Abstract
The current deluge of genomic sequences has spawned the creation of tools capable of making sense of the data. Computational and high-throughput experimental methods for generating links between proteins have recently been emerging. These methods effectively act as hypothesis machines, allowing researchers to screen large sets of data to detect interesting patterns that can then be studied in greater detail. Although the potential use of these putative links in predicting gene function has been demonstrated, a central repository for all such links for many genomes would maximize their usefulness. Here we present Predictome, a database of predicted links between the proteins of 44 genomes based on the implementation of three computational methods—chromosomal proximity, phylogenetic profiling and domain fusion—and large-scale experimental screenings of protein–protein interaction data. The combination of data from various predictive methods in one database allows for their comparison with each other, as well as visualization of their correlation with known pathway information. As a repository for such data, Predictome is an ongoing resource for the community, providing functional relationships among proteins as new genomic data emerges. Predictome is available at http://predictome.bu.edu.
INTRODUCTION
The function of a protein is perhaps best described in terms of its interactions with other proteins (1). An interaction between two proteins can be understood not only as a physical interaction, but also as an abstract association that implies some general relationship. For example, two proteins may be said to be linked if they are involved in the same metabolic pathway, or necessary for the enactment of a cellular process. Traditionally, the dominant computational method for detecting functional relationships between proteins has been database sequence similarity searches such as BLAST (2). Recently, several non-homology-based methods have been proposed for detecting such interactions, among them phylogenetic profiling (3–6), chromosomal proximity (7,8) and domain fusion (9–11), as well as high-throughput experimental methods (12–14). However, as useful as these methods are, no global database exists to perform a complementary analysis in interaction space as one does in sequence space using BLAST (2). Here we present a database of predicted links between proteins, Predictome, that is based on the implementation of published computational methods and publicly available data, to facilitate precisely such an analysis.
THE Predictome DATABASE
Several published databases exist which rely on experimental methods (15,16) or shared context (17,18) to link functionally related proteins. Similarly, the methods included in Predictome essentially serve to link one protein to another. The method of chromosomal proximity links two proteins if they are encoded close to one another along the genomic sequence, are transcribed in the same direction, and their orthologs are proximate in a number of other genomes (8). Two proteins are linked by a phylogenetic link if they share the same evolutionary pattern, such that their orthologs are either both present or absent in the genomes of known sequences (6). If two distinct proteins in one organism are encoded as one multi-domain protein in another organism, they are said to be fusion linked (9,10). The experimental detection of physical interactions between proteins by methods such as yeast two-hybrid analysis (19) provides a complementary experimental source of links to those links imputed by the sequence-based methods.
The usefulness of links between proteins to predict function has been previously demonstrated. For example, Marcotte et al. (20) were able to offer functional annotation for roughly half of the unannotated genes in Saccharomyces cerevisiae by examining the functional links which they form with the rest of the genes in that genome. These results led to the hypothesis that the predictive power of any link is increased when supported by multiple methods. Huynen et al. (21) studied the correlation of individual links predicted by different methods in Mycoplasma genitalium and found that the strength of an inference is increased when supported by multiple methods. Finally, the application of combinations of these methods has also been well reviewed (22–24).
Although the published predictive methods have been shown to be reasonably adept at detecting functional associations, their role in actually assisting protein annotation remains to be tested. The difficulty in proceeding from prediction to experimental validation may be attributed to the lack of a dedicated database that contains all of the links predicted by all of the methods. We believe that such a database will aid the scientific community in organizing and accessing the predictions and thus effectively bridge computational predictions with their experimental validation.
SOURCE DATASETS AND METHODS
The published methods for generating phylogenetic links, chromosomal proximity links and fusion links have been re-implemented to apply to the 44 microbial genomes currently available. Since a working definition of orthology is central to these three methods, we have chosen the Clusters of Orthologous Groups (COG) database, which provides a well-established model for detecting orthology, as the framework for generating these links (5,25).
