Abstract
In mycobacteria, the glucose-based disaccharide trehalose cycles between the cytoplasm, where it is a stress protectant and carbon source, and the cell envelope, where it is released as a by-product of outer mycomembrane glycan biosynthesis and turnover. Trehalose recycling via the LpqY-SugABC transporter promotes virulence, antibiotic recalcitrance, and efficient adaptation to nutrient deprivation. The source(s) of trehalose and the regulation of recycling under these and other stressors are unclear. A key technical gap in addressing these questions has been the inability to trace trehalose recycling in situ, directly from its site of liberation from the cell envelope. Here we describe a bifunctional chemical reporter that simultaneously marks mycomembrane biosynthesis and subsequent trehalose recycling with alkyne and azide groups. Using this probe, we discovered that the recycling efficiency for trehalose increases upon carbon starvation, concomitant with an increase in LpqY-SugABC expression. The ability of the bifunctional reporter to probe multiple, linked steps provides a more nuanced understanding of mycobacterial cell envelope metabolism and its plasticity under stress.
Mycobacterium is a genus that includes several notorious human and animal pathogens, among them Mycobacterium tuberculosis. These organisms have a distinctive cell envelope featuring a thick and hydrophobic outer membrane, called the mycomembrane, which not only protects against antibiotics and environmental stresses, but also harbors various immunoactive glycolipids1,2,3,4,5,6,7. Biosynthesis of the mycomembrane (Fig. 1A) is mediated by the non-mammalian disaccharide trehalose, which in the form of trehalose monomycolate (TMM) carries long-chain mycolic acids (MAs) from the cytoplasm to the periplasm. In the periplasm, TMM donates MAs to sugar acceptors, generating the major mycomembrane components trehalose dimycolate (TDM) and arabinogalactan-linked mycolate (AGM)8. These reactions, which are catalyzed by antigen 85 (Ag85) mycoloyltransferases, also produce free trehalose, which is recycled by the transporter LpqY-SugABC and then used to regenerate TMM9, 10. Trehalose is also liberated indirectly, by TDM hydrolase (Tdmh6, 10–12). Recycled trehalose can be used in central carbon metabolism, as a stress protectant, or directly, in cell surface glycoconjugates such as TMM13,14,15,16. The recycling of extracytoplasmic trehalose promotes survival of mycobacteria in macrophages and mice and in the presence of some antibiotics9, 10, 17 and promotes resilience to stress under nutrient-limited conditions10, 16. Since trehalose is released as a by-product of outer mycomembrane glycan biosynthesis or turnover, and enhanced turnover occurs under different stress conditions6, 12, 14, 18, the source of extracytoplasmic trehalose has been assumed to be turnover14. We recently demonstrated that mycomembrane remodeling in carbon-limited Mycobacterium smegmatis and M. tuberculosis comprises synthesis in addition to turnover11, a surprising result given the resource and energy requirements of catabolism. Whether mycomembrane biosynthesis can serve as a source of trehalose for recycling is an open question.
Figure 1.
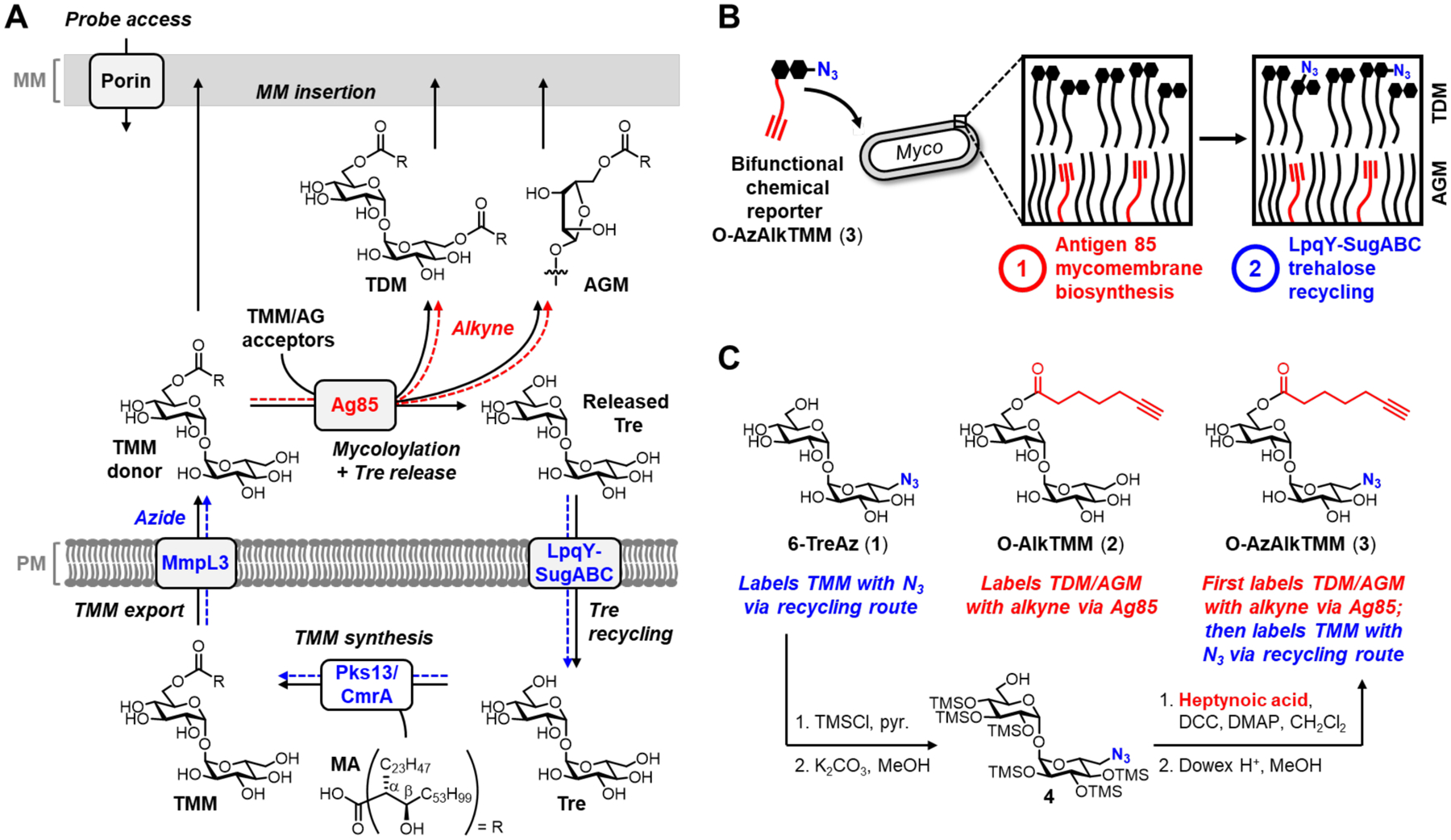
(A) Mycomembrane metabolism. Routes of probe incorporation are shown in blue and red. (B) Schematic showing simplified proposed labeling outcome when using the new bifunctional reporter O-AzAlkTMM (3). (C) Chemical reporters used in this study and synthesis of O-AzAlkTMM (3). (1, (6-TreAz)) and (2, (O-AlkTMM)) were reported previously in19 and20, respectively. Colors correlate reporter group to proposed incorporation route in (A) and major glycan target in (B). Tre, trehalose; TMM, trehalose monomycolate; MA, mycolic acid; TDM, trehalose dimycolate; AGM, arabinogalactan mycolates.
Chemical strategies for modifying the bacterial cell surface have a range of applications, including detecting and profiling bacterial cell envelope components, modulating the immunogenicity of the bacterium, and delivering diagnostic or therapeutic chemical cargo specifically to the bacterium21,22,23,24. Many of these strategies involve metabolic labeling of an envelope component (e.g., glycan, lipid, or protein) with a bioorthogonal functional group, followed by chemoselective reaction to functionally modify the cell-surface component. Chemical reporters based on biosynthetic precursors for peptidoglycan, lipopolysaccharide, teichoic acids, bacterial glycoproteins, and other components have been valuable tools for studying the cell envelopes of Gram-negative and Gram-positive bacteria25, 26.
