Abstract
Globally decreased histone 3, lysine 27 tri-methylation (H3K27me3) is a hallmark of H3K27-altered diffuse midline gliomas (DMGs) and group-A posterior fossa ependymomas (PFAs). H3K27-altered DMGs are largely characterized by lysine-to-methionine mutations in histone 3 at position 27 (H3K27M). Most PFAs overexpress EZH inhibitory protein (EZHIP), which possesses a region of similarity to the mutant H3K27M. Both H3K27M and EZHIP inhibit the function of the polycomb repressive complex 2 (PRC2) responsible for H3K27me3 deposition. These tumors often arise in neighboring regions of the brainstem and posterior fossa. In rare cases PFAs harbor H3K27M mutations, and DMGs overexpress EZHIP. These findings together raise the possibility that certain cell populations in the developing hindbrain/posterior fossa are especially sensitive to modulation of H3K27me3 states. We identified shared molecular features by comparing genomic, bulk transcriptomic, chromatin-based profiles, and single-cell RNA-sequencing (scRNA-seq) data from the two tumor classes. Our approach demonstrated that 1q gain, a key biomarker in PFAs, is prognostic in H3.1K27M, but not H3.3K27M gliomas. Conversely, Activin A Receptor Type 1 (ACVR1), which is associated with mutations in H3.1K27M gliomas, is overexpressed in a subset of PFAs with poor outcome. Despite diffuse H3K27me3 reduction, previous work shows that both tumors maintain genomic H3K27me3 deposition at select sites. We demonstrate heterogeneity in shared patterns of residual H3K27me3 for both tumors that largely segregated with inferred anatomic tumor origins and progenitor populations of tumor cells. In contrast, analysis of genes linked to H3K27 acetylation (H3K27ac)-marked enhancers showed higher expression in astrocytic-like tumor cells. Finally, common H3K27me3-marked genes mapped closely to expression patterns in the human developing hindbrain. Overall, our data demonstrate developmentally relevant molecular similarities between PFAs and H3K27M DMGs and support the overall hypothesis that deregulated mechanisms of hindbrain development are central to the biology of both tumors.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40478-023-01514-z.
Keywords: Cancer, Chromatin biology, Onco-histones, Pediatric tumors, Brain development, Neuro-oncology
Introduction
Advances in high-throughput sequencing have provided incredible windows into understanding the diversity and heterogeneity in cancers. Genomic, transcriptomic, and epigenomic studies have aided in molecular classification of tumors, elucidation of candidate driving events, and identification of potential cells of origin for many cancers, including pediatric brain tumors. Nevertheless, brain tumors remain the leading cause of mortality among all pediatric cancers [1]. Unlike in adult populations, pediatric brain tumors frequently arise in infratentorial regions [2], suggesting a spatiotemporal susceptibility to tumorigenesis. Indeed, transcriptional analysis of pediatric brain tumors including medulloblastoma, gliomas, and ependymomas suggest shared expression programs with certain developmental lineages [3–10].
Group A posterior fossa ependymomas (PFAs) and diffuse midline gliomas (DMGs), H3K27-altered, are characterized by global reduction of the repressive histone modification histone 3 lysine 27 trimethylation (H3K27me3) [11–15]. Global H3K27me3 depletion is mediated by inhibition of the function of the polycomb repressive complex-2 (PRC2), which contains the H3K27-specific methyltransferase enhancer of zeste homolog 2 (EZH2). The majority of these DMGs harbor missense mutations in histone 3-encoding genes (primarily H3-3A and H3C2) [13–15] that result in a lysine-to-methionine substitution at position 27 (H3K27M). This histone tail mutation inhibits PRC2 activity [11, 12, 16]. Most PFAs, in contrast, exhibit overexpression of EZH inhibitory protein (EZHIP) [17, 18] that similarly suppresses PRC2 activity. EZHIP contains a methionine residue at position 406 that mimics the H3K27M mutant histone [19–23]. In addition to depleting H3K27me3, each of these tumors demonstrates increased global levels of the activation-associated mark H3K27 acetylation (H3K27ac) [18, 20, 22–27].
With their epigenetic similarities, H3K27M DMGs and PFAs also share sensitivities to inhibitors targeting histone-modifying enzymes, including histone lysine demethylases, and histone deacetylases [25–30]. Although H3K27me3 levels are greatly reduced globally, specific regions of the genome retain H3K27me3, most of which are CpG islands commonly known to be canonical PRC2 binding sites [11, 20, 31–33]. Moreover, both tumor types appear to rely on residual EZH2 activity. Both pharmacologic and genetic inhibition of EZH2 reduce tumor viability in models of H3K27M DMGs and PFAs [33–35], suggesting that the tumors rely on residual PRC2 activity.
In addition to their molecular similarities, PFAs and H3K27M DMGs arise in nearby structures associated with the hindbrain. The floor of the fourth ventricle is formed by the pons and the roof by the cerebellum. Whereas the majority of H3K27M DMGs arise from the pons, most PFAs are found in association with the fourth ventricle and cerebellum [36, 37]. Moreover, it is increasingly recognized that a rare population of H3-wildtype, low-H3K27me3 DMGs overexpress EZHIP, and that a similarly small percentage of PFAs harbor H3K27M mutations [18, 38–40]. The extent to which these two tumor subtypes share biology, and how this relates to hindbrain development, remains an unanswered question. To address this gap in our knowledge, we hypothesized that shared molecular features of H3K27M DMGs and PFAs will reveal key programs within the developing human hindbrain. We addressed this hypothesis by examining genomic, transcriptomic, and epigenomic characterizations of each tumor type to characterize shared biology to better understand the origins and potential vulnerabilities of these tumors.
Methods
Copy number alteration analyses
Arm-length copy number alterations for PFAs and DMGs were obtained from supplementary data of published datasets [36, 37]. For tumor samples from the NCI, tumors were classified with the Molecular Neuropathology classifier (v11b6) [41]. Genomic Identification of Significant Targets in Cancer (GISTIC) version 2.0.23 was used to identify and score broad copy number events. Score cutoffs of ± 0.3 were used to call a gain or loss of a chromosome arm consistent with the methods employed by Mackay et al. [36].
Gene expression analyses
PFA (GSE100240, GSE64415) microarray expression data and DMG (Mackay et al. [36]) meta-analysis expression data were obtained. A limited differential expression analysis was conducted by splitting the expression datasets by the variable of interest and applying a two-sided student’s t test. Multiple testing correction was applied by the Benjamini–Hochberg method. For gene set enrichment analysis (GSEA), ranked gene lists were generated by taking the product of the negative log-10 transformation of the q-value and the difference in expression for each gene. GSEA 4.2.3 was used to run GSEA Preranked on the resulting gene lists with the following parameters: 15–500 as min–max gene set size; weighted scoring scheme; meandiv normalization; Abs_max_of_probes mode. For standard deviation analyses, the probe with the maximum average value across samples in PFAs was used. Any genes that did not appear in both gene sets were excluded from plotting.
Histone 3 lysine 27 (H3K27) trimethylation ChIP-seq analyses
For samples from Bender et al. [11], BED files with hg19 alignment were obtained from the authors. BED files aligned to hg19 from Harutyunyan et al. [32] were obtained from the Genetics and Genomics Analysis Platform (GenAP). Data aligned to hg19 from Bayliss et al. [31] (GSE89451) and Mack et al. [33] (GSE89451) were obtained from GEO. Bedtools 2.29.2 was used to intersect peaks from each dataset with a custom BED file containing genes and their promoters defined as 2 kb upstream from the transcription start site (TSS). Genes with peaks in greater than two-thirds of tumor samples for each class of tumor were defined as common H3K27me3-retaining genes and used for downstream analyses.
H3K27ac and enhancer analyses
We obtained published H3K27M DMG-specific enhancer-associated genes from Krug et al. [24] and PFA-specific enhancer-associated genes from Mack et al. [42]. We computed the overlap in genes from these two gene sets. Each of these curated gene sets was in part derived from comparison against enhancer profiles of other brain tumors. To identify enhancers that were unique to each tumor type independent of these comparisons with non-PFAs and non-H3K27M DMGs, we accessed the ChIP-seq files available from each study and reanalyzed them. We conducted Rank-ordering of Super Enhancer (ROSE) analysis to call super enhancers for each tumor. We examined if PFA-specific enhancers from the original analysis were called as super enhancers in any of the DMG ChIP-seq samples. If an enhancer appeared at least once in the DMG samples, we included it the shared enhancer list. We repeated the converse process for DMG-specific enhancers. This method allowed us to avoid biases generated by comparisons with non-PFAs and non-H3K27M DMGs. Pathway analyses of shared or unique enhancers were completed using Enrichr [43–45]. Volcano plots were generated using code generated by Appyter.
Single-cell RNA-seq analyses
Published RNA-Seq by Expectation–Maximization (RSEM) values were obtained for PFA and DMG datasets [4, 6] and loaded as Seurat objects using Seurat (4.1.1) and SeuratObject (4.1.0). Each dataset was independently normalized, and then variable features were selected. Seurat’s SelectIntegrationFeatures and FindIntegrationAnchors were sequentially applied to the datasets, and then IntegrateData was applied based on the anchors identified. The resulting integrated dataset underwent dimensional reduction and clustering. FindConservedMarkers was used to identify differentially expressed genes by cluster. Developing brain single-cell RNA-seq data from Aldinger et al. [46] were obtained from https://cbl-dev.cells.ucsc.edu and analyzed using Seurat. AddModuleScore was used to generate a score for the H3K27me3-retained gene signature. Single-cell RNA-seq data from Eze et al. [47] was obtained from https://cells-test.gi.ucsc.edu/?ds=early-brain and processed using Seurat as described in [47], and CRABP1 feature plots were generated using the Seurat ViolinPlot function, ggplot2 (3.3.5), and ggprism (1.0.3). For area under the curve measurements from the Sepp et al. [48] dataset, raw count measurements were processed with AUCell (from Bioconductor 3.16) using the H3K27me3-retained gene signature.
Allen Brain Atlas analyses
Human data were obtained from BrainSpan, and mouse data were obtained from the Allen Developing Mouse Brain map. All resulting analyses were completed with GraphPad Prism 9.1.1.
Statistical analyses
Graphs were plotted and statistical analyses were performed using Prism software (versions 9.1.1, Graphpad, La Jolla, CA). Data are represented as the means ± standard deviation (S.D.) or as violin plots. The sample size (n) along with the statistical test performed and corresponding p-values are indicated in each figure or figure legend. Progression-free and overall survival metadata, when available, was analyzed comparing survival curves utilized the Log-Rank (Mantel-Cox) test for significance. Unpaired two-tailed, two-sided, Student’s t test or one-way ANOVA followed by multiple comparisons analysis using either Ordinary one-way ANOVA or Kruskal–Wallis tests were used to analyze data. Data were considered significant if p-values, adjusted where appropriate, were below 0.05 (95% confidence intervals).
Results
H3.1K27M and EZHIP-DMGs share key copy-number alterations with PFAs
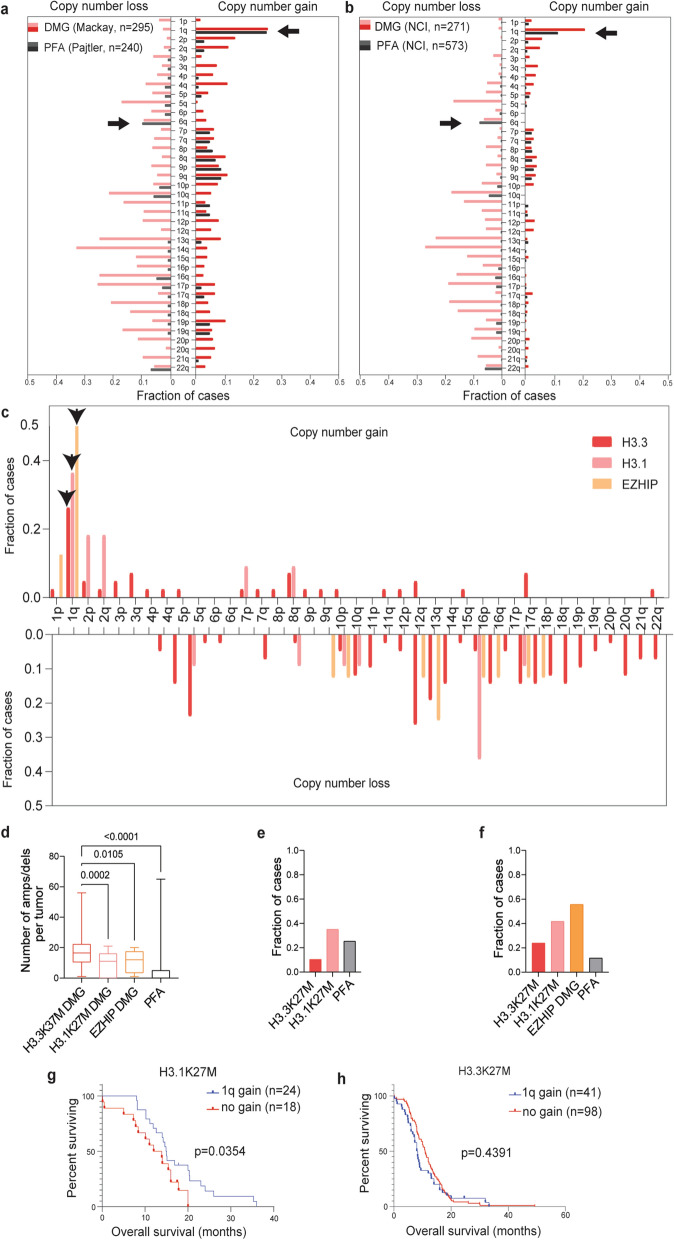
To characterize the shared features of H3K27M DMG and PFAs, we began by comparing copy number changes in two published data sets and from Mackay et al. ([36], H3K27M DMG = 295) and Pajtler et al. ([37], PFA = 240) [18, 36]. Overall, H3K27M DMGs demonstrated far greater genomic instability compared to PFAs. Nevertheless, comparison of copy number alterations demonstrated that PFAs harbor several recurrent chromosome arm alterations that are shared with H3K27M DMGs. These included gain of the long arm of chromosome 1 (1q) and loss of 6q (Fig. 1a). We validated these findings in two independent, non-overlapping tumor cohorts (PFA = 573 and DMG = 271) curated by the National Cancer Institute (NCI) (Fig. 1b).
