Abstract
Glucosinolates (GSLs) are secondary plant metabolites that are enriched in rapeseed and related Brassica species, and they play important roles in defense due to their anti-nutritive and toxic properties. Here, we conducted a genome-wide association study of six glucosinolate metabolites (mGWAS) in rapeseed, including three aliphatic glucosinolates (m145 gluconapin, m150 glucobrassicanapin and m151 progoitrin), one aromatic glucosinolate (m157 gluconasturtiin) and two indole glucosinolates (m165 indolylmethyl glucosinolate and m172 4-hydroxyglucobrassicin), respectively. We identified 113 candidate intervals significantly associated with these six glucosinolate metabolites. In the genomic regions linked to the mGWAS peaks, 187 candidate genes involved in glucosinolate biosynthesis (e.g., BnaMAM1, BnaGGP1, BnaSUR1 and BnaMYB51) and novel genes (e.g., BnaMYB44, BnaERF025, BnaE2FC, BnaNAC102 and BnaDREB1D) were predicted based on the mGWAS, combined with analysis of differentially expressed genes. Our results provide insight into the genetic basis of glucosinolate biosynthesis in rapeseed and should facilitate marker-based breeding for improved seed quality in Brassica species.
Keywords: Brassica napus, glucosinalates, mGWAS, candidate genes
1. Introduction
Glucosinolates (GSLs) are secondary metabolites comprising sulfur and nitrogen that are specially produced in Brassica species, providing these plants with their pungent odor [1,2,3]. GSLs also play important roles in plant defense against pests and in human health [4,5]. However, high levels of GSLs affect the quality of seed oil and the nutritional value of seed meal from rapeseed. In addition, some hydrolysates produced from GSLs, such as oxazolidin-2-thione, have toxic effects on human and animal health [6,7,8,9,10]. Thus, further reducing glucosinolate levels in seed meal is an important goal in rapeseed breeding.
Generally, GSLs share the same basic structure, including β-D-thiosaccharide and (Z)-n-hydroxamic sulfate, but have variable R-side chain groups due to the precursor amino acids [11]. The lengths and modifications of these variable R-side chain groups determine the chemical properties of each GSL. Correspondingly, GSLs can be divided into three types based on the source of precursor amino acids, for example, aliphatic GSLs derived from alanine (Ala), leucine (Leu), isoleucine (Ile), methionine (Met) and valine (Val); indole GSLs derived from tryptophan (Trp); and aromatic GSLs derived from phenylalanine (Phe) and tyrosine (Tyr) [12,13,14]. To date, over 200 GSLs have been discovered [15,16], which were usually synthesized through three steps. The first step is the lengthening of the progenitor amino acid side chain. This mainly occurs during the biosynthesis of aliphatic and aromatic GSLs, in which amino acids form 2-oxic acids via the action of branched-chain amino acid transaminase (BCAT) and then undergo elongation by enzymes, such as methylthinomalate synthase (MAM), isopropyl malate isomerase (IPMI) and isopropyl malate dehydrogenase (IMD). The extended 2-oxic acid molecule is converted into the corresponding amino acid by BCAT [5,17,18,19]. The second step forms the glucosinolate core structure. The extended amino acid is converted via the action of cytochrome P450 79s (CYP79s) to aldoxime, which is oxidized into the activated forms by CYP83s. These activated forms could bind to glutathione that is converted to first thiohydroxamic acid by glutathione S-transferases (GSTFs), γ-glutamyl peptide (GGP1) and C-S lyase (Super root 1 (SUR1)), and then to the GSL core structure by glucosyltransferases (UGT74s) and sulfotransferases (SOTs) [5,20,21,22,23,24]. The last step involves side chain modification of the core structure, primarily through hydroxylation, sulfation, glycosylation, desaturation and methylation of R groups [25]. The core structures of aliphatic GSLs are oxidized and hydroxylated by a series of enzymes, including flavin monooxygenase (FMOGS-OX) and alkenyl hydroxalkyl-producing proteins (AOPs) for aliphatic GSLs, 2-oxoglutarate-dependent dioxygenase (GS-OH) for aromatic GSLs and CYP81Fs and indole glucosinolate O-methyltransferases (IGMTs) for indole GSLs [26,27,28].
In Arabidopsis, the glucosinolate biosynthesis pathway is one of the best characterized specialized metabolite pathways [12]. Progress has also been made in mining many genes associated with the GSL pathway. Many catalytic enzymes that function in the biosynthesis of different GSLs have been identified, such as CYP79A2, CYP83B1, SUR1, UGT74B1, sulfotransferase (ST5A) and so on [29,30,31,32,33,34]. In addition, some MYB transcription factors have also been shown to regulate the expression of genes encoding enzymes in the GSL biosynthesis pathway. For example, AtMYB51 activates the transcription of GSL-biosynthesis-related genes (AtTSB1, AtCYP79B2, AtCYP79B3, AtCYP83B1 and AtST5a and so on), leading to the accumulation of GSLs [35]. The aliphatic GSL pathway is regulated by MYB28 and MYB29 [36,37], while indole GSL biosynthesis is jointly regulated by MYB34, MYB51 and MYB122 [38,39]. Several MYC transcription factors also function in GSL biosynthesis by regulating the jasmonic acid metabolism pathway [40]. Furthermore, MYC2, MYC3 and MYC4 interact with multiple MYB transcription factors (e.g., MYB28, MYB29, MYB34, MYB51 and MYB76), thereby contributing to GSL biosynthesis [40,41]. A recent study showed that WRKY33 not only directly targets the promoters of the gene MYB51 and the GSL biosynthesis gene CYP83B1 to regulate the de novo biosynthesis of indole GSLs, but also regulates the expression of genes involved in side chain modification (CYP81F2, IGMT1 and IGMT2), thereby increasing the biosynthesis of indole GSLs [42].
Rapeseed (Brassica napus L.) is used worldwide as an oil crop and source of edible vegetable oil and feed meal. Therefore, breeding rapeseed varieties with low GSL levels in seeds is an important breeding goal. To date, many rapeseed resources with low seed GSL contents have been developed in polyploid rapeseed [43,44,45]. However, due to the complexity of the rapeseed genome, identifying the GSL biosynthesis pathway in rapeseed and the mechanism leading to low GSL content in B. napus seeds has been challenging. In this study, we performed a genome-wide association study for six glucosinolate metabolites in 143 rapeseed accessions using 239,945 SNP markers obtained by resequencing [46,47,48]. Our goals were to identify candidate genes to enrich the metabolic regulatory network of the GSL biosynthesis pathway, and to lay a foundation for the breeding of B. napus with improved quality.
2. Results
2.1. Identification and Statistical Analysis of Glucosinolate Metabolites in B. napus
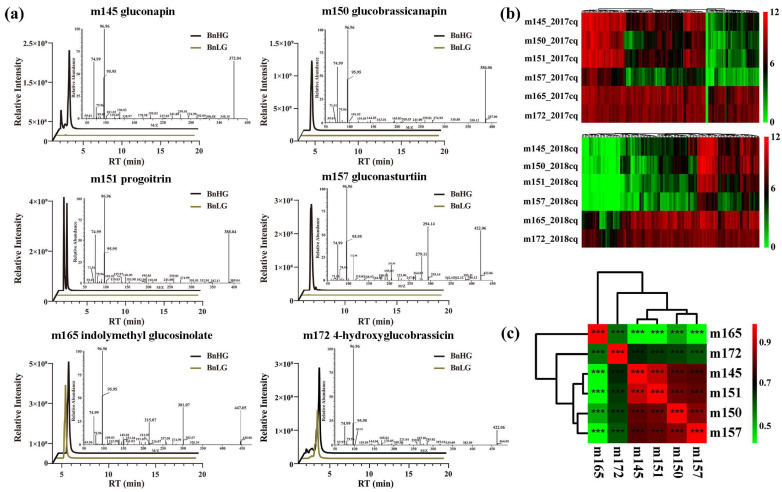
Based on ultrahigh-performance liquid chromatography-heated electrospray ionization-tandem mass spectrometry (UPLC-HESI-MS/MS) analysis, we obtained six glucosinolate metabolites with high content in rapeseed at 35 days after flowering (DAF), including three aliphatic GSLs (m145 gluconapin, m150 glucobrassicanapin and m151 progoitrin), one aromatic GSL (m157 gluconasturtiin) and two indole GSLs (m165 indolylmethyl-glucosinolate and m172 4-hydroxyglucobrassicin) (Figure 1a,b). Correlation analysis showed that the levels of three aliphatic GSLs (m145, m150 and m151) and aromatic GSL (m157) were positively correlated (|r| ≥ 0.8, p < 0.001), whereas the level of m172 showed a higher correlation with those of three aliphatic GSLs (|r| ≥ 0.6, p < 0.001) than with that of m165 (Figure 1c, Table 1 and Table S6 and Figure S4), indicating the consistency of these traits across various environments. Subsequently, we calculated the range, average, standard deviation, coefficient of variation, diversity index and heritability of the levels of the six glucosinolate metabolites (Table 1). The coefficient of variation among these metabolites ranged from 0.42 to 1.97, with an average of 1.31, and varied greatly under different years. The coefficient of variation of six glucosinolate metabolites in high glucosinolate content rapeseed was generally lower than those in low and medium glucosinolate content rapeseed (Table S7). Furthermore, genotype (G) × environment (E) interaction was also highly significant (Table 1), indicating that the accumulation levels in seeds are easily affected by environmental conditions. In addition, the order of the metabolites based on diversity index was m165 > m172 > m145 > m151 > m150 > m157, while the order based on heritability was m150 > m165 > m145 > m151 > m157 > m172 (Table 1). Therefore, all six glucosinolate metabolites not only exhibited obvious differences between BnHG and BnLG (Figure 2a), but also showed continuous variation in 2017cq and 2018cq (Figure 2b).
Figure 1.
Characterization of six glucosinolate metabolites in B. napus seeds. (a) Ion peaks of six GSLs in seeds at 35 DAP, including MS and MS/MS spectra. BnHG represents the rapeseed accession Zhongyou 821 with high total GSL contents, and BnLG represents rapeseed accession Zhongshuang 11 with low total GSL contents. (b) Heatmap showing the variable accumulation of six glucosinolate metabolites in the population at 35 DAP. (c) Correlation analysis among the levels of the six glucosinolate metabolites. The asterisks indicate significant difference (***, p < 0.001).
Table 1.
