Abstract
Disorders of the synovial joint, such as osteoarthritis and rheumatoid arthritis, afflict a substantial proportion of the global population. However, current clinical management has not been focused on fully restoring the native function of joints. Organ-on-a-chip, also called a microphysiological system, which typically accommodates multiple human cell-derived tissues/organs under physiological culture conditions, is an emerging platform that potentially overcomes the limitations of current models in developing therapeutics. Herein, the major progress in generating organs-on-chips for studying arthritis is reviewed, the challenges faced when these novel platforms enter the next phase of development and application are discussed, and the potential for organ-on-a-chip technology to investigate the pathogenesis of joint diseases and the development of efficacious therapies is presented.
Keywords: Arthritis, Joint disease, Microphysiological system, Disease modeling, Drug testing, Personalized medicine
Joint diseases: old but unsolved medical problems
Joint disorders are physically and psychologically debilitating and significantly compromise the patient’s quality of life. Joint diseases such as osteoarthritis (OA) and rheumatoid arthritis (RA) are characterized by joint inflammation and degeneration. The global prevalence of OA and RA has been estimated to be 3,754 cases and 460 cases, respectively, per 100,000 persons [1,2]. The limited sensitivity and accuracy of current diagnostic tests are major hurdles to the early diagnosis of these chronic conditions [3]. Consequently, joint diseases are only diagnosed when joint destruction has already begun. Despite decades of investigations into disease mechanisms, much remains unknown about OA and RA etiologies [4].
Humans have been afflicted with joint disorders for a long time [5]. For decades, cartilage degradation was considered the prominent, if not the only, relevant feature of OA. Nevertheless, it is now appreciated that the joint functions as an organ and that OA is a disease of the entire organ, with pathological changes observed in all joint components [6]. Besides cartilage degeneration, abnormal turnover of subchondral bone, synovium inflammation (synovitis), and altered secretome of the infrapatellar fat pad (IPFP) have all also been observed in synovial joints (see Glossary) with OA [7]. The risk factors of OA include aging, trauma, joint injury, chronic overload, obesity, genetic predisposition (see Glossary), abnormal hormone profile, and metabolic syndrome [4]. The molecular basis of OA pathogenesis has been explored extensively. However, the full map of events involved in OA, from the onset to the end stage, has not been established. Current treatment options for OA merely provide symptomatic relief and cannot halt or reverse OA progression. Thus far, because no FDA-approved disease-modifying OA drugs (DMOADs) are available, total joint arthroplasty remains the final option for end-stage disease.
Rheumatoid arthritis (RA) is a chronic and complex autoimmune disease characterized by hyperplasia of the synovial lining layer and erosive destruction of joints [8]. A prominent symptom of RA is synovial inflammation (synovitis), where the synovium experiences leukocyte infiltration and increased angiogenesis [8]. Proinflammatory cytokines are present at higher concentrations in the synovial fluid of RA joints. Pannus formation in RA ultimately leads to bone erosion and cartilage damage, causing pain and limited range of motion. RA is a highly heterogeneous group of diseases with variations in disease genotype, course, and clinical manifestations [9]. Several disease-modifying anti-rheumatic drugs (DMARDs) are available [9]. However, the efficacy of these DMARDs has been severely hampered by RA heterogeneity. Additionally, a sizable proportion of RA patients suffer from refractory disease, which is resistant (refractory) to multiple therapeutic agents with distinct structures and mechanisms of action [10].
In addition to OA and RA, which are arthritic joint disorders with high prevalence, other types of arthritis (see Glossary), such as psoriatic arthritis and septic arthritis, can also cause severe problems in the joint. Although the clinical management of these less common joint disorders has advanced dramatically, major challenges remain in their diagnosis and treatment [11,12].
To address the unmet need for efficacious treatment of joint disorders, it is critical to establish clinically relevant disease models to decipher disease mechanisms, identify novel therapeutics, and more reliably inform the selection of therapeutic options.
Lack of disease models with high fidelity for fully simulating arthritis in humans
Conventional in vitro and in vivo models: successes and setbacks
Conventional in vitro models utilized to simulate arthritis include two-dimensional (2D) cell culture, three-dimensional (3D) mono-culture and co-culture, and tissue explants (Box 1, Figure 1) [6]. These models are generally characterized by high throughput but limited physiological relevance (see Glossary). Recently, organoids have emerged as a rapidly growing biotechnology to study tissue development and pathogenesis, and their applications in studying joint pathologies are just beginning [13].
Box 1. Conventional in vitro models simulating joint diseases.
Also referred to as monolayer cultures, 2D cell cultures have been widely used in joint disease research. 2D culture is economical and highly reproducible; with their convenience and relatively low resource utilization, 2D models support high-throughput testing. However, 2D cultures represent oversimplified models. They do not provide a 3D microenvironment as observed in vivo, and the physical and chemical properties of the substrate, often polystyrene plastic, differ considerably from those of the native ECM of joint tissues. Therefore, 2D models are usually used in preliminary experiments preceding in-depth studies involving more complex and physiologically relevant models.
Models of 3D cultures can be created in scaffold-free (e.g., pellet culture of chondrocytes) form or based on 3D cell-laden scaffolds, and a key feature is the cell-cell or cell-material contact and interactions in all three dimensions, a situation observed in native tissues. Furthermore, the properties of the scaffold materials (e.g., stiffness and anchored functional biomolecules) utilized in 3D cultures can be tuned to simulate those of the native tissue ECM. Therefore, 3D culture overcomes many drawbacks inherent to 2D culture. However, the interconnections between different tissues are missing in most 3D cultures that only include a single cell type. The reciprocal cell-cell and tissue-tissue communications are critical in drug testing applications, as the response of one tissue may be affected by the presence of other tissues and their secretome [7].
Tissue explants retain the native ECM in which the cells reside, representing the most physiologically relevant in vitro samples for studying human diseases. However, it is generally recognized that the properties of the explants vary greatly with donors and harvest sites. Consequently, many tissue samples are needed to produce results of statistical significance, a requirement that is often prohibitive due to limited tissue availability and ethical considerations. Lastly, maintenance of viable explants with native features for the long term is still a challenge.
Figure 1. Different models for investigating synovial joint diseases with varying throughput and complexity.
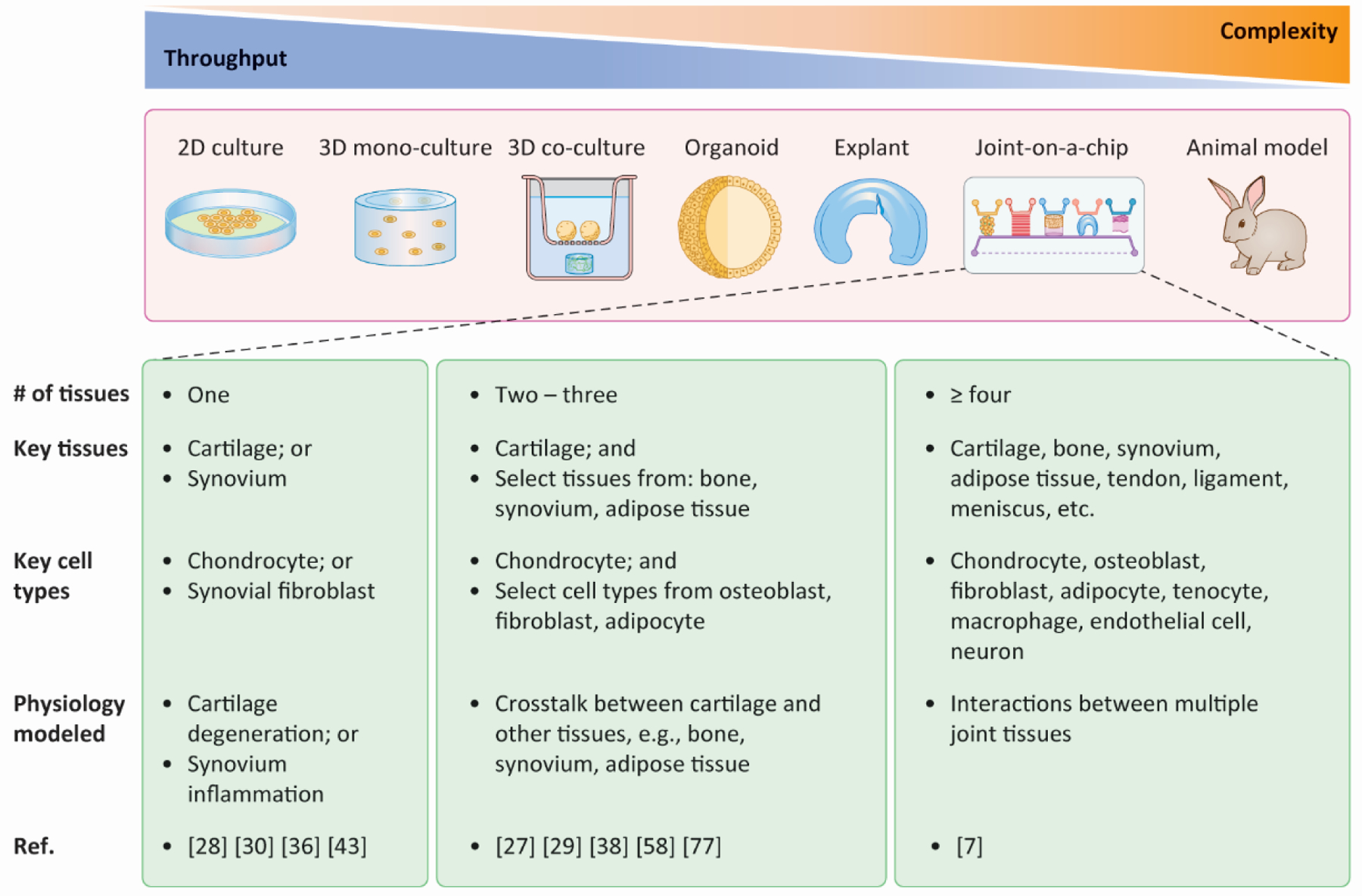
Currently, throughput and complexity/physiological relevance are, in most cases, mutually preclusive. Therefore, a trade-off between throughput and complexity is usually made when selecting a specific model. Depending on the design and application, joint-on-a-chip platforms can possess a wide range of throughput and complexity. 2D: two-dimensional; 3D: three-dimensional; Ref.: references.
