Abstract
Purpose of Review:
The Gram-negative oral pathogen Tannerella forsythia is implicated in the pathogenesis of periodontitis, an inflammatory disease characterized by progressive destruction of the tooth supporting structures affecting over 700 million people worldwide. This review highlights the basis of why and how T. forsythia interacts with Fusobacterium nucleatum, a bacterium considered to be a bridge between the early and late colonizing bacteria of the dental plaque.
Recent Findings:
The recent findings indicate that these two organisms have a strong mutualistic relationship that involves foraging by T. forsythia on F. nucleatum peptidoglycan and utilization of glucose, released by the hydrolytic activity of T. forsythia glucanase, as a nutrient by F. nucleatum. In addition, T. forsythia has the unique ability to generate a toxic and inflammogenic compound, methylglyoxal, from glucose. This compound can induce inflammation, leading to the degradation of periodontal tissues and release of host components as nutrients for bacteria to further exacerbate the disease.
Summary:
In summary, this article will present our current understanding of mechanisms underpinning T. forsythia-F. nucleatum mutualism, and how this mutualism might impact periodontal disease progression.
Keywords: periodontitis, peptidoglycan, methylglyoxal, periodontal pathogens, glucans
INTRODUCTION
My interest in Tannerella forsythia and oral microbiology in general began when I joined Dr. Robert Genco’s laboratory as a postdoctoral fellow in 1991. His laboratory at the time was studying the molecular mechanisms of the periodontal pathogen, Porphyromonas gingivalis. One of the research projects was aimed at defining the role of a subunit protein, FimA, that forms hair-like structures known as fimbriae on the surface of P. gingivalis, in bacterial colonization as well as in inflammation. As my first project in the lab, I generated the full length recombinant FimA protein and its several truncated derivatives, and subsequently with the help of colleagues in the lab delineated the functional domains of the FimA protein responsible for binding to salivary proteins [1–3]. I will never forget the ‘childlike’ excitement and enthusiasm Dr. Genco showed when he first saw the Coomassie stained protein gel with the purified recombinant FimA protein band at an expected size! I experienced the same enthusiasm, curiosity and love for science that he had that day until his last breath! His passion for oral microbiology-immunology was infectious and I feel fortunate to have had the opportunity to learn from him in the subsequent years. Dr. Genco was a pioneering figure who influenced many individuals through his work ethic and dedication. I continued my interest in P. gingivalis fimbriae and co-authored many papers on the subject with Dr. Genco, such as on the potential of FimA peptides as vaccine antigens against P. gingivalis. The studies led to the development of live vaccine vectors based on the commensal oral streptococci Streptococcus gordonii for the delivery of FimA peptide antigens [4, 5]. As a proof of principle concept for a periodontal vaccine, we showed that via oral immunization with such genetically modified vectors expressing FimA protein it was possible to significantly block the P. gingivalis-associated alveolar bone loss in a rodent model [6].
A bacterium that caught my attention while working on P. gingivalis was Bacteroides forsythus - a ‘new kid on the block’ at the time - now known as Tannerella forsythia. This bacterium was isolated by Anne Tanner at the Forsyth Institute in 1979 as a slow growing fusiform-shaped bacterium from plaque samples of periodontitis patients [7]. The organism could grow on blood agar plates when Fusobacterium nucleatum was co-streaked alongside as a helper bacterium [8]. Subsequently, Wyss (1989) [9] showed that supplementation of the culture medium with N- acetylmuramic acid (MurNAc), an amino sugar that together with N-acetylglucosamine (GlcNAc) forms the repeating disaccharide backbone of the peptidoglycan layer, it was possible to support the growth of the organism. In 2001, based on 16S rRNA gene sequencing, the organism was placed in a new genus, Tannerella, and T. forsythia became the founding member of the genus [10]. The studies have demonstrated that T. forsythia existed in the oral cavity of humans at least since the prehistoric Neolithic times, and the organism has thus adapted well to its oral niche in modern humans [11]; it is also found in the oral cavity of cats [12] and dogs [13]. It remains to be determined what adaptive advantages the loss of ability to de novo synthesize peptidoglycan amino sugars MurNAc provide to the bacterium, except for possibly reducing the genomic burden to some extent. In the genus Tannerella, species that have also generated interest in recent years include the taxa designated as Tannerella sp. BU063 (also known as human oral taxon 286 [HOT 286]) [14] and Tannerella sp. BU045 (HOT 808) [15]. Though these spp. are found frequently in periodontal disease-associated plaques, they have not been found to proliferate to high densities, and thus their role in the pathogenesis of periodontal diseases is debatable [16]. However, BU045 is more frequent in periodontitis, whereas BU063 is considered a ‘periodontal health associated’ species [17]. BU063 has recently been cultivable in the presence of helper bacteria [17] while BU045 remains to be cultivated.
