ABSTRACT
Genome analysis of Corynebacterium lactis revealed a bacteriocin gene cluster encoding a putative bacteriocin of the linaridin family of ribosomally synthesized and posttranslationally modified peptides (RiPPs). The locus harbors typical linaridin modification enzymes but lacks genes for a decarboxylase and methyltransferase, which is unusual for type B linaridins. Supernatants of Corynebacterium lactis RW3-42 showed antimicrobial activity against Corynebacterium glutamicum. Deletion of the precursor gene crdA clearly linked the antimicrobial activity of the producer strain to the identified gene cluster. Following purification, we observed potent activity of the peptide against Actinobacteria, mainly other members of the genus Corynebacterium, including the pathogenic species Corynebacterium striatum and Corynebacterium amycolatum. Also, low activity against some Firmicutes was observed, but there was no activity against Gram-negative species. The peptide is resilient towards heat but sensitive to proteolytic degradation by trypsin and proteinase K. Analysis by mass spectrometry indicates that corynaridin is processed by cleaving off the leader sequence at a conserved motif and posttranslationally modified by dehydration of all threonine and serin residues, resulting in a monoisotopic mass of 3,961.19 Da. Notably, time-kill kinetics and experiments using live biosensors to monitor membrane integrity suggest bactericidal activity that does not involve formation of pores in the cytoplasmic membrane. As Corynebacterium species are ubiquitous in nature and include important commensals and pathogens of mammalian organisms, secretion of bacteriocins by species of this genus could be a hitherto neglected trait with high relevance for intra- and interspecies competition and infection.
IMPORTANCE Bacteriocins are antimicrobial peptides produced by bacteria to fend off competitors in ecological niches and are considered to be important factors influencing the composition of microbial communities. However, bacteriocin production by bacteria of the genus Corynebacterium has been a hitherto neglected trait, although its species are ubiquitous in nature and make up large parts of the microbiome of humans and animals. In this study, we describe and characterize a novel linaridin family bacteriocin from Corynebacterium lactis and show its narrow-spectrum activity, mainly against other actinobacteria. Moreover, we were able to extend the limited knowledge on linaridin bioactivity in general and for the first time describe the bactericidal activity of such a bacteriocin. Interestingly, the peptide, which was named corynaridin, appears bactericidal, but without formation of pores in the bacterial membrane.
KEYWORDS: Corynebacterium, linaridin, antimicrobial peptides, bacteriocins
INTRODUCTION
Genome sequencing and bioinformatics have promoted the discovery of ribosomally synthesized, bioactive molecules over the past decades. The heterogeneous group of antimicrobial peptides produced by bacteria, so-called bacteriocins, gained special interest because of their potential use as food preservatives and alternatives to antibiotics (1–5). However, the primary biological function of bacteriocins is to provide the producer with a selective advantage over target organisms in a complex and competitive ecological niche (6, 7).
While bacteriocins have been extensively studied in lactic acid bacteria (LAB), comparably little knowledge is available about production of such compounds by other bacteria (e.g., Actinobacteria). Nevertheless, several studies suggest the widespread occurrence of bacteriocin gene clusters (BGCs) in non-LAB species, including the genus Corynebacterium (8–11). Species of this genus are widespread in nature, make up one of the largest groups of bacteria in the human and animal skin microbiome, and are also present in food products, including raw milk or cheese (12–14).
Besides toxicogenic Corynebacterium species (e.g., Corynebacterium diphtheriae and Corynebacterium ulcerans), many (nondiphtheritic) species of the genus have been described as commensals (13). Being a dominant bacterial group of the human skin microbiome, also nondiphtheritic corynebacteria are regularly found in infectious tissue. However, in most cases they are regarded rather as contamination from surrounding skin than the etiological agent of the infection itself (15). Nevertheless, improved methods to discriminate infection and colonization and an increasing number of reports suggest that some Corynebacterium species are important opportunistic pathogens for humans and animals (16). The clinical relevance of nondiphtheritic corynebacteria becomes even more apparent with increasing reports of multidrug-resistant strains, mostly identified in nosocomial environments (17, 18). Species like Corynebacterium striatum and Corynebacterium amycolatum have been described to cause infections in elderly, immunocompromised patients and are associated with chronic wounds (19). Also, recent studies suggest that microbe-microbe interactions of Corynebacterium species with other commensals or pathogens like Staphylococcus aureus might influence the behavior and fitness of both species (20). Interestingly, only a very few reports on bacteriocin production in corynebacteria exist (21, 22).
In general, bacteriocins of Gram-positive bacteria can be classified into small (<10 kDa) modified (class I) and unmodified (class II) as well as larger (>10 kDa) heat-labile (class III) peptides/proteins (1). Class I bacteriocins usually contain posttranslational amino acid modifications, such as dehydration, heterocycle formation, glycosylation, methylation, etc., that are often important for their biological activity (23). Thus, class I bacteriocins are also referred to as ribosomally synthesized and posttranslationally modified peptides (RiPPs) (1). Linaridins are a group of RiPPs with an overall linear structure, containing dehydrated amino acids such as dehydrobutyrine (24). So far, only five members of this family have been described in detail: i.e., cypemycin, grisemycin, legonaridin, mononaridin, and salinipeptins (24–30). Nevertheless, in silico analyses suggest that linaridin BGCs are widespread in nature and especially in Actinobacteria (8). In contrast to lanthipeptides, which also contain dehydrated amino acids (e.g., nisin), linaridin biosynthesis is considered to be essentially different from that of other RiPPs. Modification of the prototypic type A linaridin cypemycin includes dehydration of threonine residues, N-terminal methylation, and C-terminal oxidative decarboxylation of cysteine and subsequent formation of a heterocyclic S-[(Z)-2-aminovinyl]-d-cysteine (AviCys) moiety (24, 25). N-terminal methylation was shown to be crucial for the activity of cypemycin (28). In contrast, type B linaridin gene clusters do not encode decarboxylases and thus lack C-terminal modification. Instead, genes for so far uncharacterized short-chain oxidoreductases have been identified: e.g., in the gene cluster for the biosynthesis of legonaridin (27).
While extensive studies have been carried out on the structure and chemistry behind their modifications, comparably little is known about the biological functions of linaridins. The antimicrobial activity of cypemycin appears to be limited to Micrococcus luteus (29). Additionally, cypemycin possesses cytotoxic activity against mouse P388 leukemia cells (26, 29). The structurally similar salinipeptins, however, inhibit growth of a Streptococcus pyogenes strain but not that of M. luteus (30). So far, for none of the hitherto described linaridins receptors or mode of action have been proposed.
In this study, we describe the identification and partial characterization of a novel linaridin discovered in Corynebacterium lactis RW3-42, a strain isolated from raw cow’s milk (31).
RESULTS
In silico analysis of a bacteriocin gene cluster in C. lactis.
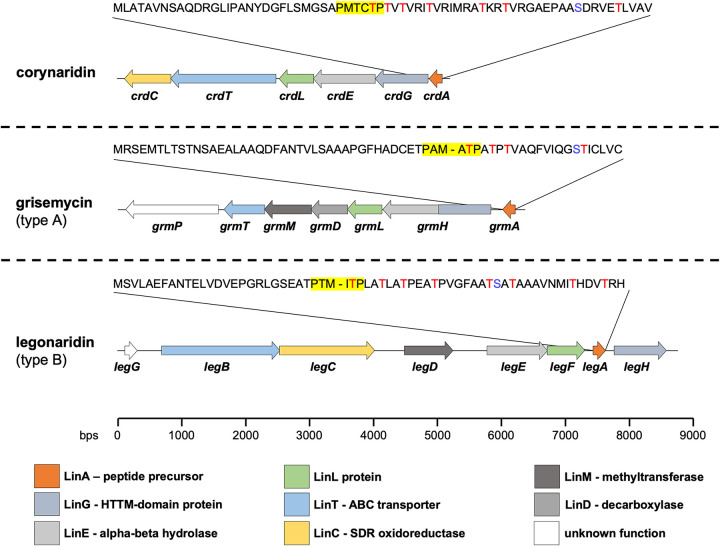
In silico analyses using the web-based tool BAGEL4 revealed several, yet undescribed bacteriocin gene clusters (BGCs) in the genus Corynebacterium (9). In this study, we closely examined one of the predicted BGCs in the genome of C. lactis RW2-5 isolated from raw cow’s milk and found an identical cluster in the strain C. lactis RW3-42 upon sequencing of the locus (31). The identified BGC consists of six genes, including a gene for a conserved LinL protein homolog (here named crdL) that was so far only associated with linaridin biosynthesis and is typically used to identify corresponding gene clusters (Fig. 1) (28). Furthermore, a hypothetical peptide precursor gene (here named crdA) was identified that encodes a peptide of 69 amino acids with a conserved hexapeptide cleavage motif, PxxxTP, at positions 29 to 34. The peptide also harbors a high number of threonine residues (seven in total) in its C-terminal part, which are often posttranslationally modified in RiPPs as shown for cypemycin or nisin (Fig. 1) (23).
