Abstract
Dolichyl monophosphates (DolPs) are essential lipids in glycosylation pathways that are highly conserved across almost all domains of life. The availability of DolP is critical for all glycosylation processes, as these lipids serve as membrane-anchored building blocks used by various types of glycosyltransferases to generate complex post-translational modifications of proteins and lipids. The analysis of DolP species by reverse-phase liquid chromatography-mass spectrometry (RPLC–MS) remains a challenge due to their very low abundance and wide range of lipophilicities. Until now, a method for the simultaneous qualitative and quantitative assessment of DolP species from biological membranes has been lacking. Here, we describe a novel approach based on simple sample preparation, rapid and efficient trimethylsilyl diazomethane-dependent phosphate methylation, and RPLC-MS analysis for quantification of DolP species with different isoprene chain lengths. We used this workflow to selectively quantify DolP species from lipid extracts derived of Saccharomyces cerevisiae, HeLa, and human skin fibroblasts from steroid 5-α-reductase 3- congenital disorders of glycosylation (SRD5A3-CDG) patients and healthy controls. Integration of this workflow with global lipidomics analyses will be a powerful tool to expand our understanding of the role of DolPs in pathophysiological alterations of metabolic pathways downstream of HMG-CoA reductase, associated with CDGs, hypercholesterolemia, neurodegeneration, and cancer.
Introduction
DolPs are lipids with a long carbon chain consisting of alpha-saturated polyisoprenoid phosphates. DolPs with 18–21 isoprene units have been described in mammals1 and are present in almost all eukaryotic membranes.2 DolP species play crucial roles as glycan carriers in cellular glycosylation reactions, where they serve as membrane-anchored acceptors for saccharides, provide sugar substrates to a broad set of different transferases, and enable the formation of complex glycan structures.2 Genetic defects in glycosylation form a large group of >165 diseases known as CDGs and result in a wide range of metabolic dysfunctions associated with defects in the process of carbohydrate attachment to proteins or lipids.3 Altered endogenous DolP levels have also been found in several other pathological and physiological conditions, including Alzheimer’s disease, dementia, and Prion disease.4−7 DolP accounts for up to 20% of the total cellular dolichol pool8,9 and is rapidly metabolized as part of the dolichol cycle. The de novo synthesis of dolichol via isoprenoid precursors in eukaryotes, archaea, and some prokaryotes is initiated by the mevalonate pathway, which is also used for the synthesis of cholesterol, heme, and ubiquinone (Figure S1). Prenylation of farnesyl diphosphate represents the committed step in the synthesis of polyprenol, which is reduced to dolichol by steroid 5-α-reductase 3 (SRD5A3). DolK then converts dolichol to DolP, which in turn serves as an acceptor for monosaccharides, including mannose. DolP-mannose (DolP-Man) forms a critical metabolic hub that channels the mannose unit into the major glycosylation pathways.10−12 Both the de novo synthesis of DolP and its recycling can adjust steady-state levels of DolP to cellular demand. DolP with 19 isoprene units has been shown to orientate perpendicularly to the plane of the lipid bilayer.13,14 Intercalation of DolP has been shown to perturb the fatty acyl chain order of phosphoglycerolipids.13,14 These biophysical properties of long-chain DolP species may facilitate membrane remodeling that occurs during the translocation of lipid-bound glycans across membranes and during membrane fusion.14,15 Furthermore, both dolichol and DolP have been reported to accumulate in membranes during aging,16 and DolP has been shown to be a rate-limiting metabolite for cellular glycosylation and thus essential for cell proliferation and development.17 Accurate identification and quantification of DolPs at the cellular level using powerful analytical techniques such as MS is therefore crucial for understanding mechanisms involved in DolP metabolism and glycosylation pathways.
So far, two different analytical platforms have been used for cellular DolP analysis, first normal-phase liquid chromatography in combination with fluorescence detection1,18 for quantification and second MS-based workflows (see, e.g., refs (18−22)) for structural characterizations. A frequently used robust quantification method for DolPs by Hennet and co-workers18 is based on the conversion of non-fluorescent DolPs into their fluorescent derivatives in a multi-step procedure, which is a labor-intensive and time-consuming process for routine analyses. Methods based on MS workflows18−22 usually involve qualitative analyses of DolPs, which are often poorly ionized and suppressed by highly abundant lipid species.23,24 To our knowledge, there are currently no methods for the simultaneous qualitative and quantitative determination of DolP species from biological membranes.
RPLC-based approaches are widely used in metabolomics and lipidomics platforms due to their versatility, robustness, and MS compatibility.25−28 RPLC-MS analysis of DolP species are, however, complicated by the following factors: (1) sensitivity; DolPs represent only a small percentage (∼0.1% in eukaryotes) of total cellular phospholipids and thus have a very low abundance; (2) chemical diversity; DolP species cover a wide range in lipophilicity with an octanol:water partition coefficient (LogP) value distribution over 14 units (from 22.4 of DolP C65 to 37.4 of DolP C105); (3) dynamic range; individual DolP concentrations vary in human cells from 10.8 μg/g in the liver to 169 μg/g in testis.29
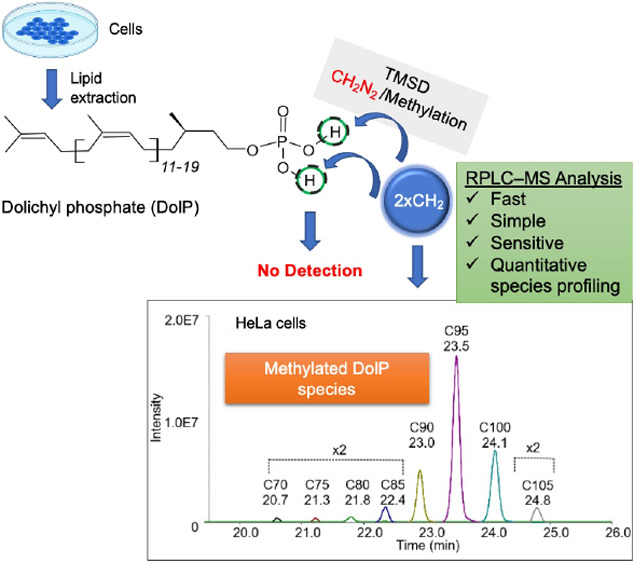
We have addressed these challenges by combining TMSD-based phosphate head group methylation with RPLC-MS analysis to simultaneously characterize and quantify DolPs from biological samples. TMSD methylation has been demonstrated to significantly increase the sensitivity of detection of various phospholipids, including the low abundance phosphoinositides or bis(monoacylglycerol-) phosphates, overcoming the previously mentioned challenges.30−37
Here, we describe for the first time that TMSD methylation of DolPs can significantly improve the sensitivity of quantification through enhanced MS ionization efficiency and specificity by RPLC separation from co-eluting peaks. The developed RPLC-MS method offers high sensitivity (LOD ∼1 pg), broad molecular coverage, and wide dynamic range. In addition, the sample preparation workflow used in this study is simple, fast, and cost- and labor-efficient and does not require additional DolP enrichment steps such as solid phase extraction columns. As a proof of principle, we applied this workflow to profile DolP from HeLa cells and S. cerevisiae and to determine DolP levels in fibroblasts from SRD5A3-CDG patients and controls.
