Abstract
Here we described PerturbSci-Kinetics, a novel combinatorial indexing method for capturing three-layer single-cell readout (i.e., whole transcriptome, nascent transcriptome, sgRNA identities) across hundreds of genetic perturbations. Through PerturbSci-Kinetics profiling of pooled CRISPR screens targeting a variety of biological processes, we were able to decipher the complexity of RNA regulations at multiple levels (e.g., synthesis, processing, degradation), and revealed key regulators involved in miRNA and mitochondrial RNA processing pathways. Our technique opens up the possibility of systematically decoding the genome-wide regulatory network underlying RNA temporal dynamics at scale and cost-effectively.
Cellular functions are determined by the expression of millions of RNA molecules, which are tightly regulated across several critical steps, including but not limited to RNA synthesis, splicing, and degradation. Dysregulated transcriptome kinetics have been linked to various diseases, including cancer1, intellectual disability2, and neurodegenerative disorders3. However, our knowledge regarding how critical molecular regulators affect genome-wide RNA kinetics is still scarce, partly due to the lack of scalable tools. For example, while single-cell transcriptome analysis coupled with pooled CRISPR screens have recently yielded fundamental insight into the gene regulatory mechanisms4–9, the readout of these methods only provides a snapshot of gene expression programs, thus is insufficient to decipher the complexity of RNA dynamics (e.g., synthesis, splicing, and degradation). To resolve this challenge, we developed PerturbSci-Kinetics, by integrating CRISPR-based pooled genetic screens, highly scalable single-cell RNA-seq by combinatorial indexing, and metabolic labeling to recover single-cell transcriptome dynamics across hundreds of genetic perturbations.
The key features of the new method include: (i) A novel combinatorial indexing strategy (referred to as ‘PerturbSci’) was developed for targeted enrichment and amplification of the sgRNA region that carries the same cellular barcode with the single-cell whole transcriptome (Fig 1a). A modified CROP-seq vector system8 was adopted in PerturbSci, enabling the direct capture of sgRNA sequences59 (Extended Data Fig 1). With extensive optimizations on primer designs and reaction conditions (Extended Data Fig 2), PerturbSci yields a high capture rate of sgRNA (i.e., up to 99.7%), comparable to previous approaches for single-cell profiling of pooled CRISPR screens4–9. Furthermore, built on an extensively improved single-cell RNA-seq by three-level combinatorial indexing (i.e., EasySci-RNA10), PerturbSci substantially reduced the library preparation costs for single-cell RNA profiling of pooled CRISPR screens (Fig 1b, Supplementary file 3). In addition, to maximize the gene knockdown efficacy, we used a multimeric fusion protein dCas9-KRAB-MeCP211, a highly potent transcriptional repressor that outperforms conventional dCas9 repressors. (ii) By integrating PerturbSci with 4-thiouridine (4sU) labeling method, PerturbSci-Kinetics exhibited an order of magnitude higher throughput than the previous single-cell metabolic profiling approaches (e.g., scEU-seq, sci-fate, scNT-seq)12–15(Fig 1a). Of note, we extensively optimized the cell fixation condition to reduce the cell loss rate during permeabilization and in-situ thiol (SH)-linked alkylation reaction16–22 (referred to as ‘chemical conversion’) (Extended Data Fig 3). Following 4sU labeling and chemical conversion, the nascent transcriptome and the whole transcriptome from the same cell can be distinguished by T to C conversion in reads mapping to mRNAs14. The kinetic rate of mRNA dynamics (e.g., synthesis and degradation) were then calculated as a multilayer readout for each genetic perturbation (Fig 1a, Methods). We further optimized the computational pipeline for nascent reads calling based on the established pipeline of sci-fate14, enabling the separation of single cell nascent transcriptomes with high accuracy (Extended Data Fig 4).
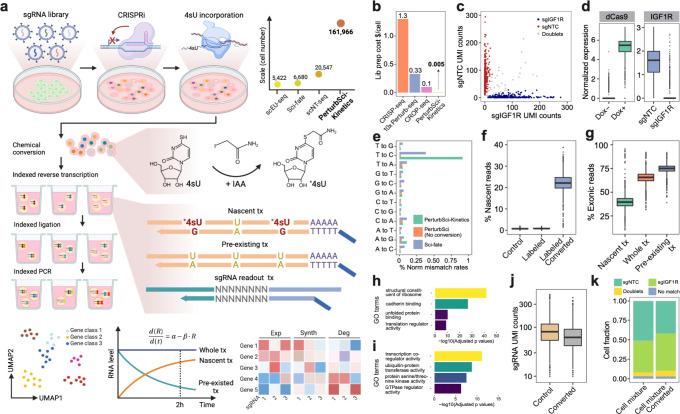
Fig. 1. PerturbSci-Kinetics enables joint profiling of transcriptome dynamics and high-throughput gene perturbations by pooled CRISPR screens.
a. Scheme of the experimental and computational strategy for PerturbSci-Kinetics. The dot plot on the upper right shows the number of cells profiled in this study for comparison with the published single-cell metabolic profiling datasets14,15,24. Scale, the highest number of cells profiled in a single experiment of each technique. IAA, iodoacetamide. *4sU, chemically modified 4sU. R, steady-state RNA level. α, RNA synthesis rate. β, RNA degradation rate. Exp, steady-state expression. Synth, synthesis rate. Deg, degradation rate. b. Bar plot showing the estimated library preparation cost for PeturbSci-Kinetics and other published techniques25,26 for single-cell transcriptome analysis coupled with CRISPR screens. c. Scatter plot showing the number of unique sgRNA transcripts detected per cell in the PerturbSci experiment for profiling cells transduced with sgNTC or sgIGF1R. d. The left box plot shows the normalized expression of dCas9-KRAB-MeCP2 in untreated or Dox-induced HEK293-idCas9 cells. The right box plot shows the normalized expression of IGF1R in Dox-induced HEK293-idCas9 cells transduced with sgNTC or sgIGF1R. Gene counts of each single cell were normalized by the total gene count, multiplied by 1e4 and then log-transformed. e. Bar plot showing the normalized percentage of all possible single base mismatches in reads from sci-fate (blue), and PerturbSci-Kinetics on chemically converted (green) or unconverted cells (orange). Normalized mismatch rates, the percentage of each type of mismatch in all sequencing bases. f. Box plot showing the fraction of recovered nascent reads in single-cell transcriptomes across conditions: no 4sU labeling + no chemical conversion, 4sU labeling + no chemical conversion, and 4sU labeling + chemical conversion. g. Box plot showing the ratio of reads mapped to exonic regions of the genome in nascent reads, pre-existing reads, and reads of the whole transcriptomes across single cells. h-i. Bar plots showing the significantly enriched Gene Ontology (GO) terms in the list of genes with low (h) or high (i) nascent reads ratio (Methods). j. Box plot showing the number of unique sgRNA transcripts detected per cell in cells with or without the chemical conversion. k. We performed PerturbSci-Kinetics experiment using converted/unconverted HEK293-idCas9 cells transduced with sgNTC/sgIGF1R. Stacked bar plot showing the fraction of converted/unconverted cells identified as sgNTC/sgIGF1R singlets, doublets, and cells with no sgRNA detected.