We and others have exploited trehalose-mediated mycomembrane construction to create trehalose-based reporters for modifying the cell surface of mycobacteria (Fig. 1C)19, 26–28. Following the publication of a fluorescent trehalose analogue that labels mycobacteria via Ag8528, azide-modified trehalose (TreAz) analogues such as 6-TreAz (1) were reported to incorporate into the mycomembrane by LpqY-SugABC-mediated uptake and subsequent elaboration to azide-modified trehalose glycolipids, which can then be functionalized via click chemistry19. More recently, we developed TMM-based reporters, including O-AlkTMM (Fig. 1C, (2)) and related structures, which deliver bioorthogonal groups to MA acceptors—predominantly the abundant glycolipid AGM, and to a lesser extent trehalose glycolipids and (in some organisms) O-mycoloylated proteins20,29,30,31. These and other reporters11, 32, 33 are specific for isolated steps of the mycomembrane assembly cycle, e.g. Ag85 activity, Tdmh activity, and recycling/TMM synthesis activity.
Here, we sought to develop a mycomembrane probe that could report multiple, linked steps of mycobacterial mycomembrane metabolism. We designed a bifunctional chemical reporter 6-azido-6-deoxy-6’-O-(6’-heptynoyl)-α,α-D-trehalose (Fig. 1C, O-AzAlkTMM (3)) that exploits multiple steps in mycomembrane construction to deliver orthogonal alkyne and azide functionalities to the cell surface. We hypothesized that the TMM-mimicking bifunctional chemical reporter O-AzAlkTMM (3) would first undergo Ag85-mediated transfer of its heptynoyl chain to MA acceptors, in the process releasing 6-TreAz (1), which would then undergo inside-out incorporation into new TMM via the LpqY-SugABC recycling pathway (Figs. 1A–B). We anticipated that in situ delivery of biosynthetically-derived trehalose, in turn, would enable us to monitor stress-induced recycling in a more precise, physiologically-relevant manner.
The target molecule O-AzAlkTMM (3) was chemically synthesized from 6-TreAz (Fig. 1C). To expose the 6-OH group of 6-TreAz for later acylation, it was subjected to per-O-trimethylsilylation followed by regioselective 6-O-monodesilylation using potassium carbonate in methanol (50% over two steps). Next, alcohol 4 was acylated with heptynoic acid in the presence of DCC and DMAP (81%). Global desilylation with H+ resin yielded 3 (99%), whose structure and purity were established by NMR and MS. 1H NMR showed downfield-shifted absorptions for the 6’-position hydrogen nuclei, confirming that esterification occurred at the desired site (Supporting Information).
We carried out initial metabolic labeling experiments in M. smegmatis, a fast-growing, non-pathogenic organism that is commonly used to model aspects of the mycobacterial cell envelope. First, we tested whether O-AzAlkTMM (3) could efficiently deliver both alkynes and azides to the cell surface. M. smegmatis was cultured in 50 μM 6-TreAz (1), O-AlkTMM (2), O-AzAlkTMM (3), or left untreated. Cells were washed, fixed, and subjected to 1) strain-promoted alkyne-azide cycloaddition (SPAAC) with a cyclooctyne-conjugated fluorophore, to detect incorporated azides, followed by 2) copper-catalyzed alkyne-azide cycloaddition (CuAAC) with an azide-conjugated fluorophore, to detect incorporated alkynes. Flow cytometry analysis of the labeled cells showed that 6-TreAz and O-AlkTMM incorporation were only detected with cyclooctyne and azide fluorophores, respectively (Fig. 2A). By contrast, incorporation of bifunctional reporter O-AzAlkTMM could be detected by both fluorophores, confirming its ability to dual-label the cell surface. Labeling by O-AzAlkTMM was ~35–40% of either 6-TreAz or O-AlkTMM alone (Fig. 2A). These data imply that (i) the azide moiety on O-AzAlkTMM decreases O-AzAlkTMM uptake relative to O-AlkTMM and/or O-AlkTMM incorporation by Ag85 but (ii) once 6-TreAz is released by Ag85 from O-AzAlkTMM, it is efficiently internalized and incorporated onto the cell surface.
Figure 2.
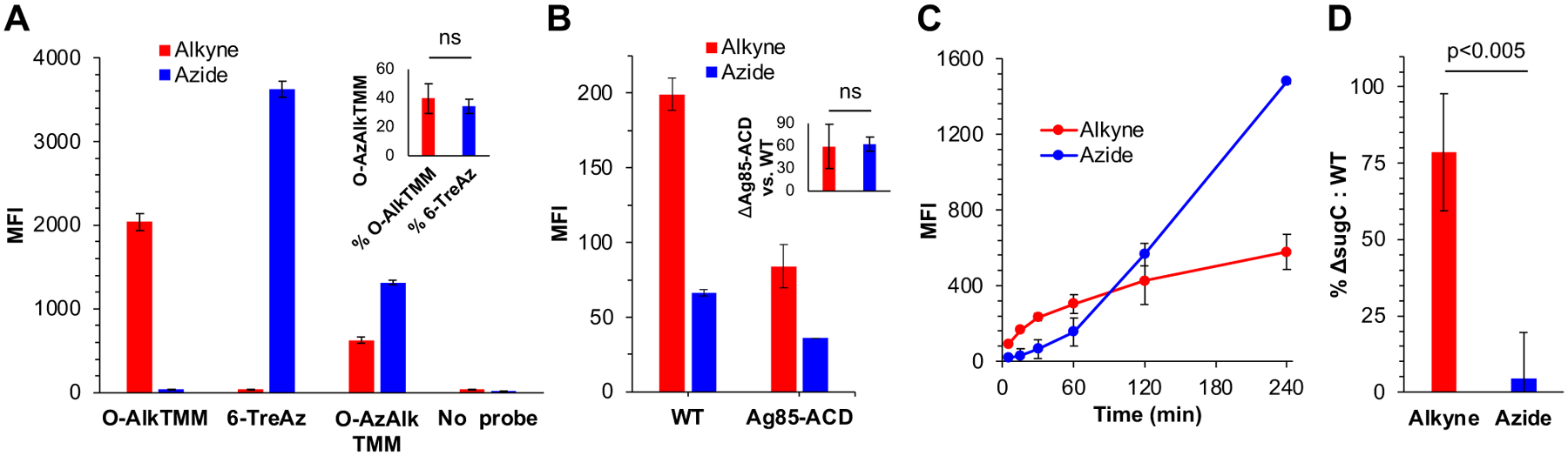
Two-step incorporation of bifunctional reporter O-AzAlkTMM into the M. smegmatis mycomembrane. (A) Labeling of wild-type (WT) M. smegmatis with 50 μM O-AlkTMM, 6-TreAz, or O-AzAlkTMM for 15 min or ~10% generation time. Fluorescence from sequential SPAAC (with DBCO Cy5, to detect azides) and CuAAC (with Carboxyrhodamine 110 Azide, to detect alkynes) was quantitated by flow cytometry. MFI, median fluorescence intensity. Representative data from five independent experiments performed in triplicate are shown. Inset, O-AzAlkTMM labeling as a % of O-AlkTMM or 6-TreAz over the five experiments. (B) Loss of Ag85ACD decreases both alkyne and azide signal from O-AzAlkTMM. WT and Δag85ACD M. smegmatis were labeled with O-AzAlkTMM as in (A), except that alkynes and azides were respectively and individually revealed by CuAAC reaction with complementary azido- or alkynyl-Carboxyrhodamine 110. Representative data from four independent experiments performed in triplicate are shown. Inset, Δag85ACD labeling as a % of wild-type (WT) over the four experiments. (C) Time-dependence of alkyne- and azide-derived labeling from O-AzAlkTMM. Wild-type M. smegmatis was labeled with O-AzAlkTMM as in Fig. 2A and aliquots were taken at the indicated time points for CuAAC as in Fig. 2B. Data from three independent experiments plotted. (D) Azide but not alkyne signal from O-AzAlkTMM is dependent on the presence of SugC. WT and ΔsugC M. smegmatis were labeled as in Fig. 2A and CuAAC performed as in Fig. 2B. Data (from three independent experiments performed in triplicate) for the mutant were normalized to WT and expressed as fold-change. Statistical significance assessed by two-tailed Student’s t test. ns, not significant (p>0.05). Error bars, standard deviation of technical or biological replicates as described above.