Fig. 1.
H3.1K27M and EZHIP-DMGs share key copy-number alterations with PFAs. a Copy-number profiles (loss left panel, gain right panel) of H3K27M DMGs (Mackay et al. 2017, n = 295) [36] and PFAs (Pajtler et al. 2015, n = 240) [37] from published DNA methylation cohorts. Arrows indicate shared recurrent alterations (more than 5% of tumors in all cohorts) in a and b with known survival associations in PFAs. X-axis = fraction of cases and Y-axis = chromosomes. b Copy-number profiles (loss left panel, gain right panel) of H3K27-altered DMGs (n = 271) and PFAs (n = 573) from the National Cancer Institute (NCI). Arm-level copy-number gain/loss calls were made using GISTIC score cutoffs of ± 0.3. c Frequency of chromosome arm-level copy number alterations (gain top panel, loss bottom panel) for H3K27-altered DMGs from b segregated by molecular alterations: H3.3 mutant (n = 42), H3.1 mutant (n = 11) and H3-wildtype (H3-WT), EZHIP-expressing (n = 8). X-axis = chromosomes and Y-axis = fraction of cases. d Quantification of the frequency of amplifications (amps) and deletions (dels) (Y-axis) in DMGs segregated by molecular alterations (X-axis) and in PFAs from the NCI cohort. Data analyzed by one-way ANOVA with 95% confidence intervals. H3.3 mutant (n = 48), H3.1 mutant (n = 12), and EZHIP (n = 8). e Frequency of 1q gain (Y-axis) for subtypes of H3K27M DMG and PFAs from published datasets depicted in a. H3.3 mutant (n = 245), H3.1 mutant (n = 49), and PFA (n = 240). DMG sample pHGG_META_0223 was excluded from as it is H3.2-mutant. f Frequency of 1q gain (Y-axis) for subtypes of H3K27-altered DMG and PFA ependymoma from the NCI cohort depicted in 1b. H3.3 mutant (n = 42), H3.1 mutant (n = 11), and EZHIP (n = 8). g Overall survival (months) analysis of 1q gain in H3.1K27M DMGs with (n = 24) or without (n = 18) 1q gain. h Overall survival (months) analysis of 1q gain in H3.3K27M DMGs with (n = 41) or without (n = 98) 1q gain. Data in g–h analyzed using Kaplan–Meier with Log-Rank test with 95% confidence intervals
Gain of 1q has previously been associated with worse overall survival outcome in PFAs in multiple studies [18, 49–52], a finding reproduced in our investigation of the NCI PFA cohort (Additional file 1: Fig. S1a). We noted that 1q gain was also the most common recurrent arm-length gain in H3K27M DMGs; however, there was no significant difference in overall survival in H3K27M DMGs with or without 1q gain from the published cohorts (Additional file 1: Fig. S1b). Previous work had demonstrated differences in the copy-number landscapes based on the type of H3 mutation with H3.3- versus (vs.) H3.1-mutant tumors exhibiting increased genomic instability [36]. We examined copy number trends with respect to H3.3 vs. H3.1 status in the NCI cohort and evaluated how H3 wildtype (H3-WT) EZHIP-expressing DMGs compared to each histone subtype. We observed that H3.1-mutant and EZHIP-expressing DMGs had fewer arm-length copy-number events (Fig. 1c) and focal amplifications and deletions (Fig. 1d) compared to H3.3 mutant DMGs. Gain of 1q was observed at a higher frequency in H3.1 vs. H3.3K27M DMGs in both the Mackay et al. (H3.1 = 35.1% vs. H3.3 = 10.5%; Fig. 1e) and the NCI cohort (H3.1 = 41.7% vs. H3.3 = 23.9%; Fig. 1f). Notably, half of EZHIP-expressing DMGs also exhibited 1q gain (Fig. 1f). Unexpectedly, 1q gain was associated with better overall survival in H3.1K27M tumors (Fig. 1g), but not in H3.3K27M-mutant tumors (Fig. 1h) or in other pediatric brain tumors—including other pediatric gliomas [36, 53], other posterior fossa ependymomas [54], and medulloblastomas [55] with the exception of a small sample-size cohort of group 4 medulloblastoma (Additional file 1: Fig. S1c–h). This intriguing finding of a 1q gain survival association in H3.1K27M tumors was unable to be assessed further in the NCI cohort as the majority of these tumors did not have sequencing information to distinguish H3.1 from H3.3 mutations.
In addition to recurrent 1q gain, the other alterations observed in at least 5% of each cohort were losses of 6q, 10q, and 22q (Fig. 1a, b). Loss of 6q has recently been associated with a subset of PFAs with poor survival rates [56]. Assessment of the 6q loss in the DMG cohort demonstrated a trend toward worse survival in 6q loss tumors (Additional file 1: Fig. S1i). In H3K27M DMGs, there was no major difference in 6q status when comparing H3.1 and H3.3-mutant cohorts (Additional file 1: Fig. S1j). Given the genomic instability of DMGs in general, we asked if 6q alterations were associated with high arm-level alterations. In every cohort, tumors with 6q loss were associated with higher levels of arm-level losses across the genome. In contrast, the frequency of arm gains in these tumors was not significantly different from non-6q loss tumors in each cohort (Additional file 1: Fig. S1k). We hypothesized that despite the relatively quiescent genome PFAs exhibit compared to H3K27M DMGs, the two tumor types might share certain alterations that permit or enhance tumorigenicity in the context of H3K27me3 dysregulation. Our findings highlight a spectrum of genomic alteration patterns that varies by driving alteration with H3.3-mutant gliomas exhibiting the greatest alteration frequencies.
ACVR1 expression is prognostic in PFAs and is associated with transcriptomic signatures observed in ACVR1-mutant DMGs
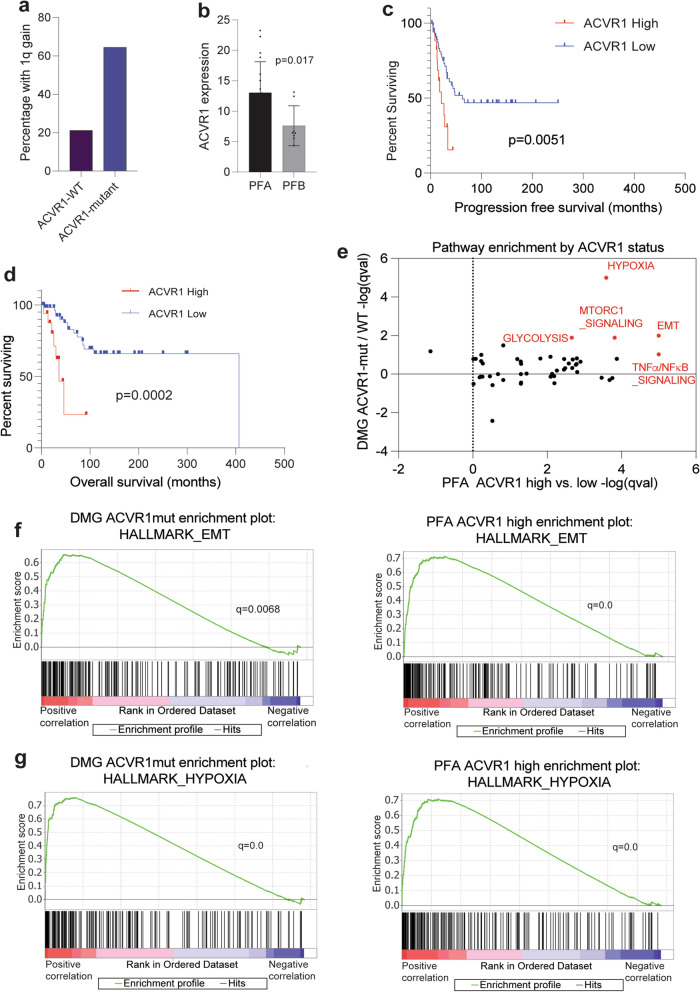
Due to our observed differences between H3.1 and H3.3 K27M DMGs and 1q status, we extended our comparative analysis to include other genomic alterations that segregate with mutant histone subtypes. Activating mutations in activin A receptor type 1 (ACVR1) occur more frequently in H3.1 vs. H3.3 DMGs [36, 57–59]. Concordant with our findings of 1q gain enrichment in H3.1 tumors, we observed increased frequency of ACVR1 mutations in 1q gain tumors vs. non-1q gain tumors (Fig. 2a).
Fig. 2.
ACVR1 expression is prognostic in PFAs and associated with transcriptomic signatures observed in ACVR1-mutant DMGs. a Frequency of 1q gain (Y-axis) in ACVR1-mutant (n = 31) vs. WT (n = 137) H3K27M DMGs from Mackay et al. [36]. b ACVR1 RNA expression levels (Y-axis) in PFAs (n = 19) compared to PFBs (n = 8) from Bayliss et al. 2016 [31]. Data analyzed by unpaired, two-tailed t test. c Progression-free survival (months) analysis comparing PFAs with ACVR1 high (the top quartile, n = 19) vs. low (lower three quartiles, n = 57) expression. d Overall survival (months) analysis comparing PFAs with ACVR1 high (the top quartile, n = 19) vs. low (lower three quartiles, n = 57) expression. Data in 2c-d analyzed using Kaplan–Meier, Log-Rank test with 95% confidence intervals. e Comparison of enriched pathways from differentially expressed genes in ACVR1-mutant vs. -wildtype H3K27M DMGs (from 2a, Y-axis) and ACVR1-high vs. -low PFAs (from 2d, X-axis). f Gene set enrichment analyses of the Hallmark epithelial-to-mesenchymal transition (EMT) pathway for genes ranked by differential expression in ACVR1-mutant DMGs or in high ACVR1 expressing PFAs. g Gene set enrichment analyses of the Hallmark hypoxia pathway for genes ranked by differential expression in ACVR1-mutant DMGs or in high ACVR1 expressing PFAs
Recent work has identified rare mutations of ACVR1 in PFA tumors [60]. These findings prompted us to investigate ACVR1 in PF ependymomas. First, we observed increased expression of ACVR1 in PFAs compared to group B posterior fossa ependymomas (PFBs), a subtype of PF ependymomas that lacks EZHIP expression [18] (Fig. 2b). We found tumors with the highest levels of ACVR1 expression in the Pajtler et al. cohort to be associated with worse progression-free and overall survival outcomes (Fig. 2c, d). We next investigated whether ACVR1-mutant H3K27M DMGs and ACVR1-high PFAs shared transcriptomic signatures. We found that both tumors demonstrated enrichment of many ACVR1-mutant [61] and H3.1K27M-enriched [62] pathways including hypoxia/HIF1-α transcription factor network and epithelial-to-mesenchymal transition, as well as glycolysis, TNFα/NFκB signaling, and mTORC1 signaling (Fig. 2e–g).
