Significance
Prostate cancer (PCa) affects over 250,000 men in the US. Androgen receptor (AR) signaling inhibitors are the mainstay therapeutics for PCa. Advanced PCa that expresses AR splice variants (AR-SVs) does not respond to current therapeutic strategies. In this manuscript, we uncovered the physicochemical properties of the intrinsically disordered transactivation domain of AR and AR-SV and developed AR-irreversible covalent antagonists (SARICA) to the transactivation domain for the treatment of advanced PCa. SARICAs were also used to identify a potential binding region in the AR and AR-SV transactivation domain.
Keywords: Androgen receptor (AR), AR splice variants (AR-SVs), castration-resistant prostate cancer (CRPC), Selective AR irreversible covalent antagonists (SARICA), liquid–liquid phase separation (LLPS)
Abstract
Androgen receptor (AR) and its splice variants (AR-SVs) promote prostate cancer (PCa) growth by orchestrating transcriptional reprogramming. Mechanisms by which the low complexity and intrinsically disordered primary transactivation domain (AF-1) of AR and AR-SVs regulate transcriptional programming in PCa remains poorly defined. Using omics, live and fixed fluorescent microscopy of cells, and purified AF-1 and AR-V7 recombinant proteins we show here that AF-1 and the AR-V7 splice variant form molecular condensates by liquid–liquid phase separation (LLPS) that exhibit disorder characteristics such as rapid intracellular mobility, coactivator interaction, and euchromatin induction. The LLPS and other disorder characteristics were reversed by a class of small-molecule-selective AR-irreversible covalent antagonists (SARICA) represented herein by UT-143 that covalently and selectively bind to C406 and C327 in the AF-1 region. Interfering with LLPS formation with UT-143 or mutagenesis resulted in chromatin condensation and dissociation of AR-V7 interactome, all culminating in a transcriptionally incompetent complex. Biochemical studies suggest that C327 and C406 in the AF-1 region are critical for condensate formation, AR-V7 function, and UT-143’s irreversible AR inhibition. Therapeutically, UT-143 possesses drug-like pharmacokinetics and metabolism properties and inhibits PCa cell proliferation and tumor growth. Our work provides critical information suggesting that clinically important AR-V7 forms transcriptionally competent molecular condensates and covalently engaging C327 and C406 in AF-1, dissolves the condensates, and inhibits its function. The work also identifies a library of AF-1-binding AR and AR-SV-selective covalent inhibitors for the treatment of PCa.
The human proteome consists of ~20,000 unmodified proteins, of which only 10 to 15% are pharmacologically targetable (1, 2). Some of the limitations that preclude discovering drugs to pharmacologically intractable proteins include absence of a binding pocket, intrinsically disordered structure, and lack of structural information (2). Transcription factors typically contain a well-defined DNA-binding domain (DBD) and a low complexity unstructured activation domain that impairs structural elucidation (3). Such regions and proteins are called intrinsically disordered regions (IDRs) and intrinsically disordered proteins (IDPs), respectively. The activation domain of many transcription factors is the primary coactivator-interacting region, and the activity depends on their disordered structure (4, 5).
Recent research suggests that transcription factors and their disordered transactivation domains form liquid–liquid phase separation (LLPS) membrane-less molecular condensates (6, 7). These LLPS molecular condensates have been correlated with forming transcriptionally active complex at superenhancers, interacting with cofactors such as mediator complex, and modulating oncogenic transcription. The condensates have been shown to play important roles in various diseases such as cancer (8, 9).
Castration-resistant prostate cancer (CRPC) is a significant clinical challenge (10). Current therapeutic strategies for advanced CRPC include androgen receptor (AR) antagonists and a CYP17A1 inhibitor that prevents the synthesis of androgens (11–13). About 30% of patients do not benefit from these drugs due to inherent resistance (10). Although AR ligand-binding domain (LBD) is the domain to which ligands bind, the activation function-1 (AF-1) region in the N-terminal domain (NTD) is the primary coactivator-interacting surface that retains over 70% of AR’s transactivation function (Fig. 1A) (14), making it an important drug discovery target. AR splice variants (AR-SVs) have a deletion of LBD, which results in a constitutively active AR. AR-V7, v567es, and other clinically identified AR-SVs appear as resistance mechanisms to existing treatments (10, 15–18). With their intrinsically disordered structure and undetectable binding pockets (using conventional methods), AR-SVs and AF-1 are important disease modifiers with little available structural information. Molecules discovered without the aid of structural information (19–24) fail to provide details about binding pockets.
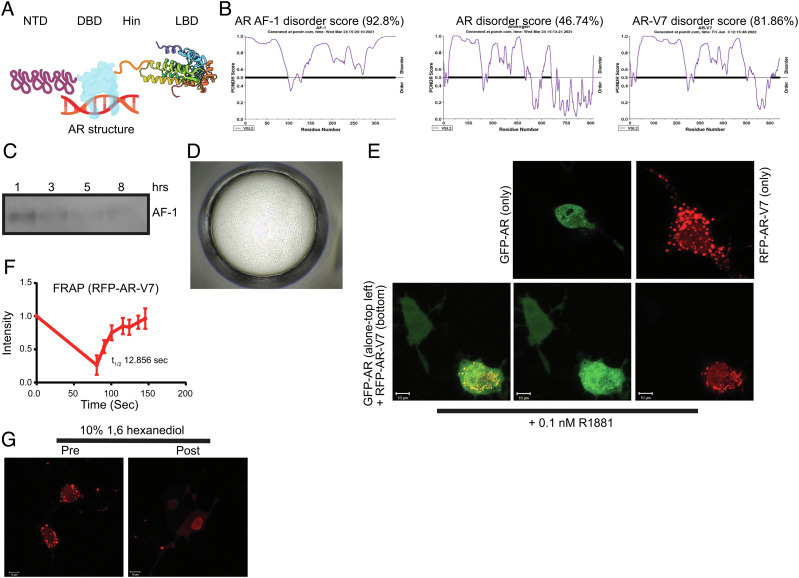
Fig. 1.
AR AF-1 and AR-V7 exhibit disordered protein characteristics. (A) AR structure (created using biorender.com; for illustration purpose only). (B) Pondr score. Pondr (www.pondr.com) disorder score for AR AF-1, AR full-length, and AR-V7. (C) Time-course for stability of recombinant-purified AR AF-1 (141 to 486). Purified recombinant AR AF-1 protein (5 ng) was incubated for the indicated time points at room temperature. Proteins were fractionated on SDS-PAGE and AR western blot was performed with an antibody that binds to the AF-1 region (AR-441, against AR fragment 302 to 318). (D) Recombinant-purified AF-1 (141 to 486) forms LLPS. Purified recombinant AR AF-1 (1 mg/ml) was seeded in a buffer (1:1 silver bullet E4 (1% w/v protamine sulfate, 0.02M HEPES pH 6.8) (HR2-096) + 30% PEG-3350 + 0.05 M HEPES pH 6.8) at 14 °C for 6 h and imaged using a phase contrast microscope. (E) AR-V7 forms molecular condensate in cells. COS7 cells were transfected with 1 µg turbo-red AR-V7 or GFP-AR or both. Twenty-four hours after transfection, the cells in DME + 5%csFBS without phenol red were treated with 0.1 nM R1881. Cells were imaged using a fluorescent confocal microscope 24 h after treatment. (F) FRAP. COS7 cells were transfected with 0.25 µg RFP-AR-V7. Twenty-four hours after transfection, the cells were fed with DME + 10% FBS. Forty-eight hours after transfection, selected regions were photobleached, and the recovery was monitored. Intensity of selected regions is provided as line graph (half-life (t1/2) is derived from an average of four independent measurements; represented as average ± SE). (G) 1,6 hexanediol dissolves the condensates. COS7 cells were transfected with RFP-AR-V7. Live cells in DME + 10% FBS were imaged before and 10 min after 10% 1,6 hexanediol addition. FRAP-fluorescence recovery after photobleaching; LLPS, liquid–liquid phase separation; NTD, N-terminal domain; DBD, DNA-binding domain; LBD, ligand-binding domain; RFP, red fluorescent protein; GFP, green fluorescent protein.
Although AR is important for the development and maintenance of the prostate, transcriptional reprogramming and differential cistrome binding makes AR oncogenic in the development of prostate carcinogenesis (25, 26). The mechanisms by which the AR, AR-SV, and their critical transactivation domain promote these changes is unknown. In addition, considering that the NTD of AR and AR-SV is intrinsically disordered and contains a powerful transactivation domain, there is a high possibility that it drives the formation of transcriptionally active molecular condensates. Previous evidence found that AR DBD and AR AF-1 in the presence of heavy water form molecular condensates (27, 28). It is intriguing that the ordered DBD region forms condensates. We utilized chemical and biological tools to unravel the mechanisms by which AR forms transcriptionally active complex, including molecular condensate by LLPS. We describe herein molecules that covalently bind to specific cysteines in AF-1 and show efficacy in models of prostate cancer (PCa). These molecules will also aid in critically advancing our knowledge of the physicochemical characteristics of AF-1 and AR-SVs.
Results
AR-AF-1 and AR-V7 Exhibit Properties of IDP and IDR.
IDPs or IDRs have several physicochemical characteristics such as temperature-dependent instability, LLPS, molecular condensate formation, and intracellular mobility, which are critical for their activity (4, 6, 29–32). We evaluated the disorder score of the AF-1 region, AR, and AR-V7 using the Pondr algorithm (www. ondr.com) (33). The Pondr algorithm uses neural networks and computational methods to predict the disordered nature of a protein sequence. While AF-1 and AR-V7 have high disorder scores of 92.8% and 81.9%, respectively, the full-length AR’s disorder score is 46.7%, suggesting that the DBD and LBD have a more ordered structure than the AF-1 region that lowers the AF-1 Pondr score (Fig. 1B). This sequence-based computational analysis is consistent with previous empirical studies suggesting an ordered DBD and LBD, and disordered NTD (34, 35).
We next evaluated the stability of purified recombinant AF-1 protein. While at 4 °C AF-1 protein was stable even after 24 h, AF-1 was unstable at room temperature with most of the protein disappearing by 3 h (Fig. 1C). Considering that the AF-1 and AR-V7 are constitutively active and disordered, they have the potential to form phase-separated molecular condensates in the presence or absence of coactivators. The catGRANULE algorithm computationally calculates the probability of a protein sequence to form LLPS condensates (36, 37). Any score greater than 0 is indicative of the protein’s propensity to form LLPS. The propensity score for AR AF-1 is 1.74, and for AR-V7 is 1.35, while for AR LBD is (−)0.836, indicating that the AF-1 domain and AR-V7 could potentially form LLPS because they are structurally disordered (SI Appendix, Fig. S1A).
