Abstract
Osteoarthritis (OA) is the most common joint disease primarily characterized by cartilage degeneration. Milk-derived extracellular vesicles (mEVs) were reported to inhibit catabolic and inflammatory processes in the cartilage of OA patients. However, the current therapies target the advanced symptoms of OA, and it is significant to develop a novel strategy to inhibit the processes driving OA pathology. In this study, we investigated the therapeutic potential of mEVs in alleviating OA in vivo. The results revealed that mEVs ameliorated cartilage degeneration by increasing hyaline cartilage thickness, decreasing histological Osteoarthritis Research Society International (OARSI) scores, enhancing matrix synthesis, and reducing the expression of cartilage destructive enzymes in the destabilization of medial meniscus (DMM) mice. In addition, the disturbed gut microbiota in DMM mice was partially improved upon treatment with mEVs. It was observed that the pro-inflammatory bacteria (Proteobacteria) were reduced and the potential beneficial bacteria (Firmicutes, Ruminococcaceae, Akkermansiaceae) were increased. mEVs could alleviate the progression of OA by restoring matrix homeostasis and reshaping the gut microbiota. These findings suggested that mEVs might be a potential therapeutic dietary supplement for the treatment of OA.
Keywords: bovine milk, extracellular vesicles, osteoarthritis, cartilage degeneration, gut microbiota
1. Introduction
Osteoarthritis (OA) is one of the most common musculoskeletal diseases characterized by cartilage degeneration [1]. Due to global aging and the increase in obesity, the OA burden poses a serious challenge to healthy aging. OA affects approximately 303.1 million people worldwide and has become the common cause of disability [2]. Currently, there are no approved disease-modifying drugs available, and the main therapies for OA are typically focused on pain control, intra-articular injections, and joint replacement, all of which simply target the symptoms of advanced disease [3,4]. Therefore, it is significant to develop effective and safe strategies to prevent or treat OA.
Milk-derived extracellular vesicles (mEVs) are nanoparticles and carry a cargo rich in proteins, lipids, and microRNAs that can be transported between cells [5,6]. It has been reported that mEVs could provide protection to the luminal cargo against harsh degrading conditions of the gut and were subsequently detected in multiple organs [6,7,8]. In recent years, there is an emerging interest in the use of mEVs as biomarkers or treatments for various diseases, such as breast cancer [6], ulcerative colitis [7], and rheumatoid arthritis [9]. However, only one study has shown that TGF-β and miR-148a, which are carried by mEVs, have the potential for cartilage protection in vitro, but the underlying mechanism of mEVs alleviating cartilage degeneration remains to be explored [10].
Gut microbiota has been regarded as an important factor in the development of OA [11,12]. There is increasing interest in the relationship between gut microbiota and OA. Alteration in normal gut microbiota has been implicated in metabolic syndrome and inflammation, which are also important components in the development of OA [13]. A recent work has suggested the role of gut microbiota in OA. For instance, the destabilization of the medial meniscus (DMM)-induced mice conventionally had more severe joint damage compared with the germ-free mice [14]. Several groups have reported that intervention of supplementation with either probiotics or prebiotics decreased the histological changes associated with OA and modified systemic inflammation profiles [15,16,17]. mEVs have been shown to regulate intestinal immunity by altering gut microbiota in both healthy and diseased mice [7,18]. In this study, the therapeutic effects of mEVs on cartilage degeneration and gut microbiota were investigated in the DMM-induced OA mice model.
2. Materials and Methods
2.1. Isolation and Identification of mEVs
Bovine milk contains a large number of non-EV proteins, milk fat, sugars, and other components. To isolate and purify mEVs, we developed an efficient method in our previous study [18,19]. Raw milk was produced from lactating Holstein cows obtained from a local dairy farm, and the samples were immediately transported under refrigeration to the laboratory within 2 h. The raw milk was incubated with 0.05% chymosin and 0.3% CaCl2 at 4 °C for 90 min. To remove milk fat globules, residual chymosin, and other debris, the sample was centrifuged at 16,500× g for 30 min at 4 °C. The supernatant was filtered by 0.22 μm filters (Millipore, Hayward, CA, USA) and ultracentrifuged at 100,000× g at 4 °C for 1 h (Hitachi, Ltd., Tokyo, Japan). After being filtered by 0.22 μm filters, the supernatant was ultracentrifuged at 135,000× g at 4 °C for 90 min. The pellets were resuspended with PBS and washed by ultracentrifugation at 100,000× g at 4 °C for 1 h. The mEVs were resuspended in PBS and transferred onto the 100 kDa filters (Millipore, Hayward, CA, USA) through 0.22 μm filters and centrifuged at 5000× g for 30 min twice. The mEVs were resuspended in PBS, and the protein concentration was determined using the BCA kit (Beyotime, Shanghai, China). The mEVs were stored at 4 °C for 1 week.
The size distribution and concentration were detected by nanoparticle tracking analysis (NTA) using ZetaView PMX 100 (Particle Metrix, Meerbusch, Germany). The size distribution and particle concentration were evaluated in three replicates. Transmission electron microscopy (TEM) was employed to observe the morphology of mEVs, and mEVs were imaged using HITACHI HT7700 Transmission Electron Microscope (Hitachi, Ltd., Tokyo, Japan). The mEV marker proteins (TSG101, CD81, CD63) and negative control (calnexin) were analyzed by Western blotting. Primary antibodies against TSG101 (ab133586), Calnexin (ab227310), and CD63 (ab134045) were purchased from Abcam (Cambridge, UK). CD81 (GTX101766) was purchased from GeneTex (Irvine, CA, USA).
2.2. Animal Experiments
A total of 57 specific pathogen-free male C57BL/6J mice (7 weeks of age) were purchased from Beijing Vita River Laboratory Animal Technology Co., Ltd. (Beijing, China). All animal experiments were in accordance with the Committee on the Ethics of Animal Experiments of Ocean University of China (permission number: spxy2021082701). Mice were housed at 22 °C with 12 h light/dark cycles. After a 1-week adaption period, mice were divided into three groups randomly: sham group (PBS), DMM + PBS group (PBS), DMM + mEVs group (mEVs, 0.2 mg/kg of body weight). Then, all groups were gavaged once daily with PBS or mEVs and kept for 12 weeks.
After 2 weeks of gavage, OA was induced by surgical destabilization of the medial meniscus (DMM) of the right knee in DMM + PBS and DMM + mEVs groups. Specifically, mice were anesthetized with 5% chloral hydrate by intraperitoneal injection, then the joint capsule was opened, and the medial meniscotibial ligament was cut to destabilize the meniscus. In the sham group, the joint capsule was surgically opened, but the medial meniscotibial ligament was intact. At 10 weeks after surgery, the mice were anesthetized, and blood was taken from the eyeballs. The serum was obtained by centrifugation at 4000× g for 40 min at 4 °C. The levels of cartilage degradation markers C-telopeptide of type Ⅱ collagen (CTX-Ⅱ) and cartilage oligomeric matrix protein (COMP) in the serum were detected using enzyme-linked immunosorbent assay (ELISA) kits according to the manufacturer’s instructions (Nanjing Jiancheng Bio, Nanjing, China).
