Abstract
The prevalence of metabolic disease has rising and affected over 1,000 million populations globally. Since the metabolic disease and its related complication are board, it has become the major health hazard of modern world. However, Long term medication of metabolic disease may cause serious side effects and risk for adverse health problems. Recently, emerging studies focus on exploring the mechanistic details of metabolic state in disease development and progression. Gut bacteria ecosystem was considered to play a pivotal role in regulating energy homeostasis and great associated with the development of metabolic disease. Accumulated evidences indicated that Akkermansia muciniphila, Faecalibacterium prausnitzii, and Roseburia hominis improve the balance of the microecology in the intestine of the host and have positive effects on enhancing nutrients absorption. Hence, the novel probiotics as therapeutic target to modify gut microbiota generally focus on improving microbiota dysbiosis, and offers new prospects for treating metabolic disease. In the present review, we discuss the significant roles and regulatory properties of specific bacterium in the context of intestinal microbial balance, explores the kinds of harmful/beneficial bacteria that were likely to act as indicator for metabolic disease. Further proposed a stepwise procedure in the basis of sequencing technology with that of innovative option to reestablish the microbial equilibrium and prevent metabolic disease.
Keywords: Disease prediction, Metabolic disease, Microbiota, Novel probiotics
1. Introduction
The term of metabolic syndrome (MetS) refers to a constellation of associated metabolic diseases states, characterized by insulin resistance (IR), dyslipidemia, hyperglycemia, and high blood pressure, which resultant type 2 diabetes mellitus (T2DM), central obesity, hypertension, and increased the risk of coronary artery disease (CAD), chronic renal failure, and malignant development. Since various diagnostic criteria of MetS are proposed, the incidence and impact are differing from countries. The reported epidemic of the syndrome was according to the consensus definitions, age, gender, and ethnicity [1]. In addition, secondary lifestyle and socioeconomic strata were suggested to influence prevalence across the aforementioned physiological factors. In generally, the global prevalence of MetS among the adult population were reported to range between 20 and 25% across countries and region [2]. Besides, the western dietary patterns were strongly correlated with developing metabolic abnormalities and a main risk factor for chronic cardiometabolic syndrome. Interestingly, the gut microbiota, via extracting energy from the human diet by which it may affect host health that was supporting the aforementioned associations was recently emphasized.
Collectively, the metabolic diseases is a multifactorial and multiorgan pathophysiologic state, which were linked to genetic architecture, healthy behavior change, and also intensely affected by gastrointestinal tract microbial ecosystem, is now been considered as a pivotal role in metabolic health and diseases, acting as a second genome [3]. Emerging studies proposed that gut microbiota contributed to a variety of physiological functions impacting human metabolic balance and disorders. The genome-wide association study have shown that gut microbiota diversity is involved in core function, including the modulation of nutrients and energy harvest efficiency, maintenance of intestinal epithelial barrier functions, stimulation of host immune response to discriminate commensals from pathogens. Two predominant bacterial phyla have been implicated in the composition of human gut microbiota: Bacteroidetes and Firmicutes. The population ration of these phyla (F/B ratio) is widely accepted as a potential indicator for maintaining gut homeostasis. The increased F/B ratio, expanding population of Firmicutes phyla and/or contracting population of Bacteroidetes phyla in individuals, which was demonstrated to characterize the dysbiosis signature of host gut microbiota. Since specific gut microbiota composition revealed high-risk rates for gastrointestinal dysfunction and leads to devastating metabolic consequence, the develop strategies for preventing perturbation of microbiota communication is now becomes a major focus. In addition, a rich diversity in gut microbiota and higher abundant in probiotics is conductive to enrich microbial consortium that are principally produced primary metabolites, of which short-chain fatty acids (SCFAs) directly or indirectly meliorated peripheral inflammation, impacted immune cells function, act through G protein-coupled-receptors signaling activation. Indeed, the balance of the intestinal microbial community is crucial to promote an overall homeostasis condition of human host achieving global health goals.
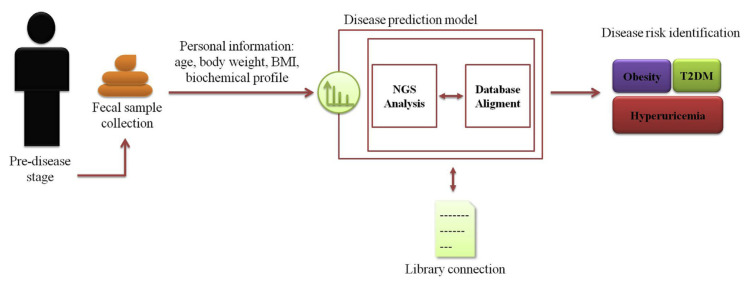
Although both scientific and commercial advances for probiotics that are commonly restricted to specific strain or species mainly include Lactobacillus and Bifidobacteria, are generally identified as traditional probiotics, other novel candidate therapeutic probiotics have yet been fully developed up to now, and which are more likely to establish the preferable mechanism for deeper investigation. Among recent evidences on metagenome sequencing based analysis, the SCFAs-producing bacteria have been referred to as potential candidates for the application of next generation to treat or avert, even cure for metabolic diseases of human beings. Besides, advanced molecular microbial profiling technologies generated massive information about the intestine ecological dynamics impact in metabolic pathological processes. In order to manage the high-dimensional data, the artificial intelligence in machine learning technology would be helpful to boost information processing. Although new nucleic acid sequencing tool and powerful algorithms enable us to deeper understand microbial community, the complex interaction between intestine microbiome and the host disease state is still a challenge. In the present review, we aim to provide an overview of recent literature that focus on intestinal ecosystem dynamics in the host with metabolic disease, and helping guide further study will clarify the potential for and support a novel predictive model for metabolic disease development to promote and achieve anti-metabolic derangement. The systemic framework based on sample collection, NGS analysis, data connection, and final metabolic diseases risk identification (Fig. 1).
Fig. 1.
Graphical abstract of the proposed working model system. The systemic framework based on sample collection, NGS analysis, data connection, and final metabolic diseases risk identification.