FIG 1.
Comparison of the BGC of C. lactis RW3-42 with BGCs for other bacteriocins of the linaridin family. Shown is the genetic organization of the corynaridin, grisemycin, and legonaridin BGCs as predicted by BAGEL4 and blastp analyses. The amino acid sequences of the precursor peptides are displayed above the gene cluster. Threonine residues are displayed in red letters. The hexapeptide cleavage site PxxxTP in the precursor peptides is highlighted in yellow. Predicted functions of modification enzymes and their corresponding genes are indicated by a color scheme according to the color key at the bottom of the figure.
The deduced amino acid sequences of the other proteins encoded in the BGC were further analyzed using blastp and checked for homologies to enzymes involved in biosynthesis of described members of the linaridin family. Similar to other linaridin BGCs, the crd locus contains no gene for a protease involved in removal of the leader peptide but genes for an N-terminal horizontally transferred transmembrane helix domain protein (HTTM domain protein, crdG) and an alpha-beta-fold hydrolase (crdE) are present (Fig. 1). These putative modification enzymes are assumed to participate in the maturation of linaridins, although their exact mechanism remains unclear (24). Interestingly, no decarboxylase gene but a putative short-chain oxidoreductase (SDR) gene (crdC) typical for biosynthetic operons of type B linaridins is part of the BGC. This observation and the absence of a C-terminal cysteine residue in the peptide precursor suggest that the BGC of C. lactis encodes a type B linaridin. Notably, we were also unable to identify a gene for a methyltransferase involved in N-terminal methylation, which was described for, e.g., legonaridin (24, 27). The ABC transporter gene (crdT) of the BGC is supposedly involved in transport and/or processing of the peptide or immunity of the host (32). Sequence alignments of the enzymes of the C. lactis BGC with those of the legonaridin biosynthesis of Streptomyces sp. strain CT34 revealed only low overall homologies (22 to 32%) (see Table S2 and Fig. S1 to S6 in the supplemental material). Overall, our in silico analyses suggest that the BGC of C. lactis RW3-42 encodes a novel type B linaridin lacking N-terminal methylation, for which we propose the designation “corynaridin.”
Furthermore, we analyzed whether the corynaridin precursor exists in other BGCs of Corynebacterium species and found a highly similar gene cluster in the genome of C. striatum 1329_CAUR, isolated from the wound of an intensive care unit patient (Fig. S7) (33). The respective peptide precursor shows 84% identity to corynaridin, with a conserved PxxxTP hexapeptide motif and seven putatively modified threonine residues at its N terminus. Interestingly, a previous, extensive bioinformatic analysis of the NCBI genome database identified 561 linaridin BGCs, mainly in Actinobacteria, including the ones in C. lactis and C. striatum plus four others in Corynebacterium spp. (8). Closer examination of these BGCs shows that also in these genomes typical linaridin biosynthesis genes are present, but the overall genetic architectures and precursor peptide sequences are significantly different (Fig. S8 and S9).
Growth, antimicrobial activity, and kinetics of secretion of corynaridin.
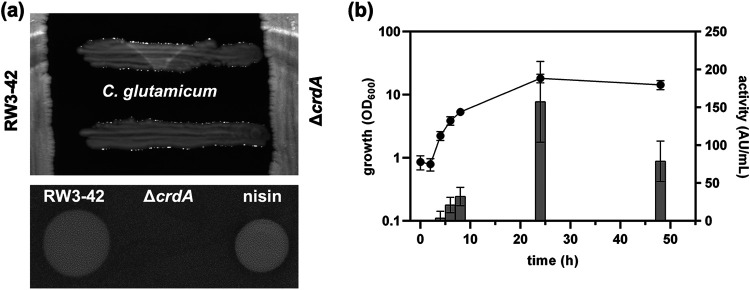
Since bacteriocins are often most effective against closely related species, we first tested if C. lactis RW3-42 is able to inhibit growth of different Corynebacterium species. Analysis of its antimicrobial capacity against Corynebacterium glutamicum ATCC 13032 revealed a clear zone of inhibition in cross-streak and spot-on-lawn assays using supernatants of C. lactis RW3-42 (Fig. 2a). Also, other Corynebacterium species, M. luteus DSM 20030, and Pediococcus acidilactici 347 were inhibited in growth (Table 1 and Table S3). In contrast, no activity of C. lactis RW3-42 was detected against Cutibacterium acnes DSM 16379, the other tested Firmicutes, or Gram-negative bacteria, like Escherichia coli MG1655 and Pseudomonas fluorescens DSM 50090, in cross-streak assays (Table 1 and Table S3). To verify that the antimicrobial activity is related to the predicted BGC (Fig. 1), we generated a C. lactis ΔcrdA mutant strain with a clean, markerless deletion of crdA, encoding the corynaridin precursor. The mutant did not show antimicrobial activity against C. glutamicum ATCC 13032 in cross-streak or spot-on-lawn assays confirming that the antimicrobial activity of C. lactis RW3-42 is related to crdA (Fig. 2a).
FIG 2.
Analysis of the antimicrobial activity produced by C. lactis RW3-42. (a) Cross-streak assay (upper panel) and spot-on-lawn assays (lower panel) indicating the secretion of an antimicrobial substance by C. lactis RW3-42 compared to the C. lactis ΔcrdA deletion mutant strain using C. glutamicum ATCC 13032 as indicator. Ten microliters of a nisin standard (250 μg/mL) was used as a positive control in the spot-on-lawn assay. (b) Growth (OD600) (left y axis; black dots) and kinetics of antimicrobial activity (BU per milliliter) (shown as arbitrary units [AU] on the right y axis; gray columns) of C. lactis RW3-42 in CLI medium containing 1% (wt/vol) glucose. Antimicrobial activity was measured with C. glutamicum ATCC 13032 as an indicator. Values are means and standard deviations from at least six biological replicates.
TABLE 1.
Inhibitory spectrum of C. lactis RW3-42 and purified corynaridin assessed with cross-streak and spot-on-lawn assays
| Strain | Phylum | Activitya |
|
|---|---|---|---|
| Cross-streak | RPC fractionb | ||
| Corynebacterium ammoniagenes DSM 20306 | Actinobacteria | + | + |
| Corynebacterium amycolatum DSM 6922 | Actinobacteria | + | + |
| Corynebacterium canis DSM 45402 | Actinobacteria | (+) | ND |
| Corynebacterium casei DSM 44701 | Actinobacteria | + | + |
| Corynebacterium efficiens DSM 44549 | Actinobacteria | + | + |
| Corynebacterium glutamicum ATCC 13032 | Actinobacteria | + | + |
| Corynebacterium lipophiloflavum DSM 44291 | Actinobacteria | + | + |
| Corynebacterium striatum DSM 20668 | Actinobacteria | + | + |
| Corynebacterium xerosis DSM 20743 | Actinobacteria | + | + |
| Cutibacterium acnes DSM 16379 | Actinobacteria | − | (+) |
| Micrococcus luteus DSM 20030 | Actinobacteria | + | + |
| Bacillus subtilis DSM 402 | Firmicutes | − | − |
| Lactobacillus plantarum DSM 1055 | Firmicutes | − | − |
| Lactococcus lactis IL1403 | Firmicutes | − | + |
| Listeria innocua LMG2785 | Firmicutes | − | (+) |
| Listeria monocytogenes EGD-e | Firmicutes | − | − |
| Pediococcus acidilactici 347 | Firmicutes | + | − |
| Staphylococcus aureus ATCC 29213 | Firmicutes | − | − |
| Staphylococcus epidermidis DSM 3269 | Firmicutes | − | − |
| Pseudomonas fluorescens DSM 50090 | Gammaproteobacteria | − | − |
| Escherichia coli K-12 MG1655 | Proteobacteria | − | − |
+, zone of inhibition; (+), diffuse zone of inhibition; −, no zone of inhibition; ND, not determined.
