Abstract
Emerging and re-emerging viruses, such as Zaire Ebola virus (EBOV), pose a global threat and require immediate countermeasures, including the rapid development of effective vaccines that are easy to manufacture. Synthetic self-amplifying RNAs (saRNAs) attend to these needs, being safe and strong immune stimulators that can be inexpensively produced in large quantities, using cell-free systems and good manufacturing practice. Here, the first goal was to develop and optimize an anti-EBOV saRNA-based vaccine in terms of its antigen composition and route of administration. Vaccinating mice with saRNAs expressing the EBOV glycoprotein (GP) alone or in combination with the nucleoprotein (NP) elicited antigen-specific immune responses. GP-specific antibodies showed neutralizing activity against EBOV. Strong CD4+ T cell response against NP and GP and CD8+ T cell response against NP were detected by ELISpot assays. Intramuscular vaccination with saRNAs conferred better immune response than intradermal. Finally, mice vaccinated in a prime-boost regimen with saRNAs encoding both GP and NP or with GP alone survived an EBOV infection. In addition, a single dose of GP and NP saRNAs was also protective against fatal EBOV infection. Overall, saRNAs expressing viral antigens represent a promising vaccine platform.
Keywords: Ebola virus, self-amplifying RNA, vaccine, combination of saRNAs, glycoprotein, nucleoprotein, lipid nanoparticles
Graphical abstract
Our results show that self-amplifying RNAs (saRNAs), which encode both the Ebola virus glycoprotein and nucleoprotein, are a promising vaccine candidate. A single intramuscular vaccination of a combination of both saRNAs formulated in lipid nanoparticles protected 100% of mice from lethal Ebola infection and drastically reduced signs of disease.
Introduction
The outbreak of Zaire Ebola virus (EBOV) in West Africa from 2013 to 2016 and the ongoing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic show how emerging and re-emerging viruses can suddenly become a global threat that requires measures of disease control to be developed in a short time frame, such as mass production of vaccines. Like EBOV and SARS-CoV-2, most emerging viruses are transmitted from wildlife to humans, which makes it difficult to predict when and where an outbreak will occur. Due to globalization and relatively easy access to remote rural areas, where a close contact with wild animals is more common than in more developed regions, the frequency and magnitude of outbreaks caused by emerging viruses appear to be increasing.1 Therefore, the capacity to rapidly develop vaccines against emerging viruses needs to be strengthened. Self-amplifying RNA (saRNA) vaccine technology as well as mRNA technology enable rapid and scalable vaccine production, requiring minimal amounts of RNA per dose. In mice, saRNAs have been shown to confer equivalent protection against influenza virus as mRNA vaccines, but at much lower doses.2 saRNA vaccines based on replicons of positive-strand RNA viruses such as alphaviruses have been developed against infectious diseases and were shown to induce robust neutralizing antibody titers and T cell responses.2,3,4 The synthetically produced RNA used for saRNA vaccines is non-infectious, has no inherent mutagenic activity, does not integrate into the cellular genome, and is rapidly degraded by natural cellular mechanisms.5 saRNAs used in this study are derived from the alphavirus Semliki Forest virus (SFV). The genome of alphaviruses is divided into two open reading frames (ORFs): the first ORF encodes proteins constituting the RNA-dependent RNA polymerase (replicase), and the second ORF encodes the viral structural proteins. In the saRNAs used here, the second ORF was either replaced by the ORF coding for the EBOV glycoprotein (GP) or the EBOV nucleoprotein (NP), while the sequence encoding the viral replicase remained intact. The viral replicase drives the intracellular amplification of the RNA replicon after injection of the vaccine.6
EBOV belongs to the family of Filoviridae and causes severe disease with an unusually high case fatality ratio, averaging about 50% (25%–90%, depending on the outbreak).7 The EBOV genome encodes for seven structural proteins. Five of the proteins, the nucleoprotein NP, VP35, VP30, VP24, and the polymerase L form the nucleocapsid, and VP40 forms a matrix layer around the nucleocapsid and is associated with the viral lipid envelope into which the surface glycoprotein GP is inserted. In this proof of concept study, the feasibility of combining two EBOV antigens, GP and NP, to generate strong humoral and T cell responses was investigated using BioNTech`s saRNA vaccine technology. Both targets, GP and NP, are included in Mvabea combined with Zabdeno, one of the two European Medicines Agency (EMA)-approved vector-based vaccines against EBOV.8,9,10 GP is the only surface protein of EBOV and is present in the viral envelope as a trimer; it has essential functions in viral attachment, fusion, and entry into host cells. In addition, GP is likely to play a major role in pathogenesis.11,12 Therefore, GP represents an important target for the development of vaccines and recombinant neutralizing monoclonal antibodies against EBOV. Yet, the T cell response of EBOV survivors is directed predominantly against NP, and therefore, EBOV NP was included in vaccines to achieve a better cellular immune response.13,14 It is already known that activated T cells, especially CD8+ T cells, play an important role in protective immunity against EBOV.13,15,16,17,18,19
In the present study, mice were immunized with saRNAs encoding either EBOV GP or NP or a combination of both. Comparative analyses showed that intramuscular (i.m.) injection of saRNAs resulted in the strongest humoral and cellular immunogenicity. Antigen-specific antibodies were detected, including GP-specific antibodies with neutralizing capacity against EBOV. In addition, CD4+ T cell responses against NP and GP and CD8+ T cell response against NP were detected. Finally, a combination of NP and GP saRNA protected mice from lethal EBOV infection after prime-boost and prime-only immunization.
Results
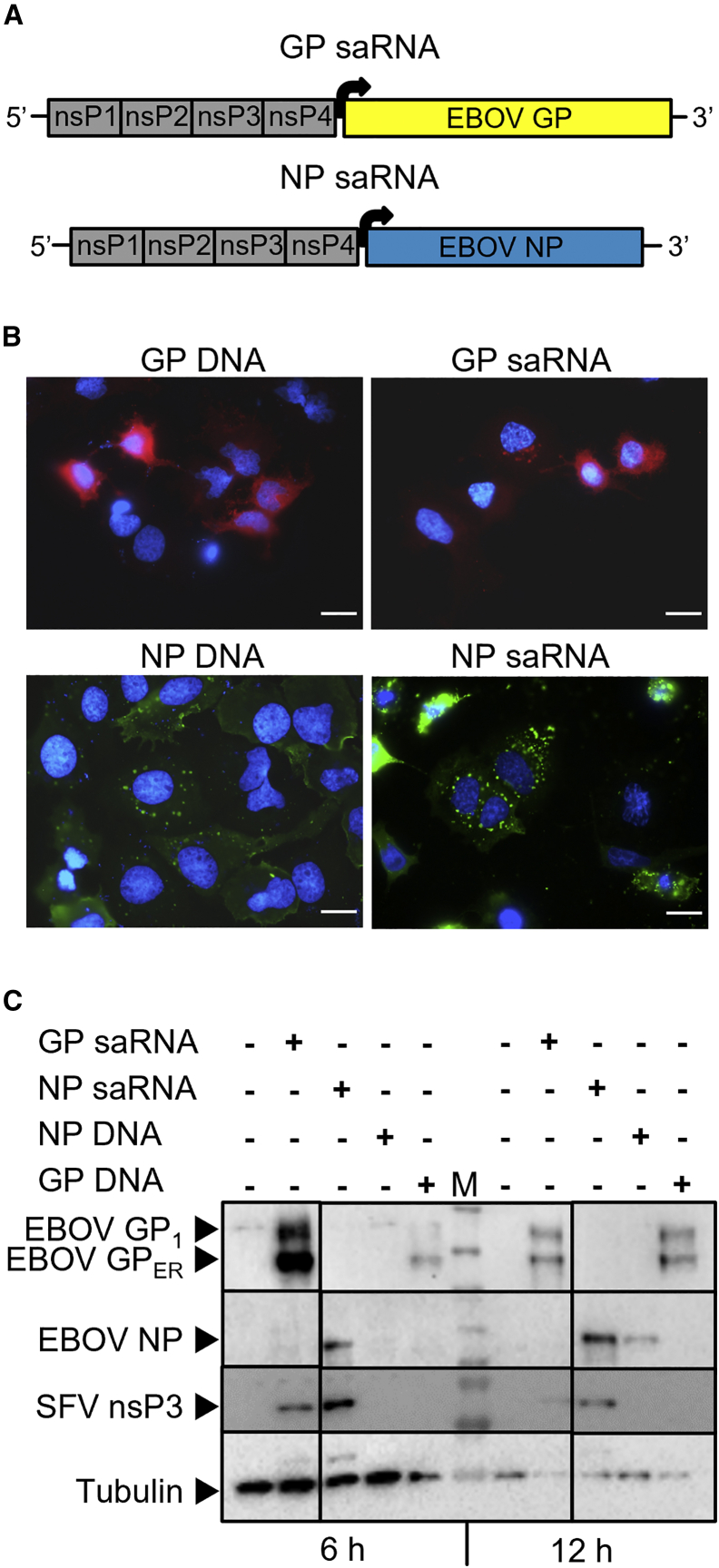
Efficient expression of EBOV NP and GP from saRNA
Two SFV-based saRNA constructs were designed to express EBOV GP (GP saRNA) or EBOV NP (NP saRNA). Both constructs encode the SFV RNA-dependent RNA polymerase, composed of the nonstructural proteins (nsP) 1 to 4, and EBOV GP or EBOV NP under control of a subgenomic promotor (Figure 1A). Subcellular localization of GP and NP was determined by immunofluorescence imaging of HuH7 cells 24 h after transfection of 2 μg of GP saRNA, NP saRNA, or DNA plasmids encoding GP or NP (Figure 1B). The expression pattern of the proteins was comparable between saRNA and plasmid DNA and similar to the distribution of NP and GP in EBOV-infected cells.20 NP (green) is present in cytoplasmic inclusions of variable size surrounding the nucleus.21 GP is localized at the plasma membrane, the site of viral budding.20,22 The expression of viral proteins was also analyzed by western blot (Figure 1C). Two bands for EBOV GP were detected, the GP1 subunit of the protein (EBOV GP1, approx. 180 kDa) and a smaller band that corresponds to the ER precursor form of GP (EBOV GPER, approx. 110 kDa).23 At 6 h after transfection, the expression of GP was higher when cells were transfected with GP saRNA than with GP-encoding plasmid DNA. At 12 h after transfection, the expression of GP was similar for GP saRNA- and GP plasmid-transfected cells. Moreover, NP expression was detected at 6 h only in cells transfected with NP saRNA, and at 12 h after transfection, NP expression was stronger in NP saRNA than in NP plasmid-transfected cells. Tubulin signals from the 6-h samples were stronger than those from the 12-h samples because twice the sample volume was loaded.
