Abstract
Hematologic malignancies are one of the most common cancers, and the incidence has been rising in recent decades. The clinical and molecular features of hematologic malignancies are highly heterogenous, and some hematologic malignancies are incurable, challenging the treatment, and prognosis of the patients. However, hematopoiesis and oncogenesis of hematologic malignancies are profoundly affected by epigenetic regulation. Studies have found that methylation-related mutations, abnormal methylation profiles of DNA, and abnormal histone deacetylase expression are recurrent in leukemia and lymphoma. Furthermore, the hypomethylating agents and histone deacetylase inhibitors are effective to treat acute myeloid leukemia and T-cell lymphomas, indicating that epigenetic regulation is indispensable to hematologic oncogenesis. Epigenetic regulation mainly includes DNA modifications, histone modifications, and noncoding RNA-mediated targeting, and regulates various DNA-based processes. This review presents the role of writers, readers, and erasers of DNA methylation and histone methylation, and acetylation in hematologic malignancies. In addition, this review provides the influence of microRNAs and long noncoding RNAs on hematologic malignancies. Furthermore, the implication of epigenetic regulation in targeted treatment is discussed. This review comprehensively presents the change and function of each epigenetic regulator in normal and oncogenic hematopoiesis and provides innovative epigenetic-targeted treatment in clinical practice.
Subject terms: Haematological cancer, Cancer genetics
Introduction
Hematologic malignancies are among the most common cancers and can involve all systems and organs. Hematologic malignancies mainly include leukemia, lymphoma, and multiple myeloma (MM), all of which are highly heterogenous in molecular characteristics, leading to severe difficulties in individualized treatment. Moreover, hematologic malignancies, especially MM, are often incurable and finally relapse or become refractory, appealing for innovative treatment strategies to increase treatment response and to improve their prognosis. Recent studies found that hematologic malignancies show recurrent methylation-related mutations, abnormal methylation profiles of DNA, and abnormal histone deacetylase expression, especially in leukemia and lymphoma. Furthermore, the hypomethylating agents and histone deacetylase inhibitors are effective to treat acute myeloid leukemia and T-cell lymphomas, indicating that epigenetic regulation is indispensable to hematologic oncogenesis. Thus, a comprehensive exploration of the change and the influence of each epigenetic mediators on hematologic oncogenesis assists to develop an innovative targeted treatment to improve the treatment response and prognosis of the patients.
Epigenetics originally referred to heritable features of a cellular phenotype that were independent of changes in DNA sequence. With the development of numerous studies and enlightened perspectives, epigenetics currently defines chromatin-based reactions that regulate DNA-templated processes. Chromatin consists of DNA and histones in a macromolecular complex that serves as a scaffold for genome packing. Histones are mainly divided into five categories. H2A, H2B, H3, and H4 are highly conserved, and an octamer consisting of two of each wrapped with 146 base pairs of DNA serves as the core of nucleosomes. H1 is variable among species and binds with internucleosome linear DNA to form a higher-level structure. According to the composition of chromatin, epigenetic regulation mainly includes alterations in DNA modifications, alterations in histone modifications, chromatin remodeling, and noncoding RNAs (ncRNAs) (Fig. 1). Epigenetic regulation plays an important role in various DNA-based processes, including DNA replication, repair, and transcription. With the development of epigenetic techniques, studies have explored the role of epigenetic regulation in hematopoiesis and have depicted the epigenome in hematologic malignancies. This review aims to highlight the influence of DNA modifications, histone modifications, and ncRNAs in hematopoiesis and their implications in targeted therapy of hematologic malignancies.
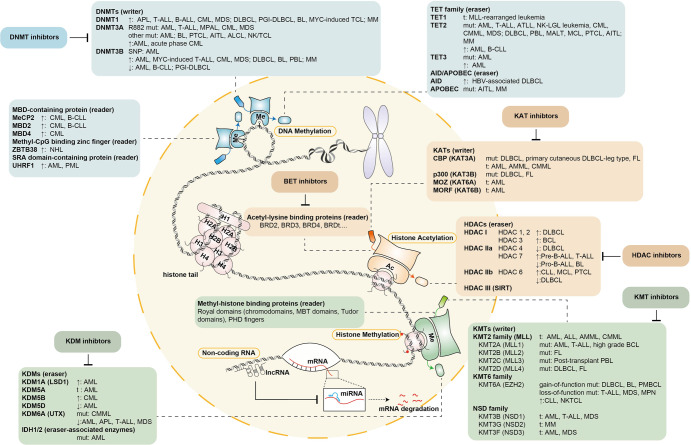
Fig. 1.
The overview of epigenetic regulation of hematologic malignancies. Epigenetic regulation of hematologic malignancies mainly includes DNA methylation, histone acetylation, histone methylation, and noncoding RNA. First, DNA methylation depends on writers, readers, and erasers. DNMTs are writers of DNA methylation, and targeted treatments focusing on DNMTs have been developed. Readers of DNA methylation include MBD-containing proteins, methyl-CpG binding zinc fingers, and SRA domain-containing proteins. Erasers of DNA methylation mainly consist of the TET family and the AID/APOBEC family. Second, histone acetylation depends on the writer KATs, the reader acetyl-lysine binding proteins, and the eraser HDACs. Correspondingly, innovative treatments targeting KATs, BETs, and HDACs are being developed. Third, histone methylation depends on the writer KMTs, the reader methyl-histone binding proteins, and the eraser KDMs. KMT inhibitors and KDM inhibitors are being explored in the treatment of hematologic malignancies. In addition, microRNAs and long noncoding RNAs also contribute to hematologic oncogenesis
DNA methylation alteration is common in hematologic malignancies
DNA methylation was first discovered from pneumococcal types when DNA was discovered as the hereditary material in mammals.1,2 DNA methylation is characterized by the carbon-5 position of the cytosine adding with a methyl group to form 5-methylcytosine (5mC).3 DNA methylation usually happens in the cytosine-guanine dinucleotides (CpG) sites by the DNA methyltransferase (DNMT) enzymes.3
DNA methylation in the normal genome
In the human genome, CpG sites spread over the whole genome, and are highly methylated, except CpG islands in normal somatic cells.4–7 Approximately 45% of the mammalian intergenic regions of the genome consist of transposable elements and viral elements, which are silenced by heavy methylation.8 However, CpG islands, consisting of 500–2000 bps GC-rich sequences, are usually unmethylated in normal cells.4,5,9 In addition, around 70% of gene promoters include CpG islands.10 While most CpG islands are unmethylated, many oncogenes have methylated promoter CpG islands in normal cells, and the function of oncogenes were repressed.11 In addition, distal regulatory regions, like enhancers, lack CpG islands and are regions with low methylation.12
DNA methylation in the overall cancer cells
However, normal epigenetic processes are disrupted in multiple diseases, including inflammatory diseases, precancerous conditions, and cancers.13–16 Generally, overall genome-wide hypomethylation or hypermethylation with regional DNA hypermethylation or hypomethylation of CpG islands can be observed in these diseases.13,17,18 A loss of DNA methylation of transposable elements and repeat elements, accompanied by aberrant expression, results in the dysregulation of pathways.19 The hypermethylation of unmethylated promoter CpG islands is related to the inhibition of tumor suppressor genes and functional genes. However, the hypomethylation of methylated promoter CpG islands is related to the activation of oncogenes, while CpG-poor regions in CpG island shores or enhancers tend to undergo methylation in cancer cells.20,21
The change of DNA methylation in hematologic malignancies
Extensive studies have reported that DNA methylation patterns in regulatory regions play an important role in the development of cell proliferation and the function of leukemic stem cells (LSCs). The change of DNA methylation in hematologic malignancies is frequently observed in two aspects, the methylation level of methyltransferase genes themselves, and the mutations of methyltransferase genes or demethylase genes. Regarding the former aspect, studies have shown that the promoter of DNMT1 gene was unmethylated in all acute promyelocytic leukemia (APL) patients.22 Tirdad et al. found that patients with B and T-acute lymphoblastic leukemia (ALL) had unmethylated promoters in the DNMT1 gene, whereas the control group showed a relatively methylated promoter.23 Abnormal hypomethylation of the DNMT3A gene was also observed in 55.3% of acute myeloid leukemia (AML) patients, and was related to an adverse prognosis in cytogenetically normal (CN)-AML patients.24 The hypomethylation of promoters or intragenic regions of methyltransferase genes probably led to DNMT hyperactivation, causing cancer cell hypermethylation in leukemia. In terms of the latter aspect, recurrent DNMT3A mutations and TET2 mutations were found in leukemia. For example, patients with DNMT3A mutations and IDH1/2 mutations exhibited a mixed DNA hydroxy-/methylation profile compared with samples from healthy controls.25 AML patients with DNMT3A mutations displayed lower levels of DNA methylation as well as fewer concurrently hypermethylated genes.26 A loss of DNMT3A caused enhancer hypomethylation in FLT3-ITD-associated leukemias and was critical for the inhibition of leukemic transformation.27 Studies identified somatic mutations by exome sequencing in AML-M5, and found that DNMT3A mutants caused decreased enzymatic activity and abnormal affinity to histone H3.28 In chronic myelomonocytic leukemia (CMML) patients with TET2 mutations, AIM2 and SP140, the non-CpG island promoters, were hypermethylated.29 TET2 was found to be transcriptionally repressed or silenced in 71% and 17% of T-ALL patients, respectively.30 Apart from the frequently observed DNA methylation alterations, the change of other methylation mediators was also discovered. For example, the high expression of the methyltransferase DNMT3B was related to poor prognosis in AML patients.31 Myelodysplastic syndrome (MDS) and AML-mesenchymal stem cells (MSCs) displayed global hypomethylation and underexpression of the methyltransferase DNMT1 and the methyl-binding protein UHRF1.32 Changes in the methylation patterns of methyl-binding proteins, MBD2 and MeCP2, were observed in B-chronic lymphocytic leukemia (CLL).33 The recurrent change in DNA methylation profile in AML and ALL could predict patients’ prognosis and treatment efficacy.34
Enzymes of DNA methylation
Various enzymes participate in DNA methylation, and can be divided into three categories based on their function. The proteins that catalyze and cause a certain modification are referred to as writers. The proteins that eliminate an existing modification are referred to as erasers. The proteins that recognize an existing modification and recruit other macromolecular complexes to the template chain are referred to as readers.
Writers: DNA methyltransferases directly catalyze DNA methylation
The DNMT family can directly catalyze DNA methylation, including DNMT1, DNMT3A, and DNMT3B.35 It has been known for many years that DNMT3A and DNMT3B, so-called de novo DNMTs, can develop a new methylation pattern in unmodified DNA.36,37 The structure and function of DNMT3A and DNMT3B are extremely similar. On the other hand, DNMT1 primarily methylates hemimethylated DNA,38,39 and can repair DNA methylation.40 In addition, DNMT1 can maintain an established pattern of DNA methylation, copying the DNA methylation pattern when DNA replicates (Fig. 2).41–43 The three DNMTs are necessary for embryonic or neonatal development.37,44 DNMT3A is also critical for hematopoietic stem cell (HSC) differentiation.45 DNMT2 and DNMT3L have no catalytic function. DNMT2 functions as an RNA methyltransferase. DNMT3L is associated with DNMT3A and DNMT3B, and promotes their methyltransferase activity.46–48
Fig. 2.
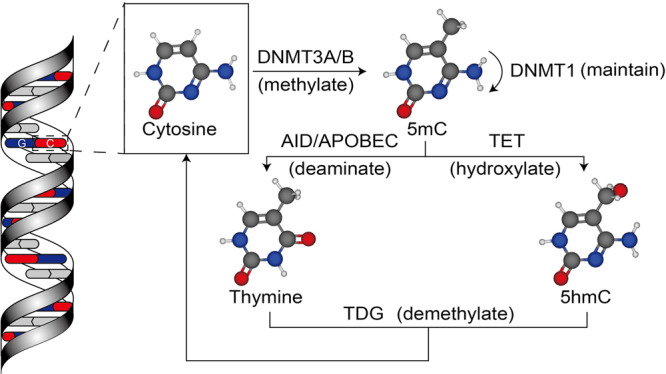
The methylation and demethylation pathways. DNMT3A and DNMT3B can methylate the cytosine (5mC) and set up a new methylation pattern to unmodified DNA. DNMT1 maintains an established pattern of DNA methylation during DNA replication. 5mC can be chemically modified at two sites: the amine group and the methyl group. The amine group of 5mC can be deaminated by AID/APOBEC to thymine (Thy). The methyl group of 5mC can be added a hydroxyl group by TET enzymes to generate 5-hydroxymethyl-cytosine (5hmC). Eventually, the products, Thy and 5hmU, can be recognized and removed by TDG
DNMT1 maintains the methylation
DNMT1 maintains an established pattern of DNA methylation. Upregulated DNMT1 could induce abnormal regional hypermethylation, and cause the pathogenesis of leukemia. The phenomena of DNMT1 alteration have been frequently observed in leukemia and lymphoma. Studies have shown that DNMT1 is overexpressed in APL,22 AML,49,50 ALL,50 MDS, and chronic myelogenous leukemia (CML) patients.49,51 The methylation of CpG islands in promoter regions is often observed in lymphomas. DNMT1 was overexpressed in Burkitt’s lymphoma (BL),52 diffuse large B-cell lymphoma (DLBCL),53 primary gastrointestinal diffuse large B-cell lymphoma (PGI-DLBCL)54 and other lymphomas.55,56 DNMT1 overexpression was identified as an independent risk factor in PGI-DLBCL.54 Furthermore, DNMT1 overexpression was associated with advanced clinical stages and resistance to treatment in DLBCL.53 DNMT1 could predict survival and Ki-67 expression in DLBCL patients received with the R-CHOP regimen.57 In addition, the expression of DNMT1 in U266 myeloma cells was higher than that in normal control cells.58
The function of DNMT1 in hematological malignancy progression has been reported in different pathways. In leukemia, conditional knockout of DNMT1 inhibited leukemia development, and DNMT1 haploinsufficiency delayed the progression of leukemogenesis and impaired LSC self-renewal without altering normal hematopoiesis.59 In AML, Wang et al. discovered that the expression level of DNMT1 in AML patients was decreased compared with that in healthy controls, and was negatively regulated by miR-148a in AML cell lines.60 In ALL, recent studies demonstrated that the level of exosomal DNMT1 mRNA transcripts was elevated in ALL patients, which might reprogram leukemia progression.61 In CML, DNMT1 was upregulated by BCR-ABLp210, and promoted tumor stem cell priming,62 and silencing the DNMT1 gene could inhibit the proliferation and promote the apoptosis of CML K562 cells.63 Regarding lymphoma, DNMT1 played a critical role in the maintenance of MYC-induced T-cell lymphomas and BL and contributed to abnormal methylation.64,65 In addition, DNMT1 was correlated with cell cycle and DNA replication gene sets in DLBCL.66 In MM, DNMT1 also promoted the methylation of SOCS-167 and TJP168 in myeloma cells, thereby decreasing their expression and regulating MM development.
DNMT3A catalyzes de novo methylation
Among the different DNMTs, the DNMT3A has been investigated and reported the most frequently in hematologic malignancies, and its change is presented below in the order of leukemia, lymphoma, and MM, respectively.
In leukemia, mutations of DNMT3A, especially in R882H, are one of the most frequent recurrent genetic changes in AML. Mutations of DNMT3A also occur in clonal hematopoiesis, MDS, ALL, CMML, mixed phenotype acute leukemia (MPAL), and pediatric AML patients.69–80 The prevalence of DNMT3A mutations was 7.4% in adult Thai AML patients,81 25% in US patients,82 19.7% in Korean patients,83 4.0–13.9% in Chinese patients,84,85 6% in Brazilian patients,86 20.9% in German patients87 and 17.9% in Egyptian AML patients.88 The majority of DNMT3A mutations have been reported to be related to increased relapse89 and poor survival in AML,82,83,90–100 CMML,101 ALL,71,75,102,103 Chinese pediatric AML,70 and Chinese pediatric ALL patients.71 In addition, DNMT3A1 and DNMT3A2V have been reported as the main variants in AML.104 In contrast to de novo AML, most mutations occurred in the methyltransferase domain other than arginine at position 882 in therapy-related and secondary AML.105
Furthermore, DNMT3A mutations were associated with several clinical characteristics in leukemia. Studies showed that DNMT3A mutation was not only related to the intermediate-risk cytogenetic group in de novo AML90,106,107 but also associated with age; the white blood cell (WBC) count, platelet (PLT) count, and blast percentage in peripheral blood; M4/M5 immunophenotype; FLT3 mutation; NPM1 mutation; IDH1/2 mutation; and CEBPA mutation in AML.87,92,97,98,107–112 In T-ALL, DNMT3A mutation was also associated with increased age, high WBC, high BM blast cell percentage, and extramedullary disease.103 Regarding cell of origin, the DNMT3A R882H mutation occurred more frequently in the T-ALL subtype than in the B-ALL subtype.71 DNMT3A mutations also showed a marked predilection for T-lineage differentiation in MPAL.93 Besides the genetic mutation of DNMT3A, overexpression of the DNMT3A protein was also reported in AML and the acute phase of CML.49,113 The expression levels in AML patients were higher than ALL patients or healthy controls,65,114 and DNMT3A expression acted as a potential prognostic biomarker.113
In addition, DNMT3A mutations, together with other mutations, were related to the prognosis of leukemia patients. For example, FLT3 and/or NPM1 mutations contributed to survival differences in DNMT3A-mutant patients.81,115 However, the IDH1/2 gene had little effect on patients’ survival with a DNMT3A mutation.116 Dose escalation of anthracycline in the induction regimen was correlated with improved survival in AML patients with DNMT3A mutations.117 Allogeneic hematopoietic stem cell transplantation (allo-HSCT) could increase the survival of CN-AML patients with DNMT3A mutations.118 In patients with complete response (CR) after complete donor chimerism allo-HSCT, no DNMT3A R882H mutation was found.119 DNMT3A mutant could always be detected during remission and did not predict prognosis in AML patients.119–122 While, DNMT3A R882/FLT3-ITD had poor prognosis in AML patients after allo-HSCT.123–125 DNMT3A variants were also associated with the progression of CML after tyrosine kinase inhibitor (TKI) therapy.126
The relationship between the type of DNMT3A mutation and clinical outcomes remained controversial in leukemia patients. One study reported that outcomes of patients with R882 and non-R882 missense mutations were similar, while patients with truncation mutations had comparable outcomes to those of patients with wild-type DNMT3A.99 However, in another study, patients with the R882 mutation and those with non-R882 mutations showed different clinical outcomes; patients with the R882 mutation had unfavorable relapse-free survival (RFS), while patients with non-R882 mutations had favorable overall survival (OS).87 In addition, the DNMT3AR882 mutations were associated with adverse prognoses in older patients, while non-R882 mutations were related to worse prognoses in younger patients.109 Mutations in DNMT3A exon 23 have also been reported to independently predict an unfavorable prognosis in older AML patients.127
Though less frequently, DNMT3A mutations were also found in lymphomas. DNMT3A mutations occurred in 11% of T-cell lymphomas,128 26.6–39% of peripheral T-cell lymphomas (PTCLs),129–131 26.1–34% of angioimmunoblastic T-cell lymphomas (AITLs),129,131–133 and were also found in NK/T-cell lymphoma/leukemia,134 breast implant-associated anaplastic large cell lymphoma (ALCL) and BL.135 However, DNMT3A mutations were infrequent in primary cutaneous CD4+ small/medium T-cell lymphoproliferative disorder (PCSMLPD),136 monomorphic epitheliotropic intestinal T-cell lymphoma (MEITL) and enteropathy-associated T-cell lymphoma (EATL).137 DNMT3A mutations were more commonly seen in patients of African ancestry compared than in those of European ancestry.138 DNMT3A mutations or expression were related to prognosis in lymphoma patients. Study findings suggested that high expression of DNMT3A was significantly related to worse OS and progression-free survival (PFS) in patients with PGI-DLBCL patients treated with R-CHOP regimen.54 DNMT3A mutation was found to be correlated with shorter PFS in AITL patients.133 Since DNMT3A mutations were significantly decreased after therapy,139 they could serve as sensitive indicators in circulating tumor DNA (ctDNA) and provide a noninvasive method of monitoring minimal residual disease in AITL.140
The evidence for the effects of DNMT3A on MM is contradictory. Study findings indicated that DNMT3A mutations were present in newly diagnosed MM patients and those treated by autologous stem cell transplantation (ASCT).141,142 Furthermore, DNMT3A overexpression was demonstrated in the cells of MM patients.58,143 Different from lymphoma, studies showed that DNMT3A underexpression was associated with worse OS in MM patients.144
DNMT3A potentiated hematologic oncogenesis in many ways. First, the DNMT3A R882 mutation stimulated the mTOR pathway145 and reactivated the leukemic transcription factor MEIS1,146 which initiated AML. DNMT3A R882 mutations also enhanced abnormal stem cell gene expression, which promoted leukemia progression.147 Studies also found that DNMT3A R882 mutations reduced AML cell apoptosis by augmenting PRDX2,148 and induced CML by disturbing DNA methylation.149 In T-cell lymphomas, the DNMT3A mutation was found in both programmed cell death 1 (PD-1) + T cells and supportive cells, indicating its role in both tumor cells and microenvironment.150,151 Second, DNMT3A was found to be a haploinsufficient tumor suppressor in various hematologic malignancies. The haploinsufficiency of DNMT3A could transform FLT3-ITD myeloproliferative neoplasm (MPN) into AML,152 and cooperate with oncogenic KRAS to promote the development of T-ALL.153 The haploinsufficient tumor suppressor effect of DNMT3A was also reported in CLL,154 CD8+ PTCL,155, and AITL.156–159 Third, DNMT3A mutations were closely related to treatment response. Studies showed that DNMT3A mutations induced anthracycline resistance in AML by impairing nucleosome remodeling,160 and evaded chemotherapy and infiltrated the central nervous system in a patient with AML.161 DNMT3AR882H-dependent AML cells were sensitive to hypomethylating agents, such as azacytidine (AZA)162 and decitabine.163 In addition, DOT1L could be a therapeutic target,164 and resistin may serve as an ancillary drug for AML patients with DNMT3A mutation.165
DNMT3B also catalyzes de novo methylation
DNMT3B caused the generation of abnormal methylation during the development of hematologic malignancies. In leukemia, studies showed that DNMT3B was overexpressed in MDS, AML and CML patients.49,51,65 The DNMT3A expression level in AML patients was higher compared with ALL patients.65 However, another study found that DNMT3B expression was decreased in AML113 and B-CLL.33 Exploring the polymorphism C46359T in the DNMT3B promoter, Li et al. found different distributions of genotypes in different races and that the CT heterozygote was related to the pathogenesis of AL.166 Furthermore, DNMT3B mutations, polymorphisms, and expression were related to AML prognosis. DNMT3B overexpression was related to adverse prognosis in older CN-AML patients.167 The G allele of rs1569686 in DNMT3B represented poor outcomes for AML, while the C allele of rs2424908 was associated with favorable outcomes.168,169
In lymphoma, DNMT3B expression was also increased in BL,52 DLBCL53, and plasmablastic lymphoma (PBL),170 and it was identified as an adverse prognostic factor.53 DNMT3B overexpression was associated with advanced clinical stages and resistance to treatment in DLBCL.53 However, studies also found that the expression of DNMT3B was lower in PGI-DLBCL patients.54 In MM, the expression of DNMT3B was increased in in U266 and RPMI8226 myeloma cells.58,171
DNMT3B mainly affected the progression of hematologic malignancies. Some studies found that the loss of DNMT3B accelerated MLL-AF9 leukemia progression.172 Others showed that high expression of DNMT3B contributed to abnormal DNA methylation and MYC-driven tumor development in T-ALL and BL.65
Readers: the methyl-binding proteins recognize DNA methylation and mediate subsequent reactions
DNA methylation can directly inhibit the binding of transcription factor and repress gene transcription when it occurred at the regulatory region of a gene.173 In addition, the methylated site recruits methyl-binding proteins (MBPs), the readers, and then attracts the members of the chromatin remodeling complex, to activate or inhibit transcription.174,175 MBPs mainly include three separate families of proteins: “methyl-CpG-binding domain (MBD)-containing proteins”, “methyl-CpG binding zinc fingers”, and “Set and RING-associated (SRA) domain-containing proteins”.176
MBD-containing proteins
This is the first identified MBP family, of which all members have the conserved MBD domains. MeCP2 was the first MBP to be identified.177 MeCP2 had an MBD domain, which comprises 70–85 amino acids, and recognizes and binds to methylated sites.178 Then, this MBD domain could recognize other proteins with methyl-binding potentials.179 According to the presence of domains excepted MBD, there were three groups of this family: (1) MeCP2-MBDs: MeCP2, MBD1, MBD2, MBD3, MBD4, MBD5, and MBD6; (2) histone methyltransferase-MBDs: SETDB1 and SETDB2; and (3) histone acetyltransferase-MBDs: BAZ2A and BAZ2B. However, the classification above is according to their structural characteristics other than methyl-binding abilities, thus not all proteins can bind to methylated DNA. The MBD domain of MeCP2,180 MBD1,181 MBD2179, and MBD4182 can bind to methylated CpG. However, MBD3,183 MBD5, MBD6,184 BAZ2A, and BAZ2B185 cannot bind to methylated DNA. Moreover, both SETDB1 and SETDB2 play a role as protein lysine methyltransferase activity.186,187
The expression of MBD2 and MeCP2 was found to be increased in CML.188 Studies found that deregulation of the epigenetic repertoire, including MBD2 and MeCP2 in B-CLL.33 The homozygous CC genotype in MBD2 (rs603097, -2176C>T) was associated with favorable outcomes of non-Hodgkin lymphoma (NHL).189 MeCP2 could epigenetically regulate SOCS5 expression in T-ALL and the SPAN-XB core promoter sequence in MM.190,191 The MeCP2/SIN3a deacetylating complex could epigenetically silence the proapoptotic gene BIM in ALCL.192 The knockdown of MBD2 inhibited apoptosis induced by B1 and overcame the resistance caused by Bcl-2 in HL60 cells.193 MBD4 was found to be overexpressed in imatinib-resistant K562 cells, and the knockdown of MBD4 expression decreased cell survival after treatment with hydrogen peroxide and doxorubicin.194
Methyl-CpG binding zinc fingers
The second family of MBPs is the “methyl-CpG binding zinc fingers” family, the members of which, including Kaiso, zinc-finger, and BTB domain containing 4 (ZBTB4), and ZBTB38. They can identify methylated and unmethylated DNA by zinc-finger motifs.195 The first member of the family was Kaiso.196 ZBTB4 and ZBTB38 are homologs of Kaiso proteins.197 Kaiso is a transcription factor and the binding partner of p120-catenin in the cytoplasm.198 It is encoded by the ZBTB33 gene which is located on the X chromosome.199 Kaiso binds to a couple of methylated CpG dinucleotides.196 ZTBT4 and ZBTB38 can bind to a single methylated CpG.197
The knockdown of Kaiso increased cell proliferation and blocked granulocytic differentiation in CML blast crisis.200 Kaiso protected human umbilical vein endothelial cells against apoptosis through the overexpression of BCL2 and decreased expression of BAX and BIK.201 HBV preferential target genes, such as ZBTB38, showed significantly altered expression levels in NHL.202
SRA domain-containing proteins
The third family of MBPs is the “SRA domain-containing proteins” family, including UHRF1 and UHRF2. They contain the SRA domain, which can bind to methylated DNA. The SRA domain usually bind to the hemimethylated regions,203,204 while the MBD/MeCP2 domains preferentially bind to the paired methylated DNA. UHRF1, also known as ICBP90 (human) or Np95 (mouse), binds to hemimethylated DNA and then attracts DNMT1 to methylate the sequence during replication.205,206 UHRF2, also known as NIRF or Np97, is involved in the regulation of the cell cycle.207
The UHRF1 level was found to be increased in aneuploid AML.208 The UHRF1 promoter was found to be hypomethylated in leukemia patients.209 UHRF1 was found to regulate the transcriptional repressor HBP1 through MIF in T-ALL, and UHRF1-knockdown mice lived longer.210 Furthermore, difluoromethylornithine (DFMO) and thymoquinone were found to downregulate UHRF1 in Jurkat cells.211,212 UHRF1 promoted ubiquitination-mediated degradation of the tumor-suppressive promyelocytic leukemia protein.213 Berberine was a novel drug for the treatment of MM via targeting UHRF1.202
Erasers: DNA demethylases remove the existing methylation
DNA demethylation is a dynamic process including passive or active pathways. Passive DNA demethylation happened in dividing cells, where newly incorporated cytosine remains unmethylated by DNMT1 inhibition or dysfunction. Active DNA demethylation can happen in both dividing and nondividing cells, which requires enzymes to remove 5mC to develop a permissive state for subsequent gene expression.214–217 Enzymes in active DNA demethylation involve the ten-eleven translocation (TET) methylcytosine dioxygenases, activation-induced cytidine deaminase/apolipoprotein B mRNA-editing enzyme complex (AID/APOBEC), and thymine DNA glycosylase (TDG).218–220 5mC can be modified at two sites, the amine group and the methyl group. The AID/APOBEC can deaminate an amine group of 5mC to thymine (Thy).221 The TET enzymes can add a hydroxyl group to the methyl group of 5mC and generate 5-hydroxymethyl cytosine (5hmC). IDH enzymes converted isocitrate to α-ketoglutarate, which is critical for TET catalytic function. Eventually, the products, Thy and 5hmC, can be recognized and removed by TDG (Fig. 2).221
TET1: oxidation of 5mC to 5-hydroxymethyl cytosine (5hmC)
The disruption of normal DNA demethylation was identified to be associated with oncogenesis. Both genetic mutations and abnormal protein expression of the TET family were reported in hematologic malignancies. TET1 was found to act as oncogene in MLL-rearranged leukemia.222 TET1 expression was decreased, whereas the expression of TET2 and TET3 was increased in AML.223,224 In refractory AML, TET1 expression was increased compared to that in treatment-responsive patients.225 Moreover, TET1, TET2, and TET3 overexpression was found to be related to poor prognosis in AML.223,224,226 The expression of TET1 and TET3 decreased in CLL, and high TET2 expression was related to longer survival in CLL.227 In addition, TET mutations could also be observed in hematological malignancies. The most frequently mutated gene was TET2, with TET2 mutations happening in 32% (10/31) of patients with human T-cell lymphotropic virus type I (HTLV-I)-induced acute adult T-cell leukemia (ATL).228 TET2 mutations also occurred in AML and T-ALL patients.229,230 In the experimental study, TET1 promoted the growth of T-ALL and could be antagonized via PARP inhibition.231
TET2: oxidation of 5mC to 5-hydroxymethyl cytosine (5hmC)
TET2 mutations were found to happen in patients with hematological malignancies, and affected DNA hypermethylation.232 In leukemia, studies found that 33% of patients had low levels of TET2-specific differentially methylated CpG (DMC) methylation, and these patients had longer OS.233 TET2 mutations occurred in 6–23% of CN-AMLs,234–236 1.4% of childhood AMLs,76 7.6% of younger adult AMLs,237 and 11.8–27.4% of AMLs82,235,238–245 and were unfavorable prognostic factors.82,236,238,239,241,246 TET2 mutations occurred in 14% of adult T-cell leukemia/lymphomas (ATLLs)247 and 32% of ATLs,228 and lower TET2 expression was associated with adult T-cell leukemia aggressiveness.248 Furthermore, TET2 mutations were also observed in 28% of chronic natural killer large granular lymphocyte (NK-LGL) leukemia cases.249 In addition, TET2 mutations were also observed in advanced phase CML,250,251 and the presence of TET2 SNP rs3442524 suggested disease progression.251 However, some studies reported TET2 mutation did not affect prognosis. TET2 mutation alone was found to have no prognostic impact on event-free survival (EFS) or OS in one study.245 Similarly, TET2 mutations were detected in 19.8–23.3% of secondary AMLs, and did not influence OS.252,253 In addition, TET2 mutations occurred in 32–65% of CMML and were favorable prognostic factors,29,254–260 but were associated with poor survival in the presence of ASXL1 mutations.261 While in another study, TET2 mutations were correlated with poor survival.262 There are different types of TET2 mutations, and patients have different clinical outcomes. Complete deletion of the TET_JBP domain (ΔJBP) of TET2 in AML patients had a lower CR rate and shorter EFS and OS.263 TET2 I1762V acted as a favorable prognostic factor in AML.264 TET2 SNP rs2454206 correlated with improved survival in childhood CN-AML.265 TET2 exon 2 splicing status might improve survival in CN-AML patients.266 TET2 mutations were reported to be associated with several clinical characteristics. TET2 was associated with a normal karyotype, high WBC count, low PLT count and high age.238,255,267 Arginase 1 overexpression was related to DNMT3A and TET2 mutations in lower-grade MDS and CMML.268 Besides the genetic alteration of TET2, the expression of TET2 protein was also different between patients with hematological malignancies and healthy people. For example, the expression of TET2 was showed to be increased in B-CLL,269 but decreased in AML270,271 and childhood ALL,272,273 and was related to poor survival.270,271
TET2 is a well-established tumor suppressor in the context of lymphomas. Studies have detected TET2 mutations in mantle cell lymphoma (MCL),274 DLBCL,275, and non-anaplastic PTCLs.276 TET2 mutations occurred in 18–92% of AITLs,129,132,247,277 5.7–12% of DLBCLs,247,278 14% of ATLLs,247 11% of PBLs,279 38–64% of PTCLs,280,281 75% of PTCLs with T-follicular helper phenotype (PTCL-TFH),129 85% of mucosa-associated lymphoid tissue (MALT) lymphomas,282 and 18% of T-cell lymphomas.128 TET2 mutations were correlated with poor survival in AITL and PTCL, not otherwise specified (PTCL-NOS) patients.281,283 Germline TET2 loss of function was found to cause childhood immunodeficiency and lymphomagenesis.284 Besides the genetic alterations, TET expression was found to increase in NHL.285 Moreover, TET2 mutations were correlated with advanced-stage disease, thrombocytopenia, and high International Prognostic Index (IPI) scores in AITL and PTCL-NOS.281 TET2 mutations were related to positive B symptoms.280 In MM, TET2 mutations were also detected,141,142,286–288 and might be related to survival and resistance to therapy at relapse.287
The recovery of TET2 function blocked abnormal self-renewal and progression of leukemia.289 MEG3 promoter hypermethylation might result from decreased TET2 activity in AML.290 TET2 mutations affected DNA methylation in non-CpG island of enhancers and transcription factor-binding sites in CMML.291 Moreover, the expression of TET2 decreased, and promoter methylation of TET2 significantly increased the risk of ALL.292 Cooperation between KDM6B overexpression and TET2 deficiency was found to initiate the pathogenesis of CMML.293 RHOA mutations were correlated with a T-follicular helper (TFH) cell phenotype and TET2 and IDH2 mutations.294 AID and TET2 cooperation modulated FANCA expression through active demethylation in DLBCL.295 Reduced TET2 function led to T-cell lymphoma with TFH-like features in mice.296
TET3: oxidation of 5mC to 5-hydroxymethyl cytosine (5hmC)
TET3 mutations are infrequently observed in AML, but the expression of TET3 in HSCs and human peripheral blood T cells has been found to decrease with age.297,298 The expression of TET3 was found to be increased in AML, which was associated with poor OS and disease-free survival (DFS).223,224,227 TET3 mutation could be detected in newly diagnosed AML patients.299
AID/APOBEC: deamination of 5mC to thymine (Thy)
The AID/APOBEC family of proteins was a group of cytidine deaminases, and played roles in multiple physiological functions.300,301 The family consists of eleven members in humans: AID and APOBEC1, APOBEC2, seven APOBEC3 proteins (APOBEC3A, APOBEC3B, APOBEC3C, APOBEC3D, APOBEC3F, APOBEC3G, APOBEC3H) and APOBEC4.302–307 APOBEC1 is hardly expressed in CLL, while APOBEC3A and APOBEC3G are both present in CLL and normal B cells.308 In a patient with two relapses, the interplay of aberrant AID/APOBEC activity was observed.309 APOBEC overactivity was also observed in primary plasma cell leukemia (pPCL).310
Furthermore, aberrant AID/APOBEC activity-associated mutations were enriched in early clonal hematopoiesis-associated mutations in AITL/PTCL-NOS.311 AID/APOBEC overactivity was also observed in HBV-associated DLBCL.312
APOBEC-associated mutations were enriched in patients with disease progression and were associated with a shorter time to progression in smoldering MM (SMM).313–315 APOBEC mutations were detected in MM316–319 and might be related to high risk, a shorter progression time, and therapy resistance.287,317,320 The APOBEC mutation was observed in 3.8% of myeloma cases and was associated with the translocation-mediated deregulation of MAF and MAFB, which was associated with poor prognosis.319 Dysregulated APOBEC3G was found to cause DNA damage and promoted genomic instability in MM.321 APOBEC3B overexpression was found to constitutively generate DNA substitutions and deletions in myeloma cells.322
TDG: demethylation of Thy and 5hmC to cytosine
TDG was a member of the uracil DNA glycosylase (UDG) superfamily. TDG is required for embryonic development.323 DNA hypermethylation of TDG in MM cell lines was found to lead to low expression of genes, and inhibit DNA repair activity during hydrogen peroxide-induced DNA damage.324 Moreover, SUMO-1 modification and colocalization with the promyelocytic leukemia protein were required in the noncovalent SUMO-1 binding activity of TDG.325
IDH1/2: conversion of isocitric acid to α-ketoglutaric acid
Isocitrate dehydrogenase (IDH) converted isocitrate to α-ketoglutarate (α-KG). The IDH family included IDH1 and IDH2, and their mutant forms transformed α-KG into 2-hydroxyglutarate (2-HG). 2-HG inhibited DNA demethylases and histone demethylases, thus increasing methylation of both DNA and histones.326 There are three conserved mutational hotspots in the IDH enzymes. The mutational hotspot of IDH1 is R132, while the mutational hotspot of IDH2 were R140 and R172.
IDH1 mutations occurred in 6–9% of AML patients, especially in patients with normal karyotype AML (8–16%).327–333 IDH1 mutations often co-occurred with normal karyotypes and NPM1 mutations,328–331,333 and were associated with wild-type CEBPA and no FLT3 mutation.331 Published results on the prognostic effects of IDH1 mutations failed to reach an agreement. Although some studies have shown that IDH mutations have no prognostic effect on OS in IDH-mutated (IDH1 and IDH2) patients or in overall patients,328–331 IDH1 mutations have a poor prognosis impact on low-risk or intermediate -risk subgroups of patients with normal karyotype AML.328,331,333 In AML patients under 60 years old in the low-risk group, 5-year DFS (42% vs. 59%) and OS rates (50% vs. 63%) of IDH-mutated patients were significantly lower than those in IDH wild-type patients. In low-risk AML patients, IDH mutations (combined IDH1 and IDH2) were correlated with lower 5-year RFS and OS rates, respectively.333 The same phenomenon was found in IDH1 or IDH2 mutations alone, although the number of patients in each subgroup was small and only the RFS analysis was statistically different.333 IDH1 mutations were also correlated with poor EFS and OS in patients with intermediate-risk normal karyotype AML.328 IDH2 mutations have been observed in 8–12% of AML patients,327–329,333,334 with a higher mutation frequency of 19% in the normal karyotype.331 In almost all cases, IDH2 and IDH1 mutations were mutually exclusive.328,329,331 IDH2 mutation could occur at R172 or R140, while R140 mutation happened more frequently.331,333,334 Interestingly, IDH2R172 mutations appeared to repel NPM1 mutations and FLT3-ITD.331,333,334 The prognostic effects of IDH2 mutations were also inconsistent. Some studies reported no prognostic value of IDH2 mutations,328,329,333 while others reported a good prognosis for IDH2-mutated patients.327,334 In one study, IDH2 mutations were associated with poor prognosis in a subpopulation of AML patients with normal karyotype and other low-risk factors.333 However, IDH2R140 mutations were correlated with increased survival in all patients as well as in a subgroup of patients with the low-risk group.327 In the latter subgroup, patients with IDH1 or IDH2 mutations had a significant increase in OS at 3 years compared to patients with NPM1 mutations without FLT3-ITD and patients without IDH1 or IDH2 mutations. These results suggested that in normal karyotype AML patients without FLT3-ITD, NPM1 mutations conferred survival benefits only when IDH mutations were present simultaneously.327 The conflicting results of these studies required further investigation.
