Abstract
CONSTANS (CO) is a master flowering-time regulator that integrates photoperiodic and circadian signals in Arabidopsis thaliana. CO is expressed in multiple tissues, including young leaves and seedling roots, but little is known about the roles and underlying mechanisms of CO in mediating physiological responses other than flowering. Here, we show that CO expression is responsive to jasmonate. CO negatively modulated jasmonate-imposed root-growth inhibition and anthocyanin accumulation. Seedlings from co mutants were more sensitive to jasmonate, whereas overexpression of CO resulted in plants with reduced sensitivity to jasmonate. Moreover, CO mediated the diurnal gating of several jasmonate-responsive genes under long-day conditions. We demonstrate that CO interacts with JASMONATE ZIM-DOMAIN (JAZ) repressors of jasmonate signaling. Genetic analyses indicated that CO functions in a CORONATINE INSENSITIVE1 (COI1)-dependent manner to modulate jasmonate responses. Furthermore, CO physically associated with the basic helix-loop-helix (bHLH) subgroup IIId transcription factors bHLH3 and bHLH17. CO acted cooperatively with bHLH17 in suppressing jasmonate signaling, but JAZ proteins interfered with their transcriptional functions and physical interaction. Collectively, our results reveal the crucial regulatory effects of CO on mediating jasmonate responses and explain the mechanism by which CO works together with JAZ and bHLH subgroup IIId factors to fine-tune jasmonate signaling.
CO is a negative regulator of jasmonate-imposed root growth inhibition and anthocyanin accumulation, and functions together with JAZ repressors and bHLH subgroup IIId factors in jasmonate signaling.
Introduction
The phytohormone jasmonate is a signaling molecule that is essential for regulating a variety of physiological processes (Chini et al., 2016; Howe et al., 2018; Guo et al., 2018a; Zhou et al., 2019; Wasternack, 2020; Cao et al., 2022). Mutations blocking jasmonate biosynthesis or signaling adversely affect several developmental programs and stress responses in Arabidopsis (Arabidopsis thaliana), such as root-growth inhibition, stamen development, trichome formation, and anthocyanin accumulation (Chini et al., 2016; Howe et al., 2018). Jasmonate is perceived by the F-box protein CORONATINE INSENSITIVE1 (COI1), which binds to ASK1 and ASK2 (Arabidopsis SKP1-like), CULLIN1, and RING-BOX1 (RBX1) to form the SCFCOI1 complex (Xie et al., 1998; Xu et al., 2002; Liu et al., 2004; Ren et al., 2005; Yan et al., 2009). After jasmonate is perceived, the SCFCOI1 complex recruits JASMONATE ZIM-DOMAIN (JAZ) proteins, which are critical repressors of jasmonate signaling, for their subsequent degradation via the 26S proteasome pathway (Chini et al., 2007; Thines et al., 2007; Sheard et al., 2010). The removal of JAZ proteins alleviates their repressive effect on downstream components to modulate various jasmonate responses (Fonseca et al., 2009; Fernández-Calvo et al., 2011; Kazan and Manners, 2013; Guo et al., 2018b).
JAZ repressors negatively regulate jasmonate signaling by interacting with and inhibiting multiple transcription factors. For example, the basic helix-loop-helix (bHLH) subgroup IIIe transcription factors MYC2, MYC3, and MYC4, which are the most extensively characterized JAZ-binding factors, control a subset of jasmonate-related processes, such as the inhibition of root elongation and stress responses (Boter et al., 2004; Lorenzo et al., 2004; Dombrecht et al., 2007; Fernández-Calvo et al., 2011; Schweizer et al., 2013; Qi et al., 2015; Liu et al., 2019; Wang et al., 2019; You et al., 2019). Several bHLH subgroup IIId transcription factors (bHLH3, bHLH13, bHLH14, and bHLH17) also physically interact with JAZ proteins and suppress jasmonate responses (Nakata et al., 2013; Sasaki-Sekimoto et al., 2013; Song et al., 2013a; Fonseca et al., 2014). Recently, we revealed that JAZ proteins directly interact with and inhibit the activities of the transcription factors ROOT HAIR DEFECTIVE 6 (RHD6) and RHD6-LIKE1 (RSL1) to regulate jasmonate-stimulated root hair development (Han et al., 2020). Furthermore, the interacting partners of JAZ repressors include several critical transcriptional regulators, such as ETHYLENE INSENSITIVE3 (EIN3), FILAMENTOUS FLOWER (FIL), TARGET OF EAT1 (TOE1), and ABSCISIC ACID INSENSITIVE5 (ABI5) (Fernández-Calvo et al., 2011; Qi et al., 2011; Song et al., 2011; Zhu et al., 2011; Hu et al., 2013; Jiang et al., 2014; Boter et al., 2015; Zhai et al., 2015; Ju et al., 2019; Pan et al., 2020). Although diverse transcription factors involved in jasmonate responses have been identified, the transcriptional regulation underlying jasmonate signaling-related processes has not been comprehensively characterized. Further clarifying the crucial JAZ-interacting regulators and elucidating the biological significance of their interactions may provide new insights into the molecular basis of the tight modulation and fine-tuning of jasmonate signaling networks.
B-box (BBX) proteins are a class of zinc-finger transcription factors that contain a conserved BBX domain in their N-terminal regions with or without a CCT (CONSTANS, CO-like, and TIMING OF CAB2 EXPRESSION1 [TOC1]) domain at the C terminus (Gangappa and Botto, 2014). Both the N-terminal and C-terminal regions of BBX proteins are involved in protein–protein interactions, DNA binding, or their transcriptional function (Gangappa and Botto, 2014). The BBX family of transcription factors controls a wide range of physiological processes, including the photoperiodic regulation of flowering, shade avoidance, seedling photomorphogenesis, and stress responses (Sarmiento, 2013; Gangappa and Botto, 2014). CONSTANS (CO, also named BBX1), which is the most well-known member of the BBX family, is a master regulator of flowering in the photoperiodic and circadian pathways (Putterill et al., 1995; Simon et al., 1996; Suárez-López et al., 2001; Takada and Goto, 2003; An et al., 2004; Song et al., 2013b, 2015; Shim et al., 2017). Under long-day conditions, CO directly stimulates the transcription of FLOWERING LOCUS T (FT), which encodes a floral induction-related long-distance signal, to determine flowering time in Arabidopsis (Samach et al., 2000; Corbesier et al., 2007; Tiwari et al., 2010; Song et al., 2013b; Shim et al., 2017). Further research revealed that CO functions in the vascular tissue (phloem) to activate FT expression and trigger photoperiodic flowering (An et al., 2004; Shim et al., 2017). The expression of CO under the control of a phloem-specific promoter was sufficient for inducing early flowering and rescuing the late-flowering phenotype of co mutants (An et al., 2004). Interestingly, spatial expression pattern analyses showed that CO is expressed in multiple tissues, including the shoot apex, young leaves, and seedling roots (Takada and Goto, 2003; An et al., 2004). These observations suggest that CO may contribute to the regulation of other physiological or developmental processes in addition to flowering.
The objective of this study was to identify additional CO regulatory functions and to characterize the molecular mechanisms by which CO mediates jasmonate signaling. We initially confirmed the spatial pattern of CO expression and detected its transcripts in young leaves and seedling roots. We also observed that the CO transcription level decreases in seedlings treated with methyl jasmonate (MeJA). Phenotypic analyses indicated that CO negatively regulates jasmonate-induced root-growth inhibition and anthocyanin accumulation. Compared to the wild-type control, loss-of-function co mutants exhibited greater MeJA-induced root-growth inhibition and anthocyanin accumulation. Conversely, CO overexpression decreased the sensitivity of transgenic seedlings to MeJA. Moreover, CO was involved in mediating the diurnal gating of several jasmonate-responsive genes under long-day conditions. Additionally, CO was able to interact with several JAZ repressors of jasmonate signaling. Genetic analyses demonstrated that the MeJA-hyposensitive phenotype of myc2-1 is attenuated by the CO mutation; however, there were no obvious differences between coi1-2 (which overaccumulates JAZ proteins) and co-1 coi1-2 mutants, suggesting that CO and MYC2 have the opposite effects and CO functions in a COI1-dependent manner to modulate jasmonate responses. Furthermore, CO physically interacted with the bHLH subgroup IIId transcription factors bHLH3 and bHLH17. Further analyses showed that CO works cooperatively with bHLH17 in repressing jasmonate signaling, and that JAZ proteins interfere with their transcriptional functions and physical association. Collectively, our results indicate that CO is a critical negative regulator of jasmonate-induced root-growth inhibition and anthocyanin accumulation. The data presented herein may be useful for clarifying how CO functions together with JAZ repressors and bHLH subgroup IIId transcription factors to precisely control jasmonate signaling.
Results
CO expression is responsive to jasmonate
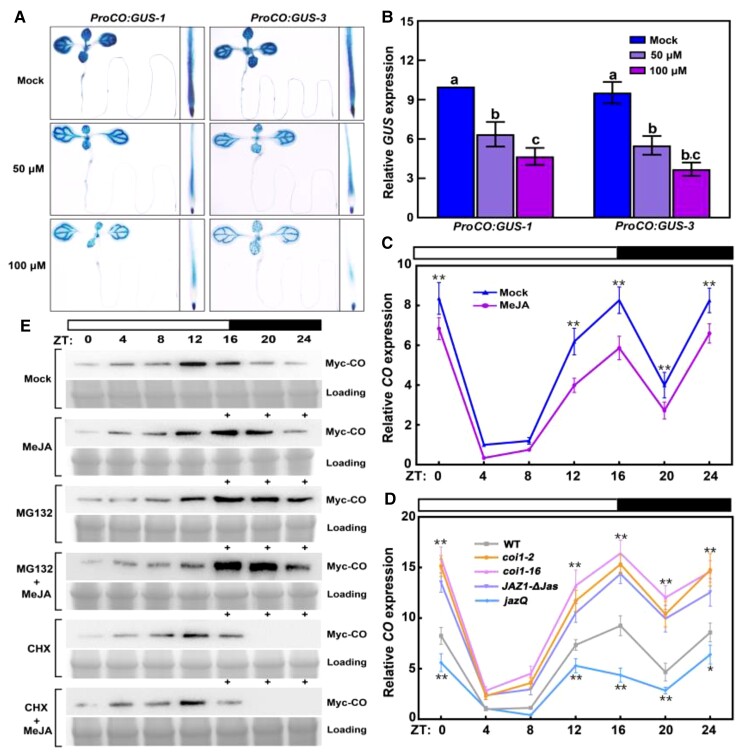
CO is a major positive regulator of flowering in the photoperiodic pathway (Putterill et al., 1995; Simon et al., 1996; Samach et al., 2000; Takada and Goto, 2003; An et al., 2004; Shim et al., 2017). More specifically, CO expression in the vascular tissue (phloem) promotes flowering (An et al., 2004). Because CO is expressed in multiple tissues, such as young leaves and seedling roots (Takada and Goto, 2003; An et al., 2004), we speculated that CO might be involved in other physiological or developmental processes besides flowering. We first determined the spatial expression pattern of CO. To this end, we cloned a CO promoter fragment (proCO; 3,576 bp) upstream of the β-glucuronidase reporter gene (GUS) to generate a reporter construct (ProCO:GUS), which we introduced into wild-type Columbia-0 (Col-0) plants. Consistent with the results of previous studies (Takada and Goto, 2003; An et al., 2004), GUS staining revealed CO promoter activity in the root and shoot apexes, young leaves, and seedling roots (Figure 1A).
Figure 1.
Expression patterns of CO in response to jasmonate. A, GUS staining of ProCO:GUS transgenic seedlings. Eight-day-old ProCO:GUS-1 and ProCO:GUS-3 seedlings grown in long days were treated with or without (mock) 50- or 100-μM MeJA for 4 h, and then the samples were harvested at ZT 16 for staining. In the mock treatment, an equal volume of 10% (v/v) ethanol was added. Experiments were performed three times with similar results. B, RT-qPCR analysis of relative GUS transcript levels in ProCO:GUS-1 and ProCO:GUS-3 seedlings. Total RNA was extracted from seedlings grown and treated as in (A). Relative GUS expression level of GUS in mock-treated ProCO:GUS-1 seedlings was set to 10. Values are means ± Sd from eight independent biological replicates (n = 8). Different lowercase letters indicate significant differences (P < 0.05) based on a two-way ANOVA (genotype × treatment interaction). C, RT-qPCR analysis of jasmonate-regulated CO expression in wild-type (WT, Col-0) seedlings. Total RNA was extracted from Col-0 seedlings grown on MS medium with or without (mock) 30-μM MeJA for 8 days under long-day conditions. The samples were harvested from ZT0 to ZT24. Time is expressed as h from dawn (ZT0). CO expression levels in mock-treated seedlings at ZT4 were set to 1. Values are means ± Sd from eight independent biological replicates (n = 8). **P < 0.01 as determined by a one-way ANOVA compared to Col-0. D, RT-qPCR analysis of CO expression in Col-0, coi1-2, coi1-16, jazQ, and JAZ1-ΔJas seedlings. Eight-day-old Col-0 and mutant seedlings grown in long days on MS medium were harvested from ZT0 to ZT24. CO expression levels in Col-0 seedlings at ZT4 were set to 1. Values are means ± Sd from eight independent biological replicates (n = 8). *P < 0.05; **P < 0.01 as analyzed by a one-way ANOVA compared to Col-0. E, Involvement of MeJA in CO abundance. Whole proteins were extracted from 8-day-old 35S:3Myc-CO-1 transgenic seedlings grown in long days (harvested from ZT0 to ZT24) with or without (mock) MeJA and/or the proteasome inhibitor (MG132) or protein synthesis inhibitor CHX treatment. For MeJA and/or MG132 or CHX treatments, seedlings were treated with 100-μM MeJA and/or 100-μM MG132 or 100-μM CHX for 4 h (+), and then the samples were harvested at ZT16, ZT20, and ZT24. CO was detected by immunoblotting with an anti-Myc antibody (1:10,000) and a secondary antibody (goat anti-mouse, 1:10,000). Experiments were performed three times with similar results.