2.3. Histology and Immunohistochemistry
The right knee joints were fixed with 4% paraformaldehyde, decalcified with 10% EDTA for 4 weeks, and then embedded in paraffin. The sections (3 μm in thickness) were cut and then stained with hematoxylin–eosin (H&E), safranin o-fast green (Safranin O), and toluidine blue. The cartilage thickness in the tibial plateau, including hyaline cartilage (HC) and calcified cartilage (CC), was measured with HE staining and determined by Image J software (National Institutes of Health, Bethesda, MD, USA). Distances between surfaces of articular cartilage and the tidemark line indicate hyaline cartilage thickness, and distances between the tidemark line and subchondral bone plate (SBP) indicate calcified cartilage thickness. The Osteoarthritis Research Society International (OARSI) scores were used to grade the cartilage degeneration of the knee joint [20]. For immunofluorescence, antigens were retrieved by boiling tissue section in sodium citrate buffer, and endogenous peroxidase activity was blocked by incubation in a 3% hydrogen peroxide solution. The tissue sections were incubated with 3% BSA for 30 min at room temperature to block non-specific binding, followed by straining with the primary antibodies overnight at 4 °C. After the secondary antibody incubation, the sections were stained with DAB and hematoxylin and visualized under a light microscope (Olympus, Tokyo, Japan).
2.4. 16S rRNA Bacterial Sequencing
Mice feces were obtained 10 weeks after surgery. Fecal pellets were freshly harvested from mice and immediately frozen in liquid nitrogen and stored at −80 °C. The genomic DNA of feces was extracted, and the bacterial 16S rRNA genes V3–V4 region was amplified using the forward primer 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and the reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′). Sample-specific 7 bp barcodes were incorporated into the primers for multiplex sequencing. PCR amplicons were purified, quantified, and pooled in equal amounts. The paired-end 2 × 250 bp sequencing was performed using the Illumina MiSeq platform with MiSeq Reagent Kit V3 at Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China). For alpha diversity analysis, the indexes of Chao1, Observed_species, Shannon, Simpson, Faith_pd, and Pielou_e were calculated using QIIME2, R software and ggplot2 package. For analyzing beta diversity, principal coordinate analysis was performed via QIIME2, R software and ape package. Linear discriminant analysis Effect Size (LEfSe) was performed to detect differentially abundant taxa across groups using Python LEfSe package, R software and ggtree package.
2.5. Statistical Analysis
All results were expressed as mean ± standard deviations (SD). One-way analysis of variance (ANOVA) with Tukey’s test for multiple comparisons was performed using GraphPad Prime 8 (GraphPad Software, San Diego, CA, USA). A p value less than 0.05 was considered to be statistically significant. All analyses and graphing of the data were carried out using GraphPad Prime 8.
3. Results
3.1. Characterization of mEVs
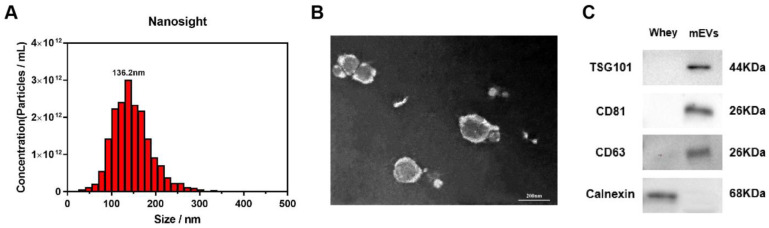
The isolated mEVs were characterized by NTA, TEM, and Western blot. The results demonstrated that the diameters of the isolated mEVs were distributed among 100–200 nm, and the mean peak of particle size was located at 136.2 nm (Figure 1A). The image in Figure 1B showed that the isolated mEVs were typically round spherical shapes with double-layer membranes. Moreover, the EV-positive markers TSG101, CD81, and CD63 were expressed in isolated mEVs, and Calnexin was not detected (Figure 1C).
Figure 1.
Characterization of mEVs. (A) The size distribution of mEVs was measured by NTA (Nanoparticle tracking analysis). The peak diameter was about 136.2 nm. (B) Transmission electron microscopy images of mEVs. Scale bars, 200 nm. (C) mEV markers and calnexin were assessed by Western blot analysis.
3.2. mEVs Reverse Cartilage Degeneration in DMM-Induced Mice
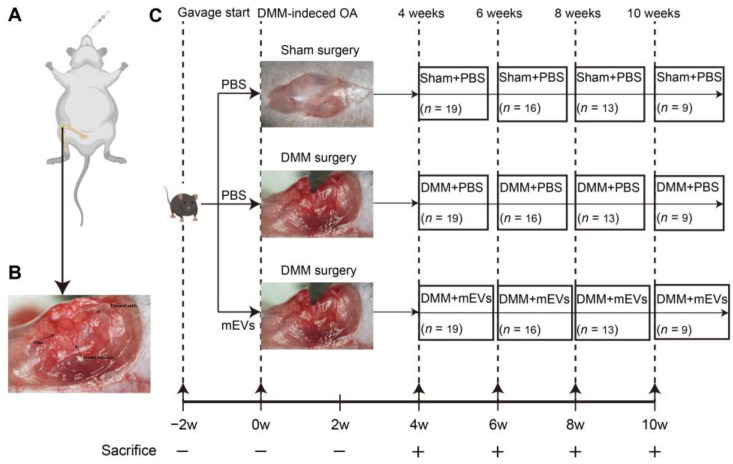
In this study, mice were gavaged with PBS or mEVs for 2 weeks before DMM surgery. To assess the therapeutic effect of mEVs in the development of OA, knee joints from each group of animals at 4, 6, 8 and 10 weeks post-surgery were harvested for histological analysis (Figure 2). To determine the severity of DMM-induced OA, the serum levels of CTX-Ⅱ and COMP were measured at different stages. The results showed that the serum CTX-Ⅱ and COMP in the DMM+PBS group were significantly higher than those of the other two groups at 8 weeks post-surgery, indicating that the OA model was successfully established and mEVs could effectively prevent the progression of OA (Supplementary Material Figures S1 and S2).
Figure 2.
Schematic illustration of in vivo experimental design. (A) Schematic diagram of animal model showing DMM-induced molded in the right hind limb and treated by oral administration. (B) Position of the femur, tibia, and medial meniscus in the knee joint. (C) Mice were pre-gavaged for 2 weeks, and then, DMM surgery was performed to establish the OA model, and tissue samples were harvested every 2 weeks from the fourth week after surgery for subsequent analysis. The number of samples in each group is expressed as an n value.