H3K27me3 commonly enriched genes in PFAs and H3K27M DMGs exhibit heterogeneity corresponding to tumor anatomic location
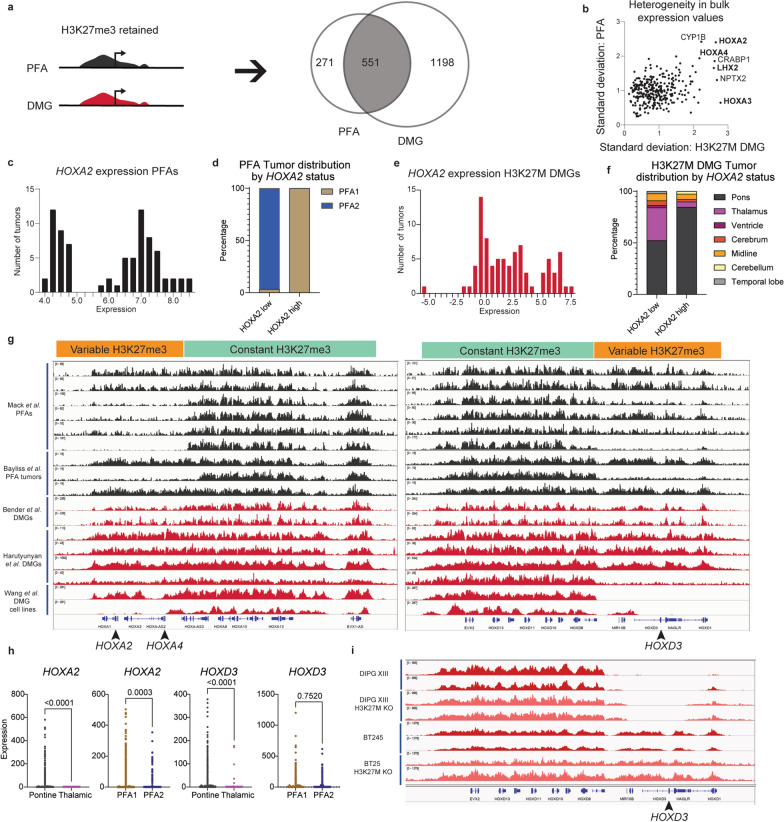
We next focused our efforts on defining common epigenetic signatures in H3K27M DMGs and PFAs. Despite global reduction of H3K27me3, multiple studies have demonstrated that both tumors retain H3K27me3 at specific genomic sites [11, 16, 21, 32, 33, 35]. Prior studies have established that the patterns of retained H3K27me3 are distinct from other pediatric brain tumors [11, 33] and have pointed to several shared sites. While specific loci such as the CDKN2A locus have been pointed out as key sites of retained H3K27me3 in data from both tumor subtypes, more global analyses of the similarities and differences in H3K27me3 landscapes in these tumors are lacking. We performed a systematic comparison of the genes that retain H3K27me3 in H3K27M DMGs and PFAs to better understand this shared chromatin landscape.
H3K27me3 chromatin-immunoprecipitation with high throughput sequencing (ChIP-seq) profiles generated by our group [31] and others [11, 32, 33] were analyzed. We examined H3K27me3 enrichment in nine PFA ependymomas and six DMGs to define genes with commonly methylated promoters and gene bodies between the two tumors. We identified 551 genes with H3K27me3 retained in both tumor sets (Fig. 3a). Because H3K27me3 is a repressive mark, the extent of repression was examined by analyzing standard deviation values of gene expression for each of these 551 genes within both H3K27M DMGs and PFAs. Analysis of bulk expression data from both tumor cohorts identified high variance in expression levels of several H3K27me3-associated genes in both PFAs and H3K27M DMGs with greatest variability in homeobox genes including HOXA2, HOXA4, and LHX2. (Fig. 3b). Examination of genomic H3K27me3 at HOX gene clusters in PFAs, H3K27M DMGs, and patient-derived H3K27M-DMG cell lines demonstrated distinct variations in enrichment (Fig. 3c). In both tumor sets, some samples showed H3K27me3 enrichment along the entire length of the HOXA and HOXD clusters, but others demonstrated variability in H3K27me3 at the HOXA1 through HOXA7 loci and the proximal HOXD locus (Fig. 3c).
Fig. 3.
Shared H3K27me3-enriched genes in PFAs and H3K27M DMGs exhibit heterogeneity corresponding to tumor anatomic location. a Workflow of H3K27me3 ChIP-seq analysis. Genes with promoter (2 kb upstream) or gene-body H3K27me3 peaks in more than two-thirds of PFAs (≥ 7/9 samples) and H3K27M DMGs (≥ 5/6) were identified as H3K27me3-retaining genes in each tumor type. The resulting gene sets were then compared to identify shared and subtype-specific H3K27me3-retaining genes. b Heterogeneity in expression of H3K27me3-retaining genes was identified by plotting the expression standard deviations of each gene identified from 3a in H3K27M DMGs (X-axis) and PFAs (Y-axis). Genes, including HOXD3, for which expression data was unavailable for both subtypes were excluded from plotting. c Histogram of HOXA2 expression values per tumor (X-axis = binned expression scores, Y-axis = number of tumors) in PFAs. d Comparison of HOXA2-high (expression > 5 units) vs. -low (expression < 5.5 units) groups based on the local minimum between modes in DNA-methylation defined, anatomically distinct PFA1 (n = 49) and PFA2 (n = 29) subtypes of PFAs. e Histogram of HOXA2 expression values per tumor (X-axis = binned expression scores, Y-axis = number of tumors) in H3K27M DMGs. f Comparison of HOXA2-high (expression > 4.5 units) vs. -low (expression < 4.5 units) groups based on the local minimum between modes with anatomic location of H3K27M DMGs (pons = 65; thalamus = 26; midline not otherwise specified (NOS) = 5; cerebellum = 1, and other = 2). g H3K27me3 ChIP-seq tracks for PFAs (black, n = 9, from Mack et al. [33], and Bayliss et al. [31]) and H3K27M DMGs (red, middle, n = 6, from Bender et al. [11] and Harutyunyan et al. [32]) tumors and patient-derived H3K27M cell lines (red, bottom, n = 2 from Wang et al. [63]) at the HOXA and HOXD clusters. Positions of proximal HOX genes associated with variable expression (HOXA2, HOXA4, HOXD3) indicated with arrowheads. Regions identified with differential patterns in H3K27me3 enrichment are labeled as variable (orange) and constant (green). Anatomic site for DMGs and PFA1/2 status were not available. h Expression patterns of HOXA2 and HOXD3 in single-cell RNA-sequencing (scRNA-seq) experiments (Filbin et al. and Gojo et al.) [4, 6]. Tumors were grouped by anatomic site (pons vs. thalamus for H3K27M DMG) or methylation-based subtype (PFA1 vs PFA2 for PFAs). i H3K27me3 ChIP-seq tracks at the HOXD locus from isogenic patient-derived H3K27M DIPG XIII and BT425 cell lines with or without H3K27M-knockdown
HOXA2, HOXA3, and HOXA4 have been identified as transcriptional markers that differentiate PFA1 and PFA2 [18]. Pajtler et al. proposed that the PFA1 tumors overexpress HOXA-family genes and hypothesized distinct anatomical origins PFA1 vs. PFA2 ependymomas [18]. Assessment of HOXA2 bulk expression revealed a bimodal expression pattern in PFAs (Fig. 3d). As expected, segregation of HOXA2 high vs. low PFAs occurred almost completely along with PFA1 vs. PFA2 status with majority of HOXA2 low tumors corresponding to PFA2 subgroup (Fig. 3e). H3K27M DMGs similarly exhibited bimodal HOXA2-high vs. low distribution (Fig. 3f). Moreover, HOXA2 expression levels revealed similar anatomic segregation with majority of thalamic tumors demonstrating low HOXA2 expression (Fig. 3g). HOXA4 expression demonstrated a similar, though less striking pattern (Additional file 1: Fig. S2a–d). HOXD3, for which expression data in PFAs was unavailable, also followed a similar bimodal distribution (Additional file 1: Fig. S2e) and segregation with pontine tumors in HOXD3-high H3K27M DMGs (Additional file 1: Fig. S2f). Evidence of HOX family expression in single-cell data from DMGs and PFAs [4, 6] further supported this anatomic pattern in expression. Tumors derived from the pons and PFA1 showed higher HOXA2 and HOXA4 expression, while HOXD3 expression was elevated in pontine vs. thalamic tumors but was not significantly different between PFA1 and PFA2 tumors (Figs. 3h and Additional file 1: Fig. S2g). This distinction in HOX gene expression patterns between DMGs and PFAs may reflect subtle regional differences in the cells of origin; however, HOXD3 expression data from additional PFA tumors will be necessary to validate this hypothesis.
Differential H3K27me3 enrichment at these loci could be due to varying effects of PRC2 inhibition by the H3K27M mutation. To test this hypothesis, we examined published ChIP-seq tracks from DIPG-XIII and BT245 H3K27M patient-derived cell lines with or without H3K27M knockdown [32]. In both cell lines, H3K27M knockdown did not alter H3K27me3 genomic distribution at these genomic loci (Fig. 3i), suggesting that differential H3K27me3 at this site is not dependent on H3K27M. Overall, both H3K27M DMGs and PFA ependymomas showed differences in H3K27me3 genomic distribution at specific gene loci consistent with gene expression differences based on tumor anatomic location. These patterns are in contrast with the initial hypothesis that H3K27M and EZHIP might drive expression patterns through modulating the degree of retained H3K27me3 at PRC2-target sites and highlight the importance of the anatomic sites of origin on this epigenetic signature.
Enhancer signatures in PFAs and H3K27M DMGs show high expression in astrocyte-like tumor cells
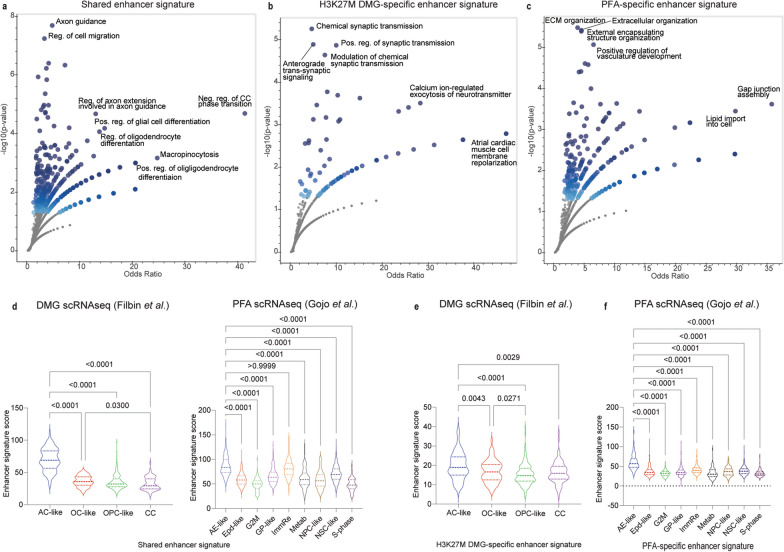
Both H3K27M mutations and EZHIP overexpression lead to a global increase in H3K27 acetylation (H3K27ac), a mark associated with active transcription [18, 22, 24–27]. Moreover, H3K27M DMGs and PFAs exhibit unique H3K27ac marked enhancer and super enhancer profiles [42, 64, 65]. We combined information from gene lists generated by Mack et al. and Krug et al. and reanalyzed publicly available H3K27ac ChIP-seq data from these studies to identify enhancers that are shared by, or unique to, each tumor (Additional file 1: Fig. S3). H3K27ac-marked gene enhancers shared by both H3K27M DMGs and PFAs (n = 533, Additional file 1: Fig. S3) were mainly associated with axonogenesis, axon guidance, and cell motility pathways, as well as differentiation of glial and oligodendrocyte lineages (Fig. 4a). DMG-specific enhancers mapped mainly to genes enriched for synaptic signaling (Fig. 4b), whereas PFA-specific enhancers were associated with genes enriched for extracellular organization and angiogenesis (Fig. 4c).
Fig. 4.