T. forsythia is a major periodontal pathogen involved in the development of periodontitis. This is corroborated by its high abundance in periodontal disease pockets, its prevalence in patients with chronic periodontitis [18–21] and the fact that the organism is able to induce experimental periodontitis in rodent models [22, 23]. T. forsythia, P. gingivalis and Treponema denticola - a triad known as ‘red- complex’ - are organisms which are strongly associated in periodontitis [20]. However, as we realize now based on 16S rDNA bacterial identification schemes, there are potentially many more species associated with the disease [24] and the disease etiology involves a synergistic and dysbiotic complex community where red-complex bacteria likely play a significant role [25].
In this review, we present recent findings of our group highlighting the survival strategies of T. forsythia in the oral cavity, depending largely on interspecies metabolic exchange between T. forsythia and its dental plaque partner F. nucleatum and how this metabolic exchange might affect microbial ecology and periodontal immunity at large.
T. forsythia – F. nucleatum synergistic partnership.
A large body of clinical evidence implicates T. forsythia in the pathogenesis of periodontitis, a disease that leads to inflammatory alveolar bone loss and is a well-established risk factor of atherosclerosis, diabetes, and low weight or preterm births [26, 27]. Targeted gene deletion strategy [28] has facilitated the identification of many virulence factors of T. forsythia (reviewed in references [26, 29]); e. g., leucine-rich repeat BspA protein [30], sialidases [31], surface (S)-layer glycoproteins [32], matrix metalloprotease-like enzyme karilysin [33] and KLIK proteases [34], and dipeptidyl aminopeptidase IV [35]. Recent studies have also implicated T. forsythia in the etiology of endodontic and dental implant infections [36, 37]. Higher levels of T. forsythia have been reported in the dental plaque samples of post-menopausal obese women [38, 39] and the bacterium has also been isolated from women with vaginosis [40]. A recent case control prospective study implicated T. forsythia in the development of esophageal cancers [41]. In experimental settings using rodent models, T. forsythia has been shown to enhance periodontal bone loss when given as a polymicrobial infection with the red-complex species P. gingivalis and T. denticola [23]. T. forsythia has been also shown to induce foam cell formation and accelerate the formation of atherosclerotic lesions in mice [42].
F. nucleatum is an opportunistic pathogen and is considered a ‘bridge bacterium’ since it facilitates the development of dental plaque due to its ability to co-aggregate with both the early-and late-colonizing bacterial species [43, 44]. Moreover, in recent years F. nucleatum has gained much notoriety and attention due to its probable role in colorectal cancer development [45, 46]. T. forsythia has been shown to co-aggregate and form synergistic cobiofilms with F. nucleatum in vitro [47], and in human dental plaque the two species coexist in close physical proximity [48]. Moreover, co-inoculation of these two organisms induces a synergistic increase in abscess formation in rabbits [49] and alveolar bone loss [50] in mice, as compared to infection with either organism alone. Thus, both in vitro and in vivo data suggest a mutualistic partnership between these two species.
Interspecies sensing and metabolite exchange.
Peptidoglycan scavenging.
As mentioned above, T. forsythia is auxotrophic for the peptidoglycan amino sugar, N-acetylmuramic acid (MurNAc). MurNAc is a unique amino sugar found only in the chemical make-up of bacteria. The peptidoglycan layer (cell-wall) is a polymer of alternating β−1,4 linked MurNAc and GlcNAc residues in which MurNAc is attached to L- and D-amino acid-containing stem pentapeptides that crosslink adjacent sugar chains [51]. The peptidoglycan layer is essential in all bacteria as it provides a rigid covering (exoskeleton) surrounding the cytoplasmic membrane, protects bacteria from osmotic shock, and determines the shape [52]. The MurNAc auxotrophy of T. forsythia is due to the lack of UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) and UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) - key enzymes involved in the de novo synthesis of MurNAc from simple sugars in bacteria [53]. In order to cope with this critical deficiency, the bacterium relies on exogenous sources for obtaining the precursors (MurNAc or muropeptides) of peptidoglycan. For this purpose, T. forsythia has evolved committed pathways for transporting and recycling MurNAc and peptidoglycan fragments from its environment - the subgingival niche, where these components are abundantly released by cohabiting bacteria as a consequence of their cell-wall breakdown/restructuring during division and growth. We have recently shown that T. forsythia has evolved a novel transport system, MurTKQ, comprising an integral membrane protein (TfMurT), a sugar kinase TfMurK), and an etherase (TfMurQ) for the transport and utilization of exogenous MurNAc [54]. However, an important fact is that the human host cannot synthesize MurNAc and the cohabiting bacteria are the sole source of free MurNAc or MurNAc containing peptidoglycan fragments (muropeptides). Conceivably, peptidoglycan fragments released by cohabiting bacteria during their cell-wall turnover or death could be readily accessible to T. forsythia in the microbial communities. We recently showed in an in vitro model that T. forsythia can utilize muropeptides derived from F. nucleatum via transport through a muropeptide permease (TfAmpG) whose expression is regulated by a unique hybrid two-component system (Tf GppX) [55]. A close physical association of these two organisms in the dental plaque biofilms [48] and the fact that these organisms show synergistic co-biofilm activity in vitro [56] and virulence in a mouse model of periodontitis [50] suggest T. forsythia’s dependence on F. nucleatum for peptidoglycan.
