Abstract
Ribosomes are abundant, large RNA-protein complexes that are the sites of all protein synthesis in cells. Defects in ribosomal proteins (RPs), including proteoforms arising from genetic variations, alternative splicing of RNA transcripts, post-translational modifications and alterations of protein expression level, have been linked to a diverse range of diseases, including cancer and aging. Comprehensive characterization of ribosomal proteoforms is challenging but important for the discovery of potential disease biomarkers or protein targets. In the present work, using E.coli 70S RPs as an example, we first developed a top-down proteomics approach on a Waters Synapt G2 Si mass spectrometry (MS) system, and then applied it to the HeLa 80S ribosome. The results were complemented by a bottom-up approach. In total, 50 out of 55 RPs were identified using the top-down approach. Among these, more than 30 RPs were found to have their N-terminal methionine removed. Additional modifications such as methylation, acetylation, and hydroxylation were also observed, and the modification sites were identified by bottom-up MS. In a HeLa 80S ribosomal sample, we identified 98 ribosomal proteoforms, among which multiple truncated 80S ribosomal proteoforms were observed, the type of information which is often overlooked by bottom-up experiments. Although their relevance to diseases is not yet known, the integration of top-down and bottom-up proteomics approaches paves the way for the discovery of proteoform-specific disease biomarkers or targets.
Keywords: Ribosomal proteins, Top-down MS, Bottom-up MS, Proteoforms
Graphical abstract
Highlights
-
•
An integrated top-down and bottom-up proteomics approach for characterizing ribosomal protein heterogeneity.
-
•
Ribosomal proteoforms arisen from genetic variations, alternative splicing, and PTMs were observed for both E.coli and Hela ribosome.
-
•
Truncated ribosomal proteoforms were frequently observed in Hela ribosome.
1. Introduction
The ribosome is a highly complex and dynamic macromolecular machine, a central regulatory hub in cells for protein quality control, orchestration of mRNA decay and stress signaling [[1], [2], [3]]. As the site of protein synthesis in the cell, it is made up of ribosomal proteins (RPs) and ribosomal RNA (rRNA). The translation of messenger RNA (mRNA) into a protein and the folding of the resulting protein into an active form are prerequisites for virtually every cellular process [4]. mRNA codons are decoded by the respective aminoacyl-tRNAs (aa-tRNAs) in the small subunit. The large subunit is responsible for the peptide bond formation [5]. Each ribosome contains 50−80 RPs, and about 2/3 of these are conserved among all kingdoms of life, while others are specific for bacteria, archaea, or eucaryotes [6]. Most RPs maintain their proper structures as specific parts of the ribosome through association with rRNA and neighboring RPs, and a few of them directly participate in the work of particular ribosomal functional sites [5]. Defects in ribosome biogenesis, translation, and the functions of individual proteins, including mutations in proteins, alternative splicing, post-translational modifications (PTMs), and alteration of protein expression level, have been linked to a diverse range of diseases, such as cancer and aging [7,8]. For example, a variety of mutations in RPs S7, S17, S19, S24, L5, L11, and L35A due to nonsense and missense mutations, small insertions and deletions, splice site defects and large deletions, and rearrangements have been found in Diamond-Blackfan anemia [9,10]. L5 and L11 mutations or deletions have also been identified in human malignancies [11]. Alternative splicing of eukaryotic transcripts is a mechanism that enables cells to generate vast protein diversity from a limited number of genes. Previous surveys of alternative splicing events engaged by the ribosome have shown that at least 75% of human exon-skipping events detected in transcripts were also detected in ribosome profiling data; thus a major fraction of splice variants is translated [12]. The abnormal splicing events contribute to tumor progression as oncogenic drivers and/or bystander factors [13]. Additionally, it has been suggested that PTMs such as phosphorylation and ubiquitylation have functional consequences for translational control and are linked to human disease [14]. Aberrant expression of RP genes has also been reported and is largely due to the changes in pre-mRNA splicing efficiency and mRNA stability [15].
Genetic variants, alternative splicing, and PTMs are the predominant factors causing proteoforms, which contribute greatly to the largely unmapped complexity of the human proteome. The term ‘proteoform’ describes all of the different molecular forms in which the protein product of a single gene can be found, including changes due to genetic variations, alternatively spliced RNA transcripts, and PTMs [16]. It is estimated that the ∼20,000 coding genes generate more than a million different proteoforms [17]. The function of proteins can be strongly modulated by mutations, alternative splicing, and PTMs, such as phosphorylation, acetylation, methylation, and other >400 known PTMs in biology [18]. One classical case is histone: histone modifications as well as histone variants function as “docking sites” and a network for the recruitment of protein factors to facilitate the chromatin remodeling and thus the regulation of gene expression [19]. As proteoforms are tightly linked to the functioning of cells and tissues that underlie complex phenotypes, their identification and quantification are critically important as a source of insights into the fundamental workings of biological systems, thereby promoting the identification of diagnostic markers and therapeutic targets. Thus, proteoform characterization, namely identifying precise molecular forms of proteins, has been considered as the next proteome currency [18].
Mass spectrometry (MS)-based proteomics has become an indispensable approach to the quantitative profiling of proteins as well as their interactions and modifications [20]. Bottom-up is the most commonly used approach due to its technical accessibility, robustness, sensitivity, and high-throughput. In bottom-up proteomics, proteins are extracted and digested by sequence-specific enzymes such as trypsin, and then these peptides are separated by liquid chromatography (LC) and analyzed by tandem MS [20]. With the rapid improvement of MS measurement speed, this technology has become widely used in biomedical fields [21], and even approached coverage of the whole proteome [22]. However, a key challenge associated with bottom-up proteomics is that it is not generally possible to identify proteoforms, as different proteoforms often share most of their peptides with one another [18,23]. Consequently, proteoforms are therefore overlooked in bottom-up proteomics [16,24]. In addition, artifacts can be introduced during sample preparation by enzymatic or chemical modification or degradation, including oxidation and chain cleavage. As a result, proteoforms can be proteolytically truncated, thereby forming new proteoforms with a loss of correlative power and relationship to their function [17]. As an alternative, the ‘top-down’ proteomic approach, in which the entire proteoform is analyzed by LC-MS/MS without prior digestion to peptides [25] can be used. In 2011, Tran et al. [26] reported the proteome-wide discovery of proteoforms with the top-down approach. Using a new four-dimensional separation system, they identified 1,043 gene products from human cells that are dispersed into more than 3,000 protein species created by PTM, RNA splicing, and proteolysis. Over the last decade, top-down proteomics has developed rapidly with the continuous advances in the technology, making it possible to identify intact proteins from complex clinical samples [27]. Ideally, the complete amino acid sequence and localized PTMs are obtained. But for especially large (>30 kDa) proteins or those harboring many PTMs, there are often ambiguities in the complete description of related proteoforms [28]. Proteoforms of low abundance can be overlooked [29] and some factors such as protein solubility, proteome complexity, and proteoform function should be considered before beginning the experiment [30].
