Abstract
The agricultural sector and environmental safety both work hand in hand to promote sustainability in important issues like soil health, plant nutrition, food safety, and security. The conventional methods have greatly harmed the environment and people’s health and caused soil fertility and quality to decline as well as deteriorate. Keeping in view the excessive exploitation and cascade of degradation events due to unsustainable farming practices, the need of the hour demands choosing an appropriate, eco-friendly strategy to restore soil health, plant nutrition, and environmental aspects. The priority highlights a need for a sustainable and environment-friendly upgradation of the present agricultural systems to utilize the beneficial aspects related to harnessing the gene-microbiome strategies which would help in the restoration and replenishment of the microbial pool. Thus, exploring the microbiome is the utmost priority which gives a deep insight into the different aspects related to soil and plant and stands out as an important contributor to plant health and productivity. “Microbes” are important drivers for the biogeochemical cycles and targets like sustainability and safety. This essential microbial bulk (soil microbiome) is greatly influenced by agricultural/farming practices. Therefore, with the help of microbiome engineering technologies like meta-transcriptomics, meta-proteomics, metabolomics, and novel gene-altering techniques, we can easily screen out the highly diverse and balanced microbial population in the bulk of soil, enhancing the soil’s health and productivity. Importantly, we need to change our cultivation strategies to attain such sustainability. There is an urgent need to revert to natural/organic systems of cultivation patterns where the microbiome hub can be properly utilized to strengthen soil health, decrease insect pest and disease incidence, reduce greenhouse gas emissions, and ultimately prevent environmental degradation. Through this article, we wish to propose a shift in the cultivation pattern from chemical to the novel, upgraded gene-assisted designed eco-friendly methodologies which can help in incorporating, exploring, and harnessing the right microbiome consortium and can further help in the progression of environmentally friendly microbiome technologies for agricultural safety and productivity.
Graphical abstract

Supplementary Information
The online version contains supplementary material available at 10.1007/s10142-023-00982-9.
Keywords: Microbiome, Farming systems, Multiomics, Gene tools, Sustainability, Environmental safety, Agricultural security
Introduction
Food security, environmental problems, and farmers’ having small farm production systems are the main causes of the increasing burden and threat to the economy (FAO 2016; FAO 2019). These economic burdens are further accentuated because of the indiscriminate and excessive incorporation of pesticides; these not only result in actual yield loss but also pose great threats in terms of soil health, air, and water pollution and ultimately negatively affect human health. Therefore, the policymakers should closely check the farmers’ health and ecological concerns while framing successive policies related to the economic efficiency and productivity of horticultural production (Lampel et al. 2008).
The increasing dependence of the farmers on chemical inputs for vegetable production and management (Table 1) indicates the lack of awareness regarding other cost-effective, eco-friendly, and safe alternatives. The present scenario demands extensive studies on the adverse effects of pesticide usage and its exposure, which can prove harmful in terms of farmers’ health and environmental issues, and also to implement and avail appropriate strategies to finally reduce the losses due to various hazards (Ranit et al. 2021).
Table 1.
The commonly used pesticides (insecticides/fungicides) by the farmers for agricultural practices and operations, their consumption patterns (MT), uses, and detrimental effects on health (source: Centre for Science and Environment, 2013, Ministry of Agriculture)
| S.NO | Most commonly preferred insecticides and fungicides | Consumption pattern (MT) | Uses in agriculture | Detrimental effects on health |
|---|---|---|---|---|
| 1. | Insecticides | |||
| a.) | Phorate | 3000–3282 | Insecticides used against insects, mites/nematodes |
a) Respiratory failure and paralysis b) Central nervous system disorders due to secretion of neurotoxins by pesticides c) Kidney failure, problems related to hormonal disorders, impacts on male semen production d) Increased incidence of dreadful and life-threatening diseases like cancer |
| b.) | Acephate | 1500–1514 | Organophosphate/soil insecticides | |
| c.) | Cypermethrin | 2000–2500 | Synthetic insecticide | |
| d.) | Methyl parathion | 2000–2837 | Organophosphate insecticides | |
| 2. | Fungicides | |||
| a.) | Mancozeb | 3000–3115 | Fungicide |
Farmers’ pattern of pesticide usage over the years is based on multiple factors like pesticide application (in terms of insect, pest, and disease management), appropriate farm size, benefit-to-cost ratio, increased quantity and yield attributes, and varying agro-ecological conditions (Schaffer et al. 2018). These contemporary methods for applying harmful fertilizers and pesticides are used to produce crops in extraordinary quantities and yields while avoiding key considerations for environmental sustainability, food security, and food quality.
The aggressive and exaggerated use of chemical fertilizers and pesticides for the control of insects, diseases, and pests harms the beneficial microflora, which causes issues with the health and quality of the soil. In turn, this upsets the harmonious relationship between plant and microbial interactions. Additionally, these disrupted interactions result in changes in major and micronutrients, decreased nutrient efficiency and uptake, modifications to the biological balance, a rise in deadly diseases like cancer, and other associated environmental dangers. Estimates of the total economic burden typically externalize and depict the costs of treating health issues and other severe environmental impacts brought on by excessive pesticide use (Kohl et al. 2019).
The literature also confirms that the extensive contamination of subterranean natural water sources and serious damage produced by the excessive nitrate leaching process via soil profile arises from the vigorous application of pesticides, especially due to the high nitrogen content (Thakur 2022).
There is a significant propensity toward alternative tactics that are safe and secure in terms of both ecological and health as a result of the growing concerns and burning issues around the excessive usage and application of pesticide doses (Thakur 2017e). The propensity for a sustainable natural approach is also a result of routinely observing the worsening imbalances in the food chain and water sources caused by the persistence of harmful residues and the loss of nutrients and flavor components. As a result, a novel method of farming known as sustainable agriculture, which has been gradually developing since 1999, has emphasized using agricultural practices that are particularly eco-friendly, consequently, preserving and stabilizing the soil ecosystem’s long-term ecological balance and encouraging effective nutrient cycling and biogeochemical processes at a stabilized scale (Leahy et al. 2014; Frederiks and Wesseler 2019).
In the current review, we concentrate on sustainable organic farming practices, particularly emphasizing the positive aspects relating to soil health, controlling the spread of insect-pest diseases, and finally highlighting microbiome-based techniques and innovations with their applications for sustainable crop farming and food safety. The main goal is to popularize and connect various microbiome-based technologies with on-farm input generation (using local resources, natural farming, and sustainable practices) so that farmers can easily access technological interventions through sustainable microbiome-based applications and methods (Thakur et al. 2022a, b).
Soil-microbiome: clustural organization of microbes
Soil stands as a consortium of many microbes including fungi, archaea, and bacterial flora. These microbes are capable of forming symbiotic associations (Kassam et al. 2009) with plant roots and are actively engaged in colonizing a specific area called the “rhizosphere.”
The assortment of microbes related to a given surrounding (both biotic and abiotic) and their aggregate genetic materials is named the microbiome. The soil microbiome can be defined as the “cluster of micro-organisms which forms the crucial part of the soil.” Examination of microbiomes of pests, plants, and regular natural assets could be utilized to foster novel management instruments for hostile insects and pests.
Symbiotic microbes (Skinner et al. 2014), an essential component of the soil microbiome, carry out various essential functions like:
Nutrient cycling, including all biogeochemical processes like the sulfur cycle, nitrogen cycle, phosphorus cycle, and carbon cycle.
Involved in minimizing threats like soil erosion, by gripping up the soil structure and subsequently leading to a successful increase in the organic matter content, which stands as an important boosting factor in enriching the fertility, health, porosity, and water holding capacities of the soil.
Rhizosphere-associated microbes contribute to activating specific types of plant defense mechanisms, which confers disease resistance to crops.
Improving the tolerance of plants towards various environmental stresses like variation in temperature and moisture content.
Initiating and acerbating processes like root growth and nutrient uptake.
Effects of intensive agricultural practices on diversity and functioning of soil microbiome
Agricultural practices based on intensive mechanisms often degrade and exploit the diversity and functioning the of soil microbiome, (Lakshmanan et al. 2014; Munster 2019) by promoting unessential microbial activity. This exploitation leads to risks associated with a reduction in crop yields, an increase in cost, and deteriorated environmental impacts (Thakur 2022).
The first problem in intensive farming practice is the (Postma et al. 2012) use of plowing methods and machinery which leads to destruction in both soil pore spaces which are generally residential areas of beneficial microbes and fungal networks — the area involved in maintaining the firm soil structure and storing the organic matter. This degradation leads to fertility and soil health issues, and water and nutrient retention problems. The second problem is faulty practices like continuous cropping methods that make the crops more sensitive to soil-borne diseases, which can directly affect the yield and can result in heavy losses to farmers (Letourneau et al. 2008). The third problem is the use of heavy doses of nitrogen fertilizers for obtaining high yields. The nitrogen cycle results in the conversion of excessive fertilizer to nitrate which can run off into various water streams causing environmental damage. On the other hand, they also produce nitrous oxide, which is a major contributor to global warming as it is a greenhouse gas (Thakur 2017a, b, c, d, e).
Nitrate leaching is a common runoff process witnessed in agricultural processes where the water contaminated with excessive nitrate leaches through the soil causing nitrogen losses from agricultural fields and contaminating the water sources in the form of algal blooms (eutrophication). The eutrophication problems, observed more in non-organic modes of farming, are generally the biggest threat to environmental degradation of the beneficial flora due to larger nitrogen inputs and higher stocking densities. It is well documented that in the case of organic practices, there is no negative impact on the ecology and environment due to the utilization of eco-friendly inputs, whereas the chemical cultivation system degrades the environment due to synthetic herbicides and pesticides (Shepherd et al. 2003). The burning global warming issues are continuously threatening the ecological processes due to harmful emissions of greenhouse gases (GHGs) from agriculture. The major reviews regarding the greenhouse emission highlight that though produced in organic cultivation methods (application, production, and storage of manures, interaction between different fodder and litter, variation in livestock breeding, etc.) are comparatively reported to have lower emissions as compared to non-organic methods (Skinner et al. 2014).
The transition from chemical cultivation pattern to eco-friendly organic strategies (in-conversion farming)
Intensive agricultural production is essential to satisfy food requirements. During 1950–1960, food production was boosted by using agrochemicals, but the physical, chemical, and biological properties of the soil were destroyed by using these chemicals causing great damage (Bonvoisin et al. 2020). They mined the power of “biological resistance” and micro-organisms beneficial to the soil thus increasing the susceptibility to pests and diseases. The consequences of excessive usage of agrochemicals are marked by the presence of toxic pesticide residues in each cropland. Toxic pesticides work as a “slow poison” for the soil and lead to problems in the quality and quantity of the crop (Jangir 2008; Barrett et al. 2017). It also results in a reduction of the helpful microbes, which leads to an imbalance in the ecology. The use of non-renewable energy chemicals often targets essential soil properties. It destroys the physio-chemical properties, reduces the friendly pool of predators, and enhances the toxic residual hazards in seed, eventually posing a great danger to human health and the environment (Braga et al. 2020).
Considering these hazardous effects, the present scenario demands quality products — the conversion of intensive chemical cropping to a safe natural organic farming system. This promises to boost crop yields and yield high profits from the foreign exchange on export. Conventional farming is also assumed to disturb the food chain and lead to a decrease in soil fertility, an increase in soil erosion, loss of biodiversity, and a decrease in the production of various species. Chemical fertilizers and pesticides are used on crops to kill and destroy harmful pests (Thakur 2017a, b, c, d, e; Kour et al. 2019). Currently, many farmers are using conventional farming to increase their cost-to-benefit ratios. They are unaware of the adverse effect of these products on human health and the environment. Many programs are being organized by the government to spread awareness about the ill effects of chemical farming on rapidly using pesticides and genetically modified organisms (GMOs) to produce crops (Brescia 2020).
From 1999 to the present, there are major modifications in agricultural practices. Farmers who were adopting conventional farming started switching to organic farming, as they recognize and saw the many associated benefits such as converting from chemical farming to organic farming (Thakur 2017a, b, c, d, e) which led to increased production as well as an increase in the cost of the products due to high nutritive value and flavor contents. This process of converting from conventional or chemical farming to organic farming is known as in-conversion farming. Farmers started using bioorganic compounds in the farming process. In-conversion farming generally adopts organic processes. This process prohibits chemicals from being used on crops. They must rely on items such as green manure for this organic production. Recently, there has been a significant increase in the number of producers under in-conversion farming. Despite lack of the support from the government in providing assistance, knowledge, and extension facilities to the farmers in India (Panneerselvam et al. 2012), in-conversion farming has only grown in popularity and acceptance. To get fully converted into organic farming the producer must be “in conversion” for at least 2 years.
Increasing threats related to erosion of soil, insect-pest management problems, and rigorous use of chemicals can be reduced by adopting environmentally friendly strategies like crop rotation and crop covering (green manure), which is a crucial part of typical organic farming. It involves the progressive increase in the addition of organic inputs in the soil, as nature does in a forest. Farmers activate the ability of the soil to effectively absorb water, reducing the impact of extra leaching or flooding. In addition, it improves the absorption and storage capacity of soils in retaining an active pool of organic carbon and important nutrients, which are required for the attainment of healthy produce, making it more resistant to pests and diseases (Reddy 2017). Therefore, organic farming promises a secure and sustainable environment, nutritionally enriched food, and good living standard by upgrading the lifestyle of small livelihoods in India. It also promises sustainable, secure, and residue-free long-term agriculture because of its associated low costs. Farmers adopting the organic method of farming are receiving better yield, and better growth, as compared to conventional farming (and earning profit from the crops (Redlich et al. 2018).