Recombinant AR AF-1 protein purified by FPLC Superdex size-exclusion column (99% pure by mass spectrometry analysis and folded and unaggregated (SI Appendix, Fig. S1 B and C)) formed LLPS condensates in the absence of cofactors such as MED1 (Fig. 1D). Condensates were formed in 6 h after plating at 14 °C.
To test if AR-V7 also forms molecular condensates, turbo-red-tagged AR-V7 plasmid was transfected into COS7 cells and the fluorescent protein in live and fixed cells was visualized by confocal microscopy (Fig. 1E). AR-V7 molecular condensates were observed both predominantly in the nucleus with fluorescence also present in the cytoplasm (Fig. 1E and SI Appendix, Fig. S1E). This is consistent with previous observation that 90% of AR-V7 is found in the nucleus and around 10% in the cytoplasm (38). While RFP-tagged AR-V7 formed condensates in cells, GFP-tagged AR in the presence of androgen R1881 had a punctate nuclear distribution (Fig. 1E and SI Appendix, Fig. S1D). RFP vector (fixed cells) and RFP-tagged AR (live cells) transfected into COS7 cells (SI Appendix, Fig. S1D) failed to form molecular condensates, suggesting that the condensates formed are characteristics of AR-V7. We also found that condensates formed at relatively low plasmid concentration of RFP-AR-V7 (SI Appendix, Fig. S1E). Since AR-V7 forms homodimers and heterodimers with other AR-SVs and AR (38, 39), we determined if AR-V7 changes the cellular distribution of AR. AR bound to androgen R1881 did not form condensates in the absence of AR-V7, but colocalized with AR-V7 as molecular condensates (Fig. 1E). Cells that express only GFP-AR showed nuclear punctate distribution, while cells that express both AR and AR-V7 together exhibited a different distribution. This suggests that AR-V7 is capable of promoting AR’s molecular condensate formation, although AR alone is incapable of forming condensates. RFP-AR-V7 mobility was determined by fluorescence-recovery after photobleaching (FRAP) in cells transfected with RFP-AR-V7 (Fig. 1F). RFP-AR-V7 that was laser photobleached recovered quickly, and this recovery was observed in the absence and presence of GFP-AR, indicating that AR-V7-AR heterodimer does not alter AR-V7 mobility.
Exposure of molecular condensates to 1,6 hexanediol results in their disruption (40). Treatment of cells expressing RFP-AR-V7 with 10% 1,6 hexanediol disrupted the condensates (Fig. 1G). Collectively, these data suggest that AR-AF-1 and AR-V7 exhibit IDP properties.
Discovery and Characterization of Selective AR-Irreversible Covalent Antagonists (SARICAs).
Chemical biology is a powerful approach that provides deeper understanding of structural and biological properties of proteins. Since many pharmacologically untargetable IDPs, including AR-SV are disease modifiers, alternate methods to target these proteins are necessary. One successful approach employed to target such proteins is to develop selective covalent binders or alkylating agents (41). About 30% of the drugs used clinically are covalent-binders that utilize electrophile–nucleophile interaction (42). Although covalent-binding molecules offer an attractive option, they also exhibit significant challenges due to cross-reactivity of the electrophile with proteins other than the intended target, resulting in off-target toxicity (42, 43).
Several of the 400 molecules synthesized in our SARD program degrade AR by binding to AF-1 (19–23). We hypothesized that covalent molecules belonging to the SARD structural series not only will potentially become next-generation therapeutics but will also increase our understanding of the AR AF-1 domain conformation, which is indispensable for AR and AR-SV function. To create the covalent molecules, we used two SARD molecules, UT-34 (20) and 21c (22) that exhibit excellent properties both in vitro and in vivo. The SARD molecules possess properties such as AR-NTD binding, degradation, long half-life, low clearance in metabolism assays, and optimum pharmacokinetic (PK) and pharmacodynamic (PD) properties. These molecules also inhibit prostate tumor xenografts in both castrated and intact animals.
Structure–activity relationship (SAR) to obtain covalent molecules with our first-generation SARDs (19, 21) failed to produce any active molecule, while SAR with our second-generation pyrazole SARDs such as UT-34 and 21c (20, 22) provided molecules that bind to AF-1 covalently. Utilizing UT-34 and 21c as starting point, we created a library of covalent molecules including UT-143 and UT-215, which were used in the following studies (Fig. 2A). We chose a carbon–carbon double bond, Michael addition, as an electrophile to form a covalent bond with nucleophilic amino acids (44, 45). The position of the carbon–carbon double bond on the molecule also required optimization and eventually found that the region that links the A-ring and B-ring is the optimum position to place the Michael addition. We synthesized ~25 molecules to obtain UT-143 and UT-215. Considering that the A and the B rings were optimized for our SARD program, the optimization was more focused on the position of the Michael addition. Since the SAR chemistry is beyond the scope of the present study, we will focus on the characterization of the two promising covalent molecules. To determine selectivity, the Michael addition electrophile was also added to an LBD-binding selective AR modulator (SARM), enobosarm (Enob; NB-enob, where NB=nonbinder), and to an intermediate chemical of SARD and SARM synthetic scheme, 153 (NB-153) (Fig. 2A) (22, 46). As exposed electrophiles are reactive, NB-153 does not contain a pyrazole ring and NB-enob where the pyrazole B ring was replaced with a 6-membered ring to serve as controls for the specificity of the Michael addition.
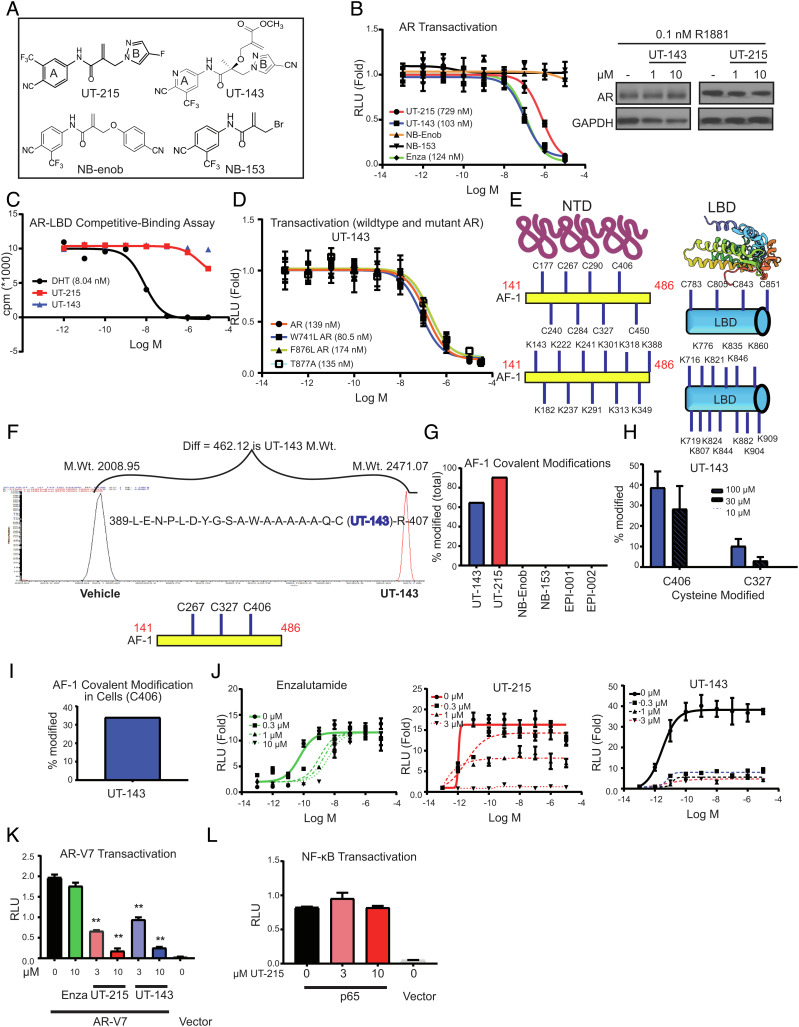
Fig. 2.
Discovery and characterization of SARICAs. (A) Chemical structures. (B) AR transactivation. COS7 cells plated in 24-well plates in DME + 5% csFBS without phenol red were transfected in quadruplicates with 0.25 µg GRE-LUC, 0.025 µg CMV-hAR, and 0.01 µg CMV-renilla-LUC. Cells in DME + 5% csFBS without phenol red were treated 24 h after transfection with a dose–response of the indicated compounds in the presence of 0.1 nM R1881. Dual luciferase assay was performed 24 h after treatment, and firefly luciferase values were normalized to renilla luciferase values. The data are average and SE of three biological replicates each performed in quadruplicates. Western blot. LNCaP cells maintained in 1% csFBS-containing medium for 2 d were treated with the compounds in the presence of 0.1 nM R1881. Cells were harvested 24 h after treatment, and the protein extracts were immunoblotted for AR and GAPDH. Representative blot is shown. (C) Competitive ligand-binding assay. Recombinant human AR-LBD protein (unpurified crude prep) was incubated with a dose–response of the indicated compounds in the presence of 6 nM 3H R1881 for 16 to 20 h. The 3H-bound protein captured by hydroxyapatite was washed to remove unbound radioactivity, and the bound 3H was counted using a scintillation counter. (D) Mutant AR transactivation. Transactivation assay was performed as indicated above with wild-type AR in the presence of 0.1 nM R1881 or mutant ARs in the presence of 0.3 nM R1881. The data are average and SE of three biological replicates each performed in quadruplicates. (E) Structure of AR NTD and LBD (for illustration purpose only) and the location of cysteines and lysines in the AF-1 and LBD (uniprot: P10275). (F) Mass Spectrometry profile and an example binding peptide sequence. Purified recombinant AR AF-1 protein (10 µg) was incubated in the presence or absence of 100 µM SARICAs for 12 to 16 h at 4 °C. Protein was digested with trypsin overnight at 4 °C, and the peptides were identified by mass spectrometry. An example peptide, its modification, and molecular weight shift by a SARICA are shown. The three modified amino acids C406 > C326 > C267 are shown below the Mass spectrometry profile. X-axis is m/z. (G) Quantification of modified AF-1. Mass spectrometry experiments were performed as indicated above with 100 µM indicated compounds. Percentage of the peptides covalently modified relative to unmodified peptides by various compounds was calculated and represented as bar graph (representative of n = 3 to 5). (H) Mass spectrometry experiment with dose–response of UT-143. Mass Spectrometry experiments were performed as indicated above with a dose–response of the compounds. Percentage of C406 and C327 modified is represented (n = 3 to 5). (I) Covalent modification in cells. COS7 cells were transfected with gal-4-AF-1. Cells were treated with 30 µM UT-143 in DME + 10%FBS for 2 h. AF-1 was immunoprecipitated with Gal-4 antibody, trypsinized, and mass spectrometry was performed. Percent-modified C406 AF-1 peptide is shown. (J) Schild plot for AR transactivation. COS7 cells were transfected in quadruplicates as indicated above. Cells in DME + 5% csFBS without phenol red were treated with a dose–response (1 pM to 10 µM) of R1881 in the presence or absence of the indicated doses of various compounds. Dual luciferase assay was performed 24 h after treatment. (K) SARICAs inhibit AR-V7 transactivation. COS7 cells in triplicates were transfected with 0.25 µg GRE-LUC, 0.025 µg pCDNA3.1 AR-V7 or vector, and 0.01 µg CMV-renilla LUC. Cells in DME + 10% FBS were treated 24 h after transfection, and luciferase assay was performed 48 h after transfection (n = 3). A representative of three independent experiments is shown. (L) The experiment was conducted as shown for AR-V7 with p65 and NFkB-LUC transfected instead of AR-V7 and GRE-LUC (n = 3). Numbers shown in graphs are IC50 values. RLU – relative light units; SARICA – selective AR irreversible covalent antagonist. AF-1 – activation function-1 domain; AR – androgen receptor; LBD- ligand-binding domain; GST – glutathione S transferase; C – cysteine; K – lysine; A – alanine. Values represented in bar graphs are average ± SE.
