Abstract
The tissue-specific profile of alternatively spliced genes (ASGs) and their involvement in reproduction processes characteristic of turkey testis, epididymis, and ductus deferens were investigated for the first time in birds. Deep sequencing of male turkey reproductive tissue RNA samples (n = 6) was performed using Illumina RNA-Seq with 2 independent methods, rMATs and SUPPA2, for differential alternative splicing (DAS) event prediction. The expression of selected ASGs was validated using quantitative real-time reverse transcriptase-polymerase chain reaction. The testis was found to be the site of the highest number of posttranscriptional splicing events within the reproductive tract, and skipping exons were the most frequently occurring class of alternative splicing (AS) among the reproductive tract. Statistical analysis revealed 86, 229, and 6 DAS events in the testis/epididymis, testis/ductus deferens, and epididymis/ductus deferens comparison, respectively. Alternative splicing was found to be a mechanism of gene expression regulation within the turkey reproduction tract. In testis, modification was observed for spermatogenesis specific genes; the changes in 5’ UTR could act as regulator of MEIG1 expression (a player during spermatocytes meiosis), and modification of 3’ UTR led to diversification of CREM mRNA (modulator of gene expression related to the structuring of mature spermatozoa). Sperm tail formation can be regulated by changes in the 5’ UTR of testicular SLC9A3R1 and gene silencing by producing dysfunctional variants of ODF2 in the testis and ATP1B3 in the epididymis. Predicted differentially ASGs in the turkey reproductive tract seem to be involved in the regulation of spermatogenesis, including acrosome formation and sperm tail formation and binding of sperm to the zona pellucida. Several ASGs were classified as cilia by actin and microtubule cytoskeleton organization. Such genes may play a role in the organization of sperm flagellum and post-testicular motility development. To our knowledge, this is the first functional investigation of alternatively spliced genes associated with tissue-specific processes in the turkey reproductive tract.
Key words: alternative splicing, spermatogenesis, sperm maturation, male reproductive tract, turkey
INTRODUCTION
Bird male anatomy and physiology are unique among the vertebrate reproductive systems. Spermatogenesis in birds is divided into an early stage in which diploid spermatogonia is proliferating into round spermatids through mitotic and meiotic division, and a late stage in which round spermatids are transformed into spermatozoa (Asano and Tajima, 2017). Avian spermatozoa undergo post-testicular maturation during passage through the male genital tract (Asano and Tajima, 2017). For functional maturation bird spermatozoa incorporate proteins secreted from epithelium into plasma membrane during epididymal transit (Ahammad et al., 2011a,b). Ductus epididymis and ductus deferens are the main sources of apocrine secretion in the bird reproductive tract (Aire and Josling, 2000). Knowledge about the detailed mechanisms of male reproduction in birds is still limited. Recently, a new comprehensive analysis method, such as next-generation sequencing (NGS), was introduced to study the specificity of bird reproduction (Liu et al., 2017 2018; Wang et al., 2017; Słowińska et al., 2020). In the turkey reproductive tract, tissue-specific processes were investigated, and potential candidate genes involved in spermatogenesis, spermiogenesis, and flagellum formation in the testis and in post-testicular sperm maturation in the epididymis and ductus deferens were identified (Słowińska et al., 2020).
During spermatogenesis, the gene expression pattern is highly dynamic, and alternative splicing (AS) seems to be one of the regulatory mechanisms of male gamet generation in mammals (Elliott and Grellscheid, 2006; Song et al., 2020). AS, common to eukaryotes, provides a versatile means of regulating gene expression using different combinations of exons from the same primary transcript, resulting in the generation of different mature transcripts and then coding the same, shorter, or even distinct proteins (Elliott and Grellscheid, 2006). Approximately 95% of human genes undergo AS (Pan et al., 2008). Seven basic modes of AS are recognized, including skipping exon (SE), retention intron (RI), alternative 5’ splice site (A5) and alternative 3’ splice site (A3), alternative first exon (AF) and alternative last exon (AL) and mutually exclusive exon (MXE) (Figure 1A). AS is particularly prevalent in the mammalian testis (Yeo et al., 2004; de la Grange et al., 2010), and several key genes undergoing AS are involved in specific stages of spermatogenesis and in the regulation of genes participating in sperm tail formation.
Figure 1.
Alternative splicing events detected in the turkey reproductive tract. (A) Common models of AS and the corresponding transcript variants. (B) Number of detected AS events and the main classes of AS events across the turkey reproductive tract. Different letters (a-c) indicate statistical significance of the total number of AS events at P-value ≤ 0.0001.
A few studies on AS have been reported in relation to bird reproduction (Zhao et al., 2007; Huang et al., 2012; Lang et al., 2020; Rogers et al., 2021, Sun et al., 2021). AS of testis-specific lactate dehydrogenase C (LDHC) and doublesex- and mab-3-related transcription factor 1 (DMRT1) were reported for chicken and pigeon (Zhao et al., 2007; Huang et al., 2012). NGS has been recently applied for the characterization of AS events during chicken male germline stem cell differentiation (Sun et al., 2021), sex-specific aspects of the stress response in rock doves (Lang et al., 2020) and the demonstration of the role of AS in the evolution of phenotypic complexity in birds (Rogers et al., 2021). Nevertheless, the splicing profiles of genes involved in reproductive processes in birds have remained unexplored thus far. Therefore, in this study, RNASeq data generated previously by Słowińska et al. (2020) were used to detect AS events in the testis, epididymis and ductus deferens of turkeys. The key genes and pathways controlled by AS were described in relation to the specificity of turkey reproduction.
MATERIALS AND METHODS
This study is based on the same RNA sequencing data as previous transcriptomic analysis of the turkey reproductive tract published by Słowińska et al. (2020). Full-length transcriptome sequences were deposited in the GEO database as the GSE142428_transcriptome_Meleagris_gallopavo.fa.gz file. Currently, using the same RNA sequencing data, we focus on differentially alternatively spliced genes (DASGs) occurring specifically in the testis, epididymis, and ductus deferens.
Birds, Housing, and Tissue Collection
Male reproductive tract tissue samples were obtained from six 38-wk-old turkeys. (British United Turkeys Big 6, Grelier, Saint-Laurent-de-la-Plaine, France) maintained under standard husbandry conditions at the Turkey Testing Farm of the Department of Poultry Science (University of Warmia and Mazury in Olsztyn, Poland) as described in detail (Słowińska et al., 2020). The sampling time was chosen within reproductive season – the 8th week of reproduction season, when a high quality semen is produced (Kotłowska et al., 2005). Only bird producing high quality semen were chosen for experiment, as shown by Słowińska et al. (2020). The tissues were immediately frozen in liquid nitrogen and designated for total RNA isolation. All experiments involving animals were conducted in accordance with national guidelines for agricultural animal care and veterinary practice, following EU Directive 2010/63/UE.
RNA Isolation and the Evaluation of RNA Integrity
RNA isolation and evaluation of RNA integrity was performed according to previously described methods (Słowińska et al., 2020) and presented in Supplementary Table 1. Briefly, total RNA was extracted from samples using a Total RNA Mini kit (A&A Biotechnology, Gdynia, Poland) according to the manufacturer's instructions. Genomic DNA was removed from RNA samples using a DNase I Amplification Grade Kit (Sigma Aldrich Co., St. Louis, Mi, MI). The RNA concentration and quality were evaluated spectrophotometrically using an ND-1000 spectrophotometer (NanoDrop Technologies LLC, Wilmington, DE). The RNA integrity was determined using a 2100 Bioanalyzer with an RNA 6000 Nano LabChip Kit (Agilent Technologies, Santa Clara, CA). Only samples with RNA integrity numbers (RINs: 28S/18S ratio) above 8.0 and rRNA ratios above 1.0 were used for NGS. The total number of samples was 18 across 3 tissues studied (testis, epididymis, and ductus deferens).
Library Preparation and Sequencing Procedures
The total RNA for each sample was prepared for library construction by the Illumina TruSeq mRNA LT Sample Prep kit (Illumina, Inc., San Diego, CA). Poly-A-containing mRNA was first purified with poly-T-attached magnetic beads. The divalent cations were then used to fragment the mRNA under increasing temperatures. First-strand cDNA was attached to RNA fragments using SuperScript II reverse transcriptase (Invitrogen, Waltham, MA). DNA Polymerase I and RNase H were used for second-strand cDNA synthesis. To prevent fragments from ligating to each other, single ‘A’ nucleotides were added to the 3′ ends. As part of the RNA-seq qualification process, the qPCR Quantification Protocol Guide (KAPA Library Quantification kits for Illumina Sequencing platforms) and TapeStation D1000 ScreenTape system (Agilent Technologies, Waldbronn, Germany) were utilized. In the next step, the prepared libraries were transferred to the NovaSeq6000 platform (Illumina), where the sequencing procedure was run as expected: 2 × 100 bp paired-end reads and assumed a number of reads per sample higher than 90 million. The raw data were submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (accessed on 20 December 2019) under accession No. PRJNA597008 and used for AS analysis.
Quality Control and Mapping Process
FastQC software (v. 0.11.7, https://bioinformatics.babraham.ac.uk) was applied to conduct the quality control of raw and trimmed reads. The reads with a Phred sequence quality score > 20 and preserved 90 bp sequence length (after adapter removal) were transferred to the following analyses. The cutting and quality screening processes were evaluated by Trimmomatic software, v. 0.38 (Bolger et al., 2014). The paired-end reads were aligned to the reference turkey genome (Meleagris_gallopavo.UMD2.dna_sm.toplevel.fa) using ENSEMBL/GENCODE database annotation and STAR software, v. 2.4 (Dobin et al., 2013). The Ensembl GTF annotation was merged with all sorted BAM files operating StringTie, version 1.3.3 (Pertea et al., 2015) to enrich annotations. Additionally, trimmed reads were mapped to the newly obtained turkey transcriptome formed by the StringTie tool and gffread script (https://github.com/gpertea/gffread). The mapping process was performed using the Salmon 0.14.1 tool (Patro et al., 2017).
Alternative Splicing Events and Differential Analysis
Alternative splicing research was detected by event-based methods described previously by Mehmood et al. (2020). Two methods with different background were used: SUPPA2, the superfast pipeline for AS analysis based on transcriptome as reference sequence (SUPPA2 v.2; Trincado et al., 2018) and rMATS, replicate multivariate analysis of transcript splicing based on genome as reference sequence (rMATS v.3.2.5; Shen et al., 2014). Only differentially splicing events confirmed by both methods were taken into consideration. Applying these miscellaneous methods allowed us to obtain stringent and robust results for the identification of splicing events. The percent of splicing inclusion (PSI) values was calculated for all AS events according to the reads mapped in the vicinity of junction sites. The differential alternative splicing (DAS) events between the turkey testis, epididymis and ductus deferens were statistically tested (FDR < 0.05). Moreover, each method assumed DAS event filtration according to ΔPSI > 0.1. The final set of differentially spliced transcripts was obtained by the intersection of DAS events from both methods according to genomic coordinates using bedtools software (Quinlan et al., 2010). The DAS events were classified into 5 subtypes (rMATS): A5, A3, MXE, RI, and SE. Furthermore, the SUPPA method extracted two additional splice subtypes: AF and AL. The results were visualized by R bioconductor packages (http://www.R- project.org/; v.4.1.1; accessed on 1 February 2021), i.a. ggplot (v.3.3.5) to draw volcano plots, GOplot (v. 1.0.2) to obtain the ontology results. The DAS events were heatmapped by Juntis software (Yang et al., 2021), and selected events were plotted by rmats2sashimiplot.py scripts (https://github.com/Xinglab/rmats2sashimiplot (accessed on 1 February 2021).
Functional Enrichment Analysis
The functional profiling of DASGs into 3 Gene Ontology (GO) categories, biological processes, cellular components, and molecular functions, was performed using ShinyGO v0.75: Gene Ontology Enrichment Analysis + more (Ge et al., 2020; turkey as the selected species) with adjusted p-value cutoff (FDR) < 0.05. The charts summarizing the 10 most significantly enriched pathways were generated automatically by ShinyGO v0.75. For additional evaluation of DASGs involved in reproduction, we used G:Profiler (Raudvere et al., 2019; Meleagris gallopavo as the reference organism) and the Ingenuity Pathway Analysis package with the same settings as previously reported (Słowińska et al., 2017). Finally, the functional prediction and the gene names of the identified DASGs were mapped to the UniProtKB database (www.uniprot.org). The possible interactions between DASGs involved in flagellum/cilium formation were established by STRING (Search Tool for Retrieval of Interacting Genes, Szklarczyk et al., 2019), with a medium confidence score cutoff of 0.4.
