Abstract
The proapoptotic BCL-2 homology (BH3)-only endoplasmic reticulum (ER)–resident protein BCL-2 interacting killer (BIK) positively regulates mitochondrial outer membrane permeabilization, the point of no return in apoptosis. It is generally accepted that BIK functions at a distance from mitochondria by binding and sequestering antiapoptotic proteins at the ER, thereby promoting ER calcium release. Although BIK is predominantly localized to the ER, we detect by fluorescence lifetime imaging microscopy–FRET microscopy, BH3 region–dependent direct binding between BIK and mitochondria-localized chimeric mutants of the antiapoptotic proteins BCL-XL and BCL-2 in both baby mouse kidney (BMK) and MCF-7 cells. Direct binding was accompanied by cell type–specific differential relocalization in response to coexpression of either BIK or one of its target binding partners, BCL-XL, when coexpressed in cells. In BMK cells with genetic deletion of both BAX and BAK (BMK-double KO), our data suggest that a fraction of BIK protein moves toward mitochondria in response to the expression of a mitochondria-localized BCL-XL mutant. In contrast, in MCF-7 cells, our data suggest that BIK is localized at both ER and mitochondria-associated ER membranes and binds to the mitochondria-localized BCL-XL mutant via relocalization of BCL-XL to ER and mitochondria-associated ER membrane. Rather than functioning at a distance, our data suggest that BIK initiates mitochondrial outer membrane permeabilization via direct interactions with ER and mitochondria-localized antiapoptotic proteins, which occur via ER–mitochondria contact sites, and/or by relocalization of either BIK or antiapoptotic proteins in cells.
Keywords: BCL-2 family, apoptosis, BCL-2 interacting killer, BIK, subcellular localization fluorescence lifetime imaging microscopy, FLIM–FRET
Abbreviations: BDEB, BIK–DAPk1–ERK1/2–BAK; BH, BCL-2 homology; BIK, BCL-2 interacting killer; BMK, baby mouse kidney; Cb5, cytochrome b5; ChPTDSS1, mCherry fused to full-length phosphatidylserine synthase-1; DAPk1, Death-Associated Protein kinase 1; DKO, double KO; DMEM, Dulbecco's modified Eagle's medium; ER, endoplasmic reticulum; ERK1/2, extracellular-regulated signal kinase 1 and 2; FHS, FLIM–hyperspectral; FLIM, fluorescence lifetime imaging microscopy; HEK293T, human embryonic kidney 293T cell line; IRES, internal ribosome entry site; MAM, mitochondria-associated ER membrane; MBR, membrane-binding region; MEM, minimum essential medium; MitoGreen, MitoTracker Green; MitoRed, MitoTrackerRed; MOM, mitochondrial outer membrane; MOMP, mitochondrial outer membrane permeabilization; qF3, quantitative Fast FLIM–FRET; ROI, region of interest; TCSPC, time-correlated single-photon counting; TMRE, tetramethylrhodamine ethyl ester
Defects in apoptosis have been linked to cancer and neurodegenerative disease (1, 2). The commitment step in apoptosis is finely controlled by a large group of semiredundant homologous “BCL-2 family” proteins. These proteins participate in primarily binary protein–protein interactions in the cell, culminating in mitochondrial outer membrane (MOM) permeabilization (MOMP) that releases proapoptotic factors into the cytoplasm, thereby committing the cell to death (reviewed in Ref. (3)). BCL-2 family proteins share one to four regions of BCL-2 homology (BH regions) and heterodimerize via the BH3 region of one protein binding to a canonical binding pocket found in multi-BH region proteins. The “direct activation” (4), “embedded together” (5, 6), “unified” (7), and “hierarchical” (8) models divide the BCL-2 family proteins into four functional groups: (1) “BH3-only activator proteins” sense apoptotic stimuli and activate (2) the “pore-forming proteins” (“effector”), BAX and BAK to oligomerize in and permeabilize the MOM. (3) “Antiapoptotic proteins” bind to and inhibit both pore-forming and BH3-only activator proteins. The resulting mutual sequestration has a prosurvival effect but renders the cells sensitive to the inhibition of antiapoptotic proteins. Finally, (4) “BH3-only sensitizer proteins” sense apoptotic stimuli and promote apoptosis indirectly by inhibiting antiapoptotic proteins and releasing BH3-only activators and activated pore-forming proteins. The embedded together model first recognized the active role of the lipid membrane in regulating BCL-2 family proteins (5, 6) and was updated in 2014 (9) to include three modes of antiapoptotic function: facilitating retrotranslocation of BAX from the MOM to cytoplasm (mode 0) (10, 11), inhibiting BH3 activators (mode 1) and active BAX/BAK (mode 2) (7). The dominant mode of action depends on multiple cellular factors, including cell type, tissue of origin, apoptotic agonist, and so on (3). Most BCL-2 family proteins have a C-terminal membrane-binding region (MBR) that determines subcellular localization and promotes proapoptotic or antiapoptotic function in cells via both membrane-dependent and membrane-independent means (5, 12). An example of the latter is the double-bolt locking to the antiapoptotic proteins BCL-XL and BCL-2 by the MBR of BIML, which prevents displacement of BIM from antiapoptotic proteins by drugs targeting BCL-2 or BCL-XL (13). Membrane binding can lead to conformational and therefore functional changes in BCL-2 family proteins, changes in interaction affinities, and alterations in the availability of binding partners (and their local concentrations), thereby imparting different modes of action depending on the subcellular location (reviewed in Ref. (3)).
Though in vitro studies clearly show a functional difference between BH3-only activators and sensitizers (4, 14, 15, 16), the biological significance of BH3-only activator function continues to be debated (17, 18). The importance of activator function is challenging to address in live cells because of the functional redundancy within the BCL-2 family. For example, because of mutual sequestration, BH3-only activators also act as sensitizers (3). Further complicating the study of BH3 activator function in cells, BH3-independent BAX/BAK autoactivation can be triggered by a variety of physical insults and by small molecules (19, 20).
The “indirect activation” model was proposed to explain the observation that apoptosis was observed in mouse embryonic fibroblast cells lacking three well-known BH3-only activators (BID, BIM, and PUMA) (20) suggesting that BAX/BAK autoactivate and can kill when BH3-only proteins neutralize antiapoptotic proteins. While this result suggests that in transformed cell lines grown in a hyperoxygenated environment on plastic that BH3-only activator function plays a minor (if any) role in cell death, it is difficult to directly extend to normal tissue. Moreover, another group observed partial resistance to apoptosis in the same triple knockdown with additional knockdown of NOXA and interpreted the result as evidence of BH3-only activator function in a “hierarchical model” (8, 21). In this model, BH3-only activators function downstream of antiapoptotic proteins and as BH3-only sensitizers but are still key players in the pathway. The embedded together model, however, sufficiently explains hierarchy by the differences in affinity of BH3-only activators for binding antiapoptotic proteins versus the pore-former proteins BAX and BAK (3, 14).
Recently, a “membrane-mediated permissive” (“membrane-mediated spontaneous BAX/BAK activation” or the “BH3-independent”) model was proposed based on gene knockout studies in HCT116 cells lacking eight known BH3-only proteins (OctaKO) (17, 18). OctaKO cells were resistant to apoptosis; however, normal rates of BAX/BAK-dependent cell death could be restored by blocking or genetically eliminating antiapoptotic proteins, BCL-XL and MCL-1 (18), suggesting spontaneous activation of BAX/BAK. The observation that in this system BAX/BAK activation was dependent on the C-terminal MBR of the proteins was interpreted as suggesting that the membrane itself was the direct BAX/BAK activator, in the absence of BH3-only proteins (18). Consistent with membrane bilayers promoting BAX activation in a liposome system (22), BH3-independent BAX/BAK activation was shown to occur in the presence of nonphysiologic concentrations of divalent cations and lipids (made with only two lipids, one cardiolipin:two phosphatidylcholines, 1 tetraoleoyl cardiolipin:2 phosphatidylcholine) at pH >8. In contrast, in experiments using isolated mitochondria (or liposomes with lipid composition similar to mitochondria), permeabilization because of spontaneous BAX/BAK activation is negligible compared with the rate observed in the presence of nanomolar levels of BH3-only activator (14, 23). Furthermore, membrane binding was not sufficient to activate the BAX mutant “S184V” that spontaneously targets membranes and retains cell killing function in cells. Rather, BAX S184V requires the addition of tBID, PUMA, or BIM (but not the sensitizer BAD) to initiate homo-oligomerization and BAX activation (24). In addition, HCT116 and mouse embryonic fibroblast cells reconstituted with BAX-S184V do not spontaneously undergo apoptosis (24, 25). Nevertheless, when BAX, BAK, and MCL-1 were knocked out in the OctaKO cell line (HCT116 Octa-MCL-1/BAX/BAK KO cells), no difference in the rate of apoptosis was observed when the cell line was reconstituted with BAX or BAK and the sensitizer BAD or a BH3 activator, BID or BIM, suggesting that in HCT116 cells the predominant role of BH3-only proteins is to inhibit antiapoptotic proteins (in HCT116 cells, MCL-1 and BCL-XL) (17). However, in primary tissues isolated from adult mouse and human patients (characterized as expressing little to no BAX, BAK, or antiapoptotic proteins and classified as “apoptosis refractory”), the addition of both recombinant BAX and a BH3-only activator was required to initiate MOMP (26). The “membrane-mediated permissive” model also fails to explain the evidence in vitro (16) and mouse models (26) demonstrating that BID preferentially activates BAK, whereas BIM activates both BAX and BAK.
As described previously, much of our understanding of BCL-2 family proteins has come from studying their interactions at the MOM (reviewed in Ref. (3)). There are still many uncertainties regarding the function of BCL-2 family proteins at the endoplasmic reticulum (ER), though the majority of the literature points toward a role in regulating Ca2+ release as a prodeath signal that is communicated to the mitochondria. Furthermore, as described by the microdomain hypothesis (27, 28), decreased mitochondria–ER distance is associated with increased efficacy of ER-to-mitochondria Ca2+ transfer (29). Mitochondrial Ca2+ uptake can lead to mitochondrial Ca2+ overload, which impairs energy production, increases the production of reactive oxygen species, and may cause mitochondrial swelling and cristae remodeling; together promoting apoptosis (30, 31, 32, 33).
BCL-2 family proteins at the ER regulate the rate of release of ER Ca2+ stores in this Ca2+-dependent apoptosis pathway (reviewed by Refs. (30, 34)). Mutual sequestration of BCL-2 family proteins at the ER also can prevent the proteins from functioning at mitochondria (35, 36). Furthermore, inhibition of antiapoptotic proteins at the ER may release proapoptotic proteins that subsequently localize to mitochondria, promoting apoptosis. Together, these data have been interpreted as supporting a “from a distance” model (36), in which some form of signal (Ca2+ or displaced BCL-2 family protein(s)) is transmitted from the ER to mitochondria to promote cell death.
For most BCL-2 family proteins, only a small fraction of the total population is located at the ER (3). However, BCL-2 interacting killer (BIK) is predominantly localized at the ER (4, 37, 38, 39, 40, 41). BIK BH3 peptide functions only as a sensitizer, and BIK is routinely referred to as a BH3-only sensitizer in the literature (4). However, when the BH3 region of BIK was used to replace the BH3 region of BID, the chimera functions as a BAX activator (42), putting into question whether full-length BIK is only a sensitizer or can also function as an activator. Here, we studied the sensitizer function of BIK by examining its interactions with antiapoptotic proteins.
Human BIK has been proposed to initiate MOMP via Ca2+-dependent (41, 43, 44, 45) and -independent pathways, which involve the displacement of proapoptotic BH3 activators and pore formers from sequestration by antiapoptotic proteins (4, 37). In studies using isolated light membranes (primarily ER microsomes) containing exogenously expressed hemagglutinin-tagged BIK, alkali extraction and protease digestion were used to establish that the C-terminal MBR of BIK transverses the membrane and anchors BIK into the ER facing the cytosol (37). Furthermore, swapping the MBR of BIK with that of the well-characterized ER-localized tail-anchor sequence from cytochrome b5 (Cb5) resulted in a chimeric protein with a proapoptotic function equal to wildtype BIK. Moreover, the coincubation of isolated light membranes containing BIK with isolated mitochondria resulted in MOMP, without BIK relocalizing to mitochondria. Evidence has since been presented for the expression of BIK in cells leading to displacement of activated BAX/BAK from antiapoptotic proteins, thereby promoting pore formation/ER Ca2+ release, and apoptosis (43, 44). These results suggest that BIK promotes cell death from its position at the ER, consistent with the “from a distance” model.
Recently, an alternative function for BIK was proposed in which BIK-induced BAK accumulation at the ER and recruited Death-Associated Protein kinase 1 (DAPk1) as part of a larger complex (45). The levels of BIK and BAK increased with interferon-γ treatment, resulting in the formation of BAK oligomers on the ER membrane and cell death that was dependent on the inhibition of BCL-2 by BIK (45). Coimmunoprecipitation experiments suggested that BIK, extracellular-regulated signal kinase 1 and 2 (ERK1/2), DAPk1 (46), and BAK may form a “BIK–DAPk1–ERK1/2–BAK” (BDEB) complex. The authors proposed a model of the BDEB complex located at sites of direct contact between the ER and mitochondria known as the “mitochondria-associated ER membranes” (MAMs). These complexes are envisioned as spanning the ER and MOM, thereby linking and reducing the distance between these membranes and increasing the efficacy of ER-to-mitochondria calcium transfer. The direct physical links of BIK to mitochondria in this model challenge the “from a distance” hypothesis of how BIK induces MOMP.
Here, we directly test both models for how BIK regulates apoptosis by examining the spatial distribution of BIK and its antiapoptotic binding partners in cells. Since the antiapoptotic proteins, BCL-XL and BCL-2, localize to both ER and mitochondria, we replaced the wildtype MBR with tail-anchor sequences from Cb5 and “ActA,” which localize proteins to the ER or mitochondria, respectively (47, 48, 49, 50). If direct binding is required for ER-localized BIK to regulate apoptosis, it would be expected to bind to and regulate only ER-localized antiapoptotic proteins. To measure direct binding between BIK and its targets, we used fluorescence lifetime imaging microscopy (FLIM) to measure the FRET between full-length human BIK and ER- or mitochondrial-localized versions of BCL-XL and BCL-2. Unexpectedly, we observed in cells that although localized at ER, BIK directly binds to antiapoptotic proteins even when the antiapoptotic protein was localized to mitochondria. We found that these interactions depend on the BH3 region of BIK and occur in cells lacking BAX and BAK, consistent with direct binding interactions but not the formation of DBEB complexes. Secondary structure predictions suggest that BIK could span the distance between the MAMs and mitochondria at ER–mitochondria contact sites. Thus, our results indicate that localization at MAMs may enable BIK to directly bind to antiapoptotic proteins at mitochondria. Our results suggest that the unexpected binding between ER-localized BIK and mitochondria-localized BCL-XL observed here results from cell type–specific relocalization of BIK or BCL-XL to ER or MAMs enabling an interaction that takes place spanning the membranes.
Results and discussion
BIK interacts with BCL-2 and BCL-XL proteins localized at the ER or mitochondria
Biophysical methods and microscopic imaging have been key to unraveling the complex BCL-2 family interaction network (51). Further delineation of how and where these interactions occur is an important step for understanding BCL-2 family protein function. When expressed in live cells, BIK with an N-terminally fused fluorescent protein remains functional (52). We recently demonstrated that in live cells a fluorescent protein fusion of BIK binds to BCL-2, BCL-XL, and BCL-W with high affinity and MCL-1 with lower affinity (52). Here, we investigate where these interactions occur in cells. Because of the low affinity of binding to MCL-1 and the restricted expression pattern of BCL-W, we focused only on the interactions between BIK and BCL-2 and BCL-XL. In cells, BIK localizes to the ER, whereas wildtype BCL-XL and BCL-2 localize to both the ER and MOM. While BCL-2 is constitutively membrane bound (53, 54, 55), a fraction of BCL-XL is soluble in the cytoplasm and the rest is peripherally bound to membranes (56, 57, 58). Previous data suggest that BIK binds BCL-XL and BCL-2 at the ER suggesting BIK inhibits only the ER-localized subset of antiapoptotic proteins. Inconsistent with this model, BIK is a very potent inducer of apoptosis.
To visualize the proteins, we used cells that expressed BIK and BCL-2 or BCL-XL proteins with fluorescence proteins fused to the amino terminus and measured binding interactions between them by FRET. As a convenient nomenclature, the fusion of mCerulean3 (FRET donor) or Venus (FRET acceptor) to the N terminus of a protein of interest is indicated by a superscript “C” or “V,” respectively. For example, CBIK indicates that mCerulean3 is fused to the N terminus of full-length human BIK protein. To avoid imaging dead/dying cells, we used baby mouse kidney (BMK) cells that have the genes encoding both BAX and BAK proteins knocked out (BMK-double KO [DKO]) for most of our experiments, as these pore-forming proteins are required for BIK to elicit cell death.
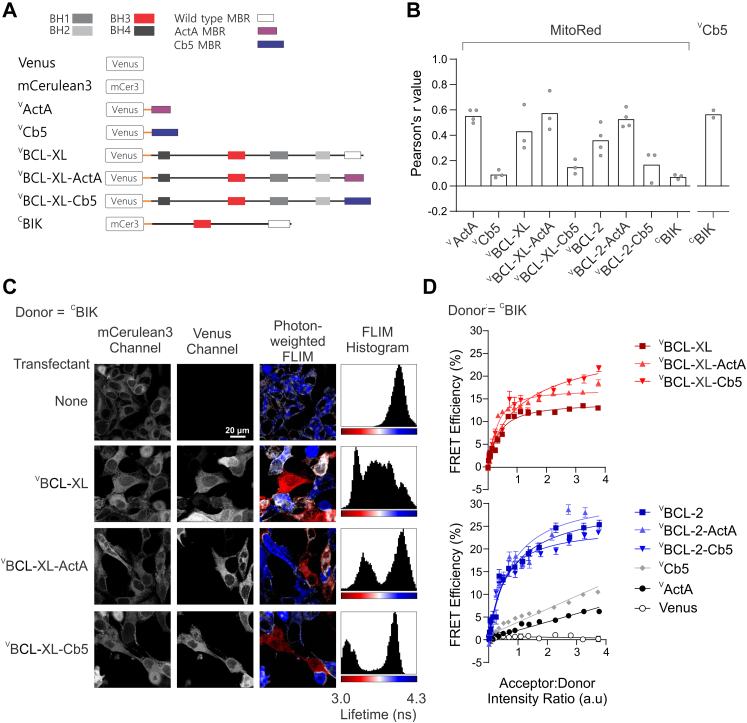
To specify the subcellular location of BIK–antiapoptotic protein interactions, we constructed mutants of VBCL-XL and VBCL-2 in which the wildtype MBR was replaced with the well-characterized tail-anchor targeting sequences of “Cb5” for ER localization or “ActA” for mitochondrial localization (47, 50, 59) (Fig. 1A). As controls localized to the same membranes, Venus was fused to the ActA and Cb5 sequences alone (VActA and VCb5). In BMK-DKO cells stained with DRAQ5 (to enable visualization of nuclei and guide cell segmentation), Pearson’s correlation was measured for individual cells for each fusion protein with either MitoTrackerRed (MitoRed), a marker of mitochondrial localization, or with VCb5, a marker for ER localization. As expected, images of VActA, VBCL-XL, VBCL-XL-ActA, and VBCL-2-ActA generated high Pearson’s r correlation values with MitoRed, whereas images of VCb5, VBCL-XL-Cb5, VBCL-2-Cb5, and CBIK generated low values (see Figs. 1B and S1 for examples). The high correlation value obtained between images of VCb5 and CBIK (Fig. 1B) confirms that in live cells CBIK is predominantly localized to the ER, as reported previously based on subcellular fractionation with the ER-resident protein, calnexin, but not mitochondria-resident protein, TOM-20 (37).
