Summary
Potatoes consist of flavonoids that provide health benefits for human consumers. To learn more about how potato tuber flavonoid accumulation and flesh pigmentation are controlled, we analyzed the transcriptomic and metabolomic profile of potato tubers from three colored potato clones at three developmental phases using an integrated approach. From the 72 flavonoids identified in pigmented flesh, differential abundance was noted for anthocyanins, flavonols, and flavones. Weighted gene co-expression network analysis further allowed modules and candidate genes that positively or negatively regulate flavonoid biosynthesis to be identified. Furthermore, an R2R3-MYB repressor StMYB3 and an R3-MYB repressor StMYBATV involved in the modulation of anthocyanin biosynthesis during tuber development were identified. Both StMYB3 and StMYBATV could interact with the cofactor StbHLH1 and repress anthocyanin biosynthesis. Our results indicate a feedback regulatory mechanism of a coordinated MYB activator-repressor network on fine-tuning of potato tuber pigmentation during tuber development.
Subject areas: Plant biology, Plant Genetics, Transcriptomics
Graphical abstract

Highlights
-
•
Seventy-two flavonoids were identified in differentially pigmented potato tuber
-
•
Candidate genes for flavonoid biosynthesis were identified
-
•
Two MYB repressors play roles in a feedback regulatory network on tuber pigmentation
Plant biology; Plant Genetics; Transcriptomics
Introduction
With potato (Solanum tuberosum L.) being a major staple crop around the world, the pigmented cultivars have attracted research interest because of substantial anthocyanin accumulation, giving both specialty and capacity for nutritional benefits to human health.1,2 Anthocyanins are classified within the flavonoid group of compounds, which are derived from the well-characterized flavonoid biosynthetic pathway in many plants.3 Flavonols, flavones, isoflavones, flavanones, anthocyanins, and condensed tannins (proanthocyanidins, PAs) are among the final products of flavonoid biosynthesis, which starts with phenylalanine.3 The phenylalanine is then converted into p-coumaroyl-CoA, with the reaction catalyzed by phenylalanine ammonia-lyase (PAL), 4-coumarate-CoA ligase (4CL), and cinnamic acid hydroxylase (C4H). Through a condensation reaction, chalcone synthase (CHS) then converts p-coumaroyl-CoA and fatty acid-derived malonyl CoA into naringenin chalcone which is subsequently catalyzed by different enzymes within the pathway to yield various flavonoid compounds. Of these, flavanone 3-hydroxylase (F3H), flavonoid 3′5′-hydroxylase (F3′5′H), flavonoid 3′-hydroxylase (F3′H), and dihydroflavonol 4-reductase (DFR) direct the route toward leucoanthocyanins. The colorless leucoanthocyanins are then converted by anthocyanidin synthase (ANS/LDOX) to colored anthocyanidins such as cyanidin, delphinidin, and pelargonidin prior to glycosylation by Uridine 5'-diphosphate (UDP)-glucose flavonoid-O-glycosyl transferase (UFGT) or other chemical modifications such as methylation and acylation. These products are transported from the cytosol to vacuoles for storage via different types of mechanisms mediated by the glutathione S-transferases (GST), ATP binding cassette (ABC) proteins, and multidrug and toxic extrusion (MATE).4,5,6 Other enzymes involved in the manufacture of flavonols, flavones, isoflavones, and PAs include flavonol synthase (FLS), flavone synthase (FNS), isoflavone synthase (IFS), anthocyanin reductase (ANR), and leucoanthocyanidin reductase (LAR).3,7,8
In addition to genes encoding flavonoid-related enzymes, transcription factors (TFs) that influence gene expression have been found in various species. For instance, gene expression within this pathway is controlled by the MYB-bHLH-WD40 (MBW) complex which basically involves interactions between MYB, the basic-helix-loop-helix (bHLH), and the WD40/WDR regulators.9,10 Within the flavonoid pathway, the different branches that yield separate end compounds are separately regulated by different MYB TFs (mainly R2R3-MYB), leading to flavonol, PAs, and anthocyanins,11,12 with 13 out of 126 R2R3-MYB members controlling flavonoid metabolism in the case of Arabidopsis.13 Flavonol biosynthesis requires regulation by MYB11, MYB12, and MYB111 of the R2R3-MYB group of TFs,14 while in the seed coat of Arabidopsis, the accumulation of PAs is regulated by a minimum of four MBW complexes TT2(MYB123)-TT8/GL3/EGL3(bHLH)-TTG1(WD40), MYB5-TT8-TTG1 with partially overlapped functions.15 The R2R3-MYB activators which take part in the process of anthocyanin synthesis are well characterized in many species, with some examples being VvMYBA2 and VvMYBA1 in grapevine (Vitis vinifera) berries,16 StAN1 in potato (S. tuberosum),17,18 and AtMYB114, AtMYB113, AtMYB90/PAP2, and AtMYB75/PAP1 in Arabidopsis19 as well as MdMYB110a, MdMYB10, MdMYBA, and MdMYB1 in apples (Malus × domestica).20,21 Similarly, in Arabidopsis, R2R3-MYB and R3-MYB repressors such as AtMYB3/4/6 and CAPRICE (CPC)/TRIPTYCHON (TRY)/AtMYBL2 are also recognized.22,23
Other TFs such as MADS-box,24 zinc finger proteins,25 AP2/ERF,26 the WRKY family,27,28 and the bZIP family29,30 also highlight the significance of transcriptional regulation for controlling flavonoid biosynthesis. They mostly co-regulate flavonoid production by interacting with members of the MBW complex; for example, AP2/ERF TFs complexed with PyMYB114 to positively co-regulate anthocyanin accumulation in pear (Pyrus bretschneideri),31 while in red-fleshed apple, the MdHY5-MdWRKY41-MdMYB complex influences the synthesis of anthocyanin and PAs.28 Other transcription factors are also recognized as upregulating key R2R3 MYB controllers. For example, several peach NAC [No apical meristem (NAM), Arabidopsis transcription activation factor (ATAF), Cup-shaped cotyledon (CUC)] TFs upregulate peach MYB10 expression in red-fleshed peaches (Prunus persica).32
Characterizing the flavonoid biosynthesis pathway in potatoes as well as associated regulatory genes has been the focus of several studies.17,18,33,34,35,36,37,38,39,40,41,42,43 In the present work, we examined potato clones developed by conventional breeding procedures from the same ancestor, which showed yellow, red, and purple pigmentation. This feature allows the identification of factors responsible for flavonoid biosynthesis without confounding the genetic background. Joint transcriptome and metabolome analyses identified potato tubers' major flavonoid compounds and corresponding genes. A total of 72 flavonoids were placed, and weighted gene co-expression network analysis (WGCNA) provided co-expressed gene modules that allowed genes which are essential for controlling flavonoid synthesis to be identified. Two MYB repressors (an R2R3-MYB and an R3-MYB) were determined engaged in feedback regulatory mechanisms underlying the biosynthesis of flavonoid accumulation in colored potatoes during tuber development. The current study will clarify the temporal accumulation of flavonoids in different colored potato tubers, expand our understanding of the regulatory mechanism of flavonoid synthesis, and facilitate research into engineering flavonoid composition in potato tubers.
Results
Metabolic differences between the three potato clones over development
Seventy-two types of flavonoids were detected in three potato clones at different developmental stages (Figure 1A) including 24 anthocyanins, 21 flavones, 18 flavonols, four flavan-3-ols, two dihydroflavones, one dihydrochalcone, one dihydroflavonol, and one isoflavone (Table 1). We screened differentially accumulated flavonoids (DAFs) using a |Log2 (fold change)| ≥ 1, a variable importance in project (VIP) value of ≥1, and a p value <0.05 as thresholds. The numbers of DAF ranged from 31 to 56, 27 to 51, and 42 to 62 between two potato clones at three developmental stages (S1, S2, and S3) and from 1 to 3, 15 to 40, and 9 to 32 between two developmental stages of Y, R, and P, respectively (Figures 1B and 1C).
Figure 1.
Tuber color and number of differentially accumulated flavonoids among three potato clones at three developmental stages
(A) Tuber colors of three potato clones at three developmental stages, CIP 302281.17, CIP 302281.25, and CIP 302281.15, were renamed as Y, R, and P according to their dominant flesh color. S1, S2, and S3 represent tuberization, tuber bulking, and tuber maturation stages, respectively. Scale bar, 1 cm.
(B) Numbers of differentially accumulated flavonoids (DAFs) among Y, R, and P at three stages.
(C) Numbers of DAFs among three developmental stages. See also Table 1 and Figure S1.
Table 1.