Phenotypic variations for six glucosinolate metabolite contents in an association panel of B. napus.
| Name | No. 1 | Env. 2 | Min. 3 | Max. 4 | Avg. 5 | SD 6 | CV 7 | Skew. 8 | Kurt. 9 | H’ 10 | Fge 11 | Her. 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aliphatic GSLs | m145 | 2017cq | 0.00 | 3007.14 | 456.59 | 584.04 | 1.28 | 1.59 | 2.33 | 1.37 | 57.46 ** | 0.63 |
| 2018cq | 0.00 | 943.99 | 125.82 | 222.02 | 1.76 | 2.21 | 4.12 | 1.07 | ||||
| mean | 0.00 | 1975.56 | 291.20 | 403.03 | 1.52 | 1.90 | 3.23 | 1.22 | ||||
| m150 | 2017cq | 0.00 | 2538.57 | 278.17 | 473.62 | 1.70 | 2.41 | 6.06 | 1.09 | 59.12 ** | 0.80 | |
| 2018cq | 0.00 | 1407.20 | 146.98 | 276.58 | 1.88 | 2.56 | 6.58 | 1.04 | ||||
| mean | 0.00 | 1972.88 | 212.58 | 375.10 | 1.79 | 2.48 | 6.32 | 1.06 | ||||
| m151 | 2017cq | 0.01 | 2418.94 | 374.82 | 534.24 | 1.43 | 1.63 | 1.94 | 1.20 | 86.47 ** | 0.52 | |
| 2018cq | 0.00 | 659.77 | 70.16 | 138.30 | 1.97 | 2.48 | 5.68 | 0.97 | ||||
| mean | 0.00 | 1539.35 | 222.49 | 336.27 | 1.70 | 2.06 | 3.81 | 1.08 | ||||
| aromatic GSL | m157 | 2017cq | 0.00 | 594.24 | 59.27 | 108.99 | 1.84 | 2.95 | 9.57 | 0.87 | 87.95 ** | 0.49 |
| 2018cq | 0.00 | 152.39 | 18.24 | 31.70 | 1.74 | 2.32 | 5.06 | 1.06 | ||||
| mean | 0.00 | 373.31 | 38.75 | 70.35 | 1.79 | 2.64 | 7.31 | 0.96 | ||||
| indole GSLs | m165 | 2017cq | 0.00 | 1071.04 | 435.89 | 231.28 | 0.53 | 0.57 | 0.23 | 2.09 | 18.36 ** | 0.66 |
| 2018cq | 1.24 | 763.78 | 248.89 | 183.86 | 0.74 | 0.54 | −0.33 | 1.88 | ||||
| mean | 0.62 | 917.41 | 342.39 | 207.57 | 0.63 | 0.56 | −0.05 | 1.98 | ||||
| m172 | 2017cq | 0.00 | 840.19 | 357.75 | 152.00 | 0.42 | 0.20 | 0.10 | 1.97 | 17.62 ** | 0.42 | |
| 2018cq | 8.66 | 370.66 | 159.13 | 74.23 | 0.47 | 0.31 | 0.11 | 1.90 | ||||
| mean | 4.33 | 605.42 | 258.44 | 113.12 | 0.45 | 0.26 | 0.10 | 1.93 |
cq, Chongqing environment; 1 No., metabolite ID; 2 Env., environment; 3 Min., minimum; 4 Max., maximum; 5 Avg., average; 6 SD, standard deviation; 7 CV, coefficient of variation; 8 Skew., Skewness; 9 Kurt., Kurtosis; 10 H’, Shannon–Wiener diversity index; 11 Fge, the F-values for G × E for glucosinolate content. Asterisks indicate significant differences (**, p < 0.01). 12 Her., heritability. Metabolite content is expressed in μg/g FW.
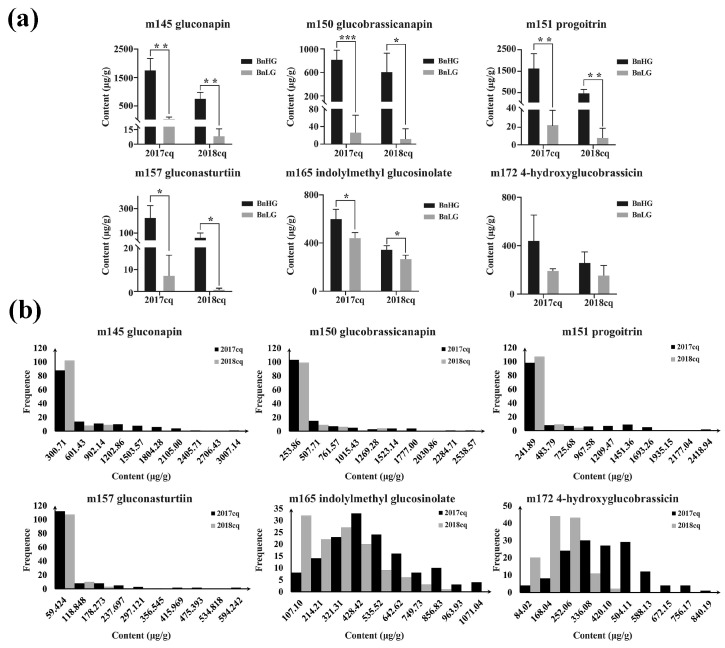
Figure 2.
Variation in the contents of six glucosinolate metabolites in the rapeseed panel in different years (2017cq and 2018cq). (a) Differences in the contents of six glucosinolate metabolites in BnHG and BnLG. Asterisks indicate significant differences (*, p < 0.05; **, p < 0.01; and ***, p < 0.001). (b) Frequency distributions of the six glucosinolate metabolites for 143 rapeseed accessions in the two years (2017cq and 2018cq).
2.2. Genome-Wide Association Study of Glucosinolate Metabolites (mGWAS)
To avoid identifying the false-positive associations in mGWAS, we selected six models to identify significant associations between phenotypes and genotypes. Based on the QQ plots of the six models (Figure S1), we performed GLM with Q model and MLM with Q+K model for the GWAS to mine more candidate genes of GSL metabolites, respectively. Herein, we used 239,945 high-quality SNPs (minor allele frequency (MAF) > 0.05 and the call frequencies <0.8) for the association analysis. The association signals were determined using a p-value < 4.17 × 10−6. The results of mGWAS for the six glucosinolate metabolites in two years (2017cq and 2018cq) and BLUP (best linear unbiased prediction) values (Figure 3, and Tables S1–S3) are summarized below.
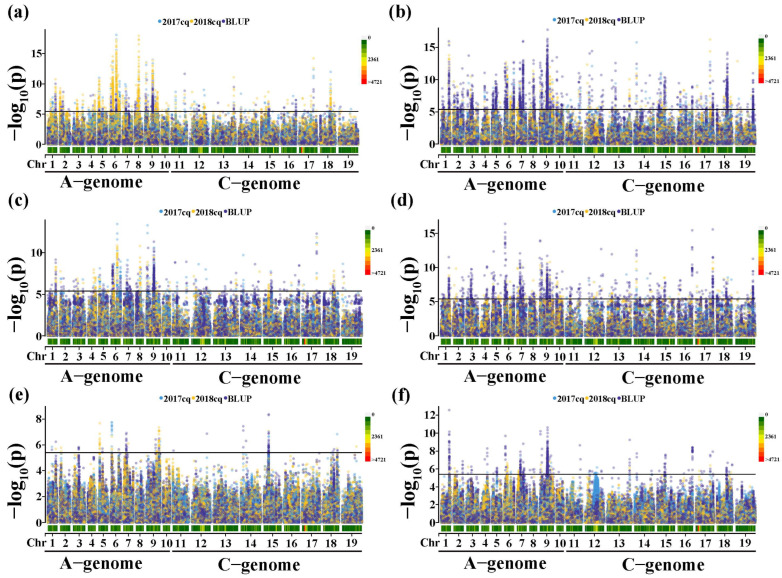
Figure 3.
Manhattan plots of association analysis for six glucosinolate metabolites. (a) Manhattan plot for m145 in 2017cq, 2018cq and BLUP values. (b) Manhattan plot for m150 in 2017cq, 2018cq and BLUP values. (c) Manhattan plot for m151 in 2017cq, 2018cq and BLUP values. (d) Manhattan plot for m157 in 2017cq, 2018cq and BLUP values. (e) Manhattan plot for m165 in 2017cq, 2018cq and BLUP values. (f) Manhattan plot for m172 in 2017cq, 2018cq and BLUP values. Different-colored spots represent different environments. The horizontal lines indicate the Bonferroni-adjusted significance threshold (4.17 × 10−6).
For the aliphatic GSL m145, we identified 1597, 2543 and 681 significantly associated SNPs based on 2017cq, 2018cqs and BLUP values, respectively, 389 SNPs of which were repeatedly detected in this study (Figure 3a, Figure S2a and Table S1). These significant SNPs primarily covered 79 candidate intervals located across the entire B. napus genome (Table S3). Importantly, two significant regions with high SNP densities were located on chromosomes A09 (17.88~22.59 Mb) and A06 (10.98~17.71 Mb) at the intervals designated qGSL-A09-5 and qGSL-A06-3, respectively. We found that the most significant SNPs (S6_16080408 and S9_18653335) could explain 49.67% and 43.75% of phenotypic variance, respectively. Therefore, we used these two intervals to predict the candidate genes to control the accumulation of the aliphatic GSL m145 in seeds (Table S3). In addition, 49 candidate regions were only associated with m145 in one or any two years and BLUP values. We believe that these remaining candidate regions represent minor-effect intervals influencing the accumulation of m145 (Table S3). These findings seem to suggest that m145 accumulation is strongly affected by environmental factors.
For the aliphatic glucosinolate, we identified m150, 2080, 2620 and 3444 significantly associated SNPs based on 2017cq, 2018cq and BLUP values, respectively, and 1431 SNPs were repeatedly detected in two years and BLUP values (Figure 3b and Figure S2b and Table S1). These significant SNPs primarily covered 94 candidate intervals across the entire B. napus genome (Table S3). Most repeated significant SNPs (1168/1431) were located in the interval qGSL-A09-5 with the most significant SNPs (S9_18653335) that explained 47.71% of phenotypic variance. In addition, the SNPs in the intervals qGSL-A01-4, qGSL-A06-2 and qGSL-C08-5 were repeatedly detected, which all explained more than 35% of the phenotypic variance (Table S1). Therefore, we believe that these regions are important interval regions for identifying the candidate genes for controlling the accumulation of m150. Furthermore, in total, 51 candidate regions are detected for m150, at least in one or any two years and BLUP values, suggesting that these remaining candidate regions are minor-effect intervals influencing the accumulation of m150 (Table S3).
For the aliphatic glucosinolate m151, we identified 977, 629 and 747 significantly associated SNPs based on 2017cq, 2018cq and BLUP values, respectively, while only 225 SNPs were repeatedly detected in this study (Figure 3c and Figure S2c and Table S1). These significant SNPs primarily covered 67 candidate intervals across the B. napus genome, except for chromosome A01 (Table S3). Among the 225 SNPs, highly significant SNPs were repeatedly detected at intervals qGSL-A06-3, qGSL-A09-1, qGSL-A09-5 and qGSL-C07-6, which all explained more than 30% of phenotypic variance. Therefore, these regions should be considered as major interval regions controlling the accumulation of m151. In addition, 46 candidate regions with minor effects are detected for m151, at least in one or any two years and BLUP values (Table S3).
For the aromatic glucosinolate m157, we identified 311, 550 and 1403 significantly associated SNPs based on 2017cq, 2018cq and BLUP values, respectively, 19 SNPs of which were repeatedly detected in this study (Figure 3d and Figure S2d and Table S1). These significant SNPs primarily covered 99 candidate intervals across the entire B. napus genome (Table S3). One significant association locus on chromosome C04 (named qGSL-C04-2) was detected, with the peak SNP S14_6056639, which explained 41.07% of phenotypic variance and was repeatedly detected in different environments (Table S3). Therefore, we used this interval region of chromosome C04 (5.85~6.27 Mb) to predict the candidate genes for controlling the accumulation of m157 in seeds. The interval region on chromosome A09 (named qGSL-A09-5) was also detected in different years, with the peak SNP S9_18294706, which explained 37.58% of phenotypic variance. Furthermore, S7_8312132 explaining 40.84% of phenotypic variance was identified and located on chromosome A07, named qGSL-A07-3. Thus, we believe that the interval regions (qGSL-A09-5, qGSL-A07-3 and qGSL-C04-2) are also closely related to the accumulation of m157 in rapeseed. Correspondingly, 79 candidate regions with minor effects were also detected for m157, at least in one or any two years and BLUP values (Table S3).