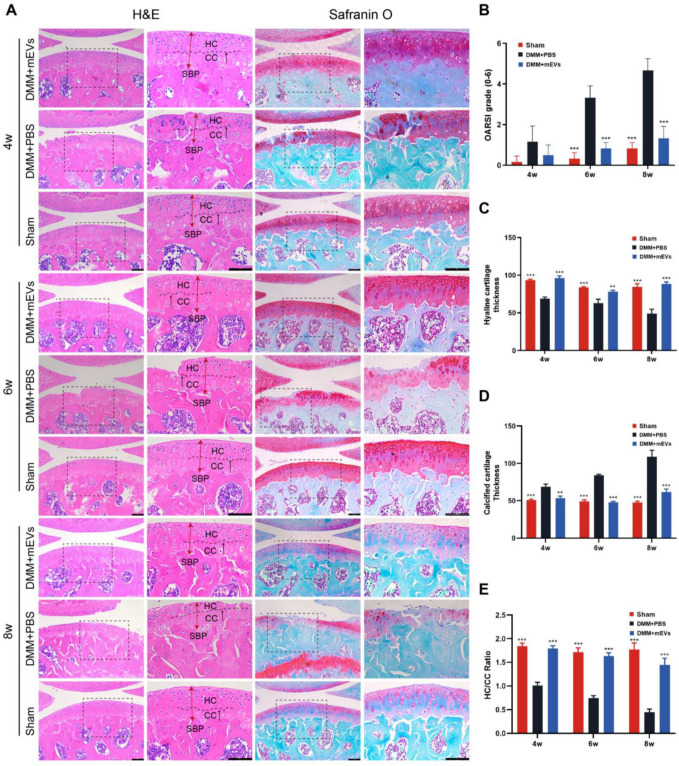
Hematoxylin–eosin and safranin o-fast green staining showed the occurrence of cartilage degeneration and cartilage proteoglycan loss after DMM surgery, and the OARSI scoring was taken to quantify the severity of cartilage damage (Figure 3 and Supplementary Material Table S1). At the early time point of 4 weeks after surgery, there were no significant differences in OARSI scores among the three groups, but the articular cartilage was locally damaged in the DMM+PBS group. Compared to the DMM+PBS group, the ratio of HC/CC was increased by 77.94% in the DMM+mEVs group. At 6 weeks post-surgery, the DMM group showed partial loss of hyaline cartilage and a significant increase in OARSI score, while the HC/CC ratio was significantly increased by 118.67% in the DMM+mEVs group in comparison to the DMM+PBS group. At 8 weeks post-surgery, the DMM+PBS group showed a significant decrease in hyaline cartilage thickness, a marked increase in OARSI scores, severe cartilage erosion, and exposure of subchondral bone. However, mEVs treatment protected against cartilage degeneration, and the HC/CC ratio increased by 219.92% compared to that of the DMM+PBS group. As described above, the articular cartilage showed local wear at 4 weeks post-surgery, and the severe cartilage tissue erosion and exposure of subchondral bone could be observed at 8 weeks post-surgery. mEVs treatment could reduce cartilage tissue damage and attenuate the development of cartilage erosion after DMM surgery.
Figure 3.
Effects of mEVs on articular cartilage structure in DMM-induced mice. (A) Hematoxylin–eosin and safranin o-fast green staining at 4, 6, and 8 weeks after DMM surgery. Scale bars: 100 μm. The tibial plateau cartilage thickness in the weight-bearing area is indicated by red double-headed arrows. The black arrow indicates the tidemark (boundary of HC and CC) of cartilage. HC, hyaline cartilage; CC, calcified cartilage; SBP, subchondral bone plate. (B) The joint lesions were graded on a scale of 0–6 using the OARSI scoring system. Data were presented as the mean ± SD (n = 5). (C–E) Quantification of cartilage thickness of all groups. All data are shown as means ± SD deviation (n = 3). ** p < 0.01, *** p < 0.001 vs. DMM+PBS group.
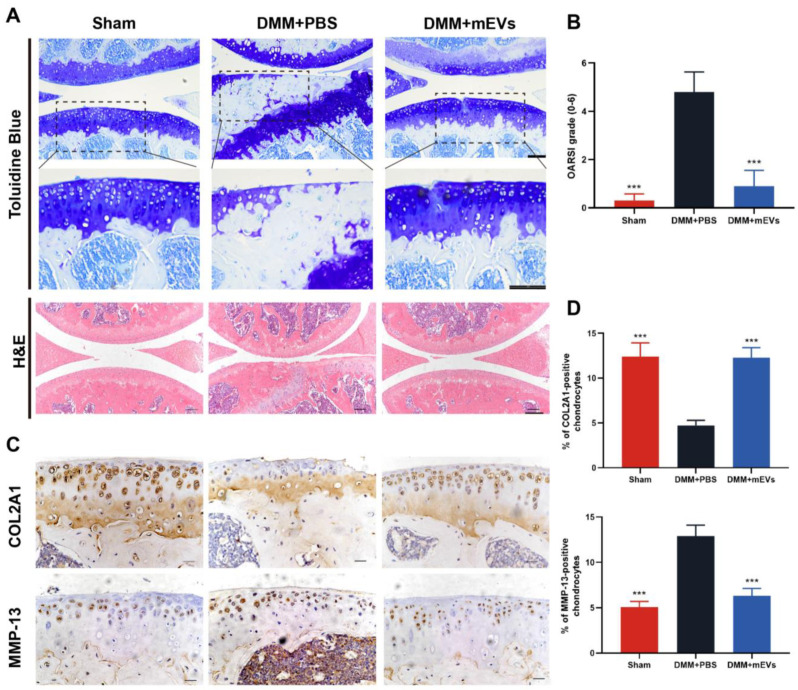
Toluidine blue and hematoxylin–eosin staining showed cartilage degeneration at 10 weeks post-surgery (Figure 4). The DMM+PBS group displayed severely calcified cartilage and extensive loss of cartilage, whereas the mEVs treatment groups showed more complete integration of cartilage with a smooth surface (Figure 4A). The OARSI scoring was taken to quantify the severity of cartilage damage, and the result showed that the mEVs treatment group significantly relieved the cartilage lesion compared with the DMM+PBS group. To further examine the effect of mEVs on cartilage degeneration, an immunohistochemical experiment was conducted to assess the change in COL2A1 and MMP-13 protein levels in articular cartilage. As shown in Figure 4C,D, the DMM-induced decrease in COL2A1 could be largely reversed by mEVs. The mEVs treatment strongly reduced the percentage of MMP-13-positive chondrocytes in DMM-induced mice compared with that of the DMM+PBS group. These results indicated that mEVs treatment in DMM-induced mice promoted chondrocyte anabolism and inhibited its catabolism effectively, which prevented erosion of the cartilage matrix after DMM surgery.