Figure 1.
BIK interacts with BCL-2 and BCL-XL at the endoplasmic reticulum and mitochondria.A, stick representations of fusion proteins indicating name (left) and location of the mCerulean3 or Venus fluorophore tethered by a flexible linker (orange). BH1–4: BCL-2 homology regions 1 to 4. B, Pearson’s r correlation for the indicated fusion protein with the mitochondrial stain, MitoRed, or with VCb5 as indicated above. A minimum of 50 cells were analyzed to determine the median for each biological replicate (gray symbols), and the mean of the biological replicates was determined (bars). See Fig. S1 for example images. C, confocal FLIM–FRET data with photon-weighted lifetime and FLIM histograms represented on the same pseudocolor scale. Cells that constitutively express CBIK were transiently transfected with constructs to express the proteins indicated at the left (transfectant). A decrease in fluorescence lifetime (shift from blue to red) indicates binding in photon-weighted FLIM images and FLIM histograms. D, FLIM–FRET binding curves measuring binding of CBIK to several Venus-fusion proteins (listed on the right) in BMK-DKO cells. Acceptor:donor intensity ratio is used to represent the relative concentrations of acceptor to donor in cells, expressed in arbitrary units of intensity. Each curve represents binned data from thousands of subcellular regions of interest. The median FRET efficiency and standard error for each binned point were plotted from a single representative experiment. Biological replicates (n = 3, Fig. S2A) were not combined as the instrument acquisition settings were not identical. While wildtype and chimera BCL-2 and BCL-XL proteins bind CBIK, the negative control acceptor proteins (gray, white, and black) do not bind. BH, BCL-2 homology; BIK, BCL-2 interacting killer; BMK, baby mouse kidney; DKO, double KO; FLIM, fluorescence lifetime imaging microscopy; MBR, membrane-binding region; MitoRed, MitoTrackerRed.
To measure protein binding by FRET, two candidate proteins were tagged with either a donor or an acceptor fluorophore and expressed together in live cells. The donor fluorophore (mCerulean3) can nonradiatively transfer energy to the acceptor (Venus) fluorophore only if they come into close proximity (1–10 nm). FRET efficiency decreases by the sixth power of the distance between the two fluorophores and is maximal only when the fluorophores are in the right orientation for FRET. As the chromophore is located inside the beta-barrel structure of the fluorescence protein, even if two fluorophores are in the closest apposition possible, they will be almost 5 nm apart. Consequently, if FRET is detected, the proteins of interest must be close enough to bring the donor and acceptor to within ∼5 nm in cells. At high ratios of acceptor–donor protein expressed in cells, the binding of the donor molecules becomes saturated. Thus, a curve that saturates indicates binding has occurred, whereas a linear increase in FRET indicates random collisions. The most accurate method for detecting FRET in cells is measuring the decrease in the fluorescence lifetime of the donor (measured in nanoseconds) that results from the energy transfer event (60, 61, 62).
Confocal images of BMK-DKO cells stably expressing CBIK (donor channel) and transiently expressing the indicated Venus-fusion protein (acceptor channel) illustrate the subcellular localization of the proteins (Fig. 1C). Photon-weighted FLIM images colored based on the calculated lifetime per pixel (from red to blue corresponding to 3.0–4.3 ns) and the intensity based on the number of photons collected in the donor image illustrate the distribution of CBIK (blue) and CBIK–VBCL-XL complexes (red). Intensity-weighted images are used for visualization only and not for any of the binding calculations. In cells expressing VBCL-XL, the red areas indicate a lower mCerulean3 lifetime as a result of the binding of VBCL-XL to CBIK, whereas blue areas indicate mCerulean3 lifetimes (average ∼3.8 ns) correlating to unbound CBIK. White areas indicate a mixture of free and bound CBIK proteins. In the FLIM histograms (right panels), the height of each peak represents the number of pixels in the image at each lifetime from 3 to 4.3 ns. As expected, in FLIM images of cells expressing only CBIK, there is a single peak in the FLIM histogram, representing the donor lifetime alone (∼3.8 ns). In cells expressing VBCL-XL and the VBCL-XL mutants with altered localization, multiple distinct populations of mCerulean3 lifetime indicate complexes with different affinities, compositions, and/or geometries. Surprisingly, for both VBCL-XL-Cb5 and VBCL-XL-Acta, there is a clear second lifetime population that appears as a second peak in the FLIM histogram and corresponds to complexes with different distributions of FRET efficiencies. The distance between these peaks reports both the extent of binding and the geometry (position of donor and acceptor fluorophores) of these protein complexes, both of which affect FRET efficiency. Interestingly, the data for VBCL-XL suggest that there may be multiple complexes and/or regions of interest (ROIs) with different proportions of bound and unbound CBIK (Fig. 1C, see multiple peaks in the FLIM histogram for BCL-XL). The latter may have arisen because of differences in expression levels, the amount of VBCL-XL in the cytoplasm, and local concentrations for VBCL-XL compared with the mutants with specific localizations. Nevertheless, CBIK bound to both VBCL-XL and ER-localized VBCL-XL-Cb5 as expected. Unexpectedly, for the mitochondria-localized protein VBCL-XL-ActA, there was an unanticipated clear second population with a pronounced decrease in lifetime indicative of direct binding to CBIK.
Quantification of the data from 100 images, each containing ∼5 to 10 cells (see Fig. 1C for examples), from multiple technical replicates (wells in a microtiter plate), resulted in thousands of subcellular ROIs for each biological replicate enabling the generation of FLIM–FRET binding curves showing CBIK bound to VBCL-XL and VBCL-2. Surprisingly, CBIK was found to bind equally well to antiapoptotic protein chimeric mutants with either the Cb5 or ActA localization signals that result in localization at the ER or mitochondria, respectively (Fig. 1D).
We recently published a quantitative Fast FLIM–FRET (qF3) method, which enables live-cell measurements of apparent Kd values (52). However, the expression levels for the fluorescence protein–tagged query proteins required for qF3 exceed the actual Kds of Bcl-2 family interactions. Thus, the apparent Kds measured by qF3 can be most effectively used to measure the effects of point mutations and small molecules on binding interactions (52). For the studies reported here, we used a simpler version of FLIM–FRET that enables determining relative binding efficiencies (62, 63). In the method used here, FRET efficiency and binding curve saturation are both indicators of binding. However, as with qF3, the binding curves generated by FLIM–FRET are a combination of collisional events plus binding; therefore, it is important to include a nonbinding mutant to correct for changes in collision frequencies that primarily depend on subcellular localization. For example, in Figure 1D, the donor mCerulean3-BIK localizes to ER membranes (Fig. 1B). Therefore, collisions with the VCb5 negative control generate a straight line with a higher slope because they are more frequent than for the mitochondria- or cytoplasm-localized collisional controls, VActA and Venus, respectively (Fig. 1D). As expected, binding to the wildtype VBCL-XL and VBCL-2, known to bind BIK in live cells (52), resulted in binding curves with higher FRET efficiency that also saturate when the acceptor:donor ratio is greater than 1. In contrast, primarily collisions were detected for the negative binding control, CBIK-L61G. Collisions were much more frequent for BIK-L61G with BCL-2 than with BCL-XL likely because both CBIK and VBCL-2 are constitutively membrane bound with a substantial fraction of Bcl-2 located at ER. In contrast, VBCL-XL localizes to the cytosol and is peripherally bound to ER and mitochondria and is, therefore, less likely to collide with ER membrane–bound CBIK. While it is unclear if the low level of collisions of CBIK and VActA represents a baseline level of mislocalization, the high efficiency of CBIK binding to VBCL-XL-ActA or VBCL-2-ActA warranted closer scrutiny to determine how and where these unexpected interactions occurred in cells.
BIK binding to mitochondria-localized BCL-XL or BCL-2 occurs via the BIK BH3 region
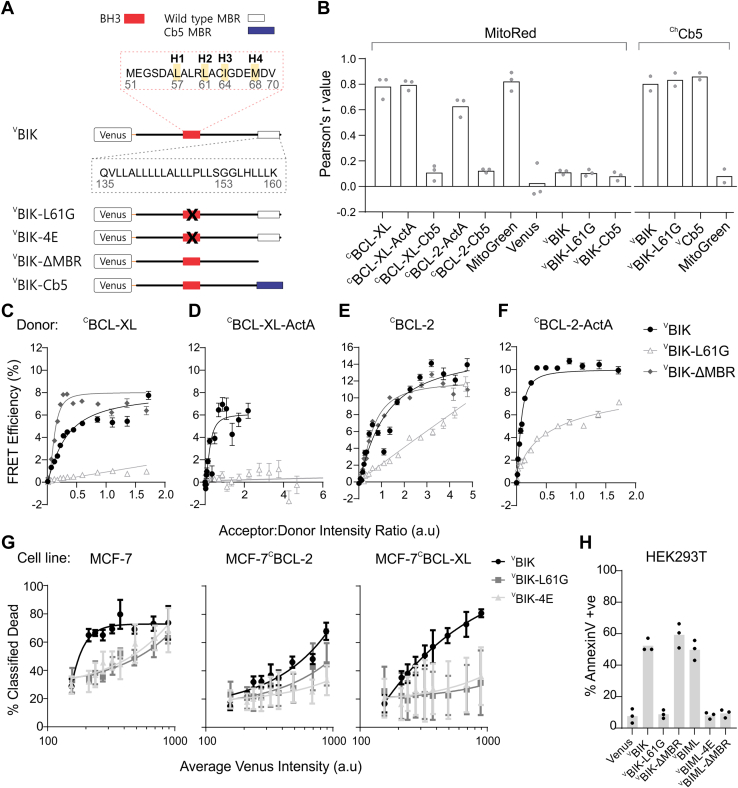
To identify which region(s) of the BIK protein is required for binding to mitochondria-localized BCL-XL and BCL-2, we created mutants of BIK predicted to disrupt binding to antiapoptotic proteins. To facilitate binding measurements in live cells, we reversed the donor and acceptor constructs and generated stable BMK-DKO cell lines expressing each of the donors CBCL-XL, CBCL-XL-ActA, CBCL-2, or CBCL-2-ActA enabling FRET measurements upon transient expression of acceptor VBIK mutants. To inactivate the BH3 region of BIK, the hydrophobic amino acid leucine (L) at position 61 (H2) was mutated to glycine (G) (VBIK-L61G, Fig. 2A) (37, 38). The MBR of BIK is a classical tail-anchor sequence of ∼20 amino acids (>60% leucine) that anchors BIK into the ER membrane (37, 64) (Fig. 2A, see BIK sequence 135–160). When the MBR of BIK was exchanged for the Cb5 sequence (BIK-Cb5), the ER-localized mutant still promoted mitochondrial cytochrome c release (37) leading to the hypothesis that BIK promotes apoptosis “from a distance” rather than directly at the MOM. In contrast, many other BCL-2 family proteins translocate to mitochondria to regulate apoptosis including BID (65), NOXA (66), and BIML (14, 67). To test the hypothesis that BIK acts at a distance, we generated two more mutants, the ER-localized “VBIK-Cb5” and the MBR-truncated “VBIK-ΔMBR,” which we expect to remain in the cytoplasm (Fig. 2A). Data for other BIK mutants, discussed briefly later, are shown in Fig. S2B.
Figure 2.
BIK binding to BCL-XL and BCL-2 at the ER and mitochondria is BH3 dependent but MBR independent.A, stick diagrams of Venus fused to BIK (VBIK) and BIK mutants, indicating the positions of the “L61G” and “4E” mutations in the BH3 region (X in red box) and the MBR (in white box). The amino acid sequence of the BH3 region of BIK (red dotted box) with positions of four key hydrophobic residues (H1–4) highlighted. The amino acid sequence of the BIK MBR (black dotted box). B, Pearson’s r value correlation coefficients for micrographs of the indicated fusion protein or the control MitoGreen dye (listed below) with MitoRed dye (n = 3) or with the ER-localized marker mCherryCb5, ChCb5 (n = 2) indicated above in BMK-DKO cells. See Fig. S1, A and B, for example images. C–F, FLIM–FRET binding curves for the mCerulean3 donor fusion protein (indicated above) stably expressed in BMK-DKO cells binding to the acceptor proteins expressed by transient transfection (legend, F). Acceptor:donor intensity ratio is used to represent the relative concentrations of acceptor to donor in cells, expressed in arbitrary units of intensity. Each curve represents binned data from thousands of subcellular regions of interest. The median FRET efficiency and standard error for each binned point were plotted from a single experiment. Data for biological replicates (n = 3) for C–F are included in Fig. S2, C–F, respectively, along with results of similar experiments performed in MCF-7 cells. G, cell death measured for Venus-positive cells based on a linear classifier incorporating AnnexinV labeling, nuclear shrinkage (small nuclei [SN]) and decreased TMRE intensity (mitochondrial transmembrane potential). The cell line is labeled above each graph. G, BH3-dependent and BIK-induced cell death is inhibited in MCF-7 and MCF-7 cells stably expressing CBCL-2 or CBCL-XL. Venus-fusion protein(s) transiently expressed in cells are indicated in the legend. The average intensity of Venus per cell was used as a measure of BIK and BIK mutant protein expression. For each replicate, data were binned by Venus intensity, and median cell death was quantified per bin. The mean and standard error of three biological replicates are displayed (symbols). H, in HEK293T cells, BIK-induced cell death depends on an intact BH3 region but is independent of the MBR. As controls, cell death measurements are reported for cells expressing VBIML, VBIML-4E (inactive BH3-4E mutant), and VBIML-ΔMBR (MBR deleted). Each point represents the percent of Venus-positive cells that were AnnexinV positive. Each point is an individual replicate with the bar representing the mean. BH, BCL-2 homology; BIK, BCL-2 interacting killer; DKO, double KO; ER, endoplasmic reticulum; FLIM, fluorescence lifetime imaging microscopy; HEK293T, human embryonic kidney 293T cell line; MBR, membrane-binding region; MitoGreen, MitoTracker Green; MitoRed, MitoTracker Red; TMRE, tetramethylrhodamine ethyl ester.
One potential explanation for our findings and previous data in support of “killing from a distance” models would be if the localization of the fluorescence protein fusions differs from our expectations. Therefore, the localization of the fusion proteins with altered MBRs was assessed in BMK-DKO cells using Pearson’s r correlation values with the mitochondrial marker MitoRed (see Figs. 2B and S1 for example images). As expected, CBCL-XL, CBCL-XL-ActA, CBCL-2-ActA, and MitoTracker Green (MitoGreen) colocalized with MitoRed, whereas CBCL-XL-Cb5, CBCL-2-Cb5, VBIK, or VBIK-L61G had much lower Pearson’s r values indicating that these ER-localized proteins have poor correlation with mitochondria. An expression plasmid encoding the mCherry fluorophore fused to the Cb5 sequence (ChCb5) was generated as a red ER landmark. As expected, ChCb5 was highly correlated with VBIK, VBIK-L61G, and VCb5 but not MitoGreen (Fig. 2B). Nevertheless, FLIM–FRET measurements revealed that VBIK binds to CBCL-XL, CBCL-2, CBCL-XL-ActA, and CBCL-2-ActA (Fig. 2, C–F). In contrast, FLIM measurements for the control mutant VBIK-L61G, with a single-point mutation in the BH3 region, resulted in a linear increase in and low % FRET efficiency with CBCL-XL or CBCL-XL-ActA indicating collisions rather than binding (Fig. 2, C and D). For unknown reasons, a much higher frequency of collisions was detected for VBIK-L61G with CBCL-2 and CBCL-2-ActA suggesting that background collisions might contribute to the higher FRET efficiency observed for VBIK binding to CBCL-2 (Fig. 2, E and F). While high rates of collisions were reproducible in BMK-DKO cells, the curvature in the line suggesting there may be some authentic binding was not routinely reproducible (Fig. S2, D–F). The reason for high rates of collisions for the two proteins is not clear as collisions for the same proteins were infrequent in MCF-7 cells (Fig. S2, D–F). Nevertheless, the remarkable effect of a single BH3 mutation reinforces the importance of the BIK BH3 region for binding to mitochondria-localized CBCL-XL-ActA and CBCL-2-ActA. Eliminating binding to ER membranes entirely by deletion of the MBR of BIK (VBIK-ΔMBR) marginally improved binding to CBCL-XL in Figure 2C but not in other replicates (Fig. S2, C and E) and did not impact binding to CBCL-2 (Figs. 2E and S2, C and E).
To test the proapoptotic function of the BIK mutants, we quantified cell death induced by transient expression in multiple cell lines. In MCF-7 cells, data were binned by arbitrary units of intensity as a measure of the relative expression of each mutant. MCF-7 cells are more sensitive to the expression of VBIK (Fig. 2G), than the VBIK-L61G and VBIK-4E mutations, consistent with the effects of the mutations on binding to CBCL-XL and CBCL-2 (Fig. 2, E and F). Unexpectedly, there was residual proapoptotic function of VBIK-L61G and VBIK-4E but only at high levels of expression (Fig. 2G). High levels of expression of Venus alone had no effect in MCF-7 cells (13), suggesting that cell death because of high expression of VBIK-L61G may be because of overexpression of a tail-anchored membrane protein, which could disrupt membrane integrity (68), or could suggest that another region of BIK outside the BH3 has minor proapoptotic activity in MCF-7 cells. Consistent with the hypothesis that overexpression of a tail-anchored membrane protein is poorly tolerated in MCF-7 cells, high levels of expression of the tail-anchored protein VBIMEL-4E also has some proapoptotic activity (13).
Stable expression of CBCL-2 or CBCL-XL in MCF-7 cells provided resistance to VBIK expression, as expected based on mutual sequestration models and our FLIM–FRET results (Fig. 2G). Therefore, the BIK binding to antiapoptotic proteins measured by FLIM–FRET is functionally relevant in apoptosis. However, at high levels of VBIK expression apoptosis resistance because of expression of CBCL-XL/CBCL-2 were overcome consistent with the binding detected by FLIM–FRET and suggesting that once the unoccupied exogenously expressed antiapoptotic proteins were saturated that BIK may function as a sensitizer.
Also in line with our FLIM–FRET data demonstrating that VBIK-ΔMBR binds BCL-2 and BCL-XL (Fig. 2, C–F) expression of VBIK, VBIK-Cb5, and VBIK-ΔMBR, each killed both BMK and HCT116 cells in a BAX- and BAK-dependent manner (Fig. S2G). Swapping the MBR for the known ER tail anchor, “Cb5,” confirmed previous results that lead to the “from a distance” model, suggesting BIK can kill cells while anchored at ER membranes (37, 44). In contrast, our results contradict data reported in Hep3B cells, suggesting that the MBR and localization at membranes are critical for the proapoptotic activity of BIK (41). Unlike results obtained for BIM, deletion of the MBR did not affect cell death induced by BIK (compare VBIK and VBIK-ΔMBR) (14) (Fig. 2H). As expected from the FLIM–FRET data, BH3 mutant, VBIK-L61G, had no proapoptotic activity in BMK, HCT116, or human embryonic kidney 293T (HEK293T) cells (Figs. 2H and S2G) consistent with previous reports in H1299 human lung carcinoma cells (43) and mouse and human airway epithelial cells (45).
