Significance
Our spatiotemporal assessment of wheat diversity using a century of varietal-use evidence for the United States provides a more nuanced view on crop diversity than the widely held views regarding genetic erosion in modern agriculture. Our research has direct implications for UN and related policy forums regarding modern science and its impact on crop biodiversity. As revealed in the win-win outcome brought about by scientifically selected crop varieties, investments in modern crop breeding can help achieve, not undercut, the UN’s SDG biodiversity goals.
Keywords: wheat, diversity, crop variety, variety adoption
Abstract
A prevalent and persistent biodiversity concern is that modern cropping systems lead to an erosion in crop genetic diversity. Although certain trait uniformity provides advantages in crop management and marketing, farmers facing risks from change in climate, pests, and markets are also incentivized to adopt new varieties to address complex and spatially variable genetics, environment, and crop management interactions to optimize crop performance. In this study, we applied phylogenetically blind and phylogenetically informed diversity metrics to reveal significant increases in both the spatial and temporal diversity of the US wheat crop over the past century. Contrary to commonly held perceptions on the negative impact of modern cropping systems on crop genetic diversity, our results demonstrated a win-win outcome where the widespread uptake of scientifically selected varieties increased both crop production and crop diversity.
Halting biodiversity loss is crucial to achieving many of the UN’s sustainable development goals and is a leading development target in its own right (1, 2). Agriculture is seen as both a key cause of the global “biodiversity crisis” (1, pp.19 and 52, 3, 4) and a principal means of addressing it (5, p. 20). The nexus between biodiversity and agriculture is complex and multidimensional. With 36.7% of the world’s land mass in cropping and animal agriculture (6), promoting sustainable agricultural productivity growth is key to feeding a large and still growing world population at affordable prices, while at the same time stalling growth in or shrinking the footprint of agriculture to return areas to wild or at least more natural, biodiverse landscapes. In 2019, more than half (51.4%) of the 8,270 quadrillion calories consumed directly by humans were sourced from just five crops—rice (18.3% of total calories), wheat (18.2%), sugar (6.6%), maize (5.4%), and soybeans (2.9%) (6). This translates into large areas of the world growing the same crop type, for example, 216 million hectares (15.0% of total harvested area) of wheat, 197 million hectares (13.8%) of corn, and 162 million hectares (11.3%) of rice in 2019 (6). Another long-standing biodiversity concern is genetic erosion within cropping agriculture (e.g., refs. 7 and 8), which Brush (9, p. 154) succinctly defined as “...the loss of variability in crop populations” (see also Khoury et al. (10) and the discussion and references therein). Within modern cropping systems, the transition from growing landraces, essentially farmer-bred crop varieties, to scientifically bred varieties is seen as a pivotal point in the narrowing of genetic diversity within agriculture (8, 11). But the subsequent decades of using varieties developed by scientists rather than farmers are perceived as a further, if not the primary, cause of a narrowing genetic variability in crop populations (12, Appendix I; 7, ch. 7).
However, there is a lack of assessment on the diversity of the cropping materials used in farmers’ fields, which matters most in terms of the environmental and economic consequences of those production systems. As Khoury et al. (10) point out, with few exceptions (e.g., refs. 13–15), most of the 105 studies they reviewed “…analyzed trends in the diversity of modern cultivars that were available, registered or bred in a given area, not the extent of their cultivation (e.g., planted area) or the varietal turnover rate” (10, p. 99). In fact, genetic erosion due to scientific selection is neither inevitable, nor necessarily the most probable outcome in farmers’ fields. Duvick (16) noted two countervailing forces regarding crop diversity. On the one hand is the substantial economic gains to be had from the lower unit costs of food production, processing, and consumption that come from uniformity in particular phenotypic traits in certain cropping systems (e.g., uniform crop emergence, flowering and harvesting times; plant, seed, or fruit size, shape or composition; and so on). On the other hand, there are other economically valuable crop traits—e.g., resilience to changes in biotic and abiotic stresses over time, or responsiveness to locational differences in the agro-ecological attributes that affect crop production—that promote the development and use of more diverse germplasm. Another factor that incentivizes more diverse cropping systems is the (time and space) variation in market demand for different quality attributes in modern wheat varieties, including their protein and oil content; test weight; and grain color, size, and shape (see, e.g., ref. 17).
From this economic framing, it naturally follows that an informative assessment of crop genetic diversity considers both the temporal and the spatial dimensions of diversity and ideally factors in the adoption not just the availability of crop genetic resources. To do this, we use a purpose-built dataset to quantify the changing spatiotemporal pattern of varietal diversity in the US wheat crop over the past century spanning the years 1919 to 2019; a period that encompasses most of the time that scientific crop breeding was informed by Mendelian methods of genetic selection (18–20). To do so, we draw on phylogenetically informed metrics of diversity developed in the ecological literature and introduce the notion of a “metacommunity” to enable the joint assessment of changes in temporal and spatial crop diversity. We used the generalized phylogenetic diversity (PD) measure proposed by Chao et al. (21) and applied the Hill numbers (or the effective number of varieties) framework to quantify the changing varietal diversity of the US wheat crop during the past 100 y. The generalized PD and Hill numbers framework incorporates both varietal relatedness and varietal abundance to allow for the joint assessment of the spatiotemporal dynamics of varietal diversity within a cropping system that is heavily reliant on scientifically selected varieties.
Results
Phylogenetically Blind Measures of Wheat Biodiversity.
The United States planted wheat area peaked in 1981 (88.3 million acres), thereafter declining steadily to just 45.5 million acres by 2019 (22). In recent years, around 70% of the acreage was planted to winter wheat, with spring wheat averaging around 25% and durum wheat less than 5% (22). Among all three classes of wheat that were commercially grown across 16 major wheat-producing states during the period 1919 to 2019, we identified a total of 1,353 unique named varieties, each accounting for at least 0.5% state-level area share by market class in the year they were grown.
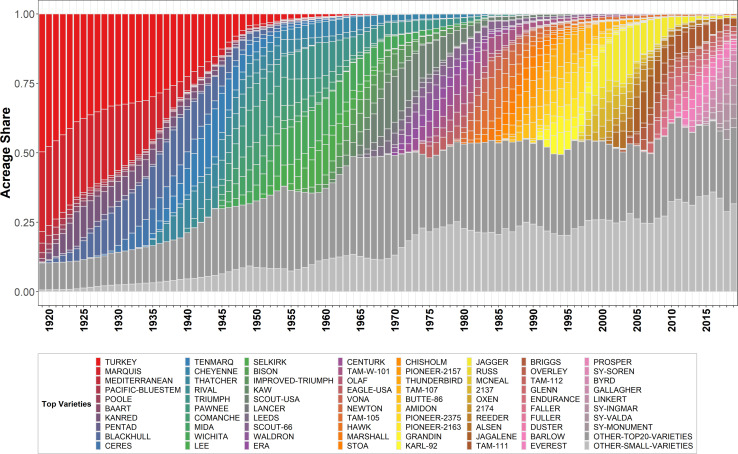
As shown in Fig. 1, the area dynamics of major varieties exhibit a strong regularity. First, the acreage shares attributed to the top wheat varieties decline over time (see decrease in overall colored areas in Fig. 1). In 1919, the top five varieties accounted for 88% of the US wheat planted acreage, by 1964 that share had shrunk to just 34%. The colored plus darker gray elements of Fig. 1 signify the share of area planted to the top 20 varieties (99% in 1919 down to 68% in 2019). Second, the temporal pattern of major variety uptake typically follows an S-shaped function (consistent with Griliches’ 1957 classic study (23) of the US adoption of hybrid corn), followed by a period of dis-adoption as newer varieties gained popularity. Finally, Fig. 1 graphically illustrates the waves of faster varietal turnover that have consistently swept through the US wheat crop over the past century. Specifically, earlier varieties (such as Turkey) that were once dominant for several decades are no longer major varieties, while in recent years the top varieties tend to completely turnover within a period of 5 y.
Fig. 1.
