Abstract
Although emerging evidence shows that gut microbiota-mediated metabolic changes regulate intestinal pathogen invasions, little is known about whether and how gut microbiota-mediated metabolites affect pathogen infection in the distal organs. In this study, untargeted metabolomics was performed to identify the metabolic changes in a subacute ruminal acidosis (SARA)-associated mastitis model, a mastitis model with increased susceptibility to Staphylococcus aureus (S. aureus). The results showed that cows with SARA had reduced cholic acid (CA) and deoxycholic acid (DCA) levels compared to healthy cows. Treatment of mice with DCA, but not CA, alleviated S. aureus-induced mastitis by improving inflammation and the blood-milk barrier integrity in mice. DCA inhibited the activation of NF-κB and NLRP3 signatures caused by S. aureus in the mouse mammary epithelial cells, which was involved in the activation of TGR5. DCA-mediated TGR5 activation inhibited the NF-κB and NLRP3 pathways and mastitis caused by S. aureus via activating cAMP and PKA. Moreover, gut-dysbiotic mice had impaired TGR5 activation and aggravated S. aureus-induced mastitis, while restoring TGR5 activation by spore-forming bacteria reversed these changes. Furthermore, supplementation of mice with secondary bile acids producer Clostridium scindens also activated TGR5 and alleviated S. aureus-induced mastitis in mice. These results suggest that impaired secondary bile acid production by gut dysbiosis facilitates the development of S. aureus-induced mastitis and highlight a potential strategy for the intervention of distal infection by regulating gut microbial metabolism.
Subject terms: Bacteriology, Pathogens
Introduction
The gut microbiota has been proven to play an important role in maintaining host physiological homeostasis and regulating disease outcomes including infectious disease1,2. Mounting evidence reveals that gut microbiota-mediated metabolites are one of the most common mechanisms of host-microbiota interaction and thus participate in the pathogenesis of pathogen invasion3. It has been shown that gut dysbiosis-derived metabolites can promote pathogen colonization in multiple manners. For example, gut microbiota-derived 1,2-propanediol regulated the metabolic pathways and increased the virulence expression of Citrobacter rodentium, enabling metabolic adaptation and pathogenicity4. Microbiota-liberated host sugars, such as sialic acid and fucose, promoted the postantibiotic expansion of Salmonella typhimurium and Clostridium difficile by serving as preferential energy sources5. Tovaglieri et al., also found that gut microbiota-derived metabolites including 4-methyl benzoic acid, 3,4-dimethylbenzoic acid, hexanoic acid and heptanoic acid aggravated enterohemorrhagic Escherichia coli-induced intestinal barrier injury by promoting the expression of flagellin6. However, it has also been shown that commensal microbiota-derived beneficial components limit pathogen invasion and subsequently improve outcomes of disease. Sun X et al. showed that commensal anaerobic microbiota-produced deoxycholic acid (DCA), a secondary bile acid derived from cholic acid (CA), reduces Campylobacter jejuni (C. jejuni)-induced colitis, while the depletion of secondary bile acids (BAs)-producing bacteria by clindamycin exacerbates this colitis7. In addition, prior gut infection increases the resistance of the microbiota to subsequent infection by increasing taurine, a metabolite derived from BA metabolism, which is then converted to sulfide and inhibits pathogen respiration8. Another well-known gut microbiota-derived metabolite that regulates infectious disease is short-chain fatty acids (SCFAs). Commensal Bacteroides-produced propionate increases intestinal colonization resistance to Salmonella by disrupting intracellular pH homeostasis9. Butyrate protects mice against Clostridium difficile-induced colitis by improving the intestinal barrier and limiting bacterial translocation by stabilizing the hypoxia-inducible factor 1α10. The regulation of gut microbiota structure by depleting commensal Bacteroides vulgatus facilitated intestinal Vibrio cholerae (V. cholerae) pathogenesis in mice by reducing V. cholerae growth-inhibitory metabolites including SCFAs but elevating metabolites that enhance V. cholerae proliferation including N-acetylglucosamine and gluconolactone11. These findings suggest that different gut microbiota-mediated metabolic profiles endow hosts with distinct susceptibilities to pathogens and subsequent outcomes of intestinal diseases. However, little is known about the effects of microbiota-derived metabolites on the pathogenesis of distal pathogen infections.
Staphylococcus aureus (S. aureus) is a formidable bacterium that can induce severe diseases and even death in humans and animals12. Mounting evidence has reported that S. aureus could be resistant to most antibiotics, which enables the appearance of new clones, including methicillin-resistant S. aureus (MRSA), which serves as a major cause of community-associated infections and lacks safe and effective prevention and control strategies13. S. aureus-associated mastitis is one of the most common diseases in lactating women and animals, which increases the risk of breast cancer and causes huge economic losses14,15. Recently, mounting evidence has indicated that host factors, such as the gut microbiota, participate in the pathogenesis of pathogen-induced mastitis. For example, it has been shown that gut-dysbiotic mice have increased inflammatory responses in the mammary gland and develop exacerbated mastitis symptoms upon bacterial infection16,17. Moreover, modification of the gut microbiota and metabolism by fecal microbiota transplantation, probiotics, and direct metabolite supplementation alleviate pathogen-induced mastitis16,17. On dairy farms, S. aureus-associated mastitis has been associated with gastrointestinal diseases, such as subacute ruminal acidosis (SARA)18. A previous study indicated that cows experiencing SARA and challenged intramammarily with lipopolysaccharide (LPS) experienced stronger metabolic disturbances and altered innate immunity19. Our previous results also showed that SARA cows had increased susceptibility to S. aureus-induced mastitis20. These suggest the increased risk of mastitis in cows with SARA, however, the underlying mechanism remains unknown. Previous studies have shown that different dietary patterns affect the microbial composition and metabolic profile in the mammary gland21,22, leading to increased susceptibility to mammary diseases22. Consumption of the Mediterranean diet increased beneficial microbiota-associated metabolites, including BAs and tryptophan-derived aryl hydrocarbon receptor (AhR) ligands, compared with the Western Diet21. These metabolites have been linked to altered susceptibility to bacterial infections and subsequent inflammatory responses7,17. However, whether microbiota-associated metabolic changes in the mammary gland of SARA cows are responsible for the increased susceptibility to S. aureus is poorly understood along with the underlying mechanism.
In this study, using untargeted metabolomics, we showed that SARA cows had distinct metabolic profiles in their milk compared with heathy cows, especially reduced CA and DCA levels. Consumption of DCA, but not CA, ameliorated S. aureus-induced mastitis in mice. The underlying mechanism was involved in the Takeda G protein-coupled receptor 5 (TGR5)-mediated cyclic adenosine monophosphate (cAMP)-protein kinase A (PKA) pathways, which inhibited the activation of proinflammatory NF-κB and NLRP3 in the mammary gland. Gut-dysbiotic mice caused by vancomycin had reduced TGR5 activation in the mammary gland, leading increased susceptibility to S. aureus-induced mastitis. Conversely, treatment of mice with spore-forming bacteria (SFB) restored TGR5 activation and alleviated S. aureus-induced mastitis. Moreover, commensal Clostridium scindens (C. scindens) with the capacity of DCA producing also attenuated mastitis caused by S. aureus in mice. These results indicate that gut microbiota-mediated DCA contributes to protecting against pathogen-induced mastitis, indicating that regulating gut microbiota and its metabolism can be a potential strategy for the intervention of distal pathogen infection.
Results
High concentrate diet-associated SARA reduces BA levels in the mammary glands of cows
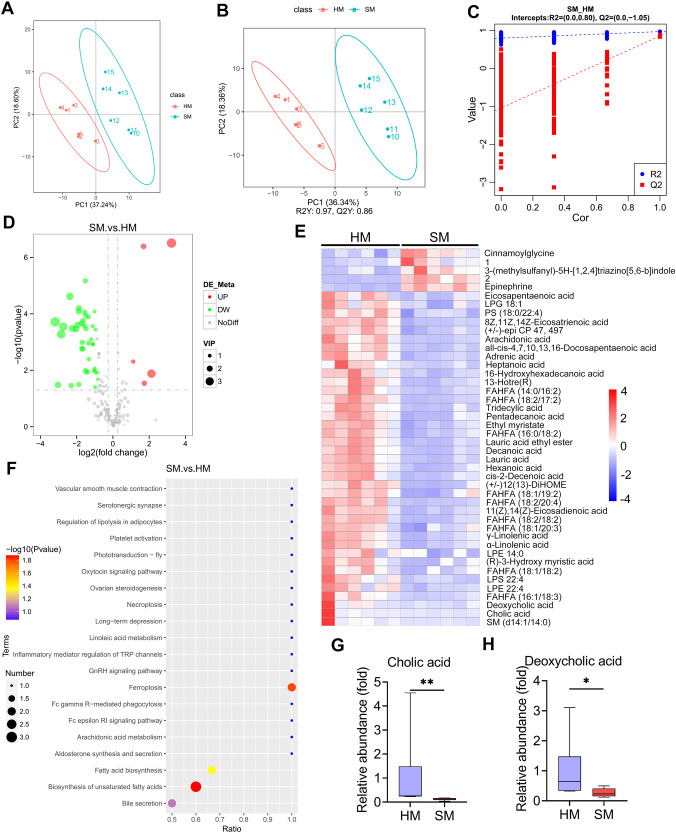
Our previous results showed that SARA cows had increased susceptibility to S. aureus-induced mastitis20. To investigate the underlying metabolic changes, untargeted metabolomics was performed on milk samples from healthy and SARA cows. To acquire stable and accurate metabolome results, data quality control (QC) was performed through the Pearson correlation analysis. The Pearson correlation of the ruminal QC samples was high, suggesting reliable data quality (Supplementary Fig. 1A). Moreover, the QC samples clustered tightly together in the principal component analysis (PCA) score plots (Supplementary Fig. 1B, C), which further confirmed quality of the data. A total of 213 annotated metabolites were identified in all the milk samples by using the HMDB annotation, Kyoto Encyclopedia of Genes and Genomes (KEGG), and LIPID MAPS (Supplementary Fig. 2A–C, Supplementary Table 1). PCA showed that the milk samples from the cows with SARA (SM) were significantly separated from the healthy cows (HM) (PC1 was 37.24% and PC2 was 18.60%, Fig. 1A). Moreover, distinct clusters from the SM compared with the HM were confirmed by the partial least squares discrimination analysis (PLS-DA) score plots (p < 0.001, Fig. 1B). The stability and reliability of the PLS-DA were confirmed by 7-fold cross-validation (R2Y:0.97, Q2Y:0.86, Fig. 1B). Furthermore, a model containing metabolic information by fitting the PLS-DA through 200 times random permutation testing was performed to confirm the ability of the PLS-DA to correctly classify new samples. The results demonstrated that the model was reliable and not overfitting, as shown by the intercepts of the goodness-of-fit (R2) being greater than the goodness-of-prediction (Q2) and the intercept of Q2 less than zero (Fig. 1C).
