Abstract
Pathologic hyperreactive inflammatory responses occur when there is excessive activation of a proinflammatory NF-κB pathway and a reduced cytoprotective NRF2 cascade. The noncytotoxic, highly selective COX-2 inhibitory flavonol-enriched butanol fraction (UaB) from Uvaria alba (U. alba) was investigated for its inflammatory modulating potential by targeting NF-κB activation and NRF2 activity. Enzyme-linked immunosorbent assay was initially performed to measure levels of proinflammatory mediators [nitric oxide (NO), prostaglandin E2, and reactive oxygen species (ROS)] and cytokines [tumor necrosis factor-alpha (TNF-α), IL-1β, and IL-6], followed by reverse transcription-polymerase chain reaction and western blotting to determine mRNA and protein expression, respectively. Using immunofluorescence staining combined with western blot analysis, the activation of NF-κB was further investigated. NRF2 activity was also measured using a luciferase reporter assay. UaB abrogated protein and mRNA expressions of inducible nitric oxide synthase (iNOS), COX-2, TNF-α, IL-1β, and IL-6 in RAW 264.7 macrophages, thereby suppressing the production of proinflammatory mediators and cytokines. This was further validated when a concentration-dependent decrease in NO and ROS production was observed in zebrafish (Danio rerio) larvae. UaB also increased NRF2 activity in HaCaT/ARE cell line and attenuated NF-κB activation by inhibiting the nuclear translocation of transcription factor p65 in RAW 264.7 macrophages. Nontargeted LC–MS analysis of UaB revealed the presence of the flavonols quercitrin (1), quercetin (2), rutin (3), kaempferol (4), and kaempferol 3-O-rutinoside (5). Molecular docking indicates that major flavonol aglycones have high affinity toward COX-2 NSAID-binding sites, TNF-α, and TNF-α converting enzyme, while the glycosylated flavonoids showed strong binding toward iNOS and IKK—all possessing dynamic stability when performing molecular dynamics simulations at 140 ns. This is the first report to have elucidated the mechanistic anti-inflammatory potential of the Philippine endemic plant U. alba.
1. Introduction
Inflammation is a physiologic response of the body to eliminate unwanted stimuli and prevent further tissue damage.1 However, the insufficient or excessive inflammatory response can be pathologic.1−3 In a bacterium-induced infection, inflammation could be mediated by lipopolysaccharide (LPS) that acts as an antigen triggering the immune system to activate macrophages and lymphocytes to produce proinflammatory mediators [prostaglandins (PGs) and nitric oxide (NO)] and cytokines [tumor necrosis factor-alpha (TNF-α), IL-1, IL-2, IL-6, IL-8, and IL-12].1,3,4 Another contributory factor to inflammation is the excessive production of reactive oxygen species (ROS), known as oxidative stress. Although ROS is required for some physiologic functions, excess ROS has damaging effects as it induces inflammatory responses and even cell death, both of which are important in chronic diseases.5,6 At the molecular level, these processes are regulated by the transcription factor NF-κB, which can either be a homodimer or heterodimer with p50, p52, or p65 subunits. The NF-κB pathway is of interest because of its role in chronic diseases, including rheumatoid arthritis, atherosclerosis, cancer, and neurodegenerative diseases.2,7 The biosynthesis of proinflammatory mediators and cytokines is regulated by the all-encompassing NF-κB signaling pathway. Inactivated NF-κB bound to its inhibitor (IκB) is found in the cytoplasm. Nuclear translocation of NF-κB is necessary for the transcription of proinflammatory mediators and cytokines. For nuclear translocation of NF-κB to occur, phosphorylation of IκB is required, which is initiated by IκB kinase (IKK) in the presence of an inflammatory stimulus. Upon activation, NF-κB proteins are expressed to serve as transcription factors for inflammatory genes initiating the cascade of inflammatory reactions, such as the production of the COX-2 enzyme required for synthesizing prostaglandin E2 (PGE2), which induces acute inflammation.4,7 Furthermore, counter-regulatory cytoprotective mechanisms also play an important role against oxidative stress to maintain redox homeostasis in cells and tissues. NRF2 and its principal negative regulator, Keap1, play a pivotal role in this function as NRF2 regulates the NF-κB signaling pathway by decreasing the intracellular ROS levels and inhibiting its nuclear translocation.8,9 Thus, efforts to target NF-κB and NRF2-Keap1 signaling cascades are appealing strategies to modulate inflammatory responses using natural products or synthetic compounds.10
Flavonols are polyphenolic compounds ubiquitously present in several food products and have demonstrated to exhibit a wide range of biological activities, including anti-inflammatory effects. Flavonols act through a number of mechanisms to mitigate and attenuate inflammatory responses and may act as potential cardioprotective, neuroprotective, and chemopreventive agents.11 For example, chlorogenic acid showed anti-inflammatory activity by inhibiting macrophage and neutrophil recruitment in vivo in zebrafish [Danio rerio (D. rerio)] models (Yang et al.).12−14 Among the plant-based flavonoid producers are species belonging to the genus Uvaria. In the Philippines, the fruit of Uvaria species is used as food by the Aetas in the Bataan and Zambales provinces of Luzon. Aside from being used as a food source, 4 of the 20 Uvaria spp. have been studied for their pharmacological potential.15−21Uvaria alba (U. alba) Merr. was found to produce cytostatic and antitubercular seco-cyclohexenes18 and reported to contain phosphodiesterase and acetylcholinesterase inhibitory constituents implicating their potential as cancer and Alzheimer’s disease-targeting agents.22 Recognizing the association of inflammatory processes with a myriad of diseases, it was of our best interest to explore the anti-inflammatory potential of the noncytotoxic, highly selective COX-2 inhibitory flavonol-enriched butanol fraction of U. alba (UaB) by attenuating the proinflammatory NF-κB pathway while increasing the cytoprotective NRF2 level.
2. Results
2.1. Screening for Selective COX-2 Inhibition
The crude extract and fractions of U. alba were screened for their potential in attenuating inflammation by enzymatic inhibition of COX-2 using enzyme-linked immunosorbent assay (ELISA) (Table 1). Only UaB showed enzymatic inhibition of COX-2 with an IC50 of 0.63 μg/mL, while the reference drug celecoxib had an IC50 of 0.34 μg/mL. To establish anti-COX-2 selectivity, the UaB fraction was further screened for inhibition of COX-1. Interestingly, UaB did not inhibit COX-1 with an IC50 value >100 μg/mL. In terms of selectivity, UaB (SI > 158.73) was more selective compared to celecoxib (SI = 7.74).
Table 1. Cytotoxicity and Enzymatic Inhibition of Cyclooxygenase Using the Crude Extract and Fractions from U. albaa.
| sample | cytotoxicity
(IC50, μg/mL) |
anti-inflammatory property
(IC50, μg/mL) |
|||
|---|---|---|---|---|---|
| HaCaT | HepG2 | COX-2 | COX-1 | selectivity index (SI) | |
| Ua | >100 | >100 | >100 | NT | NT |
| UaP | >100 | >100 | >100 | NT | NT |
| UaD | >100 | 79.7 ± 1.2 | >100 | NT | NT |
| UaB | >100 | >100 | 0.63 ± 0.5 | >100 | >158.73 |
| +control | |||||
| doxorubicin | 19.8 ± 0.8 | 11.4 ± 0.5 | NT | NT | NT |
| celecoxib | NT | 0.34 ± 0.2 | 2.63 ± 0.7 | 7.74 | |
| –control | |||||
| DMSO | NA | NA | NA | NA | NT |
Ua: Uvaria alba crude extract; UaP: U. alba petroleum ether fraction; UaB: U. alba butanol fraction; UaD: U. alba dichloromethane fraction; HepG2: human liver cancer cells; HaCaT: nontumorigenic human keratinocytes; COX-1 and -2: cyclooxygenase-1 and -2; NT: not tested; and NA: no activity.
2.2. Screening for Cytotoxicity against HaCaT and HepG2
Cytotoxicity assay on liver cancer cells (HepG2) and nontumorigenic human keratinocytes (HaCaT) was performed (Table 1). Among the samples tested, the UaD fraction was slightly cytotoxic against HepG2 with an IC50 value of 79.7 μg/mL. Fraction UaB, on the other hand, was noncytotoxic to HepG2. Additionally, the crude extract and fractions from U. alba conferred no cytotoxicity to HaCaT.
2.3. Effect of UaB on the Viability of RAW 264.7 Macrophages
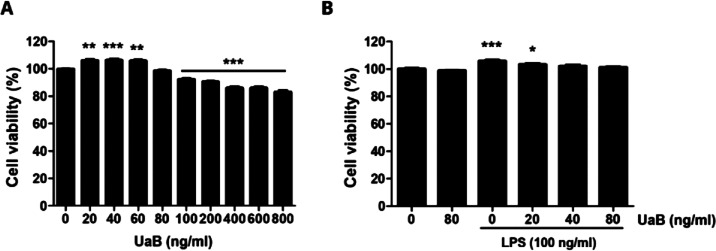
Following the identification of UaB as a noncytotoxic, highly selective COX-2 inhibitor using the enzymatic assay, we further characterized its anti-inflammatory potential in vitro. We initially determined the baseline cytotoxicity of UaB on RAW 264.7 macrophages and performed a macrophage viability assay. UaB was found to be noncytotoxic to RAW 264.7 macrophages in concentrations up to 80 ng/mL with 100% viability (Figure 1). Furthermore, upon introduction with LPS, all cells in the culture medium remained viable. Since the UaB concentration was noncytotoxic up to 80 ng/mL, this was then used as the maximum screening concentration for evaluating the inflammatory modulating effects of UaB in LPS-challenged RAW 264.7 macrophages.
Figure 1.
Effect of UaB fraction and LPS on the cell viability of RAW 264.7 macrophages. RAW 264.7 cells were stimulated with different concentrations of UaB alone for 24 h (A) or pretreated with or without UaB for 1 h before 100 ng/mL LPS stimulation for 24 h (B). Cell viability was analyzed using the MTT assay. Each value indicates the mean ± SD of three independent experiments. Significant differences among the groups were determined (*p < 0.05, **p < 0.01, and *** p < 0.0001, compared with the control cells).
2.4. Effects of UaB on LPS-Induced Production of Proinflammatory Mediators in RAW 264.7 Macrophages
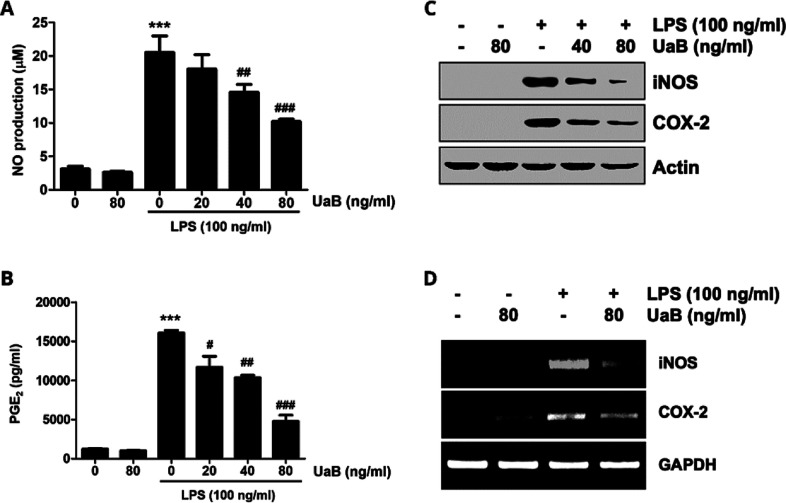
Stimulation of RAW 264.7 macrophages with LPS alone significantly increased the production of NO and PGE2 in the culture medium (Figure 2). However, upon introduction with UaB, there was a concentration-dependent decline in proinflammatory mediators (Figure 2). Additionally, western blotting analysis and reverse transcription-polymerase chain reaction (RT-PCR) were performed to determine the effects of UaB in protein and mRNA expression of iNOS and COX-2, respectively (Figure 2). Experimental data showed that there was a significant reduction in the expression of iNOS and COX-2 following the introduction of UaB as indicated by the fading of the bands at 80 ng/mL, implicating the suppression in the biosynthesis of NO and PGE2 in LPS-challenged RAW 264.7 macrophages, respectively.