As a proof-of-concept, we first tested our approach in a mouse 3T3-L1-CRISPRi cell line transduced with a non-target control (NTC) sgRNA or sgRNA targeting a Fto gene (encoding an RNA demethylase). We found that sgRNA expression was detected in over 99% of all cells, with a median of 284 sgRNA UMI detected per cell in our optimal condition (i.e., 1uM gRNA primer + 50uM dT primer in reverse transcription) (Extended Data Fig 2f). We then generated a human HEK293 cell line with the inducible expression of dCas9-KRAB-MeCP211 (HEK293-idCas9) and tested the sgRNA capture efficiency using an NTC sgRNA and a sgRNA targeting the IGF1R gene (encoding insulin-like growth factor 1 receptor). The transductions of the NTC and target sgRNAs were performed independently, such that each cell received a unique perturbation. We then carried out a PerturbSci experiment on a 1:1 mixture of cells from these two conditions. We recovered the target sgRNA expression in 96.7% of cells, of which 95.2% were annotated as sgRNA singlets with a median of 81 sgRNA UMIs detected per cell (Fig 1c). Single-cell gene expression analysis confirmed the induction of dCas9 after Dox treatment, as well as the significantly decreased IGF1R expression in cells transduced with the target sgRNA (Fig 1d). Strongly reduced IGF1R mRNA and protein levels were further validated by RT-qPCR and flow cytometry (Extended Data Fig 5), validating the high knockdown efficiency of the system.
We next sought to validate the PerturbSci-Kinetics for capturing three-layer readout (i.e., whole transcriptome, nascent transcriptome, sgRNA identities) at the single-cell level. Following 4sU labeling (200uM for two hours), we mixed HEK293-idCas9 cells transduced with NTC or IGF1R sgRNA at a 1:1 ratio for fixation and chemical conversion. We observed a significant enrichment of T to C mismatches in mapped reads of the chemical conversion group, similar to our previous study14(Fig 1e). A median of 22.1% of newly synthesized reads was recovered in labeled and chemically converted cells, compared to only 0.8% in control groups (Fig 1f). Reassuringly, the proportion of reads mapped to exonic regions was significantly lower in newly synthesized reads compared with pre-existing reads (p-value < 1e-20, Tukey’s test after ANOVA) (Fig 1g). Indeed, genes with a higher fraction of nascent reads were significantly enriched in highly dynamic biological processes such as transcription coregulator activity (FDR = 5.7e-12) and protein kinase activity (FDR = 2.6e-08)23 (Fig 1h). By contrast, genes with a lower fraction of nascent reads were strongly enriched for processes essential for cell vitality, such as the structural constituent of ribosome (FDR = 1.5e-42), unfolded protein binding (FDR = 4.5e-11), and translation regulator activity (FDR = 8.2e-10) (Fig 1i). Notably, the chemical conversion step is fully compatible with sgRNA detection at single-cell resolution: we recovered sgRNAs from 97% of chemically converted cells (a median of 62 sgRNA UMIs/cell), 92.6% of which were annotated as sgRNA singlets (Fig 1j–k). These analyses demonstrate the capacity of PerturbSci-Kinetics to profile both transcriptome dynamics and the associated perturbation identity at the single-cell level.
To dissect the impact of key genetic regulators on transcriptome kinetics, we performed a PerturbSci-Kinetics experiment on HEK293-idCas9 cells transduced with a library of 699 sgRNAs, containing 15 NTC sgRNAs and sgRNAs targeting 228 genes involved in a variety of biological processes including mRNA transcription, processing, degradation, and others (Fig 2a, Supplementary Table 1). The cloning and lentiviral packaging were carried out in a pooled fashion similar to the previous report27 (Methods). We then infected the HEK293-idCas9 cell line with the sgRNA lentiviral library at a low multiplicity of infection (MOI) (2 repeats at MOI = 0.1 and 2 repeats at MOI = 0.2) to ensure most cells received only one sgRNA. After a 5-day puromycin selection to remove non-infected cells, we harvested a fraction of cells for bulk library preparation (‘day 0’ samples). The rest of the cells were treated with Doxycycline (Dox) to induce the dCas9-KRAB-MeCP2 expression for an additional seven days. We then introduced 4sU labeling (200uM for two hours) and harvested samples for both bulk and single-cell PerturbSci-Kinetics library preparation (‘day 7’ samples). The time window for the screening period was chosen to minimize the effect of population dropout28 (Methods).
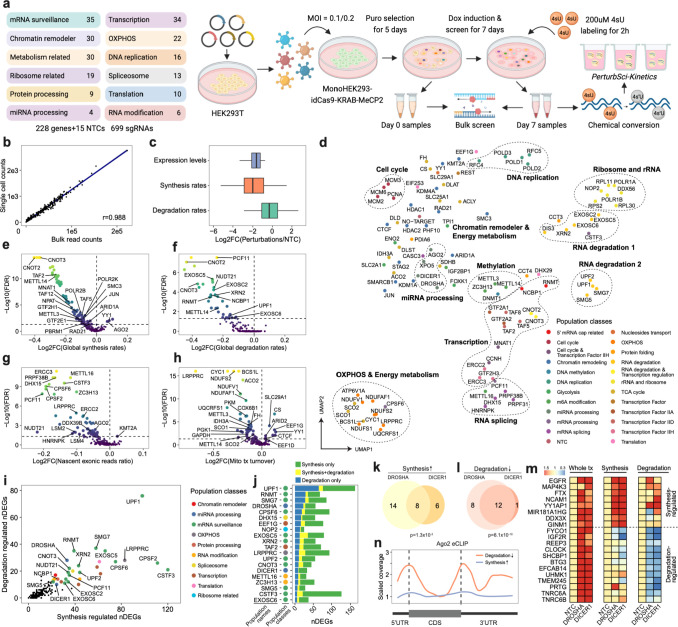
Fig 2. Characterizing the impact of genetic perturbations on gene-specific transcriptional and degradation dynamics with PerturbSci-Kinetics.
a. Scheme of the experimental design of the PerturbSci-Kinetics screen. The main steps are described in the text. b. The scatter plot shows the correlation between perturbation-associated cell count (PerturbSci-Kinetics) and sgRNA read counts (bulk screen). c. Box plot showing the log2 transformed fold change of gene expression, synthesis rates, and degradation rates of target genes across perturbations in comparison with the NTC cells. d. UMAP visualization of genetic perturbations profiled by PerturbSci-Kinetics. We aggregated single-cell transcriptomes in each perturbation, followed by dimension reduction using PCA and visualization using UMAP. Population classes, the functional categories of genes targeted in different perturbations. e-h. Scatter plots showing the extent and the significance of changes on the distributions of global synthesis (e), degradation (f), nascent exonic reads ratio (g), and mitochondrial transcriptome turnover (h) upon perturbations compared to NTC cells. The fold changes were calculated by dividing the median values of each perturbation with that of NTC cells and were log2 transformed. i. Scatter plot showing the number of synthesis/degradation-regulated DEGs from different perturbations. nDEGs: the number of DEGs. j. Top20 perturbations ordered by the number of degradation-regulated DEGs. Synthesis only, DEGs with significant changes in synthesis rates. Degradation only, DEGs with significant changes in degradation rates. Synthesis+degradation, DEGs with significant changes in both synthesis and degradation rates. k-l. Venn diagrams showing the number of merged DEGs with significantly enhanced synthesis (k) or impaired degradation (l) between DROSHA and DICER1. Based on statistical test results, merged DEGs of DROSHA and DICER1 perturbations were classified into synthesis-regulated genes (i.e., the upregulation of these genes was mainly driven by increased synthesis rates) and degradation-regulated genes (i.e., the upregulation of these genes were mainly driven by reduced degradation rates). Merged DEGs with p-value <= 0.05 on synthesis increase/degradation decrease in at least one perturbation were included in the diagram, in which genes with p-value < 0.1 on synthesis increase/degradation decrease in both perturbations were regarded as shared hits between two perturbations. m. Heatmaps showing the steady-state expression, synthesis and degradation rate changes of genes sharing the same regulatory mechanism upon DROSHA and DICER1 knockdown as shown in k, l. Tiles of each row were colored by fold changes of values of perturbations relative to NTC. n. Line plot showing the Ago2 binding patterns on the transcript regions of protein-coding genes in Figure 2n and 2o. The transcript regions of genes were assembled by merging all exons, and were divided into 5’UTR, coding sequence (CDS), and 3’UTR based on coordinates of the 5’ most start codon and the 3’ most stop codon. Single-base coverage of Ago2 eCLIP on each gene was calculated, binned, and scaled to 0–1. After merging and averaging scaled binned coverage of genes in the same group together, the lowest coverage value in the CDS was used to scale the averaged merged coverage again to visualize the Ago2/RISC binding pattern.