Inhibition of IDH1 mutant promoted cycling of LSCs.335 A genomic analysis was performed in R/R AML patients with IDH mutations who received ivosidenib (IDH1 inhibitor) treatment, and the results suggested that primary resistance to ivosidenib was correlated with RTK pathway mutations.336,337 IDH2 mutation cooperated with a NUP98-HOXD13 fusion caused early immature thymocyte precursor ALL.338
IDH2R172 mutation defined a unique subgroup of patients with AITL.158 Patients with IDH2 mutations had increased H3K27me3 and DNA hypermethylation of gene promoters. In a retrospective multicenter study, IDH2R172 mutation was found in 19–45% AITL patients and was correlated with poor survival,339 and different pathologic and clinical features.132,133,340–342 When using plasma cell-free DNA as liquid biopsy for the diagnosis of AITL, IDH2R172 hotspot mutations were detected in 15% of patients.343 Studies found that IDH2 mutation was confined in the malignant T cell of AITL, and that 2-HG was elevated in tumor tissue and serum of patients.344
IDH2 was observed overexpressed in CD138+ cells from MM, and it promoted progression and poor prognosis of MM by regulating m6A RNA methylation.345 IDH2 inhibition increased efficacy of proteasome inhibitor in MM, MCL, and BL cells.346
Histone modifications mainly include acetylation, methylation, and phosphorylation
Histone modifications in normal hematopoiesis
Posttranslational modifications of amino acids, especially lysines, at the N-terminal tail of the histone core change the chromatin configuration, ranging from the condensed repressive status to the open active status. There are at least sixteen types of histone modifications, including acetylation, methylation, phosphorylation, ubiquitylation, sumoylation, and ADP ribosylation. The histone modifications most essential for hematopoiesis and hematologic malignancy oncogenesis are acetylation, methylation, and phosphorylation.
HSCs are commonly recognized to arise from the endothelial-to-hematopoietic transition, which occurs at the early arterial endothelial cell, hemogenic endothelial cell, pre-HSC, and long-term HSC stages. Based on the results of low-input itChIP-seq and Hi-C assays, the stimulative histone modifications, H3K27-ac and H3K4me1, which regulate enhancer activation, already exist in the initiation stage of early arterial endothelial cells and hemogenic endothelial cells. During the differentiation of HSCs, the stimulative H3K27-ac gradually increases while the repressive H3K27me3 decreases, leading to the steady expression of HSC genes.347 In lineage differentiation, histone modification also plays an important role. A crucial lineage-mediating protein, BAP1, regulates the balance between lymphopoiesis and myelopoiesis by altering the histone modifications of the promoters of pro-hematopoietic factors and myelopoiesis-promoting factors. Upon BAP1 deficiency, H2AK119-ub1 and H3K27me3 are strengthened on the promoter of the pro-hematopoietic factor Scf, leading to its downregulation. In addition, H3K4-me3 and H3K27-ac are strengthened on the promoter of the myelopoiesis-promoting factor Csf3, leading to its upregulation and steering the differentiation to myeloid lineage, which causes CMML-like disease.348,349 The dynamic balance among the writers, readers, and erasers of histone modifications is essential to hematopoiesis and oncogenesis, which will be discussed below (Fig. 1).
The change of histone acetylation pattern in hematologic malignancies
Acetylation reduces the positive charge of lysines and in turn weakens the interaction between histones and negatively charged DNA, causing an open active euchromatin status, as evidenced by ChIP-seq, which reveals histone acetylation at promoters and enhancers of actively transcribed genes.350 Histone deacetylase (HDAC) erases the acetyl group from lysines in the N-terminus, which amplifies the positive charge of lysines and in turn strengthens the interaction between histones and the negatively charged DNA, leading to the condensed repressive heterochromation status.351 Histone lysine acetyltransferase (KAT, also named HAT) and HDAC counteract subtly to sustain the hematopoiesis balance. In normal erythropoiesis, erythroid-stimulating transcription factors, such as GATA-1, recruit KAT to the β-globin locus to increase the acetylation levels of H3 and H4, which in turn upregulates globin during erythropoiesis.352 When the expression of wild-type or chromosomal-fused transcription factors is abnormal due to epigenetic dysregulation, the hematopoiesis balance is disturbed and malignancies occur.353 For example, TAL1/SCL is a wild-type transcription factor steering differentiation to the erythroid and megakaryocytic lineage instead of the myeloid lineage. Upon association with the coactivating complex of p300 and PCAF, TAL1/SCL was found to be upregulated and propelled erythropoiesis leading to murine erythroleukemia. However, TAL1/SCL was found to be downregulated upon association with the corepressive complex of HDAC1 and mSin3A, impeding erythropoiesis.354 Regarding fusion proteins caused by chromosome translocation, PML-RARα, RUNXI-MTG8, and AML-ETO1 recruit abnormal histone-modifying enzymes to target genes, causing leukemogenesis.355 In lymphoma, various oncogenic mutations which were related with histone acetylation were also discovered.356
Writers: histone acetyltransferases directly catalyze histone acetylation
KATs are divided into two types. Type A KATs are mainly nuclear and consist of five families, the p300/CBP, MYST, GNAT, general transcription factor/orphan KAT, and SRC/NCoA families. Type B KATs, consisting of KAT1, are mainly cytoplasmic.357 Of the many KAT families, the p300/CBP family, and MYST family are the most essential KATs in hematologic malignancy oncogenesis, and are presented below, respectively.
The p300/CBP family includes p300, also named KAT3B or EP300, and CBP, also named KAT3A or CREBBP. CBP has been found to bind to viral oncoprotein E1A, which is associated with oncogenesis.358 CBP could also serve as a tumor suppressor, as various loss-of-function mutations have been found in lymphoma. For example, monoallelic mutations in CBP, which inactivated KAT and led to B-cell oncogenesis, were discovered in 40% of follicular lymphoma (FL) patients, up to 30% of DLBCL patients, and 26% of primary cutaneous DLBCL-leg type patients.359,360 In addition, these inactivating mutations were found to cooperate with BCL2, BCL6, and MYC to promote lymphoma development, promoting the aggressiveness of lymphoma and indicating its indispensable role as tumor suppressors.361 The chromosomal translocation of MLL-CBP, t(11;16)(q23;p13), was found to specifically drive the proliferation of granulocyte/macrophage progenitors (GMPs), leading to acute myelomonocytic leukemia (AMML) and CMML.362 Furthermore, P300 mutations were also found in 10% of FL and DLBCL patients,363 which disrupted the binding of transcription factors, such as CREB and c-Myb, and caused multilineage hematopoietic defects.364 In DLBCL, mutations of CBP/p300 could repress acetylation of H3K27, stimulate the Notch pathway, which in turn upregulated CCL2/CSF1, leading to M2 polarization of the tumor-associated macrophages and lymphoma progression.365
The MYST family includes MOZ, MORF, HBO1, TIP60, and HMOF. This family was frequently discovered to promote leukemogenesis by translocation, which consistently activated the downstream signaling pathway. For example, MOZ, also named KAT6A, was discovered to translocate with various genes. MOZ-CBP translocation, t(8;16)(p11;p13), led to consistent activation of KAT and was related to M4/M5 AML.366 MOZ-TIF2 translocation, induced by inv(8)(p11;q13), simulated aggressive leukemia by introducing stem cell activity to progenitor cells, leading to persistent self-renewal and leukemogenesis.367 MORF, also known as KAT6B, whose translocation with CBP induced by t(10;16)(q22;p13) caused a loss of monoacetylated H4K16, is a well-recognized indicator of AML transformation.352 HBO1, also known as KAT7, is the acetyltransferase of H3K14, and its loss leads to abnormal consumption and exhaustion of HSCs; therefore, HBO1 is indispensable for HSC maintenance and self-renewal.368
Readers: acetyl-lysine binding proteins recognize histone acetylation and mediate subsequent reactions
Readers of histone lysine acetylation include a highly conserved binding domain, the bromodomain. One of the most explored and targeted readers is the BET family, consisting of BRD2, BRD3, BRD4, and BRDt. However, few studies have revealed the contribution of BET family genetic alterations to the oncogenesis of hematologic malignancies. Most of the studies exploring its pathogenic influence have focused on solid tumors, for example, midline carcinoma. The translocation of the nuclear protein of testis (NUT) to BRD4, induced by t(15;19)(q14;p13.1), impeded the differentiation and promoted the proliferation of epithelial cells. On the one hand, the fusion protein stimulated p300 and amplified hyperacetylation, which recruited BRD4 and oncogenic transcription components. On the other hand, aberrant occupation of BRD4 inhibited c-fos transcription, which in turn inhibited epithelial differentiation.369 A similar oncogenic translocation to NUT was also discovered in BRD3.370,371 The evidence of histone acetylation readers contributing to hematologic malignancies are lacking and requires further investigation.
Erasers: histone deacetylases remove the existing acetylation of histones
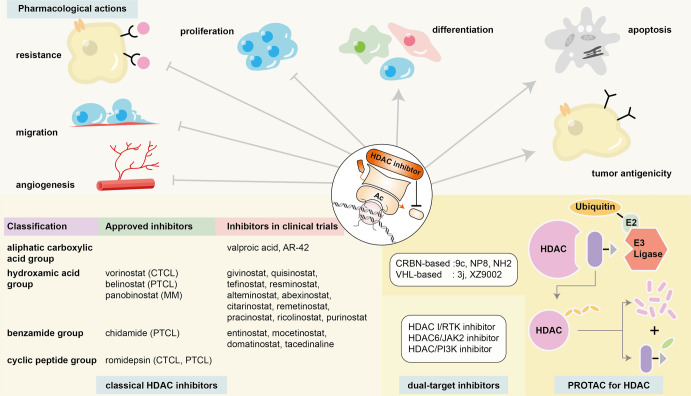
HDACs are categorized into four classes, including the zinc-dependent classic HDACs (class I, II, and IV) and the NAD+-dependent nonclassic HDACs (class III). The three categories of classic HDACs can be divided based on sequence homology to yeast deacetylases. Class I HDACs, including HDAC1, HDAC2, HDAC3, and HDAC8, are homologous to reduced potassium dependency-3 (Rpd3) and localize mainly in the nucleus; these HDACs form multiprotein complexes and regulate transcription and proliferation. Class II HDACs are homologous to histone deacetylase-1 (Hda1). Class IIa HDACs, including HDAC4, HDAC5, HDAC7, and HDAC9, move between the nucleus and cytoplasm. Class IIa HDACs have only one catalytic domain and have weaker deacetylation capability than the nucleus-localizing class IIb HDACs, including HDAC6 and HDAC10, which contain two catalytic domains. Class IV HDACs share a homologous sequence to both Rpd3 and Hda1 and reside in the nucleus. Class IV HDACs include only HDAC11, which is the smallest among the various HDACs. The nonclassic class III HDACs consist of SIRT1 to 7.
The change of HDAC manifested itself mainly through altered protein expression, instead of genetic mutations, in hematologic malignancies. Impaired expression of H3K4ac1 and H3K4ac3 was commonly observed in lymphoma.372 The overexpression of HDAC1 and HDAC2 was discovered in DLBCL compared to their expression in normal lymphoid tissues.373 The inhibition of HDAC1 and HDAC2 was found to impede Eμ-myc B-cell lymphoma development via apoptosis stimulation and proliferation repression, indicating the oncogenic role of HDAC1 and HDAC2.374 HDAC1 expression was found to be significantly related to worse prognosis in DLBCL patients and PTCL-NOS patients.375 HDAC3 overexpression was pathogenic when CBP mutations were present. In normal differentiation, the BCL6-SMRT-HDAC3 complex counteracts CBP to regulate the acetylation balance of H3K27. However, upon CBP malfunction due to mutations, the BCL6-SMRT-HDAC3 complex overacts in enhancer deacetylation of the B-cell signaling pathway without opponents, leading to B-cell lymphomagenesis.376 In contrast, HDAC4 was a tumor suppressor. It serves as a corepressor of BCL6 and represses oncogene expression upon recruitment, impeding B-cell lymphomagenesis.377 DLBCL patients with high HDAC2 expression and low HDAC4 expression were found to display significantly worse prognosis than those with low HDAC2 expression and high HDAC4 expression, also indicating the pathogenic role of HDAC2 and the protective role of HDAC4 in DLBCL.378 HDAC6 expression varies in different malignancies, and its role is controversial. In DLBCL, HDAC6 showed a tendency toward low or negative expression in most patients, and the minority of DLBCL patients who showed strong HDAC6 expression had a significantly better prognosis than the others. However, in PTCL, HDAC6 showed a tendency toward to be overexpressed, which was significantly related to worse prognosis.373 Apart from PTCL, HDAC6 was also found to play a pathogenic role in MCL, and its inhibition exerted an antitumor influence.379 HDAC6 could also regulate immunogenicity and downregulate PD-1/PD-L1 in cell subpopulations of CLL and MM.380,381 HDAC6 inhibition could decrease Treg cells and myeloid-derived suppressor cells (MDSCs) and increase the cytotoxicity of T cells, strengthening immune response of the host to counteract myeloma cells.381 In a CLL Eµ-TCL1 mice model, HDAC6 inhibition could reduce the immunosuppression caused by tumor T cells, and enhance immunomodulation of the supportive B cells,380 indicating the potential combined treatment of HDAC inhibitors and immune treatment, which is presented in the Targeted Therapy section of this review. HDAC7 was crucial to committing pro-B cells to pre-B cells by interacting with MEF2C. Thus, HDAC7 downregulation was found in pro-B-ALL and BL, while its upregulation was found in pre-B-ALL and B-ALL t(8;14).382,383 HDAC7 was also shown to be essential to the survival and TCR engagement of thymic T cells during differentiation, whose overexpression was discovered in T-ALL patients.384
The class III HDACs are the SIRT family, which has been found less associated with HDAC function. The SIRT family showed a tendency to regulate HSC aging through various pathways. For example, SIRT1 preserved aged HSCs by stimulating FOXP3 and inhibiting the mTOR pathway. Moreover, SIRT1 rendered HSCs vulnerable to stepwise mutation development in CML, while SIRT1 inhibition strengthened DNA repair and improved the sensitivity of leukemic stem cells (LSCs) to imatinib.385 The evidence of the SIRT family affecting hematologic malignancies is relatively limited. Further investigation is needed to better reveal their relationship.
The change of histone methylation pattern in hematologic malignancies
The methylation of histones mainly occurs on lysines and arginines. Lysine methylation manifests itself as mono-, di-, and trimethylation. Arginine methylation can be symmetrical or asymmetrical. Unlike acetylation, which alters chromatin status and regulates transcription by changing the charge of the lysine, methylation does not change the charge of lysine or arginine. Whether lysine methylation stimulates or inhibits transcription depends on the position and features of methylation. For example, methylation on H3K4, H3K36, and H3K79 is more often related to active euchromatin status, while methylation on H3K9, H3K27, and H4K20 usually causes repressive heterochromatin.386 Moreover, H3K9me1 is commonly seen in actively transcribed genes, while H3K9me3 is frequently discovered in repressed genes.387 Arginine methylation is commonly related to gene activation.388
Writers: histone methyltransferases directly catalyze methylation of the histone
Histone methyltransferases are mainly divided into lysine methyltransferases (KMTs) and protein arginine methyltransferases (PRMTs). KMTs mainly consist of KMT2, KMT6, NSD, and KMT1 families. Members of the KMT2 family, including KMT2A (also known as MLL1), KMT2B (also known as MLL2), KMT2C (also known as MLL3), KMT2D (also known as MLL4), KMT2F (also known as SETD1A), and KMT2G (also known as SETD1B), catalyze the methylation of H3K4. MLL is essential for proliferation and lineage differentiation during hematopoiesis. MLL can not only activate genes by methylating H3K4 and recruiting KAT but also inhibit genes by recruiting HDACs and polycomb group (PcG) proteins.389 Recurrent mutations and fusion proteins related to MLL have been reported in various hematologic malignancies. On the one hand, KMT2A mutations have been reported in AML (2.5%), T-ALL (5.6%), and high-grade B-cell lymphoma (14%).390 Recurrent KMT2B mutations were discovered in more than 90% of FLs.391 KMT2C mutations were revealed in 45.5% of posttransplant plasmablastic lymphomas.392 KMT2D mutations were reported in more than 70% of FL patients and nearly 30% of DLBCL patients, which impaired its methylation capability and steered the transformation of cancer stem cells.391,393,394 On the other hand, MLL fusion proteins caused by the translocation of chromosome 11q23 and more than 60 counterparts have been discovered in leukemia, such as the t(9;11) translocation and the t(4;11) translocation identified in de novo AML and ALL, and the t(11;16) translocation discovered in therapy-related AMML and CMML.362 In MLL fusion proteins, DNA binding domains were found to be preserved, and HOX genes were overexpressed and promoted leukemogenesis.395 Moreover, partial tandem duplication (PTD) of MLL was also found in AML patients with normal karyotypes, which occurred after DNA methylation-related mutations (TET2, DNMT3A, IDH1/2) but before kinase mutations (FLT3, RAS).396 MLL-PTD was found to be significantly related to worse prognoses in AML patients.327
The KMT6 family includes KMT6A, also named EZH2, which catalyzes the trimethylation of H3K27. EZH2 serves as an important part of the polycomb repressive complex (PRC), which negatively regulates transcription. EZH2 has a dual role, as evidenced by its oncogenic role and tumor-suppressive role in different hematologic malignancies. As oncogenes, EZH2 gain-of-function mutations (Y641, A682, and A692) in catalytic SET domains amplified H3K27 methylation, which in turn inhibited the differentiation of plasma cells, promoting oncogenesis in B-cell lymphoma.363,397 These mutations were discovered in 22% of DLBCLs, 10% of BLs, and 4% of primary mediastinal B-cell lymphomas. Excessive EZH2 expression was also found to exist in NK/T-cell lymphoma and CLL.398,399 As tumor suppressors, loss-of-function mutations of EZH2 were found in T-ALL, MDS, and MPN.400–402
Members of the NSD family, including KMT3A (also known as SETD2), KMT3B (also known as NSD1), KMT3G (also known as NSD2), and KMT3F (also known as NSD3), catalyze the methylation of H3K36 and H4K20. NSD1 was found to be fused to NUP98, a part of the nuclear core complex, caused by the translocation of t(5;11)(q35;p15.5), in 16.1% of pediatric AML with normal cytogenetics.403 The NUP98/NSD1 translocation was significantly related to FLT3-ITD variations and worse prognoses, which could be attenuated only through bone marrow transplantation.404,405 NUP98/NSD1 protein was shown to influence CD34+CD133+ hematopoietic precursor cells in MDS, AML, and T-ALL patients; transform bone marrow cells; and trigger AML in mouse models.406,407 NSD2 was found to be fused to FGFR3, caused by the translocation of t(4;14)(p16.3;q32), in 15% of MM patients. The NSD2/FGFR3 translocation was significantly related to 13q- and worse prognoses, which could not be overcome even by a high-dose regimen and ASCT.408,409 NSD3 was also found to fuse to NUP98, caused by the t(8;11)(p11.2;p15) translocation, in AML and radiation-related MDS patients.410,411
PRMT, catalyzing the mono- and demethylation of histone arginine, participates in the epigenetic regulation of leukemogenesis. PRMT4 was found to be indispensable to the onset of MLL-AF9-driven AML.412 PRMT4 could also negatively regulate CBP/p300 coactivation by methylating CBP.413 PRMT5 stimulated the self-renewal and viability of LSCs by triggering the Wnt/β-catenin pathway in CML.414 PRMT7 was shown to regulate glycine metabolism to preserve LSCs in CML, and the loss of PRMT7 downregulated glycine decarboxylase and propelled glycine metabolism to produce toxic methylglyoxal in LSCs without influencing normal hematopoiesis. PRMT7 inhibition impeded leukemogenesis based on CML mouse models and primary CD34+ cells from CML patients.415
Readers: methyl-histone binding proteins recognize histone methylation and mediate subsequent reactions
Since the methylation of lysines occurs in various sites in different forms, the readers of methylated lysines are diverse. Methyllysine readers are mainly divided into two groups based on recognition domains, the Royal domain (chromodomains, MBT domains, and Tudor domains) and PHD fingers. In NUP98-PHD (PHF23 or JARID1A) fusion AML, the PHD finger was found to read H3K4me2/3 and inhibit its removal at various lineage differentiation transcription factors, which caused persistent activation of Hox, Pbx1, and Gata3 transcription factors and led to leukemogenesis. Mutations in PHD fingers, which prevented H3K4me2/3 reading, were shown to impede leukemogenesis, indicating the essential role of methyl reading in leukemogenesis.416 In addition to hematologic malignancies, abnormal methyllysine reading was more commonly seen in solid cancers. HP1 belongs to the chromodomain family, and decreased HP1 expression was reported in breast cancer, colon cancer, ovarian cancer, and papillary thyroid carcinoma.417 ING belongs to the PHD finger family, and its mutations were discovered in melanoma.418 Further research is required to explore the influence of altered methylation readers on hematologic malignancies.
Erasers: histone demethylases remove the existing methylation of histones
The most explored lysine demethylases (KDMs) can be divided into two groups. The first group requires an amine oxidation reaction relying on flavin adenine dinucleotide (FAD) as cofactor, which can only demethylate mono- or dimethyllysine. The second group, the Jumonji demethylase, relies on oxidation and radical attack, such as a-ketoglutarate, and can methylate mono-, di-, and trimethyllysine.
LSD1, also named KDM1A, belongs to the first group. LSD1 has a dual role, as evidenced by its transcription-activating and repressing ability. On the one hand, LSD1 was found to demethylate H3K9me1/2 and repress transcription when related to androgen or estrogen receptors.419,420 On the other hand, LSD1 was shown to promote transcription after demethylating H3K4me1/2 in promoter regions. LSD1 has also been shown to serve as an essential member of transcription repressing complexes, such as CoREST, HDAC2, and ZNF217.421 Moreover, LSD1 was found to demethylate H3K4 after HDAC deacetylation, which in turn assisted in and amplified transcription inhibition, as evidenced by the impaired LSD1 function caused by HDAC inhibitors.422,423 In hematopoiesis, LSD1 was found to mediate the function of the TAL1, GATA-1, and C/EBPa transcription factors; steer erythroid differentiation,424,425 and promote erythroleukemia by inhibiting GFI1 superenhancers.426 LSD1 inhibition was demonstrated to reactivate PU.1-dependent enhancers and eradicate AML in mouse models.427
The KDM5 family, which demethylates H3K4, belongs to the second group and consists of KDM5A, KDM5B, KDM5C, and KDM5D. KDM5A translocation with NUP98 was frequently found to be pathogenic in AML patients, and KMD5A downregulation suppressed proliferation and induced the apoptosis of AML cells.428,429 The loss or inhibition of KDM5D promoted cell differentiation, impeded the growth of APL cells, and improved sensitivity to all-trans retinoic acid treatment.430 KDM5B was overexpressed in CML, and mediated myeloid differentiation and Toll-like receptors via GATA and AP-1 transcription factors. KDM5B knockdown impaired colony formation in CML cells.431 KDM5D, located on chromosome Y, encodes a demethylase of H3K4. KDM5D downregulation was associated with human and mouse AML. The contribution of mosaic loss of chromosome Y to leukemogenesis might be attributable to KDM5D loss.
KDM6A, also named UTX, plays various roles in hematopoiesis and hematologic malignancies. UTX is essential for protecting young hematopoietic stem progenitor cells (HSPCs) from aging. UTX deficiency was associated with the aggregation of reactive oxygen species, reduced DNA damage repair, and aging in HSPCs.432 Regarding hematologic malignancies, UTX mutations were found in 8% of CMML patients.74 UTX has been demonstrated to be a tumor suppressor that represses myeloid leukemogenesis and preserves drug sensitivity in MDS, AML, APL, and even T-ALL.433–437 However, studies also discovered that UTX served as a pro-oncogenic cofactor indispensable to leukemia development in TAL1-positive T-ALL, and UTX inhibition significantly impeded TAL1-positive leukemia.438
Because methylation is essential to both DNA modification and histone modification, the 2-HG caused by IDH mutants inhibited demethylases of both DNA and histones, especially the Jumonji family. Recurrent IDH1/2 mutations were found in 20% of AML patients, and these mutations transformed isocitrate into 2-HG instead of the original a-KG in the tricarboxylic acid cycle, leading to the accumulation of 2-HG and a decrease in a-KG. However, the Jumonji family relies on a-KG to exert a demethylating function, and 2-HG displays a similar orientation in the catalytic core of the JmjC domain, leading to the repression of Jumonji family function, which in turn increases the histone methylation level. The phenomena and influence of IDH mutations on hematologic malignancies have been discussed in the DNA methylation section of this review. IDH inhibitors have been approved by the Food and Drug Administration (FDA) for AML treatment and are presented in the Targeted therapy section below.439
The change of histone phosphorylation is less frequently reported in hematologic malignancies
Histone phosphorylation displays less evidence than histone acetylation and methylation in hematologic malignancies. Histone phosphorylation plays a role in crucial cellular reactions, including apoptosis, transcription, DNA repair and replication, and usually occurs on threonine, tyrosine, and serine residues. Kinases not only stimulate signal transduction but also phosphorylate histones. For example, JAK2 mutations are frequently discovered in MPN, ALL, and AML. H3Y41 phosphorylated by JAK2 was found to disturb the binding of the chromatin repressor HP1a and to stimulate the Lmo2 oncogene to promote leukemogenesis.355 In primary mediastinal B-cell lymphoma and Hodgkin lymphoma (HL), excessive IL-13 and the amplification of chromosome 9p24 were found to stimulate JAK2, which in turn phosphorylated H3Y41 and activated various oncoproteins, including MYC and JAK2 itself. Moreover, JAK2 was found to stimulate PD-L1/2 expression, which conferred immune escape of cancer cells.440–442 Furthermore, in activated B-cell-like (ABC) DLBCL, JAK1 was discovered to phosphorylate H3Y41, which in turn stimulated MYC, MYD88, and IRF4. MYD88 overexpression was shown to produce excessive IL-6 and IL-10 which in turn activated JAK1 in a positive feedback loop.443 In CML, H2AX phosphorylation was found to be stimulated by imatinib and resveratrol, which in turn triggered apoptosis via the caspase-3/Mst1 pathway.444–446
Noncoding RNAs mainly include micrornas and long noncoding RNAs to exert epigenetic regulation function
Traditionally, ncRNAs refer to RNA transcripts that do not encode proteins.447,448 Although around 75% of the human genome can be transcribed, only about 2% of it is translated into mRNAs that encode proteins.449–451 A substantial percentage of the human genome is translated into regulatory, catalytic, and structural RNAs.447,451–453 Recent studies have demonstrated that some ncRNA transcripts can also encode small peptides within 100 amino acids.454 NcRNAs participate in a variety of cellular activities, regulate gene expression and protein function, and are functionally involved in normal development, physiological functions, and the pathogenesis of illness. On the basis of RNA length, many kinds of ncRNAs can be distinguished. Currently, the most extensively researched ncRNAs are microRNAs (miRNAs) and long noncoding RNAs (lncRNAs).It has been revealed that the dysregulation of miRNAs and lncRNAs is involved in all hallmarks of cancer initiation and progression,455–457 including hematologic malignancies (Fig. 1).458 Table 1 shows the selected miRNAs and lncRNAs implicated in hematologic malignancies.
Table 1.
Selected microRNAs and long noncoding RNAs involved in hematologic malignancies
| Name | NcRNA Class | Implicated hematologic malignances | Function roles in tumorigeneses | References |
|---|---|---|---|---|
| miR-155 | miRNA | DLBCL(ABC), CLL, HL, PMBL, PTLD, pediatric BL, CTCL, AML | Oncogene | 484,731–736 |
| miR-150 | miRNA | CLL, MDS, MLL-associated leukemia | Oncogene | 737–739 |
| miR-21 | miRNA | CLL, DLBCL(ABC) | Oncogene | 491,733 |
| miR-221 | miRNA | DLBCL(ABC) | Oncogene | 740 |
| miR130b | miRNA | DLBCL | Oncogene | 499 |
| miR-29 | miRNA | CLL | Oncogene | 508,741 |
| miR-181 | miRNA | CLL, AML, APL | Oncogene/tumor suppressor | 508,742–745 |
| miR-15a/16–1 | miRNA | CLL, APL, MM | Tumor suppressor | 475,476,733,746 |
| miR-143/145 | miRNA | CLL, DLBCL, MALT, BL | Tumor suppressor | 747 |
| miRNA-193b-3p | miRNA | T-ALL | Tumor suppressor | 748 |
| miR-497/195 | miRNA | ALL | Tumor suppressor | 749 |
| miR-22 | miRNA | AML | Oncogene | 750,751 |
| miR-9 | miRNA | MLL-rearranged AML/t(8;21), EVI1+AML | Oncogene/tumor suppressor | 752–754 |
| miR-17–92 cluster | miRNA | MLL-rearranged AML | Oncogene | 755,756 |
| miR-146a | miRNA | del(5q) MDS/MDS-derived AML | Tumor suppressor | 757–760 |
| miR-125b | miRNA | AML, B-ALL | Oncogene | 761,762 |
| miR-126 | miRNA | AML | Oncogene | 763–765 |
| miR-155 | miRNA | FLT3-ITD-induced AML | Oncogene | 735,736 |
| 193a | miRNA | AML | Tumor suppressor | 766,767 |
| miR-193b | miRNA | AML | Oncogene/tumor suppressor | 768–770 |
| miR-223 | miRNA | AML | Tumor suppressor | 771,772 |
| miR-495 | miRNA | MLL-rearranged AML | Tumor suppressor | 773 |
| miR-30–5p | miRNA | MM | Tumor suppressor | 774 |
| miR-137 and miR-197 | miRNA | MM | Tumor suppressor | 775 |
| miR-214 | miRNA | MM | Tumor suppressor | 776 |
| miR-26a | miRNA | MM | Tumor suppressor | 777 |
| LUNAR1 | lncRNA | T-ALL | Oncogene | 564 |
| TCLlnc1 | lncRNA | T-ALL | Oncogene | 566 |
| DANCR | lncRNA | AML | Oncogene | 565 |
| HOXBLINC | lncRNA | NPM1-mutant AML | Oncogene | 778 |
| HOXB-AS3 | lncRNA | NPM1-mutated AML | Oncogene | 779 |
| BlackMamba | lncRNA | ALK− anaplastic large cell lymphoma | Oncogene | 780 |
| HOXB-AS3 | lncRNA | NPM1-mutated AML | Oncogene | 779 |
| MEG3 | lncRNA | AML | Tumor suppressor | 567 |
| H19 | lncRNA | CML | Tumor suppressor | 781 |
| BGL3 | lncRNA | BCR-ABL-positive CML | Tumor suppressor | 568 |
| NEAT1 | lncRNA | MM | Oncogene | 782 |
| CRNDE | lncRNA | MM | Oncogene | 783 |
| MALAT1 | lncRNA | MM | Oncogene | 784 |
ABC activated B-cell-like, ALL acute lymphoblastic leukemia, AML acute myeloid leukemia, APL acute promyelocytic leukemia, BL Burkitt’s lymphoma, CLL chronic lymphocytic leukemia, CTCL cutaneous T-cell lymphoma/leukemia, DLBCL diffuse large B-cell lymphoma, EVI1 Ecotropic viral integration site 1, FLT3 FMS-like tyrosine kinase 3 receptor, HL Hodgkin lymphoma, ITD internal tandem duplication, lncRNA long noncoding RNA, MALT mucosa-associated lymphoid tissue, MDS myelodysplastic syndrome, miRNA microRNA, MLL mixed lineage leukemia, MM multiple myeloma, ncRNA noncoding RNA, NPM1 Nnucleophosmin 1, PMBL primary mediastinal B-cell lymphoma, PTLD posttransplant lymphoproliferative disorder
The physiologic role of microRNAs
MiRNAs are small single-stranded ncRNAs that are usually approximately 22 nucleotides (nt) in length,459 and they were first identified in 1993 in studies on the development of C. elegans.460,461 MiRNAs regulate posttranscriptional gene expression predominantly via sequence-complementary binding to the 3' untranslated region (3' UTR) of target messenger RNA (mRNA), resulting in the degradation of the associated mRNA or the suppression of protein expression.462,463 Moreover, miRNAs can also interact with other targets, such as loci of the protein-coding region of mRNAs,464–466 5’ UTRs of mRNAs,467 intronic and intergenic transcripts,468,469 and other ncRNAs.470,471
The synthesis of miRNAs involves multiple processes. RNA polymerase II (Pol II) first transcribes miRNA-encoding genes in the nucleus, producing a lengthy primary transcript called pri-miRNA. The pri-miRNAs fold back on themselves to create a distinctive hairpin structure, which is recognized and cleaved by the heterotrimeric complex of Drosha endonuclease and its companion protein, DGCR8, resulting in the release of a 60-nt pre-miRNA. The pre-miRNAs are then exported to the cytoplasm by Exportin 5 and the Ran-GTP complex, where Dicer cleaves both pre-miRNA strands near the loop to form a miRNA duplex. One of the miRNA strands is chosen to serve as the guide strand, which is then loaded into an Argonaute protein to form the RNA-induced silencing complex (RISC).463,472,473 After the RISC is formed, the miRNA within the RISC pairs with the target mRNA to direct posttranscriptional repression.463 A single miRNA can target various genes, while multiple miRNAs can target a single gene.463 According to the latest miRbase (v22) data, the total number of human miRNAs identified to date includes 2654 mature miRNA molecules,474 and these miRNAs play extensive fundamental roles in normal physiologic processes and disease states.
In 2002, miRNAs' role in cancer was established for the first time, upon discovering frequent deletions and downregulation of the miR15 and miR16 genes in CLL.475 miR-15/16 have been identified as negative regulators of the BCL2 oncogene and the receptor kinase-like orphan receptor 1 (ROR1) gene.476,477 Subsequently, miRNA dysregulation has been found in almost all studied cancers, including solid tumors and hematologic malignancies.478–481 In cancer, miRNAs function mainly in two ways: as tumor suppressors or as oncogenes.482 Changes in miRNA function in cancer cells are mainly due to changed expression levels of mature or precursor miRNAs compared to related normal tissues.457 The underlying processes of miRNA expression pattern deregulation are diverse, and include miRNA locus deletions or amplifications, miRNA gene mutations, epigenetic and transcriptional regulation, posttranscriptional modification, and dysregulation of miRNA processing.473
MiRNAs can function as oncogenes in hematologic malignancies
MiRNAs can function as oncogenes in hematologic malignancies, thereby contributing to leukemia and lymphoma tumorigenesis. These microRNAs can directly suppress the activity of tumor suppressors or indirectly limit the activity of oncogene-negative regulators.482 A typical example is miR155, which is mainly involved in B-cell malignances.483–485 The copy number of miR-155 is 10- to 30-fold more in B-cell lymphomas, including DLBCL, than in normal B cells.484 Transgenic mice carrying a miR-155 transgene expressed selectively in B cells were found to exhibit a gradual process of an initiation and progression of B-cell malignancy.483 Overexpression of miR-155 in lymphoid organs resulting in disseminated lymphoma in a mouse model, whereas withdrawal of miR-155 in tumor-established mice resulted in rapid lymphoma remission.484 Furthermore, delivery of anti-miR-155 therapy as the sole treatment intervention was found to effectively suppress tumor growth.486 A negative regulator of the kinase AKT, Src homology-2 domain-containing inositol 5-phosphatase 1 (SHIP1) was targeted by MiR-155 and its expression was suppressed. Decreased SHIP1 expression was found to enhance AKT signaling, subsequently promoting cell proliferation and survival and ultimately leading to lymphoma.485 Cobomarsen, a miR-155 inhibitor, has been evaluated in multiple types of lymphoma (NCT 02580552). Another example is miR-21, which has been identified as being overexpressed in a variety of solid tumors and hematologic malignancies.478,487–489 Overexpression of MiR-21 was observed in CLL,490 DLBCL,491 AML, and HL.492,493 Overexpression of miR-21 in a mouse model was capable of the initiation, maintenance and survival of tumors, ultimately leading to a pre-B-cell lymphoma. Strikingly, the inactivation of miRNA-21 resulted in rapid and completed tumor regression in a few days, induced by the apoptosis and rapid proliferative arrest of tumor cells.494 The miR-17–92 cluster has also been demonstrated to be an oncogene. The human miR-17–92 cluster is located on chromosome 13q31.3 and comprises six miRNAs: miR-17, miR-18a, miR-19a, miR-19b-1, miR-20a, and miR-92-1.495 The transcript C13orf25, which included the miR-17–92 cluster, was first observed to have elevated expression in malignant lymphoma.496 The overexpression of the miR-17–92 cluster, in concert with the expression of the c-myc oncogene, accelerated the formation of B-cell lymphoma tumors in a mouse model, according to a subsequent study.497 C-myc might directly bind to the miR17–92 cluster locus and stimulate the miR17–92 cluster’s expression.498 However, this cluster of microRNAs inhibited E2F1, a transcription factor that promotes cell cycle progression and is also targeted by c-myc.498 In an analysis of a large cohort of 532 newly diagnosed cases of DLBCL, serum levels of the miR130b were correlated with shorter PFS and OS.499 Comprehensive research revealed a correlation between serum miR130b and tumor miR130b. In DLBCL, the activity of miR130b on the IFNAR1/p-STAT1 axis reduced lymphoma cell OX40L expression. Through the interaction of OX40L/OX40 lymphoma cells and Th17 cells, the downregulation of tumor cell OX40L expression leads to the increase of Th17 cells at the tumor site.499 MiR130b overexpression also promoted the function of Th17 cells by increasing IL17 production. Thus, miR130b contributed to the progression of DLBCL by fostering a tumor microenvironment that is immunosuppressive and hostile. Therapeutic targeting miR130b with OX40 agoniztic antibody and lipid nanoparticles (LNP)-miR130b antagomir significantly inhibited Th17 cells and miR130b-overexpressing tumor growth in vitro and in vivo, and could be candidates for immunotherapeutic strategies for treating OX40-deficient B-cell lymphoma.499
MiRNAs could also function as tumor suppressors in hematologic malignancies
MiRNAs can also function as tumor suppressors, and the prototypical examples are miR-15 and miR-16 in CLL.475 The deletion of 13q14 was the most prevalent genomic aberration detected in more than half of CLL cases.500 MiR15 and miR16 genes located within a 30-kb region of 13q14 that was lost in approximately 65% of cases of CLL. In 68 percent of CLL cases, allelic loss of this region was related with the deletion or downregulation of miR15 and miR16.475 This was the first time miRNA genes were found to be involved in cancer. The expression of miR-15 and miR-16 was then shown to be inversely associated to the expression of antiapoptotic Bcl-2, and both miR-15 and miR-16 targeted the 3' end of the BCL2 cDNA, thus adversely regulating BCL2 at the posttranscriptional stage. The loss of miR-15a and miR-16-1 caused BCL2 overexpression, which then worked as a driver of malignant transformation.476 Furthermore, miR-15 and miR-16 were also found to target the receptor tyrosine kinase-like orphan receptor 1 (ROR1) gene,477 which encodes an onco-embryonic antigen expressed on the surface of CLL cells.501–503 ROR1 is a receptor for Wnt5a,504 which could accelerate the development/progression of leukemia in a mouse model,505 and could be targeted by anti-ROR1 monoclonal antibodies for the treatment of CLL.506,507 Overall, miR-15/16 depletion enhanced the overexpression of ROR1 and BCL2, two important genes in the initiation and progression of CLL, acting as tumor suppressors in CLL.
