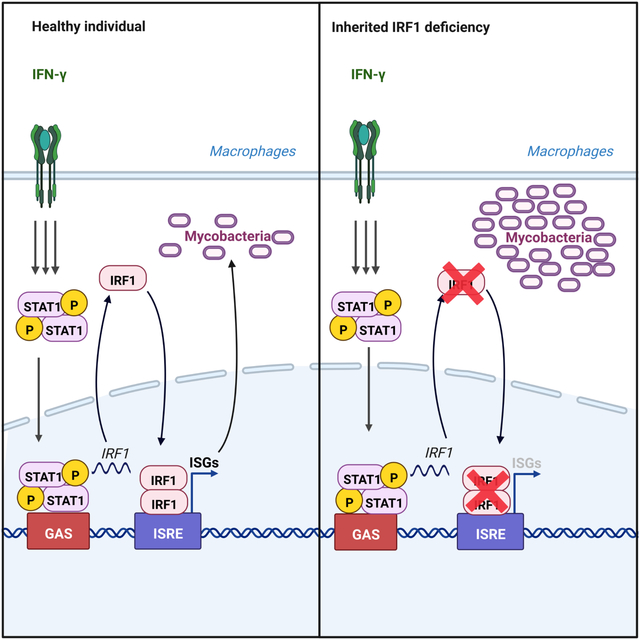
Summary
Inborn errors of human IFN-γ-dependent macrophagic immunity underlie mycobacterial diseases, whereas inborn errors of IFN-α/β-dependent intrinsic immunity underlie viral diseases. Both types of IFNs induce the transcription factor IRF1. We describe unrelated children with inherited complete IRF1 deficiency and early-onset, multiple, life-threatening diseases caused by weakly virulent mycobacteria and related intramacrophagic pathogens. These children have no history of severe viral disease, despite exposure to many viruses, including SARS-CoV-2, which is life-threatening in individuals with impaired IFN-α/β immunity. In leukocytes or fibroblasts stimulated in vitro, IRF1-dependent responses to IFN-γ are, both quantitatively and qualitatively, much stronger than those to IFN-α/β. Moreover, IRF1-deficient mononuclear phagocytes do not control mycobacteria and related pathogens normally when stimulated with IFN-γ. By contrast, IFN-α/β-dependent intrinsic immunity to nine viruses, including SARS-CoV-2, is almost normal in IRF1-deficient fibroblasts. Human IRF1 is essential for IFN-γ-dependent macrophagic immunity to mycobacteria, but largely redundant for IFN-α/β-dependent antiviral immunity.
Keywords: Inborn errors of immunity, Mycobacterium, interferon-γ, interferon-stimulated gene, IRF1, viruses
Graphical Abstract
In Brief
Studies in humans with interferon regulatory factor 1 (IRF1) deficiency reveal differences in how by type I and type II interferon immune responses protect humans against different types of pathogens.
Introduction
The discovery of inborn errors of immunity (IEI) underlying severe infectious diseases delineates the essential versus redundant functions of the corresponding human genes in host defense in natura, while clarifying the pathogenesis of these infections1–3. Mendelian susceptibility to mycobacterial disease (MSMD) is the most extensively studied monogenic susceptibility to a single type of infection in otherwise healthy individuals with apparently normal resistance to most other infections. Patients with MSMD are selectively vulnerable to weakly virulent mycobacteria — bacillus Calmette-Guérin (BCG) vaccines and environmental mycobacteria (EM) — and, in some cases, Mycobacterium tuberculosis and other intramacrophagic microorganisms4–9. MSMD is typically “isolated”, but can occasionally be “syndromic”, if associated with at least one other key infectious or non-infectious clinical phenotype4. Mutations of 19 different genes can account for MSMD: CYBB, IFNG, IFNGR1, IFNGR2, IL12B, IL12RB1, IL12RB2, IL23R, NEMO, SPPL2A, and TBX21 for isolated MSMD; ISG15, JAK1, RORC, TYK2, USP18 and ZNFX1 for syndromic MSMD; and IRF8, STAT1, and TYK2 for isolated or syndromic MSMD depending on the mutation4,6–8,10. Allelic forms at these 19 loci define 35 genetic etiologies of MSMD. Eighteen of the 19 known MSMD-causing genes encode products involved in the production of interferon-γ (IFN-γ) (IFNG, IL12B, IL12RB1, IL12RB2, IL23R, ISG15, RORC, TBX21, TYK2), cellular responses to IFN-γ (CYBB, JAK1, IFNGR1, IFNGR2, STAT1, USP18), or both (IRF8, NEMO, SPPL2A) (Figure S1A). ZNFX1 is the only gene for which the mechanism of MSMD is not yet understood6. Thus, MSMD is typically caused by IEI of IFN-γ immunity. These conditions display a high level of genetic and allelic heterogeneity, but striking physiological homogeneity11.
However, only three etiologies of isolated MSMD are truly Mendelian, i.e. with complete clinical penetrance1: autosomal recessive (AR) complete IFN-γ, IFN-γR1, and IFN-γR2 deficiencies (Figure S1B)4,5,12–15. These three disorders abolish IFN-γ activity. The corresponding patients have early-onset, disseminated, recurrent, multiple, and life-threatening mycobacterial infections4,5,12. The other etiologies of isolated MSMD have both incomplete clinical penetrance and less severe clinical features, with a later onset, narrower range of pathogens, lower rate of recurrence, and better outcome. Overall, lower penetrance and lesser severity are associated with higher levels of residual IFN-γ activity (Figure S1B). For instance, AR partial IFN-γR1 or IFN-γR2 deficiencies, which impair but do not abolish cellular responses to IFN-γ, are less severe than the complete forms12,16. AR complete IL-12Rβ1 and IL-12p40 deficiencies are also milder conditions; they abolish cellular responses to IL-12 and IL-23, reducing IFN-γ production to 1–10% of normal levels17–19. AR complete IL-12Rβ2 and IL-23R deficiencies are even milder and less penetrant, as there is a selective defect of the response to IL-12 or IL-2320,21 (Philippot et al., under revision). Despite being recessively inherited complete deficiencies, these four conditions are less penetrant and severe than recessive defects of IFN-γ or its receptor12,19,20,22. Finally, homozygosity for TYK2 p.P1104A, which impairs but does not abolish IFN-γ induction in response to IL-23 only, is associated with the lowest known penetrance of MSMD (below 0.5%)23. Thus, studies of MSMD have revealed that human antimycobacterial immunity is a genetically controlled quantitative trait: the lower the level of IFN-γ activity, the more severe the disease and the higher its penetrance16.
Less is known about the cellular basis of human immunity to mycobacteria1. Studies of peripheral leukocytes from MSMD patients have suggested that some IFN-γ-producing lymphocyte subsets, either alone or in combination, are essential for antimycobacterial immunity8,24–26,20. These essential subsets include the combination of natural killer (NK) cells, γδ T cells, and type 1 and type 2 innate lymphoid cells (ILC1 and ILC2) impaired in IL-12Rβ2 deficiency20; the invariant NK (iNKT) and mucosal-associated invariant T (MAIT) cells impaired in IL-23R20,21 (Philippot et al., under revision), RORγ/RORγT24, IL-12Rβ1, and TYK2 deficiencies20,23; the TH1* cells impaired in RORγ/RORγT, IRF8 and SPPL2a deficiencies24,25; the γδ T cells impaired in RORγ/RORγT24; the γδ2+ T cells impaired in IL-12Rβ1, and IL-23R deficiencies20 (Philippot et al., under revision); and the combination of NK, iNKT, MAIT and Vδ2+ γδ T cells impaired in T-bet deficiency8. These studies also suggested that IL-12- and IL-23-producing type 2 dendritic cells (DCs), which are impaired in both AR SPPL2a25 and autosomal dominant (AD) IRF8 deficiencies25,27, are essential for antimycobacterial immunity. Myeloid cells induce IFN-γ in lymphoid cells via at least ISG1526, IL-1220,23, and IL-2320,23, promoting the development of IFN-γ-producing cells, such as TH1* cells25. They are also activated by IFN-γ, which is commonly considered more of a macrophage-activating factor28 than an antiviral IFN. IFN-γ controls mycobacterial growth within macrophages via JAK1, STAT1 and the NAPDH oxidase complex, as revealed by studies of MSMD-causing JAK1, STAT1 and CYBB mutations9,29–34. We investigated the cellular and molecular basis of human immunity to mycobacteria further, by studying two unrelated children with a severe and unexplained form of isolated MSMD, combining diseases due to both BCG and Mycobacterium avium in early childhood.
Results
Two unrelated children with severe mycobacterial diseases
We studied two unrelated children, P1 and P2. Both of them displayed severe forms of MSMD, with not only BCG disease, but also M. avium complex disease at a very young age, with four episodes of mycobacterial disease in P1, and two episodes in P2, before the age of six years in both cases (Figures 1A–E). Phenotypes of this severity are seen almost exclusively in patients with recessive complete defects of IFN-γ, either of the receptor chains, or STAT1. Both P1 and P2 had no history of unusually severe viral illness and the only other infection observed, histoplasmosis, is caused by an intramacrophagic fungus that has already been reported in other patients with MSMD4–6,8 (see Supplemental case report). We performed whole-exome sequencing (WES) on the two patients. The ethnicity of the two patients was confirmed by principal component analysis (PCA) on the WES data35 (Figure S1C). The homozygosity rates for P1 and P2 were 2.74% and 4%, respectively, suggesting that the parents were probably first- or second-degree cousins36. The parents being healthy, we tested the hypothesis of an AR disorder and considered homozygous variants on autosomes. The prevalence of MSMD is about 10−5. We therefore considered variants with a minor allele frequency (MAF) below 0.003 in gnomAD v2.1.1. We then selected non-synonymous and essential splice-sites variants predicted to have a combined annotation depletion-dependent (CADD) score above the 99% mutation significance cut-off (MSC)37, for genes with a gene damage index (GDI) below the cutoff of 13.36 for inborn errors with AR inheritance38 (Figure 1F). We also filtered out false-positive rare variants, which were absent or rare in public databases but had a frequency above 1% in our in-house cohort (the ‘blacklist’)39. We found no homozygous candidate copy number variants (CNVs) in known MSMD genes40. We identified 26 rare homozygous single-nucleotide variants (SNVs) in 25 genes in P1, and 17 homozygous SNVs in 17 genes in P2 (Figure 1F and Table S1). In tests of the hypothesis of genetic homogeneity, we found only one common gene: IRF1. Both patients displayed homozygous transitions predicted to be nonsense in the IRF1 canonical transcript (NM_002198.241, GRCh37): c.385C>T (p.R129*) in P1, and c.103C>T (p.Q35*) in P2 (Figure 1G). Sanger sequencing confirmed that P1 was homozygous for p.R129* and P2 was homozygous for p.Q35*, whereas asymptomatic relatives were heterozygous (Figures 1A and 1H). These findings suggest that MSMD in P1 and P2 resulted from homozygosity for these rare nonsense IRF1 variants.
Figure 1 – Homozygous complete loss-of-function IRF1 variant in patients with severe MSMD.
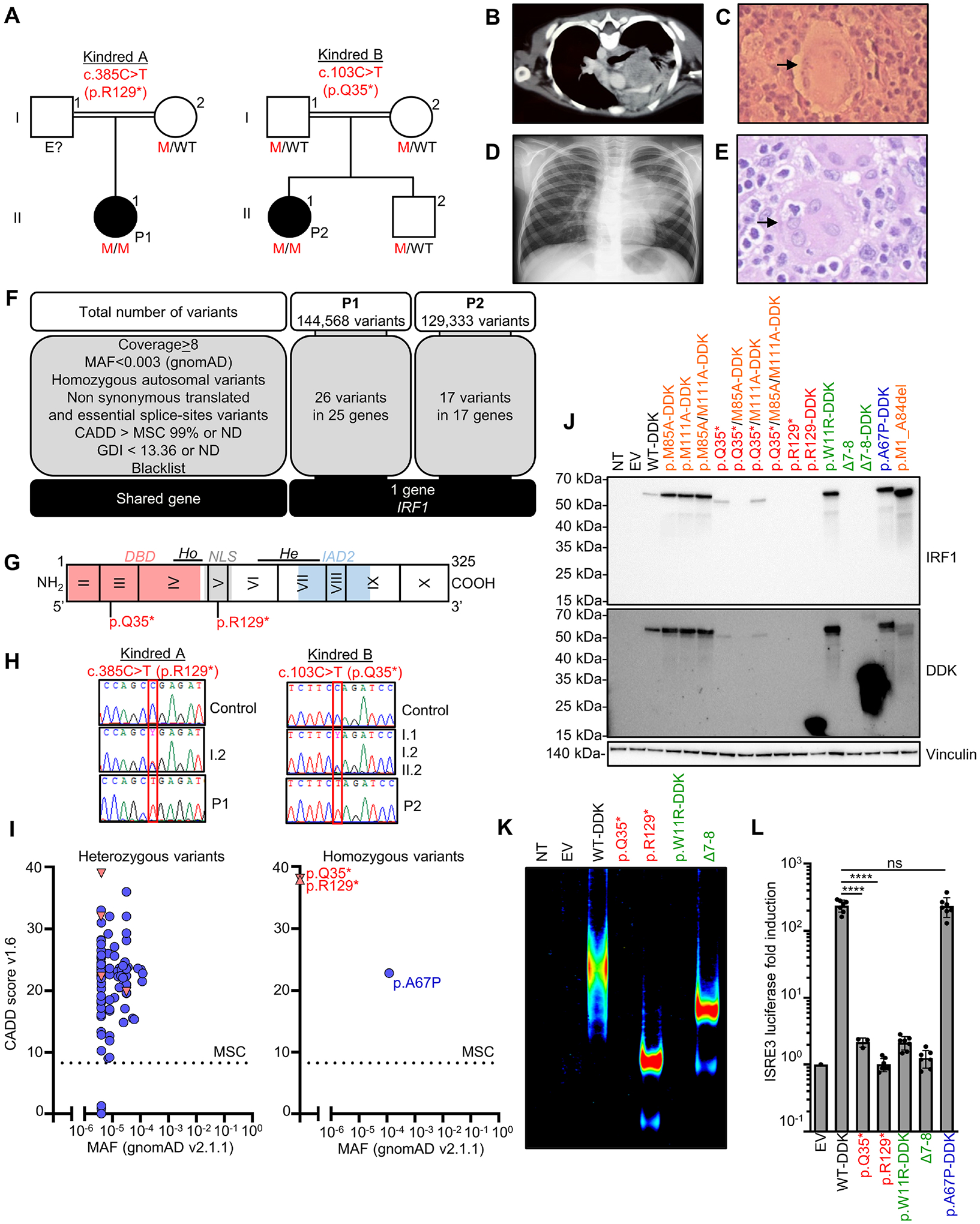
(A) Pedigrees of the two consanguineous kindreds. M=mutated; WT= wild-type; (B) Chest CT scan (P1) showing pulmonary infection. (C) Hematoxylin and eosin staining of a lymph node (P1) during M. avium and H. capsulatum infections, showing a multinucleated giant cell. (D) Chest X ray for P2 showing pulmonary M. avium infection. (E) Hematoxylin-eosin-saffron stain of a lung biopsy (P2) showing a giant cell engulfing another cell by phagocytosis (arrow). (F) WES analysis of P1 and P2. (G) IRF1 protein with the DNA-binding domain (DBD), nuclear localization sequence (NLS), and IRF-associated domain type 2 in blue (IAD2). Ho = homodimerization domain; He = heterodimerization domain. (H) Electropherograms of representative IRF1 nucleotide sequences. (I) CADD score vs. minor allele frequency (MAF) for the heterozygous state in gnomAD for variants IRF1 (left), and (right) for a homozygous variant from BRAVO/TOPmed (p.A67P), and the variants present in patients. Missense variants are indicated by blue circles and predicted loss-of-function variants are indicated by red triangles. (J) Western blotof total lysate from HEK293T cells with and without transfection with various C-DDK-tagged IRF1 cDNAs or with empty vector (EV). NT = not transfected. Representative data from two independent experiments. (K) EMSA on nuclear extract of HEK293T cells transfected with EV, WT or mutant IRF1 cDNAs, incubated with an ISRE probe. Representative data from two independent experiments. (L) Dual luciferase ISRE3 reporter activity of HEK293T cells transfected with EV or mutant IRF1 cDNAs. Data from 3–7 independent experiments performed in triplicate. Bars represent the mean and standard deviation (SD). Statistical analysis by Student’s t-test. ns = not significant, p > 0.05; ****p < 0.0001.
The overexpressed IRF1 p.Q35* and p.R129* mutants are loss-of-function
IRF1 encodes interferon regulatory factor 1 protein (IRF1), a transcription factor induced by IFN-γ, the deficiency of which in mice underlies susceptibility to various pathogens, including mycobacteria43,49,51,53,55,57,62,83,84,96–98,109,110,147 (Table S2). IRF1 is a 325-amino acid transcription factor composed of an amino-terminal DNA-binding domain (DBD) that can bind to DNA interferon-stimulated response elements (ISRE) or positive regulatory domain I (PRDI) motifs64, an intermediate segment containing a putative nuclear localization sequence (NLS), and a carboxyterminal IRF association domain 2 (IAD2), which is crucial for transcriptional activity65,66,68,70 (Figure 1G). Both the c.385C>T and c.103C>T variants are predicted to be loss-of-function (pLOF) and are rare or private in public databases (Figure 1E–I and Supplemental information). We studied the impact of the IRF1 variants by transiently transfecting human embryonic kidney (HEK)293T cells with plasmids encoding WT IRF1 (NM_002198.2), mutants p.R129* and p.Q35*, or p.A67P cDNAs. We used two previously described LOF mutants isolated from human tumors as negative controls: (i) the missense mutant p.W11R42, and (ii) a frameshift mutant resulting in the deletion of exons 7 and 844 (hereafter referred to as Δ7–8). The proteins encoded by the constructs were left untagged or were tagged with DDK at the carboxy-terminus (Figure S1E). Immunoblotting of cell extracts showed that the WT-DDK, p.W11R-DDK, and p.A67P-DDK proteins were produced at a molecular weight (MW) slightly above 50 kDa, as expected, whereas the p.R129-DDK and the Δ7–8-DDK proteins had a lower MW (Figure 1J). These results indicate that the p.R129-DDK cDNA encodes a truncated protein. Both p.R129-DDK and Δ7–8-DDK gave bands of higher intensity than the WT protein, consistent with an absence of the carboxyterminal degradation domain45. Immunoblotting with a monoclonal antibody (mAb) directed against the C-terminus of IRF1 showed no re-initiation of translation with the p.R129* cDNA (Figure 1J). However, immunoblotting of the p.Q35* protein showed this protein to have a slightly lower MW than the WT protein when probed with either the anti-IRF1 or anti-DDK mAb, suggesting that a re-initiation of translation had occurred. Two ATG codons downstream from p.Q35 and upstream from p.R129 (p.M85 and p.M111) were predicted to be potential translation re-initiation sites in analyses in silico. We mutated the corresponding methionine (ATG) codon to an alanine codon (GCG). With the WT-DDK cDNA template, the mutation of p.M85 (WT/M85A-DDK), p.M111 (WT/M111A-DDK), or both (WT/M85A/M111A-DDK) had no effect on the MW of the protein produced (Figure 1J). Conversely, with the p.Q35* cDNA template, the mutation of p.M85 (p.Q35*/M85A-DDK) abolished protein detection, whereas the mutation of p.M111 (p.Q35*/M111A-DDK) did not. The mutation of both methionine (p.Q35*/M85A/M111A-DDK) residues abolished protein production (Figure 1J). Consistently, the deletion of all amino acids upstream from M85A (p.M1_A84del) resulted in the production of a protein of the same MW as p.Q35* (Figure 1J). Overall, these results suggest that translation is re-initiated for the p.Q35* variant, resulting in the production of a protein lacking the first 84 amino-terminal amino acids of the DBD. Both the mutant proteins from the patients localized to the nucleus (Figure S1G). An EMSA with an ISRE probe found that the p.Q35* protein, like the previously described p.W11R mutant protein42, did not bind DNA, whereas the p.R129-DDK mutant, which retained the DBD, was able to bind DNA, resulting in a band of higher mobility than was observed for the WT, consistent with the lower MW of the mutant protein (Figure 1K and S1H). We then assessed the transcriptional activity of IRF1 in a dual luciferase assay with two different plasmids containing two different ISRE repeats. The p.A67P-DDK cDNA induced luciferase to WT levels, whereas the p.R129* and p.Q35* mutants and two negative controls (p.W11R-DDK42 and Δ7–844) did not (Figures 1L and S1I). Overall, these results suggest that the p.R129* (P1) and p.Q35* (P2) variants impair IRF1 production and abolish its transcriptional activity.
IRF1 protein expression is impaired in cells of the patients
IRF1 is produced ubiquitously in humans46, but its levels are highest in hematopoietic cells46,47. We used non-hematopoietic (primary fibroblasts, and simian virus 40 (SV40)-immortalized fibroblasts) and hematopoietic (Epstein-Barr virus-immortalized B lymphocytes (EBV-B cells), T-cell blasts, Herpesvirus saimiri-transformed T (HVS-T) cells, monocyte-derived macrophages (MDMs), and tissue macrophages derived from an induced pluripotent stem cell (iPSC) line48,50 (iPSC-MΦ)) to assess the impact of the p.R129* and p.Q35* IRF1 variants on endogenous IRF1 mRNA and protein levels. We performed quantitative RT-PCR (RT-qPCR) with two sets of probes and found much lower levels of IRF1 mRNA in the cells of P1 than in the corresponding cells from healthy controls, primary fibroblasts (6-fold lower; Figure 2A), SV40-fibroblasts (10-fold lower; Figure 2B), EBV-B cells (3-fold lower; Figure 2C), iPSC-MΦ (>10-fold lower; Figure 2D), and HVS-T cells (4-fold lower; Figure 2E), suggesting that the IRF1 transcript underwent nonsense-mediated mRNA decay in the cells of P1. Conversely, IRF1 mRNA levels in the cells of P2 (primary fibroblasts, SV40-fibroblasts, EBV-B cells, MDMs, and T-cell blasts) were only slightly lower than those in control cells (Figures 2A–C, and 2F–G). We then assessed IRF1 protein levels by immunoblotting and flow cytometry, with a mAb specific for the carboxy-terminus of IRF1. Primary and SV40-fibroblasts, EBV-B cells, HVS-T cells, and iPSC-MΦ from P1 contained no detectable endogenous IRF1 protein (Figures 2H–L and S2A–F). In addition, no truncated IRF1 protein was detected in SV40-fibroblasts from P1 after pretreatment with IFN-γ, even with a polyclonal antibody and prolonged exposure of the immunoblot (Figure S2F). The protein was barely detectable in primary and SV40-fibroblasts from P2 following pretreatment with IFN-γ (Figures 2H, 2K, and S2B), either on flow cytometry or after prolonged exposure of the immunoblot. The protein detected in the cells of P2 had a MW corresponding to that expected following a re-initiation of translation. Other transcription factors, such as IRF3, IRF8, IRF9, and STAT1, were produced by the cells of P1 and P2 (Figures 2H–J and S2A–F). The impaired basal levels of IRF1 protein production in the SV40-fibroblasts from both patients were corrected by stable transduction with the WT IRF1 cDNA (Figure 2L). Together, these findings suggest that homozygosity for the IRF1 p.R129* variant results in nonsense-mediated decay of the IRF1 transcript and a lack of detectable protein, whereas homozygosity for the IRF1 p.Q35* variant leads to a partial escape of mRNA nonsense-mediated decay, due to a re-initiation of translation, with low levels of production of an N-terminally truncated protein. P1 and P2 thus display an absence of IRF1 protein (P1) or the production of an abnormal IRF protein (P2) in hematopoietic and non-hematopoietic cells. These results further suggest that both patients have AR complete IRF1 deficiency.
Figure 2 – IRF1 mRNA and protein levels in cells from the two patients.
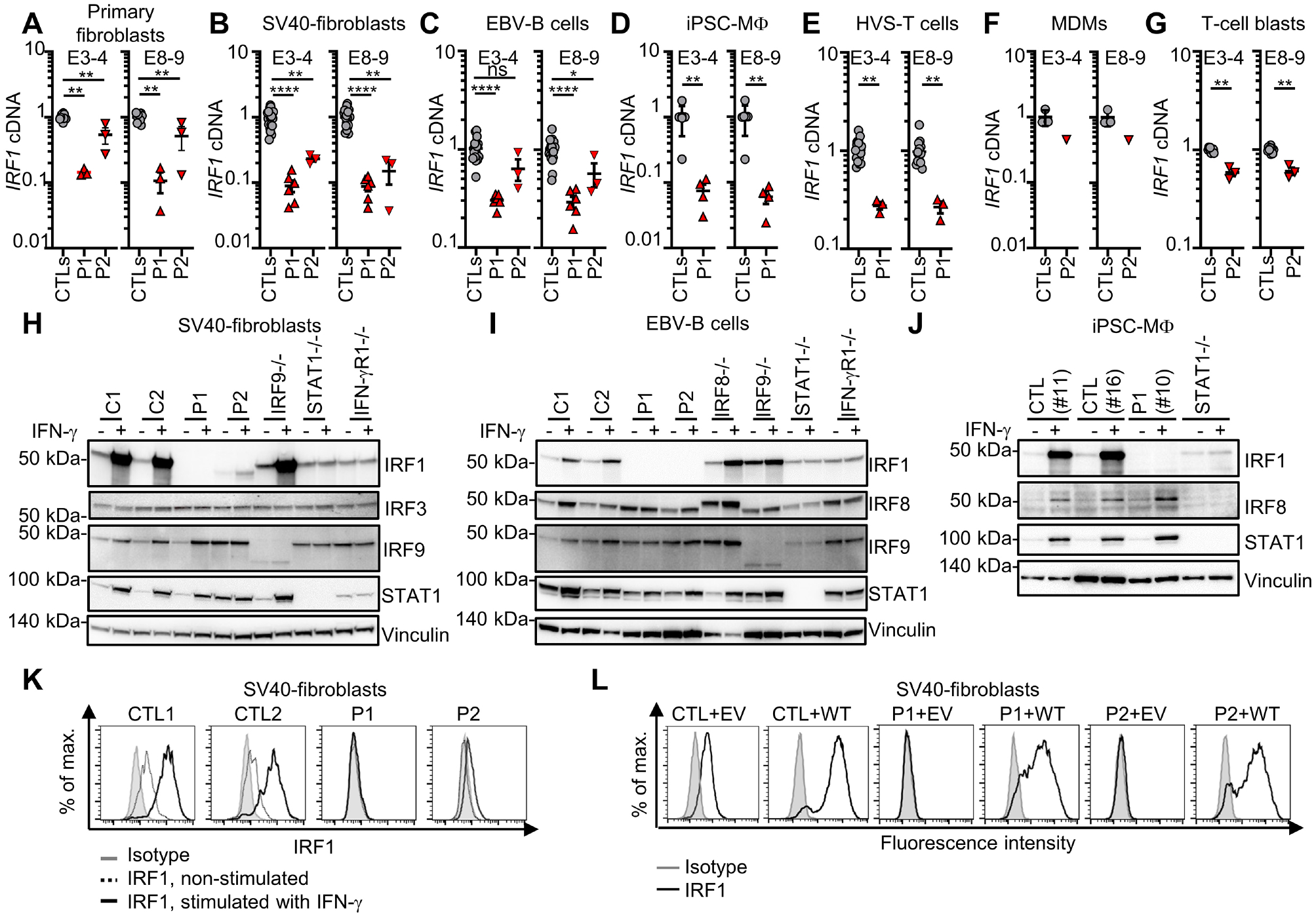
Quantitative PCR for IRF1 normalized against GUSB and the mean value for controls (CTLs) for cDNA from (A) primary fibroblasts, (B) SV40-fibroblasts, (C) EBV-B cells, (D) iPSC-derived macrophages (iPSC-MΦ cells;), (E) HSV-T cells, (F) Monocytes-derived macrophages (MDMs), and (G) T-cell blasts. Bars represent the mean and SD. Western blot for indicated protein in total lysate from (H) SV40-fibroblasts, (I) EBV-B cells, or (J) iPSC-derived MΦ, with and without IFN-γ stimulation. Data from 2–3 independent experiments are shown. (K) IRF1 staining and intracellular flow cytometry on SV40-fibroblasts with and without IFN-γ stimulation. (L) Flow cytometry with intracellular IRF1 staining on SV40-fibroblasts retrotransduced with an empty vector (EV) or WT) -IRF1 cDNA. The data shown are representative of 2–3 independent experiments. Statistical analysis by Mann-Whitney tests. ns = not significant, p > 0.05; *p < 0.05; **p < 0.01; ****p < 0.0001.
IRF1 deficiency alters the development of some innate or innate-like leukocyte subsets
We investigated the role of human IRF1 in leukocyte development, by analyzing blood cells from P1 and P2. A complete blood count (CBC) showed that both P1 and P2 had counts of polymorphonuclear neutrophils (PMN), basophils (PMB), eosinophils (PME) and peripheral blood mononuclear cells (PBMCs) that were normal for age (Figures 3A and S3A). Conventional flow cytometry also showed normal counts of monocyte subsets in P2 (Figure 3B). Both patients had low percentages and counts of circulating conventional type 1 dendritic cells (cDC1) (>10-fold decrease) and cDC2 (~5-fold decrease), whereas the counts and percentages of plasmacytoid DCs (pDCs) were at the lower end of the normal range for both patients (Figures 3C and S3B–C). NK cell counts in both patients were 80% lower than those in age-matched controls. The NK cytopenia was explained by a depletion of both CD56dim and NK CD56bright NK cells (Figures 3A, 3C and S3B). The innate lymphoid cell compartment was also affected, with a decrease in the percentages of innate lymphoid cell precursors (ILCP) (~10-fold) and type 2 innate lymphoid cells (ILC2, ~5-fold) in both patients (Figures 3C and S3D). The frequencies and counts of iNKT cells were normal in both patients, whereas the percentage of MAIT cells was normal in P1 and slightly low in P2 (Figures 3C and S3B). Counts of γδ T cells were slightly lower in P2 than in age-matched controls (Figure 3C). The frequencies of Vδ1+ and Vδ2+ T cells as a proportion of total T cells were normal and low, respectively, in both patients (Figure S3B).
Figure 3 – Phenotyping of peripheral blood leukocytes and single-cell PBMCs from IRF1-deficient patients.
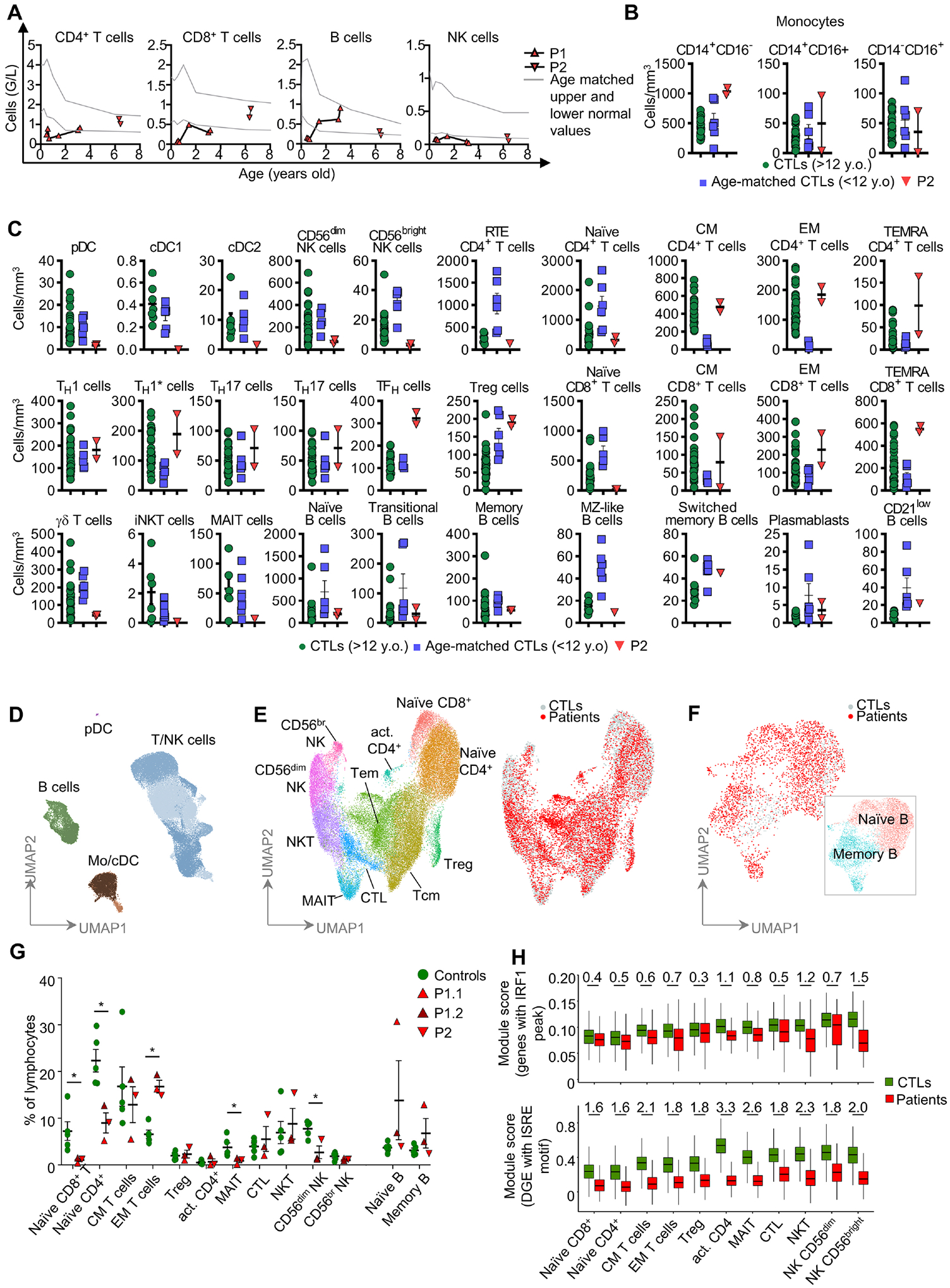
(A) Monitoring of lymphoid cell numbers in the whole blood or PBMCs of patients. (B) Counts for monocyte subsets by mass cytometry on fresh whole-blood cells. (C) Counts of myeloid and lymphoid subsets in fresh whole blood. (D) UMAP clustering for CTLs and samples from the IRF1-deficient patients (2x P1 and 1x P2) profiled by scRNA-seq or CITE-seq. General lineage populations are annotated. (E) Subclustering of T and NK cells to define cell subtypes. Overlay of 10,000 PBMCs from CTLs (gray) and 10,000 PBMCs from an IRF1-deficient patient (red) (F) As in (E), B-cell subclustering and cell-type annotation, together with an overlay of the patients’ and CTLs cells. (G) Proportions of each cell type expressed as a percentage of total lymphocytes. P1.1 corresponds to the first scRNA-seq analysis for P1 and P1.2 corresponds to the CITE-seq performed later on. (H) Module score analysis comparing the expression of genes with IRF1-binding sites within 10 kb of their TSS (top panel), and genes differentially expressed between patients and controls and predicted to have ISRE motifs in their promoters (bottom panel). Cohen’s d effect size estimates are shown for every significant variation of module expression. They were obtained by comparing the module score distribution in every cell subset in Wilcoxon signed-rank tests (q-value <= 0.05).
IRF1 deficiency alters the development of certain adaptive leukocyte subsets
Both patients also had ~95% fewer naïve CD8+ T cells than controls, and an excess of memory and T EMRA cells (Figure 3C and Table S3), consistent with the homeostatic expansion of CD8+ T cells following exposure to infectious agents. CD4+ T-cell counts were normal in both patients, but the numbers of recent thymic emigrant CD4+ T cells were only a third those in controls (Figures 3A, 3C and Table S3). Counts and percentages of TH1, TH2, TH17, TH1*, and Treg cells were also normal in both patients, whereas TFH cell counts were high in P2 (Figures 3C and S3B). B-cell counts were in the normal range in both patients, with normal frequencies of transitional B cells, memory B cells, and plasmablast cells (Figures 3A, 3C and S3B, and Table S3). The percentage of IgA+ memory B cells, was, however, higher than that in healthy controls, consistent with the abnormally high levels of plasma IgA detected in P1 (Figure S3B). However, both plasma IgA levels and IgA+ cell proportion were normal in P2. Both P1 and P2 had detectable antibodies against protein antigens and selected pneumococcal polysaccharide antigens in the serum, and normal serum levels of total IgG, and IgM (Table S4). Both patients displayed impaired development of myeloid dendritic cells (cDC1 and, to a much lesser extent, cDC2), innate lymphoid cells (ILC2 and ILCP), and IFN-γ-producing lymphoid cells (NK cells, naïve CD8+ T cells and, to a much lesser extent, naïve CD4+ T cells). Impaired development of cDC1, ILC2, NK, and CD8+ T cells were confirmed in IRF1-knockout mice (see Supplemental informations, Table S5, and Figures S3E–F).
Impairment of the lymphoid cell transcriptomes of IRF1-deficient patients
We analyzed the leukocyte development by performing single-cell RNA-seq and/or cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) on cryopreserved PBMCs. Integrated sample clustering of the various immune subsets identified 13 different major lymphoid subsets: mature (CD56dim) and immature (CD56bright) NK cells, naïve CD4+ and CD8+ T cells, activated CD4+ T cells, central memory T cells, effector memory T cells, cytotoxic T cells, MAIT and NKT cells, Treg, and naïve and memory B cells (Figures 3D–F). Consistent with the results of mass and flow cytometry, quantification of these subsets revealed low percentages of CD56dim NK cells, naïve CD4+ and CD8+ T cells, and an expansion of the population of effector memory T cells (Figures 3E–G). P1 also had an abnormally large fraction of IgA+ B cells, this feature being most pronounced at the age of three years (Figure 3G, Table S6). We performed differential gene expression analysis for each lymphoid subset, to detect changes in the transcriptional profiles of the cells of both patients. Interestingly, some genes were found to be differentially expressed in specific cells, but a sizable proportion of genes were differentially expressed in multiple subsets (Table S6). Gene ontology and pathway enrichment analysis indicated lower levels of expression for genes associated with terms such as leukocyte activation, defense response, cytokine signaling, cytotoxicity, and response to IFN in cells from the patients (Table S6). Moreover, an enrichment in genes with ISRE motifs in their promoters was observed for the genes underexpressed in patients, but not for those overexpressed in patients. We investigated the possible dysregulation of ISRE-containing gene expression, by performing a module score analysis in which we compared the expression of sets of genes between control and patient cells, and calculated effect size. We began the analysis by performing ChIP-seq on IFN-γ-stimulated mouse bone marrow-derived macrophages for the list of human orthologous genes binding IRF1. We found that IRF1-binding genes were significantly underexpressed in all subsets, this underexpression being strongest in memory and cytotoxic T cells and NK cells (Figure 3H). Underexpression was also more pronounced for differentially expressed genes with ISRE motifs in their promoters. However, IFNG expression was normal across lymphoid subsets (Table S6). Overall, these results suggest that IRF1 deficiency impairs the development of T and NK cells and the expression of target genes involved in immune activation.
Mild impairment of IFN-γ production in IRF1-deficient lymphoid cells
Given the impaired development of IFN-γ-producing lymphocytes and antigen-presenting DCs, we hypothesized that impaired IFN-γ production underlies mycobacterial disease in both these patients. We first investigated the IFN-γ secretion pathway in cell lines derived from patient cells. ISG15, the secretion of which is essential for IFN-γ production by NK cells26, was induced to a similar extent in the SV40-fibroblasts of both patients and those of healthy donors (Figure S4A). The early response to IL-12 — an inducer of IFN-γ production — which can be assessed by evaluating STAT4 phosphorylation, was also similar in the HVS-T cells of P1 and controls (Figure S4B). Moreover, IFN-γ production by HVS-T cells following stimulation with CD3/CD2/CD28 beads or PMA-ionomycin was similar for cells from P1 and controls (Figure S4C). We then studied the response to BCG of whole-blood samples from patients. IFN-γ was secreted by the peripheral leukocytes of both patients in response to BCG alone or BCG plus IL-12, and its levels in whole blood were similar to those of healthy local and travel controls (Figure 4A). Consistent with the results for whole blood, the stimulation of PBMCs from both patients with BCG in the presence or absence of IL-12 or IL-23 resulted in normal total intracellular IFN-γ production (Figure 4B). The clustering of IFN-γ-producing cells indicated that most of these cells were CD4+ and CD8+ T cells. However, the Vδ2+ cell and NK cell subsets of innate or innate-like adaptive lymphoid cells also displayed low levels of IFN-γ production in both patients (Figures 4C and S4D) consistent with their reduced circulating number. We then analyzed the in vitro differentiation of naïve CD4+ T cells from P1 after initial expansion with anti-CD2/CD3/CD28 mAb-coated beads and IL-2, followed by culture under TH0, TH1, TH2, TH9, or TH17 polarizing conditions. The production of IL-2 under TH0-polarizing conditions and the production of IFN-γ and TNF under TH1-polarizing conditions were strongly impaired in the IRF1-deficient naïve CD4+ T cells of P1 (Figures 4D and S4E). As a control, the production of IL-5 and IL-13 by IRF1-deficient naïve CD4+ T cells under TH2-polarizing conditions was normal (Figures 4D and S4E). Overall, IFN-γ production was impaired in NK and Vδ2+ cells, mostly due to their quantitative defects, and in TH1 cells upon non-mycobacterial stimulation, but the levels of production for this cytokine remained normal in CD4+ T cells, CD8+ T cells and whole blood cells stimulated with BCG.
Figure 4 – Production of IFN-γ by the lymphoid cells of IRF1-deficient patients.