Turnover of the top wheat varieties in the United States, 1919 to 2019. Notes: Colored varieties represent varieties that were among the top five in area share in at least 1 y; “OTHER-TOP20-VARIETIES” represent varieties that were among the top 20 in area share in at least 1 y (excluding the top five varieties colored already); “OTHER-SMALL-VARIETIES” represent the remaining named varieties (excluding unknown varieties).
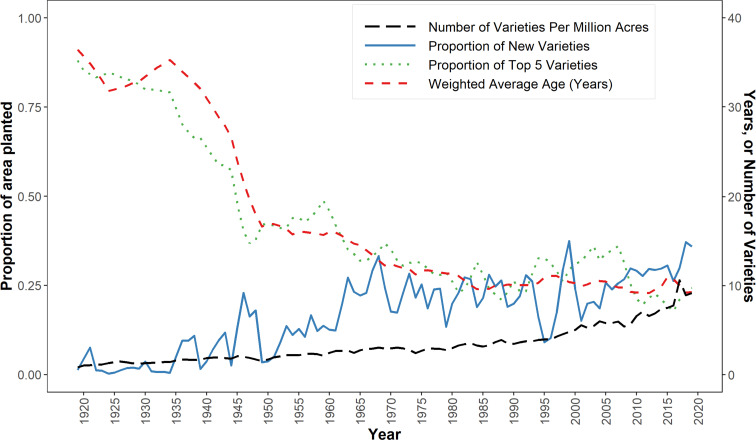
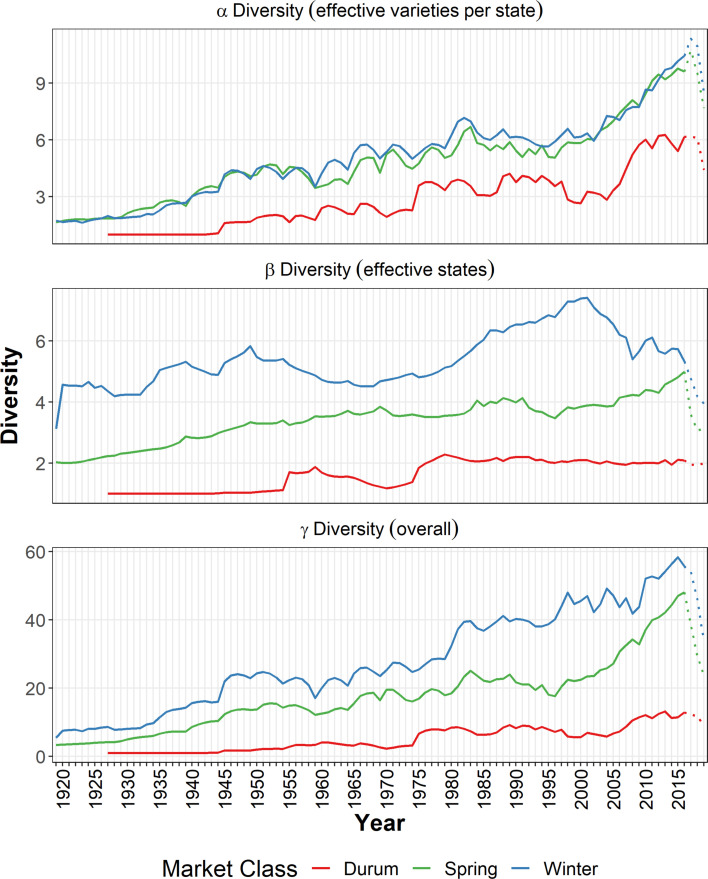
Fig. 2 plots several phylogenetically blind diversity metrics that summarize important dimensions of the varietal dynamics shown in Fig. 1. Our data reveal that the number of major commercially grown wheat varieties increased steadily over time, from just 33 in 1919 to 186 in 2019. On a per-million acre basis, the intensity of varietal use indicates an increasingly diverse spatial pattern. In 1919, varietal intensity averaged 0.8 varieties per million acres. A century later, farmers were using a much more diverse portfolio of varieties; varietal intensity had increased more than tenfold to average 9.1 varieties per million acres in 2019.
Fig. 2.
Phylogenetically blind measures of US wheat diversity, 1919 to 2019.
In 1919, just 1.3% of the planted area was sown to new varieties (i.e., <5 y old). By 2019, more than one-third (35.9%) of the US wheat area was planted to new varieties, such that the area-weighted age of commercially grown varieties declined dramatically from 36.4 y in 1919 to 16.0 y in 1960, and down to just 9.3 y in 2019. The decline in the area share of older varieties (i.e., >15 y since release) was particularly pronounced: 68.8% in 1919 and only 13.7% in 2019.
The average trends on varietal longevity mask a good deal of variation in the commercially useful life of individual varieties. For example, the winter wheat variety Cheyenne, bred at the Nebraska Agricultural Experiment Station and released for commercial use in 1933, was planted for a period of 80 y, disappearing from the varietal statistics in 2014. Its area peaked at around 2.5 million acres in 1959, falling steadily to less than 5,000 acres in 2013. In our collection, a total of 360 (26.6% of the 1,353 total) wheat varieties were long-lived (i.e., commercially grown for more than 15 y). In contrast, 146 varieties were especially short-lived, with recorded commercial use of just 1 y.
Spatiotemporal PD Patterns.
While the phylogenetically blind measures presented above unequivocally reveal increasing varietal diversity in the US wheat crop, they can be potentially misleading indicators of the extent of genetic erosion associated with the use of scientifically selected crop varieties. It is the perception of a narrowing of the genetic variation in modern cropping systems that is most closely associated with the concerns over the resilience of these systems to current and prospective climate and pest shocks. Phylogenetically blind measures of diversity will overstate the degree of genetic diversity within a given population or area extent (e.g., a field, state, or country) when the varieties in that population or locale are genetically related through shared breeding materials.
Using modern and extensive gene array technologies utilizing 4,009 unlinked polymorphic markers across 454 wheat varieties, Fradgley et al. (24) demonstrated a high correlation between pedigree- and marker-based kinship coefficients, confirming the value of using pedigree information to inform and manage wheat genetic diversity. Prior to this study, there had been numerous conflicting reports as to whether pedigree- and marker-based kinship coefficients were in agreement (e.g., refs. 25–31). However, Fradgley et al. (24) showed very clearly this earlier disagreement was almost entirely due to the artifacts of using low marker numbers (a few hundred at most, compared with 4,009 in the modern study) and erroneous pedigree or seed source data.
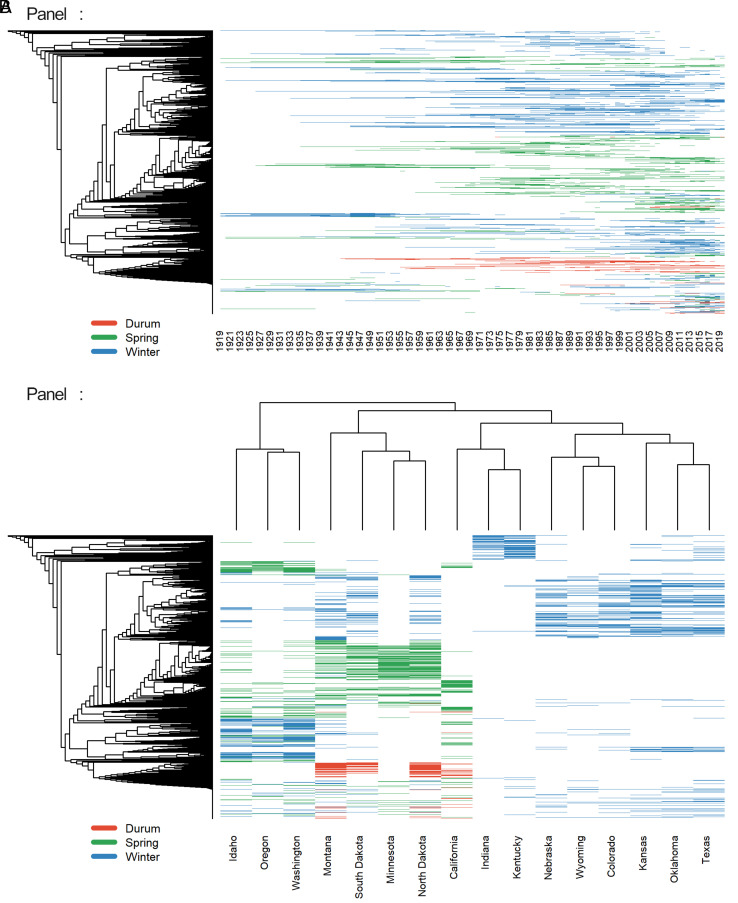
Based on the usefulness of pedigree-based diversity measures revealed in prior studies, in this study we utilize a comprehensive collection of US wheat pedigree information to infer the genetic relatedness among them (see Materials and Methods). The phylogenetic tree on the LHS of Fig. 3 graphically depicts the genetic relatedness of 1,353 commercial wheat varieties grown in the US during the period 1919 to 2019. The genetic distance between each pair of wheat varieties is represented by the pedigree branch length between them, based on their respective coefficient of parentages (COPs) that we calculated using the method described by Murphy et al. (32). The horizontal, colored lines in Fig. 3, Panel A indicate the presence or absence of each of these commercially grown wheat varieties for each of the years 1919 to 2019, while Panel B indicates the presence or absence of each of these varieties within each state during the same time period.