Flavonoid metabolites detected in three potato clones
| Metabolite Index | Compounds | Q1 (Da) | Molecular Weight (Da) | Formula | Ionization model | YS1a | RS1a | PS1a | YS2a | RS2a | PS2a | YS3a | RS3a | PS3a |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.Hmmp001656 | Pelargonin-O-rutinoside | 579 | 578 | C27H31O14 | [M + H]+ | 9 | 112000 | 47833 | 9 | 58633 | 39767 | 9 | 1038000 | 31633 |
| 2.Hmmp001174 | Pelargonin-O-Hexoside-O-rhamnoside-O-Hexoside | 741 | 740 | C33H41O19 | [M + H]+ | 9 | 133000 | 35467 | 9 | 53467 | 103733 | 9 | 1063333 | 59333 |
| 3.Zmmp001791 | Pelargonidin-3-rutinoside-5-glucoside | 741 | 741 | C33H41O19+ | [M]+ | 9 | 60867 | 18533 | 9 | 27133 | 55233 | 9 | 583000 | 32567 |
| 4.Zmmp003570 | Pelargonidin3-p-coumaroylrutinoside-5-glucoside | 887 | 887 | C42H47O21+ | [M]+ | 9 | 171000 | 56767 | 9 | 107500 | 93467 | 9 | 5163333 | 106900 |
| 5.Zmmp003304 | Pelargonidin 3-O-[2-O-Glucosyl-6-O-p-Coumaroyl-Glucoside]-5-O-(Glucoside) | 903 | 903 | C42H47O22+ | [M]+ | 9 | 4300 | 2733 | 9 | 3017 | 679 | 9 | 144333 | 4717 |
| 6.pme3392 | Pelargonidin 3-O-glucoside | 433 | 433 | C21H21O10+ | [M]+ | 9 | 82233 | 20400 | 9 | 48167 | 8877 | 9 | 693667 | 14633 |
| 7. pmb0550 | Cyanidin 3-O-glucoside (Kuromanin) | 449 | 449 | C21H21O11+ | [M]+ | 9 | 4886 | 26800 | 9 | 9 | 30833 | 9 | 9 | 26467 |
| 8.pme1773 | Cyanidin 3-O-rutinoside (Keracyanin) | 595 | 595 | C27H31O15+ | [M]+ | 9 | 110333 | 1700000 | 9 | 81633 | 2363333 | 9 | 251333 | 1286667 |
| 9.Zmmp003451 | Cyanidin-3-p-coumaroylrutinoside-5-glucoside | 903 | 903 | C42H47O22+ | [M]+ | 9 | 3043 | 110333 | 9 | 9 | 235333 | 9 | 155667 | 463667 |
| 10.Zmmp003244 | Delphinidin-3-p-coumaroylrutinoside-5-glucoside | 919 | 919 | C42H47O23+ | [M]+ | 9 | 9 | 55100 | 9 | 9 | 139667 | 9 | 9 | 357000 |
| 11.pme3256 | Delphinidin 3-O-rutinoside | 611 | 611 | C27H31O16+ | [M]+ | 9 | 9 | 1066667 | 9 | 9 | 1356667 | 9 | 5187 | 877667 |
| 12.Zmmp003722 | Peonidin-3-feruloylrutinoside-5-glucoside | 947 | 947 | C44H51O23+ | [M]+ | 9 | 3830 | 67233 | 9 | 9 | 115667 | 9 | 11733 | 74100 |
| 13.Zmmp003631 | Peonidin-3-p-coumaroylrutinoside-5-glucoside | 917 | 917 | C43H49O22+ | [M]+ | 9 | 9917 | 347667 | 9 | 1123 | 720333 | 9 | 114500 | 921333 |
| 14.pme3391 | Petunidin 3-O-glucoside | 479 | 479 | C22H23O12+ | [M]+ | 9 | 9 | 10130 | 9 | 9 | 39600 | 9 | 9 | 35433 |
| 15.Zmmp003478 | Petunidin-3-p-coumaroylrhamnoside-5-glucoside | 771 | 771 | C37H39O18+ | [M]+ | 9 | 646 | 129333 | 436 | 9 | 224333 | 9 | 2189 | 273333 |
| 16.Zmmp003779 | Petunidin-3-p-coumaroylrutinoside | 771 | 771 | C37H39O18+ | [M]+ | 9 | 9 | 146333 | 9 | 9 | 173000 | 9 | 9 | 368333 |
| 17.Zmmp001564 | Petunidin-3-rutinoside-5-glucoside | 787 | 787 | C34H43O21+ | [M]+ | 9 | 9 | 149333 | 9 | 9 | 597667 | 9 | 9 | 244000 |
| 18.Zmmp004046 | Petunidin-3-p-coumaroylrutinoside-5-glucoside | 933 | 933 | C43H49O23+ | [M]+ | 9 | 9 | 157667 | 9 | 9 | 298667 | 9 | 9 | 637333 |
| 19.Zmmp003482 | Petunidin-3-p-coumaroylrutinoside-7-glucoside | 933 | 933 | C43H49O23+ | [M]+ | 1663 | 9 | 1110000 | 9 | 9 | 2096667 | 826 | 3539 | 2780000 |
| 20.Zmmp003155 | Petunidin-3-caffeoylrutinoside-5-glucoside | 949 | 949 | C43H49O24+ | [M]+ | 9 | 9 | 77767 | 9 | 9 | 62833 | 9 | 9 | 108200 |
| 21.Zmmp003556 | Petunidin-3-feruloylrutinoside-7-glucoside | 963 | 963 | C44H51O24+ | [M]+ | 9 | 9 | 110633 | 9 | 9 | 230667 | 9 | 9 | 168000 |
| 22.Zmmp004160 | Petunidin-3-feruloylrutinoside-5-glucoside | 963 | 963 | C44H51O24+ | [M]+ | 9 | 9 | 15033 | 9 | 9 | 28200 | 9 | 9 | 28133 |
| 23.Zmmp004296 | Malvidin-3-p-coumaroylrutinoside-5-glucoside | 947 | 947 | C44H51O23+ | [M]+ | 9 | 9 | 2483 | 9 | 9 | 13033 | 9 | 9 | 23767 |
| 24.Zmmp003756 | Malvidin-3-feruloylrutinoside-5-glucoside | 977 | 977 | C45H53O24+ | [M]+ | 9 | 9 | 8267 | 9 | 9 | 19400 | 9 | 9 | 10493 |
| 25.mws2118 | Phloretin 2′-O-glucoside | 435 | 436 | C21H24O10 | [M−H]- | 1317 | 4760 | 13750 | 898 | 5590 | 34233 | 1111 | 95133 | 47367 |
| 26.Hmmn002691 | Eriodictiol-O-Hexoside-O-rhamnoside-O-Hexoside-O-rhamnoside | 903 | 904 | C39H52O24 | [M−H]- | 9 | 151000 | 48300 | 9 | 41933 | 41000 | 174 | 5836667 | 50000 |
| 27.Hmmn002379 | Eriodictiol-O-Hexoside-O-rhamnoside-O-Hexoside-O-Hexoside | 919 | 920 | C39H52O25 | [M−H]- | 9 | 9 | 132333 | 9 | 9 | 238333 | 9 | 133833 | 500667 |
| 28.mws1094 | Dihydrokaempferol | 287 | 288 | C15H12O6 | [M−H]- | 9 | 44600 | 2150 | 9 | 15800 | 9 | 9 | 106567 | 9 |
| 29.mws1422 | (−)-Epiafzelechin | 275 | 274 | C15H14O5 | [M + H]+ | 9 | 10120 | 9 | 9 | 8363 | 9 | 463 | 114733 | 4703 |
| 30.mws0034 | Epigallocatechin gallate | 457 | 458 | C22H18O11 | [M−H]- | 2236 | 9 | 5686 | 969 | 9 | 9 | 9 | 234339 | 1123 |
| 31.mws0351 | Gallate catechin gallate | 457 | 458 | C22H18O11 | [M−H]- | 9 | 9 | 9 | 8373 | 9 | 9 | 9 | 144673 | 9 |
| 32.pme3285 | Afzelechin(3,5,7,4′-Tetrahydroxyflavan) | 275 | 274 | C15H14O5 | [M + H]+ | 9 | 34400 | 886 | 9 | 23700 | 756 | 9 | 182667 | 2350 |
| 33.Hmmp002447 | Apigenin-O-rutinoside-O-Hexoside | 725 | 724 | C33H40O18 | [M + H]+ | 9 | 5706 | 9 | 9 | 11903 | 9 | 9 | 1230000 | 9 |
| 34.Hmmp002474 | Apigenin-O-Hexoside-O-rhamnoside-O-glucuronate | 755 | 754 | C33H38O20 | [M + H]+ | 9 | 49600 | 9 | 9 | 129333 | 9 | 9 | 546000 | 9 |
| 35.Hmmp002073 | Apigenin-O-Hexoside-O-rhamnoside-O-Hexoside-O-Hexoside | 903 | 902 | C39H50O24 | [M + H]+ | 9 | 7536 | 300000 | 9 | 4483 | 530667 | 9 | 442333 | 972000 |
| 36.Hmmp002266 | Apigenin-O-Hexoside-O-rhamnoside-O-rhamnoside-O-Hexoside | 887 | 886 | C39H50O23 | [M + H]+ | 9 | 366667 | 91633 | 9 | 177000 | 188000 | 3213 | 10810000 | 233667 |
| 37.pme2459 | Luteolin 7-O-glucoside(Cynaroside) | 449 | 448 | C21H20O11 | [M + H]+ | 9 | 51633 | 9 | 9 | 32767 | 9 | 439 | 509667 | 9 |
| 38.pmp000593 | Luteolin-7-O-rutinoside | 595 | 594 | C27H30O15 | [M + H]+ | 9 | 14667 | 1083 | 9 | 4527 | 439 | 9 | 130000 | 9 |
| 39.pmp001079 | Luteolin-7-O-neohesperidoside(Lonicerin) | 595 | 594 | C27H30O15 | [M + H]+ | 9 | 15267 | 9 | 9 | 5573 | 9 | 9 | 156667 | 9 |
| 40.pmp000595 | Luteolin-7,3′-Di-O-β-D-Glucoside | 611 | 610 | C27H30O16 | [M + H]+ | 9 | 9 | 19433 | 9 | 9 | 48100 | 9 | 29933 | 80933 |
| 41.Hmpp002612 | Luteolin-7-O-β-D-gentiobioside | 611 | 610 | C27H30O16 | [M + H]+ | 9 | 9 | 34567 | 9 | 9 | 57233 | 9 | 43933 | 97933 |
| 42.Hmmn003584 | Luteolin-O-rutinoside-O-rhamnoside | 739 | 740 | C33H40O19 | [M−H]- | 9 | 16033 | 9 | 9 | 4627 | 9 | 9 | 929667 | 9 |
| 43.Hmmp002169 | Luteolin-O-Hexoside-O-rhamnoside-O-rhamnoside-O-Hexoside | 903 | 902 | C39H50O24 | [M + H]+ | 9 | 8097 | 369000 | 9 | 4683 | 541000 | 9 | 457333 | 1097333 |
| 44.CWJP002289 | Luteolin-3-O-glucosyl-rhamnosyl-glucoside-glucoside | 919 | 918 | C42H46O23 | [M + H]+ | 9 | 9 | 179667 | 9 | 9 | 317667 | 9 | 1413 | 932667 |
| 45.Zmhp002897 | Isochrysoeriol C-hexosyl-O-hexoside | 625 | 624 | C28H32O16 | [M + H]+ | 9 | 9 | 57467 | 9 | 9 | 132333 | 9 | 19667 | 147000 |
| 46.mws0043 | Nobiletin | 403 | 402 | C21H22O8 | [M + H]+ | 4727 | 4740 | 5183 | 4833 | 4917 | 4380 | 4687 | 4713 | 4680 |
| 47.pmb3041 | Tricin O-saccharic acid | 521 | 522 | C23H22O14 | [M−H]- | 109800 | 210667 | 260667 | 162667 | 338333 | 176000 | 87500 | 48300 | 57733 |
| 48.Zmhp002798 | Isosaponarin | 595 | 594 | C27H30O15 | [M + H]+ | 9 | 9 | 9 | 9 | 9 | 5523 | 9 | 299333 | 7807 |
| 49.Hmmp002322 | Chrysoerio-O-Hexoside-O-rhamnoside-O-rhamnoside-O-Hexoside | 917 | 916 | C40H52O24 | [M + H]+ | 9 | 83067 | 1170000 | 1093 | 113333 | 2340000 | 9 | 1293333 | 3026667 |
| 50.Hmmp002324 | Tricin-O-Hexoside-O-rhamnoside-O-rhamnoside-O-Hexoside | 947 | 946 | C41H54O25 | [M + H]+ | 9 | 14700 | 391000 | 9 | 10393 | 716000 | 9 | 22703 | 628000 |
| 51.mws0055 | Tangeretin | 373 | 372 | C20H20O7 | [M + H]+ | 3403 | 2903 | 3227 | 3273 | 2773 | 2443 | 2963 | 2700 | 2850 |
| 52.Lmnp002584 | 5,7,4′-Trihydroxy-6-methoxyflavone-8-C-[β-D-glucosyl-(1–2)]-β-D-glucoside | 625 | 624 | C28H32O16 | [M + H]+ | 9 | 9 | 66700 | 9 | 9 | 152000 | 9 | 23533 | 153000 |
| 53.Lmnp002448 | 5,7,3′,4′-Tetrahydroxy-6-methoxyflavone-8-C-[β-D-glucosyl-(1–2)]-β-D-glucoside | 641 | 640 | C28H32O17 | [M + H]+ | 9 | 9 | 188667 | 9 | 9 | 297333 | 9 | 9 | 299000 |
| 54.Hmmp002121 | Isorhamnetin-O-gallate | 469 | 468 | C23H16O11 | [M + H]+ | 8936 | 82400 | 26533 | 949 | 110933 | 2083 | 619 | 13513 | 9 |
| 55.Hmcp001859 | Isorhamnetin-O-rutinoside | 625 | 624 | C28H32O16 | [M + H]+ | 9 | 1773 | 67033 | 9 | 9 | 164333 | 9 | 24867 | 173667 |
| 56.GQ512005 | Kaempferol-3-O-glucoside-7-O-rhamnoside | 595 | 594 | C27H30O15 | [M + H]+ | 9 | 12533 | 869 | 9 | 4627 | 653 | 9 | 159333 | 1300 |
| 57.pme0369 | Kaempferol 3-O-rutinoside(Nicotiflorin) | 593 | 594 | C27H30O15 | [M−H]- | 363 | 183667 | 7647 | 1780 | 64833 | 10727 | 1616 | 1860000 | 14920 |
| 58.Lmmp003217 | Kaempferol 3-O-β-d-(6″-O-(E)-p-coumaroyl)glucopyranoside | 595 | 594 | C30H26O13 | [M + H]+ | 1491 | 265000 | 15300 | 1293 | 77600 | 16333 | 2150 | 2850000 | 18867 |
| 59.Lmbp003230 | Kaempferol-3-O-neohesperidoside | 595 | 594 | C27H30O15 | [M + H]+ | 9 | 254000 | 10567 | 866 | 92233 | 12097 | 1733 | 3723333 | 14900 |
| 60.Lnrp002296 | Kaempferol glc-rha | 595 | 594 | C27H30O15 | [M + H]+ | 1950 | 626333 | 31233 | 3663 | 251333 | 38300 | 8850 | 6516667 | 30400 |
| 61.Lmbp002592 | Kaempferol-3,7-di-O-β-D-glucopyranoside | 611 | 610 | C27H30O16 | [M + H]+ | 9 | 9 | 34667 | 9 | 9 | 62067 | 9 | 40400 | 90900 |
| 62.CWJP002348 | Kaempferol-3-O-rhamnosyl-sophoroside-7-O-glucoside | 949 | 948 | C43H48O24 | [M + H]+ | 416 | 436 | 202000 | 332 | 1143 | 254333 | 9 | 9 | 434000 |
| 63.Lmbp003393 | Quercetin-3-O-α-L-rhamnopyranoside | 449 | 448 | C21H20O11 | [M + H]+ | 646 | 50033 | 4530 | 9 | 29333 | 5813 | 873 | 592667 | 2993 |
| 64.Lmmp002859 | Quercetin glucoside-glucoside | 627 | 626 | C27H30O17 | [M + H]+ | 2215 | 25410 | 13167 | 7437 | 8207 | 33833 | 1647 | 1283 | 52100 |
| 65.Hmmp002336 | Quercetin-O-feruloyl-Pentoside | 611 | 610 | C30H26O14 | [M + H]+ | 899 | 28267 | 44700 | 3536 | 37667 | 58933 | 893 | 52800 | 57000 |
| 66.Hmmp002041 | Quercetin-O-Hexoside-O-rhamnoside-O-rhamnoside-O-Hexoside | 919 | 918 | C39H50O25 | [M + H]+ | 9 | 9 | 179333 | 9 | 9 | 273667 | 9 | 9 | 547003 |
| 67.Lmln001801 | Myricetin-3,7-O-dirhamnoside | 625 | 626 | C27H30O17 | [M−H]- | 9 | 9 | 10110 | 9 | 9 | 23100 | 9 | 9 | 32700 |
| 68.pmp001310 | 6-Hydroxykaempferol-3,6-O-Diglucoside | 627 | 626 | C27H30O17 | [M + H]+ | 9 | 23640 | 16400 | 5723 | 7706 | 35533 | 1419 | 860 | 77467 |
| 69.Hmmp002189 | Isorhamnetin-O-Hexoside-O-rhamnoside-O-rhamnoside-O-Hexoside | 933 | 932 | C40H52O25 | [M + H]+ | 3103 | 3856 | 2903333 | 9 | 15540 | 6090000 | 9 | 9606 | 8903333 |
| 70.Hmmp002008 | Isorhamnetin-O-Hexoside-O-rhamnoside-O-Hexoside-O-Hexoside | 949 | 948 | C40H52O26 | [M + H]+ | 9 | 9 | 268667 | 9 | 9 | 301000 | 9 | 9 | 416000 |
| 71.Hmmp002240 | Isorhamnetin-O-Hexoside-O-rhamnoside-O-glucuronate-O-Hexoside | 963 | 962 | C40H50O27 | [M + H]+ | 9 | 9 | 407333 | 9 | 9 | 592000 | 9 | 1499 | 559000 |
| 72.Lmhp002800 | 2′-Hydoxy,5-methoxy Genistein-4′,7-O-diglucoside | 641 | 640 | C28H32O17 | [M + H]+ | 9 | 9 | 90067 | 9 | 9 | 247333 | 9 | 9 | 229333 |
Note: Y, R, and P represent potato clones CIP 302281.17, CIP 302281.25, and CIP 302281.15, respectively; S1, S2, and S3 represent tuberization, tuber bulking, and tuber maturation stages, respectively, as shown in Figure 1.
