Abstract
Simple Summary
Natural killer (NK) cells from patients with acute myeloid leukemia (AML), or from healthy donors and expanded with IL-15, show defective cytotoxicity and elevated levels of glycogen synthase kinase 3 beta (GSK3β), and drug inhibition of GSK3β was able to improve their cytotoxicity and maturation. To better understand its biologic role, we deleted GSK3B in NK cells and assessed their phenotype, gene expression, and function. We did not find alterations in cytotoxicity or maturation, but did observe increased metabolic function and alterations in genes related to mitochondrial metabolism. This suggests that GSK3β is a regulator of NK-cell metabolism.
Abstract
Loss of cytotoxicity and defective metabolism are linked to glycogen synthase kinase 3 beta (GSK3β) overexpression in natural killer (NK) cells from patients with acute myeloid leukemia or from healthy donors after expansion ex vivo with IL-15. Drug inhibition of GSK3β in these NK cells improves their maturation and cytotoxic activity, but the mechanisms of GSK3β-mediated dysfunction have not been well studied. Here, we show that expansion of NK cells with feeder cells expressing membrane-bound IL-21 maintained normal GSK3β levels, allowing us to study GSK3β function using CRISPR gene editing. We deleted GSK3B and expanded paired-donor knockout and wild-type (WT) NK cells and then assessed transcriptional and functional alterations induced by loss of GSK3β. Surprisingly, our data showed that deletion of GSK3B did not alter cytotoxicity, cytokine production, or maturation (as determined by CD57 expression). However, GSK3B-KO cells demonstrated significant changes in expression of genes related to rRNA processing, cell proliferation, and metabolic function, suggesting possible metabolic reprogramming. Next, we found that key genes downregulated in GSK3B-KO NK cells were upregulated in GSK3β-overexpressing NK cells from AML patients, confirming this correlation in a clinical setting. Lastly, we measured cellular energetics and observed that GSK3B-KO NK cells exhibited 150% higher spare respiratory capacity, a marker of metabolic fitness. These findings suggest a role for GSK3β in regulating NK cell metabolism.
Keywords: natural killer cells, NK cells, GSK3β, AML, Cas9/RNP, CRISPR, metabolism, OXPHOS
1. Introduction
The antitumor activity of NK cells has been widely demonstrated for multiple cancer types [1]. This antitumor role can be limited by several factors in the tumor microenvironment that mediate metabolic suppression [2,3]. As a result, chemical inhibitors, genetic, and epigenetic alterations are now being tested to overcome these suppressive mechanisms in NK cells [4]. One promising target to enhance NK cell cytotoxic activity is GSK3β, a serine threonine kinase. GSK3β has been shown to regulate multiple functions in other cell types, but its role in lymphocytes including NK cells has not been well studied. In a few reports, drug inhibition of GSK3β has been shown to improve maturation and antitumor activity of NK cells with elevated GSK3β, such as IL-15 expanded NK cells or NK cells isolated from AML patients [5,6,7].
We recently described an approach for studying human NK cell biology by combining NK cell expansion with CRISPR gene editing using Cas9 complexed with guide ribonucleoproteins (Cas9/RNP). Here, we apply this approach to studying the transcriptional and functional role of GSK3β by generating CRISPR mediated GSK3B-KO NK cells, to avoid the confounding off-target effects of small-molecule drug inhibition. To confirm the clinical relevance of the differentially expressed genes found in GSK3B-KO NK cells, we looked at expression of these genes in NK cells from patients with AML, which we previously showed to have elevated GSK3β [8,9]. Lastly, we investigated the metabolic alterations in NK cells induced by loss of GSK3β.
2. Material and Methods
2.1. Source of Primary Human NK Cells and Regulatory Approvals
Buffy coats from healthy volunteer red blood cell donations were obtained from the American Red Cross (Columbus, OH, USA). Peripheral blood was obtained from patients with untreated AML (Ohio State University Leukemia Tissue Bank). All studies and sample collections were approved through the Ohio State University Institutional Review Board, protocol number 2009C0019, or deemed IRB exempt.
2.2. Purification and Expansion of NK Cells
NK cells from AML patients and controls from normal donors were isolated by density-gradient purification followed by flow-based sorting. NK cells from both healthy donors and untreated AML patients were sorted into two subpopulations based on NK cell development (CD94+/-/NKp80+/CD16+/CD57- and CD94+/-/NKp80+/CD16+/CD57+ (stage 5 and 6, respectively)) as previously described [10,11,12,13,14,15]. NK cells for in vitro gene editing studies were isolated by negative selection using RosetteSep™ Human NK Cell Enrichment Cocktail (Stem Cell Technologies, 15065, Vancouver, BC, Canada). Purified NK cells (depleted of T cells, B cells, and monocytes and typically > 95% CD16/56+) were stimulated weekly for two weeks with irradiated CSTX002 feeder cells in AIM-V expansion medium supplemented with ICSR (CTS™AIMV™SFM/CTSTM Immune Cell SR, Thermo Fisher Scientific) [16] and 50 IU of human recombinant IL-2 (rIL-2) (Novartis). NK cell purity of WT and KO NK cells was determined after expansion and was uniformly > 98% (Supplementary Figure S1).
2.3. Tumor Cell Lines
HL60 (AML), Kasumi1 (AML), K562 (CML), DAOY (medulloblastoma), and MG63 (osteosarcoma) cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). U373 (glioblastoma) was kindly shared by Kevin Cassady (Nationwide Children’s Hospital, Columbus, OH, USA). CSTX002 feeder cells (K562 genetically modified to express 4-1BBL and membrane-bound IL-21, referred to hereafter as FC-21) was generated as previously described [16].
2.4. Generation of CRISPR-Edited NK Cells
GSK3B-KO NK cells were generated by electroporation of Cas9/RNP into NK cells at day 7 of expansion [17,18,19], targeting exon 5 of the GSK3B gene (5- CAGTATCAGGATCCAACAAG) as described previously [17,18]. Wildtype (WT) cells were electroporated in electroporation buffer without Cas9/RNP complexes as control.