UT-215 and UT-143 inhibited AR transactivation with IC50 of 700 and 150 nM, respectively, while LBD-binding antagonist enzalutamide inhibited the AR transactivation at around 150 nM (Fig. 2 B, Left). NB-enob and NB-153 failed to inhibit the AR activity, suggesting that the carbon–carbon double bond electrophile in UT-215 and UT-143 is selective to the pyrazole B-ring in our scaffold. As enob functions as an agonist, NB-enob was also tested for agonist activity and found to be inactive as an agonist. The AR antagonistic activity of UT-215 and UT-143 was elicited without an effect on AR protein levels (Fig. 2 B, Right).
UT-215 and UT-143 were also tested in glucocorticoid receptor (GR) and progesterone receptor (PR) transactivation assays. While UT-215 affected agonist-induced PR and GR transactivation, UT-143 had no such effect (SI Appendix, Fig. S2A). These data suggest that the exposed electrophile in UT-215 is reactive with other receptors, while that in UT-143 is selective for AR. We evaluated the binding of UT-215 and UT-143 to AR-LBD using a competitive ligand-binding assay (Fig. 2C). Neither molecule bound to AR-LBD, while DHT exhibited a robust binding.
Since LBD mutations can potentially result in resistance to antagonists (47), UT-143 was tested in W741L (bicalutamide-resistant mutant), T877A (hydroxyflutamide-resistant mutant and the mutant expressed in LNCaP PCa cells), and F876L (enzalutamide- and apalutamide-resistant mutant) AR transactivation assays (34, 48). UT-143 inhibited the wildtype and mutant ARs at comparable IC50s (Fig. 2D).
UT-215 and UT-143 Covalently Bind to AR-AF-1.
Covalent molecules predominantly bind to cysteines and lysines (49). While the structures of agonist-bound AR LBD and DNA-bound DBD have been resolved (34, 35, 50, 51), the structure of AR AF-1 has not been resolved. Since the precursor or parent SARDs interact with AF-1 (19, 20), the SARICAs were screened for AF-1 binding. AF-1 contains 8 cysteines and 11 lysines, while the LBD contains 4 cysteines and 13 lysines (Fig. 2E). Purified recombinant AF-1 protein (10 µg) was incubated at 4 °C with UT-215 or UT-143 overnight, trypsinized and analyzed by HPLC-mass spectrometer (LC-MS/MS). UT-215 and UT-143 bound strongly to amino acids C406 and C327, but at best weakly to amino acid C267 in the AF-1 (Fig. 2F (shows representative profile of a peptide) and Table ST1). Despite the presence of 8 cysteines and 11 lysines, the SARICAs selectively bound to C406 and C327. While UT-215 and UT-143 bound to AF-1 covalently, NB-enob and NB-153 failed to bind to AF-1 (Fig. 2G). Despite over 75% homology in the structure between UT-215, UT-143, and NB-enob, the striking difference in binding to AF-1 emphasizes the importance of the pyrazole B-ring for this scaffold’s binding to AF-1.
We then grew the AF-1 plasmid in the presence of UT-143 in bacterial cultures to determine if the molecule binds to AF-1 in vivo. AF-1 protein synthesized in the presence of UT-143 was trypsinized and LC-MS/MS was performed. UT-143 clearly bound to amino acids C406 and C327 (SI Appendix, Fig. S2B). Subsequently, we synthesized and characterized over 20 related molecules with the same electrophile. The molecules that bound to AF-1 consistently interacted with C406 and C327. EPI-001 and EPI-002, two molecules that were reported to bind covalently to AF-1 domain (52, 53), failed to bind to AF-1 (Fig. 2G), which could reflect weak binding that was unable to withstand the LC-MS/MS conditions or the requirement for higher concentrations for this scaffold to bind to AF-1. Quantification of modified residues indicated that UT-215 and UT-143 modified 60 to 80% of C406 and C327 coding peptides (and a small percent of C267) (Fig. 2G). Cross-reactivity of UT-143 with other purified recombinant proteins was also evaluated. UT-143 (100 µM) only modified 2 to 5% of purified recombinant AR-LBD and GST (SI Appendix, Fig. S2C), confirming the selectivity of UT-143 to AF-1, especially to C327 and C406.
We also found that covalent molecules could be used in developing competitive binding assays to AF-1. Recombinant AF-1 protein was preincubated with UT-34 or enzalutamide for 2 h at 4 °C and then with UT-143 overnight at 4 °C. The trypsin-digested peptides were analyzed by LC–MS/MS. UT-143-dependent C406 and C327 modifications were significantly reduced by UT-34, but not by enzalutamide (SI Appendix, Fig. S2D). This suggests that UT-34 and UT-143 create a conformation that involves C406 and C327 or a region that engages these two cysteines.
A dose response of UT-143 was performed with the purified recombinant AF-1 protein. UT-143 bound both at 30 and 100 µM to C406 and C327 and modestly at 10 µM (Fig. 2H). At concentrations lower than 100 µM, no modification of proteins (PR-LBD, GST, or AR-LBD) other than AF-1 was observed with UT-143. The binding of UT-143 to AF-1 in cells was evaluated by transfecting AF-1 into COS7 cells, treating the cells with UT-143, and immunoprecipitating AF-1. UT-143 exhibited binding to C406 (Fig. 2I). Although the C406-encoding peptide was detected, the number of peptides detected by the mass spectrometer was a limiting factor. Mutation of C406, C327, and C267 to alanine (3C-A) resulted in a complete loss of UT-143 binding to recombinant AF-1, suggesting that UT-143 does not bind to other cysteines or lysines in the absence of these three amino acids (SI Appendix, Fig. S2E).
UT-215 and UT-143 Irreversibly Inhibit AR.
Covalent antagonists can be reversible or irreversible inhibitors, which can be evaluated using a Schild plot (54). In this assay, doses of an antagonist are evaluated in the presence of increasing doses of a competing agonist. If EC50 of the agonist increases with no effect on Emax, then the antagonist is competitive. If Emax of the agonist is reduced by the antagonist, then the molecule is irreversible. AR transactivation was performed with the indicated doses of UT-215, UT-143, or enzalutamide in the presence of a dose–response of an androgen. While increasing doses of enzalutamide resulted in an increase in R1881’s EC50, UT-215 and UT-143 reduced R1881’s Emax (Fig. 2J), showing that they are SARICA.
UT-215 and UT-143 Inhibit AR-V7 Transactivation.
Since UT-215 and UT-143 bind to AF-1, we evaluated the effect of these molecules in an AR-V7 transactivation assay. UT-215 and UT-143 dose-dependently inhibited the constitutively active AR-V7 transactivation, while enzalutamide, as expected, had no effect (Fig. 2K). To determine the selectivity of these molecules, a NF-kB transactivation assay with p65-induced NF-kB-LUC, was performed with UT-215. UT-215 had no effect on p65-induced NF-kB transactivation, confirming its selectivity for AR and AR-V7 (Fig. 2L).
SARICAs Change AF-1 Conformation, Resulting in AF-1 Stabilization, Phase-Separated Molecular Condensate Disruption, and Reduced Intracellular Mobility.
We evaluated the effect of SARICAs on the various IDP properties shown in Fig. 1. Purified recombinant AF-1 protein was incubated with UT-215, UT-143, enzalutamide, and EPI-002 for 4 h at room temperature. The samples were fractionated by SDS-PAGE and immunoblotted for AF-1. UT-215 or UT-143 stabilized the AF-1 protein, while enzalutamide (LBD-binding antagonist) and EPI-002 failed to alter AF-1 stability (Fig. 3 A, Left and SI Appendix, Fig. S2F). Interestingly, UT-143 caused a pronounced shift in AF-1 molecular weight, more than its molecular weight, suggesting a stoichiometry of more than one molecule of UT-143 bound per molecule of AF-1. These results suggest that UT-215 and UT-143 bind to AF-1 covalently and change conformation, resulting in AF-1 stabilization. Subsequently, equal amounts of AF-1 protein (5 ng) synthesized in the absence or presence of UT-215 or UT-143 were incubated at room temperature for 4 h and Western blot was performed. AF-1 grown in bacteria in the presence of UT-215 or UT-143 was stable (Fig. 3 A, Right), while AF-1 grown in the absence of SARICA was not detectable, confirming that UT-215 and UT-143 covalently bound AF-1 in vitro and in vivo. Similarly, incubation of AR-V7 recombinant protein (5 ng) with UT-215 or UT-143 for 4 h at room temperature stabilized the protein, while the LBD-binding antagonist enzalutamide failed to stabilize the AR-V7 protein (Fig. 3B). An interesting observation from these studies is that while UT-143 caused a molecular weight shift of AF-1 when incubated in vitro, UT-143 for unknown reasons failed to cause this shift of AR-V7 or minimally caused the shift when AF-1 was synthesized with UT-143.