Ten-microliter RPC fraction.
Growth of C. lactis RW3-42 and kinetics of production of the antimicrobial activity in shake flask experiments indicated that the antimicrobial compound is secreted mainly during the exponential growth phase (Fig. 2b). The highest biomass of the strain was observed after 24 h (optical density at 600 nm [OD600] = 18 ± 3). Minor antimicrobial activity against C. glutamicum ATCC 13032 was first observed after 4 h and peaked at a maximum of 157 ± 53 bacteriocin units (BU)/mL after 24 h. After 48 h, a reduction of antimicrobial activity occurred, possibly due to degradation or adsorption of the peptide to biomass.
Purification of corynaridin from supernatants of C. lactis RW3-42.
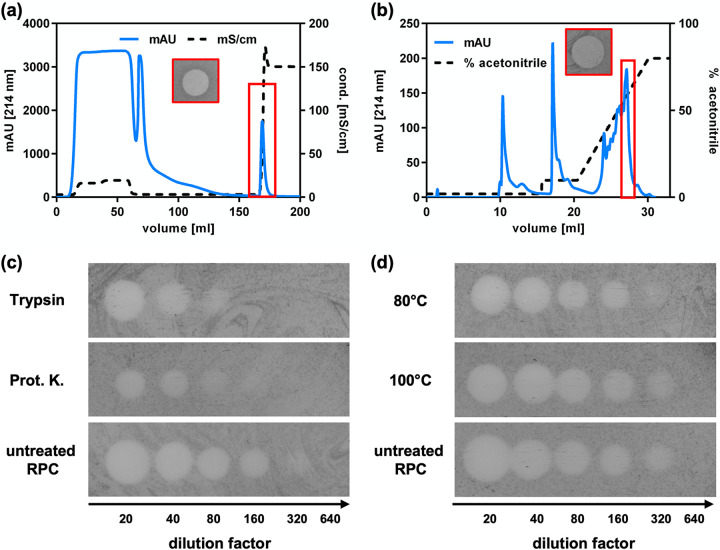
We next sought to purify the secreted antimicrobial compound for further characterization. Supernatants of 1-L cultivations of C. lactis RW3-42 were harvested, and proteins were precipitated using ammonium sulfate. The precipitate was resuspended in high-performance liquid chromatography (HPLC)-grade water, with the pH adjusted to 4, and used for cation-exchange chromatography (CIEX) (Fig. 3a). A single peak was observed following onset of elution with high-salt buffer, and the corresponding fractions exhibited activity against C. glutamicum ATCC 13032 in spot-on-lawn assays. Peak fractions were pooled for further purification via reversed-phase chromatography (RPC) using acetonitrile-H2O-trifluoroacetic acid (TFA) as the mobile phase (Fig. 3b). A combined step and linear gradient was applied and yielded several peaks, of which only one (at ~56% acetonitrile) showed activity against C. glutamicum ATCC 13032 in spot-on-lawn assays. After removal of acetonitrile and resuspension in HPLC-grade H2O, a preparation with high activity (32,000 BU/mL) was obtained.
FIG 3.
Purification of corynaridin from C. lactis RW3-42 supernatants. Supernatant proteins were precipitated by 50% (wt/vol) ammonium sulfate and analyzed by (a) cation-exchange chromatography and (b) reversed-phase chromatography. Absorbance at 214 nm is displayed as blue line, conductivity and acetonitrile concentration are displayed as dashed line. Purified corynaridin (i.e., pooled fractions of the indicated peak of reversed-phase chromatography) was analyzed for stability against (c) proteases and (d) heat treatment. For each spot, a 10-μL sample was used (RPC fraction with 606 μg/mL total protein). One representative experiment is shown for each purification step. Prot. K., proteinase K.
To assess the physicochemical properties of the purified compound, it was tested for resistance to protease and heat treatment (Fig. 3c and d). While a reduction of activity of the RPC fraction to 8,000 BU/mL (i.e., about 4-fold) was observed after incubation with trypsin or proteinase K, heat treatment at 80°C and 100°C for 10 min had no effect. This indicates that the purified antimicrobial compound is a heat-stable peptide.
Inhibitory spectrum of purified corynaridin.
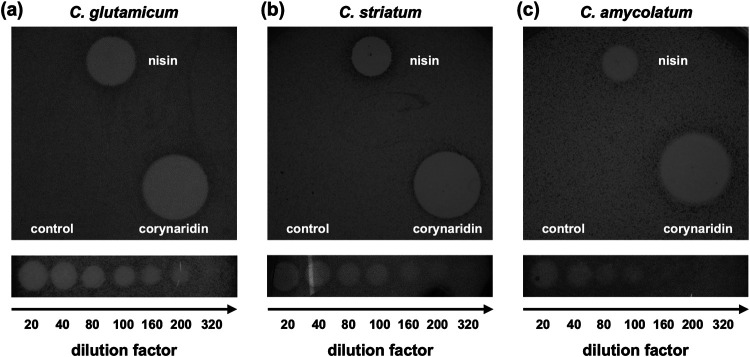
Purified corynaridin was active against all tested Corynebacterium species (Table 1 and Table S3) except C. canis DSM 45402, which could not be properly analyzed by the spot-on-lawn method. Notably, besides non-pathogenic, environmental and commensal bacteria, including C. glutamicum ATCC 13032 (Fig. 4a), emerging multiresistant pathogens like C. striatum DSM 20668 (Fig. 4b) and C. amycolatum DSM 6922 (Fig. 4c) also were inhibited by RPC-purified corynaridin. In case of P. acidilactici 347, assays with purified peptide contradicted the cross-streak results as the peptide did not inhibit growth of the strain (Table 1). L. lactis IL-1403 was the only firmicute tested that was effectively inhibited by corynaridin. For L. innocua LMG2785 and the actinobacterium C. acnes DSM 16379, low levels of inhibition were achieved only at high concentrations (>1,000 μg/mL) of the RPC fraction.
FIG 4.
Antimicrobial activity of purified corynaridin against pathogenic corynebacteria. The RPC fraction was used undiluted and in a dilution series against (a) C. glutamicum ATCC 13032, (b) C. striatum DSM 20668, and (c) C. amycolatum DSM 6922. For each spot, a 10-μL sample was used (RPC fraction with 606 μg/mL total protein). A nisin standard (250 μg/mL) and HPLC-grade H2O (control) were used as positive and negative controls, respectively.
For all other tested Gram-positive bacteria of the phylum Firmicutes, including Listeria monocytogenes EGD-e or S. aureus ATCC 29213, or Gram-negative bacteria like E. coli MG1655 or P. fluorescence DSM 50090, cross-streak and spot-on-lawn assays equally indicated that these organisms were not inhibited by corynaridin. Collectively, these findings suggest that corynaridin has a rather narrow spectrum of target organisms (Table 1).
Identification of corynaridin by LC-MS.