Organic farming mainly emphasizes food quality and human health (Andrade et al. 2011). The process of in-conversion farming involves farmers initially using organic fertilizers instead of chemical fertilizers, reducing pesticide residues. A safe and secure mode for crop protection is the beneficial use of microbial inoculants (bio-fertilizers) combined with organic manures. The new approaches, targeting the use of organic amendments in farming, have successfully proven to be beneficial in improving soil health and structure, thus increasing soil fertility and crop yields. These organic inputs provide an excellent and efficient source of plant nutrients and increase efficient nutrient uptake through managed and enriched soil. This, in turn, maintains a high load and efficient functioning of beneficial microbial populations leading to enhanced biomass and organic carbon content with increased basal respiration and metabolic rate. The successive crop yields have been witnessed with parallel improvements in soil and produce better quality products due to the incorporation of organic matter (Tonfack et al. 2009). Organic farming is further defined by various authorities: The US Department of Agriculture introduces organic farming as a natural and safe production system that strictly prohibits the use of chemical fertilizers, pesticides, artificial growth regulators, and animal feed additives. The feasibility of organic farming depends on all the synergistic strategies including green manuring, incorporation of cover crops, crop rotations, crop residues, animal manures, off-farm and on-farm organic wastes, and biological pest control strategies to maintain soil fertility and tilth. Similarly, the principles of organic cultivation highlight the sustainable management of soil through well-managed husbandry, the recycling process of agricultural residues, and reduced dependencies on external inputs. All the above strategies indicate utilizing natural methods for weed and pest control (Epelde et al. 2018). Organic farming has the potential to improve and protect the environment, protect non-renewable resources, improvement of the quality of the food, reduction in the output of foreign products, and convert agriculture into a sustainable and productive phase. Bio-controls are used in crop rotation schemes for the protection of crops from pests and diseases. Organic farming helps in the improvement of the agricultural landscape. It affects both the storage of carbon and the emission of methane and nitrous oxide in the soil. The use of compost greatly improves soil fertility.
Analysis by Durdane et al. (2011) revealed the effect of different chemical and organic fertilizers on the yield and the crop quality of the bush and dwarf type of peas. There was an obvious observable difference in yield and crop quality by using different types of inorganic and organic fertilizers. During the 2010 growing period, various types of organic fertilizers were used in different ratios (Al-Zyoud 2014). Excess use of pesticides on the crop and soil may disturb the microbial load and soil fertility. Pesticides (Table 1) are considered a threat or poison if they are misused. The government of India has officially created many acts for controlling the misuse of pesticides. Various committees were in action for controlling the excessive use of chemicals in the crop’s yield and soil. They set particular limits on the use of pesticides and banned some of the chemicals. These are termed maximum residue limits (MRLs) (Garrigou et al. 2019). Organic farming has been very important in the improvement of soil fertility, soil texture, and soil health. It also increased the life of products, which helps keep diseases away from consumers. The area under organic production has increased dramatically. Out of the total area under organic production — 3.99 M ha area is under wild organic collection; 0.54 M ha area is under organic cultivated area; and 0.18 M ha is under in-conversion phase. In 2020, 1.6% of the world’s agricultural land was organic. By region, Oceania had the highest share of organic farming (9.7%), while Europe had the lowest share (3.4%). In contrast to the global share, some countries have much higher organic shares. Liechtenstein (41.6%) and Austria (26.5%) had the highest organic shares. In 18 countries, 10% or more of the agricultural land was organic. In India, Madhya Pradesh tops the list for the maximum area under organic certification (232,887 ha). Himachal Pradesh ranks 9th with a total area of 4686 ha under organic certification (Agricultural and Processed Food Products Export Development Authority (SAPEDA 2014).
Rejuvenating soil physiology and ecology through sustainable farming system
Continuous unsustainable farming practices and rigorous agricultural intensification have caused great damage in terms of soil degradation, the decline in fertility, decreased pool of beneficial microflora, soil nutrients, and frequent soil erosion. This directly affects the ecology and provides severe consequences for the human population (Setala et al. 2014).
The natural farming strategies aim at rejuvenating and replenishing the various nutrient cycles and biogeochemical processes which are mainly driven by symbiotic associations or positive interactions of microflora. The adoption of some natural farming methods like green manuring, cover crops, incorporation of leguminous crops, intercropping crop rotations, manures, and mulches can increase the soil organic matter and can result in higher external carbon inputs (Leithold et al. 2015), increased biomass and microbial activity, soil macro and micronutrients, aggregate-bulk stability, and water-holding capacity. The breaking down of the organic matter is the major process associated with agricultural cultivation methods like crop rotation and green manuring (Scheller et al. 2008; Damalas et al. 2018). Methods related to the utilization of organic crop residues and litter have been reported to improve nutrient availability due to higher decomposition rates due to higher microbial populations and positive interaction near the soil-root zone. In comparison, in areas of utilization of non-organic methods, lower decomposition rates are observed due to excessive pesticide application which degenerates the beneficial microbial pool (Tuck et al. 2014). Therefore, the importance of sustainable and organic farming practices which are ultimately improving the soil structure and physical quality due to higher earthworm activities (vermicomposting) lowers the incidence of soil erosion (Leifeld et al. 2013).
Zero budget natural farming: re-designing the agroecosystem
The redesigning or reframing of the agroecological solutions has been trending right from the movement away from (Hill 2014) traditional farming and toward modern cultivation patterns (Palekar 2010). We are moving toward different forms of natural farming (Khadse et al. 2018; La Via Campesina 2016).
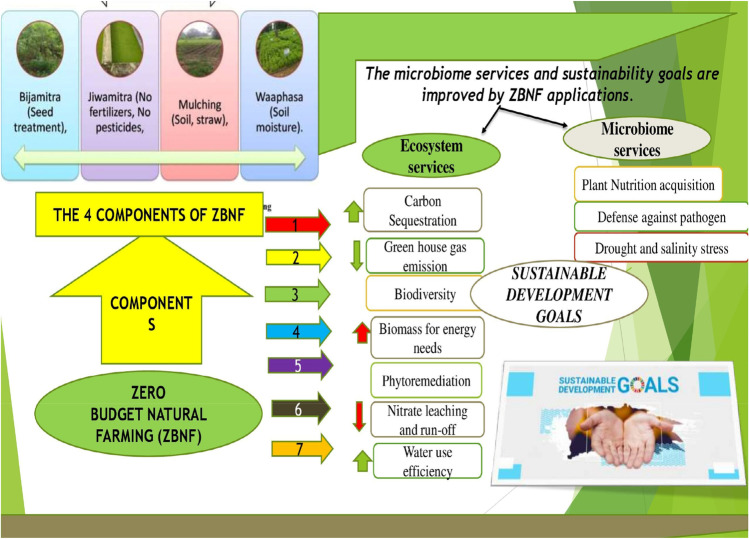
A recently framed form of natural farming is “ZBNF” (zero budget natural farming). This started as a social movement but ended as a major policy initiative. This form of farming traces its journey from Andhra Pradesh in 1980, where Subash Palekar, an agricultural scientist started (Khadse et al. 2018) explored the agricultural roots by taking the initiative of practicing the different cultivation methods (organic and chemical farming) in his farms. The expansion of the ideas has reached foreign countries to revert to the modes of natural farming and harnessing the crucial microbiome. The main aim of ZBNF is to drastically reduce the high production costs by cutting down the external dependencies on synthetic inputs and credits related to agricultural production. This natural farming concept is also called the “four-wheel concept of farming” (Fig. 1) as it depends on four essential factors (Via Campesina 2016; Pretty et al. 2018).
Fig. 1.
The different components of ZBNF (zero budget natural farming) directed towards the enhancement of microbiome services and sustainability goals
The process follows an efficient strategy to combat insects, pests, and diseases by making the farmers aware of the specific lifecycles of different pests and related antagonistic species. In addition, this green farming also acts as “substitutive farming” (Table 2) as it replaces all the chemical and synthetic fertilizers and pesticides with alternative on-farm inputs and herbs. Furthermore, all these strategies are directed toward replenishing the soil with microbial formulations and natural herbs and inputs (Williams et al. 2020).
Table 2.
The effect of practicing zero budget natural farming in community-based farming practices
| S.NO | Factors | Improvements and benefits reported by the farmers on a global scale by adopting ZBNF |
|---|---|---|
| 1. | Crop parameters |
a) The farmers reported the extended crop factors’ flexibility towards varied environmental conditions like dry climate b) The crop losses in high-value crops due to pests were stabilized by the stabilization of yields in other crop types c) Crop leftovers can be preserved as a decomposition surface as earthworms and bacteria break down organic materials to encourage slow nutrient release throughout time, in the soil d) Just as it would be in natural ecosystems, there is no weeding, no fertilizing, no soil tilting, and no plowing e) One of the most affordable farming techniques, with no external supply, totally integrates with regional biodiversity |
| 2. | Farmer’s health |
a) Reduces health hazards from chemicals, maintains agricultural ecosystems, lessens the burden on women by giving them better access to clean water and livestock feed, and decreases illnesses brought on by chemicals in food, especially in children b) Farmers reported a better recovery from bad health to good health conditions with less incidence of diseases |
| 3. | Livelihood upliftment |
a) Improvements were reported with better housing conditions and upgradation in lifestyle b) Low input, lower cultivation costs, and constant yields allow farmers to have consistent crop yields to sell in the market, enhancing the security of their livelihoods |
| 4. | Soil and Environment health |
a) Improvements in soil fertility and increased ratio of earthworms b) Moreover, processes like mulching enhanced nutrition addition through residues generated from previous crops c) Conserving the environment enables drought-prone areas to produce regular crops by enhancing soil microbial content and water retention capability d) Using fewer chemical fertilizers improves water quality and increases availability during extreme weather events by reducing runoff into rivers and wetlands e) ZBNF encourages multicultural preservation always covered in biomass. It enhances the soil microbiota as well through locally produced cow urine and bio-inoculants based on dung The soil microbiome transforms nutrients from “locked” to “bio-” in the soil f) Accessible forms, thereby producing a sustained circulation of nutrients relationship between microorganisms and plants and soil |
| 5. | Value addition and marketing |
a) The product has good quality and consumers have been observed to directly contact the producers eliminating the profits of middle man b) According to some studies, every dollar spent to encourage a farmer to use ZBNF yields an immediate advantage of 13 dollars c) The immediate advantages include cheaper cultivation costs, higher yields, lower borrowing costs, increased income from intercrops, and a little higher selling price d) Additionally, there are several social and environmental advantages, including the security of food, nutrition, and health; employment; soil and water security; restoration of coastal ecosystems; climatic resilience; biodiversity protection; and reduced risk |
| 6. | Utilization of cattle manure | ZBNF incorporates the use of cattle dung and cattle urine by farmers to maintain cattle sheds. It can be further, sold out to extended villages for creating awareness regarding natural farming |
The overall goal of ZBNF is to reach out to 6–8 million farmers and consequently convert about 8 million hectares to full ZBNF fields in the coming decades, to conserve farmers’ livelihood, ecosystem, and environment (Walker et al. 2021).
Restoring long-term agriculture sustainability: addressing food security, safety, and environmental concerns
Organic agriculture, on average, produces lower yields (Kalloo 2003) and productivities as compared to chemical cultivation methods but these gaps prominent in yield and productivity can be patched up after a successful inter-conversion period. Intensive, non-organic agriculture techniques pose a momentous threat and risk to the environment and human health. The development of pest resistance as a result of these unsustainable techniques ultimately raises concerns about the long-term sustainability of managing disease and insect pests (Ponisio et al. 2015). Thus, these unsustainable approaches do not offer a solution to our problems with food safety and security. The need of today’s era is improvement in organic strategies to increase the yield by making use of methods like crop rotation, inter and mixed cropping patterns, and reducing tillage (Niggli 2015). To minimize disadvantages and maximize the advantages of both systems, research has also recommended “partial conventionalization of organic agriculture.” This proposes alterations in field management practices to allow broader aspects of pest and plant nutrient management (Bàrberi 2015). The mixing up of various farming practices to reduce the risk of toxic residues and solve issues related to food security, yields, and environmental issues can prove a solution. Therefore, it can be concluded that organic farming practices need some modification and should be combined with other farming methods, such as integrated management practices, to ensure long-term sustainable agriculture that can feed the entire population while keeping criteria like security, quality, and ecological safety (Reganold et al. 2016). This situation demands a holistic approach where quality and safety in terms of food production or environmental issues cannot be compromised (Reddy 2010).
The increasing interest in food quality is a clear indication that organic products are safer, nutritious, and healthier (Mulet 2014, Rouphael et al. 2020). In terms of food quality and safety, consumers prefer organic produce as it ensures a high concentration of antioxidants, healthy vitamin C content, secondary metabolites, polyunsaturated fatty acids, and lower pesticide residues and cadmium concentrations (Illukpitiya and Khanal 2016).
Revival of the microbial pool and its applications
Soil health is considerably enhanced by organic farming. The fertility of the soil is often correlated with its health, which is related to a good, healthy microbiota (Doran and Zeiss 2000) with enough organic matter (Kumar et al. 2019). The activity and activation of these soil-dwelling microbial communities create the largest repository of biological diversity that continues to interact favorably with other related populations (Berendsen et al. 2012). The biologically varied microbiome, nutrient uptake and efficiency, water holding capacity, healthy soil, resilience to bad weather, and toxins make up the strongest backbone, which is one of the key characteristics of healthy soil (Spence and Bais 2013). This microbial pool plays a crucial part in controlling ecological processes, such as replenishing biogeochemical cycles an important role (Treseder et al. 2012) in the regulation of ecological processes like replenishing biogeochemical cycles, antagonistic and symbiotic plant interactions, and decomposition-transformation processes of organic material (Verheijen et al. 2015).
Harnessing the rhizosphere (plant–microbe interactions) microbiome
We aim to best utilize the benefits of the soil microbiome to achieve the goal of sustainability, (Bulgarelli et al. 2015) (through clean and green technology) (Fig. 2) for enhancing plant and crop growth, nutrient availability, uptake efficiency, abiotic tolerance, and (Delmotte et al. 2009) inducing resistance in produce against pests and diseases in agricultural production. To accomplish such integration (Bakker et al. 2012), a joint effort and strategy are required from academicians, industrial collaborators, and farmers to manage all types of plant-microbiome interactions associated with or about sustainable agricultural systems. In this regard, we must prioritize some strategies (Busby et al. 2016):
Developing a suitable crop-microbiome interaction model system integrating the right strategies and POPs (package of practices) according to specific crop and non-crop plants with related microbial genetical pools.
Defining important metagenomes related to sequencing patterns of specific beneficial microbes in the designed model systems,
Determining the functional mechanisms of environmental-microbiome interactions,
Characterizing plant genotypes by understanding the different management interactions.
Fig. 2.

Illustration highlighting the Rhizospheric plant–microbe interactions
To meet these above-set targets (Lucas et al. 2013), we should speed up our efforts to integrate and frame up effective agricultural microbiomes and sustainable strategies, which will finally strengthen the world food, supply chain (Rascovan et al. 2016), and environmental safety.
