Significance
Metamorphic proteins play an important role in both native biological processes and disease states through their shape-shifting properties, which allow them to expand their functional and dysfunctional capacity. However, the design of monomeric proteins that can shift between differently folded states remains a challenge. This paper describes a designed system that can switch reversibly between two of the most common protein topologies, 3α and α/β-plait, over a relatively narrow temperature range that is relevant to biology. The results provide mechanistic insights into how proteins can become shape-shifters, as well as adding to the protein design toolkit. The reversible 3α/αβ switch may also serve as a model system for further understanding the fundamentals of protein folding and fold switching.
Keywords: protein fold switching, metamorphic proteins, protein design, NMR, protein structure and dynamics
Abstract
Naturally occurring metamorphic proteins have the ability to interconvert from one folded state to another through either a limited set of mutations or by way of a change in the local environment. Here, we show in a designed system that it is possible to switch reversibly between two of the most common monomeric folds employing only temperature changes. We demonstrate that a latent 3α state can be unmasked from an α/β-plait topology with a single V90T amino acid substitution, populating both forms simultaneously. The equilibrium between these two states exhibits temperature dependence, such that the 3α state is predominant (>90%) at 5 °C, while the α/β-plait fold is the major species (>90%) at 30 °C. We describe the structure and dynamics of these topologies, how mutational changes affect the temperature dependence, and the energetics and kinetics of interconversion. Additionally, we demonstrate how ligand-binding function can be tightly regulated by large amplitude changes in protein structure over a relatively narrow temperature range that is relevant to biology. The 3α/αβ switch thus represents a potentially useful approach for designing proteins that alter their fold topologies in response to environmental triggers. It may also serve as a model for computational studies of temperature-dependent protein stability and fold switching.
Metamorphic proteins have malleable polypeptide chains that can adopt more than one folded topology with very little or no alteration in their amino acid sequence. Some of the earliest known examples of shape-shifting proteins, such as prions, involved formation of multimeric species that were associated with disease states (1). However, multimerization is not a necessary prerequisite for fold switching. Moreover, such large-amplitude conformational transitions can lead to new functions, not just disease states. There are now numerous examples of naturally occurring fold switches, indicating that protein fold metamorphism is a more widespread phenomenon than previously thought (2–4). Indeed, recent estimates suggest that approximately 5% of proteins in the PDB may have interconvertible topologies (5). Fold switching thus represents the ability of polypeptide chains to expand their functional capacity by adopting more than one three-dimensional structure in certain cases, providing new opportunities for the design of proteins with novel properties (6, 7).
The observation that some protein structures are metamorphic also has relevance to understanding how new folds might evolve (8). Several experimental studies have provided evidence of fold migration, where transitory amino acid sequences have been identified with topologies in between two different origin and destination structures (9, 10). Furthermore, protein design endeavors have shown that different folds may be related through a relatively small number of mutations (11, 12). In the case of the Streptococcal albumin-binding GA and IgG-binding GB domains of protein G, for example, it was demonstrated that conversion between the 3α GA topology and the 4β + α GB fold could occur with as little as a single amino acid substitution (13, 14). In addition, it was recently shown that both GA and GB can be connected with another common small fold, the α/β-plait topology of the S6 ribosomal protein, relating the three folds into a network with high-identity intersections (15). These results indicated that it was possible to derive new topologies efficiently from preexisting structures, rather than having each fold necessarily evolve as an independent entity. Computational studies further supported this notion by showing that a significant fraction of the known nonredundant folds can be connected with minimal changes to the amino acid sequences (16).
In naturally occurring metamorphic proteins characterized to date, the large-scale topology change is often caused either by an environmental factor such as pH (hemagglutinin) (17), ligand binding [Mad2 (18), KaiB (19)], redox (CLIC1) (20), proteolytic cleavage (serpins) (21), or a combination of stimuli such as salt concentration and temperature (lymphotactin) (22, 23). Here, we show how temperature alone can be used to switch reversibly between two different but very common fold topologies (24) to near-quantitative levels. We employ a designed system in which the 56-amino acid sequence for a stable, 3α-folded protein (denoted A1) is embedded within the sequence for a 95-amino acid polypeptide chain with a stable α/β-plait topology (denoted Sa1) (Fig. 1). The parent GA and S6 sequences from which A1 and Sa1 are derived have only 16% sequence identity. The A1 and Sa1 proteins were coevolved to very high sequence identity (100% over the 56-amino acid GA sequence) while maintaining their respective 3α and α/β-plait folds, as described previously (15). Briefly, this involved the following steps: 1) thread the GA sequence through the S6 fold to find alignments that minimize the most severe steric clashes; 2) design mutations to relieve unfavorable contacts and evaluate the energies computationally; and 3) maintain the parent amino acids where possible. The resulting Sa1 protein thus has an amino acid sequence that codes for both the α/β-plait and 3α folds. The competition between the embedded 3α fold and longer α/β-plait fold creates a critical state in which the latent 3α structure can be exposed with a relatively subtle valine to threonine mutation, setting up a situation in which the 3α and α/β-plait forms are populated simultaneously. The equilibrium between these two monomeric states exhibits temperature dependence such that the folds can be switched reversibly from mostly 3α (>90%) at 5 °C to mostly α/β-plait (>90%) at 30 °C. We describe the structure and dynamics of these topologies, how mutational changes affect the temperature dependence, and the energetics and kinetics of interconversion. Additionally, we demonstrate how the albumin-binding function of the 3α state can be regulated over a relatively narrow temperature range that is relevant to biology by large amplitude changes in protein structure.
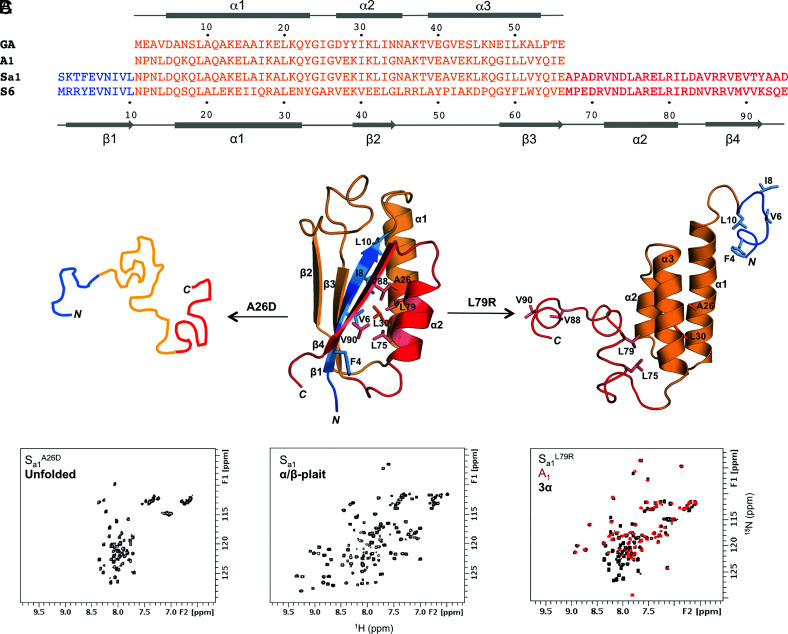
Fig. 1.
Unmasking alternative fold topologies. (A) Sequence alignment of 3α A1 and α/β-plait Sa1, with corresponding secondary structure regions at the Top and Bottom of the alignment. The parent sequences from which A1 and Sa1 are derived, GA and S6 respectively, are also shown. (B) A single amino acid mutation in Sa1 (Center) such as A26D leads to unfolding (Left), while a L79R mutation exposes the alternative 3α state (Right). Color coding is as follows: N-terminal residues 1 to 10 (blue); residues 11 to 66 aligning with the A1 amino acid sequence (orange); C-terminal residues 67 to 95 (red). (C) Representative chemical shift patterns in 2D 1H-15N HSQC spectra of the unfolded (Left), α/β-plait (Center), and 3α states (Right). The HSQC spectrum of Sa1L79R (Right, black) is overlaid with the 56-amino acid 3α-helical protein, A1 (red).