Figure 1.
In vitro characterization of saRNAs
(A) Scheme of Semliki Forest virus-based saRNA constructs, which include the RNA-dependent RNA polymerase composed of the nonstructural proteins (nsP) 1–4 and Ebola virus glycoprotein (EBOV GP) or nucleoprotein (EBOV NP) antigens, under the control of a subgenomic promotor (represented by the black arrow). The in vitro expression of saRNA-encoded EBOV GP (GP saRNA) and EBOV NP (NP saRNA) was compared with plasmid-encoded EBOV GP (GP DNA) or EBOV NP (NP DNA). HuH7 cells were transfected with 2 μg of plasmid or saRNA and analyzed after 6, 12, or 24 h. (B) Localization of EBOV GP and EBOV NP in HuH7 cells 24 h after transfection by immunofluorescence. Scale bar, 20 μm. (C) Protein expression of EBOV GP, EBOV NP (approx. 110 kDa), Semliki Forest virus nonstructural protein 3 (SFV nsP3, approx. 60 kDa), and tubulin (loading control, 55 kDa) in HuH7 lysates 6 and 12 h after transfection by western blot. EBOV GP1, GP1 subunit of EBOV GP (approx. 180 kDa), EBOV GPER, ER precursor of EBOV GP (approx. 110 kDa), M, molecular weight marker. Samples were analyzed on one membrane with irrelevant lanes cut out.
EBOV GP and NP saRNAs encapsulated within lipid nanoparticles induced strong immune responses
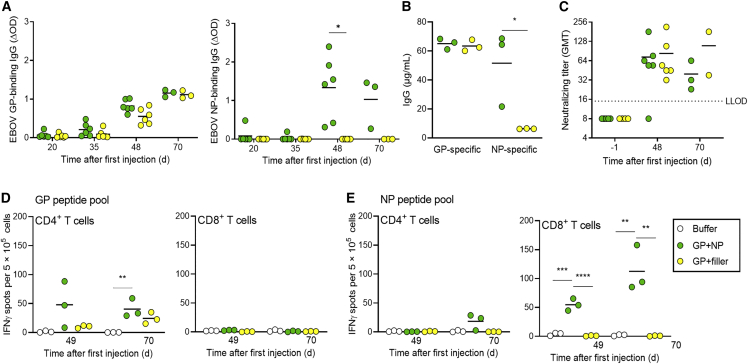
First, we wanted to investigate whether the immune responses against EBOV GP differ when used alone or in combination with NP. Lipid nanoparticles (LNP) were used as saRNA delivery vehicle as they are the most efficient and clinically most advanced delivery vehicle.24,25 5 μg of GP saRNA and 2.5 μg of NP saRNA were administered twice to groups of six C57BL/6J IFNAR−/− mice, 35 days apart. The 2:1 ratio of RNAs as well as the immunization schedule had been investigated within BioNTech SE beforehand (S.E., unpublished data). To keep the overall RNA amount equal, the GP-only group also received an saRNA encoding the SFV replicase only, without the insertion of a gene of interest (referred to as filler). Immunization with LNP-formulated GP and NP saRNAs (GP + NP and GP + filler) induced serum IgG antibodies against both GP and NP (Figures 2A and 2B). Interestingly, the humoral immune response to GP was not affected by the inclusion of NP saRNA in the vaccine, revealing no interference or competition in the immunological response to these antigens. Strong neutralizing activity against authentic EBOV was also observed in the serum of mice immunized with GP or GP + NP (Figure 2C).
Figure 2.
Immunogenicity of saRNA delivered by lipid nanoparticles
C57BL/6J IFNAR−/− mice (six per group) were intramuscularly injected on days 0 and 35 with 7.5 μg saRNA total dose (5 μg GP saRNA and 2.5 μg NP saRNA) formulated with lipid nanoparticles (LNP) (GP + NP, GP + filler). Mice in the control group were injected with formulation buffer. Serum samples were collected before immunization (day −1), after prime immunization (days 20 and 35), and after boost immunization (days 48 and 70). No IgG was detectable in serum samples before immunization. (A) Seroconversion per group over time. GP- and NP-specific IgG antibodies were measured by adding mice serum (1:100 dilution) to streptavidin plates coated with recombinant EBOV GP biotin or EBOV NP biotin fusion proteins, followed by an HRP coupled secondary antibody to allow colorimetric detection. Adsorption at 450 nm and 620 nm was measured, and the calculated ΔOD is shown. (B) GP- and NP-specific IgG concentrations on day 70 determined by four-parameter logistic fit in GraphPad Prism, against an IgG standard curve. Individual mice values are shown by the symbols; group mean values are indicated by horizontal bars. The assay’s lower limit of quantification (LLOQ) was 7.3 μg/mL. (C) Neutralizing antibody titers against authentic EBOV are shown by dots; group mean values are indicated by horizontal bars. The dotted line indicates the lower limit of detection. (D and E) Interferon γ (IFNγ) secretion was measured to assess T cell responses in splenocytes isolated on days 49 and 70, by ELISpot assay (three mice per time point). Splenocytes were stimulated for 18 h with MHC I- (CD8+) and MHC II-specific (CD4+) EBOV GP- or EBOV NP-specific peptide pools containing two to six predicted peptides and a final concentration of 1 μg/mL per peptide. Individual spot counts per mice are shown by dots (mean values of triplicate measurements); group mean values are indicated by horizontal lines. Asterisks indicate statistical significance as detailed by bars between relevant groups: ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p < 0.0001.
At day 49 and day 70 post prime vaccination, cellular immune responses were evaluated ex vivo by measuring IFNγ production by isolated splenocytes in response to EBOV GP- or NP-specific peptide pools using ELISpot assay. CD4+ T cell responses against GP and CD8+ T cell responses against NP were detected (Figures 2D and 2E), indicating that the addition of NP strengthened the vaccination-induced T cell response by additionally activating CD8+ T cells.
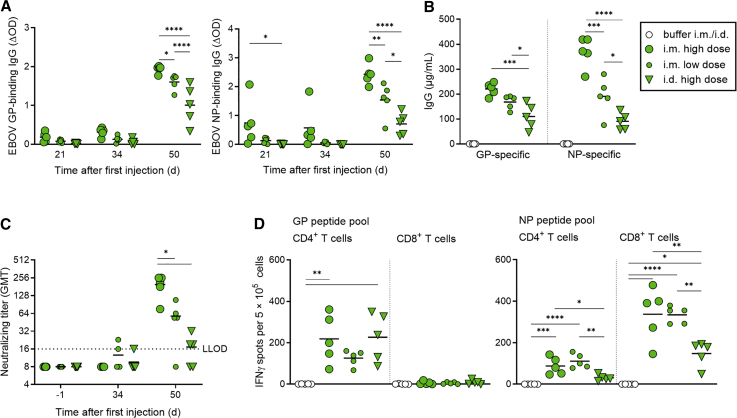
Intramuscular vaccination with EBOV-specific saRNAs was more immunogenic than intradermal vaccination
For an influenza vaccine, it was shown that intradermal (i.d.) administration of one-fifth the standard i.m. dose can elicit similar or better immune responses.26 Therefore, we wanted to compare i.d. and i.m. administration of our bivalent anti-EBOV vaccine candidate. Keeping the GP:NP saRNA ratio at 2:1, we tested the effect of 7.5 μg saRNA (high dose, i.m. and i.d.), and 1.5 μg saRNA (low dose, tested only with i.m. route). The highest GP- and NP-specific IgG antibody titers at day 50 post prime were elicited by application of the high dose i.m., followed by the low dose i.m., and the i.d. application was least effective (Figures 3A and 3B). The same was true for the neutralizing antibodies (Figure 3C). Regardless of the route of administration or dose, the highest titers of specific IgGs were detected on day 50 post prime vaccination in both cases. To investigate the T cell activation after GP + NP immunization, IFNγ ELISpot analyses were performed (Figure 3D). The previously observed induction of CD4+-specific T cell responses mainly by GP and CD8+-specific T cell responses mainly by NP could be reproduced and was independent of the route of administration of the vaccine. Interestingly, while the CD4+-specific T cell response against GP was the same after i.d. or i.m. administration, the NP-induced CD4+- and CD8+-specific T cell responses were lower in mice immunized via the i.d. route (Figure 3D).
Figure 3.
Immunogenicity of GP + NP using two routes of injection
C57BL/6J IFNAR−/− mice (five per group) were injected intramuscularly (i.m.) or intradermally (i.d.) on days 0 and 35 with LNP-saRNA vaccine candidates in two saRNA doses: high dose (7.5 μg, i.m. and i.d.) or low dose (1.5 μg, i.m. only). The ratio of GP saRNA to NP saRNA was kept at 2:1. Serum samples were collected on days −1, 21, 34, and 50. (A) Seroconversion per group over time. ELISA for EBOV GP and EBOV NP was performed as described in Figure 2. (B) Concentration of GP- and NP-specific IgG on day 50, determined by ELISA as described in Figure 2. Individual IgG concentrations are shown by dots; group mean values are indicated by lines. (C) Neutralizing antibody titers against authentic EBOV are shown by dots; group mean values are indicated by horizontal bars. The dotted line indicates the lower limit of detection (LLOD). (D) Interferon γ (IFNγ) secretion was measured to assess T cell responses in splenocytes isolated on day 50 in all animals of each group, by ELISpot assay as described in Figure 2. Individual spot counts per mice are shown by dots (mean values of triplicate measurements); group mean values are indicated by horizontal lines. Asterisks indicate statistical significance compared with buffer control: ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p < 0.0001.
