Abstract
The role of 3-dimensional (3D) genome organization in the precise regulation of gene expression is well established. Accordingly, the mechanistic connections between 3D genome alterations and disease development are becoming increasingly apparent. This opinion article provides a snapshot of our current understanding of the 3D genome alterations associated with cancers. We discuss potential connections of the 3D genome and cancer transcriptional addiction phenomenon as well as molecular mechanisms of action of 3D genome-disrupting drugs. Finally, we highlight issues and perspectives raised by the discovery of the first pharmaceutical strongly affecting 3D genome organization.
Keywords: 3D genome, transcriptional addiction, chromatin damage, CTCF, curaxins, topologically associating domains
3D genome and regulation of gene expression
Techniques exploiting the proximity ligation principle (C-methods, Box 1) have significantly improved our understanding of 3-dimensional (3D) genome organization. C-methods have confirmed the existence of chromosomal territories that are spatially compartmentalized into active and repressed chromatin domains, referred to as A and B compartments respectively (Figure 1a) [1]. Higher resolution analysis has demonstrated that these megabase-scale compartments are not uniform and represent a mosaic of smaller active or repressed compartmental domains [2]. On a sub-megabase level, the chromatin fiber is folded into topologically associating domains (TADs). The distinctive feature of TADs is that spatial contacts of remote genomic elements are more frequent within TADs than between individual TADs [3-5]. TADs are demarcated by boundaries – genomic regions that are conserved across cell types and are enriched with cohesin complex and CCCTC-binding factor (CTCF; see Glossary). The latter is a zinc finger protein that binds to DNA and, upon dimerization, tethers distant genomic regions [5]. Recent evidence suggests that TADs and compartmental domains are formed by distinct molecular mechanisms [2]. TADs are formed via dynamic DNA loop extrusion process [6, 7] and may harbour smaller contact domains, some of which are chromatin loops that mediate enhancer-promoter communications (EPCs). Chromatin loops may be generated/stabilized by specific architectural proteins, including CTCF, cohesin and Yin Yang 1 (YY1) (Figure 1a, reviewed in [8]).
Box 1. C-methods.
The current protocols to study 3D genome organization collectively referred to as C-methods are based on the Chromosome Conformation Capture (3C) procedure developed by J. Dekker [86]. To construct chromatin interaction maps reflecting the mode of chromatin packaging within the cell nucleus, a so-called proximity ligation protocol is applied. The key step of this procedure is breaking and re-ligation of DNA within the fixed nucleus. This procedure generates a certain amount of chimeric fragments due to cross-ligation of DNA fragments located far from each other on the DNA chain but close in physical space. These chimeric fragments are the spatial proximity records that can be decrypted using various analytical procedures, including sequencing. Analysis of the pools of chimeric fragments allows reconstruction of the spatial organization of the genome based on the sets of captured pairwise interactions. A number of protocols to address specific questions were developed based on the original 3C procedure (reviewed in [87]). Of special importance is Hi-C, a genome-wide protocol allowing for the analysis of the entire set of chromatin fiber spatial contacts [1]. The results of Hi-C analysis are presented in the form of 2D chromatin interaction heatmaps demonstrating the probability of interaction of any genomic segment with each other segment (Figure 1b). The resolution of maps (the size of map bin) depends on the frequency of DNA cutting by the particular restriction enzyme used and the deepness of the library sequencing and, in best cases, attends 1 kb [88]. At this resolution, one can detect both TADs and chromatin loops within TADs. The interaction frequency (P) between any two genomic fragments within the same chromosome arm depends, beside other reasons, on the distance between these fragments (s). At first approximation, the number of contacts decreases monotonically with increasing distance [1]. Abrupt changes in the curve P(s) indicate the presence of chromosomal rearrangements because the data obtained on cells with a rearranged genome are mapped on a standard genome. For example, within an inverted DNA segment, the P(s) will show an increase of contact probability instead of a decrease. Thus, Hi-C constitutes a powerful tool for detection of chromosomal rearrangements and copy number variation in human tumours [89, 90]. In some cases, it is desirable to focus analysis on a particular genome region rather than the entire genome. This aim can be achieved using various Capture-C protocols allowing selection from a Hi-C library a set of fragments complementary to a genomic region of interest.
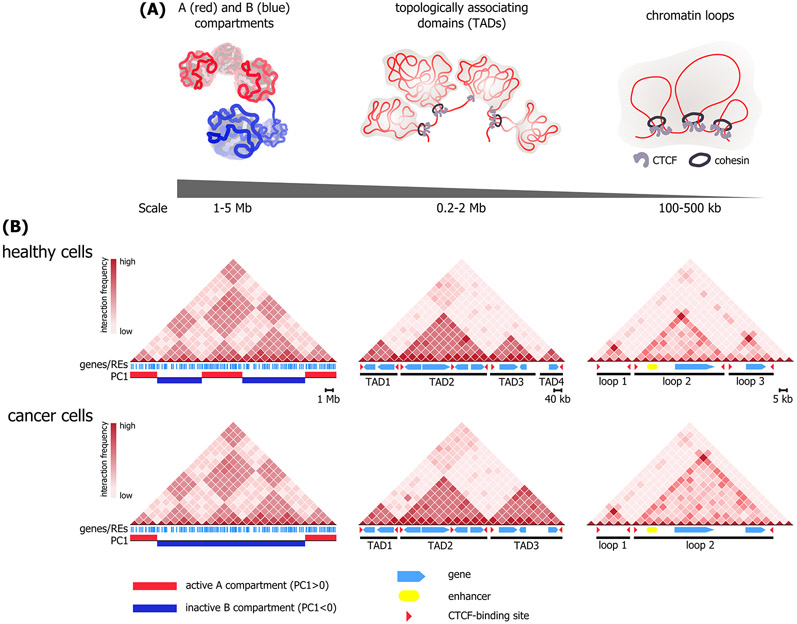
Figure 1. 3D genome organization in healthy and cancer cells.