Results
Unmasking of Alternative Fold Topologies.
The 95-amino acid protein utilized in this study, Sa1, is a version of the S6 ribosomal protein from Thermus thermophilus that has been modified to incorporate a 56-residue GA-derived sequence, A1, in its entirety (Fig. 1A) (15). Thus, while the 56-amino acid protein A1 has a 3α-helical fold, it forms a region containing the α1-helix, and β2- and β3-strands of the α/β-plait structure when it is in the context of the longer Sa1 polypeptide chain (Fig. 1B). In contrast to many native proteins, which tend to be approximated as two-state systems, single amino acid changes in Sa1 lead to three readily definable states. Depending on where the mutation is made, the outcome may be the α/β-plait fold of S6, unmasking of the alternative 3α state of GA, unfolding, or some combination of these (Fig. 1B). Generating a largely unfolded Sa1 polypeptide chain through single-site mutations requires destabilization of the core in both GA and S6 folds. There are limited options for doing this with single amino acid mutations as there are only two residues, A26 and L30, which are buried in both folds. However, more avenues exist for unmasking of the 3α fold, which requires destabilization of the α/β-plait without affecting 3α substantially. This can be done in a number of ways as the N-terminal 10 amino acids and C-terminal 29 amino acids of the Sa1 sequence are outside the folded region for the 3α state. These residues form the β1, β4, and α2 secondary structures that pack against each other in the α/β-plait. Therefore, mutations to the core contributors from β1 (F4, V6, I8, L10), β4 (V88 and V90), and α2 (L75 and L79) would be expected to perturb α/β-plait stability while having minimal effect on the stability of 3α. Lone mutations at either L75R or L79R in the α2-helix of the α/β-plait fold do in fact unmask the alternative 3α topology, giving proteins with 2D 1H-15N HSQC spectra consistent with the peak pattern seen for the 3α-fold A1 (Fig. 1C). Likewise, deletion of N-terminal residues 1 to 4 or C-terminal residues 90 to 95 is also sufficient to give NMR spectra yielding only 3α-type signals (SI Appendix, Fig. S1). Further, more subtle mutations might be expected to populate both states simultaneously. Of the possible locations outside the 3α-folded region for making such mutations, we focused on residue V90 in the β4 strand, which is at the periphery of the hydrophobic core for the α/β-plait structure (Fig. 2A). Whereas the above mutations abolished the α/β-plait fold, we postulated that a relatively small increase in polarity, from V90 to T90, might be sufficient to unmask the 3α fold without completely disrupting α/β-plait stability. Indeed, Sa1V90T gives a 2D 1H-15N HSQC spectrum with an approximately two-fold increase in the expected number of signals and a peak pattern consistent with both 3α and α/β-plait topologies being present. Assignment of resonances confirmed that the 3α and α/β-plait folds are populated simultaneously (Fig. 2B). The V90T mutation destabilizes the α/β-plait fold minimally, yet this amino acid change is enough to reveal propensity for the alternative 3α state without completely losing propensity for the α/β-plait topology.
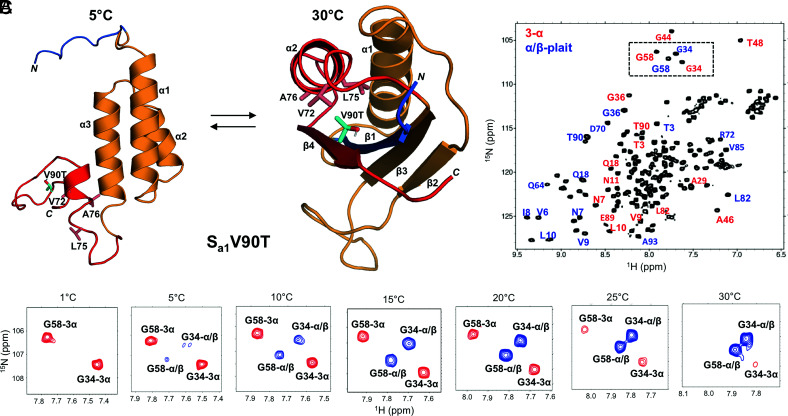
Fig. 2.
Reversible switching between 3α and α/β-plait folds with temperature. (A) Structure of Sa1V90T at 30 °C (Right) highlighting the position of V90T (cyan) on the β4-strand of the α/β-plait and its interaction with hydrophobic residues in the α2-helix. The position of V90T in the alternative 3α state at 5 °C is also shown (Left). Color coding of the main chain is as in Fig. 1. (B) 2D 1H-15N HSQC spectrum of Sa1V90T at 15 °C showing signals due to both the 3α (red labels) and α/β-plait (blue labels) states. (C) Boxed region from the HSQC spectrum in (B) shown as a function of temperature. Peaks corresponding to the 3α state (red) decrease while α/β-plait peaks (blue) increase as the temperature is raised from 1 °C to 30 °C for representative residues G34 and G58 in the Sa1V90T polypeptide chain.
Reversible Switching between 3α and α/β-Plait Folds with Temperature.
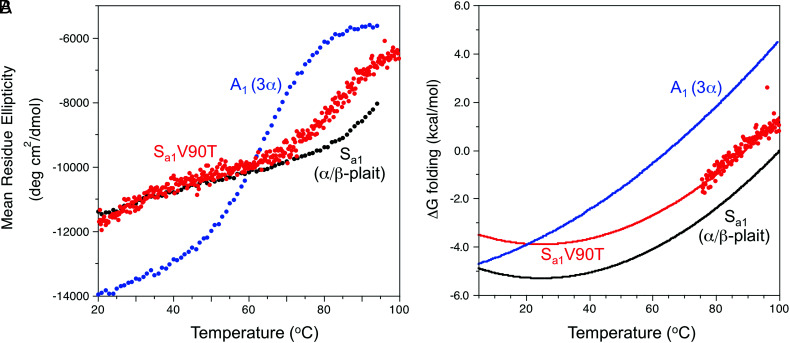
The equilibrium between the 3α and α/β-plait structures of Sa1V90T can be shifted with temperature (Fig. 2C). At 5 °C, the HSQC spectrum indicates that the major species is 3α (>90%). However, the population of 3α decreases as the temperature is raised, with a corresponding increase in signals due to the α/β-plait. By 30 °C, the major conformation is the α/β-plait fold (>90%). Moreover, the temperature-dependent switch in fold topologies is reversible (SI Appendix, Fig. S2). Thus, the 2D 1H-15N HSQC spectrum acquired at 30 °C is completely different from that recorded at 5 °C. Size exclusion chromatography combined with multi-angle static light scattering indicates that these states are monomeric at 22 °C, where the 3α and α/β-plait folds are in an approximately 30:70 equilibrium ratio (SI Appendix, Fig. S3). Temperature-dependent circular dichroism (CD) shows two partial transitions, one for the 3α to α/β-plait conversion at low temperature and another at high temperature for unfolding of the α/β-plait (Fig. 3A). The latter transition has an apparent midpoint of ~87 °C. Extrapolating a plot of ∆Gfolding versus temperature for Sa1V90T to lower temperature indicates a ∆Gfolding relative to the unfolded state of −3.9 kcal/mol at 25 °C (Fig. 3B). (The extrapolation of the high temperature transition to lower temperature estimates the equilibrium of the α/β-plait fold with the unfolded state.) In comparison, the ∆Gfolding of the parent Sa1 protein at 25 °C is −5.3 kcal/mol. Thus, the V90T mutation destabilizes the α/β-plait fold relative to the unfolded state by 1.4 kcal/mol. From HSQC analysis, the temperature midpoint (Tmid) at which the 3α and α/β-plait folds are in a 1:1 ratio is 17 °C. This is comparable with a Tmid ~ 20 °C estimated from the CD melting data where the extrapolated ΔGfolding curves for Sa1V90T and A1 intersect (Fig. 3B). The fitting method is described in Materials and Methods.