2.5. NK Cell Functional Assays
Calcein-AM was used to evaluate cytotoxicity previously described [20]. Briefly, tumor cell targets were loaded with 2 ug/mL of Calcein-AM for 30 min. Cells were washed and incubated with WT or GSK3B-KO NK cells at multiple effector/target (E:T) ratios for 4 h. For long-term killing assays, real time cell analysis—(RTCA) was performed. Briefly, the xCELLigence SP instrument (Agilent) was utilized to record cellular impedance following the previously described [21,22]. To analyze killing of target cells by NK cells, the E-Plate wells were incubated with the tethering reagent CD29 for Kasumi cell line. WT or GSK3B-KO NK cells—effector cells—were added on top of the tumor cells at 2:1 (Effector: Target) ratio. Each condition was tested in triplicate and total of three donors were evaluated. An “effector cell only” control was included to demonstrate that NK cells do not attach in presence of anti-CD29 and subtracted from the equation. Cytotoxic activities were evaluated based on the viability of the tumor cells remained attached in E-Plate surface, as reflected by Cell Index values. The xIMT software was used to plot the percentage of cytolysis. Experiment was conducted in RPMI1640 medium with 10% FBS supplemented with 50 IU of IL-2.
2.6. Metabolic Assays
Metabolic assays were performed as previously described [17]. Briefly, we used Seahorse XF Cell Mito Stress Test Kit to measure the oxygen consumption rate (OCR) (Cat# 103015-100, Agilent Technologies, Santa Clara, CA, USA) and Seahorse XF Glycolysis Stress Test Kit to measure extracellular acidification rate (ECAR) (Cat# 103020-100, Agilent Technologies, Santa Clara, CA, USA).
2.7. Antibodies
The following antibodies were used for flow cytometry: NCAM-1 (CD56) (Cat#744217—BD), CD57 (Cat#130-111-964—Miltenyi Biotec). The following antibodies were used for Western Blot: GSK3β Rabbit mAb—27C10 (Cat#9315S—Cell Signaling—1:1000), ß-Actin Mouse mAb—8H10D10 (Cat#3700S—Cell Signaling—1:1000), Anti-Rabbit IgG HRP-linked (Cat#7074S—Cell Signaling—1:5000), Anti-Mouse IgG HRP-linked (Cat#7076S—Cell Signaling—1:5000).
2.8. Cytokine Secretion
To induce cytokine secretion, NK cells at 2 × 106/mL were stimulated with 10 μg/mL PHA. After 4 h of incubation, supernatants were collected and kept in −80 °C. On the day of the assay, the supernatants were thawed and measured in duplicate with Bio-Rad Bio-plex Pro Human Immunotherapy Panel 20-Plex (Cat#12007975) according to manufacturer’s instructions. Data were acquired on Bio-rad Bio-Plex 200 system and analyzed with Bio-plex Manager software using curve-fitting with logistic regression (5PL regression) [23].
2.9. RNA-Sequencing on Non-Expanded Healthy and AML-NK
RNA-sequencing (RNA-seq) analysis was done as previously described [10]. Briefly, freshly sorted NK cell subpopulations from normal donor peripheral blood (American Red Cross; n = 3 donors) or newly diagnosed AML patients (Ohio State University Leukemia Tissue Bank; n = 5; OSU IRB# 2009C0019), were pelleted and total RNA was isolated using the Qiagen RNeasy Mini Kit (Qiagen). Directional poly-A RNA sequencing libraries were prepared and sequenced as 42-bp paired-end reads on an Illumina NextSeq 500 instrument (Illumina) to a depth of 33.2–48.0 × 106 read pairs (Active Motif). Alignment to human genome (hg19 build) was done using TopHat [24]. Transcriptome assembly and analysis was performed using Cufflinks and expression was reported as FPKM.
2.10. RNA-Sequencing on Expanded WT and GSK3B-KO NK Cells
For RNA-seq on expanded WT and GSK3B-KO NK cells, RNA libraries were prepared using the TruSeq RNA Sample Preparation Kit (Illumina Inc. San Diego, CA, USA) and sequence reads (150 bp each) were generated per library using the Illumina HiSeq4000 platform (Institute for Genomic Medicine, Nationwide Children’s Hospital). Reads were aligned and count tables were generated using Kallisto (v 0.43.1) [25]. Differential expression was then done using the Bioconductor package DeSeq2 (v 1.36.0). Volcano plots were generated using the package Glimma (v. 2.6.0) [26] within R. Gene ontology (GO) was performed using Gorillia [27] and subsequently visualized using REVIGO [28] via their online web portal.