Enhancer signatures in PFAs and H3K27M DMGs show high expression in astrocyte-like tumor cells. a Enriched gene ontology biological process (GO BP) gene sets (Y-axis Negative Log10 p value, X-axis Odds ratio) for genes associated with enhancers shared by PFAs and H3K27M DMGs. Shading of points is influenced by p-value (darker shade with lower p-values) and number of overlapping points. b Enriched gene ontology biological process (GO BP) gene sets (Y-axis Negative Log10 p value, X-axis Odds ratio) for genes associated with enhancers unique to H3K27M DMGs. c Enriched gene ontology biological process (GO BP) gene sets for genes (Y-axis Negative Log10 p value, X-axis Odds ratio) associated with enhancers unique to PFA ependymomas. d Comparison of expression of the shared enhancer signature by cell type in H3K27M DMGs [4] and PFAs [6]. Signature scores per cell were calculated by taking the mean of expression values for the genes in the signature. e Expression of the H3K27M DMG-specific enhancer signature by DMG cell type. f Expression of the PFA-specific enhancer signature by DMG cell type. Data in d–f analyzed by non-parametric Kruskal–Wallis test and 95% confidence intervals. AC-like, Astrocyte-like; OC-like, Oligodendrocyte-like; OPC-like, Oligodendrocyte precursor-like; CC- cell cycle; AE-like, Astroependymal-like; Epd-like, Ependymal-like; G2M, G to M cell cycle related; GP-like, Glialprecursor-like; ImmRe, Immune related; Metab, Metabolic; NPC-like, Neuronal precursor-like; NSC-like, Neuronal stem cell-like; S-phase, S-phase cell cycle
Due to the importance of enhancer activity in regulation of cell identity, we explored whether the enhancer signatures segregated with differentiation cell states within each tumor by assessing the single-cell RNA-seq datasets [4, 6]. Single-cell RNA-sequencing in H3K27M-DMGs and PFA ependymomas has demonstrated marked intratumoral heterogeneity [4, 6]. Both tumors contain malignant cells with varying expression of stem-cell-like, and more differentiated tumor cell signatures. H3K27M DMGs contain tumor cells bearing oligodendrocyte precursor (OPC)-like, oligodendrocyte (OC)-like and astrocyte (AC)-like transcriptional signatures [4]. Similarly, PFAs contain less differentiated neuronal stem cell (NSC)-like, glial progenitor (GP)-like, and neuronal precursor cell (NPC)-like, as well as more differentiated ependymal (Epd)-like and astroependymal (AE)-like tumor cells [6]. Mapping the expression signatures onto tumor-cell types showed the highest average expression of shared enhancer signature within astrocyte-like cells in H3K27M DMGs and astroependymal-like and immune-reactive PFA tumor cells (Fig. 4d). Evaluation of the tumor-specific enhancer signatures by single-cell type also showed increased expression in astrocytic-like lineages for both tumor types (Fig. 4d, e). We initially hypothesized that because enhancers play critical roles in regulating cell identity, and super enhancers in particular have been associated with activation of tumor-specific transcriptional programs [66], the H3K27M DMG and PFA-specific enhancer signatures might associate closely with more potent progenitor-like cell types. However, our findings demonstrate that these previously identified enhancer signatures seem to more closely align with expression programs of cells within the tumor that demonstrate astrocytic lineage features.
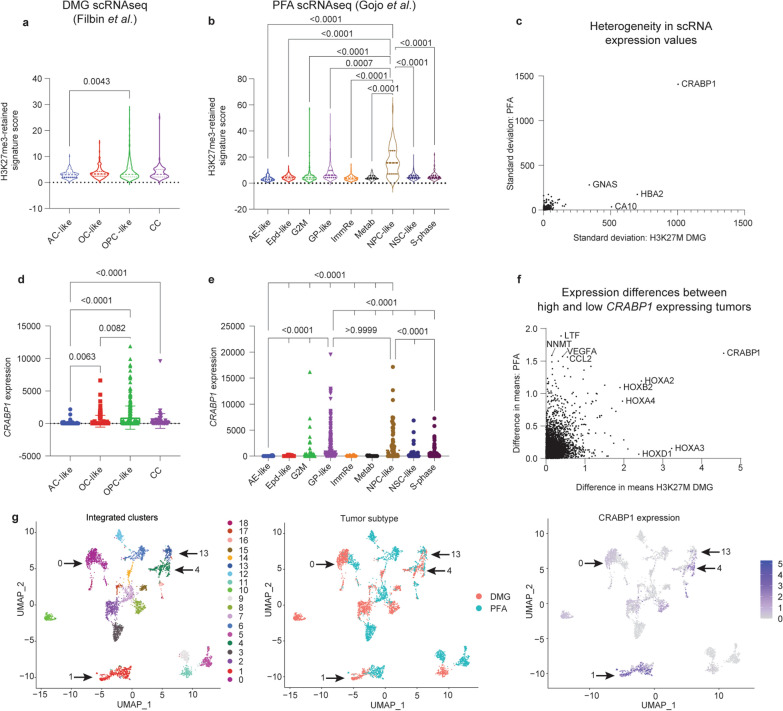
H3K27me3-enriched CRABP1 is highly expressed in progenitor populations of both tumors
In further interrogation of the single-cell datasets, we asked whether genes with retained H3K27me3 exhibited preferential expression in single-cell defined tumor cell populations. In both tumors, cells with progenitor-like states including OPC-like tumor cells in H3K27M-DMGs, and NPC-like tumor cells in PFA ependymomas, tended to have higher expression of genes in the retained H3K27me3 signature shared by H3K27M DMGs and PFAs (Fig. 5a, b).
Fig. 5.
H3K27me3-enriched CRABP1 is expressed highly in progenitor populations of both tumors. a Expression of the shared H3K27me3-retaining gene signature across cell-types in scRNA-seq data from H3K27M DMGs (Filbin et al.) [4]. Expression scores per cell were determined as the average expression of all genes in each cell. b Expression of the shared H3K27me3-retaining gene signature across cell-types in scRNA-seq data from PFA ependymomas (Gojo et al.) [6]. Only p-values for comparisons involving NPC-like cells are shown. c Comparison of the heterogeneity in expression of H3K27me3-retaining genes as measured by the standard deviation in expression of each H3K27me3-retaining gene in H3K27M DMGs (X-axis) and PFA ependymomas (Y-axis). Genes for which expression data was not available in both bulk expression data sets were excluded from plotting. 397 genes were plotted. d CRABP1 expression values per cell from scRNA-seq data in H3K27M DMGs, grouped by cell type. e CRABP1 expression values per cell from scRNA-seq data in PFAs, grouped by cell type. Only p-values for comparisons involving NPC-like and GP-like cells are shown. f Analysis of genes co-expressed with CRABP1 in bulk RNA-seq datasets by comparison of difference in means for CRABP1-high and CRABP1-low tumors in H3K27M DMGs (X-axis) and PFA ependymomas (Y-axis). A split at the mean expression of CRABP1 from the Mack et al. dataset was used to delineate H3K27M DMG CRABP1-high (n = 37) and CRABP1-low tumors (n = 46). A split in the mean expression of the highest-level CRABP1 probe from the Pajtler et al. dataset was used to delineate PFA CRABP1-high (n = 22) and -low tumors (n = 56). g Uniform Manifold Approximation and Projection (UMAP) embeddings of integrated scRNA-seq datasets of H3K27M DMG (Filbin) and PF ependymoma (Gojo). Left: identification of 20 distinct clusters. Middle: labeling by tumor of origin. Right: CRABP1 expression across single cells with color scale minimum set to 0. Kruskal–Wallis test followed by multiple comparisons analysis were used to analyze data in a–b and e–d
To further assess heterogeneity, we analyzed genes with the greatest variability in expression by comparing standard deviation values for each gene across all tumor cell types in both tumors. Expression of cellular retinoic acid binding protein 1 (CRABP1) was a clear outlier for both tumor subtypes (Fig. 5c). Moreover, CRABP1 was one of the H3K27me3 marked genes (Additional file 1: Fig. S4a) with the greatest variability in expression levels in the bulk datasets (Fig. 3b). CRABP1 is frequently dysregulated in cancer [67–71]. CRABP1 expression segregated strongly with OPC-like cells in DMGs (Fig. 5d) and both glial progenitor-like (GP-like) and neuronal precursor-like (NPC-like) in PFAs (Fig. 5e). We determined genes that correlate with CRABP1 expression in bulk expression to find that the HOX genes were among the strongest correlates in both tumor datasets (Fig. 5f). However, HOXA2 and HOXD3 did not demonstrate any distinct patterns of expression by cell type in DMGs, and the significantly different changes observed in PFAs were far less drastic (Additional file 1: Figure S4b, c).
The scRNA-seq datasets were integrated to further explore similarities in expression patterns of H3K27M DMGs and PFAs. Integrated dimensional reduction analyses identified 20 clusters, four of which (clusters 0, 1, 4, and 13) included large populations of both PFA- and DMG-derived cells (Arrows, Fig. 5g). The top marker gene in the second-largest cluster’s (cluster 1) was CRABP1 (Fig. 5g, Additional file 1: Fig. S4d). This cluster consisted primarily of progenitor and precursor cell states: OPC-like cells from the DMGs and GP-like cells from PFAs (Additional file 1: Fig. S4e). Moreover, assessment of single-cell populations from other pediatric brain tumors using a dataset with all four subgroups of medulloblastoma [72] failed to identify any populations with elevated CRABP1 expression in those tumors (Additional file 1: Fig. S4f). Overall, these data support a role for CRABP1 in specific tumor cell-states in both tumors, consistent with the hypothesis that PFA and DMG tumors may benefit from expression of similar genes to maintain progenitor states.
Common H3K27me3 signatures mirror human hindbrain brain developmental patterns
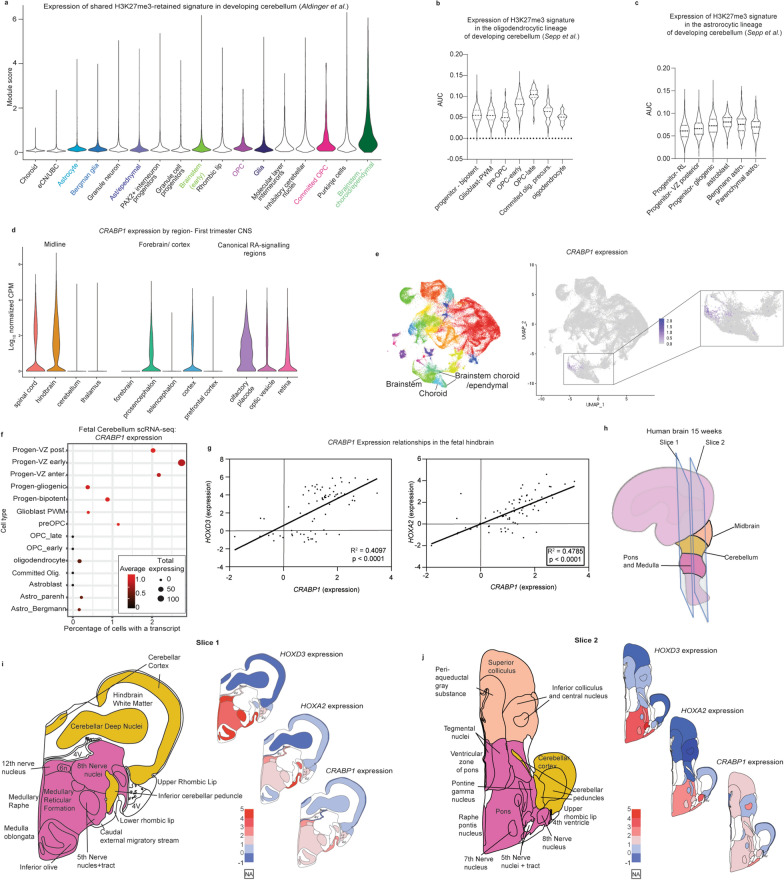
H3K27M DMGs and PFAs are proposed to arise from cells along developmental lineages in the hindbrain [4–6, 8, 10]. We first aimed to identify whether H3K27me3-associated genes exhibited specific patterns in normal human hindbrain development. We utilized scRNA-seq data from fetal human cerebellum ranging from 9–21 weeks post-conception published by Aldinger et al. [46]. Overall, average expression of H3K27me3-retained genes increased in more differentiated populations of cells associated with potential cells-of-origin for DMGs and PFAs. Committed OPCs demonstrated increased signature expression compared to OPCs, and brainstem-choroid/ependymal cells had higher levels of expression than early brainstem cells (Fig. 6a). Cells of astrocytic lineage did not show a similar pattern (Fig. 6a). Because OPCs are the leading proposed cell of origin for H3K27M DMGs [4, 65], we examined oligodendroglial lineages in more detail in a second single-cell dataset of human developing cerebellum (Sepp et al., unpublished) [48]. H3K27me3 signatures were low in progenitor cells but increased along stages of OPC differentiation with the highest expression in late-OPCs and then decreased levels in more differentiated committed OPCs and oligodendrocytes (Fig. 6b). Analysis of the astrocytic lineage cells did not yield a similar pattern (Fig. 6c).
Fig. 6.