Interestingly, a microarray-based analysis of Arabidopsis had suggested that CO transcription was repressed by jasmonate (Winter et al., 2007). To test the regulatory effect of jasmonate on CO transcription, we analyzed CO promoter activity on the basis of GUS staining and reverse transcription quantitative PCR (RT-qPCR) experiments. The GUS signal and GUS transcript levels decreased following exogenous MeJA treatment (Figure 1, A and B). We also performed RT-qPCR analyses to examine CO transcript levels in wild-type seedlings with or without MeJA treatments and observed that CO transcript levels decrease in response to MeJA treatment (Figure 1C). We then asked whether CO transcript levels are affected by critical components related to endogenous jasmonate signaling. The F-box protein COI1 is the jasmonate receptor that positively modulates jasmonate responses, whereas JAZ repressors are crucial negative regulators of jasmonate signaling (Xie et al., 1998; Chini et al., 2007; Thines et al., 2007; Yan et al., 2009). We thus examined CO transcript levels in coi1 mutants and transgenic plants overexpressing JAZ1 with a deleted Jas-encoding domain (JAZ1-ΔJas; Han et al., 2018). Relative CO transcript levels were higher in the coi1 mutants and JAZ1-ΔJas seedlings than in the wild type (Figure 1D). By contrast, CO expression levels were lower in the quintuple mutant jazQ (which lacks five JAZ repressors; Campos et al., 2016) than in the wild-type control (Figure 1D). Taken together, these results suggest that jasmonate represses CO transcription.
Previous studies have revealed that the stability of CO is strictly regulated by various signaling pathways at the post-translational level (Valverde et al., 2004; Jang et al., 2008; Liu et al., 2008; Lazaro et al., 2012; Song et al., 2012, 2014a; Zhang et al., 2015; Hayama et al., 2017). Hence, we investigated whether jasmonate also affected CO abundance. Specifically, we generated transgenic plants (35S:3Myc-CO) overexpressing a sequence encoding a fusion protein between full-length CO and the 3×Myc tag under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Pro35S; Supplemental Figure S1A). Similar to the results of earlier studies (Valverde et al., 2004; Liu et al., 2008), we detected a diurnal pattern in CO protein abundance (Figure 1E). Furthermore, we observed that CO protein accumulation clearly increases in response to MeJA (Figure 1E). Moreover, the abundance of CO further rose in seedlings treated with both MeJA and the proteasome inhibitor MG132 compared to that in seedlings treated with only MeJA or MG132 (Figure 1E), suggesting that MeJA stimulates the production of CO. However, CO protein accumulation was similar in seedlings treated with the protein synthesis inhibitor cycloheximide (CHX) with or without MeJA (Figure 1E), suggesting that MeJA may have little effect on CO degradation.
Loss of CO function stimulates jasmonate-induced root-growth inhibition and anthocyanin accumulation
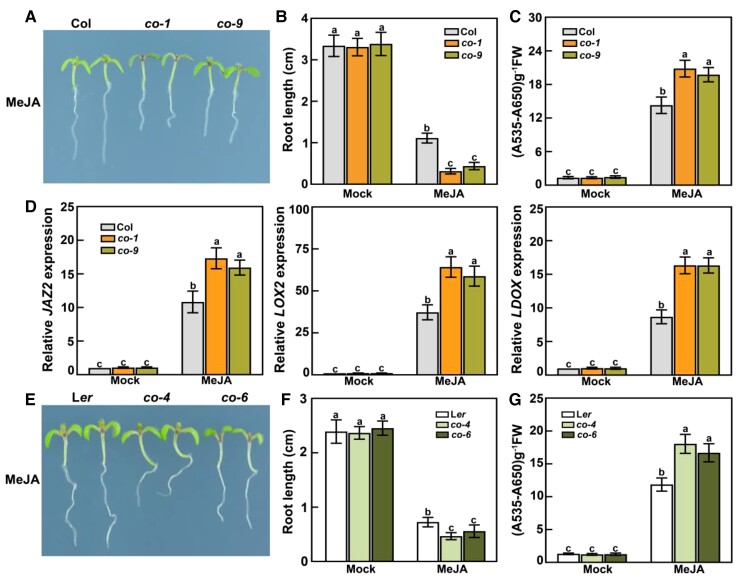
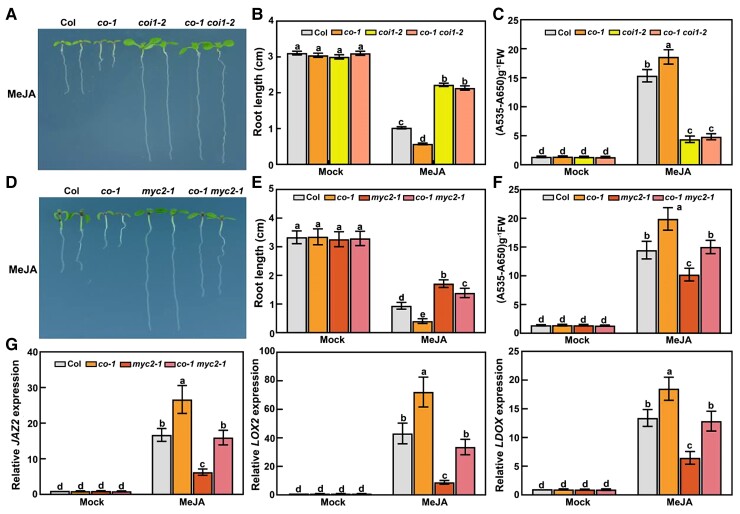
Because CO expression is responsive to jasmonate, we hypothesized that CO may help mediate responses to jasmonate. To test this possibility, we initially evaluated jasmonate-induced root-growth inhibition in the loss-of-function mutants co-1 and co-9 (Col-0 background) grown on Murashige and Skoog (MS) medium with or without 30-μM MeJA under long-day conditions (16-h light/8-h dark). The roots of co-1 and co-9 seedlings grown on MeJA-containing medium were significantly shorter than those of wild-type seedlings (Figure 2, A and B); however, in the absence of MeJA, the root lengths of the co mutants and the wild-type control were similar. In response to MeJA treatment, anthocyanin contents were higher in the co-1 and co-9 mutants than in the wild type (Figure 2C). To confirm the MeJA-induced phenotypes of the co-1 and co-9 mutants, we examined the transcript levels of several jasmonate-responsive genes in MeJA-treated co-1 and co-9 seedlings: JAZ2, LIPOXYGENASE2 (LOX2), and LEUCOANTHOCYANIDIN DIOXYGENASE (LDOX). After MeJA treatment, relative JAZ2, LOX2, and LDOX transcript levels were higher in the co-1 and co-9 seedlings than in the wild-type control (Figure 2D), indicating that CO negatively modulates the transcript levels of several jasmonate-responsive genes, possibly by modulating their transcription.
Figure 2.
Co mutants exhibit enhanced jasmonate responses under long-day conditions. A, Root phenotypes of 8-day-old wild-type (Col-0), co-1, and co-9 seedlings grown on MS medium with 30-μM MeJA under long-day conditions. B, Root length of 8-day-old seedlings grown on MS medium with or without (mock) 30-μM MeJA under long-day conditions. In the mock treatment, an equal volume of 10% (v/v) ethanol was added. The experiments were performed more than 5 times with similar results and 20 representative seedlings was measured for each repeat. Data are means ± Sd (n = 20 representative seedlings). C, Anthocyanin contents in 10-day-old Col-0, co-1, and co-9 seedlings grown on MS medium with or without (mock) 30-μM MeJA under long-day conditions. FW, fresh weight. The experiments were performed 8 times with similar results with more than 200 seedlings for each repeat. Data are means ± Sd from eight independent experiments (n = 8). D, RT-qPCR analysis of JAZ2, LOX2, and LDOX expression levels in Col-0, co-1, and co-9 seedlings. For JAZ2 and LOX2, total RNA was extracted from 8-day-old seedlings grown in long days and treated with or without (mock) 100-μM MeJA for 4 h. For LDOX, total RNA was extracted from 10-day-old seedlings grown on MS medium with or without (mock) 30-μM MeJA in long days. Relative expression levels in mock-treated Col-0 seedlings were set to 1. Values are means ± Sd from eight independent biological replicates (n = 8). E, Root phenotypes of 8-day-old Ler wild-type, co-4, and co-6 seedlings grown on MS medium with 30-μM MeJA under long-day conditions. F, Root length of 8-day-old seedlings grown on MS medium with or without (mock) 30-μM MeJA under long-day conditions. The experiments were performed more than 5 times with similar results and 20 representative seedlings were measured for each repeat. Data are means ± Sd (n = 20 representative plants). G, Anthocyanin contents in 10-day-old seedlings of Ler, co-4, and co-6 grown on MS medium with or without (mock) 30-μM MeJA under long-day conditions. FW, fresh weight. The experiments were performed 8 times with similar results with more than 200 seedlings for each repeat. Data are means ± Sd from eight independent experiments (n = 8). Different lowercase letters indicate significant differences (P < 0.05), as determined by a two-way ANOVA (genotype × treatment interaction).
To further analyze the effects of the loss of CO function on jasmonate signaling, we investigated the performances of the co-4 and co-6 mutants (Landsberg erecta [Ler] background) grown on MS medium containing 30-μM MeJA under long-day conditions. Analyses of jasmonate-regulated root growth and anthocyanin accumulation revealed the enhanced jasmonate responses of co-4 and co-6. Indeed, compared to seedlings from the wild-type Ler, the co-4 and co-6 mutants had shorter roots and accumulated more anthocyanin in the presence of MeJA (Figure 2, E–G). In response to MeJA treatment, relative JAZ2, LOX2, and LDOX transcript levels were consistently higher in the co-4 and co-6 mutants than in Ler seedlings (Supplemental Figure S2). These findings suggest that CO may negatively modulate jasmonate-induced root-growth inhibition and anthocyanin accumulation under long-day conditions.
Because jasmonate signaling is affected by photoperiod (Cagnola et al., 2018), we further analyzed the responses of co-1 and co-9 seedlings grown under short-day conditions (8-h light/16-h dark) to MeJA treatment. Seedlings from co-1, co-9, and the wild type were similarly affected by MeJA under short-day conditions (Supplemental Figure S3). Moreover, the MeJA-induced expression of LOX2 and LDOX in co-1 seedlings was similar to that in the wild-type control (Supplemental Figure S3). Accordingly, CO may not substantially affect jasmonate signaling under short-day conditions.
Overexpression of CO attenuates jasmonate-mediated root-growth inhibition and anthocyanin accumulation
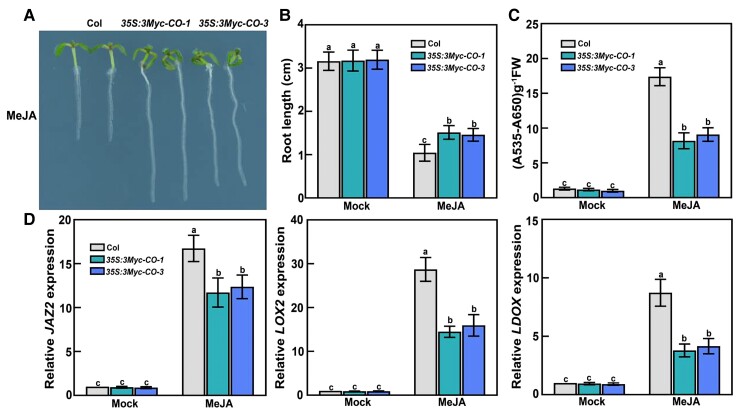
Having demonstrated that co mutants exhibit enhanced jasmonate signaling under long-day conditions, we analyzed whether CO overexpression had inhibitory effects on jasmonate responses. We selected homozygous transgenic lines overexpressing CO (35S:3Myc-CO-1 and 35S:3Myc-CO-3) for phenotypic analyses (Supplemental Figure S1A). Consistent with previous studies (Putterill et al., 1995; Simon et al., 1996; Suárez-López et al., 2001; Wang et al., 2016), we observed that the T5 progeny of 35S:3Myc-CO-1 and 35S:3Myc-CO-3 plants flowered earlier than wild-type plants. We examined jasmonate-regulated root elongation and anthocyanin accumulation in the 35S:3Myc-CO-1 and 35S:3Myc-CO-3 seedlings: their roots were much longer than those of the wild type after a 30-μM MeJA treatment under long-day conditions (Figure 3, A and B). Moreover, following MeJA treatment, anthocyanin contents were lower in the 35S:3Myc-CO-1 and 35S:3Myc-CO-3 seedlings compared to the wild type (Figure 3C). To confirm these observations, we analyzed the transcript levels of several jasmonate-responsive genes, including JAZ2, LOX2, and LDOX, in MeJA-treated 35S:3Myc-CO-1 and 35S:3Myc-CO-3 samples. The relative transcript levels of these jasmonate-inducible genes were lower in the 35S:3Myc-CO-1 and 35S:3Myc-CO-3 seedlings than in the wild-type control (Figure 3D). Thus, CO overexpression decreases the sensitivity of the transgenic seedlings to MeJA. These findings provide further evidence that CO negatively regulates jasmonate-induced root-growth inhibition and anthocyanin accumulation under long-day conditions.
Figure 3.
Overexpression of CO attenuates jasmonate responses under long-day conditions. A, Root phenotypes of 8-day-old wild-type (Col-0) and 35S:3Myc-CO transgenic seedlings grown on MS medium with 30-μM MeJA under long-day conditions. B, Root length of 8-day-old seedlings grown on MS medium with or without (mock) 30-μM MeJA under long-day conditions. In the mock treatment, an equal volume of 10% (v/v) ethanol was added. The experiments were performed more than 5 times with similar results and 20 representative seedlings was measured for each repeat. Data are means ± Sd (n = 20 representative plants). C, Anthocyanin contents in 10-day-old Col-0 and 35S:3Myc-CO seedlings grown on MS medium with or without (mock) 30-μM MeJA under long-day conditions. The experiments were performed 8 times with similar results with more than 200 seedlings for each repeat. Data are means ± Sd from eight independent experiments (n = 8). FW, fresh weight. D, RT-qPCR analysis of JAZ2, LOX2, and LDOX expression levels in Col-0 and 35S:3Myc-CO seedlings. For JAZ2 and LOX2, total RNA was extracted from 8-day-old seedlings grown in long days and treated with or without (mock) 100-μM MeJA for 4 h. For LDOX, total RNA was extracted from 10-day-old seedlings grown on MS medium with or without (mock) 30-μM MeJA under long-day conditions. Relative expression levels in mock-treated Col-0 seedlings were set to 1. Values are means ± Sd from eight independent biological replicates (n = 8). Different lowercase letters indicate significant differences (P < 0.05) as determined by a two-way ANOVA (genotype × treatment interaction).