As expected, the induction of CRISPRi significantly changed the abundance of sgRNAs in the cell population, which is consistent between replicates and the previous study29 (Extended Data Fig 6a–b, Supplementary Table 2, 3). For example, the sgRNAs targeting genes involved in essential biological functions, such as DNA replication, ribosome assembly, and rRNA processing, were strongly depleted in the screen (Extended Data Fig 6c). Reassuringly, the sgRNA abundance recovered by PerturbSci-kinetics strongly correlated with the bulk library (Pearson correlation r = 0.988, p-value < 2.2e-16) (Fig 2b). After filtering out low-quality cells, we recovered 161,966 labeled cells, 88.1% of which had matched sgRNAs. 78% of these matched cells were annotated as sgRNA singlets (Extended Data Fig 7a). Despite the relatively low (17.9%) duplication rate of sequencing, we obtained a median of 2,155 UMIs per cell. Most (698 out of 699) sgRNAs were recovered, with a median of 28 sgRNA UMIs detected per cell. We further filtered out sgRNAs with low knockdown efficiencies (<= 40% expression reduction of target genes compared with NTC) (Extended Data Fig 7b–e). Finally, 98,315 cells were retained for downstream analysis, corresponding to a median of 484 cells per gene perturbation with a median of 67.7% knockdown efficiency of target genes (Fig 2c). To further validate the impact of perturbations, we aggregated single-cell transcriptomes and generated a ‘pseudo-cell’ for each targeted gene, followed by PCA dimension reduction and UMAP visualization30. Indeed, perturbations targeting paralogous genes (e.g., EXOSC5 and EXOSC6; CNOT2 and CNOT3) or related biological processes (e.g., RNA degradation, RNA splicing, oxidative phosphorylation (OXPHOS) and energy metabolism) were readily clustered together in the low dimension space (Fig 2d).
Taking advantage of PerturbSci-Kinetics for uniquely capturing multiple layers of information, we performed differentially-expressed gene (DEG) analysis (Supplementary Table 4) and quantified gene-specific synthesis and degradation rates of DEGs in each perturbation based on an ordinary differential equation31 (Methods). As a quality control, we first examined the kinetics of genes targeted by CRISPRi, which were known to function through transcriptional repression32,33. Indeed, these genes exhibited strongly reduced synthesis rates while their degradation rates were only mildly affected (Fig 2c). We then investigated the impact of genetic perturbations on the global transcriptome dynamics (i.e., synthesis, splicing and degradation) (Methods, Supplementary Table 5, 6). As expected, the knockdown of genes involved in transcription initiation (e.g., GTF2E1, TAF2, MED21, and MNAT1), mRNA synthesis (e.g., POLR2B and POLR2K), and chromatin remodeling (e.g., SMC3, RAD21, CTCF, ARID1A) significantly downregulated the global synthesis rates but not the degradation rates (Fig 2e–f). In contrast, perturbations targeting components of critical biological processes such as DNA replication (e.g., POLA2, POLD1), ribosome synthesis and rRNA processing (e.g., POLR1A, POLR1B, RPL11, RPS15A), mRNA and protein processing (e.g., CNOT2, CNOT3, CCT3, CCT4) substantially reduced both RNA synthesis and degradation globally, indicating a compensatory mechanism for maintaining overall transcriptome homeostasis (Fig 2e–f, Extended Data Fig 8a, b). Furthermore, we observed significantly reduced fractions of exonic reads in nascent transcripts, an indicator of dysregulated splicing dynamics, following perturbations of genes involved in the main steps of RNA processing, including 5’ capping (e.g., NCBP1), RNA splicing (e.g., LSM2, LSM4, PRPF38B, HNRNPK), and 3’ cleavage/polyadenylation (e.g., CPSF2, CPSF6, NUDT21, CSTF3) (Fig 2g, Supplementary Table 7). In addition, the knockdown of genes involved in OXPHOS & energy metabolism (e.g., GAPDH, NDUFS2, ACO2) also significantly reduced the exonic reads ratio in nascent reads (Fig 2g, Extended Data Fig 8c), potentially due to the fact that the mRNA processing is highly energy-dependent34,35,36.
We next sought to investigate the regulators of mitochondrial RNA dynamics by quantifying the ratio of nascent/total read counts (referred to as “turnover rate”) mapped to mitochondrial genes (Methods). Notably, we observed a significantly downregulated turnover rate of mitochondrial-specific RNA following the perturbation of multiple metabolism-related genes (e.g., GAPDH, FH, PKM involved in glycolysis, ACO2, and IDH3A involved in the TCA cycle, NDUFS2 and COX6B1 involved in oxidative phosphorylation) (Fig 2h, Extended Data Fig 8d). Furthermore, the knockdown of LRPPRC introduced the most substantial defect in the mitochondrial turnover and the expression levels of all mitochondrial protein-coding genes (Fig 2h, Extended Data Fig 9a). Intriguingly, 5 of 13 mitochondrial protein-coding genes, including MT-CO1, MT-ATP8, MT-ND4, MT-CYB, and MT-ATP6, were regulated by both decreased transcription and increased degradation (Extended Data Fig 9a, Supplementary Table 9). This result was supported by a previous study37 (Extended Data Fig 9b) and was also consistent with the known functions of LRPPRC in regulating the life cycles of mitochondrial RNA from synthesis to degradation38–40. For comparison, the nuclear-encoded differentially expressed genes (DEGs) following LRPPRC knockdown were significantly changed mostly at the transcription level (39 out of 48 genes, Extended Data Fig 9c). Upon closer inspection of promoter regions of these synthesis-regulated genes, we observed a strong enrichment of ATF4 and CEBPG binding motifs, suggesting their potential roles as downstream transcriptional regulators of LRPPRC. Indeed, ATF4 and CEGPG have been reported as core transcriptional activators involved in stress sensing41,and both genes were substantially upregulated in LRPPRC knockdown cells (Extended Data Fig 9d–e).
Extending the above analysis, we examined the gene-specific synthesis and degradation regulation across all perturbations (Supplementary Table 10). Among all 14,618 perturbation-DEG pairs identified in the study, 22.9% of them exhibited rate changes, in which 15.1% showed significant synthesis rate changes only, 3.6% showed degradation rate changes only, and 4.2% showed both changes, suggesting complex mechanisms regulating gene expression upon perturbations42 (Extended Data Fig 10). As expected, most degradation-regulated DEGs were associated with perturbations on mRNA surveillance/processing (e.g., UPF1, UPF2, SMG5, SMG7 in nonsense-mediated mRNA decay pathway; EXOSC2, EXOSC5, EXOSC6 in RNA exosome; CSTF3, CPSF2, CPSF6, NUDT21, XRN2 for 3’ polyadenylation; RNMT, NCBP1 related to 5’ RNA capping) (Fig 2i–j). For example, the knockdown of two critical regulators in the microRNA (miRNA) pathway43 (i.e., DROSHA and DICER144,45, Extended Data Fig 11a) resulted in a group of highly overlapped DEGs(Extended Data Fig 11b). These DEGs were upregulated through decreased degradation (e.g., miRNA-mediated silencing complex (RISC) components: TNRC6A and TNRC6B46) or increased transcription (e.g., miRNA host genes: MIR181A1HG47, FTX48; genes involved in miRNA biogenesis: DDX3X49) (Fig 2k–m, Extended Data Fig 11c, Supplementary Table 11). To explore the underlying regulatory mechanisms, we examined the gene-specific binding patterns of Ago2, one of the core components in RISC for targeted mRNA binding and degradation50. Indeed, Ago2 binding was strongly enriched in the 5’ and 3’ untranslated regions (UTR) of the genes with reduced degradation, but not in genes with upregulated synthesis (Fig 2n), consistent with prior reports that miRNA induces targeted RNA degradation and translation repression mainly through binding to the UTR44,51. The analysis further demonstrates the unique capacity of PerturbSci-Kinetics for deciphering the regulatory mechanisms (degradation vs. transcription) involved in gene expression changes upon genetic perturbations.
