SUMMARY
Nutrient availability regulates the C. elegans life cycle as well as mitochondrial physiology. Food deprivation significantly reduces mitochondrial genome (mtDNA) numbers and leads to aging-related phenotypes. Here we show that the bZIP (basic leucine zipper) protein ATFS-1, a mediator of the mitochondrial unfolded protein response (UPRmt), is required to promote growth and establish a functional germline after prolonged starvation. We find that recovery of mtDNA copy numbers and development after starvation requires mitochondrion-localized ATFS-1 but not its nuclear transcription activity. We also find that the insulin-like receptor DAF-2 functions upstream of ATFS-1 to modulate mtDNA content. We show that reducing DAF-2 activity represses ATFS-1 nuclear function while causing an increase in mtDNA content, partly mediated by mitochondrion-localized ATFS-1. Our data indicate the importance of the UPRmt in recovering mitochondrial mass and suggest that atfs-1-dependent mtDNA replication precedes mitochondrial network expansion after starvation.
In brief
Uma Naresh et al. show that the mitochondrial unfolded protein response (UPRmt) is required for growth and germline proliferation after prolonged starvation. Mitochondrial localization of the bZIP protein ATFS-1 promotes mtDNA replication and is required to reestablish or regenerate the mitochondrial network depleted by autophagy.
Graphical Abstract
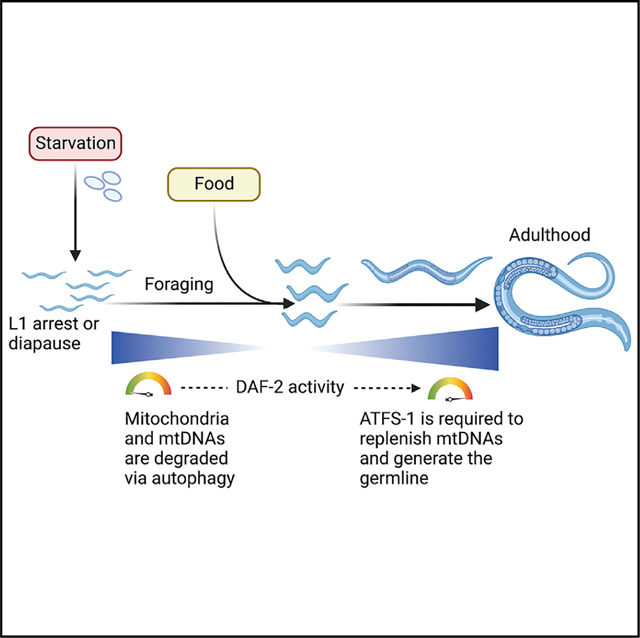
INTRODUCTION
The mitochondrial unfolded protein response (UPRmt) is a signaling pathway mediated by the bZIPprotein ATFS-1 that promotes mitochondrial network expansion by regulating transcription of mitochondrial biogenesis genes.1 The UPRmt transcriptional response has been best studied in the context of mitochondrial perturbations that slow worm development, such as mutations in genes encoding oxidative phosphorylation (OXPHOS) proteins, mtDNA heteroplasmy, or pathogen infection.2–7 During worm development, the majority of ATFS-1 is imported into mitochondria and usually degraded by the protease LONP-1.8 During mitochondrial stress or dysfunction, a fraction of ATFS-1 accumulates in the nucleus via its nuclear localization sequence (NLS), where it activates a transcription program to recover mitochondrial function.9 ATFS-1 also accumulates in dysfunctional mitochondria via its mitochondrial targeting sequence (MTS), where it binds mtDNA and promotes replication.9,10 Regulation of the nuclear transcriptional response as well as mtDNA replication by ATFS-1 during mitochondrial stress is consistent with the UPRmt promoting mitochondrial function by coordinating mitochondrion-to-nucleus communication (Figure 1A). However, little is known regarding the role of ATFS-1 in coordinating such a response when mitochondrial dysfunction is not induced by exogenous stressors or deleterious mutations.
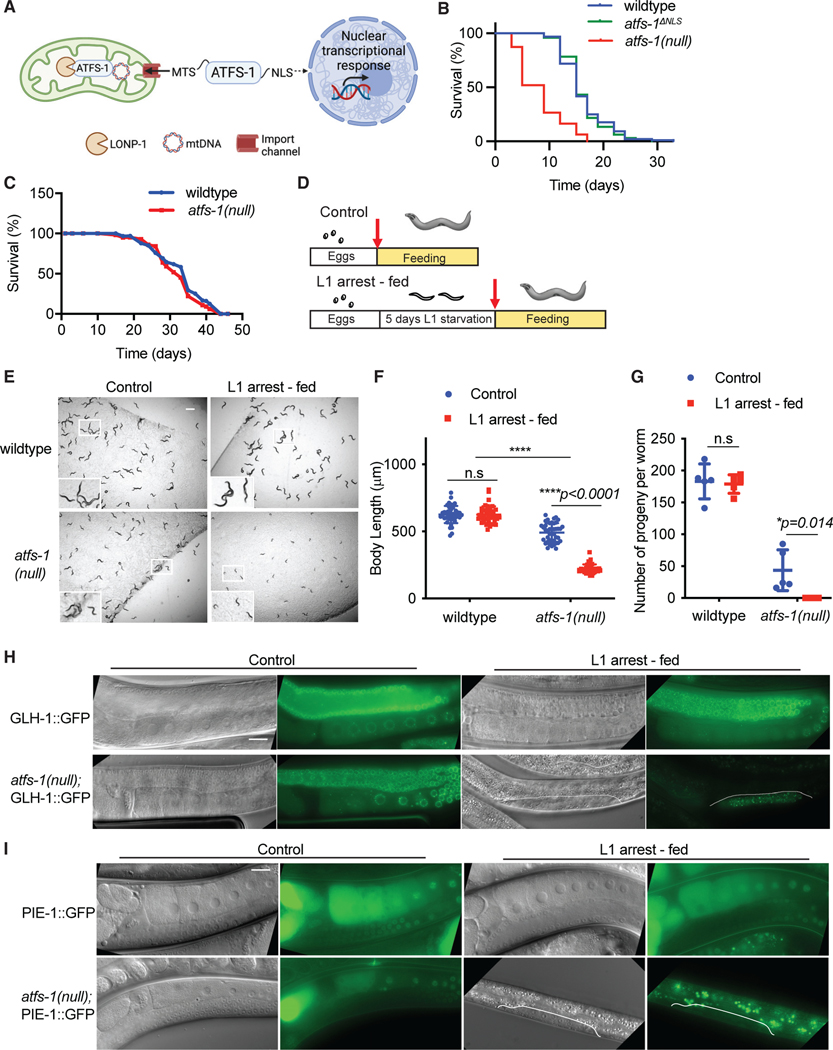
Figure 1. ATFS-1 is required for development after prolonged L1 arrest.
(A) Schematic illustrating ATFS-1-mediated UPRmt signaling.
(B) Lifespan analysis comparing wild-type, atfs-1(null), and atfs-1ΔNLS worms (statistical analyses from three independent repeats are reported in Table S3).
(C) Lifespan analysis comparing wild-type and atfs-1(null) worms during L1 arrest statistical analyses from three independent repeats are reported in Table S1).
(D) Schematic depicting the experimental protocol comparing worms that were directly hatched in the presence of bacteria (control) and worms that were maintained as arrested L1s for 5 days and then transferred to plates seeded with bacteria (L1 arrestfed).
(E) Images of wild-type and atfs-1(null) worms under control and L1 arrest-fed conditions, obtained when control worms reached the L4 stage (scale bar, 0.5 mm).
(F) Quantification of body length, comparing wild-type and atfs-1(null) worms under control and L1 arrest-fed conditions (n = 3 ± SD, ****p < 0.0001, two-way ANOVA with post-hoc Sidak’s test).
(G) Brood size quantification of wild-type and atfs-1(null) worms under control and L1 arrest-fed conditions (n = 5 ± SD, *p = 0.014, two-way ANOVA with post-hoc Sidak’s test).
(H) Images of wild-type and atfs-1(null) worms expressing GLH-1::GFP under control or L1 arrest-fed conditions, obtained when the control worms reached day 1 of adulthood (scale bar, 0.02 mm).
(I) Images of wild-type and atfs-1(null) worms expressing PIE-1::GFP under control or L1 arrest-fed conditions obtained when the control worms reached day 1 of adulthood (scale bar, 0.02 mm).