Figure 2.
Inhibition of NO and PGE2 production by UaB fraction in LPS-stimulated RAW 264.7 macrophages. RAW 264.7 cells were incubated for 1 h with the indicated concentrations of UaB prior to stimulation with 100 ng/mL LPS for 24 h (A and B). The amounts of NO (A) and PGE2 (B) in the culture supernatants were determined via the Griess reaction and a commercial ELISA kit, respectively. Values are expressed as the mean ± SD of results from three independent experiments. ***p < 0.0001 vs controls (UaB- and LPS-untreated cells); ##p < 0.001 and ###p < 0.0001 vs cells cultured with 100 ng/mL LPS. Total protein was isolated from RAW 264.7 cells pretreated with the indicated concentrations of UaB followed by treatment with 100 ng/mL LPS for 24 h and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Western blot analysis was performed using the indicated antibodies and an enhanced chemiluminescence detection system (C). RAW 264.7 cells were pretreated with various concentrations of UaB for 1 h followed by treatment with 100 ng/mL LPS for 24 h. The total RNA was isolated, and the mRNA expression of inducible nitric oxide synthase (iNOS) and COX-2 was analyzed using RT-PCR (D). The experiments were repeated three times, and similar results were obtained. Actin and GAPDH were used as internal controls for the western blot and the RT-PCR assays, respectively. NO, nitric oxide; PGE2, prostaglandin E2; RT-PCR, reverse transcription-polymerase chain reaction; LPS, lipopolysaccharide; iNOS, inducible NO synthase; and COX, cyclooxygenase.
2.5. Effects of UaB on LPS-Induced Production of Proinflammatory Cytokines in RAW 264.7 Macrophages
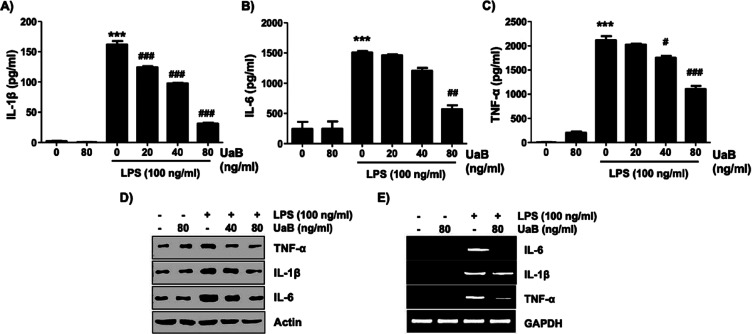
Stimulation of RAW 264.7 macrophages with LPS alone significantly increased the production of TNF-α, IL-1β, and IL-6 in the culture medium. However, following treatment with UaB, an apparent concentration-dependent decline in proinflammatory cytokines was noted (Figure 3). To further determine the effect of UaB in the protein and mRNA expression of these cytokines, western blotting and RT-PCR were performed, respectively (Figure 3). Experimental data showed a significant reduction in the expression of TNF-α, IL-1β, and IL-6 following the introduction of UaB as indicated by the fading of the bands at 80 ng/mL.
Figure 3.
Inhibition of TNF-α, IL-1β, and IL-6 production by the UaB fraction in LPS-stimulated RAW 264.7 macrophages. RAW 264.7 cells were incubated for 1 h with the indicated concentrations of UaB prior to stimulation with LPS (100 ng/mL) for 24 h (A–C). The amounts of TNF-α (A), IL-1β (B), and IL-6 (C) in the culture supernatants were determined using commercial ELISA kits. Values are expressed as the mean ± SD of results obtained from three independent experiments. ***p < 0.0001 vs controls (UaB- and LPS-untreated cells); #p < 0.01, ##p < 0.001, and ###p < 0.0001 vs cells cultured with 100 ng/mL LPS). Total protein was isolated from RAW 264.7 cells pretreated with the indicated concentrations of UaB followed by treatment with 100 ng/mL LPS for 24 h and subjected to SDS-PAGE. Western blot analyses were performed using the indicated antibodies and an enhanced chemiluminescence detection system (D). RAW 264.7 cells were pretreated with UaB for 1 h followed by treatment with 100 ng/mL LPS for 24 h, and the total RNA was isolated. The mRNA expression of TNF-α, IL-1β, and IL-6 was analyzed using RT-PCR (E). The experiments were repeated three times, and similar results were obtained. Actin and GAPDH were used as internal controls for the western blot and RT-PCR assays, respectively. IL, interleukin; TNF, tumor necrosis factor; RT-PCR, reverse transcription-polymerase chain reaction; and LPS, lipopolysaccharide.
2.6. Effects of UaB on LPS-Induced Nuclear Translocation of NF-κB in RAW 264.7 Macrophages
The effect of UaB in the NF-κB signaling pathway was also explored by immunofluorescence staining and western blot analysis. Figure 4 showed the possible impairment of the NF-κB activation posttreatment with UaB as shown by the decreased enhancement on immunofluorescence staining in comparison with the untreated LPS-induced macrophages. UaB also showed to abrogate nuclear translocation of the transcription factor p65 as indicated by western blot where a more prominent band is seen in untreated LPS-induced macrophages while a lighter band was detected in the UaB treated sample.
Figure 4.
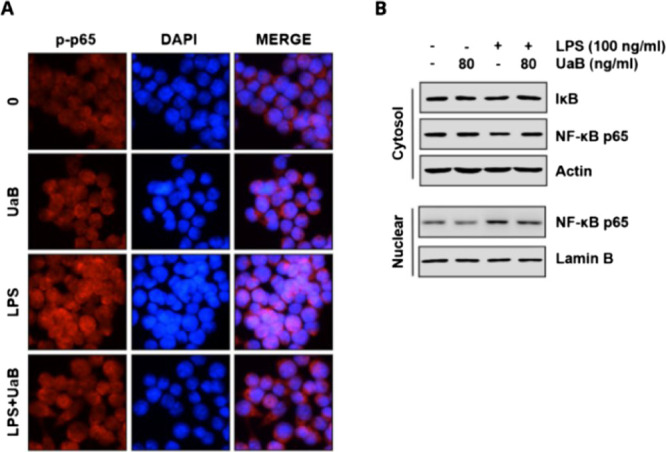
Effect of UaB fraction on LPS-induced nuclear translocation of NF-κB in RAW 264.7 macrophages. The cells were treated with 80 ng/mL UaB for 1 h prior to treatment with 100 ng/mL LPS for 30 min. Cells were pretreated with 80 ng/mL UaB for 1 h prior to stimulation with 100 ng/mL LPS for 30 min (A). Localization of NF-κB p65 was visualized following immunofluorescence staining with anti-NF-κB p65 antibody (red). The cells were also stained with DAPI for visualization of nuclei (blue). The cells were visualized using a fluorescence microscope (magnification, ×400). Cells were pretreated with 80 ng/mL UaB for 1 h prior to stimulation with 100 ng/mL LPS for 30 min (B). Nuclear and cytosolic proteins were subjected to 10% SDS-PAGE followed by western blot analysis using anti-NF-κB p65 and IκB antibodies. Lamin B and actin were used as internal controls for the nuclear and cytosolic fractions, respectively. NF, nuclear factor; IκB, inhibitor of NF-κB; and LPS, lipopolysaccharide.
2.7. Effects of UaB on NO and ROS Production in LPS-Treated Zebrafish Larvae
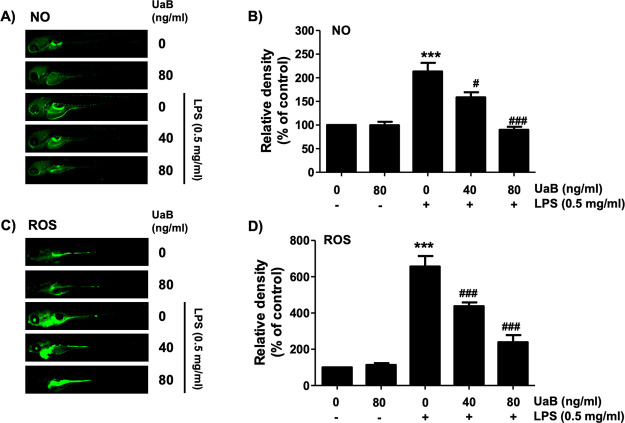
The inflammatory effect of UaB was further investigated using the zebrafish animal model. In vivo experiments showed an increase in both NO and ROS generation in LPS-treated zebrafish. However, when treated with UaB, there was a dose-dependent decrease in NO and ROS production depicted by the decreased enhancement of staining (Figure 5).
Figure 5.
Inhibition of LPS-induced NO and ROS generation by UaB in zebrafish larvae. Zebrafish at 3 dpf were microinjected with 2 nL of 0.5 mg/mL LPS and placed in E3 media containing the indicated concentrations of UaB for 24 h. The larvae were incubated with 5 μM DAF-FM-DA (A and B) or 20 μM DCF-DA (C and D) for NO and ROS detection, respectively, and visualized using the CELENAS Digital Imaging System. Relative fluorescence intensities were calculated and expressed compared to the untreated control (B and D). Each value indicates the mean ± SD and is representative of three independent experiments with 10 fish for each group. Significant differences among the groups were determined (***p < 0.001, vs LPS-unstimulated larvae; ##p < 0.01 and ###p < 0.001, vs LPS-stimulated larvae).
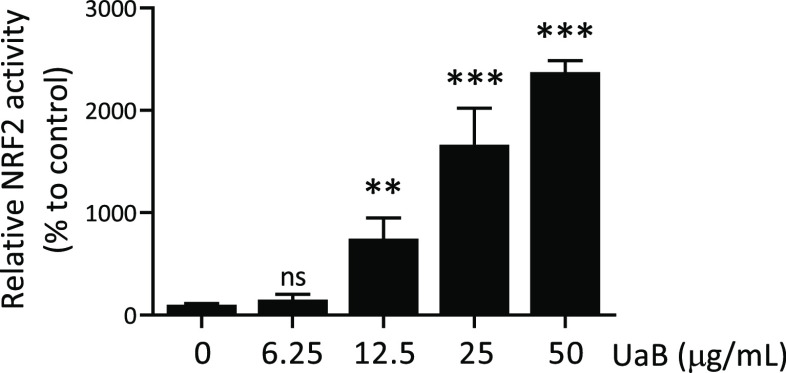
2.8. Effects of UaB on NRF2 Activity in HaCaT Keratinocytes
NRF2 is a master regulator for the expression of cytoprotective genes against oxidative stress and inflammatory responses; hence, we next determined the effects of UaB on NRF2 activity in HaCaT/ARE reporter cells. As shown in Figure 6, UaB increased NRF2 activity in a dose-dependent manner with an EC50 value of 21.6 ± 5.7 μg/mL. Therefore, UaB not only reduced proinflammatory and cytokine production following LPS exposure but also stimulated the counter-regulatory NRF2 anti-inflammatory pathway.
Figure 6.