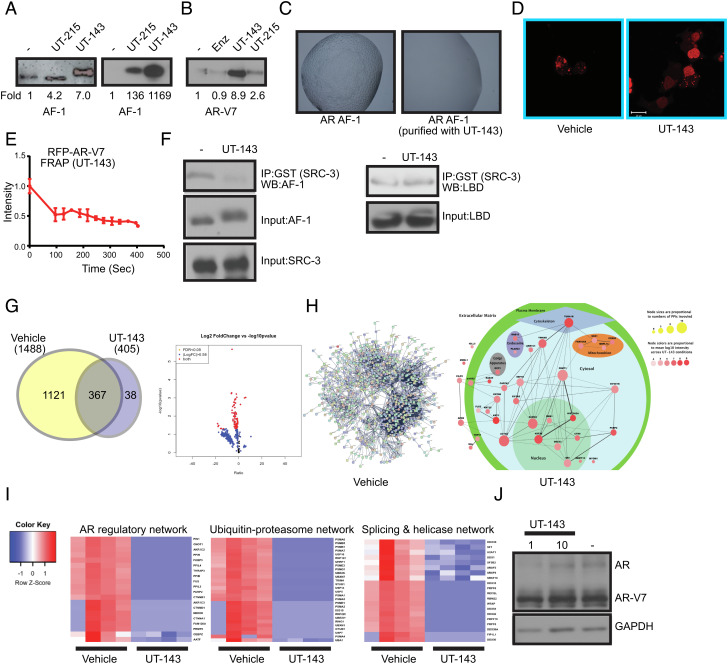
Fig. 3.
SARICA disrupts the disordered properties of AF-1 and AR-V7. (A) SARICAs stabilize AF-1 protein. (Left) Recombinant-purified AF-1 protein (5 ng) was incubated at room temperature for 4 h with DMSO or 50 µM of UT-143 and UT-215. Proteins were fractionated on an SDS-PAGE and western blot was performed with AR antibody (AR-441) that binds to the AF-1 region. The blots were quantified and expressed as fold change from vehicle controls. (Right) Bacterial cells expressing the AF-1 plasmid in the absence or presence of 30 µM UT-215 or UT-143 (added at the time of protein synthesis induction with 1 mM IPTG). Protein was purified and the purified protein (5 ng) was incubated at room temperature for 4 h. The proteins were fractionated on an SDS-PAGE and western blotted with AR-441 antibody. The blots were quantified and expressed as fold change from vehicle controls. (B) SARICAs stabilize AR-V7. Recombinant-purified AR-V7 protein (5 ng) was incubated at room temperature for 4 h in the absence or presence of 50 µM of the indicated compounds. Proteins were fractionated on an SDS-PAGE and western blot for AR-V7 was performed. The blots were quantified and expressed as fold change from vehicle controls. (C) UT-143 inhibits LLPS formation. AR AF-1 protein grown in the absence or presence of 30 µM UT-143 was incubated in the buffer shown in Fig. 1D. The LLPS formed was imaged using a microscope. (D) UT-143 dissolves AR-V7 condensates. COS7 cells were transfected with 1 µg turbo red RFP-AR-V7 and treated in DME + 10% FBS 24 h after transfection with vehicle or 10 µM UT-143 for 24 h. Cells were imaged using confocal microscope 24 h after treatment. (E) UT-143 blocks mobility of AR-V7. Photobleaching was performed in live cells that were treated with vehicle or 10 µM UT-143 and the recovery was monitored and graphed as described in Fig. 1F. Average and SE of n = 4 regions are presented. (F) UT-143 disrupts AF-1-coactivator interaction, but not AF-2-coactivator interaction. An equal amount of recombinant-purified AF-1 protein (Left) or AR LBD (Right) was incubated at 4 °C with DMSO or 100 µM UT-143 for 2 h and then incubated overnight at 4 °C with GST-tagged SRC-3-purified protein. The complex was immunoprecipitated with GST antibody and western blotted with AR-441 (for AF-1) or AR-C19 antibody (for LBD). Input controls show no change in the level of any protein. Representative experiment is presented here. (G–I) RIME assay. 22RV1 cells plated in DME + 10%FBS were treated with DMSO or 10 µM UT-143 for 16 h (n = 4/IP). Cells were fixed and the RIME assay was performed with AR-V7 of IgG antibody, and the protein complex (AR-V7 complex subtracted from IgG) detected by mass spectrometry. The data are represented as Venn diagram (G), volcano plot (G), and interacting proteins mapping (H). (I) Heatmap of selected pathways. (J) UT-143 does not change AR and AR-V7 protein expression. 22RV1 cells plated in DME + 10% FBS were treated as indicated under the RIME assay, and western blot for AR and AR-SV with an NTD-recognizing antibody was performed.
In light of these data, we hypothesized that SARICAs could change the conformation and alter the formation of LLPS condensates. LLPS molecular condensates were formed by the AF-1 protein purified in the absence of UT-143, while AF-1 synthesized and folded in the presence of UT-143 failed to form LLPS (Fig. 3C).
To test if AR-V7 condensate formation is also altered by SARICAs, COS7 cells transfected with RFP-tagged AR-V7 were treated with vehicle or UT-143. While AR-V7 formed molecular condensates in the presence of vehicle, AR-V7 was diffusely stained in the presence of UT-143 (Fig. 3D). FRAP was performed in COS7 cells transfected with RFP-AR-V7 and treated with UT-143 (Fig. 3E). UT-143 impaired the fluorescence recovery of AR-V7 after photobleaching.
UT-143-Induced AF-1 Conformation Results in Near Complete Dissociation of AR-V7-Interactome.
As IDR AF-1 is the main coactivator interacting surface of AR (6, 32), we hypothesized that a SARICA-induced conformation change and a change in its physicochemical properties will reprogram the AR and AR-V7 interactome. Purified recombinant AF-1 was incubated with recombinant GST-SRC-3 (steroid receptor coactivator 3) in the presence or absence of UT-143, and SRC-3 was immunoprecipitated and western blotted for AF-1. While the interaction was robust in vehicle-treated samples, the interaction was abrogated by UT-143 (Fig. 3 F, Left). To determine the selectivity of UT-143 to AF-1, the recombinant AR-LBD-purified protein that expresses AF-2 was incubated with GST-SRC-3 in the presence or absence of UT-143 under the same condition as in AF-1-SRC-3, SRC-3 was immunoprecipitated and western blotted for LBD. UT-143 failed to alter the LBD–SRC-3 interaction, which is further evidence for UT-143’s selectivity (Fig. 3 F, Right).
To determine the effect of UT-143 on AR-V7 interactome (55), 22RV1 PCa cells that express AR and AR-V7 were treated with vehicle or UT-143 and a rapid immunoprecipitation mass spectrometry of endogenous protein (RIME) assay was performed on AR-V7 and IgG- immunoprecipitates. AR-V7 complex (q < 0.05; ≥ 1.5-fold) with 1121 proteins was detected exclusively in the vehicle-treated samples, and 367 proteins were detected in both vehicle- and UT-143-treated samples. In contrast, AR-V7 interacted with only 38 proteins identified exclusively in UT-143-treated cells. Of the 367 proteins identified in both groups, the levels of 364 proteins were statistically significantly lower in the AR-V7 complex in UT-143-treated cells (Fig. 3 G–I). The shift in AR-V7 interactome occurred with no change in AR-V7 protein levels (Fig. 3J). Despite the dissociation of majority of the proteins from the AR-V7 complex, the gain in interaction of the 38 proteins in UT-143-treated cells is interesting. We mapped these proteins to eight subcellular localizations based on citations provided (genecards.com). The top pathways of the proteins that were dissociated from the AR-V7 complex by UT-143 include proteins that either function as AR-coactivators or that regulate AR-function, proteins belonging to the ubiquitin-proteasome pathway, and proteins involved in alternate splicing (Fig. 3I and SI Appendix, Fig. S3). U2AF1, PRPF6, SF-1, SRSF5, SF3B2, and other proteins that play a role in alternate splicing of AR were dissociated by UT-143 from the AR-V7 complex (56–59). The long-term consequence of reducing the interaction of several splicing-related proteins with AR needs further evaluation. The importance of these interacting proteins for AR and AR-V7 function will need to be evaluated in future studies. Although full-length AR was detected in both vehicle and UT-143-treated interactomes, its interaction with AR-V7 was significantly reduced by UT-143.
UT-143 Reprograms AR-V7 Chromatin Accessibility and Alters 22RV1 Transcriptome.
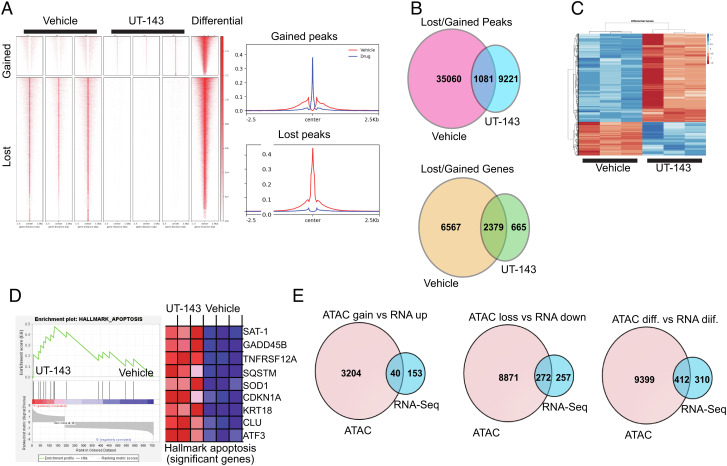
Considering the remarkable effect of UT-143 on AR-V7 interactome, we evaluated the effect of UT-143 on genome-wide chromatin accessibility by ATAC-seq in 22RV1 cells under the same conditions used for RIME (Fig. 3 G–I). UT-143 significantly reduced chromatin accessibility, indicating a marked chromatin remodeling (Fig. 4A). Vehicle-treated cells exhibited 36,141 accessible regions, while chromatin from UT-143-treated cells expressed only 10,302 accessible regions (Fig. 4B). One-thousand eighty-one (1081) regions were accessible in both conditions, with majority of them down-regulated by UT-143 (Fig. 4B). Annotating the peaks showed a dramatic reduction in the number of genes in UT-143-treated cells (Fig. 4B). No major shift in the regions the peaks were detected was observed due to UT-143 (SI Appendix, Fig. S4A). The accessible chromatin in vehicle-treated cells was enriched for SP2, ELF4, and E2F, while that in UT-143-treated cells was enriched for CCAAT enhancer-binding transcription factors, SP2, and ELK1 (SI Appendix, Fig. S4B). While SP2, ELK4, and E2F are implicated in tumor growth and metastasis, CCAAT-binding transcription factors have been found to be good prognostic indicators in cancer (60–63). The genome browser tracks are shown in SI Appendix, Fig. S4C. Gene ontology classifications are shown in SI Appendix, Fig. S4D.
