Abstract
Background
PTTG1 has been reported to be linked with the prognosis and progression of various cancers, including kidney renal clear cell carcinoma (KIRC). In this article, we mainly investigated the associations between prognosis, immunity, and PTTG1 in KIRC patients.
Method
We downloaded transcriptome data from the TCGA-KIRC database. PCR and immunohistochemistry were used, respectively, to validate the expression of PTTG1 in KIRC at the cell line and the protein levels. Survival analyses as well as univariate or multivariate Cox hazard regression analyses were used to prove whether PTTG1 alone could affect the prognosis of KIRC. The most important point was to study the relationship between PTTG1 and immunity.
Results
The results of the paper revealed that the expression levels of PTTG1 were elevated in KIRC compared with para-cancerous normal tissues, validated by PCR and immunohistochemistry at the cell line and the protein levels (P < 0.05). High PTTG1 expression was related to shorter overall survival (OS) in patients with KIRC (P < 0.05). Through univariate or multivariate regression analysis, PTTG1 was confirmed to be an independent prognostic factor for OS of KIRC (P < 0.05), and its related seven pathways were obtained through gene set enrichment analysis (GSEA; P < 0.05). Moreover, tumor mutational burden (TMB) and immunity were found to be significantly connected with PTTG1 in KIRC (P < 0.05). Correlations between PTTG1 and immunotherapy responses implied that the low-PTTG1 group was more sensitive to immunotherapy (P < 0.05).
Conclusions
PTTG1 was closely associated with TMB or immunity, and it had a superior ability to forecast the prognosis of KIRC patients.
Keywords: PTTG1, Kidney renal clear cell carcinoma, Immunity, Immunotherapy, Prognosis
1. Introduction
Renal carcinoma is a common malignant neoplasm of the urinary system, and the United States is projected to have 79,000 new cases and 13,920 deaths in 2022 [1]. The most common pathologic form of renal cell carcinoma (RCC) was kidney renal clear cell carcinoma (KIRC), which made up about 75% of all RCC [2]. KIRC was not sensitive to traditional radiotherapy or chemotherapy, and the cost of targeted therapy was expensive. Currently, the mainstay of therapy for RCC was still surgery [3]. Laparoscopic radical nephrectomy had been widely accepted by a growing number of patients and medical workers. However, patients with KIRC often lacked specific clinical manifestations in the early stages. By the time they were diagnosed, nearly one-third of patients had distant metastases, so they had missed the best time for treating [4]. In addition to surgical treatment, immunotherapy also had gained a lot of attentions. However, its effectiveness was still unsatisfactory. Therefore, in order to improve the prognosis of KIRC, looking for a meaningful biomarker was an urgent priority.
Pituitary tumor-transforming 1 (PTTG1), as a member of the PTTG family, took part in the regulation of sister chromatid separation [5,6], and it was a pituitary-derived transformation gene isolated from rat pituitary tumor cells [7]. Elevated PTTG1 expression could interfere with sister chromatid separation during mitosis, which might be a key factor in cancer [8]. A large amount of researches had confirmed that PTTG1 was an oncogenic gene and was highly expressed in various tumor tissues, including RCC, liver, prostate, and colorectal cancers [[9], [10], [11]]. As reported, PTTG1 could take part in organ development, cell proliferation, and apoptosis, being confirmed as an important gene related to tumor metastasis [[12], [13], [14], [15]]. So far, PTTG1 has been studied in many tumors, including KIRC. However, its associated roles and their relationships with immunity had not been fully elucidated. In this article, PTTG1 was selected through TCGA database mining, and the intention of the paper was to study the prognostic role of PTTG1 as well as its relationships with immunity in KIRC, offering a novel solution for the treatment of these patients in the future.
2. Method and materials
2.1. Acquisition of data
The TCGA-KIRC dataset (http://cancergenome.nih.gov/), including 539 KIRC tissues and 72 normal renal tissues, was searched for the PTTG1 transcriptome profile matrix and corresponding clinical information. The above data was standardized with the help of R software (https://www.r-project.org/). Differently expressed genes (DEG) were screened out by the usage of ‘limma’ package among KIRC and adjacent tissues, with |log2 fold change (FC)|≥1 and adjusted p values < 0.05.
2.2. Isolation of RNA, quantitative real-time PCR
HK-2, 786-O, and CAKI-1 cells, purchased from the Shanghai Institute for Biological Sciences, were employed to verify the expression of PTTG1 by PCR. HK-2, 786-O, and CAKI-1 cells were all incubated in a constant temperature incubator at 37 °C with 5% CO2. Among them, HK-2 was cultured on DMEM/F12 containing 10% FBS (Gibcol, from Life Technologies™) and 1% penicillin-streptomycin solution. 786-O was cultured on RPMI 1640 medium containing 10% FBS and 1% penicillin-streptomycin solution. CAKI-1 was incubated with McCoy’s 5 A supplemented with 10% FBS and 1% penicillin-streptomycin solution. We use Trizol reagent to extract RNA from these three types of cells, as directed by the manufacturer. Then, cDNA was obtained by using Fermentas reverse transcription kit. qRT-PCR was carried out with SYBR green reagent, and the expression of PTTG1 in renal carcinoma cells was quantified by the 2−ΔΔCT method. At the same time, GAPDH was used as an internal reference gene. The relevant primers utilized for PCR were detailed as follows: PTTG1: F: 5’- TCAGATGACGCCTATCCAG - 3’; R: 5’ - GGCACTCCACTCAAGGG - 3’. GAPDH: F: 5’- CAGGAGGCATTGCTGATGAT -3'; R: 5’- GAAGGCTGGGGCTCATTT - 3’.
