Summary
In contrast to mammals, the avian cochlea, specifically the basilar papilla, can regenerate sensory hair cells, which involves fate conversion of supporting cells to hair cells. To determine the mechanisms for converting supporting cells to hair cells, we used single-cell RNA sequencing during hair cell regeneration in explant cultures of chick basilar papillae. We identified dynamic changes in the gene expression of supporting cells, and the pseudotime trajectory analysis demonstrated the stepwise fate conversion from supporting cells to hair cells. Initially, supporting cell identity was erased and transition to the precursor state occurred. A subsequent gain in hair cell identity progressed together with downregulation of precursor-state genes. Transforming growth factor β receptor 1-mediated signaling was involved in induction of the initial step, and its inhibition resulted in suppression of hair cell regeneration. Our data provide new insights for understanding fate conversion from supporting cells to hair cells in avian basilar papillae.
Subject areas: Cell biology, Sensory neuroscience, Transcriptomics
Graphical abstract

Highlights
-
•
Chick hair cell regeneration model via direct conversion of supporting cells
-
•
Single-cell RNA sequencing reveals stepwise fate conversion of supporting cells
-
•
The intermediate precursor-like population expresses EDNRB2
-
•
Transforming growth factor β signaling is involved in the induction of conversion
Cell biology; Sensory neuroscience; Transcriptomics
Introduction
Auditory hair cells (HCs) convert sound vibration into electrical potential that stimulates the primary auditory neurons and the loss of HCs diminishes auditory function. In the mammalian cochlea, virtually no HC regeneration occurs, leading to intractable sensorineural hearing loss. In contrast, in the avian cochlea, HCs are spontaneously restored after damage resulting in the maintenance of hearing throughout life. Therefore, the avian auditory sensory epithelium, known as the basilar papilla (BP), has been used as a model to study the mechanisms of HC regeneration for decades. Avian BP HCs are regenerated through two different pathways: direct conversion of supporting cells (SCs) to HCs (SC-to-HC conversion) and SC mitosis followed by differentiation into HCs,1 with the former being the predominant mode of HC regeneration in explant cultures of chick BPs.1,2 However, the detailed molecular mechanisms underlying HC regeneration in avian BPs have not been fully elucidated. To address this question, we established an explant culture model of chick BPs for HC regeneration and performed bulk RNA sequencing (RNA-seq).3 A notable characteristic of our explant culture model is that almost all new HCs were generated through SC-to-HC conversion.3
SC-to-HC conversion is also feasible in the mammalian cochlea by forced expression of Atoh1, a basic helix-loop-helix transcription factor that is required for differentiation of HCs from precursor cells during development.4,5,6 In the neonatal mouse cochlea, forced expression of Atoh1 by pharmacological inhibition of Notch signaling induces SC-to-HC conversion.7,8 Genetic manipulation to direct forced expression of Atoh1 induces SC-to-HC conversion in the mature mammalian cochlea.9,10 However, its efficacy progressively decreases with age,11,12 suggesting that Atoh1-forced expression is not sufficient for the induction of SC-to-HC conversion in the mature mammalian cochlea. In addition, HCs generated by forced expression of Atoh1 exhibit the biophysical characteristics of HCs but fail to fully differentiate,11,12,13 indicating that additional factors are required for the generation of mature HCs in adult mammals. Recently, the efficacy of additional factors in promoting SC-to-HC conversion has been reported in neonatal or juvenile14,15 and adult mice.16,17,18 However, to the best of our knowledge, sufficient recovery of auditory function has not yet been achieved in mammals.
In contrast to the mature mammalian cochleae, SCs in mature avian BPs can spontaneously upregulate ATOH1 after HC damage. In chick BPs, ATOH1 is highly expressed in SCs shortly after HC damage, and this upregulation occurs broadly across the BP.19 Not all ATOH1-expressing SCs show a change in cell fate to become new HCs.19 In addition, in developing mouse cochleae, many sensory precursor cells initially express Atoh1 and some Atoh1-expressing cells differentiate into HCs.20 However, the distinct mechanisms by which Atoh1-expressing SCs in chick BPs or precursor cells in developing mouse cochleae acquire the HC fate are unknown. A recent review of advancements and controversies in studies of stepwise fate conversion of several somatic cells to neurons indicates the importance of characterizing immature and intermediate phenotypes during direct conversion to facilitate the elucidation of a scientifically firm footing.21 Thus, understanding the precise processes of SC-to-HC conversion in chick BPs may contribute to the achievement of more practical and sufficient SC-to-HC conversion in mature mammalian cochleae. Although it is challenging to apply findings related to HC regeneration in birds to mammals, the data generated from chick BP analyses sometimes help further understand HC regeneration in mammals.22,23
Molecular analyses of BP development have focused on major signaling pathways, including Notch,24,25,26 fibroblast growth factor,27,28 and Wnt signaling.29,30 However, information on the gene expression patterns in developing or regenerating chick BPs in comparison to those in the zebrafish lateral line31,32 was limited. The baseline data of homeostatic BPs have been recently reported using single-cell RNA-seq,33 which provides fundamental information to analyze the changes in gene expression in SCs toward HC regeneration, similar to the findings in zebrafish31,32 and mice.34 To gain insights into the processes underlying SC-to-HC conversion in chick BPs, we conducted a high-resolution transcriptional analysis of SCs during SC-to-HC conversion in our explant culture model using single-cell RNA-seq.
Results
Hair cell regeneration occurs in explant cultures of chick basilar papillae
The BPs excised from post-hatch day 1 (P1) chicks were maintained for 24 h in the control media and provided explant cultures (Figure S1A). BPs consist of two types of sensory epithelial cells: HCs and SCs (Figure S1B). HCs are divided into two phenotypes: tall and short. Tall HCs are located in the neural portion of the BP and short HCs are present in the abneural portion (Figure S1B). Several types of non-sensory cells, including homogene cells, are present in the neural and abneural areas adjacent to sensory cells (Figure S1B). These non-sensory cells are connected to epithelial cells in the roof of the cochlear duct (Figure S1B).
Half of the BPs were exposed to streptomycin (SM), an ototoxic antibiotic, for 48 h to induce HC death, and the remaining BPs were maintained in control media for 48 h (Figure S1A). Subsequently, both samples were maintained in control media for an additional 96 h (Figure S1A). At the end of the culture period, the BPs were fixed and used for histological assessment. To assess the deletion of original HCs from cultured BPs and the generation of new HCs, we performed immunostaining for MYO7A and SOX2. We observed the two layers of the surface of the 40% area from the distal end of BPs (Figure S1C) and counted the number of HCs, which were labeled with MYO7A, and the newly generated, immature HCs, which were co-stained with MYO7A and SOX2. In samples that were cultured with SM, virtually no HCs were found in the 40% area following 48-h culture (day2_SM; Figure S1D), and a certain number of HCs were identified in the same region of BPs after additional 96-h culture in the control medium (day6_SM; Figure S1D). Immunohistochemistry demonstrated the presence of newly generated immature HCs in the specimens from day6_SM (Figure S1D). Quantitative assessments ascertained the total HC loss after 48 h of exposure to SM and the occurrence of HC regeneration (Figure S1F). These findings demonstrated that HC regeneration occurred during an additional 96 h of culture after 48 h of exposure to SM.
In BP cultures without SM exposure, HCs were well maintained during the 6-day culture period (Figure S1E). However, quantitative assessments indicated modest HC loss and the generation of new HCs during the 6-day culture, although the difference was not statistically significant among experimental groups (Figures S1G). These findings suggest that HC loss and subsequent regeneration also occurred in BP explants cultured in control media, which is almost identical to the previous observation.24
Single-cell RNA sequencing of chick basilar papillae can identify clusters of supporting cells and neighboring non-sensory cells
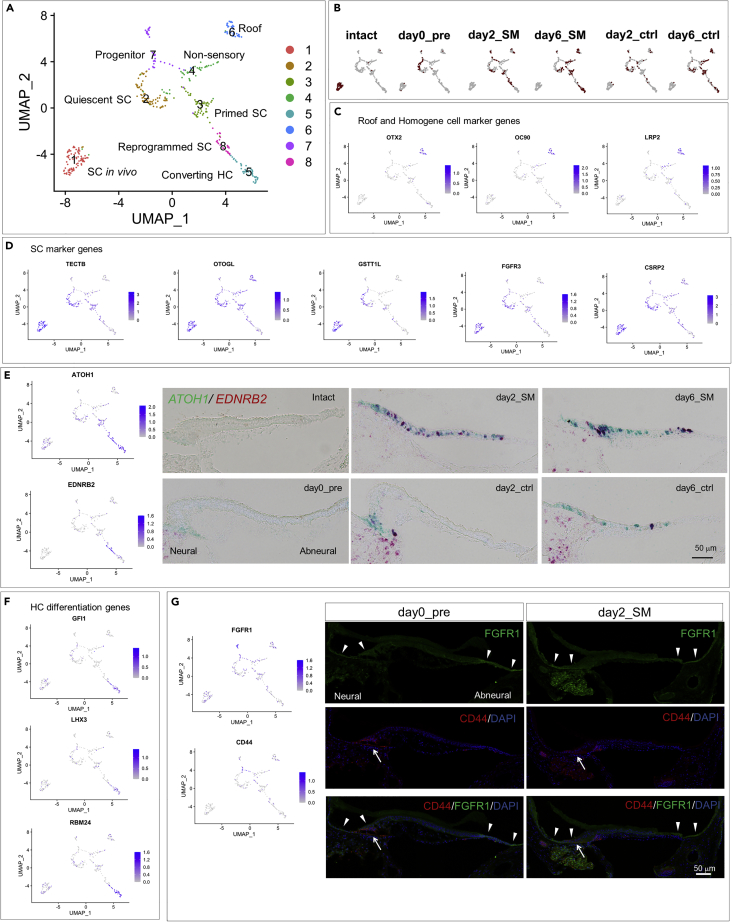
To collect SCs at different stages of HC regeneration, we prepared five experimental groups of BP explant cultures: day0_pre (before explant culture), day2_SM (after 48-h SM exposure), day6_SM (96-h culture after 48-h SM exposure), day2_Ctrl (48-h culture without SM exposure), and day6_Ctrl (144-h culture without SM exposure) (Figure 1A). Histological assessments (Figure S1) indicated that these specimens contained intact SCs in the culture condition, SCs responding to HC injury at various stages of HC regeneration, and newly generated and intact mature HCs (Figure 1A). In addition to these five experimental groups, we collected BP samples that were immediately dissected from P1 chicks, namely, the intact group, which contained HCs and SCs in vivo (Figure 1A). Consequently, we harvested single cells from the six experimental groups (15–17 BPs per group).
Figure 1.
Single-cell RNA sequencing of chick basilar papillae identifies clusters of supporting cells and neighboring non-sensory cells
(A) Experimental groups. HC: hair cell, SC: supporting cell, HG: homogene cell, NS non-sensory cell, SM: streptomycin.
(B) Uniform manifold approximation and projection (UMAP) plots of whole-cell populations (1054 cells). A circle indicates a group of clusters consisting of SCs and neighboring non-sensory cells. (C) UMAP plots for SC markers.
(D–F) UMAP plots for FGFR3 (D), AGR3 (E), and CSRP2 (F) and histological distributions (in situ hybridization for FGFR3, immunostaining for AGR3 and CSRP2) in transverse sections of the basilar papillae (BPs) before exposure to streptomycin (day0_pre). Scale bar in (F) represents 50 μm for (D–F).
(G) UMAP plots for ATOH1 and in situ hybridization for ATOH1 in transverse sections of BPs after HC injury (day2_SM and day6_SM). Scale bar represents 50 μm.
(H–K) UMAP plots for HC markers (H), a mesenchyme marker (I), a marker for neural crest-derived cells (J), and microglia markers (K). Arrowheads indicate corresponding clusters. See also Figures S1 and S2.
The capture of single cells dissociated from chick BPs and cDNA synthesis were performed using the C1 Single-Cell Auto Prep system (Fluidigm). Raw sequencing data were converted into FASTQ files and mapped to the chicken reference genome GRCg6a. After removing the cells with an RNA count of less than 20,000, detected genes less than 1,000, detected genes more than 4,000, or percentage of mitochondrial genes more than 12.5%, the remaining 1,054 cells from chick BPs were normalized. Unsupervised clustering identified 12 clusters (Figure 1B).
