ABSTRACT
Several Pseudomonas aeruginosa AmpC mutants have emerged that exhibit enhanced activity against ceftazidime and ceftolozane, while also evading inhibition by avibactam. Interestingly, P. aeruginosa strains harboring these AmpC mutations fortuitously exhibit enhanced carbapenem susceptibility. This acquired susceptibility was investigated by comparing the degradation of imipenem by wild-type and cephalosporin-resistant AmpC. We show that cephalosporin-resistant AmpC enzymes lose their efficacy for hydrolyzing imipenem and suggest that this may be due to their increased flexibility and dynamics relative to the wild type.
KEYWORDS: AmpC, Pseudomonas aeruginosa, antibiotic resistance, beta-lactamase, imipenem
INTRODUCTION
The opportunistic pathogen Pseudomonas aeruginosa often resists β-lactam action through the expression of AmpC (or Pseudomonas-derived cephalosporinase [PDC]), a broad-spectrum class C β-lactamase (1–3). Enhanced antibiotic potency is therefore ensured through the concurrent administration of β-lactamase inhibitors (BLIs) (4). Combinations such as ceftolozane-tazobactam and ceftazidime-avibactam provide effective therapies against multidrug-resistant (MDR) or extensively drug-resistant (XDR) P. aeruginosa infections, partially alleviating the urgent clinical need for new agents (5–7). However, the emergence of resistance toward these antibiotic-plus-BLI treatments is of particular concern (8).
Previously, we described AmpC-mediated ceftolozane-tazobactam resistance mechanisms, under both in vitro (9) and treatment (10) conditions. The AmpC mutations underlying these mechanisms include substitutions or deletions in (i) Ω-loop residues (11, 12) or (ii) residues in the Ω-loop vicinity interacting with it (11–15); we quantified their effects on the antibiotic ring-opening efficacy and BLI binding strength (13). Interestingly, cephalosporin-resistant P. aeruginosa strains harboring these mutations also exhibit increased susceptibility to imipenem (9, 10); however, the biochemical basis for this enhanced susceptibility remains unclear. To gain insight into imipenem susceptibility, we quantified the reduction in its degradation efficiency by wild-type (WT) AmpC and the above cephalosporin/BLI variants of the enzyme. Based on these data, we provide insights into the molecular basis for why these ampC mutations exhibit opposite effects on imipenem versus cephalosporin hydrolysis.
Cephalosporin-resistant AmpC enzymes (WT, T96I, G183D, E247K, and ΔG229-E247 [a deletion from position 229 to 247]) were expressed and purified as previously described (13). Antibiotic hydrolysis was measured using an Infinite 200 PRO plate reader (Tecan Trading AG, Switzerland) operating in absorbance mode. Nitrocefin hydrolysis was carried out as before to ensure consistency with our previous work on these variants (13). Imipenem hydrolysis was monitored at 280 nm by challenging 0.5 μM AmpC enzyme solution with imipenem concentrations ranging from 0 to 400 μM for an hour. The reaction was carried out at 25°C in far-UV-transparent plates (Greiner, Monroe, NC, USA) in pH 7 buffer (0.1 M Na2HPO4 + 0.01% [wt/wt] bovine serum albumin [BSA]). The initial rate of product (P) formation was determined from the absorbance A by linear regression, using the following relationship:
where is the change in extinction coefficient (16). All enzyme runs were performed in triplicate and analyzed using SigmaPlot (Systat Software, Inc., CA, USA). All chemicals were obtained from Sigma-Aldrich.
Table 1 lists the enzyme kinetic parameters associated with the degradation of imipenem, nitrocefin, ceftolozane, and ceftazidime for all studied variants. Nitrocefin hydrolysis was measured as a control to ensure that all mutant enzymes are active. Clearly, cephalosporin-resistant AmpC variants hydrolyze ceftolozane and ceftazidime better than wild-type enzyme ( values jump by more than an order of magnitude). In contrast, these same variants degrade imipenem much less efficiently than wild-type enzyme ( values drop to less than 10%). These contradictory mutational effects upon the β-lactam ring opening are best understood if the molecular mechanisms of antibiotic degradation are carefully considered.
TABLE 1.