Quantitative Real-Time Reverse Transcriptase PCR of Differentially Alternative Splicing Events
RNA isolation (n = 6 per group), complementary DNA (cDNA) synthesis and real-time PCR were performed as previously described (Słowińska et al., 2020; Supplementary Table 1). Briefly, total RNA was extracted using TRIzol Reagent (Invitrogen by Thermo Fisher Scientific). Genomic DNA was removed from RNA samples using a DNase I Amplification Grade Kit (Invitrogen by Thermo Fisher Scientific). Synthesis of cDNA with 1,000 ng of RNA was conducted using High-Capacity cDNA Reverse Transcription Kits with RNase Inhibitor (Applied Biosystems by Thermo Fisher Scientific) according to the manufacturer's protocol. The expression of specific mRNA splice variants of cold inducible RNA binding protein (CIRBP), T-complex protein 1 subunit theta (CCT8), neuralized E3 ubiquitin protein ligase 1 (NEURL1), and testis-expressed protein 11 (TEX11) was quantified with Custom TaqMan Gene Expression Assays (Applied Biosystems by Thermo Fisher Scientific, Pleasanton, CA) using internal oligo TaqMan probes that recognized short and long splice variants. To measure the mRNA splice variants of sodium/potassium-transporting ATPase subunit beta (ATP1B3), cell adhesion molecule 1 (CADM1), MEIG1, outer dense fiber of sperm tails 2 (ODF2), outer dense fiber of sperm tails 2 like (ODF2L) and Na(+)/H(+) exchange regulatory cofactor NHE-RF1 (SLC9A3R1), real-time PCR with SYBR Green PCR master mix (Applied Biosystems by Thermo Fisher Scientific, Foster City, CA) was used. The sequences for these genes were downloaded from the ENSEMBL database (https://www.ensembl.org). Primers for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) amplification were designed based on predicted sequences obtained from the National Center for Biotechnology Information (NCBI). The Primer3Plus online tool (https://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) was used for primer–probe and primer set design. Gene names and primer–probe set information are presented in Supplementary Table 2. Reactions were performed using an ABI ViiA7 sequence detection system (Applied Biosystems by Life Technologies, Singapore). The TaqMan-based PCR conditions were as follows: 10 min at 95°C, 45 cycles of 15 s at 95°C, and 1 min at 60°C. All results were normalized to GAPDH expression, as an endogenous control, using the PCR Miner algorithm (Zhao et al., 2005). The PCR program for the mRNA amplification in SYBR Green was as follows: initial denaturation (10 min at 95°C), followed by 40 cycles of denaturation (15 s at 95°C) and annealing (1 min at 60°C). After each PCR, melting curves were obtained by stepwise increases in temperature from 60°C to 95°C. The final volume of the reaction mixture was 10 μL and consisted of 3 μL cDNA (25 ng), 0.3 μL (300 nM) each of the forward and reverse primers, 1.4 μL nuclease-free water and 5 μL SYBR Green master mix. The specificity of the product was confirmed by electrophoresis on a 3% agarose gel. Primers were synthesized by Sigma–Aldrich Co. Primer sequences and expected PCR product lengths are reported in Supplementary Table 3.
Statistical Analysis
Gene expression of short and long variants of CCT8, CIRBP, and TEX11 in the reproductive tract was analyzed using two-way analysis of variance (ANOVA). Significant differences between means were determined by Sidak multiple comparisons tests. The differences were considered significant at P-value ≤ 0.05. Gene variants (long, short) and reproductive tract tissues (testis, epididymis, and ductus deference) were studied as separated factors and the interaction between them was taken into consideration. The Shapiro–Wilk test was used to assess data normality. RNA-Seq data were analyzed using a T test when they followed a normal distribution. Not normally distributed data were analyzed by the Mann–Whitney test (a nonparametric test). The differences were considered significant at P-value ≤ 0.05.The data are shown as the mean ± standard deviation (SD). All analyses were performed using GraphPad statistical software (GraphPad PRISM v8.4.1, GraphPad Software Inc., San Diego, CA).
RESULTS
Detection of Alternative Splicing Events and the Main Classes of AS Among the Turkey Reproductive Tract
Among the turkey reproductive tract, the highest number of AS events was detected in the testis (33,648 ± 1,381; P-value < 0.0001). The number of AS events decreased gradually from the testis to epididymis (27,996 ± 1,405), reaching the lower level of AS events in the ductus deferens (23,175 ± 1,586). The distribution of seven different types of AS events identified in the testis, epididymis, and ductus deferens is illustrated in Figure 1B. The most abundant AS type within all examined reproductive tissues was classified as A3 (22.62% - testis. 24.72% - epididymis and 25.39% - ductus deferens; Supplementary Table 4). What is worth emphasizing is that the sum of three skipping classes (AL, AF, SE) represented between 45 and 50% of all detected splicing events in each experimental tissue. The last exon skipping constituted more than 10% of spliceosome components only in the testis, whereas the other alterations accounted for approximately 7% of AL splicing. Furthermore, the MXE splice subtype comprised only 1.1 to 1.2% splicing signatures in all samples.
Differentially Alternative Splicing Patterns
Testis vs. Epididymis
To discover AS events that differ between particular parts of the male reproductive tract, 2 approaches, SUPPA2 and rMATS, were used. Only these junction events that were confirmed simultaneously by both methods were analyzed in the final results. To classify the final output, the SUPPA classification (seven types of AS events) was selected, because it is more precise in characterizing skipping exon events. Following these assumptions, the comparison between the testis and epididymis revealed 86 DAS sites (Figure 2, Supplementary Table 5). Among them, 58 were assigned to SE, 9 to A3, 6 to AL, 5 to A5, 3 to RI, 3 to AF and finally 2 to MXE events. Within the intersected results, 44 DAS events overrated PSI values in the testis samples, and 42 DAS events had increased PSI values in the epididymis. Furthermore, the splicing alterations were localized within 57 protein-coding genes. Within the top 5 genes with the highest inclusion level in the testis, the following A-kinase anchoring protein 13 (AKAP13), testis specific 10 (TSGA10), TEX11, solute carrier family 35 member C2 and RalBP1-associated Eps domain-containing protein 1 indicated skipping exon type occurrence. Interestingly, the A5 splicing event within AKAP13 was ranked in the top 5 DASGs [other four: chromatin modification-related protein MEAF6 (MEAF6), ACOT9 thioesterase, doublecortin domain-containing protein 2B, and aryl hydrocarbon receptor nuclear translocator] with the lowest ΔPSI values preferably altered in the epididymis.
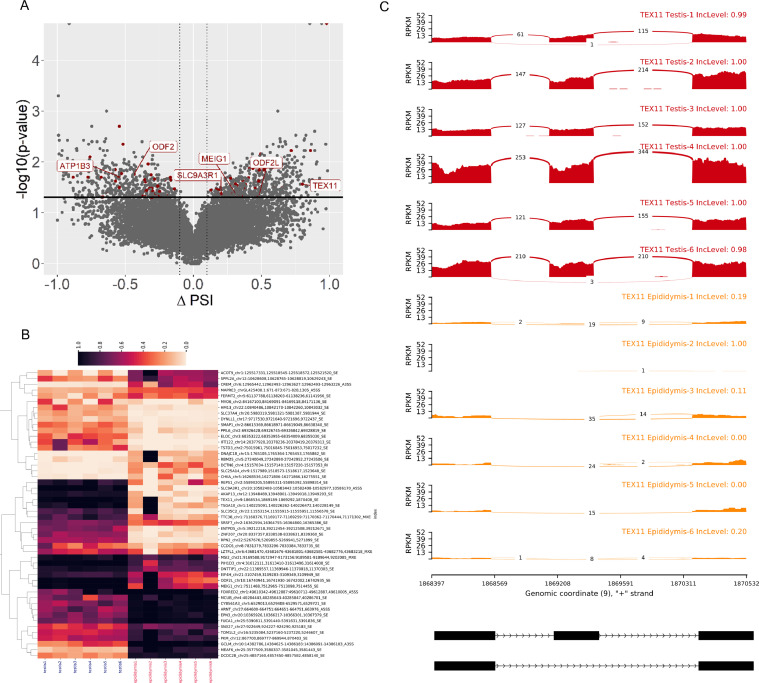
Figure 2.
Differentially expressed alternative splicing events in testis and epididymis comparison. (A) The volcano plot shows an abundance of AS events. The X-axis describes the ∆PSI values of each AS event, and the Y-axis shows the negative logarithmic adjusted P-value. The horizontal lines are equal to the negative logarithmic value of the adjusted P-value cutoff (0.05). The red dots represent the significantly different AS events, whereas gray dots are not significant. The most interesting AS events are labeled by the gene symbols. (B) Heatmap visualization of DAS events. Each track represents PSI (from 0 to 1) values of selected 50 top DAS events (rows) distributed for each RNA-seq library (columns). (C) Sashimi plot of selected AS events within the TEX11 gene. Twelve colored tracks represent biological replicates for testis (red) and epididymis (orange) libraries. Numbers on curved lines indicate reads counts engaged in each splice junction. The scale on the left presents expression values in the range of the AS regions. The PSI is presented on the right side of each sample in the upper track.
Testis vs. Ductus Deferens
The clean reads mapped in the vicinity of junction splice sites were also used to measure changes in exon inclusion/exclusion (ΔPSI) between testis and ductus deferens. Both methods identified 229 DAS events localized within 129 transcripts (Figure 3; Supplementary Table 6). The SE alteration with 145 DAS sites was the most dominant splice event. The others were classified as follows: A3 (21 events), A5 (19 events), RI (13 events), AF (11 events), MXE (10 events), and AL (10 events). The comparison of spliceosome target genes between testis and ductus deferens revealed 109 events (with ΔPSI values > 0.1) predominantly spliced in testis and 120 events (with ΔPSI values < −0.1) with preferential inclusions in ductus deferens. The five genes with the highest inclusion levels in the testis were found to have skipping exon alteration [AKAP13, gametogenetin-binding protein 2 (GGNBP2) and AP complex subunit beta], alternative 5’ splice site (exocyst complex component 7) and alternative first exon [E3 ubiquitin-protein transferase MAEA (MAEA)] occurrence. The second transcript of MAEA with alternative first exon usage (ΔPSI = −0.83) was the dominant variant in the ductus deferens. The mentioned gene together with the other four [metastasis-associated protein MTA1, MEAF6, Rab GTPase-activating protein 1-like, and spire type actin nucleation factor 1 (SPIRE1)] were classified on top of DASGs in the ductus deferens.
Figure 3.
Differentially expressed alternative splicing events in testis and ductus deferens comparison. (A) The volcano plots show an abundance of AS events. The X-axis describes the ∆PSI values of each AS event, and the Y-axis shows the negative logarithmic adjusted P-value. The horizontal lines are equal to the negative logarithmic value of the adjusted P-value cutoff (0.05). The red dots represent the significantly different AS events, whereas gray dots are not significant. The most interesting AS events are labeled by the gene symbols. (B) Heatmap visualization of DAS events. Each track represents PSI (from 0 to 1) values of selected 50 top DAS events (rows) distributed for each RNA-seq library (columns). (C). Sashimi plot of selected alternative skipping events within the SCMH1 gene. Twelve colored tracks represent biological replicates for testis (red) and ductus deferens (orange) libraries. Numbers on curved lines indicate reads counts engaged in each splice junction. The scale on the left presents expression values in the range of the AS regions. The PSI is presented on the right side of each sample in the upper track.
Epididymis vs. Ductus Deferens
Implemented stringent double verification (intersection of SUPPA2 and rMATS) permitted the identification of only 6 spliceosome changes between the epididymis and ductus deferens (Figure 4; Supplementary Table 7). Two events with ΔPSI values greater than 0.1 predominantly occurred in the epididymis, and 4 splicing events (ΔPSI < −0.1) appeared in the ductus deferens. Splicing alterations emerged within 3 protein-coding genes and 2 noncoding transcripts. There were two opposite changes in A5 splicing in torsin-4A (ΔPSI = 0.26 and ΔPSI = −0.23), which determined the predisposition of each to be transcribed in epididymis and ductus deferens, respectively. Furthermore, PDZ and LIM domain 1 (PDLIM1) showed a higher percentage of exon inclusion (ΔPSI = 0.22) in the epididymis. However, fermitin family member 2 (FERMT2) harbored an SE (ΔPSI = −0.36) event within the transcript, preferably noticed in the ductus deferens.