Fig. 3.
Changes over time in the phylogenetic variation of commercially grown US wheat varieties, 1919 to 2019. Notes: Each colored dash line indicates the presence of a specific variety within each year (Panel A) or within each state during the study period (Panel B), where red lines represent durum wheat varieties, green lines represent spring wheat varieties, and blue lines represent winter wheat varieties. Varieties are ordered along the phylogenetic tree depicted on the y-axis.
The phylogenetic variation among wheat varieties in the US increased over time, as revealed by the expanding coverage across the phylogenetic tree over time (Fig. 3, Panel A). The three market classes for wheat differ in their clustered locations within the phylogenetic tree. Most of the durum wheats (red lines) are concentrated within a closely clustered region on the phylogenetic tree. Winter wheats exhibit a much more diverse phylogenetic background than durum or spring wheats, with varieties ranging across the entire phylogenetic tree. In addition, the phylogenetic background of winter wheats constantly changes as the use of particular varieties waxes and wanes over time.
Our phylogenetic assessment of state-level data on wheat varieties in Fig. 3, Panel B leads to a natural clustering in line with the agro-climatic regions (Figs. 4 and 5). States within the Central Plains region (including Texas, Nebraska, Wyoming, Colorado, Oklahoma, and Kansas) appear to plant phylogenetically similar winter wheat varieties, which are different from the winter wheat varieties planted in the Northern Plains, Southeast, or Pacific Northwest region. States in the Northern Plains region (including Montana, North Dakota, South Dakota, and Minnesota) share phylogenetically similar spring and durum wheat varieties, which are different from the spring varieties planted in Pacific Northwest states (Washington, Oregon, and Idaho). California is unique in terms of the winter wheat varieties planted there, since many of these varieties are genetically spring-type wheats but are sown in the autumn (33). For the purposes of this study, we classified the winter wheat varieties grown in California as spring wheats given that our phylogenetic assessment in Fig. 3, Panel B shows these varieties are genetically similar to the spring wheat varieties grown in the Northern Plains.
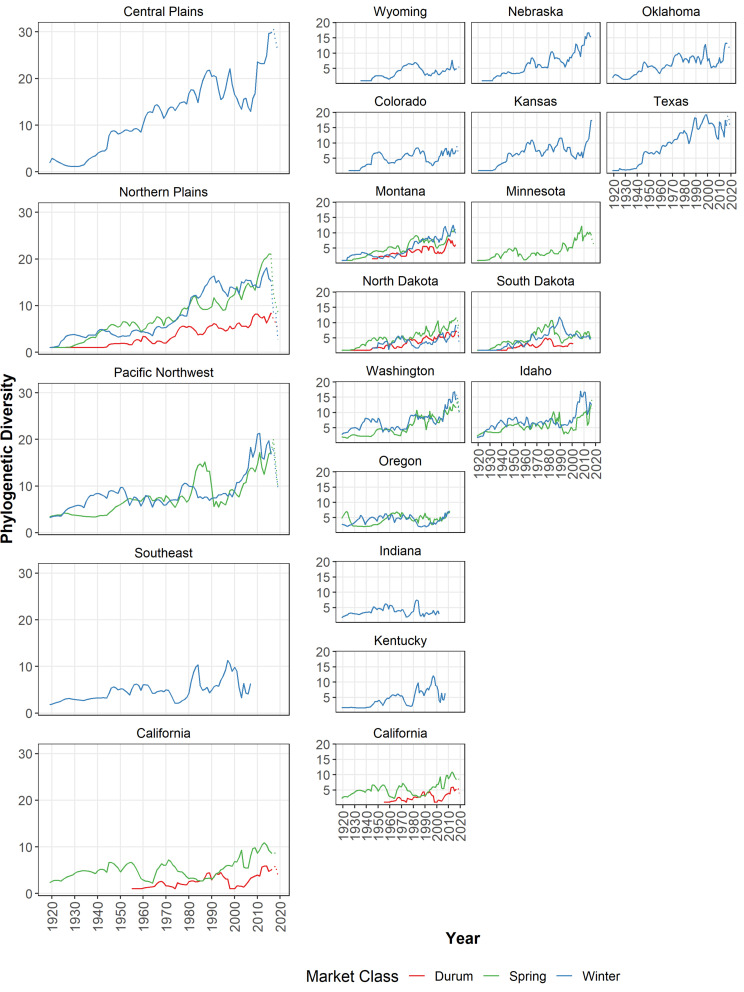
Fig. 4.
Phylogenetic diversity for major US wheat-producing regions and states, 1919 to 2019. Notes: Lines represent the phylogenetic diversity indexes of the order 1 for each region or state by different wheat market classes during 1919 to 2019. Similar trends are observed using PD indexes of order 2 (SI Appendix, Table S1).
Fig. 5.
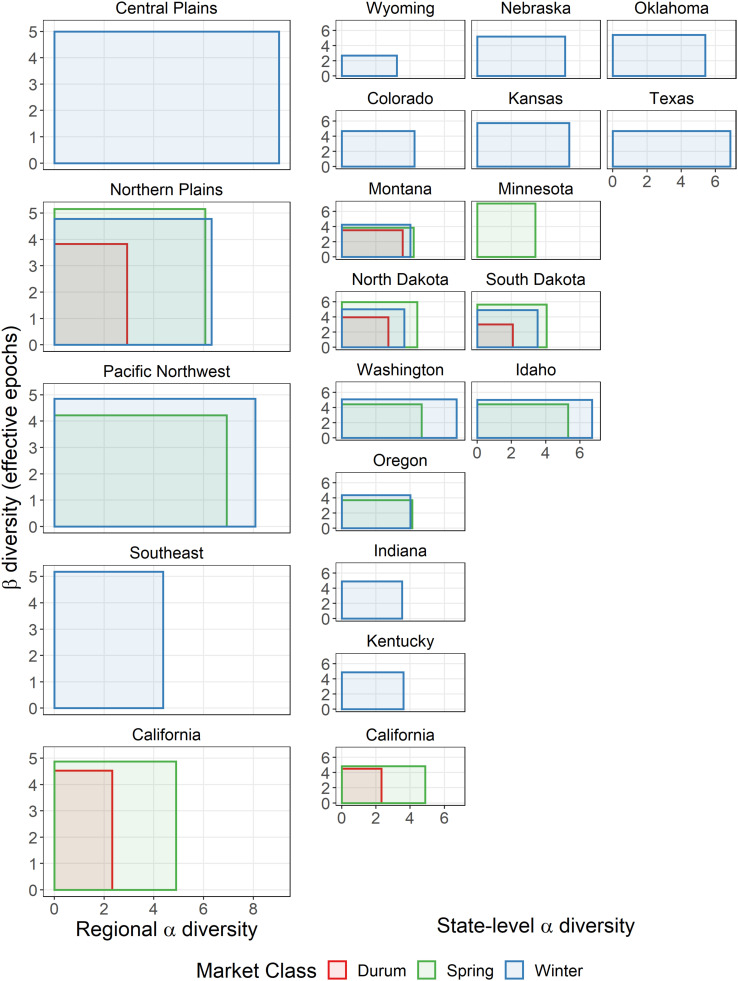
Temporal decomposition of phylogenetic diversity for major US wheat-producing regions and states, 1919 to 2019. Notes: The x-axis represents the α-diversity (effective number of varieties per epoch), and the y-axis represents temporal β-diversity (effective epochs) for each region or state, where the overall rectangle area represents the overall γ-diversity.