Here, by taking advantage of top-down MS for proteoform characterization and bottom-up MS for PTM localization and high sensitivity, we first developed and integrated the two approaches for in-depth characterization of E. coli 70S RPs on a Waters Synapt G2 Si MS system (Milford, MA, USA), which enabled the identification of 50 out of 55 70S RPs using the top-down approach. Among these, more than 30 RPs were found to have the N-terminal methionine removed. Additional modifications such as methylation, acetylation, or hydroxylation were also observed, and the modification sites were located by bottom-up MS. We then applied this approach to the characterization of HeLa 80S RPs. We identified a total of 98 ribosomal proteoforms out of 77 ribosomal genes. Among these, 79 ribosomal proteoforms of 57 genes were identified using the top-down approach, while the bottom-up approach allowed the identification of 74 or 77 RPs. Intriguingly, we observed multiple truncated ribosomal proteoforms in HeLa sample, information that is often overlooked in bottom-up experiments. Although their relevance to disease is not known, the integration of the top-down and bottom-up proteomics approaches paves the way for the discovery of proteoform-specific disease biomarkers or targets.
2. Experimental
2.1. Reagents and chemicals
Glacial acetic acid (AcOH) and hydrochloric acid (HCl) were purchased from the Guangzhou Chemical Reagent Factory (Guangzhou, China). Magnesium chloride (MgCl2), lithium chloride (LiCl), urea, dithiothreitol (DTT), iodoacetamide (IAA), and trypsin from porcine pancreas (Type IX-S, lyophilized powder, 13,000−20,000 BAEE units/mg protein) were purchased from Sigma-Aldrich (St. Louis, MO, USA). LC-MS grade acetonitrile (ACN) and formic acid (HCOOH) were purchased from Thermo Fisher Scientific Inc. (Waltham, MA, USA). Purified water was obtained using a Milli-Q system (Millipore, Bedford, MA, USA). E. coli 70S ribosomes were purchased from New England Biolabs (Ipswich, MA, USA).
2.2. Cell culture
HeLa cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and incubated overnight at 37 °C, under 5% CO2. HeLa cells were cultured in a 10-cm cell culture dish and resuspended in 1 mL of serum-containing medium using a 0.25% trypsin digestion solution (Servicebio, Wuhan, China), Cell numbers were counted by Cellometer Mini automatic cell counting instrument (Nexcelom Bioscience, Lawrence, MA, USA).
2.3. Purification of ribosomal particles
HeLa 80S ribosomes were isolated from HeLa cells by a ribosome extraction kit (Bestbio, Shanghai, China). In brief, ∼3.75 × 108 HeLa cells were centrifuged (500−1,000 g, 5 min, 4 °C) to remove medium. The collected cells were washed twice with pre-cooled phosphate buffer saline. Then 3 mL of pre-cooled reagent A was added. The cells were left on ice for 10 min and then loaded onto a Dounce homogenizer for disruption. This mixture was centrifuged for 5 min at 1,000 g. The supernatant was centrifuged for 10 min at 20,000 g and then for 4 h at 120,000 g. 2.5 mL of reagent B was added in the pellet and followed by centrifugation for 4 h at 120,000 g. The ribosome pellets were dissolved in resuspension buffer (reagent C).
2.4. Preparation of RPs
RPs and ribosome associated proteins were separated from the rRNA by glacial AcOH precipitation according to Hardy et al. [31]. To one volume of vigorously shaken ribosomal particles, 1/10 volume of 1 M MgCl2 was added. Immediately afterwards, 2 volume of glacial AcOH was added. This mixture was shaken at 4 °C for 60 min, followed by centrifugation for 10 min at 20,000 g. The supernatant was moved to a new vial and the pellet was washed with 67% glacial AcOH and re-centrifuged. The combined two supernatants contained the RP.
RPs and ribosome associated proteins were separated from the rRNA by LiCl-urea precipitation according to El-Baba et al. [32]. One volume of LiCl-urea buffer (8 M urea, 6 M LiCl, 25 mM Tris-HCl (pH 7.5), 100 mM KCl, 20 mM MgCl2, and 2 mM DTT) was added to a solution of dissociated ribosomes. This mixture was left at 2−4 °C overnight, followed by centrifugation for 10 min at 16,000 g. The supernatant containing the 30S proteins was removed from the RNA pellet. Protein concentrations were estimated by nanodrop 2000 spectrophotometer (Thermo Fisher Scientific Inc.). Protein A280 application was selected here and ammonium acetate was used for blank correction. Two microliter of the sample was pipetted onto the bottom pedestal and measured three times.
2.5. Sample preparation for bottom-up LC-MS/MS analysis
Protein solutions (∼5−10 μg) were dissolved in 8 M of urea (in 100 of mM ammonium bicarbonate, pH 7.5). Disulfide bonds were reduced using 5 mM of DTT at 37 °C for 2 h. Reduced cysteines were then alkylated using 10 of mM IAA at room temperature for 40 min in the dark. The solution was diluted to 1 M of urea with 100 mM of ammonium bicarbonate (pH 7.5) before the addition of trypsin (1:50, m/m) for overnight digestion at 37 °C. Peptides were desalted using a C18 solid phase extraction disk (3M Company, St. Paul, MN, USA) and dried on a vacuum concentrator. Desalted peptides were resolubilized in 0.1% (V/V) HCOOH in water.
2.6. Preparation of RPs for top-down LC-MS/MS
Protein solutions were concentrated and desalted using Amicon® 0.5 mL Ultra Centrifugal Filter with a 3 kDa MWCO (Millipore, Billerica, MA, USA). Samples were diluted with 200 mM ammonium acetate, and centrifuged three times at 14,000 g for 20 min.
2.7. Bottom-up LC-MS/MS analysis
Desalted peptides were loaded on a reverse-phase C18 column (75 μm × 20 cm, 1.9 μm, 120 Å) using an EASY-nLC 1200 system (Thermo Fisher Scientific Inc.). The LC-MS/MS run time was set to 60 min with flow rate of 300 nL/min. Mobile phases A (water:0.1% HCOOH, V/V) and B (ACN:0.1% HCOOH, V/V) were used for gradient elution: 5%–25% B for 28 min, 25%–40% B for 15 min, and 40%–100% B for 5 min. Samples were analyzed on Q ExactiveTM Plus Orbitrap mass spectrometer (Thermo Fisher Scientific Inc.). The mass spectrometer was operated in the positive ion mode and the spectra were acquired in the data-dependent acquisition mode. Full MS scans were acquired with 70,000 resolution and a scan mass range of m/z 355−2000. An Automatic Gain Control (AGC) target was set to 3e6 with a maximum injection time of 20 ms. A data-dependent MS/MS (dd-MS/MS) scan was acquired with 17,500 resolution. The AGC target was set to 5e4 with the maximum injection time defined as 50 ms. The data-dependent method was set to isolation and fragmentation of the 20 most intense peaks defined in the full MS scan. Parameters for isolation or fragmentation of selected ion peaks were set as follows: isolation width, 1.8 Th and higher energy collisional dissociation (HCD) normalized collision energy, 27%.