Fig. 3.
Thermal stability of Sa1V90T and comparison with parent proteins A1 (3α) and Sa1 (α/β-plait). (A) Plots of mean residue ellipticity for Sa1V90T (red), Sa1 (black), and A1 (blue) at 222 nm versus temperature. (B) Plot of ΔGfolding versus temperature for Sa1V90T (red). The data points are calculated ΔGfolding values from CD melting data, and the line represents the fitted stability profile from the Gibbs–Helmholtz equation (see Materials and Methods for details). Fitted stability profiles based on CD data are also shown for Sa1 (black) and A1 (blue).
NMR assignments of main chain resonances were made for both the high temperature and low temperature forms of Sa1V90T. At 30 °C, a three-dimensional structure was determined using a combination of chemical shift and interproton NOE restraint inputs into CS-Rosetta (Fig. 4A). The structure consists of β–strands between residues 3 and 10 (β1), residues 40 and 44 (β2), residues 60 and 66 (β3), and residues 86 and 92 (β4), and α-helices between residues 16 and 32 (α1) and residues 72 and 81 (α2). The four β-strands and two α-helices are arranged in an α/β-plait (βαββαβ) topology consistent with other S6 folds (25, 26). At 5 °C, a three-dimensional structure was also determined that has α-helices between residues 15 and 32 (α1), residues 37 and 45 (α2), and residues 49 and 63 (α3) as seen in other GA folds (Fig. 4B) (11, 27). The N-terminal amino acids 1 to 14 and C-terminal amino acids 64 to 95 are mostly disordered. However, a fourth helix (α4) is present between residues 75 and 81, which corresponds with the position of α2 in the α/β-plait. This helix is of lower stability than the other three helices and has, on average, smaller secondary ΔδCα and ΔδCO shifts (SI Appendix, Fig. S4) and fewer detectable NOEs supporting helical structure. Comparison of backbone amide chemical shift patterns for Sa1V90T at 5 °C with A1 showed that the main differences are limited to the attachment sites of the N-terminal 10 residues and the C-terminal 28 residues (Fig. 1C), indicating that the α4 helix does not interact significantly with the 3α bundle on the chemical shift timescale. Thus, the 5 °C structure of Sa1V90T closely matches that of A1 (15) over the 3α-helical region, while the 30 °C structure of Sa1V90T aligns well with the α/β-plait Sa1 structure (15) (SI Appendix, Fig. S5). The T90 side chain does not appear to form stable H-bonds in either state. Presumably, the V90T mutation decreases the stability of the α/β-plait through a decrease in hydrophobic interactions at the periphery of the core. Structures and assignments for the 5 °C and 30 °C forms of Sa1V90T have been deposited in the PDB and BMRB, respectively. Accession codes and structure statistics are provided in SI Appendix, Table S1.
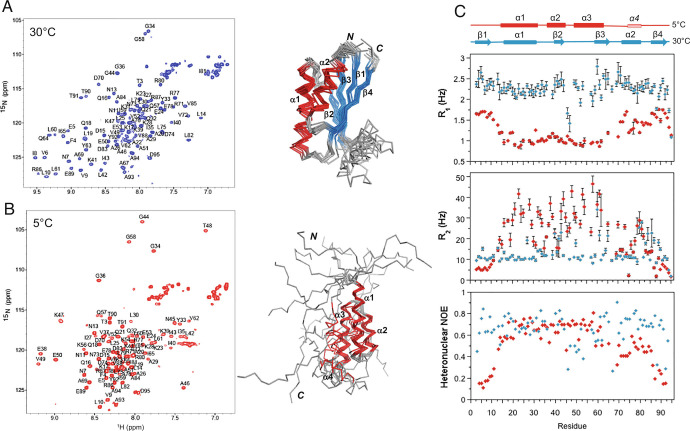
Fig. 4.
Structure and dynamics of Sa1V90T as a function of temperature. (A) 2D 1H-15N HSQC spectrum of Sa1V90T at 30 °C with backbone amide assignments (Left) and the corresponding ensemble of 10 lowest energy α/β-plait structures (right). (B) 2D 1H-15N HSQC spectrum of Sa1V90T at 5 °C with backbone amide assignments (left) and the ensemble of 10 lowest energy 3α structures (Right). (C) Plot of 15N-R1 and -R2 rates, and {1H}-15N steady-state heteronuclear NOE versus residue for Sa1V90T in the 3α state at 5 °C (red) and α/β-plait form at 30 °C (blue).
The dynamics of the Sa1V90T polypeptide chain were investigated on the nanosecond-to-picosecond timescale as a function of temperature using steady-state {1H}-15N heteronuclear NOE and 15N R1 and R2 relaxation measurements (Fig. 4C). At 5 °C, the heteronuclear NOE pattern is consistent with a well-ordered 3α-helical bundle corresponding to the structured region from residues 15 to 64, a less ordered α4 helix, and highly flexible N- and C-terminal tails from residues 1 to 14 and 82 to 95. The R1 and R2 relaxation profiles further illustrate the marked difference in the backbone dynamics of the 3α-bundle and disordered components of the 5 °C structure. Estimates of the correlation time, τC, for different regions of the polypeptide chain at 5 °C from the R1 and R2 rate constants indicated that the N-terminal tail has motions approximately three times faster than those of the 3α bundle, while the C-terminal tail tumbles about twice as fast as the 3α region (SI Appendix, Table S2). At 30 °C, in contrast, the heteronuclear NOEs and R1 and R2 values have minimal deviation from the average, consistent with an α/β-plait fold that is tumbling more uniformly over the entire length of the polypeptide chain. Notable variations correspond with loop regions, particularly the large flexible loop between the β2- and β3-strands (residues 45 to 59). At Tmid 17 °C, where the 3α and α/β-plait folds are approximately equally populated, the 3α region and α/β-plait have comparable R1 and R2 rate constants and τC, indicating that the ordered region of the 3α structure has similar fast dynamics to that of the α/β-plait (SI Appendix, Fig. S6).
How Mutations Alter the Temperature Dependence of Fold Switching.
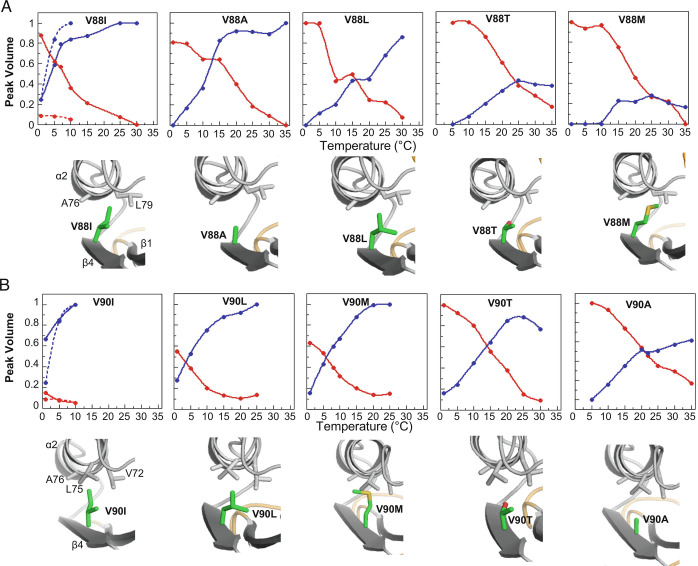
Further single amino acid substitutions were made to Sa1 at either V90 or an adjacent position in the β4-strand, V88. These were largely aliphatic mutations, altering valine to either isoleucine, leucine, alanine, methionine, or threonine. Similar to the V90T mutant, analysis of 2D 1H-15N HSQC spectra for the V88 and V90 mutants as a function of temperature demonstrated that these proteins also exhibit large amplitude fold interconversion between 1 °C and 35 °C (Fig. 5 and SI Appendix, Figs. S7 and Fig. S8). In all of these mutants, with the exception of Sa1V90I, the 3α state is populated predominantly at lower temperature and the equilibrium gradually shifts to the α/β-plait state as the temperature is raised. The temperature midpoint (Tmid) at which the 3α and α/β-plait folds are equally populated is thus highly dependent on the nature of the amino acid mutation, shifting by as much as 20 °C with relatively conservative changes. Increasing Tmid values appear to correspond approximately with decreasing α/β-plait stability as determined by temperature-dependent CD for some V88 mutants (SI Appendix, Fig. S9). This is consistent with the postulate that mutations at V88 and V90 are more likely to affect the stability of the α/β-plait than the 3α form. The residues most stabilizing to the α/β-plait at both the V88 and V90 positions are the β-branched aliphatic amino acids isoleucine and valine, which appear to provide optimal packing. In contrast, V88M is most destabilizing to the α/β-plait fold, resulting in main chain amide signal broadening in HSQC spectra at higher temperatures and precipitation of the protein (SI Appendix, Fig. S7E). Thus, relatively small changes in hydrophobicity and branching at the periphery of the α/β-plait core lead to significant shifts in the temperature dependence of the fold switch.