3. Results
3.1. Deletion of GSK3B in FC-21 Expanded NK Cells
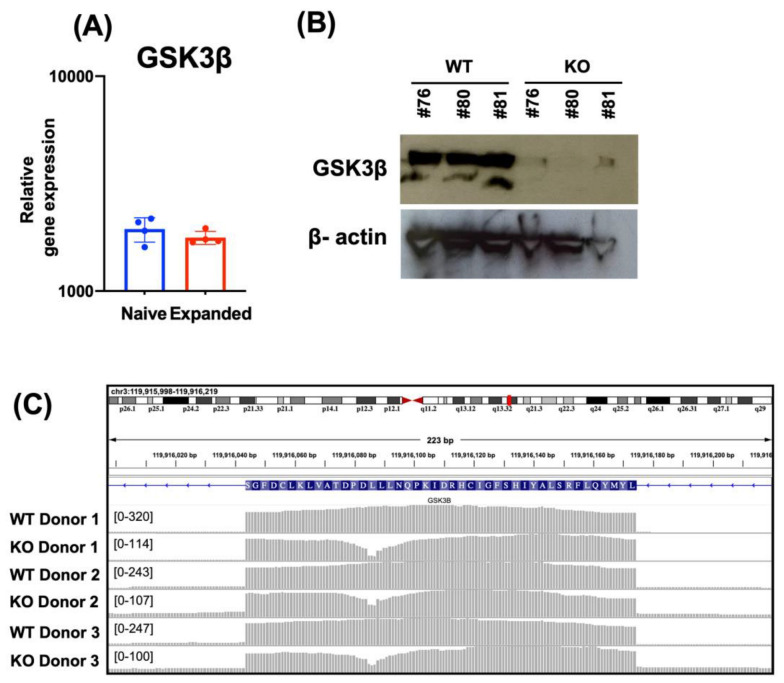
We previously described a gene editing approach for primary NK cells [17,18] in which propagation with FC-21 feeder cells enhances DNA repair machinery and expands edited cells to large numbers [19]. It was previously shown that GSK3β becomes overexpressed when NK cells are expanded in presence of soluble IL-15 and is associated with inhibition of their maturation and cytotoxic activity [5]. Therefore, we first evaluated the relative gene expression of GSK3B in both naïve and FC-21-expanded NK cells to assess the stability of GSK3β in this model system. We found that the expression level of GSK3B was similar between WT and expanded NK cells (Figure 1A).
Figure 1.
Generation of GSK3B-KO FC-21 expanded NK cells. GSK3B expression in NK cells before and after expansion on FC-21 was evaluated by RNAseq analysis. (A) The efficiency of Cas9/RNP mediated deletion of GSK3β was determined by Western blot (B) and RNA-seq (C).
To study the role of GSK3β and to avoid off-target effects of chemical inhibitors we generated GSK3B-KO NK cells and evaluated the knock-out efficiency at the protein level by Western blot (Figure 1B, uncropped blots shown in supplemental Figure S2A,B), and at the mRNA level by RNA-sequencing (Figure 1C) observed as a read drop across the region targeted by the gRNA at exon 5 of the GSK3B locus. Our data demonstrated successful GSK3β gene deletion in FC-21 expanded NK cells.
3.2. Deletion of GSK3B Does Not Alter Proliferation, Killing Potency, Cytokine Secretion, or Maturation of FC-21 Expanded NK Cells
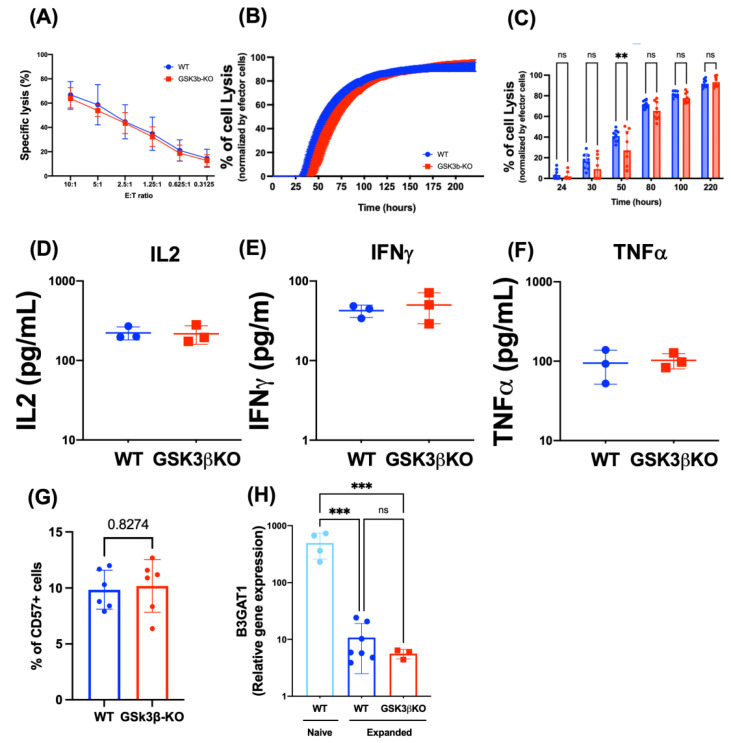
Drug inhibition of GSK3β can improve NK cell killing against AML in NK cells with elevated GSK3β, such as those expanded with IL15 [5] or from patients with AML [7]. Therefore, we next assessed the effect of knocking out GSK3B in FC-21 expanded NK cells on their anti-AML activity. Unexpectedly, the absence of GSK3β did not alter NK cell proliferation (supplementary Figure S2C,D) or killing against Kasumi1, an AML cell line, in either short (Figure 2A) or long-term (Figure 2B,C) killing assays. Additionally, GSK3B-KO NK cells showed similar killing potency as WT NK cell against HL60 (AML), K562 (CML), DAOY (medulloblastoma), and MG63 (osteosarcoma) in a standard 4 h killing assay (see supplementary Figure S3). In the previously reports, the improved killing in NK cells treated with GSK3β chemical inhibitors was also associated with increased cytokine secretion. Thus, we evaluated cytokine secretion in WT and GSK3B-KO and found that both WT and GSK3B-KO NK cells secrete high levels of IL-2, IFNγ and TNFα, and no difference was found between the two (Figure 2C).
Figure 2.
Deletion of GSK3B did not alter antitumor activity or maturation of FC-21 expanded NK cells. Both WT and GSK3B-KO NK cells were cocultured with Kasumi1 (AML tumor cell line) for 4 (A) or 220 h (B,C). “B” time course of long-term cell killing and “C” bar graph representative of indicated hours; (n = 3). (D–F) IL2, IFNγ and TNFα release was assessed after 4 h of WT and GSK3B-KO NK cells with PHA (n = 3). (G), CD57 expression was evaluated by flow cytometry on FC-21 expanded WT and GSK3B-KO NK cells (n = 6). (H), B3GAT1 expression in WT, FC-21 expanded WT, and FC-21 expanded GSK3B-KO NK cells as measured by RNA-seq. ns = non-significative, ** p ≤ 0.01, *** p ≤ 0.001. See also supplementary Figure S3 for cytotoxicity against other cancer cell lines, and supplementary Figure S4 for representative CD57 gating.