Induction of NRF2 activity by UaB fraction in HaCaT keratinocytes. HaCaT/ARE cells were treated with indicated concentrations of UaB fraction for 24 h. Luciferase activity and cell viability were measured, and relative NRF2 activity was calculated as described in the Materials and Methods section. The average of the DMSO group was used as a standard for 100%. Data are presented as mean ± SD from three independent experiments. The asterisk (*) indicates a significant difference from the solvent control cells. (ns, none significant; **p < 0.001; ***p < 0.001, and one-way ANOVA).
2.9. LC–MS Profiling of UaB
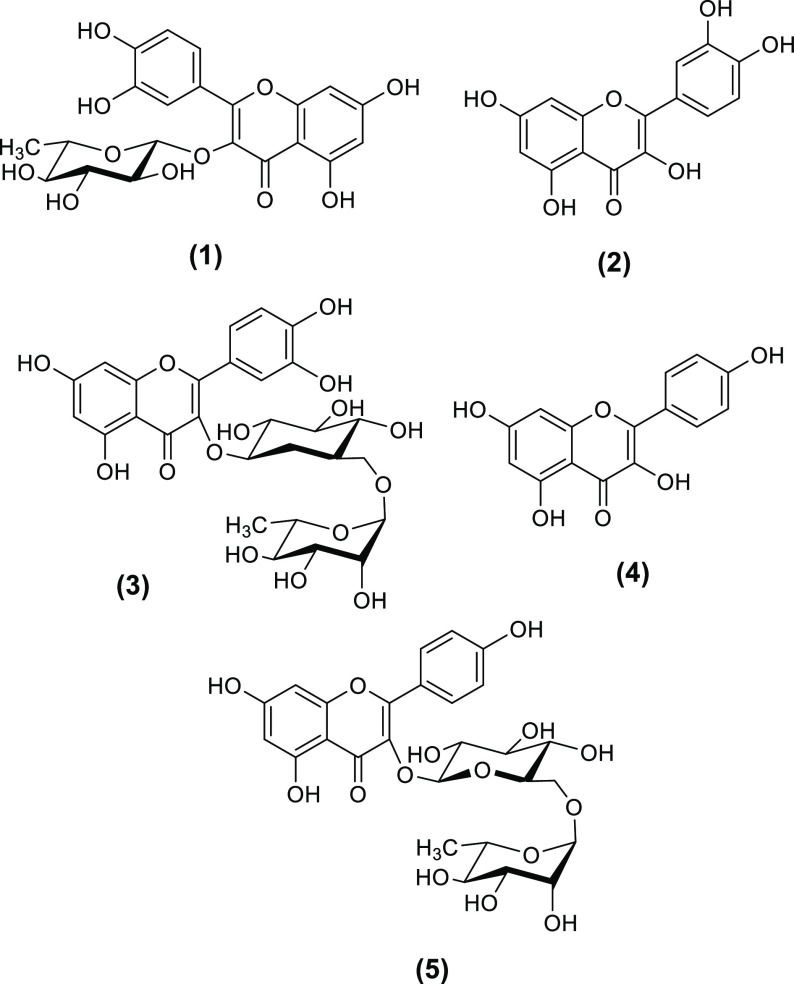
Given the selective COX-2 inhibitory activity of UaB, the secondary metabolites present in the fraction were identified through high-resolution LC–MS. The LC–MS profile (Figures S1–S9) highlighted the presence of flavonol aglycones quercetin (2) and kaempferol (4) and their glycosylated derivatives such as quercitrin (1), rutin (3), and kaempferol 3-O-rutinoside (5) (Figure 7; Table S4). Among the flavonol-containing secondary metabolites, compounds 3 and 5 were the most abundant comprising 53.1 and 23.7% in the UaB fraction.
Figure 7.
Secondary metabolites 1–5 detected in the butanol fraction of U. alba.
2.10. Molecular Docking to iNOS, COX-1/2, TACE, TNF-α, and IKK
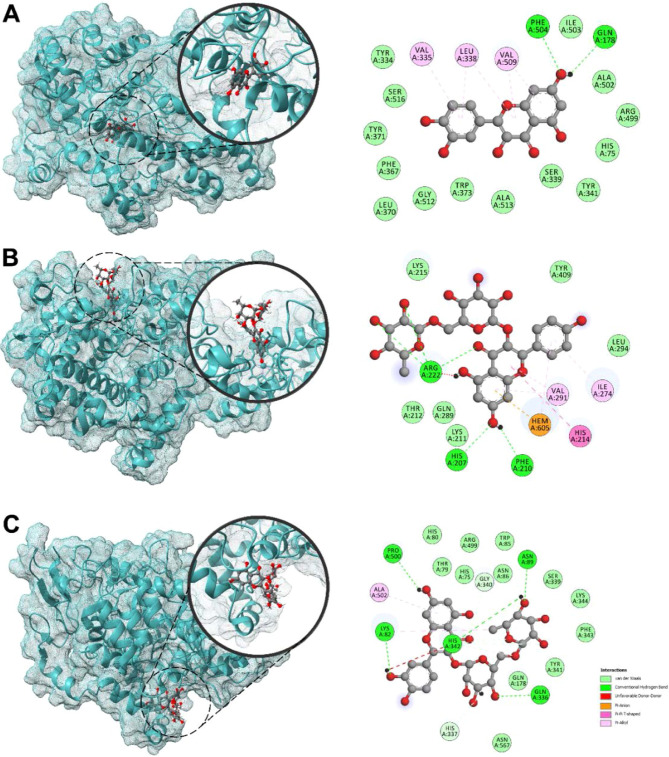
In order to further understand the potential anti-inflammatory mechanism of UaB, the structure of the putative flavonols present in the n-butanol fraction was subjected to molecular docking experiments to illustrate their binding interactions on relevant protein receptors responsible for proinflammatory responses. First, flavonols 1–5 were analyzed for their docking behavior against cyclooxygenases COX-1 and COX-2 (Table 2, Figure 8). Various binding regions of both COX-1 and COX-2 enzymes were investigated: (a) cyclooxygenase active site or the NSAID-binding site where oxygenation of arachidonic acid occurs; (b) peroxidase (POX) active site at the end of the catalytic tunnel where reduction of PGG2 to PGH2 occurs; and (c) a hydrophilic side chain that serves as entry to the catalytic tunnel. At the NSAID-binding site of COX-2, the flavonol aglycones 2 and 4 exhibited the best binding with binding energies (BEs) of −9.6 and −9.4 kcal/mol, respectively. In particular, the polycyclic core of quercetin (2) is bound onto the binding pocket via π–alkyl interactions with hydrophobic amino acids, particularly Val335, Leu338, and Val509. Two conventional hydrogen bonding with Gln178 Phe504 also played a key role in strengthening 2’s attachment to the site. Against the POX active site of COX-2, the larger glycosylated flavonols 3 and 5 showed the highest binding propensities with BEs of −9.4 and −9.5 kcal/mol, respectively. For compound 5, attachment to the POX active site is predominantly held by hydrophilic interactions between the flavonoid core and the heme group as well as several H-bonding initiated by the hydroxyl groups in C7 and the sugar moiety. At the COX-2 side pocket, 3 (−8.6 kcal/mol) and 5 (−8.5 kcal/mol) displayed the strongest binding. This binding region serves as an entrance to the narrow catalytic tunnel, which is dominated with polar residues and thus caters more to larger polyhydroxylated ligands such as 3 and 5. Molecular docking of the metabolites against COX-1 (PDB ID: 3KK6) showed that the kaempferol (4) had better binding toward the NSAID-binding site (BE = −7.9 kcal/mol); however, the compound still showed better selectivity toward COX-2.
Table 2. Binding Energies of Compounds 1–7 against COX-2 (PDB ID: 4M11) and COX-1 (PDB ID: 3KK6).
| compound | binding
energy (kcal/mol) |
|||||
|---|---|---|---|---|---|---|
| COX2-NSAID | COX2-POX | COX2-sidechain | COX1-NSAID | COX1-POX | COX1-sidechain | |
| 1 | –8.7 | –9.0 | –8.0 | –7.3 | –9.0 | –7.9 |
| 2 | –9.6 | –8.9 | –6.9 | –7.3 | –8.6 | –7.8 |
| 3 | –6.5 | –9.4 | –8.6 | –6.6 | –8.7 | –7.9 |
| 4 | –9.4 | –8.5 | –6.9 | –7.9 | –8.5 | –7.8 |
| 5 | –6.8 | –9.5 | –8.5 | –7.3 | –9 | –8.7 |
| 6 | –7.1 | –7.4 | –6.1 | –6.3 | –7.3 | –6.2 |
| 7 | –7.1 | –7.7 | –5.1 | –7.2 | –7.2 | –5.2 |
| celecoxib | –12.4 | –8.1 | –7.4 | –10.2 | –8.6 | –7.4 |
| indomethacin | –9.4 | –7.4 | –6.9 | –8.5 | –8.9 | –8.7 |
Figure 8.
3D and 2D docked poses of quercetin (2) (A) against COX-2 NSAID-binding site (PDB ID: 4M11); kaempferol 3-O-rutinoside (5) (B) against COX-2 POX-binding site (PDB ID: 4M11); and rutin (3) (C) against COX-2 side pocket (PDB ID: 4M11).
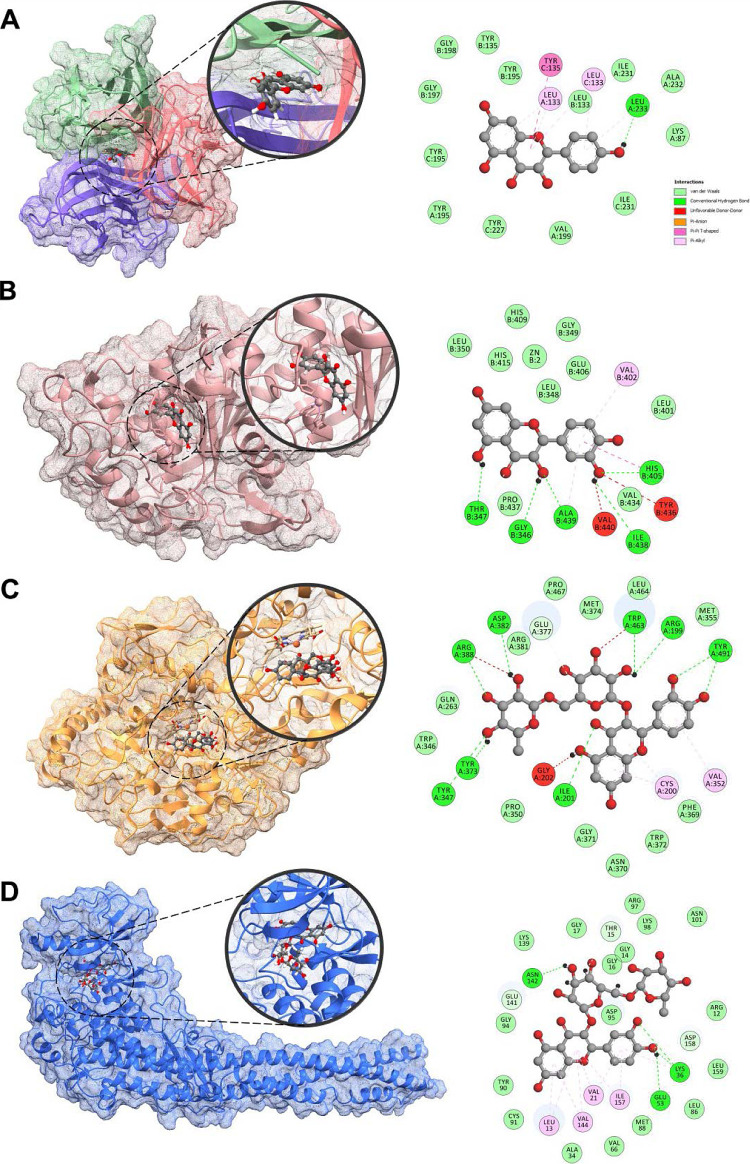
The flavonoids were also subjected to molecular docking analysis against other receptors, such as TNF-α, TNF-α converting enzyme (TACE), and iNOS (Table 3, Figure 9). Against TACE, quercetin (2) demonstrated the highest binding with a BE of −9.3 kcal/mol. Another aglycone, kaempferol (4), exhibited strong binding affinity (BE = −8.4 kcal/mol) toward the putative binding site of TNF-α trimer, which is generally held by hydrophobic interactions. The A and B rings of 4 were bound through π–π and π–alkyl interplay at the active site. Against iNOS, both rutin (3) and kaempferol 3-O-rutinoside (5) demonstrated high affinity with BEs of −10.0 kcal/mol. Both compounds are nestled to the binding site of iNOS strengthened mostly by hydrophilic interactions.