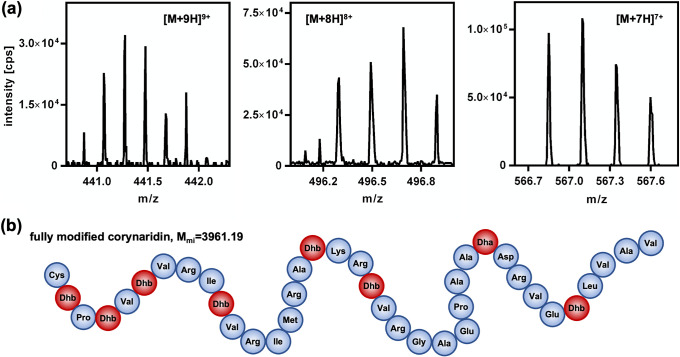
To identify the antimicrobial compound and investigate potential posttranslational modifications, corynaridin was purified from supernatants of C. lactis RW3-42 following growth in CLIV minimal medium (see Materials and Methods) with glucose as sole carbon source (Fig. S10a). Under these conditions, slow growth and strong aggregation were observed, making OD600 measurements impossible. Nevertheless, activities in supernatants were already observed during the first hours of growth, remained stable after 24 h, and levels were overall comparable to cultivation in complex medium. Supernatants harvested after 24 h of cultivation in CLIV medium were directly applied to the cation-exchange column (Fig. S10b), resulting in a higher initial purity than the previously used protocol. After subsequent RPC purification with a slightly changed protocol, a fraction was obtained that was suitable for mass spectrometry (Fig. S10c and d). The activity of the sample was 40,000 BU/mL, corresponding to 0.5 μg/mL protein (Fig. S10e). The active RPC fraction was analyzed by liquid chromatography-mass spectrometry (LC-MS), yielding rather complex time of flight (TOF) spectra. The linaridins investigated so far contain unusual postranslational modifications and were shown to carry dehydrated threonine residues (dehydrobutyrine). The corynaridin gene cluster contains the genes crdG, crdE, and crdL, which are homologues of the genes suspected to encode the enzymes responsible for threonine dehydration in linaridins (24). The primary amino acid sequence of processed corynaridin (i.e., following cleavage of the leader sequence) also contains a serine residue at position 29 that may be dehydrated in lantibiotics to dehydroalanine as shown for nisin (22). Not taking into account positional variations of dehydration and assuming processing by cleavage of the leader sequence, corynaridin may carry up to 8 dehydrated threonines/serines, resulting in 9 distinct monoisotopic masses, and each of these variants may be present in various isotopic states and carrying additional charges (protons). To identify corynaridin, the TOF spectra were filtered for signals with a minimum peak intensity of 103 cps for at least three charged peptide species with at least four isotopes. The vast majority of signals in the time-resolved spectra that matched these criteria corresponded to fully modified, processed corynaridin (dehydration of all 8 threonine/serine residues) with a monoisotopic mass of 3,961.19 Da (Fig. 5 and Table S4). However, it has to be noted that some of the signals that passed the thresholds of filtering correspond to peptide variants with lower numbers of dehydrated threonine/serine residues (Table S4).
FIG 5.
Identification of corynaridin by LC-MS. (a) Representative peaks identified in TOF spectra corresponding to fully modified, processed corynaridin with a monoisotopic mass of 3,961.19 Da carrying seven (right), eight (middle), or nine (left) positive charges (protons); (b) schematic representation of mature corynaridin with dehydration of all threonine/serine residues to dehydrobutyrine (DhB) and dehydroalanine (Dha).
Corynaridin exhibits bactericidal activity.
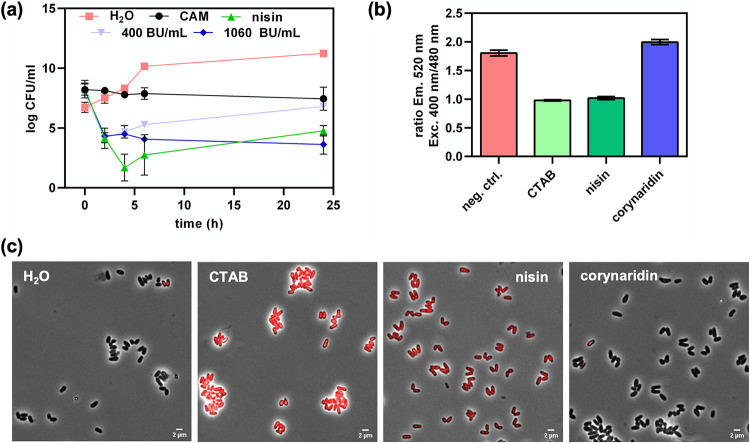
To elucidate whether corynaridin shows bactericidal or bacteriostatic activity, we employed time-kill assays with C. glutamicum ATCC 13032 as an indicator. As controls for bacteriostatic and bactericidal compounds, we used the antibiotic chloramphenicol and the pore-forming class I bacteriocin nisin, respectively (Fig. 6a). Addition of the vehicle H2O to C. glutamicum cultures had no effect on cell viability and CFU/mL steadily increased over the course of the experiment. In contrast, cultures treated with 1.25 μg/mL nisin showed a >4-log-reduced number of CFU/mL after 2 h of cultivation as a consequence of the bactericidal effect of the peptide (Fig. 6a). Addition of 6.5 μg/mL (bacteriostatic) chloramphenicol resulted in stable levels of CFU per milliliter throughout the experiment. Corynaridin-containing RPC fractions at 400 BU/mL and 1,060 BU/mL decreased the CFU per milliliter of the indicator after 2 h by 4 orders of magnitude, indicating bactericidal activity of the peptide. While cultures treated with the high concentration remained at low levels of CFU/mL, those treated with the lower concentrations showed slightly increased CFU/mL after 24 h, similar to nisin-treated samples, indicating a bactericidal mode of action of corynaridin.
FIG 6.
Bactericidal activity of corynaridin. (a) Time-kill assays with C. glutamicum ATCC 13032 as indicator, 1.25 μg/mL nisin, 6.5 μg/mL chloramphenicol, and H2O as controls, and corynaridin (RPC fractions at 400 and 1060 BU/mL). (b) Fluorescence intensity (emission at 50 nm) ratios at 400 and 480 nm of excitation for H2O (negative control [neg. ctrl.]), with CTAB, nisin, and corynaridin added to C. glutamicum ATCC 13032/pPB-pHin2Cg cells to monitor membrane damage. Values are means and standard deviations from at least three biological replicates. (c) Fluorescence microscopic pictures merged with phase-contrast images (×63) of propidium iodide-stained C. glutamicum ATCC 13032 cells after treatment with H2O, 0.05% CTAB, 1.25 μg/mL nisin, and 4,000-BU/mL RPC fraction containing corynaridin.
As bactericidal activity of bacteriocins is often related to the formation of pores in the bacterial membrane, we tested whether corynaridin causes membrane damage using a fluorescence-based whole-cell biosensor assay with C. glutamicum ATCC 13032/pPB-pHinCg (34). The strain harbors a plasmid for expression of pHluorin2, a fluorescent protein with a pH-dependent, bimodal excitation spectrum which is highly suitable to determine bacteriocin-driven pore formation (35). As expected, the detergent CTAB and pore-forming nisin led to reduced fluorescence ratios for C. glutamicum ATCC 13032/pPB-pHinCg, suggesting membrane damage (Fig. 6b). In contrast, addition of the RPC-purified corynaridin preparation at concentrations up to 10,000 BU/mL did not result in a change of fluorescence ratios. These results were further supported by fluorescence microscopy of propidium iodide-stained bacteria, which provided no evidence for compromised integrity of the bacteria (Fig. 6c). Thus, our data strongly suggest that unlike nisin and other bactericidal bacteriocins, corynaridin does not act by damaging the membrane of target cells under the tested conditions but by an as yet unknown bactericidal mode of action.
DISCUSSION
In this study, we identified a putative linaridin BGC in the genome of C. lactis RW3-42, which was isolated from raw cow’s milk (31). Deletion of the peptide precursor gene crdA in the genome of C. lactis RW3-42 completely abolished antimicrobial activity of the strain and thus confirmed the identified BGC as locus for the biosynthesis of an antimicrobial compound, which we designated corynaridin. Further in silico analyses revealed several adjacent genes for putative modification enzymes that are typically associated with the linaridin family of RiPPs (24). The absence of a decarboxylase gene in the C. lactis BGC suggests that the peptide is a type B linaridin, such as legonaridin or mononaridin (27, 36). In contrast, the corynaridin gene cluster also lacks a methyltransferase gene, which makes it unique among the hitherto described BGCs coding for peptides of the linaridin family. Mass spectrometry of the purified peptide revealed that corynaridin is processed at the predicted hexapeptide motif and displays a mass of 3,961.19 Da with all seven threonines as well as the serine dehydrated to dehydrobutyrines and dehydroalanine, respectively. The LC-MS data also suggest that the peptide is not dimethylated at its N terminus like other linardins (24). This is in line with the lack of a linM homologue for a methyltransferase in the corynaridin gene cluster. Interestingly, we also observed variants of the mature peptide with fewer threonine/serine residues dehydrated. Whether these peptides are artifacts of the sample preparation or represent true biological variation of the same precursor peptide that may also have different activity and spectra of target organisms needs to be addressed in future studies.
Purified corynaridin was stable at temperatures up to 100°C but lost activity after incubation with proteinase K or trypsin. Heat stability is a favorable trait of many bacteriocins and was also shown for, e.g., nisin (37). Interestingly, corynaridin obviously has a bactericidal mode of action against C. glutamicum ATCC 13032 in liquid cultivations but did not lead to the formation of pores as described for other class I bacteriocins such as nisin (38). As to our knowledge no receptor has been identified for the linaridins described so far, it remains to be investigated how these peptides exert their selective antimicrobial activity and, in some cases, cytotoxic activity against cancer cell lines (26).