CO mediates the diurnal gating of several jasmonate-responsive genes under long-day conditions
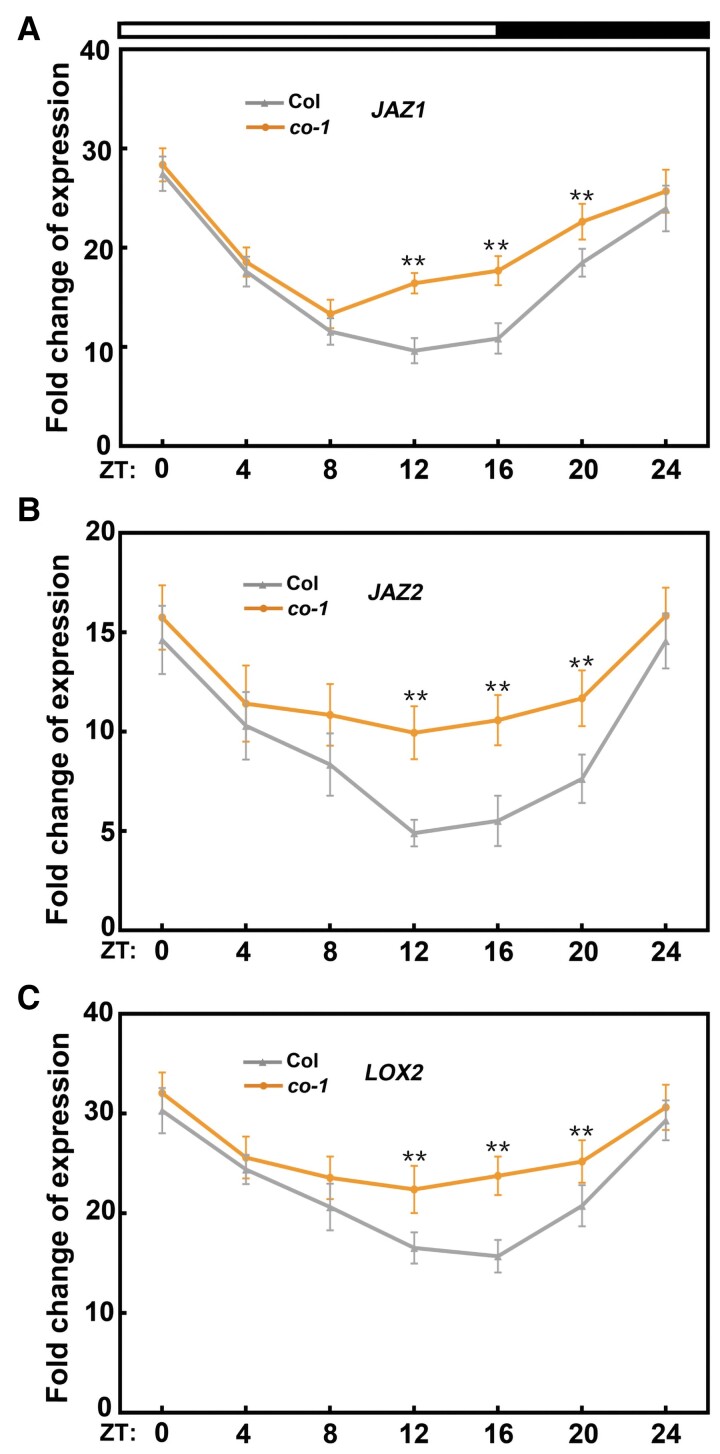
Multiple studies have confirmed that jasmonate signaling is modulated by the circadian clock (Goodspeed et al., 2012; Shin et al., 2012; Nitschke et al., 2016; Zhang et al., 2018; Thines et al., 2019). For example, Shin et al. (2012) investigated the MeJA-induced expression of several jasmonate-responsive genes throughout the day and found that the gene expression levels oscillated from dawn to dusk. On the basis of the circadian pattern of CO expression and the accumulation of the encoded protein (Suárez-López et al., 2001; Valverde et al., 2004; Liu et al., 2008), we speculated that CO might be involved in diurnal or circadian clock-mediated jasmonate signaling. To test this possibility, we examined the transcript levels of several well-known jasmonate-responsive genes (e.g. JAZ1, JAZ2, and LOX2) in MeJA-treated wild-type and co-1 seedlings throughout the day. We grew seedlings under long-day conditions and calculated the fold-change in JAZ1, JAZ2, and LOX2 transcript levels after a 1-h MeJA treatment in 4-h intervals over one diurnal cycle. In accordance with the findings of a previous study (Shin et al., 2012), we detected the rhythmic gating of MeJA responses in all three transcripts, with a peak induction at Zeitgeber time (ZT) 0 (dawn) (Figure 4, A–C). Moreover, the MeJA-induced expression of JAZ1, JAZ2, and LOX2 was significantly higher in co-1 seedlings than in the wild type at ZT12, ZT16, and ZT20 (Figure 4, A–C). These observations suggest that CO contributes to the gating of clock-mediated jasmonate signaling mainly at ZT12, ZT16, and ZT20, which corresponds to the period during which CO is expressed and the encoded protein accumulates (Suárez-López et al., 2001; Valverde et al., 2004; Liu et al., 2008).
Figure 4.
CO mediates the diurnal or circadian gating of several jasmonate-responsive genes under long-day conditions. Fold-induction of JAZ1 (A), JAZ2 (B), and LOX2 (C) expression in response to MeJA treatment. Total RNA was extracted from 8-day-old Col-0 and co-1 seedlings grown in long days and treated with or without (mock) 100-μM MeJA for 1 h at 4-h intervals over one diurnal cycle. In the mock treatment, an equal volume of 10% (v/v) ethanol was added. Gene expression levels at each time point were measured by RT-qPCR. The expression levels of each gene in mock-treated seedlings at each time point were set to 1. Fold-induction relative to the basal expression levels were then determined. Values are means ± Sd from eight independent biological replicates (n = 8). **P < 0.01 compared to Col-0, as determined by a one-way ANOVA.
CO interacts with several JAZ repressors of jasmonate signaling
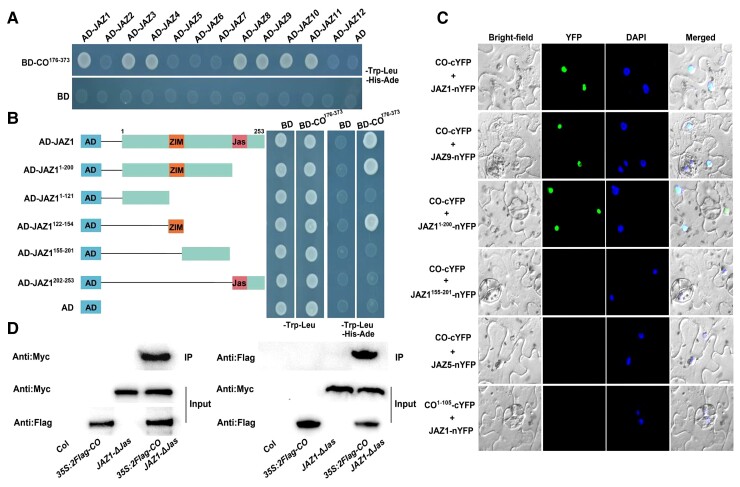
Having ascertained that CO mediates jasmonate signaling, we conducted further analyses to explore the underlying molecular mechanisms. Previous research demonstrated that JAZ proteins are repressors of jasmonate signaling and negatively regulate various responses to jasmonate (Chini et al., 2007; Thines et al., 2007). Notably, JAZ repressors interact with the COI1 receptor and transcription factors to link jasmonate perception with transcriptional changes (Chini et al., 2007; Thines et al., 2007; Kazan and Manners, 2013; Guo et al., 2018b). Because CO is responsive to exogenous jasmonate and negatively modulates jasmonate responses, we hypothesized that it might interact directly with JAZ proteins. To test this possibility, we performed yeast two-hybrid (Y2H) assays to examine the physical interactions between CO and JAZ repressors. For these analyses, we cloned the full-length JAZ coding sequences downstream of the sequence encoding the yeast (Saccharomyces cerevisiae) GAL4 activation domain in the prey vector (AD-JAZ). Additionally, we cloned the sequence encoding the C-terminal region of CO containing the CCT domain (amino acids 176–373; Gangappa and Botto, 2014) downstream of the sequence encoding the GAL4 DNA-binding domain (BD) in the bait vector (BD-CO176–373). In yeast cells, we determined that CO interacts with JAZ1, JAZ3, JAZ4, JAZ8, JAZ9, JAZ10, and JAZ11 (Figure 5A; Supplemental Figure S4). To determine which JAZ1 protein region interacts with CO, we conducted additional directed Y2H analyses, which indicated that the ZIM domain of JAZ1 (AD-JAZ1122–154) is sufficient for the interaction (Figure 5B).
Figure 5.
Physical interactions of CO with JAZ repressors. A, Yeast two-hybrid (Y2H) assays. Interactions of CO with JAZ are indicated by the ability of yeast cells to grow on SD medium lacking Leu, Trp, His, and Ade. pGBKT7 (BD) and pGADT7 (AD) vectors were used as negative controls. B, Mapping the CO-interacting domain of JAZ1 using a Y2H assay. Left, diagram of full-length and truncated JAZ1 constructs with specific regions. Right, interaction as indicated by the ability of cells to grow on SD medium lacking Leu, Trp, His, and Ade. BD and AD vectors were used as negative controls. C, BiFC assays. Fluorescence was detected in the nuclei of N. benthamiana cells co-expressing CO-cYFP and JAZ1-nYFP, JAZ9-nYFP, or JAZ11–200-nYFP. No signal was observed in the negative controls CO-cYFP and JAZ1155–201-nYFP or JAZ5-nYFP, or CO1–105-cYFP and JAZ1-nYFP. Nuclei are indicated by DAPI staining. D, Co-IP assays. Total proteins were extracted from 35S:2Flag-CO JAZ1-ΔJas transgenic Arabidopsis seedlings. Flag-tagged CO was immunoprecipitated using an anti-Flag antibody (1:250) and the co-immunoprecipitated protein was detected with an anti-Myc antibody (1:10,000, monoclonal antibody) and a secondary antibody (goat anti-mouse, 1:10,000). Similarly, Myc-tagged JAZ1 was immunoprecipitated using an anti-Myc antibody (1:250) and the co-immunoprecipitated protein was detected with an anti-Flag antibody (1:10,000). Protein input for 2Flag-CO or 3Myc-fused JAZ1 in immunoprecipitated complexes was also detected. Experiments were performed three times with similar results.
To confirm that CO interacts with JAZ repressors, we conducted bimolecular fluorescence complementation (BiFC) assays in Nicotiana benthamiana. We prepared a fusion construct encoding the C-terminal fragment of the yellow fluorescent protein (cYFP) fused to the full-length CO sequence (CO-cYFP) under the control of the 35S promoter. Additionally, we generated fusion constructs encoding the N-terminal fragment of YFP (nYFP) fused to the full-length JAZ1 or JAZ9 sequence (JAZ1-nYFP and JAZ9-nYFP). When CO-cYFP was transiently co-expressed with JAZ1-nYFP or JAZ9-nYFP in N. benthamiana leaves, we detected strong YFP fluorescence in the nucleus, as evidenced by overlap with 4′,6-diamidino-2-phenylindole (DAPI) staining (Figure 5C; Supplemental Figure S4). Moreover, we also observed the YFP signal in N. benthamiana leaves when CO-cYFP was co-expressed with the construct encoding the N-terminal amino acids (1–200) of JAZ1 fused to nYFP (JAZ11–200-nYFP; Figure 5C; Supplemental Figure S4). We detected no fluorescence in the negative controls in which CO-cYFP was co-expressed with JAZ1155–201-nYFP (amino acids 155–201 of JAZ1 fused to nYFP) or JAZ5-nYFP (JAZ5 fused to nYFP) or when CO1–105-cYFP (amino acids 1–105 of CO fused to cYFP) was co-expressed with JAZ1-nYFP (Figure 5C; Supplemental Figure S4). We also confirmed the interaction between CO and JAZ1 by conducting co-immunoprecipitation (Co-IP) assays involving transgenic Arabidopsis seedlings simultaneously overexpressing CO and JAZ1 (35S:2Flag-CO JAZ1-ΔJas; Figure 5D). These seedlings were derived from a cross between plants carrying 35S:2Flag-CO-2 (containing a 2×Flag-CO construct driven by the 35S promoter; Supplemental Figure S1B) and the previously described JAZ1-ΔJas plants (Han et al., 2018). Together, these results confirm that CO interacts with several JAZ repressors of jasmonate signaling, indicating that CO functions as a JAZ-binding factor that mediates jasmonate signaling.
CO functions in a COI1-dependent manner to mediate jasmonate responses
JAZ proteins suppress jasmonate signaling through their associations with multiple transcription factors. They accumulate to high levels in coi1 mutants, wherein they repress responses to jasmonate (Chini et al., 2007; Thines et al., 2007; Katsir et al., 2008; Browse, 2009; Fonseca et al., 2009). Because CO negatively mediates jasmonate responses and interacts with crucial components (JAZ repressors) of jasmonate signaling, we asked whether CO function is dependent on endogenous jasmonate perception and signaling. To test this possibility, we crossed co-1 with coi1-2 (a mutant with a leaky mutant allele of COI1 encoding a receptor with the missense mutation L245F; Xu et al., 2002) to obtain the co-1 coi1-2 double mutant. We then examined the MeJA-induced phenotype of the double mutant. Compared to the wild-type control, the co-1 mutant exhibited increased MeJA sensitivity, whereas the coi1-2 mutant was hyposensitive to MeJA (Figure 6, A–C; Xu et al., 2002). Similar to coi1-2, co-1 coi1-2 seedlings were hyposensitive to MeJA (Figure 6, A–C). These results suggest that the enhanced sensitivity of co-1 to jasmonate is dependent on functional COI1 and the perception of jasmonate. The MYC2 transcription factor is a central regulator of jasmonate signaling (Boter et al., 2004; Lorenzo et al., 2004). To investigate whether the relationship between CO and MYC2 may modulate jasmonate signaling, we crossed co-1 to myc2-1 to generate the co-1 myc2-1 double mutant. Genetic analyses showed that the MeJA-hyposensitive phenotype of myc2-1 is attenuated by the CO mutation (Figure 6, D–F). Additionally, relative transcript levels of jasmonate-responsive genes were higher in co-1 myc2-1 seedlings than in myc2-1 (Figure 6G). Accordingly, CO and MYC2 appear to have opposite effects on jasmonate responses.
