Summary
Aflatoxins are toxic secondary metabolites produced by some aspergilli, including Aspergillus flavus. Recently, ethanol has attracted attention as an agent for the control of aflatoxin contamination. However, as aflatoxin biosynthesis utilizes acetyl coenzyme A, ethanol may be conversely exploited for aflatoxin production. Here, we demonstrated that not only the 13C of labeled ethanol, but also that of labeled 2-propanol, was incorporated into aflatoxin B1 and B2, and that ethanol and 2-propanol upregulated aflatoxin production at low concentrations (<1% and <0.6%, respectively). In the alcohol dehydrogenase gene adh1 deletion mutant, the 13C incorporation of labeled ethanol, but not labeled 2-propanol, into aflatoxin B1 and B2 was attenuated, indicating that the alcohols have different utilization pathways. Our results show that A. flavus utilizes ethanol and 2-propanol as carbon sources for aflatoxin biosynthesis and that adh1 indirectly controls aflatoxin production by balancing ethanol production and catabolism.
Subject areas: Microbial metabolism, Mycology
Graphical abstract
Highlights
-
•
Ethanol and 2-propanol increase aflatoxin production at low concentrations
-
•
Ethanol and 2-propanol are incorporated into aflatoxin biosynthesis via acetyl-CoA
-
•
Increments in aflatoxin production by ethanol are equal to ethanol-derived aflatoxin
-
•
adh1 plays a major role in the utilization of ethanol, but not 2-propanol
Microbial metabolism; Mycology
Introduction
Aflatoxins are highly toxic fungal secondary metabolites produced by some Aspergillus species, including Aspergillus flavus and Aspergillus parasiticus. These aflatoxigenic fungi infect and contaminate crops such as maize, peanut, cotton, and tree nuts.1 In addition to posing high health risks for humans and livestock, aflatoxin contamination results in significant economic losses because of crop disposal.2 Thus, effective methods for its control are required. For this purpose, elucidation of the regulatory mechanism underlying aflatoxin production is important.
Aflatoxin is biosynthesized in at least 18 enzyme steps using 10 molecules of acetyl coenzyme A (acetyl-CoA) and 2 molecules of S-adenosylmethionine; 25 or more genes clustered in a 70-kb region on one chromosome are responsible for this process.3,4 Acetyl-CoA is produced through the oxidative decarboxylation of pyruvate via acetaldehyde and acetate, the β-oxidation of fatty acids in mitochondria and peroxisomes, and conversion from citrate.5,6 A. flavus produces mainly aflatoxin B1, the most potent naturally formed carcinogen, and a lesser amount of aflatoxin B2, a dihydro derivative of aflatoxin B1.7 Ethanol, the most familiar form of alcohol, is recognized as an inhibitor of microorganism growth and viability. High levels of ethanol increase the permeability of plasma membranes, dissipate ionic gradients across these membranes, and eliminate their electrochemical potential.8,9 These effects halt nutrient and waste exchange with the environment, leading to growth inhibition and cell death. Recently, 3.5% ethanol was reported to reduce the fungal biomass, upregulate the expression of genes related to the oxidative stress response, and downregulate the expression of aflatoxin biosynthetic cluster genes, inhibiting aflatoxin B1 production, in A. flavus strain NRRL 3357.10 These researchers found that 3–4% ethanol reduced aflatoxin B1 production and proposed that ethanol is applicable for the control of aflatoxin contamination.10
However, ethanol is also a product of alcoholic fermentation. A. flavus has enzymes involved in this fermentation pathway: pyruvate decarboxylase, which catalyzes the decarboxylation of pyruvate to acetaldehyde, and alcohol dehydrogenase, which facilitates the interconversion of acetaldehyde to ethanol.11,12 As A. flavus has genes encoding aldehyde dehydrogenase, which catalyzes the irreversible oxidation of acetaldehyde to acetate,13 it is reasonable to consider that ethanol is oxidized to acetate, which is then utilized in aflatoxin biosynthesis. Consistent with this inference, evidence suggests that ethanol is associated positively with aflatoxin production. Gupta et al.14 reported that ethanol inhibited the incorporation of [1-14C]-acetate into aflatoxin biosynthesis, likely because of the formation of acetyl-CoA from ethanol, which diluted the [1-14C]-acetate label. Bennett et al.15 reported that 10 and 100 mM ethanol increased A. parasiticus aflatoxin production in the presence of glucose.
A considerable amount of research on the incorporation of labeled acetate into aflatoxin biosynthesis has been conducted, revealing that maxima of nine and seven aflatoxin B1 carbon atoms are 13C labeled by [1-13C]- and [2-13C]-acetate, respectively.16,17,18,19 In this study, we used 13C-labeled ethanol to investigate the availability of ethanol for aflatoxin biosynthesis.
Results
Low-dose ethanol and 2-propanol increased aflatoxin production
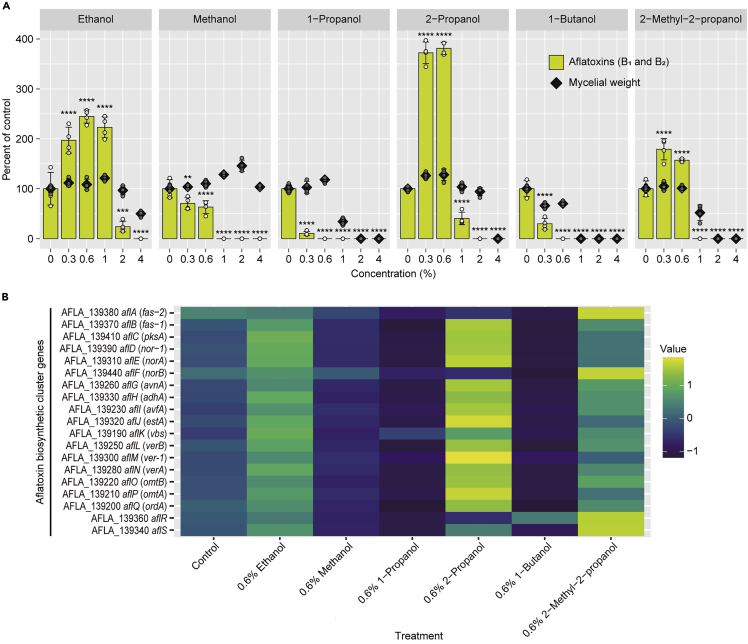
To examine the effect of low-molecular-weight alcohols on aflatoxin production, A. flavus strain IFM 47798 was cultured with the addition of 0–4% ethanol, methanol, 1-propanol, 2-propanol, 1-butanol, or 2- methyl-2-propanol for 48 h, and the aflatoxin B1 and B2 in culture supernatants and mycelial dry weights were measured (Figure 1A). Methanol, 1-propanol, and 1-butanol significantly inhibited aflatoxin production at all concentrations tested. High concentrations of ethanol, 2-propanol, and 2-methyl-2-propanol reduced aflatoxin B1 and B2 production, whereas low concentrations of these alcohols (0.3–1.0% for ethanol, 0.3–0.6% for 2-propanol, and 0.3–0.6% for 2-methyl-2-propanol) significantly increased it. At high concentrations, all alcohols except methanol reduced the mycelial weight.
Figure 1.
Effects of low-molecular-weight alcohols on the aflatoxin production and gene expression of aflatoxin biosynthetic cluster genes of Aspergillus flavus IFM 47798
(A) Effects of alcohols on aflatoxin B1 and B2 production and mycelial dry weight. Mean ± SD, n = 4. ∗∗p< 0.01, ∗∗∗p< 0.001, ∗∗∗∗p< 0.0001 versus control, ANOVA followed by Dunnett’s test.
(B) Heatmap of relative expression patterns of aflatoxin biosynthetic cluster genes with the addition of 0.6% of each alcohol, determined by RT-qPCR. mRNA levels were standardized, and averages of n = 3 are shown.
See also Figure S1.
Although to a lesser degree than in A. flavus IFM 47798, aflatoxin production in A. parasiticus NRRL 2999 was also significantly increased with 0.3% ethanol and 0.3–0.6% 2-propanol (Figure S1). 2-Methyl-2-propanol, methanol, 1-propanol, and 1-butanol showed concentration-dependent aflatoxin production inhibitory activity in this strain.
To investigate the effects of these alcohols on aflatoxin biosynthetic cluster gene expression, fungal mRNA was collected from A. flavus cultured with 0.6% of each alcohol, and the mRNA levels were determined by quantitative reverse-transcription polymerase chain reaction (RT-qPCR; Figure 1B). Ethanol, 2-propanol, and 2-methyl-2-propanol tended to increase the mRNA levels of all genes examined, whereas methanol, 1-propanol, and 1-butanol tended to decrease them, consistent with the effects of these alcohols on aflatoxin production.
