ABSTRACT
Streptococcus pneumoniae is a common bacterial pathogen that causes infections in children worldwide, even after administration of the pneumococcal conjugate vaccine. S. pneumoniae serotype 35B, especially the clonal complex 558 (CC558) lineage, has emerged globally following implementation of the 13-valent pneumococcal conjugate vaccine. Serotype 35B strains are also associated with multidrug resistance to both β-lactams and non-β-lactam drugs. In addition, a novel serotype, 35D, which is closely related to 35B and differs in polysaccharide structure, was recently reported. However, the genetic relationship among globally disseminating serotype 35B and D (35B/D) strains remains unknown. To investigate the molecular epidemiology of global serotype 35B/D strains, we conducted a genomic analysis of serotype 35B/D strains from various continents, including those from the Japanese national surveillance collection. A total of 87 isolates were identified as serotype 35B/D in the Japanese surveillance collection (n = 1,358). All the isolates were assigned to either CC558 or CC2755. Serotype 35D isolates were interspersed with serotype 35B isolates. Phylogenetic analysis revealed the formation of multiple clusters by the Japanese serotype 35B/D-CC558 isolates among the foreign isolates, which suggested multiple events of introduction of the clone into Japan. The global 35B/D-CC558 strains were found to share specific penicillin-binding protein profiles, pbp1a-4, pbp2b-7, and pbp2x-7, associated with penicillin, cephalosporin, and carbapenem nonsusceptibility. Moreover, 88.5% of the Japanese 35B/D-CC558 and 35B/D-CC2755 isolates were found to harbor the Tn916-like integrative and conjugative elements Tn2009, Tn2010, and Tn6002, associated with multidrug resistance to macrolides and tetracyclines. The results of this study imply that serotype 35B/D-CC558 strains could be frequently transmitted intercontinentally.
KEYWORDS: Streptococcus pneumoniae, pneumococcal disease, serotype 35B, serotype 35D, multidrug resistance, whole-genome sequencing
INTRODUCTION
Streptococcus pneumoniae is a common bacterial pathogen that causes infections in children worldwide despite the introduction of pneumococcal conjugate vaccines (PCVs) in many countries (1). The introduction of PCVs resulted in a universal decline in the incidence of vaccine serotype invasive pneumococcal diseases (IPDs) by over 90% (2, 3). However, replacement by non-PCV serotypes has negated the overall effect of vaccination on IPDs in several regions (4, 5). In the post-13-valent PCV (PCV13) era, non-PCV serotypes 12F, 15B and 15C (15B/C), and 35B/D are among the most prevalent, with geographical expansion across the United States, Eurasia, and Africa in a global surveillance study using whole-genome sequencing (6).
IPDs caused by serotype 35B were rare before the introduction of PCVs. In the United States, serotype 35B accounted for only 0.5% of IPDs before the introduction of PCV7 (2). However, in the PCV era, the prevalence of serotype 35B in both IPD and nasopharyngeal carriage has been increasing in the United States (3, 7), and this trend has also been observed in other countries (8). The increasing prevalence of serotype 35B is of great concern because it is associated with high rates of penicillin resistance (9). Serotype 35B is also associated with a high risk of death due to IPD (10). In particular, the 35B-clonal complex 558 (CC558) lineage was reported as one of the major contributors to the increase in the incidence of multidrug-resistant IPDs after PCV7 and PCV13 implementation in the United States (11).
This lineage also served as a donor during capsular switching of sequence type 156 (ST156) strains, resulting in the emergence of nonvaccine type and multidrug-resistant (MDR) serotype 35B-ST156 strains (12). Thus, the emergence and spread of antibiotic-resistant S. pneumoniae strains are of great concern because they are associated with poor clinical outcomes and increased health care costs; therefore, continuous monitoring of such lineages is important (13).
PCV7 and PCV13 were introduced in Japan in 2010 and 2013, respectively. An increase in the incidence of IPDs caused by non-PCV serotypes, including 35B, has also been observed after PCV introduction in Japan (14, 15). Kasahara et al. reported a 3% (19/641) prevalence of 35B among pneumococcal isolates obtained between 2002 and 2012 in Japan (16). Among these, 12/19 (63.2%) were ST558 and 7/19 (36.8%) were ST2755. After the introduction of PCV13, serotype 35B reportedly accounted for 11.0% of all pneumococcal isolates from IPD and non-IPD patients (15). CC558 isolates, all of which were penicillin resistant, accounted for 77.9% of serotype 35B isolates, whereas ST2755 accounted for 19.8% of the serotype (15). Moreover, an increase in the frequency of MDR strains resistant to β-lactams, including penicillin, and non-β-lactams, such as macrolides, tetracyclines, and trimethoprim-sulfamethoxazole, has also been reported (16–18). However, the genetic relationship between global 35B-CC558 isolates and factors attributed to the dissemination of MDR serotype 35B strains is unclear.
Recently, a newly recognized serotype, 35D, which differs from 35B polysaccharide in structure and serology, was reported (19). Serotype 35D strains lack O-acetylation of capsular polysaccharides, an antigenically dominant epitope, due to a deficient O-acetyltransferase encoded by wgiG; therefore, there is concern that these strains might escape the effect of future vaccines targeting serotype 35B (19). Furthermore, serotype 35D isolates have been reported globally, and CC558, CC156, and CC198 were the predominant CCs found in this serotype (20). Therefore, serotype 35D is speculated to have emerged sporadically from the closely related serotype 35B strains (20).
We investigated the molecular epidemiology of global serotype 35B and 35D strains, including those from a Japanese nationwide surveillance collection.
RESULTS
Whole-genome-sequencing statistics.
The whole-genome-sequencing statistics are shown in Data Set S3 in the supplemental material. The average (±standard deviation) number of contigs was 92.0 (±38.4), the N50 (shortest contig length needed to cover 50% of the genome) was 84,113 (±9,366), and the mapping depth was 121.7 (±52.5).
STs, serotypes, and antimicrobial susceptibility.