Diverse species, including vertebrates, adapt cellular responses to survive adverse environmental conditions, including prolonged starvation and extreme seasonal changes such as drought.11–14 A relatively common adaptation is when the animals enter a diapause state upon encountering severe stress, where growth is suspended but basal metabolic activities are maintained. The nematode C. elegans can undergo developmental arrest during starvation and resume development when food is encountered.15,16 When worms hatch in the absence of food, they remain arrested at the first larval stage, which is known as L1 diapause. Developmental arrest is accompanied by an increase in stress resistance and alterations in metabolic signaling pathways, including insulin signaling, transforming growth factor β (TGF-β) signaling, AMP-activated protein kinase, and fatty acid metabolism.17–21 To survive during L1 arrest, energy production requires autophagic degradation of cytosolic components, including organelles such as mitochondria.21 Sustaining a prolonged search for food during L1 arrest results in manifestation of aging-like phenotypes, including accumulation of protein aggregates and reactive oxygen species as well as fragmentation of the mitochondrial network and depletion of mtDNA.22,23 When food is encountered, the aging-like deterioration is resolved and growth resumes,23 accompanied by increased transcription and translation of components required for mitochondrial biogenesis.24–26
Nuclear chromosome replication is regulated by numerous proteins and checkpoints to ensure that genome duplication is coordinated with the cell cycle.27,28 Akin to nuclear genome replication, mtDNA replication is often followed by mitochondrial fission, presumably to ensure that each daughter mitochondrion inherits at least one mtDNA.29 However, mtDNA replication is not coordinated with the cell cycle. Although the majority of the proteins required for mtDNA replication have been identified, the upstream events that stimulate mtDNA replication remain unclear. Prolonged L1 arrest and the subsequent recovery offers a physiologically relevant as well as genetically tractable model to study the signaling pathways regulating bulk initiation of mtDNA replication and recovery of the mitochondrial network. Here, we examine the role of the UPRmt and ATFS-1 in coordinating mtDNA content recovery or expansion after prolonged starvation.
RESULTS
ATFS-1 is required for development after prolonged starvation at L1 stage
atfs-1(null) worms, which lack the entire open reading frame, have reduced mitochondrial respiration and mitochondrial function at the L4 stage as well a reduction in lifespan (Figure 1B).1,5 To survive prolonged starvation, mitochondrial function and integrity are required because loss-of-function mutations in the OXPHOS complex III component isp-1 or in the ubiquinone synthesis component clk-1 are short lived during L1 arrest.18 Thus, we hypothesized that ATFS-1 is also required for survival during starvation. atfs-1(null) worms survived as long as wild-type worms during L1 arrest (Figure 1C). These data indicate that, despite the reliance on OXPHOS, ATFS-1 is not involved in mitochondrial maintenance or survival during L1 arrest.
We next examined the growth of wild-type and atfs-1(null) worms hatched in the presence of food (referred to as “control”) compared with development upon exposure to food after 5 days of L1 arrest (referred to as “L1 arrest-fed”) (Figure 1D). The development of atfs-1(null) worms was significantly impaired after L1 arrest (Figures 1E, 1F, and S1A), whereas wild-type worms developed similarly to worms hatched on food (Figures 1E, 1F, and S1A). When raised in the presence of food, atfs-1(null) worms produce progeny, although fecundity is reduced relative to wild-type worms. However, after starvation, atfs-1(null) worms did not lay eggs or produce any viable progeny (Figures 1G and S1B). atfs-1(null) worms appeared more transparent and were smaller compared with wild-type worms of the same stage, suggestive of abnormal development (Figures S1C and S1D).
Because nearly all atfs-1(null) worms were sterile after L1 arrest, we examined the germline using transgenic reporter strains. The GLH-1::GFP fusion protein marks P granules, which are RNA-enriched compartments specific to germ cells.30 The germlines of atfs-1(null) and wild-type worms hatched in the presence of food expressed GLH-1::GFP throughout the germline (Figure 1H). However, atfs-1(null) worms that were fed after L1 arrest had only a cluster of germ cells expressing GLH-1::GFP that failed to expand (Figure 1H). We used a reporter strain expressing PIE-1::GFP to examine germline proliferation. Germ cells express PIE-1 to prevent adoption of somatic cell fates by repressing transcription.31,32 After L1 arrest, the germline in atfs-1(null) worms did not proliferate to the point where PIE-1::GFP was expressed (Figure 1I). These data indicate that the ATFS-1 is required to resume growth and establish a functional germline after prolonged L1 arrest.
ATFS-1 is required for mtDNA content recovery after L1 starvation
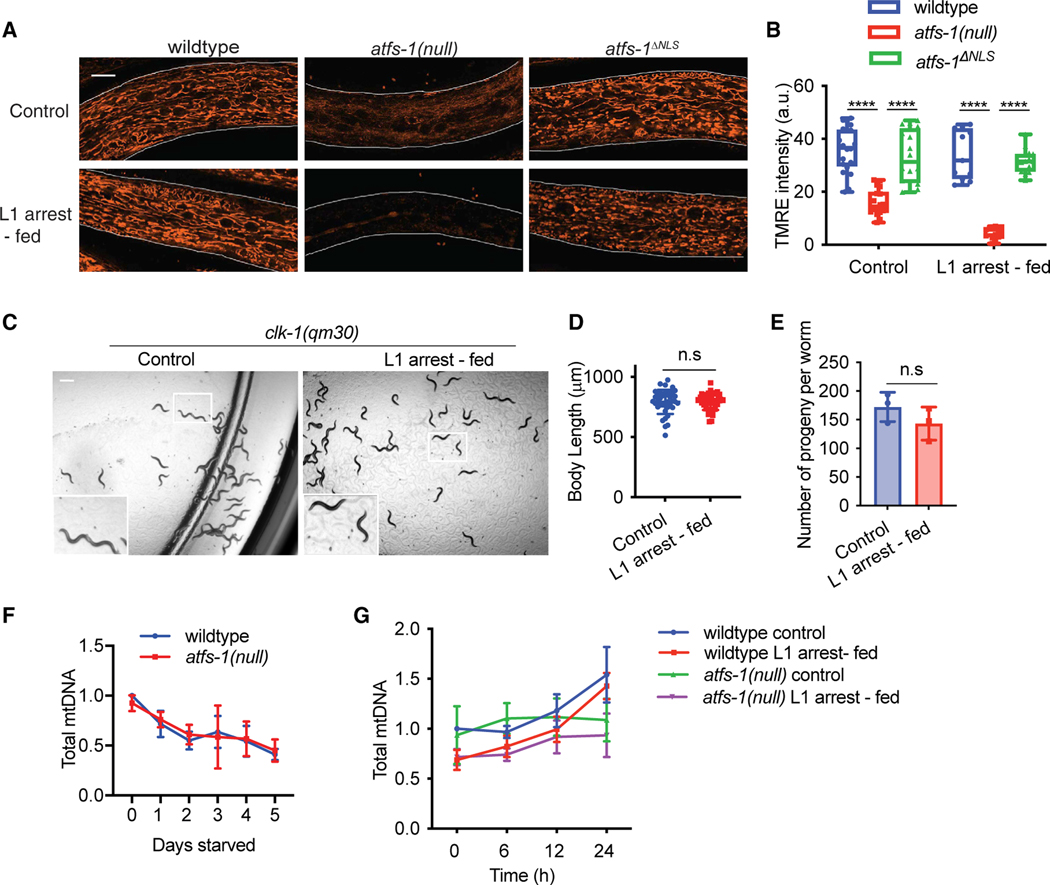
Because ATFS-1 promotes mitochondrial maintenance during growth, we examined mitochondrial network morphology using the dye TMRE (tetramethylrhodamine, ethyl ester), which accumulates in functional mitochondria. At the L4 stage, TMRE staining was comparable throughout the soma in wild-type worms fed upon hatching or after 5 days of L1 arrest (Figures 2A and 2B). As expected, atfs-1(null) worms accumulated less TMRE than wild-type worms when hatched in the presence of food, as shown previously.1 However, after 5 days of L1 arrest, TMRE staining was nearly undetectable in atfs-1(null) worms compared with worms of similar age hatched on food, indicating that these worms were unable to establish a functional mitochondrial network (Figures 2A and 2B). Because mitochondria were not detectable in atfs-1(null) worms with TMRE staining after L1 arrest, we examined mitochondrial content and morphology using a reporter in which the mitochondrial outer membrane translocase TOMM-20 was tagged with mScarlet.33 We found that, when hatched onto food, mScarlet accumulated along the mitochondrial network in atfs-1(null) mutants, similar to wild-type worms (Figure S2A). However, after L1 arrest, atfs-1(null) worms had an aberrant network morphology compared with wild-type worms (Figure S2A), suggesting that aspects of mitochondrial biogenesis occur in the absence of atfs-1, but the organelles are dysfunctional.
Figure 2. UPRmt promotes mtDNA content recovery after starvation.
(A) Images comparing TMRE staining of wild-type, atfs-1(null), and atfs-1ΔNLS worms under control or L1 arrest-fed conditions, obtained when control worms reached the L4 stage (scale bar, 10 μm; representative images from three biological replicates).
(B) Quantification of TMRE staining of wild-type, atfs-1(null), and atfs-1ΔNLS worms at the L4 stage under control or L1 arrest-fed conditions (n = 15–20 worms for each strain and condition ± SD, ****p < 0.0001, two-way ANOVA with post-hoc Sidak’s test; a.u., arbitrary unit).