2.8. Top-down LC-MS/MS analysis
Chromatographic separation of intact protein samples was conducted on a Waters Acquity ultra-performance liquid chromatography I-Class system equipped with ethylene bridged hybrid C4 column (1 mm × 100 mm, 1.7 μm). About 0.5–2 μg of material was loaded on the column heated to 80 °C. LC-MS/MS runtime was set to 70 min with a flow rate of 0.1 mL/min. Gradient elution was performed using mobile phases A (water:0.1% HCOOH, V/V) and B (ACN:0.1% formic acid, V/V): 5%–42% B for 60 min, and 42%–95% B for an additional 4 min.
All top-down MS experiments were performed on a Waters Synapt G2 Si high definition mass spectrometer instrument. The instrument was run in positive polarity and in resolution mode. The capillary voltage was set to 3 kV, and the sampling cone voltage was set to 40 V. Source temperature and desolvation temperature were set to 150 and 550 °C, respectively. Mass spectra were acquired at the m/z range of 200−2000 using fast data-dependent acquisition (DDA). The data-dependent method was set to isolation and fragmentation of the two most intense peaks. MS was calibrated using sodium formate, and Leu-enkephalin (Waters, Milford, MA, USA) was used for internal lock mass calibration.
2.9. Data analysis
Top-down MS-based proteoform identification and characterization (TopPIC) software was applied to search the top-down LC-MS/MS data of E. coli and HeLa RPs. Briefly, the RAW files were converted into mzML files using the msconvert tool. The mzML files were then processed by the top-down MS feature detection tool for spectral deconvolution. The resulted msalign files were then processed by TopPIC for database searching. The target-decoy approach was employed to evaluate the false discovery rate (FDR) of proteoform spectrum match (PrSM) and proteoform IDs. The database search results were filtered with a 1% PrSM-level FDR and a 1% proteoform-level FDR. All results were then double checked manually.
PFind (version 3.1.5) was applied for bottom-up LC-MS/MS raw data. The following parameters were used for open search: precursor ion mass tolerance, ±20 ppm; fragment tolerance, ±20 ppm. Enzymatic specificity was set to trypsin KR_C, semi-specific. Tryptic searches included maximum two missed cleavages. FDR was set to 1% at peptide and protein levels. The database containing sequences for E. coli 70S RPs and HeLa 80S RPs were downloaded from UniProt.
3. Results and discussion
3.1. Optimization of instrument parameters for top-down proteomics
Instrument parameters for top-down MS studies were first carried out using a Waters Synapt G2 Si MS system. Fast DDA was selected, and parameters related to MS/MS acquisition including scan time and collision energy ramp were optimized for better identification. As shown in Fig. 1A, the number of RPs identified increased with the increase of scan time. However, the scan time had to be balanced with LC peak width, the top n number of MS/MS acquisition, and MS/MS spectrum quality. Taken the ions centered at m/z 1014 for an example, the signal-to-noise levels with different scan time were compared. As shown in Fig. S1, the larger the number of MS/MS scans, the better the MS/MS spectrum quality was acquired. However, excessively long time would compromise the MS/MS acquired for multiple parent ions; therefore, a 5 s scan time was chosen for this study to obtain the highest number of identification as well as a significantly improved signal-to-noise level. The effect of collision energy for fragmentation was then explored. The molecular weights of RPs in E. coli were in the range of 4.3–30 kDa, and their peaks were found to be primarily concentrated between m/z 650−800 Da, which was perhaps due to their enriched positive charges. It has been previously reported that the percentage of positive residues in RPs is about twice that of the average protein, with 18.7% Lys and Arg residues for E. coli RPs [33]. Therefore, this region of the collision energy ramp was closely monitored. Fig. 1B shows a diagram of four different collision energy ramps applied in this experiment. The best fragmentation was achieved by maintaining the energy between 20 and 27 eV (Fig. 1C, high mass collision ramp energy (HME)1/low mass collision ramp energy 2–4 and HME 2) with a total identification of 38 proteoforms counted based on MS/MS data, while protein peaks with poor MS/MS spectrum quality or without MS/MS spectrum were not counted here but considered for total identification with bottom-up data integrated. The further increase of the collision energy led to diminished identification due to over fragmentation. Given the quality of the spectra offered by MS/MS fragmentation, the top two most intensive ions were selected for MS/MS fragmentation.
Fig. 1.
Parameters optimized for top-down proteomics using quadrupole time-of-flight (Q-TOF) mass spectrometry. (A) Scan time vs. number of protein identification. (B) and (C) Number of proteins identified under different collision energy conditions. LME: low mass collision ramp energy; HME: high mass collision ramp energy.
3.2. Comparison of bottom-up and top-down proteomics for the analysis of E.coli ribosomes
The 50S ribosomal subunit consists of 34 RPs (numbered L1−L36, where L8 is an artifact, and L7 and L12 are essentially identical, the only difference being that L7 is acetylated at its N-terminus whereas L12 is not) [34]. The 30S subunit consists of 21 RPs (S1–S21), and there is no difference between S20 from the small and L26 from the large subunit. In Fig. 2, we provide an overview of the top-down and bottom-up results of E.coli ribosome samples. Fig. 2A shows overlaid base peak chromatograms of three technical replicates from E. coli RPs under the top-down workflow. To achieve reliable identification of RPs by top-down MS, reproducibility had been confirmed under optimized conditions. Figs. 2B and C shows the species and numbers of identified RPs detected using both top-down and bottom-up proteomics approaches. A total of 50 RPs were identified using top-down approach, 53 RPs were determined in the bottom-up study, and 48 RPs were identified by both approaches (Fig. 2C and Table S1). 50S RP L35 (7.29 kDa) and L36 (4.36 kDa), both small proteins in the E. coli ribosome, were detected in top-down but not in the bottom-up study. This might be due to the fact that L35 has 14 Lys + 6 Arg out of 65 residues and L36 contains 12 Lys + Arg out of 38 residues, and peptides generated by trypsin digestion are therefore too small to be retained during reverse phase separation. In addition, intact L14, S1, S6, S7, and S10 were not found in top-down proteomics. Possible reasons are that these proteins are of low abundance or they are co-eluted with other proteins and suppressed during ionization. Although the bottom-up strategy detected and identified more proteins, there are still some conspicuous shortcomings associated with this method. First, a previous study of the statistics has shown that when using trypsin, 56% of all generated peptides are ≤6 residues and are thus too small to be identified by MS [35]. Therefore, the situation would be even worse for RPs with a much larger percentage of positive residues [33]. The limited sequence information inferred from small peptides is more often insufficient to assign proteins to clusters, especially to identify proteoforms, as in the case of L7/L12 (see more details in the following section). Second, some PTMs may not be detected because many of the tryptic peptides are too small to bind successfully to the stationary phases typically used in proteomics [36]. For instance, we identified methylation at Ala2 in 50S RP L33 using top-down, which is consistent with previous studies [37]; however, the relevant sequence was not fully covered in our bottom-up data (Fig. S2). Finally, we observed multiple oxidation sites (Table S2) in the bottom-up data but not in the top-down study, which is consistent with previous observations that artificial modifications can be introduced during sample preparation [38]. These artificial modifications not only complicate the data analysis, causing controversial conclusions, but also occupy a significant amount of MS instrument time and retard normal peptide identification. Overall, these results highlight the importance of multilevel proteomics for accurate characterization of the heterogeneity of proteomic samples, such as E. coli RPs.
Fig. 2.