EBOV-specific saRNA vaccines protect against fatal EBOV infection
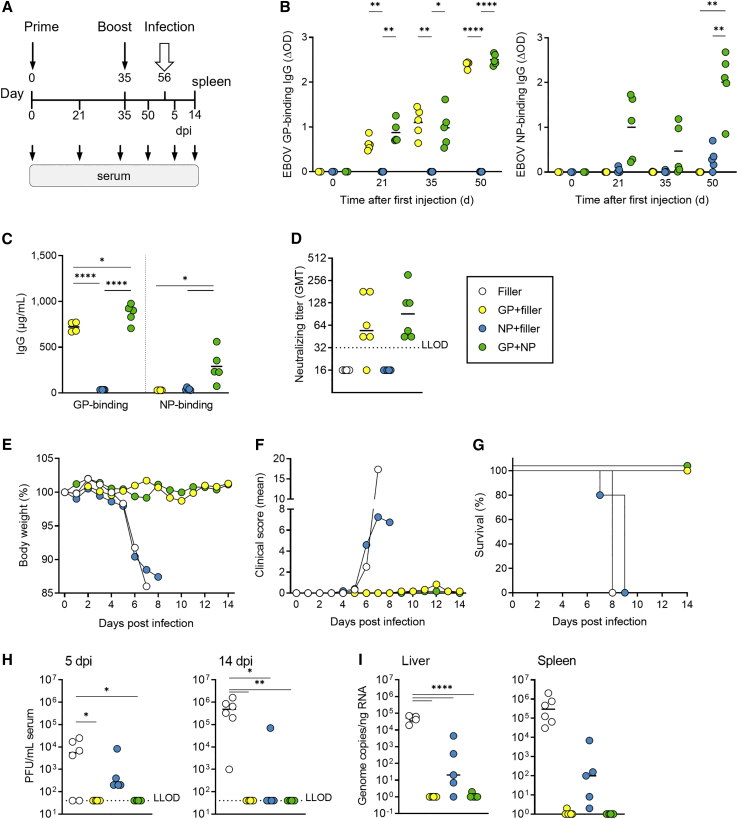
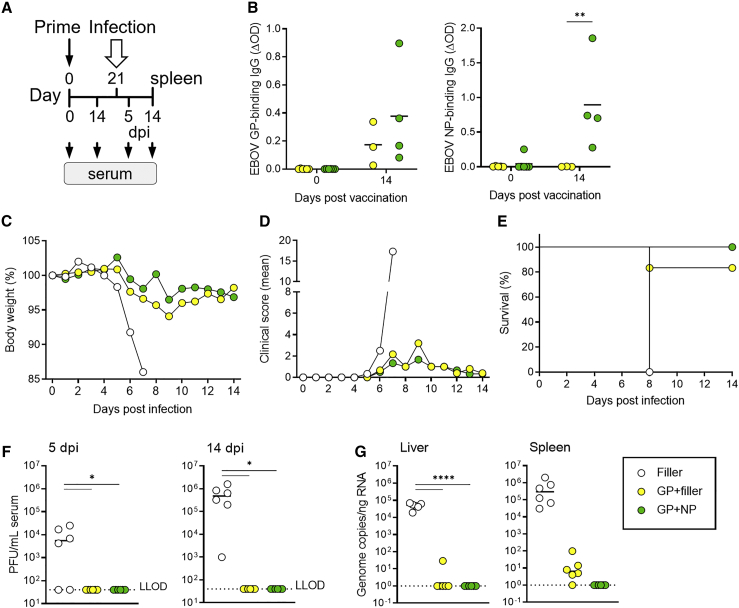
To investigate whether the vaccine-induced humoral and cellular immune responses were protective, immunized mice were infected with a lethal dose of EBOV. Mice were immunized i.m. twice (d0/d35 regimen) using the high dose of GP, NP, or GP + NP (Figure 4A), and antibody responses were analyzed. GP-specific IgG levels were comparable between the groups that received GP or GP + NP, while NP-specific IgG levels were reduced when the NP saRNA was administered alone (NP, Figures 4B and 4C). Neutralizing antibodies against EBOV were observed in the sera from mice vaccinated with GP or GP + NP. No neutralizing activity was elicited in mice vaccinated with NP (Figure 4D). On day 21 after the second vaccination, mice were infected with EBOV and monitored for 14 days. Like mice in the non-vaccinated control group, mice immunized with NP lost weight from day 5 post infection (p.i.) (Figure 4E), developed severe symptoms, and had to be euthanized between days 7 and 9 p.i. (Figures 4F and 4G). In contrast, mice that received GP or GP + NP did not lose weight and were protected from fatal EBOV infection (Figures 4E–4G).
Figure 4.
Protection against lethal EBOV infection induced by two immunizations with GP and NP or GP + NP
C57BL/6J IFNAR−/− mice (six per group) were injected with LNP-saRNA vaccine candidates on days 0 and 35. The vaccine contained 5 μg GP saRNA plus 2.5 μg saRNA encoding the replicase only (GP + filler), 2.5 μg NP saRNA plus 5 μg saRNA encoding the replicase only (NP + filler), or both (GP + NP). LNP filler contained only the replicase encoding saRNA without insertion of a gene of interest. Serum samples were collected before immunization (day 0), and after prime (days 21 and 35) and boost immunization (day 50). Mice were infected i.p. with 1,000 PFU of EBOV on day 56, and survival was recorded for 14 days (six mice per group, except NP group where only five mice were infected). (A) Schematical overview of the experimental setting. (B) Seroconversion per group over time. ELISA for EBOV GP and EBOV NP was performed of serum samples (1:100 dilution) from the indicated days, as described in Figure 2 (four or five mice per group). (C) Concentration of GP- and NP-specific IgG on day 50, determined by ELISA as described in Figure 2 (four or five mice per group). (D) Neutralizing antibody titers against authentic EBOV on day 50. The dotted line indicates the lower limit of detection (LLOD) (six mice per group). (E) Body weight curves for the different groups after EBOV infection. (F) Clinical scores of the different groups. Mice that developed severe clinical signs of infection and/or exceeded 15% of body weight loss were euthanized. (G) Survival over time. (H) Infectious EBOV particles were quantified by plaque titration in serum samples from days 5 and 14 after infection or at the day of the sacrifice. (I) EBOV genome copies in liver and spleen samples of the mice obtained at day 14 post infection or at the day of the sacrifice, by EBOV GP-specific qRT-PCR. Symbols indicate individual mouse values, with group mean values indicated by horizontal lines for all graphs, except for (E–G), where symbols represent group average values (six or five [NP] mice per group). Asterisks indicate statistical significance as detailed by bars between relevant groups: ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗∗p < 0.0001.
On days 5 and 14 p.i., no infectious virus was detected in serum samples from animals immunized with GP or GP + NP (Figure 4H). Furthermore, no viral genomes were detected in the spleen and liver of these mice (Figure 4I). In contrast, on day 5 and on the day the animals were sacrificed, we detected viral RNA and infectious virus in the serum, liver, and spleen of control mice or mice immunized with NP alone (Figures 4H and 4I).
In sum, the data show that immunization with saRNAs encoding GP alone or in combination with NP in a prime-boost regimen protected against fatal EBOV infection. Despite the observed NP-induced CD8+-specific cellular immune response, immunization with the NP saRNA alone was not sufficient to protect against EBOV infection.
A single dose of a saRNA vaccine protects against infection with EBOV
Finally, we analyzed the efficacy of a prime-only regimen of the EBOV-specific saRNA vaccine. For this purpose, mice were immunized once either with the high dose of GP or GP + NP. The antibody response was analyzed at day 14 post vaccination and showed nearly no GP-specific antibodies in the GP group and low titers in the GP + NP group (Figure 5B). At a dilution of 1:32, sera were negative for neutralizing antibodies. EBOV infection was performed at day 21 after vaccination (Figure 5A). It was surprising that all mice vaccinated with GP + NP survived the lethal EBOV infection, and only one mouse had to be euthanized after GP vaccination (Figures 5C–5E). No infectious EBOV was detected in serum samples of GP-only and GP + NP-vaccinated mice at 5 and 14 days after infection or at day 8 when one mouse of the GP group had to be sacrificed (Figure 5F). EBOV-specific genome copies were detected in the liver and spleen of control animals and, to a much lesser extent, in the GP group, while no copies were detected in the GP + NP group (Figure 5G).
Figure 5.
Protection against lethal EBOV infection induced by single immunization with GP or GP + NP saRNA
C57BL/6J IFNAR−/− mice (six per group) were injected on day 0 only. The vaccine contained 5 μg GP saRNA alone + 2.5 μg filler (GP + filler) or combined with 2.5 μg NP saRNA (GP + NP). The filler saRNA was formulated into LNPs in a total dose of 7.5 μg to keep the absolute RNA amount constant in all samples. Serum samples were collected before immunization (day 0) and on day 14 after immunization. Mice were challenged i.p. with 1,000 PFU of EBOV on day 21, and survival was recorded for 14 days (six per group). (A) Schematical overview of the experimental setting. (B) Seroconversion per group over time (three or four mice per group). ELISA for EBOV GP and EBOV NP was performed in serum samples (1:100 dilution) from the indicated days, as described in Figure 2. (C) Body weight curves for the different groups after infection. (D) Clinical scores. Mice that developed severe clinical signs of infection and/or exceeded 15% of body weight loss were euthanized. (E) Survival over time. Survival over time is shown in the graph on the right. (F) Infectious EBOV particles were quantified by plaque titration in serum samples from days 5 and 14 after infection or at the day of the sacrifice. (G) EBOV genome copies in liver and spleen samples from day 14 p.i. or from the day of the sacrifice, by EBOV GP-specific qRT-PCR. Symbols indicate individual mouse values, with group mean values indicated by horizontal lines for all graphs, except for (C–E), where symbols represent group average values of six mice per group. Asterisks indicate statistical significance as detailed by bars between relevant groups: ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗∗p < 0.0001.