Other regions of BIK have been reported to contribute to its proapoptotic activity. BIK phosphorylated at threonine (T) 33 and serine (S) 35 reportedly has greater proapoptotic activity than its unphosphorylated counterpart because of more efficient coimmunoprecipitation with BCL-2 and BCL-XL (69, 70). We created a phosphomimetic “BIK-DD” mutant where both these sites were mutated to aspartic acid (D) and, “BIK-AA,” where both residues were mutated to alanine to prevent phosphorylation in cells. In contrast to previous reports (69, 70), we observed no loss of proapoptotic function for the BIK-AA or BIK-DD mutant in either HCT116 or BMK cells (Fig. S2G). Moreover, we did not detect a difference in FRET for these mutants with CBCL-2 or CBCL-XL (Fig. S2, C and E). The activity of BIK has also been reported to be regulated by serine protease rhomboid domain–containing 1 protein cleavage of the BIK MBR at position 153 between two glycine residues, thereby destabilizing BIK in HEK293T cells (71). Thus, we hypothesized blocking this cleavage site (BIK mutant VBIK-GG153AA) or introducing a stop codon at this site (BIK mutant VBIK-G154-STOP to mimic cleavage) might change the available binding partners for BIK in cells. However, there was no difference in binding of mutants VBIK-GG153AA or VBIK-G154-STOP to wildtype or mitochondria-targeted CBCL-XL or CBCL-2 in BMK-DKO or MCF-7 cells (Fig. S2, C and E).
Altogether, these functional and binding data suggest that because of its restricted localization, BIK kills cells while anchored to the ER membrane, yet neither ER localization nor the BIK MBR is necessary for proapoptotic function. Unexpectedly, our data suggest that although the MBR localizes BIK to ER membranes, it did not prevent binding to antiapoptotic proteins localized at mitochondria. That a direct interaction is involved consistent with our observations that a functional BH3 region of BIK is required for BIK to bind to mitochondria-localized antiapoptotic proteins and to kill cells (Figs. 2 and S2). However, the localization data for the proteins were generated from cells expressing either BIK (or a BIK mutant) or one of the antiapoptotic proteins but not when BIK and the antiapoptotic proteins were coexpressed. One possible explanation is that each protein remains anchored in its target membrane (ER or mitochondria), and that binding occurs across the space between these two organelles. Another explanation for the FLIM–FRET data is that coexpression of BIK with its antiapoptotic binding partner results in a change in subcellular localization for one or both proteins.
Predicted model of ER-localized BIK binding to BCL-XL at mitochondria and ER
While BCL-XL could relocalize to the ER, it is less likely that CBCL-XL-ActA and CBCL-2-ActA would bind to ER membranes. Furthermore, as VBIK is constitutively located at ER and contains a classical ER-localized tail-anchor sequence, it is highly unlikely that it inserts into the outer mitochondrial membrane. Therefore, if the proteins remain in their target membranes, interactions must occur at regions of close apposition of the membranes. Tethering proteins (mitofusins) physically link the MAMs and MOM (45) resulting in an average distance of only 10 to 30 nm between these membrane surfaces (72, 73, 74). Recently, Mebratu et al. (45) made direct measurements of the distance and the frequency of ER–mitochondria contact sites, which suggested that expression of BIK, but not BIK-L61G, reduced the average ER–mitochondrial distance from ∼35 to ∼20 nm and increased the frequency of close ER–mito contact sites (defined as <15 nm) from 16 to 35% of the total ER surface area (45). Thus, a direct interaction between BIK and BCL-XL could occur if the proteins span the space at ER–mitochondria contact sites. To test the feasibility of this hypothesis, we modeled the putative interaction using available predictive software.
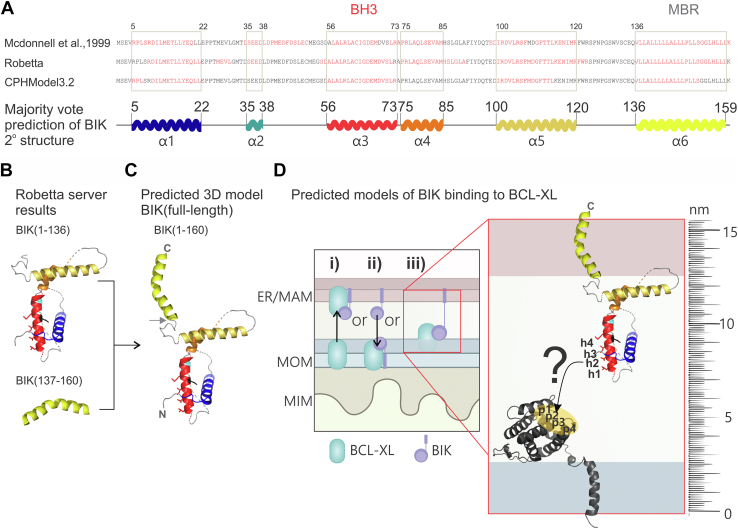
For FRET to be detected when VBIK is located primarily at the ER and MAM and CBCL-XL-ActA is located at the MOM, the proteins must be positioned such that the two fluorophores are less than 10 nm from each other. For the interaction to also be dependent on the BIK BH3 (as shown for loss of binding of BIKL61G), the BH3 sequence of BIK must be able to come in contact with the BH3-binding pocket on BCL-XL. The known structure of BCL-XL (1bxl.pdb and 6f46.pdb) suggests that the BH3-binding pocket can be located ∼2.5 nm from the MOM (see the Experimental procedures section for each measurement). As there is no published structure for BIK, we resorted to modeling in an effort to determine if interaction with CBCL-XL-ActA is feasible. There are 65 amino acids separating the BH3 sequence and the ER-anchored MBR of BIK (37). At 3.5 Ä per amino acid, a completely extended and unfolded sequence could cross ∼23 nm, suggesting that the BH3 region could theoretically reach mitochondrial BCL-XL/BCL-2 at ER–mito contact sites (defined as <15 nm (45)). However, a previous analysis of sequence hydrophobicity/net charge resulted in a prediction that BIK has three alpha-helices between the BH3 and MBR (75). To extend this observation, we predicted a secondary structure for the human BIK sequence using two available servers for 3D structure prediction (76, 77) (Figs. 3A and S3A).
Figure 3.
A predicted model of BIK binding antiapoptotic BCL-XL at mitochondrial (mito) MAM contact sites.A, secondary structure predictions from Robetta and CPHmodel3.2 servers (75). The amino acid sequence of BIK is shown where red indicates predicted α-helix. Regions selected in the boxes are predicted helical in at least two-third models (majority vote secondary [2o] structure), which was used to create our predicted model of full-length BIK. In the stick diagram shown, helices 1 to 6 in BIK are colored from blue at the N terminus, red at the BH3 region, to yellow at the C-terminal MBR. B and C, 3D models generated in PyMOL, where helix color matches (A), and within the BH3 region (red), the four conserved hydrophobic residues (h1–h4) and aspartic acid (black) are displayed as sticks. B, the predicted structure obtained from the Robetta server for residues 1 to 134 and residues 137 to 160 (submitted separately) were combined in (C) to generate a majority vote 3D model of full-length BIK. The gray arrow indicates the arbitrary point of attachment of these two fragments. N- and C-terminal residues are labeled. D, Cartoon model (left) of the possible mechanisms explaining how an interaction could occur between the known ER-anchored BIK and mito-anchored BCL-XL: (i) BCL-XL relocalizes to ER/MAMs, (ii) BIK relocalizes to mito, (iii) BIK at MAMs interacts with BCL-XL at mito. Predicted 3D model for BIK anchored in the ER membrane and BCL-XL anchored in the MOM (protein = dark gray, modeled in PyMOL to match the study by Yao et al., 2015 (103)). The position of the four hydrophobic pockets (p1–p4) on BCL-XL is highlighted to display how the corresponding residues in the BH3 region of BIK’ (h1–h4) would need to extend and rotate to bind in the right direction. Such a movement may be possible because of the four long flexible loop regions in the BIK protein to enable interaction at close ER–mito contact sites (see “ruler”). ER, endoplasmic reticulum; BH, BCL-2 homology; BIK, BCL-2 interacting killer; MAM, mitochondria-associated ER membrane; MBR, membrane-binding region; MIM, mitochondrial inner membrane; MOM, mitochondrial outer membrane.
The prediction largely agrees with that originally published by McDonnell et al. (75), except that alpha-helix 2 is considerably shortened and replaced by a loop region. This majority vote secondary structure prediction agreed best with the predicted 3D structure that resulted from submitting truncated BIK (1–136) to the Robetta server (Fig. S3B). This structure for truncated BIK is also most consistent with the MBR being constitutively embedded in the membrane (37), where it would not contribute to the structure of the cytoplasmic domain. Thus, to generate a 3D model of full-length BIK, we fused the truncated BIK (1–136) to the MBR (137–160) of BIK (analyzed separately) (Fig. 3B). The prediction suggests that BIK contains six alpha-helices with >10 amino acid long loop regions (78) between helices 1 to 2, 2 to 3, 4 to 5, and 5 to 6 (Fig. 3C), suggesting increased flexibility compared with earlier predictions (75). In this model, the BH3 region of BIK may extend up to a distance of ∼14 nm from the ER. Considering the BH3-binding pocket on BCL-XL is predicted to be ∼2.5 nm above the mitochondrial membrane, a membrane-spanning BIK–BCL-XL interaction (Fig. 3D) could engage at close ER–mito contact sites (<15 nm). The N-terminal fusions of donor and acceptor fluorophores to BIK and mitochondria-targeted BCL-XL/BCL-2 likely project further into the space between MAM and MOMs, making FRET (occurs within 10 nm) more favorable.
Deletion of the region between the BH3 and MBR of BIK has a minor impact on binding to mitochondria-anchored BCL-XL but reduces BIK proapoptotic activity in HCT116 and MCF-7 cells
Based on our structural model (iii) in Figure 3D, we hypothesized that the 65 amino acid length may allow the BH3 of BIK to extend ∼14 nm beyond the MAM surface to bind CBCL-XL-ActA at mitochondria. To test this experimentally, we generated mutant “VBIK-Δ72 to 136” where the 65 amino acids between the BH3 and the MBR in the coding region of BIK were replaced with a shorter (six amino acid) linker (see diagram in Fig. 4A). Binding of VBIK-Δ72 to 136 was assessed by FLIM–FRET with donor, CBCL-XL-ActA, expressed in BMK-DKO cells (Figs. 4B and S4A). Surprisingly, VBIK-Δ72 to 136 still binds to CBCL-XL-ActA but with somewhat reduced apparent relative affinity (Fig. 4B, compare VBIK-Δ72–136 to VBIK).
Figure 4.
Deletion of the region between the BH3 and the MBR in BIK marginally reduced the affinity of BIK binding to mitochondria-associated BCL-XL but greatly reduced proapoptotic activity.A, stick diagrams of VBIK and VBIK-Δ72 to 136’. B, FLIM–FRET binding curves for CBCL-XL-ActA expressed in BMK-DKO cells binding to the indicated acceptor proteins expressed by transient transfection. Acceptor:donor intensity ratio is used to represent the relative concentrations of acceptor to donor in cells, expressed in arbitrary units of intensity. Each curve represents binned data from thousands of subcellular regions of interest. The median FRET efficiency and standard error for each binned point were plotted from a single experiment. Data for biological replicates (n = 3) are included in Fig. S4A. VBIK-Δ72 to 136 binds with lower relative affinity than VBIK to CBCL-XL-ActA. C, example images of each Venus-fusion protein (Venus channel) labeled diagonally above, expressed in HCT116 (left set of images) or HCT116-DKO (right set of images). Cells were also stained with TMRE, AnnexinV, and DRAQ5 (nucleus). Each channel is labeled on the left and given a color that was used in generating the corresponding merged colored image below each column. Low intensities of BIK were sufficient to kill HCT116 cells. Both Venus and AnnexinV channel images were displayed on a log scale for visibility. Contrast/brightness adjustments were applied equally across all images for each channel. Original images (650 × 650 μm) were cropped (85 × 85 μm) to highlight single cells. Scale bar on top left image applies to all images. See Fig. S4B for additional Venus-fusion proteins tested in this assay. D, compared with VBIK, VBIK-Δ72 to 136 was remarkably less potent at killing HCT116 and MCF-7 cells. Cell death was quantified in Venus-positive cells based on AnnexinV intensity, nuclear shrinkage (SN [small nuclei]), and TMRE intensity using a linear classifier. The Venus-fusion proteins transiently expressed in cells are indicated in the legend in B and apply to all cell lines (indicated above the panels). The average intensity of Venus per cell was used as a measure of BIK and BIK mutant protein expression. For each replicate, data were binned by Venus intensity, and median cell death was quantified per bin. The mean and standard error of three biological replicates are displayed (symbols). Depending on the shape, data were fit to either a straight line or a sigmoidal in GraphPad Prism for visualization purposes. BH, BCL-2 homology; BIK, BCL-2 interacting killer; BMK, baby mouse kidney; DKO, double KO; FLIM, fluorescence lifetime imaging microscopy; MBR, membrane-binding region; TMRE, tetramethylrhodamine ethyl ester.
We also assessed the functional activity of VBIK-Δ72 to 136, in multiple cell lines: HCT116, HCT116 lacking BAX and BAK (HCT116-DKO) (Fig. 4, C and D), and MCF-7, and MCF-7 cells stably expressing CBCL-XL-ActA (Fig. 4D). Cell death was quantified for the indicated mutants in Figure 4D and additional mutants of BIK and BIM in Fig. S4, B and C. Residual proapoptotic activity of nonbinding BH3 mutants, VBIK-L61G and VBIK-4E, was observed in MCF-7 but not HCT-116 cells (Fig. 2). Reduced proapoptotic activity for VBIK-Δ72 to 136 in MCF-7 and HCT116 cells (Fig. 4D) is not because of protein aggregation (Fig. 4C) but did correlate with a reduction in binding (compare Fig. 4, B and D). Expression of VBIK-Δ72 to 136 did not kill HCT116-DKO, demonstrating BAX/BAK dependency of cell death. Moreover, stable expression of mitochondria-anchored CBCL-XL-ActA in MCF-7 cells had a protective effect against the expression of VBIK and VBIK-Δ72 to 136 (compare the slope of each curve in MCF-7 to MCF-7 CBCL-XL-ActA cells).
For an unknown reason, while expression of full-length VBIK eventually overcomes the protection afforded by expression of wildtype CBCL-XL (Fig. 2G), this was not observed in MCF-7 cells expressing mitochondria-anchored CBCL-XL-ActA (Fig. 4D). The same effect is observed for all full-length mutants of BIK in MCF-7-CBCL-XL-ActA cells (Fig. S4). On the other hand, expression of VBIML (BH3 activator), VBIML-ΔMBR (BH3 sensitizer), and BIK mutant lacking its MBR (VBIK–ΔMBR) killed MCF-7-CBCL-XL-ActA cells equally (Figs. 4D and S4). FLIM–FRET data consistently indicate that VBIK–ΔMBR has a slightly higher relative affinity for CBCL-XL (Fig. 2C) and CBCL-XL-ActA (Fig. S4A) than VBIK. Together, these results suggest that the enhanced proapoptotic activity of VBIK–ΔMBR compared with full-length VBIK in MCF-7-CBCL-XL-ActA cells is not likely related to potential BAX/BAK activator activity or to its enhanced affinity for CBCL-XL-ActA but rather related to the difference in localization and/or freedom of this mutant to move within cells (assessed later). Overall, the VBIK-Δ72 to 136 mutation had less effect than a BH3 mutation (L61G or 4E mutant) on binding to CBCL-XL-ActA (Fig. 4B); however, a shift in the binding curve was detected. Though these results indicate that the region between the BH3 and MBR is not required for BIK to interact with mitochondria-anchored BCL-XL-ActA, the combined FLIM–FRET and cell death results suggest that the VBIK-Δ72 to 136 deletion impairs binding, which translated to impaired efficacy. While this is not proof that “spanning the gap” is the mechanism of binding (Fig. 3D model [iii]), these results are still consistent with this model.
The observed binding between VBIK-Δ72 to 136 and CBCL-XL-ActA suggests that binding occurs in regions where the distance is less than the average reported distance in the literature between 10 and 30 nm (Naon, 2014, Rizzuto, 2009, and Vance, 1990). At present, FLIM–FRET lacks the resolving power to address these distances directly. However, contacts closer than 10 nm have been reported. For example, in RBL-2H3 cells, the average distance at close contact sites was 6 ± 1 nm (79). Closer contacts have also been reported in cells under stress (45, 80, 81) and demonstrated that expression of BIK, but not BIK-L61G, reduced the average ER–mitochondrial distance from ∼35 to ∼20 nm and increased the frequency of close ER–mito contact sites (defined as <15 nm) from 16 to 35% of the total ER surface area (45). If the interaction between the BH3 of BIK and mitochondria-associated BCL-XL further promotes such tight junctions, then a membrane-spanning interaction may still explain the interaction with VBIK-Δ72 to 136. As shown in our model, BCL-XL projects the BH3-binding site ∼2.5 nm into the space, and the six amino acid flexible linker in the VBIK-Δ72 to 136 construct can be expected to project the BH3 ∼2 nm (3.5 Å × 6 = 21 Å = 2.1 nm), which may be sufficient for binding between VBIK-Δ72 to 136 and CBCL-XL-ActA at ER–mito contact sites but would require the distance to be less than <6 nm. Thus, the simpler explanation is that VBIK and/or CBCL-XL-ActA relocalize in response to the coexpression of both proteins in cells (Fig. 2D, models [i] and [ii]).
The binding of BIK to antiapoptotic proteins occurs in mitochondrial regions of the cell
The FLIM–FRET data displayed in Figures 1 and 2 were acquired on our ISS-Alba platform, which uses nearest neighbor pixel binning to obtain sufficient counts for lifetime fitting at the expense of spatial resolution. As the binned area is a square, the averaged areas further degrade biological resolution by including pixels from both cytoplasm and membranes as well as neighboring structures in cells that may not be connected. To improve spatial resolution and determine whether the binding between BIK and mitochondria-localized BCL-XL and BCL-2 mutants (CBCL-XL-ActA and CBCL-2-ActA) is occurring at mitochondria, we performed structure-based segmentation and binning of pixels within mitochondrial ROIs before lifetime analysis (Fig. 5). For these experiments, FLIM–FRET data were collected on a custom FLIM–hyperspectral (FHS) instrument (the INO-FHS) that enables the simultaneous collection of FLIM images of the donor (mCerulean3), and intensity images of the acceptor (Venus) and MitoRed stain (52, 82) minimizing the effects of protein movement in live cells. In cells, CBCL-XL and CBCL-2 can be located at the ER and/or mitochondria, and CBCL-XL is also found in the cytoplasm; therefore, images segmented based on the intensities in the mCerulean3 channel (in Fig. 5A, called “donor image segmentation”) will contain data based on the localization of the antiapoptotic proteins irrespective of location. Mitochondrial staining with MitoRed enabled segmentation and pixel binning at mitochondria (in Fig. 5A, called “MitoRed image segmentation”) to determine if the interaction of VBIK with CBCL-XL and CBCL-2 occurs at mitochondria without using antiapoptotic mutants with foreign tail-anchor sequences. If there is a large difference in affinity of the interaction at mitochondria, then the shapes of the binding curves that result from donor image segmentation are expected to be different than the binding curves obtained by donor image segmentation, using the total mCerulean3-positive areas. However, the resulting binding curves displayed below the corresponding segmentation maps strongly suggest that in BMK-DKO cells, the binding interactions in both regions of the cell are similar (Fig. 5B). This result is consistent with ER-localized BIK binding to mitochondria-localized BCL-XL and BCL-2 with similar affinities as binding to BCL-XL and BCL-2 in other regions of the cell. This suggests that there are authentic complexes of the proteins at or in close proximity to mitochondria and at other cellular locations. In control experiments, using either ROI segmentation approach, only collisions (straight lines) were detected in images of cells expressing VBIK-4E with CBCL-XL or CBCL-2, suggesting that our measurements for VBIK represent BH3-dependent binding interactions (Fig. 5B).
Figure 5.