Fig. 1. The metabolic profiles of the milk samples from the healthy and SARA cows.
A PCA score plots for milk samples (n = 6). B PLS-DA score plots for the milk samples (n = 6). C Cross-validation plot with a permutation test repeated 200 times. The intercepts of R2 = (0.0, 0.80) and Q2 = (0.0,–1.05), indicate that the PLS-DA model is not overfitting. D Volcano plots indicate the results of the pairwise comparisons of metabolites in the healthy and SARA cows. The vertical and horizontal dashed lines indicate the threshold for the twofold abundance difference and the p = 0.05 threshold, respectively. Student’s t-test was performed for the comparison. The significant metabolites are presented in red (upregulated) or green (downregulated). E The hierarchical cluster analysis of the different milk metabolites between healthy and SARA cows. 1. 1-methyl-N-(3-methyl-5-cinnolinyl)-1H-imidazole-4-sulfonamide; 2. N-(1,1-Dioxotetrahydro-1H-1λ6-thiophen-3-yl)-4-methoxybenzamide. F The pathway enrichment analysis of significantly elevated metabolites in the SARA samples according to the KEGG pathway. The relative levels of cholic acid (G) and deoxycholic acid (H) in the healthy and SARA samples were determined. Data are presented as boxplots, with the center line representing the median, the boundary of the whiskers representing the minimum and maximum values of the dataset, and the boundary of the box representing the 25th and 75th percentile of the dataset (G and H). Mann-Whitney U test was performed (G, H). *p < 0.05, **p < 0.01. PLS-DA, partial least squares discrimination analysis; VIP, variable importance in the projection.
We identified 44 significantly differential metabolites in the SM compared with the HM (5 upregulated and 39 downregulated, Supplementary Table 1, 2, and 3) by considering the variable importance in the projection (VIP), fold change (FC), and P-value. The global distribution of different metabolites was shown by volcano plots, which indicated distinct metabolite levels between the SM and HM (Fig. 1D). Moreover, the metabolite levels in the SM were significantly different from those in the HM through differential metabolite cluster analysis by using a heatmap (Fig. 1E). KEGG pathway enrichment analysis indicated that the pathways enriched in the SM were mainly bile secretion, biosynthesis of unsaturated fatty acids, fatty acid biosynthesis and ferroptosis (Fig. 1F). Considering the role of gut microbiota in the pathogenesis of pathogen-induced mastitis16,17 and microbiota-mediated BA metabolism in regulating gut pathogen invasion7,8, we further focused on bile metabolism and showed that the secondary bile acid DCA and primary bile acid CA were significantly depleted in the SM samples compared with those in the HM group (Fig. 1G, H). Notably, our previously published rumen microbiome analysis of these SARA cows indicates that they have fewer Clostridia which might include those capable of making DCA from CA but that genus-level data is insufficient to know for sure20. Collectively, these results indicated that SARA cows had altered metabolic profiles in their milk, especially reduced BAs, which may thus promote S. aureus-induced mastitis.
The secondary bile acid DCA, but not the primary bile acid CA, alleviates S. aureus-induced mastitis in mice
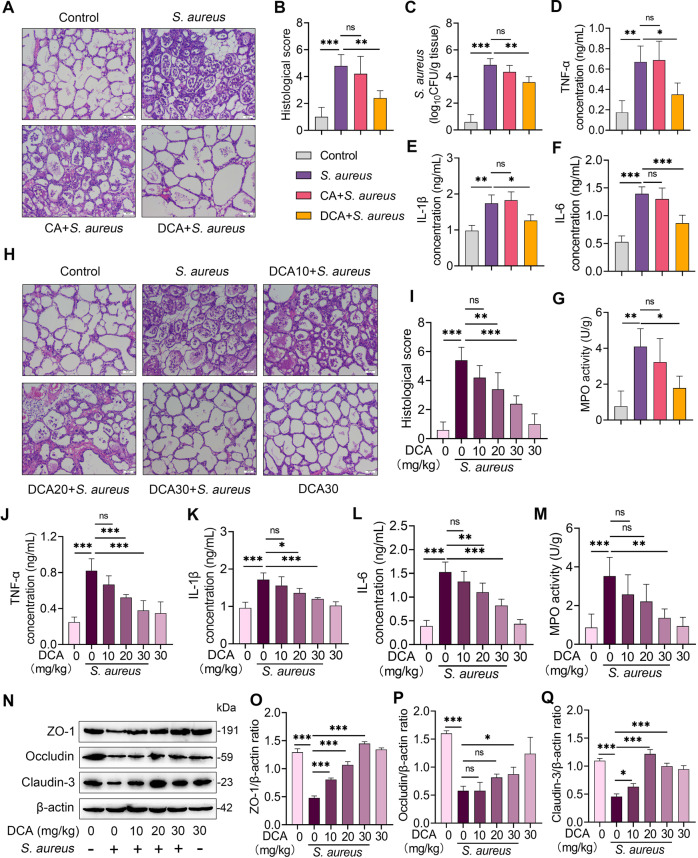
We next investigated the role of BAs in mastitis using a S. aureus-induced mouse mastitis model. The results showed that S. aureus treatment induced significant mastitis traits by increasing mammary injury and inflammatory infiltration and responses compared with the control mice (Fig. 2A–G). Pretreatment with DCA intraperitoneally, but not CA, alleviated S. aureus-induced mastitis, as evidenced by DCA improving mammary injury (Fig. 2A, B), mammary S. aureus burden (Fig. 2C), proinflammatory markers including TNF-α, IL-1β and IL-6 and MPO activity (Fig. 2D–G) compared with that of the S. aureus group. To confirm the protective effects of DCA on S. aureus-induced mastitis, mice were pretreated with different doses of DCA and the results found that DCA ameliorated S. aureus-induced mammary damage (Fig. 2H, I) and inflammatory parameters (Fig. 2J–M) in a dose-dependent manner. Increased mammary immune cell infiltration and inflammatory responses were associated with impaired blood-milk barrier integrity16,17,20. Indeed, DCA pretreatment improved the mammary barrier disruption resulting from S. aureus, as evidenced by the restoration of the tight-junction (TJ) proteins ZO-1, Occludin and Claudin-3 compared with those in the S. aureus-treated group (Fig. 2N–Q). These results suggest that the secondary bile acid DCA, but not the primary bile acid CA, improves S. aureus-induced mastitis in mice.
Fig. 2. DCA but not CA alleviates S. aureus-induced mastitis in mice.
Mice were pretreated with 30 mg/kg of CA or DCA for 2 h intraperitoneally followed by S. aureus-induced mastitis. Twenty-four hours after S. aureus infection, the mammary tissues were collected for determination. A Representative H&E-stained sections of the mammary gland from the indicated groups (scale bar, 50 μm). B Histological score for the mammary gland based on H&E-stained sections (n = 6). C Mammary S. aureus load (n = 6). Inflammatory parameters of the mammary gland from different groups, including TNF-α (D), IL-1β (E), and IL-6 (F) concentrations and MPO activity (G), were performed (n = 6). H–Q Mice were pretreated with different dose of DCA (10, 20, and 30 mg/kg) for 2 h intraperitoneally and then stimulated with S. aureus for next 24 h. H Representative mammary H&E-stained sections from the indicated mice (scale bar, 50 μm). I Histological score of the mammary gland from different treatment groups (n = 6). The inflammatory parameters of the mammary gland from different treatment groups, TNF-α (J), IL-1β (K), and IL-6 (L) concentrations and MPO activity (M), were measured (n = 6). N Represented images of ZO-1, Occludin, and Claudin-3 in the mammary glands from the indicated mice. The intensities of ZO-1, Occludin, and Claudin-3 were determined (O–Q). Data are expressed as the means ± SD (B-G, I–M, and O–Q) and one-way ANOVA was performed, followed by Tukey’s test (B–G, I-M, and O–Q). *p < 0.05, **p < 0.01, ***p < 0.001 indicate significance. ns no significance.