Relative amounts of flavonoids in potato tubers at three stages. The number 9 represents this kind of flavonoid was not detected1–24 = Anthocyanins25 = Dihydrochalcone26–27 = Dihydroflavones28 = Dihydroflavonol29–32 = Flavan-3-ols33–53 = Flavones54–71 = Flavonols72 = Isoflavone
Ten flavonoids were ubiquitously accumulated in the flesh of R and P tubers, even when the tuber flesh of R was white or yellow at S1 and S2, with most of these flavonoids being pelargonidin (Pg) derivatives. Forty-seven flavonoids were accumulated in purple flesh of P at three stages, of which 20 flavonoids were exclusively reserved in purple flesh: two malvidin (Mv) derivatives, nine petunidin (Pt) derivatives, one delphinidin (Dp), one cyanidin (Cy) derivative, four flavonols, two flavones, and one isoflavone (Figure S1 and Table S2). Twenty-three flavonoids accumulated in white, yellow, and red flesh of R at three stages, in which seven flavonoids were specifically accumulated: five flavones and two flavan-3-ols. The numbers of common and unique flavonoids in different clones at different developmental stages are shown in Venn diagrams in Figures S1A and S1B. Twenty-four flavonoids were present in red and purple tuber flesh, in which nine flavonoids were exclusively accumulated: one Dp derivative, five flavones, two flavonols, and one dihydroflavone (Figure S1A and Table S2). We further identified 40 DAFs between white/yellow flesh and red flesh of R, of which 13 flavonoids showed uniquely significant accumulation in red flesh with |Log2(fold change)| from 1.62 to 12.25: five anthocyanins (Pg, Dp, and Cy derivatives), two flavan-3-ols, four flavonols, and two flavones (Figure S1B and Table S2).
The composition or general structure of flavonoid compounds differed depending on the potato cultivar and developmental stage, indicating that the pigmentation of potato tubers could be influenced by these metabolites. Several kaempferol, apigenin, dihydrokaempferol, eriodictyol, and quercetin derivatives and all the six Pg derivatives were more abundant in red flesh. The common monomeric precursors (flavan-3-ols) of the proanthocyanidins—afzelechin and epiafzelechin—showed high accumulation in the red flesh of the red clone at S3. The accumulation of Pn, Cy, Mv, Pt, and Dp led to the dark purple appearance of the P clone during tuber development; the high accumulation of Pg derivatives and with Cy and Pn derivatives in red flesh of R led to the red coloration at the S3 stage (Figure 2).
Figure 2.
Heatmap of metabolites of flavonoid synthesis pathway in tuber flesh of three potato clones at three developmental stages
The pathway was built based on the KEGG pathway and references. Each colored cell represents the values of each compound ion normalized by converting into log10 unit according to the color scale (average value of three biological replicates×three clones×three stages, n = 9). Api, apigenin; Cy, cyanidin; Dp, delphinidin; Eri, eriodictyol; gen, gentiobioside; glu, glucoside; gluc, glucopyranoside; hex, hexoside; Kae, kaempferol; Lut, luteolin; Mv, malvidin; Nar, naringenin; neo, neohesperidoside; pen, pentoside; Pg, pelargonidin; Pn, peonidin; Que, quercetin; rha, rhamnoside; rham, rhamnopyranoside; rut, rutinoside; sop, sophoroside; PAL (phenylalanine ammonia-lyase); 4CL (4-coumarate-CoA ligase); C4H (trans-cinnamate 4-monooxygenase); CHI, chalcone isomerase; F3H, flavonoid 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; F3′5′H, flavonoid 3′5′-hydroxylase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase; FLS, flavonol synthase; FNS, flavone synthase; LAR, leucoanthocyanidin reductase; ANR, anthocyanin reductase. See also Table 1.
Transcriptome analysis of the tuber flesh of three potato clones at three stages
About 71.69%–88.58% of the clean reads were mapped to the reference genome. Out of 16,420 identified DEGs, between 548 and 4,706 were upregulated, while between 686 and 4,596 genes were downregulated (Figure S2). Differential Gene Ontology (GO) enrichment analysis focused on three major categories: molecular functions, cellular components, and biological processes, with p values <0.05. The carbohydrate derivative binding, anion binding, and ADP binding in the molecular function; thylakoid, plastid envelope, and chloroplast envelope in the cellular component; and response to stimuli, response to stress, and response to chemicals in the biological processes were the top three terms with the lowest p values in each category (Figure S3 and Table S3).
We used systems biology approach by employing weighted gene co-expression network analysis (WGCNA) to investigate the gene regulatory network of flavonoid synthesis in potato tubers.44 This analysis resulted in 22 distinct modules (Table S4). The analysis of the modules revealed that three modules (grey60, greenyellow, and royalblue) were highly and positively correlated with most of the anthocyanins, flavonols, dihydroflavonols, flavones, dihydroflavones, flavan-3-ols, and dihydrochalcones, whereas two modules (green and red) had a high negative correlation (Figures 3A–3C). The results indicated that the genes within these modules may be involved in synthesizing flavonoids in tuber flesh during development.
Figure 3.
Weighted gene co-expression network analysis (WGCNA)
(A-C) Module–flavonoid relationship. A row and column respectively indicate a module and a specific flcvonoid compound in potato clones (details in Table 1). Each cell’s value represents the correlation coefficient between a module and the flavonoid and is displayed based on the color scale on the right. p values are shown in parentheses in the center of each cell. The red or green arrows indicate the modules with higher positive or negative correlation with flavonoid compounds.
(A) module-anthocyanin relationships; (B) module-flavone relationships; (C) module-flavonol, flavan-3-ols, and dihydrochalcone relationships. See also Tables 1 and S4.
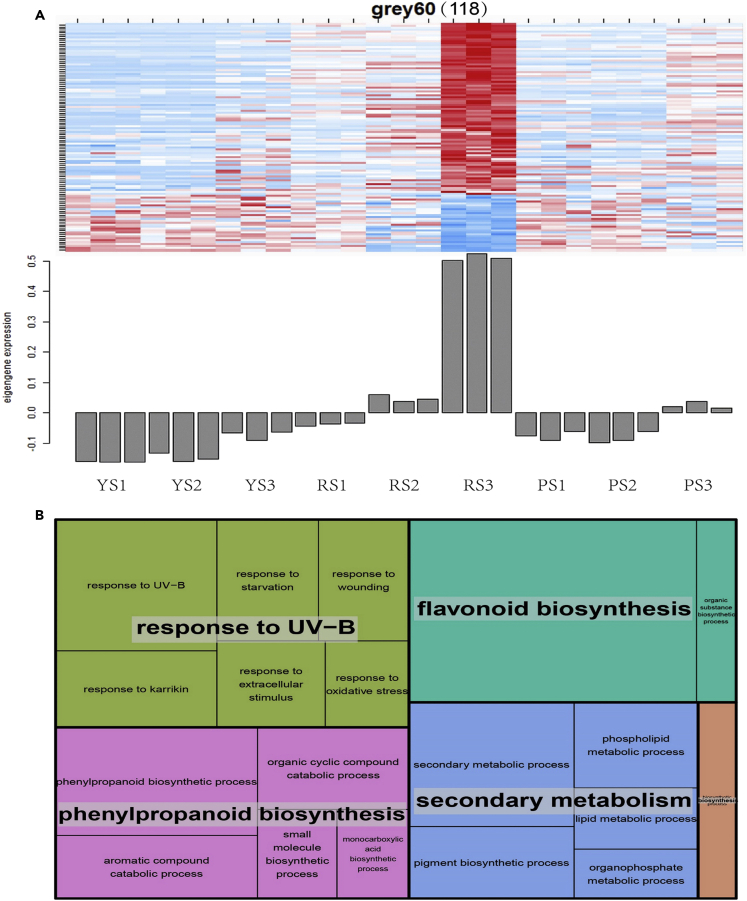
The three gene sets (grey60 modules 118 genes, greenyellow 556 genes, and royalblue modules 63 genes) were positively correlated with flavonoids; of these, a significant correlation was particularly found between the grey60 module and flavonoid metabolites in the red flesh of R (r2 > 0.86, p < 1e-08), such as all Pg derivatives, apigenin, luteolin, and kaempferol derivatives (Figure 3). These genes were highly upregulated or downregulated in the red flesh of R according to kME (eigengene-based connectivity) value (Figure 4A), whereas the genes in greenyellow module were more closely related to the flavonoid metabolites in the purple flesh of P (Figure S4A). Enrichment analysis of GO terms revealed that for genes co-expressed in the grey60 module, “response to UV-B” as well as the biosynthesis of “flavonoid” and “phenylpropanoid” was significantly enriched biological processes (Figure 4B and Table S5). In contrast, the “DNA recombination”, “organelle organization”, and “mitochondrial transmembrane transport” biological processes were significantly enriched in the greenyellow module, while in the royalblue module, significantly enriched genes were mostly involved in “heterocycle metabolism”, “regulation of developmental process”, and “organic cyclic compound metabolism” (Figures S4A and S4B, Table S5).