BIK binds BCL-XL and BCL-2 at ROIs corresponding to mitochondria in live cells.A, images of mCerulean3 and MitoRed in BMK-DKO cells expressing donor CBCL-2 or CBCL-XL (indicated above), with corresponding segmentation maps selected based on each channel below. B, FLIM–FRET binding curves were generated from data obtained from several images of BMK-DKO cells analyzed using the two ROI segmentation approaches. FLIM–FRET binding curves are shown below the corresponding segmentation schemes for CBCL-2 (left) and CBCL-XL (right) (n = 3). Positive and negative controls for binding in this experiment are included in Fig. S5. BIK, BCL-2 interacting killer; BMK, baby mouse kidney; DKO, double KO; FLIM, fluorescence lifetime imaging microscopy; MitoRed, MitoTracker Red; ROI, region of interest.
As controls for these FLIM–FRET binding curves, we used VtBID, a protein known to interact with CBCL-XL and CBCL-2 at mitochondria (3) and VCb5 and VActA as negative controls for collisions at the ER and mitochondria, respectively (Fig. S5). As expected, binding curves generated with either segmentation approach were very similar for VtBID as complexes are found overwhelmingly localized at mitochondria. Furthermore, the frequency of collisions with the constitutively membrane-bound BCL-2 were more frequent than for BCL-XL for both VCb5 and VActA, as expected (Fig. S5). Although we cannot establish subcellular localization because of the limited spatial resolution of FLIM, for FRET to be detected VBIK must be within 10 nm of CBCL-XL and CBCL-2. Thus, it appears that coexpression of the proteins results in relocalization of sufficient BIK close enough to mitochondria to bind to the mitochondrial localized antiapoptotic proteins.
In BMK cells lacking BAX and BAK, a fraction of VBIK relocalizes to and binds mitochondria-localized CBCL-XL-ActA
The subcellular localization of BCL-2 family proteins is primarily determined by the MBR (12, 83, 84). While the “ActA” and “Cb5” MBR localization sequences have well-established specificities for mitochondria and ER, respectively (47, 48, 49, 50), less is known about the localization of BIK in cells expressing interacting BCL-2 family proteins with different localizations. To test in BMK-DKO cells whether the localization of transiently expressed VBIK is different in cells stably expressing CBCL-XL, CBCL-XL-ActA, or CBCL-XL-Cb5, the cells were stained with MitoRed, and colocalization was calculated for images of three channels: mCerulean3 expression in the “blue channel” reports localization of the different BCL-XL constructs; Venus expression in the “green channel” reports localization of VBIK; and MitoRed staining in the “red channel” identifies mitochondria.
Correlation between green:red (VBIK:MitoRed), blue:green (CBCL-XL, CBCL-XL-ActA, or CBCL-XL-Cb5:VBIK, and blue:red (CBCL-XL, CBCL-XL-ActA, or CBCL-XL-Cb5: MitoRed) channels was assessed using Pearson’s r values (analysis summarized in Fig. S6). Colocalization of VBIK and VBIK-Cb5 with mitochondria (green:red channel correlation) was higher (0.34) in cells expressing CBCL-XL or CBCL-XL-ActA compared with cells expressing CBCL-XL-Cb5 (0.16). This change in colocalization was not seen for VBIK-L61G, demonstrating that binding between the proteins was required (Fig. 6A, top heatmap). Furthermore, the expression of CBCL-XL-Cb5 did not lead to an increase in colocalization of VBIK or VBIK-Cb5 with MitoRed. The increase in colocalization of VBIK and VBIK-Cb5 compared with VBIK-L61G with CBCL-XL or CBCL-XL-ActA (blue:green) was even more pronounced (Fig. 6A, middle heatmap), suggesting that BH3-dependent binding to BIK may also occur at nonmitochondrial locations in the cell, depending on the localization of the antiapoptotic proteins. In contrast, there was no change in correlation between CBCL-XL or CBCL-XL-ActA with MitoRed (blue:red) because of the expression of VBIK, suggesting the extent to which CBCL-XL and CBCL-XL-ActA localized at the MOM is unchanged (Fig. 6A, bottom heatmap). The simplest explanation for these data is that a population of VBIK and VBIK-Cb5 localized closer to mitochondria when cells stably expressed the binding partner: CBCL-XL or CBCL-XL-ActA. Nevertheless, the amount of VBIK and VBIK-Cb5 located near mitochondria in cells expressing CBCL-XL or CBCL-XL-ActA was much less than was observed for VActA localizing with mitochondria suggesting only a subpopulation of VBIK and VBIK-Cb5 relocalized.
Figure 6.
In BMK-DKO cells, a fraction of BIK relocalizes in response to binding BCL-XL.A, results of “three-way” (green:red, blue:green, and blue:red) Pearson’s correlation analyses (Fig. S6) displayed in heatmap format, where the mean Pearson’s r value from three biological replicates to the nearest two decimals is shown in each square and represented on the color scale from −0.2 to 1 shown at the right. BMK-DKO cells stably expressing CBCL-XL (n = 2), CBCL-XL-ActA (n > 3), or CBCL-XL-Cb5 (n > 3) (blue channel listed left of each row) were transfected with Venus-fusion proteins (green channel listed above each column) and then stained with MitoRed (red channel). Colocalization was measured only in cells positive for Venus expression. To the right of each heatmap, the two colored squares indicate which channels were analyzed. Note for examining the bottom heatmap, the correlation is between the stably expressed mCerulean3-fused protein (blue channel) and MitoRed (red channel) in cells expressing the protein in the green channel listed above. These results indicate that in cells expressing wildtype CBCL-XL or mitochondria-associated CBCL-XL-ActA but not ER-associated CBCL-XL-Cb5, BIK moves closer to mitochondria. B, model summarizing how BIK may bind mitochondria-associated CBCL-XL in BMK-DKO cells?: Unknowns. Results in A suggest BIK moves toward mitochondria in BMK-DKO cells to bind BCL-XL at mitochondria, which may be explained by redistribution of BIK to MAMs or by movement of BIK out of the ER and to mitochondria. BIK, BCL-2 interacting killer; BMK, baby mouse kidney; DKO, double KO; ER, endoplasmic reticulum; MAM, mitochondria-associated ER membrane; MitoRed, MitoTracker Red.
One explanation for these results is that VBIK and VBIK-Cb5 relocalized to mitochondria in cells expressing CBCL-XL or CBCL-XL-ActA (Fig. 6B). The measured correlation between VBIK/VBIK-Cb5 and MitoRed in BMK-DKO cells expressing CBCL-XL or CBCL-XL-ActA was ∼0.3, compared with a Pearson’s r of 0.7 for VActA, suggesting that only a fraction of VBIK relocalized from the ER to mitochondria (Fig. 6A, top heatmap). To ensure saturation in the FLIM–FRET binding curves, VBIK was expressed at much higher levels than CBCL-XL or CBCL-XL-ActA. Thus, relocalization of a fraction of the VBIK may have been sufficient to saturate binding to the antiapoptotic proteins at the mitochondria.
The MBR of BIK is highly hydrophobic and spans the ER membrane (37). Hence, we anticipated that after synthesis, soluble BIK would rapidly partition into the ER membrane facilitated by the proteins of the GET family responsible for localizing tail-anchor proteins at the ER (reviewed by Ref. (85)). Once embedded in the membrane, BIK is unlikely to “jump” from the ER to mitochondria. Instead, it seems likely that VBIK binds to BCL-XL at sites of close contact between the ER and mitochondria. Consistent with this view, purified light membrane fractions containing BIK induced cytochrome C release from isolated mitochondria, without any BIK relocalization detected at mitochondria (37). Moreover, BIK was recently shown to be localized to the MAM, where it was suggested to form a complex with ER-localized BAK, DAPk1, ERK1/2, and an unknown mitochondrial protein. This complex was proposed to span the distance between the ER/MAMs and mitochondria (45). An alternate possible explanation is that a fraction of BIK located in MAMs could participate in binding to BCL-XL or BCL-2 at mitochondria (Fig. 6B). Transit of BIK from ER to MAMs could explain both the FLIM–FRET and aforementioned relocalization data, as MAMs would be expected to partially colocalize with mitochondria (45, 86). To examine these possibilities in another context, we used human MCF-7 cells that contain endogenous BIK as well as BAX and BAK to examine the binding of CBCL-2 and CBCL-XL to VBIK as well as the relocalization of VBIK and CBCL-XL.
As expected from the results with BMK cells, FLIM–FRET measurements revealed that in MCF-7 cells, CBCL-XL bound to VBIK, VBIK-AA, and VBIK-DD (Fig. S2C) and CBCL-2 bound to VBIK, VBIK-AA, VBIK-DD, VBIK-GG153AA, and VBIK-GG154STOP (Fig. S2E). Moreover, CBCL-XL-ActA and CBCL-2-ActA bound directly to VBIK but not VBIK-L61G (Fig. S2, D and F, respectively). Thus, in MCF-7 cells, we again observed BH3-dependent binding of proteins located at different subcellular locations; mitochondrial localized antiapoptotic proteins binding to the ER localized the proapoptotic BH3-protein BIK.
In MCF-7 cells, a fraction of CBCL-XL moves away from mitochondria and toward the MAMs/ER in response to BH3-dependent binding to VBIK
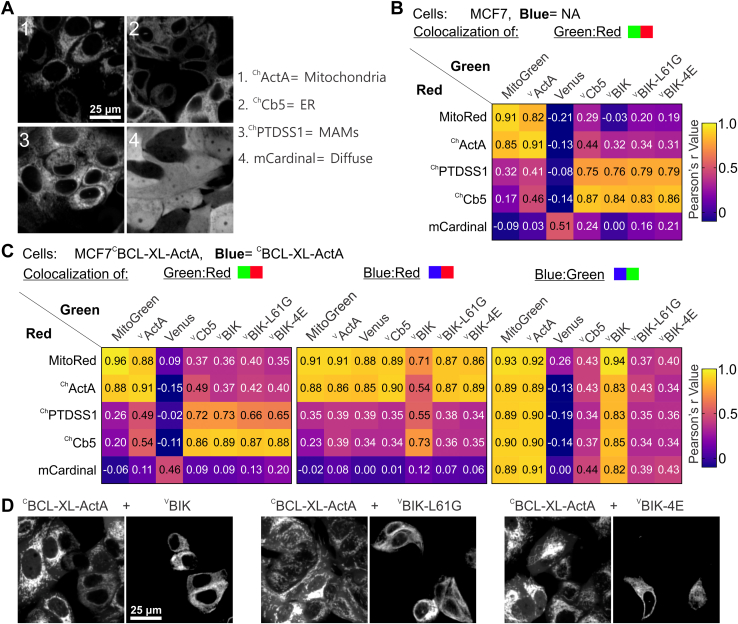
To evaluate the hypothesis that VBIK moves to MAM/mitochondria in response to the expression of its binding partner at mitochondria, mCherry-fused landmarks for MAM and other subcellular membranes were expressed individually in cells coexpressing VBIK and CBCL-XL-ActA to enable a more in-depth analysis of localization. As lentiviral infection of BMK cells was inefficient, MCF-7 and MCF-7-CBCL-XL-ActA cell lines were used (Fig. 7A). Thus, mCherry fusions to the Cb5 and ActA localization sequences, ChCb5 and ChActA, identified ER and mitochondria, respectively. In addition, mCherry fused to full-length phosphatidylserine synthase-1 (ChPTDSS1), a protein that participates in phosphatidylserine biosynthesis and is highly enriched in MAMs in mammalian cells, was used as a landmark for MAM (39, 87). Expression of the red fluorescent protein, “mCardinal” was used as a landmark for “diffuse” expression throughout the cell. As positive controls for Pearson’s r correlation, mitochondria in parental MCF-7 and MCF-7-CBCL-XL-ActA cells were stained with either MitoRed or MitoGreen. The different Venus-fusion proteins were expressed by transient transfection (Table S2), and a pan-caspase inhibitor was added to prevent expression of VBIK killing the MCF-7 cell lines (Fig. 2G). Given the previous results in BMK-DKOs (Fig. 6), we hypothesized that coexpression would result in movement of a fraction of BIK but not VBIK-L61G or VBIK-4E to mitochondria or MAMs. To quantify the effect of CBCL-XL-ActA expression on VBIK colocalization with landmarks in cells, and vice versa, three-way Pearson’s correlation was determined as aforementioned (Figs. 6 and S6) but now for Cerulean and Venus (blue:green), Cerulean and mCherry (blue:red), and Venus and mCherry (green:red) fusion proteins. To enable visual comparison of a large number of correlations, the three-way colocalization results are displayed in heatmaps where higher Pearson’s correlation is represented by a shift from purple to yellow in Figure 7, B and C.
Figure 7.
Localization ofCBCL-XL-ActA changes as a BH3-dependent response toVBIK expression in live MCF-7 cells.A, example images of each mCherry-fusion protein landmark (red channel) expressed in MCF-7 cells. Numbered images (1–4) are labeled to the right, with the name of the expressed protein: ChActA, ChCb5, ChPTDSS1, or mCardinal and expected localization. Original images (320 × 320 μm) were cropped (85 × 85 μm) to highlight localization. Scale bar on (1) applies to all images. Cells were transiently transfected with constructs to express the Venus-fusion proteins indicated in B (green channel). B and C, results of Pearson’s correlation analyses (Fig. S6) are displayed in heatmap format where the mean Pearson’s r value from three biological replicates is displayed within each square to the nearest two decimals and is represented on the color scale from −0.2 to 1 shown at the right. Landmarks are listed next to each row (red channel), and Venus-fusion proteins are above each column (green channel). Above each plot, the two colored squares indicate which channels were analyzed. B, green:red colocalization in parental MCF-7 cells that do not express a cerulean fusion protein (blue channel, not applicable [NA]). C, three-way colocalization in MCF-7 cells expressing CBCL-XL-ActA (blue). Note for examining the center heatmap, the correlation is between the stably expressed mCherry landmarks (red) with CBCL-XL-ActA (blue) in cells expressing the protein in the green channel listed above. These results indicate that only in cells expressing VBIK does CBCL-XL-ActA move toward the ER and MAM, and away from mitochondria. D, for data quantified in (C), example images (grouped: mCerulean3 and Venus channel left and right, respectively) of MCF-7 cells expressing CBCL-XL-ActA with VBIK (left), or VBIK-L61G (middle), or VBIK-4E (right). For each set of images, compare CBCL-XL-ActA localization in cells that are transfected with VBH3 (Venus channel) to CBCL-XL-ActA localization in untransfected cells. BH, BCL-2 homology; BIK, BCL-2 interacting killer; ER, endoplasmic reticulum; MAM, mitochondria-associated ER membrane.
As expected, in both MCF-7 and MCF-7-CBCL-XL-ActA cells, high green:red channel correlation (>0.8) was observed for the mitochondria landmarks (ChActA and MitoRed) with the positive controls for mitochondrial localization: VActA & MitoGreen; and for the ER localization landmark (ChCb5) with the positive control for ER localization (VCb5). In MCF-7 cells, the correlation between the MAM landmark (ChPTDSS1) and VCb5 (∼0.75) was only slightly lower than between ChCb5 and VCb5 (∼0.86), and higher than between ChPTDSS1 and either mitochondrial marker (<0.5), suggesting that in these cells MAMs are closely associated the ER (Fig. 7B). The measured correlation of VBIK, VBIK-L61G, and VBIK-4E with all landmarks was similar to those for VCb5 with the same landmarks suggesting that as expected, in cells not expressing BCL-XL-ActA that VBIK is predominantly localized to ER and/or MAMs but not to mitochondria.
Data acquired in MCF-7 CBCL-XL-ActA cells are presented in three heatmaps, one for each of the green:red, blue:red, and blue:green correlations (Fig. 7C). In contrast to our findings in BMK cells in MCF-7 cells, expression of CBCL-XL-ActA did not change the localization of VBIK as its correlation with all landmarks remained similar to those for VBIK-L61G and VBIK-4E and VCb5 (Fig. 7C, left panel). Moreover, unlike the results in BMK cells (Fig. 6B, green:red correlation), expression of CBCL-XL-ActA did not result in an increase in correlation for VBIK with the mitochondrial landmarks MitoRed or ChActA (Fig. 7C). Nevertheless, in MCF-7 cells, there was a high correlation in localization for VBIK and CBCL-XL-ActA (Fig. 7C, left panel) consistent with the two proteins binding to each other as demonstrated by FLIM–FRET (Fig. S2D). Higher colocalization of VBIK than VCb5 with ChPTDSS1 was not observed, a result difficult to interpret in terms of potential enrichment of VBIK at MAMs (Fig. 7, B and C, compare green:red colocalization) as the analysis did not differentiate ER and MAMs.
Unexpectedly, VBIK expression in MCF-7CBCL-XL-ActA cells resulted in an increase in correlation between CBCL-XL-ActA with both ChPTDSS1 and ChCb5 (Fig. 7C, compare blue:red colocalization). Furthermore, VBIK expression led to a corresponding decrease of about ∼0.2 in the correlation between CBCL-XL-ActA and the mitochondrial landmarks (MitoRed and ChActA) (Fig. 7C, compare blue:red colocalization). Taken together, the simplest explanation for the data is that a fraction of CBCL-XL-ActA changed localization from the mitochondria to the ER or MAMs. Nevertheless, CBCL-XL-ActA remained well correlated with mitochondria landmarks (0.71 and 0.54 for MitoRed and ChActA, respectively), suggesting that only a fraction of the CBCL-XL-ActA moved away from mitochondria. Consistent with the changes observed being because of BH3-dependent BCL-XL-ActA binding to BIK, expression of VBIK-L61G or VBIK-4E did not change the correlation between CBCL-XL-ActA and red channel landmarks (blue:red). Thus, the blue:red colocalization data indicate that a fraction of CBCL-XL-ActA moves away from a mitochondrial localization toward that of the MAM and/or ER when its binding partner VBIK is expressed in MCF-7 cells. Example images showing this change in blue channel localization in cells expressing VBIK are shown in Figure 7D.