Top-down and bottom-up proteomics characterization of E.coli ribosome. (A) Overlaid base peak chromatograms of three technical replicates from E.coli ribosome. (B) The number of proteins identified in 30S and 50S using top-down and bottom-up approach, respectively. (C) Venn diagram showing the overlap of top-down and bottom-up identification.
3.3. Top-down characterization of PTMs and truncations in E.coli RPs
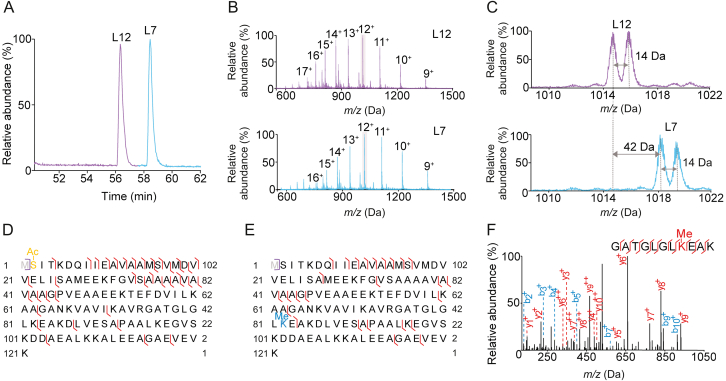
Several PTMs were observed and are listed in Table S1. The most common PTM involved the loss of the N-terminal methionine (−131.04 Da). Of the 50 RPs identified by top-down approach, 34 had the N-terminal methionine removed. The N-terminal methionine can be co-translationally cleaved by the enzyme methionine aminopeptidase, and this most usually occurs when the amino acid on the second position (P’) has a short side chain [39]. In addition, subunits L3, L7/12, L33, and S11 were found to be monomethylated, and acetylation of subunit L7, trimethylation of L11, and methyl-thiolation (D89) of S12 were also observed (Tables S1 and S2), which is consistent with previous results [[40], [41], [42]]. In the 50S ribosome, L3 forms a cluster with proteins L14 and L19 and methylation of E. coli RP L3 has been previously suggested to play a role in ribosomal assembly [43]. L11 has been previously observed to undergo N-terminal methionine cleavage and three trimethylations at the newly formed N-terminus and two internal lysines [44]. In the E. coli ribosome sample, we observed a primary intact mass peak of ∼14,870 Da, corresponding to a loss of N-terminal methionine and the addition of nine methylations. As shown in Figs. 3A and B, L7 (acetylated) and L12 (none acetylated) were both observed and well separated in the top-down proteomics MS. The expanded views of L7 and L12 peaks showed that both of L7 and L12 had a methylated proteoform (Fig. 3C). Top-down sequencing located the acetylation at the N-terminus of L7 and the methylation site of L12 amoneresidues 63−83. Bottom-up data further pinpointed the methylation to position 82 (Fig. 3D−F). Nevertheless, information about L7/L12 proteoforms cannot be differentiated in the bottom-up MS (Fig. S3). Proteins L7/L12 from 50S ribosomal subunits of E. coli form a well-defined domain and are involved in interactions with translation factors during protein biosynthesis [34], which are required for peptide chain termination [45,46]. The removal of L7/L12 reduces the rate of protein synthesis and its accuracy by an order of magnitude [47]. An increase of amounts of monomethylated L7/12 in cells grown at lower temperatures was also noted [48]. Beyond that, oxygenase-catalyzed protein hydroxylation regulates transcription in animals, and hydroxylation of the 50S RP L16 at Arg81 has been previously reported [49].
Fig. 3.
Top-down and bottom-up characterization of L12 and L7. (A) Base peak chromatograms of L12 and L7. (B) Corresponding mass spectrometry (MS) spectra of L7 and L12. (C) Expanded MS spectra of L12 and L7. Sequence maps of (D) L7 and (E) L12 proteoforms, respectively. (F) MS/MS spectrum of peptide at m/z 677.8559.
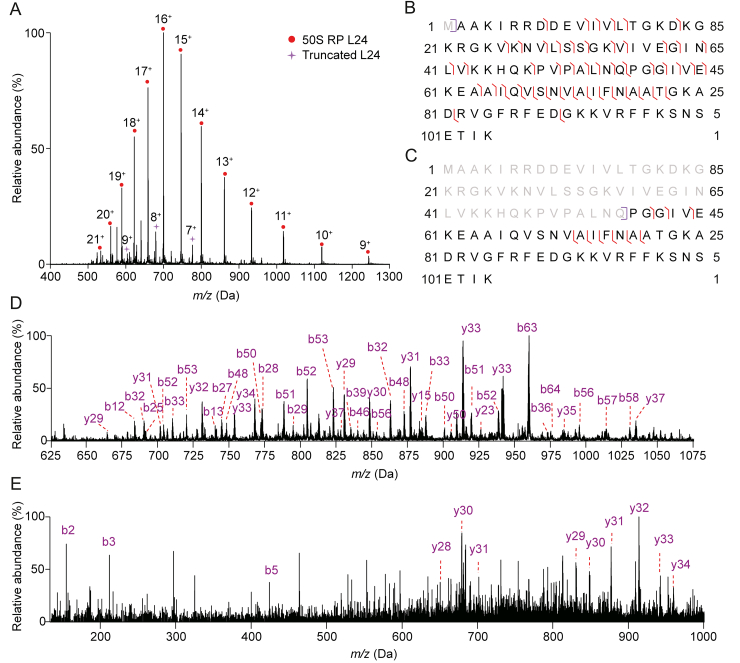
Molecular integrity is one of the most important factors which ensure the biological function of the proteins. Loss of molecular integrity, especially truncation due to alternative splicing or proteolysis, has become a major issue [50]. Two truncated proteoforms of 50S RP L24 (Fig. 4A) and 30S RP S2 were observed. Taking L24 for an example, top-down sequencing yielded a backbone cleavage efficiency of nearly 50% for L24, and located the truncation to a site between Gln54 and Pro55 (Figs. 4B−E). However, in Fig. 4, the truncated L24 proteoform and the intact L24 proteoform appeared to be co-eluted, which raised the concern that they might be caused by on-column due to acid induced hydrolysis or in source degradation. On-column degradation has been previously reported for antibody analyses, the main hydrolysis sites, including Asp-Xaa, Gly-Xaa, and Asn-Xaa, have been summarized by Vlasak et al. [51]. However, the truncated site of L24 (Gln-Pro) does not match the reported sites; it, therefore, largely rules out the cause of on-column degradation. In addition, Asp-Pro is the recognized facial in-source cleavage site [52]. Although Asp-Pro can be frequently found in RP sequences, no cleavage was observed for this site, and thus the truncated L24 proteoform is most probably biologically relevant.
Fig. 4.
Top-down mass spectrometry (MS) spectra and sequence coverage of 50S ribosomal protein (RP) L24 and truncated L24 (amino acids (AAs) 55−104): (A) MS spectrum. (B) and (D) Sequence map and MS/MS spectrum of 50S RP L24. (C) and (E) Sequence map and MS/MS spectrum of truncated 50S RP L24 (AAs 55−104).