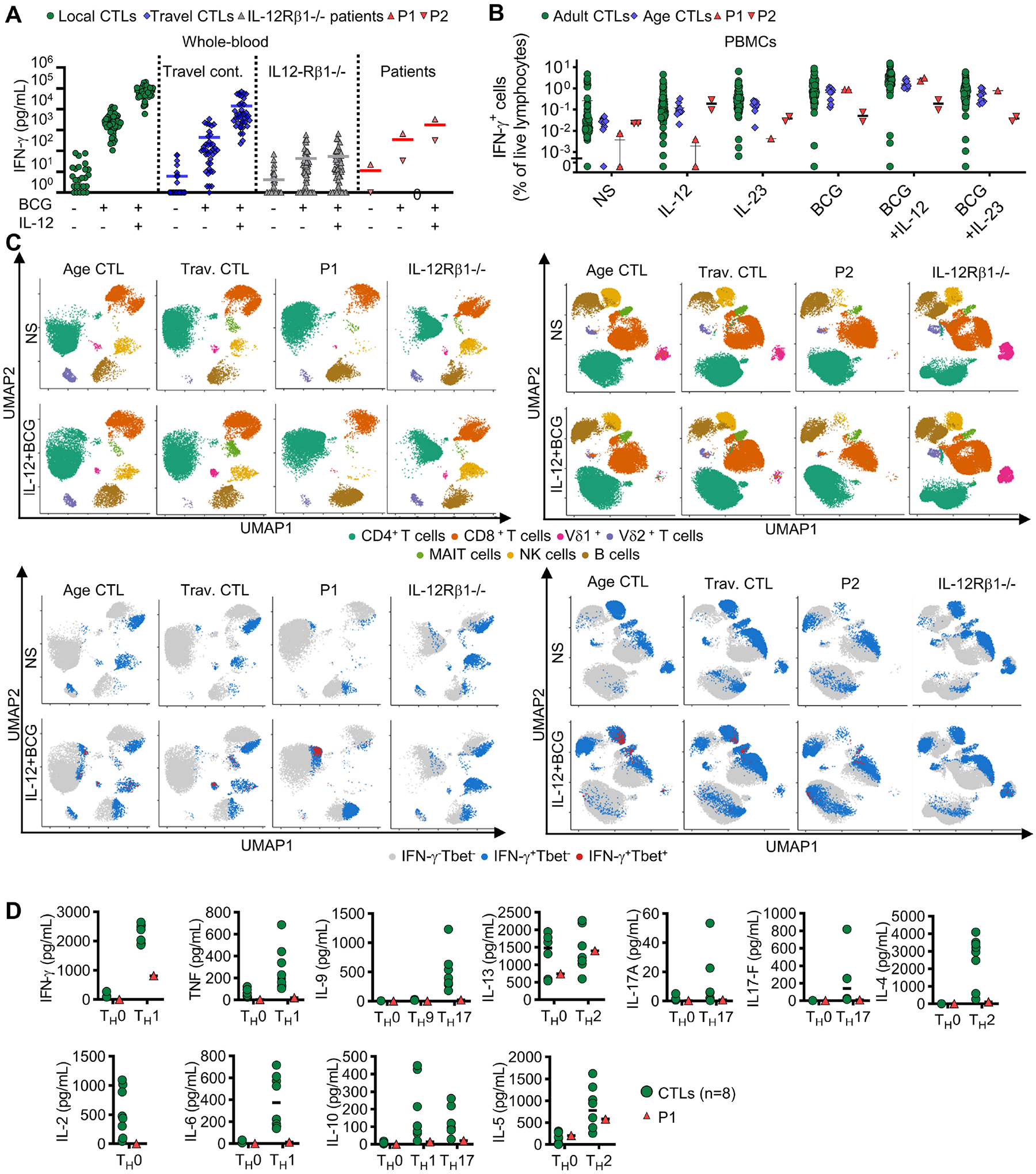
(A) Induction of IFN-γ secretion in a whole-blood assay, for controls (CTLs), IL-12Rβ1-deficient patients and patients. Bars represent the mean. (B) Intracellular flow cytometry on IFN-γ+ PBMCs after stimulation with IL-12, IL-23, or BCG. Bars represent the mean. Technical duplicates of the same experiment are shown for P1 and P2. (C) UMAP analysis of intracellular T-bet and IFN-γ expression by intracellular spectral flow cytometry across various PBMC subsets. Lymphoid subsets based on surface marker expression (upper panel), and their levels of IFN-γ and T-bet expression (lower panel). (D) Cytokine levels in the supernatant of naïve CD4+ T cells in polarizing conditions.
Impaired response to IFN-γ of IRF1-deficient fibroblasts
IRF1 is a protein found predominantly in the nucleus in the resting state52,54. The levels of production of this protein regulate its activity56,58–60. IRF1 can bind as a monomer, homodimer, or heterodimer to ISRE motifs in target genes, inducing their expression, as shown in human30,56,61,63 and mouse cells58. IFN-γ is the strongest known stimulus increasing IRF1 protein levels, through the direct induction of IRF1 mRNA synthesis30,65 and translation56,58–60, in a STAT1-dependent manner, within two to three hours61,67. We confirmed these findings for mRNA induction, translation and increases in gene expression following IFN-γ stimulation in SV40-fibroblasts (Figures S5A–C). We then investigated whether IRF1 deficiency impaired cellular responses to IFN-γ. As expected, the phosphorylation of STAT1 after 20 minutes of stimulation with IFN-γ, as determined by flow cytometry, was normal in EBV-B cells and SV40-fibroblasts from both patients (Figures S5D–E). We then comprehensively profiled the transcriptional response of the primary fibroblasts of patients after 30 minutes, 2 hours, and 8 hours of stimulation with IFN-γ, by RNA sequencing (RNA-seq). Primary fibroblasts from healthy donors displayed differential expression relative to non-stimulated conditions for 7 genes at 30 minutes, 343 genes at 2 hours, and 1,484 genes at 8 hours of stimulation, with IRF1 among the differentially regulated genes at all these timepoints (Figures S5F–G, and Table S7). We found that 20% of these genes were expressed differently between patients and controls after 8 hours. However, this percentage was significantly lower at 2 hours (13%, p-value 0.0012), and after 30 minutes of stimulation (0%). We also found that, relative to controls, the cells of both patients produced 30% and 40% less RNA for IFN-γ inducible genes at 2 hours and 8 hours, respectively (Figure S5H). Among genes differentially expressed in cells of both patients, an enrichment in the expression of genes with ISRE motifs in their proximal promoters was observed at both 2 and 8 hours (Figure 5A), but it was stronger at the 8-hour timepoint (p-value 1e-42) than at 2 hours (p-value 1e-15) (Figure 5A). The impaired induction of GBP4 transcription in response to IFN-γ was confirmed in SV40-fibroblasts from both patients (Figure 5B) and was rescued by stable transduction with a retroviral vector overexpressing WT IRF1 (Figure 5B). The amounts of protein generated for four IRF1 target genes — as demonstrated by RNA-seq (APOL3, GBP1, RARRES3, and CD274), immunoblotting or flow cytometry of SV40-fibroblasts or primary fibroblasts from patients — were also lower than those for control cells (Figures 5C and S5I–K). The impaired induction of CD274, the ligand of PD169, was corrected in patients’ SV40-fibroblasts by transduction of WT IRF1 cDNA (Figures S5J–K). We also confirmed the impaired induction of several proteins, including GBP1, GBP4, and APOL3, in the primary fibroblasts of both patients, after 24 hours of stimulation with IFN-γ, as shown by mass spectrometry (Figure 5D). The mass of inducible protein was found to be 30% smaller in the patients’ cells (Figure 5D). These results suggest that IRF1 controls the enhancement of the second wave of response to IFN-γ, downstream from STAT1.
Figure 5 – Response to IFN-γ of IRF1-deficient fibroblasts.
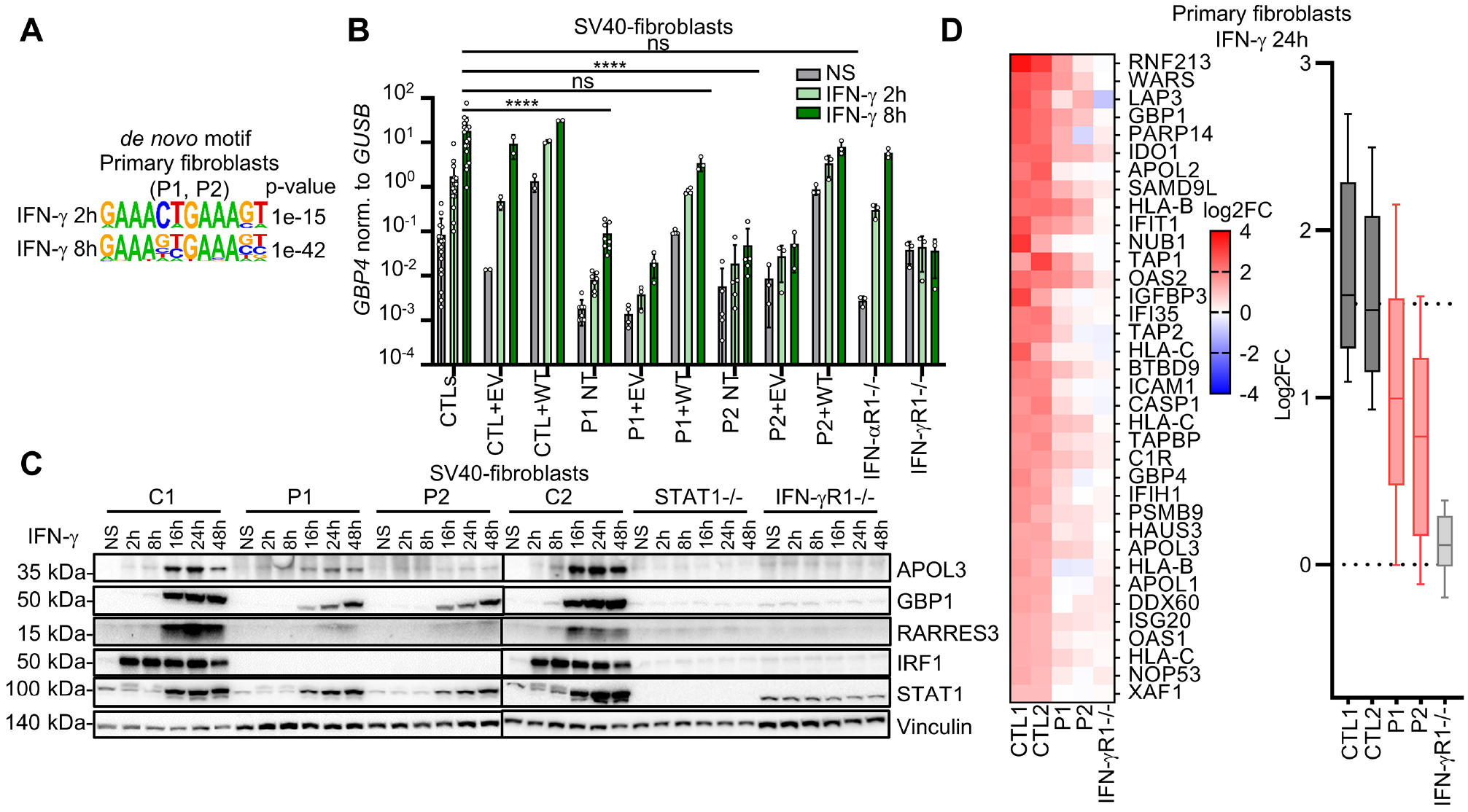
(A) HOMER de novo motif analysis of the genes differentially expressed in the primary fibroblasts after IFN-γ stimulation. (B) RT-qPCR for GBP4 (normalized against GUSB) in SV40-fibroblasts with or without retrotransduction with EV or WT IRF1 cDNA and with or without stimulation IFN-γ. Data from 2–6 independent experiments are shown. Bars represent the mean and SD. (C) Immunoblots in SV40-fibroblasts with and without stimulation with IFN-γ. (D) Mass spectrometry on lysates of primary fibroblasts with and without IFN-γ stimulation. On the right, heatmaps for proteins (i) positively induced after stimulation with a log2FC>1 over the mean in the non-stimulated state for controls (ii) and a log2FC<0.5 over the mean in the non-stimulated state for patients. On the left, 10th-90th percentiles for all proteins positively induced after stimulation with a log2FC>1 over the mean value in the non-stimulated state for controls.
Impaired response to IFN-γ in IRF1-deficient myeloid cells
We then studied cellular responses to IFN-γ in leukocytes from patients. IL-12p40 secretion into whole blood in response to BCG or BCG plus IFN-γ was in the control range for both patients (Figure S6A). By contrast, IL-12p70 induction in response to IFN-γ was impaired in both patients (Figure S6B). Impaired IL-12p70 induction probably contributed to the disseminated histoplasmosis observed in P1, as such infections are frequently reported in patients with complete IL-12Rβ1 or IL-12p40 deficiencies12,19,22. Patients with such deficiencies are also prone to isolated episodes of mycobacterial disease, but they almost never present recurrent episodes, contrasting with observations for P1 and P2. Impaired IL-12p70 production is, therefore, unlikely to explain the recurrence of mycobacterial disease in these two patients. Mononuclear myeloid cells are the best studied and classically defined effectors of IFN-γ-induced immunity28. They are also the only cells in which mycobacteria can replicate71. We therefore comprehensively profiled the transcriptome of mononuclear myeloid cells from patients after IFN-γ stimulation, by RNA-seq on iPSC-MΦ (P1) and MDMs (P2). After 8 hours of stimulation, 73 genes were found to display differential expression in the myeloid cells of both patients relative to controls, with all but one of these genes downregulated in the patients (Figures 6A, S6C–D, and Table S7). The expression of known MSMD genes was normal, except for IL12RB1 and ZNFX1, which displayed mild downregulation (~4-fold). Conversely, the genes strongly expressed in controls but with much lower levels of expression (10- to 500-fold) in both patients included several genes known to be involved in cell-autonomous defense against intracellular pathogens, such as members of the GBP family72,73 (chr1p22), members of the APOL family (chr22q12, including APOL374), IDO175, and RARRES352,76 (Figure 6A). The genes differentially expressed in the mononuclear myeloid cells of both patients were significantly enriched in (i) genes with an ISRE motif in their proximal promoter (Figure 6B) and (ii) loci to which IRF1 is known to bind, as detected by ChIP-seq in resting77 and IFN-γ-activated myeloid cells58 (Figure S6E). We confirmed the impairment of GBP4 and APOL3 induction by IFN-γ, by RT-qPCR on iPSC-MΦ derived from P1 and MDMs from P2 (Figure S6F). The impairment of APOL3, GBP1, IDO1, and RARRES3 protein production in response to IFN-γ stimulation was confirmed by western blotting on iPSC-MΦ from P1 and MDMs from P2 (Figure 6C). ZNFX1 expression was normal (Figure 6C). The oxidative burst in response to stimulation with IFN-γ and/or PMA was reduced in MDMs from P2 (Figure 6D). We then analyzed IFN-γ-dependent immunity to intramacrophagic pathogens in IRF1-deficient myeloid cells. The pretreatment with IFN-γ of WT THP1 cells reduced the intracellular growth of Salmonella Typhimurium (Figures 6E–F and S6G) and Mycobacterium abscessus relative to non-stimulated conditions (Figure 6G). By contrast, IFN-γ had no effect in THP1 IRF1-knockout (KO) cells78 or in THP1 IFN-γR1KO and STAT1KO cells (Figures 6F–G and S6G). Similar findings were obtained for the MDMs of P2 for M. abscessus infection (Figures 6H and S6H). Together, these results suggest that human IRF1 governs IFN-γ-dependent macrophage activation and resistance to intracellular pathogens.
Figure 6 – IFN-γ immunity in IRF1-deficient macrophages.

(A) Heatmaps showing genes differentially expressed (|log2(FC)| > 1 and adj. p-value < 0.05) in CTL cells after 8 hours of stimulation with 103 IU/mL IFN-γ, and differentially expressed in iPSC-MΦ (P1) and MDMs (P2) (|log2(FC)| > 1 and adj. p-value < 0.05). (B) HOMER de novo motif analysis of the genes differentially expressed in the iPSC-MΦ (P1), and in MDMs (P2); after IFN-γ stimulation. (C) Western blot of protein extracts in iPSC-MΦ, and MDMs, with and without stimulation with IFN-γ. (D) Extracellular H2O2 release for MDMs from CTLs and P2 (technical duplicates ± SD). (E) HLA-DR expression by flow cytometry, using THP1-Ф with and without IFN-γ stimulation. (F) Gentamicin protection assay performed on PMA-differentiated THP1-Ф with and without IFN-γ pretreatment and Salmonella Typhimurium-GFP (Stm-GFP) infection. Results are expressed as the proportion of Salmonella Typhimuriumhigh on Salmonella Typhi+. Representative results from 2–3 independent experiments. (G) CFU assays on PMA-differenciated THP1-Ф with and without IFN-γ pretreatment, following infection by Mycobacterium abscessus. All replicates from n=3 independent experiments are displayed. (H) Flow cytometry analyses on MDMs with and without IFN-γ pretreatment, following infection for 24 hours with M. abscessus-tdTomato.
Mildly impaired responses to IFN-α/β in IRF1-deficient cells
In addition to its known role in IFN-γ-related immunity, the antiviral role of IRF1 has also been extensively studied. The ablation52,61,79,80 and overexpression81,82 of the IRF1 gene have been shown to be associated with susceptibility and resistance to viral infections, respectively, in vivo in mice83–87 (Table S2) and in vitro in human cell lines61,79,81. Neither P1 nor P2 presented life-threatening viral infections (Supplemental Case report and Supplemental information). Phage immunoprecipitation-sequencing (PhIP-Seq) confirmed that both patients has been exposed to multiple DNA and RNA viruses (Figure 7A and Table S4). Stimulation with IFN-α/β induces IRF1, albeit at a level lower than observed after stimulation with IFN-γ59–61,65,67. We confirmed these findings in human cells and also found, surprisingly, that IFN-α/β-dependent IRF1 induction was ISGF3-independent (i.e. STAT2 and IRF9-independent) but GAS-dependent (i.e. STAT1-dependent) (Figures S7A–C). We analyzed antiviral IFN-α/β immunity in the patients’ cells by flow cytometry, RNA-seq, and in viral growth assays. The early response to IFN-α/β in SV40-fibroblasts and EBV-B cells, assessed by evaluating STAT1 phosphorylation after 20 minutes of stimulation with IFN-α2b and IFN-β, was normal in both patients (Figure S7D). We then studied the transcriptomic response of primary fibroblasts stimulated with IFN-α2b for 30 minutes, 2 hours, or 8 hours. We compared these responses to those of the cells of patients with complete deficiencies of IRF9, STAT1, or IFNAR1. Primary fibroblasts from healthy donors displayed differential expression relative to unstimulated conditions for 110 genes, 667 genes, and 1,093 genes for these three timepoints, respectively, with IRF1 significantly upregulated at all these timepoints (Figures 7B–C, and Table S7). The impaired induction of various interferon-stimulated genes (ISGs; (for 2%, 10%, and 8% of the genes induced in controls)) was observed in the patients’ cells at these three timepoints. Quantitatively, 81% and 71% of the mRNA levels of IFN-α inducible genes in controls were correctly induced in both patients, at 2 and 8 hours, respectively (Figure 7D). The corresponding percentages were 24% and 54% in IRF9-deficient cells, and 8% and 23% in STAT1-deficient cells, for the same timepoints. Most of the ISGs dysregulated in IRF1-deficient cells have an ISRE motif in their promoters (Figure 7E). A subset of these ISGs also displayed impaired induction in IRF1-deficient cells but not in IRF9-deficient cells, defining these genes as GAF-IRF1-dependent (Figure 7E, and Table S7).
Figure 7 – IFN-α and IFN-γ-driven antiviral immunity in the cells of IRF1-deficient patients.
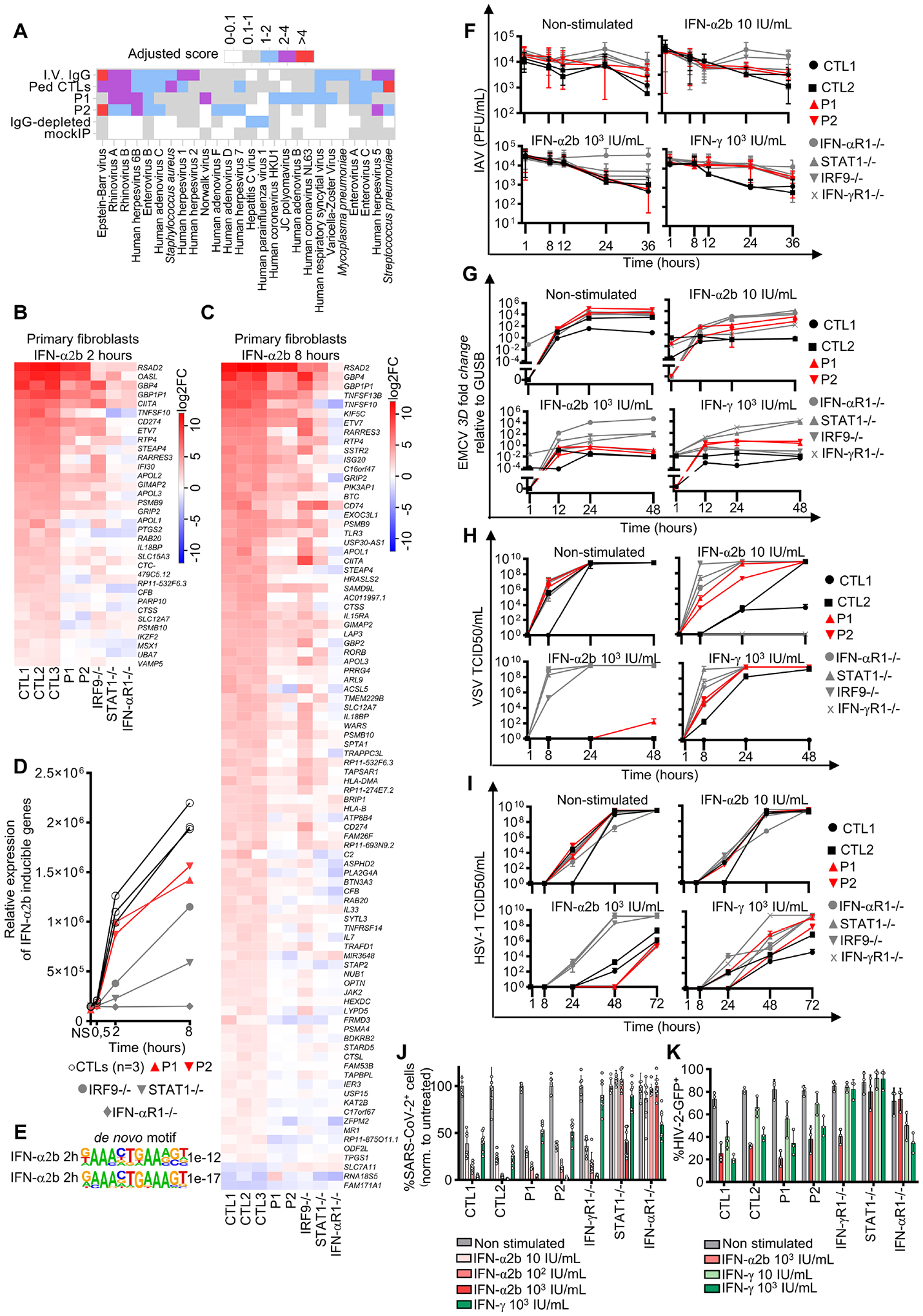
(A) Antiviral antibody responses to species for which at least one sample tested seropositive by PhIP-Seq. “IVIG” correspond to the mean response for samples from pooled patients on IVIGs and “pediatric CTLs” to pediatric controls. A hierarchical clustering of samples based on antiviral antibody levels is shown at the top. Heatmap showing genes differentially expressed (|log2(FC)| > 1 and adj. p-value < 0.05) in CTL cells after 2 hours (B) or 8 hours (C) of IFN-α2b stimulation, and differentially expressed in primary fibroblasts from P1 (|log2(FC)| > 1 and adj. p-value < 0.05) relative to CTLs. Genes differentially expressed in P1 and P2 relative to the control group at 2 and 8 hours from among those differentially expressed at both timepoints relative to non-stimulated fibroblasts in the control group, i.e., with a |log2(FC)| > 1 and adj. p-value < 0.05 after Benjamini-Hochberg correction in controls, and with a |log2(FC)| > 1 and adj. p-value < 0.05 after correction in patients relative to controls. (D) RNA-sequencing of IFN-α2b-inducible genes (logFC>2 in controls) in primary fibroblasts with and without IFN-α2b stimulation. (E) HOMER de novo motif analysis of genes differentially expressed in primary fibroblasts with IFN-α2b stimulation. (F) Influenza A virus (IAV), (G) encephalomyocarditis virus (EMCV), (H) vesicular stomatitis Indiana virus (VSV), (I) HSV-1, (J) SARS-CoV-2, and (K) HIV-2 infection of SV40-fibroblasts after pretreatment with IFN-α2b or IFN-γ. All viral infections were performed in 2–3 independent experiments.
Normal antiviral activity of IFN-α/β and IFN-γ in IRF1-deficient cells
We assessed the possible impact of these findings on the replication of several viruses in SV40-fibroblasts after pretreatment with IFN-α2b. In these conditions, the cells of the patients controlled the replication of encephalomyelitis virus (EMCV), influenza A virus (IAV), hepatitis A virus (HAV), herpes simplex virus type 1 (HSV-1), human immunodeficiency viruses type 1 and 2 (HIV-1 and HIV-2), yellow fever live-attenuated viral vaccine (YF17D-venus), SARS-CoV-2, and vesicular stomatitis virus Indiana (VSV) as effectively as control cells, whereas viral replication continued unabated in IFN-αR1−/−, IRF9−/−, STAT2−/− or STAT1−/− cells (Figures 7F–K and S7F–I), except for EMCV and VSV, for which greater susceptibility was observed with a low dose of IFN-α (Figure 7G–H). We conducted similar experiments with IFN-γ, which was initially described as an antiviral molecule88–91 before being identified as the macrophage-activating factor28. We found that IRF1-deficient cells had a lower susceptibility to infection following IFN-γ pretreatment, like control cells, for all viruses other than IAV, EMCV, and VSV, for which resistance was weaker than in control cells (Figures 7F–K and S7F–I). Collectively, these results suggest that the antiviral response of IRF1-deficient cells to IFN-α and IFN-γ is only mildly impaired and sufficient to protect IRF1-deficient fibroblasts against viral infection, consistent with the absence of severe viral illnesses in these two patients.
Discussion
We report here that AR complete IRF1 deficiency is a genetic etiology of isolated and severe MSMD. The two unrelated IRF1-deficient patients of Latin American and Turkish ancestries described here experienced recurrent early-onset life-threatening mycobacterial diseases due to multiple mycobacteria (BCG, M. avium) despite treatment with multiple antimycobacterial drugs, and even treatment with recombinant IFN-γ in the case of P1. Remarkably, IRF1 deficiency was found to be as clinically severe as AR complete deficiencies of IFN-γ, IFN-γR1, or IFN-γR24,5,12. Patients with these deficiencies display adverse reactions to BCG when vaccinated at birth, followed by relapses of BCG disease and/or disseminated disease caused by EM4,5,12. Disseminated infection with M. avium before six years of age is a hallmark of a lack of IFN-γ immunity due to inherited defects of the IFN-γ response pathway4,5,12,34, whereas M. avium disease at a later age may be due to profound deficiencies as a result of the production of anti-IFN-γ auto-antibodies92,93, or a severe progressive quantitative defect of IFN-γ myeloid target cells caused by GATA2 deficiency94,95. Given the severity of the clinical phenotype in these two unrelated patients, the clinical penetrance of inherited IRF1 deficiency for MSMD is probably complete. Consistently, Irf1−/− mice are susceptible to both BCG96 and M. tuberculosis97,98, and other intramacrophagic pathogens99–103. Myeloid cells are classically considered to be the effector cells for IFN-γ28, and this cytokine is also their most potent activator104. Our findings indicate that IRF1 governs the response to IFN-γ, downstream from STAT1, in mononuclear myeloid cells. IRF1 controls the potent induction of several genes encoding intracellular components known to be effectors of resistance to intracellular pathogens, such as a protein from the GBP family72–74,100,105,106, IDO175, RARRES3/PLAAT476, and APOL374,107. Our work extends the list of known IFN-γ-inducible IRF1-dependent genes in mice58 to human genes with no known ortholog in mice108, such as RARRES376 and APOL374,107. Inherited deficiencies of some of these IFN-γ- and IRF1-dependent effector genes may underlie MSMD or TB. IRF1 deficiency underlies severe and isolated MSMD, as in patients with AR IFN-γ, IFN-γR1, or IFN-γ2 deficiencies4,5,12 whose mononuclear myeloid cells cannot respond correctly to IFN-γ.
We also found additional quantitative and qualitative deficiencies of IFN-γ-producing lymphocytes in the two IRF1-deficient patients, consistent with reported findings for Irf1−/− mice83,101,109–113. The observed deficiencies included an impairment of the development of NK cells, and naïve αβ CD8+ T cells, and impaired IFN-γ production by residual NK cells and γδ2+ T cells. However, the leukocytes of both patients produced normal total amounts of IFN-γ upon exposure to mycobacteria in vitro, suggesting that the abnormal counts or function of IFN-γ-producing lymphoid cell subsets in peripheral blood made only a marginal contribution to mycobacterial disease. IRF1 deficiency impairs the development of myeloid DCs, predominantly that of cDC1, the counts of which were low in the blood of both IRF1-deficient patients and in the tissues of Irf1−/− mice. Our results suggest that IRF1 is essential for myeloid cell maturation, through an as yet unknown mechanism, possibly involving interaction with IRF825,27,114,115. Indeed, human IRF8 governs DC development in a gene dosage-dependent manner, as illustrated by AD and AR IRF8 deficiencies25,27,114–116. We also documented low levels of IL-12p70 induction in both patients, which is more likely to result from direct binding to the IL12A locus, as previously shown in murine myeloid cells117,118, rather than a decrease in myeloid cell number. Consistently, patients with AD PU.1 deficiency, who have low circulating myeloid cell counts, have a normal capacity to produce IL-12119. Overall, in addition to their profoundly impaired cellular responses to IFN-γ, the patients with IRF1 deficiency have a mild impairment of IFN-γ production, which may have contributed to their MSMD.
By contrast, neither patient presented any severe viral diseases, despite low levels of ILCP, NK cells, and naïve CD8+ T cells. Patients with severe combined immunodeficiency (SCID) caused by LOF mutations of IL2RG or JAK3 who undergo hematopoietic stem cell transplantation do not reconstitute a normal pool of peripheral ILC and NK cells120. They are not prone to viral infections other than HPV-driven common and flat warts, perhaps due to the persistence of deficiencies of keratinocytes or antigen-presenting cells in the skin121. Isolated deficiencies of ILCs and NK cells in patients with IEIs, such as GINS1, MCM4, or MCM10 deficiency120,122–124, can lead to various degrees of susceptibility to viral diseases, mostly caused by CMV. These IEIs are caused by genes with expression profiles not restricted to NK cells or ILCs, and the defect may be broader120,122–124. Patients with inherited CD8, TAP1, TAP2, TAPASIN, or β2-microglobulin deficiencies, all of whom have low levels of HLA-I in all cell types tested and low counts of blood CD8+ T cells, are not prone to viral infections either125–131. IEIs impairing CD8+ T-cell effector or expansion functions can underlie susceptibility to EBV132. P1 had not encountered CMV or EBV, but P2 had been exposed to CMV and had controlled its replication and tested positive for EBV by PCR. Overall, the low but non-zero counts of circulating antiviral lymphoid cell subsets in the two patients with IRF1 deficiency have so far proved sufficient to ensure immunity against the many viruses encountered in childhood. Innate and adaptive leukocytic immunity to viruses is evidently affected by complete IRF1 deficiency, albeit with no apparent clinical consequences, implying that other antiviral mechanisms can compensate for this relatively broad lymphoid deficiency.
More surprisingly, cell-intrinsic, IFN-α/β immunity is IRF1-independent for the nine viruses we have tested in vitro. The contrast between AR STAT1 and IRF1 deficiencies is striking in this respect, as the two patients with IRF1 deficiency did not suffer from the life-threatening viral diseases seen in patients with AR complete STAT1 deficiency33,34. Despite the impaired induction of a subset of target genes in fibroblasts stimulated with IFN-α/β, the patients displayed no severe viral diseases, even upon infection with SARS-CoV-2, probably the most potent known sensor of IFN-α/β deficiency133–138. Consistently, the cells of the patients controlled the replication of the seven RNA and DNA viruses tested in vitro, unlike cells from patients with AR STAT1, STAT2, IRF9, or IFNAR1 deficiency. All IEIs affecting the response to IFN-α/β underlying susceptibility to acute viral diseases (i.e. AR IFN-αR1, IFN-αR2, STAT1, STAT2, or IRF9 deficiencies) are caused by deficiencies leading to a complete abolition of ISGF3 function in response to stimulation with IFN-α/β138–145. These observations suggest that functional ISGF3 (composed of STAT1, STAT2, and IRF9) is essential for IFN-α/β intrinsic immunity, whereas IRF1 (formerly known as ISGF265,146) is largely redundant for such immunity in vivo, as demonstrated by viral replication levels. We found that human IRF1 was driven by GAF but not ISGF3 complexes in response to IFN-α/β, but the GAF/STAT1-dependent induction of IRF1, and of subsequent target ISGs, such as GBP, RARRES3, APOL3, was not required for immunity to many viruses. By contrast, Irf1-deficient mice have been reported to be susceptible to a number of viral infections147,83,84,87,86. The lack of IRF1 in mice may, nevertheless, be partly compensated by other IRFs for antiviral immunity148. IRF1 is not induced after stimulation with IFN-λ61. In addition, humans with a complete deficiency of IL-10Rβ display a complete lack of response to IFN-λ and are apparently not susceptible to infectious diseases149,150, except fulminant viral hepatitis A151. Overall, the natural course of infectious diseases in the known patients with inherited deficiencies of IRF1 (this study), IRF3152,153, IRF7138,140, or IRF9139,154 suggests that IRF1 is essential for IFN-γ-dependent myeloid antimycobacterial immunity, whereas IRF3, IRF7 and IRF9 are essential for IFN-α/β-dependent antiviral immunity.
Limitations of the Study
Our molecular, cellular, immunological, and clinical studies of two unrelated patients with inherited IRF1 deficiency do not exclude the possibility of predisposition to unusual and/or severe clinical diseases caused by viruses that have not been tested in vitro, or that have not been encountered by the patients in vivo, or both, or even such diseases caused by seemingly benign viruses in different infection conditions (e.g. high levels of inoculum, atypical route of infection, infection before vaccination), in these or other IRF1-deficient patients. Any susceptibility to viruses in these patients might be due to impaired IRF1-dependent IFN-α/β immunity, or due to the deficit of cytotoxic NK and CD8+ T cells, or both. In particular, the long-term outcome of infection with viruses capable of latency, such as those of the Herpesviridae, is unknown.
STAR METHODS
RESSOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jean-Laurent Casanova (casanova@mail.rockefeller.edu).
Materials Availability
All raw and processed data and biological materials, including immortalized cell lines from patients, are available upon request from the Lead Contact under a Material/Data Transfer Agreement with Inserm or the Rockefeller University.