Crop health and nutrition
There is a significant increase in the research associated with the increasing potential to improve parameters like crop health and nutrition, via effective plant, soil, and microbe interactions, i.e. harnessing the rhizosphere. Easy availability and utilization capacities of nutrients are determined by efficient translocation of nutrients. Therefore, the major criteria of efficient nutrient utilization by the plants and crops are linked to the group of some beneficial microflora known as the “PGPRs” (Ratnadass 2020). Their survival and sustaining capacities around the root zone with or without symbiotic colonies like VAM (vesicular arbuscular mycorrhiza) affect the overall translocation process of nutrition. They form a part of a process called “Rhizosphere Competence” where they actively participate in promoting nitrogen uptake, protecting root zone and system from the attack of pathogen microbes and pests, promoting nutrient mineralization, and supporting arbuscular mycorrhizal associations (Ratnadass et al. 2012). The arbuscular associations further promote efficient utilization of phosphorus, various micronutrients, and water supply under low-input conditions (Rothschild et al. 2002; Ratnadass and Barzman 2014).
Developing microbiome-based innovations for sustainable expansion of crop farming and food safety (plant diseases)
Under shrinking arable land and shifting climatic circumstances, cultivation frameworks are feeling the strain of meeting the needs for food and fiber for a growing global population in an environmentally responsible manner. Additionally, routine agricultural practices have caused a decline in soil fertility, and in rare instances, improper and excessive use of synthetic pesticides and composts has led to soil exploitation, which hurts both human and ecological wellness. Around the world, it is believed that a rising number of plant pests, including invertebrates, pathogens including viruses, bacteria, fungus, and weeds, are to blame for up to 40–50% of losses in agricultural productivity, quality, and yield (Peshin and Dhawan 2009). Chemical fertilizers and hazardous pesticides are routinely used as part of pest management strategies. The use of genetically modified organisms (GMOs), which are outlawed by organic farming methods and tactics, is permitted under these non-agricultural management techniques. The health and utility of plants are significantly influenced by the soil and plant microbiomes.
Additionally, because they are the primary drivers of global biogeochemical cycles, microbes are essential to sustainable agricultural methods (Fig. 3). There is growing evidence that, with the development of appropriate innovations, the plant microbiome can be regulated to possibly decrease the occurrence of plant diseases, increase resource use efficiencies, and ultimately improve the production of agricultural products while reducing the contribution of synthetic composts and pesticides and causing the decreased use of natural resources (Peshin and Zhang 2014). Conservational biological control methods in organic farming target utilizing the beneficial organisms or population pool of natural resident enemies and predators to naturally focus them to suppress the pest population and further lower the prevalence of diseases (Birkhofer et al. 2008). In addition, the use of some vegetation prominent predators can be higher under organic cultivation methods which can control the rate of animal pests in major crops. The foliar and root diseases can be controlled by following an appropriate organic management protocol incorporating suggested applications of organic pesticides or biopesticides (Geiger et al. 2010). The appropriate organic strategies may prove more eco-friendly, safe, and secure and would reduce the incidence of insects, pests, and diseases to minimal levels as compared to the non-organic strategies.
Fig. 3.
The different practices/tools for sustainable and eco-friendly farming strategies
In organic agriculture, most soil-borne (Birkhofer et al. 2016) diseases and pests can be controlled by activating the biodiversity prominently present in the soil and environment. This activation can be initiated by incorporating good soil amendments to feed the soil microflora in a balanced rotation (Harman et al. 2004). Research indicates the high suppressibility of diseases in organic soils, but more work must be done to focus on using this soil microflora as an important management tool to increase soil defenses in a balanced and controlled manner (He et al. 2010).
Advancement in sustainable microbiome-based technology, research, and industrial processing
A vital challenge to crop safety issues is the emergence of pesticide resistance, and for some insects and pests, there are no elective control techniques. Improvement of newly manufactured pesticides has gotten progressively exorbitant and testing, mostly because of the trouble of changing over a lead compound in a product that can go through exacting ecological and security guidelines. A new report assessed that in the USA, the advancement of an engineered pesticide presently costs more than US $300 million and takes almost 12 years (Jacquet et al. 2010). The need to beat the resistance issues and to get advancements in reasonable horticulture requires the disclosure of new insecticidal specialists. Lately, the push to foster novel insect poisons with negligible ecological effect has prompted a resurgence of interest in biopesticides (i.e. pesticides dependent on living organic entities or their normal items, including their qualities and metabolites). Presently, the worldwide market for biopesticides is esteemed at US $3 billion and is relied upon to develop by 15% in the following 4 years, outperforming the market development of engineered pesticidal materials by 10-folds (Damalas et al. 2018). The following points discuss sustainable microbiome-based technology and help us understand the current new advancements in this field of research.
Virus communities in the soil microbiome
There are around 107–109 virus units per gram of soil. These soil viruses are extremely unknown and quite diversified in nature because their prevalence is modest in comparison to the viruses (Williamson et al. 2017) prominent in the marine ecosystem. According to several research on marine virus populations, phages primarily regulate the concentration of nutrients by eradicating 15–45% of the marine ecosystem’s microbial population (Sime-Ngando 2014). Viral communities have grown and become more diverse due to recent advancements in the field of viral metagenomics. As a result, new viral communities have been discovered, and more knowledge has been gained regarding how these viral communities (Fig. 4) affect various microbial communities and microbial cacti activities importantly is more effective to investigate these microbial populations in interaction than to research them separately (Tshikantwa et al. 2018).
Fig. 4.
Beneficial viruses and their roles in crop growth and maintenance
Biological engineering of soil
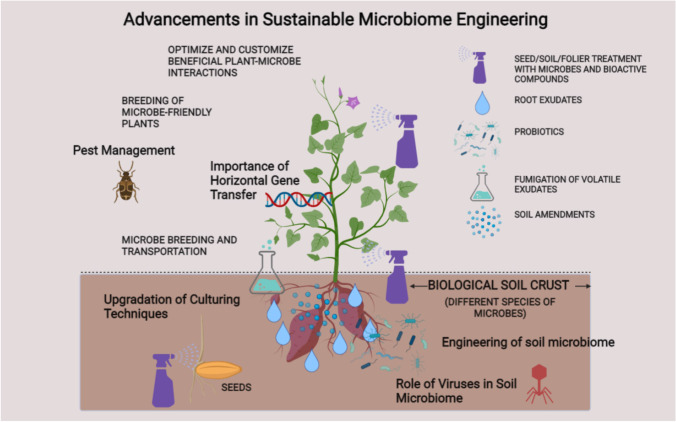
The majority of the soil’s microbial population, known as the soil microbiome, is heavily influenced by agricultural methods and cultivation patterns. A lot of information has been collected about the characterization of microbiomes. It is time to move on from the “finding phase” and into the “translational phase.” For stimulating and encouraging plant growth under varied conditions and to resist the various biotic and abiotic challenges, we must deploy these microbiome-based remedies in situ. A highly diverse and balanced microbial community in the soil can be achieved with the use of soil microbiome engineering techniques. The soil becomes healthier and more productive as a result. This technique aims to support soil microbial diversity, bio-organic cycles, and biogeochemical processes by employing the fewest chemicals necessary. Compared to chemically treated soil, eco-friendly strategies like organic farming in long run can boost soil richness and productivity (Hartmann et al. 2015), thus ensuring effective use and management. These activities aid in determining how these cutting-edge engineering methods affect the soil microbiome, to improve crop quality and productivity (Figs. 5 and 6). Here are several methods for modifying microbial communities to carry out particular tasks to improve plant performance.
Fig. 5.
New age advancements and upgradation in sustainable microbiome engineering
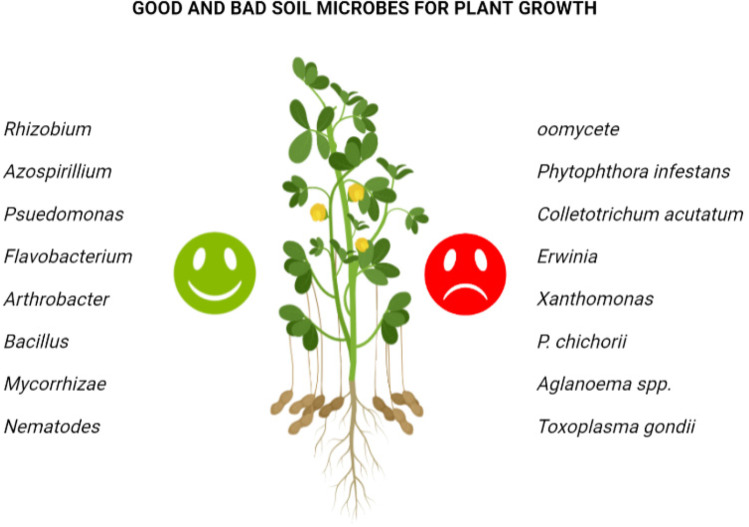
Fig. 6.
Microbes present in the soil that are good and bad for plant growth
Host-mediated engineering and optimization of the microbiome
The microbiome-based applications and methods provide an easy pathway to improve plant performance using microbiome-based solutions and also create well-adapted specific species which possess features beneficial to the host. Therefore, the most preferred criteria correspond to the screening of special microbiota that is native in nature, and well adapted to the environmental conditions including its soil types, and plant niche (e.g. rhizosphere or phyllosphere). This will further provide a better chance of establishment and the manifestation (Jochum et al. 2019) of beneficial traits compared to foreign microbiota (Gu et al. 2020). Furthermore, it is now possible to select a plant-optimized microbiome with the help of an experimental evolutionary design approach (Morella et al. 2020). These evolved microbiomes can survive for many generations on, in, and around plants providing long-term advantages and multiple benefits (Arias-Sanchez et al. 2019). For example, specialized microbial communities can be assembled through artificial selection to alter the flowering time of plants in a highly reproducible manner. Accelerating flowering time helps plants maximize the chances of reproduction under multiple stress conditions (Kazan & Lyons 2016). These techniques aim to ultimately the development of stress-tolerant phenotypes with increased productivity under changing climates like drought (Jochum et al. 2019) and salt tolerance (Skinner et al. 2014).
Plant engineering through the utilization of optimized microbiome
Through gene engineering, plants themselves can be modified/altered to perform specialized microbiome assembly or improved microbial-mediated tasks. For instance, poplar plants have undergone genetic modification to reduce the activity of cinnamoyl-CoA reductase (CCR). This affects the rhizosphere microbiome without changing the structure or function of the root endophytic population (Beckers et al. 2016). The composition of the root microbiome is affected by host genetics. Utilizing microbiome-based features for selection in plant breeding has enormous promise.
The heredity of microbiomes and their correlation with agronomic traits are both supported by recent studies. For instance, data on the rhizosphere microbiome collected during the flowering stage from 16 distinct canola genotypes showed that genetics was responsible for (Taye et al. 2020) 37–59% of the variation in the number of different microbial species. The same goes for the taxa Amycolatopsis sp. Serratiaproteamaculans, Pedobacter sp. Arthrobacter sp. Stenotrophomonas sp. Fusarium merismoides, and Fusicolla sp. which have been shown to positively affect canola yields through field research (Lay et al. 2018a, b). Breeding for disease resistance has resulted in unintended changes in rhizosphere ecosystems, including an increase in bacterial taxa that metabolize chemicals protecting against root infections, according to community-level physiological profiling and meta-transcriptomic analyses (Yi et al. 2014; Wintermans et al. 2016; Mendes et al. 2019).
The successful application of these strategies in breeding programs requires a deeper understanding of genotype interactions, notwithstanding advances in our understanding of how the microbiome influences host performance (Busby et al. 2017). Future research must concentrate on identifying plant genes that affect the composition of the microbiome. These investigations must be carried out in natural fields that are managed according to best practices and involve multiple locations and multiyear field trials to obtain the most reliable results (Wille et al. 2019). By manipulating these candidate genes, we might be able to create designer plants with specific microbiomes and the advantages that go along with them. For instance, in rice, the phyllosphere microbiome is influenced by genes involved in glucose metabolism and stress responses (Roman-Reyna et al. 2019). standing of genotype interactions, notwithstanding advances in our understanding of how the microbiome influences host performance (Busby et al. 2017). Future research must concentrate on identifying plant genes that affect the composition of the microbiome. These investigations must be carried out in natural fields that are managed according to best practices and involve multiple locations and multiyear field trials to obtain the most reliable results (Wille et al. 2019). By manipulating these candidate genes, we might be able to create designer plants with specific microbiomes and the advantages that go along with them. For instance, in rice, the phyllosphere microbiome is influenced by genes involved in glucose metabolism and stress responses (Roman-Reyna et al. 2019).
Optimization of the microbiome through various management practices
Certain effective management practices such as crop rotation, cover crops, mulching, and use of organic manure encourage the development of disease-suppressive soils. The disease’s suppressive properties can be easily passed on and transferred to the suppressive soils by mixing small portions of conducive soils with suppressive soils (Raaijmakers and Mazzola 2016). Therefore, the beneficial microbial effects of certain management practices can be transferred, thus steering the agroecosystems towards a healthier state. The legacy effect of management practices on the assembly of soil microbiomes impacts gene expression ultimately impacting plant physiology, specifically plant hormone production (Blair et al. 2018). A new study demonstrates that soil microbiomes can be designed to induce resistance against aboveground herbivorous insects through plant-soil feedback (Pineda et al. 2020). Organic farming and associated holistic practices have been shown to improve the structure and activities of the soil microbiome and soil nutrient cycling (Mendes et al. 2011). However, the management decisions are influenced and affected by various parameters ranging from the selection of varieties and site, soil health, utilization of manures/fertilizers, nutrient applications, yield and profitability, use of machinery, and the benefit-to-cost ratio analysis (Trivedi et al. 2020). Therefore, more studies are still required to trace out the holistic approach for designing and engineering more sustainable management practices based on different parameters taken into consideration and finally accessing the various impacts on soil microbiota to contribute more sustainably to agricultural production in the long run.