Discussion
The data provided in this study demonstrated the potential of saRNA-based vaccines to confer complete protection against EBOV infection.
We have optimized an saRNA-based anti-EBOV vaccine in terms of its antigen composition and route of administration. Vaccination of mice with saRNAs expressing EBOV GP alone or in combination with EBOV NP elicited antigen-specific immune responses, and higher doses had superior immunogenicity. Two factors contributed to the increased response per dose of our saRNA vaccine candidate (Figure 3). On the one hand, large amounts of antigen are produced by the transfected cell after replication of the saRNA. The finally exhausted target cells undergo cell death and lysis,25,27 thereby releasing antigen to provide an ideal constellation for continued B cell stimulation and antibody production. On the other hand, replication of the saRNA provides stimulation of innate immune sensors, which serves an adjuvant role to increase adaptive immune responses.28 Together, the intracellular amplification of coding RNA and the stimulation of innate immune signaling facilitate a strong immune response at relatively low amounts of applied RNA. This could be an advantage of saRNA over mRNA vaccines. In fact, in mice saRNAs have been shown to confer equivalent protection against influenza virus as mRNA vaccines, but at much lower doses.2 Furthermore, it was shown that an i.d. injection of an influenza vaccine can achieve an equivalent immune response as an i.m. injection of a 5-fold higher dose of vaccine, an effect attributed to the diversity and high number of immune cells in the dermis.26,29 Interestingly, our study showed that the administration of saRNAs via the i.m. route led to a higher humoral and partly also higher cellular immune response than the i.d. route (Figure 3). As recently shown by others,30 the way in which the saRNA is formulated may influence the immune response in our case as well, depending on the route of administration. In addition, the antigen itself could also affect the immune response depending on the route of administration. The CD4+-specific T cell response against GP after i.d. or i.m. administration was the same, while the NP-induced CD4+- and CD8+-specific T cell responses in mice injected via i.d. route were weaker. It was shown by Leboux et al.31 that the uptake of antigens after i.d. application depends on the antigen itself and the formulation. But there is a lack of knowledge on how different antigens, when administered together, lead to different immune responses depending on the route of administration. Overall, further studies are required to elucidate the differences in immune response after i.d. and i.m. vaccination in relation to the antigen used.
In the present study, a prime-boost and prime-only immunization regimen with the combination of a GP and NP saRNA protected all mice from lethal EBOV infection (Figures 4 and 5) despite only low levels of EBOV-specific antibodies being raised 7 days before the challenge using the prime-only setting. The protection of the prime-boost group likely results from the neutralizing activity of antibodies targeting EBOV GP, and T cell responses against both EBOV GP and NP; for the prime-only group the correlate of protection is less clear as T cell responses have not been analyzed. The efficacy of EBOV-specific RNA- or DNA-based vaccine candidates was analyzed in other preclinical studies using different vaccination schedules,32,33,34,35 but only two studies evaluated a prime-only regimen in mice.32,35 For a DNA-based vaccine expressing either GP or NP the prime-only regimen was not protective, and four doses of the vaccine were needed to ensure full protection.32 In contrast, an RNA dendrimer nanoparticle vaccine conferred protection against EBOV after just one dose of the vaccine.35 However, to achieve this, 40 μg of vaccine had to be administered, compared with the 7.5 μg of saRNAs used in our study.
One of the studies examined the efficacy of a vaccine candidate based on Venezuelan equine encephalitis virus replicon particles (VRP) encoding either GP or NP (NP-VRP or GP-VRP) of EBOV. The VRPs were analyzed in BALB/c mice and two different strains of guinea pigs using adapted EBOVs. Administration of the GP-VRP alone or in combination with the NP-VRP protected guinea pigs and mice, while the immunization with NP-VRP protected only mice.33 Compared with a replicon particle-based vaccine, manufacturing and upscaling of saRNA-based vaccines is much easier because the VRP production and purification step can be omitted.36,37 Although promising vaccine candidates in small animal models, VRPs expressing EBOV GP and NP, like other vaccines tested (inactivated virus, recombinant vaccinia virus expressing EBOV GP) failed to protect non-human primates (NHPs) from fatal EBOV infection.38 Other vaccine candidates were able to generate protection against infection with filoviruses in a small animal model and also in NHPs.39 Interestingly, Marburg virus (MARV) VRPs expressing MARV GP alone or in combination with MARV NP were shown to protect both guinea pigs and NHPs from homologous MARV infection.40,41 These examples illustrate that small animal models do not necessarily predict efficacy of vaccine candidates in NHPs, and further testing in NHPs is needed as they most closely mimic human infection.39,42 Nevertheless, small animal models have their important roles in vaccine development because they can provide proof of concept data to justify further evaluation in NHP models that are much more challenging because of ethical and practical reasons.43
In vaccinated mice we detected CD4+ T cell responses against GP and NP but only CD8+ T cell responses against NP (Figures 2 and 3). Our data suggest that the observed NP-specific CD8+ T cell response contributes to vaccine efficacy, as mice vaccinated with both GP and NP saRNAs in a prime-only regimen were fully protected, but not with GP saRNA alone. This assumption is supported by previous work showing that the adoptive transfer of EBOV NP-specific CD8+ T cells in mice led to protection from lethal EBOV infection.44,45 Additionally, a modified vaccinia virus Ankara delivering EBOV NP was recently shown to mediate protection via CD8+ T cell responses.19 Furthermore, protection of NHPs induced by an adenovirus-based vaccine appears to be mediated via CD8+ T cell responses, underscoring the role of cellular immune responses in protection against EBOV.46
Further studies on adenovirus-vectored vaccines in NHPs showed that the combination of EBOV GP and NP in one vaccine was able to protect NHPs from EBOV challenge if administered only once,47,48,49 and removal of NP from the vaccine did not affect its efficacy.47,48 In fact, the studies mentioned above did not analyze immune responses specifically for GP and NP, as we did, but determined the induced humoral and cellular immunity for both proteins at the same time (ELISA using whole viral antigen or VLPs) or only against GP.32,33,47,48,49,50,51,52 A direct comparison of GP- and NP-specific humoral and cellular immune responses with other study results is therefore not possible.
Overall, we demonstrated that a prime-boost as well as a prime-only application of an saRNA vaccine expressing EBOV GP and NP conferred complete protection against EBOV infection in IFNAR−/− mice. Both humoral and cellular immune responses contributed to the protection of the prime-boost group.
Materials and methods
Design of saRNA constructs
For in vitro synthesis of saRNA, a DNA template encoding Semliki Forest virus RNA-dependent RNA polymerase, shortly replicase (GenBank: KP699763), from naturally occurring SFV and the gene of interest replacing the viral structural proteins under the control of the natural subgenomic promotor was used. A segmented 100-nt poly(A) tail interrupted by a short linker (A30LA70, where L = GCAUAUGACU) was used to improve plasmid stability in E. coli.53
The Ebola structural GP sequence was obtained from the Ebola subtype Zaire, virus strain H. sapiens-wt/GIN/2014/Makona-Gueckedou-C07 (GenBank: KJ660347.2). The NP sequence from the Ebola subtype Zaire, virus strain H.sapiens-wt/SLE/2014/Makona-EM096 (GenBank: KM034551.1) was used.
RNA synthesis by in vitro transcription
In vitro RNA synthesis and purification were previously described using a synthetic cap analog that provides superior translational efficiencies.54 Briefly, T7 in vitro transcription was based on MEGAscript T7 Transcription Kit (Thermo Fisher, formerly Ambion, Schwerte, Germany) protocols. The general procedure was carried out similarly to as described before,54 starting with linear DNA template containing the T7 promoter, and particularly with respect to co-transcriptional capping with the synthetic cap analog beta-S-ARCA(D1) (used in 4:1 ratio regarding guanosine triphosphate). High-yielding processes qualified for our particular systems were developed; here, protocols have been modified and optimized with respect to long saRNA with up to 10,000 nucleotides.55 Concentration, purity, and integrity of the RNA were assessed by spectrophotometry (NanoDrop 2000c; PeqLab) and on-chip electrophoresis (2100 BioAnalyzer; Agilent), respectively.
In vitro expression of saRNA- and DNA-encoded EBOV GP and EBOV NP
HuH7 cells (2 x 105 cells per 6-well plate) were transfected with 2 μg of plasmid DNA (pCAGGS-EBOV-NP, pCAGGS-EBOV-GP56) or saRNA construct using either TransIT-LT1 (for DNA; Mirus Bio, Madison, Wisconsin) or Lipofectamine MessengerMAX (for saRNA; Thermo Fisher, Schwerte, Germany) transfection reagents, according to the manufacturers’ instructions. Protein expression was analyzed 6 and 12 h post transfection (p.t.) by western blot (WB) and 24 h p.t. by immunofluorescence (IF). For WB, whole-cell extracts were prepared using 1x SDS sample buffer, and proteins were separated on SDS-polyacrylamide gels and transferred to nitrocellulose membranes (Amersham Protran 0.45 NC; Amersham Biosciences Europe, Freiburg, Germany).57 Membrane blocking was performed in phosphate-buffered saline (PBS) containing 10% skim milk. Immunostaining was performed using primary antibodies diluted in PBS containing 1% (w/v) skim milk and 0.1% Tween 20: chicken anti-NP EBOV (1:1,000 dilution,58), mouse anti-GP EBOV 3B11 (1:100 dilution,59), mouse anti-alpha-tubulin (1:500 dilution, Clone DM 1A, Sigma-Aldrich), rabbit anti-SFV-nsp3 (1:5,000 dilution, kindly provided by A. Merits60), and HRP-conjugated secondary antibodies (1:30,000 dilution), and images were acquired with the ChemiDoc XRS + System (Bio-Rad; Feldkirchen, Germany). IF was performed as described by Kolesnikova et al.61 Briefly, the cells were fixed, proteins were detected using chicken anti-NP EBOV (1:100 dilution) and mouse anti-GP EBOV 3B11 (1:50 dilution) as primary antibodies and Alexa Fluor594-conjugated anti-mouse or Alexa Fluor Plus 488-conjugated anti-chicken IgY (H + L) Cross-Adsorbed secondary antibody (1:500 dilution, Thermo Fisher) as secondary antibodies. 4′,6′-diamidino-2-phenylindole (DAPI, 0.5 μg/mL) was used to stain cell nuclei. Microscopic images were acquired on a Zeiss Axiophot upright fluorescence microscope (63x objective) using a Spot inside B/W QE digital camera (Visitron Systems, Puchheim, Germany) and VisiView image acquisition software.