13C of labeled ethanol was incorporated into aflatoxin B1
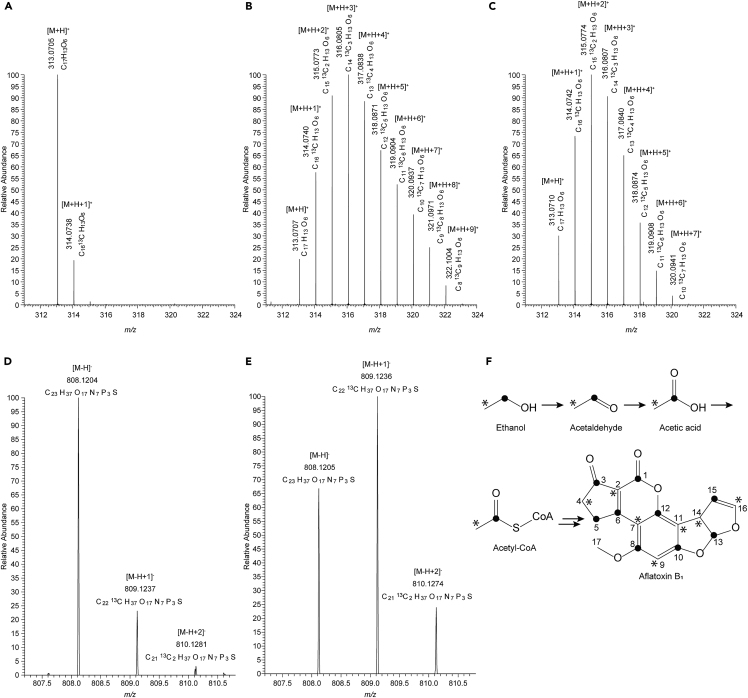
To investigate the availability of ethanol in aflatoxin biosynthesis, A. flavus IFM 47798 was cultured with [1-13C]- or [2-13C]-ethanol for 48 h, and the aflatoxin B1 and B2 produced were analyzed by liquid chromatography/mass spectrometry (LC/MS). In samples cultured with [1-13C]-ethanol, nine ion peaks from m/z 313.07 to m/z 322.10 at intervals of 1.003, corresponding to the exact mass difference between 13C and 12C, were clearly observed for aflatoxin B1 (C17H12O6); they were designated [M+H]+ to [M+H+9]+ (Figures 2A and 2B). The relative abundances of [M+H+8]+, [M+H+9]+, and the slightly detected [M+H+10]+ (m/z 323.10) were 24.9, 8.4, and 0.4, respectively (Table S1). Considering that naturally occurring 13C is present at an abundance of approximately 1.1%, the abundance of [M+H+10]+ was explainable by the natural 13C contributions from [M+H]+ to [M+H+9]+ (see Table S1 for calculation). [M+H+9]+ was much more abundant than estimated by the natural 13C contribution. These results indicate that up to nine carbon atoms of aflatoxin B1 can be 13C labeled by [1-13C]-ethanol. Similarly, in samples cultured with [1-13C]-ethanol, nine ion peaks from m/z 315.09 to m/z 324.12 at intervals of 1.003 were observed for aflatoxin B2 (C17H14O6; Figures S2A and S2B). The application of the same calculation as for aflatoxin B1 indicated that up to nine carbon atoms of aflatoxin B2 can be 13C labeled by [1-13C]-ethanol.
Figure 2.
[1-13C]-ethanol was incorporated into aflatoxin B1 biosynthesis via acetyl-CoA
(A–C) Mass spectra of aflatoxin B1 extracted from control culture and cultures supplemented with [1-13C]- and [2-13C]-ethanol, respectively.
(D and E) Mass spectra of acetyl-CoA extracted from A. flavus mycelia of the control culture and culture supplemented with [1-13C]-ethanol, respectively. The observed mass and predicted molecular formula are shown above each peak.
(F) Predicted pathway of 13C incorporation from labeled ethanol to aflatoxin B1. • and ∗ indicate 13C derived from 13C of [1-13C]- and [2-13C]-ethanol, respectively.
In samples cultured with [2-13C]-ethanol, seven ion peaks appeared for aflatoxin B1 and were designated [M+H]+ to [M+H+7]+ (Figure 2C). The relative abundances of [M+H+6]+, [M+H+7]+, and slightly detected [M+H+8]+ were 14.7, 3.9, and 0.4, respectively (Table S2). The abundance of [M+H+8]+ was reasonable, considering the natural 13C contributions of [M+H]+ to [M+H+7]+, but that of [M+H+7]+ was much greater than estimated (Table S2). For aflatoxin B2, seven ion peaks designated [M+H]+ to [M+H+7]+ were identified, and the [M+H+8]+ peak was not observed (Figure S2C). These results indicate that up to seven carbon atoms of aflatoxin B1 and B2 are 13C labeled by [2-13C]-ethanol.
Acetyl-CoA was extracted from the fungal mycelia cultured with [1-13C]-ethanol and analyzed by LC/MS. [M-H+1]- was more abundant than [M-H]-, indicating that the 13C of [1-13C]-ethanol was incorporated into the acetyl moiety of acetyl-CoA, and that the 13C-labeled acetyl-CoA was used for aflatoxin B1 and B2 biosynthesis (Figures 2D–2F and S2D).
Ethanol incorporation into acetyl-CoA and aflatoxin occurred at high rates
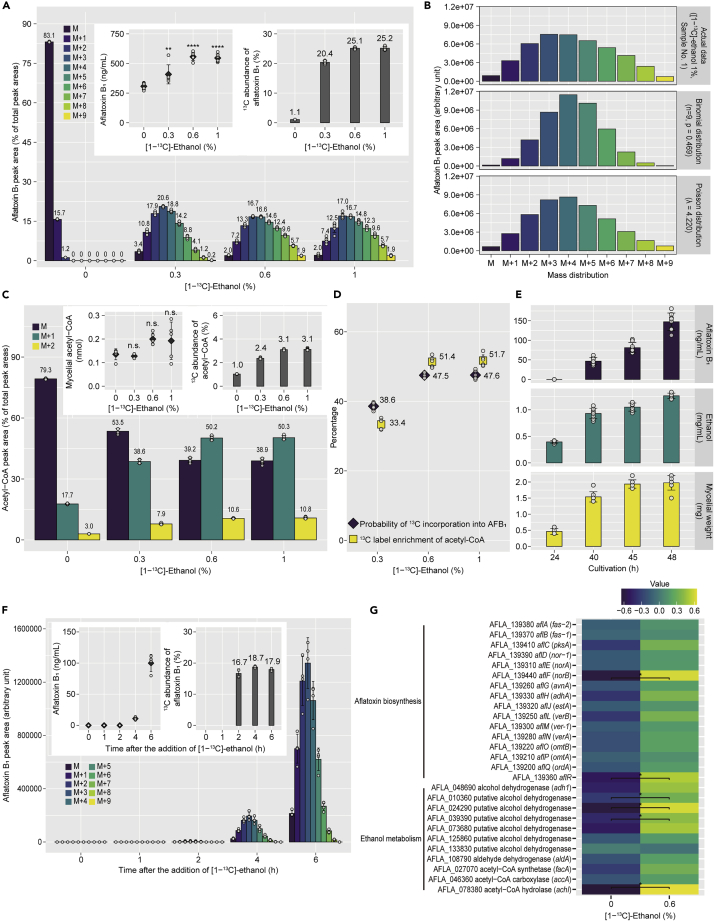
To quantitatively evaluate the incorporation of 13C of labeled ethanol into aflatoxin B1, A. flavus was cultured with 0%, 0.3%, 0.6%, and 1% [1-13C]-ethanol for 48 h, and the aflatoxin B1 produced was analyzed by LC/MS (see Table 1 for mass range used for peak area quantification). As exhibited with unlabeled ethanol (Figure 1A), 0.3–1.0% [1-13C]-ethanol increased aflatoxin B1 production (Figure 3A, upper left panel). The 13C abundance in aflatoxin B1, which contains 17 carbon atoms, was also calculated using the peak area values (Figure 3A, upper right panel). For example, when relative peak areas from [M+H]+ to [M+H+9]+ were 2.0%, 7.4%, 12.6%, 17.0%, 16.7%, 14.8%, 12.3%, 9.6%, 5.7%, and 1.9%, respectively (as for 1% [1-13C]-ethanol in Figure 3A), the 13C abundance was 25.2% [(1 × 0.074 + 2 × 0.126 + 3 × 0.17 + 4 × 0.167 + 5 × 0.148 + 6 × 0.123 + 7 × 0.096 + 8 × 0.057 + 9 × 0.019) ÷ 17 × 100 = 25.2].20 Other isotopes, such as 2H and 18O, had limited abundance. [1-13C]-Ethanol at 0.3–1.0% also increased aflatoxin B2 production; the amount of aflatoxin B2 produced was approximately one-twelfth that of aflatoxin B1 (Figure S3A, upper left panel). With 0.3–1.0% [1-13C]-ethanol, the 13C abundance in aflatoxin B2 was 22.0–24.5%, comparable to that in aflatoxin B1 (Figure S3A, upper right panel).
Table 1.