Similar to the computational methods, high-throughput experimental methods are also capable of yielding putative links between proteins. Recently, the yeast two-hybrid method has been used as a systematic tool for establishing global sets of physical interactions between proteins (12–14), and these interactions are available from publicly accessible web sites. These data sets have been compiled and integrated into Predictome.
The usefulness of this database naturally increases as the number of methods it includes grows and we expect that more methods will be added over time. For example, links based on the correlated expression of genes derived from DNA chips and microarrays would be of tremendous value. Also, we expect in librio links, based on automated literature searches for the co-occurrence of genes/proteins in the same publication (26,27), to be added in the future. In addition, users of the database have the option of submitting their own links on the submission page of the web site.
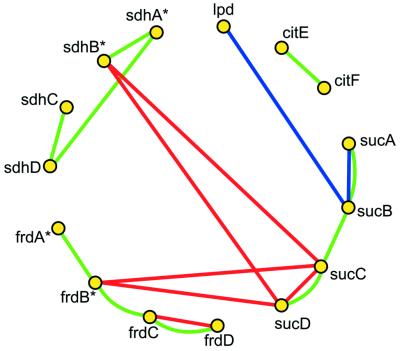
Through an analysis of the predicted inter-protein links based upon the various methods, it is possible to explore the relationships between these methods for correlation with each other and with known biological pathways and processes. Figure 1 illustrates such a comparison for a subset of 15 Escherichia coli proteins involved in the tricarboxylic acid (TCA) cycle. Since all 15 genes are in the same pathway, the predictive links among them recover existing, known associations. In order to assess the overall sensitivity of the links, we examine their correlation with three reference databases: COG (5,25), KEGG (28) and GeneQuiz (29) (Table 1). This analysis provides an evaluation of the methods used to create links, as well the selectivity of categorization in these databases. As is illustrated in Figure 1, few linked proteins are linked by more than one method. Table 2 shows the correlation between the sets of links generated by different methods. It is apparent from these results that false positives correspond to a substantial fraction of the links, typically ∼30%, and are difficult to identify given the limitations of genomic annotation. To assist users in identifying links of higher confidence, each link in Predictome is marked when the association agrees with a functional assignment in COG or pathway information in KEGG. Furthermore, users of the database can view those links produced by multiple methods, which are therefore less likely to be produced by chance.
Figure 1.
Visualization of predicted links among components of the TCA cycle in E.coli. Red, links based on phylogenetic profiling; blue, gene fusion links; green, links established by chromosomal proximity. frdB/sdhB and frdA/sdhA are paralogous pairs (indicated by *).
Table 1. Correlation of predictive methods with COG, KEGG and GeneQuiz.
| Method | Total Links | KEGG | COG | GeneQuiz | |||
|---|---|---|---|---|---|---|---|
| Qualified linksa | In same pathwayb | Qualified linksa | In same functional categoryb | Qualified linksa | In same functional categoryb | ||
| Phylogenetic profiling | 30 487 | 3905 (13%) | 79% | 18 959 (62%) | 64% | 4627 (15%) | 64% |
| Chromosomal proximity | 18 714 | 6215 (33%) | 87% | 12 456 (67%) | 72% | 5787 (15%) | 64% |
| Domain fusion | 248 472 | 4609 (2%) | 71% | 114 526 (46%) | 57% | 19 954 (8%) | 51% |
| Physical interaction | 2832 | 182 (6%) | 62% | 478 (17%) | 32% | 552 (19%) | 33% |
aQualified links are defined as links where both proteins participating in the link are present in KEGG, COG or GeneQuiz, and indicated as a percentage of the total links in parentheses. A link in KEGG refers to the participation in the same pathway and in COG and GeneQuiz to members of the same functional category.
bThe percentage of links in the same pathway or functional category is calculated using qualified links only.
Table 2. Correlation between the links generated by the different methods of Predictome.
| Physical interaction | Chromosomal proximity | Phylogenetic profiling | Domain fusion | |
|---|---|---|---|---|
| Physical interaction | 2832 | 7 | 10 | 6 |
| Chromosomal proximity | – | 18 714 | 1838 | 2143 |
| Phylogenetic profiling | – | – | 30 487 | 1687 |
| Domain fusion | – | – | – | 248 472 |
Value indicates the number of identical links between the sets.