Common H3K27me3-retained signature mirrors human hindbrain brain developmental patterns. a Expression of the shared DMG/PFA-derived H3K27me3 gene signature in scRNA-seq data of the developing human posterior fossa (Aldinger et al.) [46] grouped by cell type. Highlighted groups indicate cell types pertinent to DMGs/PFAs. b H3K27me3 gene-signature expression along the oligodendrocyte lineage in an independent scRNA-seq atlas of the developing posterior fossa (Sepp et al., unpublished) [48]. c H3K27me3 gene-signature expression along the astrocytic lineage in the scRNA-seq atlas of the developing posterior fossa used in b. d First-trimester human brain scRNA-seq [47] expression patterns of CRABP1 grouped by anatomic region. Left: midline structures, middle: forebrain and cortical structures, right: regions with canonical retinoic acid (RA) signaling. e CRABP1 expression patterns in the developing cerebellum scRNA-seq atlas from a. Left: UMAP embedding colored by cluster, middle: CRABP1 expression across all cells in UMAP embedding, right (inset): magnified depiction of CRABP1 expression patterns in a subset of cells including brainstem and brainstem-derived choroidal/ependymal cells. f CRABP1 expression patterns in astrocytic and oligodendrocyte lineages of the developing cerebellum scRNA-seq atlas from b and c. Early progenitors are grouped along the top of the Y-axis. The more differentiated oligodendrocyte and the astrocytic lineages are each grouped below along the Y-axis. The percentage of cells expressing at least one CRABP1 transcript (X-axis), the total number of cells expressing at least one CRABP1 transcript (point size) and the average expression value (color) are depicted. g Microarray-based log2 expression levels from micro-dissected fetal hindbrain tissue at 15 and 16 weeks post-conception from BrainSpan’s prenatal lateral microdissection (LMD) Microarray comparing CRABP1 expression patterns to those of HOXA2 and HOXD3. Data were analyzed with a simple linear regression and R-squared calculated with a goodness of fit test with 95% confidence intervals. h Schematic depicting sectioning of the 15–16 weeks post conception fetal brain utilized for depictions in g, I, and j. Early anatomic structures of the infratentorial brain are colored in peach (midbrain), yellow (cerebellum), and deep pink (pons and medulla). i-j Expression of HOXD3, HOXA2, and CRABP1 at 15 and 16 weeks post conception mapped to infratentorial brain at slices 1 (i) and slices 2 (j) from h. Left: schematic of brain sub-structures colored by overarching structure corresponding to key in h. Right, from top to bottom heatmaps of HOXD3, HOXA2, and CRABP1 expression in corresponding areas. Illustration adapted from reference figure from the Allen Brain Atlas
Given its notable expression pattern in the single-cell tumor datasets (Fig. 5), we examined CRABP1 expression in the developing hindbrain utilizing a scRNA-seq dataset generated from human brain samples from the first trimester [47]. Carnegie stages are used to define periods of human brain development [73]. In the hindbrain, we noted a peak of CRABP1 expression in stage 19, which corresponds to the peak in expression of H3-3A, the histone variant most commonly mutated in DMGs (Additional file 1: Fig. S5a). With respect to anatomic location, CRABP1 expression was higher in the spinal cord and hindbrain vs. thalamus and cerebellum (Fig. 6d). Known areas of high retinoic acid (RA) signaling in development, including the olfactory placode and optic pathways demonstrated enrichment of CRABP1 expression, whereas expression in forebrain/cortical-related structures was variable (Fig. 6d). Similarly, CRABP1 expression was restricted to a subset of brainstem cells in the Aldinger et al. developing cerebellum atlas (Fig. 6e). At a cellular level, CRABP1 expression was highest in the progenitor cell types compared to their more differentiated oligodendrocytic and astrocytic cells (Fig. 6f).
Data from mouse development have previously identified CRABP1 expression as specific to rhombomeres 4–6 (r4–6) [74]. The r4-6 segments give rise to the caudal portion of the basilar pons (r4) and retropontine structures (r5–6) [75, 76]. Given this spatial patterning, the Allen Brain Atlas’ Developing Human Transcriptome was examined to determine patterns of CRABP1 expression [77, 78]. Within hindbrain structures, CRABP1 expression strongly correlated with HOX genes including HOXA2 and HOXD3 (Fig. 6g). Moreover, HOXD3, HOXA2 and CRABP1 expression in the developing human hindbrain at 15-weeks gestation was mainly restricted to developing human pontine and medullary structures (Fig. 6h–j, Additional file 1: Fig. S5b), a finding supported by developing mouse brain expression patterns (Additional file 1: Fig. S5c). Altogether, these data support the positional expression patterns observed in both H3K27M DMGs and PFAs, consistent with these patterns reflecting epigenetic states from developmental lineages.
Discussion
While EZHIP-expressing PFAs and H3K27-altered DMGs (including H3.3 and H3.1K27M gliomas and EZHIP positive DMGs) have distinct natural histories and histologic features, evidence of their shared molecular features continues to mount as we profile broader samples of these tumor types. Furthermore, both tumors most commonly arise in neighboring hindbrain/posterior fossa-derived structures. This pattern suggests that developmental lineages in this region possess susceptibility to alterations that impede canonical PRC2 activity and therefore regulation of H3K27me3 state. Leveraging the existing molecular knowledge about each of these tumor types, we exploited genomic, bulk transcriptomic, scRNA-seq, and epigenomic profiles of both cancers to elucidate shared features that may improve our understanding of tumor biology.
Assessment of the genomic landscape of these tumors demonstrated that H3.1K27M DMGs showed greater copy number similarity with EZHIP-expressing DMGs than did H3.3K27M tumors, with more quiescent landscapes and higher frequencies of 1q gain. These profiles of relatively lower genomic instability may point toward greater commonalities between PFAs and H3.1K27M vs. H3.3K27M DMGs. Consistent with this pattern, we identified high ACVR1—a gene frequently altered in H3.1-mutant DMGs—expression to be associated with: (a) worse survival in PFAs, and (b) upregulation of pathways that were also enriched in ACVR1-mutated gliomas.
We additionally characterized common and unique features of H3K27M DMG and PFA chromatin landscapes based on patterns of H3K27me3/ac genomic deposition. The two tumors exhibited a considerable overlap in both H3K27me3-marked and H3K27ac-enhancer-associated genes. Exploration of the transcriptional states associated with these gene sets demonstrated inter- and intra-tumoral heterogeneity of expression patterns. Common enhancer-associated genes were enriched in cell motility and oligodendrocyte and glial differentiation pathways. Moreover, scRNA-seq demonstrated the highest levels of expression of these genes in astrocytic lineage cell types, especially in comparison to progenitor cell types. These patterns highlight the unique insights single-cell resolution transcriptomics can provide and illustrate how tumor-specific enhancers generated from bulk-sequencing methods may not fully reflect the regulatory landscapes of the tumors.
Interrogation of genes associated with residual H3K27me3 also yielded important insights. The sensitivity of H3K27M DMGs and PFAs to EZH2 inhibition [33–35] points to a potential role for H3K27me3 in maintaining tumor cell proliferative capacity. Data from developing cerebellar atlases provide some support for this model in that upregulation of genes within these signatures is seen along oligodendrocyte lineages and in brainstem choroid/ependymal cells. Furthermore, we noted distinct expression profiles of proximal vs. distal HOX genes, which correlated with inferred anatomic origins of tumor, and these genes demonstrate distinct anterior–posterior boundaries of expression in the developing human hindbrain. These data suggest that developmental epigenetic profiles and origins of PFAs and H3K27M DMGs may influence the H3K27me3 landscape and gene expression programs in these tumors.
CRABP1, like the HOX genes, has been shown to have highly specific expression patterns in mouse nervous system development [74]. In both PFAs and DMGs, its expression correlated with specific tumor cell types, with an enrichment among progenitor-like cells. While its precise role remains unclear, CRABP1 has been proposed to be a critical regulator of retinoic acid signaling, preventing canonical RA-induced transcriptional programs [79]. This role is of particular interest due to retinoic acid’s function in hindbrain patterning [80–84] and the previous identification of the retinoic acid binding protein RXRA as the top ranked super-enhancer associated gene in H3K27M DMGs [65]. Together these data suggest that CRABP1 may be associated with progenitor cells in early stages of the developing hindbrain that may be dysregulated in both PFAs and H3K27M DMGs. Further investigation is needed to elucidate whether CRABP1 contributes to maintenance of progenitor-type states in the presence of retinoids during development.
Overall, our studies further define molecular commonalities in the biology between DMGs and PFAs that express H3K27M and EZHIP and highlight similar patterns in the heterogeneity among tumors of each class. Moreover, some of these common epigenetic signatures showed specific patterns in human hindbrain development supporting the overall hypothesis that hindbrain epigenetic programs contribute to the biology of both tumors.
Supplementary Information
Additional file 1: Supplementary Figures 1–5.
Acknowledgements
The work was supported in part by grants from National Institute of Neurological Disorders and Stroke (NINDS) R01NS110572 (SV), National Cancer Institute (NCI) R01ACA261926 (SV), NCI R37CA214955-01A1 (AR), NCI P30CA046592 (AR), Chad Tough Defeat DIPG foundation (SV), Alex Lemonade Stand Foundation (SV), the Hyundai Hope on Wheel Foundation (SV), and the Rogel Cancer Center PIBS Graduate Student Scholarship (MP).
Author contributions
MP and SV conceptualized the project and wrote the manuscript. MP, DP, PRN, PKJ, LJ, BE, MC, and SV performed formal analyses. MP, PRN, PKJ, and SV created visualizations of the data. AR, KA, SP, MF, AMC, AB, and SV supervised the analyses and provided valuable feedback. All authors reviewed, edited, and approved the manuscript. All authors read and approved the final manuscript.
Availability of data and materials
Data were obtained as described in the methods section.
Declarations
Ethics approval and consent to participate
No consent to participate was required for the present study.
Consent for publication
All authors consented to publication.
Competing interests
The authors of this manuscript have no competing interests to disclose.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Curtin SC, Minino AM, Anderson RN (2016) Declines in cancer death rates among children and adolescents in the United States, 1999–2014. NCHS Data Brief 1–8 [PubMed]
