Protein fold prediction using deep-learning artificial intelligence (AI) has transformed the field of protein structure prediction (1–3). By combining physical and geometric constraints—and especially patterns extracted from the Protein Data Bank (4)—these machine learning algorithms can predict protein structures at or near atomic resolution and do so in seconds. Today, these computational methods have now solved more than 200 million protein structures, which are accessible from the AlphaFold Protein Structure Database (5) (https://alphafold.ebi.ac.uk/). This accomplishment seems all the more remarkable because few thought it possible or saw it coming. Deservedly, deep-learning AI was named Science magazine’s 2021 “breakthrough of the year” (6). Clearly, deep-learning AI represents a major advance in protein fold prediction.
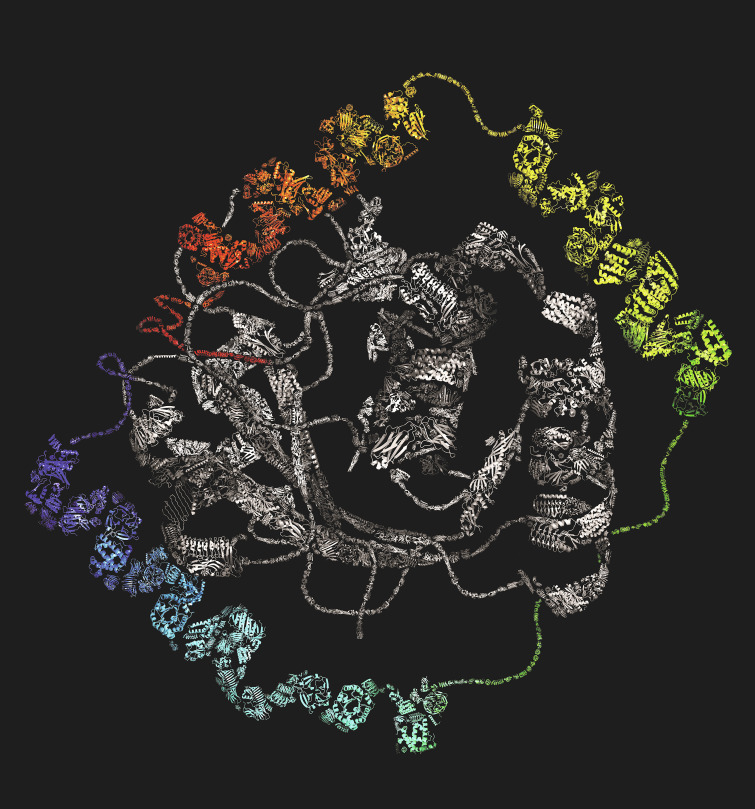
Protein domains, like those in this composite picture, are conspicuous structural units in globular proteins. Their identification has been a topic of intense biochemical interest dating back to the earliest crystal structures. Understanding the folding process of all sorts of proteins, not just their ultimate fold, should be high priority for biochemists in the coming years. Image credit: Lauren L. Porter.
But this is not folding prediction. Patterns extracted from proteins in the Protein Data Bank (PDB) provide a ready “parts list,” circumventing the folding process entirely. These patterns are “fully baked.” That is, a pattern extracted from a solved structure in the PDB is fully preorganized; any physical–chemical organizing interactions have already been realized during folding. The situation is analogous to interpreting a movie by fast-forwarding to the final scene without first watching the previous two hours; we know how it ends, but we don’t know why.
And we do need to know why. If a specific project depends solely on knowledge of a protein structure, an AI solution may be sufficient. But the burning question remains: How does that structure emerge from a linear sequence of amino acid residues in aqueous solution? Recognizing nature’s patterns has been a familiar intermediate step toward deeper understanding. Often, it takes a while. A moment’s reflection is sufficient to recall examples of phenomena that challenged smart thinkers over successive generations but, once understood, can ultimately be explained in an hour. Avogadro’s number, the number of units in one mole of any substance, is such an example. Here, we argue that moving from AI-based pattern recognition to a first principles understanding of protein folding requires an understanding of the relevant chemistry and physics.
