- 19.Banerjee K, Kolomeisky AB, Igoshin OA. 2017. Elucidating interplay of speed and accuracy in biological error correction. Proc. Natl Acad. Sci. USA 114, 5183-5188. ( 10.1073/pnas.1614838114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopfield JJ. 1974. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity (protein synthesis/DNA replication/amino-acid recognition). Proc. Natl Acad. Sci. USA 71, 4135-4139. ( 10.1073/pnas.71.10.4135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopfield JJ, Yamane T, Yue V, Coutts SM. 1976. Direct experimental evidence for kinetic proofreading in amino acylation of TRNAIle (stoichiometry of energy coupling/amino acyl TRNA synthetase/error rate in biosynthesis). Proc. Natl Acad. Sci. USA 73, 1164-1168. ( 10.1073/pnas.73.4.1164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murugan A, Huse DA, Leibler S. 2012. Speed, dissipation, and error in kinetic proofreading. Proc. Natl Acad. Sci. USA 109, 12 034-12 039. ( 10.1073/pnas.1119911109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ninio J. 1975. Kinetic amplification of enzyme discrimination. Biochimie 57, 587-595. ( 10.1016/S0300-9084(75)80139-8) [DOI] [PubMed] [Google Scholar]
- 24.Skinner MK. 2015. Environmental epigenetics and a unified theory of the molecular aspects of evolution: a neo-Lamarckian concept that facilitates neo-Darwinian evolution. Genome Biol. Evol. 7, 1296-1302. ( 10.1093/gbe/evv073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee TI, et al. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science (1979) 298, 799-804. ( 10.1126/science.1075090) [DOI] [PubMed] [Google Scholar]
- 26.Bjedov I, Tenaillon O, Gerard B, Souza V, Denamur E, Radman M, Taddei F, Matic I. 2003. Stress-induced mutagenesis in bacteria. Science (1979) 300, 1399-1404. ( 10.1126/science.1082240) [DOI] [PubMed] [Google Scholar]
- 27.Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. 2006. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science (1979) 313, 89-92. ( 10.1126/science.1127912) [DOI] [PubMed] [Google Scholar]
- 28.Kolb H. 1995. Simple anatomy of the retina. Salt Lake City, UT: University of Utah Health Sciences Center. [PubMed] [Google Scholar]
- 29.Gillespie PG, Walker RG. 2001. Molecular basis of mechanosensory transduction. Nature 413, 194-202. ( 10.1038/35093011) [DOI] [PubMed] [Google Scholar]
- 30.Bray D. 2009. Wetware: a computer in every living cell. New Haven, CT: Yale University Press. [Google Scholar]
- 31.Bray D. 1995. Protein molecules as computational elements in living cells. Nature 376, 307-312. ( 10.1038/376307a0) [DOI] [PubMed] [Google Scholar]
- 32.Crick F. 1970. Central dogma of molecular biology. Nature 227, 561-563. ( 10.1038/227561a0) [DOI] [PubMed] [Google Scholar]
- 33.Laland K, Matthews B, Feldman MW. 2016. An introduction to niche construction theory. Evol. Ecol. 30, 191-202. ( 10.1007/s10682-016-9821-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laland KN, Odling-Smee FJ, Feldman MW. 1999. Evolutionary consequences of niche construction and their implications for ecology. Proc. Natl Acad. Sci. USA 96, 10 242-10 247. ( 10.1073/pnas.96.18.10242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Odling-Smee FJ, Laland KN, Feldman MW. 1996. Niche construction. Am. Nat. 147, 641-648. ( 10.1086/285870) [DOI] [Google Scholar]