DCA attenuates S. aureus-induced mastitis through the TGR5-cAMP-PKA-NF-κB/NLRP3 pathways
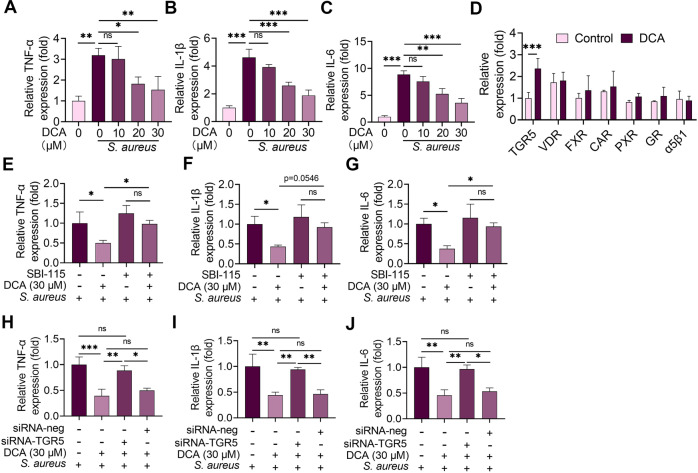
It is well known that BAs have different antimicrobial abilities and the biofilm of S. aureus contributes to its infection7,23. We, therefore, investigated whether DCA and CA regulated the growth and biofilm formation of S. aureus. The results showed that DCA (636.82 μM reduced the OD600 by almost 50%) had a better capacity to limit S. aureus growth in vitro than CA (1835.67 μM had few inhibitory effects) (Supplementary Fig. 3A–B). In addition, no significant influences on the biofilm formation of S. aureus were observed in less than 60 μM of DCA and CA treatment groups (Supplementary Fig. 3C, D). Consistent with the results found in other cell types24, a dose of DCA that did not inhibit the growth of S. aureus reduced S. aureus-induced TNF-α, IL-1β and IL-6 mRNA in the mouse mammary epithelial cells (MMECs) (Fig. 3A–C), suggesting that DCA-alleviated S. aureus-induced inflammation is not entirely dependent on the inhibition of S. aureus growth. Secondary BAs were reported by multiple receptors25–27, including the vitamin D receptor (VDR), Takeda G protein-coupled receptor 5 (TGR5), α5β1, glucocorticoid receptor (GR), constitutive androstane receptor (CAR), farnesoid X receptor (FXR), and pregnane X receptor (PXR). We next found that only TGR5 was markedly increased in the DCA-treated MMECs compared with the control group (Fig. 3D). To confirm the role of TGR5 activation in the effects of DCA, we blocked TGR5 activation with the specific inhibitor SBI-11528 or siRNA-TGR5. Both SBI-115 and siRNA-TGR5 reversed the protective effects of DCA on S. aureus-induced proinflammatory cytokine production in the MMECs (Fig. 3E–J). Although CA has also been reported to activate TGR5, we found that 30 μM of the CA treatment had no significant influence on S. aureus-induced proinflammatory cytokine production in the MMECs (Supplementary Fig. 4A–C). These results indicate that TGR5 activation is responsible for the protective effects of DCA on S. aureus-induced mastitis.
Fig. 3. DCA alleviates S. aureus-induced inflammation in MMECs by activating TGR5.
Cells were pretreated with DCA (10, 20 and 30 μM) for 2 h followed by S. aureus treatment for next 24 h, and the relative mRNA levels of proinflammatory TNF-α (A), IL-1β (B), and IL-6 (C) from the indicated group were detected by qPCR. D Cells were treated with DCA (30 μM) for 24 h and the relative mRNA levels of TGR5, VDR, FXR, CAR, PXR, GR and α5β1 were determined using qPCR. Cells were pre-treated with SBI-115 for 2 h and then treated with DCA (30 μM) for an additional 2 h followed by S. aureus treatment for the next 24 h. Relative mRNA levels of proinflammatory TNF-α (E), IL-1β (F), and IL-6 (G) from the different treatment groups were detected by qPCR. H–J Cells were pretreated with siRNA-TGR5 or siRNA-neg for 48 h followed by DCA (30 μM) and S. aureus treatment as mentioned above, and the relative mRNA levels were detected by qPCR. Data are expressed as the means ± SD (A–J) and one-way ANOVA followed by Tukey’s test (A–C and E–J) and Student’s t-test was performed (D). *p < 0.05, **p < 0.01, ***p < 0.001 indicate significance. ns no significance.
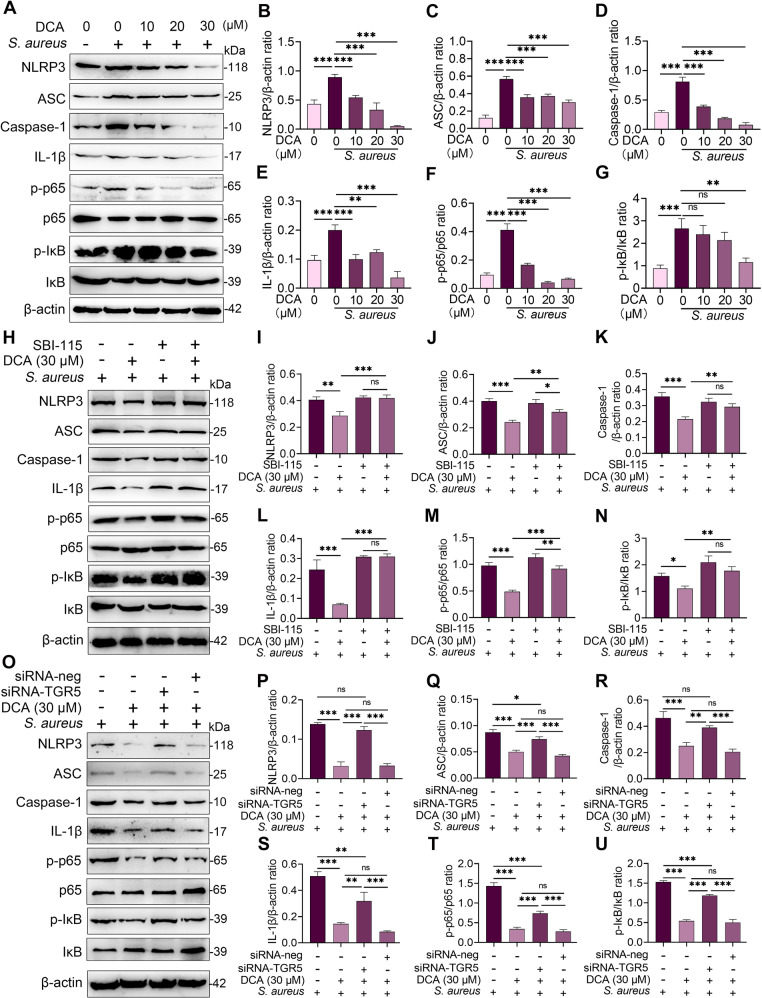
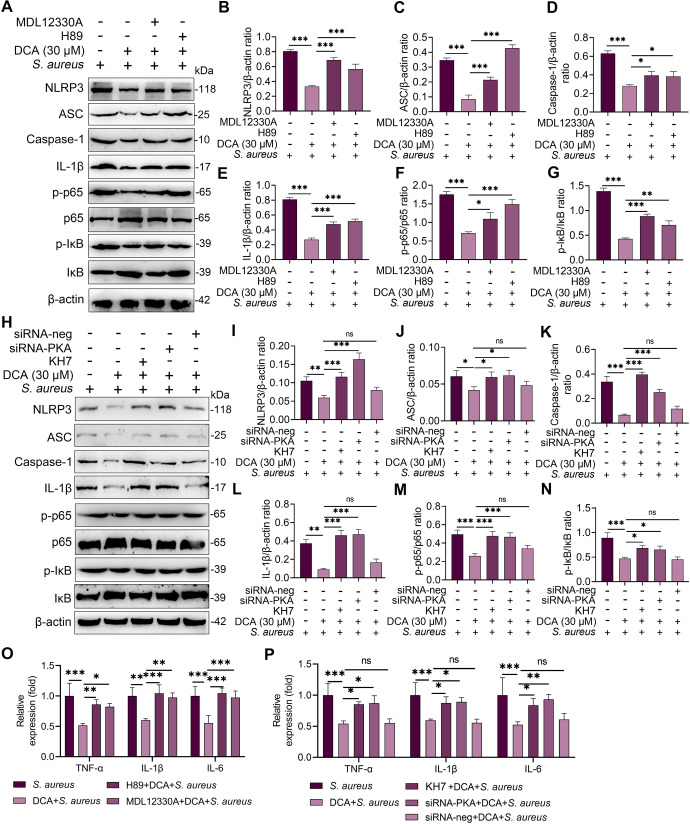
Proinflammatory cytokine secretion was attributed to the activation of proinflammatory pathways, such as NF-κB and NLRP329. As expected, DCA treatment inhibited the activation of the NF-κB and NLRP3 pathways in the S. aureus-treated MMECs in a dose-dependent manner (Fig. 4A–G). We further investigated whether these effects were mediated by TGR5 activation. The results showed that the SBI-115 and siRNA-TGR5 treatments reversed the reduction in p-p65, p-IκB, NLRP3, and IL-1β resulting from DCA treatment in the S. aureus-treated MMECs (Fig. 4H–U), leading to TGR5 activation inhibiting S. aureus-induced activation of the NF-κB and NLRP3 pathways. Previous studies have shown that TGR5 binding with ligands contributes to cAMP production, leading to the activation of PKA24,26. We, therefore, investigated whether the cAMP-PKA pathway is involved in TGR5-mediated NF-κB and NLRP3 inhibition by using the adenylate cyclase inhibitor MDL12330A and PKA inhibitor H8930,31. Pretreatment with both MDL12330A and H89 reversed the protective effect of TGR5 activation on NF-κB and NLRP3 activation (Fig. 5A–G). To confirm these results, another cAMP inhibitor KH7, and siRNA-PKA were also performed. Consistently, the KH7 and siRNA-PKA treatments reversed the decrease in NF-κB and NLRP3 activation induced by DCA in S. aureus-treated MMECs (Fig. 5H–N). Moreover, the inhibition of cAMP and PKA by inhibitor and siRNA reversed the decrease in TNF-α, IL-1β, and IL-6 expression caused by DCA in S. aureus-treated MMECs (Fig. 5O, P). Collectively, these results indicated that DCA alleviated S. aureus-induced mastitis through the TGR5-cAMP-PKA-NF-κB/NLRP3 axis.
Fig. 4. DCA inhibits S. aureus-induced NF-κB and NLRP3 activation by activating TGR5 in MMECs.
