Abstract
Glioblastoma is a heterogeneous tumor for which effective treatment options are limited and often insufficient. Studies examining the intratumoral transcriptional and proteomic heterogeneity of the glioblastoma microenvironment to characterize the spatial distribution of potential molecular and cellular therapeutic immuno-oncology targets are limited. We applied an integrated multimodal approach comprised of NanoString GeoMx® Digital Spatial Profiling (DSP), single cell RNA-seq (scRNAseq), and expert neuropathological assessment to characterize archival formalin-fixed paraffin-embedded (FFPE) glioblastoma specimens. Clustering analysis and spatial cluster maps highlighted the intratumoral heterogeneity of each specimen. Mixed cell deconvolution analysis revealed that neoplastic and vascular cells were the prominent cell types throughout each specimen, with macrophages, oligodendrocyte precursors, neurons, astrocytes, and oligodendrocytes present in lower abundance and illustrated the regional distribution of the respective cellular enrichment scores. Spatial resolution of the actionable immunotherapeutic landscape showed that robust B7H3 gene and protein expression was broadly distributed throughout each specimen and identified STING and VISTA as potential targets. Lastly, we uncovered remarkable variability in VEGFA expression and discovered unanticipated associations between VEGFA, endothelial cell markers, hypoxia, and the expression of immunoregulatory genes, indicative of regionally distinct immunosuppressive microdomains. This work provides an early demonstration of the ability of an integrated panelbased spatial biology approach to characterize and quantify the intrinsic molecular heterogeneity of the glioblastoma microenvironment.
BACKGROUND
Tumors are dynamic tissues comprised of malignant cells, various immune cell subtypes, distinct non-immune stromal cells, metabolites, soluble signaling molecules, and differential oxygenation and nutritional gradients.1,2 To truly harness the potential of immuno-oncology and identify rational therapeutic targets, a detailed molecular and cellular understanding of the tumor microenvironment is required. Spatial profiling is a novel approach that is well suited to meet this challenge by generating dozens to thousands of regionally distinct gene expression profiles from a single tissue.3-8 Glioblastoma is a challenging molecularly and cellularly heterogeneous malignancy of the central nervous system (CNS) with variable penetration of immune cells and resistant cancer stem cell subclones.9-16 We aimed to utilize spatial profiling using the NanoString GeoMx platform to characterize the intratumoral heterogeneity of this disease and reveal region-specific microenvironmental gene expression profiles. The data presented here demonstrates the utility of integrating clinical neuropathology, single cell sequencing, and spatial profiling technologies to identify and characterize the multiple region-specific tumor microenvironments within individual archived formalin-fixed paraffin embedded human glioblastoma specimens.
MATERIALS AND METHODS
Patient specimens
Archived FFPE specimens were obtained from the Department of Pathology at the Keck School of Medicine of the University of Southern California under Institutional Review Board protocol HS-11-00385 and in compliance with the Declaration of Helsinki. Specimens were obtained from adult patients with a neuropathological diagnosis of glioblastoma, IDH-wildtype, CNS World Health Organization (WHO) Grade 4.17 Representative hematoxylin and eosin (H&E) stained slides from each specimen were evaluated for the following features to determine their suitability for profiling: tissue size, percent necrosis, and hemorrhage. IDH mutation status and specimen age were also evaluated. Of the seven specimens identified, three were selected for further investigation. Serial 5μm slides were cut from each block and used for spatial RNA and protein profiling.
Protein assay slide preparation
Slides were baked in a drying oven, deparaffinized, rehydrated in an ethanol gradient, and placed in ditheyl pyrocarbonate (DEPC)-treated water. Antigen retrieval was performed with 1X citrate buffer,pH6 in a pressure cooker on high temperature setting for 15 minutes. The slides were then blocked and incubated with the primary antibody mix and the fluorescently conjugated morphology markers. The morphology markers used are as follows: Iba1-647 at 1:75 dilution (EMD Millipore, MABN92-AF647), CD3-594 at 1:50 dilution (Abcam, ab196147), GFAP-532 at 1:3000 dilution (Novus Biologicals, NBP2-33184AF532), and SYTO 13 at 1:5 dilution (Thermo Scientific S7575).
RNA assay slide preparation
Slides were prepared by baking in a drying oven and then processed using the Leica BOND platform (Leica Biosystems) as specified by the NanoString GeoMx DSP Slide Preparation User Manual. Briefly, slides were deparaffinized with xylene then rehydrated through a graded ethanol series. Targets were exposed by incubating the slides in a pressure cooker at 100°C in 1X Tris-EDTA buffer, pH9 followed by proteinase K digestion. Next, the GeoMx NGS-RNA Probe Mix (NanoString Technologies) was applied to each slide, and slides were incubated at 37°C for 16-24 hours. This probe mix contained barcoded oligonucleotide probes against 1,700 gene targets and included internal positive and negative control probes. After washing, the slides were then blocked in Buffer W (NanoString Technologies) and stained with morphology markers for 1 hour. The fluorescently conjugated morphology markers used are as follows: Iba1 at 1:75 dilution (EMD Millipore, MABN92-AF647), CD3 at 1:50 dilution (Abcam, ab196147), GFAP at 1:3000 dilution (Novus Biologicals, NBP2-33184AF532), and SYTO 13 nuclei acid stain at 1:5 dilution (Thermo Scientific S7575).
Neuropathological assessment and ROI selection criteria
Regions of interest (ROIs) in each tissue were arbitrarily selected without regard to the morphology markers in attempts to broadly sample multiple regions of each tissue. H&E-stained sections for each histologically-determined glioblastoma specimen were divided into individual ROIs. The mean ROI diameter and average nuclei/ROI are listed in Supplemental Table 1. Each ROI was reviewed by a board-certified neuropathologist (PJC) and annotated for histological features of glioblastoma according to the 2016 revised fourth edition of the World Health Organization Classification of Tumors of the Central Nervous System.17 Noted histological features included microvascular proliferation, blood vessels without microvascular proliferation, necrosis, pseudopalisading cells, hemorrhage, and tumor cellularity. Tumor cellularity was further provided as a qualitative visual estimate of cellularity (low, medium, and high) according to guidelines previously published for glioblastoma.16 Estimates of each feature were given as a percentage of ROI occupancy. Discriminating factors for ROI selection included the avoidance of areas with significant hemorrhage, necrosis, and other acellular regions that would result in a low signal-to-noise ratio. ROIs were matched as best as possible between serial sections for the RNA and protein assays.
Library preparation and sequencing
Samples were prepared for next generation sequencing (NGS) on an Illumina NextSeq 550 as specified by GeoMx – NGS Readout User Manual (MAN-10117, Nanostring Technologies). The quality of the libraries were checked using an Agilent Bioanalyzer and subsequently sequenced on an Illumina NextSeq 550 at a concentration of 1.6pM with a 5% PhiX spike-in as per Illumina manual instructions.
Normalization of GeoMx data
RNA Data: The negative control probes were used to estimate the number of background counts in each data point. This calculation was performed separately for each of the two probe pools used in this study. Expected background was subtracted from the raw counts, and negative values were reset to 0. Finally, each segment’s data was scaled such that all segments had the same 85th percentile expression value. Protein Data: Individual ROIs were examined to identify outliers with low signal and/or high background. Next, the data were normalized by dividing the antibody count value by the geometric mean of the housekeeping antibodies GAPDH and S6 as suggested by the GeoMx - NGS Data Analysis User Manual (MAN-10119-05, NanoString Technologies).
Single-sample gene set enrichment analysis
Single-sample gene set enrichment analysis (ssGSEA) was performed with the gene set variation analysis (GSVA) R library using the ssGSEA method with a Poisson kernel, applied to the normalized gene expression data described above. The fgsea R package was used to access the Reactome gene sets, and all sets with 5-500 genes were considered.