Michaelis-Menten kinetic parameters of WT and mutant AmpC enzymes for nitrocefin, imipenem, ceftazidime, and ceftolozanea
| Enzyme | Parameterb | Mean ± SD for: |
|||
|---|---|---|---|---|---|
| Ceftolozane | Ceftazidime | Nitrocefin | Imipenem | ||
| WT | kcat (s−1) | (1.0 ± 0.1) × 10−2 | (4.9 ± 0.6) × 10−3 | 50 ± 8 | (4.57 ± 0.01) × 10−3 |
| Km (μM) | 2,100 ± 300 | 350 ± 80 | 90 ± 30 | 37 ± 5 | |
| kcat/Km (μM−1 s−1) | (5 ± 1) × 10−6 | (1.4 ± 0.4) × 10−5 | (5 ± 2) × 10−1 | (1.2 ± 0.2) × 10−4 | |
| Fold change | 1 | 1 | 1 | 1 | |
| T96I | kcat (s−1) | NDc | ND | 1.4 ± 0.2 | (1.2 ± 0.2) × 10−3 |
| Km (μM) | ND | ND | 40 ± 10 | 61 ± 40 | |
| kcat/Km (μM−1 s−1) | (1.99 ± 0.04) × 10−4 | (2.27 ± 0.04) × 10−4 | (4 ± 1) × 10−2 | (2 ± 1) × 10−5 | |
| Fold change | 40 ± 2 | 17 ± 3 | 0.07 ± 0.02 | 0.16 ± 0.08 | |
| G183D | kcat (s−1) | (1.8 ± 0.2) × 10−1 | ND | 0.8 ± 0.1 | (1.8 ± 0.2) × 10−3 |
| Km (μM) | 1,200 ± 200 | ND | 24 ± 11 | 140 ± 40 | |
| kcat/Km (μM−1 s−1) | (1.5 ± 0.3) × 10−4 | (1.41 ± 0.02) × 10−4 | (3 ± 2) × 10−2 | (1.3 ± 0.4) × 10−5 | |
| Fold change | 31 ± 5 | 10.3 ± 0.1 | 0.06 ± 0.04 | 0.11 ± 0.03 | |
| E247K | kcat (s−1) | 1.5 ± 0.3 | (6.6 ± 0.4) × 10−1 | 0.9 ± 0.04 | (2.61 ± 0.18) × 10−3 |
| Km (μM) | 3,600 ± 700 | 1,020 ± 90 | 25 ± 4 | 140 ± 20 | |
| kcat/Km (μM−1 s−1) | (4 ± 1) × 10−4 | (6.5 ± 0.7) × 10−4 | (36 ± 6) × 10−3 | (1.9 ± 0.4) × 10−5 | |
| Fold change | 80 ± 20 | 47 ± 5 | 0.07 ± 0.01 | 0.16 ± 0.03 | |
| ΔG229-E247 | kcat (s−1) | ND | ND | 0.5 ± 0.05 | (3.5 ± 0.1) × 10−3 |
| Km (μM) | ND | ND | 50 ± 20 | 320 ± 20 | |
| kcat/Km (μM−1 s−1) | (1.22 ± 0.02) × 10−4 | (1.38 ± 0.02) × 10−4 | (10 ± 4) × 10−3 | (1.1 ± 0.1) × 10−5 | |
| Fold change | 25 ± 1 | 10.1 ± 0.1 | 0.019 ± 0.008 | 0.09 ± 0.01 | |
The kinetic parameters for nitrocefin and imipenem were obtained from the experimental data collected for this study (see Fig. S1 and S2 in the supplemental material); the parameters for cephalosporin were obtained from reference 7. All experiments were performed in triplicate. Reported uncertainties were obtained from regression fits.
Fold change, ratio of the mutant to WT AmpC substrate specificity constants (kcat/Km).
ND, not determined. For these enzymes, the Km values for ceftolozane and ceftazidime are significantly larger than 5 mM, and the Michaelis-Menten plots are linear not hyperbolic, precluding the determination of kcat and Km as independent parameters; the slope of the linear Michaelis-Menten plot measured at substrate concentrations close to zero, however, will be equal to , allowing kcat/Km to be determined.
β-Lactamase enzymes (E) degrade cephalosporins (C) through the following mechanism (17):
| (1) |
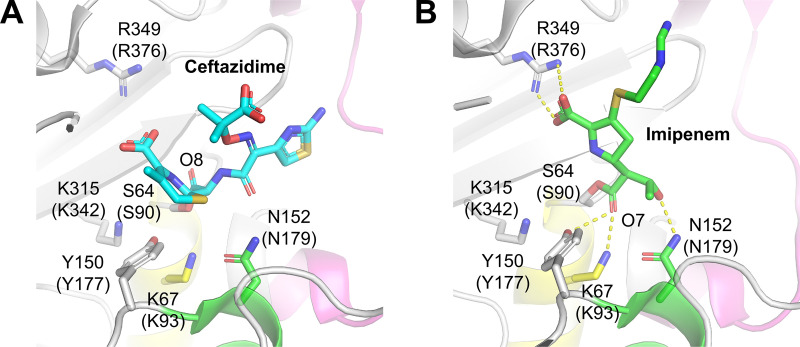
The third step of this mechanism is postulated (13, 17) to be the rate-determining step for ceftolozane and ceftazidime hydrolysis, and structural data suggest that cephalosporins bind AmpC in catalytically competent conformations (15)—the carbonyl oxygen lies in the oxyanion hole (formed by the backbone amides of S64 and A318), allowing T150 and water to activate it (Fig. 1A).
