Summary
Gene regulation is a central topic in cell biology. Advances in omics technologies and the accumulation of omics data have provided better opportunities for gene regulation studies than ever before. For this reason deep learning, as a data-driven predictive modeling approach, has been successfully applied to this field during the past decade. In this article, we aim to give a brief yet comprehensive overview of representative deep-learning methods for gene regulation. Specifically, we discuss and compare the design principles and datasets used by each method, creating a reference for researchers who wish to replicate or improve existing methods. We also discuss the common problems of existing approaches and prospectively introduce the emerging deep-learning paradigms that will potentially alleviate them. We hope that this article will provide a rich and up-to-date resource and shed light on future research directions in this area.
Keywords: deep learning, neural network, gene regulation, omics
In this review, Li et al. comprehensively discuss deep-learning applications for understanding gene regulation at various omics levels. They point out the problems and challenges of deep learning in such applications and ways to alleviate them. Emerging deep-learning methods and prospective applications are also discussed.
Introduction
Understanding gene regulation is a central topic in cell biology. Gene regulation in eukaryotes takes place at various stages of the central dogma, including the genomic level, transcriptomic level, and proteomic level. During the past two decades, advances in omics technologies, including those in genomics, transcriptomics, and proteomics, have enabled a better systematic understanding of multiple levels of gene regulation than ever before. Developments in microarray, DNA and RNA sequencing, mass spectrometry, and single-cell technologies have provided foundations for experimental techniques to study gene regulation at a greater scale and finer resolution. This includes techniques such as chromatin immunoprecipitation sequencing (ChIP-seq)1 for protein-DNA binding, cross-linking immunoprecipitation sequencing (CLIP-seq)2,3 for protein-RNA binding, DNase I hypersensitive sites sequencing (DNase-seq)4 and assay for transposase-accessible chromatin using sequencing (ATAC-seq)5 for genomic-wide chromatin accessibility, high-throughput RNA sequencing (RNA-seq) for gene expression level profiling, and 3′ region extraction and deep sequencing (3′-READS)6 for polyadenylation. In the meantime, large omics projects utilizing such techniques, such as the 1000 Genomes Project,7 Encyclopedia of DNA Elements (ENCODE),8 Roadmap Epigenomics,9 and the Genotype-Tissue Expression (GTEx) project,10 have been launched to decipher biological processes at various levels from genotype to phenotype in various individuals, species, and tissue types. Concurrently, omics data from individual studies are continuously uploaded to and collected by publicly accessible databases such as the Sequence Read Archive (SRA),11 the European Nucleotide Archive (ENA),12 and the UniProt Archive (UniParc).13 The aforementioned projects and databases have proved to be invaluable resources for omics studies because they not only support discoveries in the original studies but also enable continued analysis by independent researchers.
As a result, the requirement of analytical algorithms to process, interpret, and discover patterns in omics data has been stronger than ever before. Statistical learning-based data-mining algorithms, such as logistic regression (LR), support vector machines (SVM), and hidden Markov models, have been extensively applied in omics since its inception.14,15 Such algorithms are sometimes termed “shallow learning” algorithms because they operate on extracted features from an object of interest and run only a few inference steps as specified by a pre-determined statistical model. Although effective, such models rely heavily on how those features are engineered. A good feature engineering technique that captures a highly discriminative pattern will result in much better performance than those that overlook them. In the fields where knowledge of such patterns is limited (as is usually the case in omics), feature engineering-based machine-learning algorithms using a priori knowledge usually fail to take care of important aspects that are beyond our current understanding. Relying on feature engineering will result in degraded performance and potentially miss new discoveries. Such an approach could also result in poor model generalizability, as features used in one scenario may not be as effective in other cases. It would be very much appreciated if such discriminative features can be automatically discovered by the learning algorithms directly from the data themselves.
Since 2012, deep learning has achieved remarkable success in various other fields via a data-driven approach.16 Deep learning is a general term for machine-learning algorithms that are made up of deep neural networks (DNNs). DNNs consist of multiple artificial neural network layers, which are biologically inspired data-processing units that serve as non-linear transformation functions of their inputs. As each layer takes as input the result from the previous layer, the transformation becomes increasingly complex when the number of layers increases. Those functions are learnable in the sense that they can adjust themselves during the “training” process. Deep-learning models are usually trained by fitting themselves on the training data through the optimization of an objective function (the “loss function”) using the gradient descent algorithm.16 It is then expected to be able to perform inference tasks on data that come from the same or similar statistical distribution as the training data. Although the success of deep learning is in part due to its learning capacity, generalizability, and computational efficiency on dedicated computational architectures, the most important aspect is its representation learning ability. In contrast to the “shallow learning” algorithms, deep learning, based on DNNs, perform inference tasks with a deep and hierarchical architecture. The lower layers in the hierarchy learn the “representations,” which are highly discriminative features discovered by the algorithm using a data-driven approach. Its higher layers summarize the representations from the lower layers and produce the result of the inference. This makes deep learning especially useful in omics because it overcomes the limitations of the “shallow learning” methods and could discover patterns in biological sequences or measurements that are yet unknown. This is undoubtedly one of the reasons why many successful deep-learning applications in omics have emerged in the past decade. In addition, copious omics data take a form that is amenable to being processed by deep-learning algorithms. For example, there is a similarity between biological sequences and natural languages. Certain sequence motifs serve as regulatory codes, and the interactions between such codes serve as the regulatory grammar. This has led to numerous successful biological applications of off-the-shelf deep-learning models.17,18,19 Returning to the topic of gene regulation, it will be of great interest to find out whether deep learning in omics can decipher the regulatory code and grammar, model the regulatory process, help us understand the regulatory mechanism, and assist us in achieving the major goals of omics.
In this survey and perspective, we aim to give a brief yet systematic review of the application of deep learning in gene regulation studies with various kinds of omics data. We will cover applications of deep learning at various omics levels, including the genomic, transcriptomic, and proteomic levels (Figure 1). We will focus on the formulation of various prediction tasks addressing different biological questions that are attempted by deep learning. We will investigate the model architectures used by the studies and discuss their functionalities and design principles. In particular, we comprehensively list the datasets that are used in each study as a convenient reference for researchers willing to replicate existing methods or develop new methods in this field. Prospectively, we will discuss the application potential of various emerging deep-learning paradigms, such as self-supervised learning, meta-learning, and large-scale pre-trained models for biological sequences, that will potentially alleviate the problems of existing approaches. We also point out the trend of using the integration of structural information, multi-omics profiles, and single-cell profiles for gene regulation studies. We hope that this article will provide a rich and up-to-date resource and serve as a starting point for new researchers interested in this area.
Figure 1.
Deep-learning applications in gene regulation at various omics levels
Review of deep-learning applications in gene regulation
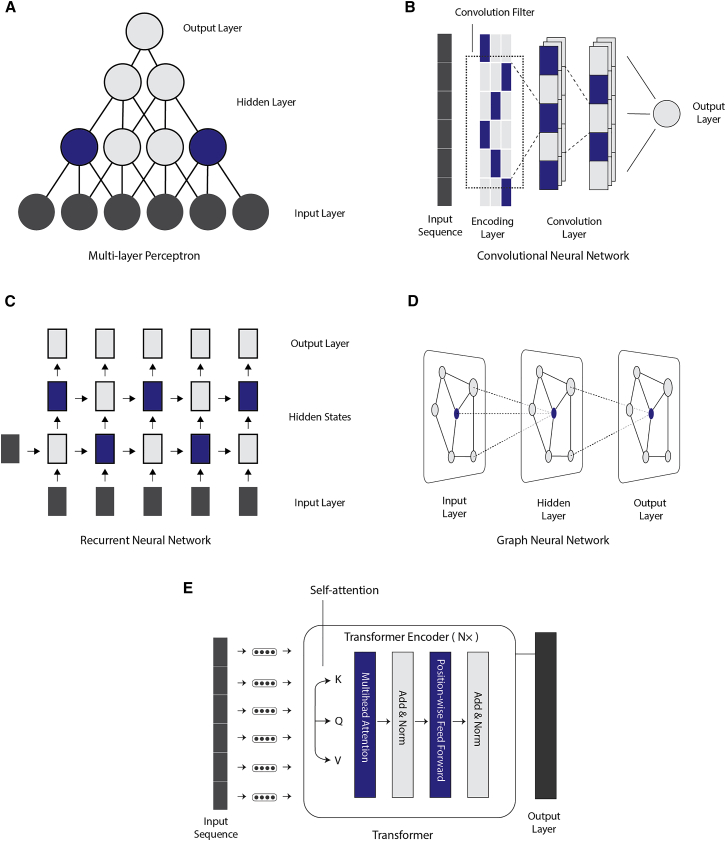
Types of neural networks used in gene regulation studies
During the past decade, the neural network models used in gene regulation studies have largely followed what has been used in computer vision and natural language processing, from which deep learning first originated. The most popular types include multi-layer perceptrons (MLPs) (Figure 2A), which are quite popular in early applications of deep learning on tabular data. The input layer of MLPs directly takes in the data values from the input dataset and is subsequently processed by one or more hidden layers. Finally, an output layer summarizes the processed information of the earlier hidden layers to produce the final prediction.
Figure 2.
Common deep-learning architectures used in gene regulation studies
(A) Multi-layer perceptron.
(B) Convolutional neural network.
(C) Recurrent neural network.
(D) Graph neural network.
(E) Transformer.
Following the success in image recognition and text classification, convolutional neural networks (CNNs)20 have proved to be very useful for handling raw biological sequence data, whether it is DNA, RNA, or protein sequences (Figure 2B). The CNNs employ convolution filters to process sequential or image data in a way that respects the spatial structure of the data. To handle long-range interactions of sequential data, recurrent neural networks (RNNs) such as gated recurrent units (GRUs)21 and long short-term memory (LSTM)22 have received particular interest in biological sequence analysis (Figure 2C). The RNNs employ hidden states in the network that will remember sequential information at earlier locations. These hidden states benefit the modeling of long-range interactions. Graph neural networks (GNNs) are designed to handle structured datasets that are represented as a graph (Figure 2D). GNNs also have input, hidden, and output layers in their architectures. In contrast to plain MLPs that handle individual data points independently, the hidden layers and output layers of GNNs respect the topological structure of the dataset. In more recent years, inspired by the success in natural language processing and understanding, Transformers19,23,24 have also received a lot of attention for biological sequence data processing (Figure 2E). Transformers are powerful learners of sequential data, partly due to their employment of the self-attention mechanism that handles the pairwise interdependencies between the sequence elements. More recently, Transformers are also popular choices for self-supervised learning on biological sequences,18 which we will discuss in subsequent sections. According to the nature of the prediction tasks formulated in each study, researchers designed deep-learning architectures utilizing one or more of the aforementioned networks with highly customized configurations to achieve higher performance, greater computational efficiency, and better biological interpretability. In this article, we do not wish to provide a comprehensive introduction to the neural network types used in deep learning. Instead, we refer readers to dedicated hands-on tutorials25,26 and introductory textbooks27 on deep learning.
Genomic-level applications
In this section, we review the most representative research works that applied deep learning to the study of genomic-level regulations. All works that we review have formulated a supervised learning problem and can predict functional genomic features from the genomic sequence. In this way, they aim to decipher the regulatory code and grammar from the genomic sequence and predict how genetic variations will affect a particular regulatory mechanism.
The relevant studies and methods are listed and summarized in Table 1. In particular, we list the functionalities each method achieves, the datasets each method uses, and the deep-learning architectures on which the methods are based. As the regulatory code and grammar of genomic sequences are interpreted differently in different organisms and tissue/cell types, most models have particular ways to provide organism- and tissue-/cell-type-specific predictions. Therefore, we particularly highlight the species and tissue/cell types that are involved in each study.
Table 1.
Genomic-level deep-learning applications
| Method name | Year | Functionalities |
Datasets | Model | Species | Tissue/cell types | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DHSa | Histonea | TFa | Varianta | DNA met.a | 3Da | ||||||
| Functional genomic models: | |||||||||||
| Deepbind17 | 2015 | ✓ | ✓ |
|
human, mouse | multiple | |||||
| DeepSEA30 | 2015 | ✓ | ✓ | ✓ | ✓ |
|
human | multiple | |||
| Basset34 | 2016 | ✓ | ✓ |
|
|
human | multiple | ||||
| DanQ35 | 2016 | ✓ | ✓ | ✓ | ✓ |
|
|
human | multiple | ||
| CpGenie36 | 2017 | ✓ | ✓ |
|
|
human, mouse | multiple lymphoblastoid cell lines | ||||
| De-Fine37 | 2018 | ✓ | ✓ |
|
|
human | K562 and GM12878 | ||||
| Basenji38 | 2018 | ✓ | ✓ | ✓ |
|
|
human | multiple | |||
| Expecto41 | 2018 | ✓ | ✓ | ✓ | ✓ |
|
|
human | multiple (>200) | ||
| Xpresso42 | 2020 | ✓ | ✓ |
|
|
human, mouse | multiple | ||||
| DeepMEL43 | 2020 | ✓ | ✓ |
|
|
trained on human, with cross-species generalizability | melanoma samples | ||||
| Enformer46 | 2021 | ✓ | ✓ | ✓ | ✓ |
|
|
human, mouse | multiple | ||
| BPNet48 | 2021 | ✓ |
|
|
mouse | mESC | |||||
| 3D genomic models: | |||||||||||
| Akita50 | 2020 | ✓ | ✓ |
|
human, mouse | multiple (multiple human and mouse cell lines; mouse neuronal tissues) | |||||
| Orca56 | 2022 | ✓ | ✓ | ✓ | ✓ |
|
human | multiple (HFF and H1 hESC cell lines) | |||
| GraphReg57 | 2022 | ✓ | ✓ | ✓ | ✓ |
|
human, mouse | multiple | |||
DHS, DNase I hypersensitivity prediction; Histone, histone modification prediction; TF, transcription factor binding prediction; Variant, effect of genetic variants prediction; DNA met, DNA methylation prediction; 3D, 3D genome-folding prediction.