The native joint tissues experience a myriad of complex, superimposing signals from both biomolecules and physical loading, which are challenging to recapitulate in the current in vitro models [4]. Therefore, animal models have been employed to study joint diseases for decades and have contributed significantly to our understanding of arthritis, including disease onset, mechanism exploration, molecular genetics, environmental factors, and therapeutic safety and efficacy [4,14–16]. Compared with conventional 2D and 3D culture models, animal models allow us to study the changes of all joint components and their crosstalk in disease progress .
However, there exist major anatomical, biomechanical, and genetic differences between animal models and humans, which may contribute to the limited translation from animal studies to clinical trials. For example, differences in size, weight, and mode of ambulation (biped vs quadruped) result in the generation of very different forces that require unique adaptations [6]. Besides, establishing animal models requires specialized techniques and is associated with high housing and maintenance costs. Although animal models of OA and RA share similarities with the human diseases, these models present differences from the human conditions and have shown varying predictive power for drug efficacy in human patients. In addition, since OA and RA affect all joint components, an in-depth understanding of joint pathophysiology requires the dissection of each tissue component’s contributions, which is costly and technically challenging, albeit feasible, in animal models [14,17,18].
Since drug candidates with sufficient efficacy in preclinical animal tests still have high attrition rates in Phase II/III clinical trials [19], there have been persistent concerns about the predictive power of animal models. Furthermore, given the cost and space required for animal research, alternative approaches capable of faithfully emulating joint pathophysiology would certainly be welcomed.
Diverse patient population requires personalized disease models
The patient population with joint disorders is highly heterogeneous, indicated by diverse structural changes, variability in the quality, intensity and features of the associated pain, and different responses to the same treatments [9,20,21]. Both intact and severely damaged cartilage can be found in the same knee joint [22], indicating the diverse degradation rates and underlying mechanisms [22]. In addition, many factors, including genetic background, age, sex, body weight, and physical exercise, are highly variable in different patients and may all influence joint health [4]. Another critical consideration in the diagnosis, prognosis, and treatment of joint disorders is that the disease stage varies from patient to patient.
Using a single-cell atlas for healthy and OA articular cartilage, Grandi and coworkers stratified OA patients in three subgroups based on the combinations of distinct chondrocyte subpopulations, offering a potential criterion for devising precision medicine tailored to specific patient subsets [21]. The heterogeneous nature of RA is shown in various aspects, such as the clinical phenotypes, genetic risk factors, and cellular and molecular synovial signatures [9]. RA patients respond drastically differently to conventional first-line treatments, i.e., small-molecule DMARDs, and respond likewise to later-line, biological DMARDs [e.g., anti-tumor necrosis factor (TNF) drugs] administered when classic RA therapeutics show inadequate efficacy [20]. Such inconsistent drug response profiles observed stem from patient heterogeneity (see Glossary), highlighting the need for patient stratification and targeted treatments.
Unfortunately, patient variability has not been sufficiently addressed in conventional joint disease models. Therefore, personalized joint disease models are highly desirable to explore patient-specific etiology and pathogenic processes and develop individualized therapies accordingly.
Organs-on-chips (OoCs): an emerging in vitro model for understanding joint diseases
The lack of clinically relevant models is a challenge to many diseases [23,24]. The concept of organ-on-a-chip (OoC, see Glossary) systems was coined about ten years ago to generate systems that could overcome the limitations of preclinical models available. Although the definition of OoC has not reached a consensus, it generally refers to miniaturized in vitro platforms that culture cells in a 3D environment with appropriate medium flow to perform specific functions of their in vivo counterparts [25]. These systems contain fundamental cellular and extracellular components of native tissues and often simulate the tissue-tissue crosstalk under physiological and pathological conditions. While there exist static OoCs, these systems lack the dynamic medium flow required to simulate drug pharmacokinetics or enable the integration of multiple OoCs [26]. It should be clarified that OoCs are not intended to emulate all biological characteristics of an organ, nor do they necessarily mimic its anatomy (although the tissue arrangement in multi-tissue joint OoCs often mimics the native organ structure) [25]. Instead, they are minimally but appropriately functional units established with relatively low consumption of cells and media to meet the requirements of envisaged applications, such as studying the biochemical crosstalk between various cell types across different tissues, disease modeling, drug evaluation, toxicity testing, and development of personalized medicine (see Glossary).
The applications of OoCs to study joint disorders started when the OoC concept was first proposed, and are gaining increasing attention in recent years (Figure 1) [7,27–29]. The key tissue components incorporated in joint OoCs are described in Box 2, with representative examples from published studies shown in Figure IA–E. Although OoCs containing a single cell type are in use [28,30], the more physiologically relevant OoC platforms feature diverse cellular components to emulate the interactions between cells and tissues [7]. Over the past few decades, a major research advance in OA and RA was achieved with the acknowledgment that they are “whole-joint diseases” [6], hence the importance of establishing multi-tissue joint OoCs.
Box 2. Key tissue components in joint OoCs.
Cartilage is the most extensively studied tissue in joint-mimicking OoCs, as cartilage degeneration is a prominent feature of joint diseases such as OA and RA. Several cartilage-on-a-chip systems have been established with primary chondrocytes as the cell source (Figure IA) [28,30,53]. Subchondral bone plays critical roles in joint homeostasis and pathogenesis [68]. However, few studies have reported bone-mimicking chips, possibly due to bone’s more complex cellular composition and architecture than cartilage. When generating bone-mimicking tissues in OoCs, a mineral component (e.g., HAp and tri-calcium phosphate) is usually incorporated into the scaffold [38,69,70]. Although not developed to investigate joint diseases, chips simulating cancer cell invasion and metastasis to an engineered bone niche [71] can provide valuable insights into the design and generation of subchondral bone-on-a-chip systems. Given the inseparable connections and well-documented biochemical and biophysical communications between articular cartilage and subchondral bone, the OC tissue ideally should be produced in situ in a single construct, generating an ECM gradient akin to that seen in the native OC tissue. Gradient tissue engineering strategies [72,73] are thus needed to form the bone-cartilage interface. Biphasic OC tissues, comprising both osseous and chondral components, have also been generated using a dual-flow chip to replicate the inseparable cartilage-subchondral bone interactions (Figure IB, C) [38,39,58].
Synovitis accompanies the pathogenesis of both OA and RA, hence the great importance of a synovium compartment in a joint OoC. The feasibility of generating synovial-like tissues has been demonstrated in several previous studies (Figure ID) [27,36]. In particular, the synovial lining only contains two types of cells, i.e., fibroblast and macrophage-like synoviocytes. Therefore, a successful generation of synovial tissues for OoCs should include both cell types [52].
The inclusion of adipose tissue in joint OoCs to emulate the IPFP is justified by the observation that altered lipid metabolism and inflammatory factor secretion are associated with OA pathogenesis and development [74]. However, most adipose tissue-on-a-chip systems established thus far (see Figure IE as an example) were intended to study metabolic disorders such as type II diabetes [75,76]. Nevertheless, these devices can serve as helpful blueprints for incorporating an adipose compartment in joint-on-a-chip systems [7]. We established an adipose tissue-on-a-chip system by culturing human adipose tissue-derived MSC (ASC)-laden hydrogels in a perfusion bioreactor; the encapsulated ASCs retained their adipogenic differentiation potential after 28 days of culture in the chip [51].
Other joint tissues, including tendon, ligament, and meniscus, have not been generated in current OoC systems. However, the inclusion of these tissues would be necessary for highly physiologically relevant joint-on-a-chip systems designed for specialized, clinical need-driven applications in future studies. Moreover, meniscal damage is increasingly appreciated as a hallmark of OA and injury to the anterior cruciate ligament and meniscus is a prominent risk factor for OA.
Figure I. Representative joint components developed in OoC or organoids.