A–G Cells were pretreated with DCA (10, 20, and 30 μM) for 2 h followed by S. aureus treatment for the next 24 h and the protein levels of the NF-κB and NLRP3 pathways from the indicated groups were determined by western blotting (A). The relative intensities of p-p65, p-IκB, NLRP3, ASC, and IL-1β were determined (B–G). H–N Cells were pretreated with SBI-115 for 2 h and then treated with DCA (30 μM) for an additional 2 h followed by S. aureus treatment for the next 24 h. The protein levels of the NF-κB and NLRP3 pathways were determined by western blotting (H), and the relative intensities of p-p65, p-IκB, NLRP3, ASC, and IL-1β were determined (I–N). O–U. Cells were pretreated with siRNA-TGR5 or siRNA-neg for 48 h followed by DCA (30 μM) and S. aureus treatment as mentioned above, and the protein levels of the NF-κB and NLRP3 pathways were determined by western blotting. Data are expressed as the means ± SD (B–G, I–N, and P–U) and one-way ANOVA was performed, followed by Tukey’s test (B–G, I–N, and P–U). *p < 0.05, **p < 0.01, ***p < 0.001 indicate significance. ns, no significance.
Fig. 5. DCA alleviates S. aureus-induced inflammation in MMECs through the cAMP-PKA- NF-Κb/NLRP3 pathways.
A–G and O. Cells were pretreated with H89 (30 μM) and MDL12330A (10 μM) for 2 h and then treated with DCA (30 μM) for an additional 2 h followed by S. aureus treatment for the next 24 h. The protein levels of NF-κB and NLRP3 pathways from the indicated groups were determined by western blotting (A). The relative intensities of p-p65, p-IκB, NLRP3, ASC, and IL-1β were determined (B–G). The relative mRNA levels of proinflammatory gene from the indicated group were detected by qPCR (O). H–N and P. Cells were pretreated with siRNA for 48 h and then treated with DCA (30 μM) and KH7 (10 μM) for an additional 2 h followed by S. aureus treatment for the next 24 h. The protein levels of NF-κB and NLRP3 pathways from the indicated groups were determined by western blotting (H) and The relative intensities of p-p65, p-IκB, NLRP3, ASC and IL-1β were determined (I–N). The relative mRNA levels of proinflammatory gene from the indicated group were detected by qPCR (P). Data are expressed as the means ± SD (B–G, I–N and O, P) and one-way ANOVA was performed, followed by Tukey’s test (B–G, I–N, and O, P). *p < 0.05, **p < 0.01, ***p < 0.001 indicate significance.
Gut microbiota-produced DCA mediates TGR5 activation and the development of S. aureus-induced mastitis in mice
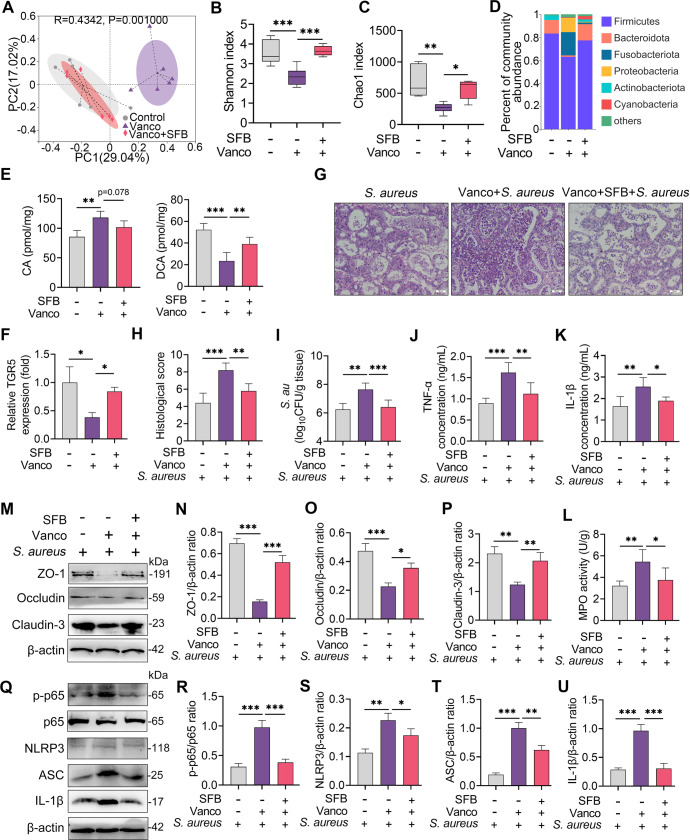
Gut microbiota plays an important role in the translation of primary into secondary BAs32,33, such as CA converting into DCA by gut microbiota-mediated enzymes. Together with the different effects of DCA and CA on S. aureus-induced mastitis, we thus investigated the role of the gut microbiota in the pathogenesis of mastitis caused by S. aureus. Vancomycin-sensitive anaerobic microbes are known as the predominant taxa for secondary BA production34,35. We then modified the gut microbiota by depleting gut anaerobic microbes using vancomycin and compensating with anaerobic SFB. Principal coordinate analysis based on the Bray-Curtis distance showed that the gut microbiota structure of the vancomycin treatment group was markedly separated from that of the control or SFB treatment groups (R = 0.4342, P = 0.001) (Fig. 6A). Venn diagram analysis found that the vancomycin-treated mice had reduced species in the gut microbiota compared with the control group but reversed after the SFB compensation (Supplementary Fig. 5A). Alpha diversity analysis, including Shannon, Chao1, ACE and Simpson indices, showed that the vancomycin-treated group had reduced richness and diversity of the gut microbiota compared with that of the control and SFB compensation groups (Fig. 6B, C and Supplementary Fig. 5B, C). At the phylum level, the vancomycin treatment reduced the abundance of the obligate anaerobic Firmicutes and Bacteroidota, but enriched the abundance of Fusobacteriota and Proteobacteria, while these changes were reversed after the SFB transplantation (Fig. 6D). At the genus level, the vancomycin-treated mice also showed distinct microbial compositions from those of the control or SFB compensation groups (Supplementary Fig. 5D). Linear discriminant analysis effect size (LEfSe) showed that Clostridium and Fusobacterium were significantly changed among different groups (Supplementary Fig. 5E). We next showed that the vancomycin treatment reduced DCA but increased CA levels in the mammary gland compared with the control and SFB treatment groups (Fig. 6E). Consistently, the vancomycin treatment reduced the expression of TGR5 in the mammary gland compared with the control group, but was restored after the SFB compensation (Fig. 6F). Moreover, the vancomycin-treated mice developed more severe mastitis than the control and SFB compensation mice upon S. aureus infection, as shown by aggravated mammary damage, bacterial burden and proinflammatory parameters (Fig. 6G–L). However, the SFB transplantation from the control mice alleviated S. aureus-induced mastitis compared with that of the vancomycin-treated mice (Fig. 6G–L). Likewise, the vancomycin-treated mice had reduced TJ expressions and increased NF-κB and NLRP3 activation compared with the control group, while the SFB treatment restored the barrier function and limited NF-κB and NLRP3 activation caused by S. aureus (Fig. 6M–U). To confirm the protective role of gut microbiota-metabolized DCA in S. aureus-induced mastitis, CA and DCA were performed orally in conventional and gut-dysbiotic mice. Interestingly, oral gavage with CA alleviated S. aureus-induced mastitis in the conventional mice but reversed after the depletion of commensal microbiota by an antibiotic cocktail, accompanied by changed DCA levels in the mammary gland (Supplementary Fig. 6A–F). Moreover, treatment with DCA in gut-dysbiotic mice still alleviated S. aureus-induced mastitis (Supplementary Fig. 6A–F). These results indicate that gut microbiota-produced DCA mediates the activation of TGR5 and the development of S. aureus-induced mastitis in mice.
Fig. 6. Gut dysbiosis impairs TGR5 activation and aggravates S. aureus-induced mastitis in mice.
Mice were treated with vancomycin and supplemented with SFB, followed by S. aureus-induced mastitis. A Principal coordinate analysis score plots indicate the separation of intestinal microbiota structure (R = 0.4342, P = 0.001) in the vancomycin-treated mice from that of control or SFB-transplanted mice based on Bray-Curtis distance (n = 6). B-C The Shannon (B) and Chao1 (C) indices showed that vancomycin reduced the gut microbial diversity and richness but was restored by SFB transplantation (n = 6). D The gut microbial composition at the phylum level from different treatment groups. E Mammary DCA and CA levels from the indicated groups were detected (n = 6). F The relative expression of TGR5 mRNA from different treatment groups (n = 6). G Representative images of H&E-stained sections from different groups (scale bar, 50 μm). H The histological score of the mammary gland from different groups (n = 6). I Mammary S. aureus load. The proinflammatory markers of TNF-α (J), IL-1β (K), and MPO activity (L) were measured (n = 6). M Representative images of ZO-1, Occludin and Claudin-3 of the mammary gland from the indicated mice. The intensities of ZO-1, Occludin and Claudin-3 were determined (N–P). Q The protein levels of the NF-κB and NLRP3 pathways in the mammary gland were detected using western blotting. The relative intensities of p-p65, NLRP3, ASC, and IL-1β in the mammary gland were detected (R–U). Data are expressed as boxplots, with the center line representing the median, the boundary of the whiskers representing the minimum and maximum values of the dataset, and the boundary of the box representing the 25th and 75th percentile of the dataset (B, C) or means ± SD (E, F, H-L, N–P, and R–U) and one-way ANOVA was performed, followed by Tukey’s test (E, F, H–L, N–P and R–U). *p < 0.05, **p < 0.01, ***p < 0.001 indicate significance.