Figure 4.
Differentially expressed alternative splicing events in epididymis and ductus deferens comparison. (A) The volcano plots show an abundance of AS events. The X-axis describes the ∆PSI values of each AS event, and the Y-axis shows the negative logarithmic adjusted Pvalue. The horizontal lines are equal to the negative logarithmic value of the adjusted P-value cutoff (0.05). The red dots represent the significantly different AS events, whereas gray dots are not significant. The most interesting AS events are labeled by the gene symbols. (B). Heatmap visualization of DAS events. Each track represents PSI (from 0 to 1) values of all significant DAS events (rows) distributed for each RNA-seq library (columns). (C). Sashimi plot of selected alternative skipping events within the DCTN6 gene. Twelve colored tracks represent biological replicates for ductus deferens (red) and epididymis (orange) libraries. Numbers on curved lines indicate counts engaged in each splice junction. The scale on the left presents expression values in the range of the AS regions. The PSI is presented on the right side of each sample in the upper track.
Functional Classification of Differentially Alternative Splicing Events
Testis vs. Epididymis
Functional analysis of 57 ASGs differentiating the testis and epididymis revealed significant enrichment of 42 pathways representing biological processes, 50 pathways representing cellular components and 11 pathways representing molecular functions (Supplementary Table 5). The cilium organization pathway was the most enriched biological process (FDR = 0.0034); cilium, sperm flagellum and 9+2 motile cilium were found to be the most enriched cellular components (FDR = 0.0025). CAMP-dependent protein kinase activity and dopamine receptor binding were found to be the most enriched molecular function pathways (FDR = 0.0455; Figure 5A, Supplementary Table 5).
Figure 5.
Gene ontology enrichment analysis of differentially alternatively spliced genes identified in testis vs. epididymis (A); testis vs. ductus deferens (B) and epididymis vs. ductus deferens comparison (C). Charts generated by Shiny GO v0.75 represent the 20 most enriched gene sets (FDR < 0.05), selected species: turkey.
Grouping of DASGs by functional categories defined by high-level GO Biology Process terms revealed genes involved in ‘reproduction’ (Supplementary Table 5). Further enrichment analysis indicated the involvement of DASGs in male reproduction processes, including spermatogenesis (FDR = 0.0062), flagellated sperm motility (FDR = 0.0333) and sperm axoneme (FDR = 0.0144) and cilium organization (FDR = 0.0352; Table 1, Supplementary Table 5). Pathways indicated by ShinyGO were enriched by an additional analysis using G:Profiler, IPA, and manual searching using the UniProt database (Table 1).
Table 1.
Involvement of DAS genes between testis and epididymis comparison in reproduction processes.
| Testis vs. Epididymis | ShinyGO/turkey | G:Profiler/turkey | IPA/mammals | UniProt |
|---|---|---|---|---|
| Spermatogenesis | ||||
| Spermatogenesis | MEIG1, TSGA10 | CREM, MEIG1, TEX11, TSGA10 | MEIG1, TEX11, TSGA10 | |
| Arrest in spermatogenesis | CREM, MEIG1 | |||
| Male gamete generation | MEIG1, TSGA10 | CREM | ||
| Spermatid differentiation | MEIG1 | |||
| Sperm flagellum and motility | ||||
| Sperm flagellum | ODF2, ATP1B3, SLC9A3R1 | DNAAF6, PIH1D3 | ||
| Flagellated sperm motility | MEIG1 | ODF2L, ATP1B3, SLC9A3R1 | ||
| Sperm motility | MEIG1 | |||
| Sperm axoneme and microtubule | ||||
| Sperm axoneme assembly | MEIG1 | DNAAF6, MEIG1 | MEIG | |
| 9+2 motile cilium | ODF2 ATP1B3, SLC9A3R1 | ODF2L, ATP1B3, SLC9A3R1 | ||
| Microtubule bundle formation | MEIG1 | DYNLL1, MAPRE3 | ||
| Microtubule-based process | ODF2, MEIG1, MAPRE3, ZNF207, DYNLL1, SLC9A3R1, PIH1D3, IFT122, UVRAG | DYNLL1, MAPRE3, ZNF207, SLC9A3R1, IFT122, ODF2 | ||
| Microtubule dynamics | APLP2, DNAAF6, DYNLL1, IFT122, MAPRE3, MYO6, ODF2, PKM, SLC9A3R1, TSGA10 | |||
| Cilium assembly | ||||
| Cilium movement | MEIG1 | ODF2, ATP1B3, SLC9A3R1 | ||
| Cillum organization | ODF2, ATP1B3, SLC9A3R1 | ODF2, IFT122 | ODF2L, ODF2, SLC9A3R1, IFT122, LZTFL1 | |
Testis vs. Ductus Deferens
Functional analysis of 129 ASGs differentiating testis and ductus deferens revealed significant enrichment of 32 pathways representing biological process, 46 pathways of cellular components and 2 pathways of molecular function (Supplementary Table 6). Macromolecule and protein localization (FDR = 0.0003 and FDR = 0.0010, respectively), positive regulation of cellular processes, and regulation of biological processes (FDR = 0.0010) were the most enriched biological processes; non–membrane-bounded organelles and intracellular non-membrane-bounded organelles were the most enriched cellular components (FDR = 2.44e-08), and protein binding was found to be the most enriched molecular function pathway FDR = 0.0009; Figure 5B, Supplementary Table 6).
Enrichment analysis of genes involved in ‘reproduction’ (Supplementary Table 6) indicated involvement of DASGs in male reproduction processes, represented by spermatogenesis (FDR= 0.0120), flagellated sperm motility (FDR = 0.0298)/sperm flagellum assembly (FDR = 0.0194), sperm axoneme (FDR = 0.0194), actin assembly (FDR = 0.0120-0.0496), and binding of sperm to zona pellucida/sperm-egg recognition (FDR = 0.0226; Table 2, Supplementary Table 6). Pathways indicated by ShinyGO were enriched by an additional analysis using IPA and manual searching using the UniProt database (Table 2).
Table 2.
Involvement of DAS genes between the testis and ductus deferens comparison in reproduction processes.
| Testis vs. ductus deferens | ShinyGO/turkey | IPA/mammals | UniProt |
|---|---|---|---|
| Spermatogenesis | |||
| Spermatogenesis | JAG2, NEURL1 | CADM 1 | JAG2, GGNBP2, SCMH1, SUN1,CADM1, CREM |
| Arrest in spermatogenesis | CADM1 | ||
| Meiotic cell cycle | SPIRE1, MLH3 | MLH3, SPIRE1, SUN1 | |
| Male gamete generation | JAG2, NEURL1, MLH3 | ||
| Attachment of spermatocytes and spermatids | CADM1 | ||
| Proliferation of spermatogonia | CIRBP | ||
| Apoptosis of spermatids | CADM1, CSNK2A2 | ||
| Sperm flagellum and motility | |||
| Flagellated sperm motility | NEURL1 | PIHD3 | |
| Sperm motility | NEURL1, DNAAF6 | ||
| Sperm binding | |||
| Zona pellucida receptor complex | CCT8 | CCT8 | |
| Sperm axoneme and microtubule | |||
| Sperm axoneme | NEURL1 | ||
| Microtubule bundle formation | MAPRE3 | KIF23, MAP2, MAPRE3, SLAIN2, CKAP5 | |
| Microtubule-based process | ZNF207, SPIRE1, KIF23, IFT122, SLAIN2, ODF2, NEURL1 | MAP2, DYNLL1, MAPRE3, SLAIN2 | ZNF207, DYNLL1 |
| Microtubule cytoskeleton | ODF2, CCT8, CEP41, PPP2R5A, KIF23, IFT122, SLAIN2 | KIF23, MACF1 | |
| Actin assembly | |||
| Parallel actin filament bundle assembly | SPIRE1, FLNB, PDLIM1, FERMT2 | ABI1, HSPB7, MYOC, PDLIM1 | ABI1, FERMT2, FHOD3, FLNB, MACF1, MAEA, SPTAN1 |
| Formin-nucleated actin cable assembly | SPIRE1 | ||
| Actin cytoskeleton | FLNB, PDLIM1, FERMT2, VCL, AKAP13 | ||
| Morphology of actin filaments | ABI1, HSPB7, MYOC, PDLIM1 | ||
| Formation of actin filaments | AKAP13, ATXN2, CDC14A, FERMT2, FHOD3, MAP2, MYOC, PDLIM1, SPIRE1 | PDLIM1, SPIRE1 | |
| Cilium assembly | |||
| Cilium movement | ENTPD5 | ||
| Cilium organization | CEP41, IFT122, ODF2, NEURL1 | CEP41 | |
Epididymis vs. Ductus Deferens
Functional analysis of 3 ASGs differentiating epididymis and ductus deferens revealed significant enrichment of 131 pathways representing biological process, 24 pathways of cellular components and 13 pathways of molecular function (Supplementary Table 7). Pathways connected with integrin activation were the most enriched biological processes (FDR = 0.0181); stress fiber and actin filament bundle were the most enriched cellular components (FDR = 1.715e-05). Transforming growth factor beta receptor binding pathways and phosphatidylinositol-3,4,5-trisphosphate binding were the most enriched molecular function pathways (FDR = 0.0164; Figure 5C, Supplementary Table 7). No genes involved in ‘reproduction’ were enriched in the epididymis vs. ductus deferens comparison.
Validation of Selected Differential Alternative Splicing Events
To validate the NGS results, 10 alternatively spliced transcripts with exon skipping were selected for qRT–PCR (CCT8, CIRBP, NEURL1, TEX11) and RT–PCR, with PCR product analysis by gel electrophoresis (ATP1B3, CADM1, MEIG1, ODF2 and ODF2L, SLC9A3R1). The DASGs were chosen for important physiological processes associated with male reproduction such as spermatogenesis (CADM1, CIRBP, MEIG1, NEURL1, TEX11), sperm flagellum and motility (ATP1B3, NEURL1, ODF2, ODF2L, SLC9A3R1) and sperm binding to the zona pellucida (CCT8). Among these 10 DASGs, the expression profiles showing inclusion (long variant, LV) and exon skipping (short variant, SV) were confirmed for 7 genes (Figure 6).
Figure 6.
Real-time validation of selected differentially alternatively spliced genes detected by RNA-Seq. Data are expressed as the mean ± SD. (A–G) The calculated percent of splicing inclusion and A’-G’) expression of T-complex protein 1 subunit theta (CCT8), cold inducible RNA binding protein (CIRBP), testis-expressed protein 11 (TEX11), cell adhesion molecule 1 (CADM1), meiosis-expressed gene 1 protein (MEIG1), outer dense fiber of sperm tails 2 (ODF2) and outer dense fiber of sperm tails 2 like (ODF2L). GAPDH, glyceraldehyde-3-phosphate dehydrogenase. Different letters indicate statistical significance at P ≤ 0.05; a,b indicate the difference between long and short variants (LV, SV) in the testis (A’) and x,y indicate the deferens between the long and shot variants in ductus deferens (A’ and B’) or epididymis (C’); * indicates the deferens between tissues for long or short variants.
The observed differences in the mRNA expression levels of CIRBP and TEX11 confirmed that long variants were expressed at higher levels in the testis (Figures 6B’-C’). For CCT8, long variants were highly expressed in the ductus deferens compared to those in the testis (Figure 6A’). Differences between long and short variants were observed in the testis for CCT8, in the ductus deferens for CCT8 and CIRBP (Figures 6A’-B’) and in the epididymis for TEX11 (Figure 6C’).
Semiquantitative PCR showed 2 products with and without an additional skipped exon, and a corresponding difference in band size of amplification products was observed (Figures 6D’-G’). The abundance of long versions of genes with the presence of skipped exons was observed in the testis for MEIG1 and ODF2L (Figures 6E’ and G’). Two products with and without an additional skipped exon (representing LV and SV, respectively) were observed in epididymis for MEIG1, ODF2, ODF2L (Figures 6E’-G’), and ductus deferens for CADM1 (Figure 6D’). The PSIs for genes are presented in Figures 6A–6G.