(A) Hierarchical structure of interphase chromatin. Chromosome territories (left) are partitioned into A- (red) and B-compartments (blue) formed by long-range spatial interactions between distant genome loci and containing transcriptionally more active or repressed genome regions, respectively. At a sub-megabase level, chromatin is folded into topologically associating domains (TADs, center), commonly interpreted as self-interacting globular structures. The internal structure of TADs is represented by arrays of chromatin loops (contact domains, right) formed by spatial contacts between CCCTC-binding factor (CTCF)/cohesin-binding sites. (B) 3D genome organization properties of healthy and cancer cells. Colour intensity on an exemplary Hi-C maps (heatmaps) reflects average interaction frequency between corresponding genomic bins; the darkest red bins correspond to most frequent interactions. Genes and gene regulatory elements (enhancers, CTCF-binding sites; if appropriate) are shown below the heatmaps. Positions of chromatin compartments, TADs and chromatin loops are depicted under the appropriate heatmap. Principal component analysis characterizes the A/B status of chromatin compartments (A, principal component 1 (PC1) > 0, red; B PC1 < 0, blue).
Spatial genome organization represents an additional level in the epigenetic regulation of gene expression. In mammals, transcription is controlled by enhancers that are typically located distantly from target promoters. Recent evidence suggests that the transcription factor (TF)-bound enhancers attract transcription machinery and serve as nucleation centres for the assembly of a liquid active environment via liquid-liquid phase separation (LLPS) [9-11]. To be activated by an enhancer, a gene should be located within this compartment. Because most enhancers are located far from the target genes, this activation can be achieved by looping out the intervening DNA segment, and recent studies demonstrate that such situations are quite common [12-14]. At the 3D genome organization level, it is also possible to unite several individual enhancers in a common activating complex, as in the case of super-enhancers (SEs), composed of several functional enhancers [15]. The number of enhancers in mammalian and Drosophila cells is at least ten times higher than the number of genes [16, 17]. The possibility of gene activation by various combinations of enhancers increases the regulatory capacity of the eukaryotic cell transcription control system. Indeed, simple reconfiguration of an extended genomic region may be sufficient for certain genes’ activation by juxtaposing them to enhancers and for repression of other genes by juxtaposing them to silencers. TAD boundaries constitute an obstacle for efficient EPC, i.e. a particular enhancer usually activates the expression of genes located within the same TAD [18, 19]. Disruption of TAD boundaries frequently impairs transcription regulation and may potentially be the cause of various diseases including cancer [20, 21]. The present study focuses mainly on our current understanding of how the 3D genome organization is altered in transformed cells and what opportunities this understanding provides in terms of treating cancer. The vulnerability of cancer cells, which is associated with the 3D genome alterations, is discussed from the transcriptional phenomenon perspective. We also invite discussion on the possibility of curing cancer by direct manipulation of cancer 3D genome.
3D genome in cancer
Spatial organization of the genome was analyzed in a number of cancer types including breast cancer, prostate cancer, gliomas and several hematologic cancers (Figure 1) [22-30]. Noteworthy, the primary patient-derived cancer cells have been investigated only in case of acute leukaemia [22, 30] while in the remaining cases cancer cell lines have been used to study cancer-associated 3D genome changes. Generally, there are no qualitative differences between 3D genomes of healthy and cancer cells. All levels of genome folding (A/B compartments, TADs and chromatin loops) can be found in cancer cells although the structural variation of 3D cancer genome is slightly more pronounced [31]. The number and locations of megabase-scale A/B chromatin compartments in cancer cells do not significantly differ from the compartments in related healthy cells [23, 27, 28, 32]. In breast cancer, multiple myeloma, B-cell lymphoma and T-cell acute lymphoblastic leukaemia, a compartment type switch (A-to-B and vice versa) was reported for up to 20 % of genomic regions (Figure 1b) [23, 28, 32]. Considering chromatin compartments’ mapping algorithms, it is more likely that this compartment switch reflects changes in gene expression in cancer cells rather than plays a causative role in cancer development.