PD Indexes.
We used the genetic relatedness among varieties based on their respective position within the phylogenetic tree, and the abundance of each variety based on its state-specific planted area shares, to calculate a PD index for each of the 16 major US wheat-producing states and their corresponding agro-climatic regions during the period 1919 to 2019 using the method described by Chao et al. (21). The overall, and dominant, trend is for the PD of wheat to increase over time in all three market classes and regions plotted in Fig. 4. Looking in more spatial detail, for winter wheat, many states—including Kansas, Kentucky, Montana, Nebraska, Oklahoma, Texas, and Washington—had substantial increases in PD, especially in the more recent decades, while some of the smaller wheat growing states (Oregon, Wyoming, and Indiana) had rather stagnant PD over the longer run. The PD indexes for spring wheat are generally similar to those for winter wheat within states growing both market classes (such as South Dakota, Montana, and Washington). In Montana and South Dakota, the PD indexes for durum wheats are almost always less than those for winter wheat, whereas in North Dakota the PD of durum wheat is comparable to that of spring and winter wheat.
Temporal Decomposition of PD.
The PD trends reported above track regional and state-level changes over time in a PD index, where that index is calculated by treating each agro-climatic region or state at each point in time as a discrete community of wheat varieties. Adopting an approach introduced by Whitakker (34) in his study of diversity changes in the vegetative cover of two forested regions, a more nuanced and more insightful measurement approach to analyzing the variation in PD among US wheat varieties is to introduce the concept of a metacommunity. In this first instance, we treat the wheat varieties grown over the entire century in each region or state as a distinct “temporal metacommunity,” such that the overall PD (aka γT-diversity) of each geographic-specific metacommunity can be decomposed into two components: an αT-diversity, which captures the average effective number of phylogenetically distinct varieties planted each year in a given geography, and a temporal βT-diversity, which measures the effective number of phylogenetically distinct epochs for each region or state. Thus, for example, geographies with higher numbers of phylogenetically distinct varieties in a year will have larger αT-diversity values, while those geographies with higher turnover rates of phylogenetically distinct varieties will have higher βT-diversity values. Fig. 5 plots the αT-diversity (number of effectively distinct varieties per year) on the x-axis and temporal βT-diversity (number of effectively distinct epochs) on the y-axis for each region or state, where the rectangle area (equal to αT x βT) represents each region or state’s overall (spatiotemporal) γT-diversity.
Regional diversity is higher than the state-level diversity within each region, reflecting the larger geographic areas encompassed by each region, which include more farmers in total and intrinsically has more diverse agro-ecologies. That in turn leads to more spatially and temporally diverse demands for varieties by crop producers at the regional versus state level. Winter wheat states differ markedly in terms of their αT-, βT-, and γT-diversities. Washington, Idaho, and Texas are the top three ranked states in terms of their overall γT-diversities, with both high αT-diversity (averaging 6.7, 6.7, and 6.9 effectively distinct varieties per year, respectively) and high temporal βT-diversity (5.1, 5.0, and 4.7 distinctly different epochs during the 101-y period spanning 1919 to 2019). In contrast, Wyoming has the lowest overall γT-diversity, with both the lowest αT-diversity (averaging just 3.2 effectively distinct varieties each year) and lowest temporal βT-diversity (with just 2.7 phylogenetically distinct epochs). Kansas is the most βT-diverse state, with 5.7 phylogenetically distinct epochs during the period 1919 to 2019, indicative of a rapid rate of varietal turnover.
For spring wheat, North Dakota has the highest overall γT-diversity with 4.4 effective varieties each year (αT-diversity) and 6.0 phylogenetically distinct years (βT-diversity). Idaho and Minnesota have the highest αT-diversity and βT -diversity, respectively. In Washington, the αT-diversity for spring wheat (4.7) is smaller than it is for winter wheat (6.7), with a difference of 2.0 effectively distinct varieties. Spring wheat in North Dakota and South Dakota has higher αT-, βT-, and γT-diversities than wheats in the other two market classes, suggesting both a higher effectively distinct number of varieties and a faster rate of replacement for spring wheat in these states.
Among the four durum wheat states, Montana planted 3.6 effectively distinct varieties each year, higher than North Dakota, South Dakota, and California which ranged from 2.0 to 2.7 effectively distinct varieties annually. California has the highest temporal βT-diversity (4.5 distinct epochs) among all four states.
Spatial Decomposition of PD.
To characterize the spatial variation of wheat varieties nationally, the collection of all major wheat growing states in the United States for each year is treated as a “spatial metacommunity.” In this instance, the γS-diversity represents the overall diversity in the United States, which can be decomposed into αS-diversity—representing the average number of effectively distinct varieties in a state—and the spatial βS-diversity—which represents the effective number of phylogenetically distinct states in the United States. For the three market classes of wheat in the United States, their overall γS-diversity and its αS- and βS-decompositions are plotted over time in Fig. 6. Generally, the diversity indexes are increasing over time for all three market classes of wheat in the United States, where winter wheat has the highest overall γS-diversity followed by spring wheat. Winter wheat and spring wheat have similar αS-diversity indexes over time, suggesting that the state-level wheat varietal diversities are similar among these two market classes. However, winter wheat has much higher spatial βS-diversities than spring wheat, consistent with the fact that winter wheat is grown in more states over larger geographic areas than spring wheat. This is also consistent with the notion that market forces are likely to drive toward the development and deployment of varieties that better align varietal genetics across the diverse, location-specific agro-ecologies in which they are grown.
Fig. 6.
Spatial decomposition of phylogenetic diversity for major US wheat-producing states, 1919 to 2019. Notes: Limitations in reported area-by-variety data since 2000 (and especially after 2016, dashed lines) account for the decline in the effective number of states for these more recent years (SI Appendix, Fig. S2 details data availability).
With only four major durum wheat states, the spatial βS-diversity and overall γS-diversity for durum are the lowest among the three market classes. The overall γS-diversities for the US wheat crop are heavily influenced by both the αS-diversity and βS-diversity, suggesting that both the within-state varietal diversity and across-state spatial variations are important for the overall diversity of the wheat population across each of the three market classes.
Discussion
Adding to a growing body of empirical evidence of wheat crop diversity (see, e.g., the compilation in SI Appendix, Table S1), our results provide further, more comprehensive, evidence that the increasingly intensive use of scientifically selected crop varieties has led to more, not less, biodiverse cropping practices, at least regarding diversity in the US wheat crop. This substantial increase in varietal diversity over the past century has been achieved in tandem with a fourfold increase in US average wheat yields. As we show in SI Appendix, Fig. S3, the preponderance of varieties planted in the United States was bred by public agencies. Wheat varietal improvement in Canada is also largely a public affair (35). And while the private sector has made significant in-roads to wheat breeding in Europe and the United Kingdom (see, e.g., ref. 36), in Australia the shift toward privately led crop improvement still involves complex collaborative research, funding and even co-ownership arrangements with public entities (37). The private presence in other crops and other countries is variable. For example, Heisey et al. (38) reported almost all US corn acreage has been planted with privately bred hybrid varieties since the 1980s, whereas Pardey et al. (39) show that the private agricultural R&D presence is still comparatively small in many low- and middle-income counties. There is much commentary (e.g., refs. 36 and 38) on the extent to which the private versus public sourcing of varietal innovations affects the varietal diversity found in farmers’ fields, but little hard evidence to the level of detail we provide here on this important issue. Moreover, the fundamental supply and demand factors we broached above as the primary drivers of varietal diversity are in play for both public and private breeders.