Production of lipid nanoparticles
LNPs were manufactured by controlled mixing of saRNA dissolved in aqueous buffer with ethanolic solution of lipids at a 3:1 volume ratio and 12 mL/min, using a NanoAssemblr (Precision Nanosystems, Vancouver, Canada). The mixture was dialyzed against 1x DPBS (GIBCO) for 2.5 h in a Slide-A-Lyser 10K MWCO dialysis cassette (Thermo Fisher Scientific, Waltham, MA, USA) and re-concentrated using Amicon 30K Ultra Centrifugal Filters (Merck, Darmstadt, Germany). For studies using the i.m. route, the composition named LNP-C12 was used (1,2-dioleyloxy-3-dimethylaminopropane [DODMA]:cholesterol [Chol]:1,2-di-(9Z-octadecenoyl)-sn-glycero-3-phosphoethanolamine [DOPE]:N-palmitoylsphingo-sine-1-succinyl-methoxypoly(ethylene-glycol)2000 [PEGcerC16]; 40:48:10:2; N/P ratio 4) and composition LNP-C09 (DODMA:Chol:1,2-distearoyl-sn-glycero-3-phosphocholine [DSPC]:PEGcerC16; 40:48:10:2; N/P ratio 2.67) was used for the i.d. application. Average LNP hydrodynamic size (Z average in nm) and polydispersity index (PDI) were determined by dynamic light scattering on a DynaPro PlateReader II and analyzed with the Dynamics v.7.8.1 software (both from Wyatt Technology, Dernbach, Germany). LNP samples were diluted to 0.01 mg/mL in PBS. All samples have been measured in duplicates, ten data points were recorded per replicate, and each measurement lasted 10 s. The data of all experiments are presented in Figure S1. saRNA concentration and encapsulation efficiency was performed by a modified Quant-iT Ribogreen Assay (Invitrogen, Carlsbad, CA). LNP samples or buffer were diluted in 1x Tris-EDTA (TE) buffer to a mRNA concentration between 2 and 5 ng/μL. Accessible saRNA was measured by diluting the LNP sample in 1x TE, and the total RNA amount was quantified by diluting the LNP sample in 2% Triton X-100 (VWR International, Darmstadt, Germany). Ribogreen reagent was added to each sample, and the fluorescent signal was quantified in an Infinite F200PRO microplate reader (Tecan, Männedorf, Switzerland).
Mice and animal experiments
C57BL/6J mice deficient for the type I interferon (IFN) α receptor (C57BL/6J IFNAR−/−)62,63 were chosen as model because of their susceptibility to infection with non-adapted EBOV. Male/female C57BL/6J IFNAR−/− mice, 7 to 9 weeks-old at study start, were bred at BioNTech SE’s animal facility (Figures 2 and 3) or at the animal facility of the Institute of Virology at the Philipps University Marburg (Figures 4 and 5). Mixed-sex mice were used for all experiments, except for the GP prime-only group, which included only male mice.
All experiments and protocols were approved by the local authorities (animal welfare committees; Marburg: Regierungspräsidium Gieβen AZ V54 –19 c 20 15 h 01 MR 20/7 Nr. G 47/2018; BioNTech SE: Landesuntersuchungsamt Koblenz AZ 23 177-07/G 18-12-100), conducted according to the recommendations of Federation of European Laboratory Animal Science Associations and Gesellschaft für Versuchstierkunde Society for Laboratory Animal Science (GV-SOLAS) and in compliance with the German animal welfare act and Directive 2010/63/EU.
Regarding BioNTech SE’s animal facility, animals were housed under specific-pathogen-free (SPF) conditions in individually ventilated cages (Sealsafe GM500 IVC Green Line, TECNIPLAST, Hohenpeiβenberg, Germany; 500 cm2) with a maximum of five animals per cage. The temperature and relative humidity in the cages and animal unit was kept at 20°C–24°C and 45%–55%, respectively, and the air change (AC) rate in the cages at 75 AC/hour. The cages with dust-free bedding made of debarked chopped aspen wood (product code: LTE E−001, Abedd LAB & VET Service, Vienna, Austria) and additional nesting material were changed weekly. Autoclaved ssniff M-Z food (product code: V1124, ssniff Spezialdiäten, Soest, Germany) and autoclaved water (tap water) were provided ad libitum and changed at least once weekly. All materials were autoclaved prior to use.
Regarding Marburg’s animal facility, mice were bred under SPF conditions. For experiments under BSL2 and BSL4 conditions, mice were housed in negative pressure cages (IsoCage N, Tecniplast, Hohenpeiβenberg, Germany) in groups of maximum five animals per cage. Only animals with an unobjectionable health status were selected for the experiments. Mice were kept at 20°C–24°C room temperature and 45%–55% humidity. Chopped aspen wood (LASbedding PG3, LASvendi, Soest, Germany) was used as bedding material and nestlets (Ancare, Bellmore, NY, USA) as nesting material as well as a mouse house (UNO BV, Zevenaar, Netherlands) were provided. Autoclaved water and regular mouse diet (1324, Altromin Spezialfutter, Lage, Germany) were supplied ad libitum. Prior to infection experiments, cups of nutritional diet gel (DietGel Boost, Clear H2O, Westbrook, ME, USA) were added to the cages to ensure sufficient calory uptake for weakened animals.
Virus
EBOV (Ebola virus Zaire, variant Mayinga, GenBank: NC_002549) was used for infections of C57BL/6J IFNAR−/− mice. All experiments with EBOV were carried out in the BSL4 laboratory of the Philipps University of Marburg, Germany.
Immunization and infection of IFNAR−/− mice
The mice were immunized as follows. For Figures 2 and 3 mice were immunized with a dose volume of 20 μL i.m. in the tibialis posterior or i.d. (dorsal back) on the study days indicated. For i.m. and i.d. injections, mice were anesthetized via inhalation (2.5% isoflurane; Abbott, Ludwigshafen, Germany) and legs were shaved. For Figures 4 and 5, mice were immunized two times i.m. in the hindlimb (quadriceps muscle) on the study days indicated with 20 μL of either the respective vaccine or filler saRNA as control. To keep the overall RNA amount in the combination vaccine experiments equal, GP or NP groups have always been filled up with an saRNA encoding the SFV replicase only, without the insertion of a gene of interest (referred to as filler). In the challenge infections, the filler RNA has also been formulated into LNPs and was used as negative control.
At the BSL4 animal facility, groups of C57BL/6J IFNAR−/−mice (n = 5–6) were infected intranasally (i.n.) according to an adapted protocol19,64 on day 21 after the only or last (prime-boost) vaccination with 30 μL of DMEM containing 1,000 plaque-forming units (PFU) of EBOV under short isoflurane anesthesia (CP-Pharma, Burgdorf, Germany). The mice were monitored daily for weight loss and clinical scoring, comprising spontaneous behavior, and general condition was performed. Blood samples to determine viral load were taken 5 days p.i. under short isoflurane anesthesia at the facial vein. All surviving animals were euthanized at day 14, and final serum samples were collected.
Endpoint of challenge experiments/termination criteria
Animals were euthanized in accordance with §4 of the German animal welfare act and the recommendation of GV-SOLAS by cervical dislocation under isoflurane anesthesia. The experiment was terminated after an observation period of 14 days after EBOV infection. Body weight losses exceeding 15% or a high severity level in any of the other categories were on their own sufficient reason for immediate euthanasia. After EBOV infection, mice were monitored at least once daily and a clinical score, which comprised weight, general condition, and spontaneous behavior, was determined. Mice were euthanized when a clinical score of 10 (e.g., weight loss >15%) or 6 on 2 consecutive days was reached. The scoring table is included in the supplemental information (Table S1).
Blood sampling
At BioNTech SE’s animal facility, blood samples for serum analysis of NP- and GP-specific IgG by ELISA were collected from the retro-orbital sinus under inhalation anesthesia. Blood (50 μL) was collected in heparin-coated serum tubes (BD Microtainer, Franklin Lakes, New Jersey) on indicated study days. In addition, blood was collected from 10% of the animals prior to the first immunization.
For Marburg’s animal facility, blood samples were taken at the depicted time points after vaccination (Figures 4A and 5A) at the facial vein to determine neutralizing and binding antibodies by virus neutralization test or ELISA, respectively.