Some controversial miRNAs function as either oncogenes or tumor suppressors
Some miRNAs can function as either oncogenes or tumor suppressors, depending on the different contexts they encounter. The previously stated miR-17–92 cluster could be both oncogenic and tumor suppressive by targeting E2F1, which induces apoptosis in some cancers. c-MYC promoted the expression of the E2F1 gene,507 whereas E2F1 was able to induce c-MYC expression through positive feedback. miR-17-5p and miR-20a are capable of targeting E2F1, reducing the reciprocal activation of MYC/E2F1, and inhibiting c-MYC-mediated cell proliferation and carcinogenesis. MiR-29 works as a tumor suppressor by targeting the expression of TCL1, an oncogene in aggressive CLL,508 but transgenic mice that overexpressed miR-29 in B cells developed indolent CLL, suggesting that miR-29 plays an oncogenic role.509
The physiologic role of long noncoding RNAs
LncRNAs are noncoding RNA transcripts longer than 200 nt that do not encode proteins.510,511 Pol II predominantly transcribes LncRNAs. Similar to mRNAs, they are frequently capped with 7-methyl guanosine (m7G) at their 5' ends, polyadenylated at their 3' ends, and posttranslationally spliced.512 LncRNAs are expressed at lower levels, are shorter, often lack an open reading frame (ORF), and are less evolutionarily conserved than protein-coding RNAs.513–517 Low expression of lncRNAs is associated with repressive histone modifications at their gene promoters,518,519 and they are also processed less efficiently than mRNAs.518,520 The majority of lncRNAs synthesized are retained in the nucleus.514,516 The remaining genes are spliced and transported to the cytoplasm.521 LncRNAs regulate gene expression at multiple levels, including epigenetic, transcriptional, and posttranscriptional levels.512 They exert biologic activity by interacting with DNA, RNA, and proteins through their functional modules which incorporate sequence motifs and secondary structures.522 They can act in cis or trans manners, and they can not only modulate their nearby chromatin state and gene expression but also leave their local transcription site and execute functions at distant cellular locations.523 LncRNAs can regulate chromatin by recruiting chromatin modifiers to the promoters of target genes, resulting in transcriptional repression or activation.524–526 Certain lncRNAs serve as decoys for chromatin modifiers by sequestering them from target gene promoters or interacting directly with chromatin.527 lncRNAs may also interact directly with DNA to produce RNA-DNA hybrids, such as triplexes528,529 or R-loops,530–533 which coordinate with chromatin modifiers or transcription factors to activate or repress the transcription of targeted genes.530–532,534 LncRNAs play a pivotal role in transcriptional regulation. By interacting with the transcription machinery, they modify recruited transcription factors or RNA polymeras,535 histone modification stations535,536 and chromatin accessibility537,538 to inhibit target gene expression. Some enhancer loci can be translated into lncRNAs, which are known as enhancer-associated lncRNAs (elncRNAs).539,540 ElncRNAs can promote target gene expression through the action of preexisting chromatin conformations541 or through the recruitment of chromatin-activating complexes to the promoters of target genes.542 They can recruit looping factors, thereby directly stimulating chromatin looping and distant gene expression.543–545 Multiple lncRNAs can form complex regulatory units to regulate the expression of target genes in concert.546 Specifically, regulatory DNA elements are just embedded in lncRNA loci and can stimulate the expression of neighboring genes.547,548 At the posttranscriptional level, lncRNAs also play important regulatory roles. They interact with proteins through their sequence motifs or structural motifs, subsequently interfering with mRNA splicing,549,550 regulating mRNA turnover,551,552 and modulating signaling pathways.553 In addition to binding with proteins, lncRNAs interact with RNAs through base pairing and then recruit proteins that regulate mRNA degradation.554–556 Furthermore, some lncRNAs are complementarily base paired with miRNAs and thus can competitively bind with miRNAs or “sponging” miRNAs, and modulate corresponding miRNA-targeted gene expression.471,557 Finally, lncRNAs are components of nuclear paraspeckle, and paraspeckles act as scaffolds for regulatory molecules.558–561
As described above, lncRNAs act extensively to regulate gene expression, and play important roles in development and various biological and disease contexts, including cancer. Accumulating evidence shows that lncRNAs are differentially expressed in tumors and are involved in malignant transformation and cancer progression.456,562,563 Similar to miRNAs, lncRNAs function as tumor suppressers and oncogenes or have dual effects.482 In hematologic malignancies, lncRNAs may influence cell proliferation, cell cycle regulation, and drug resistance, acting as tumor suppressors or oncogenes.
LncRNA could function as an oncogene in hematologic malignancies
LUNAR1 (leukemia-induced noncoding activator RNA), a NOTCH-regulated lncRNA transcript in human T-ALL, is an example of a lncRNA functioning as an oncogene in hematologic malignancies.564 LUNRAR1 is controlled by the Notch1/Rpbj activator complex and has the ability to boost IGF1R mRNA expression and maintain IGF1 signaling, hence being essential for effective in vitro and in vivo T-ALL proliferation. LncRNA DANCR has been identified as a lncRNA related with LSCs in AML. DANCR is increased in functionally validated LSC-enriched populations and maintains the self-renewal and quiescence of LSCs. In vivo knockdown of Dancr in a primary murine model of AML slowed disease progression and increased mouse survival, confirming its oncogenic function in acute leukemia.565 TCLlnc1 was a newly identified lncRNA that acted as an oncogenic driver in the progression of PTCL.566 Serum TCLlnc1 level was correlated with tumor TCLlnc1 level. TCLlnc1 could be exploited as a possible biomarker for PTCL prognosis as it aws associated with high-risk clinical characteristics and poor prognosis.566 Overexpression of TCL1nc1 enhanced PTCL cell proliferation and migration in vitro and in vivo.566 TCLlnc1 interacted with the transcription activator heterogeneous nuclear ribonucleoprotein D (HNRNPD) and the Y-box binding protein-1 (YBX1) as a modular scaffold, thereby upregulating the transcription of the TGFB2 and TGFBR1 genes and activating the tumor growth factor-β signaling pathway, leading to lymphoma progression.566
LncRNAs could also function as tumor suppressors in hematologic malignancies
LncRNAs may also function as tumor suppressors in hematologic malignancies, as illustrated by the downregulation of the lncRNA MEG3 in AML. MEG3 has been shown to inhibit leukemia cell growth via p53-dependent and p53-independent pathways, and WT1 and TET2 cooperate to upregulate MEG3 expression. MEG3 lowers cell proliferation, causes G0/G1 cell cycle arrest, and decreases AML leukemogenesis in animal models of AML.567 CML-specific LncRNA BGL3 is a critical regulator of cellular transformation mediated by Bcr-Abl. It was found that BGL3 overexpression makes leukemic cells more susceptible to apoptosis and suppresses Bcr-Abl-induced carcinogenesis. Transgenic animals expressing BGL3 were resistant to the Bcr-Abl-induced transformation of primary bone marrow, indicating that BGL3 is a tumor suppressor in CML.568
Targeted therapy based on epigenetic regulation
Targeting DNA methylation
DNA hypomethylating agents (HMAs)
Targeting abnormal DNA methylation has been explored, and DNA hypomethylating agents (HMAs) have been proposed. Two cytidine analogs, 5-azacytidine/vidaza (5-aza-CR, azacitidine) and 5-aza-2’-deoxycytidine/dacogen (5-aza-dCR, decitabine), have been approved to treat MDS by the FDA.569,570 The uridine-cytidine kinase phosphorylated azacitidine to a monophosphate derivative and then diphosphate and triphosphate forms. The triphosphate form (5-AZA-CTP) is primarily incorporated into RNA (∼80–90%) but also some into DNA (10–20%).569,571,572 Decitabine is a prodrug that undergoes a 3- step phosphorylation process intracellularly to be converted to the active moiety, decitabine triphosphate (5-AZA-dCTP), which is then incorporated into DNA by DNA polymerases. 5-AZA-dCTP forms a covalent complex with DNMTs, which leads to the trapping and degradation of the enzyme.573,574 It has been shown that cytidine deaminase (CDA) can deaminate azacytidine and decitabine to inactive aza-uradine nucleosides.575 Cedazuridine is a CDA inhibitor that prevents the degradation of decitabine when taken orally and increases the oral bioavailability of the drug. ASTX727 (decitabine/cedazuridine) has been approval by the USA and Canada for the treatment of MDS and CMML in July 2020.576 The details are shown in Fig. 3. Guadecitabine (SGI-110) is a second-generation HMA consists of a dinucleotide of decitabine and deoxyguanosine,577 which is an active metabolite of decitabine that prevents drug clearance by deamination by CDA has a longer half-life, bioavailability and greater safety.578 It is being tested in clinical trials for MDS and AML. OR-1200 and OR-2100, orally available single-compound prodrugs of decitabine, targeted aberrant DNA hypermethylation and repressed the development of CML and ATL.579,580 Myelosuppression was found to be the most common toxicity of azacitidine and decitabine, and adverse events (AEs) were generally transient, resolving during therapy.581–583 Zebularine is a DNMT-inhibiting cytosine nucleoside analog with stability in acidic environments and in aqueous solutions.584 Zebularine was shown to efficiently decrease AhR gene methylation in childhood ALL cells.585 However, zebularine required a near millimolar dose and had limited bioavailability in rodents (<7%) and primates (<1%); thus, it did not show good performance in a clinical trial.586 Although the TET family plays an essential role in DNA demethylation, TET protein inhibitors have yet to be tested for cancer treatment. The intracellular metabolism of azanucleosides is shown in Fig. 2. The completed clinical trials with results of targeting DNA methylation are concluded in Table 2.
Fig. 3.
Intracellular metabolism of azacytidine and decitabine. Azacytidine (5-AZA-CR) and decitabine (5-AZA-dCR) are modified in a 3-step phosphorylation process by different metabolic pathways. 5-AZA-CTP/5-AZA-dCTP is incorporated into RNA or DNA by RNA/DNA polymerases, respectively. CDA can deaminate azacytidine and decitabine to inactive aza-uradine nucleosides. CDA cytidine deaminase, 5-AZA-CR 5-azacitidine, 5-AZA-CMP 5-azacitidine monophosphate, 5-AZA-CDP 5-azacitidine diphosphate, 5-AZA-CTP 5-azacitidine triphosphate, 5-AZA-U aza-uridine nucleosides, 5-AZA-dCR 5-aza-2’-deoxycytidine, 5-AZA-dCMP 5-aza-2’-deoxycytidine monophosphate, 5-AZA-dCDP 5-aza-2’-deoxycytidine diphosphate, 5-AZA-dCTP 5-aza-2’-deoxycytidine triphosphate, 5-AZA-dU deoxyuridine
Table 2.
The completed clinical trials with results of targeting DNA methylation therapy
| Conditions | Interventions | Phases | Patients | NCT number |
|---|---|---|---|---|
| Azacitidine | ||||
| AML/ALL/JMML/MDS | Azacitidine | 2 | 17 | NCT02458235 |
| High-risk MDS | Azacitidine | 2 | 72 | NCT01599325 |
| AL/MDS | Azacitidine+Sorafenib | 1/2 | 60 | NCT01254890 |
| High-risk MDS/AML | Azacitidine+Sorafenib | 2 | 16 | NCT02196857 |
| High-risk MDS/AML | Azacytidine+Lenalidomide | 1/2 | 94 | NCT01038635 |
| R/R AML/AML with11q23 rearrangement | Azacitidine+Pinometostat | 1/2 | 1 | NCT03701295 |
| CMML | Azacitidine | 2 | 11 | NCT01350947 |
| High-risk MDS | Azacitidine | 2 | 16 | NCT00721214 |
| AML | Azacitidine | 2 | 24 | NCT00387647 |
| MDS/AML/MPN/CMML | Azacitidine/APR-246 | 1/2 | 55 | NCT03072043 |
| High-risk MDS | Azacitidine | NA | 25 | NCT00660400 |
| Myelofibrosis | Azacitidine | 2 | 34 | NCT00569660 |
| MDS/CMML | Azacitidine+Volasertib | 1 | 5 | NCT02201329 |
| MDS | Azacitidine | 3 | 40 | NCT01186939 |
| High-risk MDS/CMML/AML | Azacitidine+Pevonedistat | 2 | 120 | NCT02610777 |
| MDS/AML/MM/NHL/HL | Azacitidine | 1 | 31 | NCT00652626 |
| Low and intermediate-1 risk MDS | Azacitidine/Decitabine | 2 | 113 | NCT01720225 |
| High-risk MDS | Azacitidine | 4 | 44 | NCT01201811 |
| Advanced MDS | Azacytidine+Lenalidomide | 1/2 | 37 | NCT00352001 |
| Relapsed AML | Azacitidine+Gemtuzumab ozogamicin | 1/2 | 50 | NCT00766116 |
| Leukemia/AML/MDS | Azacitidine | 3 | 187 | NCT00887068 |
| MM | Azacitidine | NA | 17 | NCT01050790 |
| MDS | Azacitidine | 2 | 25 | NCT00384956 |
| AML | Azacitidine/Conventional Care Regimen | 3 | 488 | NCT01074047 |
| Poor-risk AML/MDS | Azacitidine+Sargramostim | 2 | 25 | NCT01700673 |
| AML | Azacitidine/Lenalidomide/Best Supportive Care | 2 | 88 | NCT01358734 |
| Leukemia | Azacytidine/PKC412 | 1/2 | 54 | NCT01202877 |
| AML | Azacitidine+Lenalidomide | 1/2 | 45 | NCT00890929 |
| AML/MDS | Azacitidine+Vorinostat | 2 | 110 | NCT00948064 |
| AML | Azacitidine+MLN4924 | 1 | 64 | NCT01814826 |
| AML/MDS/CMML | Azacitidine | 2 | 43 | NCT01083706 |
| R/R AML | Azacitidine+Lenalidomide | 2 | 37 | NCT01743859 |
| MDS/AML/CMML | Azacitidine+PF-04449913 (Glasdegib) | 1 | 73 | NCT02367456 |
| AML/MDS | Azacytidine | 2 | 40 | NCT02497404 |
| AML | Azacitidine+Lenalidomide | 1/2 | 31 | NCT01016600 |
| MDS/CMML/AML | Azacytidine+Panobinostat (LBH589) | 1/2 | 113 | NCT00946647 |
| MDS/AML | Azacytidine+Valproic Acid+ATRA | 2 | 34 | NCT00326170 |
| Refractory MM | Azacitidine Lenalidomide+Dexamethasone | 1/2 | 45 | NCT01155583 |
| MDS | Azacytidine | 2 | 151 | NCT00102687 |
| MDS | Azacytidine+Etanercept | 1/2 | 32 | NCT00118287 |
| AML/CMML/MDS | Azacitidine/+Entinostat | 2 | 197 | NCT00313586 |
| AML | Azacitidine+Midostaurin | 1/2 | 34 | NCT01093573 |
| R/R AML/high-risk MDS | Azacytidine+Ara-C | 1/2 | 36 | NCT00569010 |
| AML/MDS | Azacytidine+Valproic Acid+Ara-C | 2 | 11 | NCT00382590 |
| CML | Azacitidine | 2 | 24 | NCT00813124 |
| Childhood R/R ALL/AML | Azacytidine+Chemotherapy | 1 | 15 | NCT01861002 |
| High-risk MDS | Azacitidine/Conventional Care | 3 | 358 | NCT00071799 |
| R/R DLBCL | Azacitidine Vorinostat | 1/2 | 17 | NCT01120834 |
| High-risk MDS | Azacytidine+Valproic Acid+ATRA | 2 | 62 | NCT00439673 |
| R/R AML | Azacitidine+Vorinostat+Gemtuzumab Ozogamicin | 1/2 | 52 | NCT00895934 |
| DLBCL | Azacytidine+R-CHOP | 1/2 | 14 | NCT01004991 |
| Refractory lymphoma | Azacytidine -SAHA-GBM | 1/2 | 61 | NCT01983969 |
| AML | Intensive Chemotherapy/+Glasdegib/+Azacitidine/+Glasdegib | 3 | 730 | NCT03416179 |
| AML | Azacitadine+Pracinostat | 2 | 50 | NCT01912274 |
| AML/MDS | Azacitidine (CC-486) | 1/2 | 31 | NCT01835587 |
| MDS | Eltrombopag/+Hypomethylating Agent | 2 | 29 | NCT01893372 |
| Decitabine | ||||
| Relapsed AML | Decitabine+Pembrolizumab | 1/2 | 10 | NCT02996474 |
| AML/high-risk MDS | Decitabine+Vosaroxin | 1/2 | 66 | NCT01893320 |
| AML/MDS | Decitabine | 2 | 114 | NCT01687400 |
| AML | Decitabine | 2 | 74 | NCT01786343 |
| AML | Decitabine+Talacotuzumab (JNJ-56022473; anti CD123) v.s. Decitabine | 2/3 | 326 | NCT02472145 |
| MDS | Decitabine | 2 | 37 | NCT00744757 |
| AML/MDS | Decitabine | 2 | 10 | NCT00760084 |
| MDS/AML | Decitabine/+Valproic Acid | 2 | 153 | NCT00414310 |
| Low and intermediate-1 risk MDS | Decitabine v.s. Azacitidine | 2 | 113 | NCT01720225 |
| High-risk MDS | Decitabine+Clofarabine | 2 | 42 | NCT00903760 |
| AML/high-risk MDS | Decitabine+Gemtuzumab Ozogamicin | 2 | 43 | NCT00882102 |
| MDS | Decitabine+Arsenic Trioxide+Ascorbic Acid | 2 | 7 | NCT00621023 |
| AML/high-risk MDS | Decitabine+Gemtuzumab Ozogamicin | 2 | 71 | NCT00968071 |
| R/R AML/high-risk MDS | Decitabine+Mitoxantrone Hydrochloride+Etoposide+Cytarabine | 1/2 | 52 | NCT01729845 |
| Low or intermediate-1 risk MDS | Decitabine | 2 | 67 | NCT00619099 |
| MDS | Decitabine | 3 | 135 | NCT01751867 |
| AML/MDS | Decitabine | 1 | 16 | NCT01378416 |
| AML | Decitabine | 2/3 | 50 | NCT00398983 |
| AML/high-risk MDS/MPN | Decitabine+Cytarabine | NA | 12 | NCT02121418 |
| AML/MDS | Decitabine+Vorinostat | 1 | 71 | NCT00479232 |
| AML/high-risk MDS | Decitabine+GCLAM | 1/2 | 28 | NCT02921061 |
| MDS | Decitabine | 1 | 39 | NCT00796003 |
| MDS/AML | Decitabine+LBH589 | 1/2 | 52 | NCT00691938 |
| Advanced MDS | Decitabine | 2 | 99 | NCT00260065 |
| AL | Decitabine+Clofarabine+Idarubicin+Cytarabine | 1/2 | 65 | NCT01794702 |
| AML | Decitabine | 3 | 485 | NCT00260832 |
| AML/high-risk MDS | Decitabine+Clofarabine+Cytarabine | 2 | 122 | NCT00778375 |
| MDS | Decitabine | 2 | 128 | NCT00067808 |
| AML | Decitabine+Cytarabine | 2 | 44 | NCT01829503 |
| AML | Decitabine | 2 | 55 | NCT00358644 |
| High-risk MDS | Decitabine+Vorinostat+Natural killer (NK) cells | 2 | 9 | NCT01593670 |
| AML | Decitabine | 2 | 546 | NCT00416598 |
| AML | Decitabine | 2 | 55 | NCT00492401 |
| R/R AML | Decitabine+total body irradiation | 2 | 20 | NCT01707004 |
| MDS | Decitabine | 1/2 | 25 | NCT01165996 |
| AML/MDS | PF-04449913+Ara-C+Decitabine+Daunorubicin+Cytarabine | 2 | 255 | NCT01546038 |
| Guadecitabine | ||||
| MDS | SGI-110 (guadecitabine) | 2 | 22 | NCT03075826 |
| AML | SGI-110 (guadecitabine) | 3 | 815 | NCT02348489 |
| AML | SGI-110 (guadecitabine)/+Idarubicin or Cladribine | 2 | 44 | NCT02096055 |
| AML | SGI-110 (guadecitabine) | 1 | 21 | NCT02293993 |
| ASTX727 (Cedazuridine/Decitabine) | ||||
| MDS/CMML | Decitabine/Cedazuridine | 1/2 | 130 | NCT02103478 |
AL acute leukemia, ALL acute lymphoblastic leukemia, AML acute myeloid leukemia, Ara-C cytosine arabinoside, ATRA all-trans retinoic acid, CMML chronic myelomonocytic leukemia, DLBCL diffuse large B-cell lymphoma, HL Hodgkin lymphoma, JMML juvenile myelomonocytic leukemia, MDS myelodysplastic syndrome, MM multiple myeloma, MPN myeloproliferative neoplasm, NHL non-Hodgkin lymphoma, R/R relapsed/refractory
Furthermore, many non-nucleoside analogs have been reported as DNA HMAs, which are usually small-molecule inhibitors and directly target catalytic sites rather than being incorporated into DNA. GSK3685032 was a DNMT1-selective inhibitor that could inhibit DNA methylation, transcriptional activation and cancer cell proliferation inhibition in vitro.587 Epigallocatechin gallate (EGCG) was the most effective polyphenol in green tea and inhibited the proliferation of APL, AML, and CML cells by inhibiting the expression of DNMTs.588–592 EGCG also induced the demethylation and transcription of the p16 gene in the MM cell line CA46.593 Thymoquinone (TQ) was a major component of Nigella sativa seeds, inducing DNMT1 dysfunction,594 and could enhance demethylation in AML.595 TQ could also mediate apoptosis by targeting UHRF1 in Jurkat cells.212 The combination of DFMO and TQ decreased the expression of the UHRF1, DNMT1 and HDAC1 genes in Jurkat cells.211 Quercetin, an important dietary flavonoid, eliminated DNMT1 and DNMT3a expression and enhanced the apoptosis of leukemia cells.596 Harmine, a beta-carboline alkaloid derivative of Peganum harmala, could inhibit DNMT1 expression in leukemia cells.597 Curcumin, a component of the popular Indian spice turmeric, could decrease DNMT1 expression and played an anti-leukemic role in AML.598 Novel curcumin liposomes modified with hyaluronan was found to downregulate DNMT1 expression and played an anti-leukemic role in AML.599 NT1721, a novel epidithiodiketopiperazine, was shown to deplete DNMT1 protein levels, causing the re-activation of silenced tumor suppressor genes.600 Furthermore, parthenolide, a major component of the feverfew medicinal plant, reduced the expression of DNMT1 in primary effusion lymphoma models.601 Oridonin was an ent-kaurene diterpenoid extracted from the Chinese herb Rabdosia rubescens, and was shown to inhibit DNMT3AR882 mutation-driven clonal hematopoiesis and leukemia.602 RG108, a DNA methyltransferase inhibitor, could inhibit the activity of DNMT enzymes and cause demethylation.603 Emodin, an active component in the roots and rhizomes of numerous Chinese medicinal herbs, was demonstrated to inhibit human lymphoma Raji cell proliferation by UHRF1-DNMT3A-∆Np73 pathways.604 Berberine was a main constituent of optidis rhizome and an isoquinoline alkaloid, and it was reported to repress the expression of DNMT1 and DNMT3B, and to induce apoptosis in the human MM cell line U266 through p53 promoter hypomethylation.605 MG98, oligonucleotide antisense to DNMT1, displayed no pharmacodynamic or clinical activity when administered to patients with high-risk MDS.606
Combination treatment of HMAs with immune treatment
HMAs in combination with immune checkpoint inhibitors were widely used.607 CD47 was a macrophage checkpoint and can be targeted for AML and MDS,608 and its inhibitor in combination with azacitidine had increased efficacy of AML and higher-risk MDS, especially in patients with TP53 mutation.609 T-cell immunoglobulin domain and mucin domain-3 (TIM-3) was a T-cell immune checkpoint, and highly expressed in LSCs. Sabatolimab (an anti-TIM-3 monoclonal antibody) in combination with HMAs had durable responses in HR-MDS and AML patients.607 In a phase 1b trial, ipilimumab (CTLA-4) showed limited efficacy in HR-MDS patients after failing from HMAs.610 HMAs could increase the expression level of PD-1, PD-L1, PD-L2, and CTLA-4 in patients with AML and MDS, which caused HMA resistance.610 Therefore, there are clinical trials combining PD-1/PD-L1 inhibitors with HMAs in AML and MDS patients. The ORR rate and median OS were higher in azacitidine plus nivolumab plus ipilimumab than azacitidine plus nivolumab group.611 While, high-risk MDS patients after the failure of HMAs did not benefit from pembrolizumab.612 Decitabine in combinatipn with PD-1 inhibitors was also used in classical HL (cHL) treatment. Decitabine increased the efficacy of anti-PD-1 antibody in refractory cHL, even in patients with anti-PD-1 monotherapy resistance.613,614 The completed clinical trials with HMAs plus other treatments, including chemotherapy, PD-1 inhibitor, are listed in Table 2.
IDH inhibitors
The IDH1 inhibitors ivosidenib, olutasidenib, and IDH2 inhibitor enasidenib have been approved by FDA for patients with relapsed/refractory IDH1 mutant AML. In a phase I study, the ORR of ivosidenib was 41.6%, the CR and CRi was 30.4%.615 In a phase 1 clinical trial, the ORR of olutasidenib was 41% and 46% in combination with azacitidine in relapsed/refractory AML.616 The ORR of enasidenib in relapsed/refractory AML patients with IDH2 mutations was 40.3%, and the CR was 19.3% in a phase I/II study.617 The median OS for all pateints was 9.3 months, but 19.7 months for patients who achieved CR.
Furthermore, the IDH1 inhibitor ivosidenib combined with azacytoside as a first-line treatment for AML patients carrying an IDH1 mutation has been approved by the FDA. In a phase Ib clinical trial, the ORR was 78.3% and the CR was 60.9%.618 In a phase 3 study, the EFS was longer in the combination group than in the placebo-and-azacitidine group (HR = 0.33, P = 0.002). The median OS in ivosidenib and azacitidine group (24.0 months) was longer than the placebo and azacitidine group (7.9 months) (HR = 0.44, P = 0.001).619
The ongoing clinical trials are listed in Table 3. The clinical trials mainly focus on AML, and MDS. The IDH inhibitor combined with other treatments, including chemotherapys, PD-1 inhibitor, MEK inhibitor, and FLT3/AXL inhibitors, are listed in Supplementary Table 1.
Table 3.
The clinical trials of IDH inhibitors
| NCT number | Conditions | Interventions | Phase | Number enrolled |
|---|---|---|---|---|
| NCT04176393 | R/R AML | Ivosidenib | Phase 1 | 30 |
| NCT03564821 | IDH1-mutated myeloid neoplasms | Ivosidenib | Phase 1 | 18 |
| NCT03245424 | AML | Ivosidenib | NA | NA |
| NCT02074839 | R/R AML/MDS/other IDH1-mutated positive hematologic malignancies | Ivosidenib | Phase 1 | 291 |
| NCT03839771 | AML/MDS | Ivosidenib | Phase 3 | 968 |
| NCT03503409 | AML/MDS | Ivosidenib | Phase 2 | 68 |
| NCT05282459 | AML | Enasidenib | Phase 1/2 | 48 |
| NCT04203316 | R/R AML | Enasidenib | Phase 2 | 10 |
| NCT03515512 | AML/CML | Enasidenib | Phase 1 | 23 |
| NCT01915498 | Hematologic neoplasms | Enasidenib | Phase 1 | 345 |
| NCT03720366 | AML | Enasidenib | Phase 1 | 40 |
| NCT03728335 | AML | Enasidenib | Phase 1 | 15 |
| NCT03881735 | R/R AML | Enasidenib | Phase 2 | 0 |
| NCT03744390 | AML/MDS | Enasidenib | Phase 2 | 68 |
| NCT03723057 | AML | Enasidenib | NA | NA |
| NCT04522895 | AML/MDS/CML | Enasidenib | Phase 2 | 50 |
AML acute myeloid leukemia, CML chronic myeloid leukemia, MDS myelodysplastic syndrome, NA not applicable, R/R relapsed/refractory
Targeting HDAC
HDAC inhibitors not only trigger cell differentiation, apoptosis, autophagy, and cell cycle arrest but also modulate immune reactions and inhibit angiogenesis in various hematologic malignancies and some solid tumors.620 It has been hypothesized that malignant cells are more vulnerable to epigenetic treatment, rendering the specificity and selectivity of malignant cells over normal cells.351 A number of HDAC inhibitors have been developed and demonstrated effective in treating hematologic malignancies. The U.S FDA and China National Medical Products Administration (NMPA) have approved five HDAC inhibitors for the treatment of T-cell lymphoma, MM, and breast cancer. The classical inhibitors, dual-target inhibitors, and PROTAC for HDAC are discussed below (Fig. 4).
Fig. 4.
Classic HDAC inhibitors, dual-target HDAC inhibitors, and RPOTAC for HDAC. HDAC inhibitors not only inhibit angiogenesis, cell migration, resistance to treatment, and cell proliferation but also stimulate cell differentiation, apoptosis, and tumor antigenicity. The development of HDAC-targeting treatment ranges from classic HDAC inhibitors, dual-target HDAC inhibitors, and PROTAC for HDACs. Classic HDAC inhibitors are divided into four groups based on structures. The first group is aliphatic carboxylic acid group. None of the HDAC inhibitors of this group have been approved for hematologic malignancy treatment. The second group is hydroxamic acid group and includes vorinostat, belinostat, and panobinostat, which has been approved for treatment of CTCL, PTCL, and MM, respectively. The third group is benzamide group and includes chidamide, which has been approved for the treatment of PTCL. The last group is the cyclic peptide group and includes romidepsin, which has been approved for the treatment of CTCL and PTCL. Furthermore, dual-target HDAC inhibitors have been explored in hematologic malignancy treatment, which also target RTK, JAK2, and PI3K. In additions, PROTAC for HDACs has been developed recently
Classic HDAC inhibitors
Classic HDAC inhibitors can be divided into four groups based on their zinc-binding group: the aliphatic carboxylic acid group, hydroxamic acid group, benzamide group, and cyclic peptide group. In general, the aliphatic carboxylic acid group displays weak deacetylation ability and mild selectivity for class I HDACs. The hydroxamic acid group displays strong pan-deacetylation ability with an IC50 value of nM level. The benzamide group and cyclic peptide group have strong selectivity for class I HDAC.
In the aliphatic carboxylic acid group, which inhibits class I HDACs, sodium butyrate has been investigated in the chemoprevention of breast cancer and colon cancer. Valproic acid has been approved for seizure treatment, and several clinical trials are investigating its combination with chemotherapy to treat CLL, AML, MDS, and lymphoma. AR-42 resembles phenylbutyrate in structure and inhibits HDACs with an IC50 of 30 nM. In AML, AR-42 was shown to repress the NF-κB pathway, downregulate oncogenic Kit through HSP90 disturbance, and specifically trigger the apoptosis of LSCs without affecting normal HSCs.621 In MM, AR-42 could trigger cell cycle arrest and the apoptosis of MM cells, and downregulated CD44, which represents resistance to immunomodulating agents (IMiDs) and dexamethasone.622,623 However, in clinical trials, AR-42 demonstrated only mild disease control of MM and a relatively low overall response rate (ORR) (23.1%) when combined with decitabine in AML, impeding its further application in hematologic malignancy treatment.624,625
In the hydroxamic acid group, vorinostat, belinostat, and panobinostat have been approved by the FDA, while ricolinostat and citarinostat are being investigated in clinical trials. Vorinostat, also named SAHA, inhibits class I, II, and IV HDACs, and in 2006 was the first HDAC inhibitor approved by the U.S. FDA to treat cutaneous T-cell lymphoma/leukemia (CTCL). For CTCL patients who had previously received two lines of systemic treatment, the ORR of vorinostat treatment was 30%, and the median time to progression was 202 days. Serious adverse events (sAEs) of grade 3 or above included pulmonary embolism (5%) and fatigue (4%).626,627 Combination treatment of vorinostat with rituximab, proteasome inhibitors (PIs), and HMA is under investigation in lymphoma. In MM, the combination of vorinostat, PIs and IMiDs is being explored in clinical trials. Furthermore, various clinical trials exploring vorinostat combined with chemotherapy to treat AML, MDS, ALL, CML, and CLL and to prevent graft-versus-host disease (GVHD) have been completed or are ongoing (Table 4). In 2014, belinostat, also known as PXD101, which inhibits class I and II HDACs with nanomolar IC50 values, was the third HDAC inhibitor approved by the U.S. FDA to treat relapsed/refractory (RR) PTCL. For R/R PTCL patients, the ORR of belinostat was 25.8%, and the median duration of response (DoR) was 8.4 months. The sAEs of grade 3 or above were hematologic AEs (6.2–10.9%), pneumonia (5.4%), dyspnea (6.2%), and fatigue (5.4%).628 In addition to PTCL, belinostat has been investigated alone or in combination with rituximab, idarubicin, HMA, NEDD8 activating enzyme inhibitors, and PIs in the treatment of R/R aggressive B-cell lymphoma, AML, MDS, and MM in various clinical trials (Table 4). In 2015, panobinostat (also named LBH589), which inhibits class I, II, and IV HDACs, was the fourth HDAC inhibitor approved by the U.S. FDA to treat MM. In the PANORAMA1 trial, R/R MM patients received bortezomib and dexamethasone combined with panobinostat or a placebo. The panobinostat group displayed significantly longer PFS (median 11.99 months) and higher CR or near CR rate (27.6%) than the placebo group, although no significant benefit in OS was discovered.629,630 In the PANORAMA2 trial, heavily pretreated bortezomib-refractory MM patients received treatment with panobinostat, bortezomib, and dexamethasone, and the ORR was 34.5%.631 In the MUK-six trial, R/R MM patients received treatment with panobinostat, bortezomib, thalidomide and dexamethasone, and the ORR was 91%.632 However, the U.S. FDA has issued a warning regarding an increased risk of diarrhea, cardiac toxicity, and abnormal electrocardiogram manifestations with the use of panobinostat. Various clinical trials have examined panobinostat alone or combined with PIs, IMiDs, and everolimus in MM and lymphoma. In addition, AML, MDS, myelofibrosis (MF), and GVHD were also explored to confirm the safety and efficacy of panobinostat combined with corresponding standard therapy (Table 4). Apart from the approved HDAC inhibitors, various not-yet-approved inhibitors have been explored in clinical trials. Givinostat, also named ITF2357, inhibits HDAC1 and HDAC3 and has been demonstrated to regulate NFE2 and C-MYB hematopoietic transcription factors and trigger the apoptosis in JAK2V617F-positive MPN.633,634 It was also shown to trigger apoptosis, regulate the cell cycle, and promote differentiation in T-ALL and B-ALL.635–637 Several clinical trials investigated its efficacy in MPN and HL, and the ORR of givinostat in the treatment of polycythemia vera was more than 80%, accompanied by a favorable safety profile.638–640 Quisinostat, also named JNJ-26481585, inhibits HDAC1, HDAC2, and HDAC3 with nanomolar IC50 value. In MM cells, quisinostat stimulated the caspase cascade and upregulated p21, which in turn triggered apoptosis and cell cycle arrest in vitro. Quisinostat alone could also inhibit angiogenesis and significantly reduce the tumor burden in MM mouse models.641,642 Quisinostat combined with bortezomib significantly remodeled bone structure and reduced bone disease.643 When combined with bortezomib and dexamethasone, quisinostat exerted an ORR of 88.2% in R/R MM patients.644 The cutaneous response rate to quisinostat in previously treated CTCL patients was 24%.645 Tefinostat, also named CHR-2845, is a pan-HDAC inhibitor that exerts anti-leukemic activity in monocytoid-lineage leukemia in vitro.646 In a phase I trial including 18 patients with R/R hematologic malignancies, tefinostat displayed a good tolerance profile and showed early signs of response in a CMML patient and an AML-M2 patient.647 Resminostat, also named 4SC-201, inhibits HDAC1, HDAC3, and HDAC6. Resminostat repressed proliferation and induced the apoptosis and G0/G1 cell cycle arrest of MM cells in vitro. Resminostat also disturbed the Akt signaling pathway by reducing 4E-BP1 and p70S6k phosphorylation.648 In addition, resminostat exerted an antitumor effect when combined with ruxolitinib in CTCL models.649 In the phase II SAPHIRE trial, resminostat showed an ORR of 34% in R/R HL patients.650 Abexinostat, also named PCI-24781, is a pan-HDAC inhibitor that mainly targets HDAC1. Abexinostat can stimulate apoptosis via the NFκB pathway and remodel chromatin in lymphoma cells.651,652 Various clinical trials have examined abexinostat in FL, MCL, and DLBCL. The ORR of abexinostat in R/R FL patients was 64.3%, and that in R/R MCL patients was 27.3%.653 Alteminostat, also named CKD-581, is a pan-HDAC inhibitor that inhibits proliferation; downregulates c-Myc, BCL-2, BCL-6, and MCL-1; and upregulates p53, p21, and H2AX phosphorylation in MM cells and double-hit/double-expressor DLBCL cells.654,655 In a phase I trial including MM and lymphoma patients who were refractory to standard treatment, alteminostat exhibited a favorable safety profile with a relatively low PR rate of 5.6%.656 Citarinostat, also named ACY-241, inhibits HDAC6 and HDAC3 and exerts synergistic antimyeloma effects combined with pomalidomide in vitro and in MM mouse models.657 Several clinical trials have explored its safety and efficacy in MM patients. Remetinostat, also named SHP-141, inhibits HDAC1, HDAC3, and HDAC6 and was designed to exert efficacy specifically and locally in the skin.658 Clinical trials have examined remetinostat in early stage CTCL. Pracinostat, also named SB939, is a pan-HDAC inhibitor that showed synergism with JAK2 inhibitors in AML models and increased sensitivity to BCR-ABL kinase inhibitors in CML.659,660 Several clinical trials have investigated its efficacy in MDS and MF. Pracinostat showed only modest or mild efficacy in MF and showed an even lower ORR when combined with HMAs compared with HMA alone, preventing its further application to the two diseases.661–664 Ricolinostat, also named ACY-1215, is a first-in-class HDAC6 inhibitor. Ricolinostat combined with bortezomib caused sustained endoplasmic reticulum stress and apoptosis by stimulating caspase-3, caspase8, caspase 9, and poly (ADP) ribosome polymerase in MM cells in vitro. In a plasmacytoma MM model and disseminated MM model, ricolinostat combined with bortezomib significantly inhibited tumor growth and extended OS.665 It also increased sensitivity to daratumumab by upregulating CD38 expression in MM cells.666 A similar antitumor effect was also found in lymphoma.667 A combination treatment of ricolinostat with an immune checkpoint inhibitor and ibrutinib was effective in CLL and FL, respectively, in vitro and in vivo.668,669 Several clinical trials are examining its effect in R/R MM, CLL, and lymphoma. For R/R MM patients, ricolinostat combined with bortezomib and dexamethasone showed an ORR of 37%.670 and ricolinostat combined with lenalidomide and dexamethasone displayed an ORR of 55%.671 Purinostat, an HDAC I/IIb inhibitor, has demonstrated promising antitumor effects in B-ALL and DLBCL mouse models and is being explored in B-cell lymphoma and MM in China.672,673
Table 4.