The expression of known SC markers was examined to identify clusters containing SCs. Janesick et al.33 demonstrated the similarity of expressed genes in neighboring non-sensory cells to those in SCs. Therefore, in this step, we extracted clusters containing SCs and neighboring non-sensory cells. The distributions of the SC marker genes TECTB, OTOGL, ID2, and GSTTL1 indicated that a group of clusters 1, 5, 8, 9, and 11 included SCs (Figures 1B and 1C). We histologically examined the expression of FGFR3, AGR3, and CSRP2 as SC marker candidates, which were observed in SCs and neighboring non-sensory cells in day0_pre samples by in situ hybridization (ISH) or immunohistochemistry (Figures 1D–1F). These markers were also found in clusters 1, 5, 8, 9, and 11 (Figures 1D–1F). ATOH1, a key transcription factor for SC-to-HC conversion, is expressed in SCs in response to HC injury and converting HCs.35 Histologically, ATOH1 expression was broadly observed in SCs after HC injury by SM (Figure 1G), which is similar to previous observations.3,19,35 High expression of ATOH1 was found in clusters 1 and 8 (Figure 1G), indicating that clusters 1 and 8 contained SCs responding to HC injury. Altogether, we identified clusters 1, 5, 8, 9, and 11 as SC clusters that contained SCs and neighboring non-sensory cells for subsequent analyses.
To characterize other clusters, we examined the distribution of HC markers (OCM, MYO7A, and SLC34A2),33 mesenchymal cell markers (POU3F4),36,37 a neural crest-derived cell marker (FOXD),38,39 and microglial markers (LAPTM5 and C1QB).40 All HC marker genes were strongly expressed in clusters 2, 3, 6, and 7 (Figure 1H). The expression of POU3F4 was identified in cluster 4 (Figure 1I), and that of FOXD3 was selectively detected in cluster 10 (Figure 1J). Expression of LAPTM5 and C1QB was noted in cluster 12 (Figure 1K).
Identification of supporting cells at different stages toward hair cell regeneration
We extracted clusters 1, 5, 8, 9, and 11 (Figure 1B), which contained SCs and neighboring non-sensory cells, and re-clustered this dataset. Eight SC clusters were obtained (Figures 2A and S2). Figure 2B shows the distribution of the SC clusters in each experimental group. The cells derived from the intact group were clearly separated from those harvested from the cultured BP samples, which were distributed in SC cluster 1 (Figure 2A). Thus, we annotated SC cluster 1 as SCs in vivo.
Figure 2.
Identification of supporting cell clusters at different stages toward hair cell regeneration
(A) Uniform manifold approximation and projection (UMAP) plots of supporting cells and non-sensory cells (436 cells in total).
(B) UMAP plots for each experimental group.
(C, D, and F) UMAP plots for roof and homogene cell markers (C), supporting cell markers (D), and hair cell differentiation genes (F).
(E) UMAP plots for ATOH1 and EDNRB2 and in situ hybridization for ATOH1 (green) and EDNRB2 (red) in transverse sections of basilar papillae (BPs) of each experimental group. Scale bar represents 50 μm.
(G) UMAP plots for FGFR1 and CD44 and immunostaining for FGFR1 (green) and CD44 (red) in transverse sections of BP of day0_pre and day2_SM specimens. Arrowheads indicate FGFR1 expression, and arrows indicate CD44 expression. Scale bar represents 50 μm. See also Figures S3 and S4 and Tables S1 and S3.
In the hierarchical cluster analysis, SC cluster 6 was clearly separated from the other SC clusters (Figure S2). SC cluster 6 was characterized by the expression of OTX2, OC90, and LRP2 (Figures 2C, S3F, and Table S1). OTX2 is a marker for the tegmentum vasculosum of the chick cochlear duct roof,41 and OC90 is expressed in the roof of the developing mouse cochleae.42 Thus, SC cluster 6 has the characteristics of roof cells. LRP2 is a specific marker for homogene cells,33,43 and its expression is observed in the apical compartments of homogene cells adjacent to the roof of the cochlear duct.33 Altogether, SC cluster 6 may be composed of apical compartments of homogene cells and roof cells. On the basis of these findings, we annotated SC cluster 6 as roof cells (Figure 2A).
SC cluster 4 also exhibited LRP2 expression (Figure 2C), indicating that SC cluster 4 shares the identity of homogene cells. Hierarchical cluster analysis showed similarities among SC clusters 2, 3, and 4 (Figure S2). Among the three clusters, SC cluster 4 was characterized by low expression of SC marker genes (Figure 2D). In comparison with SC cluster 2 or 3, SC cluster 4 exhibited lower expression of several SC marker genes (FGFR3, TECTB, GSTT1L, and LCAT) (Figures S4A, S4B, and Table S3). On the other hand, in homeostatic chick BPs, Janesick et al.33 demonstrated the expression of SC marker genes including TECTB, ID2, and TIMP3, in the basal component of homogene cells and non-sensory cells adjacent to the sensory epithelium. CYR61 was included in the differentially expressed genes (DEGs) for SC cluster 4 (Figure S3D and Table S1), and its expression was observed in non-sensory cells adjacent to the sensory epithelium.3 On the basis of these findings, SC cluster 4 was annotated as a cluster of non-sensory cells (Figure 2A).
Intense expression of SC marker genes (Figure 2D) and faint or virtually no expression of LRP2 (Figure 2C) were characteristics of SC cluster 2. We then annotated SC cluster 2 as a quiescent SC. SC cluster 3 also showed expression of SC marker genes (Figure 2D), but in comparison with SC cluster 2, SC cluster 3 showed downregulation of several SC marker genes (CSRP2, TECTB, FGFR3, LCAT, GSTT1L, and AGR3) (Figure 3A and Table S2), indicating that while SC identity was still maintained in SC cluster 3, the erasure of SC identity had been initiated. Based on this finding, we speculated that SC cluster 3 might be involved in the process of priming SC-to-HC conversion. Thus, we annotated SC cluster 3 as primed SCs (Figure 2A).
Figure 3.
Volcano plots for comparison of expressed genes between SC clusters
Volcano plots showing highly expressed genes (FDR <0.05, log-fold-change threshold >0.5) in comparisons between SC clusters 3 and 2 (A), SC clusters 5 and 8 (B), SC clusters 8 and 3 (C), and SC clusters 8 and 2 (D). Genes of interest are marked by colors corresponding to Figure 2A. See also Table S2.
We highlighted the distribution of ATOH1 expression in SC clusters. High expression of ATOH1 was observed in SC clusters 5 and 8 (Figure 2E), and ATOH1 was included in the DEGs of SC clusters 5 and 8 (Figures S3E and S3H, and Table S1). In SC cluster 5, high expression of the HC differentiation genes GFI1,44 LHX3,45 and RBM2446,47 was also identified (Figure 2F). In comparison with SC cluster 8, SC cluster 5 showed upregulation of HC marker genes (MYO7A, GPX2, CXCL14, TMC2, and OCM) and HC differentiation genes (GFI1 and RBM24) (Figure 3B and Table S2). In contrast, SC cluster 8 exhibited higher expression of SC marker genes, including TMSB4X, TECTB, and CSRP2, than SC cluster 5 (Figure 3B and Table S2). These findings indicate that SC cluster 5 was in a more advanced stage of differentiation into HCs than SC cluster 8. Thus, we annotated SC cluster 5 as a cluster for converting HCs. Among HC marker genes, the DEGs of SC cluster 5 included one tall HC maker (CXCL14)33 but no short HC marker.
Compared to SC cluster 2 or 3, SC cluster 8 showed downregulation of SC marker genes (AGR3, TECTB, LCAT, OCM, and TIMP3) (Figures 3C and 3D, and Table S2), indicating the progression of erasure of SC identity in comparison with SC clusters 2 and 3. In addition, DLL1 was identified in the DEGs of SC cluster 8 (Figure S3H and Table S1). In comparison with SC clusters 2 or 3, DLL1 and ATOH1 were upregulated in SC cluster 8 (Figures 3B and 3C, and Table S2), indicating that SC cluster 8 cells were directed to differentiate into HCs.26 Another characteristic of SC cluster 8 was the upregulation of the cell-cycle-associated genes MCM6, STMN1, CKS1B, and CDKN1C (Figure S3H and Table S1). Wnt-associated genes FZD9 and SFRP2 (Figure S3H and Table S1), which are expressed in the precursor cells of chick BPs during development,30 were also upregulated in SC cluster 8. Altogether, SC cluster 8 was directed to erase SC identity and to reverse cell identity to a developmentally immature status. Therefore, we annotated SC cluster 8 as reprogrammed SCs (Figure 2A). The expression of EDNRB2 was noted among the DEGs in SC cluster 8 (Figure S3H and Table S1). Furthermore, EDNRB2 distribution was comparatively specific for SC cluster 8 (Figure 2E). We histologically examined the expression of EDNRB2 in BPs together with ATOH1. In specimens from day2_SM, day6_SM, and day6_ctrl, some ATOH1-expressing SCs co-expressed EDNRB2 (Figure 2E). On the basis of these results, we considered that EDNRB2 could be a marker for reprogrammed SCs.
SC cluster 7 showed unique DEGs, including PMP22, GNL3, and CD44 (Figure S3G and Table S1), which are known to play roles in the maintenance of progenitor or stem cell pools.48,49,50,51,52,53 In addition, FGFR1 expression was noted in SC cluster 7 (Figures 2G, S3G, S4C, and S4D and Table S1). FGFR1 is crucial for the generation of the progenitor pool in the auditory sensory epithelium during murine cochlear development.54,55,56 On the basis of these findings, SC cluster 7 was annotated as a cluster of progenitor pools. To examine the localization of SC cluster 7 cells, immunohistochemistry for FGFR1 and CD44 was performed using specimens from day0_pre and day2_SM. FGFR1 expression was predominantly observed in non-sensory cells adjacent to the neural or abneural edges of the sensory epithelium (Figure 2G). CD44 expression was observed in non-sensory cells and SCs at the neural edge of the sensory epithelium (Figure 2G). Some CD44-positive cells co-expressed FGFR1 (Figure 2G). These findings indicate that SC cluster 7 cells may be present in the far neural and abneural regions of BPs, which are almost identical to the previously reported localization of stem cell-like populations in chick BPs.57
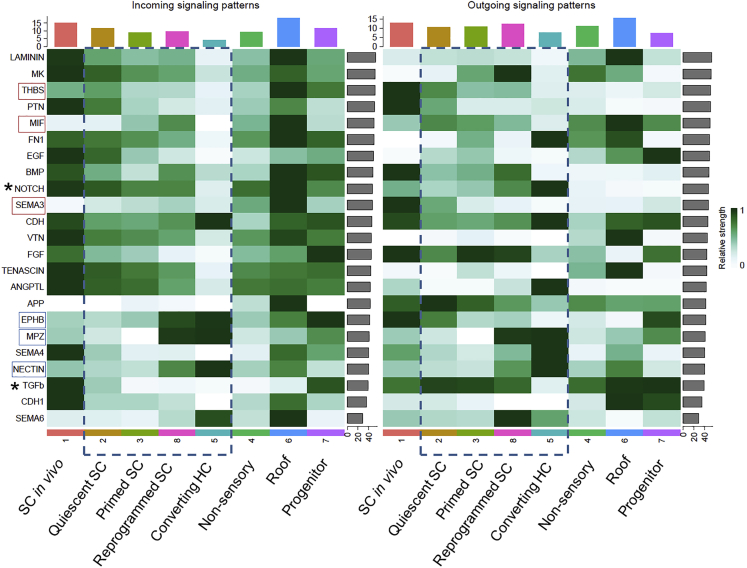
To investigate the activated signaling pathways in SC clusters, we performed a signaling pathway analysis using CellChat.58 As for NOTCH signaling, comparatively intense incoming signaling was observed in SC clusters 2, 3, and 8, whereas virtually no incoming signaling was seen in SC cluster 5 (Figure 4). Nonetheless, in outgoing signaling patterns, intense signaling was seen in SC cluster 5 (Figure 4), which is consistent with acquiring HC identity in converting HCs. While observing incoming signaling patterns of SC clusters 2, 3, 8, and 5, which contain SCs at different stages of SC-to-HC conversion, temporal activation of THBS signaling pathway was observed in SC cluster 2, and we noted a gradual upregulation of MIF and SEMA3 signaling pathways from SC cluster 2 to 8 (Figure 4). This finding suggested the involvement of these signaling pathways in the early phase of SC-to-HC conversion. Conversely, the activation of EPHB, MPZ, and NECTIN signaling pathways was noted in SC clusters 8 and 5 (Figure 4), which are known to play a key role in the regulation of adhesion molecules in the cochlear sensory epithelium.59,60,61 Particularly, EPHB and NECTIN signaling pathways play a crucial role in mosaic patterning of HCs and SCs in developing cochlear sensory epithelia.59,60 The activation of these signaling pathways is expected for the late phase of HC regeneration processes.
Figure 4.
Heatmaps of incoming signaling and outgoing signaling patterns of SC clusters
A bar plot of the x axis indicates the sum of the centrality scores for each SC cluster and that of the y axis indicates the sum of the centrality scores for each signal.