Glucan hydrolysis and methylglyoxal production.
We have recently shown that T. forsythia secretes a β-glucanase enzyme (TfGlcA) that can digest β−1,6 and β−1,3 linked β-glucans into glucose monomers and polymers [57]. Interestingly, the expression of this enzyme is induced when the bacterium comes in contact with F. nucleatum. The β-glucanase coding gene is part of an operon located immediately downstream of an operon that codes for a putative extracytoplasmic function (ECF) sigma factor and an anti-sigma factor regulatory proteins, which we predict are responsible for regulating β-glucanase expression in response to F. nucleatum and possibly other environmental stimuli [57]. Furthermore, glucose released from β-glucan hydrolysis by β-glucanase serves as a source of carbon for F. nucleatum and promotes its biomass in mixed species biofilms [57]. However, this increased glucose availability does not promote T. forsythia biomass since the bacterium is asaccharolytic. Rather, glucose can become toxic to the bacterium due to its conversion into methylglyoxal (MGO) via a methylglyoxal synthase (MgsA) catalyzed pathway in T. forsythia [58, 59]. MGO is an electrophilic di-carbonyl compound which is toxic to both bacterial and host cells. In addition, MGO can covalently modify host proteins to generate glycation adducts known as advanced glycation endproducts (AGEs) which, via activation of RAGEs (receptors for AGEs), can induce inflammatory responses [60, 61]. Potentially, therefore, dietary intake of β-glucans (rich in oats and breads) might promote dental plaque development and periodontal inflammation. Interestingly, fusobacteria can benefit from their assocaition with T. forsythia in other ways as well. For instance, fusobacteria can metabolize silaic acid released by the action of T. forsythia NanH siliadase enzyme on host silaoglycoproteins (salivary mucins and cell surface proteins) as a carbon source and/or repurpose the sugar to cloak their surface with the sugar [31].
Potential consequence of peptidoglycan scavenging and MGO production on periodontal microbiota and inflammation.
While peptidoglycan plays structural and physiological roles in bacteria, its components are immunomodulatory in nature and can orchestrate host immunity [62–65]. Specifically, peptidoglycan fragments of both Gram-negative and Gram-positive bacteria can induce inflammation by activating the NOD (nucleotide binding and oligomerization domain) receptors expressed in innate immune cells such as monocytes, macrophages, neutrophils and epithelial cells. NODs, including NOD1 and NOD2, are a family of intracellular pattern recognition receptors that recognize peptidoglycan components [66–68]. NOD2 related signaling has been strongly correlated with inflammatory diseases such as Crohn’s disease and the Blau syndrome [69–71]. The specific ligands for NOD1 and NOD2 receptors are the dipeptides D-γ-glutamyl-meso-DAP (iE-DAP) and MurNAc-L-Ala-D-isoGln (mMDP), respectively, which are released in the environment when bacteria breakdown and recycle their cell-wall during division [72, 73]. We hypothesis that T. forsythia associated muropeptide scavenging might lead to dampening of NOD2 signaling in the subgingival niche and T. forsythia in this scenario might act as a key modulator of local inflammation during periodontitis.
As mentioned above, another outcome of T. forsythia-F. nucleatum interaction is MGO production. A plausible role of T. forsythia-produced MGO in periodontitis is supported by clinical studies that show higher levels of MGO correlating with T. forsythia loads are present in gingival exudates from severely inflamed lesions of periodontitis patients [74]. Moreover, in vitro data suggest that free MGO can directly induce gingival connective tissue damage by causing apoptosis of gingival fibroblasts [75] and T. forsythia-secreted MGO can cause collagen glycation which in a RAGE-dependent manner can induce pro-inflammatory cytokine production [59]. In addition, subgingival bacteria that lack the abilty to detoxify MGO are likely to be susceptible to MGO. Taken together, MGO might be an important contributing factor in the development and severity of periodontitis by promoting both inflammation and microbial dysbiosis.