Fig. 5.
How mutations alter the temperature dependence of fold switching. (A) Relative peak volume of 3α (red) and α/β-plait (blue) conformational states observed in the 2D 1H-15N HSQC spectra of Sa1V88 mutants as temperature is raised from 1 to 35 °C. The peak volume is an average value from the main chain amide signals of 4 to 6 residues that are in the folded regions of both forms. The intersection of the plots indicates the temperature (Tmid) at which the α/β-plait and 3α states are equally populated. Pymol models of the mutations (green) are shown below each of the plots. (B) As in (A), but for Sa1V90 mutants. The temperature dependence of Sa1 is overlaid in the V88I and V90I plots as a broken line. Lines serve only as a visual guide.
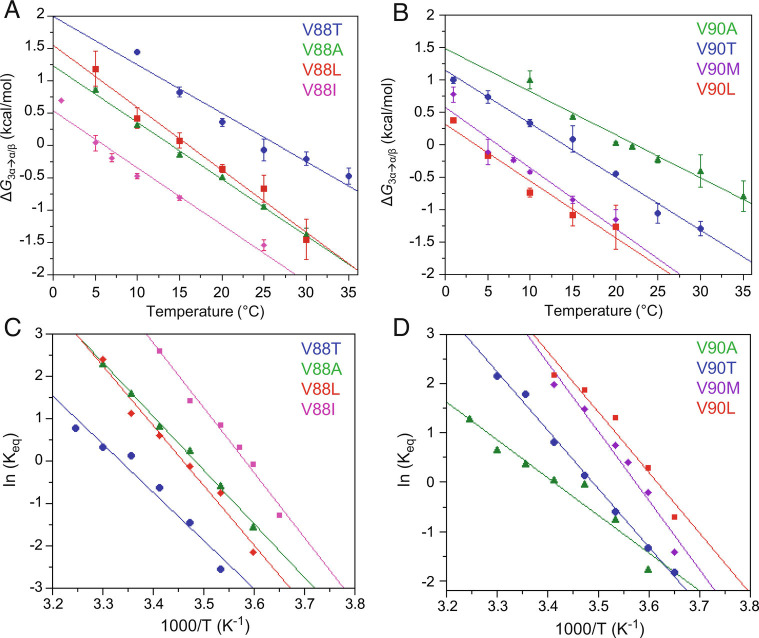
Energetics and Kinetics of Fold Interconversion.
The free energy difference between the 3α and α/β-plait folds, ΔG3α→α/β, was determined from the equilibrium constant, Keq, given by the population ratio of the 3α to α/β-plait states as a function of temperature (Fig. 6 A and B). Where the population shifts almost completely (>90%) from one state to the other within the 1 to 35 °C temperature range used (V88A, V88L, V90T, V90A), the switch corresponds with a free energy change of approximately 2 to 3 kcal mol−1. For other mutants where the Tmid is closer to one end of the temperature range (V88I, V88T, V90L, V90M), the approximately linear ΔG3α→α/β versus temperature plots and their similar slopes suggest that they likely have comparable free energy differences between their 3α and α/β-plait forms. Additionally, the changes in enthalpy, ΔH°, and entropy, ΔS°, of 3α to α/β-plait fold conversion were estimated from the Van’t Hoff equation,
Fig. 6.
Energetics of fold interconversion between 3α and α/β-plait structures. (A) ΔG3α→α/β versus temperature for the Sa1 mutants V88T (blue), V88A (green), V88L (red), and V88I (magenta). (B) ΔG3α→α/β versus temperature for Sa1 mutants V90A (green), V90T (blue), V90L (red), and V90M (purple). ΔG3α→α/β is calculated for V88 and V90 mutants from the equilibrium constant for exchange between 3α and α/β-plait conformations, K3α→α/β, at temperatures of 1 to 35 °C using the equation ΔG3α→α/β = −RTln K3α→α/β. The equilibrium constant at each temperature is an average value determined from peak volumes for main chain amide signals of 4 to 6 residues that are in the folded regions of both 3α and α/β-plait forms and have similar relaxation properties in both states. (C) Van’t Hoff plots of logKeq versus 1/T for the Sa1 mutants V88T (blue), V88A (green), V88L (red), and V88I (magenta). (D) Van’t Hoff plots for the Sa1 mutants V90A (green), V90T (blue), V90L (red), and V90M (purple). The plots in (C and D) are fitted to the linear form of the Van’t Hoff equation (Eq. 1), which is used to estimate the enthalpy and entropy of the fold conversion from the slope and intercept, respectively.
| [1] |
by relating the equilibrium constant, Keq, to the change in temperature. The negative slope of the Van’t Hoff plots for both the V88 and V90 mutants indicates an endothermic reaction which is consistent with the requirement of a temperature increase to drive the conversion from the 3α to α/β-plait state (Fig. 6 C and D). The average ΔH°3α→α/β was 26.5 ±3.3 kcal mol−1 for V88 mutants and 22.7 ± 5.4 kcal mol−1 for the V90 mutants, while the average ΔS°3α→α/β was 0.09 ± 0.01 kcal mol−1K−1 for V90 mutants and 0.08 ± 0.02 kcal mol−1K−1 for the V90 mutants.
