Significance
Mutations in epidermal growth factor receptor (EGFR) occur in 15 to 20% of lung cancers. Patients with EGFR-mutant lung cancer are typically treated with anticancer drugs called EGFR inhibitors (EGFRi), but therapy often fails due to acquired drug resistance. Here, we show that loss of the epigenetic repressor CBX5 confers EGFRi resistance through a mechanism that involves upregulation of the transcription factor E2F1 and its target, the antiapoptotic protein BIRC5 (survivin). We demonstrate that pharmacological inhibition of this CBX5-E2F1-BIRC5 axis, through either restoration of CBX5 expression or inhibition of BIRC5, represents a therapeutic approach for treating EGFRi-resistant lung cancer. Our results provide potential treatment opportunities for EGFR-mutant lung cancer patients who have failed EGFRi therapy due to the emergence of acquired resistance.
Keywords: drug resistance, transcription regulation, epigenetics, lung cancer, EGFR
Abstract
Although epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (EGFRi) are approved for treating EGFR-mutant lung adenocarcinoma (LUAD), emergence of acquired resistance limits their clinical benefits. Several mechanisms for acquired resistance to EGFRi in LUAD have been identified; however, the molecular basis for this resistance remains unknown in ~30% of LUAD. Chromatin and DNA modifiers and their regulators play important roles in determining response to anticancer therapies. Therefore, to identify nongenetic mechanisms of EGFRi resistance in LUAD, we performed an epigenome-wide shRNA screen targeting 363 human epigenetic regulator genes. This screen identified loss of the transcriptional repressor chromobox homolog 5 (CBX5) as a driver of EGFRi resistance in EGFR-mutant LUAD. Loss of CBX5 confers resistance to multiple EGFRi in both cell culture and mice. We found that CBX5 loss in EGFR-mutant LUAD cells leads to increased expression of the transcription factor E2F1, which in turn stimulates expression of the antiapoptotic gene BIRC5 (survivin). This E2F1-mediated upregulation of BIRC5 in CBX5-knockdown LUAD cells attenuates apoptosis induction following EGFRi treatment. Consistent with these results, knockdown of E2F1 or BIRC5 partly rescues CBX5-knockdown-induced EGFRi resistance in cell culture and mice. EGFRi-resistant LUAD cell lines show reduced CBX5 expression compared to parental lines; however, bromo- and extra-terminal (BET)-domain inhibitors (BETi) restore CBX5 expression in these cells and sensitize them to EGFRi/BETi combination therapy. Similarly, treatment with a BIRC5 inhibitor suppresses growth of EGFRi-resistant LUAD cells. Collectively, these studies identify CBX5 loss as a driver of EGFRi resistance and reveal therapeutic opportunities for treating EGFRi-resistant LUAD.
Lung cancer is one of the most commonly diagnosed cancers and the leading cause of death due to cancer in both men and women (1–3). Genome-scale studies have discovered several actionable oncogenic mutational subtypes of lung cancer. Among these are oncogenic mutations in the gene epidermal growth factor receptor (EGFR), which are found in approximately 15 to 20% of lung adenocarcinomas (LUADs), the most common form of non–small cell lung cancer (4–6). Oncogenic EGFR mutations result in its constitutive activation, producing cancer cells that are dependent on EGFR signaling for survival, as observed by the suppression of EGFR-mutant LUAD following inhibition of EGFR signaling (7, 8). These observations have led to the development of several highly efficacious EGFR tyrosine kinase inhibitors (EGFRi) for treating EGFR-mutant LUAD, which are currently approved for clinical use (9–11).
Initial overall response rates and disease control rates for EGFRi in patients with EGFR-mutant LUAD are generally high (12, 13). However, a significant proportion of individuals that show strong early responses to EGFRi ultimately acquire resistance to these therapies (14, 15). Multiple mechanisms for acquired resistance to EGFRi in EGFR-mutant LUAD have been identified (16–18). However, the molecular basis for acquired resistance to EGFRi remains unknown in approximately 30% of LUAD cases (19).
Epigenetic regulators, such as chromatin and DNA regulators and modifiers, have been shown to play important roles in resistance to targeted therapeutic agents, including in the context of EGFRi (20). Furthermore, many of these epigenetic regulators can be effectively targeted using small-molecule inhibitors (20). Thus, epigenetic drivers of EGFRi resistance represent promising therapeutic targets for treating drug-resistant tumors.
In this study, to comprehensively identify epigenetic drivers of EGFRi resistance in LUAD, we conducted an unbiased large-scale short-hairpin (sh)RNA screen, targeting 363 human epigenetic regulator genes. Using this strategy, we uncovered a role for chromobox homolog 5 (CBX5), also known as heterochromatin protein 1alpha, in EGFRi resistance. CBX5 is a methyl-lysine–binding protein that localizes at heterochromatin sites (21, 22) and has been shown to function in gene silencing (21, 23). Here, we found that loss of CBX5 confers resistance to multiple different EGFRi compounds. This resistance occurs, in part, due to upregulation of the transcription factor E2 promoter binding factor 1 (E2F1), leading to transcriptional activation of the antiapoptotic gene baculoviral inhibitorof apoptosis (IAP) repeat–containing 5 (BIRC5; also known as survivin) and inhibition of cancer cell apoptosis following EGFRi treatment. Consistent with these observations, we find that CBX5 is down-regulated, whereas E2F1 and BIRC5 are up-regulated in EGFRi-resistant LUAD cell lines compared to EGFRi-sensitive parental cells. Notably, however, bromo- and extra-terminal (BET)-domain inhibitors (BETi) can restore CBX5 expression in EGFRi-resistant cell lines, sensitizing cells to EGFRi/BETi combination therapy and blocking EGFRi-resistant LUAD tumor growth. Similarly, a small-molecule BIRC5 inhibitor induces apoptosis and suppresses growth of EGFRi-resistant LUAD tumor cells in vitro and in vivo. Collectively, these findings demonstrate that loss of CBX5 confers EGFRi resistance and uncover therapeutic opportunities for treating EGFRi-resistant LUAD.
Results
A Large-Scale Epigenome-Wide Human shRNA Screen Identifies CBX5 as a Modifier of EGFRi Response.