Mixed cell deconvolution
Cell type abundance was estimated using the SpatialDecon algorithm.18 The expected expression profiles for each cell type were derived using a publicly available scRNA-seq dataset derived from glioblastoma specimens.19 Cells that fell below the inflection point, were considered empty by emptyDrops, had a gene count above 2.5x the average gene count, or had a percentage of mitochondrial genes > 0.05 were removed from the data matrix.20 Genes were removed if they appeared in less than 2 cells or had low biological significance as measured by scran.21 The original cell type calls were retained with the exception of the myeloid cell cluster. Cells with the myeloid cell label were re-clustered and marker genes identified using Seurat.22 Clustered marker genes were compared to PanglaoDB markergenes (ubiquitousness index < 0.1, sensitivity > 0.6, specificity < 0.4, canonical marker).23 Cell clusters were named according to the PanglaoDB cell type with the most overlapping marker genes. Both new cell cluster names, “macrophage” and “microglia”, were manually reviewed for correctness. Each cell cluster’s profile was reported as the arithmetic mean of its cells’ expression profiles. From the matrix of expected cell type expression profiles, a subset of 432 informative genes was defined as all genes that surpassed a minimal expression level in at least one cell type and had a single cell type accounting for at least 40% of their total expression level across the columns of the cell type matrix. This yielded a reference matrix of 432 genes in 8 cell types.
Statistical Analysis
Constellation plots were generated using JMP Pro 15 (SAS). Heatmaps, correlation matrices, and dendrograms were generated using Morpheus visualization and analysis software (https://software.broadinstitute.org/morpheus/). Uniform Manifold Approximation and Projection (UMAP) plots of the processed glioblastoma scRNA-seq data from Darmanis et al. were generated using the Single Cell Expression Atlas web interface (https://www.ebi.ac.uk/gxa/sc/home).19 Stacked bar plots, dot plots, line graphs, and scatter plots were generated using Prism9 (GraphPad).
Spatial Heatmaps
The ROIs used for spatial profiling were manually applied to full slide images of the hematoxylin and eosin-stained tissues as individual vector graphics using Adobe Illustrator 2021 (Adobe). Individual ROI vector graphics were pseudo-colored according to the corresponding color profile representing the value attributed to the gene/score in question.
RESULTS
Region-specific transcriptional profiles reveal intratumoral heterogeneity within individual glioblastoma specimens.
Glioblastoma is a markedly heterogeneous malignancy, both morphologically and molecularly.14,15 To assess the intratumoral transcriptional heterogeneity within each specimen, 32 spatially distinct ROIs were randomly chosen to capture a range of regions in each specimen, avoiding hemorrhagic and acellular portions of the tissues (Figure 1A-C). After processing and normalization, 29/32 ROIs in HGG03, 29/32 ROIs in HGG05, and 31/32 ROIs in HGG06 were deemed sufficient for downstream analysis based on quality control thresholds and abundance of normalized gene counts.
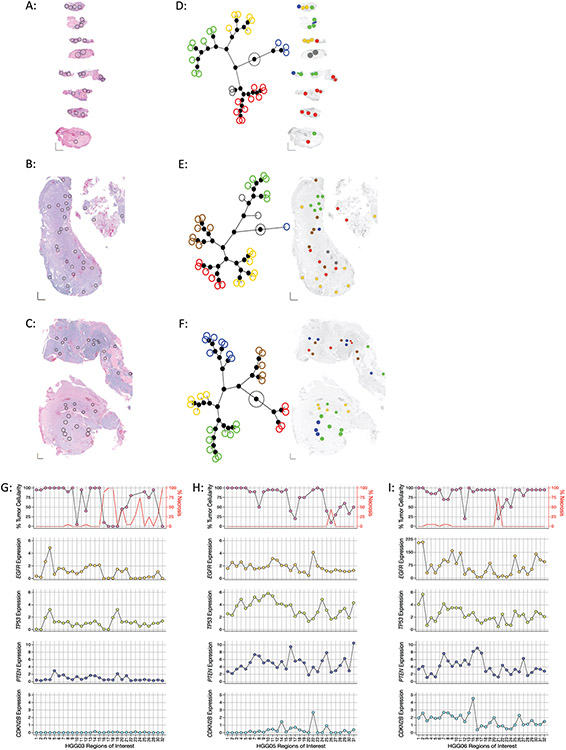
Figure 1: Regional intratumoral heterogeneity of gene expression profiles within individual glioblastoma specimens.
(A-C) Histology images of tumor samples (A) HGG03, (B) HGG05, and (C) HGG06 with spatially distinct regions of interest (ROIs) in each specimen indicated by black circles. XY scale bars in bottom left corner of each image represent 1000μm on each axis. (D-E) Left: Constellation plots depicting the unsupervised hierarchical clustering of the entire 1,700 gene panel in specimens (D) HGG03, (E) HGG05, and (F) HGG06 showing distinct transcriptionally associated clusters. Each colored circle corresponds to an individual ROI. Right: Spatial cluster maps showing the cluster identity and distribution of ROIs within each specimen where individual ROIs are colored according to their assigned clusters as indicated by the constellation plots on the left. (G-I) Regional heterogeneity in the expression of commonly altered glioblastoma tumor suppressor genes and oncogenes. Dot plots showing the percentage of viable tumor (top, left Y-axis), necrosis (top, right Y-axis) and the expression of EGFR (second row), TP53 (third row), PTEN (fourth row), and CDKN2B (bottom row) in each ROI in specimens (G) HGG03, (H) HGG05, and (I) HGG06. Percent tumor and necrosis were scored by standard neuropathological examination. ROI’s where tumor cellularity was ambiguous were not plotted.
Unsupervised hierarchical clustering was used to investigate the relatedness of the gene expression profiles for each ROI within each specimen (Figure 1D-F, left). Spatial cluster maps were generated to visualize the regional distribution of the ROIs comprising the different constellation plot clusters within each glioblastoma specimen (Figure 1D-F, right). This analysis illustrated the intrinsic transcriptional heterogeneity within individual glioblastoma specimens by showing the regional enrichment or dispersion of ROIs belonging to the same cluster assignment.
Next, we investigated the extent of intertumoral heterogeneity across the three specimens. Principal component analysis demonstrated that the ROIs in specimens HGG03 and HGG05 formed tight independent clusters with little overlap, indicating the transcriptional distinction between the two specimens (Supplemental Figure 1). The ROIs in specimen HGG06 clustered in a more diffuse manner with ROIs overlapping both the HGG03 and HGG05 clusters as well as distally related ROIs, indicating that regions of specimen HGG06 are transcriptionally similar to specimens HGG03 and HGG05 while other regions are transcriptionally divergent (Supplemental Figure 1). Together, these data demonstrate that glioblastomas are comprised of multiple regionally distinct gene expression profiles, further highlighting the intrinsic inter- and intra-tumoral heterogeneity of this complex disease.
Regional expression of genes commonly altered in glioblastoma.
EGFR, TP53, PTEN, and CDKN2B are recurrently mutated in a significant fraction of glioblastomas.14,24,25 We examined the relative expression levels of each of these genes in each ROI with respect to the percent tumor cellularity as determined by standard neuropathological assessment and observed regional variability in both tumor cellularity and gene expression values (Figure 1G-I). We observed profound differences in the percent viable tumor versus percent necrosis throughout specimen HGG03 whereas appreciable necrosis was only present in a single ROI in both specimens HGG05 and HGG06. Notably, CDKN2B expression was uniformly absent in all ROIs in specimen HGG03 and mostly absent in specimen HGG05, suggestive of widespread CDKN2B loss (Figure 1G-H, bottom panels). Whether the observed lack of CDKN2B was due to genomic deletion, point mutation, or epigenetic silencing was not determined in this study. Similarly, with little exception, low PTEN expression was also noted in the ROIs from specimen HGG03 (Figure 1G, fourth panel).
Marked variation in EGFR expression was seen in ROIs from specimen HGG06 (Figure 1I, second panel). Next, we determined if the regional alterations in EGFR expression were associated with differences in tumor cellularity. We observed lower EGFR expression and decreased tumor cellularity in only 2/32 ROIs examined (Figure 1I, first and second panels), indicating that decreased EGFR expression was not associated with decreased tumor cell content. We interpreted these data to indicate the presence of clonal glioma populations with varied EGFR gene expression. While not confirmed as part of this study, the magnitude of EGFR expression in specimen HGG06 may reflect clonal amplification of the EGFR locus in ROIs with excessive EGFR expression. Together, these data highlight the regional heterogeneity in the expression of recurrently mutated genes in glioblastoma.