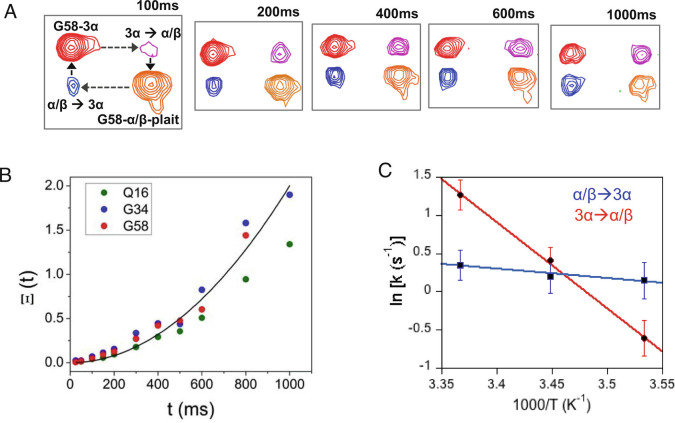
The simultaneous detection of two sets of NMR signals corresponding to the α/β-plait and 3α forms in the 2D 1H-15N HSQC spectrum of Sa1V90T indicated that the two folds are undergoing slow exchange relative to the NMR chemical shift timescale. We employed 2D longitudinal 15N ZZ-exchange spectroscopy, which measures chemical exchange on the millisecond to seconds time scale, to quantify the kinetics of the interconversion between the two folded states of Sa1V90T.(28, 29) A series of 2D 1H-15N HSQC spectra were collected with variable delays to detect buildup of exchange peaks and estimate the forward, k3α→α/β, and reverse, kα/β→3α, exchange rate constants for three residues, Q16, G34, and G58, that had well-resolved autopeaks and crosspeaks (Fig. 7 A and B). At 17 °C, where the 3α and α/β-plait conformations are close in population, the two folds exchanged with an average rate constant of 1.2 to 1.5 s−1, in accord with a slow exchange event. To determine the activation energies of the transition states for the forward and reverse reactions, the temperature dependence of the exchange rate constants was measured by repeating the ZZ-exchange analysis at 10 °C and 24 °C (SI Appendix, Table S3). While kα/β→3α was minimally affected by the change in temperature, k3α→α/β decreased to 0.6 s−1 at 10 °C and increased to 3.6 s−1 when the temperature was raised to 24 °C. The resulting activation energy, Ea, from 3α→α/β-plait was 22.0 ± 0.4 kcal/mol, while the Ea for α/β-plait→3α was 2.4 ± 0.5 kcal/mol (Fig. 7C).
Fig. 7.
Kinetics of fold interconversion between α/β-plait and 3α states using ZZ-exchange NMR. (A) Spectra showing the buildup of exchange crosspeaks (blue and magenta) and decay of autopeaks (red and orange) for G58 at several mixing times at 17 °C. (B) The time-dependent buildup of crosspeaks was determined for three residues (Q16, G34, and G58) using the composite peak intensity ratio (Ξ) fit equation (Eq. 3). The average curve fit is shown in black. (C) Arrhenius analysis of activation energy, Ea. The average exchange rate constants from residues Q16, G43, and G58 were obtained from ZZ-exchange experiments at 10, 17, and 24 °C. The logarithm of the rate constants was plotted against inverse temperature and Ea was obtained from the slope of the linear fit equation (Eq. 4).
Comparison of the change in enthalpy from the Van’t Hoff equation and the activation energy from ZZ-exchange analysis provides consistent and independently derived energetic values for 3α→α/β-plait conversion. The activation energy, Ea, of 22.0 kcal mol−1 for the 3α→α/β-plait fold switch of Sa1V90T approximates the observed change in enthalpy, ΔH°3α→α/β, of 23.5 kcal mol−1. For the endothermic 3α→α/β-plait conversion, ΔH°3α→α/β describes the heat absorbed by the reaction while Ea provides the energy required to reach the transition state. The proximity of these values suggests that the Ea of the 3α to α/β-plait switch is dominated by an enthalpic barrier.
Regulation of Fold and Function over a Narrow Temperature Range.
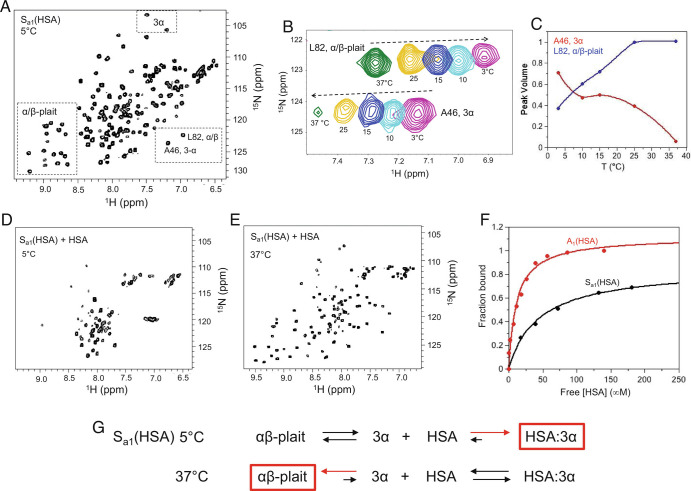
The designed 3α-helical A1 protein (Fig. 1) does not bind to human serum albumin (HSA), but HSA-binding function was engineered into the 56-amino acid sequence using five mutations (E28K, K29Y, A41G, K44S, and V52A) that are based on structural alignment to the wild-type 3α-helical GA module in complex with HSA (30) (SI Appendix, Fig. S10 A and B). The corresponding mutations were also introduced into the longer Sa1 sequence to generate a protein, Sa1(HSA), which has a destabilized α/β-state and partially populates the alternative 3α form at 5 °C (Fig. 8A). Similar to Sa1V90T and the other variants described above, the equilibrium between the 3α and α/β states in Sa1(HSA) is temperature dependent with ~55% 3α at 5 °C, ~95% α/β at 37 °C, and a Tmid ~ 7 °C (Fig. 8 B and C). The HSA-binding epitope is intact in the 3α state but is cryptic in the α/β-plait form (SI Appendix, Fig. S10C). Addition of HSA to 15N-Sa1(HSA) at 5 °C leads to loss of backbone amide signals in the HSQC spectrum from both the α/β state and the folded region of the 3α conformation (Fig. 8D). However, resonances corresponding to the disordered N- and C-terminal regions of the 3α state are detectable, consistent with the formation of an approximately 77 kDa 3α/HSA complex in which only the faster tumbling ends of the 3α species can be readily observed. When the temperature is raised to 37 °C, the narrow linewidth α/β state signals reappear and the 3α state resonances are no longer detected, indicating dissociation of the 3α/HSA complex and fold switching to the α/β-plait (Fig. 8E).
Fig. 8.
Regulation of fold and HSA-binding function over a narrow temperature range. (A) The 2D 1H-15N HSQC spectrum of Sa1(HSA) at 5 °C. Signature 3α and α/β-plait chemical shifts are boxed. (B) Overlay of the 2D 1H-15N HSQC spectra of Sa1(HSA) from 3 to 37 °C showing residue L82 from the α/β-plait fold and A46 from the 3α state. (C) Normalized peak volume of 3α residue A46 (red) and α/β-plait residue L82 (blue) as a function of temperature. (D) The 2D 1H-15N HSQC spectrum of Sa1(HSA) saturated with four molar equivalents of HSA at 5 °C showing loss of signature 3α and α/β-plait signals. The intense resonances near 8 ppm in the proton dimension correspond with disordered N- and C-terminal regions of the 3α state. (E) The 2D 1H-15N HSQC spectrum of Sa1(HSA) with four molar equivalents of HSA at 37 °C showing α/β-plait signals. (F) HSA-binding curves for A1(HSA) (red) and Sa1(HSA) (black) at 5 °C. The fraction bound is extracted from peak intensity decay of amide proton signals in 2D 1H-15N HSQC spectra of 15N-labeled protein and is plotted against the free HSA concentration. (G) HSA-binding reaction to Sa1(HSA) at 5 °C and 37 °C. The reaction equation at 5 °C is generated based on the absence of α/β-plait resonances at high concentration of HSA. The reaction equation at 37 °C is based on the appearance of α/β-plait signals at high concentration of HSA.
The dissociation constant for the binding reaction of Sa1(HSA) with HSA is ~45 μM at 5 °C, higher than for A1(HSA), which has a KD of ~12 μM at 5 °C (Fig. 8F). The weaker binding of Sa1(HSA) is due to the fact that it populates 3α and α/β almost equally at 5 °C and needs to fold switch to 3α to bind HSA completely. Therefore, at 5 °C, the free energy of fold switching from 3α to α/β for Sa1(HSA) is near 0 kcal/mol and is outweighed by the stability increase of HSA binding to the 3α state, resulting in the gain of HSA-binding function (Fig. 8G). In contrast, the free energy of fold switching to the α/β state at 37 °C is less than −2 kcal/mol based on the relative population of α/β-plait (Fig. 6). Thus, the energetic gain from fold switching to the α/β state at 37 °C outweighs the stability increase from HSA binding to 3α and leads to a >20-fold attenuation of HSA binding with effective loss of function. The competing equilibria of fold switching and HSA binding are therefore shifted using temperature changes over a relatively narrow range that regulate both fold and function.