Deepbind17 is a pioneer work dedicated to the prediction of nucleic acid-protein binding. Based on a CNN architecture, it can learn from multiple DNA-protein binding experimental profiling technologies, including protein binding microarrays60 (PBM), ChIP-seq,1 and HT-SELEX.61 DeepSEA30 is one of the seminal works that applied deep learning to whole-genome functional genomics annotations. DeepSEA uses a CNN-based architecture that takes in 1,000-bp human genomic DNA sequence and performs a multi-task prediction of DNase I hypersensitivity (DHS), transcription factor (TF) binding, and histone modification in multiple cell lines. DeepSEA was trained on 125 DHS profiles (by DNase-seq4) and 690 TF binding profiles (by ChIP-seq1 for 160 distinct TFs) from ENCODE,8 and 104 histone modification profiles (by ChIP-seq) from Roadmap Epigenomics.31 DeepSEA uses one model to predict the signals measured from 919 epigenomic profiles (including 125 DHS predictions, 690 TF binding predictions, and 104 histone modification predictions). This multi-task design allows it to share the learned genomic grammar while performing different tasks. Additionally, the authors trained a boosted logistic classifier using the predictions of DeepSEA and showed that it could prioritize functional non-coding regulatory mutations in HGMD32 and expression quantitative trait loci (eQTL) in GRASP.33
Motivated by the success of DeepSEA, follow-up works have improved and extended DeepSEA in multiple different aspects. Basset34 is also a CNN-based model particularly focused on DHS prediction. It extends DeepSEA’s DHS prediction to a total of 164 cell types (125 cell types from ENCODE8 and 39 cell types from Roadmap Epigenomics9). Basset trained cell-type-specific models by fitting one model for each cell type. Instead of using the pure CNN architecture, DanQ35 explored the effectiveness of a hybrid architecture combining CNN and bidirectional LSTM (BiLSTM). It outperformed DeepSEA even though they were trained exactly on the same dataset.
The above integrative functional genomic models do not shadow the effectiveness of dedicated predictive models. CpGenie36 predicts genomic DNA methylation status from their sequences. It was trained on restricted representation bisulfite sequencing (RRBS) and whole-genome bisulfite sequencing (WGBS) profiles from ENCODE. De-Fine37 focused on TF binding prediction and was trained specifically on TF binding profiles from the K562 and the GM12878 cell lines from ENCODE. In contrast to the previous studies, De-Fine used a cell-line-specific genome sequence instead of the human reference genome sequence, pointing out that cell-line-specific genomic variation may affect model training and prediction. It also experimented with quantitative prediction instead of binary prediction of functional genomic elements.
More recent studies further improved the network architecture and integrated genomic predictions with transcriptomic predictions, such as promoter activity and gene expression levels. Compared with the previous methods, Basenji,38 as a successor to Basset, significantly enlarged the size of genomic sequence that a CNN-based model can take in. Using the “dilated convolution”62 that exponentially increases the receptive field of high-level neurons, the model is able to process 131 kbp inputs. The model is, therefore, able to recognize motifs that have long-range dependencies. To make quantitative predictions of each genomic feature, Basenji uses Poisson regression-based log likelihood as the loss function. Expecto,41 which is an official redesign of DeepSEA by the original authors, enlarges DeepSEA to a three-stage model. The first stage is still CNN based and resembles the original DeepSEA model, but with larger-sized input (2,000 bp). The second stage is a spatial transformation module that reduces dimensionality and weighs contributions from nearby sites according to their relative distance. The third stage performs a gradient-boosted linear regression of the gene expression levels using the genomic features produced from stage 2 and 218 tissue expression profiles from GTEx,10 Roadmap Epigenomics, and ENCODE. Expecto was shown to be able to prioritize mutations related to several immunity-related diseases, such as Crohn’s disease, ulcerative colitis, inflammatory bowel disease, and Bechet’s disease. Similar to the prediction target of Expecto, Xpresso42 is also a gene expression level predictor. However, Xpresso was deliberately designed to make such predictions based purely on the genomic sequence surrounding the promoter (∼10.5 kbp). By inspecting the discrepancy between model prediction and ground-truth measurements, the researchers made interesting discoveries of several genes’ regulatory mechanisms beyond their promoter activity. This included polycomb-mediated transcriptional gene silencing,63 enhancer-mediated transcriptional gene activation, and microRNA-mediated gene repression. Enformer46 is the first to employ the CNN + Transformer hybrid architecture for gene expression level and epigenetic feature prediction. Using the same dataset as Basenji247 (an updated version of Basenji), it achieved remarkably higher accuracy than its predecessors. Enformer was trained by alternately feeding in human and mouse genomic sequences, enabling it to perform cross-species inference.
Recent works have also explored the possibility of different problem formulations and measurements from alternative experimental techniques. DeepMEL43 constructed a model similar to that of DanQ for the prediction of chromatin accessibility in melanoma cell lines. DeepMEL was trained using melanoma omniATAC-seq64 data (an improvement over plain ATAC-seq), and instead of predicting the binary chromatin accessibility per location it predicts 24 co-accessible regions as identified by cisTopic.45 This better utilizes the co-regulatory mechanism of accessible chromatin regions. BPNet,48 a 10-layer CNN with dilated convolution and residual connections, trained on ChIP-nexus49 (a high-resolution improvement of ChIP-seq) data of four pluripotency TFs (Oct4, Sox2, Nanog, and Klf4), is able to produce base-resolution binding affinity prediction to genomic sequences of all four TFs in a multi-task fashion. BPNet is able to discover interesting cooperativity between TF motifs located within 1,000 bp regions, such as the Oct4-Sox2 motif and Oct4-Oct4 motif, and the cooperation between the Nanog motif and AT-rich motif in a periodic manner.
All of the above methods regard the genome as linear sequences per chromosome. However, the cellular genome does have a three-dimensional (3D) structure. The 3D structure of the genome is under extensive regulation and is able to affect gene expression, DNA replication, and DNA repair. Several approaches have been dedicated to deciphering the regulatory code and grammar of the 3D structure of the genome from genomic sequences. Akita50 is a deep-learning method that can predict genome folding from the genomic sequences. After training on Hi-C65 and Micro-C51 profiling data from five human cell lines, one mouse cell line, and multiple mouse neuronal tissues, Akita is able to infer the genome-folding map of each cell type, which is a two-dimensional (2D) matrix representing pairwise contact between genomic regions. Akita utilizes a Basenji-like architecture as its “trunk” for processing ∼1 Mb genomic sequences. It then uses a “head” to transform the one-dimensional (1D) genomic sequence representations into 2D maps. The mean squared error between the predicted 2D map and experimental Hi-C or Micro-C data is used as the training objective. Orca,56 a very recent improvement on Akita, enables the prediction of the genome-folding map at multiple resolutions. Orca uses a multi-resolution 1D-CNN genomic sequence encoder which can take in 256 Mb, 32 Mb, or 1 Mb inputs and encodes them into 1D sequence representations. Orca then uses a cascading 2D-CNN decoder to decode sequence representations into 2D genome-folding maps. Using only the Micro-C profiles of the two human cell lines (HFF and H1 hESC) that Akita has used, Orca produces genome-folding map predictions at various scales, from 1 Mbp regions each within one chromosome to 256 Mbp regions that cover multiple chromosomes. Furthermore, Orca’s sequence encoder is simultaneously trained on the DHS and histone modification profiles of the two cell lines from ENCODE and Roadmap Epigenomics, making it an integrative and multi-purpose model. In contrast to the earlier works that have focused on sequence-based prediction of genome-folding maps, a recent model, GraphReg,57 instead utilized the 3D structure of the genome for better prediction of gene expression levels. GraphReg contains a set of two models, Epi-GraphReg and Seq-GraphReg. Epi-GraphReg infers tissue-agnostic gene expression levels based on epigenetic and 3D genomic profiles, and Seq-GraphReg infers tissue-aware gene expression levels based on genomic sequence and 3D genomic profiles. Both Epi-GraphReg and Seq-GraphReg utilize graph attention networks59 for modeling the spatial interactions between genomic locations.
Transcriptomic-level applications
The transcriptome serves as a central stage for gene regulation. The initiation of transcription requires the recognition of a promoter sequence by an RNA polymerase, binding of transcription factors to enhancers, and determination of a transcriptional start site (TSS). Such a process can be extensively regulated to control the rate of gene expression, and, if a gene has multiple promoters, the utilization of different promoters may produce RNA transcripts with different 5′ UTRs that will potentially have different translational efficiency.66 RNA splicing is also a highly regulated process and is a significant contributor to eukaryotic transcriptome diversity.67 In eukaryotes, the possible usage of multiple polyadenylation sites (PASs) produces mRNAs with different 3′ UTRs that may contain important regulatory elements.68 After the completion of the above process, the mRNA molecule is transported out of the nucleus. RNA subcellular localization controls the spatial distribution of the newly transcribed mRNAs. Post-transcriptional mRNAs may also be selectively targeted by microRNAs (miRNAs), which are able to downregulate the expression of certain genes. The 5′ UTR of mRNA has an important effect on its translational efficiency, which directly controls the rate of protein synthesis.
We summarize research works that use deep learning to model each of the aforementioned processes in Table 2. Although some of the “genomic-level” prediction methods in the previous section may also have some transcriptomic-level predictions, especially for the integrative functional genomic models such as Basenji and Expecto, we focus here on methods that are dedicated to particular aspects of transcriptomic-level regulation.
Table 2.
Transcriptomic-level deep-learning applications
| Method name | Year | Main functionalities | Datasets | Model | Species | Tissue/cell types |
|---|---|---|---|---|---|---|
| Promoter/TSS: | ||||||
| CNNProm69 | 2017 | promoter recognition |
|
human mouse Arabidopsis E. coli B. subtilis |
Non-specific | |
| DeeReCT-PromID73 | 2019 | promoter recognition on highly imbalanced dataset |
|
|
human | non-specific |
| DeeReCT-TSS74 | 2021 | promoter recognition guided by RNA-seq |
|
|
human | multiple (10 cell types) |
| Splicing: | ||||||
| Barash et al.75 | 2010 | splicing prediction of cassette exons |
|
|
mouse | multiple (27 tissues) |
| Leung et al.77 | 2014 | splicing prediction of cassette exons |
|
|
mouse | multiple (5 tissues) |
| Xiong et al.79 | 2015 | splicing prediction of cassette exons in exon triplets |
|
|
human | multiple (16 tissues) |
| DARTS80 | 2019 | splicing prediction of cassette exons guided by RNA-seq |
|
human | K562, HepG2 | |
| SpliceAI81 | 2019 | Splice sites prediction from pre-mRNA |
|
human | non-specific | |
| Pangolin83 | 2022 | splice sites prediction from pre-mRNA |
|
|
human rhesus monkey mouse rat |
multiple (4 tissues in each species) |
| Polyadenylation: | ||||||
| Leung et al., 201886 | 2018 | PAS quantification (pairwise comparison) |
|
|
human | multiple (7 tissue types) |
| DeeReCT-PolyA91 | 2019 | PAS recognition |
|
human mouse |
non-specific | |
| APARENT96 | 2019 | PAS quantification |
|
|
human | non-specific |
| DeeReCT-APA97 | 2021 | PAS quantification |
|
|
mouse (BL, SP, and BLxSP F1 hybrid) | fibroblast |
| RNA subcellular localization: | ||||||
| RNATracker98 | 2019 | subcellular localization prediction |
|
human | HepG2 and HEK293T cell lines | |
| MicroRNA targets: | ||||||
| MiRTDL102 | 2015 | microRNA targets prediction |
|
|
human mouse rat |
non-specific |
| Translation: | ||||||
| Cuperus et al.104 | 2017 | 5′ UTR translational efficiency prediction |
|
|
yeast | N/A |
| RNA-protein binding: | ||||||
| DeepBind17 | 2015 | sequence-based RNA-protein binding prediction |
|
|
multiple (24 eukaryotes) | non-specific |
| NucleicNet106 | 2019 | structure-based RNA-protein binding prediction |
|
|
multiple | non-specific |
CNNProm69 is an early deep-learning-based method for promoter sequence recognition. The model uses one to two layers of CNN for the binary classification of sequences into promoter/non-promoter sequences. Effectiveness has been demonstrated in both prokaryotes (Escherichia coli and Bacillus subtilis) and eukaryotes (human, mouse, and Arabidopsis). As a successor to CNNProm, DeeReCT-PromID73 enlarged the size of the input to 600 bp and enabled genome-wide scanning of promoters. The authors pointed out that models for promoter recognition that are trained on curated balanced datasets may not be directly applicable for genome-wide scanning. This is because the majority of genomic regions are negative examples (non-promoters) and, therefore, the tolerability of the false-positive rate should be much lower. DeeReCT-PromID employed a strategy for iteratively selecting hard negative samples to reduce the false-positive rate of the model. DeeReCT-TSS further improved on DeeReCT-PromID by inferring promoter usage in different cell lines through both promoter sequences and RNA-seq evidence. It demonstrated its functionality by training and evaluating on ten FANTOM539 cell lines, using genomic sequence and RNA-seq as input and matched CAGE-seq40 data as ground truth.
RNA splicing plays a critical role in transcriptomic-level regulation. By producing transcripts with different combinations of exons and introns, it contributes significantly to eukaryotic transcriptomic diversity. Given the complexity of different patterns of alternative splicing, early works on deep-learning-based splicing prediction particularly focused on one alternative splicing type: the cassette exons. For example, an early work of Barash et al. developed a one-layer neural network for the prediction of cassette exon differential usage across mouse tissues.75 The network takes in a 1,014-dimensional vector containing sequence features flanking the exon of interest and outputs three-class classification scores for “increased usage,” “decreased usage,” or “no change.” The model is trained on microarray profiles of 3,665 cassette exons in 27 mouse tissues.76 Leung et al. further enlarged the dataset to a total of 11,019 cassette exons in mouse and increased the dimension of sequence features to 1,393 for tissue-specific splicing pattern prediction.77 Using the same sequence features as Leung et al., Xiong et al. developed an MLP for the prediction of 10,689 human cassette exons.79 The model was trained on Bodymap 2.0 RNA-seq data (NCBI accession GEO: GSE30611) and used a Bayesian MCMC procedure to reduce overfitting. Using the predictive model, the researchers were able to examine mutations that alter splicing in genes involved in several human genetic diseases. DARTS80 provided an example of integrating deep-learning-processed sequence features and RNA-binding protein (RBP) expression levels with low-coverage RNA-seq evidence for the differential usage analysis of cassette exons between conditions. DARTS integrates MLP-based deep learning with the Bayesian hypothesis test (BHT) by asking its deep-learning module to provide a prior distribution for its BHT module. In this way, DARTS enables deep-learning-guided study of alternative splicing even when the experimental RNA-seq data are not of enough sequencing depth.