Lastly, to our knowledge, the studies described here provided the first method to quantitatively characterize the genome-wide mRNA kinetic rates (e.g., synthesis and degradation rates) across hundreds of genetic perturbations in a single experiment. We included the step-by-step protocols and the data processing pipeline as supplementary files (Supplementary file 1–4) to facilitate the broad applications of the technique. Our analysis illustrates the advantages of PerturbSci-Kinetics over conventional assays that solely profile gene expression changes. By capturing three layers of readout (e.g., whole, nascent transcriptome, and sgRNA identify) at the single-cell resolution, PerturbSci-Kinetics uniquely enables us to dissect the critical regulators of gene-specific transcription, processing, and degradation in a massive-parallel manner. Finally, PerturbSci-Kinetics is built on the recently developed EasySci-RNA10 and can be readily scaled up to profiling genome-wide perturbations (e.g., 10,000s genes or cis-regulatory elements) across millions of single cells, thus enabling the systematic characterization of cell-type-specific gene regulatory network at unprecedented scale and resolution.
Materials and Methods:
Cell culture
The 3T3-L1-CRISPRi cell line was obtained from the Tissue Culture facility at the University of California, Berkeley. The HEK293 cell line was a gift from the Scott Keeney Lab at Memorial Sloan Kettering Cancer Center. The HEK293T cell line was obtained from ATCC (CRL-3216). All cells were maintained at 37 °C and 5% CO2 in high glucose DMEM medium supplemented with L-Glutamine and Sodium Pyruvate (Gibco 11995065) and 10% Fetal Bovine Serum (FBS; Sigma F4135). When generating a monoclonal cell line, the medium was supplemented with 1% Penicillin-Streptomycin (Gibco 15140163). In the screening experiment, sgRNA-transduced HEK293-idCas9 cells were cultured in high glucose DMEM medium supplemented with L-Glutamine (Gibco 11965092) and 10% FBS, following the induction of dCas9-KRAB-MeCP2 expression by 1ug/ml Dox (Sigma D5207),
Generation of monoclonal HEK293-idCas9 cell line
To generate HEK293 with Dox-inducible dCas9-KRAB-MeCP2 expression, the lentiviral plasmid Lenti-idCas9-KRAB-MeCP2-T2A-mCherry-Neo was constructed. A dCas9-KRAB-MeCP2-T2A insert was amplified from dCas9-KRAB-MeCP2 (Addgene #110821). A T2A-mCherry Gblock was synthesized by IDT. Gibson Assembly reaction (NEB E2611S) was performed at 50 °C with a mixture of Bsp119I-digested Lenti-Neo-iCas9 (Thermo FD0124; Addgene #85400), dCas9-KRAB-MeCP2-T2A amplicon, T2A-mCherry Gblock for 60 minutes to construct a dCas9-KRAB-MeCP2-T2A-mCherry plasmid. The reaction product was transformed into NEBstable competent cells (NEB C3040H), and colonies were inoculated and amplified in LB medium (Gibco 10855001) with 50ug/ml Sodium Ampicillin (Sigma A8351) at 37 °C overnight.
After plasmid extraction (QIAGEN No.27106) and sequencing validation, the plasmid was co-transfected with psPAX2 (Addgene #12260) and pMD2.G (Addgene #12259) into low-passage HEK293T cells in a 10cm dish using Polyjet (SignaGen SL100688) for 24 hours. Cells were gently washed twice with PBS, then cultured in a medium with 10mM Sodium Butyrate (Sigma TR-1008-G) for another 24 hours. The supernatant was collected, and cell debris was cleared by spinning down (5min, 1000×g) and passed through a 0.45 μm filter. The lentivirus was concentrated 10x by the Lenti-X concentrator (TaKaRa 631231), and the virus suspension was flash frozen by Liquid Nitrogen and was stored at −80 °C.
The lentivirus titer was determined by examining the ratio of mCherry+ cells after 24 hours of transduction and 48 hours of Dox induction. Polybrene (Sigma TR-1003) at a final concentration of 8ug/ml was used to enhance the transduction efficiency. Then HEK293 cells were counted and transduced with lentivirus at MOI = 0.2 for 48 hours. Cells were treated with Dox for 48 hours, and the top 10% of cells with the strongest mCherry fluorescence were sorted to each well of a 96-well plate containing 100ul medium. After a 3-week expansion, monoclonal cells that survived were transferred to larger dishes for further expansion. We picked the clone with inducible homogeneous strong mCherry expression and normal morphology for the following experiment.
Gene Knockdown and efficacy examination
To simplify the lentiviral titer measurement, CROP-seq-opti-Puro-T2A-GFP was assembled by adding a T2A-GFP downstream of Puromycin resistant protein coding sequence on the CROP-seq-opti plasmid (Addgene #106280). Flanking MluI and CsiI digestion sites were added to the GFP Gblock (IDT) by PCR. Both amplicon and CROP-seq-opti vector were digested using MluI (Thermo, FD0564) and CsiI (Thermo, FD2114) at 37 °C for 30 minutes, and were ligated at room temperature for 20 minutes using the Blunt/TA Ligase Master Mix (NEB M0367S). Transformation, clone amplification, and sequencing validation were done as stated above.
Oligos corresponding to individual guides for ligation were ordered as standard DNA oligos from IDT with the following design:
Plus strand: 5’-CACCG[20bp sgRNA plus strand sequence]-3’
Minus strand: 5’-AAAC[20bp sgRNA minus strand sequence]C-3’
Oligos were reconstituted into 100uM and were mixed and phosphorylated using T4 PNK (NEB M0201S) by incubating at 37 °C for 30 minutes. The reaction was heated at 95 °C for 5 minutes and then ramped down to 25 °C by −0.1 °C/second to anneal oligos into a double-stranded duplex. The CROP-seq-opti-Puro-T2A-GFP was digested by Esp3I (NEB R0734L) at 37 °C for 30 minutes, then the linearized backbone and the annealed duplex were ligated at room temperature for 20 minutes using the Blunt/TA Ligase Master Mix (NEB M0367S). Transformation, clone amplification, sequencing validation, lentivirus generation, and titer measurement were done as stated above.
For the mouse 3T3-L1-CRISPRi cells, they were counted and incubated with lentivirus inserted with either non-target control (NTC) sgRNA or sgRNA targeting a Fto gene, and 8ug/ml of Polybrene. For the human HEK293-idCas9 cells, they were counted and incubated with NTC sgRNA or sgRNA targeting an IGF1R gene, and 8ug/ml of Polybrene. Transduction was then performed at MOI = 0.2 for 48 hours. Based on the results of our puromycin titration experiments, sgRNA-transduced 3T3-L1-CRISPRi cells were selected by 2.5ug/ml Puromycin for 2 days and 2ug/ml Puromycin for 3 days, and sgRNA-transduced HEK293-idCas9 cells were selected by 1.5ug/ml Puromycin for 3 days and 1ug/ml Puromycin for 2 days.