Figure 4.
Effects of mEVs on cartilage degradation in DMM-induced mice. (A) Toluidine blue and hematoxylin–eosin-stained sections of knee joints. Scale bars: 100 μm. The dashed black box indicates the magnified region. (B) The joint lesions were graded on a scale of 0–6 using the OARSI scoring system. Data were presented as the mean ± SD (n = 5). (C,D) Immunohistochemistry analysis of COL2A1 and MMP-13 in sections of tibial plateau of knee joints. Scale bar, 25 μm. All data are shown as means ± SD deviation (n = 5). *** p < 0.001 vs. DMM+PBS group.
3.3. mEVs Rectify Gut Microbiota Imbalance in DMM-Induced Mice
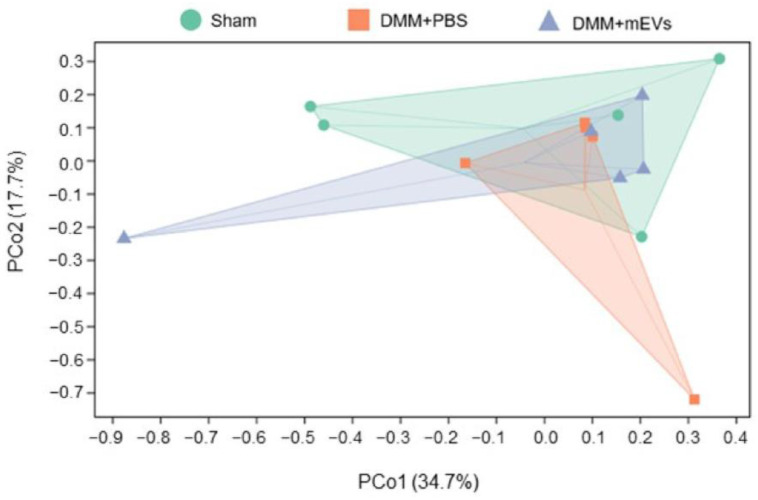
To characterize the gut microbiota in DMM-induced mice under different treatments, microbiota in fecal samples were analyzed from three groups of mice (sham group, DMM+PBS group, DMM+mEVs group) by 16S rRNA sequencing. The results showed that the sham group and DMM+PBS group had distinct intestinal microbial populations, and mEVs tended to reverse the key changes induced by DMM surgery. Specifically, a higher number of ASVs/OTU was observed in DMM-induced mice compared with that of sham-operated or mEVs-treated mice (Figure 5). As reflected by the indexes of Chao 1, Observed_species, Shannon, Simpson, Faith_pd, and Pielou_e, the microbiota in DMM-induced mice showed a higher α-diversity compared with that of the sham group and DMM+mEVs group, but there were no significant differences (p > 0.05) (Table 1). Furthermore, β-diversity was observed by the principle coordinate analysis (PCoA). As shown in Figure 6, PCoA revealed that the sham group and the DMM+mEVs group had more similar microbial community, but the differences among the three groups were not significant. These results indicated that mEVs treatment decreased the diversity of gut microbiota and thereby reversed the dysregulated microbiota community structure of DMM-induced mice.
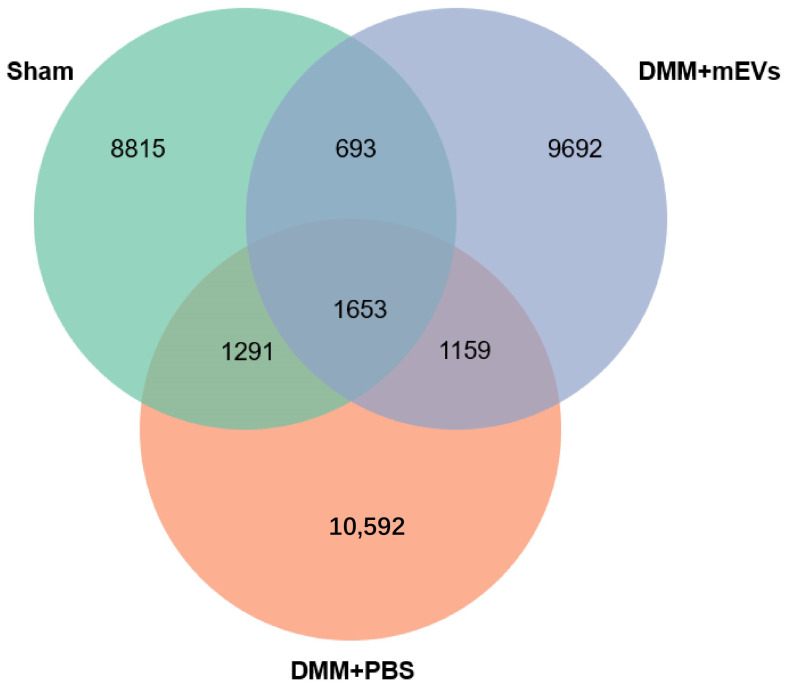
Figure 5.
Venn diagram of ASV/OUT in the feces. The numbers of ASVs/OTUs were evaluated by 16S rRNA gene V3–V4 pyrosequencing reads. ASVs, amplicon sequence variants; OTUs, operational taxonomic units.
Table 1.
Alpha diversity indexes were calculated with QIME2 according to AVS/OUT numbers of each group (n = 5).
| Alpha Diversity Indexes/Groups | Sham | DMM+PBS | DMM+mEVs |
|---|---|---|---|
| Chao1 | 3460.11 ± 908.70 | 4081.75 ± 1749.60 | 3319.48 ± 1632.58 |
| Observed_species | 3080.24 ± 75.15 | 3747.52 ± 1571.90 | 3065.48 ± 1635.47 |
| Shannon | 7.64 ± 0.38 | 8.64 ± 1.79 | 7.64 ± 2.33 |
| Simpson | 0.94 ± 0.03 | 0.96 ± 0.06 | 0.93 ± 0.09 |
| Faith_pd | 179.63 ± 36.40 | 190.00 ± 63.78 | 180.54 ± 54.26 |
| Pielou_e | 0.66 ± 0.03 | 0.73 ± 0.10 | 0.66± 0.16 |
Data are presented as the mean ± SD (n = 5).
Figure 6.
β-diversity evaluated using weighted_unifrac based PCoA (n = 5).