Masses and mass ranges used for target ion quantification.
| Compound | Formula | Ion name | Calculated mass (m/z)a | Mass range for quantification (m/z)b |
|---|---|---|---|---|
| Aflatoxin B1 | C17H12O6 | [M+H]+ | 313.0707 | 313.0691–313.0723 |
| C1613CH12O6 | [M+H+1]+ | 314.0740 | 314.0724–314.0756 | |
| C1513C2H12O6 | [M+H+2]+ | 315.0774 | 315.0758–315.0790 | |
| C1413C3H12O6 | [M+H+3]+ | 316.0807 | 316.0791–316.0823 | |
| C1313C4H12O6 | [M+H+4]+ | 317.0841 | 317.0825–317.0857 | |
| C1213C5H12O6 | [M+H+5]+ | 318.0874 | 318.0858–318.0890 | |
| C1113C6H12O6 | [M+H+6]+ | 319.0908 | 319.0892–319.0924 | |
| C1013C7H12O6 | [M+H+7]+ | 320.0941 | 320.0925–320.0957 | |
| C913C8H12O6 | [M+H+8]+ | 321.0975 | 321.0959–321.0991 | |
| C813C9H12O6 | [M+H+9]+ | 322.1009 | 322.0993–322.1025 | |
| Aflatoxin B2 | C17H14O6 | [M+H]+ | 315.0863 | 315.0847–315.0879 |
| C1613CH14O6 | [M+H+1]+ | 316.0895 | 316.0881–316.0913 | |
| C1513C2H14O6 | [M+H+2]+ | 317.0930 | 317.0914–317.0946 | |
| C1413C3H14O6 | [M+H+3]+ | 318.0964 | 318.0948–318.0980 | |
| C1313C4H14O6 | [M+H+4]+ | 319.0997 | 319.0981–319.1013 | |
| C1213C5H14O6 | [M+H+5]+ | 320.1031 | 320.1015–320.1047 | |
| C1113C6H14O6 | [M+H+6]+ | 321.1064 | 321.1048–321.1080 | |
| C1013C7H14O6 | [M+H+7]+ | 322.1098 | 322.1082–322.1114 | |
| C913C8H14O6 | [M+H+8]+ | 323.1132 | 323.1116–323.1148 | |
| C813C9H14O6 | [M+H+9]+ | 324.1165 | 324.1149–324.1181 | |
| Aflatoxin G1 | C17H12O7 | [M+H]+ | 329.0656 | 329.0640–329.0672 |
| C1613CH12O7 | [M+H+1]+ | 330.0689 | 330.0672–330.0706 | |
| Aflatoxin G2 | C17H14O7 | [M+H]+ | 331.0812 | 331.0795–331.0829 |
| C1613CH14O7 | [M+H+1]+ | 332.0846 | 332.0829–332.0863 | |
| Acetyl-CoA | C23H37O17N7P3S | [M-H]- | 808.1174 | 808.1186–808.1234 |
| C2213CH37O17N7P3S | [M-H+1]- | 809.1208 | 809.1216–809.1264 | |
| C2113C2H37O17N7P3S | [M-H+2]- | 810.1242 | 810.1254–810.1302 |
Aflatoxins were detected as hydrogen adducts in positive mode; acetyl-CoA was detected as hydrogen loss in negative mode.
Calculated using Xcalibur Qual Browser software (Thermo Fisher Scientific).
Peak areas of the target ions were quantified by setting the detection mass range in Xcalibur Qual Browser software.
Figure 3.
Added [1-13C]-ethanol was utilized for acetyl-CoA and aflatoxin B1 biosynthesis at high rates
(A) Percentages of peak areas of aflatoxin B1 produced by A. flavus cultured with [1-13C]-ethanol; aflatoxin B1 concentrations and 13C abundance are shown in the inset panels. Mean ± SD, n = 6. ∗∗p< 0.01, ∗∗∗∗p< 0.0001 versus control, ANOVA followed by Dunnett’s test.
(B) Mass distributions generated by binomial and Poisson distribution models with estimated parameters; upper, actual data from 1% [1-13C]-ethanol–treated sample #1; middle, peak areas calculated by binomial distribution with estimated p and n = 9; lower, peak areas from [M+H]+ to [M+H+9]+ calculated by Poisson distribution with estimated λ.
(C) Percentages of peak areas of acetyl-CoA extracted from A. flavus cultured with [1-13C]-ethanol; acetyl-CoA quantities and 13C abundance are shown in the inset panels. Mean ± SD, n = 6. n.s., not significant, ANOVA followed by Dunnett’s test.
(D) Probability of 13C incorporation into aflatoxin B1 and 13C label enrichment of acetyl-CoA, estimated by binomial regression and expressed as mole percent excess (MPE),21 respectively. Mean ± SD, n = 6.
(E) Time-courses of A. flavus aflatoxin B1 production, ethanol production, and mycelial growth. Mean ± SD, n = 8.
(F) Peak areas of aflatoxin B1 collected after the addition of 0.6% [1-13C]-ethanol at 24 h cultivation; aflatoxin B1 concentrations and 13C abundance are shown in the inset panels. Mean ± SD, n = 4.
(G) Heatmap of relative gene expression patterns, determined by RT-qPCR 6 h after the addition of ethanol at 24 h cultivation. mRNA levels were standardized, and averages of n = 8 are shown. ∗p< 0.05, unpaired t test followed by the two-stage step-up procedure of Benjamini, Krieger, and Yeku to control the false discovery rate at 0.1.
See also Figure S3.
When the contributions from natural isotopes are ignored and only 13C from labeled ethanol is considered, the mass distribution of aflatoxin B1 can be computed by the binomial distribution and Poisson distribution, the limiting case of binomial distribution.22 The peak area values from [M+H]+ to [M+H+9]+ of aflatoxin B1 were fitted to the binomial and Poisson distribution models, and the parameters were estimated. The mass distributions predicted by the two models with estimated parameters were strikingly close to the actual data (Figure 3B), supporting the assumption of binomial behavior of 13C incorporation from [1-13C]-ethanol into aflatoxin B1. The probability estimated with the binominal distribution model, taken as the probability of 13C incorporation into aflatoxin B1, was 38.6–47.6% (Figure 3D). Thus, this proportion of the aflatoxin B1 produced was likely [1-13C]-ethanol derived (158, 265, and 260 mg/mL for 0.3%, 0.6%, and 1% [1-13C]-ethanol, respectively). The increments in aflatoxin B1 caused by [1-13C]-ethanol (101, 250, and 239 mg/mL for 0.3%, 0.6%, and 1% [1-13C]-ethanol, respectively) were comparable to the amounts of [1-13C]-ethanol–derived aflatoxin B1, suggesting that the entire increase in aflatoxin B1 was attributable to the added ethanol.
The mycelial acetyl-CoA of A. flavus cultured with 0–1% [1-13C]-ethanol was extracted and analyzed by LC/MS.[1-13C]-ethanol did not significantly affect the amount of acetyl-CoA (Figure 3C, upper left panel), but resulted in an increase in the 13C abundance to 2.4–3.1% (Figure 3C, upper right panel). Based on the ratio of the peak area values of [M-H+1]– and [M-H]–, the 13C label enrichment in acetyl-CoA was calculated to be 33.4–51.7% MPE (Figure 3D).21 The probability of 13C incorporation into aflatoxin B1 (38.6–47.6%) was comparable to the 13C label enrichment of acetyl-CoA, indicating that the rates of 13C incorporation into intracellular acetyl-CoA and aflatoxin B1 biosynthesis were consistent (Figure 3D).
A. flavus was cultured with 0.3–1.0% [2-13C]-ethanol, and the aflatoxin B1 and mycelial acetyl-CoA produced were analyzed. The 13C abundance in aflatoxin B1 was increased to 15.4–20.5%, less than with [1-13C]-ethanol, consistent with the finding that up to seven carbon atoms can be 13C-labeled by [2-13C]-ethanol (Figure S3B). For acetyl-CoA, the increase in 13C abundance was equivalent to that observed with [1-13C]-ethanol (Figure S3C). The probability of 13C incorporation into aflatoxin B1 was 37.5–49.4%, equivalent to that with [1-13C]-ethanol and comparable to the 13C label enrichment of acetyl-CoA (Figure S3D). Thus, there was no difference between the availability of [1-13C]-ethanol and [2-13C]-ethanol for acetyl-CoA and aflatoxin production.
To investigate the incorporation of 13C in a shorter time, 0.6% [1-13C]-ethanol was added 24 h after inoculation, when aflatoxin production had not started, and the aflatoxin B1 in the culture supernatant was analyzed 0, 1, 2, 4, and 6 h later. From the onset of aflatoxin production, the 13C abundance in aflatoxin B1 reached 16.7% and remained equivalent at 4 and 6 h (Figures 3E and 3F), indicating the ethanol is used for aflatoxin biosynthesis at a constant rate immediately after its addition. Fungal mRNA was collected from A. flavus exposed to ethanol for 6 h, and gene expression was examined by RT-qPCR. The gene expression of the aflatoxin production regulator aflR, aflatoxin biosynthetic enzyme aflF, and putative alcohol dehydrogenases AFLA_010360, AFLA_024290, and AFLA_039390 was significantly increased compared with the controls (Figure 3G), suggesting that ethanol stimulates the regulatory mechanisms of ethanol metabolism and aflatoxin production within a short time.