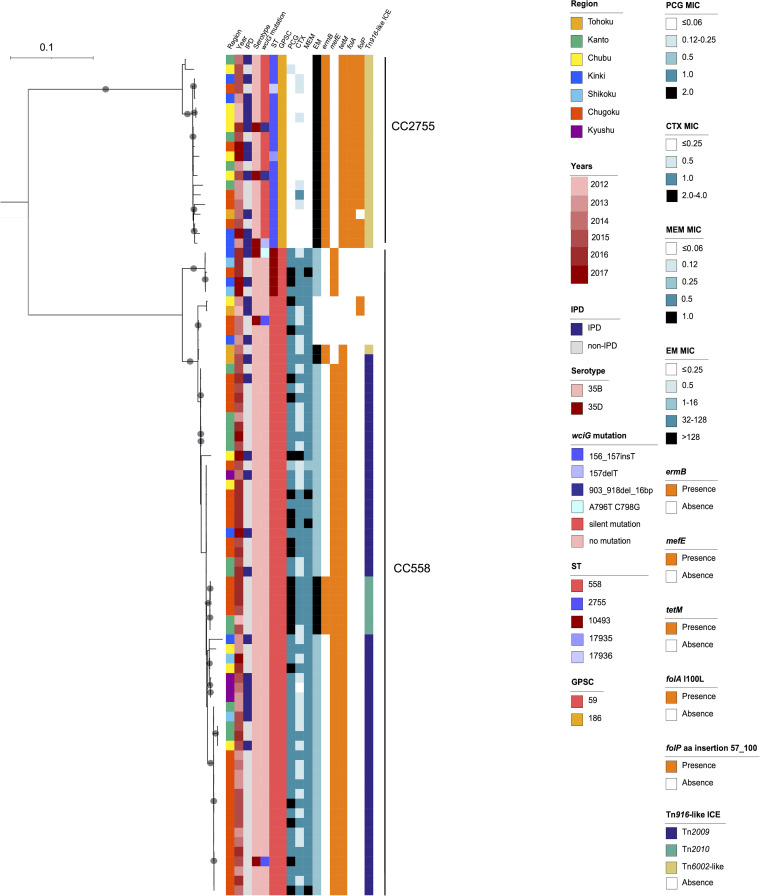
Among the 87 isolates originally typed as serotype 35B, we identified five sequence types (STs): ST558 (n = 62); ST10493 (n = 5), which is a single-locus variant (SLV) of ST558; ST2755 (n = 18); and ST17935 (n = 1) and ST17936 (n = 1), which are SLVs of ST2755. ST558 and ST10493 were assigned to CC558 and global pneumococcal sequence cluster 59 (GPSC59), whereas ST2755, ST17935, and ST17936 were assigned to CC2755 and GPSC186 (Fig. 1). Phylogenetic analysis of the Japanese serotype 35B and 35D isolates suggested a distant genetic relationship between CC558 and CC2755 (Fig. 1). Serotype 35D isolates were interspersed with serotype 35B isolates, irrespective of their CCs (Fig. 1).
FIG 1.
Phylogenetic tree of the serotype 35B and 35D isolates in Japan, generated by using RAxML-NG. Colored bars indicate characteristics of isolates including the region of isolation, year of isolation, serotype, susceptibility to penicillin, cefotaxime, meropenem, and erythromycin, presence of antimicrobial resistance determinants ermB, tetM, folA I100L mutation, and folP insertion, and types of Tn916-like integrative elements. Gray circles indicate ≥ 95% confidence by bootstrap analysis.
The wciG alignment revealed that 63 isolates (72.4%) were identical to the serotype 35B reference (accession number KX021817), 21 isolates had silent mutations (G657A [a change of G to A at position 657] or C795T), and 6 isolates (6.9%) had either frameshift or nonsense mutations and were genotypically typed as serotype 35D (Table 1). Quellung reactions confirmed the phenotype of all 81 serotype 35B isolates. Among the six isolates genotypically typed as serotype 35D, three were phenotypically typed as serotype 35D, two as serotype 35B, and one as a mixture of both serotypes (Table 1). Moreover, among the 81 serotype 35B isolates, 26 (29.9%) were collected from IPD cases, compared with 4/6 (66.7%) of the serotype 35D isolates. Furthermore, three (50%) of the six serotype 35D isolates belonged to CC558 and the other three to CC2755 (Fig. 1). Significantly more CC2755 isolates originated from IPD cases than did CC558 isolates (12/20 [60.0%] versus 18/67 [26.9%], respectively; ~ odds ratio [OR], 4.08; 95% confidence interval [CI], 1.46 to 11.38; P = 0.014). Antimicrobial susceptibility results are summarized in Table 2. Overall, the rates of nonsusceptibility to penicillin G (PCG), cefotaxime (CTX), meropenem (MEM), erythromycin (EM), and levofloxacin (LFX) were 78.2%, 40.2%, 75.9%, 94.3%, and 0%, respectively. CC558 showed significantly higher rates of nonsusceptibility to PCG, CTX, and MEM. Of 82 EM-resistant isolates, 28 (34.1%) showed extremely high MICs (>128 mg/L) for EM (Fig. 1).
TABLE 1.
Profiles of six genotypically serotype 35D S. pneumoniae isolates identified in this study
| Strain | Prefecture (region) | Yr isolated | IPD/non-IPDa | Origin | Sequence type (ST) | wciG mutation(s) | Type of mutation | Reaction to antiserumb |
Phenotypic serotypec | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pool G | Type 29 | Group 35 | fs35a | fs35b | fs35c | fs29b | fs42a | |||||||||
| PC0204 | Osaka (Kinki) | 2013 | IPD | Blood | 2755 | 157delT | Frameshift | + | + | + | + | − | + | + | − | 35B/D |
| PC0607 | Yamaguchi (Chugoku) | 2014 | Non-IPD | Middle-ear fluid | 558 | 156_157insT | Frameshift | + | + | + | + | − | + | + | − | 35B |
| PC0780 | Yamaguchi (Chugoku) | 2015 | Non-IPD | Middle-ear fluid | 558 | 156_157insT | Frameshift | + | + | + | + | − | + | + | − | 35B |
| PC0870 | Nagano (Chubu) | 2015 | IPD | Blood | 2755 | 903_918del_16bp | Frameshift | + | + | + | − | − | +/−d | + | − | 35D |
| PC1044 | Aichi (Chubu) | 2016 | IPD | CSFe | 2755 | 903_918del_16bp | Frameshift | + | + | − | − | − | + | + | − | 35D |
| PC1147 | Hyogo (Kinki) | 2016 | IPD | Blood | 10493 | A796T, C798G | Nonsense | + | + | − | − | − | + | + | − | 35D |
IPD, invasive pneumococcal disease.
Serotyping was performed using pneumococcal typing antisera (Statens Serum Institut, Copenhagen, Denmark).
Phenotypic serotypes were determined based on the criteria described by Lo et al. (20).
+/−, under the microscope, cells derived from a single-colony overnight culture showed both positive and negative effects on the antisera tested.
CSF, cerebrospinal fluid.
TABLE 2.
Antimicrobial susceptibility according to clonal complex
| Antimicrobiala | No. of nonsusceptible isolates (%) inb: |
Total (n = 87) | P value for CC558 vs CC2755c | |||||
|---|---|---|---|---|---|---|---|---|
| CC558 |
Total CC558 (n = 67) | CC2755 |
Total CC2755 (n = 20) | |||||
| ST558 (n = 62) | ST10493 (n = 5) | ST2755 (n = 18) | Others (n = 2) | |||||
| PCG | 62 (100) | 5 (100) | 67 (100) | 1 (5.6) | 0 (0) | 1 (0.5) | 68 (78.2) | <0.001 |
| CTX | 30 (48.4) | 4 (80.0) | 34 (50.7) | 1 (5.6) | 0 (0) | 1 (0.5) | 35 (40.2) | <0.001 |
| MEM | 61 (98.4) | 5 (100) | 66 (98.5) | 0 (0) | 0 (0) | 0 (0) | 66 (75.9) | <0.001 |
| EM | 57 (91.9) | 5 (100) | 62 (92.5) | 18 (100) | 2 (100) | 20 (100) | 82 (94.3) | 0.585 |
| LFX | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1.000 |
PCG, penicillin G; CTX, cefotaxime; MEM, meropenem; EM, erythromycin; LFX, levofloxacin.
