Abstract
As a class of essential enzymes in protein translation, aminoacyl–transfer RNA (tRNA) synthetases (aaRSs) are organized into two classes of 10 enzymes each, based on two conserved active site architectures. The (αβ)2 glycyl-tRNA synthetase (GlyRS) in many bacteria is an orphan aaRS whose sequence and unprecedented X-shaped structure are distinct from those of all other aaRSs, including many other bacterial and all eukaryotic GlyRSs. Here, we report a cocrystal structure to elucidate how the orphan GlyRS kingdom specifically recognizes its substrate tRNA. This structure is sharply different from those of other aaRS-tRNA complexes but conforms to the clash-free, cross-class aaRS-tRNA docking found with conventional structures and reinforces the class-reconstruction paradigm. In addition, noteworthy, the X shape of orphan GlyRS is condensed with the largest known spatial rearrangement needed by aaRSs to capture tRNAs, which suggests potential nonactive site targets for aaRS-directed antibiotics, instead of less differentiated hard-to-drug active site locations.
Cocrystal structure of orphan GlyRS-tRNA complex provides insights into aaRS classifications and antibiotics discovery.
INTRODUCTION
The functions of aminoacyl–transfer RNA (tRNA) synthetases (aaRSs) in protein biosynthesis are so essential that the members in this enzyme family are believed to have appeared before the last universal common ancestor and evolved thereafter (1, 2). On the basis of their active site architectures, aaRSs are divided into two classes of 10 enzymes each (3), which are further divided into three subclasses (a, b, and c) depending on the closeness of the evolutionary relationships (4). The early work of Rodin and Ohno (5) and further works by Carter et al. (6–8) on urzymes revealed that two classes of aaRSs might arise from opposite strands of early RNA genomes, and they were found to approach the acceptor stem of tRNA from opposite sides (9). Specially, when members of class IIa are bound to tRNA, they do so in a way that allows members of class Ia to also fit onto the same tRNA with no steric clashes. However, class IIa members cannot be paired with members of class Ib or Ic. Thus, Ia enzymes specifically pair with IIa, Ib enzymes with IIb, and Ic enzymes with IIc (4, 10). Two types (class Ib or IIb) of lysyl-tRNA synthetase (LysRS) have been found in different organisms, and they can be simultaneously docked to tRNALys without steric crashes (11, 12). These observations, and other considerations based on the sequences of active site regions, gave rise to the hypothesis that early aaRSs bound in pairs to protect each tRNA from degradation by heat or nucleases (10). This hypothesis is generally consistent with all the aaRSs whose tRNA binding modes have been clarified.
In most bacteria, the class II glycyl-tRNA synthetase (GlyRS) in charge of the synthesis of glycyl-tRNAGly is a special member of aaRS family (13). While GlyRS in eukaryotes, archaea, and some bacteria is a homodimer like most class II aaRSs (14, 15), GlyRS in most bacteria is a unique (αβ)2 heterotetramer (13, 16). Unlike other aaRSs, where the ancestral relationship between them can be clearly seen through evolution, in sequence and structure, the (αβ)2 GlyRS is distinct from any other aaRS, including other GlyRSs. Thus, it is a unique orphan aaRS.
Recently, crystal structures of Escherichia coli GlyRS and Thermanaerothrix daxensis GlyRS were solved as representative (αβ)2 orphan GlyRSs (12, 17). Orphan GlyRS adopts an unprecedented X-shaped architecture with two α subunits forming a globe at the center and two β subunits flanking the two sides of the α subunit dimer. Regarding its unusual structure, when we docked in silico the orphan GlyRS to its tRNA partner, we found that, for this docking model, there was no steric clash with the way that subclass Ia enzyme fits onto the tRNA (12). This result provided further support to the hypothesis for the origin of the aaRS classes (4, 5, 10, 18, 19). In addition, this foundation provided the basis for the present work, which was motivated by the obvious need to obtain a cocrystal of the unique GlyRS with bound tRNA/s to further test the hypothesis about the origin of the two classes of aaRSs.
Furthermore, we wanted to find at least a starting path toward using the unique X-shaped GlyRS to exploit for badly needed new antibiotics. Because of being essential proteins encoded by single genes, aaRSs have long been targets for development of antibiotics (20). Both active and editing sites have been probed for this purpose. Success was achieved with a potent leucyl-tRNA synthetase (LeuRS) editing site inhibitor that was eventually developed into a U.S. Food and Drug Administration–approved antifungal agent (tavaborole) (21). Much earlier, the isoleucyl-tRNA synthetase (IleRS) active site–directed natural product mupirocin was developed as an approved and widely used antibacterial (22). Further developments have been hindered by several factors, not the least of which is achieving species and aaRS specificity. The specificity problems are difficult because the active sites of aaRSs across evolution are built on either the well conserved class I or class II architectural framework. However, the unusual structure of the (αβ)2 orphan GlyRS in pathogenic bacteria suggested an opportunity to exploit its well-differentiated (from host GlyRS and all other aaRSs) unique structure for antibiotic development. Notably, the recognition of substrate tRNAGly by GlyRS has species specificity. Bacterial (αβ)2 GlyRS can only aminoacylate tRNAGly molecules from bacteria but not those from eukaryotes (23). The best-known discriminator base is nucleotide 73, which is conserved as U in prokaryotic tRNAGly and as A in eukaryotic tRNAGly, respectively (23, 24). The kingdom-specific GlyRS-tRNA interactions provide a valuable chance for developing bacteria-selective inhibitors, but the detailed information about how (αβ)2 GlyRS recognizes and charges bacterial tRNAGly is needed.
Here, we report a cocrystal structure of E. coli orphan GlyRS bound with two tRNAGly and two intermediate adenylate-like analogs, 2-chloro-5′-O-[N-(glycyl)sulfamoyl] adenosine (GlySA). The structure mimics the aminoacylation state in that the 3′ CCA end of tRNAGly extends with a unique conformation into the active site cavity to approach GlySA, awaiting to receive the activated glycine. Compared with the tRNA-free state, a large rotation of the HD domain (related to a hydrolase superfamily with a conserved histidine-aspartate catalytic doublet) (25), together with a large movement of the anticodon binding domain (ABD) of the β subunit, was observed upon tRNAGly binding. A loop in the HD domain directly interacts with the critical discriminator base U73. By revealing how the enzyme fits onto its tRNAs, the cocrystal structure further supports the hypothesis of the origin of the aaRS classes. Moreover, drugging a potential nonactive site to prevent the CCA end of tRNA from accessing the active site may be an alternative way to work around the difficulties associated with the more conventional active site–directed inhibitors.
RESULTS
The orphan EcGlyRS is cocrystallized with two bound tRNAGly
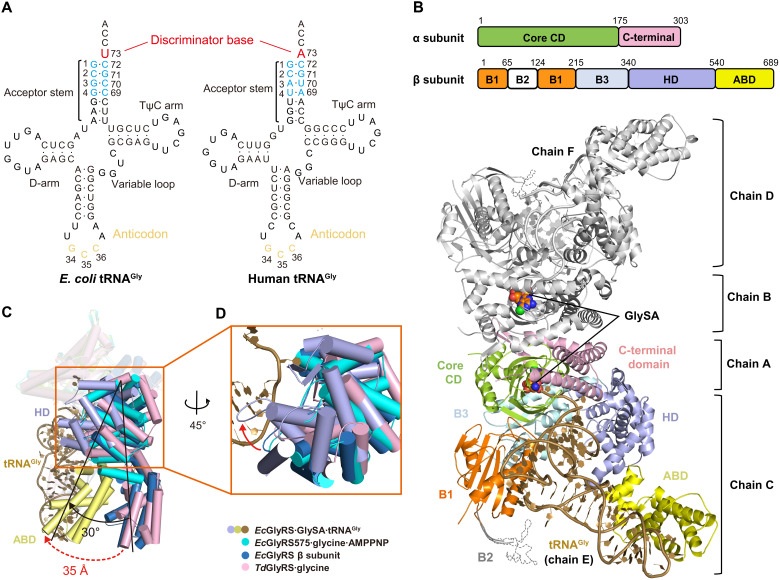
To understand how (αβ)2 heterotetrameric orphan GlyRS recognizes and charges its kingdom-specific substrate tRNAGly, we determined a cocrystal structure of E. coli GlyRS (EcGlyRS) in complex with EctRNAGly(GCC) (Fig. 1A) and an intermediate product analog GlySA (26) at 2.9 Å with R/Rfree = 0.230/0.260 (table S1). The asymmetric unit contains one EcGlyRS∙GlySA∙tRNAGly ternary complex consisting of an (αβ)2 EcGlyRS, two GlySA, and two tRNAGly molecules with a total molecular weight of about 280 kDa (Fig. 1B). EcGlyRS is organized as two protomers (one α subunit plus one β subunit), which are symmetric to each other through a noncrystallographic twofold rotation axis for both their structure and tRNA binding modes (Fig. 1B and fig. S1). Because of the better density map (fig. S2A), the protomer consisting of chains A (α subunit) and C (β subunit) and the corresponding substrate tRNAGly (chain E) were used in the following structural analysis unless otherwise indicated.
Fig. 1. The overall view of orphan EcGlyRS∙GlySA∙tRNAGly ternary complex.
(A) Cloverleaf models of E. coli and human tRNAGly(GCC). (B) Cartoon representation of the overall structure of orphan (αβ)2 EcGlyRS in complex with E. coli tRNAGly(GCC) and an intermediate product analog GlySA. The protomer consisting of chains A and C is colored the same as domain diagram above, and the bound tRNAGly (chain E) is colored in dark yellow. The other protomer (chains B and D) and its corresponding substrate tRNAGly (chain F) are colored in gray. GlySA is presented as spheres in both active site cavities. (C) Structural comparison of EcGlyRS∙GlySA∙tRNAGly complex, EcGlyRS575∙glycine∙AMPPNP complex (cyan) (PDB code 7eiv), EcGlyRS β subunit predicted by AlphaFold2 (blue) (www.alphafold.ebi.ac.uk/entry/P00961), and TdGlyRS∙glycine (pink) (PDB code 7lu4) complex reveals that major conformation changes occur at the C-terminal part of β subunit. HD and ABD rotate about 30° upon tRNA binding, and it brings ABD move inward about 35 Å to make contacts with the anticodon loop of tRNAGly. (D) A large rotation of HD domain for clamping the major groove of tRNAGly acceptor stem.
