Significance
How nanoparticle mechanical properties affect the preservation of membrane proteins on cell membrane-coated nanoparticles and the consequent bio–nano interactions remains largely unexplored. In this work, we synthesized mesenchymal stem cell membrane-coated nanoparticles and aim to answer two main questions: 1) whether cell membrane coating can translocate all membrane proteins in an intact state onto nanoparticle cores, thus preserving their structures and functions; 2) whether the property of nanoparticle cores affects the membrane protein profile and integrity after membrane coating. This article reports that nanoparticle elasticity affects the membrane-coating process resulting in a different protein density on the cell membrane-coated nanoparticles, which leads to less macrophage uptake but higher cancer cell uptake.
Keywords: nanoparticles, elasticity, cell membrane coating
Abstract
Cell membrane-coated nanoparticles are emerging as a new type of promising nanomaterials for immune evasion and targeted delivery. An underlying premise is that the unique biological functions of natural cell membranes can be conferred on the inherent physiochemical properties of nanoparticles by coating them with a cell membrane. However, the extent to which the membrane protein properties are preserved on these nanoparticles and the consequent bio–nano interactions are largely unexplored. Here, we synthesized two mesenchymal stem cell (MSC) membrane-coated silica nanoparticles (MCSNs), which have similar sizes but distinctly different stiffness values (MPa and GPa). Unexpectedly, a much lower macrophage uptake, but much higher cancer cell uptake, was found with the soft MCSNs compared with the stiff MCSNs. Intriguingly, we discovered that the soft MCSNs enabled the forming of a more protein-rich membrane coating and that coating had a high content of the MSC chemokine CXCR4 and MSC surface marker CD90. This led to the soft MCSNs enhancing cancer cell uptake mediated by the CD90/integrin receptor-mediated pathway and CXCR4/SDF-1 pathways. These findings provide a major step forward in our fundamental understanding of how the combination of nanoparticle elasticity and membrane coating may be used to facilitate bio–nano interactions and pave the way forward in the development of more effective cancer nanomedicines.
Nanoparticles (NPs) have been explored as a promising platform for drug delivery and tumor targeting (1–3). Cell membrane-coated NPs have attracted significant interest in recent years. Conventional NPs have a range of attractive intrinsic properties, including having tunable particle size and surface properties. A significant development, the coating of NPs with a cell membrane with the retention of the surface properties of the source cells, offers many additional advantages, such as a longer circulation time and better targeting (4–6). A wide variety of cell membranes have been used with the goal of making various membrane-coated NPs that retain the unique biological functions of the source cells. However, this development is still at an early stage with two key questions remaining to be addressed. First, can the cell membrane-coating process translocate all of the membrane proteins in an intact state onto NP cores, with their structures and functions being preserved? Second, what is the impact of the inherent physicochemical properties of the NP cores on the membrane protein profile and integrity after membrane coating, and how does it influence the interaction of cell membrane-coated NPs with other cells?
The preservation of membrane proteins in this coating process is critical. Several methods have been developed for characterizing the protein profile of cell membrane coating, including sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) analysis, western blot, or flow cytometry. SDS–PAGE is usually used to compare the protein profiles of cell membranes before and after coating, but it is nonspecific, qualitative, and insensitive to protein changes. Western blot and flow cytometry are often used to determine the levels of certain protein markers (7). Many studies have demonstrated that the protein profile can be well preserved after cell membrane coating, but quantitative analysis about protein quantity and levels of particular protein markers in the cell membrane-coated NPs is lacking.
On the other hand, the physicochemical properties of the NP core (e.g., charge, size and materials) can play an important role in regulating their cell membrane coating. For instance, NP charge can affect the synthesis of membrane-coated NPs. Negatively charged NPs are more favorable for controlling membrane coating as positively charged NPs often cause extensive aggregation due to the strong electrostatic interactions (8). NP size and materials have also shown to have a large impact on the integrity of the cell membrane coating (9). Further, NP elasticity or stiffness, a NP physicochemical attribute, plays a critical role in regulating cellular uptake, blood circulation, and tumor targeting (10–13). However, the impact of NP elasticity on the membrane-coating process and, especially, the composition of membrane proteins translocated onto the NP surface and associated biological functions have yet to be explored.
In this study, we studied the effect of NP elasticity on the formation of mesenchymal stem cell (MSC) membrane-coated NPs and their interactions with cells. The MSC membrane is highly attractive for NP coating due to its ease in expansion and isolation, as well as, more importantly, its specific membrane molecular composition that enables MSC chemotaxis and other processes. Moreover, MSCs exhibit tumor-tropic migratory properties, which offer great potential for tumor targeting (14, 15). These features make MSC membrane-coated NPs promising candidates for targeted cancer drug delivery (16–18). Here, we began by fabricating soft and hard MSC membrane-coated silica nanocapsules (MCSNs) with Young’s moduli of 44 MPa and 2.3 GPa, respectively. Their mechanical properties were characterized using atomic force microscopy (AFM) before and after MSC membrane coating. We then explored their interactions with macrophage cells and cancer cells. We found, and report, that the membrane-coating process was affected by nanoparticle elasticity and led to the resulting MCSNs having differing protein density in the coated cell membrane SNs, and, in turn, to a reduced macrophage uptake but with a higher cancer cell uptake. We used transmission electron microscopy (TEM) imaging and theoretical simulation to investigate changes in MCSN morphology in interacting with various cells, as well as in confirming that no significant deformation occurred during the cell internalization process. Our studies showed that the elasticity of MCSNs can have unanticipated and complex impacts in receptor-mediated targeting and deformation of the nanoparticles in the cell uptake process, and in determining their cellular uptake. The overall outcome of this work is the finding that MCSNs can be varied to promote evasion of immune cells but enhance the targeting and uptake of cancer cells.
Results
Synthesis of Soft and Hard MCSNs.
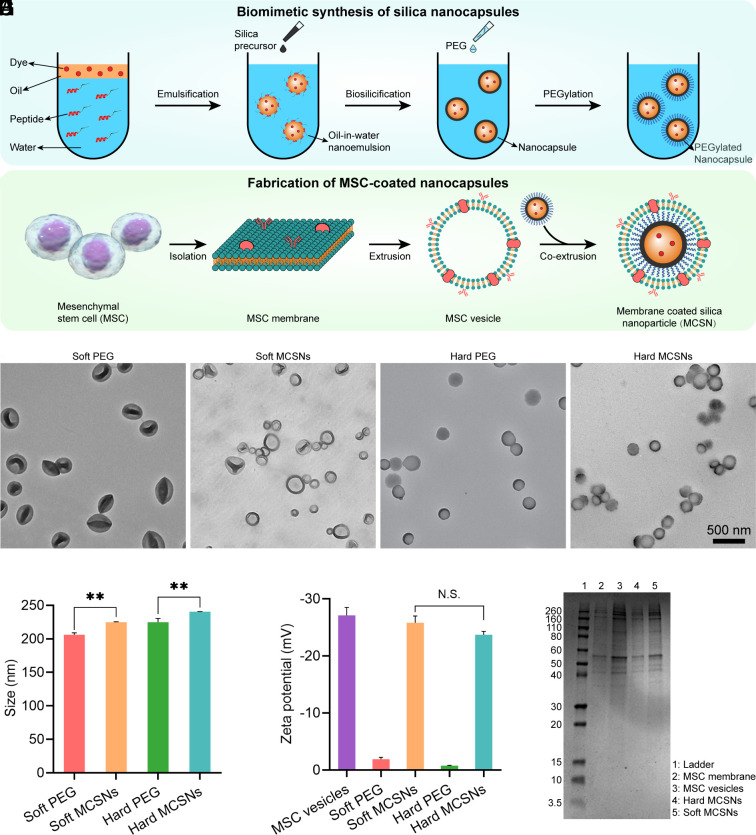
Silica nanocapsules (SNs) were synthesized as the NP core. Briefly, SNs with different stiffness were fabricated using a nanoemulsion-templated method developed in our laboratory (19). A designed peptide SurSi (Ac-MKQLAHSVSRLEHARKKRKKRKKRKKGGGY-CONH2) was used to form oil-in-water nanoemulsions. Then a silica precursor, triethoxyvinylsilane (TEVS) or tetraethoxysilane (TEOS), was added to the nanoemulsions to initiate biosilicification, allowing the formation of the oil-core silica-shell nanocapsules (Fig. 1A). The silica precursor TEVS induces the formation of the soft SNs, while TEOS allows the formation of the stiff SNs (20). The silica shell of the SNs consists of approximately 18 wt% SurSi peptide (21). These as-prepared SNs were then PEGylated to screen the surface charge because positively charged NPs would interact with negatively charged cell membranes leading to aggregation during extrusion (22). The PEG density is approximately 0.9 molecules/nm2 (20). To harvest the MSC cell membrane, stem cells were lysed in a hypotonic solution followed by disruption using sonication. Then centrifugation and extrusion were used to form the MSC membrane vesicles. At last, the PEGylated SNs were coextruded with the MSC membrane vesicles to form the MCSNs (Fig. 1B).
Fig. 1.
