Abstract
The demographic and epidemiological transitions of the past 200 years are well documented at an aggregate level. Understanding differences in individual and group risks for mortality during these transitions requires linkage between demographic data and detailed individual cause of death information. This paper describes the digitization of almost 185,000 causes of death for Ohio to supplement demographic information in the Longitudinal, Intergenerational Family Electronic Micro-database (LIFE-M). To extract causes of death, our methodology combines handwriting recognition, extensive data cleaning algorithms, and the semi-automated classification of causes of death into International Classification of Diseases (ICD) codes. Our procedures are adaptable to other collections of handwritten data, which require both handwriting recognition and semi-automated coding of the information extracted.
Keywords: Death certificate, Semi-automated classification, Cause of death
1. Introduction
How does early-life nutrition affect the development of cancer, adult hypertension, and cardiovascular disease? How does child-hood exposure to toxins such as lead affect suicide rates and the development of Alzheimer’s disease? How did the early 20th century’s public health campaigns to clean water, improve sanitation, and deliver vaccines affect later-life health and longevity? Demographic and epidemiological transitions have brought substantial improvements in human welfare over the past 200 years; however, the role of individual genetics, circumstances, local policies, and environmental conditions in shaping health is less well understood. The lack of historical individual-level data on cause of death has been a key limitation.
This paper describes the semi-automated digitization of 173,227 individual causes of death from Ohio to supplement demographic information in the Longitudinal, Intergenerational Family Electronic Micro-database (LIFE-M). Our methodology combines handwriting recognition to extract causes of death, extensive data cleaning algorithms, and classification of causes of death into International Classification of Diseases (ICD) codes. We call the process “semi-automated,” because a human field expert created a dictionary of common misspellings and corrections, and an automated computer program applied these corrections at scale.
We selected death certificates linked through the Longitudinal, Intergenerational Family Electronic Micro-database (LIFE-M), which allows us to link cause of death to vital records (birth, marriage, and death certificates) and decennial censuses over four generations (or 120 years) for two states. LIFE-M contains cohorts born, aging, and dying over the 20th century and includes crucial early-life and intergenerational information not available in other data. Digitizing cause-of-death information facilitates path-breaking research on the relationships between cause of death and genetic, demographic, socioeconomic, and early-life environmental factors for family networks across four generations.
Our approach relies on undergraduate research assistants and neural networks to segment images of handwriting. We then used two independent training datasets of multiple human transcriptions to train a handwriting recognition model to digitize causes of death. Spelling and script recognition errors were corrected using Google’s autocorrect feature, semi-automated human corrections, and a curated pattern-based dictionary. Using a combination of keywords and human review by a field expert, keywords were matched to the ICD-2 (Bertillon et al. 1910) at the broadest and finest levels in order to be consistent with medical concepts and disease classifications for the period. The resulting dataset assigns cause of death codes to 82% of all deaths and 87% of records with machine-recognizable text with 88% accuracy. The output of our pipeline is the classification of 173,227 hand-written causes of death into ICD codes.
The extracted causes of death cover well-known causes of death pre/post vaccine development (e.g., whooping cough) as well as causes of death still common today (e.g., pneumonia). We have posted complete documentation, code, and data associated with this project for public use at ICPSR Linkage Library (Project149,841 at http://doi.org/10.3886/E149841).
Our methods are adaptable to other historical sources containing semi-structured handwriting. Our contributions include methods for locating a specific piece of information in structured documents (that change format over time), standardizing terms using Google’s spelling correction tools, and mapping causes of death into a classification scheme conformable with historical language and modern death classification. Similar processes could be used for other structured sources with features that need to be classified into finite lists. Because our goal was to take production to scale, we invested in developing algorithms to parse words and phrases into causes of death and a concise and informative classification system. This paper provides a roadmap of the process including lessons learned. At key points, we highlight forks in the road and alternative routes, where other researchers could make different choices.
2. Step-by-step process to digitize and classify cause of death
Some of the most important, open questions in health and aging relate to the impact of longitudinal and, likely, intergenerational factors. Although a large body of research documents differences in health by subgroups and across time, existing micro-data limits a deeper understanding of the mechanisms behind these relationships, how they have changed over time, and how they differ between disease classes. In the United States, only a small fraction of death certificates are linked to demographic and socioeconomic data from earlier in life, and linkages are relatively recent. The situation is similar in other English-language jurisdictions. By contrast, longstanding linkages between individual death certificates and data from earlier in life exist in Scandinavian countries (Maret-Ouda et al., 2017; Thorvaldsen et al., 2015, Mandmakers and Kok 2020, Reid et al., 2002, Hall et al., 2000).
In the United States the National Longitudinal Mortality Study (NLMS) links a selection of Current Population Survey (1973–2011) and census (1980) data to death certificates, and includes 3.8 million individuals linked to 550,000 deaths. The NLMS has been widely used for research. For example, research using cause of death in the NLMS finds racial differences in breast cancer mortality by education attainment (Kim et al., 2005; Akinyemiju et al., 2013), differences in brain cancer mortality by occupation (Van Wijngaarden and Dosemeci 2006), male-female differences in lung cancer by education level (Limin et al. 2009), and differences in longevity by socioeconomic status (Du et al., 2011; Chetty et al., 2016). Similar analyses identify key differences in demographic, economic, and geographic factors associated with stroke mortality (Howard et al., 1995; Howard et al., 1997), injury mortality (Hussey 1997), suicide (Kposowa 1999; Kposowa 2001; Kposowa 2013), and heart disease (Mackenbach et al., 2000; Muntaner et al., 2001; Sorlie et al., 2004; Cooper et al., 2009; Coady et al., 2014). Because the relationship between socioeconomic factors and mortality is not stable over time (Bengtsson et al., 2020) or even across places (Bengtsson and van Poppel, 2011), documenting variation in the cause-specific antecedents of mortality in the United States is an important research focus.