The α subunit consists of a core catalytic domain (CD) and a C-terminal domain, and the β subunit could be divided into five domains named as B1, B2, B3, HD, and ABD. While the anticodon loop of tRNAGly is recognized by ABD, the acceptor stem binds to the α subunit and HD domain of the β subunit (from the same protomer). The 3′ terminal CCA end of tRNAGly extends into the active site (Fig. 1B). Notably, on the basis of structural and mutagenesis analysis, the B2 domain was previously speculated to contribute to the recognition of tRNAGly through interacting with the elbow region of tRNAGly (12). However, the electron density of the B2 domain is weak (because of its dynamics) that this interaction could not be observed in our EcGlyRS∙GlySA∙tRNAGly ternary complex structure. It is likely that the capture of tRNAGly by EcGlyRS involves multiple steps, and the B2 domain may only interact with tRNAGly at a step other than the state captured in the crystal. Human GlyRS (HsGlyRS) also has a similar domain named Ins3 that recognizes the elbow region of human tRNAGly, and a large movement of Ins3 was observed between different steps of catalysis (27). The Ins3 domain facilitating tRNA binding is coming from the other subunit of homodimeric HsGlyRS, and this cross-subunit/protomer tRNA binding manner is commonly used in class II aaRSs (28–30). In contrast, our structure indicates that the two protomers of (αβ)2 GlyRS recognize and capture their substrate tRNAGly independently.
The C-terminal part of the β subunit undergoes a large rotation and movement upon tRNAGly binding
To clarify the structural movements of EcGlyRS required for tRNA binding, the structure of the EcGlyRS∙GlySA∙tRNAGly complex was superimposed on that of the EcGlyRS575∙glycine∙AMPPNP complex [Protein Data Bank (PDB) code 7eiv] (12), TdGlyRS∙glycine complex (PDB code 7lu4) (17), and full-length EcGlyRS β subunit (predicted by AlphaFold2; www.alphafold.ebi.ac.uk/entry/P00961) (31). Compared with those GlyRS structures without tRNA binding, a large domain rotation and movement in the EcGlyRS∙GlySA∙tRNAGly complex occurs at the C-terminal part of the β subunit (HD and ABD) (Fig. 1C). Notably, the C-terminal parts of the β subunits in GlyRS structures without bound tRNAGly all have a similar orientation to each other (Fig. 1C). This observation supports that the large domain rotation of GlyRS in the existence of tRNA has a particular biological relevance. This rotation of about 30° clamps tRNA in the EcGlyRS∙GlySA∙tRNAGly complex (Fig. 1, C and D). It happens mainly at the linker between the B3 and HD domains (fig. S3A), with the HD and ABD domains rotating as a whole (fig. S3B). As a result, the ABD moves inward about 35 Å to make contact with the anticodon loop of tRNAGly in EcGlyRS∙GlySA∙tRNAGly complex (Fig. 1C).
Large domain movements upon tRNA binding occur in some other aaRSs. For class I aaRSs, the largest structural movement induced by tRNA binding is the C-terminal domain of tyrosyl-tRNA synthetase (TyrRS), which moves about 25 Å to contact and recognize the long variable loop of tRNATyr (fig. S4A) (32). For class II aaRSs, the Ins3 domain of human GlyRS and the C-Ala domain of alanyl-tRNA synthetase (AlaRS) move 30 to 40 Å to contact the elbow region of their respective tRNAs (fig. S4, B and C) (27, 33). Notably, the largest movements in these aaRSs mainly occur with small accessory domains, which play subsidiary roles in tRNA binding (such as the Ins3 and C-Ala domains). In contrast, the ABD of (αβ)2 GlyRS plays a central role in tRNA recognition, and it, together with the HD domain, constitutes almost half of the β subunit. Thus, bacterial (αβ)2 GlyRS might be the only aaRS member where a central tRNA recognition domain undergoes such a large spatial movement to capture its tRNA substrate.
Moreover, rotation of the HD domain upon tRNAGly binding also resulted in a much larger interface between the HD domain and the α subunit [608 Å2 versus 252 Å2 as calculated by PISA (www.pdbe.org/pisa/)] (fig. S3C) (34). The α and β subunits are easy to dissociate, and, as previously discussed, the two subunits fuse via a linker into a single polypeptide in some (αβ)2 GlyRSs (35). Thus, tRNA binding may facilitate the stabilization of the orphan (αβ)2 heterotetramer for catalysis.
HD domain binds to the major groove of the tRNAGly acceptor stem
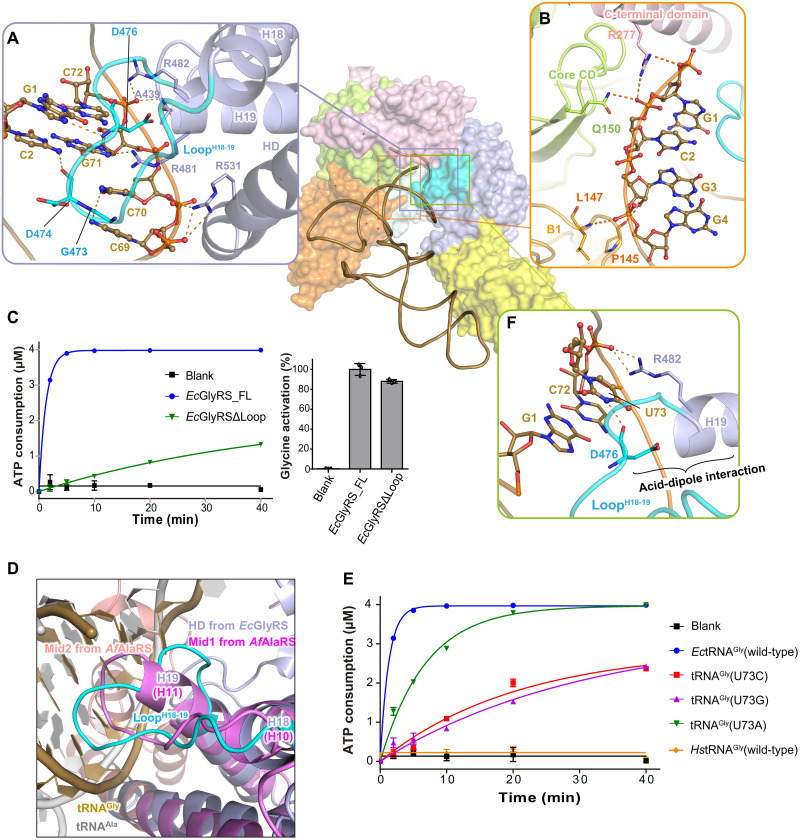
Binding of the acceptor stem of tRNAGly mainly involves the α subunit and the HD domain of the β subunit. Particularly, the loop between helices H18 and H19 (LoopH18-19) and the N-terminal part of H19 from the HD domain insert into the major groove of the tRNAGly acceptor stem and thereby interact extensively with the first four base pairs (G1∙C72, C2∙G71, G3∙C70, and G4∙C69) (Fig. 2A and fig. S2B) of that stem. The backbones of Gly473, Asp474, and Asp476 from LoopH18-19 form four base-specific hydrogen-bonding interactions with G1, C2, C69, and C70, while Arg481 at the N terminus of H19 forms a hydrogen bond (H-bond) with the base of G71 (Fig. 2A). In addition to base-specific interactions, the backbones of the first four base pairs form multiple interactions with Ala439, Arg482, and Arg531 from the HD domain of the β subunit (Fig. 2A), with Pro145 and Leu147 from the B1 domain of the β subunit, and with Gln150 and Arg277 from the α subunit (Fig. 2B).
Fig. 2. The HD domain of the β subunit of orphan GlyRS participates in the recognition of tRNAGly acceptor stem and discriminator U73.
(A) The acceptor stem of tRNAGly is mainly recognized by the α subunit and the HD domain of the β subunit. The loop (colored in cyan) between helices H18 and H19 (LoopH18-19) inserts into the major groove of tRNAGly acceptor stem and forms broad interactions with the first four base pairs of the acceptor stem. (B) The α subunit and the B1 domain of β subunit assist the binding of tRNAGly acceptor stem through backbone-mediated nonspecific interactions. (C) Deletion of the LoopH18-19 of orphan EcGlyRS markedly reduced the tRNA-dependent ATP consumption but not the tRNA-independent glycine activation, supporting an important role of LoopH18-19 in tRNAGly binding as observed in the cocrystal structure. The enzymatic assay data here and later are shown as means ± SD (n = 3). (D) Comparison of orphan EcGlyRS with Archaeoglobus fulgidus AlaRS (AfAlaRS) (PDB code 3wqy) for their similar modes on recognizing tRNA acceptor stems. Mid1 (purple) and Mid2 (salmon) subdomains of AfAlaRS and substrate tRNA (gray) are presented as cartoon. (E) The aminoacylation of E coli tRNAGly, its U73 mutants, and human tRNAGly by orphan EcGlyRS. (F) Recognition of the discriminator base U73 of tRNAGly by orphan EcGlyRS. The polar interactions between the first four base pairs and U73 of tRNAGly and the key residues of EcGlyRS are described as orange dashed lines.