2.3. Immunohistochemistry staining (IHC)
Immunohistochemistry staining was conducted as previously reported [16]. 10 pairs of KIRC tissue and its adjacent tissues samples were collected from the Affiliated Hospital of Nantong University. The tissue was first embedded in paraffin wax and then cut into 4 μm slices, followed by dewaxing and hydration. After sufficient washing with PBS, thermal antigen repair was performed with the citric acid antigen repair solution. The endogenous peroxidase activity was inactivated by incubation with 3% hydrogen peroxide for 20 min at room temperature and then closed with 3% BSA for 20 min at room temperature. The PTTG1 antibody (orb374037, Biorbyt) was used to incubate tissue section. Next, primary antibodies were added and the sections were incubated flat in a wet box at 4 °C in the refrigerator overnight. The dilution ratio of the primary antibody was 1:200. The next day, the sections were removed and then covered with secondary antibody dropwise and incubated for 30 min at room temperature. After shaking dry, DAB chromogenic solution was added dropwise and observed under the microscope for 5 min, and the color development time was controlled. Then the sections were restained in hematoxylin for 2 min and then fractionated with 1% hydrochloric acid ethanol for 10 s. Finally, the slices were dehydrated, dried, sealed with drops of neutral gum, and photographed under the microscope. We interpreted the immunohistochemical results according to the literature [17]. The number of positive cells was calculated as follows: 0 was 0, ≤ 25% was 1, 26%–50% was 2, and 51% or more was 3. Staining intensity was calculated as follows: 0 points if no coloring or coloring was not obvious, 1, 2 and 3 points if the color was yellow, brown and tawny, respectively. The scores of positive cells and staining intensity were multiplied, and the results were regarded as high expression if the score was >3 and low expression if the score was ≤3. The study was approved by the Institutional Research Ethics Committees of Affiliated Hospital of Nantong University.
2.4. Survival analysis and clinical correlation analysis
With the median of PTTG1 gene expression in the sample as the boundary, PTTG1 was divided into a high expression group and a low expression group. The Kaplan-Meier survival curve was employed to analyze the correlation between PTTG1 and the OS in KIRC patients. Univariate or multivariate Cox hazard regression analyses were used to prove whether PTTG1 alone could affect the prognosis of KIRC.
2.5. Gene set enrichment analysis (GSEA)
GSEA, as a computational method, could enrich genes in certain signaling pathways [18]. According to the expression level of PTTG1, they were divided into two groups: high and low expression groups. GSEA analysis was conducted to obtain the signaling pathways enriched by the high and low groups of PTTG1, with 1000 permutations. Pathways were screened by FDR q-value, nominal p-value, and normalized enrichment score (NES). When the nominal p value is less than 0.05, it was considered meaningful [18].
2.6. Protein–protein interaction (PPI) network analysis, microsatellite instability (MSI), tumor mutational burden (TMB) and neoantigen
We applied PPI to analyze the correlations among PTTG1 and other proteins through STRING website (https://string-db.org/). Using Spearman’s method, we investigated the relationships between PTTG1 and MSI, TMB, and neoantigen [19,20]. The above analyses were conducted using the Sangerbox tools (http://www.sangerbox.com/tool).
2.7. Correlation of PTTG1 expression with immune checkpoint molecules, immune infiltrations, immune cells pathway and tumor microenvironment
With the assistance of the estimate algorithm in the R package, we obtained the Immune, Stromal and ESTIMATEScore from the PTTG1 expression matrix, with the threshold of p < 0.01 [21]. The immune infiltration in KIRC was estimated by applying CIBERSORT, with the threshold of p < 0.01 [22]. Using Spearman’s method, we investigated the relationships between PTTG1 and immune checkpoint molecules, immune cells pathway. The above analyses were conducted using the Sangerbox tools (http://www.sangerbox.com/tool).
2.8. Prediction of immunotherapy response
TIDE (http://tide.dfci.harvard.edu/), as a computing architecture, could be used to simulate immune escape from tumors. So, we applied TIDE to predict the immunotherapy responses [23]. The Cancer Immunome Atlas (TCIA; https://tcia.at/) was a dataset of 20 solid cancers with over 8000 tumor samples. The immunophenoscore (IPS) of KIRC was obtained by using TCIA, which was also used to predict immunotherapy responses [24].
3. Results
3.1. The expression level of PTTG1 in KIRC
We downloaded PTTG1 mRNA expression data of 539 KIRC samples and 72 para-cancerous renal samples from the TCGA-KIRC database. By comparison, we discovered that the expression of PTTG1 in KIRC was remarkably up-regulated compared to that in non-cancerous tissues, which implied that PTTG1 might be an oncogenic gene (Fig. 1A, p < 0.05). Pairwise boxplot of 72 KIRC samples and 72 para-cancerous renal samples demonstrated that PTTG1 was highly expressed in the majority of KIRC tissues (Fig. 1B, p < 0.05). Boxplot of 539 KIRC samples and 72 para-cancerous renal samples showed that PTTG1 had a higher expression in tumor tissues than that in normal tissues (Fig. 1C, p < 0.05). Moreover, the receiver operating characteristic (ROC) curve was utilized to analyze one-year, three-year, and five-year survival in patients with KIRC. The areas under the ROC curve (AUC) values were 0.654, 0.626, and 0.640, indicating the low diagnostic performance (Fig. 1D). As was shown in Fig. 1E, the patients were divided into a low-PTTG1 group and a high-PTTG1 group, referring to the median mRNA expression level of PTTG1. Survival analysis showed that the low-PTTG1 group had a better prognosis (p < 0.05). Moreover, PCR further confirmed the increased expression of PTTG1 in 786-O cell lines (Fig. 1F). Through the results, we concluded PTTG1 might play oncogenic roles in KIRC.
Fig. 1.