DISCUSSION
In this study, we provided new information regarding alternatively spliced genes in the turkey reproductive tract, which is, to our knowledge, the first such study in birds. Testis was characterized by the highest number of AS events, and skipped exons were the most frequently occurring class of AS. Statistical analysis revealed 86, 229, and 6 DAS events in the testis/epididymis, testis/ductus deferens, and epididymis/ductus deferens comparison, respectively. The GO analysis of DASGs between the testis and epididymis revealed involvement in cilium organization and homeostasis. The overall enriched GO term processes encompassed macromolecule and protein localization, regulation of biological and cellular processes and non-membrane-bounded organelles in the testis vs. ductus deferens comparison. Integrin activation and stress fibers were found in epididymis vs. ductus deferens enrichment annotations. The DASGs from testis vs. epididymis and testis vs. ductus deferens were also grouped in ‘reproduction’, indicating their involvement in male reproductive processes, including spermatogenesis and sperm motility.
In this study, we focused on the reproductive tissue-specific profile of DAS events and their involvement in reproductive physiological processes characteristic of the testis, epididymis, and ductus deferens such as the regulation of spermatogenesis and sperm tail formation. It seems to be a novel strategy compared to most studies focusing on comparative analyses of testicular splicing genes and transcripts from somatic tissues such as brain, liver, heart or muscle (Yeo et al., 2004; de la Grange et al., 2010). Therefore, discussion of the AS of genes and their specific expression profiles in the reproductive tract, which can influence protein structure and function in different parts of the reproductive tract, was found to be very challenging due to a lack of similar approaches in previous studies.
Testis–The Site of Extensive Alternative Splicing in Turkeys
The available data concerning birds indicated a high number of developmentally dynamic AS in the testes and brains of chickens (Mazin et al., 2021) and an association of gonadal AS with phenotypic sexual dimorphism (Rogers et al., 2021). Our study is the first attempt to describe the whole transcriptome of the turkey reproductive tract in terms of AS. The testis was recognized as an organ with the highest amount of posttranscriptional splicing events, which gradually decreased in epididymis and ductus deferens. The reason for the abundance of AS in the testis is quite multifarious. The high level of AS events was connected with extensive developmental processes (Elliott and Grellscheid, 2006) and the contribution of splicing regulation in spermatogenesis and fertility (Gallego-Paez et al., 2017). It was also indicated that testis-specific splicing may represent “background” noise induced by high levels of cell proliferation or nonspecific fluctuation in splicing regulator expression (Elliott and Grellscheid, 2006; Gallego-Paez et al., 2017; Mazin et al., 2021). Nevertheless, it is now clear that turkey testis is characterized by a high number of AS, which likely reflects the specificity of this organ, which is likely related to spermatogenesis.
Variation in Alternative Splicing Across Turkey Reproductive Tract Tissues
In birds, SE together with MXE splicing events were found to be the most common type of autosomal splicing in turkey, duck, and guinea fowl gonads (Rogers et al., 2021). SE together with RI was also found to be the dominant class of AS in chicken testis (Mazin et al., 2021). Our study indicated that turkey reproductive tract genes identified previously by Słowińska et al. (2020) exhibit multiple different types of splicing events. Moreover, SEs, including AF and AL, are the most frequent class of AS events across the turkey reproductive tract. However, in contrast to chickens, we did not find RI to be a dominant class of AS in turkey testes. It seems that the testis-specific feature of the turkey is an increased level of AS events with a high proportion of SE splicing, which is associated with the generation of functional isoforms (Weatheritt et al., 2016). Such transcriptomic modifications may be important for proper testis development and functioning, especially during spermatogenesis in turkeys.
Alternative Splicing as a Regulator of Spermatogenesis in Turkeys
The available data describing splicing events within the reproductive system of the birds focused on 2 genes: chicken DMRT1 and pigeon LDHC (Zhao et al., 2007; Huang et al., 2012). The presence of 3 splicing events situated within the DMRT1 locus was also detected in our study; however, no significant expression changes between particular parts of the reproductive tract were recorded. Furthermore, LDHC has not been annotated thus far in the turkey genome, and we also did not detect the expression of this transcript in our research. However, for the first time, we identified several alternatively spliced genes, MEIG1, TEX11, TSGA10, CREM (testis vs. epididymis comparison) and CADM1, CIRBP, CREM, nonspecific serine/threonine protein kinase (CSNK2A2), GGNBP2, delta-like protein (JAG2), MutL homolog 3 (MLH3), NEURL1, SAM domain-containing protein (SCMH1), SPIRE1, and SUN domain-containing protein (SUN1) (testis vs. ductus deferens comparison), involved in spermatogenesis.
Among alternatively spliced genes in the turkey reproduction tract, AS has been previously confirmed in mammalian testis for the CREM, MEIG1, TSGA10, and SUN1 genes (Walker et al., 1994; Ever et al., 1999; Behnam et al., 2006; Göb et al., 2010). Moreover, none of them have been previously reported for bird reproduction. In the turkey reproductive tract, we found 8 AS events of CREM, whereas in human and rat testis, 10 AS variants of CREM were reported (Walker et al., 1994; Guo et al., 2015). CREM is known to play a role in modulation of gene expression related to the structuring of mature spermatozoa (Lalli et al., 1996; Monaco et al., 1996; Walker and Habener, 1996). As a result of AS, CREM gene expression originates different isoforms, which in turn can be divided into activators or repressors of gene expression (Foulkes et al., 1992). In turkey testis, we detected increased splicing modification within the last exon of CREM transcripts (skipping last exon with 3’ UTR, Supplementary Figure 1A). The 3’ UTR is a versatile region that is enriched for regulatory elements and can influence the polyadenylation, translation efficiency, localization, and stability of mRNA (Miyamoto et al., 1996; Takagaki et al., 1996; Edwalds-Gilbert et al., 1997; Lutz, 2008; Barrett et al., 2012). It was indicated that the 3’ UTRs of testis-specific genes in mammals become progressively shorter during spermatogenesis (Liu et al., 2007; Li et al., 2012, 2016) as a consequence of transcription termination. Therefore, the generation of 3’ UTR-alternative splicing and polyadenylation may have the potential to diversify mRNA expression patterns in bird male reproduction. It is possible that diversification of CREM mRNA in the turkey reproductive tract, especially in the testis, may be involved, similar to mammals, in the effective regulation of spermatogenesis.
MEIG1, abundantly expressed in the testis, both in somatic and germinal cells, acts in spermatocytes in the execution of meiosis during spermatogenesis (Ever et al., 1999; Salzberg et al., 2010). In murine testis, MEIG1 encodes 2 alternative transcripts that differ in the 5′ UTR due to alternative promoters (Ever et al., 1999). We are the first to report the differences in alternative splicing MEIG1 in the turkey reproductive tract. Among 3 AS events of MEIG1 detected in turkey testis, one of them indicated skipping exon changes in the 5’ UTR between the testis and epididymis (FDR = 0.042). Genes with differences in the 5′ UTR are relatively common in mammals (Araujo et al., 2012). It seems that splicing in the 5’ UTR in MEIG1 mRNA may be a common feature for mammals and birds. These variations in the 5′ UTR can function as important switches to regulate gene expression and transcript translation (Araujo et al., 2012). We identified 2 different MEIG1 transcripts derived from 2 different promoters (Supplementary Figure 1B). A shorter transcript (without changes in the 5’ UTR) was expressed in the epididymis, where it may also be efficiently translated and involved in sperm maturation, while a longer transcript (with changes in the 5’ UTR) was predominantly expressed in the testis and may be the main player during spermatogenesis.
As a result of AS, 2 isoforms of the murine TSGA10 gene have been expressed in mature testes (Behnam et al., 2006). We detected alternatively spliced transcripts of TSGA10 in the turkey reproductive tract. One of the skipping exon events was found to differentiate the testis and epididymis (FDR = 0.006). Similarly, whole exon 16 exclusion was responsible for shorter transcription of TSGA10 and production of its protein with lower molecular weight in mouse testis (Behnam et al., 2006). TSGA10 is known to be a potential key factor in the process of spermatid differentiation/maturation, playing a role in chromatin division during spindle formation in mitosis (Asgari et al., 2021). Moreover, TSGA10 is involved in the correct arrangement of the mitochondrial sheath in spermatozoa (Luo et al., 2021), and its localization was confirmed to be to the midpiece and sperm tail in spermatozoa (Behnam et al., 2006; Sha et al., 2018). Identified in this study exon skipping produces 2 transcripts of TSGA10 with different lengths (Supplementary Figure 1C). A longer transcript was predominantly expressed in the testis, similar to MEIG1, which may be a main player during spermatogenesis. A shorter transcript that is expressed in the epididymis may be involved in the post-testicular development of sperm motility described previously by Słowińska et al. (2020).
Turkey testis was also characterized by the presence of 15 alternatively spliced transcripts of SUN1, 3 of which with skipping exons showed tissue-specific expression differentiating testis and ductus deferens. There is no available information considering bird SUN1 function in spermatogenesis. In mammals, SUN1 is required for telomere attachment to the nuclear envelope and gametogenesis (Ding et al., 2007). In murine testis, seven Sun1 various splicing variants were identified (Göb et al., 2011). Interestingly, the shortest variant of the Sun1 gene was generated by deletions of exons 7 to 10, leading to complete loss of hydrophobic sequences: H1 and part of H2, belonging to the nucleoplasmic domains of SUN1 and potentially functioning as a transmembrane anchor domain (Liu et al., 2007; Göb et al., 2011). Astonishing turkey SUN1 seems to undergo similar AS modification. In detail, exons 6, 9, and 10 were deleted in short variants of turkey SUN1 predominantly expressed in the turkey testis (Supplementary Figure 1D). Following Göb et al. (2011), we observed similar modification within the SUN1 gene, which may cause N-terminal region protein modification and differences in effective inner nuclear membrane targeting and membrane retention. As a consequence, SUN1 was not identified in the nuclear envelope of spermatids but instead was localized to the acrosomal membrane system (Göb et al., 2011). The similar splicing modification of the SUN1 gene between mammals and birds is very interesting, suggesting its conserved function among vertebrates, especially related to the acrosome.
In addition to the genes discussed above, the majority of all differentially spliced genes in the turkey reproductive tract have not yet been reported to undergo AS in mammals. Longer variants (without skipping) of genes, such as CSNK2A2, GGNBP2, JAG2, MLH3, SCMH1, and TEX11, dominated in the testis, suggesting their potential role in spermatogenesis. To date, only the expression of TEX11 and CSNK2A2 has been reported for bird/turkey testis, and their function was suggested to be connected with meiosis and formation of the acrosome during vesicle trafficking, respectively (Słowińska et al., 2020). In mammals, specific engagement in meiosis was reported for MLH3 and TEX11 (Tang et al., 2011; Jung et al., 2019), chromatin condensation and modification for SCMH1 (Takada et al., 2007) and formation of head morphology for CSNK2A2 (Xu et al., 1999). Interestingly, in this study, exon skipping with intact reading frames among coding sequences was identified as a form of RNA splicing in all mentioned genes, producing shorter variants of mRNAs that encode multiple protein isoforms of functional proteins in the epididymis and ductus deferens, whereas long variants were present in the testis. In summary, AS in the turkey reproductive tract can be a mechanism causing tissue-specific differences in protein isoform activity, as previously suggested by Sharpe et al. (2017).
On the other hand, turkey testis was also characterized by the presence of dominant alternatively spliced short variations of CADM1, NEURL1, and SPIRE1. To date, only the expression of NEURL1 has been reported for birds/turkey as a part of ubiquitin-dependent regulation of spermatogenesis (Słowińska et al., 2020). In mammals, NEURL1 is required for axonemal integrity in spermatozoa (Vollrath et al., 2001) and is ontologically classified as ‘flagellated sperm motility’ (GO:0030317). In birds, CADM1 has been identified thus far only in chicken immune cell populations of the spleen, liver, and lungs (Vu Manh et al., 2014; Hu et al., 2019; Alber et al., 2020; Sutton et al., 2021), and no reports are available for SPIRE1 expression in avians. For both genes, support of germ cell development during spermatogenesis has been reported in mammals (Wakayama et al., 2009; Wen et al., 2018). The mentioned genes NEURL1, CADM1, and SPIRE1 were modulated by exon exclusion splicing. The SE was observed between exons 9 and 11 of SPIRE1, and this unannotated event has not been identified in the current turkey genomic database. The splicing analysis revealed a stronger preference, including an unannotated exon (E10), in the ductus deferens, which affected the elongation of SPIRE1 post-transcriptional products. In the testis, this skipping event reduced SPIRE1 transcripts by 174 nucleotides in the CDS (Supplementary Figure 1E). Further studies should focus on the role and consequence of exon skipping for protein function in the turkey reproductive system during spermatogenesis in the testis, epididymis, and ductus deferens, where spermatozoa develop, undergo maturation and acquire motility, respectively.