For the indole glucosinolate m165, we identified 50, 79 and 82 significantly associated SNPs based on 2017cq, 2018cq and BLUP values, respectively, containing 4 SNPs repeatedly detected in this study (Figure 3e and Figure S2e and Table S1). However, these significant SNPs primarily covered 22 candidate intervals located on chromosome A01, A02, A03, A05, A06, A07, A09, C01, C02, C04, C05, C08 and C09, respectively (Table S3). Three of these interval regions were repeatedly detected and located on chromosomes A06, A09 and C05, namely qGSL-A06-3, qGSL-A09-7 and qGSL-C05-3, respectively. The peak SNP mapped on qGSL-C05-3, qGSL-A06-3 and qGSL-A09-7 explained more than 20% of phenotypic variance (Table S3). In addition, the remaining intervals were detected for m165, at least in one or any two years and BLUP values (Table S3).
For the indole glucosinolate m172, we identified 211, 132 and 1035 significantly associated SNPs based on 2017cq, 2018cqs and BLUP values, respectively, including 47 repeatedly detected SNPs (Figure 3e and Figure S2f and Table S1). These significant SNPs primarily covered 50 candidate intervals across the B. napus genome, except the chromosome C09 (Table S3). For 47 SNPs, 6 significant SNPs were mapped on chromosome A01 (named qGSL-A01-4) with peak SNP S1_19207516, which explained 42.78% of phenotypic variance, and the peak SNP S9_18611734 was mapped on chromosome A09 (named qGSL-A09-5), explaining 38.62% of phenotypic variation. Therefore, these interval regions were used to predict the candidate genes for controlling the accumulation of m172 in seeds (Table S3). In addition, 43 candidate regions were detected for m172, at least in one or any two years and BLUP values (Table S3).
Altogether, we identified 113 candidate regions significantly associated with six glucosinolate metabolites, which were widely distributed throughout the B. napus genome. In particular, candidate interval regions qGSL-A07-3 and qGSL-A09-5 located on chromosome A07 and A09 were detected for five of the six glucosinolate metabolites (all except m165) in two years and BLUP values. In addition, qGSL-A01-4, qGSL-A02-2, qGSL-A03-3, qGSL-A05-2, qGSL-A06-1, qGSL-A06-3, qGSL-A09-6, qGSL-A09-7, qGSL-C04-2 and qGSL-C08-5 were repeatedly detected in at least one year or BLUP value. In these repeatedly detected interval regions, most of them were simultaneously associated with three glucosinolate metabolites (m145, m150 and m151) or any two of these (Table S3), perhaps because they are all aliphatic GSLs. Moreover, m145 is the precursor of m151 in the 4C pathway of aliphatic GSL biosynthesis in B. napus.
2.3. Candidate Gene Mining
Based on the physical positions of the significant SNPs, we mapped the intervals to the corresponding chromosomes of the reference genome B. napus Darmor-bzh (v4.1) [49] and searched for candidate genes for each significant locus based on the mGWAS data combined with the differentially expressed genes in BnHG vs. BnLG. In total, 187 candidate genes were identified and predicted for GSLs in this study, including genes encoding enzymes in the GSL biosynthesis pathway and their homologs, transcription factor genes and some novel candidate genes, respectively (Table S4).
Based on the known GSL biosynthesis pathway, we identified 64 candidate genes in the GSL biosynthesis pathway (including genes encoding enzymes in this pathway and their homologs) among the intervals, which showed differential expression during seed development in BnHG vs. BnLG (Figure 4 and Figure 5, Table S4).
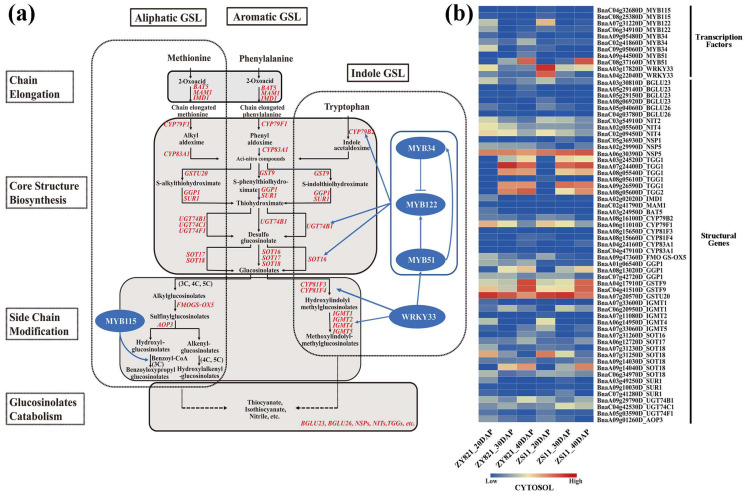
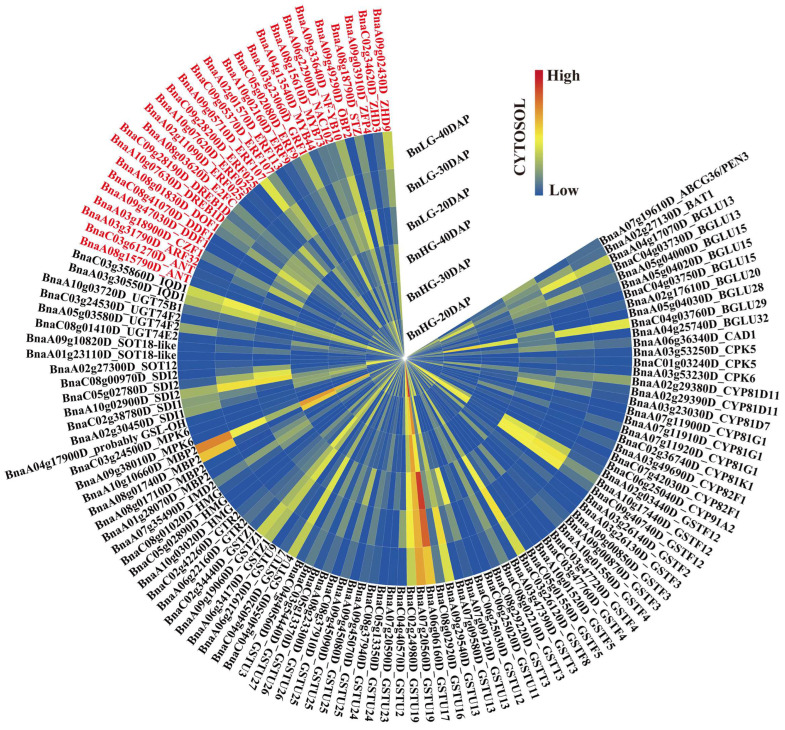
Figure 4.
The proposed GSL biosynthesis pathway (a) and expression patterns of known candidate genes in B. napus (b). The proposed pathway was constructed based on published data [33,38,42,50,51,52,53]. BAT, bile acid transporter [54]; IMD, isopropylmalate dehydrogenase [55]; MAM1, methylthioalkylmalate synthase 1 [56]; CYP, cytochrome P450 [57]; UGT, UDP-glucosyl transferase [22,58]; GST, glutathione S-transferase [59,60]; GGP1, gamma-glutamyl peptidase 1 [61]; SUR1, S-alkyl-thiohydroximate lyase [31]; SOT, sulfotransferase [62]; FMOGS-OX, flavin-monooxygenase glucosinolate S-oxygenase [26]; IGMT, indole glucosinolate O-methyltransferase [63]; AOP3, 2-oxoglutarate-dependent dioxygenase [64]; BGLU, β-glucosidase [65]; NSP, nitrile specifier protein [66]; NIT, nitrilase [67]; TGG, thioglucoside glucohydrolase [68]. Genes with red color represent the identified candidate genes for six glucosinolate metabolites (Table S4). The expression levels of the candidate genes in BnLG and BnHG are listed in Table S5. Further, 20, 30 and 40 DAP represent the seeds after 20, 30 and 40 days after pollination, respectively.
Figure 5.
Expression patterns of the 123 novel candidate genes in B. napus. Genes shown in black are structural genes and those shown in red are transcription factor genes. The expression levels of these candidate genes in BnLG and BnHG are listed in Table S5. Further, 20, 30 and 40 DAP represent the seeds after 20, 30 and 40 days after pollination, respectively.
Among these, key genes encoding enzymes that participate in the GSL biosynthesis pathway, such as genes encoding isopropyl malate dehydrogenase 1 (IMD1), methylthioalkyl malate synthase (MAM1), bile acid transporter (BAT5), cytochrome P450s (CYP79F1, CYP79B2, CYP83A1, CYP81F3 and CYP81F4), UDP-glucosyl transferases (UGT74B1, UGT74C1s and UGT74F1), glutathione S-transferases (GSTU20, GSTF9), γ-glutamyl peptidase 1 (GGP1), S-alkyl thiohydrogen oxidase lyase (SUR1), sulfotransferases (SOT16, SOT17 and SOT18), flavin-monooxygenase glucosinolate S-oxygenase 5 (FMOGS-OX5), oxoglutarate-dependent dioxygenase (AOP3) and indole-glucoside O-methyltransferases (IGMT1, IGMT2, IGMT4 and IGMT5), were identified on the following chromosomes: A01 (GGP1, BnaA01g06540D), A02 (IMD1, BnaA02g02020D), A03 (SUR1, BnaA03g49250D; and BAT5, BnaA03g24950D), A04 (CYP83A1, BnaA04g24160D; and GSTF9, BnaA04g17910D), A05 (UGT74F1, BnaA05g03590D), A06 (CYP79F1, BnaA06g11010D; IGMT4, BnaA06g14950D; and SOT17, BnaA06g12720D), A07 (GSTU20, BnaA07g20570D; IGMT1, BnaA07g33600D; IGMT2, BnaA07g11080D; IGMT5, BnaA07g33060D; SOT16, BnaA07g31260D; SOT18, BnaA07g31230D; and BnaA07g31250D), A08 (CYP79B2, BnaA08g16100D; CYP81F3, BnaA08g15650D; CYP81F4, BnaA08g15660D; and GGP1, BnaA08g13020D), A09 (FMOGS-OX, BnaA09g47360D; SOT18, BnaA09g14030D; and BnaA09g14040D; SUR1, BnaA09g10030D; UGT74B1, BnaA09g29790D; and AOP3, BnaA09g01260D), C02 (MAM1, BnaC02g41790D), C04 (CYP83A1, BnaC04g47910D; GSTF9, BnaC04g41510D; and UGT74C1, BnaC04g42530D), C06 (IGMT1, BnaC06g20950D; and SOT18, BnaC06g34970D) and C07 (GGP1, BnaC07g42720D; and SUR1, BnaC07g41280D), respectively (Figure 4, Table S4), indicating that our GWAS approach was successful in this study.