To test this presumption in more detail, we investigated the pathways by which O-AzAlkTMM incorporated into the cell surface. For these experiments (Figs. 2B–D) we subjected half of the sample to CuAAC with an alkyne-conjugated fluorophore, to detect incorporated azides, and the other half to CuAAC with an azide-conjugated fluorophore, to detect incorporated alkynes. Pre-incubation of M. smegmatis with ebselen, a cysteine-reactive inhibitor of Ag85 activity27, 34, decreased alkyne-derived signal from O-AzAlkTMM as expected (Fig. S1, Supporting Information). However, the azide-derived signal remained unaffected. Although we do not yet understand why 6-TreAz signal does not decline in the presence of ebselen, we hypothesized that there may be compensatory changes in the presence of cysteine-reactive ebselen that could enhance 6-TreAz recovery. Therefore, we compared O-AzAlkTMM labeling in wild-type and ΔAg85-ACD (ΔMSMEG_6396–6399) M. smegmatis, which lacks three out of five Ag85 enzymes32. Both azide- and alkyne-dependent labeling decreased in the mutant strain (Fig. 2B). We next monitored O-AzAlkTMM labeling kinetics in wild-type M. smegmatis and found that azide-derived signal lagged alkyne-derived signal at early time points (Fig. 2C), as expected if alkynyl fatty acid incorporation temporally precedes azido trehalose assimilation. These experiments, coupled with extensive literature data characterizing similar TMM-based reporters20,30,29,31,33,35, support the idea that transfer of the alkyne-terminated acyl chain of O-AzAlkTMM to acceptor molecules, such as arabinogalactan, is catalyzed by Ag85, which in turn liberates 6-TreAz for subsequent incorporation into the mycobacterial cell surface.
Exogenously-added 6-TreAz is recycled by the LpqY-SugABC transporter prior to M. smegmatis cell surface incorporation19. Therefore, we predicted that labeling by the 6-TreAz released upon the alkyne-terminated acyl chain transfer of O-AzAlkTMM would similarly depend on LpqY-SugABC. We compared O-AzAlkTMM labeling in wild-type and ΔsugC M. smegmatis, which lacks a functional trehalose transporter19. Only the azide-dependent labeling decreased in the mutant strain (Fig. 2D). While these data do not completely rule out alternative routes, they further support the hypothesis that O-AzAlkTMM undergoes Ag85-mediated alkyne incorporation on the cell surface, thus unmasking 6-TreAz, which then traverses the LpqY-SugABC → Pks13/CmrA → MmpL3 pathway to incorporate into surface TMM from the inside-out27.
After confirming that bifunctional reporter O-AzAlkTMM efficiently delivers alkynes and azides to the mycomembrane through the expected biosynthetic pathways, we next sought to demonstrate its utility in simultaneous visualization of mycomembrane biosynthesis, marked by its alkyne moiety, and biosynthesis-dependent trehalose recycling, marked by its azide moiety. To simultaneous detect alkyne- and azide-dependent labelling in single cells, we subjected O-AzAlkTMM-labelled M. tuberculosis (ΔleuD ΔpanCD auxotroph derivative of strain H37Rv that can be manipulated under BSL2 conditions36) to successive CuAAC with azido- and alkynyl-fluorophores (Fig. 3A), then imaged (Figs. 3B, S2) and analyzed by flow cytometry (Fig. 3C). Azide-derived fluorescence from either 6-TreAz or O-AzAlkTMM labelling was only apparent in one-quarter of M. tuberculosis cells (Figs. 3B–C, S2), suggesting that detection and/or incorporation is heterogenous in this organism. By contrast, we were able to detect alkyne-derived fluorescence in 80–90% of M. tuberculosis cells labelled with O-AlkTMM or O-AzAlkTMM (Figs. 3B–C, S2). In M. tuberculosis in which both azide- and alkyne-derived fluorescence was detectable, labelling was not spatially coincident. These data suggest that the subcellular sites of Ag85-dependent mycomembrane biosynthesis and trehalose release (marked with alkynes) are distinct from the sites of trehalose recycling-dependent TMM biosynthesis (marked with azides) and highlight the ability of O-AzAlkTMM to illuminate multiple, linked steps in mycomembrane metabolism in single M. tuberculosis cells.
Figure 3.
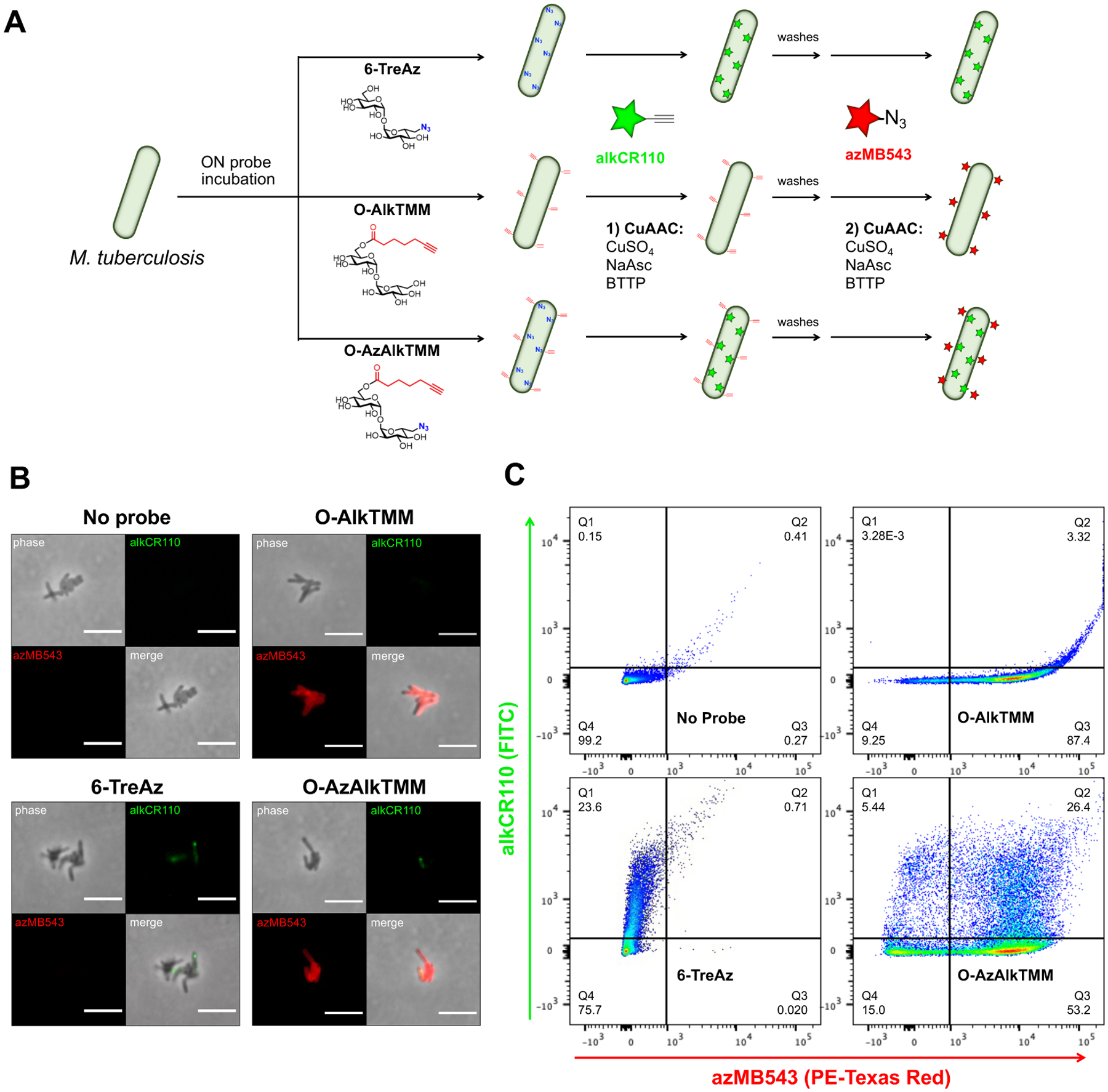
ΔleuD ΔpanCD M. tuberculosis was labeled +/− 6-TreAz, O-AlkTMM, or O-AzAlkTMM for 16–24 hrs or ~1 generation time, fixed, then subjected to successive rounds of CuAAC, the first with Carboxyrhodamine 110 alkyne (alkCR110), and the second with MB 543 Picolyl Azide (azMB543, see Methods for details). Schematic of labeling, (A); representative images (see Fig. S2 for additional images), (B); flow cytometry analysis, (C).