SC clusters of interest are indicated by squares made of blue dotted lines. Signaling pathways showing the upregulation of incoming signaling in quiescent and primed SCs are marked with red squares, and those showing the upregulation of incoming signaling in reprogrammed SCs and converting HCs are marked with blue squares. NOTCH and TGFb signaling pathways are marked using asterisks.
Consequently, SC clusters 2, 3, 8, and 5 were extracted as clusters containing SCs at different stages of SC-to-HC conversion for a subsequent pseudotime trajectory analysis.
Pseudotime trajectory analysis reveals the stepwise fate conversion of supporting cells to hair cells
To characterize the processes underlying SC-to-HC conversion, we extracted the dataset of SC clusters 2, 3, 5, and 8 and performed a pseudotime trajectory analysis using Slingshot v1.6.0.62 The uniform manifold approximation and projection plots exhibited an approximately linear distribution of SCs (Figure 5A). We examined alterations in the expression levels of the genes of interest along a pseudotime trajectory. To classify genes according to alteration patterns along a pseudotime trajectory, we identified 500 DEGs along a pseudotime trajectory by fitting a random forest regression model.63 The top 500 genes that showed alterations along a pseudotime were extracted and classified into six time-series clusters (Figures 5D–5I and S5 and Table S4). In Figures 5B–5I and S5, the x axis represents the pseudotime (0–1.0) and the y axis shows the normalized expression value of the genes of interest.
Figure 5.
Pseudotime trajectory analysis reveals the stepwise fate conversion of supporting cells to hair cells
(A) Uniform manifold approximation and projection (UMAP) plots of the SC clusters 2, 3, 5, and 8 in Figure 2A (211 cells in total). A pseudotime ordering was inferred by Slingshot v1.6.0.
(B and C) Normalized expression values of ATOH1 and EDNRB2 (B) and hair cell differentiation genes (C) along a pseudotime line. Black dotted, red dotted, and black lines indicate the timing of EDNRB2 induction, induction of hair cell differentiation genes, and ATOH1 downregulation, respectively.
(D–I) Normalized expression values of representative genes for six time-series clusters (patterns 1–6) along a pseudotime line and the top 10 genes for each cluster. See also Figures S5 and S7 and Table S4.
First, we focused on alterations in the expression of ATOH1 and EDNRB2 (Figure 5B). These genes were included in pattern 6 of the time-series clusters. ATOH1 upregulation was initiated from time point 0 on the pseudotime, and its peak was observed at approximately 0.8 (Figure 5B). The peak of ATOH1 expression is marked by a black line (Figures 5B–5I and S5), which indicates the time point for the initiation of ATOH1 downregulation. EDNRB2 upregulation was initiated at around 0.4 on the pseudotime (black dotted lines in Figures 5B–5I and S5) and reached the peak slightly earlier than ATOH1 (Figure 5B). To determine the timing of the initiation of HC differentiation, we examined the expression patterns of HC differentiation genes, including GFI1, LHX3, and RBM24 (Figure 5C). The upregulation of these genes was initiated at around 0.6 on the pseudotime trajectory (red dotted lines in Figures 5B–5I and S5), slightly later than EDNRB2 induction.
Figures 5D–5I show the representative alterations and the top 10 genes for each time-series cluster. Patterns 1–3 and 6 showed transient or continuous upregulation, and patterns 4 and 5 exhibited downregulation along a pseudotime. Pattern 1 exhibited transient upregulation up to EDNRB2 induction (Figure 5D). Pattern 2 showed transient upregulation before EDNRB2 induction (Figure 5E). The peaks of transient upregulation of pattern 3 genes appeared around the initiation of HC differentiation gene induction (Figure 5F). Pattern 6 included both transiently and continuously upregulated genes, and peaks of transiently upregulated genes were observed after the induction of HC differentiation genes (Figures 5B, 5I, and S5F). The continuously upregulated genes included HC marker (CXCL14 and OCM, Figure 5I and S5F) and nascent HC marker (NREP and CALB2, Figure S5F) genes.43 The transiently upregulated genes included ATOH1, EDNRB2, HMBG2, and UCHL1 (Figures 5B, 5I, and S5F), which are expressed by progenitor cells in various organs.20,64,65,66,67 In particular, Hao et al.66 demonstrated HMGB2 expression in the intermediate progenitor cells during neurogenesis in the adult macaque hippocampus. The induction of HC marker genes was initiated at the same time as HC differentiation gene induction (Figure 5I and S5F), while the other genes in pattern 6, including nascent HC marker genes, were upregulated after EDNRB2 induction (Figure S5F). Regarding the downregulated genes, pattern 4 genes appeared downregulated after the EDNRB2 induction (Figure 5G), whereas pattern 5 genes were downregulated before the EDNRB2 induction (Figure 5H). Many SC marker genes were included in patterns 4 (TECTB, TMSB4X, ID2, CSRP2, and TIMP3; Figures 5G and S5D) and 5 (LCAT and GSTT1L; Figure 5H).
From the point of view of SC and HC identities, the expression levels of SC marker genes were almost maintained and those of HC differentiation genes were not upregulated before EDNRB2 induction (black dotted lines in Figures 5 and S5). In contrast, the expression level of ATOH1 gradually increased before EDNRB2 induction. Thus, we annotated this period (before EDNRB2 induction) as the priming stage for SC-to-HC conversion. After EDNRB2 induction, most SC marker genes were downregulated (Figures 5G and S5D). Slightly later, upregulation of HC differentiation genes was initiated (Figure 5C). During the period between EDNRB2 induction and HC differentiation gene induction, the erasure of SC identity began, but the acquisition of HC identity was not initiated. Therefore, this period was annotated as the initial stage. After HC differentiation gene induction, the erasure of SC identity and acquisition of HC identity progressed simultaneously. The period between HC differentiation gene induction and ATOH1 downregulation was annotated as the intermediate stage. SCs in this period exhibited high expression of progenitor marker genes, including pattern 6 genes, suggesting that SCs were reprogrammed to an immature state during this period. After ATOH1 downregulation, which is a sign of initiation for the maturation process of regenerated HCs,19 the acquisition of HC identity progressed further, and progenitor marker genes were downregulated. This stage was annotated as the late stage. Thus, HC specification may occur during the intermediate stage.
Transforming growth factor β signaling is involved in the initiation of fate conversion of supporting cells to hair cells
As candidates for triggers for EDNRB2 induction, we focused on pattern 1 genes, which were upregulated up to EDNRB2 induction (Figure 5D). Among the pattern 1 genes, we highlighted LOXL1 and VIM, because these two genes exhibited remarkable upregulation in pattern 1 genes. These genes are associated with epithelial–mesenchymal transition (EMT)68,69 and known to be downstream of transforming growth factor β (TGFb) signaling.68,70,71,72,73,74 Additionally, the signaling pathway analysis indicated involvement of THBS, MIF, and SEMA3 signaling pathways in the early phase of SC-to-HC conversion (Figure 4), which are also associated with EMT and TGFb signaling.75,76,77,78 EMT is involved in the induction of regenerative processes79,80 and the gain of epithelial stem cell properties.81 Mouse utricle epithelial cells also acquire the features of prosensory cells via EMT.82 Therefore, we focused on TGFb signaling as upstream of both LOXL1 and VIM. In addition, TGFb1 application induced upregulation of LOXL1 and downregulation of LCAT in mouse astrocyte primary cultures,83 which is consistent with the present findings before EDNRB2 induction in a pseudotime trajectory analysis (Figures 5D and 5H). Furthermore, the expression of TGFBR1 was observed in SC clusters 2, 3, and 8 (Figure S6). Thus, we examined the effects of pharmacological inhibition of TGFb receptor 1 (TGFbR1) on the transition from the priming stage to the intermediate stage by assessing EDNRB2 expression.
Chick BP explants were cultured with SM alone or SM and the TGFbR1 inhibitor SB505124, which is a selective inhibitor of TGFbR1,84 at a concentration of 2 μM for 48 h (Figure 6A). In both BPs cultured with SM alone and SM and SB505124, total HC loss was confirmed. qPCR demonstrated a significant increase in EDNRB2 expression in samples incubated with SM alone compared to those before SM exposure (Figure 6B). Supplementation with SB505124 significantly reduced EDNRB2 expression 48 h after SM exposure (Figure 6B). We also examined the histological expression of EDNRB2. ISH for EDNRB2 demonstrated a trend of decreasing EDNRB2-expressing SCs in BPs following supplementation with SB505124 (Figure 6C). We quantified EDNRB2-expressing cells by counting nuclei stained with DAPI in EDNRB2-expressing cells in three transverse sections of BPs from the distal (0%–30% from the distal end), middle (30%–70%), and proximal (70%-the proximal end) portions of each sample. The numbers of EDNRB2-expressing cells in BPs treated with SM and SB505124 were significantly lower than those in BPs treated with SM alone (Figure 6C). Thus, inhibition of TGFbR1 suppressed EDNRB2 expression in BP explants after HC injury.
Figure 6.
Pharmacological inhibition of transforming growth factor β type 1 receptor suppresses fate conversion of supporting cells to hair cells
(A) Experimental design and groups. SM: streptomycin, HC: hair cell, qPCR: quantitative real-time PCR, ISH: in situ hybridization.
(B) qPCR reveals a significant increase in EDNRB2 expression levels by 48-h exposure to SM (day0_pre [n = 6] vs. day2_SM [n = 6]) and significant suppression by the TGFbR1 inhibitor SB505124 (day2_SM [n = 6] vs. day2_SM + SB [n = 5]). Exposure to SM significantly increases ATOH1 expression in qPCR (day0_pre [n = 4] vs. day2_SM [n = 4]) and supplementation of SB505124 attenuates an increase of ATOH1 expression induced by SM (day2_SM [n = 4] vs. day2_SM + SB [n = 4]). SM exposure significantly increases LOXL1 expression (day0_pre [n = 3] vs. day2_SM [n = 4]), while SB505124 shows no significant effect on LOXL1 expression levels (day2_SM [n = 4] vs. day2_SM + SB [n = 4]).
(C) In situ hybridization for EDNRB2 (red) in transverse sections of the basilar papillae (BPs) in the day2_SM and day2_SM + SB samples. The number of EDNRB2-expressing cells in day2_SM + SB samples (n = 5) is significantly lower than that in day2_SM samples (n = 4).
(D) Regenerated hair cells are labeled by MYO7A (red) and nuclear staining with DAPI (blue) in transverse sections of the basilar papillae (BPs) in day6_SM (n = 4) and day6_SM + SB samples (n = 6). The number of regenerated hair cells in day6_SM + SB samples was significantly lower than that in day6_SM samples. Scale bars in (C) and (D) represent 100 μm. Error bars in (B)–(D) represent standard errors. Welch t-test was used to calculate p values. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S6.
Next, we examined the effects of EDNRB2 suppression by a TGFbR1 inhibitor on the generation of new HCs in specimens cultured for an additional 96 h after 48 h of SM exposure (Figure 6A). The number of MYO7A-positive cells in specimens treated with SM and SB505124 (day6_SM + SB) was significantly lower than those treated with SM alone (day6_SM) (Figure 6D). Altogether, inhibition of TGFbR1 repressed the transition from the priming stage to the intermediate stage of SC-to-HC conversion, resulting in a reduction in regenerated HCs.
We also examined alterations in ATOH1 and LOXL1 expressions by supplementation of a TGFbR1 inhibitor SB505124. SM exposure induced significant increases in both ATOH1 and LOXL1 expression levels in qPCR analyses, whereas supplementation of SB505124 showed a significant attenuation in ATOH1 expression levels, but not in LOXL1 expression levels (Figure 6B). This result in qPCR was consistent with results in the pseudotime trajectory. In the pseudotime trajectory, ATOH1 expression levels increased from the priming stage to the intermediate stage of SC-to-HC conversion (Figure 5B). Conversely, LOXL1 expression levels in the intermediate stage were almost identical to that in the priming stage (Figure 5D). Hence, qPCR results in ATOH1 and LOXL1 expressions supported our hypothesis that TGFbR1 inhibition suppressed the transition from the priming stage to the intermediate stage of SC-to-HC conversion.