CIRBP is a cold-shock protein whose activation is increased in response to cold stress (Nishiyama et al., 1997). In birds, CIRBP is expressed in the testis, and its transcription is decreased after heat stress in chickens (Wang et al., 2013a; Liu et al., 2022). The role of CIRBP in the heat-stress response of bird testes is still unknown. In mammals, CIRBP plays a key function in heat stress-induced testicular spermatogenic cell injury, acting as a modulator of cyclin B1 expression, which inhibits spermatogenesis arrest (Xia et al., 2012; Liu et al., 2022). The turkey CIRBP gene was characterized by multiple splicing events, and 5 of them differentiated the testis and ductus deferens (Supplementary Figure 1F). One of them, in the testis, demonstrated a higher inclusion level of splicing exons in comparison to the ductus deferens. This indicated that a longer transcript of CIRBP may be most commonly expressed in the testis and the expression of the shorter transcript may dominate in this ductus deferens. Alternative variants of genes can encode shorter protein isoforms differing at the C-terminus end, mainly in the ductus deferens. It was indicated that both alternative transcription start and termination sites are the principal drivers of transcript isoform diversity across tissues and may underlie the majority of cell type-specific proteomes and functions (Reyes and Huber, 2018). It seems that the splicing mechanism is important for CIRBP, making this gene a potential tissue-specific reproductive marker.
On the other hand, four CIRBP sequence inclusions dominated in the ductus deferens. All of the events displayed significant AS within the 3’ UTR location. Two of them displayed significant AS, with a shared CDS end. Three of the mentioned events were classified as RI and one as AL, which occurred more frequently in the ductus deferens. In summary, our results suggest the importance of the AS mechanism for the adaptation of CIRBP to specific requirements in particular parts of the turkey reproductive system.
Acrosome
The DAS event of spectrin alpha (SPTAN1) was identified within the turkey reproductive tract. Spectrin, together with other major cytoskeletal proteins, such as actin and tubulin, is reported to be involved in the regulation of acrosome reactions in mammals (Dvoráková et al., 2005). To our knowledge, there is a lack of information regarding SPATAN1 in bird semen. Although SPTAN1 transcripts were identified in turkeys, there was no differentiation among the reproductive tract systems (Słowińska et al., 2020). We found that AS of the SPTAN1 gene caused exon 22 skipping, leading to new gene isoform production. The turkey testis was characterized by the dominance of the exon inclusion and skipping event characteristic of the ductus deferens (FDR = 0.0187; supplementary Figure 1G). Sperm acrosome formation is an important stage of spermiogenesis, and testicular transcripts participating in acrosome formation were described in detail for turkeys (Słowińska et al., 2020). This study expands this knowledge about acrosome formation in turkeys by indicating the importance of AS mechanisms in acrosome development in birds.
Alternative Splicing of Genes Involved in Sperm Tail Formation and Ciliary Function
Several genes involved in sperm tail formation and ciliary function were predicted to undergo AS in the turkey reproductive tract (Figure 7). Four genes undergoing AS in the turkey reproductive tract were classified as sperm flagellum components (ATP1B3, ODF2, SLC9A3R1, TSGA10; Figure 7). Recently, the proteins ATP1B3, ODF2, and TSGA10 were identified by Labas et al. (2015) in spermatozoa and SLC9A3R1 in seminal plasma of chickens. However, their function in avian reproduction is still unknown. In mammals, TSGA10, ODF2, and SLC9A3R1 are present in the sperm tail (Tarnasky et al., 2010; Chávez et al., 2012; Luo et al. 2021), and ATP1B3 presence was confirmed in mouse spermatozoa (Zhou et al., 2018). To date, AS of ODF2 and TSGA10 has been reported for mammalian testes (Brohmann et al., 1997; Shao et al., 1997; Turner et al., 1997; Hoyer-Fender et al., 1998; Petersen et al., 1999).
Figure 7.
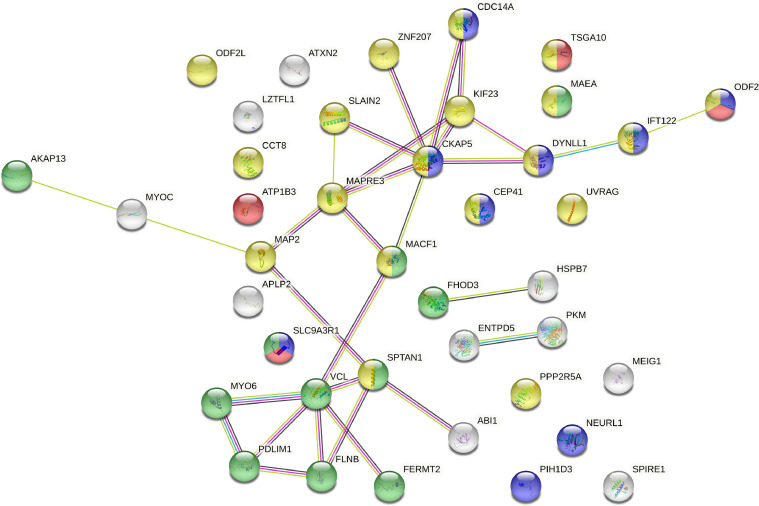
Protein–protein interaction networks for differentially alternatively spliced genes involved in sperm tail formation and ciliary function, according to an analysis by the STRING online database, version 11.5. Functional enrichment classified proteins into sperm flagellum (red color; GO:0036126; FDR = 0.0079); cilium organization (blue color; GO:0044782; FDR = 4.81e-05); actin cytoskeleton (green color; GO:0015629; FDR = 6.68e-07) and microtubule cytoskeleton (yellow color; GO: GO:0015630; FDR = 1.44e-09). There are 4 types of associations presented: experimental (purple), database (light blue), text mining (yellow), and coexpression (black) evidence.
ODF2 is the major protein component of the outer dense fibers (ODF) of the mammalian principal and midpiece sperm tail. In avian semen, ODF2 has been identified in chicken spermatozoa (Labas et al., 2015). The ODF2s seems to be involved in sperm motility; inhibition of tyrosine phosphorylation of ODF2 adversely impacts sperm motility (Mariappa et al., 2010). Two slightly different cDNAs encoding ODF2 proteins isolated from a human testis cDNA library were characterized by variable N-terminal regions (Petersen et al., 1999). In our study, we found ODF2 skipping of the first exon within variants presented exclusively in the testis (Supplementary Figure 1H). The spliced exon was localized at the beginning of the translational start codon of the turkey ODF2, contributing to the posttranscriptional and translation consequences for short variants. In the testis, the produced short variants of ODF2 may be dysfunctional. This is an unexpected result showing that in the turkey reproductive tract, the epididymis may be the main site of ODF2 protein activation, participating in sperm maturation that in birds takes place in the epididymis and ductus deferens.
Active Na+ and K+ exchange in the sperm membranes is under the control of the Na,K-ATPase pump (Jimenez et al., 2011). The ATP1B identified in our study is a part of the functional pump core and is required for its trafficking to the plasma membrane in mammals (Clausen et al., 2017). To date, the production of ATP1B3 protein has been reported in chicken spermatozoa (Labas et al., 2015). To our knowledge, no AS of ATP1B3 has been reported for vertebrates. In our study, we found inclusion of the additional exon between the annotated first and second exons, causing changes in the proper reading frame of the ATP1B3 gene (Supplementary Figure 1I). The prevalence of inclusion within ATP1B3 epididymis variants may cause the production of dysfunctional protein isoforms. This may suggest that ATP1B3 may regulate ion flow and membrane potential maintenance, processes crucial for sperm motility that take place in the testis. Presumably, ATP1B3 is silenced via AS in the epididymis, and the AS mechanism of this gene must be explored in ATP1B3-targeted research.
The SLC9A3R1 gene is important for sperm ion channel regulation in mammals (Chan et al., 2009; Chen et al., 2009; Chávez et al., 2012). The protein Na+/H+ exchanger regulatory factor 1 (NHERF1) encoded by SLC9A3R1 regulates the concentration of Cl−/HCO3− during capacitation of mammalian spermatozoa (Chávez et al., 2012). The presence of NHERF1 was reported in sperm, exactly to the midpiece (Chávez et al., 2012). In contrast to mammals, avian spermatozoa are motile during passage through the genital ductus and do not require capacitation within the hen's reproductive tract (Clulow and Jones, 1982). Interestingly, SLC9A3R1 protein has been found thus far only in avian seminal plasma (Labas et al., 2015; Słowińska et al., 2017), suggesting a low level of SLC9A3R1 in the sperm cell or secretion of this protein by reproductive tissue directly to seminal plasma, as previously described for other avian seminal plasma proteins (Słowińska et al. 2014,2019,2021). We found changes inside the SLC9A3R1 5’ UTR that differentiate turkey testis and epididymis (FDR = 0.0429, Supplementary Figure 1J). The differences in their 5′ UTRs produced 2 splicing variants. The longer first exon (proximal promoter) of SLC9A3R1 was dominant in the testis, and the shorter version was specific for the epididymis. The comparable modification was described above for the MEIG1 gene, suggesting that shorter transcripts (without changes in the 5’ UTR) were expressed in the epididymis, which could also be efficiently translated and involved in sperm maturation, while longer (with changes in the 5’ UTR) transcripts were predominantly expressed in the testis and could be the main player during spermatogenesis. Altogether, it can be suggested that variations in the 5′ UTR are an important mechanism for the regulation of gene expression and transcript translation during spermatogenesis and sperm maturation in turkeys.
We distinguished several genes undergoing AS in the turkey reproductive tract involved in ‘cilium organization’ and ‘actin and microtubule cytoskeleton’ (Figure 7). According to Olcese et al. (2017), the motile cilium of epithelia is highly similar in structure organization to the motile flagella of sperm. Therefore, we decided to discuss genes classified in the ciliary function in relation to sperm flagella. It should be emphasized that all proteins encoded by the genes involved in ciliary organization were identified both in spermatozoa and testis. Interestingly, in our study, most of the identified genes were reported for the first time to undergo AS in the male reproductive tract. Previously, somatic cells or cancer tissues have been found to be the site of AS of the mentioned genes in mammals.
The turkey spermatozoa axoneme is composed of the 9 + 2 microtubule structure (Thurston and Hess, 1987; Jamieson et al., 2007). Genes involved in spermatozoa axoneme formation have been previously described in detail for turkey testis, where the flagellar structure is primarily formed (Słowińska et al., 2020). In this study, we provided new information regarding the AS of genes involved in microtubule cytoskeleton organization in the turkey axoneme. Among them, we found a regulatory mechanism of microtubule binding [microtubule-associated protein (MAP2), microtubule-associated protein RP/EB family member], microtubule motor activity [MAP2, dynein light chain (DYNLL1)] and microtubule polymerization [MAP2, MAPRE3, SLAIN motif family member 2 (SLAIN2), cytoskeleton-associated protein 5 (CKAP5)]. The presence of MAP2, MAPRE3, CKAP5 and DYNLL1 proteins has been confirmed for mammalian spermatozoa (Peddinti et al., 2008). Among them, DYNLL1 was reported for chickens (Labas et al., 2015), suggesting a direct role in spermatozoa motility development by involvement in flagellar structure organization.
Actin filament-based processing and organization were indicated to be involved in the post-testicular development of sperm motility in turkeys (Słowińska et al., 2020). Among bird spermatozoa, the presence of actin was confirmed for quail and domestic fowl (Aire and Ozegbe, 2008). We found posttranscriptional AS modification within genes participating in actin filament processing and organization. These genes are directly involved in cytoskeleton organization [FHOD3 protein (FHOD3), filamin B (FLNB), MAEA, unconventional myosin-6 (MYO6), and PDLIM1] and actin filament binding [FERMT2, FHOD3, microtubule-actin cross-linking factor 1 (MACF1) and MYO6]. The presence of FLNB and MYO6 has been so far confirmed in chicken sperm during semen quality phenotyping (Labas et al., 2015).