Experimental Section
Reagents, Chemicals, and Lipid Standards
DolP mixture (#900201X, hereinafter referred to as DolP standard) was purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). Polyprenyl monophosphate (PolP) C60 standard was obtained from CymitQuimica (#48-62-1060, Barcelona, Spain). For reagents and chemicals, see Supporting Information.
Cell Culture
HeLa cells were cultured in alphaMEM medium (Sigma Aldrich) supplemented with 10% fetal calf serum (FCS, Bioconcept, Switzerland), 2 mM glutamine, and penicillin–streptomycin (#P4333, Sigma Aldrich) and incubated in a humidified 5% CO2 atmosphere at 37 °C. S. cerevisiae (strain W303-1B, derived from strain W30338) cells were cultured in YPD medium. Patient and control fibroblasts were cultured in Dulbecco’s modified Eagle’s medium (high glucose; Life Technologies) supplemented with 10% FCS (PAN Biotech, Aidenbach, Germany), 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells were harvested and prepared for further treatments, as described in Supporting Information.
Alkaline Hydrolysis and Lipid Extraction
For the analysis of DolP from HeLa, S. cerevisiae, and human fibroblasts, the sample preparation and extraction protocol by Haeuptle et al.18 was used with few modifications. Briefly, ∼1 × 106 HeLa or fibroblast cells were collected by trypsinization and washed with the medium (HeLa) or PBS (fibroblasts). S. cerevisiae cells were resuspended in 155 mM ammonium bicarbonate and homogenized using beads (0.5 mm; BioSpec Products). Lysates were diluted to 0.8 OD units (∼8 × 106 cells) per 200 μL. DolP extractions were performed in 13 × 100 mm Pyrex glass tubes. 20 pmol of PolP C60 was added to cell pellets as internal standard (IS), followed by 1 mL of MeOH and then 1 mL of H2O. Phosphate esters were hydrolyzed by adding 0.5 mL of 15 M KOH, followed by an incubation of the samples for 60 min at 85 °C. Phase partitioning was induced by adding 1 mL of MeOH and 4 mL of dichloromethane, and lipids were further hydrolyzed for 60 min at 40 °C. The lower phase was washed four times with 2.7 mL of dichloromethane/MeOH/H2O (3:48:47, v/v/v) and evaporated to dryness in a Turbovap evaporator chamber. In order to normalize sample amounts, phosphatidylcholine (PC) as bulk membrane lipid was extracted and analyzed as described.39 Subsequent MS analysis was typically performed immediately after extraction. If measurements had to be postponed, lipid extracts were stored as lipid films under nitrogen at −20 °C for a maximum of 3 days.
Methylation of DolPs
For methylation of DolP in lipid extracts or standards, the TMSD reagent was used with the necessary precautions. Dried samples were dissolved in 200 μL of dichloromethane: MeOH (6.5:5.2, v/v). Then, 10 μL of TMSD was added, and samples were incubated for 40 min at RT. Following this incubation, 1 μL acetic acid was added to neutralize an excess of TMSD.40 For RPLC–MS measurements, dried samples were reconstituted in 100 μL of MeOH. See Supporting Information for safety considerations for handling TMSD.
Nano-Electrospray Direct Infusion MS Analysis
DolP standard and PolP C60 were methylated separately, evaporated to dryness, and resuspended in 60 μL each of 50 mM ammonium acetate in MeOH. As a control, the un-methylated DolP standard was prepared in the same way, omitting the TMSD methylation step. Both methylated and un-methylated DolPs (each at a final concentration of 42 ng/ μL) were then mixed with methylated PolP C60 (6 pmol/μl) in 25 mM ammonium acetate in chloroform: MeOH (1:1 v/v) and transferred to a 96-well plate (Eppendorf, Hamburg, Germany). 5 μL of the abovementioned sample was infused by nano-electrospray (nESI) direct infusion (DI) using a TriVersa NanoMate (Advion Biosciences, Ithaca, NY, USA) coupled to a Q-Exactive Plus high-resolution MS (Thermo Scientific, MA, USA) (nESI-DIMS) for 7 min. The positive ionization voltage was set to 1.75 kV, and back pressure was 0.65 psi. MS analysis was performed in the range of m/z 930–1579 using a maximal injection time (max IT) of 3000 ms, automatic gain control for an ion target (AGC target) of 5 × 106, and resolution setting at R = 140,000 ( at m/z 200), and ion transfer capillary temperature was set to 200 °C. Data analysis was performed using LipidXplorer,41 and IS-based normalization of peak intensities of DolPs was performed. The methylation efficiency was calculated using the following equation
RPLC-MS Analysis
Separation of methylated DolPs was performed on a Dionex Ultimate 3000 UHPLC system coupled to a Q-Exactive Plus HRMS (RPLC-MS) operating in positive ionization mode. RPLC separation of methylated DolPs was carried out on a Waters CSH C18 column (1 × 150 mm, 1.7 μm) at a 0.1 mL/min flow rate, maintained at 55 °C. The mobile phase consisted of solvent A [acetonitrile:water (6:4, v/v)] and solvent B [isopropyl alcohol:acetonitrile (9:1, v/v)], buffered with 10 mM ammonium acetate and 0.1% formic acid. The following gradient profile was applied: 40% B at 0 min, 50% B in 3 min, 54% B in 9 min, 70% B in 9.1 min, 90% B in 17 min, 90% B for 27.5 min, 40% B in 27.6 min, and then 40% B was maintained up to 30 min. Lipids were detected with HRMS full scan MS (AGC target of 3 × 106, R = 70,000 (at m/z 200) and with parallel reaction monitoring mode (max IT = 100 ms, AGC target of 2 × 105, R = 17,500 (at m/z 200), isolation window 4.0 m/z, and NCE 50). The following MS parameters were set: spray voltage on a HESI II ion source of 3.5 kV, sheath gas 30, auxiliary gas 10, spare gas 1 unit, S-Lens 50 eV, and capillary temperature 250 °C. The data analysis and processing were performed by Thermo Qual/Quan Browser Xcalibur (v 3.1; Thermo Fisher Scientific, Waltham, MA, USA). Peak areas of monoisotopic peaks were corrected for natural 13C isotope abundance. Relative peak area ratios of DolPs to IS were converted to molar concentrations using a known IS concentration. Statistical analysis of datasets was carried out using Microsoft Excel 2016 to perform Student t-tests. All graphs were generated using GraphPad Prism version 5.0.3 (GraphPad Software, Inc.).