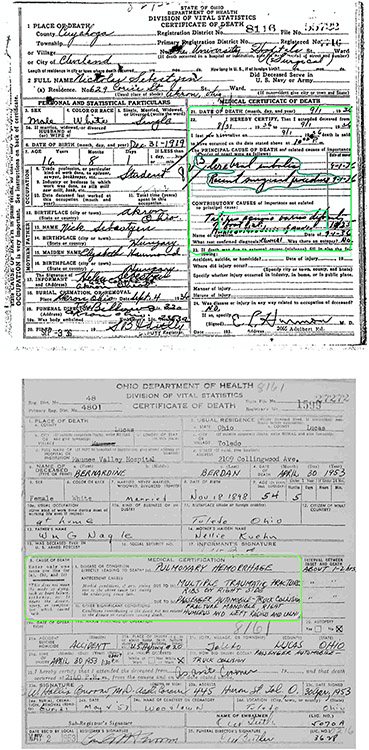
The goal of this project is to encode and classify cause of death in order to enhance the new large-scale Longitudinal, Intergenerational Family Electronic Micro-database (LIFE-M). Cause of death is usually a hand-written entry on death certificates. An example of a death certificate image is shown in Fig. 1. Digitizing cause of death has the potential to make LIFE-M more useful to researchers interested in aging and mortality by allowing them to examine relationships between a multitude of early-life and intergenerational factors, longevity, and cause of death. The following sections provide a step-by-step process for how we assigned ICD codes to hand-written causes of death.
Fig. 1.
Examples of Death Certificate Images.
2.1. Collecting images
Several dimensions of image quality are important for accurate handwriting recognition. We give a brief overview of the images we worked with, and refer readers to Appendix B1 for an extended discussion of how to collect images to facilitate accurate handwriting recognition. The death certificates we worked with had been converted from microfilm to grayscale digital images at a resolution of 200 PPI. The original physical size of the death certificates was approximately half the size of an 8.5 × 11″ page. Before proceeding with handwriting recognition, we familiarized ourselves with varying document layouts over time, and the quality of the images. The layout of the death certificates was largely consistent over time. However, the location of information on contributory causes shifted (as shown in the two examples in Fig. 1).
We also observed that while most images were aligned to the grid, some documents were scanned at a slight angle. Materials converted from microfilm lack the sharpness of modern scanned or photographed images, as some information is lost with each transformation. However, the relatively sharp color contrasts between the handwritten (and printed) text which had generally been written in black ink, and the white background page proved decisive. The death certificates we worked with had been printed and written between 1908 and 1953, and microfilmed in the late twentieth century. Thus, there was little evident fading of the ink, or yellowing of the paper, either of which would reduce contrast. Dealing with deviations from these conditions may be important with other collections. In short, we were fortunate to work with material with strong color contrast that was generally well aligned to the grid, and could focus on idiosyncrasies of the handwriting.
2.2. Create training data using multiple human transcriptions
All death images can be viewed at FamilySearch.org. We viewed the images and created training data from multiple human transcribers of 4796 death certificates. We created a system in Python that automated the process of opening death certificates and asked undergraduate research assistants (trainers) to enter causes of death. The program uses a list of URLs to access death records and, when available, displays death certificate images.
Viewing the death certificate, the trainer identified the handwritten cause of death on the record and typed it into the program terminal. Each record was transcribed twice, once each by two different trainers for a total of 9592 transcriptions. These transcriptions were split into training (80%), validation (10%), and test sets (10%).2
2.3. Automated segmentation of cause of death
Death certificates, typically written either in ledgers or on forms, are structured, meaning that handwritten information typically appears in certain parts of the document (see Fig. 1). Nevertheless, identifying this structure brings several challenges. First, cause of death is written on forms that change over time. Second, the location of the handwriting within the relevant segment changes, with responses appearing on different lines and handwriting extending outside of the given box.
To detect and segment the location of a hand-written cause of death, we leverage tremendous advancements in deep convolutional neural networks (Hafiz and Bhat 2020; He et al., 2017; Redmon et al., 2016). Neural networks are designed to handle instance segmentation, where the model detects and segments instances of objects, such as humans, cars or animals in a variety of settings. Our approach uses a deep learning-based neural network called Mask-RCNN from He et al. (2017) to identify the ‘Medical Certificate of Death’ (MCD) region and the handwritten causes of death strings from the death certificates. We use maskrcnn-benchmark,3 a pytorch implementation of Mask-RCNN which was created by Facebook. We fine-tune the pretrained ResNet-50 architecture using training data created using a tool called “LabelMe,” which labeled the bounded boxes for 600 images. We find that this model and implementation perform well, resulting in 0.969 IoU (Intersection over Union), with minimal training time (<15 h), and low memory usage. We use a modified version of the library that does not flip images or do random crops as these do not apply to our dataset scenario. However, we did use Imagemagick to deskew the images as a pre-processing step. The pytorch implementation that we used has since been replaced by another tool created by Facebook called Detectron2 (Wu et al., 2019) and is the segmentation tool used in Layout Parser (Shen et al., 2021).
