Significance
Fibrosis resulting from macrophage-mediated foreign body reactions after surgical implantation is known to be modulated by cell mechanoreception at the cell–implant interface. However, the mechanism by which macrophages recognize material surface stiffness remains unclear. Here, we demonstrate that cell membrane deformation is important for the early process of macrophage stiffness sensingleading to a heterogeneous distribution of the curvature-sensing protein Baiap2 and ultimately to cytoskeleton remodeling and inflammation inhibition. Through theoretical simulations and experiments, we propose a feedback model for stiffness sensing based on cell material properties and physical characteristics of materials, which established a link between stiffness sensing and topography sensing, offering perspectives for future material design and targeted therapies.
Keywords: foreign body reaction, biomaterial recognition, stiffness, deformation, mechanotransduction
Abstract
Implants are widely used in medical applications and yet macrophage-mediated foreign body reactions caused by implants severely impact their therapeutic effects. Although the extensive use of multiple surface modifications has been introduced to provide some mitigation of fibrosis, little is known about how macrophages recognize the stiffness of the implant and thus influence cell behaviors. Here, we demonstrated that macrophage stiffness sensing leads to differential inflammatory activation, resulting in different degrees of fibrosis. The potential mechanism for macrophage stiffness sensing in the early adhesion stages tends to involve cell membrane deformations on substrates with different stiffnesses. Combining theory and experiments, we show that macrophages exert traction stress on the substrate through adhesion and altered membrane curvature, leading to the uneven distribution of the curvature-sensing protein Baiap2, resulting in cytoskeleton remodeling and inflammation inhibition. This study introduces a physical model feedback mechanism for early cellular stiffness sensing based on cell membrane deformation, offering perspectives for future material design and targeted therapies.
Implant materials are widely used in clinical applications, including orthopedics (1), cardiology (2), and plastic surgery (3), and have shown broad application prospects in biomonitoring (4, 5). However, implants induce immune-mediated foreign body reactions (FBRs) to varying degrees, which may result in the formation of dense fibrous tissue capsules surrounding the implanted device that may affect its function (6, 7). Although the extensive use of multiple surface modifications and associated medications attempts to provide some mitigation of fibrosis, even minor reactions may lead to treatment failure and the need for secondary surgery (8, 9), resulting in a great financial burden and discomfort to the patient.
The innate immune system, particularly macrophages, is thought to be critical in the foreign body response (10, 11). Their functional activation may trigger an acute or chronic cascade of inflammation that mediates subsequent fibrosis progression (12, 13). Thus, armed with a thorough knowledge of the macrophages responding to implants, we can improve the design of implant materials to lessen the harm wrought by FBRs.
Recent studies have found that the chemical and physical characteristics of the material surface, such as topography, hydrophobicity, charge, and stiffness, play a role in regulating the phenotypes of macrophages and influence the extent of fibrous capsule formation (14, 15). Among these characteristics, stiffness is an inherent property of biomaterials, which has been actively explored in tissue engineering to regulate the migration and differentiation of stem cells (16, 17). Recent studies have also revealed the significance of microenvironmental stiffness on tumor immunity (18). Although the stiffness of implants currently approved for clinical use is well beyond the range of cellular perception, changes in stiffness at the microscopic level remain a potentially effective direction for material modification to modulate overall inflammation.
It is widely accepted that macrophages can recognize material stiffness and thus influence the success of implantation. Blakney et al. (18). and Sridharan et al. (19). demonstrated that increased stiffness promoted changes in macrophage polarization and migration patterns; conversely, another study demonstrated that stiffer substrates enhanced the anti-inflammatory polarization of macrophages (20). While the differences in the cell sources and matrix materials used by the investigators have led to various conclusions, the mechanisms by which macrophages respond to stiffness remain unexplored to date. In addition, exploring the mechanisms by which macrophages recognize stiffness may be more useful for the development of relevant therapeutics and the design of materials than determining the molecular mechanisms behind responsiveness to changes in stiffness.
Here, we investigated how macrophages recognize material stiffness by constructing hydrogel substrates with different stiffnesses and found that the recognition process starts at an early stage of cell adhesion. During the early cell adhesion phases, we observed that cell plasma membrane morphology differences were influenced by the different stiffnesses of the materials. Based on cell membrane tension and Laplace’s law, macrophages exert traction stress on the substrate through adhesion. As a result of the variations in the elastic modulus of the biomaterials, the curvature of the cellular plasma membrane changed. Furthermore, the curvature protein Baiap2 was found to assist cells in recognizing stiffness by participating in cellular deformation, providing insights into the mechanism of stiffness perception.
Results
Macrophage Recognition of Stiffness Leads to Differential Inflammatory Activation and Fibrosis.
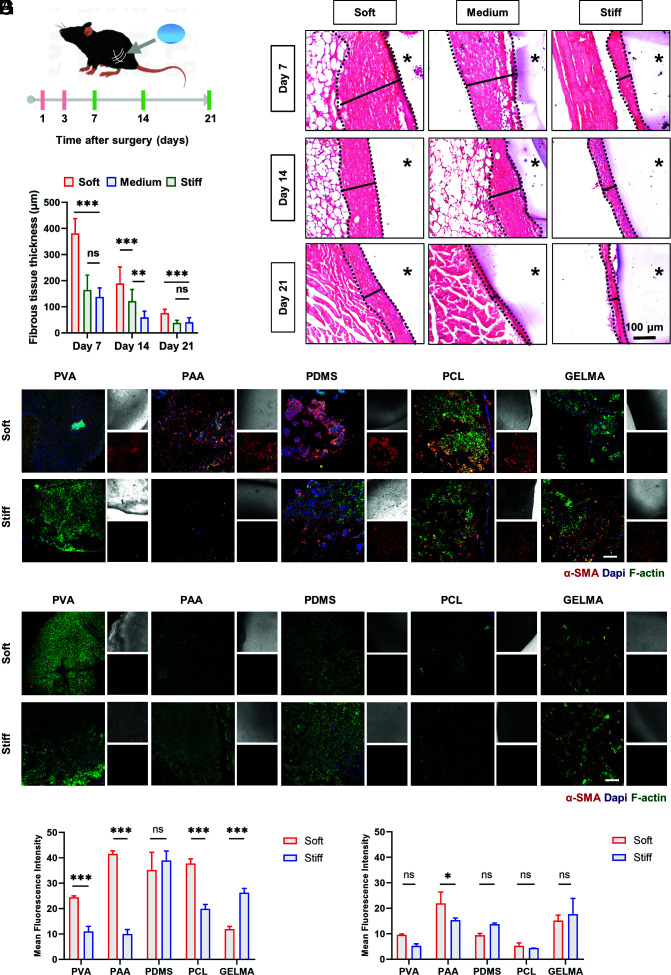
To confirm the effect of implant stiffness on fibrosis, we fabricated PVA hydrogels with stiffnesses varying from 0.5 KPa to 56 KPa. It was confirmed that no significant difference in topography, hydrophobicity, or cell viability among the hydrogels (SI Appendix, Fig. S1 A–D). These hydrogels were then buried into the bilateral thigh muscles of C57BL/6 mice, which were retained for a maximum of 21 d (Fig. 1A). Excised tissues obtained from the implantation site were analyzed by H&E staining. The images showed that the thickness of the fibrous capsule increased with decreasing implant stiffness and that fibrosis diminished over time (Fig. 1 B and C). To determine the relationship between macrophage recognition and fibrous capsule formation, we performed flow cytometry on tissues at the implantation site on Days 1 and 3 postoperatively. On Day 3, CD11b+F4/80+-labeled macrophages were recruited in large numbers and showed upregulated expression of inflammatory markers CD80+ and CD86+ around soft implants, suggesting that differences in activation generated by macrophage recognition of stiffness contributed to the different extent of fibrous encapsulation (SI Appendix, Fig. S2A).
Fig. 1.