Quantification and spatial distribution of tumor-infiltrating immune cells.
To determine the abundance and localization of infiltrating immune cells within each tissue, we examined the expression of known lymphocyte and macrophage marker genes. In each of the specimens, we found that the expression of the macrophage marker genes was more abundant than that of the lymphocyte marker genes, excluding CD4 and NCAM1 (Figure 2A-C). To further validate this finding, we mined a publicly available glioblastoma scRNA-seq dataset.19 The scRNA-seq data supported our spatial profiling data and showed abundant gene expression of macrophage markers and an absence of lymphocyte markers except for CD4 and NCAM1 (Figure 2D). The scRNA-seq data also showed that NCAM1 mapped to the neoplastic cell cluster (Figure 2D), consistent with the existing data stating that NCAM1 marks glioma cells in addition to NK cells.26,27 Correlation analysis of the spatial gene expression data showed that CD4 expression significantly correlated with the expression of various macrophage marker genes rather than lymphocyte marker genes in each of the specimens (Supplemental Figure 2), suggesting that a population of CD4+ macrophages exists within these specimens.28
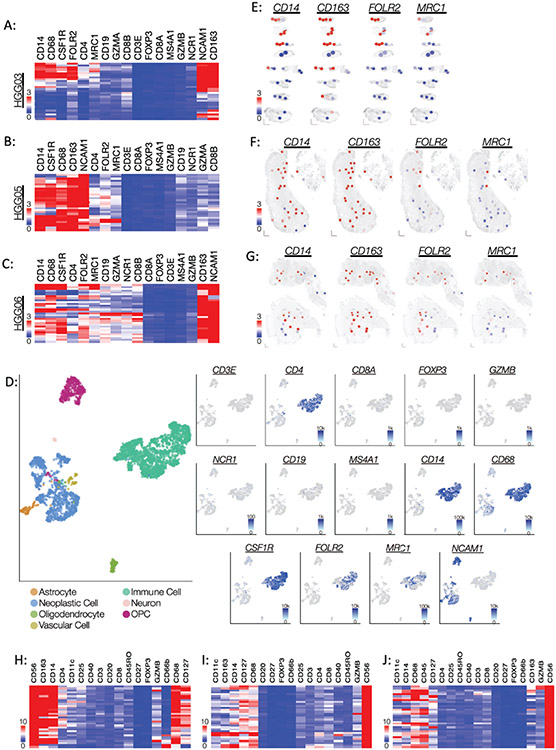
Figure 2: Regional expression of immune cell markers indicates a robust and widespread infiltration of macrophages.
(A-C) Heatmaps illustrating the unsupervised hierarchical clustering of the gene expression counts of lymphocyte and macrophage markers in specimens (A) HGG03, (B) HGG05, and (C) HGG06. Each row represents an individual ROI. Dendrograms were omitted to minimize figure congestion. (D) scRNA-seq data from glioblastoma tissue displayed as UMAP plots. Left: Cell type annotations. Right: Expression of individual lymphocyte and macrophage markers. (E-G) Spatial heatmaps demonstrating the distribution of pan-macrophage and M2-macrophage markers in specimens (D) HGG03, (E) HGG05, and (F) HGG06. (H-I) Heatmaps of the normalized antibody count data for lymphocyte and macrophage markers in specimens (H) HGG03, (I) HGG05, and (J) HGG06. The data are presented as fold change over mean IgG control value.
Spatial heatmaps were used to illustrate the regional expression of CD14 (monocyte/macrophage marker), CD163 (macrophage marker), FOLR2 (M2-tumor associated macrophage (TAM)), and MRC1 (M2-TAM) and revealed that CD163 was prominently expressed throughout most regions of the tissues (Figure 2E-G). While both FOLR2 and MRC1 mark M2-TAMs, we observed that FOLR2 gene counts were more abundant and more widely distributed than that of MRC1 in each of the specimens (Figure 2A-C, E-G). To validate the spatial gene expression data, we performed spatial proteomic profiling on serial tissue sections using molecularly barcoded antibodies against a panel of immune cell markers. Consistent with the gene expression data, this validation experiment showed that CD14, CD68, CD163, and NCAM1 proteins were highly expressed in each of the specimens (Figure 2H-J). Both granzyme-B and CD56 mark NK cells and subpopulations of cytotoxic T-cells. Spearman analysis revealed that CD56 protein expression does not positively correlate with granzyme-B protein expression, thus providing additional evidence that CD56 marks glioblastoma cells rather than NK cells and/or cytotoxic T-cells (Supplemental Figure 3). Taken together, these data suggest that macrophages are the prevailing immune cells within these glioblastoma specimens and that variations in the regional expression of the M2-TAM markers FOLR2 and MRC1 exist, highlighting the regional macrophage diversity in glioblastoma.29
Deconvolution and spatial mapping of resident glioblastoma cell types.
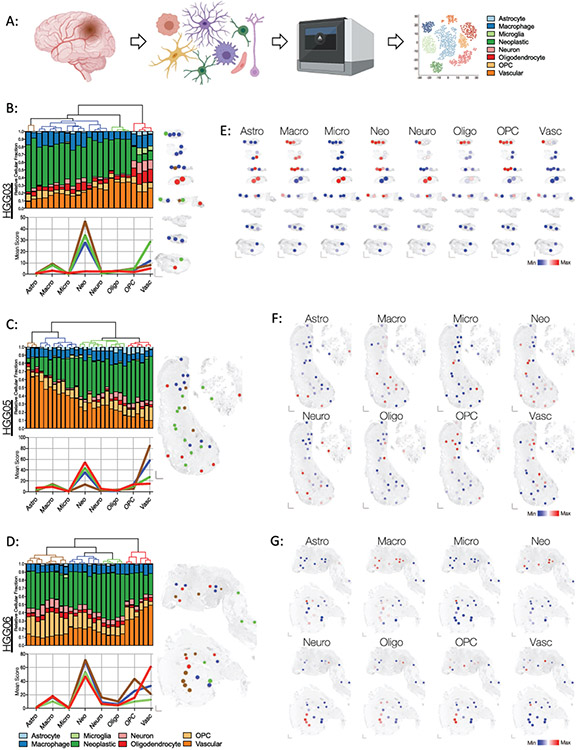
The SpatialDecon algorithm was used to compute abundance scores of resident astrocytes, macrophages, microglia, neoplastic cells, neurons, oligodendrocytes, oligodendrocyte precursor cells (OPC), and vascular cells based on glioblastoma scRNA-seq data (Figure 3A).18,19 The lack of sufficient infiltrating lymphocytes in the scRNA-seq data precluded the generation of a lymphocyte cell score. Analysis of the SpatialDecon results showed that the ROIs were predominated by neoplastic and vascular cells, albeit in varying relative proportions (Figure 3B-D). The relatedness in the cellular composition of each ROI was assessed using unsupervised hierarchical clustering and was subsequently visualized using spatial cluster maps (Figure 3B-D). Spatial heatmaps were used to localize the relative abundance of each computationally inferred cell type score within different regions of each specimen, illustrating the variable intratumoral distribution of resident glioblastoma cell types (Figure 2E-G).
Figure 3: Spatially resolved enrichment of glioblastoma cell type-specific gene signatures using SpatialDecon mixed cell deconvolution.
