Abstract
Alternative polyadenylation (APA) creates distinct transcripts from the same gene by cleaving the pre-mRNA at poly(A) sites that can lie within the 3′ untranslated region (3′UTR), introns, or exons. Most studies focus on APA within the 3′UTR; however, here, we show that CPSF6 insufficiency alters protein levels and causes a developmental syndrome by deregulating APA throughout the transcript. In neonatal humans and zebrafish larvae, CPSF6 insufficiency shifts poly(A) site usage between the 3′UTR and internal sites in a pathway-specific manner. Genes associated with neuronal function undergo mostly intronic APA, reducing their expression, while genes associated with heart and skeletal function mostly undergo 3′UTR APA and are up-regulated. This suggests that, under healthy conditions, cells toggle between internal and 3′UTR APA to modulate protein expression.
CPSF6 loss disrupts polyadenylation site choice and causes a developmental syndrome in humans and zebrafish.
INTRODUCTION
The proteome is far more diverse than the genome, with some research estimating that our cells produce hundreds of thousands of proteins from a mere 25,000 genes (1). Much of this diversification occurs at the level of mRNA precursors (pre-mRNA), which can undergo various processes before being translated into proteins (2). One process that has garnered great interest in recent years is polyadenylation. As the nascent RNA matures, the polyadenylation machinery cleaves it at a poly(A) site (PAS) in the 3′ untranslated region (UTR) and synthesizes a polyadenylate or poly(A) tail to signal that transcription should be terminated and to protect the end from degradation (3). The length of the final 3′UTR determines the number of regulatory elements available to be bound by microRNA and RNA binding proteins to direct mRNA translation, stability, localization, and the tissue-specific functions of the resulting protein (4). There is not just one PAS per mRNA, however: the 3′UTR can contain several PAS, and PAS can also reside within exons, introns, and even the 5′UTR (4, 5). By convention, the 3′UTR upstream of the proximal PAS is called the constitutive 3′UTR, while the downstream region that is present only in longer isoforms is designated the “alternative UTR” (4). The term alternative polyadenylation (APA) thus describes cleavage that uses any alternative PAS in the 3′UTR or at sites internal to the gene. When APA takes place at a PAS within the gene, it is termed “internal APA” in contradistinction to 3′UTR-APA.
The choice of PAS can alter the ratios of expressed isoforms in response to hormonal and other exogenous signals (6). For example, two different-length transcripts of the steroidogenic acute regulatory (StAR) gene are expressed at equal levels in healthy adrenal glands, but stimulation of cholesterol metabolism by the cyclic adenosine 5′-monophosphate (AMP) analog, Br–cyclic AMP, favors the formation of the less stable transcript through the use of a more distal PAS (7). Similarly, elevated glucose levels favor distal PAS usage inHGRG-14 (high-glucose-regulated gene 14) mRNA in diabetic nephropathy (6), whereas hyperactivation of mTORC1 (mechanistic target of rapamicin complex 1) favors the use of proximal PAS in RBX1 (Ring-Box 1) in the context of certain tumors (8). There are also instances in which mutations within a PAS alter the outcomes of APA, for example, favoring the retention of a long 3′UTR in HBB in b-thalassemia (9) or in GIMAP5 (GTPase, IMAP Family Member 5 ) in systemic lupus erythematosus (10). Beyond these gene-specific alterations in the context of specific conditions, however, it has been difficult to determine how PAS are selected or what consequences might follow if there were to be global changes in APA.
We had an opportunity to study the effect of PAS choice at the whole-organism level when we identified patients bearing mutations in CPSF6 (cleavage and polyadenylation specific factor 6), one of three proteins comprising the APA subcomplex called cleavage factor I (CFI). Patients with either CPSF6 deletions or loss-of-function mutations develop a previously unidentified syndrome that affects neurological, cardiovascular, and skeletal development. Examining both patient fibroblasts and zebrafish larvae, we found that CPSF6 loss deregulates APA, with consequences for protein expression, by shifting PAS choice: about three-quarters of the pre-mRNA that normally undergo 3′UTR APA in healthy controls instead underwent internal APA and vice versa. Overall, genes involved in neurodevelopment switched to internal APA, with a consequent reduction in protein expression, whereas genes involved in cardiovascular and skeletal development underwent 3′UTR APA and showed elevated protein expression.
RESULTS
Loss of CPSF6 function causes neurological, cardiac, and skeletal abnormalities
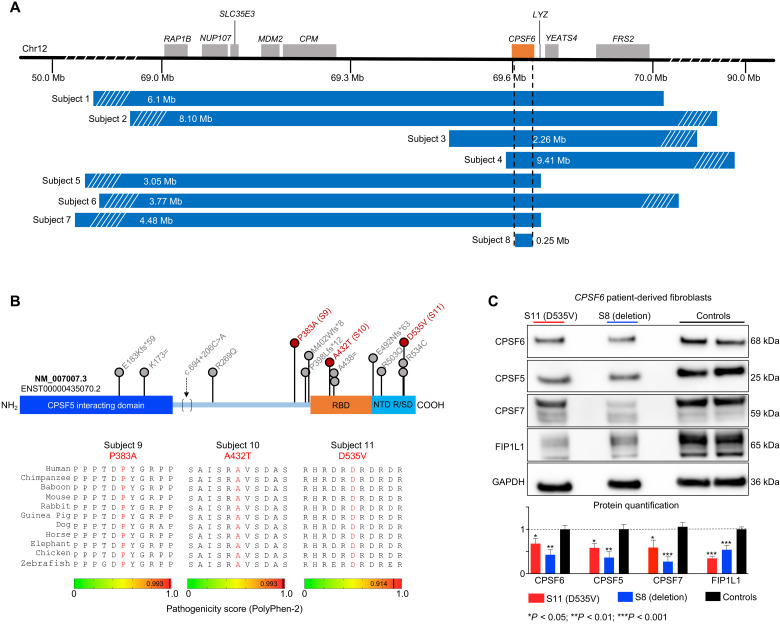
The core cleavage-and-polyadenylation machinery consists of four subcomplexes that have unhelpfully similar names: CFI, CFII, cleavage and polyadenylation specificity factor, and cleavage stimulation factor (4). We focused on the CFI complex, because it is known to be involved in PAS selection and one of its member proteins, CPSF5 (cleavage and polyadenylation specific factor 5, also known as NUDT21, nudix hydrolase 21), had previously been identified in patients with a developmental disorder (11). We therefore screened several patient databases for individuals with deletions, duplications, or variants involving CPSF6 or CPSF7 (cleavage and polyadenylation specific factor 7), the other two members of the CFI complex (see Materials and Methods). Both genes are predicted to be highly intolerant to loss of function (pLI = 1, see Materials and Methods) (12). CPSF7 did not turn up in any of the databases but CPSF6 did. There were eight subjects (S1 to S8) who bear heterozygous deletions spanning 0.25 to 9.41 megabases (Mb), within which the minimal region of overlap was CPSF6 (Fig. 1A, Table 1, and fig. S1, A and B). Of 15 individuals with a pathogenic single-nucleotide variant (SNV) (13) in CPSF6, we were able to enroll three (S9 to S11; Fig. 1B, fig. S2, Table 1, tables S1 and S2, and Materials and Methods). All three missense variants alter residues that are highly conserved down to zebrafish (phyloP, >5) and positionally intolerant to missense variation. Polyphen2 predicts all three variants to be highly damaging (Fig. 1B and table S2). One subject also had a variant in SCAF4 (SR-Related CTD Associated Factor 4), which we determined did not contribute to the phenotype (fig. S1, C to E; see Materials and Methods).
Fig. 1. Heterozygous deletion and missense mutations reduce CPSF6 levels in patients.
(A) Deletions spanning CPSF6 (orange box) on chromosome 12q15 identified in eight subjects. Dashed lines indicate the minimal overlapping region, which deleted 99% of CPSF6 (subject 8). Mb, megabases. (B) Schematic of the CPSF6 protein showing the RNA binding domain (RBP; orange), the Arg/Ser-rich domain (R/SD) within the nuclear targeting domain (NTD; turquoise), and the CPSF5-interacting domain (blue). Database searches identified 15 individuals with 13 missense variants (indicated by lollipops), which are plotted here for context, but only subjects 9 to 11 (red) were enrolled in this study. Bottom: Evolutionary alignment shows that the three variants in our subjects affect residues that are conserved from zebrafish to humans and have high pathogenicity scores. See fig. S2 for details of the three splicing variants. (C) Representative Western blot and relative quantification shows that subjects 8 and 11 (the only subjects from whom we obtained fibroblasts) have lower CPSF6, CPSF5, CPSF7, and FIP1L1 protein levels than controls. Data were normalized to GAPDH(glyceraldehyde-3-phosphate dehydrogenase) protein. Data represent means ± SEM from at least four technical and biological replicates compared to healthy age-matched fibroblasts, *P < 0.05, **P < 0.01, and ***P < 0.001.
Table 1. Summary of molecular and clinical features of 11 individuals with loss of CPSF6 function through deletion or missense mutation.
All subjects were de-identified and assigned a subject (S) number. –, no known; y, years; m, month; w, weeks; ASD, autism spectrum disorder; ADHD, attention-deficit hyperactivity disorder; CMTC, cutis marmorata telangiectatica congenita.
| Subject | S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | S10 | S11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | Male | Female | – | Female | Male | Female | Female | Female | Male | Female | Male |
| Age at last evaluation | 6y | – | – | – | 13y | 15y | 9y | <1y | 39y | 10y | 5y |
| Mutation genomic location GRCh37-Chr12 (amino acid change) | Deletion 64174854–70290866 | Deletion 67813733–75913733 | Deletion 69396710–71660633 | Deletion 69586439–78994711 | Deletion 66747385–69802582 | Deletion 67326299–71099672 | Deletion 65263944–69744248 | Deletion 69642510–6966703261-2400_*3692del | Missense 1147C>G (P383A) | Missense 1294G>A (A432T) | Missense 1604A>T (D535V) |
| Putative age at onset | Gestational (growth retardation) | 1y | – | – | <1y | 4y | 4y | Birth | <1y | 4y | 6m |
| Failure to thrive | Yes | – | – | – | – | Yes | – | – | No | No | No |
| Global delay | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Intellectual disab. | Yes | Yes | Yes | Profound | Yes | Severe | Yes | Yes | No | No | Mild |
| Speech delay | Yes | – | – | Severe | Yes | Severe | Severe | Yes | Mild | No | Mild |
| Motor delay | Yes | – | – | – | – | Yes | Yes | Yes | Yes | Yes | Yes |
| Seizures | – | – | – | – | – | – | – | – | No | Yes | Yes |
| ASD | – | – | – | – | Yes | Yes | Yes | – | No | No | No |
| Behavioral issues | Irritable, tantrums, ADHD | – | – | Tantrums, ADHD | Parallel play, fleeting eye contact, tantrums, ADHD | Panic attacks tantrums, ADHD | Tantrums | – | Some obsessive behavior | ADHD | No |
| Cardiovascular | – | CMTC | – | CMTC | – | No | – | Cardiomyopathy ventricular septal defects | Cardiomyopathy, heart block at 14y, trans-ischemic attack at 38y | No | No |
| Feeding difficulty | Yes | – | – | – | Yes | Yes | Gastrostomy dependent | Yes | No | No | No |
| Ophthalmic/ocular | – | Yes | – | – | Esotropia | Myopia | Strabismus | – | Retinal dystrophy, esotropia, cataracts | Squint & hypermetropia | Strabismus |
| Skeletal/body anomalies | Hand brachydactyly, flat feet, delayed bone age, short middle finger and distal phalanges | 2–3 toe syndactyly | Yes | Localized hirsutism, striae distensae | – | Disorganized/absent transversal palmar creases, fifth finger clinodactyly, 2–3 toe syndactyly tendon release in toes | Pes planus | Extra vertebra at C3, congenital diaphragmatic hernia | Scoliosis, asthenic body habitus, pectus excavatum | No | Broad first toes, hypermobile joints |
| Craniofacial dysmorphia | Microcephaly, broad forehead, pointed chin, down-slanting palpebral fissures, hypertelo-rism, deep-set eyes, tented lips | Microphthalmia broad forehead, convex nasal ridge, high palate, thin upper lip vermillion, low-set ears, deep set eyes, flat occiput, frontal bossing | Microcephaly | – | Short philtrum, widely spaced teeth, hypotelorism | Microcephaly, congenital left eye ptosis, micrognathia, ears slightly small, smile less broad on left; eyes slightly up-slanting at the lateral edges, wide and flat nasal bridge, long philtrum | Microcephaly, slightly short jaw, slightly up slanted palpebral fissures, narrow nose, prominent columella and nasal bridge, | – | Myoapathic face | High forehead and hypertelorism | Myopathic face, high forehead |
| Other | Recurring ear infections, fatigue | Fine hair, sparse eyelashes | – | – | Elevated AST indicating liver impairment | Hemoptysis at 5m, respiratory failure at 10y with pulmonary hemorrhage; left vocal cord paralysis, encephalopathy dysphagia, and delayed myelination; ovarian insufficiency | Intrauterine growth restriction at 20w and decreased fetal movement | Sensorineural hearing loss | Ataxia, chronic kidney disease | Hypotonia as newborn | Ataxia |
All subjects showed global developmental delays and motor delays. Deletions were associated with speech delays and intellectual disability ranging from severe to profound [by the criteria of the Diagnostic and Statistical Manual of Mental Disorders, Sixth Edition]; missense mutations were associated with mild or no delays in speech or cognition (Table 1). Two of the missense mutation subjects (S9 and S11) have had ataxia since childhood. Behavioral issues were noted in several deletion subjects as well as S9 and S10. Skeletal anomalies were frequent but variable and typically involved digital malformations; craniofacial dysmorphia were noted mostly among the subjects with deletions beyond CPSF6 (Table 1). Several subjects had ophthalmologic abnormalities: S9, at 39 years of age, exhibited retinal dystrophy, esotropia, and cataracts. S8 (with 99% of CPSF6 deleted) and S9 have cardiomyopathy, and S9 had complete heart block at 14 years of age and a transient ischemic attack at age 38. [Among the other 12 individuals with SNV whom we were unable to enroll, 5 were noted to have cardiac valve abnormalities (table S1).] This association between cardiovascular anomalies and CPSF6 variation was independently found to be statistically significant in an exome sequencing cohort of 13,218 patients [P = 0.0116; odds ratio (95% confidence interval) = 2.83 (1.21; 6.35)] (see Materials and Methods). S10 and S11 both experienced seizures, although the latter had only febrile seizures in childhood.