Figure 6.
Jasmonate responses of co-1 coi1-2 and co-1 myc2-1 double mutant seedlings. A, Root phenotypes of 8-day-old wild type (Col-0), co-1, coi1-2, and co-1 coi1-2 seedlings grown on MS medium with 30-μM MeJA under long-day conditions. B, Root length of 8-day-old seedlings grown on MS medium with or without (mock) 30-μM MeJA under long-day conditions. In the mock treatment, an equal volume of 10% (v/v) ethanol was added. The experiments were performed more than 5 times with similar results and 20 representative seedlings were measured for each repeat. Data are means ± Sd (n = 20 representative plants). C, Anthocyanin contents in 10-day-old Col-0, co-1, coi1-2, and co-1 coi1-2 seedlings grown on MS medium with or without (mock) 30-μM MeJA under long-day conditions. The experiments were performed 8 times with similar results with more than 200 seedlings for each repeat. Data are means ± Sd from eight independent experiment (n = 8). FW, fresh weight. D, Root phenotypes of 8-day-old Col-0, co-1, myc2-1, and co-1 myc2-1 seedlings grown on MS medium with 30-μM MeJA under long-day conditions. E, Root length of 8-day-old seedlings grown on MS medium with or without (mock) 30-μM MeJA. The experiments were performed more than 5 times with similar results and 20 representative seedlings were measured for each repeat. Data are means ± Sd (n = 20 representative plants). F, Anthocyanin contents in 10-day-old Col-0, co-1, myc2-1, and co-1 myc2-1 seedlings grown in long days on MS medium with or without (mock) 30-μM MeJA. The experiments were performed 8 times with similar results with more than 200 seedlings for each repeat. Values are means ± Sd from eight independent experiments (n = 8). G, RT-qPCR analysis of JAZ2, LOX2, and LDOX expression levels in Col-0, co-1, myc2-1, and co-1 myc2-1 seedlings. For JAZ2 and LOX2, total RNA was extracted from 8-day-old seedlings grown in long days and treated with or without (mock) 100-μM MeJA for 4 h. For LDOX, total RNA was extracted from 10-day-old seedlings grown in long days on MS medium with or without (mock) 30-μM MeJA. Relative expression levels in mock-treated Col-0 seedlings were set to 1. Values are means ± Sd from eight independent biological replicates (n = 8). Different lowercase letters indicate significant differences (P < 0.05) as determined by a two-way ANOVA (genotype × treatment interaction).
CO physically associates with the bHLH subgroup IIId transcription factors bHLH3 and bHLH17
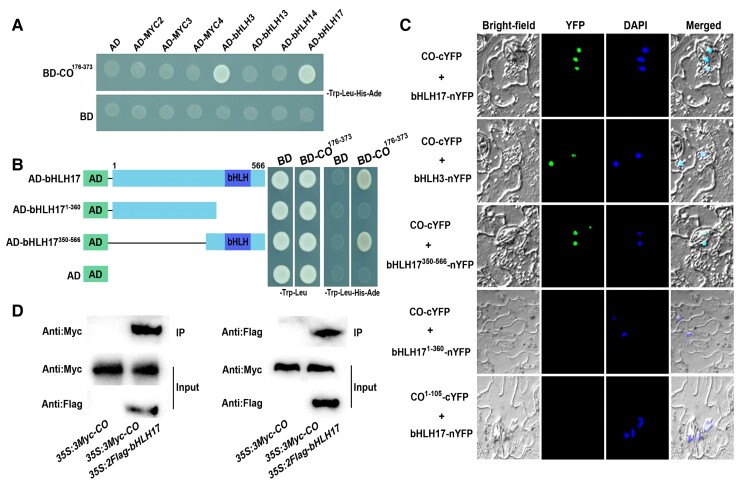
Because of the contrasting effects of CO and MYC2 on jasmonate signaling, we tested whether CO interacts with MYC2 and its homologs (MYC3 and MYC4). To this end, we generated constructs encoding full-length MYC2, MYC3, and MYC4 fused to the GAL4 activation domain in the prey vector (AD-MYC) for Y2H analyses. However, we detected no interactions between CO and MYC2, MYC3, or MYC4 in yeast cells (Figure 7A; Supplemental Figure S5). Interestingly, in addition to CO, several bHLH subgroup IIId transcription factors (bHLH3, bHLH13, bHLH14, and bHLH17) reportedly bind to JAZ repressors and negatively modulate jasmonate responses (Nakata et al., 2013; Sasaki-Sekimoto et al., 2013; Song et al., 2013a; Fonseca et al., 2014), prompting us to investigate their association with CO. We first examined their possible interactions in an Y2H assay by cloning the full-length coding sequence of each bHLH transcription factor gene downstream of the sequence of the GAL4 activation domain in the prey vector (AD-bHLH). In yeast cells, CO strongly interacted with bHLH3 and bHLH17, but not with bHLH13 or bHLH14 (Figure 7A; Supplemental Figure S5). Next, we determined which bHLH17 region is essential for interaction with CO. Specifically, we divided bHLH17 into its N-terminal fragment (amino acids 1–360) and its C-terminal fragment (amino acids 350–566) that includes the bHLH domain (Figure 7B). We determined that the C-terminal region of bHLH17 is required for interaction with CO (Figure 7B).
Figure 7.
Physical interactions of CO with bHLH3 and bHLH17. A, Y2H assays. Interactions of CO with bHLH3 and bHLH17 are indicated by the ability of yeast cells to grow on SD medium lacking Leu, Trp, His, and Ade. pGBKT7 (BD) and pGADT7 (AD) were used as negative controls. B, Mapping the CO-interacting domain of bHLH17 using a Y2H assay. Left, diagram of full-length and truncated bHLH17 constructs with specific regions. Right, interaction as indicated by the ability of cells to grow on SD medium lacking Leu, Trp, His, and Ade. BD and AD vectors were used as negative controls. C, BiFC assays. Fluorescence was detected in the nuclei of N. benthamiana cells co-expressing CO-cYFP and bHLH3-nYFP, bHLH17-nYFP, or bHLH17350–566-nYFP. No signal was observed in the negative controls co-expressing CO-cYFP and bHLH171–360-nYFP, or CO1–105-cYFP and bHLH17-nYFP. Nuclei are indicated by DAPI staining. D, Co-IP assays. Total proteins were extracted from 35S:3Myc-CO 35S:2Flag-bHLH17 transgenic seedlings. Flag-tagged bHLH17 was immunoprecipitated using an anti-Flag antibody (1:250) and the co-immunoprecipitated protein was detected with an anti-Myc antibody (1:10,000). Similarly, Myc-tagged CO was immunoprecipitated using an anti-Myc antibody (1:250) and the co-immunoprecipitated protein was detected with an anti-Flag antibody (1:10,000). Protein input for 2Flag-bHLH17 or 3Myc-CO in immunoprecipitated complexes was also detected. Experiments were performed three times with similar results.
We confirmed the interaction of CO with bHLH3 or bHLH17 by BiFC and Co-IP assays in planta. When CO-cYFP was co-expressed with bHLH3-nYFP or bHLH17-nYFP in N. benthamiana leaves, we observed YFP fluorescence in the nucleus (Figure 7C; Supplemental Figure S5). We also detected YFP signal detected in N. benthamiana leaves when CO-cYFP was co-expressed with bHLH17350–566-nYFP (bHLH17 C-terminal amino acids 350–566 fused to nYFP; Figure 7C; Supplemental Figure S5). We detected no YFP signal in the negative control in which CO-cYFP was co-expressed with bHLH171–360-nYFP (bHLH17 N-terminal amino acids 1–360 fused to cYFP) or when CO1–105-cYFP was co-expressed with bHLH17-nYFP (Figure 7C; Supplemental Figure S5). Moreover, the results of the Co-IP assays provided further in vivo evidence for the direct interaction between CO and bHLH17 (Figure 7D). These findings indicate that CO physically interacts with the transcription factors bHLH3 and bHLH17 in the nucleus of plant cells.
CO works together with bHLH17 in suppressing jasmonate signaling, whereas JAZ1 affects their transcriptional functions and physical interaction
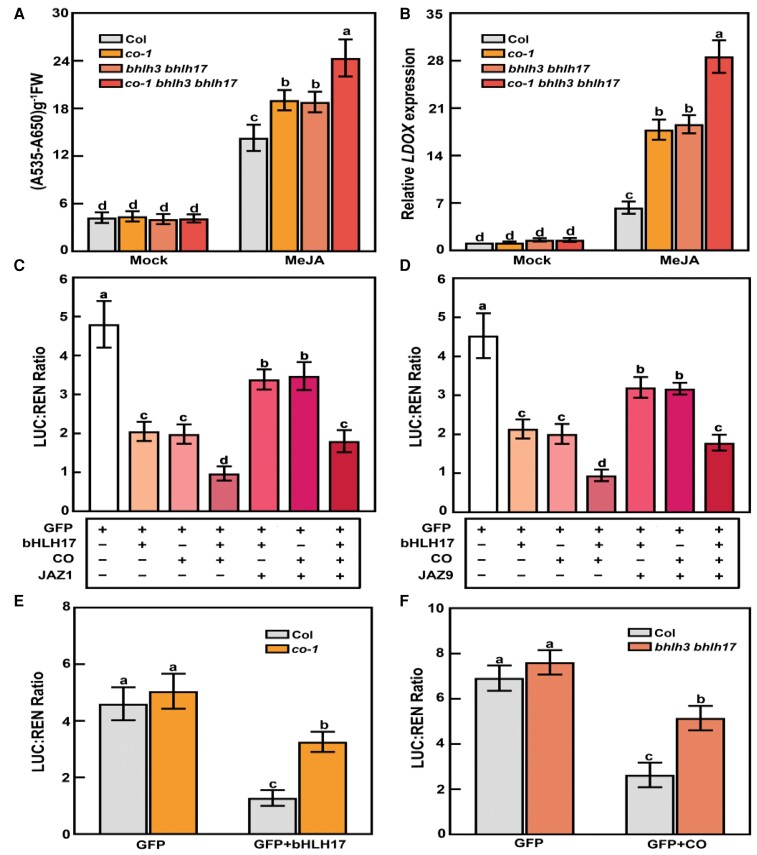
Having established that CO physically interacts with bHLH3 and bHLH17, we wondered whether they act cooperatively to mediate jasmonate signaling. Accordingly, we crossed co-1 to the bhlh3 bhlh17 double mutant to generate the co-1 bhlh3 bhlh17 triple mutant, which we then treated with MeJA for a phenotypic examination. Consistent with the results of previous studies (Sasaki-Sekimoto et al., 2013; Song et al., 2013a; Fonseca et al., 2014), bhlh3 bhlh17 seedlings accumulated more anthocyanin in the presence of MeJA than the wild type (Figure 8A). Notably, jasmonate signaling was more enhanced in the co-1 bhlh3 bhlh17 triple mutant than in co-1 and bhlh3 bhlh17. Anthocyanin levels were much higher in co-1 bhlh3 bhlh17 seedlings than in co-1 and bhlh3 bhlh17 seedlings (Figure 8A). To verify this observation, we quantitatively analyzed the expression of the jasmonate-responsive gene LDOX in MeJA-treated co-1 bhlh3 bhlh17 seedlings. Relative LDOX transcript levels were significantly higher in co-1 bhlh3 bhlh17 than in co-1 or bhlh3 bhlh17 seedlings (Figure 8B). These findings suggest that CO functions together with bHLH3 and bHLH17 to negatively regulate jasmonate signaling.
Figure 8.
CO works cooperatively with bHLH17 in suppressing jasmonate signaling. A, Anthocyanin contents in 10-day-old long-day-grown seedlings of wild type (Col), co-1, bhlh3 bhlh17, and co-1 bhlh3 bhlh17 on MS medium with or without (mock) 30-μM MeJA. In the mock treatment, an equal volume of 10% (v/v) ethanol was added. The experiments were performed 8 times with similar results with more than 200 seedlings for each repeat. Data are means ± Sd from eight independent experiments (n = 8). FW, fresh weight. Different lowercase letters indicate significant differences (P < 0.05) as determined by a two-way ANOVA (genotype × treatment interaction). B, RT-qPCR analysis of LDOX expression levels in Col, co-1, bhlh3 bhlh17, and co-1 bhlh3 bhlh17. Total RNA was extracted from 10-day-old seedlings grown in long days on MS medium with or without (mock) 30-μM MeJA. The expression level of LDOX in mock-treated Col-0 seedlings was set to 1. Data are means ± Sd from eight independent biological replicates using different batches of seedlings (n = 8). Different lowercase letters indicate significant differences (P < 0.05) as determined by a two-way ANOVA (genotype × treatment interaction). C and D, Transient dual-luciferase reporter assays showing that CO and bHLH17 function cooperatively to repress JAZ2 transcription, whereas JAZ1 and JAZ9 proteins affect their transcriptional functions. Data are means ± Sd from eight biological replicates using different batches of Col-0 protoplasts (n = 8 times). Different lowercase letters indicate significant differences (P < 0.05) as determined by a two-way ANOVA. E, Transient transcriptional activity assays showing that repression of the JAZ2 promoter by bHLH17 decreases in the co-1 mutant. Data are means ± Sd from eight biological replicates using different batches of Col-0 and co-1 protoplasts (n = 8). B Different lowercase letters indicate significant differences (P < 0.05) as determined by a two-way ANOVA (for an interaction of genotype × effector/protein expressed in the protoplasts). F, Transient transcriptional activity assays showing that repression of the JAZ2 promoter by CO decreases in the bhlh3 bhlh17 double mutant. Data are means ± Sd from eight biological replicates using different batches of Col-0 and bhlh3 bhlh17 protoplasts (n = 8). Different lowercase letters indicate significant differences (P < 0.05) as determined by a two-way ANOVA (for an interaction of genotype × effector/protein expressed in the protoplasts).