Synthesis of MCSNs and their characterization. (A) Schematic illustration of the fabrication of SNs. (B) Synthesis of MCSNs. (C–F) TEM images of PEGylated soft SNs, soft MCSNs, PEGylated hard SNs, and hard MCSNs, respectively. Samples of B and D were negatively stained with uranyl acetate. (Scale bar, 500 nm.) (G) Particle size of different NPs (n = 3; mean ± SD). (H) Zeta-potential of different NPs (n = 3; mean ± SD). (I) SDS–PAGE of the protein profile. From Left to Right were i) ladder, ii) isolated MSC membrane, iii) MSC membrane formed vesicles, iv) hard MCSNs, and v) soft MCSNs.
Fig. 1 C–F show the morphology of soft and hard SNs and MCSNs before and after membrane coating. The soft SNs are seen significantly deformed (Fig. 1C) in contrast to the spherical morphology of the stiff SNs (Fig. 1E) mainly due to their dry state under the vacuum condition of TEM. After membrane coating, the MCSNs show a less deformed morphology with a clear membrane structure observed on the particle surface of around 5 to 10 nm in thickness. The hydrodynamic diameters of the soft and stiff SNs measured using a DLS increased from 206.2 ± 2.8 nm to 224.8 ± 0.8 nm, and from 224.9 ± 0.6 nm to 240.5 ± 0.3 nm, respectively, after coating (Fig. 1G). The increase in the NP sizes agrees well with the cell membrane thickness of approximately 5 to 10 nm. The zeta potential of the soft and hard MCSNs decreases from −1.89 ± 0.33 mV to −25.8 ± mV and from 0.75 ± 0.07 mV to −23.7 ± 0.6 mV, respectively, which are similar to that of the MSC membrane-formed vesicles (−27.1 ± 1.4 mV). These data demonstrate the successful coating of the MSC membrane. The protein profiles of the isolated cell membrane, the membrane-formed vesicles, soft MCSNs, and hard MCSNs were analyzed using SDS-PAGE (Fig. 1I). The MSC membrane proteins were well retained in the vesicles and MCSNs, suggesting the cell membrane was successfully coated.
To evaluate their long-term stability, MCSNs were suspended in phosphate-buffered saline (PBS) buffer for 2 wk. As displayed in SI Appendix, Fig. S1 A and B, both soft and hard MCSNs exhibited a stable size over the 2-wk duration of the study. The MCSN stability was also investigated in Dulbecco’s Modified Eagle Medium (DMEM) with 10% Fetal bovine serum (FBS). Potential aggregations were then monitored for 8 h by ultraviolet–visible spectroscopy (UV-vis) (23). Minimal change in absorbance was observed (SI Appendix, Fig. S1C), confirming the stability of both soft and hard MCSNs in cell culture medium.
Liquid AFM was applied to measure the mechanical properties of the SNs and MCSNs. SI Appendix, Fig. S2A displays the height profiles of the soft and hard SNs. Compared with the hard SNs, the soft SNs exhibit much smaller heights on the mica surface. The reconstructed AFM-3D topographical images (SI Appendix, Fig. S2B) show that the soft SNs and MCSNs exhibit disc-like structures while the hard SNs and MCSNs have hemispherical shapes, indicating that the soft ones are easier to flatten on the mica substrates compared with the hard SNs. Young’s moduli of the SNs and MCSNs were measured in water by AFM using a contact model and calculated on the basis of the linear theory of thin elastic shells and force-indentation curves (SI Appendix, Fig. S3). The coated membrane did not change the stiffness of both soft and hard SNs significantly. The soft MCSNs display a similar Young’s modulus (44.0 MPa) as that of the soft SNs (43.7 MPa) (SI Appendix, Fig. S2C), and both the hard SNs and MCSNs have the same Young’s modulus of 2.3 GPa.
Cellular Uptake by Macrophages.
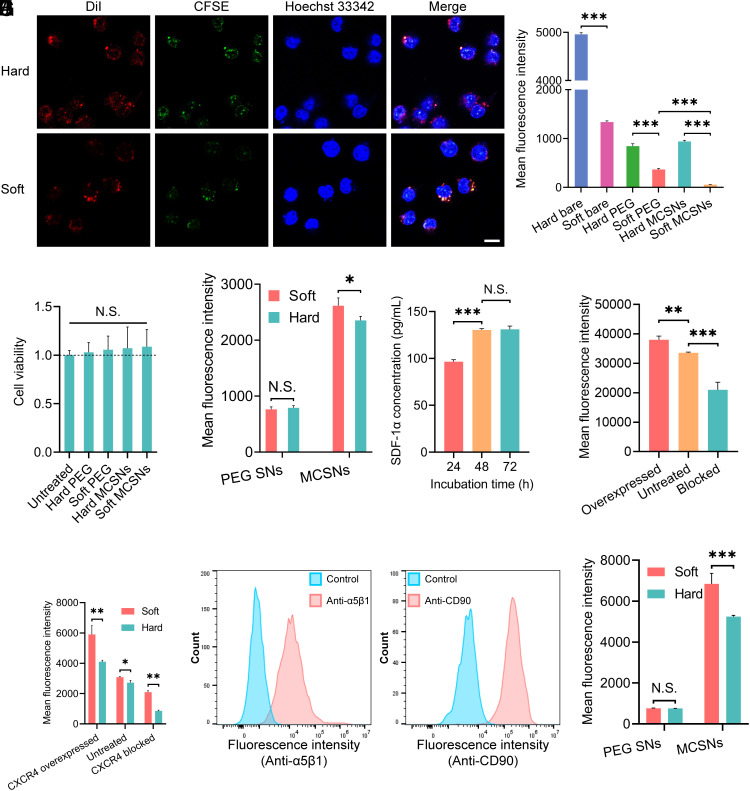
Immune evasion of NPs is critical for improving their blood circulation and tumor accumulation (24). MSC coating has been shown to be capable of preventing NPs from recognition by the immune system due to the hypoimmunogenic and low expression of major histocompatibility complex molecules (25, 26). Therefore, the MCSNs that retain the MSC membrane proteins are expected to exhibit reduced macrophage cellular uptake. To study the immune evasion effect, the cellular uptake of MCSNs by RAW264.7 macrophage cells were studied. First, the dual-fluorescent labeled MCSNs [DiI in the oil core and MSC membrane labeled with Carboxyfluorescein succinimidyl ester (CFSE)] were fabricated to elucidate the internalization process. Three fluorescent dyes, DiI, (549/565 nm), CFSE (492/517 nm), and Hoechst 33342 (343/361 nm), were employed to label the SNs, MSC cell membrane, and nucleus of macrophage cells, respectively. Then the cells were coincubated with the as-prepared MCSNs and visualized using Confocal Microscopy. The confocal images (Fig. 2A) show a high degree of overlapping fluorescent signals of DiI and CFSE, indicating that both soft and hard MCSNs remained intact during cell uptake.
Fig. 2.
The interaction between MCSNs and macrophage cells or cancer cells, and the surface properties of MCSNs. (A) CLSM images of MCSNs illustrating the colocalization of the coated membrane and hard SNs (i), the coated membrane, and soft SNs (ii). Blue: Hoechst-labeled nucleus of RAW 264.7 cells; Red: DiI-labeled SNs; Green: CFSE-labeled cell membrane. (Scale bar, 10 µm.) (B) Cell uptake of DiI-encapsulated bare SNs, PEGylated SNs, and MCSNs by macrophage-like cells RAW 267.3. Nanoparticle dose: 1.25 × 109 capsules/mL. Cell uptake was quantitatively analyzed by flow cytometry (n = 3; mean ± SD). (C) Cell viability of Huh-7 cells under various treatments (n = 8; mean ± SD). (D) Cell uptake of DiI-encapsulated PEGylated SNs and MCSNs by HCC cancer cells Huh-7. Nanoparticle dose: 1.25 × 109 capsules/mL. Cell uptake was quantitatively analyzed by flow cytometry (n = 3; mean ± SD). (E) The SDF-1α concentration in the Huh-7 cell culture medium at different incubation times. (F) Flow cytometry analysis of the presence of CXCR4 on the surface of MSCs expressed by mean fluorescence intensity. (G) Flow cytometry analysis of the presence of CXCR4 on the surface of soft and hard NPs expressed by mean fluorescence intensity. (H) Flow cytometry analysis of the presence of integrin α5β1 on the surface of Huh-7 cells expressed by overlay distribution. Cell incubated with only secondary antibody was the control groups. (I) Flow cytometry analysis of the presence of CD90 on the surface of MSCs expressed by overlay distribution. Cell incubated with isotype FITC Mouse IgG1 was the control group. (J) Flow cytometry analysis of the presence of CD90 on the surface of soft and hard NPs expressed by mean fluorescence intensity.
Next, cell uptake was quantitatively analyzed by flow cytometry. Both the PEGylated SNs and MCSNs demonstrated a much lower cell uptake compared with the corresponding bare SNs (Fig. 2B). The soft MCSNs exhibited the lowest uptake, approximately 10 times lower than that of the hard MCSNs. Interestingly, the soft MCSNs had an even lower uptake than the soft PEGylated SNs, while this was not observed in hard nanocapsules, demonstrating the intricate interplay between NP elasticity, surface property and cellular uptake. In this regard, the soft NPs in combination with MSC membrane coating resulted in minimum macrophage uptake.
Effect of Stiffness and Surface Properties of NPs on Their Cancer Cellular Uptake.