Data and Code Availability
RNA-seq, single-cell RNA-seq, and CITE-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. The mass spectrometry proteomics data have been deposited with the ProteomeXchange Consortium via the PRIDE partner repository and datasets identifier is listed in the key resource table. Original western blot images, flow cytometry data, mass spectrometry data, and microscopy data reported in this paper will be shared by the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Human IRF1 (clone D5E4) | Cell Signaling | Cat# 8478, RRID:AB_10949108 |
| Human IRF1 (rabbit polyclonal) | Proteintech | Cat# 11335-1-AP, RRID:AB_2877759 |
| Human IRF8 (goat polyclonal) | Santa Cruz | Cat# sc-6058, RRID:AB_649510 |
| Human IRF8 (clone D20D8) | Cell signaling | Cat# 5628, RRID:AB_10828231 |
| Human IRF9 (rabbit polyclonal) | Santa Cruz | Cat# sc-496, RRID:AB_2127709 |
| Human IRF9 (rabbit polyclonal) | Proteintech | Cat# 14167-1-AP, RRID:AB_2296227 |
| Human IRF3 (clone D9J5Q) | Cell Signaling | Cat# 10949, RRID:AB_2797733 |
| Human STAT1 (clone 1) | Beckton-Dickinson | Cat# 610115, RRID:AB_397521 |
| Human pSTAT1 (clone 4a) | Beckton-Dickinson | Cat# 612232, RRID:AB_399555 |
| Human STAT2 (clone B-3) | Santa Cruz | Cat# sc-514193, RRID:AB_2810271 |
| Human MX1 (polyclonal) | ProteinTech | Cat# 13750-1-AP, RRID:AB_2266768 |
| Human ISG15 (clone F-9) | Santa-Cruz | Cat# sc-166755, RRID:AB_2126308 |
| Human vinculin (clone EPR8185) | Abcam | Cat# ab129002, RRID:AB_11144129 |
| Human vinculin (clone 7F9) | Santa Cruz | Cat# sc-376248-HRP, RRID:AB_10991536 |
| Anti-mouse IgG (H + L)-HRP-conjugated | Bio-Rad | Cat# 170-6516, RRID:AB_11125547 |
| Anti-rabbit IgG (H + L)-HRP-conjugated | Bio-Rad | Cat# 170-6515, RRID:AB_11125142 |
| Human DDK-tag (clone M2) | Sigma-Aldrich | Cat# A8592, RRID:AB_439702 |
| Human DDK (clone M2) | Cell Signaling | Cat# 14793, RRID:AB_2572291 |
| Human Isotype rabbit | Cell Signaling | Cat# 2729, RRID:AB_1031062) |
| Human GBP1 (clone 1B1) | Santa-Cruz | Cat# sc-53857, RRID:AB_2109333 |
| Human APOL3 (clone EPR8238) | Abcam | Cat# ab154869 |
| Human RARRES3 (rabbit polyclonal) | ProteinTech | Cat# 12065-1-AP, RRID:AB_2175704 |
| anti-rabbit IgG Alexa Fluor 555 | ThermoFischer Scientific | Cat# A-21429, RRID:AB_2535850 |
| PE-Dazzle-594- Human PD-L1 (CD274) antibody (clone 29E.2A3) | BioLegend | Cat# 329732, RRID:AB_2616889 |
| PE-Dazzle-594-Mouse IgG2b, κ Isotype Ctrl Antibody | BioLegend | Cat# 400358 |
| PE-Human STAT1 (clone 1) | Beckton-Dickinson | Cat# 558537, RRID:AB_647231 |
| PE-Human pSTAT1 (clone 4a) | Beckton-Dickinson | Cat# 612564, RRID:AB_399855 |
| AF647-Human pSTAT1 (clone 4a) | Beckton-Dickinson | Cat# 612597, RRID:AB_399880) |
| PE Mouse IgG1, κ Isotype Control | Beckton-Dickinson | Cat# 554680, RRID:AB_395506 |
| AF647 Mouse IgG1, κ Isotype Control | Beckton-Dickinson | Cat# 565363, RRID:AB_2869665 |
| PE-conjugated goat anti-rabbit | Thermo Fisher Scientific | Cat# A10542, RRID:AB_2534042 |
| Human IFN-γ-BUV737 clone 4S.B3 | BD Horizon | Cat# 564620, RRID:AB_2869591 |
| Human TNF-PerCP clone Mab11 | BioLegend | Cat# 502924, RRID:AB_2561288 |
| Human IL-9-PE clone MH9A3 | BD Pharmingen | Cat# 560807, RRID:AB_2033985 |
| Human IL-13-BV421 clone JES10-5A2 | BD Horizon | Cat# 563580, RRID:AB_2738290 |
| Human IL-4-AF488 clone 8D4-8 | BioLegend | Cat# 500710, RRID:AB_1877131 |
| Human IL-17A-BV510 clone BL168 | BioLegend | Cat# 512330, RRID:AB_2562745 |
| Human IL-17F-BV650 clone O33-782 | BD Horizon | Cat# 564264, RRID:AB_2869555 |
| Human IL-2-BV750 clone MQ1-17H12 | BD Horizon | Cat# 566361, RRID:AB_2739710 |
| Human IL-21-eF660 clone eBio3A3-N2 | Thermo Fisher Scientific | Cat# 50-7219-42, RRID:AB_10598202 |
| Human aCD3-Alexa532 (Clone UCHT1) | Thermo Fisher Scientific | Cat# 58-0038-42, RRID:AB_11218675 |
| Human γδTCR-FITC (clone) | Thermo Fisher Scientific | Cat# 11-9959-42, RRID:AB_10669049 |
| Human Vδ2-APC-Fire750) | BioLegend | Cat# 331420, RRID:AB_2687326 |
| Human CD56-BV605 (clone 5.1H11) | BioLegend | Cat# 362538, RRID:AB_2565856 |
| Human CD4-BV750 | BD Biosciences | Cat# 566356, RRID:AB_2744426 |
| Human CD8a-BV510 (clone RPA-T8) | BioLegend | Cat# 301047, RRID:AB_2561378 |
| Human Vα7.2-BV711 (clone 3C10) | BioLegend | Cat# 351731, RRID:AB_2629679 |
| Human Vα24-Jα18-PE-Cy7 (clone 6B11) | BioLegend | Cat# 342912, RRID:AB_2562230 |
| Human Vδ1-Vioblue | Miltenyi Biotec | Cat# 30-100-555 |
| Human CD161-PE (clone HP-3G10) | BioLegend | Cat# 339938, RRID:AB_2564141 |
| Human Vβ11-APC (Miltenyi Biotec) | Miltenyi Biotec | Cat# 58-0038-42, RRID:AB_11218675 |
| Human CD1a-biotin (clone HI149) | BioLegend | Cat# 300112, RRID:AB_389344 |
| Human CD14-biotin (clone 61D3) | Invitrogen | Cat# 13-0149-82, RRID:AB_466373 |
| Human CD34-biotin (clone 4H11) | BioLegend | Cat# 316404 |
| Human CD123-biotin (clone 6H6) | BioLegend | Cat# 306004, RRID:AB_314578 |
| Human CD203c-biotin (clone FR316A11) | Miltenyi Biotec | Cat# 130-092-345, RRID:AB_615067 |
| Human CD303-biotin (clone AC144) | Miltenyi Biotec | Cat# 130-090-691, RRID:AB_244166 |
| Human FcεRIα-biotin (clone AER-37 CRA-1) | BioLegend | Cat# 334606, RRID:AB_2571885 |
| Human TCRαβ-biotin (clone IP26) | BioLegend | Cat# 306704, RRID:AB_314632 |
| Human TCRγδ-biotin (clone B1) | BD Biosciences | Cat# 555716, RRID:AB_396060 |
| Human CD4 FITC (clone OKT4) | BioLegend | Cat# 317408, RRID:AB_571951 |
| Human CD336 PerCP-eFluor710 (clone 44.189) | Thermo Fisher Scientific | Cat# 46-3369-42, RRID:AB_2573749 |
| Human EOMES PE (clone WD1928) | Thermo Fisher Scientific | Cat# 12-4877-42, RRID:AB_2572615 |
| Human CD8a PE-CF594 (clone RPA-T8) | BD Biosciences | Cat# 562282, RRID:AB_11154052 |
| Human CD127 PE-Cy7 (clone eBioRDR5) | Thermo Fisher Scientific | Cat# 25-1278-42, RRID:AB_1659672 |
| Human CD294 AF647 (clone BM16) | BD Biosciences | Cat# 558042, RRID:AB_2112699 |
| Human CD161 AF700 (clone HP-3G10) | BioLegend | Cat# 339942, RRID:AB_2565870 |
| Human CD94 APC-Fire750 (clone DX22) | BioLegend | Cat# A305-518A, RRID:AB_2773751 |
| Human CD335 BV421 (clone 9E2/NKp46) | BD Biosciences | Cat# 564065, RRID:AB_2738572 |
| Human CD45RA BV570 (clone HI100) | BioLegend | Cat# 304132, RRID:AB_2563813 |
| Human CD117 BV605 (clone 104D2) | BioLegend | Cat# 313218, RRID:AB_2562025 |
| Human CD3 BV650 (clone UCHT1) | BD Biosciences | Cat# 563852 |
| Human CD7 BV711 (clone M-T701) | BD Biosciences | Cat# 564018, RRID:AB_2738544 |
| Human T-bet BV786 (clone O4-46) | BD Biosciences | Cat# 564141, RRID:AB_2738615 |
| Human CD19 BUV395 (clone SJ25C1) | BD Biosciences | Cat# 563549, RRID:AB_2738272 |
| Human CD16 BUV496 (clone 3G8) | BD Biosciences | Cat# 564653, RRID:AB_2744294 |
| Human CD25 BUV563 (clone 2A3) | BD Biosciences | Cat# 565699, RRID:AB_2744341 |
| Human CD56 BUV737 (clone NCAM16.2) | BD Biosciences | Cat# 564447, RRID:AB_2744432 |
| Human CD45 BUV805 (clone HI30) | BD Biosciences | Cat# 612891, RRID:AB_2870179 |
| Human CD11c (S-HCL-3) | BioLegend | Cat# 371523, RRID:AB_2814332 |
| Human CD141 (M80) | BioLegend | Cat# 344127, RRID:AB_2832671 |
| Human CD161 (HP-3G10) | BioLegend | Cat# 339949, RRID:AB_2832665 |
| Human CD14 (Me5E2) | BioLegend | Cat# 301857, RRID:AB_2800735 |
| Human CD16 (3G8) | BioLegend | Cat# 302063, RRID:AB_2800737 |
| Human CD19 (HIB19) | BioLegend | Cat# 302263, RRID:AB_2800740 |
| Human CD1c (L161) | BioLegend | Cat# 331549, RRID:AB_2832653 |
| Human CD28 (CD28.2) | BioLegend | Cat# 302961, RRID:AB_2800750 |
| Human CD370 (CLEC9A) (8F9) | BioLegend | Cat# 353811, RRID:AB_2876671 |
| Human CD38 (HB-7) | BioLegend | Cat# 356639, RRID:AB_2814303 |
| Human CD3e (UCHT1) | BioLegend | Cat# 300477, RRID:AB_2800722 |
| Human CD4 (RPA-T4) | BioLegend | Cat# 300565, RRID:AB_2800724 |
| Human CD45RA (HI100) | BioLegend | Cat# 304161, RRID:AB_2800763 |
| Human CD45RO (UCHL1) | BioLegend | Cat# 304259, RRID:AB_2800766 |
| Human CD56 (NCAM) (5.1H11) | BioLegend | Cat# 362561, RRID:AB_2814309 |
| Human CD66b (6/40c) | BioLegend | Cat# 392913, RRID:AB_2832738 |
| Human CD69 (FN50) | BioLegend | Cat# 310949, RRID:AB_2800809 |
| Human CD8a (RPA-T8) | BioLegend | Cat# 301069, RRID:AB_2800729 |
| Human TCR gd (B1) | BioLegend | Cat# 331233, RRID:AB_2814200 |
| Human TCR Va7.2 (3C10) | BioLegend | Cat# 351737, RRID:AB_2819993 |
| IgG1 k Isotype Ctl (MOPC-21). | BioLegend | Cat# 400185 |
| 89 Y Human CD45 (clone HI30) PBMC panel 1 | Fluidigm | Cat# 3089003B, RRID:AB_2661851 |
| 113 In Human CD57 (clone HCD57) PBMC panel 1 | BioLegend | Cat# 322302, RRID:AB_535988 |
| 115 In Human CD11c (clone Bu15) PBMC panel 1 | BioLegend | Cat# 337202, RRID:AB_1236381 |
| 141 Pr Human CD33 (clone WM53) PBMC panel 1 | BioLegend | Cat# 303410, RRID:AB_2074243 |
| 142 Nd Human CD19 (clone HIB19) PBMC panel 1 | BioLegend | Cat# 302202, RRID:AB_314232 |
| 143 Nd Human CD45RA (clone HI100) PBMC panel 1 | BioLegend | Cat# 304102, RRID:AB_314406 |
| 144 Nd Human CD141 (clone M80) PBMC panel 1 | BioLegend | Cat# 344102, RRID:AB_2201808 |
| 145 Nd Human CD4 (clone RPA-T4) PBMC panel 1 | BioLegend | Cat# 300502, RRID:AB_314070 |
| 146 Nd Human CD8 (clone RPA-T8) PBMC panel 1 | BioLegend | Cat# 301002, RRID:AB_314120 |
| 147 Sm Human CD20 (clone 2H7) PBMC panel 1 | BioLegend | Cat# 302302, RRID:AB_314250 |
| 148 Nd Human CD16 (clone 3G8) PBMC panel 1 | BioLegend | Cat# 302014, RRID:AB_314214 |
| 149 Sm Human CD127 (clone A019D5) PBMC panel 1 | Fluidigm | Cat# 3149011B, RRID:AB_2661792 |
| 150 Nd Human CD1c (clone L161) PBMC panel 1 | BioLegend | Cat# 331502, RRID:AB_1088995 |
| 151 Eu Human CD123 (clone 6H6) PBMC panel 1 | BioLegend | Cat# 306002, RRID:AB_314576 |
| 152 Sm Human CD66b (clone G10F5) PBMC panel 1 | BioLegend | Cat# 305102, RRID:AB_314494 |
| 153 Eu Human PD-1 (clone EH12.2H7) PBMC panel 1 | BioLegend | Cat# 329926, RRID:AB_11147365 |
| 154 Sm Human CD86 (clone IT2.2) PBMC panel 1 | BioLegend | Cat# 305410, RRID:AB_314530 |
| 155 Gd Human CD27 (clone O323) PBMC panel 1 | BioLegend | Cat# 302802, RRID:AB_314294 |
| 156 Gd Human CCR5 (clone J418F1) PBMC panel 1 | BioLegend | Cat# 359102, RRID:AB_2562457 |
| 158 Gd Human CD117 (clone 104D2) PBMC panel 1 | BioLegend | Cat# 313202, RRID:AB_314981 |
| 159 Tb Human CD24 (clone ML5) PBMC panel 1 | BioLegend | Cat# 311102, RRID:AB_314851 |
| 160 Gd Human CD14 (clone M5E2) PBMC panel 1 | BioLegend | Cat# 301810, RRID:AB_314192 |
| 161 Dy Human CD56 (clone B159) PBMC panel 1 | BD Biosciences | Cat# 555513, RRID:AB_395903 |
| 162 Dy Human gdTCR (clone REA591) PBMC panel 1 | Miltenyi | Cat# 130-122-291; RRID: AB_2801872 |
| 163 Dy Human CRTh2 (clone REA598) PBMC panel 1 | Milentyi | Cat# 130-122-305, RRID:AB_2801886 |
| 164 Dy Human CLEC12A (clone 50C1) PBMC panel 1 | BioLegend | Cat# 353602, RRID:AB_10962440 |
| 165 Ho Human CCR6 (clone G034E3) PBMC panel 1 | BioLegend | Cat# 353402, RRID:AB_10918625 |
| 166 Er Human CD25 (clone M-A251) PBMC panel 1 | BioLegend | Cat# 356102, RRID:AB_2561752 |
| 167 Er Human CCR7 (clone G043H7) PBMC panel 1 | BioLegend | Cat# 353256, RRID:AB_2814291 |
| 168 Er Human CD3 (clone UCHT1) PBMC panel 1 | BioLegend | Cat# 300402, RRID:AB_314056 |
| 169 Tm Human CX3CR1 (clone 2A9-1) PBMC panel 1 | BioLegend | Cat# 341602, RRID:AB_1595422 |
| 170 Er Human CD38 (clone HB-7) PBMC panel 1 | BioLegend | Cat# 356602, RRID:AB_2561794 |
| 171 Yb Human CD161 (clone HP-3G10) PBMC panel 1 | BioLegend | Cat# 339902, RRID:AB_1501090 |
| 172 Yb Human CD209 (clone 9E9A8) PBMC panel 1 | BioLegend | Cat# 330102, RRID:AB_1134253 |
| 173 Yb Human CXCR3 (clone REA232) PBMC panel 1 | Miltenyi | Cat# 130-108-022, RRID:AB_2655743 |
| 174 Yb Human HLADR (clone L243) PBMC panel 1 | BioLegend | Cat# 307602, RRID:AB_314680 |
| 176 Yb Human CCR4 (clone 205410) PBMC panel 1 | R&DSystems | Cat# MAB1567, RRID:AB_2074395 |
| 209 Bi Human CD11b (clone ICRF44) PBMC panel 1 | Fluidigm | Cat# 3209003B, RRID:AB_2687654 |
| 89 Y Human CD45 (clone HI30) PBMC panel 2 | Fluidigm | Cat# 3089003B, RRID:AB_2661851 |
| 113 In Human HLA-ABC (clone W6/32) PBMC panel 2 | BioLegend | Cat# 311402, RRID:AB_314871 |
| 115 In Human CD11c (clone Bu15) PBMC panel 2 | BioLegend | Cat# 337202, RRID:AB_1236381 |
| 141 Pr Human CD33 (clone WM53) PBMC panel 2 | BioLegend | Cat# 303410, RRID:AB_2074243 |
| 142 Nd Human CD19 (clone HIB19) PBMC panel 2 | BioLegend | Cat# 302202, RRID:AB_314232 |
| 143 Nd Human CD45RA (clone HI100) PBMC panel 2 | BioLegend | Cat# 304102, RRID:AB_314406 |
| 144 Nd Human CD141 (clone M80) PBMC panel 2 | BioLegend | Cat# 344102, RRID:AB_2201808 |
| 145 Nd Human CD4 (clone RPA-T4) PBMC panel 2 | BioLegend | Cat# 300502, RRID:AB_314070 |
| 146 Nd Human CD8 (clone RPA-T8) PBMC panel 2 | BioLegend | Cat# 301002, RRID:AB_314120 |
| 147 Sm Human CLEC9A (clone 8F9) PBMC panel 2 | BioLegend | Cat# 353802, RRID:AB_10983070 |
| 148 Nd Human CD16 (clone 3G8) PBMC panel 2 | BioLegend | Cat# 302014, RRID:AB_314214 |
| 149 Sm Human FceRIa (clone AER-37) PBMC panel 2 | BioLegend | Cat# 334602, RRID:AB_1227649 |
| 150 Nd Human CD1c (clone L161) PBMC panel 2 | BioLegend | Cat# 331502, RRID:AB_1088995 |
| 151 Eu Human CD123 (clone 6H6) PBMC panel 2 | BioLegend | Cat# 306002, RRID:AB_314576 |
| 152 Sm Human CD66b (clone G10F5) PBMC panel 2 | BioLegend | Cat# 305102, RRID:AB_314494 |
| 153 Eu Human CD83 (clone HB15e) PBMC panel 2 | BioLegend | Cat# 305302, RRID:AB_314510 |
| 154 Sm Human CD86 (clone IT2.2) PBMC panel 2 | BioLegend | Cat# 305410, RRID:AB_314530 |
| 155 Gd Human CD27 (clone O323) PBMC panel 2 | BioLegend | Cat# 302802, RRID:AB_314294 |
| 156 Gd Human PD-L1 (clone 29E.2A3) PBMC panel 2 | BioLegend | Cat# 329711, RRID:AB_2228868 |
| 158 Gd Human CD163 (clone REA812) PBMC panel 2 | Miltenyi | Cat# 130-122-293, RRID:AB_2801874 |
| 159 Tb Human CD103 (clone Ber-Act8) PBMC panel 2 | BioLegend | Cat# 350202, RRID:AB_10639864 |
| 160 Gd Human CD14 (clone M5E2) PBMC panel 2 | BioLegend | Cat# 301810, RRID:AB_314192 |
| 161 Dy Human CD56 (clone B159) PBMC panel 2 | BD Biosciences | Cat# 555513, RRID:AB_395903 |
| 162 Dy Human CD64 (clone 10,1) PBMC panel 2 | BioLegend | Cat# 305016, RRID:AB_2103461 |
| 163 Dy Human CD172a/b (clone SE5A5) PBMC panel 2 | Fluidigm | Cat# 3163017B, RRID:AB_2864730 |
| 164 Dy Human CD40 (clone HB14) PBMC panel 2 | BioLegend | Cat# 334302, RRID:AB_1236384 |
| 166 Er Human CD169 (clone 7-239) PBMC panel 2 | BioLegend | Cat# 346002, RRID:AB_2189031 |
| 167 Er Human CD117 (clone 104D2) PBMC panel 2 | BioLegend | Cat# 313202, RRID:AB_314981 |
| 168 Er Human CD3 (clone UCHT1) PBMC panel 2 | BioLegend | Cat# 300402, RRID:AB_314056 |
| 169 Tm Human CX3CR1 (clone 2A9-1) PBMC panel 2 | BioLegend | Cat# 341602, RRID:AB_1595422 |
| 170 Er Human CD38 (clone HB-7) PBMC panel 2 | BioLegend | Cat# 356602, RRID:AB_2561794 |
| 171 Yb Human CD207 (clone 1000) PBMC panel 2 | BioLegend | Cat# 352202, RRID:AB_10898115 |
| 172 Yb Human CD206 (clone 44607) PBMC panel 2 | BioLegend | Cat# 321112, RRID:AB_571921 |
| 174 Yb Human HLADR (clone L243) PBMC panel 2 | BioLegend | Cat# 307602, RRID:AB_314680 |
| 175 Lu Human Axl (clone 108724) PBMC panel 2 | R&DSystems | Cat# MAB154, RRID:AB_2062558 |
| 176 Yb Human CD209 (clone 9E9A8) PBMC panel 2 | BioLegend | Cat# 330102, RRID:AB_1134253 |
| 209 Bi Human CD11b (clone ICRF44) PBMC panel 2 | Fluidigm | Cat# 3209003B, RRID:AB_2687654 |
| 163Dy Human CXCR3 (clone G025H7) Whole blood custom panel | Fluidigm | Cat# 3163004B, RRID:AB_2810969 |
| 152Sm Human TCRgd (clone 11F2) Whole blood custom panel | Fluidigm | Cat# 3152008B, RRID:AB_2687643 |
| 142Nd Human CD19 (clone HIB19) Whole blood custom panel | Fluidigm | Cat# 3142001B, RRID:AB_2651155 |
| 144Nd Human CD38 (clone HIT2) Whole blood custom panel | Fluidigm | Cat# 3144014B, RRID:AB_2687640 |
| 151Eu Human CD123 (clone 6H6) Whole blood custom panel | Fluidigm | Cat# 3151001B, RRID:AB_2661794 |
| 153Eu Human Va7.2 (clone 3C10) Whole blood custom panel | Fluidigm | Cat# 3153024B, RRID:AB_2891190 |
| 154Sm Human CD3 (clone UCHT1) Whole blood custom panel | Fluidigm | Cat# 3154003B, RRID:AB_2811086 |
| 155Gd Human CD45RA (clone HI100) Whole blood custom panel | Fluidigm | Cat# 3155011B, RRID:AB_2810246 |
| 158Gd Human CD27 (clone L128) Whole blood custom panel | Fluidigm | Cat# 3158010B, RRID:AB_2858231 |
| 159Tb Human CD1c (clone L161) Whole blood custom panel | Biolegend | Cat# 331502, RRID:AB_1088995 |
| 161Dy Human CLEC9A (clone 8F9) Whole blood custom panel | Fluidigm | Cat# 3161018B, RRID:AB_2810252 |
| 164Dy Human CD161 (clone HP-3G10) Whole blood custom panel | Fluidigm | Cat# 3164009B, RRID:AB_2687651 |
| 168Er Human CD8 (clone SK1) Whole blood custom panel | Fluidigm | Cat# 3168002B, RRID:AB_2892771 |
| 170Er Human iNKT (clone 6B11) Whole blood custom panel | Fluidigm | Cat# 3170015B |
| 175Lu Human CCR4 (clone L291H4) Whole blood custom panel | Fluidigm | Cat# 3175035A, RRID:AB_2921320 |
| 174Yb Human CD4 (clone RPA-T4) Whole blood custom panel | Biolegend | Cat# 300502, RRID:AB_314070 |
| 162Dy Human CD21 (clone REA940) Whole blood custom panel | Miltenyi Biotec Inc. | Cat# 130-124-315, RRID:AB_2811646 |
| 165Ho Human NKG2C (clone REA205) Whole blood custom panel | Miltenyi Biotec Inc. | Cat# 130-122-278, RRID:AB_2801859 |
| 148Nd Human CD20 (clone 2H7) Whole blood custom panel | Biolegend | Cat# 302302, RRID:AB_314250 |
| 173Yb Human HLA-DR (clone L243) Whole blood custom panel | Fluidigm | Cat# 3173005B, RRID:AB_2810248 |
| 156Gd Human CCR10 (clone REA326) Whole blood custom panel | Miltenyi Biotec Inc. | Cat# 130-122-317, RRID:AB_2801898 |
| 089Y Human CD45 (clone HI30) Whole blood custom panel | Fluidigm | Cat# 3089003B, RRID:AB_2661851 |
| 116Cd Human CD66b (clone QA17A51) Whole blood custom panel | Biolegend | Cat# 396902, RRID:AB_2814367 |
| 141Pr Human CCR6 (clone G034E3) Whole blood custom panel | Fluidigm | Cat# 3141003A, RRID:AB_2687639 |
| 143Nd Human CD127 (clone A019D5) Whole blood custom panel | Fluidigm | Cat# 3143012B, RRID:AB_2810240 |
| 147Sm Human CD11c (clone Bu15) Whole blood custom panel | Fluidigm | Cat# 3147008B, RRID:AB_2687850 |
| 149Sm Human CD25 (clone 2A3) Whole blood custom panel | Fluidigm | Cat# 3149010B, RRID:AB_2756416 |
| 150Nd Human NKVFS1 (clone NKVFS1) Whole blood custom panel | Bio-Rad | Cat# MCA2243GA, RRID:AB_323743 |
| 167Er Human CCR7 (clone G043H7) Whole blood custom panel | Fluidigm | Cat# 3167009A, RRID:AB_2858236 |
| 169Tm Human NKG2A (clone Z199) Whole blood custom panel | Fluidigm | Cat# 3169013B, RRID:AB_2756426 |
| 171Yb Human CXCR5 (clone RF8B2) Whole blood custom panel | Fluidigm | Cat# 3171014B, RRID:AB_2858239 |
| 166Er Human CD24 (clone ML5) Whole blood custom panel | Fluidigm | Cat# 3166007B, RRID:AB_2661803 |
| 145Nd Human CD31 (clone WM59) Whole blood custom panel | Fluidigm | Cat# 3145004B, RRID:AB_2737262 |
| 160Gd Human CD14 (clone M5E2) Whole blood custom panel | Fluidigm | Cat# 3160001B, RRID:AB_2687634 |
| 176Yb Human CD56 (clone NCAM16.2) Whole blood custom panel | Fluidigm | Cat# 3176008B, RRID:AB_2661813 |
| 172Yb Human CD57 (clone HNK-1) Whole blood custom panel | Biolegend | Cat# 359602, RRID:AB_2562403 |
| 150Nd Human KIR3DL1L2 (clone REA970) Whole blood custom panel | Miltenyi Biotec Inc. | Cat# 130-126-489, RRID:AB_2889458 |
| 146Nd Human IgD (clone IA6-2) Whole blood custom panel | Fluidigm | Cat# 3146005B, RRID:AB_2811082 |
| 209Bi Human CD16 (clone 3G8) Whole blood custom panel | Fluidigm | Cat# 3209002B, RRID:AB_2756431 |
| eFluor 450 Murine CD3ε (clone 1452C11) | Thermo Fisher Scientific | Cat# 48-0031-82, RRID:A14714 |
| BUV395 Murine CD4 (clone GK1.5) | BD Bioscience | Cat# 563790, RRID:AB_2738426 |
| PE Murine CD5 (clone 53-7.3) | Thermo Fisher Scientific | Cat# 12-0051-82, RRID:AB_465523 |
| eFluor 615 Murine CD8α (clone 53-6.7) | Thermo Fisher Scientific | Cat# 47-0081-82, RRID:AB_1272185 |
| Brilliant Violet 650 Murine CD11b (clone M1/70) | Thermo Fisher Scientific | Cat# 416-0112-82 |
| PE Murine CD11b (clone M1/70) | Thermo Fisher Scientific | Cat# 12-0112-82, RRID:AB_2734869 |
| eFluor 450 Murine CD11b (clone M1/70) | Thermo Fisher Scientific | Cat# 48-0112-82, RRID:AB_1582236 |
| FITC Murine CD11c (clone N418) | Thermo Fisher Scientific | Cat# 11-0114-82, RRID:AB_464940 |
| PE Murine CD11c (clone N418) | Thermo Fisher Scientific | Cat# 12-0114-82, RRID:AB_465552 |
| eFluor 450 Murine CD11c (clone N418) | Thermo Fisher Scientific | Cat# 48-0114-82, RRID:AB_1548654 |
| PE Murine CD19 (clone eBio1D3) | Thermo Fisher Scientific | Cat# 12-0193-82, RRID:AB_657659 |
| Alexa Fluor 660 Murine CD19 (clone eBio1D3) | Thermo Fisher Scientific | Cat# 606-0193-82, RRID:AB_2896251 |
| eFluor 450 Murine CD19 (clone eBio1D3) | Thermo Fisher Scientific | Cat# 48-0193-82, RRID:AB_2734905 |
| Alexa Fluor 488 Murine CD25 (clone PC61.5) | Thermo Fisher Scientific | Cat# 53-0251-82, RRID:AB_763472 |
| Brilliant Violet 711 Murine CD25 (clone PC61.5) | Thermo Fisher Scientific | Cat# 407-0251-82 |
| PerCP-Cyanine5.5 Murine CD26 (clone H194-112) | Thermo Fisher Scientific | Cat# 45-0261-82, RRID:AB_1548738 |
| Brilliant Violet 785 Murine CD44 (clone IM7) | BioLegend | Cat# 103059, RRID:AB_2571953 |
| Brilliant Violet 785 Murine CD45 (clone 30-F11) | BioLegend | Cat# 103149, RRID:AB_2564590 |
| PE/Cyanine7 Murine CD45 (clone 30-F11) | BioLegend | Cat# 103114, RRID:AB_312979 |
| Brilliant Violet 711 Murine CD45 (clone 30-F11) | BioLegend | Cat# 103147, RRID:AB_2564383 |
| Brilliant Violet 650 Murine CD45 (clone 30-F11) | BioLegend | Cat# 103151, RRID:AB_2565884 |
| PE Murine CD45R (B220) (clone RA3-6B2) | Thermo Fisher Scientific | Cat# 12-0452-82, RRID:AB_465671 |
| Brilliant Ultra Violet 395 Murine CD45R (B220) (clone RA3-6B2) | Thermo Fisher Scientific | Cat# 363-0452-82 |
| eFluor 450 Murine CD45R (B220) (clone RA3-6B2) | Thermo Fisher Scientific | Cat# 48-0452-82, RRID:AB_1548761 |
| eFluor 450 Murine CD49b (clone DX5) | Thermo Fisher Scientific | Cat# 48-5971-82, RRID:AB_10671541 |
| PE/Dazzle 594 Murine CD62L (clone MEL-14) | BioLegend | Cat# 104448, RRID:AB_2566163 |
| BV786 Murine CD64 (clone X54-5/7.1) | BD Biosciences | Cat# 741024; RRID:AB_2740644 |
| APC Murine CD86 (clone GL-1) | BioLegend | Cat# 105012, RRID:AB_493342 |
| Brilliant Violet 510 Murine CD90.2 (Thy-1.2) (clone 53-2.1) | BioLegend | Cat# 140319, RRID:AB_2561395 |
| APC Murine CD117 (c-Kit) (clone 2B8) | Thermo Fisher Scientific | Cat# 17-1171-82, RRID:AB_469430 |
| Brilliant Violet 711 Murine CD127 (clone A7R34) | BioLegend | Cat# 135035, RRID:AB_2564577 |
| Brilliant Violet 650 Murine CD127 (clone A7R34) | BioLegend | Cat# 135043, RRID:AB_2629681 |
| PE Murine CD135 (Flt3) (clone A2F10) | BioLegend | Cat# 135306, RRID:AB_1877217 |
| APC Murine CD172a (SIRPa) (clone P84) | Thermo Fisher Scientific | Cat# 17-1721-82, RRID:AB_10733158 |
| PE/Cyanine7 Murine CD317 (BST2, PDCA-1) (clone 927) | BioLegend | Cat# 348416, RRID:AB_2716221 |
| PE/Dazzle 594 Murine F4/80 (clone BM8) | BioLegend | Cat# 123146, RRID:AB_2564133 |
| PE Murine FcεR1α (clone 36951) | Thermo Fisher Scientific | Cat# 12-5898-82, RRID:AB_466028 |
| eFluor 450 Murine FcεR1α (clone 36951) | Thermo Fisher Scientific | Cat# 48-5898-82, RRID:AB_2574086 |
| FITC Murine FOXP3 (clone FJK-16s) | Thermo Fisher Scientific | Cat# 11-5773-82, RRID:AB_465243 |
| eFluor 660 Murine GATA3 (clone TWAJ) | Thermo Fisher Scientific | Cat# 50-9966-42, RRID:AB_10596663 |
| PE-Cyanine7 Murine KLRG1 (clone 2F1) | Thermo Fisher Scientific | Cat# 25-5893-82, RRID:AB_1518768 |
| Biotin Murine Integrin α4β7 (clone DATK32) | BioLegend | Cat# 120612, RRID:AB_11203892 |
| Alexa Fluor 488 Murine Ly-6A/E (Sca-1) (clone E13-161.7) | BioLegend | Cat# 122516, RRID:AB_756201 |
| Murine Ly6C (clone AL-21) | BD Biosciences | Cat# 553104, RRID:AB_394628 |
| PE Murine Ly6G/Ly6C (Gr-1) (clone RB6-8C5) | Thermo Fisher Scientific | Cat# 12-5931-82, RRID:AB_466045 |
| eFluor 450 Murine Ly6G/Ly6C (Gr-1) (clone RB6-8C5) | Thermo Fisher Scientific | Cat# 48-5931-82, RRID:AB_1548788 |
| PerCP-Cy5.5 Murine Ly6G (clone 1A8) | BD Biosciences | Cat# 560602; RRID:AB_1727563 |
| BUV805 Murine MHC Class II (I-A/I-E) (clone M5/114) | BD Biosciences | Cat# 748844; RRID:AB_2873247 |
| PE-Cyanine7 Murine NK1.1 (clone PK136) | Thermo Fisher Scientific | Cat# 25-5941-82, RRID:AB_469665 |
| eFluor 450 Murine NK1.1 (clone PK136) | Thermo Fisher Scientific | Cat# 48-5941-82, RRID:AB_2043877 |
| PerCP-Cyanine5.5 Murine NK1.1 (clone PK136) | Thermo Fisher Scientific | Cat# 45-5941-82, RRID:AB_914361 |
| PE Murine Siglec-F (clone E50-2440) | BD Biosciences | Cat# 562068; RRID:AB_394341 |
| PerCP-eFluor 710 Murine IL-33R (ST2) (clone RMST2-2) | Thermo Fisher Scientific | Cat# 46-9335-82, RRID:AB_2573883 |
| eFluor 450 Murine TCRβ (clone H57-597) | Thermo Fisher Scientific | Cat# 48-5961-82, RRID:AB_11039532 |
| PE Murine TCRβ (clone H57-597) | Thermo Fisher Scientific | Cat# 12-5961-82, RRID:AB_466066 |
| eFluor 450 Murine TCR γ/δ (clone eBioGL3) | Thermo Fisher Scientific | Cat# 48-5711-82, RRID:AB_2574071 |
| PE Murine TCR γ/δ (clone eBioGL3) | Thermo Fisher Scientific | Cat# 12-5711-82, RRID:AB_465934 |
| eFluor 450 Murine TER-119 (clone TER-119) | Thermo Fisher Scientific | Cat# 48-5921-82, RRID:AB_1518808 |
| PE-Cyanine7 Murine TER-119 (clone TER-119) | Thermo Fisher Scientific | Cat# 25-5921-82, RRID:AB_469661 |
| Murine XCR-1 (clone ZET) | Biolegend | Cat# 148204, RRID:AB_2563843 |
| Bacterial and virus strains | ||
| Salmonella enterica subsp. enterica serovar Typhimurium GFP | ATCC | 14028GFP |
| Mycobacterium abscessus sensu stricto, strain CIP104536T, smooth with pTEC27 | Bernut et al., 2014212 | N/A |
| expanded T7 Virscan phage library | S. Elledge (Brigham and Women’s Hospital and Harvard University Medical School, Boston, MA, USA) | VirScan Phage Library, version 3 |
| Vesicular stomatitis virus Indiana (VSV) | Bastard et al., 2021143 | N/A |
| Human immunodeficiency viruses-1 (HIV-1) reporter virus (NL4-3 ΔenvΔnef encoding GFP in nef) | Bhargava et al., 2021171 | N/A |
| Human immunodeficiency viruses-2 (HIV2) reporter virus (ROD9 ΔenvΔnef encoding GFP in nef) | Manel et al., 2010173 | N/A |
| Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) NYC isolate | Zhang et al., 2020138 | GenBank OM345241 |
| Yellow-fever vaccine virus-venus (YF17D-venus) reporter | Yi et al., 2011179 | N/A |
| Influenza A virus A/California/4/2009 (IAV) | Manicassamy et al., 2010213 | N/A |
| Herpes simplex virus 1 (HSV-1) | ATCC | VR-1493 |
| Hepatitis A virus (HAV) reporter virus (HM175/18f-NLuc) | Yamane et al., 201952 | N/A |
| Encephalomyocarditis virus (EMCV) | Gao et al., 2021187 | N/A |
| Biological samples | ||
| Peripheral blood mononuclear cells from indicated individuals | This manuscript | N/A |
| Plasma from indicated individuals | This manuscript | N/A |
| Biopsies from indicated individuals | This manuscript | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Protamine sulfate | Merck | Cat# P3369-10G |
| Recombinant interferon gamma-1b (Imukin) | Clinigen Healthcare France | Cat# 3400955776789 |
| Recombinant interferon alpha-2b (Introna) | MSD France | Cat# 3400934956287 |
| Aldesleukin (Proleukin) | Novartis | Cat# 3400956215867 |
| Recombinant Human IFN-beta 1a (Mammalian) Protein | Bio-Techne | Cat# 11410-2 |
| Collagenase IV | Gibco | Cat# 17104-019 |
| DpnI | New England Biolab | Cat# R0176L |
| M-CSF | Peprotech | Cat# 300-25 |
| IL-3 | Peprotech | Cat# 200-03 |
| M-CSF | # 204-IL-010 | Cat# 216-MC-010 |
| IL-4 | R&D Systems | Cat# 204-IL-010 |
| ProLong Gold with DAPI | Thermo Fisher Scientific | Cat #P36931 |
| Aqua Dead Cell Stain kit | Thermo Fisher Scientific | Cat# L34957 |
| Critical commercial assays | ||
| SureSelect Human All Exon V6 | Agilent | Cat# 5190-8864 |
| Human SNP Array 6.0 | Agilent | Cat# 901153 |
| GoTaq DNA Polymerase | Promega | Cat# M3005 |
| RNeasy Plus Mini Kit | Qiagen | Cat# 74136 |
| Quick-RNA MicroPrep Kit | Zymo | Cat# R1051 |
| Universal PCR Master Mix (2X), no AmpErase UNG | Thermo Fisher Scientific | Cat# 4352042 |
| RiboZero TruSeq Stranded Total RNA Library Prep Kit | Illumina | Cat# 20020596 |
| ELISA IL-12p40 | R&D Systems | Cat# DP400 |
| ELISA IL-12p70 HS | R&D Systems | Cat# HS120 |
| NE-PER nuclear and cytoplasmic extraction reagents | Thermo Fisher Scientific | Cat #78835 |
| RNeasy Plus Mini Kit | Qiagen | Cat# 74136 |
| Quick-RNA MicroPrep Kit | Zymo | Cat# R1051 |
| SuperScript II Reverse Transcriptase | Thermo Fisher Scientific | Cat# 18064014 |
| High-Capacity RNA-to-cDNA Kit | Applied Biosystems | Cat# 4387406 |
| TaqMan Fast Universal PCR Master Mix (2X), no AmpErase UNG | Thermo Fisher Scientific | Cat# 4352042, |
| Deposited data | ||
| RNA sequencing on primary fibroblasts | This manuscript | gene expression omnibus (GEO: GSE218033) |
| RNA sequencing on iPSC-derived macrophages | This manuscript | gene expression omnibus (GEO: GSE218033) |
| RNA sequencing on monocyte-derived macrophages | This manuscript | gene expression omnibus (GEO: GSE218033) |
| Mass-spectrometry on primary fibroblasts | This manuscript | ProteomeXchange (PXD037759) |
| scRNAseq on cryopreserved PBMCs | This manuscript | gene expression omnibus (GEO: GSE216489) |
| Experimental models: Cell lines | ||
| HEK293T cells | ATCC | Cat# CRL-11268, RRID:CVCL_1926 |
| THP1 WT | Song et al., 202178 | ATCC Cat# TIB-202, RRID:CVCL_0006 |
| THP1 IRF1KO | Song et al., 202178 | N/A |
| THP1 IFN-γR1KO clone 30 | This manuscript | N/A |
| THP1 STAT1KO clone 13 | This manuscript | N/A |
| iPSC healthy control clone 11 | Ackermann et al., 2014191 | hCD34-iPSC11 |
| iPSC healthy control clone 16 | Lachmann et al., 2014189 | hCD34-iPSC16 |
| iPSC P1 clone 10 | This manuscript | N/A |
| iPSC P2 clone 3 | This manuscript | N/A |
| iPSC STAT1 | Haake et al., 202050 | iSTAT1_compl |
| Experimental models: Organisms/strains | ||
| C57BL/6 (B6) WT | This manuscript | In-house colony established from Jax # 000664 |
| C57BL/6 (B6) IRF1−/− | This manuscript | Jax # 002762 backcrossed to a B6 background |
| Oligonucleotides | ||
| IRF1 genomic exon 3F | ThermoFischer Scientific | TGGTCTGTTTAAGCCAGCCTC |
| IRF1 genomic exon 3R | ThermoFischer Scientific | CAGAAACACAAGTCTGCCACC |
| IRF1 genomic exon 5F | ThermoFischer Scientific | TTCCACCTCTCACCAAGAACC |
| IRF1 genomic exon 5R | ThermoFischer Scientific | CAGAGAAGGTATCAGGGCTGG |
| IRD700-conjugated ISRE probe-F | Metabion | GATCGGGAAAGGGAAACCGAAACTGAA |
| IRD700-conjugated ISRE probe-R | Metabion | TCAGTTTCGGTTTCCCTTTCCCGATC |
| oligo(dT)12–18 | Thermo Fisher Scientific | Cat# 18418012 |
| IRF1 exons 3–4 qPCR probe | ThermoFischer Scientific | Cat# Hs00971960_m1 |
| IRF1 exons 8–9 | ThermoFischer Scientific | Cat# Hs00971965_m1 |
| GBP4 | ThermoFischer Scientific | Cat# Hs00364728_m1 |
| APOL3 | ThermoFischer Scientific | Cat# Hs00758274_m1 |
| GUSB | ThermoFischer Scientific | Cat# 1702016 |
| IRF1-p.M85A-DDK-F | Eurofins | CAACTTTCGCTGTGCCGCGAACTCCCT GCCAGAT |
| IRF1-p.M85A-DDK-R | Eurofins | ATCTGGCAGGGAGTTCGCGGCACAGC GAAAGTTG |
| IRF1-p.M111A-DDK-F | Eurofins | GCGAGTGTACCGGGCGCTTCCACCTCTC |
| IRF1-p.M111A-DDK-R | Eurofins | GAGAGGTGGAAGCGCCCGGTACACTCGC |
| IRF1-p.Q35*-F | Eurofins | GAGGAGATGATCTTCTAGATCCCATGG AAGC |
| IRF1-p.Q35*-R | Eurofins | GCTTCCATGGGATCTAGAAGATCATCT CCTC |
| IRF1-p.R129*-F | ThermoFischer Scientific | GTCGAAGTCCAGCTGAGATGCTAAG |
| IRF1-p.R129*-R | ThermoFischer Scientific | CTTAGCATCTCAGCTGGACTTCGAC |
| IRF1-p.R129-DDK-F | ThermoFischer Scientific | GCTGGACTTCGACTTTCTTTCTTTTCTCTG |
| IRF1-p.R129-DDK-R | ThermoFischer Scientific | ACGCGTACGCGGCCGCTCGA |
| IRF1-p.W11R-DDK-F | ThermoFischer Scientific | GCATGAGACCCCGGCTAGAGATG |
| IRF1-p.W11R-DDK-R | ThermoFischer Scientific | CATCTCTAGCCGGGGTCTCATGC |
| IRF1-Δ7-8-F | ThermoFischer Scientific | CTGGAGTCAGGGCCTGCTCC |
| IRF1-Δ7-8-R | ThermoFischer Scientific | CTCTTGGAGCAGTCGGAGTGGC |
| IRF1-Δ7-8-DDK-F | ThermoFischer Scientific | TTGAGTAGGTACCCCTTCCCATCCACGTTTG |
| IRF1-Δ7-8-DDK-R | ThermoFischer Scientific | ACGCGTACGCGGCCGCTCGA |
| IRF1-p.A67P-DDK-F | ThermoFischer Scientific | GCCGATACAAACCAGGGGAAAAG |
| IRF1-p.A67P-DDK-R | ThermoFischer Scientific | CTTTTCCCCTGGTTTGTATCGGC |
| IRF1-p.M1_A84del-F | Eurofins | ATGAACTCCCTGCCAGATAT |
| IRF1-p.M1_A84del-R | Eurofins | GGCGATCGCGGCGGCAGATC |
| IRF1 retrovirus F | ThermoFischer Scientific | GATCCATTTAAATTCGAATTCATGCCCA TCACTCGGATGCGC |
| IRF1 retrovirus R | ThermoFischer Scientific | ATCGATACCGTCGACCTCGAGTTAAAC CTTATCGTCGTCATC |
| sgRNA IFNGR1 exon 3F | Eurofins | CACCGACACATTCTACTCACCATCT |
| sgRNA IFNGR1 exon 3R | Eurofins | AAACAGATGGTGAGTAGAATGTGTA |
| sgRNA STAT1 exon 3F | Eurofins | CACCGTATTTGCAGCTCGTTTGTGG |
| sgRNA STAT1 exon 3R | Eurofins | AAACCCACAAACGAGCTGCAAATAA |
| Recombinant DNA | ||
| pCMV6-EV | Origene | Cat# RCPS100001 |
| pCMV6-IRF1-WT-DDK | Origene | Cat# RC203500 |
| pCMV6-IRF1-p.M85A-DDK | This manuscript | N/A |
| pCMV6-IRF1-p.M111A-DDK | This manuscript | N/A |
| pCMV6-IRF1-M85A/M111A-DDK | This manuscript | N/A |
| pCMV6-IRF1-p.Q35* | This manuscript | N/A |
| pCMV6-IRF1-p.Q35*/M85A-DDK | This manuscript | N/A |
| pCMV6-IRF1-p.Q35*/M111A-DDK | This manuscript | N/A |
| pCMV6-IRF1-p.Q35*/M85A/M111A-DDK | This manuscript | N/A |
| pCMV6-IRF1-p.R129* | This manuscript | N/A |
| pCMV6-IRF1-p.R129-DDK | This manuscript | N/A |
| pCMV6-IRF1-p.W11R-DDK | This manuscript | N/A |
| pCMV6-IRF1-Δ7-8 | This manuscript | N/A |
| pCMV6-IRF1-Δ7-8-DDK | This manuscript | N/A |
| pCMV6-IRF1-p.A67P-DDK | This manuscript | N/A |
| pCMV6-IRF1-p.M1_A84del | This manuscript | N/A |
| pGL4.10[luc2] backbone with three GGAAAGGGAAACCGAAACTGAA repeats | Guerin et al., 2018158 | Cat# E6651 |
| pGL4.10[luc2] backbon with five GGGAAAGTGAAACTA repeats | Hernandez et al., 2018183 | N/A |
| pRL-SV40 | Promega | Cat# E2231 |
| lentiCRISPR v2 | Addgene | Cat# 52961 |
| psPAX2 | Addgene | Cat# 12260 |
| pCMV-VSV-G | Addgene | Cat# 8454 |
| pHXB2-Env | NIH-AIDS Reagent Program | Cat# 1069 |
| Software and algorithms | ||
| phip-stat | Larman et al., 2011214 | https://github.com/lasersonlab/phip-stat |
| Bowtie for alignment of PhIP-Seq raw reads | Larman et al., 2011214 | http://bowtie-bio.sourceforge.net/index.shtml |
| R | The R Project for Statistical Computing | https://www.r-project.org |
| DESeq2 | Love et al., 2014201 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Uniform Manifold Approximation and Projection (UMAP) | Becht et al., 2018208 | v.0.3.5 |
| BVAtools | https://bitbucket.org/mugqic/bvatools | https://bitbucket.org/mugqic/bvatools |
| Cell Ranger | 10X Genomics | v3.0.1 for scRNA and v6.0.1 for CITE-seq |
| DoubletFinder package | McGinnis et al., 2019206 | v2.0.3 |
| Seurat R package | Stuart et al., 2019207 | v4.0.2 |
| MAST | Finak et al., 2015209 | https://github.com/RGLab/MAST |
| gProfiler R package | Raudvere et al., 2019210 | https://biit.cs.ut.ee/gprofiler/gost |
| FastQC | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| STAR | Dobin et al., 2013197 | v2.7.3a |
| RSeQC | Wang et al., 2012198 | v0.11.2 |
| HTSeq-count | Anders et al., 2015199 | v3.26.8 |
| edgeR package | McCarthy et al., 2012200 | N/A |
| HOMER | Heinz et al., 2010202 | v4.11 |
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.