CC, clonal complex; ST, sequence type.
A P value of <0.05 was considered statistically significant.
PBP type and antimicrobial resistance genes.
All CC558 (n = 67) isolates had penicillin-binding protein (PBP) genes pbp1a-4 and pbp2b-7, and 66 of 67 (98.5%) isolates had pbp2x-7, whereas one had pbp2x-JP46 (Fig. 2). All CC2755 (n = 20) isolates had pbp1a-0, and 19 of these had pbp2b-0, whereas one isolate had a novel type of pbp2b. Various pbp2x types were observed; JP34 (n = 5), JP36 (n = 3), and JP37 (n = 3) were the predominant types of pbp2x (Data Set S1).
FIG 2.
Recombination-free maximum-likelihood tree of the GPSC59 isolates in Japan and the previously deposited isolates in the NCBI database, rooted by the serotype 35B reference strain Utah35B (ST377, BAA-660TM [ATCC], accession no. AP025939). Colored bars indicate characteristics of isolates including the country of isolation, year of isolation, host status, serotype, sequence type, type of pbp1a, php2b, and pbp2x, and presence of antimicrobial resistance determinants ermB, tetM, folA I100L mutation, and folP insertion. Gray circles indicate ≥ 95% confidence by bootstrap analysis.
The prevalence of antimicrobial resistance determinants in serotypes 35B and 35D, according to CC, is shown in Table 3. The number of isolates with macrolide resistance mediated by ermB, which showed extremely high MICs for EM, was significantly higher in CC2755 than in CC558 (CC558, 8/67 [11.9%]; CC2755, 20/20 [100%]; P < 0.001). In contrast, macrolide resistance mediated by mefE was significantly higher in CC558 than in CC2755 (CC558, 60/67 [89.6%]; CC2755, 0/20 [0%]; P < 0.001). The folA substitution and folP insertion mutations, to which full trimethoprim-sulfamethoxazole resistance is attributed, were exclusively found in CC2755. All CC558 isolates were rrgC (type 1 pilus [pilus-1]) positive and sipA (pilus-2) negative, and all CC2755 isolates were negative for both genes.
TABLE 3.
Prevalence of antimicrobial resistance determinant genes according to serotype and clonal complex
| Antimicrobial resistance determinant(s) | No. of isolates (%) according to serotype and clonal complex |
P value fora: |
||||||
|---|---|---|---|---|---|---|---|---|
| 35B (n = 81) |
Total 35B | 35D (n = 6) |
Total 35D | 35B vs 35D | CC558 vs CC2755 | |||
| CC558 (n = 64) | CC2755 (n = 17) | CC558 (n = 3) | CC2755 (n = 3) | |||||
| ermB | 8 (12.5) | 17 (100) | 25 (30.9) | 0 (0) | 3 (100) | 3 (50.0) | 0.382 | <0.001 |
| mefE | 58 (90.6) | 0 (0) | 58 (71.6) | 2 (66.7) | 0 (0) | 2 (33.3) | 0.072 | <0.001 |
| tetM | 56 (87.5) | 17 (100) | 73 (90.1) | 2 (66.7) | 3 (100) | 5 (83.3) | 0.307 | 0.111 |
| folA I100L and folP insertion | 0 (0) | 16 (94.1) | 16 (19.8) | 0 (0) | 3 (100) | 3 (50.0) | 0.115 | <0.001 |
A P value of <0.05 was considered statistically significant.
Phylogenetic analysis of serotype 35B and 35D isolates in Japan and other countries.
The phylogenetic tree of the global ST558 and ST10493 strains in GPSC59 revealed that the Japanese isolates formed four major clusters and two minor clusters interspersed among the global strains, mainly those from the United States (Fig. 2). ST10493 isolates from Japan, which formed major cluster 3 (Fig. 2), appeared to diverge from ST558 strains from the United States and not the Japanese strains. The phylogenetic tree of GPSC186 showed that the Japanese clade was genetically distinct from the previously deposited strains from other countries, whereas all GPSC186 strains were from Asian countries, and uniformly possessed ermB, tetM, and folA substitution and folP insertion mutations (Fig. S1).
Tn916-like ICE structure and cps locus analysis.
Of the total study isolates, 77 (88.5%) had Tn916-like integrative and conjugative elements (ICEs) with tetM, which encodes tetracycline resistance (Fig. 3). Of the 67 CC558 isolates, 50 (74.6%) had Tn2009, which contains a macrolide efflux genetic assembly (MEGA) element that carries mefE. The MEGA element is inserted between open reading frame 6 (ORF6) and ORF9 of Tn916. Six (9.0%) of the CC558 isolates had Tn2010, which contains erythromycin resistance cassettes, including ermB, located between ORF20 and ORF21 of Tn2009. All ST2755 isolates and one CC558 isolate had Tn6002-like ICEs that contained erythromycin resistance cassettes, including ermB, between ORF19 and ORF20 and a hypothetical protein between ORF20 and ORF21 of Tn916, whereas mefE was not identified. None of the five ST10493 (CC558) isolates had Tn916-like ICEs, but they had the MEGA alone.
FIG 3.
Comparisons of the Tn916-like integrative conjugative elements (ICEs) of serotype 35B/35D isolates. Red bands between the sequences indicate BLASTN matches. In Tn2009, a macrolide efflux genetic assembly (MEGA) that contained mefE was inserted between ORF6 and ORF9 of Tn916. Tn2010, in addition to the insertion of the MEGA in the same spot as Tn2009, had an ermB-containing cassette that was integrated with ORF20 of Tn916. Tn6002 had an ermB-containing cassette that was integrated with ORF20 of Tn916, and mefE was not identified. The reference sequence of Tn916 was submitted as NCBI reference sequence U09422.1.
We compared the cps loci and wciG sequences of the ST558 isolate (PC0780; genotypically 35D and phenotypically 35B), the ST2755 isolate (PC0204; genotypically 35D and phenotypically 35B/D), and the reference genome of the cps locus of serotype 35B (accession number CR931705.1) (Fig. S2). The cps locus of PC0780 was consistent with that of the reference genome (accession number CR931705.1), whereas that of PC0204 contained three additional insertion sequences (ISs). We also evaluated the locations of the mutations/insertions/deletions in wciG of our isolates and previously reported serotype 35D isolates (20). The locations of the mutations/insertions/deletions were associated with the low-GC-content (AT-rich) regions in wciG (Fig. S3).