As dCas9-BFP-KRAB was constitutively expressed in 3T3-L1-CRISPRi cells, the target gene started being silenced once sgRNA lentivirus was introduced. For HEK293-idCas9 cells, Dox treatment for a minimum of 72 hours was required before examining the knockdown effect.
For RT-qPCR validation, primers targeting IGF1R were selected from PrimerBank (https://pga.mgh.harvard.edu/primerbank/) and were synthesized from IDT. Total RNA in 1e6 cells of each sample was extracted using the RNeasy Mini kit (QIAGEN 74104) and the concentration was measured by Nanodrop. 1ug total RNA was then reverse-transcribed into the first strand cDNA by SuperScript VILO Master Mix (Thermo 11755050). PowerTrack SYBR Green Master Mix (Thermo A46109) was used for RT-qPCR following the manufacturer’s instructions.
For flow cytometry validation, 1e6 cells of each sample were harvested and resuspended in 100ul of PBS-0.1% sodium azide-2% FBS. BV421 Mouse Anti-Human CD221 (BD 565966) and BV421 Mouse IgG1 k Isotype Control (BD 562438) at the final concentration of 10 ug/ml were added, and reactions were incubated at 4 °C in the dark with rotation for 30 minutes. Cells were then washed twice using PBS-0.1% sodium azide-2% FBS, and fluorescence signals were recorded.
Construction of pooled sgRNA library
Genes of interest were selected manually, considering their functions and expression levels in HEK293 cells. The sgRNA sequences targeting genes of interest with the best performances were obtained from an established optimized sgRNA library (only sgRNA set A is considered)29. Finally, 684 sgRNAs targeting 228 genes (3 sgRNAs/gene) and 15 non-targeting controls were included in the present study.
The single-stranded sgRNA library was synthesized in a pooled manner by IDT in the following format: 5’-GGCTTTATATATCTTGTGGAAAGGACGAAACACCG[20bp sgRNA plus strand sequence]GTTTAAGAGCTATGCTGGAAACAGCATAGCAAGTT-3’
100ng of oligo pool was amplified by PCR using primers targeting 5’ homology arm (HA) and 3’ HA with limited cycles (x12) to avoid introducing amplification biases. The PCR product was purified, and double-stranded library amplicons were extracted by DNA electrophoresis and gel extraction. Then the insert was cloned into Esp3I-digested CROP-seq-opti-Puro-T2A-GFP by Gibson Assembly (50 °C for 60 minutes). In parallel, a control Gibson Assembly reaction containing only the backbone was set. Both reactions were cleaned up by 0.75x AMPURE beads (Beckman Coulter A63882) and eluted in 5uL EB buffer (QIAGEN 19086), then were transformed into Endura Electrocompetent Cells (Lucigen, 602422) by electroporation (Gene Pulser Xcell Electroporation System, Bio-Rad, 1652662). After 1 hour of recovery at 250rpm, 37 °C, each reaction was spread onto an in-house 245 mm Square agarose plate (Corning, 431111) with 100ug/ml of Carbenicillin (Thermo, 10177012) and was then grown at 32 °C for 13 hours to minimize potential recombination and growth biases. All colonies from each reaction were scraped from the plate and the CROP-seq-opti-Puro-T2A-GFP-sgRNA plasmid library was extracted using ZymoPURE II Plasmid Midiprep Kit (Zymo, D4200). The lentiviral library was generated as stated above with extended virus production time. The step-by-step protocol is included in the supplementary materials.
The pooled PerturbSci-Kinetics screen experiment
For each replicate, 7e6 uninduced HEK293-idCas9 cells were seeded. After 12 hours, two replicates were transduced at MOI=0.1 (1000x coverage/sgRNA) and another two replicates were transduced at MOI=0.2 (2000x coverage/sgRNA) with 8ug/ml of Polybrene for 24 hours. Then we replaced the culture medium with the virus-free medium and culture cells for another 24 hours. Transduced cells were selected by 1.5ug/ml of Puromycin for 3 days and 1ug/ml of Puromycin for 2 days. During the selection, we passed cells every 2 or 3 days to ensure at least 1000x coverage. At the end of the drug selection, we harvested 1.4e6 cells in each replicate (2000x coverage/sgRNA) as day0 samples of the bulk screen and pellet down at 500×g, 4 °C for 5 minutes. Cell pellets were stored at −80 °C for genomic DNA extraction later. Then the dCas9-KRAB-MeCP2 expression was induced by adding Dox at the final concentration of 1ug/ml, and L-glutamine+, sodium pyruvate-, high glucose DMEM was used to sensitize cells to perturbations on energy metabolism genes. Cells were cultured in this condition for additional 7 days and were passed every other day with 4000x coverage/sgRNA. On day7, 6ml of the original media from each plate was mixed with 6uL of 200mM 4sU (Sigma T4509-25MG) dissolved in DMSO (VWR 97063-136) and was put back for nascent RNA metabolic labeling. After 2 hours of treatment, 1.4e6 cells in each replicate were harvested as day7 samples of the bulk screen, and the rest of the cells were fixed and stored for single-cell PerturbSci-Kinetics profiling (see the next section).
Genomic DNA of bulk screen samples was extracted using Quick-DNA Miniprep Plus Kit (Zymo, D4068T) following the manufacturer’s instructions and quantified by Nanodrop. All genomic DNA was used for PCR to ensure coverage. The primer targeting the U6 promoter region with P5-i5-Read1 overhang and the primer targeting the sgRNA scaffold region with P7-i7-Read2 overhang was used for generating the bulk screen libraries for sequencing.
Library preparation for the PerturbSci-Kinetics
After trypsinization, cells in each 10cm dish were collected into a 15ml falcon tube and kept on ice. Cells were spun down at 300×g for 5 minutes (4 °C) and washed once in 3ml ice-cold PBS. Cells were fixed with 5ml ice-cold 4% Paraformaldehyde (PFA) in PBS (Santa Cruz Biotechnology sc-281692) for 15 minutes on ice. PFA was then quenched by adding 250ul 2.5M Glycine (Sigma 50046-50G), and cells were pelleted at 500×g for 5 minutes (4 °C). Fixed cells were washed once with 1ml PBSR (PBS, 0.% SUPERase In (Thermo AM2696), and 10mM dithiothreitol (DTT; Thermo R0861)), and were then resuspended, permeabilized, and further fixed in 1ml PBSR-triton-BS3 (PBS, 0.1% SUPERase In, 0.2% Triton-X100 (Sigma X100-500ML), 2mM bis(sulfosuccinimidyl)suberate (BS3; Thermo, PG82083), 10mM DTT) for 5 minutes. Additional 4ml of PBS-BS3 (PBS, 2mM BS3, 10mM DTT) was then added to dilute Triton-X100 while keeping the concentration of BS3, and cells were incubated on ice for 15 minutes. Cells were pelleted at 500×g, 4 °C for 5 minutes and resuspended in 500ul nuclease-free water (Corning 46-000-CM) supplemented with 0.1% SUPERase In and 10mM DTT. 3ml of 0.05N HCl (Fisher Chemical, SA54-1) was added for further permeabilization. After 3 minutes of incubation on ice, 3.5ml Tris-HCl, pH 8.0 (Thermo 15568025), and 35ul of 10% Triton X-100 were added to each tube to neutralize the HCl. After spinning down at 4 °C, 500×g for 5 minutes, cells were finally resuspended in 400ul PSB-DTT at the concentration of ~2e6 cells/100ul (PBS, 1% SUPERase In, 1% BSA (NEB B90000S), 1mM DTT), mixed with 10% DMSO, and were slow-frozen and stored in −80 °C.