- 2.Pollack IF. Pediatric brain tumors. Semin Surg Oncol. 1999;16:73–90. doi: 10.1002/(sici)1098-2388(199903)16:2<73::aid-ssu2>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 3.Chen CCL, Deshmukh S, Jessa S, Hadjadj D, Lisi V, Andrade AF, Faury D, Jawhar W, Dali R, Suzuki H, Pathania M, Dubois F, Woodward E, Hébert S, Coutelier M, Karamchandani J, Albrecht S, Brandner S, De Jay N, Gayden T, Bajic A, Harutyunyan AS, Marchione DM, Mikael LG, Juretic N, Zeinieh M, Russo C, Maestro N, Bassenden AV, Hauser P, Virga J, Bognar L, Klekner A, Zapotocky M, Vicha A, Krskova L, Vanova K, Zamecnik J, Sumerauer D, Ekert PG, Ziegler DS, Ellezam B, Filbin MG, Blanchette M, Hansford JR, Khuong-Quang D-A, Berghuis AM, Weil AG, Garcia BA, Garzia L, Mack SC, Beroukhim R, Ligon KL, Taylor MD, Bandopadhayay P, Kramm C, Pfister SM, Korshunov A, Sturm D, Jones DTW, Salomoni P, Kleinman CL, Jabado N. Histone H3.3G34-mutant interneuron progenitors co-opt PDGFRA for gliomagenesis. Cell. 2020;183:1617–1633.e22. doi: 10.1016/j.cell.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filbin MG, Tirosh I, Hovestadt V, Shaw ML, Escalante LE, Mathewson ND, Neftel C, Frank N, Pelton K, Hebert CM, Haberler C, Yizhak K, Gojo J, Egervari K, Mount C, van Galen P, Bonal DM, Nguyen QD, Beck A, Sinai C, Czech T, Dorfer C, Goumnerova L, Lavarino C, Carcaboso AM, Mora J, Mylvaganam R, Luo CC, Peyrl A, Popovic M, Azizi A, Batchelor TT, Frosch MP, Martinez-Lage M, Kieran MW, Bandopadhayay P, Beroukhim R, Fritsch G, Getz G, Rozenblatt-Rosen O, Wucherpfennig KW, Louis DN, Monje M, Slavc I, Ligon KL, Golub TR, Regev A, Bernstein BE, Suva ML. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science (80-) 2018;360:331–335. doi: 10.1126/science.aao4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillen AE, Riemondy KA, Amani V, Griesinger AM, Gilani A, Venkataraman S, Madhavan K, Prince E, Sanford B, Hankinson TC, Handler MH, Vibhakar R, Jones KL, Mitra S, Hesselberth JR, Foreman NK, Donson AM. Single-cell RNA sequencing of childhood ependymoma reveals neoplastic cell subpopulations that impact molecular classification and etiology. Cell Rep. 2020;32:108023. doi: 10.1016/j.celrep.2020.108023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gojo J, Pavelka Z, Zapletalova D, Schmook MT, Mayr L, Madlener S, Kyr M, Vejmelkova K, Smrcka M, Czech T, Dorfer C, Skotakova J, Azizi AA, Chocholous M, Reisinger D, Lastovicka D, Valik D, Haberler C, Peyrl A, Noskova H, Pal K, Jezova M, Veselska R, Kozakova S, Slaby O, Slavc I, Sterba J. Personalized treatment of H3K27M-mutant pediatric diffuse gliomas provides improved therapeutic opportunities. Front Oncol. 2019;9:1436. doi: 10.3389/fonc.2019.01436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendrikse LD, Haldipur P, Saulnier O, Millman J, Sjoboen AH, Erickson AW, Ong W, Gordon V, Coudière-Morrison L, Mercier AL. Failure of human rhombic lip differentiation underlies medulloblastoma formation. Nature. 2022;609:1021–1028. doi: 10.1038/s41586-022-05215-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jessa S, Blanchet-Cohen A, Krug B, Vladoiu M, Coutelier M, Faury D, Poreau B, De Jay N, Hébert S, Monlong J. Stalled developmental programs at the root of pediatric brain tumors. Nat Genet. 2019;51:1702–1713. doi: 10.1038/s41588-019-0531-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith KS, Bihannic L, Gudenas BL, Haldipur P, Tao R, Gao Q, Li Y, Aldinger KA, Iskusnykh IY, Chizhikov VV, Scoggins M, Zhang S, Edwards A, Deng M, Glass IA, Overman LM, Millman J, Sjoboen AH, Hadley J, Golser J, Mankad K, Sheppard H, Onar-Thomas A, Gajjar A, Robinson GW, Hovestadt V, Orr BA, Patay Z, Millen KJ, Northcott PA. Unified rhombic lip origins of group 3 and group 4 medulloblastoma. Nature. 2022;609:1012–1020. doi: 10.1038/s41586-022-05208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vladoiu MC, El-Hamamy I, Donovan LK, Farooq H, Holgado BL, Sundaravadanam Y, Ramaswamy V, Hendrikse LD, Kumar S, Mack SC. Childhood cerebellar tumours mirror conserved fetal transcriptional programs. Nature. 2019;572:67–73. doi: 10.1038/s41586-019-1158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DTW, Kool M, Zapatka M, Northcott PA, Sturm D, Wang W, Radlwimmer B, Højfeldt JW, Truffaux N, Castel D, Schubert S, Ryzhova M, Seker-Cin H, Gronych J, Johann PD, Stark S, Meyer J, Milde T, Schuhmann M, Ebinger M, Monoranu C-M, Ponnuswami A, Chen S, Jones C, Witt O, Collins VP, von Deimling A, Jabado N, Puget S, Grill J, Helin K, Korshunov A, Lichter P, Monje M, Plass C, Cho Y-J, Pfister SM. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science (80-) 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartzentruber J, Korshunov A, Liu X-Y, Jones DTW, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang D-AK, Tönjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jäger N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Frühwald MC, Roggendorf W, Kramm C, Dürken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, Jabado N. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 14.Sturm D, Witt H, Hovestadt V, Khuong-Quang D-A, Jones DTW, Konermann C, Pfaff E, Tönjes M, Sill M, Bender S, Kool M, Zapatka M, Becker N, Zucknick M, Hielscher T, Liu X-Y, Fontebasso AM, Ryzhova M, Albrecht S, Jacob K, Wolter M, Ebinger M, Schuhmann MU, van Meter T, Frühwald MC, Hauch H, Pekrun A, Radlwimmer B, Niehues T, von Komorowski G, Dürken M, Kulozik AE, Madden J, Donson A, Foreman NK, Drissi R, Fouladi M, Scheurlen W, von Deimling A, Monoranu C, Roggendorf W, Herold-Mende C, Unterberg A, Kramm CM, Felsberg J, Hartmann C, Wiestler B, Wick W, Milde T, Witt O, Lindroth AM, Schwartzentruber J, Faury D, Fleming A, Zakrzewska M, Liberski PP, Zakrzewski K, Hauser P, Garami M, Klekner A, Bognar L, Morrissy S, Cavalli F, Taylor MD, van Sluis P, Koster J, Versteeg R, Volckmann R, Mikkelsen T, Aldape K, Reifenberger G, Collins VP, Majewski J, Korshunov A, Lichter P, Plass C, Jabado N, Pfister SM. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 15.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, Zhang J, Gajjar A, Dyer MA, Mullighan CG, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Ellison DW, Zhang J, Baker SJ. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan K-M, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, Sarkaria J, Zhang Z. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013;27:985–990. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antin C, Tauziède-Espariat A, Debily M-A, Castel D, Grill J, Pagès M, Ayrault O, Chrétien F, Gareton A, Andreiuolo F, Lechapt E, Varlet P. EZHIP is a specific diagnostic biomarker for posterior fossa ependymomas, group PFA and diffuse midline gliomas H3-WT with EZHIP overexpression. Acta Neuropathol Commun. 2020;8:183. doi: 10.1186/s40478-020-01056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pajtler KW, Wen J, Sill M, Lin T, Orisme W, Tang B, Hubner JM, Ramaswamy V, Jia S, Dalton JD, Haupfear K, Rogers HA, Punchihewa C, Lee R, Easton J, Wu G, Ritzmann TA, Chapman R, Chavez L, Boop FA, Klimo P, Sabin ND, Ogg R, Mack SC, Freibaum BD, Kim HJ, Witt H, Jones DTW, Vo B, Gajjar A, Pounds S, Onar-Thomas A, Roussel MF, Zhang J, Taylor JP, Merchant TE, Grundy R, Tatevossian RG, Taylor MD, Pfister SM, Korshunov A, Kool M, Ellison DW. Molecular heterogeneity and CXorf67 alterations in posterior fossa group A (PFA) ependymomas. Acta Neuropathol. 2018;136:211–226. doi: 10.1007/s00401-018-1877-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubner JM, Muller T, Papageorgiou DN, Mauermann M, Krijgsveld J, Russell RB, Ellison DW, Pfister SM, Pajtler KW, Kool M. EZHIP/CXorf67 mimics K27M mutated oncohistones and functions as an intrinsic inhibitor of PRC2 function in aggressive posterior fossa ependymoma. Neuro Oncol. 2019;21:878–889. doi: 10.1093/neuonc/noz058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain SU, Do TJ, Lund PJ, Rashoff AQ, Diehl KL, Cieslik M, Bajic A, Juretic N, Deshmukh S, Venneti S, Muir TW, Garcia BA, Jabado N, Lewis PW. PFA ependymoma-associated protein EZHIP inhibits PRC2 activity through a H3 K27M-like mechanism. Nat Commun. 2019;10:2146. doi: 10.1038/s41467-019-09981-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain SU, Rashoff AQ, Krabbenhoft SD, Hoelper D, Do TJ, Gibson TJ, Lundgren SM, Bondra ER, Deshmukh S, Harutyunyan AS, Juretic N, Jabado N, Harrison MM, Lewis PW. H3 K27M and EZHIP impede H3K27-methylation spreading by inhibiting allosterically stimulated PRC2. Mol Cell. 2020;80:726–735. doi: 10.1016/j.molcel.2020.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piunti A, Smith ER, Morgan MAJ, Ugarenko M, Khaltyan N, Helmin KA, Ryan CA, Murray DC, Rickels RA, Yilmaz BD, Rendleman EJ, Savas JN, Singer BD, Bulun SE, Shilatifard A. CATACOMB: An endogenous inducible gene that antagonizes H3K27 methylation activity of Polycomb repressive complex 2 via an H3K27M-like mechanism. Sci Adv. 2019;5:eaax2887. doi: 10.1126/sciadv.aax2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ragazzini R, Perez-Palacios R, Baymaz IH, Diop S, Ancelin K, Zielinski D, Michaud A, Givelet M, Borsos M, Aflaki S, Legoix P, Jansen P, Servant N, Torres-Padilla ME, Bourc’his D, Fouchet P, Vermeulen M, Margueron R. EZHIP constrains polycomb repressive complex 2 activity in germ cells. Nat Commun. 2019;10:3858. doi: 10.1038/s41467-019-11800-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krug B, De Jay N, Harutyunyan AS, Deshmukh S, Marchione DM, Guilhamon P, Bertrand KC, Mikael LG, McConechy MK, Chen CCL, Khazaei S, Koncar RF, Agnihotri S, Faury D, Ellezam B, Weil AG, Ursini-Siegel J, De Carvalho DD, Dirks PB, Lewis PW, Salomoni P, Lupien M, Arrowsmith C, Lasko PF, Garcia BA, Kleinman CL, Jabado N, Mack SC. Pervasive H3K27 acetylation leads to ERV expression and a therapeutic vulnerability in H3K27M gliomas. Cancer Cell. 2019;35:782–797.e8. doi: 10.1016/j.ccell.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagaraja S, Vitanza NA, Woo PJ, Taylor KR, Liu F, Zhang L, Li M, Meng W, Ponnuswami A, Sun W, Ma J, Hulleman E, Swigut T, Wysocka J, Tang Y, Monje M. Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell. 2017;31:635–652. doi: 10.1016/j.ccell.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panwalkar P, Tamrazi B, Dang D, Chung C, Sweha S, Natarajan SK, Pun M, Bayliss J, Ogrodzinski MP, Pratt D, Mullan B, Hawes D, Yang F, Lu C, Sabari BR, Achreja A, Heon J, Animasahun O, Cieslik M, Dunham C, Yip S, Hukin J, Phillips JJ, Bornhorst M, Griesinger AM, Donson AM, Foreman NK, Garton HJL, Heth J, Muraszko K, Nazarian J, Koschmann C, Jiang L, Filbin MG, Nagrath D, Kool M, Korshunov A, Pfister SM, Gilbertson RJ, Allis CD, Chinnaiyan AM, Lunt SY, Blüml S, Judkins AR, Venneti S. Targeting integrated epigenetic and metabolic pathways in lethal childhood PFA ependymomas. Sci Transl Med. 2021;13:eabc0497. doi: 10.1126/scitranslmed.abc0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piunti A, Hashizume R, Morgan MA, Bartom ET, Horbinski CM, Marshall SA, Rendleman EJ, Ma Q, Takahashi YH, Woodfin AR, Misharin AV, Abshiru NA, Lulla RR, Saratsis AM, Kelleher NL, James CD, Shilatifard A. Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat Med. 2017;23:493–500. doi: 10.1038/nm.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anastas JN, Zee BM, Kalin JH, Kim M, Guo R, Alexandrescu S, Blanco MA, Giera S, Gillespie SM, Das J, Wu M, Nocco S, Bonal DM, Nguyen QD, Suva ML, Bernstein BE, Alani R, Golub TR, Cole PA, Filbin MG, Shi Y. Re-programing chromatin with a bifunctional LSD1/HDAC inhibitor induces therapeutic differentiation in DIPG. Cancer Cell. 2019;36:528–544.e10. doi: 10.1016/j.ccell.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Grasso CS, Tang Y, Truffaux N, Berlow NE, Liu L, Debily M-A, Quist MJ, Davis LE, Huang EC, Woo PJ. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med. 2015;21:555–559. doi: 10.1038/nm.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hashizume R, Andor N, Ihara Y, Lerner R, Gan H, Chen X, Fang D, Huang X, Tom MW, Ngo V. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med. 2014;20:1394–1396. doi: 10.1038/nm.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bayliss J, Mukherjee P, Lu C, Jain SU, Chung C, Martinez D, Sabari B, Margol AS, Panwalkar P, Parolia A, Pekmezci M, McEachin RC, Cieslik M, Tamrazi B, Garcia BA, La Rocca G, Santi M, Lewis PW, Hawkins C, Melnick A, David Allis C, Thompson CB, Chinnaiyan AM, Judkins AR, Venneti S (2016) Lowered H3K27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. Sci Transl Med 8:366ra161. 10.1126/scitranslmed.aah6904 [DOI] [PMC free article] [PubMed]