Discussion
Single-cell RNA sequencing reveals stepwise fate conversion from supporting cells to hair cells
For single-cell RNA-seq of SCs at different stages of SC-to-HC conversion, dissociated single cells were harvested from chick BPs before and after explant cultures with or without HC damage. Unsupervised clustering of the whole-cell populations identified five clusters containing SCs and neighboring non-sensory cells. Re-clustering of this subpopulation identified four clusters consisting of SCs at different stages of SC-to-HC conversion. A pseudotime trajectory analysis of the extracted SC clusters revealed the time courses of alterations in expressed genes during SC-to-HC conversion, which indicated the stepwise conversion of SCs to nascent HCs. We divided the SC-to-HC conversion processes into the priming, initial, intermediate, and late stages according to alterations in gene expression associated with SC or HC identity along a pseudotime trajectory (Figure 7). The priming stage was characterized by gradient upregulation of ATOH1 and transient upregulation of several genes, including LOXL1 and VIM. In the initial stage, erasure of SC identity, not acquisition of HC identity, was induced. The characteristic genes for the intermediate stage, which can be associated with reversal to a precursor cell state, and HC marker genes were upregulated in the intermediate stage. In the late stage, HC differentiation progressed in parallel with the downregulation of precursor genes. In short, HC regeneration in chick BPs occurs in a stepwise fashion, and reversal to a precursor cell state may be a crucial process.
Figure 7.
Schematic drawing of stepwise conversion from supporting cells to hair cells
Alterations in gene expression associated with supporting cell (SC) and hair cell (HC) identities along a pseudotime line indicate that SC-to-HC conversion occurs in four steps, i.e., the priming, initial, intermediate, and late stages. The priming stage is characterized by a gradual increase in ATOH1 and the upregulation of LOXL1 and VIM. The initial stage is characterized by erasure of SC marker genes and induction of EDNRB2 upregulation. The intermediate stage is characterized by the initiation of HC identity acquisition and high expression of precursor-associated genes (EDNRB2 and HMGB2). The late stage is characterized by upregulation of HC marker genes and downregulation of precursor-associated genes.
Initiation of fate conversion from supporting cells to hair cells
We focused on LOXL1 and VIM as candidate switch genes for SC-to-HC conversion based on SC genetic profiles of single-cell RNA-seq and highlighted TGFb signaling as a common upstream modulator of LOXL1 and VIM. Pharmacological inhibition of TGFb signaling attenuated the expression of EDNRB2, a marker for reprogrammed SCs and subsequent HC formation in our explant culture model. This is the first report showing the involvement of TGFb signaling in the induction of SC reprogramming during spontaneous HC regeneration in chick BPs.
A recent study focusing on LIN28B and FST indicated the involvement of TGFb signaling in the induction of SC reprogramming in neonatal mouse cochleae.85 Neither LOXL1 nor VIM was identified in the dataset of bulk RNA-seq for mouse cochlear organoids.85 Besides, in the current study, LIN28B was not identified in DEGs for SC clusters or a pseudotime trajectory. FST was included in pattern 6 genes in a pseudotime trajectory, although its impact was low (Table S4). One of the key findings of Li et al.85 was the necessity for the strict regulation of TGFb signaling activity by FST; FST is a suppressor of TGFb signaling, and its expression is upregulated as a negative feedback in response to LIN28B induction. Excessive activation of TGFb signaling and EMT suppressed HC formation in mouse cochlear organoids.85 In the present study, moderate activation of several signaling pathways associated with TGFb signaling and EMT was observed during the early stage of HC regeneration in the signaling pathway analysis (Figure 4). However, the activation of incoming signaling of TGFb pathway was not apparent, while outgoing signaling was activated in SCs during the early phase of SC-to-HC conversion (Figure 4). Furthermore, alterations in TGFBR1 and TGFB2 expression levels during the early phase of SC-to-HC conversion were not found in the pseudotime trajectory. This could be associated with a strict regulation of TGFb signaling during the early phase of HC regeneration in chick BPs.
Two recent studies regarding the initiation of HC regeneration in chick BPs demonstrated the activation of the interferon/Janus kinase/signal transducer and activator of transcription (IFN/JAK/STAT) signaling pathway immediately after HC damage,3,43 suggesting that IFN/JAK/STAT signaling is an initial stimulus for SC activation toward HC regeneration. In the present dataset, the expression of IFN/JAK/STAT signaling-associated genes was not noted in DEGs for SC clusters or a pseudotime trajectory. However, some IFN/JAK/STAT signaling-associated genes were identified in DEGs for SC clusters or a pseudotime trajectory. IFI6 was included in pattern 2 genes of the pseudotime trajectory analysis (Table S4, Figure S7). Interestingly, few SCs in the priming stage expressed very high expression of IFI6 (Figure S7), which is consistent with the expression pattern of IFI6 in tissues in our previous study.3 IFIT5 was included in pattern 4 and LY6CLEL (LY6E) was included in pattern 1 (Table S4). These findings indicate that the IFN/JAK/STAT signaling plays a role in the priming of SC-to-HC conversion.
The other candidate for the initiation of SC-to-HC conversion is the MIF signaling pathway. The MIF signaling pathway exhibited upregulation of the incoming signals in SC clusters 3 and 8 and that of the outgoing signals in SC clusters 2 and 3 in the signaling pathway analysis (Figure 4). In the pseudotime trajectory, MIF signaling pathway was included in pattern 5, which showed high expression during the early phase of the priming stage (Figure S5E). The level of MIF incoming signals in SC cluster 1, normal SCs in vivo, was low in the signaling pathway analysis (Figure 4), suggesting that upregulation of its expression occurs after HC damage. MIF is a cytokine expressed in various cell types, including neuronal cells.86 The JAK/STAT signaling is included in its downstream signaling pathways.87 Further studies are needed to determine the role of MIF signaling pathway in SC-to-HC conversion.
Possible mechanisms for hair cell specification
The present results suggest that HC specification occurs during the intermediate stage of SC-to-HC conversion. Thus, highly expressed genes in the intermediate stage, namely, EDNRB2, HMGB2, and UCHL1, could contribute to the HC specification of reprogrammed SCs.
Mammals have two subtypes of endothelin receptors, EDNRA and EDNRB, while the chicken has three subtypes, EDNRA, EDNRB1, and EDNRB2.88 EDNRB2 stimulates intracellular calcium, mitogen-activated protein kinase/extracellular signal-regulated kinase, and cAMP/protein kinase A signaling pathways, similar to mammalian EDNRB.88 EDNRB modulates the differentiation of Schwann cell precursors89,90 and glial cell precursors.91 HMGB2 encodes high-mobility group box2, a chromatin-associated protein that regulates transcription, cell proliferation, and differentiation.92 HMGB2 is involved in the adipogenesis of mesenchymal stem cells93,94 and hematopoiesis in hematopoietic stem cells.95 In addition, HMGB2 regulates the neurogenic-to-gliogenic fate transition of neural stem/progenitor cells via epigenetic modifications.96 UCHL1 encodes ubiquitin C-terminal hydrolase L1, also known as PGP9.5, a deubiquitinating enzyme that is abundant in neurons97 and plays a crucial role in metabolic regulation.98,99 UCHL1 enhances the differentiation of neural progenitor cells into neurons67 and inhibits neurodegeneration by responding to energy requirements and ER stress.99 In addition, the nascent HC marker genes NREP and CALB2 also initiate upregulation in the initial stage, similar to EDNRB2, and exhibit comparatively high expression levels in the intermediate stage (Figure S5F), suggesting the possible roles of these genes in HC specification. The distinct roles of the above-mentioned molecules in HC specification should be examined in future studies.
The suppression of TGFb signaling might be involved in HC specification of reprogrammed SCs. In the late stage of SC-to-HC conversion, significant downregulation of TGFB2 expression was noted (Figure S3E). In the signaling pathway analysis, downregulation of TGFb signaling was apparent in converting HCs (Figure 4). In a mouse cochlear organoid model, exogenous TGFB2 blocked HC formation in LIN28B- and FST-overexpressing cultures.85 We, therefore, consider that downregulation of TGFb signaling may be related to induction of HC specification and/or differentiation in the late phase of SC-to-HC conversion.
Identification of reprogrammed supporting cells
A single-cell RNA-seq analysis for HC regeneration processes in P7 chick BPs in vivo revealed an exclusive set of genes associated with both responding SCs and newly generated, nascent HCs.43 Unfortunately, despite the use of single-cell RNA-seq, their dataset of unsupervised clustering did not fit for a pseudotime trajectory analysis. Instead, they demonstrated three DEGs, namely CALB2, USP18, and TRIM25 that were shared by responding SCs and nascent HCs.43 In the present dataset, CALB2 and USP18 were included in DEGs for converting HCs (Figure S3E) and CALB2 was found in DEGs for reprogrammed SCs (Figure S3H). Janesick et al.43 demonstrated that UCHL1, VIM, STMN1, and HPGDS expression was upregulated in responding SCs, in comparison with that in quiescent SCs, which exhibited high expression in the initial and/or intermediate stage of SC-to-HC conversion in the present pseudotime trajectory (Figures 5 and S5). NREP, CALB2, UCHL1, HMGB2, and EDNRB2 expression was upregulated in nascent HCs, compared with that in normal HCs,43 which are included in pattern 6 genes in the present pseudotime trajectory (Figures 5 and S5). This suggests that nascent HCs in the study conducted by Janesick et al.43 will be composed of reprogrammed SCs and converting HCs shown in the present study.
Despite the differences in the age (P1 vs P7), experimental condition (in vitro vs in vivo), and mechanism underlying HC regeneration (direct conversion alone vs direct conversion + mitotic cell division) between the present study and the study by Janesick et al.,43 the genetic profiles in both single-cell RNA-seq are compatible, suggesting the validity of the gene profiles in the present study for investigating HC regeneration processes in chick BPs. We, therefore, consider that molecules or signaling pathways that were detected in both studies will be associated with fundamental events that occur in SCs during HC regeneration in chick BPs. Unlike the study by Janesick et al.,43 the present dataset of SC clustering fitted for a pseudotime trajectory analysis, which contributed to the identification of the intermediate precursor stage or reprogrammed SCs expressing a specific marker EDNRB2 during SC-to-HC conversion.