Previously studies have identified MYB transcription factors that regulate GSL biosynthesis in various plants [36,38,50,69]. In the current study, we identified 39 transcription factors that were predicted to regulate GSL biosynthesis, including 15 AP2/ERF transcription factors (ERFs, ARF3, DREBs and ANT), seven zinc finger proteins (CZF1, STZ, ZHDs, DOF1, OBP2 and ZFP4), two WRKY transcription factors (WRKY33), one growth regulator (GRF1), one NAC family protein (NAC102), one E2F transcription factor (E2FC), one nuclear transcription factor Y subunit B (NF-YB10) and 11 MYB transcription factors (MYBs). Among these, homologous genes of MYB34, MYB51, MYB122 and WRKY33, which are involved in GSL biosynthesis [42,70], were identified on the interval regions of chromosomes A03, A04, A07, A09, C02, C06, C08 and C09, respectively (Table S4). We also identified some novel transcription factor genes (e.g., MYB44, ERF025, NF-YB10 and E2FC) associated with the significant SNPs, with obvious differences in expression in developing seeds of BnHG and BnLG (Figure 5, Table S4). These transcription factors might play an important role in the biosynthesis of GSLs in B. napus.
In addition, we identified 29 candidate genes encoding enzymes responsible for the hydrolysis of GSLs, including genes encoding three nitrile hydrases (NITs), three nitrile specifier proteins (NSPs), one glutathione gamma-glutamylcysteine transferase (PCS1), fifteen β-glucosidases (BGLUs), one ABC transporter family protein (PEN3) and six glucosidolate glucohydrolases (TGGs), respectively (Figure 4 and Figure 5, Table S4). Correspondingly, we also identified two calmodulin-binding proteins (IQD1), three structural genes (HMGs), four genes encoding myrosinase-binding proteins (MBP2) and five protein kinase genes (CPKs and MPK6) located in the interval regions for indole GSLs (Figure 5, Table S4). Among these, WRKY33, CPK5 and MPK6 were shown to mediate indole GSL biosynthesis [71], which in consistent with our results. We also identified two glucosinolate transporters (GTR2) and five TRP family proteins (SDIs) (Figure 5, Table S4).
Altogether, numerous key homologous genes, including known genes (such as IMD1, MAM1, IGMTs, MYB34, MYB51, MYB122 and so on) and novel genes (MYB44, ERF025, ARF3, NF-YB10, E2FC and so on) were identified within the confidence intervals of GSLs. Our findings not only demonstrate the reliability of the association genetics approach, but also provide new insight into elucidating the biosynthesis of these GSLs in B. napus seeds.
3. Discussion
GSLs are well-known secondary metabolites that play important roles in plant defense against diseases and insects and in human nutrition/health [1,2,3,72]. However, some glucosinolates in seed meal have deleterious effects on poultry and livestock, leading to efforts to develop low-glucosinolate Brassica crops [43,73]. Based on the functional group of amino acids, GSLs are divided into three major types, namely aliphatic GSLs, indole GSLs and aromatic GSLs, respectively [12,13,14]. Aliphatic GSLs comprise a major proportion of total GSLs in seeds [74,75,76]. In this study, we identified six major GSL metabolites in rapeseed, including three aliphatic GSLs (m145 gluconapin, m150 glucobrassicanapin and m151 progoitrin), one aromatic GSL (m157 gluconasturtiin) and two indole GSLs (m165 Indolylmethyl glucosinolate and m172 4-hydroxyglucobrassicin) (Figure 1 and Figure 2, Table 1), respectively. However, among the six GSLs examined, four showed highly significant differences in abundance in BnHG vs. BnLG seeds, whereas two indole GSLs did not (Figure 2a). It seems that aliphatic GSLs account for most of the total GSL content, comprising an average of 1539.35~1975.56 μg/g FW in both years of the study. We detected a higher positive correlation between the contents of aliphatic GSLs and aromatic GSLs vs. indole GSLs (Figure 1c), supporting the notion that aliphatic GSLs and aromatic GSLs are somewhat correlated in B. napus seeds. However, deeper knowledge of the specific GSLs in B. napus seeds is required to help breeders improve the current varieties and select plants with advantageous properties.
Unlike in the model plant Arabidopsis thaliana, many copies of homologous genes involved in the GSL biosynthesis pathway are present in the rapeseed genome, as B. napus is an allopolyploid plant with a complex genome [49]. However, numerous studies, including quantitative trait locus (QTL) mapping and candidate gene identification, have investigated the mechanisms involved in GSL production in rapeseed. For example, numerous QTL for GSLs in leaves and seeds of B. napus have been detected [77,78]. For seed GSL content, Xu et al. [79] and Wang et al. [80] also identified the significantly associated sites located on chromosomes A9, C2 and C9, respectively. We previously identified 11 significant SNPs associated with seed GSL accumulation in B. napus located on chromosomes A08, A09, C03 and C09 [81]. Tan et al. [82] recently detected 15 reliable quantitative trait loci (QTLs) for seed GSL content via a GWAS. With the rapid development of gene chip and genome sequencing technology, mGWAS combined with genomic and transcriptomic analysis is suitable for studying the metabolism, genetic characteristics and biochemical properties of plants, and has been widely used for structural and functional analysis of metabolites and functional genomics [83,84,85]. In the current study, we detected 113 candidate intervals with significant associations with glucosinolate metabolites in rapeseed via mGWAS (Figure 3, Table S3). Of these, 13 candidate intervals were significantly associated with only a single metabolite, while 100 were associated with multiple metabolites. Importantly, we repeatedly detected the significant interval regions on chromosomes A02, A08, A09, C03, C07 and C09, which is consistent with previous studies [77,78,81,82,86,87,88,89]. We also identified many new candidate intervals located on chromosomes A04, C01, C05, C06 and C08, further demonstrating the power of our approach (Figure 2, Table S3).
Producing seeds with little or no GSL represents an important breeding objective of B. napus in the past few decades. Indeed, ideal rapeseed has been developed containing high GSL levels in vegetative tissues and little or no GSL in mature seeds by allele mining [78,90]. Numerous studies have also revealed many key genes involved in GSL biosynthesis in B. napus, such as GTR2 [82], MYB28 [91], MAM1, CYP83A1, UGT74B1 [92] and LEC1 [93]. In this study, we identified two candidate genes involved in amino acid side chain extension, BnaMAM1 (BnaC02g41790D) and BnaIMD1 (BnaA02g02020D), which are located in candidate intervals qGSL-A02-1 and qGSL-C02-11, associated with three aliphogenic GSLs and one aromatic GSL (Table S4), and showed higher expression profiles in BnHG vs. BnLG (Figure 4, Table S5). Furthermore, their homologous genes in A. thaliana are involved in the biosynthesis of GSLs [56,94]. We also identified BnaFOMGS-OX5 (BnaA09g47360D) and BanGS-OH (BnaA04g17900D), which participate in the side chain modification process of aliphatic GSL biosynthesis and showed higher expression profiles in BnHG vs. BnLG (Figure 4 and Figure 5, Tables S4 and S5) [95,96]. In addition, several genes encode major enzymes that catalyze the production of the core structures of GSL, including CYP79 genes [97], CYP83 genes [20], GGP1 [43], SUR1 [31], UGT74 genes [22] and SOT genes [62]. We identified homologs of these genes, such as BnaCYP79F1 (BnaA06g11010D), BnaCYP79B2 (BnaA08g16100D), BnaCYP83A1 (BnaA04g24160D and BnaC04g47910D), BnaGGP1 (BnaA01g06540D, BnaA08g13020D and BnaC07g42720D), BnaSUR1 (BnaA09g10030D), BnaUGT74B1 (BnaA09g29790D), BnaUGT74C1 (BnaC04g42530D), BnaSOT16 (BnaA07g31260D), BnaSOT17 (BnaA06g12720D) and BnaSOT18 (BnaA07g31230D, BnaA07g31250D, BnaA09g14030D, BnaA09g14040D and BnaC06g34970D) (Figure 4, Table S4 and S5). In addition, understanding GSL metabolites is important for targeting genes in the relevant biosynthetic pathways. Here, we identified the homologs of genes involved in diverse aspects of GSL biosynthesis and accumulation in B. napus, such as BGLU genes (15), NSP genes (3), CAD1 (1), TGG genes (6) and NIT genes (3) (Figure 4, Table S4 and S5), which are essential for the catabolism and hydrolysis of GSL metabolites [66,67,69,98,99]. Importantly, some novel genes in the GSL biosynthesis pathway were also identified. For example, BnaSOT12 (BnaA02g27300D) is homologous to AtSOT12, which functions in flavonoid, brassinosteroid and salicylic acid activity, and is involved in plant responses to salt, osmotic stress and phytohormones [100,101]. The candidate genes, BnCYP81D11 (BnaA02g29380D and BnaA02g29390D), BnaCYP81D7 (BnaA03g23030D), BnaCYP81G1 (BnaA07g11900D, BnaA07g11910D and BnaA07g11920D), BnaCYP81F3 (BnaA08g15650D), BnaCYP81F4 (BnaA08g15660D) and BnaCYP81K1 (BnaC02g36740D), are located in the interval regions associated with glucosinolate metabolites and showed differential expression in BnHG vs. BnLG seeds (Figure 4 and Figure 5, Tables S4 and S5). Among them, CYP81F1 and CYP81F3 are known to be involved in indole GSL biosynthesis [102,103], but there are few reports of these genes in B. napus. In addition, 46 related transferase genes, including GSTFs, GSTU and GSTT genes along with GSTZ, showed different expression levels in BnHG vs. BnLG seeds (Figure 5, Tables S4 and S5). Similarly, some of these genes are known to be involved in GSL biosynthesis in other species [1,104].
GSLs play diverse roles in plant defense, and several transcription factors have been identified as important regulators of GSL biosynthesis. For example, WRKY33 directly regulates indolic glucosinolate (IGS) biosynthesis, specifically the production of 4-methoxyindole-3-ylmethyl glucosinolate (4MI3G), by directly activating the expression of CYP81F2, IGMT1 and IGMT2 [42], while the R2R3-MYB transcription factors, MYB51, MYB34, MYB122 and MYB115, regulate aliphatic and indole GSL biosynthesis [35,50,69,70,105]. In addition, the zinc finger protein OBP2 has been implicated in indole GSL biosynthesis in A. thaliana [106]. In this study, we identified genes encoding various transcription factors, such as MYBs (MYB34, MYB51, MYB122 and MYB115), AP2/ERFs (ERF9), WRKY (WRKY33) and Zinc finger protein (OBP2), located in the interval regions for GSL metabolites (Figure 4 and Figure 5, Tables S4 and S5). In addition, we identified some new transcription factors, including MYB44 (BnaA04g13540D), DREB1D (BnaA10g07630D and BnaC09g28190D), NAC102 (BnaA06g22900D), NF-YB10 (BnaA09g33640D) and E2FC (BnaA08g03620D), that showed differential gene expression profiles in BnHG vs. BnLG, suggesting they might be involved in regulating GSL biosynthesis in B. napus (Figure 5, Tables S4 and S5). Our findings suggest that the different candidate homologous genes play conserved roles in GSL biosynthesis, although their biological functions in B. napus must be confirmed through careful analysis.
In conclusion, we identified 113 interval regions for six glucosinolate metabolites in B. napus seeds (Figure 3, Table S3). We also identified 187 candidate genes for GSL biosynthesis and accumulation. Our results enrich our knowledge of the GSL biosynthesis pathway and provide candidate genes for improving the compositions of specific GSLs in rapeseed, which could be precisely modified by gene editing in the future.