(C) Images comparing development of clk-1(qm30) worms under control and L1 arrest-fed conditions on day 1 of adulthood (scale bar, 0.5 mm).
(D) Body length quantification of clk-1(qm30) worms under control and L1 arrest-fed conditions, measured on day 1 of adulthood (n = 3 ± SD, Student’s t test).
(E) Brood size quantification of clk-1(qm30) worms under control and L1 arrest-fed conditions (n = 3 ± SD, 10–15 worms each replicate, Student’s t test).
(F) mtDNA quantification of wild-type and atfs-1(null) starved L1 worms over a period of 5 days as determined by qPCR (n = 3 ± SD, one-way ANOVA; wild-type worms: ***p = 0.0007, post hoc Dunnett’s test shows significant difference between 0-day starved worms and 1- to 5-day starved worms; atfs-1(null) worms: *p = 0.0191, post-hoc Dunnett’s test shows significant difference between 0-day starved worms and 3- to 5-day starved worms).
(G) Time course of mtDNA content quantification over 24 h as determined by qPCR comparing wild-type and atfs-1(null) worms under control and L1 arrest-fed conditions (n = 6 for wild-type and n = 3 for atfs-1(null) ± SD, **p = 0.0039 comparing L1 arrest-fed wild-type and atfs-1(null) mtDNA content after 24 h of feeding, two-way ANOVA with post hoc Tukey’s multiple comparisons test).
Because atfs-1(null) worms harbored dysfunctional mitochondria and were developmentally impaired after prolonged L1 arrest, we hypothesized that worms with impaired OXPHOS dysfunction may also have similar defects when exiting starvation. However, we found that clk-1(qm30) and complex II-defective mev-1(kn-1) mutant worms developed at the same rate and had similar brood sizes regardless of whether they were hatched in the presence of food or underwent L1 arrest prior to feeding (Figures 2C–2E and S2B–S2D). OXPHOS is reduced in clk-1(qm30) and mev-1(kn-1) worms but not completely impaired.34 These data suggest that the presence of functional ATFS-1 in clk-1 and mev-1 mutants allows OXPHOS dysfunction mutants to cope during development after starvation, in contrast to atfs-1(null) worms, which lack UPRmt-mediated processes.
We have shown previously that, concomitant with reduced membrane potential, atfs-1(null) worms harbor fewer mtDNAs than wild-type worms.1 Thus, we examined the mtDNA content of atfs-1(null) worms during L1 arrest and after food introduction. A previous study found that mtDNAs were depleted during L1 arrest by autophagy.22 Similarly, mtDNA content was reduced by ~50% in wild-type worms after 5 days of L1 arrest (Figure 2F). atfs-1(null) worms hatched with similar quantities of mtDNA as wild-type worms, which was also reduced by 50% after 5 days of L1 arrest (Figure 2F). As expected, mtDNA content increased in wild-type worms upon food introduction. However, mtDNA content remained low in atfs-1(null) worms (Figure 2G). These data suggest that, when L1 worms begin to feed, ATFS-1 is required to increase or recover the mtDNA content that was degraded during L1 arrest (Figures 2F and 2G).
Wild-type and atfs-1(null) worms that are directly hatched onto food contain similar quantities of mtDNA (Figure 2G) and develop into fertile adults, suggesting that atfs-1 does not regulate mtDNA content in embryos prior to feeding. During starvation, although wild-type and atfs-1(null) worms undergo a reduction in mtDNA content, after food introduction, mtDNA increases more slowly in atfs-1(null) worms compared with wild-type worms (Figures 2F and 2G). These findings suggest that, for normal development after prolonged L1 arrest, ATFS-1 is required to increase mtDNA content to the level that L1 wild-type and atfs-1(null) harbor at the time that they hatch. These findings also emphasize the challenges in mediating the metabolic transition that must occur when the worm goes from a catabolic state to fuel the search for food to an anabolic state required for growth and development when feeding begins.
Mitophagy does not impair mtDNA accumulation in atfs-1(null) worms after L1 arrest
Given the reduced mtDNA accumulation in atfs-1(null) worms after L1 arrest, we hypothesized that this may be due to impaired mtDNA replication or increased mitophagy-mediated clearance as a result of accumulating mitochondrial damage in the absence of the UPRmt. To determine the effect of mitophagy, we examined recovery from L1 arrest in atfs-1(null);pdr-1(tm598) worms, which contain a loss-of-function mutation in the ubiquitin ligase Parkin, which is required for mitophagy in C. elegans.6,35 Although atfs-1(null);pdr-1(tm598) worms developed slower than either single mutant, they are able to develop to adulthood when hatched in the presence of food without prolonged starvation (Figures S3A and S3B). However, similar to atfs-1(null) worms, the development and recovery of mtDNA content after L1 arrest was impaired in atfs-1(null);pdr-1(tm598) worms (Figures S3A–S3D). These findings suggest that the impaired recovery of mtDNA content in atfs-1(null) worms is not due to mitophagy.
ATFS-1-dependent nuclear transcription is not required for mtDNA recovery
Our findings suggest that recovery of mtDNA content after prolonged L1 arrest could be due to ATFS-1 directly promoting mtDNA replication in mitochondria or regulating nuclear transcription of genes that promote mtDNA replication. To examine the role of ATFS-1-dependent nuclear transcription, we compared wild-type, atfs-1(null), and mutant worms in which the NLS in ATFS-1 was mutated.1,10 Consistent with impaired nuclear activity of atfs-1ΔNLS worms, previous studies have shown that the increase in atfs-1-dependent transcription of the nuclear gene hsp-6 was reduced in atfs-1ΔNLS worms raised during mitochondrial stress.1,10 ATFS-1ΔNLS is imported into mitochondria and processed similarly to wild-type ATFS-1.10 atfs-1DNLS worms had similar lifespans to wild-type worms, whereas the lifespan of atfs-1(null) worms was considerably reduced (Figure 1B). We generated clk-1(qm30);atfs-1DNLS worms to examine the effect of reduced atfs-1-dependent nuclear transcription during mitochondrial dysfunction. We found that clk-1(qm30);atfs-1ΔNLS worms developed considerably more slowly and produced fewer progeny than clk-1(qm30) worms, indicating that the nuclear activity of ATFS-1 is required for growth during OXPHOS perturbation (Figures S4A and S4B).
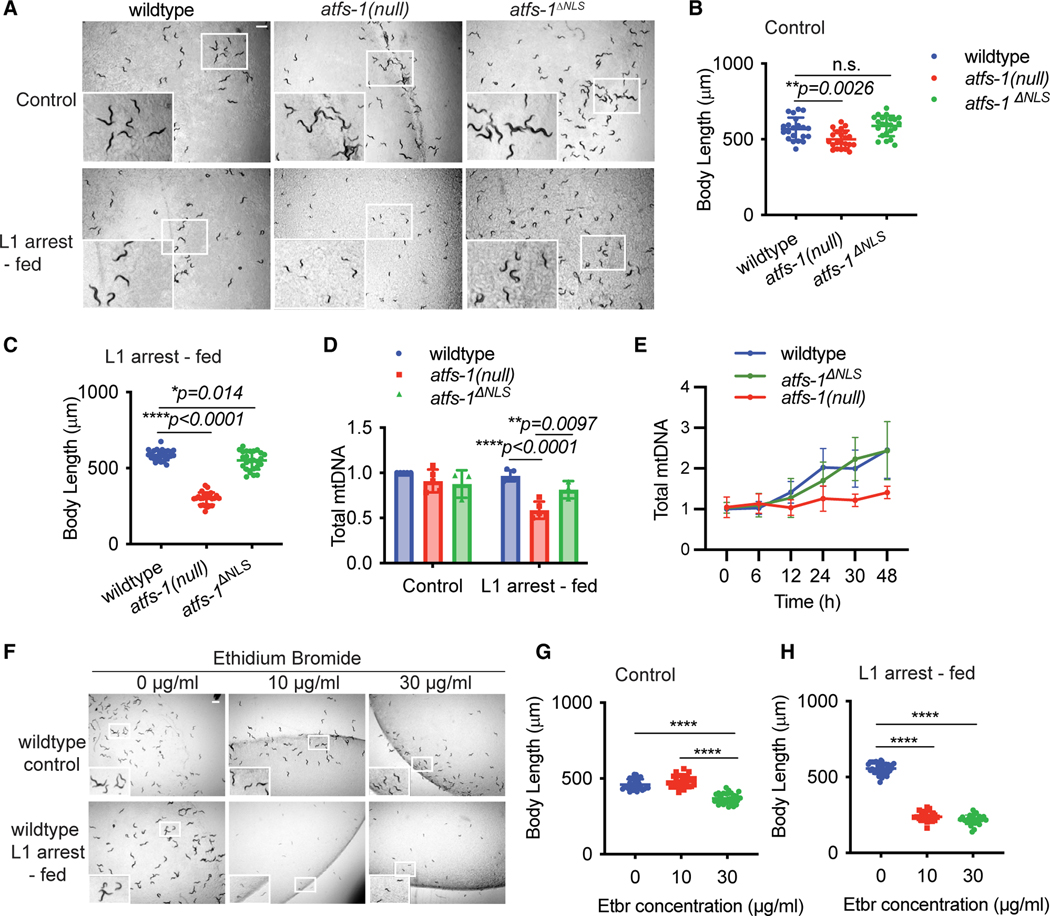
We next examined the role of ATFS-1 nuclear activity during development after prolonged L1 arrest. atfs-1ΔNLS worms developed much faster than atfs-1(null) worms (Figures 3A–3C). Upon feeding after prolonged L1 arrest, mtDNA content increased at similar rates in wild-type and atfs-1ΔNLS worms but was impaired in atfs-1(null) worms (Figures 3D and 3E). Unlike atfs-1(null) worms, atfs-1ΔNLS worms established a functional mitochondrial network upon feeding, as determined by TMRE staining (Figures 2A and 2B). However, the mitochondrial network was more fragmented in atfs-1ΔNLS worms relative to wild-type worms (Figure S4C), indicating that atfs-1-dependent transcription is required to maintain optimal mitochondrial function after prolonged starvation.