EBOV GP- and NP-specific IgG ELISA
GP- and NP-specific IgG antibodies were detected in serum samples using ELISA. Recombinant proteins from EBOV (strain H.sapiens-wt/GIN/2014/Kissidougou-C15) produced in E.coli or baculovirus-infected insect cells were used. Recombinant EBOV GP protein (GenBank: AHX24649.1; Met1-Gln650; 69.3 kDa; cat. no.: 40442-V08B1) or recombinant EBOV NP protein (GenBank: AHX24646.1; His630-Gln739; 15.6 kDa; cat. no. 40443-V07 × 101; both from Sino Biological via LSZ Life Sciences, Beijing, P.R. China) were biotinylated using the EZ-Link Sulfo-NHS-LC-Biotinylation Kit (cat. no.: 21435, Thermo Fisher Scientific, Rockford, Illinois), following manufacturer’s instructions, to enable high affinity binding to streptavidin-precoated 96-well plates (cat. no.: 734–1284; Nunc Immobilizer, Thermo Fisher Scientific, Waltham, Massachusetts). Biotinylation of the recombinant protein stocks was directly assessed using an HABA/Avidin assay (Pierce Biotin Quantitation Kit, cat. no.: 28005, Thermo Fisher Scientific, Rockford, IL). Streptavidin-precoated plate wells with 100 ng/100 μL (1 μg/mL) biotinylated recombinant protein or with serial dilutions (1:100 to 1:3,200) of a biotin-conjugated mouse IgG isotype (0.5 mg/mL stock; cat. no.: 0107-08; Southern Biotech, Birmingham, Alabama) were incubated overnight at 4°C. After plate washing and blocking of unspecific binding sites (casein blocking buffer, cat. no.: B6429, Sigma-Aldrich, Saint Louis, MO), diluted serum samples from immunized mice were added to coated wells and incubated for 1 h at 37°C on a shaker. For positive control wells, human anti-EBOV GP (1:1,000 dilution, clone KZ52, cat. no.: 0260-001, IBT Bioservices, Rockville, MD) or rabbit anti-EBOV NP (1:100 dilution, cat. no.: 0301-012, IBT Bioservices) was added to wells. To detect bound antibody, horseradish peroxidase (HRP)-conjugated secondary antibody was added to wells: goat anti-mouse IgG (1:15,000 dilution, cat. no.: 115-035-071), goat anti-human IgG (1:5,000 dilution, cat. no.: 109-035-098, both from Jackson ImmunoResearch West Grove, PA), or goat anti-rabbit IgG (1:10,000 dilution, cat. no.: A0545, Merck, Darmstadt, Germany). HRP substrate TMB one (cat. no.: 4380; Kementec, Taastrup, Denmark) was added to wells, and the enzymatic reaction ran for 8 min at RT. Reaction was stopped by addition of sulfuric acid (cat. no.: 1.007.161.000, Merck, Darmstadt, Germany), and the absorbance from wells was measured at 450 nm and 620 nm (Epoch microplate spectrophotometer, BioTek, Winooski, VT). IgG concentration was determined using four-parameter logistic (4-PL) fit (GraphPad Prism 8 Software, La Jolla, CA) against the mouse IgG standard curve analyzed in parallel.
EBOV neutralization assay
EBOV neutralization assay was performed as described by Ehrhardt et al.65 Briefly, mouse sera were serially diluted and incubated with 100 TCID50 units of EBOV Mayinga (GenBank: NC_002549). Following incubation at 37°C for 1 h, Vero C1008 cells (ATCC CRL-1586) were added. Cytopathic effects were evaluated at day 7 p.i. Neutralization was defined as absence of CPE in serum dilutions. Neutralization titers of four replicates were calculated as geometric mean titers for sera (reciprocal value). The lower limit of detection (LLOD) of the assay is determined by the first dilution of the respective serum.
Virus titration by plaque assay
Vero C1008 cells were cultured to 100% confluence and infected with 10-fold serial dilutions of mouse sera starting at a dilution of 1:20 or 1:100. After 1 h the inoculum was replaced by an overlay consisting of 2% carboxymethylcellulose (cat. no.: C-5678, Sigma-Aldrich) in 1X Minimum Essential Medium (cat. no.: 51200-046, Thermo Fisher Scientific) supplemented with 2% fetal bovine serum, penicillin (50 U/mL), streptomycin (50 μg/mL), and glutamine (2 mM). At day 5 p.i. cells were fixed with 4% paraformaldehyde for 2 days. Cells were washed three times with PBS and permeabilized with PBS containing 0.1% Triton X-100 for 10 min. After three washes with PBS, cells were incubated with 100 mM glycine in PBS for 10 min. After one wash with PBS, the cells were incubated in blocking solution (BS, 2% BSA, 0.2% Tween 20, 5% glycerol in PBS). The virus-induced plaques were stained with an EBOV-specific goat serum (1:200 dilution) and an AlexaFluor488 secondary antibody from rabbit (1:500 dilution, cat. no. A27012, Thermo Fisher Scientific). Plaques were counted using a Nikon TS100 FL microscope, and PFU/mL were calculated.
T cell epitope prediction
The peptides for stimulation of splenocytes (Table S2) were selected based on a prediction of immunodominant peptides via database research (Immune Epitope Database and Analysis Resource, IEDB). In general, epitope prediction is based on, e.g., amphipathicity profile and recognized sequence motifs. The prediction method utilized by IEDB uses input protein amino acid sequences to identify binding cores, binding affinities, and residues flanking peptides based on large-scale systematic evaluation. Specificity for MHC I (processed by CD8+ T cells) or MHC II (processed by CD4+ T cells) was predicted by the length of the synthesized peptides (8–11 mers for MHC I and 13–17 mers for MHC II).
IFNγ secretion by splenocytes: ELISpot assay
ELISpot assay was performed using the Mabtech Mouse IFNγ ELISpotPLUS kit (cat. no. 3321-4APT-2, Mabtech, Nacka Strand, Sweden). Total isolated splenocytes (5 × 105) were seeded on pre-coated ELISpot plates and stimulated with the indicated peptide pools (1 μg/mL final concentration per peptide) overnight (18 h) in a humidified incubator at 37°C. Splenocytes were stimulated with MHC I- and MHC II-specific GP- or NP-specific peptide pools, with each pool containing six predicted peptides as described in the T cell epitope prediction section. Control measurements were performed using an irrelevant peptide pool (6 mg/mL murine leukemia virus envelope glycoprotein 70 peptide AH1; SPSYVYHQF66), medium only, or concanavalin A (2 mg/mL, cat. no. C0412, Sigma-Aldrich, Saint Louis, MO). IFNγ secretion was measured to assess T cell responses. Spots were visualized with a biotin-conjugated anti-IFNγ antibody followed by incubation with streptavidin-alkaline phosphatase and 5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium substrate. Plates were scanned using a CTL ImmunoSpot Analyzer and analyzed with the ImmunoCapture V6.3 software. All tests were performed in triplicate, and spot counts were summarized as median values for each triplicate.
Quantitative real-time RT-PCR analysis of virus load in mouse tissue samples
Tissue samples from liver and spleen of immunized and challenged mice were excised and homogenized in 1 mL DMEM with ceramic and glass beads (Lysing Matrix H 2-mL tubes, MP Biomedicals) in a Mixer Mill MM 400 (Retsch, Germany) instrument three times for 5 min. Homogenates were centrifuged for 5 min at 2,400 rpm in a Mikro 200R centrifuge (Hettich Lab Technology, Germany) to remove tissue debris. Aliquots of 100 μL of supernatants were used for RNA isolation with the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. The RNA amount of organ homogenates was measured by spectrophotometry with the NanoDrop ND-100. Total RNA was reverse transcribed and quantified by the means of a standard curve based on a real-time RT-PCR protocol that has been previously published to differentiate between EBOV virus subtypes Sudan and Zaire67 and adapted to our lab. Briefly, the One Step RT Kit (Qiagen) was used in combination with the primer pair (forward TGGGCTGAAAAYTGCTACAATC, reverse CTTTGTGMACATASCGGCAC) and probes (6FAM-TTACCCCCACCGCCGGATG-BHQ1, 6FAM-CTACCAGCAGCGCCAGACGG-BHQ1) on an ABI StepOnePlus Real-Time PCR System (Life Technologies Instruments). Cycling steps were as follows: 30 min at 50°C, 15 min at 95°C, followed by 45 cycles at 95°C for 15 s and 58°C for 30 s.
Statistical analysis
GraphPad Prism 8 Software (La Jolla, California) was used for statistical analysis and figure generation. Comparisons among groups were done for each measurement day by mixed effects analysis (Figures 2A, 3A, 3C, 4B, 4C, and 5B) or one-way ANOVA test (Figures 2B–2E, 3B, 3D, 4H, 4I, 5F, and 5G) with Tukey’s multiple comparison post-test. In all figures, asterisks indicate statistical significance as detailed by bars between relevant groups: ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p < 0.0001.
Acknowledgments
We thank Arianne Plaschke, Jonathan Mottl, Klara Zwadlo, Janina Vogt, Saskia Krapp, Denis Lutz, Imke Biermann, Bernadette Jesionek, Diana Schneider, Nele Brüne, Astrid Herwig, Dirk Becker, Katharina Kowalski, and Jörg Schmidt for excellent technical assistance. Special thanks to Andres Merits (University of Tartu, Estonia) for providing an SFV nsP3 antiserum. We thank Tim Beiβert for critically proofreading the manuscript. This work was supported by the German Center for Infection Research (DZIF), section Emergency Vaccines (FKZ 8033801805). The study was funded in part by BioNTech SE, who was involved in all stages of the study conduct and analysis.
Author contributions
Conceptualization, S.E., V.K., S.B., U.S.; methodology, S.E., V.K., A.K.; formal analysis: S.E., V.K., L.AS. investigation, S.E., V.K., S.N., J.M.H., H.B., K.W., A.K., E.D., S.H., C.R., L.S.; visualization, S.E., V.K.; resources, S.W., S.N.; writing – original draft, S.E., V.K., L.A.S., S.B.; supervision, U.S., S.B.; writing – review & editing, all authors.
Declaration of interests
S.E., K.W., H.B., J.M.H., and H.H. are employees and shareholders of BioNTech SE. L.A.S. is an employee of BioNTech SE. S.N. is a former employee of BioNTech SE and is currently employed by Merck KGaA. U.S. is a management board member and shareholder at BioNTech SE. S.E., J.M.H., H.H., S.B., and V.K. have a patent related to this work with the International Patent Application No. PCT/EP2022/061735 “REPLICON COMPOSITIONS AND METHODS OF USING SAME FOR THE TREATMENT OF DISEASES” BioNTech SE, TrOn GmbH, DZIF e.V., Our Ref.: 1405/PABION-167. S.H. and E.D. are former employees of the Philipps University of Marburg and are currently employed by GlaxoSmithKline.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2022.10.011.