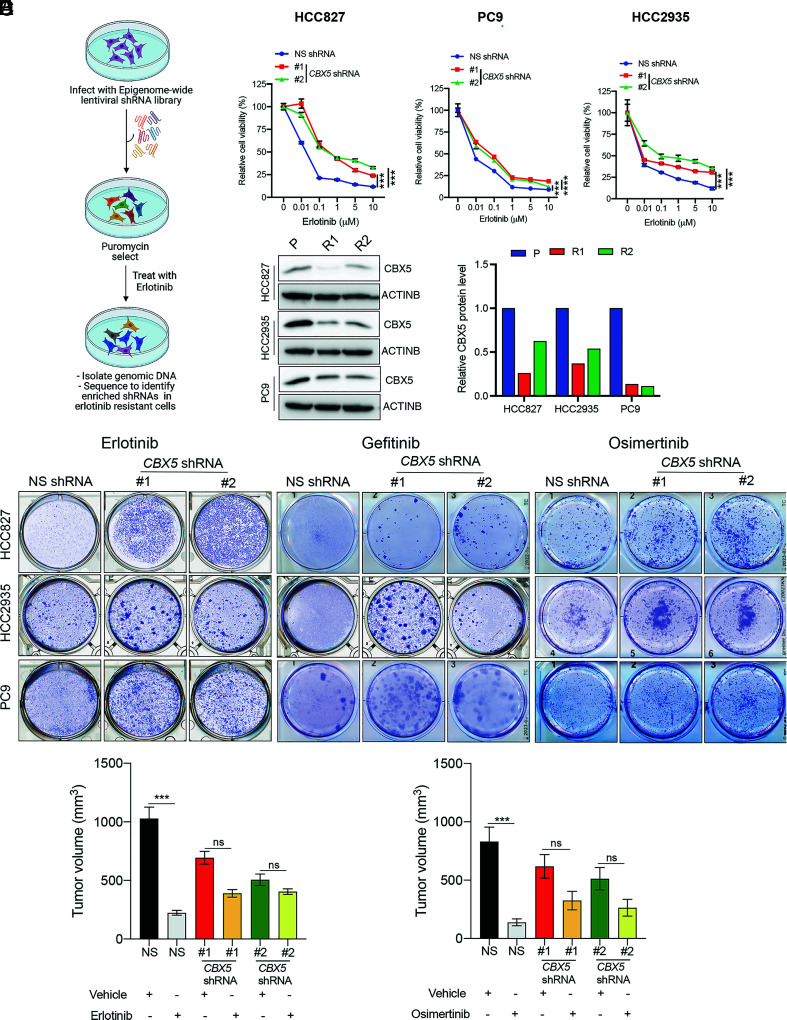
The role of epigenetic regulators in conferring resistance to EGFRi therapies is not fully known, and only a few studies thus far have identified epigenetic mechanisms that drive EGFRi resistance (24–26). To comprehensively identify additional epigenetic regulators that confer resistance to EGFRi, we performed a large-scale unbiased epigenome-wide shRNA screen. To this end, we assembled a library of 1,862 shRNAs targeting 363 known and predicted epigenetic regulators (Dataset S1). We then infected EGFR-mutant, EGFRi-sensitive HCC827 LUAD cells with lentiviral particles containing the shRNA library at 0.2 multiplicity of infection (MOI) to ensure that each cell received no more than one shRNA and selected with puromycin to enrich for shRNA-containing cells. After selection, cells were treated for 4 wk with the EGFRi erlotinib, which binds competitively and reversibly to the ATP-binding site of the EGFR kinase domain (27), and shRNA-expressing colonies that survived treatment were harvested and subjected to genomic DNA sequencing to identify integrated shRNAs (Fig. 1A). From this analysis, we identified shRNAs corresponding to six different epigenetic regulators (SI Appendix, Table S1).
Fig. 1.
A large-scale epigenome-wide human shRNA screen identifies CBX5 as a modifier of EGFRi response. (A) Schematic overview of the large-scale epigenome-wide shRNA screen used to identify epigenetic regulators involved in erlotinib resistance. (B) MTT assay monitoring relative cell viability (%) of HCC827, PC9, and HCC2935 cells expressing an NSshRNA or one of two CBX5 shRNAs and treated for 5 d with the indicated concentrations of erlotinib or dimethyl sulfoxide (DMSO) control (0). (C, Left) Immunoblot analysis measuring CBX5 expression in the indicated EGFRi-sensitive (P) and EGFRi-resistant (R1 or R2) EGFR-mutant LUAD cell lines. ACTINB was used as the loading control. (C, Right) Bar graph showing quantification of the immunoblots. (D) The indicated LUAD cell lines expressing an NS shRNA or CBX5 shRNAs were treated with erlotinib (100 nM), gefitinib (100 nM), osimertinib (20 nM for HCC827; 20 nM for PC9; 100 nM for HCC2935), or DMSO control, for 2 wk, and survival was measured in clonogenic assays. Representative wells for cells grown under the indication conditions are shown. (E) NSG mice (n = 5) were subcutaneously injected with 5 × 106 PC9 cells expressing an NS or CBX5 shRNA and treated with either vehicle (0.5% methylcellulose) or erlotinib (25 mg/kg) 3 d per week until the end of the experiment. Average tumor volumes at the end of the experiment are shown. (F) NSG mice (n = 5) were subcutaneously injected with PC9 cells expressing an NS shRNA or CBX5 shRNA and treated with either vehicle (0.5% carboxy-propyl cellulose and 0.1% Tween 80) or osimertinib (2.5 mg/kg) 3 d per week until the end of the experiment. Average tumor volumes at the end of the experiment are shown. Data are presented as the mean ± SEM. ns = not significant; ***P < 0.001, and ****P < 0.0001.
To confirm these candidates in secondary validation experiments, we knocked down expression of each gene individually in HCC827 cells, using two independent, knockdown-validated shRNAs (SI Appendix, Fig. S1 A and B and Materials and Methods). Cells expressing either the gene-specific shRNAs or a nonspecific (NS) control shRNA were then treated with erlotinib, and survival was measured. We found that knockdown of all genes except sirtuin 4 (SIRT4) confers resistance to erlotinib in HCC827 cells (Fig. 1B and SI Appendix, Fig. S1C). To further generalize our findings and determine whether these genes broadly act as drivers of erlotinib resistance, we individually knocked down expression of all five candidates in two additional EGFR-mutant, EGFRi-sensitive LUAD cell lines (PC9 and HCC2935) (SI Appendix, Fig. S2A) and measured cell viability in the presence of erlotinib. We found that out of the five genes tested, only knockdown of CBX5 and myeloid/lymphoid or mixed‐lineage leukemia translocated to 6 (MLLT6) confers resistance to erlotinib in PC9 and HCC2935 cells (Fig. 1B and SI Appendix, Fig. S2B).