It is well known that the treatment of ribosomal subunits with monovalent cations such as Li+ at a high concentration leads to dissociation of the RPs [53]. To exclude the potential cause of truncation during the sample preparation step of RP extraction, we changed the glacial AcOH method to LiCl-urea precipitation, and found that the truncated proteoforms remained (Figs. S4 and S5). This result suggests that these truncated proteoforms are most probably biologically relevant. In E. coli, transcription and translation are coupled and ribosomes can directly contact RNA polymerase. The contacts are formed between the RNA exit site of the RNA polymerase and RPs S2, S3, or S4 on the ribosome. S2 has been suggested to act as an anchor for the polymerase during coupling of transcription and translation [54]; such an effect might potentially be affected by the truncation of S2 (193−241).
3.4. Top-down and bottom-up characterization of HeLa 80S ribosomal proteoforms
After the development of the top-down proteomics method using an E. coli ribosome sample, we tested our method using a HeLa 80S ribosome. The eukaryotic 80S ribosome consists of a small 40S subunit and a large 60S subunit. The small 40S subunit comprises 33 S-proteins, whereas the large 60S subunit consists of 47 L-proteins [55]. In total, we identified 98 ribosomal proteoforms out of 77 ribosomal genes. Among these, 79 ribosomal proteoforms of 57 genes were identified using top-down approach, while bottom-up approach allowed the identification of 74 or 77 RPs (Tables S3 and S4, Fig. S6). In contrast to the most common modification of methionine cleavage in the E. coli ribosome, methionine cleavage accompanied by N-terminal acetylation is a prevalent protein modification in eukaryotes (Fig. 5A). In addition, dimethylation for 40S RP S10 and S25, hydroxyproline for 40S RP S23, and hydroxyhistidine for 60S RP L8 were observed by the top-down, but not by the bottom-up approach. It is intriguing that the majority of the proteoforms observed in top-down were in truncated forms. In total, we observed truncated RP proteoforms for 40S including S11, S19, and S28, and 19 RP proteoforms for large 60S including L4, L7a, L13, L13a, L19, L21, L26, L28, L34, L35, L36, and L37a, with L28 having the maximum number of 4 (Fig. 5 and Table S3). Recently, van de Waterbeemd et al. [56] reported a combination of top-down, bottom-up, and native MS for the dissection of the ribosome complex in four distinct ribosomal particles. Apart from the observation of ribosomal proteoforms due to PTMs and truncations, assembly and stoichiometry of individual RPs were also revealed. Protein-truncating proteoforms can originate from nonsense single-nucleotide variants, frameshift insertions or deletions, large structural variants, or alternative splicing, which often causes loss of protein function though it is also possible to observe gain-of-function effects [57,58].
Fig. 5.
HeLa 80S ribosome structure and ribosomal proteoforms identified using a top-down proteomics approach. (A) HeLa 80S ribosomal proteoforms identified using a top-down proteomics approach. (B) 60S RP L19 structure under pre- and post-translocation. (C) Observed ribosomal proteoforms are highlighted on the structure of 80S ribosome. PTM: post-translational modification; DiMe: dimethylation; Ac: acetylation; Hydroxy: hydroxylation; Me: methylation; Met: methionine.
RPs are the key components in the basic assembly of ribosomes, which not only promotes the folding of rRNAs to form a functional three-dimensional structure during rRNA processing, but also stabilizes the final spatial conformation of the ribosome and collaborates with rRNA to catalyze the process of protein synthesis [59]. For example, RP L19 is involved in the major rearrangements in structure during the transition from a pre- to a post-translocation state of the ribosome [55]. RP L19 is a long protein helix that protrudes from the 60S subunit and crosses over to the 40S subunit. The C-terminal helix of L19 is linked in the pre-translocation state, but it dynamically changes its conformation upon subunit rotation and becomes linear in the post-translocation state (Fig. 5B). This conformational transition results in the formation of salt bridges between the positively charged Arg172 and Arg176 side chains of protein L19 and the phosphate moieties of 18S rRNA nucleotides G909 and G910 [55]. We also observed a truncated L19 proteoform without the C-terminal part (amino acids (AAs) 2−16) (Fig. S7).
Apart from their core role in the cell translation and protein synthesis, RPs have also been found to have extra-ribosomal functions involved in cell proliferation, differentiation, apoptosis, DNA repair, modulation of cell migration and invasion, and other cellular processes [11,[60], [61], [62], [63], [64], [65]]. Many of the RPs that we observed in truncated form (Fig. 5C) have been previously linked to the development and progression of hematological, metabolic, and cardiovascular diseases, and cancer [7,66]. For example, the RP L19 gene is highly overexpressed in prostate cancer cell lines and has been considered as a potential target for therapeutic intervention [67]. We also observed two truncated proteoforms of RP L26 (Figs. S8 and S9) and four truncated proteoforms of RP L28 (Figs. S10−S13), which have been suggested as potential therapeutic markers/targets for pancreatic cancer [68,69]. RP L28 is the one with the largest number of truncated proteoforms in our study, including AAs 36−87, AAs 68−137, AAs 2−11, and AAs 2−35 (Figs. S10−S13). Several truncated proteoforms of L28 have also been reported previously [70], although correlation between their level of expression and the severity of disease has yet to be found. Labriet et al. [71] found that germline variability and tumor expression level of RP gene L28 are associated with survival of metastatic colorectal cancer patients. Additionally, abnormal expressions of an array of RPs including L13, L13a, L19, L26, L28, L34, L35a, L36, S8, S11, S19, and S20 have been reported for cancers such as hepatocellular carcinoma, colon and neoplastic colorectal cancer, and pancreatic cancer tissues and cells [7,8,61,62].
Furthermore, in the bottom-up experiments, many more proteins were detected than in the top-down approach, including proteins associated with the ribosome as well as those nonspecifically attached (Table S4). Such significant contrast can come from several sources. One is due to the inherent sensitivity of the bottom-up approach compared to that of top-down, and peptides are more easily sprayed, transferred, and fragmented than intact proteins. Secondly, nanoLC system was used for bottom-up while microflow LC system was used for top-down. Thirdly, the experiments were performed on two different platforms, i.e., an orbitrap mass spectrometer used for bottom-up and a quadrupole time-of-flight (Q-TOF) for top-down. When the condition permits, performing top-down on an orbitrap can achieve better identification due to its higher resolving power and better fragmentation efficiency of HCD. Last but not least, apart from the RPs identified by top-down and shown in Table S3, there are a percentage of protein peaks that have not yet been successfully assigned due to factors such as poor signal-to-noise ratio, poor MS/MS spectrum quality, absence of an MS/MS spectrum due to low top n acquisition number of MS/MS or low peak intensity, limited fragmentation of large proteins, and limited instrument resolving power for correct charge state assignment or overlapping peak separation. However, this is also positive and leaves room for improvement. For example, with the continuous advances of instrument design, apart from the well-known high resolution mass spectrometers for top-down experiments, such as orbitrap and Fourier transform ion cyclotron resonance, some TOF instruments with mass resolving power >100,000 have been reported [72,73] and can potentially offer a more affordable top-down platform. In addition, the integration of new fragmentation devices such as an electromagnetostatic electron capture dissociation cell for electron-based fragmentation can further enhance gas-phase fragmentation of proteoforms [74]. Recently, Mehaffey et al. [75] implemented a multistage MS/MS approach to identify unique proteoforms from E. coli RPs, including 22 protein-metal complexes and 10 protein-protein complexes. Additionally, mapping metal-bound holo fragment ions resulting from ultraviolet photodissociation of protein-metal complexes offers insights into the metal-binding sites.