Figure 4.
Expression profiles, functions of genes in grey60 module, which is positively correlated with flavonoid compounds
(A) Eigengene expression values and heatmap based on FPKM of each gene of grey60 module. The eigengene value of the module or gene is shown on the y axis; the x axis indicates the type of potato clones (Y, R, and P) and the three developmental stages (S1, S2, and S3). The number of genes in grey60 module is indicated at the top.
(B) Results of the analysis of enriched GO terms for the grey60 module as visualized by the “TreeMap” view of REVIGO. Common colors represent groupings based on parent GO terms, and each rectangle is a single cluster representative for the non-redundant GO term. The representatives are joined into “superclusters” of loosely related terms, visualized with different colors. The size of the rectangles reflect the p value. See also Tables S4 and S5.
Gene networks were further constructed and visualized by WGCNA and Cytoscape for identifying key hub regulatory genes within these modules.45 In this case, an edge weight ≥0.15 was selected, with each node and the connecting lines (edges) representing a gene and co-expression relationships, respectively. For the analysis, it was assumed that key regulatory genes were more likely to be found in hub genes having most connections within the network.
In the grey60 module network, most genes were flavonoid-related pathway genes, while StCHI (Soltu. DM.05G022280) was the hub gene having most edges (26), followed by StPAL (Soltu. DM.05G026870) with 23 edges. In green-yellow module network, Soltu. DM.10G014550, of unknown function, had most edges (113), followed by a DNA repair protein (Soltu. DM.07G017450) with 103 edges. In royalblue module network, histone deacetylase HDT2-like gene Soltu. DM.04G014010 and myosin-11-like gene Soltu.DM.06G001190 are the hub genes with the highest number of edges (9 edges and 8 edges, respectively) (Table S6).
The green module (1237 genes) was strongly negatively correlated with most of the flavonoids, and the red module was also negatively correlated with most flavonoids (Figure 3). The StAN1 (Soltu. DM.10G020850)—anthocyanin biosynthesis activator of the potato tuber—and the cofactor StbHLH1 (Soltu. DM.09G019660)18 were assigned into the red and green modules, with kME value −0.8 and −0.9 (Table S4). The F3′5′H (Soltu. DM.11G020990) and DFR (Soltu.DM.02G024900) genes within the anthocyanin biosynthesis pathway were assigned into the green module, with kME value −0.9, showing they were highly upregulated in the colored flesh of R and P (Table S4). The anthocyanin cofactor StbHLH1 was also in the green module with a strong negative correlation, suggesting the expression of F3′5′H and DFR pathway genes was more likely to be associated with StbHLH1, which was only highly upregulated in colored flesh. The expression level was consistent with an anthocyanin accumulation trend.
In the red module network, high expression of genes with positive kME values was only observed for white and yellow tuber flesh of Y, whereas the genes with negative kME values were highly upregulated in the flesh of R and P at three stages. “DNA recombination” and “ribosome biogenesis” represented the main biological processes in which these genes were mainly involved (Figure S4). The anthocyanin activator StAN1 was assigned to the red module with a significantly negative correlation (kME = − 0.8) (Table S4). Unlike F3′5′H, DFR and StbHLH1 expression levels, StAN1 was highly upregulated in the flesh of R and P at three stages (Table S4), suggesting StAN1 was activated at the initial developmental stage of the tuber in colored cultivars, to participate in regulating flavonoid biosynthesis.
Expression of flavonoid synthesis-related genes in three potato clones
We identified 8 differentially expressed PALs: 2 of them (Soltu. DM.09G005690 and Soltu. DM.05G026870), named PAL-1 and PAL-2, were in module grey60, which were highly expressed in colored flesh, suggesting the two PALs were involved in flavonoid biosynthesis (Figure 5). Five 4CLs and 3 C4Hs were identified in our dataset; the relatively high expression of 4CL-4, C4H-2, and C4H-3 in tuber flesh ensured the production of precursor compounds for the phenolic and flavonoid biosynthesis. CHS and CHI are key enzymes controlling flavonoid biosynthesis. In the present study, we identified two CHSs and two CHIs, belonging to module grey60, which were highly expressed in tuber flesh of R and P at three developmental stages, and the highest expression level was present in red flesh of R at S3. In RS3, a consistently high expression of F3H results in the production of dihydrokaempferol. DFR and FLS use dihydrokaempferol, dihydroquercetin, or dihydromyricetin as the substrate for respectively producing colored anthocyanins and colorless flavonols such as kaempferol, quercetin, or myricetin, with the level of FLS and DFR expression influencing the amount of both compounds.46 The kaempferol, quercetin, and myricetin derivatives in RS3 and PS3 were highly accumulated in our datasets (Figure 2 and Table 1); however, the FLS transcription levels identified did not correlate with flavonol content. The high expression of UFGT, ANS, and DFR in RS3 led to a high accumulation of Pg derivatives. In addition, the F3′5′H also showed high expression in RS3, leading to the production of Cy derivatives, thus the flesh red color. We also found the monomeric precursors—flavan 3-ols (in this case afzelechin and epifzelechin)—were highly accumulated in RS3, which were formed via the reduction of leucopelargonidin by LAR or via the reduction of pelargonidin by ANR.47 2 ANR genes were identified, but no LAR was detected in our work, suggesting the other genes were involved.
Figure 5.
The expression profile of major flavonoid-related genes in tuber flesh of Y, R, and P potato clones at three developmental stages
Each row and column represent a gene and a library, respectively. The blue to red color of cells represent the log2-transformed FPKMs normalized by row scale. The original FPKM values are shown in the middle of each cell. Gene IDs were presented in parentheses and “Soltu.DM.” was omitted for clarity. ∗PAL (phenylalanine ammonia-lyase); 4CL (4-coumarate-CoA ligase); C4H (trans-cinnamate 4-monooxygenase); CHS (chalcone synthase); CHI (chalcone isomerase); F3H (naringenin 3-dioxygenase); DFR (dihydroflavonol 4-reductase); F3′H (flavonoid 3′-monooxygenase); F3′5′H (flavonoid 3′, 5′-hydroxylase); AOMT (flavonoid 3′,5′-methyltransferase); ANS (anthocyanidin synthase); UFGT (UDP-glucose flavonoid 3-O glucosyltransferase); GST (glutathione transferase); MATE (Multidrug and toxic extrusion); ABC transporter (ATP-binding cassette transporter); FLS (flavonol synthase); ANR (anthocyanidin reductase).
In purple flesh, the F3′H was weakly expressed, but high luteolin, eriodictyol, myricetin, quercetin, Dp, Pt, Mv, and Pt derivatives contents were accumulated, suggesting the highly expressed F3H and F3′5′H genes, coupled with the high expression of UFGT, ANS, DFR, and AOMT, were mainly responsible for affecting flavonoid biosynthesis by routing flux through dihydromyricetin, dihydroquercetin, and luteolin. This led to the increased accumulation of Dp, Pt, Mv, and Pt derivatives, finally resulting in the dark-purple color in tuber flesh of P at stage S3. GSTs, MATEs, and the ABC proteins act as carrier proteins for sequestering anthocyanin and proanthocyanidin (PA) within vacuoles.7 We found two GSTs, one MATE, and two ABC transporters in module grey60 which could very likely participate in flavonoid synthesis. Also, a plasma membrane H + -ATPase energizes TT12—the MATE transporter for the import of PAs into the vacuole.48 In our dataset, we identified an F-type H+-ATPase (Soltu. DM. 02G026440) in the grey60 module highly expressed in tuber flesh of R at stage three, which could be responsible for this function (Figure 5).
Flavonoid synthesis regulated by transcription factors
In the grey60 module, one R2R3-MYB (Soltu.DM.05G004700) and one MYB-related (Soltu.DM.12G023200) TF showed high intra-modular connectivity with kME value > 0.9, highly expressed in colored flesh. In the greenyellow module, 22 genes were identified as TFs or transcriptional regulators (TRs), and six of them showed higher absolute values of kME (>-0.8), including zinc finger TFs (C2C2 and C3H), STK, SNF2, and ARID families. In the green module, 36 genes were assigned as TFs or TRs, and 12 of them showed high absolute values of kME (>0.8), of which two C2C2-GATA TFs and three SET TRs were in the positive kME sets. They were all downregulated in colored flesh, whereas seven TFs, including bHLH1, C2H2, BPC, MYB-related, HB TFs, and PHD, GNAT TRs, were all upregulated in colored flesh. In the red module, 14 out of 42 TFs or TRs showed a high absolute value of kME (>0.8). In the positive kME sets, TFs upregulated in white or yellow flesh of Y included a bHLH showing high expression with the Fragments Per Kilobase of transcript per Million fragments mapped (FPKM) >100; one MADS-MIKC (AtAGL1-like) and one RWP-RK were highly expressed with an FPKM >20 (Figure 6). The anthocyanin activator StAN1 was highly expressed in tuber flesh of R and P at three stages. The HB-other TF (Soltu.DM.04G013310) displayed an overlapping expression pattern with StAN1, but the FPKM was relatively low (between 1 and 4) (Figure 6). Thus, apart from the reported anthocyanin-related StAN1 and StbHLH1 TFs, other TFs in the same module displayed similar expression patterns to those of StAN1 and StbHLH1, thereby suggesting their potential involvement in regulating the synthesis of flavonoids in potato tuber flesh.
Figure 6.
The expression profile of transcription factors in tuber flesh of Y, R, and P potato clones at three developmental stages in grey60, greenyellow, royalblue, green, and red modules
The blue to red color of cells represent the log2-transformed FPKMs. “Soltu.DM.” in Gene ID was omitted for clarity. The TFs StAN1 (Soltu.DM.10G020850) in red module and StbHLH1 (Soltu.DM.09G019660) in green module were shown in red color. See also Table S4.
MYBs identified that repress anthocyanin biosynthesis
The grey60 module was the main one associated with the flavonoid compounds (highest positive correlation; Figure 3). Within this module, co-expressed genes were significantly enriched for “flavonoid biosynthesis” (Figure 4B), and most of the flavonoid-related genes were classified into the module grey60 (Figure 5 and Table S4). Thus, the genes in grey60 module were further analyzed, with results indicating that 34 genes were correlated with an edge weight ≥0.15 (Table S6). Only two TFs—an R2R3-MYB TF (Soltu.DM.05G004700) and an MYB-related (Soltu.DM.12G023200) TF—were included in this network, with kMEs of 0.92 and 0.91, respectively, and a high degree of connectivity suggesting they are the hub genes in this module (Figure 7A). Based on phylogenetic analysis, Soltu.DM.05G004700 clustered with AtMYB3 and AtMYB4, which belong to group 4 of the MYB proteins in Arabidopsis49; therefore it was annotated as StMYB3. The deduced protein of Soltu. DM.05G004700 shares high identity with PhMYB27, FaMYB1, and AtMYB3. Two conserved domains in the C terminus of the subgroup 4 type MYB TFs, C1() and C2/EAR motif (LxLxL or DLNxxP), which are required for their repressive activity, were found in StMYB3 (Figures 7B and 7C). By BLAST analysis of Soltu.DM.12G023200, it was completely identical with the reported StMYBATV, which was characterized as an anthocyanin negative regulator in potato cell culture,42,50 and therefore Soltu.DM.12G023200 was annotated as StMYBATV.
Figure 7.
Network of correlated genes within the grey60 module and the sequence analysis of StMYB3
(A) Network was visualized by Cytoscape. The hub TFs StMYB3 and StMYBATV are shown by large circles with red arrows. The green to red color of nodes represents the degree of connectivity, the green to red color of edges represents the weight above a threshold of 0.1.
(B and C) Phylogenetic tree and amino acid sequence alignment of StMYB3, AtMYB3, AtMYB4, PhMYB27, FaMYB1, AmMYB308, SlTHM27, and PhMYB4; the StMYB3 shares high identity with PhMYB27 and FaMYB1. A black color indicates conserved and partially conserved residues while grey and greyish white boxes respectively show the R2 and R3 domains. The red boxes show the conserved motifs C1 and C2 in the C-terminus. The following GenBank accession numbers are used: AmMYB308 (P81393.1), AtMYB3 (NP 564176.2), AtMYB4 (NP 195574.1), PhMYB4 (F1B281.1), SlTHM27 (NP_001233975.1), FaMYB1 (AAK84064.1), PhMYB27 (AHX24372.1), and StMYB3 (MW768000). See also Table S6.