Discussion
Here, we describe the reversible interconversion between two common folds, the 3α helix bundle and the α/β-plait, using only temperature. The stabilities of the 3α and α/β-plait topologies of the Sa1V90T polypeptide chain are similar over the temperature range used, with small free energy differences, ΔG3α→α/β, of approximately 2 to 3 kcal/mol needed to shift the population from more than 90% 3α at 5 °C to more than 90% α/β-plait at 30 °C (Fig. 6). One likely contributing factor to the destabilization of the α/β-plait fold relative to the 3α fold at 5 °C is that hydrophobic forces decrease at lower temperatures (31, 32). Because the α/β-plait has a larger hydrophobic core (~3,200 Å2) than the 3α topology (~1,700 Å2), destabilizing effects of the temperature-dependent decrease in hydrophobicity for the α/β-plait fold may outweigh those for the 3α structure sufficiently to get the small observed difference in free energy. This would lead to the more conformationally expanded 3α state being favored at lower temperature. While this may seem counterintuitive, it has analogy to the phenomenon of cold denaturation, which is also driven by weakening of the hydrophobic effect with decreasing temperature (33, 34). The results highlight and reinforce a common feature of fold switching, which is that the environmental trigger necessarily induces a decrease in the stability of one state relative to the other.
The results obtained from ZZ-exchange data suggest that the pathways of the forward and reverse reactions are significantly different energetically. For the 3α→α/β conversion, the Ea of 22.0 kcal/mol is comparable to that required for the global unfolding and refolding of similar sized proteins (ubiquitin, lymphotactin), (35, 36) which has previously been posited as a mechanism for fold switching of lymphotactin. In contrast, the α/β→3α transition has a significantly lower Ea value of 2.4 kcal/mol. Although the reasons for these differences are not well understood, one contributing factor may come from the possible energetic similarities of this transition to cold denaturating events, as mentioned above. The α/β→3α transition requires the well-ordered α/β-plait structure to convert to a 3α fold with N- and C-terminal disordered ends (Fig. 2A), thus having the net effect of unfolding for approximately 40% of the polypeptide chain as the temperature is lowered. While heat denaturation is endothermic, cold unfolding is an exothermic process (37). The putative heat release from this partial unfolding transition may therefore offset the energy of activation requirements for α/β→3α conversion, leading to a lower value.
The temperature dependence of the 3α/αβ switch is exquisitely sensitive to mutation (Fig. 5). The Sa1 parent protein, which has an α/β-plait topology, does not show any evidence of an alternative fold at 25 °C. Yet a single relatively subtle amino acid substitution of a residue at the periphery of the core, V90T, destabilizes the α/β-plait fold by 1.4 kcal/mol relative to the unfolded state (Fig. 3B) and is enough to reveal the temperature-dependent fold switch. Moreover, other single-site mutations at either V88 or V90 shift the Tmid from less than 5 °C to 25 °C by altering the stability of the α/β-plait, while likely having minimal impact on the 3α state stability. The fact that nearly all of these V88X or V90X mutations, with the exception of X = Thr, are aliphatic substitutions (X = I, L, V, M, or A) on the outside of the core demonstrates the delicate balance of the system. With coding for both the 3α and α/β-plait folds present in a single polypeptide chain, small changes in amino acid composition can alter the relative stabilities of the two competing states, leading to changes in the temperature-dependent profiles.
In almost all of the mutants investigated, the temperature dependence is completely reversible, indicating that neither fold is kinetically trapped. The exception is Sa1V88M, where the protein is stable at lower temperatures (1 to 20 °C) in the 3α topology, but begins to precipitate and presumably aggregate at higher temperatures (25 to 35 °C) once it switches to mostly α/β-plait. The structure of Sa1V90T indicates that V88 is more buried than T90. In models, branched aliphatic side chains can be accommodated at both sites (Fig. 5), whereas the unbranched methionine introduces more steric clashes at position 88 than 90. This may destabilize the α/β-plait form of Sa1V88M and increase the propensity to form aggregated states. This characteristic is reminiscent of a prion, where fold switching from a benign α-helical state to a β-stranded form is presumed to precede oligomerization to fibrils, protein deposition, and disease states (38). Here, however, the biophysical properties of fold switching between the monomeric 3α and α/β-plait forms of the Sa1V90T mutant can be analyzed, because switching is decoupled from complicating oligomerization events. The present results in a designed system therefore provide a useful framework for understanding the structural, energetic, and kinetic factors controlling fold switching. Moreover, they indicate that common topologies can undergo fold switching without the need to progress to multimeric states.
In Sa1V90T, the N- and C-termini are mostly disordered in the 3α state (Fig. 4). Nonetheless, they exhibit some propensity for the secondary structured regions present in the alternative state (SI Appendix, Fig. S4), providing nucleating sites for folding to the α/β-plait. The conformational propensities of these regions in the 3α state do not increase as the temperature is raised and as the population of the α/β-plait form increases, consistent with switching from the 3α to α/β-plait structures being a highly cooperative process. Involvement of disordered regions, which are also observed in a number of natural fold switches, permits flexibility of phi/psi-angles in at least one of the two states (3). This relaxes the relatively tight structural constraint of populating two different folds with a single polypeptide chain. In addition, these types of large-scale intramolecular topological changes have some analogy at the intermolecular level. For example, addition of an unstructured peptide in trans to the globular folded protein superoxide dismutase promotes switching from an immunoglobulin-like β-sandwich to an alternative structure with amyloid characteristics (39). Taken together, these observations indicate that disordered regions can play an important role in the remodeling of ordered states.
In summary, our results show that temperature alone can be used to switch from one common fold topology to another and that subtle changes in amino acid composition modulate the relative stabilities of the two competing states over a reasonably narrow temperature range (5 to 37 °C) that is relevant to biology. Moreover, the HSA-binding function can be tightly regulated over this temperature range by shifting the equilibrium between the 3α state, which binds HSA, and the α/β-plait form, which does not bind HSA (Fig. 8). Such studies therefore provide insights into how new folds and functions could potentially evolve. They also add to the new strategies being developed for the design of proteins with novel properties (40, 41). Since the parameters of temperature and single-site mutation can be simulated in silico, the GA/S6 system may serve as a useful model for further computational studies of temperature-dependent protein stability and fold switching. Underlying these efforts, the physicochemical principles determined from designed protein fold switches will also be applicable to natural shape shifters.
Materials and Methods
Sample Preparation.
Site-directed mutants were prepared using mutagenic primers and a Q5 site-directed mutagenesis kit (BioLabs). Sa1 and variants were cloned into an eXact tag pH0720 vector, including an engineered subtilisin prodomain and an HSA-binding GA sequence as N-terminal tags that were removed during purification (42). BL21DE3 E. coli cells were transformed with the vector and grown in M9 minimal media for 15N- and 13C/15N-labeling. Cells were grown at 37 °C to a density of 0.6 to 0.8 OD600, and protein expression was induced with 1 mM IPTG at 25 °C for 18 h. The cells were harvested by centrifugation, resuspended in 100 mM potassium phosphate (KPi) buffer (pH 7.0) containing 150 mM NaCl and a protease inhibitor cocktail tablet (Pierce), and lysed by sonication. Soluble cell extract of prodomain-GA fusion protein was loaded onto a 5 mL subtilisin column. The column was washed with five column volumes of 100 mM KPi (pH 7.0), then 20 column volumes of 500 mM NaCl, 100 mM KPi (pH 7.0) to remove impurities, followed by five column volumes of 100 mM KPi. To cleave and elute the purified protein, 6 mL of 3.5 mM imidazole, 100 mM KPi (pH 7.0) was injected at 1 mL/min. The eluent was passed through an HSA column to remove uncleaved fusion protein and the cleaved target protein was collected in the flow through. The purified protein was concentrated to 0.2 to 0.3 mM in 100 mM KPi buffer (pH 7.0) containing 5% D2O for NMR analysis. The oligomeric state of the Sa1V90T protein was assessed by static light scattering using analytical size-exclusion high-performance liquid chromatography in combination with a Minidawn Treos (Wyatt Technologies) multiangle light scattering instrument.