Finally, the integration of top-down and bottom-up approaches can be greatly critical for analyzing challenging systems, such as the identification of low abundance phosphorylated proteoforms. Using this approach, Brown et al. [76] successfully mapped five novel sites of phosphorylation and confirmed the sixth site of potassium channel (Kir) 2.1, an integral membrane protein critical for the maintenance of the resting membrane potential and phase-3 repolarization of the cardiac action potential in the heart. While for in-depth analyses of phosphorylated proteoforms or modified proteoforms, enrichment is often essential [77]. Beyond that, the integration of top-down and bottom-up data can be a rate-limiting step. In our study, we applied PFind for bottom-up proteomic data and TopPIC for top-down proteomic data, respectively, and integrated them manually. To improve the throughput of data integration and visualization, Schaffer et al. [78] augmented the software program Proteoform Suite by Smith and co-workers [79] and ProteoCombiner by Chamot-Rooke and co-workers [80] to enable the large-scale integration of bottom-up and top-down proteomics data and data visualization to improve proteoform assessment. This software program has greatly improved and extended the proteomic analysis. We expect that more new tools will emerge in the future to facilitate large-scale integration of bottom-up and top-down proteomics data.
4. Conclusions
Accumulating evidence emphasizes that the heterogeneity of RP expression is usually accompanied by the occurrence and development of tumors. Specifically, defects in ribosomal proteoforms have been linked to a range of diseases. Comprehensive characterization of ribosomal proteoforms is the prerequisite for the discovery of potential disease biomarkers or protein targets. In the present work, we developed top-down proteomics approach to in-depth characterization of ribosomal proteoforms using a Waters Synapt G2 Si Q-TOF MS system, and then further integrated a bottom-up approach for locating PTM sites. Interestingly, apart from proteoforms with a various type of PTMs, a number of truncated proteoforms were identified in our study, while bottom-up experiments alone often failed to spot this type of information. Although the relevance of these truncated ribosomal proteoforms requires in-depth digging, it offers an advanced way to pinpoint disease-specific proteoform biomarkers or targets.
CRediT author statement
Ying Zhang: Methodology, Validation, Investigation, Formal analysis, Data curation, Writing - Original draft preparation, Reviewing and Editing, Visualization; Qinghua Cai: Resources; Yuxiang Luo: Visualization; Yu Zhang: Resources, Visualization; Huilin Li: Conceptualization, Methodology, Resources, Writing - Reviewing and Editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China (Grant Nos.: 91953102 and 81872836), Natural Science Foundation of Guangdong Province, China (Grant Nos.: 2019A1515011265 and 2022A1515010965), the Fundamental Research Funds for Sun Yat-sen University, China (Grant No.: 19ykzd26), and Open Project Funding of the State Key Laboratory of Crop Stress Adaptation and Improvement (Grant No.: 2020KF05). Huilin Li would like to thank the Pearl River Talent Recruitment Program for support.
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2022.11.003.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Pechmann S., Willmund F., Frydman J. The ribosome as a hub for protein quality control. Mol. Cell. 2013;49:411–421. doi: 10.1016/j.molcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikeuchi K., Izawa T., Inada T. Recent progress on the molecular mechanism of quality controls induced by ribosome stalling. Front. Genet. 2019;9:743. doi: 10.3389/fgene.2018.00743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowman J.C., Petrov A.S., Frenkel-Pinter M., et al. Root of the tree: The significance, evolution, and origins of the ribosome. Chem. Rev. 2020;120:4848–4878. doi: 10.1021/acs.chemrev.9b00742. [DOI] [PubMed] [Google Scholar]
- 4.Peña C., Hurt E., Panse V.G. Eukaryotic ribosome assembly, transport and quality control. Nat. Struct. Mol. Biol. 2017;24:689–699. doi: 10.1038/nsmb.3454. [DOI] [PubMed] [Google Scholar]
- 5.Petrov A.S., Gulen B., Norris A.M., et al. History of the ribosome and the origin of translation. Proc. Natl. Acad. Sci. U S A. 2015;112:15396–15401. doi: 10.1073/pnas.1509761112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korobeinikova A.V., Garber M.B., Gongadze G.M. Ribosomal proteins: Structure, function, and evolution. Biochemistry (Mosc.) 2012;77:562–574. doi: 10.1134/S0006297912060028. [DOI] [PubMed] [Google Scholar]
- 7.Kang J., Brajanovski N., Chan K.T., et al. Ribosomal proteins and human diseases: Molecular mechanisms and targeted therapy. Signal Transduct. Target. Ther. 2021;6:323. doi: 10.1038/s41392-021-00728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turi Z., Lacey M., Mistrik M., et al. Impaired ribosome biogenesis: Mechanisms and relevance to cancer and aging. Aging. 2019;11:2512–2540. doi: 10.18632/aging.101922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boria I., Quarello P., Avondo F., et al. A new database for ribosomal protein genes which are mutated in Diamond-Blackfan anemia. Hum. Mutat. 2008;29:E263−E270. doi: 10.1002/humu.20864. [DOI] [PubMed] [Google Scholar]
- 10.Gazda H.T., Sheen M.R., Vlachos A., et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am. J. Hum. Genet. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oršolić I., Bursać S., Jurada D., et al. Cancer-associated mutations in the ribosomal protein L5 gene dysregulate the HDM2/p53-mediated ribosome biogenesis checkpoint. Oncogene. 2020;39:3443–3457. doi: 10.1038/s41388-020-1231-6. [DOI] [PubMed] [Google Scholar]
- 12.Weatheritt R.J., Sterne-Weiler T., Blencowe B.J. The ribosome-engaged landscape of alternative splicing. Nat. Struct. Mol. Biol. 2016;23:1117–1123. doi: 10.1038/nsmb.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y., Qian J., Gu C., et al. Alternative splicing and cancer: A systematic review. Signal Transduct. Target. Ther. 2021;6:78. doi: 10.1038/s41392-021-00486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simsek D., Barna M. An emerging role for the ribosome as a nexus for post-translational modifications. Curr. Opin. Cell Biol. 2017;45:92–101. doi: 10.1016/j.ceb.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petibon C., Ghulam M.M., Catala M., et al. Regulation of ribosomal protein genes: An ordered anarchy. Wiley Interdiscip. Rev. RNA. 2021;12:e1632. doi: 10.1002/wrna.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith L.M., Kelleher N.L. The Consortium for Top Down Proteomics, Proteoform: A single term describing protein complexity. Nat. Methods. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aebersold R., Agar J.N., Amster I.J., et al. How many human proteoforms are there? Nat. Chem. Biol. 2018;14:206–214. doi: 10.1038/nchembio.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith L.M., Kelleher N.L. Proteoforms as the next proteomics currency. Science. 2018;359:1106–1107. doi: 10.1126/science.aat1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Millán-Zambrano G., Burton A., Bannister A.J., et al. Histone post-translational modifications − cause and consequence of genome function. Nat. Rev. Genet. 2022;23:563–580. doi: 10.1038/s41576-022-00468-7. [DOI] [PubMed] [Google Scholar]