Divergence time estimation.
We next performed Bayesian analysis to estimate the time to the most recent common ancestor (TMRCA) of the ST558 and ST10493 strains with serotype 35B/D, using BEAST2 (21). The TMRCAs of the four major clusters (containing ≥ 5 isolates) of these strains in Japan (major clusters 1 to 4) (Fig. 2 and 4), estimated using corresponding global GPSC59 strains, were 1983 (major cluster 1; 95% highest posterior density [HPD], 1979 to 1987), 1992 (major cluster 2; 95% HPD, 1989 to 2006), 2001 (major cluster 3; 95% HPD, 1999 to 2003), and 1992 (major cluster 4; 95% HPD, 1989 to 1994) (Fig. 4).
FIG 4.
Bayesian phylogenetic reconstruction of ST558 and ST10493 isolates in GPSC59 from Japan and other countries, based on isolates. Major clusters (≥ 5 isolates) and minor clusters (< 5 isolates) containing only Japanese isolates are highlighted in gray. The estimated times of the most recent common ancestor of each major cluster and its 95% HPD are displayed.
DISCUSSION
Serotype 35B has become one of the most prevalent pneumococcal serotypes globally after the introduction of PCV13 (3, 6, 8). In addition, CC558 strains detected in various regions worldwide have exhibited multidrug resistance (12, 16, 17). This motivated us to analyze the genetic data of serotype 35B strains to understand their global spread after the introduction of PCV7 and -13, using whole-genome sequencing of serotype 35B strains isolated from four countries, the United States, Israel, Qatar, and South Africa (22–26), and compare them with those of strains isolated in Japan through a 6-year nationwide pneumococcal surveillance study. In addition, we assessed the phenotypic and genetic basis of the newly recognized serotype 35D, a serotype 35B variant, which is reportedly distributed globally, and suggested that its invasive potential was conferred by the loss of O-acetylation in the pneumococcal capsular polysaccharides (20).
Similar to the foreign serotype 35B-CC558 strains, CC558 strains in Japan were found to be associated with multidrug resistance, especially to a broad range of β-lactams, such as penicillin, cephalosporin, and carbapenem. This β-lactam resistance of serotype 35B-CC558 was caused by specific PBP profiles, pbp1a-4, pbp2b-7, and pbp2x-7, shared by global MDR serotype 35B-CC558 isolates (12, 26). Considering the identical CC and PBP profiles, we believe that they share an ancestor and have recently diverged from each other. Our analysis showed that the TMRCA of the strains in Japan was between 1983 and 2001. In particular, our phylogenetic analysis revealed multiple clade formation of the ST558 and ST10493 strains in Japan among the foreign strains, indicating multiple events of introduction of the clone into Japan from foreign countries and vice versa. Our previous studies on global serotype 15A-ST63 (27) and serotype 19A-ST320 (28) and other studies on serotype 3-ST180 (29) and serotype 23F-ST81 (30) demonstrated that pneumococcal clades in phylogenetic trees tended to generate phylogenetic clades based on geographic location. This was especially true in Japan for geopolitical reasons; the country is surrounded by water and foreign migration events are rare. Previous studies suggested that CC558 is associated with multidrug resistance (17, 31) and pili, a pneumococcal virulence factor that facilitates colonization (15, 32, 33). Serotype 35B strains show high potential for biofilm formation in vitro (34). These findings suggest that the 35B-CC558 lineage is likely to have a high potential for colonizing the human nasopharynx and being resistant to clearance, which might be associated with a longer duration of carriage and consequent international transmission. Although further studies are needed to support characteristics like the duration of colonization, ability of adhesion, and fitness cost in colonizers, the trend of this clone should be monitored because serotype 35B is not covered by any of the current conjugate vaccines, polysaccharide vaccines, or the upcoming PCV20.
In a previous study, seroconversion to 35D from 35B was suggested to occur independently via the variable point mutations and indels in wciG (20). Additionally, in our study, two pairs of the 35D isolates possessed the same mutation/indels in wciG and were derived from the same regions, which suggested clonal expansion of the 35D isolates. As discussed above, the 35B-CC558 lineage is suspected to have a high potential for colonizing the human nasopharynx and being resistant to clearance. A recent study showed that within-host microevolution of S. pneumoniae was rapid and adaptive during natural colonization (35), which might facilitate the seroconversion. Considering this result, a longer duration of carriage can also accelerate the nucleotide substitution rate within the host and might lead to seroconversion to 35D. In a previous study, wciG-mediated O-acetylation seemed to yield an antigenically dominant epitope for serotype 35B, and seroconversion to 35D was associated with the loss of O-acetylation, resulting in escape from the acquired immune system response targeting serotype 35B (19). The immune escape might lead to the clonal expansion of the 35D isolates. These findings imply that serotype conversion from 35B to 35D might occur frequently and expand clonally, especially under selective pressure, such as a vaccine covering serotype 35B or immune pressure during colonization. Because limited analysis of 35D isolates was conducted due to the small sample size, further studies are needed to investigate the mechanisms of the serotype conversion.
In addition to the global epidemic CC558 lineage, CC2755 is another prevalent lineage in Japan and was reported predominately in the country (15, 16). CC2755 isolates were associated with multidrug resistance to macrolides, tetracyclines, and trimethoprim-sulfamethoxazole. Macrolide resistance in S. pneumoniae is a major clinical problem because macrolides are commonly prescribed for treating bacterial infections in the upper and lower respiratory tracts, including community-acquired infections (36). In Japan, the proportion of macrolide consumption to total antibiotic consumption in outpatient settings is higher than those in European and North American countries (37). The incidence of macrolide-resistant IPDs was reported to have decreased following PCV introduction in the United States and globally (38, 39). However, in Japan, the rate of macrolide resistance in S. pneumoniae remains very high, >80% in the nationwide surveillance in 2021, even after PCV13 implementation in 2013 (40). In this study, 94.3% of the 35B/D isolates were resistant to erythromycin, and most of the resistance was mediated by ermB and/or mefE carried on the MEGA in ST10493 (CC558) and on Tn916-like ICEs Tn2009 and Tn2010 in ST558 (CC558) and Tn6002 in CC2755. Previously, we found that the predominant ICEs associated with macrolide resistance in 15A-CC63 and 19A-CC3111 strains in Japan were Tn6002 and Tn2017, respectively (41, 42). Recent studies have suggested that Tn916-like ICEs, such as Tn2009, Tn2010, Tn3872, Tn6002, and the MEGA, are also commonly found in viridans group streptococci colonizing the human throat, mainly Streptococcus mitis, Streptococcus oralis, and Streptococcus parasanguinis (43). Another recent study revealed that high macrolide consumption in the community induced an increase in the prevalence of nasopharyngeal ermB and mefA/-E resistomes in preschool children (44). These findings imply that the acquisition of macrolide resistance determinants like ermB and mefE through intra- and interspecies transfer of Tn916-like ICEs or MEGAs accelerates the clonal expansion of the strains under the selective pressure of high macrolide consumption. Further studies are needed to elucidate the roles of and relationships between ICEs and the MEGA in macrolide resistance in S. pneumoniae and other commensal bacteria in the nasopharynx.