2. Type-2 diabetes mellitus
The resistance of peripheral tissue including muscle, liver, and adipose tissue to insulin actions, which forced islet β cell to increase the secretion of insulin was mainly observed in T2DM. The detailed mechanistic pathway and major risk factors for T2DM is well established. Currently, scientific research aims to verify the significance of other factors involved, such as gut microbiota and its secondary metabolites in T2DM development. The first human study by Larsen et al. demonstrated a significant difference across healthy and T2DM subjects in terms of the composition of intestinal microbiome [4]. Furthermore, faecal microbiota composition profiles from 18 individuals with/or without T2DM, characterized by a substantial reduction of Firmicutes phyla and increasing the relative abundance of Bacteroidetes phyla and Proteobacteria phyla in T2DM patients. Although meta-genome-wide association analysis have determined the Bacteroidetes and Firmicutes were major phyla in human gastrointestinal tract, the gut dysbiosis caused by other phyla such as Actinobacteria, Proteobacteria, and Fusobacteria promote susceptibility to insulin resistance, typical pathological condition of diabetes [5]. In the context, emerging human data and animal’s model focus on characterizing properties of the microbiome in T2DM, and further evaluate the relative abundance of specific bacteria taxa to identify their relationship with this metabolic condition. Recently, Chatelier and the colleagues described low gene count of Roseburia intestinalis and Faecalibacterium prausnitzii were founded in individuals with T2DM, as well as elevated oxidative stress, inflammatory status, and gut leakage. Deeper, microbiom taxonomic and functional profiles from 784 gut metagenomes showed that non-diabetic treated T2D patients (n = 106) exhibited significantly decreased in butyrate-producing species (Roseburia spp., Subdoligranulum spp., and Clostridiales spp.) as compared to non-diabetic participants (n = 554). According to meta-analysis pipeline and validation in all datasets, the abundance of genus Akkermansia was 3-fold increased after metformin treatment, which was in consisted with genus Escherichia [6]. Table 1 summarizes the findings of gut dysbiosis in the abundance and richness from both preclinical animal models and human trail of T2DM.
Table 1.
Main findings of gut microbial populations changing associated with T2DM.
| Subjects’ Characteristics | Implicated microbiota | References |
|---|---|---|
| Female participants with T2DM (Denmark) | phylum Firmicutes ↓/class Clostridia ↓/class Betaproteobacteria ↑/genus Roseburia ↓ | [4] |
| Male participants with T2DM (European) | Clostridium clostridioforme ↑/Clostridium hathewayi ↑/Bacteroides intestinalis ↓ | [41] |
| Participants with T2DM (Chinese) | Bacteroides caccae ↑/Clostridium hathewayi ↑/Clostridium ramosum ↑/Clostridium symbiosum ↑/Eggerthella lenta ↑/Escherichia coli ↑/Clostridiales sp. SS3/4 ↓/Eubacterium rectale ↓/Faecalibacterium prausnitzii ↓/Roseburia intestinalis ↓/Roseburia inulinivorans ↓ | [30] |
| Male db/db mice (8-week-old) | family Bacteroidaceae ↓/family Prevotellaceae ↓/genus Clostridium ↓/phylum Verrucomicrobia ↑/species Lactobacillus reuteri ↑ | [42] |
| Tsumura Suzuki obese diabetes mice (12-week-old) | Clostridium ruminantiun ↑/Clostridium celatum ↑/Ruminococcus callidus ↑/Clostridium colinum ↓ | [43] |
| Participants with pre-DM/T2DM (Chinese) | Akkermansia muciniphila ATCCBAA-835 ↓/Faecalibacterium prausnitzii L2-6 ↓/Verrucomicrobiae ↓ | [44] |
| Participants with T2DM (Poland) | genus Roseburia ↓/family Clostridiaceae ↓/genus Ruminococcus ↑/family Enterobacteriaceae ↑ | [45] |
3. Obesity
Western dietary habit and sedentary lifestyle invariably cause rapid induction of IR and hyperglycemia, as well as obesity. The resident microbiota was extensively considered as an indispensable enteroendocrine organ in the host functioning with properties of energy harvest, especially for the context to develop overweight and obesity. Preliminary evidence from animal studies, the intestinal microbiota was able to affect weight gain/or lose, and associated with body composition. Based on studies of germ-free mice, even microbial inoculation for functional microbiome research has linked the association between intestinal microbiota and obesity. Colonization of germ-free C57BL/6 mice with conventional microbiota harvested from cecum as a consequence of IR and total body fat elevated approximately to the level of donor subjects. Intriguingly, the increased body fat components were in consist with it reflected increased energy output and decreased consumption despite markedly less food intake. As discussed suggested, the intestinal microbiota may served as mediators in energy metabolism [7]. Excepted for vitamin synthesis, saccharolytic fermentation carried out by the intestinal microbiota produced a variety of microbial metabolites such as SCFA, mediated by predominant species including Roseburia, Lactobacillus, Bifidobacterium, and Fecalibacterium. Concentration of these microbial-produced metabolites was recently recognized to correlate with elevated fat content and obesity. Remely and the colleagues reported that obese participants underwent nutrients counseling and extra glucagon-like peptide intervention exhibited an increased trend in abundance of Faecalibacterium prausnitzii and epigenetic methylation of FFAR3 and LINE-1 [8]. The results were consisted with other comparative study between obese and lean participants, and showed that Faecalibacterium prausnitzii level and systemic inflammatory markers such as fecal calprotectin and plasma C-reactive protein were increased in obese [9]. Similar to animal model of diet-induced obesity, a significant reduction in butyrate-producing probiotics related to Roseburia app. and Eubacterum rectale in relation to energy harvested ability of subjects with obesity when compared to lean ones [10]. Moreover, the prebiotic properties of Roseburia spp. was demonstrated to along with reduced IL-6, MCP-1, subcutaneous adipose fat, and downregulated genes expression involved in hepatic cholesterol synthesis (C/EBPα, FAT/CD36, aP2, and LPL) [11]. Overall, the main characteristic of microbiota composition is emphasized by the depletion and repletion resultant. The vast majority of those phenomenons strongly indicated that obese phenotype exhibit a significant shift in the richness of helpful and potentially pathogenic bacteria in compared with lean phenotype (Table 2).