C. scindens with the capacity of secondary BA production alleviates S. aureus-induced mastitis in mice
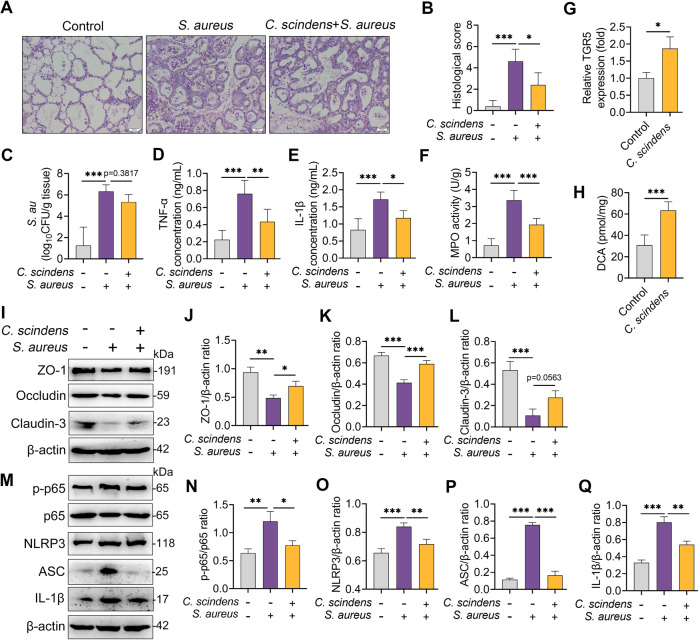
We next investigated the effects of C. scindens, a commensal microbe known to convert primary BAs into secondary BAs26,36, on S. aureus-induced mastitis in mice. As expected, colonization with C. scindens alleviated S. aureus-induced mammary damage (Fig. 7A, B), the S. aureus burden (Fig. 7C), and proinflammatory marker expressions (Fig. 7D–F). Notably, the C. scindens-colonized mice had increased TGR5 gene expression and DCA levels in their mammary glands compared with the control mice (Fig. 7G, H). Moreover, C. scindens supplementation alleviated S. aureus-induced blood-milk barrier damage, as evidenced by restored TJ proteins including ZO-1, Occludin, and Claudin-3 compared with the S. aureus treatment group (Fig. 7I–L). In addition, treating with C. scindens ameliorated S. aureus-induced NF-κB and NLRP3 activation in the mammary glands (Fig. 7M–Q). These results suggest that secondary BA producer improved S. aureus-induced mastitis through the activation of TGR5 in mice.
Fig. 7. Clostridium scindens with DCA-producing capacity alleviates S. aureus-induced mastitis in mice.
A Representative images of the H&E-stained sections from different groups (scale bar, 50 μm). B The histological score of the mammary gland from different groups (n = 6). C Mammary S. aureus load (n = 6). The proinflammatory markers of TNF-α (D), IL-1β (E), and MPO activity (F) were measured (n = 6). G Relative expression of TGR5 mRNA from different treated groups (n = 6). H Mammary DCA levels from the indicated mice (n = 6). I Representative images of ZO-1, Occludin, and Claudin-3 of the mammary gland from the indicated mice. Intensities of ZO-1, Occludin, and Claudin-3 were determined (J–L). M The protein levels of NF-κB and NLRP3 pathways of the mammary glands were detected using western blotting. The relative intensities of p-p65, NLRP3, ASC, and IL-1β of the mammary glands were detected (N–Q). Data are expressed as the means ± SD (B–H, J–L, and N–Q) and one-way ANOVA was performed, followed by Tukey’s test (B–H, J–L, and N–Q). *p < 0.05, **p < 0.01, ***p < 0.001 indicate significance.
Discussion
MRSA-associated infections are of significant concern for humans and animals13. Mounting evidence suggests that an alternated host’s state endows different susceptibilities to pathogens including MRSA6,16,17, which may be attributed to distinct metabolic profiles caused by the host and gut microbiota3,4,11. However, whether and how gut microbiota-derived metabolic changes contribute to the development of MRSA-associated diseases in distal organs remains unknown. In this study, we identified significantly different metabolic characteristics in the mammary glands of SARA cows, which may be involved in the changed susceptibility to S. aureus-induced mastitis20. Among the metabolites associated with gut microbiota is the BA metabolism characterized by reduced CA and DCA in the SARA cows. Supplementation of the mice with DCA, but not CA, attenuated S. aureus-associated mastitis by limiting inflammatory responses and improving the blood-milk barrier disruption in which the underlying mechanism was involved in TGR5-mediated NF-κB and NLRP3 inhibition through the cAMP-PKA pathways. The activation of mammary TGR5 was regulated by gut microbiota, enabling different responses and development to S. aureus-induced mastitis, as evidenced by gut dysbiosis caused by vancomycin-impaired mammary TGR5 expression but restored after SFB compensation. Moreover, treatment mice with secondary BA producer also activated mammary TGR5 and alleviated S. aureus-induced mastitis in mice.
Host- or microbiota-mediated metabolic changes have been proven to play a central role in the development and outcomes of infectious diseases3,9,37. We then performed untargeted metabolomics to identify potential metabolites that affected the susceptibility of pathogen-induced mastitis. The results showed that SARA cows had metabolic profiles in their milk that were distinct from those of healthy cows. Previous studies indicated that SARA increased systemic inflammatory responses and changed metabolites in the circulation38, which was associated with reduced milk yield and fat synthesis. Consistently, our results showed that SARA cows had changed metabolites enriched in fatty acid biosynthesis. A recent study also showed that cows with mastitis had different metabolic traits in their serum39, leading that mastitis was associated with changed host homeostasis. Of note, gut microbiota-associated BAs, especially CA and DCA, were reduced in SARA cows, which act as important factors for regulating host function and the outcomes of diseases7,25,40. Consistently, a previous study indicated that consumption of a fiber-enriched diet increased serum secondary bile acids levels in cows with subclinical mastitis, including DCA and lithocholic acid41. Another study showed that mammary glands from Mediterranean diet-fed animals had higher levels of bile acid metabolites, including cholate, glycocholate, taurocholate, deoxycholate and chenodeoxycholate21, which suggests that dietary components are a main determinant of secondary bile acids metabolism. In clinical and subclinical mastitis cows, lower serum secondary bile acids including DCA and 12-ketolithocholic acid were also detected42. Interestingly, Thomas et al. found a dynamic change in BA concentrations in milk after intramammary challenge with Streptococcus uberis43, implying the potential role of BAs in pathogen-induced mastitis. In addition, bile salts have been reported to regulate zinc uptake and capsule synthesis in a mastitis-associated pathogenic E. coli44, suggesting that different bile acids may have different effects on the host or pathogens.
In our study, treatment with DCA, but not CA, alleviated S. aureus-induced mastitis in mice. Similar results were found in other disease models in which DCA, but not CA, improved the outcome of autoimmune uveitis in mice26. We also confirmed the role of DCA in regulating host responses but not inhibiting bacterial growth since non-antimicrobial dose of DCA also reduced S. aureus-induced inflammation. Our results agree with previous findings that a non-antimicrobial dose of DCA has the capacity to limit LPS- or pathogen-induced inflammatory responses in macrophages24. DCA treatment restored the decrease in TJ proteins caused by S. aureus, suggesting improved blood-milk barrier integrity. In intestinal epithelial cells, DCA was also found to regulate TJ levels45. Impaired barrier function is a common response to increased inflammatory responses that are attributed to activated inflammatory signatures, such as the NF-κB and NLRP3 pathways. S. aureus invasion has been known to activate the NF-κB and NLRP3 pathways and these signatures play an essential role in the pathogenesis of mastitis16. However, DCA depressed the activation of NF-κB and NLRP3 in the MMECs, which agrees with the finding that DCA can inhibit the activation of proinflammatory pathways24. DCA can work through multiple host receptors, including TGR5, FXR, and VDR26. We then determined that TGR5 was significantly increased after DCA treatment in the mammary gland. In return, blocking TGR5 with SBI-115 reversed the protective effects of DCA. Another study also showed that SBI-115 reversed the effects of DCA or INT777, a specific TGR5 agonist, on autoimmune uveitis26. Moreover, increased TGR5 activation regulates the expression of cAMP and subsequent PKA24,26. Blocking cAMP and PKA with MDL12330A and H89 impaired the effects of DCA on NF-κB and NLRP3 and subsequent proinflammatory cytokines in the MMECs. These results showed that TGR5-mediated cAMP-PKA pathways contributed to the effects of DCA on S. aureus-induced mastitis in mice.
Considering the role of DCA and CA in S. aureus-induced mastitis in mice and the gut microbiota in BA metabolism and mastitis7,25, we next studied gut microbiota-mediated TGR5 activation in the pathogenesis of mastitis. Mice treated with vancomycin had reduced DCA levels and TGR5 activation and developed more serious mastitis than the control mice. Similar results were also found in a previous study in which depletion of gut obligate anaerobic microbes by clindamycin aggravated C jejuni-induced colitis in specific pathogen-free (SPF) IL10−/− mice by reducing intestinal DCA levels7. Hu et al. also showed that impairment of the secondary BA reservoir by antibiotics also exacerbated autoimmune uveitis in mice26. Supplementation with SFB restored the DCA and TGR5 levels in the mammary glands and ameliorated S. aureus-induced mastitis in mice, supporting the essential role of spore-forming Clostridium in the production of secondary BAs25,35. C. scindens increased mammary TGR5 activation compared with the control mice and reduced S. aureus-induced mastitis. Another study also showed that C. scindens mirrored the role of DCA or TGR5 activation in uveitis26. These results suggest that gut microbiota-mediated secondary BA production contributes to protecting against S. aureus-induced mastitis in mice. Notably, our results did not allow us to rule out the role of other metabolites in the pathogenesis of S. aureus-induced mastitis, as well as gut microbiota-derived host immune changes. In addition, whether the protective effects of gut microbiota-mediated secondary BAs on S. aureus are conservative in other organs needs to be proven. Future studies also need to explore the effects and potential mechanisms of other types of primary and secondary BAs on S. aureus infection.