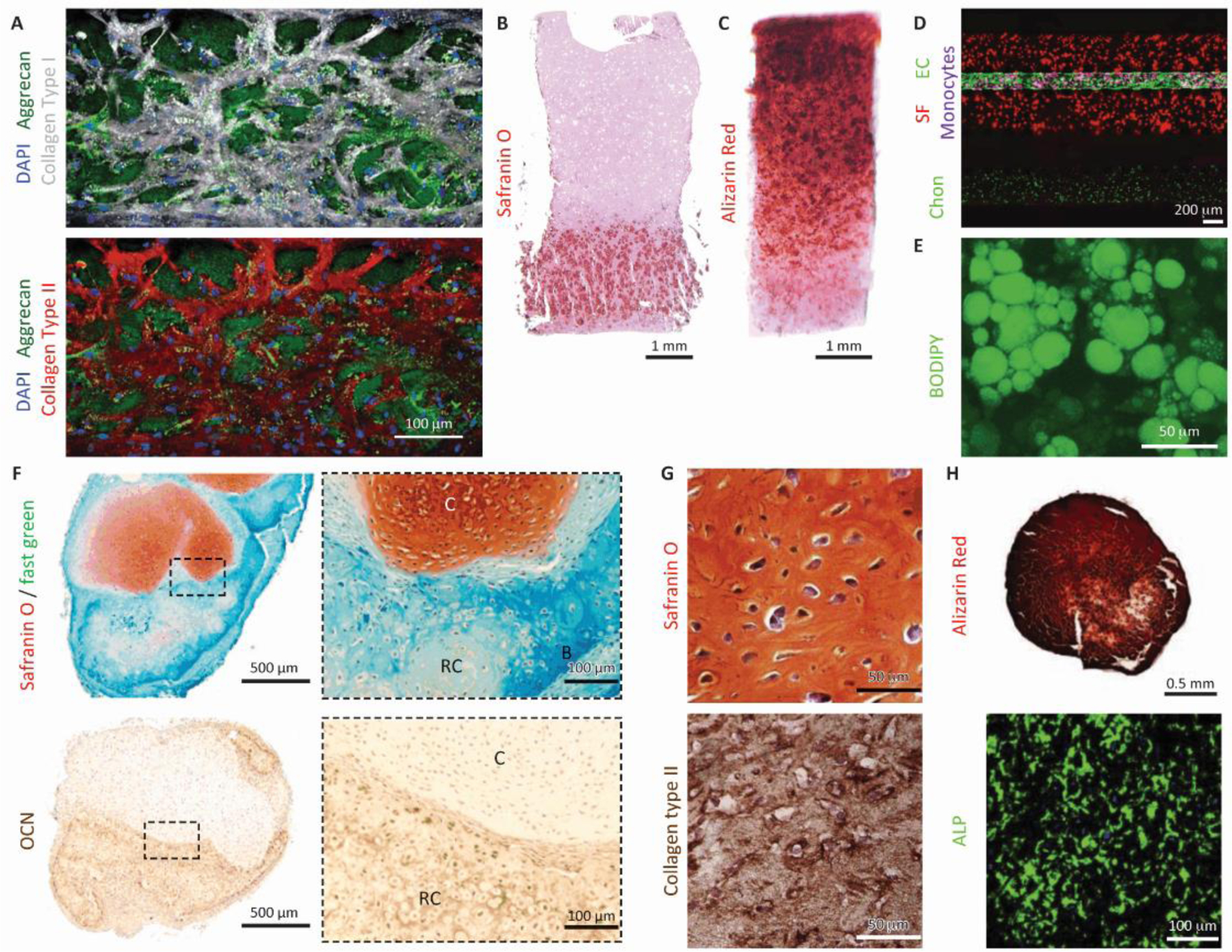
(A) Cartilage tissue created from human chondrocytes in a microfluidic chip. scale bar = 100 µm. Reproduced with permission from [28]. (B,C) Osteochondral tissue engineered from human MSCs in a dual-flow chip. Scale bar = 1 mm. Reproduced with permission from [35]. (D) Cartilage-synovium (with endothelial channel) chip. SF: synovial fibroblast, EC: endothelial cell, Chon: chondrocytes. Scale bar = 200 µm. Reproduced with permission from [27]. (E) Adipose tissue engineered from adipose-derived stem cells in a microfluidic chip. Scale bar = 50 µm. Reproduced with permission from [45]. (F) An osteochondral organoid formed after four weeks of subcutaneous implantation, with the cartilage layer from human iPSC (cartilage layer) and the bone layer from human periosteum-derived cells. OCN: osteocalcin. C: cartilage, RC: remodeling cartilage, B: bone. Scale bar = 500 μm (overview images) or 100 μm (magnified view indicated by dashed borders). Reproduced with permission from [91]. (G) Cartilage organoids derived from human MSCs. Scale bar = 50μm. Reproduced with permission from [89]. (H) Bone organoids derived from human MSCs. ALP: alkaline phosphatase. Scale bar= 0.5 mm (Alizarin Red) or 100 μm (ALP immunostaining). Reproduced with permission from [89].
For multi-tissue OoCs, a key challenge is the development of medium compositions that allows the maturation and maintenance of tissue phenotypes and, at the same time, enable the active, reciprocal crosstalk between the tissue components [23]. In a joint-mimicking OoC, such a medium is likely a “simulated synovial fluid” (SSF) that emulates the key functions of the synovial fluid in the articular cavity [7]. The SSF can be shared by tissues such as the cartilage, synovium, IPFP, and meniscus in the chip, resembling the synovial fluid bathing these tissues in an articular joint. Tissue-secreted biomolecules, including various growth factors, cytokines and chemokines, are transported by SSF to other tissues, providing biochemical signals involved in tissue crosstalk.
Furthermore, many studies have revealed the correlations between joint health and other tissues/organs in the human body, highlighting the necessity of looking beyond the joint organ to research joint pathophysiology [14,15,31]. For example, the interconnections between the gut microbiome and joint diseases such as OA and RA have been unveiled in several recent studies [15,31]. Collins and colleagues found that mice with lipodystrophy (with no adipose tissue) were protected from post-traumatic and spontaneous knee OA, indicating that tissues outside the knee joint also affect joint health, possibly through systemic effects [14]. Therefore, investigations into integrating joint-mimicking chips with OoCs emulating other organs, which have not been reported thus far, would be highly valuable. A human body-on-a-chip system [23] can be established to comprehensively evaluate the interconnections between joints and other organs and assess the local and systemic effects of potential DMOADs and DMARDs.
Joint-on-a-chip Systems
Fabrication of joint OoCs
The important considerations for engineering tissues, fabricating chip devices, and establishing single-tissue and multi-tissue joint OoCs are presented in Figure 2.
Figure 2. Fabrication of synovial joint-mimicking OoCs and key considerations.
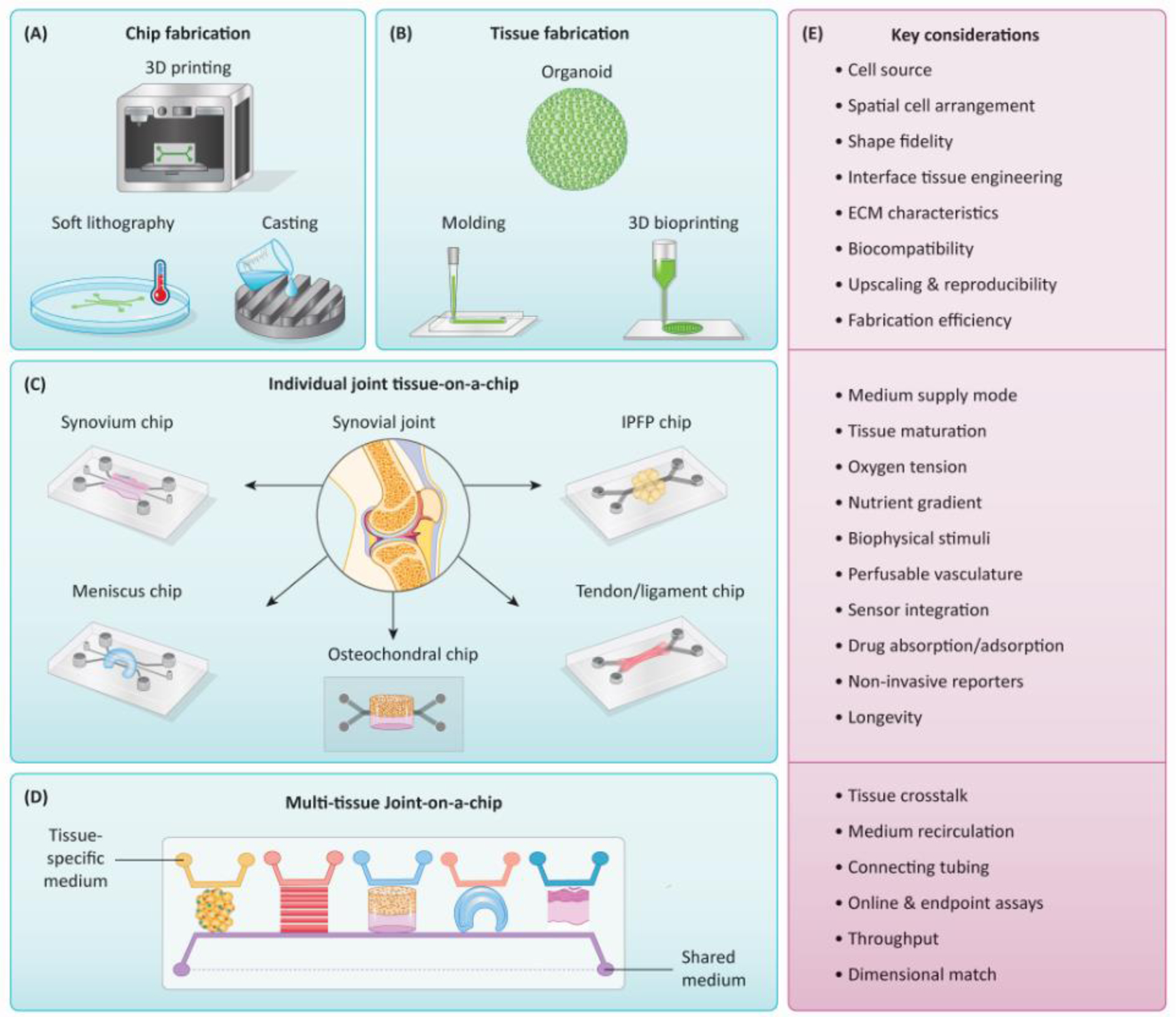
(A, B) The chip devices and tissues are usually fabricated separately using various methods, although 3D printing can potentially prepare tissue-containing devices in a single step. (C) Single-tissue chips can emulate individual tissues in the synovial joint. (D) Multi-tissue joint-on-a-chip systems can be established by integrating single-tissue chips to achieve higher complexity and physiological relevance. To facilitate tissue-tissue crosstalk, a shared medium (purple stream, with dashed line indicating recirculation) can be used to simulate the synovial fluid or blood flow. (E) The key considerations in creating joint OoCs revolve around recapitulating the environmental cues (e.g., oxygen tension, pH, compression, shear stress, biochemical factor gradient, etc.) experienced by native joint tissues. To assess the tissue health state, conventional off-line analysis and characterization can be used after manual sample collection, which however needs to terminate the culture, thus are incompatible with the continuous, real-time, and on-chip monitoring. Therefore, as a critical step toward the automatic and continuous examination of functional readouts, integrating different kinds of sensors is particularly important to capture sensitive, transient cellular responses or collect biophysical and biochemical signals over extended durations of OoC cultures. 3D: three-dimensional. IPFP: infrapatellar fat pad.
Typical OoCs are constructed using microfluidic devices (see Glossary) fabricated from the transparent, elastic, and gas-permeable polymer polydimethylsiloxane (PDMS) using soft lithography (Figure 2) [28]. A major drawback of the PDMS materials is their absorption and adsorption of small hydrophobic molecules, which represents a severe limitation for drug testing applications [32]. Alternative materials, therefore, are being explored for the fabrication of OoC devices. In addition, PDMS-based devices often require manual casting, punching, and assembly, thus significantly decreasing the reproducibility and yield of the fabrication process. Many OoCs have been fabricated by 3D printing (Figure 2) [33,34], which requires minimal manual handling, offers more freedom in chip design, and generates virtually unlimited shape and geometry. By casting OoC parts using 3D printed molds, Novak and coworkers realized large-scale production of OoC devices [35].