We then rationalized that if a gene is involved in erlotinib resistance, it might be down-regulated in EGFRi-resistant LUAD cells. To test this hypothesis, we generated erlotinib-resistant cell lines by treating EGFR-mutant LUAD cells (HCC827, PC9, and HCC2935) with erlotinib. As expected, we found that the resulting cells showed significant resistance to erlotinib compared to parental cells (SI Appendix, Fig. S3 A and B). We next measured expression of CBX5 and MLLT6 in all EGFRi-resistant and parental LUAD cell lines and found that, as expected, expression of the CBX5 protein was substantially down-regulated in EGFRi-resistant cells compared to parental cells (Fig. 1C). In contrast, expression of MLLT6 was unchanged in resistant cells (SI Appendix, Fig. S3C). Based on these collective results, we focused on CBX5 for our subsequent studies.
CBX5 Loss Confers Resistance to EGFRi in Long-Term clonogenic Assays and Mouse Models with EGFR-Mutant Human LUAD Xenografts.
Patients in the clinic are treated for an extended period with EGFRi drugs. Therefore, to better mirror this clinical scenario, we determined whether CBX5 loss also confers resistance to erlotinib in long-term clonogenic assays. Consistent with the results from our short-term cell survival assays, we found that shRNA-mediated CBX5 knockdown leads to erlotinib resistance in clonogenic assays (Fig. 1D and SI Appendix, Fig. S4). We then tested whether CBX5 loss also drives resistance to other EGFRi compounds, such as gefitinib and osimertinib. Both erlotinib and gefitinib reversibly bind to the ATP-binding site of the EGFR kinase domain (27). In contrast, osimertinib is a third-generation, covalent, irreversible, and mutant-selective EGFRi that shows superior efficacy to previous-generation drugs, such as erlotinib and gefitinib, and is also effective against LUAD with the EGFR T790M gatekeeper mutation (12, 28, 29). We found that similar to erlotinib, CBX5 knockdown results in resistance to both gefitinib and osimertinib, as observed by a significantly increased number of colonies produced by LUAD cells expressing CBX5 shRNAs, compared to those expressing a negative-control NS shRNA, in clonogenic assays (Fig. 1D and SI Appendix, Fig. S4).
The above results establish a role for CBX5 loss in EGFRi resistance for cells in culture. To determine whether CBX5 loss also confers EGFRi resistance in vivo, we used a mouse xenograft model of human EGFR-mutant LUAD. We injected EGFR-mutant PC9 cells expressing either one of two independent, knockdown-validated CBX5 shRNAs or an NS shRNA subcutaneously into the flanks of NOD scid gamma (NSG) mice. Once tumors became palpable (~100 mm3), mice were treated with erlotinib three times a week, and tumor volumes were measured weekly. Consistent with the results from cell culture experiments, we found that CBX5 shRNA-expressing PC9 tumor Xenografts display significantly higher resistance to erlotinib compared to NS shRNA-expressing tumors (Fig. 1E). We then performed analogous experiments to determine whether CBX5 knockdown in PC9 Xenografts also induces resistance to osimertinib. Consistent with results in erlotinib-treated mice, we found that tumors produced from PC9 cells expressing CBX5 shRNAs also display increased resistance to osimertinib, as compared to those generated by NS shRNA-expressing cells (Fig. 1F). Collectively, these findings demonstrate that CBX5 loss drives resistance to different EGFRi in both cell culture and mice.
Loss of CBX5 Confers Resistance to EGFRi via Up-regulating the Transcription Factor E2F1.
Previous studies have shown that activation of mitogen-activated protein kinase and the phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT) pathways by various mechanisms can promote resistance to EGFRi in LUAD (30, 31). Therefore, we tested whether CBX5 loss induces activation of EGFR or other key downstream antiapoptotic signaling pathways, such as PI3K/AKT and/or MEK/ERK, by measuring the activation of these pathways in LUAD cell lines expressing CBX5 shRNAs or a control NS shRNA. However, we did not detect any consistent or substantial pathway alterations in CBX5-knockdown EGFR-mutant cells (SI Appendix, Fig. S5), indicating that CBX5-loss–driven resistance to EGFRi occurs independently of these oncogenic signaling pathways.
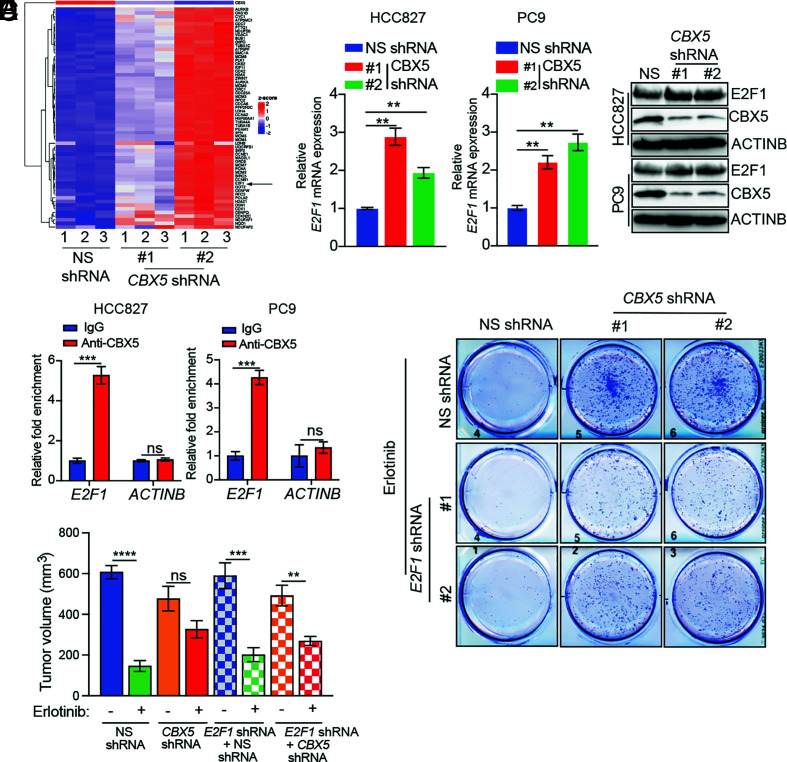
To further explore the mechanism of CBX5-loss–induced EGFRi resistance, and based on the role of CBX5 as a known transcriptional repressor (32), we performed transcriptome-wide RNA-sequencing (RNA-seq) to measure changes in mRNA expression resulting from CBX5 knockdown in HCC827 cells. The results from this analysis revealed a significant increase in expression of the transcription factor E2F1, as well as enrichment of E2F1 as a major upstream regulator of genes altered in response to CBX5 knockdown by Ingenuity Pathway Analysis (Fig. 2A, SI Appendix, Fig. S6, and Datasets S2 and S3). Consistent with these findings, we further found that CBX5 knockdown in EGFR-mutant LUAD cells (HCC827 and PC9) leads to increased expression of E2F1 (Fig. 2 B and C). Next, to determine whether E2F1 is a direct target of CBX5, we measured the association between CBX5 and the E2F1 promoter using a Cleavage Under Targets and Release using Nuclease (CUT&RUN) assay. The results from this assay revealed enrichment of CBX5 at the E2F1 promoter, indicating that CBX5 directly promotes transcriptional repression of E2F1 (Fig. 2D).