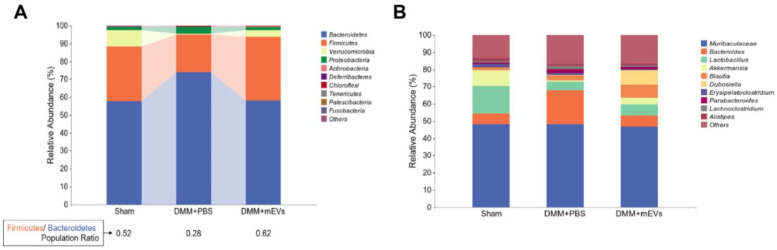
To further investigate the changes in intestinal flora composition in DMM-induced mice with or without mEVs treatment, taxonomic composition analysis was used to determine the differences in bacterial abundance at different levels. We first observed the changes in bacterial abundance in different treatment groups at the phylum and genus levels. It has been reported that OA patients have a higher relative abundance of Bacteroides and a lower relative abundance of Firmicutes compared to healthy people [19]. As shown in Figure 7A, it was observed that Bacteroidetes and Firmicutes were the main phylum in mice, Bacteroidetes were increased and Firmicutes were decreased in DMM-induced mice, whereas mEVs treatment reversed those changes. Furthermore, mEVs treatment increased the Firmicutes/Bacteroidetes (F/B) ratio in DMM-induced mice. DMM surgery resulted in an increase in Proteobacteria and a decrease in Firmicutes at the phylum. At the genus level, the relative abundance of Bacteroides was highest in DMM+PBS group among three groups, while the DMM+PBS group had the lowest relative abundance of Lactobacillus and Akkermansia among the three groups, but there were no significant differences in relative abundance of Muribaculaceae among the three groups (Figure 7B).
Figure 7.
The relative abundance of phylum/genus in each experimental group. (A) The relative abundance of phylum in each experimental group. The ratio of the relative abundance of Firmicutes to Bacteroidetes for each group was calculated. (B) The relative abundance of genus in each experimental group.
At the family and genus level, the ratio of certain flora was also altered in DMM-induced mice. The relative abundance of Ruminococcaceae and Akkermansiaceae was decreased, and Bacteroidaceae and Enterobacteriaceae were increased at the family level in DMM-induced mice. Additionally, it has been demonstrated that the abundance of Enterobacter increased as arthritis progressed in CIA-susceptible mice [21]. In DMM-induced mice, Enterobacteriaceae was observed to be richer compared to the sham group and DMM+mEVs group. At the genus level, DMM-induced mice displayed a reduction of Lactobacillus, Akkermansia, and Lachnospiraceae_NK4A136_group, and the abundance of Bacteroides increased in DMM-induced mice, which were partially reversed by mEVs treatment (Table 2). In addition, Akkermansia_muciniphila and Muribaculaceae_mouse_gut decreased in the feces of DMM-induced mice (Supplementary Material Figure S3).
Table 2.
Relative abundance of several bacteria at the family/genus level (n = 5).
| Family/Genus | Strains | Sham (%) | DMM+PBS (%) | DMM+mEVs (%) |
|---|---|---|---|---|
| Family | Bacteroidaceae | 0.06 ± 0.07 | 0.20 ± 0.29 | 0.06 ± 0.04 |
| Ruminococcaceae | 0.03 ± 0.03 | 0.02 ± 0.01 | 0.03 ± 0.03 | |
| Akkermansiaceae | 0.09 ± 0.16 | 0.01 ± 0.01 | 0.04 ± 0.06 | |
| Enterobacteriaceae | 0.00 ± 0.00 | 0.02 ± 0.04 | 0.00 ± 0.01 | |
| Genus | Bacteroides | 0.06 ± 0.07 | 0.20 ± 0.29 | 0.06 ± 0.04 |
| Lactobacillus | 0.16 ± 0.21 | 0.05 ± 0.06 | 0.07 ± 0.12 | |
| Akkermansia | 0.09 ± 0.16 | 0.01 ± 0.01 | 0.04 ± 0.06 | |
| Lachnospiraceae_NK4A136_group | 0.02 ± 0.03 | 0.00 ± 0.00 | 0.02 ± 0.02 |
Data are presented as the mean ± SD (n = 5).
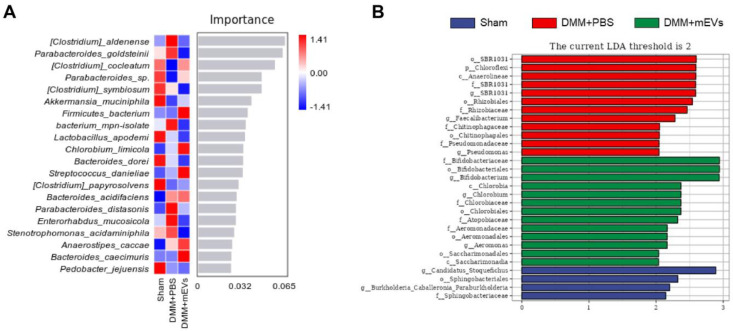
In addition, random forest analysis and line discriminant analysis (LDA) can detect species that differ significantly in abundance among groups (Figure 8). Random forest analysis was applied to screen out the indicative species. It was observed that Clostridium_aldenense, Parabacteroides_goldsteinii, Clostridium_cocleatum and Parabacteroides_sp. had high importance that differed in abundance among groups. Clostridium_aldenense and Parabacteroides_goldsteinii were more abundant in the DMM+PBS group, while Clostridium_cocleatum and Parabacteroides_sp. were more abundant in the sham group and DMM+mEVs group (Figure 8A). LDA analysis revealed that key phylotypes were determined as distinguished biomarkers. It was found that some bacteria were overrepresented in the DMM+PBS group, such as SBR1031 order, Chloroflexi phylum, and Anaerolineae class, which might be the potential biomarkers and intervention targets of OA therapy (Figure 8B and Figure S4). Furthermore, Bifidobacteriaceae family, Bifidobacteriales order, and Bifidobacterium genus were overrepresented in the DMM+mEVs group, and Candidatus_Stoquefichus genus was overrepresented in the sham group (Figure 8B).
Figure 8.
Specific species were altered in each experimental group. (A) Bar plot species importance at the species level performed by random forest analysis. (B) Differentially enriched gut microbiota in each group of mice at the species level by linear discriminant analysis (LDA).
4. Discussion
In the present study, we investigated the therapeutic effects of mEVs on OA in DMM-induced mice. EVs can be considered regulators of cell communication to transfer bioactive cargos (lipids, miRNAs, proteins) to elicit diverse cellular responses in recipient cells [22]. To date, mEVs have been used in many diseases, but only one study has reported its protection on cartilage from OA patients. It was demonstrated that mEVs had a potential therapy on human osteoarthritic joints by reducing the expression of cartilage-destructive enzymes and pro-inflammatory cytokines in vitro [10]. In our initial observation, it was found that oral administration of mEVs alleviated the progress of cartilage degeneration effectively in the DMM-induced OA model. In addition, a single dose of mEVs was used to interfere with DMM-induced mice in this study; more doses should be selected to fully elucidate the inhibitory effect of mEVs on cartilage degeneration in subsequent studies.