Implant material stiffness modulates macrophage-mediated foreign body response. (A) Schematic of implant surgery of different stiffness hydrogels implanted in the quadriceps muscle group of mice and the experimental timeline for investigating the correlation between implant material stiffness and foreign body response. (B) Representative images of H&E staining of the implantation site at 7, 14, and 21 d after PVA hydrogel implant surgery. The asterisk represents implanted hydrogels. (Scale bar, 100 μm.) (C) Quantification of the variation in fibrous capsule thickness with time and material stiffness by H&E staining (n = 15; **P = 0.003). One-way ANOVA with Tukey’s posttest. (D–G) Z-stack confocal images of multiple materials with different stiffnesses recovered 14 d after implantation surgery, which were stained with DAPI (nuclei, blue), phalloidin (F-actin, green), and α-SMA (myofibroblast cells, red). And the analysis of the fluorescence intensity of α-SMA. (D and F) Mice were injected with blank control liposomes every 2 d. (E and G) Mice were injected with clodronate liposomes every 2 d. (Scale bar, 200 µm.) N = 3; *P = 0.0261, two-way ANOVA. *P < 0.033, **P < 0.002, ***P < 0.001; ns, no significant difference.
To clarify whether this stiffness-modulated fibrosis phenomenon would extend to other materials, we implanted a series of materials with different stiffnesses, including PDMS, polyacrylamide (PAA) hydrogels, methacrylate-gelatin (GelMA) hydrogels, and polycaprolactone (PCL). After 14 d, confocal images from the retrieved materials suggested that differences in stiffness, except in PDMS, led to differences in the thickness of the fibrotic capsule and cell deposition (Fig. 1 D and F). Besides, the clearance of macrophages reduced the differences to some extent (Fig. 1 E and G and SI Appendix, Fig. S2 B–D). Although the trend of the stiffness-modulated fibrosis phenomenon varies depending on the material types, macrophages ultimately influenced the stiffness-modulated fibrosis of the implants. In the case of PDMS, the most widely used prosthetic implant material which has viscoelastic qualities that distinguish it from the other elastic materials mentioned above, the mechanism involved postimplantation was different (21).
Macrophage Stiffness Sensing Is an Early Process.
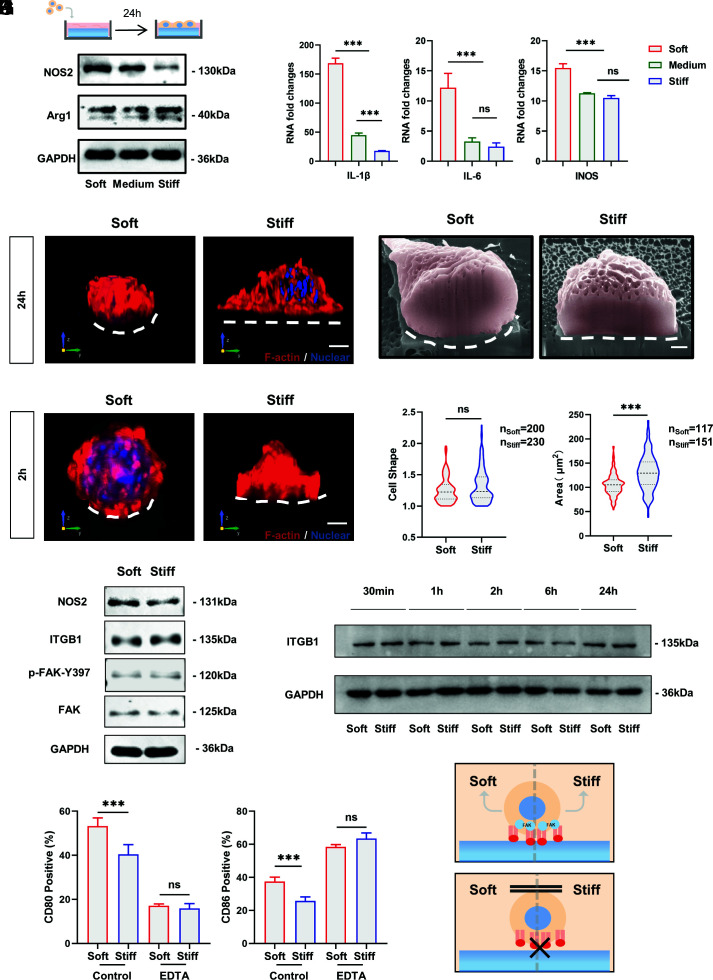
Cellular mechanoreception consists of three stages: mechanosensing, mechanotransduction, and mechanoresponse (22–24). To investigate the mechanism of macrophage stiffness sensing, we examined the response of macrophages to substrate stiffness in vitro. Bone marrow-derived macrophages (BMDMs) and the stable macrophage cell line RAW264.7 were cultured on PVA hydrogels coated with 10 μg/mL RGD (SI Appendix, Fig. S1E). BMDMs and RAW264.7 cells showed stronger inflammatory activation on the soft substrate (SI Appendix, Fig. S3 A and B). Western blot and real-time qPCR also indicated the presence of relatively higher secretion of inflammatory factors by cells on the soft substrate, consistent with the in vivo results (Fig. 2 A and B). Similarly, the flow cytometry results of PAA and PDMS in vitro matched the results in vivo (SI Appendix, Fig. S3 C and D).
Fig. 2.
Adhesion is required for macrophage stiffness sensing, and macrophage inflammatory activation is independent of integrin-mediated FAK phosphorylation. (A and B) RAW264.7 cells were cultured on RGD-coated PVA hydrogel for 24 h using Western blotting and RT–PCR to detect the expression of inflammatory markers and factors. (C) Representative 3D rendering images of RAW264.7 cells cultured on soft and stiff PVA hydrogels for 24 h. F-actin is red, and nuclei are blue. (Scale bar, 3 μm.) (D) Representative FIB-SEM images of RAW264.7 cells after 24 h of culturing on soft and stiff PVA hydrogels. The white dashed line marks the ventral shape of the cell. (Scale bar, 1 µm.) (E) Representative 3D rendering images of RAW264.7 cells cultured on soft and stiff PVA hydrogels for 2 h. F-actin is red, and nuclei are blue. (Scale bar, 3 μm.) (F and G) Quantification of cell shape (ratio of maximum cell diameter to minimum cell diameter) and cell area on soft and stiff substrates. Data were analyzed using a nonparametric test. (H) To clarify whether macrophages show stiffness-related functional alterations at early stages and whether the classical pathway of integrins is involved, Western blotting was performed to examine protein expression after 2 h of culturing on hydrogels. (I) Expression of ITGB1 over time on substrates with different stiffnesses. (J) Inflammatory marker expression of RAW264.7 cells on substrates with different stiffnesses by flow cytometry after EDTA inhibition of integrin function (n = 3). (K) Schematic illustration. Macrophage stiffness sensing requires integrin-mediated adhesion. The data presented were analyzed by one-way ANOVA with Tukey’s posttest unless otherwise stated. *P < 0.033, **P < 0.002, ***P < 0.001; ns, no significant difference.
In addition to functional changes, we observed changes in cell morphology. In addition to the observation of greater cell spreading on stiff substrates, which is consistent with the literature (18, 20), there were differences in the three-dimensional morphology of the cells. The curvature of the plasma membrane ventral to the cells decreased with increasing matrix stiffness (Fig. 2C). To exclude possible fluorescence artifacts due to the Z-stack resolution of confocal microscopy, we used focused ion beam scanning electron microscopy (FIB-SEM) to section cells in situ to confirm these results (Fig. 2D). The change in the plasma membrane curvature and the difference in cell spreading persisted at the early stage of macrophage adhesion, which suggests that macrophage recognition of stiffness and corresponding behavioral changes are early and transient processes (Fig. 2 E–G).
Macrophage Stiffness Sensing Requires Adhesion.
Integrins are now well acknowledged as facilitators of mechanotransduction, transmitting extracellular physical signals into the cell, and converting them into biochemical signals that regulate cell behaviors (25, 26). Therefore, the integrin-FAK pathway of macrophages at 2 h was examined by Western blot (Fig. 2H). Differences in cellular NOS2 expression existed, while no differences in FAK phosphorylation levels or ITGB1 expression were observed in the early stage, indicating that integrin–ligand binding does not differ significantly on the surface of different substrates. Furthermore, it could be observed by immunofluorescence and Western blot that for macrophages, integrins are mainly involved in the formation of podosomes rather than conventional focal adhesion, and their expression did not differ significantly over time on soft and stiff substrates (Fig. 2I and SI Appendix, Fig. S3E).
However, integrins remain involved in the recognition of matrix stiffness by macrophages. After the inactivation of integrins by depletion of cations with EDTA, the difference in activation of stiffness sensing disappeared (Fig. 2J). Thus, early-stage macrophage stiffness sensing requires the establishment of initial adhesion and is independent of the difference in the degree of FAK phosphorylation (Fig. 2K).