The expression level of PTTG1 in KIRC from TCGA database. a The expression level of PTTG1 in different cancers. B Pairwise boxplot of PTTG1 expression between the KIRC and normal tissues in TCGA dataset (T = 72, N = 72). c Boxplot of PTTG1expression between KIRC and normal tissues in TCGA database (T = 539, N = 72). d ROC curve of PTTG1. e K–M survival analysis of PTTG1. f The PCR results of PTTG1 expression in normal renal tubular epithelial cells and KIRC cells. *P < 0.05; **P < 0.01; ***P < 0.001.
3.2. Immunohistochemistry staining and relationship between PTTG1 and clinical clinicopathologic factors
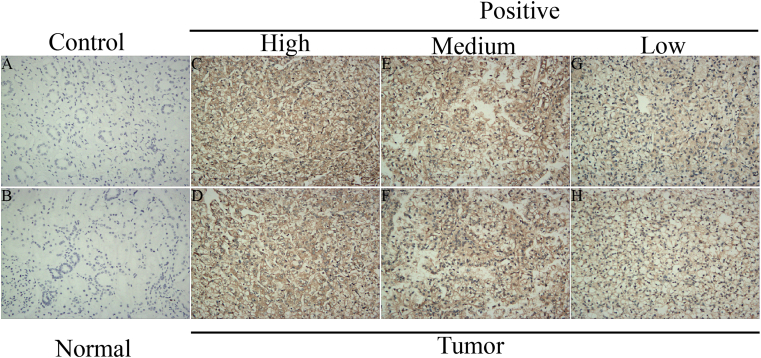
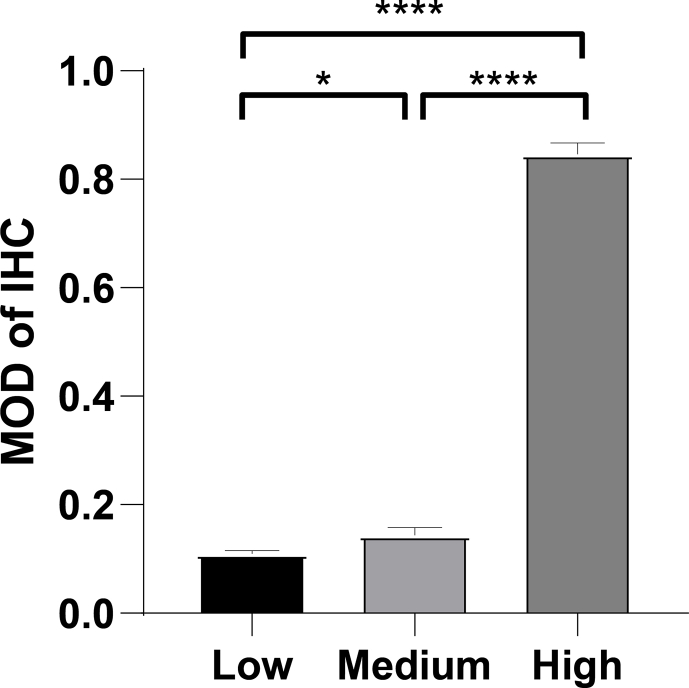
We verified the expression of PTTG1 at the protein level by using immunohistochemistry staining (Fig. 2A-2H). As displayed in Fig. S1, PTTG1 showed low, medium, and high protein expression levels in KIRC tissues, while showing negative expression in paraneoplastic tissues, indicating a higher expression in tumor tissues. As shown in Fig. 3A-3H, elevated PTTG1 expression was closely associated with gender, higher grade, M stage, T stage, and N stage (p < 0.05). Meanwhile, PTTG1 showed a significant difference between the white and African races (p < 0.05; Fig. 3F). In contrast, the relationship between PTTG1 and age was not statistically significant (Fig. 3A).
Fig. 2.
Immunohistochemistry staining of KIRC tissues. a-h The results of immunohistochemistry staining indicated that PTTG1 was highly expressed in KIRC tissues.
Fig. 3.
The relationships between the expression level of PTTG1 and clinicopathologic characteristics. a Age. B Gender. c Grade. d M. e N. f Race. g Stage. h T.
3.3. Univariate/multivariate cox hazard regression analysis
Univariate or multivariate Cox hazard regression analyses were used to prove whether PTTG1 alone could affect the OS prognosis of KIRC (Fig. 4 and Table 1). Based on the univariate regression analysis, age, grade, stage, T, M, and PTTG1 expression were closely associated with the OS prognosis of KIRC (Fig. 4A). Meanwhile, multivariate regression analysis showed that age, grade, stage, and PTTG1 expression were independent prognostic factors of KIRC (Fig. 4B). Combined with the above results, we drew the conclusion that PTTG1 alone could affect the OS prognosis of KIRC.
Fig. 4.
PTTG1 could serve as an independent prognostic factor in KIRC. a Univariate Cox hazard regression analysis of clinicopathologic variables and PTTG1 in KIRC. b Multivariate Cox hazard regression analysis of clinicopathologic variables and PTTG1 in KIRC.
Table 1.