- 32.Harutyunyan AS, Krug B, Chen H, Papillon-Cavanagh S, Zeinieh M, De Jay N, Deshmukh S, Chen CCL, Belle J, Mikael LG, Marchione DM, Li R, Nikbakht H, Hu B, Cagnone G, Cheung WA, Mohammadnia A, Bechet D, Faury D, McConechy MK, Pathania M, Jain SU, Ellezam B, Weil AG, Montpetit A, Salomoni P, Pastinen T, Lu C, Lewis PW, Garcia BA, Kleinman CL, Jabado N, Majewski J. H3K27M induces defective chromatin spread of PRC2-mediated repressive H3K27me2/me3 and is essential for glioma tumorigenesis. Nat Commun. 2019;10:1262. doi: 10.1038/s41467-019-09140-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mack SC, Witt H, Piro RM, Gu L, Zuyderduyn S, Stütz AM, Wang X, Gallo M, Garzia L, Zayne K, Zhang X, Ramaswamy V, Jäger N, Jones DTW, Sill M, Pugh TJ, Ryzhova M, Wani KM, Shih DJH, Head R, Remke M, Bailey SD, Zichner T, Faria CC, Barszczyk M, Stark S, Seker-Cin H, Hutter S, Johann P, Bender S, Hovestadt V, Tzaridis T, Dubuc AM, Northcott PA, Peacock J, Bertrand KC, Agnihotri S, Cavalli FMG, Clarke I, Nethery-Brokx K, Creasy CL, Verma SK, Koster J, Wu X, Yao Y, Milde T, Sin-Chan P, Zuccaro J, Lau L, Pereira S, Castelo-Branco P, Hirst M, Marra MA, Roberts SS, Fults D, Massimi L, Cho YJ, Van Meter T, Grajkowska W, Lach B, Kulozik AE, von Deimling A, Witt O, Scherer SW, Fan X, Muraszko KM, Kool M, Pomeroy SL, Gupta N, Phillips J, Huang A, Tabori U, Hawkins C, Malkin D, Kongkham PN, Weiss WA, Jabado N, Rutka JT, Bouffet E, Korbel JO, Lupien M, Aldape KD, Bader GD, Eils R, Lichter P, Dirks PB, Pfister SM, Korshunov A, Taylor MD. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506:445–450. doi: 10.1038/nature13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michealraj KA, Kumar SA, Kim LJY, Cavalli FMG, Przelicki D, Wojcik JB, Delaidelli A, Bajic A, Saulnier O, MacLeod G, Vellanki RN, Vladoiu MC, Guilhamon P, Ong W, Lee JJY, Jiang Y, Holgado BL, Rasnitsyn A, Malik AA, Tsai R, Richman CM, Juraschka K, Haapasalo J, Wang EY, De Antonellis P, Suzuki H, Farooq H, Balin P, Kharas K, Van Ommeren R, Sirbu O, Rastan A, Krumholtz SL, Ly M, Ahmadi M, Deblois G, Srikanthan D, Luu B, Loukides J, Wu X, Garzia L, Ramaswamy V, Kanshin E, Sánchez-Osuna M, El-Hamamy I, Coutinho FJ, Prinos P, Singh S, Donovan LK, Daniels C, Schramek D, Tyers M, Weiss S, Stein LD, Lupien M, Wouters BG, Garcia BA, Arrowsmith CH, Sorensen PH, Angers S, Jabado N, Dirks PB, Mack SC, Agnihotri S, Rich JN, Taylor MD. Metabolic regulation of the epigenome drives lethal infantile ependymoma. Cell. 2020 doi: 10.1016/j.cell.2020.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohammad F, Weissmann S, Leblanc B, Pandey DP, Hojfeldt JW, Comet I, Zheng C, Johansen JV, Rapin N, Porse BT, Tvardovskiy A, Jensen ON, Olaciregui NG, Lavarino C, Sunol M, de Torres C, Mora J, Carcaboso AM, Helin K. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med. 2017;23:483–492. doi: 10.1038/nm.4293. [DOI] [PubMed] [Google Scholar]
- 36.Mackay A, Burford A, Carvalho D, Izquierdo E, Fazal-Salom J, Taylor KR, Bjerke L, Clarke M, Vinci M, Nandhabalan M, Temelso S, Popov S, Molinari V, Raman P, Waanders AJ, Han HJ, Gupta S, Marshall L, Zacharoulis S, Vaidya S, Mandeville HC, Bridges LR, Martin AJ, Al-Sarraj S, Chandler C, Ng H-K, Li X, Mu K, Trabelsi S, Brahim DH-B, Kisljakov AN, Konovalov DM, Moore AS, Carcaboso AM, Sunol M, de Torres C, Cruz O, Mora J, Shats LI, Stavale JN, Bidinotto LT, Reis RM, Entz-Werle N, Farrell M, Cryan J, Crimmins D, Caird J, Pears J, Monje M, Debily M-A, Castel D, Grill J, Hawkins C, Nikbakht H, Jabado N, Baker SJ, Pfister SM, Jones DTW, Fouladi M, von Bueren AO, Baudis M, Resnick A, Jones C. Integrated molecular meta-analysis of 1000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017;32:520–537.e5. doi: 10.1016/j.ccell.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pajtler KW, Witt H, Sill M, Jones DTW, Hovestadt V, Kratochwil F, Wani K, Tatevossian R, Punchihewa C, Johann P, Reimand J, Warnatz H-J, Ryzhova M, Mack S, Ramaswamy V, Capper D, Schweizer L, Sieber L, Wittmann A, Huang Z, van Sluis P, Volckmann R, Koster J, Versteeg R, Fults D, Toledano H, Avigad S, Hoffman LM, Donson AM, Foreman N, Hewer E, Zitterbart K, Gilbert M, Armstrong TS, Gupta N, Allen JC, Karajannis MA, Zagzag D, Hasselblatt M, Kulozik AE, Witt O, Collins VP, von Hoff K, Rutkowski S, Pietsch T, Bader G, Yaspo M-L, von Deimling A, Lichter P, Taylor MD, Gilbertson R, Ellison DW, Aldape K, Korshunov A, Kool M, Pfister SM. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27:728–743. doi: 10.1016/j.ccell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castel D, Kergrohen T, Tauziede-Espariat A, Mackay A, Ghermaoui S, Lechapt E, Pfister SM, Kramm CM, Boddaert N, Blauwblomme T, Puget S, Beccaria K, Jones C, Jones DTW, Varlet P, Grill J, Debily MA. Histone H3 wild-type DIPG/DMG overexpressing EZHIP extend the spectrum diffuse midline gliomas with PRC2 inhibition beyond H3–K27M mutation. Acta Neuropathol. 2020 doi: 10.1007/s00401-020-02142-w. [DOI] [PubMed] [Google Scholar]
- 39.Mariet C, Castel D, Grill J, Saffroy R, Dangouloff-Ros V, Boddaert N, Llamas-Guttierrez F, Chappé C, Puget S, Hasty L. Posterior fossa ependymoma H3 K27-mutant: an integrated radiological and histomolecular tumor analysis. Acta Neuropathol Commun. 2022;10:1–13. doi: 10.1186/s40478-022-01442-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pratt D, Quezado M, Abdullaev Z, Hawes D, Yang F, Garton HJL, Judkins AR, Mody R, Chinnaiyan A, Aldape K, Koschmann C, Venneti S (2020) Diffuse intrinsic pontine glioma-like tumor with EZHIP expression and molecular features of PFA ependymoma. In: Acta Neuropathol Commun. Laboratory of Brain Tumor Metabolism and Epigenetics, Department of Pathology, University of Michigan, 3520E MSRB 1, 1150 W. Medical Center, Ann Arbor, MI, 48109, USA. Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, Bethesd, p 37 [DOI] [PMC free article] [PubMed]
- 41.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, Koelsche C, Sahm F, Chavez L, Reuss DE, Kratz A, Wefers AK, Huang K, Pajtler KW, Schweizer L, Stichel D, Olar A, Engel NW, Lindenberg K, Harter PN, Braczynski AK, Plate KH, Dohmen H, Garvalov BK, Coras R, Hölsken A, Hewer E, Bewerunge-Hudler M, Schick M, Fischer R, Beschorner R, Schittenhelm J, Staszewski O, Wani K, Varlet P, Pages M, Temming P, Lohmann D, Selt F, Witt H, Milde T, Witt O, Aronica E, Giangaspero F, Rushing E, Scheurlen W, Geisenberger C, Rodriguez FJ, Becker A, Preusser M, Haberler C, Bjerkvig R, Cryan J, Farrell M, Deckert M, Hench J, Frank S, Serrano J, Kannan K, Tsirigos A, Brück W, Hofer S, Brehmer S, Seiz-Rosenhagen M, Hänggi D, Hans V, Rozsnoki S, Hansford JR, Kohlhof P, Kristensen BW, Lechner M, Lopes B, Mawrin C, Ketter R, Kulozik A, Khatib Z, Heppner F, Koch A, Jouvet A, Keohane C, Mühleisen H, Mueller W, Pohl U, Prinz M, Benner A, Zapatka M, Gottardo NG, Driever PH, Kramm CM, Müller HL, Rutkowski S, von Hoff K, Frühwald MC, Gnekow A, Fleischhack G, Tippelt S, Calaminus G, Monoranu C-M, Perry A, Jones C, Jacques TS, Radlwimmer B, Gessi M, Pietsch T, Schramm J, Schackert G, Westphal M, Reifenberger G, Wesseling P, Weller M, Collins VP, Blümcke I, Bendszus M, Debus J, Huang A, Jabado N, Northcott PA, Paulus W, Gajjar A, Robinson GW, Taylor MD, Jaunmuktane Z, Ryzhova M, Platten M, Unterberg A, Wick W, Karajannis MA, Mittelbronn M, Acker T, Hartmann C, Aldape K, Schüller U, Buslei R, Lichter P, Kool M, Herold-Mende C, Ellison DW, Hasselblatt M, Snuderl M, Brandner S, Korshunov A, von Deimling A, Pfister SM. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mack SC, Pajtler KW, Chavez L, Okonechnikov K, Bertrand KC, Wang X, Erkek S, Federation A, Song A, Lee C, Wang X, McDonald L, Morrow JJ, Saiakhova A, Sin-Chan P, Wu Q, Michaelraj KA, Miller TE, Hubert CG, Ryzhova M, Garzia L, Donovan L, Dombrowski S, Factor DC, Luu B, Valentim CLL, Gimple RC, Morton A, Kim L, Prager BC, Lee JJY, Wu X, Zuccaro J, Thompson Y, Holgado BL, Reimand J, Ke SQ, Tropper A, Lai S, Vijayarajah S, Doan S, Mahadev V, Miñan AF, Gröbner SN, Lienhard M, Zapatka M, Huang Z, Aldape KD, Carcaboso AM, Houghton PJ, Keir ST, Milde T, Witt H, Li Y, Li C-J, Bian X-W, Jones DTW, Scott I, Singh SK, Huang A, Dirks PB, Bouffet E, Bradner JE, Ramaswamy V, Jabado N, Rutka JT, Northcott PA, Lupien M, Lichter P, Korshunov A, Scacheri PC, Pfister SM, Kool M, Taylor MD, Rich JN. Therapeutic targeting of ependymoma as informed by oncogenic enhancer profiling. Nature. 2018;553:101–105. doi: 10.1038/nature25169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie Z, Bailey A, Kuleshov M V, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, Jeon M, Ma’ayan A (2021) Gene set knowledge discovery with Enrichr. Curr Protoc 1:e90. 10.1002/cpz1.90 [DOI] [PMC free article] [PubMed]
- 46.Aldinger KA, Thomson Z, Phelps IG, Haldipur P, Deng M, Timms AE, Hirano M, Santpere G, Roco C, Rosenberg AB, Lorente-Galdos B, Gulden FO, O’Day D, Overman LM, Lisgo SN, Alexandre P, Sestan N, Doherty D, Dobyns WB, Seelig G, Glass IA, Millen KJ. Spatial and cell type transcriptional landscape of human cerebellar development. Nat Neurosci. 2021;24:1163–1175. doi: 10.1038/s41593-021-00872-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eze UC, Bhaduri A, Haeussler M, Nowakowski TJ, Kriegstein AR. Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia. Nat Neurosci. 2021;24:584–594. doi: 10.1038/s41593-020-00794-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sepp M, Leiss K, Sarropoulos I, Murat F, Okonechnikov K, Joshi P, Leushkin E, Mbengue N, Schneider C, Schmidt J (2021) Cellular development and evolution of the mammalian cerebellum. bioRxiv [DOI] [PMC free article] [PubMed]
- 49.Godfraind C, Kaczmarska JM, Kocak M, Dalton J, Wright KD, Sanford RA, Boop FA, Gajjar A, Merchant TE, Ellison DW. Distinct disease-risk groups in pediatric supratentorial and posterior fossa ependymomas. Acta Neuropathol. 2012;124:247–257. doi: 10.1007/s00401-012-0981-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mendrzyk F, Korshunov A, Benner A, Toedt G, Pfister S, Radlwimmer B, Lichter P. Identification of gains on 1q and epidermal growth factor receptor overexpression as independent prognostic markers in intracranial ependymoma. Clin Cancer Res Off J Am Assoc Cancer Res. 2006;12:2070–2079. doi: 10.1158/1078-0432.CCR-05-2363. [DOI] [PubMed] [Google Scholar]
- 51.Rajeshwari M, Sharma MC, Kakkar A, Nambirajan A, Suri V, Sarkar C, Singh M, Saran RK, Gupta RK. Evaluation of chromosome 1q gain in intracranial ependymomas. J Neurooncol. 2016;127:271–278. doi: 10.1007/s11060-015-2047-z. [DOI] [PubMed] [Google Scholar]