Limitations of the study
In the present study, single-cell RNA-seq revealed the stepwise conversion from SCs to HCs through reversal to precursor status in chick BP explant cultures. EDNRB2 is specifically expressed in SCs reprogrammed for HC regeneration. TGFb signaling is involved in the induction of SC reprogramming. Whether the same mechanism works in in vivo settings of chick BPs or mammalian cochleae remains to be explored. Our data provide new insights into the mechanisms of SC-to-HC conversion in chick BPs and will contribute to the exploration of critical signaling pathways for HC regeneration through SC-to-HC conversion in mammals in future studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| mouse anti-MYO7A | Developmental Studies Hybridoma Bank | catalog #138-1; RRID:AB_2282417 |
| rabbit anti-SOX2 | Sigma–Aldrich | catalog #AB5603; RRID:AB_2286686 |
| rabbit anti-ARG3 | Proteintech Group | catalog #11967-1-AP; RRID:AB_2877809 |
| rabbit anti-CSRP2 | MyBioSource | catalog #MBS9204419 |
| mouse anti-CD44 | Thermo Fisher Scientific | catalog #MA528693; RRID:AB_2745652 |
| rabbit anti-FGFR1 | Honda A et al., 2018100 | N/A |
| Alexa Fluor 594 goat anti-mouse | Thermo Fisher Scientific | catalog # A11005; RRID:AB_141372 |
| Alexa Fluor 568 goat anti-mouse | Thermo Fisher Scientific | catalog # A11004; RRID:AB_2534072 |
| Alexa Fluor 488 goat anti-rabbit | Thermo Fisher Scientific | catalog # A11034; RRID:AB_2576217 |
| Alexa Fluor 546 goat anti-rabbit | Thermo Fisher Scientific | catalog #A11010; RRID:AB_2534077 |
| Chemicals, peptides, and recombinant proteins | ||
| medium 199 | Thermo Fisher Scientific | catalog #11043023 |
| DMEM-high glucose | Nacalai Tesque | catalog #08459-35 |
| HyClone fetal bovine serum | Thermo Fisher Scientific | catalog #12389802 |
| penicillin/streptomycin | Sigma–Aldrich | catalog #P4333 |
| Thermolysin | Sigma–Aldrich | catalog #T7902 |
| Accutase | Nacalai Tesque | catalg #12679-54 |
| TrypLE Select enzyme | Thermo Fisher Scientific | catalog# 12563011 |
| DAPI | Thermo Fisher Scientific | catalog # D1306 |
| Reagent kit v2 | Fluidigm | catalog #SKU: 101-3473 |
| Nextera XT DNA Library Prep Kit | Illumina | catalog #FC-131-1024 |
| RNeasy Plus Micro kit | QIAGEN | catalog # 74004 |
| SuperScript III First-Strand Synthesis System | Thermo Fisher Scientific | catalog #N8080234 |
| Power SYBR Green PCR Master Mix | Thermo Fisher Scientific | catalog #4367659 |
| TaqMan Fast Advanced Master Mix | Thermo Fisher Scientific | catalog #4444557 |
| SB505124 | Sigma–Aldrich | catalog #S4696-5MG |
| Critical commercial assays | ||
| RNAscope 2.5 HD Duplex Detection Kit | Advanced Cell Diagnostics Inc | catalog #322430 |
| C1 Single-Cell mRNA seq HT IFC | Fluidigm | catalog #SKU: 101-4981 |
| Deposited data | ||
| Sequencing data | This paper | GEO accession number: GSE209791 |
| Experimental models: Organisms/strains | ||
| Post-hatch day 1 male Momiji (Gallus domesticus) chicken | Goto Furan-Jo (Gifu, Japan) | N/A |
| Oligonucleotides | ||
| ATOH1 | Advanced Cell Diagnostics Inc | catalog #574081-C1 or C2 for ggATOH1 |
| FGFR3 | Advanced Cell Diagnostics Inc | catalog #574091-C2 for ggFGFR3 |
| EDNRB2 | Advanced Cell Diagnostics Inc | catalog #1065931-C2 for ggEDNRB2 |
| Hprt1 forward, 5ʹ-GCACTATGACTCTACCGACTATTGC-3ʹ | Eurofins Genomics K.K. | N/A |
| Hprt1 reverse, 5ʹ-CAGTTCTGGGTTGATGAGGTT-3ʹ | Eurofins Genomics K.K. | N/A |
| Ednrb2 forward, 5ʹ-GCAGTCAGTTTTGCCCTTCG-3ʹ | Eurofins Genomics K.K. | N/A |
| Ednrb2 reverse, 5ʹ-ATGAGAACATTCGGCCCGTT-3ʹ | Eurofins Genomics K.K. | N/A |
| Atoh1 forward, 5′-AACAACGACAAGAAGCTCTCCAA-3′ | Eurofins Genomics K.K. | N/A |
| Atoh1 reverse, 5′-GCGAGGGCGCTGATGTAG-3′ | Eurofins Genomics K.K. | N/A |
| ID: Gg07187773_s1, Gene:LOXL1 | Taqman assay gene expression | Cat.no. 4351370 |
| ID: Gg03338898_m1, Gene:HPRT1 | Taqman assay gene expression | Cat.no. 4448485 |
| Software and algorithms | ||
| FASTP v0.19.6 | Chen et al., 2018101 | https://github.com/OpenGene/fastp |
| GRCg6a | NCBI | https://www.ncbi.nlm.nih.gov/assembly/GCF_000002315.6 |
| STAR v2.7.0a | Dobin et al., 2013102 | http://code.google.com/p/rna-star/ |
| SUBREAD v1.6.3 | Liao et al., 2013103 | https://subread.sourceforge.net/ |
| Seurat v4.0.3 R package | Hao et al., 2021104; Stuart et al., 2019105 | https://satijalab.org/seurat/ |
| R v4.0.3 | R Project for Statistical Computing_ |
https://www.R-project.org/ |
| quminorm v0.1.0 R package | Townes et αl., 2020106 | https://github.com/willtownes/quminorm/ |
| sctransform v0.3.3 R package | Hafemeister and Satija, 2019107 | https://cran.r-project.org/web/packages/sctransform/index.html |
| Slingshot v1.6.0 R package | Street et al., 201862 | https://github.com/kstreet13/slingshot |
| ranger v0.13.1 R package | Wright and Ziegler, 201763 | https://cran.r-project.org/web/packages/ranger/index.html |
| dtwclust v5.5.10 R package | Sardá-Espinosa, 2019108 | https://github.com/asardaes/dtwclust/releases/tag/v5.5.10 |
| CellChat v1.5.0 | Jin et al., 202158 | https://github.com/sqjin/CellChat/releases/tag/v1.5.0 |
| Rmagic v2.0.3 | van Dijk et al., 2018109 | https://github.com/cran/Rmagic |
| GraphPad Prism7 | GraphPad Software | https://www.graphpad.com/ |
| Adobe Photoshop 2020 | Adobe Systems | https://www.adobe.com/jp/products/photoshop.html |
| Real-Time PCR System Software v2.0 | Thermo Fisher Scientific | https://www.thermofisher.com/order/catalog/product/jp/ja/4443420 |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Other | ||
| TC20 automated cell counter | Bio-Rad Laboratories | catalog #1450101J1 |
| HiSeq2500 | Illumina | catalog #SY–401–2501 |
| TCS-SPE confocal microscope | Leica Microsystems | https://www.leica-microsystems.com/products/confocal-microscopes/p/leica-tcs-spe/downloads/ |
| BX50 microscope | Olympus | https://www.olympus-lifescience.com/ja/technology/museum/micro/1993_02/ |
| StepOnePlus Real-Time PCR system | Thermo Fisher Scientific | catalog #4376598 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Takayuki Nakagawa (tnakagawa@ent.kuhp.kyoto-u.ac.jp).
Materials availability
The study did not generate new unique reagents.
Experimental model and subject details
Explant culture of chick basilar papillae
Post-hatch day 1 (P1) male Momiji (Gallus domesticus) chicks were purchased from Goto Furan-Jo (Gifu, Japan) and placed for less than 3 h in the transport box in the experimental room, which was maintained at approximately 23°C. All animal procedures were performed following the National Institutes of Health (NIH; Bethesda, MD, USA) Guide for the Care and Use of Laboratory Animals (NIH Publications No. 8023, revised 1978) and were approved by the Animal Research Committee of Kyoto University Graduate School of Medicine (MedKyo21123).
After euthanasia by CO2 inhalation followed by decapitation, the middle ear cavity was opened, followed by the extirpation of the cochlear duct. Cochlear ducts were placed in ice-cold sterile medium 199. The tegmentum vasculosum was carefully removed from the cochlear duct. The remaining tissue containing the basilar papilla (BP) was cultured in a free-floating manner (submerged in the medium) in 500 μl control medium consisting of Dulbecco’s modified Eagle’s medium with 4.5 g/l glucose supplemented with 1% fetal bovine serum (FBS) in 48-well plates (Iwaki Co. Limited, Tokyo, Japan) for 24 h. Thereafter, BP specimens were provided for explant cultures. To damage hair cells (HCs), BP explants were maintained with the culture media containing penicillin/streptomycin (SM; 200 penicillin units per ml and 78 μM streptomycin). After the SM exposure for 48 h, BP was washed out by changing the medium twice, and then cultured in DMEM-high glucose with 1% FBS and penicillin (control medium). The control specimens were incubated in the control media without exposure to SM. Culture media were changed every other day.
Methods details
Single cell preparation and RNA-sequencing
The following samples were provided for the single cell RNA sequencing (RNA-seq) analysis: day0_pre (before SM exposure, n = 17), day2_SM (immediately after 48-h SM exposure, n = 16), day6_SM (96 h after SM exposure, n = 15), day2_Ctrl (48-h culture in the control medium, n = 16), day6_Ctrl (144-h culture in the control medium, n = 15) and intact BPs that were harvested immediately after the dissection from the temporal bones (n = 17). For enzymatic dissociation of BPs for single cell RNA-seq, specimens were incubated in prewarmed 500 μg/ml thermolysin in medium 199 at 37°C for 30 min. To stop enzymatic dissociation, specimens were transferred into medium 199 supplemented with 5% FBS. Afterwards, the BP, the sensory epithelium was separated from the surrounding tissues under the stereomicroscope. Then, the samples were transferred into an Eppendorf tube by a pipette. After centrifugation (800g x 5 min) at room temperature, the supernatant was removed. Samples were incubated in an enzyme cocktail containing 50% Accutase and 50% TrypLE Select enzyme (1X) (1:1) for 7 min at 37°C. After gently pipetting 10-20 times, samples were incubated for additional 4 min at 37°C. After inactivation of enzymatic activity with medium 199 supplemented with 5% FBS, further mechanical dissociation by pipetting was performed. Dissociated cells were collected after centrifugation (800g x 5 min) and resuspended in medium 199 supplemented with 5% FBS. We sampled 1 μl of a cell suspension and quantified the number of living cells by a TC20 automated cell counter. Finally, the cell concentration was adjusted to 500 cells per μl.
Capture of the single cells and cDNA synthesis were conducted using C1 Single-Cell mRNA seq HT IFC for medium cell size (10-17 μm) and Reagent kit v2 according to manufacturer’s instruction. Briefly, 12,000 cells of each sample were loaded onto HT IFC, and 400 cells were captured in the IFC. Sequencing libraries were prepared by transposase-assisted tagmentation and enrichment of 3’end of genes using Nextera XT DNA Library Prep Kit. The libraries were sequenced on the HiSeq2500 for 25 and 75 cycles paired-end reads. The raw sequencing data in bcl format were converted into FASTQ files using the Illumina bcl2fastq2 software.
Data preprocessing
We applied C1 mRNA Seq HT Demultiplex Script v2.0 (Fluidigm) to demultiplex the FASTQ files. We performed quality check and trimming using FASTP v0.19.6101 with default settings otherwise additional polyG and polyX tail trimming. Trimmed reads were mapped to the chicken reference genome GRCg6a (https://www.ncbi.nlm.nih.gov/assembly/GCF_000002315.6) using STAR v2.7.0a102 with default settings. The generated BAM files were transformed into gene expression matrix of 2,400 cells using featureCounts function from SUBREAD v1.6.3 package.103 We performed a downstream analysis using Seurat 4.0.3 library104,105 on R 4.0.3 (R Core Team (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.). After removing the cells of which RNA count was less than 20,000, detected genes was less than 1,000, detected gene was more than 4,000, or percentage of mitochondrial genes was more than 12.5%, 1,054 cells were analyzed. The dataset was normalized using transcripts per million,110 quasi UMI normalization106 and R package sctransform,107 which included the selection of variable genes. We performed principal component analysis (PCA) to reduce dimensionality on the normalized data matrix. For PCA, we used the top 3,000 most highly variable genes and kept the first 10 principal components (PCs). For 2-dimentional visualization, PC embedding was passed into the uniform manifold approximation and projection (UMAP).111 The parameters for UMAP were as follows: min dist = 0.01, n (the number of neighbors) = 30. We applied the Louvain clustering algorithm for community detection. The Seurat FindMarkers function was used to explore marker genes for the clusters (Wilcoxon rank-sum test). The clusters were annotated based on established marker genes. During analyses of differentially expressed genes, we set a log-fold-change threshold > 0.5 and FDR < 0.05. P-value was adjusted by Bonferroni correction.
To calculate communication networks in the supporting cell subset, we used R software CellChat v1.5.0.58 We imputed the data using Rmagic v2.0.3109 with default parameters before the CellChat analysis. We inferred the communication network, including signaling pathway and ligand-receptor pairs information using their default mean method, trimean. We visualized incoming and outgoing signaling patterns.
Reconstruction of the pseudotime trajectory was performed by Slingshot v1.6.0.62 In order to find genes that are differentially expressed along the pseudotime, we built a random forest regression model that predicts the value of the pseudotime from the expression value of each gene. We built the model using R ranger package.63 The parameters for the regression model were as follows: mtry (randomly selected predictors) = 800, trees (number of trees) = 1,000, min_n (minimal node seize) = 15. Using this model, we calculated the importance of the genes at the pseudo-time and selected 500 most important genes. We classified the 500 selected genes into six clusters using dtwclust package.108 We performed time series clustering based on Euclidean distances. Volcano plots were made in GraphPad Prism7 (GraphPad Software).
Immunohistochemistry
Immunostaining was carried out on whole mount preparations or frozen sections of the chick cochlea duct. After the fixation with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 15 min at room temperature, PBS wash was done for two times. The dehydration was performed for the frozen section from 15% sucrose with 0.2mM ethylenediaminetetraacetic acid (EDTA) in 1×PBS at 4 °C overnight to 30% sucrose with 0.2mM EDTA next night. Specimens were embedded in Tissue-Tek O.C.T. Compound (Sakura Fine technical Co., Ltd., Tokyo, Japan) and then, cryostat-cut sections (10 μm) were mounted directly onto MAS-coated slides (Matsunami Glass Ind. Ltd, Osaka, Japan). The ensemble of sections collected from 2-3 BPs was distributed through almost 20 slides (7-9 sections per slide) for each experiment. And each slide contains a reliable representation of the whole length of the BP in a serial manner (first tissue section on slide numbered 1 and the following tissue section on slide number 2 and so on) which provided uniformity of treatment for cross-sections throughout the cochlear length.