Fig. 2.
Upregulation of the transcription factor E2F1 resulting from loss of CBX5 leads to increased EGFRi resistance in vitro and in vivo. (A) Heatmap showing the top 50 genes up-regulated in HCC827 cells expressing CBX5 shRNAs compared to cells expressing an NS shRNA, as measured by RNA-seq. (B) Expression of E2F1 mRNA in the indicated LUAD cell lines expressing a CBX5 shRNAs or NS shRNA, measured by qRT-PCR. Data were normalized to ACTINB, and the results with NS shRNA were set to 1. (C) Immunoblot analysis measuring expression of E2F1 and CBX5 in the indicated LUAD cell lines expressing an NS or CBX5 shRNA. (D) Relative enrichment of CBX5 on the E2F1 and ACTINB promoters, as measured by CUT&RUN. (E) PC9 cells expressing an NS or CBX5 shRNA alone, or in combination with an E2F1 shRNA, were treated with erlotinib (100 nM) or DMSO, and survival was measured in clonogenic assays. Representative wells for cells grown under the indicated conditions are shown. (F) PC9 cells expressing an NS or CBX5 shRNA alone, or in combination with an E2F1 shRNA, were injected subcutaneously into the flanks of NSG mice (n = 5). Mice were treated with either vehicle (0.5% methylcellulose) or erlotinib (25 mg/kg) 3 d per week until the end of the experiment. Average tumor volumes at the end of the experiment are shown. Data are presented as the mean ± SEM. ns = not significant; **P < 0.01, ***P < 0.001, and ****P < 0.0001.
We then determined whether E2F1 upregulation drives resistance to EGFRi following CBX5 knockdown. We first asked whether ectopic expression of E2F1, similar to CBX5 knockdown, confers resistance to EGFRi. We found that ectopic E2F1 expression in EGFR-mutant LUAD cells (HCC827, PC9, and HCC2935) (SI Appendix, Fig. S7A) promotes EGFRi resistance, as evidenced by reduced levels of cleaved caspase 3 (CASP3) and increased colony formation following erlotinib treatment (SI Appendix, Fig. S7 A and B).
We then determined whether E2F1 knockdown could restore EGFRi sensitivity to CBX5-knockdown cells in culture. We simultaneously knocked down expression of E2F1 and CBX5 in LUAD cells and conducted clonogenic assays to assess whether double knockdown resensitizes cells to EGFRi. Our results show that E2F1 knockdown partly rescues CBX5-loss–induced resistance to EGFRi, as observed by a reduced number of colonies produced by E2F1/CBX5-double-knockdown cells relative to CBX5-knockdown cells (Fig. 2E and SI Appendix, Fig. S8 A and B). Based on this finding, we then determined whether simultaneous knockdown of CBX5 and E2F1 could also restore sensitivity to EGFRi in vivo. We injected PC9 cells expressing both CBX5 and E2F1 shRNAs into NSG mice. The results from this experiment showed that simultaneous knockdown of E2F1 and CBX5 partly restored tumor sensitivity to EGFRi (Fig. 2F). Collectively, these results demonstrate that CBX5-loss-induced upregulation of E2F1 mediates, at least in part, the observed increase in EGFRi resistance following CBX5 knockdown.
E2F1 Upregulates Expression of the Antiapoptotic Gene BIRC5 to Promote EGFRi Resistance in CBX5-Depleted LUAD Cells.