Substitution of the first base pair by other nucleotides has been reported to cause an 11- to 43-fold decrease in tRNAGly aminoacylation (36). Replacement of the second base pair lowered aminoacylation by 5- to 10-fold, and mutation of the third base pair resulted in a smaller but still notable decrease (one- to fourfold) (36). Thus, these biochemical data confirmed the structural observation that the first four base pairs are important determinants of tRNAGly recognition. Consistently, sequence analysis using the tRNAviz program (37) showed that the first four base pairs are highly conserved among the bacterial tRNAGly molecules (fig. S5). On the other side, the key residues on LoopH18-19 and helix H19 of the HD domain that is involved in interacting with the first four base pairs of tRNAGly are also highly conserved among the aligned (αβ)2 GlyRSs (fig. S6). In addition, in EcGlyRS, deletion of LoopH18-19 largely decreased its activity on the tRNAGly-dependent step of aminoacylation as shown in the adenosine 5′-triphosphate (ATP) consumption assay but not glycine activation (Fig. 2C).
In many class II aaRSs, protein sequences outside the aminoacylation domain (e.g., insertion domains or editing domain) facilitate recognition of the acceptor stems and the 3′ termini of their cognate tRNAs (fig. S7). For example, in HsGlyRS, the Ins1 and WHEP domains contact the minor groove of the acceptor stem and 3′ terminus of tRNA, respectively (fig. S7A) (38). The N2 domain (editing domain) of threonyl-tRNA synthetase (ThrRS) contacts the minor groove of the acceptor stem (fig. S7B) (28). In addition, the insertion domains of histidyl-tRNA synthetase (HisRS) and aspartyl-tRNA synthetase (AspRS) participate in recognizing the 3′ termini of their cognate tRNAs (fig. S5, C and D) (39, 40). In our analysis, AlaRS shows the highest similarity with orphan (αβ)2 GlyRS on how the tRNA acceptor stem is recognized (Fig. 2D). While its Mid2 subdomain clamps tRNA from the minor groove side of the tRNAAla acceptor stem, AlaRS inserts a loop between helices H10 and H11 (LoopH10-11) from the Mid1 subdomain into the major groove of the acceptor stem, forming multiple interactions with the bases and backbones of the first three base pairs of tRNAAla (Fig. 2D and fig. S7E) (33). The similar tRNA acceptor stem binding mode between LoopH10-11 of the Mid1 subdomain of AlaRS and LoopH18-19 of the HD domain of (αβ)2 GlyRS (Fig. 2D and fig. S7, E and F) supports the potential evolutionary relationship between these two aaRSs, which has been discussed by others (12, 17, 41).
HD domain directly contacts the discriminator base U73
Orphan (αβ)2 GlyRS was reported not to glycylate eukaryotic tRNAGly (Fig. 2E) (23). U73 is absolutely conserved in prokaryotic tRNAGly and as A73 in eukaryotic tRNAGly (Fig. 1A and fig. S5). Nucleotide 73 is considered as the major determinant for kingdom-specific glycylation of tRNAGly (23, 42). Our ATP consumption assay confirmed that U73A substitution decreased the aminoacylation of EctRNAGly by EcGlyRS and that the U73C and U73G mutants decreased aminoacylation even more strongly than U73A (Fig. 2E).
In our structure of the EcGlyRS∙GlySA∙tRNAGly ternary complex, the pyrimidine ring of U73 points inward and forms stacks with C72, while Arg482 from helix H19 of the HD domain interacts with the backbone phosphate of U73 (Fig. 2F). Because of the insertion of LoopH18-19 into the major groove of the acceptor stem, the main chain oxygen of Asp476 can form a base-specific H-bonding interaction with N3 of the U73 pyrimidine ring (Fig. 2F). Notably, the local conformation of LoopH18-19 is well stabilized by an intramolecular acid–α helix dipole interaction between the β-carboxyl of the conserved Asp476 and the N terminus of helix H19 (Fig. 2F and fig. S6). Thus, substitution of U73 by other nucleotides would result in loss of the Asp476 H-bond, and, with larger purine bases, such a substitution might also cause a clash with LoopH18-19.
The preference for U73 in bacterial tRNAGly may also have other reasons beyond GlyRS recognition. For instance, bacterial d-aminoacyl-tRNA deacylase (DTD) prefers A73 to U73 (43). Thus, the cognate bacterial Gly-tRNAGly (U73) could escape from unexpected deaminoacylation by DTD, while the noncognate Gly-tRNAAla (A73) could be efficiently cleaved. Similarly, eukaryotic DTD prefers U73, so that the cognate eukaryotic Gly-tRNAGly (A73) can be avoided (43).
tRNAGly CCA end binds to the active site cavity of orphan GlyRS at the aminoacylation mimicking state
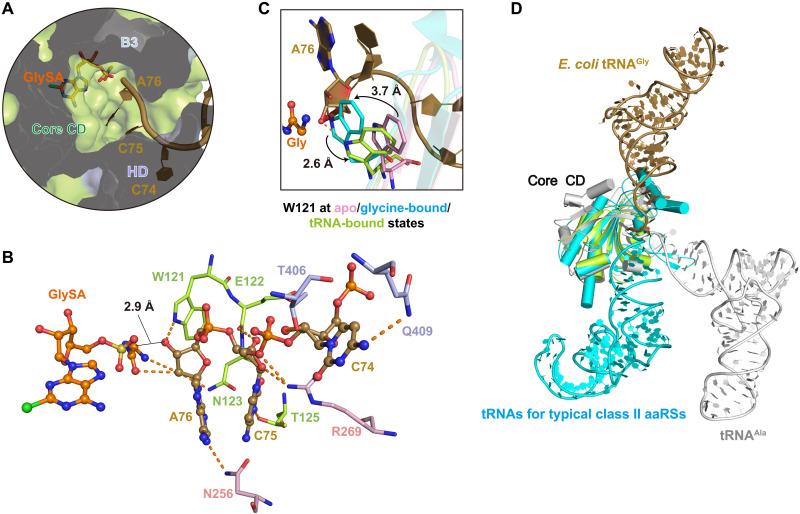
The active cleft on the α subunit is partially covered by the B1, B3, and HD domains of the β subunit, resulting in an active site cavity deeper than that of other typical class II aaRSs (12). Therefore, the 3′ CCA end of tRNAGly adopts an extended conformation to reach the glycyl group of the intermediate product glycyl adenylate (Fig. 3A and fig. S2, D and E), and this conformation of the CCA end was stabilized by forming extensive interactions with residues from the α subunit and HD domain of the β subunit (Fig. 3A).
Fig. 3. tRNAGly CCAend binds to orphan EcGlyRS active site cavity at the aminoacylation mimicking state.
(A) The 3′ CCA end of tRNAGly extends into the deep aminoacylation pocket of orphan EcGlyRS. EcGlyRS, GlySA and substrate tRNAGly are presented as surface, sticks, and cartoons, respectively. (B) The interactions of CCA end of tRNAGly with orphan EcGlyRS and intermediate product analog GlySA. The polar interactions are described as orange dashed lines. The distance between the carbonyl group of glycine moiety of GlySA and the 3′-OH of the ribose moiety of A76 is about 2.9 Å. (C) Structural movements of Trp121 on the ɑ subunit upon substrate binding. Trp121 at apo state [α subunit of Campylobacter jejuni GlyRS (CjGlyRS); PDB code 3rf1], glycine-bound state (EcGlyRS·glycine·AMPPNP complex; PDB code 7eiv), and tRNA-bound state are colored in pink, cyan, and green, respectively. (D) Structural superimposition of aminoacyl-tRNA synthetases (aaRSs)–tRNA complexes according to their core catalytic domain reveals that E. coli tRNAGly and tRNAAla locate at positions, which is different from tRNA substrates of other typical class II synthetases including human GlyRS. E. coli tRNAGly, tRNAAla, and a representative tRNA for typical class II synthetases [HsGlyRS (PDB code 5e6m), ThrRS (PDB code 1qf6), HisRS (PDB code 4rdx), ProRS (PDB code 1h4s), SerRS (PDB code 1ser), AspRS (PDB code 1c0a), and phenylalanyl-tRNA synthetase (PheRS; PDB code 1eiy)] are colored in dark yellow, gray, and cyan, respectively.
The cytosine group of C74 is oriented toward the HD domain of the EcGlyRS β subunit and forms an H-bond with the side chain of Gln409 (Fig. 3, A and B). In addition, the sugar of C74 interacts with Thr406 from the HD domain of the β subunit (Fig. 3B). The cytosine ring of C75 H-bonds with the side chain of Thr125 from the α subunit, and the backbone of C75 contacts the side chain of Arg269 and the main chain of Glu122 from the α subunit (Fig. 3B). The adenine ring of A76 H-bonds with the side chain of Asn256 from the α subunit, and its sugar ring also H-bonds with the side chain of Trp121 from the α subunit (Fig. 3B).
Notably, structural movement of Trp121 on the α subunit was observed upon substrate binding. Compared with GlyRS in the apo state (PDB code 3rf1), the binding of glycine and an ATP analog caused a 3.7-Å movement of Trp121 to coordinate glycine by forming a cation-π interaction (12). With tRNAGly bound, Trp121 moves back about 2.6 Å to allow A76 to approach the glycyl moiety of the intermediate analog (Fig. 3C). In this state, the 2′-OH of A76 sugar forms two H-bonds with the carbonyl oxygen and amine group of GlySA, respectively (Fig. 3B). The distance between the carbonyl group of glycine moiety and the 3′-OH of A76 is about 2.9 Å (Fig. 3B), suggesting that our complex structure mimics the aminoacylation state in that the 3′-OH of nucleotide A76 of tRNAGly is positioned to receive the glycyl moiety from the intermediate product, consistent with past work that class II aaRSs fuse the amino acid to the 3′ position (44). Except for Thr406 in the HD domain (interacts with tRNAGly CCA end through its backbone) and Thr125 in the α subunit (changed to Ser in some GlyRSs), all other residues mentioned above for CCA end interactions are absolutely conserved in the aligned (αβ)2 GlyRSs (figs. S6 and S8).