Table 3. Binding Energies of Compounds 1–7 against iNOS (PDB ID: 4NOS), TNF-α (PDB ID: 1TNF), TACE (PDB ID: 3KME), and IKK (PDB ID: 4KIK).
| compound | binding
energy (kcal/mol) |
|||
|---|---|---|---|---|
| iNOS | TNF-α | TACE | IKK | |
| 1 | –9.0 | –8.4 | –8.5 | –10.2 |
| 2 | –8.4 | –8.3 | –9.3 | –9.8 |
| 3 | –10.0 | –5.2 | –8.2 | –10.7 |
| 4 | –8.9 | –8.4 | –8.7 | –9.5 |
| 5 | –10.0 | –6.1 | –8.3 | –10.4 |
| 6 | –7.5 | –7.0 | –6.8 | –7.5 |
| 7 | –7.1 | –7.9 | –7.7 | –6.9 |
| tilarginine | –6.5 | |||
| pentoxifylline | –8.6 | |||
| apratastat | –6.8 | |||
| MLN120B | –9.6 | |||
Figure 9.
3D and 2D docked poses of kaempferol (4) (A) against TNF-α (PDB ID: 1TNF); quercetin (2) (B) against TACE (PDB ID: 3KME); rutin (3) (C) against iNOS (PDB ID: 4NOS); and rutin (3) (D) against IKK (PDB ID: 4KIK).
Finally, flavonols 1–5 were docked against IKK. Among the metabolites, rutin (3) demonstrated a strong binding toward the IKK (BE = −10.7 kcal/mol). The flavonoid core of 3 is affixed to the binding pocket through π–alkyl interactions and conventional hydrogen bonding in the B ring. The presence of a sugar moiety on 3 strengthened the interactions through H-bonding and C–H bonding.
2.11. MD Simulation of Top-Ranked Ligands in Complex with iNOS, COX-1/2, TACE, TNF-α, and IKK
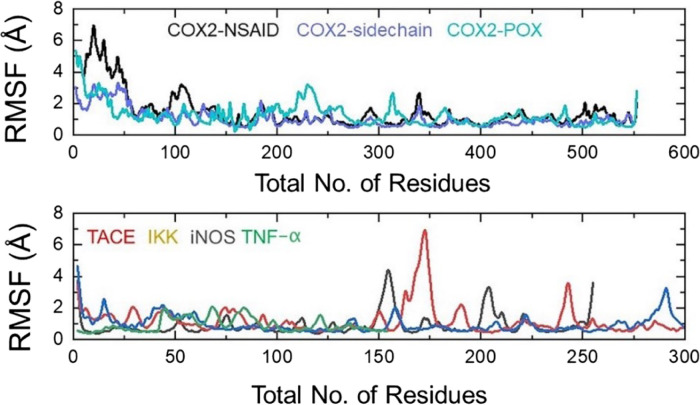
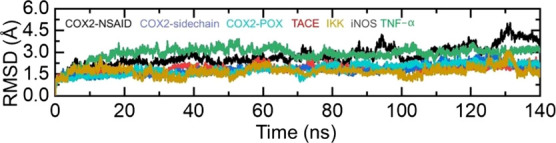
Understanding the structural dynamic features is an important parameter to reveal the binding of ligands to a specific receptor. Herein, using the MD simulation approach, we performed structural stability and residual flexibility analysis to understand the binding stability of each ligand. Structural stability calculated as the root mean square deviation (RMSD) as a function of time revealed that all the ligands bind stably in the binding cavity except for some minor deviations in a particular complex. In the case of COX2-NSAID_2, the structure demonstrated a little unstable behavior between 100 and 140 ns, while the average RMSD was calculated to be 2.5 Å. Moreover, for all the complexes remained lower comparatively and demonstrated average RMSD between 2.0 and 2.5 Å, respectively. This shows the stable binding of these ligands into the binding cavity of the respective receptors. The RMSD graphs of all the complexes are given in Figure 10.
Figure 10.
Dynamics stability investigation of ligands inside the binding cavity of enzymes over 140 ns simulation time for the following complexes: COX2-NSAID_2, COX2-sidechain_3, COX2-POX_5, TACE_2; TNF-α_4, iNOS_3; and IKK_3.
To understand the residual flexibility that confers an important role in biological processes and demonstrates the strength of binding, we used the root mean square fluctuation (RMSF) approach to calculate the flexibility. As displayed in Figure 11, the COX2-NSAID_2, COX2-sidechain_3, and COX2-POX_5 complexes demonstrated a more similar pattern of flexibility. Moreover, the RMSF for other complexes including TACE_2, IKK_3, iNOS_3, and TNF-α_4 revealed acceptable fluctuation of the residues’ flexibility by minimizing the flexibility of the residues thus showing the stable binding and alteration of internal dynamics. Like the RMSD, the RMSF plot, showing a relatively minimal fluctuation pattern, demonstrates similar behavior consequently favoring the stable binding of ligands. The RMSF of each complex is shown in Figure 11.
Figure 11.
Residual flexibility investigation of ligands inside the binding cavity of enzymes over 140 ns simulation time for the following complexes: COX2-NSAID_2, COX2-sidechain_3, COX2-POX_5, TACE_2; TNF-α_4, iNOS_3; and IKK_3.
Binding free energy prediction of small molecules to a larger biological macromolecule by MM/GBSA is arguably the most widely used approach to re-investigate docking conformation, determining structural stability and predicting interacting hotspots and binding affinities. The aforementioned method is computationally less expensive than the extensive alchemical free energy methods and is categorized as more accurate than conventional scoring functions. Taking into account the high significance of this method, we calculate the total binding free energy for each complex. For each complex COX2-NSAID_2 (−25.23), COX2-sidechain_3 (−22.74), COX2-POX_5 (−26.53), TACE_2 (−29.08), IKK_3 (−23.24), iNOS_3 (−22.11), and TNF-α_4 reported −35.96 kcal/mol. This shows that for these ligands, particularly TNF-α, the binding was robust. The other energy terms such as van der Waals (vdW), electrostatic, surface accessible (SA), and generalized born (GB) are also given in Table 4.
Table 4. Binding Free Energy Calculation Results Calculated in kcal/mola.
| complexes | vdW | electrostatic | SA | GB | total |
|---|---|---|---|---|---|
| COX2-NSAID_2 | –16.47 | –25.58 | –6.33 | 23.15 | –25.23 |
| COX2-sidechain_3 | –13.23 | –29.92 | –4.87 | 25.28 | –22.74 |
| COX2-POX_5 | –17.66 | –35.38 | –3.37 | 29.88 | –26.53 |
| TACE_2 | –18.28 | –17.62 | –7.41 | 14.23 | –29.08 |
| IKK_3 | –26.36 | –21.77 | –5.32 | 30.21 | –23.24 |
| iNOS_3 | –31.52 | –17.1 | –2.11 | 28.62 | –22.11 |
| TNF-α_4 | –25.28 | –21.22 | –5.47 | 16.01 | –35.96 |
vdW: van der Waals; SA: surface accessible; GB: generalized born.
3. Discussion
Inflammation is the body’s natural defense against invading pathogens, toxins, and physical trauma.1 However, a hyperactive inflammatory response is also associated with autoimmune and neurodegenerative diseases, anaphylactic reactions, and neoplastic development.1 Thus, there have been efforts to discover and design immunomodulatory compounds to overcome the unwanted sequelae of an overwhelming inflammatory cascade.10 In this study, we investigated the inflammatory modulating properties of the butanol fraction from U. alba (UaB). Utilizing an enzyme-based assay, our initial screening revealed that UaB selectively inhibited COX-2, one of the key enzymes mediating the conversion of arachidonic acid to PGs.23 To further confirm this result, we investigated in vitro the potential of UaB in attenuating proinflammatory mediators and cytokines in LPS-challenged RAW 264.7 macrophages. Our results revealed that UaB suppressed the synthesis of NO and PGE2 by downregulating protein and mRNA expression of iNOS and COX-2, respectively. These proinflammatory mediators induce oxidative stress and play an integral part in the progression of chronic inflammatory diseases.24 Thus, NO and PGE2 are potential therapeutic targets for modulating inflammation-associated pathologies.25−28
In addition, our data also reported the significant decline of proinflammatory cytokines in LPS-challenged macrophages by downregulating protein and mRNA expression of TNF-α, IL-1β, and IL-6. Similar to proinflammatory mediators, these cytokines have chemotactic and vasoactive properties.24 TNF-α, in particular, upregulates proinflammatory cytokines, promotes angiogenesis, activates NF-κB, and stimulates NO production. IL-1β, together with TNF-α, induces fever, causes coagulation associated with inflammatory response, and induces severe inflammatory cascade through autocrine and paracrine mechanisms.1 Meanwhile, IL-6, on the other hand, is associated with chronic inflammation and serves as an inducer of acute phase reactants, B cells, and T lymphocytes.1,29 Inhibition of cytokines could attenuate a severe and potentially fatal inflammatory cascade.30,31 Additionally, we tested the effects of UaB on the production of NO and ROS in LPS-challenged zebrafish in vivo. Results showed a concentration-dependent decrease in both NO production and ROS accumulation posttreatment with UaB, thereby validating the observed anti-inflammatory potential in vitro.
The biosynthesis of proinflammatory mediators and cytokines is regulated by the all-encompassing NF-κB signaling pathway. In this study, we demonstrated the potential of UaB in suppressing NF-κB activation. Nuclear translocation of NF-κB is necessary for the transcription of proinflammatory mediators and cytokines. In the cytosol, NF-κB is inactive until IκB is phosphorylated through the initiation of IKK.4,7 Hence, the role of UaB in suppressing nuclear translocation of NF-κB p65 is an indication of its mechanistic anti-inflammatory properties (Figure 10). Several polyphenolic natural products have been shown to suppress inflammatory responses through this mechanism.10,32 Anthocyanins from black rice and carvacrol, a phenolic monoterpene present in plant essential oil of the Lamiaceae family, both abrogated nuclear translocation of NF-κB.29,33
We also explored the NRF2 activity of flavonol-enriched UaB. The NRF2-Keap1-ARE pathway is the main modulator of antioxidant and phase II detoxification genes. The activation of this pathway upregulates the expression of antioxidant and cytoprotective proteins, protecting cells against infections, free radicals, and pathologic inflammatory cascade. The increased activity of NRF2 in HaCaT/ARE reporter cells clearly showed that flavonols from UaB may play a role in activating counter-regulatory defense mechanisms against inflammatory processes.