In conclusion, we conducted whole-genome sequencing to investigate the molecular epidemiology of Japanese serotype 35B/D S. pneumoniae isolates and compared the sequences to previously reported global ones. We revealed multiple events of introduction of the MDR 35B/D-CC558 lineage into Japan, suggesting frequent intercontinental transmission of this lineage. In addition, serotype 35D isolates showed clonality, which suggested potential clonal expansion of the 35D isolates. Our study also revealed that 35B/D isolates in Japan are highly associated with multidrug resistance, including broad-spectrum β-lactam resistance in CC558 and macrolide, tetracycline, and clindamycin resistance via transferable elements, the MEGA, and Tn916-like ICEs, in both CCs. Global molecular surveillance utilizing whole-genome-sequencing methods will help to understand the dissemination patterns of the global epidemic clones.
MATERIALS AND METHODS
Bacterial isolates.
This study was part of a nationwide prospective surveillance study of pediatric IPDs and non-IPDs in Japan between January 2012 and December 2017 (31, 45). A total of 1,358 pneumococcal isolates were collected from 249 medical institutions across Japan and included 87 serotype 35B and 35D isolates obtained from 30 IPD and 57 non-IPD patients. Among those from the IPD patients, 23 isolates were obtained from blood, 6 from cerebrospinal fluid, and 1 from joint fluid. Non-IPD cases included 50 patients with otitis media and 7 with pneumonia. The detailed characteristics of the isolates are shown in Data Set S1.
Antimicrobial susceptibility tests.
Susceptibility tests for penicillin G (PCG), erythromycin (EM), cefotaxime (CTX), meropenem (MEM), and levofloxacin (LFX) were performed using the broth microdilution method following the Clinical and Laboratory Standards Institute (CLSI) guidelines (46). The 2015 CLSI standard for categorization (46) was used to determine susceptibility, and isolates identified as having intermediate or full resistance were grouped together as nonsusceptible. We used the CLSI criteria for meningitis for all isolates to allow comparison with previous reports. The following breakpoints were applied for susceptibility, intermediate resistance, resistance, and high resistance, respectively: PCG, ≤0.06, 0.12, 1.0, and ≥2 μg/mL; CTX, ≤0.5, 1.0, 2.0, and >2 μg/mL; MEM, ≤0.25, 0.5, 1.0, and >1 μg/mL; and EM, ≤0.25, 0.5, 16, and >16 μg/mL.
Whole-genome sequencing and genome analyses.
Whole-genome sequencing was performed on 79/87 of the 35B and 35D isolates in the current study, whereas sequencing data for the other 8 isolates were obtained previously (27). We extracted total genomic DNA using the QIAamp DNA minikit (Qiagen, Hilden, Germany) and prepared sequencing libraries using the Nextera XT DNA library preparation kit (Illumina, San Diego, CA, USA) (27, 41). The details of genome analyses are shown in the supplemental material.
In silico and phenotypic serotyping and wciG locus comparison.
We performed in silico serotyping using Pathogenwatch (https://pathogen.watch). We performed multilocus sequence typing (MLST) to determine the exact matches for the seven loci (aroE, gdh, gki, recP, spi, xpt, and ddl) and assigned clonal complexes (CCs) in agreement with six of the seven loci, with the most predominant sequence type (ST) representing a CC. We extracted sequences of the wciG locus from the assembled contigs using BLAST+ version 2.9.0 (47) and the serotype 35B wciG reference locus (GenBank accession number KX021817). We performed phenotypic serotyping via the Quellung reaction using pneumococcal typing antisera (Statens Serum Institut, Copenhagen, Denmark) on an overnight culture derived from a single colony. We interpreted the Quellung results as previously described (20).
PBP typing, antimicrobial resistance gene detection, and GPSC identification.
We extracted the penicillin-binding protein 1A (PBP1A), -2B, and -2X transpeptidase domain sequences of the isolates and assigned PBP transpeptidase domain types using the U.S. Centers for Disease Control and Prevention PBP-type database (https://www.cdc.gov/streplab/pneumococcus/mic.html). PBP types that had not been previously published in the database (last accessed on 1 April 2021) were labeled with the prefix JP (e.g., “pbp1a-JP1”). Some of these unassigned PBP types from Japan have been reported (27, 28, 41, 42, 48). Furthermore, we detected ermB, ermTR, mefA, mefE, tetM, and tetO genes and searched for point mutations within the folA and folP genes in the assembled contigs (41). In addition, we assigned global pneumococcal sequence cluster (GPSC) numbers and detected tet(S/M) using Pathogenwatch (https://pathogen.watch).
Tn916-like ICE analysis and cps locus comparison.
We extracted the sequences of Tn916-like integrative and conjugative elements (ICEs) from the assembled contigs using BLAST+ (47) and an Enterococcus faecalis Tn916 reference sequence (GenBank accession no. U09422.1). The analyzed sequences were annotated using Prokka version 1.14.6 (49), and the structures of the regions were analyzed manually using the Artemis Comparison Tool (ACT) (50). We also extracted sequences of the cps locus from the assembled contigs using BLAST+ and manually analyzed them using ACT.
Single nucleotide polymorphism (SNP) and phylogenetic analyses.
The core genomes of the 87 serotype 35B and 35D isolates were identified using Prokka version 1.14.6 (49) and Roary version 3.13.0 (51), with standard parameters. A maximum-likelihood phylogenetic tree was generated from the core genome alignment using RAxML-NG version 1.0.3 with a GTR+Γ DNA substitution model (52). To investigate the genetic relationship between the strains previously deposited as the same GPSCs, we obtained sequence data from the European Nucleotide Archive (https://www.ebi.ac.uk/ena/browser/home). A total of 88 strains (GPSC59, 75 strains, and GPSC186, 13 strains) were included for further analysis (Data Set S2).
Bayesian analysis.
We reconstructed a maximum-credibility clade tree and obtained the dates of ancestors or nodes of the ST558 and ST10493 clades using the Bayesian Markov chain Monte Carlo framework. For this analysis, we performed recombination prediction using Gubbins version 2.4.1 (53). The final SNP alignments, without recombination regions, were used as the input data set for BEAST2 version 2.6.6 (21).
The details of genome analyses are shown in the supplemental material.
Statistical analysis.
Categorical variables were compared using the χ2 and Fisher’s exact tests, as appropriate. A two-sided P value of <0.05 was considered to indicate statistical significance. All statistical analyses were performed using R version 3.6.0.
Ethics statement.
This study was reviewed and approved by the Ethics Committee of Mie Hospital (acceptance number, 23-8). Informed consent for collection and use of patient information and specimens was obtained from each parent/guardian by a primary physician.
Data availability.