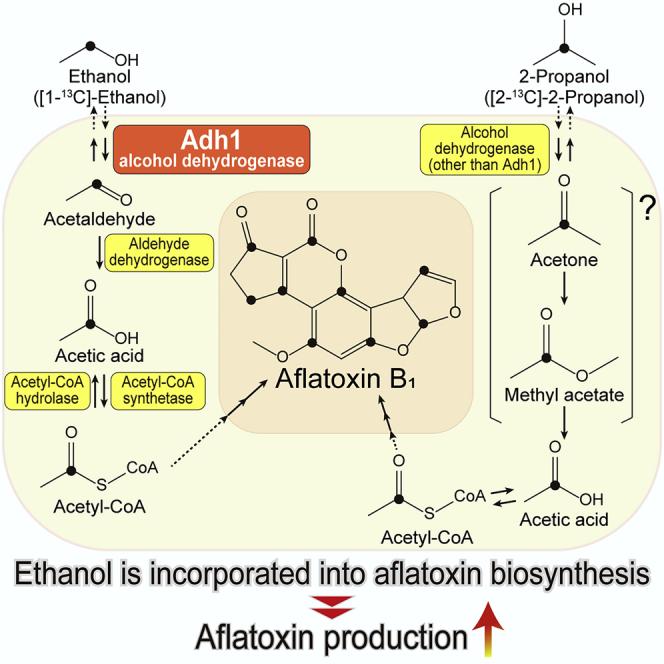
Alcohol dehydrogenase adh1 was involved in ethanol incorporation and aflatoxin production
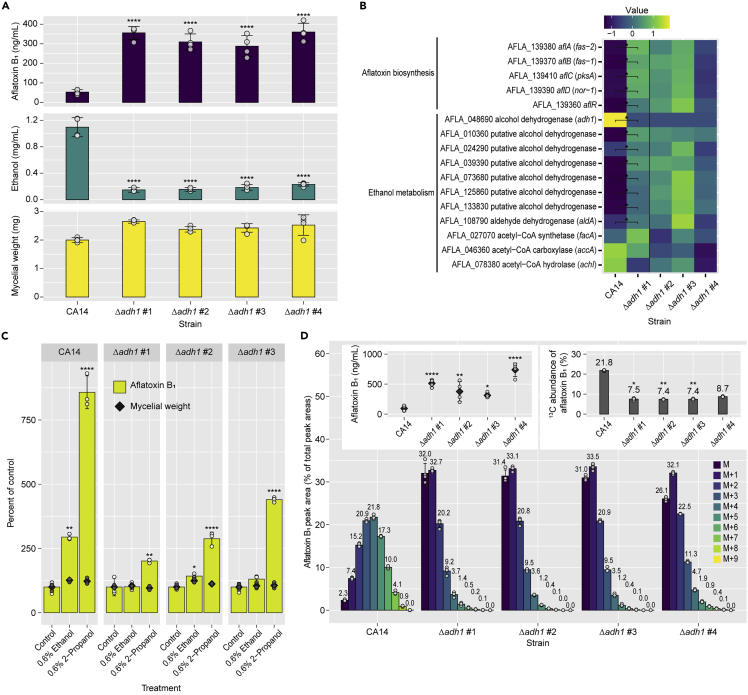
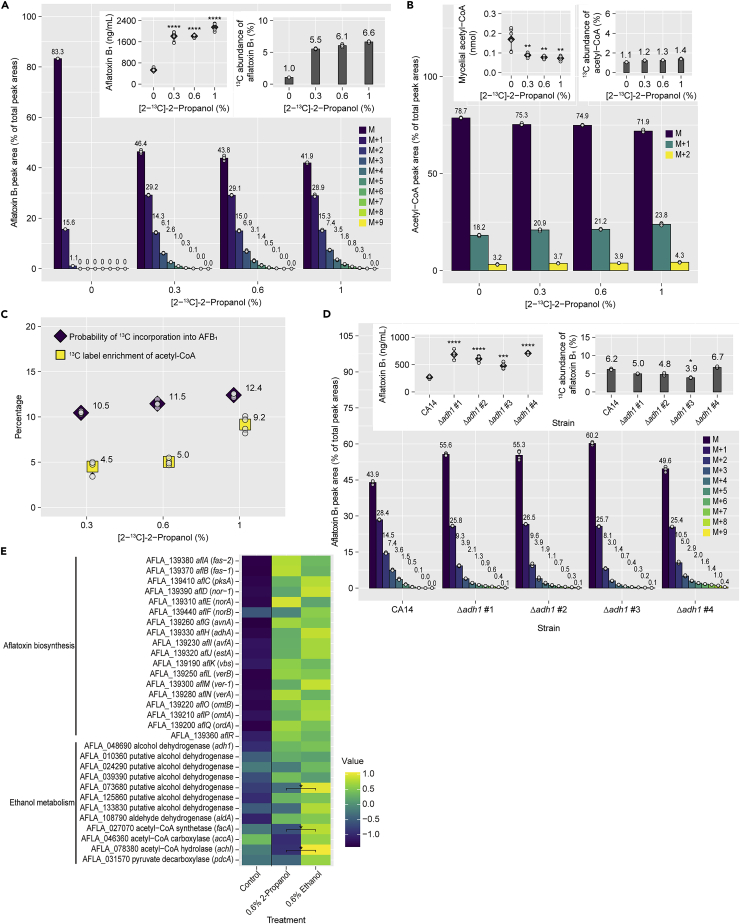
Among the putative alcohol dehydrogenase genes in A. flavus, adh1 encoding AFLA_048690 has the highest sequence identity (57% identity, 88% similarity) with yeast adh1, which plays a primary role in ethanol fermentation in yeast (Table S3).23 Thus, we prepared adh1 deletion mutants using the A. flavus CA14 (Δku70ΔpyrG) strain, whose aflatoxin production was increased with the addition of 0.6% and 1% ethanol, as with the IFM 47798 strain (Figures S4A–S4D). The mycelial weights of Δadh1 strains cultured for 48 h were comparable to that of the parental strain, but the ethanol produced in the supernatant was decreased (Figure 4A). Aflatoxin B1 production was significantly increased in the Δadh1 strains. Gene expression in these strains was investigated by RT-qPCR, and was comparable to that in the parental strain (Figure 4B). Consistent with the increase in aflatoxin production, the expression of aflatoxin biosynthetic cluster genes was upregulated. All putative alcohol dehydrogenase genes apart from adh1 and aldehyde dehydrogenase gene aldA (AFLA_108790) were also upregulated.
Figure 4.
The alcohol dehydrogenase gene adh1 plays primary roles in ethanol incorporation and aflatoxin production
(A) Amounts of aflatoxin B1 and ethanol in the culture supernatant and mycelial dry weight of Δadh1 strains after 48 h cultivation. Mean ± SD, n = 6. ∗∗∗∗p< 0.0001 versus CA14, ANOVA followed by Dunnett’s test.
(B) Heatmap of relative gene expression patterns, determined by RT-qPCR. Gene expression levels were standardized, and averages of n = 4 are shown. ∗p< 0.05, unpaired t test followed by the two-stage step-up procedure of Benjamini, Krieger, and Yeku to control the false discovery rate at 0.1.
(C) Effects of ethanol and 2-propanol on the aflatoxin B1 production and mycelial growth of Δadh1 strains. The control values were set to 100% for each strain. Mean ± SD, n = 3. ∗p< 0.05, ∗∗p< 0.01, ∗∗∗∗p< 0.0001 versus each control, ANOVA followed by Dunnett’s test.
(D) Percentages of peak areas of aflatoxin B1 produced by Δadh1 strains supplemented with [1-13C]-ethanol; aflatoxin B1 concentrations and 13C abundance are shown in the inset panels. Mean ± SD, n = 4. ∗p< 0.05, ∗∗p< 0.01, ∗∗∗∗p< 0.0001 versus CA14, ANOVA followed by Dunnett’s test (aflatoxin B1 concentration) or Kruskal–Wallis test followed by Dunn’s test (13C abundance).
See also Figure S4.
Δadh1 strains were incubated with 0.6% ethanol or 2-propanol, and aflatoxin B1 accumulation was quantified. Ethanol did not significantly affect aflatoxin production of Δadh1 strains #1 or #3. It increased the aflatoxin production of Δadh1 strain #2 by 1.4-fold, less than in the parental strain (2.9-fold). 2-Propanol increased aflatoxin production in the parental strain and all Δadh1 strains (Figure 4C). To confirm the involvement of adh1 in the incorporation of ethanol into aflatoxin biosynthesis, CA14 and Δadh1 strains were cultured with 0.3% [1-13C]-ethanol and the aflatoxin B1 and B2 produced were analyzed. Aflatoxin B1 and B2 production remained higher in Δadh1 strains than in parental strains (Figure 4D and S4E, upper left panel), as was the case without ethanol (Figure 4A). The mass distribution patterns of aflatoxin B1 and B2 differed markedly between the parental and Δadh1 strains and the 13C abundance in aflatoxin B1 and B2 was significantly decreased in Δadh1 strains #1–3 compared with that in the parental strain (Figure 4D and Figure S4E, upper right panel), indicating that 13C incorporation into aflatoxin B1 and B2 from [1-13C]-ethanol was impaired by adh1 gene deletion.