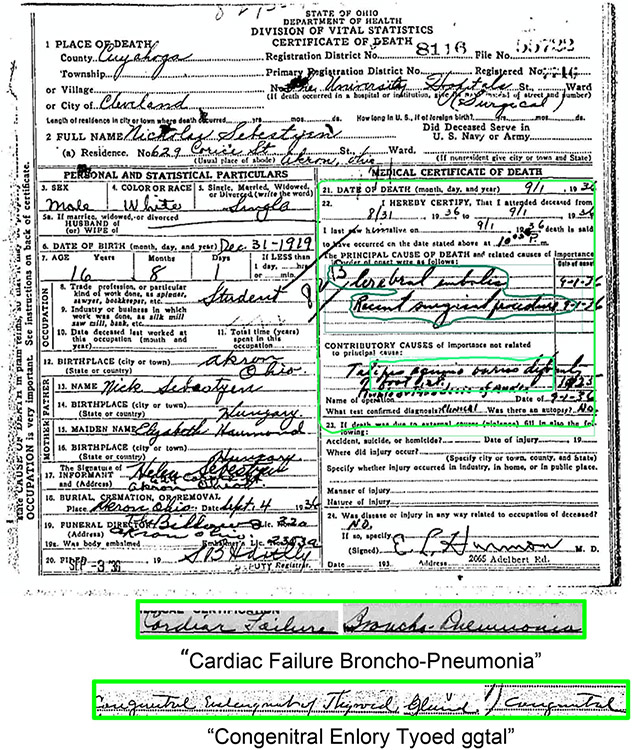
The immediate and contributory causes of death are then extracted and concatenated into a single line for ingestion and text prediction by the handwriting recognition model as shown in Fig. 2.
Fig. 2.
Example Output of the Mask-RCNN Segmentation Model that Identifies ’Medical Certificate of Death’ (MCD) Region and Handwritten Responses, “Cardiac Failure Broncho-Pneumonia”, “Congenitral Enlory Tyoed ggtal”.
2.4. Handwriting recognition
The next step in the process is to recognize and digitize handwriting strings extracted from the last step. This step involves both image recognition and sequential character prediction. One of the most popular approaches for such a task is the one proposed by Shi et al. (2015). It uses a combination of Convolutional Neural Network (CNN) and long-short term memory (LSTM), where the CNN extracts feature maps from the input image, and the LSTM maps them to a sequential output that predicts each character of the text in the input image. Since the invention of the Transformer model (Vaswani et al., 2017), work in this area has shifted from recurrent neural networks to pure attention based models. Recently, Kang et al. (2022) adapted the transformer model to handwriting recognition where they further pushed the accuracy frontier on the IAM benchmark dataset. For our work, we adopt a CNN-LSTM architecture as in Shi et al. (2015). We fine-tuned the handwriting recognition module from Start-Follow-Read 4 from Wigington et al. (2018) using our own training data. Notably, our retrained MaskRCNN with a ResNet-50 backend mentioned in the previous section performed better than the original Start-of-Line detector used in the original paper to find lines of interest due to having a much deeper network.
From this process, researchers should expect transcription output to be messy, and have relatively high character error rates. For instance, our handwriting recognition performance had a character error rate (CER) of 0.28. This is considerably higher than the character error rate achieved by some of the most advanced methods using commonly used training data like the RIMES dataset which can achieve character error rates less than 0.03. Nevertheless, human inspection of the data shows that the transcriptions are recognizably close to correct words or phrases. Consider, for example, how “bacillary disentoy” may easily be recognized as “bacillary dysentery” by a human even though the CER is close to what we observed in our data on average. With cursive or script handwriting, internal letters that are linked to others are more likely to be incorrectly transcribed by humans and software. However, the pattern of the letters within the word, and the visual similarity of the “o” and “e” mean many readers will see “bacillary disentoy” and map it quickly to “bacillary dysentery.”
We used three approaches to reduce spelling errors and variations in our output. First, BYU developed a system where rapid feedback can be acquired by presenting users with the segmented causes of death images and a transcription of the image. We found that transcription correction is faster, more accurate and easier for users when compared to de novo transcription generation. This system is called reverse indexing and was used on any cause of death that was a single word. This system can be accessed at https://indexing.familytech.byu.edu.
A second, fully automated approach is to use Google’s search algorithm, which easily fixes spelling errors. For instance, Google translated misspellings in strings like “Prmature Bith” and “Ifluenza” to “premature birth” and “influenza.” In addition, Google’s experience with misspellings identified harder to detect errors, for example correcting the phrase, “Heulet Feve,r” to “scarlet fever.” We wrote a script to search for all digitized strings in Google and retrieve the corresponding misspelling-corrected strings from Google’s suggestions. In total, Google corrected a large number of character errors, resulting in updates to 71% out of 199,876 strings. We also conducted a systematic analysis of Google corrections to determine their accuracy and helpfulness, which is reported in later sections.