Some concerns remain about the potential of MSCs in promoting the growth of cancer cells due to the activation of integrins. A WST assay was used to evaluate the effect of MCSNs on the cell viability and proliferation of Huh-7 liver cancer cells and immortalized human hepatocyte (IHH) cells. The model cell Huh-7 is a typical hepatocellular carcinoma (HCC) cell line from human. After a 48-h incubation, their growth was not affected confirming no promotion of cancer cell growth, and no significant toxicity was detected for these two cells (Fig. 2C and SI Appendix, Fig. S4).
The impact of NP elasticity on NP–cell interactions was investigated using the soft and hard PEGylated SNs, and MCSNs (44 MPa and 2.3 GPa) (Fig. 2D). Both soft and hard PEGylated SNs exhibited a similar Huh-7 cancer cell uptake due to the nonspecific NP–cell interactions, demonstrating that a minimum effect of NP stiffness on their nonspecific cell uptake, which agrees well with our previous studies (3, 20). In contrast, both the soft and hard MCSNs exhibited significantly higher Huh-7 cell uptake compared with the PEGylated SNs, confirming that MSC coating could enhance cancer cell uptake. Moreover, the soft MCSNs showed higher uptake than that of the hard ones. Based on these results, we further explore the surface properties of the soft and hard MCSNs.
The surface properties of MSCs and MCSNs relating to their targeting effect were investigated. It has been reported that the CXCR4/SDF-1 interaction plays an important role in enhancing cancer cell targeting of MSC membrane-coated NPs (27, 28). First, the secretion of SDF-1α from Huh-7 cells was measured using an enzyme-linked immunosorbent assay (ELISA), showing that the concentration of SDF-1α increased significantly between 24 and 48 h, then remained unchanged from 48 to 72 h (Fig. 2E and SI Appendix, Fig. S5). Furthermore, the expression level of the CXCR4 receptor on MSCs was controlled by either inducing the CXCR4 overexpression using hypoxia treatment of MSCs (29, 30), or blocking the CXCR4 function using a CXCR4 antagonist AMD3100. The CXCR4 expression level of MSCs was assessed by flow cytometry, showing that the CXCR4 was up-regulated or blocked successfully (Fig. 2F). Then, soft and hard NPs were coated with the CXCR4-up-regulated or blocked membrane, and the CXCR4 levels of an equal number of the corresponding MCSNs were confirmed by flow cytometry (Fig. 2G). We found that the relative density of CXCR4 on MCSNs changed accordingly when the CXCR4 of MSCs membrane was regulated (Table 1). Intriguingly, the soft MCSNs always showed significantly higher CXCR4 receptors on the NP surface compared with their counterpart hard ones regardless of the CXCR4 upregulation or blocking (Table 1).
Table 1.
Properties including hydrodynamic size, Zeta potential, surface CXCR4 and CD90 density and Young’s modulus of SNs and MCSNs
| PEGylated soft SNs | PEGylated hard SNs | Soft MCSNs | Hard MCSNs | |
|---|---|---|---|---|
| Hydrodynamic size (Diameter, nm) | 206.2 | 224.9 | 224.8 | 240.5 |
| Zeta potential (mV) | −1.89 | −0.75 | −25.8 | −23.7 |
| Normalized CXCR4 density | N/A | N/A | 1.134 | 1 |
| Normalized CD90 density | N/A | N/A | 1.236 | 1 |
| Young’s modulus (GPa) | 0.0437 | 2.3 | 0.0440 | 2.3 |
Another important surface marker is CD90, a glycoprotein expressed on the surface of the cell membrane and a typical marker of MSCs, which can associate with multiple cell surface molecules and form multiprotein complexes (31, 32). Notably, CD90 can specifically bind to various kinds of integrins, including α5β1, αvβ3, αMβ2, and αxβ2 (33, 34). The model in this study, HCC cell line, Huh-7, has been reported to have a high expression of α5β1 integrin” (35). The expression of α5β1 integrin on the surface of Huh-7 cancer cells and IHH cells were determined (Fig. 2H and SI Appendix, Fig. S6), showing that the α5β1 integrin is overexpressed on Huh-7 compared with a human hepatocyte cell line IHH. The level of CD90 markers on both MSCs and MCSNs was also confirmed (Fig. 3 I and J), again with a significantly higher concentration of CD90 on the soft MCSNs than that on the hard MCSNs, which agrees with our finding about a higher CXCR4 concentration on the soft MCSNs. We further quantified the whole protein concentration of the MCSNs, and found that the soft MCSNs consist of a much higher amount of membrane proteins than that of the hard MCSNs (SI Appendix, Fig. S7). Considering the similar size and charge of the SN particle core (Table 1), the only difference of the MCSN lies in their elasticity. Therefore, the high concentration of CXCR4 and CD90 receptors on the soft MCSNs is attributable to the NP stiffness, suggesting its critical role in regulating the membrane coating process and consequently the final membrane protein compositions on the cell membrane-coated NPs.
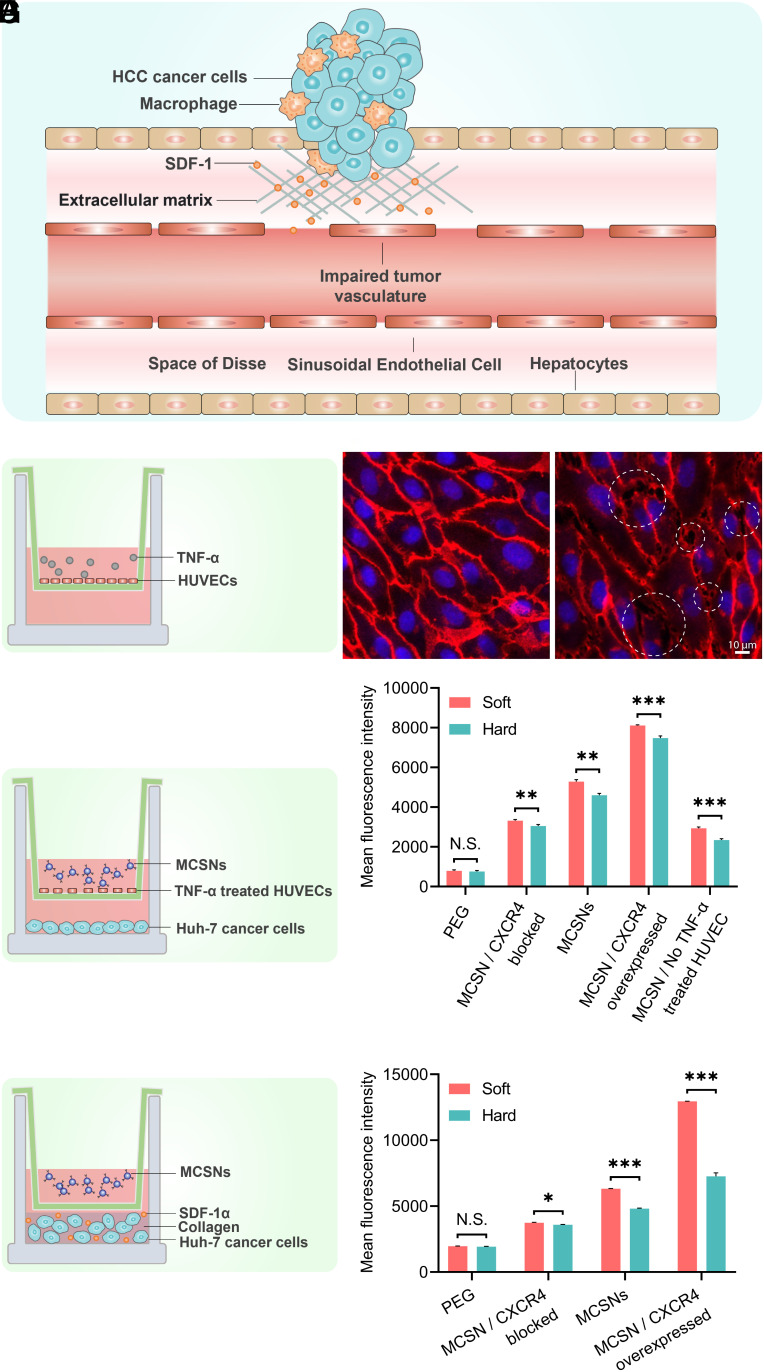
Fig. 3.
(A) Schematic diagram of the characteristics of HCC tumor, including leaky tumor vasculature and dense ECM. (B) Schematic representation of an impaired vasculature model using a transwell insert with a TNF-α-treated HUVECs layer and (C) The Confocal Laser Scanning Microscopy (CLMS) images of HUVECs with or without TNF-α treated. (Scale bar, 10 µm.) (D) Schematic representation of a transwell model with a HUVEC layer and Huh-7 cells (E) Cell uptake of DiI-encapsulated PEGylated SNs and MCSNs by Huh-7 liver cancer cells analyzed by flow cytometry of (D) model. (F) Schematic representation of a transwell model with Huh-7 cells dispersed in collagen (G) Cell uptake of DiI-encapsulated PEGylated SNs and MCSNs by Huh-7 liver cancer cells analyzed by flow cytometry of (F) model.