A. flavus incorporated 2-propanol into acetyl-CoA and aflatoxin biosynthesis
To examine whether 2-propanol, like ethanol, is incorporated into aflatoxin biosynthesis, A. flavus was cultured with [2-13C]-2-propanol and the aflatoxin B1 and B2 and mycelial acetyl-CoA produced were analyzed by LC/MS.
The significant increase in aflatoxin B1 production attributable to [2-13C]-2-propanol was confirmed (Figure 5A, upper left panel). The mass distributions indicated that the 13C of [2-13C]-2-propanol was incorporated into aflatoxin B1, and the 13C abundance in the labeled aflatoxin B1 was 5.5–6.6% (Figure 5A, upper right panel). Aflatoxin B2 production was also increased by [2-13C]-2-propanol, and the 13C abundance in aflatoxin B2 was 5.5–7.0%, consistent with that in aflatoxin B1 (Figure S5A). Unexpectedly, [2-13C]-2-propanol decreased the amount of mycelial acetyl-CoA produced significantly (Figure 5B, upper left panel). The 13C abundance in acetyl-CoA increased to 1.2–1.4% with the addition of [2-13C]-2-propanol (Figure 5B, upper right panel), indicating that the 13C of [2-13C]-2-propanol was incorporated to acetyl-CoA. The probability of 13C incorporation into aflatoxin B1 and the 13C label enrichment of acetyl-CoA were comparable to each other, but much lower than those observed for [1-13C]-ethanol (Figure 5C).
Figure 5.
2-Propanol is inefficiently incorporated into acetyl-CoA and aflatoxin biosynthesis through a different pathway from that of ethanol
(A) Percentages of peak areas of aflatoxin B1 from the culture supplemented with [2-13C]-2-propanol; aflatoxin B1 concentrations and 13C abundance are shown in the inset panels. Mean ± SD, n = 6. ∗∗∗∗p<0.0001 versus control, ANOVA followed by Dunnett’s test.
(B) Percentages of peak areas of acetyl-CoA extracted from A. flavus cultured with [2-13C]-2-propanol; acetyl-CoA concentrations and 13C abundance are shown in the inset panels. Mean ± SD, n = 6. ∗∗p< 0.01 versus control, ANOVA followed by Dunnett’s test.
(C) Probability of 13C incorporation into aflatoxin B1 and 13C label enrichment of fungal acetyl-CoA. Mean ± SD, n = 6.
(D) Percentages of peak areas of aflatoxin B1 collected from the cultures of Δadh1 strains with 0.3% [2-13C]-2-propanol; aflatoxin B1 concentrations and 13C abundance are shown in the inset panels. Mean ± SD, n = 4. ∗p< 0.05, ∗∗∗p< 0.001, ∗∗∗∗p< 0.0001 versus CA14, ANOVA followed by Dunnett’s test (aflatoxin B1 concentrations) or Kruskal–Wallis test followed by Dunn’s test (13C abundance).
(E) Heatmap of relative gene expression patterns, determined by RT-qPCR. Gene expression levels were standardized, and averages of n = 6 are shown. ∗p< 0.05, unpaired t test followed by the two-stage step-up procedure of Benjamini, Krieger, and Yeku to control the false discovery rate at 0.1.
See also Figure S5.
To examine the involvement of adh1 in the incorporation of 2-propanol into aflatoxin biosynthesis, Δadh1 strains were cultured with 0.3% [2-13C]-2-propanol and the aflatoxin B1 and B2 produced were analyzed (Figures 5D and S5B). The mass distributions of aflatoxin B1 and B2 in the Δadh1 strains resembled those in the parental strain. Consistent with this result, the 13C abundance in aflatoxin B1 and B2 did not differ between the parental and Δadh1 strains apart from Δadh1 #3, in which13C was slightly less abundant (Figures 5D and S5B, upper right panel). These results suggest that one or more alcohol dehydrogenases other than adh1 is responsible for the incorporation of 2-propanol into the metabolic pathway of aflatoxin biosynthesis.
The effects of 2-propanol and ethanol on gene expression were compared using RT-qPCR. The aflatoxin biosynthetic cluster gene expression levels did not differ between the alcohols. The expression of a putative alcohol dehydrogenase gene, acetyl-CoA synthetase gene, and acetyl-CoA hydrolase gene was upregulated in ethanol (Figure 5E).
Discussion
The use of alcohols for the development of an economical and effective method of preventing aflatoxin contamination of food and feed is promising.10 All low-molecular-weight alcohols tested in this study inhibited aflatoxin production at concentrations >2%, but ethanol, 2-propanol, and 2-methyl-2-propanol increased the aflatoxin B1 and B2 production of A. flavusIFM47798 at low concentrations. In accordance with the effects on aflatoxin production, 0.6% ethanol, 2-propanol, and 2-methyl-2-propanol increased the expression of most aflatoxin biosynthetic cluster genes. Thus, the application of these alcohols to prevent aflatoxin contamination may be counterproductive, as their non-uniform distribution on target crops could cause topical aflatoxin accumulation. We plan to investigate the applicability of methanol, 1-propanol, and 1-butanol as aflatoxin-controlling agents.
Consistent with previous reports on 13C- and 14C-labeled acetate incorporation,18,19 the C-1 and C-2 atoms of ethanol were incorporated into up to nine and seven carbon atoms, respectively, of aflatoxin B1 and B2. Considering that the branching point of aflatoxin B1 and B2 in the biosynthetic pathway is versicolorin B desaturation, and that the pathways before and after this desaturation are identical,24 the equivalence of the aflatoxin B1 and B213C incorporation patterns is reasonable. Surprisingly, the addition of 0.6% and 1% [1-13C]-ethanol increased the 13C label enrichment of acetyl-CoA to levels exceeding 50%, meaning that >50% of the intracellular acetyl-CoA was ethanol derived, despite the synthesis of acetyl-CoA in various pathways in multiple organelles, such as β-oxidation in the mitochondria and peroxisome.25 Given the biosynthetic pathway from ethanol to aflatoxin B1, the 13C label enrichment of acetyl-CoA directed to aflatoxin biosynthesis should be equal to the probability of 13C incorporation into aflatoxin B1, as the amount of aflatoxin B2 was negligible compared with that of aflatoxin B1. Although the conversion of ethanol to acetyl-CoA may occur in the cytosol and the early steps of aflatoxin biosynthesis (acetyl-CoA conversion to norsolorinic acid) have been proposed to occur in the peroxisome,25,26,27 the 13C label enrichment of acetyl-CoA in whole cell extracts was equivalent to the probability of 13C incorporation into aflatoxin B1. This finding suggests that ethanol-derived acetyl-CoA is transported uniformly throughout the cell, including to the peroxisome, and is directed in an unbiased manner to aflatoxin biosynthesis. Furthermore, the 13C abundance in aflatoxin B1 reached nearly 17%, even with the addition of [1-13C]-ethanol just 2 h before the onset of aflatoxin accumulation, suggesting that ethanol-derived acetyl-CoA spreads in the cell immediately after the addition of ethanol, and that the biosynthesis of the aflatoxin B1 molecule from acetyl-CoA is accomplished within 2 h. To elucidate the intracellular source of acetyl-CoA and the kinetics of its utilization in aflatoxin biosynthesis, it would be beneficial to visualize and continuously observe the subcellular localization of 13C-labeled acetyl-CoA derived from 13C-labeled ethanol.
In Δadh1 strains, the effects of exogenous ethanol on the increases in the amount and 13C abundance in aflatoxin B1 and B2 were largely reduced, indicating that adh1 is responsible for ethanol catabolism to acetyl-CoA and aflatoxin. Moreover, whereas ethanol production was ceased, aflatoxin biosynthetic cluster gene expression and aflatoxin B1 production were upregulated strongly in the Δadh1 strains. These results demonstrate the existence of a metabolic link between aflatoxin production and alcohol fermentation, for whichadh1 is primarily responsible. adh1 may regulate this metabolic balance by determining the fate of acetaldehyde (i.e., whether it is converted to ethanol or acetate used for aflatoxin production). adh1 gene deletion may cause the accumulation of acetate and nicotinamide adenine dinucleotide hydride (NADH), the reduced cofactor of alcohol dehydrogenases. As aflatoxin biosynthesis consumes many reducing agents in the form of nicotinamide adenine dinucleotide phosphate,3 the accumulation of NADH and acetate may lead to increased aflatoxin production. Considering that adh1 gene is expressed under conditions conducive to aflatoxin production,12 and that ethanol production occurs before aflatoxin production, A. flavus may produce ethanol via adh1 in advance, and metabolize it after the nutrient is depleted for use in aflatoxin biosynthesis.