We recently demonstrated that non-replicating, carbon-starved M. smegmatis and M. tuberculosis continue to synthesize AGM10. We hypothesized that this final, Ag85-mediated step of mycomembrane catabolism serves as a source of trehalose under these conditions, likely in addition to TDM turnover that we and others have shown occurs in response to various stressors6, 10, 12, 14, 16, 18. To test this hypothesis, we cultured M. smegmatis in high or low carbon media and briefly labeled with O-AzAlkTMM. The proportion of azide-dependent signal (6-TreAz/alkyne-fluorophore-marked trehalose recycling) compared to alkyne-dependent signal (O-AlkTMM/azide-fluorophore-marked AGM biosynthesis) was elevated under carbon-limited conditions (Figs. 4A). This experiment suggested that a greater proportion of the trehalose liberated from Ag85 activity was recycled upon adaptation to carbon limitation. We next investigated the kinetics of adaptation by transferring M. smegmatis from high glucose medium to medium lacking glucose and monitoring azide- and alkyne-derived fluorescence over time. We used M. smegmatis pre-adapted to low glucose as a control. Compared to that of pre-adapted M. smegmatis, the azide:alkyne ratio of M. smegmatis that had been cultured in high glucose prior to transfer increased in the first hour after transfer and plateaued thereafter (Fig. 4B). These data suggest that the efficiency of trehalose capture from AGM biosynthesis increases as M. smegmatis adapts to carbon-limited conditions.
Figure 4.
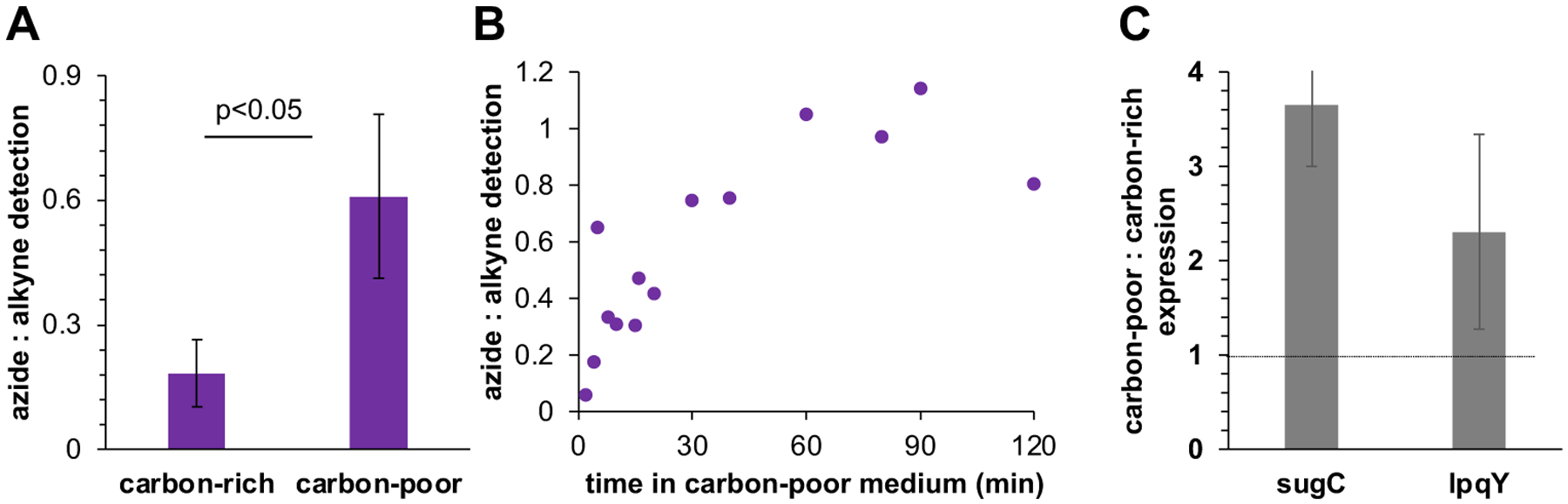
Increased efficiency of trehalose capture from mycomembrane synthesis under glucose limitation. (A) M. smegmatis cultured in 0.02% (carbon-poor) or 2% (carbon-rich) glucose as in10 was labeled with O-AzAlkTMM as in Fig. 2A except that the bacteria were transferred to no-glucose medium for labeling10 (see Methods). Data for the azide-derived fluorescence were normalized to alkyne-derived fluorescence and expressed as fold-change from four independent experiments performed in duplicate or triplicate. (B) Rapid increase in trehalose recycling upon adaptation to carbon limitation. M. smegmatis cultured in low or high glucose as in Fig. 4A was washed, transferred to no-glucose medium, and labeled with O-AzAlkTMM at indicated time points. Data for the azide-derived fluorescence were normalized to alkyne-derived fluorescence, and data for trehalose capture from carbon-rich M. smegmatis were further normalized to M. smegmatis that had been pre-adapted, i.e., cultured in carbon-poor medium prior to transfer. Data combined from five independent experiments. Each time point is from 1, 2, or 3 of the independent experiments. (C) The expression of trehalose transporter genes sugC and lpqY is higher in carbon-poor conditions. Expression monitored by qRT-PCR for M. smegmatis cultured in low or high glucose as in Fig. 4A. Expression data for sugC, lpqY and control gene sigA from carbon-limited M. smegmatis were normalized to expression data for M. smegmatis cultured in high glucose. sugC and lpqY ratios were further normalized to sigA ratio. Error bars, standard deviations for five independent experiments.
Increased recycling could reflect changes in the efficiency or abundance of the LpqY-SugABC transporter. While we have not ruled out the former, the timing of adaptation suggested a role for enhanced transcription/translation of the transporter genes17,37. Accordingly, we monitored the mRNA levels of trehalose transporter genes sugC and lpqY by qRT-PCR. Consistent with our hypothesis, we found that the relative expression of trehalose transporter increases under carbon limitation (Fig. 4C). As expression of bacterial carbohydrate transporters can be induced by their substrates38,39, enhanced expression of LpqY-SugABC is also consistent with mycomembrane turnover, which we previously showed occurs in response to carbon limitation and liberates trehalose10. Taken together, these data support a model in which mycobacteria make mycomembrane biosynthesis more efficient under carbon limitation by increasing the expression of LpqY-SugABC and extracting a greater proportion of the trehalose by-product.
M. tuberculosis is a pathogen that survives for decades in the hostile, carbohydrate-poor environment of its human host. Illuminating the mechanisms by which mycobacteria survive under stressful conditions has the potential to identify new targets for treatment. Trehalose recycling is known to support mycobacterial survival under stress9, 10, 17, but the source of extracellular trehalose in what are presumably slow or non-growing organisms has not been directly characterized, e.g. Ag85-mediated biosynthesis or Tdmh-dependent turnover of the mycomembrane. While metabolic labeling can report on trehalose recycling19, direct delivery of trehalose probes may or may not recapitulate native flux through the pathway. As well, such strategies do not provide information on the provenance of the free trehalose. Here we addressed this technical gap by synthesizing O-AzAlkTMM, a bifunctional probe that reports trehalose recycling directly from one of its potential sources, as a by-product of mycomembrane biosynthesis. To our knowledge, this is the first example of a probe that marks multiple metabolic processes with distinct functional groups, i.e., alkyne for cell envelope biosynthesis and azide for cell envelope recycling. Using O-AzAlkTMM, we showed that recycling of biosynthesis-derived trehalose increases under carbon starvation and is likely enabled by increased expression of the transporter. The biosynthetic origin of the recycled trehalose is surprising as de novo expansion of the bacterial surface is usually equated with cell replication. While we do not yet know why carbon-limited M. smegmatis and M. tuberculosis continue to synthesize mycomembrane, we previously showed that synthesis is part of an overall program of remodeling that also includes turnover and correlates with decreased permeability10. Enhanced capture of remodeling by-products enables mycobacteria to “waste not” under unfavorable conditions.