The clinical trials of approved HDAC inhibitors
| Conditions | Interventions | Phase | N | NCT number | Status |
|---|---|---|---|---|---|
| Vorinostat | |||||
| CTCL | Vorinostat | 1 | 10 | NCT00771472 | Completed |
| Advanced CTCL | Vorinostat | 2 | 74 | NCT00091559 | Completed |
| ND T-cell lymphoma | Vorinostat+CHOP | 1/2 | 14 | NCT00787527 | Completed |
| ND T-cell lymphoma, MCL, R/R other types of lymphoma | Vorinostat+Rituximab+Ifosfamide+Carboplatin+Etoposide | 1/2 | 29 | NCT00601718 | Completed |
| ENKT | Vorinostat+Azacitidine | 1 | 18 | NCT00336063 | Active, not recruiting |
| Low-grade NHL | Vorinostat | 2 | 37 | NCT00253630 | Completed |
| Indolent NHL | Vorinostat+Rituximab | 2 | 30 | NCT00720876 | Completed |
| R/R NHL, AML, ALL, CML | Vorinostat+Decitabine | 1 | 80 | NCT00275080 | Completed |
| Relapsed NHL | Vorinostat+Combination Chemotherapy | 2 | 30 | NCT04220008 | Not yet recruiting |
| Indolent B-cell NHL, MCL | Vorinostat | 2 | 56 | NCT00875056 | Completed |
| R/R MCL, DLBCL | Vorinostat+Bortezomib | 2 | 65 | NCT00703664 | Completed |
| MCL, R/R B-NHL, R/R CLL | Vorinostat+Cladribine+Rituximab | 2 | 57 | NCT00764517 | Completed |
| ND stage II-IV DLBCL | Vorinostat+Rituximab+Combination Chemotherapy | 1/2 | 83 | NCT00972478 | Active, not recruiting |
| R/R DLBCL, FL, HL | Vorinostat+Pembrolizumab | 1 | 52 | NCT03150329 | Completed |
| R/R DLBCL | Vorinostat+Azacitidine | 1/2 | 17 | NCT01120834 | Completed |
| Relapsed DLBCL | Vorinostat | 2 | 18 | NCT00097929 | Completed |
| Elderly relapsed DLBCL | Vorinostat+Cyclophosphamide, Etoposide, Prednisone and Rituximab | 1/2 | 30 | NCT00667615 | Completed |
| R/R B-cell lymphoma | Vorinostat+Carfilzomib | 1 | 20 | NCT01276717 | Completed |
| R/R advanced HL | Vorinostat | 2 | 27 | NCT00132028 | Completed |
| HIV-related DLBCL, other aggressive B-cell lymphoma | Vorinostat+Rituximab+Combination Chemotherapy | 1/2 | 107 | NCT01193842 | Completed |
| Lymphoma | Vorinostat+Niacinamide+Etoposide | 1 | 40 | NCT00691210 | Completed |
| High-risk lymphoma | Vorinostat after stem cell transplantation | 1 | 23 | NCT00561418 | Completed |
| Relapsed lymphoma | Vorinostat | 1 | 10 | NCT00127140 | Completed |
| R/R lymphoma | Vorinostat+Gemcitabine+Busulfan+ Melphalan+Stem Cell Transplant | 1 | 78 | NCT01421173 | Completed |
| R/R lymphoma | Vorinostat+Alisertib | 1 | 34 | NCT01567709 | Completed |
| NHL after ASCT | Vorinostat+Bortezomib | 2 | 27 | NCT00992446 | Completed |
| ND MM | Vorinostat+Bortezomib+Lenalidomide+Dexamethasone | 1 | 30 | NCT01038388 | Completed |
| ND MM in maintenance treatment | Vorinostat+Lenalinomide | 3 | 4420 | NCT01554852 | Active, not recruiting |
| R-resistant MM | Vorinostat+Lenalinomide+Dexamethasone | 1/2 | 25 | NCT01502085 | Completed |
| R/R MM | Vorinostat+Bortezomib | 1 | 34 | NCT00111813 | Completed |
| R/R MM | Vorinostat+Bortezomib | 1 | 9 | NCT00858234 | Completed |
| R/R MM | Vorinostat+Bortezomib | 1 | 40 | NCT00310024 | Completed |
| R/R MM | Vorinostat+Bortezomib | 2 | 143 | NCT00773838 | Completed |
| R/R MM | Vorinostat+Lenalinomide+Dexamethasone | 1 | 31 | NCT00642954 | Completed |
| R/R MM | Vorinostat+Bortezomib+Doxorubicin+Dexamethasone | 1/2 | 34 | NCT01394354 | Completed |
| R/R MM | Vorinostat+Carfilzomib+Lenalidomide+Dexamethasone | 1/2 | 17 | NCT01297764 | Active, not recruiting |
| MM after ASCT | Vorinostat+Lenalinomide | 1 | 19 | NCT00729118 | Completed |
| MM after ASCT | Vorinostat+Bortezomib | 2 | 31 | NCT00839956 | Completed |
| MM | Vorinostat+Bortezomib | 3 | 637 | NCT00773747 | Completed |
| MM | Vorinostat+Bortezomib+ Dexamethasone | 2 | 16 | NCT01720875 | Completed |
| Young ND AML | Vorinostat/+Idarubicin and Cytarabine v.s. Cytarabine+Daunorubicin Hydrochloride | 3 | 754 | NCT01802333 | Completed |
| ND AML, MDS | Vorinostat+Azacitidine | 2 | 110 | NCT00948064 | Completed |
| R/R AML | Vorinostat+Temozolomide | 2 | 23 | NCT01550224 | Completed |
| R/R AML, MDS | Vorinostat+Decitabine+Cytarabine | 1 | 17 | NCT01130506 | Completed |
| R/R AML, ALL, CML | Vorinostat+Flavopiridol | 1 | 24 | NCT00278330 | Completed |
| R/R AML | Vorinostat+Decitabine+Fludarabine+Cytarabine+Filgrastim (G-CSF) | 1 | 37 | NCT03263936 | Completed |
| R/R or poor prognosis AML, MDS, CML, ALL | Vorinostat+Decitabine | 1 | 50 | NCT00357708 | Completed |
| R/R AML | Vorinostat+venetocla+azacitidine+Cytarabine+Fludarabine+Filgrastim | 1 | 40 | NCT05317403 | Not yet recruiting |
| Elder R/R AML | Vorinostat+Azacitidine+Gemtuzumab Ozogamicin | 1/2 | 52 | NCT00895934 | Completed |
| Poor-risk AML, high-risk MDS | Vorinostat+Sorafenib | 1 | 15 | NCT00875745 | Completed |
| Pediatric or young adult AML, MDS | Vorinostat+Azacytidine after alloSCT | 1 | 15 | NCT03843528 | Recruiting |
| Complex/poor-risk cytogenetics AML or FLT3-ITD | Vorinostat+Sorafenib+Bortezomib | 1/2 | 37 | NCT01534260 | Completed |
| AML | Vorinostat | 2 | 37 | NCT00305773 | Completed |
| AML, MDS | Vorinostat+Decitabine | 1 | 71 | NCT00479232 | Completed |
| AML, MDS | Vorinostat+Azacitidine | 1/2 | 135 | NCT00392353 | Active, not recruiting |
| AML, high-risk MDS | Vorinostat/+Azacitidine | 2 | 260 | NCT01617226 | Completed |
| AML, high-risk MDS | Vorinostat+Idarubicin+Cytarabine | 2 | 106 | NCT00656617 | Completed |
| R/R leukemia, MDS | Vorinostat+Idarubicin | 1 | 40 | NCT00331513 | Completed |
| AML before alloSCT | Vorinostat+Fludarabine Phosphate+Clofarabine+Busulfan | 1 | 70 | NCT02083250 | Completed |
| High-risk MDS | Vorinostat+Low Dose Cytarabine | 1/2 | 52 | NCT00776503 | Completed |
| High-risk MDS before CD3-/CD19- NK Cells Infusion | Vorinostat+Decitabine | 2 | 9 | NCT01593670 | Completed |
| High-risk MDS, CMML | Vorinostat/Lenalidomide/- +Zzacitidine | 2 | 282 | NCT01522976 | Completed |
| Acute leukemia, MDS, MPN | Vorinostat+Cytarabine+Etoposide | 1 | 25 | NCT00357305 | Completed |
| Infant ALL | Vorinostat+Bortezomib+Chemotherapy | 1/2 | 50 | NCT02553460 | Active, not recruiting |
| Accelerated phase or blastic phase CML, ALL | Vorinostat+Dasatinib | 1 | 5 | NCT00816283 | Completed |
| ND CLL/SLL | Vorinostat+Fludarabine+Cyclophosphamide+Rituximab | 1/2 | 40 | NCT00918723 | Completed |
| Reduced intensity, related donor stem cell transplant | Vorinostat+Tacrolimus+Mycophenolate | 1/2 | 61 | NCT00810602 | Completed |
| Unrelated stem cell transplant | Vorinostat+Tacrolimus+Methotrexate | 2 | 26 | NCT01790568 | Completed |
| AlloSCT | Vorinostat+Tacrolimus+Methotrexate+Mycophenolate Mofetil+Cyclophosphamide | 1/2 | 49 | NCT03842696 | Recruiting |
| Stem cell transplant | Vorinostat+Tacrolimus+Methotrexate | 2 | 12 | NCT01789255 | Completed |
| Belinostat | |||||
| PTCL | Belinostat+CHOP | 1 | 23 | NCT01839097 | Completed |
| R/R PTCL | Belinostat | 2 | 129 | NCT00865969 | Completed |
| R/R BL, DLBCL | Belinostat | 2 | 22 | NCT00303953 | Completed |
| R/R NHL | Belinostat+Carfilzomib | 1 | 19 | NCT02142530 | Completed |
| Relapsed aggressive high-risk lymphoma | Belinostat+Rituximab+Yttrium Y 90 Ibritumomab Tiuxetan | 2 | 5 | NCT01686165 | Completed |
| Adult T-cell leukemia/lymphoma | Belinostat+Zidovudine | 2 | 20 | NCT02737046 | Recruiting |
| R/R AML | Belinostat+Pevonedistat | 1 | 30 | NCT03772925 | Recruiting |
| AML unsuitable for standard intensive therapy | Belinostat+Idarubicin | 1/2 | 41 | NCT00878722 | Completed |
| AML | Belinostat | 2 | 12 | NCT00357032 | Completed |
| R/R acute leukemia, MDS | Belinostat+Bortezomib | 1 | 41 | NCT01075425 | Completed |
| AML, ALL, CML, MDS | Belinostat+Azacitidine | 1 | 56 | NCT00351975 | Completed |
| MDS | Belinostat | 2 | 21 | NCT00357162 | Completed |
| Advanced MM | Belinostat+Dexamethsone | 2 | 25 | NCT00131261 | Completed |
| Panobinostat | |||||
| R/R MM | Panobinostat+Bortezomib+Dexamethasone | 3 | 767 | NCT01023308 | Completed |
| R/R MM | Panobinostat+Bortezomib+Dexamethasone | 2 | 249 | NCT02654990 | Active, not recruiting |
| R/R MM | Panobinostat+Bortezomib+Dexamethasone | 2 | 31 | NCT02290431 | Completed |
| Relapsed and bortezomib-refractory MM | Panobinostat+Bortezomib+Dexamethasone | 2 | 55 | NCT01083602 | Completed |
| R/R MM | Panobinostat+Bortezomib | 1 | 62 | NCT00532389 | Completed |
| R/R MM | Panobinostat+Carfilzomib | 1/2 | 80 | NCT01496118 | Completed |
| R/R MM | Panobinostat+Carfilzomib | 1 | 32 | NCT01549431 | Completed |
| R/R MM | Panobinostat+Carfilzomib | 1 | 46 | NCT01301807 | Active, not recruiting |
| R/R MM | Panobinostat+Lenalidomide+Dexamethasone | 2 | 32 | NCT01651039 | Completed |
| R/R MM | Panobinostat+Bortezomib+Lenalidomide+Dexamethasone | 1 | 21 | NCT01965353 | Completed |
| R/R MM | Panobinostat+Daratumumab+Bortezomib+Dexamethasone | 1 | 27 | NCT04956302 | Recruiting |
| Relapsed MM | Panobinostat+Melphalan | 1/2 | 40 | NCT00743288 | Completed |
| Adult MM | Panobinostat+Lenalidomide+Dexamethasone | 1 | 46 | NCT00532675 | Completed |
| ND transplant-eligible MM | Panobinostat+Bortezomib+Lenalidomide+Dexamethasone | 1 | 77 | NCT01440582 | Completed |
| R/R MM before autoSCT | Panobinostat+Gemcitabine+Busulfan+Melphalan | 2 | 83 | NCT02506959 | Active, not recruiting |
| MM after autoSCT | Panobinostat | 2 | 30 | NCT02722941 | Active, not recruiting |
| Recurrent MM, lymphoma | Panobinostat+Everolimus | 1/2 | 124 | NCT00918333 | Completed |
| R/R MM, lymphoma | Panobinostat+Everolimus | 1 | 11 | NCT00962507 | Completed |
| Refractory CTCL | Panobinostat | 2 | 139 | NCT00425555 | Completed |
| R/R PTCL, ENKT | Panobinostat+Bortezomib | 2 | 25 | NCT00901147 | Completed |
| R/R MCL | Panobinostat+Bortezomib | 1 | 3 | NCT01504776 | Completed |
| R/R DLBCL | Panobinostat | 2 | 35 | NCT01523834 | Completed |
| R/R WM | Panobinostat | 2 | 39 | NCT00936611 | Completed |
| R/R NHL | Panobinostat | 2 | 41 | NCT01261247 | Completed |
| R/R classical HL | Panobinostat | 2 | 129 | NCT00742027 | Completed |
| R/R HL | Panobinostat+Lenalidomide | 2 | 24 | NCT01460940 | Completed |
| Relapsed HL | Panobinostat/+Ifosfamide+Carboplatin+Etoposide | 1/2 | 62 | NCT01169636 | Completed |
| HL maintenance | Panobinostat | 3 | 41 | NCT01034163 | Completed |
| R/R lymphoma | Panobinostat+Everolimus | 1/2 | 31 | NCT00967044 | Completed |
| Pediatric lymphoma, AML, ALL | Panobinostat for lymphoma, Panobinostat+Cytarabine for Leukemia | 1 | 30 | NCT01321346 | Completed |
| High-risk AML, MDS after HSCT | Panobinostat | 3 | 52 | NCT04326764 | Active, not recruiting |
| AML younger than 65 years old | Panobinostat+Chemotherapy | 1 | 40 | NCT01242774 | Completed |
| ND AML older than 65 years old | Panobinostat+Idarubicin+Cytarabine | 1/2 | 46 | NCT00840346 | Completed |
| ND AML, advanced MDS | Panobinostat+Daunorubicin+Cytarabine | 1 | 29 | NCT01463046 | Completed |
| Refractory AML | Panobinostat | 2 | 59 | NCT00880269 | Completed |
| R/R AML | Panobinostat+Azacitidine+Mitoxantrone | 1 | 59 | NCT01055483 | Completed |
| AML, MDS | Panobinostat+Decitabine | 1/2 | 52 | NCT00691938 | Completed |
| AML, MDS, CMML | Panobinostat+Azacitidine | 1 | 10 | NCT01613976 | Completed |
| AML, MDS, CMML | Panobinostat+Azacitidine | 1/2 | 113 | NCT00946647 | Completed |
| Previously treated chronic phase CML | Panobinostat+Imatinib | 1 | 9 | NCT00686218 | Completed |
| MF | Panobinostat+Ruxolitinib | 1 | 61 | NCT01433445 | Completed |
| MF | Panobinostat+Ruxolitinib | 1/2 | 20 | NCT01693601 | Completed |
| MF, PV, GVHD, AML | Panobinostat/+Ruxolitinib | 4 | 298 | NCT02386800 | Recruiting |
| Advanced hematologic malignancies | Panobinostat | 1/2 | 175 | NCT00621244 | Completed |
| AlloSCT | Panobinostat+Sirolimus+Tacrolimus | 2 | 42 | NCT02588339 | Completed |
| AlloSCT | Panobinostat+Corticosteroids | 1/2 | 22 | NCT01111526 | Completed |
| Chidamide | |||||
| ND PTCL | Chidamide+CHOP | 1 | 30 | NCT02809573 | Completed |
| ND PTCL | Chidamide+CHOEP | 1/2 | 100 | NCT02987244 | Recruiting |
| ND PTCL | Chidamide+Azacitidine/+CHOP | 3 | 107 | NCT05075460 | Not yet recruiting |
| ND PTCL unfit for conventional chemotherapy | Chidamide+Azacitidine | 2 | 28 | NCT04480125 | Recruiting |
| R/R PTCL | Chidamide+Parsaclisib | 1/2 | 28 | NCT05083208 | Not yet recruiting |
| R/R PTCL | Chidamide+Lenalidomide | 2 | 44 | NCT04329130 | Recruiting |
| R/R PTCL | Chidamide+Sintilimab | 2 | 51 | NCT04512534 | Recruiting |
| R/R PTCL | Chidamide+Sintilimab+Azacitidine | 2 | 30 | NCT04052659 | Not yet recruiting |
| R/R PTCL | Chidamide v.s. Mitoxantrone Hydrochloride Liposome Injection | 3 | 190 | NCT04668690 | Not yet recruiting |
| ND AITL | Chidamide+CHOP | 2 | 23 | NCT03853044 | Active, not recruiting |
| R/R ATIL | Chidamide+Azacitidine | 2 | 20 | NCT05179213 | Not yet recruiting |
| R/R AITL | Chidamide+Sintilimab | 2 | 83 | NCT04831710 | Not yet recruiting |
| R/R AITL | Chidamide+Rituximab+Lenalidomide | NA | 26 | NCT04319601 | Recruiting |
| ND ENKT | Chidamide+Sintilimab | 2 | 30 | NCT04994210 | Recruiting |
| R/R ENKT | Chidamide+Sintilimab | 1/2 | 40 | NCT03820596 | Completed |
| Stage I/II ENKT | Chidamide/+Gemcitabine+Cisplatin+Dexamethasone+Radiotherapy | NA | 76 | NCT04511351 | Recruiting |
| R/R CTCL | Chidamide+Sintilimab | 2 | 52 | NCT04296786 | Recruiting |
| T-NHL before autoSCT | Chidamide+Carmustine+Etoposide+Cytarabine+Melphalan | 2 | 23 | NCT05367856 | Not yet recruiting |
| ND double-expressor DLBCL | Chidamide/+RCHOP | 3 | 418 | NCT04231448 | Recruiting |
| R/R DLBCL | Chidamide+Rituximab+Anti-PD-1 Antibody | 2 | 27 | NCT05115409 | Not yet recruiting |
| R/R transplant-ineligible DLBCL | Chidamide+Rituximab+Gemcitabine+Oxaliplatin | 2 | 54 | NCT04022005 | Recruiting |
| R/R B-NHL | Chidamide | 2 | 100 | NCT03245905 | Recruiting |
| R/R B-NHL before CAR-T | Chidamide/+Fludarabine+Cyclophosphamide | 1/2 | 120 | NCT05370547 | Recruiting |
| Aggressive R/R B-NHL undergoing decitabine-primed tandem CD19/CD20 CAR-T | Chidamide v.s. Decitabine v.s. Chidamide+Decitabine | 1/2 | 80 | NCT04553393 | Recruiting |
| R/R NHL | Chidamide+Chiauranib | 1/2 | 9 | NCT03974243 | Completed |
| R/R NHL | Chidamide+Decitabine+Immune checkpoint inhibitors | 1/2 | 100 | NCT05320640 | Recruiting |
| NHL who relapsed after CAR-T | Chidamide+Decitabine | 1/2 | 100 | NCT04337606 | Recruiting |
| NHL | Chidamide | 1 | 13 | NCT02697552 | Completed |
| ND primary central nervous system lymphoma | Chidamide+Rituximab+Methotrexate | 2 | 51 | NCT04516655 | Not yet recruiting |
| R/R classical HL | Chidamide+Decitabine+Camrelizumab | 2 | 100 | NCT04233294 | Recruiting |
| Anti-PD-1 antibody-resistant classical HL | Chidamide/+Decitabine+Camrelizumab | 2 | 200 | NCT04514081 | Recruiting |
| Aggressive lymphoma | Chidamide+BEAC+autoSCT | 2 | 69 | NCT03629873 | Active, not recruiting |
| R/R MM | Chidamide+Lenalidomide+Dexamethasone | 2 | 25 | NCT03605056 | Not yet recruiting |
| Primary high-risk MM | Chidamide/+Bortezomib+Lenalidomide+Dexamethasone | 1/2 | 50 | NCT04025450 | Recruiting |
| CBF AML | Chidamide v.s. Cytarabine | 1/2 | 250 | NCT03031262 | Recruiting |
| R/R AML | Chidamide+Azacitidine+Venetoclax | 2 | 30 | NCT05305859 | Not yet recruiting |
| R/R AML | Chidamide+Cladribine | 2 | 31 | NCT05330364 | Recruiting |
| R/R AML | Chidamide+Decitabine+Priming IAG | 2 | 40 | NCT03985007 | Completed |
| R/R AML | Chidamide/+Azacitidine+HAG | 2 | 210 | NCT05029141 | Recruiting |
| AML after transplant | Chidamide+Azacitidine | 1/2 | 20 | NCT05270200 | Recruiting |
| T-ALL | Chidamide+Ruxolitinib | 1/2 | 50 | NCT05075681 | Recruiting |
| B-ALL undergoing haploidentical alloSCT | Chidamide+Ruxolitinib | 2 | 50 | NCT05088226 | Recruiting |
| Adult Ph-like ALL | Chidamide v.s. Dasatinib | 2/3 | 120 | NCT03564470 | Recruiting |
| HLH | Chidamide+VP-16+Methylprednisolone | NA | 20 | NCT05137522 | Recruiting |
| ENKT-HLH | Chidamide+Sintilimab v.s Azacitidine+Sintilimab v.s. L-DEP | 2 | 37 | NCT05008666 | Not yet recruiting |
| Steroid-resistant/steroid-dependent Severe cGVHD | Chidamide | 1/2 | 20 | NCT05140616 | Recruiting |
| Romidepsin | |||||
| Conditions | Interventions | Phase | Patients | NCT number | Status |
| ND PTCL | Romidepsin/+CHOP | 3 | 421 | NCT01796002 | Active, not recruiting |
| ND PTCL | Romidepsin+Lenalidomide | 2 | 35 | NCT02232516 | Active, not recruiting |
| ND PTCL | Romidepsin+CHOP | 1/2 | 37 | NCT01280526 | Completed |
| ND young nodal PTCL before HSCT | Romidepsin+CHOEP | 1/2 | 89 | NCT02223208 | Active, not recruiting |
| R/R PTCL | Romidepsin+Ixazomib | 1/2 | 11 | NCT03547700 | Active, not recruiting |
| R/R PTCL | Romidepsin+Carfilzomib | 1/2 | 50 | NCT03141203 | Completed |
| R/R PTCL | Romidepsin+Gemcitabine | 2 | 20 | NCT01822886 | Completed |
| R/R PTCL | Romidepsin+Pembrolizumab | 1/2 | 39 | NCT03278782 | Recruiting |
| R/R PTCL | Romidepsin+Ifosfamide+Carboplatin+Etoposide | 1 | 22 | NCT01590732 | Completed |
| Progressive or relapsed PTCL | Romidepsin | 2 | 131 | NCT00426764 | Completed |
| Progressive or relapsed PTCL | Romidepsin | 1/2 | 51 | NCT01456039 | Completed |
| PTCL, CTCL | Romidepsin | 2 | 131 | NCT00007345 | Completed |
| R/R mature TCL | Romidepsin+Venetoclax | 2 | 9 | NCT03534180 | Active, not recruiting |
| R/R TCL | Romidepsin+Tenalisib | 1/2 | 33 | NCT03770000 | Completed |
| R/R TCL | Romidepsin+Parsaclisib | 1 | 20 | NCT04774068 | Recruiting |
| R/R TCL | Romidepsin+Duvelisib v.s. Bortezomib+Duvelisib | 1 | 114 | NCT02783625 | Active, not recruiting |
| R/R TCL | Romidepsin+Azacitidine+Lenalidomide+Dexamethasone | 1 | 30 | NCT04447027 | Recruiting |
| R/R AITL | Romidepsin+Sintilimab | 2 | 83 | NCT04831710 | Not yet recruiting |
| R/R ENKT | Romidepsin | 1 | 16 | NCT01913119 | Completed |
| TCL | Romidepsin+Azacitidine+Duvelisib | 1 | 60 | NCT04639843 | Not yet recruiting |
| TCL, T-ALL after alloSCT | Romidepsin | 1 | 10 | NCT02512497 | Recruiting |
| Previously treated PTCL, DLBCL | Romidepsin+Gemcitabine+Dexamethasone+Cisplatin | 1 | 21 | NCT01846390 | Completed |
| CTCL | Romidepsin+Brentuximab Vedotin | 1 | 27 | NCT02616965 | Recruiting |
| R/R CTCL | Romidepsin+doxorubicin HCl liposomal | 1 | 24 | NCT01902225 | Completed |
| R/R CTCL | Romidepsin | 2 | 102 | NCT00106431 | Completed |
| T-NHL after autoSCT | Romidepsin maintenance | 2 | 47 | NCT01908777 | Active, not recruiting |
| R/R NHL | Romidepsin | 2 | 35 | NCT00077194 | Completed |
| R/R NHL | Romidepsin+Alisertib | 1 | 26 | NCT01897012 | Completed |
| R/R NHL | Romidepsin+Carfilzomib+Lenalidomide | 1/2 | 31 | NCT02341014 | Active, not recruiting |
| R/R aggressive lymphoma | Romidepsin+Gemcitabine+Oxaliplatin+Dexamethasone | 1 | 24 | NCT02181218 | Completed |
| R/R lymphoma, myeloma | Romidepsin+Lenalidomide | 1/2 | 62 | NCT01755975 | Active, not recruiting |
| R/R MM | Romidepsin | 2 | 50 | NCT00066638 | Completed |
| R/R AML | Romidepsin | 2 | 47 | NCT00062075 | Completed |
| R/R leukemia, MDS, MPN | Romidepsin+Decitabine | 1 | 36 | NCT00114257 | Completed |
| R/R CLL/SLL | Romidepsin+Bortezomib | 1 | 18 | NCT00963274 | Completed |
AITL angioimmunoblastic T-cell lymphoma, ALL acute lymphoblastic leukemia, alloSCT allogenic stem cell transplantation, AML acute myeloid leukemia, ASCT autologous stem cell transplantation, BL Burkitt’s lymphoma, CLL chronic lymphocytic leukemia, CML chronic myeloid leukemia, CTCL cutaneous T-cell lymphoma/leukemia, DLBCL diffuse large B-cell lymphoma, ENKT extranodal NK/T-cell lymphoma, FL follicular lymphoma, GVHD graft-versus-host disease, HL Hodgkin lymphoma, ITD internal tandem duplication, MCL mantle cell lymphoma, MDS myelodysplastic syndrome, MF myelofibrosis, MM multiple myeloma, ND newly diagnosed, NHL non-Hodgkin lymphoma, PV polycythemia vera, PTCL peripheral T-cell lymphoma, R rituximab, R/R relapsed/refractory, SLL small lymphocytic lymphoma
In the benzamide group, chidamide, also named CS055 or tucidinostat, inhibits HDAC1, HDAC2, HDAC3, and HDAC10, and has been approved by the China NMPA and EMA to treat R/R PTCL.674,675 For R/R PTCL patients, the ORR of chidamide was 28%, and the median OS was 21.4 months, with good tolerance in clinical trials.676 In a real-world study, the ORR of chidamide monotherapy was 39.06%, and that of chidamide combined with chemotherapy was 51.18% in R/R PTCL.677 In addition, there are numerous ongoing clinical trials exploring its efficacy in NHL (mostly T-cell lymphoma), AML, ALL, MM, and hemophagocytic lymphohistiocytosis.678 In addition to the approved chidamide, other benzamide-based HDAC inhibitors are being explored in clinical trials. Entinostat, also known as MS275, inhibits HDAC1, HDAC3, and HDAC8. Preclinical studies revealed that entinostat relieved the epigenetic silencing of LAT2 caused by AML1/ETO, and inhibited leukemic maintenance.679–681 Entinostat stimulated apoptosis alone and synergized with rituximab in B-cell lymphoma, with BCL2 inhibitors in HL, and with bendamustine in MM.682–684 Various clinical trials are investigating its efficacy in MDS, AML, CMML, and lymphoma. However, bendamustine combined with azacytidine displayed worse ORR and OS in AML and MDS compared with azacytidine treatment alone, indicating antagonism and higher toxicity of the two agents in this setting.685,686 Mocetinostat, also named MGCD0103, inhibits HDAC1, HDAC2, HDAC3, and HDAC11. Mocetinostat triggered H3 and H4 acetylation and exerted significant antitumor effects in various cancer cells.351 Various clinical trials have explored its effect on R/R lymphoma and AML. Domatinostat, also named 4SC-202, inhibits HDAC1, HDAC2, and HDAC3, and displayed a manageable safety profile with signs of antitumor effects in a phase I trial enrolling patients with advanced hematological malignancies.687 Tacedinaline, also named CI-994, is a selective HDAC1 inhibitor and has been explored in advanced MM patients.
In the cyclic peptide group, romidepsin, also named FK228 or depsipeptide, inhibits HDAC1 and HDAC2 and was the second HDAC inhibitor approved by the U.S. FDA to treat CTCL and PTCL. Romidepsin inhibited proliferation and angiogenesis and promoted cell cycle arrest and apoptosis in various cancer cells.688 For previously treated or refractory CTCL patients, romidepsin monotherapy displayed an ORR of 34%.689,690 For R/R PTCL patients, romidepsin monotherapy displayed an ORR of 38%.691 Numerous ongoing clinical trials are investigating romidepsin combined with anti-PD-1 monoclonal antibody, PIs, PI3K inhibitors, BCL2 inhibitors, IMiDs, and chemotherapy in R/R T-cell NHL.
Dual-target HDAC inhibitors
In addition to the classic HDAC inhibitors, dual-target HDAC inhibitors have also demonstrated favorable antitumor effects in preclinical models. CUDC-101, an HDAC I/receptor tyrosine kinase (RTK) bifunctional inhibitor, inhibits HDAC I, EGFR, and HER2 and has been demonstrated effective in preclinical models of gemcitabine-treated lymphoma, APL, and several solid tumors.692,693 Several clinical trials have investigated its role in solid tumors, while no trials on hematologic malignancies have been conducted. An HDAC6/JAK2 inhibitor was designed based on vorinostat and pacritinib, and showed preclinical efficacy in AML and ALL.694 CUDC-907, an HDAC/PI3K inhibitor, exerted an ORR of 37% in R/R DLBCL patients, including those with MYC-alterations.695 HDAC/LSD1 inhibitors and HDAC I/tubulin inhibitors display favorable efficacy in solid tumors, such as colon cancer and breast cancer, but few studies have explored their application to hematologic malignancies.696,697
PROTACs for HDACs
The reversibility of classic small-molecule HDAC inhibitors requires sustainable exposure to a high in vivo drug concentration to maintain sufficient inhibition. However, maintaining a high in vivo drug concentration is sometimes challenging. Proteolysis-targeting chimeras (PROTACs) provide an innovative strategy for degrading HDACs more persistently. A PROTAC is a small bifunctional compound consisting of a ligand for the target protein, an E3 ligase recognition moiety, and a linker. After the ligand binds with the target protein on one side, the E3 ligase on the other side mediates the ubiquitination of the target protein by the ubiquitin-conjugating E2 enzyme. The target protein labeled with ubiquitin is recognized and degraded by proteasomes. The ubiquitination and degradation process are highly efficient and recyclable, making PROTACs a promising strategy to degrade HDACs. Several HDAC-targeted PROTACs have been developed in recent years.
According to the E3 ligase complex, these PROTACs are mainly divided into cereblon (CRBN)-based PROTACs and Von Hippel-Lindau (VHL)-based PROTACs. The first-in-class HDAC6-targeted PROTAC, compound 9c, was developed by conjugating a pan-HDAC nonselective inhibitor to thalidomide as CRBN. The concentration of half-maximal degradation (DC50) for HDAC6 was 34 nM, and HDAC6 was significantly degraded by 9c at a concentration of 80 nM in multiple myeloma MM.1S cells. However, other HDACs were also inhibited by 9c, indicating the need for HDAC6-targeted PROTACs with higher selectivity and specificity.698 Later, HDAC6-targeted PROTACs with higher selectivity were developed by conjugating a selective HDAC6 inhibitor, nexturastat A, to pomalidomide as CRBN. The degrader, NP8, exerted efficient HDAC6 degradation in MM.1S cells with a DC50 of 3.8 nM without affecting other types of HDACs.699 Another degrader, NH2, exerted similarly efficient HDAC6 degradation in MM.1S cells with a DC50 of 3.2 nM without affecting other types of HDACs. Moreover, the onset of degradation occurred within an hour and peaked at 6–8 h, and HDAC6 rapidly recovered three hours after washout, indicating its efficient and reversible capability to degrade HDAC6.700
In addition to CRBN-based PROTACs, VHL-based PROTACs targeting HDAC have also been developed. An HDAC6 degrader, compound 3j, was designed by conjugating nexturastat A to VHL, which showed robust HDAC6 degradation with a DC50 of 7.1 nM in MM.1S cells. Furthermore, the maximal degradation (Dmax) of HDAC6 reached up to 90% in MM.1S cells.701 An HDAC3 degrader, XZ9002, was developed by conjugating a selective HDAC I inhibitor, SR-5228, to VHL. The DC50 of HDAC3 in breast cancer MDA-MB-468 cells was 42 nM.702
Combination treatment of HDAC inhibitors with HMAs
Inspired by the encouraging efficacy of HMAs in treating leukemia, numbers of clinical trials have explored the combination treatment of HMAs and HDAC inhibitors in AML and ALL. The majority of clinical trials focused on AML. However, most results were disappointing. A phase I study explored the combination of the HDAC inhibitor AR-42 with decitabine in 13 newly diagnosed or relapsed/refractory AML patients. The ORR was only 23.1% with dose-limiting toxicities occurring at the third dosage. The results were not satisfying, probably due to the small sample size, high-risk baseline clinical features of participants, and the relatively mild HDAC inhibition capability of AR-42.624 A phase I study investigated decitabine combined with the HDAC inhibitor valproic acid or not in 25 AML patients. The ORR of decitabine monotherapy group was similar to that in combination treatment group. The addition of valproic acid to decitabine showed little treatment effect bonus, but caused encephalopathy. The evident effect of valproic acid on central nervous system, as evidenced by its approved indications as seizures, narrowed its application in combination treatment in AML patients.703 The addition of valproate to decitabine in AML treatment was proved insignificant again in a phase II trial with larger sample size.704 Moreover, the combination treatment of decitabine even with the commonly recognized effective HDAC inhibitor vorinostat still exerted limited ORR (23%) in AML patients.705 In relapsed ALL, combined treatment of vorinostat with decitabine exerted an ORR of 39–46.2%, which was higher than that in AML. However, treatment-related infections were common.706,707 The failure of combination treatment of HDAC inhibitors with HMAs could be attributable to the potential epigenetic antagonism. Several clinical trials are still ongoing in PTCL and DLBCL. Extensive studies are required to thoroughly reveal the exact mechanism and interaction between the two epigenetic therapies.
Combination treatment of HDAC inhibitors with immune treatment
HDACs can regulate both innate and adaptive immune response. In innate immune response, HDACs could promote or inhibit Toll-like receptor (TLR) signaling pathways determined by the types of HDACs and diseases, leading to either an increase or decrease in cytokine and chemokine secretion. Regarding adaptive immune response, HDAC inhibitors could upregulate MHC-I and promote antigen processing, which amplified the antitumor reaction.708 Panobinostat has been reported to activate Notch pathway and enhance the anti-leukemic influence of human γδT cells in vitro,709 and could exert synergistic effect with interferon-α in mouse models.710 Trichostatin A, an HDAC inhibitor, has also been proved to induce dendritic cell differentiation from AML leukemic blasts in vitro.711 These encouraging preclinical findings indicate the promising application of combination treatment of HDAC inhibitors and immune treatment in hematologic malignancies. The number of clinical trials are investigating the combined treatment of HDAC inhibitors with rituximab in B-cell lymphoma, with lenalidomide in MM, and with brentuximab vedotin in CTCL patients. Various ongoing clinical trials are also exploring the combined treatment of HDAC inhibitors with PD-1 monoclonal antibodies in the treatment of HL, DLBCL, FL, PTCL, CTCL, AITL, ENKT, and HLH secondary to ENKT (Table 4). Their results are widely anticipated, and will better reveal the optimal individualized treatment of hematologic malignancies.
Targeting other histone modification agents
Although HDAC inhibitors are one of the most explored and applied epigenetic regimens, agents targeting other histone modification agents are also being developed (Fig. 1).
Regarding histone acetylation, agents targeting writers mainly include CBP/P300 inhibitors. CCS1477, a CBP inhibitor, is being explored to treat NHL, MM, AML, and MDS in a phase I/IIa trial (NCT04068597).712 Agents targeting readers include BET inhibitors. OTX015 inhibited proliferation and stimulated cell cycle arrest and apoptosis in leukemia cells. It also downregulated BRD2, BRD4, and MYC in vitro.713 In lymphoma, OTX015 stimulated the apoptosis of DLBCL cells with mutations in MYD88, CD79B, or CARD11, and repressed MYC- and E2F1-related expression.714 OTX015 showed mild efficacy in acute leukemia and lymphoma patients, while no efficacy was found in MM patients.715 CPI-0610, GSK525762, RO6870810, and FT-1101 are being explored in hematologic malignancies in clinical trials.716 Apart from the classic BET inhibitors, BET-targeted PROTACs have been reported. ARV-825 was developed by conjugating the BRD4 inhibitor OTX015 to pomalidomide as CRBN. ARV0825 exerted rapid, efficient and sustained degradation of BRD4 and lethal activity compared to BET inhibitors in Burkitt lymphoma cells, mantle cell lymphoma cells, post-MPN secondary AML cells, and T-cell ALL cells.717–721 dBET1 was generated by conjugating the BRD4 inhibitor, JQ1, to phthalimide as CRBN. The efficient and specific degradation of BRD2, BRD3, and BRD4 was accompanied by superior apoptosis induction in AML cells and xenograft models.722
For histone methylation, agents targeting writers include EZH2 inhibitors and DOT1L inhibitors. EZH2 inhibitors in the clinical stage include tazemetostat, CPI-1205, SHR2554, and PF-06821497. Tazemetostat has been approved by the U.S. FDA to treat metastatic or advanced epithelioid sarcoma. Although most of the EZH2 inhibitor clinical trials focus on solid tumors, several trials are investigating its effect on R/R B-NHL.723 The DOT1L inhibitor, pinometostat, also named EPZ5676, has been demonstrated to impair H3K79 methylation and moderately effective in treating MLL-rearranged leukemia.724 Agents targeting histone methylation erasers mainly include LSD1 inhibitors, also named KDM1A inhibitors. ORY-1001, TCP, and GSK2879552 are being investigated in the treatment of R/R AML in clinical trials. INCB059872 and IMG-7289 have been explored in MPN trials. CC-90011 was investigated in R/R NHL.725
In terms of histone phosphorylation, the JAK2 inhibitors (ruxolitinib, fedratinib, and pacritinib) have been approved by the U.S FDA to treat MPN.726 Various clinical trials have been investigating their role in CMML, CLL, ALL, and post-MPN AML.727,728 In addition, aurora inhibitors have been developed and have shown synergism with docetaxel in apoptosis stimulation to inhibit lymphoma.729,730
Conclusion
In conclusion, epigenetic regulation plays a fundamental role in hematopoiesis and oncogenesis of hematologic malignancies. Abnormal DNA methylation profiles, including genome-wide hypomethylation and aberrant hypermethylation or hypomethylation of CpG islands, are frequently observed in hematologic malignancies. Upregulation or mutations of DNA methylation writers (DNMT1, DNMT3A, and DNMT3B) are pathogenic in AML. Readers of DNA methylation mainly consist of MBD-containing proteins, methyl-CpG binding zing fingers, and SRA domain-containing proteins, whose upregulation is frequently identified in CML, NHL, and AML, respectively. DNA methylation erasers mainly include the TET family, whose mutations, translocation, and upregulation are relatively common in AML. Correspondingly, HMAs have been developed to target aberrant DNA methylation profiles and have been demonstrated effective to treat MDS, AML, and CMML. Furthermore, histone acetylation and methylation are involved in hematologic oncogenesis. KATs are histone acetylation writers, whose mutations are detected in DLBCL and translocations are identified in AML. HDACs are histone acetylation erasers, whose aberrant expression has been demonstrated in lymphoma. Correspondingly, HDAC inhibitors have been proposed and demonstrated effective in treatment of CTCL, PTCL, and MM. Mutations, translocations, and aberrant expression of histone methylation writers, KMTs, and histone methylation erasers, KDMs, are also found in AML and other hematologic malignancies. Dysregulation of miRNAs and lncRNAs also contributes to hematologic oncogenesis. In addition to DNMT inhibitors and HDAC inhibitors, innovative epigenetic treatment targeting KATs, BETs, KMTs, KDMs, and ncRNAs are emerging and will provide novel treatment strategies in hematologic malignancies.