SpliceAI81 is the first deep-learning-based splice site predictor for all splicing types. SpliceAI simulates the in vivo pre-mRNA processing machinery and directly predicts splice sites from raw pre-mRNA sequences. SpliceAI takes in long input (5,000 nt) to handle large chunks of pre-mRNA sequences and performs three-class classification (no splice site, donor site, and acceptor site) per each pre-mRNA location. SpliceAI also utilized the dilated convolution62 and residual block108 for increased receptive field of high-level neurons. Pangolin83 further extends SpliceAI in a multi-task fashion for the detection of splice sites in a total of four tissue types (heart, liver, brain, and testis) from four species (human, rhesus monkey, mouse, and rat).
The termination of eukaryotic Pol II transcription in eukaryotic cells requires cleavage at the 3′ end of the transcript and an addition of a poly(A) tail, a process called polyadenylation. Similar to promoters that determine TSSs, PASs determine transcription termination sites. A gene may have multiple competing PASs, and cells from different tissue types or conditions may preferentially use each of them. Such alternative polyadenylation (APA) could modify the 3′ UTRs of transcripts and could strongly affect mRNA stability109 and cellular localization,110 and is involved in various human diseases. DeeReCT-PolyA91 is one of the first deep-learning methods for recognizing PASs. Using a CNN with group normalization95 to increase robustness, DeeReCT-PolyA takes in 200 nt sequences and predicts whether they contain a PAS or not. The model achieved state-of-the-art performance and substantially outperformed non-deep-learning methods on two human polyadenylation datasets, the Dragon human poly(A) dataset92 and the Omni human poly(A) dataset,93 and one mouse polyadenylation dataset from Xiao et al.94 Instead of tackling the PAS recognition problem, Leung et al. were the first to address the PAS quantification problem by applying a two-branch CNN for pairwise comparison of competing PAS of a gene.86 Leung et al. assembled a reference of PASs in the human genome using four different reference databases and used 3′-seq data from Lianoglou et al.90 for quantification. Instead of casting the PAS quantification problem into pairwise comparison problems, DeeReCT-APA97 handles variable numbers of PASs per gene using a combined CNN and BiLSTM architecture. DeeReCT-APA is able to model the interactions between competing PASs and achieves better performance than the model from Leung et al.86 Instead of training models only on endogenous PAS sequences, APARENT96 trained a two-layer CNN on 3 million synthesized massively parallel reporter assay (MPRA) sequences of APA. The MPRA is able to measure hundreds of thousands of synthesized PAS sequences’ regulatory activities in parallel. APARENT’s CNN was trained to predict the measured regulatory activity given the PAS sequence. Using a gradient-based optimization of input sequences, the APARENT model is able to engineer PAS sequences to have desired levels of regulatory activity.
Transcriptomic-level regulation can also be carried out through other post-transcriptional mechanisms. mRNA subcellular localization controls gene expression both spatially (by transporting mRNA into different subcellular structures) and quantitatively (by modulating the accessibility of mRNA to ribosomes). RNATracker98 is a deep-learning tool that predicts such localization patterns of mRNAs. Utilizing mRNA sequence and RNA secondary structure predicted from RNAplfold,99 RNATracker is able to classify mRNAs into their plausible subcellular localizations. The version of RNATracker trained on CeFra-Seq data100 classifies mRNA localizations into cytosol, nuclear, membranes, and insoluble, while the version trained on APEX-RIP data101 classifies localizations into ER mitochondrial, cytosol, and nuclear. mRNAs can also be targeted with miRNAs that could silence mRNA expression. MiRTDL102 is a CNN-based tool that is able to predict potential miRNA-mRNA interactions. The 5′ UTR of an mRNA greatly affects ribosomal translational efficiency and is under frequent regulation. Similar to the motivation of the APARENT model for the prediction of PAS strength, Cuperus et al. developed a 3-layer CNN to predict 5′ UTR translational efficiency by training it on 489,348 synthesized 5′ UTRs in yeast.104 The translational efficiency of each synthesized 50-nt-long 5′ UTR was measured by a massively parallel growth selection experiment and was used as the CNN’s prediction target.
Under the hood of all the transcriptomic-level gene regulations are complex interactions between RNA and various other types of biomolecules. RNA-protein binding is certainly one of the most important, as most post-transcriptional regulations are mediated through RBPs. Besides predicting DNA-protein binding, DeepBind17 is also able to predict RNA-protein bindings. After being trained on the RNAcompete assay data,105 it is able to predict the binding preference of an RNA molecule to 207 distinct RBPs from 24 eukaryotes based on the RNA sequence. NucleicNet106 pursued a path different from DeepBind. Instead of performing sequence-based RNA-protein binding predictions, it makes RBP-centric predictions based on their structures. Trained on 158 ribonucleoprotein structures from the PDB,107 it is able to predict an RBP’s binding sites for RNAs as well as each binding site’s preference of each type of RNA constituents. Readers are referred to Wei et al.111 for a detailed survey of deep-learning applications in RNA-protein binding predictions.
Proteomic-level applications
After a protein is translated from its mRNA template, regulation can take place at the proteomic level via a number of mechanisms. Protein post-translational modifications (PTMs) contain a family of such regulatory processes that covalently modify a protein after it is translated. For example, the serine (Ser), threonine (Thr), and tyrosine (Tyr) residues can be modified by phosphorylation, which plays an important role in intracellular signal transduction. Furthermore, the lysine (Lys) residues can be modified through ubiquitination, which can mark a protein for degradation. Protein subcellular localization determines the cellular compartments where a protein resides and exerts its functions, which will substantially affect its function and activities.
We summarize these proteomic-level deep-learning applications in Table 3 (upper half). Similarly, we highlight their functionalities, training datasets, and model architectures to facilitate future explorations in this area.
Table 3.
Proteomic- and phenotype-level deep-learning applications
| Method name | Year | Main functionalities | Datasets | Model | Species |
|---|---|---|---|---|---|
| Post-translational modification (PTM): | |||||
| DeepPhos112 | 2019 | phosphorylation site prediction (general/residual-specific/kinase-specific) |
|
|
human |
| DeepUbi120 | 2019 | ubiquitination prediction |
|
|
multiple (176 species) |
| MusiteDeep122,123,124 | 2017-2020 | multiple PTM prediction |
|
|
multiple animal species |
| Protein-subcellular localization: | |||||
| DeepLoc126 | 2017 | subcellular localization prediction |
|
|
multiple eukaryotes |
| Genotype-to-phenotype inference in animal species: | |||||
| Zhou et al.128 | 2019 | prediction of the effect of non-coding variants to autism spectrum disorder |
|
|
human |
| DeepWAS129 | 2020 | using genomic deep-learning model to enhance genome-wide association studies |
|
human | |
| Genotype-to-phenotype inference in plant species: | |||||
| DeepGP133 | 2020 | multiple phenotype prediction in polyploid outcrossing species |
|
|
strawberry blueberry |
| Shook et al.137 | 2021 | crop yield prediction based on genotype and environmental factors |
|
|
soybean |
Deep-learning models have been developed for the prediction of PTMs from protein sequences. DeepPhos112 is a densely connected CNN architecture119 that predicts phosphorylation sites from protein sequences. DeepPhos112 formulates three different phosphorylation prediction tasks. The general prediction involves predicting whether an amino acid position is a phosphorylation site. The residual-specific prediction requires a model to predict in which amino acid type phosphorylation occurs. The kinase-specific prediction requires a model to predict which kinase is responsible for the phosphorylation event. DeepUbi120 is a CNN architecture that predicts ubiquitination from protein sequences, achieving an area under the curve of 0.9 in a total of 176 species. MusiteDeep122,123,124 is a series of works for multiple PTM type prediction. Its latest version uses an ensemble of multi-layer CNN and Capsule Network125 that is able to handle 13 different PTM types. MusiteDeep updates its predictions for UniProt protein sequences every 3 months, and its prediction results are available at https://www.musite.net.
DeepLoc126 is a deep-learning-based prediction tool that is able to infer protein subcellular localization from their sequence. DeepLoc is designed as a combined CNN and LSTM architecture. It uses attention-based decoding to identify sequence regions with high predictive power. It organizes the ten subcellular locations in a hierarchical manner and uses hierarchical tree classification likelihood to train the model. In this way, without using information from homology sequences, it is able to achieve 78% accuracy for a total of ten subcellular locations.
Phenotypic-level applications in animal and plant species
The aforementioned deep-learning applications in gene regulation have been mainly at the microscopic level of biological processes. However, bridging the gap between the genotype and the phenotype represents one of the ultimate goals of gene regulation studies. In this section, we summarize deep-learning models that have been developed to assist in genotype-to-phenotype and phenotype-to-genotype inferences in animal and plant species (Table 3, lower half).
Following the development of DeepSEA, Zhou et al. further developed DeepSEA-based models for the prediction of the effect of non-coding variants on autism spectrum disorder (ASD).128 In this work, two DeepSEA-based models were trained to predict transcriptional and post-transcriptional regulatory effects separately. The resulting predictions were summarized into disease impact scores through training an LR model on top of the model predictions using known disease-associated mutations. Using the disease impact scores, it was then able to prioritize disease-associated mutations observed in 1,790 ASD-affected families. DeepWAS129 introduces a deep-learning-assisted genome-wide association study (GWAS) pipeline. DeepWAS utilizes the pre-trained DeepSEA model to produce a list of candidate variants for a GWAS. In this way, it reduces the number of candidates for GWAS and increased its statistical power. The authors demonstrated its effectiveness by improving three existing GWASs for multiple sclerosis,130 major depressive disorder,131 and body height.132
In plant species, deep learning has also been applied in multiple plant phenotype prediction tasks. For example, DeepGP133 applied a CNN-based model for phenotype prediction in two polyploid outcrossing species: strawberry and blueberry. Five strawberry fruit quality traits were predicted for strawberry individuals based on microarray genotypes, and five blueberry fruit quality traits were predicted for blueberry individuals based on genotypes obtained from Rapid Genomics Capture-seq.136 Shook et al. studied the possibility of predicting crop yield based on genotype and environmental factors. Using the Uni-form Soybean Tests data,138 which contains soybean yields in United States and Canada during 2003–2015, the authors separately built LSTM and temporal attention139 models for soybean crop yield prediction. At each time step, the model considers the crop’s genotype and seven weather variables during its growth period and forecasts the yield during harvest seasons. Such deep-learning applications in plant phenotype prediction tasks will provide valuable insights for plant breeding.
Problems and limitations of current deep-learning applications
Challenges in training and interpreting deep-learning models
In this section we discuss challenges that are common in the applications of deep learning, especially those in model training and model interpretation.
Deep-learning models are known to be difficult to train because their high non-linearity makes the optimization of the objective function difficult. Typically, deep-learning models are trained using stochastic gradient descent (SGD), as it is an efficient first-order optimization algorithm and its stochasticity allows it to jump out from local minima.27 However, the size of each gradient descent step (the “learning rate”) can be difficult to configure. A small learning rate can result in slow training progress and becoming stuck in a local minimum; a high learning rate will make the algorithm fail to converge. The direction of an SGD update, i.e., the direction toward which the objective function decreases the fastest, could also alternate too rapidly, making the optimization trajectory oscillate around a local minimum.140
Therefore, several improvements to SGD have been made to make the algorithm more efficient and stable, as summarized in Table 4. Adagrad141 adaptively modulates the learning rate for each model parameter based on its magnitude. RMSprop142 also adaptively modulates the learning rates, but it is based on the exponential moving averages of the parameters’ magnitude. Improvements to the direction of parameter update are also available, such as momentum and Nesterov momentum.156 Adam140 introduces the momentum update to RMSprop and has been effective in the optimization of large CNNs. More recently, several successors to Adam have become more and more popular, including NAdam143 (Adam with Nesterov momentum), AdamW144 (Adam with decoupled weight decay), and RAdam145 (Adam with more stabilized adaptive learning rate in the warm-up process). To boost model performance, researchers are always encouraged to apply these algorithms and their variants in real-world practice.
Table 4.
Common problems in deep learning and their solutions
| Method name | Year | Features |
|---|---|---|
| Improvements to SGD optimization: | ||
| Adagrad141 | 2011 | adaptive learning rate |
| RMSprop142 | 2013 | adaptive learning rate |
| Adam140 | 2014 | adaptive learning rate, momentum update |
| NAdam143 | 2016 | adaptive learning rate, Nesterov momentum update |
| AdamW144 | 2017 | adaptive learning rate, decoupled weight decay |
| RAdam145 | 2019 | adaptive learning rate, momentum update |
| Tools for hyperparameter tuning: | ||
| Raytune146 | 2018 | Tensorflow,147 Pytorch148 |
| KerasTuner149 | 2019 | Keras150 |
| Model interpretation: | ||
| Example-based methods | explain models using data points themselves application examples: Alipanahi et al., Bogard et al.17,96 |
|
| Perturbation-based methods | application examples: Alipanahi et al., Avsec et al.17,48 | |
| Attribution-based methods | application examples: Avsec et al., Janssens et al.48,151 | |
| Integrated gradients152 | 2017 | |
| SHAP153 | 2017 | |
| DeepLIFT154 | 2018 | |
| Captum155 | 2020 | |
| Model-based methods | encourage model interpretability through model design application examples: Ji et al., Zhou et al.19,41 |
|
Another challenge in training deep-learning models concerns hyperparameter selection. The selection of hyperparameters can substantially affect training stability and model performance. As the model grows larger, the space of hyperparameter combinations increases exponentially. Therefore, it is necessary to employ heuristic search strategies in hyperparameter tuning. Techniques such as random search,157 coordinate descent, and Bayesian optimization158 are common choices in practice. To facilitate the hyperparameter tuning process, software libraries with integrated hyperparameter selection algorithms such as Raytune146 and KerasTuner149 can be applied to existing projects with minimal modifications to the existing source code.
Another challenge of deep learning is the difficulty in its interpretation. Unlike shallow models such as linear models, decision trees, and SVMs, deep-learning models have complex hierarchical architectures and their hidden states cannot be interpreted in easy-to-understand terminologies. However, existing deep-learning studies in gene regulation have employed various kinds of methods to improve model interpretability. We categorize such methods into four general categories, which are summarized in Table 4.