Figure 3. The increase in mtDNA content after L1 arrest does not require atfs-1-dependent nuclear transcription.
(A) Images of wild-type, atfs-1(null), and atfs-1ΔNLS worms under control and L1 arrest-fed conditions, obtained when the control condition worms reached the L4 stage (scale bar, 0.5 mm).
(B) Quantification of body length of wild-type, atfs-1(null), and atfs-1ΔNLS worms under control conditions when the worms reached the L4 stage (n = 3 ± SD, **p = 0.0026, one-way ANOVA with post hoc Dunnett’s test).
(C) Quantification of body length of wild-type, atfs-1(null), and atfs-1ΔNLS worms under L1 arrest-fed conditions when the control worms reached the L4 stage (n = 3 ± SD, *p = 0.014 comparing wild-type and atfs-1DNLS worms and ****p < 0.0001 comparing wild-type and atfs-1(null) worms, one-way ANOVA with post hoc Dunnett’s test).
(D) mtDNA quantification of wild-type, atfs-1(null), and atfs-1ΔNLS worms when the control condition worms reached the L4 stage (n = 3–5 ± SD, ****p < 0.0001 comparing wild-type and atfs-1(null) worms and **p = 0.0097 comparing atfs-1(null) and atfs-1ΔNLS L1 arrest-fed worms, two-way ANOVA with post hoc Tukey’s multiple comparisons test).
(E) Time course of mtDNA quantification by qPCR comparing wild-type, atfs-1(null), and atfs-1ΔNLS worms fed for 48 h after L1 arrest (n = 4 ± SD for wild-type *p = 0.029 comparing wild-type and atfs-1(null) worm mtDNA at 48 h, *p = 0.035 comparing atfs-1(null) and atfs-1ΔNLS worm mtDNA at 48 h, two-way ANOVA with post hoc Tukey’s multiple comparisons test).
(F) Images of wild-type control and L1 arrest-fed worms treated with 0, 10, and 30 μg/mL of EtBr when the control worms reached the L4 stage (scale bar, 0.5 mm).
(G) Quantification of body length of wild-type control worms treated with 0, 10, and 30 μg/mL of EtBr, measured when worms treated with 0 mg/mL EtBr reached the L4 stage (n = 3, ****p < 0.0001, one-way ANOVA with post hoc Tukey’s test).
(H) Quantification of body length of wild-type L1 arrest-fed worms treated with 0, 10, and 30 μg/mL of EtBr when the worms treated with 0 μg/mL EtBr reached the L4 stage (n = 3, ****p < 0.0001, one-way ANOVA with post hoc Tukey’s test).
To examine the role of mitochondrion-localized ATFS-1 in recovery from L1 arrest, we evaluated the mitochondrial network in atfs-1R/R worms in which the strength of the ATFS-1 MTS was increased by substituting the threonine and aspartic acid residues at positions 10 and 24 with arginine residues. Consistent with increased import of ATFS-1R/R into mitochondria, atfs-1R/R worms have an impaired nuclear response during mitochondrial stress and a fragmented mitochondrial network, as shown previously.1 Similar to atfs-1ΔNLS worms, L1 arrested atfs-1R/R worms developed to adulthood upon feeding (Figure S4D). The mtDNA content in atfs-1R/R worms was also higher than in atfs-1(null) worms exiting prolonged L1 arrest (Figure S4E). These findings indicate that mitochondrion-localized ATFS-1 is required to resume growth and recover mtDNA content after prolonged L1 arrest.
We found previously that ATFS-1 binds directly to mtDNA in dysfunctional mitochondria and that inhibition of atfs-1 during mitochondrial stress results in an aberrant increase of mtDNA-encoded OXPHOS transcripts.9 We hypothesized that, in addition to promoting mtDNA recovery, mitochondrion-localized ATFS-1 might also limit mtDNA-encoded transcript accumulation to promote development after starvation. We found that several mtDNA-encoded mRNAs were increased in atfs-1(null) worms but not nuclear genome-encoded OXPHOS genes after introduction of food after starvation (Figures S4F and S4G). These findings suggest that, in addition to promoting mtDNA replication, mitochondrion-localized ATFS-1 represses or limits transcription of mtDNA-encoded OXPHOS transcripts during growth upon feeding after prolonged L1 starvation, potentially to facilitate efficient OXPHOS complex assembly.9
We next sought to determine whether the atfs-1-mediated recovery of mtDNA content is required for recovery from prolonged L1 arrest. Thus, we exposed L1-arrested wild-type worms to ethidium bromide (EtBr), which intercalates in mtDNA and inhibits replication2,36 and transcription.37 Exposure to 10 or 30 mg/mL EtBr modestly affected development of wild-type worms hatched directly onto food (Figures 3F and 3G). The same doses severely impaired development of worms after prolonged L1 arrest (Figures 3F and 3H). These data suggest that recovery of mtDNA copy numbers mediated by mitochondrion-localized ATFS-1 is required for growth and germline development after prolonged L1 arrest, but it is difficult to determine the effect of aberrant mtDNA transcription.
daf-2 inhibition promotes atfs-1-dependent mtDNA replication
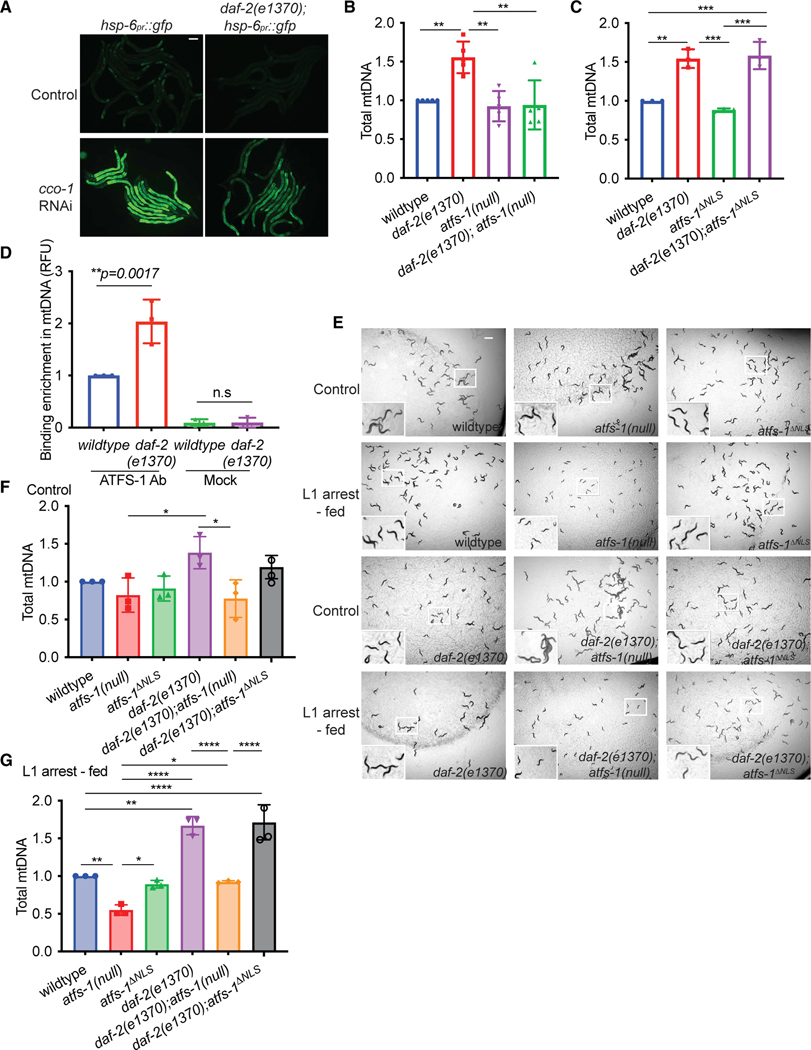
Last, we sought to gain insight into the upstream events regulating atfs-1-dependent mtDNA replication that occur after prolonged L1 arrest. We and others found previously that inhibition of the insulin-like receptor DAF-2 using a hypomorphic mutant (daf-2(e1370)) impairs atfs-1-dependent induction of hsp-6pr::gfp during mitochondrial stress (Figure 4A), suggesting that DAF-2 activity promotes nuclear accumulation of ATFS-1.1,38 daf-2(e1370) worms hatched in the presence of food harbored nearly 1.5-fold more mtDNAs than wild-type worms at the L4 stage, suggesting that modest inhibition of DAF-2 promotes mtDNA replication39 (Figure 4B). We found that the increase in mtDNAs was abrogated in daf-2(e1370);atfs-1(null) mutants (Figure 4B). daf-2(e1370);atfs-1ΔNLS worms harbored similar amounts of mtDNAs as daf-2(e1370) worms (Figure 4C), suggesting that mitochondrion-localized ATFS-1 promotes mtDNA replication when daf-2 is inhibited. Consistent with ATFS-1 localizing to mitochondria, a chromatin precipitation (ChIP) assay using ATFS-1-specific antibodies9 indicated that ATFS-1 interacts with 2-fold more mtDNAs upon DAF-2 inhibition (Figure 4D). These data indicate that decreased DAF-2 activity reduces atfs-1-dependent transcription in the nucleus and allows mitochondrion-localized ATFS-1 to increase mtDNA content.