Both whole-mount samples and section samples were incubated in blocking solution (1% bovine serum albumin and 5% normal goat serum in PBS) for 30 min at room temperature. The samples were incubated with primary antibodies in a blocking solution overnight at 4 °C, followed by incubation with the corresponding secondary antibodies for 1 h at room temperature. Nuclear staining was done by 4',6-diamidino-2-phenylindole (DAPI). After several washes in 1xPBS, samples were mounted using FluoromountG (catalog # 00-4958-02, Southern Biotech). All fluorescence images were obtained with a TCS-SPE confocal microscope (equipped with 40×/1.15 OIL CS. objective, Leica Microsystems, Wetzlar, Germany). As for whole mount samples, each image taken by the BX50 microscope (Olympus, Tokyo, Japan) was identified at first by immunostaining for MYO7A and nuclear staining with DAPI, and the total BP length between the proximal portion and the distal portion was calculated by ImageJ software. Around 40% of the distal end of each BP was used for quantitative analyses in whole-mount samples. Optical sections in the xy-field (“z-sections”) were imaged and recorded at 4-μm intervals with the span adjusted to include the HC layer and SC layer in the xy-field of view. As for transverse sections, 3 sections for each image were z-stack projection. Image processing for figures was accomplished in Adobe Photoshop 2020. Images presented are representative of three independent experiments. We counted numbers of MYO7A-expressing cells with DAPI nuclear staining as hair cell numbers and those of MYO7A and SOX2 co-expressing cells as immature hair cell numbers. Welch t-test for the two groups was calculated in GraphPad Prism 7.
In situ hybridization
The preparation of the frozen-section sample for the in situ hybridization (ISH) was almost the same as immunostaining protocols except for the longer fixation (at 4 °C for 3-4 h), higher EDTA concentration (333mM) in 30% sucrose and thicker sections (14 μm). All ISH experiments were carried out with the RNAscope 2.5 HD Duplex Detection Kit on frozen section samples according to the manufacture’s instruction. Briefly, slides were baked at 60 °C for 15 min and washed with 1xPBS. The specimens were then post-fixed in pre-chilled 4% PFA for 15 min, washed in 2 changes of double distilled water (DDW) for 1 min each before dehydration through 50%, 70%, 100% and 100% ethanol for 5 min each. The slides were air-dried for 5 min before hydrogen peroxide treatment for 10 min, washed with DDW for a brief time twice. Slides were boiled in the target retrieval solution for 3 min and washed with DDW and then changed for 100% ethanol. Slides were dried up again with baking at 60 °C for 10 min and then allowed to put into the Protease III Solution at 40 °C for 5 min, and the hybridization with the pre-warmed probe was performed at 40 °C for 120 min. After washing with wash buffer and 5xSSC solution overnight, the specimens were stained according to the manufacturer’s instructions. On each individual section, three target mRNAs were examined: ATOH1 (#574081-C1 or C2 for ggATOH1, Advanced Cell Diagnostics Inc.), FGFR3 (574091-C2 for ggFGFR3, Advanced Cell Diagnostics Inc.), and EDNRB2 (1065931-C2 for ggEDNRB2, Advanced Cell Diagnostics Inc.). Both positive and negative control probes (#453961-C2 for ggUBC [ubiquitin C] or #320751, Advanced Cell Diagnostics Inc.) were also labeled. The duplex negative control probe (#320751, Advanced Cell Diagnostics Inc.) was used on one section per slide for control purposes. Probes in channel 2 were labeled with AP enzyme and a red substrate. DAPI was used to mark cell nuclei. Slides were then imaged using a BX50 microscope to take 20x, 40x bright-field and fluorescence with DAPI filter from more than 3 BP samples for each time point, and a representative image was cited in the figures. Image processing for figures was accomplished in Adobe Photoshop 2020. Images presented are representative of three independent experiments.
Quantitative real-time PCR
Total RNA for each sample was extracted pooled 10 BPs using the RNeasy Plus Micro kit (catalog # 74004, QIAGEN, Venlo, Netherlands) according to the manufacturer’s protocol. DNase I treatment was performed using spin columns. RNA was reverse-transcribed using the SuperScript III First-Strand Synthesis System. Quantitative real-time polymerase chain reaction (qPCR) was performed using a StepOnePlus Real-Time PCR system. cDNA was amplified using the Power SYBR Green PCR Master Mix or TaqMan™ Fast Advanced Master Mix. The experiment was performed in triplicate. Target gene expression was normalized to hypoxanthine phosphoribosyltransferase 1 (Hprt1).112 cDNA from the brain tissue of post-hatched 1-day-old chickens was used to generate standard curves for each gene. Relative quantification was performed using Real-Time PCR System Software v2.0, with the 2−ΔΔCT method (Thermo Fisher Scientific). The primers used are shown in the key resources table.
Pharmacological inhibition of transforming growth factor β type 1 receptors
Chick BP explants were cultured with SM (78 μM) alone (day2_SM) or SM and a transforming growth factor β type 1 receptor inhibitor, SB505124 (catalog #S4696-5MG, Sigma-Aldrich) at a concentration of 2μM (day2_SM+SB) for 48 h. To validate EDNRB2 expression, we harvested BP explants after 48-h culture and performed qPCR and ISH. For qPCR assessment related to Figure 5B, we used 10 to 11BPs for each experimental group per experiment. BPs before SM exposure were used as controls (day0_pre). For quantification of EDNRB2-expressing cells related to Figure 5C, we counted EDNRB2-expressing cells with DAPI staining in three transverse sections of BPs from the distal (0-30% from the distal end), middle (30-70%) and proximal (70%-the proximal end) portions in each sample. The sum of three sections was defined as the number of EDNRB2-expressing cells for each sample. To examine effects of pharmacological inhibition of transforming growth factor β type 1 receptor on hair cell regeneration, we used BP explants that were cultured with SM or SM+SB505124 for 48 h followed additional 96-h culture in control media, and HC count was done in transverse sections using immunohistochemistry for MYO7A and nuclear staining with DAPI.
Quantification and statistical analysis
Differential expression analysis between single cell RNA-seq clusters was performed with Seurat 4.0.3 library104,105 on R 4.0.3 (R Core Team (2020). We set a log-fold-change threshold > 0.5 and false discovery rate (FDR) < 0.05. P-value was adjusted by Bonferroni correction. Differentially expression analysis along the pseudotime was performed using R ranger package.63 Statistical analyses were conducted using Welch t-test for the two groups (Figure 5) or one-way ANOVA with Tukey’s correction for the three groups (Figure S1) in GraphPad Prism 7. Additional details are provided in the figure legends.
Acknowledgments
We thank J.S. Stone and her lab members (Univ. of Washington) for technical advice regarding explant cultures of chick basilar papillae, S. Heller (Stanford Univ.) for practical advice regarding tissue preparation for in situ hybridization of chick basilar papillae, A. Watanabe and S. Sakamoto (Kyoto Univ.) for performing single-cell RNA sequencing, and S. Suzuki, C. Long, and H. Doods (Boehringer Ingelheim Pharma GmbH) for discussions regarding bioinformatics analyses. The study was partly supported by KAKENHI (Grants-in-Aid for Scientific Research, 20K09708 to T.N., Grant-in-Aid for Young Scientists [Start-up], 21K20964 to M.M., Grant-in-Aid for Young Scientists [B], 22K16899 to M.M.) from the Japan Society for the Promotion of Science, by Start-up FY2021 to M.M. from Kyoto University Research Administration Office, by AMED (Japan Agency for Medical Research and Development) under Grant (20lm0203013j0002) to T.O., and by Boehringer Ingelheim Pharma GmbH to T.N.
Author contributions
Conceptualization, M.M. and T.N.; Methodology, R.Y., T.K., M.M., and T.N.; Formal analysis, M.M., R.Y., T.K., and T.N.; Investigation, M.M., R.Y., T.K., and H.O.; Resources, M.M., R.Y., T.O., N.Y., K.O., and T.N.; Data curation, R.Y., T.K., M.M., and T.N.; Writing – original draft, M.M., R.Y., T.K., and T.N.; Writing – review & editing, T.O., N.Y., K.O., and T.N.; Visualization, M.M., R.Y., and T.N.; Supervision, N.Y., K.O., and T.N.; Funding acquisition, M.M., T.O., and T.N.
Declaration of interests
The authors declare no competing interests.
Published: January 25, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106046.
Supplemental information
Data and code availability
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Stone J.S., Cotanche D.A. Hair cell regeneration in the avian auditory epithelium. Int. J. Dev. Biol. 2007;51:633–647. doi: 10.1387/ijdb.072408js. [DOI] [PubMed] [Google Scholar]
- 2.Shang J., Cafaro J., Nehmer R., Stone J. Supporting cell division is not required for regeneration of auditory hair cells after ototoxic injury in vitro. J. Assoc. Res. Otolaryngol. 2010;11:203–222. doi: 10.1007/s10162-009-0206-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsunaga M., Kita T., Yamamoto R., Yamamoto N., Okano T., Omori K., Sakamoto S., Nakagawa T. Initiation of supporting cell activation for hair cell regeneration in the avian auditory epithelium: an explant culture model. Front. Cell. Neurosci. 2020;14 doi: 10.3389/fncel.2020.583994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bermingham N.A., Hassan B.A., Price S.D., Vollrath M.A., Ben-Arie N., Eatock R.A., Bellen H.J., Lysakowski A., Zoghbi H.Y. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- 5.Chen P., Johnson J.E., Zoghbi H.Y., Segil N. The role of Math1 in inner ear development: uncoupling the establishment of the sensory primordium from hair cell fate determination. Development. 2002;129:2495–2505. doi: 10.1242/dev.129.10.2495. [DOI] [PubMed] [Google Scholar]
- 6.Woods C., Montcouquiol M., Kelley M.W. Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat. Neurosci. 2004;7:1310–1318. doi: 10.1038/nn1349. [DOI] [PubMed] [Google Scholar]
- 7.Doetzlhofer A., Basch M.L., Ohyama T., Gessler M., Groves A.K., Segil N. Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev. Cell. 2009;16:58–69. doi: 10.1016/j.devcel.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamamoto N., Tanigaki K., Tsuji M., Yabe D., Ito J., Honjo T. Inhibition of Notch/RBP-J signaling induces hair cell formation in neonate mouse cochleas. J. Mol. Med. 2006;84:37–45. doi: 10.1007/s00109-005-0706-9. [DOI] [PubMed] [Google Scholar]
- 9.Izumikawa M., Minoda R., Kawamoto K., Abrashkin K.A., Swiderski D.L., Dolan D.F., Brough D.E., Raphael Y. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat. Med. 2005;11:271–276. doi: 10.1038/nm1193. [DOI] [PubMed] [Google Scholar]
- 10.Kawamoto K., Ishimoto S.I., Minoda R., Brough D.E., Raphael Y. Math1 gene transfer generates new cochlear hair cells in mature Guinea pigs in vivo. J. Neurosci. 2003;23:4395–4400. doi: 10.1523/JNEUROSCI.23-11-04395.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly M.C., Chang Q., Pan A., Lin X., Chen P. Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. J. Neurosci. 2012;32:6699–6710. doi: 10.1523/JNEUROSCI.5420-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z., Dearman J.A., Cox B.C., Walters B.J., Zhang L., Ayrault O., Zindy F., Gan L., Roussel M.F., Zuo J. Age-dependent in vivo conversion of mouse cochlear pillar and Deiters' cells to immature hair cells by Atoh1 ectopic expression. J. Neurosci. 2012;32:6600–6610. doi: 10.1523/JNEUROSCI.0818-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu L.M., Zhao L.P., Wu L.J., Guo L., Li W.Y., Chen Y. Characterization of the transcriptomes of Atoh1-induced hair cells in the mouse cochlea. Am. J. Stem Cells. 2020;9:1–15. [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y., Gu Y., Li Y., Li G.L., Chai R., Li W., Li H. Generation of mature and functional hair cells by co-expression of Gfi1, Pou4f3, and Atoh1 in the postnatal mouse cochlea. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109016. [DOI] [PubMed] [Google Scholar]