FIG 1.
Active site structures of AmpC with ceftazidime (PDB accession number 1IEL) (A) and imipenem (1LL5) (B). Residues are labeled with numbers corresponding to E. coli AmpC; numbers in parentheses correspond to Pseudomonas aeruginosa PAO1. α-Helix residues (91 to 106) is shown in yellow, α-helix residues (154 to 165) appears in green, and Ω-loop residues (229 to 247) is indicated in magenta.
Unfortunately, we were unable to crystalize the P. aeruginosa AmpC mutant proteins in this study. However, the structures of wild-type P. aeruginosa and Escherichia coli AmpC proteins are very similar (the root mean square deviation [RMSD] of alpha carbon is ~1.49 Å) (18, 19). Based on the X-ray crystal structures of E. coli AmpC mutant enzymes solved by Thomas et al. (20), we speculate that the effects of the amino acid changes (T96I, G183D, E247K, and ΔG229-E247) upon the structure of P. aeruginosa mutant proteins are as follows (see Fig. S3 in the supplemental material). In wild-type AmpC, the threonine (T96) hydroxyl can form a hydrogen bond with the backbone carbonyl of E247, which stabilizes the Ω-loop. Replacing threonine with isoleucine will prevent this hydrogen bond from forming. Similarly, the E247K replacement precludes the formation of a side chain-backbone hydrogen bond. On the other hand, the G183D replacement exchanges a small hydrogen atom for a bulky negatively charged group that can clash with other nearby moieties, such as E247. These three replacements may reduce favorable interactions between the Ω-loop and the (91 to 106) and (154 to 165) helices, thereby increasing the Ω-loop dynamics. In fact, Thomas et al. saw both loop structure perturbation and a reduction in protein stability as characterized by the melting point when these mutants were introduced (20). Based upon Fig. S3, the deletion mutation of the Ω-loop (ΔG229-E247) may also affect the folding free energy of the (91 to 106) and (154 to 165) helices.
Indeed, we measured the melting temperature of the P. aeruginosa AmpC mutant enzymes (T96I, G183D, E247K, and ΔG229-E247) in our previous work (13), and all the proteins exhibited a reduction in melting point relative to the wild type. This reduction in protein stability causes an increase in the enzyme dynamics (13, 21), allowing water molecules to access the protein interior. We postulate that this increase in dynamics and water access is the likely reason why the magnitude of khydrolysis for ceftolozane and ceftazidime is greater for the mutant enzymes than for wild-type enzyme (13).
Conversely, the crystal structure showed that the thermodynamically stable imipenem-AmpC complex has the imipenem carbonyl oxygen O7 outside the oxyanion hole (Fig. 1B) (14, 22). The activation of the carbonyl oxygen is required for β-lactamase catalysis, which led Maveyraud et al. to suggest (22) the existence of a catalytically active “canonical” acyl-enzyme complex structure having the carbonyl oxygen in optimal geometry. This canonical structure is a high-energy conformer because of the unique torsional strain it requires—namely, the strain that the 6α-1R-hydroxyethyl substituent applies to the acyl-serine bond (22). Therefore, we suggest the following mechanism of imipenem (I) ring opening to be more realistic than the scheme in equation 1:
| (2) |
In this mechanism, the hydrolysis step is still the slow rate-determining step, allowing all prior steps to be represented as equilibria. After substrate binding, the first necessary step for catalysis is the activation of its carbonyl oxygen through the displacement of water from the oxyanion hole and S64 acylation (22). Next, once the complex is formed, it equilibrates between two conformations (22): (i) the canonical form, in which the ester carbonyl is held in the oxyanion hole by two hydrogen bonds, and (ii) the “lower energy form,” similar to the crystal structure. Finally, hydrolysis of the acyl bond occurs, which requires the complex to be in the canonical form prior to deacylation (22).