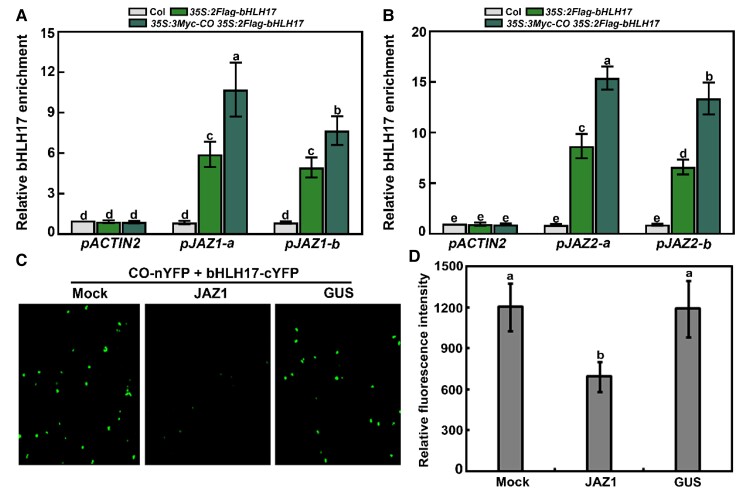
We also investigated the mechanisms underlying the additive effects of CO and bHLH17 on jasmonate signaling. More specifically, we conducted dual-luciferase (LUC) reporter assays in Arabidopsis mesophyll protoplasts (Yoo et al., 2007) to assess whether CO enhances the effect of bHLH17 on transcription. The effectors consisted of CO, bHLH17, or GFP (green fluorescent protein) under the control of the 35S promoter (Supplemental Figure S6). We also constructed a reporter construct comprising the LUC gene driven by the JAZ2 promoter (Fonseca et al., 2014). Consistent with the findings of a previous study (Fonseca et al., 2014), expressing bHLH17 decreased relative LUC activity from the ProJAZ2:LUC reporter in wild-type protoplasts (relative to the effects of GFP alone) (Figure 8, C and D; Supplemental Figure S7). We obtained similar results when CO was expressed in this assay with the same reporter (Figure 8, C and D; Supplemental Figure S7). Moreover, relative LUC activity decreased more in the presence of CO and bHLH17 than in the presence of GFP and bHLH17 (Figure 8, C and D; Supplemental Figure S7), suggesting that CO and bHLH17 function cooperatively to repress JAZ2 transcription. To assess whether the loss of CO function would affect the transcriptional function of bHLH17, we analyzed the ability of bHLH17 to suppress JAZ2 transcription in mesophyll protoplasts prepared from the wild-type control and the co-1 mutant. We measured lower relative LUC activity from the ProJAZ2:LUC reporter in bHLH17-expressing wild-type protoplasts than in co-1 protoplasts (Figure 8E; Supplemental Figure S7). Similarly, relative LUC activity from the ProJAZ2:LUC reporter was lower in CO-expressing wild-type protoplasts than in bhlh3 bhlh17 protoplasts (Figure 8F; Supplemental Figure S7). These findings support the notion that CO and bHLH17 reciprocally enhance their repressive effects on transcription. To further elucidate the regulatory effect of CO on bHLH17, we explored whether CO stimulates the enrichment of bHLH17 to the promoter regions of its downstream genes in vivo by chromatin immunoprecipitation (ChIP) assay. We observed that the enrichment of bHLH17 to the JAZ1 and JAZ2 promoter regions (pJAZ1-a, pJAZ1-b, pJAZ2-a, and pJAZ2-b; Supplemental Table S1) is greater in 35S:3Myc-CO 35S:2Flag-bHLH17 seedlings than in 35S:2Flag-bHLH17 seedlings (Figure 9, A and B). Thus, CO appears to enhance the enrichment of bHLH17 to the promoter regions of downstream jasmonate-responsive genes in plants.
Figure 9.
CO enhances the enrichment of bHLH17 at the promoter of target genes, and JAZ1 affects the CO-bHLH17 interaction. A and B, CO enhances the enrichment of bHLH17 at the promoter regions of JAZ1 (pJAZ1-a and pJAZ1-b) and JAZ2 (pJAZ2-a and pJAZ2-b). Eight-d-old wild-type (Col-0), 35S:2Flag-bHLH17, and 35S:3Myc-CO 35S:2Flag-bHLH17 seedlings grown in long days and treated with 100-μM MeJA for 1 h and harvested at ZT16 were used in ChIP assays with an anti-Flag antibody (or IgG antibody as a negative control). The enrichment (nonspecific binding) level of bHLH17 at the untranslated region of ACTIN2 (pACTIN2) in Col-0 seedlings was set to 1. Data are means ± Sd from eight independent biological replicates using different batches of seedlings (n = 8). Different lowercase letters indicate significant differences (P < 0.05) as determined by a two-way ANOVA (for an interaction of genotype × DNA/promoter region). C, BiFC analyses showing that JAZ1 attenuates the interaction between CO and bHLH17. Fluorescence was detected 48 h after co-expression of CO-cYFP + bHLH17-nYFP (mock), JAZ1 CO-cYFP + bHLH17-nYFP (+JAZ1), or GUS + CO-cYFP or bHLH17-nYFP (+GUS). D, Quantitative analysis of YFP fluorescence intensity in (C). Fifty independent fluorescent spots were assessed for fluorescence intensity. Data are means ± Sd (n = 50). One representative fluorescent spot resulting from the co-expression of CO-cYFP + bHLH17-nYFP (mock) was used as the control and its relative intensity value was set to 1,200. Experiments were performed more than three times with similar results. Different lowercase letters indicate significant differences (P < 0.05) as determined by a two-way ANOVA.
Because JAZ repressors interact with CO and bHLH subgroup IIId transcription factors, we investigated the modulatory effects of JAZ proteins on these transcription factors in dual-luciferase reporter assays conducted using Arabidopsis mesophyll protoplasts (Yoo et al., 2007). We expressed JAZ1 (or JAZ9) with CO and bHLH17 in protoplasts derived from the wild-type control. Relative LUC activity from the ProJAZ2:LUC reporter was higher in protoplasts expressing JAZ1 (or JAZ9) with CO and bHLH17 than in protoplasts expressing only CO and bHLH17 (Figure 8, C and D). These results suggest that JAZ1 and JAZ9 proteins antagonize the regulatory effects of CO and bHLH17 to modulate the transcription of downstream target genes. To further clarify the regulatory effects of JAZ proteins on CO and bHLH17, we performed BiFC assays to analyze the interaction between CO and bHLH17 in the presence of JAZ1. When JAZ1 was co-expressed with CO-cYFP and bHLH17-nYFP in N. benthamiana leaves, the YFP signal decreased (Figure 9, C and D). In the negative control, the expression of GUS with CO-cYFP and bHLH17-nYFP did not affect fluorescence intensity (Figure 9, C and D). These results suggest that JAZ1 interferes with the physical association between CO and bHLH17.
Discussion
CO is a critical negative regulator of jasmonate-imposed root-growth inhibition and anthocyanin accumulation
The BBX member transcription factor CO is a major regulator of flowering in the photoperiodic pathway. Previous studies have highlighted its regulatory effects on FT expression and flowering induction under long-day conditions (Putterill et al., 1995; Simon et al., 1996; Samach et al., 2000; Suárez-López et al., 2001; Takada and Goto, 2003; An et al., 2004; Corbesier et al., 2007; Tiwari et al., 2010; Shim et al., 2017). CO is widely expressed in multiple tissues, and its expression in the vascular tissue (phloem) is sufficient to stimulate FT transcription and trigger photoperiodic flowering (Takada and Goto, 2003; An et al., 2004; Shim et al., 2017). Although there has been considerable progress in recent years, other potential functions of CO and their underlying molecular mechanisms remain unknown. Investigating the direct involvement of CO in other biological processes and thoroughly elucidating the associated regulatory mechanisms will advance our understanding of CO-mediated signaling networks. In this study, we confirmed that CO was expressed in the shoot apex, young leaves, and roots of seedlings (Figure 1A), which is in accordance with previous studies (Takada and Goto, 2003; An et al., 2004). Intriguingly, we also revealed that CO transcript and CO protein levels were responsive to the phytohormone jasmonate (e.g. MeJA; Figure 1, A–E). Phenotypic analyses showed that loss-of-function co mutants were more sensitive to jasmonate than wild-type seedlings (Figure 2, A–G). Additionally, compared to control seedlings, co mutant seedlings had shorter roots and accumulated more anthocyanin following jasmonate treatment (Figure 2, A–G). By contrast, CO overexpression rendered the transgenic seedlings less sensitive to jasmonate than the wild type (Figure 3, A–D). These findings suggest that CO negatively regulates jasmonate-induced root-growth inhibition and anthocyanin accumulation in Arabidopsis seedlings.
Jasmonate is an essential signaling compound that controls multiple developmental processes and stress responses in plants, such as root elongation, trichome formation, stamen development, and anthocyanin accumulation (Chini et al., 2016; Hu et al., 2017; Huang et al., 2017; Zhang et al., 2017; Howe et al., 2018; Guo et al., 2018a). Jasmonate signal transduction involves profound changes in the cellular gene expression profile associated with the complex interplay between negative and positive components (e.g. JAZ proteins and downstream transcriptional regulators). Additionally, JAZ proteins inhibit jasmonate signaling by physically associating with and attenuating multiple transcription factors. For example, recent studies have revealed that JAZ proteins interact with the transcription factors ABI5 and ABI3 to modulate abscisic acid signaling and seed germination (Ju et al., 2019; Pan et al., 2020). Other targets of JAZ proteins include MYC transcription factors, essential components of the WD-repeat/bHLH/MYB transcriptional complexes, EIN3, FIL, TOE1, MYB21, RHD6, WRKY57, and INDUCER OF CBF EXPRESSION1 (ICE1), all of which regulate diverse aspects of jasmonate responses (Chini et al., 2007; Fernández-Calvo et al., 2011; Qi et al., 2011; Song et al., 2011; Zhu et al., 2011; Hu et al., 2013; Jiang et al., 2014; Boter et al., 2015; Zhai et al., 2015; Han et al., 2020). Two repressors of jasmonate signaling in apple (Malus domestica), MdJAZ1 and MdJAZ2, were recently reported to interact with MdBBX37 to repress the transcriptional activation of C-repeat binding factor genes by MdBBX37 (An et al., 2021). The results of the present study suggested that CO also physically associated with several JAZ repressors and that the C-terminal region with the CCT domain of CO was sufficient for these interactions (Figure 5, A–D). As the C-terminal fragment of MdBBX37 lacks a CCT domain and shares low similarity with that of CO, it is possible that the MdJAZ–MdBBX37 and JAZ–CO interactions do not occur through a conserved domain. Interestingly, previous studies have revealed that JAZ proteins exhibit physical associations with bHLH transcription factors (e.g. MYC2 and ICE1) also through different domains (Fernández-Calvo et al., 2011; Hu et al., 2013). These findings underscore the involvement of different domains from transcription factors in their interactions with JAZ proteins, and future elucidations may shed light on the distinct and complex transcriptional mechanisms of jasmonate signaling. Our further investigation demonstrated that JAZ proteins interfered with the transcriptional function of CO and the interaction of CO with bHLH17 in mediating jasmonate signaling (Figures 8 and 9). Based on our genetic analyses, we propose that CO functions in a COI1-dependent manner and CO and MYC2 have opposite effects on jasmonate responses (Figure 6, A–G). These findings indicate that CO is a JAZ-binding factor that participates in jasmonate signaling via direct protein–protein interactions.
Previous investigations have shown that the bHLH family subgroup IIId transcription factors bHLH3, bHLH13, bHLH14, and bHLH17 interact with JAZ repressors and negatively modulate jasmonate responses (Nakata et al., 2013; Sasaki-Sekimoto et al., 2013; Song et al., 2013a; Fonseca et al., 2014). The quadruple mutant bhlh3 bhlh13 bhlh14 bhlh17 exhibits severe jasmonate-repressed root growth and jasmonate-induced anthocyanin accumulation (Song et al., 2013a). Interestingly, we observed that CO also interacted with bHLH3 and bHLH17 and acted cooperatively with these two transcription factors to suppress jasmonate signaling (Figures 7 and 8). Through their physical interaction, CO enhanced the transcriptional function of bHLH17 and its enrichment at the promoter regions of downstream jasmonate-responsive genes (Figures 8 and 9). Moreover, our results suggested that JAZ proteins affected the transcriptional activity of CO, while also inhibiting the CO–bHLH17 interaction associated with jasmonate signaling (Figures 8 and 9). Consistent with these findings, we detected an interaction between the C-terminal region of CO with the CCT domain crucial for its transcriptional function (Gangappa and Botto, 2014) and bHLH3/bHLH17 and JAZ proteins (Figures 5 and 7). Thus, we uncovered a critical signaling module in which, CO acts together with bHLH subgroup IIId transcription factors (bHLH3 and bHLH17) to negatively regulate jasmonate signaling, but their transcriptional functions and physical interactions are repressed by JAZ proteins. Unlike CO and bHLH subgroup IIId transcription factors, the transcription factors MYC2, MYC3, and MYC4 stimulate jasmonate-mediated root-growth inhibition and anthocyanin accumulation (Boter et al., 2004; Lorenzo et al., 2004; Dombrecht et al., 2007; Fernández-Calvo et al., 2011). Phenotypic and biochemical analyses showed that CO/bHLH subgroup IIId transcription factors and MYC2 have opposite effects on jasmonate signaling (Figure 6, D–G; Nakata et al., 2013; Sasaki-Sekimoto et al., 2013; Song et al., 2013a; Fonseca et al., 2014), which may be explained by the competitive inhibition of MYC by CO/bHLH subgroup IIId transcription factors. Because a direct interaction between MYC and CO/bHLH subgroup IIId transcription factors has not been detected (Figure 7A; Song et al., 2013a), further research is required to explore the molecular mechanisms underlying the opposite effects of MYC and CO/bHLH subgroup IIId transcription factors.