Scientific history since Galileo and Newton has taught us that once the principles are understood, more accurate solutions, unanticipated insights, and revealing predictions are likely to follow quickly. Indeed, the ultimate aim of science is to rationalize recurrent patterns by formulating first principles. By analogy, protein structure prediction using AI-assisted pattern recognition is comparable with Mendeleev’s compilation of the periodic table of the elements before its eventual derivation from quantum mechanics—first pattern recognition, then first principles. Accordingly, it is crucial to support ongoing research. The literature suggests numerous fertile approaches are already in play, including one we discuss here.
Into the Fold
A little background is needed to fully appreciate the significance of the protein fold breakthrough. The protein folding problem was first articulated in the 1930s (7). To this day, a mechanistic understanding of the folding reaction remains a challenge, perhaps the most significant unsolved problem at the chemistry–biology interface.
For proteins, function follows form (i.e., the three-dimensional structure of the protein is responsible for its biological function). At present, the three-dimensional structures of almost 200,000 proteins solved by X-ray crystallography, nuclear magnetic resonance (NMR), and cryo-electron microscopy can be accessed in the PDB (4), a freely available, government-supported repository (https://www.rcsb.org/).
Remarkably, proteins can self-assemble spontaneously and reversibly into their unique native three-dimensional structure under suitable physiological conditions. Here, “spontaneous” means that no external energy source such as ATP hydrolysis is required. This chemistry was established 60 years ago by Anfinsen and Haber, who showed that purified ribonuclease can self-assemble spontaneously in salty water (8), and many subsequent experiments with other proteins confirmed its generality (9). Successful self-assembly of purified ribonuclease—free of cellular components—proved that the information needed to determine the protein’s native state is encoded solely within its amino acid sequence. In essence, protein folding is physical chemistry, not cell biology, and sequence alone determines structure.
The reversible folding reaction, U(nfolded)⇌N(ative), differs from an ordinary chemical reaction in that no covalent bonds are made or broken when a protein folds (although some proteins are stabilized by covalently formed disulfide bonds); the population just re-equilibrates in response to changed chemical and/or physical conditions that either disfavor or favor the folded state. Some larger proteins are apt to get “stuck” during folding and require helper proteins called chaperones, which can liberate the incompletely folded conformer, and shift it toward the U state to try again, iteratively if necessary.
The underlying physical chemistry responsible for spontaneous self-assembly is at the root of macromolecular-based life on Earth (10). With this overall perspective in mind, the following sections seek to place the current success of AI-based protein structure prediction within a broader scientific framework.
The success of deep-learning AI is, in effect, an existence proof that an essentially complete set of patterns is embedded in these structures. This approach solves the fold problem, at least in part, but the fundamental question remains: How does the relevant physical chemistry select the native structure from a protein’s amino acid sequence? This is the classic protein folding problem. Protein folding links linear sequences of amino acid residues to the three-dimensional world of the cell, a spontaneous transition under suitable physiological conditions (8), although some larger proteins may necessitate chaperones, as mentioned above.
Stepping Stones
So where do we go from here? Surely there is much that remains to be learned by using AI-based approaches. However, the ultimate goal is to move beyond empirical pattern recognition to the underlying physical chemistry responsible for determining the protein’s three-dimensional structure. Many years of research have been directed toward this ultimate goal: See, for example (11) and references therein, or (12), developed along quite different lines. Here, we invoke the simplifying realization that, of thermodynamic necessity, globular proteins are built on scaffolds of repetitive secondary structure (α-helices and strands of β-sheet) (13), and this thermodynamic imperative imposes a stringent limit on the number of viable folds for small proteins the size of ribonuclease.
Ongoing AI research offers an expanding modeling toolkit to the community. A natural direction is physics-informed AI in which existing physical models can be transformed into descriptors within a machine learning framework (14). Such human–machine collaboration represents one promising route to capture “fundamental laws” for protein structure.