To evaluate the molecular effects of the identified variants, we developed cell lines from fibroblasts obtained from S8 (CPSF6-only deletion) and S11 (p.D535V de novo missense mutation) shortly after birth. As expected, cell lines from S8 had half the quantities of CPSF6 mRNA and protein as found in three healthy age-matched controls; cell lines from S11 showed no change in CPSF6 mRNA (fig. S1F) but had 65% of the levels of CPSF6 protein found in the healthy controls (Fig. 1C). Reduced expression of CPSF6 down-regulates the other members of the CFIm complex: CPSF5 and CPSF7 protein levels were reduced in S8 and S11 to similar extents (Fig. 1C). The abundance of FIP1L1 (factor interacting with PAPOLA and CPSF1), another APA factor that interacts with the CFI complex (14), was also reduced by 50%. This is consistent with previous work showing that the stoichiometry of the complexes is carefully balanced (15).
Loss of CPSF6 function affects polyadenylation globally by switching PAS site usage
Depletion of CPSF6 has been suggested to enhance cleavage at proximal PAS in the 3′UTR (4). To understand whether this is the case in patients, we used poly(A)-ClickSeq (PAC-seq) (16) to sequence polyA+ mRNA in cell lines derived from S8, S11, and their age-matched controls and then analyzed the resulting data with PolyA-miner (17), which allows for simultaneous measurement of mRNA expression and PAS site selection genome wide. Principal components analysis (PCA) assessing global poly(A) distribution cleanly separated the subjects and controls, confirming the reproducibility of the PAC-seq analysis (fig. S3A).
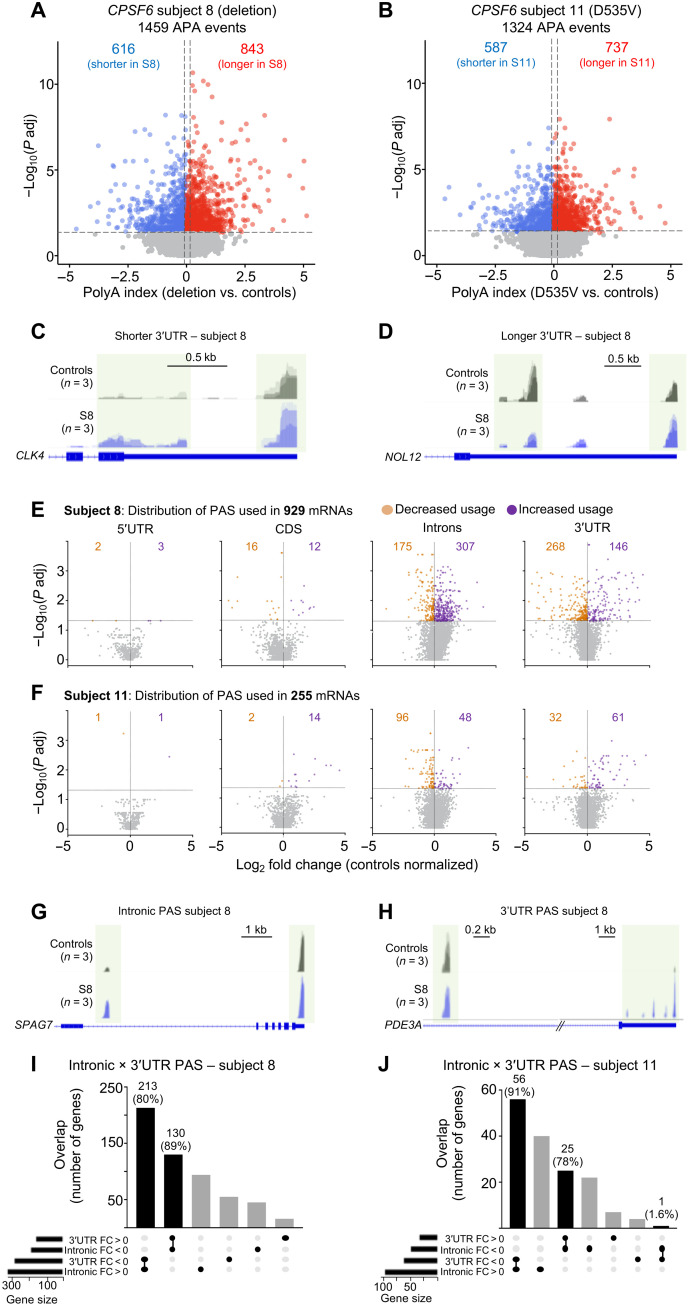
S8 and S11 exhibited a similar number of total APA events (Fig. 2, A to D, and tables S3 and S4). One-quarter to one-third of transcripts that are shorter or longer than the same transcript in controls were the same in both subjects (fig. S3B). For transcripts with APA events occurring only in the 3′UTR, there were a similar number of shorter and longer 3′UTRs (831 in S8 and 754 in S11; fig. S3, C and D, and tables S5 and S6). We then analyzed the locations of PAS used throughout the whole transcript, i.e., from the 5′UTR through coding sequences (CDS), introns, and 3′UTR, to identify mRNA that showed statistically significant changes in the location of PAS favored for APA compared to healthy controls (Fig. 2, E to H, and tables S7 to S14). Although far fewer mRNA met this criterion in S11 than in S8 fibroblasts (only 255 events, or 73% of the number in S8), the two cell lines were similar in that APA events were least likely to involve PAS in the 5′UTRs or CDSs and most likely to involve those in introns and 3′UTRs. More than half of the APA events involved intronic PAS. It is worth noting that these PAS were dispersed throughout the transcripts (i.e., not concentrated in the last intron before the 3′UTR). APA events in S8 not only relied on intronic PAS, they tended to use more of candidate PAS available within those introns (Fig. 2, E and F). Conversely, despite having nearly as many APA events involving the 3′UTR, S8 cells tended to use fewer of the available PAS in that region. In S11, APA events used a greater variety of PAS in coding regions and the 3′UTR but fewer in introns.
Fig. 2. Loss of CPSF6 function globally affects APA.
(A and B) Differences in APA between subjects 8 (A) and 11 (B) compared to controls. The horizontal dashed lines indicate the −log10 (P adjusted) ≥ 1.325 (P adjusted ≤ 0.05), and the vertical dashed lines indicate polyA index ≥ +0.1 and ≤ −0.1, with positive values indicating longer transcripts and negative values indicating shorter transcripts. (C and D) Representative IGV tracks from subject 8 (S8) showing an example of longer and shorter 3′UTR. (C) CLK4 has a shorter 3′UTR than controls, and (D) NOL12 has a longer 3′UTR than controls. (E and F). Locations of PAS (5′UTR, CDS, introns, and 3′UTR) from subjects 8 and 11 fibroblasts relative to controls. Purple [log2 fold change (FC) > 0] and orange (log2 FC < 0) dots represent PAS usage in that location (n = 3 replicates for each subject). The horizontal and vertical dashed lines in (E) and (F) indicate the −log10 (P adjusted) ≥ 1.325 (P adjusted ≤ 0.05) and the log2 FC, respectively. (G and H) Representative IGV tracks from subject 8 (S8) showing SPAG7 switches from 3′UTR to intronic PAS usage, and PDE3A does the reverse. (I and J) Upset plots shows switching between internal and 3′UTR APA. Fold change compared to healthy controls. PAC-seq was performed on three independent biological samples from subjects 8 and 11 and their controls, each in triplicate.
The most notable pattern among significant APA events, however, was that those using 3′UTR PAS in healthy controls switched to intronic PAS in subjects 8 and 11 (80 and 91%, respectively; Fig. 2, I and J). At the same time, the majority of events using intronic PAS in healthy controls shifted to 3′UTR PAS in subjects 8 and 11 (Fig. 2, I and J). It thus appears that CPSF6 insufficiency deregulates PAS selection. For the rest of the paper, we will refer to transcripts that undergo APA using distal PAS in the 3′UTR as “longer” transcripts and those that undergo APA using either proximal 3′UTR or internal PAS as “shorter.”
Shifts between internal and 3′UTR PAS usage affect mRNA and protein abundance
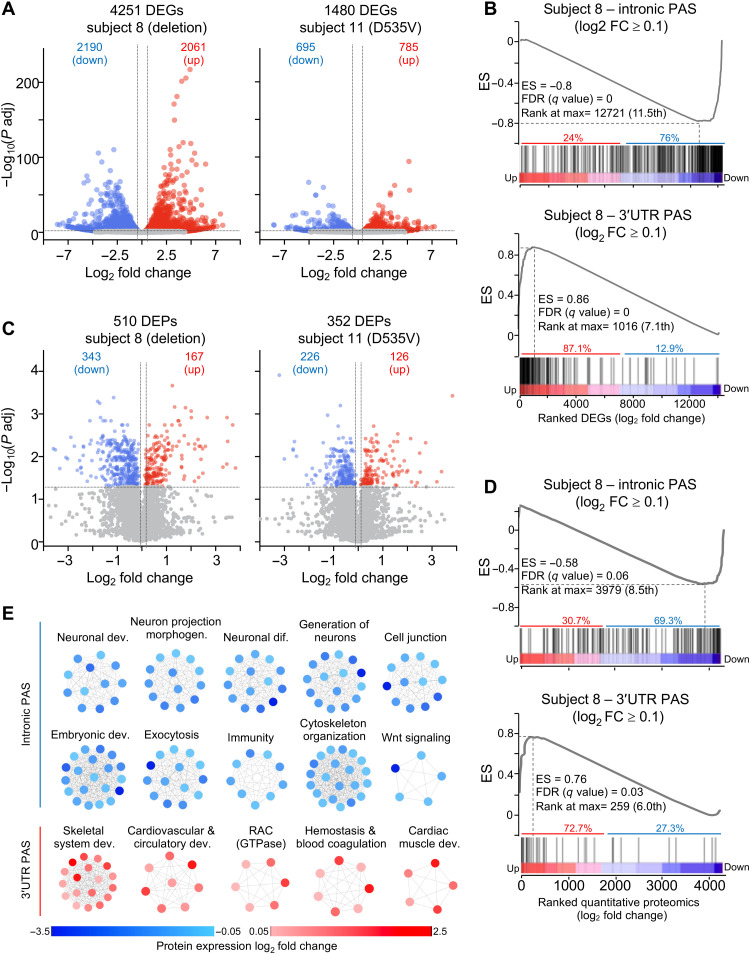
We next analyzed our PAC-seq dataset for differentially expressed genes (DEGs) (16). PCA again readily distinguished three groups (fig. S4A). PolyA-miner analysis identified more DEGs in subject 8 than in 11 (Fig. 3A and tables S15 and S16). The DEGs were fairly evenly split between up- and down-regulated genes, with substantial but incomplete overlap in the genes affected in the two subjects (fig. S4B). We found that majority of longer transcripts were up-regulated (fig. S4, C and D). Shorter transcripts, however, were equally likely to be up- or down-regulated; even the length of the 3′UTR itself did not correlate with higher or lower levels of mRNA (fig.. S4, E and F). This runs counter to expectation (18, 19), so we examined the usage of intronic and 3′UTR PAS among the DEGs.
Fig. 3. Intronic and 3′UTR PAS usage tracks with gene and protein expression change.
(A) Volcano plots of DEGs in fibroblasts from subjects 8 and 11 relative to age-matched controls (B) GSEA of preranked mRNA with intronic (top) or 3′UTR PAS usage (bottom) intersected with DEGs from subject 8 ranked from the most up-regulated to the most down-regulated gene. (C) DEPs in subjects 8 and 11 compared to three healthy age-matched controls, each in triplicate. (D) Preranked GSEA of mRNA with intronic PAS usage (top) and 3′UTR PAS usage (bottom) intersected with quantitative proteomics from subject 8 ranked from the most up-regulated to the most down-regulated protein. ES, GSEA enrichment score. FDR and rank at max are calculated by GSEA (see table S18). (E) Enriched biological categories of transcripts with intronic or 3′UTR PAS in subject 8. Notably, all mRNA undergoing intronic APA use were down-regulated (blue), whereas those undergoing 3′UTR APA resulted in up-regulated (red) protein expression. Each dot represents a protein belonging to a specific biological category. In (A) and (C), the vertical dashed lines indicate the log2 FC ≥ 0.263 (1.2 FC) for the up-regulated genes (A) or proteins (C) and log2 FC ≤ −0.263 (−1.2 FC) for the down-regulated genes (A) or proteins (C). The horizontal dashed line indicates the −log10 (P adjusted) ≥ 1.325 (P adjusted ≤ 0.05).
We performed gene set enrichment analysis (GSEA) (20) to see how transcripts with significantly greater intronic PAS usage in S8 and S11, compared to controls, were distributed among the DEGs and found that 76% were strongly down-regulated (Fig. 3B, top, and fig. S4G). Conversely, 87% of transcripts that predominantly used intronic PAS in healthy controls and switched to 3′UTR PAS usage in S8 and S11 were among the most strongly up-regulated (Fig. 3B, bottom, and fig. S4H). CPSF6 insufficiency thus has a bidirectional effect: Where it increases intronic APA, it produces unstable mRNA, but where it favors 3′UTR APA, it augments production of stable mRNA.
To probe the effects of CPSF6 loss of function at the protein level, we performed quantitative liquid chromatography–tandem mass spectrometry (LC-MS/MS) (see Materials and Methods). PCA separated S8, S11, and the healthy controls (fig. S4I). As expected, there were more differentially expressed proteins (DEPs) in S8 than in S11 (Fig. 3C and tables S17 and S18), and the DEGs from RNA sequencing (RNA-seq) strongly correlated with the DEPs (fig. S4, J and K). Most of the transcripts that switched from 3′UTR usage in healthy controls to intronic PAS in S8 were strongly down-regulated at the protein level (Fig. 3D, top). Because S11 had fewer DEPs, GSEA did not find statistically significant enrichment, but of 21 transcripts that underwent intronic APA, three-quarters exhibited lower protein expression (table S19). Thus, increased intronic APA typically reduced mRNA and protein expression, most likely by prematurely terminating transcription (21).
Conversely, three-quarters of the mRNA that underwent internal APA in healthy controls but switched to 3′UTR APA in S8 led to strongly up-regulated proteins (Fig. 3D, bottom). In S11, 11 of 14 genes with aberrant 3′UTR PAS usage were up-regulated (table S19). Preferential use of 3′UTR PAS thus typically increased protein expression.
Last, we examined the biological pathways to which the DEPs belonged. The down-regulated proteins from mRNA that had shifted to internal APA in S8 (there were too few to study in S11) were enriched in functions related to neuronal development and differentiation, as well as exocytosis and cytoskeletal organization (Fig. 3E). The up-regulated proteins expressed from the mRNAs with high 3′UTR PAS usage were enriched in functions related to cardiovascular, circulatory, and skeletal development (Fig. 3E and table S20). These enrichments are consistent with phenotypic features noted in the subjects and reveal that the disrupted APA profile is more complex than initially thought based on knocking down CPSF6 in cell culture models (22).