Methods
Chemical synthesis.
Materials and reagents were obtained from commercial sources without further purification unless otherwise noted. Anhydrous solvents were obtained either commercially or from an alumina column solvent purification system. All reactions were carried out in oven-dried glassware under inert gas unless otherwise noted. Analytical TLC was performed on glass-backed silica gel 60 Å plates (thickness 250 μm) and detected by charring with 5% H2SO4 in EtOH for detection of sugar. Column chromatography was performed using flash-grade silica gel 32–63 μm (230–400 mesh). 1H NMR and 13C NMR spectra were recorded at 500 MHz with chemical shifts in ppm (δ) referenced to TMS or solvent peaks. NMR spectra were obtained on a Varian Inova 500 instrument. Coupling constants (J) are reported in hertz (Hz). Electrospray ionization (ESI) mass spectra were obtained using a Waters LCT Premier XE.
6’-azido-6’-deoxy-2,3,4,2’,3’,4’-hexakis-O-(trimethylsilyl)-α,α-D-trehalose (4).
To a stirring solution of 6’-azido-6’-deoxy-α,α-D-trehalose19 (576 mg, 1.57 mmol) in anhydrous CH2Cl2 (60 mL) was added Et3N (30 mL). The solution was cooled to 0 °C and trimethylsilyl chloride (5.0 mL, 39.4 mmol) was added dropwise. After stirring overnight, TLC (hexanes/ethyl acetate 10:1 with 1% Et3N) indicated the generation of a major product (Rf = 0.94). The reaction mixture was concentrated by rotary evaporation, re-suspended in CH2Cl2, and washed once with water in a separatory funnel. The organic layer was dried over anhydrous Na2SO4, filtered, and concentrated by rotary evaporation to give the per-trimethylsilylated intermediate (1.22 g), which was directly taken forward to the next step. To a solution of the intermediate in CH3OH and CH2Cl2 (7 mL, 3:1) stirring at 0 °C was added K2CO3 (0.010 g, 0.072 mmol). The reaction was left stirring at 0 °C and monitored by TLC (hexanes/ethyl acetate 10:1 with 1% Et3N). After 5 hours, TLC showed conversion to a major product (Rf = 0.44) and the reaction mixture was filtered, concentrated by rotary evaporation, and subjected to silica gel column chromatography (hexanes/ethyl acetate 15:1 → 10:1 containing 1% Et3N) to give 4 (0.63 g, 50% over two steps) as a syrup. 1H NMR (500 MHz, CDCl3): 4.93 (d, J = 3.0 Hz, 1 H, H-1 or H-1’), 4.92 (d, J = 3.5 Hz, 1 H, H-1 or H-1’), 3.99−3.94 (m, 1 H, H-5’), 3.91−3.83 (m, 3 H, H-3, H-3’, H-5), 3.74−3.63 (m, 2 H, H-6a and H-6b), 3.52−3.33 (m, 6 H, H-4, H-4’, H-2, H-2’, H-6a’, H-6b’), 1.75 (dd, J = 5.4, 7.4 Hz, 1 H, 6-OH), 0.23−0.08 (m, 54 H, TMS CH3s). 13C24 NMR (125 MHz, CDCl3): 94.9, 94.6, 73.40, 74.38, 73.1, 72.9, 72.8, 72.6, 72.5, 71.5, 61.7, 51.7, 1.2, 1.13, 1.09, 1.0, 0.23, 0.22. MS (ESI-TOF) m/z: [M+Na]+ Calcd for C30H69N3O10Si6Na 822.3496; Found 822.3484.
6-azido-6-deoxy-6’-O-(6’-heptynoyl)-2,3,4,2’,3’,4’-hexakis-O-(trimethylsilyl)-α,α-D-trehalose (5).
An oven-dried round bottom flask was charged with DCC (0.120 g, 0.582 mmol) and DMAP (0.018 g, 0.15 mmol). A suspension of the reagents was made by the addition of anhydrous CH2Cl2 (1.5 mL), followed by the dropwise addition of 6-heptynoic acid (55 μL, 0.44 mmol). The mixture was then cooled to 0 °C. To the stirring solution was added a freshly prepared solution of compound 4 (0.233 g, 0.291 mmol) in CH2Cl2 (2 mL). The reaction mixture was stirred and gradually allowed to warm to room temperature. After stirring overnight, TLC (hexanes/ethyl acetate 4:1) showed the generation of the presumed ester product (Rf = 0.5). The reaction mixture was filtered and the crude product was concentrated by rotary evaporation and purified by silica gel column chromatography (hexanes/ethyl acetate 10:1 → 8:1 containing 1% Et3N) to give the pure monoester intermediate 5 (0.215 g, 81%) as a syrup. 1H NMR (500 MHz, CDCl3): 4.95 (d, J = 1.5 Hz, 1 H, H-1 or H-1’), 4.94 (d, J = 3 Hz, 1 H, H-1 or H-1’), 4.31 (dd, J = 2.0, 12 Hz, 1 H, H-6a’ or H-6b’), 4.08 (dd, J = 4.5, 12.5 Hz, 1 H, H-6a’ or H-6b’), 4.02 (ddd, J = 2.5, 3.5, 9.0 Hz, 1 H, H-5’), 3.96 (ddd, J = 3.0, 4.5, 9.5 Hz, 1 H, H-5), 3.91 (t, J = 9.0 Hz, 1 H, H-3’), 3.87 (t, J = 9.5 Hz, 1 H, H-3), 3.49 (t, J = 9.5 Hz, 1 H, H-4’), 3.48−3.39 (m, 4 H, H-2, H-2’, H-4, and H-6a or H-6b), 3.35 (dd, J = 5.0, 12.5 Hz, 1 H, H-6a or H-6b), 2.42−2.33 (m, 2 H, α-CH2), 2.21 (dt, J = 2.5, 7, 14.5 Hz, 2 H, propargylic CH2), 1.96 (t, J = 2.5 Hz, 1 H, terminal alkyne H), 1.79−73 (m, 2 H, β-CH2), 1.60−1.57 (m, γ-CH2), 0.16−0.14 (m, 54, TMS CH3s). 13C19, 24 NMR (125 MHz, CDCl3): 173.4, 94.7, 94.6, 84.0, 73.5, 73.4, 72.8, 72.7, 72.52, 72.50, 72.0, 70.9, 68.8, 63.5, 51.6, 33.7, 27.9, 24.0, 18.3, 1.17, 1.16, 1.1, 1.0, 0.30, 0.26. MS (ESI-TOF) m/z: [M+Na]+ Calcd for C30H69N3O10Si6Na 930.4071; Found 930.3744.
6-azido-6-deoxy-6’-O-(6’-heptynoyl)-α,α-D-trehalose (O-AzAlkTMM, 3).