Characterization of corynaridin revealed antimicrobial activity against several other actinobacteria comprising pathogenic and commensal Corynebacterium species as well as M. luteus and C. acnes, but it showed only low or no activity against a selection of Firmicutes and Gram-negative bacteria. C. lactis and other Corynebacterium species are frequently found in raw milk (14, 31), and some isolates of Corynebacterium bovis and C. amycolatum are causative agents of mastitis in dairy cows (39, 40). C. lactis was eventually linked to infections in companion animals, but it is not considered a pathogen so far and the determinants of infection remain unclear (41, 42). Interestingly, a putative BGC similar to that of corynaridin exists in the genome of an isolate of C. striatum from an intensive care unit (33). C. striatum is a ubiquitous species and part of the human microbiome but can also lead to infections in immunocompromised patients (43, 44). Moreover, Georgiou et al. predicted linaridin BGCs in six Corynebacterium spp., including the ones in C. lactis and C. striatum but also in a C. diphtheriae strain (8). These bacteria are not only phylogenetically related but might also be competitors in their respective habitats. Thus, secretion of bacteriocins by corynebacteria might be a hitherto largely neglected mechanism for intraspecies competition as described for other bacterial groups (e.g., lactic acid bacteria) and might have an impact on the composition of (actino)bacterial communities in general (20, 45).
In conclusion, we identified a novel, heat-stable bacteriocin produced by C. lactis RW3-42 that exerts narrow-spectrum bactericidal activity against other Actinobacteria—mainly Corynebacterium species. Our experiments suggest that corynaridin has a yet undescribed, bactericidal mode of action that does not involve pore formation. As other linaridins also showed growth-suppressing activity against cancer cell lines, they might be promising candidates for biotechnological exploitation.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
The bacterial strains used in this study (Table 2) were cultivated in GM17 medium (L. lactis IL-1403), MRS medium (Lactiplantibacillus plantarum DSM1055, P. acidilactici 347) or brain heart infusion (BHI) medium (all others) at 37°C (E. coli MG1655, Listeria innocua LMG2785, L. monocytogenes EGD-e, S. aureus ATCC 29213, Staphylococcus epidermidis DSM 3269, C. acnes DSM 16379, C. canis DSM 45402, Corynebacterium efficiens DSM 44569, C. amycolatum DSM 6922, C. striatum DSM 20668, Corynebacterium lipophiloflavum DSM 44291) for the indicated bacteria listed in parentheses or 30°C for all others, respectively. Solidified medium was prepared by addition of 16 g agar per L to the medium. For growth characterization and production, C. lactis strains were cultivated in Corynebacterium lactis (CL) medium I (CLI) (21 g/L MOPS [morpholinepropanesulfonic acid], 1 g/L K2HPO4, 1 g/L KH2PO4, 16 g/L tryptone, 10 g/L yeast extract, 0.25 g/L MgSO4, 0.01 g/L CaCl2, 0.2 mg/L biotin [pH adjusted to 7]) or CLIV [21 g/L MOPS, 1 g/L KH2PO4, 1 g/L K2HPO4, 10 g/L (NH4)2SO4, 0.1 g/L glucose, 0.2% Tween 80, SL10 trace elements, 0.2 mg/L biotin 0.01 g/L CaCl2, 0.25 g/L MgSO4 (pH adjusted to 7)]. For time-kill kinetics, 2× TY medium (16 g/L tryptone, 10 g/L yeast extract, 5 g/L NaCl) was used for the cultivation of C. glutamicum ATCC 13032. Cultivation medium of strains carrying plasmids additionally contained kanamycin (25 μg/mL) or chloramphenicol (15 μg/mL). Growth was monitored photometrically by measuring the optical density at 600 nm (OD600) at the indicated time points.
TABLE 2.
Bacterial strains and plasmids used in this study
| Species or plasmid | Strain or plasmid characteristic | Source or referencea |
|---|---|---|
| Species | ||
| Bacillus subtilis | DSM 402 | DSMZ |
| Corynebacterium lactis | RW3-42 | 31 |
| RW3-42 ΔcrdA | This study | |
| Corynebacterium ammoniagenes | DSM 20306 | DSMZ |
| Corynebacterium amycolatum | DSM 6922 | DSMZ |
| Corynebacterium canis | DSM 45402 | DSMZ |
| Corynebacterium casei | DSM 44701 | DSMZ |
| Corynebacterium efficiens | DSM 44549 | DSMZ |
| Corynebacterium glutamicum | ATCC 13032 | ATCC |
| Corynebacterium lipophiloflavum | DSM 44291 | DSMZ |
| Corynebacterium striatum | DSM 20668 | DSMZ |
| Corynebacterium xerosis | DSM 20743 | DSMZ |
| Cutibacterium acnes | DSM 16379 | DSMZ |
| Escherichia coli | MG1655 | 52 |
| Lactiplantibacillus plantarum | DSM 1055 | DSMZ |
| Lactococcus lactis | IL1403 | 53 |
| Listeria innocua | LMG2785 | 54 |
| Listeria monocytogenes | EGD-e | 55 |
| Micrococcus luteus | DSM 20030 | 56 |
| Pediococcus acidilactici | 347 | 57 |
| Pseudomonas fluorescens | DSM 50090 | DSMZ |
| Staphylococcus aureus | ATCC 29213 | ATCC |
| Staphylococcus epidermidis | DSM 3269 | 58 |
| Plasmids | ||
| pk19mobsacB | Kmr mobilizable (oriT); oriV | 59 |
| pk19mobsacB_del-crdA | pK19 derivative with 750-bp flanking regions of crdA gene of C. lactis RW3-42 genome | This study |
| pPB-pHin2Cg | pPBEx2 derivative with codon-optimized pHluorin2 gene under control of Ptuf; Kmr; pMB1 origin; pBL1 origin | 34 |
| pBAD33 | Cmr; pACYC184/p15A origin; araC | 60 |
| pBAD33_crd | pBAD33 derivative with crd gene cluster under control of PBAD | This study |
DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen; ATCC, American Type Culture Collection.
Molecular biology procedures.
Construction of pk19mobsacB_del-crdA and pBAD33_crd was carried out with standard reagents and according to protocols of the manufacturers. Oligonucleotides are listed in Table S1 in the supplemental material and were obtained from Eurofins Genomics (Ebersberg, Germany). PCR was performed in a C100 thermocycler (Bio-Rad Laboratories, Munich, Germany) using Q5 high-fidelity polymerase (New England Biolabs, Ipswich, USA) and nucleotides from Bio-Budget (Krefeld, Germany). The up- and downstream regions of the crdA gene and the crd gene cluster were amplified using oligonucleotides creating overlapping ends for Gibson assembly. The empty vector pK19mobsacB was linearized by restriction endonucleases EcoRI and SalI. The empty vector pBAD33 was linearized by restriction endonucleases KpnI and SalI. The final plasmids were verified by sequencing (Eurofins Genomics, Ebersberg, Germany). All plasmids and their relevant characteristics are listed in Table 2. For transformation, C. lactis was rendered electrocompetent and transformed as described previously for C. glutamicum (46).
In silico analyses.
Prediction of bacteriocin gene clusters (BGCs) in the genome of C. lactis RW2-5 was carried out using BAGEL4 (47). Subsequently, blastp analysis (48) was performed using the deduced proteins with standard parameters (BLOSUM62; gap existence costs, 11; gap extension cost, 1) and assigned to putative functions based on sequence similarities with published linaridin biosynthesis genes (24). The BGC of C. lactis RW3-42 was PCR amplified and cloned into pBAD33, followed by sequencing (Eurofins Genomics, Ebersberg, Germany). The resulting sequences were then aligned to the BGCs of C. lactis RW2-5 and the legonaridin biosynthesis genes. Sequence alignments were carried out using ClustalW (49) and visualized using Jalview (50).
Purification of corynaridin.