We wondered whether this observation might indicate that neuronal genes are longer than genes involved in cardiovascular and skeletal development. It is frequently claimed that neuronal genes are long, but much of this length is due to their elaborate 3′UTRs. We therefore examined intron length in both categories of genes and found that introns in neuronal genes are, on average, around 6000 nucleotides or about one-third the total average length (~20,000 nucleotides) of heart and skeletal genes (fig. S4L). This could explain why, under healthy conditions, the latter are more likely to use intronic PAS. The average number of introns was the same in neuronal and heart/skeletal genes.
Cpsf6-deficient zebrafish show skeletal, cardiac, and neurological defects
To investigate CPSF6 deficiency in a model organism, we used a previously uncharacterized zebrafish line carrying a nonsense mutation in the ortholog cpsf6 (fig. S5A; see Materials and Methods). To determine the degree of cpsf6 expression, we used primers amplifying both the regions upstream and downstream from the mutation and found that the homozygous mutant larvae expressed only 30% of the wild-type mRNA level at 6 days postfertilization (dpf) (fig. S5B). Because zebrafish express cpsf6 even at the one- to two-cell stage (23), when gene expression from zygotic DNA is still repressed, this suggested that cpsf6 mRNA is maternally inherited (23). We immunostained for cpsf6 starting from 11 hours postfertilization (hpf) to 6 dpf. At 11 hpf, cpsf6 was expressed at similar levels in cpsf6+/− and cpsf6−/− larvae and their wild-type siblings (fig. S5C). Only a few cells showed residual cpsf6 expression at 24 hpf in cpsf6−/−, and these showed no signal at all by 2 and 6 dpf (fig. S5C), indicating active nonsense-mediated decay of the mutant cpsf6 mRNA. These data confirm that cpsf6 mRNA in the zebrafish embryo is maternally inherited and that its expression supports embryogenesis for the first 24 hours of larval development. When we intercrossed cpsf6+/− animals, all of the cpsf6−/− offspring died between 7 and 15 dpf (fig. S5D). We therefore performed all the behavioral and pathological characterization no later than 6 dpf.
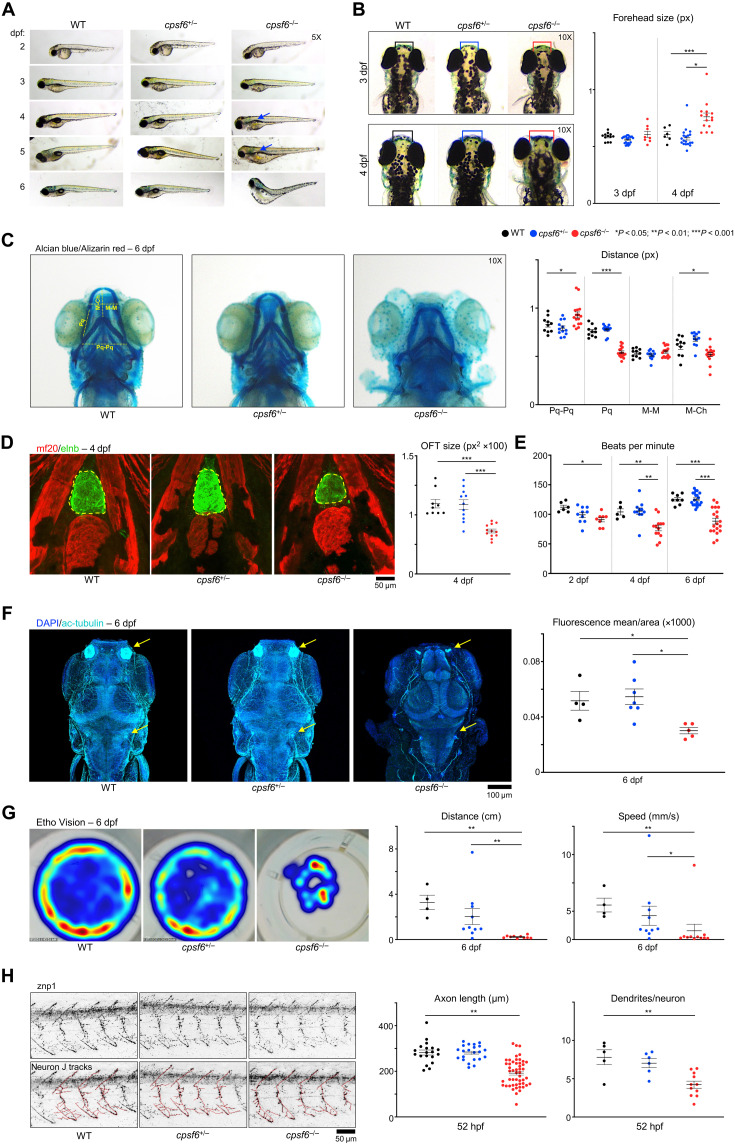
The cpsf6+/− and cpsf6−/− were morphologically indistinguishable from wild-type larvae at 2 and 3 dpf (Fig. 4A). Starting from 4 dpf, however, cpsf6−/− mutants showed extensive edema; by 5 dpf, the swim bladder has still not inflated (Fig. 4A). Skull deformities were most notable: At 4 dpf, cpsf6−/− larvae had a 25% broader forehead than their wild-type and cpsf6+/− siblings (Fig. 4B). Alcian blue and Alizarin red, which stain cartilage and bone, respectively (24), revealed a jaw defect at 6 dpf: The palatoquadrates (Pq) were shorter, and the distance between the two palatoquadrates (Pq-Pq) was longer in cpsf6−/− compared to wild-type siblings (Fig. 4C). The ventral mandibular arch, known as Meckel’s cartilage (M), appeared unaffected, but the distance between Meckel’s cartilage and the Ceratohyal cartilage at the back of the jaw (M-Ch) was shortened.
Fig. 4. Cpsf6 deficiency in zebrafish produces skeletal, neurological, and cardiac defects.
(A) Larval development of wild-type (WT), cpsf6+/−, and cpsf6−/− animals. Arrows point to where the missing swim bladder should be. Images acquired in brightfield with a 5× objective lens. (B) Dorsal images at 3 and 4 dpf, with the regions quantified at right in boxes. Images acquired in brightfield with 10× objective. (C) Ventral views of larvae at 6 dpf stained with Alcian blue and Alizarin red (24) with relative quantification of cartilage length. Pq, palatoquadrate; Pq-Pq, distance between right and left Pq; M-M, Meckel’s cartilage; M-Ch, distance between M and the Ceratohyal (78). (D) Immunofluorescence (IF) confocal microscopy with MF20 (79) (red, ventricular cardiomyocytes) and elnb (80) (green, outflow tract). Yellow dashed line encircles Outflow tract (OFT), quantified at the right. (E) Heart rate quantification (see also movies S1 to S9). (F) IF confocal images with staining for acetylated tubulin (81) and relative quantification of mean ac-tubulin fluorescence normalized by the analyzed area. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). (G) Density map of the free-swimming test acquired with EthoVision, quantifying distance and speed. (H) Confocal images of the znp1 pMN marker (75), with relative quantification of axonal length and dendritic density. Each dot represents one animal in (B) to (G) and (H) (dendrites/neuron graph); each dot in axon length graph [also in (H)] represents one neuron (total of 84, from 5 WT, 6 cpsf6+/−, and 12 cpsf6−/− animals). Data represent mean ± SEM. One-way analysis of variance (ANOVA); *P < 0.05, **P < 0.01, ***P < 0.001.
Cardiac defects were prominent. Staining with elastin b (enlb) showed a smaller cardiac outflow tract (Fig. 4D). The cpsf6−/− fish showed progressive bradycardia starting from 2 dpf, with longer pauses and shorter beats per minute at 6 dpf; some of the larvae showed sporadic episodes of heart block (Fig. 4E and movies S1 to S9). We speculate that these cardiac defects came from abnormalities in the secondary heart field cardiomyocytes (25, 26), since the atrium and ventricle, which were grossly normal, derive from the primary heart field cardiomyocytes, which develop earlier (while there is still maternal cpsf6 expression) (fig. S5E) (27).
To assess neuronal morphology and function, we used acetylated tubulin to mark axonal tracts (28). The cpsf6−/− larvae showed much less tubulin staining, particularly in the olfactory bulb and projections from the sensory lateral line organs that are important for spatial locomotion (Fig. 4F) (29). Swimming behavior at 6 dpf was terribly impaired, with cpsf6−/− barely able to move and even the heterozygous larvae having severe difficulties (Fig. 4G and movies S10 to S12). To better understand the possible origin of the swimming impairment, we traced the development and maintenance of primary motor neurons (pMNs). The pMNs are classified into three subtypes according to their location within the spinal cord: caudal primary, rostral primary, and middle primary (30). Staining with anti-synaptotagmin (znp1), which recognizes pMN, revealed shorter axons and lower dendritic density from 52 hpf on (Fig. 4H; see Materials and Methods); there was less znp1 signal and branching was impaired through 6 dpf (fig. S5F).
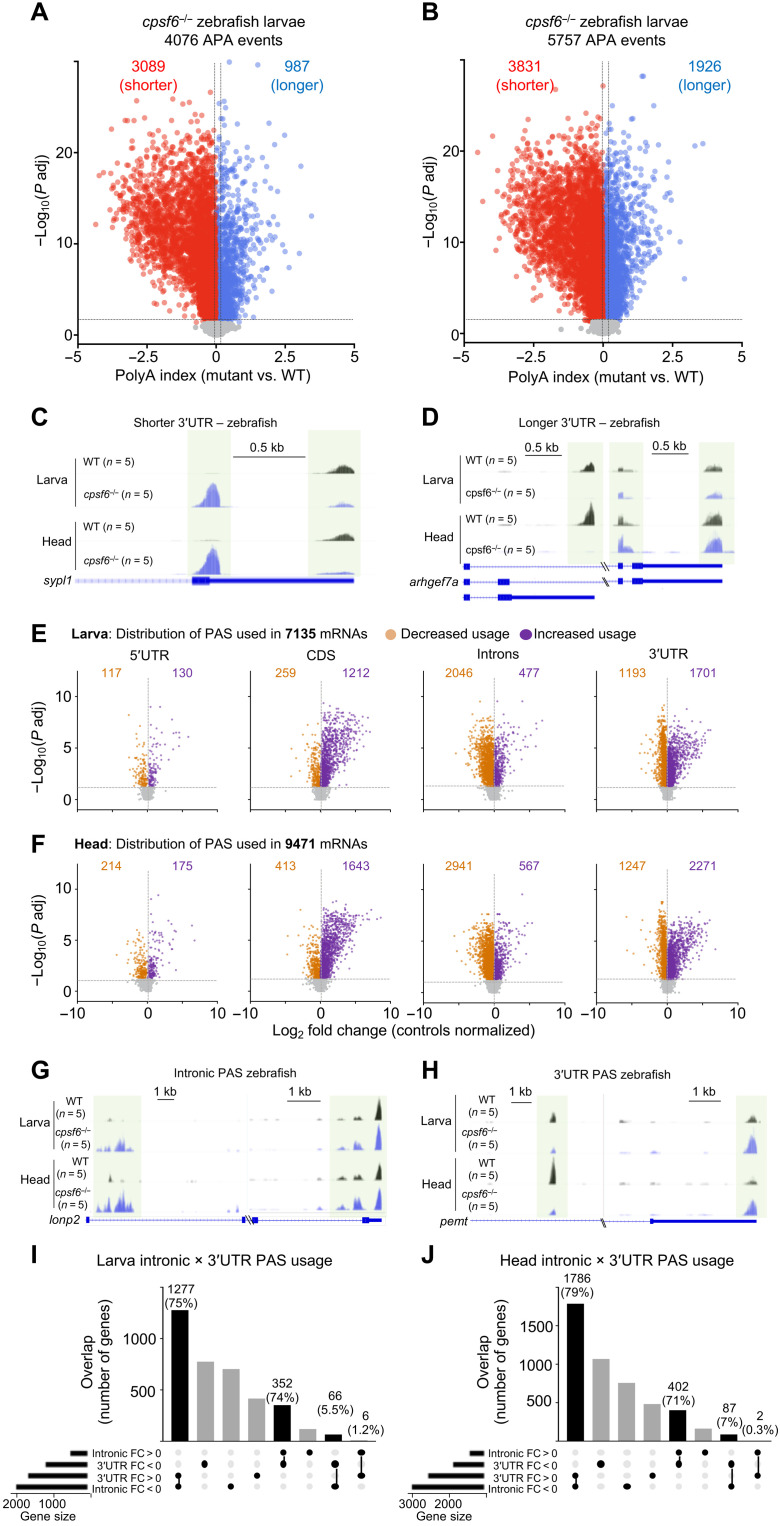
Zebrafish lacking cpsf6 show shifts between internal and 3′UTR APA
To determine whether orthologous genes underlie the similar defects in humans and zebrafish, we performed PAC-seq on polyA+ mRNA from cpsf6 homozygous mutant larvae. Staining with cpsf6 antibody revealed no differences between heterozygous and wild-type animals (fig. S5C), so we focused on the homozygotes. Since the brains of zebrafish larvae are too small to be easily extracted, we studied heads and whole larvae (i.e., the entire body, including the head) at 6 dpf along with their stage-matched wild-type controls. PCA of the APA analysis separated wild-type and cpsf6−/− in both whole-larvae and head samples (fig. S6, A and B). PAC-seq identified thousands of significant APA events from larvae and heads, which both had over twice as many short as long isoforms compared to controls (Fig. 5, A to D, and tables S21 and S22). APA in larval and head samples overlapped, with 69% of shorter and 53% of longer mRNAs belonging to the same genes. There was minimal overlap between genes with shorter mRNA in larvae and longer mRNA in the head and vice versa (fig. S6C). A scatterplot of genes expressed in both larvae and head samples showed a strong positive correlation (r = 0.73 and P < 0.0001) (fig. S6D). Despite this overlap, it is worth noting that the number of shorter isoforms in the head alone is higher than in the larvae, likely because the brain has a preponderance of genes with long 3′UTRs (31, 32).
Fig. 5. Cpsf6−/− zebrafish show altered PAS usage in larvae and head.