The chemical conversion was performed before the library preparation. Cells were thawed with shaking in the 37 °C water bath and spun down, then were washed once with 400ul PSB without DTT. Next, cells were resuspended in 100ul PSB, mixed with 40ul Sodium Phosphate buffer (PH 8.0, 500mM), 40ul IAA (100mM, Sigma I1149-5G), 20ul nuclease-free water, and 200ul DMSO with the order. The reaction was incubated at 50 °C for 15 minutes and was quenched by adding 8ul 1M DTT. Then cells were washed with PBS and were filtered through a 20um strainer (Pluriselect 43-10020-60). Cells were finally resuspended in 100ul PSB.
For library preparation, a step-by-step protocol is included as a supplementary file.
Reads processing
For bulk screen libraries, bcl files were demultiplexed into fastq files based on index 7 barcodes. Reads for each sample were further extracted by index 5 barcode matching. Then every read pair was matched against two constant sequences (Read1: 11–25bp, Read2: 11–25bp) to remove reads generated from the PCR by-product. For all matching steps, a maximum of 1 mismatch was allowed. Finally, sgRNA sequences were extracted from filtered read pairs (at 26–45bp of R1), assigned to sgRNA identities with no mismatch allowed, and read counts matrices at sgRNA and gene levels were quantified.
For PerturbSci-Kinetics transcriptome reads processing and whole-transcriptome/nascent transcriptome gene counting, the pipeline was developed based on EasySci10 and Sci-fate14 with minor modifications. After demultiplexing on index 7, Read1 were matched against a constant sequence on the sgRNA capture primer to remove unspecific priming, and cell barcodes and UMI sequences sequenced in Read1 were added to the headers of the fastq files of Read2, which were retained for further processing. After potential polyA sequences and low-quality bases were trimmed from Read2 by Trim Galore61, reads were aligned to a customized reference genome consisting of a complete hg38 reference genome and the dCas9-KRAB-MeCP2 sequence from Lenti-idCas9-KRAB-MECP2-T2A-mCherry-Neo using STAR62. Unmapped reads and reads with mapping score < 30 were filtered by samtools63. Then deduplication at the single-cell level was performed based on the UMI sequences and the alignment location, and retained reads were split into SAM files per cell. These single-cell sam files were converted into alignment tsv files using the sam2tsv function in jvarkit64. Only reads with FLAG values of 0 or 16 and high-quality mismatches with QUAL scores > 45 and CIGAR of M in them were maintained. Mutations were further filtered against background SNPs called by VarScan using our in-house EasySci data on HEK293 cells. Reads in which at least 30% of mutations were T to C mismatches were identified as nascent reads, and the list of reads were extracted from single-cell whole transcriptome sam files by Picard65. Finally, single-cell whole transcriptome gene x cell count matrix and nascent transcriptome gene x cell count matrix were constructed by assigning reads to genes if the aligned coordinates overlapped with the gene locations on the genome. At the same time, single cell exonic/intronic read numbers were also counted by checking whether reads were mapped to the exonic or the intronic regions of genes. To quantify dCas9-KRAB-MECP2 expression, a customized gtf file consisting of the complete hg38 genomic annotations and additional annotations for dCas9 was used in this step.
Read1 and read2 of PerturbSci-Kinetics sgRNA libraries were matched against constant sequences respectively, allowing a maximum of 1 mismatch. For each filtered read pair, cell barcode, sgRNA sequence, and UMI were extracted from designed positions. Extracted sgRNA sequences with a maximum of 1 mismatch from the sgRNA library were accepted and corrected, and the corresponding UMI was used for deduplication. De-duplication was performed by collapsing identical UMI sequences of each individual corrected sgRNA under a unique cell barcode. Cells with overall sgRNA UMI counts higher than 10 were maintained and the sgRNA x cell count matrix was constructed.
Bulk screen sgRNA counts analysis
For each bulk screen library, read counts of sgRNAs were normalized first by the sum of total counts to remove the biases from sequencing depth, and then the abundance of each sgRNA relative to the sum of sgNTC was calculated, assuming the NTC cells had no selection pressure during the screen. The Pearson correlations across replicates were calculated based on the relative abundances. Then the fraction changes (After vs. before the CRISPRi induction) of sgRNAs were calculated within each replicate, and the mean fold changes across replicates were log2 transformed. The raw counts of another external bulk CRISPRi screen dataset29 was processed as stated above and the log2 mean relative abundance was compared to the current study.
sgRNA singlets identification and off-target sgRNA removal
In the cell mixture experiments, cells with at least 200 whole transcriptome UMIs and 200 genes detected, and unannotated reads ratio < 40% were kept. If the count of the most abundant sgRNA was at least 3-fold of the second most abundant sgRNA within this single cell, then this cell was identified as a sgRNA singlet.
In the screen dataset, cells with at least 300 whole transcriptome UMIs and 200 genes detected, and unannotated reads ratio < 40% were kept. sgRNA identities of cells were assigned and doublets were removed based on the following criteria: the cell is assigned to a single sgRNA if the most abundant sgRNA in the cell took >= 60% of total sgRNA counts and was at least 3-fold of the second most abundant sgRNA. Then whole transcriptomes and sgRNA profiles of single cells were integrated with the matched nascent transcriptomes.
Target genes with the number of cells perturbed >= 50 were kept for further filtering. The knockdown efficiency was calculated at the individual sgRNA level to remove potential off-target or inefficient sgRNAs: whole transcriptome counts of all cells receiving the same sgRNA were merged, normalized by the total counts, and scaled using 1e6 as the scale factor, then the fold changes of the target gene expressions were calculated by comparing the normalized expression levels between corresponding perturbations and NTC. sgRNAs with >= 40% of target gene expression reduction relative to NTC were regarded as “effective sgRNAs”, and singlets receiving these sgRNAs were kept as “on-target cells”. Downstream analyses were done at the target gene level by analyzing all cells receiving different sgRNAs targeting the same gene together.
Gene Ontology analysis of genes with high or low nascent reads ratio
To validate the specificity of 4sU labeling and the computational identification of nascent reads, we identified features of gene groups with different turnover rates. Single cells were split into nascent transcriptomes and pre-existing transcriptomes, and were loaded into Seurat30. Nascent transcriptomes and pre-existing transcriptomes were normalized, scaled independently, and DEGs between the two groups were identified by FindMarkers function30 with default parameters. Then GO enrichment analyses were performed using ClusterProfiler66 on upregulated genes (genes with significantly higher fraction of nascent counts, FDR of 0.05) and downregulated genes (genes with significantly lower fraction of nascent counts, FDR of 0.05) respectively.
UMAP embedding on pseudo-cells
The count matrix of the “on-target” cells described above was loaded into Seurat30, and DEGs of each perturbation (compared to NTC) were retrieved by FindMarkers function30 with default parameters. Cells from perturbations with over one DEGs (by FindMarkers function30) were selected. We also included cells from genetic perturbations involved in similar pathways of the top perturbations. The fold changes of the normalized gene expression between perturbations and NTC were calculated, and were binned based on the gene-specific expression levels in NTC. The top 3% of genes showing the highest fold changes within each bin were selected and merged as features for Principal Component Analysis (PCA). The top 9 PCs were used as input for Uniform Manifold Approximation and Projection (UMAP) embedding (min.dist = 0.3, n.neighbors = 10).
Differential expression analysis
Pairwise differential expression analyses between each perturbation and NTC cells were performed by the differentialGeneTest() function of Monocle 267. To identify DEGs with rate changes, we selected significant hits (FDR of 5%, likelihood) with a >= 1.5-fold expression difference and counts per million (CPM) >= 5 in at least one of the tested cell pairs. To showcase LRPPRC and miRNA pathway perturbations, more stringent criteria were used to obtain DEGs with high confidence: significant hits (FDR of 5%, likelihood) with a >= 1.5-fold expression difference and CPM >= 50 in at least one of the tested cell pairs were kept.