At the level of TADs, distinctions between healthy and cancer cells can be traced more clearly, although different cancer types are characterized by mutually exclusive changes. For some cancer types (breast and prostate cancers, multiple myeloma), there have been reported acquisition of new TAD boundaries, usually accompanied by the corresponding increase in TAD number and the decrease in their size (Figure 1b) [23, 24, 28, 29]. The extent, to which the amount of TADs is increased in cancers, can vary substantially for a given pair of healthy and cancer cells. While in multiple myeloma the number of TADs is increased by 25 % compared to normal B cells, 2-3-fold increase in the amount of TADs was reported in prostate cancer cell lines vs normal prostate epithelial cells [23, 24]. For other cancer types—gliomas, acute lymphoblastic leukaemias—weakening and/or disappearance of TAD boundaries are more typical [22, 25-27]. In B-cell lymphomas, there are no differences in the number and structure of TADs [32].
Formation of excessive cancer-specific TAD boundaries often occurs in the copy number variation (CNV) regions and correlates with changes in the epigenome and gene expression [23, 24, 29]. However, this correlation does not demonstrate the causative role of altered spatial genome organization in cancer development. Nonetheless, such an expansion in TAD number may make the cancer 3D genome and, subsequently, cancer-specific gene expression, vulnerable to 3D genome-disrupting agents.
The contribution of genome-wide and/or loci-specific disappearance of TAD boundaries to cell transformation is clearer [22, 25-27]. Loss of TAD boundaries in cancer is usually a consequence of mutation or downregulation of CTCF and other chromosome architecture proteins, or cancer-specific genetic alterations (mutations, deletions, duplications) of their binding sites [22, 25-27, 33]. These perturbations can lead to disruption of chromosome insulated neighbourhoods—CTCF/cohesin-mediated DNA loops that limit the action of gene regulatory elements like enhancers and SEs [25, 26, 34, 35]. This, in turn, can result in transcriptional activation of oncogenes. The direct contribution of this mechanism to cancer development was demonstrated in vitro using C-methods and the CRISPR/Cas9 technique [25, 26]. Cancer-promoting pleiotropic effects of other chromatin structural proteins–YY1 and BORIS (Brother Of the Regulator of Imprinted Sites, also known as CTCFL)–may be explained by their ability to form new cancer-specific chromatin loops leading to transcriptional dysregulation (particularly, to oncogenes activation and/or repression of tumour suppressors) [36, 37]. YY1 is overexpressed in a wide range of cancers; however, the proposed mechanism of its contribution to cell malignization remains speculative [36]. BORIS, the germ-cell-specific paralogue of CTCF, is overexpressed in several cancer types [37, 38]. Recently, it was demonstrated that BORIS upregulation promotes the establishment of new chromatin interactions in ALK-mutated neuroblastoma cells [37]. BORIS-mediated chromatin looping stimulates SE-driven transcription of a set of proneural TFs that results in acquired chemoresistance of the cells [37]. It seems possible to selectively eliminate cancer cells by modulating the activity of particular chromatin architecture proteins like YY1 and BORIS. Moreover, cancer-specific changes in the 3D genome can promote transcriptional addiction of cancer cells (see below) and make them sensitive to various agents targeting high-level transcription of oncogenes.
Chemotherapeutics exploiting cancer transcriptional addiction
Transcriptional addiction concept describes the state when, in order to support high proliferation rates and other needs, cancer cells become strongly dependent on transcriptional regulators, such as TFs, chromatin regulators and even basal transcription machinery [39]. It is well established that in many cancers, the malignant phenotype is based on the aberrant high-level expression of oncogenes, many of which represent master TFs [39]. Several conditions sustain high-level transcription of oncogenes (Figure 2). Contribution of distant SEs and typical enhancers to oncogene transcription makes it dependent on high levels of transcriptional co-activators (such as bromodomain and the extraterminal (BET) family of proteins and certain cyclin-dependent kinases (CDKs) [15] and chromatin-associated factors [40], on the 3D genome-mediated EPCs [41], and on the formation of phase-separated condensates [9, 11]. These dependencies make high-level expression of oncogenes more vulnerable for targeting abovementioned aspects of transcription, and provide an opportunity for selective elimination of cancer cells. Particularly, it was shown in vitro and in vivo that BRD4 inhibitors selectively downregulate the expression of cancer-promoting genes by compromising SEs and, thus, stimulate cancer cell death [42-44]; many of the BET inhibitors are currently undergoing clinical trials (reviewed in [45]). Pharmacological inhibition of cyclin-dependent kinase (CDK) 7 (with or without inhibition of CDK12/13) also alters SE function and suppresses cancer-specific transcription [46-50]. The histone chaperone FACT (FAcilitates Chromatin Transcription) is almost exclusively expressed in cancer cells [51, 52] and is involved in the regulation of transcription in cancer cells [40]. Depletion of active FACT by using nucleosome-destabilizing small molecules contributes to selective elimination of cancer cells [53, 54]. These examples demonstrate that the factors contributing to high-level oncogene transcription are promising molecular targets for anti-cancer chemotherapeutics. It is not clear whether disruption of phase-separated compartments (condensates), necessary for SE function, can be clinically exploited. Until now, there was only one small molecule, aliphatic alcohol 1,6-hexanediol, to alter LLPS in living cells, but it was non-specific and extremely toxic [55]. Cellular image-based compound screening identified lipoamide and lipoic acid as the non-toxic molecules that efficiently prevent LLPS-mediated stress granule formation [56]. These compounds prevent regression of amyotrophic lateral sclerosis patient-derived (FUS mutant) motor neuron axons in culture and recover motor defects in Drosophila melanogaster expressing FUS mutants [56]. The question of whether the 3D genome can be considered as a target for anti-cancer therapy is discussed below.