Nucleotide sequence data obtained in this study have been submitted to GenBank/ENA/DDBJ under BioProject accession number PRJDB13244 (DRR356353 to DRR356452) and PRJDB13326 (AP025936 to AP025940, BRKZ01000001 to BRKZ01000009, and BRLA01000001 to BRLA01000013).
ACKNOWLEDGMENTS
Satoshi Nakano was supported by AMED under grant number JP21fk0108147, by JSPS KAKENHI under grants number 19K16637 and 21K15432, and by a research grant from Pfizer, Inc., given to his previous institution. Takao Fujisawa was supported by an unconditional grant from Pfizer through his institution. Bin Chang was supported by AMED under grants number JP21fk0108139 and JP21fk0108099.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Wahl B, O’Brien KL, Greenbaum A, Majumder A, Liu L, Chu Y, Lukšić I, Nair H, McAllister DA, Campbell H, Rudan I, Black R, Knoll MD. 2018. Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: global, regional, and national estimates for 2000–15. Lancet Glob Health 6:e744–e757. 10.1016/S2214-109X(18)30247-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pilishvili T, Lexau C, Farley MM, Hadler J, Harrison LH, Bennett NM, Reingold A, Thomas A, Schaffner W, Craig AS, Smith PJ, Beall BW, Whitney CG, Moore MR. Active Bacterial Core Surveillance/Emerging Infections Program Network. 2010. Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J Infect Dis 201:32–41. 10.1086/648593. [DOI] [PubMed] [Google Scholar]
- 3.Moore MR, Link-Gelles R, Schaffner W, Lynfield R, Lexau C, Bennett NM, Petit S, Zansky SM, Harrison LH, Reingold A, Miller L, Scherzinger K, Thomas A, Farley MM, Zell ER, Taylor TH, Pondo T, Rodgers L, McGee L, Beall B, Jorgensen JH, Whitney CG. 2015. Effect of use of 13-valent pneumococcal conjugate vaccine in children on invasive pneumococcal disease in children and adults in the USA: analysis of multisite, population-based surveillance. Lancet Infect Dis 15:301–309. 10.1016/S1473-3099(14)71081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ladhani SN, Collins S, Djennad A, Sheppard CL, Borrow R, Fry NK, Andrews NJ, Miller E, Ramsay ME. 2018. Rapid increase in non-vaccine serotypes causing invasive pneumococcal disease in England and Wales, 2000–17: a prospective national observational cohort study. Lancet Infect Dis 18:441–451. 10.1016/S1473-3099(18)30052-5. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Shimol S, Givon-Lavi N, Grisaru-Soen G, Megged O, Greenberg D, Dagan R. Israel Bacteremia and Meningitis Active Surveillance Group. 2018. Comparative incidence dynamics and serotypes of meningitis, bacteremic pneumonia and other-IPD in young children in the PCV era: insights from Israeli surveillance studies. Vaccine 36:5477–5484. 10.1016/j.vaccine.2017.05.059. [DOI] [PubMed] [Google Scholar]
- 6.Lo SW, Gladstone RA, van Tonder AJ, Lees JA, Du Plessis M, Benisty R, Givon-Lavi N, Hawkins PA, Cornick JE, Kwambana-Adams B, Law PY, Ho PL, Antonio M, Everett DB, Dagan R, von Gottberg A, Klugman KP, McGee L, Breiman RF, Bentley SD, Brooks AW, Corso A, Davydov A, Maguire A, Pollard A, Kiran A, Skoczynska A, Moiane B, Beall B, Sigauque B, Aanensen D, Lehmann D, Faccone D, Foster-Nyarko E, Bojang E, Egorova E, Voropaeva E, Sampane-Donkor E, Sadowy E, Bigogo G, Mucavele H, Belabbès H, Diawara I, Moïsi J, Verani J, Keenan J, Bhai JNNT, Ndlangisa KM, Zerouali K, Ravikumar KL, et al. 2019. Pneumococcal lineages associated with serotype replacement and antibiotic resistance in childhood invasive pneumococcal disease in the post-PCV13 era: an international whole-genome sequencing study. Lancet Infect Dis 19:759–769. 10.1016/S1473-3099(19)30297-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma D, Baughman W, Holst A, Thomas S, Jackson D, da Gloria Carvalho M, Beall B, Satola S, Jerris R, Jain S, Farley MM, Nuorti JP. 2013. Pneumococcal carriage and invasive disease in children before introduction of the 13-valent conjugate vaccine: comparison with the era before 7-valent conjugate vaccine. Pediatr Infect Dis J 32:e45–e53. 10.1097/INF.0b013e3182788fdd. [DOI] [PubMed] [Google Scholar]
- 8.Cohen R, Varon E, Doit C, Schlemmer C, Romain O, Thollot F, Béchet S, Bonacorsi S, Levy C. 2015. A 13-year survey of pneumococcal nasopharyngeal carriage in children with acute otitis media following PCV7 and PCV13 implementation. Vaccine 33:5118–5126. 10.1016/j.vaccine.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Beall B, McEllistrem MC, Gertz RE, Jr, Boxrud DJ, Besser JM, Harrison LH, Jorgensen JH, Whitney CG. Active Bacterial Core Surveillance/Emerging Infections Program Network. 2002. Emergence of a novel penicillin-nonsusceptible, invasive serotype 35B clone of Streptococcus pneumoniae within the United States. J Infect Dis 186:118–122. 10.1086/341072. [DOI] [PubMed] [Google Scholar]
- 10.Navarro-Torné A, Dias JG, Hruba F, Lopalco PL, Pastore-Celentano L, Gauci AJA. Invasive Pneumococcal Disease Study Group. 2015. Risk factors for death from invasive pneumococcal disease, Europe, 2010. Emerg Infect Dis 21:417–425. 10.3201/eid2103.140634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomczyk S, Lynfield R, Schaffner W, Reingold A, Miller L, Petit S, Holtzman C, Zansky SM, Thomas A, Baumbach J, Harrison LH, Farley MM, Beall B, McGee L, Gierke R, Pondo T, Kim L. 2016. Prevention of antibiotic-nonsusceptible invasive pneumococcal disease with the 13-valent pneumococcal conjugate vaccine. Clin Infect Dis 62:1119–1125. 10.1093/cid/ciw067. [DOI] [PubMed] [Google Scholar]
- 12.Chochua S, Metcalf BJ, Li Z, Walker H, Tran T, McGee L, Beall B. 2017. Invasive serotype 35B pneumococci including an expanding serotype switch lineage, United States, 2015–2016. Emerg Infect Dis 23:922–930. 10.3201/eid2306.170071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.CDC. 2019. Antibiotic resistance threats in the United States, 2019. Centers for Disease Control and Prevention, Atlanta, GA.