The last approach used human review. After the first two processes were complete, a field expert examined and identified terms for which Google did not suggest a spelling correction. When the field expert found misspellings or alternative phrasings, she recorded them in a correction dictionary (sometimes called a “tidy list”). A computer algorithm used this dictionary to clean other strings that had the same spelling error. For instance, words containing misspelling patterns of “apperdiatise” or “appendicit” were corrected to “appendicitis.” Another example is that the expert identified nearly 80 different misspelling patterns for “stillborn.”
For researchers interested in applying these methods, we emphasize two key points. First, handwriting recognition is likely to produce messier transcriptions on a larger and faster scale, compared to the slow, but more accurate work of human transcription. But “close” is good enough in cases such as ours, where additional technology such as Google’s suggestions may be used to correct transcription errors. Second, domain knowledge of the text being transcribed is important in determining how close is good enough. The level of character error that was tolerable for causes of death may be different for different variables, depending on the true underlying number of distinct values. Less error can be tolerated when there are more distinct values in the underlying concept of interest, because it increases the chance that a spelling error will lead to misclassification.
2.5. Classifying corrected strings into ICD codes
For each corrected string, the final step was to determine the associated ICD code. This is complicated by the fact that many strings contained words or diseases or contributing causes that could be classified into different ICD classes. Our approach relied on a highly trained expert in historical disease classification to assist in the development of computer algorithms to classify strings as diseases.
A first step identified and extracted up to three keywords and three general words for each cause of death string, which we used to identify the associated ICD code. Keywords are those that contain a specific disease (e.g., “tuberculous meningitis” and “smallpox”), while general words are those that contain either a body part (e.g., “lungs”, “skull”) or a general cause of death (e.g. “hemorrhage”). For instance, the string, “probable skill fractured prushed right chest and hemorrhage-upper lung,” yielded the keyword, “fracture" and general words, “chest,” “hemorrhage,” and “lungs”. The string, “bronchopneumonia in malnutrition” yielded the keywords, “bronchopneumonia” and “malnutrition,” and the general word, “bronchial.” Words appearing at the beginning of the string are usually the primary cause of death, whereas those that appear later are typically the contributory causes of death. To preserve this information, we order our keywords and general words by their position in the string. In assigning an ICD code, we prioritized contagious diseases and external causes regardless of their position in the string. We note that the grammar of causes of death is similar to that of occupations (Roberts et al., 2003) where the first words generally denote the primary information with subsidiary information or secondary causes appearing later. Other concepts have a different grammar: addresses typically begin with the most specific piece of information and end with the broadest entity, such as a city, state or country. For those keywords and general words not yet classified, our field expert examined lists of unigrams and bigrams sorted by frequency and manually identified and grouped terms that could be categorized as either keywords or general words. For example, “congenital malformation of the heart” and “malformation of heart” are both standardized to “cardiac malformation” as a keyword and “pulmonary” and “tuberculosis” were combined into one standardized keyword “pulmonary tuberculosis.” We repeated this process until we had extracted at least one keyword from most of our non-empty and non-gibberish strings. At present, there are nearly 500 keywords and 70 general words, which can be scaled at low marginal cost to more death records in the future.
As a second step, we created a mapping between keywords and general words and ICD codes. Information about age and sex were used for many causes of death referring to conditions of pregnancy and birth that could either have been applied to the mother or the infant (e.g., “delivery”, “miscarriage”).5 For our context, we created a 26-category system based on ICD-2 (Bertillon et al. 1910)—the second revision of the ICD—which we call ICD2-H. (“H” here stands for historical, similar to the HISCO adaptation of occupational codes.) The categories of ICD2-H were aligned with the ICD classes in use for the cohorts covered in the early years of our data (General Diseases, Diseases of the Nervous System and the Organs of Special Sense, etc.) with a few additional categories, non-bolded in Table 1. These categories reflect causes that were important over the time period (e.g., typhoid, cancer) or are of particular interest to historical researchers. The categories also group together causes easily mistaken for one another in practice, for which reliable diagnostic tools were not yet in place. For instance, pneumonia, bronchopneumonia, and bronchitis were included in one category. Although later versions of the ICD identified disease classes with more detail, the strings in our data do not have the level of detail to map to more current versions.6 More details on ICD classification are included in Appendix A.
Table 1.
ICD2-H Classifications.