Various cell sources and scaffold materials are used to create joint OoCs (Box 3). The tissue constructs in OoCs are commonly prepared by molding [28], organoid culture [36], and 3D bioprinting [37] (Box 4).
Box 3. Cell sources and scaffold materials for joint OoCs.
Joint-replicating OoC systems should ideally comprise primary cells, including osteoblasts, chondrocytes, synovial fibroblasts, synovial macrophages, adipocytes, tenocytes, ligament fibroblasts, and endothelial cells, or subsets of these cell types, as well as sensory and sympathetic innervation (Figure 1). These cells, however, can reside in difficult-to-access tissues in the human body. In addition, given the limited availability and low expansion potential of primary cells, in particular neurons followed by chondrocytes, high-throughput applications would necessarily require multiple attempts to harvest sufficient cells from different donors, which significantly increases the technical difficulty and associated cost and introduces additional inconsistencies due to the difference in genetic background.
These drawbacks can be largely overcome by utilizing stem cells. MSCs, for example, possess multipotency and can be differentiated into various musculoskeletal lineages [77,78]. Furthermore, MSCs can be isolated from many tissues throughout the body and expanded extensively in vitro, thus minimizing the number of donors required. Another potential, increasingly popular cell source is iPSCs, generated by re-programming of somatic cells [79], which eliminate the process of isolating primary cells from multiple tissue harvesting. The theoretically unlimited expansion capacity of iPSCs is desirable to OoC applications in high-throughput drug screening.
The cells cultured within OoCs are induced to deposit nascent ECM similar to that found in native tissues. This may take place in the presence or absence of a scaffold, and in the latter case, the cells are seeded as a monolayer or introduced as spheroids. A cell-laden scaffold is typically used when a 3D tissue is incorporated. The physical and chemical properties of scaffold materials utilized to engineer OoC tissue components are critical parameters in modulating the phenotypes and behavior of the seeded cells, such as the redifferentiation of 2D expanded human chondrocytes [80]. Common scaffold materials used in OoC platforms are hydrogels, such as polyethyleneglycol [28], gelatin [39], fibrin [30], and collagen [81]. The popularity of hydrogels as tissue scaffolds in OoCs is mainly attributed to their ability to crosslink in situ and their high water content, which allows sufficient nutrient diffusion and timely waste removal during the culture process.
Box 4. Common approaches to creating tissues for joint OoCs.
The most widely utilized approach to generating tissues for joint OoCs is molding, typically via injecting and crosslinking cell-laden hydrogels in compartmented chambers. When molded tissue constructs are cultured in OoC devices, the dynamic, intricately modulated microenvironment of the system contributes to tissue maturation and maintenance [7]. OoCs thus have advantages over conventional cell culture platforms in the engineering of various tissues, particularly complex tissues, although the end-use of OoCs is not in tissue engineering. As the tissue compartments in microfluid chips are as small as tens to hundreds of microns in dimension, the cell, medium, and biomaterial consumptions are significantly lower than that typically required in conventional tissue cultures.
While OoC systems generate a precisely engineered microenvironment that attempts to recapitulate the complex in vivo conditions, organoids represent another rapidly developing biotechnology to engineer human tissues and organs. Organoids are generated following developmental biology principles. While organoid technology has garnered much success in modeling the development and disease of a long list of organs [82–86], a relatively small number of studies have been dedicated to studying joint biology and pathophysiology (Box 2, Figure IF-H) [36,87–91]. Organoids resembling different joint tissues can be derived from self-organizing stem cells [89,91,92], and common methods employed to generate articular tissue-mimicking organoids include microwell plate culture [13,22,91] and hanging drop culture [93]. Although organoid and OoC have fundamental differences, they can be synergistically combined to bring out the best of both worlds to establish joint-mimicking organoid-on-a-chip platforms. These systems are expected to faithfully recapitulate joint physiological functions and thus hold enormous potential in disease modeling, drug testing, and personalized medicine [94].
A prominent advantage of 3D bioprinting is the defined spatial arrangement of cells and biomaterials. With the capacity to print heterogeneous structures using multiple materials (inks) with or without encapsulated cells, 3D printing as a bottom-up approach can be employed to establish the entire OoC in a single step [95], leading to a streamlined fabrication process with higher efficiency. With the rapid development of biomimetic 4D printing technology [96], tissue constructs that enable spatiotemporal control over their geometry with external stimuli can be potentially printed in OoC systems to simulate tissue morphological changes in vivo.
Joint OoCs modeling inflammation
Inflammation is a key characteristic of almost all joint diseases, including OA and RA [7,8]. Most joint-mimicking OoCs developed thus simulated inflammation. For example, Ertl and colleagues have developed a cartilage-on-a-chip model [30], a synovium-on-a-chip platform [36], and a chondral-synovial co-culture chip model [29] to recapitulate joint inflammation.
We also modeled inflammation-mediated joint degeneration using osteochondral (OC) chips derived from human mesenchymal stem cells (MSCs, see Glossary) [38] and induced pluripotent stem cells (iPSCs, see Glossary) [39]. Building upon our success in creating OC chips, we recently established a four-tissue OoC named miniJoint, which incorporates the largest number of tissues among all joint-mimicking OoCs developed thus far [7]. It was found that IL-1β treatment of the synovial-like fibrous tissue (SFT) induced pathological changes locally and in all other tissues, indicating active tissue interactions. Furthermore, the miniJoint allows drug testing via either “systemic” or “intraarticular” administration by introducing compounds into all medium streams or only the synovial fluid-simulating SSF.
Joint OoCs simulating mechanical stimuli
Mechanical stimulation has long been employed to promote stem cell differentiation into various musculoskeletal lineages in OoCs [40]. In addition, fluid flow-induced shear stress has been reported to substantially enhance vascularization when endothelial cells are used to generate vascularized joint tissues [41]. On the flip side, chronic overloading and mechanical injury are major OA risk factors [4].
A cartilage-on-a-chip model was established by Occhetta and colleagues, and for the first time, OA traits were induced in an OoC through mechanical injury [28]. This novel device holds promise for the screening of potential DMOADs [42]. Lee and coworkers developed a pneumatic microfluidic chip containing an array of PDMS balloons located under chondrocyte-laden alginate constructs [43], which enabled investigations into the responses of chondrocytes to compressive strains of different magnitudes. A mechanical activator was recently built to simultaneously apply cyclic physiological or supra-physiological loading to up to 24 cartilage samples [44]. Yang and colleagues utilized human adipose-derived stromal cells embedded in fibrin to engineer 3D adipose tissues in a microfluidic chip, in which the engineered tissue showed interstitial shear stress-dependent function [45].
OoC devices established thus far can generate only uniaxial forces [28], while the biomechanical environment in the native joint is much more complex. For example, articular cartilage in the synovial joint is simultaneously exposed to compressive, hydrostatic, and shear stresses. Vainieri and colleagues [46], applying compressive and shear stresses, demonstrated the importance of both stress types in OoC cultures of cartilage and OC tissues. In OoC systems, the small tissue and chamber dimensions complicate the application of multi-axial mechanical forces. One strategy to circumvent external physical forces is applying biochemical agents to biologically regulate mechanotransduction pathways [47–49].
Joint OoCs modeling immune response
Immune cells are present in the subchondral bone, synovium, IPFP, and vasculature of the synovial joint [50–52]. To provide joint OoCs with an immune response requires the introduction of relevant cell types. For example, human monocytes used in a synovium-cartilage chip could cross the endothelial channel within the synovium tissue in response to a chemokine mix and OA synovial fluid [27]. In another study, human chondrocytes and THP-1 monocytes were seeded in two lateral channels separated by a central, hydrogel-filled channel in a cartilage chip [53]. When the monocytes were treated with lipopolysaccharide (LPS) for 5 h to simulate an inflammatory condition, degradation of collagen type II was observed in the chondrocyte culture. Recently, we developed an immune-responsive microphysiological bone model by co-encapsulating human macrophages, MSCs, and LPS-contaminated polyethylene particles in 3D gelatin scaffolds in a perfusion bioreactor [54]. This platform allowed us to investigate the modulation of bone healing by immune cells under inflammatory conditions. In a microfluidic model established by Ma and coworkers, human synovium SW982 cells in the center channel migrated to the side channels containing RAW264.7 mouse macrophages and mouse bone marrow MSCs, reproducing features of bone erosion mediated by fibroblast-like synoviocytes in RA [55].
Joint OoCs for studying genetic predisposition
An expanding repertoire of genetic, genomic and epigenetic factors associated with joint diseases has been identified in recent studies [4,56]. Two broad applications of OoCs in deciphering genetic predispositions to OA can be envisaged. First, gene function may be studied by creating “transgenic” OoCs, which will significantly reduce the use of animals. After introducing specific mutations to source cells, OoCs can be employed to culture joint tissues derived from the genetically modified cells to understand the role of the mutation(s) of interest in tissue development and function and interactions between the tissues. For this purpose, CRISPR/Cas9 gene editing can be performed on patient-specific iPSCs [57]. Second, comparative transcriptomics (see Glossary) of OoC tissues generated with cells from different patients and disease subgroups can help identify unknown genetic predispositions to joint disorders. Using primary human osteogenic cells and chondrocytes from four OA patients, Tuerlings and colleageus developed an OC tissue chip [58]; they further carried out RNA sequencing (RNA-seq) for preserved cartilage and subchondral bone from 15 additional OA patients to identify potential OA risk genes [59].