4. Materials and Methods
4.1. Plant Materials
In total, 143 B. napus accessions were collected from major breeding institutes across China, including 70 low-glucosinolate content (<45 μmol/g), 46 medium-glucosinolate content (45~100 μmol/g) and 27 high-glucosinolate content (>100 μmol/g), respectively (Table S6). All accessions were grown in the growing seasons of 2017–2018 and 2018–2019 (namely 2017Cq and 2018Cq, respectively) in Chongqing, China. The field experiments are designed as in our previous research [107]. In brief, all field experiments were carried out by a randomized complete block design, with three biological replications across each environment. Each accession was grown in a plot with three rows, singling 10–12 plants each row. At the stage of flower initiation, the individual flowers were marked on each plant to ensure that seeds are at the same stage of development. The fresh seeds at 35 days after flowering (DAF) were sampled and pooled from five or more individuals, and quickly frozen in liquid nitrogen. The representative B. napus accessions with high (Zhongyou 821, BnHG) and low (Zhongshuang 11, BnLG) seed glucosinolate content were selected from 143 B. napus accessions for transcriptome sequencing (RNA-Seq), respectively. The seeds of 20, 30 and 40 DAF were also collected and pooled from five or more individuals in BnHG and BnLG plants. All samples were stored at −80 °C until further analysis.
The total RNA was extracted from the seeds of 20, 30 and 40 DAF using an EZ-10 DNAaway RNA Mini-Preps kit (Sangon Biotech, Shanghai, China) following the manufacturer instructions. Then, the qualified RNA samples were used for libraries construction and sequenced on Illumina Hiseq 2000 platform with 150 bp paired-end reads (Tianjin Novogene Bioinformatic Technology Co., Ltd., Tianjin, China). The gene expression profiles were evaluated using FPKM (fragments per kilo base of exon model per million) values. Genes were considered as differentially expressed genes (DEGs) with a minimum 2-fold difference in expression (|log2FC| ≥ 1).
4.2. Glucosinolate Matbolite Extraction
The raw metabolites were extracted from fresh seeds described in our previous research [52], with minor modifications. In short, about 100 mg of fresh seeds was crushed into powder using high-throughput tissue grinder (Tissuelyser-192, Shanghai, China). Subsequently, we added the 500 μL extract solution (80% aqueous methanol with 0.1% formic acid) and homogenized by vortex for 10 s. The homogenized extraction buffer was extracted using sonication (KQ-100E, Kunshan, China) at 4 °C for 1 h, followed by centrifugation at 12,000× g at 4 °C for 10 min. Then, the residues were repeatedly extracted. Eventually, the mixed liquid supernatants were used for UPLC-HESI-MS/MS analysis after being filtered by a 0.22 μm nylon filter. All experiments were performed in at least three replicates for each accession.
4.3. UPLC-HESI-MS/MS Analysis
The UPLC-HESI-MS/MS was performed using Dionex UltiMateTM 3000 UHPLC system (Thermo Fisher Scientific, Waltham, MA, USA) coupled to a Thermo Scientifific Q-Exactive System equipped with an S-Lens ionizer source (Thermo Scientifific, Waltham, MA, USA), including the precolumn (pore size: 1.7 μm, 2.1 × 5 mm, Waters, Wexford, Ireland) and Acquity UPLCBEH C18 column (pore size: 1.7 μ m, 2.1 mm × 150mm, Waters, Wexford, Ireland). The parameters were as follows: the mobile phases A (0.1% formic acid) and B (0.1% acetonitrile aqueous solution), 37 °C column temperature, 0.3 mL/min flow rate and 10 μL injection volume, respectively. The mobile phase gradients are 0–2 min, 5% B–10% B; 2–10 min, 10–25% B; 10–13 min, 25–95% B; 13–16 min, 95% B; 16–16.5 min, 95–5% B; 16.5–21 min, 5% B. The mass spectrometry was detected in negative ion mode with a scanning range of 100 to 1200 (m/z), 3.5 kV ion source voltage, 350 °C capillary temperature, 35 sheath gas, 10 auxiliary gas and 0 backblow air, respectively.
4.4. Data Processing and Glucosinolate Metabolite Identification
Raw data of UPLC-HESI-MS/MS were firstly treated with MS-DAIL ver4.1 MSP negative database (http://prime.psc.riken.jp/compms/msdial/main.htmL#MSP, accessed on 2 April 2022), and automatically converted by ABF (Analysis Base File) converter (http://www.reifycs.com/AbfConverter/index.html, accessed on 12 March 2020) [108]. Correspondingly, the glucosinolate metabolites were identified using Xcalibur ver4.1 based on retention time (RT), MS and MS/MS spectral data, referring to the published information [109,110]. Furthermore, the amounts of six glucosinolate metabolites were quantified by drawing standard curve of sinigrin [110,111], and the calibration curve was constructed using eight points generated from the sinigrin concentration gradients, ranging from 0.001 to 2 mg L−1 (0.001, 0.005, 0.01, 0.05, 0.20, 0.50, 1.0 and 2.0 mg L−1).
The glucosinolate metabolites were detected in two consecutive years (2017 and 2018) with replications; we further obtained the best linear unbiased prediction (BLUP) of six glucosinolate metabolites per accession using a linear model using an R script (http://www.eXtension.org/pages/61006, accessed on 14 October 2020), respectively. The content of six glucosinolate metabolites in the 2017 and 2018 growing seasons and resulting values of BLUP were used as phenotypes for GWAS, respectively. The heritability was calculated by using the multi-year repeated model. In addition, the quantitative data of glucosinolate metabolites were divided into 10 grades, and Shannon–Wiener Diversity Index was used to calculate metabolites [112,113,114]. The Pearson correlation coefficients among six glucosinolate metabolites were calculated by the R language psych package and significance tests (Student t-test) were performed [115]. Statistical analysis of glucosinolate metabolites was performed using Microsoft Office Excel 2009. The G × E analysis of glucosinolate metabolites was performed using AMMI model by R package “agricolae”.
4.5. GWAS of Glucosinolate Metabolites
Detailed methods used for SNP genotyping and mapping were previously described [46,47,48]. In brief, DNA libraries with a mean insert size of 350 bp were constructed, and 125 bp paired-end reads were generated using an Illumina HiSeq 4000 instrument at the Biomarker Technologies Corporation (Beijing, China). Low-quality bases from paired-end reads were trimmed using Trimmomatic (version 0.33) and mapped to the rapeseed genome ‘Darmor-bzh’ [49] using the Burrows–Wheeler Aligner (version 0.7.10-r789). Then, we used Picard (release 2.0.1, http://broadinstitute.github.io/picard/, accessed on 26 July 2018) and GATK (version 3.2) to process local realignment and base quality detection for the alignment results, sequentially. Further, we then used AMtools mpileup (version 0.1.19–44428cd) and GATK to perform SNP calling. In total, 239,945 high-quality SNPs with a minor allele frequency (MAF) <5% were used for further analysis. The six models, including naïve, Q, K, PCA, K + Q and K + PCA model, were applied to determine the statistical associations between phenotypes and genotypes. Quantile–quantile (QQ) plots were used for false-positive correction for association analyses. In this study, genome-wide association analysis for six glucosinolate metabolites was carried out using the GLM with Q model and MLM with Q + K model by TASSEL 5.2.1 software. The population structure Q matrix was completed by admixture_linux-1.3.0 software [116]. The significant signal of the associations between SNPs and the glucosinolate metabolites was assessed based on the threshold P < P = 1/N (where N is 239,945 SNPs in this study), and the threshold of significance was set to p < 4.17 × 10−6.
4.6. Annotation of Candidate Genes
The significant interval regions were mapped to the Darmor-bzh reference genome (version 4.1, http://www.genoscope.cns.fr/brassicanapus/data/, accessed on 26 October 2022) [49], which were anchored by the physical position of significant SNPs. Correspondingly, 200 kb flanking region of association SNPs was used as the candidate interval region. Then, the annotated genes in the interval regions were used for screening the candidate genes, and further confirmed by gene expression analysis. Herein, the gene expression levels were calculated as FPKM (Fragments Per Kilobase of transcript sequence per Millions base pairs) with the featureCounts tool in Subread [117]. Finally, correlations between the GSL phenotype and the gene expression profiles were detected to determine the candidate genes that were associated with the glucosinolate metabolites.