Figure 4. daf-2 inhibition promotes atfs-1-dependent mtDNA replication.
(A) Fluorescence images of hsp-6pr::gfp and daf-2(e1370);hsp-6pr::gfp worms raised on control or cco-1(RNAi) at the L4 stage (scale bar, 0.1 mm).
(B) mtDNA quantification of wild-type, atfs-1(null), daf-2(e1370), and daf-2(e1370);atfs-1(null) worms, determined at the L4 stage (n = 5 ± SD, ****p < 0.0001, one way ANOVA with post hoc Tukey’s multiple comparisons test).
(C) mtDNA quantification of wild-type, atfs-1ΔNLS, daf-2(e1370), and daf-2(e1370);atfs-1DNLS worms determined at the L4 stage (n = 3 ± SD, ****p < 0.0001, one-way ANOVA with post hoc Tukey’s multiple comparison test).
(D) mtDNA quantification after ATFS-1 or control ChIP in wild-type and daf-2(e1370) worms at the L4 stage (n = 3 ± SD, **p = 0.0017, one-way ANOVA with post hoc Tukey’s multiple comparisons test).
(E) Images of wild-type, atfs-1(null), atfs-1ΔNLS, daf-2(e1370), daf-2(e1370);atfs-1(null), and daf-2(e1370);atfs-1ΔNLS worms under control and L1 arrest-fed conditions, obtained when control worms reached the L4 stage (scale bar, 0.5 mm).
(F) mtDNA quantification of wild-type, atfs-1(null), atfs-1ΔNLS, daf-2(e1370), daf-2(e1370);atfs-1(null), and daf-2(e1370);atfs-1ΔNLS worms under control conditions, as determined by qPCR at the L4 stage (n = 3 ± SD, *p = 0.132, one-way ANOVA with post hoc Tukey’s multiple comparisons test).
(G) mtDNA quantification of wild-type, atfs-1(null), atfs-1ΔNLS, daf-2(e1370), daf-2(e1370);atfs-1(null), and daf-2(e1370);atfs-1ΔNLS worms under L1 arrest conditions, as determined by qPCR when control worms reached the L4 stage (n = 3 ± SD, ****p < 0.0001, one-way ANOVA with post hoc Tukey’s multiple comparisons test).
Next we examined the effect of L1 arrest on the mtDNA content of daf-2(e1370) mutants. Although daf-2(e1370) worms hatched with increased mtDNA content relative to wild-type worms, mtDNA content declined at similar rates in both strains during L1 arrest (Figure S5A). We hypothesized that the increased mtDNA content in daf-2(e1370) worms throughout L1 arrest can be advantageous for growth or recovery after prolonged L1 arrest. As expected, growth and mtDNA levels were impaired in wild-type worms exposed to EtBr, whereas daf-2(e1370) worms developed to adulthood and had increased mtDNA content after prolonged L1 arrest (Figures S5B–S5D), suggesting that the lack of mtDNA is a rate-limiting factor for growth after starvation.
We next examined the role of ATFS-1 in recovery of daf-2(e1370) worms after prolonged starvation. daf-2(e1370);atfs-1(null) worms did not develop into mature adults (Figure 4E). However, daf-2(e1370);atfs-1ΔNLS worms developed into fertile adults, similar to wild-type worms (Figure 4E). daf-2(e1370);atfs-1(null) worms harbor fewer mtDNAs relative to daf-2(e1370) and daf-2(e1370);atfs-1ΔNLS worms after starvation, suggesting that ATFS-1 functions downstream of DAF-2 inhibition to increase the mtDNA content (Figures 4F and 4G) required for development after prolonged L1 arrest. However, it remains unclear why daf-2(e1370);atfs-1(null) worms fail to develop after prolonged L1 arrest because they harbor similar quantities of mtDNA as wild-type and atfs-1ΔNLS worms, which recover efficiently from L1 arrest.
The FOXO-like transcription factor DAF-16 is the canonical downstream effector activated upon DAF-2 inhibition. DAF-16 promotes L1 survival by regulating transcription of numerous genes, including the cyclin-dependent kinase inhibitor, and by altering metabolic flux to promote trehalose synthesis during L1 arrest.20,40 daf-16 loss-of-function mutant worms are unable to develop after prolonged L1 arrest.41 daf-2(e1370);daf-16(mu86) worms had fewer mtDNAs than daf-2(e1370) worms, consistent with DAF-16 also promoting mtDNA accumulation when daf-2 is inhibited (Figure S5E). Thus, we sought to understand the relationship between daf-16 and atfs-1, using the partial deletion mutant daf-16(mu86). daf-2(e1370);daf16(mu86);atfs-1(null) worms had a slight but not significant reduction in mtDNAs compared with either single deletion, suggesting that both ATFS-1 and DAF-16 function downstream of DAF-2 to regulate mtDNA content. However, it remains unclear whether ATFS-1 and DAF-16 function in parallel pathways or in the same pathway (Figure S5F). Because daf-16-mutants are unable to maintain and survive prolonged L1 arrest,20,40 the role of DAF-16 in increasing mtDNA content upon feeding remains unclear. Based on these data, we conclude that a reduction in DAF-2 activity leads to increased mtDNA content, partially dependent on mitochondrial ATFS-1, which promotes growth and mitochondrial recovery after prolonged starvation.
DISCUSSION
C. elegans mature from egg to reproductive adult within 3 days. However, during development, worms can enter a diapause-like state at different developmental stages to endure prolonged stress, including starvation.42 Worms that hatch in the absence of food remain developmentally arrested at the L1 stage and can survive for over 30 days (Figure 1C).23 To sustain the search for food, cytosolic components, including mitochondria, are degraded by autophagy, which is used to generate ATP via OX-PHOS.21,22 When food is encountered, a major challenge to resumption of development is repairing the cellular and tissue damage incurred in the absence of food.23
Here, we find that, after 5 days of L1 arrest, ATFS-1 is required for development and to establish a functional germline. A previous study found a strong correlation between mutations that extend L1 survival and mutations that extend the adult lifespan.43 In contrast, we found that, although atfs-1(null) worms are relatively short-lived when hatched in presence of food, the absence of ATFS-1 does not affect L1 survival. ATFS-1 does not appear to have any role in the mtDNA accumulation that occurs during embryogenesis through the L1 stage. During prolonged L1 arrest, mtDNAs, and presumably whole mitochondria, are degraded via autophagy.22 Our findings indicate that mtDNA depletion via autophagy occurs independent of ATFS-1 during L1 arrest. However, to mature into reproductive adults after introduction of food, ATFS-1 is essential to increase or recover the mtDNA content depleted during L1 arrest to a level comparable with that of newly hatched worms. The NLS in ATFS-1 required for atfs-1-dependent nuclear transcription was not required to recover mtDNA content or establish a functional mitochondrial network or functional germline. However, atfs-1ΔNLS worms have perturbed mitochondrial morphology at the L4 stage, consistent with ATFS-1 coordinating mitochondrial network expansion with the increased levels of protein synthesis and growth that occur upon feeding.1
Lastly, our findings suggest that the activity of mitochondrion-localized ATFS-1 is required to increase mtDNA content upon inhibition of the insulin receptor-like protein DAF-2. We found previously that daf-2-inhibition impairs atfs-1-dependent transcription, potentially by impairing S6 kinase and reducing general rates of protein synthesis, which result in the majority of ATFS-1 trafficking to mitochondria.1 Here, we find that a reduction in DAF-2 function causes an increase in mtDNA content, which requires ATFS-1, but not ATFS-1-dependent, nuclear transcription, suggesting that a reduction in DAF-2 activity promotes mtDNA replication via mitochondrion-localized ATFS-1. We propose that this “toggle switch”-like activity of DAF-2 ensures that synthesis of mtDNAs occurs prior to activation of the transcription program required for expansion of the mitochondrial network driven by TORC1-dependent protein synthesis during worm development. These findings are consistent with our previous work indicating that mitochondrion-localized ATFS-1 promotes mtDNA replication in dysfunctional mitochondria, which can contribute to a replicative advantage of deleterious mtDNAs in a disease model.6,10 Our finding that ATFS-1 is required to increase mtDNA replication and recover mtDNAs degraded during starvation suggest a reason why such a pathway may have evolved because recovery of mtDNA content after L1 arrest is essential for development and C. elegans reproduction.