Univariate and multivariate analyses of PTTG1 and clinicopathologic factors in KIRC.
| Univariate analysis |
Multivariate analysis |
|||||||
|---|---|---|---|---|---|---|---|---|
| Clinical characteristics | HR | HR.95 L | HR.95H | pvalue | HR | HR.95 L | HR.95H | pvalue |
| age | 1.033274 | 1.019678 | 1.047052 | 1.28E-06 | 1.03580842 | 1.02074701 | 1.05109206 | 2.51E-06 |
| gender | 0.933298 | 0.679691 | 1.281529 | 0.669603 | 0.89843384 | 0.64579494 | 1.24990661 | 0.52491918 |
| race | 1.193075 | 0.715959 | 1.988138 | 0.498058 | 1.17179724 | 0.67911878 | 2.02189779 | 0.56893102 |
| grade | 1.966884 | 1.638835 | 2.360598 | 3.69E-13 | 1.37654845 | 1.10246962 | 1.7187645 | 0.0047856 |
| stage | 1.855626 | 1.643637 | 2.094956 | 1.71E-23 | 1.77271054 | 1.26676129 | 2.48073784 | 0.00084045 |
| T | 1.997582 | 1.689051 | 2.362469 | 6.29E-16 | 1.07185509 | 0.81340611 | 1.41242279 | 0.62207097 |
| M | 2.099647 | 1.660681 | 2.654644 | 5.69E-10 | 0.79820147 | 0.43322866 | 1.47064506 | 0.46973755 |
| N | 0.862971 | 0.73887 | 1.007916 | 0.062826 | 0.85616293 | 0.72858741 | 1.0060769 | 0.05924498 |
| PTTG1 | 1.061266 | 1.042659 | 1.080205 | 4.43E-11 | 1.0395034 | 1.01690132 | 1.06260784 | 0.0005518 |
3.4. Identification of PTTG1-related signaling pathways by GSEA
GSEA was used to look for significant enrichment pathways in either the high- or low-expression type of PTTG1. As displayed in Fig. 5A-5H and Table 2, the high expression type was closely related to two signaling pathways, including cell cycle pathway and p53 signaling pathway. The low expression type was closely related to five signaling pathways, containing Erbb pathway, Gnrh pathway, Insulin pathway, mTOR pathway, and TGF Beta pathway. All in all, PTTG1-related pathways in KIRC were revealed by us.
Fig. 5.
Gene set enrichment analysis (GSEA). a Cell cycle pathway. B Erbb signaling pathway. c Gnrh signaling pathway. d Insulin signaling pathway. e mTOR signaling pathway. f P53 signaling pathway. g TGF Beta signaling pathway. h seven PTTG1-related signaling pathways.
Table 2.
Gene set enrichment analysis (GSEA) of PTTG1 in KIRC.
| GeneSet name | NES | Nominal p-value | FDR q-value |
|---|---|---|---|
| CELL_CYCLE | 1.7488136 | 0.048484847 | 0.070737466 |
| ERBB_SIGNALING_PATHWAY | −1.920799 | 0.003976143 | 0.046424486 |
| GNRH_SIGNALING_PATHWAY | −1.957752 | 0.001919386 | 0.053977773 |
| INSULIN_SIGNALING_PATHWAY | −2.170076 | 0 | 0.025261166 |
| MTOR_SIGNALING_PATHWAY | −2.225056 | 0 | 0.028832098 |
| P53_SIGNALING_PATHWAY | 1.7120552 | 0.02414487 | 0.08312161 |
| TGF_BETA_SIGNALING_PATHWAY | −2.058136 | 0 | 0.03348911 |
3.5. Construction of PPI and correlations between PTTG1 expression and MSI, TMB, neoantigen
We applied PPI to study the interactions between PTTG1 and other proteins through the STRING website. As displayed in Fig. 6A, PTTG1 was closely related to the following ten proteins (PLK1, ANAPC11, ESPL1, CDKI, UBE2C, AURKB, ANAPC2, AURKA, CDC20, and FZR1). In order to investigate the associations between PTTG1 and MSI, TMB, neoantigens, we conducted the correlation analysis. As displayed in Fig. 6B and C, PTTG1 was not related to MSI and neoantigen (p = 0.18, p = 0.77). However, PTTG1 was significantly relevant to TMB (p = 0.014, Fig. 6D).
Fig. 6.
Relationships between PTTG1 and PPI, MSI, TMB, Neoantigen in KIRC. a Protein–protein interaction (PPI) network. B Correlations between PTTG1 and MSI. c Correlations between PTTG1 and neoantigen. d Correlations between PTTG1 and TMB.
3.6. Correlations between PTTG1 and tumor microenvironment, tumor immune infiltration, immune cells pathway, immune checkpoint molecules
In terms of immune checkpoint molecules, there were 31 molecules significantly linked to PTTG1, including LAG3, CD274, PDCD1, etc (Fig. 7A, all p < 0.05). Moreover, PTTG1 was markedly associated with immune cells including activated CD4 T cell, MDSC, Type 2 T helper cell, and so on (Fig. 7B). In order to further investigate the association between PTTG1 and immunity, we performed the tumor microenvironment and tumor immune infiltration analyses. In the aspect of tumor immune infiltration, PTTG1 was closely connected with B cell, CD8+ T cell, neutrophil, and dendritic cell infiltration (Fig. 7C, p < 0.01). As for the tumor microenvironment, ImmuneScore, ESTIMATEScore, and StromalScore were all strongly linked to PTTG1 (Fig. 7D, all p < 0.01). The above results all indicated that PTTG1 had a close relationship with immunity.
Fig. 7.
Relationships between PTTG1 and an immune checkpoint molecules; b immune cells pathway; c tumor immune infiltration; and d tumor microenvironment in KIRC.