Results and Discussion
Increased Sensitivity of DolP Detection through Methylation
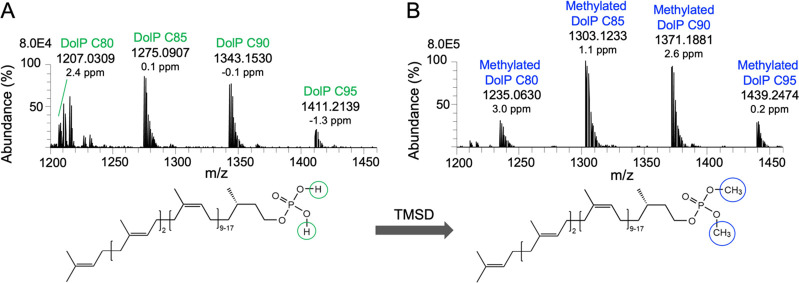
TMSD methylation has been shown to be a powerful approach to improve the ionization efficiency of anionic phospholipids in positive ion mode by eliminating the negative charge of the phosphate group.30,32 To test whether methylation of DolP species increases the sensitivity of MS detection, we used a commercial DolP mixture with chain lengths from C65-C105 as a reference standard. Since the un-methylated DolP standard cannot be analyzed by RPLC-MS, we used nESI-DIMS analysis of both un-methylated and methylated DolPs to evaluate their ionization and methylation efficiencies. As observed for anionic phospholipids, nESI-DIMS analyses showed that un-methylated ions were poorly ionized, while methylation of DolP species resulted in a significant increase in ion intensities compared to the un-methylated standard (Figure S2). However, with nESI-DIMS, we could only detect and quantify four abundant DolP species C80-C95, while other species, although present in the DolP standard, could not be detected. The nESI-DIMS analysis of the DolP standard before TMSD methylation showed four [M + NH4]+ ions, that is, DolP C80 at m/z 1207.0309, DolP C85 at m/z 1275.0907, DolP C90 at m/z 1343.1530, and DolP C95 at m/z 1411.2139 (Figure 1A). After methylation, the ion cluster was shifted to m/z 1235.0630, 1303.1233, 1371.1881, and 1439.2474, respectively (Figure 1B). The mass addition of ∼28.0333 Da indicates di-methylation (addition of C2H4; exact mass = 28.0313 Da) of the phosphate residue in the DolP species. After methylation, un-methylated or mono-methylated species were not detected (Figure S2; theoretical m/z values of DolP ions are given in Tables 1, S1, and S2).
Figure 1.
Representative positive ion nESI-DIMS spectra of DolP standard before and after methylation. nESI-DIMS spectra of un-methylated (A) and TMSD-methylated DolP standards (B). Ammonium adducts of DolP species ranging from C80 to C95 are indicated.
Table 1. DolP and PolP Molecular Compositions and Positive Ion Mode RPLC-MS Characteristics of Annotated Di-Methylated Analyte Species in This Study.
| analyte species | molecular formula | theoretical m/za | mass error (ppm) | RT (min) |
|---|---|---|---|---|
| PolP C60 | C62H103O4P | 960.7932 | –0.8 | 19.1 |
| DolP C65 | C67H113O4P | 1030.8715 | –0.4 | 20.0 |
| DolP C70 | C72H121O4P | 1098.9341 | 1.7 | 20.6 |
| DolP C75 | C77H129O4P | 1166.9967 | 0.8 | 21.2 |
| DolP C80 | C82H137O4P | 1235.0593 | 1.6 | 21.8 |
| DolP C85 | C87H145O4P | 1303.1219 | 0.8 | 22.3 |
| DolP C90 | C92H153O4P | 1371.1845 | 0.7 | 22.9 |
| DolP C95 | C97H161O4P | 1439.2471 | 0.3 | 23.5 |
| DolP C100 | C102H169O4P | 1507.3097 | 2.0 | 24.1 |
| DolP C105 | C107H177O4P | 1575.3720 | 0.3 | 24.7 |
Theoretical m/z values of [M + NH4]+ ions.
MS-based lipid quantification typically relies on internal lipid standards, that is, lipid species that are not present in the biological samples and are added before the extraction procedure. Ideally, these standards resemble the analytes in their physico-chemical properties and behave similarly in derivatization, extraction, chromatographic behavior, and MS analysis. Isotopically labeled counterparts to endogenous lipids are the molecules of choice, as standards can be used that are very close to the m/z range of the target lipid species. Since such standards are not commercially available for DolPs, we tested analytes that are very similar to DolPs but whose m/z values do not overlap those of endogenous DolPs. PolPs are structural analogues of DolP that are used in bacterial N-glycosylation and are also found in plants.2 DolP and PolP share the phosphate head group structure and have similar linear isoprene linkage, except for the α-isoprene unit, which is unsaturated in PolPs. To establish the quantitative analysis of DolPs, the commercially available PolP C60 species is a suitable standard, as it can be used as an IS in a wide range of cellular matrices, including yeast, mammals, and archaea.42
We performed the TMSD-based methylation analysis of PolP C60 as described for DolP. As observed for DolP, methylation of PolP resulted in a complete shift toward di-methylated PolP species (Figure S3), and methylated PolP showed similar RPLC-MS properties as methylated DolPs. The extracted ion chromatogram (EIC) for the [M + NH4]+ ion of methylated PolP C60 showed a chromatographic elution at RT ∼19.37 min (Figure S4A), and a protonated adduct of the methylated phosphate head group with theoretical m/z 127.0155 was observed in MS/MS experiments, which is a common characteristic of (di-)methylated phospholipids (Figure S4B). To assess the TMSD methylation efficiency for DolPs, we compared the IS-normalized peak intensities of un-methylated and TMSD-methylated C80, C85, C90, and C95 species in the DolP standard with nESI-DIMS measurements. Our analyses demonstrate that the efficiency of TMSD derivatization was ≥99% for all DolP species investigated, with only di-methylated species being formed under the given reaction conditions (Table S3). We found that the DolP species were methylated to the same relative extent in the three independent replicates, as indicated by the low relative standard deviation (RSD <1%). These results demonstrate that TMSD efficiently and selectively methylates the hydroxyl groups of phosphate residue of DolPs to di-methylated DolP species.
Methylation of DolP Improves Chromatographic Performance
Coupling of LC separation with MS detection provides better sensitivity, selectivity, and peak capacity and reduces the ion suppression observed in nESI-DIMS.43 In addition, LC-based separation of analytes reduces the complexity of biological lipid extracts and allows for enrichment of target analytes according to their physico-chemical properties. These advantages of LC-MS analysis are particularly important when analyzing analytes present as minor components in a complex mixture, as is the case for DolP species in most biological samples. Among the LC separation methods commonly used for complex lipid mixtures, RPLC is the prevalent analytical technique.26 RPLC is, however, unsuitable for the analysis of unmodified DolP species due to their wide range of lipophilicity (xlog P > 20) values. Methylation of the phosphate groups of lipids by derivatization reduces the polarity of phospholipids, suggesting that methylation is a suitable method for RPLC analyses of DolPs. We, therefore, tested whether methylation of DolP species makes these analytes amenable to RPLC separation, as has been shown previously for other anionic lipids.36,44 For the chromatographic separation of methylated DolP species, we used a conventional C18 column prior to MS detection. We found that methylation of DolPs significantly improved the ability of their RP chromatographic retention and resolution and resulted in a narrower peak shape of methylated DolPs compared to their unmodified counterparts (Figure S5). Importantly, no carryover was observed for methylated DolPs in the RPLC system. The improved chromatographic elution characteristics observed for the methylated DolP species are comparable to those described for the analysis of methylated phospholipids using supercritical fluid chromatography-MS.40 TMSD methylation in combination with RPLC-MS analysis has also been shown to be a powerful approach for the detection and quantification of phosphatidylinositol-(3,4,5)-trisphosphate species, which, comparable to DolPs, are present at very low levels in cellular extracts.31 After methylation, we detected the DolP species C70-C105 (see Tables S4 and S5 for species distributions) in the DolP standard by RPLC-MS analysis. A C65 species indicated by the manufacturer was not detectable in the standard. A list of all annotated methylated DolP species characterized in this study, their molecular formulae, and theoretical m/z values and retention times (RTs) are provided in Table 1 (see Table S1 for an extended version).