Protein corona plays a critical role in regulating nano–bio interactions (36, 37). To investigate the formation of protein corona, we first quantified the total amount of proteins on the PEGylated SNs and MCSNs in the presence and absence of FBS using the BCA assay (SI Appendix, Fig. S8). We can see that the amount of the protein corona of both the soft and hard MCSNs is much less than those PEGylated SNs, which agrees with others’ studies that coating NPs with cell membrane reduces protein adsorption thus less amount of protein corona (38). Moreover, the soft MCSNs have slightly higher amount of protein corona than that of the hard one, which corresponds well with our previous study (21). Then we examined whether the presence of protein corona affected the availability of CXCR4 on the surface of soft and hard MCSNs, and found that the available CXCR4 to bind with the antibody decreased slightly after forming a protein corona, and the CXCR4 on the soft MCSNs remained higher than the hard ones (SI Appendix, Fig. S9).
Endothelial Barrier Model for Studying MCSN Translocation.
To mimic the HCC tumor microenvironment (Fig. 3A) and to explore how the MSC coating on the nanocapsule surface affects the cancer cell uptake of MCSNs, we designed two transwell-based models. First, to study the translocation of MCSNs through the tumor endothelial barrier, an impaired vasculature model was established by culturing human umbilical vein endothelial cells (HUVECs) on the membrane of a transwell insert, which was then treated with TNF-α (39, 40) to induce the damage of hepatic sinusoidal endothelial thus increased size of fenestrations (Fig. 3B). VE-cadherin was immunostained to investigate the state of intercellular junction. After stimulation, gaps (a few micrometers) started to appear (Fig. 3C), which were similar in size to those observed in vivo (41). We then built a model with an impaired endothelial barrier on the membrane of the transwell insert and a monolayer of Huh-7 cells on the bottom of wells to explore whether the MSC coating promotes transendothelial transport of MCSNs (Fig. 3D). In this model, an equal amount of DiI-labeled soft or hard SNs and MCSNs were added into the insert and incubated for 4 h. To probe how CXCR4 affects the cell uptake, cell membrane of MSCs with different levels of CXCR4 expression (CXCR4 blocked, normal CXCR4 expression, and CXCR4 overexpressed) were used to generate different MCSNs. The quantitative cell uptake results measured by flow cytometry indicated that after blocking or overexpressing CXCR4, the MCSN cellular uptake decreased or increased proportionally to the concentration of CXCR4 on the corresponding cell membranes, suggesting that the CXCR4/SDF-1 axis plays an important role in transendothelial transport of NPs. It should also be noted that even for the CXCR4 blocked one, its cellular uptake was still significantly higher than that of the PEGylated SNs. This is probably due to the other surface marker, CD90, which increased the cellular uptake via a specific receptor-mediated uptake pathway. In addition, the NP cellular uptake through the non-TNF-α-treated HUVEC layer was two times lower than that of the TNF-α stimulated group, indicating the impaired vascular with intercellular gaps caused by tumors contributed to the enhanced translocation of NPs from blood vessel to the tumor site.
Extracellular Matrix (ECM) Barrier Model for Studying Role of the CXCR4/SDF-1 Axis in Regulating MCSN Cell Uptake.
Another transwell model was set up to study the NP–cell interaction in the presence of the ECM barrier (Fig. 3F). Huh-7 cells were dispersed in dense ECM-mimicking collagen with 120 pg/mL SDF-1α, which was added to mimic the higher concentration of SDF-1 α in the tumor microenvironment. Then the same number of different types of DiI-loaded SNs or MCSNs was added to the insert. After full digestion of the ECM mimicking collagen, Huh-7 cells were collected and analyzed using flow cytometry. The results showed that Huh-7 cell uptake increased or decreased in proportion to the overexpression or blocking of CXCR4, suggesting again the important role of the CXCR4/SDF-1 axis in regulating NP cell uptake through the ECM of tumors. More CXCR4 on the MCSNs facilitated their higher uptake. Furthermore, the cell uptakes of all types of NPs in this ECM mimicking model were significantly higher than those in the HUVEC endothelial barrier model, indicating the endothelial layer presents a major barrier in blocking the translocation of MCSNs from blood circulation to tumor sites. Based on these studies, it is clear that the CXCR4 on the MCSNs contribute to their enhanced cellular uptake by the Huh-7 liver cancer cells via the CXCR4/SDF-1 axis mechanism.
TEM Visualization of MCSN Morphology during Cell Uptake.
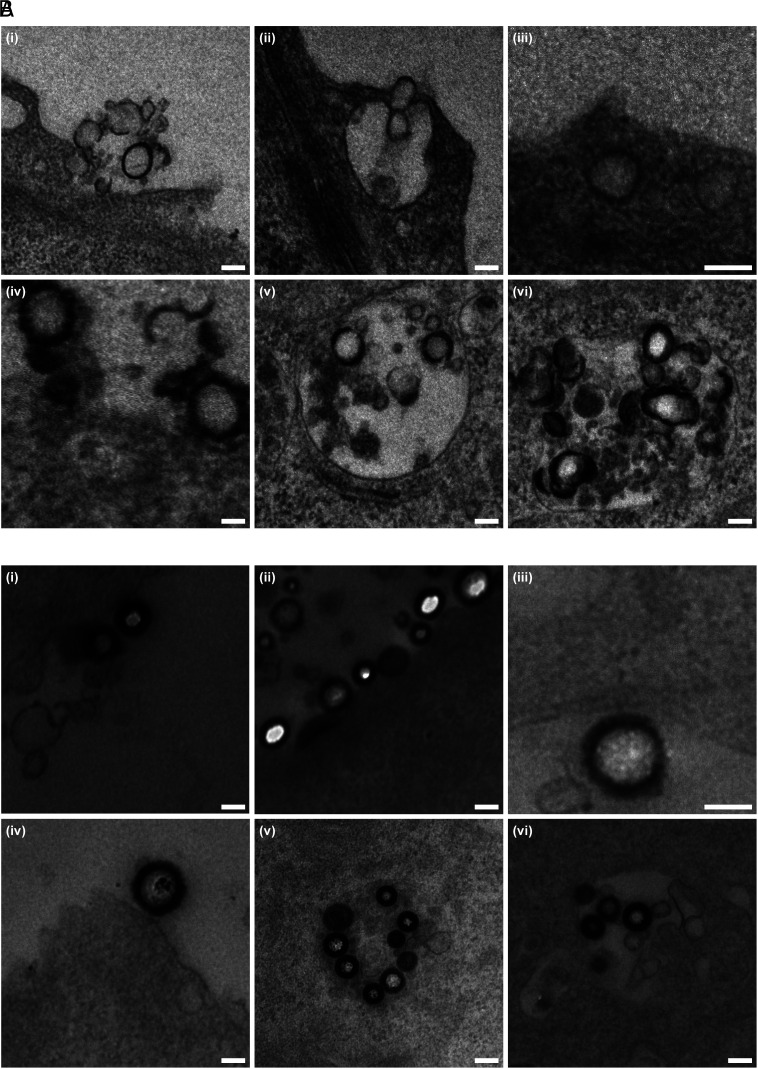
To further investigate the mechanism of the cell uptake process, the morphology of MCSNs during cell uptake was visualized by preparing 50-nm resin sections and imaged using TEM. Fig. 4 displays both soft and hard MCSNs on the cell surface or in the endosome. More debris has been found near the soft MCSNs indicating that the soft ones were easier to be peeled off during cell uptake. The particular white parts of MCSNs (e.g., Fig. 4 B, ii) were due to the holes created by the resin cutting process. To evaluate the deformation, ten MCSNs on the cell membrane were chosen, and the dimension ratio of the major axis to the minor axis was calculated (SI Appendix, Fig. S10). Similar shape transformations (1.092 for hard MCSNs and 1.119 for soft MCSNs) were observed. The cellular uptake of MCSNs by macrophages was also investigated using the same method (SI Appendix, Fig. S11). No significant deformation was observed for both the soft and hard MCSNs. Notably, not many soft MCSNs were found because their cellular uptake was minimum.
Fig. 4.
TEM images of MCSNs coincubated with Huh-7 liver cancer cells. (A) Soft MCSNs: (i–iv) on cell surface; (v and vi) in endosome. (B) Hard MCSNs: (i–iv) on cell surface; (v and vi) in endosome (Scale bar, 100 nm.)
Theoretical Studies.