The intermediate 5 (0.194 g, 0.214 mmol) was suspended in anhydrous CH3OH (60 mL) and placed under a nitrogen atmosphere. Dowex 50WX8-400 H+ ion exchange resin (1 g) was added and the reaction was left stirring at room temperature. After 30 min, TLC showed that the reaction was yet to be completed so an extra 1 g of Dowex 50WX8-400 H+ was added to the stirring reaction. After another 30 min, TLC (CH2Cl2/CH3OH 6:1) indicated that the reaction was complete (Rf = 0.44). After the ion-exchange resin was filtered off, the filtrate was concentrated by rotary evaporation to give the O-AzAlkTMM (3) (.101 g, quantitative yield) as an off-white solid. 1H NMR (500 MHz, D2O): 5.15 (d, J = 4.0 Hz, 1 H, H-1 or H-1’), 5.14 (d, J = 4.0 Hz, 1 H, H-1 or H-1), 4.42 (dd, J = 2.0, 12 Hz, 1 H, H-6a’ or H-6b’), 4.27 (dd, J = 5.5, 12.5 Hz, 1 H, H-6a’ or H-6b’), 4.00 (ddd, J = 2.0, 5.5, 10 Hz, 1 H, H-5’) 3.93 (ddd, J = 2.0, 5.5, 10 Hz, 1 H, H-5), 3.82 (t, J = 9.5 Hz, 1 H, H-3’), 3.80 (t, J = 9.5 Hz, 1 H, H-3), 3.66−3.62 (m, 3 H, H-2, H-2’, H-6a or H-6b), 3.52 (dd, J = 5.5, 13.5 Hz, 1 H, H-6a or H-6b), 3.47 (t, J = 9.5 Hz, 1 H, H-4’), 3.42 (t, J = 9.0 Hz, 1 H, H-4), 2.44 (t, J = 7.5 Hz, 2 H, α-CH2), 2.34 (t, J = 2.5 Hz, 1 H, terminal alkyne H), 2.22 (dt, J = 2.5, 7.5 Hz, 2 H, propargylic CH2), 1.71 (pent, J = 8.0 Hz, 2 H, β-CH2), 1.53 (pent, J = 7.5 Hz, 2 H, γ-CH2). 13C NMR (125 MHz, D2O, referenced to internal CH3OH standard at 49.50 ppm): 176.9, 94.24, 94.21, 86.3, 73.04, 73.02, 71.8, 71.6, 71.5, 71.1, 70.6, 70.4, 70.0, 63.7, 51.5, 33.9, 27.7, 24.2, 17.9. MS (ESI-TOF) m/z: [M+Na]+ Calcd for C19H29N3O11Na 498.1699; Found 498.1722.
Bacterial strains and culture conditions.
M. smegmatis mc2155 was grown shaking in Middlebrook 7H9 growth medium (BD Difco, Franklin Lakes, NJ) supplemented either with glucose (2% or 0.02%) or albumin-dextrose-catalase (ADC) (10%) and Tween-80 at 37 °C (7H9T). The experiment with M. smegmatis Δag85ACD strain40 (provided by Dr. Rainer Kalscheuer, University of Düsseldorf) was performed in Sauton’s medium (Fig. 2B). For carbon starvation experiments, two-day-old cultures of M. smegmatis grown in 7H9T-ADC were normalized to an OD600 of 0.1 in fresh 7H9T supplemented with 2% or 0.02% glucose and allowed to grow for 24 h. In all other experiments including endpoint labeling, ebselen treatment, and kinetic studies, two-day-old cultures were normalized to an OD of 0.1 and allowed to grow overnight. For Fig. 3, M. tuberculosis (ΔleuD ΔpanCD auxotroph derivative of H37Rv that can be manipulated under BSL2 conditions36 and was provided by Dr. Yasu Morita, University of Massachusetts Amherst) was grown shaking in Middlebrook 7H9 growth medium (BD Difco, Franklin Lakes, NJ) supplemented with OADC (BD BBL, Sparks, MD), Tween-80, 50 μg/ml leucine and 24 μg/ml pantothenic acid. For Fig. 4, M. smegmatis were cultured in 0.02% or 2% glucose then transferred briefly to no-glucose medium to avoid artifactual suppression of O-AlkTMM incorporation into AGM10.
M. smegmatis cell envelope labeling.
6-TreAz (1, 50 μM)19, O-AlkTMM (2, 50 μM)27, and bifunctional reporter O-AzAlkTMM (3, 50 μM) probes were used in this study. M. smegmatis labeling was performed mainly as described10, 41. Briefly, the OD600 was normalized to 1. Cultures were shaken in the presence of probes for 15 min at 37 °C unless otherwise stated. After incubation, the cultures were washed twice with PBST (phosphate-buffered saline and Tween-80, 0.05%) and fixed with 2% formaldehyde at room temperature for 10 min. After fixation, cultures were washed with PBST and probe incorporation was detected by CuAAC reaction with azide- and/or alkyne-fluorophores (Click Chemistry Tools, Scottsdale, AZ) as described in individual figure legends and in10, 41. Except for Fig. 2A, bacteria were resuspended in half of the volume of the starting culture of the reaction mix (1 mM CuSO4, 128 μM TBTA, 1.2 mM Sodium Ascorbate [60 mM stock solution fresh prepared in water], 20 μM azido- or alkynyl-fluorophore, in PBS) and incubated in shaking at room temperature for 30 min. Cultures were washed thrice with PBST, and either subjected to flow cytometry analysis or imaged on agar pads by fluorescence microscope. For Fig. 2A, fixed cells were resuspended in half of the volume of the starting culture in PBS containing 10 μM Cy5 DBCO (to detect cell envelope-installed azides [Click Chemistry Tools]) and incubated shaking at room temperature for 1 hour. The cells were washed twice with PBST then subjected to a CuAAC reaction with Carboxyrhodamine 110 Azide to detect cell envelope-installed alkynes, as described above.
M. tuberculosis cell envelope labeling.
Early log-phase M. tuberculosis (OD600 = 0.1) was grown for 48 hours, incubated +/− 6-TreAz (1, 500 μM)19, O-AlkTMM (2, 500 μM)27, or O-AzAlkTMM (3, 500 μM) probes for 16–24 hours, washed twice with PBST supplemented with 0.05% BSA (PBSTB), and finally fixed with freshly-prepared 4% formaldehyde in PBS at room temperature for 2 hours. After fixation, cultures were washed with PBSTB and subjected to the first CuAAC reaction to detect cell envelope-installed azides. In brief, bacteria were resuspended in half of the volume of the starting culture in reaction mix (1.5 mM CuSO4, 1.5 mM BTTP, 10 mM Sodium Ascorbate [60 mM stock solution freshly prepared in PBS], 10 μM Carboxyrhodamine 110 Alkyne [alk-CR110, Click Chemistry Tools, Scottsdale, AZ], in PBS) and incubated shaking at 30 °C for 2 hours. The cells were washed twice with PBSTB and subjected to a second CuAAC reaction using the same conditions but with MB 543 Picolyl Azide (azMB543, Click Chemistry Tools) to detect cell envelope-installed alkynes. Cells were finally washed three times with PBSTB and either subjected to flow cytometry or imaged by fluorescence microscopy.
Fluorescence microscopy.
Bacteria prepared on agar pad were imaged with a Nikon Eclipse E600 fluorescent microscope using a 100x objective and the FITC-HYO (EX 460–500, DM 505, BA 510–560) or G-2E/C TRITC (EX 528–553, DM 565, BA 600–660) filters. Images were rendered in FIJI.42
Flow cytometry analysis.
After fixation and washing, bacteria were resuspended in PBS and analyzed with a BD Biosciences Fortessa X20 SORP using three different channels: FITC (488 nm excitation laser, emission detection 530/30 band pass filter), PE-Texas Red (561 nm excitation, emission 610/20), and APC (640 nm excitation, emission 670/30). Filters are coupled with a photomultiplier tube detector using default voltage settings as determined by the cytometer set up and tracking (CS&T) software add-on, which is the quality control protocol established by BD Biosciences. Data were analyzed by FlowJo software.
qRT-PCR.