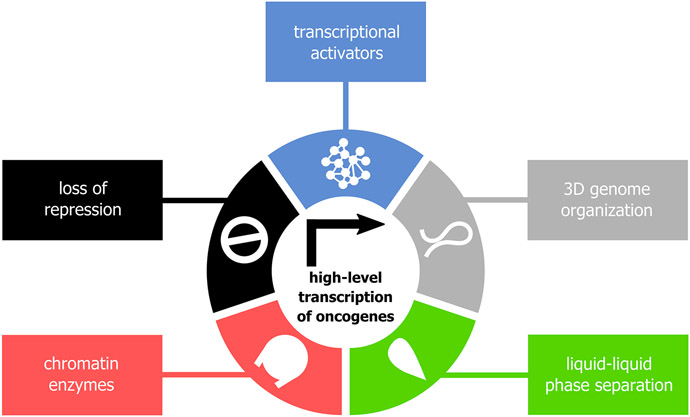
Figure 2. Factors sustaining high-level transcription of oncogenes.
High-level transcription of oncogenes often is driven by enhancers and super-enhancers. This makes it strongly dependent on several conditions: (1) high levels of transcriptional co-activators (blue), (2) the 3D genome-mediated enhancer-promoter communications (gray), (3) the formation of phase-separated condensates (green), (4) chromatin-associated factors (chromatin remodelers, histone chaperones and histone-modifying enzymes; red), and (5) the loss of transcriptional repression (black). These dependencies make possible targeting oncogenes’ transcription with minimal effects on basal transcription.
Approaches to modulate 3D genome
Most of the known methods for the large-scale interference with the spatial genome organization in living cells are genetic methods. It is well-established that 3D genome is mostly maintained by several proteins in interphase cells: CTCF, ring-shaped cohesin complex (SMC3, SMC1, RAD21 and STAG1/SA1 or STAG2/SA2) and factors that govern cohesin loading onto chromatin (NIPBL-MAU2), and its release from chromatin (WAPL) [57]. Downregulation of these factors inevitably affects 3D genome, although in different ways. It was demonstrated that depletion of CTCF, cohesin and cohesin-loading factor NIPBL in cultured cells compromises TADs and chromatin loops but does not significantly affect segregation of the genome into A and B compartments [58-60]. Downregulation of cohesin-unloading factor WAPL and its binding partners weakened compartments but increased the size of TADs and loops [61, 62]. Physical stresses such as hyperosmotic shock or heat shock can also induce rapid reversible changes in 3D genome in living cells [63-65]. However, the described approaches are not easily suitable for clinical applications.
The DNA-binding small molecules remain the mainstream of anti-cancer therapy. Their mechanism of action is traditionally attributed to DNA damage induction that leads to apoptosis [66]. Binding of small molecules to DNA not only compromises DNA integrity but also alters physical properties of the double helix, such as flexibility, charge and characteristics of the major and minor grooves [67-69]. These changes inevitably affect DNA interactions with histones and other chromatin-associated proteins [53, 70]. Nevertheless, the effects of DNA-binding compounds on the spatial genome organization had not been studied until recently [71].
3D genome as a target for anti-cancer therapy
It is now evident that 3D genome alterations can contribute to disease development. Recent analyses demonstrate that many single-nucleotide polymorphisms (SNPs), which have been identified by genome-wide association studies (GWAS) and have been linked to disease traits, are located within distant enhancers regulating gene expression through 3D genome-mediated EPCs [72, 73]. The insulated neighbourhoods concept also clarifies the impact of the 3D genome on disease development [74]. It was demonstrated in vitro that the deletion of prostate cancer risk-associated CTCF anchor regions, which created a local repressive regulatory environment (insulated neighbourhood), resulted in ~100-fold increase in expression level of genes promoting prostate cancer [35]. However, it was questionable whether 3D genome organization could be manipulated by suitable for clinical use chemical compounds in order to modulate disease-associated gene expression. Recently, a mechanism by which a small molecule—curaxin CBL0137—affects 3D genome organization and compromises long-range interactions of cis-regulatory elements has been reported [71]. Curaxins intercalate into DNA by protruding symmetrical side chains of the molecule (the structure of CBL0137 is shown in Figure 3) into the major groove of DNA and by filling the minor groove with its carbazole N-side chain [53]. Curaxins have unique features that distinguish them from most of the DNA-binding small molecules. Although curaxins are capable of inhibiting both topoisomerases I and II, they do not induce DNA damage [53, 54]. Curaxin binding to DNA results in genome-wide nucleosome destabilization [53, 75], which, in turn, generates superhelical tension that cannot be relieved due to topoisomerase inhibition and stimulates B- to Z-DNA transitions [53]. Indirect inhibition of FACT histone chaperone is the most studied activity of curaxins. It was shown in vitro and ex vivo that upon curaxin treatment histone chaperone FACT binds (1) to the multiple epitopes within the destabilized nucleosomes, and (2) to Z-DNA regions, that exhausts the cellular pool of active FACT [53, 54, 75].