Well and good, but even at best, empirical pattern recognition is a familiar intermediate in the usual course of scientific discovery, a stepping-stone in an ongoing stream. The ultimate goal of scientific understanding is to explain complex phenomena with a compact description, a model, preferably one in which the description has physical meaning and predictive power. For example, Tycho Brahe’s copious observations of planetary motions were reduced to Kepler’s three compact laws, an empirical mathematical description that was transformed into physics by Newton. This progression, from empirical data to abstract representation and then to a physical model, illustrates the ongoing, accretive process by which we learn.
Five centuries of progress in science has typically followed this familiar path:
observation → pattern recognition → theory/models (e.g., Tycho Brahe → Kepler → Newton).
That history of fundamental scientific discoveries abounds with such examples. For example:
(i) Relativity: observations of Michelson-Morley, then “empirical” Lorentz transformations, and finally Einstein’s theory.
(ii) Quantum mechanics: observation of spectral lines, then Lyman’s discovery of empirical regularities in the series of spectral lines, and finally quantum mechanics.
Thus far, protein folding is tracking this progression closely, with half a century of observation encapsulated in the PDB and breakthrough success in pattern recognition using deep learning AI. But the next step in this paradigm is still in the offing.
Commenting on AI-based fold prediction in a recent letter to Science, Moore et al. opined:
“Others, including us, feel that solving the protein-folding problem means making accurate predictions of structures from amino acid sequences starting from first principles based on the underlying physics and chemistry” (15).
Count us, the authors of the present article, among these stalwarts.
A successful physical–chemical theory of protein folding would likely provide deep insights into dynamics, mechanism, function, and the origins of protein-based life on Earth. Furthermore, if the past is any indication, there would also be additional payoffs we cannot yet imagine. Indeed, all the above-mentioned theories, once developed, went far beyond simply reproducing the empirical observations that spawned them.
First Principles
Basic research has provided countless practical applications of immense value. But let us not lose sight of the inner directive that draws us to basic research and the persisting search for first principles—that’s what we do because that’s who we are. Aristotle’s perception still rings true: “All, by nature, desire to know.”
In the most creative minds, this ineffable drive has led to the law of universal gravitation, Maxwell’s equations, E = mc2, etc. All are models. We tend to gloss over the realization that a durable model is nevertheless just a model of reality, not reality per se. Newtonian gravitation (published in 1686) is typically taught as a Kantian “thing-in-itself” (Ding an sich), an unmindful conflation of phenomenon and noumenon stemming from the remarkable effectiveness and apparent singularity of the model over the course of centuries. It’s an operational model: “Gravitation works that way, never mind why.” Although familiarity conditions intuition, we still today regard it as a weird model, and so did Newton in the 17th century. A stunning realization that Newtonian gravitation is just a model came almost three centuries later with Einstein’s general theory of relativity (1915), a superseding model that is both more far-reaching and more intuitively satisfying.
It is no accident that the examples of first principles mentioned above are from physics. Biology has lagged behind because, unlike physics, it is self-modifying and therefore more complex—far more complex. Biological experiments involve many parameters, and conclusions are meaningless in the absence of suitable controls. Physics experiments are typically simpler: Assuming accurate measurements, controls are foreign concepts. For example, the speed of light in a vacuum is a constant in any experiment.
Biological complexity notwithstanding, there is now good reason to anticipate that an authentic physical–chemical theory for protein folding is within reach. For simple proteins, the set of AI-evolved patterns is akin to the basis set of a vector space or the grammar of a language, where a set of primitives or rules can generate an open-ended set of syntactically correct constructs. In proteins, the analogous primitives would be patterns or building blocks.
Recently, it has been shown that some more complex proteins switch folds by remodeling their secondary structures α-helices and β-strands) in response to cellular stimuli (16), a radical departure from the classical Anfinsen paradigm (8) in which a given amino acid sequence gives rise to a unique three-dimensional structure under suitable folding conditions. For example, fold switching has been documented in the NusG transcription factor family (17), a large superfamily of transcriptional regulators known to be conserved from bacteria to humans. In an analogous grammar, fold-switching proteins would correspond to a context-dependent language.