We found that 2-propanol is also incorporated into aflatoxin biosynthesis via acetyl-CoA. As [2-13C]-2-propanol increased the 13C abundance in aflatoxin B1 and B2 in Δadh1 strains, adh1 is not primarily responsible for 2-propanol catabolism; secondary alcohol dehydrogenases appear to be responsible for the oxidation of 2-propanol to acetone in A. flavus.28 Acetone is converted to methyl acetate by fungal Baeyer-Villiger–type monooxygenase29 and then decomposed into acetate, which is used for aflatoxin biosynthesis. Consistent with this view, Bennett et al.15 reported that 10 and 100 mM acetone increased the aflatoxin production of A. parasiticus. Although the probability of 13C incorporation into aflatoxin B1 and 13C label enrichment of acetyl-CoA were consistent for each [2-13C]-2-propanol concentration, these values were smaller than those for ethanol. Thus, although the ethanol-triggered increase in aflatoxin production may have resulted from the exploitation of ethanol as a carbon source, the hypothesized exploitation of 2-propanol as a carbon source does not explain the much greater increase in aflatoxin production caused by 2-propanol. Furthermore, unlike ethanol, 2-propanol decreased the amount of acetyl-CoA. Takasaki et al.30 reported that acetyl-CoA synthetase is required for the ethanol utilization pathway to acetyl-CoA via acetate in Aspergillus nidulans. Thus, the reduced expression of the acetyl-CoA synthetase gene in 2-propanol–treated A. flavus may be related to the decreased acetyl-CoA level. Further studies are needed to determine why 2-propanol promoted aflatoxin production more strongly than did ethanol, despite the reduced amount of acetyl-CoA and similarity of aflatoxin biosynthetic cluster gene expression between the alcohols.
Whether 2-methyl-2-propanol is also incorporated into aflatoxin biosynthesis in A. flavus, and whether ethanol and 2-propanol are incorporated into aflatoxin biosynthesis in A. parasiticus, remain elusive. Additional research is needed to investigate commonalities and differences in mechanisms of action among alcohols and aflatoxigenic fungi.
Limitations of the study
In this study, we computed the probability of 13C incorporation into aflatoxin B1 by fitting the mass distribution of aflatoxin B1 to a binomial distribution model, although the Poisson distribution better approximated the real mass distribution data. This approach may have led to the oversimplification of the multitude of biosynthetic reactions that occur simultaneously in the cell. The neglect of the contribution of naturally occurring 13C in model fitting may have led to the overestimation of the probability of incorporation. The data we provide on the 13C label enrichment of acetyl-CoA e are approximate; to increase accuracy, the skew correction factor for natural 13C should be considered in the calculation of the sample tracer/tracee ratio. To examine the role of adh1 in ethanol and 2-propanol utilization, we prepared Δadh1 strains. However, the increased expression of other putative alcohol dehydrogenase genes in these strains suggests that these putative alcohol dehydrogenases complement the function of adh1. Thus, the precise determination of adh1’s function may require the combined application of other methods.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| [1-13C]-Ethanol | Cambridge Isotope Laboratories, Inc. | Cat#CLM-344 |
| [2-13C]-Ethanol | Cambridge Isotope Laboratories, Inc. | Cat#CLM-130 |
| [2-13C]-2-Propanol | Cambridge Isotope Laboratories, Inc. | Cat#CLM-4714 |
| Aflatoxins Mixture Standard Solution(B1, B2, G1, G2 each 25 μg/ml Acetonitrile Solution) | FUJIFILM Wako Pure Chemical | Cat#018-24341 |
| Alcohol Dehydrogenase | Sigma-Aldrich | Cat#A3263 |
| Aldehyde Dehydrogenase | Sigma-Aldrich | Cat#A6338 |
| Deposited data | ||
| All data and original code | Mendeley Data | https://doi.org/10.17632/gpnjw5xxm7.1 |
| Experimental models: Organisms/strains | ||
| Aspergillus flavus: IFM47798 strain | Medical Mycology Research Center, Chiba University | N/A |
| Aspergillus flavus: MAFF111229 strain | The Research Center of Genetic Resources, National Agriculture and Food Research Organization | N/A |
| Aspergillus parasiticus: NRRL2999 strain | Biological Resource Center, National Institute of Technology and Evaluation | N/A |
| Aspergillus flavus: CA14 strain: Δku70ΔpyrG | Fungal Genetics Stock Center | N/A |
| Aspergillus flavus: CA14 Δadh1 strain: Δku70ΔpyrGΔadh1::pyrG | This Paper | N/A |
| Oligonucleotides | ||
| RT-PCR primers | See Table S3 | N/A |
| Primers used for generation and verification of mutant strains | See Table S4 | N/A |
| Software and algorithms | ||
| GraphPad Prism version 9.3.1 | GraphPad software | https://www.graphpad.com/ |
| RStudio Version 1.4.1106 | RStudio, PBC | https://www.rstudio.com/products/rstudio/ |
| Xcalibur Qual Browser | Thermo Fisher Scientific | https://www.thermofisher.com/order/catalog/product/OPTON-30965 |
Resource availability
Lead contact
Further information and requests for data and resources should be directed to and will be fulfilled by the Lead Contact, Tomohiro Furukawa (furukawat795@affrc.go.jp).
Materials availability
Fungal strains generated in this study will be provided from the lead contact and may require a completed Materials Transfer Agreements.
Experimental model and subject details
Fungal strains and culture conditions
The A. flavus strain IFM 47798 (Medical Mycology Research Center, Chiba University, Chiba, Japan) was used as the experimental model throughout this study. The A. parasiticus strain NRRL 2999 (Biological Resource Center, National Institute of Technology and Evaluation, Tokyo, Japan) was used to investigate the effect of alcohol on aflatoxin production. The A. flavus strain CA14 (Δku70 ΔpyrG; Fungal Genetics Stock Center, Manhattan, KS, USA) was used as the parental strain in the construction of adh1 gene deletion mutants (Δku70 ΔpyrG Δadh1::pyrG). Spores of these strains were collected from a 1-week-old culture plate, suspended in 30% glycerol solution, and stored at −80°C. The spore suspension was inoculated into potato dextrose broth liquid medium (Difco, Sparks, MD, USA) in a 12-well microplate (2 mL/well) at a density of 5 × 104 spores/mL, and the microplate was placed at 28°C in the dark for the desired cultivation period. The incubation of the CA14 strain was conducted with uracil and uridine supplementation (1 mg/mL). Where indicated, alcohols were supplemented at the desired concentrations and cultivation timepoints. After cultivation, the culture broth was centrifuged to separate the supernatant and mycelia for subsequent analysis.
Method details
Aflatoxin and acetyl-CoA extraction
To extract the aflatoxins produced, 0.5 mL culture supernatant was mixed with an equal amount of chloroform, and the chloroform solution was collected and evaporated by air drying. The remaining residue was dissolved in 1 mL 90% aqueous acetonitrile and subjected to LC/MS analysis. For acetyl-CoA extraction, the harvested fungal mycelia were washed and lyophilized. The dried mycelia were transferred to a Lysing matrix C tube (MP Biomedicals, Irvine, CA, USA) and ground (FastPrep-24; MP Biomedicals). Trifluoroacetic acid (5%; 200 μL) was added to the cell debris, with vigorous mixing at 4°C. After centrifugation at 10,000 × g, 20 μL 25% ammonia aqueous solution was added and mixed to neutralize, and then centrifuged at 15,000 × g. The supernatant was filtered and subjected to LC/MS analysis.
LC/MS analysis
The detection and quantification of the aflatoxins and acetyl-CoA were performed with an LC-Orbitrap MS Exactive system (Thermo Fisher Scientific, Waltham, MA, USA). The system was operated in accurate-mass/high-resolution full-scan mode at ultra-high resolution (100,000 full width at half maximum at m/z 200), in positive ion mode for aflatoxins and negative mode for acetyl-CoA. Parameters with the heated electrospray interface (ESI) were: sheath gas/aux gas/sweep gas, 30/5/0 arbitrary units; capillary temperature/heater temperature, 250°C/250°C; and spray voltage, 4.0 kV (positive) or −4.0 kV (negative). The capillary, tube lens, and skimmer voltages were set with the auto-tuning function for each run sequence. Mass calibration of the instrument was performed before each run sequence using calibration solutions [positive, Pierce LTQ Velos ESI Positive Ion Calibration Solution (Thermo Fisher Scientific); negative, Pierce Negative Ion Calibration Solution (Thermo Fisher Scientific)]. Chromatographic separation was performed on a 250 × 4.6 mm i.d. Capcell pak C18 UG120 column (Osaka Soda, Osaka, Japan) at 40°C. For the aflatoxins, the column was eluted with carrier solvents consisting of 0.1% formic acid (A) and acetonitrile (B), at the flow rate of 0.45 mL/min with a linear gradient of 5–95% B to 22 min. The retention times for aflatoxins B1, B2, G1, and G2 were 18.1, 17.6, 17.5, and 16.9 min, respectively. For acetyl-CoA, the solvents were composed of 5 mM hexylamine (pH 6.3; A) and 90% methanol/10% 10 mM ammonium acetate buffer (pH 8.5; B). Elution was conducted at the flow rate of 0.45 mL/min with a linear gradient of 5–95% B to 30 min. The retention time was 23.9 min. Table 1 shows the analyte ion masses and mass range used for peak area quantification for the target ions. The total of peak area values from [M+H]+ to [M+H+9]+ was used to calculate aflatoxin concentrations. Calibration curves were determined using an Aflatoxins Mixture Standard (containing 25 μg/mL B1, B2, G1, and G2; FUJIFILM Wako Pure Chemical, Osaka, Japan).