For purification of corynaridin, proteins of supernatants (1 L) collected after 24 h of cultivation of C. lactis RW3-42 in CLI containing 1% (wt/vol) glucose were precipitated using ammonium sulfate (50% [wt/vol] saturation) at 4°C overnight (16 h). The precipitate was collected by centrifugation (60 min, 10,000 × g, 4°C) and resuspended in 50 mL H2O, and pH was adjusted to 4.0 using 2 M HCl. An additional centrifugation step (10 min, 10,000 × g, 4°C) was performed to remove insoluble particles. All of the following chromatographic steps were carried out with an ÄKTA pure chromatography system (Cytiva). The solution containing the peptide was applied to a HiPrep SP FF 16/10 column (GE Healthcare Life Sciences) equilibrated with 20 mM sodium phosphate buffer at pH 3.9. Unbound proteins were washed out by 5 column volumes (CVs) of 20 mM sodium phosphate buffer at pH 6.9 (buffer A). The remaining bound peptides/proteins were then eluted in with 5 CVs of 20 mM sodium phosphate buffer at pH 6.9 with 2 M NaCl (buffer B). The eluate fractions containing the bacteriocin were identified by activity assays (described below) and directly applied to reversed-phase chromatography (RPC) using a 1-mL Resource RPC column (Cytiva) or stored at −20°C until further use. To remove weakly bound proteins, a washing step was carried out with 5 CVs of 2% acetonitrile in H2O plus 0.065% TFA. Elution was performed with an initial step to 15% acetonitrile for 5 CVs followed by a linear gradient up to 80% acetonitrile. To obtain corynaridin in a higher purity for liquid chromatography-mass spectrometry (LC-MS), C. lactis was cultivated in CLIV medium with 1% (wt/vol) glucose for 24 h and supernatants were harvested as described above. After adjustment of the pH of the solution to 4 and filtration through a 0.2-μm-pore filter, the solution was directly applied to a HiPrep SP FF 16/10 column (GE Healthcare Life Sciences) equilibrated with H2O at pH 4.3 (adjusted with HCl). Unbound proteins were washed out by 2 CVs of buffer A followed by a step to 2% buffer B for 3 CVs. Elution started with a step to 5% buffer B followed by a step to 30% buffer B and a final elution at 100% buffer B. Fractions with antimicrobial activity were identified by activity assays (see below), dried using a vacuum concentrator at 60°C (Eppendorf, Hamburg, Germany), and resuspended in HPLC-grade H2O. Protein concentrations of the purification fractions were estimated using the Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific) according to the manufacturer‘s protocol.
Radial streak.
Bioprospecting for antimicrobial activity was performed using a modified cross-streak method (51). Briefly, C. lactis RW3-42 was inoculated from an overnight culture as a single streak with an inoculation loop in the center of an BHI medim agar plate and incubated aerobically for at least 3 days at 30°C. Indicator bacteria (Table 2) were cultivated overnight in 5 mL BHI medium and streaked in a line from the border of the plate toward C. lactis. The plates were then incubated for 24 to 48 h at 30 to 37°C, depending on the indicator bacteria. In the case of C. acnes, incubation was carried out in an anaerobic jar (Merck KGaA, Darmstadt, Germany) containing an AnaeroGen anaerobic incubation system (Thermo Fisher Scientific).
Determination of antimicrobial activity.
Antimicrobial activity was determined using a spot-on-lawn assay. Overnight cultures of the respective strains were inoculated with an OD600 of 0.01 into hand-warmed agar medium (16 g/L agar) and poured into sterile petri dishes. After solidification, surfaces of agar plates were air dried at room temperature under a sterile hood. Supernatants and purification fractions were serially diluted and spotted onto agar plates. Plates were incubated under the preferred conditions of the embedded bacteria until growth was visible. Volumetric bacteriocin activity (bacteriocin units [BU] per milliliter) was determined by dividing the last dilution factor resulting in a visible zone of inhibition by the volume spotted.
Liquid chromatography-mass spectrometry.
Following RPC, purified protein fractions were concentrated to ~1 mg mL−1 with LC-MS-grade H2O prior to LC-MS measurements. LC-MS was performed on an Agilent 1260 Infinity system (Agilent Technologies, Waldbronn, Germany) coupled to a quadrupole time-of-flight (QTOF) mass spectrometer (TripleTOF6600; AB Sciex, Darmstadt, Germany). LC was performed with an Ascentis Express peptide ES-C18, 2.7-μm HPLC column (53307-U; Merck, Darmstadt, Germany) with a flow rate of 200 μL min−1 and the mobile phases A (0.1% formic acid in water) and B (acetonitrile). Ultraperformance liquid chromatography (UPLC)-MS-grade 0.1% formic acid in H2O and acetonitrile were obtained from Biosolve BV (Valkenswaard, Netherlands). LC-MS-grade H2O was obtained from a Milli-Q water purification system (Merck Millipore, Burlington, MA, USA). The elution gradient was as follows: 0–70 min: linear gradient from 3 to 40% B, 70–78 min: constant 40% B, 78–79 min: step from 40 to 60% B, 79–89 min: constant 60% B, 89–90 min: step from 60 to 3% B followed by a 12-min equilibration time between injections. The column temperature was set to 21°C and injection volume to 10 μL. MS was conducted with a TurboV ion source operated in positive-ionization mode. The ion spray voltage was set to 5.5 kV, source temperature to 450°C, curtain gas to 35 lb/in2, and the support gases GS1/GS2 to 50 lb/in2/50 lb/in2. All gases were nitrogen. The QTOF mass spectrometer was operated in TOF scan mode with a dwell time of 250 ms. The declustering potential was set to 120 V and mass tolerance to 25 ppm.
Acquired mass spectra were analyzed with Python 3.9.7 and the packages pyopenms 2.7.0, pandas 1.3.4, numpy 1.21.3, pathlib 1.0.1 and matplotlib 3.4.3. TOF spectra were centroided with pyopenms and subsequently filtered for potential peptide mass-to-charge (m/z) ratios. Corynaridin peptide variants with different numbers of dehydrated serine or threonine residues were considered identified when m/z peaks had a minimum signal intensity of 103 cps for at least three charged peptide species with at least four isotopes.
Time-kill kinetics.
Fresh overnight cultures of C. glutamicum ATCC 13032 were used to inoculate 5 mL 2× TY in glass tubes with a starting OD600 of 0.5 (i.e., ~107 CFU/mL). Samples of interest were added at the indicated concentrations prior to inoculation of the medium. Cultures were then incubated on a rotary shaker at 130 rpm for 24 h at 30°C. Samples were collected at the indicated time points, diluted (10−1 to 10−8), and plated on 2× TY agar. CFU per milliliter were determined after 24 to 48 h of incubation of the plates at 30°C by counting the colonies for the respective dilution.
pHluorin assay.
For detection of membrane damage, a pHluorin assay was conducted as described earlier (34). In particular, a 5-mL BHI overnight culture containing kanamycin (50 μg/mL) of the sensor strain C. glutamicum ATCC 13032/pPB-pHin2Cg was harvested by centrifugation and resuspended to an OD600 of 3 in Listeria minimal buffer [LMB: 100 mM MOPS, 4.82 mM KH2PO4, 11.52 mM Na2HPO4, 1.7 mM MgSO4, 0.6 g/L (NH4)2SO4, 55 mM glucose (pH 6.2)]. Serial 2-fold dilutions of samples were prepared in black 96-well microtiter plates (Sarsted, Nümbrecht, Germany) with a final volume of 100 μL in each well. Subsequently, 100 μL of the sensor strain suspension was added and the plate was incubated at room temperature in the dark for 30 min. Then, pHluorin2 fluorescence was measured at 520 nm with excitation at the distinct maxima at 400 and 480 nm using an infinite M200 plate reader (Tecan, Männedorf, Switzerland).
Fluorescence microscopy.
A fresh culture of C. glutamicum ATCC 13032 was washed once in phosphate-buffered saline (PBS), and bacteria were resuspended in saline (0.9% [wt/vol] NaCl) at an OD600 of 1. An 87.5-μL concentration of the cell suspension was mixed with nisin, cetyltrimethylammonium bromide (CTAB), H2O, or the RPC fraction containing corynaridin to the indicated concentrations and incubated for 10 to 30 min in the dark. Then, the bacteria were stained using 12.5 μL propidium iodide (25 μg/mL) (Invitrogen, Darmstadt, Germany) and again incubated for 15 min in the dark. Samples were imaged using a Axio Observer Z1 (Zeiss, Oberkochen, Germany) in bright-field and fluorescence mode with a filter set for propidium iodide (excitation at 575 to 625 nm, emission at 660 to 710 nm). Images were acquired with a 63× lens objective and analyzed using the Zen software (version 2.3 SP1; Zeiss).
ACKNOWLEDGMENTS
This study was partially funded by a grant of the German Ministry for Education and Research to C.U.R. and B.J.E. within the AMPLIFY consortium (grant no. 031B0826A). The funding bodies had no role in the design of the study, analysis of the data, or writing of the manuscript.