Macrophages Recognize and Respond to Stiffness through Plasma Membrane Deformation.
Cell morphological changes have been reported to alter cell function (27, 28). However, previous studies have focused more on two-dimensional cell morphology changes than on three-dimensional cell morphology. Therefore, based on the previously observed differences in cytoplasmic membrane deformation on the substrate, we propose the hypothesis that macrophages can perceive stiffness through plasma membrane deformation.
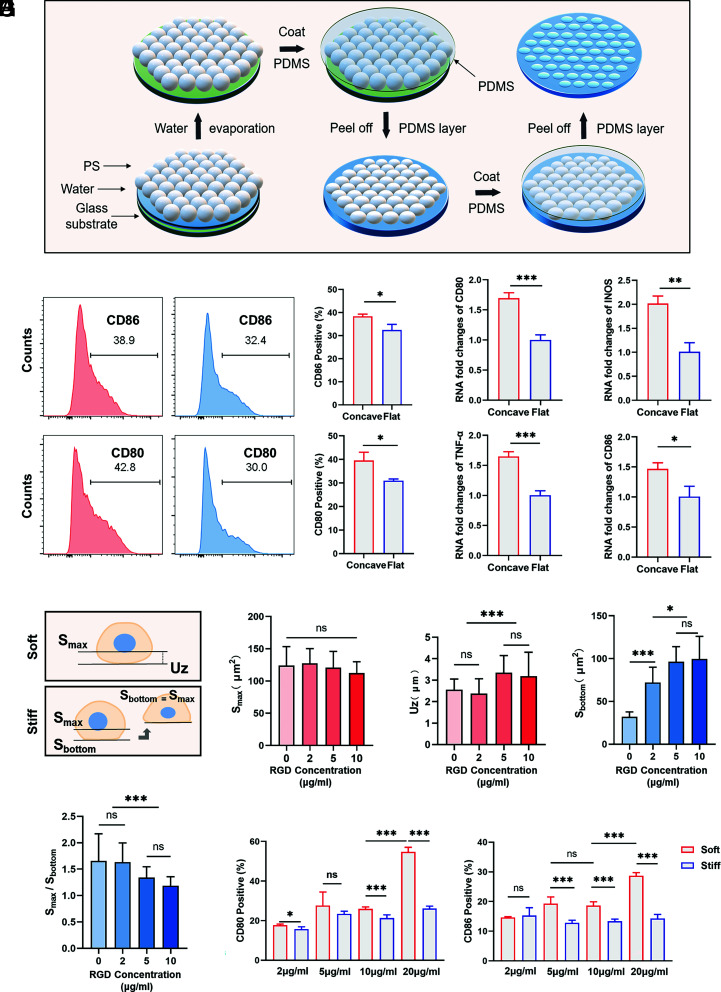
To test this, we prepared a micron-scale concave and convex substrate by the self-assembly of polystyrene (PS) microspheres and nanoimprinting techniques, on which cells had three-dimensional plasma membrane deformation (Fig. 3A). SEM and bright-field microscopy confirmed the expected concave topography (SI Appendix, Fig. S4 A–C). Compared to flat surfaces, cells had different plasma membrane deformations on concave substrates with the same stiffness (SI Appendix, Fig. S4D). The degree of macrophage activation was then examined by flow cytometry, real-time qPCR, and immunofluorescence (Fig. 3 B and C and SI Appendix, Fig. S4D), which showed higher inflammatory activation on the concave surface, consistent with the soft substrate. However, on the convex substrates, the cells tended to be distributed in the gap region between two or three convexities, rather than being located on the convex surface, showing more complex curvature changes and nuclear deformation (SI Appendix, Fig. S4F). For further determination, PDMS with different surfaces was implanted into mice. Elevated macrophage inflammation as well as increased fibrosis thickness were observed at the concave surface (SI Appendix, Fig. S4 G and H). Therefore, the mechanism of cell stiffness sensing is similar to cell topography sensing, both of which are based on the perception of the external environment through plasma membrane deformation leading to functional changes in response to mechanical clues.
Fig. 3.
Plasma membrane deformation affects macrophage function, and macrophages show differences in membrane deformation on soft and stiff substrates. (A) Schematic of the concave substrate preparation process. (B and C) RAW264.7 cells were cultured on concave and flat substrates for 24 h. Flow cytometry (n = 3; *P = 0.0146, *P = 0.0180) and RT–PCR (n = 3; *P = 0.016, **P = 0.002) were performed to examine the expression of inflammatory markers and factors. Two-tailed Student’s t test. (D) Schematic illustration. On soft substrates, because it is difficult to calculate the bottom adhesion area, the vertical distance between the plane of maximum circumference on the z-axis and the lowest point of cell adhesion is defined as Uz. On stiff substrates, the area of maximum cell circumference and the area of cell bottom adhesion were recorded as Smax and Sbottom. (E–H) The concentration of ligands on the hydrogel surface was changed to quantify Uz (n = 20) and Smax on the soft substrates (n = 20) and Smax and Sbottom on the stiff substrates (n = 24). (I) Flow cytometry analysis of macrophage responses to soft and stiff hydrogel substrates when the concentration of ligands on the hydrogel surface is changed (n = 5; *P = 0.03). Unless otherwise stated, the data presented were analyzed by one-way ANOVA with Tukey’s posttest. *P < 0.033, **P < 0.002, ***P < 0.001; ns, no significant difference.
To facilitate the discussion and analysis of deformation, the distance from the lowest point of cell–material contact to the initial plane was defined as Uz. In addition, the maximum circumference area (Smax) versus the bottom adhesion area (Sbottom) was recorded (Fig. 3D). Although macrophages do not rely on the difference of FAK phosphorylation for stiffness sensing on soft and stiff substrates, integrins still participate in the stiffness mechanoreception process. It has been found that the formation of integrin clusters is correlated with ligand distribution. Thus, we prepared PVA hydrogels coated with RGD at various concentrations. As the ligand density increased from 0 μg/mL to 10 μg/mL, the Uz of the cells on the soft substrate increased, while the Smax did not change significantly (Fig. 3 E and F). On the stiff substrate, Sbottom gradually increased, and the ratio of Smax to Sbottom converged to 1 with increasing ligand concentration, demonstrating the enhanced adhesion of the cells with increasing ligand concentration (Fig. 3 G and H and SI Appendix, Fig. S5 A–D). The difference in plasma membrane deformation on substrates increased with increasing ligand concentration. Although ligands promoted macrophage activation, as detected by flow cytometry, the difference in activation of cells on soft and stiff substrates increased with elevated ligand concentration, consistent with the observation of morphological differences (Fig. 3I and SI Appendix, Fig. S5E). Thus, integrin-mediated adhesion leads to cellular plasma membrane deformation for stiffness sensing. However, whether the deformation behavior is an actively produced adaptive change or a passive behavior of the cell remains to be discussed.
Macrophages Exert Traction Stress on the Substrate to Produce Differential Plasma Membrane Deformation.
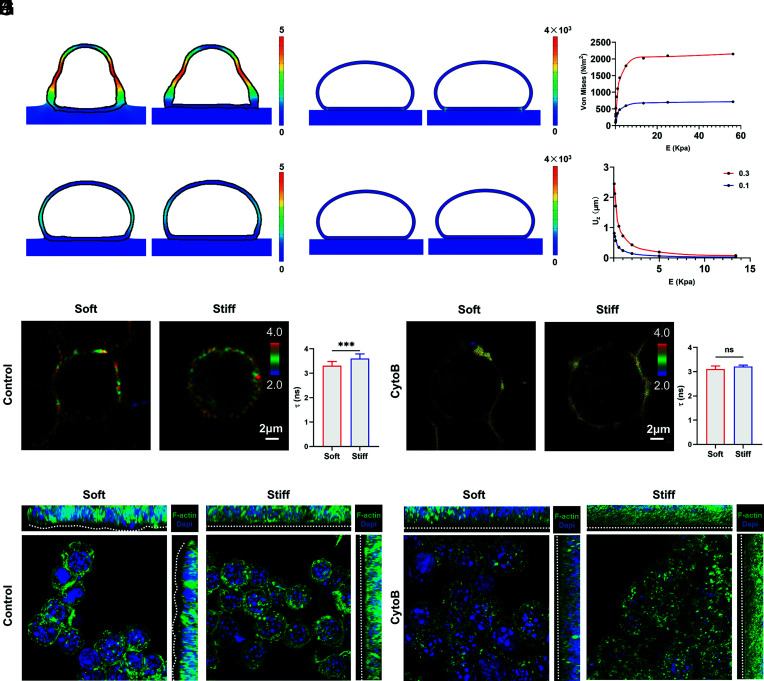
Previous studies have observed the deformation of liposomes on hydrogel substrates and found that liposome spreading generates traction stresses on the substrate (29). However, it is not yet known whether cell membrane surface tension also exists on macrophages and maintains the deformation changes of the cell membrane as well as the deformation of the region in contact with the substrate. To investigate the mechanism by which macrophages appear to differ in curvature on soft and stiff substrates, we used a finite element model (FEM) to generate stress curves and displacement curves between PVA hydrogels with different elastic moduli (Fig. 4 A and B and see Methods for details). We chose a hemispherical geometry to represent the cell (SI Appendix, Fig. S6B).