(A) Schematic of SpatialDecon workflow and the cell types identified by this process. The color key for each inferred cell type is consistent throughout the figure. (B-D) Cellular composition of each ROI in specimens (B) HGG03, (C) HGG05, and (D) HGG06. Top: Unsupervised hierarchical clustering dendrograms and associated stacked bar graphs showing relative fraction of each inferred cell type score within each ROI (column). Bottom: Mean score for each inferred cell type in each of the clusters identified above. The graph lines are colored in accordance with their assigned clusters designated by the dendrogram above. Right: Spatial cluster maps showing the cluster identity and distribution of ROIs within each specimen. Individual ROIs are colored according to their assigned clusters as indicated by the dendrograms to the left. (E-G) Spatial heatmaps showing the relative inferred cell score in each of the ROIs within specimens (E) HGG03, (F) HGG05, and (G) HGG06. Abbreviations are as follows: Astro, astrocyte; Macro, macrophage; Micro, microglia; Neo, neoplastic cells; Neuro, Neuron, Oligo, oligodendrocyte; OPC, oligodendrocyte precursor cells; Vasc, vasculature.
Comparison between computationally inferred cell type scores and neuropathological assessment.
To validate the utility and fidelity of our integrated approach, we compared the computationally inferred neoplastic and vascular cell type scores to the tumor cellularity and vascularity scores obtained by neuropathological examination (Supplemental Table 2). We observed a significant positive correlation between the inferred neoplastic score and the histological determination of percent tumor cellularity (Supplemental Table 3). Significant positive correlations between the inferred vascular score and vessels with microvascular proliferation were noted in specimens HGG05 and HGG06, whereas the computationally inferred vascular score correlated with vessels lacking microvascular proliferation in specimen HGG03. Together, these data validate the deconvolution method used in this study.
Divergent correlations between VEGFA, immunomodulators, and vascular abundance.
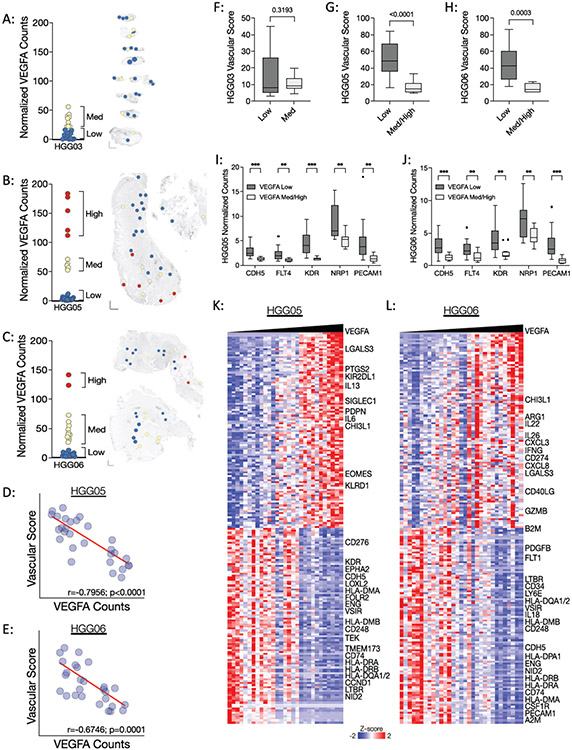
With little variation, our SpatialDecon data identified the vasculature as the most abundant non-malignant cellular component in these specimens (Figure 3B-D), prompting our examination of VEGFA expression. In comparison to HGG05 and HGG06, specimen HGG03 had lower VEGFA expression (Figure 4A-C). Unexpectedly, specimens HGG05 and HGG06 self-organized into VEGFA-High, VEGFA-Med, and VEGFA-Low ROIs, warranting deeper investigation of these specimens. Further analysis revealed significant inverse correlations between the vascular score and VEGFA expression in specimens HGG05 and HGG06 (Figure 4D-E), whereas no such correlation was apparent in specimen HGG03 (data not shown). Accordingly, we observed significantly higher vascular scores in the VEGFA-Low ROIs in specimens HGG05 and HGG06 (Figure 4F-H). The vascular score does not readily distinguish between blood and lymphatic vessels. To determine if the observed VEGFA-associated differences in the vascular score were specific to blood versus lymphatic endothelial cells, we compared the expression CDH5, FLT4, KDR, NRP1, and PECAM1 between VEGFA-Med/High ROIs and VEGFA-Low ROIs in specimens HGG05 and HGG06. This analysis showed a significantly increased expression of each endothelial cell marker in the VEGFA-Low ROIs from both specimens (Figure 4I-J).
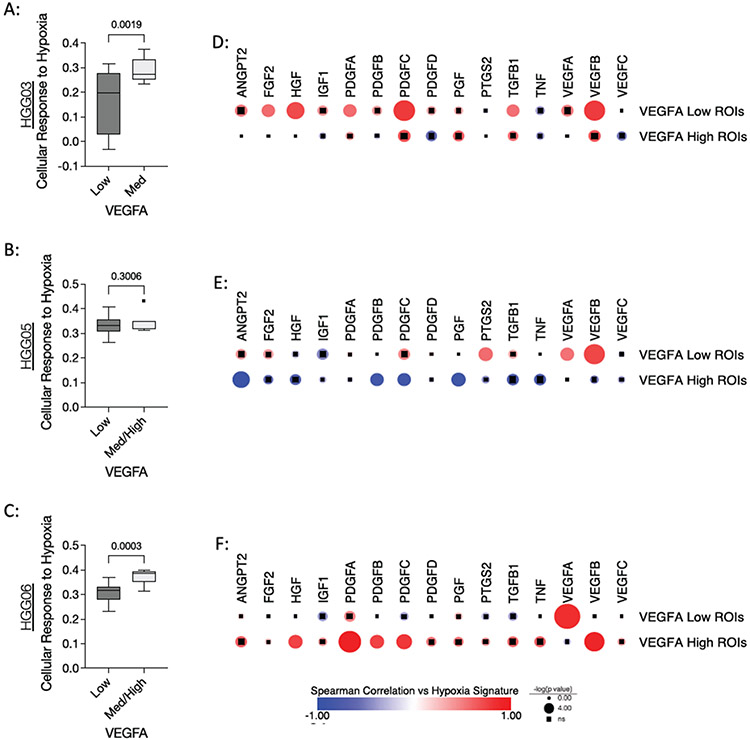
Figure 4: Regional heterogeneity in VEGFA gene expression inversely correlates with vascular abundance in specimens HGG05 and HGG06.
(A-C) Left: Dot plots of VEGFA expression in specimens (A) HGG03, (B), HGG05, and (C) HGG06. Right: Spatial heatmaps showing regions of low (blue ROIs), medium (yellow ROIs), and high (red ROIs) VEGFA expression in each corresponding specimen. (D-E) Spearman correlations between normalized VEGFA gene counts and the inferred vascular score in specimens (D) HGG05 and (E) HGG06. Correlation and significance values are listed along the X-axis of each scatter plot. (F-H) Box plots showing the vascular score in VEGFA-Low and VEGFA-Med/High ROIs in specimens (F) HGG03, (G) HGG05, and (H) HGG06. (I-J) Box plots showing the normalized gene counts of endothelial cell markers in VEGFA-Low and VEGFA-Med/High ROIs in specimens (I) HGG05 and (J) HGG06. Box plot p-values were calculated using an unpaired t-test with Welch’s correction where p < 0.05 is considered statistically significant. (K-L) Heatmaps showing the results of VEGFA nearest neighbors analysis in specimens (K) HGG05 and (L) HGG06.
Next we investigated which genes in this 1,700-gene panel correlate with the expression of VEGFA in specimens HGG05 and HGG06. Nearest neighbors analysis uncovered positive correlations between VEGFA expression and the expression of immunomodulatory genes, immune checkpoint genes, and various markers of T-cell exhaustion in both HGG05 and HGG06 (Figure 4K-L). To further contextualize the data, we performed an additional nearest neighbors analysis using GSVA scores to determine which biological pathways positively and negatively correlated with VEGFA expression in specimens HGG05 and HGG06 (Supplemental Figure 4). This analysis showed that VEGFA expression positively correlated with transcription, cell cycle progression, and receptor tyrosine kinase signaling and inversely correlated with processes related to immune responses and extracellular matrix remodeling. Additional analyses revealed significant positive and negative correlations between various endothelial cell markers and pro-angiogenic genes in a specimen-dependent manner suggesting that VEGFA-independent mechanisms support the vascularization of these tumors (Supplemental Figure 5). Taken together, these data propose a non-angiogenic function associated with excessive VEGFA expression that corresponds with the increased expression of immunomodulatory genes, thus promoting regionally distinct microenvironments in specimens HGG05 and HGG06.