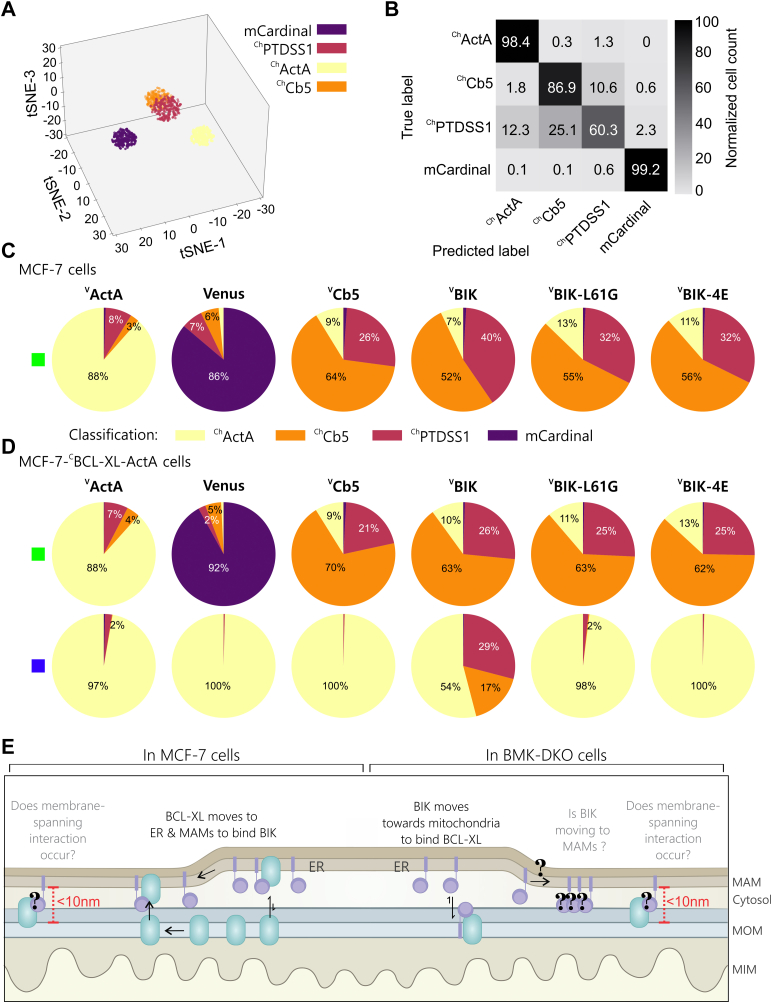
Colocalization is a single parametric measure of intensity changes in the cell and was not sufficient to resolve ER and MAMs in MCF-7 cells. Given the size of the MCF-7 cells dataset, it was possible to reanalyze the data using a Gabor-based convolutional neural network and contrastive learning to examine localization based on texture, morphology, and intensity features within the cell (see Experimental procedures section and Fig. S7, A and B). To identify features characteristic of the subcellular localization of the landmarks (ChActA, ChCb5, ChPTDSS1, and mCardinal), 116,651 single-cell images were used for contrastive learning. A t-distributed stochastic neighbor embedding plot in Figure 8A (see Fig. S7C for raw data) was used to reduce the high-dimensional feature space and permit visualization of the extent to which the learned features separated the landmarks. While the landmarks mCardinal and ChActA were clearly separated from each other and the other two landmarks, there was some overlap of ChCb5 and ChPTDSS1 within the t-distributed stochastic neighbor embedding landscape. In this representation, the MAM landmark (ChPTDSS1) fell between the ER (ChCb5) and mitochondria (ChActA) landmarks, consistent with the known biology of MAMs. Thus, the learned features were used to train a classifier to output normalized probabilities of the likelihood that a given cell belongs to each landmark. The landmark with the highest probability was then chosen as the class label for that cell. Thus, the classification accuracy does not reflect the confidence of the classifier in determining the protein distribution but rather reflects the fraction of instances that the highest probability was assigned to a particular landmark. The accuracy of the assigned classification was tested using 26,560 cell images that were not used in training to generate a normalized confusion matrix (Figs. 8B, see S7D for cell counts). The overall accuracy of the classifier was 88%. As expected from the known localization and functions of MAMs, there was some misclassification for the MAM landmark (ChPTDSS1) as either the ER (∼25% ChCb5) or mitochondria (∼12% ChActA) landmarks (Fig. 8B). Nevertheless, MAMs were better separated from ER than we observed using Pearson’s correlation. The trained classifier was then used to examine the localization of proteins expressed in MCF-7 and MCF-7CBCL-XL-ActA cells in the green, red, and blue channels with the results displayed in pie chart format (Figs. 8, C and D, see S7E for example of single-cell image data, and Fig. S7, F and G for the predicted and ground truth classification of red channel data).
Figure 8.
In MCF-7 cells, exogenous expression of BIK results in relocalization of a fraction of BCL-XL from mitochondria to mitochondria-associated ER membranes and the ER.A, visualization of multidimensional red channel MCF-7 image data after dimension reduction using the t-SNE algorithm. Each point represents 1 of the 3000 cells randomly selected from the 143,211 total cells for each of the four landmarks (listed in legend). For visualization, the 300 cells closest to the centroid were displayed. B, confusion matrix after classification of MCF-7 cell mCherry landmark images indicating the accuracy of the classifier trained on learned representations. Data were normalized to the total number of cells from the true label of each landmark and represented numerically within the grid and in grayscale (right). C and D,CBCL-XL-ActA moves toward the ER and MAM, and away from mitochondria in cells expressing VBIK. C, MCF-7 and (D) MCF-7-CBCL-XL-ActA single-cell image data from three biological replicates were combined and classified. The Venus-fusion protein expressed is indicated above in bold. The channel analyzed is indicated by the colored square to the left of each row of pie charts: Venus, green; mCerulean3, blue. Corresponding red channel data (mCherry) are included in Fig. S7F. Classification results for each channel are shown as pie charts, colored according to the landmark assignments (see legend). Images of more than 20,000 transfected cells were analyzed for each pie chart. There is no protein expressed in the blue channel for MCF-7 parental cells. Localization effects because of the expression of the indicated Venus fusion proteins can be appreciated by comparing the blue channel row-wise. The green channel rows illustrate the localization of the different Venus fusion proteins in (C) MCF-7 and (D) MCF-7-CBCL-XL-ActA cells. Blue channel,CBCL-XL-ActA, classification changed only in cells expressing VBIK. See associated analysis, supporting data, and additional BIK mutants in Figs. S7 and S8. E, Illustration of potential localizations and interactions for BIK (purple) and BCL-XL (green).
As expected, the learned representations were not channel specific, as control Venus-fusion transfectants were assigned to their expected landmarks: for example, VActA, Venus, and VCb5 were classified 88%, 86%, and 64% as ChActA, cardinal, and ChCb5, respectively, in MCF-7 cells; and similarly, 88%, 92%, and 70% as ChActA, cardinal and ChCb5, respectively, in MCF-7-CBCL-XL-ActA cells (Fig. 8, C and D, green channel). In the Pearson’s correlation analysis (Figs. 7 and S6), we assumed that the expression and localization of landmarks (red channel) were unaffected by transient transfection (green channel) or stable expression of mCerulean3-fusion proteins (blue channel). Here, we demonstrate that the frequency distribution of assigned landmark classification (red channel) is unchanged across all transfectants in MCF-7 and MCF-7-CBCL-XL-ActA cells (Fig. 8, C and D, red channel). The proportion changed by no more than 4% for any of the landmarks across all the experiments, thus we conclude that the expression of the Venus-fusion proteins did not affect either the localization of the landmarks or classification accuracy. Corroborating this conclusion, the frequency distribution of the known localizations of the red channel landmarks (ground truth) was unchanged across transfectants in both MCF-7 and MCF-7-CBCL-XL-ActA cells (compare Fig. S7, F and G for frequency distribution). Moreover, as expected from the confusion matrix (Fig. 8B), the MAM landmark was under-represented in the predicted red channel classification (Fig. S7F) compared with the red channel ground truth (Fig. S7G). Thus, the classification algorithm consistently underestimated the MAM fraction and overestimated the fractions at the mitochondria and ER. For this experiment, we asked whether the expression of VBIK and CBCL-XL-ActA together in MCF-7 cells caused any relative change in localization of the green channel (Fig. 8, C and D, compare distribution in MCF-7 to MCF-7-CBCL-XL-ActA cells) or blue channel (Fig. 8D, compare across transfectants for MCF-7-CBCL-XL-ActA cells). Though it does underestimate the fraction in the MAM, the algorithm is consistent in its classification (Fig. 8, C and D, red channel); therefore, relative changes >8% (twice the deviation in the red channel) in this distribution for the green and blue channels are meaningful.
As expected, in most cases, >90% of the blue channel images of cells expressing CBCL-XL-ActA were assigned to the mitochondria class. However, when VBIK was expressed in the cells, CBCL-XL-ActA was classified as ∼54% mitochondria, 29% MAM, and 17% ER (Fig. 8D, blue channel). This change clearly demonstrates that VBIK expression changed the localization of CBCL-XL-ActA. Furthermore, the data underestimate the extent of relocalization because for the classification to change, the subcellular features must have changed sufficiently that the highest probability class for the cell image changed. That there were no changes in localization detected for these cells in the red channel indicates that the difference in CBCL-XL-ActA (blue channel) classification upon VBIK expression was not because of morphology changes in caspase inhibitor–treated but still dying cells as such changes in morphology would be expected to be detected in the red channel as well. Finally, the expression of VBIK-L61G and VBIK-4E did not induce the relocalization of CBCL-XL-ActA. Thus, relocalization of CBCL-XL-ActA toward the MAM and ER appears to be due to BH3-dependent binding to VBIK.
To test if movement of BCL-XL-ActA to ER/MAMs in response to BIK expression is dependent on the membrane localization of BIK, we repeated the experiment analyzed in Figures 7 and 8 with mutant VBIK-ΔMBR (Fig. S8). When VBIK-ΔMBR was expressed in MCF-7 cells, it was mostly expressed diffusely throughout the cell (classified as 81% Venus, 14% ER, and 4% MAMs), as expected. In contrast, when VBIK-ΔMBR was expressed in MCF-7 cells stably expressing CBCL-XL-ActA, the truncated VBIK-ΔMBR protein partially relocalized to mitochondria, MAM, and ER membrane (classified as 24% Venus, 26% ER, 23% MAMs, and 27% mitochondria). As expected, we again observed a change in the localization of CBCL-XL-ActA (blue channel classification), in cells transiently expressing VBIK and VBIK-Δ72 to 136, which both contain an intact BH3 and MBR, but not VBIK-L61G or VBIK-4E. As supporting evidence for Figure 4, mutant VBIK-Δ72 to 136 had a similar subcellular distribution as wildtype BIK (Fig. S8, compare green channel pie charts) and relocalized CBCL-XL-ActA to ER/MAMs in MCF-7 cells (blue channel), suggesting that observed binding to CBCL-XL-ActA and reducted proapoptotic activity of this mutant is not because of mistargeting of VBIK-Δ72 to 136 in cells. Consistent with the MBR directing the localization of CBCL-XL-ActA to MAMs/ER in response to binding BIK, we did not observe the change in classification of the blue channel in cells expressing VBIK-ΔMBR (Fig. S8).
Consistent with the data obtained using Pearson’s correlation in MCF-7 cells (Fig. 7), there was no increase in the assignment of VBIK or the BH3 mutants, VBIK-L61G and VBIK-4E, to mitochondria or MAM landmarks in cells expressing CBCL-XL-ActA (Fig. 8, C and D, green channel, compare MCF-7 to MCF-7CBCL-XL-ActA). Taken together, the results in Figures 7 and 8 demonstrate that CBCL-XL-ActA changed localization in response to VBIK expression in MCF-7 cells. Furthermore, the movement of BCL-XL depended on the affinity of the BH3 region of BIK to bind BCL-XL and localization to ER/MAMs by the MBR of BIK (Fig. S8). In contrast to observations in BMK-DKO cells (Fig. 6), the expression of CBCL-XL-ActA did not have a measurable effect on the localization of the Venus-fused BIK. Though there were differences in which protein relocalized in the two cell lines studied, in each case, VBIK and CBCL-XL-ActA ended up with increasingly overlapped distributions consistent with the direct binding observed by FLIM–FRET (Figure 1, Figure 2, Figure 4 and 8E).
Conclusions
As a BH3-only sensitizer protein (4), BIK cannot activate BAX or BAK directly and instead promotes apoptosis by directly binding to and inhibiting antiapoptotic proteins, including BCL-2 and BCL-XL. While the importance of BH3 proteins activating BAX and BAK continues to be debated (8, 17); B (4, 5, 7, 18, 20) the current consensus is that BIK functions as a sensitizer. Therefore, here we examined the interactions of BIK with antiapoptotic proteins and the ensuing changes in localization and function in cells. BIK is unique among BH3-only proteins, as it contains a canonical transmembrane tail anchor (MBR) that localizes BIK to the ER in cells (37, 38, 40, 41) where it promotes apoptosis without binding to mitochondrial membranes (37). Further, it has been proposed that BIK promotes apoptosis from its position at the ER by binding to ER-localized antiapoptotic proteins, thereby promoting apoptosis at mitochondria via regulation of ER to mitochondria calcium signaling (37, 38, 43, 44) (referred to here as the “from a distance” model of BIK function). By analyzing both direct binding interactions using FLIM–FRET and subcellular localization using Pearson’s correlation and neural network–based contrastive learning of image features, our data suggest that rather than acting at a distance, BIK promotes apoptosis by directly binding to mitochondria-localized BCL-2 and BCL-XL. Furthermore, it appears that the interactions between proteins believed to be located in different membranes results in part from cell type–specific relocalization of the proteins in cells expressing both binding partners and from localization at ER–mitochondrial contact sites via MAMs. According to our predicted 3D model, a direct interaction between ER-anchored BIK and mitochondria-anchored antiapoptotic protein(s) may occur at sites where the distance between the MAM/ER and MOM is <15 nm. Indeed, our demonstration that VBIK-Δ72 to 136 binds albeit with lower relative affinity than VBIK to CBCL-XL-ActA suggests that binding likely occurs at close-contact sites where the intermembrane distance is less than 10 nm. Binding to mitochondria-localized BCL-XL at such sites may promote accumulation of BIK at the MAMs in BMK-DKO cells and/or movement of mitochondria toward ER membranes to increase close-contact sites to explain our colocalization data. In some cell types, ER/MAM-anchored BIK may bind antiapoptotic BCL-XL redirecting localization to the ER. Interestingly, recent evidence in yeast suggests that TOM20 and MAMs may play an important role in mediating BCL-2 relocalization from ER to mitochondria and vice versa during apoptosis (88). Moreover, MAMs have also been reported to regulate BAX localization and function in yeast (89). It remains to be determined how these findings translate to mammalian cells, or whether the MAMs somehow facilitate relocalization of BIK to mitochondria. Regardless of the mechanism, both relocalization events require BH3-dependent binding of BIK to mitochondria-localized antiapoptotic proteins and are more consistent with a “from a close distance” model of BIK function including ER and mitochondrial membrane close contacts at MAMs.
Recently, BIK has been shown to function at MAMs in the “BDEB” complex (45), directly participating in decreasing the ER–mitochondria distance and increasing the frequency of close-contact sites. Since our FLIM–FRET binding curves were collected in BMK cells lacking BAK (a critical component of this complex), we conclude that either BIK interacts with mitochondria-targeted antiapoptotic proteins independent of the BDEB complex, or in some cells, neither BAK nor BAX are required for BDEB complex formation. Presently, the mechanism by which the BDEB complex contacts mitochondria remains obscure (45). We speculate that a BH3-dependent interaction between BIK and a mitochondria-targeted antiapoptotic protein could tether a version of a BDEB-type complex to mitochondria.
The average concentration of CBCL-2 and CBCL-XL stably expressed in our BMK-DKO specifically is less than 2 μM at membranes (Fig. 3H of Ref. (52)). Lindner et al. (90) measured endogenous expression levels of BCL-XL expressed in whole-cell lysates of 26 matching normal (1–717 nM), and tumor (0–522 nM) tissue samples, as well as several cell lines (109–994 nM) (90). The same study reported levels of BCL-2 expressed in whole-cell lysates of 26 matching normal (78–5836 nM) and tumor (31–4504 nM) tissue samples, as well as several cell lines (22–653 nM). We expect local concentrations of endogenous BCL-2/BCL-XL at membranes to be higher than those reported in whole-cell lysates; thus, low micromolar expression levels of BCL-2/BCL-XL expressed in our stable cell lines should be reasonably close to the expression at membranes in cancer cells. While there are no reports measuring endogenous levels of BIK, BH3-only proteins are often observed to be induced/or activated in response to cell death stimuli, when measured relatively by immunoblotting. For example, BIK expression is induced in response to estrogen deprivation in MCF-7 cells (91). To overcome antiapoptotic signaling in these conditions, we, therefore, expect expression levels of BIK to be in the micromolar range. However, levels of acceptor-tagged proteins required for FLIM–FRET binding curve saturation will likely be above physiological levels required to induce cell death, given positive feedback contributes to the activation and BAX/BAK pore formation (3); thus displacing only a fraction of activator/pore former proteins may be sufficient to initiate MOMP.
Here the BMK-DKO cell lines stably expressing mCerulean3-tagged BCL-XL-ActA/BCL-2-ActA mutants were gated for similar expression levels as the wildtype protein by cell sorting. Higher expression levels of each protein are expected in MCF-7 cell lines (based on observed relative intensities but were not quantified here). Nevertheless, the FLIM–FRET binding curves collected in MCF-7 cells were consistent with those acquired in BMK-DKO cells (Fig. S2). Nevertheless, it remains a potential caveat that both FLIM–FRET and localization studies required the introduction of exogenous stable and transiently expressed fluorescent-tagged proteins of interest. Whether endogenous BIK, BCL-2, and BCL-XL proteins act in the same way cannot be determined at present as the imaging techniques are not sensitive enough to yield useful information from cells in which the fluorescence proteins are knocked into the endogenous gene locus. It is unlikely that overexpression accounts for the observed “retargeting” of BIK as an in-depth localization study used the MBR of BIK as an ER landmark (39). To address the concern that forced expression of BIK may overcome the normal constraints of its natural subcellular localization, the data were divided into expression quartiles based on average pixel intensity. No significant changes in localization results from the lowest to highest quartile were detected, suggesting that exogenously expressed BIK will localize correctly irrespective of expression level.
Our analyses suggest that in BMK cells direct binding of BIK to mitochondria-localized BCL-XL is facilitated by BIK relocalization toward mitochondria, whereas in MCF-7 cells, BCL-XL relocalized to MAMs and ER. The simplest explanation for our results is that in both cases there can be an accumulation of BIK and the antiapoptotic proteins at close contact sites with BIK in MAMs and BCL-XL in both ER and mitochondria. However, the wildtype antiapoptotic proteins can be located at both mitochondrial and ER membranes; thus, direct interaction with BIK may not occur exclusively at contact sites. The binding of BIK to antiapoptotic protein is dependent on its BH3 region as a single-point mutation to the BH3 region (BIK-L61G) abolished binding (Fig. 2). Given the changes in localization that accompany binding, it was unexpected that neither localization nor the MBR of BIK was required for its proapoptotic function that requires direct binding interactions with BCL-2 and BCL-XL (Fig. 2H, compare VBIK and VBIK-ΔMBR). Surprisingly, a 65 amino acid region in BIK outside the BH3 region was required for the maximum proapoptotic activity of BIK (Fig. 4). Deletion of this region resulted in decreased affinity of BIK-Δ72 to 136 for BCL-XL, but it is unknown whether this alone can account for the reduced activity (Fig. 4). BIK-induced relocalization of mitochondria-associated BCL-XL toward ER/MAMs in MCF-7 cells depended on the BH3 region for binding BCL-XL and the MBR of BIK for directing BCL-XL to the ER/MAMs (Fig. S8). Thus, while our results demonstrate that BIK regulates apoptosis by binding to antiapoptotic proteins directly, it remains to be determined why BIK has a tail-anchor sequence that constitutively localizes the protein to the ER and MAMs. We speculate that the normally highly restricted localization of BIK in human cells may be required for an as yet undiscovered BIK function unrelated to constitutive proapoptotic activity via BH3-region binding to antiapoptotic proteins.
Experimental procedures
Constructs
All DNA constructs have been assigned an Addgene accession number (Table S1). The pEGFP-C3 plasmid backbone includes a cytomegalovirus promoter and kanamycin/neomycin resistance. The s2193 plasmid backbone contains a human ferritin heavy chain (hFerh) promoter with its 5′UTR replaced with that of elongation factor-1α to eliminate iron regulation and an internal ribosome entry site (IRES) downstream of the multicloning site allows for separate translation of the blasticidin resistance gene. The pLVX-EF1a-IRES-Puromycin (Clontech) vector backbone was used for mammalian expression of mCherry landmark fusion proteins by lentiviral infection and contains an ampicillin resistance gene. An elongation factor-1α promoter controls the expression of the gene of interest, followed by the IRES sequence and puromycin resistance gene.
The DNA sequence encoding full-length human PTDSS1 was described in our recent publication ((39), Addgene ID 134667). The human BIK sequence aligns with National Center for Biotechnology Information Gene ID: 638. In human BCL-2- and BCL-XL-coding sequences, the sequence encoding the MBR was replaced with the targeting sequence for “Cb5” (ER specific) or “ActA” (mitochondria specific) as we previously described for BCL-2 (50) and BCL-XL (59). Here, we refer to these chimeras as BCL-2-Cb5, BCL-2-ActA, BCL-XL-Cb5, and BCL-XL-ActA. The “ActA” mitochondrial targeting sequence is the coding region of amino acids 613 to 639 of Listeria ActA (49, 50). The “Cb5” ER-targeting sequence is the coding region of the hydrophobic tail sequence (amino acids 100–134) of rat hepatic ER Cb5 (48, 50).