Fig. 4.
Macrophages generate plasma membrane deformation by applying stress to the substrate through adhesion. (A) Displacement of the cell membrane with different stiffnesses of the PVA substrate, with the color scale illustrating the displacement size (μm). (B) Von Mises equivalent strains of the cell membrane periphery with different stiffnesses of the PVA substrate, with the color scale illustrating the stress magnitude (N/m2). (C) Displacement of the cell membrane under a relaxed state with different stiffnesses of the PVA substrate, tension coefficient τ = 0.1 mN/m. Color scale illustrating the displacement size (μm). (D) Von Mises equivalent strains of the cell membrane periphery under a relaxed state with different stiffness of the PVA substrate, tension coefficient τ = 0.1 mN/m. Color scale illustrating the stress magnitude (N/m2). (E) The z-axis displacement of the bottom center of the cell membrane under the condition of substrate stiffness gradient. (red means τ = 0.3 mN/m, blue means τ = 0.1 mN/m) (F) The magnitude of von Mises equivalent strains at the periphery of the cell membrane in contact with the substrate under the condition of a substrate stiffness gradient (red means τ = 0.3 mN/m, blue means τ = 0.1 mN/m). (G and H) RAW264.7-cell membrane tension was detected by the probe Flipper-TR 2 h after cell culture. (G) Cells were treated with DMSO as control groups (soft: n = 23; stiff: n = 16), and (H) cells were treated with 20 μM cytochalasin B (n = 6). The fluorescence lifetime range used by Flipper-TR for statistics is 2 to 4 ns. (Scale bar, 2 μm.) Two-tailed Student’s t test. (I and J) Z-stack confocal images of RAW264.7 cells on soft and stiff substrates. F-actin is green, and nuclei are blue. (Scale bar, 10 μm.) (I) Cells treated with DMSO as control groups had dense and clear cytoskeletons with membrane deformation differences on different substrates. (J) Cells treated with 20 μM cytochalasin B had broken and discontinuous cytoskeletons with reduced membrane deformation variation on different substrates. *P < 0.033, **P < 0.002, ***P < 0.001; ns, no significant difference.
The deformation of the hydrogel arises from the vertical force exerted by the macrophages on the soft substrate. We assume a stable state of adhesion between the cell and the substrate, which means that the traction force of the substrate in contact with the macrophage brings the system into equilibrium. Laplace’s law can be applied to the free membrane of macrophages:
where P is the pressure outside the cell membrane and R is the radius of curvature of the membrane. Then, we simulated the gradient stiffness, and the results demonstrated that the displacement of the cell bottom gradually decreased as the substrate stiffness increased, which is consistent with our aforementioned confocal images (Fig. 4 A–E). Furthermore, on the surface of stiff substrates, the local stress in the outermost cell membrane at the cell–substrate junction is much greater than the local stress in the cell membrane on soft substrates because it is more difficult to produce displacement against stiff substrates (Fig. 4 B–F). Thus, the cells are more likely to achieve a state of contact surface bending upon contact with the soft substrate, which is consistent with our aforementioned experiments.
In contrast, we simulated cells on PDMS, a viscoelastic material with different elastic moduli, for comparison. Fitting Laplace’s law to the same model species and plotting the displacement profile (SI Appendix, Fig. S6C), the bottom of the cell tends to a flatter state regardless of the substrate stiffness (SI Appendix, Fig. S6D). This is also in agreement with the results we observed by confocal microscopy on PDMS with different stiffnesses (SI Appendix, Fig. S6A).
According to Laplace’s law, the tensile force of the cell membrane on a substrate is strongly related to the tension coefficient τ of the cell membrane surface. To verify this property, we reduced the cell membrane tension coefficient τ to 0.1 mN/m and performed the gradient stiffness simulation again (Fig. 4 C and D). The results show that under the condition of a reduced cell membrane tension coefficient, the cell membrane is more difficult to displace under all conditions of substrate stiffness, and the stress at the cell periphery is reduced (Fig. 4 E and F). Thus, the deformation depends on the substrate’s mechanical properties and cell material properties. To verify the simulation results, we used the membrane tension probe Flipper-TR. Membrane tension increased with increasing matrix stiffness after initial macrophage adhesion (Fig. 4G). It is known that cell membrane tension is mainly derived from membrane components and cortical cytoskeleton attachment (30). Thus, in addition to larger stresses, cells also exhibit more cortical cytoskeletal aggregation on stiff substrates. When cells were pretreated with 20 µM the actin inhibitor cytochalasin B before seeding, the membrane tension decreased, and the difference was diminished (Fig. 4H). In addition, the difference in cytoplasmic membrane deformation was reduced after inhibitor treatment, consistent with the simulation results (Fig. 4 I–J).
The above results demonstrated that cells apply pulling forces on substrates through initial adhesion, which results in differential deformation due to differences in material stiffness. In turn, the cytoskeleton is differentially regulated in this process, further enhancing the stabilization of the differential deformation and leading to subsequent differences in response.
Plasma Membrane Deformation Regulates the Spatial Distribution of Baiap2, Affecting the Cytoskeleton.
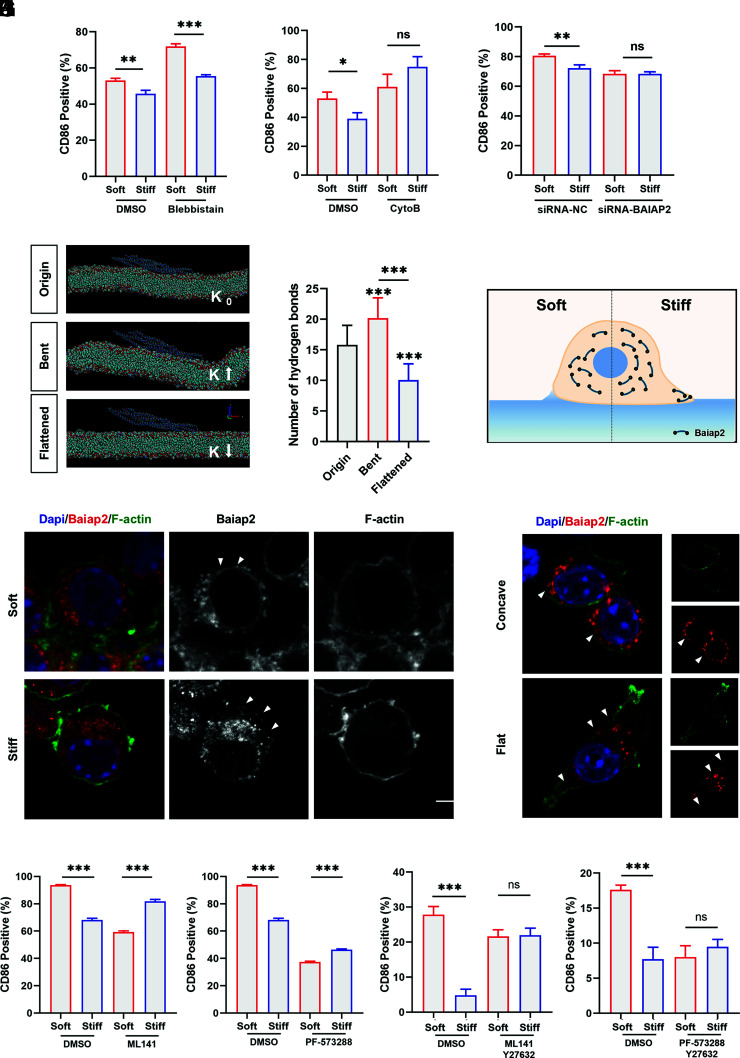
Numerous studies have suggested a relationship between the cytoskeleton and inflammatory activation (31). Flow cytometry demonstrated an increase in cellular inflammation activation after treatment of cells with cytochalasin B or the myosin inhibitor blebbistatin, suggesting that cytoskeletal remodeling facilitates the inhibition of inflammatory activation (SI Appendix, Fig. S7A). However, differences in the cell stiffness response disappeared after inhibition of actin aggregation, while differences persisted after myosin inhibition (Fig. 5 A and B). This suggests that macrophages recognize and respond to stiffness primarily through the modulation of cellular actin. Considering previous results, there exists another mechanism based on plasma membrane deformation that regulates actin aggregation in response to stiffness.