Circular Dichroism.
CD measurements were made on a Chirascan spectrometer (Applied Photophysics) using protein concentrations of 5 μM in 100 mM KPi buffer (pH 7.0) with a 1-mm path length cuvette. For thermal denaturation, ellipticities at 222 nm were continuously monitored at a scanning rate of 1 °C per min from 5 to 90 °C. The temperature unfolding profiles were converted to an apparent ∆Gfolding and fit to a theoretical curve calculated using the Gibbs–Helmholtz equation: ∆Gfolding = ∆Ho − T∆So + ∆Cp(T − To − TlnT/To), where To = 298 °K. (15, 43)
NMR Spectroscopy.
NMR spectra were collected on Bruker AVANCE III 600 and 900 MHz spectrometers fitted with a z-axis gradient 1H/13C/15N triple-resonance cryoprobe. For all multidimensional NMR spectra, NMRPipe was used for data processing, and analysis was done with Sparky (44, 45). Backbone resonances were assigned at 5 °C and 30 °C for Sa1V90T using the following triple-resonance experiments: HNCACB, CBCA(CO)NH, HNCA, HNCO, HN(CA)CO, HNHA, and HN(CA)NNH. Interproton distance restraints were obtained from 3D 15N- and 13C-edited NOESY spectra in 5% D2O with mixing times of 100 and 150 ms, respectively. The 30 °C three-dimensional structure of Sa1V90T was calculated using a standard CS-Rosetta3.2 protocol based on backbone N, HN, Hα Cα, Cβ, and CO chemical shifts as well as backbone NOE data (46). The 5 °C structure of Sa1V90T was also calculated using CS-Rosetta3.2 based on backbone N, HN, Hα Cα, Cβ, and CO chemical shifts, where TALOS (47)-predicted flexible N- and C-terminal regions (residues 2 to 14, 64 to 73, and 82 to 95) were predefined as flexible. One thousand CS-Rosetta structures were calculated, from which the 10 lowest energy structures were chosen. Protein structures were displayed and analyzed using PROCHECK-NMR (48), MOLMOL (49), and PyMol (Schrodinger).
Secondary Cα and CO chemical shifts were determined by subtracting calculated random coil values from experimental values (50). Steady-state {1H}-15N heteronuclear NOE experiments were recorded at 5 °C and 30 °C on a 600-MHz spectrometer with a relaxation delay of 3.6 s. 15N-T1 and -T2 measurements were made using 2D 15N HSQC experiments at 5, 17, and 30 °C on a 600-MHz spectrometer with a relaxation delay of 3 and 2 s, respectively. For T1 experiments at 5 and 17 °C, 12 variable delays were used (10, 50, 100, 200, 300, 500, 700, 1,000, 1,200, 1,500, 2,000, and 2,500 ms), whereas 10 variable delays were employed at 30 °C (10, 50, 100, 200, 300, 500, 700, 1,000, 1,200, and 1,500 ms). For T2 experiments at 5 and 17 °C, 11 variable delays were used (8, 16, 30, 50, 80, 100, 130, 160, 200, 300, and 500 ms), while 11 variable delays were utilized at 30 °C (8, 16, 30, 50, 80, 100, 130, 160, 200, 240, and 300 ms). T1 and T2 values were calculated from fitting an exponential decay with error estimates using Sparky. The rotational correlation time, τC, was calculated using Eq. 2,
| [2] |
where is the 15N resonance frequency (51).
To determine fold interconversion rate constants, 2D ZZ-exchange 15N HSQC experiments were recorded at 10, 17, and 24 °C on a 900-MHz spectrometer (28, 29). A range of variable delay times was used at 10 °C (100, 200, 300, 400, 600, 800, 1,000, 1,300, 1,500 ms), 17 °C (25, 50, 100, 150, 200, 300, 400, 500, 600, 800, 1,000 ms), and 24 °C (20, 50, 100, 150, 250, 300, 400, 500 ms). Changes of auto- and cross-peak intensities as a function of mixing time were analyzed using the composite peak intensity ratio method, shown in Eq. 3,
| [3] |
where IAA(t) and IBB(t) are intensities of autopeaks and IAB(t) and IBA(t) are intensities of crosspeaks, and the data were fitted using a linear least squares fit (52). Exchange rate constants were calculated from the quadratic fit function, Ξ(t), obtained from each residue. The equilibrium constant, Keq, was determined from a fully relaxed 15N HSQC spectrum. The exchange rates at each temperature were then used to calculate the activation energy, Ea, by fitting to the Arrhenius equation
| [4] |
where A is the preexponential factor and R is the gas constant.
Dissociation constants were determined as described previously (14). Purified 15N-labeled A1(HSA) and Sa1(HSA) protein samples were concentrated to 50 μM in 100 mM KPi (pH 7.0). Lyophilized HSA (Sigma Aldrich) was solubilized to 50 mg/mL in 100 mM KPi (pH 7.0). Binding experiments were performed by titrating 0.1 to 4.0 molar equivalents of HSA into a 50-μM solution of A1(HSA) or Sa1(HSA) and monitoring changes by NMR.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by NIH Grant GM62154 (to P.N.B. and J.O.). The NMR facility is supported by the University of Maryland, the National Institute of Standards and Technology, and a grant from the W. M. Keck Foundation. We would also like to thank Dr. Dorothy Beckett for useful discussions. Mention of commercial products does not imply recommendation or endorsement by NIST.
Author contributions
T.L.S., P.N.B., and J.O. designed research; T.L.S., Y.H., N.S., Y.C., and D.T.G. performed research; T.L.S., Y.H., P.N.B., and J.O. analyzed data; and T.L.S., P.N.B., and J.O. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Philip N. Bryan, Email: pbryan@potomac-affinity-proteins.com.
John Orban, Email: jorban@umd.edu.