The CDS of StMYB3 was cloned and transformed into Nicotiana tabacum by transient Agrobacterium infiltration assays. Based on transient color assays, it was found that anthocyanin production was not induced in tobacco leaves after infiltration of StMYB3 alone, but an intense red pigmentation was observed following infiltration with StAN1-R1 as an anthocyanin activator.18 On the other hand, co-infiltration of StAN1-R1 with StMYB3 resulted in the absence of pigmentation after 4 days (Figure 8A). After 7 days, accumulated anthocyanin with dark red coloration was observed in the leaf infiltrated with StAN1-R1, while very light red was seen in leaf tissue infiltrated with combined StAN1-R1 and StMYB3 (Figure 8A). These results indicate that StMYB3 was able to suppress anthocyanin accumulation. To further verify the suppressive ability of StMYB3 to block anthocyanin biosynthesis, StMYB3 was infiltrated into the young leaves of transgenic tobacco that constitutively overexpresses the anthocyanin activator StAN1-R1. The results showed that anthocyanin accumulation was repressed in transgenic tobacco leaves that transiently overexpressed StMYB3 (Figure 8B).
Figure 8.
Functional analysis of the potato StMYB3 and StMYBATV TFs
(A) Infiltration with StMYB3 only, a combination of StAN1-R1 and StMYB3 or a combination of StAN1-R1 with an empty vector (EV) was carried out on the upper left side, on the lower left side and on the right side of a tobacco leaf respectively; Pictures were taken four days (i) and seven days after infiltration (ii).
(B) Transgenic tobacco constitutively overexpressing StAN1-R1 was used to transiently overexpress StMYB3 (right side of the tobacco leaf) and EV was used as control (left side of the tobacco leaf), the infiltrated tissues were indicated by white arrow.
(C) Co-infiltration of StAN1-R1 and StMYB3 (left side of tobacco leaf) and StAN1-R1 (right side of the tobacco leaf).
(D) StMYBATV and StMYB3 prevent the activation of the promoter StDFR-LUC, by co-filtration with StAN-R1 and StbHLH1 using dual luciferase assay. Error bars (SE) are provided for four biological replicates. Data are represented as mean ± SE. One-way ANOVA was used to determine the statistical significance of results, with means compared by Least Significant Difference (LSD) to identify significant differences at p < 0.05. Different letters (a, b, c, etc.) above the bar indicate significantly different means.
(E) StbHLH interacts with StMYB3 and StMYBATV in yeast. Yeast cells were grown on -2SD (-Leu-Trp) or -4SD (-Ade-His-Leu-Trp) medium.
(F) Bimolecular fluorescence complementation assays show the interactions between StMYB3/StMYBATV and StbHLH1 proteins in epidermal cells of N. benthamiana, respectively. (i) Positive and negative controls. (ii) The interaction between StMYB3 and StbHLH1 proteins. (iii) The interaction between StMYBATV and StbHLH1 proteins. Scale bars, 20 μM.
Using cDNA of tuber flesh of R at the S3 stage, the CDS of StMYBATV was cloned. Based on phylogenetic analysis, it was found that StMYBATV belongs to the R3-MYB group, which included TRICHOMELESS (TLC1), ENHANCER OF TRY AND CPC (ETC), TRIPTYCHON (TRY), AtMYBL2, and CAPRICE (CPC) that negatively influence anthocyanin synthesis.51 To further confirm the repressive function of StMYBATV, it was transformed into N. tabacum by transient assays as well. The transient color assays in tobacco leaves showed accumulated anthocyanin, with dark red observed in the leaf infiltrated with StAN1-R1, while light red was seen in leaf tissue infiltrated with combined StAN1-R1 and StMYBATV after 7 days of infiltration (Figure 8C).
To assess how StMYB3 and StMYBATV influenced gene expression in the anthocyanin pathway, prom-3-StDFR, a promoter of the DFR gene in potato, was fused with the luciferase reporter.18 Co-infiltration of StAN1-R1 with StbHLH1 activated the promoter, while co-infiltration of StMYB3 or StMYBATV inhibited the gene activity. The repressive function of either StMYB3 or StMYBATV alone or a combination of StMYB3 and StMYBATV showed no significant difference (Figure 8D). These results further confirmed that StMYB3 and StMYBATV negatively regulate the synthesis of anthocyanins.
It was previously reported that in potato tubers, StAN1 acts together with StbHLH1 for regulating anthocyanin biosynthesis.18 The bHLH binding domain was also found in StMYB3 and StMYBATV. To further investigate whether StMYB3 and StMYBATV interacted with StbHLH1, the StMYB3/StMYBATV and StbHLH1 recombination vectors were constructed and used for yeast two-hybrid and BiFC assays. The results showed that on selective medium, only cells expressing both MYB repressors and the StbHLH1 factor showed growth (Figure 8E). Via Agrobacterium-mediated transformation of Nicotiana benthamiana leaves, the epidermal cells of StMYB3-StbHLH1 and StMYBATV-StbHLH1 combinations in N. benthamiana displayed fluorescence signals (Figure 8F).The result suggests that both repressors StMYB3 and StMYBATV can inhibit the synthesis of anthocyanin by interacting with StbHLH1 to compete with anthocyanin activator StAN1during tuber development (Figure 9).
Figure 9.
Schematic diagram of the anthocyanin regulation network in potato tuber flesh
(A) In red clone R, the R2R3-MYB activator—StAN1—was activated at S1 stage, the bHLH1 was activated at S3 stage and combined with StAN1 to activate the expression of the anthocyanin biosynthesis genes. Feedback inhibition is provided by an R2R3-MYB repressor StMYB3 and an R3-MYB repressor StMYBATV, which were also activated at S3 stage and inhibit gene expression (such as DFR) by combining with bHLH1 to prevent the synthesis of anthocyanins. MBS indicates MYB-binding sites.
(B) In yellow clone Y, TFs StAN1, StbHLH1, StMYB3, and StMYBATV were not activated at three stages.
qRT-PCR and RNA-seq datasets correlations
The RNA sequencing (RNA-seq) expression profile was validated by qPCR using gene-specific primers of eight differentially expressed structural genes and TFs. The results showed that despite variations between RNA-seq data and qPCR results in terms of relative expression of selected genes, linear regression analysis (y = 0.9782x −0.3605) suggested a high correlation (R2 = 0.8838), thus highlighting that the two analytical methods were consistent (Figure S5).
Discussion
Anthocyanins are responsible for the red to purple pigmentation of potatoes, which is attractive to consumers. They not only confer resistance against biotic and abiotic stresses but also offer potential health benefits to humans, with some examples being intervention in several chronic diseases and protection against cardiovascular diseases as well as some cancers.1,52 As a result, flavonoid production in colored potatoes has attracted increased interest, especially for understanding the underlying regulatory mechanism. The regulatory network involved in sprout pigmentation from three colored potato cultivars has been investigated based on integrated analysis of transcriptome and metabolome, and 119 genes that strongly correlated with 22 anthocyanins in sprouts were identified.39 In our current study, metabolomic and transcriptomic differences among potato tubers of three different colored potato clones at three developmental stages were comprehensively analzyed, seeking to identify candidate genes that may be involved in flavonoid biosynthesis in potato tuber during tuber development phases.
Based on WGCNA analysis, three modules (grey60, greenyellow, and royalblue) were identified to be positively correlated with flavonoid biosynthesis and further analyzed, whereas several other modules were found to have a negative correlation with flavonoid biosynthesis. There were 118 genes in grey60, which showed a higher correlation (r2 > 0.9) with Pg derivatives in the anthocyanin class; apigenin, luteolin, eriodictyol derivatives, and isosaponarin in the flavone class; and dihydrokaempferol, kaempferol, Quercetin-3-O-α-L-rhamnopyranoside, (−)-epiafzelechin, afzelechin, and phloretin 2′-O-glucoside in the flavonol and flavan-3-ol classes. In grey60, we found 13 kinds of biosynthetic pathway genes, such as two PALs, two CHSs, two CHIs, one F3H, one ANS, one AOMT, one UFGT, two GSTs, and one MATE. One ATPase and two ABC transporters had the highest expression levels in the red flesh of R, as well as increased expression levels in purple flesh of P at three stages, suggesting they are more likely to be responsible for the red pigmentation (Pg derivatives) and other flavonoid compound biosynthesis mentioned above. The identified FLS and ANR showed very low expression in the red flesh of R, at the S3 stage (FPKM 1 and 3–5, respectively), suggesting other candidate genes were involved because high accumulations of kaempferol, (−)-epiafzelechin, and afzelechin were detected in red flesh (Figure 2 and Table 1).
The transcriptional control of genes within the anthocyanin pathway by the MBW complex as well as the latter’s influence on anthocyanin production and distribution has been widely studied.12,53 The MBW complex is encoded by the TFs StAN1, StbHLH1, and StWD40 in potato tubers and leaves.17,18,41,54
StAN1 is the key regulatory gene regulating anthocyanin production in potatoes. However, diagnostic markers generated for functioning StAN1 alleles (StAN1-777 and StAN1-816) cannot be used to predict potato pigmentation variations successfully. The StAN1 promoter sequence organization is likely critical for anthocyanin synthesis control, necessitating the creation of additional diagnostic markers.55 Increased expression of StAN1-like and PAL was associated with deeper colors of the potato, with StAN1-like being predominantly expressed in potato tubers, stems, and roots; its expression was however dramatically enhanced in Purple Meigui 2 and Red Meigui 3 potato tubers.56,57 The anthocyanin activator StAN1 was assigned to the red module with a negative relationship (kME > - 0.8). Interestingly, high expression of StAN1 was observed in white/yellow flesh and colored flesh of R and P at three stages. The anthocyanin pathway genes F3′5′H, DFR, and TF StbHLH1 were classified into the green module with a negative elationship (absolute kME >0.88); they were highly expressed in all colored flesh and present in the highest expression in purple flesh of P at S3, which, combined with their relationship with anthocyanin metabolite analysis, suggest the two pathway genes and StbHLH1 may be crucial for regulating late biosynthetic genes (LBGs) to affect anthocyanin biosynthesis, by regulating the flux through purple-blue delphinidin.
Soltu.DM.10G025870, annotated as serine/threonine-protein kinase SRPK3 by the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, showed a high correlation with StAN1 in this module (with edge weight ≥0.15). It has been reported that, as a plant-specific serine/threonine protein kinase, the sucrose non-fermenting 1-related kinase (SnRK) is involved in abscisic acid (ABA)-dependent and ABA-independent signaling pathways that regulate plant adaptation to abiotic stress58; the relationship between ABA and flavonoid biosynthesis in potato tuber is worthy of further investigation.