In conclusion, our results show that the SARA cows have distinct metabolic changes compared with healthy cows. Among the metabolites potentially affecting the development of mastitis is DCA. Treatment with DCA alleviates S. aureus-induced mastitis in mice and the underlying mechanism is involved in the activation of TGR5, which inhibits the activation of NF-κB and NLRP3 signaling by cAMP and PKA. Gut dysbiosis impairs DCA production and TGR5 activation, which aggravates S. aureus-induced mastitis in mice, but restoring microbiota by SFB or commensal C. scindens ameliorates S. aureus-induced mastitis in mice. Our results suggest that gut microbiota-mediated secondary acid production is a key regulator in the pathogenesis of S. aureus-associated mastitis and acts as a basis for regulating gut microbiota-mediated metabolism to treat mastitis and other diseases.
Methods
Ethical statement
All animal experiments were subject to approval by the Institutional Animal Care and Use Committee (IACUC) of Jilin University (China). The full proposal was considered by the IACUC ethics committee, which approved the animal care and use permit license. All experiments complied with the manual of the care and use of laboratory animals published by the US National Institutes of Health.
Materials
Deoxycholic acid (DCA), cholic acid (CA), and vancomycin were bought from Sigma Aldrich (St. Louis, MO, USA). SBI-115, MDL-12330A, and H89 were purchased from MedChemExpress (MCE, USA). The primary antibodies, including phosphorylation-p65 (p-p65, #AF2006; RRID: AB_2834435), p-65 (#AF5006; RRID: AB_2834847), p-IκB (#AF2002; RRID: AB_2834433), IκB (#AF5002; RRID: AB_2834792), Occludin (#DF7504; RRID: AB_2841004), ZO-1 (#AF5145; RRID: AB_2837631), Claudin-3 (#AF0129; RRID: AB_2833313) and β-actin (#AF7018; RRID: AB_2839420) were obtained from Affinity Biosciences (OH, USA). NLRP3 (#15101), ASC (#67824), and IL-1β (#12242) were bought from Cell Signaling Technology (CST, Boston, USA). Goat anti-rabbit or Rabbit anti-mouse secondary antibodies were bought from ImmunoWay Biotechnology Company. Mouse TNF-α (Cat #430915) and IL-1β (Cat #432615) ELISA assay kits were obtained from Biolegend (CA, USA). Myeloperoxidase (MPO) assay kit was bought from Nanjing Jiancheng Bioengineering Institute (Nanjing, China).
Cow treatment and samples collection
A total of 12 Holstein cows (4–6 years, averaging ~600 kg of weight) were obtained from a farm in Qingzhou, Shangdong Province, China, and none of the animals had diseases or were treated with antibiotics or drugs within 6 months. Cows were randomly separated into healthy group and SARA group. The healthy group was treated with a standard diet of grass-legume hay and the SARA group was treated with a high-concentrate diet (70% grain diet)20,46. All cows met the daily nutrient requirements for lactation. After eight weeks of treatment, milk samples from the different treatment groups were harvested and stored in liquid nitrogen until the metabolomics analysis. The ruminal pH was used to diagnose SARA47.
Untargeted metabolomics
The milk samples (100 mL) were mixed with prechilled methanol (400 μL) by vortexing. The samples were incubated on ice for 5 min and then centrifuged at 15,000 × g and 4 °C for 5 min. The supernatant was diluted to a final concentration containing 53% methanol with LC-MS grade water. The samples were subsequently transferred to a fresh Eppendorf tube and then were centrifuged at 15,000 × g and 4 °C for 10 min. Finally, the supernatant was injected into the LC-MS/MS system for analysis.
The LC-MS/MS analyses were performed using a Vanquish UHPLC system (Thermo Fisher) coupled with an Orbitrap Q Exactive series mass spectrometer (Thermo Fisher). Samples were injected onto a Hyperil Gold column (100 × 2.1 mm, 1.9 μm) using a 16 min linear gradient at a flow rate of 0.2 mL/min. The raw data files generated by the UHPLC-MS/MS were processed using Compound Discoverer 3.1 (Thermo Fisher) to perform peak alignment, peak picking, and quantitation for each metabolite. The main parameters were set as follows: retention time tolerance, 0.2 min; actual mass tolerance, 5 ppm; signal intensity tolerance, 30%; signal/noise ratio, 3; and minimum intensity, 100000. After the peak intensities were normalized to the total spectral intensity. The normalized data were used to predict the molecular formula based on additive ions, molecular ion peaks, and fragment ions. Then, peaks were matched with the mzCloud (https://www.mzcloud.org/), mzVault, and MassList databases to obtain accurate qualitative and relative quantitative results. Statistical analyses were performed using the statistical software R (R version R-3.4.3), Python (Python 2.7.6 version) and CentOS (CentOS release 6.6), When data were not normally distributed, normal transformations were attempted using the area normalization method.
These metabolites were annotated using the KEGG database (http://www.genome.jp/kegg/) and Lipidmaps database (http://www.lipidmaps.org/). The PCA and PLS‐DA were performed at metaX. Pearson correlation analysis was used to assess the correlation of QC samples to determine the stability and accuracy of the metabolome. We applied univariate analysis (t-test) to calculate the statistical significance (P-value). Metabolites with a VIP > 1 and P-value < 0.05 and fold change ≥2 or FC ≤ 0.5 were considered to be differential metabolites. Volcano plots were used to filter metabolites of interest based on the Log2 (FC) and -log10 (P-value) of the metabolites. For the clustering heat maps, the data were normalized using z-scores of the intensity areas of differential metabolites and were plotted by the Pheatmap package in R language. The functions of these metabolites and metabolic pathways were studied using the KEGG database. The metabolic pathway enrichment of the differential metabolites was performed. When the ratio was satisfied by x/n > y/N, the metabolic pathways were considered enriched. When the P-value of metabolic pathway < 0.05, the metabolic pathways were considered as significantly enriched.
Mouse and treatments
All the SPF-grade BABL/c mice (6–8 weeks, 22–24 g) were obtained from Liaoning Changsheng Biotecnology Co., Ltd. (Benxi, China). The mice were raised with enough food and water in SPF-grade feeding conditions with 12 h light and 12 h dark daily for a week. After adapting to the feeding environment, these mice were mixed at a ratio of three females to one male in separated cages with the same feeding conditions. After confirming pregnancy by the observation of vaginal spermatozoa, the male mice were removed.
For the DCA supplementation experiment, mice 7 days after delivery were pretreated with DCA (10, 20, and 30 mg/kg) for 2 h intraperitoneally before the induction of the S. aureus-induced mastitis model24. To confirm the role of gut microbiota in converting CA to DCA and regulating mastitis pathogenesis, mice were treated with CA (30 mg/kg) or DCA (30 mg/kg) orally with or without antibiotics treatment (200 mg/kg ampicillin, neomycin and metronidazole, 100 mg/kg vancomycin) for a week. For the vancomycin treatment experiment, mice were treated with vancomycin (0.5 g/L) in the drinking water for three weeks. For the C. scindens treatment, mice were treated with antibiotics (200 mg/kg ampicillin, neomycin, and metronidazole, 100 mg/kg vancomycin) for 5 days to deplete the commensal microbiota48 and C. scindens (109 CFU/mouse) was supplemented by oral gavage for 7 consecutive days26.
Bacterial spore preparation and transplantation
For SFB transplantation experiment, the cecal contents were harvested from the conventionally fed mice under sterile conditions and the cecal contents were added to a 1.5 mL microcentrifuge tube on ice. The cecal contents were diluted 1:10 in PBS (w/v) and chloroform was added at a final concentration of 3% (v/v). The prepared cecal contents were next incubated by shaking at 200 × g at 37 °C for 30 min. After allowing the chloroform to settle to the bottom of the tube at room temperature (approximately 20 min), the top aqueous layer was removed and the SFB was resuspended with 200 μL PBS in a sterile 1.5 mL tube35,49. For transplantation, mice were pretreated with vancomycin for 14 days and then treated with 200 μL SFB daily for consecutive 7 days.
S. aureus-induced mastitis model
Lactating mice seven days after delivery were separated from the offspring for three hours and subsequently anesthetized using urethane (100 mg/kg). Next, these mice were treated with S. aureus USA300 (107 CFU/50 μL) by intraductal injection at the fourth nipple16,50. Twenty-four hours later, the mammary tissues were harvested and stored at −80 °C until determination.
Bacterial culture and growth assay
The S. aureus USA300 was cultured in a tryptone soybean broth (TSB) medium at 37 °C. The C. scindens ATCC35704 was purchased from American Type Culture Collection (ATCC) and cultured under anaerobic conditions. To investigate the effect of DCA and CA on bacterial growth, the S. aureus USA300 was incubated in TSB medium supplemented with DCA (0, 25.47, 63.68, 127.36, 191.04, 254.73, 636.82, 1273.65, 1910.48 and 2547.31 μM) or CA (0, 24.47, 61.18, 122.37, 183.56, 244.75, 611.89, 1223.78, 1835.67 and 2447.56 μM) and bacterial intensities were measured at 600 nm optical density (OD600) after incubation at 37 °C for 24 h.
Biofilm assay
To investigate whether DCA affects the formation of S. aureus biofilms, S. aureus was incubated in a TSB medium in 96-well plates supplemented with different concentrations of DCA (0, 25.47, 63.68, 127.36, 191.04, 254.73, 636.82, 1273.65, 1910.48 and 2547.31 μM) or CA (0, 24.47, 61.18, 122.37, 183.56, 244.75, 611.89, 1223.78, 1835.67 and 2447.56 μM) at 37 °C for 24 h without shaking. After incubation, the supernatants were removed carefully and the biofilm in the plate was stained with 1% crystal violet solution for 10 min. After washing with PBS three times every 2 min, the plate was dried at 72 °C for 2 h. Finally, 200 μL ethanol (75%) was added to each well and bacterial biofilm was detected at OD57051.