CONCLUSIONS
T. forsythia displays a unique affinity for F. nucleatum and the evidence suggests that this association is mutualistic and might be critical for the development of dental plaque, and hence in the progression of periodontal disease (summarized in a model in Figure 1). This mutualism is due to the following reasons. T. forsythia, is auxotrophic for the aminosugar N-acetylmuramic acid (MurNAc) which, along with N-acetylglucosamine (GlcNAc), forms the repeating disaccharide unit of the peptidoglycan backbone in most bacteria. To circumvent this deficiency, the bacterium must harvest peptidoglycan precursors from the environment for its cell wall biosynthesis and survival. The bacteriium has evolved well-committed pathways for the transport and recycling of MurNAc and peptidoglycan fragments released by the cohabiting bacteria during their cell wall turnover. In this regard, we believe F. nucleatum is a crucial partner of T. forsythia in the dental plaque. This relationship appears to be mutualistic and strategic as F. nucleatum can provide peptidoglycan fragments to T. forythia in return for glucose as a nutrient released by T. forythia β-glucanase action on glucans. Moreover, T. forsythia is a unique pathogen with an ability to produce methylglyoxal (MGO), an electrophilic di-carbonyl compound that can covalently modify host proteins to generate inflammogenic adducts known as advanced glycation endproducts (AGEs). MGO is also toxic to bacteria that lack detoxification systems, and thus MGO may aid in the release of peptidoglycan from such organisms. We hypothesis that MGO, by promoting the formation of AGEs and its killing activity against susceptible organisms, is involved in the microbial dysbiosis process. It would be interesting to determine how T. forsythia’s peptidoglycan scavenging activities and MGO production orchestrate a state of innate immune dysregulation and microbial dysbiosis in the oral cavity.
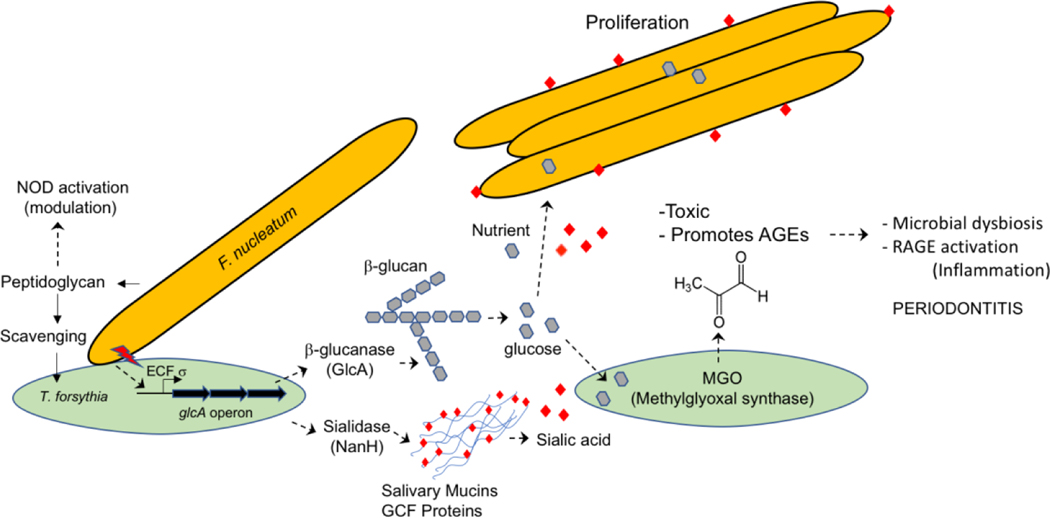
Figure 1. Model of T. forsythia - F. nucleatum interbacterial interactions in dental biofilm and their impact on periodontal ecology and inflammation.
T. forsythia utilzes peptidoglycan fragments released by F. nucleatum and possibly other bacteria for its survival. This is expected to reduce the levels of free peptidoglycan in the subgingival niche and dampen NOD (nucleotide bnding oligomerization domain containing intracellular pattern recognition proteins) - mediated inflammatory responses of the host. F. nucleatum triggers the secretion of β-glucanase from T. forsythia via an ECF σ-factor (extracytoplasmic function sigma factor) dependent induction of the glcA operon in T. forsythia. β-glucanase thus secreted can digest dietary β-glucans into glucose residues, which serve as a source of carbon for F. nucleatum and a precursor for the generation of methylglyoxal (MGO) by T. forsythia. MGO covalently modifies host proteins to generate AGEs (glycation adducts known as advanced glycation endproducts), which can trigger inflammation via the activation of RAGEs (receptor for AGEs). Moreover, sialic acid released from salivary mucins by T. forsythia-secreted sialidase enzyme NanH might be utilized by F. nucleatum to decorate its cell surface with the sugar. Figure keys: red lightning bolt. F. nucleatum stimulating ECF σ-factor in T. forsythia; red diamonds, sialic acid; grey hexagons, glucose.
ACKNOWLEDGEMENTS
I thank Dr. Karen Falkner for critical reading of this article and her helpful suggestions. The work cited from the author’s laboratory was supported by grants (DE014749 and DE022870) from the NIDCR.
Footnotes
Human and Animal Rights and Informed Consent: This article does not contain any primary research studies with human or animal subjects performed by any of the authors.