Altered microbiomes
Genetically modified microbes
Numerous genes in microbes that mediate plant–microbe interactions have been discovered using genome-based techniques (Levy et al. 2018). We must manipulate microbial genes to promote advantageous features if we are to create microbial inoculants which ensure superiority and safety. A recent development is the genetic modification of the symbiotic bee gut bacterium Snodgrassella alvi to generate double-stranded RNA that results in profuse RNA interference (RNAi) in bee hosts). The modified bacterium’s activation of RNAi shields honeybees from the infections generated by some viruses and mites (Leonard et al. 2020). Similarly, endophytic engineering providing an activated plant defence mechanism against pathogens and pests is also possible using similar upgrading microbiome strategies. The options for editing the genome have substantially expanded thanks to the very effective clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein CRISPR/Cas systems (site-directed nucleases) (Uniyal et al. 2019). Additionally, it has substantially increased the possibilities for editing genes and genomes that can be used to improve the advantageous characteristics of plant-associated bacteria. With the aid of these technologies, it is possible to create regulatory circuits (inducible in nature) that regulate the induction of advantageous features in response (Farrar et al. 2014) to the signals generated in the signaling process operating between the target organisms and the plant host (in the case of biocontrol). With the use of synthetic biology tools and biosensors, significant work is being done to redesign networks of the microbial genes for effective utilization in agriculture (Goold et al. 2018). One item, called Banana Guard, was created and has a Pseudomonas putida strain that has been genetically altered to prevent the (Kemal et al. 2016) Fusarium oxysporum infection that causes Panama disease in bananas. After detecting fusaric acid produced by Fusarium oxysporum, these modified bacteria release fungal growth inhibitors. It also possesses a kill/automatic killing switch that causes automatic destruction/self-destruction when the fungus is no longer recognized. Recently, naturally occurring cereal crop endophytic or epiphytic microorganisms were modified to facilitate nitrogen fixation). Further modifications were made to the engineered nitrogen-fixing strains to cause them to express N2-fixing genes in response to chemicals found in the rhizosphere and seeds, which are especially reported to alter the microbiomes of plants (Ryu et al. 2020). Even the development of special designer circuits for genes in microbes that are receptive to signals derived from plants for optimizing microbe interactions is now achievable with the help of modern targeted genome editing techniques. These new tools not only provide the methodologies for altering and modifying a complex microbiome but also provide excellent opportunities to even mark modifications in the genomes of the specifically cultured bacteria (Sheth et al. 2016). Technologies related to gene editing can be used to create efficient microbial inoculants or alter local microbiomes. If the use of these technologies will, in most situations, lessen agriculture’s environmental impact, the public may be far more receptive to them. Numerous initiatives are being carried out to boost nitrogen fixation in crop-associated microorganisms, which, if successful, will lessen fertilizer consumption and contemporaneous run-off that has a negative influence on waterways (Goold et al. 2018). For example, the native microbial and plant biodiversity may be impacted by the inoculation of exotic microbial strains (Schwartz et al. 2006). To accurately pinpoint the drawbacks and benefits of these methods, more research must be done.
Targeted plant microbiome engineering
The main factor influencing a plant’s growth and development is its root microbiome. It helps with nutrient uptake, the control of this microbiome, and protection against biotic stressors (Arif et al. 2020). Several microorganisms can interact in the rhizosphere with one another through functional consortia (Dubey et al. 2019). For instance, in a model grassland, PGPR and arbuscular mycorrhizal (AM) fungi complemented one another with different limiting nutrients and ecological roles (Vyas et al. 2018). To finally captivate and allure the desired microbiota, the manipulation can be incorporated into the rhizosphere microbiome with the aid of soil conditioning using some conventional/traditional soil amendments or by the activation of specific substrates and molecules in the signaling pathway (including root exudates). Numerous research has been conducted in this regard to promote the use of particular exudates/secretions and substrates in field trials (Liu et al. 2019a, b; Danish and Zafar-ul-Hye 2019; Pascale et al. 2020).
Specific microbial strains can also be injected into the soil to alter and modify the makeup of different microbial communities (del Carmen Orozco-Mosqueda et al. 2018). Some examples include:
The application of a powerful bacterial and fungal consortium (Pseudomonas libanensis TR1(bacterial part) + (Arbuscular mycorrhiza) AM fungus, Claroideoglomus claroideum BEG210 (fungal part) into the rhizosphere section of sunflower resulted in profuse development of the plant, even under heavy metal and salt stress (Ma et al. 2019).
Using bacterial cultures of Acinetobacter sp, (UQO2), Bacillus amyloliquefaciens, and Bacillus velezensis, on the roots of growing chili plants increased plant development and, consequently, the plant’s capacity to survive soil-borne Phytophthora capsicum infection (Syed-Ab-Rahman et al. 2019).
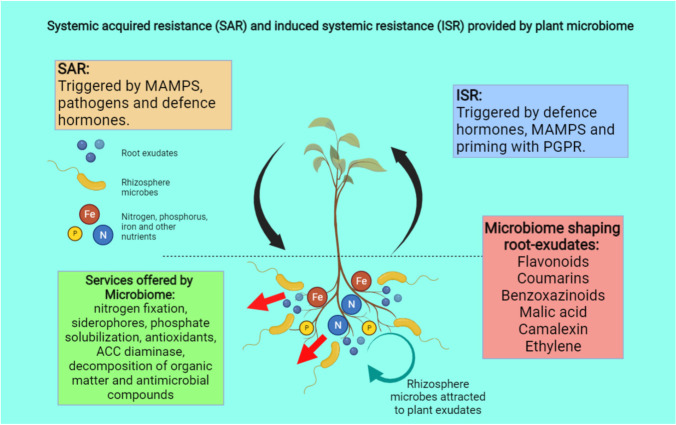
Beans grown with Agrobacterium sp. 10C2 produced more phosphorus, flavonoids as well as high antioxidants in the harvested pods as well as more nodules overall and plant biomass (Chihaoui et al. 2015). After 15 days of inoculation, numerous (plant growth promoting rhizobacteria) PGPR species, including Actinomycetes spp. and Brevibacterium spp. began to colonize the soil near the plant. The seed microbiome can be passed down across generations and has been demonstrated to enhance germination, plant survival, and performance (Tallapragada and Seshachla, 2020). Co-inoculating tomato seedlings’ rhizospheres with Pseudomonas stutzeri and Stenotrophomonas maltophilia accelerated plant growth and caused both organisms to release substances that are active against the Botrytis cinerea leaf disease (dimethyl disulfide, for example). The bacteria that were included as endophytes may multiply and populate the following generation. Roots serve as crucial gatekeepers to choose beneficial microorganisms that may enter the plant as endophytes, much to how root exudates enhance beneficial microbes in the rhizosphere. It is interesting to note that bacteria are also released by plants into the rhizosphere (Mendes et al. 2018), although how microbes enter and leave the body is unknown. In addition, roots can consume associated microbes directly and use them as a nitrogen source (Panke-Buisse et al. 2017), suggesting that the presence of microbial biomass in the rhizosphere (Fig. 7) also contributes to plant growth in an unspecific manner. Further studies will be necessary to determine whether specific microbes are preferred by plants for consumption. Recent plant breeding initiatives attempt to create crops that are more resilient and able to withstand attacks from pests and pathogens. However, because the corresponding pathways are frequently antagonistic, plants are typically unable to fight themselves against both biotic and abiotic stress at the same time (He et al. 2010).
Fig. 7.
Systemic acquired resistance (SAR) and induced systemic resistance (ISR) techniques employed against plant pathogens, which can be initiated via microbe associated molecular patterns (MAMPs) (Wintermans et al. 2016)
Multi-omics: a genetic perspective to breach into microbiome composition, interactions, and activities
Different facets of microbiomes are being revealed through integrated multi-omics methods.
Our understanding of plant-microbiome associations and interactions with host plants has expanded because of these tools (Fig. 8). These combined approaches give us a thorough grasp of how specific bacteria and microbial communities encourage plants-microbiomes synergistic interactions to assure plant health and resistance to environmental challenges.
Fig. 8.
The different techniques for studying the network of interaction, composition, and functions related to microbiome
The microbiome has been observed and examined from different angles, including (Tkacz et al. 2020a):
Microbiome composition (through advanced sequencing methods and metagenomics)
Activities (through meta-transcriptomics, metagenomics, and meta-proteomics)
The interaction networks in which they take part (through metabolomics)
Techniques are as followed:
High-throughput sequencing of the marker gene (Amplicon sequencing) tags permits for the investigation of microbial characteristics, distribution, composition, and profiling (Illumina amplicon tag (iTAG). We have compiled a thorough census of the microbiota inhabiting various crops by focusing on marker gene tags like 16S rRNA for bacteria and archaea, internal transcribed spacer (ITS) or 18S rRNA for fungi, and functional genes like ammonia monooxygenase (amoA) for nitrifying bacteria and nitrogenase gene (nif H) for nitrogen-fixing bacteria. Some specific crop microbiota studies include rice (Edwards et al. 2015), soyean (Mendes et al. 2019), corn (Walters et al. 2018), barley (Bulgarelli et al. 2012, 2013), pea (Tkacz et al. 2020b), sugarcane (Hamonts et al. 2018), citrus (Xu et al. 2018), and grapevine (Marasco et al. 2018). Plant-associated microbiota has a very unique and conserved phylogenetic structure with immense overlap between subsets of microbial communities in different plant compartments (Hamonts et al. 2018). Considering the bulk soil, most plant species house a variety of bacterial taxa belonging to the phyla Proteobacteria, Bacteroidetes, Firmicutes, and Actinobacteria; by contrast, Acidobacteria, Verrucomicrobia, and Gemmatimonadetes are observed in much less quantity in plant-associated environments compared to bulk soils.
Comprehensive sequencing: Although iTAG-based profiling enables the characterization of microorganisms, it does not offer comprehensive insights into how plant-associated microbiomes function. Shotgun sequencing has discovered microbial genomic characteristics connected to plant-microbiome colonization and interaction patterns by providing information on total DNA (Ofek-Lalzar et al. 2014).
The microbial activities and functions start from cellular mobility, toxin secretion, response to various stress levels, chemotaxis, and utilization of carbon molecules that are favorably used to improve the plant-soil ecosystem (Mitter et al. 2017). In plant-associated microbiomes, improved genes involved in transcriptional maintenance, regulation, and communication enable highly specific and coextensive responses (biocontrol activities, iron-chelation processes, nitrogen, and phosphorus mobilization, and solubilization) (Clouse and Wagner 2021). The collection and curation of comprehensive genomic data from plant/soil settings are now possible thanks to recent advances in genome curation techniques (Bulgarelli et al. 2015). This will make it possible to have a comprehensive understanding of the history and significance of the microbiomes linked with plants. We will gain a deeper knowledge of the expression of these features and microbial activity by combining and directing metagenomic approaches with other advance high-throughput methods (proteomics and metabolomics).
-
iii)
Meta-transcriptomics, meta-proteomics, and metabolomics (the omics triad): In response to drought stress, the dynamic rhizospheric microbiomes of oat, pea, and wheat showed kingdom-level differences (Bhattacharyya et al. 2021); the root-associated vigorous microbiome of sorghum showed escalating transcriptional activity related to the genes concerned with the metabolic activities of carbohydrate and amino acid (due to regular switching of actinobacterial function and activity); and the development of microbial communities as biosensors for bearing the stress generated during the drought periods (Delmotte et al. 2009). In-depth understandings of the molecular levels of phenotypes related to the microbial communities from the phyllosphere and rhizosphere have been obtained by metaproteomic investigations. Significant stability was shown between the predominant microbiome members and the related proteins found in several plant-allied environments. Metaproteomic revealed metabolic pathways that permit précised colonization and adaption in the rhizosphere sections as compared to the bulk soil, utilizing inferences from in situ physiology of the microbial communities (e.g. regulation of many primary metabolic processes was higher and faster in the rhizosphere; Bona et al. 2019). This demonstrates the necessity of shared adaptation methodologies and strategies for flourishing colonization. Although metabolomic methods are widely employed to diagnose plant diseases and the etiological agents behind them, microbiome science has yet to benefit significantly from their usage (Adeniji et al. 2020). Recently, complicated connections between different characteristics, metabolites, microorganisms, and minerals in an agroecosystem were clarified using multi-omics (phenome, ionome, metabolome, and microbiome) and integrated/synergistic informatics (multi-omics approaches will advance more quickly with improved sample preparation (e.g. elimination of host sequences for transcriptomics, universal protein isolation and extraction for proteomics), expanded databases, specific algorithms development, and specific computational methods/tools for the identification at metabolite, gene, and protein at various hierarchy levels (Ichihashi et al. 2020).
-
iv)
The art of culturomics: To create commercial microbiome inoculants and as experimental documentation to offer specific bacterial genome sequences as reference for a better comprehension of metagenome datasets and functional studies, bacterial cultures are necessary (Forster et al. 2019). A sizable portion of the microbes in plant-associated microbiomes can be cultured, notably bacteria. In addition, these important collections have allowed correlative genomics from huge/large isolate collections to find homologs related to the known bacterial genes specifically involved in plant nutrition, pathogenicity, or colonization (Levy et al. 2018). The identification and recognition of the specific protein domains in bacteria associated with plants that resemble and mimic plant-related protein domains was an intriguing finding in this area (Levy et al. 2018). The creation of impartial culture collections sequenced and obtained from a variety of crops/plants and soil types, as well as tools to compute, exchange, and standardize metadata related to specific strains, will be necessary for future studies (Finkel et al. 2017). High-throughput culture platforms can improve isolation efficiency by using microdroplets and microfluidic technology (Kaminski et al. 2016). This will enable extensive microbial isolate separation, genome sequencing, and functional screening.
Novel ways to alter the plant microbiome in situ
There are currently many methods being formulated and tested to control the entire microbiome associated with the plant without the requirement for culture (utilize transgenics or more regular methodologies). Several strategies, including the use of transgenic or more traditional methods, are now being researched and tested to mobilize the entire microbiome without the need for its respective culture. Considering this model, utilizing designed plants with root attributes that invigorate helpful organisms, for example, the active nitrogen fixers; the symbiotic associators: mycorrhiza; the active zinc, potassium, and phosphorus solubilizers; siderophore producers, and phytohormone-creating microorganisms can greatly affect agricultural productivities in a positive manner (Macdonald and Singh 2014).
Unfortunately, the use of this proposition for crops is still limited due to public perception of transgenic plants. Consideration is being given to other non-transgenic methods that can steer and direct the microbiome in situ. Some of these approaches are discussed here:
Making use of molecules active in plant-microbial signal transduction (exploiting plant-microbial communication): Plants as well as microorganisms both produce various signaling atoms to convey their prerequisites to their accomplices. For instance, phosphorus-starved plants release signal particles, which cause rhizosphere organisms to upregulate their phosphorus-assembling genes in response (Zhang et al. 2019). Such signaling molecules are also released by the plant when it is being attacked by pathogens and pests or when it needs to communicate with other plants via volatile organic compounds (VOCs) to encourage the colonization of healthy microorganisms. Cultivation, detection, and identification for utilization of these compounds may prove to be an important tool with great benefits but is difficult due to the low quantities of these compounds formed.
Utilization of microbes mixed potions (cocktails), which do not show any direct valuable impacts on plants, can expand the movement of active native beneficial microflora. These mixed potions essentially contain microorganisms with high measures of signaling molecules.