Inter- and intra-tumoral variability in the hypoxic response.
It is well established that hypoxia drives angiogenesis by upregulating the expression of VEGFA and other pro-angiogenic molecules.30 Given our unanticipated finding that VEGFA inversely correlates with vascular markers in specimens HGG05 and HGG06 (Figure 4), we sought to investigate the relationship between VEGFA expression and hypoxia. GSVA was performed using the “Cellular Response to Hypoxia” gene set from the Reactome database to quantify hypoxia in each ROI. A modest, yet statistically significant decrease of the hypoxic response gene signature was observed in the VEGFA-Low ROIs from specimens HGG03 and HGG06, whereas this signature was not differentially enriched in specimen HGG05 (Figure 5A-C). Correlation analysis was performed to investigate the relationship between the hypoxic response and various pro-angiogenic genes in VEGFA-Low and VEGFA-Med/High ROIs. This analysis showed that VEGFA gene expression positively correlated with the hypoxic response in VEGFA-Low ROIs (Figure 5D-F). Moreover, the data shows that the hypoxic response gene signature correlates with the expression of different pro-angiogenic genes in a specimen-specific manner. Together, these data illustrate regional differences in the hypoxic response and that this response is associated with angiokines other than VEGFA in regions with high VEGFA expression.
Figure 5: Associations between hypoxic response gene signature and VEGFA expression.
(A-C) Box plots showing the “Cellular Response to Hypoxia” gene set enrichment score in VEGFA-Low and VEGFA-Med/High ROIs in specimens (A) HGG03, (B) HGG05, and (C) HGG06. P-values were calculated using an unpaired t-test with Welch’s correction where p < 0.05 is considered statistically significant. (D-F) Spearman correlation matrix showing the relationships between genes encoding pro-angiogenic molecules and the “Cellular Response to Hypoxia” gene set enrichment score in VEGFA -Low and VEGFA -Med/High ROIs in specimens (D) HGG03, (E) HGG05, and (F) HGG06. Circle size indicates p value where circles with inlaid black squares indicating failure to reach statistical significance (p<0.05). Color bar indicates Spearman’s rho.
Regional expression of actionable immuno-oncology targets.
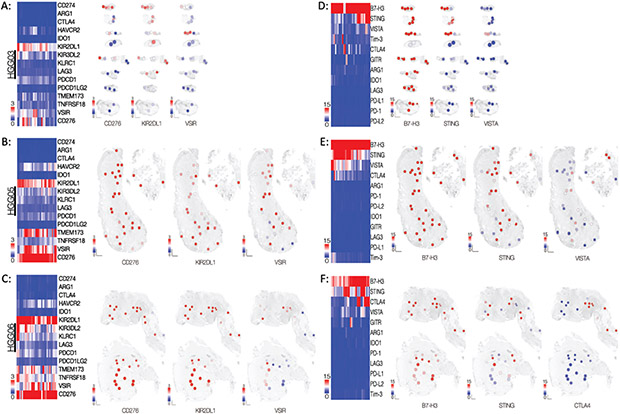
To extend our spatial analysis of the glioblastoma microenvironment, we examined the relative expression of genes encoding various actionable immuno-oncology targets. Of these targets, KIR2DL1, VSIR, and CD276 were consistently expressed in most of the ROIs in each of the three specimens (Figure 6A-C). Spatial heatmaps illustrated the regional variability of KIR2DL1, VSIR, and CD276 expression in specimen HGG03 (Figure 6A). Widespread and robust expression of CD276 and KIR2DL1 was observed throughout specimens HGG05 and HGG06 whereas regional differences in VSIR expression was noted (Figure 6B-C).
Figure 6: Immunotherapeutic targets are expressed in spatially distinct regions throughout the tissues.
(A-C, left) Heatmaps of the normalized count data for genes encoding immunotherapeutic targets in specimens (A) HGG03, (B) HGG05, and (C) HGG06. (A-C, right) Spatial heatmaps of specimens of each specimen depicting the normalized count data corresponding to the three genes with the highest median count value. (D-F, left) Heatmaps of the normalized antibody count data for immunotherapeutic targets in specimens (D) HGG03, (E) HGG05, and (F) HGG06. The data are presented as fold change over mean IgG control value. (D-E, right) Spatial heatmaps of each specimen showing the distribution of the normalized antibody count data for the three protein targets with the highest median value.
To further validate these data, we used serial sections from each specimen and applied the same ROIs to spatially profile the protein expression of B7-H3, STING, VISTA, TIM-3, CTLA4, ARG1, IDO1, PD-1, PD-L1, PD-L2, LAG3, and GITR using barcoded antibodies against each target. Antibodies against KIR2DL1 were not included in the antibody panel used for these experiments. In each specimen, high expression of B7-H3 was observed in almost all ROIs. (Figure 6D-F). Notably, regional variability in the expression of STING, VISTA, and CTLA4 was observed in each specimen whereas various other immuno-oncology targets such as PD-1, PD-L1, PD-L2, TIM3, IDO1, LAG3, ARG1, and GITR were minimally or not expressed (Figure 6D-F). The correlation between the antibody and gene expression counts for any given target in these specimens demonstrated varying degrees of statistical significance, suggesting an uncoupling of gene transcription from protein translation and/or post-translational regulation of protein stability. Nonetheless, the protein data largely agree with the gene expression data consistently showing moderate to high level expression of CD276 (encoding B7-H3) and TMEM173 (encoding STING) in each of the three specimens.
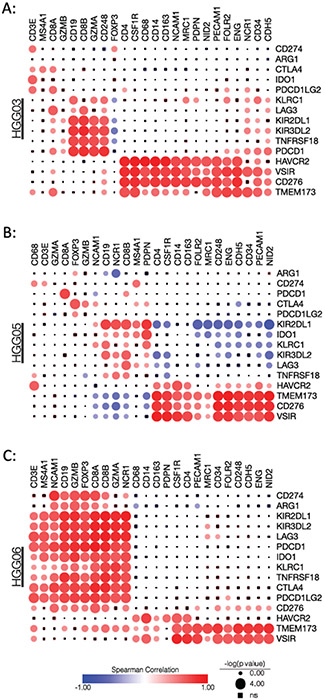
Next, we investigated the correlation between the expression of immunotherapeutic targets and the expression of different cell type marker genes. The marked differences observed in the correlation matrices between the three specimens shows the differential relationships between the expression of cell type-specific marker genes and actionable immunotherapeutic targets (Figure 7A-C). Despite the different correlation patterns observed in the specimens, CD276, VSIR, TMEM173, and, to a lesser extent, HAVCR2, consistently correlated with macrophage and vascular markers in each specimen (Figure 7A-C). Additionally, although the lymphocyte abundance in each specimen is low, the significant positive correlations between lymphocyte marker genes and genes associated with lymphocyte exhaustion suggest that the few lymphocytes present within these tumors may be exhausted. Taken together, these data, combined with the SpatialDecon data showing the regional differences in cellular composition, highlight the regional heterogeneity of the cellular and molecular immuno-oncology landscape both within and between individual glioblastoma specimens.
Figure 7: Correlations between immunotherapeutic targets and different cell type markers.
Spearman correlation matrix showing the relationships between genes encoding immunotherapeutic targets and marker genes for lymphocytes, monocytes/macrophages, endothelial cells, and glioma cells in specimens (A) HGG03, (B) HGG05, and (C) HGG06. Circle size indicates p value where circles with inlaid black squares indicating failure to reach statistical significance (p<0.05). Color bar indicates Spearman’s rho.