| ICD-2 Class | ICD2-H | Description | % of Deaths |
|---|---|---|---|
| 1 | General diseases | 19.93 | |
| 101 | Typhoid and typhus | 0.36 | |
| 102 | Tuberculosis | 4.20 | |
| 103 | Venereal disease | 0.33 | |
| 104 | Cancer/neoplasm | 2.30 | |
| 105 | Tumor without mention of cancer | 0.16 | |
| 106 | Diseases of malnutrition | 0.02 | |
| 107 | Alcoholism | 0.22 | |
| 108 | Other general diseases | 12.34 | |
| 2 | Diseases of the nervous system and the organs of special sense | 8.70 | |
| 209 | Encephalitis/meningitis/brain fever | 4.50 | |
| 210 | Other diseases of the nervous system and the organs of special sense (excepting encephalitis, meningitis and brain fever) | 4.20 | |
| 3 | 311 | Diseases of the circulatory system | 7.67 |
| 4 | Diseases of the respiratory system | 17.11 | |
| 412 | Pneumonia/bronchopneumonia/influenza/bronchitis | 15.12 | |
| 413 | Other diseases of the respiratory system (excepting pneumonia, bronchopneumonia, bronchitis and influenza) | 1.99 | |
| 5 | Diseases of the digestive system | 12.21 | |
| 514 | Diarrhea and enteritis, gastritis | 8.24 | |
| 515 | Other diseases of the digestive system (excepting diarrhea, enteritis and gastritis) | 3.97 | |
| 6 | Diseases of the genito-urinary apparatus | 2.29 | |
| 616 | Nephritis and Bright’s | 2.03 | |
| 617 | Other diseases of the genito-urinary apparatus (excepting nephritis and Bright’s disease) | 0.26 | |
| 7 | 718 | Puerperal state (maternal mortality) | 0.23 |
| 8 | 819 | Diseases of the skin and cellular tissue | 0.44 |
| 9 | 920 | Diseases of the bones and of the organs of locomotion | 0.36 |
| 10 | 1021 | Malformations | 2.94 |
| 11 | 1122 | Infancy | 10.45 |
| 12 | 1200 | Old age | 0.00 |
| 13 | External causes | 10.58 | |
| 1323 | Suicide | 0.25 | |
| 1324 | Accidents, injuries, poisoning | 10.33 | |
| 14 | 1425 | Ill-defined causes (including unknown) | 6.03 |
| 15 | 1527 | Stillbirth | 1.07 |
After creating this mapping system, each keyword and general word was classified to one of these ICD2-H codes, so that any given cause of death could have up to six classifications (3 based on keywords and 3 based on general words). As we refined classifications, we updated our output iteratively, working with the data for a time and then applying the keyword mapping and classification processes again to identify and correctly classify as many causes of death as possible. When all keywords were mapped to the same ICD2-H classification, we used the ICD classification as the final ICD2-H without further scrutiny. For example, the corrected string “carcinoma of breast with metastasis chest” resulted in keywords “carcinoma” and “metastasis," both coded to the ICD2-H “cancer/neoplasm” category. For strings with multiple ICD2-H values (101,270 cases), we assign the final ICD2-H based on the first keyword’s classification with a few modifications. Modifications took account of the fact that some words take priority, regardless of an ordering in the string (e.g. “suicide”, “accidents”). An example is when conditions arising after an accident were listed first, for example, “pulmonary hemorrhage," “accidental death” and “automobile accident.” The death would be classified as an accident in final ICD2-H.
We also differentiate between “well-defined ” and “ill-defined” causes. We use the ill-defined diseases category to differentiate between causes where no keywords or general words were extracted vs. cases where keywords or general words were extracted but are not precise enough to determine a specific cause. For the cause “inanition found head in bal”, the keyword “inanition” and general word “head” were extracted, but they are not precise enough to be assigned to any well-defined category, and thus were assigned to the ill-defined diseases category. Ill-defined is a standalone ICD2-H category while “well-defined” categories are those other than ill-defined and unknown. We prioritize well-defined over ill-defined keywords regardless of their position. (See Supplementary Appendix for more details regarding classification rules and procedures.)
The fidelity of our coding can be partly evaluated by the percentage of cases in each class that were classified using general words only. Overall, about 18% were coded solely from general words, which mostly identify a body part or system, state, or event (e.g., pregnancy, birth, cardiac). Some states and body systems are precise enough to classify to the correct ICD2-H class with high confidence (e.g., we classify the general word “valvular” to “diseases of the circulatory system”), while for other cases classification could only be made to ill-defined diseases (e.g., “chest,” “insufficiency,” “organic ”) in the absence of other more informative words. Twenty percent of all the cases classified using only general words are stillbirths (ICD2-H 1527), all based on the general word “still” which was the part of the word that our processes recognized (the full word, “stillbirth,” is a keyword in our dictionary). Roughly 20% of the cases assigned to circulatory diseases (ICD2-H 311) were classified using only general words; the general words are themselves precise to the category (e.g., “heart,” “mitral,” “carditis”). We also assigned the more finely detailed ICD2 titles to all possible causes of death, assigning causes to titles based on the instructions in the ICD2 manual for use in the U.S. Keywords and general words that did not appear in the manual, and did not have a synonym in the manual, were coded to the ill-defined category. (See also Supplementary Appendix).
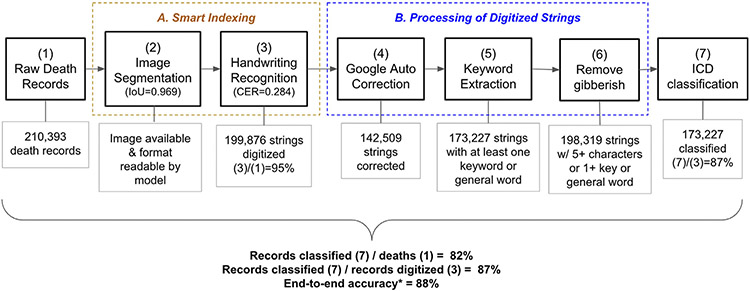
2.6. End-to-End pipeline performance evaluation
Fig. 3 summarizes our pipeline and evaluation metrics for each stage as well as an end-to-end evaluation metric. The Smart-indexing process began with a set of 210,393 Ohio death records we attempted to classify (1). Only 199,876 had images available or had images in a format that can be understood by our segmentation model (3), as the formats of death certificates change over time and the segmentation model was not able to read all variations in death certificate formats.