- 52.Varela C, Denis JA, Polentes J, Feyeux M, Aubert S, Champon B, Piétu G, Peschanski M, Lefort N. Recurrent genomic instability of chromosome 1q in neural derivatives of human embryonic stem cells. J Clin Invest. 2012;122:569–574. doi: 10.1172/JCI46268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mackay A, Burford A, Molinari V, Jones DTW, Izquierdo E, Brouwer-Visser J, Giangaspero F, Haberler C, Pietsch T, Jacques TS, Figarella-Branger D, Rodriguez D, Morgan PS, Raman P, Waanders AJ, Resnick AC, Massimino M, Garrè ML, Smith H, Capper D, Pfister SM, Würdinger T, Tam R, Garcia J, Das TM, Vassal G, Grill J, Jaspan T, Varlet P, Jones C. Molecular, pathological, radiological, and immune profiling of non-brainstem pediatric high-grade glioma from the HERBY phase II randomized trial. Cancer Cell. 2018;33:829–842. doi: 10.1016/j.ccell.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cavalli FMG, Hübner J-M, Sharma T, Luu B, Sill M, Zapotocky M, Mack SC, Witt H, Lin T, Shih DJH, Ho B, Santi M, Emery L, Hukin J, Dunham C, McLendon RE, Lipp ES, Gururangan S, Grossbach A, French P, Kros JM, van Veelen M-LC, Rao AAN, Giannini C, Leary S, Jung S, Faria CC, Mora J, Schüller U, Alonso MM, Chan JA, Klekner A, Chambless LB, Hwang EI, Massimino M, Eberhart CG, Karajannis MA, Lu B, Liau LM, Zollo M, Ferrucci V, Carlotti C, Tirapelli DPC, Tabori U, Bouffet E, Ryzhova M, Ellison DW, Merchant TE, Gilbert MR, Armstrong TS, Korshunov A, Pfister SM, Taylor MD, Aldape K, Pajtler KW, Kool M, Ramaswamy V. Heterogeneity within the PF-EPN-B ependymoma subgroup. Acta Neuropathol. 2018;136:227–237. doi: 10.1007/s00401-018-1888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B, Garzia L, Torchia J, Nor C, Morrissy AS, Agnihotri S, Thompson YY, Kuzan-Fischer CM, Farooq H, Isaev K, Daniels C, Cho B-K, Kim S-K, Wang K-C, Lee JY, Grajkowska WA, Perek-Polnik M, Vasiljevic A, Faure-Conter C, Jouvet A, Giannini C, Nageswara Rao AA, Li KKW, Ng H-K, Eberhart CG, Pollack IF, Hamilton RL, Gillespie GY, Olson JM, Leary S, Weiss WA, Lach B, Chambless LB, Thompson RC, Cooper MK, Vibhakar R, Hauser P, van Veelen M-LC, Kros JM, French PJ, Ra YS, Kumabe T, López-Aguilar E, Zitterbart K, Sterba J, Finocchiaro G, Massimino M, Van Meir EG, Osuka S, Shofuda T, Klekner A, Zollo M, Leonard JR, Rubin JB, Jabado N, Albrecht S, Mora J, Van Meter TE, Jung S, Moore AS, Hallahan AR, Chan JA, Tirapelli DPC, Carlotti CG, Fouladi M, Pimentel J, Faria CC, Saad AG, Massimi L, Liau LM, Wheeler H, Nakamura H, Elbabaa SK, Perezpeña-Diazconti M, Ponce C, de León F, Robinson S, Zapotocky M, Lassaletta A, Huang A, Hawkins CE, Tabori U, Bouffet E, Bartels U, Dirks PB, Rutka JT, Bader GD, Reimand J, Goldenberg A, Ramaswamy V, Taylor MD. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell. 2017;31:737–754.e6. doi: 10.1016/j.ccell.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baroni LV, Sundaresan L, Heled A, Coltin H, Pajtler KW, Lin T, Merchant TE, McLendon R, Faria C, Buntine M, White CL, Pfister SM, Gilbert MR, Armstrong TS, Bouffet E, Kumar S, Taylor MD, Aldape KD, Ellison DW, Gottardo NG, Kool M, Korshunov A, Hansford JR, Ramaswamy V. Ultra high-risk PFA ependymoma is characterized by loss of chromosome 6q. Neuro Oncol. 2021;23:1360–1370. doi: 10.1093/neuonc/noab034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buczkowicz P, Bartels U, Bouffet E, Becher O, Hawkins C. Histopathological spectrum of paediatric diffuse intrinsic pontine glioma: diagnostic and therapeutic implications. Acta Neuropathol. 2014;128:573–581. doi: 10.1007/s00401-014-1319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Castel D, Philippe C, Kergrohen T, Sill M, Merlevede J, Barret E, Puget S, Sainte-Rose C, Kramm CM, Jones C, Varlet P, Pfister SM, Grill J, Jones DTW, Debily MA. Transcriptomic and epigenetic profiling of “diffuse midline gliomas, H3 K27M-mutant” discriminate two subgroups based on the type of histone H3 mutated and not supratentorial or infratentorial location. Acta Neuropathol Commun. 2018;6:117. doi: 10.1186/s40478-018-0614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fontebasso AM, Papillon-Cavanagh S, Schwartzentruber J, Nikbakht H, Gerges N, Fiset P-O, Bechet D, Faury D, De Jay N, Ramkissoon LA, Corcoran A, Jones DTW, Sturm D, Johann P, Tomita T, Goldman S, Nagib M, Bendel A, Goumnerova L, Bowers DC, Leonard JR, Rubin JB, Alden T, Browd S, Geyer JR, Leary S, Jallo G, Cohen K, Gupta N, Prados MD, Carret A-S, Ellezam B, Crevier L, Klekner A, Bognar L, Hauser P, Garami M, Myseros J, Dong Z, Siegel PM, Malkin H, Ligon AH, Albrecht S, Pfister SM, Ligon KL, Majewski J, Jabado N, Kieran MW. Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat Genet. 2014;46:462–466. doi: 10.1038/ng.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pratt D, Lucas C-HG, Selvam PP, Abdullaev Z, Ketchum C, Quezado M, Armstrong TS, Gilbert MR, Papanicolau-Sengos A, Raffeld M, Choo-Wosoba H, Chan P, Whipple N, Nasrallah M, Santi M, Ramaswamy V, Giannini C, Ritzmann TA, Grundy RG, Burford A, Jones C, Hawkins C, Venneti S, Solomon DA, Aldape K. Recurrent ACVR1 mutations in posterior fossa ependymoma. Acta Neuropathol. 2022;144:373–376. doi: 10.1007/s00401-022-02435-2. [DOI] [PubMed] [Google Scholar]
- 61.Hoeman CM, Cordero FJ, Hu G, Misuraca K, Romero MM, Cardona HJ, Nazarian J, Hashizume R, McLendon R, Yu P, Procissi D, Gadd S, Becher OJ. ACVR1 R206H cooperates with H3.1K27M in promoting diffuse intrinsic pontine glioma pathogenesis. Nat Commun. 2019;10:1023. doi: 10.1038/s41467-019-08823-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Castel D, Philippe C, Calmon R, Le Dret L, Truffaux N, Boddaert N, Pages M, Taylor KR, Saulnier P, Lacroix L, Mackay A, Jones C, Sainte-Rose C, Blauwblomme T, Andreiuolo F, Puget S, Grill J, Varlet P, Debily MA. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 2015;130:815–827. doi: 10.1007/s00401-015-1478-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J, Huang TY-T, Hou Y, Bartom E, Lu X, Shilatifard A, Yue F, Saratsis A (2021) Epigenomic landscape and 3D genome structure in pediatric high-grade glioma. Sci Adv 7. 10.1126/sciadv.abg4126 [DOI] [PMC free article] [PubMed]
- 64.Krug B, De Jay N, Harutyunyan AS, Deshmukh S, Marchione DM, Guilhamon P, Bertrand KC, Mikael LG, McConechy MK, Chen CCL, Khazaei S, Koncar RF, Agnihotri S, Faury D, Ellezam B, Weil AG, Ursini-Siegel J, De Carvalho DD, Dirks PB, Lewis PW, Salomoni P, Lupien M, Arrowsmith C, Lasko PF, Garcia BA, Kleinman CL, Jabado N, Mack SC. Pervasive H3K27 acetylation leads to ERV expression and a therapeutic vulnerability in H3K27M gliomas. Cancer Cell. 2019;36:338–339. doi: 10.1016/j.ccell.2019.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nagaraja S, Quezada MA, Gillespie SM, Arzt M, Lennon JJ, Woo PJ, Hovestadt V, Kambhampati M, Filbin MG, Suva ML, Nazarian J, Monje M. Histone variant and cell context determine H3K27M reprogramming of the enhancer landscape and oncogenic state. Mol Cell. 2019;76:965–980.e12. doi: 10.1016/j.molcel.2019.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA (2013) Super-Enhancers in the Control of Cell Identity and Disease. Cell 155:934–947. 10.1016/j.cell.2013.09.053 [DOI] [PMC free article] [PubMed]
- 67.Favorskaya I, Kainov Y, Chemeris G, Komelkov A, Zborovskaya I, Tchevkina E. Expression and clinical significance of CRABP1 and CRABP2 in non-small cell lung cancer. Tumor Biol. 2014;35:10295–10300. doi: 10.1007/s13277-014-2348-4. [DOI] [PubMed] [Google Scholar]
- 68.Lind GE, Kleivi K, Meling GI, Teixeira MR, Thiis-Evensen E, Rognum TO, Lothe RA. ADAMTS1, CRABP1, and NR3C1 identified as epigenetically deregulated genes in colorectal tumorigenesis. Anal Cell Pathol. 2006;28:259–272. doi: 10.1155/2006/949506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu R-Z, Garcia E, Glubrecht DD, Poon HY, Mackey JR, Godbout R. CRABP1 is associated with a poor prognosis in breast cancer: adding to the complexity of breast cancer cell response to retinoic acid. Mol Cancer. 2015;14:1–16. doi: 10.1186/s12943-015-0380-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tanaka K, Imoto I, Inoue J, Kozaki K, Tsuda H, Shimada Y, Aiko S, Yoshizumi Y, Iwai T, Kawano T. Frequent methylation-associated silencing of a candidate tumor-suppressor, CRABP1, in esophageal squamous-cell carcinoma. Oncogene. 2007;26:6456–6468. doi: 10.1038/sj.onc.1210459. [DOI] [PubMed] [Google Scholar]
- 71.Wu Q, Lothe RA, Ahlquist T, Silins I, Tropé CG, Micci F, Nesland JM, Suo Z, Lind GE. DNA methylation profiling of ovarian carcinomas and their in vitro models identifies HOXA9, HOXB5, SCGB3A1, and CRABP1 as novel targets. Mol Cancer. 2007;6:1–11. doi: 10.1186/1476-4598-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Riemondy KA, Venkataraman S, Willard N, Nellan A, Sanford B, Griesinger AM, Amani V, Mitra S, Hankinson TC, Handler MH, Sill M, Ocasio J, Weir SJ, Malawsky DS, Gershon TR, Garancher A, Wechsler-Reya RJ, Hesselberth JR, Foreman NK, Donson AM, Vibhakar R. Neoplastic and immune single-cell transcriptomics define subgroup-specific intra-tumoral heterogeneity of childhood medulloblastoma. Neuro Oncol. 2022;24:273–286. doi: 10.1093/neuonc/noab135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.O’Rahilly R, Müller F. Developmental stages in human embryos: revised and new measurements. Cells Tissues Organs. 2010;192:73–84. doi: 10.1159/000289817. [DOI] [PubMed] [Google Scholar]
- 74.Maden M, Horton C, Graham A, Leonard L, Pizzey J, Siegenthaler G, Lumsden A, Eriksson U. Domains of cellular retinoic acid-binding protein I (CRABP I) expression in the hindbrain and neural crest of the mouse embryo. Mech Dev. 1992;37:13–23. doi: 10.1016/0925-4773(92)90011-8. [DOI] [PubMed] [Google Scholar]
- 75.Watson C, Bartholomaeus C, Puelles L. Time for radical changes in brain stem nomenclature—applying the lessons from developmental gene patterns. Front Neuroanat. 2019;13:10. doi: 10.3389/fnana.2019.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watson C, Ullmann J (2015) The Brain Stem. In: Toga AWBT-BM (ed). Academic Press, Waltham, pp 251–259
- 77.Allen Institute for Brain Science (2010) Allen developing human brain atlas: microarray
- 78.Miller JA, Ding S-L, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nhieu J, Lin Y-L, Wei L-N. CRABP1 in non-canonical activities of retinoic acid in health and diseases. Nutrients. 2022;14:1528. doi: 10.3390/nu14071528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Begemann G, Meyer A. Hindbrain patterning revisited: timing and effects of retinoic acid signalling. BioEssays. 2001;23:981–986. doi: 10.1002/bies.1142. [DOI] [PubMed] [Google Scholar]
- 81.Glover JC, Renaud J, Rijli FM. Retinoic acid and hindbrain patterning. J Neurobiol. 2006;66:705–725. doi: 10.1002/neu.20272. [DOI] [PubMed] [Google Scholar]
- 82.Hernandez RE, Putzke AP, Myers JP, Margaretha L, Moens CB (2007) Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development [DOI] [PMC free article] [PubMed]
- 83.Marshall H, Nonchev S, Sham MH, Muchamore I, Lumsden A, Krumlauf R. Retinoic acid alters hindbrain Hox code and induces transformation of rhombomeres 2/3 into a 4/5 identity. Nature. 1992;360:737–741. doi: 10.1038/360737a0. [DOI] [PubMed] [Google Scholar]
- 84.Niederreither K, Vermot J, Schuhbaur B, Chambon P, Dollé P. Retinoic acid synthesis and hindbrain patterning in the mouse embryo. Development. 2000;127:75–85. doi: 10.1242/dev.127.1.75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Figures 1–5.
Data Availability Statement
Data were obtained as described in the methods section.