Perspectives
Validation of clinical relevance of joint OoCs
The increasing adoption of OoCs will undoubtedly refine and reduce the use of animal models in biomedical research [6]. It is, however, unlikely for OoCs to fully replace animal models in the short term as investigators struggle to adequately capture the complex physiology in the native knee joint, nor in the long term given the growing appreciation of the influence that even distant structures, such as the gastrointestinal tract, can have on joint [31]. Besides, it is not possible to replicate the joint pain-related behaviors in joint OoCs. Instead, depending on the specific application, a combinatorial strategy utilizing in vivo and select in vitro models is probably the most effective approach to mirror the complexity and heterogeneity of joint diseases. Given that the use of OoCs for joint disease modeling and drug testing is still in its early stage, it is paramount to cross-validate the data from OoC platforms with those collected from human patients (Figure 3A) [7]. Particularly, patient heterogeneity represents a key consideration in the validation of joint disease models. Biomarker-based patient stratification is particularly critical for a deeper understanding of the etiology and pathology specific to the patient subsets, thus enabling more accurate modeling of the pathological changes in OoCs than via conventiaonl one-size-fits-all approaches (Figure 4, Key Figure). The validation of these patient subgroup-specific models will also benefit from the advancement in disease modeling methods and our understanding of disease mechanisms.
Figure 3. Cross-validation of in vitro and in vivo models facilitates investigations into joint disease mechanisms and the development of potential treatments.
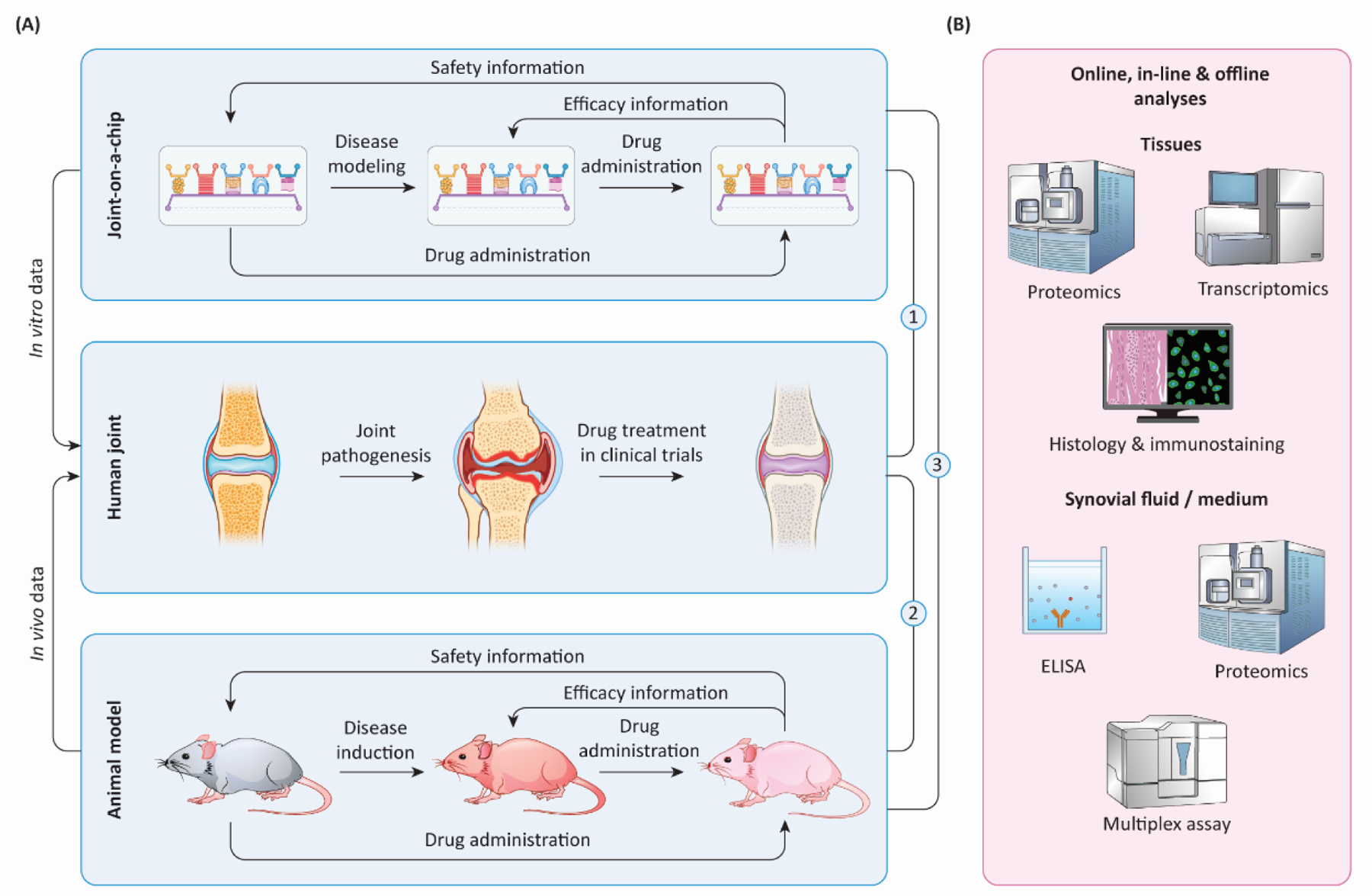
(A) Through simulating different aspects of arthritis in humans, both joint-on-a-chip (in vitro) and animal models (in vivo) provide valuable information on predicting drug safety and efficacy in humans. Before joint-on-a-chip can be used as a preclinical model, cross-check of treatment outcomes from three systems, indicated by ① ② ③, with clinically tested drugs, is essential. (B) To define the fidelity of joint-on-a-chip and animal models in simulating human arthritis and response to treatments, harvested tissues and fluids can be analyzed with different methods. ELISA: enzyme-linked immunosorbent assay.
Figure 4, Key Figure. Application of joint OoCs for precision medicine.
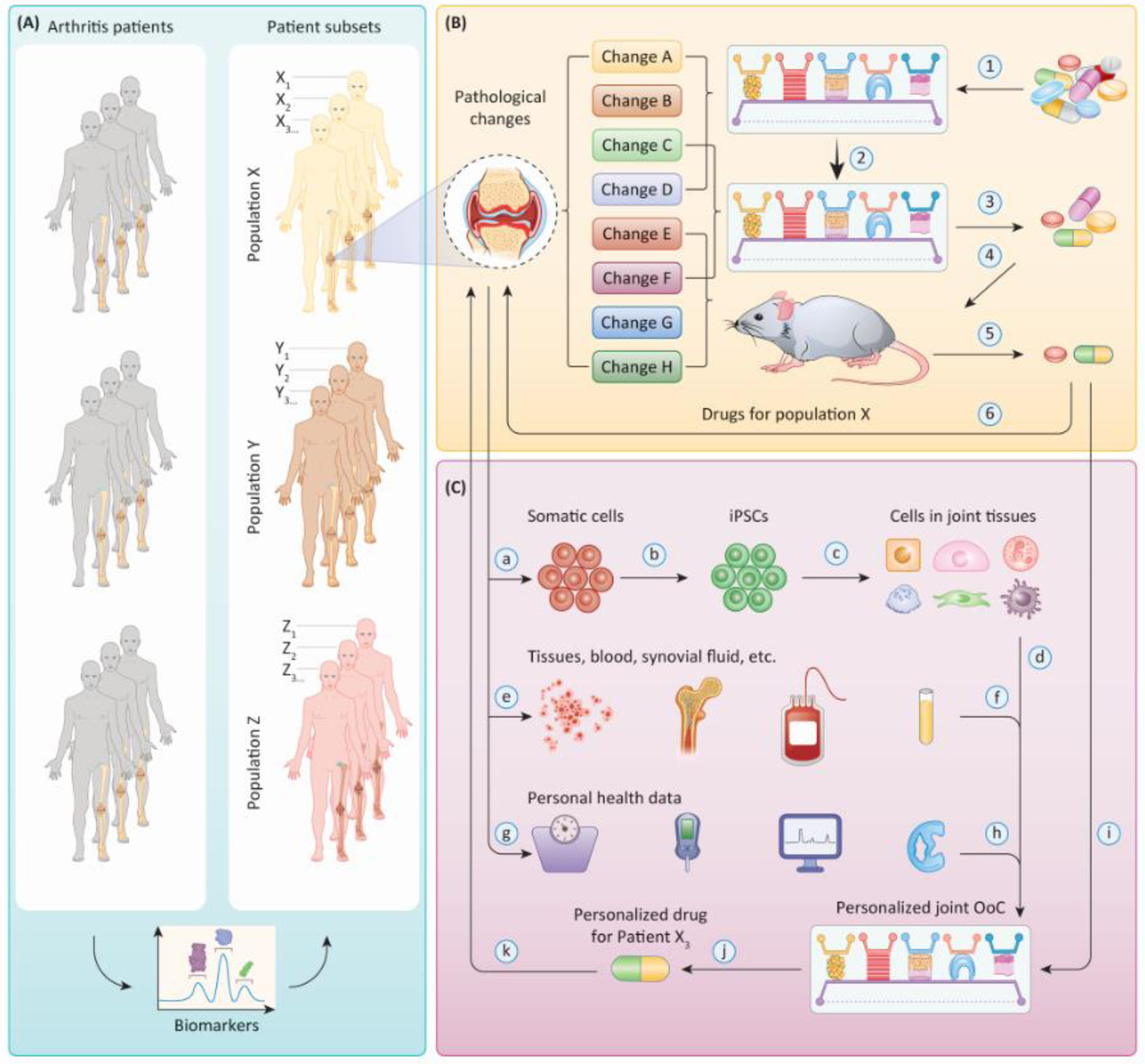
(A) Patient stratification. Arthritis patients can be stratified based on their biomarker profiles into different subsets (e.g., Population X, Y, Z). (B) Modeling of pathological changes. Joint OoCs and animal models can be established to recapitulate certain aspects of the pathological changes observed in a specific patient subpopulation (e.g., Population X) and utilized to evaluate the safety and efficacy of candidate drugs for this particular patient subset. Since not all features of the disease can be simulated in one type of OoC platform or animal model, the use of multiple models is expected. Then a drug library will undergo a series of screening processes using different models, and the candidates(s) with the best therapeutic potential are defined (Steps ① – ⑥ ). (C) Development of personalized medicine. Somatic cells can be collected from a specific individual (e.g., Patient X3) and reprogrammed into iPSCs to generate personalized joint OoC carrying the genetic background of that patient (Steps ⓐ–ⓓ). Samples of tissues, synovial fluid, and blood from the same patient can be cultured/tested in this personalized joint OoC (Steps ⓔ–ⓕ), and biochemical and biomechanical stimuli can be applied according to patient-specific health data and clinical manifestations (Steps ⓖ–ⓗ). Disease models established with such an OoC possess high patient specificity and clinical relevance, and can serve as a robust platform to identify efficacious personalized therapeutics (Steps ⓘ–ⓚ).