Table 2.
Main findings of gut microbial populations changing associated with obesity.
| Subjects’ Characteristics | Implicated microbiota | References |
|---|---|---|
| Pregnant Participants with obesity (Finland) | Bacteroides group ↑/Staphylococcus group ↑ | [46] |
| Participants with obesity (Germany) | Bifidobacterium ↓/Clostridium leptum ↓/Methanobrevibacter ↓/Firmicutes ↓/Bacteroidetes ↑ | [47] |
| Participants with obesity (Chinese) | Clostridium perfringens ↓/Bacteroides ↓ | [48] |
| Children participants with obesity (India) | Faecalibacterium prausnitzii ↑ | [49] |
| Children participants with obesity (Belgium) | Bacteroides vulgates ↓/Lactobacillus spp. ↑ | [50] |
| Adolescents participants with overweight/ and obesity (Spain) | Bacteroides fragilis ↑/Lactobacillus groups ↑/Clostridium coccoides ↓/ Bifidobacterium longum ↓/Bifidibacterium adolescentis ↓ | [51] |
| Children participants with obesity (Italy) | family Ruminococcaceae ↑/family Bacteroidaceae ↓ | [52] |
| Participants with obesity (Chinese) | Akkermansia muciniphila ↓/Fecalibacterum prausnitzii ↓/Bacteroides uniformis ↓/Bacteroides ovatus ↓/Ruminoccoccus torques ↑/Fusobacterium ulcerans ↑ | [53] |
| Female participants with obesity (Chinese) | Roseburia spp. ↓/Lachnospira spp. ↓/Clostridiales spp. ↓/Faecalibacterium spp. ↓/family Lachnospiraceae ↓ | [54] |
| Participants with obesity (Germany) | genus Akkermansia ↓/genus Dialister ↓/genus Prevotella ↑/genus Megamonas ↑/genus Phascolarctobacterium ↑ | [55] |
| Male Sprague-Dawley rats (HFD-induced obesity) | phylum Firmicutes ↑/phylum Proteobacteria ↑/phylum Actinobacteria ↑/phylum Bacteroites ↓ | [56] |
| Leptin-deficient ob/ob mice (C57BL/6J) | Bacteroides ↓/Firmicutes ↑ | [57] |
| Leptin-deficient ob/ob mice (C57BL/6J) | genus Akkermansia ↓/genus Dubosiella ↓/genus Muribaculaceae ↓/genus Turicibacter ↑/genus Lactobacillus ↑/genus Coriobacteriaceae ↑ | [58] |
Increased Bacteroides fragilis group (P = 0.001) and Lactobacillus group (P = 0.030) counts, and to decreased Clostridium coccoides group (P = 0.028), Bifidobacterium longum (P = 0.031), and Bifidobacterium adolescentis (P = 0.044) counts.
4. Hyperuricemia
Although T2DM and obesity were most prevalent metabolic diseases and main health concerns of modern life nowadays. The common Western dietary pattern is characterized by highly saturated fatty acid and sugar, especially fructose with the form of corn syrup in foods and beverages. Accumulated evidences demonstrated that high fructose consumption eventually leads to elevation in incidence and prevalence rate of obesity, metabolic dysregulation, hepatic steatosis, and renal disease. In addition, increased dietary intake of fructose was recently reported to induce inflammatory response and upregulated fructose metabolic pathway which resulted chronically elevated uric acid level in blood, also known as hyperuricemia. In recent years, emerging studies further explored novel pathogenic pathway of hyperuricemia is mainly intestinal flora, which highlighted it’s the role in purine and uric acid metabolism generated by xanthine oxidase [12]. Reversely, researches also revealed that elevated soluble serum uric acid may affect intestinal bacterial community and gut mucosal barrier stability. Recent evidence of gosling with visceral gout have reported existence of intestinal dysbiosis with higher abundant of specific germ-negative strain Proteobacteria, reflected adverse action through increasing intestinal epithelial permeability, elevated systemic lipopolysaccharide (LPS) level, and stimulate inflammatory pathway and produce kidney injury. Similarly, increased blood serum uric acid level was also observed in enteropathogenic Escherichia coli infected rabbit ileal loop model, suggesting a significant role of gut-bacteria in the development of hyperuricemia [13]. The authors founded that enteropathogenic and Shiga-toxigenic E. coli infection resulted in xanthine oxidase released into intestinal tissue and fluids and subsequently, leaded to significantly increased in uric acid level both in T84 cells and rabbit models of infection. The LPS produced by gram-negative bacteria might participate uric acid metabolism in intestine in which suggested affecting hyperuricemia. A therapeutic property for purine-induced hyperuricemic rats was contributed to fecal transplantation from normal rats. After three-week intervention, levels of uric acid, genera Vallitalea, genera Christensenella, and genera Insolitispirillum of recipient hyperuricemic rats were shown to decrease and close to normal rats [14]. Although the detail mechanism of increased uric acid content remain unfully investigated, the correlation between intestinal dysbiosis and uric acid level are well worthy of focus and further studies (Table 3).
Table 3.