Conflict of Interest: Dr. Sharma declares no conflict of interest.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
REFERENCES
Papers of particular interest have been highlighted as:
* Of imporance
** Of major importance
- 1.Sharma A, Sojar HT, Lee JY, Genco RJ. Expression of a functional Porphyromonas gingivalis fimbrillin polypeptide in Escherichia coli: purification, physicochemical and immunochemical characterization, and binding characteristics. Infect Immun. 1993;61(8):3570–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amano A, Sojar HT, Lee JY, Sharma A, Levine MJ, Genco RJ. Salivary receptors for recombinant fimbrillin of Porphyromonas gingivalis. Infect Immun. 1994;62(8):3372–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagata H, Sharma A, Sojar HT, Amano A, Levine MJ, Genco RJ. Role of the carboxyl-terminal region of Porphyromonas gingivalis fimbrillin in binding to salivary proteins. Infect Immun. 1997;65(2):422–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma A, Honma K, Sojar HT, Hruby DE, Kuramitsu HK, Genco RJ. Expression of saliva-binding epitopes of the Porphyromonas gingivalis FimA protein on the surface of Streptococcus gordonii. Biochem Biophys Res Commun. 1999;258(1):222–6. [DOI] [PubMed] [Google Scholar]
- 5.Sharma A, Nagata H, Hamada N, Sojar HT, Hruby DE, Kuramitsu HK et al. Expression of functional Porphyromonas gingivalis fimbrillin polypeptide domains on the surface of Streptococcus gordonii. Appl Environ Microbiol. 1996;62(11):3933–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma A, Honma K, Evans RT, Hruby DE, Genco RJ. Oral immunization with recombinant Streptococcus gordonii expressing Porphyromonas gingivalis FimA domains. Infect Immun. 2001;69(5):2928–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanner AC, Haffer C, Bratthall GT, Visconti RA, Socransky SS. A study of the bacteria associated with advancing periodontitis in man. J Clin Periodontol. 1979;6(5):278–307. [DOI] [PubMed] [Google Scholar]
- 8.Tanner ACR, Listgarten MA, Ebersole JL, Strzempko MN. Bacteroides forsythus sp. nov., a slow growing, fusiform Bacteroides sp. from the human oral cavity. Int J Syst Bacteriol. 1986;36:213–21. [Google Scholar]
- 9. Wyss C. Dependence of proliferation of Bacteroides forsythus on exogenous N-acetylmuramic acid. Infect Immun. 1989;57(6):1757–9. *This study showed that peptidoglycan amino sugar MurNAc is essential for the cultivation of T. forsythia.
- 10.Sakamoto M, Suzuki M, Umeda M, Ishikawa L, Benno Y. Reclassification of Bacteroides forsythus (Tanner et al. 1986) as Tannerella forsythensis corrig., gen. nov., comb. nov. Int J Syst Evol Microbiol. 2002;52(Pt 3):841–9. [DOI] [PubMed] [Google Scholar]
- 11.Warinner C, Rodrigues JF, Vyas R, Trachsel C, Shved N, Grossmann J et al. Pathogens and host immunity in the ancient human oral cavity. Nat Genet. 2014;46(4):336–44. doi: 10.1038/ng.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Booij-Vrieling HE, van der Reijden WA, Houwers DJ, de Wit WE, Bosch-Tijhof CJ, Penning LC et al. Comparison of periodontal pathogens between cats and their owners. Vet Microbiol. 2010;144:147–52. [DOI] [PubMed] [Google Scholar]
- 13.Oh C, Lee K, Cheong Y, Lee SW, Park SY, Song CS et al. Comparison of the Oral Microbiomes of Canines and Their Owners Using Next-Generation Sequencing. PloS one. 2015;10(7):e0131468. doi: 10.1371/journal.pone.0131468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beall CJ, Campbell AG, Dayeh DM, Griffen AL, Podar M, Leys EJ. Single Cell Genomics of Uncultured, Health-Associated Tannerella BU063 (Oral Taxon 286) and Comparison to the Closely Related Pathogen Tannerella forsythia. PloS one. 2014;9(2):e89398. doi: 10.1371/journal.pone.0089398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beall CJ, Campbell AG, Griffen AL, Podar M, Leys EJ. Genomics of the Uncultivated, Periodontitis-Associated Bacterium Tannerella sp. BU045 (Oral Taxon 808). mSystems. 2018;3(3). doi: 10.1128/mSystems.00018-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuger J, Luthi-Schaller H, Gmur R. Uncultivated Tannerella BU045 and BU063 are slim segmented filamentous rods of high prevalence but low abundance in inflammatory disease-associated dental plaques. Microbiology. 2007;153(Pt 11):3809–16. doi: 10.1099/mic.0.2007/010926-0. [DOI] [PubMed] [Google Scholar]
- 17.Vartoukian SR, Moazzez RV, Paster BJ, Dewhirst FE, Wade WG. First Cultivation of Health-Associated Tannerella sp. HOT-286 (BU063). J Dent Res. 2016;95:1308–13. doi: 10.1177/0022034516651078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camelo-Castillo A, Novoa L, Balsa-Castro C, Blanco J, Mira A, Tomas I. Relationship between periodontitis-associated subgingival microbiota and clinical inflammation by 16S pyrosequencing. J Clin Periodontol. 2015;42(12):1074–82. doi: 10.1111/jcpe.12470. [DOI] [PubMed] [Google Scholar]