A range of techniques can be employed to collect data from in-line/online and end-point analyses of joint OoCs for validation purposes (Figure 3B) [60]. Tissue transcriptome revealed by bulk and single-cell RNA-seq provides a wealth of information on the pathophysiology of tissues of interest [7]. Currently, RNA-seq data on joint diseases were obtained mainly from the articular cartilage of animal models and human patients [16,61]. This is because of the critical role of articular cartilage in joint health and its relatively simple cellular composition (i.e., with chondrocytes being the only one cell type) compared to more complex and diverse cellular populations in tissues such as subchondral bone and synovium. Besides RNA-seq, proteomics (see Glossary) has also been carried out to analyze chondrocytes at the single-cell level [21]. As an example of cross-comparison, mass spectroscopy-based proteomic/phosphoproteomic profiling strategies were recently employed to analyze tissues derived from human patients, animal models, and OoC systems [62]. Histology and immunostaining are also among the commonly utilized methods for biomarker identification (Figure 3B). In addition, biomarkers of joint diseases are also frequently identified by examining the synovial fluid with methods such as immunoassays (e.g., enzyme-linked immunosorbent assay and Multiplex assay) [63] and mass spectrometry (Figure 3B) [64]. These techniques can also be utilized to examine the SSF in joint OoCs [7]. Comparative analyses of the compositional changes in SSF and the synovial fluid of human patients offer valuable insights into the clinical relevance (see Glossary) of OoC disease models. Similarly, by benchmarking the results from joint OoC platforms and animal models against human patient data, valuable information can be gained to gauge the relative physiological relevance of these models [7,62]. One challenge, however, is that donor tissues from human joints, already of low availability, are primarily derived from patients with advanced joint diseases [22]. The scarcity of joint tissues from patients without or with early-stage joint diseases presents a major hurdle to validating OoCs mimicking healthy joints or studying disease onset.
As discussed above, biomarkers of joint disorders come under many different guises. Thus, it is important that molecular signatures comprising a panel of biomarkers are generated to guide clinical decision-making. With virtually unlimited “biopsies” and culture medium that OoCs can offer, we can envision accelerated discoveries of joint disease markers, paving the way for more accurate delineation of disease mechanisms and patient stratification. Furthermore, the biomarkers unearthed in OoC models can be potentially translated to clinical diagnosis, thus contributing to timely therapeutic interventions without missing the optimum treatment window [3].
Drug evaluation in joint OoCs via different administration routes
Drugs targeting joint diseases can be administered via systemic or intraarticular routes [7]. Those administered by oral administration or intravascular injection enter the circulation and exert systemic effects. Some medications are not suitable for oral delivery because of their poor oral bioavailability or concerns of toxicity to other tissues, such as the gastrointestinal system. Such drugs are generally delivered by intraarticular injection, which achieves high concentrations of the locally administered compounds. Challenges facing the intraarticular delivery route include the rapid turnover of synovial fluid (hence fast drug clearance) and the biological barrier of a dense, negatively charged cartilage extracellular matrix (ECM). Most OA drugs are designed to act on chondrocytes, meaning that they need to penetrate through the cartilage ECM, which possesses an average pore size of only ~3–4 nm (spacing between adjacent glycosaminoglycans within the collagen network) [65]. Nevertheless, since OA affects all joint tissues, other cell types in the joint, such as fibroblasts and macrophages in the synovium, are also possible targets [66].
A joint-on-a-chip platform can be designed to accommodate different drug administration routes. To simulate intraarticular injection, to-be-tested compounds can be introduced to the shared “synovial fluid-simulating” stream without being directly exposed to the subchondral bone [7]. The flow rate can be tuned to match the synovial fluid turnover rate in the native joint. To simulate systemic administration, the therapeutic agents can be added in all medium streams to reach all joint tissues, including the subchondral bone. Furthermore, the systemic drug effects can be extended via administration to OoCs that mimic other organs integrated with the joint OoC via microfluidic connection [23]. These fluidically linked systems can be employed to simulate the pharmacokinetics and pharmacodynamics of drugs and potentially predict the compounds’ safety and efficacy in human patients [26].
Physiological relevance or throughput: a need-dependent choice
An ideal OoC platform is physiologically relevant and possesses the capacity for high-throughput screening [26]. However, these two characteristics are often mutually exclusive (Figure 1). Highly physiologically relevant OoCs generally comprise multiple tissues to allow their crosstalk and thus require complicated design and increased resource consumption, making it uneconomical and unrealistic to upscale the manufacture and maintenance. However, high physiological relevance is not always the priority. Given that many compounds are removed from further consideration due to overt adverse events in the early stage of drug screening, drug testing tools with a quick turnaround at a low cost are desirable. Owing to the significance of cartilage in joint disease treatment [4], an OoC platform containing solely cartilage might suffice at the early stages. This obviates the need for multiple medium types or sophisticated tissue arrangements usually needed in a multi-tissue system [7]. When the number of candidate drugs has been reduced to, e.g., several hundred, given the active communications between articular cartilage and the subchondral bone [39], an osseous component can be introduced to establish an OC chip for the next phase of selection. With an even smaller number (e.g., less than 100) of compounds, additional tissues such as the synovium and IPFP can then be incorporated [7]. By now, the principal objective of drug testing has shifted from safety evaluation to efficacy assessment on a molecular basis for establishing the mechanism of action. Finally, with further downselection of candidate drugs, tissue compartments mimicking the tendon, ligament, and meniscus may be included to comprehensively examine the effects of each compound on all joint tissues in the context of the active interplay between them.
Personalized joint OoCs for precision medicine
Like other joint disease models, an OoC alone cannot capture all facets of the disease [6,25]. Considering the heterogeneity of joint diseases and the need for personalized treatment, it is important to use representative OA/RA models with different etiologies and pathological phenotypes in OoCs (Figure 4A, B). Patient-specific primary cells and iPSCs can be used to engineer tissue components of joint OoCs that carry the genetic backgrounds of specific patients and patient subpopulations (Figure 4C) [24]. As aging is a prominent risk factor for OA development, aging/senescent cells generated by, for example, induction with biochemical stress or extensive in vitro expansion [22], can be employed to generate joint OoCs with aging phenotypes for comparative studies on “young” and “old” joints in OoCs. Samples of tissues, synovial fluid, and blood can also be directly used in joint OoCs to reflect donor-specific physiology (Figure 4C). The availability of these samples, however, is limited and thus should probably be resorted to only in the final stages of developing personalized therapeutics.
Besides cell and tissue sources, the various biophysical and biochemical aspects of the OoC microenvironment are also major parameters that can be tuned in accordance with clinical manifestations and personal health data (Figure 4C) [67]. For example, to model post-traumatic OA caused by different injurious events, mechanical insults can be introduced to ligament, meniscus, or cartilage compartments in a joint OoC [28]. Obesity-associated OA can be recapitulated by incorporating obese adipose tissue in joint OoCs to study, for example, the roles of low-grade inflammation and diabetes mellitus in OA onset and progression [14].
Once validated, personalized joint OoCs can provide valuable patient-specific readouts that have rarely been obtainable from traditional in vitro and in vivo models [4]. Such systems can then be used to test precision medicine tailored to individual physiology and disease state (Figure 4C). Furthermore, by integrating a joint OoC with OoCs derived from the same patient (patient subset) and recapitulating other organs, a personalized body-on-a-chip system can be generated to study patient-specific comorbidities in joint disorders and test the systemic effects of therapeutics [23,26].
Concluding Remarks
The past few years have witnessed the rapid development of joint-mimicking OoCs, and exciting progress has been made in the fabrication and application of these novel systems. Joint organ-on-a-chip systems established so far have led to exciting findings on several key facets of joint disorders, including inflammation and mechanical injuries. However, there is still a paucity of multi-tissue joint OoCs with sufficient complexity and physiological relevance required in mechanistic studies. In addition, a good track record of predictive power is yet to be established for joint OoCs (see Outstanding Questions). The convergence of OoCs and other biotechnologies, including 3D printing, organoid, iPSC, and gene editing, is just beginning. The immense potential of joint OoC platforms is yet to be unleashed, which is expected to generate new insights into heterogeneous disease mechanisms, bring about a new paradigm for drug development and toxicity testing, and accelerate the clinical applications of precision medicine and personalized therapies for joint diseases.
Outstanding Questions.
Can a universal medium, or blood mimetic, be developed for all joint tissues to maintain their respective phenotypes and undergo active, reciprocal crosstalk in a joint organ-on-a-chip (OoC)?
To enable high-throughput screening, what technologies should we adopt to scale up OoC production and assess tissue status in a non-invasive manner?
How can we validate the clinical relevance of joint OoCs and demonstrate their predictive power and accuracy in preclinical evaluations of drugs?
Will “transgenic” joint (OoC) systems enable us to understand genetic predispositions to arthritis?
Besides using patient-specific cells, what other components are needed to establish personalized joint OoCs to advance precision medicine?
Highlights.
A recently developed human stem cell-derived, joint-mimicking organ-on-a-chip (OoC) comprised of four three-dimensional tissues demonstrated its capability in modeling joint inflammation and testing potential disease-modifying drugs.
Joint OoCs developed thus far fall into four broad categories, including those modeling inflammation, simulating mechanical stimuli, recapitulating immune response, and studying genetic disposition.
Accumulating evidence reveals the active, bidirectional crosstalk between different joint elements, highlighting the importance of incorporating multiple tissue components in joint OoCs.
Recent advances in induced pluripotent stem cell-based tissue engineering pave the way for building personalized joint OoC to address patient heterogeneity.
Acknowledgments
The authors are grateful to their lab members who contributed to joint OoC-related research over the past five years. This work was supported by funding from the National Institutes of Health (UG3/UH3TR002136 to RST (UG3 phase) and HL (UH3 phase), UG3/UH3TR003090 to MSG and HL), the U.S. Department of Defense (W81XWH2010902), and the Pitt Momentum Funds from the University of Pittsburgh (Teaming Grants to HL). RST is supported by the Lee Quo Wei and Lee Yik Hoi Lun Professorship in Tissue Engineering and Regenerative Medicine. Figure 1, Figure 2, Figure 3, and Figure 4 were created with elements from Biorender.com.