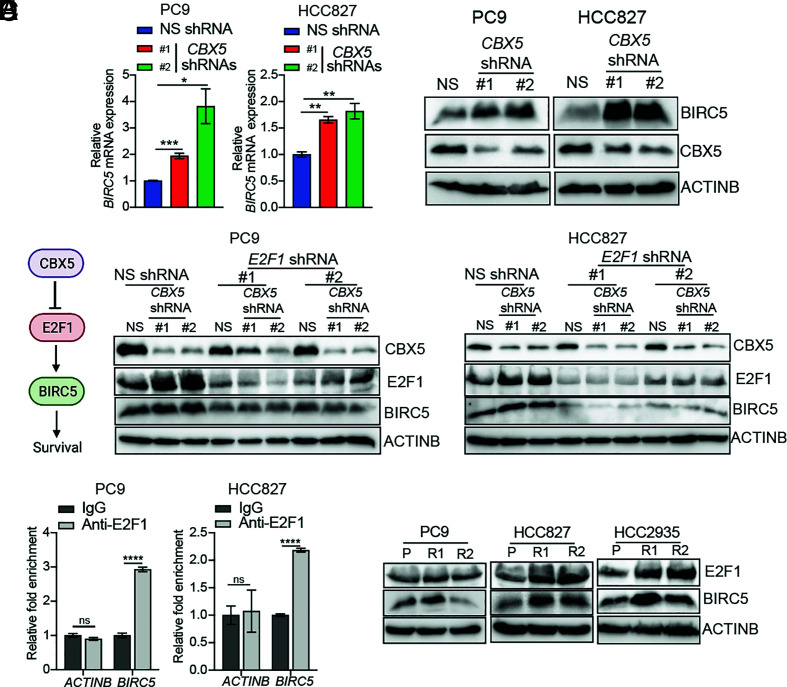
To further explore the mechanism by which E2F1 upregulation resulting from loss of CBX5 promotes EGFRi resistance, we reanalyzed our RNA-seq data, specifically focusing on genes that might be involved in apoptotic or antiapoptotic responses. We found that BIRC5, a well-known antiapoptotic gene (33, 34), is up-regulated in LUAD cells following CBX5 knockdown, as confirmed by quantitative reverse transcription (qRT)-PCR and immunoblot analysis (Fig. 3 A and B). We, therefore, determined whether E2F1 is necessary for BIRC5 upregulation downstream of CBX5 depletion. We measured expression of BIRC5 in EGFR-mutant LUAD cells expressing shRNAs targeting both CBX5 and E2F1. Consistent with a role for E2F1 in up-regulating BIRC5 after CBX5 knockdown, we detected decreased expression of BIRC5 in CBX5 and E2F1 double-knockdown EGFR-mutant LUAD cells, as compared to cells knocked down for CBX5 alone (Fig. 3C). Next, to test whether E2F1 directly regulates BIRC5 expression, we performed a CUT&RUN assay and found that E2F1 directly associates with the BIRC5 promoter (Fig. 3D). We then measured expression of E2F1 and BIRC5 in EGFRi-resistant LUAD cell lines and found that consistent with our above data, expression of both proteins was up-regulated in EGFRi-resistant LUAD cells compared to parental cells (Fig. 3E). Collectively, these results suggest a key role for the CBX5-E2F1-BIRC5 axis in conferring EGFRi resistance in LUAD.
Fig. 3.
CBX5 loss results in E2F1-induced transcriptional activation of the BIRC5 gene. (A) Relative expression of BIRC5 mRNA in the indicated LUAD cell lines expressing an NS or CBX5 shRNA, as measured by qRT-PCR. (B) Immunoblot analysis measuring expression of BIRC5 and CBX5 in the indicated LUAD cell lines expressing an NS or CBX5 shRNA. (C) Immunoblot analysis measuring expression of CBX5, E2F1, and BIRC5 in the indicated LUAD cell lines expressing an NS or CBX5 shRNA alone, or in combination with an NS or E2F1 shRNA. (D) Relative enrichment of E2F1 on the BIRC5 or ACTINB promoters measured by the CUT&RUN assay. (E) Immunoblot analysis measuring expression of E2F1 and BIRC5 in the indicated parental or EGFRi-resistant LUAD cell lines. Data are presented as the mean ± SEM. ns = not significant; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
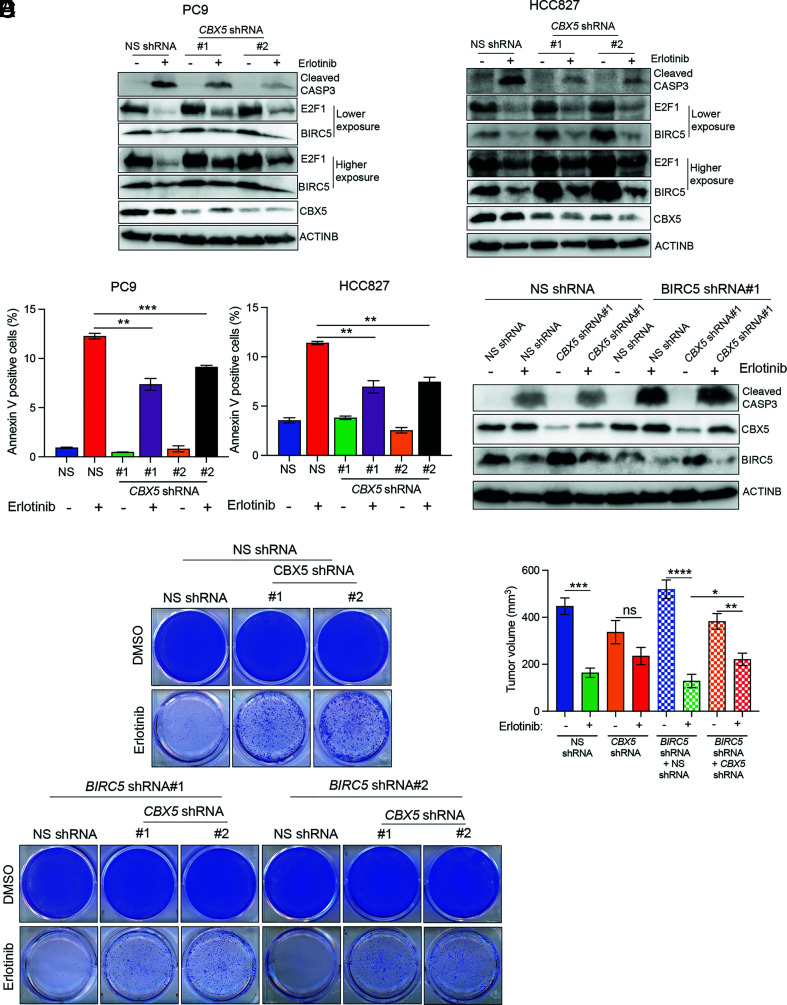
Based on our data showing that loss of CBX5 leads to increased E2F1 expression, which in turn upregulates expression of the antiapoptotic protein BIRC5, we hypothesized that CBX5 depletion promotes EGFRi resistance by conferring protection from apoptosis following EGFRi treatment. To test this possibility, we measured levels of cleaved CASP3 and annexin V in CBX5-knockdown cells following erlotinib treatment. As expected, we detected reduced levels of cleaved CASP3 following erlotinib treatment in CBX5-knockdown EGFR-mutant LUAD cells compared to control cells expressing an NS shRNA (Fig. 4A). Similarly, knockdown of CBX5 reduced the percentage of annexin V-positive cells after erlotinib treatment compared to NS shRNA control cells (Fig. 4B). Thus, our data demonstrate that CBX5 knockdown protects LUAD cells from apoptosis following EGFRi treatment.