RNA was extracted from M. smegmatis grown either in 0.02% glucose or 2% glucose medium for 24 hours. Cells were harvested by centrifugation for 4000 rpm, 5 min at RT and the obtained pellets were re-suspended in 1 mL of TRIzol reagent (Invitrogen, Carlsbad, CA) prior to bead-beating (MP Biochemicals Lysing Matrix B) at 4 °C. After bead-beating, 300 μL chloroform was added to each tube and mixed slowly. The tubes were centrifuged at 14,000 rpm for 15 min at 4 °C to separate the upper aqueous layer. The aqueous layer was then immersed in 600 μL isopropanol in a fresh tube. The tube was kept at −20°C for 1–2 h and then centrifuged for 20 min at 4 °C, 14,000 rpm to precipitate the RNA. The precipitate RNA was washed once with 75% ethanol by centrifugation for 5 min at 4 °C, 14,000 rpm and dried at RT. The RNA pellet was re-suspended in RNase-free H2O. 20 μg of RNA was treated with 2.5 μL DNase (TURBO™ DNase, Ambion, Carlsbad, CA) in a final volume of 100 μL. The reaction was incubated for 2 h at 37 °C with shaking. The RNA was cleaned up by the Qiagen RNeasy Mini Kit (Qiagen), as per the manufacturer’s instructions. cDNA synthesis was carried out using 5 μg of the cleaned-up RNA following the manufacturer’s instructions for SuperScript IV Reverse Transcriptase (Invitrogen). The cDNA was then used for qRT-PCR reactions (iTaq Universal SYBR Green Supermix, BioRad, Hercules, CA). We used the sigA gene as our internal control. Primers used in this study are as follows:
| gene | primer | sequence |
|---|---|---|
| sigA | forward | gggctacaagttctcgacct |
| reverse | ccgagcttgttgatcacctc | |
| sugC | forward | gccgtcaaagagttctcgat |
| reverse | caatcatgttgagcgtggtc | |
| lpqY | forward | atgtggagaaccagcgctac |
| reverse | cgggtacttcttctggaacg |
Supplementary Material
Acknowledgements
This work was supported by NIH DP2 AI138238 (M.S.S.), NSF CAREER Award 1654408 (B.M.S.), Camille and Henry Dreyfus Foundation Henry Dreyfus Teacher-Scholar Award TH-17-034 (B.M.S.), University of Massachusetts Amherst Institute for Applied Life Sciences Midigrant (I.L.), and the University of Massachusetts Amherst Chemistry-Biology Interface Program NIH T32 GM008515 (R.A.G.). We thank Dr. Amy Burnside, director of the University of Massachusetts Amherst Flow Cytometry Core facility, for technical support.
Footnotes
Supporting Information
Supplementary figures, experimental details, and NMR spectra for synthetic compounds.
References
- (1).Ishikawa E; Mori D; Yamasaki S Recognition of Mycobacterial Lipids by Immune Receptors. Trends Immunol 2017, 38 (1), 66–76. DOI: 10.1016/j.it.2016.10.009. [DOI] [PubMed] [Google Scholar]
- (2).Daffé M; Marrakchi H Unraveling the Structure of the Mycobacterial Envelope. Microbiol Spectr 2019, 7 (4). DOI: 10.1128/microbiolspec.GPP3-0027-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Viswanathan G; Joshi SV; Sridhar A; Dutta S; Raghunand TR Identifying novel mycobacterial stress associated genes using a random mutagenesis screen in Mycobacterium smegmatis. Gene 2015, 574 (1), 20–27. DOI: 10.1016/j.gene.2015.07.063. [DOI] [PubMed] [Google Scholar]
- (4).Sambandan D; Dao DN; Weinrick BC; Vilchèze C; Gurcha SS; Ojha A; Kremer L; Besra GS; Hatfull GF; Jacobs WR Keto-mycolic acid-dependent pellicle formation confers tolerance to drug-sensitive Mycobacterium tuberculosis. mBio 2013, 4 (3), e00222–00213. DOI: 10.1128/mBio.00222-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Lee WB; Kang JS; Choi WY; Zhang Q; Kim CH; Choi UY; Kim-Ha J; Kim YJ Mincle-mediated translational regulation is required for strong nitric oxide production and inflammation resolution. Nat Commun 2016, 7, 11322. DOI: 10.1038/ncomms11322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Yang Y; Kulka K; Montelaro RC; Reinhart TA; Sissons J; Aderem A; Ojha AK A hydrolase of trehalose dimycolate induces nutrient influx and stress sensitivity to balance intracellular growth of Mycobacterium tuberculosis. Cell Host Microbe 2014, 15 (2), 153–163. DOI: 10.1016/j.chom.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Gebhardt H; Meniche X; Tropis M; Krämer R; Daffé M; Morbach S The key role of the mycolic acid content in the functionality of the cell wall permeability barrier in Corynebacterineae. Microbiology 2007, 153 (Pt 5), 1424–1434. DOI: 10.1099/mic.0.2006/003541-0. [DOI] [PubMed] [Google Scholar]
- (8).Dulberger CL; Rubin EJ; Boutte CC The mycobacterial cell envelope - a moving target. Nat Rev Microbiol 2020, 18 (1), 47–59. DOI: 10.1038/s41579-019-0273-7. [DOI] [PubMed] [Google Scholar]
- (9).Kalscheuer R; Weinrick B; Veeraraghavan U; Besra GS; Jacobs WR Jr. Trehalose-recycling ABC transporter LpqY-SugA-SugB-SugC is essential for virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 2010, 107 (50), 21761–21766. DOI: 10.1073/pnas.1014642108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Pohane AA; Carr CR; Garhyan J; Swarts BM; Siegrist MS Trehalose Recycling Promotes Energy-Efficient Biosynthesis of the Mycobacterial Cell Envelope. mBio 2021, 12 (1). DOI: 10.1128/mBio.02801-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Holmes NJ; Kavunja HW; Yang Y; Vannest BD; Ramsey CN; Gepford DM; Banahene N; Poston AW; Piligian BF; Ronning DR; Ojha AK; Swarts BM A FRET-Based Fluorogenic Trehalose Dimycolate Analogue for Probing Mycomembrane-Remodeling Enzymes of Mycobacteria. ACS Omega 2019, 4 (2), 4348–4359. DOI: 10.1021/acsomega.9b00130.\ [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ojha AK; Trivelli X; Guerardel Y; Kremer L; Hatfull GF Enzymatic hydrolysis of trehalose dimycolate releases free mycolic acids during mycobacterial growth in biofilms. J Biol Chem 2010, 285 (23), 17380–17389. DOI: 10.1074/jbc.M110.112813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Nobre A; Alarico S; Maranha A; Mendes V; Empadinhas N The molecular biology of mycobacterial trehalose in the quest for advanced tuberculosis therapies. Microbiology 2014, 160 (Pt 8), 1547–1570. DOI: 10.1099/mic.0.075895-0. [DOI] [PubMed] [Google Scholar]
- (14).Eoh H; Wang Z; Layre E; Rath P; Morris R; Branch Moody D; Rhee KY Metabolic anticipation in Mycobacterium tuberculosis. Nat Microbiol 2017, 2, 17084. DOI: 10.1038/nmicrobiol.2017.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Shleeva MO; Trutneva KA; Demina GR; Zinin AI; Sorokoumova GM; Laptinskaya PK; Shumkova ES; Kaprelyants AS Free Trehalose Accumulation in Dormant. Front Microbiol 2017, 8, 524. DOI: 10.3389/fmicb.2017.00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lee JJ; Lee SK; Song N; Nathan TO; Swarts BM; Eum SY; Ehrt S; Cho SN; Eoh H Transient drug-tolerance and permanent drug-resistance rely on the trehalose-catalytic shift in Mycobacterium tuberculosis. Nat Commun 2019, 10 (1), 2928. DOI: 10.1038/s41467-019-10975-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Danelishvili L; Shulzhenko N; Chinison JJJ; Babrak L; Hu J; Morgun A; Burrows G; Bermudez LE Mycobacterium tuberculosis Proteome Response to Antituberculosis Compounds Reveals Metabolic “Escape” Pathways That Prolong Bacterial Survival. Antimicrob Agents Chemother 2017, 61 (7). DOI: 10.1128/AAC.00430-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Galagan JE; Minch K; Peterson M; Lyubetskaya A; Azizi E; Sweet L; Gomes A; Rustad T; Dolganov G; Glotova I; Abeel T; Mahwinney C; Kennedy AD; Allard R; Brabant W; Krueger A; Jaini S; Honda B; Yu W-H; Hickey MJ; Zucker J; Garay C; Weiner B; Sisk P; Stolte C; Winkler JK; Van de Peer Y; Iazzetti P; Camacho Diogo; Dreyfuss J; Liu Y; Dorhoi A; Mollenkopf H-J; Drogaris P; Lamontagne J; Zhou Y; Piquenot J; Park ST; Raman S; Kaufman SHE; Mohney RP; Chelsky D; Moody DB; Sherman DR; Schoolnik GK The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 2013, 499 (7457), 178–183. DOI: 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Swarts BM; Holsclaw CM; Jewett JC; Alber M; Fox DM; Siegrist MS; Leary JA; Kalscheuer R; Bertozzi CR Probing the mycobacterial trehalome with bioorthogonal chemistry. J Am Chem Soc 2012, 134 (39), 16123–16126. DOI: 10.1021/ja3062419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Foley HN; Stewart JA; Kavunja HW; Rundell SR; Swarts BM Bioorthogonal chemical reporters for selective in situ probing of mycomembrane components in mycobacteria. Angew. Chem. Int. Edit 2016, 55 (6), 2053–2057. DOI: 10.1002/anie.201509216. [DOI] [PubMed] [Google Scholar]