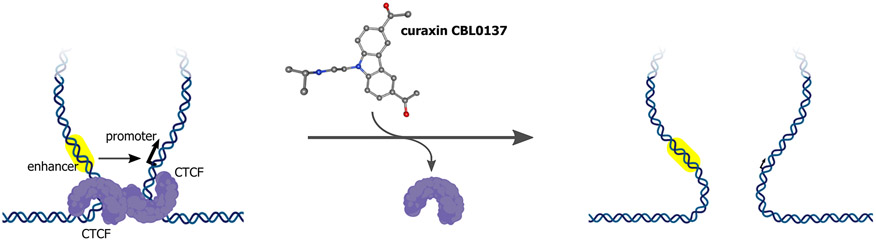
Figure 3. Mode of action of 3D genome destabilizing pharmaceuticals (curaxin CBL0137).
Curaxins intercalate into DNA and, thus, alter DNA topology, leading to the inability of CCCTC-binding factor (CTCF) to bind to its cognate DNA sites. This inability results in partial disruption of chromatin loops and in large-scale perturbations to 3D genome organization that affects enhancer-promoter communications and leads to preferential downregulation of enhancer- and super-enhancer-driven transcription of oncogenes.
Intercalation of the curaxins into DNA results in the inability of CTCF to bind to its cognate genomic sites [71]. This, in turn, stimulates partial disruption of chromatin loops and large-scale perturbations to 3D genome organization that affect EPCs and lead to preferential downregulation of enhancer and SE-driven transcription of oncogenes (Figure 3). The selective cytotoxicity of curaxins towards melanoma, neuroblastoma, glioblastoma, MLL-rearranged leukaemia and small-cell lung cancer cells was reported in a number of preclinical studies [76-81]. At least in part, the efficacy of curaxins in preclinical studies can be explained by their 3D genome-modulating properties as evidenced by the fact that curaxins downregulate MYC expression in cancer cells that could be a result of disruption of 3D genome structure. Nonetheless, it is clear that, used beyond their therapeutic window, curaxins would be toxic for healthy cells as well.
CTCF is the key 3D genome-maintaining factor, and its careful targeting may be considered as an advantageous therapeutic strategy for downregulating high-level oncogene expression. CTCF binding sites are not equal–some of them are not involved in maintaining general 3D genome organization but are rather positioned in promotor and enhancer regions that directly involve CTCF in transcriptional regulation [82]. Recent evidence suggests that CTCF’s binding sites are located in a subset of SEs and that CTCF plays a critical role in SE function [83]. Further analysis has to be performed to ascertain what kind of CTCF-binding sites lack this factor upon cell treatment with curaxins. Although DNA methylation-based molecular tools for loci-specific disruption of CTCF binding sites have been reported [84], small molecule compounds specifically inhibiting DNA-binding or dimerization (oligomerization) activities of CTCF have yet to be discovered. Such inhibitors (as well as curaxins) may constitute a good choice for treatment of cancers that overexpress BORIS, the germ line-specific CTCF paralogue, which can bind the same genomic sites, can form homodimers or heterodimers with CTCF and can stabilize new cancer-specific chromatin interactions [37].
Concluding remarks
Many anti-cancer epigenetic drugs, targeting different aspects of gene expression regulation, have been invented and are undergoing clinical trials now. The most recent of them exploit the cancer transcriptional addiction phenomenon (i.e. specifically target high-level oncogene transcription that usually depends on SE function) (reviewed in [85]). It is essentially clear how inhibitors of BET proteins and CDK7 implement selective cancer cell elimination [42, 47]. However, the exact mode of action and clinical efficiency of 3D genome-disrupting drugs remain elusive. Only one chemotherapeutic compound (curaxin CBL0137), targeting dysregulated gene expression in cancer cells via 3D genome modulation, has been reported [71]. It is intriguing to consider whether this is the beginning of a new direction in drug discovery. Proteins that maintain the 3D genome architecture and, in particular, that mediate EPC (CTCF, YY1, BORIS) may be considered as promising targets for treatment of cancers that strongly depend on SE-driven transcription. However, several basic issues should be addressed alongside possible development of the 3D genome-modulating agents (see Outstanding Questions). The most important questions are (1) whether healthy cells will be sufficiently tolerant to these kind of drugs, and (2) what is the reason for such tolerance? In other words, what does distinguish healthy and particular cancer cells in terms of chromatin structure and 3D genome organization? Although the therapeutic significance of 3D genome-modulating effects of curaxins is currently unclear (see Clinician’s Corner), the findings described here provide new insights into spatial genome organization and its potential use as a target for anti-cancer therapy.
Outstanding questions.
What are the most crucial distinctions of cancer cells in terms of chromatin structure and 3D genome organization? Is cancer cell heterogeneity manifested in their 3D genome?
Are normal cells sufficiently tolerant to 3D genome-modulating drugs? What does exactly underlie such tolerance?