Fig. 4.
BIRC5 inhibits EGFRi-induced apoptosis in the context of CBX5 loss in EGFR-mutant LUAD. (A) Immunoblot analysis measuring levels of CASP3 in LUAD cell lines expressing an NS or CBX5 shRNA and treated with erlotinib (50 nM) or DMSO for 24 h. (B) The indicated LUAD cell lines expressing an NS or CBX5 shRNA were treated with erlotinib (50 nM) or DMSO for 24 h and then analyzed by fluorescence-activated cell sorting (FACS)-based annexin V-PE staining. (C) Immunoblot analysis measuring levels of cleaved CASP3 in PC9 cells expressing an NS or CBX5 shRNA alone, or in conjunction with an NS shRNA or BIRC5 shRNA, and treated with erlotinib (50 nM) or DMSO for 24 h. (D) PC9 cells expressing an NS or CBX5 shRNA alone, or in conjunction with an NS or BIRC5 shRNA, were treated with erlotinib (100 nM) or DMSO, and survival was measured in clonogenic assays. Representative wells for cells grown under the indicated conditions are shown. (E) PC9 cells expressing an NS or CBX5 shRNA alone, or in conjunction with an NS or BIRC5 shRNA, were injected subcutaneously into the flanks of NSG mice (n = 5). Mice were treated with either vehicle (0.5% methylcellulose) or erlotinib (25 mg/kg) 3 d per week until the end of the experiment. Average tumor volumes at the end of the experiment are shown. Data are presented as the mean ± SEM. ns = not significant; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Collectively, the above findings indicate that the antiapoptotic gene BIRC5, a downstream target of E2F1 that is up-regulated in CBX5-knockdown cells, may mediate resistance to apoptosis in response to CBX5 loss. To test this possibility, we first determined whether ectopic BIRC5 expression, similar to CBX5 loss, could block apoptosis induction following EGFRi treatment. The results showed that ectopic expression of BIRC5 in EGFR-mutant LUAD cells leads to reduced levels of cleaved CASP3 following erlotinib treatment (SI Appendix, Fig. S9A). Consistent with these data, we further found that ectopic BIRC5 expression promotes EGFRi resistance, as evidenced by increased colony formation following EGFRi treatment (SI Appendix, Fig. S9B).
Next, to test whether BIRC5 upregulation downstream of E2F1 in CBX5-knockdown cells directly leads to reduced induction of apoptosis following EGFRi treatment, we simultaneously knocked down expression of both CBX5 and BIRC5 and measured markers of apoptosis. We found that simultaneous knockdown of CBX5 and BIRC5 results in elevated levels of cleaved CASP3 and increased numbers of annexin V-positive cells following treatment with EGFRi relative to CBX5 knockdown alone (Fig. 4C and SI Appendix, Fig. S9 C–E). These findings are consistent with a direct role for BIRC5 in EGFRi resistance downstream of CBX5.
We then determined whether BIRC5 knockdown could restore EGFRi sensitivity to CBX5-knockdown cells in culture and our mouse xenograft model of LUAD. We first performed clonogenic assays with LUAD cells expressing a CBX5 shRNA or shRNAs targeting both CBX5 and BIRC5. Consistent with our above data suggesting that BIRC5 protects CBX5-knockdown EGFR-mutant cells from EGFRi-induced apoptosis, we found that simultaneous knockdown of CBX5 and BIRC5 resensitizes these cells to erlotinib (Fig. 4D). We then tested the role of BIRC5 in vivo by injecting NSG mice with PC9 cells expressing an NS shRNA, a CBX5 shRNA, or both CBX5 and BIRC5 shRNAs. Animals were treated with erlotinib, and tumor volumes were measured weekly to monitor the response rate of each experimental group. In complete agreement with our cell culture studies, we found that simultaneous knockdown of CBX5 and BIRC5 resensitizes tumors derived from EGFR-mutant LUAD cells to erlotinib (Fig. 4E). Collectively, these results show that E2F1-mediated transcriptional activation of BIRC5 promotes resistance to EGFRi in EGFR-mutant LUAD cells.
BETi Restore CBX5 Levels and EGFRi Sensitivity in EGFRi-Resistant Cells.
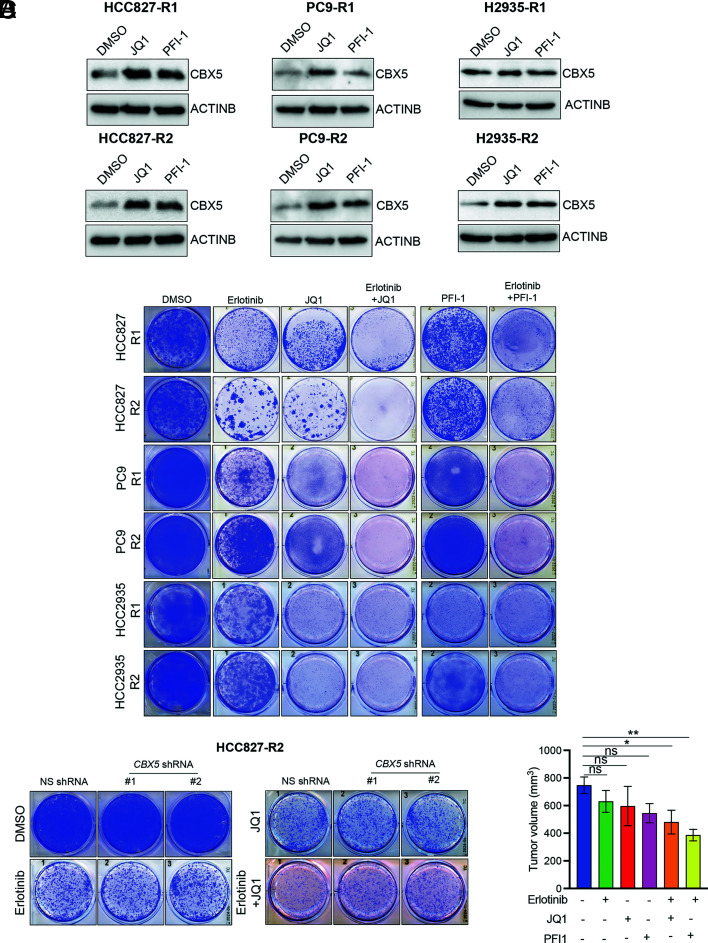
Based on our finding that loss of CBX5 results in EGFRi resistance and the observation that CBX5 levels are significantly reduced in EGFRi-resistant LAUD cells, we next tested whether pharmacological restoration of CBX5 expression could resensitize EGFRi-resistant cells to EGFRi. To identify compounds that could restore CBX5 expression, we treated EGFRi-resistant PC9 cells with a library of 33 small-molecule inhibitors targeting 25 different epigenetic regulators, which is available from the Structural Genome Consortium (SI Appendix, Table S2), and measured CBX5 expression. We found that two BETi, JQ1 and PFI-1, restored CBX5 expression in EGFRi-resistant PC9 cells and other EGFRi-resistant, EGFR-mutant LUAD cell lines (Fig. 5A and SI Appendix, Fig. S10). BET proteins are a family of transcriptional regulators characterized by the presence of two acetyl-lysine (Kac)-binding bromodomains (35–37). The BETi JQ1 and PFI act as Kac mimetics and competitively bind to the Kac-binding pocket of BET proteins, thereby displacing them from chromatin (35–37).