- (21).Siegrist MS; Swarts BM; Fox DM; Lim SA; Bertozzi CR Illumination of growth, division and secretion by metabolic labeling of the bacterial cell surface. FEMS Microbiol Rev 2015, 39 (2), 184–202. DOI: 10.1093/femsre/fuu012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Behren S; Westerlind U Glycopeptides and -Mimetics to Detect, Monitor and Inhibit Bacterial and Viral Infections: Recent Advances and Perspectives. Molecules 2019, 24 (6). DOI: 10.3390/molecules24061004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Dutta AK; Choudhary E; Wang X; Záhorszka M; Forbak M; Lohner P; Jessen HJ; Agarwal N; Korduláková J; Jessen-Trefzer C Trehalose Conjugation Enhances Toxicity of Photosensitizers against Mycobacteria. ACS Cent Sci 2019, 5 (4), 644–650. DOI: 10.1021/acscentsci.8b00962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Jayawardana KW; Jayawardena HS; Wijesundera SA; De Zoysa T; Sundhoro M; Yan M Selective targeting of Mycobacterium smegmatis with trehalose-functionalized nanoparticles. Chem Commun 2015, 51 (60), 12028–12031. DOI: 10.1039/c5cc04251h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zhang ZJ; Wang YC; Yang X; Hang HC Chemical Reporters for Exploring Microbiology and Microbiota Mechanisms. Chembiochem 2020, 21 (1–2), 19–32. DOI: 10.1002/cbic.201900535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Banahene N; Kavunja HW; Swarts BM Chemical Reporters for Bacterial Glycans: Development and Applications. Chem. Rev 2022, 122 (3), 3336–3413. DOI: 10.1021/acs.chemrev.1c00729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Foley HN; Stewart JA; Kavunja HW; Rundell SR; Swarts BM Bioorthogonal Chemical Reporters for Selective In Situ Probing of Mycomembrane Components in Mycobacteria. Angew Chem Int Ed Engl 2016, 55 (6), 2053–2057. DOI: 10.1002/anie.201509216. [DOI] [PubMed] [Google Scholar]
- (28).Backus KM; Boshoff HI; Barry CS; Boutureira O; Patel MK; D’Hooge F; Lee SS; Via LE; Tahlan K; Barry CE 3rd; Davis BD Uptake of unnatural trehalose analogs as a reporter for Mycobacterium tuberculosis. Nat Chem Biol 2011, 7 (4), 228–235. DOI: 10.1038/nchembio.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Fiolek TJ; Banahene N; Kavunja HW; Holmes NJ; Rylski AK; Pohane AA; Siegrist MS; Swarts BM Engineering the Mycomembrane of Live Mycobacteria with an Expanded Set of Trehalose Monomycolate Analogues. ChemBioChem 2019, 20 (10), 1282–1291. DOI: 10.1002/cbic.201800687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kavunja HW; Piligian BF; Fiolek TJ; Foley HN; Nathan TO; Swarts BM A chemical reporter strategy for detecting and identifying O-mycoloylated proteins in Corynebacterium. Chem. Commun 2016, 52 (95), 13795–13798. DOI: 10.1039/C6CC07143K. [DOI] [PubMed] [Google Scholar]
- (31).Kavunja HW; Biegas KJ; Banahene N; Stewart JA; Piligian BF; Groenevelt JM; Sein CE; Morita YS; Niederweis M; Siegrist MS; Swarts BM Photoactivatable Glycolipid Probes for Identifying Mycolate-Protein Interactions in Live Mycobacteria. J Am Chem Soc 2020, 142 (17), 7725–7731. DOI: 10.1021/jacs.0c01065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Kamariza M; Shieh P; Ealand CS; Peters JS; Chu B; Rodriguez-Rivera FP; Babu Sait MR; Treuren WV; Martinson N; Kalscheuer R; Kana BD; Bertozzi CR Rapid detection of Mycobacterium tuberculosis in sputum with a solvatochromic trehalose probe. Sci Transl Med 2018, 10 (430). DOI: 10.1126/scitranslmed.aam6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Hodges HL; Brown RA; Crooks JA; Weibel DB; Kiessling LL Imaging mycobacterial growth and division with a fluorogenic probe. Proc Natl Acad Sci U S A 2018, 115 (20), 5271–5276. DOI: 10.1073/pnas.1720996115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Favrot L; Grzegorzewicz AE; Lajiness DH; Marvin RK; Boucau J; Isailovic D; Jackson M; Ronning DR Mechanism of inhibition of Mycobacterium tuberculosis antigen 85 by ebselen. Nat Commun 2013, 4, 2748. DOI: 10.1038/ncomms3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Zhou X; Rodriguez-Rivera FP; Lim HC; Bell JC; Bernhardt TG; Bertozzi CR; Theriot JA Sequential assembly of the septal cell envelope prior to V snapping in Corynebacterium glutamicum. Nat Chem Biol 2019, 15 (3), 221–231. DOI: 10.1038/s41589-018-0206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Sampson SL; Dascher CC; Sambandamurthy VK; Russell RG; Jacobs WR; Bloom BR; Hondalus MK Protection elicited by a double leucine and pantothenate auxotroph of Mycobacterium tuberculosis in guinea pigs. Infect Immun 2004, 72 (5), 3031–3037. DOI: 10.1128/IAI.72.5.3031-3037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Betts JC; Lukey PT; Robb LC; McAdam RA; Duncan K Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol 2002, 43 (3), 717–731. DOI: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- (38).Titgemeyer F; Amon J; Parche S; Mahfoud M; Bail J; Schlicht M; Rehm N; Hillmann D; Stephan J; Walter B; Burkovski A; Niederweis M A genomic view of sugar transport in Mycobacterium smegmatis and Mycobacterium tuberculosis. J Bacteriol 2007, 189 (16), 5903–5915. DOI: 10.1128/JB.00257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Li M; Müller C; Fröhlich K; Gorka O; Zhang L; Groß O; Schilling O; Einsle O; Jessen-Trefzer C Detection and Characterization of a Mycobacterial L-Arabinofuranose ABC Transporter Identified with a Rapid Lipoproteomics Protocol. Cell Chem Biol 2019, 26 (6), 852–862.e856. DOI: 10.1016/j.chembiol.2019.03.002. [DOI] [PubMed] [Google Scholar]
- (40).Kamariza M; Shieh P; Ealand CS; Peters JS; Chu B; Rodriguez-Rivera FP; Babu Sait MR; Treuren WV; Martinson N; Kalscheuer R; et al. Rapid detection of. Sci Transl Med 2018, 10 (430). DOI: 10.1126/scitranslmed.aam6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).García-Heredia A; Pohane AA; Melzer ES; Carr CR; Fiolek TJ; Rundell SR; Lim HC; Wagner JC; Morita YS; Swarts BM; et al. Peptidoglycan precursor synthesis along the sidewall of pole-growing mycobacteria. Elife 2018, 7. DOI: 10.7554/eLife.37243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Schindelin J; Arganda-Carreras I; Frise E; Kaynig V; Longair M; Pietzsch T; Preibisch S; Rueden C; Saalfeld S; Schmid B; et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012, 9 (7), 676–682. DOI: 10.1038/nmeth.2019 From NLM Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