What are molecular biomarkers for the primary usage of 3D genome-modulating drugs over conventional chemotherapeutics?
Can 3D genome-modulating drugs be exploited to treat other diseases besides cancer?
Clinician’s Corner Box.
3D genome organization shows 1) how DNA is packed in the cell nucleus, and 2) how distant genomic regions, including cis-regulatory DNA elements, are interconnected in the nuclear space. 3D genome organization is an additional level in epigenetic regulation of gene expression.
3D genome organization of cancer cells differs from that of healthy cells. However, it remains elusive whether changes in the cancer 3D genome underlie dysregulated gene expression in cancer cells or that perhaps changes in gene expression provoke 3D genome alterations.
Many cancer types are addicted to high-level transcription of oncogenes (i.e. survival, proliferation and further malignization strongly depend on the production of oncogene proteins). High-level transcription, in particular, requires high levels of transcriptional co-activators, 3D genome-mediated EPCs and specific chromatin-associated factors. There are several chemotherapeutic drugs targeting transcriptional co-activators that selectively kill cancer cells.
A new group of epigenetic drugs (curaxins) that target the 3D genome organization has been reported recently. They modulate the 3D genome and, thus, alter EPCs in living cells and lead to preferential downregulation of enhancer and SE-driven transcription.
Highlights.
Changes in spatial genome organization (3D genome) of cancer cells mostly occur on the level of topologically associating domains and chromatin loops.
Many cancers are addicted to high-level transcription of oncogenes which requires involvement of transcriptional co-activators, 3D genome-mediated enhancer-promotor communication (EPCs) and specific chromatin-associated factors. There are several chemotherapeutic drugs targeting transcriptional co-activators that selectively kill cancer cells.
A new group of epigenetic drugs targets the 3D genome organization via DNA-intercalating small molecules that alter DNA topology. This leads to the inability of CCCTC-binding factor (CTCF) to bind efficiently to its cognate DNA sites, results in large-scale perturbations in the 3D genome, and leads to preferential downregulation of enhancer and super-enhancer-driven transcription.
Acknowledgements
This study was supported by Russian Science Foundation (17-74-20030 to O.L.K.). The work of V.M.S. was supported by National Institute of General Medical Sciences (R01GM119398); the work of K.V.G. was supported by National Cancer Institute (R01CA197967-04).
Glossary
- CCCTC-binding factor (CTCF)
the critical 3D genome-maintaining protein in the interphase nucleus. It is responsible for the formation of chromatin loops.
- Curaxins
carbazole-based small-molecule compounds with broad anti-cancer activity that intercalate into DNA.
- DNA loop extrusion
a molecular process that underlies TAD formation. In this process, loop-extruding factors (cohesins) form progressively larger loops that are limited by boundary elements (CTCF bound at TAD boundaries).
- Enhancer-promoter communication (EPC)
long-range interactions of gene regulatory elements and their target promoters mediated by 3D genome organization.
- Insulated neighbourhoods
chromosomal loop structures formed by two CTCF/cohesin-binding sites that limit physical interactions of particular gene regulatory elements. Certain insulated neighbourhoods are compromised in cancer, leading to dysregulated gene expression.
- Liquid-liquid phase separation (LLPS)
a process in which a protein solution is separated into protein-dense and light phases. It is critical in the formation of intracellular membrane-less compartments such as stress granules, heterochromatin foci and transcription activating condensates.
- Super-enhancer (SE)
large regulatory elements that enable cell-type-specific gene regulation. SEs regulate cancer cell proliferation and survival.
Footnotes
Disclaimer Statement
K.V.G. obtained a research grant and consulting payments from Incuron, Inc. The remaining authors declare no competing interests.