- 36.Wong ML, Bartlett S, Chen S, Tierney L. 2022. Searching for life, mindful of lyfe's possibilities. Life 12, 783. ( 10.3390/life12060783) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker SI. 2014. Top-down causation and the rise of information in the emergence of life. Information (Switzerland) 5, 424-439. ( 10.3390/info5030424) [DOI] [Google Scholar]
- 38.Walker SI, Davies PCW. 2013. The algorithmic origins of life. J. R. Soc. Interface 10, 20120869. ( 10.1098/rsif.2012.0869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higgs PG, Lehman N. 2015. The RNA world: molecular cooperation at the origins of life. Nat. Rev. Genet. 16, 7-17. ( 10.1038/nrg3841) [DOI] [PubMed] [Google Scholar]
- 40.Maury CPJ. 2009. Self-propagating β-sheet polypeptide structures as prebiotic informational molecular entities: the amyloid world. Origins Life Evol. Biospheres 39, 141-150. ( 10.1007/s11084-009-9165-6) [DOI] [PubMed] [Google Scholar]
- 41.Cairns-Smith AG. 1966. The origin of life and the nature of the primitive gene. J. Theor. Biol. 10, 53-88. ( 10.1016/0022-5193(66)90178-0) [DOI] [PubMed] [Google Scholar]
- 42.Russell M. 2018. Green rust: the simple organizing ‘seed’ of all life? Life 8, 35. ( 10.3390/life8030035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartlett S, Louapre D. 2022. Provenance of life: chemical autonomous agents surviving through associative learning. Phys. Rev. E 106, 034401. ( 10.1103/PhysRevE.106.034401) [DOI] [PubMed] [Google Scholar]
- 44.Bedau MA, Packard NH. 2003. Evolution of evolvability via adaptation of mutation rates. Biosystems 69, 143-162. ( 10.1016/S0303-2647(02)00137-5) [DOI] [PubMed] [Google Scholar]
- 45.Crombach A, Hogeweg P. 2008. Evolution of evolvability in gene regulatory networks. PLoS Comput. Biol. 4, e1000112. ( 10.1371/journal.pcbi.1000112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gould SJ. 2002. The structure of evolutionary theory. Cambridge, UK: Harvard University Press. [Google Scholar]
- 47.Pigliucci M. 2008. Is evolvability evolvable? Nat. Rev. Genet. 9, 75-82. ( 10.1038/nrg2278) [DOI] [PubMed] [Google Scholar]
- 48.Valiant L. 2013. Probably approximately correct: nature's algorithms for learning and prospering in a complex world. New York, NY: Basic Books. [Google Scholar]
- 49.Wilder B, Stanley K. 2015. Reconciling explanations for the evolution of evolvability. Adapt. Behav. 23, 171-179. ( 10.1177/1059712315584166) [DOI] [Google Scholar]
- 50.Szathmáry E, Maynard Smith J. 1995. The major evolutionary transitions. Nature 374, 227-232. ( 10.1038/374227a0) [DOI] [PubMed] [Google Scholar]
- 51.Pearl J, Mackenzie D. 2018. The book of why: the new science of cause and effect. New York, NY: Basic Books. [Google Scholar]
- 52.Scharf C. 2021. The ascent of information: books, bits, genes, machines, and life's unending algorithm. New York, NY: Riverhead Books. [Google Scholar]
- 53.Levin M, Dennett DC. 2020. Cognition all the way down. Aeon Essays. See https://aeon.co/essays/how-to-understand-cells-tissues-and-organisms-as-agents-with-agendas (accessed 7 November 2022).