Although orphan (αβ)2 GlyRS has some structural similarity to other class II aaRSs on their core catalytic domains, their substrate tRNAs extend their CCA ends to the active site cavities of the cognate aaRSs from different directions (Fig. 3D). When the core catalytic domains of orphan EcGlyRS and other class II aaRSs were superimposed, tRNAGly located at the position opposite that of most other tRNAs, with only tRNAAla located between tRNAGly and other tRNAs (Fig. 3D). In typical class II aaRSs, including eukaryotic GlyRSs, class II signature motif 2 contributes to stabilizing the 3′ CCA end of tRNA by forming charge and hydrogen bonding interactions (fig. S9A). However, this stabilization effect was observed in neither EcGlyRS nor AlaRS (PDB code 3wqy) because of their different modes of tRNA binding (fig. S9B) (33, 45).
Orphan GlyRS has a unique anticodon binding mode
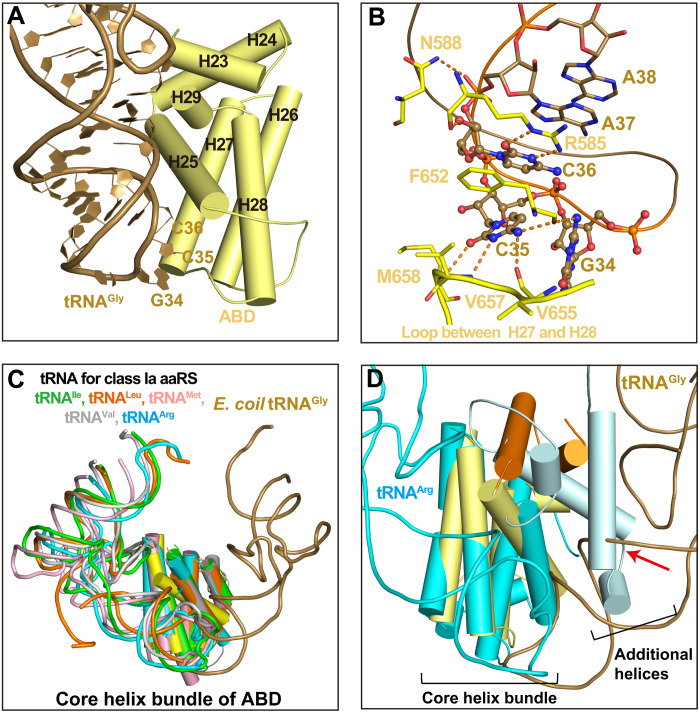
Three anticodons (with G, C, or U at the variable position 34 and C at positions 35 and 36) were used to pair the four glycine codons during protein translation (46). In the structure of the orphan EcGlyRS∙GlySA∙tRNAGly ternary complex, the ABD of the β subunit folds into an all-α structure. The ABD consists of a core four-helix bundle and an additional helix-loop-helix motif, and it specifically recognizes the anticodon triplet of tRNAGly from the minor groove side of the anticodon stem (Fig. 4A). The G34 of tRNAGly (GCC) contributes only one H-bond with the backbone of Phe652 (Fig. 4B). In contrast, C35 is surrounded by residues mainly from the loop between helices H27 and H28 (LoopH27-28), and its cytosine ring forms an H-bonding interaction network with the backbones of Phe652, Val655, Val657, and Met658. These four residues are highly conserved in aligned bacterial (αβ)2 GlyRSs (Fig. 4B and fig. S6). Furthermore, C36 forms three H-bonds with the conserved Arg585 through its cytosine ring and ribose (Fig. 4B and fig. S6).
Fig. 4. The recognition mode of tRNA anticodon loop by orphan (αβ)2 GlyRS.
(A) The ABD of EcGlyRS, consisting of a core four-helix bundle and a helix-loop-helix motif, approaches the anticodon loop from minor groove side of E. coli tRNAGly anticodon stem. (B) A zoomed-in view for anticodon-ABD interactions. The anticodon triplets (G34, C35, and C36) are presented as a ball-and-stick model, while other nucleotides and EcGlyRS residues are shown as sticks. (C) Superimposition of the ABD’s core helix bundle of EcGlyRS (yellow) with that of class Ia aaRSs [IleRS (PDB code 1ffy), LeuRS (PDB code 4aq7), MetRS (PDB code 2ct8), ValRS (PDB code 1gax), and ArgRS (PDB code 1f7u)] showed that tRNAGly binds to the ABD through a direction opposite to other tRNAs. For clarity, only ABDs’ core helix bundles and substrate tRNAs of the aaRS-tRNA complexes are shown. (D) With the unique binding mode of tRNAGly, its anticodon stem would cause clashes (indicated with a red arrow) with the accessory helices additional to the core helix bundle of ArgRS’s ABD.
These observations suggest that C35 plays a vital role in recognizing tRNAGly by bacterial GlyRSs. Consistently, a C35 mutant of E. coli tRNAGly was reported to reduce charging activity by 105- to 1500-fold, while substitution of C36 by other nucleotides damaged activity by 21- to 41-fold (36, 47). In addition to the anticodon triplet, the phosphate backbone of A37 also contributes to an interaction with Asn588. It is worth noting that the stacking interactions among nucleotides C36, A37, and A38 may stabilize the conformation of the tRNA anticodon loop (Fig. 4B).
AaRSs in different subclasses have structurally distinct ABDs. Class IIa aaRSs [ThrRS, HisRS, prolyl-tRNA synthetase (ProRS), and eukaryotic GlyRS] use an α/β-fold ABD at their C termini (27, 28, 40, 48). The all-α structure ABD of EcGlyRS is unique to class IIa aaRSs but, interestingly, similar to that of class Ia aaRSs (17, 25). We superimposed the ABD of EcGlyRS with that of the tRNA complex structures of arginyl-tRNA synthetase (ArgRS; PDB code 1f7u), valyl-tRNA synthetase (ValRS; PDB code 1gax), methionyl-tRNA synthetase (MetRS; PDB code 2ct8), LeuRS (PDB code 4aq7), and IleRS (PDB code 1ffy) (22, 49–52). EcGlyRS ABD overlaid well with the ABDs of these class Ia aaRSs, particularly for their core four-helix bundles (Fig. 4C). However, the anticodon loops of the different tRNAs approached the anticodon binding site (located at the distal end of the core four-helix bundles) from different sides (Fig. 4C). We took ArgRS as an example, whose ABD showed the highest sequence and structural similarity to that of orphan (αβ)2 GlyRS (17, 25). If tRNAGly binds to the ArgRS ABD as it does in the EcGlyRS∙GlySA∙tRNAGly ternary complex, then the α helices additional to the core four-helix bundles in the ABD of ArgRS would cause strong clashes with the anticodon stem of tRNAGly (Fig. 4D). Thus, although orphan (αβ)2 GlyRS has a class Ia–like ABD, its tRNA recognition mode is very different from that of class Ia aaRSs.
The acceptor stem–binding mode of orphan GlyRS further supports the origin of aaRS classifications
While the catalytic domain of most class I aaRSs approaches tRNA from the minor groove side of the acceptor stem, the catalytic domain of typical class II aaRSs binds the tRNA acceptor stem from the major groove side (9, 10). We aligned the tRNAs in EcGlyRS∙GlySA∙tRNAGly complex and some representative aaRS-tRNA complexes, and the catalytic domain of EcGlyRS was found to bind to the acceptor stem of tRNA from a direction substantially different from other aaRSs, such as HsGlyRS (a representative typical class IIa aaRSs) and AlaRS (has a special relationship with orphan GlyRS) (Fig. 5A). The position of the core catalytic domain of EcGlyRS is between class I and typical class II aaRSs (Fig. 5B).
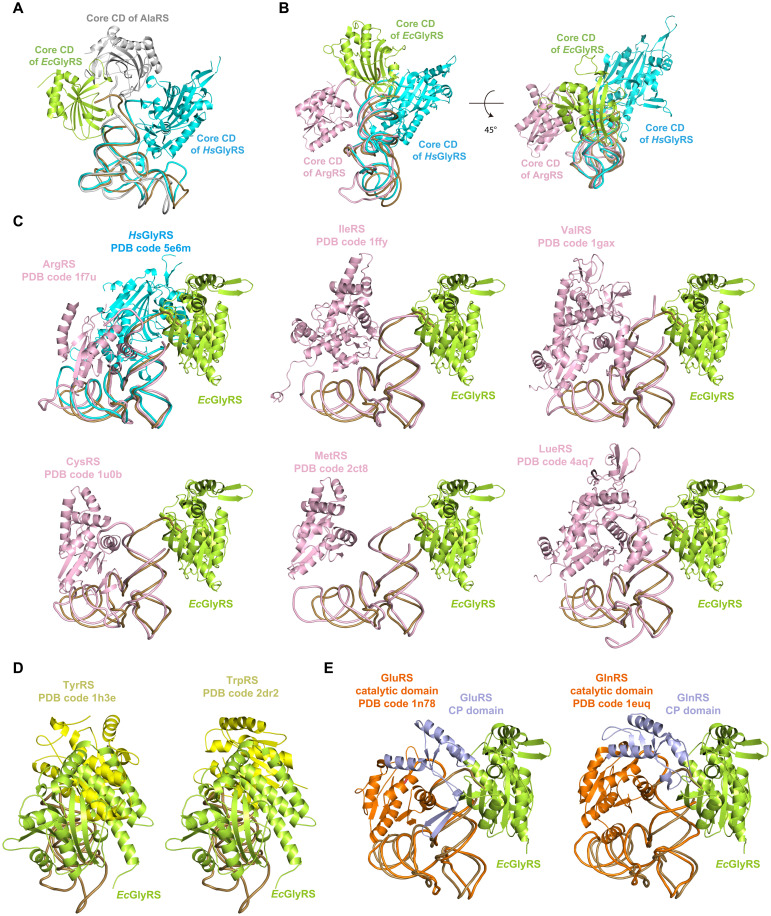
Fig. 5. The unusual tRNA binding mode of orphan (αβ)2 GlyRS supports subclass-specific pairwise docking of aaRSs on tRNA acceptor stems.