Main findings of gut microbial populations changing associated with hyperuricemia.
| Subjects’ Characteristics | Implicated microbiota | References |
|---|---|---|
| Participants with gout (Chinese) | Bacteroides caccae ↑/Bacteroides xylanisolvens ↑/Faecalibacterium prausnitzii ↓/ Bifidobacterium pseudocatenulatum ↓ | [12] |
| Male participants with gout (Ukraine) | Bifidobacterium spp. ↓/Eubacterium spp. ↓/Fusobacterium spp. ↑/Veilonella spp. ↑/Peptostreptococcus spp. ↑/Bacteroides spp. ↑ | [59] |
| Male Wistar rats (high-purine-induced hyperuricemia) | genus Vallitalea ↑/genus Christensenella ↑/genus Insolitispirillum ↑/genus Prevotella ↓/genus Anaerovibrio ↓/genus Alloprevotella ↓/genus Barnesiella ↓ | [14] |
| Uox-knockout hyperuricemia mice (C57BL/6J) | Firmicutes ↓/Bacteroides ↑/Akkermansia ↓/Ruminococcus ↓ | [60] |
| Male Sprague-Dawley rats (HFD-induced hyperuricemia) | genus Bacteroides ↑/genus Lactococcus ↑/genus Dorea ↑/genus Proteus ↑/genus Morganella ↑/genus Allobaculm ↑/genus Prevotella ↓/genus Lactobacillus ↓/genus Streptococcus ↓/genus Clostridium ↓/genus Ruminococcus ↓/genus Anaeroplasma ↓ | [61] |
5. Coronary artery disease
Generally, CAD is indicated strongly linking to various risk factors such as obesity, hyperglycemia, aging, hyperlipidemia, and hypertension, and has recently been considered to be affected by intestinal dysbiosis. The observation from patients with atherosclerosis showed that frequent bacterial signature in atherosclerotic lesions and high diversity of bacteria DNA detected in the atherosclerotic plaque area that positively correlated to leukocytes levels. Intriguingly, specific bacteria species presents in the plaque, oral cavity, and intestine share same phylotypes in the same patient which believed to involve in the development of CAD [15,16]. A case-control study enrolled 53 participants with advanced CAD and 53 healthy controls to analyze the intestinal microbial alternation between the two groups. The results showed that Chao-1 index, Shannon diversity, and observed number of operational taxonomic units (OTUs) were founded to lower in CAD fecal samples. An alternation was observed in the abundance of Lachnospiraceae NK4B4, Ruminooccus Gauvreauii were significant lower, while Ruminooccus gnavus was significant higher, and suggested to correlate with CAD progression [17]. Previously, comparative cohort study conducted in 218 participants with atherosclerosis and 187 healthy participants revealed a characteristic alteration in intestinal microbial composition and metabolic functions. The results exhibit a significant reduction in the relative abundance of novel probiotics, including Roseburia intestinals and Faecalibacterium cf. prausnitzii in participants with atherosclerosis, as well as other functional modules correlated to atherosclerosis [18]. Trimethylamine N-oxide (TMAO), one toxic metabolite produced by bacteria, is pro-atherogenic in rodent and humans [19]. The intestinal microbial-mediated TMAO production was discovered in genus level including Clostridium, Enterococcus, Acinetobacter, Citrobacter, Anaerococcus, Desulfitobacter, Streptococcus, Desulfovibrio, Enterobacteria, Escherichia, Klebsiella, Proteus, and Pseudomonas [20]. Notably, higher Akkermansia level was indicated to associated with TMAO production [21]. Although TMAO is widely accepted as marker to the pathogenesis of CAD, other microbial products such as SCFA, LPS, and secondary bile acid along the TMAO cascade are of great interest. The intestinal microbiome alteration that involved in the onset of CAD were summarized in Table 4.
Table 4.
Main findings of gut microbial populations changing associated with coronary artery disease.
| Subjects’ Characteristics | Implicated microbiota | References |
|---|---|---|
| Participants with CAD (Japan) | order Lactobacillales ↑/phylum Bacteroidetes ↓ | [62] |
| Participants with CAD (Chinese) | Escherichia Shigella ↑/Enterococcus ↑/Faecalibacterium ↓/ Subdoligranulum ↓/Roseburia ↓/Eubacterium | [63] |
| Participants with CAD (USA) | Lachnospiraceae NK484 ↓/Ruminococcus Gauvreauii ↓/Ruminococcus gnavus ↑ | [17] |
| Participants with CAD (Chinese) | phylum Bacteroidetes ↓/phylum Firmicutes ↓ | [64] |
| Participants with CAD (Japan) | Bacteroides vulgatus ↓/Bacteroides dorei ↓ | [65] |
6. Novel potential probiotics: SCFAs-producing bacteria
Metabolic diseases including T2DM, obesity, and hyperuricemia have achieved high proportions that constitute multiple public health concerns confront the human population. Moreover, recent progresses in these metabolic disorders were reported linking to the dramatic change in human intestinal ecosystem diversity. Indeed, the intestinal microbial quantity and richness is considered as a significant marker in host health condition and confronting disease. The metabolic diseases are originated from low-grade inflammation which implicated in a main triggering factor, the bacteria-produced LPS, as its translocation was restricted to gut epithelial barrier. However, the disruption of gut microbial bio-network leads to downregulation of occluding, claudins, and zonula occludens, proteins that compose epithelial tight-junction, and which eventually causes intestinal mucosal barrier leaking and results in the release of LPS into the systemic circulation. The impaired intestinal permeability promotes LPS translocation that may link to early development of IR and chronic inflammation in germ-free mice model and human subjects [22,23]. The fermented microbial production such as SCFAs, mainly acetate, propionate, and butyrate, were recently demonstrated to exert an anti-inflammatory property, regulated carbohydrates, and fatty acid metabolism [24]. Absorption of SCFAs by intestinal epithelium through specific receptors including monocarboxylate transporters and sodium-coupled monocarboxylate transporters that acts as modulators for maintaining intestinal barrier function. Besides, SCFAs interacts with metabolite-sensing G-protein coupled