APPLICATIONS AND FEATURES
Predictome often reveals many links for each protein, each potentially implying a different functional relationship. Thus, it is imperative to have computational tools for condensing the information derived from these links into a coherent format which can integrate functional annotation from various sources. Predictome has two such tools to assist in interpreting the output of the database, both based on condensing the annotation of protein groups.
The first tool uses the structured vocabulary of the Gene Ontology (GO) project (30). GO is a quickly expanding source of systematic gene annotation, where functional terms exist in a descending hierarchy of increasing specificity. When the linked partners of a given protein are mapped onto the structured vocabulary of GO, the result can be represented in graphical form. The GO analysis tool gives a quick overview of the annotation relationships among the proteins, in hierarchical format.
Another analytical tool on the site compares different text annotations for proteins, using a phrase-building algorithm to construct concise functional summaries from a variety of sources. This allows the user to quickly view the ‘consensus’ annotation for a given set. The tool provides compact summaries for proteins in genomes where there are otherwise low levels of annotation.
DATABASE IMPLEMENTATION
Predictome is implemented as a web-accessible relational database using the PostgreSQL RDBMS. The schema and instructions for use of this database can be viewed from the database web page http://predictome.bu.edu. Users can browse the database by entering gene names or keywords, and navigate through the network of predicted links. An optional Java-based applet allows for the visualization of small sections of the network. The complete list of protein links and supporting data, as well as the technical specifications of the database system are publicly accessible through the home page.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank R. Berwick for support and advice in the development of this database. This work was supported in part by a National Science Foundation Integrative Graduate Education and Research Traineeship Program grant (to J.C.M., J.M. and K.H.C.). I.Y. is supported by a Whitaker Fellowship.
REFERENCES
- 1.Eisenberg D., Marcotte,E.M., Xenarios,I. and Yeates,T.O. (2000) Protein function in the post-genomic era. Nature, 405, 823–826. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 3.Gaasterland T. and Ragan,M.A. (1998) Constructing multigenome views of whole microbial genomes. Microb. Comp. Genomics, 3, 177–192. [DOI] [PubMed] [Google Scholar]
- 4.Huynen M.A. and Bork,P. (1998) Measuring genome evolution. Proc. Natl Acad. Sci. USA, 95, 5849–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tatusov R.L., Koonin,E.V. and Lipman,D.J. (1997) A genomic perspective on protein families. Science, 278, 631–637. [DOI] [PubMed] [Google Scholar]
- 6.Pellegrini M., Marcotte,E.M., Thompson,M.J., Eisenberg,D. and Yeates,T.O. (1999) Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA, 96, 4285–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dandekar T., Snel,B., Huynen,M. and Bork,P. (1998) Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci., 23, 324–328. [DOI] [PubMed] [Google Scholar]
- 8.Overbeek R., Fonstein,M., D’Souza,M., Pusch,G.D. and Maltsev,N. (1999) The use of gene clusters to infer functional coupling. Proc. Natl Acad. Sci. USA, 96, 2896–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcotte E.M., Pellegrini,M., Ng,H.L., Rice,D.W., Yeates,T.O. and Eisenberg,D. (1999) Detecting protein function and protein–protein interactions from genome sequences. Science, 285, 751–753. [DOI] [PubMed] [Google Scholar]
- 10.Enright A.J., Iliopoulos,I., Kyrpides,N.C. and Ouzounis,C.A. (1999) Protein interaction maps for complete genomes based on gene fusion events. Nature, 402, 86–90. [DOI] [PubMed] [Google Scholar]