Fig. 4.
UT-143 alters AR-V7 chromatin accessibility and transcriptome. (A and B) ATAC-Seq. 22RV1 cells (n = 3/group) plated in RPMI + 10%FBS were treated with vehicle or 10 µM UT-143 as indicated for the RIME assay, and ATAC-seq was performed. Heatmap (A) and lost and gained peaks and annotated genes (B) are shown. (C) RNA-seq was performed in 22RV1 cells (n = 3/group) under conditions described for ATAC-seq. Heatmap (C) and GSEA plot and the corresponding heatmap for the hallmark apoptosis pathway (D) are shown. (E) Venn diagram for shared and distinct ATAC-seq annotated peaks and RNA-seq genes are shown.
Overlaying the RIME data with ATAC-Seq produced a 50% overlap between the annotated genes and proteins that were lost due to UT-143 treatment (SI Appendix, Fig. S5B; Only 911 proteins were converted into gene IDs). Of the over 400 genes shared between ATAC and RIME, 389 genes associated with 2425 high-confidence interactions. These nodes are represented in the Euclidian heatmap (SI Appendix, Fig. S5C). The group of 389 genes was ranked along X and Y axes. Each spot (X, Y) in the matrix represents the confidence score for the PPI connecting genes X and Y, and only PPIs with confidence score>0.7 were recorded. Hierarchical clustering using the Euclidean distance was carried out along both rows and columns of the matrix, clustering highly connected genes into small groups. Two groups are highlighted. Group 1 was highlighted by a sky blue box in the heatmap, consisting of 48 genes with 644 PPIs and enriched with RNA handling genes, while Group 2 is represented by a dark green box in the heatmap, including 21 genes with 117 PPIs and enriched with pathways like TCA cycle (SI Appendix, Fig. S5A).
To determine if the change in the chromatin accessibility translates into a change in the expression of genes, an RNA-sequencing was performed under the same condition as in ATAC-seq (Fig. 4C). UT-143 up-regulated 193 unique genes and down-regulated 529 unique genes (q < 0.05; fold >=1.5). Genes belonging to DNA replication and repair, and spliceosomes were highly significantly altered by UT-143 (SI Appendix, Fig. S6 A–C). AR-target genes, including FKBP5 and NKX3.1 are some of the top down-regulated genes by UT-143. GSEA analysis showed that hallmark apoptosis and p53 pathways were the top two pathways (SI Appendix, Fig. S6B). All significantly altered genes in the hallmark apoptosis pathways were up-regulated by UT-143 (Fig. 4D). Interestingly the ATAC-seq annotated genes and RNA-seq genes showed that more than 50% of the lost/down-regulated and differentially regulated genes and one-third of the up-regulated genes overlapped, indicating a good concordance between the chromatin accessibility and gene expression (Fig. 4E).
To determine the effect of UT-143 on chromatin accessibility in a cell line that expresses AR, but not AR-SV, LNCaP cells were treated with vehicle or UT-143 under the same condition as in 22RV1 cells and an ATAC-seq was performed (SI Appendix, Fig. S7 A–E). UT-143-treated cells lost 52,272 peaks and gained 25,020 peaks. UT-143 caused a downregulation of 20,166 chromatin regions and an upregulation of 6,097 chromatin regions. Unlike 22RV1 chromatin accessibility where 70% of the peaks were located to promoter region, in LNCaP cells only 10 to 15% of the peaks were mapped to the promoter region. Also, one of the top down-regulated and lost motifs in LNCaP cells is GRE/PRE, demonstrating an effect on AR consensus sequence sites. IGV tracks for AR-regulated FKBP5 shows the loss of chromatin accessibility in UT-143-treated cells. Overlaying the 22RV1 and LNCaP ATAC-seq annotated genes, shows a significant overlap of over 30 to 50% of genes between the two cell lines, demonstrating a consistency in UT-143’s action.
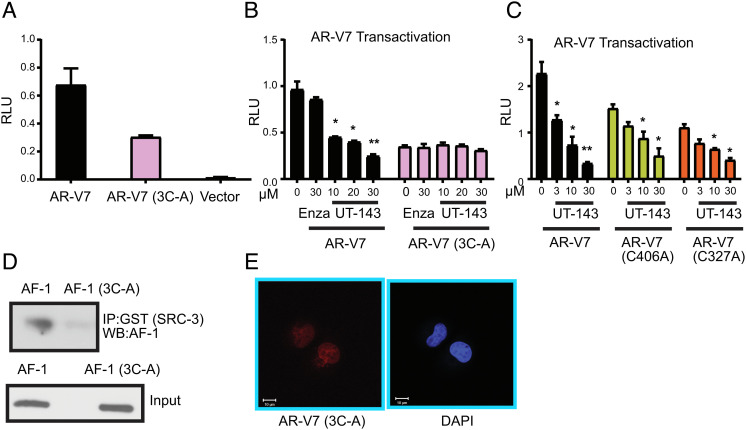
C406, C327, and C267 Are Important for AR-V7 Stability, Molecular Condensate Formation, and Function.
Selective binding of UT-143 to C406, C327, and C267 and the resulting conformation change, and inhibition of AR and AR-V7 function suggest the importance of these amino acids and this region for AR and AR-V7 function. The three amino acids were mutated to alanine (3C-A), and the effect of mutation on AR-V7 conformation, function, and molecular condensate formation was evaluated. Mutation of the three cysteines in AR-V7 exhibited a 50 to 60% decrease in AR-V7 activity (Fig. 5A). Consistent with this result and the lack of binding to 3C-A AF-1 (SI Appendix, Fig. S2E), UT-143 antagonized AR-V7, but did not affect AR-V7 3C-A transactivation (Fig. 5B). This result suggests that binding to these amino acids is critical for AR-V7 inhibitory activity of SARICAs.
Fig. 5.
Characterization of mutant AR-V7. (A) AR-V7 3C-A transactivation. COS7 cells were transfected with 0.25 µg GRE-LUC, 0.025 µg AR-V7, AR-V7 3C-A, or vector, and cmv-renilla-LUC. Cells were maintained in DME + 10% FBS and were harvested 48 h after transfection and dual luciferase assay was performed (n = 3). (B) UT-143 inhibits AR-V7, but not AR-V7 3C-A transactivation. COS7 cells were transfected with AR-V7 or AR-V7 3C-A mutant and treated in DME + 10% FBS as indicated in the figure. Dual luciferase assay was performed 24 h after treatment (n = 3). (C) Single-point mutation of cysteines in AF-1 inhibits AR-V7 transactivation but does not reverse UT-143 antagonism. COS7 cells were transfected with AR-V7 wildtype, AR-V7 C406A, or AR-V7 C327A and treated with UT-143 in DME + 10% FBS. Dual luciferase assay was performed 24 h after treatment (n = 3). (D) Mutating three cysteines in AF-1 results in its failure to interact with SRC-3. AF-1 or AF-1 3C-A-purified recombinant protein was incubated with recombinant GST-SRC-3 at 4 °C overnight. SRC-3 was immunoprecipitated with GST antibody, and western blot for AF-1 was performed. (E) AR-V7 3C-A fails to form molecular condensate. AR-V7 3C-A was transfected into COS7 cells. Cells were fixed, immunostained with AR-V7 antibody, and images obtained using confocal microscope. Experiments were performed at least three independent times. AR – androgen receptor; V7 3C-A – AR-V7 with C267, C327, and C406 mutated to alanine; C406A – Cysteine 406 mutated to alanine; C327A – Cysteine 327 mutated to alanine; C267A – Cysteine 267 mutated to alanine.
Since the triple C-A mutation decreased AR-V7 transactivation, we created single-point mutations of C406 and C327 and performed AR-V7 transactivation assays. Both C406A and C327A mutations reduced AR-V7 transactivation by ~40 to 50% (Fig. 5C). However, UT-143 retained its antagonistic effects, suggesting that the molecule can function if one of the binding cysteines is available. AR or 3C-A AR Schild plot showed that UT-143 inhibited R1881 Emax in AR, but not in 3C-A AR (SI Appendix, Fig. S8), confirming the importance of the three cysteines for SARICA’s effect.
We reasoned that if these three cysteines are important for AF-1 conformation, then they should be critical for AF-1 interaction with cofactors. Purified recombinant AF-1 or AF-1 3C-A was incubated with purified recombinant GST-SRC-3 overnight at 4 °C (AF-1 and AF-1 3C-A are stable at 4 °C), SRC-3 was immunoprecipitated with GST antibody, and western blot for AF-1 was performed (Fig. 5D). As expected, AF-1 interacted robustly with SRC-3, but mutating the three cysteines to alanine resulted in lack of interaction with SRC-3. AR-V7 3C-A mutant failed to form molecular condensates (Fig. 5E), suggesting the importance of these three amino acids for molecular condensate LLPS formation.