receptors, thereby stimulating intracellular anti-oxidative and anti-inflammatory pathways. Importantly, a significant observation in both human subject and rodent model with metabolic disease that revealed reduced abundance of dominant SCFAs-producing species, such as Faecalibacterium prausnitzii and Roseburia hominis in gastrointestinal tract and fecal samples, as well as the SCFAs concentration appear to be reduced when compared with health phenotype. Roseburia spp. metabolized complex non-digestible carbohydrate and dominantly produced butyrate during growth under fermentation. Previously, a large clinical trial analyzed the main composition of intestinal microbiota between ulcerative colitis and health participants by using denature gradient gel electrophoresis technique. Interestingly, relative abundance of butyrate-producing bacteria Roseburia hominis was founded to significant lower in ulcerative colitis group, and was inversely associated with intestinal inflammation. Base on this observation, Patterson et al. propose that mono-colonization of Roseburia hominis may regulate host-microbe cross-talk in germ-free mice. The presents of Roseburia hominis in gut result in increase in intestinal barrier integrity, anti-pathogenic activity, and T cell biology that may serve as an anti-inflammatory commensal bacterium [25]. Moreover, depletion of Roseburia hominis and Faecalibacterium prausnitzii has also been noted in ulcerative colitis and Crohn’s disease [26]. To note predominant producer of butyrate in human intestine is Faecalibacterium prausnitzii, which has been consistently mentioned as a special degrader of non-digestible dietary substrates. Oral butyrate supplementation significantly attenuated TNF-α and IL-1β production as well as macrophage chemoattractant protein-1 expression in white adipose tissue, against HFD-induced obesity and IR in C57BL/6 mice model [27]. In comparison to lean control, obese and T2DM participants has lower abundance of Faecalibacterium prausnitzii, while nutrient counseling over 3 month could increase quantity of Faecalibacterium prausnitzii and epigenetically upregulate GPR41 and long interspersed nuclear element 1 promoter activity [8]. Verdam et al. reported that increased abundant of Faecalibacterium prausnitzii in health subjects and revealed potential proinflammatory change in microbiota diversity in obese cluster [9]. Others study also showed a lower level of Faecalibacterium prausnitzii species in obese cases with T2DM is directly linked to chronic inflammation. Previously, oral administration of commensal bacterium Faecalibacterium prausnitzii markedly improved intestinal inflammation, partly attributed to it’s metabolites production that inhibited NF-κB-mediated inflammatory cascade, and suggesting a potential probiotics in Crohn’s disease treatment [28]. A research reported that Faecalibacterium prausnitzii supernatant exerted anti-inflammatory activity and improved intestinal barrier function, and was recently identified by Quévrain and colleagues that a 15 kDa protein in the culture supernatant may contributed to attenuate the severity of dinitrobenzene sulfonic acid-induced colitis in mice [29]. In addition, Faecalibacterium prausnitzii also shown different feature of modulating diabetes progression through butyrate induced regulatory pathway. Several cohort studies have confirmed the compositional changes in Faecalibacterium prausnitzii are greatly correlated with T2DM [30]. An instance, a significant inverse association between low count of Faecalibacterium prausnitzii and hemoglobin A1c (HbA1c) level founded in T2DM patients [31]. A therapeutic effect of Faecalibacterium prausnitzii as a potent probiotic supplement has been proposed for preventing diabetes in mice model [31]. Another bacteria indicator of maintaining intestinal health, Akkermansia muciniphila is recently attracted great interest. Akkermansia muciniphila was first isolated in 2004 and characterized by its mucin-degrading and SCFAs-producing abilities. Notably, Akkermansia muciniphila also played a critical role in maintaining intestinal health and it’s depletion was founded to inversely associated with several gastrointestinal-related disorders and metabolic disease. The fecal concentration of Akkermansia muciniphila and was shown to reduced several fold in ulcerative colitis and Crohn’s disease patients [32]. Also, in patients of appendicitis, the abundance of Akkermansia muciniphila was significant reduced. Karlsson et al. demonstrated that the level of Akkermansia muciniphila-like bacteria were lower in preschool children with overweight and obesity [33]. Compared to normal weight gain pregnant women, Akkermansia muciniphila numbers were lower in women with excessive weight gain [34]. Moreover, the probiotic potential of Akkermansia muciniphila was proposed and further investigated in rodent models. Colonization of Akkermansia muciniphila by oral supplementation normalized HFD-induced metabolic disorders such as endotoxemia, inflammation, and IR in obese and T2DM mice. Other studies conducted gnotobiotic mice model by colonizing 14 synthetic human intestinal bacteria species in fiber-free diet feed Swiss Webster mice, resulting in recovering intestinal community and colonic mucus barrier lesion [35]. Similarity, it was reported that participant with higher abundance of Akkermansia muciniphila exhibited great efficiencies in improving metabolic parameters, especially in fasting blood glucose value and body mass index [36]. Furthermore, a pili-like transmembrane protein named Amuc_1100 was identified from Akkermansia muciniphila, which is involved in benefiting immune function and intestinal mucosa integrity [37]. The aforementioned studies suggested that specific bacteria species, particularly of Roseburia hominis, Faecalibacterium prausnitzii, and Akkermansia muciniphila were referred to as next generation probiotics which could be served as a indicator and therapeutic tool for metabolic disease.