Limitations of the study
In the current study, we demonstrated a requirement for ATFS-1 in worm development after prolonged L1 arrest. Our findings suggest that ATFS-1 is required to recover the mtDNA content depleted during L1 arrest because of autophagy. To examine mitochondrial function, we utilized the functional dye TMRE and mScarlet fused to TOMM-20, which accumulates on the mitochondrial membrane independent of mitochondrial function. These tools enable assessment of mitochondrial function and morphology. However, we did not perform higher-resolution imaging such as electron microscopy, which may provide further insight into the mitochondrial defects that occur in the absence of ATFS-1 after L1 arrest. We found that the development of wild-type worms after L1 arrest was impaired by exposure to EtBr, whereas worms that hatched in the presence of food developed relatively normally with low EtBr concentrations. Because EtBr is a known inhibitor of mtDNA replication,44 we interpreted these findings to suggest that mtDNA replication is essential for recovery from prolonged L1 arrest. However, because ATFS-1 is also a negative regulator of transcription of mtDNA-encoded genes, we cannot rule out a role of aberrant transcription in impaired recovery from L1 arrest. Lastly, we suggest a role of DAF-2 in regulating the ATFS-1-dependent increase in mtDNA content after L1 arrest. The increased mtDNA content in daf-2(e1270) worms relative to wild-type worms partially requires ATFS-1, and the development of daf-2(e1270) worms after L1 arrest also requires ATFS-1. However, daf-2(e1270);atfs-1(null) worms have similar levels of mtDNA as atfs-1ΔNLS worms, which are able to develop after L1 arrest. The underlying biology remains unclear but may involve other downstream regulators of DAF-2. Because metabolism and growth are considerably altered in daf-2(e1270) worms, their reliance on mitochondrial function and mtDNA for development may also be altered.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents or strains must be directed to and will be fulfilled by the lead contact, Cole Haynes (cole.haynes@umassmed.edu).
Materials availability
This study did not generate new unique materials.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data deposited in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans strains
C. elegans strains were fed and cultured on HT115 Escherichia Coli strain at 20°C unless otherwise specified. The reporter strain hsp-6pr::gfp was used for visualizing UPRmt activation.2 N2 (wildtype), clk-1(qm30), daf-2(e1370), daf-16(mu86) strains were obtained from the CGC. PIE:GFP and GLH-1:GFP were gifts from Dr. Craig Mello. pdr-1(tm598) was obtained from the National BioResource project (Tokyo, Japan). atfs-1(null), atfs-1ΔNLS, and the atfs-1R/R strains were generated using CRISPR-Cas9 as described.1,5,10 vha-6p:tomm-20(1–54):mScarlet strain was a gift from Dr. Gary Ruvkun 33.
METHOD DETAILS
Starvation plates and recovery from prolonged L1 arrest
For L1 arrest, worms were synchronized by bleaching and transferred to NGM plates that lacked cholesterol and peptone for 5 days.46 Following L1 arrest, the worms were transferred to NGM plates seeded with HT115 bacteria (L1 arrest-fed condition). Control condition worms were synchronized by bleaching and allowed to hatch directly on NGM plates seeded with HT115 bacteria. All strains were maintained at 20°C throughout all experiments. For images of worm development, the L1 arrest-fed worms of each strain were compared to control worms of the same developmental stage.
Brood size quantification
Three L4 stage nematodes were randomly selected from each condition and transferred to a fresh plate and incubated at 20°C. Once worms reached adulthood, the number of progenies were quantified until the worms stopped laying eggs. Brood size was determined as the average number of progenies per worm on each plate based on the sum of total eggs. Five biological replicates per strain per condition were used and 10–15 worms were used for quantification of eggs from each replicate.
mtDNA quantification
mtDNA was measured using quantitative PCR (qPCR) similar to previously published methods.1,6,47 30–40 worms were collected per sample in lysis buffer containing 50 mM KCl, 10 mM Tris-HCl pH 8.3, 2.5 mM MgCl2, 0.45% NP-40, 0.45% Tween 20, 0.01% gelatin, fresh stock of 200 mg/mL proteinase K) and immediately frozen at −80°C for at least 15 min followed by lysis at 65C for 80 min 2μL of lysate was used per reaction, and they were performed in biological and technical triplicates using iQ SYBR Green Supermix and the Biorad qPCR CFX96 (Bio-Rad Laboratories). The average Ct (threshold cycle) of technical replicate values were obtained for mtDNA and was normalized using the 2−ΔΔCt method. Primers that specifically amplify mtDNA are listed in Table S2. Primers that amplify a non-coding region near the nuclear-encoded ges-1 gene were used as a control. Student’s t-test or one-way ANOVA was used to determine the statistical significance where applicable.
For measuring mtDNA recovery in Figure 2H over time course of 24 h, control worms were synchronized to L1s overnight prior to transferring on bacteria seeded plates and L1 arrest-fed worms were starved for 5 days prior to transferring on bacteria seeded plates.
RNA isolation and qRT-PCR
Worms were harvested from plates into tubes and washed 3 times using S. basal to remove bacteria. 1 mL of TRIzol Reagent (Invitrogen) was added to the worm pellet and the tubes were flash frozen using liquid nitrogen. Total RNA was isolated from worm pellets using the TRIzol Reagent and chloroform. cDNA was then synthesized from total RNA using the iScript cDNA Synthesis Kit (Bio-Rad). qPCR was performed using iQ SYBR Green Supermix. Primer sequences are listed in Table S2. Relative expression of target genes was normalized to the house keeping gene act-3. Fold changes in gene expression were calculated.
For qPCR of L1 worms, liquid cultures of different worm strains were grown for at least two generations with food and then prepped for eggs using bleach solution to obtain synchronized L1s. For L1 arrest-fed condition, eggs were hatched onto starvation plates for 5 days and then fed up to L2s before harvesting for RNA extraction. For control condition, eggs directly hatched on food were grown up to L2s were collected and used for RNA extraction.
Body length measurements
Body length quantifications were obtained at the L4 stage. However, as the clk-1(qm30) and clk-1(qm30); atfs-1ΔNLS strains developed slowly, body length measurements were obtained on day1 of egg lay. Body length measurements were obtained using ImageJ measuring from nose tip to worm tail as previously described.48
Microscopy
C. elegans were imaged using either a Zeiss AxioCam 506 mono camera mounted on a Zeiss Axio Imager Z2 microscope, a Zeiss AxioCam MRc camera mounted on a Zeiss SteREO Discovery V12 stereoscope or a ZEISS LSM800 confocal microscope. Exposure times were the same for each sample in each experiment.
L1 starvation survival
L1 survival assay was adapted from protocol previously described.18 Worms were grown for at least two generations at 20°C in liquid with food and bleached to collect eggs, which hatched overnight on a shaker incubator in S-basal without cholesterol. Synchronized L1s were maintained in 25- or 125-mL flasks at a concentration of ~1 worm/ml in 10–12 mL of S-basal without cholesterol and without antibiotics. Flasks were shaken at 200 rpm in a shaker at 20°C except when scoring. Viability was assessed by aliquoting 100 to 200 μL of worms to fresh plates and monitoring the number of worms that were alive or dead after 2 days post plating. The worms were scored every other day and the survival percent was calculated based on a ratio of the number of worms survived to total number of plated worms on each scoring day. The graph shows data collected from one biological replicate representative of 3 independent replicates. Summary statistics of 3 independent biological replicates are reported in Table S1. Lifespan variability between experiments may be due to variation in population density between biological replicates.49
Adult lifespan assay
Animals were raised on seeded NGM plates at 20°C until the L4 stage and ~100 L4 worms were transferred to fresh bacterial plates (10 worms per plate). Worms were scored every 2–3 days and those that were unresponsive to touch of the worm pick were marked as dead. Worms that bagged or crawled off the plate were censored and not included in the study. The figure shows data collected from a single experiment. Each experiment was repeated in 3 independent biological replicates (Table S3).