Example-based methods
For example-based methods, the deep-learning models are explained by training or testing examples in the dataset. To interpret a specific layer or a hidden state neuron, examples that result in their high activation are selected (e.g., top 5% among all examples). The commonality among those examples can be used as an interpretation. For example, the subsequences that result in high activation of a specific convolution filter can be collapsed into position weight matrices (PFMs) and visualized by sequence logos. In this way, the regulatory motifs that the model is “looking at” can be revealed. This technique is commonly used by sequence-based models, such as described by Alipahani et al.17,96 and Bogard et al.17,96
Perturbation-based methods
Another way to interpret model predictions is based on perturbation. This is done by modifying (“perturbing”) the model’s input and inspecting the changes in the output. It is expected that the model’s prediction will substantially decrease if the most discriminative part of the input example is perturbed. For example, by performing in silico pointwise mutations for a biological sequence, we can produce a so-called mutation map of the sequence by asking the model to predict a score for each of the mutated sequences. This method has been systematically investigated by DeepBind to confirm the putative sequence motifs that are recognized by its convolution filters.17
Attribution-based methods
In contrast to example-based and perturbation-based methods which still treat the deep-learning model as a black box, attribution-based methods open up the black box by attributing a model’s intermediate network value to the model’s input. Such methods compute an attribution score for each element of the input. The magnitude of the score indicates the amount of its contribution, and the sign of the score shows whether the contribution is positive or negative. Saliency map159 is one the simplest attribution-based methods. It is defined just as the model’s gradients with respect to the input. In recent years more theoretically guaranteed approaches have been developed, including Integrated Gradients,152 SHAP,153 and DeepLIFT.154 For instance, BPNet extensively utilized DeepLIFT when inspecting the model’s binding affinity predictions to the TFs. Attribution-based methods seem to have a higher sensitivity than example-based methods. In the BPNet paper, the researchers systematically discussed the complex sequence motifs that could only be discovered by DeepLIFT but not by PFMs, such as the helical periodicity patterns of Nanog.48
Model-based methods
Instead of making post hoc interpretations of the model, it is better to consider interpretability during the model’s development. There are building blocks of deep-learning models that are inherently interpretable, such as attention modules.23 Attention scores can provide the location of regions to which the model is paying attention. Dividing models into different stages and producing interpretable results at the end of each stage is also a common strategy. For example, Expecto predicts gene expression prediction in three stages.41 In the first stage, it transforms genomics sequences into epigenomic features, which consists of 2,002 genome-wide histone marks; in the second stage, it aggregates epigenomic features produced in the second step based on spatial closeness; in the third stage, it predicts gene expression level from the aggregated features. In this way, the transparency at each stage provides interpretability for the whole pipeline.
Limitations of existing deep-learning applications in gene regulation
Apart from the aforementioned general problems of deep-learning algorithms, there are limitations specific to gene regulation that will potentially challenge their application potential.
The problem of overfitting
Deep-learning models are well known for the overfitting issue. The apparent high performance on a benchmark dataset does not always imply successful generalization to other unseen examples. This is particularly true for applications in gene regulation, due to three problems that are not trivial to overcome.
-
1.
The limitation of data volume in biological studies may hinder machine-learning model development. Unlike fields such as computer vision and natural language processing, where it is easy to collect terabytes or even petabytes of training data from the internet or from crowd-sourcing platforms,160 biological data have to be generated from biological experiments. If a particular regulatory mechanism cannot be studied by an established experimental technique that generates a large enough number of training examples, it will be impossible to study them using machine-learning methods. Even though such experimental techniques are available, the unavailability of such data may also arise from financial constraints or privacy concerns.
-
2.
The biological and technical variations across experimental conditions may limit the model’s generalization performance. For example, in the Basenji paper, the authors observed, on average, a Pearson correlation of 0.479 between biological replicates,38 even though they are from the same consortium. It is therefore difficult to tell whether a model with a high performance score is generalizable to other examples or is simply overfitting to those random variations.
-
3.
The use of endogenous sequences does not always imply the model’s generalizability to unseen cases. Most models take in biological sequences as input for their prediction, but most of them only use endogenous sequences for training. For example, DeepSEA,30 Basset,34 and Expecto41 were trained solely on the human reference genome GRCh37. It remains elusive how well those models are generalizable to genetic variations that may have a different “regulatory grammar” from those that are in the observed endogenous sequences. Furthermore, the number of such sequences pertinent to a particular regulatory event may comprise only a tiny fraction of the organism’s genome, transcriptome, or proteome. For example, the promoter sequences that regulate TSS only consist of genomic sequences at the beginning of each gene. This could further limit the diversity of training data. As we have introduced in previous sections, only APARENT96 for polyadenylation and the Cuperus et al.104 model for 5′ UTR translation efficiency utilized measurements of synthesized exogenous sequences from MPRA data for model training and evaluation.
The limitation of sequence-only models
Most gene regulation is a concerted effect of both cis-acting sequence motifs and trans-acting binding molecules (mainly binding proteins) residing in a cellular environment. Most of the aforementioned deep-learning models take nucleotide or amino acid sequences containing only cis-acting information as input. Some methods modeled the trans-acting effects implicitly by making tissue-specific predictions. For example, DeepSEA, Basset, and Basenji38 perform multi-task prediction across multiple tissue types, and in Leung et al.,78 the researchers trained separate models for each tissue type. For some methods, such trans-acting effects are ignored completely (e.g., in CNNProm69 and APARENT96). For all those models with tissue-specific predictions, the trans-acting environment is assumed to be static, and the models always produce the same results for a tissue type even though gene regulation is dynamic with respect to internal and external conditions. When prediction in a new tissue type is needed, the model needs to be retrained using experimental profiles coming from that tissue, which may not always be available. The only model that explicitly models trans-acting effects is DARTS,80 which considers the expression levels of 1,498 RBPs for splicing prediction.
Not enough consideration of interactions among regulatory events
The multiple layers of gene regulation do not happen independently. A proteomic-level regulation of one protein may affect the transcriptomic-level regulation of another gene. However, most methods developed so far have only considered regulatory events independently. Even for the multi-task models that predict multiple genomic features simultaneously, the interactions between those predicted events are not explicitly taken into consideration. DeeReCT-APA97 considers the interactions among multiple PASs; however, the interaction with the regulatory events of other types, e.g., splicing, is beyond its reach.
New deep-learning methods and perspectives
In the following sections, we discuss several promising new paradigms in deep learning that will potentially overcome the limitations already described (Figure 3). We list related works in Table 5 as examples for each new paradigm and hope that they can shed light on new deep-learning-based gene regulation studies.
Figure 3.
New deep-learning paradigms for gene regulation studies
(A) Self-supervised pre-trained models.
(B) Few-shot and meta-learning models.
(C) Incorporation of structural information.
(D) Multi-omics models.
(E) Single-cell omics models.
Table 5.
New methods and perspectives
| Method name | Year | Datasets | Model | Functionalities |
|---|---|---|---|---|
| Self-supervised pre-trained models: | ||||
| ESM-1b Transformer18 | 2020 | 250 million protein sequences from UniProt Archive (UniParc)161 collected by Uniref. 50129 | Transformer (33 layers, 650M params) | pre-train task: protein masked language modeling with amino acid sequencedownstream tasks: contact map prediction, secondary structure prediction |
| DNABERT19 | 2021 | k-mers of the human genome | BERT-base162 (12 layers, 110M params) | DNA masked language modeling; downstream fine-tuning achieves strong performance on: promoter recognition, TF binding sites prediction, splice sites prediction, functional genetic variants classification, cross-species transfer learning |
| MSA Transformer24 | 2021 | 260 million MSAs from UniProt collected by UniClust30163 | Transformer adapted to MSA (12 layers, 100M params) | protein masked language modeling with MSA; downstream tasks: unsupervised and supervised contact map prediction, 8-class secondary structure prediction |
| Few-shot/meta- learning: | ||||
| DeeReCT-TSS74 | 2021 | FANTOM539 | Reptile algorithm164 for meta-learning | the Reptile meta-learning algorithm allows fast adaptation to new tissue types |
| MIMML165 | 2022 | starPepDB166 BIOPEP-UWM167 |
Prototypical Network168 for metric-based meta-learning (few-shot learning) mutual information maximization loss |
few-shot bioactive peptide function prediction |
| Incorporation of structural information: | ||||
| NucleicNet106 | 2019 | 483 RNA-protein complexes from the PDB107 and de-duplicated to 158 ribonucleoprotein structures | CNN with ResNet-like architecture | structure-based RNA-protein binding prediction |
| MaSIF169 | 2020 | PDB | geodesic convolutional neural networks170 | protein pocket classification (MaSIF-ligand) protein interface prediction (MaSIF-site)protein-protein interaction (PPI) search (MaSIF-search) |
| dMaSIF171 | 2021 | PDB | quasi-geodesic convolution on point cloud representation of protein surfaces | protein interface prediction PPI search |
| Multi-omics models: | ||||
| MOMA172 | 2016 | the study curated the dataset “Ecomics,” which has integrated data of the transcriptome, proteome, metabolome, fluxome, and phenome of E. coli under different experimental conditions (available from http://prokaryomics.com) | combination of RNN-based deep learning and LASSO regression | using a layer-by-layer approach, the model predicts multi-omics quantities (transcriptomic, proteomic, metabolomic, fluxomic, and phenomic) |
| DSPN173 | 2018 | the study curated the dataset resource “PsychENCODE,” which includes comprehensive functional genomic data (genotype, bulk transcriptome, chromatin, and Hi-C profiles) of the brain of 1,866 individuals | conditional deep Boltzmann machine174 | the model predicts brain phenotypes in an interpretable and generative way and is able to impute intermediate “molecular phenotypes” |
| deepManReg175 | 2022 | the Patch-seq176 multi-omics transcriptomic and electrophysiological data for neuron phenotype classification177 | DNN with manifold alignment178 | multi-modal alignment of multi-omics data |
| Chaudhary et al.179 | 2018 | 230 samples from TCGA with RNA-seq data, microRNA-seq data, and DNA methylation profiles | autoencoder-based dimensionality reduction,180 feature selection, and integration of multi-omics data | the multi-omics model is able to cluster patients into different survival groups, and survival-correlated autoencoder features have verified predictive performance on independent datasets |
| Utilizing single-cell profiles: | ||||
| Pseudobulk level: | ||||
| Cusanovich et al., 2018181 | 2018 | scATAC-seq of 100,000 cells from 13 tissues of adult mice | Basset model trained to predict chromatin accessibility in each of the 13 tissues |
|
| DeepFlyBrain151 | 2022 | scATAC-seq profiling of 240,919 cells of Drosophila whole brain | CNN + LSTM model used in DeepMEL | prediction of co-accessible regions in three cell subtypes: Kenyon cell, T neurons, and glia |
| Single-cell level: | ||||
| DeepCpG182 | 2017 |
|
CNN + bidirectional GRU | imputation of methylation states at the single-cell level |
| CNNC185 | 2019 |
|
CNN | TF target gene prediction, disease-related genes prediction, and causality inference between genes |
| SCALE190 | 2019 | variational autoencoder197 with Gaussian mixture model | clustering, batch effect removal, and imputation of scATAC-seq data | |
| scFAN198 | 2020 |
|
3-layer CNN | inferring TF binding activity of scATAC-seq using TF binding model pre-trained on bulk data |
| scGNN199 | 2021 |
|
GNN-based autoencoder | clustering, scRNA-seq data imputation |
| scBasset200 | 2022 |
|
6-layer CNN (1,344 bp input) | sequence-aware scATAC-seq imputation, de-noising, and cell clustering |
| scTenifoldKnk202 | 2022 |
|
quasi-manifold alignment206 | virtual gene knockout prediction |
Pre-trained self-supervised models could alleviate the problem of data insufficiency
In recent years, pre-trained models have achieved great success in processing and understanding the natural language. Pre-trained Transformer models such as BERT162 and GPT207,208,209 perform self-supervised learning on massive corpora, aiming to predict randomly masked tokens from their context (the “masked language modeling” task) or the next token given the previous tokens (the “causal language modeling” task). The pre-trained models then display strong transfer learning ability. After fine-tuning on a very small amount of data from some downstream tasks, the model achieves state-of-the-art performance (Figure 3A).
Pre-trained models for biological sequences have been developed in parallel. For example, Rives et al.18 developed a protein sequence model, ESM-1b, which is a 33-layer Transformer architecture with 650 million parameters. ESM-1b performs BERT-like masked language modeling and is trained on 250 million protein sequences from Uniref. 50,210 which contains clusters from the UniProt Archive with 50% sequence similarity. Taking the network representations from ESM-1b, downstream classifiers trained on small datasets perform quite well on protein secondary structure and protein contact map prediction. DNABERT19 is a DNA sequence model based on a 12-layer BERT-base162 Transformer architecture with 110 million parameters pre-trained on the k-mer representation of the human genome for genomic sequence modeling. DNABERT is trained with the masked language modeling task by tokenizing the human genome into k-mers. The model showed similar or even better performance on several sequence classification tasks such as promoter recognition, TF binding site prediction, splice site prediction, and functional genetic variants classification. The model also showed cross-species transfer learning ability through the prediction of mouse TF binding sites. Instead of performing the pre-training task on one amino acid sequence only, the MSA Transformer24 extended the Transformer model to handle multiple sequence alignments (MSAs) of amino acid sequences to better utilize contextual information both within sequences and across homologous sequences. The MSA Transformer showed even superior performance on downstream protein secondary structure prediction and protein contact map prediction than ESM-1b.
Through a language modeling objective, the pre-trained models can utilize a massive amount of unlabeled biological sequence data that are not specific to one species or one prediction task. In this way, it is able to discover regulatory grammars across multiple genomic regions or from multiple species. It will be of great interest to see whether such pre-trained models are systematically beneficial to downstream prediction tasks of gene regulation, especially when the size of the downstream task datasets is not enough to train deep-learning models from scratch.
Few-shot and meta-learning mechanisms produce data-efficient deep-learning models
Another trend in the deep-learning community to tackle the problem of data insufficiency is to develop deep-learning models that utilize data efficiently. In particular, “few-shot learning” is aimed at solving a prediction task with only a few training examples (Figure 3B). This challenging problem is usually tacked by “meta-learning,” whereby a “meta-model” is trained that is easily generalizable across a set of similar tasks. When it is required to perform a specific task, it is able to quickly adapt itself to it with a few provided training examples from the task.
Such methods have already been applied in the classification of biological sequences. For example, the previously introduced DeeReCT-TSS74 applied a gradient-based meta-learning algorithm, Reptile,164 for the fast adaptation of the TSS prediction model to a total of ten cell types. The authors discovered that using ∼20% of data from each cell type to pre-train a meta-model and then adapt it to a specific cell type using the rest of the data benefited model performance. MIMML165 is a newly proposed meta-learning framework for bioactive peptide function prediction. MIMML is based on the Prototypical Network,168 which performs few-shot classification by measuring the distance from a query example to a few exemplars of each class. MIMML is able to perform few-shot prediction of a total of 16 peptide functions. With the above successful applications, we expect meta-learning to have a greater impact, especially on prediction tasks, with many related classes but only a few training examples for each of them.