However, the relationship of cancer epigenetics with interdependent mechanisms like cancer immunology or metabolism should be explored further. In the future, more accurate diagnostic and prognostic information detected by high-throughput genomic technologies can provide more precise and individualized therapeutic options. Moreover, the ncRNA could possibly serve as diagnostic and prognostic molecular biomarkers in the future clinical settings. With the rapid growth of associated scientific research methods and technologies in this academic sector, ncRNA-targeted therapy may soon be a realistic treatment option for patients with hematologic malignancies.
Supplementary information
Acknowledgements
This work is supported by the National Natural Science Foundation of China (82204490 to A.Z.), the Translational Research Grant of NCRCH (2021WWB03 to T.N.), Achievement Transformation Project (CGZH21001 to T.N.), and 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (ZYJC21007 to T.N.).
Author contributions
A.Z., H.Z., and J.Y. prepared the manuscript. A.Z., H.Z., J.Y., and M.L. wrote the main parts of the article. M.L., A.Z., and T.N. produced graphics. A.Z. and T.N. performed critical editing to the manuscript. T.N. drafted the final version of the manuscript. All authors have read and approved the article.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Ailin Zhao, Hui Zhou, Jinrong Yang
Supplementary information
The online version contains supplementary material available at 10.1038/s41392-023-01342-6.
References
- 1.Avery OT, Macleod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarty M, Avery OT. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: II. Effect of desoxyribonuclease on the biological activity of the transforming substance. J. Exp. Med. 1946;83:89–96. doi: 10.1084/jem.83.2.89. [DOI] [PubMed] [Google Scholar]
- 3.Bestor TH. Cloning of a mammalian DNA methyltransferase. Gene. 1988;74:9–12. doi: 10.1016/0378-1119(88)90238-7. [DOI] [PubMed] [Google Scholar]
- 4.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 5.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl Acad. Sci. USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maunakea AK, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bird A, Taggart M, Frommer M, Miller OJ, Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40:91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 8.Schulz WA, Steinhoff C, Florl AR. Methylation of endogenous human retroelements in health and disease. Curr. Top. Microbiol. Immunol. 2006;310:211–250. doi: 10.1007/3-540-31181-5_11. [DOI] [PubMed] [Google Scholar]
- 9.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl Acad. Sci. USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 12.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 13.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karatzas PS, Mantzaris GJ, Safioleas M, Gazouli M. DNA methylation profile of genes involved in inflammation and autoimmunity in inflammatory bowel disease. Medicine. 2014;93:e309. doi: 10.1097/MD.0000000000000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Easwaran H, Tsai HC, Baylin SB. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol. Cell. 2014;54:716–727. doi: 10.1016/j.molcel.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulis M, Esteller M. DNA methylation and cancer. Adv. Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 17.Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nat. Rev. Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 19.Laird PW, Jaenisch R. DNA methylation and cancer. Hum. Mol. Genet. 1994;3(Spec No):1487–1495. doi: 10.1093/hmg/3.suppl_1.1487. [DOI] [PubMed] [Google Scholar]
- 20.Aran D, Sabato S, Hellman A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol. 2013;14:R21. doi: 10.1186/gb-2013-14-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taberlay PC, et al. Three-dimensional disorganization of the cancer genome occurs coincident with long-range genetic and epigenetic alterations. Genome Res. 2016;26:719–731. doi: 10.1101/gr.201517.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zebardast S, et al. The gene expression profile and DNA methylation pattern of cdh1 and DNMT1 genes in acute promyelocytic leukemia (APL) Rep. Biochem. Mol. Biol. 2020;8:454–457. [PMC free article] [PubMed] [Google Scholar]
- 23.Rahmani T, et al. Patterns of DNMT1 promoter methylation in patients with acute lymphoblastic leukemia. Int. J. Hematol. Oncol. Stem Cell Res. 2017;11:172–177. [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang YY, et al. DNMT3A intragenic hypomethylation is associated with adverse prognosis in acute myeloid leukemia. Leuk. Res. 2015;39:1041–1047. doi: 10.1016/j.leukres.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 25.Šestáková Š, et al. DNA methylation and hydroxymethylation patterns in acute myeloid leukemia patients with mutations in DNMT3A and IDH1/2 and their combinations. Cancer Biomark. 2019;25:43–51. doi: 10.3233/CBM-182176. [DOI] [PubMed] [Google Scholar]
- 26.Hajkova H, et al. Decreased DNA methylation in acute myeloid leukemia patients with DNMT3A mutations and prognostic implications of DNA methylation. Leuk. Res. 2012;36:1128–1133. doi: 10.1016/j.leukres.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 27.Yang L, et al. DNMT3A loss drives enhancer hypomethylation in FLT3-ITD-associated leukemias. Cancer Cell. 2016;30:363–365. doi: 10.1016/j.ccell.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 28.Yan XJ, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat. Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 29.Yamazaki J, et al. Effects of TET2 mutations on DNA methylation in chronic myelomonocytic leukemia. Epigenetics. 2012;7:201–207. doi: 10.4161/epi.7.2.19015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bensberg M, et al. TET2 as a tumor suppressor and therapeutic target in T-cell acute lymphoblastic leukemia. Proc. Natl Acad. Sci. USA. 2021;118:e2110758118. doi: 10.1073/pnas.2110758118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayette S, et al. High DNA methyltransferase DNMT3B levels: a poor prognostic marker in acute myeloid leukemia. PLoS ONE. 2012;7:e51527. doi: 10.1371/journal.pone.0051527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roux B, et al. Aberrant DNA methylation impacts HOX genes expression in bone marrow mesenchymal stromal cells of myelodysplastic syndromes and de novo acute myeloid leukemia. Cancer Gene Ther. 2022;29:1263–1275. doi: 10.1038/s41417-022-00441-w. [DOI] [PubMed] [Google Scholar]
- 33.Kn H, Bassal S, Tikellis C, El-Osta A. Expression analysis of the epigenetic methyltransferases and methyl-CpG binding protein families in the normal B-cell and B-cell chronic lymphocytic leukemia (CLL) Cancer Biol. Ther. 2004;3:989–994. doi: 10.4161/cbt.3.10.1137. [DOI] [PubMed] [Google Scholar]
- 34.Jiang H, et al. DNA methylation markers in the diagnosis and prognosis of common leukemias. Signal Transduct. Target Ther. 2020;5:3. doi: 10.1038/s41392-019-0090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 36.Ooi SK, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 38.Pradhan S, Bacolla A, Wells RD, Roberts RJ. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem. 1999;274:33002–33010. doi: 10.1074/jbc.274.46.33002. [DOI] [PubMed] [Google Scholar]
- 39.Ramsahoye BH, et al. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl Acad. Sci. USA. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mortusewicz O, Schermelleh L, Walter J, Cardoso MC, Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl Acad. Sci. USA. 2005;102:8905–8909. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-P. [DOI] [PubMed] [Google Scholar]
- 43.Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 2004;279:48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- 44.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- 45.Challen GA, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 2011;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 47.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 48.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mizuno S, et al. Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood. 2001;97:1172–1179. doi: 10.1182/blood.V97.5.1172. [DOI] [PubMed] [Google Scholar]
- 50.Li Y, et al. The expression of DNA methyltransferase DNMT1, 3A and 3B in acute leukemia and myelodysplastic syndrome. Zhonghua Nei Ke Za Zhi. 2003;42:688–691. [PubMed] [Google Scholar]
- 51.Li YH, et al. Expression and clinical significance of DNMT in patients with chronic myeloid leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2015;23:1547–1550. doi: 10.7534/j.issn.1009-2137.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 52.Robaina MC, et al. Deregulation of DNMT1, DNMT3B and miR-29s in Burkitt lymphoma suggests novel contribution for disease pathogenesis. Exp. Mol. Pathol. 2015;98:200–207. doi: 10.1016/j.yexmp.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 53.Amara K, et al. DNA methyltransferase DNMT3b protein overexpression as a prognostic factor in patients with diffuse large B-cell lymphomas. Cancer Sci. 2010;101:1722–1730. doi: 10.1111/j.1349-7006.2010.01569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao H, et al. Overexpression of DNA methyltransferase 1 as a negative independent prognostic factor in primary gastrointestinal diffuse large B-cell lymphoma treated with CHOP-like regimen and rituximab. Oncol. Lett. 2015;9:2307–2312. doi: 10.3892/ol.2015.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qayum I, Ashraf M. Increased DNA methyltransferase 1 (DNMT1) gene expression in human lymphomas by fluorescent in situ hybridization. J. Ayub Med Coll. Abbottabad. 2004;16:1–6. [PubMed] [Google Scholar]
- 56.Qayum I, Ashraf M. Dna methyltransferase 1 (DNMT1) gene activity in human lymphomas correlates with aberrant p53 gene expression. J. Ayub Med Coll. Abbottabad. 2006;18:1–6. [PubMed] [Google Scholar]
- 57.Loo SK, et al. DNMT1 is predictive of survival and associated with Ki-67 expression in R-CHOP-treated diffuse large B-cell lymphomas. Pathology. 2017;49:731–739. doi: 10.1016/j.pathol.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 58.Lu QY, Zhang ZC, Hong XL. Expression of DNA methyltransferase in myeloma U266 cells and its significance. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2011;19:1429–1431. [PubMed] [Google Scholar]
- 59.Trowbridge JJ, et al. Haploinsufficiency of Dnmt1 impairs leukemia stem cell function through derepression of bivalent chromatin domains. Genes Dev. 2012;26:344–349. doi: 10.1101/gad.184341.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang XX, Zhang H, Li Y. Preliminary study on the role of miR-148a and DNMT1 in the pathogenesis of acute myeloid leukemia. Mol. Med Rep. 2019;19:2943–2952. doi: 10.3892/mmr.2019.9913. [DOI] [PubMed] [Google Scholar]
- 61.Haque S, Vaiselbuh SR. Exosomal DNMT1 mRNA transcript is elevated in acute lymphoblastic leukemia which might reprograms leukemia progression. Cancer Genet. 2022;260–261:57–64. doi: 10.1016/j.cancergen.2021.07.004. [DOI] [PubMed] [Google Scholar]
- 62.Vicente-Duenas C, et al. Dnmt1 links BCR-ABLp210 to epigenetic tumor stem cell priming in myeloid leukemia. Leukemia. 2019;33:249–278. doi: 10.1038/s41375-018-0192-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu K, et al. Silencing DNMT1 attenuates the effect of WIF-1 gene promoter methylation on the biological behavior of chronic myeloid leukemia K562 cells. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2021;29:1768–1774. doi: 10.19746/j.cnki.issn.1009-2137.2021.06.013. [DOI] [PubMed] [Google Scholar]
- 64.Peters SL, et al. Essential role for Dnmt1 in the prevention and maintenance of MYC-induced T-cell lymphomas. Mol. Cell Biol. 2013;33:4321–4333. doi: 10.1128/MCB.00776-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poole CJ, et al. DNMT3B overexpression contributes to aberrant DNA methylation and MYC-driven tumor maintenance in T-ALL and Burkitt’s lymphoma. Oncotarget. 2017;8:76898–76920. doi: 10.18632/oncotarget.20176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loo SK, Ab Hamid SS, Musa M, Wong KK. DNMT1 is associated with cell cycle and DNA replication gene sets in diffuse large B-cell lymphoma. Pathol. Res. Pr. 2018;214:134–143. doi: 10.1016/j.prp.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 67.Wang Z, Niu XQ, Zhou WW, Lu QY. Effects of DNMT1 gene silencing on methylation of SOCS-1 gene in myeloma cells. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2015;23:713–717. doi: 10.7534/j.issn.1009-2137.2015.03.022. [DOI] [PubMed] [Google Scholar]
- 68.Li M, et al. Methylation of the promoter region of the tight junction protein-1 by DNMT1 induces EMT-like features in multiple myeloma. Mol. Ther. Oncolytics. 2020;19:197–207. doi: 10.1016/j.omto.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim MS, Kim YR, Yoo NJ, Lee SH. Mutational analysis of DNMT3A gene in acute leukemias and common solid cancers. APMIS. 2013;121:85–94. doi: 10.1111/j.1600-0463.2012.02940.x. [DOI] [PubMed] [Google Scholar]
- 70.Li W, et al. DNMT3A mutations in Chinese childhood acute myeloid leukemia. Medicine. 2017;96:e7620. doi: 10.1097/MD.0000000000007620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu YN, et al. DNMT3A mutation analysis in adult patients with acute lymphoblastic leukemia. J. Huazhong Univ. Sci. Technol. Med. Sci. 2015;35:337–342. doi: 10.1007/s11596-015-1434-1. [DOI] [PubMed] [Google Scholar]
- 72.El Ghannam D, Taalab MM, Ghazy HF, Eneen AF. DNMT3A R882 mutations in patients with cytogenetically normal acute myeloid leukemia and myelodysplastic syndrome. Blood Cells Mol. Dis. 2014;53:61–66. doi: 10.1016/j.bcmd.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 73.Lin J, et al. Recurrent DNMT3A R882 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS ONE. 2011;6:e26906. doi: 10.1371/journal.pone.0026906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jankowska AM, et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood. 2011;118:3932–3941. doi: 10.1182/blood-2010-10-311019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grossmann V, et al. The molecular profile of adult T-cell acute lymphoblastic leukemia: mutations in RUNX1 and DNMT3A are associated with poor prognosis in T-ALL. Genes Chromosomes Cancer. 2013;52:410–422. doi: 10.1002/gcc.22039. [DOI] [PubMed] [Google Scholar]
- 76.Liang DC, et al. Cooperating gene mutations in childhood acute myeloid leukemia with special reference on mutations of ASXL1, TET2, IDH1, IDH2, and DNMT3A. Blood. 2013;121:2988–2995. doi: 10.1182/blood-2012-06-436782. [DOI] [PubMed] [Google Scholar]
- 77.Zhou JF, et al. Analysis of DNMT3a mutation in childhood acute myeloid leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2012;20:1297–1301. [PubMed] [Google Scholar]
- 78.Ewalt M, et al. DNMT3a mutations in high-risk myelodysplastic syndrome parallel those found in acute myeloid leukemia. Blood Cancer J. 2011;1:e9. doi: 10.1038/bcj.2011.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thol F, et al. DNMT3A mutations are rare in childhood acute myeloid leukemia. Haematologica. 2011;96:1238–1240. doi: 10.3324/haematol.2011.046839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eckstein OS, et al. Mixed-phenotype acute leukemia (MPAL) exhibits frequent mutations in DNMT3A and activated signaling genes. Exp. Hematol. 2016;44:740–744. doi: 10.1016/j.exphem.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sirirat T, et al. Mutation analysis of isocitrate dehydrogenase (IDH1/2) and DNA methyltransferase 3A (DNMT3A) in Thai patients with newly diagnosed acute myeloid leukemia. Asian Pac. J. Cancer Prev. 2017;18:413–420. doi: 10.22034/APJCP.2017.18.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sasaki K, et al. Impact of the variant allele frequency of ASXL1, DNMT3A, JAK2, TET2, TP53, and NPM1 on the outcomes of patients with newly diagnosed acute myeloid leukemia. Cancer. 2020;126:765–774. doi: 10.1002/cncr.32566. [DOI] [PubMed] [Google Scholar]
- 83.Park DJ, et al. Characteristics of DNMT3A mutations in acute myeloid leukemia. Blood Res. 2020;55:17–26. doi: 10.5045/br.2020.55.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lu Q, Chen Y, Wang H, Li Z. DNMT3A mutations and clinical features in Chinese patients with acute myeloid leukemia. Cancer Cell Int. 2013;13:1. doi: 10.1186/1475-2867-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gou H, et al. The prevalence and clinical profiles of FLT3-ITD, FLT3-TKD, NPM1, C-KIT, DNMT3A, and CEBPA mutations in a cohort of patients with de novo acute myeloid leukemia from southwest China. Tumour Biol. 2016;37:7357–7370. doi: 10.1007/s13277-015-4601-x. [DOI] [PubMed] [Google Scholar]
- 86.Pezzi A, et al. DNMT3A mutations in patients with acute myeloid leukemia in South Brazil. Adv. Hematol. 2012;2012:697691. doi: 10.1155/2012/697691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gaidzik VI, et al. Clinical impact of DNMT3A mutations in younger adult patients with acute myeloid leukemia: results of the AML Study Group (AMLSG) Blood. 2013;121:4769–4777. doi: 10.1182/blood-2012-10-461624. [DOI] [PubMed] [Google Scholar]
- 88.El Gammal MM, et al. Clinical effect of combined mutations in DNMT3A, FLT3-ITD, and NPM1 among Egyptian acute myeloid leukemia patients. Clin. Lymphoma Myeloma Leuk. 2019;19:e281–e290. doi: 10.1016/j.clml.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 89.Markova J, et al. Prognostic impact of DNMT3A mutations in patients with intermediate cytogenetic risk profile acute myeloid leukemia. Eur. J. Haematol. 2012;88:128–135. doi: 10.1111/j.1600-0609.2011.01716.x. [DOI] [PubMed] [Google Scholar]
- 90.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Elrhman H, El-Meligui YM, Elalawi SM. Prognostic impact of concurrent DNMT3A, FLT3 and NPM1 gene mutations in acute myeloid leukemia patients. Clin. Lymphoma Myeloma Leuk. 2021;21:e960–e969. doi: 10.1016/j.clml.2021.07.011. [DOI] [PubMed] [Google Scholar]
- 92.Chen S, et al. Bioinformatics analysis identifies key genes and pathways in acute myeloid leukemia associated with DNMT3A mutation. Biomed. Res. Int. 2020;2020:9321630. doi: 10.1155/2020/9321630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kumar D, Mehta A, Panigrahi MK, Nath S, Saikia KK. DNMT3A (R882) mutation features and prognostic effect in acute myeloid leukemia in Coexistent with NPM1 and FLT3 mutations. Hematol. Oncol. Stem Cell Ther. 2018;11:82–89. doi: 10.1016/j.hemonc.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 94.Zhang Q, Wu X, Cao J, Gao F, Huang K. Association between increased mutation rates in DNMT3A and FLT3-ITD and poor prognosis of patients with acute myeloid leukemia. Exp. Ther. Med. 2019;18:3117–3124. doi: 10.3892/etm.2019.7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thol F, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J. Clin. Oncol. 2011;29:2889–2896. doi: 10.1200/JCO.2011.35.4894. [DOI] [PubMed] [Google Scholar]
- 96.Saygin C, et al. Mutations in DNMT3A, U2AF1, and EZH2 identify intermediate-risk acute myeloid leukemia patients with poor outcome after CR1. Blood Cancer J. 2018;8:4. doi: 10.1038/s41408-017-0040-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hou HA, et al. DNMT3A mutations in acute myeloid leukemia: stability during disease evolution and clinical implications. Blood. 2012;119:559–568. doi: 10.1182/blood-2011-07-369934. [DOI] [PubMed] [Google Scholar]
- 98.Loghavi S, et al. Clinical features of de novo acute myeloid leukemia with concurrent DNMT3A, FLT3 and NPM1 mutations. J. Hematol. Oncol. 2014;7:74. doi: 10.1186/s13045-014-0074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gale RE, et al. Simpson’s paradox and the impact of different DNMT3A mutations on outcome in younger adults with acute myeloid leukemia. J. Clin. Oncol. 2015;33:2072–2083. doi: 10.1200/JCO.2014.59.2022. [DOI] [PubMed] [Google Scholar]
- 100.Bullinger L, Döhner K, Döhner H. Genomics of acute myeloid leukemia diagnosis and pathways. J. Clin. Oncol. 2017;35:934–946. doi: 10.1200/JCO.2016.71.2208. [DOI] [PubMed] [Google Scholar]
- 101.Patnaik MM, et al. DNMT3A mutations are associated with inferior overall and leukemia-free survival in chronic myelomonocytic leukemia. Am. J. Hematol. 2017;92:56–61. doi: 10.1002/ajh.24581. [DOI] [PubMed] [Google Scholar]
- 102.Bond J, et al. DNMT3A mutation is associated with increased age and adverse outcome in adult T-cell acute lymphoblastic leukemia. Haematologica. 2019;104:1617–1625. doi: 10.3324/haematol.2018.197848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Aref S, El Menshawy N, El-Ghonemy MS, Zeid TA, El-Baiomy MA. Clinicopathologic effect of DNMT3A mutation in adult T-cell acute lymphoblastic leukemia. Clin. Lymphoma Myeloma Leuk. 2016;16:43–48. doi: 10.1016/j.clml.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 104.Lin N, et al. Biologico-clinical significance of DNMT3A variants expression in acute myeloid leukemia. Biochem Biophys. Res. Commun. 2017;494:270–277. doi: 10.1016/j.bbrc.2017.10.041. [DOI] [PubMed] [Google Scholar]
- 105.Fried I, et al. Frequency, onset and clinical impact of somatic DNMT3A mutations in therapy-related and secondary acute myeloid leukemia. Haematologica. 2012;97:246–250. doi: 10.3324/haematol.2011.051581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jost E, et al. Epimutations mimic genomic mutations of DNMT3A in acute myeloid leukemia. Leukemia. 2014;28:1227–1234. doi: 10.1038/leu.2013.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ibrahem L, Mahfouz R, Elhelw L, Abdsalam EM. & Soliman, R. Prognostic significance of DNMT3A mutations in patients with acute myeloid leukemia. Blood Cells Mol. Dis. 2015;54:84–89. doi: 10.1016/j.bcmd.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 108.Ahmad F, Mohota R, Sanap S, Mandava S, Das BR. Molecular evaluation of DNMT3A and IDH1/2 gene mutation: frequency, distribution pattern and associations with additional molecular markers in normal karyotype Indian acute myeloid leukemia patients. Asian Pac. J. Cancer Prev. 2014;15:1247–1253. doi: 10.7314/APJCP.2014.15.3.1247. [DOI] [PubMed] [Google Scholar]
- 109.Marcucci G, et al. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J. Clin. Oncol. 2012;30:742–750. doi: 10.1200/JCO.2011.39.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wakita S, et al. Mutations of the epigenetics-modifying gene (DNMT3a, TET2, IDH1/2) at diagnosis may induce FLT3-ITD at relapse in de novo acute myeloid leukemia. Leukemia. 2013;27:1044–1052. doi: 10.1038/leu.2012.317. [DOI] [PubMed] [Google Scholar]
- 111.Masuda S. DNMT3A mutations in acute myeloid leukemia: impact on low-risk patients with CEBPA mutations. J. Clin. Oncol. 2011;29:4592–4593. doi: 10.1200/JCO.2011.38.2127. [DOI] [PubMed] [Google Scholar]
- 112.Badar T, Atallah E. Do histone deacytelase inhibitors and azacitidine combination hold potential as an effective treatment for high/very-high risk myelodysplastic syndromes? Expert Opin. Investig. Drugs. 2021;30:665–673. doi: 10.1080/13543784.2021.1915986. [DOI] [PubMed] [Google Scholar]
- 113.Zhang TJ, Zhang LC, Xu ZJ, Zhou JD. Expression and prognosis analysis of DNMT family in acute myeloid leukemia. Aging. 2020;12:14677–14690. doi: 10.18632/aging.103520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Huang X, et al. Gene expression profiling of the DNMT3A R882 mutation in acute leukemia. Oncol. Lett. 2013;6:268–274. doi: 10.3892/ol.2013.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lauber C, et al. Survival differences and associated molecular signatures of DNMT3A-mutant acute myeloid leukemia patients. Sci. Rep. 2020;10:12761. doi: 10.1038/s41598-020-69691-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang X, et al. Clinical and biological implications of IDH1/2 in acute myeloid leukemia with DNMT3A(mut) Cancer Manag Res. 2018;10:2457–2466. doi: 10.2147/CMAR.S157632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sehgal AR, et al. DNMT3A mutational status affects the results of dose-escalated induction therapy in acute myelogenous leukemia. Clin. Cancer Res. 2015;21:1614–1620. doi: 10.1158/1078-0432.CCR-14-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xu Y, et al. Allogeneic hematopoietic stem cell transplantation could improve survival of cytogenetically normal adult acute myeloid leukemia patients with DNMT3A mutations. Am. J. Hematol. 2015;90:992–997. doi: 10.1002/ajh.24135. [DOI] [PubMed] [Google Scholar]
- 119.Berenstein R, et al. Quantitative detection of DNMT3A R882H mutation in acute myeloid leukemia. J. Exp. Clin. Cancer Res. 2015;34:55. doi: 10.1186/s13046-015-0173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gaidzik VI, et al. DNMT3A mutant transcript levels persist in remission and do not predict outcome in patients with acute myeloid leukemia. Leukemia. 2018;32:30–37. doi: 10.1038/leu.2017.200. [DOI] [PubMed] [Google Scholar]
- 121.Sun Y, et al. Persistent DNMT3A mutation burden in DNMT3A mutated adult cytogenetically normal acute myeloid leukemia patients in long-term remission. Leuk. Res. 2016;49:102–107. doi: 10.1016/j.leukres.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 122.Ottone T, et al. Longitudinal detection of DNMT3A(R882H) transcripts in patients with acute myeloid leukemia. Am. J. Hematol. 2018;93:E120–e123. doi: 10.1002/ajh.25061. [DOI] [PubMed] [Google Scholar]
- 123.Ardestani MT, et al. FLT3-ITD compared with DNMT3A R882 mutation is a more powerful independent inferior prognostic factor in adult acute myeloid leukemia patients after allogeneic hematopoietic stem cell transplantation: a retrospective cohort study. Turk. J. Haematol. 2018;35:158–167. doi: 10.4274/tjh.2018.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tang SH, et al. Effect of FLT3-ITD with DNMT3A R882 double-mutation on the prognosis of acute myeloid leukemia after allogeneic hematopoietic stem cell transplantation. Zhonghua Xue Ye Xue Za Zhi. 2018;39:552–557. doi: 10.3760/cma.j.issn.0253-2727.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ahn JS, et al. DNMT3A R882 mutation with FLT3-ITD Positivity is an extremely poor prognostic factor in patients with normal-karyotype acute myeloid leukemia after allogeneic hematopoietic cell transplantation. Biol. Blood Marrow Transpl. 2016;22:61–70. doi: 10.1016/j.bbmt.2015.07.030. [DOI] [PubMed] [Google Scholar]
- 126.Kim T, et al. Exome sequencing reveals DNMT3A and ASXL1 variants associate with progression of chronic myeloid leukemia after tyrosine kinase inhibitor therapy. Leuk. Res. 2017;59:142–148. doi: 10.1016/j.leukres.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 127.Ostronoff F, et al. Mutations in the DNMT3A exon 23 independently predict poor outcome in older patients with acute myeloid leukemia: a SWOG report. Leukemia. 2013;27:238–241. doi: 10.1038/leu.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Couronne L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N. Engl. J. Med. 2012;366:95–96. doi: 10.1056/NEJMc1111708. [DOI] [PubMed] [Google Scholar]
- 129.Yao WQ, et al. Angioimmunoblastic T-cell lymphoma contains multiple clonal T-cell populations derived from a common TET2 mutant progenitor cell. J. Pathol. 2020;250:346–357. doi: 10.1002/path.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nicolae A, et al. Nodal cytotoxic peripheral T-cell lymphoma occurs frequently in the clinical setting of immunodysregulation and is associated with recurrent epigenetic alterations. Mod. Pathol. 2022;35:1126–1136. doi: 10.1038/s41379-022-01022-w. [DOI] [PubMed] [Google Scholar]
- 131.Sakata-Yanagimoto M, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat. Genet. 2014;46:171–175. doi: 10.1038/ng.2872. [DOI] [PubMed] [Google Scholar]
- 132.Odejide O, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123:1293–1296. doi: 10.1182/blood-2013-10-531509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lemonnier F, et al. Integrative analysis of a phase 2 trial combining lenalidomide with CHOP in angioimmunoblastic T-cell lymphoma. Blood Adv. 2021;5:539–548. doi: 10.1182/bloodadvances.2020003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mondejar R, et al. Molecular basis of targeted therapy in T/NK-cell lymphoma/leukemia: a comprehensive genomic and immunohistochemical analysis of a panel of 33 cell lines. PLoS ONE. 2017;12:e0177524. doi: 10.1371/journal.pone.0177524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang J, et al. Identification of clinical molecular targets for childhood Burkitt lymphoma. Transl. Oncol. 2020;13:100855. doi: 10.1016/j.tranon.2020.100855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Beltzung F, et al. Primary cutaneous CD4+ small/medium T-cell lymphoproliferative disorders: a clinical, pathologic, and molecular study of 60 cases presenting with a single lesion: a multicenter study of the french cutaneous lymphoma study group. Am. J. Surg. Pathol. 2020;44:862–872. doi: 10.1097/PAS.0000000000001470. [DOI] [PubMed] [Google Scholar]
- 137.Mutzbauer G, et al. SYK expression in monomorphic epitheliotropic intestinal T-cell lymphoma. Mod. Pathol. 2018;31:505–516. doi: 10.1038/modpathol.2017.145. [DOI] [PubMed] [Google Scholar]
- 138.Lee MJ, et al. Genome-defined African ancestry is associated with distinct mutations and worse survival in patients with diffuse large B-cell lymphoma. Cancer. 2020;126:3493–3503. doi: 10.1002/cncr.32866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Suehara Y, et al. Mutations found in cell-free DNAs of patients with malignant lymphoma at remission can derive from clonal hematopoiesis. Cancer Sci. 2019;110:3375–3381. doi: 10.1111/cas.14176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sakata-Yanagimoto M, et al. Detection of the circulating tumor DNAs in angioimmunoblastic T- cell lymphoma. Ann. Hematol. 2017;96:1471–1475. doi: 10.1007/s00277-017-3038-2. [DOI] [PubMed] [Google Scholar]
- 141.Song J, et al. The application of NextGen sequencing in the diagnosis of myeloid neoplasms in myeloma patients with cytopenia. Clin. Lymphoma Myeloma Leuk. 2022;22:e414–e426. doi: 10.1016/j.clml.2021.12.008. [DOI] [PubMed] [Google Scholar]
- 142.Mouhieddine TH, et al. Clonal hematopoiesis is associated with adverse outcomes in multiple myeloma patients undergoing transplant. Nat. Commun. 2020;11:2996. doi: 10.1038/s41467-020-16805-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Luzna P, et al. Global DNA methylation and increased DNMT3A expression in multiple myeloma patients. Biomed. Pap. 2022 doi: 10.5507/bp.2022.006. [DOI] [PubMed] [Google Scholar]
- 144.Heuck CJ, et al. Myeloma is characterized by stage-specific alterations in DNA methylation that occur early during myelomagenesis. J. Immunol. 2013;190:2966–2975. doi: 10.4049/jimmunol.1202493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dai YJ, et al. Conditional knockin of Dnmt3a R878H initiates acute myeloid leukemia with mTOR pathway involvement. Proc. Natl Acad. Sci. USA. 2017;114:5237–5242. doi: 10.1073/pnas.1703476114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ferreira HJ, et al. DNMT3A mutations mediate the epigenetic reactivation of the leukemogenic factor MEIS1 in acute myeloid leukemia. Oncogene. 2016;35:3079–3082. doi: 10.1038/onc.2015.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lu R, et al. Epigenetic perturbations by Arg882-mutated DNMT3A potentiate aberrant stem cell gene-expression program and acute leukemia development. Cancer Cell. 2016;30:92–107. doi: 10.1016/j.ccell.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bera R, et al. Genetic and epigenetic perturbations by DNMT3A-R882 mutants impaired apoptosis through augmentation of PRDX2 in myeloid leukemia cells. Neoplasia. 2018;20:1106–1120. doi: 10.1016/j.neo.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xu J, et al. DNMT3A Arg882 mutation drives chronic myelomonocytic leukemia through disturbing gene expression/DNA methylation in hematopoietic cells. Proc. Natl Acad. Sci. USA. 2014;111:2620–2625. doi: 10.1073/pnas.1400150111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nguyen TB, et al. Identification of cell-type-specific mutations in nodal T-cell lymphomas. Blood Cancer J. 2017;7:e516. doi: 10.1038/bcj.2016.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ohshima K, Miyoshi H, Yamada K. The microenvironment and immune status of peripheral T-cell lymphoma: focusing on AITL and ATLL. Rinsho Ketsueki. 2018;59:574–587. doi: 10.11406/rinketsu.59.574. [DOI] [PubMed] [Google Scholar]
- 152.Meyer SE, et al. DNMT3A haploinsufficiency transforms FLT3ITD myeloproliferative disease into a rapid, spontaneous, and fully penetrant acute myeloid leukemia. Cancer Disco. 2016;6:501–515. doi: 10.1158/2159-8290.CD-16-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chang YI, et al. Dnmt3a haploinsufficiency cooperates with oncogenic Kras to promote an early-onset T-cell acute lymphoblastic leukemia. Am. J. Transl. Res. 2017;9:1326–1334. [PMC free article] [PubMed] [Google Scholar]
- 154.Haney SL, et al. Promoter hypomethylation and expression is conserved in mouse chronic lymphocytic leukemia induced by decreased or inactivated Dnmt3a. Cell Rep. 2016;15:1190–1201. doi: 10.1016/j.celrep.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Haney SL, et al. Dnmt3a Is a haploinsufficient tumor suppressor in CD8+ peripheral T cell lymphoma. PLoS Genet. 2016;12:e1006334. doi: 10.1371/journal.pgen.1006334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang X, et al. Dnmt3a loss and Idh2 neomorphic mutations mutually potentiate malignant hematopoiesis. Blood. 2020;135:845–856. doi: 10.1182/blood.2019003330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lewis NE, et al. Clonal hematopoiesis in angioimmunoblastic T-cell lymphoma with divergent evolution to myeloid neoplasms. Blood Adv. 2020;4:2261–2271. doi: 10.1182/bloodadvances.2020001636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang C, et al. IDH2R172 mutations define a unique subgroup of patients with angioimmunoblastic T-cell lymphoma. Blood. 2015;126:1741–1752. doi: 10.1182/blood-2015-05-644591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Laginestra MA, et al. Whole exome sequencing reveals mutations in FAT1 tumor suppressor gene clinically impacting on peripheral T-cell lymphoma not otherwise specified. Mod. Pathol. 2020;33:179–187. doi: 10.1038/s41379-019-0279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Guryanova OA, et al. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat. Med. 2016;22:1488–1495. doi: 10.1038/nm.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chen YY, et al. Mutant DNMT3A clone evading chemotherapy and infiltrating central nervous system in a patient with molecularly good-risk acute myeloid leukemia. Ann. Hematol. 2014;93:1441–1442. doi: 10.1007/s00277-013-1985-9. [DOI] [PubMed] [Google Scholar]
- 162.Scheller M, et al. Hotspot DNMT3A mutations in clonal hematopoiesis and acute myeloid leukemia sensitize cells to azacytidine via viral mimicry response. Nat. Cancer. 2021;2:527–544. doi: 10.1038/s43018-021-00213-9. [DOI] [PubMed] [Google Scholar]
- 163.Metzeler KH, et al. DNMT3A mutations and response to the hypomethylating agent decitabine in acute myeloid leukemia. Leukemia. 2012;26:1106–1107. doi: 10.1038/leu.2011.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rau RE, et al. DOT1L as a therapeutic target for the treatment of DNMT3A-mutant acute myeloid leukemia. Blood. 2016;128:971–981. doi: 10.1182/blood-2015-11-684225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Que Y, et al. Study on the immune escape mechanism of acute myeloid leukemia with DNMT3A mutation. Front. Immunol. 2021;12:653030. doi: 10.3389/fimmu.2021.653030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li Y, et al. The C46359T polymorphism of DNMT3B promoter gene and pathogenesis of acute leukemia. Zhonghua Nei Ke Za Zhi. 2005;44:588–591. [PubMed] [Google Scholar]
- 167.Niederwieser C, et al. Prognostic and biologic significance of DNMT3B expression in older patients with cytogenetically normal primary acute myeloid leukemia. Leukemia. 2015;29:567–575. doi: 10.1038/leu.2014.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zheng Q, et al. Association between DNA methyltransferases 3B gene polymorphisms and the susceptibility to acute myeloid leukemia in Chinese Han population. PLoS ONE. 2013;8:e74626. doi: 10.1371/journal.pone.0074626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ait Boujmia OK, Nadifi S, Dehbi H, Lamchahab M, Quessar A. The influence of DNMT3A and DNMT3B gene polymorphisms on acute myeloid leukemia risk in a Moroccan population. Curr. Res. Transl. Med. 2020;68:191–195. doi: 10.1016/j.retram.2020.08.001. [DOI] [PubMed] [Google Scholar]
- 170.Morscio J, et al. Clinicopathologic comparison of plasmablastic lymphoma in HIV-positive, immunocompetent, and posttransplant patients: single-center series of 25 cases and meta-analysis of 277 reported cases. Am. J. Surg. Pathol. 2014;38:875–886. doi: 10.1097/PAS.0000000000000234. [DOI] [PubMed] [Google Scholar]
- 171.Wang G, et al. Synergetic effects of DNA methylation and histone modification during mouse induced pluripotent stem cell generation. Sci. Rep. 2017;7:39527. doi: 10.1038/srep39527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zheng Y, et al. Loss of Dnmt3b accelerates MLL-AF9 leukemia progression. Leukemia. 2016;30:2373–2384. doi: 10.1038/leu.2016.112. [DOI] [PubMed] [Google Scholar]
- 173.Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev. 1993;3:226–231. doi: 10.1016/0959-437X(93)90027-M. [DOI] [PubMed] [Google Scholar]
- 174.Nan X, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 175.Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 176.Mahmood N, Rabbani SA. DNA methylation readers and cancer: mechanistic and therapeutic applications. Front. Oncol. 2019;9:489. doi: 10.3389/fonc.2019.00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 178.Nan X, Meehan RR, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell Biol. 1998;18:6538–6547. doi: 10.1128/MCB.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Klose RJ, et al. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol. Cell. 2005;19:667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 181.Lopez-Serra L, Esteller M. Proteins that bind methylated DNA and human cancer: reading the wrong words. Br. J. Cancer. 2008;98:1881–1885. doi: 10.1038/sj.bjc.6604374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hashimoto H, et al. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40:4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yildirim O, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Laget S, et al. The human proteins MBD5 and MBD6 associate with heterochromatin but they do not bind methylated DNA. PLoS ONE. 2010;5:e11982. doi: 10.1371/journal.pone.0011982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Parry L, Clarke AR. The roles of the methyl-CpG binding proteins in cancer. Genes Cancer. 2011;2:618–630. doi: 10.1177/1947601911418499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Falandry C, et al. CLLD8/KMT1F is a lysine methyltransferase that is important for chromosome segregation. J. Biol. Chem. 2010;285:20234–20241. doi: 10.1074/jbc.M109.052399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Li ZY, et al. Corrigendum to "the long noncoding RNA MEG3 and its target miR-147 regulate JAK/STAT pathway in advanced chronic myeloid leukemia". EBioMedicine. 2018;37:569. doi: 10.1016/j.ebiom.2018.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Li Q, et al. Role of one-carbon metabolizing pathway genes and gene-nutrient interaction in the risk of non-Hodgkin lymphoma. Cancer Causes Control. 2013;24:1875–1884. doi: 10.1007/s10552-013-0264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sharma ND, et al. Epigenetic silencing of SOCS5 potentiates JAK-STAT signaling and progression of T-cell acute lymphoblastic leukemia. Cancer Sci. 2019;110:1931–1946. doi: 10.1111/cas.14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wang Z, Zhang J, Zhang Y, Srivenugopal KS, Lim SH. SPAN-XB core promoter sequence is regulated in myeloma cells by specific CpG dinucleotides associated with the MeCP2 protein. Int. J. Cancer. 2006;119:2878–2884. doi: 10.1002/ijc.22259. [DOI] [PubMed] [Google Scholar]