When modifying the entire microbiome in situ, it is crucial to identify and screen the microbiota hub of crop species and understand their involvement in microbiome assembly and activity.
Synthetic biology, a branch of industrial science, offers yet another important tool for creating crop probiotics with innovative and predictable properties that, when incorporated and implemented on a plant can boost up the activities of the helpful organisms in a predetermined manner.
The designing techniques like in situ genome designing can be used to directly engineer and finally alter/modify the in situ microbiome’s genome (Sheth et al. 2016). Here, transportable hereditary components, such as plasmids, can be introduced to native microflora, assisting in the development of desired and directional capabilities.
Rhizosphere microbiome, plant-microorganism cooperation, structural makeup, and functional range are governed by micro-RNA (miRNA), which is present in plants. This has been used to reestablish sound digestive systems (Liu and Weiner 2016) and could be a huge device to target recipient organisms for improved yielding of crops.
Use of microfluidics-based technologies allows us a unique insight into the fascinating root-microenvironment for observing the plant-microbiome interactions by providing dynamic interaction imaging. Using this technology, we will be able to accelerate the ongoing discoveries in the field of research related to the microbiome and its effective management strategies (Bashiardes et al. 2016; Nicholson et al. 2005)
The utilization of ecological technologies and techniques in combination with the above-mentioned upgraded technologies (such as crop rotation and non-tillage practices integrated with the breeding techniques with a specific microbiome) have immense potential to alter the host microbiomes, reducing plant and soil diseases and finally accelerating sustainable productivity. However, these technologies are still very new and need further research to be fully efficient and usable to avoid negative outcomes.
The advent of computational models: methods and methodology for microbiome study
The research in the field of the microbiome is highly interdisciplinary and has various important applications and techniques to study it. Here, we will discuss some of these important methods of computational origin and models that can be used for data collection and methods to analyze this data for further understanding. Microbiome study design and methods include:
Data collection: Sequencing techniques referred to as the next generation techniques are used for the identification of genetic material of microbial origin and other techniques, like meta-proteomics for proteins, meta-transcriptomics for gene expression, and metabolomics for small molecules, are used for the identification of microbial functional products (Hettich et al. 2013; Bashiardes et al. 2016; Nicholson et al. 2005). A large amount of data is gathered and analyzed using these techniques which reveals a very distinct aspect of the microbiome and microbial population.
Statistical analysis models: Surveys of marker genes of the microbiome are employed often for a broad understanding of the microbial population. Under these studies, sample processing is done via sequencing and bioinformatic pathways. Then, the collected data is organized in tables as operational taxonomic unit (OTU) counts. Generally, the statistical analysis begins through the OTU table, which is scarce and highly definite. Throughout the sample, it displays theatrical inconsistency in the library size (total number of counts). On the other hand, distributions of Dirichlet Multinomial can be taken as practical statistics and might be employed for standardizing the counts throughout the samples (Holmes et al. 2012). As not many zeros are there in the counts for gene sets of microbes, methods like reads per kilobase million (RPKM) can be used for standardization.
Models based on patterns: Models that are based on patterns are generally free of any conventions. Under intricate ecosystems (Coyte et al. 2015), these models are focused on discovering important and adequate data to replicate the results and planned patterns (Zeng et al. 2015). These models are steady and expressive with the potential of giving collective probabilistic associations showing the universal dynamics of multifaceted ecosystems like microbiome dependent patterns of biodiversity and environmental interactions. Metacommunity modeling is based on these patterns.
Association graphs: This technique is used for the discovery of co-occurrence: In the field of informatics, association graphs are used extensively for the discovery (Jayadevaprakash et al. 2005) and understanding of relationships between vital agents which form the compound system (Bartoli et al. 2000). In the past, these association graphs were employed for the retrieval of text-based data and the study of hierarchical patterns (Balaneshinkordan and Kotov 2016). These graphs were also helpful in interpreting data models and lately, in applications like-varied array of multi-variate data informatics (Hosseinkhani et al. 2012; Date et al. 2013; Luo et al. 2017).
Network inference models: Several network inference-based models are present nowadays which can deliver value in microbiome studies (Balaneshinkordan and Kotov 2016). For example, data theoretical representations might be employed in conjecturing systems of microbial communication (Li et al. 2018). The basis of these models is a probable description of a plentiful number of species with their growth over time and in a particular surrounding, instead of specific guesses regarding the dynamics of microbes. Moreover, since these models are based on entropies, extraction of relevant data is possible from the raw microbial information (Servadio et al. 2018); (Fig. 9).
Fig. 9.
A graphical sketch establishing recent trends in Micro-Biome Based Green & Clean Technology (2015–2022)
Major breakthrough genome mechanism: a robust gene editing methodology
Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas) presents an important breakthrough channel through gene editing methodologies. It further enhances the researcher’s ability to incorporate editing and altering processes in DNA sequences, thus modifying gene functioning in animals and plants (Xu et al. 2019). CRISPR technique is becoming a powerful choice for introducing gene of interest, desired traits (disease resistance) in wide range of economically important crops. In addition, various research demonstrates successful integration of gene traits through CRISPR mediated engineered resistance, also leading to a broad-spectrum action against a wide variety of pathogen groups (Oliva et al. 2019). Moreover, the researches and literature also support the commercialization of CRISPR crop varieties which are not only limited to protected cultivation criteria or laboratory experimentations. The commercialization and marketing of C. sativa (CRISPR-Crop) with enriched omega-3 oil is hitting the market records in the USA (Waltz 2018). This provides a clear indication that CRISPR ENGINEERED CROPS are the FUTURE CROPS and will soon reach the consumers with exciting applications (Kaur et al. 2021). CRISPR technology not only provides a disease resistance property to the crop but also helps in de novo induction of meristem, thus reducing tissue culture tedious/ time-consuming steps for the production of dicotyledonous plants (Maher et al. 2020). Moreover, CRISPR/LbCas12a, a temperature tolerant gene, has enabled insertion of large DNA ranging up to 2 kb, thus increasing the efficiency and target methodologies (Merker et al. 2020; Lu et al. 2020). Similarly, CRISPR heat inducible makeover has resulted in increasing and accerlating the gene targeting mechanism in maize (Barone et al. 2020). Thus, CRISPR is providing an exciting platform for chromosome engineering, gene restructuring, and somatic chromosomal engineering for sorting out the genetic linkages (Kim et al. 2018). In order to raise crop tolerance to different diseases, decrease the use of chemical pesticides, and safeguard the environment, it is possible to manipulate susceptibility genes (also known as S-genes) in agricultural techniques. For breeding novel cultivars with high tolerance or even resistance to specific diseases, inhibiting the expression of susceptibility genes is a fantastic method and also ensures a smart move from traditional plant breeding to precision breeding systems. Additionally, CRISPR/Cas genome editing is being used to enhance post-harvest quality, crop storage, and developing tolerance against many environmental stress (Zhang et al. 2021). Furthermore, certain fruits, like the tomato and peach, have long-standing issues with shipping and storage. Fruits that have reached full maturity and have a superb flavor become mushy and challenging to carry and store for a long time. Many researchers employed CRISPR/Cas genome editing technology to replace and mutate the tomato ALC gene. CRISPR-homology-directed repair mediated gene substitution is significantly more difficult than CRISPR/Cas knock-out mutagenesis, just like in other plant species (Yu et al. 2017). Without influencing other agronomic variables like plant size and fruit firmness, the CRISPR-edited tomato showed improved storage ability and long shelf life (Yu et al. 2017). Currently, agrobacterium-mediated gene transformation, which is restricted to a few plant species, is the most effective method for obtaining genome editing events. Although some kinds of plants, including agriculturally significant crops, can regenerate, this is only possible in a small subset of genotypes or cultivars. Therefore, it is urgently necessary to either design a new transformation approach that does not involve plant regeneration or a highly effective genotype-independent plant tissue culture and plant regeneration system. By experimenting with various explants, plant growth hormones/regulators, active compounds, and combinations of these effectors, it is feasible to increase the ability of plants to regenerate. As a result, these developments would ensure the elite gene transfer to the cultivars (Li et al. 2021). However, CRISPR-mediated geminivirus resistance techniques are utilizing the direct methods for DNA targets and a wide variety of important genes have been identified which can be potentially used in case of plant virus diseases (Zaidi et al. 2020). Moreover, these technologies need several years to get well equipped with the standard regulatory guidelines and protocols to remove the bottlenecks related to these upcoming tools and techniques (Yang et al. 2019). Furthermore, CRISPR promises that it will help to address many of the major problems of the twenty-first century, from food and agricultural production to medical and health problems.
Sustainable development goals: the genetic march towards zero hunger strategy
Global economic downturns brought on by the COVID-19 pandemic and climatic changes are preventing the globe from meeting the sustainable development goals (SDGs), which have as their ultimate objective the eradication of hunger by 2030 (Veillet et al. 2019). Although the usage of pesticides, fungicides, and herbicides has been considerably decreased due to transgenic and traditionally cross-bred crop types, they still require close monitoring to prevent the spread of weeds and insect pests through natural selection (Qin et al. 2020). We must boost food production responsibly and make sure that it is distributed fairly if we want to end poverty. Agriculture production must overcome a number of obstacles, such as sustaining yield while maintaining sustainability, enhancing grain quality and nutritional value, and developing resilience to biotic and abiotic pressures (Cai et al. 2020). Scientists bear a heavy responsibility for encouraging sustainable agriculture, boosting nutrition, and eradicating hunger by the year 2030. Creating sustainable crops that can adapt to shifting environmental conditions is one strategy to assure food security. Additionally, concerns with transgenic crops’ affordability, effectiveness, bioethics, regulation, and public acceptance limit their potential for use. The second sustainable development goal (SDG2) established by the United Nations in 2015 is to end hunger worldwide by 2030 (Xing et al. 2021). To feed the world’s population with healthy food and achieve the goal of ending hunger by 2030 will require greater and more reliable agricultural production that meets the population's nutritional needs. Many staple food crops around the world lacks important nutrients. Gene/genome editing technologies (such zinc finger nucleases (ZFNs; transcription activator-like effector nucleases (TALENs), CRISPR/Cas systems, etc.) have the potential to greatly enhance the nutrient density, attractiveness, and post-harvest quality of crops to assist reach the 2030 goals (Malabarba et al. 2021).
Conclusions and future prospects
The advantages of these sustainable practices include efficient uptake and supply of minerals, improved crop resistance to diseases, reduced nitrate leaching, better soil structure, and porosity and water infiltration. Organic farmers have increased in numbers dramatically. According to data from the national center of organic farming, a total of 246,576 farmers have switched to organic farming as they are under in-conversion farming. The plant-microbe interactions formed may be treated as harmful or beneficial to the associated host. The studies, however, on these interactions have been carried out in isolation as such we need to know the experimental evidence which focuses on the role of root-microbiome Furthermore, scientific attempts need to be executed to expand the molecular analysis and studies regarding strengthening the plant-microbiome interaction to direct it towards attaining crop productivity. There is an alarming need to investigate, understand, and explore the parameters related to soil diversity and crop productivity, to stabilize crop yields, and soil health by reducing inputs of chemical fertilizers. The needs to harness crop–microbe associations are of utmost priority for combating ecological and environmental issues. The diverse nature of the microbial community needs to be better understood to both explore its function and develop specifically tagged biomarkers that are directed to monitor soil parameters. More advancement in genome analysis is needed to enable microbiologists to grab an understanding of different microbe-plant interactions. This may also lead to the identification of some novel microbes which can be used to produce useful compounds like antibiotics, biopesticides, and biofertilizers. These advancements would lead to the raising of new crop varieties that selectively favor beneficial crop-microbe interactions. This will help the small-scale farmers in raising their income by introducing on-farm input generation, which involves minimal costs for the inputs; this can be integrated with the ZBNF concept. This will also help the farmer in a long way to reduce the cost of cultivation and improve the environment, the soil health, and provide safe food to humans, which is the most important need of the day. The purchase costs of chemical fertilizers and pesticides are increasing every year and seem to continue in the future. This economic burden has resulted in poor returns for the farmers. The toxic residue in the food is also posing threat to human health and is resulting in emerging many dreadful diseases to the human folk as such we need to switch over to a cultivation system that helps in sustaining the ecology and reduces the cost of cultivation of the commercially cultivated crops like tomato, cauliflower, and capsicum, etc. The microbial population enhances sustainability in agricultural practices by decreasing the use of chemical fertilizers and pesticides for fighting against plant diseases and elevating crop yields. The properties of the microbiome lead to better plant growth, low environmental stress, and good crop health. Several microbial consortia are designed for successful and high crop growth as they possess numerous beneficial traits. For better plant growth, novel steps should be taken and new agricultural practices must be adopted. A complete investigation of the key microbial communities is required which can help us in the expansion of microbiome studies and related fields for acquiring better benefits. The information acquired will also help us improve plant disease resistance, nutrient cycle enhancement, and crop output. In addition, tying microbiome-based innovations to other sustainable development goals (zero hunger, industry, innovation, and infrastructure, clean water and sanitation, responsible consumption and production, and sustainable cities and communities) will point us in the direction of developing new strategies for a better and more effective application of microbiome engineering to help us realize our goal of sustainable and secure farming.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank Researchers Supporting Project (RSP2023R118), King Saud University, Riyadh, Saudi Arabia.
Author contribution
N.T. formulated the whole idea and brought this issue as this is the need of the hour, proof read the manuscript, added figures and tables. M.N. N.A.M. R.C. H.O.E. and S.A.K added various sections, figures, and methodology to the article. C.M.H. S.K.S. and A.A.S added the concepts related to the sustainability and novel gene techniques.
Funding
Researchers Supporting Project (RSP2023R118), King Saud University.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article.
Declarations
Ethics approval and consent to participate
“Not applicable”.
Consent for publication
“Not applicable”.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adeniji AA, Babalola OO, Loots DT. Metabolomic applications for understanding complex tripartite plant–microbes’ interactions: strategies and perspectives. Biotechnology Reports. 2020;25:00425. doi: 10.1016/j.btre.2020.e00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Zyoud F. Adoption range of integrated pest management (IPM) techniques by greenhouse vegetable growers in Jordan. Jordan J Agric Sci. 2014;10:504–525. [Google Scholar]
- Andrade GC, Freguglia RM, Furlani RP, Torres NH, Tornisielo VL. Determination of pesticide residues in tomato using dispersive solid-phase extraction and gas chromatography/ion trap mass spectrometry. J Braz Chem Soc. 2011;22:1701–1708. [Google Scholar]
- APEDA RN (2014) Agricultural and Processed Food Products Export Development Authority.