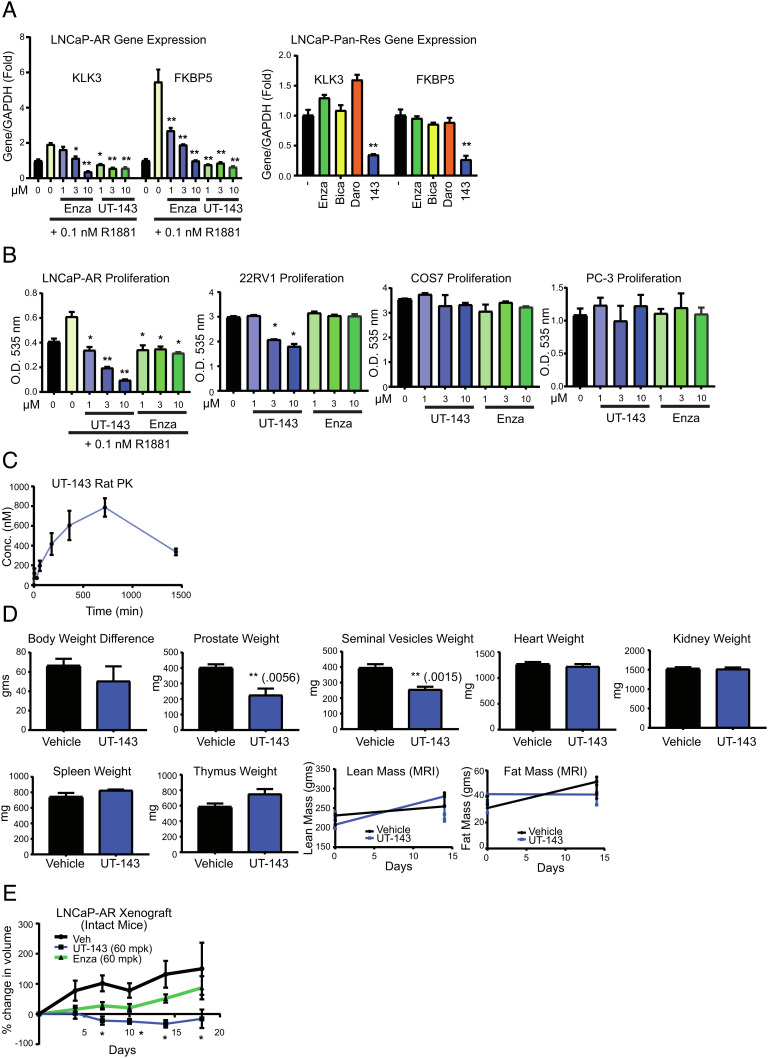
UT-143 Inhibits AR-Target Gene Expression and Proliferation of PCa.
Due to covalent binding of AF-1 and robust inhibition of AR and AR-V7 function, these molecules will be appropriate to be tested in PCa models. UT-143 and enzalutamide dose-dependently inhibited the R1881-induced expression of PSA (KLK3) and FKBP5 in LNCaP PCa cell line (Fig. 6 A, Left). Long-term culture of LNCaP cells sequentially with 10 µM enzalutamide, bicalutamide, and darolutamide created a multidrug-resistant LNCaP cells. Incubation of these cells with various compounds showed that only UT-143 inhibited the expression of AR-target genes, while LBD-binding antagonists failed to inhibit the expression of these genes (Fig. 6 A, Right). PCa cell proliferation results are consistent with the gene expression results with UT-143 inhibiting the proliferation of AR-positive LNCaP-AR and AR- and AR-SV-positive 22RV1 cells, but not AR-negative PCa PC-3 cells and non-PCa COS7 cells (Fig. 6B).
Fig. 6.
UT-143 inhibits AR function, PCa cell proliferation and tumor growth. (A) UT-143 inhibits AR-target gene expression in PCa cells. LNCaP cells (Left) maintained in RPMI + 1% csFBS without phenol red or LNCaP cells that are resistant to AR antagonists (LNCaP pan-res; Right) maintained in RPMI + 10% FBS for 2 d were treated with various compounds (10 µM for LNCaP-pan resistant) as indicated in the presence or absence of 0.1 nM R1881. RNA was extracted and expression of KLK3 and FKBP5 was measured by real-time PCR and normalized to GAPDH expression (n = 4). (B) UT-143 selectively inhibits PCa cell proliferation. AR-positive (LNCaP and 22RV1), AR-negative (PC-3) PCa cells, and non-cancerous (COS7) cells were treated in various media (LNCaP in 1% csFBS-containing medium; 22RV1, PC-3, and COS7 cells in 10% FBS-containing medium) for 6 d with medium change and re-treated after 3 d. Cells were fixed and SRB assay was performed (n = 4). (C) PK properties of UT-143. Male Sprague–Dawley rats (n = 6) were treated once orally with 20 mg/kg UT-143 (20% DMSO + 80% PEG-300). Blood was collected at the indicated time points and serum concentration of UT-143 was measured using LC–MS/MS (n = 3/time point). (D) Hershberger PD assay with UT-143. Male Sprague–Dawley rats (n = 5/group) were treated orally with vehicle (20% DMSO + 80% PEG-300) or 20 mg/kg UT-143 for 13 d. Body weight, lean mass, and fat mass (EchoMRI) were recorded at study initiation and at killing. Animals were killed on day 14, and prostate, seminal vesicles, and indicated organs were weighed. (E) UT-143 inhibits LNCaP-AR xenograft in intact mice. LNCaP-AR cells were implanted subcutaneously in male NSG mice (n = 8 to 10/group). Once the tumors grow to 100 to 300 mm3, the animals were randomized and treated orally with vehicle (40% DMSO + 80% PEG-300), 60 mg/kg/day enzalutamide or UT-143. Tumor volume was measured twice weekly and represented as % change from day 0 of treatment initiation. Values are expressed as average ± SE. Enza-enzalutamide; pan-res-pan antagonist resistant; MRI-magnetic resonance imaging.
PK and PD Effects of UT-143.
Male Sprague–Dawley rats orally received a single dose of 20 mg/kg UT-143, and blood was collected at different time points to determine the PK properties. Serum was separated, and concentration of UT-143 was quantified using the LC-MS/MS method. UT-143 was orally bioavailable with a half-life of greater than 12 h (Fig. 6C). We also performed a Hershberger PD assay in which the effect of AR-targeting molecules on androgen-dependent tissues such as prostate and seminal vesicles are evaluated. Male Sprague–Dawley rats (n = 5/group) were treated with vehicle or 20 mg/kg UT-143. Body weight and composition to determine lean and fat mass were recorded on day 1 and at killing on day 14. Animals were killed after 13 d of treatment, and weights of androgen-target tissues prostate and seminal vesicles were recorded. To determine potential off-target toxicity, the weights of the heart, kidney, spleen, and thymus were determined. UT-143 had no effect on body weight or composition or weights of the kidney, heart, spleen, and thymus, while it significantly inhibited the weights of the androgen-target prostate and seminal vesicles (Fig. 6D). The experiment was repeated to compare UT-143 with one of the precursor SARDs, UT-34, in the Hershberger assay. The effect of UT-143 was comparable to the effect observed with UT-34 (SI Appendix, Fig. S9A).
In Vivo Effects of UT-143.
Tumor xenograft studies were performed in immunocompromised mice. LNCaP-AR cells were implanted subcutaneously in male NSG mice. Once the tumors grew to 100 to 300 mm3, the animals were randomized and treated orally with vehicle, 60 mg/kg/day UT-143 or enzalutamide for 18 d. Since this study was performed in intact noncastrated animals, enzalutamide was ineffective potentially due to its inability to compete with circulating androgen excess. UT-143 effectively inhibited the tumor growth with greater than 95% tumor growth inhibition (Fig. 6E). UT-143 at 60 mg/kg/day was also tested in the AR-negative PC-3 cell line tumor xenograft model (SI Appendix, Fig. S9B). UT-143 did not affect the tumor growth, suggesting that the inhibition of tumor growth is selective to AR-positive tumors.
Discussion
The results herein advance our understanding of the AF-1 IDR in AR-SVs and AR, which are important drug targets. First, SARICAs clearly demonstrate the importance of the pyrazole ring in our scaffold for AF-1 binding, while other molecules with a six-membered ring or no B-ring were unable to bind to this region. The direct evidence provided by mass spectrometry demonstrate small molecule AR AF-1-binding SARICAs are a class of targeted molecules for AR. The compounds bound to already synthesized protein as well as protein actively being synthesized, in part through amino acids C406 and C327 of AF-1. We found that small molecule covalent binders of IDPs can change physicochemical properties such as LLPS and molecular condensate formation. The approach shown in the manuscript could be applied to other IDPs to elucidate structural information.
Covalent-binding molecules offer several advantages over competitive antagonists or inhibitors (64). The efficacy of competitive antagonists can be reduced by increasing substrate or agonist levels, thereby displacing the antagonists by breaking the weak noncovalent hydrophobic and hydrogen bonds (42, 64). This competition between antagonists and agonists will result in a dynamic equilibrium, which could provide diseases, especially proliferative diseases such as cancer, an opportunity to develop alternate pathways to displace the antagonists. In contrast, covalent irreversible antagonists permanently bind to a protein with tenfold higher energy than hydrogen bonds, which can be displaced only by recycling the protein (42, 64).
Binding of all the active covalent compounds in our library to the same amino acids provides proof for a binding region in AF-1. For a covalent molecule to be effective and selective, it first has to bind noncovalently to a binding region and then form a covalent bond with an appropriate nucleophile (65). To achieve selectivity, the noncovalent binding of a molecule to the region should be strong to promote selective covalent binding to the nearby nucleophile. The results observed with UT-143 clearly suggest that the SARD backbone of the molecule binds to a binding region noncovalently with high affinity, before engaging the nucleophiles C327 and C406 for covalent binding. Results with NB-enob and NB-153 indicate that the pyrazole B-ring of the SARDs is critical for burying the SARICAs to a binding region in the AF-1 domain, resulting in the carbon–carbon double bond Michael addition with C406 and C327.
The results herein: 1) provide first-hand evidence for the existence of a binding region in the AR NTD that has only been speculated using indirect biophysical analysis; 2) indicate that the apo conformation of AR-NTD and AR-V7 has an exposed binding region that facilitates binding of SARICAs to C327 and C406 for Michael addition; 3) show that the pyrazole B-ring in our scaffold is important for AF-1 binding and covalent bond formation, since NB-enob and NB-153 failed to demonstrate any binding; and D) show that C327 and C406 are important for the function of AR-SV.
Cysteine is a rare amino acid, constituting only 2 to 3% of the human proteome (66, 67), while lysine is one of the most abundant amino acids, constituting 6 to 7% of the human proteome (67, 68). Since disulfide bonds are critical for the integrity of proteins, cysteines are usually buried in folded proteins, making it difficult for covalent molecules to gain access. SARICAs failed to interact with any of the 11 lysines in AF-1 and interacted with only two specific cysteines, C327 and C406. Also, of the 8 cysteines in AF-1, the molecules interacted selectively with C327 and C406. The binding to these two cysteines was not unique to UT-215 and UT-143, as over 10 molecules belonging to this scaffold bound precisely to them. These results suggest that binding to the AF-1 region only exposes C327 and C406, facilitating their interaction with the molecules.