To the extent our results are generalizable, they can profoundly reframe the policy and practical implications of a sizable body of literature and policy debate that ignores phylogenetic (within-crop species) diversity when assessing the implications of intensified cropping systems on agricultural biodiversity (e.g., ref. 40). While landraces (or farmer-bred varieties) may exhibit considerable spatial diversity at any point in time (41), their comparatively slow rates of varietal turnover (see. e.g., refs. 42 and 43) increase the vulnerability of these stands to unprecedented (in nature and magnitude) biotic or abiotic shocks occasioned by relative rapid changes in climate and human- or naturally mediated invasions of foreign pests and diseases. In contrast, the relatively rapid turnover of crop varieties in more intensive agricultural systems that use scientifically selected material (9, ch. 7, 44, 45) opens up prospects for more rapid, and thereby more valuable, genetic responses to changing climate, pest, and disease circumstances. In addition, scientifically selected varieties that optimize their performance in locationally variable agro-ecological environments mean that market forces are also likely to spur more spatially diverse seed development and deployments to achieve better-performing G x E (genetics-by-environment) matchups.
Conserving or enhancing (within species) crop biodiversity in and of itself is a multi-instrument, multi-objective problem, where the crop biodiversity outcomes are envisaged as a means (instrument) to other ends (e.g., yield growth; resilience to a multitude of climate or pest and disease shocks; and various food security, access, and equity outcomes). Beyond the genetic diversity of the crop per se, there is above and below ground (e.g., plant, microbe, and insect) biodiversity within cropping systems, the biodiversity inherent in dual animal-livestock production systems, and the much more encompassing notions of managed, natural, or wild systems of biodiversity involving land outside of agriculture to be considered. All of these factors enrich the biological diversity of the larger ecological and agricultural setting.
Setting policies, and subsequently putting them into practice, to achieve desired outcomes (in this instance agricultural biodiversity) requires close and careful alignment of policy instruments with targets (46–48). If enhancing crop (within species) diversity at consequential temporal and spatial scales is the target, our results show that the intelligent intensification of cropping systems using scientifically selected crop varieties can be an especially effective instrument. Moreover, in this particular instance, this same instrument has multiple other desired outcomes. The higher yields associated with more-intensive cropping systems clearly have positive food security implications by increasing crop output, lowering the unit costs of production, or both. This in turn lowers the price of food, with especially equitable impacts on poorer people who spend a substantially larger share of their meager incomes on food (49). If our results are generalizable such that increased yields and crop genetic diversity can both be achieved under intensive cropping systems, they are also likely to be impactful on biodiversity more generally by shrinking the footprint of cropping agriculture. In the spirit of Waggoner (50) and Borlaug (51), Stevenson et al. (52), for example, carried out a counterfactual simulation to assess the land use consequences of global agriculture absent the Green Revolution yield gains associated with genetic improvement in wheat, rice, coarse grains, and other crops (cassava, lentils, beans, and potatoes) over the period 1961 to 2004. They concluded that “… the total crop area in 2004 would have been between 17.9 and 26.7 million hectares larger in a world that had not benefited from crop germplasm improvement since 1965. Of these hectares, 12.0 to 17.7 million would have been in developing countries, displacing pastures and resulting in an estimated 2 million hectares of additional deforestation (52, p. 8363).” In fact, US wheat production increased 3.3-fold over the past century (22), while planted area declined by 41%, with a fourfold increase in average yield (from 867.5 kg/ha in 1919 to 3476.9 kg/ha in 2019). Similarly, the 2019 global area in wheat was roughly comparable to the acreage in the late 1960s, so that output grew by threefold in-line with the increase in average global wheat yields (6). The choice of targets and instruments is naturally context sensitive. In poorer parts of the world, the priority policy target may be to improve the well-being of poor people, especially by way of increasing crop productivity (thus expanding the supply and lowering the price of food, with equitable implications for poorer consumers given larger shares of their incomes are typically spent on food). In this instance, our results indicate that an efficient instrument to cost-effectively achieve well-being targets for poorer people would be to intelligently intensify wheat production by way of expanding the use of scientifically selected wheat varieties; a strategy that calls for doubling down on the science and seed systems that at present often underserve farmers in poorer countries. Based on our study, this particular configuration of policy instruments and targets is also likely to improve biodiversity outcomes, both directly by increasing the spatiotemporal diversity of wheat varieties in use and indirectly by stalling the expansion or even shrinking the footprint of unnatural agricultural landscapes in favor of increasing areas in more natural environments.
Materials and Methods
Data on Wheat Varieties in the United States.
Data on planted area-by-variety were collected for the US wheat crop for the period 1919 to 2019 by the authors and colleagues at the International Science and Technology Practice and Policy center and the GEMS Informatics Center, University of Minnesota. From 1919 to 1984, acreage-by-variety data are reported quinquennially for a total of 42 states by USDA’s Statistical Bulletins on the Distribution of the Varieties and Classes of Wheat in the United States. Thereafter, we sourced the required data from state-specific agricultural statistical services, which dropped to 16 states in 2000 and the years following. These 16 states, as listed in Figs. 4 and 5, accounted for 89% of total US wheat area in 2019. Based on end-use purpose and growth habit, US wheat varieties can be categorized into six market classes; hard red winter, soft red winter, hard white, soft white, hard red spring, and durum (53, 54). Unfortunately, the area-by-variety data that are integral to the diversity metrics we developed are consistently reported for only three market classes based on growth habit—specifically, winter, spring (excluding durum), and durum—and so these are the class classifications used in this study.
The states for which we have area-by-variety data account for around 97% of total US durum acreage in 2019, 100% of spring acreage, and 84% of winter acreage. However, even within the 16 major wheat-growing states, not all states report area-by-variety data for all years. To construct a complete state-level panel on wheat variety areas, we filled in the missing years’ variety area information using linear interpolation based on years before and after any missing years. We used each variety’s year of release and last year of reported use nationally to determine the beginning and end year of each variety’s use when interpolating their state-specific areas. Information on each variety’s year of release was collected from multiple sources, including the Genetic Resource Information System (GRIS) for Wheat and Triticale (55), the Germplasm Resources Information Network (GRIN) (56), the GrainGenes database (57), crop registration narratives from scientific journals such as Crop Science, Journal of the American Society of Agronomy, and Journal of Plant Registration, the Plant Variety Protection Office (58), and searches from elsewhere such as private company websites and university websites.
Additionally, information on each variety’s name, market class, and crop pedigree was also collected. Varieties often have aliases or different spellings depending on the time and location they were marketed and adopted. To avoid double counting the same varieties with different names across states and over time, we reconciled the aliases of different varieties and standardized the names across all the reported wheat varieties grown commercially in the United States from 1919 to 2019. Multiple sources of wheat genetic and pedigree information were used to consolidate varietal names and their pedigrees, including the GRIS for Wheat and Triticale (55), the GRIN (56), the GrainGenes database (57), and the Plant Variety Protection Office (58). Where possible, the entire pedigree of each variety and their parents were traced back to either a landrace, a wild accession, or a local variety. The pedigree information for all reported commercially grown varieties in the United States was collected and processed. Inconsistencies among reported pedigrees (and varietal name) were reconciled by PedTools, a Python library developed by the GEMS Informatics Center at the University of Minnesota that maps input varieties for a crop to aliases, standardizes naming protocols and recursively reconstructs pedigrees in principle back to landraces through repeated observations of parent–child relationships found in the literature. To process the 1,353 commercially grown varieties included in this study, PedTools was applied to 2,597 varieties (including non-commercially grown crossing materials) for the final pedigree analysis.
Diversity Measures.