Glossary
- Arthritis
a disease characterized by joint pain, swelling, stiffness, and decreased range of motion. There are many types of arthritis, with osteoarthritis and rheumatoid arthritis being the most common
- Clinical relevance
a measurement of the effectiveness of a particular treatment based on whether there is a noticeable change in a patient’s clinical status. It is also referred to as clinical significance
- Genetic predisposition
an increased likelihood of developing a certain disease based on genetic characteristics. It is also referred to as genetic susceptibility
- Induced pluripotent stem cell (iPSC)
pluripotent stem cells that are generated by genetically reprogramming adult somatic cells, which can self-renew and differentiate into cell types from all three germ layers
- Mesenchymal stem cell (MSC)
non-haematopoietic adult stromal cells that can self-renew, adhere to tissue culture plastic, and differentiate into multiple lineages such as osteocytes, chondrocytes, and adipocytes
- Microfluidic device
an instrument containing a set of microchannels (typically submillimeter in size) etched or molded into a material (glass, silicon, or polymer such as polydimethylsiloxane) for processing low volumes of fluids. It is often characterized by a high surface-to-volume ratio
- Organ-on-a-chip (OoC)
a miniaturized, in vitro cell culture platform that establishes a minimally but appropriately functional unit to recapitulate specific functions of their in vivo counterparts under healthy or pathological conditions. It is typically established in a microfluidic device, but the OoCs at millimeter scale are also seen
- Organoid
a self-organizing, self-renewing three-dimensional cell aggregate produced in vitro from primary tissues or stem cells, which is a miniaturized and simplified version of an organ and exhibits organ functionality
- Patient heterogeneity
variation between patients that can be explained by certain patient characteristics, such as age, sex, disease stage, disease history, genetic profile, etc
- Personalized medicine
practice of medicine tailored to individual patients based on their genetic profile and other biomarker information to guide clinical decision-making to achieve optimal outcomes. It is also referred to as precision medicine
- Physiological relevance
the characteristic of normal or healthy biological functioning
- Proteomics
a branch of biotechnology concerned with the systemic, large-scale identification and analysis of the entire set of proteins present in a cell, tissue, or organism
- Synovial joint
joints characterized by the presence of a fluid-filled joint cavity/space, the walls of which are formed by the articular capsule lined by the synovium. It is the most common and mobile joint type in the human body
- Transcriptomics
comprehensive analysis of the whole set of RNA transcripts for a cell, tissue, or organism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Safiri S et al. (2020) Global, regional and national burden of osteoarthritis 1990–2017: a systematic analysis of the Global Burden of Disease Study 2017. Ann. Rheum. Dis 79, 819–828 [DOI] [PubMed] [Google Scholar]
- 2.Almutairi K et al. (2021) The global prevalence of rheumatoid arthritis: a meta-analysis based on a systematic review. Rheumatol. Int 41, 863–877 [DOI] [PubMed] [Google Scholar]
- 3.Camacho-Encina M et al. (2019) Discovery of an autoantibody signature for the early diagnosis of knee osteoarthritis: data from the Osteoarthritis Initiative. Annals of the Rheumatic Diseases 78, 1699–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He Y et al. (2020) Pathogenesis of osteoarthritis: risk factors, regulatory pathways in chondrocytes, and experimental models. Biology 9, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Matteo B et al. (2021) Osteoarthritis: an ancient disease, an unsolved conundrum. Int. Orthop 45, 313–317 [DOI] [PubMed] [Google Scholar]
- 6.Makarczyk MJ et al. (2021) Current models for development of disease-modifying osteoarthritis drugs. Tissue Eng. Part C Methods 27, 124–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z et al. (2022) Human mesenchymal stem cell-derived miniature joint system for disease modeling and drug testing. Adv. Sci, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nygaard G and Firestein GS (2020) Restoring synovial homeostasis in rheumatoid arthritis by targeting fibroblast-like synoviocytes. Nat. Rev. Rheumatol 16, 316–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humby F et al. (2019) Synovial cellular and molecular signatures stratify clinical response to csDMARD therapy and predict radiographic progression in early rheumatoid arthritis patients. Ann. Rheum. Dis 78, 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buch MH (2018) Defining refractory rheumatoid arthritis. Ann. Rheum. Dis 77, 966–969 [DOI] [PubMed] [Google Scholar]
- 11.Van den Bosch F and Coates L (2018) Clinical management of psoriatic arthritis. Lancet 391, 2285–2294 [DOI] [PubMed] [Google Scholar]
- 12.Masters EA et al. (2022) Skeletal infections: microbial pathogenesis, immunity and clinical management. Nat. Rev. Microbiol, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Y et al. (2021) Generating 3D-cultured organoids for pre-clinical modeling and treatment of degenerative joint disease. Signal Transduct. Target Ther 6, 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins KH et al. (2021) Adipose tissue is a critical regulator of osteoarthritis. Proc. Natl. Acad. Sci. U. S. A 118, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schott EM et al. (2018) Targeting the gut microbiome to treat the osteoarthritis of obesity. JCI Insight 3, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smeriglio P et al. (2020) Inhibition of TET1 prevents the development of osteoarthritis and reveals the 5hmC landscape that orchestrates pathogenesis. Sci. Transl. Med 12, eaax2332. [DOI] [PubMed] [Google Scholar]
- 17.Doyran B et al. (2017) Nanoindentation modulus of murine cartilage: a sensitive indicator of the initiation and progression of post-traumatic osteoarthritis. Osteoarthritis Cartilage 25, 108–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao L et al. (2020) Acute synovitis after trauma precedes and is associated with osteoarthritis onset and progression. Int J Biol Sci 16, 970–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He H et al. (2019) Survey of clinical translation of cancer nanomedicines—lessons learned from successes and failures. Acc. Chem. Res 52, 2445–2461 [DOI] [PubMed] [Google Scholar]
- 20.Guan Y et al. (2019) Machine learning to predict anti–tumor necrosis factor drug responses of rheumatoid arthritis patients by integrating clinical and genetic markers. Arthritis Rheum 71, 1987–1996 [DOI] [PubMed] [Google Scholar]
- 21.Grandi FC et al. (2020) Single-cell mass cytometry reveals cross-talk between inflammation-dampening and inflammation-amplifying cells in osteoarthritic cartilage. Sci. Adv 6, eaay5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang N et al. (2022) Engineering osteoarthritic cartilage model through differentiating senescent human mesenchymal stem cells for testing disease-modifying drugs. Sci. China Life Sci 65, 309–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jalili-Firoozinezhad S et al. (2021) Modeling the human body on microfluidic chips. Trends Biotechnol 39, 838–852 [DOI] [PubMed] [Google Scholar]
- 24.Liu C et al. (2018) Modeling human diseases with induced pluripotent stem cells: from 2D to 3D and beyond. Development 145, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vunjak-Novakovic G et al. (2021) Organs-on-a-chip models for biological research. Cell 184, 4597–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novak R et al. (2020) Robotic fluidic coupling and interrogation of multiple vascularized organ chips. Nat. Biomed. Eng 4, 407–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mondadori C et al. (2021) Recapitulating monocyte extravasation to the synovium in an organotypic microfluidic model of the articular joint. Biofabrication 13, 045001. [DOI] [PubMed] [Google Scholar]
- 28.Occhetta P et al. (2019) Hyperphysiological compression of articular cartilage induces an osteoarthritic phenotype in a cartilage-on-a-chip model. Nat. Biomed. Eng 3, 545–557 [DOI] [PubMed] [Google Scholar]
- 29.Rothbauer M et al. (2021) Establishment of a human three-dimensional chip-based chondro-synovial coculture joint model for reciprocal cross talk studies in arthritis research. Lab Chip 21, 4128–4143 [DOI] [PubMed] [Google Scholar]
- 30.Rosser J et al. (2019) Microfluidic nutrient gradient–based three-dimensional chondrocyte culture-on-a-chip as an in vitro equine arthritis model. Mater. Today Bio 4, 100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Artacho A et al. (2021) The pretreatment gut microbiome is associated with lack of response to methotrexate in new-onset rheumatoid arthritis. Arthritis Rheum 73, 931–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell SB et al. (2021) Beyond polydimethylsiloxane: Alternative materials for fabrication of organ-on-a-chip devices and microphysiological systems. ACS Biomater. Sci. Eng 7, 2880–2899 [DOI] [PubMed] [Google Scholar]
- 33.Matai I et al. (2020) Progress in 3D bioprinting technology for tissue/organ regenerative engineering. Biomaterials 226, 119536. [DOI] [PubMed] [Google Scholar]
- 34.Weisgrab G et al. (2019) Functional 3D printing for microfluidic chips. Adv. Mater. Technol 4, 1900275 [Google Scholar]
- 35.Novak R et al. (2018) Scalable fabrication of stretchable, dual channel, microfluidic organ chips. JoVE, e58151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothbauer M et al. (2020) Monitoring tissue-level remodelling during inflammatory arthritis using a three-dimensional synovium-on-a-chip with non-invasive light scattering biosensing. Lab Chip 20, 1461–1471 [DOI] [PubMed] [Google Scholar]
- 37.Homan KA et al. (2016) Bioprinting of 3D convoluted renal proximal tubules on perfusable chips. Sci. Rep 6, 34845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin H et al. (2014) Stem cell-based microphysiological osteochondral system to model tissue response to interleukin-1β. Mol. Pharm 11, 2203–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin Z et al. (2019) Osteochondral tissue chip derived from iPSCs: modeling OA pathologies and testing drugs. Front. Bioeng. Biotechnol 7, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee J et al. (2020) Combinatorial screening of biochemical and physical signals for phenotypic regulation of stem cell–based cartilage tissue engineering. Sci. Adv 6, eaaz5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park D et al. (2020) Integrating organs-on-chips: Multiplexing, scaling, vascularization, and innervation. Trends Biotechnol 38, 99–112 [DOI] [PubMed] [Google Scholar]