Additionally, Cichocki et al., previously reported that GSK3β drug inhibition drove maturation of IL-15 expanded NK cell, evidenced by CD57 expression [5]. Thus, we analyzed CD57 expression on both FC-21 expanded WT and GSK3B-KO NK cells by flow cytometry and found no difference between them (Figure 2D). We then investigated FC-21 expanded NK cells for their expression of B3GAT1, the gene encoding the enzyme responsible for catalyzing the CD57 carbohydrate epitope. Interestingly, we found that B3GAT1 was 30-fold downregulated in FC-21 expanded NK cells compared to naïve NK cells, and that this low expression was not altered by GSK3B deletion (Figure 2E). This explains the low CD57 surface expression in both WT and gene edited cells, and also suggests that B3GAT1 is not directly regulated by GSK3β and that other elements may be involved in regulating the expression of CD57 in NK cells.
3.3. Transcriptional Changes in GSK3B-KO NK Cells
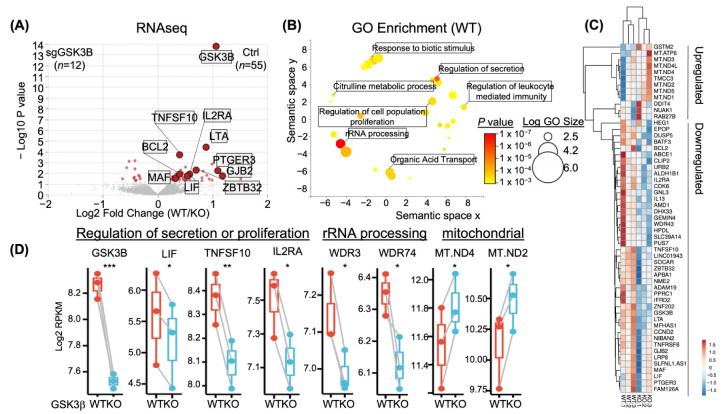
To study the role of GSK3β in NK cells, we next performed bulk RNA-seq on WT or GSK3B-KO NK cells after expansion with FC-21. Differential gene expression analysis via Deseq2 revealed 55 genes (12 upregulated and 43 downregulated) with significantly altered expression (adjusted P value < 0.05, Paired Deseq2 test) in WT NK cells compared to GSK3B-KO cells. The most significant of these differentially expressed genes was GSK3B (P = 1.59 × 10−14), thus the majority of changes detected were of loss of expression concurrent with the loss of GSK3β expression (Figure 3A,D).
Figure 3.
GSK3B-KO in NK resulted in transcriptomic changes, RNA-seq analysis was performed on NK cells from FC-21 expanded WT and GSK3B-KO. (A) volcano plot representing DEG (WT/KO). (B) GO enrichment (WT). Heatmap (C) and RNA-seq boxplots (D) showing the top downregulated and top upregulated genes affected by GSK3B deletion. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001. For (C), rows are centered; unit variance scaling is applied to rows. Both rows and columns are clustered using correlation distance and average linkage.
We next used gene ontology (GO) analysis of the 55 WT-specific transcripts to determine what expression programs were regulated by GSK3β. GO results were then visualized via REVIGO, which clusters redundant categories for biological interpretation (Figure 3B). Unique GO clusters included those for ribosomal RNA (rRNA) processing, secretion, metabolic processes, and proliferation (Figure 3B). For example, GSK3B-KO lost transcription of genes involved within NK cell homeostatic renewal, such as the high affinity IL2 trimeric receptor, encoded by IL2RA, and the survival gene BCL2 (Figure 3C). We likewise observed downregulation of genes that encode for TNFα effectors, namely leukemia inhibitory factor (LIF) and TNFSF10 (encoding TRAIL), within GSK3B-KO. Additional expression analysis suggested deregulation of global biosynthesis as some genes involved in ribosomal RNA processing were lost (e.g., WDR3, WDR74, see Figure 3C) and those involved in mitochondrial function were gained (e.g., MT-ND4 and MT-ND2, see Figure 3C). Together, transcriptome analysis suggested a role of GSK3β in regulating NK cell homeostasis, effector function, and metabolism.
3.4. Deletion of GSK3B Leads to Metabolic Reprogramming of NK Cells
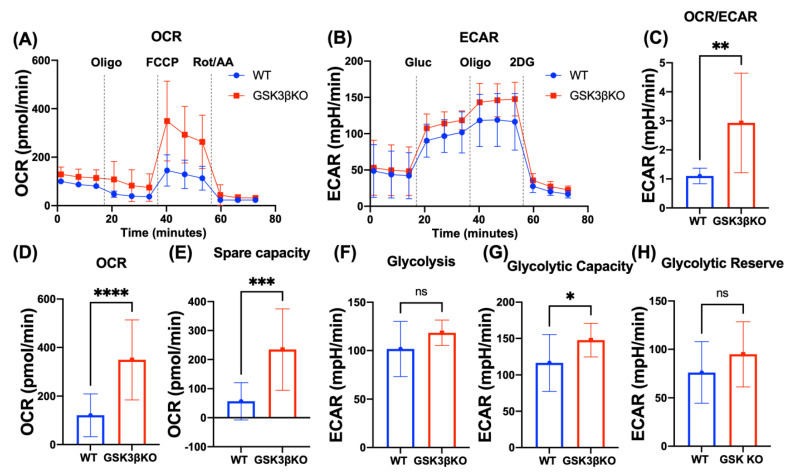
To explore the effect of upregulation of the mitochondrial genes identified by RNA-seq, we next examined the cellular metabolism of WT and GSK3B-KO NK cells by assessing both mitochondrial (Figure 4A,D,E) and glycolytic stress (Figure 4B,F–H). We found that GSK3B-KO NK cells presented an overall shift towards oxidative metabolism with higher OCR and spare respiratory capacity (Figure 4A–E). Spare respiratory capacity was previously described as pivotal for memory T cell formation and as a reliable indicator of metabolic fitness [29,30]. Additionally, GSK3B-KO NK cells also showed modestly higher ECAR, glycolytic capacity, and glycolytic reserve than WT NK cells, indicating that the increase in oxidative phosphorylation was not compensatory for decreased glycolysis. Together this suggests that deletion of GSK3B induces NK cells to enhance overall metabolic capacity, which could be beneficial for sustaining antitumor activity.