A large number of biodiversity measures have been proposed in the disciplines of ecology, genetics, economics, information theory, and other sciences (59–62). Among them, the most commonly used are phylogenetically blind in the sense that they tally the distributions of each type of entity, regardless of each entity type’s taxonomic, genetic, or functional similarities. Such indexes include an entity richness index, the Shannon entropy index (61), and the Gini-Simpson index (63), all of which are special cases of the generalized Tsallis entropy measures (64, 65), also known as HCDT entropy indexes (66–68). HCDT indexes can be converted into so-called true diversity measures. Such measures, also known as the effective number of species or “Hill numbers,” represent the hypothetical number of equally abundant entities that would give the same diversity index value as was actually observed (65, 69). “Hill numbers” do not depend on the functional form of the index and satisfy the replication principle, whereby the index value doubles if each entity grouping was divided into two equal new groups (65, 70). Thus, Hill numbers allow for a unified and intuitive interpretation of diversity across locations.
For wheat, new varieties are typically developed by genetic crosses among existing varieties, and thus, the contribution of each new variety to the overall crop diversity depends on their relatedness to existing varieties. Commonly used phylogenetically blind biodiversity measures such as species richness and Shannon entropy are unsuitable to differentiate areas growing many genetically similar, but nonetheless differentiated (by name), crop varieties from those areas with many genetically distant crop varieties. Thus, characterizing the biodiversity of a crop species in modern agricultural landscapes requires a phylogenetically informed approach, which by construction incorporates the notion of varietal relatedness into the measure of diversity.
A growing number of phylogenetically informed biodiversity measures have been proposed to account for taxonomic, functional, or phylogenetic similarities among species within a community, such as Rao’s quadratic entropy (71), taxonomic cladistics diversity (CD) (72), PD (73), pure diversity measure (60, 74), functional diversity (75), and many others (e.g., refs. 21 and 56–59). Among these alternatives, one common approach to account for the genetic relatedness among biological individuals is the PD measure proposed by Faith (73). This diversity measure is defined as the sum of all the phylogenetic branches along the minimum spanning path to quantify the evolutionary history shared among individuals. Weitzman (76 showed that a community’s diversity value can be represented by the branch length of the hypothetical phylogenetic tree.
PD measures typically focus on the presence or absence of a species to measure the overall genetic variation within a community, without taking into account the relative abundance of each species. However, species abundance provides crucial additional information regarding the composition of the community, especially for agro-ecosystems where a few popular crop varieties may dominate the majority of the landscape, while numerous other varieties account for comparatively small portions of the overall cropped area. To incorporate both species abundance and species phylogenetic distances, Chao et al. (21) generalized the traditional phylogenetic measure and proposed a PD measure based on Hill numbers that quantifies “the mean effective number of species” and in so doing unified many of the existing measures of biodiversity.
To calculate a generalized PD, a phylogenetic tree is first constructed based on distances between species of the community using the UPGMA method (unweighted pair group method with arithmetic mean). Both molecular markers and pedigree information have been used in major crop genetic diversity studies to derive genetic distances among crop varieties. For this study, we use the COP concept to infer genetic relatedness from pedigree information on all named US wheat varieties planted during the period 1919 to 2019. Following Murphy et al. (32), COP calculates the proportion of shared genetic material among varieties based on their respective pedigrees under the following assumptions: 1) A cultivar inherits half of its genes from each parent; 2) all parental lines are homozygous and homogeneous; and 3) all landraces are unrelated to each other. Defining a pair-wise dissimilarity index between variety and variety as , we can obtain a pair-wise dissimilarity matrix for the collection of all wheat varieties over the entire study period in the United States. Based on the pair-wise dissimilarity matrix, a phylogenetic tree can be constructed for all US wheat varieties. Using the phylogenetic tree, the generalized PD (denoted as ) for a community of wheat varieties can then be calculated using Chao et al.’s (21) method as:
where is the number of branch segments in the tree, denotes the length of branch (), denotes the branch abundance (sum of relative abundance of all species descended from branch ), and denotes the exponent value (i.e., order) given to the branch abundance normalized by the mean branch length, which is defined as . For special cases of the order spanning the entire age of the phylogenetic tree, it is shown that becomes the total branch length, which is the traditional Faith’s PD; can be linked to a generalization of Shannon entropy to incorporate phylogenetic distances; and can be linked to Rao’s quadratic entropy (21, 77). The phylogenetic Hill number is then calculated as:
For a state with equally common species that are completely distinct from each other along the phylogenetic tree, the diversity measure always gives exactly . Thus, the phylogenetic Hill number can be interpreted as the effective number of maximally distinct lineages with equal relative abundance (21).
Spatial and Temporal Diversity Decomposition.
The generalized PD indexes introduced above are static measures of biodiversity for a single crop (in this instance wheat) community (e.g., a US state, agro-climatic region, or the United States as a whole). However, variation in both the spatial and temporal dimensions of an ever-changing mix of crop varieties is one of the most fundamental features of modern agricultural systems. Thus, decomposing biodiversity into its spatial and temporal components provides a means of characterizing the dynamic changes over space and time in agricultural biodiversity. A growing number of long-term datasets have been used to examine the spatial and temporal patterns of biodiversity change within ecological systems (e.g., ref. 78). This study incorporates both spatial and temporal decompositions into an assessment of the diversity dynamics of a major crop species.
Whittaker (34) first proposed the decomposition of the overall diversity ( -diversity) into within-community ( -diversity) and between-community ( -diversity) components, using either an additive or multiplicative rule. For spatial decomposition within each time period, each agroclimatic region or US state can be treated as a separate crop community, where the variation among different regions or states reflects the spatial diversity across the landscape. For temporal decomposition with each region or US state, each year can be treated as a separate crop community where the variation across years reflects the temporal diversity within each state.
Here we define a crop community as the collection of varieties planted within a given region or state for a given year for a given crop. Then a “spatial metacommunity” is defined as the collection of all communities within a single year (i.e., all states in the United States within a given year), and a “temporal metacommunity” is defined as the collection of communities over multiple years within the same geography (i.e., all years within a given region or state). With these definitions of spatial and temporal metacommunities, following Marcon et al. (79), the total species neutral HCDT entropy ( -entropy) for a metacommunity can be decomposed as:
where - and -entropies for the metacommunity (i.e., and ) are the weighted sums of local entropies within each community (i.e., and ). The weight adjusts for sample size differences among communities, which is commonly defined as where is the number of individuals (here, crop varieties) in a local community and is the total number of individuals for a m.etacommunity. The - and -entropies for a community are calculated as:
where
Similarly, as a linear transformation of generalized entropy, the generalized PD for the metacommunity can be decomposed as:
where
The corresponding decomposition of the diversity index , also known as the phylogenetic Hill number, is then obtained as:
where
Here is the transformed exponential defined as
Depending on the grouping of communities into a given “spatial metacommunity” or a “temporal metacommunity,” the above formula allows us to decompose diversity into its respective spatial or temporal dimensions. Essentially, for a metacommunity (i.e., a collection of local communities), the overall -diversity is decomposed into an average local community diversity ( -diversity) and a measure of the effective number of communities ( -diversity). Intuitively, for a given year in the United States with an overall PD index value , the -diversity component of this “spatial metacommunity” indicates the average diversity of wheat varieties growing within a state (i.e., the average effectively distinct number of varieties from maximally distinct lineages with equal relative abundance in a state), while the -diversity component of this “spatial metacommunity” indicates the effectively distinct number of states (i.e., the equivalent number of states that each has effective number of varieties that are distinct from each other). Similarly, for a “temporal metacommunity” (i.e., multiple periods or epochs for a given geography) with an overall PD index value , the -component indicates the average diversity of wheat varieties growing in each epoch in each region or state, while the -component indicates the effectively distinct number of epochs (i.e., the equivalent number of epochs where each has effectively distinct number of varieties). Using a unique long-run panel dataset for a major crop species, such spatial and temporal decompositions allow us to better understand the impact of crop varietal turnover on modern agricultural genetic diversity and address key questions concerning 1) the overall trends in the PD of the US wheat crop and 2) changes in either the spatial or temporal dimensions of diversity in the US wheat crop over the past century.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We acknowledge the research assistance of Connie Chan-Kang from the GEMS Informatics Center, University of Minnesota. We also acknowledge the Minnesota Supercomputing Institute for computational resources.