Based on high-resolution LCMS profiling, the majority of the putatively identified metabolites from UaB are flavonol derivatives. Flavonols are known to exhibit anti-inflammatory properties via a variety of mechanisms, including inhibition of regulatory enzymes and transcription factors, both of which play essential roles in the regulation of mediators implicated in inflammation. The more abundant metabolites in the UaB fraction are quercetin (2), rutin (3), kaempferol (4), and kaempferol 3-O-rutinoside (5), which individually have been shown to be anti-inflammatory agents via in vitro and in vivo experiments.34−37
To further evaluate the role of UaB fraction as an anti-inflammatory, we investigated the binding behavior of the putative metabolites of UaB through molecular docking and molecular dynamic simulation methodologies against proinflammatory enzymes. In the docking analysis of the metabolites against COX-2, the compounds were bound to various active sites that probably inhibit specific enzymatic activities of COX-2. In particular, the smaller flavonol aglycone 2 bound strongly to the NSAID-binding site situated at the middle of a narrow tunnel, while the larger glycosylated5,6 flavonols 5 and 3 preferred binding at the peroxidase binding site and at the hydrophilic side chain, an entry to the cyclooxygenase tunnel. The various mechanisms of attachment at different catalytic sites of flavonols 1–5 suggest a potentially strong COX-2-inhibitory activity of the fraction, which corroborates with the in vitro data. The flavonoids also show selectivity toward COX-2 over COX-1 (Tables 1 and 2). Moreover, molecular docking analysis revealed that the aglycones 2 and 4 exhibited high affinity toward TACE and TNF-α with BEs of −9.3 and −8.4 kcal/mol, respectively, while the glycosylated flavonoid 3 showed high affinity toward iNOS and IKK with BEs of −10.0 and −10.7 kcal/mol. The top protein–ligand complexes of these enzymes and respective flavonoids were also found to be dynamically stable at 140 ns when molecular dynamics simulations were performed (Figures 10 and 11).
These computational data corroborate with in vitro and in vivo experiments, suggesting the profound anti-inflammatory potential of the UaB fraction. As such, this may help in understanding the key roles of dietary flavonoids in the regulation of inflammatory responses, which are valuable in the discovery of drug leads for the prevention, mitigation, and treatment of acute chronic inflammatory diseases.20,38,39
4. Conclusions
Compelling evidence showed that the flavonol-enriched U. alba butanol fraction (UaB) ameliorates inflammatory responses in LPS-challenged RAW 264.7 macrophages by attenuating the production of proinflammatory mediators (NO and PGE2) and cytokines (TNF-α, IL-1β, and IL-6) through the downregulation of their protein and mRNA expression. There was also a concentration-dependent decrease in the production and accumulation of NO and ROS in LPS-challenged zebrafish treated with UaB. The anti-inflammatory effects of UaB are attributed to its potential of increasing the cytoprotective NRF2 activity while attenuating the NF-κB pathway by inhibiting the nuclear translocation of transcription factor p65. Through nontargeted LC–MS analysis, molecular docking, and molecular dynamics simulations, the aglycone and glycosylated flavonols identified in UaB exhibited strong binding and form stable complexes with various active sites of COX-2 as well as with the binding pocket of TNF-α, TACE, iNOS, and IKK.
This is the first report to have elucidated the mechanistic anti-inflammatory properties of the Philippine endemic plant, U. alba. As such, isolation and synergistic studies of biologically active flavonoids from U. alba are highly encouraged to create a potent cocktail of anti-inflammatory agents. This cocktail can then be assayed using in vivo mouse models for inflammatory diseases to further establish its potential in ameliorating pathologic inflammatory disease mechanisms.
5. Materials and Methods
5.1. Plant Collection and Identification
The leaves of U. alba were collected in the lowlands of Palauig, Zambales, Luzon, Philippines (15°43′ N, 119°91′ E) in May 2020. These were authenticated with voucher specimens (USTH 1631) deposited at the University of Santo Tomas Herbarium and at the Philippine National Herbarium, Manila, Philippines.
5.2. Crude Extraction and Fractionation
The ground air-dried leaves (2.1 kg) were extracted with technical-grade (1,1) DCM–MeOH and concentrated in vacuo at 45 °C. 349 g of crude DCM–MeOH extract (Ua) was yielded. Ua crude extract was suspended in distilled water (20 L) and partitioned against increasing polarity of solvents, yielding three fractions, namely, petroleum ether (UaP), DCM (UaD, 92.6 g), and n-butanol (UaB, 137.5 g) fractions at room temperature. The fractions were concentrated in vacuo using a Buchi R-215 (Sankt Gallen, Switzerland) and tested for biological activities.17,18,22,40
5.3. High-Resolution Liquid Chromatography–Mass Spectrometry
UaB was chemically profiled to identify its putative secondary metabolites using untargeted LC-HRMS analysis. This was performed on a Shimadzu LC-20 AD apparatus equipped with an autosampler (SIL-20A, Shimadzu), a diode array detector (SPD-M20AV, Shimadzu), and coupled with a microToF II (Bruker Daltonics) ESI-QToF mass spectrometer. High-performance liquid chromatography column Chromolith Performance RP-18e (2.0 × 100 mm2 i.d.) was used for the analysis. The eluents were acetonitrile and water with 0.1% acetic acid. After injecting 5 μL of the UaB fraction, flow elution was set at 0.2 mL/min. The peaks were monitored at 350 nm. The mass spectra were recorded in the mass range m/z 50–2000. Bruker DataAnalysis 4.3 software (Bruker, Germany) was used for data acquisition and analysis. Individual components were identified by comparison of their m/z values in the total ion count profile with those compounds described in the literature or by matching their MS/MS spectra with those reported in a public repository of mass spectral data called MassBank.
5.4. ELISA-Based Cyclooxygenase Inhibition Assay
The potential anti-inflammatory property of U. alba fractions was evaluated based on its inhibition of COX-1 and COX-2 isoenzymes. Following the manufacturer’s protocol (cat. nos. 701070 and 701080; Cayman Chemical Co., Ann Arbor, MI, USA), an ELISA was performed with minor modifications (Garcia et al.32). U. alba samples were dissolved in DMSO and the 96-well plates were read using a Glomax microplate reader (Promega, Madison, Wisconsin) at 450 nm. The concentration–response curve in triplicate measurements was then used to estimate IC50 values GraphPad PRISM 5 software (GraphPad Software Inc., CA, USA). Selectivity index of the samples against COX-1 and COX-2 enzymes was determined using the formula: IC50 value COX-1/IC50 value COX-2.41
5.5. Cytotoxicity Assay
U. alba crude extract and fractions were assayed for cytotoxicity against nontumorigenic human keratinocytes (HaCaT) and human liver cancer cells (HepG2) (Leibniz Institute DSMZ, Braunschweig, Germany) using the CellTiter-Blue assay (Promega, Mannheim, Germany). Both HaCaT and HepG2 cells were maintained in an RPMI 1640 medium (Life Technologies, Darmstadt, Germany) supplemented with 10% fetal bovine serum (Life Technologies, Darmstadt, Germany), 100 U/mL penicillin, and 100 mg/mL streptomycin (Promocell, Heidelberg, Germany). Cultures were incubated at 37 °C under a humidified atmosphere and 5% CO2. The cell lines were subcultured every 3–4 days.29,30
Cells were seeded in 96-well plates at concentration of 1 × 104 cells/well. After 24 h incubation, the medium was removed, and the test samples were added to the cells. The reference chemotherapeutic drug was doxorubicin. Following the addition of the samples, the plates were cultured for 24 h at 37 °C. Hereafter, 20 μL of the CellTiter-Blue reagent was dispensed to each well and the cells were further incubated for 2 h at 37 °C. Fluorescence was measured using the microplate reader Infinite M200 (Tecan Trading AG, Switzerland) at excitation of 560 nm and emission of 590 nm. The IC50 values were computed using GraphPad PRISM 5 software (GraphPad Software Inc.).
5.6. Macrophage Culture and LPS Stimulation
UaB having a selective COX-2 inhibitory activity was further tested for its anti-inflammatory properties on LPS-challenged murine macrophages. RAW 264.7 cell line (ATCC, Manassas, VA, USA) was maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum, 2 mM l-glutamine, 100 U/mL penicillin, and 100 U/mL streptomycin (WelGENE Inc., Daegu, Korea). Cell cultures were incubated at 37 °C in a humidified atmosphere containing 5% CO2 and 95% air.
UaB was dissolved in DMSO, and final concentrations were adjusted by dilution with a complete culture medium. To stimulate the cells, the medium was replaced with fresh DMEM. LPS (E. coli Serotype 055:B5; cat. no. L2880; Sigma-Aldrich, Merck KGaA) was added in the presence or absence of the UaB fraction for the indicated periods.
5.7. Assessment of Macrophage Viability
To evaluate cytotoxicity of the UaB on murine macrophages, RAW 264.7 cells were seeded in 96-well plates at a density of 1 × 103 cells/well. Cells were treated with various concentrations of the UaB fraction for 1 h prior to incubation with 100 ng/mL LPS for 24 h. Subsequently, 3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyl tetrazolium bromide (MTT) (Sigma-Aldrich, Merck KGaA) was added to each well at 0.5 mg/mL, followed by incubation for 3 h at 37 °C in the dark. MTT solution was removed and 200 μL of 5% DMSO was added to dissolve the crystals. The viable cells were detected by reading the absorbance of formazan at 540 nm using an ELISA microplate reader (Dynatech Laboratories, Chantilly, VA, USA). The optical density of formazan formed in the untreated cells was considered to represent 100% viability.
5.8. Measurement of NO and PGE2 Production
RAW 264.7 cells were pretreated with UaB for 1 h; subsequently, they were stimulated with LPS for 24 h. Controls were maintained under the same culture conditions; however, they were not pre-incubated or stimulated. NO levels were indirectly determined by measuring the stable NO catabolite nitrite in the medium utilizing the Griess reaction. In brief, the conditioned medium (100 μL) was mixed with the same volume of Griess reagent (Sigma-Aldrich, Merck KGaA) and incubated for 10 min at room temperature. The optical density at 540 nm was measured using an ELISA microplate reader, and the nitrite concentration was calculated according to a standard curve generated from known concentrations of sodium nitrite. The PGE2 concentration in the conditioned medium was measured using a commercial PGE2 ELISA kit (cat. no. 514010; Cayman Chemical Co., Ann Arbor, MI, USA) following the manufacturer’s instructions.
5.9. ELISA for Proinflammatory Cytokines
The generation of proinflammatory cytokines TNF-α, IL-1β, and IL-6 was measured using ELISA kits (Table S5). RAW 264.7 cells were pre-incubated with the UaB for 1 h, followed by LPS stimulation for 24 h, and cytokine contents in the cell-free supernatants were measured using cytokine sandwich ELISA kits (cat. nos. MTA00B and MLB00C; R&D Systems, Minneapolis, MN, USA) according to the manufacturer’s protocols.