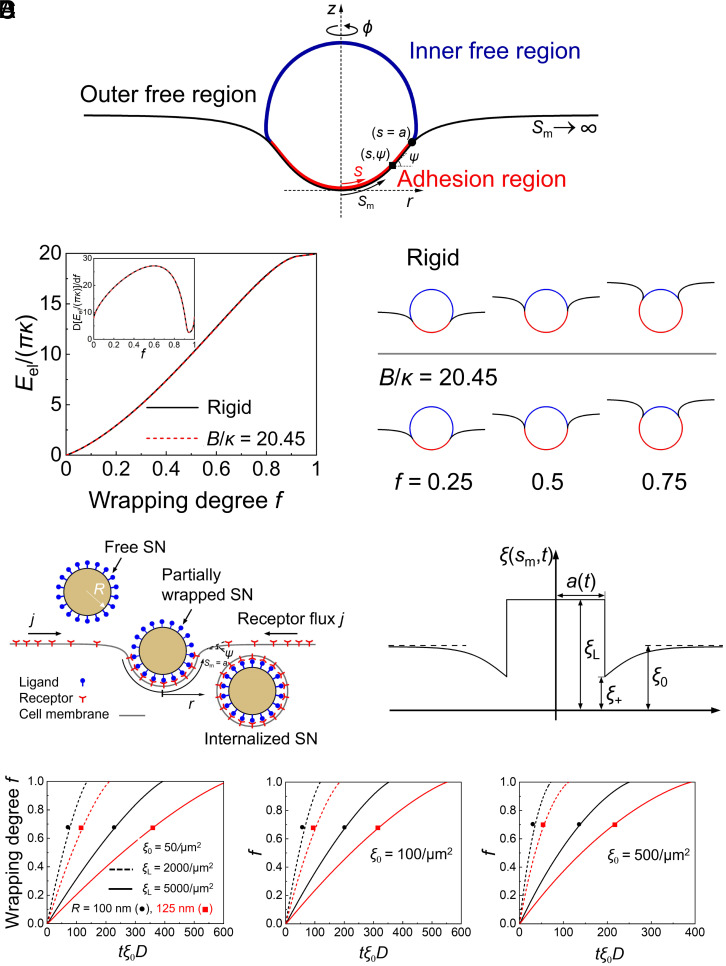
Theoretical simulations were conducted to elucidate how stiffness and specific binding affect the cellular uptake of the soft and hard SNs. Based on the force-indentation depth curves from the experiments (SI Appendix, Fig. S3) and the Reissner’s formula on thin shell deformation (42, 43), Young’s moduli of the nanocapsule materials were determined. The SNs are assumed to undergo an axisymmetric deformation during the adhesive wrapping by the cell membrane (Fig. 5A). Taking the cell membrane bending rigidity κ and membrane tension σ of representative values, the current theoretical results, consistent with our previous theoretical studies (44), show that the lipid membrane alone can barely deform the solid thin-shell nanocapsule during the simple membrane wrapping process (Fig. 5B). Soft and hard SNs show infinitesimal difference in the wrapping energy and configurations, indicating that the mechanical behaviors of wrapping around spherical soft and hard SNs can be modeled as the wrapping around spherical rigid SNs, consistent with the results observed by TEM.
Fig. 5.
Theoretical studies. (A) Schematic of membrane wrapping around an elastic thin-shelled spherical nanocapsule in the adopted cylindrical coordinate (r, φ, z). Arclengths s and sm of the nanocapsule and membrane are measured from the bottom pole, respectively. (B) Left: Elastic energy Eel versus the wrapping degree f at the normalized membrane tension σR2/κ = 3 and adhesion energy γ = 0. Inset, energy derivative d[Eel/(πκ)]/df versus f. Right: Selected wrapping configurations at σR2/κ = 3 and γR2/κ = 7 for hard and soft (B/κ = 20.45) nanocapsules. (C) Schematic of receptor-mediated endocytosis of a spherical hard nanocapsule of radius R (Left). During the wrapping process, the receptor density distribution is nonuniform with receptors depleted near the binding region. The resulting gradient of receptor density ξ in turn induces global receptor diffusion toward the binding site (Right). (D) Wrapping degree f versus the normalized time tξ0D for different values of the nanocapsule radius R and ligand density ξL at the binding energy per receptor–ligand bond of 15 kBT, D = 0.1 μm2/s, κ = 20 kBT, σR2/κ = 3 and ξ0 = 50/μm2, 100/μm2, and 500/μm2. The parameters used in the theoretical model are also summarized in SI Appendix, Table S1.
The dynamic wrapping process (Fig. 5C) can be described with a mechanical model taking into account both the cell membrane deformation and the evolution of receptor density ξ(sm,t) as a function of the membrane arclength sm and time t, as proposed in refs. 45 and 46 and confirmed by molecular dynamics simulations (47). As a result, the wrapping degree f, a ratio between the area of the SN-membrane contact region and the total surface area of the undeformed SN, correlates with the normalized time tξ0D for different values of the nanocapsule radius R and the density ξL of ligands on the SN surfaces, D being the receptor diffusivity (Fig. 5D). The initial density of the receptors on the cell membrane is ξ0. As ξ0 increases, the wrapping time decreases. Compared with the hard MCSNs, the soft ones have more receptors (CD90) for specific binding so that the wrapping time is shorter. Moreover, the nanocapsule of a larger radius R or a lower ligand density ξL has a longer period of wrapping time for the MCSNs to be fully wrapped by the cell membrane, as more receptors are required to diffuse toward the wrapped nanocapsules.
Discussion
In our previous studies, we observed a complex trade-off between NP stiffness and nano–bio interactions. Soft NPs exhibit better immune evasion as evidenced by the very low macrophage uptake thus longer circulation, while stiff NPs demonstrate better targeting effect as suggested by the enhanced uptake of receptor-positive tumor cells and higher accumulation at the tumor tissue (48). It would be ideal to design a NP with an optimal stiffness so that it can achieve not only good immune evasion but also enhanced tumor accumulation. In this work, we demonstrated that we were able to fabricate MSC-functionalized nanocapsules, which are superior in both tumor targeting and immune evasion.
We systematically studied the impact of mechanical properties and surface proteins on MCSNs’ immune evasion and tumor targeting. Two nanocapsules were synthesized with Young’s Moduli of 44 MPa and 2.3 GPa, respectively. Intriguingly, we found that the NP stiffness not only affected their interactions with cells, but also impacted the protein coating of MSC cell membrane. The soft MCSNs formed the membrane coating with a higher amount of proteins, leading to a higher concentration of membrane protein receptors per unit surface area of MCSNs, such as CD90 and CXCR4, two important receptors on MSC cell membranes. This is probably due to the deformation of soft SNs during the MCSN fabrication process. When the MCSN were fabricated through extrusion in the presence of MSC cell membrane, they became deformed thus a bigger surface area compared with their spherical counterpart. Although the cell membrane amount per area remains the same for both the deformed nanocapsules and spherical nanocapsules, the total amount of membrane proteins on the deformed MCSN is higher due to its bigger surface area. Then when they relaxed to spherical shape, the membrane protein density on the spherical surface becomes higher. In contrast, the stiff MCSNs remain spherical through the whole fabrication process, the cell membrane coating and density do not change. Consequently, the soft MCSN exhibited much higher cancer cell uptake via the receptor-mediated pathway than the stiff MCSNs. This finding agrees with our previous work, demonstrating that the total amount of proteins adsorbed onto nanocapsules increased as their stiffness decreased. Also, the protein corona profile varied with the NP stiffness, showing that the hard nanocapsules had a much higher amount of complement protein (Complement C3) and immunoglobulin proteins in their protein corona (21).
The CXCR4/SDF-1 axis also plays an important role in regulating the NP–cell interactions (30, 49). Ma et al. fabricated neural stem cell membrane–coated NPs for targeting SDF-1-enriched ischemic brain (50). The enhanced accumulation of cell membrane–coated NPs at the ischemic region in the mice model is attributed to the high level of CXCR4 on NPs. Bose et al. (27) designed a poly(lactic-co-glycolic acid) (PLGA) NPs coated with cell membrane isolated from CXCR4 overexpressed human adipose-derived stem cells. They showed that the CXCR4-overexpressed PLGA exhibit better targeting toward the ischemic tissue compared with the nonengineered stem cell-coated PLGA in their mouse experiments. They also demonstrated the CXCR4-SDF-1 axis is linked with endothelial barrier penetration. Zhang et al. (51) synthesized PLGA NPs coated with CXCR4-overexpressed MSCs and found that these NPs exhibit an osteoporosis-targeting ability responding to the SDF-1 gradient in an ovariectomized rat model. Their animal experiments demonstrated the role of CXCR4 in regulating NP targeting effect, but how the chemokine–ligand interaction affects the targeting process remain unexplored. To systematically address this question, we developed two in vitro models to study the effect of CXCR4 on the translocation of NPs across the endothelial barrier as well as on NP penetration through the ECM. First, we confirmed that the surface protein CXCR4 expression level promoted the translocation of MCSNs across the inflamed endothelium, which is consistent with previous work (27, 52). Then we designed an SDF-1α-enriched collagen ECM model and confirmed that both the CXCR4 and CD90 contributed to tumor targeting. These results agree well with those reported animal experiment results (16, 51). Our simulation results further demonstrated no significant deformation of both the soft and hard MCSNs, which corresponds well the TEM results, and the enhanced uptake of the soft MCSNs is mainly due to the high density of CXCR4 and CD90 receptors on the soft MCSN surface, which enhance NP targeting and binding to cell membrane, thus a rapid wrapping.
In summary, we report the critical role of NP stiffness in regulating the formation of cell membrane-coated NPs, the protein profile of the coating membrane, consequently the NP–cell interactions. Soft MCSNs tended to form a membrane coating with more proteins thus higher density of particular receptors (CXCR4 and CD90 in our study), leading to a higher cancer cell uptake than that of the hard MCSNs. We developed two in vitro models to confirm the important role of CXCR4/SDF-1 axis in the enhancement of cancer targeting through the endothelial barrier and ECM. Our finding provides valuable insights into the properties of cell membrane-coated NPs as well as the fundamental understanding of how MSC membrane coating contributes to cancer cell uptake. In this work, we primarily focused on in vitro bio–nano interactions. These nanocapsules can be loaded with hydrophobic payloads in the oil core by dissolving them in the oil phase before making the oil-in-water nanoemulsions, and could be potentially used for cancer treatment, and future systematic in vivo studies including circulation time, biodistribution, pharmacokinetics, pharmacodynamics, toxicity, and clearance will be essential for developing effective cell membrane-coated NPs for targeted cancer therapy.