Incorporation of structural information benefits modeling
We have previously pointed out the limitation of sequence-only models for not explicitly considering trans-acting factors. At the molecular level, such factors are constantly dependent on the interactions between protein-protein interactions or nucleic acid-protein interactions. Therefore, the accurate modeling of trans-acting factors in gene regulation requires the incorporation of structural information from the cis- and trans-acting counterparts. Recent breakthroughs in de novo protein structure prediction by AlphaFold2 have greatly enriched our resource for protein structures.211 Prediction of 3D structures of the genome50,56 and secondary structures of RNA has also experienced significant progress.212,213 Therefore, systematically incorporating structural information for deep-learning models in gene regulation is closer to reality than ever before (Figure 3C).
Recent works that perform deep learning on protein 3D structures are able to inspire future works that aim to incorporate such information into deep-learning models. MaSIF169 enabled deep learning on protein surfaces. Using a geodesic convolutional neural network,170 MaSIF is able to predict the binding of common ligands to protein interfaces and search for protein surfaces involved in PPIs. dMaSIF,214 as a successor to MaSIF, reduced the computational complexity of MaSIF by replacing geodesic convolution with quasi-geodesic convolution. Both methods produce deep representations of a protein surface that can be easily reused by other downstream deep-learning predictors. They could serve as a starting point for incorporating structural information in proteomic-level regulation prediction tasks, where protein-protein interaction is abundant. NucleicNet,106 introduced in the section “transcriptomic-level applications,” could serve as an example for transcriptomic-level models that aim to factor in such structural information. NucleicNet represents the binding protein’s 3D structure as a 3D grid with physicochemical properties. Using a CNN with residual connections,108 it is able to predict binding specificities of each type of the constituents of RNA. It will be of great interest as to whether such structural information and binding preference of RBPs, encoded in the deep representations of NucleicNet, can be explicitly utilized in modeling transcriptomic-level gene regulation in order to make such kinds of predictions more reliable and convincing.
Development of multi-omics models
Biologists usually rely on multiple experimental techniques for the confirmation of certain discoveries. Utilizing multiple data sources will also provide more evidence for predictive deep-learning models (Figure 3D). There are already existing methods that integrate multi-omics data from multiple sources for clinical predictive modeling. For example, Chaudhary et al.179 developed a multi-omics deep-learning model for the prediction of patient survival in hepatocellular carcinoma. The model is trained using 230 samples from The Cancer Genome Atlas (TCGA) with DNA methylation profiles, RNA-seq data, and microRNA-seq data. Their strategy to aggregate multi-omics data was to use autoencoder-based dimensionality reduction180 and feature selection in each data type. The selected features are then concatenated for downstream multi-omics clustering and discriminative prediction.
The developments of multi-omics models can be further inspired by multi-modal machine learning.215 Processing multi-omics data is essentially dealing with data coming from multiple modalities. The concept of multi-modal fusion strategies for multi-modal inputs, e.g., early fusion versus late fusion, is also applicable to multi-omics models. Early fusion, i.e., data integration at the networks’ early processing stage, may favor two omics profiles that are similar in the technology of measurement, and late fusion, i.e., data integration at the networks’ late processing stage, may favor two omics profiles that are similar in their subject of measurement but with very different technologies. Production of the so-called joint representations215 by mapping data points from multiple omics data sources into a same semantic space may also be beneficial for unified downstream network processing and analysis. DeepManReg175 shows itself as such an example. Designed as a DNN with a manifold alignment objective, it performs multi-modal alignment of transcriptomic and electrophysiological data in a Patch-seq multi-omics experiment176 and has been effective in neuron phenotype classification.
Although most existing deep-learning methods independently consider each regulatory event, gene regulation itself is a holistic cellular process. Future deep-learning models for gene regulation modeling should not only integrate multi-omics data sources as input but also consider the relationship between multi-omics quantities in their output (Figure 3D). For this, the Multi-Omics Model and Analytics (MOMA)172 provided such an example in E. coli. The MOMA model predicts multi-omics quantities of E. coli given their different growth conditions. Using RNN-based deep learning and LASSO regression, MOMA adopts a layer-by-layer approach to predict transcriptomic, proteomic, metabolomic, fluxomic, and phenomic quantities one after another, specifically taking into consideration the effect on an omics quantity by the quantities from previous omics layers. Similarly, the Deep Structured Phenotype Network (DSPN)173 predicts brain phenotypes from multiple functional genomic data modalities based on a hierarchical conditional deep Boltzmann machine (DBM) architecture.174 The DBMs are also arranged in a layer-by-layer fashion by first predicting the “intermediate molecular phenotypes” and then the brain phenotypes. This makes DSPN a generative model that is more interpretable than the common discriminative models in deep learning. These could shed light on future gene regulation modeling works that aim to simulate the underlying biological processes more realistically.
Utilization of single-cell profiles
Nearly all of the aforementioned deep-learning models for gene regulation have been trained on bulk sequencing profiles. In recent years, single-cell omics profiling technologies have improved substantially. This includes single-cell RNA-seq (scRNA-seq) for gene expression level profiling, single-cell ATAC-seq (scATAC-seq)192,216 for chromatin accessibility profiling, single-cell bisulfite sequencing (scBS-seq),183 and single-cell reduced representation bisulfite sequencing (scRRBS-seq)217 for methylation profiling, single-cell ChIP-seq (scChIP-seq)218 for protein-DNA binding profiling, and Smart-seq219 for full-length transcriptome profiling. Therefore, more and more data at single-cell resolution have accumulated. Single-cell profiles distinguish themselves from bulk profiles in their high dimensionality, high dropout rate (sparsity), and low sequencing quality and coverage. This not only introduces new challenges in data processing, analysis, and interpretation but also for the development of gene regulation models that utilize them.
Current deep-learning-based gene regulation models generally utilize single-cell profiles in two different ways (Figure 3E). The first operates at the pseudobulk level. The model aggregates single-cell measurements within each cell cluster into one rofile. The model then utilizes the aggregated pseudobulk profiles in the same way as bulk omics profiles. Despite information loss during the aggregation process, the utilization of pseudobulk profiles still has an advantage over real bulk omics profiles because they represent measurements from pure cell types without interference from others. As an example, Cusanovich et al. performed scATAC-seq on ∼100,000 somatic cells from adult mice.181 The researchers developed a model based on the architecture of Basset to predict chromatin accessibility in each of the 85 identified cell types in a multi-task fashion. The model was trained on the aggregated pseudobulk profiles within each cell cluster. Very recently Janssens et al. developed the DeepFlyBrain model, based on DeepMEL, for the prediction of chromatin co-accessible regions in the Drosophila brain.151 Similarly, the authors trained the DeepFlyBrain model on the aggregated pseudobulk profiles of three cell types, namely Kenyon cell, T neurons, and glia.
The other way is to utilize single-cell profiles at the genuine single-cell level. As single-cell profiles are well known for their sparsity, much research has been dedicated to applying deep learning for the imputation and inference on single-cell profiles. For example, DeepCpG,182 trained on the scBS-seq and scRRBS-seq profiles of multiple human and mouse tissues, uses a CNN + bidirectional GRU architecture and can impute methylation status for low-coverage single-cell methylation profiles. scGNN199 is a GNN-based autoencoder model for scRNA-seq data enhancement. scGNN utilizes multiple GNN and autoencoders that are effective in producing relationship-aware cell embeddings. The authors demonstrated that scGNN was effective in improving cell clustering and data imputation among four independent publicly available scRNA-seq datasets. SCALE190 is a variational autoencoder and Gaussian mixture model-based deep-learning model that performs imputation for low-coverage scATAC-seq profiles. Additionally, SCALE’s latent embedding of each cell was shown to be effective in scATAC-seq cell clustering and batch effect removal. scBasset is a recent model for scATAC-seq profile imputation. It improves upon SCALE by guiding imputation with the underlying genomic sequence. This is achieved by processing the genomic sequences into deep representations with a 6-layer CNN and incorporating them at the imputation step. scFAN198 is able to infer single-cell TF binding activity from scATAC-seq profiles. scFAN utilizes a sequence-based TF binding model that was trained on bulk TF binding profiles. scFAN then infers the per-cell TF binding activity by asking the model to predict the TF binding affinity to the chromatin-accessible regions of each cell as reported by the scATAC-seq profile.
Deep learning has also been effective in making inferences on gene regulation networks using scRNA-seq data. For example, CNNC185 infers the causality between two genes, e.g., gene A and gene B, in a gene regulatory network. CNNC uses a CNN to analyze the 2D expression level histogram between the two genes as if it were a 2D image and predicts whether there is an interaction between gene A and gene B, and, if so, whether gene A causally influences gene B or vice versa. scTenifoldKnk202 is a model for the in silico prediction of the gene knockout (KO) effects based on scRNA-seq data and gene regulatory networks. scTenifoldKnk first constructs a gene regulatory network based on a given scRNA-seq dataset. It then performs an in silico KO experiment by modifying the edges of the target genes in the network. scTenifoldKnk then performs a quasi-manifold alignment of the network before and after KO to predict its influence on the gene expression levels of all genes in the network.
As more and more single-cell profiling techniques are emerging and maturing, it is expected that more deep-learning applications for the imputation and inference in those data modalities are going to emerge. With the accumulation of evidence provided by single-cell gene regulation profiles, future gene regulation models will certainly better capture the gene regulation heterogeneity among cells.
Conclusions
To conclude, deep learning has certainly had successful applications in gene regulation. Being a data-driven approach, deep-learning-based methods have successfully modeled regulatory processes at various omics levels with high accuracy. With further improvement in deep-learning paradigms, ongoing development in omics technologies, and accumulation of omics data, deep-learning models are expected to be more accurate and make breakthroughs by providing biologically insightful predictions. We believe that in the foreseeable future, deep-learning-based predictive models for gene regulation will become indispensable tools that will aid biologists in solving real-world biological problems.
Acknowledgments
This work was supported by Office of Research Administration (ORA) at KAUST under award numbers FCC/1/1976-44-01, FCC/1/1976-45-01, URF/1/4098-01-01, URF/1/4352-01-01, REI/1/5202-01-01, REI/1/4940-01-01, RGC/3/4816-01-01, and REI/1/0018-01-01.