Figure 4.
Deletion of GSK3B enhances metabolic capacity of NK cells. (A,B—mitochondrial and glycolytic stress, respectively) summarized data of metabolic analysis of paired WT and GSK3B-KO NK cells (n = 3; mean ± SD). (C) OCR/ECAR ratio. (D) Maximum respiratory capacity. (E) reserve capacity derived from (A). (F) Glycolysis, (G) glycolytic capacity, and (H) glycolytic reserve derived from (B). n = 4, ns = non-significant, * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001.
3.5. GSK3β Is Highly Expressed in NK Cells at Stage 5 and 6 of Maturation from AML Patients
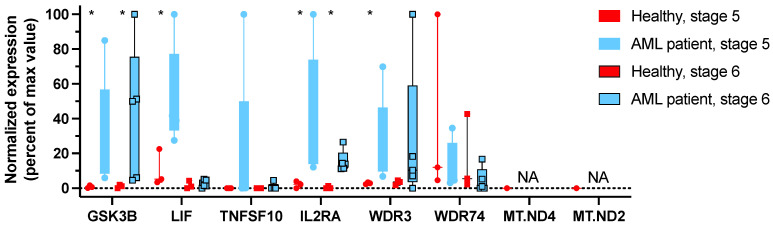
To further study the role of GSK3β in NK cells, we studied transcriptional changes in NK cells from AML patients which have been shown to overexpress GSK3β, to validate key genes identified as differentially expressed in GSK3B-KO NK cells. To correct for confounding by differences in NK cell maturation, we separately studied the expression of level of these genes in NK maturation stages 5 and 6. First, we confirmed that GSK3β was statistically elevated in NK cells from AML patients when compared to healthy donor NK cells, for both stage 5 and stage 6 cells. We then evaluated the relative expression of the key genes identified in GSK3B-KO NK cells (Figure 5) and found strong concordance of downregulated genes in GSK3B-KO NK cells being upregulated in GSK3β-overexpressing NK cells from patients with AML. LIF was only overexpressed in stage 5 but not in stage 6. IL2RA expression was significantly increased in both AML-NK5 and AML-NK6 when compared to healthy NK cells. No difference was observed in expression level of WDR74, but WDR3 was significantly increased in AML-NK5. We did not identify any mRNA transcripts for MT-ND4 and MT-ND2 in any of these samples, despite finding MT-ND4 and MT-ND2 transcripts at high levels in the GSK3B-KO experiments. Since these mtDNA transcripts are not recovered efficiently by all purification methods, this may be explained differences in RNA isolation and sequencing methods in the GSK3B-KO and AML-NK experiments. Although the reverse correlation between the differentially expressed genes in GSK3B-KO NK cells versus AML-NK cells suggests a possible role of GSK3β on NK-cell activity in these patients, further studies would be beneficial.
Figure 5.
The downregulated genes identified in GSK3B-KO NK cells were mostly upregulated in GSK3β-overexpressing NK cells from patients with AML. RNA-seq analysis was performed on NK cells at different stages of maturation isolated by sorting from healthy donors and patients with AML. * p ≤ 0.05. (Healthy donors, n = 3; AML patient, n = 5).
4. Discussion
NK cells play an important role in immunosurveillance and preventing tumor development and progression. NK cells from patients with AML exhibit great defects in both numbers and functional activities, unable to control AML development, progression and relapse [31,32]. Even though phenotypic changes have been well described, specific molecular explanations for these dysfunctions are still needed [33]. It was previously reported that the high expression of GSK3β on AML NK cells profoundly impacted NK cells killing ability [7]. Here, we further demonstrated that the enhanced expression of GSK3β is present in NK cells at both stage 5 and 6 of development in patients with AML. There was previously limited evidence on the biologic mechanisms of GSK3β-mediated dysfunction in NK cells. Here, we used Cas9/RNP gene editing to generate GSK3B-KO NK cells [17,18,19] to enable precise gene-level understanding of GSK3β effects on primary human NK cells, which revealed novel information on the impact of GSK3β on transcriptional regulation of homeostasis, effector function, and metabolism.
We showed that expansion of NK cells on FC-21 does not increase GSK3β expression levels as was observed in IL15-expanded NK cells, which may reduce the need for GSK3β inhibition in adoptive NK cell therapy.
Deletion of GSK3B resulted in increased mitochondrial respiratory capacity that was associated with an increase in Complex 1 genes, MT-ND2 and MT-ND4, which favor OXPHOS metabolism. Defined as “The Most Complex Complex” [34], Complex 1, also known as NADH dehydrogenase, is composed of 46 subunits, seven of which (including MT-ND2 and MT-ND4) are encoded in the mitochondrial genome. Complex 1 catalyzes electrons transfer from NADH through the respiratory chain, using ubiquinone as an electron acceptor [35]. Furthermore, the expression of mitochondrial respiratory complex 1 was previously associated with gain in mitochondrial oxidative phosphorylation (OXPHOS) activity. Additionally, targeting OXPHOS with a complex I inhibitor decreased OXPHOS in pancreatic cancer cells reinforcing the correlation between OXPHOS and Complex 1 [36]. In line with previously published evidence, our data showed that GSK3B deletion resulted in increased OXPHOS, with higher maximum and spare respiratory capacities. The edited cells also shift their metabolic profile more towards OCR than ECAR which can improve ATP generation and provide more energy to the cells essential for effector function. Previously Bou-Tayeh et al. [37], demonstrated that NK cells from both leukemic mice and patients with AML displayed similar metabolic defects.