(A and B) Volcano plots show APA change in cpsf6−/− larvae (A) and heads (B) compared to WT age-matched siblings. The horizontal and vertical dashed lines in (A) and (B) indicate the −log10 (P adjusted) ≥ 1.325 (P adjusted ≤ 0.05) and polyA index ≥+0.1 and ≤−0.1, respectively. (C and D) Representative IGV tracks for shorter (C) and longer (D) 3′UTRs. Light green boxes highlight the polyA site used. (E and F) Volcano plots showing the polyA read distribution in 5′UTR, CDS, introns, and 3′UTR from cpsf6−/− larvae (E) and heads (F), compared to WT stage-matched larvae and heads, respectively. Each dot represents a transcript with greater (purple, log2 FC > 0) or lesser (orange, log2 FC < 0) PAS usage in a certain location. The horizontal and vertical dashed lines in (E) and (F) indicate the −log10 (P adjusted) ≥ 1.325 (P adjusted ≤ 0.05) and the log2 FC, respectively. (G and H) Representative IGV tracks for intronic PAS usage (G) and 3′UTR PAS usage (H). (I) In cpsf6−/− larvae, 75% of the mRNA undergoing 3′UTR APA in mutant underwent intronic APA WT, while 74% switch in the other direction. (J) In cpsf6−/− heads, 79% of mRNA that had 3′UTR APA underwent intronic APA in controls, and 71% switch from intronic to 3′UTR APA in controls. PAC-seq was performed on 10 (five larvae and five heads) independently collected groups of cpsf6−/− compared to 10 WT stage-matched animals. Each group of animals was composed of 30 to 40 larvae or 40 to 50 heads.
To visualize PAS usage, we analyzed all the polyA read distributions in the 5′UTR, exon, intron, and 3′UTR from larvae and heads compared to their stage-matched controls (Fig. 5, E to H, and tables S23 to S30). PAS usage was distributed fairly evenly across the CDSs, introns, and 3′UTR (21, 35, and 40% in larvae; 22, 37, and 37% in the head). The distribution of APA events is thus almost random across those three regions. This suggests that a complete loss of cpsf6 function (as opposed to the 35 or 50% loss of function in S11 and S8) relieves the selectivity of binding and/or cleavage.
Comparing just intronic and 3′UTR APA, roughly three-quarters of genes undergoing 3′UTR APA in the larvae or heads underwent intronic APA in controls (Fig. 5, I and J). The converse was also true, as it had been in human fibroblasts: 74% of genes subject to intronic PAS usage in cpsf6−/− larvae and 71% of them in the head overlapped with genes undergoing 3′UTR PAS usage in wild type (Fig. 5, I and J). Loss of CPSF6 therefore causes pathogenic shifts between internal and 3′UTR APA in both humans and zebrafish.
Loss of cpsf6 suppresses neuronal genes and up-regulates cardiovascular and skeletal genes
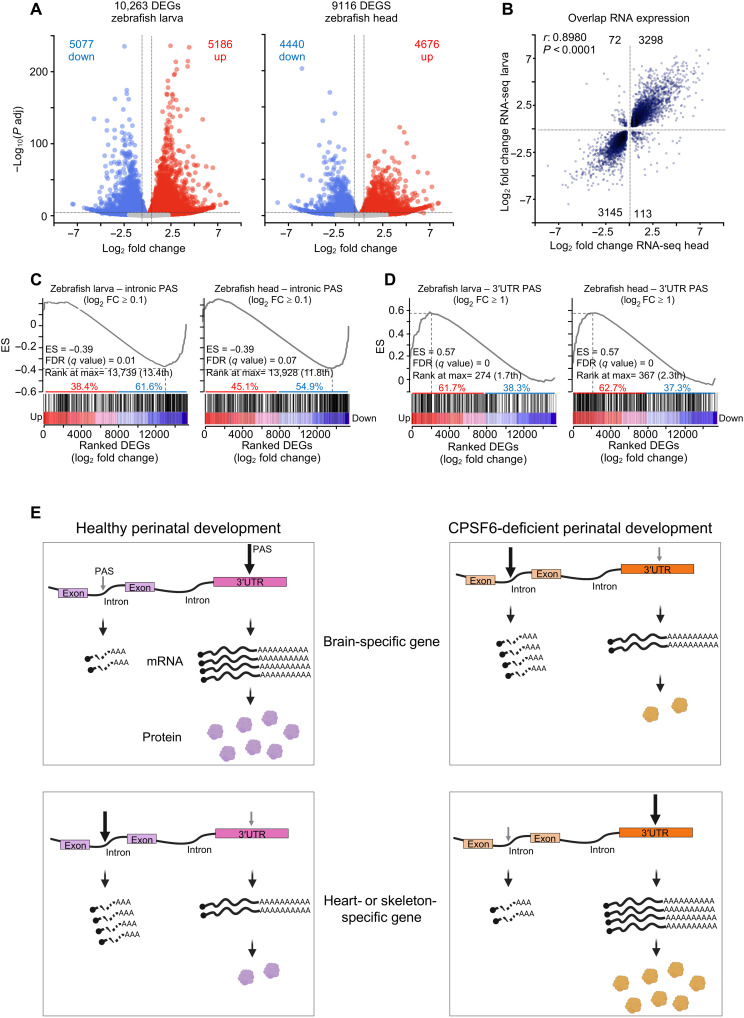
Following the same analysis we used for patient-derived fibroblasts, we analyzed mRNA to identify transcripts that were differentially expressed between wild-type and mutant fish, separating larvae and head samples. PCA neatly separated cpsf6−/− and wild-type controls (fig. S6, E and F). As in human subject cells, roughly half the DEGs in larvae and heads were down-regulated and half up-regulated (Fig. 6A and tables S31 and S32). The majority of DEGs overlapped between larval and head samples (97%; r = 0.8980) (Fig. 6B). Most of the transcripts, whether short or long, were equally likely to be up- or down-regulated (fig. S6, G and H).
Fig. 6. Gene expression changes correspond with PAS selection across species.
(A) Volcano plots of DEGs from cpsf6−/− larvae and heads compared to their WT stage-matched animals. The blue and red dots represent down- and up-regulated genes, respectively. DEGs are defined as having P adjusted < 0.05 and log2 FC > 0.263 (1.2 FC). (B) Scatterplot showing 97% correlation (r = 0.8980, P < 0.0001) between DEGs from cpsf6−/− larvae and heads, compared to their respective WT stage-matched animals. (C and D) Preranked GSEA of genes with intronic PAS usage (C) or 3′UTR PAS usage (D) intersected with DEGs from larvae and heads, ranked from the most up- to the most down-regulated gene. ES, enrichment score; the FDR and rank at max are calculated by GSEA. (E) Model of how cells may use APA to toggle between suppressing and augmenting protein abundance. In healthy perinatal development, a subset of genes involved in neuronal functions undergoes APA within the 3′UTR, which stabilizes the mRNA and up-regulates the resulting protein levels. A different subset of genes enriched in cardiac and skeletal development tend to undergo internal APA, generating short and unstable transcripts that result in down-regulation of protein levels. Loss of CPSF6 function causes these trends to switch.
As with the human samples, GSEA revealed that the majority of transcripts with high internal APA in cpsf6−/− larvae/head were strongly down-regulated (Fig. 6C). Conversely, the transcripts that switched to 3′UTR-APA in fish were strongly up-regulated (fig. S6, G and H). Because so many genes used 3′UTR PAS, we performed an additional GSEA using a more stringent cutoff; this analysis confirmed the strong up-regulation (Fig. 6D).
We next asked whether the DEGs in zebrafish overlap with those of the human subjects. S8 shared 704 DEGs (429 up-regulated and 275 down-regulated), and S11 shared 282 DEGs (178 up-regulated, 104 down-regulated) with the mutant zebrafish larvae (fig. S7A). We then performed DAVID GO (33) followed by a clustering analysis using REVIGO (34) on these genes (tables S33 to S38) and showed that the down-regulated genes are enriched in biological processes related to neuronal development and maintenance, such as cranial ganglion development, synaptic vesicle development, neuronal projection, axogenesis, and regulation of heart contraction [fig. S7, B, C (red circles), and D]. Genes involved in organ development, heart and outflow tract morphogenesis, circulatory system development, skeletal and chondrocyte differentiation, and kidney development tended to be up-regulated [fig. S7, B, C (blue circles), and D].
Loss of function of CPSF6 in both humans and zebrafish thus causes a syndrome involving impairments in neurological, cardiovascular, and skeletal development by causing the majority of transcripts that normally undergo internal APA to instead undergo 3′UTR APA and vice versa (Fig. 6E). The implication is that even under healthy conditions, internal APA, particularly intronic APA, plays a substantial role in regulating protein expression during early development.
DISCUSSION
In this study, we find that loss of CPSF6 function exerts a bidirectional effect on PAS choice and protein expression. Our findings differ from those of previous studies that studied knockout of CPSF6 in human embryonic kidney (HEK) 293T and C2C12 cells and found enrichment in shorter RNA isoforms (22, 35). There are several possible explanations for these differences. APA is cell type specific, and human fibroblasts could differ from HEK293T and C2C12 cells. This does not explain, however, why we found the same trends in humans and zebrafish without distinguishing cell types. Another possibility is that different results derive from different degrees of knockdown; however, in this study, we examined cells from a patient with 65% of wild-type levels of CPSF6, one with 50% of wild-type CPSF6, and zebrafish that were functionally null, all from a similar period of early development. We propose that our results differ simply because we looked at PAS selection throughout the entire transcript rather than just within the 3′UTR. The fact that cpsf6-deficient zebrafish echo the phenotypic features and APA patterns observed in human CPSF6 deficiency provides a compelling case that CPSF6 governs PAS site selection in ways that are highly conserved across vertebrate species, with clear consequences for mRNA and protein expression during development.
Previous work examining only 3′UTR APA has been unable to resolve the question of whether APA affects protein expression. Some early studies had found that cancer cells tend to have mRNA isoforms with shorter 3′UTR and up-regulated protein expression, at least for the several genes examined (36, 37). Yet one study looking at 13,000 genes showed that 3′UTR length had only a modest effect on translational efficiency (19) (they did not examine protein abundance), while another study examined several hundred proteins and concurred that 3′UTR length had little effect on expression overall (38). Even studies on intronic polyadenylation, which truncates and down-regulates mRNA, often look at only a handful of proteins (39, 40). To our knowledge, only one study has examined how APA across the entire transcript (in exonic, intronic, and 3′UTR regions) influences gene expression: LaForce et al. (41) found that loss of CLP1 (cleavage factor polyribonucleotide kinase subunit 1) function produced some APA changes that correlated with gene expression but did not explore effects at the protein level.
Our results make it clear that APA plays an important role in modulating protein levels during development, but they also reveal gaps in our current understanding of CPSF6 function. It has been proposed that a subset of mRNAs are enriched in UGUA enhancers at the distal PAS, which, in theory, would help recruit the APA machinery (14, 42). In the context of diminished expression of the CFIm complex, recall that reduction of CPSF6 also reduced CPSF5 and CPSF7, there would be too little CPSF5/6/7 to strengthen binding at the distal PAS, so the proximal PAS would be used (14, 43). Because most studies on PAS usage confine their investigation to different PAS within the 3′UTR, they cannot help us explain why loss of CPSF6 would cause different tissues or pathways to switch between 3′UTR APA and internal APA. Differences in PAS strength would explain our observation that below-normal concentrations of CPSF6 increased the use of intronic PASs in transcripts involved in neuronal function, but it would not explain why other transcripts associated with cardiovascular or skeletal function switch to 3′UTR-APA. Nor would it explain what appears to be a loss of selectivity for PAS, as we observed in subject 8 for intronic PAS, in subject 11 for CDS and 3′UTR PAS, and in the mutant zebrafish, in which APA events took place almost randomly throughout the transcripts. It is possible that the PAS that are selected share specific features or that CPSF6 sterically hinders binding at certain PAS so that its absence relieves this restriction. It is also plausible that the choice of PAS reflects a response to signals (e.g., hormones) from outside the cell rather than any intrinsic feature of the PAS.
Recent studies of the CFI complex have sought to map its function in various cell lines and tissues. Knockdown of CPSF5 (NUDT21) has been reported to bias HeLa cells and healthy human lung fibroblasts toward shorter 3′UTR isoforms (44, 45). In mice, loss of one copy of CPSF5 caused learning deficits with almost no observed changes in APA, but knockdown in neurons appeared to favor shorter 3′UTRs (46). Conversely, knockdown of FIP1L1 caused enrichment in longer 3′UTR (22). In none of these studies, however, was there a correlation between the length of the 3′UTRs and mRNA or protein concentrations (47). It was only when we examined APA events throughout the entire transcript that we found that switching between internal and 3′UTR-APA led to reliable changes in mRNA and protein expression within specific tissues.
Very recent work showing sequential polyadenylation in retained transcripts (48) suggests that APA is even more complex than previously appreciated, occurring at different PAS in two phases. This study found that CPSF6 and FIP1L1 bind distal PAS in poly(A)+ mRNA in the nuclear matrix, which is enriched with mRNAs with partially spliced introns or with longer 3′UTR. These nascent mRNAs are retained in the nuclear matrix so that they might undergo posttranscriptional processing (phase two) using the proximal PAS before being exported from the nucleus. This is reminiscent of how neurons use intron retention to keep a readily releasable pool of fully transcribed, partially spliced transcripts to respond quickly to neural activity (49). It will take further investigation to determine how insufficient expression of CPSF6 or FIP1L1 affects cotranscriptional APA, posttranscriptional APA, or both, and how the cellular context influences the choice of internal or 3′UTR PAS.
MATERIALS AND METHODS
Ethics
The study was conducted in accordance with the Declaration of Helsinki. Informed consent was obtained from all family members. Subject 11 approval was granted by the Tartu University Ethics Committee (certificate nos. 259/T-2, 263/M-16, 283/M-10, and 287/M-15). All study procedures were defined under protocol #AAAR7750 (V.A.G.) and #AAAJ8651 (W.K.C.), approved by the Institutional Review Board at Columbia University Irving Medical Center (V.A.G.); the Dell Children’s Medical Group, Austin (TX); Stanford School of Medicine, Stanford (CA); University of Rochester Medical Center, Rochester (NY); Baylor Genetics Laboratories, Houston, TX; Monroe Carell Jr. Children’s Hospital at Vanderbilt, Nashville (TN); Sørlandet Sykehus (Hospital), Kristiansand, Norway; Tartu University Hospital, Tartu, Estonia. The study also adhered to tenets outlined by the Declaration of Helsinki.
Sequencing and genetic analyses
Before undergoing trio or singleton exome sequencing, most patients underwent genetic screening that included karyotyping, chromosomal microarray, CAG and CGG repeat expansion analysis for selected genes, and metabolic testing, without detection of significant findings. Chromosomal array plots of oligonucleotide arrays have been obtained for subject 8, while Sanger sequencing results have been obtained for subject 11 (fig. S1, A and B).