Fig. 3.
Summary of Processing Pipeline and Performance Metrics.
The next component of the pipeline involved processing these digitized strings. For the 199,876 records with digitized strings, Google suggested auto corrections in around 71% of cases (4). To assess the quality of the Google corrections, we drew a random sample of cases where Google suggested at least one correction to our raw digitized strings. 44% of Google’s corrections aided in determining the correct cause of death. For the remaining 56% of cases, Google corrected a word that was not used in ICD classification. For instance, the relevant keyword was already correctly spelled in the raw digitized string. Importantly, a careful manual inspection of a sample of records revealed that none of the reviewed cases showed that Google auto-correction led to an incorrect ICD classification.
Finally, our pattern-based keyword extraction program extracted at least one keyword or general word for 173,227 strings (5). Next, we removed gibberish cases where the digitized strings contain fewer than 5 characters and no keyword or general word was extracted so that strings that are short but meaningful will remain in our dataset (6). For instance, “flu” is retained, but gibberish short strings like “pki” are removed.
Our final dataset contains 173,227 cause-of death classifications (7). The final output resulted in 82% of the initial 210,393 records and 87% of the 199,876 digitized strings being assigned to an ICD code, which we present as part of our end-to-end performance metric in the final three rows of the figure.
As a complement to these measures, we also created an end-to-end metric of classification accuracy. Because we have no ground truth, this metric was developed using the following steps. First, we took a subsample of around 500 death record images with legible causes of death. Second, we asked three authors of this paper to review these images alongside the assigned ICD code to determine if it is correct. Third, we used these classifications to compute end-to-end error rates for our ICD assignments. We found that 88% (452/511) of sampled cases were in agreement with the ICD code assigned by our semi-automated process. Among the 12% of cases determined to be incorrect, around two thirds were due to issues with the smart indexing process and one third due to errors in the post-cleaning and assignment process.
3. Conclusion and applications
Handwriting was and remains a common method of data collection. Many primary sources of great use to economic historians are handwritten, and handwriting persists today as a data collection tool, because of its low cost and flexibility in the field.
As handwritten records are digitized, the challenges of handwriting recognition have become salient. Lowering the costs of digitizing these sources offers the potential to transform the research horizon, allowing quantitative history to reach previously inaccessible areas and topics.
This paper shows how we undertook the work of digitizing cause of death from death records and points to where researchers may depart from our choices. Our seven-step pipeline shows how we divided tasks, but future work should continue to make progress to integrate them. The boundaries between these steps are porous: cleaning and keyword identification overlap with coding and classification. It is useful for researchers to assess at the beginning and end of each stage what kind of data they are trying to construct and the precision needed in different variables. We were able to accept, and work with, a high character error rate, because misspelled medical terms can still be recognized as distinct causes of death and assigned to a coarse classification scheme. Researchers whose key variables of interest are proper names (people, streets, geographic points at fine precision) or numbers would require much lower character error rates.
The approach we took is most applicable to large collections of administrative records. Our techniques could be adapted to extract structured data from tabular material in ledgers and bound volumes where the records of multiple entities are arrayed in a spreadsheet style display. Correctly placing material in the right rows and columns is critical. We recommend researchers examine the logical and physical structure of such documents for visible cues to the structure of the data that can be recognized by a computer. Human and computer methods work well together, and different humans may notice different aspects of document structure. Thus, we see promise in using citizen science tools such as the Zooniverse Project Builder as an alternative to the research assistants used in this project to gather data on the physical layout of documents as well as validate transcriptions.
In conclusion, future work could apply our methods to civil and vital registration documents, as they are in a format similar to the Ohio certificates. We also suggest applying these methods to human resource, personnel records, health forms, and property records. There are many more examples. We hope that more researchers undertake such efforts and continue to improve upon the methods used in this project.
Supplementary Material
Acknowledgments
This project is generously supported by the National Science Foundation (SMA-1,539,228), the National Institute on Aging (R21 AG056912), and the University of California, Los Angeles. Supplemental funding from the National Institute on Aging (R01 AG057704) allowed us to add cause of death for records in Ohio. Pilot work on this project was generously supported by the University of Michigan’s Department of Economics Michigan Institute for Teaching and Research in Economics (MITER) grant, the University of Michigan’s Population Studies Center Small Grant (R24 HD041028), and the Michigan Center for the Demography of Aging (MiCDA) grant (P30 AG012846–21). We gratefully acknowledge the use of the services and facilities of the Population Studies Center at the University of Michigan (R24 HD041028 and P2C HD041028), the California Center for Population Research at UCLA (P2C HD041022), and the Minnesota Population Center (P2C HD041023).
Footnotes
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.eeh.2022.101474.
Online access: https://ucla.app.box.com/s/s67qo1e6zbxa4ofzc3ij64kgge23wxx1.