5.10. RT-PCR Assay
RAW 264.7 cells were pretreated with UaB for 1 h, followed by treatment with 100 ng/mL LPS for 24 h. Controls were maintained under the same culture conditions but were not pre-incubated or stimulated. Total RNA was extracted using TRIzol reagent (Invitrogen, Thermo Fisher Scientific Inc., Waltham, MA, USA), according to the manufacturer’s instructions. The complementary (c)DNA of each sample was prepared using 2 μg RNA, 1 μL of Moloney’s murine leukemia virus reverse transcriptase, 1 mM deoxynucleoside triphosphate, and 1 μL of oligo(dT) according to the manufacturer’s protocol. DNA amplification was performed in an AccuPower PCR PreMix (Bioneer Corp., Daejeon, Korea). iNOS, COX-2, TNF-α, IL-1β, and IL-6 genes were amplified from the cDNA using PCR (Eppendorf, Hamburg, Germany). After amplification, the PCR products were electrophoresed on 1% agarose gels using a 1 Kb Plus DNA Ladder (0787018, Thermo Fisher Scientific, Waltham, MA, USA) and visualized following staining with ethidium bromide (Sigma-Aldrich, Merck KGaA) for 10 min at room temperature under ultraviolet irradiation using the gel documentation system (CHEMI-SMART 2026M.WL, Vilber Lourmat, Marne-la-Valle, France). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. The PCR primers are as follows: iNOS forward, 5′-ATG TCC GAA GCA AAC ATCAC-3′ and reverse, 5′-TAA TGT CCA GGA AGT AGG TG-3′; COX-2 forward, 5′-CAG CAA ATC CTT GCT GTT CC-3′ and reverse, 5′-TGG GCA AAG AAT GCA AAC ATC-3′; TNF-α forward, 5′-TCT CAT CAG TTC TAT GGC CC-3′ and reverse, 5′-GGG AGT AGA CAA GGT ACA AC-3′; IL-1β forward, 5′-GGG CTG CTT CCA AAC CTT TG-3′ and reverse, 5′-GCT TGG GAT CCA CAC TCT CC-3′, IL-6 forward, 5′-AAG TGC ATC ATC GTT GTT TTCA-3′ and reverse, 5′-GAG GAT ACC ACT CCC AAC AG-3′; and GAPDH forward, 5′-AGG CCG GTG CTG AGT ATG TC-3′ and reverse, 5′-TGC CTG CTT CAC CAC CTT CT-3′ (Bioneer Corp.). The PCR reaction was initiated at 94 °C for 2 min, followed by 25 cycles of 94 °C for 30 s, annealing temperature for 30 s and 72 °C for 30 s, and a final extension step at 72 °C for 5 min. The annealing temperatures were 63 °C for iNOS, COX-2, TNF-α, IL-1β, and IL-6, and 61 °C for GAPDH.
5.11. Protein Isolation and Western Blot Analysis
RAW 264.7 cells were incubated with the UaB at the indicated concentrations for 1 h prior to stimulation with 100 ng/mL LPS for 24 h. As described previously by Park et al. 2021,6 the cells were collected, lysed with a cell lysis buffer, and the protein concentration was determined using the Bradford Protein assay kit (Bio-Rad Laboratories, Hercules, CA, USA). In a parallel experiment, cytoplasmic and nuclear extracts were prepared using an NE-PER Nuclear and Cytoplasmic Extraction reagents kit (Pierce, Thermo Fisher Scientific, Inc.) following the manufacturer’s instructions. For western blotting, equal amounts of protein samples (30 μg/lane) were subjected to 10–13% SDS-PAGE, and then electrophoretically transferred onto polyvinylidene difluoride membranes (Schleicher & Schuell, Keene, NH, USA). Subsequently, the membranes were blocked with 5% nonfat dry milk in tris-buffered saline containing 0.1% triton X-100 (TBST) for 1 h and probed with specific primary antibodies at 4 °C overnight.42 After washing with TBST, the membranes were incubated with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies (dilution, 1:500; cat. no. sc-2004, goat anti-rabbit IgG-HRP; sc-2005, goat anti-mouse IgG-HRP; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) for 2 h at room temperature. The protein bands were detected on X-ray film using an enhanced chemiluminescence kit (cat. no. RPN 2232; GE Healthcare Life Sciences, Little Chalfont, UK) following the manufacturer’s instructions.
5.12. Immunofluorescence Staining
The effect of UaB on LPS-induced nuclear translocation of NF-κB was assessed using immunofluorescence microscopy. RAW 264.7 cells were initially grown on glass coverslips for 24 h at 37 °C in a humidified atmosphere containing 5% CO2 and subsequently incubated with 80 ng/mL UaB for 1 h prior to treatment with 100 ng/mL LPS for 30 min in the same culture conditions. The cells were fixed in 3.7% paraformaldehyde for 15 min, permeabilized with 0.2% triton X-100 in phosphate-buffered saline (PBS) for 15 min, and blocked for 10 min at room temperature with PBS containing 5% bovine serum albumin (Sigma-Aldrich, Merck KGaA). The cells were then stained with the primary antibody from Santa Cruz Biotechnology, Inc. (sc-8008, Dallas, TX, USA) against NF-κB p65 (1:100 dilution) overnight at 4 °C. Subsequently, cells were incubated with a fluorescein-conjugated anti-rat immunoglobulin G (1100 dilution; cat. no. 31629; Molecular Probes, Thermo Fisher Scientific, Inc.) in the dark for 40 min at 37 °C. Nuclei were sequentially stained with 2.5 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) solution (Sigma-Aldrich, Merck KGaA). The slides were then mounted, and fluorescence images were captured using a fluorescence microscope (Carl Zeiss, Oberkochen, Germany).
5.13. Zebrafish Maintenance and LPS Microinjection
Zebrafish (D. rerio) are often used for in vitro assays because they are small and easy to maintain, have a short lifespan and reproductive cycle, and are transparent, allowing for easy visualization of their internal structures. Additionally, previous genome sequencing studies revealed that 70% of human genes have a zebrafish homologue, making them suitable models for studying biological processes. They are relatively inexpensive and are not protected by animal welfare laws, making them a convenient choice for research.43−45 In this study, the AB strain zebrafish were provided by the laboratory of Dr. C.H. Kang (Nakdong National Institute of Biological Resources, Sangju, Republic of Korea) and were maintained at 28.5 °C with a 14/10 h light/dark cycle according to the standard guidelines of the Animal Care and Use Committee of Jeju National University (Approval no.: 2019–0053, Jeju, Republic of Korea). Fertilized embryos were collected after natural spawning as previously described and cultured in 2 mg/L methylene blue containing E3 embryo media at 28.5 °C.46 Three days postfertilized (dpf) zebrafish larvae were anesthetized using 0.04% tricaine (Sigma-Aldrich Chemical Co.), and LPS (0.5 mg/mL, 2 nL in each larva) was microinjected into the yolk using a Drummond NANOJECT III injector (Drummond Scientific, Broomall, PA, USA). The negative control group was injected with PBS. The larvae were washed three times after LPS microinjection and immediately placed in E3 media containing the indicated concentrations of UaB. Each group of larvae was cultured at 28.5 °C for 24 h.
5.14. NO and ROS Staining in Zebrafish Larvae
The production of NO and ROS in zebrafish larvae was visualized using 4-amino-5-methylamino-2′7′-difluorofluores-cein diacetate (DAF-FM-DA, Sigma-Aldrich Chemical Co.) and DCF-DA, respectively, 24 h after chemical treatment as previously described.46 In brief, zebrafish embryos (4 dpf) were transferred to 24-well plates and incubated with 5 μM DAF-FM-DA and 20 μM dichlorodihydrofluorescein diacetate(DCF-DA) for 30 min and visualized using the CELENAS Digital Imaging System (Logos Biosystems, Anyang, Gyeonggido, Republic of Korea). Fluorescence intensities were calculated using ImageJ software (Wayne Rasband, National Institute of Health, Bethesda, MD, USA) and expressed as a percentage compared to the untreated control.
5.15. NRF2 Reporter Assay
HaCaT/ARE cell line was cultured as described in a previous publication.47 The luciferase reporter assay and the cell viability assay were also performed based on methods described in the same paper. Relative luciferase activity was calculated by normalizing luciferase activity to cell viability. The average relative luciferase activity of DMSO wells was defined as the control and attributed a relative NRF2 activity of 100%.
5.16. Statistical Analyses
Each in vitro experiment was performed in triplicate and values are expressed as the mean ± standard deviation. Statistical analysis was performed using GraphPad PRISM 5 software (GraphPad Software Inc.). Differences between groups were assessed using analysis of variance followed by Tukey’s post hoc test or unpaired Student’s t-test. p < 0.05 was deemed statistically significant.
5.17. Molecular Docking
Molecular docking methodologies are used for predicting the binding behavior of ligands onto various receptors.20,48−50 In this study, all molecular docking experiments were performed on the University of California San Francisco (UCSF) Chimera platform.51 The three-dimensional structures of the proteins were retrieved from the RCSB protein data bank (RCSB.org) added to the docking platform in PDB format. Each protein crystal structure was processed by removing existing co-crystallized ligands and water molecules. Meanwhile, the ligands were added to the docking platform, rendered from SMILES notation or added as an SYBYL mol2 file. Minimization and docking preparation of ligand and protein structures were done by adding the missing hydrogen atoms and appropriate charges to the structures employing the Gasteiger charge method computed using Amber’s Antechamber module.52 The docking procedure was done using a flexible ligand into a flexible active site protocol, where the ligand was allowed to be flexible and torsion within a grid box encompassing the ligand-binding cavity of each enzyme. A grid box was set around the bound co-crystallized ligand of the enzyme. With all docking parameters maintained at default values (number of binding modes = 10 at maximum exhaustiveness search), molecular docking simulation was performed following the Broyden–Fletcher–Goldfarb–Shanoo algorithm of AutoDock Vina (version 1.1.2).53 After each docking experiment, AutoDock Vina provides a set of docking poses for each ligand and corresponding binding affinities in which the docking pose with the best affinity was further subjected to postdock analysis. Visualization and analysis of the enzyme–ligand complex conformation were carried out using ChimeraX and Biovia Discovery Studios (version 4.1).54 Validation of the docking protocol was done by extracting the bound co-crystallized ligand and re-docking it to the set grid.20,40,54,55
5.18. Molecular Dynamics Simulation and Binding Free Energy Calculations
Using the AMBER20 tool, the dynamics and binding of each ligand in their respective pockets were performed by employing the FF19SB force field.56−58 For solvation purposes, an optimal point charge water box module was used while sodium and chlorine ions were added for system neutralization. Using steepest descent and conjugate gradient algorithms, gentle minimization was achieved followed by equilibration using both weak and without restraint and heating at 300 K and constant pressure.59,60 PMEMD.CUDA enabled simulation for 140 ns each.61 The trajectories were processed using CPPTRAJ and PTRAJ modules.62
Using MM/GBSA, the binding free energy for each complex was calculated, which is used by large-scale studies such as protein–protein, protein–ligand, and protein–RNA/DNA interactions.63−66 To estimate the total binding energy, such as electrostatic, GB, SA, and vdW, the MMGBSA.py script was utilized.67 The following equation was used for energy calculation:
The above-mentioned component of the total free energy was calculated using the equation below:
Acknowledgments
This work was supported in part by the International Foundation for Science (Grant no. F/5376-1).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c06451.
LC chemical profile of Uvaria alba dichloromethane (DCM) fraction (negative-ion mode MS detection); mass spectra of compounds 1, 3, 5 & 6; LC chemical profile of Uvaria alba DCM fraction (positive-ion mode MS detection); mass spectra of compounds 2, 4 & 7; secondary metabolites 1–7 detected in the n-butanol fraction of U. alba; the gene primer sequence; PCR reaction data; data of RNA quantification; secondary metabolites detected in the n-butanol fraction of U. alba (UaB); and the list of antibodies used in this study (PDF)
Author Contributions
K.I.R.N.: conceptualization, visualization, methodology, validation, formal analysis, data curation, writing—original draft, and writing—review & editing; M.T.J.Q., I.T.M., A.K., A.M.P., H.C.P.A., D.Y.P. I.V., S.M.M.T., A.W.S., S.Y.J., and G.Y.K.: methodology, validation, formal analysis, data curation, writing—original draft, and writing—review & editing; O.B.V., D.Q.W., and D.T.: visualization and validation; Y.H.C. and A.P.G.M.: conceptualization, visualization, methodology, validation, and writing—review & editing.
The authors declare no competing financial interest.