References
- 1.Lieberman-Aiden E et al. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rowley MJ and Corces VG (2018) Organizational principles of 3D genome architecture. Nat. Rev. Genet 19, 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sexton T et al. (2012) Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 [DOI] [PubMed] [Google Scholar]
- 4.Nora EP et al. (2012) Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon JR et al. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanborn AL et al. (2015) Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. USA 112, E6456–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fudenberg G et al. (2016) Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 15, 2038–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szabo Q et al. (2019) Principles of genome folding into topologically associating domains. Sci. Adv 5, eaaw1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabari BR et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hahn S (2018) Phase Separation, Protein Disorder, and Enhancer Function. Cell 175, 1723–1725 [DOI] [PubMed] [Google Scholar]
- 11.Cho WK et al. (2018) Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phanstiel DH et al. (2017) Static and Dynamic DNA Loops form AP-1-Bound Activation Hubs during Macrophage Development. Mol. Cell 67, 1037–1048 e1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vernimmen D et al. (2009) Chromosome looping at the human alpha-globin locus is mediated via the major upstream regulatory element (HS −40). Blood 114, 4253–4260 [DOI] [PubMed] [Google Scholar]
- 14.de Laat W and Grosveld F (2003) Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 11, 447–459 [DOI] [PubMed] [Google Scholar]
- 15.Hnisz D et al. (2017) A Phase Separation Model for Transcriptional Control. Cell 169, 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Consortium EP (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnold CD et al. (2013) Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339, 1074–1077 [DOI] [PubMed] [Google Scholar]
- 18.Symmons O et al. (2014) Functional and topological characteristics of mammalian regulatory domains. Genome Res. 24, 390–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Symmons O et al. (2016) The Shh Topological Domain Facilitates the Action of Remote Enhancers by Reducing the Effects of Genomic Distances. Dev. Cell 39, 529–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valton AL and Dekker J (2016) TAD disruption as oncogenic driver. Curr. Opin. Genet. Dev 36, 34–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lupianez DG et al. (2015) Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang M et al. (2019) Proteogenomics and Hi-C reveal transcriptional dysregulation in high hyperdiploid childhood acute lymphoblastic leukemia. Nat. Commun 10, 1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu P et al. (2017) 3D genome of multiple myeloma reveals spatial genome disorganization associated with copy number variations. Nat. Commun 8, 1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taberlay PC et al. (2016) Three-dimensional disorganization of the cancer genome occurs coincident with long-range genetic and epigenetic alterations. Genome Res. 26, 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hnisz D et al. (2016) Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351, 1454–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flavahan WA et al. (2016) Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braun R et al. (2019) Single Chromosome Aneuploidy Induces Genome-Wide Perturbation of Nuclear Organization and Gene Expression. Neoplasia 21, 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barutcu AR et al. (2015) Chromatin interaction analysis reveals changes in small chromosome and telomere clustering between epithelial and breast cancer cells. Genome Biol. 16, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Achinger-Kawecka J et al. (2016) Alterations in Three-Dimensional Organization of the Cancer Genome and Epigenome. Cold Spring Harb. Symp. Quant. Biol 81, 41–51 [DOI] [PubMed] [Google Scholar]
- 30.Kloetgen A et al. (2019) Dynamic 3D chromosomal landscapes in acute leukemia. bioRxiv, doi: 10.1101/724427 [DOI] [Google Scholar]
- 31.Sauerwald N and Kingsford C (2018) Quantifying the similarity of topological domains across normal and cancer human cell types. Bioinformatics 34, i475–i483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagai LAE et al. (2019) Analyzing the 3D chromatin organization coordinating with gene expression regulation in B-cell lymphoma. BMC Med. Genomics 11, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corces MR and Corces VG (2016) The three-dimensional cancer genome. Curr. Opin. Genet. Dev 36, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katainen R et al. (2015) CTCF/cohesin-binding sites are frequently mutated in cancer. Nat. Genet 47, 818–821 [DOI] [PubMed] [Google Scholar]
- 35.Guo Y et al. (2018) CRISPR-mediated deletion of prostate cancer risk-associated CTCF loop anchors identifies repressive chromatin loops. Genome Biol. 19, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weintraub AS et al. (2017) YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 171, 1573–1588 e1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Debruyne DN et al. (2019) BORIS promotes chromatin regulatory interactions in treatment-resistant cancer cells. Nature 572, 676–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garikapati KR et al. (2017) Down-regulation of BORIS/CTCFL efficiently regulates cancer stemness and metastasis in MYCN amplified neuroblastoma cell line by modulating Wnt/beta-catenin signaling pathway. Biochem. Biophys. Res. Commun 484, 93–99 [DOI] [PubMed] [Google Scholar]
- 39.Bradner JE et al. (2017) Transcriptional Addiction in Cancer. Cell 168, 629–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandlesh P et al. (2018) Prevention of chromatin destabilization by FACT is crucial for malignant transformation. bioRxiv, doi: 10.1101/499376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yokoshi M and Fukaya T (2019) Dynamics of transcriptional enhancers and chromosome topology in gene regulation. Dev. Growth Differ 61, 343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loven J et al. (2013) Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chapuy B et al. (2013) Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 24, 777–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asangani IA et al. (2014) Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 510, 278–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y and Vakoc CR (2017) Targeting Cancer Cells with BET Bromodomain Inhibitors. Cold Spring Harb. Perspect. Med 7, a026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng M et al. (2018) Targeting MYC dependency in ovarian cancer through inhibition of CDK7 and CDK12/13. Elife 7, e39030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y et al. (2015) CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell 163, 174–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwiatkowski N et al. (2014) Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 511, 616–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Christensen