Minor allele frequencies were obtained from the gnomAD (https://gnomad.broadinstitute.org/gene/ENSG00000111605?dataset=gnomad_r2_1) and BRAVO (https://bravo.sph.umich.edu/freeze8/hg38/gene/snv/CPSF6) databases (assessed May 2022). Variant annotation was performed using ANNOVAR (https://annovar.openbioinformatics.org/en/latest/) with pathogenicity prediction scores from the dbnsfp 4.2a dataset (50).
Subjects with CPSF6 deletions or missense variants
We searched the following public databases for anonymized individuals harboring CPSF6 variants (13): ClinVar (https://ncbi.nlm.nih.gov/clinvar/), DECIPHER (51), GeneMatcher (52), MIRACA Baylor College of Medicine Medical Genetics Laboratory, and the Diaphragmatic Hernia Research and Exploration; and Advancing Molecular Science study (53). We found 15 individuals carrying a missense variant in CPSF6 and were able to enroll three of them (Fig. 1 and fig. S2). Two of the three missense variants, p.A432T (S10) and p.D535V (S11), are de novo and absent in the general population (gnomAD, 141,456 individuals; BRAVO database, 62,784 individuals), but p.P383A (S9) was detected in two alleles of presumably unaffected individuals of European descent. The 12 individuals that we were not able to enroll have novel variants that have either a) not been found in gnomAD or BRAVO, or b) have been reported in one allele according to gnomAD (subjects 17 and 18) (table S2).
The statistical association between a cardiac phenotype and CPSF6 variant was determined by screening a large cohort of 13,218 patients (Baylor Genetics) for whom diagnostic exome sequencing and phenotypic descriptions were available from the Baylor College of Medicine. Of 13,218 patients, 2350 were identified as exhibiting a cardiac phenotype, defined by one or several of the following keywords within their clinical descriptions: “cardiac,” “VSD,” “CHD,” “cutis marmorata,” “ischemic attack,” “atrioventricular,” “aortic,” “septal,” “mitral,” “cardio,” and “heart.” Of these 2350 individuals, 11 harbored putatively pathogenic variation in CPSF6. There were 18 cases of 10,850 harboring putatively pathogenic variation in CPSF6 without cardiac features being noted. A two-sided Fisher’s exact test was performed against the null hypothesis that no association exists within the 2 × 2 contingency table (Table 2).
Table 2. Contingency table (2 × 2) of cardiac phenotype and CPSF6 variants.
Cases identified as having one (or several) cardiac-related features had a greater likelihood of harboring a CPSF6 variant (0.47%) relative to cases without reported cardiac phenotype (0.17%; P < 0.005). Fisher’s exact test P value = 0.0116; odds ratio (95% confidence interval) = 2.83 (1.21; 6.35).
| Cardiac-related keyword | No cardiac-related keyword | |
|---|---|---|
| CPSF6 variants identified | 11 | 18 |
| No CPSF6 variants identified | 2339 | 10,850 |
For copy number variations, we limited our search to those <20 megabases. Subjects were enrolled for the study (i.e., we acquired clinical data for further analysis) only after obtaining written informed consent/assent for each study subject from their respective parents. All patient-related study procedures were conducted according to the respective ethics committees of each participating institution. Each patient underwent a full clinical examination by a neurologist and/or medical geneticist. Clinical data were directly abstracted from medical records provided by the referring clinician(s). Clinical data from subjects 2, 3, and 4 were obtained from DECIPHER. When possible, standardized assessment of impairment in cognitive domains in each subject was made in accordance with the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition, which classifies intellectual disability as mild, moderate, severe, or profound.
Genomic analysis of subject 8 found an additional mutation, a de novo c.73A > G/p.K25E missense variant in SCAF4. To determine whether this mutation contributed substantially to the phenotype, we first performed Western blot analysis on fibroblasts from S8 and three healthy age-matched controls and found no difference between them in SCAF4 protein expression (fig. S1C). Second, because the absence of SCAF4 in cell lines promotes transcriptional readthrough (54), we checked the transcriptional readthrough of two of the top SCAF4 target genes, ATG5 and SMAD2 (54), and found no differences by either quantitative real-time polymerase chain reaction (qPCR) or RNA-seq between the patient-derived cells line and the controls (fig. S1, D and E).
Fibroblast cells from CPSF6 subjects
We isolated primary fibroblasts from skin biopsies taken from the subjects during the first year of life using standard methodology (55). Briefly, the skin had been placed in a transport medium (Ham’s F10, Thermo Fisher Scientific, catalog no. 11550043), from which we removed the skin specimen using a sterile technique (in a class II biohazard cabinet). We transferred the specimen to a sterile petri dish and cut it into small pieces (<0.5 mm) using sterile scalpel blades. We transferred these pieces to the lower surface of a 25-cm2 culture flask (six to eight pieces per flask), which had been premoistened with 1 to 2 ml of AmnioMAX Complete Medium (Thermo Fisher Scientific, catalog no. 17001074) supplemented with 1% penicillin/streptomycin. Patient-derived cell lines and three healthy age-matched controls (see below) were maintained at 37°C in a humidified incubator supplemented with 5% CO2 in Dulbecco’s modified Eagle’s medium (GenDepot, Katy, TX, catalog no. CM002-320) supplemented with 10% heat-inactivated fetal bovine serum (GenDepot, Katy, TX, catalog no. F0901-050), 1% penicillin/streptomycin (GenDepot, Katy, TX, catalog no. CA005-010), and 1% l-glutamine (Hyclone Laboratories, Logan, UT, catalog no. SH30034). The medium was renewed every 3 to 4 days until ready for subculturing.
We purchased three healthy control infant fibroblast lines: CCD-1064Sk, CCD-1070Sk, and CCD-1079Sk (American Type Culture Collection, Manassas, VA). Because only male infant fibroblasts are available on the market, our controls are age-matched for both subjects but sex-matched only for S11; the controls and the two subjects are all matched for ethnicity (Caucasian). To allow for as much biological variation as possible, we performed all human cell–based experiments at least three times, using a different batch of cells (at a similar number of passages) for each of the subjects and controls.
Preparation of human cells for Western blot analysis
We collected fibroblasts from CPSF6 subjects and controls at 6 × 106 cell confluence and processed them for protein extraction. Cell pellets were subsequently lysed with radioimmunoprecipitation assay buffer consisting of 25 mM tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, and 0.1% SDS completed with 1× Xpert Protease and 1× Phosphatase Inhibitor Cocktail Solutions (GenDepot, catalog nos. P3100-100 and P3200-020). We lysed cells by pipetting them up and down with a p200 tip and then placed them on ice for 20 min. Last, we cleared the lysate of cellular debris by centrifuging 20 min at 21,000g at 4°C.
Proteins were quantified by the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, catalog no. PI23225), and the absorbance was measured by NanoDrop OneC (Thermo Fisher Scientific). Consequently, proteins were resolved by high-resolution NuPAGE 4 to 12% bis-tris gel (Thermo Fisher Scientific, catalog no. NP0335BOX) according to the manufacturer’s instructions. Antibodies used for all the Western blot experiments were as follows: rabbit α-CPSF6 [1:1000 (Bethyl Laboratories, TX, catalog no. A301-356A)], mouse α-NUDT21 (2203C3) [1:500 (Santa Cruz, TX, catalog no. sc-81109)], mouse α-CPSF7 [1:500 (Santa Cruz, TX, catalog no. sc-393880)], rabbit α-FIP1L1 [1:500 (A301-461A, Bethyl Laboratories, TX)], rabbit α-SCAF4 [1:1000 (Bethyl Laboratories, TX, catalog no. A303-951A)], and mouse α–glyceraldehyde-3-phosphate dehydrogenase [1:10,000 (Millipore, catalog no. CB1001)].
qPCR in human cells and zebrafish
For human cells, we collected cells as described above for Western blot. For zebrafish, we anesthetized cpsf6−/− larvae at 6 dpf and their wild-type stage-matched controls with 0.0168% tricaine (Sigma-Aldrich, catalog no. MS-222, E1052). RNA was extracted using the miRNeasy kit (QIAGEN, Hilden, Germany, catalog no. 217004) according to the manufacturer’s instructions.
RNA was quantified using NanoDrop OneC (Thermo Fisher Scientific). cDNA was synthesized using Quantitect Reverse Transcription kit (QIAGEN, Hilden, Germany, catalog no. 205313) starting from 1 μg of RNA. Quantitative reverse transcription PCR experiments were performed using the CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA) with PowerUP SYBR Green Master Mix (Thermo Fisher Scientific, catalog no. A25743). Real-time PCR runs were analyzed using the comparative Ct method normalized against the housekeeping human gene GAPDH (56) or zebrafish actin beta 1 (actb1). To evaluate gene expression and unambiguously distinguish cDNA from genomic DNA, deoxyribonuclease treatment was performed during RNA extraction, and specific exon-exon spanning primers were designed to amplify across introns of the gene tested.
PAC-seq in human fibroblasts and zebrafish
Total RNA was extracted from human fibroblasts using the miRNeasy kit (QIAGEN, Hilden, Germany, catalog no. 217004) according to the manufacturer’s instructions. Zebrafish cpsf6−/− larvae were harvested as described above, at 6 dpf (before there is sexual differentiation), along with their wild-type stage-matched controls, and larvae or heads were collected for RNA extraction. Five samples were used from each genotype for both larvae and heads for a total of 20 samples: five cpsf6−/− larvae, five stage-matched larvae, five cpsf6−/− heads, and five heads stage-matched controls. Forty and 60 larvae and heads were combined and homogenized for each sample, respectively.
The PAS-seq library prep and sequencing protocol was adapted from (16). We added 1 to 2 μg of total RNA to 2 μl of a1:5 AzVTP:dNTP mixture at 5 mM (AzATP, AzCTP, AzGTP, AzTTP, and baseclick) and then added 1 μl of a 100 μM stock of the 3′Illumina_4N_21T_VN primer (5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNTTTTTTTTTTTTTTTTTTTTTVN-3′) to achieve a final volume of 13 μl. The reaction was incubated at 65° for 5 min, followed by snap cooling on ice for >1 min. We then added 7 μl of a master-mix containing the following reagents to make a 20-μl final reaction (on ice): 4 μl of 5× Superscript First Strand Buffer (Thermo Fisher Scientific), 1 μl of 0.1 M dithiothreitol, 1 μl of RNase OUT Recombinant Ribonuclease Inhibitor (Thermo Fisher Scientific), and 1 μl of Superscript III Reverse Transcriptase (Thermo Fisher Scientific). The reaction was incubated at 25°C for 10 min, 50°C for 40 min, and 75°C for 15 min. Last, we added 1 μl of RNase H (1 U/μl, diluted from higher concentration by H2O) (Thermo Fisher Scientific) and incubated the mixture at 37°C for 20 min and then 80°C for 10 min.
To clean the RT reaction, we added 37.8 μl of AMPure XP beads (1:1.8, pipette mix) and incubated the mixture for 5 min at room temperature. Beads were pelleted using a magnet for 2 min at room temperature, and the supernatant was discarded. Beads were washed twice with 200 μl of 80% ethanol and then pelleted using the magnet. Beads were then allowed to dry before being resuspended in 12 μl of 50 mM Hepes (pH 7.2) and incubated for 2 min at room temperature. Last, beads were pelleted by a magnet, and 10 μl of the supernatant was transferred to new tubes.
For the click ligation step, 20 μl of dimethyl sulfoxide was mixed with 3 μl of UMI-click-adapter (5 μM) 5′-Hexynyl-NNNNNNNNNNNNAGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT-3′ and 10 μl of the cleaned cDNA. In a separate tube at room temperature, 0.4 μl of vitamin C (50 mM) was mixed with 2 μl of Cu(II)-{Tris[(1-benzyl-4-triazolyl)methyl]amine} (TBTA) (10 mM) (Lumiprobe, #21050). The color was allowed to change from blue to clear before we added 2.4 μl of this mix to the cDNA and incubated it at room temperature for 30 min. The click ligation step was performed a second time by adding another 2.4 μl of the vitamin C/Cu(II)-TBTA to the cDNA and incubated again for 30 min.
To clean the click-ligated cDNA, we added 68 μl of AMPure XP beads (1:1.8, pipette mix) and incubated the mix at room temperature for 5 min. We pelleted beads using the magnet and discarded the supernatant. We washed the beads twice with 200 μl of 80% ethanol and pelleted them using the magnet and then allowed them to briefly dry before resuspending them in 22 μl of 10 mM tris (pH 7.4), mixing with a pipette, and incubating them for 2 min at room temperature. Last, beads were pelleted using the magnet, and 20 μl of the supernatant was transferred to a new tube.
To PCR amplify the click-ligated cDNA, a 50-μl reaction was assembled that contained 10 μl of clean click-ligated cDNA, 2 μl of a 5 μM 5′ and 3′ indexing primers mix (Integrated DNA Technologies), 13 μl of H2O, and 25 μl of 2× OneTaq Standard Buffer Master Mix (New England Biobiolabs, #M0482). The PCR reaction was then carried out for 15 to 17 cycles under the following conditions: 94°C for 4 min, 53°C for 30 s, 68°C for 10 min, 94°C for 30 s, 54°C for 30 s, and 68°C for 2 min.
To purify PAC-seq libraries before sequencing, we added 45 μl (50-μl PCR volume × 0.9) of AMPure XP beads to the PCR reaction, mixed them with a pipette, and incubated them for 5 min at room temperature. We pelleted the beads with a magnet and transferred the supernatant to new tubes. We then added 10 μl (50-μl PCR volume × 0.2) AMPure XP beads to the supernatant, which we incubated for 10 min at room temperature. We discarded the supernatant and pelleted the beads, which we washed twice with 200 μl of 85% ethanol before drying them briefly. The beads were then resuspended in 12 μl of 10 mM tris (pH 7.4), pipette mixed, and incubated further for 5 min at room temperature. Last, beads were pelleted using the magnet, and 10 μl of the supernatant was collected and subjected to quantification and quality control analysis.
Libraries derived from zebrafish RNA were prepared as described here and were sequenced at the URMC Genomics Core on a Novaseq SP100 cycle Flowcell. Libraries derived from human RNA were prepared as described here but with two exceptions: Only one barcode primer was used in PCR amplification, and the libraries were sequenced at the Genomics Core located at The University of Texas Medical Branch on a NextSeq High Output Flowcell.