- 15.Yamashita T., Zheng F., Finkelstein D., Kellard Z., Carter R., Rosencrance C.D., Sugino K., Easton J., Gawad C., Zuo J. High-resolution transcriptional dissection of in vivo Atoh1-mediated hair cell conversion in mature cochleae identifies Isl1 as a co-reprogramming factor. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S., Song J.J., Beyer L.A., Swiderski D.L., Prieskorn D.M., Acar M., Jen H.I., Groves A.K., Raphael Y. Combinatorial Atoh1 and Gfi1 induction enhances hair cell regeneration in the adult cochlea. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-78167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun S., Li S., Luo Z., Ren M., He S., Wang G., Liu Z. Dual expression of Atoh1 and Ikzf2 promotes transformation of adult cochlear supporting cells into outer hair cells. Elife. 2021;10 doi: 10.7554/eLife.66547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walters B.J., Coak E., Dearman J., Bailey G., Yamashita T., Kuo B., Zuo J. In Vivo interplay between p27Kip1, GATA3, ATOH1, and POU4F3 converts non-sensory cells to hair cells in adult mice. Cell Rep. 2017;19:307–320. doi: 10.1016/j.celrep.2017.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cafaro J., Lee G.S., Stone J.S. Atoh1 expression defines activated progenitors and differentiating hair cells during avian hair cell regeneration. Dev. Dynam. 2007;236:156–170. doi: 10.1002/dvdy.21023. [DOI] [PubMed] [Google Scholar]
- 20.Driver E.C., Sillers L., Coate T.M., Rose M.F., Kelley M.W. The Atoh1-lineage gives rise to hair cells and supporting cells within the mammalian cochlea. Dev. Biol. 2013;376:86–98. doi: 10.1016/j.ydbio.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leaman S., Marichal N., Berninger B. Reprogramming cellular identity in vivo. Development. 2022;149 doi: 10.1242/dev.200433. [DOI] [PubMed] [Google Scholar]
- 22.Lee S.G., Huang M., Obholzer N.D., Sun S., Li W., Petrillo M., Dai P., Zhou Y., Cotanche D.A., Megason S.G., et al. Myc and fgf are required for zebrafish neuromast hair cell regeneration. PLoS One. 2016;11 doi: 10.1371/journal.pone.0157768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shu Y., Li W., Huang M., Quan Y.Z., Scheffer D., Tian C., Tao Y., Liu X., Hochedlinger K., Indzhykulian A.A., et al. Renewed proliferation in adult mouse cochlea and regeneration of hair cells. Nat. Commun. 2019;10:5530. doi: 10.1038/s41467-019-13157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daudet N., Gibson R., Shang J., Bernard A., Lewis J., Stone J. Notch regulation of progenitor cell behavior in quiescent and regenerating auditory epithelium of mature birds. Dev. Biol. 2009;326:86–100. doi: 10.1016/j.ydbio.2008.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daudet N., Lewis J. Two contrasting roles for Notch activity in chick inner ear development: specification of prosensory patches and lateral inhibition of hair-cell differentiation. Development. 2005;132:541–551. doi: 10.1242/dev.01589. [DOI] [PubMed] [Google Scholar]
- 26.Petrovic J., Formosa-Jordan P., Luna-Escalante J.C., Abelló G., Ibañes M., Neves J., Giraldez F. Ligand-dependent Notch signaling strength orchestrates lateral induction and lateral inhibition in the developing inner ear. Development. 2014;141:2313–2324. doi: 10.1242/dev.108100. [DOI] [PubMed] [Google Scholar]
- 27.Bermingham-McDonogh O., Stone J.S., Reh T.A., Rubel E.W. FGFR3 expression during development and regeneration of the chick inner ear sensory epithelia. Dev. Biol. 2001;238:247–259. doi: 10.1006/dbio.2001.0412. [DOI] [PubMed] [Google Scholar]
- 28.Jacques B.E., Dabdoub A., Kelley M.W. Fgf signaling regulates development and transdifferentiation of hair cells and supporting cells in the basilar papilla. Hear. Res. 2012;289:27–39. doi: 10.1016/j.heares.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munnamalai V., Sienknecht U.J., Duncan R.K., Scott M.K., Thawani A., Fantetti K.N., Atallah N.M., Biesemeier D.J., Song K.H., Luethy K., et al. Wnt9a can influence cell fates and neural connectivity across the radial axis of the developing cochlea. J. Neurosci. 2017;37:8975–8988. doi: 10.1523/JNEUROSCI.1554-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sienknecht U.J., Fekete D.M. Comprehensive Wnt-related gene expression during cochlear duct development in chicken. J. Comp. Neurol. 2008;510:378–395. doi: 10.1002/cne.21791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baek S., Tran N.T.T., Diaz D.C., Tsai Y.Y., Acedo J.N., Lush M.E., Piotrowski T. Single-cell transcriptome analysis reveals three sequential phases of gene expression during zebrafish sensory hair cell regeneration. Dev. Cell. 2022;57:799–819.e6. doi: 10.1016/j.devcel.2022.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lush M.E., Diaz D.C., Koenecke N., Baek S., Boldt H., St Peter M.K., Gaitan-Escudero T., Romero-Carvajal A., Busch-Nentwich E.M., Perera A.G., et al. scRNA-Seq reveals distinct stem cell populations that drive hair cell regeneration after loss of Fgf and Notch signaling. Elife. 2019;8 doi: 10.7554/eLife.44431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janesick A., Scheibinger M., Benkafadar N., Kirti S., Ellwanger D.C., Heller S. Cell-type identity of the avian cochlea. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108900. [DOI] [PubMed] [Google Scholar]
- 34.Kolla L., Kelly M.C., Mann Z.F., Anaya-Rocha A., Ellis K., Lemons A., Palermo A.T., So K.S., Mays J.C., Orvis J., et al. Characterization of the development of the mouse cochlear epithelium at the single cell level. Nat. Commun. 2020;11:2389. doi: 10.1038/s41467-020-16113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis R.M., Hume C.R., Stone J.S. Atoh1 expression and function during auditory hair cell regeneration in post-hatch chickens. Hear. Res. 2012;289:74–85. doi: 10.1016/j.heares.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brooks P.M., Rose K.P., MacRae M.L., Rangoussis K.M., Gurjar M., Hertzano R., Coate T.M. Pou3f4-expressing otic mesenchyme cells promote spiral ganglion neuron survival in the postnatal mouse cochlea. J. Comp. Neurol. 2020;528:1967–1985. doi: 10.1002/cne.24867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coate T.M., Raft S., Zhao X., Ryan A.K., Crenshaw E.B., Kelley M.W. Otic mesenchyme cells regulate spiral ganglion axon fasciculation through a Pou3f4/EphA4 signaling pathway. Neuron. 2012;73:49–63. doi: 10.1016/j.neuron.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lukoseviciute M., Gavriouchkina D., Williams R.M., Hochgreb-Hagele T., Senanayake U., Chong-Morrison V., Thongjuea S., Repapi E., Mead A., Sauka-Spengler T. From pioneer to repressor: bimodal foxd3 activity dynamically remodels neural crest regulatory landscape in vivo. Dev. Cell. 2018;47:608–628.e6. doi: 10.1016/j.devcel.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mundell N.A., Labosky P.A. Neural crest stem cell multipotency requires Foxd3 to maintain neural potential and repress mesenchymal fates. Development. 2011;138:641–652. doi: 10.1242/dev.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonham L.W., Sirkis D.W., Yokoyama J.S. The transcriptional landscape of microglial genes in aging and neurodegenerative disease. Front. Immunol. 2019;10:1170. doi: 10.3389/fimmu.2019.01170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sánchez-Calderón H., Martín-Partido G., Hidalgo-Sánchez M. Otx2, Gbx2, and Fgf8 expression patterns in the chick developing inner ear and their possible roles in otic specification and early innervation. Gene Expr. Patterns. 2004;4:659–669. doi: 10.1016/j.modgep.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 42.Hartman B.H., Durruthy-Durruthy R., Laske R.D., Losorelli S., Heller S. Identification and characterization of mouse otic sensory lineage genes. Front. Cell. Neurosci. 2015;9:79. doi: 10.3389/fncel.2015.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janesick A.S., Scheibinger M., Benkafadar N., Kirti S., Heller S. Avian auditory hair cell regeneration is accompanied by JAK/STAT-dependent expression of immune-related genes in supporting cells. Development. 2022;149 doi: 10.1242/dev.200113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matern M.S., Milon B., Lipford E.L., McMurray M., Ogawa Y., Tkaczuk A., Song Y., Elkon R., Hertzano R. GFI1 functions to repress neuronal gene expression in the developing inner ear hair cells. Development. 2020;147 doi: 10.1242/dev.186015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hertzano R., Dror A.A., Montcouquiol M., Ahmed Z.M., Ellsworth B., Camper S., Friedman T.B., Kelley M.W., Avraham K.B. Lhx3, a LIM domain transcription factor, is regulated by Pou4f3 in the auditory but not in the vestibular system. Eur. J. Neurosci. 2007;25:999–1005. doi: 10.1111/j.1460-9568.2007.05332.x. [DOI] [PubMed] [Google Scholar]
- 46.Cheng X., Zhang J.J., Shi D.L. Loss of Rbm24a causes defective hair cell development in the zebrafish inner ear and neuromasts. J. Genet. Genom. 2020;47:403–406. doi: 10.1016/j.jgg.2020.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Wang G., Li C., He S., Liu Z. Mosaic CRISPR-stop enables rapid phenotyping of nonsense mutations in essential genes. Development. 2021;148:dev196899. doi: 10.1242/dev.196899. [DOI] [PubMed] [Google Scholar]
- 48.Cai W., Chen G., Luo Q., Liu J., Guo X., Zhang T., Ma F., Yuan L., Li B., Cai J. PMP22 regulates self-renewal and chemoresistance of gastric cancer cells. Mol. Cancer Therapeut. 2017;16:1187–1198. doi: 10.1158/1535-7163.MCT-16-0750. [DOI] [PubMed] [Google Scholar]
- 49.Chen C., Zhao S., Karnad A., Freeman J.W. The biology and role of CD44 in cancer progression: therapeutic implications. J. Hematol. Oncol. 2018;11:64. doi: 10.1186/s13045-018-0605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hagedorn L., Suter U., Sommer L. P0 and PMP22 mark a multipotent neural crest-derived cell type that displays community effects in response to TGF-beta family factors. Development. 1999;126:3781–3794. doi: 10.1242/dev.126.17.3781. [DOI] [PubMed] [Google Scholar]
- 51.Tin A.S., Park A.H., Sundar S.N., Firestone G.L. Essential role of the cancer stem/progenitor cell marker nucleostemin for indole-3-carbinol anti-proliferative responsiveness in human breast cancer cells. BMC Biol. 2014;12:72. doi: 10.1186/s12915-014-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsai R.Y.L. Turning a new page on nucleostemin and self-renewal. J. Cell Sci. 2014;127:3885–3891. doi: 10.1242/jcs.154054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu Y., Wang T., Gu J., Huang K., Zhang T., Zhang Z., Liu H., Tang J., Mai Y., Zhang Y., et al. Characterization and generation of human definitive multipotent hematopoietic stem/progenitor cells. Cell Discov. 2020;6:89. doi: 10.1038/s41421-020-00213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayashi T., Ray C.A., Younkins C., Bermingham-McDonogh O. Expression patterns of FGF receptors in the developing mammalian cochlea. Dev. Dynam. 2010;239:1019–1026. doi: 10.1002/dvdy.22236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pirvola U., Ylikoski J., Trokovic R., Hébert J.M., McConnell S.K., Partanen J. FGFR1 is required for the development of the auditory sensory epithelium. Neuron. 2002;35:671–680. doi: 10.1016/s0896-6273(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 56.Yang L.M., Cheah K.S.E., Huh S.H., Ornitz D.M. Sox2 and FGF20 interact to regulate organ of Corti hair cell and supporting cell development in a spatially-graded manner. PLoS Genet. 2019;15 doi: 10.1371/journal.pgen.1008254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Janesick A.S., Heller S. Stem cells and the bird cochlea-Where is everybody? Cold Spring Harb. Perspect. Med. 2019;9 doi: 10.1101/cshperspect.a033183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.H., Myung P., Plikus M.V., Nie Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Defourny J. Eph/ephrin signalling in the development and function of the mammalian cochlea. Dev. Biol. 2019;449:35–40. doi: 10.1016/j.ydbio.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 60.Togashi H., Kominami K., Waseda M., Komura H., Miyoshi J., Takeichi M., Takai Y. Nectins establish a checkerboard-like cellular pattern in the auditory epithelium. Science. 2011;333:1144–1147. doi: 10.1126/science.1208467. [DOI] [PubMed] [Google Scholar]
- 61.Wesdorp M., Murillo-Cuesta S., Peters T., Celaya A.M., Oonk A., Schraders M., Oostrik J., Gomez-Rosas E., Beynon A.J., Hartel B.P., et al. MPZL2, encoding the epithelial junctional protein myelin protein zero-like 2, is essential for hearing in man and mouse. Am. J. Hum. Genet. 2018;103:74–88. doi: 10.1016/j.ajhg.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Street K., Risso D., Fletcher R.B., Das D., Ngai J., Yosef N., Purdom E., Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genom. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wright M.N., Ziegler A. ranger: a fast implementation of random forests for high dimensional data in C++ and R. J. Stat. Software. 2017;77:1–17. doi: 10.18637/jss.v077.i01. [DOI] [Google Scholar]