Fig. 5.
BETi upregulate CBX5 expression and sensitize EGFRi-resistant LUAD cell lines to erlotinib. (A) Immunoblot analysis measuring expression of CBX5 in the indicated EGFRi-resistant LUAD cell lines treated with the BETi JQ1 (2 μM) or PFI-1 (2 μM) for 48 h. (B) The indicated EGFRi-resistant LUAD cell lines were treated with erlotinib (100 nm), JQ1 (1 μM), or PFI-1 (1 μM) alone, or with a combination of erlotinib (100 nM) + JQ1 (1 μM) or erlotinib (100 nM) + PFI-1 (1 μM), and survival was measured in clonogenic assays. Representative wells for cells grown under the indicated conditions are shown. (C) HCC827-R2 cells expressing an NS or CBX5 shRNA were treated with DMSO, erlotinib (25 nM), or JQ1 (0.125 μM) alone, or with a combination of erlotinib (25 nM) + JQ1 (0.125 μM), and survival was measured in clonogenic assays. Representative wells for cells grown under the indicated conditions are shown. (D) EGFRi-resistant PC9 cells (PC9-R2) were injected subcutaneously into the flank of NSG mice (n = 5), and mice were treated with vehicle (0.5% methylcellulose/5% dextrose), erlotinib (25 mg/kg), JQ1 (50 mg/kg), or erlotinib (25 mg/kg) + JQ1 (50 mg/kg). Average tumor volumes at the end of the experiment are shown. Data are presented as the mean ± SEM. ns = not significant; *P < 0.05, **P < 0.01.
Because the loss of CBX5 expression is associated with increased resistance to EGFRi, we then determined whether the elevated CBX5 levels observed in BETi-treated EGFRi-resistant cells could restore EGFRi sensitivity. To this end, we treated EGFRi-resistant LUAD cells with either erlotinib alone or erlotinib in combination with BETi (JQ1 or PFI-1) and performed clonogenic assays. The results show that combination treatment with erlotinib and BETi resensitizes EGFRi-resistant cells, whereas no resensitization is observed with EGFRi or BETi alone (Fig. 5B). We then tested whether this effect is mediated by CBX5 by measuring resensitization in CBX5-knockdown EGFRi-resistant cells (SI Appendix, Fig. S11A). Our results show that CBX5 knockdown partially impairs the ability of BETi to resensitize resistant cells to EGFRi (Fig. 5C and SI Appendix, Fig. S11B), indicating a key role for CBX5 in this process. Notably, we further found that simultaneous treatment with BETi and EGFRi forestalls the emergence of EGFRi resistance in LUAD cells (SI Appendix, Fig. S12A).
We then tested the effect of treatment with BETi in combination with EGFRi in mice containing xenograft tumors derived from EGFRi-resistant LUAD cells. We injected mice with EGFRi-resistant PC9 cells and treated the animals with erlotinib, JQ1, or PFI-1 alone, as well as with each BETi in combination with erlotinib. We found that combination treatment with BETi and erlotinib results in significant tumor inhibition, as compared to treatment with erlotinib, JQ1, or PFI-1 alone (Fig. 5D). Thus, these findings indicate that combination treatment with BETi and EGFRi resensitizes EGFRi-resistant cells in both cell culture and in mice.
BIRC5 Inhibitors Show Tumor Suppressive Effects against EGFRi-Resistant LUAD Cells.