DISCUSSION
We have demonstrated the utility of performing spatial transcriptional profiling on archived glioblastoma specimens to reveal the molecular heterogeneity within individual patient tissues. Because of the minimal amount of archival tissue required to generate a robust dataset, spatial profiling is an ideal platform to enable in depth molecular characterization of patient specimens that are in limited supply, such as those associated with clinical trial correlative biology studies, rare tumors, and pediatric tumors. While our gene panel used in this study was fixed at 1,700 genes, recent advances in the technology has allowed for spatially-resolved profiling of >18,000 genes, covering almost the entire coding transcriptome.31-33 Moreover, the NanoString GeoMx platform adds another dimension to spatial analysis by allowing for highly multiplexed digital antibody-based protein profiling on serial sections for an integrated multianalyte approach. Lastly, by combining clinical neuropathological examination, single cell sequencing data, and spatial transcriptomic and proteomic profiling, we demonstrate the applicability of an integrated multiparametric approach to characterize the tumor microenvironment of archived FFPE tissues on a per-specimen basis.
Using deconvolution and marker gene methods to infer cell type is a common approach to reveal the cellular composition of the tumor microenvironment from RNA sequencing and gene expression datasets, yet each method has its limitation.34 The genes used to classify these cell types are not unique and can be expressed by other tissue-specific resident cell types in both pathologic and non-diseased tissues. For example, NCAM1, encoding CD56, is one of the primary genes used to describe natural killer cells yet this gene is highly expressed in non-diseased brain tissue and variably expressed in glial malignancies. 26,27 Additionally, microglia are CNS-resident macrophages yet these cells are not captured using popular methods such as XCell and CiberSort.35-37 For this reason, we used the SpatialDecon algorithm to derive resident glioblastoma cell type scores to describe the cellular composition of these specimens while retaining the contextual relevance.18 While the resultant data from the SpatialDecon algorithm significantly correlated with the data obtained upon expert neuropathological examination, this comparison exposed a shortcoming in our approach. Computationally, we were unable to discern between vessels with and without microvascular proliferation, whereas these differences were evident by neuropathological examination. We attribute this to the fact that the 1,700 gene panel used for this study was not designed to distinguish between these vascular phenotypes, especially given the recently discovered transcriptional heterogeneity between different endothelial cell populations.38-42 To differentially interrogate the region-specific transcriptional programs associated with these vascular phenotypes additional studies using high resolution, spatially-resolved whole transcriptome profiling will be needed.
B7-H3 is a cell surface protein that is highly expressed in glioblastoma along with several other CNS and solid tumors.43-45 Currently, targeting B7-H3 using chimeric antigen receptor T-cells and antibody drug conjugates is an active area of research.45-50 Our data is consistent with the published data showing that B7-H3 is the most highly expressed immunotherapy target in our panel across all specimens. Although the ROI density used in this study is low, our data shows that B7-H3 expression is widely distributed throughout the tissues at both the protein and RNA levels, further highlighting the utility of B7-H3 as a ubiquitous therapeutic target.
We observed significant positive correlations between macrophage and/or endothelial cell markers and TMEM173, VSIR, and CD276, suggesting a context for combinatorial immunotherapeutic targeting of both macrophages and the tumor vasculature. As stated above, studies involving B7-H3 (encoded by CD276) are already underway. STING (encoded by TMEM173) is part of the cytosolic DNA sensing pathway where its activation induces a potent inflammatory response primarily driven by type I interferons. STING agonists are currently in development and preclinical studies indicate that STING is an attractive immunotherapeutic target.51-54 VISTA (encoded by VSIR) promotes quiescence of myeloid cells and lymphocytes.55,56 The mechanisms by which VISTA achieves immunosuppression are quite complex yet studies have demonstrated that blocking VISTA can enhance the antitumor immune response.56-58 Conceptually, combinatorial targeting of these three immunoregulatory molecules would allow for localized inflammation via STING activation, relieve the inhibitory effects of VISTA, and target B7-H3 expressing glioma cells, TAMs, and tumor vasculature. Preclinical studies will need to be performed to test this hypothesis and evaluate the therapeutic efficacy of this combinatorial immunotherapeutic approach.
VEGFA is a known driver of tumor angiogenesis and an important therapeutic target in glioblastoma.59 Our data has revealed that there are substantial regional differences in VEGFA expression in specimens HGG05 and HGG06 and that regions with high expression of VEGFA inversely correlate with vascularity. VEGFA is subject to alternative splicing, resulting in the generation of various protein isoforms with both pro- and anti-angiogenic functions.60 The gene panel used in this study was composed of multiple exon-specific probes per gene to quantify RNA levels, potentially allowing for the examination of exon skipping events for certain genes. Unfortunately, we were unable to assess exon usage to infer alternative splicing as probes targeting the most commonly alternatively spliced exons of VEGFA were not present in this panel. Our data also show strong positive correlations between various angiokines and several vascular markers, indicating that alternative pathways promote and/or maintain the tumor vasculature in the absence of angiogenic VEGFA signaling in specimens HGG05 and HGG06.61 Importantly, neuropathological examination of the tissues determined that specimens HGG05 and HGG06 exhibited microvascular proliferation whereas specimen HGG03 did not and this may contribute to the differences described in this study.
Vascular mimicry is a phenomenon whereby tumor cells adopt transcriptional and phenotypic characteristics reminiscent of endothelial cells to establish a network of dysfunctional vascular-like structures to supply the tumor.62 Yao et al. demonstrated that VEGFA-VEGFR2 signaling in glioma stem-like cells promotes vascular mimicry.63 Hypoxia is also an established driver of vascular mimicry.62 Our finding of increased enrichment of the hypoxic response gene signature and concomitant decreased expression of endothelial cell markers/vascular score in ROIs with excessive VEGFA expression may indicate region-specific vascular mimicry. As stated above, the content of this gene panel limits the depth of interrogation of complex biological processes. Thus, a plausible alternative interpretation of these results is that the hypoxic response in these regions is driven by a VEGFA-independent mechanism that cannot be readily elucidated by our approach.
In addition to its notable role in vascular biology, VEGFA exerts immunomodulatory effects within the tumor microenvironment, including the promotion of T-cell exhaustion, recruitment of suppressive myeloid cells, and the polarization of TAMs towards an M2 phenotype.64-66 Targeting the immunosuppressive functions of VEGFA is currently an active area of research and clinical trials investigating the therapeutic efficacy of combining VEGFA antagonists with immune checkpoint inhibitors are currently ongoing.65,67-69 Here we show that, in addition to differential vascular abundance, increased VEGFA expression is associated with the expression of immunomodulatory genes in distinct regions of the specimens. Functional studies are needed to determine whether VEGFA orchestrates the immunosuppressive tissue niche or if increased VEGFA is a consequence of a preexisting localized suppressive environment. Utilizing high resolution spatial profiling technologies combined with multiplexed immunofluorescent staining will be pivotal in such studies.
In conclusion, we have successfully achieved our goal of demonstrating the applicability of spatially-resolved transcriptional profiling to reveal novel biologic insights using archived FFPE material. Currently, various spatial biology technologies exist and each platform has its strengths and limitations.8 Nonetheless, integrated multimodal investigations can offset the platform-specific weaknesses to enable robust downstream analyses that inform the biological interpretation of the data. While we have highlighted the utility of this pre-production assay for discovery-based/hypothesis generating studies, the commercial NanoString GeoMx whole transcriptome assay is extremely well suited for hypothesis driven research where ROIs are selected based on specific markers or histological features of interest and subsequently profiled using a highly multiplexed panel of approximately 18,000 gene targets.
Supplementary Material
Acknowledgements
We would like to thank Frances Chow for critically reading this manuscript and for the insightful comments. This project was conceptualized while TAM was affiliated with the University of Southern California and was completed in its entirety at the National Cancer Institute.
Funding
TAM and JG are supported by the Intramural Research Program of the NIH, NCI, Center for Cancer Research. The views and opinions contained within this article do not necessarily reflect those of the NIH or the US Department of Health and Human Services. The mention of trade names and/or commercialized products does not indicate endorsement by the US government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Ethics Approval
Specimens were obtained under the University of Southern California Institutional Review Board protocol HS-11-00385 and in compliance with the Declaration of Helsinki.
Conflicts of Interest
YM, PD, SW, and JMB are employees of NanoString Technologies. All other authors declare no competing financial interests as it pertains to the data presented in this manuscript.
Data Availability
The normalized data files are available upon request.