EXPERIMENT MODEL AND SUBJECT DETAILS
Patient 1 (P1, kindred A) is an eight-year-old girl born to consanguineous parents originating from and living in Argentina (Figure 1A). She was vaccinated with BCG at birth. At the age of five months, she was hospitalized for lymphadenitis and hepatosplenomegaly. BCG was detected in a lymph node biopsy and bone marrow aspirate by culture, attesting to disseminated BCG disease (BCG-osis). P1 was treated with a combination of amikacin, rifampicin, isoniazid, ethambutol, and levofloxacin for two years. At the age of three years, she had fever and abdominal pain. Computed tomography (CT) revealed hepatomegaly, and mediastinal and abdominal lymphadenopathies (Figure 1B). Mediastinal lymph node biopsy revealed granulomatous lymphadenitis (Figure 1C), and culture yielded M. avium. During this episode, PCR also detected Histoplasma sp. in the blood. P1 was treated with rifampicin, isoniazid, ethambutol, levofloxacin for two years for mycobacterial disease, and with amphotericin B followed by voriconazole for prophylaxis. At the age of four years, IFN-γ treatment was initiated following a suspected reactivation of mycobacterial disease. At the age of five years, P1 was hospitalized for pneumonia. M. avium was again identified in a sputum sample. Clarithromycin was added to the patient’s IFN-γ and antimycobacterial treatment. At the age of seven years, P1 had rhinitis causing upper respiratory obstruction. M. intracellulare subsp. chimaera was documented on a nasal swab. During this episode, P1 also had hepatosplenomegaly and pancytopenia. Bone marrow biopsy showed epithelioid granulomas with giant multinucleated cells. P1 was treated with liposomal amphotericin, levofloxacin, clarithromycin, and ethambutol. She is now seven years old and clinically well on immunoglobulins, recombinant IFN-γ, levofloxacin, clarithromycin, cotrimoxazole, and itraconazole.
Patient 2 (P2, kindred B) is a seven-year-old girl born to consanguineous parents originating from and living in Turkey (Figure 1A). She was vaccinated with BCG at two months of age. She presented with left axillary lymphadenitis at five months old. BCG was grown from the abscess, attesting to local-regional BCG-infection (BCG-itis). P2 was successfully treated with isoniazid, rifampicin, and ethambutol for six months. At the age of five years, she presented fever, digital clubbing, and rash. A chest computed tomography (CT)-scan revealed multiple mediastinal and hilar lymphadenopathies, and a lung mass (Figure 1D). Biopsy specimens from the lymphadenopathies and the lung mass contained multinucleated giant cells and spindle-shaped histiocytes (Figure 1E). M. avium was cultured from these lesions. P2 has since been treated with azithromycin, ethambutol, and rifabutin.
Mice used in this study (C57BL/6 (B6) mice and Irf1−/− mice were all male.
METHOD DETAILS
Patients
P1 is resident in Argentina and P2 is resident in Turkey. Informed consent was obtained in Argentina and Turkey, respectively, in accordance with local regulations and with institutional review board (IRB) approval. Experiments were conducted in Australia, Canada, France, Qatar, and the United States of America, in accordance with local regulations and with the approval of the IRB of the Rockefeller University and INSERM, for the United States of America and France, respectively. A detailed clinical case report is provided below. Healthy controls were recruited in Argentina, France, Turkey, and the United States of America.
Case reports for P1 and P2
The patient from kindred A (P1, II.1) is a girl born in 2014 and living in Tucuman, in the North East of Argentina (Figure 1A). She was vaccinated with BCG at birth. At the age of one month, she presented impetigo and a local abscess at the site of vaccination with BCG. At the age of five months, she was hospitalized with fever, hepatosplenomegaly, and intra-abdominal and mediastinal adenomegalies. M. bovis-BCG was cultured from lymph node and bone marrow aspiration fluids. P1 also had digestive symptoms, with diarrhea and intestinal subocclusion, and a cutaneous eruption. BCG-osis was successfully treated with a combination of amikacin, rifampicin, isoniazid, ethambutol, and levofloxacin. At the age of eight months, during hospitalization for intestinal malrotation surgery, P1 was found to have vesicular lesions on the scalp, trunk, perianal region, vulva, and lower limbs, but no signs of meningoencephalitis or pneumonia. The Tzanck test was positive, suggesting infection with varicella-zoster virus (VZV). Infection with this virus was confirmed by positive PhIP-Seq (Figure 7A) and serological testing for VZV (Table S4). Given that VZV was hospital-acquired in a young patient with a suspected IEI, and despite the absence of clinical signs of severity (including an absence of both pneumonia and encephalitis), P1 was pre-emptively treated with intravenous acyclovir, leading to resolution of the varicella virus infection. At the age of 11 months, P1 had a lower respiratory tract infection caused by parainfluenza virus 3 (PIV3), documented by PCR. She also experienced another lower respiratory tract infection caused by Chlamydia pneumoniae, and various other respiratory tract infections presumed to be viral. She did not require intubation for any of these respiratory tract infections. At the age of three years, she presented simultaneous disseminated infections due to M. avium and Histoplasma capsulatum. These infections were treated with amikacin, rifampicin, isoniazid, ethambutol, levofloxacin, and liposomal amphotericin B. During the episode of BCG-osis, P1 also displayed leukopenia, anemia, and thrombocytopenia. At the age of four years she presented with vesicular lesions of the right upper limb. However, PCR tests for VZV and HSV were negative. At the age of five years, P1 presented pneumonia due to M. avium intracellulare. Clarithromycin was added to the antimycobacterial treatment regimen. At the age of six years the patient suffered rhinosinusitis leading to nasal obstruction. Acinetobacter baumanii and M. intracellulare subsp. chimaera were documented on culture of a sinus sample. During this episode, the patient displayed hepatosplenomegaly and pancytopenia associated, on bone marrow biopsy, with epithelioid granuloma giant multinucleated cells without hemophagocytic lymphohistiocytosis (HLH). The patient was treated with liposomal amphotericin, levofloxacin, clarithromycin, and ethambutol. She suffered failure to thrive, from infancy (−2 to −3 SD for weight and height). Between the ages of two and four years, P1 suffered recurrent abdominal pain, with an enlarged liver and abdominal distention. Digestive tract endoscopy results were normal. Abdominal and pelvic CT-scan results were normal, with the exception of organomegaly. P1 had ventricular septal defects and café au lait spots on the skin, from birth. In addition to BCG, she was vaccinated against diphtheria, tetanus, hepatitis A, hepatitis B, Haemophilus influenzae type b, seasonal flu, pneumococcus (Prevenar 13), poliomyelitis (Salk), and rotavirus (Rotarix), and had received one dose of measles-mumps-rubella vaccine. Serological tests for HIV were negative. Chromosome 22q11.2 deletion syndrome was ruled out by fluorescence in situ hybridization. Blood karyotype was normal. The primary fibroblasts of P1 displayed normal sensitivity to mitomycin C and gave normal results in FANCD2-monoubiquitination assays155,156, ruling out Fanconi anemia. Plasma alpha-feto protein concentration was in the normal range. The oxidative burst in PNM cells (dihydrorhodamine test), and ADA and PNP activities in plasma were normal. A brain CT-scan at the age of four years was normal. P1 is currently being treated with subcutaneous recombinant IFN-γ (Imukin®), polyvalent subcutaneous immunoglobulin, levofloxacin, ethambutol, clarithromycin, trimethoprim-sulfamethoxazole, iron, folic acid, vitamin D and calcium. She is well on this treatment. The mother of P1 had no history of unusually severe infectious diseases. She did not display adverse reaction to BCG vaccination.
The patient from kindred B (P2, II.1) is a girl born in 2015 and living in Turkey (Figure 1A). She was vaccinated with BCG at birth, and presented left axillary lymphadenitis (BCG-itis) without organomegaly at the age of five months. She was successfully treated for six months with isoniazid, rifampicin, and ethambutol. At the age of one year, she presented respiratory infection due to influenza B virus that did not require oxygen support. At the age of five years, P2 was referred to hospital for intermittent fever and a rash of three months’ duration. She was treated with oral antibiotics for upper respiratory tract infection, and with colchicine for suspected familial Mediterranean fever. She had been on intramuscular antibiotics for two weeks when she was diagnosed with pneumonia, without identification of the causal microbe. On admission, she had tachypnea, but was in good general condition with no fever. Lymphadenopathies were observed in both axillary regions on clinical examination. A chest CT-scan revealed multiple lymphadenopathies in both axillary regions, in the mediastinal and hilar regions, and a mass-like lesion in the left lower lobe of the lungs. Abdominal CT-scan was normal, with no organomegaly. Antibiotic treatment with ceftriaxone and clindamycin was continued for pneumonia. A peripheral smear and bone marrow aspiration were performed; the results excluded leukemia and HLH. A left shift and leukocytosis with neutrophilia were found. Cultures of blood and nasopharyngeal fluid were negative. Three fasting gastric fluid aspiration specimens were sent for staining for acid-fast bacilli (AFB) and mycobacterial culture, but the results were negative on all three occasions. A tuberculin skin test (TST) gave a 17 mm induration, and the Quantiferon test was negative. Due to a lack of improvement, antituberculosis treatment was initiated with moxifloxacin, rifampicin, isoniazid, pyrazinamide and ethambutol. Samples of the lung mass and mediastinal lymphadenopathies were sent for histological analysis and mycobacterial culture. Cultures revealed M. avium intracellulare. The histopathology report showed a few giant cells and spindle-shaped histiocytes. P2 was treated with azithromycin, ethambutol, and rifabutin. The patient had papular hyperemic rashes on her hands and feet during clinical follow-up and was referred to a dermatologist. The cutaneous biopsy results obtained from the rashes were consistent with acute generalized exanthematous pustulosis. At the age of five years, the patient was infected with SARS-CoV-2, causing mild cold-like symptoms with no need for hospitalization. This patient has never been vaccinated against COVID-19. P2 is currently doing well. The father, mother, and brother of P2 had no history of unusually severe infectious diseases, including adverse effects of live vaccines.
Both IRF1 variants are predicted to be deleterious
IRF1 encodes interferon regulatory factor 1 protein (IRF1), a transcription factor induced by IFN-γ, the deficiency of which in mice underlies susceptibility to various pathogens, including mycobacteria43,49,51,53,55,57,62,83,84,96–98,109,110,147 (Table S2). IRF1 is a 325-amino acid transcription factor composed of an amino-terminal DNA-binding domain (DBD) that can bind to DNA interferon-stimulated response elements (ISRE) or positive regulatory domain I (PRDI) motifs64, an intermediate segment containing a putative nuclear localization sequence (NLS), and a carboxyterminal IRF association domain 2 (IAD2), which is crucial for transcriptional activity65,66,68,70 (Figure 1G). Both the c.385C>T and c.103C>T variants are predicted to be loss-of-function (pLOF). Indeed, both result in the creation of a premature stop codon, p.R129* and p.Q35*, respectively (Figure 1G). The CADD score of the two variants is 38, well above the 99% MSC37,166 of IRF1 at 8.2 (Figure 1I). An analysis of public databases, including ExAC, gnomAD v2.1.1 or v3.1.1168, BRAVO/TOPmed freeze 8170, UK Biobank172, ATAV174, the Greater Middle East variome176, and the Turkish variome178 containing WES or whole-genome sequencing (WGS) data from more than 250,000 individuals showed that the p.Q35* variant is private, whereas p.R129* is found in the heterozygous state in two individuals. All the non-synonymous coding or structural variants of IRF1 listed in public WES and WGS databases are present in the heterozygous state, with the exception of one homozygous missense variant (c.199G>C, p.A67P, rs137993322) in the TOPmed170 database with a gnomAD MAF of 1.2e-4 and a CADD score below that of p.R129* and p.Q35* (Figure 1I). We identified no other homozygous or compound heterozygous non-synonymous IRF1 coding variants in our in-house database of 20,000 WES or WGS from patients with severe infectious diseases. The cumulative frequency of heterozygous pLOF variants in gnomAD is also low (4.3×10−5) (Figure 1I). Consistently, the consensus negative selection (CoNeS) score of IRF1 is low, suggesting that this gene is under negative selection, consistent with either recessive or dominant inheritance180 (Figure S1D). These findings further suggest that homozygosity for the pLOF private IRF1 variants identified here was causal for MSMD in P1 and P2.
Impaired development of lymphoid and myeloid cells in Irf1−/− mice
IRF1-knockout mice (Irf1−/−) were first described in 1993112. Consistent with the results obtained for IRF1-deficient patients, Irf1−/− mice with various genetic backgrounds (C57BL/6 and 129/Sv) display impaired development of NK cells, and TH1 cells, together with a predominance of naïve cells among CD8+ T cells (Table S5)43,109,110,182,184. However, other leukocyte subsets do not seem to have been analyzed in these mice. We studied the immunophenotypes of Irf1−/− mice with a C57BL/6 background by isolating leukocytes from the bone marrow, spleen, and lung. We replicated the reported decrease in the absolute numbers of NK and CD8+ T cells in C57BL/6 Irf1−/− mice (Figures S3E–F). The development of myeloid DCs has been little studied, and with only rudimentary markers, in mice, but the available results suggest an impaired maturation of myeloid DCs186, whereas ILC2 maturation has not been studied. We found that cDC1 counts were low in the spleen and lungs of Irf1−/− mice, whereas cDC2 counts in the lungs of Irf1−/− mice were unaffected, and those in the spleen were only slightly lower than those of WT mice (Figures S3E–F). Spleen pDC counts were normal in Irf1−/− mice (Figure S3F). The number of mature ILC2 in the lungs was low, whereas the number of immature ILC2 progenitors in the bone marrow was normal in Irf1−/− mice (Figure S3G). Overall, IRF1 deficiency appears to impede the development of the same lymphoid and myeloid cell subsets in humans and mice, resulting in deficiencies of cDC1, ILC2, NK cells, and CD8+ T cells.
Lack of severe viral infections in IRF1-deficient patients
In addition to its known role in IFN-γ-related immunity, the antiviral role of IRF1 has also been extensively studied. The ablation52,61,79,80 and overexpression81,82 of the IRF1 gene have been shown to be associated with susceptibility and resistance to viral infections, respectively, in vivo in mice83–87 (Table S2) and in vitro in human cell lines61,79,81. Indeed, human IRF1 is induced by IFN-α/β61,188, but not by IFN-λ61. Inborn errors of IFN-α/β immunity underlie various severe viral diseases in humans2,3,138–143,154,190,192. However, neither P1 nor P2 presented life-threatening viral infections (see Supplemental Case report). The patients had experienced benign upper respiratory infections caused by Parainfluenza virus 3 (P1), influenza virus (P2), and SARS-CoV-2 (P2). The last of these infections is of particular importance as even subtle defects of type IFN-α/β immunity underlie critical COVID-19 pneumonia3,134–138. One of the major mechanisms of SARS-CoV-2 virulence depends on the ability of this virus to induce only small amounts of IFN-α/β, rendering the amounts of this cytokine produced all the more important to the infected individual3,194,196. Phage immunoprecipitation-sequencing (PhIP-Seq) confirmed that both patients has been exposed to multiple DNA and RNA viruses (Figure 7A and Table S4). Thus, both these patients with IRF1 deficiency controlled many common, and even virulent viruses well.
Genetics
P1 and P2 were genotyped from DNA extracted from whole blood, with the Genome-Wide Human SNP Array 6.0 and/or WES with SureSelect Human All Exon V6 from Agilent. IRF1 exon 3 was amplified from genomic DNA with the following primers (5’-TGGTCTGTTTAAGCCAGCCTC-3’ and 5’-CAGAAACACAAGTCTGCCACC-3’), and exon 5 with the following primers (5’-TTCCACCTCTCACCAAGAACC-3’ and 5’-CAGAGAAGGTATCAGGGCTGG-3’) both at a Tm of 60°C, with the GoTaq DNA Polymerase (#M3005, Promega). Amplicons were then sequenced by the Sanger sequencing method with Big Dye Terminator v3.1 (Thermo Fisher Scientific), and subjected to capillary electrophoresis (#A30469, Applied Biosystems 3500xL system, Thermo Fisher Scientific). The genotype of P1 was checked with DNA extracted from whole blood, granulocytes, EBV-B cells, HVS-T cells, SV40-fibroblasts, primary fibroblasts, and induced pluripotent stem cells (iPSCs). The genotype of her mother was checked with DNA extracted from whole blood and granulocytes. The genotype of P2 was checked with DNA extracted from whole blood, granulocytes, primary fibroblasts, SV40-fibroblasts and MDMs, and the genotypes of her relatives were checked with DNA extracted from whole blood.
Cell culture and stimulation
HEK293T cells and SV40-fibroblasts were cultured in Dulbecco/Vogt modified Eagle’s minimal essential medium (DMEM, #61965059, Gibco) supplemented with decomplemented 10% fetal bovine serum (FBS, #10270098, Gibco), and EBV-B cells were cultured in Roswell Park Memorial Institute medium (RPMI 1640, # 61870044, Gibco) supplemented with 10% decomplemented FBS. HVS-T cells were cultured with an equal mixture of RPMI and Panserin 401 (#P04–710401, Pan Biotech) supplemented with 10% decomplemented FBS, 1.2% Glutamax (#35050061, Gibco), gentamycin, and 100 IU/mL recombinant interleukin 2 (Aldesleukin, Novartis). T-cell blasts were cultured in ImmunoCult-XF T Cell Exp Medium (#10981, Stemcell) in the presence of IL-2 and primed every two weeks with ImmunoCult Human CD3/CD28/CD2 T-Cell Activator (#10970, Stemcell). All cells were grown at 37°C, under an atmosphere containing 5% CO2. HEK293T cells were plated at a density of 600,000 cells per well, in six-well plates. For CD274 (PD-L1) induction, SV40-fibroblasts were plated at a density of 200,000 cells per well, in six-well plates, with 2 mL DMEM-10% FBS per well, and were left unstimulated or were stimulated the following day with 102, 103, or 104 IU/mL recombinant IFN-γ (IFN-γ, Imukin, Horizon Pharma). Forty-eight hours after stimulation, cells were harvested with trypsin, and stained as described below. For p-STAT1 induction, SV40-fibroblasts were starved overnight in DMEM-1% FBS medium, and were left unstimulated or were stimulated for 20 minutes with 103 IU/mL recombinant IFN-γ (Imukin, Horizon Pharma), IFN-2αb (Introna, MSD), or 1 ng/mL IFN-β (Miltenyi Biotec). The same protocol was used for p-STAT1 induction in EBV-B cells, except that the cells were starved in RPMI-1% FBS medium and stimulated with 105 IU/mL IFN-γ or IFN-α2b, or 10 ng/mL IFN-β (Miltenyi Biotec). The reaction was stopped by adding cold PBS, and the cells were harvested with trypsin and directly stained, as described below.
Site-directed mutagenesis, transient and stable transfection
Empty vector (EV) and a plasmid containing the DDK-tagged IRF1 cDNA were obtained from a commercial source (#RCPS100001 and #RC203500, respectively, Origene). Constructs carrying single-nucleotide mutant alleles were generated from this plasmid by mutagenesis with appropriate primers, with the Pfu Ultra II Fusion HS DNA (#600674, Agilent) polymerase, followed by digestion with DpnI (#R0176L, New England Biolab). For assessments of the reinitiation of translation, methionine codons were mutated to alanine codons (ATG>GCG). Plasmids were amplified in competent E. coli cells (#C3019H, New England Biolab) and purified with a maxiprep kit (#12663, Qiagen). HEK293T cells were transiently transfected with the various constructs at a concentration of 2.5 μg/mL, with the Lipofectamine LTX kit (#15338100, Thermo Fisher Scientific) in accordance with the manufacturer’s instructions. Retroviral plasmids and vectors were prepared as previously described, with primers for site-directed mutagenesis or deletion, and were produced in Phoenix cells157.
Western blotting and EMSA
Total protein extracts were prepared by mixing cells with modified radioimmunoprecipitation assay buffer supplemented with protease inhibitors (EDTA-free Complete, Roche) and phosphatase inhibitor cocktail (PhosphoStop, Roche), 0.1 mM dithiothreitol (DTT; Life Technologies), 10−3 mM Na3VO4, and 1 mM PMSF, and incubating for 40 minutes on ice. The cytoplasmic and nuclear contents of the cells were separated with NE-PER nuclear and cytoplasmic extraction reagents (#78835, Thermo Fisher Scientific). Equal amounts of protein, according to a Bradford protein assay (#5000002, Biorad), were resolved by SDS-PAGE in a Criterion TGX 10% or 12% precast gel (Biorad) and the bands obtained were transferred to a nitrocellulose membrane (#1704159 and #1704157, Biorad). Membranes were probed with antibodies directed against IRF1 (unconjugated, clone D5E4, #8478, Cell Signaling; or unconjugated, polyclonal, #11335–1-AP, Proteintech), IRF8 (unconjugated, goat polyclonal, #sc-6058, Santa Cruz; or unconjugated, clone D20D8, #5628, Cell Signaling), IRF9 (unconjugated, rabbit polyclonal, #sc-496, Santa Cruz; or unconjugated, rabbit polyclonal, #14167–1-AP, ProteinTech), IRF3 (unconjugated, clone D9J5Q, #10949, Cell Signaling), STAT1 (unconjugated, clone 1, #610115, Beckton-Dickinson), pSTAT1 (unconjugated, clone 4a, #612232, Beckton-Dickinson), STAT2 (unconjugated, clone B-3, #sc-514193, Santa Cruz), MX1 (unconjugated, polyclonal, #13750–1-AP, ProteinTech), ISG15 (HRP-conjugated, clone F-9, sc-166755, Santa-Cruz), DDK-tag (HRP-conjugated, clone M2, #A8592, Sigma-Aldrich), GBP1 (unconjugated, clone 1B1, #sc-53857, Santa-Cruz, #A8592, Sigma-Aldrich), APOL3 (unconjugated, clone EPR8238(2), #ab154869, Abcam), RARRES3 (unconjugated, rabbit polyclonal, #12065–1-AP, ProteinTech), vinculin (unconjugated, clone EPR8185, #ab129002, Abcam; or HRP-conjugated, clone 7F9, #sc-73614-HRP, Santa Cruz), and lamin A/C (HRP-conjugated, clone E-1, #sc-376248-HRP, Santa Cruz). Unconjugated antibodies were detected by incubation with goat anti-mouse or rabbit IgG (H + L)-HRP-conjugated antibodies (#1706516 or #1706515, respectively, Biorad). Binding was detected by incubation with the Clarity Western ECL substrate (Biorad, #1705061) or SuperSignal West Femto (Thermo Fisher Scientific, #34096) with ChemiDoc MP (Biorad). The Spectra Multicolor Broad Range Protein Ladder (#26623, Thermo Fisher Scientific) or the Chameleon Duo Prestained Protein Ladder (#928–6000, Licor) was used to provide molecular weight markers. Membranes were stripped in Restore Western Blot Stripping Buffer (#21063, Thermo Fisher Scientific).
EMSA was performed by incubating 10 μg of nuclear protein lysate on ice for 30 minutes with an IRD700-conjugated ISRE probe (5′-GATCGGGAAAGGGAAACCGAAACTGAA-3’) designed on the basis of the ISRE motif from the ISG15 promoter. For supershift assays, nuclear protein lysates were incubated for 30 minutes on ice with 3 μg anti-DDK antibody (clone M2, #14793S, Cell Signaling) or the corresponding isotype (#2729S, Cell Signaling). Protein/oligonucleotide mixtures were then subjected to electrophoresis in 12.5% acrylamide/bis-acrylamide 37.5:1 gels in 0.5% TBE migration buffer for ~120 minutes at 200 mA and 4°C in the dark. Binding was detected with the Licor Odyssey CLx system (Li-Cor, Lincoln). Images were analyzed with Imagine Lab 6.0.1 build 34 (Bio-Rad Laboratories).
Confocal microscopy
SV40-fibroblasts were plated on chambered coverslips (#80826, iBidi). The following day, they were left unstimulated or were stimulated for the indicated times with 103 IU/mL IFN-γ (Imukin, Horizon Pharma). Cells were fixed by incubation for 15 minutes in 4% formaldehyde in phosphate-buffered saline (PBS), pH 7.4 at 37°C. The cells were then incubated overnight at 4°C with primary antibody (unconjugated, clone D5E4, #8478, Cell Signaling). They were washed three times in PBS, stained by incubation with secondary antibodies for one hour at room temperature (goat anti-rabbit IgG Alexa Fluor 555, #A21429), and left in ProLong Gold with DAPI (#P36931, Thermo Fisher Scientific). Cells were then visualized by confocal microscopy (×63 oil immersion lens, SP8 gSTED, Leica). Images were analyzed with Fiji software.
Luciferase assay
We used two different ISRE-luciferase reporter assays: (i) an assay using the ISRE3 reporter plasmid (pGL4.10[luc2] backbone, Promega #E6651), which contains, as previously described158, three repeats of the ISRE sequence (5’-GGAAAGGGAAACCGAAACTGAA-3’) separated by spacers designed on the basis of the ISRE motif from the ISG15 promoter, (ii) an assay using the ISRE5 reporter plasmid, which contains, as previously described139, five repeats of the ISRE sequence 5’-GGGAAAGTGAAACTA-3’. HEK293T cells were transiently transfected, in 96-well plates, with the (ISRE) reporter plasmid (100 ng/well and 100 μL DMEM-10% FBS medium), the pRL-SV40 vector (Promega, #E2231, 40 ng/well) and the IRF1 WT or mutant p.CMV6 plasmid (100 ng/well), with the Lipofectamine LTX kit (Thermo Fisher Scientific, #15338–100), according to the manufacturer’s instructions. Cells were used for the ISRE assay with the Dual-Luciferase system kit (Promega #E1980), according to the manufacturer’s protocol, 24 hours after transfection. Signal intensity was determined with a Victor X4 plate reader (Perkin Elmer). Experiments were performed in triplicate, and dual reporter activity is expressed as the fold-induction relative to cells transfected with the empty vector.
RT-qPCR and cDNA
RNA was extracted with the RNeasy Plus Mini Kit (#74136, Qiagen) or Quick-RNA MicroPrep Kit (#R1051, Zymo). Any remaining genomic DNA was removed by extraction on a column or by DNase digestion. RNA was reverse-transcribed with the SuperScript II Reverse Transcriptase (#18064014, Thermo Fisher Scientific) and oligo(dT)12–18 (#18418012, Thermo Fisher Scientific) or with the High-Capacity RNA-to-cDNA Kit (#4387406, Applied Biosystems), according to the manufacturer’s protocol. qPCR was performed on cDNA with TaqMan Fast Universal PCR Master Mix (2X), no AmpErase UNG (#4352042, Thermo Fisher Scientific) on a 7500 Real-Time PCR System (Applied Biosystems) or Taqman ViiA7, with the following probes, all from Thermo Fisher Scientific: IRF1 exons 3–4 (#Hs00971960_m1), IRF1 exons 8–9 (#Hs00971965_m1), GBP4 (#Hs00364728_m1), APOL3 (#Hs00758274_m1), and GUSB (#1702016).
Flow cytometry on cell lines
SV40-fibroblasts were incubated with the PE-Dazzle-594-PD-L1 (CD274) antibody (clone 29E.2A3, #329732, BioLegend), or the corresponding isotype (#400358, BioLegend). For pSTAT1 staining, the cells were starved overnight in DMEM-1% FBS. For intracellular staining, 106 cells were washed with PBS-2% FBS- 2 mM EDTA buffer, fixed by incubation for 10 minutes at 37°C with Fix Buffer I (#557870, Beckton Dickinson,) and permeabilized by incubation for 20 minutes at 4°C with Phosflow Perm Buffer III (#558050, Beckton Dickinson). Cells were then incubated with PE-coupled STAT1 (clone 1, #558537, Beckton Dickinson) or PE- or AF647-conjugated anti-pSTAT1 antibody (clone 4a, # 612564 or #612597, Beckton Dickinson), the corresponding isotype (#554680 and #565363, respectively, Beckton Dickinson), or unconjugated IRF1 (clone D5E4, #8478, Cell Signaling) or the corresponding isotype, for detection with PE-conjugated goat anti-rabbit antibody (#A10542, Thermo Fisher Scientific). All non-fibroblastic cells were also stained with the Aqua Dead Cell Stain kit (#L34957, Thermo Fisher Scientific). Cells were acquired on a Beckman Coulter Gallios flow cytometer and analyzed with FlowJo Software.
Mass spectrometry on primary fibroblasts
Primary fibroblasts were plated at a density of 180,000 cells/mL in 12 mL DMEM supplemented with 10% FCS in T75 flasks. The following day, they were starved overnight and incubated for 24 hours with or without 103 IU/mL IFN-γ (Imukin, Horizon Pharma). The following day, cells were harvested with trypsin, washed once in PBS, and total protein was extracted by incubation for 40 minutes on ice (with vortexing for 15 seconds every 10 minutes) in the following SDS-free RIPA buffer: 150 mM NaCl, 1% NP40 (#85124, Thermo Fisher Scientific), 50 mM Tris, 1 mM EDTA, and EDTA-free protease inhibitor (#0469332001, Roche). A volume of 80 to 120 μL buffer was used, depending on the size of the pellet. The mixture was then centrifuged at 16,800 g for 10 minutes at 4°C and the protein-containing supernatant was collected. The concentration of protein in the supernatant was determined with the Pierce BCA Protein Assay Kit (#23225, Thermo Fisher Scientific) and the standard test tube protocol. Protein integrity was assessed with a colloidal blue staining kit (#LC6025, Thermo Fisher Scientific). Samples were then stored at −80°C before further processing.
The cysteine residues were then reduced and alkylated with DTT and IAA. Proteins were precipitated in ice-cold acetone and the resulting pellet was dissolved in 200 mM EPPs pH 8.5 supplemented with 1 μg sequencing grade trypsin. Samples were digested by incubation overnight at room temperature. An additional 1 μg of trypsin was then added and the temperature was increased to 50°C for 1 hour. Peptides were labeled by incubation with TMTpro reagent for 1 hour at room temperature. Stoichiometry and labeling efficiency were checked, hydroxylamine was added to quench the reaction and the peptides were pooled. TMTpro peptides were first separated into two fractions by custom-made SCX column. Each of these two fractions was then further separated into eight fractions on high-pH reversed-phase spin columns, and the eight fractions were then concatenated to obtain a total of 12 fractions. The peptide fractions were separated by HPLC for two hours with a linear gradient on a 250 mm*75 μm EasySpray column connected to an Easy nLC 1200 HPLC machine, with an orbitrap Fusion Lumos mass spectrometer operating in DDA MS2 positive mode used for analysis. Spectra were queried against the human proteome (downloaded from uniprot.org on 2019/02/12) at a FDR of 1% with Sequest HT through Proteome Discoverer v. 2.5. A spectral purity of 75% was required to ensure adequate confidence in quantification. Protein abundances were processed further in the Perseus statistical software environment. Abundances were log2-transformed and normalized against the median intensity for each sample. The mass spectrometry proteomics data have been deposited with the ProteomeXchange Consortium via the PRIDE159 partner repository with the dataset identifier PXD037759.
Deep flow cytometry phenotyping and ex vivo naïve CD4+ T-cell polarization experiments
Cryopreserved PBMCs and their subpopulations were analyzed with a 28-color flow cytometry panel, as previously described160. PBMCs were collected from P1 at the age of five years and from P2 at the age of six years. Both patients were receiving broad-spectrum antimycobacterial drugs and P1 was also receiving recombinant IFN-γ. Cells were also labeled with anti-CD4 (APC-Cy7, RPA-T4, #557871, BD Pharmingen), anti-CD45RA (BV605, HI100, #562886, BD Horizon), and anti-CCR7 (AF700, 100503, #561143, BD Pharmingen) antibodies, and naïve (defined as CD45RA+CCR7+CD4+) T cells were isolated (>98% purity) with a FACS Aria cell sorter (BD Biosciences). Isolated cells were then cultured with T-cell activation and expansion beads (TAE; anti-CD2/CD3/CD28; Miltenyi Biotec) + IL-2 (50 IU/mL, Millipore) to allow proliferation to occur, over a period of seven days. The cells were then subcultured with TAE beads alone (TH0) or under TH1 (IL-12 [50 ng/mL; R&D Systems]), TH2 (IL-4 [1 IU/mL; Thermo Fisher Scientific], TH9 ([100 IU/mL; Thermo Fisher Scientific], TGF-β [2.5 ng/mL; R&D Systems]), or TH17 (TGF-β [2.5 ng/mL; R&D Systems], IL-1β [50 ng/mL; Peprotech], IL-6 [50 ng/mL; PeproTech], IL-21 [50 ng/mL; PeproTech], IL-23 [50 ng/mL; Thermo Fisher Scientific]) polarizing conditions. After five days of culture, the supernatant was used for assessments of the secretion of IL-2, IL-4, IL-5, IL-6, IL-9, IL-10, IL-13, IL-17A, IL-17F, IFN-γ, and TNF with a cytometric bead array (BD Biosciences). Once the supernatant had been collected, the cells were stimulated with PMA (100 ng/mL)-ionomycin (750 ng/mL) for six hours, with brefeldin A (10 μg/mL) added after the first two hours of incubation. For the assessment of intracellular cytokine production, cells were stained for IFN-γ (BUV737, 4S.B3, #564620, BD Horizon), TNF (PerCP, Mab11, #502924, BioLegend), IL-9 (PE, MH9A3, #560807, BD Pharmingen), IL-13 (BV421, JES10–5A2, #563580, BD Horizon), IL-4 (AF488, 8D4–8, #500710, BioLegend), IL-17A (BV510, BL168, #512330, BioLegend), IL-17F (BV650, O33–782, #562264, BD Horizon), IL-2 (BV750, MQ1–17H12, #566361, BD Horizon), and IL-21 (eF660, eBio3A3-N2, #50-7219-42, Thermo Fisher Scientific), and analyzed by FACS (BD FACSymphony High-Speed Cell Analyzer A3). A previously described gating strategy was used160.
Immunophenotyping of MAIT, iNKT, and γδ T cells
The immunophenotyping of MAIT, iNKT, and γδ T cells was performed as previously described8 on cryopreserved PBMCs from P1 prepared from a sample collected at the age of three years; and as previously described10 on cryopreserved PBMCs from P2 prepared from a sample collected at the age of six years. Both patients were receiving broad-spectrum antimycobacterial drugs. Briefly, staining was performed in the presence of Fcblock (Miltenyi Biotec), with Zombie-NIR live-dead exclusion dye (#423105, BioLegend), anti-CD3-Alexa532 (Clone UCHT1, # 58-0038-42, Thermo Fisher Scientific), anti-γδTCR-FITC (#11-9959-42, Thermo Fisher Scientific), anti-Vδ2-APC-Fire750 (#331420, BioLegend), anti-CD56-BV605 (clone 5.1H11, #362538, BioLegend), anti-CD4-BV750 (#5663656, BD Biosciences), anti-CD8a-BV510 (clone RPA-T8, #301047, BioLegend), anti-Vα7.2-BV711 (clone 3C10, #351731, BioLegend), anti-Vα24-Jα18-PE-Cy7 (clone 6B11, #342912, BioLegend), anti-Vδ1-Vioblue (#30-100-555, Miltenyi Biotec), anti-CD161-PE (clone HP-3G10, #339938, BioLegend) and anti-Vβ11-APC (Miltenyi Biotec) antibodies. Cells were analyzed with an Aurora cytometer (Cytek). The gating strategy for MAIT cells, iNKT cells, γδ1+ T cells, and γδ2+ T cells has been described elsewhere8,69.
ILC immunophenotyping
ILC, T, B, and NK cells were immunophenotyped on cryopreserved PBMCs from P1 and P2 prepared from samples collected at the ages of four and six years, respectively. Both patients were receiving broad-spectrum antimycobacterial drugs and P1 was also receiving recombinant IFN-γ. Briefly, biotinylated anti-human CD1a (biotin, HI149, #300112, BioLegend), CD14 (biotin, 61D3, #13–0149-82, Invitrogen), CD34 (biotin, 4H11, #316404, BioLegend), CD123 (biotin, 6H6, #306004, BioLegend), CD203c (biotin, FR3–16A11, #130-092-345, Miltenyi Biotec), CD303 (biotin, AC144, #130-090-691, Miltenyi Biotec), FcεRIα (biotin, AER-37(CRA-1), #334606, BioLegend), TCRαβ (biotin, IP26, #306704, BioLegend) and TCRγδ (biotin, B1, #555716, BD Biosciences) antibodies were used, in combination with streptavidin BUV661 (#565081, BD Biosciences), for lineage staining, along with anti-human CD4 FITC (OKT4, #317408, BioLegend), CD336 PerCP-eFluor710 (NKp44, 44.189, #46-3369-42), EOMES PE (WD1928, #12-4877-42, Invitrogen), CD8a PE-CF594 (RPA-T8, #562282, BD Biosciences), CD127 PE-Cy7 (eBioRDR5, #25-1278-42, Invitrogen), CD294 (CRTh2) AF647 (BM16, #558042, BD Biosciences), CD161 AF700 (HP-3G10, #339942, BD Biosciences), CD94 APC-Fire750 (DX22, #305518, BioLegend), CD335 (NKp46) BV421 (9E2/NKp46, #564065, BD Biosciences), CD45RA BV570 (HI100, #304132, BioLegend), CD117 BV605 (104D2, #313218, BioLegend), CD3 BV650 (UCHT1, #563852, BD Biosciences), CD7 BV711 (M-T701, #564018, BD Biosciences), T-bet BV786 (O4-46, #564141, BD Biosciences), CD19 BUV395 (SJ25C1, #563549, BD Biosciences), CD16 BUV496 (3G8, #564653, BD Biosciences), CD25 BUV563 (2A3, #565699, BD Biosciences), CD56 BUV737 (NCAM16.2, #564447, BD Biosciences) and CD45 BUV805 (HI30, #612891, BD Biosciences) antibodies. Human IgG from serum (Sigma-Aldrich) was used to block Fc receptors before staining. Extracellular staining was performed in Brilliant Stain Buffer (BD Biosciences). Dead cells were excluded with the fixable viability dye eFluor 506 (Invitrogen). Transcription factors were stained with the FoxP3 staining buffer set (Invitrogen), in accordance with the manufacturer’s instructions. Samples were acquired on a Symphony A5 cytometer (BD Biosciences) with FACSDiva 8 software and were analyzed with FlowJo v.10 (BD Biosciences).
Mass cytometry on fresh whole blood and cryopreserved PBMCs
Whole-blood mass cytometry was performed with two different panels, on two different blood samples from P2 collected two months apart, at the age of six years. P2 was receiving broad-spectrum antimycobacterial drugs. The first panel was that of the Maxpar Direct Immune Profiling Assay (#201325, Fluidigm), used according to the manufacturer’s instructions. The second was a custom-produced panel, the content of which is detailed in STAR Methods. Marked cells were frozen at −80°C after overnight dead-cell staining, and acquisition was performed on a Helios machine (Fluidigm). All the samples were processed within 24 hours of sampling. Data analysis was performed with OMIQ software. Mass cytometry on cryopreserved PBMCs was performed as previously described8 on two different samples collected at the ages of three and five years. The antibodies indicated in STAR Methods were used. The gating strategy was as previously described8.
Whole-blood activation ELISA for cytokines
Venous blood samples from healthy controls, P1 (aged 3 years), and P2 (aged 6 years) were collected in tubes containing heparin161,162. Both patients were receiving broad-spectrum antimycobacterial drugs. These samples were diluted 1:2 in RPMI 1640 (Gibco) supplemented with 100 IU/mL penicillin and 100 μg/mL streptomycin (Gibco). We then dispensed 1 mL of each diluted blood sample into each of five wells (1 mL/well) of a 48-well plate (Nunc). These samples were incubated for 48 hours at 37°C, under an atmosphere containing 5% CO2/95% air, and under various activation conditions: with medium alone, with live BCG (M. bovis-BCG, Pasteur substrain) at a MOI of 20 BCG cells/leukocyte, or with BCG plus recombinant (rh) IL-12 (20 ng/mL; R&D Systems), or BCG plus IFN-γ (Imukin, Horizon Pharma). The supernatants were then collected and subjected to ELISA.
ELISA
Supernatants from whole-blood stimulation experiments were used for determinations of IL-12p40 (#DP400, R&D Systems), IL-12p70 (#HS120, R&D Systems), and IFN-γ (#DIF50, R&D Systems), in accordance with the manufacturer’s protocol.
Phage immunoprecipitation-sequencing (PhIP-Seq)
A plasma sample was collected from P1 at the age of three years (before immunoglobulin administration) and from P2 at the age of six years. For antibody profiling by phage immunoprecipitation-sequencing (PhIP-Seq)163, plasma samples from both patients and controls were assayed and data were analyzed as previously described5,164, but with the following modifications. We calculated species-specific significance cutoff values to estimate the minimum number of enriched, non-homologous peptides required to consider a sample seropositive (as previously described163) with an in-house dataset and a generalized linear model. For each sample, we calculated virus-specific scores by dividing the counts of enriched, non-homologous peptides by the estimated cutoff score. These adjusted virus scores were used for the heatmap plot (Figure 7A). In addition to studying the patients reported here, we also calculated and plotted the mean antibody responses for a pediatric control cohort of lean individuals without infectious or immunological disease (n=111; age range: 7 to 15 years; median age: 11.0 years) described in a previous study165,167. Pooled human plasma for IVIg (Privigen® CSL Behring AG) and human IgG-depleted serum (Molecular Innovations, Inc.) served as additional controls. All research on human subjects was performed after informed written consent had been obtained or after the samples had been rendered anonymous. The procedures were approved by the Institutional Research Ethics Boards of Sidra Medicine.
Viral infection experiments
In viral infection experiments (as indicated), SV40-fibroblasts were subjected to pretreatment for 16 hours with the indicated doses of IFN-α2b (Introna, MSD) or IFN-γ (Imukin, Horizon Pharma).