An alternative approach to using research assistants to coordinate locations is creating a project on the Zooniverse citizen science platform. Zooniverse began as a research tool for physics and ecology research where identifying single or multiple coordinates (points, or two-dimensional shapes) on images was a fundamental research task. Recent (2018) updates to the Zooniverse platform allowed coordinate identification tasks to be associated with one or more subsidiary tasks such as transcribing handwriting in the bounded area. A web-based platform to identify coordinates and transcribe text can be deployed in less than a day, including the time required to learn the web-based “Project Builder” software. Coordinate identification and transcription tasks can then be performed by a private or public audience. Results are exported in a JSON format that can be reformatted into a dataset for analysis in R, Python, Stata, or other analysis tools. Each image is identified by a unique identifier, so that the multiple independent transcriptions and location coordinates can be compared. Standard string comparison metrics can be used to compare the similarity of transcriptions.
Keywords and general words that are mapped to puerperal state (age>14 & sex=Female), infancy (age<2), and stillbirth (age<2) use additional age and sex constraints indicated in the parenthesis to map to ICD codes. Age at death is inferred by subtracting year of birth from year of death on the death certificate, and the sex is obtained directly from the death certificate. For infancy and stillbirth, we chose the age threshold to be 2 (not 1) due to the delayed filing of many death records.
For example, for many diseases one needs to know whether the cause was bacterial or viral, which is not contained in our records. Although germ theory had been established in the late 19th century, it was not universally accepted in practice until well into our time period. The Flexner Report (1910) enumerated many different approaches to medical care being taught in the nation’s medical schools.
Data Availability
We have published the data in ICPSR Linkage Library.
References
- Akinyemiju Tomi F, Soliman Amr S, Johnson Norman J, Altekruse Sean F, Welch Kathy, Banerjee Mousumi, Schwartz Kendra, Merajver Sofia, 2013. Individual and Neighborhood Socioeconomic Status and Healthcare Resources in Relation to Black-White Breast Cancer Survival Disparities. J. Cancer Epidemiol doi: 10.1155/2013/490472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson Tommy, Dribe Martin, Helgertz Jonas, 2020. When Did the Health Gradient Emerge? Income, Social Class and Mortality, Sweden 1813-2015. Demography 57 (3), 953–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson Tommy, van Poppel Frans, 2011. Socioeconomic Inequalities in Death from Past to Present: an Introduction. Explor Econ. Hist 48 (3), 343–356. [Google Scholar]
- Bertillon J, 1910. Commission internationale chargée de reviser les nomenclatures nosologiques. International Classification of Causes of Sickness and Death. Government Printing Office, Washington, D.C.. [Google Scholar]
- Chetty Raj, Michael Stepner, Sarah Abraham, Shelby Lin, Benjamin Scuderi, Nicholas Turner, Augustin Bergeron, Cutler David, 2016. The Association between Income and Life Expectancy in the United States. 2001-2014. JAMA 315 (16), 1750–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coady Sean A, Johnson Norman J, Hakes Jahn K, Sorlie Paul D, 2014. Individual Education, Area Income, and Mortality and Recurrence of Myocardial Infarction in a Medicare Cohort: the National Longitudinal Mortality Study. BMC Public Health 14 (1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper Anna R, Van Wijngaarden Edwin, Fisher Susan G, Jacob Adams, M., Yost Michael G, Bowman Joseph D, 2009. A Population-Based Cohort Study of Occupational Exposure to Magnetic Fields and Cardiovascular Disease Mortality. Ann. Epidemiol 19 (1), 42–48. [DOI] [PubMed] [Google Scholar]
- Du Xianglin L, Lin Charles C, Johnson Norman J, Altekruse Sean, 2011. Effects of Individual-Level Socioeconomic Factors on Racial Disparities in Cancer Treatment and Survival: findings from the National Longitudinal Mortality Study. 1979-2003. Cancer 117 (14), 3242–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafiz Abdul Mueed, Bhat Ghulam Mohiuddin, 2020. A survey on instance segmentation: state of the art. Int J Multimed Inf Retr 9. doi: 10.1007/s13735-020-00195-x. [DOI] [Google Scholar]
- Hall Patricia Kelly, McCaa Robert, Thorvaldsen Gunnar, 2000. Handbook of International Historical Microdata for Population Research. Minnesota Population Center, Minneapolis. [Google Scholar]
- He Kaiming, Gkioxari Georgia, Dollar Piotr, Girshick Ross, 2017. Mask R-CNN. In: IEEE International Conference on Computer Vision (ICCV), pp. 2980–2988. doi: 10.1109/ICCV.2017.322. [DOI] [Google Scholar]
- Howard George, Russell Gregory B, Anderson Roger, Evans Gregory W, Morgan Timothy, Howard Virginia J, Burke Gregory L, 1995. In: Role of Social Class in Excess Black Stroke Mortality, 26. Stroke, pp. 1759–1763. [DOI] [PubMed] [Google Scholar]