- 192.Piazza R, et al. Epigenetic silencing of the proapoptotic gene BIM in anaplastic large cell lymphoma through an MeCP2/SIN3a deacetylating complex. Neoplasia. 2013;15:511–522. doi: 10.1593/neo.121784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Liang X, Xu Y, Xu K, Liu J, Qian X. B1, a novel amonafide analogue, overcomes the resistance conferred by Bcl-2 in human promyelocytic leukemia HL60 cells. Mol. Cancer Res. 2010;8:1619–1632. doi: 10.1158/1541-7786.MCR-10-0341. [DOI] [PubMed] [Google Scholar]
- 194.Dinis J, et al. DNA damage response in imatinib resistant chronic myeloid leukemia K562 cells. Leuk. Lymphoma. 2012;53:2004–2014. doi: 10.3109/10428194.2012.681654. [DOI] [PubMed] [Google Scholar]
- 195.Hudson NO, Buck-Koehntop BA. Zinc finger readers of methylated DNA. Molecules. 2018;23:2555. doi: 10.3390/molecules23102555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Prokhortchouk A, et al. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 2001;15:1613–1618. doi: 10.1101/gad.198501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Filion GJ, et al. A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol. Cell Biol. 2006;26:169–181. doi: 10.1128/MCB.26.1.169-181.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Daniel JM, Reynolds AB. The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol. Cell Biol. 1999;19:3614–3623. doi: 10.1128/MCB.19.5.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Daniel JM, Spring CM, Crawford HC, Reynolds AB, Baig A. The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 2002;30:2911–2919. doi: 10.1093/nar/gkf398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Cofre J, Menezes JR, Pizzatti L, Abdelhay E. Knock-down of Kaiso induces proliferation and blocks granulocytic differentiation in blast crisis of chronic myeloid leukemia. Cancer Cell Int. 2012;12:28. doi: 10.1186/1475-2867-12-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Xue X, Zhang J, Lan H, Xu Y, Wang H. Kaiso protects human umbilical vein endothelial cells against apoptosis by differentially regulating the expression of B-cell CLL/lymphoma 2 family members. Sci. Rep. 2017;7:7116. doi: 10.1038/s41598-017-07559-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Gu C, et al. Identification of berberine as a novel drug for the treatment of multiple myeloma via targeting UHRF1. BMC Biol. 2020;18:33. doi: 10.1186/s12915-020-00766-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 204.Avvakumov GV, et al. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 205.Bostick M, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 206.Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 207.Mori T, Li Y, Hata H, Ono K, Kochi H. NIRF, a novel RING finger protein, is involved in cell-cycle regulation. Biochem Biophys. Res. Commun. 2002;296:530–536. doi: 10.1016/S0006-291X(02)00890-2. [DOI] [PubMed] [Google Scholar]
- 208.Simonetti G, et al. Aneuploid acute myeloid leukemia exhibits a signature of genomic alterations in the cell cycle and protein degradation machinery. Cancer. 2019;125:712–725. doi: 10.1002/cncr.31837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Kim KB, et al. H3K9 methyltransferase G9a negatively regulates UHRF1 transcription during leukemia cell differentiation. Nucleic Acids Res. 2015;43:3509–3523. doi: 10.1093/nar/gkv183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Yao J, Luo Y, Zeng C, He H, Zhang X. UHRF1 regulates the transcriptional repressor HBP1 through MIF in T acute lymphoblastic leukemia. Oncol. Rep. 2021;46:131. doi: 10.3892/or.2021.8082. [DOI] [PubMed] [Google Scholar]
- 211.Alhosin M, et al. Thymoquinone and difluoromethylornithine (DFMO) Synergistically induce apoptosis of human acute T lymphoblastic leukemia Jurkat cells through the modulation of epigenetic pathways. Technol. Cancer Res. Treat. 2020;19:1533033820947489. doi: 10.1177/1533033820947489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Alhosin M, et al. Induction of apoptosis by thymoquinone in lymphoblastic leukemia Jurkat cells is mediated by a p73-dependent pathway which targets the epigenetic integrator UHRF1. Biochem. Pharm. 2010;79:1251–1260. doi: 10.1016/j.bcp.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 213.Guan D, Factor D, Liu Y, Wang Z, Kao HY. The epigenetic regulator UHRF1 promotes ubiquitination-mediated degradation of the tumor-suppressor protein promyelocytic leukemia protein. Oncogene. 2013;32:3819–3828. doi: 10.1038/onc.2012.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 215.Oswald J, et al. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 2000;10:475–478. doi: 10.1016/S0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 216.Paroush Z, Keshet I, Yisraeli J, Cedar H. Dynamics of demethylation and activation of the alpha-actin gene in myoblasts. Cell. 1990;63:1229–1237. doi: 10.1016/0092-8674(90)90418-E. [DOI] [PubMed] [Google Scholar]
- 217.Zhang F, Pomerantz JH, Sen G, Palermo AT, Blau HM. Active tissue-specific DNA demethylation conferred by somatic cell nuclei in stable heterokaryons. Proc. Natl Acad. Sci. USA. 2007;104:4395–4400. doi: 10.1073/pnas.0700181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bhutani N, et al. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Iqbal K, Jin SG, Pfeifer GP, Szabó PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl Acad. Sci. USA. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Huang H, et al. TET1 plays an essential oncogenic role in MLL-rearranged leukemia. Proc. Natl Acad. Sci. USA. 2013;110:11994–11999. doi: 10.1073/pnas.1310656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zhang T, Zhao Y, Zhao Y, Zhou J. Expression and prognosis analysis of TET family in acute myeloid leukemia. Aging. 2020;12:5031–5047. doi: 10.18632/aging.102928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Huang Y, et al. Comprehensively analyze the expression and prognostic role for ten-eleven translocations (TETs) in acute myeloid leukemia. Transl. Cancer Res. 2020;9:7259–7283. doi: 10.21037/tcr-20-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Vitkeviciene A, Skliute G, Zucenka A, Borutinskaite V, Navakauskiene R. Potential Prognostic Markers for Relapsed/Refractory vs. Responsive Acute Myeloid Leukemia. Cancers. 2022;14:2752. doi: 10.3390/cancers14112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wang J, et al. High expression of TET1 predicts poor survival in cytogenetically normal acute myeloid leukemia from two cohorts. EBioMedicine. 2018;28:90–96. doi: 10.1016/j.ebiom.2018.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Van Damme M, et al. Characterization of TET and IDH gene expression in chronic lymphocytic leukemia: comparison with normal B cells and prognostic significance. Clin. Epigenet. 2016;8:132. doi: 10.1186/s13148-016-0298-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Yeh CH, et al. Erratum to: ‘Mutation of epigenetic regulators TET2 and MLL3 in patients with HTLV-I-induced acute adult T-cell leukemia. Mol. Cancer. 2016;15:20. doi: 10.1186/s12943-016-0504-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Dolnik A, et al. Commonly altered genomic regions in acute myeloid leukemia are enriched for somatic mutations involved in chromatin remodeling and splicing. Blood. 2012;120:e83–e92. doi: 10.1182/blood-2011-12-401471. [DOI] [PubMed] [Google Scholar]
- 230.Kalender Atak Z, et al. High accuracy mutation detection in leukemia on a selected panel of cancer genes. PLoS ONE. 2012;7:e38463. doi: 10.1371/journal.pone.0038463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Bamezai S, et al. TET1 promotes growth of T-cell acute lymphoblastic leukemia and can be antagonized via PARP inhibition. Leukemia. 2021;35:389–403. doi: 10.1038/s41375-020-0864-3. [DOI] [PubMed] [Google Scholar]
- 232.Wang J, et al. Analysis of TET2 and EZH2 gene functions in chromosome instability in acute myeloid leukemia. Sci. Rep. 2020;10:2706. doi: 10.1038/s41598-020-59365-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Yamazaki J, et al. Hypomethylation of TET2 target genes identifies a curable subset of acute myeloid leukemia. J. Natl Cancer Inst. 2015;108:djv323. doi: 10.1093/jnci/djv323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Damm F, et al. TET2 mutations in cytogenetically normal acute myeloid leukemia: clinical implications and evolutionary patterns. Genes Chromosomes Cancer. 2014;53:824–832. doi: 10.1002/gcc.22191. [DOI] [PubMed] [Google Scholar]
- 235.Metzeler KH, et al. TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: a cancer and leukemia group B study. J. Clin. Oncol. 2011;29:1373–1381. doi: 10.1200/JCO.2010.32.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Tian X, et al. TET2 gene mutation is unfavorable prognostic factor in cytogenetically normal acute myeloid leukemia patients with NPM1+ and FLT3-ITD - mutations. Int J. Hematol. 2014;100:96–104. doi: 10.1007/s12185-014-1595-x. [DOI] [PubMed] [Google Scholar]
- 237.Gaidzik VI, et al. TET2 mutations in acute myeloid leukemia (AML): results from a comprehensive genetic and clinical analysis of the AML study group. J. Clin. Oncol. 2012;30:1350–1357. doi: 10.1200/JCO.2011.39.2886. [DOI] [PubMed] [Google Scholar]
- 238.Weissmann S, et al. Landscape of TET2 mutations in acute myeloid leukemia. Leukemia. 2012;26:934–942. doi: 10.1038/leu.2011.326. [DOI] [PubMed] [Google Scholar]
- 239.Ponciano-Gomez A, et al. Mutations in TET2 and DNMT3A genes are associated with changes in global and gene-specific methylation in acute myeloid leukemia. Tumour Biol. 2017;39:1010428317732181. doi: 10.1177/1010428317732181. [DOI] [PubMed] [Google Scholar]
- 240.Shaikh ARK, et al. TET2 mutations in acute myeloid leukemia: a comprehensive study in patients of Sindh, Pakistan. PeerJ. 2021;9:e10678. doi: 10.7717/peerj.10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Wei JF, et al. The incidence of TET2 gene mutation and its clinical significance in acute myeloid leukemia patients. Zhonghua Xue Ye Xue Za Zhi. 2011;32:304–307. [PubMed] [Google Scholar]
- 242.Ohgami RS, et al. Next-generation sequencing of acute myeloid leukemia identifies the significance of TP53, U2AF1, ASXL1, and TET2 mutations. Mod. Pathol. 2015;28:706–714. doi: 10.1038/modpathol.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Nibourel O, et al. Incidence and prognostic value of TET2 alterations in de novo acute myeloid leukemia achieving complete remission. Blood. 2010;116:1132–1135. doi: 10.1182/blood-2009-07-234484. [DOI] [PubMed] [Google Scholar]
- 244.Chehreghani Z, et al. Detection of TET2 mutation in patients with de novo acute myeloid leukemia: a mutation analysis of 51 Iranian patients. Asian Pac. J. Cancer Prev. 2022;23:803–806. doi: 10.31557/APJCP.2022.23.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ahn JS, et al. Adverse prognostic effect of homozygous TET2 mutation on the relapse risk of acute myeloid leukemia in patients of normal karyotype. Haematologica. 2015;100:e351–e353. doi: 10.3324/haematol.2015.126227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Chou WC, et al. TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood. 2011;118:3803–3810. doi: 10.1182/blood-2011-02-339747. [DOI] [PubMed] [Google Scholar]
- 247.Shimoda K, et al. TET2 mutation in adult T-cell leukemia/lymphoma. J. Clin. Exp. Hematop. 2015;55:145–149. doi: 10.3960/jslrt.55.145. [DOI] [PubMed] [Google Scholar]
- 248.Marçais A, et al. Adult T cell leukemia aggressivenness correlates with loss of both 5-hydroxymethylcytosine and TET2 expression. Oncotarget. 2017;8:52256–52268. doi: 10.18632/oncotarget.13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Olson TL, et al. Frequent somatic TET2 mutations in chronic NK-LGL leukemia with distinct patterns of cytopenias. Blood. 2021;138:662–673. doi: 10.1182/blood.2020005831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Makishima H, et al. CBL, CBLB, TET2, ASXL1, and IDH1/2 mutations and additional chromosomal aberrations constitute molecular events in chronic myelogenous leukemia. Blood. 2011;117:e198–e206. doi: 10.1182/blood-2010-06-292433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Hamed NA, Elhalawani NA, Kassem HS, Ayad MW, Dammag EA. The prognostic significance of TET2 single nucleotide polymorphism in Egyptian chronic myeloid leukemia. Mediterr. J. Hematol. Infect. Dis. 2020;12:e2020004. doi: 10.4084/mjhid.2020.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Kosmider O, et al. TET2 mutations in secondary acute myeloid leukemias: a French retrospective study. Haematologica. 2011;96:1059–1063. doi: 10.3324/haematol.2011.040840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Konstandin N, et al. Genomic 5-hydroxymethylcytosine levels correlate with TET2 mutations and a distinct global gene expression pattern in secondary acute myeloid leukemia. Leukemia. 2011;25:1649–1652. doi: 10.1038/leu.2011.134. [DOI] [PubMed] [Google Scholar]
- 254.Patnaik MM, et al. Prognostic interaction between ASXL1 and TET2 mutations in chronic myelomonocytic leukemia. Blood Cancer J. 2016;6:e385. doi: 10.1038/bcj.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Coltro G, et al. Clinical, molecular, and prognostic correlates of number, type, and functional localization of TET2 mutations in chronic myelomonocytic leukemia (CMML)-a study of 1084 patients. Leukemia. 2020;34:1407–1421. doi: 10.1038/s41375-019-0690-7. [DOI] [PubMed] [Google Scholar]
- 256.Cui Y, et al. Impact of TET2, SRSF2, ASXL1 and SETBP1 mutations on survival of patients with chronic myelomonocytic leukemia. Exp. Hematol. Oncol. 2015;4:14. doi: 10.1186/s40164-015-0009-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Perez C, et al. TET2 mutations are associated with specific 5-methylcytosine and 5-hydroxymethylcytosine profiles in patients with chronic myelomonocytic leukemia. PLoS ONE. 2012;7:e31605. doi: 10.1371/journal.pone.0031605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Zhao W, et al. The prognostic value of the interaction between ASXL1 and TET2 gene mutations in patients with chronic myelomonocytic leukemia: a meta-analysis. Hematology. 2022;27:367–378. doi: 10.1080/16078454.2021.1958486. [DOI] [PubMed] [Google Scholar]
- 259.Patnaik MM, et al. Number and type of TET2 mutations in chronic myelomonocytic leukemia and their clinical relevance. Blood Cancer J. 2016;6:e472. doi: 10.1038/bcj.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Grossmann V, et al. Molecular profiling of chronic myelomonocytic leukemia reveals diverse mutations in >80% of patients with TET2 and EZH2 being of high prognostic relevance. Leukemia. 2011;25:877–879. doi: 10.1038/leu.2011.10. [DOI] [PubMed] [Google Scholar]
- 261.Cui Y, et al. TET2 mutations were predictive of inferior prognosis in the presence of ASXL1 mutations in patients with chronic myelomonocytic leukemia. Stem Cell Investig. 2016;3:50. doi: 10.21037/sci.2016.09.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Kosmider O, et al. TET2 gene mutation is a frequent and adverse event in chronic myelomonocytic leukemia. Haematologica. 2009;94:1676–1681. doi: 10.3324/haematol.2009.011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Yang YC, et al. Clinical significance of truncated mutant ΔJBP of TET2 gene in patients with acute myeloid leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2021;29:1011–1018. doi: 10.19746/j.cnki.issn.1009-2137.2021.04.001. [DOI] [PubMed] [Google Scholar]
- 264.Li YW, et al. Analysis of clinical significance and prognostic impact of TET2 single nucleotide polymorphism I1762V in patients with acute myeloid leukemia. Zhonghua Xue Ye Xue Za Zhi. 2022;43:241–246. doi: 10.3760/cma.j.issn.0253-2727.2022.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Wang X, et al. Correlation of TET2 SNP rs2454206 with improved survival in children with acute myeloid leukemia featuring intermediate-risk cytogenetics. Genes Chromosomes Cancer. 2018;57:379–386. doi: 10.1002/gcc.22540. [DOI] [PubMed] [Google Scholar]
- 266.Mohamed AM, et al. TET2 exon 2 skipping is an independent favorable prognostic factor for cytogenetically normal acute myelogenous leukemia (AML): TET2 exon 2 skipping in AML. Leuk. Res. 2017;56:21–28. doi: 10.1016/j.leukres.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 267.Itzykson R, et al. Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia. 2011;25:1147–1152. doi: 10.1038/leu.2011.71. [DOI] [PubMed] [Google Scholar]
- 268.Cull AH, et al. Overexpression of Arginase 1 is linked to DNMT3A and TET2 mutations in lower-grade myelodysplastic syndromes and chronic myelomonocytic leukemia. Leuk. Res. 2018;65:5–13. doi: 10.1016/j.leukres.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 269.Hernández-Sánchez M, et al. TET2 overexpression in chronic lymphocytic leukemia is unrelated to the presence of TET2 variations. Biomed. Res. Int. 2014;2014:814294. doi: 10.1155/2014/814294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Zhang TJ, et al. TET2 expression is a potential prognostic and predictive biomarker in cytogenetically normal acute myeloid leukemia. J. Cell Physiol. 2018;233:5838–5846. doi: 10.1002/jcp.26373. [DOI] [PubMed] [Google Scholar]
- 271.Zhu Z, et al. Clinical significance of TET2 gene expression in 157 adult acute myeloid leukemia patients with normal cytogenetics. Zhonghua Xue Ye Xue Za Zhi. 2014;35:802–807. doi: 10.3760/cma.j.issn.0253-2727.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 272.Musialik E, Bujko M, Wypych A, Matysiak M, Siedlecki JA. TET2 promoter DNA methylation and expression analysis in pediatric B-cell acute lymphoblastic leukemia. Hematol. Rep. 2014;6:5333. doi: 10.4081/hr.2014.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Zhang P, et al. Low expression of TET2 gene in pediatric acute lymphoblastic leukemia is associated with poor clinical outcome. Int. J. Lab Hematol. 2019;41:702–709. doi: 10.1111/ijlh.13099. [DOI] [PubMed] [Google Scholar]
- 274.Hill HA, et al. Genetic mutations and features of mantle cell lymphoma: a systematic review and meta-analysis. Blood Adv. 2020;4:2927–2938. doi: 10.1182/bloodadvances.2019001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Kubuki Y, et al. TET2 mutation in diffuse large B-cell lymphoma. J. Clin. Exp. Hematop. 2017;56:145–149. doi: 10.3960/jslrt.56.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Au-Yeung RKH, et al. Molecular features of non-anaplastic peripheral T-cell lymphoma in children and adolescents. Pediatr. Blood Cancer. 2021;68:e29285. doi: 10.1002/pbc.29285. [DOI] [PubMed] [Google Scholar]
- 277.Schwartz FH, et al. TET2 mutations in B cells of patients affected by angioimmunoblastic T-cell lymphoma. J. Pathol. 2017;242:129–133. doi: 10.1002/path.4898. [DOI] [PubMed] [Google Scholar]
- 278.Asmar F, et al. Genome-wide profiling identifies a DNA methylation signature that associates with TET2 mutations in diffuse large B-cell lymphoma. Haematologica. 2013;98:1912–1920. doi: 10.3324/haematol.2013.088740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Ramis-Zaldivar JE, et al. MAPK and JAK-STAT pathways dysregulation in plasmablastic lymphoma. Haematologica. 2021;106:2682–2693. doi: 10.3324/haematol.2020.271957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Ye Y, et al. Correlation of mutational landscape and survival outcome of peripheral T-cell lymphomas. Exp. Hematol. Oncol. 2021;10:9. doi: 10.1186/s40164-021-00200-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Lemonnier F, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120:1466–1469. doi: 10.1182/blood-2012-02-408542. [DOI] [PubMed] [Google Scholar]
- 282.Wu F, et al. Thyroid MALT lymphoma: self-harm to gain potential T-cell help. Leukemia. 2021;35:3497–3508. doi: 10.1038/s41375-021-01289-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Guo YM, et al. Angioimmunoblastic T-cell lymphoma: histopathological grading and prognosis. Zhonghua Bing. Li Xue Za Zhi. 2019;48:784–790. doi: 10.3760/cma.j.issn.0529-5807.2019.10.007. [DOI] [PubMed] [Google Scholar]
- 284.Stremenova Spegarova J, et al. Germline TET2 loss of function causes childhood immunodeficiency and lymphoma. Blood. 2020;136:1055–1066. doi: 10.1182/blood.2020005844. [DOI] [PubMed] [Google Scholar]
- 285.Elliott EK, et al. Epigenetic regulation of miR-92a and TET2 and their association in non-Hodgkin lymphoma. Front. Genet. 2021;12:768913. doi: 10.3389/fgene.2021.768913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Maia C, et al. Biological and clinical significance of dysplastic hematopoiesis in patients with newly diagnosed multiple myeloma. Blood. 2020;135:2375–2387. doi: 10.1182/blood.2019003382. [DOI] [PubMed] [Google Scholar]
- 287.Hoang PH, et al. An enhanced genetic model of relapsed IGH-translocated multiple myeloma evolutionary dynamics. Blood Cancer J. 2020;10:101. doi: 10.1038/s41408-020-00367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Ryland GL, et al. Novel genomic findings in multiple myeloma identified through routine diagnostic sequencing. J. Clin. Pathol. 2018;71:895–899. doi: 10.1136/jclinpath-2018-205195. [DOI] [PubMed] [Google Scholar]
- 289.Cimmino L, et al. Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell. 2017;170:1079–1095 e1020. doi: 10.1016/j.cell.2017.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Yao H, et al. TET2 and MEG3 promoter methylation is associated with acute myeloid leukemia in a Hainan population. Oncotarget. 2017;8:18337–18347. doi: 10.18632/oncotarget.15440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Yamazaki J, et al. TET2 mutations affect non-CpG island DNA methylation at enhancers and transcription factor-binding sites in chronic myelomonocytic leukemia. Cancer Res. 2015;75:2833–2843. doi: 10.1158/0008-5472.CAN-14-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Bahari G, Hashemi M, Naderi M, Taheri M. TET2 promoter DNA methylation and expression in childhood acute lymphoblastic leukemia. Asian Pac. J. Cancer Prev. 2016;17:3959–3962. [PubMed] [Google Scholar]
- 293.Wei Y, et al. Cooperation between KDM6B overexpression and TET2 deficiency in the pathogenesis of chronic myelomonocytic leukemia. Leukemia. 2022;36:2097–2107. doi: 10.1038/s41375-022-01605-1. [DOI] [PubMed] [Google Scholar]
- 294.Nguyen PN, et al. Clinicopathological implications of RHOA mutations in angioimmunoblastic T-cell lymphoma: a meta-analysis: RHOA mutations in AITL. Clin. Lymphoma Myeloma Leuk. 2021;21:431–438. doi: 10.1016/j.clml.2021.03.002. [DOI] [PubMed] [Google Scholar]
- 295.Jiao J, et al. AID and TET2 co-operation modulates FANCA expression by active demethylation in diffuse large B cell lymphoma. Clin. Exp. Immunol. 2019;195:190–201. doi: 10.1111/cei.13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Muto H, et al. Reduced TET2 function leads to T-cell lymphoma with follicular helper T-cell-like features in mice. Blood Cancer J. 2014;4:e264. doi: 10.1038/bcj.2014.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Sun D, et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 2014;14:673–688. doi: 10.1016/j.stem.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Truong TP, et al. Age-dependent decrease of DNA hydroxymethylation in human T cells. J. Clin. Exp. Hematop. 2015;55:1–6. doi: 10.3960/jslrt.55.1. [DOI] [PubMed] [Google Scholar]
- 299.Togasaki E, et al. Frequent somatic mutations in epigenetic regulators in newly diagnosed chronic myeloid leukemia. Blood Cancer J. 2017;7:e559. doi: 10.1038/bcj.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 301.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 302.Muto T, Muramatsu M, Taniwaki M, Kinoshita K, Honjo T. Isolation, tissue distribution, and chromosomal localization of the human activation-induced cytidine deaminase (AID) gene. Genomics. 2000;68:85–88. doi: 10.1006/geno.2000.6268. [DOI] [PubMed] [Google Scholar]
- 303.Reaves SK, et al. Regulation of intestinal apolipoprotein B mRNA editing levels by a zinc-deficient diet and cDNA cloning of editing protein in hamsters. J. Nutr. 2000;130:2166–2173. doi: 10.1093/jn/130.9.2166. [DOI] [PubMed] [Google Scholar]
- 304.Liao W, et al. APOBEC-2, a cardiac- and skeletal muscle-specific member of the cytidine deaminase supergene family. Biochem Biophys. Res. Commun. 1999;260:398–404. doi: 10.1006/bbrc.1999.0925. [DOI] [PubMed] [Google Scholar]
- 305.Jarmuz A, et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 306.Rogozin IB, Basu MK, Jordan IK, Pavlov YI, Koonin EV. APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis. Cell Cycle. 2005;4:1281–1285. doi: 10.4161/cc.4.9.1994. [DOI] [PubMed] [Google Scholar]
- 307.Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Ferreira PG, et al. Transcriptome characterization by RNA sequencing identifies a major molecular and clinical subdivision in chronic lymphocytic leukemia. Genome Res. 2014;24:212–226. doi: 10.1101/gr.152132.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Antic Z, et al. Unravelling the sequential interplay of mutational mechanisms during clonal evolution in relapsed pediatric acute lymphoblastic leukemia. Genes. 2021;12:214. doi: 10.3390/genes12020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Cifola I, et al. Whole-exome sequencing of primary plasma cell leukemia discloses heterogeneous mutational patterns. Oncotarget. 2015;6:17543–17558. doi: 10.18632/oncotarget.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Cheng S, Zhang W, Inghirami G, Tam W. Mutation analysis links angioimmunoblastic T-cell lymphoma to clonal hematopoiesis and smoking. eLife. 2021;10:e66395. doi: 10.7554/eLife.66395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Ren W, et al. Genetic landscape of hepatitis B virus-associated diffuse large B-cell lymphoma. Blood. 2018;131:2670–2681. doi: 10.1182/blood-2017-11-817601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Bustoros M, et al. Genomic profiling of smoldering multiple myeloma identifies patients at a high risk of disease progression. J. Clin. Oncol. 2020;38:2380–2389. doi: 10.1200/JCO.20.00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Oben B, et al. Whole-genome sequencing reveals progressive versus stable myeloma precursor conditions as two distinct entities. Nat. Commun. 2021;12:1861. doi: 10.1038/s41467-021-22140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Bolli N, et al. Genomic patterns of progression in smoldering multiple myeloma. Nat. Commun. 2018;9:3363. doi: 10.1038/s41467-018-05058-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Rustad EH, et al. Timing the initiation of multiple myeloma. Nat. Commun. 2020;11:1917. doi: 10.1038/s41467-020-15740-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Hoang PH, Cornish AJ, Dobbins SE, Kaiser M, Houlston RS. Mutational processes contributing to the development of multiple myeloma. Blood Cancer J. 2019;9:60. doi: 10.1038/s41408-019-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Samur MK, et al. Genome-wide somatic alterations in multiple myeloma reveal a superior outcome group. J. Clin. Oncol. 2020;38:3107–3118. doi: 10.1200/JCO.20.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Walker BA, et al. APOBEC family mutational signatures are associated with poor prognosis translocations in multiple myeloma. Nat. Commun. 2015;6:6997. doi: 10.1038/ncomms7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Maura F, et al. Biological and prognostic impact of APOBEC-induced mutations in the spectrum of plasma cell dyscrasias and multiple myeloma cell lines. Leukemia. 2018;32:1044–1048. doi: 10.1038/leu.2017.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Talluri S, et al. Dysregulated APOBEC3G causes DNA damage and promotes genomic instability in multiple myeloma. Blood Cancer J. 2021;11:166. doi: 10.1038/s41408-021-00554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Yamazaki H, et al. Endogenous APOBEC3B overexpression constitutively generates DNA substitutions and deletions in myeloma cells. Sci. Rep. 2019;9:7122. doi: 10.1038/s41598-019-43575-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Cortazar D, et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 324.Peng B, Hurt EM, Hodge DR, Thomas SB, Farrar WL. DNA hypermethylation and partial gene silencing of human thymine- DNA glycosylase in multiple myeloma cell lines. Epigenetics. 2006;1:138–145. doi: 10.4161/epi.1.3.2938. [DOI] [PubMed] [Google Scholar]
- 325.Takahashi H, Hatakeyama S, Saitoh H, Nakayama KI. Noncovalent SUMO-1 binding activity of thymine DNA glycosylase (TDG) is required for its SUMO-1 modification and colocalization with the promyelocytic leukemia protein. J. Biol. Chem. 2005;280:5611–5621. doi: 10.1074/jbc.M408130200. [DOI] [PubMed] [Google Scholar]
- 326.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Patel JP, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Abbas S, et al. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: prevalence and prognostic value. Blood. 2010;116:2122–2126. doi: 10.1182/blood-2009-11-250878. [DOI] [PubMed] [Google Scholar]
- 329.Chotirat S, Thongnoppakhun W, Promsuwicha O, Boonthimat C, Auewarakul CU. Molecular alterations of isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) metabolic genes and additional genetic mutations in newly diagnosed acute myeloid leukemia patients. J. Hematol. Oncol. 2012;5:5. doi: 10.1186/1756-8722-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Chou WC, et al. Distinct clinical and biologic characteristics in adult acute myeloid leukemia bearing the isocitrate dehydrogenase 1 mutation. Blood. 2010;115:2749–2754. doi: 10.1182/blood-2009-11-253070. [DOI] [PubMed] [Google Scholar]
- 331.Marcucci G, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J. Clin. Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Paschka P, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J. Clin. Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 334.Chou WC, et al. The prognostic impact and stability of Isocitrate dehydrogenase 2 mutation in adult patients with acute myeloid leukemia. Leukemia. 2011;25:246–253. doi: 10.1038/leu.2010.267. [DOI] [PubMed] [Google Scholar]
- 335.Gruber E, et al. Inhibition of mutant IDH1 promotes cycling of acute myeloid leukemia stem cells. Cell Rep. 2022;40:111182. doi: 10.1016/j.celrep.2022.111182. [DOI] [PubMed] [Google Scholar]
- 336.Choe S, et al. Molecular mechanisms mediating relapse following ivosidenib monotherapy in IDH1-mutant relapsed or refractory AML. Blood Adv. 2020;4:1894–1905. doi: 10.1182/bloodadvances.2020001503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Amatangelo MD, et al. Enasidenib induces acute myeloid leukemia cell differentiation to promote clinical response. Blood. 2017;130:732–741. doi: 10.1182/blood-2017-04-779447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Goldberg L, et al. Mutant Idh2 cooperates with a NUP98-HOXD13 fusion to induce early immature thymocyte precursor ALL. Cancer Res. 2021;81:5033–5046. doi: 10.1158/0008-5472.CAN-21-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Riva M, et al. IDH2(R172) mutation in angioimmunoblastic T-cell lymphoma: a retrospective multicenter case series. Eur. J. Haematol. 2022;110:217–220. doi: 10.1111/ejh.13885. [DOI] [PubMed] [Google Scholar]
- 340.Steinhilber J, et al. The pathological features of angioimmunoblastic T-cell lymphomas with IDH2(R172) mutations. Mod. Pathol. 2019;32:1123–1134. doi: 10.1038/s41379-019-0254-4. [DOI] [PubMed] [Google Scholar]
- 341.Leclaire Alirkilicarslan A, et al. Expression of TFH markers and detection of RHOA p.G17V and IDH2 p.R172K/S mutations in cutaneous localizations of angioimmunoblastic T-cell lymphomas. Am. J. Surg. Pathol. 2017;41:1581–1592. doi: 10.1097/PAS.0000000000000956. [DOI] [PubMed] [Google Scholar]
- 342.Cairns RA, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012;119:1901–1903. doi: 10.1182/blood-2011-11-391748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Hayashida M, et al. Combination of multicolor flow cytometry for circulating lymphoma cells and tests for the RHOA(G17V) and IDH2(R172) hot-spot mutations in plasma cell-free DNA as liquid biopsy for the diagnosis of angioimmunoblastic T-cell lymphoma. Leuk. Lymphoma. 2020;61:2389–2398. doi: 10.1080/10428194.2020.1768382. [DOI] [PubMed] [Google Scholar]
- 344.Lemonnier F, et al. The IDH2 R172K mutation associated with angioimmunoblastic T-cell lymphoma produces 2HG in T cells and impacts lymphoid development. Proc. Natl Acad. Sci. USA. 2016;113:15084–15089. doi: 10.1073/pnas.1617929114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Song S, et al. IDH2 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in multiple myeloma. Oncogene. 2021;40:5393–5402. doi: 10.1038/s41388-021-01939-7. [DOI] [PubMed] [Google Scholar]
- 346.Bergaggio E, et al. IDH2 inhibition enhances proteasome inhibitor responsiveness in hematological malignancies. Blood. 2019;133:156–167. doi: 10.1182/blood-2018-05-850826. [DOI] [PubMed] [Google Scholar]
- 347.Li CC, et al. Pre-configuring chromatin architecture with histone modifications guides hematopoietic stem cell formation in mouse embryos. Nat. Commun. 2022;13:346. doi: 10.1038/s41467-022-28018-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Jeong J, et al. BAP1 shapes the bone marrow niche for lymphopoiesis by fine-tuning epigenetic profiles in endosteal mesenchymal stromal cells. Cell Death Differ. 2022;29:2151–2162. doi: 10.1038/s41418-022-01006-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Dey A, et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 2012;337:1541–1546. doi: 10.1126/science.1221711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 351.Ruzic D, et al. Targeting histone deacetylases: opportunities for cancer treatment and chemoprevention. Pharmaceutics. 2022;14:209. doi: 10.3390/pharmaceutics14010209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Rice KL, Hormaeche I, Licht JD. Epigenetic regulation of normal and malignant hematopoiesis. Oncogene. 2007;26:6697–6714. doi: 10.1038/sj.onc.1210755. [DOI] [PubMed] [Google Scholar]
- 353.Ji MM, et al. Histone modifier gene mutations in peripheral T-cell lymphoma not otherwise specified. Haematologica. 2018;103:679–687. doi: 10.3324/haematol.2017.182444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Huang S, Brandt SJ. mSin3A regulates murine erythroleukemia cell differentiation through association with the TAL1 (or SCL) transcription factor. Mol. Cell Biol. 2000;20:2248–2259. doi: 10.1128/MCB.20.6.2248-2259.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 356.Zhu Y, et al. Oncogenic mutations and tumor microenvironment alterations of older patients with diffuse large B-cell lymphoma. Front. Immunol. 2022;13:842439. doi: 10.3389/fimmu.2022.842439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Brown JA. Patent spotlight: small-molecule lysine acetyltransferase inhibitors (KATi) Pharm. Pat. Anal. 2020;9:17–28. doi: 10.4155/ppa-2019-0025. [DOI] [PubMed] [Google Scholar]
- 358.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 359.Schmitz R, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N. Engl. J. Med. 2018;378:1396–1407. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Mareschal S, et al. Identification of somatic mutations in primary cutaneous diffuse large B-cell lymphoma, leg type by massive parallel sequencing. J. Investig. Dermatol. 2017;137:1984–1994. doi: 10.1016/j.jid.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 361.García-Ramírez I, et al. Crebbp loss cooperates with Bcl2 overexpression to promote lymphoma in mice. Blood. 2017;129:2645–2656. doi: 10.1182/blood-2016-08-733469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Wang J, et al. Conditional MLL-CBP targets GMP and models therapy-related myeloproliferative disease. EMBO J. 2005;24:368–381. doi: 10.1038/sj.emboj.7600521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Pasqualucci L, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Kasper LH, et al. A transcription-factor-binding surface of coactivator p300 is required for haematopoiesis. Nature. 2002;419:738–743. doi: 10.1038/nature01062. [DOI] [PubMed] [Google Scholar]
- 365.Huang YH, et al. CREBBP/EP300 mutations promoted tumor progression in diffuse large B-cell lymphoma through altering tumor-associated macrophage polarization via FBXW7-NOTCH-CCL2/CSF1 axis. Signal Transduct. Target Ther. 2021;6:10. doi: 10.1038/s41392-020-00437-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Iyer NG, Ozdag H, Caldas C. p300/CBP and cancer. Oncogene. 2004;23:4225–4231. doi: 10.1038/sj.onc.1207118. [DOI] [PubMed] [Google Scholar]
- 367.Zhuravleva J, et al. MOZ/TIF2-induced acute myeloid leukaemia in transgenic fish. Br. J. Haematol. 2008;143:378–382. doi: 10.1111/j.1365-2141.2008.07362.x. [DOI] [PubMed] [Google Scholar]
- 368.Yang Y, et al. The histone lysine acetyltransferase HBO1 (KAT7) regulates hematopoietic stem cell quiescence and self-renewal. Blood. 2022;139:845–858. doi: 10.1182/blood.2021013954. [DOI] [PubMed] [Google Scholar]
- 369.Yan J, Diaz J, Jiao J, Wang R, You J. Perturbation of BRD4 protein function by BRD4-NUT protein abrogates cellular differentiation in NUT midline carcinoma. J. Biol. Chem. 2011;286:27663–27675. doi: 10.1074/jbc.M111.246975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Chaidos A, Caputo V, Karadimitris A. Inhibition of bromodomain and extra-terminal proteins (BET) as a potential therapeutic approach in haematological malignancies: emerging preclinical and clinical evidence. Ther. Adv. Hematol. 2015;6:128–141. doi: 10.1177/2040620715576662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.French CA, et al. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene. 2008;27:2237–2242. doi: 10.1038/sj.onc.1210852. [DOI] [PubMed] [Google Scholar]
- 372.Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 373.Marquard L, et al. Histone deacetylase 1, 2, 6 and acetylated histone H4 in B- and T-cell lymphomas. Histopathology. 2009;54:688–698. doi: 10.1111/j.1365-2559.2009.03290.x. [DOI] [PubMed] [Google Scholar]
- 374.Pillonel V, et al. Histone deacetylase 1 plays a predominant pro-oncogenic role in Eμ-myc driven B cell lymphoma. Sci. Rep. 2016;6:37772. doi: 10.1038/srep37772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Min SK, et al. Expression of HAT1 and HDAC1, 2, 3 in diffuse large B-cell lymphomas, peripheral T-cell lymphomas, and NK/T-cell lymphomas. Korean J. Pathol. 2012;46:142–150. doi: 10.4132/KoreanJPathol.2012.46.2.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Jiang Y, et al. CREBBP Inactivation promotes the development of HDAC3-dependent lymphomas. Cancer Disco. 2017;7:38–53. doi: 10.1158/2159-8290.CD-16-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Lemercier C, et al. Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor. J. Biol. Chem. 2002;277:22045–22052. doi: 10.1074/jbc.M201736200. [DOI] [PubMed] [Google Scholar]