Acknowledgments
We thank Xinfu Xu and Rui Wang from Southwest University for providing important plant materials.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants12030639/s1, Figure S1: The QQ plots of 6 glucosinolate metabolites; Figure S2: Manhattan plots of association analysis for six glucosinolates using Q+K model; Figure S3: The significant SNPs associated with 6 glucosinolates in different environments; Figure S4: Correlation analysis of six glucosinolate metabolites in 2017cq and 2018cq; Table S1: Summary of repeatedly detected SNPs for 6 glucosinolate metabolites by mGWAS; Table S2: Summary of interval regions for 6 glucosinolate metabolites in B. napus; Table S3: Candidate interval regions for 6 glucosinolate metabolites in B. napus; Table S4: Candidate genes for 6 glucosinolate metabolites in B. napus; Table S5: The expression levels of the candidate genes in BnHG and BnLG of B. napus; Table S6: The list of 143 B. napus accessions used in this study; Table S7: Statistical analysis of 6 glucosinolate metabolites in high-, medium- and low-glucosinolate-content rapeseed
Author Contributions
Conceptualization, N.Y. and C.Q.; methodology, Y.T. (Yunshan Tang) and G.Z.; software, Y.T. (Yunshan Tang), S.S. and M.G.; validation, Y.T. (Yunshan Tang), N.Y. and C.Q.; formal analysis, Y.T. (Yunshan Tang), X.J. and Y.T. (Yuhan Tang); resources, H.Z. and J.L.; data curation, F.S., R.H. and S.C.; writing—original draft preparation, Y.T. (Yunshan Tang), N.Y. and C.Q.; writing—review and editing, K.L., N.Y. and C.Q.; supervision, C.Q. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
All other datasets supporting the results of this article are included within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was funded by the National Natural Science Foundation of China (32272150, 32072093), the Innovation and Entrepreneurship Training Program for graduates of Chongqing (CYS21124), Innovation and Entrepreneurship Training Program for Undergraduates (S202210635329), the China Agriculture Research System of MOF and MARA and the 111 Project (B12006).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Grubb C.D., Abel S. Glucosinolate metabolism and its control. Trends Plant Sci. 2006;11:89–100. doi: 10.1016/j.tplants.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Prieto M., López C.J., Simal-Gandara J. Glucosinolates: Molecular structure, breakdown, genetic, bioavailability, properties and healthy and adverse effects. Adv. Food Nutr. Res. 2019;90:305–350. doi: 10.1016/bs.afnr.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Bischoff K.L. Glucosinolates. In: Gupta R., Lall R., Srivastava A., editors. Nutraceuticals, Effificacy, Safety and Toxicity. Academic Press; Cambridge, MA, USA: 2021. pp. 903–909. [Google Scholar]
- 4.Jeschke V., Burow M. Glucosinolates. In: John W., Sons L., editors. eLS. 2018. [(accessed on 2 April 2022)]. pp. 1–8. Available online: https://onlinelibrary.wiley.com/doi/full/10.1002/9780470015902.a0027968. [Google Scholar]
- 5.Chhajed S., Mostafa I., He Y., Abou-Hashem M., El-Domiaty M., Chen S. Glucosinolate biosynthesis and the glucosinolate–myrosinase system in plant defense. Agronomy. 2020;10:1786. doi: 10.3390/agronomy10111786. [DOI] [Google Scholar]
- 6.Smolinska U., Morra M., Knudsen G., Brown P. Toxicity of glucosinolate degradation products from Brassica napus seed meal toward Aphanomyces euteiches f. sp. pisi. Phytopathology. 1997;87:77–82. doi: 10.1094/PHYTO.1997.87.1.77. [DOI] [PubMed] [Google Scholar]
- 7.Hasegawa T., Yamada K., Kosemura S., Yamamura S., Hasegawa K. Phototropic stimulation induces the conversion of glucosinolate to phototropism-regulating substances of radish hypocotyls. Phytochemistry. 2000;54:275–279. doi: 10.1016/S0031-9422(00)00080-7. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt R., Bancroft I. Genetics and Genomics of the Brassicaceae. Springer; New York, NY, USA: 2011. pp. 585–596. [Google Scholar]
- 9.Velasco P., Rodríguez V.M., Francisco M., Cartea M.E., Soengas P. Genetics and breeding of Brassica crops. In: Mérillon J.M., Ramawat K., editors. Glucosinolates, Reference Series in Phytochemistry. Springer; Cham, Switzerland: 2017. pp. 61–86. [Google Scholar]
- 10.Bisht N.C., Augustine R. Development of Brassica Oilseed Crops with Low Antinutritional Glucosinolates and Rich in Anticancer Glucosinolates. In: Jaiwal P., Chhillar A., Chaudhary D., Jaiwal R., editors. Nutritional Quality Improvement in Plants. Springer; Cham, Switzerland: 2019. pp. 271–287. [Google Scholar]
- 11.Bell L., Oloyede O.O., Lignou S., Wagstaff C., Methven L. Taste and flavor perceptions of glucosinolates, isothiocyanates, and related compounds. Mol. Nutr. Food Res. 2018;62:e1700990. doi: 10.1002/mnfr.201700990. [DOI] [PubMed] [Google Scholar]
- 12.Sønderby I.E., Geu-Flores F., Halkier B.A. Biosynthesis of glucosinolates–gene discovery and beyond. Trends Plant Sci. 2010;15:283–290. doi: 10.1016/j.tplants.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Blažević I., Montaut S., Burčul F., Olsen C.E., Burow M., Rollin P., Agerbirk N. Glucosinolate structural diversity, identification, chemical synthesis and metabolism in plants. Phytochemistry. 2020;169:112100. doi: 10.1016/j.phytochem.2019.112100. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen V.T., Stewart J., Lopez M., Ioannou I., Allais F. Glucosinolates: Natural occurrence, biosynthesis, accessibility, isolation, structures, and biological activities. Molecules. 2020;25:4537. doi: 10.3390/molecules25194537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fahey J.W., Zalcmann A.T., Talalay P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry. 2001;56:5–51. doi: 10.1016/S0031-9422(00)00316-2. [DOI] [PubMed] [Google Scholar]
- 16.Clarke D.B. Glucosinolates, structures and analysis in food. Anal. Methods. 2010;2:310–325. doi: 10.1039/b9ay00280d. [DOI] [Google Scholar]
- 17.Knill T., Reichelt M., Paetz C., Gershenzon J., Binder S. Arabidopsis thaliana encodes a bacterial-type heterodimeric isopropylmalate isomerase involved in both Leu biosynthesis and the Met chain elongation pathway of glucosinolate formation. Plant Mol. Biol. 2009;71:227–239. doi: 10.1007/s11103-009-9519-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z., Hammerlindl J., Keller W., McVetty P.B., Daayf F., Quiros C.F., Li G. MAM gene silencing leads to the induction of C3 and reduction of C4 and C5 side-chain aliphatic glucosinolates in Brassica napus. Mol. Breed. 2011;27:467–478. doi: 10.1007/s11032-010-9444-y. [DOI] [Google Scholar]
- 19.Lächler K., Imhof J., Reichelt M., Gershenzon J., Binder S. The cytosolic branched-chain aminotransferases of Arabidopsis thaliana influence methionine supply, salvage and glucosinolate metabolism. Plant Mol. Biol. 2015;88:119–131. doi: 10.1007/s11103-015-0312-3. [DOI] [PubMed] [Google Scholar]
- 20.Bak S., Feyereisen R. The involvement of two P450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol. 2001;127:108–118. doi: 10.1104/pp.127.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikkelsen M.D., Petersen B., Olsen C., Halkier B. Biosynthesis and metabolic engineering of glucosinolates. Amino Acids. 2002;22:279–295. doi: 10.1007/s007260200014. [DOI] [PubMed] [Google Scholar]
- 22.Grubb C.D., Zipp B.J., Kopycki J., Schubert M., Quint M., Lim E.K., Bowles D.J., Pedras M.S.C., Abel S. Comparative analysis of A. rabidopsis UGT 74 glucosyltransferases reveals a special role of UGT 74C1 in glucosinolate biosynthesis. Plant J. 2014;79:92–105. doi: 10.1111/tpj.12541. [DOI] [PubMed] [Google Scholar]
- 23.Klein A.P., Sattely E.S. Biosynthesis of cabbage phytoalexins from indole glucosinolate. Proc. Natl. Acad. Sci. USA. 2017;114:1910–1915. doi: 10.1073/pnas.1615625114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitainda V., Jez J.M. Structural Studies of Aliphatic Glucosinolate Chain-Elongation Enzymes. Antioxidants. 2021;10:1500. doi: 10.3390/antiox10091500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hopkins R.J., van Dam N.M., van Loon J.J. Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annu. Rev. Entomol. 2009;54:57–83. doi: 10.1146/annurev.ento.54.110807.090623. [DOI] [PubMed] [Google Scholar]
- 26.Li J., Hansen B.G., Ober J.A., Kliebenstein D.J., Halkier B.A. Subclade of flavin-monooxygenases involved in aliphatic glucosinolate biosynthesis. Plant Physiol. 2008;148:1721–1733. doi: 10.1104/pp.108.125757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li G., Riaz A., Goyal S., Abel S., Quiros C. Inheritance of three major genes involved in the synthesis of aliphatic glucosinolates in Brassica oleracea. J. Am. Soc. Hort. Sci. 2001;126:427–431. doi: 10.21273/JASHS.126.4.427. [DOI] [Google Scholar]
- 28.Yi G.-E., Robin A.H.K., Yang K., Park J.-I., Hwang B.H., Nou I.-S. Exogenous methyl jasmonate and salicylic acid induce subspecies-specific patterns of glucosinolate accumulation and gene expression in Brassica oleracea L. Molecules. 2016;21:1417. doi: 10.3390/molecules21101417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez V.C., Dai R., Block A.K., Kim J. Metabolite analysis of Arabidopsis CYP79A2 overexpression lines reveals turnover of benzyl glucosinolate and an additive effect of different aldoximes on phenylpropanoid repression. Plant Signal. Behav. 2021;16:1966586. doi: 10.1080/15592324.2021.1966586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naur P., Petersen B.L., Mikkelsen M.D., Bak S., Rasmussen H., Olsen C.E., Halkier B.A. CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol. 2003;133:63–72. doi: 10.1104/pp.102.019240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikkelsen M.D., Naur P., Halkier B.A. Arabidopsis mutants in the C–S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 2004;37:770–777. doi: 10.1111/j.1365-313X.2004.02002.x. [DOI] [PubMed] [Google Scholar]
- 32.Douglas Grubb C., Zipp B.J., Ludwig-Müller J., Masuno M.N., Molinski T.F., Abel S. Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 2004;40:893–908. doi: 10.1111/j.1365-313X.2004.02261.x. [DOI] [PubMed] [Google Scholar]
- 33.Liu S., Liu Y., Yang X., Tong C., Edwards D., Parkin I.A., Zhao M., Ma J., Yu J., Huang S. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014;5:3930. doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yi G.-E., Robin A.H.K., Yang K., Park J.-I., Kang J.-G., Yang T.-J., Nou I.-S. Identification and expression analysis of glucosinolate biosynthetic genes and estimation of glucosinolate contents in edible organs of Brassica oleracea subspecies. Molecules. 2015;20:13089–13111. doi: 10.3390/molecules200713089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gigolashvili T., Berger B., Mock H.P., Müller C., Weisshaar B., Flügge U.I. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007;50:886–901. doi: 10.1111/j.1365-313X.2007.03099.x. [DOI] [PubMed] [Google Scholar]
- 36.Beekwilder J., Van Leeuwen W., Van Dam N.M., Bertossi M., Grandi V., Mizzi L., Soloviev M., Szabados L., Molthoff J.W., Schipper B. The impact of the absence of aliphatic glucosinolates on insect herbivory in Arabidopsis. PLoS ONE. 2008;3:e2068. doi: 10.1371/journal.pone.0002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller R., De Vos M., Sun J.Y., Sønderby I.E., Halkier B.A., Wittstock U., Jander G. Differential effects of indole and aliphatic glucosinolates on lepidopteran herbivores. J. Chem. Ecol. 2010;36:905–913. doi: 10.1007/s10886-010-9825-z. [DOI] [PubMed] [Google Scholar]