- 64.Abraham A.B., Bronstein R., Reddy A.S., Maletic-Savatic M., Aguirre A., Tsirka S.E. Aberrant neural stem cell proliferation and increased adult neurogenesis in mice lacking chromatin protein HMGB2. PLoS One. 2013;8 doi: 10.1371/journal.pone.0084838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bondurand N., Dufour S., Pingault V. News from the endothelin-3/EDNRB signaling pathway: role during enteric nervous system development and involvement in neural crest-associated disorders. Dev. Biol. 2018;444:S156–S169. doi: 10.1016/j.ydbio.2018.08.014. [DOI] [PubMed] [Google Scholar]
- 66.Hao Z.Z., Wei J.R., Xiao D., Liu R., Xu N., Tang L., Huang M., Shen Y., Xing C., Huang W., et al. Single-cell transcriptomics of adult macaque hippocampus reveals neural precursor cell populations. Nat. Neurosci. 2022;25:805–817. doi: 10.1038/s41593-022-01073-x. [DOI] [PubMed] [Google Scholar]
- 67.Sakurai M., Ayukawa K., Setsuie R., Nishikawa K., Hara Y., Ohashi H., Nishimoto M., Abe T., Kudo Y., Sekiguchi M., et al. Ubiquitin C-terminal hydrolase L1 regulates the morphology of neural progenitor cells and modulates their differentiation. J. Cell Sci. 2006;119:162–171. doi: 10.1242/jcs.02716. [DOI] [PubMed] [Google Scholar]
- 68.Cheng F., Shen Y., Mohanasundaram P., Lindström M., Ivaska J., Ny T., Eriksson J.E. Vimentin coordinates fibroblast proliferation and keratinocyte differentiation in wound healing via TGF-β-Slug signaling. Proc. Natl. Acad. Sci. USA. 2016;113:E4320–E4327. doi: 10.1073/pnas.1519197113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu Q., Masuda T., Kuramitsu S., Tobo T., Sato K., Kidogami S., Nambara S., Ueda M., Tsuruda Y., Kuroda Y., et al. Potential association of LOXL1 with peritoneal dissemination in gastric cancer possibly via promotion of EMT. PLoS One. 2020;15 doi: 10.1371/journal.pone.0241140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma L., Liu J., Xiao E., Ning H., Li K., Shang J., Kang Y. MiR-15b and miR-16 suppress TGF-β1-induced proliferation and fibrogenesis by regulating LOXL1 in hepatic stellate cells. Life Sci. 2021;270 doi: 10.1016/j.lfs.2021.119144. [DOI] [PubMed] [Google Scholar]
- 71.Ma L., Zeng Y., Wei J., Yang D., Ding G., Liu J., Shang J., Kang Y., Ji X. Knockdown of LOXL1 inhibits TGF-β1-induced proliferation and fibrogenesis of hepatic stellate cells by inhibition of Smad2/3 phosphorylation. Biomed. Pharmacother. 2018;107:1728–1735. doi: 10.1016/j.biopha.2018.08.156. [DOI] [PubMed] [Google Scholar]
- 72.Vervoort S.J., Lourenço A.R., van Boxtel R., Coffer P.J. SOX4 mediates TGF-β-induced expression of mesenchymal markers during mammary cell epithelial to mesenchymal transition. PLoS One. 2013;8 doi: 10.1371/journal.pone.0053238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Voloshenyuk T.G., Hart A.D., Khoutorova E., Gardner J.D. TNF-α increases cardiac fibroblast lysyl oxidase expression through TGF-β and PI3Kinase signaling pathways. Biochem. Biophys. Res. Commun. 2011;413:370–375. doi: 10.1016/j.bbrc.2011.08.109. [DOI] [PubMed] [Google Scholar]
- 74.Xie J., Wang C., Huang D.Y., Zhang Y., Xu J., Kolesnikov S.S., Sung K.L.P., Zhao H. TGF-beta1 induces the different expressions of lysyl oxidases and matrix metalloproteinases in anterior cruciate ligament and medial collateral ligament fibroblasts after mechanical injury. J. Biomech. 2013;46:890–898. doi: 10.1016/j.jbiomech.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 75.Havis E., Bonnin M.A., Esteves de Lima J., Charvet B., Milet C., Duprez D. TGFβ and FGF promote tendon progenitor fate and act downstream of muscle contraction to regulate tendon differentiation during chick limb development. Development. 2016;143:3839–3851. doi: 10.1242/dev.136242. [DOI] [PubMed] [Google Scholar]
- 76.Jäger B., Klatt D., Plappert L., Golpon H., Lienenklaus S., Barbosa P.D., Schambach A., Prasse A. CXCR4/MIF axis amplifies tumor growth and epithelial-mesenchymal interaction in non-small cell lung cancer. Cell. Signal. 2020;73 doi: 10.1016/j.cellsig.2020.109672. [DOI] [PubMed] [Google Scholar]
- 77.Lai Y.J., Tsai F.C., Chang G.J., Chang S.H., Huang C.C., Chen W.J., Yeh Y.H. miR-181b targets semaphorin 3A to mediate TGF-β-induced endothelial-mesenchymal transition related to atrial fibrillation. J. Clin. Invest. 2022;132 doi: 10.1172/JCI142548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Murphy-Ullrich J.E., Suto M.J. Thrombospondin-1 regulation of latent TGF-β activation: a therapeutic target for fibrotic disease. Matrix Biol. 2018;68–69:28–43. doi: 10.1016/j.matbio.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oh S.H., Swiderska-Syn M., Jewell M.L., Premont R.T., Diehl A.M. Liver regeneration requires Yap1-TGFβ-dependent epithelial-mesenchymal transition in hepatocytes. J. Hepatol. 2018;69:359–367. doi: 10.1016/j.jhep.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marconi G.D., Fonticoli L., Rajan T.S., Pierdomenico S.D., Trubiani O., Pizzicannella J., Diomede F. Epithelial-Mesenchymal Transition (EMT): the type-2 EMT in wound healing, tissue regeneration and organ fibrosis. Cells. 2021 doi: 10.3390/cells10071587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang L., Hu Z. Sensory epithelial cells acquire features of prosensory cells via epithelial to mesenchymal transition. Stem Cell. Dev. 2012;21:1812–1821. doi: 10.1089/scd.2011.0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hamby M.E., Coppola G., Ao Y., Geschwind D.H., Khakh B.S., Sofroniew M.V. Inflammatory mediators alter the astrocyte transcriptome and calcium signaling elicited by multiple G-protein-coupled receptors. J. Neurosci. 2012;32:14489–14510. doi: 10.1523/JNEUROSCI.1256-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.DaCosta Byfield S., Major C., Laping N.J., Roberts A.B. SB-505124 is a selective inhibitor of transforming growth factor-beta type I receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2004;65:744–752. doi: 10.1124/mol.65.3.744. [DOI] [PubMed] [Google Scholar]
- 85.Li X.J., Morgan C., Goff L.A., Doetzlhofer A. Follistatin promotes LIN28B-mediated supporting cell reprogramming and hair cell regeneration in the murine cochlea. Sci. Adv. 2022;8:eabj7651. doi: 10.1126/sciadv.abj7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jankauskas S.S., Wong D.W.L., Bucala R., Djudjaj S., Boor P. Evolving complexity of MIF signaling. Cell. Signal. 2019;57:76–88. doi: 10.1016/j.cellsig.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 87.Yang M., Wu S., Cai W., Ming X., Zhou Y., Chen X. Hypoxia-induced MIF induces dysregulation of lipid metabolism in Hep2 laryngocarcinoma through the IL-6/JAK-STAT pathway. Lipids Health Dis. 2022;21:82. doi: 10.1186/s12944-022-01693-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu H., Luo Q., Zhang J., Mo C., Wang Y., Li J. Endothelins (EDN1, EDN2, EDN3) and their receptors (EDNRA, EDNRB, EDNRB2) in chickens: functional analysis and tissue distribution. Gen. Comp. Endocrinol. 2019;283 doi: 10.1016/j.ygcen.2019.113231. [DOI] [PubMed] [Google Scholar]
- 89.Brennan A., Dean C.H., Zhang A.L., Cass D.T., Mirsky R., Jessen K.R. Endothelins control the timing of Schwann cell generation in vitro and in vivo. Dev. Biol. 2000;227:545–557. doi: 10.1006/dbio.2000.9887. [DOI] [PubMed] [Google Scholar]
- 90.Quintes S., Brinkmann B.G., Ebert M., Fröb F., Kungl T., Arlt F.A., Tarabykin V., Huylebroeck D., Meijer D., Suter U., et al. Zeb2 is essential for Schwann cell differentiation, myelination and nerve repair. Nat. Neurosci. 2016;19:1050–1059. doi: 10.1038/nn.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hammond T.R., McEllin B., Morton P.D., Raymond M., Dupree J., Gallo V. Endothelin-B receptor activation in astrocytes regulates the rate of oligodendrocyte regeneration during remyelination. Cell Rep. 2015;13:2090–2097. doi: 10.1016/j.celrep.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 92.Yanai H., Ban T., Wang Z., Choi M.K., Kawamura T., Negishi H., Nakasato M., Lu Y., Hangai S., Koshiba R., et al. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature. 2009;462:99–103. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]
- 93.Lee D., Taniguchi N., Sato K., Choijookhuu N., Hishikawa Y., Kataoka H., Morinaga H., Lotz M., Chosa E. HMGB2 is a novel adipogenic factor that regulates ectopic fat infiltration in skeletal muscles. Sci. Rep. 2018;8:9601. doi: 10.1038/s41598-018-28023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen K., Zhang J., Liang F., Zhu Q., Cai S., Tong X., He Z., Liu X., Chen Y., Mo D. HMGB2 orchestrates mitotic clonal expansion by binding to the promoter of C/EBPβ to facilitate adipogenesis. Cell Death Dis. 2021;12:666. doi: 10.1038/s41419-021-03959-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang C., Fondufe-Mittendorf Y.N., Wang C., Chen J., Cheng Q., Zhou D., Zheng Y., Geiger H., Liang Y. Latexin regulation by HMGB2 is required for hematopoietic stem cell maintenance. Haematologica. 2020;105:573–584. doi: 10.3324/haematol.2018.207092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bronstein R., Kyle J., Abraham A.B., Tsirka S.E. Neurogenic to gliogenic fate transition perturbed by loss of HMGB2. Front. Mol. Neurosci. 2017;10:153. doi: 10.3389/fnmol.2017.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wilkinson K.D., Lee K.M., Deshpande S., Duerksen-Hughes P., Boss J.M., Pohl J. The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase. Science. 1989;246:670–673. doi: 10.1126/science.2530630. [DOI] [PubMed] [Google Scholar]
- 98.Alpaugh W.F., Voigt A.L., Dardari R., Su L., Al Khatib I., Shin W., Goldsmith T.M., Coyle K.M., Tang L.A., Shutt T.E., et al. Loss of ubiquitin carboxy-terminal hydrolase l1 impairs long-term differentiation competence and metabolic regulation in murine spermatogonial stem cells. Cells. 2021 doi: 10.3390/cells10092265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reinicke A.T., Laban K., Sachs M., Kraus V., Walden M., Damme M., Sachs W., Reichelt J., Schweizer M., Janiesch P.C., et al. Ubiquitin C-terminal hydrolase L1 (UCH-L1) loss causes neurodegeneration by altering protein turnover in the first postnatal weeks. Proc. Natl. Acad. Sci. USA. 2019;116:7963–7972. doi: 10.1073/pnas.1812413116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Honda A., Kita T., Seshadri S.V., Misaki K., Ahmed Z., Ladbury J.E., Richardson G.P., Yonemura S., Ladher R.K. FGFR1-mediated protocadherin-15 loading mediates cargo specificity during intraflagellar transport in inner ear hair-cell kinocilia. Proc. Natl. Acad. Sci. USA. 2018;115:8388–8393. doi: 10.1073/pnas.1719861115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liao Y., Smyth G.K., Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Townes F.W., Irizarry R.A. Quantile normalization of single-cell RNA-seq read counts without unique molecular identifiers. Genome Biol. 2020;21:160. doi: 10.1186/s13059-020-02078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sardá-Espinosa A. Time-series clustering in R using the dtwclust package. Rice J. 2019;11:22–43. doi: 10.32614/RJ-2019-023. [DOI] [Google Scholar]
- 109.van Dijk D., Sharma R., Nainys J., Yim K., Kathail P., Carr A.J., Burdziak C., Moon K.R., Chaffer C.L., Pattabiraman D., et al. Recovering gene interactions from single-cell data using data diffusion. Cell. 2018;174:716–729.e27. doi: 10.1016/j.cell.2018.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wagner G.P., Kin K., Lynch V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theor. Biosci. 2012;131:281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 111.McInnes L., Healy J., Saul N., Großberger L. UMAP: uniform manifold approximation and projection. J. Open Source Softw. 2018;3:861. doi: 10.21105/joss.00861. [DOI] [Google Scholar]
- 112.Hassanpour H., Aghajani Z., Bahadoran S., Farhadi N., Nazari H., Kaewduangta W. Identification of reliable reference genes for quantitative real-time PCR in ovary and uterus of laying hens under heat stress. Stress. 2019;22:387–394. doi: 10.1080/10253890.2019.1574294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.