Synthesis and degradation rates calculation
After the induction of CRISPRi for 7 days, we assumed new transcriptomic steady states had been established at the perturbation level before the 4sU labeling, and the labeling didn’t disturb these new transcriptomic steady states. The following RNA dynamics differential equation is used for synthesis and degradation rates calculation similar to the previous study31:
In which R is the mRNA abundance of each gene, α is the synthesis rate of this gene, and β is the degradation rate of this gene. Since the RNA synthesis follows the zero-order kinetics and RNA degradation follows the first-order kinetics in cells, is determined by α and R · β.
As steady states had been established, the mRNA level of each gene didn’t change. We can get:
Under the assumption that the labeling efficiency was 100%, all nascent RNA were labeled during the 4sU incubation, and pre-existing RNA would only degrade. So, for nascent RNA (Rn), Rn(t = 0) = 0 and αn = α. For pre-existing RNA (Rp), Rp(t = 0) = R = α/β and αp = 0. Based on these boundary conditions, we could further solve the differential equation above on nascent RNA and pre-existing RNA of each gene.
As PerturbSci-Kinetics directly measured whole transcriptome gene expression levels and nascent transcriptome gene expression levels, pre-existing gene expression levels could be obtained by subtracting nascent transcriptome expressions from the whole transcriptome expressions. As cells were labeled by 4sU for 2 hours (t = 2), and β of each gene could be calculated based on the equations above.
Due to the shallow sequencing and the sparsity of the single cell expression data, synthesis and degradation rates of DEGs were calculated at the pseudo-cell level. We aggregated the expression profiles of all cells with the same target gene knockdown, normalized the expressions of genes by the sum of gene counts, and scaled the size of the total counts to 1e6. Synthesis and degradation rates of DEGs in the corresponding perturbed pseudo-cell were calculated as stated above. DEGs with only nascent counts or degradation counts were excluded from further examination since their rates couldn’t be estimated.
To examine the significance of synthesis and degradation rate changes upon perturbation, regarding the different cell sizes across different perturbations and NTC, which could affect the robustness of rate calculation, randomization tests were adopted. Only perturbations with cell number >= 50 were examined. For each DEG belonging to each perturbation, background distributions of the synthesis and degradation rate were generated: a subset of cells with the same size as the corresponding perturbed cells was randomly sampled from a mixed pool consisting of corresponding perturbed cells and NTC cells, then these cells were aggregated into a background pseudo-cell, and synthesis and degradation rates of the gene for testing were calculated as stated above, and the process was repeated for 500 times. Rates = 0 were assigned if only nascent counts or degradation counts were sampled during the process (referred to as invalid samplings), but only genes with less than 50 (10%) “invalid samplings” were kept for p-value calculation. The two-sided empirical p-values for the synthesis and degradation rate changes were calculated respectively by examining the occurrence of extreme values in background distributions compared to the rates from perturbed pseudo-cell. Rate changes with p-value <= 0.05 were regarded as significant, and the directions of the rate changes were determined by comparing the rates from the perturbed pseudo-cell with the background mean values. The fold changes of rates for each significant gene were calculated as follows: only NTC cells were sampled at the same size as perturbed cells and aggregated, and the background rates were calculated at the pseudo-cell level. After resampling for 200 times, these gene-specific rates were averaged. Fold changes of the rates = rates in perturbed pseudo-cell / mean rates from the NTC-only background.
Global changes of key statistics upon perturbations
For global synthesis and degradation rate changes, considering the noise from lowly-expressed genes, we selected top1000 highly-expressed genes from NTC cells, then calculated their synthesis rates and degradation rates in NTC cells and all perturbations with cell number >= 50. KS tests were performed to compare rate distributions between each perturbation and NTC cells.
During the reads processing, the number of reads aligned to exonic/intronic regions were counted at the single cell level. Then the distributions of exonic reads percentage in nascent reads from single cells with the same target gene knockdown and NTC cells were compared using the KS tests to identify genes affecting RNA processing.
The ratio of nascent mitochondrial read counts to total mitochondrial read counts was calculated in each single cell, and the distributions of the ratio from single cells with the same target gene knockdown and NTC cells were compared using the KS tests to identify the master regulator of mitochondrial mRNA dynamics.
In all global statistics examinations, the p-values were corrected from multiple comparisons, and comparisons with FDR <= 0.05 were considered as significant. The median value from each perturbation and NTC cells were compared to determine the direction of significant changes.
Ago2 eCLIP coverage analysis
To identify the potential different RISC binding patterns between synthesis/degradation-regulated DEGs in DROSHA and DICER1 perturbations, we reprocessed the raw data of Ago2 eCLIP obtained from Hela cells (two replicates, SRR7240709 and SRR7240710) from Zhang, K et, al68. Potential adapters at 3’ ends of reads were trimmed by Cutadapt69, and the first 6-base UMI were extracted and attached to headers of the reads. After STAR alignment62 and samtools filtering63, only uniquely aligned reads were kept and deduplication was performed based on the UMI and mapping coordinates using UMI-tools70. Then bam files were transformed to the single-base coverage by BEDtools71. The transcript regions of genes-of-interest were reconstructed based on the hg38 genome annotation gtf file from GENCODE. Briefly, for each gene, the exonic regions were extracted and were redivided into 5’UTR, CDS, and 3’UTR by the 5’most start codon and the 3’most stop codon annotated in the gtf. The Ago2 binding coverages of these designated regions were obtained by intersection and were binned. A small background (0.1/base) was added for smoothing. The gene-specific signal in each bin was normalized by the number of bases in each bin, and the binned coverage of each gene was scaled to be within 0–1. After aggregating scaled coverages of synthesis/degradation-regulated genes respectively, the second scaling was performed to visualize the relative enrichment of Ago2 binding at UTR compared to the CDS: fold changes of the scaled binned coverage relative to the lowest coverage value in the CDS along the aggregated transcript were calculated.
Supplementary Material
Acknowledgments:
We thank all members of the Cao lab for helpful discussions and feedback. We thank Dr. R. Satija (New York Genome Center) for insightful feedback related to this work. We thank the Tissue Culture facility of the University of California, Berkeley for the 3T3 cell line, and the Scott Keeney Lab at Memorial Sloan Kettering Cancer Center for the HEK293 cell line. We thank members of the Rockefeller University Flow Cytometry Resource Center and the Rockefeller University Genomics Resource Center for their extensive help with FACS sorting and sequencing experiments. We also thank members of the Information Technology and High-Performance Computing team at Rockefeller University, especially J. Banfelder and B. Jayaraman for the great support. We acknowledge that the research resulting in this publication was supported, in part, by The G. Harold and Leila Y. Mathers Charitable Foundation.
Funding:
This work was funded by grants from the NIH (1DP2HG012522, 1R01AG076932 and RM1HG011014) and the Mathers Foundation to J.C..
Footnotes
Competing interests statement: J.C., W.Z., and Z.X. are inventors on pending patent applications related to PerturbSci-Kinetics. Other authors declare no competing interests.
Code Availability
The computation scripts for processing PerturbSci-Kinetics were included as supplementary files.
Data Availability
The data generated by this study can be downloaded in raw and processed forms from the NCBI Gene Expression Omnibus (GSE218566, reviewers’ token: itqlgacczrgxpmb).