Analysis of LC/MS data
See the Quantification and statistical analysis section below.
RT-qPCR
The sequences of the genes related to ethanol metabolism and production in A. flavus were determined from the JCVI-afl1-v2.0 genome assembly for A. flavus NRRL 3357 with database version 105.2 (http://fungi.ensembl.org/Aspergillus_flavus/Info/Index). A search for homologous protein sequences was conducted using the Saccharomyces cerevisiae proteins registered in the Uniprot database (https://www.uniprot.org/) as queries. The search was performed with Genetyx (Tokyo, Japan) software. The sequences of aflatoxin biosynthetic cluster and 18S rRNA genes were obtained from the database. Primers for RT-qPCR were designed using Primer Express software (Thermo Fisher Scientific); their sequences are provided in Table S3.
For the preparation of cDNA, lyophilized A. flavus mycelia were ground using the FastPrep-24 instrument as described above. Total RNA was extracted using Trizol reagent (Thermo Fisher Scientific) and purified with the PureLink RNA Mini Kit (Thermo Fisher Scientific). cDNA was synthesized with ReverTra Ace qPCR RT Master Mix (TOYOBO, Osaka, Japan). qPCR was conducted using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) in a final volume of 25 μL for each reaction in an QuantStudio 12K Flex Real-Time PCR system (Thermo Fisher Scientific). The mRNA levels for each gene were normalized to those of control 18S rRNA genes for each sample. Then, the mRNA levels were standardized so that the average value was 0 and the variance was 1 in all samples for each gene. Using these standardized values, a heatmap was created.
Generation of adh1 gene deletion mutants
The Δadh1 strains were prepared using the split-marker approach as described previously.31,32 The orotidine-5′-monophosphate decarboxylase (pyrG) gene, cloned from the genome of A. flavus strain IFM 47798, was introduced into the adh1 locus of the genome of A. flavus strain CA14 (Δku70 ΔpyrG) for transformant selection based on uracil/uridine auxotrophy of the CA14 strain. Figure S3C is a schematic diagram of gene deletion. Sequences of the primers used for the preparation of replacement constructs and verification of gene deletion are listed in Table S4. To generate replacement constructs, the 5′and-3′ flanking regions of the adh1 gene were amplified in a first-round PCR with the primer pairs Del_1F/Del_2R and Del_3F/Del_4R, respectively, and named amplicons 1 and 2. Next, the pyrG gene in the genome of A. flavus strain IFM 47798 was amplified with the primer pair pyrGP5F/pyrG P6R and named amplicon 3. The PCR products were purified using the QIAquick PCR purification kit (QIAGEN, Venlo, The Netherlands). In a second-round PCR, replacement constructs 1 and 2 were generated with the primer pair Del_1F/YR-R using amplicons 1 and 3 as templates, and with the primer pair PY-F/Del_4R using amplicons 2 and 3 as templates. The resulting replacement constructs 1 and 2 were introduced into the protoplasts of A. flavus strain CA14, and transformation mixture was spread onto selection plates without uracil or uridine. Candidate adh1 deletion strains were screened based on uracil or uridine autotrophy of transformants. The desired gene deletion was verified by PCR with the primer pairs Check_1F/Check_1R and Check_2F/Check 2R (Figures S3D and S3E).
Ethanol quantification
The supernatant obtained by A. flavus culture centrifugation was diluted 10-fold in water. The diluted solution (10 μL) was transferred to a tube containing 250 μL enzyme solution [0.8 mg/mL β-nicotinamide adenine dinucleotide (Sigma-Aldrich), 0.06 mg/mL alcohol dehydrogenase (Sigma-Aldrich), 0.03 mg/mL aldehyde dehydrogenase (Sigma-Aldrich), 20 mM potassium phosphate (pH 8.0)]. The oxidation of ethanol to acetic acid was accompanied by the reduction of NAD+ to NADH, which has absorption at 340 nm. Following incubation at room temperature for 30 min, absorbance at 340 nm was measured. Ethanol concentrations were determined from the standard curve of an ethanol dilution series.
Quantification and statistical analysis
Estimation of the probability of 13C incorporation into aflatoxin B1 from LC/MS data
For estimation, the presences of stable isotopes other than 13C and natural 13C was ignored due to the limited abundance of these isotopes relative to that of incorporated 13C from labeled alcohol. When the contribution of 13C from 13C-labeled alcohols alone is taken into account, the mass distribution of aflatoxin B1 should be computed using the binomial distribution formula , where parameter n is the maximum number of carbon atoms that may be 13C labeled, k is the number of 13C-labeled carbon atoms, and parameter p is the probability of success.22 The parameter n was set to 9 for the [1-13C]-ethanol– and [2-13C]-2-propanol–treated groups and to 7 for the [2-13C]-ethanol–treated group. If the probability p is small, n is large, and np is constant, the binomial distribution is approximated by Poisson distribution: , where k is the number of 13C-labeled carbon atoms, parameter λ is the average number of 13C-labeled carbon atoms. The λ is approximately equal to np. Peak area values of aflatoxin B1 from [M+H]+ to [M+H+9]+ for [1-13C]-ethanol and [2-13C]-2-propanol and from [M+H]+ to [M+H+7]+ for [2-13C]-ethanol were subjected to binomial logistic regression using the GLM function in the R statistical platform (https://www.r-project.org). The binomial probability of success (p) was estimated using the maximum likelihood method and regarded as the probability of 13C incorporation into aflatoxin B1 in this study. Poisson regression was also performed using the GLM function in R to estimate the parameter λ.
Estimation of the 13C label enrichment of acetyl-CoA from LC/MS data
The 13C label enrichment of acetyl-CoA (in MPE) was calculated using the peak area values of [M-H]– and [M-H+1]– using an equation published previously.21,33 Briefly, the background tracer/tracee ratio (TTR; [M-H+1]–/[M-H]–) was calculated from control (no 13C-labeled alcohol treatment) peak area data. Similarly, the sample TTR was calculated as [M-H+1]–/[M-H]– from 13C-labeled alcohol-treated sample peak area data. Then, TTR was calculated as TTR = sample TTR – average of background TTR. Finally, the MPE was calculated as MPE = TTR/(1 + TTR) × 100. MPE values reflect molecular enrichment, namely the percentage of molecules containing a labeled atom,33 and thus were regarded as reflecting the 13C label enrichment of acetyl-CoA.
Statistical analysis and graph creation
The data are presented as means ± standard deviations. For all quantitative data, the numbers of biological replicates (ns) used are provided in the relevant figure legends. GraphPad Prism ver. 9.3.1 (GraphPad Software, San Diego, CA, USA) was used to perform all statistical tests. Aflatoxin and ethanol quantities were compared between groups using the two-tailed Welch’s t test, and among more than two groups using one-way ANOVA with the post-hoc two-tailed Dunnett’s test. The 13C abundance in aflatoxin was compared using the nonparametric Kruskal–Wallis test with the post-hoc Dunn’s multiple comparisons test. Values of p< 0.05 were considered to be significant. Significant differences in gene expression between groups were determined using the multiple unpaired t test and the false discovery rate approach; the two-stage step-up procedure of Benjamini, Krieger, and Yeku was used with the false discovery rate set at 0.1. RStudio ver. 1.4.1106 (https://www.rstudio.com/products/rstudio/) and ggplot2 package (https://ggplot2.tidyverse.org/) were used for graph and heatmap creation. For preparation and arrangement of visual presentations, Adobe Illustrator ver.26.0.1 (https://www.adobe.com/jp/products/illustrator.html) was used.
Acknowledgments
This work was supported by JSPS KAKENHI grant number JP19K15760. The LC/MS measurements were performed at the Research Center for Advanced Analysis of NARO.
Author contributions
Conceptualization, T.F. and S.S.; Methodology, T.F., H.N., and H.E.; Software, T.F.; Formal Analysis, T.F. and H.E.; Investigation, T.F.; Resources, H.N., M.K., and S.S.; Writing – Original Draft, T.F.; Writing – Review and Editing, M.K. and S.S.; Visualization, T.F.; Supervision, M.K. and S.S.; Funding Acquisition, T.F. and S.S.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: January 23, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106051.