The surgical model of DMM was used to explore the effect of mEVs on OA in vivo. Most notably, it is the first study, to the best of our knowledge, to evaluate the prevention effect of mEVs in the DMM-induced OA model. One of the important findings in our study is that oral administration of mEVs prevents the progress of cartilage degeneration effectively in the DMM-induced OA model. Hematoxylin–eosin, safranin o-fast green and toluidine blue staining revealed that mEVs significantly inhibited the extracellular matrix loss and improved cartilage destruction. By the end of the 12-week treatment, mEV-treated joints showed remarkable restoration of cartilage structure destruction and cartilage matrix degradation that were comparable to that of the sham group.
mEVs not only protect cartilage from damage, but also reshape and restore the composition of gut microbiota in DMM-induced mice. The microbiota is a potential driver of chronic innate activation in OA, but few studies have examined gut microbiota perturbations in OA. In a population-based cohort, OA was found to be associated with a large number of gut microbiota features, and the abundance of gut microbes was negatively associated with the prevalence of OA [23]. A recent paper reported that four bacterial clades were associated with knee pain, including Bacilli class, Lactobacillales order, Streptococcaceae family, and Streptococcus genus [24]. Furthermore, the abundant diversity of bacterial nucleic acids has been identified in the synovial fluid and cartilage samples from knee OA patients, such as Lanchospiraeae family and Coriobacteriales order [25,26]. These evidences indicated that gut microbiota might influence joint biology in OA. In line with previous reports, our findings suggested that the composition of gut microbiota was disturbed in the DMM-induced OA model, but mEVs could inhibit the development of OA by reshaping gut microbiota.
There was an interesting finding that the DMM+PBS group showed a higher α-diversity compared with that of the sham group and DMM+mEVs group. Similarly, the number of observed species was slightly higher in the bacteriome of OA patients compared with that in healthy controls via whole-metagenome shotgun sequencing, but there were no significant differences [27]. Another research study also reported that α-diversity was higher in the OA rats than that in control rats [28]. The mechanism of this phenomenon needs to be further explored. The DMM+PBS group had higher relative abundance of Proteobacteria and lower relative abundance of Firmicutes and Ruminococcaceae compared to the other groups. It has been reported that the enrichment of Proteobacteria or decrease in Firmicutes is associated with the plasma levels of pro-inflammatory cytokines (e.g., IL-6, IL-8) [29], and Ruminococcaceae has been proven to be negatively correlated with the OARSI scores and pro-inflammatory factors (e.g., IL-6, IL-1β) [30]. Moreover, the F/B ratio of DMM+PBS group was the lowest among three groups, while The F/B ratio has been considered to be associated with maintaining normal intestinal homeostasis, and changes in this ratio can lead to a variety of pathologies, such as obesity and bowel inflammation [31]. Lactobacillus has been reported to be mainly enriched in OA monkeys [32]. In contrast, in this study, Lactobacillus decreased in the feces of DMM-induced mice. Collectively, these results suggested that oral gavage of mEVs could restore the taxonomic imbalance of gut microbiota induced by DMM surgery. However, there were no significant differences among the three groups, likely due to small group sizes, and the exact mechanisms need further study.
There was some emerging evidence suggesting the correlation between microbiome and the pathogenesis of OA, and the ‘two-hit’ model of OA pathogenesis and enhancement was proposed. One ’hit’ was provided by activation of the adverse gut microbiome of innate immunity, and the other ’hit’ was underlying joint damage [33]. It emphasized that the initiation and perpetuation of OA were associated with innate immune activity, including macrophage-associated inflammatory responses, TLR activation, and complement activation [34,35,36]. TLR2 and TLR4, upregulated in OA cartilage, are stimulated by lipopolysaccharide (LPS) from an outer membrane component of Gram-negative bacteria [37]. In human OA cartilage, an increased fraction of Gram-negative constituent bacterial DNA was detected as well as increased Kyoto Encyclopedia of Gene and Genomes (KEGG) pathways associated with LPS biosynthesis in OA [26]. Bacteroidetes are a phylum of Gram-negative bacteria that impact digestion, immunity, and resistance to infection [38]. In agreement with these studies, our data demonstrated an increased abundance of Bacteroidetes in the DMM+PBS group, while oral administration of mEVs restored its abundance to a normal level. The abundance of Enterobacteriaceae (Gram-negative bacteria) increased in DMM-induced mice, which is positively correlated with the progression of arthritis [21]. Based on the above, we hypothesize that exacerbation of OA may be associated with increased LPS produced by abundant Gram-negative bacteria. In this hypothesis, mEVs may inhibit the production of LPS by altering the composition of gut microbiota. It has not been tested in our study and needs to be explored in further experiments.
Moreover, oral administration of mEVs upregulated the abundance of some beneficial gut microbes, such as Ruminococcaceae, Lachnospiraceae_NK4A136_group, and Akkermansia_muciniphila (A. muciniphila). It has been demonstrated that Ruminococcaceae was negatively correlated with the OARSI scores, and we found that the DMM+PBS group had a lower abundance of Ruminococcaceae with higher OARSI scores. A. muciniphila is a promising probiotic that can stimulate mucosal immunity to prevent pathogenic invasion and promote the thickening of the colonic mucosa [39]. The results showed that the special bacteria were affected by mEVs, but how mEVs affect OA progression via regulating gut microbial metabolism remains to be elucidated.
At present, few studies have focused on the effect of mEVs on OA. Our research initially demonstrates that oral administration of mEVs can inhibit cartilage degeneration and restore disordered gut microbiota in DMM-induced mice, but the mechanism needs to be further investigated. Meanwhile, mEV contains abundant functional cargo, such as microRNAs and proteins. It is necessary to explore which substances play the main role in alleviating OA.
5. Conclusions
This was the first report that mEVs can attenuate cartilage degeneration by suppressing the expression of catabolism, enhancing matrix synthesis, and reshaping the gut microbiota in DMM-induced OA mice. Oral administration of mEVs might be a potential approach in the treatment of OA.
Acknowledgments
Schematic illustration was created with Adobe illustrator and Medpeer.com.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/nu15030747/s1, Table S1: The OARSI scoring system; Figure S1: The serum CTX-Ⅱ levels were determined by ELISA (n = 3); Figure S2: The serum COMP levels were determined by ELISA (n = 3); Figure S3: The relative abundance of species in each experimental group; Figure S4: Cladogram based on linear discriminant analysis effect size (LEfSe) analysis showing community composition of the gut microbiota in mice.