In each plasmid, a gene encoding a fluorophore (i.e., mCerulean3, Venus, or mCherry) was subcloned at the 5′ end of the coding region for each protein of interest (i.e., BCL-XL, BCL-2, BIK, etc.). The resulting N-terminal fusion proteins expressed in cells are named with a superscript C, V, or Ch for mCerulean3, Venus, and mCherry, respectively: that is, VBIK denotes Venus fused to the N terminus of BIK protein. This notation is also used to indicate which fusion protein is encoded in each plasmid. For example, the “CBCL-XL-ActA-pEGFP-C3” plasmid contains the mCerulean3 gene cloned at the 5′ end of the coding region of the BCL-XL-ActA chimera in a pEGFP-C3 vector backbone. We also use the same format to name cell lines stably expressing the genes of one or more fusion proteins, that is, “MCF-7CBCL-XL-ActA” refers to MCF-7 cells stably expressing CBCL-XL-ActA fusion protein. Furthermore, “MCF-7CBCL-XL-ActA_ChPTDSSI” indicates additional expression of landmark, ChPTDSSI, in the MCF-7CBCL-XL-ActA cell line. VBIK mutants listed in Figures 2A and 4A and supplementary 2B were generated by PCR and/or restriction enzyme digest, as described on Addgene (see Table S1 for accession numbers). “Δ” or “del” are used to indicate deleted regions in BIK. For example, the MBR (amino acids 136–160) of BIK was deleted in “BIK-ΔMBR” and replaced by a stop codon. To make “BIK-Δ72 to 136,” the 65 amino acid region following the BH3 and up to the MBR (amino acids 72–136) was deleted and replaced by a short six amino acid (SRSGEQ) linker. Other point mutations in BIK were introduced by PCR only. For example, the “L61G” mutation was introduced to the H2 position in the BH3 of BIK, whereas the “4E” mutation was made to positions H1–H4 (Fig. 2A).
Cell culture
Cell lines were tested regularly for mycoplasma by a PCR-based test (92). All cell lines were incubated at 37 °C with 5% CO2. BMK cells with BAX and BAK gene knockout (BMK-DKO) were gifts from Eileen White (Rutgers University) (93). Human breast cancer MCF-7 cells (94) were gifted by Linda Penn (University of Toronto). Human colon carcinoma, HCT116, and HCT116-DKO cells were a gift from Dr Bert Vogelstein (The Johns Hopkins Kimmel Cancer Center) (95, 96). HEK293 cells (293T) were a gift from Dr Frank Graham (McMaster University), the originator of the cell line (97). MCF-7 cells were cultured in alpha minimum essential medium (MEM) supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin (pH 7.2). All other cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 1% MEM nonessential amino acids, and 1% penicillin–streptomycin (pH 7.2). In 2020, MCF-7, HCT116, and HEK293 cells were authenticated at the hospital for Sick Children facility in Toronto by short tandem repeat genotyping (14). Also, BMK cells were submitted to American Type Culture Collection for mouse short tandem repeat profiling of 18 loci to confirm murine origin, though no reference was available to validate the BMK cell line. BAX and BAK were not detected by Western blot for HCT116 DKO (96) and BMK DKO (93) cell lysates.
The MCF-7-CBCL-XL-ActA cell line was generated by transfection of the CBCL-XL-ActA-pEGFP-C3 plasmid, followed by selection with G418 (500 μg/ml) and fluorescence-activated cell sorting for stable gene expression as described for MCF-7-CBCL-XL (13). Cell pools stably expressing: BMK-DKOCBCL-XL and BMK-DKOCBCL-2 (13) and BMK-DKO cells stably expressing genes encoding CBCL-XL-ActA, CBCL-2-ActA, CBCL-XL-Cb5, or CBCL-2-Cb5 were created by transfection of plasmids containing genes encoding CBCL-XL or CBCL-2 in the s2193 backbone, followed by blasticidin (5 μg/ml) treatment for >2 weeks and sorting for cells positive for mCerulean3 expression by flow cytometry. MCF-7 parental and MCF-7-CBCL-XL-ActA cells stably expressing mCherry landmark fusion proteins: ChPTDSSI, ChCb5, ChActA, or mCardinal, were generated via lentiviral production and transduction as recently described in detail for MCF10A cells (39). Cells (listed in Table S1) were expanded for 1 week following infection and then sorted by flow cytometry. Compared with MCF-7 stable cell lines, BMK-DKO stable cell lines have lower expression of the mCerulean3-fusion protein because of the different promoters (cytomegalovirus versus human ferritin) in plasmids used for generating these cell lines.
Transfection of live cells in 384-well plate
At ∼60 to 80% confluency, cells were trypsinized, counted, diluted to the desired density, and seeded in a 25 μl volume into a CellCarrier-384 Ultra Microplate well plate. Cells were left to settle for 15 min before returning the plate to the incubator. The absorbance for transfection the following day is ∼60%. Thus, the number of cells seeded per was determined by doubling time: 2000 for HEK293 cells, 3000 for BMK cell lines, 5000 for MCF-7s, and 3500 for HCT116. After 12 to 24 h, cells were transfected using TransIT-X2 reagent (Mirus) following the manufacturer’s instructions. To transfect two wells of a 384-well plate, 50 ng of the DNA of interest, 50 ng pSPUTK, to 9 μl Opti-MEM and 0.4 μl TransIT-X2 reagent are mixed by vortexing and centrifuged to spin down the mixture from the sides of the tube and let stand at room temperature for 15 to 30 min. DMEM complete cell media (50 μl) were added per reaction and mixed gently by pipetting. To transfect cells, 25 μl of the transfection reaction was added on top of the 25 μl volume of seeded cells from the previous day. Control, “untransfected” wells were handled similarly but did not include the DNA in the transfection reaction. Plates were incubated for 3 to 5 h, then fresh DMEM complete media were to cells. Sample plates were incubated for 12 to 18 h and then imaged directly. Note that pSPUTK DNA was added to all single transfection reactions to maintain the required amount of DNA for efficient transfection while controlling the overexpression of the protein of interest. For cotransfections, the second construct was added in place of the 50 ng pSPUTK DNA.
For FLIM–FRET experiments, BMK-DKO cells stably expressing a mCerulean3-fusion protein (indicated on top of each graph as “donor”) were transfected with plasmids encoding the Venus-fusion protein of interest (indicated in the legend of each graph). Plasmids encoding the appropriate collisional control were included in each experiment and one well was left untransfected; details are provided in Table S2.
Image-based quantification of cell death
Cell death was quantified for cells transfected to express the indicated proteins as described (13). In brief, untransfected cells, cells transfected to express Venus, and cells treated with 200 nM staurosporine were included with each experiment as negative and positive controls for death. After incubation for 12 to 24 h, the cells were stained half an hour with 5 μM DRAQ5 and ∼0.3 ng/ml purified AnnexinV protein labeled with rhodamine and imaged on the Opera Phenix automated confocal microscope (PerkinElmer) with four channels collected simultaneously: blue (excitation: 425 nm, emission: 435–480 nm), green (excitation: 488 nm, emission: 500–550 nm), red (excitation: 561, emission: 570–630), and far red (excitation: 640 nm, emission 650–760 nm). For each experiment, mCerulean3 protein expression in stable cell lines (i.e., MCF-7-CBCL-XL or MCF-7-CBCL-2) was captured in the blue channel; transient expression of the indicated Venus-fusion proteins in the green channel; AnnexinV–rhodamine signal in the red channel; and DRAQ5 stained nuclei in the far red channel. Analysis was carried out using Harmony software (PerkinElmer) as described (13). DRAQ5 nuclear stain images were used for cell segmentation to identify nuclear and cytoplasmic areas. The average intensity of AnnexinV and nuclear area were quantified for each cell. “AnnexinV positive” (apoptotic) was defined as a cell that has a measured non-nuclear AnnexinV signal that is at least two standard deviations above the average in untreated cells. Shrinkage of the nuclear area (defined here as at least two standard deviations below the average in untreated cells) is another morphological feature of apoptosis (98). Average Venus and mCerulean3 intensities were also measured per cell. A “Venus positive” cell was defined as a cell that has a Venus intensity of at least two standard deviations above the average in untransfected cells. To quantify the proapoptotic function of Venus-fusion proteins, cells were first identified as Venus positive, and then the percentage of those cells with small nuclear area and/or AnnexinV positivity was recorded. For each of the experiments in Figures 2H and S2G, four technical replicates were included for each biological replicate (n = 3). And 1000 to >12,000 Venus-positive cells for each transfectant were analyzed per biological replicate.
To measure % cell death as a function of increasing expression of VBIK or VBIK mutants in MCF-7 (Fig. 2G) and MCF-7CBCL-XL-ActA (Fig. 4D) cells, a linear classifier was trained using the Harmony software, and a MATLAB script was used to bin single-cell data by Venus intensity. Instead of AnnexinV–rhodamine, MCF-7 cell lines were stained with 10 nM tetramethylrhodamine ethyl ester (TMRE) to assess mitochondrial inner membrane transmembrane potential. MOMP leads to depolarization of mitochondria, which results in a decrease in TMRE intensity. The classifier was trained on the red (TMRE intensity) and far red (DRAQ5 nuclear and cell morphology features) for at least 100 cells manually identified as “dead” and 100 cells identified as “alive.” Venus and mCerulean3 intensities per cell were measured after the classification of each cell as live/dead. Object-level data (measurements reported per cell) were exported from Harmony in “.CSV” format. In MATLAB, object-level data were binned by Venus intensity to calculate the percent of cells classified as dead per bin, for each biological replicate. Relatively low-intensity bins (0, 100, 200, 225, 250, 300, 350, 400, 600, 800, and 1000) arbitrary intensity units were selected to exclude cells with supraphysiologic expression of some proteins after transient transfection. Venus intensity versus % classified dead curves (a minimum of 10 points per bin, mean, and SE) were exported for each sample, for three biological replicates (n = 3) (Figs. 2G and 4D). Similarly, percent of cell death was assessed for HCT116 and HCT116-DKO cells (Fig. 4D), except the blue channel was otherwise unoccupied (no expression of mCerulean3-fusion protein), and the cells were also stained with AnnexinV as an additional marker of apoptosis. Thus, cells transfected with VBIK mutants (green channel) were stained with Hoechst (nuclear marker, blue channel), TMRE (red channel), and AnnexinV-Alexa647 (far red channel), and the latter three channels were used to train the classifier to recognize dead cells.
FLIM–FRET data acquisition on the ISS-Alba
FLIM–FRET data were acquired and analyzed as described (62). The 384-well sample plate was incubated at 25 °C during image acquisition. The pulsed excitation is interleaved allowing for the simultaneous acquisition of two channels by time-correlated single-photon counting. We used a 1 ms pixel dwell time to collect only one frame per image to minimize effects from the movement of proteins of interest during imaging. Altogether, 100 fields of view (100 μm × 100 μm) were acquired per well. For each figure, the details about cell line(s), DNA constructs, and stains added are listed in Table S2.
Watershed segmentation analysis of FLIM–FRET data acquired on ISS-Alba platform
The ISS-Alba FLIM data were acquired, and lifetimes were calculated using VistaVision software (version 4.1.011.0) (62). In brief, nearest neighbor pixel binning was used to obtain sufficient photon counts for fitting to a single exponential decay for each pixel of the confocal image. Pixels below a specified threshold level were not included. The confocal image of the donor mCerulean3 channel was used to identify ROIs; pixels were binned within ROIs to export the average donor, acceptor, and lifetime per ROI.
For analysis, two MATLAB scripts were run in succession, described in detail and available at: https://github.com/DWALab/Osterlund_et_al_2021_BIK_Localization.
The first script, “LoopThroughMultipleDataFolders.m” uses a Watershed segmentation algorithm to select ROIs, extract average donor and acceptor intensities, and average lifetime per ROI to a file entitled, “Measurements.csv” in each sample subfolder.
Where <IAcceptor> and <IDonor> are the mean acceptor and donor intensities, respectively. On the ISS-Alba, interleaved excitation and time-gated detection for both donor and acceptor channels eliminates bleed-through. The second MATLAB script is used to calculate % FRET efficiency per ROI and bin the resulting binding curves. Importantly, an untransfected (donor alone) sample must be included for each experiment to define lower and upper intensity thresholds based on the 5% and 95% confidence intervals of the donor intensity distribution and to calculate the average lifetime of the donor alone. The thresholds were applied to the analysis of all transfected wells. The FRET efficiency calculated for each ROI is as:
Where <τD> is the average lifetime of the donor alone and <τDA> is the average lifetime of the donor in the presence of the acceptor. Binding curves were constructed by binning raw measurements with similar acceptor:donor intensity ratios. The user-defined bin sizes were kept consistent for all samples. Median acceptor:donor intensity ratio, percent of FRET efficiency, number of points, standard deviation, and standard error per bin were exported to generate final binding curves named, “Binned_Results.csv” for each sample. Final binding curves displayed in Figure 1, Figure 2 and 4 (and Figs. S2 and S4) were plotted in GraphPad Prism 8 (GraphPad Software, Inc) and fit to a one-site specific Hill slope equation. Collisional control data could generally be fit to a straight line; otherwise, if curvature was observed, a one-site specific Hill slope equation was used where the Bmax was fixed to that determined for the positive control. A standard was not used to normalize the settings for data acquisition between independent replicates; therefore, the data cannot be combined. As a result, data from a single replicate are displayed with additional replicates included in Figs. S2 and S4.
FLIM–FRET with mitochondrial segmentation on INO-FHS platform
BMK-DKO cells stably expressing CBCL-XL or CBCL-2 were transfected in a 384-well plate, as described previously, with plasmids to express VBIK, VBIK-4E, VtBID, VCb5, or VActA (Table S2). Before imaging, cells were stained with 400 nM MitoRed and 5 μM DRAQ5. The INO-FHS microscope was operated as described (99). During each confocal scan, the INO-FHS microscope collects time-correlated single-photon counting (TCSPC) FLIM and 64-channel hyperspectral data. Environmental controls were set to 25 °C and 5% CO2 during image acquisition. Two INO-FHS acquisition configurations were used: configuration 1 excited Venus (detected in FLIM channel) MitoRed and DRAQ5 and configuration 2 excited mCerulean3 (detected in FLIM channel) and Venus (Table 1). We then created a Job in Nikon Elements to rapidly switch between these two imaging configurations at each field of view.
Table 1.
Microscope configurations for imaging MitoRed and DRAQ5 while acquiring mCerulean3 and Venus FLIM–FRET data for detection of binding in mitochondria-like ROIs
| Microscope component/setting | Configuration 1 | Configuration 2 |
|---|---|---|
| ExEmDichroic (D1) | BP_442/20-514/26-LP_561 | BP_442/20-514/26-LP_561 |
| DirectFilter (F1 = excite donor) | BP_510/20 | BP_433/24 |
| InterleaveFilter (F2= excite acceptor/other) | BP_563/9 | BP_510/10 |
| DetectionDichroic (D2) (splits emitted signal between TCSPC FLIM [F3] and Hyperspectral [F4]) | LP_573 | LP_520 |
| TCSPCDetectionFilter (F3) | BP_540/15 | BP_482/25 |
| SpectralDetectionFilter (F4) | LP_579 | LP_526 |
| Pinhole | 150 μm | 150 μm |
| LineFillFraction | 0.85 | 0.85 |
| Frame rate | 0.07 | 0.055 |
| Number of pixels | 600 × 600 | 600 × 600 |
| Pixel size | 335 nm | 335 nm |
| Detected in TCSPC FLIM channel | Venus | mCerulean3 |
| Detected in 64 channel Hyperspectral | MitoRed & DRAQ5 | Venus |
With these two configurations, we acquired four fields of view (200 μm × 200 μm) per well. A standard 10 nM fluorescein sample (in 0.1 M NaOH) was used to adjust the acquisition settings to match intensities acquired on the first replicate. With this standard acquired, all three biological replicates could be combined (Fig. 5).
Analysis of INO-FHS image data with mitochondrial ROI selection
The image of the donor-fusion proteins was constructed by integrating the photon counts along the temporal axis, for each pixel, in the FLIM cube. Intensity images in the hyperspectral channel were generated by summing the spectral measurements for a specified range of wavelengths for individual pixels. Images of untransfected cells expressing the donor alone were used to calculate and subtract bleedthrough of mCerulean3 emission at 525 to 560 nm by simple linear unmixing for configuration 2 hyperspectral data before generating the Venus intensity image (52, 82). The DRAQ5 data acquired in configuration 1 were not used. The Venus lifetime images acquired in configuration 1 were used to check the alignment with the corresponding image of Venus acquired in configuration 2. Once image registration between configurations 1 and 2 was confirmed, ROIs selected from configuration 1 hyperspectral channels were applied to the corresponding images acquired in configuration 2 to measure mCerulean3-bound fraction within each.
At the excitation settings used in configuration 1, the observed emission from Venus at 599 nm was negligible, thus spectral unmixing was not necessary. The Venus intensity image was generated by summing the spectral intensity from 525 to 560 nm, whereas the mitochondrial image was constructed by summing the spectral intensity from 585 to 640 nm. ROI selection was performed using a multiseed watershed segmentation algorithm applied to either the donor (mCerulean3 in TCSPC FLIM channel, configuration 1) or MitoRed image (summed intensity from spectral intensities 585–640 nm, configuration 2) (Fig. 5A). The TCSPC arrival time histograms for pixels within each ROI were combined to generate a fluorescence decay curve associated with that ROI. These decay curve values are Fourier transformed, and their phasor coordinates were used to determine fluorescence lifetime, as described (82).
FLIM–FRET binding curves were generated from configuration 2 data as described previously for data acquired on the ISS-Alba. Average lifetime was used to calculate percent FRET efficiency per ROI. ROI data for all four images acquired within the same well were exported, combined, and binned by the acceptor:donor ratio. Since acquisition settings were normalized to a standard fluorescein sample, all three biological replicates could be plotted on the same graph. Binned data from three biological replicates were combined and fit to a Hill equation for each segmentation approach (Fig. 5B).
Sample preparation and data acquisition for colocalization experiments
All colocalization experiments were performed as follows. Cells were seeded and then transfected with DNA plasmids encoding Venus fusion proteins of interest as described earlier. For each figure, the details about cell line(s), DNA constructs, and stains added per well are listed in Table S2. At the point when we normally change to fresh media 3 to 5 h after transfection, 10 μM of QVD-OPH (pan-caspase inhibitor) was added to prevent cell death in MCF-7 and MCF-7 stable cell lines only. Each experiment included an untransfected control treated with transfection reagent alone for each cell line. Cells were incubated for 12 h and then stained for 30 min before imaging on the Opera Phenix. All wells were stained with 2.5 μM of the nuclear dye DRAQ5. Depending on which proteins were expressed in the cell, cells were also stained with 200 nM MitoRed and/or 100 nM MitoGreen depending on whether the cell expressed a Venus or mCherry fluorescence protein, respectively.
Sample plates were imaged on the Opera Phenix high-content screening microscope using the 40× water immersion objective (numerical aperture: 1.1) while incubated at 37 °C with 5% CO2. Four confocal channels were collected independently, where (excitation and emission) wavelengths are indicated in nanometers: red channel (561, 570–630); blue channel (425, 435–515); green channel (488, 500–550); and far red channel (640, 650–760). Laser power/exposure settings were fixed for all three biological replicates. For MCF-7 landmark data (Figs. 7 and 8), three biological replicates, each containing at least two technical replicates and 48 fields of view per well, were collected. For colocalization data in BMK-DKO cells (Figure 1, Figure 2, Figure 6), the number of independent biological replicates (n) is specified in each figure.