Author contributions
Y.C. and P.G.P. designed research; Y.C. performed research; Y.C. and K.A.T.S. analyzed data; and Y.C., P.G.P., and K.A.T.S. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Some study data available (The state-level area-by-variety data generated and analyzed during the current study are the property of the GEMS Informatics Center at the University of Minnesota. They are maintained in the GEMS Informatics Platform (https://gems.agroinformatics.org/) from where they can be requested from the corresponding author YC for use to replicate the findings of this study. The R codes for generating the results and figures reported in the current study are available at GitHub (https://github.com/y-chai/US-Wheat-Diversity.git). PedTools used in the current study for wheat pedigree processing is a python package developed by the GEMS Informatics Center at the University of Minnesota, which is not publicly available and can be requested for use to replicate the findings of this study).
Supporting Information
References
- 1.UN (United Nations), "Sustainable development report 2021" (Cambridge University Press, Cambridge, U.K., 2021; ). [Google Scholar]
- 2.Delabre I., et al. , Actions on sustainable food production and consumption for the post-2020 global biodiversity framework. Sci. Adv. 7, eabc8259 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.UNEP (United Nations Environment Programme), "Food systems and natural resources. A report of the working group on food systems of the international resource panel", Westhoek H., Ingram J., Van Berkum S., Özay L., Hajer M., Eds. (UNESCO, Paris, 2016). [Google Scholar]
- 4.Lu Y., Bullock J. M., Biodiversity conservation in a changing environment beyond 2020. Sci. Adv. 7, eabl8162 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.UN-SCBD (Secretariat of the Convention on Biological Diversity), Global Biodiversity Outlook 5. Montreal, 2020. Online at https://www.cbd.int/gbo5.
- 6.FAO (Food and Agriculture of the United Nations), FAOSTAT database (FAO, Rome, 2022). Available at: http://www.fao.org/faostat/en/#data. [Google Scholar]
- 7.Miller J., Genetic erosion: Crop plants threatened by government neglect. Science 182, 1231–1233 (1973), 10.1126/science.182.4118.1231. [DOI] [PubMed] [Google Scholar]
- 8.Harlan J. R., Our vanishing genetic resources. Science 188, 618–621 (1975). [DOI] [PubMed] [Google Scholar]
- 9.Brush S. B., Farmers’ Bounty: Locating Crop Diversity in Contemporary World (Yale University Press, 2004). [Google Scholar]
- 10.Khoury C. K., et al. , Crop genetic erosion: Understanding and responding to loss of crop diversity. New Phytol. 233, 84–118 (2022). [DOI] [PubMed] [Google Scholar]
- 11.Frankel O. H., Genetic dangers in the green revolution. World Agric. 19, 9–13 (1970). [Google Scholar]
- 12.Fowler C., Unnatural Selection: Technology, Politics and Plant Evolution (Gordon and Breach, Switzerland, 1994). [Google Scholar]
- 13.Souza E., Fox P. N., Byerlee D., Skovmand B., Spring wheat diversity in irrigated areas of two developing Countries. Crop Sci. 34, 774–783 (1994), 10.2135/cropsci1994.0011183X003400030031x. [DOI] [Google Scholar]
- 14.Brennan J. P., Fox P. N., Impact of CIMMYT varieties on the genetic diversity of wheat in Australia, 1973–1993. Aust. J. Agric. Res. 49, 175–178 (1998). [Google Scholar]
- 15.Bowman D. T., May O. L., Creech J. B., Genetic uniformity of the U.S. Upland cotton crop since the introduction of transgenic cottons. Crop. Sci. 43, 515–518 (2003). [Google Scholar]
- 16.Duvick D. N., Genetic diversity in major farm crops on the farm and in reserve. Econ. Botany 38, 161–178 (1984). [Google Scholar]
- 17.Vitale J., Adam B., Vitale P., Economics of wheat breeding strategies: Focusing on Oklahoma hard red winter wheat. Agronomy 10, 238 (2020). [Google Scholar]
- 18.Biffen R. H., Mendel’s law of inheritance and wheat breeding. J. Agric. Sci. 1, 4–48 (1905). [Google Scholar]
- 19.Ball C. R., The history of American wheat improvement. Agric. History 4, 48–71 (1930). [Google Scholar]
- 20.Smýkal P., et al. , From Mendel’s discovery on pea to today’s plant genetics and breeding. Theor. Appl. Genet. 29, 2267–2280 (2016), 10.1007/s00122-016-2803-2. [DOI] [PubMed] [Google Scholar]
- 21.Chao A., Chiu C. H., Jost L., Phylogenetic diversity measures based on Hill numbers. Philos. Trans. R Soc. London B Biol. Sci. 365, 3599–3609 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.USDA-NASS, "United States Department of agriculture national agricultural statistics service: Quick stats" (2021). Available at: https://quickstats.nass.usda.gov/.
- 23.Griliches Z., Hybrid corn: An exploration in the economics of technological change. Econometrica 254, 501–522 (1957). [Google Scholar]
- 24.Fradgley N., et al. , A large-scale pedigree resource of wheat reveals evidence for adaptation and selection by breeders. PLoS Biol. 17, e3000071 (2019), 10.1371/journal.pbio.3000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corbellini M., Perenzin M., Accerbi M., Vaccino P., Borghi B., Genetic diversity in bread wheat, as revealed by coefficient of parentage and molecular markers, and its relationship to hybrid performance. Euphytica 123, 273–285 (2002). [Google Scholar]
- 26.Barrett B. A., Kidwell K. K., Fox P. N., Comparison of AFLP and pedigree-based genetic diversity assessment methods using wheat cultivars from the Pacific Northwest. Crop. Sci. 38, 1271–1278 (1998), 10.2135/cropsci1998.0011183X003800050026x. [DOI] [Google Scholar]
- 27.Kim H. S., Ward R. W., Genetic diversity in Eastern U.S. soft winter wheat (Triticum aestivum L. em. Thell.) based on RFLPs and coefficients of parentage. Theor. Appl. Genet. 94, 472–479 (1997). [Google Scholar]
- 28.Almanza-Pinzón M. I., Khairallah M., Fox P. N., Warburton M. L., Comparison of molecular markers and coefficients of parentage for the analysis of genetic diversity among spring bread wheat accessions. Euphytica 130, 77–86 (2003). [Google Scholar]
- 29.Marić S., Bolarić S., Martinčić J., Pejić I., Kozumplik V., Genetic diversity of hexaploid wheat cultivars estimated by RAPD markers, morphological traits and coefficients of parentage. Plant Breed. 123, 366–369 (2004). [Google Scholar]
- 30.Fufa H., et al. , Comparison of phenotypic and molecular marker-based classifications of hard red winter wheat cultivars. Euphytica 145, 133–146 (2005). [Google Scholar]
- 31.Barbosa-Neto J. F., Sorrells M. E., Cisar G., Prediction of heterosis in wheat using coefficient of parentage and RFLP-based estimates of genetic relationship. Genome 39, 1142–1149 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Murphy J. P., Cox T. S., Rodgers D. M., Cluster analysis of red winter wheat cultivars based upon coefficients of parentage. Crop Sci. 26, 672–676 (1986). [Google Scholar]
- 33.Jackson L., Fenandez B., Meister H., Spiller M., “Importance of small grain crops in California agriculture” in Small Grain Production Manual, Part 1, University of California Division of Agriculture and Natural Rerefsources, Ed. (2006).