- Howard George, Anderson Roger, Johnson Norman J, Sorlie Paul, Russell Gregory, Howard Virginia J, 1997. Evaluation of Social Status as a Contributing Factor to the Stroke Belt Region of the United States. Stroke 28 (5), 936–940. [DOI] [PubMed] [Google Scholar]
- Hussey Jon M., 1997. The Effects of Race, Socioeconomic Status, and Household Structure on Injury Mortality in Children and Young Adults. Matern. Child Health J. 1 (4), 217–227. [DOI] [PubMed] [Google Scholar]
- Kang Lei, Pau Riba, Marçal Rusiñol, Alicia Fornés, Mauricio Villegas, 2022. Pay attention to what you read: non-recurrent handwritten text-line recognition. Pattern Recognit. 129, 108766. doi: 10.1016/j.patcog.2022.108766. [DOI] [Google Scholar]
- Kim Catherine, Eby Elizabeth, Piette John D., 2005. Is Education Associated with Mortality for Breast Cancer and Cardiovascular Disease among Black and White Women? Gend Med 2 (1), 13–18. doi: 10.1016/S1550-8579(05)80005-1. [DOI] [PubMed] [Google Scholar]
- Kposowa Augustine J., 1999. Suicide Mortality in the United States: differentials by Industrial and Occupational Groups. Am. J. Ind. Med 36 (6), 645–652. [DOI] [PubMed] [Google Scholar]
- Kposowa Augustine J., 2001. Unemployment and Suicide: a Cohort Analysis of Social Factors Predicting Suicide in the US National Longitudinal Mortality Study. Psychol. Med 31 (1), 127–138. [DOI] [PubMed] [Google Scholar]
- Kposowa Augustine J., 2013. Association of Suicide Rates, Gun Ownership, Conservatism and Individual Suicide Risk. Soc. Psychiatry Psychiatr. Epidemiol 48 (9), 1467–1479. [DOI] [PubMed] [Google Scholar]
- Maret-Ouda John, Wenjing Tao, Karl Wahlin, Jesper Lagergren, 2017. Nordic registry-based cohort studies: possibilities and pitfalls when combining Nordic registry data. Scand. J. Public Health 45 (17), 14–19. [DOI] [PubMed] [Google Scholar]
- Mackenbach Johan P, Cavelaars AEJM, Kunst Anton E, Groenhof Feikje, 2000. Socioeconomic Inequalities in Cardiovascular Disease Mortality. An International Study. Eur. Heart J 21 (14), 1141–1151. [DOI] [PubMed] [Google Scholar]
- Muntaner Carles, Sorlie Paul, O’Campo Patricia, Johnson Norman, Backlund Eric, 2001. Occupational Hierarchy, Economic Sector, and Mortality from Cardiovascular Disease among Men and Women: findings from the National Longitudinal Mortality Study. Ann. Epidemiol 11 (3), 194–201. [DOI] [PubMed] [Google Scholar]
- Redmon Joseph, Divvala Santosh, Girshick Ross, Farhadi Ali, 2016. You Only Look Once: unified, Real-Time Object Detection. In: IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 779–788. doi: 10.1109/CVPR. [DOI] [Google Scholar]
- Reid Alice, Davies Ros, Garrett Eilidh., 2002. Nineteenth-century Scottish demography from linked censuses and civil registers: a ‘sets of related individuals’ approach. Hist. Comput 14 (1–2), 61–86. [Google Scholar]
- Roberts Evan, Woollard Matthew, Ronnander Chad, Dillon Lisa Y., Thorvaldsen Gunnar, 2003. Occupational Classification in the North Atlantic Population Project. Hist Methods 36 (2), 89–96. [Google Scholar]
- Shen Zejiang, Zhang Ruochen, Dell Melissa, Lee Benjamin Charles Germain, Carlson Jacob, Li Weining, 2021. Layoutparser: a Unified Toolkit for Deep Learning Based Document Image Analysis. In: International Conference on Document Analysis and Recognition. Springer, pp. 131–146. [Google Scholar]
- Shi Baoguang, Bai Xiang, Yao Cong, 2015. An End-to-End Trainable Neural Network for Image-Based Sequence Recognition and Its Application to Scene Text Recognition. IEEE Trans. Pattern Anal. Mach. Intell 2298–2304. doi: 10.1109/TPAMI.2016.2646371. [DOI] [PubMed] [Google Scholar]
- Sorlie Paul D, Coady Sean, Lin Charles, Arias Elizabeth, 2004. Factors Associated with out-of-Hospital Coronary Heart Disease Death: the National Longitudinal Mortality Study. Ann. Epidemiol 14 (7), 447–452. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen G, Andersen T, Sommerseth HL, 2015. Record linkage in the historical population register for Norway. In: Population Reconstruction. Springer, Cham, pp. 155–171. [Google Scholar]
- Van Wijngaarden Edwin, Dosemeci Mustafa, 2006. Brain Cancer Mortality and Potential Occupational Exposure to Lead: findings from the National Longitudinal Mortality Study. 1979–1989. Int. J. Cancer 119 (5), 1136–1144. [DOI] [PubMed] [Google Scholar]
- Vaswani Ashish, Shazeer Noam, Parmar Niki, Uszkoreit Jakob, Jones Llion, Gomez Aidan N., Kaiser Łukasz, Polosukhin Illia, 2017. Attention is all you need. Advances in Neural Information Processing Systems. Curran Associates Inc. [Google Scholar]
- Wigington Curtis, Tensmeyer Chris, Davis Brian, Barrett William, Price Brian, Cohen Scott, 2018. Start, Follow, Read: end-to-End Full-Page Handwriting Recognition. In: European Conference on Computer Vision. Springer, pp. 367–383. [Google Scholar]
- Wu Y, Kirillov A, Massa F, Lo WY, Girshick R 2019. Detectron2. https://github.com/facebookresearch/detectron2. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We have published the data in ICPSR Linkage Library.