Binding of Sperm to Zona Pellucida
The CCT8 gene undergoing AS in the turkey reproductive tract is known to be involved in the binding of sperm to the zona pellucida in mammals (Arangasamy et al., 2011; Staicu et al., 2021). In our study, we found 2 DAS events of CCT8 in the turkey reproductive tract. One of them differentiating the testis and ductus deferens (FDR = 0.012) was characterized by the inclusion of an additional exon between the 2 last exons, giving rise to changes within the terminal part of the CCT8 coding sequence in the ductus deferens (Supplementary Figure 1K). According to Ye et al. (2014), both transcript variants from the skipping event site, that is, in which the exon is either included (inclusion isoform) or excluded (skipping isoform), are typically present in one cell, and the prevailing conditions dictate which isoform is dominantly expressed. Our study indicates that the CCT8 variant with exon inclusion is dominantly expressed in the ductus deferens. Biological factors that may influence the regulation of CCT8 variant expression remain unknown. It can be suggested that AS of genes involved in sperm-egg binding in turkeys may intentionally produce additional protein isoforms for successful fertilization, as was previously indicated for the redundant form of acrosin in turkeys’ spermatozoa (Słowińska et al., 2012).
Interestingly, results concerning genetic mutation, particularly single nucleotide polymorphisms (SNPs) agree with results of this study in terms of heterogeneity and expression site. In birds, SNPs has been studied mainly in the relation to performance traits. A huge number of SNPs markers were identified in bird blood (Kerstens et al., 2009; Adams et al., 2021). Among them, novel SNPs in chicken and turkey genes were found to be associated with bird growth (ADIPOR2, MYF5, MYOG and PRDM16; Han et al., 2012; Wang et al., 2015; Wei et al., 2016) and muscle formation (MYL1 and RYR1; Chaves et al., 2003; Droval et al., 2012). In duck, SNP screening and genotyping has been also analysed in relation to egg production (Wang et al., 2013b) and eggshell quality (Tan et al., 2021). However, there is lack of knowledge concerning SNP of genes connected directly with reproductive performance in turkey males. Further studies should focus on searching of novel SNP in genes participating directly in spermatogenesis and post-testicular sperm maturation in birds.
CONCLUSIONS
In conclusion, AS in the turkey reproductive tract seems to be a mechanism leading to tissue-specific gene diversification. Testis was found to be the site of the highest number of posttranscriptional splicing events within the reproductive tract, and SE was the most frequently occurring class of AS. ASGs seem to be involved in the regulation of spermatogenesis, including acrosome and sperm tail formation and binding of sperm to the zona pellucida. Several ASGs were classified as cilia by actin and microtubule cytoskeleton organization. Such genes may play a role during the development of post-testicular motility. Further studies are crucial to research the expression and transmission of alternative splicing over 2 or 3 generations.
ACKNOWLEDGEMENTS
This work was supported by the National Science Centre, Poland (2016/21/B/NZ9/03583).
DISCLOSURES
The authors have declared that they have no conflicts of interest.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2023.102484.
Appendix. Supplementary materials
Supplementary Table 1. Steps of RNA preparation for gene expression analysis.
Supplementary Table 2. Primer – probe sequences for TaqMan-based real-time PCR.
Supplementary Table 3. Primer sequences for real-time PCR.
Supplementary Table 4. Number of detected AS events and the main classes of AS events across the turkey reproductive tract.
Supplementary Table 5. The list of differentially alternative splicing (DAS) events between testis and epididymis discovered by SUPPA and rMATS tools and gene ontology analysis by ShinyGO v0.75.
Supplementary Table 6. The list of differentially alternative splicing (DAS) events between testis and ductus deferens discovered by SUPPA and rMATS tools and gene ontology analysis by ShinyGO v0.75.
Supplementary Table 7. The list of differentially alternative splicing (DAS) events between epididymis and ductus deferens discovered by SUPPA and rMATS tools and gene ontology analysis by ShinyGO v0.75.
Supplementary Figure 1. Selected differentially alternative splicing events described by changes in percentage splicing inclusion (PSI).
REFERENCES
- Adams S.M., Derks M.F.L., Makanjuola B.O., Marras G., Wood B.J., Baes C.F. Investigating inbreeding in the turkey (Meleagris gallopavo) genome. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahammad M.U., Nishino C., Tatemoto H., Okura N., Kawamoto Y., Okamoto S., Nakada T. Maturational changes in motility, acrosomal proteolytic activity, and penetrability of the inner perivitelline layer of fowl sperm, during their passage through the male genital tract. Theriogenology. 2011;76:1100–1109. doi: 10.1016/j.theriogenology.2011.05.017. [DOI] [PubMed] [Google Scholar]
- Ahammad M.U., Nishino C., Tatemoto H., Okura N., Kawamoto Y., Okamoto S., Nakada T. Maturational changes in the survivability and fertility of fowl sperm during their passage through the male reproductive tract. Anim. Reprod. Sci. 2011;128:129–136. doi: 10.1016/j.anireprosci.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Aire T.A., Josling D. Ultrastructural study of the luminal surface of the ducts of the epididymis of gallinaceous birds. Onderstepoort J. Vet. Res. 2000;67:191–199. [PubMed] [Google Scholar]
- Aire T.A., Ozegbe P.C. Immunohistochemistry of the cytoskeleton in the excurrent ducts of the testis in birds of the Galloanserae monophyly. Cell Tissue Res. 2008;333:311–321. doi: 10.1007/s00441-008-0631-7. [DOI] [PubMed] [Google Scholar]
- Alber A., Morris K.M., Bryson K.J., Sutton K.M., Monson M.S., Chintoan-Uta C., Borowska D., Lamont S.J., Schouler C., Kaiser P., Stevens M.P., Vervelde L. Avian pathogenic Escherichia coli (APEC) strain-dependent immunomodulation ofrespiratory granulocytes and mononuclear phagocytes in CSF1R- reporter transgenic chickens. Front. Immunol. 2020;10:3055. doi: 10.3389/fimmu.2019.03055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arangasamy A., Kasimanickam V.R., DeJarnette J.M., Kasimanickam R.K. Association of CRISP2, CCT8, PEBP1 mRNA abundance in sperm and sire conception rate in Holstein bulls. Theriogenology. 2011;76:570–577. doi: 10.1016/j.theriogenology.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Araujo P.R., Yoon K., Ko D., Smith A.D., Qiao M., Suresh U., Burns S.C., Penalva L.O. Before it gets started: regulating translation at the 5′ UTR. Comp. Funct. Genomics. 2012;2012 doi: 10.1155/2012/475731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano A., Tajima A. In: Pages 59–73 in Avian Reproduction, Advances in Experimental Medicine and Biology. Sasanami T., editor. Springer Nature Singapore Pte Ltd; Singapore: 2017. Development and preservation of avian sperm. [DOI] [PubMed] [Google Scholar]
- Asgari R., Bakhtiari M., Rezazadeh D., Yarani R., Esmaeili F., Mansouri K. TSGA10 as a potential key factor in the process of spermatid differentiation/maturation: deciphering its association with autophagy pathway. Reprod. Sci. 2021;28:3228–3240. doi: 10.1007/s43032-021-00648-6. [DOI] [PubMed] [Google Scholar]
- Barrett L.W., Fletcher S., Wilton S.D. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell. Mol. Life Sci. 2012;69:3613–3634. doi: 10.1007/s00018-012-0990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnam B., Modarressi M.H., Conti V., Taylor K.E., Puliti A., Wolfe J. Expression of Tsga10 sperm tail protein in embryogenesis and neural development: from cilium to cell division. Biochem. Biophys. Res. Commun. 2006;344:1102–1110. doi: 10.1016/j.bbrc.2006.03.240. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohmann H., Pinnecke S., Hoyer-Fender S. Identification and characterization of new cDNAs encoding outer dense fiber proteins of rat sperm. J. Biol. Chem. 1997;272:10327–10332. doi: 10.1074/jbc.272.15.10327. [DOI] [PubMed] [Google Scholar]
- Chan H.C., Ruan Y.C., He Q., Chen M.H., Chen H., Xu W.M., Chen W.Y., Xie C., Zhang X.H., Zhou Z. The cystic fibrosis transmembrane conductance regulator in reproductive health and disease. J. Physiol. 2009;587:2187–2195. doi: 10.1113/jphysiol.2008.164970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves L.D., Ostroski B.J., Reed K.M. Myosin light chain genes in the turkey (Meleagris gallopavo) Cytogenet. Genome. Res. 2003;102:340–346. doi: 10.1159/000075772. [DOI] [PubMed] [Google Scholar]
- Chávez J.C., Hernández-González E.O., Wertheimer E., Visconti P.E., Darszon A., Treviño C.L. Participation of the Cl-/HCO(3)- exchangers SLC26A3 and SLC26A6, the Cl- channel CFTR, and the regulatory factor SLC9A3R1 in mouse sperm capacitation. Biol. Reprod. 2012;86:1–14. doi: 10.1095/biolreprod.111.094037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.Y., Xu W.M., Chen Z.H., Ni Y., Yuan Y.Y., Zhou S.C., Zhou W.W., Tsang L.L., Chung Y.W., Hoglund P., Chan H.C., Shi Q.X. Cl-is required forHCO3 entry necessary for sperm capacitation in guinea pig: involvement of a Cl/HCO3 exchanger (SLC26A3) and CFTR. Biol. Reprod. 2009;80:115–123. doi: 10.1095/biolreprod.108.068528. [DOI] [PubMed] [Google Scholar]
- Clausen M.V., Hilbers F., Poulsen H. The structure and function of the Na, K-ATPase isoforms in health and disease. Front. Physiol. 2017;8:371. doi: 10.3389/fphys.2017.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clulow J., Jones R.C. Production, transport, maturation, storage and survival of spermatozoa in the male Japanese quail, Coturnix coturnix. J. Reprod. Fertil. 1982;64:259–266. doi: 10.1530/jrf.0.0640259. [DOI] [PubMed] [Google Scholar]
- de la Grange P., Gratadou L., Delord M., Dutertre M., Auboeuf D. Splicing factor and exon profiling across human tissues. Nucleic Acids Res. 2010;38:2825–2838. doi: 10.1093/nar/gkq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Xu R., Yu J., Xu T., Zhuang Y., Han M. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell. 2007;12:863–872. doi: 10.1016/j.devcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droval A.A., Binneck E., Marin S.R., Paião F.G., Oba A., Nepomuceno A.L., Shimokomaki M. A new single nucleotide polymorphism in the ryanodine gene of chicken skeletal muscle. Genet. Mol. Res. 2012;11:821–829. doi: 10.4238/2012.April.3.4. [DOI] [PubMed] [Google Scholar]
- Dvoráková K., Moore H.D., Sebková N., Palecek J. Cytoskeleton localization in the sperm head prior to fertilization. Reproduction. 2005;130:61–69. doi: 10.1530/rep.1.00549. [DOI] [PubMed] [Google Scholar]
- Edwalds-Gilbert G., Veraldi K.L., Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott D.J., Grellscheid S.N. Alternative RNA splicing regulation in the testis. Reproduction. 2006;132:811–819. doi: 10.1530/REP-06-0147. [DOI] [PubMed] [Google Scholar]
- Ever L., Steiner R., Shalom S., Don J. Two alternatively spliced Meig1 messenger RNA species are differentially expressed in the somatic and in the germ-cell compartments of the testis. Cell Growth Differ. 1999;10:19–26. [PubMed] [Google Scholar]