7. Novel predict tool for metabolic disease
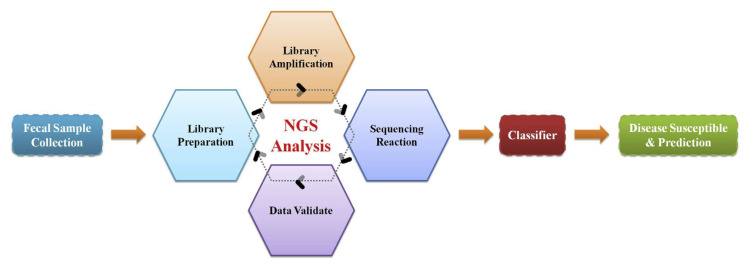
With the fast grow in collecting healthcare records, big data analysis is expected to become popular in the future. Given that the term disease prediction is recently regard as a novel concept globally with major reason of it’s application to prevent or forecast disease progression of human beings. The intestinal microbiota is recognized an essential metabolic organ response for dietary utilization and regulate host energy metabolism. It seem that referring to novel probiotic bacteria species, we have mentioned and a powerful analysis tool, next-generation sequencing (NGS) for big data construction that may suitable for extracting information and generating a disease prediction model (Fig. 2). To assess the linkage between diabetes and host gut microbiota dysbiosis, compositional and functional change of microbiota during sub-clinical state of diabetes was analysis in 36 fecal spacemen of Korea twin participants using metagenomic shogun sequencing technology. A next generation probiotic, Akkermansia muciniphilia was indicated playing a crucial marker that precede disease onset, and functional alteration associated with low-grade inflammation in consist with previous observation [38]. These results suggested that Akkermansia muciniphilia served as an early diagnostic biomarker of diabetes and applicable for microbial-based metabolic therapy. Although studies have suggested specific bacterial taxa might potentially used as a predictor/biomarker for the early diagnosis of MetS, global complexity of human intestinal microbiome and individual host properties limits the development of significant means for optimizing personal microbial-related references and disease model. To overcome the difficulty of intestinal microbiota complexity and diversity over country, ethnicity, and region, it still needs deeper and comprehensive studies to clarify the role of specific-species bacteria/ novel potential probiotics in metabolic disease and determined a new disease risk algorithm or personalized based medicine. Emerging sequencing technology, high throughput analysis such as next-generation sequencing and shotgun metagenomic methods enables generation of thousands or even millions of sequence reads from specimens and help profiling intestinal microbial diversity. The massive databases of phylogenetic and functional diversity have provided preliminary information of microbial communities between healthy and metabolic impairments. Global collaborative efforts such as International Census of Marine Microbes project and European Metagenomics of the Human Intestinal Trace project have provided preliminary microbial communities dataset and thus, necessitates methods to acquire meaningful data for multidimensional analysis [39]. Recently, machine learning methods exert great potential to process large data and applied to exploit microbial community datasets. Supervised machine learning algorithms such as support vector machine (SVM), random forest (RF), and gradient boosting (GB), can help developing multi-layer perceptrons, which might potentially applied to extract microbiome data [40]. Therefore, the consistently growing in vivo and in vitro investigations with metabolic associated diseases requires an emergency advance from interventional evaluation to predictive, preventive, and precision medicine. The flow chart caters to corresponding disease following a standardized protocol through NGS analysis, data extraction, and classification.
Fig. 2.
Schematic representation of disease prediction flow chart. The flow chart caters to corresponding disease following a standardized protocol through next-generation sequencing (NGS) analysis, data extraction, and classification.
8. Conclusion
To date, the intestinal microbiota has been proven to greatly associate with host metabolic status and disease development. Modern sequencing technology has provided a fast and convenient way to identified specific alteration of the intestinal microbiota between disease and health. However, the present review summarized results from human clinical trials and animal studies, and gives a perspective of intestinal microbiota balance and diversity modulate metabolic diseases which emphasized the commercial development and clinical application. Although there is a substantial heterogeneity in microbial composition between species and needs further confirmed. Finally, we also proposed a disease predicted model in performance of NGS based detection and suggested the futurity of this proposed model.
Acknowledgments
All persons who have made substantial contributions to the work reported in the manuscript (e.g., technical help, writing and editing assistance, general support), but who do not meet the criteria for authorship, are named in the Acknowledgments and have given us their written permission to be named. If we have not included an Acknowledgments in our manuscript, then that indicates that we have not received substantial contributions from non-authors.
Funding Statement
This research work was supported by the grant MOST 107-2320-B-040-019-MY3 from the Ministry of Science and Technology (MOST), Taiwan.
Footnotes
Conflict of interest
The authors declare that no conflicts of interest exist.
Funding
This research work was supported by the grant MOST 107-2320-B-040-019-MY3 from the Ministry of Science and Technology (MOST), Taiwan.
References
- 1.Sun SS, Liang R, Huang TT, Daniels SR, Arslanian S, Liu K, et al. Childhood obesity predicts adult metabolic syndrome: the Fels Longitudinal Study. J Pediatr. 2008;152:191–200. doi: 10.1016/j.jpeds.2007.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grundy SM. Metabolic syndrome pandemic. Arterioscler Thromb Vasc Biol. 2008;28:629–36. doi: 10.1161/ATVBAHA.107.151092. [DOI] [PubMed] [Google Scholar]
- 3.Boulange CL, Neves AL, Chilloux J, Nicholson JK, Dumas ME. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016;8:42. doi: 10.1186/s13073-016-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petruzzelli M, Moschetta A. Intestinal ecology in the metabolic syndrome. Cell Metabol. 2010;11:345–6. doi: 10.1016/j.cmet.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 6.Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–6. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–23. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Remely M, Aumueller E, Merold C, Dworzak S, Hippe B, Zanner J, et al. Effects of short chain fatty acid producing bacteria on epigenetic regulation of FFAR3 in type 2 diabetes and obesity. Gene. 2014;537:85–92. doi: 10.1016/j.gene.2013.11.081. [DOI] [PubMed] [Google Scholar]