The MBW complex is reported to be sensitive to feedback repression by MYB repressors.59 FaMYB1, the first anthocyanin-related R2R3-MYB repressor was identified in strawberry. Although it is highly expressed during the red ripening stages of the fruit, it may actually repress transcription during later fruit maturation stages to balance anthocyanin content.60 A conserved network of transcriptional activators (DPL, PHZ, and AN1) and repressors (MYB27 and MYBx) regulate anthocyanin pigmentation in eudicots. PhMYB27 is orthologous to FaMYB1 and was shown to be repressed under bright light but highly expressed under shaded conditions. Furthermore, being part of the MBW complex, it acts on PhAN1 as well as genes involved in anthocyanin biosynthesis through its C-terminal EAR motif, thereby suppressing transcription. PhAN1 is also targeted PhMYBx, competitive R3-MYB repressor which inhibits PhAN1 activity; overall, the ability of the MBW complex to inhibit and activate PhAN1, PhMYB27, and PhMYBx provides both reinforcement and feedback regulation.59,61 Similarly, in tomato, the R3-MYB repressor SlMYBL2 was significantly upregulated in Pro35S: BrTT8 seedlings by the MBW complex itself under high light. However, at the same time, it could also bind competitively with the MYB interaction domain of BrTT8 to inhibit anthocyanin biosynthesis, leading to restricted anthocyanin production.62
Identification of these repressors and elucidation of the network of activators and repressors regulating the synthesis of flavonoids in other plant species have raised renewed interest. In our work, StMYB3 and StMYBATV were identified as highly correlated with flavonoid accumulation, and they are the orthologous genes of PhMYB27 and PhMYBx. It was assumed that they may act in the negative feedback regulation mechanism on anthocyanin biosynthesis as it is the case for PhMYB27 and PhMYBx.59 StMYB3 and StMYBATV are highly expressed at the tuber pigmentation stage (Figure 6); this is consistent with what was observed for MYB repressors involved in anthocyanin biosynthesis in other species. In contrast, while potato white skin displays high expression of StMYB113, StMYBA1, and StAN1, the bHLH partners' expression levels appeared to be constrained, thus indicating that bHLHs could be critical for regulating the synthesis of anthocyanins in potato.57 We hypothesize that StMYB3 and StMYBATV could form complexes with StbHLH1 to prevent the MBW complex from being formed, thus preventing it from interacting with the R2R3-MYB activator, leading to restricted anthocyanin production (Figure 9). This hypothesis is in line with our results that the interaction of StMYB3 and StMYBATV with the anthocyanin promoting StbHLH1, the inhibition of anthocyanin production in tobacco (N. tabacum), and the inhibition of DFR promotor activation in Nicotiana benthemiana leaves where StMYB3, StMYBATV, and StAN1 with StbHLH1 were transiently co-expressed (Figure 8). The StMYB3 and StMYBATV may be activated by the MBW complex itself and/or other unknown proteins under certain developmental stages. There was no significant difference of StAN1 expression in the flesh of R at three stages, but the StbHLH1 transcript was significantly higher in red flesh of R at the S3 stage than that in yellow flesh of R at the S2 stage, suggesting StMYB3 and StMYBATV might be activated by StbHLH1 or other unknown proteins, rather than StAN1, which requires further investigation.
In conclusion, we explored the regulatory network connected to flavonoid biosynthesis using targeted metabolome and transcriptome analysis in different potato clones during development. Five modules were identified that positively or negatively correlated with flavonoid accumulation. The provided candidate genes for potato anthocyanin, flavone, and flavonol accumulation can guide future functional studies. Two MYB repressors involved in flavonoid biosynthesis during potato tuber pigmentation were functionally identified, suggesting a feedback regulatory mechanism underlying the biosynthesis of flavonoid accumulation in colored potatoes during tuber development.
Limitations of the study
This work revealed a dynamic regulatory mechanism of potato tuber pigmentation: (1) a total of 72 flavonoids were identified in pigmented flesh; (2) the candidate genes that positively or negatively regulate flavonoid biosynthesis were identified based on WGCNA analysis; (3) two MYB repressors (an R2R3-MYB StMYB3 and an R3-MYB StMYBATV) were determined engaged in feedback regulatory mechanisms. Although the function and regulatory mechanism of StMYB3 and StMYBATV on anthocyanin biosynthesis have been preliminarily determined based on yeast two-hybrid assay, transient assay, and BiFC assay, further verification of their function in potato tuber via stable genetic transformation of potato using overexpression, RNAi, and CRISPR/Cas9 systems is needed. The detailed mechanism needs to be further explored.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Agrobacterium tumefaciens strains GV3101 | Shanghai Weidi Biotechnology | AC1030S |
| Biological samples | ||
| Solanum tuberosum genotypes:CIP 302281.17, CIP 302281.25, CIP 302281.15 | This study | N/A |
| Critical commercial assays | ||
| PureLink Plant RNA Reagent Kit | Invitrogen | CAT#12183018A |
| Platinum Taq DNA Polymerase High Fidelity | Invitrogen | CAT#10966018 |
| SuperReal Pre-Mix Plus | TIANGEN | CAT#FP205 |
| Dual-Luciferase Reporter Assay System | Promega | CAT#E1910 |
| Deposited data | ||
| KEGG database | KEGG | https://www.genome.jp/kegg/pathway.html |
| Raw Data for RNA-seq | This study | GenBank: PRJNA782081 |
| Oligonucleotides | ||
| Gene specific primers for qRT-PCR analysis and full length cloning, see Table S1 | This study | N/A |
| Recombinant DNA | ||
| pNC-Cam2304-MCS35S | (Yan et al., 2019)63 | N/A |
| pGreenII 0800-LUC-prom-3-StDFR | (Liu et al., 2016)18 | N/A |
| pSAK277-StAN1-R1 | (Liu et al., 2016)18 | N/A |
| pSAK277-StbHLH1 | (Liu et al., 2016)18 | N/A |
| pNC-Cam2304-MCS35S-MYBATV | This study | N/A |
| pNC-Cam2304-MCS35S-MYB3 | This study | N/A |
| pNC-GADT7 (AD-EV, AD-StAN1, AD-StMYB3, and AD-StMYBATV) | This study | N/A |
| pNC-GBKT7 (BD-StbHLH1) | This study | N/A |
| pNC-BiFC-Enn-(StAN1/StMYB3/StMYBATV) | This study | N/A |
| pNC-BiFC-Ecn-StbHLH1 | This study | N/A |
| Software and algorithms | ||
| R v 3.6.3 | R Core Team 2020 | https://www.r-project.org/ |
| DESeq2 v1.34.0 | (Love et al., 2014)64 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| TBtools | (Chen et al., 2020)65 | https://github.com/CJ-Chen/TBtools |
| Cytoscape | (Shannon et al., 2003)45 | https://cytoscape.org |
| Other | ||
| CFX96 Touch Real-Time PCR Detection System | Bio-Rad | CFX96 |
| HiSeq 4000 Sequencing System | Metware Biotechnology | N/A |
| UPLC-ESI-MS/MS | Metware Biotechnology | N/A |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact Jiangping Bai (baijp@gsau.edu.cn).
Materials availability
Plasmids generated in this study are available from the lead contact upon request.
Experimental model and subject details
Plant material and sampling
Three potato clones (yellow clone – CIP 302281.17, red clone – CIP 302281.25, and purple clone – CIP 302281.15) derived from the same parentage were acquired from Hebei North University and grown in field conditions at Dingxi Academy of Agricultural Science, Gansu Province. The dominant flesh color of the yellow clone transformed from white to pale yellow and then to yellow over development. On the other hand, the red and purple clones, changed from white/pale yellow/red to pale purple/purple/dark purple during the three developmental stages. These were abbreviated to Y, R, and P. The tuber flesh of the three potato clones was collected at the tuberization stage (S1), tuber bulking stage (S2), and tuber maturation stage (S3) (100, 130, 170 days after sowing). The flesh tissue inside the vascular ring of five tubers was collected from each potato clone and at each time point before being pooled, with samples collected from three individual plants used as three biological replicates. After collection, liquid nitrogen was used to immediately freeze samples prior to storage at −80°C.
Method details
Widely targeted metabolic profiling and data analysis
Sample preparation and extraction
Metabolomic analyses were carried out by Metware Biotechnology Co., Ltd (Wuhan, China), as described before.66,67 In brief, after freeze-drying (Scientz - 100F, Scientz, China), each potato tuber was ground into powder at 30 Hz and for 1.5 min with the help of a mixer mill (MM 400, Retsch, Germany) containing zirconia beads. The resulting powder (100 mg) was dissolved in 1.2 mL of 70% aqueous methanol, vortexed six times (once every half an hour for 30 s) and stored overnight at 4°C. This was followed by centrifugation for 10 min at 12000 rpm and after filtration (SCAA-104, 0.22 μm pore size; ANPEL, Shanghai, China), samples were analyzed by UPLC-MS/MS.
UPLC conditions
For sample analysis with an UPLC-ESI-MS/MS system (UPLC, SHIMADZU NexeraX2; MS, Applied Biosystems 4500 Q TRAP), an Agilent SB-C18 column (1.8 μm, 2.1 mm∗100 mm) was used, with mobile phases A and B consisting of pure water with 0.1% of formic acid and acetonitrile with 0.1% of formic acid (v/v) respectively. A temperature of 40°C, a flow rate of 0.35 mL/min as well as an injection volume of 4 μL were also selected as additional parameters. A gradient elution was applied, starting with 95% of A and 5% of B. Within 9 min, 5% of A and 95% of B was selected as a linear gradient and maintained for 1min before switching to 95% of A and 5% of B within 1.1 min. This final gradient was maintained for 2.9 min. Analysis of eluted samples was subsequently performed with an ESI-triple quadrupole-linear ion trap (QTRAP) - MS.
ESI-Q TRAP-MS/MS
A triple quadruple-linear ion trap mass spectrometer (QTRAP, AB4500 Q TRAP UPLC/MS/MS System) to which an ESI Turbo Ion-Spray interface was connected, was used to acquire linear ion trap (LIT) and triple quadruple (QQQ) scans. The spray, controlled by the Analyst 1.6.3 software (ABSciex), was operated in positive and negative ion modes and under the following conditions: a source temperature of 550°C; turbo spray; ion source; a high collisionally-activated dissociation (CAD); pressures of 25, 50 and 60 psi for curtain gas (CUR) as well as ion source gases I (GSI) and II (GSII), respectively; an ion spray voltage (IS) of 5500V and −4500V for positive ion and negative ion modes respectively. Mass calibration and instrument tuning was carried out with polypropylene glycol solutions at concentrations of 10 and 100 μmol/L in QQQ and LIT modes respectively. QQQ scans were acquired as multiple reaction monitoring (MRM) experiments with nitrogen, as the collision gas, set to medium. Collision energy (CE) and Declustering potential (DP) for individual MRM transitions was done after additional optimization of CE and DP. A specific set of MRM transitions was monitored for each period depending on the eluents within this period.
Metabolomics analysis
As described in other studies, the metabolic profile was determined using the self-built database MWDB (Metware biotechnology Co., Ltd. Wuhan, China).66,67 MS data were qualitatively analyzed by comparing fragmentation patterns, retention times (RTs) as well as precursor ion (Q1) and production (Q3) values with the results obtained when using standards (Sigma-Aldrich,USA) under the same conditions. Moreover, the MRM mode was used for quantifying metabolites. For this purpose, signal strengths were first acquired by screening the ions for each metabolite through QQQ MS, with the area under chromatographic peaks subsequently indicating relative metabolite content. In addition, metabolites that were differentially produced between samples were screened and quantified using metaX software (http://metax.genomics.cn/), with an absolute log2FC (fold change) ≥ 1 and a VIP (variable importance in project) value ≥ 1 selected as thresholds for classifying metabolites as being differentially regulated between groups. VIP values were extracted from OPLS-DA result, which also include permutation plots and score plots, was obtained by using R package MetaboAnalystR. For the identified metabolites, principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) were then performed in R (www.r-project.org/) before mapping these diffierential metabolites onto the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.kegg.jp/kegg/pathway.html). All experiments were carried out with triplicate samples.
RNA extraction, library construction and sequencing
The PureLink Plant RNA Reagent Kit (Invitrogen, USA) was used as specified by the manufacturer for total RNA extraction from the tuber flesh of three potato clones at three developmental stages. RNA quality and quantity were then determined by electrophoresis (1% agarose gel) as well as with a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, USA) and a Qubit 2.0 Fluorometer (Life Technologies, USA). Using the RNA Nano 6000 Assay Kit, the integrity of the extracted RNA was also assessed on an Agilent Bioanalyzer 2100 (Agilent Technologies, USA).
Messenger RNAs (mRNAs), purified from the total RNA with the help of a Dynabeads mRNA Purification Kit (Invitrogen, USA), were fragmented before using random hexamer primers to synthesize a first cDNA strand by reverse-transciption. This was followd by the synthesis of a second strand using DNA Polymerase I, dNTPs and buffer. Double-stranded cDNAs, purified with AMPure XP beads, were repaired at the ends, poly-A-tails as well as adaptors were ligated, and fragment size selection was carried out through AMPure XP beads. Finally, cDNA libraries were generated by PCR enrichment prior to sequencing on an Illumina HiSeq platform by MetWare Biotechnology Co. Ltd (Wuhan, China).