PAC-seq analysis
Data processing
Human samples were sequenced to a depth of ~21 million per sample with 150–base pair (bp)–long single-end reads, and zebrafish samples were sequenced to a depth of ~24 million per sample with 121-bp-long single-end reads. For each sample, PCR duplicates were identified using Unique molecular identifiers (UMI) bar codes. UMI tools extract (57) was used to extract the UMI nucleotides from the reads and append them to the read names. Initial quality control was performed using fastp (58). First four nucleotides and the reads shorter than 40 nucleotides were also filtered using the -f and -l options, respectively. Adapter contamination (AGATCGGAAGAGC) was trimmed using the -a option. Reference human genome and annotations of the build GRCh38 release 33 were downloaded from the GENCODE portal. Reference zebrafish genome and annotations of the build GRCz11 were downloaded from the Lawson Laboratory (https://umassmed.edu/lawson-lab/reagents/zebrafish-transcriptome/) and Ensemble data portal, respectively. UMI marked and trimmed reads were aligned to respective reference genomes using Bowtie2 (59) version 2.2.6 with parameters -D 20 R 3 N 0 L 20 -i S,1,0.50 (very-sensitive-local). Sample-wise alignments were saved as Sequence Alignment Map (SAM) files. SAMtools (60) V0.1.19 “view,” “sort” and “index” modules were used to convert the SAM files to Binary Alignment Maps files, sort by chromosomal coordinates, and index, respectively. Mapped reads are deduplicated using UMI tools dedup (57) based on the UMI marked read names and the mapping coordinates.
We analyzed APA events at two levels. First, we calculated the overall effect of APA for a transcript [shorter versus longer, represented by the poly(A) index; see Figs. 2, A and B, and 5, A and B]. Then, we analyzed the locations of all of the PAS used in the APA events for that transcript (intronic/3′UTR/5′UTR/CDS; see Figs. 2, E and F, and 5, E and F). We provide details for each of these analyses below.
APA analysis
We used PolyA-miner (17) to identify shifts or changes at specific PAS. Each PAS is subjected to the same denoising filters and a β-binomial test. We combined individual PAS P values to infer the gene-level-APA P value. We computed the overall poly(A) index using vector projections to determine whether the overall effect was to create shorter- or longer-than-normal transcripts. Human samples were processed with the following parameters: -pa_p 0.6 -pa_a 5 -pa_m 3 -ip_u 30 -ip_d 40 -a 0.65 -novel_d 5000 -expNovel 1 -t BB. A detailed description of the pipeline and the parameters is given in (61). We extended the PolyA-miner v1.0 with additional upstream (-ip_u) and downstream (-ip_d) mispriming window parameters. Each polyA site is scanned with a window of -ip_u and -ip_d for mispriming/genomic stretches of adenosine (A) nucleotides. We also updated the PolyA-miner to selectively filter specific gene features (CDS, intronic, 3′UTR, and 5′UTR) from the APA analysis. Annotated polyadenylation sites from PolyA_DB3 (62) were converted to GRCh38 coordinate system using liftOver from the UCSC genome browser and used as reference. PolyA-miner v1.31 was used in the Python 3.8 environment. We analyzed 3′UTR-APA by excluding the polyA sites mapped to other genomic regions with the parameter -ignore UTR5, CDS, Intron, UN. Zebrafish samples were processed using the following parameters -p 20 -pa_p 0.6 -pa_a 5 -pa_m 5 -ip_u 30 -ip_d 40 -a 0.65 -outPrefix ZF_Body_ALL -novel_d 5000 -expNovel 1 -t BB and no reference polyadenylation annotations with PolyA-miner v1.3 in the Python 3.9 environment. PCA on the PolyA cleavage site counts was performed to confirm genotypes separated (63).
PAS usage analysis
We added up the read counts of all PAS mapped to each region of interest—CDS, introns, the 5′UTR, and the 3′UTR—to determine whether the number and variety of PAS used in each region was greater or lesser relative to controls. CDS, 5′UTR, and 3′UTR were extracted in bed format. Features from multiple isoforms of a transcript were merged. Intronic features were obtained by subtracting the merged exonic regions from the gene coordinates. PolyA sites were extracted in de novo mode and cleaned for potential mispriming as done in APA in analysis above. Read counts of the cleaned polyA sites were extracted using featureCounts (64). Reads mapping to multiple gene features (Eon-Intron/Intron-Exon/Intron-UTR/Exon-UTR junctions) were assigned to the feature with the read 3′ end. Each polyA site is mapped to one of the four features (CDS, intron, 3′UTR, and 5′UTR). Any multimapped polyA features were marked unknown. Total reads per feature were computed by adding all individual polyA site counts mapped to respective features, and total gene counts were computed as the sum of all individual feature polyA counts. A β-binomial test (65) was performed using the proportions of individual feature counts to the total gene counts to infer differential polyA usage changes. Genes with less than 10 reads mapped to a specific feature were ignored, respectively.
DEG analysis
We computed gene levels counts as the sum total of reads mapped to the polyA sites of a gene. We excluded genes with an average read count of less than two across the samples. We then normalized raw read counts and tested for differential expression using DESeq2 (66) as previously described (63).
Mass spectrometry
Total proteins were extracted from subject and control fibroblasts using urea lysis buffer [8 M urea, 75 mM NaCl, 50 mM tris/HCl (pH 8.0), and 1 mM EDTA]. Protein concentrations of all samples were determined by Pierce BCA assay. Twenty micrograms of total protein per sample was processed further. Disulfide bonds were reduced with 5 mM dithiothreitol for 45 min at room temperature, and cysteines were subsequently alkylated with 10 mM iodoacetamide in the dark for 45 min at room temperature. Proteins were precipitated onto magnetic SP3 beads as described in (67) by adding ethanol to the samples, resulting in a sample that was 50% organic solvent, and by shaking for 8 min at room temperature. Beads were washed three times with 1.5 ml of 80% ethanol and reconstituted in 100 μl of ammonium bicarbonate. Samples were then digested off the beads using Promega sequencing grade modified trypsin in an enzyme-to-substrate ratio of 1:50. After 16 hours of digestion, samples were acidified to a final concentration of 1% formic acid. Tryptic peptides were taken off the beads and evaporated to dryness in a vacuum concentrator. Desalted peptides were then labeled with the TMTpro mass tag labeling reagent. TMTpro reagent (0.1 U) was used per 20 μg of sample. (TMT allows for multiplexing of samples so that there is greater quantification precision during mass spectrometry analysis.) Peptides were dissolved in 30 μl of 50 mM Hepes (pH 8.5) solution, and the TMTpro reagent was added in 12.3 μl of MeCN. After 1-hour incubation, the reaction was stopped with 4 μl of 5% hydroxylamine for 20 min at 25°C. Differentially labeled peptides were mixed, desalted on C18 StageTips according to (68), evaporated to dryness in a vacuum concentrator, and reconstituted in 100 μl of 4.5 mM ammonium formate (pH 10) in 2% acetonitrile for high-performance liquid chromatography (HPLC) fractionation.
Channels 131C through 134 were left out of downstream comparisons (Table 3). We performed mass spectrometry on all three healthy controls, but because we had used 1079 for PAC-seq comparative analysis, we used it for the mass spectrometry analysis as well. The mixed, desalted sample was fractionated into 12 fractions via basic reversed-phase chromatography as per (69). The sample was separated on the basis of hydrophobicity using an Agilent Zorbax 300 Extend-C18 15-cm column on an Agilent 1260 Series HPLC into a 96 deep-well plate. Solvent A and solvent B were 4.5 mM ammonium formate in 2% acetonitrile and 4.5 mM ammonium formate in 90% acetonitrile, respectively. The gradient mimics that in (69) but with a flow rate of 0.2 ml/min. The collected wells were then concatenated into 12 fractions in an alternating pattern to evenly distribute each part of the gradient into each concatenated fraction.
Table 3. Proteomics labeling scheme.
Tandem mass tag (TMT) labeling was used to label different samples. The name of the reporter ion channel is listed in the table with the corresponding sample name and replicate number. The channel 127N was not used.
| TMTpro channel | Sample | Replicate |
|---|---|---|
| 126 | Subject 11 | 1 |
| 127N | Empty | – |
| 127C | Subject 11 | 2 |
| 128N | Subject 11 | 3 |
| 128C | Subject 8 | 1 |
| 129N | Subject 8 | 2 |
| 129C | Subject 8 | 3 |
| 130N | Control - 1079 | 1 |
| 130C | Control - 1079 | 2 |
| 131N | Control - 1079 | 3 |
| 131C | Unrelated sample | 1 |
| 132N | Unrelated sample | 2 |
| 132C | Unrelated sample | 3 |
| 133N | Unrelated sample | 1 |
| 133C | Unrelated sample | 2 |
| 134 | Unrelated sample | 3 |
LC-MS/MS analysis on a Q-Exactive HF
About 1 μg of total peptides was analyzed on a Waters M-Class UPLC using a 25-cm Ionopticks Aurora column coupled to a benchtop Thermo Fisher Scientific Orbitrap Q Exactive HF mass spectrometer. Peptides were separated at a flow rate of 400 nl/min with a 160-min gradient, including sample loading and column equilibration times. MS1 spectra were measured with a resolution of 120,000, an AGC target of 3 × 106, and a mass range from 300 to 1800 mass/charge ratio (m/z). Up to 12 MS2 spectra per duty cycle were triggered at a resolution of 60,000, an AGC target of 1 × 105, an isolation window of 0.8 m/z, a normalized collision energy of 28, a scan range of 200 to 2000 m/z, and a fixed first mass of 110 m/z.
Proteomics: Quantification and statistical analysis
All raw data were analyzed with MaxQuant software version 1.6.10.43 (70) using a UniProt database (Homo sapiens, UP000005640), and MS/MS searches were performed with the following parameters: TMTpro-16plex labeling on the MS2 level, oxidation of methionine and protein N-terminal acetylation as variable modifications; carbamidomethylation as fixed modification; trypsin/P as the digestion enzyme; precursor ion mass tolerances of 20 parts per million (ppm) for the first search (used for nonlinear mass recalibration) and 7 ppm for the main search; and a fragment ion mass tolerance of 20 ppm. For identification, we applied a maximum false discovery rate (FDR) of 1% separately on protein and peptide levels. We required 1 or more unique/razor peptides for protein identification and at least two MS/MS spectra ratio counts for quantification for each TMT channel. This gave us a total of 4358 quantified protein groups.
First, protein group intensities were normalized for the number of observable peptides in each protein group using the iBAQ value for a given protein group (71). iBAQ value refers to the sum of all peptide intensities of a protein group divided by the number of observable peptides in that protein group. To normalize, a protein group intensity in a given channel was divided by the sum of intensities across the TMT plex for that protein group and multiplied by the iBAQ value of that protein group.
Next, we normalized the corrected TMT MS2 intensity such that, at each channel’s intensity, values were added up to exactly 1,000,000; therefore, each protein group value can be regarded as a normalized microshare. A pseudo count of 1 was added to all zero values to allow for log2 transformation of the data. Once in log2 space, the data were then used for downstream differential expression analysis.
Zebrafish mutation and genotype
All zebrafish work followed protocols approved by Columbia University Irving Medical Center’s Institutional Animal Care and Use Committee. We obtained cpsf6sa9322 from the Zebrafish International Resource Center (ZIRC) (72). This allele has a point mutation (C>T) creating a premature stop codon in exon 4. To genotype adult fish, we extracted genomic DNA from the fin; for paraformaldehyde (PFA)–fixed larvae, we used whole larva for genotyping. Fins or larvae were dissolved in 50 or 20 μl of 50 mM NaOH, respectively, incubated for 20 min at 95°C neutralized with tris-HCl (pH 7.5; 1:10). We performed PCR with EconoTaq PLUS GREEN (Lucigen, Middleton, WI, catalog no. 30033-1) according to the manufacturer’s instructions, using between 2 and 5 μl of genomic DNA and the following primers: cpsf6-fw 5′-GCCTTCCCCTCTCCTGACAT-3′ and cpsf6-rv 5′-ATCTCTCTGTGGCCCTCACC-3′. Two microliters of the 666-bp PCR product was digested with 0.3 μl of Bsph I (New England Biolabs, catalog no. R0517). The C>T mutation creates a Bsph I restriction site; the PCR product from wild-type zebrafish will result in one DNA band (666 bp), cpsf6+/− will result in three bands (666, 466, and 200 bp), and cpsf6−/− will result in two bands (466 and 200 bp).
Depigmentation of embryos
When necessary, embryos were depigmented after overnight fixation in 4% PFA at 4°C. They were washed twice in PBT (phosphate-buffered saline–0.1% Tween) and then incubated in 3% H2O2, 0.5% KOH, and 0.1% Tween for 35 min at 6 dpf. Subsequently, the embryos were washed twice again in PBT.
Zebrafish imaging and video experiments
Bright-field acquisition
We anesthetized zebrafish from 2 to 6 dpf with 0.0168% tricaine (Sigma-Aldrich, catalog no. MS-222, E1052) and acquired images in brightfield using the Zeiss stereomicroscope at 5× (Fig. 4A) and 10× (Fig. 4, B and C). We recorded videos of the hearts beating for 30 s per zebrafish with the Zeiss stereomicroscope at 10× (movies S1 to S9). Beats were counted for 30× and doubled to achieve beats per minute.
Immunofluorescence
Whole-mount immunofluorescence for heart and axonal projection were performed as previously described (73) for the following antibodies: mouse monoclonal antibodies against sarcomeric myosin heavy chain, α-MF20 (1:200; Developmental Studies Hybridoma Bank); mouse monoclonal atrial myosin heavy chain, α-S46 (1:20; Developmental Studies Hybridoma Bank); rabbit polyclonal anti–elastin b, α-elnb (1:500); mouse monoclonal anti-acetylated tubulin, α-ac-tubulin (1:250; Millipore Sigma, catalog no. T7451); and rabbit α-CFIm68 (1:200; Bethyl Laboratories, TX, catalog no. A301-356A). MF20 and S46 were obtained from the Developmental Studies Hybridoma Bank maintained by the Department of Biological Sciences, University of Iowa, under contract NO1-HD-2-3144 from the National Institute of Child Health and Human Development. Elnb was generated in collaboration by the Waxman lab and YenZym (www.YenZym.com) from the PGAGYQQQYPGFGGPGAGGPGS (amino acids 1958 to 1979) (74).
Whole-mount immunofluorescence for pMNs was performed as previously described (75) using mouse monoclonal anti-synaptotagmin 2, α-znp1 (1:100; Zebrafish International Resource Center, ZIRC, catalog no. AB_10013783). The following secondary antibodies were used: goat anti-mouse immunoglobulin G2b (IgG2b) Alexa Fluor 568 (Thermo Fisher scientific, catalog no. A-21144), goat anti-mouse IgG1 Alexa Fluor 488 (Thermo Fisher scientific, catalog no. A-21121), and goat anti-mouse IgG2a Alexa Fluor 568 (Thermo Fisher scientific, catalog no. A-21134). All the secondary antibodies were used at a dilution of 1:200. Larvae stained for MF20/S46 were placed in a glass-well plate with concave wells and imaged using a fluorescence stereomicroscope. For all other staining, we mounted larvae in 0.1% low-melting agarose and acquired images with a Zeiss LSM-800 confocal microscope. Z-stack images were processed with Fiji software (76); we quantified axonal length with NeuronJ (77), a Fiji plugin (76).