VSV (Indiana strain) replication experiments were performed as previously described143. Briefly, 1.25 × 105 SV40-fibroblasts per well were plated in 24-well plates, in DMEM supplemented with 10% FBS. Cells were infected with VSV at a multiplicity of infection (MOI) of 0.1, in DMEM supplemented with 2% FBS. Cells and supernatants were obtained at various time points (1, 8, 24 and 48 hours) and frozen. VSV titers were determined by calculating the 50% end point (TCID50), as described by Reed and Muench169, following the inoculation of Vero cell cultures in 96-well plates.
For HIV infections, the HIV-1 reporter virus HIV-GFP env-nef- was NL4–3 ΔenvΔnef encoding GFP in nef171 and the HIV-2 reporter virus was ROD9 ΔenvΔnef encoding GFP in nef173. Viral particles were produced by transfecting 293FT cells in six-well plates with 3 μg DNA and 8 μL TransIT-293 (Mirus Bio) per well: for VSV-G-pseudotyped HIV-1, 0.2 μg CMV-VSVG, 0.2 μg HXB2175 and 2.6 μg HIV-GFP env-nef-; for VSV-G-pseudotyped HIV-2, 0.4 μg CMV-VSVG and 2.6 μg HIV-2ROD9ΔenvΔnef GFP. One day after transfection, the medium was removed, and fresh medium was added. Viral supernatants were harvested the following day, passed through a filter with 0.45 μm pores, and frozen at −80°C. SV40-fibroblasts were plated in a 96-well plate at a density of 2.5 × 104/mL in 200 μL of DMEM supplemented with 10% FCS per well. The following day, the cells were left unstimulated or were stimulated by overnight incubation with 103 IU/mL IFN-α2a (#11343506, Immunotools), or 10 or 103 IU/mL of IFN-γ (Imukin, Horizon Pharma). Cells were washed and then incubated with infected viral supernatants in the presence of 1 μg/mL protamine. Two days later, the GFP expression of living cells was assessed on a BD FACSVerse flow cytometer.
The SARS-CoV-2 NYC isolate (GenBank OM345241) was obtained from a de-identified patient in July 2020. The virus isolate was amplified though six- to seven-day passages in Caco-2 cells at 37°C. After each passage, virus-containing supernatant was harvested, clarified by centrifugation (3,000 × g for 10 minutes), and filtered through a 0.22 μm-mesh disposable vacuum filter system. The passage 3 stock, used in this study, had a titer of 3.4 × 106 PFU/mL, as determined on Vero E6 cells with a 1% methylcellulose overlay, as previously described177. SV40-fibroblasts stably transduced with ACE2 were used to seed 96-well plates at a density of 7,000 cells per well in the presence or absence of the indicated doses of IFN-α2b (Introna; MSD) or IFN-γ. The cells were infected with SARS-CoV-2 16 hours later, by adding the 0.1 μL of viral inoculum to the media (final volume 110 μl) and centrifuging the cells for 5 minutes at 500 × g at room temperature. The infections were conducted in four replicates (separate wells). The cells were fixed, 24 hours post-infection, with neutral buffered formalin at a final concentration of 10%, stained for SARS-CoV-2 with an anti-N antibody at a dilution of 1:3000 (#GTX135357; GeneTex), then with an Alexa Fluor 647-conjugated secondary antibody (#A-21245; Invitrogen) and 1 μg/mL Hoechst 33342 (# H3570; Invitrogen). Plates were imaged with ImageXpress micro XL and analyzed with MetaXpress (Molecular Devices).
The YF17D-venus reporter virus expressing the Venus fluorescent protein was generated as described by Yi et al.179; the titer of this virus was 1.8 × 106 TCID50/mL on Huh-7.5 cells. SV40-fibroblasts were used to seed 96-well plates at a density of 8,000 cells/well in the presence or absence of 10 or 103 IU/mL IFN-α2b (Introna; MSD), or 103 IU/mL of IFN-γ (Imukin, Horizon Pharma). After incubation for 16 hours, the medium was removed and the cells were inoculated with 50 μL YF17D-venus diluted in Opti-MEM (#51985–091; Gibco). The dilution used was determined empirically (1/32) so as to obtain > 90% YF17D-venus positive IFNAR1-deficient cells at 3 dpi (MOI ≈ 0.3; titer determined on Huh-7.5 cells). After inoculation for 1 hour, the inoculum was removed and 100 μL phenol-free medium (FluoroBrite DMEM; #A1896701; Gibco) supplemented with 10% FBS was added. Infections were performed with six replicates (separate wells). Three hours post-infection, the nuclei of the cells were stained by incubation with 1 μg/mL Hoechst 33342 (#H3570; Invitrogen). Live imaging and quantification were performed with a BioTek Cytation 7 microscope.
For hepatitis A virus assays, SV40-fibroblasts were used to seed 12-well plates. They were incubated at 37°C for 24 h, and were then treated with IFN-α2b (103 IU/mL) or IFN-γ (103 IU/mL), or left untreated for 16 hours before infection with the HM175/18f-NLuc reporter virus (6 ×103 genome equivalents/well)52. The cells were incubated for another 72 hours, then harvested and assayed for nanoluciferase (NLuc) activity, as previously described52. The replication of influenza A virus (IAV) was assessed as previously described at a MOI of 10, with the A/California/4/2009 strain181. The HSV-1 replication experiment was performed as previously described139,143,183 with a MOI of 0.001.
The EMCV replication experiment was performed on SV-40 fibroblasts plated at a density of 5×104 cells per well in 48-well plates. The cells were subjected to pretreated with the indicated concentrations of IFN-α2b or IFN-γ, or were left untreated, for 16 hours before infection. Cells were then incubated with EMCV at a MOI of 0.01 for 1 hour, washed twice in PBS, and then transferred to fresh complete DMEM only, or complete DMEM supplemented with the indicated concentrations of IFN-α2b or IFN-γ. Virus samples were collected one hour post-infection (hpi), 12 hpi, 24 hpi, and 48 hpi. Total RNA was extracted from the mixture of cells and supernatant with the Quick-RNA MicroPrep Kit (#R1051, Zymo Research), according to the manufacturer’s protocol. Reverse transcriptase-PCR was performed with random hexamers (#18080–051, Invitrogen). The viral titer was determined with SYBR Green qPCR methods (#4385612, Applied Biosystems) for EMCV 3D, using the previously described primers185,187, with β-glucuronidase (GUSB) as the housekeeping gene for normalization. The results are expressed according to the ΔΔCt method, as recommended by the manufacturer.
iPSC-derived macrophages (iPSC-MΦ) and MDMs
The iPSC clones C16 and C11 derived from a healthy control, and the iPSC clones derived from a patient with complete STAT1 deficiency complete have been described elsewhere50,189,191. We obtained iPSCs from P1 by reprogramming primary fibroblasts with a “4-in-1” 3rd generation SIN lentiviral vector containing the original Yamanaka factors, as previously described193. Cells were maintained on irradiated CF1 mouse embryonic fibroblasts (#A34180 or #A34181) cultured in KnockOut DMEM (#10829018) supplemented with 20% KnockOut Serum Replacement medium (#10828028), GlutaMAX Supplement (#35050061), penicillin-streptomycin (#15140122), MEM non-essential amino acids solution (#11140050), and 2-mercaptoethanol (#31350010), all from Thermo Fisher Scientific. Cultures were split weekly with collagenase IV (#17104–019, Gibco). Pluripotency was confirmed by an embryonic stem cell-like morphology, and the expression of SSEA-A, TRA1–60 and alkaline phosphatase. The karyotype was shown to be normal by R-banding and SNP array techniques. We then obtained macrophages from the iPSCs (iPSC-MΦ) as follows: the medium was depleted of bFGF for seven days, and embryonic bodies were allowed to form by incubation on an orbital shaker in the presence of ROCK inhibitor (#1254, Biotechne)189,191. After five days, EBs were collected and transferred to differentiation medium composed of STEMdiff APEL2 Medium (#5275, Stem Cell Technologies) supplemented with 50 ng/L recombinant human M-CSF and 25 ng/mL IL-3 (Peprotech)189,191. The medium was replaced weekly, and it was possible to harvest monocytic cells from the supernatant after 21 days, for terminal differentiation in RPMI supplemented with 10% FBS, 1% PS and 50 ng/mL M-CSF for 10 days189,191.
MDMs were prepared as previously described195. Briefly CD14+ cells were isolated from PBMCs by positive selection with anti-CD14 MicroBeads (Miltenyi Biotec). Cells were cultured in 12-well plates, in RPMI 1640 containing 10% FBS and M-CSF (50 ng/mL, R&D Systems). On day 7, IL-4 (50 ng/mL, R&D Systems) was added, and the cells were incubated for a further seven days for the completion of MDM differentiation. Primary fibroblasts, iPSC-MΦ, or MDMs were left unstimulated or were stimulated with 103 IU/mL recombinant IFN-γ (Imukin, Horizon Pharma).
RNA-seq
Primary fibroblasts were plated at a density of 700,000 cells per well in six-well plates, in 2 mL DMEM supplemented with 10% FBS. They were starved overnight in DMEM with 0.3% BSA (#A1595, Sigma-Aldrich). Terminally differentiated iPSC-MФ were positively selected with anti-CD14 MicroBeads (Miltenyi Biotec) to ensure that a pure population was obtained and >50,000 cells per well were plated in 12-well plates, in RPMI supplemented with 10% FBS and 100 ng/mL M-CSF. Primary fibroblasts, iPSC-MФ, and MDMs were left unstimulated or were stimulated for 2 or 8 hours with 103 IU/mL recombinant IFN-γ (Imukin, Horizon Pharma). Primary fibroblasts were left unstimulated or were stimulated with 103 IU/mL recombinant IFN-α2b (Introna, MSD). Total RNA was extracted from cells with the Quick-RNA MicroPrep kit (#R1051, Zymo Research), and treated with DNAse (Zymo) to remove residual genomic DNA. Ribosomal RNA (rRNA) and mitochondrial RNA (mRNA) were depleted. RNA-seq libraries were prepared with the Illumina RiboZero TruSeq Stranded Total RNA Library Prep Kit (Illumina) and sequenced on a NovaSeq machine in the 100- or 150-nt, paired-end configuration. The RNA-seq FASTQ files were first inspected with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to ensure that the raw data were of high quality. For each subject, the two FASTQ files generated were then mapped onto the human reference genome (Ensembl GRCh37 release 75) with STAR v.2.7.3a, in the two-pass mode197. The mapping quality of each BAM file was then evaluated with RSeQC198. Reads were quantified to generate gene-level feature counts from the read mapping, with HTSeq-count v.0.11.2199. We normalized the datasets with the functions DGEList and calcNormFactors from the edgeR version 3.26.8 package200 implemented in R v.3.6.3. We retained only genes with at least 1 count-per-million (CPM) in at least two samples. We considered a gene to be differentially expressed between two sets of conditions if the log2-fold-change between the two sets of conditions was greater than 1 (absolute value) and the adjusted p-value was below 0.05, according to the calculations made with the DESeq function of the DESeq2 package version 1.24.0201 implemented in R. Differential gene expression data were plotted on heatmaps with heatmap.2 implemented in R. We used the findMotifs.pl script from HOMER v4.11202 with the human v.6.3 database and the parameter -len 12.
Generation of THP1 cells with knockouts of IFN-γR1 or STAT1
THP1KO cells were prepared as previously described8. Briefly, the plasmid was digested with BsmbI. Annealed forward and reverse sgRNA for the target sequence (IFNGR1 exon 3 or STAT1 exon 3) was inserted into the pLentiCRISPRv2 plasmid digested with BsmbI. The resulting plasmid was used to transduce HEK293T cells in the presence of pCMV-VSV-G (#8454, Addgene), pHXB2 (#1069, NIH-AIDS Reagent Program), psPAX2 (#12260, Addgene), and pLentiCRISPRv2 (#52961, Addgene) with inserted sgRNA. After 24 hours, the supernatant of HEK293T cells was filtered through a membrane with 45 μm pores, protamine was added to a final concentration of 8 μg/mL, and 100 μL of the viral supernatant was added to 200,000 THP1 WT cells in a 96-well round-bottomed plate. Spinoculation was performed for 2 hours at 1200 × g. Two days later, selection on 5 ng/mL puromycin was initiated for five days. The cells were then cultured under single-clone conditions. After expansion IFN-γR1KO or STAT1KO clones completely deficient for the molecules concerned were identified on the basis of their (i) on complete lack of expression of IFN-γR1 or STAT1, respectively, on flow cytometry; (ii) complete lack of induction of HLA-DR after 24 hours of stimulation with 104 IU/mL IFN-γ (Imukin, Horizon Pharma).
Infection of macrophages with bacteria and mycobacteria
THP1 WT cells, the previously described IRF1KO cells78, or custom-generated IFN-γR1KO or STAT-1KO cells were cultured in FCS-supplemented RPMI. Theydifferentiated into adherent macrophages with 48 hours of PMA treatment (20 ng/mL, #P8139–1MG, Sigma Aldrich). The cells were then left unstimulated or were stimulated for 24 hours with IFN-γ at a concentration of 103 IU/mL (Imukin, Horizon Pharma). Cells were then infected as previously described74 with Salmonella enterica subsp. enterica serovar Typhimurium GFP (14028GFP, ATCC), in a gentamicin protection assay. Briefly, Salmonella Typhimurium-GFP was cultured overnight in LB broth supplemented with 100 μg/mL ampicillin. The overnight culture was then diluted 1:33 in the same medium and cultured for another three hours until it reached exponential growth phase. The bacteria were then washed once with 1X PBS and used to infect cells at a MOI of 5–10. After 45 minutes, the medium was changed and the cells were incubated with gentamicin (#15750060, Thermo Fisher Scientific) at a concentration of 100 μg/mL for 1 hour. The medium was then removed; the cells were gently washed three times with 1X PBS and incubated with RPMI supplemented with 10% FCS and 20 μg/mL gentamicin with or without 103 IU/mL IFN-γ for the indicated infection time. The cells were then treated with trypsin, and fixed by incubation with 4% paraformaldehyde (#30525-89-4, Santa Cruz) for 20 minutes at room temperature. Cells were acquired on a Beckman Coulter Gallios flow cytometer and analyzed with FlowJo Software. The infection of MDMs (differentiation with M-CSF and IL-4 (#216-MC-010 and #204-IL-010, respectively, R&D Systems)) with M. abscessus-tdTomato203 and intracellular CFU determination were performed as previously described204. Infected MDMs were acquired on a Beckton Dickinson Fortessa flow cytometer and analyzed with FlowJo Software.
CITE-seq
Single-cell RNA-seq profiling was performed on a first PBMC sample from P1 (P1.1 sampled at the age of three years), and cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) was performed on PBMCs obtained from P1 (P1.2, sample obtained at the age of five years), P2 (sample obtained at the age of six years), one pediatric control and two other healthy controls. Both patients were receiving broad-spectrum antimycobacterial drugs. P1 was also receiving recombinant IFN-γ at the age of five years but not at the age of three years. The frozen PBMCs were quickly thawed at 37°C and gently resuspended by serial additions of DMEM + 10% heat-inactivated FBS, to obtain a final volume of 14 mL. The cell suspension was centrifuged at 300 × g for 5 minutes, and the cells were washed twice with 5 mL DMEM + 10% HI-FBS each to remove cell debris. Cells were counted and viability was assessed with the LIVE/DEAD Viability kit (Thermo Fisher Scientific), according to the manufacturer’s guidelines. Residual red blood cells were removed with 25 μL of the antibody cocktail and beads from the StemCell EasySep PBMC isolation kit. One million cells were resuspended in 1 mL Biolegend Cell Staining Buffer and passed through a 40 μm-mesh Flowmi cell strainer (Sigma) to remove aggregates. Cells were then centrifuged at 400 × g for 5 minutes and resuspended in 45 μL Biolegend Cell Staining Buffer supplemented with 5 μL Human TruStain FcX Fc block (Biolegend). Cells were then incubated for 30 minutes at 4°C with a cocktail of DNA-barcoded TotalSeq-B antibodies (Biolegend): CD11c (S-HCL-3), CD141 (M80), CD161 (HP-3G10), CD14 (Me5E2), CD16 (3G8), CD19 (HIB19), CD1c (L161), CD28 (CD28.2), CD370 (CLEC9A) (8F9), CD38 (HB-7), CD3e (UCHT1), CD4 (RPA-T4), CD45RA (HI100), CD45RO (UCHL1), CD56 (NCAM) (5.1H11), CD66b (6/40c), CD69 (FN50), CD8a (RPA-T8), TCR gd (B1), TCR Va7.2 (3C10), IgG1 k Isotype control (MOPC-21). Cells were washed twice in cell staining buffer, once in PBS + 0.04% BSA, and resuspended at a concentration of 1000 cells/μL in PBS + 0.04% BSA. The samples were loaded onto a 10X Genomics Chromium G chip and reverse transcription and library preparation were performed with the Chromium Single-Cell 3′ Reagent Kits (v2 for P1.1, and v3.1 for the others), in accordance with the manufacturer’s instructions. The quality of the cDNA and feature barcode library was assessed with a TapeStation (Agilent), before sequencing on the S4 flow cells of an Illumina NovaSeq 6000 sequencer.
Two additional control PBMC scRNA-seq datasets were obtained from another study that we performed205. For the analysis, we included scRNA-seq for P1.1 and 2 controls, and CITE-seq for 3 controls and the P1.2 and P2 samples. The analysis was performed as previously described205. Briefly, sequence read quality was assessed with BVAtools (https://bitbucket.org/mugqic/bvatools). Cell Ranger (v3.0.1 for scRNA and v6.0.1 for CITE-seq) was used to map reads to the hg38 human reference genome assembly, to perform filtering, and to count barcodes and unique molecular indices (UMIs). Genes that were not expressed in any of the datasets were discarded. Cells with >20% mitochondrial genes were excluded. We filtered out low-quality cells and doublets, by excluding cells falling outside the [−1SD;+2.5SD] interval from the UMI and gene count distributions. We identified and removed any remaining doublets by manually removing cells that co-expressed markers for different cell types (CD14, CD79A, TRBC1, HBA2 and LILRA4). We then used DoubletFinder package v2.0.3 to identify and filter out any remaining cell doublets206. Once dead cells and doublets had been removed, the patient and control samples were analyzed together with the Seurat v4.0.2 R package207 and cell clustering was performed by the Uniform Manifold Approximation and Projection dimension reduction method208 applied to the most variable genes, but excluding mitochondrial and ribosomal protein genes, together with sex-related genes as both patients were female. Based on the clustering pattern obtained, we performed subclustering for the large immune cell lineages separately (myeloid/pDC, T/NK, and B cells) to identify any remaining doublets. Small residual cell clusters expressing multiple cell type-specific markers were, therefore, excluded. A final UMAP208 clustering identified 14 different cells, including 11 T cells and 2 B cells; myeloid cells were excluded from further analyses because of the small numbers of these cells in the patient samples, probably due to the poor survival of these cells during sample freezing and shipping. The protein markers identified with CITE-seq were used to assign a cell-type identity to each cluster, in conjunction with the MAST approach209 to identify the marker genes for each cluster. Differential gene expression analysis was performed with MAST for each lymphoid cell cluster, by comparing the RNA levels in P1.2 and P2 with those in five healthy controls. Adjusted p-values were estimated with the Benjamini-Hochberg (BH) procedure. Every significant differentially expressed gene (absolute FC >=1.5 and adj-p-value ≤0.001) was classified as down- or upregulated, extracted and used for downstream analysis. All the differential expression results are provided in Table S6. Enrichment analyses for GO biological processes, KEGG and Reactome pathways and Transfac motifs were performed separately on up- and downregulated differentially expressed genes and for each cell subpopulation, with the gProfiler R package and the g:SCS threshold method210; all the enrichment results are provided in Table S6.
We evaluated the effects on IRF1-binding genes in the cells of the patients by estimating module scores with Seurat’s AddModuleScore function, using the default settings. Module score was determined for each cell by calculating the mean transcriptional activity of the genes from the module and then subtracting a mean expression value for a set of control genes. The list of IRF1-binding genes was obtained as follows: IRF1 binding sites were identified by ChIP-seq in IFN-γ-treated (3 h) mouse bone marrow-derived macrophages obtained in a previous study58, we then retain genes with IRF1 sites within 10 kb on either side of the transcription start site and performed a conversion to human orthologs. Control genes were randomly sampled from bins defined based on the observed level of expression for the genes in the module. The significance of differences between expression in the patients and control module expression was then assessed by comparing the corresponding module score distributions for each cell subset in a Wilcoxon signed rank-test approach (q-value <= 0.05). We obtained a sample size-free evaluation of the difference in expression by calculating Cohen’s d effect size for every significant variant.
The scRNA-seq and CITE-seq datasets have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE216489.
Generation of single-cell suspensions from murine blood and tissues for flow cytometry
All mice were kept in specific pathogen-free conditions and handled according to the guidelines and regulations of the Canadian Council on Animal Care. Experimental protocols were approved by the McGill University Institutional Animal Care Committee (protocol number 2018–8014). Groups of male C57BL/6 (B6) and Irf1−/− mice (originally from The Jackson Laboratory, reared in-house) were killed, and their blood was collected by cardiac puncture and transferred to heparin-containing tubes (Sarstedt). Blood was diluted 1:5 in PBS, centrifuged in a benchtop centrifuge and the red blood cells in the pellet were lysed with Red Blood Cell Lysing Buffer Hybri-Max (Sigma-Aldrich) according to the manufacturer’s instructions. Cells were washed twice with FACS buffer (PBS supplemented with 2% heat-inactivated FBS) and resuspended in FACS buffer. Mouse lungs were processed as previously described211 but with minor modifications. Briefly, lungs were perfused with 5 mL ice-cold PBS, isolated and finely minced. Lung pieces were digested by incubation in 2.5 mL lung digestion buffer (RPMI-1640 supplemented with 5% FBS, 0.2 mg/mL Liberase [Roche] and 0.1 mg/mL DNase I [Roche]) for 50 minutes at 37°C. Digests were homogenized by passage through a syringe (18 G needle) and a cell strainer with 70 μM pores. Cells were washed with PBS and red blood cells were lysed with Red Blood Cell Lysing Buffer Hybri-Max (Sigma-Aldrich). Cells were washed twice and resuspended in FACS buffer. For the generation of single-cell splenocyte suspensions, spleens were isolated, gently homogenized between the frosted ends of glass slides and digested by incubation in 3 mL spleen digestion buffer (HBSS supplemented with 10 mM HEPES, 150 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 1 mg/mL collagenase D [Roche] and 0.2 mg/mL DNase I [Roche]) for 30 minutes at 37°C. Cell suspensions were further homogenized by pipetting and incubated for an additional 15 minutes at 37°C. EDTA was added to a final concentration of 1 mM and the suspensions were incubated for another 10 minutes at room temperature. Splenocytes were collected by centrifugation. The pellet was washed with PBS and red blood cells were lysed with Red Blood Cell Lysing Buffer Hybri-Max (Sigma-Aldrich). Cells were washed twice and resuspended in FACS buffer. Bone marrow was isolated from femurs and tibiae and red blood cells were lysed with Red Blood Cell Lysing Buffer Hybri-Max (Sigma-Aldrich). Cells were washed twice and resuspended in FACS buffer.
Flow cytometry on mouse cells
Pelleted cells were resuspended in 2.4G2 hybridoma supernatant diluted in FACS buffer and the resulting suspensions were incubated for 15 minutes on ice to block Fcγ receptors. Surface markers were then stained by incubation with the corresponding fluorochrome-labeled antibody dilutions in FACS buffer for 30 minutes on ice. Dead cells were excluded by staining with Fixable Viability Dye eFluor 780 (Thermo Fisher Scientific) according to the manufacturer’s instructions. Intracellular staining was performed with the FoxP3/Transcription factor staining kit (Thermo Fisher Scientific), in accordance with the manufacturer’s protocol. Stained cell suspensions were acquired on a BD LSRFortessa Cell Analyzer (BD Biosciences) and data were analyzed with FlowJo v.10 software (BD Biosciences). All the antibodies used for murine flow cytometry analyses are listed in STAR Methods.
QUANTIFICATION AND STATISTICAL ANALYSIS
Unless otherwise indicated in the figure legends, the statistics provided correspond to independent experiments. Mann-Whitney or Student’s t-tests were performed. In the relevant figures, n.s. indicates not significant, p>0.05; **** p<0.0001; ***p < 0.001; **p < 0.01; and *p < 0.05. Analyses were performed with GraphPad software.
Supplementary Material
Figure S1 – Identification of homozygous complete loss-of-function IRF1 variants in patients with severe MSMD, Related to Figure 1. (A) Cells involved in the production of and response to IFN-γ. Proteins for which a mutation of the corresponding gene has been recognized to cause isolated MSMD are depicted in blue, those responsible for syndromic MSMD are shown in red, and those that can cause either isolated MSMD or syndromic MSMD are depicted with crossed lines. (B) Relative clinical severity of isolated MSMD forms vs. relative residual IFN-γ activity. (C) Principal component analysis of WES data from the two patients and our in-house WES database. (D) Consensus negative selection (CoNeS) of IRF1. (E) Diagram of the various IRF1 cDNA constructs used for transient overexpression experiments. (F) RT-qPCR on HEK293T cells without (NT) and with transfection with empty vector (EV) or various IRF1 cDNAs. The bars represent the mean. Data from two to four independent experiments are shown. (G) Western blot for DDK, IRF1, lamin A/C, and vinculin on cytoplasmic (C) and nuclear (N) extracts from HEK293T cells not transfected (NT) or transfected with EV or various IRF1 cDNAs. Data representative of three independent experiments are shown. (H) EMSA and supershift of the nuclear extract of HEK293T cells transfected with EV or various IRF1 cDNAs with DDK tagging of the C-terminus of the protein, incubated with a fluorescent ISRE probe. Representative data from two independent experiments are shown. (I) Dual luciferase ISRE5 reporter activity of HEK293T cells transfected with EV or various IRF1 cDNAs. Bars represent the mean and SD. Data from three to six independent experiments are shown. Statistical analysis was performed with Student’s t-test. ns = not significant, p > 0.05; ****p < 0.0001.
Figure S2 – IRF1 protein levels in cells from IRF1-deficient patients, Related to Figure 2. (A) Western blot for IRF1, IRF3, IRF9, STAT1, and vinculin on total lysate from HVS-T cells from three healthy controls (CTLs), P1, and a patient with complete IRF9 deficiency. Data from a single experiment are shown. (B) IRF1 expression in primary fibroblasts from three CTLs, P1, P2, and a patient with complete IFN-γR1 deficiency, with and without 16 hours of stimulation with 103 IU/mL IFN-γ. Intracellular IRF1 (upper panel) and STAT1 (lower panel) levels, assessed by flow cytometry on (C) HVS-T cells (from a CTL and P1), (D) SV40-fibroblasts (from a CTL, P1, P2, and from a patient with complete STAT1 deficiency), or (E) EBV-B cells (from a CTL, P1, P2, and from a patient with complete STAT1 deficiency). (F) IRF1 and STAT1 expression in SV40-fibroblasts from three CTLs, P1, and a patient with complete IFN-γR1 deficiency, with and without 16 hours of stimulation with 103 IU/mL IFN-γ, or in HEK293T cells transfected with p.R129-DDK IRF1 cDNA. Data representative of three independent experiments are shown.
Figure S3 – Leukocyte development in IRF1-deficient patients and in IRF1-deficient mice, Related to Figure 3. (A) Monitoring of counts of leukocytes, monocytes and lymphocytes, polymorphonuclear neutrophils (PMN), polymorphonuclear eosinophils (PME), and polymorphonuclear basophils (PMB) in the blood of the patients, in red. (B) Mass or flow cytometry analysis of cryopreserved PBMC subsets from CTLs, P1, and P2. (C) Flow cytometry analysis of dendritic cell (DC) subsets in fresh PBMCs from a CTL, P1, and P2. (D) Flow cytometry immunophenotyping of live CD45+ lymphocytes from PBMCs, for ILCP (CD117+CRTh2−) and ILC2 (CRTh2+), in CTLs, P1, and P2. (E) Conventional flow cytometry counts of cDC1, cDC2, NK cells, ILC2, CD4+ T cells, Treg and CD8+ T cells, in the lungs of WT mice (n=5), and IRF1-deficient mice (n=5). (F) Conventional flow cytometry counts of Ly6C+ monocytes, cDC1, cDC2, pDCs, NK cells, CD4+ T cells, Treg and CD8+ T cells in the spleen of WT mice (n=5), and IRF1-deficient mice (n=5). (G) Conventional flow cytometry counts of CLP, CHILP, and ILC2P in the bone marrow of WT mice (n=5), and IRF1-deficient mice (n=5). Unpaired two-tailed t-tests were used for the statistical analysis. *p < 0.05; ***p < 0.001.
Figure S4 – IFN-γ production pathway in P1 and P2, Related to Figure 4. (A) Western blot analysis of ISG15 and MX1 induction in the SV40-fibroblasts of three CTLs, P1, P2, one patient with complete ISG15-deficiency, and one patient with complete IFN-αR1 deficiency. (B) pSTAT4 levels in HVS-T cells from two controls (CTLs), P1, and a patient with complete IL-12Rβ1 deficiency without stimulation (gray), and after 15 minutes of stimulation with 50 ng/mL IL-12 (solid black line), or 105 IU/mL IFN-α2b (dotted black line). Data representative of two independent experiments are shown. (C) Production of IFN-γ by the HVS-T cells of controls (CTLs), P1, and a patient with complete IFN-γ deficiency with and without 2 hours of stimulation with brefeldin, anti-CD3/CD2/CD28 mAb-coated beads, or with PMA-ionomycin. Bars represent the mean and SD. Data from one experiment are shown. (D) Analysis of IFN-γ+ cells on intracellular flow cytometry for various PBMC subsets, following 48 hours of stimulation with IL-12, IL-23, or BCG. Bars represent the mean and SD. Technical duplicates of the same experiment are shown for P1 and P2. (E) Intracellular flow cytometry analysis of IFN-γ, TNF, IL-9, IL-13, IL-4, IL-17A, IL-17F, IL-2, and IL-21 levels in naïve CD4+ T cells from P1 and controls after expansion for 5 days with anti-CD2/CD3/CD28 mAb-coated beads and IL-2, followed by culture for 7 days in polarizing conditions, and stimulation with PMA-ionomycin in the presence of brefeldin for 6 hours.
Figure S5 – Response to IFN-γ in non-hematopoietic cells from P1 and P2, Related to Figure 5. IRF1 RT-qPCR (A) and western blot for IRF1, pSTAT1, STAT1 and vinculin (B) on SV40-fibroblasts with and without stimulation with 103 IU/mL IFN-γ. Data from one experiment are shown. (C) Confocal microscopy of IRF1 in SV40-fibroblasts of a CTL, P1, and a STAT1-deficient patient stimulated for the indicated times. Phosphorylation of STAT1, analyzed by staining and flow cytometry after 20 minutes of incubation with or without 105 IU/mL IFN-γ, in EBV-B cells (D), or with or without 103 IU/mL IFN-γ in SV40-fibroblasts (E) from controls (CTLs), the patients (P1 and P2), or patients with complete IFN-γR1 or STAT1 deficiency. Data representative of two independent experiments are shown. Heatmaps of genes differentially expressed (log2FC) in primary fibroblasts from controls or patients, after (F) 2 or (G) 8 hours of stimulation with 103 IU/mL IFN-γ. We show the genes differentially expressed in patients relative to the control group at both time points (2 or 8 hours), for the genes differentially expressed at both time points relative to non-stimulated fibroblasts in the control group, i.e., with a |log2(FC)| > 1 and adj. p-value < 0.05 in controls, and with a |log2(FC)| > 1 and adj. p-value < 0.05 in patients relative to controls at the corresponding time point. We used the IFN-γR1−/− patient as a negative control. (H) Amount of mRNA, assessed by RNA sequencing for IFN-α2b-inducible genes (logFC>2 in controls) in primary fibroblasts from the indicated individuals with and without 30 minutes, 2 hours, and 8 hours of stimulation with IFN-γ. (I) and (J) Flow cytometry analysis of CD274 (PD-L1) expression with and without stimulation for 48 hours with 102, 103, or 104 IU/mL IFN-γ in SV40-fibroblasts with and without stable retrotransduction with EV or WT IRF1 cDNA. (K) Flow cytometry analysis of the CD274 expression of primary fibroblasts with and without 48 hours of stimulation with 102 or 103 IU/mL IFN-γ.
Figure S6 – Response to IFN-γ in myeloid cells from P1 and P2, Related to Figure 6. Induction of IL-12p40 (A) and IL-12p70 (B) secretion in whole-blood assays for local and travel controls (CTLs), an IL-12p40-deficient patient, P1, and P2, following stimulation with live BCG alone or in combination with IFN-γ (5,000 IU/mL). Bars represent the mean. Heatmaps of genes differentially expressed (log2(FC)) in iPSC-derived macrophages (iPSC-MΦ), and monocytes-derived macrophages (MDMs) from controls (CTL) or patients, after (C) 2 or (D) 8 hours of stimulation with 103 IU/mL IFN-γ. We show the genes differentially expressed in patients relative to the control group at both time points (2 or 8 hours), for genes differentially expressed at both time points relative to non-stimulated cells in the control group, i.e., with a |log2(FC)| > 1 and adj. p-value < 0.05 after Benjamini-Hochberg correction in controls, and with a |log2(FC)| > 1 and adj. p-value < 0.05 after correction in patients relative to controls at the corresponding time point. (E) Fold-enrichment in dysregulated genes, as determined by RNA-seq, for the myeloid cells of both patients relative to IRF1 ChIP-seq results for resting human monocytes77 and IFN-γ-activated murine bone marrow-derived macrophages58. (F) RT-qPCR for APOL3 and GBP4 in iPSC-MΦ or MDMs with and without stimulation for 2 and 8 hours with 103 IU/mL IFN-γ. The bars represent the mean. (G) Examples of flow cytometry dot plots obtained for gentamicin protection assay performed on PMA-differentiated WT or IRF1KO, IFN-γR1KO, or STAT1KO THP1-derived macrophages (THP1-Ф) with and without 24 hours of pretreatment with 103 IU/mL IFN-γ and infection with Salmonella Typhimurium-GFP (Stm-GFP) at a MOI of 6 for 8 hours. The percent Salmonella Typhimurium+ cells is displayed in black and the percent Salmonella Typhimuriumhigh cells is displayed in red. (H) Examples of flow cytometry dot plots obtained for Figure 6H.
Figure S7 – Antiviral IFN-α/β and IFN-γ cell-intrinsic immunity of IRF1-deficient fibroblasts, Related to Figure 7. (A) RT-qPCR for IRF1 and MX1 in SV40-fibroblasts from the indicated controls or patients after 2 hours of stimulation with 103 IU/mL IFN-α2a or IFN-γ. (B) IRF1 levels determined by RNA sequencing, in primary fibroblasts stimulated for 30 minutes, 2 hours, or 8 hours with IFN-α2b or IFN-γ. Results are normalized against non-stimulated conditions. (C) Western blot of IRF1, MX1, STAT1, STAT2, IRF9, and vinculin in SV40-fibroblasts from controls (CTLs), the patients (P1, P2), or patients with complete STAT1, STAT2, IRF9, IFN-αR1, or IFN-γR1 deficiencies stimulated for 4 hours with IFN-γ or IFN-α2β. (D) Phosphorylation of STAT1, as assessed by staining and flow cytometry, after 20 minutes of incubation with or without 103 IU/mL IFN-α2b or 10 ng/mL IFN-β, in SV40-fibroblasts or EBV-B cells from two controls (CTLs), the patients (P1, P2), and patients with complete IFN-αR1 or STAT1 deficiency. Data representative of two to three experiments are shown. (E) Venn diagram of genes positively induced in primary fibroblasts of CTLs (log2FC>1) but less strongly induced (log2FC<−1) in IRF1-, STAT1-, or IRF9-deficient primary fibroblasts than in controls after 2 or 8 hours of stimulation with IFN-α2b. (F) Hepatitis A virus (HAV), (G) human immunodeficiency virus-1 (HIV-1), and (H) yellow fever vaccine virus-venus (YF17D-venus) replication in SV40-fibroblasts for the indicated controls or patients (P1 and P2), after pretreatment for 16 hours with the indicated doses of IFN-α2b or IFN-γ. All viral infections were performed in two or three independent experiments.
Table S6 –CITE-seq data, Related to Figure 3. Differential gene expression analysis for each scRNA-seq lymphoid cell cluster and enrichment analysis for genes significantly up- and downregulated in scRNA-seq analyses of patient cells. Log2FC and adjusted p values for the comparison of gene expression of P1.2 and P2 cells with healthy controls cells; an absolute FC ≥1.5 and adj. p value ≤0.001 were considered significant. Gene ontology (biological process), pathway and Transfac motif-enriched genes are analyzed.
Table S7 –RNA sequencing and mass-spectrometry data, Related to Figures 5, 6, and 7. – (Sheet 1) log2FC and adj. p-value for RNA-seq on primary fibroblasts from three controls, both IRF1-deficient patients (P1 and P2), and patients with autosomal recessive deficiencies of IRF9, STAT1, IFN-αR1, IFN-γR1, or IFN-γR2, with and without stimulation for 30 minutes, 2 hours, or 8 hours with 103 IU/mL IFN-γ or IFN-α2b. (Sheet 2) – log2FC and adj. p-value for RNA-seq on iPSC-MΦ from two clones from controls (#11 and #16), P1 (#10), and a patient with autosomal recessive STAT1 deficiency, with and without stimulation for 2 or 8 hours with 103 IU/mL IFN-γ. (Sheet 3) – log2FC and adj. p-value for RNA-seq on MDMs from three controls and P2, with and without stimulation for 2 or 8 hours with 103 IU/mL IFN-γ. (Sheet 4) – Mass spectrometry quantification of protein extracted from primary fibroblasts from the indicated individuals with and without prior stimulation for 24 hours. Values were log2-transformed and normalized against the median.
Highlights.
Inherited complete human IRF1 deficiency underlies severe mycobacterial disease.
Human IRF1 is essential for IFN-γ-dependent macrophagic immunity to mycobacteria.
Human IRF1 is essential for IFN-γ- and STAT1-dependent immunity to mycobacteria.
Human IRF1 is largely redundant for IFN-α/β-dependent antiviral intrinsic immunity.
Acknowledgments
We thank the patients and their families, all members of the laboratory, Stephen Elledge (for the PhIP-Seq phage display viral library), and Jose Mauricio Ayala, Véronique Cholette and Rebecca Bellworthy. The Laboratory of Human Genetics of Infectious Diseases is supported in part by grants from Inserm, Paris Cité University, the St. Giles Foundation, The Rockefeller University, the Center for Clinical and Translational Science (UL1TR001866), the National Center for Research Resources and the National Center for Advancing Sciences, the National Institutes of Health (NIH), (R01AI095983, R01AI088364, R01AI163029, and U19AI162568), the National Institute of Allergy and Infectious Diseases, the French National Research Agency (ANR) under the “Investments for the future” program (ANR-10-IAHU-01), the Integrative Biology of Emerging Infectious Diseases Laboratory of Excellence (ANR-10-LABX-62-IBEID), the French Foundation for Medical Research (FRM) (EQU201903007798), the ANRS Nord-Sud (ANRS-COV05), ANRS (ECTZ170784-ANRS0073), GENVIR (ANR-20-CE93-003), GENMSMD (ANR-16-CE17-0005-01), AABIFNCOV (ANR-20-CO11-0001), GenMIS-C (ANR-21-COVR-0039), SUNLIVE (ANR-19-CE15-0012-01), MAFMACRO (ANR-22-CE92-0008)grants, Ecos-NORD (ECOS N°C19S01), the Fisher Center for Alzheimer’s Research Foundation, the Meyer Foundation, the JPB Foundation, the European Union’s Horizon 2020 research and innovation program (824110, EASI-Genomics), the Square Foundation, Grandir–Fonds de solidarité pour l’Enfance, the Fondation du Souffle, the SCOR Corporate Foundation for Science, the French Ministry of Higher Education, Research, and Innovation (MESRI-COVID-19), and REACTing-INSERM. The Laboratory of Virology and Infectious Disease was supported in part by the NIH (R01AI091707-10 to C.M.R.). J.L.P. was supported by the Francois Wallace Monahan Postdoctoral Fellowship at The Rockefeller University and the European Molecular Biology Organization Long-Term Fellowship (ALTF 380-2018). N.Mar. was supported by Sidra Medicine and the Qatar National Research Fund (NPRP9-251-3-045). The Yale Center for Mendelian Genomics (UM1HG006504) was funded by the National Human Genome Research Institute, the Yale GSP Coordinating Center (U24 HG008956), and the Yale High-Performance Computing Center (S10OD018521). This research was partly supported by Calcul Québec, Compute Canada Canada Institute of Health Research (CIHR) Project Grant to D.La. (#168959) and a CIHR Foundation Grant (to P.G.). D.La. was also supported by an FRQS Chercheur-Boursier Junior 1 Award and the Canada Foundation for Innovation John R. Evans Leaders Fund. P.G. is supported by a Distinguished James McGill Professorship award from McGill University. S.M.L is supported by the NIH: R01-AI103083 and R01-AI150095. J.R. was supported by poste d’accueil Inserm”. J.R., P.B., and T.L.V were supported by the MD-PhD program of the Imagine Institute by the Bettencourt-Schueller Foundation. N.L received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (852178 grant), German Research Foundation, (DFG) under Germany’s Excellence Strategy—EXC 2155—project number 390874280 and REBIRTH “Förderung aus Mitteln des Niedersächsischen Vorab”. A.-L.N. was supported by the international PhD program of the Imagine Institute, by the Bettencourt-Schueller Foundation and the fin de thèse FRM program (FDT202204015102). R.Y. was supported by the Sackler Center for Biomedicine and Nutrition, the Shapiro-Silverberg Fund for the Advancement of Translational Research at the Center for Clinical and Translational Science of the Rockefeller University, and the Research Grant Program from the Immune Deficiency Foundation. D.Le. was supported by FRMfellowship (FDM202006011282). C.S.M was supported by an Early-Mid Career Research Fellowship from the Department of Health of the New South Wales Government of Australia. S.G.T was supported by an NHMRC Leadership 3 Investigator Grant (1176665) and NHMRC grant (1113904). M.O. was supported by the David Rockefeller Graduate Program, the Funai Foundation for Information Technology, the Honjo International Scholarship Foundation, and the New York Hideyo Noguchi Memorial Society. This work was supported by grants from ANRS (ECTZ118797), Sidaction (20-2-AEQ-12822-2) and FRM (EQU202103012774) to N.Man., and H.I. was supported by fellowships from Institut Curie, Séneca Foundation (20941/PD/18), and ANRS (ECTZ171453). A.N.S. was supported in part by the European Union’s Horizon 2020 research and innovation program (789645 Marie Sklodowska-Curie grant). Y.-H.C. is supported by an A*STAR International Fellowship. J. Bo. is an EMBO postdoctoral fellow. We thank the NIH Tetramer Core Facility (NTCF) for providing the 5-OP-RU-loaded MR1 tetramer, which was developed jointly with Dr. James McCluskey, Dr. Jamie Rossjohn, and Dr. David Fairlie.