CO modulates the diurnal gating of jasmonate signaling
The circadian clock is an endogenous biological oscillator that provides an adaptive advantage to land plants (Dunlap, 1999; Pruneda-Paz and Kay, 2010; Hsu and Harmer, 2014; Webb et al., 2019; Yang et al., 2021). Several recent studies have revealed that jasmonate accumulation and signaling are controlled by the circadian clock (Goodspeed et al., 2012; Shin et al., 2012; Nitschke et al., 2016; Zhang et al., 2018; Thines et al., 2019). More specifically, Goodspeed et al. (2012) detected a circadian pattern in the accumulation of jasmonate, with peak levels in the middle of the day. Moreover, Shin et al. (2012) analyzed MeJA-induced expression of several jasmonate-responsive genes at different times over diurnal cycles and found that these genes were rhythmically expressed, with peak expression levels at dawn. Moreover, the circadian clock component TIME FOR COFFEE (TIC) physically interacts with MYC2 and prevents MYC2 from accumulating to negatively modulate jasmonate signaling. Similarly, we detected a rhythmic diurnal pattern in MeJA-induced expression of JAZ1, JAZ2, and LOX2, with the highest expression levels at dawn (Figure 4, A–C). Furthermore, relative JAZ1, JAZ2, and LOX2 transcript levels were significantly higher in co-1 seedlings than in wild-type seedlings at ZT12, ZT16, and ZT20 (Figure 4, A–C). These results indicate that CO is involved in the gating of circadian clock-mediated jasmonate signaling when CO contents increase (Suárez-López et al., 2001; Valverde et al., 2004; Liu et al., 2008). Because bHLH3 and bHLH17 interact with CO (Figure 7), we also analyzed their diurnal expression patterns in MeJA-treated wild-type seedlings. The bHLH3 and bHLH17 transcript levels did not show a diurnal pattern in response to MeJA (Supplemental Figure S8). Nevertheless, the precise biochemical mechanisms underlying the circadian clock-mediated gating of jasmonate signaling remain to be characterized. Elucidating the potential relationships between circadian clock components and key regulators of jasmonate signaling will enhance our understanding of circadian clock-mediated gating of jasmonate signaling networks.
The circadian clock and jasmonate signaling pathways are both involved in the regulation of plant growth and stress responses by synchronizing internal physiological processes with surrounding environmental changes. Considering that jasmonate accumulation and signaling are rhythmically regulated (Figure 4; Goodspeed et al., 2012; Shin et al., 2012; Nitschke et al., 2016; Zhang et al., 2018; Thines et al., 2019), we speculate that the circadian gating of jasmonate responses may contribute to maintaining an appropriate temporal balance so that plant growth and stress responsiveness are optimized for the prevailing conditions. Because jasmonate activates stress responses at the cost of energy for plant growth and development, such a gating mechanism underlying jasmonate signaling may allow plants to promote growth in the evening and restrict growth in the morning. Consistently, when jasmonate signaling is relatively less active at night, daily plant growth and development are promoted, which are also modulated by rhythmic gibberellin (GA) and auxin signaling (Figure 4; Covington and Harmer, 2007; Nozue et al., 2007; Rawat et al., 2009; Arana et al., 2011; Goodspeed et al., 2012; Shin et al., 2012; Thines et al., 2019). In terms of defense responses, jasmonate is vital for the establishment of resistance against necrotrophic pathogen infection and wounding by feeding insects. Previous studies have highlighted that jasmonate-mediated plant defense against necrotrophic fungal pathogens is modulated by the circadian clock and differs with a time of day (Hevia et al., 2015; Ingle et al., 2015; Zhang et al., 2019a, 2019b). Moreover, the circadian clock also gates jasmonate-regulated resistance to insect herbivory (Goodspeed et al., 2012; Sharma and Bhatt, 2015). As jasmonate signaling is stimulated in the morning, it is possible that this regulation of jasmonate helps plants establish the appropriate defense to anticipate particular pathogen infections and insect attacks during the morning phase. Despite recent advances, in-depth analyses of the direct implication of crucial regulators that integrate the circadian clock and jasmonate signaling pathways may shed light on the molecular basis of the balance between plant growth and stress tolerance.
A network of transcriptional regulators mediates jasmonate signaling
The COI1/JAZ-mediated jasmonate signaling pathway delays flowering in Arabidopsis (Robson et al., 2010; Yang et al., 2012; Zhai et al., 2015). Under long-day conditions, coi1-2 and JAZ1-overexpressing plants flower earlier than wild-type plants (Zhai et al., 2015). Detailed analyses of the responsible mechanism revealed that a subset of JAZ proteins interacts with and inhibits the transcription factors TOE1 and TOE2, which directly repress FT transcription and flowering (Zhai et al., 2015). Similarly, the bHLH subgroup IIIe transcription factors (MYC2, MYC3, and MYC4) are required for jasmonate-mediated inhibition of flowering (Wang et al., 2017). The myc2 myc3 myc4 triple mutant accumulates more FT transcripts and flowers earlier than the wild type (Wang et al., 2017). The results of these studies provided insights into how COI1/JAZ-mediated jasmonate signaling negatively modulates flowering time. In this study, we identified CO as a JAZ-binding protein that helps mediate jasmonate responses (Figure 5, A–D). Zhang et al. (2015) showed that TOE proteins interact with CO and transmit a photoperiodic signal with inhibitory effects on CO. The bhlh3 bhlh13 bhlh14 bhlh17 quadruple mutant exhibits a late-flowering phenotype, indicating that these bHLH subgroup IIId transcription factors also stimulate flowering (Song et al., 2013a). The effects of bHLH subgroup IIId transcription factors on flowering time may involve antagonism of the repressive effects of bHLH subgroup IIIe transcription factors. These findings collectively suggest that jasmonate-regulated flowering may require a network of transcriptional regulators that combine to delay flowering.
Considering that CO and bHLH subgroup IIId transcription factors function oppositely (or antagonistically) to MYC and TOE proteins in mediating jasmonate-regulated processes, we speculate that the effects of different JAZ-binding factors on jasmonate signaling are balanced to maintain appropriate jasmonate signaling levels. This fine-tuning of jasmonate responses may optimize the balance between establishing stress tolerance and plant growth and development. Similar dual modulations have been described in other situations. For example, jasmonate-activated EIN3 inhibits MYC2 function to regulate jasmonate-induced apical hook curvature and defense responses to the necrotrophic fungal pathogen Botrytis cinerea (Berrocal-Lobo et al., 2002; Lorenzo et al., 2004; Zhu et al., 2011; Zhang et al., 2014; Song et al., 2014b). In this study, we found that exogenous application of jasmonate (i.e. MeJA) down-regulated CO transcript levels in wild-type seedlings (Figure 1, A–C). Additionally, CO transcript levels increased in the coi1 mutant and JAZ1-ΔJas seedlings (Figure 1D), indicative of the suppressive effects of jasmonate on CO transcription. By contrast, CO increased in abundance in the presence of jasmonate (Figure 1E). Considering these findings along with the fact that JAZ repressors interact with and inhibit the transcriptional function of CO (Figures 5 and 8), it is possible that jasmonate has dual (negative and positive) regulatory effects on CO that strictly control jasmonate signaling. Further analyses of the regulatory mechanisms underlying the dual effects of jasmonate on CO transcription and the accumulation of the encoded protein will help understand how CO precisely controls jasmonate signaling.
The transcription of CO and the stability of the encoded protein are strictly controlled by multiple regulators and signaling pathways. More specifically, GIGANTEA (GI) and FLAVIN-BINDING, KELCH REPEAT, F-BOX1 (FKF1) play a major role in maintaining appropriate CO expression levels (Imaizumi et al., 2005; Sawa et al., 2007; Song et al., 2012, 2014a). Previous studies have demonstrated that GI physically associates with FKF1 to form a complex that recruits CYCLING DOF FACTOR (CDF) proteins, which are repressors of CO transcription, for degradation (Imaizumi et al., 2005; Sawa et al., 2007; Fornara et al., 2009). In the cdf1 cdf2 cdf3 cdf5 quadruple mutant, CO transcript levels are high in the morning, regardless of photoperiod or daylength changes (Fornara et al., 2009). Moreover, several critical light-signaling components modulate CO abundance. For example, the RING finger E3 ubiquitin ligase CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) and its interacting partner SUPPRESSOR OF PHYA-105 (SPA) facilitate CO protein degradation at night (Laubinger et al., 2006; Jang et al., 2008). Another RING finger E3 ubiquitin ligase, HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 (HOS1), works synergistically with COP1 to precisely regulate the timing of CO accumulation (Lazaro et al., 2012). Additionally, photoreceptors (e.g. phytochromes and cryptochromes), ZEITLUPE (ZTL), PSEUDO-RESPONSE REGULATORs (PRRs), GI, and TOE modulate CO stability (Valverde et al., 2004; Zuo et al., 2011; Song et al., 2012, 2014a; Hayama et al., 2017). Because jasmonate also affects CO transcription (Figure 1, A–D), further research is needed to determine whether jasmonate operates through the known upstream CO regulators listed above. Furthermore, whether the crucial regulators of CO also participate in jasmonate signaling should be investigated. Interestingly, the results presented in Figure 1E showed that CO accumulation increased in response to MeJA, and its accumulation further rose in seedlings treated with both MeJA and MG132 compared to seedlings treated with only MeJA or MG132. These observations suggest that the production of CO is activated by MeJA. It is possible that jasmonate affects CO mRNA stability and influences the subsequent translation and/or it affects how efficiently CO is translated. The underlying mechanisms should be clarified in future investigations.
Based on our results and those of other studies, we propose the following simplified model to explain the molecular mechanism underlying CO-mediated jasmonate signaling in Arabidopsis. When the jasmonate concentration is low, JAZ proteins interact with CO and bHLH3/bHLH17, interfere with CO–bHLH3/bHLH17 interactions, and repress transcription factor functions (Figures 5, 8, and 9). When jasmonate concentrations increase, COI1 perceives jasmonate and targets JAZ proteins for degradation via the SCFCOI1–26S proteasome pathway (Chini et al., 2007; Thines et al., 2007; Yan et al., 2009; Sheard et al., 2010). The degradation of JAZ repressors subsequently releases CO and bHLH3/bHLH17 to form a transcriptional complex that negatively regulates jasmonate-induced root-growth inhibition and anthocyanin accumulation (Figures 2–7; Nakata et al., 2013; Sasaki-Sekimoto et al., 2013; Song et al., 2013a; Fonseca et al., 2014). Additionally, CO accumulates in response to jasmonate (Figure 1E), which also contributes to the repressive effects of CO on jasmonate signaling.
Materials and methods
Materials and plant growth conditions
Taq DNA polymerases were obtained from Takara Biotechnology (Dalian, China); MeJA was purchased from Sigma-Aldrich. Other common chemicals were obtained from Sangon Biotech (Shanghai, China). The wild-type and mutant Arabidopsis plants used in this study were in the Col-0 or Landsberg erecta (Ler) genetic background. The co-1 (CS3325), co-4 (CS177), co-6 (CS179), co-9 (CS870084), bhlh3 (CS871836), and bhlh17 (CS874647) mutants were obtained from the Arabidopsis Biological Resource Center at Ohio State University (http://abrc.osu.edu). The coi1-2, coi1-16, jazQ (jaz1 jaz3 jaz4 jaz9 jaz10), and myc2-1 mutants have been described (Ellis and Turner, 2002; Xu et al., 2002; Campos et al., 2016; Wang et al., 2017). To generate transgenic plants 35S:3Myc-CO, 35S:2Flag-CO, or 35S:2Flag-bHLH17, the full-length coding sequence of CO or bHLH17 was cloned into the binary vector pOCA30 in the sense orientation under the control of the CaMV 35S promoter (Han et al., 2020). The transgenic plants overexpressing JAZ1 with a deleted Jas domain (JAZ1-ΔJas) were described previously (Han et al., 2018). The co-1 coi1-2, co-1 myc2-1, bhlh3 bhlh17, and co-1 bhlh3 bhlh17 mutant lines were generated via standard crossing. Plants were grown in an artificial growth chamber at 22°C under a 16-h light (100 mE m−2 s−1, white fluorescent bulbs, full light wavelength range)/8-h dark long-day photoperiod or an 8-h light/16-h dark short-day photoperiod. MeJA was dissolved in 10% (v/v) ethanol as a 10-mM stock solution. In the mock treatment, an equal volume of 10% (v/v) ethanol was added to the medium.
GUS staining
The putative CO promoter sequence (proCO; 3,576 bp) was amplified from Col-0 genomic DNA using gene-specific primers (Takada and Goto, 2003; Supplemental Data Set S1). The proCO:GUS construct was cloned into the pOCA28 binary vector and introduced into wild-type (Col-0) plants using the floral dip method (Clough and Bent, 1998). T5 seedlings from proCO:GUS transgenic lines were treated with 50- or 100-μM MeJA for 4 h and then samples were harvested at ZT 16 for staining. The histochemical detection of GUS activity was performed as previously described (Chen et al., 2010). The primers used for cloning are listed in Supplemental Data Set S1.
RNA extraction and RT-qPCR
Total RNA was extracted from seedlings of the indicated age that had been treated with or without MeJA using Trizol reagent (Invitrogen) for RT-qPCR analysis, which was performed as previously described (Han et al., 2020). Briefly, 1.0-μg DNase-treated total RNA was reverse transcribed in a 20-μL reaction volume using the oligo-(dT)18 primer and Moloney murine leukemia virus reverse transcriptase (Fermentas, Hanover, MD, USA). The cDNA was diluted 1:1 prior to use. Each qPCR analysis was completed using 1.0-μL cDNA, a SYBR Premix Ex Taq kit (Takara Biotechnology), and a LightCycler 480 real-time PCR instrument (Roche) according to the manufacturer's instructions. ACTIN2 (At3g18780) was used as the internal control. The gene-specific qPCR primers are listed in Supplemental Table S1.