3.7. Prediction of immunotherapy response
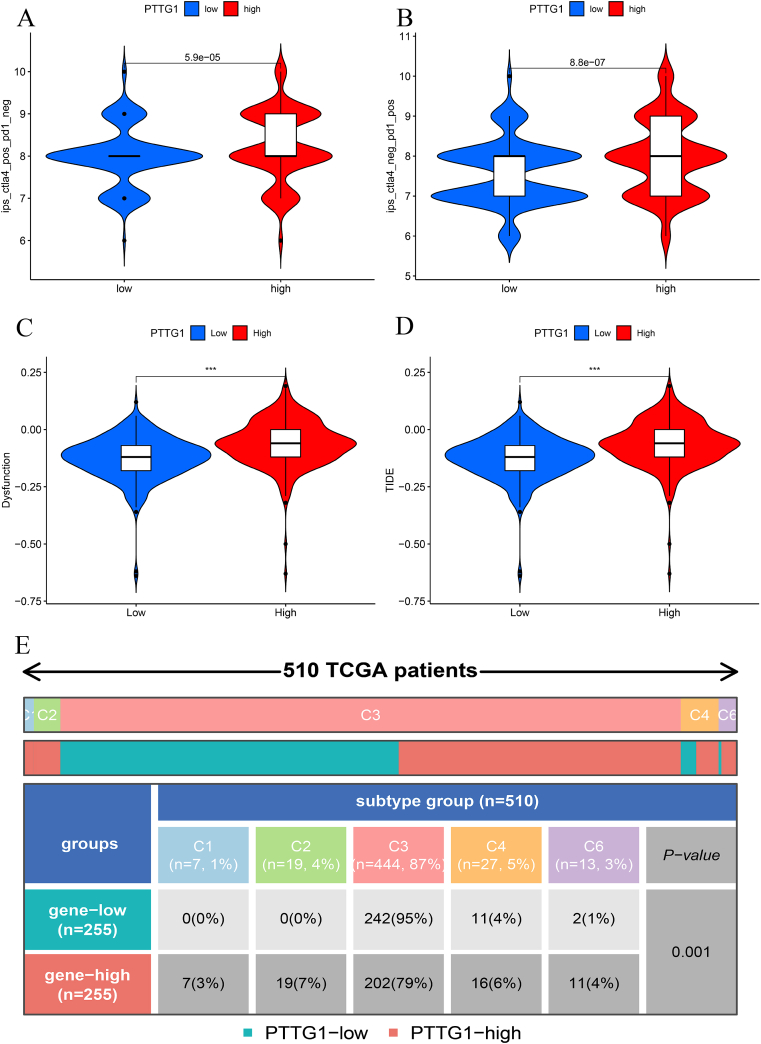
We applied TIDE and TCIA algorithms to explore the relationships between PTTG1 and the immunotherapy responses. As displayed in Fig. 8A-B, we explored the relationships between IPS and PTTG1 expression. Obviously, there were differences in IPS scores of anti-CTLA-4 or anti-PD-1 treatment between high- and low-expression groups of PTTG1 (p < 0.05). As presented in Fig. 8C-D, KIRC patients with low expression of PTTG1 had less immune dysfunction and a greater response to immunotherapy. The above results indicated that the group with low PTTG1 expression was more sensitive to immune checkpoint inhibitors. Besides, the distributions of PTTG1 expression in pan-cancer immune subtypes were also significantly different (Fig. 8E).
Fig. 8.
Predictions of the immunotherapy response in KIRC patients. a Associations between PTTG1 and IPS of anti-CTLA-4 or anti-PD-1 treatment; b Associations between PTTG1 and IPS of anti-CTLA-4 or anti-PD-1 treatment; c Immune dysfunction scores in high and low PTTG1 expression groups by TIDE; d TIDE scores in high and low PTTG1 expression groups by TIDE; e The distributions of PTTG1 expression in pan-cancer immune subtypes; Figure S1. Mean optical density of the immunohistochemistry results.
4. Discussion
PTTG1 was a cellular transforming factor with little or no expression in normal tissues [5]. However, aberrantly high PTTG1 expression had been found in several malignancies, including adrenocortical carcinoma, prostate cancer, and oral squamous cell carcinoma, playing vital roles in tumor cell proliferation, invasion, and metastasis [[25], [26], [27]]. PTTG1 had been found to be involved in microtubule nucleation and cell polarity formation, and its knockdown could lead to severe polarity and motility defects in cells [28]. Emanuela et al. found that PTTG1 could promote the invasion of seminoma and predict the prognosis of seminoma [29]. Studies in KIRC proposed that PTTG1 might participate in the process of epithelial-mesenchymal transition through the AKT/MMP pathway [30]. Moreover, previous research also demonstrated that PTTG1 could affect the prognosis of KIRC [31]. Nevertheless, the relationship between PTTG1 and immunity remained unclear, which was the key content of this study.
In this paper, bioinformatics analysis was used to gather data on KIRC from TCGA database. Through database information, we concluded that PTTG1 expression was up-regulated in KIRC. The high expression of PTTG1 in KIRC was further confirmed by immunohistochemistry and PCR. The survival analysis implied that elevated PTTG1 was closely connected to a poorer OS prognosis in patients with KIRC. Moreover, the PTTG1 expression was closely associated with gender, higher grade, M stage, T stage, and N stage. Univariate/multivariate regression analysis implied that PTTG1 was an independent prognostic factor for KIRC. Combining the above results, we came to the conclusion that PTTG1 could be a better predictor of KIRC. GSEA was applied to analyze PTTG1-related signaling pathways in KIRC, and we finally found seven signaling pathways, including cell cycle, Erbb, Gnrh, Insulin, mTOR, p53 and TGF beta signaling pathways. The above signaling pathways would be beneficial to illustrate the pathogenesis of KIRC.
In addition, the associations among PTTG1 and MSI, TMB, and neoantigen were also analyzed. From the above results, we found that PTTG1 was positively correlated with TMB. However, no relationships were found between PTTG1 and MSI or neoantigens. TMB represented the number of mutations per megabyte in the gene region examined in a tumor sample [32]. TMB indirectly reflected the ability of tumors to produce neoantigens and predicted the effect of immunotherapy on different tumors [[33], [34], [35]]. Studies had shown that higher TMB was associated with better OS and could have a better response to immune checkpoint inhibitors (ICIs) [36,37]. Moreover, TMB, PD-L1, and DNA mismatch repair defects could be used as biomarkers of ICIs [[38], [39], [40]]. Since PTTG1 was positively correlated with TMB, it remained to be discussed whether PTTG1 could be used as a biomarker to predict the ICIs’ responses.