- 14.Ubukata K, Takata M, Morozumi M, Chiba N, Wajima T, Hanada S, Shouji M, Sakuma M, Iwata S. Invasive Pneumococcal Diseases Surveillance Study Group. 2018. Effects of pneumococcal conjugate vaccine on genotypic penicillin resistance and serotype changes, Japan, 2010–2017. Emerg Infect Dis 24:2010–2020. 10.3201/eid2411.180326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyazaki H, Shibuya R, Chang B, Inukai T, Miyazaki Y, Ubukata K, Nakamura S, Matsumoto T. 2020. Genetic characteristics of piliated Streptococcus pneumoniae serotype 35B, increased after introduction of pneumococcal vaccines in Japan. J Infect Chemother 26:1198–1204. 10.1016/j.jiac.2020.06.016. [DOI] [PubMed] [Google Scholar]
- 16.Kasahara K, Komatsu Y, Koizumi A, Chang B, Ohnishi M, Muratani T, Mikasa K. 2014. Serotype 35B Streptococcus pneumoniae, Japan, 2002–2012. J Infect Chemother 20:228–230. 10.1016/j.jiac.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 17.Olarte L, Kaplan SL, Barson WJ, Romero JR, Lin PL, Tan TQ, Hoffman JA, Bradley JS, Givner LB, Mason EO, Hultén KG. 2017. Emergence of multidrug-resistant pneumococcal serotype 35B among children in the United States. J Clin Microbiol 55:724–734. 10.1128/JCM.01778-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richter SS, Diekema DJ, Heilmann KP, Dohrn CL, Riahi F, Doern GV. 2014. Changes in pneumococcal serotypes and antimicrobial resistance after introduction of the 13-valent conjugate vaccine in the United States. Antimicrob Agents Chemother 58:6484–6489. 10.1128/AAC.03344-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geno KA, Saad JS, Nahm MH. 2017. Discovery of novel pneumococcal serotype 35D, a natural WciG-deficient variant of serotype 35B. J Clin Microbiol 55:1416–1425. 10.1128/JCM.00054-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo SW, Gladstone RA, van Tonder AJ, Hawkins PA, Kwambana-Adams B, Cornick JE, Madhi SA, Nzenze SA, Du Plessis M, Kandasamy R, Carter PE, Eser ÖK, Ho PL, Elmdaghri N, Shakoor S, Clarke SC, Antonio M, Everett DB, von Gottberg A, Klugman KP, McGee L, Breiman RF, Bentley SD. 2018. Global distribution of invasive serotype 35D Streptococcus pneumoniae isolates following introduction of 13-valent pneumococcal conjugate vaccine. J Clin Microbiol 56:e00228-18. 10.1128/JCM.00228-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, Heled J, Jones G, Kühnert D, De Maio N, Matschiner M, Mendes FK, Müller NF, Ogilvie HA, Du Plessis L, Popinga A, Rambaut A, Rasmussen D, Siveroni I, Suchard MA, Wu C-H, Xie D, Zhang C, Stadler T, Drummond AJ. 2019. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol 15:e1006650. 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Croucher NJ, Kagedan L, Thompson CM, Parkhill J, Bentley SD, Finkelstein JA, Lipsitch M, Hanage WP. 2015. Selective and genetic constraints on pneumococcal serotype switching. PLoS Genet 11:e1005095. 10.1371/journal.pgen.1005095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chewapreecha C, Marttinen P, Croucher NJ, Salter SJ, Harris SR, Mather AE, Hanage WP, Goldblatt D, Nosten FH, Turner C, Turner P, Bentley SD, Parkhill J. 2014. Comprehensive identification of single nucleotide polymorphisms associated with beta-lactam resistance within pneumococcal mosaic genes. PLoS Genet 10:e1004547. 10.1371/journal.pgen.1004547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Croucher NJ, Finkelstein JA, Pelton SI, Parkhill J, Bentley SD, Lipsitch M, Hanage WP. 2015. Population genomic datasets describing the post-vaccine evolutionary epidemiology of Streptococcus pneumoniae. Sci Data 2:150058. 10.1038/sdata.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Croucher NJ, Finkelstein JA, Pelton SI, Mitchell PK, Lee GM, Parkhill J, Bentley SD, Hanage WP, Lipsitch M. 2013. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat Genet 45:656–663. 10.1038/ng.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metcalf BJ, Gertz RE, Gladstone RA, Walker H, Sherwood LK, Jackson D, Li Z, Law C, Hawkins PA, Chochua S, Sheth M, Rayamajhi N, Bentley SD, Kim L, Whitney CG, McGee L, Beall B. Active Bacterial Core Surveillance Team. 2016. Strain features and distributions in pneumococci from children with invasive disease before and after 13-valent conjugate vaccine implementation in the USA. Clin Microbiol Infect 22:60.e9–60.e29. 10.1016/j.cmi.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakano S, Fujisawa T, Ito Y, Chang B, Matsumura Y, Yamamoto M, Nagao M, Suga S, Ohnishi M, Ichiyama S. 2018. Spread of meropenem-resistant Streptococcus pneumoniae serotype 15A-ST63 clone in Japan, 2012–2014. Emerg Infect Dis 24:275–283. 10.3201/eid2402.171268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano S, Fujisawa T, Chang B, Ito Y, Akeda H, Fujita J, Matsumura Y, Yamamoto M, Suga S, Oishi K, Nagao M, Ohnishi M. 2022. Whole-genome analysis-based phylogeographic investigation of Streptococcus pneumoniae serotype 19A sequence type 320 isolates in Japan. Antimicrob Agents Chemother 66:e01395-21. 10.1128/AAC.01395-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Azarian T, Mitchell PK, Georgieva M, Thompson CM, Ghouila A, Pollard AJ, von Gottberg A, Du Plessis M, Antonio M, Kwambana-Adams BA, Clarke SC, Everett D, Cornick J, Sadowy E, Hryniewicz W, Skoczynska A, Moïsi JC, McGee L, Beall B, Metcalf BJ, Breiman RF, Ho PL, Reid R, O’Brien KL, Gladstone RA, Bentley SD, Hanage WP. 2018. Global emergence and population dynamics of divergent serotype 3 CC180 pneumococci. PLoS Pathog 14:e1007438. 10.1371/journal.ppat.1007438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434. 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakano S, Fujisawa T, Ito Y, Chang B, Matsumura Y, Yamamoto M, Suga S, Ohnishi M, Nagao M. 2020. Nationwide surveillance of paediatric invasive and non-invasive pneumococcal disease in Japan after the introduction of the 13-valent conjugated vaccine, 2015–2017. Vaccine 38:1818–1824. 10.1016/j.vaccine.2019.12.022. [DOI] [PubMed] [Google Scholar]
- 32.Golden AR, Adam HJ, Gilmour MW, Baxter MR, Martin I, Nichol KA, Demczuk WHB, Hoban DJ, Zhanel GG. 2015. Assessment of multidrug resistance, clonality and virulence in non-PCV-13 Streptococcus pneumoniae serotypes in Canada, 2011–13. J Antimicrob Chemother 70:1960–1964. 10.1093/jac/dkv061. [DOI] [PubMed] [Google Scholar]
- 33.Regev-Yochay G, Hanage WP, Trzcinski K, Rifas-Shiman SL, Lee G, Bessolo A, Huang SS, Pelton SI, McAdam AJ, Finkelstein JA, Lipsitch M, Malley R. 2010. Re-emergence of the type 1 pilus among Streptococcus pneumoniae isolates in Massachusetts, USA. Vaccine 28:4842–4846. 10.1016/j.vaccine.2010.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Domenech M, Damián D, Ardanuy C, Liñares J, Fenoll A, García E. 2015. Emerging, non-PCV13 serotypes 11A and 35B of Streptococcus pneumoniae show high potential for biofilm formation in vitro. PLoS One 10:e0125636. 10.1371/journal.pone.0125636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaguza C, Senghore M, Bojang E, Gladstone RA, Lo SW, Tientcheu P-E, Bancroft RE, Worwui A, Foster-Nyarko E, Ceesay F, Okoi C, McGee L, Klugman KP, Breiman RF, Barer MR, Adegbola RA, Antonio M, Bentley SD, Kwambana-Adams BA. 2020. Within-host microevolution of Streptococcus pneumoniae is rapid and adaptive during natural colonisation. Nat Commun 11:3442. 10.1038/s41467-020-17327-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metlay JP, Waterer GW, Long AC, Anzueto A, Brozek J, Crothers K, Cooley LA, Dean NC, Fine MJ, Flanders SA, Griffin MR, Metersky ML, Musher DM, Restrepo MI, Whitney CG. 2019. Diagnosis and treatment of adults with community-acquired pneumonia. An official clinical practice guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med 200:e45–e67. 10.1164/rccm.201908-1581ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.The Government of Japan. 2016. Japan: National action plan on antimicrobial resistance (AMR) 2016–2020. https://www.who.int/publications/m/item/japan-national-action-plan-on-antimicrobial-resistance-(amr).