(A) The catalytic domain of orphan EcGlyRS binds to the acceptor stem of tRNA from a direction different to human GlyRS (a representative class IIa aaRS) and AlaRS [the aaRS member has a special evolutionary relationship with (αβ)2 GlyRS] when aligning their tRNA molecules. (B) The catalytic domains of most class I aaRSs (such as ArgRS) approach tRNA from the minor groove side of acceptor stem, and the catalytic domains of typical class II aaRSs (such as human GlyRS) bind tRNA acceptor stem from the major groove side. In comparison, the core catalytic domain of orphan EcGlyRS locates between class I aaRSs and typical class II aaRSs and at the top of tRNA acceptor stem. (C) The catalytic domain of orphan EcGlyRS could be paired with that of class Ia aaRSs on the acceptor stem of tRNA without clashes. (D) The catalytic domain of orphan EcGlyRS would cause clashes with that of class Ic aaRSs when aligning their tRNAs. (E) The catalytic domain of orphan EcGlyRS would cause clashes with the insertion sequences (e.g., CP domain) of the catalytic domains of class Ib aaRSs. For clarity, only the α subunit of EcGlyRS (green) and catalytic domains of class Ia (pink), Ib (orange), and Ic (yellow) aaRSs and their respective tRNAs are shown. CP domains of class Ib synthetases are colored in light blue.
The catalytic domains of aaRSs in the same subclasses but different classes can be paired without clashes in a highly specific way (Ia paired with IIa, Ib paired with IIb, and Ic paired with IIc), which probably protected the acceptor stems from degradation in the early evolution of life forms (10). The cocrystal structure is a bit different from the docking mode of the EcGlyRS-tRNA complex that we proposed previously, where the enzyme bound from the side of the stem (12). This difference is due to the unforeseen structural movements and rotations. In the present structure, the orphan GlyRS catalytic domain is at the top of the tRNA acceptor stem (Fig. 5B). When tRNA molecules were aligned, structure superposition revealed that the catalytic domain of orphan EcGlyRS could still fit onto the acceptor stem without clashes with the catalytic domain of class Ia aaRSs (Fig. 5C) but not with the catalytic domain of class Ic aaRSs (Fig. 5D) and the inserted connecting peptide (CP) in the catalytic domain of class Ib aaRSs, which is responsible for the recognition of tRNA acceptor stem and is called “acceptor binding domain” (Fig. 5E) (53). Thus, the idiosyncratic structure of the complex provided a fresh test of the “origin of classes” hypothesis, showing that, regardless of its unusual overall structure and tRNA binding mode, orphan (αβ)2 GlyRS could still be paired with class Ia aaRSs. This result further supports the hypothesis that the two classes of aaRSs can be defined by how they copair on tRNA.
Structure of orphan (αβ)2 bacterial GlyRS suggests a unique approach to aaRS antibiotics
As 20 essential enzymes, aaRSs have long been considered as ideal targets for developing antibiotics (20). The unusual orphan (αβ)2 infectious bacterial GlyRS suggested an opportunity to exploit that well-differentiated structure (from host GlyRS and all other aaRSs) for antibiotic development. Infectious bacterial organisms that have orphan (αβ)2 GlyRSs include Streptococcus pneumoniae, Helicobacter pylori, Chlamydia trachomatis, Pseudomonas aeruginosa, among others.
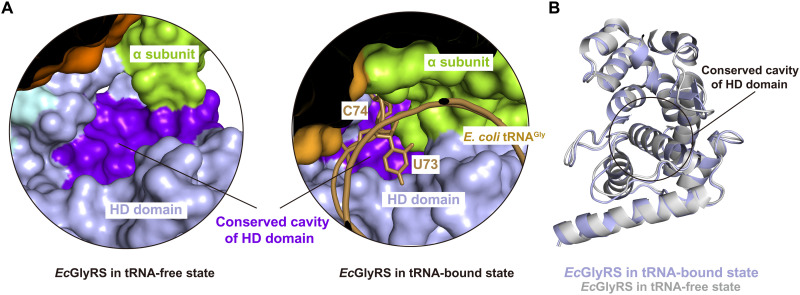
Our previous work identified a highly conserved cavity on the HD domain of orphan GlyRSs, which is absent in eukaryotic GlyRSs. Mutation of residues Lys364, Arg367, Lys394, and Asp459 around this bacterial-specific cavity reduced the aminoacylation activity to bacterial tRNAGly, but the mechanism was unclear (12). tRNA binding requires a large rotation of the C-terminal HD and ABD domains of the β subunit. While this cavity on HD is exposed in the structure of the tRNA-free state enzyme (Fig. 6A), it gets largely covered by the ɑ subunit and closely contacts nucleotides U73 and C74 of the acceptor arm of bound tRNAGly in the EcGlyRS∙GlySA∙tRNAGly complex (Fig. 6A). However, the structure itself of this cavity is not reshaped by this rotation (Fig. 6B). Likely, chemicals binding to this cavity could disrupt the 3′ CCA end of bacterial tRNAGly from accessing the active site. Thus, this conserved cavity on the HD domain, which is idiosyncratic to the orphan GlyRSs, presents a unique opportunity to go beyond the highly conserved active sites to find and develop antibiotics.
Fig. 6. The conserved cavity on HD domain is largely covered by the ɑ subunit and nucleotides U73 and C74 of tRNA acceptor arm after tRNA binding.
(A) The rotation of the C-terminal part of β subunit upon tRNA binding makes the conserved cavity on HD domain, which is exposed to surface in tRNA-free state, largely covered by α subunit and nucleotides U73 and C74 of the tRNA acceptor arm. (B) The structure of HD domain and its conserved cavity have almost no changes upon tRNA binding.
DISCUSSION
Eukaryotic GlyRS, like class IIa ThrRS, HisRS, and ProRS, has an α/β-fold ABD at the C terminus and recognizes the anticodon triplet of tRNAGly from the major groove side of the anticodon stem (38). In contrast, the ABD in orphan (αβ)2 GlyRS is an all-α structure, and it binds anticodon triplets from the minor groove side of the stem (Fig. 4A). Studies have suggested that orphan GlyRS is related to the ancestor of AlaRS (12, 41), a special class IIa aaRS that lacks an ABD and instead recognizes tRNAAla through the G3∙U70 identity base pair (33). Possibly, the preorphan GlyRS did not have an ABD and recognized tRNAGly mainly on the basis of U73 and the first four base pairs. When preorphan GlyRS evolved to more accurately and effectively capture tRNAGly by integrating an ABD, the ABD was not obtained from other class IIa aaRSs but from ArgRS, because the ABD of ArgRS has multiple specific interactions with C35 of tRNAArg. For other tRNAs with C at position 35, their corresponding aaRSs recognize tRNA either through the shape of the cognate tRNA [e.g., seryl-tRNA synthetase (SerRS) and cysteinyl-tRNA synthetase (CysRS)] or by forming more interactions with another nucleotide in the anticodon loop [e.g., trytophanyl-tRNA synthetase (TrpRS)] (29, 54–56).
A minihelix comprising the acceptor-TpsiC stem of tRNAGly is charged by eukaryotic and prokaryotic GlyRSs (42). The first two base pairs of the acceptor stem are well conserved as G1∙C72 and C2∙G71 in tRNAGly molecules from all three kingdoms of life (fig. S5). Mutations of either base pair strongly reduced the charging activity of bacterial and eukaryotic tRNAGly by their cognate GlyRSs (36). Both HsGlyRS and EcGlyRS have direct base-specific interactions with the first two base pairs of their respective tRNAGly (Fig. 2A) (38). However, different mechanisms are used. For example, for HsGlyRS, Arg283 from the catalytic domain forms three H-bonds with G1 and G71 (38). In contrast, Asp474, Asp476, and Arg481 from the HD domain of orphan EcGlyRS form three H-bonds with C2, G1, and G71, respectively (Fig. 2A). The use of distinct domains to retain the acceptor stem recognition of the first two base pairs strongly suggests the cross-kingdom evolutionary pressure, which focuses on the same base pairs for a critical discrimination of tRNAGly. However, orphan GlyRS charges only tRNAGly from bacteria but not tRNAGly from eukaryotes. N73 (conserved as U in bacteria and A in eukaryotic tRNAGly) is considered as a key discriminator for kingdom specificity of tRNAGly glycylation. Thus, despite common cross-kingdom recognition of the first two base pairs of the acceptor stem, N73 has ostensibly evolved to block cross-kingdom charging of tRNAGly. However, substitution of U73 by A in E. coli tRNAGly decreased but did not completely abolish the aminoacylation activity in our assay (Fig. 2E), consistent with a previous report that E. coli tRNAGly U73A mutant caused only an 11-fold decrease in Vmax/Km (35). Thus, other discriminators beside U73 may contribute to the kingdom specificity of tRNAGly, which requires further studies.
While traditional aaRS inhibitors are active site–targeted competitors of amino acid and/or ATP binding, other studies have revealed that blocking aaRS-tRNA interactions might be a viable alternative strategy for drugging aaRSs (57–62). The first representative cocrystal structure of orphan (αβ)2 GlyRS in complex with tRNAGly presented here clarifies the mechanisms of how the unique structure of orphan (αβ)2 GlyRS recognizes its substrate tRNAGly. Our structural study not only reinforces the paradigm of the origin of aaRS classifications but also shows bacterial-specific large conformational changes and nonactive site regions associated with the binding of tRNAGly, which informs on a potential approach to the development of bacteria-specific antimicrobials.