Footnotes
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
J.-L.C. serves on the scientific advisory boards of ADMA Biologics Inc., Kymera Therapeutics, and Elixiron Immunotherapeutics. All other authors declare no competing interests.
References
- 1.Casanova J-L, and Abel L (2021). Lethal Infectious Diseases as Inborn Errors of Immunity: Toward a Synthesis of the Germ and Genetic Theories. Annu. Rev. Pathol 16, 23–50. 10.1146/annurev-pathol-031920-101429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casanova J-L, and Abel L (2021). Mechanisms of viral inflammation and disease in humans. Science 374, 1080–1086. 10.1126/science.abj7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Q, Bastard P, COVID Human Genetic Effort, Cobat A, and Casanova J-L (2022). Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature 10.1038/s41586-022-04447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bustamante J (2020). Mendelian susceptibility to mycobacterial disease: recent discoveries. Hum. Genet 139, 993–1000. 10.1007/s00439-020-02120-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kerner G, Rosain J, Guerin A, Al-Khabaz A, Oleaga-Quintas C, Rapaport F, Massaad MJ, Ding JY, Khan T, Ali FA, et al. (2020). Inherited human IFN-gamma deficiency underlies mycobacterial disease. J. Clin. Invest 130, 3158–3171. 10.1172/JCI135460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Voyer T, Neehus A-L, Yang R, Ogishi M, Rosain J, Alroqi F, Alshalan M, Blumental S, Ali FA, Khan T, et al. (2021). Inherited deficiency of stress granule ZNFX1 in patients with monocytosis and mycobacterial disease. Proc. Natl. Acad. Sci. U. S. A 118. 10.1073/pnas.2102804118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin-Fernandez M, Buta S, Le Voyer T, Li Z, Dynesen LT, Vuillier F, Franklin L, Ailal F, Muglia Amancio A, Malle L, et al. (2022). A partial form of inherited human USP18 deficiency underlies infection and inflammation. J. Exp. Med 219, e20211273. 10.1084/jem.20211273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang R, Mele F, Worley L, Langlais D, Rosain J, Benhsaien I, Elarabi H, Croft CA, Doisne J-M, Zhang P, et al. (2020). Human T-bet Governs Innate and Innate-like Adaptive IFN-γ Immunity against Mycobacteria. Cell 10.1016/j.cell.2020.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daza-Cajigal V, Albuquerque AS, Young DF, Ciancanelli MJ, Moulding D, Angulo I, Jeanne-Julien V, Rosain J, Minskaia E, Casanova J-L, et al. (2022). Partial human Janus kinase 1 deficiency predominantly impairs responses to interferon gamma and intracellular control of mycobacteria. Front. Immunol 13, 888427. 10.3389/fimmu.2022.888427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogishi M, Arias AA, Yang R, Han JE, Zhang P, Rinchai D, Halpern J, Mulwa J, Keating N, Chrabieh M, et al. (2022). Impaired IL-23-dependent induction of IFN-γ underlies mycobacterial disease in patients with inherited TYK2 deficiency. J. Exp. Med 219, e20220094. 10.1084/jem.20220094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casanova JL, and Abel L (2020). The human genetic determinism of life-threatening infectious diseases: genetic heterogeneity and physiological homogeneity? Hum. Genet 139, 681–694. 10.1007/s00439-020-02184-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bustamante J, Boisson-Dupuis S, Abel L, and Casanova JL (2014). Mendelian susceptibility to mycobacterial disease: genetic, immunological, and clinical features of inborn errors of IFN-gamma immunity. Semin. Immunol 26, 454–470. 10.1016/j.smim.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newport MJ, Huxley CM, Huston S, Hawrylowicz CM, Oostra BA, Williamson R, and Levin M (1996). A mutation in the interferon-gamma-receptor gene and susceptibility to mycobacterial infection. N. Engl. J. Med 335, 1941–1949. 10.1056/NEJM199612263352602. [DOI] [PubMed] [Google Scholar]
- 14.Jouanguy E, Altare F, Lamhamedi S, Revy P, Emile JF, Newport M, Levin M, Blanche S, Seboun E, Fischer A, et al. (1996). Interferon-gamma-receptor deficiency in an infant with fatal bacille Calmette-Guérin infection. N. Engl. J. Med 335, 1956–1961. 10.1056/NEJM199612263352604. [DOI] [PubMed] [Google Scholar]
- 15.Dorman SE, and Holland SM (1998). Mutation in the signal-transducing chain of the interferon-gamma receptor and susceptibility to mycobacterial infection. J. Clin. Invest 101, 2364–2369. 10.1172/JCI2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupuis S, Döffinger R, Picard C, Fieschi C, Altare F, Jouanguy E, Abel L, and Casanova JL (2000). Human interferon-gamma-mediated immunity is a genetically controlled continuous trait that determines the outcome of mycobacterial invasion. Immunol. Rev 178, 129–137. 10.1034/j.1600-065x.2000.17810.x. [DOI] [PubMed] [Google Scholar]
- 17.Prando C, Samarina A, Bustamante J, Boisson-Dupuis S, Cobat A, Picard C, AlSum Z, Al-Jumaah S, Al-Hajjar S, Frayha H, et al. (2013). Inherited IL-12p40 Deficiency. Medicine (Baltimore) 92, 109–122. 10.1097/MD.0b013e31828a01f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fieschi C, Dupuis S, Catherinot E, Feinberg J, Bustamante J, Breiman A, Altare F, Baretto R, Le Deist F, Kayal S, et al. (2003). Low penetrance, broad resistance, and favorable outcome of interleukin 12 receptor beta1 deficiency: medical and immunological implications. J. Exp. Med 197, 527–535. 10.1084/jem.20021769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Beaucoudrey L, Samarina A, Bustamante J, Cobat A, Boisson-Dupuis S, Feinberg J, Al-Muhsen S, Jannière L, Rose Y, de Suremain M, et al. (2010). Revisiting human IL-12Rβ1 deficiency: a survey of 141 patients from 30 countries. Medicine (Baltimore) 89, 381–402. 10.1097/MD.0b013e3181fdd832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Barricarte R, Markle JG, Ma CS, Deenick EK, Ramirez-Alejo N, Mele F, Latorre D, Mahdaviani SA, Aytekin C, Mansouri D, et al. (2018). Human IFN-gamma immunity to mycobacteria is governed by both IL-12 and IL-23. Sci. Immunol 3, eaau6759. 10.1126/sciimmunol.aau6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staels F, Lorenzetti F, De Keukeleere K, Willemsen M, Gerbaux M, Neumann J, Tousseyn T, Pasciuto E, De Munter P, Bossuyt X, et al. (2022). A Novel Homozygous Stop Mutation in IL23R Causes Mendelian Susceptibility to Mycobacterial Disease. J. Clin. Immunol 10.1007/s10875-022-01320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prando C, Samarina A, Bustamante J, Boisson-Dupuis S, Cobat A, Picard C, AlSum Z, Al-Jumaah S, Al-Hajjar S, Frayha H, et al. (2013). Inherited IL-12p40 deficiency: genetic, immunologic, and clinical features of 49 patients from 30 kindreds. Medicine (Baltimore) 92, 109–122. 10.1097/MD.0b013e31828a01f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boisson-Dupuis S, Ramirez-Alejo N, Li Z, Patin E, Rao G, Kerner G, Lim CK, Krementsov DN, Hernandez N, Ma CS, et al. (2018). Tuberculosis and impaired IL-23-dependent IFN-γ immunity in humans homozygous for a common TYK2 missense variant. Sci. Immunol 3, eaau8714. 10.1126/sciimmunol.aau8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okada S, Markle JG, Deenick EK, Mele F, Averbuch D, Lagos M, Alzahrani M, Al-Muhsen S, Halwani R, Ma CS, et al. (2015). IMMUNODEFICIENCIES. Impairment of immunity to Candida and Mycobacterium in humans with bi-allelic RORC mutations. Science 349, 606–613. 10.1126/science.aaa4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong XF, Martinez-Barricarte R, Kennedy J, Mele F, Lazarov T, Deenick EK, Ma CS, Breton G, Lucero KB, Langlais D, et al. (2018). Disruption of an antimycobacterial circuit between dendritic and helper T cells in human SPPL2a deficiency. Nat. Immunol 19, 973–985. 10.1038/s41590-018-0178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bogunovic D, Byun M, Durfee LA, Abhyankar A, Sanal O, Mansouri D, Salem S, Radovanovic I, Grant AV, Adimi P, et al. (2012). Mycobacterial disease and impaired IFN-γ immunity in humans with inherited ISG15 deficiency. Science 337, 1684–1688. 10.1126/science.1224026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hambleton S, Salem S, Bustamante J, Bigley V, Boisson-Dupuis S, Azevedo J, Fortin A, Haniffa M, Ceron-Gutierrez L, Bacon CM, et al. (2011). IRF8 mutations and human dendritic-cell immunodeficiency. N. Engl. J. Med 365, 127–138. 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nathan CF, Murray HW, Wiebe ME, and Rubin BY (1983). Identification of interferon-gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J. Exp. Med 158, 670–689. 10.1084/jem.158.3.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bustamante J, Arias AA, Vogt G, Picard C, Galicia LB, Prando C, Grant AV, Marchal CC, Hubeau M, Chapgier A, et al. (2011). Germline CYBB mutations that selectively affect macrophages in kindreds with X-linked predisposition to tuberculous mycobacterial disease. Nat. Immunol 12, 213–221. 10.1038/ni.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chapgier A, Boisson-Dupuis S, Jouanguy E, Vogt G, Feinberg J, Prochnicka-Chalufour A, Casrouge A, Yang K, Soudais C, Fieschi C, et al. (2006). Novel STAT1 alleles in otherwise healthy patients with mycobacterial disease. PLOS Genet 2, e131. 10.1371/journal.pgen.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conti F, Aragão Filho WC, Prando C, Deswarte C, Hubeau M, Newburger PE, Casanova J-L, Bustamante J, and Condino-Neto A (2015). Phagocyte nicotinamide adenine dinucleotide phosphate oxidase activity in patients with inherited IFN-γR1 or IFN-γR2 deficiency. J. Allergy Clin. Immunol 135, 1393–1395.e1. 10.1016/j.jaci.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dupuis S, Dargemont C, Fieschi C, Thomassin N, Rosenzweig S, Harris J, Holland SM, Schreiber RD, and Casanova JL (2001). Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation. Science 293, 300–303. 10.1126/science.1061154. [DOI] [PubMed] [Google Scholar]
- 33.Dupuis S, Jouanguy E, Al-Hajjar S, Fieschi C, Al-Mohsen IZ, Al-Jumaah S, Yang K, Chapgier A, Eidenschenk C, Eid P, et al. (2003). Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet 33, 388–391. 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- 34.Le Voyer T, Sakata S, Tsumura M, Khan T, Esteve-Sole A, Al-Saud BK, Gungor HE, Taur P, Jeanne-Julien V, Christiansen M, et al. (2021). Genetic, Immunological, and Clinical Features of 32 Patients with Autosomal Recessive STAT1 Deficiency. J. Immunol, ji2001451. 10.4049/jimmunol.2001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belkadi A, Pedergnana V, Cobat A, Itan Y, Vincent QB, Abhyankar A, Shang L, El Baghdadi J, Bousfiha A, Exome/Array, C., et al. (2016). Whole-exome sequencing to analyze population structure, parental inbreeding, and familial linkage. Proc. Natl. Acad. Sci. U. S. A 113, 6713–6718. 10.1073/pnas.1606460113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fareed M, and Afzal M (2017). Genetics of consanguinity and inbreeding in health and disease. Ann. Hum. Biol 44, 99–107. 10.1080/03014460.2016.1265148. [DOI] [PubMed] [Google Scholar]
- 37.Itan Y, Shang L, Boisson B, Ciancanelli MJ, Markle JG, Martinez-Barricarte R, Scott E, Shah I, Stenson PD, Gleeson J, et al. (2016). The mutation significance cutoff: gene-level thresholds for variant predictions. Nat. Methods 13, 109–110. 10.1038/nmeth.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Itan Y, Shang L, Boisson B, Patin E, Bolze A, Moncada-Velez M, Scott E, Ciancanelli MJ, Lafaille FG, Markle JG, et al. (2015). The human gene damage index as a gene-level approach to prioritizing exome variants. Proc. Natl. Acad. Sci. U. S. A 112, 13615–13620. 10.1073/pnas.1518646112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maffucci P, Bigio B, Rapaport F, Cobat A, Borghesi A, Lopez M, Patin E, Bolze A, Shang L, Bendavid M, et al. (2019). Blacklisting variants common in private cohorts but not in public databases optimizes human exome analysis. Proc. Natl. Acad. Sci. U. S. A 116, 950–959. 10.1073/pnas.1808403116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bigio B, Seeleuthner Y, Kerner G, Migaud M, Rosain J, Boisson B, Nasca C, Puel A, Bustamante J, Casanova J-L, et al. (2021). Detection of homozygous and hemizygous complete or partial exon deletions by whole-exome sequencing. NAR Genomics Bioinforma 3, lqab037. 10.1093/nargab/lqab037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maruyama M, Fujita T, and Taniguchi T (1989). Sequence of a cDNA coding for human IRF-1. Nucleic Acids Res 17, 3292. 10.1093/nar/17.8.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eason DD, Shepherd AT, and Blanck G (1999). Interferon regulatory factor 1 tryptophan 11 to arginine point mutation abolishes DNA binding. Biochim. Biophys. Acta 1446, 140–144. 10.1016/s0167-4781(99)00078-0. [DOI] [PubMed] [Google Scholar]
- 43.Ohteki T, Yoshida H, Matsuyama T, Duncan GS, Mak TW, and Ohashi PS (1998). The transcription factor interferon regulatory factor 1 (IRF-1) is important during the maturation of natural killer 1.1+ T cell receptor-alpha/beta+ (NK1+ T) cells, natural killer cells, and intestinal intraepithelial T cells. J. Exp. Med 187, 967–972. 10.1084/jem.187.6.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee EJ, Jo M, Park J, Zhang W, and Lee JH (2006). Alternative splicing variants of IRF-1 lacking exons 7, 8, and 9 in cervical cancer. Biochem. Biophys. Res. Commun 347, 882–888. 10.1016/j.bbrc.2006.06.145. [DOI] [PubMed] [Google Scholar]
- 45.Watarai H, Nakagawa R, Omori-Miyake M, Dashtsoodol N, and Taniguchi M (2008). Methods for detection, isolation and culture of mouse and human invariant NKT cells. Nat. Protoc 3, 70–78. 10.1038/nprot.2007.515. [DOI] [PubMed] [Google Scholar]
- 46.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 47.Kroger A, Koster M, Schroeder K, Hauser H, and Mueller PP (2002). Activities of IRF-1. J. Interferon Cytokine Res 22, 5–14. 10.1089/107999002753452610. [DOI] [PubMed] [Google Scholar]
- 48.Neehus A-L, Lam J, Haake K, Merkert S, Schmidt N, Mucci A, Ackermann M, Schubert M, Happle C, Kühnel MP, et al. (2018). Impaired IFNγ-Signaling and Mycobacterial Clearance in IFNγR1-Deficient Human iPSC-Derived Macrophages. Stem Cell Rep 10, 7–16. 10.1016/j.stemcr.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taniguchi T, Ogasawara K, Takaoka A, and Tanaka N (2001). IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol 19, 623–655. 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 50.Haake K, Neehus A-L, Buchegger T, Kühnel MP, Blank P, Philipp F, Oleaga-Quintas C, Schulz A, Grimley M, Goethe R, et al. (2020). Patient iPSC-Derived Macrophages to Study Inborn Errors of the IFN-γ Responsive Pathway. Cells 9, 483. 10.3390/cells9020483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamura T, Yanai H, Savitsky D, and Taniguchi T (2008). The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol 26, 535–584. 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 52.Yamane D, Feng H, Rivera-Serrano EE, Selitsky SR, Hirai-Yuki A, Das A, McKnight KL, Misumi I, Hensley L, Lovell W, et al. (2019). Basal expression of interferon regulatory factor 1 drives intrinsic hepatocyte resistance to multiple RNA viruses. Nat. Microbiol 4, 1096–1104. 10.1038/s41564-019-0425-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ikushima H, Negishi H, and Taniguchi T (2013). The IRF family transcription factors at the interface of innate and adaptive immune responses. Cold Spring Harb. Symp. Quant. Biol 78, 105–116. 10.1101/sqb.2013.78.020321. [DOI] [PubMed] [Google Scholar]
- 54.Maarifi G, Martin M-F, Zebboudj A, Boulay A, Nouaux P, Fernandez J, Lagisquet J, Garcin D, Gaudin R, Arhel NJ, et al. (2022). Identifying enhancers of innate immune signaling as broad-spectrum antivirals active against emerging viruses. Cell Chem. Biol 29, 1113–1125.e6. 10.1016/j.chembiol.2022.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Honda K, and Taniguchi T (2006). IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol 6, 644–658. 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 56.Frontini M, Vijayakumar M, Garvin A, and Clarke N (2009). A ChIP-chip approach reveals a novel role for transcription factor IRF1 in the DNA damage response. Nucleic Acids Res 37, 1073–1085. 10.1093/nar/gkn1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Minegishi Y, Saito M, Morio T, Watanabe K, Agematsu K, Tsuchiya S, Takada H, Hara T, Kawamura N, Ariga T, et al. (2006). Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity 25, 745–755. 10.1016/j.immuni.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 58.Langlais D, Barreiro LB, and Gros P (2016). The macrophage IRF8/IRF1 regulome is required for protection against infections and is associated with chronic inflammation. J. Exp. Med 213, 585–603. 10.1084/jem.20151764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shao L, Hou W, Scharping NE, Vendetti FP, Srivastava R, Roy CN, Menk AV, Wang Y, Chauvin JM, Karukonda P, et al. (2019). IRF1 Inhibits Antitumor Immunity through the Upregulation of PD-L1 in the Tumor Cell. Cancer Immunol. Res 7, 1258–1266. 10.1158/23266066.CIR-18-0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, Zaretsky JM, Sun L, Hugo W, Wang X, et al. (2017). Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep 19, 1189–1201. 10.1016/j.celrep.2017.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Forero A, Ozarkar S, Li H, Lee CH, Hemann EA, Nadjsombati MS, Hendricks MR, So L, Green R, Roy CN, et al. (2019). Differential Activation of the Transcription Factor IRF1 Underlies the Distinct Immune Responses Elicited by Type I and Type III Interferons. Immunity 51, 451–464.e6. 10.1016/j.immuni.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aly S, Mages J, Reiling N, Kalinke U, Decker T, Lang R, and Ehlers S (2009). Mycobacteria-induced granuloma necrosis depends on IRF-1. J. Cell. Mol. Med 13, 2069–2082. 10.1111/j.1582-4934.2008.00470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rettino A, and Clarke NM (2013). Genome-wide Identification of IRF1 Binding Sites Reveals Extensive Occupancy at Cell Death Associated Genes. J. Carcinog. Mutagen 10.4172/21572518.S6-009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Escalante CR, Yie J, Thanos D, and Aggarwal AK (1998). Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature 391, 103–106. 10.1038/34224. [DOI] [PubMed] [Google Scholar]
- 65.Pine R, Decker T, Kessler DS, Levy DE, and Darnell JE (1990). Purification and cloning of interferon-stimulated gene factor 2 (ISGF2): ISGF2 (IRF-1) can bind to the promoters of both beta interferon- and interferon-stimulated genes but is not a primary transcriptional activator of either. Mol. Cell. Biol 10, 2448–2457. 10.1128/mcb.10.6.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meraro D, Hashmueli S, Koren B, Azriel A, Oumard A, Kirchhoff S, Hauser H, Nagulapalli S, Atchison ML, and Levi BZ (1999). Protein-protein and DNA-protein interactions affect the activity of lymphoid-specific IFN regulatory factors. J. Immunol 163, 6468–6478. 10.4049/jimmunol.163.12.6468. [DOI] [PubMed] [Google Scholar]
- 67.Chapgier A, Wynn RF, Jouanguy E, Filipe-Santos O, Zhang S, Feinberg J, Hawkins K, Casanova JL, and Arkwright PD (2006). Human complete Stat-1 deficiency is associated with defective type I and II IFN responses in vitro but immunity to some low virulence viruses in vivo. J. Immunol 176, 5078–5083. 10.4049/jimmunol.176.8.5078. [DOI] [PubMed] [Google Scholar]
- 68.Schaper F, Kirchhoff S, Posern G, Koster M, Oumard A, Sharf R, Levi BZ, and Hauser H (1998). Functional domains of interferon regulatory factor I (IRF-1). Biochem. J 335 (Pt 1), 147–157. 10.1042/bj3350147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ogishi M, Yang R, Aytekin C, Langlais D, Bourgey M, Khan T, Ali FA, Rahman M, Delmonte OM, Chrabieh M, et al. (2021). Inherited PD-1 deficiency underlies tuberculosis and autoimmunity in a child. Nat. Med 27, 1646–1654. 10.1038/s41591-021-01388-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, and Taniguchi T (1988). Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-beta gene regulatory elements. Cell 54, 903–913. 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- 71.Emile JF, Patey N, Altare F, Lamhamedi S, Jouanguy E, Boman F, Quillard J, Lecomte-Houcke M, Verola O, Mousnier JF, et al. (1997). Correlation of granuloma structure with clinical outcome defines two types of idiopathic disseminated BCG infection. J. Pathol 181, 25–30. 10.1002/(SICI)1096-9896(199701)181:1. [DOI] [PubMed] [Google Scholar]
- 72.Kim B-H, Shenoy AR, Kumar P, Das R, Tiwari S, and MacMicking JD (2011). A family of IFN-γ-inducible 65-kD GTPases protects against bacterial infection. Science 332, 717–721. 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- 73.Wandel MP, Kim B-H, Park E-S, Boyle KB, Nayak K, Lagrange B, Herod A, Henry T, Zilbauer M, Rohde J, et al. (2020). Guanylate-binding proteins convert cytosolic bacteria into caspase-4 signaling platforms. Nat. Immunol 21, 880–891. 10.1038/s41590-020-0697-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gaudet RG, Zhu S, Halder A, Kim B-H, Bradfield CJ, Huang S, Xu D, Mamiñska A, Nguyen TN, Lazarou M, et al. (2021). A human apolipoprotein L with detergent-like activity kills intracellular pathogens. Science 373, eabf8113. 10.1126/science.abf8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carlin JM, Borden EC, and Byrne GI (1989). Interferon-induced indoleamine 2,3-dioxygenase activity inhibits Chlamydia psittaci replication in human macrophages. J. Interferon Res 9, 329–337. 10.1089/jir.1989.9.329. [DOI] [PubMed] [Google Scholar]
- 76.Rinkenberger N, Abrams ME, Matta SK, Schoggins JW, Alto NM, and Sibley LD (2021). Over-expression screen of interferon-stimulated genes identifies RARRES3 as a restrictor of Toxoplasma gondii infection. eLife 10, e73137. 10.7554/eLife.73137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shi L, Perin JC, Leipzig J, Zhang Z, and Sullivan KE (2011). Genome-wide analysis of interferon regulatory factor I binding in primary human monocytes. Gene 487. 10.1016/j.gene.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song R, Gao Y, Dozmorov I, Malladi V, Saha I, McDaniel MM, Parameswaran S, Liang C, Arana C, Zhang B, et al. (2021). IRF1 governs the differential interferon-stimulated gene responses in human monocytes and macrophages by regulating chromatin accessibility. Cell Rep 34, 108891. 10.1016/j.celrep.2021.108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Panda D, Gjinaj E, Bachu M, Squire E, Novatt H, Ozato K, and Rabin RL (2019). IRF1 Maintains Optimal Constitutive Expression of Antiviral Genes and Regulates the Early Antiviral Response. Front. Immunol 10, 1019. 10.3389/fimmu.2019.01019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sen N, Sung P, Panda A, and Arvin AM (2018). Distinctive Roles for Type I and Type II Interferons and Interferon Regulatory Factors in the Host Cell Defense against Varicella-Zoster Virus. J. Virol 92, e01151–18. 10.1128/JVI.01151-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, and Rice CM (2011). A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485. 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schoggins JW, and Rice CM (2011). Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol 1, 519–525. 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brien JD, Daffis S, Lazear HM, Cho H, Suthar MS, Gale M, and Diamond MS (2011). Interferon regulatory factor-1 (IRF-1) shapes both innate and CD8(+) T cell immune responses against West Nile virus infection. PLOS Pathog 7, e1002230. 10.1371/journal.ppat.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nair S, Michaelsen-Preusse K, Finsterbusch K, Stegemann-Koniszewski S, Bruder D, Grashoff M, Korte M, Koster M, Kalinke U, Hauser H, et al. (2014). Interferon regulatory factor-1 protects from fatal neurotropic infection with vesicular stomatitis virus by specific inhibition of viral replication in neurons. PLOS Pathog 10, e1003999. 10.1371/journal.ppat.1003999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang P, Gamero AM, and Jensen LE (2019). IL-36 promotes anti-viral immunity by boosting sensitivity to IFN-α/β in IRF1 dependent and independent manners. Nat. Commun 10, 4700. 10.1038/s41467-019-12318-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nair S, Poddar S, Shimak RM, and Diamond MS (2017). Interferon Regulatory Factor 1 Protects against Chikungunya Virus-Induced Immunopathology by Restricting Infection in Muscle Cells. J. Virol 91, e01419–17. 10.1128/JVI.01419-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mboko WP, Rekow MM, Ledwith MP, Lange PT, Schmitz KE, Anderson S, and Tarakanova VL (2017). Interferon Regulatory Factor 1 and Type I Interferon Cooperate To Control Acute Gammaherpesvirus Infection. J. Virol 91, e01444–16. 10.1128/JVI.01444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wheelock EF (1965). Interferon-Like Virus-Inhibitor Induced in Human Leukocytes by Phytohemagglutinin. Science 149, 310–311. 10.1126/science.149.3681.310. [DOI] [PubMed] [Google Scholar]
- 89.Kornbluth RS, Oh PS, Munis JR, Cleveland PH, and Richman DD (1989). Interferons and bacterial lipopolysaccharide protect macrophages from productive infection by human immunodeficiency virus in vitro. J. Exp. Med 169, 1137–1151. 10.1084/jem.169.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Karupiah G, Xie QW, Buller RM, Nathan C, Duarte C, and MacMicking JD (1993). Inhibition of viral replication by interferon-gamma-induced nitric oxide synthase. Science 261, 1445–1448. 10.1126/science.7690156. [DOI] [PubMed] [Google Scholar]
- 91.Sancéau J, Sondermeyer P, Béranger F, Falcoff R, and Vaquero C (1987). Intracellular human gamma-interferon triggers an antiviral state in transformed murine L cells. Proc. Natl. Acad. Sci. U. S. A 84, 2906–2910. 10.1073/pnas.84.9.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Puel A, Bastard P, Bustamante J, and Casanova J-L (2022). Human autoantibodies underlying infectious diseases. J. Exp. Med 219, e20211387. 10.1084/jem.20211387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shih H-P, Ding J-Y, Yeh C-F, Chi C-Y, and Ku C-L (2021). Anti-interferon-γ autoantibody-associated immunodeficiency. Curr. Opin. Immunol 72, 206–214. 10.1016/j.coi.2021.05.007. [DOI] [PubMed] [Google Scholar]
- 94.Oleaga-Quintas C, de Oliveira-Júnior EB, Rosain J, Rapaport F, Deswarte C, Guérin A, Sajjath SM, Zhou YJ, Marot S, Lozano C, et al. (2021). Inherited GATA2 Deficiency Is Dominant by Haploinsufficiency and Displays Incomplete Clinical Penetrance. J. Clin. Immunol 41, 639–657. 10.1007/s10875-020-00930-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Spinner MA, Sanchez LA, Hsu AP, Shaw PA, Zerbe CS, Calvo KR, Arthur DC, Gu W, Gould CM, Brewer CC, et al. (2014). GATA2 deficiency: a protean disorder of hematopoiesis, lymphatics, and immunity. Blood 123, 809–821. 10.1182/blood-2013-07-515528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kamijo R, Harada H, Matsuyama T, Bosland M, Gerecitano J, Shapiro D, Le J, Koh SI, Kimura T, and Green SJ (1994). Requirement for transcription factor IRF-1 in NO synthase induction in macrophages. Science 263, 1612–1615. 10.1126/science.7510419. [DOI] [PubMed] [Google Scholar]
- 97.Cooper AM, Pearl JE, Brooks JV, Ehlers S, and Orme IM (2000). Expression of the nitric oxide synthase 2 gene is not essential for early control of Mycobacterium tuberculosis in the murine lung. Infect. Immun 68, 6879–6882. 10.1128/iai.68.12.6879-6882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yamada H, Mizuno S, and Sugawara I (2002). Interferon regulatory factor 1 in mycobacterial infection. Microbiol. Immunol 46, 751–760. 10.1111/j.1348-0421.2002.tb02760.x. [DOI] [PubMed] [Google Scholar]
- 99.Ko J, Gendron-Fitzpatrick A, and Splitter GA (2002). Susceptibility of IFN regulatory factor-1 and IFN consensus sequence binding protein-deficient mice to brucellosis. J. Immunol 168, 2433–2440. 10.4049/jimmunol.168.5.2433. [DOI] [PubMed] [Google Scholar]
- 100.Man SM, Karki R, Malireddi RK, Neale G, Vogel P, Yamamoto M, Lamkanfi M, and Kanneganti TD (2015). The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat. Immunol 16, 467–475. 10.1038/ni.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lohoff M, Ferrick D, Mittrucker HW, Duncan GS, Bischof S, Rollinghoff M, and Mak TW (1997). Interferon regulatory factor-1 is required for a T helper 1 immune response in vivo. Immunity 6, 681–689. 10.1016/s1074-7613(00)80444-6. [DOI] [PubMed] [Google Scholar]
- 102.Fehr T, Schoedon G, Odermatt B, Holtschke T, Schneemann M, Bachmann MF, Mak TW, Horak I, and Zinkernagel RM (1997). Crucial role of interferon consensus sequence binding protein, but neither of interferon regulatory factor 1 nor of nitric oxide synthesis for protection against murine listeriosis. J. Exp. Med 185, 921–931. 10.1084/jem.185.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Khan IA, Matsuura T, Fonseka S, and Kasper LH (1996). Production of nitric oxide (NO) is not essential for protection against acute Toxoplasma gondii infection in IRF-1−/− mice. J Immunol 156, 636–643. 10.4049/jimmunol.156.2.636. [DOI] [PubMed] [Google Scholar]
- 104.Nathan CF, Prendergast TJ, Wiebe ME, Stanley ER, Platzer E, Remold HG, Welte K, Rubin BY, and Murray HW (1984). Activation of human macrophages. Comparison of other cytokines with interferon-gamma. J. Exp. Med 160, 600–605. 10.1084/jem.160.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huang S, Zhu S, Kumar P, and MacMicking JD (2021). A phase-separated nuclear GBPL circuit controls immunity in plants. Nature 594, 424–429. 10.1038/s41586-021-03572-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim JH, Castroverde CDM, Huang S, Li C, Hilleary R, Seroka A, Sohrabi R, Medina-Yerena D, Huot B, Wang J, et al. (2022). Increasing the resilience of plant immunity to a warming climate. Nature 607, 339–344. 10.1038/s41586-022-04902-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nathan C (2021). Rethinking immunology. Science 373, 276–277. 10.1126/science.abj5637. [DOI] [PubMed] [Google Scholar]
- 108.Medetgul-Ernar K, and Davis MM (2022). Standing on the shoulders of mice. Immunity 55, 1343–1353. 10.1016/j.immuni.2022.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ogasawara K, Hida S, Azimi N, Tagaya Y, Sato T, Yokochi-Fukuda T, Waldmann TA, Taniguchi T, and Taki S (1998). Requirement for IRF-1 in the microenvironment supporting development of natural killer cells. Nature 391, 700–703. 10.1038/35636. [DOI] [PubMed] [Google Scholar]
- 110.Penninger JM, Sirard C, Mittrucker HW, Chidgey A, Kozieradzki I, Nghiem M, Hakem A, Kimura T, Timms E, Boyd R, et al. (1997). The interferon regulatory transcription factor IRF-1 controls positive and negative selection of CD8+ thymocytes. Immunity 7, 243–254. 10.1016/s1074-7613(00)80527-0. [DOI] [PubMed] [Google Scholar]
- 111.Kano S, Sato K, Morishita Y, Vollstedt S, Kim S, Bishop K, Honda K, Kubo M, and Taniguchi T (2008). The contribution of transcription factor IRF1 to the interferon-gamma-interleukin 12 signaling axis and TH1 versus TH-17 differentiation of CD4+ T cells. Nat. Immunol 9, 34–41. 10.1038/ni1538. [DOI] [PubMed] [Google Scholar]
- 112.Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, Kundig TM, Amakawa R, Kishihara K, Wakeham A, et al. (1993). Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell 75, 83–97. 10.1093/nar/23.12.2127. [DOI] [PubMed] [Google Scholar]
- 113.Taki S, Sato T, Ogasawara K, Fukuda T, Sato M, Hida S, Suzuki G, Mitsuyama M, Shin EH, Kojima S, et al. (1997). Multistage regulation of Th1-type immune responses by the transcription factor IRF-1. Immunity 6, 673–679. 10.1016/s1074-7613(00)80443-4. [DOI] [PubMed] [Google Scholar]
- 114.Bigley V, Maisuria S, Cytlak U, Jardine L, Care MA, Green K, Gunawan M, Milne P, Dickinson R, Wiscombe S, et al. (2018). Biallelic interferon regulatory factor 8 mutation: A complex immunodeficiency syndrome with dendritic cell deficiency, monocytopenia, and immune dysregulation. J. Allergy Clin. Immunol 141, 2234–2248. 10.1016/j.jaci.2017.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cytlak U, Resteu A, Pagan S, Green K, Milne P, Maisuria S, McDonald D, Hulme G, Filby A, Carpenter B, et al. (2020). Differential IRF8 Transcription Factor Requirement Defines Two Pathways of Dendritic Cell Development in Humans. Immunity 53, 353–370.e8. 10.1016/j.immuni.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rosain J, Bernasconi A, Prieto E, Caputi L, Le Voyer T, Buda G, Marti M, Bohlen J, Neehus A-L, Castaños C, et al. (2022). Pulmonary Alveolar Proteinosis and Multiple Infectious Diseases in a Child with Autosomal Recessive Complete IRF8 Deficiency. J. Clin. Immunol 10.1007/s10875-022-01250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu J, Cao S, Herman LM, and Ma X (2003). Differential regulation of interleukin (IL)-12 p35 and p40 gene expression and interferon (IFN)-gamma-primed IL-12 production by IFN regulatory factor 1. J. Exp. Med 198, 1265–1276. 10.1084/jem.20030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Salkowski CA, Kopydlowski K, Blanco J, Cody MJ, McNally R, and Vogel SN (1999). IL-12 is dysregulated in macrophages from IRF-1 and IRF-2 knockout mice. J. Immunol 163, 1529–1536. 10.4049/jimmunol.163.3.1529. [DOI] [PubMed] [Google Scholar]
- 119.Le Coz C, Nguyen DN, Su C, Nolan BE, Albrecht AV, Xhani S, Sun D, Demaree B, Pillarisetti P, Khanna C, et al. (2021). Constrained chromatin accessibility in PU.1-mutated agammaglobulinemia patients. J. Exp. Med 218, e20201750. 10.1084/jem.20201750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vély F, Barlogis V, Vallentin B, Neven B, Piperoglou C, Ebbo M, Perchet T, Petit M, Yessaad N, Touzot F, et al. (2016). Evidence of innate lymphoid cell redundancy in humans. Nat. Immunol 17, 1291–1299. 10.1038/ni.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Béziat V (2020). Human genetic dissection of papillomavirus-driven diseases: new insight into their pathogenesis. Hum. Genet 139, 919–939. 10.1007/s00439-020-02183-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jouanguy E, Gineau L, Cottineau J, Béziat V, Vivier E, and Casanova J-L (2013). Inborn errors of the development of human natural killer cells. Curr. Opin. Allergy Clin. Immunol 13, 589–595. 10.1097/ACI.0000000000000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cottineau J, Kottemann MC, Lach FP, Kang Y-H, Vély F, Deenick EK, Lazarov T, Gineau L, Wang Y, Farina A, et al. (2017). Inherited GINS1 deficiency underlies growth retardation along with neutropenia and NK cell deficiency. J. Clin. Invest 127, 1991–2006. 10.1172/JCI90727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mace EM, Paust S, Conte MI, Baxley RM, Schmit MM, Patil SL, Guilz NC, Mukherjee M, Pezzi AE, Chmielowiec J, et al. (2020). Human NK cell deficiency as a result of biallelic mutations in MCM10. J. Clin. Invest 130, 5272–5286. 10.1172/JCI134966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Beziat V, Sleiman M, Goodridge JP, Kaarbo M, Liu LL, Rollag H, Ljunggren HG, Zimmer J, and Malmberg KJ (2015). Polyclonal Expansion of NKG2C(+) NK Cells in TAP-Deficient Patients. Front. Immunol 6, 507. 10.3389/fimmu.2015.00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ardeniz Ö, Unger S, Onay H, Ammann S, Keck C, Cianga C, Gerçeker B, Martin B, Fuchs I, Salzer U, et al. (2015). β2-Microglobulin deficiency causes a complex immunodeficiency of the innate and adaptive immune system. J. Allergy Clin. Immunol 136, 392–401. 10.1016/j.jaci.2014.12.1937. [DOI] [PubMed] [Google Scholar]