- 11.Yanai I., Derti,A. and DeLisi,C. (2001) Genes linked by fusion events are generally of the same functional category: a systematic analysis of 30 microbial genomes. Proc. Natl Acad. Sci. USA, 98, 7940–7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uetz P., Giot,L., Cagney,G., Mansfield,T.A., Judson,R.S., Knight,J.R., Lockshon,D., Narayan,V., Srinivasan,M., Pochart,P. et al. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- 13.Ito T., Chiba,T., Ozawa,R., Yoshida,M., Hattori,M. and Sakaki,Y. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA, 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rain J.C., Selig,L., De Reuse,H., Battaglia,V., Reverdy,C., Simon,S., Lenzen,G., Petel,F., Wojcik,J., Schachter,V. et al. (2001) The protein–protein interaction map of Helicobacter pylori. Nature, 409, 211–215. [DOI] [PubMed] [Google Scholar]
- 15.Xenarios I., Rice,D.W., Salwinski,L., Baron,M.K., Marcotte,E.M. and Eisenberg,D. (2000) DIP: the database of interacting proteins. Nucleic Acids Res., 28, 289–291. Updated article in this issue: Nucleic Acids Res. (2002), 30, 303–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bader G.D., Donaldson,I., Wolting,C., Ouellette,B.F., Pawson,T. and Hogue,C.W. (2001) BIND—the Biomolecular Interaction Network Database. Nucleic Acids Res., 29, 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snel B., Lehmann,G., Bork,P. and Huynen,M.A. (2000) STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res., 28, 3442–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nitschke P., Guerdoux-Jamet,P., Chiapello,H., Faroux,G., Henaut,C., Henaut,A. and Danchin,A. (1998) Indigo: a World-Wide-Web review of genomes and gene functions. FEMS Microbiol. Rev., 22, 207–227. [DOI] [PubMed] [Google Scholar]
- 19.Fields S. and Song,O. (1989) A novel genetic system to detect protein–protein interactions. Nature, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- 20.Marcotte E.M., Pellegrini,M., Thompson,M.J., Yeates,T.O. and Eisenberg,D.A (1999) Combined algorithm for genome-wide prediction of protein function. Nature, 402, 83–86. [DOI] [PubMed] [Google Scholar]
- 21.Huynen M., Snel,B., Lathe,W.,III and Bork,P. (2000) Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res., 10, 1204–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galperin M.Y. and Koonin,E.V. (2000) Who’s your neighbor? New computational approaches for functional genomics. Nat. Biotechnol., 18, 609–613. [DOI] [PubMed] [Google Scholar]
- 23.Huynen M., Snel,B., Lathe,W. and Bork,P. (2000) Exploitation of gene context. Curr. Opin. Struct. Biol., 10, 366–370. [DOI] [PubMed] [Google Scholar]
- 24.Marcotte E.M. (2000) Computational genetics: finding protein function by nonhomology methods. Curr. Opin. Struct. Biol., 10, 359–365. [DOI] [PubMed] [Google Scholar]
- 25.Tatusov R.L., Natale,D.A., Garkavtsev,I.V., Tatusova,T.A., Shankavaram,U.T., Rao,B.S., Kiryutin,B., Galperin,M.Y., Fedorova,N.D. and Koonin,E.V. (2001) The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res., 29, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcotte E.M., Xenarios,I. and Eisenberg,D. (2001) Mining literature for protein–protein interactions. Bioinformatics, 17, 359–363. [DOI] [PubMed] [Google Scholar]
- 27.Andrade M.A. and Bork,P. (2000) Automated extraction of information in molecular biology. FEBS Lett., 476, 12–17. [DOI] [PubMed] [Google Scholar]
- 28.Kanehisa M. and Goto,S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res., 28, 27–30. Updated article in this issue: Nucleic Acids Res. (2002), 30, 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrade M.A., Brown,N.P., Leroy,C., Hoersch,S., de Daruvar,A., Reich,C., Franchini,A., Tamames,J., Valencia,A., Ouzounis,C. et al. (1999) Automated genome sequence analysis and annotation. Bioinformatics, 15, 391–412. [DOI] [PubMed] [Google Scholar]
- 30.The Gene Ontology Consortium (2001) Creating the gene ontology resource: design and implementation. Genome Res., 11, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]