Fig. 5.
Plasma membrane deformation mediates inflammatory responses by affecting Baiap2 to regulate the cytoskeleton. (A and B) RAW264.7 cells were treated with inhibitors 30 min before cell culture and for the next 24 h after being cultured on substrates. Flow cytometry was performed to determine the importance of myosin (A) and actin (B) in macrophage stiffness sensing by examining the expression of the inflammatory marker CD86 (n = 3; *P = 0.01, **P = 0.005). (C) The role of Baiap2 in stiffness perception was examined by flow cytometry after the knockdown of Baiap2 expression in RAW264.7 cells (n = 3; **P = 0.002). (D) Representative images of the state of the I-BAR domain when the membrane curvature K changes. (E) Analysis and evolution of the number of hydrogen bonds between the protein and membrane after an initial simulation of 20 ns and a conditional simulation of 2 ns (n = 20). (F) Schematic diagram of Baiap2 distribution. (G) Representative immunofluorescence images of the distribution of Baiap2 when Raw264.7 cells were cultured on PVA hydrogels for 2 h. F-actin is green, nuclei are blue, and Baiap2 is red. Triangular symbols indicate differences in the local distribution of Baiap2 on the inner side of the membrane. (Scale bar, 5 μm.) (H) Representative immunofluorescence images of the distribution of Baiap2 when Raw264.7 cells were cultured on processed surfaces for 2 h. F-actin is green, nuclei are blue, and Baiap2 is red. Triangular symbols indicate differences in the local distribution of Baiap2 on the inner side of the membrane. (Scale bar, 2 μm.) (I–L) Flow cytometry analysis of inflammatory differences in RAW264.7 cells cultured on hydrogels of different stiffnesses after the use of inhibitions. (n = 3). Unless otherwise stated, the data presented were analyzed by one-way ANOVA with Tukey’s posttest. *P < 0.033, **P < 0.002, ***P < 0.001; ns, no significant difference.
The Bin–Amphiphysin–Rvs (BAR) domain proteins are a family of proteins that sense and generate curvature consisting of helical bundles that associate in an antiparallel manner to form dimers of different sizes and curvatures (32). They interact with membranes via nonspecific electrostatic interactions and specific curvatures determined by their protein structures, which can bind directly to cytoskeleton assembly factors and/or kinesins.
In this way, we used molecular dynamics to simulate the interaction between three subfamily domains of curvature-sensing proteins and flat cell membranes. Our results screen and identify the inverse BAR (I-BAR) domain, which has a flatter negative curvature that facilitates binding to the membrane (SI Appendix, Fig. S7 B–D and see Methods for details). To examine the significance of I-BAR in the process of macrophage recognition to surface stiffness, we knockdown representative I-BAR protein Baiap2 via siRNA silencing, and the activation differences diminished, which indicated that Baiap2 is essential for curvature-based stiffness sensing (Fig. 5C and SI Appendix, Fig. S7F). To explore the mechanisms involved, we constructed a bending and stretching membrane model by molecular dynamics. The I-BAR domain binds to the membrane after 20 ns of simulation and causes negative curvature of the membrane (SI Appendix, Fig. S7 G and H). Then, the membrane is bent by squeezing and flattened by stretching for 2 ns (Fig. 5D). Binding is enhanced by an increase in the number of hydrogen bonds between the protein and the membrane when the membrane is bent. When the membrane is flattened, the number of hydrogen bonds is reduced, and the binding is lost (Fig. 5E). It has been reported that BAR domain proteins have autoinhibitory intramolecular interactions and the interactions can be alleviated by binding to other proteins or membranes (32). The adaptability of the protein structure to the membrane curvature in the simulations suggests possible localization changes when membrane deformation occurs. Therefore, we hypothesize that membrane deformation affects the binding of Baiap2 to the membrane and thus its regulatory function on the cytoskeleton.
The distribution of Baiap2 was observed by immunofluorescence (Fig. 5G). Consistent with the simulation results, Baiap2 aggregates more in the regions with large curvature than in those with small curvature. On soft substrates, Baiap2 is uniformly distributed on the inner side of the membrane due to the relatively gentle curvature. On stiff substrates, Baiap2 is unevenly distributed within the cell. Due to the near-zero curvature at the bottom and greater curvature at the rim, some of the proteins are scattered and distributed, and some are bound to the limbal membrane, which leads to the elongation of actin filaments and subsequent spreading. Similarly, Baiap2 has an altered distribution on the morphological substrate (Fig. 5H). The Baiap2 distribution was consistent with pits due to the presence of intrinsic topography. Moreover, it has been reported that an increase in the local density of BAR domain proteins can promote their linear arrangement to stabilize local membrane deformations, while an increase in membrane curvature can also promote an increase in linear aggregation events (33). Therefore, changes based on adhesion-mediated membrane deformations can affect the distribution and function of Baiap2 (Fig. 5F).
Studies have demonstrated that the self-inhibited state of Baiap2 slows the growth of actin filament ends. Activation of Cdc42 by FAK phosphorylation also alleviates the self-inhibition by binding to the Baiap2 auxiliary structural domain SH3, which promotes progressive actin filament elongation (34–36). And the results of FAK inhibitors and Cdc42 inhibitors validate this (Fig. 5 I–J). However, the difference in inflammatory activation between soft and stiff substrates reverted, indicating other potential pathways also involved in the regulation of stiffness sensing. Rho-associated protein kinases (ROCKs) are a key regulator of the actin cytoskeleton and have been reported to be relevant for stiffness sensing (37, 38). The trend of inflammatory activation was unchanged when ROCK was inhibited, while the inflammation difference on substrates of different stiffness disappeared when ROCK was inhibited along with FAK or Cdc42 (Fig. 5 K and L and SI Appendix, Fig. S7K).
Based on the above results, it can be assumed that ligand–integrin mediated adhesion leads to cytoplasmic membrane deformation that affects the distribution and function of Baiap2. On a stiff surface, Baiap2 binds to the membrane periphery and promotes actin aggregation to inhibit inflammation, whereas, on a soft surface, the uniform density and intracellular distribution of Baiap2 inhibit cytoskeleton remodeling, leading to relatively higher inflammatory activation.
Discussion
Matrix stiffness has been demonstrated to have implications in embryonic development, stem cell differentiation, and tumor microenvironment immunity for subsequent disease treatment and drug development. Recent work has demonstrated the effects of biomaterial stiffness on macrophage-immune responses, but the mechanism of macrophage stiffness sensing remains unclear (18–20). In this study, we demonstrated that macrophages can influence intracellular curvature-sensing proteins through an adhesion-mediated plasma membrane deformation mechanism, which in turn regulates cytoskeletal remodeling in response to stiffness.
Many studies have shown that biological behavior can be explained by the laws of physics (39, 40). Surface tension refers to the tension acting on any boundary along the surface of the liquid surface layer due to the unbalanced molecular attraction. Surface tension exists in many phenomena in living organisms, for example, Laplace’s law is often used to describe the recoil force of alveoli (41). Murrell et al. revealed the liposome adhesion behaviors on deformable substrates and that indentation is generated by an applied pressure through membrane tension elevated by adhesion to the substrate (29). On this basis, we hypothesized that recognition of substrate stiffness by macrophages is also a physical behavior-mediated process. The interaction of integrins and ligands provides the basis for a theoretical model based on Laplace’s law, leading to further membrane deformation. To investigate the potential relationship between cell membrane deformation and stiffness sensing, a concave surface and a flat surface with the same stiffness were prepared to produce similar three-dimensional plasma membrane deformation in cells, which led to the same functional changes as cells on different stiffness surfaces and confirmed the speculation.