To investigate the relationship between PTTG1 and immunity, immune checkpoint molecules, immune cell pathways, tumor immune infiltration, and tumor microenvironment were applied. The term "tumor microenvironment" referred to the surrounding microenvironment, in which tumor cells existed, including surrounding blood vessels, immune cells, fibroblasts, and so on. The existence of tumor microenvironment could improve the proliferation and migration of tumor cells and their immune escape ability [41]. An increasing number of studies demonstrated that tumor microenvironment had a great impact on the development and treatment of KIRC [42]. In this paper, PTTG1 was significantly related to ImmuneScore, ESTIMATEScore and StromalScore. Furthermore, PTTG1 had direct relevance to checkpoint molecules (PDCD1, CD274, CTLA4, and LAG3). Immune checkpoints were a kind of immunosuppressive molecule expressed on immune cells that was used to regulate immune activation. In this case, PD-1 (programmed cell death protein 1) was a typical example of an immune checkpoint. PD-1 inhibited the autoimmune response by binding PD-L1 or PD-L2 and inhibiting the function of T lymphocytes. Both PD-1 and PD-L1 have been found to be expressed in tumor cells, and the PD-1/PD-L2 axis has been shown to inhibit tumor growth via the AKT and ERK1/2 pathways [43]. CTLA-4 acted as a transmembrane receptor on T cells that bound to receptors on the surface of antigen cells to terminate the immune response [44]. As to lymphocyte activation gene-3 (LAG3), it may be a better immunotherapy target than PD-1 or CTLA-4. Because the antibodies of PD-1 and CTLA-4 could only activate effector T cells, but not inhibit the activity of regulatory T cells (Tregs) [45]. Given the relationships between PTTG1 and immune checkpoints, we could speculate on the expression of immune checkpoints in KIRC through the expression of PTTG1.
Tumor genomic changes, PD-L1 expression, and immune microenvironment infiltration all had a significant impact on immunotherapy efficacy [46]. In recent years, the mechanism of tumor immunosuppression and escape mediated by immune checkpoints had become a hot spot in the research of immunotherapy for advanced tumors, such as CTLA-1, PD-1, and PD-L1 [47,48]. Due to the complexity and heterogeneity of KIRC, the effect of immunotherapy showed obvious individual differences, and patients have different degrees of drug resistance [49]. Therefore, it was of great clinical significance to determine biomarkers that could predict the therapeutic effects. Current candidate predictive biomarkers include tumor antigens (MSI, TMB, neoantigen), inflammatory cells (tumor-infiltrating lymphocytes, genetic markers, tumor-associated immune cells expressing PD-L1 protein, etc.), tumor immunosuppressive cells or proteins (PD-L1, CTLA-4, regulatory T cells, bone marrow-derived suppressor cells), and host factors (microorganisms, single nucleotide polymorphisms, etc.) [50]. Alborelli et al. mentioned that a tumor may respond better to immunotherapy if it presented a higher TMB and more neoantigens [51]. By sequencing exons and their matching normal DNA from two groups of non-small cell lung cancer patients treated with Pembrolizumab, the study by Rizvi et al. found that higher TMB in tumors was strongly associated with prolonged PFS in these patients [40]. It had been suggested that PTTG1 might be a potential predictor of ICB response in papillary renal cell carcinoma patients [52]. In this paper, we further explored the relationship between PTTG1 and the immunotherapy responses in KIRC. Aiming at PD-1 and CTLA-4, two popular targets of immunotherapy, IPS was used to compare the response of the high and low PTTG1 expression groups to immunotherapy. Immune checkpoint inhibitors blocked the inhibitory effect of tumor cells on immune cells by binding to immune checkpoints. Compared to the high PTTG1 expression group, the low PTTG1 expression group responded better to immunotherapy and was less likely to develop immune dysfunction, which might explain the poor prognosis in the group with high PTTG1 expression. However, the results of these immunotherapy reactions were all based on the information in the database. Due to the limited amount of patient information in the database, more patient information needed to be collected to further verify the specific efficacy.
According to the above results, PTTG1 could be used as an independent factor affecting the prognosis of KIRC and was closely related to immunity. In addition, in this study, we found that PTTG1 was closely related to the expression levels of multiple immune checkpoints, TMB, and IPS in KIRC, indicating the potential value of PTTG1 as a new therapeutic target and a predictive marker of the responses to ICIs for KIRC. But this study also had some limitations. The limited information in the TCGA database and the small sample size of normal kidney tissue might influence our conclusion. More clinical information and basic trials were needed to confirm the relationship between PTTG1 and immunotherapy response.
5. Conclusions
Overall, our study suggested that PTTG1 might serve as a marker to help clinicians better predict the OS prognosis of KIRC. GSEA analysis revealed that PTTG1 might affect the development of KIRC through the cell cycle, ErbB, GnrH, Insulin, mTOR, p53 and TGF beta signaling pathways. Moreover, PTTG1 was also related to TMB, immunity, and immunotherapy responses. More clinical patient data and deeper studies were needed to further validate our findings.
6. Ethics approval and consent to participate
The study was approved by the Institutional Research Ethics Committees of Affiliated Hospital of Nantong University.
Declarations
Author contribution statement
Xinyu Zhang: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Hao Ji: Analyzed and interpreted the data; Wrote the paper.
Yeqing Huang: Performed the experiments.
Bingye Zhu: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Qianwei Xing: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Funding statement
Yeqing Huang was supported by National Natural Science Foundation of China (81802580).
Qianwei Xing was supported by Nantong Science and Technology Planning Project (JC 2021183).
Data availability statement
Data associated with this study has been deposited at The RNA-sequencing data and corresponding clinical information were downloaded from the Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). The IPS values are downloaded from The Cancer Immunome Atlas (TCIA, https://tcia.at/home).