REFERENCES
- 1.Labani-Motlagh A, Ashja-Mahdavi M, Loskog A. The Tumor Microenvironment: A Milieu Hindering and Obstructing Antitumor Immune Responses. Front Immunol. 2020;11:940. doi: 10.3389/fimmu.2020.00940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. Nov 2013;19(11):1423–37. doi: 10.1038/nm.3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merritt CR, Ong GT, Church SE, et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat Biotechnol. May 2020;38(5):586–599. doi: 10.1038/s41587-020-0472-9 [DOI] [PubMed] [Google Scholar]
- 4.Rodriques SG, Stickels RR, Goeva A, et al. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science. Mar 29 2019;363(6434):1463–1467. doi: 10.1126/science.aaw1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salmen F, Stahl PL, Mollbrink A, et al. Barcoded solid-phase RNA capture for Spatial Transcriptomics profiling in mammalian tissue sections. Nat Protoc. Nov 2018;13(11):2501–2534. doi: 10.1038/s41596-018-0045-2 [DOI] [PubMed] [Google Scholar]
- 6.Stahl PL, Salmen F, Vickovic S, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. Jul 1 2016;353(6294):78–82. doi: 10.1126/science.aaf2403 [DOI] [PubMed] [Google Scholar]
- 7.Villacampa EG, Larsson L, Kvastad L, Andersson A, Carlson J, Lundeberg J. Genome-wide Spatial Expression Profiling in FFPE Tissues. bioRxiv. 2020:2020.07.24.219758. doi: 10.1101/2020.07.24.219758 [DOI] [Google Scholar]
- 8.Bassiouni R, Gibbs LD, Craig DW, Carpten JD, McEachron TA. Applicability of spatial transcriptional profiling to cancer research. Mol Cell. April 15 2021;81(8):1631–1639. doi: 10.1016/j.molcel.2021.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rutledge WC, Kong J, Gao J, et al. Tumor-infiltrating lymphocytes in glioblastoma are associated with specific genomic alterations and related to transcriptional class. Clin Cancer Res. Sep 15 2013;19(18):4951–60. doi: 10.1158/1078-0432.CCR-13-0551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salmaggi A, Boiardi A, Gelati M, et al. Glioblastoma-derived tumorospheres identify a population of tumor stem-like cells with angiogenic potential and enhanced multidrug resistance phenotype. Glia. Dec 2006;54(8):850–60. doi: 10.1002/glia.20414 [DOI] [PubMed] [Google Scholar]
- 11.Kumar A, Boyle EA, Tokita M, et al. Deep sequencing of multiple regions of glial tumors reveals spatial heterogeneity for mutations in clinically relevant genes. Genome Biol. Dec 03 2014;15(12):530. doi: 10.1186/s13059-014-0530-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manini I, Caponnetto F, Dalla E, et al. Heterogeneity Matters: Different Regions of Glioblastoma Are Characterized by Distinctive Tumor-Supporting Pathways. Cancers (Basel). October 13 2020;12(10)doi: 10.3390/cancers12102960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer M, Reimand J, Lan X, et al. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc Natl Acad Sci U S A. Jan 20 2015;112(3):851–6. doi: 10.1073/pnas.1320611111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. Jan 19 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. Mar 2006;9(3):157–73. doi: 10.1016/j.ccr.2006.02.019 [DOI] [PubMed] [Google Scholar]
- 16.Puchalski RB, Shah N, Miller J, et al. An anatomic transcriptional atlas of human glioblastoma. Science. May 11 2018;360(6389):660–663. doi: 10.1126/science.aaf2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. June 2016;131(6):803–20. doi: 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 18.Danaher P, Kim Y, Nelson B, et al. Advances in mixed cell deconvolution enable quantification of cell types in spatial transcriptomic data. Nat Commun. Jan 19 2022;13(1):385. doi: 10.1038/s41467-022-28020-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darmanis S, Sloan SA, Croote D, et al. Single-Cell RNA-Seq Analysis of Infiltrating Neoplastic Cells at the Migrating Front of Human Glioblastoma. Cell Rep. October 31 2017;21(5):1399–1410. doi: 10.1016/j.celrep.2017.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lun ATL, Riesenfeld S, Andrews T, et al. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. March 22 2019;20(1):63. doi: 10.1186/s13059-019-1662-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lun AT, Bach K, Marioni JC. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. Apr 27 2016;17:75. doi: 10.1186/s13059-016-0947-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stuart T, Butler A, Hoffman P, et al. Comprehensive Integration of Single-Cell Data. Cell. June 13 2019;177(7):1888–1902.e21. doi: 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franzén O, Gan LM, Björkegren JLM. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database (Oxford). January 01 2019;2019 doi: 10.1093/database/baz046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. Oct 23 2008;455(7216):1061–8. doi: 10.1038/nature07385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. Oct 10 2013;155(2):462–77. doi: 10.1016/j.cell.2013.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gingras MC, Roussel E, Bruner JM, Branch CD, Moser RP. Comparison of cell adhesion molecule expression between glioblastoma multiforme and autologous normal brain tissue. J Neuroimmunol. Mar 1995;57(1-2):143–53. doi: 10.1016/0165-5728(94)00178-q [DOI] [PubMed] [Google Scholar]
- 27.Todaro L, Christiansen S, Varela M, et al. Alteration of serum and tumoral neural cell adhesion molecule (NCAM) isoforms in patients with brain tumors. J Neurooncol. Jun 2007;83(2):135–44. doi: 10.1007/s11060-006-9312-0 [DOI] [PubMed] [Google Scholar]
- 28.Shaw TN, Houston SA, Wemyss K, et al. Tissue-resident macrophages in the intestine are long lived and defined by Tim-4 and CD4 expression. J Exp Med. June 04 2018;215(6):1507–1518. doi: 10.1084/jem.20180019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landry AP, Balas M, Alli S, Spears J, Zador Z. Distinct regional ontogeny and activation of tumor associated macrophages in human glioblastoma. Sci Rep. November 11 2020;10(1):19542. doi: 10.1038/s41598-020-76657-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krock BL, Skuli N, Simon MC. Hypoxia-induced angiogenesis: good and evil. Genes Cancer. Dec 2011;2(12):1117–33. doi: 10.1177/1947601911423654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan M, Yoo SJ, Clijsters M, et al. Visualizing in deceased COVID-19 patients how SARS-CoV-2 attacks the respiratory and olfactory mucosae but spares the olfactory bulb. Cell. November 24 2021;184(24):5932–5949.e15. doi: 10.1016/j.cell.2021.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salem F, Perin L, Sedrakyan S, et al. The spatially resolved transcriptional profile of acute T cell-mediated rejection in a kidney allograft. Kidney Int. Jan 2022;101(1):131–136. doi: 10.1016/j.kint.2021.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delorey TM, Ziegler CGK, Heimberg G, et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature. July 2021;595(7865):107–113. doi: 10.1038/s41586-021-03570-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sturm G, Finotello F, Petitprez F, et al. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. July 15 2019;35(14):i436–i445. doi: 10.1093/bioinformatics/btz363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. Nov 2017;18(1):220. doi: 10.1186/s13059-017-1349-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. July 2019;37(7):773–782. doi: 10.1038/s41587-019-0114-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. May 2015;12(5):453–7. doi: 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalucka J, de Rooij LPMH, Goveia J, et al. Single-Cell Transcriptome Atlas of Murine Endothelial Cells. Cell. February 20 2020;180(4):764–779.e20. doi: 10.1016/j.cell.2020.01.015 [DOI] [PubMed] [Google Scholar]