- 9.Verdam FJ, Fuentes S, de Jonge C, Zoetendal EG, Erbil R, Greve JW, et al. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity (Silver Spring) 2013;21:E607–15. doi: 10.1002/oby.20466. [DOI] [PubMed] [Google Scholar]
- 10.Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol. 2007;73:1073–8. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neyrinck AM, Possemiers S, Druart C, Van de Wiele T, De Backer F, Cani PD, et al. Prebiotic effects of wheat arabinoxylan related to the increase in bifidobacteria, Roseburia and Bacteroides/Prevotella in diet-induced obese mice. PLoS One. 2011;6:e20944. doi: 10.1371/journal.pone.0020944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo Z, Zhang J, Wang Z, Ang KY, Huang S, Hou Q, et al. Intestinal microbiota distinguish gout patients from healthy humans. Sci Rep. 2016;6:20602. doi: 10.1038/srep20602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crane JK, Naeher TM, Broome JE, Boedeker EC. Role of host xanthine oxidase in infection due to enteropathogenic and Shiga-toxigenic Escherichia coli. Infect Immun. 2013;81:1129–39. doi: 10.1128/IAI.01124-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Lv Q, Ren H, Gao L, Zhao P, Yang X, et al. The altered gut microbiota of high-purine-induced hyperuricemia rats and its correlation with hyperuricemia. PeerJ. 2020;8:e8664. doi: 10.7717/peerj.8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ott SJ, El Mokhtari NE, Musfeldt M, Hellmig S, Freitag S, Rehman A, et al. Detection of diverse bacterial signatures in atherosclerotic lesions of patients with coronary heart disease. Circulation. 2006;113:929–37. doi: 10.1161/CIRCULATIONAHA.105.579979. [DOI] [PubMed] [Google Scholar]
- 16.Koren O, Spor A, Felin J, Fak F, Stombaugh J, Tremaroli V, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4592–8. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toya T, Corban MT, Marrietta E, Horwath IE, Lerman LO, Murray JA, et al. Coronary artery disease is associated with an altered gut microbiome composition. PLoS One. 2020;15:e0227147. doi: 10.1371/journal.pone.0227147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jie Z, Xia H, Zhong SL, Feng Q, Li S, Liang S, et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. 2017;8:845. doi: 10.1038/s41467-017-00900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang WH, Hazen SL. The contributory role of gut microbiota in cardiovascular disease. J Clin Invest. 2014;124:4204–11. doi: 10.1172/JCI72331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Obaide MAI, Singh R, Datta P, Rewers-Felkins KA, Salguero MV, Al-Obaidi I, et al. Gut microbiota-dependent trimethylamine-N-oxide and serum biomarkers in patients with T2DM and advanced CKD. J Clin Med. 2017:6. doi: 10.3390/jcm6090086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falony G, Vieira-Silva S, Raes J. Microbiology meets big data: the case of gut microbiota-derived trimethylamine. Annu Rev Microbiol. 2015;69:305–21. doi: 10.1146/annurev-micro-091014-104422. [DOI] [PubMed] [Google Scholar]
- 22.Burcelin R, Garidou L, Pomie C. Immuno-microbiota cross and talk: the new paradigm of metabolic diseases. Semin Immunol. 2012;24:67–74. doi: 10.1016/j.smim.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 23.Hormannsperger G, Schaubeck M, Haller D. Intestinal microbiota in animal models of inflammatory diseases. ILAR J. 2015;56:179–91. doi: 10.1093/ilar/ilv019. [DOI] [PubMed] [Google Scholar]
- 24.Byrne CS, Chambers ES, Morrison DJ, Frost G. The role of short chain fatty acids in appetite regulation and energy homeostasis. Int J Obes (Lond) 2015;39:1331–8. doi: 10.1038/ijo.2015.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patterson AM, Mulder IE, Travis AJ, Lan A, Cerf-Bensussan N, Gaboriau-Routhiau V, et al. Human gut symbiont Roseburia hominis promotes and regulates innate immunity. Front Immunol. 2017;8:1166. doi: 10.3389/fimmu.2017.01166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tilg H, Danese S. Roseburia hominis: a novel guilty player in ulcerative colitis pathogenesis? Gut. 2014;63:1204–5. doi: 10.1136/gutjnl-2013-305799. [DOI] [PubMed] [Google Scholar]
- 27.Vinolo MA, Rodrigues HG, Festuccia WT, Crisma AR, Alves VS, Martins AR, et al. Tributyrin attenuates obesity-associated inflammation and insulin resistance in high-fat-fed mice. Am J Physiol Endocrinol Metab. 2012;303:E272–82. doi: 10.1152/ajpendo.00053.2012. [DOI] [PubMed] [Google Scholar]
- 28.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–6. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carlsson AH, Yakymenko O, Olivier I, Hakansson F, Postma E, Keita AV, et al. Faecalibacterium prausnitzii supernatant improves intestinal barrier function in mice DSS colitis. Scand J Gastroenterol. 2013;48:1136–44. doi: 10.3109/00365521.2013.828773. [DOI] [PubMed] [Google Scholar]
- 30.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 31.Xu J, Lian F, Zhao L, Zhao Y, Chen X, Zhang X, et al. Structural modulation of gut microbiota during alleviation of type 2 diabetes with a Chinese herbal formula. ISME J. 2015;9:552–62. doi: 10.1038/ismej.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–8. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 33.Karlsson CL, Onnerfalt J, Xu J, Molin G, Ahrne S, Thorngren-Jerneck K. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity (Silver Spring) 2012;20:2257–61. doi: 10.1038/oby.2012.110. [DOI] [PubMed] [Google Scholar]
- 34.Santacruz A, Collado MC, Garcia-Valdes L, Segura MT, Martin-Lagos JA, Anjos T, et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br J Nutr. 2010;104:83–92. doi: 10.1017/S0007114510000176. [DOI] [PubMed] [Google Scholar]
- 35.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016;167:1339–1353 e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barton W, Penney NC, Cronin O, Garcia-Perez I, Molloy MG, Holmes E, et al. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut. 2018;67:625–33. doi: 10.1136/gutjnl-2016-313627. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Xu W, Wang R, Cheng R, Tang Z, Zhang M. The outer membrane protein Amuc_1100 of Akkermansia muciniphila promotes intestinal 5-HT biosynthesis and extracellular availability through TLR2 signalling. Food Funct. 2021;12:3597–610. doi: 10.1039/d1fo00115a. [DOI] [PubMed] [Google Scholar]