- Arias-Sanchez FI, Vessman B, Mitri S. Artificially selecting microbial communities: if we can breed dogs, why not microbiomes? PLoS Biol. 2019;17:3000356. doi: 10.1371/journal.pbio.3000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif I, Batool M, Schenk PM. Plant microbiome engineering: expected benefits for improved crop growth and resilience. Trends Biotechnol. 2020;38(12):1385–1396. doi: 10.1016/j.tibtech.2020.04.015. [DOI] [PubMed] [Google Scholar]
- Bakker MG, Manter DK, Sheflin AM, Weir TL, Vivanco JM. Harnessing the rhizosphere microbiome through plant breeding and agricultural management. Plant Soil. 2012;360:1–3. [Google Scholar]
- Balaneshinkordan S, Kotov A (2016) An empirical comparison of term association and knowledge graphs for query expansion. In Advances in Information Retrieval: 38th European Conference on IR Research, ECIR 2016, Padua, Italy, March 20–23, 2016. Proceedings 38. Springer International Publishing, pp 761–767
- Bàrberi P. Functional biodiversity in organic systems: the way forward. Sust Agric Res. 2015;4:37937. [Google Scholar]
- Barone P, Wu E, Lenderts B, Anand A, Gordon-Kamm W, Svitashev S, Kumar S. Efficient gene targeting in maize using inducible CRISPR-Cas9 and marker-free donor template. Mol Plant. 2020;13(8):1219–1227. doi: 10.1016/j.molp.2020.06.008. [DOI] [PubMed] [Google Scholar]
- Bartoli M, Pelillo M, Siddiqi K, Zucker SW. Attributed tree homomorphism using association graphs. In Proceedings 5th International Conference on Pattern Recognition. ICPR. 2000;2:133–136. [Google Scholar]
- Bashiardes S, Zilberman-Schapira G, Elinav E (2016) Use of metatranscriptomics in microbiome research. Bioinform Biol Insights 4610 [DOI] [PMC free article] [PubMed]
- Barrett CB, Christian P, Shiferaw BA. The structural transformation of African agriculture and rural spaces: Introduction to a special section. Agric Econ. 2017;48(S1):5–10. [Google Scholar]
- Bhattacharyya A, Pablo CH, Mavrodi OV, Weller DM, Thomashow LS, Mavrodi DV (2021) Rhizosphere plant-microbe interactions under water stress. In Advances in applied microbiology, vol 115. Academic Press, pp 65–113 [DOI] [PMC free article] [PubMed]
- Beckers B, Op De Beeck M, Weyens N, Van Acker R, Van Montagu M, Boerjan W, Vangronsveld J. Lignin engineering in field-grown poplar trees affects the endosphere bacterial microbiome. Proc Nat Acad Sci, USA. 2016;113:2312–2317. doi: 10.1073/pnas.1523264113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen RL, Pieterse CM, Bakker PA. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Birkhofer K, Bezemer TM, Bloem J, Bonkowski M, Christensen S, Dubois D, Ekelund F, Fließbach A, Gunst L, Hedlund K, Mäder P. Long-term organic farming fosters below and above ground biota: implications for soil quality, biological control and productivity. Soil Biol Biochem. 2008;40:2297–2308. [Google Scholar]
- Birkhofer K, Arvidsson F, Ehlers D, Mader VL, Bengtsson J, Smith HG. Organic farming affects the biological control of hemipteran pests and yields in spring barely independent of landscape complexity. Landsc Ecol. 2016;31:567–579. [Google Scholar]
- Blair PM, Land ML, Piatek MJ, Jacobson DA, Lu TS, Doktycz MJ, Pelletier DA. Exploration of the biosynthetic potential of the Populus microbiome. mSystems. 2018;3:00045–18. doi: 10.1128/mSystems.00045-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bona E, Massa N, Novello G, Boatti L, Cesaro P, Todeschini V, Magnelli V, Manfredi M, Marengo E, Mignone F, et al. Metaproteomic characterization of the Vitis vinifera rhizosphere. FEMS Microbiol Ecol. 2019;95:204. doi: 10.1093/femsec/fiy204. [DOI] [PubMed] [Google Scholar]
- Bonvoisin T, Utyasheva L, Knipe D, Gunnel D, Eddleston M. Suicide by pesticide poisoning in India: a review of pesticide regulations and their impact on suicide trends. BMC Public Health. 2020;20:251. doi: 10.1186/s12889-020-8339-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga ARC, de Rosso VV, Harayashiki CAY, Jimenez PC, Castro ÍB. Global health risks from pesticide use in Brazil. Nature Food. 2020;1:312–314. doi: 10.1038/s43016-020-0100-3. [DOI] [PubMed] [Google Scholar]
- Brescia S. From crisis to healthy farming and food systems. Agric Hum Values. 2020;37:633–634. doi: 10.1007/s10460-020-10106-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D, Rott M, Schlaeppi K, Loren V, van Themaat E, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- Bulgarelli D, Schlaeppi K, Spaepen S, Van Themaat EV, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- Bulgarelli D, Garrido-Other R, Münch PC, Weiman A, Dröge J, Pan Y, McHardy AC, Schulze-Lefert P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe. 2015;17:392–403. doi: 10.1016/j.chom.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby PE, Peay KG, Newcombe G. Common foliar fungi of Populus trichocarpa modify Melampsora rust disease severity. New Phytol. 2016;209:1681–1692. doi: 10.1111/nph.13742. [DOI] [PubMed] [Google Scholar]
- Busby PE, Soman C, Wagner MR, Friesen ML, Kremer J, Bennett A, Morsy M, Eisen JA, Leach JE, Dangl JL. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 2017;15:2001793. doi: 10.1371/journal.pbio.2001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Chen L, Zhang Y, Yuan S, Su Q, Sun S, Wu C, Yao W, Han T, Hou W. Target base editing in soybean using a modified CRISPR/Cas9 system. Plant Biotechnol J. 2020;18:1996. doi: 10.1111/pbi.13386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihaoui SA, Trabelsi D, Jdey A, Mhadhbi H, Mhamdi R. Inoculation of Phaseolus vulgaris with the nodule-endophyte Agrobacterium sp. 10C2 affects the richness and structure of rhizosphere bacterial communities and enhances nodulation and growth. Arch Microbiol. 2015;197:805–813. doi: 10.1007/s00203-015-1118-z. [DOI] [PubMed] [Google Scholar]
- Clouse KM, Wagner MR. Plant genetics as a tool for manipulating crop microbiomes: Opportunities and challenges. Frontiers in Bioengineering and Biotechnology. 2021;9:567548. doi: 10.3389/fbioe.2021.567548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: networks, competition, and stability. Science. 2015;350:663–666. doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- Damalas CA, Koutroubas SD. Current status and recent developments in biopesticide use. Agriculture. 2018;8:13. [Google Scholar]
- Danish S, Zafar-ul-Hye M. Co-application of ACC-deaminase producing PGPR and timber-waste biochar improves pigments formation, growth, and yield of wheat under drought stress. Sci Rep. 2019;9:1–3. doi: 10.1038/s41598-019-42374-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date K, Gross GA, Khopkar S, Nagi R, Sambhoos K (2013) Data association and graph analytical processing of hard and soft intelligence data. In Proceedings of the 16th international conference on information fusion. IEEE, pp 404–411
- del Carmen Orozco-Mosqueda M, del Carmen Rocha Granados M, Glick BR, Santoyo G (2018) Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms. Microbiol Res 208:25–31 [DOI] [PubMed]
- Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, Schlapbach R, von Mering C, Vorholt JA. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc Natl Acad Sci. 2009;106:16428–16433. doi: 10.1073/pnas.0905240106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran JW, Zeiss MR. Soil health and sustainability: managing the biotic component of soil quality. Appl Soil Ecol. 2000;15:3–11. [Google Scholar]
- Dubey A, Malla MA, Khan F, Chowdhary K, Yadav S, Kumar A, Sharma S, Khare PK, Khan ML. Soil microbiome: a key player for conservation of soil health under changing climate. Biodivers Conserv. 2019;28:2405–2429. [Google Scholar]
- Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci. 2015;112:911–920. doi: 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epelde L, Jauregi L, Urra J, Ibarretxe L, Romo J, Goikoetxea I, Garbisu C. Characterization of composted organic amendments for agricultural use. Front Sustain Food Syst. 2018;2:44. [Google Scholar]
- FAO (2016) The FAO action plan on antimicrobial resistance 2016–2020. FAO, Roma
- Food and Agriculture Organization of the United Nations (FAO) and World Health Organization (WHO), Global Situation of Pesticide Management in Agriculture and Public Health (2019).
- Farrar K, Bryant D, Cope-Selby N. Understanding and engineering beneficial plant-microbe interactions: plant growth promotion in energy crops. Plant Biotechnol J. 2014;12:1193–1206. doi: 10.1111/pbi.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel OM, Castrillo G, Herrera Paredes S, Salas Gonzalez I, Dangl JL. Understanding and exploiting plant beneficial microbes. Curr Opin Plant Biol. 2017;38:155–163. doi: 10.1016/j.pbi.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster SC, Kumar N, Anonye BO, Almeida A, Viciani E, Stares MD, Dunn M, Mkandawire TT, Zhu A, Shao Y, et al. A human gut bacterial genome and culture collection for improved metagenomic analyses. Nat Biotechnol. 2019;37:186–192. doi: 10.1038/s41587-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederiks C, Wesseler JH. A comparison of the EU and US regulatory frameworks for the active substance registration of microbial biological control agents. Pest Manag Sci. 2019;75:87–103. doi: 10.1002/ps.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigou A, Laurent C, Berthet A, Colosio C, Jas N, Daubas-Letourneux V, Jackson Filho JM, Jouzel JN, Samuel O, Baldi I, Lebailly P, Galey L, Goutille F. Judon N Critical review of the role of PPE in the prevention of risks related to agricultural pesticide use. Saf Sci. 2019;123:104527. [Google Scholar]
- Geiger F, Bengtsson J, Berendse F, Weisser WW, Emmerson M, Morales MB, Ceryngier P, Liira J, Tscharntke T, Winqvist C, Eggers S. Persistent negative effects of pesticides on biodiversity and biological control potential on European farmland. Basic Appl Ecol. 2010;11:97–105. [Google Scholar]
- Goold HD, Wright P, Hailstones D. Emerging opportunities for synthetic biology in agriculture. Genes. 2018;9:341. doi: 10.3390/genes9070341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Dong K, Geisen S, Yang W, Yan Y, Gu D, Liu N, Borisjuk N, Luo Y, Friman VP. The effect of microbial inoculant origin on the rhizosphere bacterial community composition and plant growth-promotion. Plant Soil. 2020;452:105–117. [Google Scholar]
- Hamonts K, Trivedi P, Garg A, Janitz C, Grinyer J, Holford P, Botha FC, Anderson IC, Singh BK. Field study reveals core plant microbiota and relative importance of their drivers. Environ Microbiol. 2018;20:124–140. doi: 10.1111/1462-2920.14031. [DOI] [PubMed] [Google Scholar]
- Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. Trichoderma species—opportunistic, avirulent plant symbionts. Nat Rev Microbiol. 2004;2:43–56. doi: 10.1038/nrmicro797. [DOI] [PubMed] [Google Scholar]
- Hartmann M, Frey B, Mayer J, Mäder P, Widmer F. Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 2015;9:1177–1194. doi: 10.1038/ismej.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Yang Y, Yang G, Yu L. Studies on antibacterial activity and antibacterial mechanism of a novel polysaccharide from Streptomyces Virginia H03. Food Control. 2010;21:1257–1262. [Google Scholar]
- Hettich RL, Pan C, Chourey K, Giannone RJ. Metaproteomics: harnessing the power of high-performance mass spectrometry to identify the suite of proteins that control metabolic activities in microbial communities. Anal Chem. 2013;85:4203–4214. doi: 10.1021/ac303053e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SB (2014) Considerations for enabling the ecological redesign of organic and conventional agriculture: a social ecology and psychosocial perspective. Organic Farming, Prototype for Sustainable Agricultures: Prototype for Sustainable Agricultures 401–422
- Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS ONE. 2012;7:30126. doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseinkhani J, Chaprut S, Taherdoost H (2012) Criminal network mining by web structure and content mining. In 11th WSEAS International Conference on Information Security and Privacy (ISP'12), Prague, Czech Republic September, pp 24–26
- Ichihashi Y, Date Y, Shino A, Shimizu T, Shibata A, Kumaishi K, Funahashi F, Wakayama K, Yamazaki K, Umezawa A, et al. Multi-omics analysis on an agroecosystem reveals the significant role of organic nitrogen to increase agricultural crop yield. Proc Nat Acad Sci, USA. 2020;117:14552–14560. doi: 10.1073/pnas.1917259117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illukpitiya P, Khanal P (2016) Consumer perception of organic food and product marketing. Organic Farming for Sustainable Agriculture 315–324
- Jacquet S, Miki T, Noble R, Peduzzi P, Wilhelm S. Viruses in aquatic ecosystems: important advancements of the last 20 years and prospects for the future in the field of microbial oceanography and limnology. AIOL Journal. 2010;1:97–141. [Google Scholar]
- Jangir RP, Rathore MS, Bisnoi HR, Sundria MM (2008) National workshop on spices and aromatic plants. Agricultural Research Station 6–7.
- Jayadevaprakash N, Mukhopadhyay S, Palakal M (2005) Generating association graphs of non-cooccurring text objects using transitive methods. In Proceedings of the 2005 ACM symposium on Applied computing 141–145.
- Jochum MD, McWilliams KL, Pierson EA, Jo YK. Host-mediated microbiome engineering (HMME) of drought tolerance in the wheat rhizosphere. PLoS ONE. 2019;14:e0225933. doi: 10.1371/journal.pone.0225933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalloo K (2003) Research and extension activities on organic agriculture in India. Organic farming in horticulture for sustainable production 29–30.