Materials and Methods
Materials.
Peptide SurSi was custom synthesized by GenScript Corporation (Piscataway) with a purity ≥95%. Miglyol 812 oil was purchased from Caesar & Loretz GmbH (Hilden) and passed through heat-activated silica gel (Sigma-Aldrich) prior to use. Water having resistivity >18.2 MΩ·cm was obtained from a Milli-Q system (Millipore) equipped with a 0.22 μm filter. Methoxy poly (ethylene glycol) succinimidyl carbonate ester (mPEG-NHS, MW 5000) was obtained from Nanocs (New York, NY, USA). DiI stains were purchased from Thermo Fisher Scientific (Scoresby). Anti-CXCR4 antibody was obtained from Thermo Fisher Scientific (Scoresby). FITC-Anti-CD90 antibody, anti-integrin α5β1 antibody, human SDF-1 alpha ELISA kit and Alexa Fluor 594 Goat Anti-Mouse (IgG) secondary antibody was purchased from Abcam (Cambridge). High concentration type I collagen gel was purchased from Corning (New York, NY, USA). Other reagents and chemicals were of analytical grade purchased from Sigma-Aldrich and used as received unless otherwise stated.
Cell Culture.
Human MSCs obtained from A/Prof Michael R Doran (Queensland University of Technology, Brisbane, Australia) were isolated from bone marrow aspirates and characterized as previously reported (53). Bone marrow aspirates were collected from fully informed healthy human volunteer donors who provided written consent. Healthy volunteer donors were recruited from Mater Private Hospital, Brisbane, Australia. Ethical approval was granted through the Mater Health Services Human Research Ethics Committee and ratified by the Queensland University of Technology Human Ethics Committee (number: 1000000938).
MSCs were cultured in DMEM with 10% FBS and 10 ng/mL basic fibroblast growth factor (FGF-2). RAW264.7 (murine macrophage cells) and Huh-7 cells (human HCC cells) were cultured in DMEM supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco). IHH cells were cultured in DMEM: Ham’s F12 medium (v/v), supplemented with ITS, 40 ng/mL dexamethasone (DEX), and 10% FBS. HUVECs were cultured in EGM-2 endothelial cell growth medium with a growth factor kit (SingleQuots Supplements). All the cells were incubated at 37 °C in a humidified atmosphere with 5% CO2.
Isolation of Cell Membrane from MSCs.
When the MSC cells in the T175 flask achieved 80 to 90% confluence, they were washed twice using ice-cold PBS buffer and were harvested in 5 mL 0.25% Trypsin-EDTA. After 5-min incubation, 10 mL cell culture medium was added, and the cell suspension was centrifuged at 1,200 g for 3 min. The cell pellet was then washed by PBS buffer and centrifuged at 1,200 g for 3 min. The washed cell pellet was resuspended in the 1 mL hypotonic lysing buffer consisting of 10 mM Tris, 10 mM MgCl2, and 1X EDTA-free protease inhibitor and incubated in ice overnight with shaking. Then the cell dispersed in the lysing buffer was sonicated using a Sonifier 450 ultrasonicator (Branson Ultrasonics) for 10 times 5 s each time. During each break, the samples emerged in ice for 30 s. Then the as-prepared solution was centrifuged at 3,200 g for 5 min at 4 °C to remove the cell nucleus and intact cells. The resulting supernatant was collected and then centrifuged at 7,000 g for 10 min to discard mitochondrial. Then the supernatant was centrifuged at 15,000 g for 60 min at 4 °C. Finally, the isolated membrane pellet was washed by 1 mL PBS buffer twice and then stored in the same buffer at 4 °C.
Preparation of SNs.
Nanoemulsions were prepared using a nanoemulsion template method. Briefly, peptide SurSi solution (400 μM) and ZnCl2 (800 μM) were mixed in HEPES buffer (25 mM, pH 7). To make a nanoemulsion, SurSi solution and Miglyol 812 oil with the volume ratio of 50:1 were mixed and sonicated using a Sonifier 450 ultrasonicator (Branson Ultrasonics). The nanoemulsion was dialyzed in HEPES buffer (25 mM, pH 7.5) using a dialysis membrane (cutoff molecular weight: 10 kDa, Thermo Fisher Scientific) for 20 h at 4 °C to remove the excess peptide molecules in the bulk solution. To make SNs, the nanoemulsion was diluted one time with HEPES buffer (25 mM, pH 7.5), and was then mixed with either TEVS, 80 mM or TEOS, 40 mM. The reaction was conducted under stirring at room temperature for 30 h (TEOS) or 50 h (TEVS). To purify SNs, suspensions were dialyzed in HEPES buffer (25 mM, pH 7.5) at 4 °C for 20 h. To make PEGylated SNs, 3-aminopropyltri-ethoxysilane (APTES) was added into the as-prepared nanocapsule solution at a final concentration of 4 mM, and stirred for 4 h at room temperature. Then mPEG-NHS was added into the solution at a final concentration of 8 mM, and the suspension was then stirred for 4 h at room temperature. The PEGylated SNs were washed twice by centrifugation (15,000 g, 5 min) and resuspended in PBS buffer.
MCSNs Synthesis.
To coat the MSC cell membrane, an extrusion method was employed. All the filters, as well as syringes, were wetted with PBS buffer. For each membrane-coating process, 3 × 105 MSCs were used for membrane isolation and resuspended in 0.5 mL PBS, and the membrane protein in solution was determined by Bicinchoninic acid (BCA) assay (212.75 μg/mL). The isolated membrane solution was then extruded through a 400-nm polycarbonate membrane 21 times using a mini-extruder (Avanti Polar Lipids) by gently pushing the plunger of the filled syringe. After this step, the cell membrane vesicles were prepared. Then 100 μL of SNs was added to the membrane vesicle solution, and the mixture was extruded through a 400-nm polycarbonate membrane 13 times. The resulting particles were washed three times by centrifuging at 15,000 g for 3 min. Then these MCSNs were resuspended in PBS buffer for further use.
Protein Characterization.
The protein concentrations of MSC whole-cell lysate, MSC membrane, MSC membrane-formed vesicles, hard MCSNs, and soft MCSNs were measured using BCA assay (BCA Protein Assay Kit, Thermofisher Scientific). Then the samples with the equivalent volume of solution were loaded on Bio-Rad’s Mini-PROTEAN TGX gels, and the protein profiles were examined by Coomassie blue staining. The protein pattern in SDS-PAGE gel was imagined by a ChemiDox (BioRad).
Protein Corona Quantification Using BCA Assay.
To investigate the protein corona formation, PEGylated SNs or MCSNs were mixed with (1:1 v/v) FBS and incubated for 4 h. After incubation, the samples were washed three times using PBS and resuspended in PBS. Then the protein concentration was measured using the BCA assay.
Immunofluorescence Staining for Flow Cytometry.
Surface CXCR4 expression of different treated MSCs was verified by indirect immunostaining and assessed by flow cytometry. Briefly, 1 × 106 MSCs were collected, washed, and fixed by 4% formaldehyde. After washing, the cells were then resuspended in 2% Bovine Serum Albumin (BSA) to block nonspecific antibody binding for 30 min. Next, the cells were washed and coincubated with 4 μg/mL anti-CXCR4 (# 35-8800, Thermofisher Scientific) for 30 min. After this, the cells were washed three times, resuspended in PBS, and coincubated with Alexa Fluor 594 Goat Anti-Mouse (IgG) secondary antibody for 30 min (ab150116, Abcam). Then the MSCs were washed three times, resuspended in 0.5 mL PBS. The cells were finally collected and measured by a CytoFLEX flow cytometer (Beckman Coulter). For the indirect immunostaining of integrin α5β1, anti-integrin α5β1 antibody (ab275977) and TRITC Anti-Rabbit (IgG) secondary antibody were used. Surface CD90 expression of MSCs was verified by direct immunostaining in a similar method. The cells were coincubated with FITC anti-CD90 (ab124527, Abcam) for 30 min after fixation and blocking. After washing, the cells were ready for analysis. The surface receptors of MCSNs were verified using the same method.
The effect of protein corona on the presence of CXCR4 was measured using flow cytometry. To prepare MCSNs with corona, MCSNs were mixed with (1:1 v/v) FBS and incubated for 4 h before a 3-time washing using PBS. Then MCSNs and MCSNs with corona were resuspended in 2% BSA solution for 30 min. After blocking, these nanoparticles were centrifuged and coincubated with 20 μg/mL anti-CXCR4 for 30 min. Then after washing with PBS, MCSNs and MCSNs with corona were coincubated with Alexa Fluor 594 Goat Anti-Mouse (IgG) secondary antibody for 30 min. After washing, the samples were used to measure the presence of CXCR4 on the MCSN particle surface using flow cytometry.
Dynamic Light Scattering (DLS).
Hydrodynamic sizes and zeta-potentials of all the nanoparticles were measured using a Malvern Zetasizer Nano ZS (Malvern Instruments). All the samples are 1,000 times diluted before measurement.
TEM.