Supplemental information
References
- 1.Murray K.A., Preston N., Allen T., Zambrana-Torrelio C., Hosseini P.R., Daszak P. Global biogeography of human infectious diseases. Proc. Natl. Acad. Sci. USA. 2015;112:12746–12751. doi: 10.1073/pnas.1507442112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogel A.B., Lambert L., Kinnear E., Busse D., Erbar S., Reuter K.C., Wicke L., Perkovic M., Beissert T., Haas H., et al. Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol. Ther. 2018;26:446–455. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brazzoli M., Magini D., Bonci A., Buccato S., Giovani C., Kratzer R., Zurli V., Mangiavacchi S., Casini D., Brito L.M., et al. Induction of broad-based immunity and protective efficacy by self-amplifying mRNA vaccines encoding influenza virus hemagglutinin. J. Virol. 2016;90:332–344. doi: 10.1128/JVI.01786-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moyo N., Vogel A.B., Buus S., Erbar S., Wee E.G., Sahin U., Hanke T. Efficient induction of T cells against conserved HIV-1 regions by mosaic vaccines delivered as self-amplifying mRNA. Mol. Ther. Methods Clin. Dev. 2019;12:32–46. doi: 10.1016/j.omtm.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloom K., van den Berg F., Arbuthnot P. Self-amplifying RNA vaccines for infectious diseases. Gene Ther. 2021;28:117–129. doi: 10.1038/s41434-020-00204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou X., Berglund P., Rhodes G., Parker S.E., Jondal M., Liljeström P. Self-replicating Semliki Forest virus RNA as recombinant vaccine. Vaccine. 1994;12:1510–1514. doi: 10.1016/0264-410x(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization Ebola Virus Disease: WHO Response. https://www.who.int/news-room/fact-sheets/detail/ebola-virus-disease
- 8.Matz K.M., Marzi A., Feldmann H. Ebola vaccine trials: progress in vaccine safety and immunogenicity. Expert Rev. Vaccin. 2019;18:1229–1242. doi: 10.1080/14760584.2019.1698952. [DOI] [PubMed] [Google Scholar]
- 9.Pollard A.J., Launay O., Lelievre J.-D., Lacabaratz C., Grande S., Goldstein N., Robinson C., Gaddah A., Bockstal V., Wiedemann A., et al. Safety and immunogenicity of a two-dose heterologous Ad26.ZEBOV and MVA-BN-Filo Ebola vaccine regimen in adults in Europe (EBOVAC2): a randomised, observer-blind, participant-blind, placebo-controlled, phase 2 trial. Lancet Infect. Dis. 2021;21:493–506. doi: 10.1016/S1473-3099(20)30476-X. [DOI] [PubMed] [Google Scholar]
- 10.Henao-Restrepo A.M., Camacho A., Longini I.M., Watson C.H., Edmunds W.J., Egger M., Carroll M.W., Dean N.E., Diatta I., Doumbia M., et al. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ça Suffit!) Lancet. 2017;389:505–518. doi: 10.1016/S0140-6736(16)32621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furuyama W., Shifflett K., Feldmann H., Marzi A. The Ebola virus soluble glycoprotein contributes to viral pathogenesis by activating the MAP kinase signaling pathway. Plos Pathog. 2021;17:e1009937. doi: 10.1371/journal.ppat.1009937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jain S., Martynova E., Rizvanov A., Khaiboullina S., Baranwal M. Structural and functional aspects of ebola virus proteins. Pathogens. 2021;10:1330. doi: 10.3390/pathogens10101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herst C.V., Burkholz S., Sidney J., Sette A., Harris P.E., Massey S., Brasel T., Cunha-Neto E., Rosa D.S., Chao W.C.H., et al. An effective CTL peptide vaccine for Ebola Zaire Based on Survivors' CD8+ targeting of a particular nucleocapsid protein epitope with potential implications for COVID-19 vaccine design. Vaccine. 2020;38:4464–4475. doi: 10.1016/j.vaccine.2020.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakabe S., Sullivan B.M., Hartnett J.N., Robles-Sikisaka R., Gangavarapu K., Cubitt B., Ware B.C., Kotliar D., Branco L.M., Goba A., et al. Analysis of CD8+ T cell response during the 2013-2016 ebola epidemic in West Africa. Proc. Natl. Acad. Sci. USA. 2018;115:E7578–E7586. doi: 10.1073/pnas.1806200115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin J.E., Sullivan N.J., Enama M.E., Gordon I.J., Roederer M., Koup R.A., Bailer R.T., Chakrabarti B.K., Bailey M.A., Gomez P.L., et al. A DNA vaccine for Ebola virus is safe and immunogenic in a phase I clinical trial. Clin. Vaccin. Immunol. 2006;13:1267–1277. doi: 10.1128/CVI.00162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marzi A., Feldmann H. Ebola virus vaccines: an overview of current approaches. Expert Rev. Vaccin. 2014;13:521–531. doi: 10.1586/14760584.2014.885841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sridhar S. Clinical development of Ebola vaccines. Ther. Adv. Vaccin. 2015;3:125–138. doi: 10.1177/2051013615611017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren S., Wei Q., Cai L., Yang X., Xing C., Tan F., Leavenworth J.W., Liang S., Liu W. Alphavirus replicon DNA vectors expressing ebola GP and VP40 antigens induce humoral and cellular immune responses in mice. Front. Microbiol. 2017;8:2662. doi: 10.3389/fmicb.2017.02662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kupke A., Volz A., Dietzel E., Freudenstein A., Schmidt J., Shams-Eldin H., Jany S., Sauerhering L., Krähling V., Gellhorn Serra M., et al. Protective CD8+ T cell response induced by modified vaccinia virus ankara delivering ebola virus nucleoprotein. Vaccines (Basel) 2022;10 doi: 10.3390/vaccines10040533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nanbo A., Watanabe S., Halfmann P., Kawaoka Y. The spatio-temporal distribution dynamics of Ebola virus proteins and RNA in infected cells. Sci. Rep. 2013;3:1206. doi: 10.1038/srep01206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoenen T., Shabman R.S., Groseth A., Herwig A., Weber M., Schudt G., Dolnik O., Basler C.F., Becker S., Feldmann H. Inclusion bodies are a site of ebolavirus replication. J. Virol. 2012;86:11779–11788. doi: 10.1128/JVI.01525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhattacharyya S., Hope T.J. Full-length Ebola glycoprotein accumulates in the endoplasmic reticulum. Virol. J. 2011;8:11. doi: 10.1186/1743-422X-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Escudero-Pérez B., Volchkova V.A., Dolnik O., Lawrence P., Volchkov V.E. Shed GP of Ebola virus triggers immune activation and increased vascular permeability. PLoS Pathog. 2014;10:e1004509. doi: 10.1371/journal.ppat.1004509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R., Zhou J., Bouton C.R., Rogers P., Polra K., et al. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 2020;11:3523. doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blakney A.K., Ip S., Geall A.J. An Update on Self-Amplifying mRNA Vaccine Development. Vaccines (Basel) 2021;9 doi: 10.3390/vaccines9020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kenney R.T., Frech S.A., Muenz L.R., Villar C.P., Glenn G.M. Dose sparing with intradermal injection of influenza vaccine. N. Engl. J. Med. 2004;351:2295–2301. doi: 10.1056/NEJMoa043540. [DOI] [PubMed] [Google Scholar]
- 27.Pepini T., Pulichino A.-M., Carsillo T., Carlson A.L., Sari-Sarraf F., Ramsauer K., Debasitis J.C., Maruggi G., Otten G.R., Geall A.J., et al. Induction of an IFN-mediated antiviral response by a self-amplifying RNA vaccine: implications for vaccine design. J. Immunol. 2017;198:4012–4024. doi: 10.4049/jimmunol.1601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwasaki A., Medzhitov R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015;16:343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belshe R.B., Newman F.K., Cannon J., Duane C., Treanor J., van Hoecke C., Howe B.J., Dubin G. Serum antibody responses after intradermal vaccination against influenza. N. Engl. J. Med. 2004;351:2286–2294. doi: 10.1056/NEJMoa043555. [DOI] [PubMed] [Google Scholar]
- 30.Anderluzzi G., Lou G., Woods S., Schmidt S.T., Gallorini S., Brazzoli M., Johnson R., Roberts C.W., O'Hagan D.T., Baudner B.C., Perrie Y. The role of nanoparticle format and route of administration on self-amplifying mRNA vaccine potency. J. Control. Release. 2022;342:388–399. doi: 10.1016/j.jconrel.2021.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leboux R.J.T., Schipper P., van Capel T.M.M., Kong L., van der Maaden K., Kros A., Jiskoot W., de Jong E.C., Bouwstra J.A. Antigen uptake after intradermal microinjection depends on antigen nature and formulation, but not on injection depth. Front. Allergy. 2021;2:642788. doi: 10.3389/falgy.2021.642788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanderzanden L., Bray M., Fuller D., Roberts T., Custer D., Spik K., Jahrling P., Huggins J., Schmaljohn A., Schmaljohn C. DNA vaccines expressing either the GP or NP genes of Ebola virus protect mice from lethal challenge. Virology. 1998;246:134–144. doi: 10.1006/viro.1998.9176. [DOI] [PubMed] [Google Scholar]
- 33.Pushko P., Bray M., Ludwig G.V., Parker M., Schmaljohn A., Sanchez A., Jahrling P.B., Smith J.F. Recombinant RNA replicons derived from attenuated Venezuelan equine encephalitis virus protect Guinea pigs and mice from Ebola hemorrhagic fever virus. Vaccine. 2000;19:142–153. doi: 10.1016/s0264-410x(00)00113-4. [DOI] [PubMed] [Google Scholar]