- 127.Hanalioglu D, Ayvaz DC, Ozgur TT, van der Burg M, Sanal O, and Tezcan I (2017). A novel mutation in TAP1 gene leading to MHC class I deficiency: Report of two cases and review of the literature. Clin. Immunol 178, 74–78. 10.1016/j.clim.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 128.Yabe T, Kawamura S, Sato M, Kashiwase K, Tanaka H, Ishikawa Y, Asao Y, Oyama J, Tsuruta K, Tokunaga K, et al. (2002). A subject with a novel type I bare lymphocyte syndrome has tapasin deficiency due to deletion of 4 exons by Alu-mediated recombination. Blood 100, 1496–1498. 10.1182/blood-2001-12-0252. [DOI] [PubMed] [Google Scholar]
- 129.Dumontet E, Osman J, Guillemont-Lambert N, Cros G, Moshous D, and Picard C (2015). Recurrent Respiratory Infections Revealing CD8α Deficiency. J. Clin. Immunol 35, 692–695. 10.1007/s10875-015-0213-x. [DOI] [PubMed] [Google Scholar]
- 130.de la Calle-Martin O, Hernandez M, Ordi J, Casamitjana N, Arostegui JI, Caragol I, Ferrando M, Labrador M, Rodriguez-Sanchez JL, and Espanol T (2001). Familial CD8 deficiency due to a mutation in the CD8 alpha gene. J. Clin. Invest 108, 117–123. 10.1172/JCI10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mancebo E, Moreno-Pelayo MA, Mencía A, de la Calle-Martín O, Allende LM, Sivadorai P, Kalaydjieva L, Bertranpetit J, Coto E, Calleja-Antolín S, et al. (2008). Gly111Ser mutation in CD8A gene causing CD8 immunodeficiency is found in Spanish Gypsies. Mol. Immunol 45, 479–484. 10.1016/j.molimm.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 132.Tangye SG, and Latour S (2020). Primary immunodeficiencies reveal the molecular requirements for effective host defense against EBV infection. Blood 135, 644–655. 10.1182/blood.2019000928. [DOI] [PubMed] [Google Scholar]
- 133.Abolhassani H, Landegren N, Bastard P, Materna M, Modaresi M, Du L, Aranda-Guillén M, Sardh F, Zuo F, Zhang P, et al. (2022). Inherited IFNAR1 Deficiency in a Child with Both Critical COVID-19 Pneumonia and Multisystem Inflammatory Syndrome. J. Clin. Immunol 10.1007/s10875-022-01215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Asano T, Boisson B, Onodi F, Matuozzo D, Moncada-Velez M, Maglorius Renkilaraj MRL, Zhang P, Meertens L, Bolze A, Materna M, et al. (2021). X-linked recessive TLR7 deficiency in ~1% of men under 60 years old with life-threatening COVID-19. Sci. Immunol 6, eabl4348. 10.1126/sciimmunol.abl4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann H-H, Zhang Y, Dorgham K, Philippot Q, Rosain J, Béziat V, et al. (2020). Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370. 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bastard P, Gervais A, Le Voyer T, Rosain J, Philippot Q, Manry J, Michailidis E, Hoffmann H-H, Eto S, Garcia-Prat M, et al. (2021). Autoantibodies neutralizing type I IFNs are present in ~4% of uninfected individuals over 70 years old and account for ~20% of COVID-19 deaths. Sci. Immunol 6, eabl4340. 10.1126/sciimmunol.abl4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Khanmohammadi S, Rezaei N, Khazaei M, and Shirkani A (2021). A Case of Autosomal Recessive Interferon Alpha/Beta Receptor Alpha Chain (IFNAR1) Deficiency with Severe COVID-19. J. Clin. Immunol 10.1007/s10875-021-01166-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, Ogishi M, Sabli IKD, Hodeib S, Korol C, et al. (2020). Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370, eabd4570. 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hernandez N, Melki I, Jing H, Habib T, Huang SSY, Danielson J, Kula T, Drutman S, Belkaya S, Rattina V, et al. (2018). Life-threatening influenza pneumonitis in a child with inherited IRF9 deficiency. J. Exp. Med 215, 2567–2585. 10.1084/jem.20180628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ciancanelli MJ, Huang SX, Luthra P, Garner H, Itan Y, Volpi S, Lafaille FG, Trouillet C, Schmolke M, Albrecht RA, et al. (2015). Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science 348, 448–453. 10.1126/science.aaa1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hambleton S, Goodbourn S, Young DF, Dickinson P, Mohamad SMB, Valappil M, McGovern N, Cant AJ, Hackett SJ, Ghazal P, et al. (2013). STAT2 deficiency and susceptibility to viral illness in humans. Proc. Natl. Acad. Sci. U. S. A 110, 3053–3058. 10.1073/pnas.1220098110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Duncan CJA, Mohamad SMB, Young DF, Skelton AJ, Leahy TR, Munday DC, Butler KM, Morfopoulou S, Brown JR, Hubank M, et al. (2015). Human IFNAR2 deficiency: Lessons for antiviral immunity. Sci. Transl. Med 7, 307ra154. 10.1126/scitranslmed.aac4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bastard P, Manry J, Chen J, Rosain J, Seeleuthner Y, AbuZaitun O, Lorenzo L, Khan T, Hasek M, Hernandez N, et al. (2021). Herpes simplex encephalitis in a patient with a distinctive form of inherited IFNAR1 deficiency. J. Clin. Invest 131, e139980. 10.1172/JCI139980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Moens L, Van Eyck L, Jochmans D, Mitera T, Frans G, Bossuyt X, Matthys P, Neyts J, Ciancanelli M, Zhang S-Y, et al. (2017). A novel kindred with inherited STAT2 deficiency and severe viral illness. J. Allergy Clin. Immunol 139, 1995–1997.e9. 10.1016/j.jaci.2016.10.033. [DOI] [PubMed] [Google Scholar]
- 145.Gothe F, Stremenova Spegarova J, Hatton CF, Griffin H, Sargent T, Cowley SA, James W, Roppelt A, Shcherbina A, Hauck F, et al. (2022). Aberrant inflammatory responses to type I interferon in STAT2 or IRF9 deficiency. J. Allergy Clin. Immunol, S0091–6749(22)00185–3. 10.1016/j.jaci.2022.01.026. [DOI] [PubMed] [Google Scholar]
- 146.Kessler DS, Levy DE, and Darnell JE (1988). Two interferon-induced nuclear factors bind a single promoter element in interferon-stimulated genes. Proc. Natl. Acad. Sci. U. S. A 85, 8521–8525. 10.1073/pnas.85.22.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kimura T, Nakayama K, Penninger J, Kitagawa M, Harada H, Matsuyama T, Tanaka N, Kamijo R, Vilcek J, Mak TW, et al. (1994). Involvement of the IRF-1 transcription factor in antiviral responses to interferons. Science 264, 1921–1924. 10.1126/science.8009222. [DOI] [PubMed] [Google Scholar]
- 148.Carlin AF, Plummer EM, Vizcarra EA, Sheets N, Joo Y, Tang W, Day J, Greenbaum J, Glass CK, Diamond MS, et al. (2017). An IRF-3-, IRF-5-, and IRF-7-Independent Pathway of Dengue Viral Resistance Utilizes IRF-1 to Stimulate Type I and II Interferon Responses. Cell Rep 21, 1600–1612. 10.1016/j.celrep.2017.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Charbit-Henrion F, Bègue B, Sierra A, Hanein S, Stolzenberg M-C, Li Z, Pellegrini S, Garcelon N, Jeanpierre M, Neven B, et al. (2018). Copy number variations and founder effect underlying complete IL-10Rβ deficiency in Portuguese kindreds. PLOS One 13, e0205826. 10.1371/journal.pone.0205826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang Q (2020). Human genetics of life-threatening influenza pneumonitis. Hum. Genet 139, 941–948. 10.1007/s00439-019-02108-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Korol CB, Belkaya S, Alsohime F, Lorenzo L, Boisson-Dupuis S, Brancale J, Neehus A-L, Vilarinho S, Zobaida A, Halwani R, et al. (2022). Fulminant Viral Hepatitis in Two Siblings with Inherited IL-10RB Deficiency. J. Clin. Immunol 10.1007/s10875-022-01376-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Andersen LL, Mork N, Reinert LS, Kofod-Olsen E, Narita R, Jorgensen SE, Skipper KA, Honing K, Gad HH, Ostergaard L, et al. (2015). Functional IRF3 deficiency in a patient with herpes simplex encephalitis. J. Exp. Med 212, 1371–1379. 10.1084/jem.20142274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lévy R, Bastard P, Lanternier F, Lecuit M, Zhang S-Y, and Casanova J-L (2021). IFN-α2a Therapy in Two Patients with Inborn Errors of TLR3 and IRF3 Infected with SARS-CoV-2. J. Clin. Immunol 41, 26–27. 10.1007/s10875-020-00933-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bravo Garcia-Morato M, Calvo Apalategi A, Bravo-Gallego LY, Blazquez Moreno A, Simon-Fuentes M, Garmendia JV, Mendez Echevarria A, Del Rosal Rabes T, Dominguez-Soto A, Lopez-Granados E, et al. (2019). Impaired control of multiple viral infections in a family with complete IRF9 deficiency. J. Allergy Clin. Immunol 144, 309–312.e10. 10.1016/j.jaci.2019.02.019. [DOI] [PubMed] [Google Scholar]
- 155.Pinto FO, Leblanc T, Chamousset D, Le Roux G, Brethon B, Cassinat B, Larghero J, de Villartay J-P, Stoppa-Lyonnet D, Baruchel A, et al. (2009). Diagnosis of Fanconi anemia in patients with bone marrow failure. Haematologica 94, 487–495. 10.3324/haematol.13592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Soulier J (2011). Fanconi anemia. Hematol. Am. Soc. Hematol. Educ. Program 2011, 492–497. 10.1182/asheducation-2011.1.492. [DOI] [PubMed] [Google Scholar]
- 157.Martínez-Barricarte R, de Jong SJ, Markle J, de Paus R, Boisson-Dupuis S, Bustamante J, van de Vosse E, Fleckenstein B, and Casanova J-L (2016). Transduction of Herpesvirus saimiri-Transformed T Cells with Exogenous Genes of Interest. Curr. Protoc. Immunol 115, 7.21C.1–7.21C.12. 10.1002/cpim.15. [DOI] [PubMed] [Google Scholar]
- 158.Guerin A, Kerner G, Marr N, Markle JG, Fenollar F, Wong N, Boughorbel S, Avery DT, Ma CS, Bougarn S, et al. (2018). IRF4 haploinsufficiency in a family with Whipple’s disease. Elife 7, e32340. 10.7554/eLife.32340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Perez-Riverol Y, Bai J, Bandla C, García-Seisdedos D, Hewapathirana S, Kamatchinathan S, Kundu DJ, Prakash A, Frericks-Zipper A, Eisenacher M, et al. (2022). The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res 50, D543–D552. 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Payne K, Li W, Salomon R, and Ma CS (2020). OMIP-063: 28-Color Flow Cytometry Panel for Broad Human Immunophenotyping. Cytometry A 97, 777–781. 10.1002/cyto.a.24018. [DOI] [PubMed] [Google Scholar]
- 161.Feinberg J, Fieschi C, Doffinger R, Feinberg M, Leclerc T, Boisson-Dupuis S, Picard C, Bustamante J, Chapgier A, Filipe-Santos O, et al. (2004). Bacillus Calmette Guerin triggers the IL-12/IFN-gamma axis by an IRAK-4- and NEMO-dependent, non-cognate interaction between monocytes, NK, and T lymphocytes. Eur. J. Immunol 34, 3276–3284. 10.1002/eji.200425221. [DOI] [PubMed] [Google Scholar]
- 162.Esteve-Solé A, Sologuren I, Martínez-Saavedra MT, Deyà-Martínez À, Oleaga-Quintas C, Martinez-Barricarte R, Martin-Nalda A, Juan M, Casanova J-L, Rodriguez-Gallego C, et al. (2018). Laboratory evaluation of the IFN-γ circuit for the molecular diagnosis of Mendelian susceptibility to mycobacterial disease. Crit. Rev. Clin. Lab. Sci 55, 184–204. 10.1080/10408363.2018.1444580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xu GJ, Kula T, Xu Q, Li MZ, Vernon SD, Ndung’u T, Ruxrungtham K, Sanchez J, Brander C, Chung RT, et al. (2015). Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science 348, aaa0698. 10.1126/science.aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Drutman SB, Mansouri D, Mahdaviani SA, Neehus A-L, Hum D, Bryk R, Hernandez N, Belkaya S, Rapaport F, Bigio B, et al. (2020). Fatal Cytomegalovirus Infection in an Adult with Inherited NOS2 Deficiency. N. Engl. J. Med 382, 437–445. 10.1056/NEJMoa1910640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hasan MR, Rahman M, Khan T, Saeed A, Sundararaju S, Flores A, Hawken P, Rawat A, Elkum N, Hussain K, et al. (2021). Virome-wide serological profiling reveals association of herpesviruses with obesity. Sci. Rep 11, 2562. 10.1038/s41598-021-82213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang P, Bigio B, Rapaport F, Zhang S-Y, Casanova J-L, Abel L, Boisson B, and Itan Y (2018). PopViz: a webserver for visualizing minor allele frequencies and damage prediction scores of human genetic variations. Bioinformatics 34, 4307–4309. 10.1093/bioinformatics/bty536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Khan T, Rahman M, Ali FA, Huang SSY, Ata M, Zhang Q, Bastard P, Liu Z, Jouanguy E, Béziat V, et al. (2021). Distinct antibody repertoires against endemic human coronaviruses in children and adults. JCI Insight 6, e144499. 10.1172/jci.insight.144499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443. 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.REED LJ, and MUENCH H (1938). A SIMPLE METHOD OF ESTIMATING FIFTY PER CENT ENDPOINTS. Am. J. Epidemiol 27, 493–497. 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 170.Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, Taliun SAG, Corvelo A, Gogarten SM, Kang HM, et al. (2021). Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590, 290–299. 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bhargava A, Williart A, Maurin M, Davidson PM, Jouve M, Piel M, Lahaye X, and Manel N (2021). Inhibition of HIV infection by structural proteins of the inner nuclear membrane is associated with reduced chromatin dynamics. Cell Rep 36, 109763. 10.1016/j.celrep.2021.109763. [DOI] [PubMed] [Google Scholar]
- 172.Halldorsson BV, Eggertsson HP, Moore KHS, Hauswedell H, Eiriksson O, Ulfarsson MO, Palsson G, Hardarson MT, Oddsson A, Jensson BO, et al. (2022). The sequences of 150,119 genomes in the UK Biobank. Nature 607, 732–740. 10.1038/s41586-022-04965-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Manel N, Hogstad B, Wang Y, Levy DE, Unutmaz D, and Littman DR (2010). A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature 467, 214–217. 10.1038/nature09337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ren Z, Povysil G, Hostyk JA, Cui H, Bhardwaj N, and Goldstein DB (2021). ATAV: a comprehensive platform for population-scale genomic analyses. BMC Bioinformatics 22, 149. 10.1186/s12859-021-04071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lahaye X, Satoh T, Gentili M, Cerboni S, Silvin A, Conrad C, Ahmed-Belkacem A, Rodriguez EC, Guichou J-F, Bosquet N, et al. (2016). Nuclear Envelope Protein SUN2 Promotes Cyclophilin-A-Dependent Steps of HIV Replication. Cell Rep 15, 879–892. 10.1016/j.celrep.2016.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Scott EM, Halees A, Itan Y, Spencer EG, He Y, Azab MA, Gabriel SB, Belkadi A, Boisson B, Abel L, et al. (2016). Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat. Genet 48, 1071–1076. 10.1038/ng.3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mendoza EJ, Manguiat K, Wood H, and Drebot M (2020). Two Detailed Plaque Assay Protocols for the Quantification of Infectious SARS-CoV-2. Curr. Protoc. Microbiol 57, ecpmc105. 10.1002/cpmc.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kars ME, Başak AN, Onat OE, Bilguvar K, Choi J, Itan Y, Çağlar C, Palvadeau R, Casanova J-L, Cooper DN, et al. (2021). The genetic structure of the Turkish population reveals high levels of variation and admixture. Proc. Natl. Acad. Sci. U. S. A 118, e2026076118. 10.1073/pnas.2026076118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yi Z, Sperzel L, Nürnberger C, Bredenbeek PJ, Lubick KJ, Best SM, Stoyanov CT, Law LMJ, Yuan Z, Rice CM, et al. (2011). Identification and characterization of the host protein DNAJC14 as a broadly active flavivirus replication modulator. PLOS Pathog 7, e1001255. 10.1371/journal.ppat.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Rapaport F, Boisson B, Gregor A, Béziat V, Boisson-Dupuis S, Bustamante J, Jouanguy E, Puel A, Rosain J, Zhang Q, et al. (2021). Negative selection on human genes underlying inborn errors depends on disease outcome and both the mode and mechanism of inheritance. Proc. Natl. Acad. Sci. U. S. A 118. 10.1073/pnas.2001248118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lim HK, Huang SXL, Chen J, Kerner G, Gilliaux O, Bastard P, Dobbs K, Hernandez N, Goudin N, Hasek ML, et al. (2019). Severe influenza pneumonitis in children with inherited TLR3 deficiency. J. Exp. Med 216, 2038–2056. 10.1084/jem.20181621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Testa U, Stellacci E, Pelosi E, Sestili P, Venditti M, Orsatti R, Fragale A, Petrucci E, Pasquini L, Belardelli F, et al. (2004). Impaired myelopoiesis in mice devoid of interferon regulatory factor 1. Leukemia 18, 1864–1871. 10.1038/sj.leu.2403472. [DOI] [PubMed] [Google Scholar]
- 183.Hernandez N, Bucciol G, Moens L, Le Pen J, Shahrooei M, Goudouris E, Shirkani A, Changi-Ashtiani M, Rokni-Zadeh H, Sayar EH, et al. (2019). Inherited IFNAR1 deficiency in otherwise healthy patients with adverse reaction to measles and yellow fever live vaccines. J. Exp. Med 216, 2057–2070. 10.1084/jem.20182295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.McElligott DL, Phillips JA, Stillman CA, Koch RJ, Mosier DE, and Hobbs MV (1997). CD4+ T cells from IRF-1-deficient mice exhibit altered patterns of cytokine expression and cell subset homeostasis. J. Immunol 159, 4180–4186. 10.4049/jimmunol.159.9.4180. [DOI] [PubMed] [Google Scholar]
- 185.Wang Z, Liu Y, Lin W, and Cui S (2012). A real-time PCR to detect and analyze virulent EMCV loads in sows and piglets. Mol. Biol. Rep 39, 10013–10017. 10.1007/s11033-012-1870-y. [DOI] [PubMed] [Google Scholar]
- 186.Gabriele L, Fragale A, Borghi P, Sestili P, Stellacci E, Venditti M, Schiavoni G, Sanchez M, Belardelli F, and Battistini A (2006). IRF-1 deficiency skews the differentiation of dendritic cells toward plasmacytoid and tolerogenic features. J. Leukoc. Biol 80, 1500–1511. 10.1189/jlb.0406246. [DOI] [PubMed] [Google Scholar]
- 187.Gao D, Ciancanelli MJ, Zhang P, Harschnitz O, Bondet V, Hasek M, Chen J, Mu X, Itan Y, Cobat A, et al. (2021). TLR3 controls constitutive IFN-β antiviral immunity in human fibroblasts and cortical neurons. J. Clin. Invest 131, 134529. 10.1172/JCI134529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wüst S, Schad P, Burkart S, and Binder M (2021). Comparative Analysis of Six IRF Family Members in Alveolar Epithelial Cell-Intrinsic Antiviral Responses. Cells 10, 2600. 10.3390/cells10102600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lachmann N, Happle C, Ackermann M, Lüttge D, Wetzke M, Merkert S, Hetzel M, Kensah G, Jara-Avaca M, Mucci A, et al. (2014). Gene correction of human induced pluripotent stem cells repairs the cellular phenotype in pulmonary alveolar proteinosis. Am. J. Respir. Crit. Care Med 189, 167–182. 10.1164/rccm.201306-1012OC. [DOI] [PubMed] [Google Scholar]
- 190.Bastard P, Michailidis E, Hoffmann H-H, Chbihi M, Le Voyer T, Rosain J, Philippot Q, Seeleuthner Y, Gervais A, Materna M, et al. (2021). Auto-antibodies to type I IFNs can underlie adverse reactions to yellow fever live attenuated vaccine. J. Exp. Med 218, e20202486. 10.1084/jem.20202486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ackermann M, Lachmann N, Hartung S, Eggenschwiler R, Pfaff N, Happle C, Mucci A, Göhring G, Niemann H, Hansen G, et al. (2014). Promoter and lineage independent anti-silencing activity of the A2 ubiquitous chromatin opening element for optimized human pluripotent stem cell-based gene therapy. Biomaterials 35, 1531–1542. 10.1016/j.biomaterials.2013.11.024. [DOI] [PubMed] [Google Scholar]
- 192.Casanova J-L, and Abel L (2022). From rare disorders of immunity to common determinants of infection: Following the mechanistic thread. Cell 185, 3086–3103. 10.1016/j.cell.2022.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Warlich E, Kuehle J, Cantz T, Brugman MH, Maetzig T, Galla M, Filipczyk AA, Halle S, Klump H, Schöler HR, et al. (2011). Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol. Ther. J. Am. Soc. Gene Ther 19, 782–789. 10.1038/mt.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Blanco-Melo D, Nilsson-Payant BE, Liu W-C, Uhl S, Hoagland D, Møller R, Jordan TX, Oishi K, Panis M, Sachs D, et al. (2020). Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 181, 1036–1045.e9. 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Neehus A-L, Moriya K, Nieto-Patlán A, Le Voyer T, Lévy R, Özen A, Karakoc-Aydiner E, Baris S, Yildiran A, Altundag E, et al. (2021). Impaired respiratory burst contributes to infections in PKCδ-deficient patients. J. Exp. Med 218, e20210501. 10.1084/jem.20210501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Thorne LG, Bouhaddou M, Reuschl A-K, Zuliani-Alvarez L, Polacco B, Pelin A, Batra J, Whelan MVX, Hosmillo M, Fossati A, et al. (2022). Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature 602, 487–495. 10.1038/s41586-021-04352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wang L, Wang S, and Li W (2012). RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185. 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 199.Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.McCarthy DJ, Chen Y, and Smyth GK (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288–4297. 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Takaki K, Davis JM, Winglee K, and Ramakrishnan L (2013). Evaluation of the pathogenesis and treatment of Mycobacterium marinum infection in zebrafish. Nat. Protoc 8, 1114–1124. 10.1038/nprot.2013.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Daher W, Leclercq L-D, Johansen MD, Hamela C, Karam J, Trivelli X, Nigou J, Guérardel Y, and Kremer L (2022). Glycopeptidolipid glycosylation controls surface properties and pathogenicity in Mycobacterium abscessus. Cell Chem. Biol 29, 910–924.e7. 10.1016/j.chembiol.2022.03.008. [DOI] [PubMed] [Google Scholar]
- 205.Lévy R, Langlais D, Béziat V, Rapaport F, Rao G, Lazarov T, Bourgey M, Zhou YJ, Briand C, Moriya K, et al. (2021). Inherited human c-Rel deficiency disrupts myeloid and lymphoid immunity to multiple infectious agents. J. Clin. Invest 131. 10.1172/JCI150143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.McGinnis CS, Murrow LM, and Gartner ZJ (2019). DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst 8, 329–337.e4. 10.1016/J.CELS.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2019). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol 37, 38–44. 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 209.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 16, 278. 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, and Vilo J (2019). g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 47, W191–W198. 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Duerr CU, McCarthy CDA, Mindt BC, Rubio M, Meli AP, Pothlichet J, Eva MM, Gauchat J-F, Qureshi ST, Mazer BD, et al. (2016). Type I interferon restricts type 2 immunopathology through the regulation of group 2 innate lymphoid cells. Nat. Immunol 17, 65–75. 10.1038/ni.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Bernut A, Herrmann J-L, Kissa K, Dubremetz J-F, Gaillard J-L, Lutfalla G, and Kremer L (2014). Mycobacterium abscessus cording prevents phagocytosis and promotes abscess formation. Proc. Natl. Acad. Sci. U. S. A 111, E943–952. 10.1073/pnas.1321390111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Manicassamy B, Medina RA, Hai R, Tsibane T, Stertz S, Nistal-Villán E, Palese P, Basler CF, and García-Sastre A (2010). Protection of mice against lethal challenge with 2009 H1N1 influenza A virus by 1918-like and classical swine H1N1 based vaccines. PLOS Pathog 6, e1000745. 10.1371/journal.ppat.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Larman HB, Zhao Z, Laserson U, Li MZ, Ciccia A, Gakidis MAM, Church GM, Kesari S, Leproust EM, Solimini NL, et al. (2011). Autoantigen discovery with a synthetic human peptidome. Nat. Biotechnol 29, 535–541. 10.1038/nbt.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 – Identification of homozygous complete loss-of-function IRF1 variants in patients with severe MSMD, Related to Figure 1. (A) Cells involved in the production of and response to IFN-γ. Proteins for which a mutation of the corresponding gene has been recognized to cause isolated MSMD are depicted in blue, those responsible for syndromic MSMD are shown in red, and those that can cause either isolated MSMD or syndromic MSMD are depicted with crossed lines. (B) Relative clinical severity of isolated MSMD forms vs. relative residual IFN-γ activity. (C) Principal component analysis of WES data from the two patients and our in-house WES database. (D) Consensus negative selection (CoNeS) of IRF1. (E) Diagram of the various IRF1 cDNA constructs used for transient overexpression experiments. (F) RT-qPCR on HEK293T cells without (NT) and with transfection with empty vector (EV) or various IRF1 cDNAs. The bars represent the mean. Data from two to four independent experiments are shown. (G) Western blot for DDK, IRF1, lamin A/C, and vinculin on cytoplasmic (C) and nuclear (N) extracts from HEK293T cells not transfected (NT) or transfected with EV or various IRF1 cDNAs. Data representative of three independent experiments are shown. (H) EMSA and supershift of the nuclear extract of HEK293T cells transfected with EV or various IRF1 cDNAs with DDK tagging of the C-terminus of the protein, incubated with a fluorescent ISRE probe. Representative data from two independent experiments are shown. (I) Dual luciferase ISRE5 reporter activity of HEK293T cells transfected with EV or various IRF1 cDNAs. Bars represent the mean and SD. Data from three to six independent experiments are shown. Statistical analysis was performed with Student’s t-test. ns = not significant, p > 0.05; ****p < 0.0001.
Figure S2 – IRF1 protein levels in cells from IRF1-deficient patients, Related to Figure 2. (A) Western blot for IRF1, IRF3, IRF9, STAT1, and vinculin on total lysate from HVS-T cells from three healthy controls (CTLs), P1, and a patient with complete IRF9 deficiency. Data from a single experiment are shown. (B) IRF1 expression in primary fibroblasts from three CTLs, P1, P2, and a patient with complete IFN-γR1 deficiency, with and without 16 hours of stimulation with 103 IU/mL IFN-γ. Intracellular IRF1 (upper panel) and STAT1 (lower panel) levels, assessed by flow cytometry on (C) HVS-T cells (from a CTL and P1), (D) SV40-fibroblasts (from a CTL, P1, P2, and from a patient with complete STAT1 deficiency), or (E) EBV-B cells (from a CTL, P1, P2, and from a patient with complete STAT1 deficiency). (F) IRF1 and STAT1 expression in SV40-fibroblasts from three CTLs, P1, and a patient with complete IFN-γR1 deficiency, with and without 16 hours of stimulation with 103 IU/mL IFN-γ, or in HEK293T cells transfected with p.R129-DDK IRF1 cDNA. Data representative of three independent experiments are shown.
Figure S3 – Leukocyte development in IRF1-deficient patients and in IRF1-deficient mice, Related to Figure 3. (A) Monitoring of counts of leukocytes, monocytes and lymphocytes, polymorphonuclear neutrophils (PMN), polymorphonuclear eosinophils (PME), and polymorphonuclear basophils (PMB) in the blood of the patients, in red. (B) Mass or flow cytometry analysis of cryopreserved PBMC subsets from CTLs, P1, and P2. (C) Flow cytometry analysis of dendritic cell (DC) subsets in fresh PBMCs from a CTL, P1, and P2. (D) Flow cytometry immunophenotyping of live CD45+ lymphocytes from PBMCs, for ILCP (CD117+CRTh2−) and ILC2 (CRTh2+), in CTLs, P1, and P2. (E) Conventional flow cytometry counts of cDC1, cDC2, NK cells, ILC2, CD4+ T cells, Treg and CD8+ T cells, in the lungs of WT mice (n=5), and IRF1-deficient mice (n=5). (F) Conventional flow cytometry counts of Ly6C+ monocytes, cDC1, cDC2, pDCs, NK cells, CD4+ T cells, Treg and CD8+ T cells in the spleen of WT mice (n=5), and IRF1-deficient mice (n=5). (G) Conventional flow cytometry counts of CLP, CHILP, and ILC2P in the bone marrow of WT mice (n=5), and IRF1-deficient mice (n=5). Unpaired two-tailed t-tests were used for the statistical analysis. *p < 0.05; ***p < 0.001.
Figure S4 – IFN-γ production pathway in P1 and P2, Related to Figure 4. (A) Western blot analysis of ISG15 and MX1 induction in the SV40-fibroblasts of three CTLs, P1, P2, one patient with complete ISG15-deficiency, and one patient with complete IFN-αR1 deficiency. (B) pSTAT4 levels in HVS-T cells from two controls (CTLs), P1, and a patient with complete IL-12Rβ1 deficiency without stimulation (gray), and after 15 minutes of stimulation with 50 ng/mL IL-12 (solid black line), or 105 IU/mL IFN-α2b (dotted black line). Data representative of two independent experiments are shown. (C) Production of IFN-γ by the HVS-T cells of controls (CTLs), P1, and a patient with complete IFN-γ deficiency with and without 2 hours of stimulation with brefeldin, anti-CD3/CD2/CD28 mAb-coated beads, or with PMA-ionomycin. Bars represent the mean and SD. Data from one experiment are shown. (D) Analysis of IFN-γ+ cells on intracellular flow cytometry for various PBMC subsets, following 48 hours of stimulation with IL-12, IL-23, or BCG. Bars represent the mean and SD. Technical duplicates of the same experiment are shown for P1 and P2. (E) Intracellular flow cytometry analysis of IFN-γ, TNF, IL-9, IL-13, IL-4, IL-17A, IL-17F, IL-2, and IL-21 levels in naïve CD4+ T cells from P1 and controls after expansion for 5 days with anti-CD2/CD3/CD28 mAb-coated beads and IL-2, followed by culture for 7 days in polarizing conditions, and stimulation with PMA-ionomycin in the presence of brefeldin for 6 hours.
Figure S5 – Response to IFN-γ in non-hematopoietic cells from P1 and P2, Related to Figure 5. IRF1 RT-qPCR (A) and western blot for IRF1, pSTAT1, STAT1 and vinculin (B) on SV40-fibroblasts with and without stimulation with 103 IU/mL IFN-γ. Data from one experiment are shown. (C) Confocal microscopy of IRF1 in SV40-fibroblasts of a CTL, P1, and a STAT1-deficient patient stimulated for the indicated times. Phosphorylation of STAT1, analyzed by staining and flow cytometry after 20 minutes of incubation with or without 105 IU/mL IFN-γ, in EBV-B cells (D), or with or without 103 IU/mL IFN-γ in SV40-fibroblasts (E) from controls (CTLs), the patients (P1 and P2), or patients with complete IFN-γR1 or STAT1 deficiency. Data representative of two independent experiments are shown. Heatmaps of genes differentially expressed (log2FC) in primary fibroblasts from controls or patients, after (F) 2 or (G) 8 hours of stimulation with 103 IU/mL IFN-γ. We show the genes differentially expressed in patients relative to the control group at both time points (2 or 8 hours), for the genes differentially expressed at both time points relative to non-stimulated fibroblasts in the control group, i.e., with a |log2(FC)| > 1 and adj. p-value < 0.05 in controls, and with a |log2(FC)| > 1 and adj. p-value < 0.05 in patients relative to controls at the corresponding time point. We used the IFN-γR1−/− patient as a negative control. (H) Amount of mRNA, assessed by RNA sequencing for IFN-α2b-inducible genes (logFC>2 in controls) in primary fibroblasts from the indicated individuals with and without 30 minutes, 2 hours, and 8 hours of stimulation with IFN-γ. (I) and (J) Flow cytometry analysis of CD274 (PD-L1) expression with and without stimulation for 48 hours with 102, 103, or 104 IU/mL IFN-γ in SV40-fibroblasts with and without stable retrotransduction with EV or WT IRF1 cDNA. (K) Flow cytometry analysis of the CD274 expression of primary fibroblasts with and without 48 hours of stimulation with 102 or 103 IU/mL IFN-γ.
Figure S6 – Response to IFN-γ in myeloid cells from P1 and P2, Related to Figure 6. Induction of IL-12p40 (A) and IL-12p70 (B) secretion in whole-blood assays for local and travel controls (CTLs), an IL-12p40-deficient patient, P1, and P2, following stimulation with live BCG alone or in combination with IFN-γ (5,000 IU/mL). Bars represent the mean. Heatmaps of genes differentially expressed (log2(FC)) in iPSC-derived macrophages (iPSC-MΦ), and monocytes-derived macrophages (MDMs) from controls (CTL) or patients, after (C) 2 or (D) 8 hours of stimulation with 103 IU/mL IFN-γ. We show the genes differentially expressed in patients relative to the control group at both time points (2 or 8 hours), for genes differentially expressed at both time points relative to non-stimulated cells in the control group, i.e., with a |log2(FC)| > 1 and adj. p-value < 0.05 after Benjamini-Hochberg correction in controls, and with a |log2(FC)| > 1 and adj. p-value < 0.05 after correction in patients relative to controls at the corresponding time point. (E) Fold-enrichment in dysregulated genes, as determined by RNA-seq, for the myeloid cells of both patients relative to IRF1 ChIP-seq results for resting human monocytes77 and IFN-γ-activated murine bone marrow-derived macrophages58. (F) RT-qPCR for APOL3 and GBP4 in iPSC-MΦ or MDMs with and without stimulation for 2 and 8 hours with 103 IU/mL IFN-γ. The bars represent the mean. (G) Examples of flow cytometry dot plots obtained for gentamicin protection assay performed on PMA-differentiated WT or IRF1KO, IFN-γR1KO, or STAT1KO THP1-derived macrophages (THP1-Ф) with and without 24 hours of pretreatment with 103 IU/mL IFN-γ and infection with Salmonella Typhimurium-GFP (Stm-GFP) at a MOI of 6 for 8 hours. The percent Salmonella Typhimurium+ cells is displayed in black and the percent Salmonella Typhimuriumhigh cells is displayed in red. (H) Examples of flow cytometry dot plots obtained for Figure 6H.
Figure S7 – Antiviral IFN-α/β and IFN-γ cell-intrinsic immunity of IRF1-deficient fibroblasts, Related to Figure 7. (A) RT-qPCR for IRF1 and MX1 in SV40-fibroblasts from the indicated controls or patients after 2 hours of stimulation with 103 IU/mL IFN-α2a or IFN-γ. (B) IRF1 levels determined by RNA sequencing, in primary fibroblasts stimulated for 30 minutes, 2 hours, or 8 hours with IFN-α2b or IFN-γ. Results are normalized against non-stimulated conditions. (C) Western blot of IRF1, MX1, STAT1, STAT2, IRF9, and vinculin in SV40-fibroblasts from controls (CTLs), the patients (P1, P2), or patients with complete STAT1, STAT2, IRF9, IFN-αR1, or IFN-γR1 deficiencies stimulated for 4 hours with IFN-γ or IFN-α2β. (D) Phosphorylation of STAT1, as assessed by staining and flow cytometry, after 20 minutes of incubation with or without 103 IU/mL IFN-α2b or 10 ng/mL IFN-β, in SV40-fibroblasts or EBV-B cells from two controls (CTLs), the patients (P1, P2), and patients with complete IFN-αR1 or STAT1 deficiency. Data representative of two to three experiments are shown. (E) Venn diagram of genes positively induced in primary fibroblasts of CTLs (log2FC>1) but less strongly induced (log2FC<−1) in IRF1-, STAT1-, or IRF9-deficient primary fibroblasts than in controls after 2 or 8 hours of stimulation with IFN-α2b. (F) Hepatitis A virus (HAV), (G) human immunodeficiency virus-1 (HIV-1), and (H) yellow fever vaccine virus-venus (YF17D-venus) replication in SV40-fibroblasts for the indicated controls or patients (P1 and P2), after pretreatment for 16 hours with the indicated doses of IFN-α2b or IFN-γ. All viral infections were performed in two or three independent experiments.
Table S6 –CITE-seq data, Related to Figure 3. Differential gene expression analysis for each scRNA-seq lymphoid cell cluster and enrichment analysis for genes significantly up- and downregulated in scRNA-seq analyses of patient cells. Log2FC and adjusted p values for the comparison of gene expression of P1.2 and P2 cells with healthy controls cells; an absolute FC ≥1.5 and adj. p value ≤0.001 were considered significant. Gene ontology (biological process), pathway and Transfac motif-enriched genes are analyzed.
Table S7 –RNA sequencing and mass-spectrometry data, Related to Figures 5, 6, and 7. – (Sheet 1) log2FC and adj. p-value for RNA-seq on primary fibroblasts from three controls, both IRF1-deficient patients (P1 and P2), and patients with autosomal recessive deficiencies of IRF9, STAT1, IFN-αR1, IFN-γR1, or IFN-γR2, with and without stimulation for 30 minutes, 2 hours, or 8 hours with 103 IU/mL IFN-γ or IFN-α2b. (Sheet 2) – log2FC and adj. p-value for RNA-seq on iPSC-MΦ from two clones from controls (#11 and #16), P1 (#10), and a patient with autosomal recessive STAT1 deficiency, with and without stimulation for 2 or 8 hours with 103 IU/mL IFN-γ. (Sheet 3) – log2FC and adj. p-value for RNA-seq on MDMs from three controls and P2, with and without stimulation for 2 or 8 hours with 103 IU/mL IFN-γ. (Sheet 4) – Mass spectrometry quantification of protein extracted from primary fibroblasts from the indicated individuals with and without prior stimulation for 24 hours. Values were log2-transformed and normalized against the median.