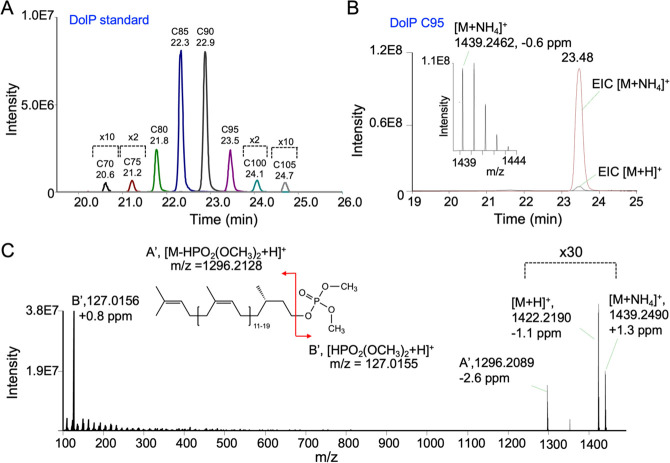
An overlay of EICs of different DolP species obtained after RPLC-MS analysis of the methylated DolP standard demonstrates baseline resolution of DolP species with different chain lengths within a RT range of ∼20 to 25 min (Figure 2A, Table 1). Increasing isoprene units (with increased degree of unsaturation and carbon length) resulted in higher RTs of methylated DolP species (Table 1). This increase of RTs is attributed to an increase in the hydrophobic interactions of DolP species with the stationary phase.45 The isotopologues of the [M + NH4]+ adducts of DolP species were fully resolved at R = 70,000 (at m/z 200). For DolP C90, C95, C100, and C105 species, the M + 1 isotope peak was more abundant than the monoisotopic M + 0 peak (see Figure S2 for DolP C90 and DolP C95), as is expected due to the increased contribution of 13C isotopes.
Figure 2.
RPLC-MS characteristics of di-methylated DolP C95 species in DolP standard. (A) Representative overlay of EICs of di-methylated [M + NH4]+ ions of DolP species. For each EIC peak, DolP species and RTs are indicated. (B) Overlay of EICs (extraction window of ±5 ppm) of [M + NH4]+ and [M + H]+ ions, with the corresponding MS spectrum of the [M + NH4]+ ion. m/z(theoretical) of [M + NH4]+ ion = 1439.2471 and of [M + H]+ ion = 1422.2205. (C) MS/MS spectrum resulting from collision-induced dissociation (CID, NCE 10%) of [M + NH4]+, showing the intact [M + NH4]+ and [M + H]+ ions and the methylated phosphate head group ion. Capital letters A’ and B′ refer to the illustration of the fragmentation pattern shown in the simplified linear structure displayed as inset.
Having demonstrated the baseline separation of DolP species by MS analysis of [M + NH4]+ ions, we performed MS/MS analyses of the individual DolP species for further characterization, as exemplified for the DolP species C95 present in the DolP standard (Figure 2B,C). Following RPLC separation and MS detection, the EICs of DolP C95, detected as [M + NH4]+ and [M + H]+ ions, show that the abundance of the [M + H]+ ion was typically ∼30-fold lower than the corresponding [M + NH4]+ ion (Figure 2B). The [M + NH4]+ adducts were therefore used for the mass spectrometric characterization of DolPs.
CID of the precursor [M + NH4]+ ion at low normalized collision energy (NCE 10%) resulted in the appearance of a highly abundant head group fragment ion in the MS/MS spectrum, corresponding to a protonated dimethylphosphate [HPO2(OCH3)2 + H]+ ion (Figure 2B,C). This head group fragment ion was found in all MS/MS spectra of methylated DolP species, independent of the number of isoprene units, suggesting phosphodiester bond cleavage as the major fragmentation pathway. This type of fragmentation has already been demonstrated for sphingosine-1-phosphates with di-methylated phosphate head groups46 and may provide a useful approach to scan for phospholipids, including yet unidentified species. In addition to the main fragment ion, other less abundant ions corresponding to the loss of ammonia and subsequent neutral loss of a dimethyl phosphate were also observed (Figure 2C).
The DolP species present in the DolP standard have varying degrees of hydrocarbon chain unsaturation (13–21 double bonds per lipid) and range in length from 67 to 107 C-atoms. Their different physico-chemical properties (such as polarity or pKa) could consequently influence the derivatization and RP-chromatographic behavior of DolPs. We, therefore, determined the relative distribution of the major DolP species C80, C85, C90, and C95, present in the DolP standard, comparing the profiles before and after methylation and with and without RPLC separation prior to MS detection. Our data demonstrate that the profile of DolP species is independent of the methylation reaction, and similar results are obtained for methylated DolPs in the presence or absence of RPLC separation (Table S4).
Method Validation
The optimized RPLC-MS method was validated according to ICH guidelines (see Supporting Information for experimental details and results). First, we tested the RPLC-MS method for LOQs and determined the LOD and the linearity range (Table S5, Figure S6). We found that the LOD for all DolP species was ≤2 pg on the column. Our method showed excellent linearity over an extensive range with correlation coefficient (r2) values of 0.99–1.00 for all DolP species. The linear range of 0.05–800 pg/μL was sufficiently low to cover the quantification of endogenous DolP species at the cellular level. The LOQ of ≤10 pg (on column) is, to our knowledge, the most sensitive method for quantitative analysis of DolP species reported to date. Further, the performance of the method was evaluated at three quality control (QC) levels, low (5), medium (20), and high (50) concentrations (in pg/μL) of the methylated DolP standard spiked into methylated HeLa cells (matrix blanks). The specificity and selectivity of the method was tested by comparing matrix blanks (HeLa cells extracts) with and without methylation and by comparing matrix blanks with and without spiked in methylated DolP/PolP C60 standards. For extraction recovery, QCs were spiked in blank solvent. The intra- and inter-day precision expressed as % CV (n = 3) was in the range of 1.1–7.9 and 0.5–11.2%, respectively, with intra- and inter-day accuracy ranging from 100.1 to 110.4% and 101.2 to 110.3%, respectively (Table S6). For all DolP species, an extraction efficiency of ≥95% was found, except for DolP C105, which was only recovered in the high concentration sample (Table S7). Post-preparative autosampler stability results are summarized in Table S7 and indicate that there was no significant degradation of DolPs in these samples in the autosampler at 4 °C for over 20 h (Table S7). No background interference for methylated ions was found. Overall, the validated method is accurate, precise, and acceptable for the analysis of biological samples. Taken together, TMSD methylation of DolPs prior to RPLC-MS offers several advantages, for example, the simple pre-column derivatization procedure, the higher stability of derivatized samples, and the enhanced sensitivity of detection due to masking of the phosphate head groups and improved ionization of the methylated analytes.