Several groups showed that NK cells from AML patients or expanded with IL15 have high levels of GSK3β and that chemical inhibition of GSK3β can improve their antitumor activity through an increase in NFKB signaling molecules (RELA, RELB, c-REL, NFKIBA) [5] However, our data showed that FC21 expansion of NK cells does not increase GSK3β levels, and therefore its deletion did not alter their cytotoxicity, in both short and long-term killing assays we used here. In vivo experiments or other approaches that mimic the harsh tumor microenvironment may be able to show the effect of these metabolic improvements on anti-tumor activity of GSK3B-KO NK cells.
NK cells with high GSK3β have been shown to express low levels of CD57. A benefit of using GSK inhibitors in these NK cells was to increase CD57 expression level suggesting maturation to an adaptive memory-like phenotype. This protein is recognized as a maturation/senescence marker and its frequency increases with age [36] and it has been reported to be missing or dim on fetal and infant NK cells [38]. The CD57 expression, denoting terminal differentiation of NK cells, should be beneficial from an immunotherapy standpoint, as several clinical studies reported that high expression of CD57 on NK cells is associated with better clinical outcome [5,38,39]. It has been shown that drug inhibition of GSK3β resulted in increased CD57 expression. Of note, CD57 is not a DNA-encoded protein, but rather a carbohydrate epitope catalyzed by B3GAT1 [40]. We showed that B3GAT1 levels decrease in FC-21 expanded NK cells, but that both CD57 and B3GAT1 remain unchanged after the deletion of GSK3B. This suggests that GSK3β does not directly regulate B3GAT1, or therefor CD57, implying other regulators and/or pathways control the CD57 expression associated with maturation and memory. We also note that there was no correlation between GSK3B expression and Stage 5 or 6 NK cells, for AML-NK or healthy donor NK cells, despite Stage 5 and 6 being defined by the expression of CD57.
5. Conclusions
Using CRISPR gene deletion of GSK3B in human primary NK cells we showed that the GSK3B does not any affect in cytotoxicity or maturation of the cells, but has a role in metabolic function in NK cells. Taken together, we show that GSK3β is a negative metabolic regulator in NK cells.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15030705/s1, Figure S1: Flow cytometry gating strategy to evaluate CD56 expression on NK cells; Figure S2: Protein expression on NK cells after CRISPR/Casp9; Figure S3: WT and GSK3B-KO NK cells showed similar killing potency; Figure S4: Flow cytometry gating strategy to evaluate CD57+ cells.
Author Contributions
Conceptualization, M.N.K., M.S.F.P. and D.A.L.; methodology, M.N.K., M.S.F.P., K.S., Y.S., A.T., C.M., P.L.C. and B.L.M.-B.; validation, M.N.K., M.S.F.P., P.L.C. and B.L.M.-B.; formal analysis, M.N.K., M.S.F.P., P.L.C., B.L.M.-B. and D.A.L.; investigation, M.S.F.P., M.N.K. and D.A.L.; resources, M.N.K. and D.A.L.; data curation, M.S.F.P., M.N.K., P.L.C., B.L.M.-B. and D.A.L.; writing—original draft preparation, M.S.F.P., M.N.K. and D.A.L.; writing—review and editing, M.N.K., M.S.F.P. and D.A.L.; visualization, M.S.F.P., M.N.K. and D.A.L.; supervision, M.N.K. and D.A.L.; funding acquisition, M.N.K. and D.A.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of THE Ohio State University—protocol 2009C0019 from 25 July 2022.
Informed Consent Statement
IRB protocol 2009C0019 from 25 July 2022 approved and waiver of informed consent from the patient for this protocol.
Data Availability Statement
All data reported in this paper will be shared by the correspondence authors upon request.
Conflicts of Interest
M.S.F.P.: reports licensing and royalty fees from Kiadis Pharma, and intellectual property interest under provisional application 63/439,426. A.T.: Licensing and royalty fees from Kiadis Pharma. B.L.M.B., have consulting with Radiant Biotherapeutics and receive laboratory funding from Ikena Oncology. M.N.K. reports consulting, licensing and royalty fees from Kiadis Pharma, and intellectual property interest under provisional application 63/439,426. D.A.L.: Scientific advisory board and consulting, licensing and royalty fees from Kiadis Pharma, and intellectual property interest under provisional application 63/439,426. K.S., Y.S., C.M. and P.L.C. reported none.