MATERIALS AND METHODS
Protein expression and purification
The coding sequence for the full-length α [UniProt identifier (ID): P00960] and β (UniProt ID: P00961) subunits of EcGlyRS were amplified from the genomic DNA of E. coli strain K12 with polymerase chain reaction (PCR) and inserted into the plasmids pET20b and pET28a, respectively. The β subunit has a hexahistidine tag at the C terminus for protein purification. E. coli BL21(DE3) cells cotransformed with two plasmids were grown at 37°C in LB medium supplemented with ampicillin (100 mg/liter) and kanamycin (50 mg/liter) until optical density at 600 nm reached 0.6. Coexpression of the α and β subunits of EcGlyRS was induced by adding 0.2 mM isopropyl-β-d-thiogalactopyranoside. After growth at 20°C for 20 hours, E. coli cells were harvested by centrifugation at 5000 revolutions per min (rpm) for 30 min. The cell pellets were resuspended and sonicated in lysis buffer [50 mM tris-HCl (pH 8.0), 400 mM NaCl, and 20 mM imidazole]. The cell lysate was centrifuged at 18,000 rpm for 30 min at 4°C, and the supernatant was loaded onto a Ni–nitrilotriacetic acid (NTA) column preequilibrated with lysis buffer. The Ni-NTA column was washed with 20 column volumes of lysis buffer to remove impurities, and the target protein was eluted with elution buffer [50 mM tris-HCl (pH 8.0), 400 mM NaCl, and 200 mM imidazole]. The protein in elution fraction was concentrated to 15 to 20 mg/ml using a 50-kDa Ultra-15 centrifugal filter device and then further purified using a HiLoad 16/60 Superdex 200 pg column with the running buffer [200 mM NaCl and 20 mM tris-HCl (pH 8.0)]. Protein purity was assessed with SDS–polyacrylamide gel electrophoresis (SDS-PAGE). The purified (αβ)2 EcGlyRS protein was concentrated in the storage buffer [100 mM NaCl and 10 mM tris-HCl (pH 7.0)] and frozen at −80°C before use.
In vitro transcription of tRNAGly
E. coli tRNAGly (GCC), the most favorable tRNAGly molecule in E. coli cells (63, 64), was produced with in vitro T7 polymerase transcription. The initial DNA template was generated by PCR using primer1 (5′-TAATACGACTCACTATAGCGGGAATAGCTCAGTTGGTAGAGCACGACCTTGCCAAG-3′) and primer2 (5′-TGGAGCGGGAAACGAGACTCGAACTCGCGACCCCGACCTTGGCAAGGTCGTGCTC-3′). These two primers covered the full sequence of tRNAGly (GCC) and partially overlapped with each other (underlined nucleotides), and primer1 also contains a T7 promoter sequence (nucleotides in bold). The PCR product was further amplified by the second round of PCR with primer3 (5′-TAATACGACTCACTATAGCGGGAATAGC-3′) and primer4 (5′-UGGAGCGGGAAACGAGACTCG-3′), and the product was then used as the DNA template for in vitro T7 transcription assay without additional purification. The first two nucleotides at the 5′ terminus of primer4 (nucleotides in italics) were methylated at their 2′-hydroxyl groups to reduce nontemplated nucleotide addition by the T7 RNA polymerase (65). The T7 transcription reaction consisted of 200 mM tris-HCl (pH 8.0), 2 mM spermidine, 10 mM dithiothreitol (DTT), 20 mM MgCl2, DNA template (50 ng/μl), 4 mM ATP, 4 mM uridine 5′-triphosphate, 4 mM guanosine 5′-triphosphate, 4 mM cytidine 5′-triphosphate, 20 mM guanosine 5′-monophosphate, and 2 μM T7 polymerase. After incubation at 37°C for 3 to 4 hours, the transcripts were desaturated at 95°C for 5 min. The transcripts were purified by 12% PAGE supplemented with 8 M urea. The target tRNA transcripts were extracted from gel with 0.5 M ammonium acetate, precipitated by ethanol at −20°C overnight, collected by centrifugation, and redissolved in a buffer consisting of 20 mM tris-HCl (pH 8.0) and 1 mM EDTA. The tRNA product was heated at 65°C for 5 min and then refolded by slowly cooling to room temperature after the addition of 10 mM MgCl2. The refolded tRNA was concentrated with a 3-kDa Ultra-4 centrifugal filter device (Millipore), and its final concentration was determined by the ultraviolet absorbance at the wavelength of 260 nm. The tRNA product was stored at −80°C before use.
Crystallography
The sitting-drop vapor diffusion method was used to crystallize the EcGlyRS∙GlySA∙tRNAGly complex. In brief, the full-length (αβ)2 EcGlyRS (22 mg/ml) was mixed with tRNAGly(GCC) (in vitro transcript, 7.5 mg/ml) and 2 mM GlySA. GlySA was synthesized as described in scheme S1. After an incubation for 30 min on ice, crystallization drops were set up by mixing 0.5 μl of protein complex solution with 0.5 μl of reservoir solution [0.15 M MgCl2, 0.1 M NaCl, 0.1 M tris-HCl (pH 8.5), 30% polyethylene glycol, molecular weight 300]. Crystallization drops were equilibrated against 60 μl of reservoir solution at 8°C. Large crystals appeared after 4 to 7 days and were directly flash-frozen in liquid nitrogen without additional cryoprotectant solution. Diffraction data were collected using a single crystal at 100 K with a wavelength of 0.979 Å at beamline BL02U1 of the Shanghai Synchrotron Radiation Facility, China. The oscillation angle was 0.5° for each frame, and the whole dataset contained 360 frames.
The diffraction data were automatically processed with Aquarium (66), which used the autoPROC (67). The structure was solved using the molecular replacement method in the program MOLREP (68) with structures of EcGlyRS575∙glycine∙AMPPNP complex (PDB code 7eiv), full-length EcGlyRS β subunit predicted by AlphaFold2 (www.alphafold.ebi.ac.uk/entry/P00961) (31), and free Geobacillus kaustophilus tRNAGly (PDB code 4mgm) as search models. Iterative refinement of the structure model was carried out using Coot (69) and Refmac5 (70). The stereochemical quality of the final model was assessed using MolProbity (71). The statistics for the data collection and structural refinements are listed in table S1. The structure was analyzed in PyMOL (www.pymol.org), which was also used to create the figures.
ATP consumption assay
An ATP consumption assay was used to evaluate the aminoacylation activity of EcGlyRS on tRNAGly as described (12). In brief, the 30 μl of mixture containing 50 nM EcGlyRS, 4 μM ATP, 500 μM glycine, E. coli tRNAGly (GCC) (0.25 mg/ml), 30 mM Hepes (pH 7.5), 150 mM NaCl, 30 mM KCl, 40 mM MgCl2, 1 mM DTT, and 0.1% bovine serum albumin was prepared and incubated at room temperature. Five microliters of the mixture was collected from the reaction at different time points (2, 5, 10, 20, and 40 min) and mixed with 5 μl of Kinase-Glo Reagent (Promega) in a 384-well microplate to stop the reaction. After 15 min, the luminescence which reflects the concentration of the remaining ATP in the well was read on a Synergy H1 microplate reader (BioTek). The reaction without adding tRNAGly was used as the control. Last, the ATP consumption was calculated as the difference between the luminescence in the well with RNAGly and the well without tRNAGly. Each reaction was repeated three times, and the results are expressed as the means ± SD (n = 3). Statistical analyses were performed with GraphPad Prism 6.0 software, and a one-phase association equation was used for the curve fitting of the ATP consumption assay.
Measurement of glycine activation activity of EcGlyRS
Glycine activation by EcGlyRS_FL and EcGlyRS∆Loop was detected by using a coupled assay (12). EcGlyRS could catalyze an adenylyl-imidodiphosphate (AMP-PNP) and a glycine to generate a molecule of glycyl-AMP, which is attacked by a pyrophosphate to form a glycine and one ATP. Hexokinase consumes one ATP to phosphorylate glucose to glucose 6-phosphate, from which glucose 6-phosphate dehydrogenase then consumes one nicotinamide adenine dinucleotide phosphate (NADP+) to produce one 6-phosphogluconate and one NADPH (reduced form of NADP+). The experiment was conducted at room temperature in a clear 96-well microplate (Corning). Ninety microliters of reaction buffer consisting of 50 mM Hepes (pH 7.5), 10 mM MgCl2, 50 mM KCl, 1 mM DTT, 3 mM AMP-PNP, 10 mM glycine, 10 mM d-glucose, 2.5 mM sodium pyrophosphate, 0.5 mM NADP+, 2.5 U of yeast hexokinase, and 2.5 U of glucose 6-phosphate dehydrogenase was placed in the wells of the microplate. The reactions were started by adding 10 μl of EcGlyRS_FL or EcGlyRS∆Loop (final enzyme concentrations were 200 nM) to each well. The reduction of NADP+ to NADPH in the coupled assay was continuously recorded for the first 10 min by monitoring the absorbance at 340 nm using a Synergy H1 microplate reader (BioTek), and the reactions without adding inorganic pyrophosphate were used as blank controls. The slope of NADPH formation in the first 10 min (equal to the glycine activation rate) of wild-type EcGlyRS_FL was defined as 100%. The results were from three independent assays, and the error bars represent SDs.
Data analysis and figure preparation
Figures were created using the PyMOL. Protein sequence alignment analysis was performed using Clustal Omega program (72). The conservation scores were calculated by program Jalview (73). tRNA sequence analysis was performed using the tRNAviz program (37). Statistical analysis was performed using GraphPad Prism 6.0 software, each enzymatic activity assay was repeated three times, and the results are expressed as the means ± SD (n = 3).
Acknowledgments
We thank the staff of beamline BL02U1 at Shanghai Synchrotron Radiation Facility (SSRF) for assistance during data collection.
Funding: This work was supported by the National Natural Science Foundation of China (nos. 22177140, 22207133), the Guangdong Basic and Applied Basic Research Foundation (no. 2019A1515011571), Program for Guangdong Introducing Innovative and Entrepreneurial Teams (no. 2016ZT06Y337), Guangdong Provincial Key Laboratory of Chiral Molecule and Drug Discovery (no. 2019B030301005), and the National Foundation for Cancer Research.