Declaration of interest’s statement
The authors declare no conflict of interest.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.heliyon.2023.e13201.
Contributor Information
Bingye Zhu, Email: zhubingye1995@163.com.
Qianwei Xing, Email: xingqianwei@ntu.edu.cn.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
FigS1.
References
- 1.Siegel R.L., et al. Cancer statistics. CA A Cancer J. Clin. 2022;72(1):7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Yang F.Q., et al. Foxl1 inhibits tumor invasion and predicts outcome in human renal cancer. Int. J. Clin. Exp. Pathol. 2014;7(1):110–122. [PMC free article] [PubMed] [Google Scholar]
- 3.Russo P. Renal cell carcinoma: presentation, staging, and surgical treatment. Semin. Oncol. 2000;27(2):160–176. [PubMed] [Google Scholar]
- 4.Morris M.R., Latif F. The epigenetic landscape of renal cancer. Nat. Rev. Nephrol. 2017;13(1):47–60. doi: 10.1038/nrneph.2016.168. [DOI] [PubMed] [Google Scholar]
- 5.Chen L., et al. Identification of the human pituitary tumor transforming gene (hPTTG) family: molecular structure, expression, and chromosomal localization. Gene. 2000;248(1–2):41–50. doi: 10.1016/s0378-1119(00)00096-2. [DOI] [PubMed] [Google Scholar]
- 6.Zou H., et al. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285(5426):418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- 7.Pei L., Melmed S. Isolation and characterization of a pituitary tumor-transforming gene (PTTG) Mol. Endocrinol. 1997;11(4):433–441. doi: 10.1210/mend.11.4.9911. [DOI] [PubMed] [Google Scholar]
- 8.Jallepalli P.V., et al. Securin is required for chromosomal stability in human cells. Cell. 2001;105(4):445–457. doi: 10.1016/s0092-8674(01)00340-3. [DOI] [PubMed] [Google Scholar]
- 9.Fraune C., et al. Upregulation of PTTG1 is associated with poor prognosis in prostate cancer. Pathol. Int. 2020;70(7):441–451. doi: 10.1111/pin.12938. [DOI] [PubMed] [Google Scholar]
- 10.Gao S., et al. Computational analysis for identification of early diagnostic biomarkers and prognostic biomarkers of liver cancer based on GEO and TCGA databases and studies on pathways and biological functions affecting the survival time of liver cancer. BMC Cancer. 2021;21(1):791. doi: 10.1186/s12885-021-08520-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang S., et al. Distinct expression pattern and prognostic values of pituitary tumor transforming gene family genes in non-small cell lung cancer. Oncol. Lett. 2019;18(5):4481–4494. doi: 10.3892/ol.2019.10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramaswamy S., et al. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 2003;33(1):49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 13.Romero F., et al. Human securin, hPTTG, is associated with Ku heterodimer, the regulatory subunit of the DNA-dependent protein kinase. Nucleic Acids Res. 2001;29(6):1300–1307. doi: 10.1093/nar/29.6.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vlotides G., Eigler T., Melmed S. Pituitary tumor-transforming gene: physiology and implications for tumorigenesis. Endocr. Rev. 2007;28(2):165–186. doi: 10.1210/er.2006-0042. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z., Yu R., Melmed S. Mice lacking pituitary tumor transforming gene show testicular and splenic hypoplasia, thymic hyperplasia, thrombocytopenia, aberrant cell cycle progression, and premature centromere division. Mol. Endocrinol. 2001;15(11):1870–1879. doi: 10.1210/mend.15.11.0729. [DOI] [PubMed] [Google Scholar]
- 16.Wei C., et al. High expression of FER tyrosine kinase predicts poor prognosis in clear cell renal cell carcinoma. Oncol. Lett. 2013;5(2):473–478. doi: 10.3892/ol.2012.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soh J.W., et al. Protein kinase G activates the JNK1 pathway via phosphorylation of MEKK1. J. Biol. Chem. 2001;276(19):16406–16410. doi: 10.1074/jbc.C100079200. [DOI] [PubMed] [Google Scholar]
- 18.Subramanian A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hundal J., et al. pVAC-Seq: a genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 2016;8(1):11. doi: 10.1186/s13073-016-0264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao Y., et al. Prognostic value and immunological role of PDCD1 gene in pan-cancer. Int. Immunopharm. 2020;89(Pt B) doi: 10.1016/j.intimp.2020.107080. [DOI] [PubMed] [Google Scholar]
- 21.Yoshihara K., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L., et al. NUPR1 imparts oncogenic potential in bladder cancer. Cancer Med. 2022 doi: 10.1002/cam4.5518. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y., et al. Survival prognosis, tumor immune landscape, and immune responses of PPP1R18 in kidney renal clear cell carcinoma and its potentially double mechanisms. World J. Oncol. 2022;13(1):27–37. doi: 10.14740/wjon1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang L., et al. DLX2 is a potential immune-related prognostic indicator associated with remodeling of tumor microenvironment in lung squamous cell carcinoma: an integrated bioinformatical analysis. Dis. Markers. 2022;2022 doi: 10.1155/2022/6512300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao X.L., et al. [Downregulation of PTTG1 expression inhibits the proliferation and invasiveness and promotes the apoptosis of human prostate cancer LNCaP-AI cells] Zhonghua Nan ke Xue. 2017;23(7):589–597. [PubMed] [Google Scholar]
- 26.Romero Arenas M.A., et al. Protein expression of PTTG1 as a diagnostic biomarker in adrenocortical carcinoma. Ann. Surg Oncol. 2018;25(3):801–807. doi: 10.1245/s10434-017-6297-1. [DOI] [PubMed] [Google Scholar]
- 27.Zhang E., et al. Pituitary tumor-transforming gene 1 (PTTG1) is overexpressed in oral squamous cell carcinoma (OSCC) and promotes migration, invasion and epithelial-mesenchymal transition (EMT) in SCC15 cells. Tumour Biol. 2014;35(9):8801–8811. doi: 10.1007/s13277-014-2143-2. [DOI] [PubMed] [Google Scholar]