- 42.Collison J (2019) Cartilage-on-a-chip to aid OA drug development. Nat. Rev. Rheumatol 15, 511. [DOI] [PubMed] [Google Scholar]
- 43.Lee D et al. (2018) Pneumatic microfluidic cell compression device for high-throughput study of chondrocyte mechanobiology. Lab Chip 18, 2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capuana E et al. (2021) A high-throughput mechanical activator for cartilage engineering enables rapid screening of in vitro response of tissue models to physiological and supra-physiological loads. Cells Tissues Organs, 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang F et al. (2021) A 3D human adipose tissue model within a microfluidic device. Lab Chip 21, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vainieri ML et al. (2018) Mechanically stimulated osteochondral organ culture for evaluation of biomaterials in cartilage repair studies. Acta Biomater 81, 256–266 [DOI] [PubMed] [Google Scholar]
- 47.López-Jiménez C et al. (2022) TRPV4 activation enhances compressive properties and glycosaminoglycan deposition of equine neocartilage sheets. Osteoarthr. Cartil. Open 4, 100263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee W et al. (2014) Synergy between Piezo1 and Piezo2 channels confers high-strain mechanosensitivity to articular cartilage. Proceedings of the National Academy of Sciences 111, E5114–E5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee W et al. (2021) Inflammatory signaling sensitizes Piezo1 mechanotransduction in articular chondrocytes as a pathogenic feed-forward mechanism in osteoarthritis. Proceedings of the National Academy of Sciences 118, e2001611118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romero-López M et al. (2020) Macrophage effects on mesenchymal stem cell osteogenesis in a three-dimensional in vitro bone model. Tissue Eng. Part A 26, 1099–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Donnell , B.T. et al. (2020) Adipose tissue-derived stem cells retain their adipocyte differentiation potential in three-dimensional hydrogels and bioreactors. Biomolecules 10, 1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao Q et al. (2021) Macrophages modulate the function of MSC- and iPSC-derived fibroblasts in the presence of polyethylene particles. Int. J. Mol. Sci 22, 12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oliveira IM et al. (2021) Modulation of inflammation by anti-TNF α mAb-dendrimer nanoparticles loaded in tyramine-modified gellan gum hydrogels in a cartilage-on-a-chip model. J. Mater. Chem. B 9, 4211–4218 [DOI] [PubMed] [Google Scholar]
- 54.Gao Q et al. (2021) The effects of macrophage phenotype on osteogenic differentiation of MSCs in the presence of polyethylene particles. Biomedicines 9, 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma H-P et al. (2018) A microfluidic chip-based co-culture of fibroblast-like synoviocytes with osteoblasts and osteoclasts to test bone erosion and drug evaluation. R. Soc. Open Sci 5, 180528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ratneswaran A and Kapoor M (2021) Osteoarthritis year in review: genetics, genomics, epigenetics. Osteoarthr. Cartil 29, 151–160 [DOI] [PubMed] [Google Scholar]
- 57.Lilianty J et al. (2021) Generation of a heterozygous COL2A1 (p.G1113C) hypochondrogenesis mutation iPSC line, MCRIi019-A-7, using CRISPR/Cas9 gene editing. Stem Cell Res 56, 102515. [DOI] [PubMed] [Google Scholar]
- 58.Tuerlings M et al. (2022) Capturing essential physiological aspects of interacting cartilage and bone tissue with osteoarthritis pathophysiology: A human osteochondral unit-on-a-chip model. Adv. Mater. Technol. n/a, 2101310 [Google Scholar]
- 59.Tuerlings M et al. (2021) Development of a human osteochondral construct on a microfluidic chip–to advance functional studies of osteoarthritis risk genes. Osteoarthr. Cartil 29, S108–S109 [Google Scholar]
- 60.Marrero D et al. (2021) Gut-on-a-chip: Mimicking and monitoring the human intestine. Biosensors and Bioelectronics 181, 113156. [DOI] [PubMed] [Google Scholar]
- 61.Sebastian A et al. (2021) Single-Cell RNA-Seq Reveals Transcriptomic Heterogeneity and Post-Traumatic Osteoarthritis-Associated Early Molecular Changes in Mouse Articular Chondrocytes. Cells 10, 1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuzmanov U et al. (2020) Mapping signalling perturbations in myocardial fibrosis via the integrative phosphoproteomic profiling of tissue from diverse sources. Nat. Biomed. Eng 4, 889–900 [DOI] [PubMed] [Google Scholar]
- 63.Tang R et al. (2020) Gene therapy for follistatin mitigates systemic metabolic inflammation and post-traumatic arthritis in high-fat diet–induced obesity. Sci. Adv 6, eaaz7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carlson AK et al. (2019) Characterization of synovial fluid metabolomic phenotypes of cartilage morphological changes associated with osteoarthritis. Osteoarthr. Cartil 27, 1174–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ng L et al. (2003) Individual cartilage aggrecan macromolecules and their constituent glycosaminoglycans visualized via atomic force microscopy. Journal of Structural Biology 143, 242–257 [DOI] [PubMed] [Google Scholar]
- 66.Fan Y et al. (2022) Exosomes in the Pathogenesis, Progression, and Treatment of Osteoarthritis. Bioengineering 9, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Den Berg A et al. (2019) Personalised organs-on-chips: functional testing for precision medicine. Lab Chip 19, 198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu W et al. (2021) Microenvironment in subchondral bone: predominant regulator for the treatment of osteoarthritis. Ann. Rheum. Dis 80, 413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jusoh N et al. (2015) Microfluidic vascularized bone tissue model with hydroxyapatite-incorporated extracellular matrix. Lab Chip 15, 3984–3988 [DOI] [PubMed] [Google Scholar]
- 70.Pirosa A et al. (2021) An in vitro chondro-osteo-vascular triphasic model of the osteochondral complex. Biomaterials 272, 120773. [DOI] [PubMed] [Google Scholar]
- 71.Marturano-Kruik A et al. (2018) Human bone perivascular niche-on-a-chip for studying metastatic colonization. Proc. Natl. Acad. Sci. U.S.A 115, 1256–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li C et al. (2019) Buoyancy-driven gradients for biomaterial fabrication and tissue engineering. Adv. Mater 31, 1900291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sant S et al. (2010) Biomimetic gradient hydrogels for tissue engineering. The Canadian Journal of Chemical Engineering 88, 899–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robinson WH et al. (2016) Low-grade inflammation as a key mediator of the pathogenesis of osteoarthritis. Nat. Rev. Rheumatol 12, 580–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Loskill P et al. (2017) WAT-on-a-chip: a physiologically relevant microfluidic system incorporating white adipose tissue. Lab Chip 17, 1645–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tanataweethum N et al. (2018) Establishment and characterization of a primary murine adipose tissue-chip. Biotechnology and Bioengineering 115, 1979–1987 [DOI] [PubMed] [Google Scholar]
- 77.Lin H et al. (2019) Bone marrow mesenchymal stem cells: aging and tissue engineering applications to enhance bone healing. Biomaterials 203, 96–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Z et al. (2021) Graphene oxide-functionalized nanocomposites promote osteogenesis of human mesenchymal stem cells via enhancement of BMP-SMAD1/5 signaling pathway. Biomaterials 277, 121082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamanaka S (2012) Induced pluripotent stem cells: past, present, and future. Cell Stem Cell 10, 678–684 [DOI] [PubMed] [Google Scholar]
- 80.Bachmann B et al. (2020) Stiffness matters: Fine-tuned hydrogel elasticity alters chondrogenic redifferentiation. Front. Bioeng. Biotechnol 8, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Varone A et al. (2021) A novel organ-chip system emulates three-dimensional architecture of the human epithelia and the mechanical forces acting on it. Biomaterials 275, 120957. [DOI] [PubMed] [Google Scholar]
- 82.Qian X et al. (2019) Brain organoids: advances, applications and challenges. Development 146, dev166074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miller AJ et al. (2019) Generation of lung organoids from human pluripotent stem cells in vitro. Nat. Protoc 14, 518–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Phipson B et al. (2019) Evaluation of variability in human kidney organoids. Nat. Methods 16, 79–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Broutier L et al. (2016) Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat. Protoc 11, 1724–1743 [DOI] [PubMed] [Google Scholar]
- 86.Puschhof J et al. (2021) Intestinal organoid cocultures with microbes. Nat. Protoc 16, 4633–4649 [DOI] [PubMed] [Google Scholar]
- 87.Strobel HA et al. (2021) Vascularized adipocyte organoid model using isolated human microvessel fragments. Biofabrication 13, 035022. [DOI] [PubMed] [Google Scholar]
- 88.Chen S et al. (2022) The horizon of bone organoid: A perspective on construction and application. Bioact. Mater 18, 15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu Y et al. (2019) Robust bone regeneration through endochondral ossification of human mesenchymal stem cells within their own extracellular matrix. Biomaterials 218, 119336. [DOI] [PubMed] [Google Scholar]
- 90.Nilsson Hall G et al. (2020) Developmentally engineered callus organoid bioassemblies exhibit predictive in vivo long bone healing. Adv. Sci 7, 1902295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hall GN et al. (2021) Patterned, organoid-based cartilaginous implants exhibit zone specific functionality forming osteochondral-like tissues in vivo. Biomaterials 273, 120820. [DOI] [PubMed] [Google Scholar]
- 92.O’Connor SK et al. (2021) Formation of osteochondral organoids from murine induced pluripotent stem cells. Tissue Eng Part A 27, 1099–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klingelhutz AJ et al. (2018) Scaffold-free generation of uniform adipose spheroids for metabolism research and drug discovery. Sci. Rep 8, 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Takebe T et al. (2017) Synergistic engineering: Organoids meet organs-on-a-chip. Cell Stem Cell 21, 297–300 [DOI] [PubMed] [Google Scholar]
- 95.Lee H and Cho D-W (2016) One-step fabrication of an organ-on-a-chip with spatial heterogeneity using a 3D bioprinting technology. Lab Chip 16, 2618–2625 [DOI] [PubMed] [Google Scholar]
- 96.Hu Y et al. (2020) Botanical-inspired 4D printing of hydrogel at the microscale. Adv. Funct. Mater 30, 1907377 [Google Scholar]