Supplementary Material
References
- Hung Y. L.; Suzuki K. The Pattern Recognition Receptors and Lipopolysaccharides (LPS)-Induced Systemic Inflammation. Int. J. Res. Stud. Med. Health Sci. 2017, 2, 1–7. [Google Scholar]
- Biswas R.; Bagchi A. NFkB Pathway and Inhibition: An Overview. Comput. Mol. Biol. 2016, 6, 1–20. 10.5376/cmb.2016.06.0001. [DOI] [Google Scholar]
- Fujiwara N.; Kobayashi K. Macrophages in Inflammation. Curr. Drug Targets: Inflammation Allergy 2005, 4, 281–286. 10.2174/1568010054022024. [DOI] [PubMed] [Google Scholar]
- Liu T.; Zhang L.; Joo D.; Sun S. C. NF-κB Signaling in Inflammation. Signal Transduction Targeted Ther. 2017, 2, 17023. 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; Ren Z.; Zhang J.; Chuang C. C.; Kandaswamy E.; Zhou T.; Zuo L. Role of ROS and Nutritional Antioxidants in Human Diseases. Front. Physiol 2018, 9, 477. 10.3389/fphys.2018.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.; Lee H.; Hong S.; Molagoda I. M. N.; Jeong J. W.; Jin C. Y.; Kim G. Y.; Choi S. H.; Hong S. H.; Choi Y. H. Inhibition of Lipopolysaccharide-Induced Inflammatory and Oxidative Responses by Trans-Cinnamaldehyde in C2C12 Myoblasts. Int. J. Med. Sci. 2021, 18, 2480. 10.7150/IJMS.59169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak P. P.; Firestein G. S. NF-ΚB: A Key Role in Inflammatory Diseases. J. Clin. Invest. 2001, 107, 7–11. 10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S.; Buttari B.; Panieri E.; Profumo E.; Saso L. An Overview of Nrf2 Signaling Pathway and Its Role in Inflammation. Molecules 2020, 25, 5474. 10.3390/molecules25225474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares M. P.; Seldon M. P.; Gregoire I. P.; Vassilevskaia T.; Berberat P. O.; Yu J.; Tsui T.-Y.; Bach F. H. Heme Oxygenase-1 Modulates the Expression of Adhesion Molecules Associated with Endothelial Cell Activation. J. Immunol. 2004, 172, 3553–3563. 10.4049/jimmunol.172.6.3553. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y.; Gaynor R. Role of the NF-KB Pathway in the Pathogenesis of Human Disease States. Curr. Mol. Med. 2001, 1, 287–296. 10.2174/1566524013363816. [DOI] [PubMed] [Google Scholar]
- Tresserra-Rimbau A.; Lamuela-Raventos R. M.; Moreno J. J. Polyphenols, Food and Pharma. Current Knowledge and Directions for Future Research. Biochem. Pharmacol. 2018, 156, 186–195. 10.1016/j.bcp.2018.07.050. [DOI] [PubMed] [Google Scholar]
- Yang L.; Zhou X.; Huang W.; Fang Q.; Hu J.; Yu L.; Ma N.; Zhang W. Protective Effect of Phillyrin on Lethal LPS-Induced Neutrophil Inflammation in Zebrafish. Cell. Physiol. Biochem. 2018, 43, 2074–2087. 10.1159/000484192. [DOI] [PubMed] [Google Scholar]
- Yang L. L.; Wang G. Q.; Yang L. M.; Huang Z. B.; Zhang W. Q.; Yu L. Z. Endotoxin Molecule Lipopolysaccharide-Induced Zebrafish Inflammation Model: A Novel Screening Method for Anti-Inflammatory Drugs. Molecules 2014, 19, 2390–2409. 10.3390/molecules19022390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q.; Zhu J.; Cao F.; Chen F. Anti-Inflammatory Properties of Extracts from Chimonanthus Nitens Oliv. Leaf. PLoS One 2017, 12, e0181094 10.1371/journal.pone.0181094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macabeo A. P. G.; Tudla F. A.; Alejandro G. J. D.; Kouam S. F.; Hussain H.; Krohn K. Benzoylated Derivatives from Uvaria rufa. Biochem. Syst. Ecol. 2010, 38, 857–860. 10.1016/j.bse.2010.08.007. [DOI] [Google Scholar]
- Macabeo A. P. G.; Tudla F. A.; Krohn K.; Franzblau S. G. Antitubercular Activity of the Semi-Polar Extractives of Uvaria rufa. Asian Pac. J. Trop. Med. 2012, 5, 777–780. 10.1016/S1995-7645(12)60142-4. [DOI] [PubMed] [Google Scholar]
- Macabeo A. P. G.; Martinez F. P. A.; Kurtán T.; Tóth L.; Mándi A.; Schmidt S.; Heilmann J.; Alejandro G. J. D.; Knorn M.; Dahse H. M.; Franzblau S. G. Tetrahydroxanthene-1,3(2 H)-Dione Derivatives from Uvaria valderramensis. J. Nat. Prod. 2014, 77, 2711–2715. 10.1021/np500538c. [DOI] [PubMed] [Google Scholar]
- Macabeo A. P. G.; Letada A. G.; Budde S.; Faderl C.; Dahse H. M.; Franzblau S. G.; Alejandro G. J. D.; Pierens G. K.; Garson M. J. Antitubercular and Cytotoxic Chlorinated Seco-Cyclohexenes from Uvaria alba. J. Nat. Prod. 2017, 80, 3319–3323. 10.1021/acs.jnatprod.7b00679. [DOI] [PubMed] [Google Scholar]
- Notarte K. I. R.; Devanadera M. K. P.; Mayor A. B. R.; Cada M. C. A.; Pecundo M. H.; Macabeo A. P. G. Toxicity, Antibacterial, and Antioxidant Activities of Fungal Endophytes Colletotrichum and Nigrospora Spp. Isolated from Uvaria grandiflora. Philipp. J. Sci. 2019, 148, 505–512. [Google Scholar]
- Quimque M. T. J.; Notarte K. I. R.; Fernandez R. A. T.; Mendoza M. A. O.; Liman R. A. D.; Lim J. A. K.; Pilapil L. A. E.; Ong J. K. H.; Pastrana A. M.; Khan A.; Wei D.-Q.; Macabeo A. P. G. Virtual Screening-Driven Drug Discovery of SARS-CoV2 Enzyme Inhibitors Targeting Viral Attachment, Replication, Post-Translational Modification and Host Immunity Evasion Infection Mechanisms. J. Biomol. Struct. Dyn. 2021, 39, 4316–4333. 10.1080/07391102.2020.1776639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudla F. A.; Aguinaldo A. M.; Krohn K.; Hussain H.; Macabeo A. P. G. Highly Oxygenated Cyclohexene Metabolites from Uvaria rufa. Biochem. Syst. Ecol. 2007, 35, 45–47. 10.1016/j.bse.2006.08.006. [DOI] [Google Scholar]
- Quimque M. T.; Notarte K. I.; Letada A.; Fernandez R. A.; Pilapil D. Y.; Pueblos K. R.; Agbay J. C.; Dahse H. M.; Wenzel-Storjohann A.; Tasdemir D.; Khan A.; Wei D. Q.; Gose Macabeo A. P. Potential Cancer- And Alzheimer’s Disease-Targeting Phosphodiesterase Inhibitors from Uvaria alba: Insights from In Vitro and Consensus Virtual Screening. ACS Omega 2021, 6, 8403–8417. 10.1021/acsomega.1c00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.; Deng H.; Cui H.; Fang J.; Zuo Z.; Deng J.; Li Y.; Wang X.; Zhao L. Inflammatory Responses and Inflammation-Associated Diseases in Organs. Oncotarget 2018, 9, 7204–7218. 10.18632/oncotarget.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniandy K.; Gothai S.; Badran K. M. H.; Kumar S. S.; Esa N. M.; Arulselvan P. Suppression of Proinflammatory Cytokines and Mediators in LPS-Induced RAW 264.7 Macrophages by Stem Extract of Alternanthera sessilis via the Inhibition of the NF-ΚB Pathway. J. Immunol. Res. 2018, 2018, 3430684 10.1155/2018/3430684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. H.; Kim G. Y.; Lee H. H. Anti-Inflammatory Effects of Cordycepin in Lipopolysaccharide-Stimulated RAW 264.7 Macrophages through Toll-like Receptor 4-Mediated Suppression of Mitogen-Activated Protein Kinases and NF-ΚB Signaling Pathways. Drug Des. Dev. Ther. 2014, 8, 1941–1953. 10.2147/DDDT.S71957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.; Cha H. J.; Lee H.; Kim G. Y.; Choi Y. H. The Regulation of the TLR4/NF-ΚB and Nrf2/HO-1 Signaling Pathways Is Involved in the Inhibition of Lipopolysaccharide-Induced Inflammation and Oxidative Reactions by Morroniside in RAW 264.7 Macrophages. Arch. Biochem. Biophys. 2021, 706, 108926 10.1016/j.abb.2021.108926. [DOI] [PubMed] [Google Scholar]
- Martinez J.; Sanchez T.; Moreno J. J. Regulation of Prostaglandin E2 Production by the Superoxide Radical and Nitric Oxide in Mouse Peritoneal Macrophages. Free Rad. Res. 2000, 32, 303. 10.1080/10715760000300301. [DOI] [PubMed] [Google Scholar]
- Vivancos M.; Moreno J. J. Role of Ca2+-Independent Phospholipase A2 and Cyclooxygenase/Lipoxygenase Pathways in the Nitric Oxide Production by Murine Macrophages Stimulated by Lipopolysaccharides. Nitric Oxide 2002, 6, 255–262. 10.1006/niox.2001.0410. [DOI] [PubMed] [Google Scholar]
- Limtrakul P.; Yodkeeree S.; Pitchakarn P.; Punfa W. Suppression of Inflammatory Responses by Black Rice Extract in RAW 264. 7 Macrophage Cells via Downregulation of NF- kB and AP-1 Signaling Pathways. Asian Pac. J. Cancer Prev. 2015, 16, 4277–4283. 10.7314/APJCP.2015.16.10.4277. [DOI] [PubMed] [Google Scholar]
- Fang Y.; Yang L.; He J. Plantanone C Attenuates LPS-Stimulated Inflammation by Inhibiting NF-ΚB/INOS/COX-2/MAPKs/Akt Pathways in RAW 264.7 Macrophages. Biomed. Pharmacother. 2021, 143, 112104 10.1016/J.BIOPHA.2021.112104. [DOI] [PubMed] [Google Scholar]
- Novilla A.; Djamhuri D. S.; Nurhayati B.; Rihibiha D. D.; Afifah E.; Widowati W. Anti-Inflammatory Properties of Oolong Tea (Camellia Sinensis) Ethanol Extract and Epigallocatechin Gallate in LPS-Induced RAW 264. 7 Cells. Asian Pac. J. Trop. Biomed. 2017, 7, 1005–1009. 10.1016/J.APJTB.2017.10.002. [DOI] [Google Scholar]
- Garcia K. Y. M.; Quimque M. T. J.; Primahana G.; Ratzenböck A.; Cano M. J. B.; Llaguno J. F. A.; Dahse H. M.; Phukhamsakda C.; Surup F.; Stadler M.; Macabeo A. P. G. COX Inhibitory and Cytotoxic Naphthoketal-Bearing Polyketides from Sparticola junci. Int. J. Mol. Sci. 2021, 22, 12379. 10.3390/ijms222212379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somensi N.; Rabelo T. K.; Guimarães A. G.; Quintans-Junior L. J.; de Souza Araújo A. A.; Moreira J. C. F.; Gelain D. P. Carvacrol Suppresses LPS-Induced pro-Inflammatory Activation in RAW 264.7 Macrophages through ERK1/2 and NF-KB Pathway. Int. Immunopharmacol. 2019, 75, 105743 10.1016/j.intimp.2019.105743. [DOI] [PubMed] [Google Scholar]