We express the experimentally determined enzyme specificity constant of imipenem hydrolysis in terms of the microscopic equilibrium/rate constants seen in the scheme in equation 2 (see the supplemental material):
| (3) |
Equation 3 allows us to rationalize the observed the loss of imipenem degradation activity seen in AmpC mutants. Because these mutations presumably increase protein flexibility/dynamics, all enzyme species depicted in the scheme in equation 2 (except E − Icanonical, which has to be “locked” in a catalytically competent state) gain additional degrees of freedom. This increases the entropic penalty required for accessing the E − Icanonical conformation as represented by the constant Kring opening (23). The net effect of this penalty is an increase in the apparent activation barrier, which leads to an increase in Km and a decrease in the measured values for imipenem seen in the mutant AmpCs.
In conclusion, here, we compared the imipenem degradation efficiency of wild-type AmpC with mutants of the enzyme that evade avibactam and have enhanced activity toward cephalosporins (13). We showed that in contrast to cephalosporins, these mutants exhibit significantly smaller imipenem substrate specificity than the wild type. We rationalized this by considering the structural requirements of the catalytic cycle and the effects these point mutations have upon the AmpC dynamics. Cephalosporin resistance-conferring AmpC mutations cause a decrease in protein thermal stability (13), which increases the frequency of dynamic fluctuations (entropy) (21). We suggest that in contrast to cephalosporin degradation, this entropy gain lowers the imipenem degradation efficacy of AmpC enzymes. This efficiency loss leads to higher steady-state concentrations of imipenem inside the bacterium (13), increasing the susceptibility of cephalosporin-resistant P. aeruginosa strains toward imipenem.
ACKNOWLEDGMENTS
This research was supported by Natural Sciences and Engineering Research Council (NSERC) of Canada grants to M.K. (grant number RGPIN-2017-05935) and B.L.M. (grant number RGPIN-2020-05682).
B.L.M., G.C., K.K., and M.K. contributed to the writing of the manuscript. Kinetic assays were performed by G.C. and M.K. Analysis of the kinetic data was performed by G.C. and M.K. B.L.M. and A.O. provided the plasmids encoding WT ampC and the E247K, T96I, and ΔG229-E247 ampC mutants. This research was conceived by A.O., B.L.M., and M.K.
Footnotes
Supplemental material is available online only.
Contributor Information
Brian L. Mark, Email: brian.mark@umanitoba.ca.
Antonio Oliver, Email: antonio.oliver@ssib.es.
Mazdak Khajehpour, Email: mazdak.khajehpour@umanitoba.ca.
REFERENCES
- 1.Poole K. 2011. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2:65. 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park JT, Uehara T. 2008. How bacteria consume their own exoskeletons (turnover, and recycling of cell wall peptidoglycan). Microbiol Mol Biol Rev 72:211–227. 10.1128/MMBR.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moya B, Dötsch A, Juan C, Blázquez J, Zamorano L, Haussler S, Oliver A. 2009. Beta-lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog 5:e1000353. 10.1371/journal.ppat.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drawz SM, Bonomo RA. 2010. Three decades of beta-lactamase inhibitors. Clin Microbiol Rev 23:160–201. 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moya B, Zamorano L, Juan C, Pérez JL, Ge Y, Oliver A. 2010. Activity of a new cephalosporin, CXA-101 (FR264205), against beta-lactam-resistant Pseudomonas aeruginosa mutants selected in vitro and after antipseudomonal treatment of intensive care unit patients. Antimicrob Agents Chemother 54:1213–1217. 10.1128/AAC.01104-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torrens G, Cabot G, Ocampo-Sosa AA, Conejo MC, Zamorano L, Navarro F, Pascual Á, Martínez-Martínez L, Oliver A. 2016. Activity of ceftazidime-avibactam against clinical and isogenic laboratory Pseudomonas aeruginosa isolates expressing combinations of most relevant β-lactam resistance mechanisms. Antimicrob Agents Chemother 60:6407–6410. 10.1128/AAC.01282-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Duin D, Bonomo RA. 2016. Ceftazidime/avibactam and ceftolozane/tazobactam: second-generation β-lactam/β-lactamase inhibitor combinations. Clin Infect Dis 63:234–241. 10.1093/cid/ciw243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horcajada JP, Montero M, Oliver A, Sorlí L, Luque S, Gómez-Zorrilla S, Benito N, Grau S. 2019. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev 32:e00031-19. 10.1128/CMR.00031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabot G, Bruchmann S, Mulet X, Zamorano L, Moyà B, Juan C, Haussler S, Oliver A. 2014. Pseudomonas aeruginosa ceftolozane-tazobactam resistance development requires multiple mutations leading to overexpression and structural modification of AmpC. Antimicrob Agents Chemother 58:3091–3099. 10.1128/AAC.02462-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraile-Ribot PA, Cabot G, Mulet X, Periañez L, Martín-Pena ML, Juan C, Pérez JL, Oliver A. 2018. Mechanisms leading to in vivo ceftolozane/tazobactam resistance development during the treatment of infections caused by MDR Pseudomonas aeruginosa. J Antimicrob Chemother 73:658–663. 10.1093/jac/dkx424. [DOI] [PubMed] [Google Scholar]
- 11.MacVane SH, Pandey R, Steed LL, Kreiswirth BN, Chen L. 2017. Emergence of ceftolozane-tazobactam-resistant Pseudomonas aeruginosa during treatment is mediated by a single AmpC structural mutation. Antimicrob Agents Chemother 61:e01183-17. 10.1128/AAC.01183-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lahiri SD, Walkup GK, Whiteaker JD, Palmer T, McCormack K, Tanudra MA, Nash TJ, Thresher J, Johnstone MR, Hajec L, Livchak S, McLaughlin RE, Alm RA. 2015. Selection and molecular characterization of ceftazidime/avibactam-resistant mutants in Pseudomonas aeruginosa strains containing derepressed AmpC. J Antimicrob Chemother 70:1650–1658. 10.1093/jac/dkv004. [DOI] [PubMed] [Google Scholar]
- 13.Slater CL, Winogrodzki J, Fraile-Ribot PA, Oliver A, Khajehpour M, Mark BL. 2020. Adding insult to injury: mechanistic basis for how AmpC mutations allow Pseudomonas aeruginosa to accelerate cephalosporin hydrolysis and evade avibactam. Antimicrob Agents Chemother 64:e00894-20. 10.1128/AAC.00894-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beadle BM, Shoichet BK. 2002. Structural basis for imipenem inhibition of class C beta-lactamases. Antimicrob Agents Chemother 46:3978–3980. 10.1128/AAC.46.12.3978-3980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powers RA, Caselli E, Focia PJ, Prati F, Shoichet BK. 2001. Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: implications for resistance mutations and inhibitor design. Biochemistry 40:9207–9214. 10.1021/bi0109358. [DOI] [PubMed] [Google Scholar]
- 16.Takeuchi D, Akeda Y, Sugawara Y, Sakamoto N, Yamamoto N, Shanmugakani RK, Ishihara T, Shintani A, Tomono K, Hamada S. 2018. Establishment of a dual-wavelength spectrophotometric method for analysing and detecting carbapenemase-producing Enterobacteriaceae. Sci Rep 8:15689. 10.1038/s41598-018-33883-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chow C, Xu H, Blanchard JS. 2013. Kinetic characterization of hydrolysis of nitrocefin, cefoxitin, and meropenem by β-lactamase from Mycobacterium tuberculosis. Biochemistry 52:4097–4104. 10.1021/bi400177y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powers RA, Shoichet BK. 2002. Structure-based approach for binding site identification on AmpC β-lactamase. J Med Chem 45:3222–3234. 10.1021/jm020002p. [DOI] [PubMed] [Google Scholar]
- 19.Lahiri SD, Mangani S, Durand-Reville T, Benvenuti M, Luca FD, Sanyal G, Docquier J-D. 2013. Structural insight into potent broad-spectrum inhibition with reversible recyclization mechanism: avibactam in complex with CTX-M-15 and Pseudomonas aeruginosa AmpC β-lactamases. Antimicrob Agents Chemother 57:2496–2505. 10.1128/AAC.02247-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas VL, McReynolds AC, Shoichet BK. 2010. Structural bases for stability-function tradeoffs in antibiotic resistance. J Mol Biol 396:47–59. 10.1016/j.jmb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Závodszky P, Kardos J, Svingor Á, Petsko GA. 1998. Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proc Natl Acad Sci USA 95:7406–7411. 10.1073/pnas.95.13.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maveyraud L, Mourey L, Kotra LP, Pedelacq J-D, Guillet V, Mobashery S, Samama J-P. 1998. Structural basis for clinical longevity of carbapenem antibiotics in the face of challenge by the common class A β-lactamases from the antibiotic-resistant bacteria. J Am Chem Soc 120:9748–9752. 10.1021/ja9818001. [DOI] [Google Scholar]
- 23.Åqvist J, Kazemi M, Isaksen GV, Brandsdal BO. 2017. Entropy and enzyme catalysis. Acc Chem Res 50:199–207. 10.1021/acs.accounts.6b00321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aac.01409-22-s0001.pdf, PDF file, 0.6 MB (637KB, pdf)