- 38.Schroeder MR, Chancey ST, Thomas S, Kuo W-H, Satola SW, Farley MM, Stephens DS. 2017. A population-based assessment of the impact of 7- and 13-valent pneumococcal conjugate vaccines on macrolide-resistant invasive pneumococcal disease: emergence and decline of Streptococcus pneumoniae serotype 19A (CC320) with dual macrolide resistance mechanisms. Clin Infect Dis 65:990–998. 10.1093/cid/cix446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andrejko K, Ratnasiri B, Hausdorff WP, Laxminarayan R, Lewnard JA. 2021. Antimicrobial resistance in paediatric Streptococcus pneumoniae isolates amid global implementation of pneumococcal conjugate vaccines: a systematic review and meta-regression analysis. Lancet Microbe 2:e450–e460. 10.1016/S2666-5247(21)00064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Japan Nosocomial Infections Surveillance (JANIS). 2022. Annual open report 2021. https://janis.mhlw.go.jp/english/report/open_report/2021/3/1/ken_Open_Report_Eng_202100_clsi2012.pdf.
- 41.Nakano S, Fujisawa T, Ito Y, Chang B, Matsumura Y, Yamamoto M, Suga S, Ohnishi M, Nagao M. 2019. Penicillin-binding protein typing, antibiotic resistance gene identification, and molecular phylogenetic analysis of meropenem-resistant Streptococcus pneumoniae serotype 19A-CC3111 strains in Japan. Antimicrob Agents Chemother 63:e00711-19. 10.1128/AAC.00711-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakano S, Fujisawa T, Ito Y, Chang B, Matsumura Y, Yamamoto M, Suga S, Ohnishi M, Nagao M. 2019. Whole-genome sequencing analysis of multidrug-resistant serotype 15A Streptococcus pneumoniae in Japan and the emergence of a highly resistant serotype 15A-ST9084 clone. Antimicrob Agents Chemother 63:e02579-18. 10.1128/AAC.02579-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brenciani A, Tiberi E, Tili E, Mingoia M, Palmieri C, Varaldo PE, Giovanetti E. 2014. Genetic determinants and elements associated with antibiotic resistance in viridans group streptococci. J Antimicrob Chemother 69:1197–1204. 10.1093/jac/dkt495. [DOI] [PubMed] [Google Scholar]
- 44.Keenan JD, Chin SA, Amza A, Kadri B, Nassirou B, Cevallos V, Cotter SY, Zhou Z, West SK, Bailey RL, Porco TC, Lietman TM. Rapid Elimination of Trachoma (PRET) Study Group. 2018. The effect of antibiotic selection pressure on the nasopharyngeal macrolide resistome: a cluster-randomized trial. Clin Infect Dis 67:1736–1742. [DOI] [PubMed] [Google Scholar]
- 45.Nakano S, Fujisawa T, Ito Y, Chang B, Suga S, Noguchi T, Yamamoto M, Matsumura Y, Nagao M, Takakura S, Ohnishi M, Ihara T, Ichiyama S. 2016. Serotypes, antimicrobial susceptibility, and molecular epidemiology of invasive and non-invasive Streptococcus pneumoniae isolates in paediatric patients after the introduction of 13-valent conjugate vaccine in a nationwide surveillance study conducted in Japan in 2012–2014. Vaccine 34:67–76. 10.1016/j.vaccine.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 46.CLSI. 2022. Performance standards for antimicrobial susceptibility testing, 32nd ed. M100-Ed32. Clinical & Laboratory Standards Institute, Wayne, PA. https://clsi.org/standards/products/microbiology/documents/m100/.
- 47.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 48.Nakano S, Fujisawa T, Ito Y, Chang B, Matsumura Y, Yamamoto M, Suga S, Ohnishi M, Nagao M. 2020. Streptococcus pneumoniae serotype 12F-CC4846 and invasive pneumococcal disease after introduction of 13-valent pneumococcal conjugate vaccine, Japan, 2015–2017. Emerg Infect Dis 26:2660–2668. 10.3201/eid2611.200087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 50.Carver TJ, Rutherford KM, Berriman M, Rajandream M-A, Barrell BG, Parkhill J. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422–3423. 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 51.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35:4453–4455. 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43:e15. 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Set S1. Download aac.01083-22-s0001.xlsx, XLSX file, 0.02 MB (21.3KB, xlsx)
Data Set S2. Download aac.01083-22-s0002.xlsx, XLSX file, 0.01 MB (12.1KB, xlsx)
Data Set S3. Download aac.01083-22-s0003.xlsx, XLSX file, 0.01 MB (12.8KB, xlsx)
Data Set S4. Download aac.01083-22-s0004.pdf, PDF file, 0.97 MB (978KB, pdf)
Data Availability Statement
Nucleotide sequence data obtained in this study have been submitted to GenBank/ENA/DDBJ under BioProject accession number PRJDB13244 (DRR356353 to DRR356452) and PRJDB13326 (AP025936 to AP025940, BRKZ01000001 to BRKZ01000009, and BRLA01000001 to BRLA01000013).