- 378.Lee SH, et al. Expression of histone deacetylases in diffuse large B-cell lymphoma and its clinical significance. Int. J. Med. Sci. 2014;11:994–1000. doi: 10.7150/ijms.8522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Pérez-Salvia M, et al. In vitro and in vivo activity of a new small-molecule inhibitor of HDAC6 in mantle cell lymphoma. Haematologica. 2018;103:e537–e540. doi: 10.3324/haematol.2018.189241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Maharaj K, et al. HDAC6 inhibition alleviates CLL-induced T-cell dysfunction and enhances immune checkpoint blockade efficacy in the Eμ-TCL1 model. Front. Immunol. 2020;11:590072. doi: 10.3389/fimmu.2020.590072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Bae J, et al. Histone deacetylase (HDAC) inhibitor ACY241 enhances anti-tumor activities of antigen-specific central memory cytotoxic T lymphocytes against multiple myeloma and solid tumors. Leukemia. 2018;32:1932–1947. doi: 10.1038/s41375-018-0062-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Azagra A, et al. In vivo conditional deletion of HDAC7 reveals its requirement to establish proper B lymphocyte identity and development. J. Exp. Med. 2016;213:2591–2601. doi: 10.1084/jem.20150821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Barneda-Zahonero B, et al. The transcriptional repressor HDAC7 promotes apoptosis and c-Myc downregulation in particular types of leukemia and lymphoma. Cell Death Dis. 2015;6:e1635. doi: 10.1038/cddis.2014.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Kasler HG, et al. Histone deacetylase 7 regulates cell survival and TCR signaling in CD4/CD8 double-positive thymocytes. J. Immunol. 2011;186:4782–4793. doi: 10.4049/jimmunol.1001179. [DOI] [PubMed] [Google Scholar]
- 385.Wang Z, et al. SIRT1 deacetylase promotes acquisition of genetic mutations for drug resistance in CML cells. Oncogene. 2013;32:589–598. doi: 10.1038/onc.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Ojaimi MA, et al. Disorders of histone methylation: molecular basis and clinical syndromes. Clin. Genet. 2022;102:169–181. doi: 10.1111/cge.14181. [DOI] [PubMed] [Google Scholar]
- 387.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 388.Liu MK, et al. Methylation alterations and advance of treatment in lymphoma. Front. Biosci. 2021;26:602–613. doi: 10.52586/4970. [DOI] [PubMed] [Google Scholar]
- 389.Xia ZB, Anderson M, Diaz MO, Zeleznik-Le NJ. MLL repression domain interacts with histone deacetylases, the polycomb group proteins HPC2 and BMI-1, and the corepressor C-terminal-binding protein. Proc. Natl Acad. Sci. USA. 2003;100:8342–8347. doi: 10.1073/pnas.1436338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Castiglioni S, et al. KMT2A: umbrella gene for multiple diseases. Genes. 2022;13:514. doi: 10.3390/genes13030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Leeman-Neill RJ, et al. Phenogenomic heterogeneity of post-transplant plasmablastic lymphomas. Haematologica. 2022;107:201–210. doi: 10.3324/haematol.2020.267294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Zhang J, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat. Med. 2015;21:1190–1198. doi: 10.1038/nm.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Green MR, et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood. 2013;121:1604–1611. doi: 10.1182/blood-2012-09-457283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Sun QY, et al. Ordering of mutations in acute myeloid leukemia with partial tandem duplication of MLL (MLL-PTD) Leukemia. 2017;31:1–10. doi: 10.1038/leu.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Yap DB, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Papakonstantinou N, et al. The histone methyltransferase EZH2 as a novel prosurvival factor in clinically aggressive chronic lymphocytic leukemia. Oncotarget. 2016;7:35946–35959. doi: 10.18632/oncotarget.9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Yan J, et al. EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood. 2013;121:4512–4520. doi: 10.1182/blood-2012-08-450494. [DOI] [PubMed] [Google Scholar]
- 400.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Nikoloski G, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat. Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 402.Ernst T, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 403.Hollink IH, et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood. 2011;118:3645–3656. doi: 10.1182/blood-2011-04-346643. [DOI] [PubMed] [Google Scholar]
- 404.Struski S, et al. NUP98 is rearranged in 3.8% of pediatric AML forming a clinical and molecular homogenous group with a poor prognosis. Leukemia. 2017;31:565–572. doi: 10.1038/leu.2016.267. [DOI] [PubMed] [Google Scholar]
- 405.Shiba N, et al. NUP98-NSD1 gene fusion and its related gene expression signature are strongly associated with a poor prognosis in pediatric acute myeloid leukemia. Genes Chromosomes Cancer. 2013;52:683–693. doi: 10.1002/gcc.22064. [DOI] [PubMed] [Google Scholar]
- 406.Mohanty S, et al. Targeted inhibition of the NUP98-NSD1 fusion oncogene in acute myeloid leukemia. Cancers. 2020;12:2766. doi: 10.3390/cancers12102766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Crescenzi B, et al. NUP98/11p15 translocations affect CD34+ cells in myeloid and T lymphoid leukemias. Leuk. Res. 2015;39:769–772. doi: 10.1016/j.leukres.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 408.Avet-Loiseau H, et al. Bortezomib plus dexamethasone induction improves outcome of patients with t(4;14) myeloma but not outcome of patients with del(17p) J. Clin. Oncol. 2010;28:4630–4634. doi: 10.1200/JCO.2010.28.3945. [DOI] [PubMed] [Google Scholar]
- 409.Gertz MA, et al. Clinical implications of t(11;14)(q13;q32), t(4;14)(p16.3;q32), and -17p13 in myeloma patients treated with high-dose therapy. Blood. 2005;106:2837–2840. doi: 10.1182/blood-2005-04-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Taketani T, et al. NUP98-NSD3 fusion gene in radiation-associated myelodysplastic syndrome with t(8;11)(p11;p15) and expression pattern of NSD family genes. Cancer Genet. Cytogenet. 2009;190:108–112. doi: 10.1016/j.cancergencyto.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 411.Rosati R, et al. NUP98 is fused to the NSD3 gene in acute myeloid leukemia associated with t(8;11)(p11.2;p15) Blood. 2002;99:3857–3860. doi: 10.1182/blood.V99.10.3857. [DOI] [PubMed] [Google Scholar]
- 412.Greenblatt SM, et al. CARM1 is essential for myeloid leukemogenesis but dispensable for normal hematopoiesis. Cancer Cell. 2018;33:1111–1127.e1115. doi: 10.1016/j.ccell.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Chevillard-Briet M, Trouche D, Vandel L. Control of CBP co-activating activity by arginine methylation. EMBO J. 2002;21:5457–5466. doi: 10.1093/emboj/cdf548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Jin Y, et al. Targeting methyltransferase PRMT5 eliminates leukemia stem cells in chronic myelogenous leukemia. J. Clin. Investig. 2016;126:3961–3980. doi: 10.1172/JCI85239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Liu C, et al. Loss of PRMT7 reprograms glycine metabolism to selectively eradicate leukemia stem cells in CML. Cell Metab. 2022;34:818–835.e817. doi: 10.1016/j.cmet.2022.04.004. [DOI] [PubMed] [Google Scholar]
- 416.Wang GG, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Dialynas GK, Vitalini MW, Wallrath LL. Linking heterochromatin protein 1 (HP1) to cancer progression. Mutat. Res. 2008;647:13–20. doi: 10.1016/j.mrfmmm.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Coles AH, Jones SN. The ING. gene Fam. Regul. Cell growth tumorigenesis. J. Cell Physiol. 2009;218:45–57. doi: 10.1002/jcp.21583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Garcia-Bassets I, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Metzger E, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 421.Cowger JJ, Zhao Q, Isovic M, Torchia J. Biochemical characterization of the zinc-finger protein 217 transcriptional repressor complex: identification of a ZNF217 consensus recognition sequence. Oncogene. 2007;26:3378–3386. doi: 10.1038/sj.onc.1210126. [DOI] [PubMed] [Google Scholar]
- 422.Lee MG, et al. Functional interplay between histone demethylase and deacetylase enzymes. Mol. Cell Biol. 2006;26:6395–6402. doi: 10.1128/MCB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Forneris F, et al. A highly specific mechanism of histone H3-K4 recognition by histone demethylase LSD1. J. Biol. Chem. 2006;281:35289–35295. doi: 10.1074/jbc.M607411200. [DOI] [PubMed] [Google Scholar]
- 424.Kohrogi K, et al. LSD1 defines erythroleukemia metabolism by controlling the lineage-specific transcription factors GATA1 and C/EBPα. Blood Adv. 2021;5:2305–2318. doi: 10.1182/bloodadvances.2020003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Hu X, et al. LSD1-mediated epigenetic modification is required for TAL1 function and hematopoiesis. Proc. Natl Acad. Sci. USA. 2009;106:10141–10146. doi: 10.1073/pnas.0900437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Tatsumi G, et al. LSD1-mediated repression of GFI1 super-enhancer plays an essential role in erythroleukemia. Leukemia. 2020;34:746–758. doi: 10.1038/s41375-019-0614-6. [DOI] [PubMed] [Google Scholar]
- 427.Zhang S, Liu M, Yao Y, Yu B, Liu H. Targeting LSD1 for acute myeloid leukemia (AML) treatment. Pharm. Res. 2021;164:105335. doi: 10.1016/j.phrs.2020.105335. [DOI] [PubMed] [Google Scholar]
- 428.Noort S, et al. The clinical and biological characteristics of NUP98-KDM5A in pediatric acute myeloid leukemia. Haematologica. 2021;106:630–634. doi: 10.3324/haematol.2019.236745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Shokri G, Doudi S, Fathi-Roudsari M, Kouhkan F, Sanati MH. Targeting histone demethylases KDM5A and KDM5B in AML cancer cells: a comparative view. Leuk. Res. 2018;68:105–111. doi: 10.1016/j.leukres.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 430.Xu S, et al. KDM5A suppresses PML-RARα target gene expression and APL differentiation through repressing H3K4me2. Blood Adv. 2021;5:3241–3253. doi: 10.1182/bloodadvances.2020002819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Xue S, et al. Histone lysine demethylase KDM5B maintains chronic myeloid leukemia via multiple epigenetic actions. Exp. Hematol. 2020;82:53–65. doi: 10.1016/j.exphem.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 432.Sera Y, et al. UTX maintains the functional integrity of the murine hematopoietic system by globally regulating aging-associated genes. Blood. 2021;137:908–922. doi: 10.1182/blood.2019001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Tian L, et al. Kdm6a deficiency restricted to mouse hematopoietic cells causes an age- and sex-dependent myelodysplastic syndrome-like phenotype. PLoS ONE. 2021;16:e0255706. doi: 10.1371/journal.pone.0255706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Stief SM, et al. Loss of KDM6A confers drug resistance in acute myeloid leukemia. Leukemia. 2020;34:50–62. doi: 10.1038/s41375-019-0497-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Zheng L, et al. Utx loss causes myeloid transformation. Leukemia. 2018;32:1458–1465. doi: 10.1038/s41375-018-0011-6. [DOI] [PubMed] [Google Scholar]
- 436.Gozdecka M, et al. UTX-mediated enhancer and chromatin remodeling suppresses myeloid leukemogenesis through noncatalytic inverse regulation of ETS and GATA programs. Nat. Genet. 2018;50:883–894. doi: 10.1038/s41588-018-0114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Rocha-Viegas L, et al. Role of UTX in retinoic acid receptor-mediated gene regulation in leukemia. Mol. Cell Biol. 2014;34:3765–3775. doi: 10.1128/MCB.00839-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Benyoucef A, et al. UTX inhibition as selective epigenetic therapy against TAL1-driven T-cell acute lymphoblastic leukemia. Genes Dev. 2016;30:508–521. doi: 10.1101/gad.276790.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Montalban-Bravo G, DiNardo CD. The role of IDH mutations in acute myeloid leukemia. Future Oncol. 2018;14:979–993. doi: 10.2217/fon-2017-0523. [DOI] [PubMed] [Google Scholar]
- 440.Drennan AC, Rui L. HiJAKing the epigenome in leukemia and lymphoma. Leuk. Lymphoma. 2017;58:2540–2547. doi: 10.1080/10428194.2017.1312370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Rui L, et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell. 2010;18:590–605. doi: 10.1016/j.ccr.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Rosenwald A, Staudt LM. Gene expression profiling of diffuse large B-cell lymphoma. Leuk. Lymphoma. 2003;44(Suppl 3):S41–S47. doi: 10.1080/10428190310001623775. [DOI] [PubMed] [Google Scholar]
- 443.Rui L, et al. Epigenetic gene regulation by Janus kinase 1 in diffuse large B-cell lymphoma. Proc. Natl Acad. Sci. USA. 2016;113:E7260–e7267. doi: 10.1073/pnas.1610970113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Wu XP, et al. Resveratrol induces apoptosis of human chronic myelogenous leukemia cells in vitro through p38 and JNK-regulated H2AX phosphorylation. Acta Pharm. Sin. 2015;36:353–361. doi: 10.1038/aps.2014.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Dong Y, et al. H2AX phosphorylation regulated by p38 is involved in Bim expression and apoptosis in chronic myelogenous leukemia cells induced by imatinib. Apoptosis. 2014;19:1281–1292. doi: 10.1007/s10495-014-0997-9. [DOI] [PubMed] [Google Scholar]
- 446.Zhang YJ, et al. Imatinib induces H2AX phosphorylation and apoptosis in chronic myelogenous leukemia cells in vitro via caspase-3/Mst1 pathway. Acta Pharm. Sin. Sin. 2012;33:551–557. doi: 10.1038/aps.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Eddy SR. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 448.Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2:986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 450.Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 451.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 454.Dragomir MP, et al. FuncPEP: A database of functional peptides encoded by non-coding RNAs. Noncoding RNA. 2020;6:41. doi: 10.3390/ncrna6040041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 456.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 458.Calin GA, Croce CM. Investigation of microRNA alterations in leukemias and lymphomas. Methods Enzymol. 2007;427:193–213. doi: 10.1016/S0076-6879(07)27011-9. [DOI] [PubMed] [Google Scholar]
- 459.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 460.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 461.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 462.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 463.Bartel DP. Metazoan microRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Rigoutsos I. New tricks for animal microRNAS: targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Res. 2009;69:3245–3248. doi: 10.1158/0008-5472.CAN-09-0352. [DOI] [PubMed] [Google Scholar]
- 465.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 466.Schnall-Levin M, et al. Unusually effective microRNA targeting within repeat-rich coding regions of mammalian mRNAs. Genome Res. 2011;21:1395–1403. doi: 10.1101/gr.121210.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Zhou H, Rigoutsos I. MiR-103a-3p targets the 5' UTR of GPRC5A in pancreatic cells. Rna. 2014;20:1431–1439. doi: 10.1261/rna.045757.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Zisoulis DG, et al. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Leung AK, et al. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat. Struct. Mol. Biol. 2011;18:237–244. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Cesana M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Anastasiadou E, Faggioni A, Trivedi P, Slack FJ. The nefarious nexus of noncoding RNAs in cancer. Int. J. Mol. Sci. 2018;19:2072. doi: 10.3390/ijms19072072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Dragomir MP, Knutsen E, Calin GA. Classical and noncanonical functions of miRNAs in cancers. Trends Genet. 2022;38:379–394. doi: 10.1016/j.tig.2021.10.002. [DOI] [PubMed] [Google Scholar]
- 474.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–d162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Calin GA, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Rassenti LZ, et al. MicroRNA dysregulation to identify therapeutic target combinations for chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2017;114:10731–10736. doi: 10.1073/pnas.1708264114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 480.Ramkissoon SH, et al. Hematopoietic-specific microRNA expression in human cells. Leuk. Res. 2006;30:643–647. doi: 10.1016/j.leukres.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 481.Fabbri M, et al. MicroRNAs and noncoding RNAs in hematological malignancies: molecular, clinical and therapeutic implications. Leukemia. 2008;22:1095–1105. doi: 10.1038/leu.2008.30. [DOI] [PubMed] [Google Scholar]
- 482.Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell. 2019;179:1033–1055. doi: 10.1016/j.cell.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl Acad. Sci. USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Eis PS, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl Acad. Sci. USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl Acad. Sci. USA. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Cheng CJ, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518:107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Si ML, et al. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 488.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 489.Schetter AJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. J. Am. Med. Assoc. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Fulci V, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–4951. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- 491.Lawrie CH, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int. J. Cancer. 2007;121:1156–1161. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- 492.Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Löwenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111:5078–5085. doi: 10.1182/blood-2008-01-133355. [DOI] [PubMed] [Google Scholar]
- 493.Navarro A, et al. MicroRNA expression profiling in classic Hodgkin lymphoma. Blood. 2008;111:2825–2832. doi: 10.1182/blood-2007-06-096784. [DOI] [PubMed] [Google Scholar]
- 494.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 495.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Ota A, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.CAN-03-3773. [DOI] [PubMed] [Google Scholar]
- 497.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 499.Sun R, et al. Therapeutic targeting miR130b counteracts diffuse large B-cell lymphoma progression via OX40/OX40L-mediated interaction with Th17 cells. Signal Transduct. Target Ther. 2022;7:80. doi: 10.1038/s41392-022-00895-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Döhner H, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N. Engl. J. Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 501.Baskar S, et al. Unique cell surface expression of receptor tyrosine kinase ROR1 in human B-cell chronic lymphocytic leukemia. Clin. Cancer Res. 2008;14:396–404. doi: 10.1158/1078-0432.CCR-07-1823. [DOI] [PubMed] [Google Scholar]
- 502.Broome HE, Rassenti LZ, Wang HY, Meyer LM, Kipps TJ. ROR1 is expressed on hematogones (non-neoplastic human B-lymphocyte precursors) and a minority of precursor-B acute lymphoblastic leukemia. Leuk. Res. 2011;35:1390–1394. doi: 10.1016/j.leukres.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Cui B, et al. High-level ROR1 associates with accelerated disease progression in chronic lymphocytic leukemia. Blood. 2016;128:2931–2940. doi: 10.1182/blood-2016-04-712562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Fukuda T, et al. Antisera induced by infusions of autologous Ad-CD154-leukemia B cells identify ROR1 as an oncofetal antigen and receptor for Wnt5a. Proc. Natl Acad. Sci. USA. 2008;105:3047–3052. doi: 10.1073/pnas.0712148105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Widhopf GF, 2nd, et al. ROR1 can interact with TCL1 and enhance leukemogenesis in Eμ-TCL1 transgenic mice. Proc. Natl Acad. Sci. USA. 2014;111:793–798. doi: 10.1073/pnas.1308374111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Choi MY, et al. Phase I trial: cirmtuzumab inhibits ROR1 signaling and stemness signatures in patients with chronic lymphocytic leukemia. Cell Stem Cell. 2018;22:951–959.e953. doi: 10.1016/j.stem.2018.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Chen Y, et al. Cirmtuzumab blocks Wnt5a/ROR1 stimulation of NF-κB to repress autocrine STAT3 activation in chronic lymphocytic leukemia. Blood. 2019;134:1084–1094. doi: 10.1182/blood.2019001366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Pekarsky Y, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 509.Santanam U, et al. Chronic lymphocytic leukemia modeled in mouse by targeted miR-29 expression. Proc. Natl Acad. Sci. USA. 2010;107:12210–12215. doi: 10.1073/pnas.1007186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 512.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021;22:96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Iyer MK, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Guo CJ, et al. Distinct Processing of lncRNAs Contributes to Non-conserved Functions in Stem Cells. Cell. 2020;181:621–636.e622. doi: 10.1016/j.cell.2020.03.006. [DOI] [PubMed] [Google Scholar]
- 517.Quinn JJ, et al. Rapid evolutionary turnover underlies conserved lncRNA-genome interactions. Genes Dev. 2016;30:191–207. doi: 10.1101/gad.272187.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Melé M, et al. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017;27:27–37. doi: 10.1101/gr.214205.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Lagarde J, et al. High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat. Genet. 2017;49:1731–1740. doi: 10.1038/ng.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Zuckerman B, Ulitsky I. Predictive models of subcellular localization of long RNAs. Rna. 2019;25:557–572. doi: 10.1261/rna.068288.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Zuckerman B, Ron M, Mikl M, Segal E, Ulitsky I. Gene architecture and sequence composition underpin selective dependency of nuclear export of long RNAs on NXF1 and the TREX complex. Mol. Cell. 2020;79:251–267.e256. doi: 10.1016/j.molcel.2020.05.013. [DOI] [PubMed] [Google Scholar]
- 522.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat. Rev. Cancer. 2018;18:5–18. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Csorba T, Questa JI, Sun Q, Dean C. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl Acad. Sci. USA. 2014;111:16160–16165. doi: 10.1073/pnas.1419030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Rosa S, Duncan S, Dean C. Mutually exclusive sense-antisense transcription at FLC facilitates environmentally induced gene repression. Nat. Commun. 2016;7:13031. doi: 10.1038/ncomms13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Jain AK, et al. LncPRESS1 is a p53-regulated lncRNA that safeguards pluripotency by disrupting SIRT6-mediated de-acetylation of histone H3K56. Mol. Cell. 2016;64:967–981. doi: 10.1016/j.molcel.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Postepska-Igielska A, et al. LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol. Cell. 2015;60:626–636. doi: 10.1016/j.molcel.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 529.Blank-Giwojna A, Postepska-Igielska A, Grummt I. lncRNA KHPS1 activates a poised enhancer by triplex-dependent recruitment of epigenomic regulators. Cell Rep. 2019;26:2904–2915.e2904. doi: 10.1016/j.celrep.2019.02.059. [DOI] [PubMed] [Google Scholar]
- 530.Boque-Sastre R, et al. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc. Natl Acad. Sci. USA. 2015;112:5785–5790. doi: 10.1073/pnas.1421197112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Arab K, et al. GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nat. Genet. 2019;51:217–223. doi: 10.1038/s41588-018-0306-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Ariel F, et al. R-loop mediated trans action of the APOLO long noncoding RNA. Mol. Cell. 2020;77:1055–1065.e1054. doi: 10.1016/j.molcel.2019.12.015. [DOI] [PubMed] [Google Scholar]
- 533.Niehrs C, Luke B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 2020;21:167–178. doi: 10.1038/s41580-019-0206-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Beckedorff FC, et al. The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation. PLoS Genet. 2013;9:e1003705. doi: 10.1371/journal.pgen.1003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Latos PA, et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 536.Stojic L, et al. Transcriptional silencing of long noncoding RNA GNG12-AS1 uncouples its transcriptional and product-related functions. Nat. Commun. 2016;7:10406. doi: 10.1038/ncomms10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Thebault P, et al. Transcription regulation by the noncoding RNA SRG1 requires Spt2-dependent chromatin deposition in the wake of RNA polymerase II. Mol. Cell Biol. 2011;31:1288–1300. doi: 10.1128/MCB.01083-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Rom A, et al. Regulation of CHD2 expression by the Chaserr long noncoding RNA gene is essential for viability. Nat. Commun. 2019;10:5092. doi: 10.1038/s41467-019-13075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Hon CC, et al. An atlas of human long non-coding RNAs with accurate 5' ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.Melo CA, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol. Cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 542.Grossi E, et al. A lncRNA-SWI/SNF complex crosstalk controls transcriptional activation at specific promoter regions. Nat. Commun. 2020;11:936. doi: 10.1038/s41467-020-14623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Isoda T, et al. Non-coding transcription instructs chromatin folding and compartmentalization to dictate enhancer-promoter communication and T cell fate. Cell. 2017;171:103–119.e118. doi: 10.1016/j.cell.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Cai Z, et al. RIC-seq for global in situ profiling of RNA-RNA spatial interactions. Nature. 2020;582:432–437. doi: 10.1038/s41586-020-2249-1. [DOI] [PubMed] [Google Scholar]
- 545.Xiang JF, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Anderson KM, et al. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539:433–436. doi: 10.1038/nature20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Dao LTM, et al. Genome-wide characterization of mammalian promoters with distal enhancer functions. Nat. Genet. 2017;49:1073–1081. doi: 10.1038/ng.3884. [DOI] [PubMed] [Google Scholar]
- 548.Engreitz JM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Yap K, et al. A short tandem repeat-enriched RNA assembles a nuclear compartment to control alternative splicing and promote cell survival. Mol. Cell. 2018;72:525–540.e513. doi: 10.1016/j.molcel.2018.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 550.Wu H, et al. Unusual Processing generates SPA lncRNAs that sequester multiple RNA binding proteins. Mol. Cell. 2016;64:534–548. doi: 10.1016/j.molcel.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 551.Lee S, et al. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell. 2016;164:69–80. doi: 10.1016/j.cell.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Tichon A, Perry RB, Stojic L, Ulitsky I. SAM68 is required for regulation of Pumilio by the NORAD long noncoding RNA. Genes Dev. 2018;32:70–78. doi: 10.1101/gad.309138.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Liu B, et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 554.Kretz M, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555.Gong C, Maquat L. E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Wang J, Gong C, Maquat LE. Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev. 2013;27:793–804. doi: 10.1101/gad.212639.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Clemson CM, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.Tripathi V, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Yamazaki T, et al. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell. 2018;70:1038–1053.e1037. doi: 10.1016/j.molcel.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 561.Lin Y, Schmidt BF, Bruchez MP, McManus CJ. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res. 2018;46:3742–3752. doi: 10.1093/nar/gky046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 563.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Trimarchi T, et al. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158:593–606. doi: 10.1016/j.cell.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Bill M, et al. Expression and functional relevance of long non-coding RNAs in acute myeloid leukemia stem cells. Leukemia. 2019;33:2169–2182. doi: 10.1038/s41375-019-0429-5. [DOI] [PubMed] [Google Scholar]
- 566.Zhao P, et al. A novel lncRNA TCLlnc1 promotes peripheral T cell lymphoma progression through acting as a modular scaffold of HNRNPD and YBX1 complexes. Cell Death Dis. 2021;12:321. doi: 10.1038/s41419-021-03594-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Lyu Y, et al. Dysfunction of the WT1-MEG3 signaling promotes AML leukemogenesis via p53-dependent and -independent pathways. Leukemia. 2017;31:2543–2551. doi: 10.1038/leu.2017.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Guo G, et al. A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene. 2015;34:1768–1779. doi: 10.1038/onc.2014.131. [DOI] [PubMed] [Google Scholar]
- 569.Raj K, Mufti GJ. Azacytidine (Vidaza(R)) in the treatment of myelodysplastic syndromes. Ther. Clin. Risk Manag. 2006;2:377–388. doi: 10.2147/tcrm.2006.2.4.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Santos FP, Kantarjian H, Garcia-Manero G, Issa JP, Ravandi F. Decitabine in the treatment of myelodysplastic syndromes. Expert Rev. Anticancer Ther. 2010;10:9–22. doi: 10.1586/era.09.164. [DOI] [PubMed] [Google Scholar]
- 571.Kaminskas E, Farrell AT, Wang YC, Sridhara R, Pazdur R. FDA drug approval summary: azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist. 2005;10:176–182. doi: 10.1634/theoncologist.10-3-176. [DOI] [PubMed] [Google Scholar]
- 572.Hollenbach PW, et al. A comparison of azacitidine and decitabine activities in acute myeloid leukemia cell lines. PLoS ONE. 2010;5:e9001. doi: 10.1371/journal.pone.0009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Chilakala S, et al. Tracking decitabine incorporation into malignant myeloid cell DNA in vitro and in vivo by LC-MS/MS with enzymatic digestion. Sci. Rep. 2019;9:4558. doi: 10.1038/s41598-019-41070-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574.Chen J, Odenike O, Rowley JD. Leukaemogenesis: more than mutant genes. Nat. Rev. Cancer. 2010;10:23–36. doi: 10.1038/nrc2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Chabot GG, Bouchard J, Momparler RL. Kinetics of deamination of 5-aza-2'-deoxycytidine and cytosine arabinoside by human liver cytidine deaminase and its inhibition by 3-deazauridine, thymidine or uracil arabinoside. Biochem Pharm. 1983;32:1327–1328. doi: 10.1016/0006-2952(83)90293-9. [DOI] [PubMed] [Google Scholar]
- 576.Dhillon S. Decitabine/cedazuridine: first approval. Drugs. 2020;80:1373–1378. doi: 10.1007/s40265-020-01389-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577.Griffiths EA, et al. SGI-110: DNA methyltransferase inhibitor oncolytic. Drugs Future. 2013;38:535–543. [PMC free article] [PubMed] [Google Scholar]
- 578.Issa JJ, et al. Safety and tolerability of guadecitabine (SGI-110) in patients with myelodysplastic syndrome and acute myeloid leukaemia: a multicentre, randomised, dose-escalation phase 1 study. Lancet Oncol. 2015;16:1099–1110. doi: 10.1016/S1470-2045(15)00038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579.Kamachi K, et al. Targeting DNMT1 by demethylating agent OR-2100 increases tyrosine kinase inhibitors-sensitivity and depletes leukemic stem cells in chronic myeloid leukemia. Cancer Lett. 2022;526:273–283. doi: 10.1016/j.canlet.2021.11.032. [DOI] [PubMed] [Google Scholar]
- 580.Watanabe T, et al. Targeting aberrant DNA hypermethylation as a driver of ATL leukemogenesis by using the new oral demethylating agent OR-2100. Blood. 2020;136:871–884. doi: 10.1182/blood.2019003084. [DOI] [PubMed] [Google Scholar]
- 581.Silverman LR, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J. Clin. Oncol. 2002;20:2429–2440. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- 582.Fenaux P, et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 2009;10:223–232. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583.Wu D, et al. Decitabine for treatment of myelodysplastic syndromes in Chinese patients: an open-label, phase-3b study. Adv. Ther. 2015;32:1140–1159. doi: 10.1007/s12325-015-0263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Cheng JC, et al. Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J. Natl Cancer Inst. 2003;95:399–409. doi: 10.1093/jnci/95.5.399. [DOI] [PubMed] [Google Scholar]
- 585.Andrade AF, et al. Zebularine induces chemosensitization to methotrexate and efficiently decreases AhR gene methylation in childhood acute lymphoblastic leukemia cells. Anticancer Drugs. 2014;25:72–81. doi: 10.1097/CAD.0000000000000028. [DOI] [PubMed] [Google Scholar]
- 586.Holleran JL, et al. Plasma pharmacokinetics, oral bioavailability, and interspecies scaling of the DNA methyltransferase inhibitor, zebularine. Clin. Cancer Res. 2005;11:3862–3868. doi: 10.1158/1078-0432.CCR-04-2406. [DOI] [PubMed] [Google Scholar]
- 587.Pappalardi MB, et al. Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia. Nat. Cancer. 2021;2:1002–1017. doi: 10.1038/s43018-021-00249-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 588.Vitkeviciene A, Baksiene S, Borutinskaite V, Navakauskiene R. Epigallocatechin-3-gallate and BIX-01294 have different impact on epigenetics and senescence modulation in acute and chronic myeloid leukemia cells. Eur. J. Pharm. 2018;838:32–40. doi: 10.1016/j.ejphar.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 589.Shi X, Gao HY, Yan W, He XW, Yang W. Effects of EGCG on proliferation, cell cycle and DAPK1 gene methylation of acute promyelocytic leukemia NB4 cell line. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2018;26:1288–1293. doi: 10.7534/j.issn.1009-2137.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 590.Wu M, et al. Epigallocatechin gallate induces CHD5 gene demethylation to promote acute myeloid leukemia cell apoptosis in vitro by regulating p19(Arf)-p53-p21(Cip1) signaling pathway. Nan Fang. Yi Ke Da Xue Xue Bao. 2020;40:1230–1238. doi: 10.12122/j.issn.1673-4254.2020.09.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Borutinskaite V, Virksaite A, Gudelyte G, Navakauskiene R. Green tea polyphenol EGCG causes anti-cancerous epigenetic modulations in acute promyelocytic leukemia cells. Leuk. Lymphoma. 2018;59:469–478. doi: 10.1080/10428194.2017.1339881. [DOI] [PubMed] [Google Scholar]
- 592.Fan LP, et al. Effect of epigallocatechin-3-galate on human acute monocytic leukemia cell line U937 and its relevant mechanism. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2010;18:286–290. [PubMed] [Google Scholar]
- 593.Yu AF, Shen JZ, Chen ZZ, Fan LP, Lin FA. Demethylation and transcription of p16 gene in malignant lymphoma cell line CA46 induced by EGCG. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2008;16:1073–1078. [PubMed] [Google Scholar]
- 594.Pang J, et al. Thymoquinone exerts potent growth-suppressive activity on leukemia through DNA hypermethylation reversal in leukemia cells. Oncotarget. 2017;8:34453–34467. doi: 10.18632/oncotarget.16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595.Al-Rawashde FA, et al. Thymoquinone inhibits growth of acute myeloid leukemia cells through reversal SHP-1 and SOCS-3 hypermethylation: in vitro and in silico evaluation. Pharmaceuticals. 2021;14:1287. doi: 10.3390/ph14121287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596.Alvarez MC, Maso V, Torello CO, Ferro KP, Saad STO. The polyphenol quercetin induces cell death in leukemia by targeting epigenetic regulators of pro-apoptotic genes. Clin. Epigenet. 2018;10:139. doi: 10.1186/s13148-018-0563-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 597.Oodi A, Norouzi H, Amirizadeh N, Nikougoftar M, Vafaie Z. Harmine, a novel DNA methyltransferase 1 inhibitor in the leukemia cell line. Indian J. Hematol. Blood Transfus. 2017;33:509–515. doi: 10.1007/s12288-016-0770-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598.Yu J, et al. Curcumin down-regulates DNA methyltransferase 1 and plays an anti-leukemic role in acute myeloid leukemia. PLoS ONE. 2013;8:e55934. doi: 10.1371/journal.pone.0055934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599.Sun D, et al. Novel curcumin liposome modified with hyaluronan targeting CD44 plays an anti-leukemic role in acute myeloid leukemia in vitro and in vivo. ACS Appl. Mater. Interfaces. 2017;9:16857–16868. doi: 10.1021/acsami.7b02863. [DOI] [PubMed] [Google Scholar]
- 600.Kowolik CM, Lin M, Xie J, Overman LE, Horne DA. NT1721, a novel epidithiodiketopiperazine, exhibits potent in vitro and in vivo efficacy against acute myeloid leukemia. Oncotarget. 2016;7:86186–86197. doi: 10.18632/oncotarget.13364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601.Karam L, et al. Anticancer activities of parthenolide in primary effusion lymphoma preclinical models. Mol. Carcinog. 2021;60:567–581. doi: 10.1002/mc.23324. [DOI] [PubMed] [Google Scholar]
- 602.Liao M, et al. Oridonin inhibits DNMT3A R882 mutation-driven clonal hematopoiesis and leukemia by inducing apoptosis and necroptosis. Cell Death Disco. 2021;7:297. doi: 10.1038/s41420-021-00697-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 603.Brueckner B, et al. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65:6305–6311. doi: 10.1158/0008-5472.CAN-04-2957. [DOI] [PubMed] [Google Scholar]
- 604.Lin Y, Chen W, Wang Z, Cai P. Emodin promotes the arrest of human lymphoma Raji cell proliferation through the UHRF1-DNMT3A-∆Np73 pathways. Mol. Med. Rep. 2017;16:6544–6551. doi: 10.3892/mmr.2017.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 605.Qing Y, et al. Berberine induces apoptosis in human multiple myeloma cell line U266 through hypomethylation of p53 promoter. Cell Biol. Int. 2014;38:563–570. doi: 10.1002/cbin.10206. [DOI] [PubMed] [Google Scholar]
- 606.Klisovic RB, et al. A phase I biological study of MG98, an oligodeoxynucleotide antisense to DNA methyltransferase 1, in patients with high-risk myelodysplasia and acute myeloid leukemia. Clin. Cancer Res. 2008;14:2444–2449. doi: 10.1158/1078-0432.CCR-07-1320. [DOI] [PubMed] [Google Scholar]
- 607.Abaza Y, Zeidan AM. Immune checkpoint inhibition in acute myeloid leukemia and myelodysplastic syndromes. Cells. 2022;11:2249. doi: 10.3390/cells11142249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 608.Majeti R, et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138:286–299. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609.Sallman DA, et al. The first-in-class anti-CD47 antibody magrolimab (5F9) in combination with azacitidine is effective in MDS and AML patients: ongoing phase 1b results. Blood. 2019;134:569. doi: 10.1182/blood-2019-126271. [DOI] [Google Scholar]
- 610.Zeidan AM, et al. A multi-center phase I trial of ipilimumab in patients with myelodysplastic syndromes following hypomethylating agent failure. Clin. Cancer Res. 2018;24:3519–3527. doi: 10.1158/1078-0432.CCR-17-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 611.Daver N, et al. Azacitidine (AZA) with Nivolumab (Nivo), and AZA with Nivo + Ipilimumab (Ipi) in relapsed/refractory acute myeloid leukemia: a non-randomized, prospective, phase 2 study. Blood. 2019;134:830–830. doi: 10.1182/blood-2019-131494. [DOI] [Google Scholar]
- 612.Chien KS, et al. Phase II study of azacitidine with pembrolizumab in patients with intermediate-1 or higher-risk myelodysplastic syndrome. Br. J. Haematol. 2021;195:378–387. doi: 10.1111/bjh.17689. [DOI] [PubMed] [Google Scholar]
- 613.Nie J, et al. Addition of low-dose decitabine to anti-PD-1 antibody camrelizumab in relapsed/refractory classical Hodgkin lymphoma. J. Clin. Oncol. 2019;37:1479–1489. doi: 10.1200/JCO.18.02151. [DOI] [PubMed] [Google Scholar]
- 614.Gourd E. New treatment for relapsed or refractory Hodgkin’s lymphoma. Lancet Oncol. 2019;20:e298. doi: 10.1016/S1470-2045(19)30289-X. [DOI] [PubMed] [Google Scholar]
- 615.DiNardo CD, et al. Durable remissions with ivosidenib in IDH1-mutated relapsed or refractory AML. N. Engl. J. Med. 2018;378:2386–2398. doi: 10.1056/NEJMoa1716984. [DOI] [PubMed] [Google Scholar]