- 54.Frank A, Grinspoon D, Walker S. 2022. Intelligence as a planetary scale process. Int. J. Astrobiol. 21, 47-61. ( 10.1017/S147355042100029X) [DOI] [Google Scholar]
- 55.Grinspoon D. 2016. Earth in human hands: shaping our planet's future. New York, NY: Grand Central Publishing. [Google Scholar]
- 56.Wong ML, Bartlett S. 2022. Asymptotic burnout and homeostatic awakening: a possible solution to the Fermi paradox? J. R. Soc. Interface 19, 20220029. ( 10.1098/rsif.2022.0029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brabazon A, O'neill M, Mcgarraghy S. 2015. Natural computing algorithms. Berlin, Germany: Springer-Verlag. [Google Scholar]
- 58.Rozenberg G, Bäck T, Kok JN. 2012. Handbook of natural computing, 1st edn. Berlin, Germany: Springer Berlin. [Google Scholar]
- 59.Hopfield JJ. 1988. Artificial neural networks. IEEE Circuits Devices Mag. 4, 3-10. ( 10.1109/101.8118) [DOI] [Google Scholar]
- 60.Priddy KL, Keller PE. 2005. Artificial neural networks: an introduction. Bellingham, WA: SPIE Press. [Google Scholar]
- 61.Mitchell M. 1996. An introduction to genetic algorithms. Cambridge, MA: MIT Press. [Google Scholar]
- 62.Whitley D. 2001. An overview of evolutionary algorithms: practical issues and common pitfalls. Inf. Softw. Technol 43, 817-831. ( 10.1016/S0950-5849(01)00188-4) [DOI] [Google Scholar]
- 63.Beni G, Wang J. 1993. Swarm intelligence in cellular robotic systems. In Robots and biological systems: towards a new bionics? (eds Dario P, Sandini G, Aebischer P), pp. 703-712. Berlin, Germany: Berlin, Germany: Springer Berlin Heidelberg. [Google Scholar]
- 64.Chakraborty A, Kar AK. 2017. Swarm intelligence: a review of algorithms. In Nature-inspired computing and optimization: theory and applications (eds Patnaik S, Yang X-S, Nakamatsu K), pp. 475-494. Cham, Switzerland: Springer International Publishing. [Google Scholar]
- 65.Hunt JE, Cooke DE. 1996. Learning using an artificial immune system. J. Netw. Comp. Appl. 19, 189-212. ( 10.1006/jnca.1996.0014) [DOI] [Google Scholar]
- 66.Timmis J, Neal M, Hunt J. 2000. An artificial immune system for data analysis. Biosystems 55, 143-150. ( 10.1016/S0303-2647(99)00092-1) [DOI] [PubMed] [Google Scholar]
- 67.Cheng S-M, Karyotis V, Chen PY, Chen KC, Papavassiliou S. 2013. Diffusion models for information dissemination dynamics in wireless complex communication networks. J. Complex Syst. 2013, 1-13. ( 10.1155/2013/972352) [DOI] [Google Scholar]
- 68.Burchett JN, Elek O, Tejos N, Prochaska JX, Tripp TM, Bordoloi R, Forbes AG. 2020. Revealing the dark threads of the cosmic web. Astrophys. J. 891, L35. ( 10.3847/2041-8213/ab700c) [DOI] [Google Scholar]
- 69.Noor MAF. 2022. Thinking outside Earth's box—how might heredity and evolution differ on other worlds? Evolution 15, 13. ( 10.1186/s12052-022-00172-4) [DOI] [Google Scholar]
- 70.Hazen RM, Griffin PL, Carothers JM, Szostak JW. 2007. Functional information and the emergence of biocomplexity. Proc. Natl Acad. Sci. USA 104(suppl. 1), 8574-8581. ( 10.1073/pnas.0701744104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kolchinsky A, Wolpert DH. 2018. Semantic information, autonomous agency and non-equilibrium statistical physics. Interface Focus 8, 20180041. ( 10.1098/rsfs.2018.0041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dooley KJ. 1997. A complex adaptive systems model of organization change. Nonlinear Dynamics Psychol. Life Sci. 1, 69-97. [DOI] [PubMed] [Google Scholar]
- 73.Hazen RM. 2009. The emergence of patterning in life's origin and evolution. Int. J. Dev. Biol. 53, 683-692. ( 10.1387/ijdb.092936rh) [DOI] [PubMed] [Google Scholar]
- 74.Hazen RM, Eldredge N. 2010. Themes and variations in complex systems. Elements 6, 43-46. ( 10.2113/gselements.6.1.43) [DOI] [Google Scholar]
- 75.Holland JH. 1995. Hidden order: how adaptation builds complexity. New York, NY: Basic Books. [Google Scholar]