Alcian blue–Alizarin red staining
We performed Alcian blue and Alizarin red staining as previously described (24). Briefly, we fixed up to 100 larvae in 4% PFA for 2 hours at room temperature. We then dehydrated the larvae with 50% (v/v) ethanol for 10 min at room temperature and stained them with an acid-free solution of Alcian blue–Alizarin red overnight (also at room temperature). The next day, we washed the larvae with water and bleached them for 30 min at room temperature with a solution of 3% H2O2 and 2% KOH. After bleaching, we resuspended the larvae in 20% (v/v) glycerol and 0.25% KOH and took images using a bright-field stereomicroscope.
Free swimming assay on zebrafish larvae
We used 6-dpf larvae for the swimming assay. We placed individual larvae in 24-well plates placed on top of a light pad to provide backlighting. We video-recorded the fish for 10 min using the Supereyes B003+ camera (at 18 frames/s) and iSpy software. Analysis was performed using EthoVision XT (Noldus).
DAVID gene ontology and REVIGO clustering analysis
We performed DAVID gene ontology as previously described (33). Biological processes were obtained as an output file, and significant gene ontology terms (FDR < 0.25) were used for clustering using REVIGO (34). Both DAVID GO (33) and PubMed (https://pubmed.ncbi.nlm.nih.gov/) were used to infer the biological pathways in Fig. 6 (E and F).
Gene set enrichment analysis
Preranked GSEA was performed as previously described (20) using the ranked lists of RNA-seq or quantitative mass spectrometry and the list of genes with intronic or 3′UTR PAS usage as input. The output file was used to graph the enrichment score.
Primers used in this manuscript
The following primers were used: Hs_CPSF6, 5′-GATGTCGGCGAAGAGTTCA-3′ (forward) and 5′-CTTCTGGGGCATCTCCATTA-3′ (reverse); Hs_GAPDH, 5′-CGACCACTTTGTCAAGCTCA-3′ (forward) and 5′-TTACTCCTTGGAGGCCATGT-3′ (reverse); Zf_cpsf6 exon 2, 5′-AAAGGGGCTCCAGCTAATGT-3′ (forward); Zf_cpsf6 exon 3, 5′-TATTGAGCGAATGGCATCTG-3′ (reverse); Zf_cpsf6 exon 9, 5′-AGCTCACTGCAGGACTGCTT-3′ (forward) and 5′-TTCTCACGAGAGCGACTGTG-3′ (reverse); Zf_actb1, 5′-TCTCTTGCTCCTTCCACCAT-3′ (forward) and 5′-GGGCCAGACTCATCGTACTC-3′ (reverse); Zf_cpsf6 genotype, 5′-GCCTTCCCCTCTCCTGACAT-3′ (forward) and 5′-ATCTCTCTGTGGCCCTCACC-3′ (reverse).
Quantification and statistical analysis
Experimental design
At every stage of the study, the experimenter was blinded to the identity of the cell lines. For example, experimenter #1 made a list of samples and controls to be tested, and experimenter #2 randomized this list and relabeled the tubes; experimenter #2 was the only person with the key to identify the samples. These samples were then distributed to experimenter #3 to culture the cells, then to experimenter #1 to perform Western blots, and lastly, experimenters #1 and #4 analyzed the data. Only then was the key applied to identify the samples. For behavioral assays and immunofluorescence, the experimenter was blinded to the genotype of the animals, as described in the preceding paragraph.
Software and statistical analysis
Unless otherwise specified, all experimental statistics and graphs were analyzed using GraphPad Prism 9 (https://graphpad.com/scientific-software/prism/) and Excel Software (Microsoft). Statistical details and numbers of replicates for each experiment can be found in figures and legends.
Acknowledgments
We thank the families who participated in this study and all the clinicians who provided clinical information. We thank M. Shirasu-Hiza and S. J. Tener for the use of the recording free swimming in zebrafish (see Materials and Methods) and J. Waxman for the anti-elnb antibody (74). We also thank members of the Gennarino laboratory for helpful discussions.
Funding: This work was supported by National Institute of Neurological Disorders and Stroke (NINDS; R01NS109858 to V.A.G.); National Human Genome Research Institute (NHGRI; R01 HG012216 to M.J.); National Institute on Aging (NIA; R01 AG071869 to M.J.); National Institute of General Medical Science (NIGM; R35 GM128802 to M.J.); National Heart, Lung, and Blood Institute (NHLBI; R01 HL131438-01A1 to K. L.T.); Paul A. Marks Scholar Program, Columbia University Vagelos College of Physicians and Surgeons (V.A.G.); TIGER Award, Taub Institute, Columbia University Irving Medical Center (V.A.G.); Columbia Stem Cell Initiative (CSCI), Columbia University Irving Medical Center (V.A.G. and N.d.P.); National Science Foundation Graduate Research Fellowship Program, NSF-GRFP (L.C.T.); Estonian Research Council; grant PRG471 (K.Õ. and K.R.); United States Department of Agriculture (USDA/ARS) under Cooperative Agreement No. 58-3092-0-001 (H.K.Y.); Duncan NRI Zoghbi Scholar Award (H.K.Y.); and Gulf Coast Consortia on the NLM Training Program in Biomedical Informatics and Data Science (T15 LM0070943 to V.S.J.).
Author contributions: Conceptualization: N.d.P., V.B., M.J., K. L.T., H.K.Y., E.J.W., and V.A.G. Methodology: N.d.P., M.J., K. L.T., H.K.Y., E.J.W., and V.A.G. Software: N.D.E., L.C.T., V.S.J., H.K.Y., and E.J.W. Validation: N.d.P., C.F., N.D.E., M.J., H.K.Y., E.J.W., and V.A.G. Formal analysis: N.d.P., N.D.E., W.L., L.C.T., V.S.J., M.J., H.K.Y., E.J.W., and V.A.G. Investigation: N.d.P., C.F., N.D.E., W.L., L.C.T., K.-L.H., A.L., P.J., L.B., M.C., S.B., and V.A.G. Resources: L.B., K.Õ., K.R., M.H.W., J.A.R., W.B., K.T., T.P., T.G., A.S., C.-T.F., J.K.G.-A., C.A.B., A.H.-K., J.A.B., A.A.N., W.K.C., M.J., K. L. T., H.K.Y., E.J.W., and V.A.G. Data curation: N.d.P., N.D.E., V.S.J., K.Õ., K.R., M.H.W., J.A.R., W.B., K.T., T.P., T.G., A.S., C.-T.F., J.K.G.-A., C.A.B., A.H.-K., J.A.B., A.N., W.K.C., M.J., H.K.Y., E.J.W., and V.A.G. Writing—original draft: N.d.P. and VAG. Writing—review and editing: N.d.P., M.C., W.K.C., V.B., M.J., K. L.T., H.K.Y., E.J.W., and V.A.G. Visualization: N.d.P., C.F., W.L., V.B., H.K.Y., and V.A.G. Supervision: V.A.G. Project administration: V.A.G. Funding acquisition: V.A.G.
Competing interests: W.B. and J.A.R. work with The Department of Molecular and Human Genetics at Baylor College of Medicine, which receives revenue from clinical genetic testing completed at Baylor Genetics Laboratories. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The patient-derived fibroblasts from subject 8 and subject 11 can be provided by the corresponding author (V.A.G.) pending scientific review and a completed material transfer agreement (MTA). Requests should be submitted to vag2138@cumc.columbia.edu. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, V.A.G. (vag2138@cumc.columbia.edu). All unique/stable reagents generated in this study are available from the lead contact. Data and code used in this study are freely available. The PAC-seq data are available in the NCBI Gene Expression Omnibus (GEO), accession number: GSE206559 GSE human and zebrafish (https://ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE206559). The LC-MS/MS data are available in the Mass Spectrometry Interactive Virtual Environment (MASSIVE) at ftp://massive.ucsd.edu/MSV000090759/.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S7
Legends for tables S1 to S38
Legends for movies S1 to S12
References
Other Supplementary Material for this : manuscript includes the following:
Tables S1 to S38
Movies S1 to S12
REFERENCES AND NOTES
- 1.E. A. Ponomarenko, E. V. Poverennaya, E. V. Ilgisonis, M. A. Pyatnitskiy, A. T. Kopylov, V. G. Zgoda, A. V. Lisitsa, A. I. Archakov, The size of the human proteome: The width and depth. Int. J. Anal. Chem. 2016, 7436849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.S. N. Floor, J. A. Doudna, Tunable protein synthesis by transcript isoforms in human cells. ELife 5, e10921 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.N. J. Proudfoot, A. Furger, M. J. Dye, Integrating mRNA processing with transcription. Cell 108, 501–512 (2002). [DOI] [PubMed] [Google Scholar]
- 4.B. Tian, J. L. Manley, Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 18, 18–30 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Y. Zhang, L. Liu, Q. Qiu, Q. Zhou, J. Ding, Y. Lu, P. Liu, Alternative polyadenylation: Methods, mechanism, function, and role in cancer. J. Exp. Clin. Cancer Res. 40, 51 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.N. Abdel Wahab, J. Gibbs, R. M. Mason, Regulation of gene expression by alternative polyadenylation and mRNA instability in hyperglycaemic mesangial cells. Biochem. J. 336 (Pt 2), 405–411 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.H. Duan, N. Cherradi, J. J. Feige, C. Jefcoate, cAMP-dependent posttranscriptional regulation of steroidogenic acute regulatory (STAR) protein by the zinc finger protein ZFP36L1/TIS11b. Mol. Endocrinol. 23, 497–509 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.J.-W. Chang, W. Zhang, H.-S. Yeh, E. P. de Jong, S. Jun, K.-H. Kim, S. S. Bae, K. Beckman, T. H. Hwang, K.-S. Kim, D.-H. Kim, T. J. Griffin, R. Kuang, J. Yong, mRNA 3′-UTR shortening is a molecular signature of mTORC1 activation. Nat. Commun. 6, 7218 (2015). [DOI] [PubMed] [Google Scholar]
- 9.D. Rund, C. Dowling, K. Najjar, E. A. Rachmilewitz, H. H. Kazazian Jr., A. Oppenheim, Two mutations in the beta-globin polyadenylylation signal reveal extended transcripts and new RNA polyadenylylation sites. Proc. Natl. Acad. Sci. U.S.A. 89, 4324–4328 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.A. Hellquist, M. Zucchelli, K. Kivinen, U. Saarialho-Kere, S. Koskenmies, E. Widen, H. Julkunen, A. Wong, M.-L. Karjalainen-Lindsberg, T. Skoog, J. Vendelin, D. S. Cunninghame-Graham, T. J. Vyse, J. Kere, C. M. Lindgren, The human GIMAP5 gene has a common polyadenylation polymorphism increasing risk to systemic lupus erythematosus. J. Med. Genet. 44, 314–321 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.V. A. Gennarino, C. E. Alcott, C.-A. Chen, A. Chaudhury, M. A. Gillentine, J. A. Rosenfeld, S. Parikh, J. W. Wheless, E. R. Roeder, D. D. G. Horovitz, E. K. Roney, J. L. Smith, S. W. Cheung, W. Li, J. R. Neilson, C. P. Schaaf, H. Y. Zoghbi, NUDT21-spanning CNVs lead to neuropsychiatric disease and altered MeCP2 abundance via alternative polyadenylation. eLife 4, e10782 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.K. J. Karczewski, L. C. Francioli, G. Tiao, B. B. Cummings, J. Alföldi, Q. Wang, R. L. Collins, K. M. Laricchia, A. Ganna, D. P. Birnbaum, L. D. Gauthier, H. Brand, M. Solomonson, N. A. Watts, D. Rhodes, M. Singer-Berk, E. M. England, E. G. Seaby, J. A. Kosmicki, R. K. Walters, K. Tashman, Y. Farjoun, E. Banks, T. Poterba, A. Wang, C. Seed, N. Whiffin, J. X. Chong, K. E. Samocha, E. Pierce-Hoffman, Z. Zappala, A. H. O’Donnell-Luria, E. V. Minikel, B. Weisburd, M. Lek, J. S. Ware, C. Vittal, I. M. Armean, L. Bergelson, K. Cibulskis, K. M. Connolly, M. Covarrubias, S. Donnelly, S. Ferriera, S. Gabriel, J. Gentry, N. Gupta, T. Jeandet, D. Kaplan, C. Llanwarne, R. Munshi, S. Novod, N. Petrillo, D. Roazen, V. Ruano-Rubio, A. Saltzman, M. Schleicher, J. Soto, K. Tibbetts, C. Tolonen, G. Wade, M. E. Talkowski; Genome Aggregation Database Consortium, B. M. Neale, M. J. Daly, D. G. MacArthur, The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.W. Lee, N. de Prisco, V. A. Gennarino, S. Buttery, How to expand the method details in your Cell Press paper with step-by-step STAR protocols. STAR Protoc. 3, 101550 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Y. Zhu, X. Wang, E. Forouzmand, J. Jeong, F. Qiao, G. A. Sowd, A. N. Engelman, X. Xie, K. J. Hertel, Y. Shi, Molecular mechanisms for CFIm-mediated regulation of mRNA alternative polyadenylation. Mol. Cell 69, 62–74.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D. C. Di Giammartino, K. Nishida, J. L. Manley, Mechanisms and consequences of alternative polyadenylation. Mol. Cell 43, 853–866 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.N. D. Elrod, E. A. Jaworski, P. Ji, E. J. Wagner, A. Routh, Development of Poly(A)-ClickSeq as a tool enabling simultaneous genome-wide poly(A)-site identification and differential expression analysis. Methods 155, 20–29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.H. K. Yalamanchili, C. E. Alcott, P. Ji, E. J. Wagner, H. Y. Zoghbi, Z. Liu, PolyA-miner: Accurate assessment of differential alternative poly-adenylation from 3′Seq data using vector projections and non-negative matrix factorization. Nucleic Acids Res. 48, e69 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.C. Mayr, What are 3′UTRs doing? Cold Spring Harb. Perspect. Biol. 11, a034728 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.N. Spies, C. B. Burge, D. P. Bartel, 3′UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res. 23, 2078–2090 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.A. Subramanian, P. Tamayo, V. K. Mootha, S. Mukherjee, B. L. Ebert, M. A. Gillette, A. Paulovich, S. L. Pomeroy, T. R. Golub, E. S. Lander, J. P. Mesirov, Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.A. Kishor, S. E. Fritz, N. Haque, Z. Ge, I. Tunc, W. Yang, J. Zhu, J. R. Hogg, Activation and inhibition of nonsense-mediated mRNA decay control the abundance of alternative polyadenylation products. Nucleic Acids Res. 48, 7468–7482 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.W. Li, B. You, M. Hoque, D. Zheng, W. Luo, Z. Ji, J. Y. Park, S. I. Gunderson, A. Kalsotra, J. L. Manley, B. Tian, Systematic profiling of poly(A)+ transcripts modulated by core 3′ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLOS Genet. 11, e1005166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.C. M. Borini Etichetti, A. Tenaglia, M. N. Arroyo, J. E. Girardini, Expression of zebrafish cpsf6 in embryogenesis and role of protein domains on subcellular localization. Gene Expr. Patterns 36, 119114 (2020). [DOI] [PubMed] [Google Scholar]