Data Availability Statement
RNA-seq, single-cell RNA-seq, and CITE-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. The mass spectrometry proteomics data have been deposited with the ProteomeXchange Consortium via the PRIDE partner repository and datasets identifier is listed in the key resource table. Original western blot images, flow cytometry data, mass spectrometry data, and microscopy data reported in this paper will be shared by the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Human IRF1 (clone D5E4) | Cell Signaling | Cat# 8478, RRID:AB_10949108 |
| Human IRF1 (rabbit polyclonal) | Proteintech | Cat# 11335-1-AP, RRID:AB_2877759 |
| Human IRF8 (goat polyclonal) | Santa Cruz | Cat# sc-6058, RRID:AB_649510 |
| Human IRF8 (clone D20D8) | Cell signaling | Cat# 5628, RRID:AB_10828231 |
| Human IRF9 (rabbit polyclonal) | Santa Cruz | Cat# sc-496, RRID:AB_2127709 |
| Human IRF9 (rabbit polyclonal) | Proteintech | Cat# 14167-1-AP, RRID:AB_2296227 |
| Human IRF3 (clone D9J5Q) | Cell Signaling | Cat# 10949, RRID:AB_2797733 |
| Human STAT1 (clone 1) | Beckton-Dickinson | Cat# 610115, RRID:AB_397521 |
| Human pSTAT1 (clone 4a) | Beckton-Dickinson | Cat# 612232, RRID:AB_399555 |
| Human STAT2 (clone B-3) | Santa Cruz | Cat# sc-514193, RRID:AB_2810271 |
| Human MX1 (polyclonal) | ProteinTech | Cat# 13750-1-AP, RRID:AB_2266768 |
| Human ISG15 (clone F-9) | Santa-Cruz | Cat# sc-166755, RRID:AB_2126308 |
| Human vinculin (clone EPR8185) | Abcam | Cat# ab129002, RRID:AB_11144129 |
| Human vinculin (clone 7F9) | Santa Cruz | Cat# sc-376248-HRP, RRID:AB_10991536 |
| Anti-mouse IgG (H + L)-HRP-conjugated | Bio-Rad | Cat# 170-6516, RRID:AB_11125547 |
| Anti-rabbit IgG (H + L)-HRP-conjugated | Bio-Rad | Cat# 170-6515, RRID:AB_11125142 |
| Human DDK-tag (clone M2) | Sigma-Aldrich | Cat# A8592, RRID:AB_439702 |
| Human DDK (clone M2) | Cell Signaling | Cat# 14793, RRID:AB_2572291 |
| Human Isotype rabbit | Cell Signaling | Cat# 2729, RRID:AB_1031062) |
| Human GBP1 (clone 1B1) | Santa-Cruz | Cat# sc-53857, RRID:AB_2109333 |
| Human APOL3 (clone EPR8238) | Abcam | Cat# ab154869 |
| Human RARRES3 (rabbit polyclonal) | ProteinTech | Cat# 12065-1-AP, RRID:AB_2175704 |
| anti-rabbit IgG Alexa Fluor 555 | ThermoFischer Scientific | Cat# A-21429, RRID:AB_2535850 |
| PE-Dazzle-594- Human PD-L1 (CD274) antibody (clone 29E.2A3) | BioLegend | Cat# 329732, RRID:AB_2616889 |
| PE-Dazzle-594-Mouse IgG2b, κ Isotype Ctrl Antibody | BioLegend | Cat# 400358 |
| PE-Human STAT1 (clone 1) | Beckton-Dickinson | Cat# 558537, RRID:AB_647231 |
| PE-Human pSTAT1 (clone 4a) | Beckton-Dickinson | Cat# 612564, RRID:AB_399855 |
| AF647-Human pSTAT1 (clone 4a) | Beckton-Dickinson | Cat# 612597, RRID:AB_399880) |
| PE Mouse IgG1, κ Isotype Control | Beckton-Dickinson | Cat# 554680, RRID:AB_395506 |
| AF647 Mouse IgG1, κ Isotype Control | Beckton-Dickinson | Cat# 565363, RRID:AB_2869665 |
| PE-conjugated goat anti-rabbit | Thermo Fisher Scientific | Cat# A10542, RRID:AB_2534042 |
| Human IFN-γ-BUV737 clone 4S.B3 | BD Horizon | Cat# 564620, RRID:AB_2869591 |
| Human TNF-PerCP clone Mab11 | BioLegend | Cat# 502924, RRID:AB_2561288 |
| Human IL-9-PE clone MH9A3 | BD Pharmingen | Cat# 560807, RRID:AB_2033985 |
| Human IL-13-BV421 clone JES10-5A2 | BD Horizon | Cat# 563580, RRID:AB_2738290 |
| Human IL-4-AF488 clone 8D4-8 | BioLegend | Cat# 500710, RRID:AB_1877131 |
| Human IL-17A-BV510 clone BL168 | BioLegend | Cat# 512330, RRID:AB_2562745 |
| Human IL-17F-BV650 clone O33-782 | BD Horizon | Cat# 564264, RRID:AB_2869555 |
| Human IL-2-BV750 clone MQ1-17H12 | BD Horizon | Cat# 566361, RRID:AB_2739710 |
| Human IL-21-eF660 clone eBio3A3-N2 | Thermo Fisher Scientific | Cat# 50-7219-42, RRID:AB_10598202 |
| Human aCD3-Alexa532 (Clone UCHT1) | Thermo Fisher Scientific | Cat# 58-0038-42, RRID:AB_11218675 |
| Human γδTCR-FITC (clone) | Thermo Fisher Scientific | Cat# 11-9959-42, RRID:AB_10669049 |
| Human Vδ2-APC-Fire750) | BioLegend | Cat# 331420, RRID:AB_2687326 |
| Human CD56-BV605 (clone 5.1H11) | BioLegend | Cat# 362538, RRID:AB_2565856 |
| Human CD4-BV750 | BD Biosciences | Cat# 566356, RRID:AB_2744426 |
| Human CD8a-BV510 (clone RPA-T8) | BioLegend | Cat# 301047, RRID:AB_2561378 |
| Human Vα7.2-BV711 (clone 3C10) | BioLegend | Cat# 351731, RRID:AB_2629679 |
| Human Vα24-Jα18-PE-Cy7 (clone 6B11) | BioLegend | Cat# 342912, RRID:AB_2562230 |
| Human Vδ1-Vioblue | Miltenyi Biotec | Cat# 30-100-555 |
| Human CD161-PE (clone HP-3G10) | BioLegend | Cat# 339938, RRID:AB_2564141 |
| Human Vβ11-APC (Miltenyi Biotec) | Miltenyi Biotec | Cat# 58-0038-42, RRID:AB_11218675 |
| Human CD1a-biotin (clone HI149) | BioLegend | Cat# 300112, RRID:AB_389344 |
| Human CD14-biotin (clone 61D3) | Invitrogen | Cat# 13-0149-82, RRID:AB_466373 |
| Human CD34-biotin (clone 4H11) | BioLegend | Cat# 316404 |
| Human CD123-biotin (clone 6H6) | BioLegend | Cat# 306004, RRID:AB_314578 |
| Human CD203c-biotin (clone FR316A11) | Miltenyi Biotec | Cat# 130-092-345, RRID:AB_615067 |
| Human CD303-biotin (clone AC144) | Miltenyi Biotec | Cat# 130-090-691, RRID:AB_244166 |
| Human FcεRIα-biotin (clone AER-37 CRA-1) | BioLegend | Cat# 334606, RRID:AB_2571885 |
| Human TCRαβ-biotin (clone IP26) | BioLegend | Cat# 306704, RRID:AB_314632 |
| Human TCRγδ-biotin (clone B1) | BD Biosciences | Cat# 555716, RRID:AB_396060 |
| Human CD4 FITC (clone OKT4) | BioLegend | Cat# 317408, RRID:AB_571951 |
| Human CD336 PerCP-eFluor710 (clone 44.189) | Thermo Fisher Scientific | Cat# 46-3369-42, RRID:AB_2573749 |
| Human EOMES PE (clone WD1928) | Thermo Fisher Scientific | Cat# 12-4877-42, RRID:AB_2572615 |
| Human CD8a PE-CF594 (clone RPA-T8) | BD Biosciences | Cat# 562282, RRID:AB_11154052 |
| Human CD127 PE-Cy7 (clone eBioRDR5) | Thermo Fisher Scientific | Cat# 25-1278-42, RRID:AB_1659672 |
| Human CD294 AF647 (clone BM16) | BD Biosciences | Cat# 558042, RRID:AB_2112699 |
| Human CD161 AF700 (clone HP-3G10) | BioLegend | Cat# 339942, RRID:AB_2565870 |
| Human CD94 APC-Fire750 (clone DX22) | BioLegend | Cat# A305-518A, RRID:AB_2773751 |
| Human CD335 BV421 (clone 9E2/NKp46) | BD Biosciences | Cat# 564065, RRID:AB_2738572 |
| Human CD45RA BV570 (clone HI100) | BioLegend | Cat# 304132, RRID:AB_2563813 |
| Human CD117 BV605 (clone 104D2) | BioLegend | Cat# 313218, RRID:AB_2562025 |
| Human CD3 BV650 (clone UCHT1) | BD Biosciences | Cat# 563852 |
| Human CD7 BV711 (clone M-T701) | BD Biosciences | Cat# 564018, RRID:AB_2738544 |
| Human T-bet BV786 (clone O4-46) | BD Biosciences | Cat# 564141, RRID:AB_2738615 |
| Human CD19 BUV395 (clone SJ25C1) | BD Biosciences | Cat# 563549, RRID:AB_2738272 |
| Human CD16 BUV496 (clone 3G8) | BD Biosciences | Cat# 564653, RRID:AB_2744294 |
| Human CD25 BUV563 (clone 2A3) | BD Biosciences | Cat# 565699, RRID:AB_2744341 |
| Human CD56 BUV737 (clone NCAM16.2) | BD Biosciences | Cat# 564447, RRID:AB_2744432 |
| Human CD45 BUV805 (clone HI30) | BD Biosciences | Cat# 612891, RRID:AB_2870179 |
| Human CD11c (S-HCL-3) | BioLegend | Cat# 371523, RRID:AB_2814332 |
| Human CD141 (M80) | BioLegend | Cat# 344127, RRID:AB_2832671 |
| Human CD161 (HP-3G10) | BioLegend | Cat# 339949, RRID:AB_2832665 |
| Human CD14 (Me5E2) | BioLegend | Cat# 301857, RRID:AB_2800735 |
| Human CD16 (3G8) | BioLegend | Cat# 302063, RRID:AB_2800737 |
| Human CD19 (HIB19) | BioLegend | Cat# 302263, RRID:AB_2800740 |
| Human CD1c (L161) | BioLegend | Cat# 331549, RRID:AB_2832653 |
| Human CD28 (CD28.2) | BioLegend | Cat# 302961, RRID:AB_2800750 |
| Human CD370 (CLEC9A) (8F9) | BioLegend | Cat# 353811, RRID:AB_2876671 |
| Human CD38 (HB-7) | BioLegend | Cat# 356639, RRID:AB_2814303 |
| Human CD3e (UCHT1) | BioLegend | Cat# 300477, RRID:AB_2800722 |
| Human CD4 (RPA-T4) | BioLegend | Cat# 300565, RRID:AB_2800724 |
| Human CD45RA (HI100) | BioLegend | Cat# 304161, RRID:AB_2800763 |
| Human CD45RO (UCHL1) | BioLegend | Cat# 304259, RRID:AB_2800766 |
| Human CD56 (NCAM) (5.1H11) | BioLegend | Cat# 362561, RRID:AB_2814309 |
| Human CD66b (6/40c) | BioLegend | Cat# 392913, RRID:AB_2832738 |
| Human CD69 (FN50) | BioLegend | Cat# 310949, RRID:AB_2800809 |
| Human CD8a (RPA-T8) | BioLegend | Cat# 301069, RRID:AB_2800729 |
| Human TCR gd (B1) | BioLegend | Cat# 331233, RRID:AB_2814200 |
| Human TCR Va7.2 (3C10) | BioLegend | Cat# 351737, RRID:AB_2819993 |
| IgG1 k Isotype Ctl (MOPC-21). | BioLegend | Cat# 400185 |
| 89 Y Human CD45 (clone HI30) PBMC panel 1 | Fluidigm | Cat# 3089003B, RRID:AB_2661851 |
| 113 In Human CD57 (clone HCD57) PBMC panel 1 | BioLegend | Cat# 322302, RRID:AB_535988 |
| 115 In Human CD11c (clone Bu15) PBMC panel 1 | BioLegend | Cat# 337202, RRID:AB_1236381 |
| 141 Pr Human CD33 (clone WM53) PBMC panel 1 | BioLegend | Cat# 303410, RRID:AB_2074243 |
| 142 Nd Human CD19 (clone HIB19) PBMC panel 1 | BioLegend | Cat# 302202, RRID:AB_314232 |
| 143 Nd Human CD45RA (clone HI100) PBMC panel 1 | BioLegend | Cat# 304102, RRID:AB_314406 |
| 144 Nd Human CD141 (clone M80) PBMC panel 1 | BioLegend | Cat# 344102, RRID:AB_2201808 |
| 145 Nd Human CD4 (clone RPA-T4) PBMC panel 1 | BioLegend | Cat# 300502, RRID:AB_314070 |
| 146 Nd Human CD8 (clone RPA-T8) PBMC panel 1 | BioLegend | Cat# 301002, RRID:AB_314120 |
| 147 Sm Human CD20 (clone 2H7) PBMC panel 1 | BioLegend | Cat# 302302, RRID:AB_314250 |
| 148 Nd Human CD16 (clone 3G8) PBMC panel 1 | BioLegend | Cat# 302014, RRID:AB_314214 |
| 149 Sm Human CD127 (clone A019D5) PBMC panel 1 | Fluidigm | Cat# 3149011B, RRID:AB_2661792 |
| 150 Nd Human CD1c (clone L161) PBMC panel 1 | BioLegend | Cat# 331502, RRID:AB_1088995 |
| 151 Eu Human CD123 (clone 6H6) PBMC panel 1 | BioLegend | Cat# 306002, RRID:AB_314576 |
| 152 Sm Human CD66b (clone G10F5) PBMC panel 1 | BioLegend | Cat# 305102, RRID:AB_314494 |
| 153 Eu Human PD-1 (clone EH12.2H7) PBMC panel 1 | BioLegend | Cat# 329926, RRID:AB_11147365 |
| 154 Sm Human CD86 (clone IT2.2) PBMC panel 1 | BioLegend | Cat# 305410, RRID:AB_314530 |
| 155 Gd Human CD27 (clone O323) PBMC panel 1 | BioLegend | Cat# 302802, RRID:AB_314294 |
| 156 Gd Human CCR5 (clone J418F1) PBMC panel 1 | BioLegend | Cat# 359102, RRID:AB_2562457 |
| 158 Gd Human CD117 (clone 104D2) PBMC panel 1 | BioLegend | Cat# 313202, RRID:AB_314981 |
| 159 Tb Human CD24 (clone ML5) PBMC panel 1 | BioLegend | Cat# 311102, RRID:AB_314851 |
| 160 Gd Human CD14 (clone M5E2) PBMC panel 1 | BioLegend | Cat# 301810, RRID:AB_314192 |
| 161 Dy Human CD56 (clone B159) PBMC panel 1 | BD Biosciences | Cat# 555513, RRID:AB_395903 |
| 162 Dy Human gdTCR (clone REA591) PBMC panel 1 | Miltenyi | Cat# 130-122-291; RRID: AB_2801872 |
| 163 Dy Human CRTh2 (clone REA598) PBMC panel 1 | Milentyi | Cat# 130-122-305, RRID:AB_2801886 |
| 164 Dy Human CLEC12A (clone 50C1) PBMC panel 1 | BioLegend | Cat# 353602, RRID:AB_10962440 |
| 165 Ho Human CCR6 (clone G034E3) PBMC panel 1 | BioLegend | Cat# 353402, RRID:AB_10918625 |
| 166 Er Human CD25 (clone M-A251) PBMC panel 1 | BioLegend | Cat# 356102, RRID:AB_2561752 |
| 167 Er Human CCR7 (clone G043H7) PBMC panel 1 | BioLegend | Cat# 353256, RRID:AB_2814291 |
| 168 Er Human CD3 (clone UCHT1) PBMC panel 1 | BioLegend | Cat# 300402, RRID:AB_314056 |
| 169 Tm Human CX3CR1 (clone 2A9-1) PBMC panel 1 | BioLegend | Cat# 341602, RRID:AB_1595422 |
| 170 Er Human CD38 (clone HB-7) PBMC panel 1 | BioLegend | Cat# 356602, RRID:AB_2561794 |
| 171 Yb Human CD161 (clone HP-3G10) PBMC panel 1 | BioLegend | Cat# 339902, RRID:AB_1501090 |
| 172 Yb Human CD209 (clone 9E9A8) PBMC panel 1 | BioLegend | Cat# 330102, RRID:AB_1134253 |
| 173 Yb Human CXCR3 (clone REA232) PBMC panel 1 | Miltenyi | Cat# 130-108-022, RRID:AB_2655743 |
| 174 Yb Human HLADR (clone L243) PBMC panel 1 | BioLegend | Cat# 307602, RRID:AB_314680 |
| 176 Yb Human CCR4 (clone 205410) PBMC panel 1 | R&DSystems | Cat# MAB1567, RRID:AB_2074395 |
| 209 Bi Human CD11b (clone ICRF44) PBMC panel 1 | Fluidigm | Cat# 3209003B, RRID:AB_2687654 |
| 89 Y Human CD45 (clone HI30) PBMC panel 2 | Fluidigm | Cat# 3089003B, RRID:AB_2661851 |
| 113 In Human HLA-ABC (clone W6/32) PBMC panel 2 | BioLegend | Cat# 311402, RRID:AB_314871 |
| 115 In Human CD11c (clone Bu15) PBMC panel 2 | BioLegend | Cat# 337202, RRID:AB_1236381 |
| 141 Pr Human CD33 (clone WM53) PBMC panel 2 | BioLegend | Cat# 303410, RRID:AB_2074243 |
| 142 Nd Human CD19 (clone HIB19) PBMC panel 2 | BioLegend | Cat# 302202, RRID:AB_314232 |
| 143 Nd Human CD45RA (clone HI100) PBMC panel 2 | BioLegend | Cat# 304102, RRID:AB_314406 |
| 144 Nd Human CD141 (clone M80) PBMC panel 2 | BioLegend | Cat# 344102, RRID:AB_2201808 |
| 145 Nd Human CD4 (clone RPA-T4) PBMC panel 2 | BioLegend | Cat# 300502, RRID:AB_314070 |
| 146 Nd Human CD8 (clone RPA-T8) PBMC panel 2 | BioLegend | Cat# 301002, RRID:AB_314120 |
| 147 Sm Human CLEC9A (clone 8F9) PBMC panel 2 | BioLegend | Cat# 353802, RRID:AB_10983070 |
| 148 Nd Human CD16 (clone 3G8) PBMC panel 2 | BioLegend | Cat# 302014, RRID:AB_314214 |
| 149 Sm Human FceRIa (clone AER-37) PBMC panel 2 | BioLegend | Cat# 334602, RRID:AB_1227649 |
| 150 Nd Human CD1c (clone L161) PBMC panel 2 | BioLegend | Cat# 331502, RRID:AB_1088995 |
| 151 Eu Human CD123 (clone 6H6) PBMC panel 2 | BioLegend | Cat# 306002, RRID:AB_314576 |
| 152 Sm Human CD66b (clone G10F5) PBMC panel 2 | BioLegend | Cat# 305102, RRID:AB_314494 |
| 153 Eu Human CD83 (clone HB15e) PBMC panel 2 | BioLegend | Cat# 305302, RRID:AB_314510 |
| 154 Sm Human CD86 (clone IT2.2) PBMC panel 2 | BioLegend | Cat# 305410, RRID:AB_314530 |
| 155 Gd Human CD27 (clone O323) PBMC panel 2 | BioLegend | Cat# 302802, RRID:AB_314294 |
| 156 Gd Human PD-L1 (clone 29E.2A3) PBMC panel 2 | BioLegend | Cat# 329711, RRID:AB_2228868 |
| 158 Gd Human CD163 (clone REA812) PBMC panel 2 | Miltenyi | Cat# 130-122-293, RRID:AB_2801874 |
| 159 Tb Human CD103 (clone Ber-Act8) PBMC panel 2 | BioLegend | Cat# 350202, RRID:AB_10639864 |
| 160 Gd Human CD14 (clone M5E2) PBMC panel 2 | BioLegend | Cat# 301810, RRID:AB_314192 |
| 161 Dy Human CD56 (clone B159) PBMC panel 2 | BD Biosciences | Cat# 555513, RRID:AB_395903 |
| 162 Dy Human CD64 (clone 10,1) PBMC panel 2 | BioLegend | Cat# 305016, RRID:AB_2103461 |
| 163 Dy Human CD172a/b (clone SE5A5) PBMC panel 2 | Fluidigm | Cat# 3163017B, RRID:AB_2864730 |
| 164 Dy Human CD40 (clone HB14) PBMC panel 2 | BioLegend | Cat# 334302, RRID:AB_1236384 |
| 166 Er Human CD169 (clone 7-239) PBMC panel 2 | BioLegend | Cat# 346002, RRID:AB_2189031 |
| 167 Er Human CD117 (clone 104D2) PBMC panel 2 | BioLegend | Cat# 313202, RRID:AB_314981 |
| 168 Er Human CD3 (clone UCHT1) PBMC panel 2 | BioLegend | Cat# 300402, RRID:AB_314056 |
| 169 Tm Human CX3CR1 (clone 2A9-1) PBMC panel 2 | BioLegend | Cat# 341602, RRID:AB_1595422 |
| 170 Er Human CD38 (clone HB-7) PBMC panel 2 | BioLegend | Cat# 356602, RRID:AB_2561794 |
| 171 Yb Human CD207 (clone 1000) PBMC panel 2 | BioLegend | Cat# 352202, RRID:AB_10898115 |
| 172 Yb Human CD206 (clone 44607) PBMC panel 2 | BioLegend | Cat# 321112, RRID:AB_571921 |
| 174 Yb Human HLADR (clone L243) PBMC panel 2 | BioLegend | Cat# 307602, RRID:AB_314680 |
| 175 Lu Human Axl (clone 108724) PBMC panel 2 | R&DSystems | Cat# MAB154, RRID:AB_2062558 |
| 176 Yb Human CD209 (clone 9E9A8) PBMC panel 2 | BioLegend | Cat# 330102, RRID:AB_1134253 |
| 209 Bi Human CD11b (clone ICRF44) PBMC panel 2 | Fluidigm | Cat# 3209003B, RRID:AB_2687654 |
| 163Dy Human CXCR3 (clone G025H7) Whole blood custom panel | Fluidigm | Cat# 3163004B, RRID:AB_2810969 |
| 152Sm Human TCRgd (clone 11F2) Whole blood custom panel | Fluidigm | Cat# 3152008B, RRID:AB_2687643 |
| 142Nd Human CD19 (clone HIB19) Whole blood custom panel | Fluidigm | Cat# 3142001B, RRID:AB_2651155 |
| 144Nd Human CD38 (clone HIT2) Whole blood custom panel | Fluidigm | Cat# 3144014B, RRID:AB_2687640 |
| 151Eu Human CD123 (clone 6H6) Whole blood custom panel | Fluidigm | Cat# 3151001B, RRID:AB_2661794 |
| 153Eu Human Va7.2 (clone 3C10) Whole blood custom panel | Fluidigm | Cat# 3153024B, RRID:AB_2891190 |
| 154Sm Human CD3 (clone UCHT1) Whole blood custom panel | Fluidigm | Cat# 3154003B, RRID:AB_2811086 |
| 155Gd Human CD45RA (clone HI100) Whole blood custom panel | Fluidigm | Cat# 3155011B, RRID:AB_2810246 |
| 158Gd Human CD27 (clone L128) Whole blood custom panel | Fluidigm | Cat# 3158010B, RRID:AB_2858231 |
| 159Tb Human CD1c (clone L161) Whole blood custom panel | Biolegend | Cat# 331502, RRID:AB_1088995 |
| 161Dy Human CLEC9A (clone 8F9) Whole blood custom panel | Fluidigm | Cat# 3161018B, RRID:AB_2810252 |
| 164Dy Human CD161 (clone HP-3G10) Whole blood custom panel | Fluidigm | Cat# 3164009B, RRID:AB_2687651 |
| 168Er Human CD8 (clone SK1) Whole blood custom panel | Fluidigm | Cat# 3168002B, RRID:AB_2892771 |
| 170Er Human iNKT (clone 6B11) Whole blood custom panel | Fluidigm | Cat# 3170015B |
| 175Lu Human CCR4 (clone L291H4) Whole blood custom panel | Fluidigm | Cat# 3175035A, RRID:AB_2921320 |
| 174Yb Human CD4 (clone RPA-T4) Whole blood custom panel | Biolegend | Cat# 300502, RRID:AB_314070 |
| 162Dy Human CD21 (clone REA940) Whole blood custom panel | Miltenyi Biotec Inc. | Cat# 130-124-315, RRID:AB_2811646 |
| 165Ho Human NKG2C (clone REA205) Whole blood custom panel | Miltenyi Biotec Inc. | Cat# 130-122-278, RRID:AB_2801859 |
| 148Nd Human CD20 (clone 2H7) Whole blood custom panel | Biolegend | Cat# 302302, RRID:AB_314250 |
| 173Yb Human HLA-DR (clone L243) Whole blood custom panel | Fluidigm | Cat# 3173005B, RRID:AB_2810248 |
| 156Gd Human CCR10 (clone REA326) Whole blood custom panel | Miltenyi Biotec Inc. | Cat# 130-122-317, RRID:AB_2801898 |
| 089Y Human CD45 (clone HI30) Whole blood custom panel | Fluidigm | Cat# 3089003B, RRID:AB_2661851 |
| 116Cd Human CD66b (clone QA17A51) Whole blood custom panel | Biolegend | Cat# 396902, RRID:AB_2814367 |
| 141Pr Human CCR6 (clone G034E3) Whole blood custom panel | Fluidigm | Cat# 3141003A, RRID:AB_2687639 |
| 143Nd Human CD127 (clone A019D5) Whole blood custom panel | Fluidigm | Cat# 3143012B, RRID:AB_2810240 |
| 147Sm Human CD11c (clone Bu15) Whole blood custom panel | Fluidigm | Cat# 3147008B, RRID:AB_2687850 |
| 149Sm Human CD25 (clone 2A3) Whole blood custom panel | Fluidigm | Cat# 3149010B, RRID:AB_2756416 |
| 150Nd Human NKVFS1 (clone NKVFS1) Whole blood custom panel | Bio-Rad | Cat# MCA2243GA, RRID:AB_323743 |
| 167Er Human CCR7 (clone G043H7) Whole blood custom panel | Fluidigm | Cat# 3167009A, RRID:AB_2858236 |
| 169Tm Human NKG2A (clone Z199) Whole blood custom panel | Fluidigm | Cat# 3169013B, RRID:AB_2756426 |
| 171Yb Human CXCR5 (clone RF8B2) Whole blood custom panel | Fluidigm | Cat# 3171014B, RRID:AB_2858239 |
| 166Er Human CD24 (clone ML5) Whole blood custom panel | Fluidigm | Cat# 3166007B, RRID:AB_2661803 |
| 145Nd Human CD31 (clone WM59) Whole blood custom panel | Fluidigm | Cat# 3145004B, RRID:AB_2737262 |
| 160Gd Human CD14 (clone M5E2) Whole blood custom panel | Fluidigm | Cat# 3160001B, RRID:AB_2687634 |
| 176Yb Human CD56 (clone NCAM16.2) Whole blood custom panel | Fluidigm | Cat# 3176008B, RRID:AB_2661813 |
| 172Yb Human CD57 (clone HNK-1) Whole blood custom panel | Biolegend | Cat# 359602, RRID:AB_2562403 |
| 150Nd Human KIR3DL1L2 (clone REA970) Whole blood custom panel | Miltenyi Biotec Inc. | Cat# 130-126-489, RRID:AB_2889458 |
| 146Nd Human IgD (clone IA6-2) Whole blood custom panel | Fluidigm | Cat# 3146005B, RRID:AB_2811082 |
| 209Bi Human CD16 (clone 3G8) Whole blood custom panel | Fluidigm | Cat# 3209002B, RRID:AB_2756431 |
| eFluor 450 Murine CD3ε (clone 1452C11) | Thermo Fisher Scientific | Cat# 48-0031-82, RRID:A14714 |
| BUV395 Murine CD4 (clone GK1.5) | BD Bioscience | Cat# 563790, RRID:AB_2738426 |
| PE Murine CD5 (clone 53-7.3) | Thermo Fisher Scientific | Cat# 12-0051-82, RRID:AB_465523 |
| eFluor 615 Murine CD8α (clone 53-6.7) | Thermo Fisher Scientific | Cat# 47-0081-82, RRID:AB_1272185 |
| Brilliant Violet 650 Murine CD11b (clone M1/70) | Thermo Fisher Scientific | Cat# 416-0112-82 |
| PE Murine CD11b (clone M1/70) | Thermo Fisher Scientific | Cat# 12-0112-82, RRID:AB_2734869 |
| eFluor 450 Murine CD11b (clone M1/70) | Thermo Fisher Scientific | Cat# 48-0112-82, RRID:AB_1582236 |
| FITC Murine CD11c (clone N418) | Thermo Fisher Scientific | Cat# 11-0114-82, RRID:AB_464940 |
| PE Murine CD11c (clone N418) | Thermo Fisher Scientific | Cat# 12-0114-82, RRID:AB_465552 |
| eFluor 450 Murine CD11c (clone N418) | Thermo Fisher Scientific | Cat# 48-0114-82, RRID:AB_1548654 |
| PE Murine CD19 (clone eBio1D3) | Thermo Fisher Scientific | Cat# 12-0193-82, RRID:AB_657659 |
| Alexa Fluor 660 Murine CD19 (clone eBio1D3) | Thermo Fisher Scientific | Cat# 606-0193-82, RRID:AB_2896251 |
| eFluor 450 Murine CD19 (clone eBio1D3) | Thermo Fisher Scientific | Cat# 48-0193-82, RRID:AB_2734905 |
| Alexa Fluor 488 Murine CD25 (clone PC61.5) | Thermo Fisher Scientific | Cat# 53-0251-82, RRID:AB_763472 |
| Brilliant Violet 711 Murine CD25 (clone PC61.5) | Thermo Fisher Scientific | Cat# 407-0251-82 |
| PerCP-Cyanine5.5 Murine CD26 (clone H194-112) | Thermo Fisher Scientific | Cat# 45-0261-82, RRID:AB_1548738 |
| Brilliant Violet 785 Murine CD44 (clone IM7) | BioLegend | Cat# 103059, RRID:AB_2571953 |
| Brilliant Violet 785 Murine CD45 (clone 30-F11) | BioLegend | Cat# 103149, RRID:AB_2564590 |
| PE/Cyanine7 Murine CD45 (clone 30-F11) | BioLegend | Cat# 103114, RRID:AB_312979 |
| Brilliant Violet 711 Murine CD45 (clone 30-F11) | BioLegend | Cat# 103147, RRID:AB_2564383 |
| Brilliant Violet 650 Murine CD45 (clone 30-F11) | BioLegend | Cat# 103151, RRID:AB_2565884 |
| PE Murine CD45R (B220) (clone RA3-6B2) | Thermo Fisher Scientific | Cat# 12-0452-82, RRID:AB_465671 |
| Brilliant Ultra Violet 395 Murine CD45R (B220) (clone RA3-6B2) | Thermo Fisher Scientific | Cat# 363-0452-82 |
| eFluor 450 Murine CD45R (B220) (clone RA3-6B2) | Thermo Fisher Scientific | Cat# 48-0452-82, RRID:AB_1548761 |
| eFluor 450 Murine CD49b (clone DX5) | Thermo Fisher Scientific | Cat# 48-5971-82, RRID:AB_10671541 |
| PE/Dazzle 594 Murine CD62L (clone MEL-14) | BioLegend | Cat# 104448, RRID:AB_2566163 |
| BV786 Murine CD64 (clone X54-5/7.1) | BD Biosciences | Cat# 741024; RRID:AB_2740644 |
| APC Murine CD86 (clone GL-1) | BioLegend | Cat# 105012, RRID:AB_493342 |
| Brilliant Violet 510 Murine CD90.2 (Thy-1.2) (clone 53-2.1) | BioLegend | Cat# 140319, RRID:AB_2561395 |
| APC Murine CD117 (c-Kit) (clone 2B8) | Thermo Fisher Scientific | Cat# 17-1171-82, RRID:AB_469430 |
| Brilliant Violet 711 Murine CD127 (clone A7R34) | BioLegend | Cat# 135035, RRID:AB_2564577 |
| Brilliant Violet 650 Murine CD127 (clone A7R34) | BioLegend | Cat# 135043, RRID:AB_2629681 |
| PE Murine CD135 (Flt3) (clone A2F10) | BioLegend | Cat# 135306, RRID:AB_1877217 |
| APC Murine CD172a (SIRPa) (clone P84) | Thermo Fisher Scientific | Cat# 17-1721-82, RRID:AB_10733158 |
| PE/Cyanine7 Murine CD317 (BST2, PDCA-1) (clone 927) | BioLegend | Cat# 348416, RRID:AB_2716221 |
| PE/Dazzle 594 Murine F4/80 (clone BM8) | BioLegend | Cat# 123146, RRID:AB_2564133 |
| PE Murine FcεR1α (clone 36951) | Thermo Fisher Scientific | Cat# 12-5898-82, RRID:AB_466028 |
| eFluor 450 Murine FcεR1α (clone 36951) | Thermo Fisher Scientific | Cat# 48-5898-82, RRID:AB_2574086 |
| FITC Murine FOXP3 (clone FJK-16s) | Thermo Fisher Scientific | Cat# 11-5773-82, RRID:AB_465243 |
| eFluor 660 Murine GATA3 (clone TWAJ) | Thermo Fisher Scientific | Cat# 50-9966-42, RRID:AB_10596663 |
| PE-Cyanine7 Murine KLRG1 (clone 2F1) | Thermo Fisher Scientific | Cat# 25-5893-82, RRID:AB_1518768 |
| Biotin Murine Integrin α4β7 (clone DATK32) | BioLegend | Cat# 120612, RRID:AB_11203892 |
| Alexa Fluor 488 Murine Ly-6A/E (Sca-1) (clone E13-161.7) | BioLegend | Cat# 122516, RRID:AB_756201 |
| Murine Ly6C (clone AL-21) | BD Biosciences | Cat# 553104, RRID:AB_394628 |
| PE Murine Ly6G/Ly6C (Gr-1) (clone RB6-8C5) | Thermo Fisher Scientific | Cat# 12-5931-82, RRID:AB_466045 |
| eFluor 450 Murine Ly6G/Ly6C (Gr-1) (clone RB6-8C5) | Thermo Fisher Scientific | Cat# 48-5931-82, RRID:AB_1548788 |
| PerCP-Cy5.5 Murine Ly6G (clone 1A8) | BD Biosciences | Cat# 560602; RRID:AB_1727563 |
| BUV805 Murine MHC Class II (I-A/I-E) (clone M5/114) | BD Biosciences | Cat# 748844; RRID:AB_2873247 |
| PE-Cyanine7 Murine NK1.1 (clone PK136) | Thermo Fisher Scientific | Cat# 25-5941-82, RRID:AB_469665 |
| eFluor 450 Murine NK1.1 (clone PK136) | Thermo Fisher Scientific | Cat# 48-5941-82, RRID:AB_2043877 |
| PerCP-Cyanine5.5 Murine NK1.1 (clone PK136) | Thermo Fisher Scientific | Cat# 45-5941-82, RRID:AB_914361 |
| PE Murine Siglec-F (clone E50-2440) | BD Biosciences | Cat# 562068; RRID:AB_394341 |
| PerCP-eFluor 710 Murine IL-33R (ST2) (clone RMST2-2) | Thermo Fisher Scientific | Cat# 46-9335-82, RRID:AB_2573883 |
| eFluor 450 Murine TCRβ (clone H57-597) | Thermo Fisher Scientific | Cat# 48-5961-82, RRID:AB_11039532 |
| PE Murine TCRβ (clone H57-597) | Thermo Fisher Scientific | Cat# 12-5961-82, RRID:AB_466066 |
| eFluor 450 Murine TCR γ/δ (clone eBioGL3) | Thermo Fisher Scientific | Cat# 48-5711-82, RRID:AB_2574071 |
| PE Murine TCR γ/δ (clone eBioGL3) | Thermo Fisher Scientific | Cat# 12-5711-82, RRID:AB_465934 |
| eFluor 450 Murine TER-119 (clone TER-119) | Thermo Fisher Scientific | Cat# 48-5921-82, RRID:AB_1518808 |
| PE-Cyanine7 Murine TER-119 (clone TER-119) | Thermo Fisher Scientific | Cat# 25-5921-82, RRID:AB_469661 |
| Murine XCR-1 (clone ZET) | Biolegend | Cat# 148204, RRID:AB_2563843 |
| Bacterial and virus strains | ||
| Salmonella enterica subsp. enterica serovar Typhimurium GFP | ATCC | 14028GFP |
| Mycobacterium abscessus sensu stricto, strain CIP104536T, smooth with pTEC27 | Bernut et al., 2014212 | N/A |
| expanded T7 Virscan phage library | S. Elledge (Brigham and Women’s Hospital and Harvard University Medical School, Boston, MA, USA) | VirScan Phage Library, version 3 |
| Vesicular stomatitis virus Indiana (VSV) | Bastard et al., 2021143 | N/A |
| Human immunodeficiency viruses-1 (HIV-1) reporter virus (NL4-3 ΔenvΔnef encoding GFP in nef) | Bhargava et al., 2021171 | N/A |
| Human immunodeficiency viruses-2 (HIV2) reporter virus (ROD9 ΔenvΔnef encoding GFP in nef) | Manel et al., 2010173 | N/A |
| Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) NYC isolate | Zhang et al., 2020138 | GenBank OM345241 |
| Yellow-fever vaccine virus-venus (YF17D-venus) reporter | Yi et al., 2011179 | N/A |
| Influenza A virus A/California/4/2009 (IAV) | Manicassamy et al., 2010213 | N/A |
| Herpes simplex virus 1 (HSV-1) | ATCC | VR-1493 |
| Hepatitis A virus (HAV) reporter virus (HM175/18f-NLuc) | Yamane et al., 201952 | N/A |
| Encephalomyocarditis virus (EMCV) | Gao et al., 2021187 | N/A |
| Biological samples | ||
| Peripheral blood mononuclear cells from indicated individuals | This manuscript | N/A |
| Plasma from indicated individuals | This manuscript | N/A |
| Biopsies from indicated individuals | This manuscript | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Protamine sulfate | Merck | Cat# P3369-10G |
| Recombinant interferon gamma-1b (Imukin) | Clinigen Healthcare France | Cat# 3400955776789 |
| Recombinant interferon alpha-2b (Introna) | MSD France | Cat# 3400934956287 |
| Aldesleukin (Proleukin) | Novartis | Cat# 3400956215867 |
| Recombinant Human IFN-beta 1a (Mammalian) Protein | Bio-Techne | Cat# 11410-2 |
| Collagenase IV | Gibco | Cat# 17104-019 |
| DpnI | New England Biolab | Cat# R0176L |
| M-CSF | Peprotech | Cat# 300-25 |
| IL-3 | Peprotech | Cat# 200-03 |
| M-CSF | # 204-IL-010 | Cat# 216-MC-010 |
| IL-4 | R&D Systems | Cat# 204-IL-010 |
| ProLong Gold with DAPI | Thermo Fisher Scientific | Cat #P36931 |
| Aqua Dead Cell Stain kit | Thermo Fisher Scientific | Cat# L34957 |
| Critical commercial assays | ||
| SureSelect Human All Exon V6 | Agilent | Cat# 5190-8864 |
| Human SNP Array 6.0 | Agilent | Cat# 901153 |
| GoTaq DNA Polymerase | Promega | Cat# M3005 |
| RNeasy Plus Mini Kit | Qiagen | Cat# 74136 |
| Quick-RNA MicroPrep Kit | Zymo | Cat# R1051 |
| Universal PCR Master Mix (2X), no AmpErase UNG | Thermo Fisher Scientific | Cat# 4352042 |
| RiboZero TruSeq Stranded Total RNA Library Prep Kit | Illumina | Cat# 20020596 |
| ELISA IL-12p40 | R&D Systems | Cat# DP400 |
| ELISA IL-12p70 HS | R&D Systems | Cat# HS120 |
| NE-PER nuclear and cytoplasmic extraction reagents | Thermo Fisher Scientific | Cat #78835 |
| RNeasy Plus Mini Kit | Qiagen | Cat# 74136 |
| Quick-RNA MicroPrep Kit | Zymo | Cat# R1051 |
| SuperScript II Reverse Transcriptase | Thermo Fisher Scientific | Cat# 18064014 |
| High-Capacity RNA-to-cDNA Kit | Applied Biosystems | Cat# 4387406 |
| TaqMan Fast Universal PCR Master Mix (2X), no AmpErase UNG | Thermo Fisher Scientific | Cat# 4352042, |
| Deposited data | ||
| RNA sequencing on primary fibroblasts | This manuscript | gene expression omnibus (GEO: GSE218033) |
| RNA sequencing on iPSC-derived macrophages | This manuscript | gene expression omnibus (GEO: GSE218033) |
| RNA sequencing on monocyte-derived macrophages | This manuscript | gene expression omnibus (GEO: GSE218033) |
| Mass-spectrometry on primary fibroblasts | This manuscript | ProteomeXchange (PXD037759) |
| scRNAseq on cryopreserved PBMCs | This manuscript | gene expression omnibus (GEO: GSE216489) |
| Experimental models: Cell lines | ||
| HEK293T cells | ATCC | Cat# CRL-11268, RRID:CVCL_1926 |
| THP1 WT | Song et al., 202178 | ATCC Cat# TIB-202, RRID:CVCL_0006 |
| THP1 IRF1KO | Song et al., 202178 | N/A |
| THP1 IFN-γR1KO clone 30 | This manuscript | N/A |
| THP1 STAT1KO clone 13 | This manuscript | N/A |
| iPSC healthy control clone 11 | Ackermann et al., 2014191 | hCD34-iPSC11 |
| iPSC healthy control clone 16 | Lachmann et al., 2014189 | hCD34-iPSC16 |
| iPSC P1 clone 10 | This manuscript | N/A |
| iPSC P2 clone 3 | This manuscript | N/A |
| iPSC STAT1 | Haake et al., 202050 | iSTAT1_compl |
| Experimental models: Organisms/strains | ||
| C57BL/6 (B6) WT | This manuscript | In-house colony established from Jax # 000664 |
| C57BL/6 (B6) IRF1−/− | This manuscript | Jax # 002762 backcrossed to a B6 background |
| Oligonucleotides | ||
| IRF1 genomic exon 3F | ThermoFischer Scientific | TGGTCTGTTTAAGCCAGCCTC |
| IRF1 genomic exon 3R | ThermoFischer Scientific | CAGAAACACAAGTCTGCCACC |
| IRF1 genomic exon 5F | ThermoFischer Scientific | TTCCACCTCTCACCAAGAACC |
| IRF1 genomic exon 5R | ThermoFischer Scientific | CAGAGAAGGTATCAGGGCTGG |
| IRD700-conjugated ISRE probe-F | Metabion | GATCGGGAAAGGGAAACCGAAACTGAA |
| IRD700-conjugated ISRE probe-R | Metabion | TCAGTTTCGGTTTCCCTTTCCCGATC |
| oligo(dT)12–18 | Thermo Fisher Scientific | Cat# 18418012 |
| IRF1 exons 3–4 qPCR probe | ThermoFischer Scientific | Cat# Hs00971960_m1 |
| IRF1 exons 8–9 | ThermoFischer Scientific | Cat# Hs00971965_m1 |
| GBP4 | ThermoFischer Scientific | Cat# Hs00364728_m1 |
| APOL3 | ThermoFischer Scientific | Cat# Hs00758274_m1 |
| GUSB | ThermoFischer Scientific | Cat# 1702016 |
| IRF1-p.M85A-DDK-F | Eurofins | CAACTTTCGCTGTGCCGCGAACTCCCT GCCAGAT |
| IRF1-p.M85A-DDK-R | Eurofins | ATCTGGCAGGGAGTTCGCGGCACAGC GAAAGTTG |
| IRF1-p.M111A-DDK-F | Eurofins | GCGAGTGTACCGGGCGCTTCCACCTCTC |
| IRF1-p.M111A-DDK-R | Eurofins | GAGAGGTGGAAGCGCCCGGTACACTCGC |
| IRF1-p.Q35*-F | Eurofins | GAGGAGATGATCTTCTAGATCCCATGG AAGC |
| IRF1-p.Q35*-R | Eurofins | GCTTCCATGGGATCTAGAAGATCATCT CCTC |
| IRF1-p.R129*-F | ThermoFischer Scientific | GTCGAAGTCCAGCTGAGATGCTAAG |
| IRF1-p.R129*-R | ThermoFischer Scientific | CTTAGCATCTCAGCTGGACTTCGAC |
| IRF1-p.R129-DDK-F | ThermoFischer Scientific | GCTGGACTTCGACTTTCTTTCTTTTCTCTG |
| IRF1-p.R129-DDK-R | ThermoFischer Scientific | ACGCGTACGCGGCCGCTCGA |
| IRF1-p.W11R-DDK-F | ThermoFischer Scientific | GCATGAGACCCCGGCTAGAGATG |
| IRF1-p.W11R-DDK-R | ThermoFischer Scientific | CATCTCTAGCCGGGGTCTCATGC |
| IRF1-Δ7-8-F | ThermoFischer Scientific | CTGGAGTCAGGGCCTGCTCC |
| IRF1-Δ7-8-R | ThermoFischer Scientific | CTCTTGGAGCAGTCGGAGTGGC |
| IRF1-Δ7-8-DDK-F | ThermoFischer Scientific | TTGAGTAGGTACCCCTTCCCATCCACGTTTG |
| IRF1-Δ7-8-DDK-R | ThermoFischer Scientific | ACGCGTACGCGGCCGCTCGA |
| IRF1-p.A67P-DDK-F | ThermoFischer Scientific | GCCGATACAAACCAGGGGAAAAG |
| IRF1-p.A67P-DDK-R | ThermoFischer Scientific | CTTTTCCCCTGGTTTGTATCGGC |
| IRF1-p.M1_A84del-F | Eurofins | ATGAACTCCCTGCCAGATAT |
| IRF1-p.M1_A84del-R | Eurofins | GGCGATCGCGGCGGCAGATC |
| IRF1 retrovirus F | ThermoFischer Scientific | GATCCATTTAAATTCGAATTCATGCCCA TCACTCGGATGCGC |
| IRF1 retrovirus R | ThermoFischer Scientific | ATCGATACCGTCGACCTCGAGTTAAAC CTTATCGTCGTCATC |
| sgRNA IFNGR1 exon 3F | Eurofins | CACCGACACATTCTACTCACCATCT |
| sgRNA IFNGR1 exon 3R | Eurofins | AAACAGATGGTGAGTAGAATGTGTA |
| sgRNA STAT1 exon 3F | Eurofins | CACCGTATTTGCAGCTCGTTTGTGG |
| sgRNA STAT1 exon 3R | Eurofins | AAACCCACAAACGAGCTGCAAATAA |
| Recombinant DNA | ||
| pCMV6-EV | Origene | Cat# RCPS100001 |
| pCMV6-IRF1-WT-DDK | Origene | Cat# RC203500 |
| pCMV6-IRF1-p.M85A-DDK | This manuscript | N/A |
| pCMV6-IRF1-p.M111A-DDK | This manuscript | N/A |
| pCMV6-IRF1-M85A/M111A-DDK | This manuscript | N/A |
| pCMV6-IRF1-p.Q35* | This manuscript | N/A |
| pCMV6-IRF1-p.Q35*/M85A-DDK | This manuscript | N/A |
| pCMV6-IRF1-p.Q35*/M111A-DDK | This manuscript | N/A |
| pCMV6-IRF1-p.Q35*/M85A/M111A-DDK | This manuscript | N/A |
| pCMV6-IRF1-p.R129* | This manuscript | N/A |
| pCMV6-IRF1-p.R129-DDK | This manuscript | N/A |
| pCMV6-IRF1-p.W11R-DDK | This manuscript | N/A |
| pCMV6-IRF1-Δ7-8 | This manuscript | N/A |
| pCMV6-IRF1-Δ7-8-DDK | This manuscript | N/A |
| pCMV6-IRF1-p.A67P-DDK | This manuscript | N/A |
| pCMV6-IRF1-p.M1_A84del | This manuscript | N/A |
| pGL4.10[luc2] backbone with three GGAAAGGGAAACCGAAACTGAA repeats | Guerin et al., 2018158 | Cat# E6651 |
| pGL4.10[luc2] backbon with five GGGAAAGTGAAACTA repeats | Hernandez et al., 2018183 | N/A |
| pRL-SV40 | Promega | Cat# E2231 |
| lentiCRISPR v2 | Addgene | Cat# 52961 |
| psPAX2 | Addgene | Cat# 12260 |
| pCMV-VSV-G | Addgene | Cat# 8454 |
| pHXB2-Env | NIH-AIDS Reagent Program | Cat# 1069 |
| Software and algorithms | ||
| phip-stat | Larman et al., 2011214 | https://github.com/lasersonlab/phip-stat |
| Bowtie for alignment of PhIP-Seq raw reads | Larman et al., 2011214 | http://bowtie-bio.sourceforge.net/index.shtml |
| R | The R Project for Statistical Computing | https://www.r-project.org |
| DESeq2 | Love et al., 2014201 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Uniform Manifold Approximation and Projection (UMAP) | Becht et al., 2018208 | v.0.3.5 |
| BVAtools | https://bitbucket.org/mugqic/bvatools | https://bitbucket.org/mugqic/bvatools |
| Cell Ranger | 10X Genomics | v3.0.1 for scRNA and v6.0.1 for CITE-seq |
| DoubletFinder package | McGinnis et al., 2019206 | v2.0.3 |
| Seurat R package | Stuart et al., 2019207 | v4.0.2 |
| MAST | Finak et al., 2015209 | https://github.com/RGLab/MAST |
| gProfiler R package | Raudvere et al., 2019210 | https://biit.cs.ut.ee/gprofiler/gost |
| FastQC | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| STAR | Dobin et al., 2013197 | v2.7.3a |
| RSeQC | Wang et al., 2012198 | v0.11.2 |
| HTSeq-count | Anders et al., 2015199 | v3.26.8 |
| edgeR package | McCarthy et al., 2012200 | N/A |
| HOMER | Heinz et al., 2010202 | v4.11 |
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.