Carrying the analogy further, AlphaFold has provided an exhaustive list of sentences in the language of proteins (5), and we are now poised to learn the grammar. That grammar is governed by the laws of physics and chemistry (18), especially thermodynamics, as described next.
Extreme adaptability is built into globular proteins by the thermodynamics of self-assembly. Of thermodynamic necessity, folded globular proteins are typically built on scaffolds of hydrogen-bonded α-helix and/or strands of β-sheet (13), enabling side chains to respond to external constraints without perturbing backbone integrity. Consistent with this thermodynamic imperative, proteins in extremophiles (thermophiles, psychrophiles, halophiles) that function successfully under extremes of pressure, temperature, pH, and ionic strength are found to retain the same overall backbone structure as their counterparts in mesophiles. Differing cellular microenvironments (cytoplasm, membrane, ribosomes, organelles) can be accommodated similarly. Such adaptability resembles Darwinian evolution at the molecular level, selecting for the “fittest” sequence that can function successfully within a given environment while keeping the overall structure intact.
Clearly, adoption of the native state during the folding reaction, U(nfolded) ⇌ N(ative), comes at an entropic price. Paying this price, the thermodynamic requirement for backbone hydrogen bonding implies that only a limited number of possible scaffold arrangements for a protein domain is possible, no more than ~10,000 (19–23). In detail, a single-domain protein like hen egg lysozyme (129 residues) has approximately 10 scaffold elements. With 10 segments of either α-helix or β-strand, there are 210 possible scaffolds, multiplied by the complexity introduced from interconnecting turns and loops, which are typically short and therefore conformationally restrictive. Thus, most of the entropic cost is prepaid on forming the hydrogen-bonded backbone scaffold, an inescapable thermodynamic requirement in both natural proteins and designed proteins (10, 24).
A possible objection to the preceding explanation is that AlphaFold (1) has had limited success with the class of intrinsically disordered proteins (25), which, by definition, lack persisting structure until paired with a cognate molecule, or again with allosteric proteins (26), regulatory proteins that involve populations rather than single structures. Additionally, AlphaFold2 stumbles on fold-switching proteins (27), as mentioned previously. Nevertheless, to date, there is no evidence that once folded, novel patterns will be found in these refractory cases. The same basic AI patterns seem likely to cover any protein in all cases.
Progress on open questions of greater complexity is ongoing. Much of our current knowledge comes from decades of work on purified proteins studied in vitro, and its applicability to folding within the complex microenvironment of a living cell remains an ongoing concern (28). Unlike in vitro denaturation studies, proteins in cells are synthesized N to C terminus, and nascent peptides remain bound to ribosomes when folding begins. To what degree, if any, does this difference affect the folding pathway? Again, some proteins require chaperones, others do not. Can we distinguish between these two classes? And, is in vivo folding controlled kinetically (29), again unlike in vitro studies of proteins at equilibrium?
In short, it seems likely that a physical–chemical theory of protein folding, one that covers the full spectrum of inquiry—conformation, dynamics, pathways, fluctuations, binding, allostery, etc.—is within our grasp. Now is not the time to halt the search!
Acknowledgments
Author contributions
S.C., M.H., R.J., K.J., D.K., A.K., S.K., D.K., J.L., A.L., S.M., J.M., C.M., J.M., S.M., R.N., K.O., D.P., J.S., T.S., G.S., I.V., X.Z., and G.R. designed research; G.R. wrote the paper; S.C., M.H., R.J., K.J., D.K., A.K., S.K., D.K., J.L., A.L., S.M., J.M., C.M., J.M., S.M., R.N., K.O., D.P., J.S., T.S., G.S., I.V., and X.Z. the ideas presented here emerged during discussions at a 2022 Telluride Science Research Center Coarse-Grained Modeling workshop. All authors were active participants.
Competing interest
The authors declare no competing interest.