Cell culture and inflammatory response assay
Mouse mammary epithelial cells (MMECs, HC11 cells) were obtained from the American Type Culture Collection (ATCC, CRL-3062) and cultured in a DMEM supplemented with 10% fetal bovine serum and 1% ampicillin and streptomycin at 37 °C with 5% CO2. For the BA treatment experiment, ampicillin and streptomycin were removed from the cells (106 cells/mL) and incubated in 6-well plates for 24 h. The prepared cells were then treated with SBI115 (100 μM), KH7 (10 μM), MDL12330A (10 μM), or H89 (30 μM) for 2 h prior to DCA. For the DCA treatment, DCA was performed 2 h prior to S. aureus treatment with a final concentration of 10, 20, or 30 μM according to the experimental demand. For the siRNA transfection experiment, TGR5 siRNA, cAMP siRNA, PKA siRNA, and the negative control siRNA were transfected using Opti-MEM and Lipofectamine52. All siRNAs were diluted in RNase-free water to a final concentration of 40 nM. The cells were incubated with the DMEM without antibiotics and serum for 24 h followed by a complete medium incubation for the next 24 h. DCA was added 48 h post-siRNA infection. The prepared cells were further treated with S. aureus (MOI 100:1) for 24 h according to the results of the preliminary experiments and the cells were harvested for determination.
Histological analysis
All samples used for the histological analysis were fixed with 4% paraformaldehyde for more than 48 h and embedded in paraffin to prepare 5-μm paraffin sections. All sections were stained with hematoxylin and eosin (H&E) after dewaxing and hydration. The histological changes in the mammary glands were performed by using an optical microscope (Olympus, Tokyo, Japan) and the histological score was assessed17.
Cytokine detection
ELISA assay was performed to determine the predominant proinflammatory cytokines of TNF-α and IL-1β in the serum, mammary gland, and cultured cell supernatant. For the mammary samples, 10% tissue homogenates were prepared using PBS and the supernatants were collected by centrifugation at 12,000 × g for 10 min. The concentrations of TNF-α and IL-1β the mammary gland were calculated according to the manufacturer’s instructions (Biolegend, CA, USA).
MPO activity assay
To determine the MPO levels, 10% tissue homogenates were prepared using a MPO buffer and detected by a MPO assay kit (A044-1-1) according to the manufacturer’s instructions (Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
DCA and CA analysis
To detect the levels of mammary CA and DCA, mammary samples were pre-weighted in lysis tubes containing ceramic beads and added Methanol (MeOH)-containing internal standard. MeOH-extracted samples were centrifuged, diluted 1:1 in 50% MeOH:water, and injected into the UPLC–MS52. CA and DCA levels were determined using commercially available standards purchased from Sigma.
Total bacterial DNA extraction and sequencing
For the gut microbiome analysis, fecal samples were collected from the individual mice. Microbial genomic DNA was extracted by using the FastDNA® Spin Kit for Soil (MP Biomedicals, U.S.). 16 S rRNA gene libraries were constructed using primers specific to the V3-V4 region (338 F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806 R (5′-GGACTACHVGGGTWTCTAAT-3′)) by an ABI GeneAmp® 9700 PCR thermocycler (ABI, CA, USA) and performed paired-end sequencing on an Illumina MiSeq PE300 platform/NovaSeq PE250 platform (Illumina, San Diego, USA) by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). Operational taxonomic units (OTUs) with a 97% similarity cutoff were clustered using UPARSE version 7.1, and chimeric sequences were identified and removed. The taxonomy of each OTU representative sequence was analyzed by RDP Classifier version 2.2 against the 16S rRNA database using a confidence threshold of 0.7. The principal coordinate analysis was performed to identify the separation of gut microbiota among the different treatment groups based on the Bray-Curtis distance and significance was analyzed by ANOSIM. LEfSe was used to identify the differential bacterial taxa in the different treatment groups (LDA score (log10) > 3).
Total RNA extraction and quantitative RT-PCR
The total RNA of the mammary tissues was extracted by Trizol (Invitrogen, CA, USA)17. Briefly, 100 mg of tissues or collected cells were extracted with 1 mL Trizol and subjected to chloroform, isopropanol, and 75% ethyl alcohol treatment under RNase-free conditions. After reverse transcription using TransStart Tip Green qPCR SuperMix (TransGen Biotech, Beijing, China), the cDNA was reacted with specific primers using a FastStart Universal SYBR Green Master Mix (ROX) (Roche, Switzerland, Basel) in a Step One Plus apparatus (Applied Biosystems, Foster City, CA, USA). The reaction conditions were performed as previously mentioned17. The primers used in this study are presented in Supplementary Table 4, and GAPDH served as an endogenous control. The 2−ΔΔCt method was performed to calculate the relative expression of genes by calibration with the control group.
Western blotting
The total protein of the mammary gland was harvested using a tissue protein extraction kit (Thermo Fisher Scientific, USA)17. Ten percent or 15% SDS-PAGE was used to separate proteins, which were then bound to 0.45 μm PVDF membranes. After blocking in 5% skim milk, the prepared PVDF membranes were incubated at 4 °C overnight with specific primary antibodies, including p-p65 (#AF2006; 1:1000), p-65 (#AF5006; 1:1000), p-IκB (#AF2002; 1:1000), IκB (#AF5002; 1:1000), Occludin (#DF7504; 1:1000), ZO-1 (#AF5145; 1:1000), Claudin-3 (#AF0129; 1:1000), NLRP3 (#15101; 1:1000), ASC (#67824; 1:1000), IL-1β (#12242; 1:1000) and β-actin (#AF7018; 1:1000). After washing three times with TBST, the PVDF membranes were treated with goat anti-rabbit or rabbit anti-mouse IgG (1:20000) for 2 h at room temperature. Finally, the proteins were detected using the ECL plus western blotting Detection System (Tanon, China). All blots or gels derive from the same experiment and that were processed in parallel. Original blots are provided in Supplementary Information.
Statistical analysis
All data were analyzed using GraphPad Prism 8 (San Diego, CA, USA) and expressed as boxplots or the means ± SD. The Student’s t test (parametric) and Mann-Whitney test (nonparametric) were performed for the comparison of two groups. One-way analysis of variance (ANOVA) was performed for the comparison of more than two groups, followed by Tukey’s test to determine the differences among groups. A p < 0.05 indicated statistical significance. Other special analyses are stated in the legends.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
The authors thank all constructive suggestions from Zhang’s Lab for the study. We would also like to acknowledge the National Natural Science Foundation of China (32122087, 32102738, and 31972749) and Natural Science Foundation of Jilin Province (20220101298JC).
Author contributions
C.Z., Y.F. and N.Z. designed the study. C.Z. and K.W. performed mouse animal experiments, metabolomics, and all statistical analysis. L.B., H.H., and Y.Z. performed in vitro experiments. X.H. performed cow experiment and samples collection. M.Q., Z.H., and Y.H. assisted with animal experiments and experimental parameter determinations. N.Z., X.H., and Y.F. obtained funding. C.Z. wrote the manuscript and all authors revised and approved the manuscript. C.Z. and K.W. contributed equally to this study.
Data availability
The 16S rRNA gene sequencing data are available in NCBI Sequence Read Archive (SRA) repository under accession number PRJNA892063.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Caijun Zhao, Keyi Wu.
Contributor Information
Xiaoyu Hu, Email: hxiaoyu@yeah.net.
Yunhe Fu, Email: fuyunhesky@sina.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41522-023-00374-8.