To examine the morphology of nanoparticles, 5 μL of the nanoparticle suspensions were placed on Formvar-coated copper grids (XXBR Technology Co., Ltd) and dried in air for 5 min. For the observation of cell membrane-coated nanoparticles, grids were then negatively stained with 5 μL of 1% uranyl acetate three times (Sigma-Aldrich). The excess solution was wicked away with absorbent paper. A JEOL 1010 transmission electron microscope (JEOL Ltd.) was used in this project. The cellular uptake process of cell membrane-coated NPs and their morphologies were also investigated by TEM at 80 kV. First, Huh-7 cells were seeded in a glass-bottom petri dish with a 20-mm bottom well (In Vitro Technologies) at a concentration of 2.5 × 105 cells/mL and cultured for 1 d. Then soft or hard MCSNs (5 × 109 capsules/mL) were added into cell culture medium for 12 h at 37 °C. Then the cells were coincubated with MCSNs (5 × 109 capsules/mL) for 2 h at 4 °C for receptor binding. Finally, the cells were incubated at 37 °C for 10 min before fixation. Then samples were fixed with glutaraldehyde and OsO4 immediately. After washing, cells were then dehydrated in a graded ethanol series (50%, 70%, 90%, 100%), infiltrated using Durcupan resin, and then incubated at 60 °C for 48 h. Finally, these cell resin samples were sliced into sections (50 nm) and stained before imaging.
AFM.
To measure the mechanical properties of nanoparticles, an AFM method was applied using a Cypher AFM (Asylum Research). First, all the nanoparticles were dialyzed in water for 20 h to remove the salt from buffer solution. Then, 20 μL nanoparticle solution was placed on a freshly cleaved mica sheet and air-dried overnight. Before the AFM experiment, nitrogen was used to clean the mica surface, and then 100 μL Milli-Q water was added on the mica gently. For imaging, cantilevers (Etalon, Tipsnano) with a nominal spring constant of 6 N/m were used. For force measurement, samples were measured in water using cantilevers with a nominal spring constant of 3.5 N/m for hard NPs and 0.06 N/m for soft NPs. For each sample, 10 different nanoparticles were chosen for analysis. Before each experiment, cantilevers were calibrated on a clean glass surface.
Nanoparticle Stability.
To study the in vitro stability of the MCSNs in physiological conditions. 500 µL aliquots of the MCSN suspensions were mixed with an equal volume of Dulbecco’s modified Eagle’s medium (DMEM, supplemented with 10% FBS). The mixtures were then incubated at 37 °C with shaking at 100 rpm. Aggregation was monitored by measurement of the absorbance at 560 nm by UV-vis.
Colocalization Study.
Three fluorescent dyes DiI, (549/565 nm), CFSE (492/517 nm), and Hoechst 33342 (343/361 nm) were used to label the SNs, the coated cell membrane, and nucleus of macrophage cells, respectively. RAW 264.7 cells at a density of 2 × 105 cells/well were seeded in 24-well plates containing coverslips (diameter of 12 mm) which have been cleaned with Piranha Solution in every well and cultured for 24 h. Then the cells were coincubated with DiI-loaded nanocapsules for 4 h. After this, the cells were washed with PBS three times, and the Hoechst 33342 was added by medium exchange with 25-min incubation. Colocalization was observed using CLSM (Zeiss 710, Jena, Germany). Because the cellular uptake of soft MCSNs was very low, the setting of laser intensity was different for soft and hard MCSNs.
Quantitative Cellular Uptake Study Using Flow Cytometry.
Cellular uptake of the nanocapsules by RAW264.7 cells or Huh-7 cancer cells was studied using a 2D cell monolayer model and quantitated by flow cytometry. Cells were seeded in 24-well plates (2.5 × 106 cells per well) and cultured for 24 h. Before each experiment, the fluorescence intensity of DiI loaded NPs were measured using a plate reader to ensure that the fluorescence intensity of each group is the same. Cells were then cocultured with DiI-loaded different types of nanocapsules for 4 h. To prepare samples, the cells were then treated with 0.25% trypsin, washed three times carefully, and then resuspended in 500 µL PBS before measurement. Nontreated cells were set as the control group. A CytoFLEX flow cytometer (Beckman Coulter) was used to measure the fluorescence intensity with excitation and emission wavelengths at 549 and 565 nm, respectively. Forward versus side scatter (FSC versus SSC) gating method was used.
Transwell Model Study.
To explore the permeability of MCSNs from injury vascular to tumor cells and the effect of CXCR4-SDF-1 axis, a transwell kit (0.4-μm pore size, 24-well plate, Costar) was used. A monolayer of HUVECs (2 × 105 per insert) was seeded on the insert. The HUVECs were cultured until confluence, and then they were treated with 20 ng/μL TNF-α for 2 h to mimic the leaky sinusoidal vasculature around the tumor. Huh-7 cancer cells were seeded in the bottom chamber. After washing the HUVECs to remove redundant TNF-α, HUVECs were incubated with Huh-7 cells with fresh medium and cocultured with DiI-loaded different types of SNs for 4 h to study the translocation and targeting from HUVEVs layer to tumor cells.
To investigate the intercellular adherens of HUVECs, vascular endothelial cadherin and nucleus were stained. Briefly, cells were fixed by 4% formaldehyde and permeabilized with 0.1% Triton X for 5 min. After incubation with 2% BSA for 1 h, HUVEC were stained with Alexa Fluor647 VE-cadherin antibody (SC-9989, Santa Cruz Biotechnology) for another 1 h. Finally, the Hoechst 33342 was added and stained for 20 min before imaging.
For the mimicking of cancer ECM, collagen gel solution (4 mg/mL) was applied and freshly prepared on ice before using. 10×PBS buffer, Type 1 collagen gel, 1 M NaOH, and Huh-7 cells dispersed in culture medium were gently mixed, and the final concentration of cells was 5 × 105/mL. 120 pg/mL of SDF-1α was premixed with culture medium. For each well of transwell, 0.5 mL collagen gel solution was added. After a 30-min incubation at 37°C, different types of NPs were added on the insert and incubated for 4 h.
To digest the collagen, collagenase was used. Collagen gel was washed with PBS buffer three times, and one gel volume, 1 mg/mL collagenase in PBS was added. Then the collagen samples were incubated at 37°C for 2 h with pipetting up and down every 15 min. When the digestion completed, an equal volume of culture medium was added and centrifuged the cell suspension at 250 g for 5 min. Finally, the cells were resuspended in 500 µL PBS for flow cytometry analysis.
Cytokine Measurement.
Quantitative analysis the secretion of SDF-1α (CXCL12α) from Huh-7 cancer cells in conditioned medium was studied by the ELISA method according to the manufacturer’s recommendation. To prepare the conditioned medium, Huh-7 cells were seeded in T25 cell flasks. When the confluence was 80%, culture medium was aspirated. After gently washing with warm PBS for twice, 5 mL FBS-free medium was added, and incubated for 24 to 72 h. Finally, the conditioned medium was centrifuged at 300 g for 10 min at 4°C and ready to use.
WST Assay.
Cell vitality was assessed using cell proliferation reagent WST-1. Each type of cells (Huh-7 and IHH) was seeded in a 96-well plate at a density of 2,000 cells/well. Then, cells were cultured in the presence of different nanoparticles by medium changing for 48 h. Next, cells were incubated with 10% WST-1 reagent for 2 h. Finally, the plate was shaken thoroughly, and the absorbance intensity at 450 nm was measured using a microplate reader Tecan Infinite 200 PRO (Tecan).
Statistical Analysis.
The two-tailed unpaired Student’s t tests were used for statistical significance between groups, with P ≥ 0.05, P < 0.05, P < 0.01, and P < 0.001 denoted as N.S., *, **, and ***, respectively. P < 0.05 was considered statistically significant. Data are represented as mean ± SD. Values for n, P, and the specific statistical test performed for each experiment are included in the appropriate figure legend or in the main text.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was performed in part at the Queensland node of the Australian National Fabrication Facility. A company established under the National Collaborative Research Infrastructure Strategy to provide nano and microfabrication facilities for Australia’s researchers. We acknowledge the facilities, and the scientific and technical assistance, of the Australian Microscopy & Microanalysis Research Facility at the Centre for Microscopy and Microanalysis, The University of Queensland. We also acknowledge A/Prof Michael R Doran for providing human MSCs and support in stem cell biology; National Health and Medical Research Council of Australia APP1126091 and APP1141121 (X.L, M.S.R, H.W and Z.P.X); National Health and Medical Research Council projects of Australia APP2008698 (C.-X.Z); Australian Research Council projects DP200101238 and DP210103079 (C.-X.Z); and National Natural Science Foundation of China, Grant nos. 11988102 and 12022207 (X.Y.).
Author contributions
D.Z., M.S.R., H.G., and C.-X.Z. designed research; D.Z., Z.W., X.Y., and X.L. performed research; J.T., H.F.W., A.B., H.L.W., and C.-X.Z. contributed new reagents/analytic tools; D.Z., Z.W., X.Y., Y.H., G.Y., Y.L., X.L., Z.P.X., H.G., and C.-X.Z. analyzed data; Z.P.X. supervision and discussion; and D.Z., X.Y., and C.-X.Z. wrote the paper.