Anthocyanin content measurement
Arabidopsis seeds were surface-sterilized with 5% [v/v] NaClO solution for 15 min and washed with sterile water three times, then incubated in the dark at 4°C for 2 days for stratification. Arabidopsis seedlings were grown on MS medium with or without 30-µM MeJA for 10 days before measuring their anthocyanin content as previously described (Qi et al., 2011). The anthocyanin content is expressed as (A535−A650) per gram fresh weight. All experiments were performed at least eight times with similar results by analyzing seedlings of different batches.
Root measurements
For jasmonate-mediated root-growth inhibition assays, the root length of 8-day-old seedlings grown on MS medium with or without 30- μM MeJA (Sigma-Aldrich) was measured. All experiments were performed at least five times with similar results by analyzing seedlings of different batches. For each sample, the root length of 20 representative seedlings was measured (n = 20). Values represent means ± Sd. Comparisons between different lines and the wild type were done by a two-way analysis of variance (ANOVA).
Yeast two-hybrid (Y2H) assays
The coding sequence encoding the C-terminal half of CO (amino acids 176–373) was cloned into pGBKT7 (Clontech) to generate the bait vector (BD-CO176–373) containing the GAL4 DNA-BD sequence. The full-length coding sequence of each target (JAZ, MYC2, MYC3, MYC4, bHLH3, bHLH13, bHLH14, or bHLH17) was individually cloned into pGADT7 vector (Clontech) to produce the prey vectors (AD-JAZ, AD-MYC, or AD-bHLH) containing the sequence encoding the GAL4 activation domain (AD). To identify specific regions critical for protein interactions, multiple truncated JAZ1 and bHLH17 sequences were cloned into pGADT7. The Y2H assays were performed as previously described (Hu et al., 2013). Yeast strain AH109 was co-transformed with the bait and prey vectors and then protein interactions were evaluated based on the ability of the cells to grow on synthetic defined (SD) medium lacking Leu, Trp, His, and Ade after 4 days of growth at 28°C. The primers used for cloning are listed in Supplemental Data Set S1.
BiFC assays
The coding sequences encoding the YFP C-terminal (64 amino acids) region (cYFP) and the YFP N-terminal (173 amino acids) region (nYFP) were inserted into separate pFGC5941 plasmids behind the FLAG or MYC tag sequences under the control of the CaMV 35S promoter to generate pFGC-cYFP and pFGC-nYFP, respectively (Kim et al., 2008). The full-length or truncated CO coding sequence was cloned into pFGC-cYFP to generate CO-cYFP or CO1–105-cYFP. Similarly, the full-length coding sequences or truncated forms of JAZ1, JAZ5, JAZ9, or bHLH17 were cloned upstream of the nYFP sequence to generate JAZ1-nYFP, JAZ5-nYFP, JAZ9-nYFP, JAZ11–200-nYFP, JAZ1155–201-nYFP, bHLH171–360-nYFP, and bHLH17350–566-nYFP. The resulting plasmids were introduced into Agrobacterium (Agrobacterium tumefaciens) strain EHA105 for transient infiltration of N. benthamiana leaves as previously described (Han et al., 2020). The experiments were conducted at least three times using different batches of N. benthamiana plants. For each replicate, more than 12 N. benthamiana plants were infiltrated and more than 600 cells were analyzed. The fluorescence of YFP in DAPI-stained leaves was examined 48 h after infiltration using a confocal laser-scanning microscope (Olympus, Tokyo, Japan). The primers used for cloning are listed in Supplemental Data Set S1.
Protein extraction, immunoblots, and Co-IP assays
Eight-day-old Arabidopsis seedlings were ground in liquid nitrogen and homogenized in extraction buffer containing 50-mM Tris–HCl (pH 7.4), 1-mM EDTA, 150-mM NaCl, 10% (v/v) glycerol, 0.1% (v/v) Triton X-100, 1-mM PMSF, and 1× Roche Protease Inhibitor Cocktail. The extracts were incubated for 3 h at 4°C and the supernatant was collected by centrifugation (12,000g for 10 min at 4°C). For immunoblotting analysis, seedlings were harvested, frozen in liquid nitrogen, and homogenized in 100 μL of 1× Laemmli SDS-PAGE protein loading buffer. The extracts were boiled at 95°C for 5 min. After centrifugation (12,000g for 10 min at room temperature), the supernatant was collected. For immunoblotting, a 20-μL volume of each sample was run on 10% SDS-PAGE gels, transferred to nitrocellulose membrane (Bio-Rad), blocked for 1 h in 5% (w/v) reconstituted nonfat skim milk powder in TBST at room temperature, and incubated with an anti-Myc antibody (1:10,000; catalog no. M4439, mouse, monoclonal antibody, Sigma-Aldrich) or an anti-Flag antibody (1:10,000; catalog no. F3165, mouse, monoclonal antibody, Sigma-Aldrich). HRP-conjugated secondary antibody (goat anti-mouse, 1:10,000; D110087) was from Sangon Biotech (Shanghai, China). The luminescence was detected with Luminata Forte Western HRP substrate (Millipore).
For Co-IP assays, total proteins were extracted from 35S:2Flag-CO 35S:3Myc-JAZ1 transgenic Arabidopsis seedlings or from 35S:3Myc-CO 35S:2Flag-bHLH17 seedlings. Immunoprecipitation was performed with protein A/G Plus-agarose beads following the manufacturer's protocol. In brief, cell lysates were precleared with protein A/G Plus-agarose beads and incubated with an anti-Myc antibody (1:250) or an anti-Flag antibody (1:250) and protein A/G Plus-agarose beads at 4°C overnight in extraction buffer. The beads were washed twice extensively with extraction buffer and the co-immunoprecipitated protein was then detected by immunoblotting using an anti-Flag antibody (1:10,000) or anti-Myc antibody (1:10,000).
ChIP analyses
The ChIP assay was performed essentially as previously described (Mukhopadhyay et al., 2008; Hu et al., 2019). Eight-day-old seedlings (with or without 100-μM MeJA treatment for 1 h; harvested at ZT0 or ZT16) of the wild type, 35S:2Flag-bHLH17, or 35S:Myc-CO 35S:2Flag-bHLH17 were crosslinked in 1% (w/v) formaldehyde and their chromatin was isolated. The anti-Flag antibody or IgG antibody (the negative control) was used to immunoprecipitate the protein–DNA (target promoter) complex, and the precipitated DNA was purified using a PCR purification kit (Qiagen) for qPCR analysis. To quantitatively assess bHLH17–DNA binding, qPCR analysis was performed as described previously (Mukhopadhyay et al., 2008) with the ACTIN2 untranslated region sequence (pACTIN2) as an endogenous control. The relative quantity value was calculated by the 2 (−DDCt) method (Mukhopadhyay et al., 2008) and presented as the DNA-binding ratio. The results presented were obtained from at least eight independent experiments. The primers used for ChIP assays are listed in Supplemental Document S1.
Transient transactivation assays
The putative promoter JAZ2 sequence (proJAZ2, 2,000 bp) was amplified from Col-0 genomic DNA and cloned into the pGreenII 0800-LUC vector to generate the ProJAZ2:LUC reporter construct (Hellens et al., 2005). The full-length coding sequences of CO, JAZ1, JAZ9, bHLH17, and GFP were individually amplified and cloned into separate pGreenII 62-SK vectors under the control of the CaMV 35S promoter as effectors (Hellens et al., 2005). Different combinations of plasmids were transfected into Arabidopsis leaf mesophyll protoplasts as previously described (Sheen, 2001). Transfected cells were cultured for 16 h in the light and then relative LUC activity was detected using a Dual-Luciferase Reporter Assay system (Promega), which measured the activities of firefly LUC and that of the internal control Renilla reniformis LUC (REN). The primers used for constructs are listed in Supplemental Data Set S1.
Statistical analysis
Statistical analyses were performed as ANOVA using Tukey's honest significant difference as a post-hoc test. Statistically significant differences were defined as those with P < 0.05. Different lowercase letters indicate significant differences (P < 0.05). Asterisks in Figures 1 and 4 also represent differences that are statistically significant (*P < 0.05) or highly significant (**P < 0.01) at the indicated times., All statistical analyses were performed using GraphPad Prism version 8.0. The results are shown in Supplemental Data Set S2.
Accession numbers
Arabidopsis Genome Initiative numbers for the genes discussed in this article are as follows: CO (At5g15840); COI1 (At2g39940); JAZ1 (At1g19180); JAZ2 (At1g74950); JAZ3 (At3g17860); JAZ4 (At1g48500); JAZ5 (At1g17380); JAZ6 (At1g72450); JAZ7 (At2g34600); JAZ8 (At1g30135); JAZ9 (At1g70700); JAZ10 (At5g13220); JAZ11 (At3g43440); JAZ12 (At5g20900); bHLH3 (At4g16430); bHLH13 (At1G01260); bHLH14 (At4G00870); bHLH17 (At2g46510); MYC2 (At1g32640); and ACTIN2 (At3g18780).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . RT-qPCR analyses of CO expression in 35S:3Myc-CO and 35S:2Flag-CO transgenic plants.
Supplemental Figure S2 . RT-qPCR analysis of relative JAZ2, LOX2, and LDOX expression levels in co-4 and co-6 mutants.
Supplemental Figure S3 . Jasmonate responses of co mutant plants under short-day conditions.
Supplemental Figure S4 . Negative controls for the interactions between CO and JAZ proteins in yeast and N. benthamiana.
Supplemental Figure S5 . Negative controls for the interactions between CO and bHLH transcription factors in yeast and N. benthamiana.
Supplemental Figure S6 . Schematic diagrams of the effectors and reporters used in the transient transactivation assays.
Supplemental Figure S7 . Accumulation of CO and bHLH17 in Arabidopsis leaf mesophyll protoplasts.
Supplemental Figure S8 . Induced expression of bHLH3 and bHLH17 in response to MeJA.
Supplemental Document S1. Information for bHLH17-binding sequences in the JAZ1 and JAZ2 promoters (for ChIP assays).
Supplemental Table S1 . Primers used for RT-qPCR analyses.
Supplemental Data Set S1. Primers used for cloning.
Supplemental Data Set S2. ANOVA tables.
Supplementary Material
Acknowledgments
We thank Drs. Daoxin Xie (Tsinghua University), Gregg A. Howe (Michigan State University), Xiaoya Chen (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences), and Zhixiang Chen (Purdue University) for sharing research materials. We thank the Arabidopsis Resource Center at Ohio State University for co, bhlh3, and bhlh17 seeds. We also thank the Central Laboratory of Xishuangbanna Tropical Botanical Garden and Kunming Institute of Botany, Chinese Academy of Sciences for technical support.
Contributor Information
Xiao Han, CAS Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; Center of Economic Botany, Core Botanical Gardens, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
Mengyi Kui, CAS Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; Center of Economic Botany, Core Botanical Gardens, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Tingting Xu, CAS Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; Center of Economic Botany, Core Botanical Gardens, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Jingwen Ye, CAS Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230027, China.
Jiancan Du, CAS Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; Center of Economic Botany, Core Botanical Gardens, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
Milian Yang, CAS Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; Center of Economic Botany, Core Botanical Gardens, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
Yanjuan Jiang, CAS Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; Center of Economic Botany, Core Botanical Gardens, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
Yanru Hu, CAS Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; Center of Economic Botany, Core Botanical Gardens, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
Funding
This work was supported by the National Natural Science Foundation of China (32100445, 31922009, 31870259, and 32270613), the Applied Basic Research Project of Yunnan Province (2019FI006, 202001AV070009, 202001AT070118, and 202101AW070005), the CAS “Light of West China” Program (to X.H.), the Youth Innovation Promotion Association of the of Chinese Academy of Sciences (Y201973 and 2022399), and “Yunnan Revitalization Talent Support Program” in Yunnan Province (YNWR-QNBJ-2018-075).