- 28.Moreno-Mateos M.A., et al. PTTG1/securin modulates microtubule nucleation and cell migration. Mol. Biol. Cell. 2011;22(22):4302–4311. doi: 10.1091/mbc.E10-10-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teveroni E., et al. Nuclear localization of PTTG1 promotes migration and invasion of seminoma tumor through activation of MMP-2. Cancers. 2021;13(2) doi: 10.3390/cancers13020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wondergem B., et al. Expression of the PTTG1 oncogene is associated with aggressive clear cell renal cell carcinoma. Cancer Res. 2012;72(17):4361–4371. doi: 10.1158/0008-5472.CAN-11-2330. [DOI] [PubMed] [Google Scholar]
- 31.Wei C., et al. High expression of pituitary tumor-transforming gene-1 predicts poor prognosis in clear cell renal cell carcinoma. Mol Clin Oncol. 2015;3(2):387–391. doi: 10.3892/mco.2014.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Addeo A., et al. TMB or not TMB as a biomarker: that is the question. Crit. Rev. Oncol. Hematol. 2021;163 doi: 10.1016/j.critrevonc.2021.103374. [DOI] [PubMed] [Google Scholar]
- 33.Friedlaender A., et al. Tissue-plasma TMB comparison and plasma TMB monitoring in patients with metastatic non-small cell lung cancer receiving immune checkpoint inhibitors. Front. Oncol. 2020;10:142. doi: 10.3389/fonc.2020.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halbert B., Einstein D.J. Hot or not: tumor mutational burden (TMB) as a biomarker of immunotherapy response in genitourinary cancers. Urology. 2021;147:119–126. doi: 10.1016/j.urology.2020.10.030. [DOI] [PubMed] [Google Scholar]
- 35.Ravaioli S., et al. Are we ready to use TMB in breast cancer clinical practice? Cancer Immunol. Immunother. 2020;69(10):1943–1945. doi: 10.1007/s00262-020-02682-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai H., et al. Mutational landscape of gastric cancer and clinical application of genomic profiling based on target next-generation sequencing. J. Transl. Med. 2019;17(1):189. doi: 10.1186/s12967-019-1941-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H., et al. Tumor mutational burden as a biomarker for advanced biliary tract cancer. Technol. Cancer Res. Treat. 2021;20 doi: 10.1177/15330338211062324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asaoka Y., Ijichi H., Koike K. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015;373(20):1979. doi: 10.1056/NEJMc1510353. [DOI] [PubMed] [Google Scholar]
- 39.Patel S.P., Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol. Cancer Therapeut. 2015;14(4):847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 40.Rizvi N.A., et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khurana A., Ansell S.M. Role of microenvironment in non-hodgkin lymphoma: understanding the composition and biology. Cancer J. 2020;26(3):206–216. doi: 10.1097/PPO.0000000000000446. [DOI] [PubMed] [Google Scholar]
- 42.Şenbabaoğlu Y., et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol. 2016;17(1):231. doi: 10.1186/s13059-016-1092-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng H., et al. New insights into the important roles of tumor cell-intrinsic PD-1. Int. J. Biol. Sci. 2021;17(10):2537–2547. doi: 10.7150/ijbs.60114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buchbinder E.I., Desai A. CTLA-4 and PD-1 pathways: similarities, differences, and implications of their inhibition. Am. J. Clin. Oncol. 2016;39(1):98–106. doi: 10.1097/COC.0000000000000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang C.T., et al. Role of LAG-3 in regulatory T cells. Immunity. 2004;21(4):503–513. doi: 10.1016/j.immuni.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 46.Angell H.K., et al. The immunoscore: colon cancer and beyond. Clin. Cancer Res. 2020;26(2):332–339. doi: 10.1158/1078-0432.CCR-18-1851. [DOI] [PubMed] [Google Scholar]
- 47.Andrews L.P., Yano H., Vignali D.A.A. Inhibitory receptors and ligands beyond PD-1, PD-L1 and CTLA-4: breakthroughs or backups. Nat. Immunol. 2019;20(11):1425–1434. doi: 10.1038/s41590-019-0512-0. [DOI] [PubMed] [Google Scholar]
- 48.Carosella E.D., et al. A systematic review of immunotherapy in urologic cancer: evolving roles for targeting of CTLA-4, PD-1/PD-L1, and HLA-G. Eur. Urol. 2015;68(2):267–279. doi: 10.1016/j.eururo.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 49.Roviello G., et al. Results from a meta-analysis of immune checkpoint inhibitors in first-line renal cancer patients: does PD-L1 matter? Ther. Adv. Med. Oncol. 2019;11 doi: 10.1177/1758835919861905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Früh M., Peters S. Genomic features of response to combination immunotherapy in lung cancer. Cancer Cell. 2018;33(5):791–793. doi: 10.1016/j.ccell.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 51.Alborelli I., et al. Tumor mutational burden assessed by targeted NGS predicts clinical benefit from immune checkpoint inhibitors in non-small cell lung cancer. J. Pathol. 2020;250(1):19–29. doi: 10.1002/path.5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian X., et al. Identification of prognostic biomarkers in papillary renal cell carcinoma and PTTG1 may serve as a biomarker for predicting immunotherapy response. Ann. Med. 2022;54(1):211–226. doi: 10.1080/07853890.2021.2011956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data associated with this study has been deposited at The RNA-sequencing data and corresponding clinical information were downloaded from the Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). The IPS values are downloaded from The Cancer Immunome Atlas (TCIA, https://tcia.at/home).