The BAR protein family plays an important role in vesicle transport, cytokinesis, and pseudopodium formation due to their specific structural properties (32). Recent studies have shown that BAR proteins play an active role in the cellular response to nanomorphology (28). Membrane deformation caused by nanomorphology not only occurs on the biomaterial surface but also occurs due to the physical interactions of the above-mentioned cell–substrate interface. Here, we observed the altered distribution of the I-BAR protein Baiap2 and identified its important role in stiffness recognition. And the important role of BAR proteins in stiffness perception also provides strategies to target macrophages to regulate inflammatory response and fibrosis.
Our results reveal a mechanism of macrophage stiffness recognition of elastic materials. Macrophage membrane deformation, which is mainly related to the materialistic properties of substrates and cells, can modulate cytoskeletal remodeling to respond to material stiffness. Although the stiffness of most implants currently used in clinical applications far exceeds cellular recognition, the emergence of surface coating technologies has found a certain balance between macroscopic biomechanics and local cellular stiffness sensing (7, 8, 18). The design of material topography may also be inspired by stiffness sensing (42). Future studies will help reveal how this mechanism can be applied to better design materials and avoid the occurrence of adverse FBRs.
Materials and Methods
Preparation of Implants.
PVA hydrogels were synthesized by preparing a mixture of 10% (w/v) PVA (Aladdin) with different concentrations of 50% (m/v) glutaraldehyde (Aladdin) at pH 3 ~ 4. It was poured into prefabricated molds or Petri dishes to polymerize overnight at 37 °C. The hydrogels were repeatedly washed with deionized water at 98 °C to remove the residual crosslinker. The tensile modulus of elasticity is obtained by a universal tensile tester. Soft, medium, and stiff PVA hydrogels have a modulus of elasticity of 0.56 kPa, 13.42 kPa, and 56.25 kPa, respectively. PDMS was fabricated using Sylgard 184 PDMS (Dow Corning) silicone elastomer mixed with the curing agent at ratios of 10:1 (w/w, E = 2,151 kPa) (43) and 50:1 (w/w, E = 15 kPa) (44). PAA hydrogels with a modulus of elasticity of 2.8 kPa and 55 kPa were prepared by adjusting the ratio of 40% acrylamide (Aladdin) and 2% bis-acrylamide (Aladdin) according to a well-established protocol (45, 46). GelMA hydrogels with a modulus of elasticity of 2 kPa and 29 kPa were prepared as previously reported by adjusting the GelMA concentration (47). PCL films with a modulus of elasticity of 330 kPa and 1,000 kPa were fabricated using PCL (average Mn = 80,000, MACKLIN) (48, 49). All materials were exposed to UV light and immersed in 75% ethanol for 1 h to ensure sterility before use. For animal experiments, all materials were cut into small discs of 5 mm diameter and 1 mm thickness (except for the thickness of PCL films). To culture cells in vitro, PVA hydrogels were cut into blocks of a size appropriate for the well plate. The PVA hydrogels were incubated with 10 μg/mL RGD solution overnight before cell culture. PAA hydrogels and PDMS were prepared on 24 mm × 24 mm diameter coverslips (CITOGLAS); PAA hydrogels were also incubated with 10 μg/mL RGD solution overnight at 4 °C according to the protocol.
Fabrication of Substrate Surface Topography.
The concave microarrays were fabricated based on a previous study (50). Coverslips (24 mm × 24 mm) were used as substrates, on which PS microspheres (20 μm) self-assembled at the PS-water/ethanol-air interface and formed uniform PS arrays after liquid evaporation. Liquid PDMS (10:1, w/w) was spin-coated on coverslips at 3,000 rpm for 5 s and heated at 95 °C for 30 s. Then, coverslips with self-assembled PS spheres on the surface were stamped on the PDMS-coated coverslips. After heat curing at 95 °C for 2 h, the coverslips originally with PS spheres on the surface were carefully removed to obtain PDMS films with PS arrays embedded on the surface. Then, liquid PDMS (10:1, w/w) was spin-coated on the PDMS films at 200 rpm for 10 s, and the films were heated at 95 °C for 2 h. Finally, the two PDMS layers were separated, and PDMS films with concave surfaces were obtained. The flat substrate is produced from coverslips with PDMS spin-coated on the surface. To obtain the convex substrate, the concave substrate was used as a mold. A layer of 100-nm-thickness Ti film was deposited on the surface.
Cell Culture and Treatment.
After euthanasia of C57Bl/6 mice, the marrow cavities were exposed and washed with DMEM (HyClone) consisting of 10% fetal bovine serum (FBS, Gibco) and 1% penicillin-streptomycin (Gibco) to obtain a cell suspension. The cell suspension was cultured overnight to remove the adherent cells. Then, the suspended cells were transferred to a culture dish and induced to differentiate in the modified cell culture medium containing M-CSF (50 ng/mL; Proteintech) for 5 d. BMDMs were then obtained. The macrophage cell line RAW264.7 (CTCC) was cultured in DMEM containing 10% FBS. To investigate the cellular response to substrate stiffness, cells were cultured on the hydrogel surface. For Baiap2 knockdown, RAW264.7 cells were transfected with 30 nM siRNA (Sangon Biotechnology) by use of PepMute (SiganGen) for 48 h before being cultured on hydrogels. All cells were cultured at 37 °C in a humidified 5% CO2 atmosphere. To study the contribution of different proteins, the 20 µM actin polymers inhibitor cytochalasin B (HY-16928; MCE), 10 µM myosin inhibitor blebbistatin (HY-13441; MCE), 10 µM FAK inhibitor PF-573288 (HY-10461; MCE), 20 µM ROCK inhibitor Y27632(HY-10071; MCE), and 20 µM Cdc42 inhibitor ML141 (HY-12755; MCE) were added to the cell culture medium 30 min before experiments. Cells were then cultured with the modified cell culture medium in subsequent experiments.
Water Contact Angle Measurement.
A contact angle tester (Bruker & Kjaer) was used to measure the hydrophobicity of PVA hydrogels. Deionized water was loaded. The volume of each droplet was 10 µL. Photographs were taken after the droplet had settled on the surface (time >15 s) for further analysis.
Scanning Electron Microscopy and Focused Ion Beam Scanning Electron Microscopy.
SEM (Zeiss) was employed in examining the surface topography of the PVA hydrogels. The PVA hydrogels were cut into cubes and dried by two standard methods to determine the pore size. One method is to freeze the hydrogels in liquid nitrogen followed by freeze-drying them in a vacuum freeze-dryer for 24 h. The other is to freeze-dry the hydrogels in a vacuum freeze-dryer overnight until they meet the requirements for SEM sample preparation. Fib-SEM (TESCAN) was used to observe cell cross sections in situ. Cells cultured on hydrogels were fixed in 2.5% glutaraldehyde for 10 min and then dehydrated in a gradient of alcohol from 30 to 100%. Samples were sprayed with gold for 90 s under 30 mA.
Material Biocompatibility Assays.
PVA hydrogels with different stiffnesses were prepared in 96-well cell culture plates before the same number of cells (5 × 103) culturing on each substrate for a maximum of 72 h, and cell proliferation was analyzed by the use of Cell Counting Kit-8 (Beyotime Biotechnology) concerning the manufacturer’s instructions. In addition, a BeyoClick™ EdU Cell Proliferation Kit with Alexa Fluor 488 (Beyotime Biotechnology) was also used. Cells were cultured on substrates prepared in 12-well plates and collected by flow cytometry.
Implant Surgery.
Female C57Bl/6 mice at 8 wk old were obtained from the animal center of Wuhan University. All the procedures of the experiments were approved by Wuhan University and conducted with reference to the guidelines of laboratory animal care and use. The mice were grouped according to the time of retrieval and the type of implant materials, and each group consisted of three mice. After anesthesia, the lower limbs were shaved, and the surgery area was sterilized with 0.5% iodophor. A 1-cm incision was made in the skin and quadriceps muscle groups. The prepared implants were gently placed in the wound of the quadriceps muscle groups, one implant per leg. The wound was carefully closed by suturing. After the euthanasia of the mice by CO2 administration, the implant and the fibrous capsule were separated from the surrounding adherent tissue. For the macrophage clearance groups, mice were injected with 200 µL of clodronate liposomes (Yeason) in the tail vein 1 d before surgery and 200 μL of clodronate liposomes in the tail vein every 2 d afterward.