DAPI staining
Animals were grown on control or L1 arrest-fed condition until day 1 adulthood. Then, the worms were washed off the plates and placed in microcentrifuge tube containing 0.01% Tween-PBS. Worms were pelleted by low-speed centrifugation and washed 2–3 times with 0.01% Tween PBS. Next, 1 mL of pre-chilled methanol from −20°C was added to the whole worm pellet and tubes were immediately placed in the freezer at −20°C for exactly 5 min. The worms were pellet again and rinsed 2–3 times with 0.1% Tween-PBS. 0.2 μL of DAPI solution was added and the tube was allowed in sit in the dark for 5 min and after, rinsed with 0.1% Tween-PBS. Finally, 30 μL of 75% glycerol solution was added to worm pellet and were ready to be imaged.
TMRE staining
Freshly seeded HT115 bacterial plates were coated with TMRE to make a final concentration of 30 μm in each plate. The worms were either synchronized by bleaching and plated onto TMRE plates either from egg stage (control) or starved in L1 stage for 5 days using starvation plates and then transferred to TMRE plates (L1 arrest-fed). Three independent biological repeats were performed. The graph was generated by quantifying of 15–25 individual worms from a single experimental repeat that is representative of all three biological replicates. TMRE quantification was done as previously described.1 In brief, the average pixel intensity values were calculated by sampling images of different worms. The average pixel intensity for each animal was calculated using ImageJ (http://rsb.info.nih.gov/ij/ ). Mean values were compared using two-way ANOVA.
Mitochondrial morphology quantification
For morphology analysis, 3–4 worms were placed on a fresh 4% agarose pad on a single slide and scored within 5 min by imaging using a ZEISS LSM800 confocal microscope (minimal time was spent for each slide as prolonged exposure may induce network fission). All visible cells in each worm were scored on based 3 levels of classification as tubular, intermediate and fragmented as previously described.23 Three biological replicates were analyzed with ~10–15 worms scored in each replicate.
Ethidium bromide experiments
Control worms were bleached directly onto plates containing 0, 10 or 30 mg/mL ethidium bromide with HT115 bacteria. For starved-fed condition, worms were first bleached onto starvation plates for 5 days and then transferred onto ethidium bromide plates as described above. Plates were imaged when at the L4 stage or when they became developmentally arrested.
ChIP-mtDNA
ATFS-1 ChIP was performed as previously described.10 Synchronized worms were cultured on plates and harvested at the L4 stage. The worms were lysed via Teflon homogenizer in chilled PBS with protease inhibitors (Roche). Cross-linking of DNA and protein was performed by treating worms with 1.85% formaldehyde containing protease inhibitors for 15 min. Glycine was added to a final concentration of 125 mM and incubated for 5 min at room temperature to quench the formaldehyde. The tubes were cold centrifuged at 3000 rpm for 2 min and then, pellets were resuspended twice in cold PBS with protease inhibitors. Samples were treated with lysis buffer containing 0.1% Sarkosyl, protease inhibitors in FA Buffer (50 mM HEPES/KOH pH 7.5, 1mM EDTA, 1% Triton X-100, 0.1% Sodium deoxycholate, 150 mM NaCl). The supernatant was precleaned with pre-blocked ChIP-grade Pierce magnetic protein A/G beads (Thermo Scientific) and then incubated with Monoclonal Mouse mAb IgG1 Isotype Control (Cell Signaling Technology, G3A1) rotating overnight at 4C. The antibody-DNA complex was precipitated with protein A/G magnetic beads or protein A Sepharose beads (Invitrogen). After washing, the crosslinks were reversed by incubation at 65°C overnight. The samples were then treated with RNaseA at 37°C for 1.5 h followed by proteinase K treatment at 55°C for 2 h. Lastly, the immunoprecipitated and input DNA were purified with ChIP DNA Clean & Concentrator (Zymo Research, D5205) and used as templates for qPCR.
QUANTIFICATION AND STATISTICAL ANALYSIS
All experiments were performed at least three times and all the represented graphs are based on biologically independent replicates. “n” represents number of biological replicates in figure legends. L1 starvation survival and adult life span are graphs from a single biological replicate representative of three independent biological repeats. The summary statistics from three biological replicates are reported in Tables S1 and S3. For lifespan analysis, statistics were performed using OASIS2 software45 and values were calculated with the log rank/Mantel Cox test. p values were calculated by the two-tailed Student’s t-test, one-way ANOVA or two-way ANOVA with post-hoc analyses. The statistical tests were chosen based on previous studies with similar methodologies and analysis performed using Graphpad Prism 7. Experiments were not blinded, and randomization was not used. For all figures, the mean ± SD is represented unless otherwise noted.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Ethidium Bromide | Invitrogen | 15585011 |
| TMRE | Thermo Fisher | T669 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| C. elegans: MQ130 (clk-1(qm30)) | Caenorhabditis Genetics Center (CGC) | RRID: WB Cat#: WBStrain00026639 |
| C. elegans: CB1370 (daf-2(e1370)) | Caenorhabditis Genetics Center (CGC) | RRID: WB Cat#: WBStrain00004309 |
| C. elegans: CB1038 (daf-16(mu86)) | Caenorhabditis Genetics Center (CGC) | RRID: WB Cat#: WBStrain00004840 |
| atfs-1 ΔNLS | Shpilka et al., 2021,1 Yang et al., 202210 | N/A |
| atfs-1 R/R | Shpilka et al., 20211 | N/A |
| pdr-1(tm598) | National BioResource Project (Tokyo, Japan) |
N/A |
| atfs-1(null) | Deng et al., 20195 | N/A |
| PIE-1::GFP | Laboratory of Dr. Craig C. Mello | N/A |
| GLH-1::GFP | Laboratory of Dr. Craig C. Mello | N/A |
| vha-6p::tomm-20(1–54)::mScarlet | Laboratory of Dr. Gary Ruvkun | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| Full list of qRT-PCR primers is presented in Table S2 |
This paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Fiji ImageJ | ImageJ | RRID:SCR_002285 |
| Prism | Graphpad | RRID:SCR_002798 |
| OASIS2 | Han et al., 201645 | https://sbi.postech.ac.kr/oasis2/surv/ |
| Biorender | Biorender | https://biorender.com/ |
Highlights.
The mitochondrial UPR is required for C. elegans growth after prolonged starvation
UPRmt is required to increase mtDNA replication after starvation-induced mtDNA decline
Mitochondrial localization of the bZIP protein ATFS-1 promotes mtDNA content recovery
The insulin-like receptor DAF-2 coordinates ATFS-1-mediated mtDNA replication and growth
ACKNOWLEDGMENTS
We thank the Caenorhabditis Genetics Center for providing C. elegans strains, funded by NIH Office of Research 362 Infrastructure Programs (P40 OD010440). This work was supported by HHMI and National Institutes of Health grants (R37AG047182-08 and R56AG075204) to C.M.H.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111875.