- 34.Whittaker R. H., Vegetation of the Siskiyou Mountains, Oregon and California. Ecol. Monogr. 30, 279–338 (1960). [Google Scholar]
- 35.Gray R. S., Kingwell R. S., Galushko V., Bolek K., Intellectual property rights and Canadian wheat breeding for the 21st century. Can. J. Agri. Econ. 65, 667–691 (2017). [Google Scholar]
- 36.Galushko V., Gray R., Twenty five years of private wheat breeding in the UK: Lessons for other countries. Sci. Public Policy 41, 765–779 (2014). [Google Scholar]
- 37.Alston J. M., Gray R. S., Wheat research funding in Australia: The rise of public–private–producer partnerships. EuroChoices 12, 30–35 (2013). [Google Scholar]
- 38.Heisey P. W., Srinivasan C. S., Thirtle C., “Public sector plant breeding in a privatizing World” (Resource Economics Division, USDA-ERS. Agriculture Information Bulletin No. 772, 2001). [Google Scholar]
- 39.Pardey P. G., Chan-Kang C., Dehmer S. P., Beddow J. M., Agricultural R&D is on the move. Nat. News 537, 301 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Dainese M., et al. , A global synthesis reveals biodiversity-mediated benefits for crop production. Sci. Adv. 5, eaax0121 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng J. H., Sun D., Nevo E., Domestication evolution, genetics and genomics in wheat. Mol. Breed. 28, 281–301 (2011), 10.1007/s11032-011-9608-4. [DOI] [Google Scholar]
- 42.Hillman G. C., Davies M. S., Measured domestication rates in wild wheats and barley under primitive cultivation, and their archaeological implications. J. World Prehist. 4, 157–222 (1990). [Google Scholar]
- 43.Honne B. I., Heun M., On the domestication genetics of self-fertilizing plants. Veg. Hist. Archaeobotany 18, 269–272 (2009). [Google Scholar]
- 44.Brennan J. P., Byerlee D., The rate of crop varietal replacement on farms: Measures and empirical results for wheat. Int. J. Plant Var. Seeds 4, 99–106 (1991). [Google Scholar]
- 45.Meng E. C. H., Smale M., Bellon M., Grimanelli D., "Definition and measurement of crop diversity for economic analysis" in Farmers, Gene Banks and Crop Breeding: Economic analyses of Diversity in Wheat, Maize and Rice, Smale M., Ed. (Kluwer Academic Publisher, Boston, 1998). [Google Scholar]
- 46.Arrow K. J., Tinbergen on economic policy. J. Am. Stats. Assoc. 53, 89–97 (1958). [Google Scholar]
- 47.Corden W. M., Trade Policy and Economic Welfare (Oxford University Press, Oxford, 1974). [Google Scholar]
- 48.Alston J. M., Pardey P. G., "The economics of agricultural innovation" in Handbook of Agricultural Economics, Barrett C. B., Just D. R., Eds. (Elsevier, Amsterdam, 2021), vol. 5. [Google Scholar]
- 49.Muhammad A., Seale J. L. Jr., Meade B., Regmi A., International Evidence on Food Consumption Patterns: An Update Using 2005 International Comparison Program Data (U.S. Dept. of Agriculture, Econ. Res. Serv., Washington, D.C., 2011). [Google Scholar]
- 50.Waggoner P. E., How much land can ten billion people spare for nature? Daedalus 125, 73–93 (1996). [Google Scholar]
- 51.Borlaug N., Feeding a hungry world. Science 318, 359 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Stevenson J. R., Villoria N., Byerlee D., Kelley T., Maredia M., Green Revolution research saved an estimated 18 to 27 million hectares from being brought into agricultural production. Proc. Natl. Acad. Sci. U.S.A. 110, 8363–8368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ek C., "Wheat classification" (Congressional Research Service Report for Congress. 88–54ENR, 1988). [Google Scholar]
- 54.U.S. Wheat Associates, Wheat classes. https://www.uswheat.org/working-with-buyers/wheat-classes/. Accessed 14 September 2022.
- 55.CIMMYT, Genetic resource information system (GRIS) for wheat and triticale. http://wheatpedigree.net. Accessed 5 December 2017.
- 56.USDA-NGRP (National Genetic Resources Program), The germplasm resources information network (GRIN). https://www.ars-grin.gov/. Accessed 1 October 2021.
- 57.USDA-ARS, GrainGenes, a database for Triticeae and Avena. https://wheat.pw.usda.gov/GG3/. Accessed 16 September 2021.
- 58.USDA-PVPO (Plant Variety Protection Office), Plant variety database. https://www.ams.usda.gov/datasets/plant-variety. Accessed 21 September 2021.
- 59.Fisher R. A., Corbet A. S., Williams C. B., The relation between the number of species and the number of individuals in a random sample of an animal population. J. Animal Ecol. 12, 42–58 (1943). [Google Scholar]
- 60.Solow A., Polasky S., Broadus J., On the measurement of biological diversity. J. Environ. Econ. Manag. 24, 60–68 (1993). [Google Scholar]
- 61.Shannon C. E., A mathematical theory of communication. SIGMOBILE Mob. Comput. Commun. Rev. 5, 3–55 (2001). [Google Scholar]
- 62.Chao A., Chiu C. H., Jost L., Unifying species diversity, phylogenetic diversity, functional diversity, and related similarity and differentiation measures through hill numbers. Annu. Rev. Ecol. Evol. Syst. 45, 297–324 (2014). [Google Scholar]
- 63.Simpson E. H., Measurement of diversity. Nature 163, 688 (1949). [Google Scholar]
- 64.Keylock C. J., Simpson diversity and the Shannon-Wiener index as special cases of a generalized entropy. Oikos 109, 203–207 (2005). [Google Scholar]
- 65.Jost L., Entropy and diversity. Oikos 113, 363–375 (2006). [Google Scholar]
- 66.Havrda J., Charvát F., Quantification method of classification processes. Concept of structural a-entropy. Kybernetika 3, 30–35 (1967). [Google Scholar]
- 67.Daróczy Z., Generalized information functions. Inf. Control 16, 36–51 (1970). [Google Scholar]
- 68.Tsallis C., Possible generalization of Boltzmann-Gibbs statistics. J. Stat. Phys. 52, 479–487 (1988). [Google Scholar]
- 69.Hill M. O., Diversity and evenness: A unifying notation and its consequences. Ecology 54, 427–432 (1973). [Google Scholar]
- 70.Tuomisto H., An updated consumer’s guide to evenness and related indices. Oikos 121, 1203–1218 (2012). [Google Scholar]
- 71.Rao C. R., Diversity and dissimilarity coefficients: A unified approach. Theor. Popul. Biol. 21, 24–43 (1982). [Google Scholar]
- 72.Vane-Wright R. I., Humphries C. J., Williams P. H., What to protect?—Systematics and the agony of choice. Biol. Conserv. 55, 235–254 (1991). [Google Scholar]
- 73.Faith D. P., Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10 (1992). [Google Scholar]
- 74.Solow A. R., Polasky S., Measuring biological diversity. Environ. Ecol. Stat. 1, 95–103 (1994). [Google Scholar]
- 75.Tilman D., Functional diversity. Encycl. Biodiversity 3, 109–120 (2001). [Google Scholar]
- 76.Weitzman M. L., On diversity. Q. J. Econ. 107, 363–405 (1992). [Google Scholar]
- 77.Warwick R. M., Clarke K. R., New ‘biodiversity’ measures reveal a decrease in taxonomic distinctness with increasing stress. Mar. Ecol. Prog. Ser. 129, 301–305 (1995). [Google Scholar]
- 78.Magurran A. E., et al. , Long-term datasets in biodiversity research and monitoring: Assessing change in ecological communities through time. Trends Ecol. Evol. 25, 574–582 (2010). [DOI] [PubMed] [Google Scholar]
- 79.Marcon E., Scotti I., Hérault B., Rossi V., Lang G., Generalization of the partitioning of Shannon diversity. PLoS One 9, e90289 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Some study data available (The state-level area-by-variety data generated and analyzed during the current study are the property of the GEMS Informatics Center at the University of Minnesota. They are maintained in the GEMS Informatics Platform (https://gems.agroinformatics.org/) from where they can be requested from the corresponding author YC for use to replicate the findings of this study. The R codes for generating the results and figures reported in the current study are available at GitHub (https://github.com/y-chai/US-Wheat-Diversity.git). PedTools used in the current study for wheat pedigree processing is a python package developed by the GEMS Informatics Center at the University of Minnesota, which is not publicly available and can be requested for use to replicate the findings of this study).