Author contributions
Z.L., E.G., and X.G. conceived the project. Z.L. and E.G. collected and reviewed relevant articles. Z.L. and E.G. drafted major parts of the manuscript. J.Z. contributed to the design of figure illustrations. W.H. and X.X. contributed to the design of tables. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
References
- 1.Kharchenko P.V., Tolstorukov M.Y., Park P.J. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ule J., Jensen K., Mele A., Darnell R.B. CLIP: a method for identifying protein–RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 3.Licatalosi D.D., Mele A., Fak J.J., Ule J., Kayikci M., Chi S.W., Clark T.A., Schweitzer A.C., Blume J.E., Wang X., et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song L., Crawford G.E. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb. Protoc. 2010;2010 doi: 10.1101/pdb.prot5384. pdb.prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoque M., Ji Z., Zheng D., Luo W., Li W., You B., Park J.Y., Yehia G., Tian B. Analysis of alternative cleavage and polyadenylation by 3' region extraction and deep sequencing. Nat. Methods. 2013;10:133–139. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siva N. 1000 Genomes project. Nat. Biotechnol. 2008;26:256–257. doi: 10.1038/nbt0308-256b. [DOI] [PubMed] [Google Scholar]
- 8.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roadmap Epigenomics Consortium. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., et al. The genotype-tissue expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leinonen R., Sugawara H., Shumway M., International Nucleotide Sequence Database Collaboration The sequence read archive. Nucleic Acids Res. 2011;39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leinonen R., Akhtar R., Birney E., Bower L., Cerdeno-Tárraga A., Cheng Y., Cleland I., Faruque N., Goodgame N., Gibson R., et al. The European nucleotide archive. Nucleic Acids Res. 2011;39:D28–D31. doi: 10.1093/nar/gkq967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eddy S.R. Hidden Markov models. Curr. Opin. Struct. Biol. 1996;6:361–365. doi: 10.1016/S0959-440X(96)80056-X. [DOI] [PubMed] [Google Scholar]
- 15.Zeng I.S.L., Lumley T. Review of statistical learning methods in integrated omics studies (an integrated information science) Bioinform. Biol. Insights. 2018;12 doi: 10.1177/1177932218759292. 1177932218759292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 17.Alipanahi B., Delong A., Weirauch M.T., Frey B.J. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 2015;33:831–838. doi: 10.1038/nbt.3300. http://www.nature.com/nbt/journal/v33/n8/abs/nbt.3300 [DOI] [PubMed] [Google Scholar]
- 18.Rives A., Meier J., Sercu T., Goyal S., Lin Z., Liu J., Guo D., Ott M., Zitnick C.L., Ma J., Fergus R. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2016239118. e2016239118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji Y., Zhou Z., Liu H., Davuluri R.V. DNABERT: pre-trained bidirectional encoder representations from Transformers model for DNA-language in genome. Bioinformatics. 2021;37:2112–2120. doi: 10.1093/bioinformatics/btab083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lecun Y., Bottou L., Bengio Y., Haffner P. Gradient-based learning applied to document recognition. Proc. IEEE. 1998;86:2278–2324. doi: 10.1109/5.726791. [DOI] [Google Scholar]
- 21.Cho K., Van Merriënboer B., Bahdanau D., Bengio Y. On the properties of neural machine translation: encoder-decoder approaches. Preprint at arXiv. 2014 doi: 10.48550/arXiv.1409.1259. [DOI] [Google Scholar]
- 22.Hochreiter S., Schmidhuber J. Long short-term memory. Neural Comput. 1997;9:1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 23.Vaswani A., Shazeer N., Parmar N., Uszkoreit J., Jones L., Gomez A.N., Kaiser Ł., Polosukhin I. Attention is all you need. Preprint at arXiv. 2017 doi: 10.48550/arXiv.1706.03762. [DOI] [Google Scholar]
- 24.Rao R.M., Liu J., Verkuil R., Meier J., Canny J., Abbeel P., Sercu T., Rives A. In: Proceedings of the 38th International Conference on Machine Learning. Marina M., Tong Z., editors. PMLR; 2021. MSA transformer. [Google Scholar]
- 25.Zou J., Huss M., Abid A., Mohammadi P., Torkamani A., Telenti A. A primer on deep learning in genomics. Nat. Genet. 2019;51:12–18. doi: 10.1038/s41588-018-0295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y., Huang C., Ding L., Li Z., Pan Y., Gao X. Deep learning in bioinformatics: introduction, application, and perspective in the big data era. Methods. 2019;166:4–21. doi: 10.1016/j.ymeth.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 27.Goodfellow I., Bengio Y., Courville A. MIT press; 2016. Deep Learning. [Google Scholar]
- 28.Weirauch M.T., Cote A., Norel R., Annala M., Zhao Y., Riley T.R., Saez-Rodriguez J., Cokelaer T., Vedenko A., Talukder S., et al. Evaluation of methods for modeling transcription factor sequence specificity. Nat. Biotechnol. 2013;31:126–134. doi: 10.1038/nbt.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jolma A., Yan J., Whitington T., Toivonen J., Nitta K.R., Rastas P., Morgunova E., Enge M., Taipale M., Wei G., et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 30.Zhou J., Troyanskaya O.G. Predicting effects of noncoding variants with deep learning–based sequence model. Nat. Methods. 2015;12:931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M.A., Beaudet A.L., Ecker J.R., et al. The NIH roadmap epigenomics mapping consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S.T., Abeysinghe S., Krawczak M., Cooper D.N. Human gene mutation database (HGMD®): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 33.Leslie R., O’Donnell C.J., Johnson A.D. GRASP: analysis of genotype–phenotype results from 1390 genome-wide association studies and corresponding open access database. Bioinformatics. 2014;30:i185–i194. doi: 10.1093/bioinformatics/btu273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelley D.R., Snoek J., Rinn J.L. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 2016;26:990–999. doi: 10.1101/gr.200535.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quang D., Xie X. DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res. 2016;44:e107. doi: 10.1093/nar/gkw226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng H., Gifford D.K. Predicting the impact of non-coding variants on DNA methylation. Nucleic Acids Res. 2017;45:e99. doi: 10.1093/nar/gkx177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang M., Tai C., E W., Wei L. DeFine: deep convolutional neural networks accurately quantify intensities of transcription factor-DNA binding and facilitate evaluation of functional non-coding variants. Nucleic Acids Res. 2018;46:e69. doi: 10.1093/nar/gky215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelley D.R., Reshef Y.A., Bileschi M., Belanger D., McLean C.Y., Snoek J. Sequential regulatory activity prediction across chromosomes with convolutional neural networks. Genome Res. 2018;28:739–750. doi: 10.1101/gr.227819.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noguchi S., Arakawa T., Fukuda S., Furuno M., Hasegawa A., Hori F., Ishikawa-Kato S., Kaida K., Kaiho A., Kanamori-Katayama M., et al. FANTOM5 CAGE profiles of human and mouse samples. Sci. Data. 2017;4:170112. doi: 10.1038/sdata.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Itoh M., Kojima M., Nagao-Sato S., Saijo E., Lassmann T., Kanamori-Katayama M., Kaiho A., Lizio M., Kawaji H., Carninci P., et al. Automated workflow for preparation of cDNA for cap analysis of gene expression on a single molecule sequencer. PLoS One. 2012;7:e30809. doi: 10.1371/journal.pone.0030809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou J., Theesfeld C.L., Yao K., Chen K.M., Wong A.K., Troyanskaya O.G. Deep learning sequence-based ab initio prediction of variant effects on expression and disease risk. Nat. Genet. 2018;50:1171–1179. doi: 10.1038/s41588-018-0160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agarwal V., Shendure J. Predicting mRNA abundance directly from genomic sequence using deep convolutional neural networks. Cell Rep. 2020;31:107663. doi: 10.1016/j.celrep.2020.107663. [DOI] [PubMed] [Google Scholar]
- 43.Minnoye L., Taskiran I.I., Mauduit D., Fazio M., Van Aerschot L., Hulselmans G., Christiaens V., Makhzami S., Seltenhammer M., Karras P., et al. Cross-species analysis of enhancer logic using deep learning. Genome Res. 2020;30:1815–1834. doi: 10.1101/gr.260844.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wouters J., Kalender-Atak Z., Minnoye L., Spanier K.I., De Waegeneer M., Bravo González-Blas C., Mauduit D., Davie K., Hulselmans G., Najem A., et al. Robust gene expression programs underlie recurrent cell states and phenotype switching in melanoma. Nat. Cell Biol. 2020;22:986–998. doi: 10.1038/s41556-020-0547-3. [DOI] [PubMed] [Google Scholar]
- 45.Bravo González-Blas C., Minnoye L., Papasokrati D., Aibar S., Hulselmans G., Christiaens V., Davie K., Wouters J., Aerts S. cisTopic: cis-regulatory topic modeling on single-cell ATAC-seq data. Nat. Methods. 2019;16:397–400. doi: 10.1038/s41592-019-0367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Avsec Ž., Agarwal V., Visentin D., Ledsam J.R., Grabska-Barwinska A., Taylor K.R., Assael Y., Jumper J., Kohli P., Kelley D.R. Effective gene expression prediction from sequence by integrating long-range interactions. Nat. Methods. 2021;18:1196–1203. doi: 10.1038/s41592-021-01252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelley D.R. Cross-species regulatory sequence activity prediction. PLoS Comput. Biol. 2020;16:e1008050. doi: 10.1371/journal.pcbi.1008050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avsec Ž., Weilert M., Shrikumar A., Krueger S., Alexandari A., Dalal K., Fropf R., McAnany C., Gagneur J., Kundaje A., Zeitlinger J. Base-resolution models of transcription-factor binding reveal soft motif syntax. Nat. Genet. 2021;53:354–366. doi: 10.1038/s41588-021-00782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He Q., Johnston J., Zeitlinger J. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat. Biotechnol. 2015;33:395–401. doi: 10.1038/nbt.3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fudenberg G., Kelley D.R., Pollard K.S. Predicting 3D genome folding from DNA sequence with Akita. Nat. Methods. 2020;17:1111–1117. doi: 10.1038/s41592-020-0958-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krietenstein N., Abraham S., Venev S.V., Abdennur N., Gibcus J., Hsieh T.-H.S., Parsi K.M., Yang L., Maehr R., Mirny L.A., et al. Ultrastructural details of mammalian chromosome architecture. Mol. Cell. 2020;78:554–565.e7. doi: 10.1016/j.molcel.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rao S.S.P., Huang S.-C., Glenn St Hilaire B., Engreitz J.M., Perez E.M., Kieffer-Kwon K.-R., Sanborn A.L., Johnstone S.E., Bascom G.D., Bochkov I.D., et al. Cohesin loss eliminates all loop domains. Cell. 2017;171:305–320.e24. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsieh T.-H.S., Cattoglio C., Slobodyanyuk E., Hansen A.S., Rando O.J., Tjian R., Darzacq X. Resolving the 3D landscape of transcription-linked mammalian chromatin folding. Mol. Cell. 2020;78:539–553.e8. doi: 10.1016/j.molcel.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonev B., Mendelson Cohen N., Szabo Q., Fritsch L., Papadopoulos G.L., Lubling Y., Xu X., Lv X., Hugnot J.-P., Tanay A., Cavalli G. Multiscale 3D genome rewiring during mouse neural development. Cell. 2017;171:557–572.e24. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou J. Sequence-based modeling of three-dimensional genome architecture from kilobase to chromosome scale. Nat. Genet. 2022;54:725–734. doi: 10.1038/s41588-022-01065-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karbalayghareh A., Sahin M., Leslie C.S. Chromatin interaction–aware gene regulatory modeling with graph attention networks. Genome Res. 2022;32:930–944. doi: 10.1101/gr.275870.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reiff S.B., Schroeder A.J., Kırlı K., Cosolo A., Bakker C., Mercado L., Lee S., Veit A.D., Balashov A.K., Vitzthum C., et al. The 4D Nucleome Data Portal as a resource for searching and visualizing curated nucleomics data. Nat. Commun. 2022;13:2365. doi: 10.1038/s41467-022-29697-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Velickovic P., Cucurull G., Casanova A., Romero A., Lio P., Bengio Y. Graph attention networks. Preprint at arXiv. 2017 doi: 10.48550/arXiv.1710.10903. [DOI] [Google Scholar]
- 60.Mukherjee S., Berger M.F., Jona G., Wang X.S., Muzzey D., Snyder M., Young R.A., Bulyk M.L. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jolma A., Kivioja T., Toivonen J., Cheng L., Wei G., Enge M., Taipale M., Vaquerizas J.M., Yan J., Sillanpää M.J., et al. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 2010;20:861–873. doi: 10.1101/gr.100552.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu F., Koltun V. Multi-scale context aggregation by dilated convolutions. Preprint at arXiv. 2016 doi: 10.48550/arXiv.1511.07122. [DOI] [Google Scholar]
- 63.Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 64.Corces M.R., Trevino A.E., Hamilton E.G., Greenside P.G., Sinnott-Armstrong N.A., Vesuna S., Satpathy A.T., Rubin A.J., Montine K.S., Wu B., et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods. 2017;14:959–962. doi: 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O., et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davuluri R.V., Suzuki Y., Sugano S., Plass C., Huang T.H.M. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 2008;24:167–177. doi: 10.1016/j.tig.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 67.Witten J.T., Ule J. Understanding splicing regulation through RNA splicing maps. Trends Genet. 2011;27:89–97. doi: 10.1016/j.tig.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Elkon R., Ugalde A.P., Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet. 2013;14:496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 69.Umarov R.K., Solovyev V.V. Recognition of prokaryotic and eukaryotic promoters using convolutional deep learning neural networks. PLoS One. 2017;12:e0171410. doi: 10.1371/journal.pone.0171410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dreos R., Ambrosini G., Cavin Périer R., Bucher P. EPD and EPDnew, high-quality promoter resources in the next-generation sequencing era. Nucleic Acids Res. 2013;41:D157–D164. doi: 10.1093/nar/gks1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gama-Castro S., Salgado H., Santos-Zavaleta A., Ledezma-Tejeida D., Muñiz-Rascado L., García-Sotelo J.S., Alquicira-Hernández K., Martínez-Flores I., Pannier L., Castro-Mondragón J.A., et al. RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 2016;44:D133–D143. doi: 10.1093/nar/gkv1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ishii T., Yoshida K., Terai G., Fujita Y., Nakai K. DBTBS: a database of Bacillus subtilis promoters and transcription factors. Nucleic Acids Res. 2001;29:278–280. doi: 10.1093/nar/29.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Umarov R., Kuwahara H., Li Y., Gao X., Solovyev V. Promoter analysis and prediction in the human genome using sequence-based deep learning models. Bioinformatics. 2019;35:2730–2737. doi: 10.1093/bioinformatics/bty1068. [DOI] [PubMed] [Google Scholar]
- 74.Zhou J., zhang b., Li H., Zhou L., Li Z., Long Y., Han W., Wang M., Cui H., Chen W., Gao X. DeeReCT-TSS: a novel meta-learning-based method annotates TSS in multiple cell types based on DNA sequences and RNA-seq data. Preprint at bioRxiv. 2021 doi: 10.1101/2021.07.14.452328. [DOI] [Google Scholar]