CL et al. (2014) Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell 26, 909–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chipumuro E et al. (2014) CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 159, 1126–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia H et al. (2013) Facilitates chromatin transcription complex is an "accelerator" of tumor transformation and potential marker and target of aggressive cancers. Cell Rep. 4, 159–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garcia H et al. (2011) Expression of FACT in mammalian tissues suggests its role in maintaining of undifferentiated state of cells. Oncotarget 2, 783–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Safina A et al. (2017) FACT is a sensor of DNA torsional stress in eukaryotic cells. Nucleic Acids Res. 45, 1925–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nesher E et al. (2018) Role of Chromatin Damage and Chromatin Trapping of FACT in Mediating the Anticancer Cytotoxicity of DNA-Binding Small-Molecule Drugs. Cancer Res. 78, 1431–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cermakova K and Hodges HC (2018) Next-Generation Drugs and Probes for Chromatin Biology: From Targeted Protein Degradation to Phase Separation. Molecules 23, E1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wheeler RJ et al. (2019) Small molecules for modulating protein driven liquid-liquid phase separation in treating neurodegenerative disease. bioRxiv, doi: 10.1101/721001 [DOI] [Google Scholar]
- 57.Zuin J et al. (2014) Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. USA 111, 996–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwarzer W et al. (2017) Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rao SSP et al. (2017) Cohesin Loss Eliminates All Loop Domains. Cell 171, 305–320 e324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nora EP et al. (2017) Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 169, 930–944 e922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wutz G et al. (2017) Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36, 3573–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haarhuis JHI et al. (2017) The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell 169, 693–707 e614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lyu X et al. (2018) Architectural Proteins and Pluripotency Factors Cooperate to Orchestrate the Transcriptional Response of hESCs to Temperature Stress. Mol. Cell 71, 940–955 e947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li L et al. (2015) Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol. Cell 58, 216–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amat R et al. (2019) Rapid reversible changes in compartments and local chromatin organization revealed by hyperosmotic shock. Genome Res. 29, 18–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferguson LR and Denny WA (2007) Genotoxicity of non-covalent interactions: DNA intercalators. Mutat. Res 623, 14–23 [DOI] [PubMed] [Google Scholar]
- 67.Almaqwashi AA et al. (2016) Mechanisms of small molecule-DNA interactions probed by single-molecule force spectroscopy. Nucleic Acids Res. 44, 3971–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hossain M et al. (2008) DNA intercalation by quinacrine and methylene blue: a comparative binding and thermodynamic characterization study. DNA Cell Biol. 27, 81–90 [DOI] [PubMed] [Google Scholar]
- 69.Boer DR et al. (2009) DNA-binding drugs caught in action: the latest 3D pictures of drug-DNA complexes. Dalton Trans. 3, 399–414 [DOI] [PubMed] [Google Scholar]
- 70.Pang B et al. (2013) Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin. Nat. Commun 4, 1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kantidze OL et al. (2019) The anti-cancer drugs curaxins target spatial genome organization. Nat. Commun 10, 1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Arensbergen J et al. (2019) High-throughput identification of human SNPs affecting regulatory element activity. Nat. Genet 51, 1160–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sadowski M et al. (2018) Spatial Chromatin Architecture Alteration by Structural Variations in Human Genomes at Population Scale. bioRxiv, doi: 10.1101/266981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun F et al. (2019) Promoter-Enhancer Communication Occurs Primarily within Insulated Neighborhoods. Mol. Cell 73, 250–263 e255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chang HW et al. (2018) Mechanism of FACT removal from transcribed genes by anticancer drugs curaxins. Sci. Adv 4, eaav2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim M et al. (2016) Preclinical Validation of a Single-Treatment Infusion Modality That Can Eradicate Extremity Melanomas. Cancer Res. 76, 6620–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dermawan JK et al. (2016) Pharmacological Targeting of the Histone Chaperone Complex FACT Preferentially Eliminates Glioblastoma Stem Cells and Prolongs Survival in Preclinical Models. Cancer Res. 76, 2432–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carter DR et al. (2015) Therapeutic targeting of the MYC signal by inhibition of histone chaperone FACT in neuroblastoma. Sci. Transl. Med 7, 312ra176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang X et al. (2017) Critical Role for GAB2 in Neuroblastoma Pathogenesis through the Promotion of SHP2/MYCN Cooperation. Cell Rep. 18, 2932–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Somers K et al. (2019) Potent antileukemic activity of curaxin CBL0137 against MLL-rearranged leukemia. Int. J. Cancer, doi: 10.1002/ijc.32582 [DOI] [PubMed] [Google Scholar]
- 81.De S et al. (2018) The FACT inhibitor CBL0137 Synergizes with Cisplatin in Small-Cell Lung Cancer by Increasing NOTCH1 Expression and Targeting Tumor-Initiating Cells. Cancer Res. 78, 2396–2406 [DOI] [PubMed] [Google Scholar]
- 82.Ohlsson R et al. (2001) CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 17, 520–527 [DOI] [PubMed] [Google Scholar]
- 83.Huang J et al. (2018) Dissecting super-enhancer hierarchy based on chromatin interactions. Nat. Commun 9, 943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu XS et al. (2016) Editing DNA Methylation in the Mammalian Genome. Cell 167, 233–247 e217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mohammad HP et al. (2019) Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat. Med 25, 403–418 [DOI] [PubMed] [Google Scholar]
- 86.Dekker J et al. (2002) Capturing chromosome conformation. Science 295, 1306–1311 [DOI] [PubMed] [Google Scholar]
- 87.Denker A and de Laat W (2016) The second decade of 3C technologies: detailed insights into nuclear organization. Genes Dev. 30, 1357–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rao SS et al. (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Harewood L et al. (2017) Hi-C as a tool for precise detection and characterisation of chromosomal rearrangements and copy number variation in human tumours. Genome Biol. 18, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chakraborty A and Ay F (2017) Identification of copy number variations and translocations in cancer cells from Hi-C data. Bioinformatics 34, 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]