- 34.Meyer M., Huang E., Yuzhakov O., Ramanathan P., Ciaramella G., Bukreyev A. Modified mRNA-based vaccines elicit robust immune responses and protect Guinea pigs from ebola virus disease. J. Infect. Dis. 2018;217:451–455. doi: 10.1093/infdis/jix592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chahal J.S., Khan O.F., Cooper C.L., McPartlan J.S., Tsosie J.K., Tilley L.D., Sidik S.M., Lourido S., Langer R., Bavari S., et al. Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc. Natl. Acad. Sci. USA. 2016;113:E4133–E4142. doi: 10.1073/pnas.1600299113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ballesteros-Briones M.C., Silva-Pilipich N., Herrador-Cañete G., Vanrell L., Smerdou C. A new generation of vaccines based on alphavirus self-amplifying RNA. Curr. Opin. Virol. 2020;44:145–153. doi: 10.1016/j.coviro.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouazzaoui A., Abdellatif A.A.H., Al-Allaf F.A., Bogari N.M., Al-Dehlawi S., Qari S.H. Strategies for vaccination: conventional vaccine approaches versus new-generation strategies in combination with adjuvants. Pharmaceutics. 2021;13:140. doi: 10.3390/pharmaceutics13020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geisbert T.W., Pushko P., Anderson K., Smith J., Davis K.J., Jahrling P.B. Evaluation in nonhuman primates of vaccines against Ebola virus. Emerg. Infect. Dis. 2002;8:503–507. doi: 10.3201/eid0805.010284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakayama E., Saijo M. Animal models for Ebola and Marburg virus infections. Front. Microbiol. 2013;4:267. doi: 10.3389/fmicb.2013.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hevey M., Negley D., Pushko P., Smith J., Schmaljohn A. Marburg virus vaccines based upon alphavirus replicons protect Guinea pigs and nonhuman primates. Virology. 1998;251:28–37. doi: 10.1006/viro.1998.9367. [DOI] [PubMed] [Google Scholar]
- 41.Falzarano D., Geisbert T.W., Feldmann H. Progress in filovirus vaccine development: evaluating the potential for clinical use. Expert Rev. Vaccin. 2011;10:63–77. doi: 10.1586/erv.10.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.St Claire M.C., Ragland D.R., Bollinger L., Jahrling P.B. Animal models of ebolavirus infection. Comp. Med. 2017;67:253–262. [PMC free article] [PubMed] [Google Scholar]
- 43.Safronetz D., Geisbert T.W., Feldmann H. Animal models for highly pathogenic emerging viruses. Curr. Opin. Virol. 2013;3:205–209. doi: 10.1016/j.coviro.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olinger G.G., Bailey M.A., Dye J.M., Bakken R., Kuehne A., Kondig J., Wilson J., Hogan R.J., Hart M.K. Protective cytotoxic T-cell responses induced by venezuelan equine encephalitis virus replicons expressing Ebola virus proteins. J. Virol. 2005;79:14189–14196. doi: 10.1128/JVI.79.22.14189-14196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson J.A., Hart M.K. Protection from Ebola virus mediated by cytotoxic T lymphocytes specific for the viral nucleoprotein. J. Virol. 2001;75:2660–2664. doi: 10.1128/JVI.75.6.2660-2664.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sullivan N.J., Hensley L., Asiedu C., Geisbert T.W., Stanley D., Johnson J., Honko A., Olinger G., Bailey M., Geisbert J.B., et al. CD8+ cellular immunity mediates rAd5 vaccine protection against Ebola virus infection of nonhuman primates. Nat. Med. 2011;17:1128–1131. doi: 10.1038/nm.2447. [DOI] [PubMed] [Google Scholar]
- 47.Sullivan N.J., Geisbert T.W., Geisbert J.B., Shedlock D.J., Xu L., Lamoreaux L., Custers J.H.H.V., Popernack P.M., Yang Z.-Y., Pau M.G., et al. Immune protection of nonhuman primates against Ebola virus with single low-dose adenovirus vectors encoding modified GPs. PLoS Med. 2006;3:e177. doi: 10.1371/journal.pmed.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sullivan N.J., Geisbert T.W., Geisbert J.B., Xu L., Yang Z.-Y., Roederer M., Koup R.A., Jahrling P.B., Nabel G.J. Accelerated vaccination for Ebola virus haemorrhagic fever in non-human primates. Nature. 2003;424:681–684. doi: 10.1038/nature01876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swenson D.L., Wang D., Luo M., Warfield K.L., Woraratanadharm J., Holman D.H., Dong J.Y., Pratt W.D. Vaccine to confer to nonhuman primates complete protection against multistrain Ebola and Marburg virus infections. Clin. Vaccin. Immunol. 2008;15:460–467. doi: 10.1128/CVI.00431-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bukreyev A., Rollin P.E., Tate M.K., Yang L., Zaki S.R., Shieh W.-J., Murphy B.R., Collins P.L., Sanchez A. Successful topical respiratory tract immunization of primates against Ebola virus. J. Virol. 2007;81:6379–6388. doi: 10.1128/JVI.00105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bukreyev A., Yang L., Zaki S.R., Shieh W.-J., Rollin P.E., Murphy B.R., Collins P.L., Sanchez A. A single intranasal inoculation with a paramyxovirus-vectored vaccine protects Guinea pigs against a lethal-dose Ebola virus challenge. J. Virol. 2006;80:2267–2279. doi: 10.1128/JVI.80.5.2267-2279.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marzi A., Ebihara H., Callison J., Groseth A., Williams K.J., Geisbert T.W., Feldmann H. Vesicular stomatitis virus-based Ebola vaccines with improved cross-protective efficacy. J. Infect. Dis. 2011;204:S1066–S1074. doi: 10.1093/infdis/jir348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orlandini von Niessen A.G., Poleganov M.A., Rechner C., Plaschke A., Kranz L.M., Fesser S., Diken M., Löwer M., Vallazza B., Beissert T., et al. Improving mRNA-based therapeutic gene delivery by expression-augmenting 3' UTRs identified by cellular library screening. Mol. Ther. 2019;27:824–836. doi: 10.1016/j.ymthe.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuhn A.N., Diken M., Kreiter S., Selmi A., Kowalska J., Jemielity J., Darzynkiewicz E., Huber C., Türeci O., Sahin U. Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther. 2010;17:961–971. doi: 10.1038/gt.2010.52. [DOI] [PubMed] [Google Scholar]
- 55.Pokrovskaya I.D., Gurevich V.V. In vitro transcription: preparative RNA yields in analytical scale reactions. Anal. Biochem. 1994;220:420–423. doi: 10.1006/abio.1994.1360. [DOI] [PubMed] [Google Scholar]
- 56.Hoenen T., Groseth A., Kolesnikova L., Theriault S., Ebihara H., Hartlieb B., Bamberg S., Feldmann H., Ströher U., Becker S. Infection of naive target cells with virus-like particles: implications for the function of ebola virus VP24. J. Virol. 2006;80:7260–7264. doi: 10.1128/JVI.00051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krähling V., Stein D.A., Spiegel M., Weber F., Mühlberger E. Severe acute respiratory syndrome coronavirus triggers apoptosis via protein kinase R but is resistant to its antiviral activity. J. Virol. 2009;83:2298–2309. doi: 10.1128/JVI.01245-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krähling V., Becker D., Rohde C., Eickmann M., Eroğlu Y., Herwig A., Kerber R., Kowalski K., Vergara-Alert J., Becker S., European Mobile Laboratory Consortium Development of an antibody capture ELISA using inactivated Ebola Zaire Makona virus. Med. Microbiol. Immunol. 2016;205:173–183. doi: 10.1007/s00430-015-0438-6. [DOI] [PubMed] [Google Scholar]
- 59.Lucht A., Grunow R., Otterbein C., Möller P., Feldmann H., Becker S. Production of monoclonal antibodies and development of an antigen capture ELISA directed against the envelope glycoprotein GP of Ebola virus. Med. Microbiol. Immunol. 2004;193:181–187. doi: 10.1007/s00430-003-0204-z. [DOI] [PubMed] [Google Scholar]
- 60.Saul S., Ferguson M., Cordonin C., Fragkoudis R., Ool M., Tamberg N., Sherwood K., Fazakerley J.K., Merits A. Differences in processing determinants of nonstructural polyprotein and in the sequence of nonstructural protein 3 affect neurovirulence of Semliki forest virus. J. Virol. 2015;89:11030–11045. doi: 10.1128/JVI.01186-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kolesnikova L., Berghöfer B., Bamberg S., Becker S. Multivesicular bodies as a platform for formation of the Marburg virus envelope. J. Virol. 2004;78:12277–12287. doi: 10.1128/JVI.78.22.12277-12287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schaupp L., Muth S., Rogell L., Kofoed-Branzk M., Melchior F., Lienenklaus S., Ganal-Vonarburg S.C., Klein M., Guendel F., Hain T., et al. Microbiota-induced type I interferons instruct a poised basal state of dendritic cells. Cell. 2020;181:1080–1096.e19. doi: 10.1016/j.cell.2020.04.022. [DOI] [PubMed] [Google Scholar]
- 63.Müller U., Steinhoff U., Reis L.F., Hemmi S., Pavlovic J., Zinkernagel R.M., Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 64.Oestereich L., Lüdtke A., Wurr S., Rieger T., Muñoz-Fontela C., Günther S. Successful treatment of advanced Ebola virus infection with T-705 (favipiravir) in a small animal model. Antivir. Res. 2014;105:17–21. doi: 10.1016/j.antiviral.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 65.Ehrhardt S.A., Zehner M., Krähling V., Cohen-Dvashi H., Kreer C., Elad N., Gruell H., Ercanoglu M.S., Schommers P., Gieselmann L., et al. Polyclonal and convergent antibody response to Ebola virus vaccine rVSV-ZEBOV. Nat. Med. 2019;25:1589–1600. doi: 10.1038/s41591-019-0602-4. [DOI] [PubMed] [Google Scholar]
- 66.Slansky J.E., Rattis F.M., Boyd L.F., Fahmy T., Jaffee E.M., Schneck J.P., Margulies D.H., Pardoll D.M. Enhanced antigen-specific antitumor immunity with altered peptide ligands that stabilize the MHC-peptide-TCR complex. Immunity. 2000;13:529–538. doi: 10.1016/s1074-7613(00)00052-2. [DOI] [PubMed] [Google Scholar]
- 67.Gibb T.R., Norwood D.A., Woollen N., Henchal E.A. Development and evaluation of a fluorogenic 5' nuclease assay to detect and differentiate between Ebola virus subtypes Zaire and Sudan. J. Clin. Microbiol. 2001;39:4125–4130. doi: 10.1128/JCM.39.11.4125-4130.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.