Funding Statement
This study was supported by funding from Office of technology commercialization at the Abagail Wexner Research Institute (TDF-2018-021, M.N.K); The Hyundai Hope on Wheels Foundation (D.A.L.); the NIH -NCI (U54-CA232561, D.A.L.) and NIH -NIAID (R21-AI156411, P.L.C., B.L.M.-B). We thank the Institute for Genomic Medicine at Nationwide Children’s Hospital for their assistance with nucleic acid sequencing.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Lee D.A. Cellular therapy: Adoptive immunotherapy with expanded natural killer cells. Immunol. Rev. 2019;290:85–99. doi: 10.1111/imr.12793. [DOI] [PubMed] [Google Scholar]
- 2.Loftus R.M., Finlay D.K. Immunometabolism: Cellular Metabolism Turns Immune Regulator. J. Biol. Chem. 2016;291:1–10. doi: 10.1074/jbc.R115.693903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poznanski S.M., Singh K., Ritchie T.M., Aguiar J.A., Fan I.Y., Portillo A.L., Rojas E.A., Vahedi F., El-Sayes A., Xing S., et al. Metabolic flexibility determines human NK cell functional fate in the tumor microenvironment. Cell Metab. 2021;33:1205–1220.e5. doi: 10.1016/j.cmet.2021.03.023. [DOI] [PubMed] [Google Scholar]
- 4.Naeimi Kararoudi M., Tullius B.P., Chakravarti N., Pomeroy E.J., Moriarity B.S., Beland K., Colamartino A.B.L., Haddad E., Chu Y., Cairo M.S., et al. Genetic and epigenetic modification of human primary NK cells for enhanced antitumor activity. Semin Hematol. 2020;57:201–212. doi: 10.1053/j.seminhematol.2020.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cichocki F., Valamehr B., Bjordahl R., Zhang B., Rezner B., Rogers P., Gaidarova S., Moreno S., Tuininga K., Dougherty P., et al. GSK3 Inhibition Drives Maturation of NK Cells and Enhances Their Antitumor Activity. Cancer Res. 2017;77:5664–5675. doi: 10.1158/0008-5472.CAN-17-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson J.K., Miller J.S. Current strategies exploiting NK-cell therapy to treat haematologic malignancies. Int. J. Immunogenet. 2018;45:237–246. doi: 10.1111/iji.12387. [DOI] [PubMed] [Google Scholar]
- 7.Parameswaran R., Ramakrishnan P., Moreton S.A., Xia Z., Hou Y., Lee D.A., Gupta K., deLima M., Beck R.C., Wald D.N. Repression of GSK3 restores NK cell cytotoxicity in AML patients. Nat. Commun. 2016;7:11154. doi: 10.1038/ncomms11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chretien A.S., Fauriat C., Orlanducci F., Galseran C., Rey J., Bouvier Borg G., Gautherot E., Granjeaud S., Hamel-Broza J.F., Demerle C., et al. Natural Killer Defective Maturation Is Associated with Adverse Clinical Outcome in Patients with Acute Myeloid Leukemia. Front. Immunol. 2017;8:573. doi: 10.3389/fimmu.2017.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chretien A.S., Devillier R., Granjeaud S., Cordier C., Demerle C., Salem N., Wlosik J., Orlanducci F., Gorvel L., Fattori S., et al. High-dimensional mass cytometry analysis of NK cell alterations in AML identifies a subgroup with adverse clinical outcome. Proc. Natl. Acad. Sci. USA. 2021;118:e2020459118. doi: 10.1073/pnas.2020459118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mundy-Bosse B.L., Weigel C., Wu Y.Z., Abdelbaky S., Youssef Y., Casas S.B., Polley N., Ernst G., Young K.A., McConnell K.K., et al. Identification and Targeting of the Developmental Blockade in Extranodal Natural Killer/T-cell Lymphoma. Blood Cancer Discov. 2022;3:154–169. doi: 10.1158/2643-3230.BCD-21-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freud A.G., Becknell B., Roychowdhury S., Mao H.C., Ferketich A.K., Nuovo G.J., Hughes T.L., Marburger T.B., Sung J., Baiocchi R.A., et al. A human CD34(+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity. 2005;22:295–304. doi: 10.1016/j.immuni.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Freud A.G., Yokohama A., Becknell B., Lee M.T., Mao H.C., Ferketich A.K., Caligiuri M.A. Evidence for discrete stages of human natural killer cell differentiation in vivo. J. Exp. Med. 2006;203:1033–1043. doi: 10.1084/jem.20052507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L., Youssef Y., Robinson C., Ernst G.F., Carson M.Y., Young K.A., Scoville S.D., Zhang X., Harris R., Sekhri P., et al. CD56 Expression Marks Human Group 2 Innate Lymphoid Cell Divergence from a Shared NK Cell and Group 3 Innate Lymphoid Cell Developmental Pathway. Immunity. 2018;49:464–476.e464. doi: 10.1016/j.immuni.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scoville S.D., Mundy-Bosse B.L., Zhang M.H., Chen L., Zhang X., Keller K.A., Hughes T., Chen L., Cheng S., Bergin S.M., et al. A Progenitor Cell Expressing Transcription Factor RORgammat Generates All Human Innate Lymphoid Cell Subsets. Immunity. 2016;44:1140–1150. doi: 10.1016/j.immuni.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freud A.G., Keller K.A., Scoville S.D., Mundy-Bosse B.L., Cheng S., Youssef Y., Hughes T., Zhang X., Mo X., Porcu P., et al. NKp80 Defines a Critical Step during Human Natural Killer Cell Development. Cell Rep. 2016;16:379–391. doi: 10.1016/j.celrep.2016.05.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moseman J.E., Foltz J.A., Sorathia K., Heipertz E.L., Lee D.A. Evaluation of serum-free media formulations in feeder cell-stimulated expansion of natural killer cells. Cytotherapy. 2020;22:322–328. doi: 10.1016/j.jcyt.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Naeimi Kararoudi M., Nagai Y., Elmas E., de Souza Fernandes Pereira M., Ali S.A., Imus P.H., Wethington D., Borrello I.M., Lee D.A., Ghiaur G. CD38 deletion of human primary NK cells eliminates daratumumab-induced fratricide and boosts their effector activity. Blood. 