- Kong L.; Luo C.; Li X.; Zhou Y.; He H. The Anti-Inflammatory Effect of Kaempferol on Early Atherosclerosis in High Cholesterol Fed Rabbits. Lipids Health Dis. 2013, 12, 115. 10.1186/1476-511X-12-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Yao J.; Han C.; Yang J.; Chaudhry M. T.; Wang S.; Liu H.; Yin Y. Quercetin Inflammation and Immunity. Nutrients 2016, 8, 167. 10.3390/nu8030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyagbemi A. A.; Omobowale T. O.; Ola-Davies O. E.; Asenuga E. R.; Ajibade T. O.; Adejumobi O. A.; Afolabi J. M.; Ogunpolu B. S.; Falayi O. O.; Ayodeji F.; Hassan F. O.; Saba A. B.; Adedapo A. A.; Yakubu M. A. Ameliorative Effect of Rutin on Sodium Fluoride-Induced Hypertension through Modulation of Kim-1/NF-ΚB/Nrf2 Signaling Pathway in Rats. Environ. Toxicol. 2018, 33, 1284–1297. 10.1002/tox.22636. [DOI] [PubMed] [Google Scholar]
- Wang J.; Fang X.; Ge L.; Cao F.; Zhao L.; Wang Z.; Xiao W. Antitumor, Antioxidant and Anti-Inflammatory Activities of Kaempferol and Its Corresponding Glycosides and the Enzymatic Preparation of Kaempferol. PLoS One 2018, 13, e0197563 10.1371/journal.pone.0197563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleki S. J.; Crespo J. F.; Cabanillas B. Anti-Inflammatory Effects of Flavonoids. Food Chem. 2019, 299, 125124 10.1016/j.foodchem.2019.125124. [DOI] [PubMed] [Google Scholar]
- Pan M. H.; Lai C. S.; Ho C. T. Anti-Inflammatory Activity of Natural Dietary Flavonoids. Food Funct. 2010, 1, 15–31. 10.1039/c0fo00103a. [DOI] [PubMed] [Google Scholar]
- Santos J.; Quimque M. T.; Liman R. A.; Agbay J. C.; Macabeo A. P. G.; Corpuz M. J.-A.; Wang Y.-M.; Lu T.-T.; Lin C.-H.; Villaflores O. B. Computational and Experimental Assessments of Magnolol as a Neuroprotective Agent and Utilization of UiO-66(Zr) as Its Drug Delivery System. ACS Omega 2021, 6, 24382–24396. 10.1021/acsomega.1c02555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan K. C.; Walters M. J.; Musee J.; Harp J. M.; Kiefer J. R.; Oates J. A.; Marnett L. J. Molecular Basis for Cyclooxygenase Inhibition by the Non-Steroidal Anti-Inflammatory Drug Naproxen. J. Biol. Chem. 2010, 285, 34950–34959. 10.1074/jbc.M110.162982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon D. H.; Cha H. J.; Choi E. O.; Leem S. H.; Kim G. Y.; Moon S. K.; Chang Y. C.; Yun S. J.; Hwang H. J.; Kim B. W.; Kim W. J.; Choi Y. H. Schisandrin A Suppresses Lipopolysaccharide-Induced Inflammation and Oxidative Stress in RAW 264.7 Macrophages by Suppressing the NF-kB, MAPKs and PI3K/Akt Pathways and Activating Nrf2/HO-1 Signaling. Int. J. Mol. Med 2018, 41, 264–274. 10.3892/ijmm.2017.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahardehi A. M.; Arsad H.; Lim V. Zebrafish as a Successful Animal Model for Screening Toxicity of Medicinal Plants. Plants 2020, 9, 1345. 10.3390/plants9101345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassar S.; Adatto I.; Freeman J. L.; Gamse J. T.; Iturria I.; Lawrence C.; Muriana A.; Peterson R. T.; van Cruchten S.; Zon L. I. Use of Zebrafish in Drug Discovery Toxicology. Chem. Res. Toxicol. 2020, 33, 95–118. 10.1021/acs.chemrestox.9b00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y.; Meijer A. H.; Schaaf M. J. M. Modeling Inflammation in Zebrafish for the Development of Anti-Inflammatory Drugs. Front. Cell Dev. Biol. 2020, 8, 620984 10.3389/fcell.2020.620984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J. W.; Cha H. J.; Han M. H.; Hwang S. J.; Lee D. S.; Yoo J. S.; Choi I. W.; Kim S.; Kim H. S.; Kim G. Y.; Hong S. H.; Park C.; Lee H. J.; Choi Y. H. Spermidine Protects against Oxidative Stress in Inflammation Models Using Macrophages and Zebrafish. Biomol. Ther. 2018, 26, 146. 10.4062/BIOMOLTHER.2016.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. S.; Chang H. S.; Hsiao H. H.; Chen Y. F.; Kuo Y. P.; Yen F. L.; Yen C. H. Identification of Beilschmiedia Tsangii Root Extract as a Liver Cancer Cell–Normal Keratinocyte Dual-Selective Nrf2 Regulator. Antioxidants 2021, 10, 544. 10.3390/antiox10040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogi S.; Quimque M. T.; Notarte K. I.; Africa J. G.; Hernandez J. B.; Tan S. M.; Calderone V.; Macabeo A. P. Virtual Combinatorial Library Screening of Quinadoline B Derivatives against SARS-CoV-2 RNA-Dependent RNA Polymerase. Computation 2022, 10, 7. 10.3390/COMPUTATION10010007. [DOI] [Google Scholar]
- de Leon V. N. O.; Manzano J. A. H.; Pilapil D. Y. H.; Fernandez R. A. T.; Ching J. K. A. R.; Quimque M. T. J.; Agbay J. C. M.; Notarte K. I. R.; Macabeo A. P. G. Anti-HIV Reverse Transcriptase Plant Polyphenolic Natural Products with in Silico Inhibitory Properties on Seven Non-Structural Proteins Vital in SARS-CoV-2 Pathogenesis. J. Genet. Eng. Biotechnol. 2021, 19, 104. 10.1186/s43141-021-00206-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez R. A.; Quimque M. T.; Notarte K. I.; Manzano J. A.; Pilapil D. Y.; de Leon V. N.; San Jose J. J.; Villalobos O.; Muralidharan N. H.; Gromiha M. M.; Brogi S.; Macabeo A. P. G. Myxobacterial Depsipeptide Chondramides Interrupt SARS-CoV-2 Entry by Targeting Its Broad, Cell Tropic Spike Protein. J. Biomol. Struct. Dyn. 2022, 40, 12209–12220. 10.1080/07391102.2021.1969281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF Chimera - A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Wang J.; Wang W.; Kollman P. A.; Case D. A. Automatic Atom Type and Bond Type Perception in Molecular Mechanical Calculations. J. Mol. Graphics Modell. 2006, 25, 247–260. 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. Software News and Update AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quimque M. T.; Notarte K. I.; Adviento X. A.; Cabunoc M. H.; de Leon V. N.; Delos Reyes F. S. L.; Lugtu E. J.; Manzano J. A.; Monton S. N.; Muñoz J. E.; Ong K. D.; Pilapil D. Y.; Roque V.; Tan S. M.; Lim J. A.; Macabeo A. P. Polyphenolic Natural Products Active In Silico against SARS-CoV-2 Spike Receptor Binding Domains and Non-Structural Proteins – A Review. Comb. Chem. High Throughput Screen. 2023, 26, 459–488. 10.2174/1386207325666210917113207. [DOI] [PubMed] [Google Scholar]
- Quimque M. T.; Notarte K. I.; de Leon V. N. O.; Manzano J. A.; Muñoz J. E.; Pilapil D. Y.; Lim J. A.; Macabeo A. P.. Computationally Repurposed Natural Products Targeting SARS-CoV-2 Attachment and Entry Mechanisms. In Frontiers of COVID-19; Springer: Cham, 2022; pp. 505–537. [Google Scholar]
- Case D. A.; Cheatham T. E.; Darden T.; Gohlke H.; Luo R.; Merz K. M.; Onufriev A.; Simmerling C.; Wang B.; Woods R. J. The Amber Biomolecular Simulation Programs. J. Comput. Chem. 2005, 26, 1668–1688. 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearlman D. A.; Case D. A.; Caldwell J. W.; Ross W. S.; Cheatham T. E.; DeBolt S.; Ferguson D.; Seibel G.; Kollman P. AMBER, a Package of Computer Programs for Applying Molecular Mechanics, Normal Mode Analysis, Molecular Dynamics and Free Energy Calculations to Simulate the Structural and Energetic Properties of Molecules. Comput. Phys. Commun. 1995, 91, 1–41. 10.1016/0010-4655(95)00041-D. [DOI] [Google Scholar]
- Salomon-Ferrer R.; Götz A. W.; Poole D.; le Grand S.; Walker R. C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- Meza J. C. Steepest Descent. WIREs Comput. Stat. 2010, 2, 719–722. 10.1002/wics.117. [DOI] [Google Scholar]
- Watowich S. J.; Meyer E. S.; Hagstrom R.; Josephs R. A Stable, Rapidly Converging Conjugate Gradient Method for Energy Minimization. J. Comput. Chem. 1988, 9, 650–661. 10.1002/jcc.540090611. [DOI] [Google Scholar]
- Salomon-Ferrer R.; Case D. A.; Walker R. C. An Overview of the Amber Biomolecular Simulation Package. WIREs Comput. Mol. Sci. 2013, 3, 198–210. 10.1002/wcms.1121. [DOI] [Google Scholar]
- Roe D. R.; Cheatham T. E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- Ali A.; Khan M. T.; Khan A.; Ali S.; Chinnasamy S.; Akhtar K.; Shafiq A.; Wei D.-Q. Pyrazinamide Resistance of Novel Mutations in PncA and Their Dynamic Behavior. RSC Adv. 2020, 10, 35565–35573. 10.1039/D0RA06072K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A.; Tahir Khan M.; Saleem S.; Junaid M.; Ali A.; Shujait Ali S.; Khan M.; Wei D.-Q. Structural Insights into the Mechanism of RNA Recognition by the N-Terminal RNA-Binding Domain of the SARS-CoV-2 Nucleocapsid Phosphoprotein. Comput. Struct. Biotechnol. J. 2020, 18, 2174–2184. 10.1016/j.csbj.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir Ul Qamar M.; Ahmad S.; Khan A.; Mirza M. U.; Ahmad S.; Abro A.; Chen L.-L.; Almatroudi A.; Wei D.-Q. Structural Probing of HapR to Identify Potent Phytochemicals to Control Vibrio Cholera through Integrated Computational Approaches. Comput. Biol. Med. 2021, 138, 104929 10.1016/j.compbiomed.2021.104929. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Khan A.; Chandra Kaushik A.; Junaid M.; Zhang X.; Wei D. Q. The Systematic Modeling Studies and Free Energy Calculations of the Phenazine Compounds as Anti-Tuberculosis Agents. J. Biomol. Struct. Dyn. 2019, 37, 4051–4069. 10.1080/07391102.2018.1537896. [DOI] [PubMed] [Google Scholar]
- Sun H.; Li Y.; Tian S.; Xu L.; Hou T. Assessing the Performance of MM/PBSA and MM/GBSA Methods. 4. Accuracies of MM/PBSA and MM/GBSA Methodologies Evaluated by Various Simulation Protocols Using PDBbind Data Set. Phys. Chem. Chem. Phys. 2014, 16, 16719–16729. 10.1039/c4cp01388c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.