We declare no conflict of interest.
E.P. performed experimental work and was involved in writing, review, and editing of the manuscript. S.J.R. performed the pHluorin experiments and analysis and was involved in writing, review, and editing of the manuscript. A.R. performed mass spectrometry, analyzed data, and was involved in review and editing of the manuscript. D.W. was involved in establishing the activity measurements and was involved in writing, review, and editing of the manuscript. B.J.E. was involved in acquisition of funding, conceptualization of the study, and writing, review, and editing of the manuscript. M.O. provided access to mass spectrometry equipment, analyzed data, and was involved in review and editing of the manuscript. C.U.R. was involved in acquisition of funding, conceptualization of the study and writing, review, and editing of the manuscript. O.G. performed experimental work, analyzed and visualized all data, and was involved in conceptualization of the study, writing, review, and editing of the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Christian U. Riedel, Email: christian.riedel@uni-ulm.de.
Oliver Goldbeck, Email: oliver.goldbeck@uni-ulm.de.
Jan Claesen, Lerner Research Institute.
REFERENCES
- 1.Alvarez-Sieiro P, Montalbán-López M, Mu D, Kuipers OP. 2016. Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol 100:2939–2951. doi: 10.1007/s00253-016-7343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chikindas ML, Weeks R, Drider D, Chistyakov VA, Dicks LM. 2018. Functions and emerging applications of bacteriocins. Curr Opin Biotechnol 49:23–28. doi: 10.1016/j.copbio.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins—a viable alternative to antibiotics? Nat Rev Microbiol 11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 4.Field D, Ross RP, Hill C. 2018. Developing bacteriocins of lactic acid bacteria into next generation biopreservatives. Curr Opin Food Sci 20:1–6. doi: 10.1016/j.cofs.2018.02.004. [DOI] [Google Scholar]
- 5.Soltani S, Hammami R, Cotter PD, Rebuffat S, Said LB, Gaudreau H, Bédard F, Biron E, Drider D, Fliss I. 2021. Bacteriocins as a new generation of antimicrobials: toxicity aspects and regulations. FEMS Microbiol Rev 45:fuaa039. doi: 10.1093/femsre/fuaa039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riley MA, Wertz JE. 2002. Bacteriocins: evolution, ecology, and application. Annu Rev Microbiol 56:117–137. doi: 10.1146/annurev.micro.56.012302.161024. [DOI] [PubMed] [Google Scholar]
- 7.Heilbronner S, Krismer B, Brötz-Oesterhelt H, Peschel A. 2021. The microbiome-shaping roles of bacteriocins. Nat Rev Microbiol 19:726–739. doi: 10.1038/s41579-021-00569-w. [DOI] [PubMed] [Google Scholar]
- 8.Georgiou MA, Dommaraju SR, Guo X, Mast DH, Mitchell DA. 2020. Bioinformatic and reactivity-based discovery of linaridins. ACS Chem Biol 15:2976–2985. doi: 10.1021/acschembio.0c00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldbeck O, Weixler D, Eikmanns BJ, Riedel CU. 2021. In silico prediction and analysis of unusual lantibiotic resistance operons in the genus Corynebacterium. Microorganisms 9:646–618. doi: 10.3390/microorganisms9030646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomes KM, Duarte RS, Bastos MdCdF. 2017. Lantibiotics produced by Actinobacteria and their potential applications (a review). Microbiology (Reading) 163:109–121. doi: 10.1099/mic.0.000397. [DOI] [PubMed] [Google Scholar]
- 11.Poorinmohammad N, Bagheban-Shemirani R, Hamedi J. 2019. Genome mining for ribosomally synthesised and post-translationally modified peptides (RiPPs) reveals undiscovered bioactive potentials of actinobacteria. Antonie Van Leeuwenhoek 112:1477–1499. doi: 10.1007/s10482-019-01276-6. [DOI] [PubMed] [Google Scholar]
- 12.Brennan NM, Brown R, Goodfellow M, Ward AC, Beresford TP, Simpson PJ, Fox PF, Cogan TM. 2001. Corynebacterium mooreparkense sp. nov. and Corynebacterium casei sp. nov., isolated from the surface of a smear-ripened cheese. Int J Syst Evol Microbiol 51:843–852. doi: 10.1099/00207713-51-3-843. [DOI] [PubMed] [Google Scholar]
- 13.Byrd AL, Belkaid Y, Segre JA. 2018. The human skin microbiome. Nat Rev Microbiol 16:143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 14.Hahne J, Kloster T, Rathmann S, Weber M, Lipski A. 2018. Isolation and characterization of Corynebacterium spp. from bulk tank raw cow’s milk of different dairy farms in Germany. PLoS One 13:e0194365-16. doi: 10.1371/journal.pone.0194365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernard K. 2012. The genus Corynebacterium and other medically relevant coryneform-like bacteria. J Clin Microbiol 50:3152–3158. doi: 10.1128/JCM.00796-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alibi S, Ferjani A, Boukadida J, Cano ME, Fernández-Martínez M, Martínez-Martínez L, Navas J. 2017. Occurrence of Corynebacterium striatum as an emerging antibiotic-resistant nosocomial pathogen in a Tunisian hospital. Sci Rep 7:9704. doi: 10.1038/s41598-017-10081-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cazanave C, Greenwood-Quaintance KE, Hanssen AD, Patel R. 2012. Corynebacterium prosthetic joint infection. J Clin Microbiol 50:1518–1523. doi: 10.1128/JCM.06439-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otsuka Y, Ohkusu K, Kawamura Y, Baba S, Ezaki T, Kimura S. 2006. Emergence of multidrug-resistant Corynebacterium striatum as a nosocomial pathogen in long-term hospitalized patients with underlying diseases. Diagn Microbiol Infect Dis 54:109–114. doi: 10.1016/j.diagmicrobio.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Silva-Santana G, Silva CMF, Olivella JGB, Silva IF, Fernandes LMO, Sued-Karam BR, Santos CS, Souza C, Mattos-Guaraldi AL. 2021. Worldwide survey of Corynebacterium striatum increasingly associated with human invasive infections, nosocomial outbreak, and antimicrobial multidrug-resistance, 1976–2020. Arch Microbiol 203:1863–1880. doi: 10.1007/s00203-021-02246-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP. 2016. Staphylococcus aureus shifts toward commensalism in response to Corynebacterium species. Front Microbiol 7:1230. doi: 10.3389/fmicb.2016.01230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abrehem K, Zamiri I. 1983. Production of a bacteriocin, ulceracin 378, by Corynebacterium ulcerans. Antimicrob Agents Chemother 24:262–267. doi: 10.1128/AAC.24.2.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pátek M, Hochmannová J, Nešvera J, Stránský J. 1986. Glutamicin CBII, a bacteriocin-like substance produced by Corynebacterium glutamicum. Antonie Van Leeuwenhoek 52:129–140. doi: 10.1007/BF00429316. [DOI] [PubMed] [Google Scholar]
- 23.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl HG, Schmidt EW, Selsted ME, et al. 2013. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma S, Zhang Q. 2020. Linaridin natural products. Nat Prod Rep 37:1152–1163. doi: 10.1039/c9np00074g. [DOI] [PubMed] [Google Scholar]
- 25.Claesen J, Bibb MJ. 2011. Biosynthesis and regulation of grisemycin, a new member of the linaridin family of ribosomally synthesized peptides produced by Streptomyces griseus IFO 13350. J Bacteriol 193:2510–2516. doi: 10.1128/JB.00171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minami Y, Yoshida K, Azuma R, Urakawa A, Kawauchi T, Otani T, Komiyama K, Omura S. 1994. Structure of cypemycin, a new peptide antibiotic. Tetrahedron Lett 35:8001–8004. doi: 10.1016/S0040-4039(00)78407-3. [DOI] [Google Scholar]
- 27.Rateb ME, Zhai Y, Ehrner E, Rath CM, Wang X, Tabudravu J, Ebel R, Bibb M, Kyeremeh K, Dorrestein PC, Hong K, Jaspars M, Deng H. 2015. Legonaridin, a new member of linaridin RiPP from a Ghanaian Streptomyces isolate. Org Biomol Chem 13:9585–9592. doi: 10.1039/c5ob01269d. [DOI] [PubMed] [Google Scholar]