- 19.Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. 2005;43(8):3944–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr., Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25(2):134–44. [DOI] [PubMed] [Google Scholar]
- 21.Tanner AC, Izard J. Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontol 2000. 2006;42:88–113. [DOI] [PubMed] [Google Scholar]
- 22.Myneni SR, Settem RP, Connell TD, Keegan AD, Gaffen SL, Sharma A. TLR2 signaling and Th2 responses drive Tannerella forsythia-induced periodontal bone loss. J Immunol. 2011;187(1):501–9. doi: 10.4049/jimmunol.1100683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kesavalu L, Sathishkumar S, Bakthavatchalu V, Matthews C, Dawson D, Steffen M et al. Rat model of polymicrobial infection, immunity, and alveolar bone resorption in periodontal disease. Infect Immun. 2007;75(4):1704–12. doi: 10.1128/IAI.00733-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. The ISME journal. 2011;6:1176. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27:409–19. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma A. Virulence mechanisms of Tannerella forsythia. Periodontol 2000. 2010;54(1):106–16. doi: 10.1111/j.1600-0757.2009.00332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanner AC, Izard J. Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontol 2000. 2006;42:88–113. doi: 10.1111/j.1600-0757.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- 28.Honma K, Kuramitsu HK, Genco RJ, Sharma A. Development of a gene inactivation system for Bacteroides forsythus: construction and characterization of a BspA mutant. Infect Immun. 2001;69(7):4686–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma A. Genome functions of Tannerella forsythia in bacterial communities. In: Kolenbrander PE, editor. Oral Microbial Communities: Genome inquiry and interspecies communication. Washington, D. C.: American Society for Microbiology; 2011. p. 135. [Google Scholar]
- 30.Sharma A, Sojar HT, Glurich I, Honma K, Kuramitsu HK, Genco RJ. Cloning, expression, and sequencing of a cell surface antigen containing a leucine-rich repeat motif from Bacteroides forsythus ATCC 43037. Infect Immun. 1998;66(12):5703–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stafford G, Roy S, Honma K, Sharma A. Sialic acid, periodontal pathogens and Tannerella forsythia: stick around and enjoy the feast! Mol Oral Microbiol. 2012;27(1):11–22. doi: 10.1111/j.2041-1014.2011.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Settem RP, Honma K, Stafford GP, Sharma A. Protein-linked glycans in periodontal bacteria: prevalence and role at the immune interface. Front Microbiol. 2013;4:PMID: 24146665 (open access). doi: 10.3389/fmicb.2013.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karim AY, Kulczycka M, Kantyka T, Dubin G, Jabaiah A, Daugherty PS et al. A novel matrix metalloprotease-like enzyme (karilysin) of the periodontal pathogen Tannerella forsythia ATCC 43037. Biol Chem. 2010;391(1):105–17. doi: 10.1515/BC.2010.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ksiazek M, Mizgalska D, Eick S, Thogersen IB, Enghild JJ, Potempa J. KLIKK proteases of Tannerella forsythia: putative virulence factors with a unique domain structure. Front Microbiol. 2015;6:312. doi: 10.3389/fmicb.2015.00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yost S, Duran-Pinedo AE. The contribution of Tannerella forsythia dipeptidyl aminopeptidase IV in the breakdown of collagen. Mol Oral Microbiol. 2018;33(6):407–19. doi: 10.1111/omi.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ribeiro AC, Matarazzo F, Faveri M, Zezell DM, Mayer MP. Exploring Bacterial Diversity of Endodontic Microbiota by Cloning and Sequencing 16S rRNA. J Endod. 2011;37(7):922–6. doi: 10.1016/j.joen.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 37.Rocas IN, Alves FR, Santos AL, Rosado AS, Siqueira JF Jr., Apical Root Canal Microbiota as Determined by Reverse-capture Checkerboard Analysis of Cryogenically Ground Root Samples from Teeth with Apical Periodontitis. J Endod. 2010;36(10):1617–21. doi: 10.1016/j.joen.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Brennan RM, Genco RJ, Wilding GE, Hovey KM, Trevisan M, Wactawski-Wende J. Bacterial species in subgingival plaque and oral bone loss in postmenopausal women. J Periodontol. 2007;78(6):1051–61. doi: 10.1902/jop.2007.060436. [DOI] [PubMed] [Google Scholar]
- 39.Haffajee AD, Socransky SS. Relation of body mass index, periodontitis and Tannerella forsythia. J Clin Periodontolol. 2009;36(2):89–99. [DOI] [PubMed] [Google Scholar]