- 39.Xie Y, He L, Lugano R, et al. Key molecular alterations in endothelial cells in human glioblastoma uncovered through single-cell RNA sequencing. JCI Insight. August 09 2021;6(15)doi: 10.1172/jci.insight.150861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Q, Eichten A, Parveen A, et al. Single-Cell Transcriptome Analyses Reveal Endothelial Cell Heterogeneity in Tumors and Changes following Antiangiogenic Treatment. Cancer Res. May 01 2018;78(9):2370–2382. doi: 10.1158/0008-5472.CAN-17-2728 [DOI] [PubMed] [Google Scholar]
- 41.Feng W, Chen L, Nguyen PK, Wu SM, Li G. Single Cell Analysis of Endothelial Cells Identified Organ-Specific Molecular Signatures and Heart-Specific Cell Populations and Molecular Features. Front Cardiovasc Med. 2019;6:165. doi: 10.3389/fcvm.2019.00165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jambusaria A, Hong Z, Zhang L, et al. Endothelial heterogeneity across distinct vascular beds during homeostasis and inflammation. Elife. January 16 2020;9doi: 10.7554/eLife.51413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kraan J, van den Broek P, Verhoef C, et al. Endothelial CD276 (B7-H3) expression is increased in human malignancies and distinguishes between normal and tumour-derived circulating endothelial cells. British Journal of Cancer. 2014/July// 2014;111(1):149–156. doi: 10.1038/bjc.2014.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Picarda E, Ohaegbulam KC, Zang X. Molecular Pathways: Targeting B7-H3 (CD276) for Human Cancer Immunotherapy. Clinical Cancer Research. 2016/July/15/ 2016;22(14):3425–3431. doi: 10.1158/1078-0432.CCR-15-2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seaman S, Zhu Z, Saha S, et al. Eradication of Tumors through Simultaneous Ablation of CD276/B7-H3-Positive Tumor Cells and Tumor Vasculature. Cancer Cell. 2017/April// 2017;31(4):501–515.e8. doi: 10.1016/j.ccell.2017.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du H, Hirabayashi K, Ahn S, et al. Antitumor Responses in the Absence of Toxicity in Solid Tumors by Targeting B7-H3 via Chimeric Antigen Receptor T Cells. Cancer Cell. February 11 2019;35(2):221–237.e8. doi: 10.1016/j.ccell.2019.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haydar D, Houke H, Chiang J, et al. Cell-surface antigen profiling of pediatric brain tumors: B7-H3 is consistently expressed and can be targeted via local or systemic CAR T-cell delivery. Neuro Oncol. June 01 2021;23(6):999–1011. doi: 10.1093/neuonc/noaa278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kendsersky NM, Lindsay J, Kolb EA, et al. The B7-H3-Targeting Antibody-Drug Conjugate m276-SL-PBD Is Potently Effective Against Pediatric Cancer Preclinical Solid Tumor Models. Clin Cancer Res. May 15 2021;27(10):2938–2946. doi: 10.1158/1078-0432.CCR-20-4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Majzner RG, Theruvath JL, Nellan A, et al. CAR T Cells Targeting B7-H3, a Pan-Cancer Antigen, Demonstrate Potent Preclinical Activity Against Pediatric Solid Tumors and Brain Tumors. Clin Cancer Res. April 15 2019;25(8):2560–2574. doi: 10.1158/1078-0432.CCR-18-0432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nehama D, Di Ianni N, Musio S, et al. B7-H3-redirected chimeric antigen receptor T cells target glioblastoma and neurospheres. EBioMedicine. Sep 2019;47:33–43. doi: 10.1016/j.ebiom.2019.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chin EN, Yu C, Vartabedian VF, et al. Antitumor activity of a systemic STING-activating non-nucleotide cGAMP mimetic. Science. Aug 21 2020;369(6506):993–999. doi: 10.1126/science.abb4255 [DOI] [PubMed] [Google Scholar]
- 52.Demaria O, De Gassart A, Coso S, et al. STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proc Natl Acad Sci U S A. Dec 15 2015;112(50):15408–13. doi: 10.1073/pnas.1512832112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perera SA, Kopinja JE, Ma Y, et al. STimulator of INterferon Genes Agonism Accelerates Anti-tumor Activity in Poorly Immunogenic Tumors. Mol Cancer Ther. Nov 23 2021;doi: 10.1158/1535-7163.MCT-21-0136 [DOI] [PubMed] [Google Scholar]
- 54.Wang-Bishop L, Wehbe M, Shae D, et al. Potent STING activation stimulates immunogenic cell death to enhance antitumor immunity in neuroblastoma. J Immunother Cancer. March 2020;8(1)doi: 10.1136/jitc-2019-000282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.ElTanbouly MA, Zhao Y, Nowak E, et al. VISTA is a checkpoint regulator for naïve T cell quiescence and peripheral tolerance. Science. January 17 2020;367(6475)doi: 10.1126/science.aay0524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan L, Tatineni J, Mahoney KM, Freeman GJ. VISTA: A Mediator of Quiescence and a Promising Target in Cancer Immunotherapy. Trends Immunol. March 2021;42(3):209–227. doi: 10.1016/j.it.2020.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li TT, Jiang JW, Qie CX, et al. Identification of active small-molecule modulators targeting the novel immune checkpoint VISTA. BMC Immunol. August 11 2021;22(1):55. doi: 10.1186/s12865-021-00446-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan J, Chen Y, Zhang Q, et al. Inhibition of lung tumorigenesis by a small molecule CA170 targeting the immune checkpoint protein VISTA. Commun Biol. July 23 2021;4(1):906. doi: 10.1038/s42003-021-02381-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng SY, Huang HJ, Nagane M, et al. Suppression of glioblastoma angiogenicity and tumorigenicity by inhibition of endogenous expression of vascular endothelial growth factor. Proc Natl Acad Sci U S A. Aug 06 1996;93(16):8502–7. doi: 10.1073/pnas.93.16.8502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mamer SB, Wittenkeller A, Imoukhuede PI. VEGF-A splice variants bind VEGFRs with differential affinities. Sci Rep. September 02 2020;10(1):14413. doi: 10.1038/s41598-020-71484-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nomura M, Yamagishi S, Harada S, Yamashima T, Yamashita J, Yamamoto H. Placenta growth factor (PlGF) mRNA expression in brain tumors. J Neurooncol. Nov 1998;40(2):123–30. doi: 10.1023/a:1006198422718 [DOI] [PubMed] [Google Scholar]
- 62.Wei X, Chen Y, Jiang X, et al. Mechanisms of vasculogenic mimicry in hypoxic tumor microenvironments. Mol Cancer. January 04 2021;20(1):7. doi: 10.1186/s12943-020-01288-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yao X, Ping Y, Liu Y, et al. Vascular endothelial growth factor receptor 2 (VEGFR-2) plays a key role in vasculogenic mimicry formation, neovascularization and tumor initiation by Glioma stem-like cells. PLoS One. 2013;8(3):e57188. doi: 10.1371/journal.pone.0057188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Apte RS, Chen DS, Ferrara N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell. March 07 2019;176(6):1248–1264. doi: 10.1016/j.cell.2019.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khan KA, Kerbel RS. Improving immunotherapy outcomes with anti-angiogenic treatments and vice versa. Nat Rev Clin Oncol. May 2018;15(5):310–324. doi: 10.1038/nrclinonc.2018.9 [DOI] [PubMed] [Google Scholar]
- 66.Yang J, Yan J, Liu B. Targeting VEGF/VEGFR to Modulate Antitumor Immunity. Front Immunol. 2018;9:978. doi: 10.3389/fimmu.2018.00978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fukumura D, Kloepper J, Amoozgar Z, Duda DG, Jain RK. Enhancing cancer immunotherapy using antiangiogenics: opportunities and challenges. Nat Rev Clin Oncol. May 2018;15(5):325–340. doi: 10.1038/nrclinonc.2018.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wallin JJ, Bendell JC, Funke R, et al. Atezolizumab in combination with bevacizumab enhances antigen-specific T-cell migration in metastatic renal cell carcinoma. Nat Commun. August 2016;7:12624. doi: 10.1038/ncomms12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu X, Giobbie-Hurder A, Liao X, et al. VEGF Neutralization Plus CTLA-4 Blockade Alters Soluble and Cellular Factors Associated with Enhancing Lymphocyte Infiltration and Humoral Recognition in Melanoma. Cancer Immunol Res. October 2016;4(10):858–868. doi: 10.1158/2326-6066.CIR-16-0084 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The normalized data files are available upon request.