- Foulkes N.S., Mellström B., Benusiglio E., Sassone-Corsi P. Developmental switch of CREM function during spermatogenesis: from antagonist to activator. Nature. 1992;355:80–84. doi: 10.1038/355080a0. [DOI] [PubMed] [Google Scholar]
- Gallego-Paez L.M., Bordone M.C., Leote A.C., Saraiva-Agostinho N., Ascensão-Ferreira M., Barbosa-Morais N.L. Alternative splicing: the pledge, the turn, and the prestige: the key role of alternative splicing in human biological systems. Hum. Genet. 2017;136:1015–1042. doi: 10.1007/s00439-017-1790-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S.X., Jung D., Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göb E., Meyer-Natus E., Benavente R., Alsheimer M. Expression of individual mammalian Sun1 isoforms depends on the cell type. Commun. Integr. Biol. 2011;4:440–442. doi: 10.4161/cib.4.4.15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göb E., Schmitt J., Benavente R., Alsheimer M. Mammalian sperm head formation involves different polarization of two novel LINC complexes. PLoS One. 2010;5:e12072. doi: 10.1371/journal.pone.0012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q., Chen X., Du Y., Guo J., Su Y. Cyclic AMP-responsive element modulator α polymorphisms are potential genetic risks for systemic lupus erythematosus. J. Immunol. Res. 2015;2015 doi: 10.1155/2015/906086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han R., Wei Y., Kang X., Chen H., Sun G., Li G., Bai Y., Tian Y., Huang Y. Novel SNPs in the PRDM16 gene and their associations with performance traits in chickens. Mol. Biol. Rep. 2012;39:3153–3160. doi: 10.1007/s11033-011-1081-y. [DOI] [PubMed] [Google Scholar]
- Hoyer-Fender S., Petersen C., Brohmann H., Rhee K., Wolgemuth D.J. Mouse Odf2 cDNAs consist of evolutionarily conserved as well as highly variable sequences and encode outer dense fiber proteins of the sperm tail. Mol. Reprod. Dev. 1998;51:167–175. doi: 10.1002/(SICI)1098-2795(199810)51:2<167::AID-MRD6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Hu T., Wu Z., Bush S.J., Freem L., Vervelde L., Summers K.M., Hume D.A., Balic A., Kaiser P. Characterization of subpopulations of chicken mononuclear phagocytes that express TIM4 and CSF1R. J. Immunol. 2019;202:1186–1199. doi: 10.4049/jimmunol.1800504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Lin Y., Jin S., Liu W., Xu Y., Zheng Y. Alternative splicing of testis-specific lactate dehydrogenase C gene in mammals and pigeon. Anim. Biotechnol. 2012;23:114–123. doi: 10.1080/10495398.2011.652325. [DOI] [PubMed] [Google Scholar]
- Jamieson B.G.M. In: Reproductive Biology and Phylogeny of Birds. Jamiesson B.G.M., editor. Science Publishers; Enfield, NH: 2007. Avian spermatozoa: structure and phylogeny. Pages 349–511. Pages 349–511. [Google Scholar]
- Jimenez T., McDermott J.P., Sánchez G., Blanco G. Na,K-ATPase alpha4 isoform is essential for sperm fertility. Proc. Natl. Acad. Sci. U. S. A. 2011;108:644–649. doi: 10.1073/pnas.1016902108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M., Wells D., Rusch J., Ahmad S., Marchini J., Myers S.R., Conrad D.F. Unified single-cell analysis of testis gene regulation and pathology in five mouse strains. Elife. 2019;8:e43966. doi: 10.7554/eLife.43966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstens H.H., Crooijmans R.P., Veenendaal A., Dibbits B.W., Chin-A-Woeng T.F., den Dunnen J.T., Groenen M.A. Large scale single nucleotide polymorphism discovery in unsequenced genomes using second generation high throughput sequencing technology: applied to turkey. BMC Genomics. 2009;10:479. doi: 10.1186/1471-2164-10-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotłowska M., Glogowski J., Dietrich G.J., Kozłowski K., Faruga A., Jankowski J., Ciereszko A. Biochemical characteristics and sperm production of turkey semen in relation to strain and age of the males. Poult. Sci. 2005;84:1763–1768. doi: 10.1093/ps/84.11.1763. [DOI] [PubMed] [Google Scholar]
- Labas V., Grasseau I., Cahier K., Gargaros A., Harichaux G., Teixeira-Gomes A.P., Alves S., Bourin M., Gérard N., Blesbois E. Qualitative and quantitative peptidomic and proteomic approaches to phenotyping chicken semen. J. Proteomics. 2015;112:313–335. doi: 10.1016/j.jprot.2014.07.024. [DOI] [PubMed] [Google Scholar]
- Lalli E., Lee J.S., Lamas M., Tamai K., Zazopoulos E., Nantel F., Penna L., Foulkes N.S., Sassone-Corsi P. The nuclear response to cAMP: role of transcription factor CREM. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1996;351:201–209. doi: 10.1098/rstb.1996.0017. [DOI] [PubMed] [Google Scholar]
- Lang A.S., Austin S.H., Harris R.M., Calisi R.M., MacManes M.D. Stress-mediated convergence of splicing landscapes in male and female rock doves. BMC Genomics. 2020;21:251. doi: 10.1186/s12864-020-6600-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Park J.Y., Zheng D., Hoque M., Yehia G., Tian B. Alternative cleavage and polyadenylation in spermatogenesis connects chromatin regulation with post-transcriptional control. BMC Biol. 2016;14:6. doi: 10.1186/s12915-016-0229-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Yeh H.J., Shankarling G.S., Ji Z., Tian B., MacDonald C.C. The τCstF64 polyadenylation protein controls genome expression in testis. PLoS One. 2012;7:e48373. doi: 10.1371/journal.pone.0048373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Brockman J.M., Dass B., Hutchins L.N., Singh P., McCarrey J.R., MacDonald C.C., Graber J.H. Systematic variation in mRNA 3processing signals during mouse spermatogenesis. Nucleic Acids Res. 2007;35:234–246. doi: 10.1093/nar/gkl919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Xu C., Bao M., Huang J., Zou L., Fan X., Zhu C., Xia W. Cold-inducible RNA-binding protein regulates cyclin B1 against spermatogenesis arrest caused by heat stress. Andrology. 2022;10:392–403. doi: 10.1111/andr.13115. [DOI] [PubMed] [Google Scholar]
- Liu Q., Pante N., Misteli T., Elsagga M., Crisp M., Hodzic D., Burke B., Roux K.J. Functional association of Sun1 with nuclear pore complexes. J. Cell. Biol. 2007;178:785–798. doi: 10.1083/jcb.200704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Sun Y., Li Y., Bai H., Xu S., Xu H., Ni A., Yang N., Chen J. Identification and differential expression of microRNAs in the testis of chicken with high and low sperm motility. Theriogenology. 2018;122:94–101. doi: 10.1016/j.theriogenology.2018.09.010. [DOI] [PubMed] [Google Scholar]
- Liu Y., Sun Y., Li Y., Bai H., Xue F., Xu S., Xu H., Shi L., Yang N., Chen J. Analyses of long non-coding RNA and mRNA profiling using RNA sequencing in chicken testis with extreme sperm motility. Sci. Rep. 2017;7:9055. doi: 10.1038/s41598-017-08738-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G., Hou M., Wang B., Liu Z., Liu W., Han T., Zhang D., Zhou X., Jia W., Tan Y., Wu Y., Wang J., Zhang X. Tsga10 is essential for arrangement of mitochondrial sheath and male fertility in mice. Andrology. 2021;9:368–375. doi: 10.1111/andr.12889. [DOI] [PubMed] [Google Scholar]
- Lutz C.S. Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chem. Biol. 2008;3:609–617. doi: 10.1021/cb800138w. [DOI] [PubMed] [Google Scholar]
- Mariappa D., Aladakatti R.H., Dasari S.K., Sreekumar A., Wolkowicz M., van der Hoorn F., Seshagiri P.B. Inhibition of tyrosine phosphorylation of sperm flagellar proteins, outer dense fiber protein-2 and tektin-2, is associated with impaired motility during capacitation of hamster spermatozoa. Mol. Reprod. Dev. 2010;77:182–193. doi: 10.1002/mrd.21131. [DOI] [PubMed] [Google Scholar]
- Mazin P.V., Khaitovich P., Cardoso-Moreira M., Kaessmann H. Alternative splicing during mammalian organ development. Nat. Genet. 2021;53:925–934. doi: 10.1038/s41588-021-00851-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehmood A., Laiho A., Venäläinen M.S., McGlinchey A.J., Wang N., Elo L.L. Systematic evaluation of differential splicing tools for RNA-seq studies. Brief. Bioinform. 2020;21:2052–2065. doi: 10.1093/bib/bbz126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S., Chiorini J.A., Urcelay E., Safer B. Regulation of gene expression for translation initiation factor eIF-2 alpha: importance of the 3′ untranslated region. Biochem. J. 1996;315:791–798. doi: 10.1042/bj3150791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco L., Nantel F., Foulkes N.S., Sassone-Corsi P. In: Pages 69–94 in Signal Transduction in Testicular Cells. Hansson V., Levy F.O., Tasken K., editors. Springer Verlag; Berlin, Germany: 1996. CREM: a transcriptional master switch governing the cAMP response in the testis. [Google Scholar]
- Nishiyama H., Higashitsuji H., Yokoi H., Itoh K., Danno S., Matsuda T., Fujita J. Cloning and characterization of human CIRP (cold-inducible RNA-binding protein) cDNA and chromosomal assignment of the gene. Gene. 1997;204:115–120. doi: 10.1016/s0378-1119(97)00530-1. [DOI] [PubMed] [Google Scholar]
- Olcese C., Patel M.P., Shoemark A., Kiviluoto S., Legendre M., Williams H.J., Vaughan C.K., Hayward J., Goldenberg A., Emes R.D., Munye M.M., Dyer L., Cahill T., Bevillard J., Gehrig C., Guipponi M., Chantot S., Duquesnoy P., Thomas L., Jeanson L., Copin B., Tamalet A., Thauvin-Robinet C., Papon J.F., Garin A., Pin I., Vera G., Aurora P., Fassad M.R., Jenkins L., Boustred C., Cullup T., Dixon M., Onoufriadis A., Bush A., Chung E.M., Antonarakis S.E., Loebinger M.R., Wilson R., Armengot M., Escudier E., Hogg C., UK10K Rare Group. Amselem S., Sun Z., Bartoloni L., Blouin J.L., Mitchison H.M. X-linked primary ciliary dyskinesia due to mutations in the cytoplasmic axonemal dynein assembly factor PIH1D3. Nat. Commun. 2017;8:14279. doi: 10.1038/ncomms14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peddinti D., Nanduri B., Kaya A., Feugang J.M., Burgess S.C., Memili E. Comprehensive proteomic analysis of bovine spermatozoa of varying fertility rates and identification of biomarkers associated with fertility. BMC Syst. Biol. 2008;2:19. doi: 10.1186/1752-0509-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C., Füzesi L., Hoyer-Fender S. Outer dense fibre proteins from human sperm tail: molecular cloning and expression analyses of two cDNA transcripts encoding proteins of 70 kDa. Mol. Hum. Reprod. 1999;5:627–635. doi: 10.1093/molehr/5.7.627. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A., Huber W. Alternative start and termination sites of transcription drive most transcript isoform differences across human tissues. Nucleic Acids Res. 2018;46:582–592. doi: 10.1093/nar/gkx1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Palmer D.H., Wright A.E. Sex-specific selection drives the evolution of alternative splicing in birds. Mol. Biol. Evol. 2021;38:519–530. doi: 10.1093/molbev/msaa242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg Y., Eldar T., Karminsky O.D., Itach S.B., Pietrokovski S., Don J. Meig1 deficiency causes a severe defect in mouse spermatogenesis. Dev. Biol. 2010;338:158–167. doi: 10.1016/j.ydbio.2009.11.028. [DOI] [PubMed] [Google Scholar]
- Sha Y.W., Sha Y.K., Ji Z.Y., Mei L.B., Ding L., Zhang Q., Qiu P.P., Lin S.B., Wang X., Li P., Xu X., Li L. TSGA10 is a novel candidate gene associated with acephalic spermatozoa. Clin. Genet. 2018;93:776–783. doi: 10.1111/cge.13140. [DOI] [PubMed] [Google Scholar]
- Shao X., Tarnasky H.A., Schalles U., Oko R., van der Hoorn F.A. Interactional cloning of the 84-kDa major outer dense fiber protein Odf84. J. Biol. Chem. 1997;272:6105–6113. doi: 10.1074/jbc.272.10.6105. [DOI] [PubMed] [Google Scholar]