Three-way colocalization analysis
The CellProfiler analysis pipeline for colocalization analysis is publicly available on GitHub (13). We used this pipeline for the analysis of two-channel colocalization in BMK-DKO cells (Figs. 1B and 2B) and three-channel data in Figures 6 and 7. Pearson’s r correlation measurements were calculated for all pairs of channels: (1) blue:green, (2) green:red, and (3) blue:red (three-way colocalization, Fig. S6).
For this analysis, images were exported from PerkinElmer Harmony software as “.Tiff” files for all four channels: red channel (mCherry signal or MitoRed); blue channel (mCerulean3 signal); green channel (Venus signal or MitoGreen); and far red channel (DRAQ5 signal). Images were analyzed in CellProfiler (Table S1, “Step1_CellProfilerAnalysis”). Analysis was customized for each dataset using the “test mode” in CellProfiler to check that segmentation appropriately identified primary and secondary objects. The script applies a log filter to the DRAQ5 channel, identifies nuclei as primary objects, and then propagates these seeds outward to identify secondary objects (the total cell area). Subtracting the nuclear area from the total cell area creates a cytoplasmic mask, which is defined as our ROI. A minimum median DRAQ5 intensity and a maximum size/area were used to identify and remove images of cells that were segmented improperly. For acceptable ROIs, the mean and median intensities of each channel, and Pearson’s r values for pairs of channels: (1) blue:green, (2) green:red, (3) blue:red were exported along with Image ID and Object ID for identification. CellProfiler exports the image directory path, which includes the row and column labels, in a separate file. A second MATLAB script was used to match the Image ID from the filename to the object-level dataset with row/column identification (Table S1, “Step2_Get_RowColumn”) and export the data as “colocalization.csv”.
A third MATLAB script uses the “colocalization.csv” data (Table S1, “Step3_FilterAndPlotData”) to apply a minimum intensity threshold for our green, red, and or blue channels to eliminate images of cells with insufficient protein expression for analysis. This script also plots Pearson’s r values for filtered ROI data, for example, Fig. S6, see left graph. For each cell line tested, a separate plot was exported for colocalization of (1) blue:green, (2) green:red, and (3) blue:red channels along with median of ROI data (Fig. S6, orange +), mode of ROI data (red +), and number of ROIs (column data) exported for each technical replicate (well transfection ID = row data) to an excel sheet. Replicate datasheets were combined using the Microsoft Excel RDBMerge plugin (author: Ron De Bruin, available publicly). When running RDBMerge in Microsoft Excel, the user must check the box to include the filename to maintain the sample identifiers included in each filename. Since the distribution of the raw ROI data was skewed (Fig. S6, see left graph), we found the mode to best represent the Pearson’s r value for each replicate. A minimum of 50 cytoplasmic ROIs was required per technical replicate for median or mode values to be included in final plots (see example Fig. S6, middle and right graphs). Finally, the mean (thick black line) of these technical replicates was used for plotting heatmaps.
Contrastive supervised learning for classifying landmark protein subcellular localizations from single-cell images
All code mentioned here has been made publicly available on our GitHub repository linked to in Table S1. To reanalyze the image dataset used for mCherry-fusion landmark colocalization analysis, we used a deep learning approach.
Our analysis pipeline was generally based on our previous work (39), with modifications. Here, we used CellProfiler 4.1.3 to locate the bounding box of each cell where cells are segmented as done for the colocalization analysis aforementioned, and the bounding box for each cell was exported into a MySQL database (db) file. The database contains tables for segmented single-cell measurements. A Python script, “ExportCellROIsToTFRecords.py” cropped the multichannel single-cell images using the bounding box coordinates. This script was also used to identify in-focus cells defined as having a measured variance of the DRAQ5 channel Laplacian greater than 30 (Fig. S7A). In-focus images have sharper edges, which results in a larger variance in the Laplacian image. Then each multichannel image was automatically resized to 100 × 100 pixels, rescaled from 0 to 1 (minimum/maximum normalization), and saved to tensor flow records (a file format used for storing large sequences of binary records). From three biological repetitions (each containing at least two technical replicates), we identified 143,211 in-focus single cells (red channel). This image dataset was then used to train a Gabor-based convolutional neural network (100, 101). Specifically, 116,651 of the red channel images (mCherry landmarks stably expressed in MCF-7 cells) including images of four subcellular locations: ER (ChCb5), mitochondria (ChActA), MAMs (ChPTDSS1), and diffuse expression (mCardinal) were used as the training dataset.
Model architecture is described in Fig. S7B. Our convolutional neural network was implemented, trained, and deployed using TensorFlow 2.3 (see our “Jupyter Notebook” on GitHub). The encoder output was flattened to produce a new image representation r (vector size of 1152), then transformed into a projection, ‘z’ using a single perceptron layer (vector size 128). The encoder and projector were trained using a supervised contrastive loss function, as described (102) and included later
where the symbol denotes the inner (dot) product, τ a scalar temperature parameter, and are the class label within each pair, is the number of samples with the same label as the anchor, and 2N is the number of samples in the minibatch.
The encoder projector network was trained for 100 epochs using Adam optimizer with a learning rate of 0.001, and τ was set to 0.05. During training, image flipping and rotation augmentations were used. Within each training batch, a single projection, “zi,” is automatically selected to be an anchor. The remaining samples were used to reconstruct positive and negative pairs; a projection pair (zi, zj) is said to be positive if the images belong to the same class and negative if the images belong to different classes (102). As the training proceeds, the encoder learns features that bring data for positive pairs closer in the projected space, while keeping projection for negative pairs further apart. After training was complete (100 epochs), the encoder was frozen, and the projector network was discarded. The trained encoder can be thought of as a transformation function from the image space into a new compressed representation used to train a classifier. The classifier consists of two fully connected layers (dropout regularization = 0.5): ReLU activation was used for the first layer, whereas a SoftMax activation was used for the second (output) layer. The classifier was trained for 10 epochs using Adam optimizer (learning rate also 0.001). Red channel classification accuracy plateaued at ∼88%. The Gabor-based convolutional encoder and classifier were combined into a single model to classify any single cell image to a given landmark.
The user-defined Master Platemap (previously used for colocalization analysis) was then used for the code to track the cell line and transfection/stain status based on the well ID of each cell. MCF-7 parental cells have no mCerulean3 expression (no blue channel to classify). In untransfected MCF-7-CBCL-XL-ActA cells, all cells were passed to classify CBCL-XL-ActA localization from the blue channel. In transfected MCF-7 and MCF-7-CBCL-XL-ActA cells, a minimum green channel filter (at least 40% of the pixels in the single-cell image must be above the mean background intensity measured in untransfected cells) was applied to select only images of transfected cells to pass into the final classification and analyzed for each channel. By this means, we examined the change in classified localization of the blue and green channels in the presence versus absence of coexpression of another protein (Fig. 8D). Expression of either CBCL-XL-ActA or any transfectant did not change the classification of the red channel landmarks in cells (Fig. S7F, red channel) “predicted labels” for the performance of the algorithm across cell lines/transfectants and compare with ground truth data in Fig. S7G).
Predictive BIK structure models
To calculate unstructured lengths, we used 3.5 Å as the average length of an amino acid in an unstructured peptide.
The Robetta server from the University of Washington’s Institute for Protein Design (77) and the CPHmodel server from the Technical University of Denmark, Department of Bio and Health Informatics (76) were used to predict BIK structure. The CPHmodel server generated a single predicted structure per submission, whereas the Robetta server generated five potential structures ranked by energy minimization, of which only model 1 (most minimized potential energy of inter-residue interactions) was used here. All resulting model.pdb files are available for download (Table S1). To each server, we submitted the full-length sequence of human BIK. However, the MBR is a known transmembrane helix that anchors into ER membranes (37) that we expect would not be exposed in solution under physiologically relevant conditions. Hence, we also submitted truncated human BIK (1–136) lacking the MBR to examine the predicted fold for the portion of BIK that faces the cytoplasm in cells alone.
We exported the sequence included in each structural model and colored amino acids according to whether they were unstructured (black), within an ɑ helix (red), or a β sheet (blue) and aligned results with the prediction of McDonnell et al. (75) (Fig. 3A). For these three sequences, we came to a “majority vote” secondary structure prediction where an amino acid was considered part of an ɑ helix in our final prediction if at least two-third models predicted it was part of an ɑ helix (Fig. 3A). Finally, we compared our predicted majority vote secondary structure to the 3D models of BIK and concluded that the Robetta server prediction BIK(1–136) matched best (Supplementary text and models in Supplementary Fig. S3A). We also exported the C-terminal MBR (BIK 137–160) from the Robetta Server prediction for full-length BIK at the same scale as BIK (1–136) (Fig. 3B). For visualization purposes only, we then generated a model of full-length BIK fusing the MBR (BIK137–160) to the C terminus of BIK (1–136) (point of attachment indicated by the gray arrow in Fig. 3C). Notably, only the portion of BIK (1–136) that faces the cytoplasm in cells was needed to measure the distance of the BH3 region of BIK may extend from the ER membrane. We measured the length of the helices within the region between the BH3 (helix 3) and the MBR (helix 6) in PyMOL. Helix 4 measures ∼14Å, and helix 5 is ∼28Å. Finally, to estimate the distance between the BH3 and MBR of BIK, we combine these data with the maximum span of the flexible loop regions between helices 3 and 4 (count 1 amino acid), between 4 and 5 (count 11 amino acids), and between 5 and 6 (count 15 amino acids). As before, using 3.5 Å as the average length of an amino acid, these loop regions could span (1 ∗ 3.5 + 11 ∗ 3.5 + 15 ∗ 3.5) Å = (3.5 + 38.5 + 52.5) Å= 94.5 Å. Together, this results in a potential span of maximally (14 + 28 + 94.5) Å= 136 Å = ∼14 nm between the BH3 and MBR.
In Figure 3D, we then generated a model of the structure of BCL-XL binding to BIK. The conformation of BCL-XL integrated into the membrane was created as previously described (103): structure of truncated BCL-XL (1bxl.pdb is soluble BCL-XL in complex with BAK peptide, hide BAK peptide in PyMOL) was merged with the structure of the C-terminal tail of BCL-XL in lipid nanodiscs (6f46.pdb) and displayed on the membrane. There is no membrane in either structure; however, based on the model by Yao et al., (103), we made measurements in PyMOL on 1bxl.pdb from an amino acid (lysine 20) on BCL-XL that was close to the membrane in their model to the BH3 region of 27.4, 24.1, and 19.7Å, giving our approximate distance of ∼2.5 nm. To approximate the distance between the N terminus of BCL-XL and the membrane, based on the model by Yao et al. (103) and using structure 1bxl.pdb in PyMOL, we measured the distance (12.7 Å = ∼1 nm) between another amino acid close to the membrane in their model (proline 38) and methionine at the first position in BCL-XL. The N terminus of BCL-XL is ∼1 nm from the membrane, but given, we added a flexible seven amino acid linker between mCerulean3 and the N terminus of BCL-XL, this projects mCerulean3 an additional ∼2.5 nm (at 3.5Ä per amino acid = 24.5 Å) from the membrane, for a distance ∼3.5 nm. Furthermore, the chromophore lies within the center of the β-barrel of the fluorophore, ∼2.5 nm from the C terminus (measurement approximated from 4AR7.pdb, mTurquoise, a fluorophore similar to mCerulean3).
Resource availability
Our lead contact is David W. Andrews (David.Andrews@SRI.utoronto.ca). All DNA constructs have been deposited with Addgene. Sources for DNA constructs, reagents, and cell lines used in this article are listed in Table S1. Table S2 lists the cell line(s) used for each experiment, DNA constructs transfected, stains added, and expected signal in each channel acquired, organized by figure identification.
Data availability
All data are available in the article or its supporting information and publicly available on Dataverse (see Table S1 for links). All original code and instructions have been made publicly available on Github (see Table S1 for links).
Supporting information
This article contains supporting information. References in the supporting information (3, 65, 75, 76, 77, 100, 101, 100, 101) are included with main references.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
E. J. O. and D. W. A. conceptualization; E. J. O. and N. H. methodology; N. H. software; D. N. validation; E. J. O. and N. H. formal analysis; E. J. O. investigation; D. W. A. resources; N. H. data curation; E. J. O. writing–original draft; E. J. O., N. H., D. N., J. M. P., and D. W. A. writing–review & editing; E. J. O. visualization; Q. F. and D. W. A. supervision; D. W. A. project administration; Q. F. and D. W. A. funding acquisition.
Funding and additional information
This work was supported by grant FDN143312 from the Canadian Institutes of Health Research to D. W. A. D. W. A. holds Tier 1 Canada Research Chair in Membrane Biogenesis.
Biography

Elizabeth J. Osterlund As a Ph.D. student at the University of Toronto, Elizabeth Osterlund used high-content analyses to detect, in live cells, direct interactions between the reported ER-localized pro-apoptotic protein, BIK, and anti-apoptotic protein(s) targeted to mitochondria. In this article, fluorescence techniques probed how this interaction occurs, and the results challenge existing models of BIK function. As a Post Doctoral Researcher at McMaster University, Elizabeth now studies the mechanisms behind Huntington’s disease.
Edited by Elizabeth Coulson
Supporting information
References
- 1.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Pemberton J.M., Pogmore J.P., Andrews D.W. Neuronal cell life, death, and axonal degeneration as regulated by the BCL-2 family proteins. Cell Death Differ. 2021;28:108–122. doi: 10.1038/s41418-020-00654-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kale J., Osterlund E.J., Andrews D.W. BCL-2 family proteins: changing partners in the dance towards death. Cell Death Differ. 2018;25:65–80. doi: 10.1038/cdd.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Letai A., Bassik M.C., Walensky L.D., Sorcinelli M.D., Weiler S., Korsmeyer S.J. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 5.Leber B., Lin J., Andrews D.W. Still embedded together binding to membranes regulates Bcl-2 protein interactions. Oncogene. 2010;29:5221–5230. doi: 10.1038/onc.2010.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leber B., Lin J., Andrews D.W. Embedded together: the life and death consequences of interaction of the bcl-2 family with membranes. Apoptosis. 2007;12:897–911. doi: 10.1007/s10495-007-0746-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Llambi F., Moldoveanu T., Tait S.W.G., Bouchier-Hayes L., Temirov J., McCormick L.L., et al. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol. Cell. 2011;44:517–531. doi: 10.1016/j.molcel.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H.-C., Kanai M., Inoue-Yamauchi A., Tu H.-C., Huang Y., Ren D., et al. An interconnected hierarchical model of cell death regulation by the BCL-2 family. Nat. Cell Biol. 2015;17:1270–1281. doi: 10.1038/ncb3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi X., Kale J., Leber B., Andrews D.W. Regulating cell death at, on, and in membranes. Biochim. Biophys. Acta. 2014;1843:2100–2113. doi: 10.1016/j.bbamcr.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Edlich F., Banerjee S., Suzuki M., Cleland M.M., Arnoult D., Wang C., et al. Bcl-x(L) retrotranslocates Bax from the mitochondria into the cytosol. Cell. 2011;145:104–116. doi: 10.1016/j.cell.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Todt F., Cakir Z., Reichenbach F., Youle R.J., Edlich F. The C-terminal helix of Bcl-x(L) mediates Bax retrotranslocation from the mitochondria. Cell Death Differ. 2013;20:333–342. doi: 10.1038/cdd.2012.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torrecillas A., Martínez-Senac M.M., Ausili A., Corbalán-García S., Gómez-Fernández J.C. Interaction of the C-terminal domain of Bcl-2 family proteins with model membranes. Biochim. Biophys. Acta. 2007;1768:2931–2939. doi: 10.1016/j.bbamem.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Liu Q., Osterlund E.J., Chi X., Pogmore J., Leber B., Andrews D.W. Bim escapes displacement by BH3-mimetic anti-cancer drugs by double-bolt locking both Bcl-XL and Bcl-2. ELife. 2019;8 doi: 10.7554/eLife.37689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chi X., Nguyen D., Pemberton J.M., Osterlund E.J., Liu Q., Brahmbhatt H., et al. The carboxyl-terminal sequence of bim enables bax activation and killing of unprimed cells. ELife. 2020;9 doi: 10.7554/eLife.44525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kale J., Chi X., Leber B., Andrews D. Examining the molecular mechanism of bcl-2 family proteins at membranes by fluorescence spectroscopy. Met. Enzymol. 2014;544:1–23. doi: 10.1016/B978-0-12-417158-9.00001-7. [DOI] [PubMed] [Google Scholar]
- 16.Sarosiek K.A., Chi X., Bachman J.A., Sims J.J., Montero J., Patel L., et al. BID preferentially activates BAK while BIM preferentially activates BAX, affecting chemotherapy response. Mol. Cell. 2013;51:751–765. doi: 10.1016/j.molcel.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang K., O’Neill K.L., Li J., Zhou W., Han N., Pang X., et al. BH3-only proteins target BCL-xL/MCL-1, not BAX/BAK, to initiate apoptosis. Cell Res. 2019;29:942–952. doi: 10.1038/s41422-019-0231-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Neill K.L., Huang K., Zhang J., Chen Y., Luo X. Inactivation of prosurvival Bcl-2 proteins activates Bax/Bak through the outer mitochondrial membrane. Genes Dev. 2016;30:973–988. doi: 10.1101/gad.276725.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brahmbhatt H., Uehling D., Al-Awar R., Leber B., Andrews D. Small molecules reveal an alternative mechanism of Bax activation. Biochem. J. 2016;473:1073–1083. doi: 10.1042/BCJ20160118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willis S.N., Fletcher J.I., Kaufmann T., van Delft M.F., Chen L., Czabotar P.E., et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science (New York, N.Y.) 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 21.Kim H., Rafiuddin-Shah M., Tu H.-C., Jeffers J.R., Zambetti G.P., Hsieh J.J.D., et al. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat. Cell Biol. 2006;8:1348–1358. doi: 10.1038/ncb1499. [DOI] [PubMed] [Google Scholar]