Using methods such as LLPS and protein stability helped us to demonstrate the potential change in AF-1 conformation by SARICAs. Previous studies have not demonstrated ligand-dependent disruption of LLPS. LLPS condensates form at superenhancer sites and are typically formed by IDPs in complex with coactivators such as MED1 (6). Many of the deregulated processes in cancer occur in LLPS condensates (7). Disruption of LLPS condensates by SARICAs is an indication of change in conformation, resulting in dissociation of coactivators and inactivation of the protein. Interestingly, recombinant AR AF-1 formed LLPS condensates in the absence of any coactivator or DNA, suggesting that the highly disordered AF-1 region (>90% disorder score) forms the LLPS condensates without requiring any other interacting protein. Most importantly, the SARICAs serve as an excellent molecular probe to confirm the biophysical data and solidify our pyrazole scaffold as AF-1-binding small molecules.
The conformation change of the AF-1 region in AR-V7 forced by UT-143 resulted in a dramatic dissociation of AR-V7 interactome, transcriptome, and chromatin accessibility in 22RV1 and chromatin accessibility in LNCaP cells. The downstream effect of conformational change and condensate disruption has far-reaching consequences on AR-V7 function.
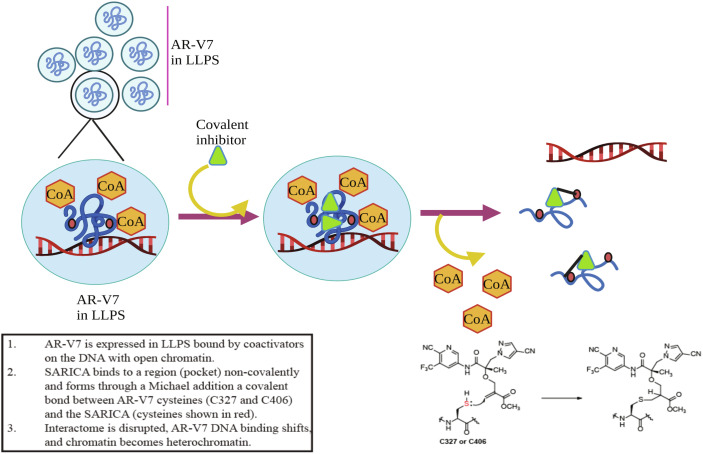
Based on these observations, we propose a model that rationalizes the binding of covalent molecules selectively to AR AF-1 or AR-V7. AF-1 exists in biomolecular condensates with the binding region exposed for SARICAs to bind (Fig. 7). Binding of SARICAs to AF-1 changes conformation and the physicochemical properties, dissociates interactome, and changes chromatin landscape. Subsequently, the electrophile binds to exposed nucleophiles C327 and C406 by Michael addition, leading to the inhibition of AF-1 function.
Fig. 7.
Model. AR AF-1 or AR-V7 is maintained in a disordered conformation in molecular condensates in complex with coactivators with a binding region exposed and the C327 and C406 (shown in red dots) buried inside. SARICA binding to the binding region leads to conformation change and dissolution of the condensates. This leads to dissociation of coactivators, exposure of C327 and C406, reorganization of DNA-binding, and change in DNA from euchromatin to heterochromatin. These sequence of events results in inhibition of PCa cell proliferation and tumor growth. SARICA binds to the two cysteines by Michael addition, resulting in inactivation of the region. CoA – coactivator, LLPS – liquid–liquid phase separation.
Binding of the SARICAs to both C327 and C406 might provide a fail-safe therapeutic opportunity as mutating two sites in a protein region in advanced cancer to overcome inhibition is rare. Our data suggest that SARICA still retains its AR-V7 antagonism, when one site is mutated.
The studies presented in this manuscript demonstrate the development of AF-1-binding AR-selective covalent binders and provide evidence for the existence of a binding region in the AF-1 domain that could be utilized to inhibit AR-SVs and other aggressive cancers that express AR-SVs. Considering that IDPs have no stable structure, this potential binding region could be unique to the SARICA scaffold and not universal. This hypothesis must be tested with other molecules belonging to other scaffolds that bind to this region, and the data need to be validated in clinical setting to determine if C406 and C327 are exposed in PCa patients.
Materials and Methods
All animal studies were performed under protocols approved by the institutional animal care and use committee (IACUC) of the University of Tennessee Health Science Center (UTHSC). Animals were quarantined for 3 to 5 d before initiation of any procedure. Animals were provided food and water ad libitum and were maintained in 12-h light–dark cycle.
Materials.
Details of the materials are provided in SI Appendix, Table ST2. Data from RNA-seq and ATAC-seq are deposited in GEO database. The GEO accession number is GSE215335.
Cell Culture.
LNCaP, COS7, and 22RV1 cells were procured from American Type Tissue Culture (ATCC, Manassas, VA). Cells were cultured as recommended by ATCC. Cells were authenticated by short terminal DNA repeat assay and were frequently checked for mycoplasma contamination. LNCaP-AR cells were provided by Dr. Charles Sawyers (Memorial Sloan Kettering Cancer Center, New York, NY) (11). LNCaP cells resistant to enzalutamide, bicalutamide, and darolutamide were obtained by culturing the cells with the three AR antagonists sequentially until the cells become resistant to all three antagonists.
Plasmids.
Several of the plasmids (GRE-LUC, CMV-renilla LUC, AR, W741L AR, F876L AR, T877A, PR, GR, AR AF-1, and AR NTD) used in this manuscript were described previously (19, 20). pCDNA3.1 AR-V7, turbo-red AR-V7, and His AR-V7 were kindly provided by Dr. Yan Dong (Tulane University, New Orleans) (38). PGEX 6p (AR AF-1, SRC-3, AR-V7, AR-LBD, AR-NTD, RFP-AR) and various mutant plasmids were created in-house.
Transactivation Assay, Protein Purification, Cell Proliferation, Western Blot, RNA Extraction, and Real-Time PCR.
were all described previously (19, 20). Mass spectrometry conditions are provided in the SI Appendix, Supplemental Methods section.
Fluorescent Microscopy and FRAP.
Microscopy was performed in live cells or in cells that were fixed and counter-stained with DAPI using Zeiss 710 confocal microscope. For FRAP, selected regions were photobleached until 70 to 90% of the signal was bleached, and the recovery was monitored.
LLPS.
AR AF-1 protein (1 mg/ml) purified in the presence or absence of 100 µM UT-143 was incubated in the respective buffer for 6 h. LLPS formation was photographed.
PK Assay.
Male Sprague–Dawley rats (n = 6) were weighed and dosed orally with 20 mg/kg of compound. Blood was collected at various time points (n = 3/time point), and serum separated. Sample preparation and LC–MS/MS condition are provided in the SI Appendix, Supplement Methods.
Hershberger Assay.
Male Sprague–Dawley rats were dosed orally with vehicle or the indicated compounds. Animals were weighed and body composition (EchoMRI) recorded at the start of the experiment and on day 14 before killing. The prostate, seminal vesicles, and other organs were isolated and weighed.
Xenograft.
Cells (1:1 mixture in growth medium and Matrigel) were implanted subcutaneously in male NSG mice. Once the tumors grow to 100 to 300 mm3, animals were randomized and treated orally with vehicle (40% DMSO + PEG-300) or the indicated doses of the compounds. Tumor volume was recorded twice weekly. Body weights were recorded at the beginning and at the end of the study. Animals were killed, tumors isolated, and were stored for future analysis.
RIME assay, ATAC and RNA-seq, and competitive ligand-binding assay methods are provided in supplement material. All experiments were repeated at least three independent times. Animal experiments were conducted with n = 5 to 10 animals/group, depending on the type of experiment. Data are represented as mean ± SE. Experiments with two groups were analyzed by t test, while experiments with more than two groups were analyzed using a one-way ANOVA, followed by TUKEY post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001.
Data, Materials, and Software Availability
ATAC-Seq, RNA-seq data have been deposited in Geo (GSE215335). All study data are included in the article and/or SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Drs. T.J. Hollingworth and Esther Marquez-Wilkins, and Ms. Michelle Sims (deceased) for their help with fluorescent microscopy and FRAP. We thank the UTHSC neuroscience imaging core for extending the microscope facility for our studies. We thank the UTHSC institutional proteomics core and its director, Dr. David Kakhniashvili for help with the covalent binding studies and the UTHSC MRC and its Director Dr. William Taylor for sequencing. We thank Dr. Ronald N. Laribee for his productive discussion and useful comments on the project.
Author contributions
E.F.P., D.D.M., and R.N. designed research; T.T., S.P., D.-J.H., Y.H., S.A., K.L.Y., V.B., W.Z., A.K.J., D.D.M., and R.N. performed research; D.-J.H., Y.H., L.M.P., and D.D.M. contributed new reagents/analytic tools; T.T., S.A., D.L.J., A.K.J., Z.Y., and R.N. analyzed data; and L.M.P., D.D.M., and R.N. wrote the paper.
Competing interest
The authors declare a competing interest. The authors have organizational affiliations to disclose, R.N. is a paid consultant to Oncternal therapeutics Inc., San Diego, CA, the licensee of the SARICA program described in the manuscript. Yes, the authors have patent filings to disclose, S.P., D.-J.H., Y.H., D.D.M., and R.N. are inventors in the patent filings related to SARICAs. Yes, the authors have research support to disclose. This work was supported by research grants from the National Cancer Institute (NCI) R01-CA229164, R01-CA229164-02S1, R01-CA229164-02S2, and R01-CA253329, Department of Defense (DOD) W81XWH-21-1-0055, and Muirhead Chair endowment.
Footnotes
This article is a PNAS Direct Submission.