- 76.Holland JH. 1992. Complex adaptive systems. Daedalus 121, 17-30. [Google Scholar]
- 77.Levin SA. 1998. Ecosystems and the biosphere as complex adaptive systems. Ecosystems 1, 431-436. ( 10.1007/s100219900037) [DOI] [Google Scholar]
- 78.Morowitz HJ. 2002. The emergence of everything: how the world became complex. Oxford, UK: Oxford University Press. [Google Scholar]
- 79.Ehlmann BL, Mustard JF, Murchie SL, Bibring JP, Meunier A, Fraeman AA, Langevin Y. 2011. Subsurface water and clay mineral formation during the early history of Mars. Nature 479, 53-60. ( 10.1038/nature10582) [DOI] [PubMed] [Google Scholar]
- 80.García-Ruiz JM, Otálora F, Sanchez-Navas A, Higes-Rolando FJ. 1994. The formation of manganese dendrites as the mineral record of flow structures. In Fractals and dynamic systems in geoscience (eds Kruhl JH), pp. 307-318. Berlin, Germany: Springer Berlin Heidelberg. [Google Scholar]
- 81.Hazen RM, Papineau D, Bleeker W, Downs RT, Ferry JM, Mccoy TJ, Sverjensky DA, Yang H. 2008. Mineral evolution. Am. Mineral. 93, 1693-1720. ( 10.2138/am.2008.2955) [DOI] [Google Scholar]
- 82.Hazen RM, Ferry JM. 2010. Mineral evolution: mineralogy in the fourth dimension. Elements 6, 9-12. ( 10.2113/gselements.6.1.9) [DOI] [Google Scholar]
- 83.Hazen RM, Morrison SM. 2022. On the paragenetic modes of minerals: a mineral evolution perspective. Am. Mineral. 107, 1262-1287. ( 10.2138/am-2022-8099) [DOI] [Google Scholar]
- 84.Hopkins M, Harrison TM, Manning CE. 2008. Low heat flow inferred from >4 Gyr zircons suggests Hadean plate boundary interactions. Nature 456, 493-496. ( 10.1038/nature07465) [DOI] [PubMed] [Google Scholar]
- 85.Hopkins MD, Harrison TM, Manning CE. 2010. Constraints on Hadean geodynamics from mineral inclusions in >4Ga zircons. Earth Planet. Sci. Lett. 298, 367-376. ( 10.1016/j.epsl.2010.08.010) [DOI] [Google Scholar]
- 86.Kastner M. 1999. Oceanic minerals: their origin, nature of their environment, and significance. Proc. Natl Acad. Sci. USA 96, 3380-3387. ( 10.1073/pnas.96.7.3380) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Keller WD. 1956. Clay minerals as influenced by environments of their formation. Am. Assoc. Pet. Geol. Bull. 40, 2689-2710. ( 10.1306/5CEAE5CE-16BB-11D7-8645000102C1865D) [DOI] [Google Scholar]
- 88.Krivovichev S. 2013. Structural complexity of minerals: information storage and processing in the mineral world. Mineral. Mag. 77, 275-326. ( 10.1180/minmag.2013.077.3.05) [DOI] [Google Scholar]
- 89.Mojzsis SJ, Harrison TM, Pidgeon RT. 2001. Oxygen-isotope evidence from ancient zircons for liquid water at the Earth's surface 4,300 Myr ago. Nature 409, 178-181. ( 10.1038/35051557) [DOI] [PubMed] [Google Scholar]
- 90.Taylor KG, Macquaker JHS. 2011. Iron minerals in marine sediments record chemical environments. Elements 7, 113-118. ( 10.2113/gselements.7.2.113) [DOI] [Google Scholar]
- 91.Cleland CE, Hazen RM, Morrison SM. 2021. Historical natural kinds and mineralogy: systematizing contingency in the context of necessity. Proc. Natl Acad. Sci. USA 118, e2015370118. ( 10.1073/PNAS.2015370118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Prabhu A, Morrison SM, Wong ML, Fox P, Hazen R. 2022. Pursuing big science questions with information-rich minerals. Goldschmidt Conf., Honolulu, HI, 10–15 July 2022. [Google Scholar]
- 93.Solé R, Munteanu A. 2004. The large-scale organization of chemical reaction networks in astrophysics. Europhys. Lett. 68, 170-176. ( 10.1209/epl/i2004-10241-3) [DOI] [Google Scholar]
- 94.Yung YL, Wong ML, Gaidos EJ. 2014. Evolution of Earth's atmosphere. In Encyclopedia of atmospheric sciences (eds North GR, Pyle J, Zhang F), vol. 5, pp. 163-167. Cambridge, MA: Academic Press. [Google Scholar]
- 95.Yung YL, Demore WB. 1999. Photochemistry of planetary atmospheres. Oxford, UK: Oxford University Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.