- Sharpe J.J., Cooper T.A. Unexpected consequences: exon skipping caused by CRISPR-generated mutations. Genome Biol. 2017;18:109. doi: 10.1186/s13059-017-1240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Słowińska M., Ciereszko A. Identification of the second form of acrosin in turkey spermatozoa. Reprod. Domest. Anim. 2012;47:849–855. doi: 10.1111/j.1439-0531.2011.01981.x. [DOI] [PubMed] [Google Scholar]
- Słowińska M., Hejmej A., Bukowska J., Liszewska E., Bilińska B., Hliwa P., Kozłowski K., Jankowski J., Ciereszko A. Expression and secretion of albumin in male turkey (Meleagris gallopavo) reproductive tract in relation to yellow semen syndrome. Poult. Sci. 2019;98:1872–1882. doi: 10.3382/ps/pey490. [DOI] [PubMed] [Google Scholar]
- Słowińska M., Liszewska E., Nynca J., Bukowska J., Hejmej A., Bilińska B., Szubstarski J., Kozłowski K., Jankowski J., Ciereszko A. Isolation and characterization of an ovoinhibitor, a multidomain Kazal-like inhibitor from turkey (Meleagris gallopavo) seminal plasma. Biol. Reprod. 2014;91:108. doi: 10.1095/biolreprod.114.118836. [DOI] [PubMed] [Google Scholar]
- Słowińska M., Nynca J., Arnold G.J., Fröhlich T., Jankowski J., Kozłowski K., Mostek A., Ciereszko A. Proteomic identification of turkey (Meleagris gallopavo) seminal plasma proteins. Poult. Sci. 2017;96:3422–3435. doi: 10.3382/ps/pex132. [DOI] [PubMed] [Google Scholar]
- Słowińska M., Pardyak L., Liszewska E., Judycka S., Bukowska J., Dietrich M.A., Paukszto Ł., Jastrzębski J., Kozłowski K., Kowalczyk A., Jankowski J., Bilińska B., Ciereszko A. Characterization and biological role of cysteine-rich venom protein belonging to CRISPs from turkey seminal plasma. Biol. Reprod. 2021;104:1302–1321. doi: 10.1093/biolre/ioab032. [DOI] [PubMed] [Google Scholar]
- Słowińska M., Paukszto Ł., Jastrzębski J.P., Bukowska J., Kozłowski K., Jankowski J., Ciereszko A. Transcriptome analysis of turkey (Meleagris gallopavo) reproductive tract revealed key pathways regulating spermatogenesis and post-testicular sperm maturation. Poult. Sci. 2020;99:6094–6118. doi: 10.1016/j.psj.2020.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Wang L., Chen D., Li F. The function of Pre-mRNA alternative splicing in mammal spermatogenesis. Int. J. Biol. Sci. 2020;16:38–48. doi: 10.7150/ijbs.34422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staicu F.D., Martínez-Soto J.C., Canovas S., Matás C. Nitric oxide-targeted protein phosphorylation during human sperm capacitation. Sci. Rep. 2021;11:20979. doi: 10.1038/s41598-021-00494-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Jin K., Zuo Q., Sun H., Song J., Zhang Y., Chen G., Li B. Characterization of alternative splicing (AS) events during chicken (Gallus gallus) male germ-line stem cell differentiation with single-sell RNA-seq. Animals (Basel) 2021;11:1469. doi: 10.3390/ani11051469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton K.M., Morris K.M., Borowska D., Sang H., Kaiser P., Balic A., Vervelde L. Characterization of conventional dendritic cells and macrophages in the spleen using the CSF1R-reporter transgenic chickens. Front. Immunol. 2021;12 [Google Scholar]
- Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L., Mering C. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada Y., Isono K., Shinga J., Turner J.M., Kitamura H., Ohara O., Watanabe G., Singh P.B., Kamijo T., Jenuwein T., Burgoyne P.S., Koseki H. Mammalian Polycomb Scmh1 mediates exclusion of Polycomb complexes from the XY body in the pachytene spermatocytes. Development. 2007;134:579–590. doi: 10.1242/dev.02747. [DOI] [PubMed] [Google Scholar]
- Takagaki Y., Seipelt R.L., Peterson M.L., Manley J.L. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- Tan G.H., Li J .Z., Zhang Y.Y., You M.F., Liao C.M., Zhang Y.G. Association of PRKCA expression and polymorphisms with layer duck eggshell quality. Br. Poult. Sci. 2021;62:8–16. doi: 10.1080/00071668.2020.1817329. [DOI] [PubMed] [Google Scholar]
- Tang L., Zeng W., Clark R.K., Dobrinski I. Characterization of the porcine testis-expressed gene 11 (Tex11) Spermatogenesis. 2011;1:147–151. doi: 10.4161/spmg.1.2.16680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnasky H., Cheng M., Ou Y., Thundathil J.C., Oko R., van der Hoorn F.A. Gene trap mutation of murine outer dense fiber protein-2 gene can result in sperm tail abnormalities in mice with high percentage chimaerism. BMC Dev. Biol. 2010;10:67. doi: 10.1186/1471-213X-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston R.J., Hess R.A. Ultrastructure of spermatozoa from domesticated birds: comparative study of turkey, chicken and guinea fowl. Scanning Microsc. 1987;1:1829–1838. [PubMed] [Google Scholar]
- Trincado J.L., Entizne J.C., Hysenaj G., Singh B., Skalic M., Elliott D.J., Eyras E. SUPPA2: fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 2018;19:40. doi: 10.1186/s13059-018-1417-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner K.J., Sharpe R.M., Gaughan J., Millar M.R., Foster P.M.D., Saunders P.T.K. Expression cloning of a rat testicular transcript abundant in germ cells which contains two leucine zipper motifs. Biol. Reprod. 1997;57:1223–1232. doi: 10.1095/biolreprod57.5.1223. [DOI] [PubMed] [Google Scholar]
- Vollrath B., Pudney J., Asa S., Leder P., Fitzgerald K. Isolation of a murine homologue of the Drosophila neuralized gene, a gene required for axonemal integrity in spermatozoa and terminal maturation of the mammary gland. Mol. Cell. Biol. 2001;21:7481–7494. doi: 10.1128/MCB.21.21.7481-7494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu Manh T.P., Marty H., Sibille P., Vern Y.Le, Kaspers B., Dalod M., Schwartz-Cornil I., Quéré P. Existence of conventional dendritic cells in Gallus gallus revealed by comparative gene expression profiling. J. Immunol. 2014;192:4510–4517. doi: 10.4049/jimmunol.1303405. [DOI] [PubMed] [Google Scholar]
- Wakayama T., Iseki S. Role of the spermatogenic-Sertoli cell interaction through cell adhesion molecule-1 (CADM1) in spermatogenesis. Anatz. Sci. Int. 2009;84:112–121. doi: 10.1007/s12565-009-0034-1. [DOI] [PubMed] [Google Scholar]
- Walker W.H., Habener J.F. Role of transcription factors CREB and CREM in cAMP-regulated transcription during spermatogenesis. Trends Endocrinol. Metab. 1996;7:133–138. doi: 10.1016/1043-2760(96)00035-5. [DOI] [PubMed] [Google Scholar]
- Walker W.H., Sanborn B.M., Habener J.F. An isoform of transcription factor CREM expressed during spermatogenesis lacks the phosphorylation domain and represses cAMP-induced transcription. Proc. Natl. Acad. Sci. U. S. A. 1994;91:12423–12427. doi: 10.1073/pnas.91.26.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li S.J., Li C., Yu G.H., Feng Y.P., Peng X.L., Gong Y.Z. Molecular cloning, expression and association study with reproductive traits of the duck LRP8 gene. Br. Poult. Sci. 2013;54:567–574. doi: 10.1080/00071668.2013.819488. [DOI] [PubMed] [Google Scholar]
- Wang L., Tian Y., Mei X., Han R., Li G., Kang X. SNPs in the adiponectin receptor 2 gene and their associations with chicken performance traits. Anim. Biotechnol. 2015;26:1–7. doi: 10.1080/10495398.2013.862254. [DOI] [PubMed] [Google Scholar]
- Wang S.H., Cheng C.Y., Tang P.C., Chen C.F., Chen H.H., Lee Y.P., Huang S.Y. Differential gene expressions in testes of L2 strain Taiwan country chicken in response to acute heat stress. Theriogenology. 2013;79 doi: 10.1016/j.theriogenology.2012.10.010. 374-82.e1-7. [DOI] [PubMed] [Google Scholar]
- Wang Y., Li J., Feng C., Zhao Y., Hu X., Li N. Transcriptome analysis of comb and testis from Rose-comb Silky chicken (R1/R1) and Beijing Fatty wild type chicken (r/r) Poult. Sci. 2017;96:1866–1873. doi: 10.3382/ps/pew447. [DOI] [PubMed] [Google Scholar]
- Weatheritt R.J., Sterne-Weiler T., Blencowe B.J. The ribosome-engaged landscape of alternative splicing. Nat. Struct. Mol. Biol. 2016;23:1117–1123. doi: 10.1038/nsmb.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Zhang G.X., Zhang T., Wang J.Y., Fan Q.C., Tang Y., Ding F.X., Zhang L. Myf5 and MyoG gene SNPs associated with Bian chicken growth trait. Genet. Mol. Res. 2016;5:15. doi: 10.4238/gmr.15037043. [DOI] [PubMed] [Google Scholar]
- Wen Q., Li N., Xiao X., Lui W.Y., Chu D.S., Wong C.K.C., Lian Q., Ge R., Lee W.M., Silvestrini B., Cheng C.Y. Actin nucleator Spire 1 is a regulator of ectoplasmic specialization in the testis. Cell Death Dis. 2018;9:208. doi: 10.1038/s41419-017-0201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z., Zheng X., Zheng H., Liu X., Yang Z., Wang X. Cold-inducible RNA-binding protein (CIRP) regulates target mRNA stabilization in the mouse testis. FEBS Lett. 2012;586:3299–3308. doi: 10.1016/j.febslet.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Xu X., Toselli P.A., Russell L.D., Seldin D.C. Globozoospermia in mice lacking the casein kinase II alpha' catalytic subunit. Nat. Genet. 1999;23:118–121. doi: 10.1038/12729. [DOI] [PubMed] [Google Scholar]
- Yang G., Cope L., He Z., Florea L. Jutils: a visualization toolkit for differential alternative splicing events. Bioinformatics. 2021;37:4272–4274. doi: 10.1093/bioinformatics/btab401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z., Chen Z., Lan X., Hara S., Sunkel B., Huang T.H., Elnitski L., Wang Q., Jin V.X. Computational analysis reveals a correlation of exon-skipping events with splicing, transcription and epigenetic factors. Nucleic Acids Res. 2014;42:2856–2869. doi: 10.1093/nar/gkt1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G., Holste D., Kreiman G., Burge C.B. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Fernald R.D. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J. Comput. Biol. 2005;12:1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Lu H., Yu H., Cheng H., Zhou R. Multiple alternative splicing in gonads of chicken DMRT1. Dev. Genes Evol. 2007;217:119–126. doi: 10.1007/s00427-006-0117-0. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Wu F., Zhang M., Xiong Z., Yin Q., Ru Y., Shi H., Li J., Mao S., Li Y., Cao X., Hu R., Liew C.W., Ding Q., Wang X., Zhang Y. EMC10 governs male fertility via maintaining sperm ion balance. J. Mol. Cell Biol. 2018;10:503–514. doi: 10.1093/jmcb/mjy024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Steps of RNA preparation for gene expression analysis.
Supplementary Table 2. Primer – probe sequences for TaqMan-based real-time PCR.
Supplementary Table 3. Primer sequences for real-time PCR.
Supplementary Table 4. Number of detected AS events and the main classes of AS events across the turkey reproductive tract.
Supplementary Table 5. The list of differentially alternative splicing (DAS) events between testis and epididymis discovered by SUPPA and rMATS tools and gene ontology analysis by ShinyGO v0.75.
Supplementary Table 6. The list of differentially alternative splicing (DAS) events between testis and ductus deferens discovered by SUPPA and rMATS tools and gene ontology analysis by ShinyGO v0.75.
Supplementary Table 7. The list of differentially alternative splicing (DAS) events between epididymis and ductus deferens discovered by SUPPA and rMATS tools and gene ontology analysis by ShinyGO v0.75.
Supplementary Figure 1. Selected differentially alternative splicing events described by changes in percentage splicing inclusion (PSI).