Histological Processing for H&E.
Tissues containing implants were fixed for 48 h in 4% PFA Fix Solution (Beyotime Biotechnology) and then dehydrated and processed for paraffin wax. Five-micrometer sections were rehydrated before hematoxylin and eosin staining (Beyotime Biotechnology).
Immunofluorescent Imaging.
Retrieved materials and cells cultured on hydrogels were fixed in 4% PFA Fix Solution. After being blocked with 2% bovine serum albumin (BSA, Roche) containing 0.1% Triton X-100 (Beyotime Biotechnology) at room temperature for 1 h, samples were incubated with primary antibodies overnight at 4 °C and then labeled with secondary antibody (Abclonal; 1:200). The primary antibodies used included αSMA (ABclonal; 1:150), NOS2 (ABclonal; 1:150), Baiap2 (ABclonal; 1:150), Cdc42 (ABclonal; 1:150), ITGB1 (ABclonal; 1:150), and Vinculin (Boster; 1:200). FITC-phalloidin (Yeasen; 1:150) and DAPI (Beyotime Biotechnology; 1:10) were then used. Images were captured by a Zeiss LSM880 confocal microscope and analyzed by ImageJ.
Flow Cytometry.
Tissues at the implant site were cut into pieces and treated with collagenase (Sigma–Aldrich) to obtain single-cell suspensions. BMDMs were marked by F4/80 (Biolegend; 1:400) and CD11b (Biolegend; 1:200). For macrophage function assays, cells were stained with CD80 (Biolegend; 1:200) and CD86 (Biolegend; 1:200) at 4 °C for 15 min. Cells were then collected by an LSRFortessa X-20 flow cytometer (Becton Dickinson) and analyzed by FlowJo software.
Real-Time qPCR.
Total RNA was obtained by isolation and extraction with the use of TRIzol reagent (Takara). cDNA was obtained by reverse transcription of total RNA with the use of HiScript II Q RT SuperMix (Vazyme) by referring to the manufacturer’s instructions. Amplification reactions were carried out with the ChamQ SYBR qPCR master mix (Vazyme) in a CFX Connect RT–PCR detection system (Bio-Rad). The results were analyzed by the 2-ΔΔCt method, normalizing to GAPDH and calibrating to the control group.
Western Blot.
Total intracellular protein was obtained with RIPA lysis buffer (Beyotime Biotechnology), and a BCA protein assay kit (Beyotime Biotechnology) was used to determine the protein concentration. After being denatured at 96 °C for 10 min with 5× loading buffer (Beyotime Biotechnology), samples were separated by 10% SDS–PAGE gels and then transferred to PVDF membranes (Roche). Blocking PVDF membranes with 5% skim milk for 1 h at room temperature and incubated with primary antibodies overnight at 4 °C. Membranes were then washed three times before incubating with secondary antibodies labeled with HRP (ABclonal; 1:10,000) for 1 h at room temperature. Signals were examined using the enhanced chemiluminescence (ECL, Advansta). The primary antibodies used included: anti-Arg1 (ABclonal; 1:1,000), anti-NOS2 (ABclonal; 1:1,000), anti-FAK (ABclonal; 1:1,000), anti-ITGB1 (ABclonal; 1:1,000), anti-p-FAK-Y397 (ABclonal; 1:1,000), anti-Baiap2 (ABclonal; 1:1,000), anti-Cdc42 (Proteintech; 1:800), and anti-GAPDH (ABclonal; 1:5,000).
Membrane Tension Assay.
Cells were stained with the membrane tension probe Flipper-TR (Spirochrome) according to the manufacturer’s instructions. FLIM images were obtained using a 488-nm pulsed laser for excitation and collecting photons through a 600/50-nm filter. The lifetime ranges from 2.8 to 7.0 ns. A longer lifetime means more membrane tension.
Molecular Dynamics Simulation.
The molecular dynamics simulation was performed by the GROMACS 2019.2 package with a charmm36 force field. The 3D structures of BAR domains were obtained from the Protein Data Bank (I-BAR, PDB ID: 1Y2O; F-BAR, PDB ID: 2EFL; BAR, PDB ID: 2ELB). The membrane containing 390 DOPSs and 910 DOPCs is established by CHARMM-GUI. The distance from the center of mass to the membrane was initially set to 1 nm for all proteins. The models were put into a 40*11*13 nm3 box and neutralized with 0.15 M CaCl2. The steepest descent method was adopted for energy minimization. Systems were heated to 323 K using the V-rescale thermostat in the NVT ensemble for 100 ps and equilibrated for 1 ns in NPT. For systems of three different domains, a 10 ns molecular dynamics simulation for domain–membrane interactions was performed with the constraint of the membrane. For I-BAR simulations, a 20 ns molecular dynamics simulation for domain–membrane interactions was performed without the constraint of the membrane. The I-BAR domain bends the membrane, and then a 2 ns simulation was performed to straighten or bend the membrane to make it more curved. The number of hydrogen bonds between the protein and the membrane was obtained to evaluate the interaction. The rmsds of proteins or systems were calculated to determine the simulation stability. VMD software was used for molecular dynamics simulations trajectory visualization and analysis.
Deformation Model.
To simplify the model, the FEM model of the cell consists of the cell membrane cortex and cytoplasm. In the process of previous experiments, we found cells with a length of 14.64 ± 1.81 μm and a height of 6.80 ± 1.19 μm (n = 16). Thus, the length and thickness of the cell in the model were set to 15 and 7 μm to be consistent with cell experiments (50). Parameters for cell were obtained from a previous publication. Young’s moduli of the cell membrane cortex and cytoplasm were set to 1 and 0.1 kPa, respectively; Poisson’s ratios of the cell membrane cortex and cytoplasm were set to 0.3 and 0.37, respectively (51). The elastic modulus of the PVA substrate was set to 0.566 and 56.252 kPa, consistent with the experimentally measured values. Additional stiffness gradients were simulated by 50, 100, 200, 566, 1,000, 2,000, 5,000, 13,421, 25,000, and 56,252 Pa. The PDMS substrate was set to 15 and 2,151 kPa (43, 44). The thickness of the cell membrane cortex was set to 0.25 μm (52). The cell membrane tension coefficient was set to 0.3 mN/m, derived from a previous study (29). To simulate conditions where membrane tension is suppressed, the membrane tension coefficient was set to 0.1 mN/m. The adhesion of the cell membrane cortex was simulated by adhesive contact interactions. During cell membrane tension loading, stable adhesion between the cell and the substrate was assumed, which was considered a fixed connection. The von Mises equivalent strains of the cell membrane were calculated as well as the displacement on the z-axis in the mode of surface tension.
Statistical Analysis.
Statistical analysis of the experimental results was performed using GraphPad Prism 8.0; data are presented as the mean ± SD. For data conforming to a normal distribution, one-way ANOVA was used for multiple groups, and a two-tailed Student’s t test was performed for two groups of samples. When the data did not have a normal distribution, they were analyzed by nonparametric tests. Differences were considered significant at P < 0.05. *P < 0.033, **P < 0.02, ***P < 0.001.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by the National Key R&D Program of China (2021YFC2400405) and the National Natural Science Foundation of China (No. 82025011).