DolP Species Profiling in Biological Samples
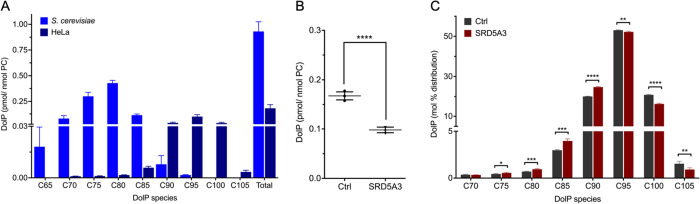
To determine the DolP content of HeLa cells, ∼1 × 106 cells were subjected to alkaline hydrolysis, extracted, methylated, and subjected to RPLC-MS analysis. The alkaline hydrolysis step is essential to ensure optimal recovery of DolP from biological samples. At the same time, oligosaccharides are released from dolichyl carriers that are converted into DolPs.47 Cellular DolP species were annotated based on accurate mass, similarity matching of precursor m/z, RPLC RTs, presence of the head group fragment in the MS/MS spectrum, and similarity matching of isotope patterns of endogenous DolP species and standards. A representative RPLC-MS chromatogram, MS, and MS/MS analysis of endogenous DolP C95 from methylated HeLa cells extracts are shown in Figure S7. Typically, PolP species are absent in Eukarya.2 If PolP standards are used that have the same number of C-atoms as the DolP species to be quantified, then the PolP M + 2 and DolP M+ isotopologues cannot be resolved at the selected resolution. In such a case, the contribution of the PolP M + 2 ions to the M+ ions of neighboring DolP species has to be taken into account. In total, we detected eight DolP species in HeLa extracts, ranging from C70 to C105 (Figure 3A), which were baseline-separated in their EIC profiles (Figure S8A). DolP C95 was the most abundant DolP species, accounting for 54.1 ± 0.2% of the total DolP pool, which is in agreement with previous reports.42,48,49 Due to the high sensitivity of our method, we were also able to identify DolP C70, C75, and C80 as minor species for the first time, accounting for 0.4 ± 0.1, 0.5 ± 1, and 0.7 ± 0.1% of total DolP species, respectively. Normalization of DolP to PC, which contributes ∼33 mol % to membrane lipids in HeLa cells,50 revealed that the DolP content of HeLa membranes is in the range of ∼0.006% of membrane lipids and thus only present in catalytic amounts.
Figure 3.
DolP species profiles determined in cellular extracts. (A) HeLa and S. cerevisiae cells were subjected to alkaline hydrolysis and lipid extraction, methylation, and RPLC-MS analysis. DolP amounts were normalized to PC as bulk membrane lipid. Data are presented as mean ± SD of three biological replicates. (B, C) Patient fibroblasts (SRD5A3-CDG) and healthy control cells (Ctrl) were subjected to lipid extraction in the presence of IS, methylation, and RPLC-MS analysis. (B) Total DolP amounts were normalized to PC. (C) Distribution of DolPs ranging from C70 to C105 displayed as a percentage of all DolP species. Data are presented as mean ± SD of four biological replicates.
Similarly, we have characterized the DolP profile of S. cerevisiae cells. In contrast to HeLa cells, DolP species in S. cerevisiae range from DolP C65 to DolP C95, with DolP C75 and DolP C80 being the dominant lipid species (Figures 3A and S8B). DolP C75 and C80 together formed 31.7 ± 0.9 and 45.5 ± 2.9% of total DolPs, respectively, and their distribution mirrors that of the DolP precursor dolichol.51
Monitoring DolPs in SRD5A3-CDG Patient Fibroblasts
Having validated the DolP quantification in cellular extracts, we determined DolP levels in fibroblasts from patients suffering from a mutation in the steroid 5-α-reductase type 3 (SRD5A3) gene.51 The gene encodes polyprenol reductase, which catalyzes the reduction of the alpha-isoprene of polyprenol in the final step of dolichol synthesis (Figure S1). SRD5A3-CDG is associated with severe visual impairment, variable ocular anomalies (e.g., iris and optic nerve coloboma, congenital cataract, and glaucoma), intellectual disability, cerebellar abnormalities, hypotonia, ataxia, ichthyosiform skin lesions, kyphosis, congenital heart defects, hypertrichosis, and abnormal coagulation, affecting children and adults (e.g., see refs (51−56)) and reflecting the central role of dolichol in cellular glycosylation. Fibroblasts from a SRD5A3-CDG patient presenting a severe phenotype were analyzed for their DolP content. Compared to healthy control fibroblasts, the DolP content was significantly decreased (41.5%) in the patient’s fibroblasts (Figure 3B), supporting a key role of SRD5A3 in maintaining cellular DolP levels. The presence of a substantial residual amount of DolP in the patient’s fibroblasts is in line with a recent report suggesting a yet unidentified alternative pathway of dolichol synthesis.57
We also observed a shift toward the shorter DolP C75–DolP C90 species, mainly at the expense of DolP C100 and DolP C105 (Figure 3C). Inhibition of the DolP synthesis pathway by the small molecular drug lovastatin also led to a decrease in DolP species chain length in liver tissue of lovastatin treated mice58 and in hepatocellular carcinoma samples in comparison to healthy human liver.59 In all cases, the underlying molecular mechanisms are unclear, highlighting the importance of DolP species profiling in the study of DolP homeostasis in health and disease.
Conclusions
Here, we describe a method for the highly sensitive quantitative analysis of DolP species in biological samples. DolP profilings have not yet been implemented in comprehensive lipidomics analyses, mainly due to their very low abundance. By combining phosphate methylation with RPLC-MS separation, we have developed a workflow to quantify DolP species from as few as 106 HeLa cells. Including the TMSD-based methylation enabled the quantification of DolP species over a wide range of lipophilicities. Due to its unprecedented sensitivity, our method now offers comprehensive coverage of DolP species ranging from C70 to C105, including minor species that were not detected in previous studies. Sample preparation has been simplified by implementing a rapid three-step procedure, including hydrolysis, extraction, and methylation. Phosphate methylation of DolP species resulted in improved peak shapes in RPLC runs. A limitation of our approach is the lack of commercially available standards that co-ionize with the DolP analytes, that is, ideally isotope-labeled ISs. Since PolPs are generally not found in eukaryotic samples, PolP mixtures with isoprene units of similar range to eukaryotic DolP species could also be considered as standards in the future. Although our approach entails a substantially reduced turnaround time, we assume that this can be further optimized, for example, through shorter gradient runs. In summary, implementation of DolP species analysis into standard lipidomics workflows will help to detect alterations in DolP steady state levels, which are intricately linked with a number of lipophilic metabolites, including cholesterol and ubiquinone, via the mevalonate pathway, thus connecting glycosylation processes with membrane homeostasis and cellular energy states. This approach also represents a promising diagnostic and/or therapeutic application for pathophysiologies such as Alzheimer’s disease that are associated with altered DolP levels in body fluids, including urine, plasma, or liquor.