- 75.Barash Y., Calarco J.A., Gao W., Pan Q., Wang X., Shai O., Blencowe B.J., Frey B.J. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. http://www.nature.com/nature/journal/v465/n7294/suppinfo/nature09000_S1.html [DOI] [PubMed] [Google Scholar]
- 76.Fagnani M., Barash Y., Ip J.Y., Misquitta C., Pan Q., Saltzman A.L., Shai O., Lee L., Rozenhek A., Mohammad N., et al. Functional coordination of alternative splicing in the mammalian central nervous system. Genome Biol. 2007;8:R108. doi: 10.1186/gb-2007-8-6-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leung M.K.K., Xiong H.Y., Lee L.J., Frey B.J. Deep learning of the tissue-regulated splicing code. Bioinformatics. 2014;30:i121–i129. doi: 10.1093/bioinformatics/btu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brawand D., Soumillon M., Necsulea A., Julien P., Csárdi G., Harrigan P., Weier M., Liechti A., Aximu-Petri A., Kircher M., et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 79.Xiong H.Y., Alipanahi B., Lee L.J., Bretschneider H., Merico D., Yuen R.K.C., Hua Y., Gueroussov S., Najafabadi H.S., Hughes T.R., et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347:1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Z., Pan Z., Ying Y., Xie Z., Adhikari S., Phillips J., Carstens R.P., Black D.L., Wu Y., Xing Y. Deep-learning augmented RNA-seq analysis of transcript splicing. Nat. Methods. 2019;16:307–310. doi: 10.1038/s41592-019-0351-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B., et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 82.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., et al. GENCODE: the reference human genome annotation for the ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zeng T., Li Y.I. Predicting RNA splicing from DNA sequence using Pangolin. Genome Biol. 2022;23:103. doi: 10.1186/s13059-022-02664-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hubbard T., Barker D., Birney E., Cameron G., Chen Y., Clark L., Cox T., Cuff J., Curwen V., Down T., et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cardoso-Moreira M., Halbert J., Valloton D., Velten B., Chen C., Shao Y., Liechti A., Ascenção K., Rummel C., Ovchinnikova S., et al. Gene expression across mammalian organ development. Nature. 2019;571:505–509. doi: 10.1038/s41586-019-1338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leung M.K.K., Delong A., Frey B.J. Inference of the human polyadenylation code. Bioinformatics. 2018;34:2889–2898. doi: 10.1093/bioinformatics/bty211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee J.Y., Yeh I., Park J.Y., Tian B. PolyA_DB 2: mRNA polyadenylation sites in vertebrate genes. Nucleic Acids Res. 2007;35:D165–D168. doi: 10.1093/nar/gkl870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Müller S., Rycak L., Afonso-Grunz F., Winter P., Zawada A.M., Damrath E., Scheider J., Schmäh J., Koch I., Kahl G. APADB: a database for alternative polyadenylation and microRNA regulation events. Database. 2014;2014:bau076. doi: 10.1093/database/bau076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Derti A., Garrett-Engele P., Macisaac K.D., Stevens R.C., Sriram S., Chen R., Rohl C.A., Johnson J.M., Babak T. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lianoglou S., Garg V., Yang J.L., Leslie C.S., Mayr C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 2013;27:2380–2396. doi: 10.1101/gad.229328.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xia Z., Li Y., Zhang B., Li Z., Hu Y., Chen W., Gao X. DeeReCT-PolyA: a robust and generic deep learning method for PAS identification. Bioinformatics. 2019;35:2371–2379. doi: 10.1093/bioinformatics/bty991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kalkatawi M., Rangkuti F., Schramm M., Jankovic B.R., Kamau A., Chowdhary R., Archer J.A.C., Bajic V.B. Dragon PolyA Spotter: predictor of poly(A) motifs within human genomic DNA sequences. Bioinformatics. 2012;28:127–129. doi: 10.1093/bioinformatics/btr602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Magana-Mora A., Kalkatawi M., Bajic V.B. Omni-PolyA: a method and tool for accurate recognition of Poly(A) signals in human genomic DNA. BMC Genom. 2017;18:620. doi: 10.1186/s12864-017-4033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xiao M.S., Zhang B., Li Y.S., Gao Q., Sun W., Chen W. Global analysis of regulatory divergence in the evolution of mouse alternative polyadenylation. Mol. Syst. Biol. 2016;12:890. doi: 10.15252/msb.20167375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu Y., He K. Group normalization. Preprint at arXiv. 2018 doi: 10.48550/arXiv.1803.08494. [DOI] [Google Scholar]
- 96.Bogard N., Linder J., Rosenberg A.B., Seelig G. A deep neural network for predicting and engineering alternative polyadenylation. Cell. 2019;178:91–106.e23. doi: 10.1016/j.cell.2019.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li Z., Li Y., Zhang B., Li Y., Long Y., Zhou J., Zou X., Zhang M., Hu Y., Chen W., Gao X. DeeReCT-APA: prediction of alternative polyadenylation site usage through deep learning. Genom. Proteom. Bioinform. 2021 doi: 10.1016/j.gpb.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yan Z., Lécuyer E., Blanchette M. Prediction of mRNA subcellular localization using deep recurrent neural networks. Bioinformatics. 2019;35:i333–i342. doi: 10.1093/bioinformatics/btz337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bernhart S.H., Hofacker I.L., Stadler P.F. Local RNA base pairing probabilities in large sequences. Bioinformatics. 2006;22:614–615. doi: 10.1093/bioinformatics/btk014. [DOI] [PubMed] [Google Scholar]
- 100.Benoit Bouvrette L.P., Cody N.A.L., Bergalet J., Lefebvre F.A., Diot C., Wang X., Blanchette M., Lécuyer E. CeFra-seq reveals broad asymmetric mRNA and noncoding RNA distribution profiles in Drosophila and human cells. RNA. 2018;24:98–113. doi: 10.1261/rna.063172.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kaewsapsak P., Shechner D.M., Mallard W., Rinn J.L., Ting A.Y. Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking. Elife. 2017;6:e29224. doi: 10.7554/eLife.29224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cheng S., Guo M., Wang C., Liu X., Liu Y., Wu X. MiRTDL: a deep learning approach for miRNA target prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 2016;13:1161–1169. doi: 10.1109/TCBB.2015.2510002. [DOI] [PubMed] [Google Scholar]
- 103.Vlachos I.S., Paraskevopoulou M.D., Karagkouni D., Georgakilas G., Vergoulis T., Kanellos I., Anastasopoulos I.-L., Maniou S., Karathanou K., Kalfakakou D., et al. DIANA-TarBase v7. 0: indexing more than half a million experimentally supported miRNA: mRNA interactions. Nucleic Acids Res. 2015;43:D153–D159. doi: 10.1093/nar/gku1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cuperus J.T., Groves B., Kuchina A., Rosenberg A.B., Jojic N., Fields S., Seelig G. Deep learning of the regulatory grammar of yeast 5' untranslated regions from 500, 000 random sequences. Genome Res. 2017;27:2015–2024. doi: 10.1101/gr.224964.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ray D., Kazan H., Chan E.T., Peña Castillo L., Chaudhry S., Talukder S., Blencowe B.J., Morris Q., Hughes T.R. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 2009;27:667–670. doi: 10.1038/nbt.1550. [DOI] [PubMed] [Google Scholar]
- 106.Lam J.H., Li Y., Zhu L., Umarov R., Jiang H., Héliou A., Sheong F.K., Liu T., Long Y., Li Y., et al. A deep learning framework to predict binding preference of RNA constituents on protein surface. Nat. Commun. 2019;10:4941. doi: 10.1038/s41467-019-12920-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.He K., Zhang X., Ren S., Sun J. Deep residual learning for image recognition. Preprint at arXiv. 2015 doi: 10.48550/arXiv.1512.03385. [DOI] [Google Scholar]
- 109.Barreau C., Paillard L., Osborne H.B. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bertrand E., Chartrand P., Schaefer M., Shenoy S.M., Singer R.H., Long R.M. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 111.Wei J., Chen S., Zong L., Gao X., Li Y. Protein–RNA interaction prediction with deep learning: structure matters. Brief. Bioinform. 2022;23:bbab540. doi: 10.1093/bib/bbab540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Luo F., Wang M., Liu Y., Zhao X.-M., Li A. DeepPhos: prediction of protein phosphorylation sites with deep learning. Bioinformatics. 2019;35:2766–2773. doi: 10.1093/bioinformatics/bty1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Diella F., Cameron S., Gemünd C., Linding R., Via A., Kuster B., Sicheritz-Pontén T., Blom N., Gibson T.J. Phospho. ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinform. 2004;5:1–5. doi: 10.1186/1471-2105-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hornbeck P.V., Kornhauser J.M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Peri S., Navarro J.D., Kristiansen T.Z., Amanchy R., Surendranath V., Muthusamy B., Gandhi T.K.B., Chandrika K.N., Deshpande N., Suresh S., et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004;32:D497–D501. doi: 10.1093/nar/gkh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lu C.-T., Huang K.-Y., Su M.-G., Lee T.-Y., Bretaña N.A., Chang W.-C., Chen Y.-J., Chen Y.-J., Huang H.-D. DbPTM 3.0: an informative resource for investigating substrate site specificity and functional association of protein post-translational modifications. Nucleic Acids Res. 2013;41:D295–D305. doi: 10.1093/nar/gks1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li H., Xing X., Ding G., Li Q., Wang C., Xie L., Zeng R., Li Y. SysPTM: a systematic resource for proteomic research on post-translational modifications. Mol. Cell. Proteomics. 2009;8:1839–1849. doi: 10.1074/mcp.M900030-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Huang Y., Niu B., Gao Y., Fu L., Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Densely connected convolutional networks. Preprint at arXiv. 2017 doi: 10.48550/arXiv.1608.06993. [DOI] [Google Scholar]
- 120.Fu H., Yang Y., Wang X., Wang H., Xu Y. DeepUbi: a deep learning framework for prediction of ubiquitination sites in proteins. BMC Bioinform. 2019;20:86. doi: 10.1186/s12859-019-2677-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xu H., Zhou J., Lin S., Deng W., Zhang Y., Xue Y. PLMD: an updated data resource of protein lysine modifications. J. Genet. Genomics. 2017;44:243–250. doi: 10.1016/j.jgg.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 122.Wang D., Zeng S., Xu C., Qiu W., Liang Y., Joshi T., Xu D. MusiteDeep: a deep-learning framework for general and kinase-specific phosphorylation site prediction. Bioinformatics. 2017;33:3909–3916. doi: 10.1093/bioinformatics/btx496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang D., Liang Y., Xu D. Capsule network for protein post-translational modification site prediction. Bioinformatics. 2019;35:2386–2394. doi: 10.1093/bioinformatics/bty977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang D., Liu D., Yuchi J., He F., Jiang Y., Cai S., Li J., Xu D. MusiteDeep: a deep-learning based webserver for protein post-translational modification site prediction and visualization. Nucleic Acids Res. 2020;48:W140–W146. doi: 10.1093/nar/gkaa275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sabour S., Frosst N., Hinton G.E. Dynamic routing between capsules. Preprint at arXiv. 2017 doi: 10.48550/arXiv.1710.09829. [DOI] [Google Scholar]
- 126.Almagro Armenteros J.J., Sønderby C.K., Sønderby S.K., Nielsen H., Winther O. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics. 2017;33:3387–3395. doi: 10.1093/bioinformatics/btx431. [DOI] [PubMed] [Google Scholar]
- 127.Höglund A., Dönnes P., Blum T., Adolph H.-W., Kohlbacher O. MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics. 2006;22:1158–1165. doi: 10.1093/bioinformatics/btl002. [DOI] [PubMed] [Google Scholar]
- 128.Zhou J., Park C.Y., Theesfeld C.L., Wong A.K., Yuan Y., Scheckel C., Fak J.J., Funk J., Yao K., Tajima Y., et al. Whole-genome deep-learning analysis identifies contribution of noncoding mutations to autism risk. Nat. Genet. 2019;51:973–980. doi: 10.1038/s41588-019-0420-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Arloth J., Eraslan G., Andlauer T.F.M., Martins J., Iurato S., Kühnel B., Waldenberger M., Frank J., Gold R., Hemmer B., et al. DeepWAS: multivariate genotype-phenotype associations by directly integrating regulatory information using deep learning. PLoS Comput. Biol. 2020;16:e1007616. doi: 10.1371/journal.pcbi.1007616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Andlauer T.F.M., Buck D., Antony G., Bayas A., Bechmann L., Berthele A., Chan A., Gasperi C., Gold R., Graetz C., et al. Novel multiple sclerosis susceptibility loci implicated in epigenetic regulation. Sci. Adv. 2016;2:e1501678. doi: 10.1126/sciadv.1501678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Muglia P., Tozzi F., Galwey N.W., Francks C., Upmanyu R., Kong X.Q., Antoniades A., Domenici E., Perry J., Rothen S., et al. Genome-wide association study of recurrent major depressive disorder in two European case–control cohorts. Mol. Psychiatry. 2010;15:589–601. doi: 10.1038/mp.2008.131. [DOI] [PubMed] [Google Scholar]
- 132.Wichmann H.-E., Gieger C., Illig T. KORA-gen-resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2005;67:26–30. doi: 10.1055/s-2005-858226. MONICA/KORA Study Group [DOI] [PubMed] [Google Scholar]
- 133.Zingaretti L.M., Gezan S.A., Ferrão L.F.V., Osorio L.F., Monfort A., Muñoz P.R., Whitaker V.M., Pérez-Enciso M. Exploring deep learning for complex trait genomic prediction in polyploid outcrossing species. Front. Plant Sci. 2020;11:25. doi: 10.3389/fpls.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gezan S.A., Osorio L.F., Verma S., Whitaker V.M. An experimental validation of genomic selection in octoploid strawberry. Hortic. Res. 2017;4:16070. doi: 10.1038/hortres.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.de Bem Oliveira I., Resende M.F.R., Jr., Ferrão L.F.V., Amadeu R.R., Endelman J.B., Kirst M., Coelho A.S.G., Munoz P.R. Genomic prediction of autotetraploids; influence of relationship matrices, allele dosage, and continuous genotyping calls in phenotype prediction. G3. 2019;9:1189–1198. doi: 10.1534/g3.119.400059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Benevenuto J., Ferrão L.F.V., Amadeu R.R., Munoz P. How can a high-quality genome assembly help plant breeders? Gigascience. 2019;8:giz068. doi: 10.1093/gigascience/giz068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shook J., Gangopadhyay T., Wu L., Ganapathysubramanian B., Sarkar S., Singh A.K. Crop yield prediction integrating genotype and weather variables using deep learning. PLoS One. 2021;16:e0252402. doi: 10.1371/journal.pone.0252402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Abney T.S., Crochet W.D. 2005. The Uniform Soybean Tests: Northern Region 2005. [Google Scholar]
- 139.Bahdanau D., Cho K., Bengio Y. Neural machine translation by jointly learning to align and translate. Preprint at arXiv. 2014 doi: 10.48550/arXiv.1409.0473. [DOI] [Google Scholar]
- 140.Kingma D.P., Ba J. Adam: a method for stochastic optimization. Preprint at arXiv. 2014 doi: 10.48550/arXiv.1409.0473. [DOI] [Google Scholar]
- 141.Duchi J., Hazan E., Singer Y. Adaptive subgradient methods for online learning and stochastic optimization. J. Mach. Learn. Res. 2011;12 [Google Scholar]
- 142.Graves A. Generating sequences with recurrent neural networks. Preprint at arXiv. 2013 doi: 10.48550/arXiv.1308.0850. [DOI] [Google Scholar]
- 143.Dozat T. 2016. Incorporating Nesterov Momentum into Adam. [Google Scholar]
- 144.Loshchilov I., Hutter F. Decoupled weight decay regularization. Preprint at arXiv. 2017 doi: 10.48550/arXiv.1711.05101. [DOI] [Google Scholar]