Competing interest
The authors have patent filings to disclose. C.-X.Z. is an inventor on a patent related to the fabrication of SNs using peptides and proteins filed by The University of Queensland (European Patent Office: EP3046870A1; Australia: AU2014321161B2; United States: US20160193581A1; and Canada: CA2959201A1).
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Michael S. Roberts, Email: m.roberts@uq.edu.au.
Huajian Gao, Email: huajian.gao@ntu.edu.sg.
Chun-Xia Zhao, Email: chunxia.zhao@adelaide.edu.au.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Blanco E., Shen H., Ferrari M., Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 33, 941–951 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Izci M., Maksoudian C., Manshian B. B., Soenen S. J., The use of alternative strategies for enhanced nanoparticle delivery to solid tumors. Chem. Rev. 121, 1746–1803 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hui Y., et al. , Understanding the effects of nanocapsular mechanical property on passive and active tumor targeting. ACS Nano 12, 2846–2857 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Liu W. L., et al. , Recent advances of cell membrane-coated nanomaterials for biomedical applications. Adv. Funct. Mater. 30, 2003559 (2020). [Google Scholar]
- 5.Zou D., et al. , Artificial cells for the treatment of liver diseases. Acta Biomater. 130, 98–114 (2021), 10.1016/j.actbio.2021.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Fang R. H., Kroll A. V., Gao W., Zhang L., Cell membrane coating nanotechnology. Adv. Mater. 30, e1706759 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin J. F., et al. , Human cancer cell membrane-coated biomimetic nanoparticles reduce fibroblast-mediated invasion and metastasis and induce T-cells. ACS Appl. Mater. Inter. 11, 7850–7861 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luk B. T., et al. , Interfacial interactions between natural RBC membranes and synthetic polymeric nanoparticles. Nanoscale 6, 2730–2737 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu L. Z., et al. , Cell membrane coating integrity affects the internalization mechanism of biomimetic nanoparticles. Nat. Commun. 12, 5726 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi X., Shi X., Gao H., Cellular uptake of elastic nanoparticles. Phys. Rev. Lett. 107, 098101 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Kong S. M., Costa D. F., Jagielska A., Van Vliet K. J., Hammond P. T., Stiffness of targeted layer-by-layer nanoparticles impacts elimination half-life, tumor accumulation, and tumor penetration. Proc. Natl. Acad. Sci. U.S.A. 118, e2104826118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang Q., et al. , The softness of tumour-cell-derived microparticles regulates their drug-delivery efficiency. Nat. Biomed. Eng. 3, 729–740 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Yu M., et al. , Rapid transport of deformation-tuned nanoparticles across biological hydrogels and cellular barriers. Nat. Commun. 9, 2607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golinelli G., et al. , Arming mesenchymal stromal/stem cells against cancer: Has the time come? Front. Pharmacol. 11, 529921 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy O., Shattering barriers toward clinically meaningful MSC therapies. Sci. Adv. 6, eaba6884 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun L., et al. , Mesenchymal stem cells functionalized sonodynamic treatment for improving therapeutic efficacy and compliance of orthotopic oral cancer. Adv. Mater. 32, e2005295 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Yang N., et al. , Surface functionalization of polymeric nanoparticles with umbilical cord-derived mesenchymal stem cell membrane for tumor-targeted therapy. ACS Appl. Mater. Interfaces 10, 22963–22973 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Gao C., et al. , Stem cell membrane-coated nanogels for highly efficient in vivo tumor targeted drug delivery. Small 12, 4056–4062 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Wibowo D., Zhao C. X., Middelberg A. P., Emulsion-templated silica nanocapsules formed using bio-inspired silicification. Chem. Commun. 50, 11325–11328 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Hui Y., et al. , Nanoparticle elasticity regulates phagocytosis and cancer cell uptake. Sci. Adv. 6, eaaz4316 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tengjisi, et al. , Influence of nanoparticle mechanical property on protein corona formation. J. Colloid Interface Sci. 606, 1737–1744 (2021), 10.1016/j.jcis.2021.08.148. [DOI] [PubMed] [Google Scholar]
- 22.Guo P., et al. , Nanomaterial preparation by extrusion through nanoporous membranes. Small 14, e1703493 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Liu Y., et al. , Stable polymer nanoparticles with exceptionally high drug loading by sequential nanoprecipitation. Angew. Chem. Int. Ed. 59, 4720–4728 (2020). [Google Scholar]
- 24.Rennick J. J., Johnston A. P. R., Parton R. G., Key principles and methods for studying the endocytosis of biological and nanoparticle therapeutics. Nat. Nanotechnol. 16, 266–276 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Stevens H. Y., Bowles A. C., Yeago C., Roy K., Molecular crosstalk between macrophages and mesenchymal stromal cells. Front. Cell Dev. Biol. 8, 600160 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Najar M., Martel-Pelletier J., Pelletier J. P., Fahmi H., Mesenchymal stromal cell immunology for efficient and safe treatment of osteoarthritis. Front. Cell Dev. Biol. 8, 567813 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bose R. J., et al. , Bioengineered stem cell membrane functionalized nanocarriers for therapeutic targeting of severe hindlimb ischemia. Biomaterials 185, 360–370 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Wang M., et al. , Recent advances in mesenchymal stem cell membrane-coated nanoparticles for enhanced drug delivery. Biomater. Sci. 9, 1088–1103 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Liu H., et al. , The role of SDF-1-CXCR4/CXCR7 axis in the therapeutic effects of hypoxia-preconditioned mesenchymal stem cells for renal ischemia/reperfusion injury. PLoS One 7, e34608 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang X., Wang C., Fitch S., Yang F., Targeting tumor hypoxia using nanoparticle-engineered CXCR4-overexpressing adipose-derived stem cells. Theranostics 8, 1350–1360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leyton L., et al. , Thy-1/CD90 a bidirectional and lateral signaling scaffold. Front. Cell Dev. Biol. 7, 132 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lv F. J., Tuan R. S., Cheung K. M., Leung V. Y., Concise review: The surface markers and identity of human mesenchymal stem cells. Stem Cells 32, 1408–1419 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Wu Y., Qiao X., Qiao S., Yu L., Targeting integrins in hepatocellular carcinoma. Expert Opin. Ther. Targets 15, 421–437 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Sokeland G., Schumacher U., The functional role of integrins during intra- and extravasation within the metastatic cascade. Mol. Cancer 18, 12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nejjari M., et al. , Expression, regulation, and function of alpha V integrins in hepatocellular carcinoma: An in vivo and in vitro study. Hepatology 36, 418–426 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Corbo C., et al. , Unveiling the in vivo protein corona of circulating leukocyte-like carriers. ACS Nano 11, 3262–3273 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Cox A., et al. , Evolution of nanoparticle protein corona across the blood-brain barrier. ACS Nano 12, 7292–7300 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Ferreira-Faria I., et al. , Stem cell membrane-coated abiotic nanomaterials for biomedical applications. J. Control Release 351, 174–197 (2022). [DOI] [PubMed] [Google Scholar]
- 39.Wang H. F., et al. , Tumor-vasculature-on-a-chip for investigating nanoparticle extravasation and tumor accumulation. ACS Nano 12, 11600–11609 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Aird W. C., Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ. Res. 100, 158–173 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Vekemans K., et al. , CC531s colon carcinoma cells induce apoptosis in rat hepatic endothelial cells by the Fas/FasL-mediated pathway. Liver Int. 23, 283–293 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Berry J. D., Mettu S., Dagastine R. R., Precise measurements of capsule mechanical properties using indentation. Soft Matter. 13, 1943–1947 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Reissner E., Stresses and small displacements of shallow spherical shells. I. J. Math. Phys. 25, 80–85 (1946). [Google Scholar]
- 44.Yi X., Gao H., Cell membrane wrapping of a spherical thin elastic shell. Soft Matter 11, 1107–1115 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Gao H., Shi W., Freund L. B., Mechanics of receptor-mediated endocytosis. Proc. Natl. Acad. Sci. U.S.A. 102, 9469 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yi X., Gao H., Kinetics of receptor-mediated endocytosis of elastic nanoparticles. Nanoscale 9, 454–463 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Shen Z., Ye H., Yi X., Li Y., Membrane wrapping efficiency of elastic nanoparticles during endocytosis: Size and shape matter. ACS Nano 13, 215–228 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Hui Y., et al. , Role of nanoparticle mechanical properties in cancer drug delivery. ACS Nano 13, 7410–7424 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Luo L., et al. , Bioengineering CXCR4-overexpressing cell membrane functionalized ROS-responsive nanotherapeutics for targeting cerebral ischemia-reperfusion injury. Theranostics 11, 8043–8056 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma J., et al. , Targeted drug delivery to stroke via chemotactic recruitment of nanoparticles coated with membrane of engineered neural stem cells. Small 15, e1902011 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang C., et al. , Nanoparticles functionalized with stem cell secretome and CXCR4-overexpressing endothelial membrane for targeted osteoporosis therapy. J. Nanobiotechnol. 20, 35 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parodi A., et al. , Synthetic nanoparticles functionalized with biomimetic leukocyte membranes possess cell-like functions. Nat. Nanotechnol. 8, 61–68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Squillaro T., Peluso G., Galderisi U., Clinical trials with mesenchymal stem cells: An update. Cell Transplant. 25, 829–848 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.