References
- 1.Schnupf P, Gaboriau-Routhiau V, Cerf-Bensussan N. Modulation of the gut microbiota to improve innate resistance. Curr. Opin. Immunol. 2018;54:137–144. doi: 10.1016/j.coi.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Monedero V, Collado MC, Rodríguez-Díaz J. Therapeutic opportunities in intestinal microbiota-virus Interactions. Trends Biotechnol. 2018;36:645–648. doi: 10.1016/j.tibtech.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 3.Troha K, Ayres JS. Metabolic adaptations to infections at the organismal level. Trends Immunol. 2020;41:113–125. doi: 10.1016/j.it.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connolly JPR, et al. Host-associated niche metabolism controls enteric infection through fine-tuning the regulation of type 3 secretion. Nat. Commun. 2018;9:4187. doi: 10.1038/s41467-018-06701-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng KM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tovaglieri A, et al. Species-specific enhancement of enterohemorrhagic E. coli pathogenesis mediated by microbiome metabolites. Microbiome. 2019;7:43. doi: 10.1186/s40168-019-0650-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun X, et al. Microbiota-derived metabolic factors reduce campylobacteriosis in mice. Gastroenterology. 2018;154:1751–1763.e1752. doi: 10.1053/j.gastro.2018.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stacy A, et al. Infection trains the host for microbiota-enhanced resistance to pathogens. Cell. 2021;184:615–627.e617. doi: 10.1016/j.cell.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobson A, et al. A gut commensal-produced metabolite mediates colonization resistance to Salmonella Infection. Cell Host Microbe. 2018;24:296–307.e297. doi: 10.1016/j.chom.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fachi JL, et al. Butyrate protects mice from Clostridium difficile-Induced Colitis through an HIF-1-Dependent mechanism. Cell Rep. 2019;27:750–761.e757. doi: 10.1016/j.celrep.2019.03.054. [DOI] [PubMed] [Google Scholar]
- 11.You JS, et al. Commensal-derived metabolites govern Vibrio cholerae pathogenesis in host intestine. Microbiome. 2019;7:132. doi: 10.1186/s40168-019-0746-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park, S. & Ronholm, J. Staphylococcus aureus in Agriculture: Lessons in Evolution from a Multispecies Pathogen. Clin. Microbiol. Rev.34, 10.1128/cmr.00182-20 (2021). [DOI] [PMC free article] [PubMed]
- 13.Lakhundi, S. & Zhang, K. Methicillin-resistant staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev.31, 10.1128/cmr.00020-18 (2018). [DOI] [PMC free article] [PubMed]
- 14.Holmes MA, Zadoks RN. Methicillin resistant S. aureus in human and bovine mastitis. J. Mammary gland Biol. Neoplasia. 2011;16:373–382. doi: 10.1007/s10911-011-9237-x. [DOI] [PubMed] [Google Scholar]
- 15.Ding ST, et al. Lactational breast abscesses caused by methicillin-resistant or methicillin-sensitive staphylococcus aureus infection and therapeutic effect of ultrasound-guided aspiration. Breastfeed. Med.: Off. J. Acad. Breastfeed. Med. 2020;15:471–474. doi: 10.1089/bfm.2020.0003. [DOI] [PubMed] [Google Scholar]
- 16.Hu X, et al. The gut microbiota contributes to the development of Staphylococcus aureus-induced mastitis in mice. ISMEJ. 2020;14:1897–1910. doi: 10.1038/s41396-020-0651-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao C, et al. Aryl hydrocarbon receptor activation by Lactobacillus reuteri tryptophan metabolism alleviates Escherichia coli-induced mastitis in mice. PLoS Pathog. 2021;17:e1009774. doi: 10.1371/journal.ppat.1009774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, et al. Rumen microbiome structure and metabolites activity in dairy cows with clinical and subclinical mastitis. J. Anim. Sci. Biotechnol. 2021;12:36. doi: 10.1186/s40104-020-00543-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Humer E, Aditya S, Zebeli Q. Innate immunity and metabolomic responses in dairy cows challenged intramammarily with lipopolysaccharide after subacute ruminal acidosis. Animal. 2018;12:2551–2560. doi: 10.1017/S1751731118000411. [DOI] [PubMed] [Google Scholar]
- 20.Hu X, et al. The Rumen Microbiota contributes to the development of mastitis in dairy cows. Microbiol. Spectr. 2022;10:e0251221. doi: 10.1128/spectrum.02512-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shively CA, et al. Consumption of Mediterranean versus Western Diet leads to distinct mammary gland microbiome populations. Cell Rep. 2018;25:47–56.e43. doi: 10.1016/j.celrep.2018.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soto-Pantoja DR, et al. Diet alters entero-mammary signaling to regulate the breast microbiome and tumorigenesis. Cancer Res. 2021;81:3890–3904. doi: 10.1158/0008-5472.CAN-20-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S, et al. Diet-induced remission in chronic enteropathy is associated with altered microbial community structure and synthesis of secondary bile acids. Microbiome. 2019;7:126. doi: 10.1186/s40168-019-0740-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo C, et al. Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity. 2016;45:802–816. doi: 10.1016/j.immuni.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 25.Long SL, Gahan CGM, Joyce SA. Interactions between gut bacteria and bile in health and disease. Mol. Asp. Med. 2017;56:54–65. doi: 10.1016/j.mam.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Hu J, et al. Gut microbiota-mediated secondary bile acids regulate dendritic cells to attenuate autoimmune uveitis through TGR5 signaling. Cell Rep. 2021;36:109726. doi: 10.1016/j.celrep.2021.109726. [DOI] [PubMed] [Google Scholar]
- 27.Sorrentino G, et al. Bile acids signal via TGR5 to activate intestinal stem cells and epithelial regeneration. Gastroenterology. 2020;159:956–968.e958. doi: 10.1053/j.gastro.2020.05.067. [DOI] [PubMed] [Google Scholar]
- 28.Masyuk TV, et al. TGR5 contributes to hepatic cystogenesis in rodents with polycystic liver diseases through cyclic adenosine monophosphate/Gαs signaling. Hepatology. 2017;66:1197–1218. doi: 10.1002/hep.29284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu S, Liu X, Yu D, Changyong E, Yang J. Morin protects LPS-induced mastitis via inhibiting NLRP3 Inflammasome and NF-κB signaling pathways. Inflammation. 2020;43:1293–1303. doi: 10.1007/s10753-020-01208-x. [DOI] [PubMed] [Google Scholar]
- 30.Ye J, et al. Scutellarin inhibits caspase-11 activation and pyroptosis in macrophages via regulating PKA signaling. Acta Pharm. Sin. B. 2021;11:112–126. doi: 10.1016/j.apsb.2020.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marunaka Y, Niisato N. H89, an inhibitor of protein kinase A (PKA), stimulates Na+ transport by translocating an epithelial Na+ channel (ENaC) in fetal rat alveolar type II epithelium. Biochem Pharm. 2003;66:1083–1089. doi: 10.1016/s0006-2952(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, et al. Review article: insights into the bile acid-gut microbiota axis in intestinal failure-associated liver disease-redefining the treatment approach. Aliment. Pharmacol. Ther. 2022;55:49–63. doi: 10.1111/apt.16676. [DOI] [PubMed] [Google Scholar]
- 33.Ma J, et al. Gut microbiota remodeling reverses aging-associated inflammation and dysregulation of systemic bile acid homeostasis in mice sex-specifically. Gut Microbes. 2020;11:1450–1474. doi: 10.1080/19490976.2020.1763770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vrieze A, et al. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J. Hepatol. 2014;60:824–831. doi: 10.1016/j.jhep.2013.11.034. [DOI] [PubMed] [Google Scholar]
- 35.Grizotte-Lake M, et al. Commensals suppress intestinal epithelial cell retinoic acid synthesis to regulate Interleukin-22 Activity and prevent microbial dysbiosis. Immunity. 2018;49:1103–1115.e1106. doi: 10.1016/j.immuni.2018.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Studer N, et al. Functional Intestinal Bile Acid 7α-Dehydroxylation by Clostridium scindens Associated with Protection from Clostridium difficile Infection in a Gnotobiotic Mouse Model. Front. Cell. Infect. Microbiol. 2016;6:191. doi: 10.3389/fcimb.2016.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Debnath N, Kumar R, Kumar A, Mehta PK, Yadav AK. Gut-microbiota derived bioactive metabolites and their functions in host physiology. Biotechnol. Genet. Eng. Rev. 2021;37:105–153. doi: 10.1080/02648725.2021.1989847. [DOI] [PubMed] [Google Scholar]
- 38.Zhang T, et al. Responsive changes of rumen microbiome and metabolome in dairy cows with different susceptibility to subacute ruminal acidosis. Anim. Nutr. 2022;8:331–340. doi: 10.1016/j.aninu.2021.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang G, et al. Identification of serum-predictive biomarkers for subclinical mastitis in dairy cows and new insights into the pathobiology of the disease. J. Agric Food Chem. 2022;70:1724–1746. doi: 10.1021/acs.jafc.1c07281. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, et al. DCA-TGR5 signaling activation alleviates inflammatory response and improves cardiac function in myocardial infarction. J. Mol. Cell. Cardiol. 2021;151:3–14. doi: 10.1016/j.yjmcc.2020.10.014. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, et al. Changes in the Profile of fecal microbiota and metabolites as well as serum metabolites and proteome after dietary inulin supplementation in dairy cows with subclinical mastitis. Front. Microbiol. 2022;13:809139. doi: 10.3389/fmicb.2022.809139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, et al. Discrepancies among healthy, subclinical mastitic, and clinical mastitic cows in fecal microbiome and metabolome and serum metabolome. J. Dairy Sci. 2022;105:7668–7688. doi: 10.3168/jds.2021-21654. [DOI] [PubMed] [Google Scholar]
- 43.Thomas FC, et al. Mastitomics, the integrated omics of bovine milk in an experimental model of Streptococcus uberis mastitis: 3. Untargeted metabolomics. Mol. Biosyst. 2016;12:2762–2769. doi: 10.1039/c6mb00289g. [DOI] [PubMed] [Google Scholar]
- 44.Olson MA, et al. Bile salts regulate zinc uptake and capsule synthesis in a mastitis-associated extraintestinal pathogenic Escherichia coli strain. Infect. Immun. 2021;89:e0035721. doi: 10.1128/IAI.00357-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raimondi F, et al. Bile acids modulate tight junction structure and barrier function of Caco-2 monolayers via EGFR activation. Am. J. Physiol. Gastrointest. Liver Physiol. 2008;294:G906–G913. doi: 10.1152/ajpgi.00043.2007. [DOI] [PubMed] [Google Scholar]
- 46.Orton T, Rohn K, Breves G, Brede M. Alterations in fermentation parameters during and after induction of a subacute rumen acidosis in the rumen simulation technique. J. Anim. Physiol. Anim. Nutr. 2020;104:1678–1689. doi: 10.1111/jpn.13412. [DOI] [PubMed] [Google Scholar]
- 47.Eger M, Riede S, Breves G. Induction of a transient acidosis in the rumen simulation technique. J. Anim. Physiol. Anim. Nutr. 2018;102:94–102. doi: 10.1111/jpn.12662. [DOI] [PubMed] [Google Scholar]
- 48.Chen X, et al. Gut dysbiosis induces the development of pre-eclampsia through bacterial translocation. Gut. 2020;69:513–522. doi: 10.1136/gutjnl-2019-319101. [DOI] [PubMed] [Google Scholar]
- 49.Velazquez, E. M., Rivera-Chávez, F. & Bäumler, A. J. Spore preparation protocol for enrichment of clostridia from murine intestine. Bio-protocol7, 10.21769/BioProtoc.2296 (2017). [DOI] [PMC free article] [PubMed]
- 50.Jiang KF, et al. Polydatin ameliorates Staphylococcus aureus-induced mastitis in mice via inhibiting TLR2-mediated activation of the p38 MAPK/NF-κB pathway. Acta Pharmacol. Sin. 2017;38:211–222. doi: 10.1038/aps.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu, M. et al. Commensal Bacillus subtilis from cow milk inhibits Staphylococcus aureus biofilm formation and mastitis in mice. FEMS Microbiol. Ecol.98, 10.1093/femsec/fiac065 (2022). [DOI] [PubMed]
- 52.Chaudhari SN, et al. Bariatric surgery reveals a gut-restricted TGR5 agonist with anti-diabetic effects. Nat. Chem. Biol. 2021;17:20–29. doi: 10.1038/s41589-020-0604-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA gene sequencing data are available in NCBI Sequence Read Archive (SRA) repository under accession number PRJNA892063.