- Kaminski TS, Scheler O, Garstecki P. Droplet microfluidics for microbiology: techniques, applications and challenges. Lab Chip. 2016;16:2168–2187. doi: 10.1039/c6lc00367b. [DOI] [PubMed] [Google Scholar]
- Kassam A, Friedrich T, Shaxson F, et al. The spread of conservation agriculture: justification, sustainability and uptake. Int J Agric Sustain. 2009;7:292–320. [Google Scholar]
- Kaur H, Pandey DK, Goutam U, Kumar V. CRISPR/Cas9-mediated genome editing is revolutionizing the improvement of horticultural crops: Recent advances and future prospects. Sci Hortic. 2021;289:110476. [Google Scholar]
- Kazan K, Lyons R. The link between flowering time and stress tolerance. J Exp Bot. 2016;67:47–60. doi: 10.1093/jxb/erv441. [DOI] [PubMed] [Google Scholar]
- Kemal RA, Islamaiah PWN, Sadah M, Lusiany T. Synthetic biology for biocontrol: a mini-review. Proc 6th Int Confer Global Res Conserv. 2016;6:92–94. [Google Scholar]
- Khadse A, Rosset PM. Zero budget natural farming in India–from inception to institutionalization. Agroecol Sustain Food Syst. 2019;43:848–871. [Google Scholar]
- Khadse A, Rosset PM, Morales H, Ferguson BG. Taking agroecology to scale: the zero-budget natural farming peasant movement in Karnataka, India. J Peasant Stud. 2018;45(1):192–219. [Google Scholar]
- Kim D, Alptekin B, Budak H. CRISPR/Cas9 genome editing in wheat. Functional & Integrative Genomics. 2018;18:31–41. doi: 10.1007/s10142-017-0572-x. [DOI] [PubMed] [Google Scholar]
- Köhl J, Kolnaar R, Ravensberg WJ (2019) Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Front Plant Sci 845. 10.3389/fpls.2019.00845 [DOI] [PMC free article] [PubMed]
- Kour D, Rana KL, Kaur T, Singh B, Chauhan VS, Kumar A, Gupta VK (2019) Extremophiles for hydrolytic enzymes productions: biodiversity and potential biotechnological applications. Bioprocessing for biomolecules production, pp 321–372
- Kumar R, Kumar S, Yashavanth BS, Meena PC. Natural Farming practices in India: Its adoption and impact on crop yield and farmers’ income. Indian J Agric Econ. 2019;74:420–432. [Google Scholar]
- Lakshmanan V, Selvaraj G, Bais HP. Functional soil microbiome: belowground solutions to an aboveground problem. Plant Physiol. 2014;166:689–700. doi: 10.1104/pp.114.245811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampel J, Scarbrough H, Macmillan S (2008) Managing through projects in knowledge-based environments: special issue introduction by the guest editors. Long Range Plan 41:7–16. 10.1016/j.lrp.2007.11.007
- Lay CY, Bell TH, Hamel C, Harker KN, Mohr R, Greer CW, Yergeau E, St- Arnaud M. Canola root-associated microbiomes in the Canadian prairies. Front Microbiol. 2018;9:1188. doi: 10.3389/fmicb.2018.01188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay CY, Bell TH, Hamel C, Harker KN, Mohr R, Greer CW, Yergeau E, St-Arnaud M. Canola root-associated microbiomes in the Canadian prairies. Front Microbiol. 2018;9:1188. doi: 10.3389/fmicb.2018.01188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy J, Mendelsohn M, Kough J, Jones R, Berckes N (2014) Biopesticide oversight and registration at the US Environmental Protection Agency. Biopesticides: state of the art and future opportunities, pp 3–18
- Leifeld J, Angers DA, Chenu C, Fuhrer J, Kätterer T, Powlson DS. Organic farming gives no climate change benefit through soil carbon sequestration. Proc Natl Acad Sci. 2013;110:984–984. doi: 10.1073/pnas.1220724110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leithold G, Hülsbergen KJ, Brock C. Organic matter returns to soils must be higher under organic compared to conventional farming. J Plant Nutr Soil Sci. 2015;178:4–12. [Google Scholar]
- Leonard SP, Powell JE, Perutka J, Geng P, Heckmann LC, Horak RD, Davies BW, Ellington AD, Barrick JE, Moran NA. Engineered symbionts activate honey bee immunity and limit pathogens. Science. 2020;367:573–576. doi: 10.1126/science.aax9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneau DK, Bothwell SG. Comparison of organic and conventional farms: challenging ecologists to make biodiversity functional. Front Ecol Environ. 2008;6:430–438. [Google Scholar]
- Levy A, Conway JM, Dangl JL, Woyke T. Elucidating bacterial gene functions in the plant microbiome. Cell Host Microbe. 2018;24:475–485. doi: 10.1016/j.chom.2018.09.005. [DOI] [PubMed] [Google Scholar]
- Li C, Brant E, Budak H, Zhang B. CRISPR/Cas: a Nobel Prize award-winning precise genome editing technology for gene therapy and crop improvement. J Zhejiang Univ Sci B. 2021;22(4):253–284. doi: 10.1631/jzus.B2100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Jing XY, Zhu X. Progress on approaches to software defect prediction. IET Softw. 2018;12(3):161–175. [Google Scholar]
- Liu F, Hewezi T, Lebeis SL, Pantalone V, Grewal PS, Staton ME. Soil indigenous microbiome and plant genotypes cooperatively modify soybean rhizosphere microbiome assembly. BMC Microbiol. 2019;19:1–19. doi: 10.1186/s12866-019-1572-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Khan MY, Carvalhais LC, Delgado-Baquerizo M, Yan L, Crawford M, Schenk PM. Soil amendments with ethylene precursor alleviate negative impacts of salinity on soil microbial properties and productivity. Sci Rep. 2019;9:1–13. doi: 10.1038/s41598-019-43305-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Weiner HL. Control of the gut microbiome by fecal microRNA. Microbial Cell. 2016;3(4):176. doi: 10.15698/mic2016.04.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Tian Y, Shen R, Yao Q, Wang M, Chen M, Dong J, Zhang T, Li F, Lei M, Zhu JK. Targeted, efficient sequence insertion and replacement in rice. Nat Biotechnol. 2020;38(12):1402–1407. doi: 10.1038/s41587-020-0581-5. [DOI] [PubMed] [Google Scholar]
- Lucas JA, García-Villaraco A, Ramos B, García-Cristobal J, Algar E, Gutierrez-Manero J. Structural and functional study in the rhizosphere of Oryza sativa L. plants growing under biotic and abiotic stress. J Appl Microbiol. 2013;115:218–235. doi: 10.1111/jam.12225. [DOI] [PubMed] [Google Scholar]
- Luo Y, Uzuner Ö, Szolovits P. Bridging semantics and syntax with graph algorithms—state-of-the-art of extracting biomedical relations. Brief Bioinform. 2017;18:160–178. doi: 10.1093/bib/bbw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Rajkumar M, Oliveira RS, Zhang C, Freitas H. Potential of plant beneficial bacteria and arbuscular mycorrhizal fungi in phytoremediation of metal-contaminated saline soils. J Hazard Mater. 2019;379:120813. doi: 10.1016/j.jhazmat.2019.120813. [DOI] [PubMed] [Google Scholar]
- Macdonald C, Singh B. Harnessing plant-microbe interactions for enhancing farm productivity. Bioengineered. 2014;5(1):5–9. doi: 10.4161/bioe.25320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher MF, Nasti RA, Vollbrecht M, Starker CG, Clark MD, Voytas DF. Plant gene editing through de novo induction of meristems. Nat Biotechnol. 2020;38(1):84–89. doi: 10.1038/s41587-019-0337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malabarba J, Chevreau E, Dousset N, Veillet F, Moizan J, Vergne E. New strategies to overcome present CRISPR/Cas9 limitations in apple and pear: efficient dechimerization and base editing. Int J Mol Sci. 2021;22:319. doi: 10.3390/ijms22010319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasco R, Rolli E, Fusi M, Michoud G, Daffonchio D. Grapevine rootstocks shape underground bacterial microbiome and networking but not potential functionality. Microbiome. 2018;6:1–17. doi: 10.1186/s40168-017-0391-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes R, Kruijt M, De Bruijn I, Dekkers E, Van Der Voort M, Schneider JH, Raaijmakers JM. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- Mendes LW, de Chaves MG, Fonseca MC, Mendes R, Raaijmakers JM, Tsai SM. Resistance breeding of common bean shapes the physiology of the rhizosphere microbiome. Front Microbiol. 2019;10:2252. doi: 10.3389/fmicb.2019.02252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes LW, Mendes R, Raaijmakers JM, Tsai SM. Breeding for soil-borne pathogen resistance impacts active rhizosphere microbiome of common bean. The ISME Journal. 2018;12(12):3038–3042. doi: 10.1038/s41396-018-0234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker L, Schindele P, Huang TK, Wolter F, Puchta H. Enhancing in planta gene targeting efficiencies in Arabidopsis using temperature-tolerant CRISPR/LbCas12a. Plant Biotechnol J. 2020;18(12):2382–2384. doi: 10.1111/pbi.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitter B, Pfaffenbichler N, Flavell R, Compant S, Antonielli L, Petric A, Sessitsch A. A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds. Front Microbiol. 2017;8:11. doi: 10.3389/fmicb.2017.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morella NM, Weng FC, Joubert PM, Metcalf CJE, Lindow S, Koskella B. Successive passaging of a plant-associated microbiome reveals robust habitat and host genotype-dependent selection. Proc Nat Acad Sci, USA. 2020;117:1148–1159. doi: 10.1073/pnas.1908600116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulet JM. Should we recommend organic crop foods on the basis of health benefits? Letter to the editor regarding the article by Barańskiet al. Br J Nutr. 2014;112:1745–1747. doi: 10.1017/S0007114514002645. [DOI] [PubMed] [Google Scholar]
- Munster D (2019) Affect beyond the human: Indian agriculture in a multispecies world. In Engaging transculturality. Routledge, pp 398–412
- Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- Niggli U. Sustainability of organic food production: challenges and innovations. Proc Nutr Soc. 2015;74:83–88. doi: 10.1017/S0029665114001438. [DOI] [PubMed] [Google Scholar]
- Ofek-Lalzar M, Sela N, Goldman-Voronov M, Green SJ, Hadar Y, Minz D. Niche and host-associated functional signatures of the root surface microbiome. Nat Commun. 2014;5:4950. doi: 10.1038/ncomms5950. [DOI] [PubMed] [Google Scholar]
- Oliva R, Ji C, Atienza-Grande G, Huguet-Tapia JC, Perez-Quintero A, Li T, ... Yang B (2019) Broadspectrum resistance to bacterial blight in rice using genome editing. Nat Biotechnol 37(11):1344–1350 [DOI] [PMC free article] [PubMed]
- Palekar S (2010) The philosophy of spiritual farming: zero budget natural farming–part 1, 5th revised renewal edn. Amravati: Zero Budget Natural Farming Research. Development and Extension Movement
- Panke-Buisse K, Lee S, Kao-Kniffin J. Cultivated sub-populations of soil microbiomes retain early flowering plant trait. Microb Ecol. 2017;73:394–403. doi: 10.1007/s00248-016-0846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerselvam P, Halberg N, Vaarst M, Hermansen JE. Indian farmers’ experience with and perceptions of organic farming. Renew Agric Food Syst. 2012;27:157–169. [Google Scholar]
- Pascale A, Proietti S, Pantelides IS, Stringlis IA. Modulation of the root microbiome by plant molecules: the basis for targeted disease suppression and plant growth promotion. Front Plant Sci. 2020;10:1741. doi: 10.3389/fpls.2019.01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peshin R, Dhawan AK (2009) Integrated pest management: volume 1: innovation-development process. Springer, Science & Business Media.