Supplemental information
Data and code availability
-
•
All data have been deposited at Mendeley Data and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
All original R code has been deposited at Mendeley Data and is publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Bennett J.W., Klich M. Mycotoxins. Clin. Microbiol. Rev. 2003;16:497–516. doi: 10.1128/CMR.16.3.497-516.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell N.J., Bowers E., Hurburgh C., Wu F. Potential economic losses to the US corn industry from aflatoxin contamination. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2016;33:540–550. doi: 10.1080/19440049.2016.1138545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yabe K., Nakajima H. Enzyme reactions and genes in aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 2004;64:745–755. doi: 10.1007/s00253-004-1566-x. [DOI] [PubMed] [Google Scholar]
- 4.Yu J., Chang P.-K., Ehrlich K.C., Cary J.W., Bhatnagar D., Cleveland T.E., Payne G.A., Linz J.E., Woloshuk C.P., Bennett J.W. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2004;70:1253–1262. doi: 10.1128/AEM.70.3.1253-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chanda A., Roze L.V., Kang S., Artymovich K.A., Hicks G.R., Raikhel N.V., Calvo A.M., Linz J.E. A key role for vesicles in fungal secondary metabolism. Proc. Natl. Acad. Sci. USA. 2009;106:19533–19538. doi: 10.1073/pnas.0907416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheridan K.J., Dolan S.K., Doyle S. Endogenous cross-talk of fungal metabolites. Front. Microbiol. 2014;5:732. doi: 10.3389/fmicb.2014.00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klich M.A. Aspergillus flavus: the major producer of aflatoxin. Mol. Plant Pathol. 2007;8:713–722. doi: 10.1111/j.1364-3703.2007.00436.x. [DOI] [PubMed] [Google Scholar]
- 8.Lam F.H., Ghaderi A., Fink G.R., Stephanopoulos G. Engineering alcohol tolerance in yeast. Science. 2014;346:71–75. doi: 10.1126/science.1257859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma M., Liu Z.L. Mechanisms of ethanol tolerance in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2010;87:829–845. doi: 10.1007/s00253-010-2594-3. [DOI] [PubMed] [Google Scholar]
- 10.Ren Y., Jin J., Zheng M., Yang Q., Xing F. Ethanol inhibits aflatoxin B1 biosynthesis in Aspergillus flavus by up-regulating oxidative stress-related genes. Front. Microbiol. 2019;10:2946. doi: 10.3389/fmicb.2019.02946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchis V., Vinas I., Roberts I.N., Jeenes D.J., Watson A.J., Archer D.B. A pyruvate decarboxylase gene from Aspergillus parasiticus. FEMS Microbiol. Lett. 1994;117:207–210. doi: 10.1111/j.1574-6968.1994.tb06766.x. [DOI] [PubMed] [Google Scholar]
- 12.Woloshuk C.P., Payne G.A. The alcohol dehydrogenase gene adh1 is induced in Aspergillus flavus grown on medium conducive to aflatoxin biosynthesis. Appl. Environ. Microbiol. 1994;60:670–676. doi: 10.1128/aem.60.2.670-676.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flipphi M., Mathieu M., Cirpus I., Panozzo C., Felenbok B. Regulation of the aldehyde dehydrogenase gene (aldA) and its role in the control of the coinducer level necessary for induction of the ethanol utilization pathway in Aspergillus nidulans. J. Biol. Chem. 2001;276:6950–6958. doi: 10.1074/jbc.M005769200. [DOI] [PubMed] [Google Scholar]
- 14.Gupta S.R., Prasanna H.R., Viswanathan L., Venkitasubramanian T.A. The effect of inorganic salts and some biologically important compounds on the incorporation of 1—14C acetate into aflatoxins by resting mycelia of Aspergillus parasiticus. Z. Lebensm. Unters. Forsch. 1975;157:19–22. doi: 10.1007/BF01785723. [DOI] [PubMed] [Google Scholar]
- 15.Bennett J.W., Lee L.S., Gaar G.G. Effect of acetone on production of aflatoxins and versicolorin pigments by resting cell cultures of aspergillus parasiticus. Mycopathologia. 1976;58:9–12. doi: 10.1007/BF00493586. [DOI] [PubMed] [Google Scholar]
- 16.Biollaz M., Büchi G., Milne G. Biosynthesis of the aflatoxins. J. Am. Chem. Soc. 1970;92:1035–1043. doi: 10.1021/ja00707a050. [DOI] [PubMed] [Google Scholar]
- 17.Cox R. Oxidative rearrangements during fungal biosynthesis. Nat. Prod. Rep. 2014;31:1405–1424. doi: 10.1039/C4NP00059E. [DOI] [PubMed] [Google Scholar]
- 18.Maggon K.K., Gupta S.K., Venkitasubramanian T.A. Biosynthesis of aflatoxins. Bacteriol. Rev. 1977;41:822–855. doi: 10.1128/br.41.4.822-855.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pachler K.G.R., Steyn P.S., Vleggaar R., Wessels P.L., Scott D.B. Carbon-13 nuclear magnetic resonance assignments and biosynthesis of aflatoxin B1 and sterigmatocystin. J. Chem. Soc. Perkin 1. 1976;1:1182–1189. doi: 10.1039/p19760001182. [DOI] [PubMed] [Google Scholar]
- 20.Lee W.N., Byerley L.O., Bergner E.A., Edmond J. Mass isotopomer analysis: theoretical and practical considerations. Biol. Mass Spectrom. 1991;20:451–458. doi: 10.1002/bms.1200200804. [DOI] [PubMed] [Google Scholar]
- 21.Kim I.-Y., Suh S.-H., Lee I.-K., Wolfe R.R. Applications of stable, nonradioactive isotope tracers in in vivo human metabolic research. Exp. Mol. Med. 2016;48:e203. doi: 10.1038/emm.2015.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadygov R.G. Poisson model to generate isotope distribution for biomolecules. J. Proteome Res. 2018;17:751–758. doi: 10.1021/acs.jproteome.7b00807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roze L.V., Chanda A., Laivenieks M., Beaudry R.M., Artymovich K.A., Koptina A.V., Awad D.W., Valeeva D., Jones A.D., Linz J.E. Volatile profiling reveals intracellular metabolic changes in Aspergillus parasiticus: veA regulates branched chain amino acid and ethanol metabolism. BMC Biochem. 2010;11:33. doi: 10.1186/1471-2091-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yabe K., Nakamura Y., Nakajima H., Ando Y., Hamasaki T. Enzymatic conversion of norsolorinic acid to averufin in aflatoxin biosynthesis. Appl. Environ. Microbiol. 1991;57:1340–1345. doi: 10.1128/aem.57.5.1340-1345.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roze L.V., Chanda A., Linz J.E. Compartmentalization and molecular traffic in secondary metabolism: a new understanding of established cellular processes. Fungal Genet. Biol. 2011;48:35–48. doi: 10.1016/j.fgb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boubekeur S., Camougrand N., Bunoust O., Rigoulet M., Guérin B. Participation of acetaldehyde dehydrogenases in ethanol and pyruvate metabolism of the yeast Saccharomyces cerevisiae: role of yeast ACDHs. Eur. J. Biochem. 2001;268:5057–5065. doi: 10.1046/j.1432-1033.2001.02418.x. [DOI] [PubMed] [Google Scholar]
- 27.Maggio-Hall L.A., Wilson R.A., Keller N.P. Fundamental contribution of β-oxidation to polyketide mycotoxin production in planta. Mol. Plant Microbe Interact. 2005;18:783–793. doi: 10.1094/MPMI-18-0783. [DOI] [PubMed] [Google Scholar]
- 28.Sealy-Lewis H.M., Fairhurst V. Substrate specificity of nine NAD+-dependent alcohol dehydrogenases in Aspergillus nidulans. Microbiology. 1995;141:2295–2300. doi: 10.1099/13500872-141-9-2295. [DOI] [PubMed] [Google Scholar]
- 29.Ferroni F.M., Tolmie C., Smit M.S., Opperman D.J. Structural and catalytic characterization of a fungal Baeyer-Villiger monooxygenase. PLoS One. 2016;11:e0160186. doi: 10.1371/journal.pone.0160186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takasaki K., Shoun H., Yamaguchi M., Takeo K., Nakamura A., Hoshino T., Takaya N. Fungal ammonia fermentation, a novel metabolic mechanism that couples the dissimilatory and assimilatory pathways of both nitrate and ethanol. J. Biol. Chem. 2004;279:12414–12420. doi: 10.1074/jbc.M313761200. [DOI] [PubMed] [Google Scholar]
- 31.Furukawa T., Katayama H., Oikawa A., Negishi L., Ichikawa T., Suzuki M., Murase K., Takayama S., Sakuda S. Dioctatin activates ClpP to Degrade mitochondrial components and inhibits aflatoxin production. Cell Chem. Biol. 2020;27:1396–1409.e10. doi: 10.1016/j.chembiol.2020.08.006. [DOI] [PubMed] [Google Scholar]
- 32.Gravelat F.N., Askew D.S., Sheppard D.C. In: Host-Fungus Interactions Methods in Molecular Biology. Brand A.C., MacCallum D.M., editors. Humana Press; 2012. Targeted gene deletion in Aspergillus fumigatus using the hygromycin-resistance split-marker approach; pp. 119–130. [DOI] [PubMed] [Google Scholar]
- 33.Wolfe R.R., Chinkes D.L. 2nd edition. Wiley-Liss; 2004. Isotope Tracers in Metabolic Research. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data have been deposited at Mendeley Data and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
All original R code has been deposited at Mendeley Data and is publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.