- 38.Yassour M, Lim MY, Yun HS, Tickle TL, Sung J, Song YM, et al. Sub-clinical detection of gut microbial biomarkers of obesity and type 2 diabetes. Genome Med. 2016;8:17. doi: 10.1186/s13073-016-0271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, et al. Ocean plankton. Structure and function of the global ocean microbiome. Science. 2015;348:1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- 40.Iadanza E, Fabbri R, Bašić-ČiČak D, Amedei A, Telalovic JH. Gut microbiota and artificial intelligence approaches: a scoping review. Health Technol. 2020:1–16. [Google Scholar]
- 41.Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 42.Yu F, Han W, Zhan G, Li S, Jiang X, Wang L, et al. Abnormal gut microbiota composition contributes to the development of type 2 diabetes mellitus in db/db mice. Aging (Albany NY) 2019;11:10454–67. doi: 10.18632/aging.102469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horie M, Miura T, Hirakata S, Hosoyama A, Sugino S, Umeno A, et al. Comparative analysis of the intestinal flora in type 2 diabetes and nondiabetic mice. Exp Anim. 2017;66:405–16. doi: 10.1538/expanim.17-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Shen D, Fang Z, Jie Z, Qiu X, Zhang C, et al. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS One. 2013;8:e71108. doi: 10.1371/journal.pone.0071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salamon D, Sroka-Oleksiak A, Kapusta P, Szopa M, Mrozinska S, Ludwig-Slomczynska AH, et al. Characteristics of gut microbiota in adult patients with type 1 and type 2 diabetes based on nextgeneration sequencing of the 16S rRNA gene fragment. Pol Arch Intern Med. 2018;128:336–43. doi: 10.20452/pamw.4246. [DOI] [PubMed] [Google Scholar]
- 46.Collado MC, Isolauri E, Laitinen K, Salminen S. Distinct composition of gutmicrobiota during pregnancy in overweight and normal-weight women. Am J Clin Nutr. 2008;88:894–9. doi: 10.1093/ajcn/88.4.894. [DOI] [PubMed] [Google Scholar]
- 47.Schwiertz A, Taras D, Schafer K, Beijer S, Bos NA, Donus C, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 2010;18:190–5. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 48.Zuo HJ, Xie ZM, Zhang WW, Li YR, Wang W, Ding XB, et al. Gut bacteria alteration in obese people and its relationship with gene polymorphism. World J Gastroenterol. 2011;17:1076–81. doi: 10.3748/wjg.v17.i8.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balamurugan R, George G, Kabeerdoss J, Hepsiba J, Chandragunasekaran AM, Ramakrishna BS. Quantitative differences in intestinal Faecalibacterium prausnitzii in obese Indian children. Br J Nutr. 2010;103:335–8. doi: 10.1017/S0007114509992182. [DOI] [PubMed] [Google Scholar]
- 50.Bervoets L, Van Hoorenbeeck K, Kortleven I, Van Noten C, Hens N, Vael C, et al. Differences in gut microbiota composition between obese and lean children: a cross-sectional study. Gut Pathog. 2013;5:10. doi: 10.1186/1757-4749-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santacruz A, Marcos A, Warnberg J, Marti A, Martin-Matillas M, Campoy C, et al. Interplay between weight loss and gut microbiota composition in overweight adolescents. Obesity (Silver Spring) 2009;17:1906–15. doi: 10.1038/oby.2009.112. [DOI] [PubMed] [Google Scholar]
- 52.Riva A, Borgo F, Lassandro C, Verduci E, Morace G, Borghi E, et al. Pediatric obesity is associated with an altered gut microbiota and discordant shifts in Firmicutes populations. Environ Microbiol. 2017;19:95–105. doi: 10.1111/1462-2920.13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y, et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med. 2017;23:859–68. doi: 10.1038/nm.4358. [DOI] [PubMed] [Google Scholar]
- 54.Allen JM, Mailing LJ, Niemiro GM, Moore R, Cook MD, White BA, et al. Exercise alters gut microbiota composition and function in lean and obese humans. Med Sci Sports Exerc. 2018;50:747–57. doi: 10.1249/MSS.0000000000001495. [DOI] [PubMed] [Google Scholar]
- 55.Louis S, Tappu RM, Damms-Machado A, Huson DH, Bischoff SC. Characterization of the gut microbial community of obese patients following a weight-loss intervention using whole metagenome shotgun sequencing. PLoS One. 2016;11:e0149564. doi: 10.1371/journal.pone.0149564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang H, Wang Q, Liang C, Su M, Wang X, Li H, et al. Acupuncture regulating gut microbiota in abdominal obese rats induced by high-fat diet. Evid Based Compl Alternat Med. 2019;2019:4958294. doi: 10.1155/2019/4958294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–5. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schroeder BO, Birchenough GMH, Pradhan M, Nystrom EEL, Henricsson M, Hansson GC, et al. Obesity-associated microbiota contributes to mucus layer defects in genetically obese mice. J Biol Chem. 2020;295:15712–26. doi: 10.1074/jbc.RA120.015771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kondratiuk VE, Tarasenko OM, Karmazina OM, Taranchuk VV. Impact of the synbiotics and urate-lowering therapy on gut microbiota and cytokine profile in patients with chronic gouty arthritis. J Med Life. 2020;13:490–8. doi: 10.25122/jml-2020-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo Y, Yu Y, Li H, Ding X, Li X, Jing X, et al. Inulin supplementation ameliorates hyperuricemia and modulates gut microbiota in Uox-knockout mice. Eur J Nutr. 2020;4:2217–30. doi: 10.1007/s00394-020-02414-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu Y, Liu Q, Li H, Wen C, He Z. Alterations of the gut microbiome associated with the treatment of hyperuricaemia in male rats. Front Microbiol. 2018;9:2233. doi: 10.3389/fmicb.2018.02233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsai HF, Hsu PN. Modulation of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis by Helicobacter pylori in immune pathogenesis of gastric mucosal damage. J Microbiol Immunol Infect. 2017;50:4–9. doi: 10.1016/j.jmii.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 63.Emoto T, Yamashita T, Sasaki N, Hirota Y, Hayashi T, So A, et al. Analysis of gut microbiota in coronary artery disease patients: a possible link between gut microbiota and coronary artery disease. J Atherosclerosis Thromb. 2016;23:908–21. doi: 10.5551/jat.32672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cui L, Zhao T, Hu H, Zhang W, Hua X. Association study of gut flora in coronary heart disease through high-throughput sequencing. BioMed Res Int. 2017;2017:3796359. doi: 10.1155/2017/3796359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoshida N, Emoto T, Yamashita T, Watanabe H, Hayashi T, Tabata T, et al. Bacteroides vulgatus and Bacteroides dorei reduce gut microbial lipopolysaccharide production and inhibit atherosclerosis. Circulation. 2018;138:2486–98. doi: 10.1161/CIRCULATIONAHA.118.033714. [DOI] [PubMed] [Google Scholar]