References
- An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, Piñeiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, et al. (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131(15): 3615–3626 [DOI] [PubMed] [Google Scholar]
- An J, Wang X, Zhang X, You C, Hao Y (2021) Apple B-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytol 229(5): 2707–2729 [DOI] [PubMed] [Google Scholar]
- Arana MV, Marín-de la Rosa N, Maloof JN, Blázquez MA, Alabadí D (2011) Circadian oscillation of gibberellin signaling in Arabidopsis. Proc Natl Acad Sci USA 108(22): 9292–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29(1): 23–32 [DOI] [PubMed] [Google Scholar]
- Boter M, Golz JF, Giménez-lbañez S, Fernandez-Barbero-Zorrilla JM, Solana R (2015) FILAMENTOUS FLOWER is a direct target of JAZ3 and modulates responses to jasmonate. Plant Cell 27(11): 3160–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M, Ruíz-Rivero O, Abdeen A, Prat S (2004) Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 18(13): 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J (2009) Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Cagnola JI, Cerdán PD, Pacín M, Andrade A, Rodriguez V, Zurbriggen MD, Legris M, Buchovsky S, Carrillo N, Chory J, et al. (2018) Long-day photoperiod enhances jasmonic acid-related plant defense. Plant Physiol 178(1): 163–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos ML, Yoshida Y, Major IT, de Oliveira Ferreira D, Weraduwage SM, Froehlich JE, Johnson BF, Kramer DM, Jander G, Sharkey TD, et al. (2016) Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat Commun 30(7): 12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Liu L, Ma K, Wang W, Lv H, Gao M, Wang X, Zhang X, Ren S, Zhang N, Guo Y-D (2022) The jasmonate-induced bHLH gene SlJIG functions in terpene biosynthesis and resistance to insects and fungus. J Integr Plant Biol 64(5): 1102–1115 [DOI] [PubMed] [Google Scholar]
- Chen L, Zhang L, Yu D (2010) Wounding-induced WRKY8 is involved in basal defense in Arabidopsis. Mol Plant Microbe Interact 23(5): 558–565 [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signaling. Nature 448(7154): 666–671 [DOI] [PubMed] [Google Scholar]
- Chini A, Gimenez-Ibanez S, Goossens A, Solano R (2016) Redundancy and specificity in jasmonate signalling. Curr Opin Plant Biol 33: 147–156 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6): 735–743 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316(5827): 1030–1033 [DOI] [PubMed] [Google Scholar]
- Covington MF, Harmer SL (2007) The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol 5(8): e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19(7): 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap J (1999) Molecular bases for circadian clocks. Cell 96(2): 271–290 [DOI] [PubMed] [Google Scholar]
- Ellis C, Turner JG (2002) A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215(4): 549–556 [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico J-M, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23(2): 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S, Chico JM, Solano R (2009) The jasmonate pathway: the ligand, the receptor and the core signaling module. Curr Opin Plant Biol 12(5): 539–547 [DOI] [PubMed] [Google Scholar]
- Fonseca S, Fernández-Calvo P, Fernández GM, Díez-Díaz M, Gimenez-Ibanez S, López-Vidriero I, Godoy M, Fernández-Barbero G, Van Leene J, De Jaeger Get al. (2014) bHLH003, bHLH013 and bHLH017 are new targets of JAZ repressors negatively regulating JA responses. PLoS One 9(1): e86182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupland G (2009) Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17(1): 75–86 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF (2014) The BBX family of plant transcription factors. Trends Plant Sci 19(7): 460–470 [DOI] [PubMed] [Google Scholar]
- Goodspeed D, Chehab EW, Min-Venditti A, Braam J, Covington MF (2012) Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc Natl Acad Sci USA 109(12): 4674–4677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Major IT, Howe GA (2018a) Resolution of growth-defense conflict: mechanistic insights from jasmonate signaling. Curr Opin Plant Biol 44: 72–81 [DOI] [PubMed] [Google Scholar]
- Guo Q, Yoshida Y, Major IT, Wang K, Sugimoto K, Kapali G, Havko NE, Benning C, Howe GA (2018b) JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc Natl Acad Sci USA 115(45): 10768–10777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Hu Y, Zhang G, Jiang Y, Chen X, Yu D (2018) Jasmonate negatively regulates stomatal development in Arabidopsis cotyledons. Plant Physiol 176(4): 2871–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Zhang M, Yang M, Hu Y (2020) Arabidopsis JAZ proteins interact with and suppress RHD6 transcription factor to regulate jasmonate-stimulated root hair development. Plant Cell 32(4): 1049–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Sarid-Krebs L, Richter R, Fernández V, Jang S, Coupland G (2017) PSEUDO RESPONSE REGULATORs stabilize CONSTANS protein to promote flowering in response to day length. EMBO J 36(7): 904–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 18(1): 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevia MA, Canessa P, Müller-Esparza H, Larrondo LF (2015) A circadian oscillator in the fungus Botrytis cinerea regulates virulence when infecting Arabidopsis thaliana. Proc Natl Acad Sci USA 112(28): 8744–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Major IT, Koo AJ (2018) Modularity in jasmonate signaling for multistress resilience. Annu Rev Plant Biol 26(69): 387–415 [DOI] [PubMed] [Google Scholar]
- Hsu P, Harmer S (2014) Wheels within wheels: the plant circadian system. Trends Plant Sci 19(4): 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Han X, Yang M, Zhang M, Pan J, Yu D (2019) The transcription factor INDUCER OF CBF EXPRESSION1 interacts with ABSCISIC ACID INSENSITIVE5 and DELLA proteins to fine-tune abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 31(7): 1520–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Jiang L, Wang F, Yu D (2013) Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25(8): 2907–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Jiang Y, Han X, Wang H, Pan J, Yu D (2017) Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J Exp Bot 68(6): 1361–1369 [DOI] [PubMed] [Google Scholar]
- Huang H, Liu B, Liu L, Song S (2017) Jasmonate action in plant growth and development. J Exp Bot 68(6): 1349–1359 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309(5732): 293–297 [DOI] [PubMed] [Google Scholar]
- Ingle RA, Stoker C, Stone W, Adams N, Smith R, Grant M, Carré I, Roden LC, Denby KJ (2015) Jasmonate signalling drives time-of-day differences in susceptibility of Arabidopsis to the fungal pathogen Botrytis cinerea. Plant J 84(5): 937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27(8): 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Liang G, Yang S, Yu D (2014) Arabidopsis WRKY57 functions as node of convergence for jasmonic acid-and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 26(1): 230–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju L, Jing Y, Shi P, Liu J, Chen J, Yan J, Chu J, Chen KM, Sun J (2019) JAZ proteins modulate seed germination through interacting with ABI5 in bread wheat and Arabidopsis. New Phytol 223(1): 246–260 [DOI] [PubMed] [Google Scholar]
- Katsir L, Chung HS, Koo AJ, Howe GA (2008) Jasmonate signaling: a conserved mechanism of hormone sensing. Curr Opin Plant Biol 11(4): 428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM (2013) MYC2: the master in action. Mol Plant 6(3): 686–703 [DOI] [PubMed] [Google Scholar]
- Kim KC, Lai Z, Fan B, Chen Z (2008) Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20(9): 2357–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Marchal V, Le Gourrierec J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U (2006) Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133(16): 3213–3222 [DOI] [PubMed] [Google Scholar]
- Lazaro A, Valverde F, Piñeiro M, Jarillo JA (2012) The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24(3): 982–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Ni W, Griffith ME, Huang Z, Chang C, Peng W, Ma H, Xie D (2004) The ASK1 and ASK2 genes are essential for Arabidopsis early development. Plant Cell 16(1): 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang Y, Li Q, Sang Y, Mao J, Lian H, Wang L, Yang H (2008). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20(2): 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Du M, Deng L, Shen J, Fang M, Chen Q, Lu Y, Wang Q, Li C, Zhai Q (2019) MYC2 regulates the termination of jasmonate signaling via an autoregulatory negative feedback loop. Plant Cell 31(1): 106–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16(7): 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A, Deplancke B, Walhout AJM, Tissenbaum HA (2008) Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat Protoc 3(4): 698–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M, Mitsuda N, Herde M, Koo AJ, Moreno JE, Suzuki K, Howe GA, Ohme-Takagi M (2013) A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant Cell 25(5): 1641–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschke S, Cortleven A, Iven T, Feussner I, Havaux M, Riefler M, Schmülling T (2016) Circadian stress regimes affect the circadian clock and cause jasmonic acid-dependent cell death in cytokinin-deficient Arabidopsis plants. Plant Cell 28(7): 1616–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448(7151): 358–361 [DOI] [PubMed] [Google Scholar]
- Pan J, Hu Y, Wang H, Guo Q, Chen Y, Howe GA, Yu D (2020) Molecular mechanism underlying the synergetic effect of jasmonate on abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 32(12): 3846–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz J, Kay S (2010) An expanding universe of circadian networks in higher plants. Trends Plant Sci 15(5): 259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80(6): 847–857 [DOI] [PubMed] [Google Scholar]
- Qi T, Huang H, Song S, Xie D (2015) Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell 27(6): 1620–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D (2011) The jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23(5): 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat R, Schwartz J, Jones MA, Sairanen I, Cheng Y, Andersson CR, Zhao Y, Ljung K, Harmer SL (2009). REVEILLE1, A Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc Natl Acad Sci USA 106(39): 16883–16888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Pan J, Peng W, Genschik P, Hobbie L, Hellmann H, Estelle M, Gao B, Peng J, Sun C, et al. (2005) Point mutations in Arabidopsis Cullin1 reveal its essential role in jasmonate response. Plant J 42(4): 514–524 [DOI] [PubMed] [Google Scholar]
- Robson F, Okamoto H, Patrick E, Harris SR, Wasternack C, Brearley C, Turner JG (2010) Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22(4): 1143–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288(5471): 1613–1616 [DOI] [PubMed] [Google Scholar]
- Sarmiento F (2013) The BBX subfamily IV: additional cogs and sprockets to fine-tune light-dependent development. Plant Signal Behav 8(4): e23831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y, Jikumaru Y, Obayashi T, Saito H, Masuda S, Kamiya Y, Ohta H, Shirasu K (2013) Basic helix-loop-helix transcription factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate responses in Arabidopsis. Plant Physiol 163(1): 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318(5848): 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer F, Fernández-Calvo P, Zander M, Diez-Diaz M, Fonseca S, Glauser G, Lewsey MG, Ecker JR, Solano R, Reymond P (2013) Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25(8): 3117–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Bhatt D (2015) The circadian clock and defense signaling in plants. Mol Plant Pathol 16(2): 210–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu F-F, Sharon M, Browse J, et al. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468(7322): 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127(4): 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Shim JS, Kubota A, Imaizumi T (2017) Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol 173(1): 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Heidrich K, Sanchez-Villarreal A, Parker JE, Davis SJ (2012) TIME FOR COFFEE represses accumulation of the MYC2 transcription factor to provide time-of-day regulation of jasmonate signaling in Arabidopsis. Plant Cell 24(6): 2470–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Igeño MI, Coupland G (1996) Activation of floral meristem identity genes in Arabidopsis. Nature 384(6604): 59–62 [DOI] [PubMed] [Google Scholar]
- Song S, Huang H, Gao H, Wang J, Wu D, Liu X, Yang S, Zhai Q, Li C, Qi T, et al. (2014b) Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26(1): 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Fan M, Zhang X, Gao H, Huang H, Wu D, Guo H, Xie D (2013a) The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet 9(7): e1003653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D (2011) The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23(3): 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Estrada DA, Johnson RS, Kim SK, Lee SY, MacCoss MJ, Imaizumi T (2014a) Distinct roles of FKF1, Gigantea, and Zeitlupe proteins in the regulation of Constans stability in Arabidopsis photoperiodic flowering. Proc Natl Acad Sci USA 111(49): 17672–17677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Ito S, Imaizumi T (2013b) Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci 18(10): 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T (2015) Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol 66: 441–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T (2012) FKF1 Conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336(6084): 1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410(6832): 1116–1120 [DOI] [PubMed] [Google Scholar]
- Takada S, Goto K (2003) TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15(12): 2856–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448(7154): 661–665 [DOI] [PubMed] [Google Scholar]
- Thines B, Parlan EV, Fulton EC (2019) Circadian network interactions with jasmonate signaling and defense. Plants (Basel) 8(8): 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Shen Y, Chang H-C, Hou Y, Harris A, Ma SF, McPartland M, Hymus GJ, Adam L, Marion C, et al. (2010) The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol 187(1): 57–66 [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303(5660): 1003–1006 [DOI] [PubMed] [Google Scholar]
- Wang H, Li S, Li Y, Xu Y, Wang Y, Zhang R, Sun W, Chen Q, Wang X-J, Li C, et al. (2019) MED25 connects enhancer-promoter looping and MYC2-dependent activation of jasmonate signaling. Nat Plants 5(6): 616–625 [DOI] [PubMed] [Google Scholar]
- Wang H, Li Y, Pan J, Lou D, Hu Y, Yu D (2017) The bHLH transcription factors MYC2, MYC3, and MYC4 are required for jasmonate-mediated inhibition of flowering in Arabidopsis. Mol Plant 10(11): 1461–1464 [DOI] [PubMed] [Google Scholar]
- Wang H, Pan J, Li Y, Lou D, Hu Y, Yu D (2016) The DELLA-CONSTANS cascade integrates gibberellin and photoperiod signaling to regulate flowering in Arabidopsis. Plant Physiol 172(1): 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C (2020) Determination of sex by jasmonate. J Integr Plant Biol 62(2): 162–164 [DOI] [PubMed] [Google Scholar]
- Webb A, Seki M, Satake A, Caldana C (2019) Continuous dynamic adjustment of the plant circadian oscillator. Nat Commun 10(1): 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2(8): e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280(5366): 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D (2002) The SCF(COI1) ubiqutin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14(8): 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Zhang C, Gu M, Bai Z, Zhang W, Qi T, Cheng Z, Peng W, Luo H, Nan F, et al. (2009) The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21(8): 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D-L, Yao J, Mei C-S, Tong X-H, Zeng L-J, Li Q, Xiao L-T, Sun T-P, Li J, Deng X-W, et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109(19): E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Han X, Yang J, Jiang Y, Hu Y (2021) The Arabidopsis circadian clock protein PRR5 interacts with and stimulates ABI5 to modulate abscisic acid signaling during seed germination. Plant Cell 33(9): 3022–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2(7): 1565–1572 [DOI] [PubMed] [Google Scholar]
- You Y, Zhai Q, An C, Li C (2019) LEUNIG-HOMOLOG mediates MYC2-dependent transcriptional activation in cooperation with the coactivators HAC1 and MED25. Plant Cell 31(9): 2187–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Zhang X, Wu F, Feng H, Deng L, Xu L, Zhang M, Wang Q, Li C (2015) Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. Plant Cell 27(10): 2814–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang L, Zeng L, Zhang C, Ma H (2015) Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev 29(9): 975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Gao M, Seitz NC, Angel W, Hallworth A, Wiratan L, Darwish O, Alkharouf N, Dawit T, Lin D, et al. (2019a) LUX ARRHYTHMO mediates crosstalk between the circadian clock and defense in Arabidopsis. Nat Commun 10(1): 2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ren Z, Zhou Y, Ma Z, Ma Y, Hou D, Xu Z, Huang X (2019b) NPR1 And redox rhythmx: connections, between circadian clock and plant immunity. Int J Mol Sci 20(5): 1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang F, Melotto M, Yao J, He SY (2017) Jasmonate signaling and manipulation by pathogens and insects. J Exp Bot 68(6): 1371–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhu Z, An F, Hao D, Li P, Song J, Yi C, Guo H (2014) Jasmonate-activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. Plant Cell 26(3): 1105–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang Y, Wei H, Li N, Tian W, Chong K, Wang L (2018) Circadian evening complex represses jasmonate-induced leaf senescence in Arabidopsis. Mol Plant 11(2): 326–337 [DOI] [PubMed] [Google Scholar]
- Zhou W, Lozano-Torres JL, Blilou I, Zhang X, Zhai Q, Smant G, Li C, Scheres B (2019) A jasmonate signaling network activates root stem cells and promotes regeneration. Cell 177(4): 942–956 [DOI] [PubMed] [Google Scholar]
- Zhu Z, An F, Feng Y (2011) Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA 108(30): 12539–12544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21(10): 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.