Supporting Information
References
- 1.C. International Human Genome Sequencing, Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Hajduk P. J., Huth J. R., Tse C., Predicting protein druggability. Drug. Discov. Today 10, 1675–1682 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Mitchell P. J., Tjian R., Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 245, 371–378 (1989). [DOI] [PubMed] [Google Scholar]
- 4.Wright P. E., Dyson H. J., Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 293, 321–331 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Dunker A. K., Cortese M. S., Romero P., Iakoucheva L. M., Uversky V. N., Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 272, 5129–5148 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Boija A., et al. , Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855 e1816 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boija A., Klein I. A., Young R. A., Biomolecular condensates and cancer. Cancer Cell 39, 174–192 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alberti S., Gladfelter A., Mittag T., Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176, 419–434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J., et al. , Emerging roles of liquid-liquid phase separation in cancer: From protein aggregation to immune-associated signaling. Front. Cell Dev Biol. 9, 631486 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antonarakis E. S., et al. , AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 371, 1028–1038 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran C., et al. , Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 324, 787–790 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scher H. I., et al. , Antitumour activity of MDV3100 in castration-resistant prostate cancer: A phase 1–2 study. Lancet 375, 1437–1446 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryan C. J., et al. , Abiraterone in metastatic prostate cancer without previous chemotherapy. N. Engl. J. Med. 368, 138–148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chamberlain N. L., Whitacre D. C., Miesfeld R. L., Delineation of two distinct type 1 activation functions in the androgen receptor amino-terminal domain. J. Biol. Chem. 271, 26772–26778 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Armstrong A. J., et al. , Prospective multicenter validation of androgen receptor splice variant 7 and hormone therapy resistance in high-risk castration-resistant prostate cancer: The PROPHECY study. J Clin. Oncol. 37, 1120–1129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Armstrong A. J., et al. , Prospective multicenter study of circulating tumor cell AR-V7 and taxane versus hormonal treatment outcomes in metastatic castration-resistant prostate cancer. JCO Precis Oncol. 4, PO.20.00200 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hornberg E., et al. , Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS One 6, e19059 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiliccioglu I., Bilen C. Y., Sozen S., Konac E., Upregulation of potential regulatory signaling molecules correlate with androgen receptor splice variants AR-V7 and AR-V567es in prostate cancer metastasis. Gene 772, 145377 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Ponnusamy S., et al. , Novel selective agents for the degradation of androgen receptor variants to treat castration-resistant prostate cancer. Cancer Res. 77, 6282–6298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ponnusamy S., et al. , Orally-bioavailable androgen receptor degrader, A potential next-generation therapeutic for enzalutamide-resistant prostate cancer. Clin. Cancer Res. (2019), 10.1158/1078-0432.CCR-19-1458. [DOI] [PubMed] [Google Scholar]
- 21.Hwang D. J., et al. , New generation of selective androgen receptor degraders: Our initial design, synthesis, and biological evaluation of new compounds with enzalutamide-resistant prostate cancer activity. J. Med. Chem. 62, 491–511 (2019). [DOI] [PubMed] [Google Scholar]
- 22.He Y., et al. , Pyrazol-1-yl-propanamides as SARD and pan-antagonists for the treatment of enzalutamide-resistant prostate cancer. J. Med. Chem. 63, 12642–12665 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Y., et al. , Exploration and biological evaluation of basic heteromonocyclic propanamide derivatives as SARDs for the treatment of enzalutamide-resistant prostate cancer. J. Med. Chem. 64, 11045–11062 (2021), 10.1021/acs.jmedchem.1c00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersen R. J., et al. , Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell 17, 535–546 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Pomerantz M. M., et al. , Prostate cancer reactivates developmental epigenomic programs during metastatic progression. Nat. Genet. 52, 790–799 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baca S. C., et al. , Reprogramming of the FOXA1 cistrome in treatment-emergent neuroendocrine prostate cancer. Nat. Commun. 12, 1979 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmed J., Meszaros A., Lazar T., Tompa P., DNA-binding domain as the minimal region driving RNA-dependent liquid-liquid phase separation of androgen receptor. Protein. Sci. 30, 1380–1392 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bielskute S., et al. , Low amounts of heavy water increase the phase separation propensity of a fragment of the androgen receptor activation domain. Protein. Sci. 30, 1427–1437 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suskiewicz M. J., Sussman J. L., Silman I., Shaul Y., Context-dependent resistance to proteolysis of intrinsically disordered proteins. Protein. Sci. 20, 1285–1297 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsvetkov P., et al. , Operational definition of intrinsically unstructured protein sequences based on susceptibility to the 20S proteasome. Proteins 70, 1357–1366 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Sabari B. R., et al. , Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu X., et al. , Structural insights of transcriptionally active, full-length androgen receptor coactivator complexes. Mol. Cell 79, 812–823 e814 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xue B., Dunbrack R. L., Williams R. W., Dunker A. K., Uversky V. N., PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta. 1804, 996–1010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bohl C. E., Miller D. D., Chen J., Bell C. E., Dalton J. T., Structural basis for accommodation of nonsteroidal ligands in the androgen receptor. J. Biol. Chem. 280, 37747–37754 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaffer P. L., Jivan A., Dollins D. E., Claessens F., Gewirth D. T., Structural basis of androgen receptor binding to selective androgen response elements. Proc. Natl. Acad. Sci. U.S.A. 101, 4758–4763 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolognesi B., et al. , A concentration-dependent liquid phase separation can cause toxicity upon increased protein expression. Cell Rep. 16, 222–231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell S. F., Jain S., She M., Parker R., Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 20, 127–133 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhan Y., et al. , Interplay between cytoplasmic and nuclear androgen receptor splice variants mediates castration resistance. Mol. Cancer Res. 15, 59–68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma T., et al. , Increased transcription and high translation efficiency lead to accumulation of androgen receptor splice variant after androgen deprivation therapy. Cancer Lett. 504, 37–48 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahn J. H., et al. , Phase separation drives aberrant chromatin looping and cancer development. Nature 595, 591–595 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carrico I. S., Chemoselective modification of proteins: Hitting the target. Chem. Soc. Rev. 37, 1423–1431 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Mah R., Thomas J. R., Shafer C. M., Drug discovery considerations in the development of covalent inhibitors. Bioorg. Med. Chem. Lett. 24, 33–39 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Baillie T. A., Approaches to mitigate the risk of serious adverse reactions in covalent drug design. Expert. Opin. Drug. Discov. 16, 275–287 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Roskoski R. Jr., Orally effective FDA-approved protein kinase targeted covalent inhibitors (TCIs). Pharmacol. Res. 165, 105422 (2021), 10.1016/j.phrs.2021.105422. [DOI] [PubMed] [Google Scholar]
- 45.Jackson P. A., Widen J. C., Harki D. A., Brummond K. M., Covalent modifiers: A chemical perspective on the reactivity of alpha, beta-unsaturated carbonyls with thiols via hetero-michael addition reactions. J. Med. Chem. 60, 839–885 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crawford J., et al. , Study design and rationale for the phase 3 clinical development program of enobosarm, a selective androgen receptor modulator, for the prevention and treatment of muscle wasting in cancer patients (POWER trials). Curr. Oncol. Rep. 18, 37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joseph J. D., et al. , A clinically relevant androgen receptor mutation confers resistance to second-generation antiandrogens enzalutamide and ARN-509. Cancer Discov. 3, 1020–1029 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Balbas M. D., et al. , Overcoming mutation-based resistance to antiandrogens with rational drug design. Elife 2, e00499 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin J. S., MacKenzie C. J., Fletcher D., Gilbert I. H., Characterising covalent warhead reactivity. Bioorg. Med. Chem. 27, 2066–2074 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nadal M., et al. , Structure of the homodimeric androgen receptor ligand-binding domain. Nat Commun 8, 14388 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He B., et al. , Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance. Mol. Cell 16, 425–438 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Banuelos C. A., et al. , Characterization of niphatenones that inhibit androgen receptor N-terminal domain. PLoS One 9, e107991(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Myung J. K., et al. , An androgen receptor N-terminal domain antagonist for treating prostate cancer. J. Clin. Invest. 123, 2948–2960 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schild H. O., pAx and competitive drug antagonism. Br J. Pharmacol. Chemother. 4, 277–280 (1949). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao Y., et al. , Targeting GRP78-dependent AR-V7 protein degradation overcomes castration-resistance in prostate cancer therapy. Theranostics 10, 3366–3381 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao H., Wang D., Gao R., Chen L., Feng Y., Down regulation of U2AF1 promotes ARV7 splicing and prostate cancer progression. Biochem. Biophys. Res. Commun. 541, 56–62 (2021). [DOI] [PubMed] [Google Scholar]
- 57.Liu W., et al. , PRPF6 promotes androgen receptor/androgen receptor-variant 7 actions in castration-resistant prostate cancer cells. Int. J. Biol. Sci. 17, 188–203 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu L. L., et al. , Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene 33, 3140–3150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawamura N., et al. , SF3B2-mediated RNA splicing drives human prostate cancer progression. Cancer Res. 79, 5204–5217 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Terrados G., et al. , Genome-wide localization and expression profiling establish Sp2 as a sequence-specific transcription factor regulating vitally important genes. Nucleic Acids Res. 40, 7844–7857 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mao C., et al. , betaKlotho inhibits cell proliferation by downregulating ELK4 and predicts favorable prognosis in prostate cancer. Cancer Manag Res. 13, 6377–6387 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pang B., et al. , Inhibition of lipogenesis and induction of apoptosis by valproic acid in prostate cancer cells via the C/EBPalpha/SREBP-1 pathway. Acta. Biochim. Biophys. Sin. (Shanghai) 53, 354–364 (2021). [DOI] [PubMed] [Google Scholar]
- 63.Nagamatsu T., Suzuki Y., Antinephritic effect of prostaglandin E1 on serum sickness nephritis in rats (5). Effect of PGE1 on disposal of heat-aggregated bovine serum albumin in the glomerulus. Jpn. J. Pharmacol. 46, 397–402 (1988). [DOI] [PubMed] [Google Scholar]
- 64.Sutanto F., Konstantinidou M., Domling A., Covalent inhibitors: A rational approach to drug discovery. RSC Med. Chem. 11, 876–884 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh J., Petter R. C., Baillie T. A., Whitty A., The resurgence of covalent drugs. Nat. Rev. Drug Discov. 10, 307–317 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Miseta A., Csutora P., Relationship between the occurrence of cysteine in proteins and the complexity of organisms. Mol. Biol. Evol. 17, 1232–1239 (2000). [DOI] [PubMed] [Google Scholar]
- 67.Rosen C. B., Francis M. B., Targeting the N terminus for site-selective protein modification. Nat. Chem. Biol. 13, 697–705 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Tekaia F., Yeramian E., Dujon B., Amino acid composition of genomes, lifestyles of organisms, and evolutionary trends: A global picture with correspondence analysis. Gene 297, 51–60 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
ATAC-Seq, RNA-seq data have been deposited in Geo (GSE215335). All study data are included in the article and/or SI Appendix.