Author contributions: Study design: L.H., P.S., and H.Z. Preparation of protein and tRNA samples: L.H., Y.J., Z.L., and B.C. Crystallization: L.H. and Y.J. Structure determination: L.H. and H.Z. Synthesis of the chemical: Z.L. and T.Z. Data analysis and experiments design: J.W., J.X., Q.G., and X.-L.Y. Supervision: P.S. and H.Z. Writing: L.H., Z.L., P.S., and H.Z.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The coordinates and structure factors for the reported crystal structure have been deposited in the Protein Data Bank under the accession code 7YSE. All materials are available from the corresponding authors upon request.
Supplementary Materials
This PDF file includes:
Supplementary Method
Scheme S1
Figs. S1 to S9
Table S1
References
REFERENCES AND NOTES
- 1.L. Lei, Z. F. Burton, Evolution of life on Earth: tRNA, aminoacyl-tRNA synthetases and the genetic code. Life (Basel) 10, 21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.L. Ribas de Pouplana, The evolution of aminoacyl-tRNA synthetases: From dawn to LUCA. Enzymes 48, 11–37 (2020). [DOI] [PubMed] [Google Scholar]
- 3.G. Eriani, M. Delarue, O. Poch, J. Gangloff, D. Moras, Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 347, 203–206 (1990). [DOI] [PubMed] [Google Scholar]
- 4.L. Ribas de Pouplana, P. Schimmel, Aminoacyl-tRNA synthetases: Potential markers of genetic code development. Trends Biochem. Sci. 26, 591–596 (2001). [DOI] [PubMed] [Google Scholar]
- 5.S. N. Rodin, S. Ohno, Two types of aminoacyl-tRNA synthetases could be originally encoded by complementary strands of the same nucleic acid. Orig. Life Evol. Biosph. 25, 565–589 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Y. Pham, L. Li, A. Kim, O. Erdogan, V. Weinreb, G. L. Butterfoss, B. Kuhlman, C. W. Carter Jr., A minimal TrpRS catalytic domain supports sense/antisense ancestry of class I and II aminoacyl-tRNA synthetases. Mol. Cell 25, 851–862 (2007). [DOI] [PubMed] [Google Scholar]
- 7.S. N. Chandrasekaran, G. G. Yardimci, O. Erdogan, J. Roach, C. W. Carter Jr., Statistical evaluation of the Rodin-Ohno hypothesis: Sense/antisense coding of ancestral class I and II aminoacyl-tRNA synthetases. Mol. Biol. Evol. 30, 1588–1604 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.L. Martinez-Rodriguez, O. Erdogan, M. Jimenez-Rodriguez, K. Gonzalez-Rivera, T. Williams, L. Li, V. Weinreb, M. Collier, S. N. Chandrasekaran, X. Ambroggio, B. Kuhlman, C. W. Carter, Functional class I and II amino acid-activating enzymes can be coded by opposite strands of the same gene. J. Biol. Chem. 291, 23830–23831 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.C. W. Carter Jr., Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu. Rev. Biochem. 62, 715–748 (1993). [DOI] [PubMed] [Google Scholar]
- 10.L. Ribas de Pouplana, P. Schimmel, Two classes of tRNA synthetases suggested by sterically compatible dockings on tRNA acceptor stem. Cell 104, 191–193 (2001). [DOI] [PubMed] [Google Scholar]
- 11.T. Terada, O. Nureki, R. Ishitani, A. Ambrogelly, M. Ibba, D. Soll, S. Yokoyama, Functional convergence of two lysyl-tRNA synthetases with unrelated topologies. Nat. Struct. Biol. 9, 257–262 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Y. Ju, L. Han, B. Chen, Z. Luo, Q. Gu, J. Xu, X.-L. Yang, P. Schimmel, H. Zhou, X-shaped structure of bacterial heterotetrameric tRNA synthetase suggests cryptic prokaryote functions and a rationale for synthetase classifications. Nucleic Acids Res. 49, 10106–10119 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.W. Freist, D. T. Logan, D. H. Gauss, Glycyl-tRNA synthetase. Biol. Chem. Hoppe Seyler 377, 343–356 (1996). [PubMed] [Google Scholar]
- 14.W. Xie, L. A. Nangle, W. Zhang, P. Schimmel, X.-L. Yang, Long-range structural effects of a Charcot-Marie-Tooth disease-causing mutation in human glycyl-tRNA synthetase. Proc. Natl. Acad. Sci. U.S.A. 104, 9976–9981 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D. T. Logan, M. H. Mazauric, D. Kern, D. Moras, Crystal structure of glycyl-tRNA synthetase from Thermus thermophilus. EMBO J. 14, 4156–4167 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D. L. Ostrem, P. Berg, Glycyl transfer ribonucleic acid synthetase from Escherichia coli: Purification, properties, and substrate binding. Biochemistry 13, 1338–1348 (1974). [DOI] [PubMed] [Google Scholar]
- 17.J.-U. Dimas-Torres, A. Rodríguez-Hernández, M. I. Valencia-Sánchez, E. Campos-Chávez, V. Godínez-López, D.-E. Rodríguez-Chamorro, M. Grøtli, C. Fleming, A. Hernández-González, M. Arciniega, A. Torres-Larios, Bacterial glycyl tRNA synthetase offers glimpses of ancestral protein topologies (2021); 10.1101/2021.08.20.456953v6. [DOI]
- 18.S. N. Rodin, A. S. Rodin, On the origin of the genetic code: Signatures of its primordial complementarity in tRNAs and aminoacyl-tRNA synthetases. Heredity (Edinb) 100, 341–355 (2008). [DOI] [PubMed] [Google Scholar]
- 19.C. W. Carter Jr., P. R. Wills, Hierarchical groove discrimination by class I and II aminoacyl-tRNA synthetases reveals a palimpsest of the operational RNA code in the tRNA acceptor-stem bases. Nucleic Acids Res. 46, 9667–9683 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.N. H. Kwon, P. L. Fox, S. Kim, Aminoacyl-tRNA synthetases as therapeutic targets. Nat. Rev. Drug Discov. 18, 629–650 (2019). [DOI] [PubMed] [Google Scholar]
- 21.F. L. Rock, W. Mao, A. Yaremchuk, M. Tukalo, T. Crepin, H. Zhou, Y.-K. Zhang, V. Hernandez, T. Akama, S. J. Baker, J. J. Plattner, L. Shapiro, S. A. Martinis, S. J. Benkovic, S. Cusack, M. R. K. Alley, An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science 316, 1759–1761 (2007). [DOI] [PubMed] [Google Scholar]
- 22.L. F. Silvian, J. Wang, T. A. Steitz, Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin. Science 285, 1074–1077 (1999). [PubMed] [Google Scholar]
- 23.K. Shiba, P. Schimmel, H. Motegi, T. Noda, Human glycyl-tRNA synthetase. Wide divergence of primary structure from bacterial counterpart and species-specific aminoacylation. J. Biol. Chem. 269, 30049–30055 (1994). [PubMed] [Google Scholar]
- 24.F. Juhling, M. Morl, R. K. Hartmann, M. Sprinzl, P. F. Stadler, J. Putz, tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 37, D159–D162 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Y. I. Wolf, L. Aravind, N. V. Grishin, E. V. Koonin, Evolution of aminoacyl-tRNA synthetases–Analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 9, 689–710 (1999). [PubMed] [Google Scholar]
- 26.J. Castropichel, M. T. Garcialopez, F. G. Delasheras, A facile synthesis of ascamycin and related analogues. Tetrahedron 43, 383–389 (1987). [Google Scholar]
- 27.X. Deng, X. Qin, L. Chen, Q. Jia, Y. Zhang, Z. Zhang, D. Lei, G. Ren, Z. Zhou, Z. Wang, Q. Li, W. Xie, Large conformational changes of insertion 3 in human glycyl-tRNA synthetase (hGlyRS) during catalysis. J. Biol. Chem. 291, 5740–5752 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R. Sankaranarayanan, A. C. Dock-Bregeon, P. Romby, J. Caillet, M. Springer, B. Rees, C. Ehresmann, B. Ehresmann, D. Moras, The structure of threonyl-tRNA synthetase-tRNA(Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell 97, 371–381 (1999). [DOI] [PubMed] [Google Scholar]
- 29.V. Biou, A. Yaremchuk, M. Tukalo, S. Cusack, The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser). Science 263, 1404–1410 (1994). [DOI] [PubMed] [Google Scholar]
- 30.N. Moor, O. Kotik-Kogan, D. Tworowski, M. Sukhanova, M. Safro, The crystal structure of the ternary complex of phenylalanyl-tRNA synthetase with tRNAPhe and a phenylalanyl-adenylate analogue reveals a conformational switch of the CCA end. Biochemistry 45, 10572–10583 (2006). [DOI] [PubMed] [Google Scholar]
- 31.M. Varadi, S. Anyango, M. Deshpande, S. Nair, C. Natassia, G. Yordanova, D. Yuan, O. Stroe, G. Wood, A. Laydon, A. Zidek, T. Green, K. Tunyasuvunakool, S. Petersen, J. Jumper, E. Clancy, R. Green, A. Vora, M. Lutfi, M. Figurnov, A. Cowie, N. Hobbs, P. Kohli, G. Kleywegt, E. Birney, D. Hassabis, S. Velankar, AlphaFold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.A. Yaremchuk, I. Kriklivyi, M. Tukalo, S. Cusack, Class I tyrosyl-tRNA synthetase has a class II mode of cognate tRNA recognition. EMBO J. 21, 3829–3840 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.M. Naganuma, S. Sekine, Y. E. Chong, M. Guo, X.-L. Yang, H. Gamper, Y. M. Hou, P. Schimmel, S. Yokoyama, The selective tRNA aminoacylation mechanism based on a single G*U pair. Nature 510, 507–511 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.E. Krissinel, K. Henrick, Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 35.E. A. Wagar, M. J. Giese, B. Yasin, M. Pang, The glycyl-tRNA synthetase of Chlamydia trachomatis. J. Bacteriol. 177, 5179–5185 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.N. Nameki, K. Tamura, H. Asahara, T. Hasegawa, Recognition of tRNA(Gly) by three widely diverged glycyl-tRNA synthetases. J. Mol. Biol. 268, 640–647 (1997). [DOI] [PubMed] [Google Scholar]