Analysis of RNA-seq data
Adapter sequences, reads of low quality (Q ≤ 20) and those with ambiguous bases (multiple “N”) were removed to obtain clean raw reads which were subsequently used for alignment with PGSC_DM_v6.1 gene models available at Solanaceae Genomics Resource at Michigan State University (http://solanaceae.plantbiology.msu.edu/pgsc_download.shtml)68 using HISAT2 (v2.0.1). DESeq2 was then used for analyzing differentially expressed genes (DEGs) between two samples based on the raw counts, with an absolute value of log2FC (log2 fold change) > 1 and a false discovery rate (FDR) < 0.05 selected as thresholds to consider genes as being diffierentially expressed.63 DEGs were then annotated against NR (Non-Redundant Protein Sequence Database), SwissProt/UniProt Plant Proteins, COG/KOG (Cluster of Orthologous Groups of proteins), Gene Ontology (GO) and KEGG. In this case, after enrichment analysis, p values <0.05 were taken as indicative of significantly enriched KEGG pathways and GO functions. The heatmap of gene expression profile was visualized by TBtools software.65
Module construction by analyzing co-expression networks
The “weighted gene co-expression network analysis (WGCNA)” package in R44 was used and it allowed 20110 genes, with Fragments Per Kilobase of transcript per Million mapped reads (FPKM) values ≥2, to be identified from the 27 samples used for performing the co-expression network analysis. The eigengene values were calculated for each module to identify associated links with flavonoid compounds, with visualization of the interaction network conducted with Cytoscape v3.7.1 44.
Construction of expression vectors
The whole coding sequences of StMYB3 (Soltu.DM.05G004700) and StMYBATV (Soltu.DM.12G023200) were amplified using cDNAs as templates, which was obtained from the red flesh of R at the S3 stage as well as the following two pairs of designed primers: MYB3F: 5′- ATGAGAAAGCCTTGTTGTGATAACA-3′, MYB3R: 5′- CTATGGAAGTGAATTGAGATCAAGCAA-3'; MYBATVF: 5′- ATGGCTGATTTGGATAGTTCAAGCA-3′, MYBATVR: 5′- TTATTGGCTGGTGGAATTTCTTGAGT-3'. Gene amplification was achieved by PCR using Platinum Taq DNA Polymerase High Fidelity (Invitrogen, USA), with full-length fragments of both genes cloned into the plant expression vector pNC-Cam2304-MCS35S by nimble cloning.63
Transient assays to assess gene functions
Transient or dual-luciferase assays, were carried out in tobacco (N. benthamiana or N. tabacum).69,70 For this assay, four different vectors were used: a pSAK277 vector containing StAN1-R1 and StbHLH1,18 the pNC-Cam2304-MCS35S vector with StMYB3 and StMYBATV being under the control of the CaMV 35S promoter, a pGreenII 0800-LUC vector containing prom-3-StDFR, a promoter of the DFR gene in potato18 and an empty vector which acted as the control. Agrobacterium tumefaciens strains GV3101 were then transformed with each of the above contructs. Agrobacterium cultures containing the reporter cassette and cassettes containing StMYB3/StMYBATV, StAN1-R1 and StbHLH1 were mixed with a ratio of 1:3:3:3 for infiltration experiments. N. benthamiana plants were grown in greenhouse until four to six leaves were available for infiltration with Agrobacterium. Approximately 300 μL of Agrobacterium culture containing genes of interest was infiltrated into a young leaf. At three days post infiltration, leaf discs were placed into a 96-well-plate containing 50 μL of PBS (PBS) with 4 replicates from each plant. LUC and REN activities were measured with a Varioskan Flash Multimode Microplate Reader (Thermo Fisher Scientific, USA).
In order to determine the functions of StMYB3 and StMYBATV, a separate color assay was performed using two-week-old seedlings of N. tabacum. Young leaves of the seedlings were injected with activated Agrobacterium cultures (OD600 = 0.8–1) consisting of either a mixture of equivalent doses of StAN1-R1 and StMYB3/StMYBATV, or StMYB3, StMYBATV alone. Meanwhile, Agrobacterium cultures containing StMYB3 and empty vector were agroinfiltrated in fully expanded leaves of 2-week-old transgenic N. tabacum plants overexpressing the activator StAN1-R1 as previously obtained (Liu et al.18). Photographs were taken seven days after the infiltration process.
Yeast two-hybrid assays
Full-length StAN1, StMYB3, and StMYBATV were cloned into pNC-GADT7, and StbHLH1 was cloned into pNC-GBKT7. The combination of the indicated pNC-GADT7 (AD-EV, AD-StAN1, AD-StMYB3, and AD-StMYBATV) constructs with pNC-GBKT7 (BD-StbHLH1) was co-transformed into the AH109 yeast strain, and yeast cells were grown on -2SD (-Leu-Trp) medium for 5 days at 30°C. Transformed colonies were plated onto -4SD (-Leu-Trp-His-Ade) medium and grown at 30°C for 5 days to test the possible interactions. The plasmid combination of AD-StAN1 and BD-StbHLH1 was the positive control.
Bimolecular fluorescence complementation (BiFC) assays
In order to investigate planta interactions, BiFC assays were used to build constructs in the pNC-BiFC-Enn and pNC-BiFC-Ecn vectors.63 The OFR of StAN1, StMYB3, StMYBATV were cloned into pNC-BiFC-Enn, and the StbHLH1 was cloned into pNC-BiFC-Ecn. Electroporation was performed to transform these constructs into the Agrobacterium GV3101 strain (Primers list for plasmid synthesis in Table S1). Mixed Agrobacterium strains were used for infiltrating N. benthamiana leaves, with fluorescence signals detected at 488 nm using a confocal laser-scanning microscope (LSCM 800, Carl Zeiss, Germany) 48 h after infiltration.
Quantitative analysis of gene expression
For qRT-PCR analysis, eight genes related to tuber color were selected, with the primers listed in Table S1 and StEF-1α (AB061263) selected as a reference gene for template normalization.71 SuperReal Pre-Mix Plus (SYBRGreen FP205; Tiangen, Beijing, China) was used as specified by the manufacturer and qPCR reactions were performed as follows: initial denaturation at 95°C for 15 min, followed by 40 cycles, each with denaturation at 95°C for 10 s and annealing at 60°C for 30 s. Finally, melting curve analysis was carried out in a temperature range of 65–95°C. The standard curve of a cDNA serial dilution analyzed the qPCR efficiency. The quantitative data were calculated from triplicate samples using the comparative Ct (2−ΔΔ Ct) method.
Quantification and statistical analysis
In the case of qPCR and dual luciferase promoter activation assays, data from four biological replicates were provided as means (±Standard Errors). Means were compared by one-way ANOVA and differences were tested for statistical significance using the least significant difference (LSD) at 5% significance level.
Acknowledgments
The Research Program was sponsored by the National Natural Science Foundation of China (31860398), the Key Program of Gansu Natural Science Foundation (22JR5RA834), the Major S&T Special Projects of Gansu Province (22ZD6NA010), the Fuxi Talent Project of Gansu Agricultural University (Gaufx-02Y04), and the Earmarked Fund for China Agriculture Research System (CARS-09-P14). We would like to thank Pro. Richard V. Espley and Pro. Andrew C. Allan (The New Zealand Institute for Plant and Food Research Limited, New Zealand) for the suggestions and help on the paper. We also would like to thank Pro. Pu Yan (Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Sciences, China) for having provided plasmids and technical advice.
Author contributions
YL and ZL: devising the experiment, statistical analysis, and writing; YL, LW, and JZ: data collection; KLW: data analysis and revision; ZB and CS: conceptualization; JZ and JB: supervision. All authors reviewed and edited the final manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: December 29, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105903.
Supplemental information
Data and code availability
The datasets generated in this study have been deposited in the Sequence Read Archive (SRA), the accession number is listed in the key resources table.Other data supporting our findings are available within the paper and its Supplementary Information files.
References
- 1.He J., Giusti M.M. Anthocyanins: natural colorants with health-promoting properties. Annu. Rev. Food Sci. Technol. 2010;1:163–187. doi: 10.1146/annurev.food.080708.100754. [DOI] [PubMed] [Google Scholar]
- 2.Jansen G., Flamme W. Coloured potatoes (Solanum Tuberosum L.) - anthocyanin content and tuber quality. Genet. Resour. Crop Evol. 2006;53:1321–1331. doi: 10.1007/s10722-005-3880-2. [DOI] [Google Scholar]
- 3.Jaakola L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013;18:477–483. doi: 10.1016/j.tplants.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Goodman C.D., Casati P., Walbot V. A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays. Plant Cell. 2004;16:1812–1826. doi: 10.1105/tpc.022574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marinova K., Pourcel L., Weder B., Schwarz M., Barron D., Routaboul J.M., Debeaujon I., Klein M. The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+ -antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell. 2007;19:2023–2038. doi: 10.1105/tpc.106.046029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mueller L.A., Goodman C.D., Silady R.A., Walbot V. AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol. 2000;123:1561–1570. doi: 10.1104/pp.123.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao J. Flavonoid transport mechanisms: how to go, and with whom. Trends Plant Sci. 2015;20:576–585. doi: 10.1016/j.tplants.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Koes R., Verweij W., Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005;10:236–242. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Patra B., Schluttenhofer C., Wu Y., Pattanaik S., Yuan L. Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim. Biophys. Acta. 2013;1829:1236–1247. doi: 10.1016/j.bbagrm.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Hichri I., Barrieu F., Bogs J., Kappel C., Delrot S., Lauvergeat V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011;62:2465–2483. doi: 10.1093/jxb/erq442. [DOI] [PubMed] [Google Scholar]
- 11.Stracke R., Ishihara H., Huep G., Barsch A., Mehrtens F., Niehaus K., Weisshaar B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007;50:660–677. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu W., Dubos C., Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015;20:176–185. doi: 10.1016/j.tplants.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Stracke R., Jahns O., Keck M., Tohge T., Niehaus K., Fernie A.R., Weisshaar B. Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-MYB12-and MYB111-independent flavonol glycoside accumulation. New Phytol. 2010;188:985–1000. doi: 10.1111/j.1469-8137.2010.03421.x. [DOI] [PubMed] [Google Scholar]
- 15.Xu W., Grain D., Bobet S., Le Gourrierec J., Thévenin J., Kelemen Z., Lepiniec L., Dubos C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014;202:132–144. doi: 10.1111/nph.12620. [DOI] [PubMed] [Google Scholar]
- 16.Azuma A., Kobayashi S., Mitani N., Shiraishi M., Yamada M., Ueno T., Kono A., Yakushiji H., Koshita Y. Genomic and genetic analysis of Myb-related genes that regulate anthocyanin biosynthesis in grape berry skin. Theor. Appl. Genet. 2008;117:1009–1019. doi: 10.1007/s00122-008-0840-1. [DOI] [PubMed] [Google Scholar]
- 17.Jung C.S., Griffiths H.M., De Jong D.M., Cheng S., Bodis M., Kim T.S., De Jong W.S. The potato developer (D) locus encodes an R2R3 MYB transcription factor that regulates expression of multiple anthocyanin structural genes in tuber skin. Theor. Appl. Genet. 2009;120:45–57. doi: 10.1007/s00122-009-1158-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y., Lin-Wang K., Espley R.V., Wang L., Yang H., Yu B., Dare A., Varkonyi-Gasic E., Wang J., Zhang J., et al. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 2016;67:2159–2176. doi: 10.1093/jxb/erw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez A., Zhao M., Leavitt J.M., Lloyd A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- 20.Espley R.V., Hellens R.P., Putterill J., Stevenson D.E., Kutty-Amma S., Allan A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007;49:414–427. doi: 10.1111/j.1365-313X.2006.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chagné D., Lin-Wang K., Espley R.V., Volz R.K., How N.M., Rouse S., Brendolise C., Carlisle C.M., Kumar S., De Silva N., et al. An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 2013;161:225–239. doi: 10.1104/pp.112.206771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin H., Cominelli E., Bailey P., Parr A., Mehrtens F., Jones J., Tonelli C., Weisshaar B., Martin C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000;19:6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubos C., Le Gourrierec J., Baudry A., Huep G., Lanet E., Debeaujon I., Routaboul J.M., Alboresi A., Weisshaar B., Lepiniec L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008;55:940–953. doi: 10.1111/j.1365-313X.2008.03564.x. [DOI] [PubMed] [Google Scholar]