Footnotes
Any opinions, findings, conclusions, or recommendations expressed in this work are those of the authors and have not been endorsed by the National Academy of Sciences
References
- 1.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tunyasuvunakool K., et al. , Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baek M., et al. , Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berman H. M., et al. , The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varadi M., et al. , AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thorp H. H., Proteins, proteins everywhere. Science 374, 1415 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Mirsky A. E., Pauling L., On the structure of native, denatured, and coagulated proteins. Proc. Natl. Acad. Sci. U.S.A. 22, 439–447 (1936). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haber E., Anfinsen C. B., Regeneration of enzyme activity by air oxidation of reduced subtilisin-modified ribonuclease. J. Biol. Chem. 236, 422–424 (1961). [PubMed] [Google Scholar]
- 9.Sosnick T. R., Barrick D., The folding of single domain proteins–have we reached a consensus? Curr. Opin. Struct. Biol. 21, 12–24 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rose G. D., Reframing the protein folding problem: Entropy as organizer. Biochemistry 60, 3753–3761 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Nassar R., Dignon G. L., Razban R. M., Dill K. A., The protein folding problem: The role of theory. J. Mol. Biol. 433, 167126. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Škrbić T., Maritan A., Giacometti A., Rose G. D., Banavar J. R., Building blocks of protein structures: Physics meets biology. Physical Rev. E 104, 014402. (2021). [DOI] [PubMed] [Google Scholar]
- 13.Rose G. D., Fleming P. J., Banavar J. R., Maritan A., A backbone-based theory of protein folding. Proc. Natl. Acad. Sci. U.S.A. 103, 16623–16633 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu X., Ericksen S. S., Mitchell J. C., DBSI: DNA-binding site identifier. Nucleic Acids Res. 41, e160. (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore P. B., Hendrickson W. A., Henderson R., Brunger A. T., The protein-folding problem: Not yet solved. Science 375, 507 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Porter L. L., Looger L. L., Extant fold-switching proteins are widespread. Proc. Natl. Acad. Sci. U.S.A. 115, 5968–5973 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porter L. L., et al. , Many dissimilar NusG protein domains switch between alpha-helix and beta-sheet folds. Nat. Commun. 13, 3802 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liwo A., et al. , Scale-consistent approach to the derivation of coarse-grained force fields for simulating structure, dynamics, and thermodynamics of biopolymers. Prog. Mol. Biol. Transl. Sci. 170, 73–122 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Chothia C., Proteins. One thousand families for the molecular biologist. Nature 357, 543–544 (1992). [DOI] [PubMed] [Google Scholar]
- 20.Przytycka T., Aurora R., Rose G. D., A protein taxonomy based on secondary structure. Nat. Struct. Biol. 6, 672–682 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Koonin E. V., Wolf Y. I., Karev G. P., The structure of the protein universe and genome evolution. Nature 420, 218–223 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Hubner I. A., Arakaki A. K., Shakhnovich E., Skolnick J., On the origin and highly likely completeness of single-domain protein structures. Proc. Natl. Acad. Sci. U.S.A. 103, 2605–2610 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmermann M., Towfic F., Jernigan R. L., Kloczkowski A., Short paths in protein structure space originate in graph structure. Proc. Natl. Acad. Sci. U.S.A. 106, E137; author reply E138 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J., et al. , Scaffolding protein functional sites using deep learning. Science 377, 387–394 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruff K. M., Pappu R. V., AlphaFold and implications for intrinsically disordered proteins. J. Mol. Biol. 433, 167208. (2021). [DOI] [PubMed] [Google Scholar]
- 26.Nussinov R., Zhang M., Liu Y., Jang H., AlphaFold, artificial intelligence (AI), and allostery. J. Phys Chem. B 126, 6372–6383 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakravarty D., Porter L. L., AlphaFold2 fails to predict protein fold switching. Protein Sci. 31, e4353. (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monteith W. B., Pielak G. J., Residue level quantification of protein stability in living cells. Proc. Natl. Acad. Sci. U.S.A. 111, 11335–11340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chattopadhyay G., et al. , Mechanistic insights into global suppressors of protein folding defects. PLoS Genet. 18, e1010334. (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]