2020;136:2416–2427. doi: 10.1182/blood.2020006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naeimi Kararoudi M., Dolatshad H., Trikha P., Hussain S.A., Elmas E., Foltz J.A., Moseman J.E., Thakkar A., Nakkula R.J., Lamb M., et al. Generation of Knock-out Primary and Expanded Human NK Cells Using Cas9 Ribonucleoproteins. J. Vis. Exp. 2018:e58237. doi: 10.3791/58237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kararoudi M.N., Likhite S., Elmas E., Yamamoto K., Schwartz M., Sorathia K., Pereira M.D.S.F., Sezgin Y., Devine R.D., Lyberger J.M., et al. Optimization and validation of CAR transduction into human primary NK cells using CRISPR and AAV. Cell Rep. Methods. 2022;2:100236. doi: 10.1016/j.crmeth.2022.100236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Somanchi S.S., McCulley K.J., Somanchi A., Chan L.L., Lee D.A. A Novel Method for Assessment of Natural Killer Cell Cytotoxicity Using Image Cytometry. PLoS ONE. 2015;10:e0141074. doi: 10.1371/journal.pone.0141074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerignoli F., Abassi Y.A., Lamarche B.J., Guenther G., Santa Ana D., Guimet D., Zhang W., Zhang J., Xi B. In vitro immunotherapy potency assays using real-time cell analysis. PLoS ONE. 2018;13:e0193498. doi: 10.1371/journal.pone.0193498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naeimi Kararoudi M., Likhite S., Elmas E., Schwartz M., Sorathia K., Yamamoto K., Chakravarti N., Moriarity B.S., Meyer K., Lee D.A. CD33 Targeting Primary CAR-NK Cells Generated By CRISPR Mediated Gene Insertion Show Enhanced Anti-AML Activity. Blood. 2020;136:3. doi: 10.1182/blood-2020-142494. [DOI] [Google Scholar]
- 23.Foltz J.A., Moseman J.E., Thakkar A., Chakravarti N., Lee D.A. TGFbeta Imprinting During Activation Promotes Natural Killer Cell Cytokine Hypersecretion. Cancers. 2018;10:423. doi: 10.3390/cancers10110423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C., Pachter L., Salzberg S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 26.Su S., Law C.W., Ah-Cann C., Asselin-Labat M.L., Blewitt M.E., Ritchie M.E. Glimma: Interactive graphics for gene expression analysis. Bioinformatics. 2017;33:2050–2052. doi: 10.1093/bioinformatics/btx094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Supek F., Bosnjak M., Skunca N., Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearce E.L., Poffenberger M.C., Chang C.H., Jones R.G. Fueling immunity: Insights into metabolism and lymphocyte function. Science. 2013;342:1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu H., Blum R.H., Bernareggi D., Ask E.H., Wu Z., Hoel H.J., Meng Z., Wu C., Guan K.L., Malmberg K.J., et al. Metabolic Reprograming via Deletion of CISH in Human iPSC-Derived NK Cells Promotes In Vivo Persistence and Enhances Anti-tumor Activity. Cell Stem Cell. 2020;27:224–237.e226. doi: 10.1016/j.stem.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lichtenegger F.S., Lorenz R., Gellhaus K., Hiddemann W., Beck B., Subklewe M. Impaired NK cells and increased T regulatory cell numbers during cytotoxic maintenance therapy in AML. Leuk. Res. 2014;38:964–969. doi: 10.1016/j.leukres.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 32.Khaznadar Z., Henry G., Setterblad N., Agaugue S., Raffoux E., Boissel N., Dombret H., Toubert A., Dulphy N. Acute myeloid leukemia impairs natural killer cells through the formation of a deficient cytotoxic immunological synapse. Eur. J. Immunol. 2014;44:3068–3080. doi: 10.1002/eji.201444500. [DOI] [PubMed] [Google Scholar]
- 33.Costello R.T., Sivori S., Marcenaro E., Lafage-Pochitaloff M., Mozziconacci M.J., Reviron D., Gastaut J.A., Pende D., Olive D., Moretta A. Defective expression and function of natural killer cell-triggering receptors in patients with acute myeloid leukemia. Blood. 2002;99:3661–3667. doi: 10.1182/blood.V99.10.3661. [DOI] [PubMed] [Google Scholar]
- 34.Ohnishi T. NADH-quinone oxidoreductase, the most complex complex. J. Bioenerg. Biomembr. 1993;25:325–329. doi: 10.1007/BF00762457. [DOI] [PubMed] [Google Scholar]
- 35.Ugalde C., Hinttala R., Timal S., Smeets R., Rodenburg R.J., Uusimaa J., van Heuvel L.P., Nijtmans L.G., Majamaa K., Smeitink J.A. Mutated ND2 impairs mitochondrial complex I assembly and leads to Leigh syndrome. Mol. Genet. Metab. 2007;90:10–14. doi: 10.1016/j.ymgme.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Masoud R., Reyes-Castellanos G., Lac S., Garcia J., Dou S., Shintu L., Abdel Hadi N., Gicquel T., El Kaoutari A., Dieme B., et al. Targeting Mitochondrial Complex I Overcomes Chemoresistance in High OXPHOS Pancreatic Cancer. Cell Rep. Med. 2020;1:100143. doi: 10.1016/j.xcrm.2020.100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bou-Tayeh B., Laletin V., Salem N., Just-Landi S., Fares J., Leblanc R., Balzano M., Kerdiles Y.M., Bidaut G., Herault O., et al. Chronic IL-15 Stimulation and Impaired mTOR Signaling and Metabolism in Natural Killer Cells During Acute Myeloid Leukemia. Front. Immunol. 2021;12:730970. doi: 10.3389/fimmu.2021.730970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nielsen C.M., White M.J., Goodier M.R., Riley E.M. Functional Significance of CD57 Expression on Human NK Cells and Relevance to Disease. Front. Immunol. 2013;4:422. doi: 10.3389/fimmu.2013.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cichocki F., Cooley S., Davis Z., DeFor T.E., Schlums H., Zhang B., Brunstein C.G., Blazar B.R., Wagner J., Diamond D.J., et al. CD56dimCD57+NKG2C+ NK cell expansion is associated with reduced leukemia relapse after reduced intensity HCT. Leukemia. 2016;30:456–463. doi: 10.1038/leu.2015.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gorska A., Gruchala-Niedoszytko M., Niedoszytko M., Maciejewska A., Chelminska M., Skrzypski M., Wasag B., Kaczkan M., Lange M., Nedoszytko B., et al. The Role of TRAF4 and B3GAT1 Gene Expression in the Food Hypersensitivity and Insect Venom Allergy in Mastocytosis. Arch. Immunol. Ther. Exp. 2016;64:497–503. doi: 10.1007/s00005-016-0397-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the correspondence authors upon request.