- 22.Vasquez-Montes V., Rodnin M.V., Kyrychenko A., Ladokhin A.S. Lipids modulate the BH3-independent membrane targeting and activation of BAX and Bcl-xL. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2025834118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lovell J.F., Billen L.P., Bindner S., Shamas-Din A., Fradin C., Leber B., et al. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135:1074–1084. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 24.Kim H., Tu H.-C., Ren D., Takeuchi O., Jeffers J.R., Zambetti G.P., et al. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol. Cell. 2009;36:487–499. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng R., Tong J.S., Li H., Yue B., Zou F., Yu J., et al. Targeting Bax interaction sites reveals that only homo-oligomerization sites are essential for its activation. Cell Death Differ. 2013;20:744. doi: 10.1038/cdd.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarosiek K.A., Fraser C., Muthalagu N., Bhola P.D., Chang W., McBrayer S.K., et al. Developmental regulation of mitochondrial apoptosis by c-myc governs age- and tissue-specific sensitivity to cancer therapeutics. Cancer Cell. 2017;31:142–156. doi: 10.1016/j.ccell.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rizzuto R., Brini M., Murgia M., Pozzan T. Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science (New York, N.Y.) 1993;262:744–747. doi: 10.1126/science.8235595. [DOI] [PubMed] [Google Scholar]
- 28.Rizzuto R., Pinton P., Carrington W., Fay F.S., Fogarty K.E., Lifshitz L.M., et al. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science (New York, N.Y.) 1998;280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 29.Korzeniowski M.K., Szanda G., Balla T., Spät A. Store-operated Ca2+ influx and subplasmalemmal mitochondria. Cell Calcium. 2009;46:49–55. doi: 10.1016/j.ceca.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Contreras L., Drago I., Zampese E., Pozzan T. Mitochondria: the calcium connection. Biochim. Biophys. Acta. 2010;1797:607–618. doi: 10.1016/j.bbabio.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 31.Gottschalk B., Klec C., Waldeck-Weiermair M., Malli R., Graier W.F. Intracellular Ca2+ release decelerates mitochondrial cristae dynamics within the junctions to the endoplasmic reticulum. Pflugers Archi. 2018;470:1193–1203. doi: 10.1007/s00424-018-2133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemasters J.J., Theruvath T.P., Zhong Z., Nieminen A.-L. Mitochondrial calcium and the permeability transition in cell death. Biochim. Biophys. Acta. 2009;1787:1395–1401. doi: 10.1016/j.bbabio.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang D., Lu C., Whiteman M., Chance B., Armstrong J.S. The mitochondrial permeability transition regulates cytochrome c release for apoptosis during endoplasmic reticulum stress by remodeling the cristae junction. J. Biol. Chem. 2008;283:3476–3486. doi: 10.1074/jbc.M707528200. [DOI] [PubMed] [Google Scholar]
- 34.Vervliet T., Clerix E., Seitaj B., Ivanova H., Monaco G., Bultynck G. Modulation of Ca2+ signaling by anti-apoptotic B-cell lymphoma 2 proteins at the endoplasmic reticulum-mitochondrial interface. Front. Oncol. 2017;7:75. doi: 10.3389/fonc.2017.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pihán P., Carreras-Sureda A., Hetz C. BCL-2 family: integrating stress responses at the ER to control cell demise. Cell Death Differ. 2017;24:1478–1487. doi: 10.1038/cdd.2017.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomenius M.J., Distelhorst C.W. Bcl-2 on the endoplasmic reticulum: Protecting the mitochondria from a distance. J. Cell Sci. 2003;116:4493–4499. doi: 10.1242/jcs.00829. [DOI] [PubMed] [Google Scholar]
- 37.Germain M., Mathai J.P., Shore G.C. BH-3-only BIK functions at the endoplasmic reticulum to stimulate cytochrome c release from mitochondria. J. Biol. Chem. 2002;277:18053–18060. doi: 10.1074/jbc.M201235200. [DOI] [PubMed] [Google Scholar]
- 38.Mathai J.P., Germain M., Marcellus R.C., Shore G.C. Induction and endoplasmic reticulum location of BIK/NBK in response to apoptotic signaling by E1A and p53. Oncogene. 2002;21:2534–2544. doi: 10.1038/sj.onc.1205340. [DOI] [PubMed] [Google Scholar]
- 39.Schormann W., Hariharan S., Andrews D.W. A reference library for assigning protein subcellular localizations by image-based machine learning. J. Cell Biol. 2020;219 doi: 10.1083/jcb.201904090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilfling F., Weber A., Potthoff S., Vögtle F.-N., Meisinger C., Paschen S.A., et al. BH3-only proteins are tail-anchored in the outer mitochondrial membrane and can initiate the activation of Bax. Cell Death Differ. 2012;19:1328–1336. doi: 10.1038/cdd.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao X., Wang L., Sun Y., Ye L., Lu J., Yuan Y., et al. The endoplasmic reticulum (ER)-target protein Bik induces Hep3B cells apoptosis by the depletion of the ER Ca2+ stores. Mol. Cell Biochem. 2008;312:33–38. doi: 10.1007/s11010-008-9718-4. [DOI] [PubMed] [Google Scholar]
- 42.Hockings C., Anwari K., Ninnis R.L., Brouwer J., O’Hely M., Evangelista M., et al. Bid chimeras indicate that most BH3-only proteins can directly activate Bak and Bax, and show no preference for Bak versus Bax. Cell Death Dis. 2015;6:e1735. doi: 10.1038/cddis.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Germain M., Mathai J.P., McBride H.M., Shore G.C. Endoplasmic reticulum BIK initiates DRP1-regulated remodelling of mitochondrial cristae during apoptosis. EMBO J. 2005;24:1546–1556. doi: 10.1038/sj.emboj.7600592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mathai J.P., Germain M., Shore G.C. BH3-only BIK regulates BAX,BAK-dependent release of Ca2+ from endoplasmic reticulum stores and mitochondrial apoptosis during stress-induced cell death. J. Biol. Chem. 2005;280:23829–23836. doi: 10.1074/jbc.M500800200. [DOI] [PubMed] [Google Scholar]
- 45.Mebratu Y.A., Leyva-Baca I., Wathelet M.G., Lacey N., Chand H.S., Choi A.M.K., et al. Bik reduces hyperplastic cells by increasing Bak and activating DAPk1 to juxtapose ER and mitochondria. Nat. Commun. 2017;8:803. doi: 10.1038/s41467-017-00975-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mebratu Y.A., Dickey B.F., Evans C., Tesfaigzi Y. The BH3-only protein BikJBlkJNbk inhibits nuclear translocation of activated ERK1J2 to mediate IFNγ-induced cell death. J. Cell Biol. 2008;183:429–439. doi: 10.1083/jcb.200801186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Annis M.G., Zamzami N., Zhu W., Penn L.Z., Kroemer G., Leber B., et al. Endoplasmic reticulum localized Bcl-2 prevents apoptosis when redistribution of cytochrome c is a late event. Oncogene. 2001;20:1939–1952. doi: 10.1038/sj.onc.1204288. [DOI] [PubMed] [Google Scholar]
- 48.Mitoma J., Ito A. The carboxy-terminal 10 amino acid residues of cytochrome b5 are necessary for its targeting to the endoplasmic reticulum. EMBO J. 1992;11:4197–4203. doi: 10.1002/j.1460-2075.1992.tb05513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pistor S., Chakraborty T., Walter U., Wehland J. The bacterial actin nucleator protein ActA of Listeria monocytogenes contains multiple binding sites for host microfilament proteins. Curr. Biol. 1995;5:517–525. doi: 10.1016/s0960-9822(95)00104-7. [DOI] [PubMed] [Google Scholar]
- 50.Zhu W., Cowie A., Wasfy G.W., Penn L.Z., Leber B., Andrews D.W. Bcl-2 mutants with restricted subcellular location reveal spatially distinct pathways for apoptosis in different cell types. EMBO J. 1996;15:4130–4141. [PMC free article] [PubMed] [Google Scholar]
- 51.Bleicken S., Hantusch A., Das K.K., Frickey T., Garcia-Saez A.J. Quantitative interactome of a membrane Bcl-2 network identifies a hierarchy of complexes for apoptosis regulation. Nat. Commun. 2017;8:73. doi: 10.1038/s41467-017-00086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osterlund E.J., Hirmiz N., Pemberton J.M., Nougarède A., Liu Q., Leber B., et al. Efficacy and specificity of inhibitors of BCL-2 family protein interactions assessed by affinity measurements in live cells. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abm7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dlugosz P.J., Billen L.P., Annis M.G., Zhu W., Zhang Z., Lin J., et al. Bcl-2 changes conformation to inhibit Bax oligomerization. EMBO J. 2006;25:2287–2296. doi: 10.1038/sj.emboj.7601126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Janiak F., Leber B., Andrews D.W. Assembly of Bcl-2 into microsomal and outer mitochondrial membranes. J. Biol. Chem. 1994;269:9842–9849. [PubMed] [Google Scholar]
- 55.Nguyen M., Millar D.G., Yong V.W., Korsmeyer S.J., Shore G.C. Targeting of Bcl-2 to the mitochondrial outer membrane by a COOH-terminal signal anchor sequence. J. Biol. Chem. 1993;268:25265–25268. [PubMed] [Google Scholar]
- 56.Billen L.P., Kokoski C.L., Lovell J.F., Leber B., Andrews D.W. Bcl-XL inhibits membrane permeabilization by Competing with bax. PLoS Biol. 2008;6:e147. doi: 10.1371/journal.pbio.0060147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hsu Y.T., Wolter K.G., Youle R.J. Cytosol-to-membrane redistribution of bax and bcl-X(L) during apoptosis. Proc. Natl. Acad. Sci. U. S. A. 1997;94:3668–3672. doi: 10.1073/pnas.94.8.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaufmann T., Schlipf S., Sanz J., Neubert K., Stein R., Borner C. Characterization of the signal that directs Bcl-x(L), but not Bcl-2, to the mitochondrial outer membrane. J. Cell Biol. 2003;160:53–64. doi: 10.1083/jcb.200210084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fiebig A.A., Zhu W., Hollerbach C., Leber B., Andrews D.W. Bcl-XL is qualitatively different from and ten times more effective than Bcl-2 when expressed in a breast cancer cell line. BMC Cancer. 2006;6:213. doi: 10.1186/1471-2407-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Algar W.R., Hildebrandt N., Vogel S.S., Medintz I.L. FRET as a biomolecular research tool — understanding its potential while avoiding pitfalls. Nat. Met. 2019;16:815–829. doi: 10.1038/s41592-019-0530-8. [DOI] [PubMed] [Google Scholar]
- 61.Day R.N., Schaufele F. Imaging molecular interactions in living cells. Mol. Endocrinol. (Baltimore, Md. 2005;19:1675–1686. doi: 10.1210/me.2005-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Osterlund E.J., Liu Q., Andrews D.W. In: Mitochondrial Medicine. Methods in Molecular Biology. Weissig V., Edeas M., editors. Vol. 1264. Humana Press; New York, NY: 2015. The use of FLIM-FRET for the detection of mitochondria-associated protein interactions. [DOI] [PubMed] [Google Scholar]
- 63.Liu Q., Leber B., Andrews D.W. Interactions of pro-apoptotic BH3 proteins with anti-apoptotic Bcl-2 family proteins measured in live MCF-7 cells using FLIM FRET. Cell Cycle (Georgetown, Tex.) 2012;11:3536–3542. doi: 10.4161/cc.21462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chinnadurai G., Vijayalingam S., Rashmi R. BIK, the founding member of the BH3-only family proteins: mechanisms of cell death and role in cancer and pathogenic processes. Oncogene. 2008;27:S20–S29. doi: 10.1038/onc.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shamas-Din A., Bindner S., Zhu W., Zaltsman Y., Campbell C., Gross A., et al. tBid undergoes multiple conformational changes at the membrane required for Bax activation. J. Biol. Chem. 2013;288:22111–22127. doi: 10.1074/jbc.M113.482109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oda E., Ohki R., Murasawa H., Nemoto J., Shibue T., Yamashita T., et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science (New York, N.Y.) 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 67.Wang X., Wu S. Real-time monitoring BimL interacting with Bcl-xL during UV-induced apoptosis. Seventh Int. Conf. Photon. Imaging Biol. Med. 2008;7280 doi: 10.1117/12.822629. [DOI] [Google Scholar]
- 68.Andreu-Fernández V., Genoves A., Lee T.-H., Stellato M., Lucantoni F., Orzáez M., et al. Peptides derived from the transmembrane domain of Bcl-2 proteins as potential mitochondrial priming tools. ACS Chem. Biol. 2014;9:1799–1811. doi: 10.1021/cb5002679. [DOI] [PubMed] [Google Scholar]
- 69.Li Y.M., Wen Y., Zhou B.P., Kuo H.-P., Ding Q., Hung M.-C. Enhancement of Bik antitumor effect by Bik mutants. Cancer Res. 2003;63:7630–7633. [PubMed] [Google Scholar]
- 70.Verma S., Zhao L.J., Chinnadurai G. Phosphorylation of the pro-apoptotic protein BIK: mapping of phosphorylation sites and effect on apoptosis. J. Biol. Chem. 2001;276:4671–4676. doi: 10.1074/jbc.M008983200. [DOI] [PubMed] [Google Scholar]
- 71.Wang Y., Guan X., Fok K.L., Li S., Zhang X., Miao S., et al. A novel member of the Rhomboid family, RHBDD1, regulates BIK-mediated apoptosis. Cell Mol. Life Sci. 2008;65:3822–3829. doi: 10.1007/s00018-008-8452-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Naon D., Scorrano L. At the right distance: ER-mitochondria juxtaposition in cell life and death. Biochim Biophys Acta. 2014;1843:2184–2194. doi: 10.1016/j.bbamcr.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 73.Rizzuto R., Marchi S., Bonora M., Aguiari P., Bononi A., De Stefani D., et al. Ca(2+) transfer from the ER to mitochondria: when, how and why. Biochim Biophys Acta. 2009;1787:1342–1351. doi: 10.1016/j.bbabio.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vance J.E. Phospholipid synthesis in a membrane fraction associated with mitochondria. J Biol Chem. 1990;265:7248–7256. [PubMed] [Google Scholar]
- 75.McDonnell J.M., Fushman D., Milliman C.L., Korsmeyer S.J., Cowburn D. Solution structure of the proapoptotic molecule BID: a structural basis for apoptotic agonists and antagonists. Cell. 1999;96:625–634. doi: 10.1016/s0092-8674(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 76.Nielsen M., Lundegaard C., Lund O., Petersen T.N. CPHmodels-3.0-remote homology modeling using structure-guided sequence profiles. Nucl. Acids Res. 2010;38:W576–W581. doi: 10.1093/nar/gkq535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang J., Anishchenko I., Park H., Peng Z., Ovchinnikov S., Baker D. Improved protein structure prediction using predicted interresidue orientations. Proc. Natl. Acad. Sci. U. S. A. 2020;117:1496–1503. doi: 10.1073/pnas.1914677117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dhar J., Chakrabarti P. Defining the loop structures in proteins based on composite β-turn mimics. Protein Eng. Des. Select. 2015;28:153–161. doi: 10.1093/protein/gzv017. [DOI] [PubMed] [Google Scholar]
- 79.Csordás G., Renken C., Várnai P., Walter L., Weaver D., Buttle K.F., et al. Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 2006;174:915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bravo R., Vicencio J.M., Parra V., Troncoso R., Munoz J.P., Bui M., et al. Increased ER-mitochondrial coupling promotes mitochondrial respiration and bioenergetics during early phases of ER stress. J. Cell Sci. 2011;124:2143–2152. doi: 10.1242/jcs.080762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Giacomello M., Pellegrini L. The coming of age of the mitochondria–ER contact: a matter of thickness. Cell Death Differ. 2016;23:1417. doi: 10.1038/cdd.2016.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Osterlund E.J., Hirmiz N., Pemberton J.M., Fang Q., Andrews D.W. Automated, quantitative Fast FLIM-FRET (qF3): a step-by-step protocol to measure dissociation constants for protein-protein interactions in live-cell screening applications. Protoc. Exchange. 2022 doi: 10.21203/RS.3.PEX-1354/V1. [DOI] [Google Scholar]
- 83.Andreu-Fernández V., García-Murria M.J., Bañó-Polo M., Martin J., Monticelli L., Orzáez M., et al. The C-terminal domains of apoptotic BH3-only proteins mediate their insertion into distinct biological membranes. J. Biol. Chem. 2016;291:25207–25216. doi: 10.1074/jbc.M116.733634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gómez-Fernández J.C. Functions of the C-terminal domains of apoptosis-related proteins of the Bcl-2 family. Chem. Phys. Lipids. 2014;183:77–90. doi: 10.1016/j.chemphyslip.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 85.Denic V. A portrait of the GET pathway as a surprisingly complicated young man. Trends Biochem. Sci. 2012;37:411–417. doi: 10.1016/j.tibs.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sakai S., Watanabe S., Komine O., Sobue A., Yamanaka K. Novel reporters of mitochondria-associated membranes (MAM), MAMtrackers, demonstrate MAM disruption as a common pathological feature in amyotrophic lateral sclerosis. FASEB J.. 2021;35 doi: 10.1096/fj.202100137R. [DOI] [PubMed] [Google Scholar]
- 87.Stone S.J., Vance J.E. Phosphatidylserine synthase-1 and -2 are localized to mitochondria-associated membranes. J. Biol. Chem. 2000;275:34534–34540. doi: 10.1074/jbc.M002865200. [DOI] [PubMed] [Google Scholar]
- 88.Lalier L., Mignard V., Joalland M.-P., Lanoé D., Cartron P.-F., Manon S., et al. TOM20-mediated transfer of Bcl2 from ER to MAM and mitochondria upon induction of apoptosis. Cell Death Dis. 2021;12:182. doi: 10.1038/s41419-021-03471-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Légiot A., Céré C., Dupoiron T., Kaabouni M., Camougrand N., Manon S. Mitochondria-Associated Membranes (MAMs) are involved in Bax mitochondrial localization and cytochrome c release. Microb. Cell (Graz, Austria) 2019;6:257–266. doi: 10.15698/mic2019.05.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lindner A.U., Concannon C.G., Boukes G.J., Cannon M.D., Llambi F., Ryan D., et al. Systems analysis of BCL2 protein family interactions establishes a model to predict responses to chemotherapy. Cancer Res. 2013;73:519–528. doi: 10.1158/0008-5472.CAN-12-2269. [DOI] [PubMed] [Google Scholar]
- 91.Hur J., Chesnes J., Coser K.R., Lee R.S., Geck P., Isselbacher K.J., et al. The Bik BH3-only protein is induced in estrogen-starved and antiestrogen-exposed breast cancer cells and provokes apoptosis. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2351–2356. doi: 10.1073/pnas.0307337101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hopert A., Uphoff C.C., Wirth M., Hauser H., Drexler H.G. Mycoplasma detection by pcr analysis. In Vitro Cell Dev. Biol. Anim. 1993;29A:819–821. doi: 10.1007/BF02634350. [DOI] [PubMed] [Google Scholar]
- 93.Degenhardt K., Sundararajan R., Lindsten T., Thompson C., White E. Bax and Bak independently promote cytochrome C release from mitochondria. J. Biol. Chem. 2002;277:14127–14134. doi: 10.1074/jbc.M109939200. [DOI] [PubMed] [Google Scholar]
- 94.Graham K.A., Trent J.M., Osborne C.K., McGrath C.M., Minden M.D., Buick R.N. The use of restriction fragment polymorphisms to identify the cell line MCF-7. Breast Cancer Res. Treat. 1986;8:29–34. doi: 10.1007/BF01805922. [DOI] [PubMed] [Google Scholar]
- 95.Lengauer C., Kinzler K.W., Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 96.Zhang L., Yu J., Park B., Kinzler K., Vogelstein B. Role of BAX in the apoptotic response to anticancer agents. Science (New York, N.Y.) 2000;290:989–992. doi: 10.1126/science.290.5493.989. [DOI] [PubMed] [Google Scholar]
- 97.Graham F., Smiley J., Russell W., Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 98.Hacker G. The morphology of apoptosis. Cell Tissue Res. 2000;30:5–17. doi: 10.1007/s004410000193. [DOI] [PubMed] [Google Scholar]
- 99.Osterlund E.J., Hirmiz N., Tardif C., Andrews D.W. Rapid imaging of BCL-2 family interactions in live cells using FLIM-FRET. Met. Mol. Biol. 2019;1877:305–335. doi: 10.1007/978-1-4939-8861-7_19. [DOI] [PubMed] [Google Scholar]
- 100.Molaei S., Shiri Ahmad Abadi M.E. Maintaining filter structure: a gabor-based convolutional neural network for image analysis. Appl. Soft Comput. 2020;88 [Google Scholar]
- 101.Pérez J.C., Alfarra M., Jeanneret G., Bibi A., Thabet A., Ghanem B., et al. In: Computer Vision – ECCV 2020. ECCV 2020. Lecture Notes in Computer Science. Vedaldi A., Bischof H., Brox T., Frahm J.M., editors. Vol. 12354. Springer; Cham: 2020. Gabor Layers Enhance Network Robustness. [Google Scholar]
- 102.Khosla P., Teterwak P., Wang C., Sarna A., Tian Y., Isola P., et al. arXiv; 2020. Supervised Contrastive Learning. [Google Scholar]
- 103.Yao Y., Fujimoto L.M., Hirshman N., Bobkov A.A., Antignani A., Youle R.J., et al. Conformation of BCL-XL upon membrane-integration. J. Mol. Biol. 2015;427:2262–2270. doi: 10.1016/j.jmb.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the article or its supporting information and publicly available on Dataverse (see Table S1 for links). All original code and instructions have been made publicly available on Github (see Table S1 for links).