Acknowledgments
This project was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Project-ID 289991887 – Forschungsgruppe FOR 2509, project 02, – Project-ID 112927078 – Transregio TRR83, project 01, and – Project-ID 278001972 – TRR 186, project Z04. The authors gratefully acknowledge the data storage service SDS@hd supported by the Ministry of Science, Research, and the Arts Baden-Württemberg (MWK) and the DFG through grant INST 35/1314-1 FUGG and INST 35/1503-1 FUGG. We thank Timo Sachsenheimer for PC measurements, Iris Leibrecht and Alexia Herrmann for technical assistance, and Ingo Amm for providing S. cerevisiae cells.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.2c03623.
Additional experimental details, methods, and results (PDF)
Accession Codes
All lipidomics data have been deposited to the Metabolights database, where they are available under accession number MTBLS4095 for RPLC-MS data and MTBLS4228 for nESI-DIMS data
Author Contributions
D.K. and B.B.: Conceptualization, methodology, investigation, writing—original draft preparation, review, and editing. D.K.: Formal analysis, data curation, and visualization. B.B.: Supervision and funding acquisition. P.P.: Writing—Review and editing. F.K., L.B., and C.T.: Cell culture preparation of biological samples. C.T.: Funding acquisition. All authors contributed to and approved the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Haeuptle M. A.; Welti M.; Troxler H.; Hulsmeier A. J.; Imbach T.; Hennet T. Improvement of dolichol-linked oligosaccharide biosynthesis by the squalene synthase inhibitor zaragozic acid. J. Biol. Chem. 2011, 286, 6085–6091. 10.1074/jbc.M110.165795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler J.; Imperiali B. Stereochemical Divergence of Polyprenol Phosphate Glycosyltransferases. Trends Biochem. Sci. 2018, 43, 10–17. 10.1016/j.tibs.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefeber D. J.; Freeze H. H.; Steet R.; Kinoshita T.. Congenital Disorders of Glycosylation. In Essentials of Glycobiology, Varki A., Cummings R. D., Esko J. D., Stanley P., Hart G. W., Aebi M., Mohnen D., Kinoshita T., Packer N. H., Prestegard J. H., Schnaar R. L., Seeberger P. H. Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor (NY), 2022; pp. 599–614. [PubMed] [Google Scholar]
- Guan Z.; Söderberg M.; Sindelar P.; Prusiner S. B.; Kristensson K.; Dallner G. Lipid composition in scrapie-infected mouse brain: prion infection increases the levels of dolichyl phosphate and ubiquinone. J. Neurochem. Lond. 1996, 66, 277–285. 10.1046/j.1471-4159.1996.66010277.x. [DOI] [PubMed] [Google Scholar]
- Hall N. A.; Patrick A. D. Dolichol and phosphorylated dolichol content of tissues in ceroid-lipofuscinosis. J. Inherit. Metab. Dis. 1985, 8, 178–183. 10.1007/BF01805431. [DOI] [PubMed] [Google Scholar]
- Söderberg M.; Edlund C.; Alafuzoff I.; Kristensson K.; Dallner G. Lipid composition in different regions of the brain in Alzheimer’s disease/senile dementia of Alzheimer’s type. J. Neurochem. 1992, 59, 1646–1653. 10.1111/j.1471-4159.1992.tb10994.x. [DOI] [PubMed] [Google Scholar]
- Spiro M. J.; Spiro R. G. Control of N-linked carbohydrate unit synthesis in thyroid endoplasmic reticulum by membrane organization and dolichyl phosphate availability. J. Biol. Chem. 1986, 261, 14725–14732. 10.1016/S0021-9258(18)66931-5. [DOI] [PubMed] [Google Scholar]
- Cavallini G.; Sgarbossa A.; Parentini I.; Bizzarri R.; Donati A.; Lenci F.; Bergamini E. Dolichol: A Component of the Cellular Antioxidant Machinery. Lipids 2016, 51, 477–486. 10.1007/s11745-016-4137-x. [DOI] [PubMed] [Google Scholar]
- Tollbom O.; Dallner G. Dolichol and dolichyl phosphate in human tissues. Br. J. Exp. Pathol. 1986, 67, 757–764. [PMC free article] [PubMed] [Google Scholar]
- Kornfeld R.; Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 1985, 54, 631–664. 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger S.; Gentzsch M.; Tanner W. Protein O-mannosylation. Biochim. Biophys. Acta 1999, 1426, 297–307. 10.1016/S0304-4165(98)00131-7. [DOI] [PubMed] [Google Scholar]
- Kinoshita T.; Ohishi K.; Takeda J. GPI-anchor synthesis in mammalian cells: genes, their products, and a deficiency. J. Biochem. 1997, 122, 251–257. 10.1093/oxfordjournals.jbchem.a021746. [DOI] [PubMed] [Google Scholar]
- Vigo C.; Grossman S. H.; Drost-Hansen W. Interaction of dolichol and dolichyl phosphate with phospholipid bilayers. Biochim. Biophys. Acta 1984, 774, 221–226. 10.1016/0005-2736(84)90295-5. [DOI] [PubMed] [Google Scholar]
- Zhou G. P.; Troy F. A. 2nd. NMR study of the preferred membrane orientation of polyisoprenols (dolichol) and the impact of their complex with polyisoprenyl recognition sequence peptides on membrane structure. Glycobiology 2005, 15, 347–359. 10.1093/glycob/cwi016. [DOI] [PubMed] [Google Scholar]
- van Duijn G.; Valtersson C.; Chojnacki T.; Verkleij A. J.; Dallner G.; de Kruijff B. Dolichyl phosphate induces non-bilayer structures, vesicle fusion and transbilayer movement of lipids: a model membrane study. Biochim. Biophys. Acta 1986, 861, 211–223. 10.1016/0005-2736(86)90423-2. [DOI] [PubMed] [Google Scholar]
- Andersson M.; Appelkvist E. L.; Kristensson K.; Dallner G.; Gustafsson P. Age-dependent changes in the levels of dolichol and dolichyl phosphates in human brain. Acta Chem. Scand. B 1987, 41, 144–146. 10.3891/acta.chem.scand.41b-0144. [DOI] [PubMed] [Google Scholar]
- Dricu A.; Wang M.; Hjertman M.; Malec M.; Blegen H.; Wejde J.; Carlberg M.; Larsson O. Mevalonate-regulated mechanisms in cell growth control: role of dolichyl phosphate in expression of the insulin-like growth factor-1 receptor (IGF-1R) in comparison to Ras prenylation and expression of c-myc. Glycobiology 1997, 7, 625–633. 10.1093/glycob/7.5.625. [DOI] [PubMed] [Google Scholar]