- Peshin R, Zhang W (2014) Integrated pest management and pesticide use. Springer, Netherlands, pp 1–46
- Pineda A, Kaplan I, Hannula SE, Ghanem W, Bezemer TM. Conditioning the soil microbiome through plant–soil feedbacks suppress an aboveground insect pest. New Phytol. 2020;226:595–608. doi: 10.1111/nph.16385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponisio LC, M'Gonigle LK, Mace KC, Palomino J, De Valpine P, Kremen C. Diversification practices reduce organic to conventional yield gap. Proc Royal Soc B Biol Sci. 2015;282:20141396. doi: 10.1098/rspb.2014.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma-Blaauw MB, De Goede RG, Bloem J, Faber JH, Brussaard L. Agricultural intensification and de-intensification differentially affect taxonomic diversity of predatory mites, earthworms, enchytraeids, nematodes and bacteria. Appl Soil Ecol. 2012;57:39–49. [Google Scholar]
- Pretty J, Benton TG, Bharucha ZP, Dicks LV, Flora CB, Godfray HCJ, Goulson D, Hartley S, Lampkin N, Morris C, et al. Global assessment of agricultural system redesign for sustainable intensification. Nat Sustain. 2018;1(8):441–446. [Google Scholar]
- Qin L, Li J, Wang Q, Xu Z, Sun L, Alariqi M, Manghwar H, Wang G, Li B, Ding X, Rui H. High-efficient and precise base editing of C• G to T• A in the allotetraploid cotton (Gossypium hirsu-tum) genome using a modified CRISPR/Cas9 system. Plant Biotechnol J. 2020;18:45–56. doi: 10.1111/pbi.13168.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JM, Mazzola M. Soil immune responses. Science. 2016;352(6292):1392–1393. doi: 10.1126/science.aaf3252. [DOI] [PubMed] [Google Scholar]
- Rani L, Thapa K, Kanojia N, Sharma N, Singh S, Grewal AS, ... Kaushal J (2021) An extensive review on the consequences of chemical pesticides on human health and environment. J Clean Prod 283:124657. 10.1016/j.jclepro.2020.124657
- Rascovan N, Carbonetto B, Perrig D, Díaz M, Canciani W, Abalo M, Vazquez MP. Integrated analysis of root microbiomes of soybean and wheat from agricultural fields. Sci Rep. 2016;6:1–12. doi: 10.1038/srep28084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnadass A. Crop protection for agricultural intensification systems in Sub-Saharan Africa. Sustain Agric Rev. 2020;39:1–34. doi: 10.1007/978-3-030-38881-2_1. [DOI] [Google Scholar]
- Ratnadass A, Barzman M. Ecological intensification for crop protection. Sustain Agric Rev. 2014;14:53–81. doi: 10.1007/978-3-319-06016-3_3. [DOI] [Google Scholar]
- Ratnadass A, Fernandes P, Avelino J, Habib R. Plant species diversity for sustainable management of crop pests and diseases in agroecosystems: a review. Agron Sustain Dev. 2012;32:273–303. doi: 10.1007/s13593-011-0022-4. [DOI] [Google Scholar]
- Reddy BS. Organic farming: status, issues and prospects–a review. Agric Econ Res Rev. 2010;23:343–358. [Google Scholar]
- Reddy PP (2017) Agro-ecological approaches to pest management for sustainable agriculture. Springer, Singapore, pp 29–41
- Redlich S, Martin EA, Steffan-Dewenter I. Landscape-level crop diversity benefits biological pest control. J Appl Ecol. 2018;55:2419–2428. doi: 10.1111/1365-2664.13126. [DOI] [Google Scholar]
- Reganold JP, Wachter JM. Organic agriculture in the twenty-first century. Nat Plants. 2016;2:1–8. doi: 10.1038/nplants.2015.221. [DOI] [PubMed] [Google Scholar]
- Roman-Reyna V, Pinili D, Borjaa FN, Quibod I, Groen SC, Mulyaningsih ES, Rachmat A, Slamet-Loedin IH, Alexandrov N, Mauleon R, et al. The rice leaf microbiome has a conserved community structure controlled by complex host–microbe interactions. Cell Host Microbe. 2019;19:00340. doi: 10.2139/ssrn.3382544. [DOI] [Google Scholar]
- Rothschild N, Novotný Č, Šašek V, Dosoretz CG. Ligninolytic enzymes of the fungus Irpex lacteus (Polyporus tulipiferae): isolation and characterization of lignin peroxidase. Enzyme Microb Technol. 2002;31:627–633. [Google Scholar]
- Rouphael Y, Colla G. Editorial: biostimulants in agriculture. Front Plant Sci. 2020;11:40. doi: 10.3389/fpls.2020.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu MH, Zhang J, Toth T, Khokhani D, Geddes BA, Mus F, Garcia-Costas A, Peters JW, Poole PS, Ane JM, et al. Control of nitrogen fixation in bacteria that associate with cereals. Nat Microbiol. 2020;5:314–330. doi: 10.1038/s41564-019-0631-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäffer A, Filser J, Frische T, Gessner M, Köck W, Kratz W, ... Scheringer M (2018) The Silent Spring: On the need for sustainable plant protection. Deutsche Akademie der Naturforscher Leopoldina eV-Nationale Akademie der Wissenschaften (German National Academy of Sciences) Leopoldin
- Scheller E, Joergensen RG. Decomposition of wheat straw differing in nitrogen content in soils under conventional and organic farming management. J Plant Nutr Soil Sci. 2008;171:886–892. [Google Scholar]
- Schwartz MW, Hoeksema JD, Gehring CA, Johnson NC, Klironomos JN, Abbott LK, Pringle A. The promise and the potential consequences of the global transport of mycorrhizal fungal inoculum. Ecological Letters. 2006;9:501–515. doi: 10.1111/j.1461-0248.2006.00910.x. [DOI] [PubMed] [Google Scholar]
- Servadio JL, Convertino M. Optimal information networks: application for data-driven integrated health in populations. Sci Adv. 2018;4:1701088. doi: 10.1126/sciadv.1701088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setala H, Bardgett RD, Birkhofer K, Brady M, Byrne L, De Ruiter PC, Van der Putten WH. Urban and agricultural soils: conflicts and trade-offs in the optimization of ecosystem services. Urban Ecosyst. 2014;17:239–253. [Google Scholar]
- Shepherd M, Pearce B, Cormack B, Philipps L, Cuttle S, Bhogal A, Unwin R (2003) An assessment of the environmental impacts of organic farming. A review for DEFRA-funded Project OF0405
- Sheth RU, Cabral V, Chen SP, Wang HH. Manipulating bacterial communities by in situ microbiome engineering. Trends Genet. 2016;32:189–200. doi: 10.1016/j.tig.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sime-Ngando T. Environmental bacteriophages: viruses of microbes in aquatic ecosystems. Front Microbiol. 2014;5:355. doi: 10.3389/fmicb.2014.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner C, Gattinger A, Muller A, Mäder P, Flieβbach A, Stolze M, Niggli U. Greenhouse gas fluxes from agricultural soils under organic and non-organic management—a global meta-analysis. Sci Total Environ. 2014;468:553–563. doi: 10.1016/j.scitotenv.2013.08.098. [DOI] [PubMed] [Google Scholar]
- Spence C, Bais H. Probiotics for plants: rhizospheric microbiome and plant fitness. Mol Microb Ecol Rhizosphere. 2013;1:713–721. [Google Scholar]
- Syed-Ab-Rahman SF, Xiao Y, Carvalhais LC, Ferguson BJ, Schenk PM. Suppression of Phytophthora capsici infection and promotion of tomato growth by soil bacteria. Rhizosphere. 2019;9:72–75. [Google Scholar]
- Tallapragada P, Seshachla U (2020) Seed microbiome and its implication in plant growth promotion and health. In The plant microbiome in sustainable agriculture, pp 47–64
- Taye ZM, Helgason BL, Bell JK, Norris CE, Vail S, Robinson SJ, Parkin IA, Arcand M, Mamet S, Links MG, et al. Core and differentially abundant bacterial taxa in the rhizosphere of field grown Brassica napus genotypes: implications for canola breeding. Front Microbiol. 2020;10:3007. doi: 10.3389/fmicb.2019.03007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur N (2017a) Increased soil-microbial-eco-physiological interactions and microbial food safety in tomato under organic strategies. Probiotics and plant health, pp 215–232
- Thakur N (2017b) Increased soil-microbial-eco-physiological interactions and microbial food safety in tomato under organic strategies. In: Kumar, V. Kumar, M. Sharma, S. Prasad, R. (eds) Probiotics and Plant Health. Springer, Singapore. 10.1007/978-981-10-3473-2_9
- Thakur N (2017c) Organic farming, food quality, and human health: a trisection of sustainability and a move from pesticides to eco-friendly biofertilizers. Probiotics in agroecosystem, pp 491–515
- Thakur N (2017d). Integrated approach for the management of differential patterns of diseases and pest incidence in lychee. In: Kumar, M. Kumar, V. Bhalla-Sarin, N. Varma, A. (eds) Lychee Disease Management. Springer, Singapore. 10.1007/978-981-10-4247-8_16
- Thakur N (2017e) Organic farming, food quality, and human health: a trisection of sustainability and a move from pesticides to eco-friendly biofertilizers. In: V Kumar, Kumar M, Sharma S, Prasad R (eds) Probiotics in Agroecosystem. Springer, Singapore. 10.1007/978-981-10-4059-7_26
- Thakur N. Physicochemical analysis and studies on soil profile of Prunus Armeniaca collected from mid-hill of Himachal Pradesh. Natl Acad Sci Lett. 2022;45:541–548. [Google Scholar]
- Thakur N, Nigam M, Tewary R, Rajvanshi K, Kumar M, Shukla SK, Mahmoud G-E, Gupta S. Drivers for the behavioural receptiveness and non-receptiveness of farmers towards organic cultivation system. J King Saud Univ - Sci. 2022;34(5):1018–3647. [Google Scholar]
- Thakur N, Sharma P, Govender PP, Shukla SK. Analysis and extraction of curcumin at mid and late phase harvested Curcuma Longa samples collected from Western Himalayan regions. Chemistry Africa. 2022;5:1733–1742. [Google Scholar]
- Tkacz A, Bestion E, Bo Z, Hortala M, Poole PS. Influence of plant fraction, soil, and plant species on microbiota: a multi-kingdom comparison. Mol Bio. 2020;11:02785–2819. doi: 10.1128/mBio.02785-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkacz A, Pini F, Turner TR, Bestion E, Simmonds J, Howell P, Greenland A, Cheema J, Emms DM, Uauy C, et al. Agricultural selection of wheat has been shaped by plant–microbe interactions. Front Microbiol. 2020;11:132. doi: 10.3389/fmicb.2020.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonfack LB, Bernadac A, Youmbi E, Mbouapouognigni VP, Ngueguim M, Akoa A. Impact of organic and inorganic fertilizers on tomato vigor, yield and fruit composition under tropical andosol soil conditions. Fruits. 2009;64:167–177. [Google Scholar]
- Treseder KK, Balser TC, Bradford MA, Brodie EL, Dubinsky EA, Eviner VT, Waldrop MP. Integrating microbial ecology into ecosystem models: challenges and priorities. Biogeochemistry. 2012;109:7–18. [Google Scholar]
- Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK. Plant–microbiome interactions: from community assembly to plant health. Nat Rev Microbiol. 2020;18:607–621. doi: 10.1038/s41579-020-0412-1. [DOI] [PubMed] [Google Scholar]
- Tshikantwa TS, Ullah MW, He F, Yang G. Current trends and potential applications of microbial interactions for human welfare. Front Microbiol. 2018;9:1156. doi: 10.3389/fmicb.2018.01156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck SL, Winqvist C, Mota F, Ahnström J, Turnbull LA, Bengtsson J. Land-use intensity and the effects of organic farming on biodiversity: a hierarchical meta-analysis. J Appl Ecol. 2014;51:746–755. doi: 10.1111/1365-2664.12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uniyal AP, Mansotra K, Yadav SK, Kumar V (2019) An overview of designing and selection of sgRNAs for precise genome editing by the CRISPR-Cas9 system in plants. 3 Biotech 9(6):223 [DOI] [PMC free article] [PubMed]
- Veillet F, Perrot L, Chauvin L, Kermarrec MP, Guyon-Debast A, Chauvin JE, Nogué F, Mazier M. Transgene-free genome editing in tomato and potato plants using agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int J Mol Sci. 2019;20:402. doi: 10.3390/ijms20020402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheijen LM, Aerts R, Brovkin V, Cavender-Bares J, Cornelissen JH, Kattge J, Van Bodegom PM. Inclusion of ecologically based trait variation in plant functional types reduces the projected land carbon sink in an earth system model. Glob Chang Biol. 2015;21:3074–3086. doi: 10.1111/gcb.12871. [DOI] [PubMed] [Google Scholar]
- Via Campesina L (2016) Zero budget natural farming in India. Family Farming Knowledge Platform of Fao
- Vyas P, Kumar D, Dubey A, Kumar A. Screening and characterization of Achromobacter xylosoxidans isolated from the rhizosphere of Jatropha curcas L. (energy crop) for plant-growth-promoting traits. J Adv Res Biotechnol. 2018;3:1–8. [Google Scholar]
- Walker G, Osbahr H, Cardey S. Thematic collages in participatory photography: a process for understanding the adoption of zero budget natural farming in India. Int J Qual Methods. 2021;20:1–13. [Google Scholar]
- Walters WA, Jin Z, Youngblut N, Wallace JG, Sutter J, Zhang W, Gonzalez-Pena A, Peiffer J, Koren O, Shi Q, et al. Large-scale replicated field study of maize rhizosphere identifies heritable microbes. Pro Nat Acad Sci, USA. 2018;115:7368–7373. doi: 10.1073/pnas.1800918115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltz E. With a free pass, CRISPR-edited plants reach market in record time. Nat Biotechnol. 2018;36(1):6–7. doi: 10.1038/nbt0118-6b. [DOI] [PubMed] [Google Scholar]
- Wille L, Messmer MM, Studer B, Hohmann P. Insights to plant–microbe interactions provide opportunities to improve resistance breeding against root diseases in grain legumes. Plant, Cell Environ. 2019;42:20–40. doi: 10.1111/pce.13214. [DOI] [PubMed] [Google Scholar]
- Williams PA, Sikutshwa L, Shackleton S. Acknowledging indigenous and local knowledge to facilitate collaboration in landscape approaches—lessons from a systematic review. Land. 2020;9(9):331. [Google Scholar]
- Williamson KE, Fuhrmann JJ, Wommack KE, Radosevich M. Viruses in soil ecosystems: an unknown quantity within an unexplored territory. Annu Rev Virol. 2017;4:201–219. doi: 10.1146/annurev-virology-101416-041639. [DOI] [PubMed] [Google Scholar]
- Wintermans PC, Bakker PA, Pieterse CM. Natural genetic variation in Arabidopsis for responsiveness to plant growth-promoting rhizobacteria. Plant Mol Biol. 2016;90:623–634. doi: 10.1007/s11103-016-0442-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Z, Li L, Wang W (2021) Evaluation for education of the sustainable development of engineering college students base on NLP method. In 2021 2nd International Conference on Information Science and Education (ICISE-IE). IEEE, pp 705–709
- Xu J, Zhang Y, Zhang P, Trivedi P, Riera N, Wang Y, Liu X, Fan G, Tang J, Coletta-Filho HD, et al. The structure and function of the global citrus rhizosphere microbiome. Nat Commun. 2018;9:4894. doi: 10.1038/s41467-018-07343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Xu X, Gong Q, Li Z, Li Y, Wang S, Yang Y, Ma W, Liu L, Zhu B, Zou L, Chen G. Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol Plant. 2019;12(11):1434–1446. doi: 10.1016/j.molp.2019.08.006. [DOI] [PubMed] [Google Scholar]
- Yanar D, Naif G, Yusuf Y, Mine A, Perihan C. Effect of different organic fertilizers on yield and fruit quality of indeterminate tomato. Sci Rese Essays. 2011;6:3623–3628. [Google Scholar]
- Yang X, Guo W, Li F, Sunter G, Zhou X. Geminivirus-associated betasatellites: exploiting chinks in the antiviral arsenal of plants. Trends Plant Sci. 2019;24(6):519–529. doi: 10.1016/j.tplants.2019.03.010. [DOI] [PubMed] [Google Scholar]
- Yi HY, Chowdhury M, Huang YD, Yu XQ. Insect antimicrobial peptides and their applications. Appl Microbiol Biotechnol. 2014;98:5807–5822. doi: 10.1007/s00253-014-5792-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu QH, Wang B, Li N, Tang Y, Yang S, Yang T, Xu J, Guo C, Yan P, Wang Q, Asmutola P. CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci Rep. 2017;7(1):11874. doi: 10.1038/s41598-017-12262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SSEA, Mahas A, Vanderschuren H, Mahfouz MM. Engineering crops of the future: CRISPR approaches to develop climate-resilient and disease-resistant plants. Genome Biol. 2020;21(1):1–19. doi: 10.1186/s13059-020-02204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q, Sukumaran J, Wu S, Rodrigo A. Neutral models of microbiome evolution. PLoS Comput Biol. 2015;11:1004365. doi: 10.1371/journal.pcbi.1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zhang Z, Unver T, Zhang B. CRISPR/Cas: a powerful tool for gene function study and crop improvement. J Adv Res. 2021;21(29):207–221. doi: 10.1016/j.jare.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu YX, Zhang N, Hu B, Jin T, Xu H, ... Bai Y (2019) NRT1. 1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat Biotechnol 37(6):676–684 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.