- 38.Frerigmann H., Gigolashvili T. MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol. Plant. 2014;7:814–828. doi: 10.1093/mp/ssu004. [DOI] [PubMed] [Google Scholar]
- 39.Frerigmann H., Gigolashvili T. Update on the role of R2R3-MYBs in the regulation of glucosinolates upon sulfur deficiency. Front. Plant Sci. 2014;5:626. doi: 10.3389/fpls.2014.00626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schweizer F., Fernández-Calvo P., Zander M., Diez-Diaz M., Fonseca S., Glauser G., Lewsey M.G., Ecker J.R., Solano R., Reymond P. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell. 2013;25:3117–3132. doi: 10.1105/tpc.113.115139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frerigmann H. Glucosinolate regulation in a complex relationship–MYC and MYB–no one can act without each other. Adv. Bot. Res. 2016;80:57–97. [Google Scholar]
- 42.Tao H., Miao H., Chen L., Wang M., Xia C., Zeng W., Sun B., Zhang F., Zhang S., Li C. WRKY33-mediated indolic glucosinolate metabolic pathway confers resistance against Alternaria brassicicola in Arabidopsis and Brassica crops. J. Integr. Plant Biol. 2022;64:1007–1019. doi: 10.1111/jipb.13245. [DOI] [PubMed] [Google Scholar]
- 43.Agnihotri A., Prem D., Gupta K. The chronicles of oil and meal quality improvement in oilseed rape. Adv. Bot. Res. 2007;45:49–97. [Google Scholar]
- 44.Gupta S., Pratap A. History, origin, and evolution. Adv. Bot. Res. 2007;45:1–20. [Google Scholar]
- 45.He Y., Yang Z., Tang M., Yang Q.-Y., Zhang Y., Liu S. Enhancing canola breeding by editing a glucosinolate transporter gene lacking natural variation. Plant Physiol. 2022;188:1848–1851. doi: 10.1093/plphys/kiac021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu K., Wei L., Li X., Wang Y., Wu J., Liu M., Zhang C., Chen Z., Xiao Z., Jian H. Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 2019;10:1154. doi: 10.1038/s41467-019-09134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang T., Wei L., Wang J., Xie L., Li Y.Y., Ran S., Ren L., Lu K., Li J., Timko M.P. Integrating GWAS, linkage mapping and gene expression analyses reveals the genetic control of growth period traits in rapeseed (Brassica napus L.) Biotechnol. Biofuels. 2020;13:134. doi: 10.1186/s13068-020-01774-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qian M., Fan Y., Li Y., Liu M., Sun W., Duan H., Yu M., Chang W., Niu Y., Li X. Genome-wide association study and transcriptome comparison reveal novel QTL and candidate genes that control petal size in rapeseed. J. Exp. Bot. 2021;72:3597–3610. doi: 10.1093/jxb/erab105. [DOI] [PubMed] [Google Scholar]
- 49.Chalhoub B., Denoeud F., Liu S., Parkin I.A., Tang H., Wang X., Chiquet J., Belcram H., Tong C., Samans B. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345:950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y., Li B., Huai D., Zhou Y., Kliebenstein D.J. The conserved transcription factors, MYB115 and MYB118, control expression of the newly evolved benzoyloxy glucosinolate pathway in Arabidopsis thaliana. Front. Plant Sci. 2015;6:343. doi: 10.3389/fpls.2015.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Augustine R., Bisht N.C. Biofortification of oilseed Brassica juncea with the anti-cancer compound glucoraphanin by suppressing GSL-ALK gene family. Sci Rep. 2015;5:18005. doi: 10.1038/srep18005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shang G., Zhao H., Tong L., Yin N., Hu R., Jiang H., Kamal F., Zhao Z., Xu L., Lu K. Genome-Wide Association Study of Phenylalanine Derived Glucosinolates in Brassica rapa. Plants. 2022;11:1274. doi: 10.3390/plants11091274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robin A.H.K., Yi G.-E., Laila R., Yang K., Park J.-I., Kim H.R., Nou I.-S. Expression profiling of glucosinolate biosynthetic genes in Brassica oleracea L. var. capitata inbred lines reveals their association with glucosinolate content. Molecules. 2016;21:787. doi: 10.3390/molecules21060787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gigolashvili T., Yatusevich R., Rollwitz I., Humphry M., Gershenzon J., Flügge U.-I. The plastidic bile acid transporter 5 is required for the biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana. Plant Cell. 2009;21:1813–1829. doi: 10.1105/tpc.109.066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He Y., Mawhinney T.P., Preuss M.L., Schroeder A.C., Chen B., Abraham L., Jez J.M., Chen S. A redox-active isopropylmalate dehydrogenase functions in the biosynthesis of glucosinolates and leucine in Arabidopsis. Plant J. 2009;60:679–690. doi: 10.1111/j.1365-313X.2009.03990.x. [DOI] [PubMed] [Google Scholar]
- 56.Redovniković I.R., Textor S., Lisnić B., Gershenzon J. Expression pattern of the glucosinolate side chain biosynthetic genes MAM1 and MAM3 of Arabidopsis thaliana in different organs and developmental stages. Plant Physiol. Biochem. 2012;53:77–83. doi: 10.1016/j.plaphy.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 57.Zang Y.-X., Kim J.-H., Park Y.-D., Kim D.-H., Hong S.-B. Metabolic engineering of aliphatic glucosinolates in Chinese cabbage plants expressing Arabidopsis MAM1, CYP79F1, and CYP83A1. BMB Rep. 2008;41:472–478. doi: 10.5483/BMBRep.2008.41.6.472. [DOI] [PubMed] [Google Scholar]
- 58.Zheng H., Zhang C., Wang Y., Zhou W., Chen J., Yan X., Li Z., Huang S., Li M., Tang Y. Overexpression of the glucosyltransferase gene BoaUGT74B1 enhances the accumulation of indole glucosinolates in Chinese kale. Sci. Horti. 2021;288:110302. doi: 10.1016/j.scienta.2021.110302. [DOI] [Google Scholar]
- 59.Czerniawski P., Bednarek P. Glutathione S-transferases in the biosynthesis of sulfur-containing secondary metabolites in Brassicaceae plants. Front. Plant Sci. 2018;9:1639. doi: 10.3389/fpls.2018.01639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang A., Luo R., Li J., Miao R., An H., Yan X., Pang Q. Arabidopsis Glutathione-S-Transferases GSTF11 and GSTU20 Function in Aliphatic Glucosinolate Biosynthesis. Front. Plant Sci. 2022;12:816233. doi: 10.3389/fpls.2021.816233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Geu-Flores F., Møldrup M.E., Böttcher C., Olsen C.E., Scheel D., Halkier B.A. Cytosolic γ-glutamyl peptidases process glutathione conjugates in the biosynthesis of glucosinolates and camalexin in Arabidopsis. Plant Cell. 2011;23:2456–2469. doi: 10.1105/tpc.111.083998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klein M., Papenbrock J. Sulfotransferases and their role in glucosinolate biosynthesis. In: Khan N.A., Singh S., Umar S., editors. Sulfur Assimilation and Abiotic Stress in Plants. Springer; Berlin/Heidelberg, Germany: 2008. pp. 149–166. [Google Scholar]
- 63.Pfalz M., Mukhaimar M., Perreau F., Kirk J., Hansen C.I.C., Olsen C.E., Agerbirk N., Kroymann J. Methyl transfer in glucosinolate biosynthesis mediated by indole glucosinolate O-methyltransferase 5. Plant Physiol. 2016;172:2190–2203. doi: 10.1104/pp.16.01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jensen L.M., Kliebenstein D.J., Burow M. Investigation of the multifunctional gene AOP3 expands the regulatory network fine-tuning glucosinolate production in Arabidopsis. Front. Plant Sci. 2015;6:762. doi: 10.3389/fpls.2015.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meier K., Ehbrecht M.D., Wittstock U. Glucosinolate content in dormant and germinating Arabidopsis thaliana seeds is affected by non-functional alleles of classical myrosinase and nitrile-specifier protein genes. Front. Plant Sci. 2019;10:1549. doi: 10.3389/fpls.2019.01549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wittstock U., Meier K., Dörr F., Ravindran B.M. NSP-dependent simple nitrile formation dominates upon breakdown of major aliphatic glucosinolates in roots, seeds, and seedlings of Arabidopsis thaliana Columbia-0. Front. Plant Sci. 2016;7:1821. doi: 10.3389/fpls.2016.01821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Piotrowski M., Schönfelder S., Weiler E.W. The Arabidopsis thaliana isogene NIT4 and its orthologs in tobacco encode β-cyano-L-alanine hydratase/nitrilase. J. Biol. Chem. 2001;276:2616–2621. doi: 10.1074/jbc.M007890200. [DOI] [PubMed] [Google Scholar]
- 68.Barth C., Jander G. Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 2006;46:549–562. doi: 10.1111/j.1365-313X.2006.02716.x. [DOI] [PubMed] [Google Scholar]
- 69.Rask L., Andréasson E., Ekbom B., Eriksson S., Pontoppidan B., Meijer J. Myrosinase: Gene family evolution and herbivore defense in Brassicaceae. Plant Mol. Biol. 2000;42:93–114. doi: 10.1023/A:1006380021658. [DOI] [PubMed] [Google Scholar]
- 70.Frerigmann H., Glawischnig E., Gigolashvili T. The role of MYB34, MYB51 and MYB122 in the regulation of camalexin biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 2015;6:654. doi: 10.3389/fpls.2015.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang L., Zhang Y., Guan R., Li S., Xu X., Zhang S., Xu J. Co-regulation of indole glucosinolates and camalexin biosynthesis by CPK5/CPK6 and MPK3/MPK6 signaling pathways. J. Integr. Plant Biol. 2020;62:1780–1796. doi: 10.1111/jipb.12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hasan M., Friedt W., Pons-Kühnemann J., Freitag N., Link K., Snowdon R. Association of gene-linked SSR markers to seed glucosinolate content in oilseed rape (Brassica napus ssp. napus) Theor. Appl. Genet. 2008;116:1035–1049. doi: 10.1007/s00122-008-0733-3. [DOI] [PubMed] [Google Scholar]
- 73.Kondra Z., Stefansson B. Inheritance of the major glucosinolates of rapeseed (Brassica napus) meal. Can. J. Plant Sci. 1970;50:643–647. doi: 10.4141/cjps70-122. [DOI] [Google Scholar]
- 74.Kim Y.B., Li X., Kim S.-J., Kim H.H., Lee J., Kim H., Park S.U. MYB transcription factors regulate glucosinolate biosynthesis in different organs of Chinese cabbage (Brassica rapa ssp. pekinensis) Molecules. 2013;18:8682–8695. doi: 10.3390/molecules18078682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cartea M.E., de Haro A., Obregón S., Soengas P., Velasco P. Glucosinolate variation in leaves of Brassica rapa crops. Plant Foods Hum. Nutr. 2012;67:283–288. doi: 10.1007/s11130-012-0300-6. [DOI] [PubMed] [Google Scholar]
- 76.Li P., Zhao Y., Zhang W., Ding X., Yang M., Wang X., Xie C., Fu T. Analysis of Glucosinolate Components and Profiles in Brassica napus. Zhongguo Nong Ye Ke Xue. 2005;38:1346–1352. [Google Scholar]
- 77.Feng J., Long Y., Shi L., Shi J., Barker G., Meng J. Characterization of metabolite quantitative trait loci and metabolic networks that control glucosinolate concentration in the seeds and leaves of Brassica napus. New Phytol. 2012;193:96–108. doi: 10.1111/j.1469-8137.2011.03890.x. [DOI] [PubMed] [Google Scholar]
- 78.Liu S., Huang H., Yi X., Zhang Y., Yang Q., Zhang C., Fan C., Zhou Y. Dissection of genetic architecture for glucosinolate accumulations in leaves and seeds of Brassica napus by genome-wide association study. Plant Biotechnol. J. 2020;18:1472–1484. doi: 10.1111/pbi.13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu J., Long Y., Wu J., Xu H., Zhao Z., Wen J., Meng J., Shi C. QTL identification on two genetic systems for rapeseed glucosinolate and erucic acid contents over two seasons. Euphytica. 2015;205:647–657. doi: 10.1007/s10681-015-1379-2. [DOI] [Google Scholar]