Author contributions
Y.N., H.Q., and Y.Z. designed research; Y.N., H.Q., F.Z., S.J., Q.T., and W.M. performed research; Y.N., H.Q., and W.C. contributed new reagents/analytic tools; Y.N., H.Q., and W.C. analyzed data; and Y.N., H.Q., and R.J.M. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Rankin E. A., AJRR: Becoming a National US joint registry. Orthopedics 36, 175–176 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Overtchouk P., et al. , Advances in transcatheter mitral and tricuspid therapies. BMC Cardiovasc. Disord. 20, 1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel K., Brandstetter K., Solid implants in facial plastic surgery: Potential complications and how to prevent them. Facial Plast. Surg. 32, 520–531 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Li Z., Cui Y., Zhong J., Recent advances in nanogenerators-based flexible electronics for electromechanical biomonitoring. Biosens. Bioelectron. 186, 113290 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Wang D., Tan J., Zhu H., Mei Y., Liu X., Biomedical implants with charge-transfer monitoring and regulating abilities. Adv. Sci. (Weinh) 8, 2004393 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson J. M., Rodriguez A., Chang D. T., Foreign body reaction to biomaterials. Semin. Immunol. 20, 86–100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carnicer-Lombarte A., Chen S.-T., Malliaras G. G., Barone D. G., Foreign body reaction to implanted biomaterials and its impact in nerve neuroprosthetics. Front. Bioeng. Biotechnol. 9, 622524 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kastellorizios M., et al. , Foreign body reaction to subcutaneous implants. Adv. Exp. Med. Biol. 865, 93–108 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Kharbikar B. N., Chendke G. S., Desai T. A., Modulating the foreign body response of implants for diabetes treatment. Adv. Drug. Delivery Rev. 174, 87–113 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zdolsek J., Eaton J. W., Tang L., Histamine release and fibrinogen adsorption mediate acute inflammatory responses to biomaterial implants in humans. J. Transl. Med. 5, 31 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorbet M. B., Sefton M. V., Biomaterial-associated thrombosis: Roles of coagulation factors, complement, platelets and leukocytes. Biomaterials 25, 5681–5703 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Chu C., et al. , Modulation of foreign body reaction and macrophage phenotypes concerning microenvironment. J. Biomed. Mater. Res. A. 108, 127–135 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Doloff J. C., et al. , Colony stimulating factor-1 receptor is a central component of the foreign body response to biomaterial implants in rodents and non-human primates. Nat. Mater. 16, 671–680 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thevenot P., Hu W., Tang L., Surface chemistry influences implant biocompatibility. Curr. Top. Med. Chem. 8, 270–280 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veiseh O., et al. , Size- and shape-dependent foreign body immune response to materials implanted in rodents and non-human primates. Nat. Mater. 14, 643–651 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hadden W. J., et al. , Stem cell migration and mechanotransduction on linear stiffness gradient hydrogels. Proc. Natl. Acad. Sci. U.S.A. 114, 5647–5652 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z., et al. , Heterogeneous responses to mechanical force of prostate cancer cells inducing different metastasis patterns. Adv. Sci. (Weinh) 7, 1903583 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blakney A. K., Swartzlander M. D., Bryant S. J., The effects of substrate stiffness on the in vitro activation of macrophages and in vivo host response to poly(ethylene glycol)-based hydrogels. J. Biomed. Mater. Res. A. 100, 1375–1386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sridharan R., Cavanagh B., Cameron A. R., Kelly D. J., O’Brien F. J., Material stiffness influences the polarization state, function and migration mode of macrophages. Acta Biomater. 89, 47–59 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Chen M., et al. , Substrate stiffness modulates bone marrow-derived macrophage polarization through NF-κB signaling pathway. Bioact. Mater 5, 880–890 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhuri O., Cooper-White J., Janmey P. A., Mooney D. J., Shenoy V. B., The impact of extracellular matrix viscoelasticity on cellular behavior. Nature 584, 535–546 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maurer M., Lammerding J., The driving force: Nuclear mechanotransduction in cellular function, fate, and disease. Annu. Rev. Biomed. Eng. 21, 443–468 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Z., Guo S. S., Fässler R., Integrin-mediated mechanotransduction. J. Cell Biol. 215, 445–456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martino F., Perestrelo A. R., Vinarský V., Pagliari S., Forte G., Cellular mechanotransduction: From tension to function. Front. Physiol. 9, 824 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng B., et al. , Nanoscale integrin cluster dynamics controls cellular mechanosensing via FAKY397 phosphorylation. Sci. Adv. 6, eaax1909 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kechagia J. Z., Ivaska J., Roca-Cusachs P., Integrins as biomechanical sensors of the microenvironment. Nat. Rev. Mol. Cell Biol. 20, 457–473 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Zheng X., et al. , Near-infrared-triggered dynamic surface topography for sequential modulation of macrophage phenotypes. ACS Appl. Mater. Interfaces 11, 43689–43697 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Lou H.-Y., et al. , Membrane curvature underlies actin reorganization in response to nanoscale surface topography. Proc. Natl. Acad. Sci. U.S.A. 116, 23143–23151 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murrell M. P., et al. , Liposome adhesion generates traction stress. Nat. Phys. 10, 163–169 (2014). [Google Scholar]
- 30.Sitarska E., Diz-Muñoz A., Pay attention to membrane tension: Mechanobiology of the cell surface. Curr. Opin. Cell Biol. 66, 11–18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McWhorter F. Y., Wang T., Nguyen P., Chung T., Liu W. F., Modulation of macrophage phenotype by cell shape. Proc. Natl. Acad. Sci. U.S.A. 110, 17253–17258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishimura T., Morone N., Suetsugu S., Membrane re-modelling by BAR domain superfamily proteins via molecular and non-molecular factors. Biochem. Soc. Trans. 46, 379–389 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Jarin Z., et al. , Unusual organization of I-BAR proteins on tubular and vesicular membranes. Biophys. J. 117, 553–562 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Disanza A., et al. , CDC42 switches IRSp53 from inhibition of actin growth to elongation by clustering of VASP. EMBO J. 32, 2735–2750 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kast D. J., et al. , Mechanism of IRSp53 inhibition and combinatorial activation by Cdc42 and downstream effectors. Nat. Struct. Mol. Biol. 21, 413–422 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berthet J., Varelas X., Integrin-FAK-CDC42-PP1A signaling gnaws at YAP/TAZ activity to control incisor stem cells. Bioessays 39, 10.1002/bies.201700116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kümper S., Marshall C. J., ROCK-driven actomyosin contractility induces tissue stiffness and tumor growth. Cancer Cell 19, 695–697 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Sridharan R., Cavanagh B., Cameron A. R., Kelly D. J., O’Brien F. J., Material stiffness influences the polarization state, function and migration mode of macrophages. Acta Biomater. 89, 47–59 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Reversat A., et al. , Cellular locomotion using environmental topography. Nature 582, 582–585 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Luciano M., et al. , Cell monolayers sense curvature by exploiting active mechanics and nuclear mechanoadaptation. Nat. Phys. 17, 1382–1390 (2021). [Google Scholar]
- 41.Bernhard W., Lung surfactant: Function and composition in the context of development and respiratory physiology. Ann. Anat. 208, 146–150 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Chen L., et al. , Nanostructural surfaces with different elastic moduli regulate the immune response by stretching macrophages. Nano. Lett. 19, 3480–3489 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Noskovicova N., et al. , Suppression of the fibrotic encapsulation of silicone implants by inhibiting the mechanical activation of pro-fibrotic TGF-β. Nat. Biomed Eng. 5, 1437–1456 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Balaban N. Q., et al. , Force and focal adhesion assembly: A close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 3, 466–472 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Aratyn-Schaus Y., Oakes P. W., Stricker J., Winter S. P., Gardel M. L., Preparation of complaint matrices for quantifying cellular contraction. J. Vis. Exp. 2173 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tse J. R., Engler A. J., Preparation of hydrogel substrates with tunable mechanical properties. Curr. Protoc. Cell Biol. Chapter 10, Unit 10.16 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Zhuang Z., et al. , Control of matrix stiffness using methacrylate-gelatin hydrogels for a macrophage-mediated inflammatory response. ACS Biomater Sci. Eng. 6, 3091–3102 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Khampieng T., Yamassatien V., Ekabutr P., Pavasant P., Supaphol P., Protein adsorption and cell behaviors on polycaprolactone film: The effect of surface topography. Adv. Polymer Technol. 37, 2030–2042 (2018). [Google Scholar]
- 49.Das A., Adhikary S., Chowdhury A. R., Barui A., Substrate-dependent control of the chiral orientation of mesenchymal stem cells: Image-based quantitative profiling. Biomed. Mater. 16, 034102 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Xiong Y., et al. , A flexible, ultra-highly sensitive and stable capacitive pressure sensor with convex microarrays for motion and health monitoring. Nano. Energy 70, 104436 (2020). [Google Scholar]
- 51.Wang L., Wang L., Xu L., Chen W., Finite element modelling of single cell based on atomic force microscope indentation method. Comput. Math. Methods Med. 2019, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barreto S., Clausen C. H., Perrault C. M., Fletcher D. A., Lacroix D., A multi-structural single cell model of force-induced interactions of cytoskeletal components. Biomaterials 34, 6119–6126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.