- 24.M. B. Walker, C. B. Kimmel, A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 82, 23–28 (2007). [DOI] [PubMed] [Google Scholar]
- 25.E. de Pater, L. Clijsters, S. R. Marques, Y.-F. Lin, Z. V. Garavito-Aguilar, D. Yelon, J. Bakkers, Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development 136, 1633–1641 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.F. Tessadori, J. H. van Weerd, S. B. Burkhard, A. O. Verkerk, E. de Pater, B. J. Boukens, A. Vink, V. M. Christoffels, J. Bakkers, Identification and functional characterization of cardiac pacemaker cells in zebrafish. PLOS ONE 7, e47644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.M. Buckingham, S. Meilhac, S. Zaffran, Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 6, 826–835 (2005). [DOI] [PubMed] [Google Scholar]
- 28.C. A. Devine, B. Key, Identifying axon guidance defects in the embryonic zebrafish brain. Methods Cell Sci. 25, 33–37 (2003). [DOI] [PubMed] [Google Scholar]
- 29.E. D. Thomas, I. A. Cruz, D. W. Hailey, D. W. Raible, There and back again: Development and regeneration of the zebrafish lateral line system. Wiley Interdiscip. Rev. Dev. Biol. 4, 1–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.P. Z. Myers, J. S. Eisen, M. Westerfield, Development and axonal outgrowth of identified motoneurons in the zebrafish. J. Neurosci. 6, 2278–2289 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.P. Smibert, P. Miura, J. O. Westholm, S. Shenker, G. May, M. O. Duff, D. Zhang, B. D. Eads, J. Carlson, J. B. Brown, R. C. Eisman, J. Andrews, T. Kaufman, P. Cherbas, S. E. Celniker, B. R. Graveley, E. C. Lai, Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep. 1, 277–289 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.P. Miura, S. Shenker, C. Andreu-Agullo, J. O. Westholm, E. C. Lai, Widespread and extensive lengthening of 3′UTRs in the mammalian brain. Genome Res. 23, 812–825 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D. W. Huang, B. T. Sherman, R. A. Lempicki, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 34.F. Supek, M. Bošnjak, N. Škunca, T. Šmuc, REVIGO summarizes and visualizes long lists of gene ontology terms. PLOS ONE 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.H.-W. Tseng, A. Mota-Sydor, R. Leventis, I. Topisirovic, T. F. Duchaine, Distinct, opposing functions for CFIm59 and CFIm68 in mRNA alternative polyadenylation of Pten and in the PI3K/Akt signalling cascade. Nucleic Acids Res. 50, 9397–9412 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.C. Mayr, D. P. Bartel, Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–684 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R. Sandberg, J. R. Neilson, A. Sarma, P. A. Sharp, C. B. Burge, Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320, 1643–1647 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.A. R. Gruber, G. Martin, P. Müller, A. Schmidt, A. J. Gruber, R. Gumienny, N. Mittal, R. Jayachandran, J. Pieters, W. Keller, E. van Nimwegen, M. Zavolan, Global 3′UTR shortening has a limited effect on protein abundance in proliferating T cells. Nat. Commun. 5, 5465 (2014). [DOI] [PubMed] [Google Scholar]
- 39.S. J. Dubbury, P. L. Boutz, P. A. Sharp, CDK12 regulates DNA repair genes by suppressing intronic polyadenylation. Nature 564, 141–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.S.-H. Lee, I. Singh, S. Tisdale, O. Abdel-Wahab, C. S. Leslie, C. Mayr, Widespread intronic polyadenylation inactivates tumour suppressor genes in leukaemia. Nature 561, 127–131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.G. R. LaForce, J. S. Farr, J. Liu, C. Akesson, E. Gumus, O. Pinkard, H. C. Miranda, K. Johnson, T. J. Sweet, P. Ji, A. Lin, J. Coller, P. Philippidou, E. J. Wagner, A. E. Schaffer, Suppression of premature transcription termination leads to reduced mRNA isoform diversity and neurodegeneration. Neuron 110, 1340–1357.e7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R. Elkon, A. P. Ugalde, R. Agami, Alternative cleavage and polyadenylation: Extent, regulation and function. Nat. Rev. Genet. 14, 496–506 (2013). [DOI] [PubMed] [Google Scholar]
- 43.G. Martin, A. R. Gruber, W. Keller, M. Zavolan, Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′UTR length. Cell Rep. 1, 753–763 (2012). [DOI] [PubMed] [Google Scholar]
- 44.C. P. Masamha, Z. Xia, J. Yang, T. R. Albrecht, M. Li, A.-B. Shyu, W. Li, E. J. Wagner, CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 510, 412–416 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.T. Weng, J. Ko, C. P. Masamha, Z. Xia, Y. Xiang, N. Y. Chen, J. G. Molina, S. Collum, T. C. Mertens, F. Luo, K. Philip, J. Davies, J. Huang, C. Wilson, R. A. Thandavarayan, B. A. Bruckner, S. S. Jyothula, K. A. Volcik, L. Li, L. Han, W. Li, S. Assassi, H. Karmouty-Quintana, E. J. Wagner, M. R. Blackburn, Cleavage factor 25 deregulation contributes to pulmonary fibrosis through alternative polyadenylation. J. Clin. Invest. 129, 1984–1999 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.C. E. Alcott, H. K. Yalamanchili, P. Ji, M. E. van der Heijden, A. Saltzman, N. Elrod, A. Lin, M. Leng, B. Bhatt, S. Hao, Q. Wang, A. Saliba, J. Tang, A. Malovannaya, E. J. Wagner, Z. Liu, H. Y. Zoghbi, Partial loss of CFIm25 causes learning deficits and aberrant neuronal alternative polyadenylation. eLife 9, e50895 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.N. K. Mohanan, F. Shaji, G. R. Koshre, R. S. Laishram, Alternative polyadenylation: An enigma of transcript length variation in health and disease. Wiley Interdiscip. Rev. RNA 13, e1692 (2022). [DOI] [PubMed] [Google Scholar]
- 48.P. Tang, Y. Yang, G. Li, L. Huang, M. Wen, W. Ruan, X. Guo, C. Zhang, X. Zuo, D. Luo, Y. Xu, X.-D. Fu, Y. Zhou, Alternative polyadenylation by sequential activation of distal and proximal PolyA sites. Nat. Struct. Mol. Biol. 29, 21–31 (2022). [DOI] [PubMed] [Google Scholar]
- 49.O. Mauger, F. Lemoine, P. Scheiffele, Targeted intron retention and excision for rapid gene regulation in response to neuronal activity. Neuron 92, 1266–1278 (2016). [DOI] [PubMed] [Google Scholar]
- 50.X. Liu, X. Jian, E. Boerwinkle, dbNSFP: A lightweight database of human nonsynonymous SNPs and their functional predictions. Hum. Mutat. 32, 894–899 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.H. V. Firth, S. M. Richards, A. P. Bevan, S. Clayton, M. Corpas, D. Rajan, S. V. Vooren, Y. Moreau, R. M. Pettett, N. P. Carter, DECIPHER: Database of chromosomal imbalance and phenotype in humans using ensembl resources. Am. J. Hum. Genet. 84, 524–533 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.N. Sobreira, F. Schiettecatte, D. Valle, A. Hamosh, GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 36, 928–930 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.L. Yu, J. Wynn, L. Ma, S. Guha, G. B. Mychaliska, T. M. Crombleholme, K. S. Azarow, F. Y. Lim, D. H. Chung, D. Potoka, B. W. Warner, B. Bucher, C. A. LeDuc, K. Costa, C. Stolar, G. Aspelund, M. S. Arkovitz, W. K. Chung, De novo copy number variants are associated with congenital diaphragmatic hernia. J. Med. Genet. 49, 650–659 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.L. H. Gregersen, R. Mitter, A. P. Ugalde, T. Nojima, N. J. Proudfoot, R. Agami, A. Stewart, J. Q. Svejstrup, SCAF4 and SCAF8, mRNA anti-terminator proteins. Cell 177, 1797–1813.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.M. J. Barch, Association of Cytogenetic Technology, The ACT Cytogenetics Laboratory Manual, Second Edition. (Raven Press, 1991).
- 56.J. Vandesompele, K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, F. Speleman, Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, research0034.1 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.T. Smith, A. Heger, I. Sudbery, UMI-tools: Modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res. 27, 491–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.S. Chen, Y. Zhou, Y. Chen, J. Gu, fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.B. Langmead, S. L. Salzberg, Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.H. Li, B. Handsaker, A. Wysoker, T. Fennell, J. Ruan, N. Homer, G. Marth, G. Abecasis, R. Durbin; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.H. K. Yalamanchili, N. D. Elrod, M. K. Jensen, P. Ji, A. Lin, E. J. Wagner, Z. Liu, A computational pipeline to infer alternative poly-adenylation from 3′ sequencing data. Methods Enzymol. 655, 185–204 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.R. Wang, R. Nambiar, D. Zheng, B. Tian, PolyA_DB 3 catalogs cleavage and polyadenylation sites identified by deep sequencing in multiple genomes. Nucleic Acids Res. 46, D315–D319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.H. K. Yalamanchili, Y. W. Wan, Z. Liu, Data analysis pipeline for RNA-seq experiments: From differential expression to cryptic splicing. Curr. Protoc. Bioinformatics 59, 11.15.1–11.15.21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Y. Liao, G. K. Smyth, W. Shi, featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Q. Tan, H. K. Yalamanchili, J. Park, A. de Maio, H.-C. Lu, Y.-W. Wan, J. J. White, V. V. Bondar, L. S. Sayegh, X. Liu, Y. Gao, R. V. Sillitoe, H. T. Orr, Z. Liu, H. Y. Zoghbi, Extensive cryptic splicing upon loss of RBM17 and TDP43 in neurodegeneration models. Hum. Mol. Genet. 25, 5083–5093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.M. I. Love, W. Huber, S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.C. S. Hughes, S. Moggridge, T. Müller, P. H. Sorensen, G. B. Morin, J. Krijgsveld, Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 14, 68–85 (2019). [DOI] [PubMed] [Google Scholar]
- 68.J. Rappsilber, M. Mann, Y. Ishihama, Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007). [DOI] [PubMed] [Google Scholar]
- 69.J. Li, J. G. van Vranken, L. Pontano Vaites, D. K. Schweppe, E. L. Huttlin, C. Etienne, P. Nandhikonda, R. Viner, A. M. Robitaille, A. H. Thompson, K. Kuhn, I. Pike, R. D. Bomgarden, J. C. Rogers, S. P. Gygi, J. A. Paulo, TMTpro reagents: A set of isobaric labeling mass tags enables simultaneous proteome-wide measurements across 16 samples. Nat. Methods 17, 399–404 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.J. Cox, M. Mann, MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 71.B. Schwanhäusser, D. Busse, N. Li, G. Dittmar, J. Schuchhardt, J. Wolf, W. Chen, M. Selbach, Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 72.R. N. W. Kettleborough, E. M. Busch-Nentwich, S. A. Harvey, C. M. Dooley, E. de Bruijn, F. van Eeden, I. Sealy, R. J. White, C. Herd, I. J. Nijman, F. Fényes, S. Mehroke, C. Scahill, R. Gibbons, N. Wali, S. Carruthers, A. Hall, J. Yen, E. Cuppen, D. L. Stemple, A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature 496, 494–497 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.J. Alexander, D. Y. Stainier, D. Yelon, Screening mosaic F1 females for mutations affecting zebrafish heart induction and patterning. Dev. Genet. 22, 288–299 (1998). [DOI] [PubMed] [Google Scholar]
- 74.Y. C. Song, T. E. Dohn, A. B. Rydeen, A. V. Nechiporuk, J. S. Waxman, HDAC1-mediated repression of the retinoic acid-responsive gene ripply3 promotes second heart field development. PLOS Genet. 15, e1008165 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Y. Arribat, K. S. Mysiak, L. Lescouzères, A. Boizot, M. Ruiz, M. Rossel, P. Bomont, Sonic Hedgehog repression underlies gigaxonin mutation-induced motor deficits in giant axonal neuropathy. J. Clin. Invest. 129, 5312–5326 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak, A. Cardona, Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.E. Meijering, M. Jacob, J.-C. F. Sarria, P. Steiner, H. Hirling, M. Unser, Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A 58A, 167–176 (2004). [DOI] [PubMed] [Google Scholar]
- 78.Y. C. M. Staal, J. Meijer, R. J. C. van der Kris, A. C. de Bruijn, A. Y. Boersma, E. R. Gremmer, E. P. Zwart, P. K. Beekhof, W. Slob, L. T. M. van der Ven, Head skeleton malformations in zebrafish (Danio rerio) to assess adverse effects of mixtures of compounds. Arch. Toxicol. 92, 3549–3564 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.D. Bader, T. Masaki, D. A. Fischman, Immunochemical analysis of myosin heavy chain during avian myogenesis in vivo and in vitro. J. Cell Biol. 95, 763–770 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.M. Miao, A. E. Bruce, T. Bhanji, E. C. Davis, F. W. Keeley, Differential expression of two tropoelastin genes in zebrafish. Matrix Biol. 26, 115–124 (2007). [DOI] [PubMed] [Google Scholar]
- 81.G. Piperno, M. T. Fuller, Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J. Cell Biol. 101, 2085–2094 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.S. Rohr, C. Otten, S. Abdelilah-Seyfried, Asymmetric involution of the myocardial field drives heart tube formation in zebrafish. Circ. Res. 102, e12–e19 (2008). [DOI] [PubMed] [Google Scholar]
- 83.A. Khan, A. Mathelier, Intervene: A tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinformatics 18, 287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S7
Legends for tables S1 to S38
Legends for movies S1 to S12
References
Tables S1 to S38
Movies S1 to S12