- 616.Watts JM, et al. Olutasidenib alone or with azacitidine in IDH1-mutated acute myeloid leukaemia and myelodysplastic syndrome: phase 1 results of a phase 1/2 trial. Lancet Haematol. 2022;10:e46–e58. doi: 10.1016/S2352-3026(22)00292-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 617.Stein EM, et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood. 2017;130:722–731. doi: 10.1182/blood-2017-04-779405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 618.DiNardo CD, et al. Mutant isocitrate dehydrogenase 1 inhibitor ivosidenib in combination with azacitidine for newly diagnosed acute myeloid leukemia. J. Clin. Oncol. 2021;39:57–65. doi: 10.1200/JCO.20.01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619.Montesinos P, et al. Ivosidenib and azacitidine in IDH1-mutated acute myeloid leukemia. N. Engl. J. Med. 2022;386:1519–1531. doi: 10.1056/NEJMoa2117344. [DOI] [PubMed] [Google Scholar]
- 620.Shanmugam G, Rakshit S, Sarkar K. HDAC inhibitors: Targets for tumor therapy, immune modulation and lung diseases. Transl. Oncol. 2022;16:101312. doi: 10.1016/j.tranon.2021.101312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 621.Guzman ML, et al. Selective activity of the histone deacetylase inhibitor AR-42 against leukemia stem cells: a novel potential strategy in acute myelogenous leukemia. Mol. Cancer Ther. 2014;13:1979–1990. doi: 10.1158/1535-7163.MCT-13-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 622.Canella A, et al. HDAC inhibitor AR-42 decreases CD44 expression and sensitizes myeloma cells to lenalidomide. Oncotarget. 2015;6:31134–31150. doi: 10.18632/oncotarget.5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Zhang S, et al. The novel histone deacetylase inhibitor, AR-42, inhibits gp130/Stat3 pathway and induces apoptosis and cell cycle arrest in multiple myeloma cells. Int. J. Cancer. 2011;129:204–213. doi: 10.1002/ijc.25660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 624.Liva SG, et al. Phase I study of AR-42 and decitabine in acute myeloid leukemia. Leuk. Lymphoma. 2020;61:1484–1492. doi: 10.1080/10428194.2020.1719095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 625.Sborov DW, et al. A phase 1 trial of the HDAC inhibitor AR-42 in patients with multiple myeloma and T- and B-cell lymphomas. Leuk. Lymphoma. 2017;58:2310–2318. doi: 10.1080/10428194.2017.1298751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 626.Olsen EA, et al. Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma. J. Clin. Oncol. 2007;25:3109–3115. doi: 10.1200/JCO.2006.10.2434. [DOI] [PubMed] [Google Scholar]
- 627.Duvic M, et al. Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL) Blood. 2007;109:31–39. doi: 10.1182/blood-2006-06-025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 628.O'Connor OA, et al. Belinostat in patients with relapsed or refractory peripheral T-cell lymphoma: results of the pivotal phase II BELIEF (CLN-19) study. J. Clin. Oncol. 2015;33:2492–2499. doi: 10.1200/JCO.2014.59.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 629.San-Miguel JF, et al. Overall survival of patients with relapsed multiple myeloma treated with panobinostat or placebo plus bortezomib and dexamethasone (the PANORAMA 1 trial): a randomised, placebo-controlled, phase 3 trial. Lancet Haematol. 2016;3:e506–e515. doi: 10.1016/S2352-3026(16)30147-8. [DOI] [PubMed] [Google Scholar]
- 630.San-Miguel JF, et al. Panobinostat plus bortezomib and dexamethasone versus placebo plus bortezomib and dexamethasone in patients with relapsed or relapsed and refractory multiple myeloma: a multicentre, randomised, double-blind phase 3 trial. Lancet Oncol. 2014;15:1195–1206. doi: 10.1016/S1470-2045(14)70440-1. [DOI] [PubMed] [Google Scholar]
- 631.Richardson PG, et al. PANORAMA 2: panobinostat in combination with bortezomib and dexamethasone in patients with relapsed and bortezomib-refractory myeloma. Blood. 2013;122:2331–2337. doi: 10.1182/blood-2013-01-481325. [DOI] [PubMed] [Google Scholar]
- 632.Popat R, et al. Bortezomib, thalidomide, dexamethasone, and panobinostat for patients with relapsed multiple myeloma (MUK-six): a multicentre, open-label, phase 1/2 trial. Lancet Haematol. 2016;3:e572–e580. doi: 10.1016/S2352-3026(16)30165-X. [DOI] [PubMed] [Google Scholar]
- 633.Amaru Calzada A, et al. Givinostat and hydroxyurea synergize in vitro to induce apoptosis of cells from JAK2(V617F) myeloproliferative neoplasm patients. Exp. Hematol. 2013;41:253–260.e252. doi: 10.1016/j.exphem.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 634.Amaru Calzada A, et al. The HDAC inhibitor Givinostat modulates the hematopoietic transcription factors NFE2 and C-MYB in JAK2(V617F) myeloproliferative neoplasm cells. Exp. Hematol. 2012;40:634–645.e610. doi: 10.1016/j.exphem.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 635.Yao C, et al. Potent induction of apoptosis by givinostat in BCR-ABL1-positive and BCR-ABL1-negative precursor B-cell acute lymphoblastic leukemia cell lines. Leuk. Res. 2017;60:129–134. doi: 10.1016/j.leukres.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 636.Savino AM, et al. The histone deacetylase inhibitor givinostat (ITF2357) exhibits potent anti-tumor activity against CRLF2-rearranged BCP-ALL. Leukemia. 2017;31:2365–2375. doi: 10.1038/leu.2017.93. [DOI] [PubMed] [Google Scholar]
- 637.Pinazza M, et al. An immediate transcriptional signature associated with response to the histone deacetylase inhibitor Givinostat in T acute lymphoblastic leukemia xenografts. Cell Death Dis. 2016;6:e2047. doi: 10.1038/cddis.2015.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638.Rambaldi A, et al. Long-term safety and efficacy of givinostat in polycythemia vera: 4-year mean follow up of three phase 1/2 studies and a compassionate use program. Blood Cancer J. 2021;11:53. doi: 10.1038/s41408-021-00445-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 639.Rambaldi A, et al. Safety and efficacy of the maximum tolerated dose of givinostat in polycythemia vera: a two-part Phase Ib/II study. Leukemia. 2020;34:2234–2237. doi: 10.1038/s41375-020-0735-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 640.Finazzi G, et al. A phase II study of Givinostat in combination with hydroxycarbamide in patients with polycythaemia vera unresponsive to hydroxycarbamide monotherapy. Br. J. Haematol. 2013;161:688–694. doi: 10.1111/bjh.12332. [DOI] [PubMed] [Google Scholar]
- 641.Stühmer T, et al. Preclinical anti-myeloma activity of the novel HDAC-inhibitor JNJ-26481585. Br. J. Haematol. 2010;149:529–536. doi: 10.1111/j.1365-2141.2010.08126.x. [DOI] [PubMed] [Google Scholar]
- 642.Deleu S, et al. The effects of JNJ-26481585, a novel hydroxamate-based histone deacetylase inhibitor, on the development of multiple myeloma in the 5T2MM and 5T33MM murine models. Leukemia. 2009;23:1894–1903. doi: 10.1038/leu.2009.121. [DOI] [PubMed] [Google Scholar]
- 643.Deleu S, et al. Bortezomib alone or in combination with the histone deacetylase inhibitor JNJ-26481585: effect on myeloma bone disease in the 5T2MM murine model of myeloma. Cancer Res. 2009;69:5307–5311. doi: 10.1158/0008-5472.CAN-08-4472. [DOI] [PubMed] [Google Scholar]
- 644.Moreau P, et al. Quisinostat, bortezomib, and dexamethasone combination therapy for relapsed multiple myeloma. Leuk. Lymphoma. 2016;57:1546–1559. doi: 10.3109/10428194.2015.1117611. [DOI] [PubMed] [Google Scholar]
- 645.Child F, et al. Phase II multicentre trial of oral quisinostat, a histone deacetylase inhibitor, in patients with previously treated stage IB-IVA mycosis fungoides/Sézary syndrome. Br. J. Dermatol. 2016;175:80–88. doi: 10.1111/bjd.14427. [DOI] [PubMed] [Google Scholar]
- 646.Zabkiewicz J, et al. The targeted histone deacetylase inhibitor tefinostat (CHR-2845) shows selective in vitro efficacy in monocytoid-lineage leukaemias. Oncotarget. 2016;7:16650–16662. doi: 10.18632/oncotarget.7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 647.Ossenkoppele GJ, et al. A phase I first-in-human study with tefinostat - a monocyte/macrophage targeted histone deacetylase inhibitor - in patients with advanced haematological malignancies. Br. J. Haematol. 2013;162:191–201. doi: 10.1111/bjh.12359. [DOI] [PubMed] [Google Scholar]
- 648.Mandl-Weber S, et al. The novel inhibitor of histone deacetylase resminostat (RAS2410) inhibits proliferation and induces apoptosis in multiple myeloma (MM) cells. Br. J. Haematol. 2010;149:518–528. doi: 10.1111/j.1365-2141.2010.08124.x. [DOI] [PubMed] [Google Scholar]
- 649.Karagianni F, et al. Combination of resminostat with ruxolitinib exerts antitumor effects in the chick embryo chorioallantoic membrane model for cutaneous T cell lymphoma. Cancers. 2022;14:1070. doi: 10.3390/cancers14041070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 650.Walewski J, et al. Resminostat in patients with relapsed or refractory Hodgkin lymphoma: results of the phase II SAPHIRE study. Leuk. Lymphoma. 2019;60:675–684. doi: 10.1080/10428194.2018.1492122. [DOI] [PubMed] [Google Scholar]
- 651.Markozashvili D, et al. Histone deacetylase inhibitor abexinostat affects chromatin organization and gene transcription in normal B cells and in mantle cell lymphoma. Gene. 2016;580:134–143. doi: 10.1016/j.gene.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 652.Bhalla S, et al. PCI-24781 induces caspase and reactive oxygen species-dependent apoptosis through NF-kappaB mechanisms and is synergistic with bortezomib in lymphoma cells. Clin. Cancer Res. 2009;15:3354–3365. doi: 10.1158/1078-0432.CCR-08-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 653.Evens AM, et al. A phase I/II multicenter, open-label study of the oral histone deacetylase inhibitor abexinostat in relapsed/refractory lymphoma. Clin. Cancer Res. 2016;22:1059–1066. doi: 10.1158/1078-0432.CCR-15-0624. [DOI] [PubMed] [Google Scholar]
- 654.Kim SJ, Kim S, Choi YJ, Kim UJ, Kang KW. CKD-581 downregulates Wnt/β-catenin pathway by DACT3 induction in hematologic malignancy. Biomol. Ther. 2022;30:435–446. doi: 10.4062/biomolther.2022.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 655.Kim SJ, Kim UJ, Yoo HY, Choi YJ, Kang KW. Anti-cancer effects of CKD-581, a potent histone deacetylase inhibitor against diffuse large B-Cell lymphoma. Int. J. Mol. Sci. 2020;21:4377. doi: 10.3390/ijms21124377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 656.Cho H, et al. Phase I study of CKD-581, a pan-histone deacetylase inhibitor, in patients with lymphoma or multiple myeloma refractory to standard therapy. Invest. N. Drugs. 2018;36:877–885. doi: 10.1007/s10637-018-0582-0. [DOI] [PubMed] [Google Scholar]
- 657.North BJ, et al. Enhancement of pomalidomide anti-tumor response with ACY-241, a selective HDAC6 inhibitor. PLoS ONE. 2017;12:e0173507. doi: 10.1371/journal.pone.0173507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 658.Kilgour JM, et al. Phase II open-label, single-arm trial to investigate the efficacy and safety of topical remetinostat gel in patients with basal cell carcinoma. Clin. Cancer Res. 2021;27:4717–4725. doi: 10.1158/1078-0432.CCR-21-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 659.Rauzan M, Chuah CT, Ko TK, Ong ST. The HDAC inhibitor SB939 overcomes resistance to BCR-ABL kinase Inhibitors conferred by the BIM deletion polymorphism in chronic myeloid leukemia. PLoS ONE. 2017;12:e0174107. doi: 10.1371/journal.pone.0174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 660.Novotny-Diermayr V, et al. The oral HDAC inhibitor pracinostat (SB939) is efficacious and synergistic with the JAK2 inhibitor pacritinib (SB1518) in preclinical models of AML. Blood Cancer J. 2012;2:e69. doi: 10.1038/bcj.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 661.Yalniz FF, et al. A phase II study of addition of pracinostat to a hypomethylating agent in patients with myelodysplastic syndromes who have not responded to previous hypomethylating agent therapy. Br. J. Haematol. 2020;188:404–412. doi: 10.1111/bjh.16173. [DOI] [PubMed] [Google Scholar]
- 662.Bose P, et al. A phase 2 study of pracinostat combined with ruxolitinib in patients with myelofibrosis. Leuk. Lymphoma. 2019;60:1767–1774. doi: 10.1080/10428194.2018.1543876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 663.Garcia-Manero G, et al. Phase 2, randomized, double-blind study of pracinostat in combination with azacitidine in patients with untreated, higher-risk myelodysplastic syndromes. Cancer. 2017;123:994–1002. doi: 10.1002/cncr.30533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 664.Quintás-Cardama A, et al. Therapy with the histone deacetylase inhibitor pracinostat for patients with myelofibrosis. Leuk. Res. 2012;36:1124–1127. doi: 10.1016/j.leukres.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 665.Santo L, et al. Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood. 2012;119:2579–2589. doi: 10.1182/blood-2011-10-387365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 666.García-Guerrero E, et al. Upregulation of CD38 expression on multiple myeloma cells by novel HDAC6 inhibitors is a class effect and augments the efficacy of daratumumab. Leukemia. 2021;35:201–214. doi: 10.1038/s41375-020-0840-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 667.Amengual JE, et al. Dual targeting of protein degradation pathways with the selective HDAC6 inhibitor ACY-1215 and bortezomib is synergistic in lymphoma. Clin. Cancer Res. 2015;21:4663–4675. doi: 10.1158/1078-0432.CCR-14-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 668.Vekaria PH, et al. Functional cooperativity of p97 and histone deacetylase 6 in mediating DNA repair in mantle cell lymphoma cells. Leukemia. 2019;33:1675–1686. doi: 10.1038/s41375-018-0355-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 669.Lee DH, Kim GW, Kwon SH. The HDAC6-selective inhibitor is effective against non-Hodgkin lymphoma and synergizes with ibrutinib in follicular lymphoma. Mol. Carcinog. 2019;58:944–956. doi: 10.1002/mc.22983. [DOI] [PubMed] [Google Scholar]
- 670.Vogl DT, et al. Ricolinostat, the first selective histone deacetylase 6 inhibitor, in combination with bortezomib and dexamethasone for relapsed or refractory multiple myeloma. Clin. Cancer Res. 2017;23:3307–3315. doi: 10.1158/1078-0432.CCR-16-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 671.Yee AJ, et al. Ricolinostat plus lenalidomide, and dexamethasone in relapsed or refractory multiple myeloma: a multicentre phase 1b trial. Lancet Oncol. 2016;17:1569–1578. doi: 10.1016/S1470-2045(16)30375-8. [DOI] [PubMed] [Google Scholar]
- 672.Zhu Z, et al. Therapeutic efficacy of an injectable formulation of purinostat mesylate in SU-DHL-6 tumour model. Ann. Med. 2022;54:743–753. doi: 10.1080/07853890.2022.2045347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 673.Yang L, et al. Purinostat Mesylate is a uniquely potent and selective inhibitor of HDACs for the treatment of BCR-ABL-induced B-cell acute lymphoblastic leukemia. Clin. Cancer Res. 2019;25:7527–7539. doi: 10.1158/1078-0432.CCR-19-0516. [DOI] [PubMed] [Google Scholar]
- 674.Lu X, Ning Z, Li Z, Cao H, Wang X. Development of chidamide for peripheral T-cell lymphoma, the first orphan drug approved in China. Intractable Rare Dis. Res. 2016;5:185–191. doi: 10.5582/irdr.2016.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 675.Ning ZQ, et al. Chidamide (CS055/HBI-8000): a new histone deacetylase inhibitor of the benzamide class with antitumor activity and the ability to enhance immune cell-mediated tumor cell cytotoxicity. Cancer Chemother. Pharm. 2012;69:901–909. doi: 10.1007/s00280-011-1766-x. [DOI] [PubMed] [Google Scholar]
- 676.Shi Y, et al. Results from a multicenter, open-label, pivotal phase II study of chidamide in relapsed or refractory peripheral T-cell lymphoma. Ann. Oncol. 2015;26:1766–1771. doi: 10.1093/annonc/mdv237. [DOI] [PubMed] [Google Scholar]
- 677.Shi Y, et al. Chidamide in relapsed or refractory peripheral T cell lymphoma: a multicenter real-world study in China. J. Hematol. Oncol. 2017;10:69. doi: 10.1186/s13045-017-0439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 678.Zhang MC, et al. Clinical efficacy and molecular biomarkers in a phase II study of tucidinostat plus R-CHOP in elderly patients with newly diagnosed diffuse large B-cell lymphoma. Clin. Epigenet. 2020;12:160. doi: 10.1186/s13148-020-00948-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 679.Zhou L, et al. HDAC inhibition by SNDX-275 (Entinostat) restores expression of silenced leukemia-associated transcription factors Nur77 and Nor1 and of key pro-apoptotic proteins in AML. Leukemia. 2013;27:1358–1368. doi: 10.1038/leu.2012.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 680.Ramsey JM, et al. Entinostat prevents leukemia maintenance in a collaborating oncogene-dependent model of cytogenetically normal acute myeloid leukemia. Stem Cells. 2013;31:1434–1445. doi: 10.1002/stem.1398. [DOI] [PubMed] [Google Scholar]
- 681.Duque-Afonso J, et al. The HDAC class I-specific inhibitor entinostat (MS-275) effectively relieves epigenetic silencing of the LAT2 gene mediated by AML1/ETO. Oncogene. 2011;30:3062–3072. doi: 10.1038/onc.2011.32. [DOI] [PubMed] [Google Scholar]
- 682.Frys S, et al. Entinostat, a novel histone deacetylase inhibitor is active in B-cell lymphoma and enhances the anti-tumour activity of rituximab and chemotherapy agents. Br. J. Haematol. 2015;169:506–519. doi: 10.1111/bjh.13318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 683.Cai B, et al. Combination of bendamustine and entinostat synergistically inhibits proliferation of multiple myeloma cells via induction of apoptosis and DNA damage response. Cancer Lett. 2013;335:343–350. doi: 10.1016/j.canlet.2013.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 684.Jóna A, et al. The histone deacetylase inhibitor entinostat (SNDX-275) induces apoptosis in Hodgkin lymphoma cells and synergizes with Bcl-2 family inhibitors. Exp. Hematol. 2011;39:1007–1017.e1001. doi: 10.1016/j.exphem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 685.Prebet T, et al. Azacitidine with or without Entinostat for the treatment of therapy-related myeloid neoplasm: further results of the E1905 North American Leukemia Intergroup study. Br. J. Haematol. 2016;172:384–391. doi: 10.1111/bjh.13832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 686.Prebet T, et al. Prolonged administration of azacitidine with or without entinostat for myelodysplastic syndrome and acute myeloid leukemia with myelodysplasia-related changes: results of the US Leukemia Intergroup trial E1905. J. Clin. Oncol. 2014;32:1242–1248. doi: 10.1200/JCO.2013.50.3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 687.von Tresckow B, et al. Phase I study of domatinostat (4SC-202), a class I histone deacetylase inhibitor in patients with advanced hematological malignancies. Eur. J. Haematol. 2019;102:163–173. doi: 10.1111/ejh.13188. [DOI] [PubMed] [Google Scholar]
- 688.Smolewski P, Robak T. The discovery and development of romidepsin for the treatment of T-cell lymphoma. Expert Opin. Drug Discov. 2017;12:859–873. doi: 10.1080/17460441.2017.1341487. [DOI] [PubMed] [Google Scholar]
- 689.Whittaker SJ, et al. Final results from a multicenter, international, pivotal study of romidepsin in refractory cutaneous T-cell lymphoma. J. Clin. Oncol. 2010;28:4485–4491. doi: 10.1200/JCO.2010.28.9066. [DOI] [PubMed] [Google Scholar]
- 690.Piekarz RL, et al. Phase II multi-institutional trial of the histone deacetylase inhibitor romidepsin as monotherapy for patients with cutaneous T-cell lymphoma. J. Clin. Oncol. 2009;27:5410–5417. doi: 10.1200/JCO.2008.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 691.Piekarz RL, et al. Phase 2 trial of romidepsin in patients with peripheral T-cell lymphoma. Blood. 2011;117:5827–5834. doi: 10.1182/blood-2010-10-312603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 692.Li H, Cui R, Ji M, Jin SY. CUDC-101 enhances the chemosensitivity of gemcitabine-treated lymphoma cells. Leuk. Res. 2021;106:106575. doi: 10.1016/j.leukres.2021.106575. [DOI] [PubMed] [Google Scholar]
- 693.Zhang T, et al. CUDC-101 overcomes arsenic trioxide resistance via caspase-dependent promyelocytic leukemia-retinoic acid receptor alpha degradation in acute promyelocytic leukemia. Anticancer Drugs. 2020;31:158–168. doi: 10.1097/CAD.0000000000000847. [DOI] [PubMed] [Google Scholar]
- 694.Yang EG, et al. Design and synthesis of Janus kinase 2 (JAK2) and histone deacetlyase (HDAC) bispecific inhibitors based on pacritinib and evidence of dual pathway inhibition in hematological cell Lines. J. Med Chem. 2016;59:8233–8262. doi: 10.1021/acs.jmedchem.6b00157. [DOI] [PubMed] [Google Scholar]
- 695.Oki Y, et al. CUDC-907 in relapsed/refractory diffuse large B-cell lymphoma, including patients with MYC-alterations: results from an expanded phase I trial. Haematologica. 2017;102:1923–1930. doi: 10.3324/haematol.2017.172882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 696.Duan YC, et al. Design and synthesis of tranylcypromine derivatives as novel LSD1/HDACs dual inhibitors for cancer treatment. Eur. J. Med. Chem. 2017;140:392–402. doi: 10.1016/j.ejmech.2017.09.038. [DOI] [PubMed] [Google Scholar]
- 697.Zhang X, et al. Design, synthesis and biological evaluation of colchicine derivatives as novel tubulin and histone deacetylase dual inhibitors. Eur. J. Med. Chem. 2015;95:127–135. doi: 10.1016/j.ejmech.2015.03.035. [DOI] [PubMed] [Google Scholar]
- 698.Yang K, et al. Development of the first small molecule histone deacetylase 6 (HDAC6) degraders. Bioorg. Med. Chem. Lett. 2018;28:2493–2497. doi: 10.1016/j.bmcl.2018.05.057. [DOI] [PubMed] [Google Scholar]
- 699.An Z, Lv W, Su S, Wu W, Rao Y. Developing potent PROTACs tools for selective degradation of HDAC6 protein. Protein Cell. 2019;10:606–609. doi: 10.1007/s13238-018-0602-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 700.Yang H, et al. Plasticity in designing PROTACs for selective and potent degradation of HDAC6. Chem. Commun. 2019;55:14848–14851. doi: 10.1039/C9CC08509B. [DOI] [PubMed] [Google Scholar]
- 701.Yang K, et al. Development of selective histone deacetylase 6 (HDAC6) degraders recruiting Von Hippel-Lindau (VHL) E3 ubiquitin ligase. ACS Med Chem. Lett. 2020;11:575–581. doi: 10.1021/acsmedchemlett.0c00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 702.Xiao Y, et al. Discovery of histone deacetylase 3 (HDAC3)-specific PROTACs. Chem. Commun. 2020;56:9866–9869. doi: 10.1039/D0CC03243C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 703.Blum W, et al. Phase I study of decitabine alone or in combination with valproic acid in acute myeloid leukemia. J. Clin. Oncol. 2007;25:3884–3891. doi: 10.1200/JCO.2006.09.4169. [DOI] [PubMed] [Google Scholar]
- 704.Lübbert M, et al. Valproate and retinoic acid in combination with decitabine in elderly nonfit patients with acute myeloid leukemia: results of a multicenter, randomized, 2 × 2, phase II trial. J. Clin. Oncol. 2020;38:257–270. doi: 10.1200/JCO.19.01053. [DOI] [PubMed] [Google Scholar]
- 705.How J, et al. A phase I trial of two sequence-specific schedules of decitabine and vorinostat in patients with acute myeloid leukemia. Leuk. Lymphoma. 2015;56:2793–2802. doi: 10.3109/10428194.2015.1018248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 706.Burke MJ, et al. Decitabine and vorinostat with chemotherapy in relapsed pediatric acute lymphoblastic leukemia: a TACL pilot study. Clin. Cancer Res. 2020;26:2297–2307. doi: 10.1158/1078-0432.CCR-19-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 707.Burke MJ, et al. A therapeutic trial of decitabine and vorinostat in combination with chemotherapy for relapsed/refractory acute lymphoblastic leukemia. Am. J. Hematol. 2014;89:889–895. doi: 10.1002/ajh.23778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 708.Borcoman E, et al. HDAC inhibition to prime immune checkpoint inhibitors. Cancers. 2021;14:66. doi: 10.3390/cancers14010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 709.He Y, et al. HDAC inhibitor LBH589 suppresses the proliferation but enhances the antileukemic effect of human γδT cells. Mol. Ther. Oncolytics. 2020;18:623–630. doi: 10.1016/j.omto.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 710.Salmon JM, et al. Epigenetic activation of plasmacytoid DCs drives IFNAR-dependent therapeutic differentiation of AML. Cancer Discov. 2022;12:1560–1579. doi: 10.1158/2159-8290.CD-20-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 711.Moldenhauer A, et al. Histone deacetylase inhibition improves dendritic cell differentiation of leukemic blasts with AML1-containing fusion proteins. J. Leukoc. Biol. 2004;76:623–633. doi: 10.1189/jlb.1103581. [DOI] [PubMed] [Google Scholar]
- 712.He ZX, et al. Current development of CBP/p300 inhibitors in the last decade. Eur. J. Med. Chem. 2021;209:112861. doi: 10.1016/j.ejmech.2020.112861. [DOI] [PubMed] [Google Scholar]
- 713.Coudé MM, et al. BET inhibitor OTX015 targets BRD2 and BRD4 and decreases c-MYC in acute leukemia cells. Oncotarget. 2015;6:17698–17712. doi: 10.18632/oncotarget.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 714.Boi M, et al. The BET bromodomain inhibitor OTX015 affects pathogenetic pathways in preclinical B-cell tumor models and synergizes with targeted drugs. Clin. Cancer Res. 2015;21:1628–1638. doi: 10.1158/1078-0432.CCR-14-1561. [DOI] [PubMed] [Google Scholar]
- 715.Amorim S, et al. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: a dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol. 2016;3:e196–e204. doi: 10.1016/S2352-3026(16)00021-1. [DOI] [PubMed] [Google Scholar]
- 716.Stathis A, Bertoni F. BET proteins as targets for anticancer treatment. Cancer Disco. 2018;8:24–36. doi: 10.1158/2159-8290.CD-17-0605. [DOI] [PubMed] [Google Scholar]
- 717.Wu S, et al. BRD4 PROTAC degrader ARV-825 inhibits T-cell acute lymphoblastic leukemia by targeting 'Undruggable' Myc-pathway genes. Cancer Cell Int. 2021;21:230. doi: 10.1186/s12935-021-01908-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 718.Saenz DT, et al. Targeting nuclear β-catenin as therapy for post-myeloproliferative neoplasm secondary AML. Leukemia. 2019;33:1373–1386. doi: 10.1038/s41375-018-0334-3. [DOI] [PubMed] [Google Scholar]
- 719.Sun B, et al. BET protein proteolysis targeting chimera (PROTAC) exerts potent lethal activity against mantle cell lymphoma cells. Leukemia. 2018;32:343–352. doi: 10.1038/leu.2017.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 720.Saenz DT, et al. Novel BET protein proteolysis-targeting chimera exerts superior lethal activity than bromodomain inhibitor (BETi) against post-myeloproliferative neoplasm secondary (s) AML cells. Leukemia. 2017;31:1951–1961. doi: 10.1038/leu.2016.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 721.Lu J, et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chemistry. Biol. 2015;22:755–763. doi: 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 722.Winter GE, et al. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 723.Duan R, Du W, Guo W. EZH2: a novel target for cancer treatment. J. Hematol. Oncol. 2020;13:104. doi: 10.1186/s13045-020-00937-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 724.Stein EM, et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood. 2018;131:2661–2669. doi: 10.1182/blood-2017-12-818948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 725.Fang Y, Liao G, Yu B. LSD1/KDM1A inhibitors in clinical trials: advances and prospects. J. Hematol. Oncol. 2019;12:129. doi: 10.1186/s13045-019-0811-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 726.Pandey G, Kuykendall AT, Reuther GW. JAK2 inhibitor persistence in MPN: uncovering a central role of ERK activation. Blood. Cancer J. 2022;12:13. doi: 10.1038/s41408-022-00609-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 727.Dao KT, et al. Efficacy of ruxolitinib in patients with chronic neutrophilic leukemia and atypical chronic myeloid leukemia. J. Clin. Oncol. 2020;38:1006–1018. doi: 10.1200/JCO.19.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 728.Bose P, et al. A phase 1/2 study of ruxolitinib and decitabine in patients with post-myeloproliferative neoplasm acute myeloid leukemia. Leukemia. 2020;34:2489–2492. doi: 10.1038/s41375-020-0778-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 729.Qi W, et al. AT9283, a novel aurora kinase inhibitor, suppresses tumor growth in aggressive B-cell lymphomas. Int. J. Cancer. 2012;130:2997–3005. doi: 10.1002/ijc.26324. [DOI] [PubMed] [Google Scholar]
- 730.Qi W, et al. Aurora inhibitor MLN8237 in combination with docetaxel enhances apoptosis and anti-tumor activity in mantle cell lymphoma. Biochem. Pharm. 2011;81:881–890. doi: 10.1016/j.bcp.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 731.Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;39:167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 732.Kluiver J, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J. Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 733.Calin GA, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl Acad. Sci. USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 734.Kluiver J, et al. Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes Chromosomes Cancer. 2006;45:147–153. doi: 10.1002/gcc.20273. [DOI] [PubMed] [Google Scholar]
- 735.Gerloff D, et al. NF-κB/STAT5/miR-155 network targets PU.1 in FLT3-ITD-driven acute myeloid leukemia. Leukemia. 2015;29:535–547. doi: 10.1038/leu.2014.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 736.Wallace JA, et al. miR-155 promotes FLT3-ITD-induced myeloproliferative disease through inhibition of the interferon response. Blood. 2017;129:3074–3086. doi: 10.1182/blood-2016-09-740209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 737.Wang M, et al. miRNA analysis in B-cell chronic lymphocytic leukaemia: proliferation centres characterized by low miR-150 and high BIC/miR-155 expression. J. Pathol. 2008;215:13–20. doi: 10.1002/path.2333. [DOI] [PubMed] [Google Scholar]
- 738.Hussein K, et al. Significant inverse correlation of microRNA-150/MYB and microRNA-222/p27 in myelodysplastic syndrome. Leuk. Res. 2010;34:328–334. doi: 10.1016/j.leukres.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 739.Jiang X, et al. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell. 2012;22:524–535. doi: 10.1016/j.ccr.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 740.Felli N, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc. Natl Acad. Sci. USA. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 741.Herling M, et al. TCL1 shows a regulated expression pattern in chronic lymphocytic leukemia that correlates with molecular subtypes and proliferative state. Leukemia. 2006;20:280–285. doi: 10.1038/sj.leu.2404017. [DOI] [PubMed] [Google Scholar]
- 742.Calin GA, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 743.Debernardi S, et al. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21:912–916. doi: 10.1038/sj.leu.2404605. [DOI] [PubMed] [Google Scholar]
- 744.Kitada S, et al. Expression of apoptosis-regulating proteins in chronic lymphocytic leukemia: correlations with In vitro and In vivo chemoresponses. Blood. 1998;91:3379–3389. doi: 10.1182/blood.V91.9.3379. [DOI] [PubMed] [Google Scholar]
- 745.Naguibneva I, et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat. Cell Biol. 2006;8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 746.Roccaro AM, et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood. 2009;113:6669–6680. doi: 10.1182/blood-2009-01-198408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 747.Akao Y, Nakagawa Y, Kitade Y, Kinoshita T, Naoe T. Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci. 2007;98:1914–1920. doi: 10.1111/j.1349-7006.2007.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 748.Mets E, et al. MicroRNA-193b-3p acts as a tumor suppressor by targeting the MYB oncogene in T-cell acute lymphoblastic leukemia. Leukemia. 2015;29:798–806. doi: 10.1038/leu.2014.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 749.Boldrin E, et al. MicroRNA-497/195 is tumor suppressive and cooperates with CDKN2A/B in pediatric acute lymphoblastic leukemia. Blood. 2021;138:1953–1965. doi: 10.1182/blood.2020007591. [DOI] [PubMed] [Google Scholar]
- 750.Jiang X, et al. miR-22 has a potent anti-tumour role with therapeutic potential in acute myeloid leukaemia. Nat. Commun. 2016;7:11452. doi: 10.1038/ncomms11452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 751.Shen C, et al. The PU.1-modulated microRNA-22 Is a regulator of monocyte/macrophage differentiation and acute myeloid leukemia. PLoS Genet. 2016;12:e1006259. doi: 10.1371/journal.pgen.1006259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 752.Chen P, et al. miR-9 is an essential oncogenic microRNA specifically overexpressed in mixed lineage leukemia-rearranged leukemia. Proc. Natl Acad. Sci. USA. 2013;110:11511–11516. doi: 10.1073/pnas.1310144110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 753.Senyuk V, et al. Critical role of miR-9 in myelopoiesis and EVI1-induced leukemogenesis. Proc. Natl Acad. Sci. USA. 2013;110:5594–5599. doi: 10.1073/pnas.1302645110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 754.Emmrich S, et al. miR-9 is a tumor suppressor in pediatric AML with t(8;21) Leukemia. 2014;28:1022–1032. doi: 10.1038/leu.2013.357. [DOI] [PubMed] [Google Scholar]
- 755.Mi S, et al. Aberrant overexpression and function of the miR-17-92 cluster in MLL-rearranged acute leukemia. Proc. Natl Acad. Sci. USA. 2010;107:3710–3715. doi: 10.1073/pnas.0914900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 756.Wong P, et al. The miR-17-92 microRNA polycistron regulates MLL leukemia stem cell potential by modulating p21 expression. Cancer Res. 2010;70:3833–3842. doi: 10.1158/0008-5472.CAN-09-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 757.Starczynowski DT, et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat. Med. 2010;16:49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 758.Zhao JL, et al. NF-kappaB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc. Natl Acad. Sci. USA. 2011;108:9184–9189. doi: 10.1073/pnas.1105398108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 759.Boldin MP, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 2011;208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 760.Su YL, et al. Myeloid cell-targeted miR-146a mimic inhibits NF-κB-driven inflammation and leukemia progression in vivo. Blood. 2020;135:167–180. doi: 10.1182/blood.2019002045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 761.Bousquet M, et al. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J. Exp. Med. 2008;205:2499–2506. doi: 10.1084/jem.20080285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 762.So AY, et al. Dual mechanisms by which miR-125b represses IRF4 to induce myeloid and B-cell leukemias. Blood. 2014;124:1502–1512. doi: 10.1182/blood-2014-02-553842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 763.Li Z, et al. Overexpression and knockout of miR-126 both promote leukemogenesis. Blood. 2015;126:2005–2015. doi: 10.1182/blood-2015-04-639062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 764.de Leeuw DC, et al. Attenuation of microRNA-126 expression that drives CD34+38- stem/progenitor cells in acute myeloid leukemia leads to tumor eradication. Cancer Res. 2014;74:2094–2105. doi: 10.1158/0008-5472.CAN-13-1733. [DOI] [PubMed] [Google Scholar]
- 765.Lechman ER, et al. miR-126 regulates distinct self-renewal outcomes in normal and malignant hematopoietic stem cells. Cancer Cell. 2016;29:214–228. doi: 10.1016/j.ccell.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 766.Gao XN, et al. MicroRNA-193a represses c-kit expression and functions as a methylation-silenced tumor suppressor in acute myeloid leukemia. Oncogene. 2011;30:3416–3428. doi: 10.1038/onc.2011.62. [DOI] [PubMed] [Google Scholar]
- 767.Li Y, et al. Epigenetic silencing of microRNA-193a contributes to leukemogenesis in t(8;21) acute myeloid leukemia by activating the PTEN/PI3K signal pathway. Blood. 2013;121:499–509. doi: 10.1182/blood-2012-07-444729. [DOI] [PubMed] [Google Scholar]
- 768.Popovic R, et al. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;113:3314–3322. doi: 10.1182/blood-2008-04-154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 769.Li Z, et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat. Commun. 2012;3:688. doi: 10.1038/ncomms1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 770.Bhayadia R, et al. Endogenous tumor suppressor microRNA-193b: therapeutic and prognostic value in acute myeloid leukemia. J. Clin. Oncol. 2018;36:1007–1016. doi: 10.1200/JCO.2017.75.2204. [DOI] [PubMed] [Google Scholar]
- 771.Fazi F, et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12:457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 772.Pulikkan JA, et al. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115:1768–1778. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 773.Jiang X, et al. MiR-495 is a tumor-suppressor microRNA down-regulated in MLL-rearranged leukemia. Proc. Natl Acad. Sci. USA. 2012;109:19397–19402. doi: 10.1073/pnas.1217519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 774.Zhao JJ, et al. miR-30-5p functions as a tumor suppressor and novel therapeutic tool by targeting the oncogenic Wnt/β-catenin/BCL9 pathway. Cancer Res. 2014;74:1801–1813. doi: 10.1158/0008-5472.CAN-13-3311-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 775.Yang Y, et al. miR-137 and miR-197 induce apoptosis and suppress tumorigenicity by targeting MCL-1 in multiple myeloma. Clin. Cancer Res. 2015;21:2399–2411. doi: 10.1158/1078-0432.CCR-14-1437. [DOI] [PubMed] [Google Scholar]
- 776.Misiewicz-Krzeminska I, et al. Restoration of microRNA-214 expression reduces growth of myeloma cells through positive regulation of P53 and inhibition of DNA replication. Haematologica. 2013;98:640–648. doi: 10.3324/haematol.2012.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 777.Hu Y, et al. Targeting of CD38 by the tumor suppressor miR-26a serves as a novel potential therapeutic agent in multiple myeloma. Cancer Res. 2020;80:2031–2044. doi: 10.1158/0008-5472.CAN-19-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 778.Zhu G, et al. HOXBLINC long non-coding RNA activation promotes leukemogenesis in NPM1-mutant acute myeloid leukemia. Nat. Commun. 2021;12:1956. doi: 10.1038/s41467-021-22095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 779.Papaioannou D, et al. The long non-coding RNA HOXB-AS3 regulates ribosomal RNA transcription in NPM1-mutated acute myeloid leukemia. Nat. Commun. 2019;10:5351. doi: 10.1038/s41467-019-13259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 780.Fragliasso V, et al. The novel lncRNA BlackMamba controls the neoplastic phenotype of ALK(-) anaplastic large cell lymphoma by regulating the DNA helicase HELLS. Leukemia. 2020;34:2964–2980. doi: 10.1038/s41375-020-0754-8. [DOI] [PubMed] [Google Scholar]
- 781.Yang J, et al. The identification of long non-coding RNA H19 target and its function in chronic myeloid leukemia. Mol. Ther. Nucleic Acids. 2020;19:1368–1378. doi: 10.1016/j.omtn.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 782.Taiana E, et al. Long non-coding RNA NEAT1 targeting impairs the DNA repair machinery and triggers anti-tumor activity in multiple myeloma. Leukemia. 2020;34:234–244. doi: 10.1038/s41375-019-0542-5. [DOI] [PubMed] [Google Scholar]
- 783.David A, et al. The long non-coding RNA CRNDE regulates growth of multiple myeloma cells via an effect on IL6 signalling. Leukemia. 2021;35:1710–1721. doi: 10.1038/s41375-020-01034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 784.Hu Y, et al. Targeting the MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in multiple myeloma. Leukemia. 2018;32:2250–2262. doi: 10.1038/s41375-018-0104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.