REFERENCES
- 1.Shpilka T, Du Y, Yang Q, Melber A, Uma Naresh N, Lavelle J, Kim S, Liu P, Weidberg H, Li R, et al. (2021). UPR(mt) scales mitochondrial network expansion with protein synthesis via mitochondrial import in Caenorhabditis elegans. Nat. Commun 12, 479. 10.1038/s41467-020-20784-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, and Ron D. (2004). Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J. Cell Sci 117, 4055–4066. 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- 3.Durieux J, Wolff S, and Dillin A. (2011). The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144, 79–91. 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moullan N, Mouchiroud L, Wang X, Ryu D, Williams EG, Mottis A,Jovaisaite V, Frochaux MV, Quiros PM, Deplancke B, et al. (2015). Tetracyclines disturb mitochondrial function across eukaryotic models: a call for caution in biomedical research. Cell Rep. 10, 1681–1691. 10.1016/j.celrep.2015.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng P, Uma Naresh N, Du Y, Lamech LT, Yu J, Zhu LJ, PukkilaWorley R, and Haynes CM (2019). Mitochondrial UPR repression during Pseudomonas aeruginosa infection requires the bZIP protein ZIP-3. Proc. Natl. Acad. Sci. USA 116, 6146–6151. 10.1073/pnas.1817259116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin YF, Schulz AM, Pellegrino MW, Lu Y, Shaham S, and Haynes CM (2016). Maintenance and propagation of a deleterious mitochondrial genome by the mitochondrial unfolded protein response. Nature 533, 416–419. 10.1038/nature17989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gitschlag BL, Kirby CS, Samuels DC, Gangula RD, Mallal SA,and Patel MR (2016). Homeostatic responses regulate selfish mitochondrial genome dynamics in C. elegans. Cell Metab. 24, 91–103. 10.1016/j.cmet.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, and Haynes CM (2012). Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337, 587–590. 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, and Haynes CM (2015). Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt). Mol.Cell 58, 123–133. 10.1016/j.molcel.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Q, Liu P, Anderson NS, Shpilka T, Du Y, Naresh NU, Li R,Zhu LJ, Luk K, Lavelle J, et al. (2022). LONP-1 and ATFS-1 sustain deleterious heteroplasmy by promoting mtDNA replication in dysfunctional mitochondria. Nat. Cell Biol 24, 181–193. 10.1038/s41556-021-00840-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu CK, Wang W, Brind’Amour J, Singh PP, Reeves GA, Lorincz MC, Alvarado AS, and Brunet A. (2020). Vertebrate diapause preserves organisms long term through Polycomb complex members. Science 367, 870–874. 10.1126/science.aaw2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Gilst M. (2020). A time to grow and a time to pause. Science 367, 851–852. 10.1126/science.aba8064. [DOI] [PubMed] [Google Scholar]
- 13.Hu CK, and Brunet A. (2018). The African turquoise killifish: a researchorganism to study vertebrate aging and diapause. Aging Cell 17, e12757. 10.1111/acel.12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hand SC, Denlinger DL, Podrabsky JE, and Roy R. (2016). Mechanisms of animal diapause: recent developments from nematodes, crustaceans, insects, and fish. Am. J. Physiol. Regul. Integr. Comp. Physiol 310, R1193–R1211. 10.1152/ajpregu.00250.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baugh LR (2013). To grow or not to grow: nutritional control of development during Caenorhabditis elegans L1 arrest. Genetics 194, 539–555. 10.1534/genetics.113.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carranza-García E, and Navarro RE (2020). Insights into the hypometabolic stage caused by prolonged starvation in L4-adult Caenorhabditis elegans hermaphrodites. Front. Cell Dev. Biol 8, 124. 10.3389/fcell.2020.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan REW, Chen Y, Moore BT, Jordan JM, Maxwell CS, Schindler AJ, and Baugh LR (2015). dbl-1/TGF-beta and daf-12/NHR signaling mediate cell-nonautonomous effects of daf-16/FOXO on starvation-induced developmental arrest. PLoS Genet. 11, e1005731. 10.1371/journal.pgen.1005731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee BH, and Ashrafi K. (2008). A TRPV channel modulates C. elegansneurosecretion, larval starvation survival, and adult lifespan. PLoS Genet. 4, e1000213. 10.1371/journal.pgen.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demoinet E, Li S, and Roy R. (2017). AMPK blocks starvation-inducibletransgenerational defects in Caenorhabditis elegans. Proc. Natl. Acad.Sci. USA 114, E2689–E2698. 10.1073/pnas.1616171114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baugh LR, and Sternberg PW (2006). DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr. Biol 16, 780–785. 10.1016/j.cub.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 21.Kang C, You YJ, and Avery L. (2007). Dual roles of autophagy in thesurvival of Caenorhabditis elegans during starvation. Genes Dev. 21, 2161–2171. 10.1101/gad.1573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hibshman JD, Leuthner TC, Shoben C, Mello DF, Sherwood DR,Meyer JN, and Baugh LR (2018). Nonselective autophagy reduces mitochondrial content during starvation in Caenorhabditis elegans. Am. J. Physiol. Cell Physiol 315, C781–C792. 10.1152/ajpcell.00109.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roux AE, Langhans K, Huynh W, and Kenyon C. (2016). Reversible-age-related phenotypes induced during larval Quiescence in C. elegans.Cell Metab. 23, 1113–1126. 10.1016/j.cmet.2016.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baugh LR, Demodena J, and Sternberg PW (2009). RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science 324, 92–94. 10.1126/science.1169628. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, and Kim SK (2003). Global analysis of dauer gene expressionin Caenorhabditis elegans. Development 130, 1621–1634. 10.1242/dev.00363. [DOI] [PubMed] [Google Scholar]
- 26.Stadler M, and Fire A. (2013). Conserved translatome remodeling innematode species executing a shared developmental transition. PLoS Genet. 9, e1003739. 10.1371/journal.pgen.1003739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vermeulen K, Van Bockstaele DR, and Berneman ZN (2003). The cellcycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif 36, 131–149. 10.1046/j.1365-2184.2003.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sclafani RA, and Holzen TM (2007). Cell cycle regulation of DNA replication. Annu. Rev. Genet 41, 237–280. 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis SC, Uchiyama LF, and Nunnari J. (2016). ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science 353, aaf5549. 10.1126/science.aaf5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seydoux G. (2018). The P granules of C. elegans: a genetic model for thestudy of RNA-protein condensates. J. Mol. Biol 430, 4702–4710. 10.1016/j.jmb.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mello CC, Schubert C, Draper B, Zhang W, Lobel R, and Priess JR (1996). The PIE-1 protein and germline specification in C. elegans embryos. Nature 382, 710–712. 10.1038/382710a0. [DOI] [PubMed] [Google Scholar]
- 32.Reese KJ, Dunn MA, Waddle JA, and Seydoux G. (2000). Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol.Cell 6, 445–455. 10.1016/s1097-2765(00)00043-5. [DOI] [PubMed] [Google Scholar]
- 33.Mao K, Ji F, Breen P, Sewell A, Han M, Sadreyev R, and Ruvkun G(2019). Mitochondrial dysfunction in C. elegans activates mitochondrial relocalization and nuclear hormone receptor-dependent detoxification genes. Cell Metab. 29, 1182–1191.e4. 10.1016/j.cmet.2019.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Felkai S, Ewbank JJ, Lemieux J, Labbé JC, Brown GG, and Hekimi S. (1999). CLK-1 controls respiration, behavior and aging in the nematode Caenorhabditis elegans. EMBO J. 18, 1783–1792. 10.1093/emboj/18.7.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narendra D, Tanaka A, Suen DF, and Youle RJ (2008). Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol 183, 795–803. 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinus RD, Garth GP, Webster TL, Cartwright P, Naylor DJ, Høj PB, and Hoogenraad NJ (1996). Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. Eur. J. Biochem 240, 98–103. 10.1111/j.1432-1033.1996.0098h.x. [DOI] [PubMed] [Google Scholar]
- 37.Zylber E, Vesco C, and Penman S. (1969). Selective inhibition of thesynthesis of mitochondria-associated RNA by ethidium bromide. J. Mol.Biol 44, 195–204. 10.1016/0022-2836(69)90414-8. [DOI] [PubMed] [Google Scholar]
- 38.Gatsi R, Schulze B, Rodrıģuez-Palero MJ, Hernando-Rodrıģuez B, Baumeister R, and Artal-Sanz M. (2014). Prohibitin-mediated lifespan and mitochondrial stress implicate SGK-1, insulin/IGF and mTORC2 in C. elegans. PLoS One 9, e107671. 10.1371/journal.pone.0107671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haroon S, Li A, Weinert JL, Fritsch C, Ericson NG, AlexanderFloyd J, Braeckman BP, Haynes CM, Bielas JH, Gidalevitz T, and Vermulst M. (2018). Multiple molecular mechanisms rescue mtDNA disease in C. elegans. Cell Rep. 22, 3115–3125. 10.1016/j.celrep.2018.02.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hibshman JD, Doan AE, Moore BT, Kaplan RE, Hung A, Webster AK, Bhatt DP, Chitrakar R, Hirschey MD, and Baugh LR (2017). daf-16/FoxO promotes gluconeogenesis and trehalose synthesis during starvation to support survival. Elife 6, e30057. 10.7554/eLife.30057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olmedo M, Mata-Cabana A, Rodriguez-Palero MJ, Garcia-Sanchez S, Fernandez-Yanez A, Merrow M, et al. (2020). Prolonged quiescence delays somatic stem cell-like divisions in Caenorhabditis elegans and is controlled by insulin signaling. Aging Cell 19, e13085. 10.1111/acel.13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baugh LR, and Hu PJ (2020). Starvation responses throughout theCaenorhabditis elegans life cycle. Genetics 216, 837–878. 10.1534/genetics.120.303565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muñoz MJ, and Riddle DL (2003). Positive selection of Caenorhabditis elegans mutants with increased stress resistance and longevity. Genetics 163, 171–180. 10.1093/genetics/163.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsang WY, and Lemire BD (2002). Mitochondrial genome content isregulated during nematode development. Biochem. Biophys. Res. Commun 291, 8–16. 10.1006/bbrc.2002.6394. [DOI] [PubMed] [Google Scholar]
- 45.Han SK, Lee D, Lee H, Kim D, Son HG, Yang JS, Lee SJV, andKim S. (2016). Oasis 2: online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget 7, 56147–56152. 10.18632/oncotarget.11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stiernagle T. (2006). Maintenance of C. elegans (WormBook), pp. 1–11. 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed]
- 47.Valenci I, Yonai L, Bar-Yaacov D, Mishmar D, and Ben-Zvi A. (2015). Parkin modulates heteroplasmy of truncated mtDNA in Caenorhabditis elegans. Mitochondrion 20, 64–70. 10.1016/j.mito.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 48.Mörck C, and Pilon M. (2006). C. elegans feeding defective mutants have shorter body lengths and increased autophagy. BMC Dev. Biol 6, 39. 10.1186/1471-213X-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Artyukhin AB, Schroeder FC, and Avery L. (2013). Density dependence in Caenorhabditis larval starvation. Sci. Rep 3, 2777. 10.1038/srep02777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data deposited in this paper is available from the lead contact upon request.