Reference:
- 1.Huang H. et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurosaki T., Popp M. W. & Maquat L. E. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 20, 406–420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weskamp K. & Barmada S. J. RNA Degradation in Neurodegenerative Disease. Adv Neurobiol 20, 103–142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaitin D. A. et al. Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell 167, 1883–1896.e15 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Adamson B. et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell 167, 1867–1882.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixit A. et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 167, 1853–1866.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie S., Duan J., Li B., Zhou P. & Hon G. C. Multiplexed Engineering and Analysis of Combinatorial Enhancer Activity in Single Cells. Mol. Cell 66, 285–299.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Datlinger P. et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill A. J. et al. On the design of CRISPR-based single-cell molecular screens. Nat. Methods 15, 271–274 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sziraki A. et al. A global view of aging and Alzheimer’s pathogenesis-associated cell population dynamics and molecular signatures in the human and mouse brains. Preprint at 10.1101/2022.09.28.509825. [DOI] [PMC free article] [PubMed]
- 11.Yeo N. C. et al. An enhanced CRISPR repressor for targeted mammalian gene regulation. Nat. Methods 15, 611–616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erhard F. et al. scSLAM-seq reveals core features of transcription dynamics in single cells. Nature 571, 419–423 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Hendriks G.-J. et al. NASC-seq monitors RNA synthesis in single cells. Nat. Commun. 10, 3138 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao J., Zhou W., Steemers F., Trapnell C. & Shendure J. Sci-fate characterizes the dynamics of gene expression in single cells. Nat. Biotechnol. 38, 980–988 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu Q. et al. Massively parallel and time-resolved RNA sequencing in single cells with scNT-seq. Nat. Methods 17, 991–1001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cleary M. D., Meiering C. D., Jan E., Guymon R. & Boothroyd J. C. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat. Biotechnol. 23, 232–237 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Dolken L. et al. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA 14, 1959–1972 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller C. et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol. Syst. Biol. 7, 458–458 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duffy E. E. et al. Tracking Distinct RNA Populations Using Efficient and Reversible Covalent Chemistry. Mol. Cell 59, 858–866 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwalb B. et al. TT-seq maps the human transient transcriptome. Science 352, 1225–1228 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Rabani M. et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 29, 436–442 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller M. R., Robinson K. J., Cleary M. D. & Doe C. Q. TU-tagging: cell type–specific RNA isolation from intact complex tissues. Nat. Methods 6, 439–441 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawata K. et al. Metabolic labeling of RNA using multiple ribonucleoside analogs enables the simultaneous evaluation of RNA synthesis and degradation rates. Genome Res. 30, 1481–1491 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Battich N. et al. Sequencing metabolically labeled transcripts in single cells reveals mRNA turnover strategies. Science 367, 1151–1156 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Ziegenhain C. et al. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol. Cell 65, 631–643.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Ding J. et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol. 38, 737–746 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joung J. et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Replogle J. M. et al. Mapping information-rich genotype-phenotype landscapes with genome-scale Perturb-seq. Cell 185, 2559–2575.e28 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanson K. R. et al. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat. Commun. 9, 5416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stuart T. et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu X. et al. Mapping transcriptomic vector fields of single cells. Cell 185, 690–711.e45 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones P. L. et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19, 187–191 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Dominguez A. A., Lim W. A. & Qi L. S. Beyond editing: repurposing CRISPR–Cas9 for precision genome regulation and interrogation. Nature Reviews Molecular Cell Biology vol. 17 5–15 Preprint at 10.1038/nrm.2015.2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim S. H. & Lin R. J. Pre-mRNA splicing within an assembled yeast spliceosome requires an RNA-dependent ATPase and ATP hydrolysis. Proc. Natl. Acad. Sci. U. S. A. 90, 888–892 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colgan D. F. & Manley J. L. Mechanism and regulation of mRNA polyadenylation. Genes & Development vol. 11 2755–2766 Preprint at 10.1101/gad.11.21.2755 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Kikkawa S. et al. Conversion of GDP into GTP by nucleoside diphosphate kinase on the GTP-binding proteins. J. Biol. Chem. 265, 21536–21540 (1990). [PubMed] [Google Scholar]
- 37.Siira S. J. et al. LRPPRC-mediated folding of the mitochondrial transcriptome. Nat. Commun. 8, 1532 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruzzenente B. et al. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 31, 443–456 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu L. et al. LRP130 protein remodels mitochondria and stimulates fatty acid oxidation. J. Biol. Chem. 286, 41253–41264 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pajak A. et al. Defects of mitochondrial RNA turnover lead to the accumulation of double-stranded RNA in vivo. PLoS Genet. 15, e1008240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pakos-Zebrucka K. et al. The integrated stress response. EMBO Rep. 17, 1374–1395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buccitelli C. & Selbach M. mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet. 21, 630–644 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Chipman L. B. & Pasquinelli A. E. miRNA Targeting: Growing beyond the Seed. Trends Genet. 35, 215–222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Treiber T., Treiber N. & Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 20, 5–20 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Kim Y.-K., Kim B. & Kim V. N. Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc. Natl. Acad. Sci. U. S. A. 113, E1881–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park M. S. et al. Multidomain Convergence of Argonaute during RISC Assembly Correlates with the Formation of Internal Water Clusters. Mol. Cell 75, 725–740.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu B., Shyr Y., Cai J. & Liu Q. Interplay between miRNAs and host genes and their role in cancer. Brief. Funct. Genomics 18, 255–266 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chureau C. et al. Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Hum. Mol. Genet. 20, 705–718 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Zhao L., Mao Y., Zhao Y. & He Y. DDX3X promotes the biogenesis of a subset of miRNAs and the potential roles they played in cancer development. Scientific Reports vol. 6 Preprint at 10.1038/srep32739 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heinrichs A. A slice of the action. Nat. Rev. Mol. Cell Biol. 5, 677–677 (2004). [Google Scholar]
- 51.Lytle J. R., Yario T. A. & Steitz J. A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proc. Natl. Acad. Sci. U. S. A. 104, 9667–9672 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Replogle J. M. et al. Combinatorial single-cell CRISPR screens by direct guide RNA capture and targeted sequencing. Nat. Biotechnol. 38, 954–961 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liscovitch-Brauer N. et al. Profiling the genetic determinants of chromatin accessibility with scalable single-cell CRISPR screens. Nat. Biotechnol. 39, 1270–1277 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mimitou E. P. et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat. Methods 16, 409–412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scarpulla R. C., Vega R. B. & Kelly D. P. Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol. Metab. 23, 459–466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aibar S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.La Torre A., Georgi S. & Reh T. A. Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proc. Natl. Acad. Sci. U. S. A. 110, E2362–70 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Janowski B. A. et al. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat. Struct. Mol. Biol. 13, 787–792 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Griffin K. N. et al. Widespread association of the Argonaute protein AGO2 with meiotic chromatin suggests a distinct nuclear function in mammalian male reproduction. Genome Res. 32, 1655–1668 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moshkovich N. et al. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev. 25, 1686–1701 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krueger F. A wrapper around Cutadapt and FastQC to consistently apply adapter and quality trimming to FastQ files, with extra functionality for RRBS data. TrimGalore (accessed on 27 August 2019). [Google Scholar]
- 62.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Danecek P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lindenbaum P. JVarkit: java-based utilities for Bioinformatics. (2015) doi: 10.6084/m9.figshare.1425030.v1. [DOI] [Google Scholar]
- 65.Picard. https://broadinstitute.github.io/picard/.
- 66.Yu Guangchuang, Wang Li-Gen, GiovanniDall’Olio (formula interface of compareCluster). clusterProfiler. (Bioconductor, 2017). doi: 10.18129/B9.BIOC.CLUSTERPROFILER. [DOI] [Google Scholar]
- 67.Qiu X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang K. et al. A novel class of microRNA-recognition elements that function only within open reading frames. Nat. Struct. Mol. Biol. 25, 1019–1027 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011). [Google Scholar]
- 70.Smith T., Heger A. & Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Research vol. 27 491–499 Preprint at 10.1101/gr.209601.116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quinlan A. R. & Hall I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated by this study can be downloaded in raw and processed forms from the NCBI Gene Expression Omnibus (GSE218566, reviewers’ token: itqlgacczrgxpmb).