- 145.Liu L., Jiang H., He P., Chen W., Liu X., Gao J., Han J. On the variance of the adaptive learning rate and beyond. Preprint at arXiv. 2019 doi: 10.48550/arXiv.1908.03265. [DOI] [Google Scholar]
- 146.Liaw R., Liang E., Nishihara R., Moritz P., Gonzalez J.E., Stoica I. Tune: a research platform for distributed model selection and training. Preprint at arXiv. 2018 doi: 10.48550/arXiv.1807.05118. [DOI] [Google Scholar]
- 147.Abadi M. 2016. TensorFlow: Learning Functions at Scale. [Google Scholar]
- 148.Paszke A., Gross S., Massa F., Lerer A., Bradbury J., Chanan G., Killeen T., Lin Z., Gimelshein N., Antiga L. PyTorch: an imperative style, high-performance deep learning library. Preprint at arXiv. 2019 doi: 10.48550/arXiv.1912.01703. [DOI] [Google Scholar]
- 149.O'Malley T.B., Elie, Long J., Chollet F., Jin H., Invernizzi L. 2019. KerasTuner.https://github.com/keras-team/keras-tuner [Google Scholar]
- 150.Chollet F.a.o. 2015. Keras.https://keras.io [Google Scholar]
- 151.Janssens J., Aibar S., Taskiran I.I., Ismail J.N., Gomez A.E., Aughey G., Spanier K.I., De Rop F.V., González-Blas C.B., Dionne M., et al. Decoding gene regulation in the fly brain. Nature. 2022;601:630–636. doi: 10.1038/s41586-021-04262-z. [DOI] [PubMed] [Google Scholar]
- 152.Sundararajan M., Taly A., Yan Q. PMLR); 2017. Axiomatic Attribution for Deep Networks; pp. 3319–3328. [Google Scholar]
- 153.Lundberg S.M., Lee S.-I. A unified approach to interpreting model predictions. Preprint at arXiv. 2017 doi: 10.48550/arXiv.1705.07874. [DOI] [Google Scholar]
- 154.Shrikumar A., Greenside P., Kundaje A. PMLR); 2017. Learning Important Features through Propagating Activation Differences; pp. 3145–3153. [Google Scholar]
- 155.Kokhlikyan N., Miglani V., Martin M., Wang E., Alsallakh B., Reynolds J., Melnikov A., Kliushkina N., Araya C., Yan S. Captum: a unified and generic model interpretability library for pytorch. Preprint at arXiv. 2020 doi: 10.48550/arXiv.2009.07896. [DOI] [Google Scholar]
- 156.Nesterov Y.E. 1983. A Method for Solving the Convex Programming Problem with Convergence Rate O(1/k2) pp. 543–547. [Google Scholar]
- 157.Bergstra J., Bengio Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 2012;13:281–305. [Google Scholar]
- 158.Shahriari B., Swersky K., Wang Z., Adams R.P., De Freitas N. Taking the human out of the loop: a review of Bayesian optimization. Proc. IEEE. 2016;104:148–175. [Google Scholar]
- 159.Simonyan K., Vedaldi A., Zisserman A. Deep inside convolutional networks: visualising image classification models and saliency maps. Preprint at arXiv. 2013 doi: 10.48550/arXiv.1312.6034. [DOI] [Google Scholar]
- 160.Paolacci G., Chandler J., Ipeirotis P.G. Running experiments on amazon mechanical turk. Judgment and Decision making. 2010;5:411–419. [Google Scholar]
- 161.UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Devlin J., Chang M.-W., Lee K., Toutanova K. Association for Computational Linguistics; Minneapolis, Minnesota: 2019. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding; pp. 4171–4186. [Google Scholar]
- 163.Mirdita M., Von Den Driesch L., Galiez C., Martin M.J., Söding J., Steinegger M. Uniclust databases of clustered and deeply annotated protein sequences and alignments. Nucleic Acids Res. 2017;45:D170–D176. doi: 10.1093/nar/gkw1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nichol A., Achiam J., Schulman J. On first-order meta-learning algorithms. Preprint at arXiv. 2018 doi: 10.48550/arXiv.1803.02999. [DOI] [Google Scholar]
- 165.He W., Jiang Y., Jin J., Li Z., Zhao J., Manavalan B., Su R., Gao X., Wei L. Accelerating bioactive peptide discovery via mutual information-based meta-learning. Brief. Bioinform. 2022;23:bbab499. doi: 10.1093/bib/bbab499. [DOI] [PubMed] [Google Scholar]
- 166.Aguilera-Mendoza L., Marrero-Ponce Y., Beltran J.A., Tellez Ibarra R., Guillen-Ramirez H.A., Brizuela C.A. Graph-based data integration from bioactive peptide databases of pharmaceutical interest: toward an organized collection enabling visual network analysis. Bioinformatics. 2019;35:4739–4747. doi: 10.1093/bioinformatics/btz260. [DOI] [PubMed] [Google Scholar]
- 167.Minkiewicz P., Iwaniak A., Darewicz M. BIOPEP-UWM database of bioactive peptides: current opportunities. Int. J. Mol. Sci. 2019;20:5978. doi: 10.3390/ijms20235978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Snell J., Swersky K., Zemel R.S. Prototypical networks for few-shot learning. Preprint at arXiv. 2017 doi: 10.48550/arXiv.1703.05175. [DOI] [Google Scholar]
- 169.Gainza P., Sverrisson F., Monti F., Rodolà E., Boscaini D., Bronstein M.M., Correia B.E. Deciphering interaction fingerprints from protein molecular surfaces using geometric deep learning. Nat. Methods. 2020;17:184–192. doi: 10.1038/s41592-019-0666-6. [DOI] [PubMed] [Google Scholar]
- 170.Masci J., Boscaini D., Bronstein M., Vandergheynst P. Geodesic convolutional neural networks on riemannian manifolds. Preprint at arXiv. 2015 doi: 10.48550/arXiv.1501.06297. [DOI] [Google Scholar]
- 171.Sverrisson F., Feydy J., Correia B.E., Bronstein M.M. Fast end-to-end learning on protein surfaces. Preprint at bioRxiv. 2021 doi: 10.1101/2020.12.28.424589. [DOI] [Google Scholar]
- 172.Kim M., Rai N., Zorraquino V., Tagkopoulos I. Multi-omics integration accurately predicts cellular state in unexplored conditions for Escherichia coli. Nat. Commun. 2016;7:13090. doi: 10.1038/ncomms13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang D., Liu S., Warrell J., Won H., Shi X., Navarro F.C.P., Clarke D., Gu M., Emani P., Yang Y.T., et al. Comprehensive functional genomic resource and integrative model for the human brain. Science. 2018;362:eaat8464. doi: 10.1126/science.aat8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Salakhutdinov R., Hinton G. In: Proceedings of the Twelth International Conference on Artificial Intelligence and Statistics. David van D., Max W., editors. PMLR; 2009. Deep Boltzmann machines. [Google Scholar]
- 175.Nguyen N.D., Huang J., Wang D. A deep manifold-regularized learning model for improving phenotype prediction from multi-modal data. Nat. Comput. Sci. 2022;2:38–46. doi: 10.1038/s43588-021-00185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Cadwell C.R., Scala F., Li S., Livrizzi G., Shen S., Sandberg R., Jiang X., Tolias A.S. Multimodal profiling of single-cell morphology, electrophysiology, and gene expression using Patch-seq. Nat. Protoc. 2017;12:2531–2553. doi: 10.1038/nprot.2017.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gouwens N.W., Sorensen S.A., Baftizadeh F., Budzillo A., Lee B.R., Jarsky T., Alfiler L., Baker K., Barkan E., Berry K., et al. Integrated morphoelectric and transcriptomic classification of cortical GABAergic cells. Cell. 2020;183:935–953.e19. doi: 10.1016/j.cell.2020.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Nguyen N.D., Blaby I.K., Wang D. ManiNetCluster: a novel manifold learning approach to reveal the functional links between gene networks. BMC Genom. 2019;20:1003. doi: 10.1186/s12864-019-6329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Chaudhary K., Poirion O.B., Lu L., Garmire L.X. Deep learning–based multi-omics integration robustly predicts survival in liver CancerUsing deep learning to predict liver cancer prognosis. Clin. Cancer Res. 2018;24:1248–1259. doi: 10.1158/1078-0432.CCR-17-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hinton G.E., Salakhutdinov R.R. Reducing the dimensionality of data with neural networks. Science. 2006;313:504–507. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- 181.Cusanovich D.A., Hill A.J., Aghamirzaie D., Daza R.M., Pliner H.A., Berletch J.B., Filippova G.N., Huang X., Christiansen L., DeWitt W.S., et al. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell. 2018;174:1309–1324.e18. doi: 10.1016/j.cell.2018.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Angermueller C., Lee H.J., Reik W., Stegle O. DeepCpG: accurate prediction of single-cell DNA methylation states using deep learning. Genome Biol. 2017;18:67. doi: 10.1186/s13059-017-1189-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Smallwood S.A., Lee H.J., Angermueller C., Krueger F., Saadeh H., Peat J., Andrews S.R., Stegle O., Reik W., Kelsey G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hou Y., Guo H., Cao C., Li X., Hu B., Zhu P., Wu X., Wen L., Tang F., Huang Y., Peng J. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016;26:304–319. doi: 10.1038/cr.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yuan Y., Bar-Joseph Z. Deep learning for inferring gene relationships from single-cell expression data. Proc. Natl. Acad. Sci. USA. 2019;116:27151–27158. doi: 10.1073/pnas.1911536116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Alavi A., Ruffalo M., Parvangada A., Huang Z., Bar-Joseph Z. A web server for comparative analysis of single-cell RNA-seq data. Nat. Commun. 2018;9:4768. doi: 10.1038/s41467-018-07165-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yevshin I., Sharipov R., Valeev T., Kel A., Kolpakov F. GTRD: a database of transcription factor binding sites identified by ChIP-seq experiments. Nucleic Acids Res. 2016:gkw951. doi: 10.1093/nar/gkw951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B., et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Xiong L., Xu K., Tian K., Shao Y., Tang L., Gao G., Zhang M., Jiang T., Zhang Q.C. SCALE method for single-cell ATAC-seq analysis via latent feature extraction. Nat. Commun. 2019;10:4576. doi: 10.1038/s41467-019-12630-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Corces M.R., Buenrostro J.D., Wu B., Greenside P.G., Chan S.M., Koenig J.L., Snyder M.P., Pritchard J.K., Kundaje A., Greenleaf W.J., et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 2016;48:1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Buenrostro J.D., Wu B., Litzenburger U.M., Ruff D., Gonzales M.L., Snyder M.P., Chang H.Y., Greenleaf W.J. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Li W.V., Li J.J. An accurate and robust imputation method scImpute for single-cell RNA-seq data. Nat. Commun. 2018;9:997. doi: 10.1038/s41467-018-03405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chen X., Miragaia R.J., Natarajan K.N., Teichmann S.A. A rapid and robust method for single cell chromatin accessibility profiling. Nat. Commun. 2018;9:5345. doi: 10.1038/s41467-018-07771-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Preissl S., Fang R., Huang H., Zhao Y., Raviram R., Gorkin D.U., Zhang Y., Sos B.C., Afzal V., Dickel D.E., et al. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat. Neurosci. 2018;21:432–439. doi: 10.1038/s41593-018-0079-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Chen X., Litzenburger U.M., Wei Y., Schep A.N., LaGory E.L., Choudhry H., Giaccia A.J., Greenleaf W.J., Chang H.Y. Joint single-cell DNA accessibility and protein epitope profiling reveals environmental regulation of epigenomic heterogeneity. Nat. Commun. 2018;9:4590. doi: 10.1038/s41467-018-07115-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kingma D.P., Welling M. Auto-encoding variational bayes. Preprint at arXiv. 2013 doi: 10.48550/arXiv.1312.6114. [DOI] [Google Scholar]
- 198.Fu L., Zhang L., Dollinger E., Peng Q., Nie Q., Xie X. Predicting transcription factor binding in single cells through deep learning. Sci. Adv. 2020;6:eaba9031. doi: 10.1126/sciadv.aba9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wang J., Ma A., Chang Y., Gong J., Jiang Y., Qi R., Wang C., Fu H., Ma Q., Xu D. scGNN is a novel graph neural network framework for single-cell RNA-Seq analyses. Nat. Commun. 2021;12:1882. doi: 10.1038/s41467-021-22197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Yuan H., Kelley D.R. scBasset: sequence-based modeling of single-cell ATAC-seq using convolutional neural networks. Nat. Methods. 2022;19:1088–1096. doi: 10.1038/s41592-022-01562-8. [DOI] [PubMed] [Google Scholar]
- 201.Buenrostro J.D., Corces M.R., Lareau C.A., Wu B., Schep A.N., Aryee M.J., Majeti R., Chang H.Y., Greenleaf W.J. Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell. 2018;173:1535–1548.e16. doi: 10.1016/j.cell.2018.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Osorio D., Zhong Y., Li G., Xu Q., Yang Y., Tian Y., Chapkin R.S., Huang J.Z., Cai J.J. scTenifoldKnk: an efficient virtual knockout tool for gene function predictions via single-cell gene regulatory network perturbation. Patterns. 2022;3:100434. doi: 10.1016/j.patter.2022.100434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Little D.R., Gerner-Mauro K.N., Flodby P., Crandall E.D., Borok Z., Akiyama H., Kimura S., Ostrin E.J., Chen J. Transcriptional control of lung alveolar type 1 cell development and maintenance by NK homeobox 2-1. Proc. Natl. Acad. Sci. USA. 2019;116:20545–20555. doi: 10.1073/pnas.1906663116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Nugent A.A., Lin K., Van Lengerich B., Lianoglou S., Przybyla L., Davis S.S., Llapashtica C., Wang J., Kim D.J., Xia D., et al. TREM2 regulates microglial cholesterol metabolism upon chronic phagocytic challenge. Neuron. 2020;105:837–854.e9. doi: 10.1016/j.neuron.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 205.Chen L., Toke N.H., Luo S., Vasoya R.P., Fullem R.L., Parthasarathy A., Perekatt A.O., Verzi M.P. A reinforcing HNF4–SMAD4 feed-forward module stabilizes enterocyte identity. Nat. Genet. 2019;51:777–785. doi: 10.1038/s41588-019-0384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wang C., Mahadevan S. 2009. A General Framework for Manifold Alignment. [Google Scholar]
- 207.Radford A., Narasimhan K., Salimans T., Sutskever I. 2018. Improving Language Understanding by Generative Pre-training. [Google Scholar]
- 208.Radford A., Wu J., Child R., Luan D., Amodei D., Sutskever I. Language models are unsupervised multitask learners. OpenAI Blog. 2019;1:9. [Google Scholar]
- 209.Brown T.B., Mann B., Ryder N., Subbiah M., Kaplan J., Dhariwal P., Neelakantan A., Shyam P., Sastry G., Askell A., et al. Language models are few-shot learners. Preprint at arXiv. 2020 doi: 10.48550/arXiv.2005.14165. [DOI] [Google Scholar]
- 210.Suzek B.E., Wang Y., Huang H., McGarvey P.B., Wu C.H., UniProt Consortium UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015;31:926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Townshend R.J.L., Eismann S., Watkins A.M., Rangan R., Karelina M., Das R., Dror R.O. Geometric deep learning of RNA structure. Science. 2021;373:1047–1051. doi: 10.1126/science.abe5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Xinshi Chen Y.L., Umarov R., Gao X., Song L. ICLR; 2020. RNA Secondary Structure Prediction by Learning Unrolled Algorithms. [Google Scholar]
- 214.Sverrisson F., Feydy J., Correia B.E., Bronstein M.M. Fast end-to-end learning on protein surfaces. Preprint at bioRxiv. 2021 doi: 10.1101/2020.12.28.424589. [DOI] [Google Scholar]
- 215.Baltrusaitis T., Ahuja C., Morency L.-P. Multimodal machine learning: a survey and taxonomy. IEEE Trans. Pattern Anal. Mach. Intell. 2019;41:423–443. doi: 10.1109/tpami.2018.2798607. [DOI] [PubMed] [Google Scholar]
- 216.Cusanovich D.A., Daza R., Adey A., Pliner H.A., Christiansen L., Gunderson K.L., Steemers F.J., Trapnell C., Shendure J. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Farlik M., Sheffield N.C., Nuzzo A., Datlinger P., Schönegger A., Klughammer J., Bock C. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 2015;10:1386–1397. doi: 10.1016/j.celrep.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Rotem A., Ram O., Shoresh N., Sperling R.A., Goren A., Weitz D.A., Bernstein B.E. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol. 2015;33:1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Picelli S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]