- 20.Aebersold R., Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 21.Yates J.R., Ruse C.I., Nakorchevsky A. Proteomics by mass spectrometry: Approaches, advances, and applications. Annu. Rev. Biomed. Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 22.Müller J.B., Geyer P.E., Colaço A.R., et al. The proteome landscape of the kingdoms of life. Nature. 2020;582:592–596. doi: 10.1038/s41586-020-2402-x. [DOI] [PubMed] [Google Scholar]
- 23.Aebersold R., Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 24.Chait B.T. Mass spectrometry: Bottom-up or top-down? Science. 2006;314:65–66. doi: 10.1126/science.1133987. [DOI] [PubMed] [Google Scholar]
- 25.Kelleher N.L. Peer reviewed: Top-down proteomics. Anal. Chem. 2004;76:196A−203A. [PubMed] [Google Scholar]
- 26.Tran J.C., Zamdborg L., Ahlf D.R., et al. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature. 2011;480:254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toby T.K., Fornelli L., Kelleher N.L. Progress in top-down proteomics and the analysis of proteoforms. Annu. Rev. Anal. Chem. (Palo Alto Calif.) 2016;9:499–519. doi: 10.1146/annurev-anchem-071015-041550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith L.M., Agar J.N., Chamot-Rooke J., et al. The human proteoform project: Defining the human proteome. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abk0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Catherman A.D., Skinner O.S., Kelleher N.L. Top down proteomics: Facts and perspectives. Biochem. Biophys. Res. Commun. 2014;445:683–693. doi: 10.1016/j.bbrc.2014.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melby S.J.S., Roberts D.S., Larson E.J., et al. Novel strategies to address the challenges in top-down proteomics. J. Am. Soc. Mass Spectrom. 2021;32:1278–1294. doi: 10.1021/jasms.1c00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardy S.J.S., Kurland C.G., Voynow P., et al. The ribosomal proteins of Escherichia coli. I. purification of the 30S ribosomal proteins. Biochemistry. 1969;8:2897–2905. doi: 10.1021/bi00835a031. [DOI] [PubMed] [Google Scholar]
- 32.El-Baba T.J., Raab S.A., Buckley R.P., et al. Thermal analysis of a mixture of ribosomal proteins by vT-ESI-MS: Toward a parallel approach for characterizing the stabilitome. Anal. Chem. 2021;93:8484–8492. doi: 10.1021/acs.analchem.1c00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burton B., Zimmermann M.T., Jernigan R.L., et al. A computational investigation on the connection between dynamics properties of ribosomal proteins and ribosome assembly. PLoS Comput. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gudkov A.T. The L7/L12 ribosomal domain of the ribosome: Structural and functional studies. FEBS Lett. 1997;407:253–256. doi: 10.1016/s0014-5793(97)00361-x. [DOI] [PubMed] [Google Scholar]
- 35.Tsiatsiani L., Heck A.J.R. Proteomics beyond trypsin. FEBS J. 2015;282:2612–2626. doi: 10.1111/febs.13287. [DOI] [PubMed] [Google Scholar]
- 36.Chabanet C., Yvon M. Prediction of peptide retention time in reversed-phase high-performance liquid chromatography. J. Chromatogr. 1992;599:211–225. doi: 10.1016/0021-9673(92)85475-9. [DOI] [PubMed] [Google Scholar]
- 37.Chang C.N., Schwartz M., Chang F.N. Identification and characterization of a new methylated amino acid in ribosomal protein L33 of Escherichia coli. Biochem. Biophys. Res. Commun. 1976;73:233–239. doi: 10.1016/0006-291x(76)90698-7. [DOI] [PubMed] [Google Scholar]
- 38.Dupree E.J., Jayathirtha M., Yorkey H., et al. A critical review of bottom-up proteomics: The good, the bad, and the future of this field. Proteomes. 2020;8:14. doi: 10.3390/proteomes8030014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wingfield P.T. N-terminal methionine processing. Curr. Protoc. Protein Sci. 2017;88 doi: 10.1002/cpps.29. 6.14.1–6.14.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Demirci H., Gregory S.T., Dahlberg A.E., et al. Multiple-site trimethylation of ribosomal protein L11 by the PrmA methyltransferase. Structure. 2008;16:1059–1066. doi: 10.1016/j.str.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suh M.J., Hamburg D.M., Gregory S.T., et al. Extending ribosomal protein identifications to unsequenced bacterial strains using matrix-assisted laser desorption/ionization mass spectrometry. Proteomics. 2005;5:4818–4831. doi: 10.1002/pmic.200402111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Running W.E., Ravipaty S., Karty J.A., et al. A top-down/bottom-up study of the ribosomal proteins of Caulobacter crescentus. J. Proteome Res. 2007;6:337–347. doi: 10.1021/pr060306q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lhoest J., Colson C. Cold-sensitive ribosome assembly in an Escherichia coli mutant lacking a single methyl group in ribosomal protein L3. Eur. J. Biochem. 1981;121:33–37. doi: 10.1111/j.1432-1033.1981.tb06425.x. [DOI] [PubMed] [Google Scholar]
- 44.Cameron D.M., Gregory S.T., Thompson J., et al. Thermus thermophilus L11 methyltransferase, PrmA, is dispensable for growth and preferentially modifies free ribosomal protein L11 prior to ribosome assembly. J. Bacteriol. 2004;186:5819–5825. doi: 10.1128/JB.186.17.5819-5825.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brot N., Tate W.P., Caskey C.T., et al. The requirement for ribosomal proteins L7 and L12 in peptide-chain termination. Proc. Natl. Acad. Sci. U S A. 1974;71:89–92. doi: 10.1073/pnas.71.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oleinikov A.V., Jokhadze G.G., Traut R.R. A single-headed dimer of Escherichia coli ribosomal protein L7/L12 supports protein synthesis. Proc. Natl. Acad. Sci. U S A. 1998;95:4215–4218. doi: 10.1073/pnas.95.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pettersson I., Kurland C.G. Ribosomal protein L7/L12 is required for optimal translation. Proc. Natl. Acad. Sci. U S A. 1980;77:4007–4010. doi: 10.1073/pnas.77.7.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang F.N. Temperature-dependent variation in the extent of methylation of ribosomal proteins L7 and L12 in Escherichia coli. J. Bacteriol. 1978;135:1165–1166. doi: 10.1128/jb.135.3.1165-1166.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ge W., Wolf A., Feng T., et al. Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans. Nat. Chem. Biol. 2012;8:960–962. doi: 10.1038/nchembio.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeBoever C., Tanigawa Y., Lindholm M.E., et al. Medical relevance of protein-truncating variants across 337,205 individuals in the UK Biobank study. Nat. Commun. 2018;9:1612. doi: 10.1038/s41467-018-03910-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vlasak J., Ionescu R. Fragmentation of monoclonal antibodies. mAbs. 2011;3:253–263. doi: 10.4161/mabs.3.3.15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu W., Vath J.E., Huberty M.C., et al. Identification of the facile gas-phase cleavage of the Asp-Pro and Asp-Xxx peptide bonds in matrix-assisted laser desorption time-of-flight mass spectrometry. Anal. Chem. 1993;65:3015–3023. doi: 10.1021/ac00069a014. [DOI] [PubMed] [Google Scholar]