- 37.B. Y. Lin, P. P. Chan, T. M. Lowe, tRNAviz: Explore and visualize tRNA sequence features. Nucleic Acids Res. 47, W542–W547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.X. Qin, X. Deng, L. Chen, W. Xie, Crystal structure of the wild-type human GlyRS bound with tRNA(Gly) in a productive conformation. J. Mol. Biol. 428, 3603–3614 (2016). [DOI] [PubMed] [Google Scholar]
- 39.S. Eiler, A. Dock-Bregeon, L. Moulinier, J. C. Thierry, D. Moras, Synthesis of aspartyl-tRNA(Asp) in Escherichia coli–A snapshot of the second step. EMBO J. 18, 6532–6541 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Q. Tian, C. Wang, Y. Liu, W. Xie, Structural basis for recognition of G-1-containing tRNA by histidyl-tRNA synthetase. Nucleic Acids Res. 43, 2980–2990 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.M. I. Valencia-Sanchez, A. Rodriguez-Hernandez, R. Ferreira, H. A. Santamaria-Suarez, M. Arciniega, A. C. Dock-Bregeon, D. Moras, B. Beinsteiner, H. Mertens, D. Svergun, L. G. Brieba, M. Grotli, A. Torres-Larios, Structural insights into the polyphyletic origins of glycyl tRNA synthetases. J. Biol. Chem. 291, 14430–14446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D. Hipps, K. Shiba, B. Henderson, P. Schimmel, Operational RNA code for amino acids: Species-specific aminoacylation of minihelices switched by a single nucleotide. Proc. Natl. Acad. Sci. U.S.A. 92, 5550–5552 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.P. Kumar, A. Bhatnagar, R. Sankaranarayanan, Chiral proofreading during protein biosynthesis and its evolutionary implications. FEBS Lett. 596, 1615–1627 (2022). [DOI] [PubMed] [Google Scholar]
- 44.M. Sprinzl, F. Cramer, Site of aminoacylation of tRNAs from Escherichia coli with respect to the 2′- or 3′-hydroxyl group of the terminal adenosine. Proc. Natl. Acad. Sci. U.S.A. 72, 3049–3053 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.C. W. Carter Jr., P. R. Wills, Class I and II aminoacyl-tRNA synthetase tRNA groove discrimination created the first synthetase-tRNA cognate pairs and was therefore essential to the origin of genetic coding. IUBMB Life 71, 1088–1098 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.L. E. Leiva, A. Pincheira, S. Elgamal, S. D. Kienast, V. Bravo, J. Leufken, D. Gutierrez, S. A. Leidel, M. Ibba, A. Katz, Modulation of Escherichia coli translation by the specific inactivation of tRNA(Gly) under oxidative stress. Front. Genet. 11, 856 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.M. H. Mazauric, H. Roy, D. Kern, tRNA glycylation system from Thermus thermophilus. tRNAGly identity and functional interrelation with the glycylation systems from other phylae. Biochemistry 38, 13094–13105 (1999). [DOI] [PubMed] [Google Scholar]
- 48.A. Yaremchuk, M. Tukalo, M. Grotli, S. Cusack, A succession of substrate induced conformational changes ensures the amino acid specificity of Thermus thermophilus prolyl-tRNA synthetase: Comparison with histidyl-tRNA synthetase. J. Mol. Biol. 309, 989–1002 (2001). [DOI] [PubMed] [Google Scholar]
- 49.B. Delagoutte, D. Moras, J. Cavarelli, tRNA aminoacylation by arginyl-tRNA synthetase: Induced conformations during substrates binding. EMBO J. 19, 5599–5610 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.S. Fukai, O. Nureki, S. Sekine, A. Shimada, J. Tao, D. G. Vassylyev, S. Yokoyama, Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNA(Val) and valyl-tRNA synthetase. Cell 103, 793–803 (2000). [DOI] [PubMed] [Google Scholar]
- 51.K. Nakanishi, Y. Ogiso, T. Nakama, S. Fukai, O. Nureki, Structural basis for anticodon recognition by methionyl-tRNA synthetase. Nat. Struct. Mol. Biol. 12, 931–932 (2005). [DOI] [PubMed] [Google Scholar]
- 52.A. Palencia, T. Crepin, M. T. Vu, T. L. Lincecum Jr., S. A. Martinis, S. Cusack, Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase. Nat. Struct. Mol. Biol. 19, 677–684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.M. A. Rould, J. J. Perona, D. Söll, T. A. Steitz, Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science 246, 1135–1142 (1989). [DOI] [PubMed] [Google Scholar]
- 54.S. Hauenstein, C. M. Zhang, Y. M. Hou, J. J. Perona, Shape-selective RNA recognition by cysteinyl-tRNA synthetase. Nat. Struct. Mol. Biol. 11, 1134–1141 (2004). [DOI] [PubMed] [Google Scholar]
- 55.N. Shen, L. Guo, B. Yang, Y. Jin, J. Ding, Structure of human tryptophanyl-tRNA synthetase in complex with tRNATrp reveals the molecular basis of tRNA recognition and specificity. Nucleic Acids Res. 34, 3246–3258 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.B. Kuhle, M. Hirschi, L. K. Doerfel, G. C. Lander, P. Schimmel, Structural basis for shape-selective recognition and aminoacylation of a D-armless human mitochondrial tRNA. Nat. Commun. 13, 5100 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.H. Zhou, L. Sun, X.-L. Yang, P. Schimmel, ATP-directed capture of bioactive herbal-based medicine on human tRNA synthetase. Nature 494, 121–124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.J. Guo, B. Chen, Y. Yu, B. Cheng, Y. Cheng, Y. Ju, Q. Gu, J. Xu, H. Zhou, Discovery of novel tRNA-amino acid dual-site inhibitors against threonyl-tRNA synthetase by fragment-based target hopping. Eur. J. Med. Chem. 187, 111941 (2020). [DOI] [PubMed] [Google Scholar]
- 59.B. Chen, S. Luo, S. Zhang, Y. Ju, Q. Gu, J. Xu, X.-L. Yang, H. Zhou, Inhibitory mechanism of reveromycin A at the tRNA binding site of a class I synthetase. Nat. Commun. 12, 1616 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Z. Cai, B. Chen, Y. Yu, J. Guo, Z. Luo, B. Cheng, J. Xu, Q. Gu, H. Zhou, Design, synthesis, and proof-of-concept of triple-site inhibitors against aminoacyl-tRNA synthetases. J. Med. Chem. 65, 5800–5820 (2022). [DOI] [PubMed] [Google Scholar]
- 61.Y. Manickam, N. Malhotra, S. Mishra, P. Babbar, A. Dusane, B. Laleu, V. Bellini, M. A. Hakimi, A. Bougdour, A. Sharma, Double drugging of prolyl-tRNA synthetase provides a new paradigm for anti-infective drug development. PLOS Pathog. 18, e1010363 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.J. Zhou, Z. Huang, L. Zheng, Z. Hei, Z. Wang, B. Yu, L. Jiang, J. Wang, P. Fang, Inhibition of plasmodium falciparum lysyl-tRNA synthetase via an anaplastic lymphoma kinase inhibitor. Nucleic Acids Res. 48, 11566–11576 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Y. Nakamura, T. Gojobori, T. Ikemura, Codon usage tabulated from international DNA sequence databases: Status for the year 2000. Nucleic Acids Res. 28, 292 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.P. P. Chan, T. M. Lowe, GtRNAdb: A database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 37, D93–D97 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.C. Kao, M. Zheng, S. Rudisser, A simple and efficient method to reduce nontemplated nucleotide addition at the 3′ terminus of RNAs transcribed by T7 RNA polymerase. RNA 5, 1268–1272 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.F. Yu, Q. Wang, M. Li, H. Zhou, K. Liu, K. Zhang, Z. Wang, Q. Xu, C. Xu, Q. Pan, J. He, Aquarium: An automatic data-processing and experiment information management system for biological macromolecular crystallography beamlines. J. Appl. Cryst. 52, 472–477 (2019). [Google Scholar]
- 67.C. Vonrhein, C. Flensburg, P. Keller, A. Sharff, O. Smart, W. Paciorek, T. Womack, G. Bricogne, Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.A. Vagin, A. Teplyakov, Molecular replacement with MOLREP. Acta Crystallogr. D 66, 22–25 (2010). [DOI] [PubMed] [Google Scholar]
- 69.P. Emsley, B. Lohkamp, W. G. Scott, K. Cowtan, Features and development of coot. Acta Crystallogr. D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.G. N. Murshudov, A. A. Vagin, E. J. Dodson, Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 71.V. B. Chen, W. B. Arendall 3rd, J. J. Headd, D. A. Keedy, R. M. Immormino, G. J. Kapral, L. W. Murray, J. S. Richardson, D. C. Richardson, MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.F. Sievers, A. Wilm, D. Dineen, T. J. Gibson, K. Karplus, W. Li, R. Lopez, H. McWilliam, M. Remmert, J. Soding, J. D. Thompson, D. G. Higgins, Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.A. M. Waterhouse, J. B. Procter, D. M. Martin, M. Clamp, G. J. Barton, Jalview Version 2–A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.R. Cain, R. Salimraj, A. S. Punekar, D. Bellini, C. W. G. Fishwick, L. Czaplewski, D. J. Scott, G. Harris, C. G. Dowson, A. J. Lloyd, D. I. Roper, Structure-guided enhancement of selectivity of chemical probe inhibitors targeting bacterial seryl-tRNA synthetase. J. Med. Chem. 62, 9703–9717 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Method
Scheme S1
Figs. S1 to S9
Table S1
References