- 28.Claesen J, Bibb M. 2010. Genome mining and genetic analysis of cypemycin biosynthesis reveal an unusual class of posttranslationally modified peptides. Proc Natl Acad Sci USA 107:16297–16302. doi: 10.1073/pnas.1008608107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanki K, Kazuhiko O, Toshiaki S, Kazuro S, Hong Y, Yoko T, Masahiko H, Toshio O. 1993. New antibiotic, cypemycin taxonomy, fermentation, isolation and biological characteristics. J Antibiot (Tokyo) 46:1666–1671. [DOI] [PubMed] [Google Scholar]
- 30.Shang Z, Winter JM, Kauffman CA, Yang I, Fenical W. 2019. Salinipeptins: integrated genomic and chemical approaches reveal unusual d-amino acid-containing ribosomally synthesized and post-translationally modified peptides (RiPPs) from a Great Salt Lake Streptomyces sp. ACS Chem Biol 14:415–425. doi: 10.1021/acschembio.8b01058. [DOI] [PubMed] [Google Scholar]
- 31.Wiertz R, Schulz SC, Müller U, Kämpfer P, Lipski A. 2013. Corynebacterium frankenforstense sp. nov. and Corynebacterium lactis sp. nov., isolated from raw cow milk. Int J Syst Evol Microbiol 63:4495–4501. doi: 10.1099/ijs.0.050757-0. [DOI] [PubMed] [Google Scholar]
- 32.Mo T, Liu WQ, Ji W, Zhao J, Chen T, Ding W, Yu S, Zhang Q. 2017. Biosynthetic insights into linaridin natural products from genome mining and precursor peptide mutagenesis. ACS Chem Biol 12:1484–1488. doi: 10.1021/acschembio.7b00262. [DOI] [PubMed] [Google Scholar]
- 33.Roach DJ, Burton JN, Lee C, Stackhouse B, Butler-Wu SM, Cookson BT, Shendure J, Salipante SJ. 2015. A year of infection in the intensive care unit: prospective whole genome sequencing of bacterial clinical isolates reveals cryptic transmissions and novel microbiota. PLoS Genet 11:e1005413–21. doi: 10.1371/journal.pgen.1005413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weixler D, Berghoff M, Ovchinnikov KV, Reich S, Goldbeck O, Seibold GM, Wittmann C, Bar NS, Eikmanns BJ, Diep DB, Riedel CU. 2022. Recombinant production of the lantibiotic nisin using Corynebacterium glutamicum in a two-step process. Microb Cell Fact 21:11. doi: 10.1186/s12934-022-01739-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crauwels P, Schäfer L, Weixler D, Bar NS, Diep DB, Riedel CU, Seibold GM. 2018. Intracellular phluorin as sensor for easy assessment of bacteriocin-induced membrane-damage in Listeria monocytogenes. Front Microbiol 9:3038. doi: 10.3389/fmicb.2018.03038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang F, Wei W, Zhao J, Mo T, Wang X, Huang X, Ma S, Wang S, Deng Z, Ding W, Liang Y, Zhang Q. 2021. Genome mining and biosynthesis study of a type B linaridin reveals a highly versatile α-N-methyltransferase. CCS Chem 3:1049–1057. doi: 10.31635/ccschem.020.202000247. [DOI] [Google Scholar]
- 37.Rollema HS, Kuipers OP, Both P, De Vos WM, Siezen RJ. 1995. Improvement of solubility and stability of the antimicrobial peptide nisin by protein engineering. Appl Environ Microbiol 61:2873–2878. doi: 10.1128/aem.61.8.2873-2878.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bierbaum G, Sahl H-G. 2009. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr Pharm Biotechnol 10:2–18. doi: 10.2174/138920109787048616. [DOI] [PubMed] [Google Scholar]
- 39.Hommez J, Devriese LA, Vaneechoutte M, Riegel P, Butaye P, Haesebrouck F. 1999. Identification of nonlipophilic corynebacteria isolated from dairy cows with mastitis. J Clin Microbiol 37:954–957. doi: 10.1128/JCM.37.4.954-957.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watts JL, Lowery DE, Teel JF, Rossbach S. 2000. Identification of Corynebacterium bovis and other coryneforms isolated from bovine mammary glands. J Dairy Sci 83:2373–2379. doi: 10.3168/jds.S0022-0302(00)75126-5. [DOI] [PubMed] [Google Scholar]
- 41.Antunes JdP, Ribeiro MG, Demoner LdC, Ramos JN, Baio PVP, Simpson-Louredo L, Santos CS, Hirata R, Ferioli RB, Romera ARC, Vieira VV, Mattos-Guaraldi AL. 2015. Cutaneous abscess caused by Corynebacterium lactis in a companion dog. Vet Microbiol 178:163–166. doi: 10.1016/j.vetmic.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 42.Lim FS, Loong SK, Khoo JJ, Tan KK, Zainal N, Abdullah MF, Khor CS, AbuBakar S. 2018. Identification and characterization of Corynebacterium lactis isolated from Amblyomma testudinarium of Sus scrofa in Malaysia. System Appl Acarol 23:1838–1844. doi: 10.11158/saa.23.9.10. [DOI] [Google Scholar]
- 43.Lee PP, Ferguson DA, Sarubbi FA. 2005. Corynebacterium striatum: an underappreciated community and nosocomial pathogen. J Infect 50:338–343. doi: 10.1016/j.jinf.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 44.McMullen AR, Anderson N, Wallace MA, Shupe A, Burnham CA. 2017. When good bugs go bad: epidemiology and antimicrobial resistance profiles of Corynebacterium striatum, an emerging multidrug-resistant, opportunistic pathogen. Antimicrob Agents Chemother 61:e01111-17. doi: 10.1128/AAC.01111-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kommineni S, Bretl DJ, Lam V, Chakraborty R, Hayward M, Simpson P, Cao Y, Bousounis P, Kristich CJ, Salzman NH. 2015. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 526:719–722. doi: 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Rest ME, Lange C, Molenaar D. 1999. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl Microbiol Biotechnol 52:541–545. doi: 10.1007/s002530051557. [DOI] [PubMed] [Google Scholar]
- 47.Van Heel AJ, De Jong A, Song C, Viel JH, Kok J, Kuipers OP. 2018. BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res 46:W278–W281. doi: 10.1093/nar/gky383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 50.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haber M, Ilan M. 2014. Diversity and antibacterial activity of bacteria cultured from Mediterranean Axinella spp. sponges. J Appl Microbiol 116:519–532. doi: 10.1111/jam.12401. [DOI] [PubMed] [Google Scholar]
- 52.Bachmann BJ. 1972. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev 36:525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, Ehrlich SD, Sorokin A. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res 11:731–753. doi: 10.1101/gr.gr-1697r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nilsen T, Nes IF, Holo H. 2003. Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl Environ Microbiol 69:2975–2984. doi: 10.1128/AEM.69.5.2975-2984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, Charbit A, Chetouani F, Couvé E, de Daruvar A, Dehoux P, Domann E, Domínguez-Bernal G, Duchaud E, Durant L, Dussurget O, Entian KD, Fsihi H, García-del Portillo F, Garrido P, Gautier L, Goebel W, Gómez-López N, Hain T, Hauf J, Jackson D, Jones LM, Kaerst U, Kreft J, Kuhn M, Kunst F, Kurapkat G, Madueno E, Maitournam A, Vicente JM, Ng E, Nedjari H, Nordsiek G, Novella S, de Pablos B, Pérez-Diaz JC, Purcell R, Remmel B, Rose M, Schlueter T, Simoes N, et al. 2001. Comparative genomics of Listeria species. Science 294:849–852. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- 56.Cohn F. 1872. Untersuchungen über Bakterien. Beitr Biol Pflanz 1:127–224. [Google Scholar]
- 57.Rodríguez JM, Cintas LM, Casaus P, Martínez MI, Suárez A, Hernández PE. 1997. Detection of pediocin PA-1-producing pediococci by rapid molecular biology techniques. Food Microbiol 14:363–371. doi: 10.1006/fmic.1996.0084. [DOI] [Google Scholar]
- 58.Peters G, Locci R, Pulverer G. 1982. Adherence and growth of coagulase-negative staphylococci on surfaces of intravenous catheters. J Infect Dis 146:479–482. doi: 10.1093/infdis/146.4.479. [DOI] [PubMed] [Google Scholar]
- 59.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. pK18mobsacB. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 60.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.01756-22-s0001.pdf, PDF file, 2.2 MB (2.2MB, pdf)