Data, Materials, and Software Availability
[structure coordinates; NMR assignments] data have been deposited in [PDB/PDBDev; BMRB](8E6Y/00000132; 51338, 51339) (53–56). All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Uversky V. N., Intrinsic disorder in proteins associated with neurodegenerative diseases. Front. Biosci. 14, 5188–238 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Bryan P. N., Orban J., Proteins that switch folds. Curr. Opin. Struct. Biol. 20, 482–488 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kulkarni P., et al. , Structural metamorphism and polymorphism in proteins on the brink of thermodynamic stability. Protein Sci. 27, 1557–1567 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murzin A. G., Biochemistry - Metamorphic proteins. Science 320, 1725–1726 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Porter L. L., Looger L. L., Extant fold-switching proteins are widespread. Proc. Natl. Acad. Sci. U.S.A. 115, 5968–5973 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambroggio X. I., Kuhlman B., Computational design of a single amino acid sequence that can switch between two distinct protein folds. J. Am. Chem. Soc. 128, 1154–1161 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Ambroggio X. I., Kuhlman B., Design of protein conformational switches. Curr. Opin. Struct. Biol. 16, 525–530 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Dishman A. F., et al. , Evolution of fold switching in a metamorphic protein. Science 371, 86–90 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian P., Louis J. M., Baber J. L., Aniana A., Best R. B., Co-Evolutionary fitness landscapes for sequence design. Angew. Chem. Int. Ed. Engl. 57, 5674–5678 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cordes M. H., Burton R. E., Walsh N. P., McKnight C. J., Sauer R. T., An evolutionary bridge to a new protein fold. Nat. Struct. Biol. 7, 1129–1132 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Alexander P. A., He Y., Chen Y., Orban J., Bryan P. N., The design and characterization of two proteins with 88% sequence identity but different structure and function. Proc. Natl. Acad. Sci. U.S.A. 104, 11963–11968 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porter L. L., He Y., Chen Y., Orban J., Bryan P. N., Subdomain interactions foster the design of two protein pairs with ∼80% sequence identity but different folds. Biophys. J. 108, 154–162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexander P. A., He Y., Chen Y., Orban J., Bryan P. N., A minimal sequence code for switching protein structure and function. Proc. Natl. Acad. Sci. U.S.A. 106, 21149–21154 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Y., Chen Y., Alexander P. A., Bryan P. N., Orban J., Mutational tipping points for switching protein folds and functions. Structure 20, 283–291 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y., Rules for designing protein fold switches and their implications for the folding code. bioRxiv [Preprint] (2021). 10.1101/2021.05.18.444643 (Accessed 18 May 2021). [DOI]
- 16.Cao B., Elber R., Computational exploration of the network of sequence flow between protein structures. Proteins 78, 985–1003 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bullough P. A., Hughson F. M., Skehel J. J., Wiley D. C., Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371, 37 (1994). [DOI] [PubMed] [Google Scholar]
- 18.Luo X., et al. , The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat. Struct. Mol. Biol. 11, 338–345 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Tseng R., et al. , Structural basis of the day-night transition in a bacterial circadian clock. Science 355, 1174–1180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Littler D. R., et al. , The intracellular chloride ion channel protein CLIC1 undergoes a redox-controlled structural transition. J. Biol. Chem. 279, 9298–9305 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Gettins P. G. W., Serpin structure, mechanism, and function. Chem. Rev. 102, 4751–4803 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Kuloğlu E. S., McCaslin D. R., Markley J. L., Volkman B. F., Structural rearrangement of human lymphotactin, a C chemokine, under physiological solution conditions. J. Biol. Chem. 277, 17863–17870 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuinstra R. L., et al. , Interconversion between two unrelated protein folds in the lymphotactin native state. Proc. Natl. Acad. Sci. U.S.A. 105, 5057–5062 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Day R., Beck D. A., Armen R. S., Daggett V., A consensus view of fold space: Combining SCOP, CATH, and the dali domain dictionary. Protein Sci. 12, 2150–2160 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindahl M., et al. , Crystal structure of the ribosomal protein S6 from Thermus thermophiius. EMBO. J. 13, 1249–1254 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haglund E., et al. , The HD-exchange motions of ribosomal protein S6 are insensitive to reversal of the protein-folding pathway. Proc. Natl. Acad. Sci. U.S.A. 106, 21619–21624 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Y., et al. , Structure, dynamics, and stability variation in bacterial albumin binding modules: Implications for species specificity. Biochemistry 45, 10102–10109 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Farrow N. A., Zhang O., Forman-Kay J. D., Kay L. E., A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J. Biomol. NMR 4, 727–34 (1994). [DOI] [PubMed] [Google Scholar]
- 29.Montelione G. T., Wagner G., 2D chemical exchange NMR spectroscopy by proton-detected heteronuclear correlation. J. Am. Chem. Soc. 111, 3096–3098 (1989). [Google Scholar]
- 30.Lejon S., Frick I.-M., Bjorck L., Wikstrom M., Svensson S., Crystal structure and biological implications of a bacterial albumin binding module in complex with human serum albumin. J. Biol. Chem. 279, 42924–42928 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Baldwin R. L., Temperature dependence of the hydrophobic interaction in protein folding. Proc. Natl. Acad. Sci. U.S.A. 83, 8069–8072 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Dijk E., Hoogeveen A., Abeln S., The hydrophobic temperature dependence of amino acids directly calculated from protein structures. PLoS Comput. Biol. 11, e1004277 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaremko M., et al. , Cold denaturation of a protein dimer monitored at atomic resolution. Nat. Chem. Biol. 9, 264–270 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dias C. L., Ala-Nissila T., Karttunen M., Vattulainen I., Grant M., Microscopic mechanism for cold denaturation. Phys. Rev. Lett. 100, 118101 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Charlier C., et al. , Study of protein folding under native conditions by rapidly switching the hydrostatic pressure inside an NMR sample cell. Proc. Natl. Acad. Sci. U.S.A. 115, E4169–E4178 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tyler R. C., Murray N. J., Peterson F. C., Volkman B. F., Native-state interconversion of a metamorphic protein requires global unfolding. Biochemistry 50, 7077–7079 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim S. B., Palmer J. C., Debenedetti P. G., Computational investigation of cold denaturation in the Trp-cage miniprotein. Proc. Natl. Acad. Sci. U.S.A. 113, 8991–8996 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prusiner S. B., Prions. Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ivanova M. I., et al. , Aggregation-triggering segments of SOD1 fibril formation support a common pathway for familial and sporadic ALS. Proc. Natl. Acad. Sci. U.S.A. 111, 197–201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davey J. A., Damry A. M., Goto N. K., Chica R. A., Rational design of proteins that exchange on functional timescales. Nat. Chem. Biol. 13, 1280–1285 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Dishman A. F., Volkman B. F., Design and discovery of metamorphic proteins. Curr. Opin. Struct. Biol. 74, 102380 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruan B., Fisher K. E., Alexander P. A., Doroshko V., Bryan P. N., Engineering subtilisin into a fluoride-triggered processing protease useful for one-step protein purification. Biochemistry 43, 14539–14546 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Becktel W. J., Schellman J. A., Protein stability curves. Biopolymers 26, 1859–1877 (1987). [DOI] [PubMed] [Google Scholar]
- 44.Delaglio F., et al. , NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 45.Lee W., Tonelli M., Markley J. L., NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen Y., et al. , Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl. Acad. Sci. U.S.A. 105, 4685–4690 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen Y., Bax A., Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 56, 227–241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M., AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 (1996). [DOI] [PubMed] [Google Scholar]
- 49.Koradi R., Billeter M., Wuthrich K., MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 29–32 (1996). [DOI] [PubMed] [Google Scholar]
- 50.Kjaergaard M., Poulsen F. M., Sequence correction of random coil chemical shifts: Correlation between neighbor correction factors and changes in the Ramachandran distribution. J. Biomol. NMR 50, 157–165 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Kay L. E., Torchia D. A., Bax A., Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry 28, 8972–8979 (1989). [DOI] [PubMed] [Google Scholar]
- 52.Miloushev V. Z., et al. , Dynamic properties of a type II cadherin adhesive domain: Implications for the mechanism of strand-swapping of classical cadherins. Structure 16, 1195–1205 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Solomon T. L., Orban J., NMR structure of Sa1_V90T at 30 degrees Celsius. PDB. 10.2210/pdb8E6Y/pdb. Accessed 23 August 2022. [DOI]
- 54.Solomon T. L., Orban J., NMR structure of Sa1_V90T at 5 degrees Celsius. PDBDev. https://pdb-dev.wwpdb.org/entry.html?PDBDEV_00000132. Accessed 30 June 2022.
- 55.Solomon T. L., He Y., Orban J., Chemical shift assignment of Sa1_V90T at 5 degrees Celsius. BMRB. https://bmrb.io/data_library/summary/index.php?bmrbId=51338. Accessed 22 February 2022.
- 56.Solomon T. L., He Y., Orban J., Chemical shift assignment of Sa1_V90T at 30 degrees Celsius. BMRB. https://bmrb.io/data_library/summary/index.php?bmrbId=51339. Accessed 22 February 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
[structure coordinates; NMR assignments] data have been deposited in [PDB/PDBDev; BMRB](8E6Y/00000132; 51338, 51339) (53–56). All study data are included in the article and/or SI Appendix.