Author Contributions
Conceptualization, Q.L. and H.Y.; methodology, Q.L.; Software, Q.L., J.L. and H.H.; validation, Q.L., H.H., J.L. and T.Z.; formal analysis, Q.L. and H.H.; resources, H.Y.; data curation, Q.L., T.Z. and Y.Y.; writing—original draft preparation, Q.L.; writing—review and editing, Y.Y., X.T. and Z.Z.; visualization, X.T.; supervision, Z.Z. and H.Y.; funding acquisition, H.Y. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
All animal experiments were approved by the Committee on the Ethics of Animal Experiments of Ocean University of China (permission number: spxy2021082701). All procedures were performed under deep anesthesia. After deep anesthesia, mice were sacrificed by cervical dislocation after eyeball blood was collected, and then, knee joints were harvested.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest. All authors consent to the publication of this manuscript.
Funding Statement
This work was supported by the Key Program of Natural Science Foundation of Shandong Province in China (no. ZR2020KC009).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Robinson W.H., Lepus C.M., Wang Q., Raghu H., Mao R., Lindstrom T.M., Sokolove J. Low-grade inflammation as a key mediator of the pathogenesis of osteoarthritis. Nat. Rev. Rheumatol. 2016;12:580–592. doi: 10.1038/nrrheum.2016.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Safiri S., Kolahi A.A., Smith E., Hill C., Bettampadi D., Mansournia M.A., Hoy D., Ashrafi-Asgarabad A., Sepidarkish M., Almasi-Hashiani A., et al. Global, regional and national burden of osteoarthritis 1990–2017: A systematic analysis of the Global Burden of Disease Study 2017. Ann. Rheum. Dis. 2020;79:819–828. doi: 10.1136/annrheumdis-2019-216515. [DOI] [PubMed] [Google Scholar]
- 3.Bennell K.L., Paterson K.L., Metcalf B.R., Duong V., Eyles J., Kasza J., Wang Y., Cicuttini F., Buchbinder R., Forbes A., et al. Effect of Intra-articular Platelet-Rich Plasma vs Placebo Injection on Pain and Medial Tibial Cartilage Volume in Patients With Knee Osteoarthritis: The RESTORE Randomized Clinical Trial. Jama. 2021;326:2021–2030. doi: 10.1001/jama.2021.19415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang Y., Xu X., Li X., Xiong J., Li B., Duan L., Wang D., Xia J. Chondrocyte-Targeted MicroRNA Delivery by Engineered Exosomes toward a Cell-Free Osteoarthritis Therapy. ACS Appl. Mater. Interfaces. 2020;12:36938–36947. doi: 10.1021/acsami.0c10458. [DOI] [PubMed] [Google Scholar]
- 5.Zhong J., Xia B., Shan S., Zheng A., Zhang S., Chen J., Liang X.-J. High-quality milk exosomes as oral drug delivery system. Biomaterials. 2021;277:121126. doi: 10.1016/j.biomaterials.2021.121126. [DOI] [PubMed] [Google Scholar]
- 6.Samuel M., Fonseka P., Sanwlani R., Gangoda L., Chee S.H., Keerthikumar S., Spurling A., Chitti S.V., Zanker D., Ang C.-S., et al. Oral administration of bovine milk-derived extracellular vesicles induces senescence in the primary tumor but accelerates cancer metastasis. Nat. Commun. 2021;12:3950. doi: 10.1038/s41467-021-24273-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tong L., Hao H., Zhang Z., Lv Y., Liang X., Liu Q., Liu T., Gong P., Zhang L., Cao F., et al. Milk-derived extracellular vesicles alleviate ulcerative colitis by regulating the gut immunity and reshaping the gut microbiota. Theranostics. 2021;11:8570–8586. doi: 10.7150/thno.62046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiklander O.P., Nordin J.Z., O’Loughlin A., Gustafsson Y., Corso G., Mäger I., Vader P., Lee Y., Sork H., Seow Y., et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J. Extracell. Vesicles. 2015;4:26316. doi: 10.3402/jev.v4.26316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arntz O.J., Pieters B.C., Oliveira M.C., Broeren M.G., Bennink M.B., de Vries M., van Lent P.L., Koenders M.I., van den Berg W.B., van der Kraan P.M., et al. Oral administration of bovine milk derived extracellular vesicles attenuates arthritis in two mouse models. Mol. Nutr. Food Res. 2015;59:1701–1712. doi: 10.1002/mnfr.201500222. [DOI] [PubMed] [Google Scholar]
- 10.Pieters B.C.H., Arntz O.J., Aarts J., Feitsma A.L., van Neerven R.J.J., van der Kraan P.M., Oliveira M.C., van de Loo F.A.J. Bovine Milk-Derived Extracellular Vesicles Inhibit Catabolic and Inflammatory Processes in Cartilage from Osteoarthritis Patients. Mol. Nutr. Food Res. 2022;66:e2100764. doi: 10.1002/mnfr.202100764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu X.H., Yang Y.Q., Cao R.R., Bo L., Lei S.F. The causal role of gut microbiota in development of osteoarthritis. Osteoarthr. Cartil. 2021;29:1741–1750. doi: 10.1016/j.joca.2021.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Ling Z., Xiao H., Chen W. Gut Microbiome: The Cornerstone of Life and Health. Adv. Gut Microbiome Res. 2022;2022:9894812. doi: 10.1155/2022/9894812. [DOI] [Google Scholar]
- 13.Morris J.L., Letson H.L., Gillman R., Hazratwala K., Wilkinson M., McEwen P., Dobson G.P. The CNS theory of osteoarthritis: Opportunities beyond the joint. Semin. Arthritis Rheum. 2019;49:331–336. doi: 10.1016/j.semarthrit.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Ulici V., Kelley K.L., Azcarate-Peril M.A., Cleveland R.J., Sartor R.B., Schwartz T.A., Loeser R.F. Osteoarthritis induced by destabilization of the medial meniscus is reduced in germ-free mice. Osteoarthr. Cartil. 2018;26:1098–1109. doi: 10.1016/j.joca.2018.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O-Sullivan I., Natarajan Anbazhagan A., Singh G., Ma K., Green S.J., Singhal M., Wang J., Kumar A., Dudeja P.K., Unterman T.G., et al. Lactobacillus acidophilus Mitigates Osteoarthritis-Associated Pain, Cartilage Disintegration and Gut Microbiota Dysbiosis in an Experimental Murine OA Model. Biomedicines. 2022;10:1298. doi: 10.3390/biomedicines10061298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rios J.L., Bomhof M.R., Reimer R.A., Hart D.A., Collins K.H., Herzog W. Protective effect of prebiotic and exercise intervention on knee health in a rat model of diet-induced obesity. Sci. Rep. 2019;9:3893. doi: 10.1038/s41598-019-40601-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schott E.M., Farnsworth C.W., Grier A., Lillis J.A., Soniwala S., Dadourian G.H., Bell R.D., Doolittle M.L., Villani D.A., Awad H., et al. Targeting the gut microbiome to treat the osteoarthritis of obesity. JCI Insight. 2018;3:e95997. doi: 10.1172/jci.insight.95997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tong L., Hao H., Zhang X., Zhang Z., Lv Y., Zhang L., Yi H. Oral Administration of Bovine Milk-Derived Extracellular Vesicles Alters the Gut Microbiota and Enhances Intestinal Immunity in Mice. Mol. Nutr. Food Res. 2020;64:e1901251. doi: 10.1002/mnfr.201901251. [DOI] [PubMed] [Google Scholar]