- 40.Africa CW, Nel J, Stemmet M. Anaerobes and bacterial vaginosis in pregnancy: virulence factors contributing to vaginal colonisation. Int J Environ Res Public Health. 2014;11(7):6979–7000. doi: 10.3390/ijerph110706979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peters BA, Wu J, Pei Z, Yang L, Purdue MP, Freedman ND et al. Oral Microbiome Composition Reflects Prospective Risk for Esophageal Cancers. Cancer Res. 2017;77(23):6777–87. doi: 10.1158/0008-5472.CAN-17-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee HR, Jun HK, Choi BK. Tannerella forsythia BspA increases the risk factors for atherosclerosis in ApoE(−/−) mice. Oral Dis. 2014;20(8):803–8. doi: 10.1111/odi.12214. [DOI] [PubMed] [Google Scholar]
- 43.Al-Ahmad A, Wunder A, Auschill TM, Follo M, Braun G, Hellwig E et al. The in vivo dynamics of Streptococcus spp., Actinomyces naeslundii, Fusobacterium nucleatum and Veillonella spp. in dental plaque biofilm as analysed by five-colour multiplex fluorescence in situ hybridization. J Med Microbiol. 2007;56(Pt 5):681–7. doi: 10.1099/jmm.0.47094-0. [DOI] [PubMed] [Google Scholar]
- 44.Han YW. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol. 2015;23:141–7. doi: 10.1016/j.mib.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rubinstein MR, Baik JE, Lagana SM, Han RP, Raab WJ, Sahoo D et al. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/beta-catenin modulator Annexin A1. EMBO Rep. 2019;20(4). doi: 10.15252/embr.201847638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65(12):1973–80. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma A, Inagaki S, Sigurdson W, Kuramitsu HK. Synergy between Tannerella forsythia and Fusobacterium nucleatum in biofilm formation. Oral Microbiol Immunol. 2005;20(1):39–42. doi: 10.1111/j.1399-302X.2004.00175.x. [DOI] [PubMed] [Google Scholar]
- 48.Zijnge V, van Leeuwen MB, Degener JE, Abbas F, Thurnheer T, Gmur R et al. Oral biofilm architecture on natural teeth. PloS one. 2010;5(2):e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takemoto T, Kurihara H, Dahlen G. Characterization of Bacteroides forsythus isolates. J Clin Microbiol. 1997;35(6):1378–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Settem RP, El-Hassan AT, Honma K, Stafford GP, Sharma A. Fusobacterium nucleatum and Tannerella forsythia Induce Synergistic Alveolar Bone Loss in a Mouse Periodontitis Model. Infect Immun. 2012;80(7):2436–43. doi: 10.1128/iai.06276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park JT. Murein syntheisis. In: Neidhardt JL, Ingraham KB, Low B, Magasanik M, Schaechter, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D. C.: American Society for MIcrobiology; 1987. p. 663–7. [Google Scholar]
- 52.Vollmer W, Seligman SJ. Architecture of peptidoglycan: more data and more models. Trends Microbiol.18(2):59–66. doi: 10.1016/j.tim.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 53.Ruscitto A, Sharma A. Peptidoglycan synthesis in Tannerella forsythia: Scavenging is the modus operandi. Mol Oral Microbiol. 2018;33:125–32. doi: 10.1111/omi.12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruscitto A, Hottmann I, Stafford GP, Schaffer C, Mayer C, Sharma A. Identification of a Novel N-Acetylmuramic Acid Transporter in Tannerella forsythia. J Bacteriol. 2016;198(22):3119–25. doi: 10.1128/JB.00473-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruscitto A, Honma K, Veeramachineni VM, Nishikawa K, Stafford GP, Sharma A. Regulation and Molecular Basis of Environmental Muropeptide Uptake and Utilization in Fastidious Oral Anaerobe Tannerella forsythia. Front Microbiol. 2017;8:648. doi: 10.3389/fmicb.2017.00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma A, Inagaki S, Sigurdson W, Kuramitsu HK. Synergy between Tannerella forsythia and Fusobacterium nucleatum in biofilm formation. Oral Microbiol Immunol. 2005;20(1):39–42. [DOI] [PubMed] [Google Scholar]
- 57. Honma K, Ruscitto A, Sharma A. beta-Glucanase Activity of the Oral Bacterium Tannerella forsythia Contributes to the Growth of a Partner Species, Fusobacterium nucleatum, in Cobiofilms. Appl Environ Microbiol. 2018;84(1). doi: 10.1128/AEM.01759-17. **This study showed that T. forsythia contact with F. nucleatum leads to the induction of β-glucanse
- 58.Maiden MFJ, Pham C, Kashket S. Glucose toxicity effect and accumulation of methylgloxal by the periodontal pathogen Bacteroides forsythus. Anaerobe. 2004;10:27–32. [DOI] [PubMed] [Google Scholar]
- 59. Settem RP, Honma K, Shankar M, Li M, LaMonte M, Xu D et al. Tannerella forsythia produced methylglyoxal causes advanced glycation endproducts (AGEs) accumulation to trigger cytokine secretion in human monocytes. Mol Oral Microbiol. 2018;33:292–9. doi: 10.1111/omi.12224. *This study showed that T. forsythia promotes formation of AGEs via production of methylglyoxal.