- 53.Lerman M.I., Spirin A.S., Gavrilova L.P., et al. Studies on the structure of ribosomes: II. Stepwise dissociation of protein from ribosomes by caesium chloride and the re-assembly of ribosome-like particles. J. Mol. Biol. 1966;15:268–281. doi: 10.1016/s0022-2836(66)80226-7. [DOI] [PubMed] [Google Scholar]
- 54.Blaha G.M., Diggs S., Tam T.K., et al. 2022. The effects of ribosomal proteins uS2, uS3, and uS4 on transcription, FASEB J. [DOI] [Google Scholar]
- 55.Khatter H., Myasnikov A.G., Natchiar S.K., et al. Structure of the human 80S ribosome. Nature. 2015;520:640–645. doi: 10.1038/nature14427. [DOI] [PubMed] [Google Scholar]
- 56.van de Waterbeemd M., Tamara S., Fort K.L., et al. Dissecting ribosomal particles throughout the kingdoms of life using advanced hybrid mass spectrometry methods. Nat. Commun. 2018;9:2493. doi: 10.1038/s41467-018-04853-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rivas M.A., Pirinen M., Conrad D.F., et al. Human genomics. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science. 2015;348:666–669. doi: 10.1126/science.1261877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neverov A.D., Artamonova I.I., Nurtdinov R.N., et al. Alternative splicing and protein function. BMC Bioinformatics. 2005;6:266. doi: 10.1186/1471-2105-6-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie X., Guo P., Yu H., et al. Ribosomal proteins: Insight into molecular roles and functions in hepatocellular carcinoma. Oncogene. 2018;37:277–285. doi: 10.1038/onc.2017.343. [DOI] [PubMed] [Google Scholar]
- 60.Wang W., Nag S., Zhang X., et al. Ribosomal proteins and human diseases: Pathogenesis, molecular mechanisms, and therapeutic implications. Med. Res. Rev. 2015;35:225–285. doi: 10.1002/med.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Challa S., Khulpateea B.R., Nandu T., et al. Ribosome ADP-ribosylation inhibits translation and maintains proteostasis in cancers. Cell. 2021;184:4531–4546. doi: 10.1016/j.cell.2021.07.005. e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pecoraro A., Pagano M., Russo G., et al. Ribosome biogenesis and cancer: Overview on ribosomal proteins. Int. J. Mol. Sci. 2021;22:5496. doi: 10.3390/ijms22115496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xie J., Zhang W., Liang X., et al. RPL32 promotes lung cancer progression by facilitating p53 degradation. Mol. Ther. Nucleic Acids. 2020;21:75–85. doi: 10.1016/j.omtn.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li C., Ge M., Chen D., et al. RPL21 siRNA blocks proliferation in pancreatic cancer cells by inhibiting DNA replication and inducing G1 arrest and apoptosis. Front. Oncol. 2020;10:1730. doi: 10.3389/fonc.2020.01730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ebright R.Y., Lee S., Wittner B.S., et al. Deregulation of ribosomal protein expression and translation promotes breast cancer metastasis. Science. 2020;367:1468–1473. doi: 10.1126/science.aay0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Slimane S.N., Marcel V., Fenouil T., et al. Ribosome biogenesis alterations in colorectal cancer. Cells. 2020;9:2361. doi: 10.3390/cells9112361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bee A., Ke Y.Q., Forootan S., et al. Ribosomal protein L19 is a prognostic marker for human prostate cancer. Clin. Cancer Res. 2006;12:2061–2065. doi: 10.1158/1078-0432.CCR-05-2445. [DOI] [PubMed] [Google Scholar]
- 68.Muro S., Miyake Y., Kato H., et al. Serum anti-60S ribosomal protein L29 antibody as a novel prognostic marker for unresectable pancreatic cancer. Digestion. 2015;91:164–173. doi: 10.1159/000371545. [DOI] [PubMed] [Google Scholar]
- 69.Li C., Ge M., Yin Y., et al. Silencing expression of ribosomal protein L26 and L29 by RNA interfering inhibits proliferation of human pancreatic cancer PANC-1 cells. Mol. Cell. Biochem. 2012;370:127–139. doi: 10.1007/s11010-012-1404-x. [DOI] [PubMed] [Google Scholar]
- 70.Ota T., Suzuki Y., Nishikawa T., et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 71.Labriet A., Lévesque É., Cecchin E., et al. Germline variability and tumor expression level of ribosomal protein gene RPL28 are associated with survival of metastatic colorectal cancer patients. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-49477-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yavor M.I., Pomozov T.V., Kirillov S.N., et al. High performance gridless ion mirrors for multi-reflection time-of-flight and electrostatic trap mass analyzers. Int. J. Mass Spectrom. 2018;426:1–11. [Google Scholar]
- 73.Richardson K., Hoyes J. A novel multipass oa-TOF mass spectrometer. Int. J. Mass Spectrom. 2015;377:309–315. [Google Scholar]
- 74.Shen X., Xu T., Hakkila B., et al. Capillary zone electrophoresis-electron-capture collision-induced dissociation on a quadrupole time-of-flight mass spectrometer for top-down characterization of intact proteins. J. Am. Soc. Mass Spectrom. 2021;32:1361–1369. doi: 10.1021/jasms.0c00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mehaffey M.R., Xia Q., Brodbelt J.S. Uniting native capillary electrophoresis and multistage ultraviolet photodissociation mass spectrometry for online separation and characterization of Escherichia coli ribosomal proteins and protein complexes. Anal. Chem. 2020;92:15202–15211. doi: 10.1021/acs.analchem.0c03784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brown K.A., Anderson C., Reilly L., et al. Proteomic analysis of the functional inward rectifier potassium channel (kir) 2.1 reveals several novel phosphorylation sites. Biochemistry. 2021;60:3292–3301. doi: 10.1021/acs.biochem.1c00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roberts D.S., Chen B., Tiambeng T.N., et al. Reproducible large-scale synthesis of surface silanized nanoparticles as an enabling nanoproteomics platform: Enrichment of the human heart phosphoproteome. Nano Res. 2019;12:1473–1481. doi: 10.1007/s12274-019-2418-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schaffer L.V., Millikin R.J., Shortreed M.R., et al. Improving proteoform identifications in complex systems through integration of bottom-up and top-down data. J. Proteome Res. 2020;19:3510–3517. doi: 10.1021/acs.jproteome.0c00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cesnik A.J., Shortreed M.R., Schaffer L.V., et al. Proteoform Suite: Software for constructing, quantifying, and visualizing proteoform families. J. Proteome Res. 2018;17:568–578. doi: 10.1021/acs.jproteome.7b00685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lima D.B., Dupré M., Duchateau M., et al. ProteoCombiner: Integrating bottom-up with top-down proteomics data for improved proteoform assessment. Bioinformatics. 2021;37:2206–2208. doi: 10.1093/bioinformatics/btaa958. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.