- 24.Nesi N., Debeaujon I., Jond C., Stewart A.J., Jenkins G.I., Caboche M., Lepiniec L. The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell. 2002;14:2463–2479. doi: 10.1105/tpc.004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sagasser M., Lu G.H., Hahlbrock K., Weisshaar B. A. thaliana TRANSPARENT TESTA 1 is involved in seed coat development and defines the WIP subfamily of plant zinc finger proteins. Genes Dev. 2002;16:138–149. doi: 10.1101/gad.212702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng Y., Liu L., Yuan C., Guan J. Molecular characterization of ethylene-regulated anthocyanin biosynthesis in plums during fruit ripening. Plant Mol. Biol. Rep. 2016;34:777–785. doi: 10.1007/s11105-015-0963-x. [DOI] [Google Scholar]
- 27.Ishida T., Hattori S., Sano R., Inoue K., Shirano Y., Hayashi H., Shibata D., Sato S., Kato T., Tabata S., et al. Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell. 2007;19:2531–2543. doi: 10.1105/tpc.107.052274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao Z., Jiang H., Wang S., Wang Y., Yu L., Zou Q., Liu W., Jiang S., Wang N., Zhang Z., Chen X. The MdHY5-MdWRKY41-MdMYB transcription factor cascade regulates the anthocyanin and proanthocyanidin biosynthesis in red-fleshed apple. Plant Sci. 2021;306:110848. doi: 10.1016/j.plantsci.2021.110848. [DOI] [PubMed] [Google Scholar]
- 29.An J.P., Qu F.J., Yao J.F., Wang X.N., You C.X., Wang X.F., Hao Y.J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017;4:17056. doi: 10.1038/hortres.2017.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartmann U., Sagasser M., Mehrtens F., Stracke R., Weisshaar B. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 2005;57:155–171. doi: 10.1007/s11103-004-6910-0. [DOI] [PubMed] [Google Scholar]
- 31.Yao G., Ming M., Allan A.C., Gu C., Li L., Wu X., Wang R., Chang Y., Qi K., Zhang S., Wu J. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 2017;92:437–451. doi: 10.1111/tpj.13666. [DOI] [PubMed] [Google Scholar]
- 32.Zhou H., Lin-Wang K., Wang H., Gu C., Dare A.P., Espley R.V., He H., Allan A.C., Han Y. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 2015;82:105–121. doi: 10.1111/tpj.12792. [DOI] [PubMed] [Google Scholar]
- 33.Jung C.S., Griffiths H.M., De Jong D.M., Cheng S., Bodis M., De Jong W.S. The potato P locus codes for flavonoid 3', 5'-hydroxylase. Theor. Appl. Genet. 2005;110:269–275. doi: 10.1007/s00122-004-1829-z. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y., Cheng S., De Jong D., Griffiths H., Halitschke R., De Jong W. The potato R locus codes for dihydroflavonol 4-reductase. Theor. Appl. Genet. 2009;119:931–937. doi: 10.1007/s00122-009-1100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Jung C.S., De Jong W.S. Genetic analysis of pigmented tuber flesh in potato. Theor. Appl. Genet. 2009;119:143–150. doi: 10.1007/s00122-009-1024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Jong H. Inheritance of pigmented tuber flesh in cultivated diploid potatoes. Am. Potato J. 1987;64:337–343. doi: 10.1007/bf02853595. [DOI] [Google Scholar]
- 37.Rommens C.M., Richael C.M., Yan H., Navarre D.A., Ye J., Krucker M., Swords K. Engineered native pathways for high kaempferol and caffeoylquinate production in potato. Plant Biotechnol. J. 2008;6:870–886. doi: 10.1111/j.1467-7652.2008.00362.x. [DOI] [PubMed] [Google Scholar]
- 38.Stushnoff C., Ducreux L.J.M., Hancock R.D., Hedley P.E., Holm D.G., Mcdougall G.J., Mcnicol J.W., Morris J., Morris W.L., Sungurtas J.A., et al. Flavonoid profiling and transcriptome analysis reveals new gene-metabolite correlations in tubers of Solanum tuberosum L. J. Exp. Bot. 2010;61:1225–1238. doi: 10.1093/jxb/erp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho K., Cho K.S., Sohn H.B., Ha I.J., Hong S.Y., Lee H., Kim Y.M., Nam M.H. Network analysis of the metabolome and transcriptome reveals novel regulation of potato pigmentation. J. Exp. Bot. 2016;67:1519–1533. doi: 10.1093/jxb/erv549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonar N., Liney M., Zhang R., Austin C., Dessoly J., Davidson D., Stephens J., McDougall G., Taylor M., Bryan G.J., Hornyik C. Potato miR828 is associated with purple tuber skin and flesh color. Front. Plant Sci. 2018;9:1742. doi: 10.3389/fpls.2018.01742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D'Amelia V., Aversano R., Batelli G., Caruso I., Castellano Moreno M., Castro-Sanz A.B., Chiaiese P., Fasano C., Palomba F., Carputo D. High AN1 variability and interaction with basic helix-loop-helix co-factors related to anthocyanin biosynthesis in potato leaves. Plant J. 2014;80:527–540. doi: 10.1111/tpj.12653. [DOI] [PubMed] [Google Scholar]
- 42.D'Amelia V., Villano C., Batelli G., Çobanoğlu Ö., Carucci F., Melito S., Chessa M., Chiaiese P., Aversano R., Carputo D. Genetic and epigenetic dynamics affecting anthocyanin biosynthesis in potato cell culture. Plant Sci. 2020;298:110597. doi: 10.1016/j.plantsci.2020.110597. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y., Lin-Wang K., Espley R.V., Wang L., Li Y., Liu Z., Zhou P., Zeng L., Zhang X., Zhang J., Allan A.C. StMYB44 negatively regulates anthocyanin biosynthesis at high temperatures in tuber flesh of potato. J. Exp. Bot. 2019;70:3809–3824. doi: 10.1093/jxb/erz194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian J., Han Z.Y., Zhang J., Hu Y., Song T., Yao Y. The balance of expression of dihydroflavonol 4-reductase and flavonol synthase regulates flavonoid biosynthesis and red foliage coloration in crabapples. Sci. Rep. 2015;5:12228. doi: 10.1038/srep12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hammerbacher A., Paetz C., Wright L.P., Fischer T.C., Bohlmann J., Davis A.J., Fenning T.M., Gershenzon J., Schmidt A. Flavan-3-ols in Norway Spruce: biosynthesis, accumulation, and function in response to attack by the bark beetle-associated fungus Ceratocystis polonica. Plant Physiol. 2014;164:2107–2122. doi: 10.1104/pp.113.232389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baxter I.R., Young J.C., Armstrong G., Foster N., Bogenschutz N., Cordova T., Peer W.A., Hazen S.P., Murphy A.S., Harper J.F. A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2005;102:2649–2654. doi: 10.1073/pnas.0406377102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stracke R., Werber M., Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- 50.D'Amelia V., Staiti A., D'Orso F., Maisto M., Piccolo V., Aversano R., Carputo D. Targeted mutagenesis of StISAC stabilizes the production of anthocyanins in potato cell culture. Plant Direct. 2022;6:e433. doi: 10.1002/pld3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang S., Chen J.G. Regulation of cell fate determination by single-repeat R3 MYB transcription factors in Arabidopsis. Front. Plant Sci. 2014;5:133. doi: 10.3389/fpls.2014.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crowe F.L., Roddam A.W., Key T.J., Appleby P.N., Overvad K., Jakobsen M.U., Tjønneland A., Hansen L., Boeing H., Weikert C., et al. Fruit and vegetable intake and mortality from ischaemic heart disease: results from the European Prospective Investigation into Cancer and Nutrition (EPIC)-Heart study. Eur. Heart J. 2011;32:1235–1243. doi: 10.1093/eurheartj/ehq465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramsay N.A., Glover B.J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 54.Liu Y., Tikunov Y., Schouten R.E., Marcelis L.F.M., Visser R.G.F., Bovy A. Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables: a review. Front. Chem. 2018;6:52. doi: 10.3389/fchem.2018.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strygina K.V., Kochetov A.V., Khlestkina E.K. Genetic control of anthocyanin pigmentation of potato tissues. BMC Genet. 2019;20:27–43. doi: 10.1186/s12863-019-0728-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tengkun N., Dongdong W., Xiaohui M., Yue C., Qin C. Analysis of key genes involved in potato anthocyanin biosynthesis based on genomics and transcriptomics data. Front. Plant Sci. 2019;10:603. doi: 10.3389/fpls.2019.00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Payyavula R.S., Singh R.K., Navarre D.A. Transcription factors, sucrose, and sucrose metabolic genes interact to regulate potato phenylpropanoid metabolism. J. Exp. Bot. 2013;64:5115–5131. doi: 10.1093/jxb/ert303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dey A., Samanta M.K., Gayen S., Maiti M.K. The sucrose non-fermenting 1-related kinase 2 gene SAPK9 improves drought tolerance and grain yield in rice by modulating cellular osmotic potential, stomatal closure and stress-responsive gene expression. BMC Plant Biol. 2016;16:158. doi: 10.1186/s12870-016-0845-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Albert N.W., Davies K.M., Lewis D.H., Zhang H., Montefiori M., Brendolise C., Boase M.R., Ngo H., Jameson P.E., Schwinn K.E. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell. 2014;26:962–980. doi: 10.1105/tpc.113.122069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aharoni A., De Vos C.H., Wein M., Sun Z., Greco R., Kroon A., Mol J.N., O'Connell A.P. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 2001;28:319–332. doi: 10.1046/j.1365-313x.2001.01154.x. [DOI] [PubMed] [Google Scholar]
- 61.Albert N.W., Lewis D.H., Zhang H., Schwinn K.E., Jameson P.E., Davies K.M. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 2011;65:771–784. doi: 10.1111/j.1365-313X.2010.04465.x. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y., Li Y., Li W., Hu Z., Yu X., Tu Y., Zhang M., Huang J., Chen G. Metabolic and molecular analysis of nonuniform anthocyanin pigmentation in tomato fruit under high light. Hortic. Res. 2019;6:56. doi: 10.1038/s41438-019-0138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yan P., Zeng Y., Shen W., Tuo D., Li X., Zhou P. Nimble cloning: a simple, versatile, and efficient system for standardized molecular cloning. Front. Bioeng. Biotechnol. 2019;7:460. doi: 10.3389/fbioe.2019.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 66.Chen W., Gong L., Guo Z., Wang W., Zhang H., Liu X., Yu S., Xiong L., Luo J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol. Plant. 2013;6:1769–1780. doi: 10.1093/mp/sst080. [DOI] [PubMed] [Google Scholar]
- 67.Wang A., Li R., Ren L., Gao X., Zhang Y., Ma Z., Ma D., Luo Y. A comparative metabolomics study of flavonoids in sweet potato with different flesh colors (Ipomoea batatas (L.) Lam) Food Chem. 2018;260:124–134. doi: 10.1016/j.foodchem.2018.03.125. [DOI] [PubMed] [Google Scholar]
- 68.Pham G.M., Hamilton J.P., Wood J.C., Burke J.T., Zhao H., Vaillancourt B., Ou S., Jiang J., Buell C.R. Construction of a chromosome-scale long-read reference genome assembly for potato. GigaScience. 2020;9:giaa100. doi: 10.1093/gigascience/giaa100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Espley R.V., Brendolise C., Chagné D., Kutty-Amma S., Green S., Volz R., Putterill J., Schouten H.J., Gardiner S.E., Hellens R.P., Allan A.C. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell. 2009;21:168–183. doi: 10.1105/tpc.108.059329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1:13. doi: 10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tang X., Zhang N., Si H., Calderón-Urrea A. Selection and validation of reference genes for RT-qPCR analysis in potato under abiotic stress. Plant Methods. 2017;13:85. doi: 10.1186/s13007-017-0238-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated in this study have been deposited in the Sequence Read Archive (SRA), the accession number is listed in the key resources table.Other data supporting our findings are available within the paper and its Supplementary Information files.