- 19.Liu S., Li G., Zhu Y., Xu C., Yang Q., Xiong A., Weng J., Yu F., Zeng H. Analysis of gut microbiome composition, function, and phenotype in patients with osteoarthritis. Front. Microbiol. 2022;13:980591. doi: 10.3389/fmicb.2022.980591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glasson S.S., Chambers M.G., Van Den Berg W.B., Little C.B. The OARSI histopathology initiative-recommendations for histological assessments of osteoarthritis in the mouse. Osteoarthr. Cartil. 2010;18:S17–S23. doi: 10.1016/j.joca.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 21.Liu X., Zeng B., Zhang J., Li W., Mou F., Wang H., Zou Q., Zhong B., Wu L., Wei H., et al. Role of the Gut Microbiome in Modulating Arthritis Progression in Mice. Sci. Rep. 2016;6:30594. doi: 10.1038/srep30594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Somiya M., Yoshioka Y., Ochiya T. Biocompatibility of highly purified bovine milk-derived extracellular vesicles. J. Extracell. Vesicles. 2018;7:1440132. doi: 10.1080/20013078.2018.1440132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson M.A., Verdi S., Maxan M.E., Shin C.M., Zierer J., Bowyer R.C.E., Martin T., Williams F.M.K., Menni C., Bell J.T., et al. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat. Commun. 2018;9:2655. doi: 10.1038/s41467-018-05184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boer C.G., Radjabzadeh D., Medina-Gomez C., Garmaeva S., Schiphof D., Arp P., Koet T., Kurilshikov A., Fu J., Ikram M.A., et al. Intestinal microbiome composition and its relation to joint pain and inflammation. Nat. Commun. 2019;10:4881. doi: 10.1038/s41467-019-12873-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y., Chen B., Li S., Yang L., Zhu D., Wang Y., Wang H., Wang T., Shi B., Gai Z., et al. Detection and characterization of bacterial nucleic acids in culture-negative synovial tissue and fluid samples from rheumatoid arthritis or osteoarthritis patients. Sci. Rep. 2018;8:14305. doi: 10.1038/s41598-018-32675-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunn C.M., Velasco C., Rivas A., Andrews M., Garman C., Jacob P.B., Jeffries M.A. Identification of Cartilage Microbial DNA Signatures and Associations With Knee and Hip Osteoarthritis. Arthritis Rheumatol. 2020;72:1111–1122. doi: 10.1002/art.41210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C., Zhang Y., Yao X., Li S., Wang G., Huang Y., Yang Y., Zhang A., Liu C., Zhu D., et al. Characterizations of the Gut Bacteriome, Mycobiome, and Virome in Patients with Osteoarthritis. Microbiol. Spectr. 2022. online ahead of print . [DOI] [PMC free article] [PubMed]
- 28.Lan H., Hong W., Qian D., Peng F., Li H., Liang C., Du M., Gu J., Mai J., Bai B., et al. Quercetin modulates the gut microbiota as well as the metabolome in a rat model of osteoarthritis. Bioengineered. 2021;12:6240–6250. doi: 10.1080/21655979.2021.1969194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmed N., Biagi E., Nylund L., Candela M., Ostan R., Bucci L., Pini E., Nikkïla J., Monti D., Satokari R., et al. Through Ageing, and Beyond: Gut Microbiota and Inflammatory Status in Seniors and Centenarians. PLoS ONE. 2010;5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Z., Chen J., Li B., Zeng B., Chou C.-H., Zheng X., Xie J., Li H., Hao Y., Chen G., et al. Faecal microbiota transplantation from metabolically compromised human donors accelerates osteoarthritis in mice. Ann. Rheum. Dis. 2020;79:646–656. doi: 10.1136/annrheumdis-2019-216471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stojanov S., Berlec A., Štrukelj B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel disease. Microorganisms. 2020;8:1715. doi: 10.3390/microorganisms8111715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan Y., Yi X., Duan Y., Jiang B., Huang T., Inglis B.M., Zheng B., Si W. Alteration of the gut microbiota in rhesus monkey with spontaneous osteoarthritis. BMC Microbiol. 2021;21:328. doi: 10.1186/s12866-021-02390-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Z., Kraus V.B. Does lipopolysaccharide-mediated inflammation have a role in OA? Nat. Rev. Rheumatol. 2016;12:123–129. doi: 10.1038/nrrheum.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scanzello C.R., Plaas A., Crow M.K. Innate immune system activation in osteoarthritis: Is osteoarthritis a chronic wound? Curr. Opin. Rheumatol. 2008;20:565–572. doi: 10.1097/BOR.0b013e32830aba34. [DOI] [PubMed] [Google Scholar]
- 35.Bondeson J., Wainwright S.D., Lauder S., Amos N., Hughes C.E. The role of synovial macrophages and macrophage-produced cytokines in driving aggrecanases, matrix metalloproteinases, and other destructive and inflammatory responses in osteoarthritis. Arthritis Res. Ther. 2006;8:R187. doi: 10.1186/ar2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Q., Rozelle A.L., Lepus C.M., Scanzello C.R., Song J.J., Larsen D.M., Crish J.F., Bebek G., Ritter S.Y., Lindstrom T.M., et al. Identification of a central role for complement in osteoarthritis. Nat. Med. 2011;17:1674–1679. doi: 10.1038/nm.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H.A., Cho M.-L., Choi H.Y., Yoon C.S., Jhun J.Y., Oh H.J., Kim H.-Y. The catabolic pathway mediated by Toll-like receptors in human osteoarthritic chondrocytes. Arthritis Rheum. Off. J. Am. Coll. Rheumatol. 2006;54:2152–2163. doi: 10.1002/art.21951. [DOI] [PubMed] [Google Scholar]
- 38.Russell A.B., Wexler A.G., Harding B.N., Whitney J.C., Bohn A.J., Goo Y.A., Tran B.Q., Barry N.A., Zheng H., Peterson S.B., et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe. 2014;16:227–236. doi: 10.1016/j.chom.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Vos W.M. Microbe Profile: Akkermansia muciniphila: A conserved intestinal symbiont that acts as the gatekeeper of our mucosa. Microbiology. 2017;163:646–648. doi: 10.1099/mic.0.000444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.