- 80.Wang B., Wu Z., Li Z., Zhang Q., Hu J., Xiao Y., Cai D., Wu J., King G.J., Li H. Dissection of the genetic architecture of three seed-quality traits and consequences for breeding in Brassica napus. Plant Biotechnol. J. 2018;16:1336–1348. doi: 10.1111/pbi.12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qu C.-M., Li S.-M., Duan X.-J., Fan J.-H., Jia L.-D., Zhao H.-Y., Lu K., Li J.-N., Xu X.-F., Wang R. Identification of candidate genes for seed glucosinolate content using association mapping in Brassica napus L. Genes. 2015;6:1215–1229. doi: 10.3390/genes6041215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tan Z., Xie Z., Dai L., Zhang Y., Zhao H., Tang S., Wan L., Yao X., Guo L., Hong D. Genome-and transcriptome-wide association studies reveal the genetic basis and the breeding history of seed glucosinolate content in Brassica napus. Plant Biotechnol. J. 2022;20:211–225. doi: 10.1111/pbi.13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen W., Gao Y., Xie W., Gong L., Lu K., Wang W., Li Y., Liu X., Zhang H., Dong H. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat. Genet. 2014;46:714–721. doi: 10.1038/ng.3007. [DOI] [PubMed] [Google Scholar]
- 84.Slaten M.L., Yobi A., Bagaza C., Chan Y.O., Shrestha V., Holden S., Katz E., Kanstrup C., Lipka A.E., Kliebenstein D.J. mGWAS uncovers Gln-Glucosinolate seed-specific interaction and its role in metabolic homeostasis. Plant Physiol. 2020;183:483–500. doi: 10.1104/pp.20.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen J., Hu X., Shi T., Yin H., Sun D., Hao Y., Xia X., Luo J., Fernie A.R., He Z. Metabolite-based genome-wide association study enables dissection of the flavonoid decoration pathway of wheat kernels. Plant Biotechnol. J. 2020;18:1722–1735. doi: 10.1111/pbi.13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li F., Chen B., Xu K., Wu J., Song W., Bancroft I., Harper A.L., Trick M., Liu S., Gao G. Genome-wide association study dissects the genetic architecture of seed weight and seed quality in rapeseed (Brassica napus L.) DNA Res. 2014;21:355–367. doi: 10.1093/dnares/dsu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wei D., Cui Y., Mei J., Qian L., Lu K., Wang Z.M., Li J., Tang Q., Qian W. Genome-wide identification of loci affecting seed glucosinolate contents in Brassica napus L. J. Integr. Plant Biol. 2019;61:611–623. doi: 10.1111/jipb.12717. [DOI] [PubMed] [Google Scholar]
- 88.Jan H.U., Guan M., Yao M., Liu W., Wei D., Abbadi A., Zheng M., He X., Chen H., Guan C. Genome-wide haplotype analysis improves trait predictions in Brassica napus hybrids. Plant Sci. 2019;283:157–164. doi: 10.1016/j.plantsci.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 89.Gao C., Zhang F., Hu Y., Song L., Tang L., Zhang X., Wang A., Wu X. Dissecting the genetic architecture of glucosinolate compounds for quality improvement in flowering stalk tissues of Brassica napus. Hortic. Plant J. 2022 doi: 10.1016/j.hpj.2022.09.001. [DOI] [Google Scholar]
- 90.Nour-Eldin H.H., Madsen S.R., Engelen S., Jørgensen M.E., Olsen C.E., Andersen J.S., Seynnaeve D., Verhoye T., Fulawka R., Denolf P. Reduction of antinutritional glucosinolates in Brassica oilseeds by mutation of genes encoding transporters. Nat. Biotechnol. 2017;35:377–382. doi: 10.1038/nbt.3823. [DOI] [PubMed] [Google Scholar]
- 91.Zhou X., Zhang H., Xie Z., Liu Y., Wang P., Dai L., Zhang X., Wang Z., Wang Z., Wan L. Natural variation and artificial selection at the BnaC2. MYB28 locus modulate Brassica napus seed glucosinolate. Plant Physiol. 2023;191:352–368. doi: 10.1093/plphys/kiac463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Y., Huai D., Yang Q., Cheng Y., Ma M., Kliebenstein D.J., Zhou Y. Overexpression of three glucosinolate biosynthesis genes in Brassica napus identifies enhanced resistance to Sclerotinia sclerotiorum and Botrytis cinerea. PLoS ONE. 2015;10:e0140491. doi: 10.1371/journal.pone.0140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Elahi N., Duncan R.W., Stasolla C. Modification of oil and glucosinolate content in canola seeds with altered expression of Brassica napus Leafy Cotyledon1. Plant Physiol. Biochem. 2016;100:52–63. doi: 10.1016/j.plaphy.2015.12.022. [DOI] [PubMed] [Google Scholar]
- 94.He Y., Galant A., Pang Q., Strul J.M., Balogun S.F., Jez J.M., Chen S. Structural and functional evolution of isopropylmalate dehydrogenases in the leucine and glucosinolate pathways of Arabidopsis thaliana. J. Biol. Chem. 2011;286:28794–28801. doi: 10.1074/jbc.M111.262519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kong W., Li J., Yu Q., Cang W., Xu R., Wang Y., Ji W. Two novel flavin-containing monooxygenases involved in biosynthesis of aliphatic glucosinolates. Front. Plant Sci. 2016;7:1292. doi: 10.3389/fpls.2016.01292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Halkier B.A., Du L. The biosynthesis of glucosinolates. Trends Plant Sci. 1997;2:425–431. doi: 10.1016/S1360-1385(97)90026-1. [DOI] [PubMed] [Google Scholar]
- 97.Mikkelsen M.D., Petersen B.L., Glawischnig E., Jensen A.B., Andreasson E., Halkier B.A. Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol. 2003;131:298–308. doi: 10.1104/pp.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang L., Kawaguchi R., Morikawa-Ichinose T., Allahham A., Kim S.-J., Maruyama-Nakashita A. Sulfur deficiency-induced glucosinolate catabolism attributed to two β-glucosidases, BGLU28 and BGLU30, is required for plant growth maintenance under sulfur deficiency. Plant Cell Physiol. 2020;61:803–813. doi: 10.1093/pcp/pcaa006. [DOI] [PubMed] [Google Scholar]
- 99.De Benedictis M., Brunetti C., Brauer E.K., Andreucci A., Popescu S.C., Commisso M., Guzzo F., Sofo A., Ruffini Castiglione M., Vatamaniuk O.K. The Arabidopsis thaliana knockout mutant for phytochelatin synthase1 (cad1-3) is defective in callose deposition, bacterial pathogen defense and auxin content, but shows an increased stem lignification. Front. Plant Sci. 2018;9:19. doi: 10.3389/fpls.2018.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marsolais F., Boyd J., Paredes Y., Schinas A.-M., Garcia M., Elzein S., Varin L. Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a (At2g14920) and AtST1 (At2g03760) Planta. 2007;225:1233–1244. doi: 10.1007/s00425-006-0413-y. [DOI] [PubMed] [Google Scholar]
- 101.Lacomme C., Roby D. Molecular cloning of a sulfotransferase in Arabidopsis thaliana and regulation during development and in response to infection with pathogenic bacteria. Plant Mol. Biol. 1996;30:995–1008. doi: 10.1007/BF00020810. [DOI] [PubMed] [Google Scholar]
- 102.Pfalz M., Mikkelsen M.D., Bednarek P., Olsen C.E., Halkier B.A., Kroymann J. Metabolic engineering in Nicotiana benthamiana reveals key enzyme functions in Arabidopsis indole glucosinolate modification. Plant Cell. 2011;23:716–729. doi: 10.1105/tpc.110.081711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhao L., Wang C., Zhu F., Li Y. Mild osmotic stress promotes 4-methoxy indolyl-3-methyl glucosinolate biosynthesis mediated by the MKK9–MPK3/MPK6 cascade in Arabidopsis. Plant Cell Rep. 2017;36:543–555. doi: 10.1007/s00299-017-2101-8. [DOI] [PubMed] [Google Scholar]
- 104.Piślewska-Bednarek M., Nakano R.T., Hiruma K., Pastorczyk M., Sanchez-Vallet A., Singkaravanit-Ogawa S., Ciesiołka D., Takano Y., Molina A., Schulze-Lefert P. Glutathione transferase U13 functions in pathogen-triggered glucosinolate metabolism. Plant Physiol. 2018;176:538–551. doi: 10.1104/pp.17.01455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Malitsky S., Blum E., Less H., Venger I., Elbaz M., Morin S., Eshed Y., Aharoni A. The transcript and metabolite networks affected by the two clades of Arabidopsis glucosinolate biosynthesis regulators. Plant Physiol. 2008;148:2021–2049. doi: 10.1104/pp.108.124784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Skirycz A., Reichelt M., Burow M., Birkemeyer C., Rolcik J., Kopka J., Zanor M.I., Gershenzon J., Strnad M., Szopa J. DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 2006;47:10–24. doi: 10.1111/j.1365-313X.2006.02767.x. [DOI] [PubMed] [Google Scholar]
- 107.Qu C., Jia L., Fu F., Zhao H., Lu K., Wei L., Xu X., Liang Y., Li S., Wang R. Genome-wide association mapping and Identification of candidate genes for fatty acid composition in Brassica napus L. using SNP markers. BMC Genomics. 2017;18:232. doi: 10.1186/s12864-017-3607-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tsugawa H., Ikeda K., Takahashi M., Satoh A., Mori Y., Uchino H., Okahashi N., Yamada Y., Tada I., Bonini P. MS-DIAL 4: Accelerating lipidomics using an MS/MS, CCS, and retention time atlas. BioRxiv. 2020 doi: 10.1101/2020.02.11.944900. [DOI] [PubMed] [Google Scholar]
- 109.Hwang I.M., Park B., Dang Y.M., Kim S.-Y., Seo H.Y. Simultaneous direct determination of 15 glucosinolates in eight Brassica species by UHPLC-Q-Orbitrap-MS. Food Chem. 2019;282:127–133. doi: 10.1016/j.foodchem.2018.12.036. [DOI] [PubMed] [Google Scholar]
- 110.Qu C., Yin N., Chen S., Wang S., Chen X., Zhao H., Shen S., Fu F., Zhou B., Xu X. Comparative analysis of the metabolic profiles of yellow-versus black-seeded rapeseed using UPLC–HESI–MS/MS and transcriptome analysis. J. Agric. Food Chem. 2020;68:3033–3049. doi: 10.1021/acs.jafc.9b07173. [DOI] [PubMed] [Google Scholar]
- 111.Yin N.-W., Wang S.-X., Jia L.-D., Zhu M.-C., Yang J., Zhou B.-J., Yin J.-M., Lu K., Wang R., Li J.-N. Identification and characterization of major constituents in different-colored rapeseed petals by UPLC–HESI-MS/MS. J. Agric. Food Chem. 2019;67:11053–11065. doi: 10.1021/acs.jafc.9b05046. [DOI] [PubMed] [Google Scholar]
- 112.Fang K., Xia Z., Li H., Jiang X., Qin D., Wang Q., Wang Q., Pan C., Li B., Wu H. Genome-wide association analysis identified molecular markers associated with important tea flavor-related metabolites. Hortic. Res. 2021;8:42. doi: 10.1038/s41438-021-00477-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kuznetsova A., Brockhoff P.B., Christensen R.H. lmerTest package: Tests in linear mixed effects models. J. Stat. Softw. 2017;82:1–26. doi: 10.18637/jss.v082.i13. [DOI] [Google Scholar]
- 114.Bates D. lme4: Linear Mixed-Effects Models Using S4 Classes. 2010. R Package Version 1.1–30. [(accessed on 14 October 2022)]. Available online: http://CRAN.R-project.org/package=lme4.
- 115.Revelle W., Maechler M., Bolker B., Walker W. Psych: Procedures for Psychological, Psychometric, and Personality Research. 2015. R Rackage Version 2.2.9. [(accessed on 14 October 2022)]. Available online: https://CRAN.R-project.org/package=psych.
- 116.Alexander D.H., Shringarpure S.S., Novembre J., Lange K. Admixture 1.3 Software Manual. 2020. Software Version 1.3.0. [(accessed on 15 September 2022)]. Available online: https://dalexander.github.io/admixture/admixture-manual.pdf.
- 117.Liao Y., Smyth G.K., Shi W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All other datasets supporting the results of this article are included within the article and Supplementary Materials.