- 60.Bierhaus A, Humpert PM, Morcos M, Wendt T, Chavakis T, Arnold B et al. Understanding RAGE, the receptor for advanced glycation end products. J Mol Med (Berl). 2005;83(11):876–86. doi: 10.1007/s00109-005-0688-7. [DOI] [PubMed] [Google Scholar]
- 61.Ramasamy R, Vannucci SJ, Yan SS, Herold K, Yan SF, Schmidt AM. Advanced glycation end products and RAGE: a common thread in aging, diabetes, neurodegeneration, and inflammation. Glycobiology. 2005;15(7):16R–28R. doi: 10.1093/glycob/cwi053. [DOI] [PubMed] [Google Scholar]
- 62.Bertsche U, Mayer C, Gotz F, Gust AA. Peptidoglycan perception--sensing bacteria by their common envelope structure. Int J Med Microbiol. 2015;305(2):217–23. doi: 10.1016/j.ijmm.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 63.Boneca IG. The role of peptidoglycan in pathogenesis. Curr Opin Microbiol. 2005;8(1):46–53. doi: 10.1016/j.mib.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 64.McDonald C, Inohara N, Nunez G. Peptidoglycan signaling in innate immunity and inflammatory disease. J Biol Chem. 2005;280(21):20177–80. doi: 10.1074/jbc.R500001200. [DOI] [PubMed] [Google Scholar]
- 65.Stewart-Tull DE. The immunological activities of bacterial peptidoglycans. Annu Rev Microbiol. 1980;34:311–40. doi: 10.1146/annurev.mi.34.100180.001523. [DOI] [PubMed] [Google Scholar]
- 66.Caruso R, Warner N, Inohara N, Nunez G. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity. 2014;41(6):898–908. doi: 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nigro G, Fazio LL, Martino MC, Rossi G, Tattoli I, Liparoti V et al. Muramylpeptide shedding modulates cell sensing of Shigella flexneri. Cellular microbiology. 2008;10(3):682–95. doi: 10.1111/j.1462-5822.2007.01075.x. [DOI] [PubMed] [Google Scholar]
- 68.Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nat Immunol. 2006;7(12):1250–7. doi: 10.1038/ni1412. [DOI] [PubMed] [Google Scholar]
- 69.Radford-Smith G, Pandeya N. Associations between NOD2/CARD15 genotype and phenotype in Crohn’s disease--Are we there yet? World J Gastroenterol. 2006;12(44):7097–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim TH, Payne U, Zhang X, Iwanaga Y, Davey MP, Rosenbaum JT et al. Altered host:pathogen interactions conferred by the Blau syndrome mutation of NOD2. Rheumatol Int. 2007;27(3):257–62. doi: 10.1007/s00296-006-0250-0. [DOI] [PubMed] [Google Scholar]
- 71.Chamaillard M, Philpott D, Girardin SE, Zouali H, Lesage S, Chareyre F et al. Gene-environment interaction modulated by allelic heterogeneity in inflammatory diseases. Proc Natl Acad Sci U S A. 2003;100(6):3455–60. doi: 10.1073/pnas.0530276100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4(7):702–7. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 73.Girardin SE, Travassos LH, Herve M, Blanot D, Boneca IG, Philpott DJ et al. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J Biol Chem. 2003;278(43):41702–8. doi: 10.1074/jbc.M307198200. [DOI] [PubMed] [Google Scholar]
- 74.Kashket S, Maiden MF, Haffajee AD, Kashket ER. Accumulation of methylglyoxal in the gingival crevicular fluid of chronic periodontitis patients. J Clin Periodontol. 2003;30(4):364–7. doi:322 [pii]. [DOI] [PubMed] [Google Scholar]
- 75.Retamal IN, Hernandez R, Gonzalez-Rivas C, Caceres M, Arancibia R, Romero A et al. Methylglyoxal and methylglyoxal-modified collagen as inducers of cellular injury in gingival connective tissue cells. J Periodontal Res. 2016;51(6):812–21. [DOI] [PubMed] [Google Scholar]