- Haeuptle M. A.; Hülsmeier A. J.; Hennet T. HPLC and mass spectrometry analysis of dolichol-phosphates at the cell culture scale. Anal. Biochem. 2010, 396, 133–138. 10.1016/j.ab.2009.09.020. [DOI] [PubMed] [Google Scholar]
- Löw P.; Dallner G.; Mayor S.; Cohen S.; Chait B.; Menon A. J. T. J. The mevalonate pathway in the bloodstream form of Trypanosoma brucei. Identification of dolichols containing 11 and 12 isoprene residues. J. Biol. Chem. 1991, 266, 19250–19257. 10.1016/S0021-9258(18)54990-5. [DOI] [PubMed] [Google Scholar]
- Wolucka B. A.; Rozenberg R.; de Hoffmann E.; Chojnacki T. Desorption chemical ionization tandem mass spectrometry of polyprenyl and dolichyl phosphates. J. Am. Soc. Mass Spectrom. 1996, 7, 958–964. 10.1016/1044-0305(96)80514-9. [DOI] [PubMed] [Google Scholar]
- Guan Z.; Naparstek S.; Kaminski L.; Konrad Z.; Eichler J. Distinct glycan-charged phosphodolichol carriers are required for the assembly of the pentasaccharide N-linked to the Haloferax volcanii S-layer glycoprotein. Mol. Microbiol. 2010, 78, 1294–1303. 10.1111/j.1365-2958.2010.07405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi Y.; Fujinami D.; Kohda D. Comparative Analysis of Archaeal Lipid-linked Oligosaccharides That Serve as Oligosaccharide Donors for Asn Glycosylation. J. Biol. Chem. 2016, 291, 11042–11054. 10.1074/jbc.M115.713156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech N. B.; Enke C. G. Practical implications of some recent studies in electrospray ionization fundamentals. Mass Spectrom. Rev. 2001, 20, 362–387. 10.1002/mas.10008. [DOI] [PubMed] [Google Scholar]
- Antignac J.-P.; de Wasch K.; Monteau F.; De Brabander H.; Andre F.; Le Bizec B. The ion suppression phenomenon in liquid chromatography–mass spectrometry and its consequences in the field of residue analysis. Anal. Chim. Acta 2005, 529, 129–136. 10.1016/j.aca.2004.08.055. [DOI] [Google Scholar]
- Yabré M.; Ferey L.; Somé I. T.; Gaudin K. Greening Reversed-Phase Liquid Chromatography Methods Using Alternative Solvents for Pharmaceutical Analysis. Molecules 2018, 23, 1065. 10.3390/molecules23051065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajka T.; Fiehn O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. TrAC, Trends Anal. Chem. 2014, 61, 192–206. 10.1016/j.trac.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete S.; Veuthey J.-L.; Guillarme D. New trends in reversed-phase liquid chromatographic separations of therapeutic peptides and proteins: Theory and applications. J. Pharm. Biomed. Anal. 2012, 69, 9–27. 10.1016/j.jpba.2012.03.024. [DOI] [PubMed] [Google Scholar]
- Kochling J.; Wu W.; Hua Y.; Guan Q.; Castaneda-Merced J. A platform analytical quality by design (AQbD) approach for multiple UHPLC-UV and UHPLC–MS methods development for protein analysis. J. Pharm. Biomed. Anal. 2016, 125, 130–139. 10.1016/j.jpba.2016.03.031. [DOI] [PubMed] [Google Scholar]
- Hartley M. D.; Imperiali B. At the membrane frontier: a prospectus on the remarkable evolutionary conservation of polyprenols and polyprenyl-phosphates. Arch. Biochem. Biophys. 2012, 517, 83–97. 10.1016/j.abb.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T.; Shu Q.; Liu P.; Niu L.; Guo X.; Ding X.; Xue P.; Xie Z.; Wang J.; Zhu N.; Wu P.; Niu L.; Yang F. Characterization and relative quantification of phospholipids based on methylation and stable isotopic labeling. J. Lipid Res. 2016, 57, 388–397. 10.1194/jlr.M063024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J.; Anderson K. E.; Juvin V.; Smith T. S.; Karpe F.; Wakelam M. J. O.; Stephens L. R.; Hawkins P. T. Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat. Methods 2011, 8, 267–272. 10.1038/nmeth.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. C.; Byeon S. K.; Moon M. H. Relative Quantification of Phospholipids Based on Isotope-Labeled Methylation by Nanoflow Ultrahigh Performance Liquid Chromatography-Tandem Mass Spectrometry: Enhancement in Cardiolipin Profiling. Anal. Chem. 2017, 89, 4969–4977. 10.1021/acs.analchem.7b00297. [DOI] [PubMed] [Google Scholar]
- Wei F.; Wang X.; Ma H. F.; Lv X.; Dong X. Y.; Chen H. Rapid profiling and quantification of phospholipid molecular species in human plasma based on chemical derivatization coupled with electrospray ionization tandem mass spectrometry. Anal. Chim. Acta 2018, 1024, 101–111. 10.1016/j.aca.2018.04.012. [DOI] [PubMed] [Google Scholar]
- Xia T.; Ren H.; Zhang W.; Xia Y. Lipidome-wide characterization of phosphatidylinositols and phosphatidylglycerols on CC location level. Anal. Chim. Acta 2020, 1128, 107–115. 10.1016/j.aca.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T.; Shu Q.; Hou J.; Liu P.; Niu L.; Guo X.; Liu C. C.; Yang F. Profiling and relative quantitation of phosphoinositides by multiple precursor ion scanning based on phosphate methylation and isotopic labeling. Anal. Chem. 2015, 87, 513–521. 10.1021/ac503224j. [DOI] [PubMed] [Google Scholar]
- Pan M.; Qin C.; Han X. Quantitative Analysis of Polyphosphoinositide, Bis(monoacylglycero)phosphate, and Phosphatidylglycerol Species by Shotgun Lipidomics After Methylation. Methods Mol. Biol. 2021, 2306, 77–91. 10.1007/978-1-0716-1410-5_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; Palavicini J. P.; Cseresznye A.; Han X. Strategy for Quantitative Analysis of Isomeric Bis(monoacylglycero)phosphate and Phosphatidylglycerol Species by Shotgun Lipidomics after One-Step Methylation. Anal. Chem. 2017, 89, 8490–8495. 10.1021/acs.analchem.7b02058. [DOI] [PubMed] [Google Scholar]
- Thomas B. J.; Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell 1989, 56, 619–630. 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Ozbalci C.; Sachsenheimer T.; Brugger B.. Quantitative analysis of cellular lipids by nano-electrospray ionization mass spectrometry. In Membrane Biogenesis; Methods in Molecular Biology; Humana Press, Totowa, NJ, 2013; vol. 1033, pp. 3 −20. [DOI] [PubMed] [Google Scholar]
- Lee J. W.; Nishiumi S.; Yoshida M.; Fukusaki E.; Bamba T. Simultaneous profiling of polar lipids by supercritical fluid chromatography/tandem mass spectrometry with methylation. J. Chromatogr. A 2013, 1279, 98–107. 10.1016/j.chroma.2013.01.020. [DOI] [PubMed] [Google Scholar]
- Herzog R.; Schwudke D.; Shevchenko A. LipidXplorer: Software for Quantitative Shotgun Lipidomics Compatible with Multiple Mass Spectrometry Platforms. Curr. Protoc. Bioinform. 2013, 43, 14.12.11–14.12.30. 10.1002/0471250953.bi1412s43. [DOI] [PubMed] [Google Scholar]
- Schenk B.; Fernández F.; Waechter C. J. J. G.. The ins(ide) and out(side) of dolichyl phosphate biosynthesis and recycling in the endoplasmic reticulum. 2001, 11 ( (5), ), 61R–70R, 10.1093/glycob/11.5.61r. [DOI] [PubMed]
- García-Cañaveras J. C.; Donato M. T.; Castell J. V.; Lahoz A. A Comprehensive Untargeted Metabonomic Analysis of Human Steatotic Liver Tissue by RP and HILIC Chromatography Coupled to Mass Spectrometry Reveals Important Metabolic Alterations. J. Proteome Res. 2011, 10, 4825–4834. 10.1021/pr200629p. [DOI] [PubMed] [Google Scholar]
- Li P.; Gawaz M.; Chatterjee M.; Lämmerhofer M. Targeted Profiling of Short-, Medium-, and Long-Chain Fatty Acyl-Coenzyme As in Biological Samples by Phosphate Methylation Coupled to Liquid Chromatography–Tandem Mass Spectrometry. Anal. Chem. 2021, 93, 4342–4350. 10.1021/acs.analchem.1c00664. [DOI] [PubMed] [Google Scholar]
- Smith M.; Jungalwala F. B. Reversed- phase high performance liquid chromatography of phosphatidylcholine: a simple method for determining relative hydrophobic interaction of various molecular species. J. Lipid Res. 1981, 22, 697–704. 10.1016/S0022-2275(20)37381-8. [DOI] [PubMed] [Google Scholar]
- Narayanaswamy P.; Shinde S.; Sulc R.; Kraut R.; Staples G.; Thiam C. H.; Grimm R.; Sellergren B.; Torta F.; Wenk M. R. Lipidomic “Deep Profiling”: An Enhanced Workflow to Reveal New Molecular Species of Signaling Lipids. Anal. Chem. 2014, 86, 3043–3047. 10.1021/ac4039652. [DOI] [PubMed] [Google Scholar]
- Elmberger P. G.; Eggens I.; Dallner G. Conditions for quantitation of dolichyl phosphate, dolichol, ubiquinone and cholesterol by HPLC. Biomed. Chromatogr. 1989, 3, 20–28. 10.1002/bmc.1130030106. [DOI] [PubMed] [Google Scholar]
- Chang M. M.; Imperiali B.; Eichler J.; Guan Z. N-Linked Glycans Are Assembled on Highly Reduced Dolichol Phosphate Carriers in the Hyperthermophilic Archaea Pyrococcus furiosus. PLoS One 2015, 10, e0130482 10.1371/journal.pone.0130482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewska E.; Danikiewicz W. Polyisoprenoids: Structure, biosynthesis and function. Prog. Lipid Res. 2005, 44, 235–258. 10.1016/j.plipres.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Gerl M. J.; Bittl V.; Kirchner S.; Sachsenheimer T.; Brunner H. L.; Luchtenborg C.; Ozbalci C.; Wiedemann H.; Wegehingel S.; Nickel W.; Haberkant P.; Schultz C.; Kruger M.; Brugger B. Sphingosine-1-Phosphate Lyase Deficient Cells as a Tool to Study Protein Lipid Interactions. PLoS One 2016, 11, e0153009 10.1371/journal.pone.0153009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantagrel V.; Lefeber D. J.; Ng B. G.; Guan Z.; Silhavy J. L.; Bielas S. L.; Lehle L.; Hombauer H.; Adamowicz M.; Swiezewska E.; De Brouwer A. P. SRD5A3 is required for converting polyprenol to dolichol and is mutated in a congenital glycosylation disorder. Cell 2010, 142, 203–217. 10.1016/j.cell.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeken J.; Lefeber D. J.; Matthijs G. SRD5A3 defective congenital disorder of glycosylation: clinical utility gene card. Eur. J. Hum. Genet. 2020, 28, 1297–1300. 10.1038/s41431-020-0647-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamarus Jaman N.; Rehsi P.; Henderson R. H.; Lobel U.; Mankad K.; Grunewald S. SRD5A3-CDG: Emerging Phenotypic Features of an Ultrarare CDG Subtype. Front. Genet. 2021, 12, 737094 10.3389/fgene.2021.737094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kara B.; Ayhan O.; Gokcay G.; Basbogaoglu N.; Tolun A. Adult phenotype and further phenotypic variability in SRD5A3-CDG. BMC Med. Genet. 2014, 15, 10. 10.1186/1471-2350-15-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger M.; Turk M.; Kraus C.; Uebe S.; Ekici A. B.; Krumbiegel M.; Huchzermeyer C.; Reis A.; Thiel C. SRD5A3-CDG: Twins with an intragenic tandem duplication. Eur. J. Med. Genet. 2022, 65, 104492 10.1016/j.ejmg.2022.104492. [DOI] [PubMed] [Google Scholar]
- Wheeler P. G.; Ng B. G.; Sanford L.; Sutton V. R.; Bartholomew D. W.; Pastore M. T.; Bamshad M. J.; Kircher M.; Buckingham K. J.; Nickerson D. A.; Shendure J.; Freeze H. H. SRD5A3-CDG: Expanding the phenotype of a congenital disorder of glycosylation with emphasis on adult onset features. Am. J. Med. Genet. A 2016, 170, 3165–3171. 10.1002/ajmg.a.37875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundahl J. E.; Guan Z.; Rust S.; Reunert J.; Muller B.; Du Chesne I.; Zerres K.; Rudnik-Schoneborn S.; Ortiz-Bruchle N.; Hausler M. G.; Siedlecka J.; Swiezewska E.; Raetz C. R.; Marquardt T. Life with too much polyprenol: polyprenol reductase deficiency. Mol. Genet. Metab. 2012, 105, 642–651. 10.1016/j.ymgme.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low P.; Andersson M.; Edlund C.; Dallner G. Effects of mevinolin treatment on tissue dolichol and ubiquinone levels in the rat. Biochim. Biophys. Acta 1992, 1165, 102–109. 10.1016/0005-2760(92)90081-6. [DOI] [PubMed] [Google Scholar]
- Eggens I.; Elmberger P. G.; Low P. Polyisoprenoid, cholesterol and ubiquinone levels in human hepatocellular carcinomas. Br. J. Exp. Pathol. 1989, 70, 83–92. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.