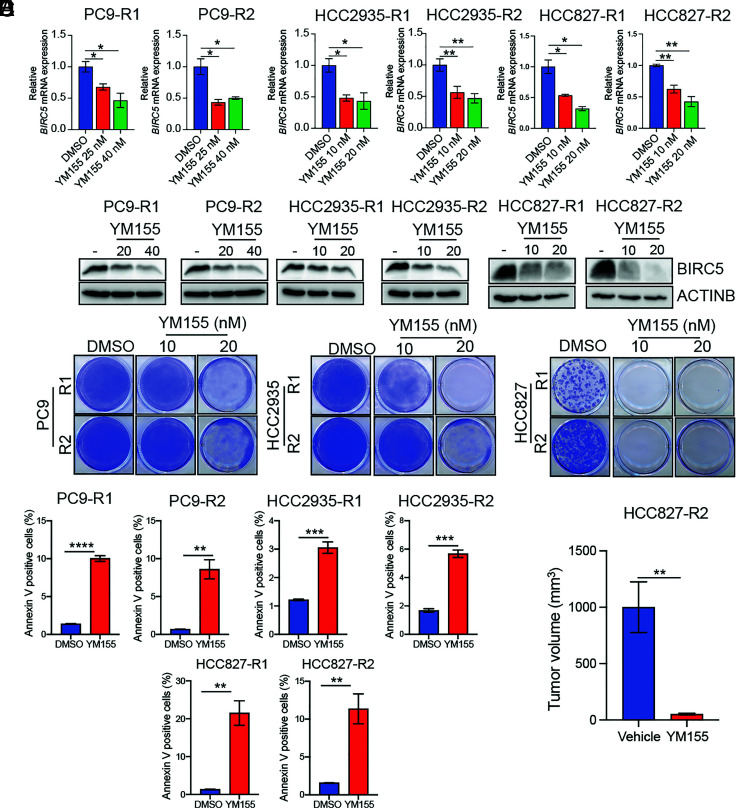
Our above studies have shown that increased BIRC5 expression downstream of CBX5 suppression mediates EGFRi resistance in EGFR-mutant LUAD cells. We, therefore, aimed to test the feasibility of BIRC5 suppression as a therapeutic approach for treating EGFRi-resistant LUAD. For these experiments, we used YM155, a small-molecule inhibitor that functions by transcriptionally repressing BIRC5 (38). As expected, we found that treatment with YM155 represses BIRC5 expression in EGFRi-resistant LUAD cells (Fig. 6 A and B) and blocks their growth in clonogenic assays (Fig. 6C). Furthermore, combination treatment with YM155 and EGFRi forestalls development of drug-resistant LUAD cells (SI Appendix, Fig. S12B), and as expected, treatment with YM155 induces apoptosis of EGFR-mutant LUAD cells (Fig. 6D). Lastly, to determine whether BIRC5 inhibition suppresses EGFRi-resistant LUAD tumor growth in vivo, we injected mice with EGFRi-resistant HCC827 cells and treated tumor-bearing animals with YM155 or vehicle. Consistent with the results from our cell culture experiments, we found that treatment with YM155 potently suppresses growth of EGFRi-resistant LUAD tumors in mice (Fig. 6E). These results further support a key role for the CBX5→E2F1→BIRC5 pathway in conferring resistance to EGFRi in LUAD and identify BIRC5 suppression as a strategy for treating EGFRi-resistant LUAD.
Fig. 6.
The BIRC5 inhibitor YM155 blocks growth of EGFRi-resistant LUAD cells in cell culture and mice. (A) Relative expression of BIRC5 mRNA in the indicated EGFRi-resistant LUAD cell lines treated with DMSO or BIRC5 inhibitor YM155 (10 and 20 nm for HCC827-R1, HCC827-R2, HCC2935-R1, and HCC2935-R2; 20 and 40 nm for PC9-R1 and PC9-R2) for 48 h, as measured by qRT-PCR. (B) Immunoblot analysis measuring expression of BIRC5 in the indicated EGFRi-resistant LUAD cell lines treated with DMSO or YM155 (10 and 20 nm for HCC827-R1, HCC827-R2, HCC2935-R1, and HCC2935-R2; 20 and 40 nm for PC9-R1 and PC9-R2) for 48 h. (C) The indicated EGFRi-resistant LUAD cell lines were treated with DMSO or YM155 (10 and 20 nm), and survival was measured in clonogenic assays. Representative wells for cells grown under the indicated conditions are shown. (D) The indicated LUAD EGFRi-resistant cell lines were treated with YM155 (20 nM for HCC827-R1, HCC827-R2, HCC2935-R1, and HCC2935-R2; 50 nM for PC9-R1 and PC9-R2) or DMSO control for 48 h and then analyzed by FACS-based annexin V-PE staining. (E) EGFRi-resistant HCC827 cells (HCC827-R2) were injected subcutaneously into the flanks of NSG mice (n = 5), and animals were treated with vehicle (0.9% saline) or YM155 (3.5 mg/kg). Average tumor volumes at the end of the experiment are shown. Data are presented as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Discussion
Successful therapeutic targeting of EGFR-mutant LUAD with EGFRi is a major accomplishment of modern-day cancer medicine. However, acquired resistance to EGFRi is frequent and, in most instances, inevitable (15, 21), which poses a significant hurdle to achieving long-term clinical benefits in patients with EGFR-mutant LUAD. One approach to overcoming this limitation involves identifying drivers of EGFRi resistance and utilizing this information to develop improved treatments for EGFRi-resistant tumors. Previous studies have documented the success of such approaches. For example, the third-generation EGFRi osimertinib is effective against LUAD with the EGFR T790M mutation, which is insensitive to other EGFRi compounds, such as erlotinib and gefitinib (11, 12). Combination therapy using drugs that work by different mechanisms may decrease the likelihood that resistant cancer cells will develop, and thus, there is great interest in testing the efficacy of EGFRi combination therapies in preclinical studies and clinical trials (27, 39).
A number of nongenetic mechanisms, including epigenetic alterations, have been shown to promote development of acquired drug resistance (20). In particular, several previous studies have identified important roles for epigenetic regulatory proteins and epigenetic mechanisms in EGFRi resistance (24–26, 40) In this study, we conducted a comprehensive epigenome-wide screen and identified CBX5 as a protein whose loss drives resistance to EGFRi in LUAD. CBX5 is a methyl-lysine–binding protein that localizes at heterochromatin sites and is implicated in both gene silencing and supranucleosomal chromatin structure (21). This protein is targeted to specific loci by the histone lysine methyltransferase SUV39H1, which selectively methylates histone H3 on lysine-9 to generate a binding site for CBX5, leading to gene repression (41). Notably, aberrant CBX5-mediated gene regulation has been implicated in multiple pathological conditions, including lung fibrosis (32) and cancer (42–44). Notably, we observed substantial reduction in CBX5 protein levels in EGFRi-resistant LUAD cells in vitro, suggesting that loss of CBX5 may underlie EGFRi resistance in LUAD patients. In the future, it would be important to determine whether EGFR-mutant LUAD patients treated with EGFRi show similar reduction in CBX5 when they acquire resistance to this therapy.
E2Fs are a large family of transcription factors that directly associate with a retinoblastoma (RB) protein, or other RB-related pocket proteins, such as p107 and p130, and together, are corecruited to E2F target promoters (45–47). Loss of RB leads to deregulation of E2F transcription factor activity, resulting in cell proliferation and/or cell death (48–50). E2F1 is a member of the E2F transcription factor family that functions as a transcriptional activator (51). Here, we show that loss of CBX5 results in upregulation of E2F1 and consequently, an E2F1-mediated increase in expression of the antiapoptotic protein BIRC5 (Fig. 7). We also establish a role for E2F1 in EGFRi resistance downstream of CBX5 by showing that E2F1 knockdown partially restores EGFRi sensitivity in cells lacking CBX5.
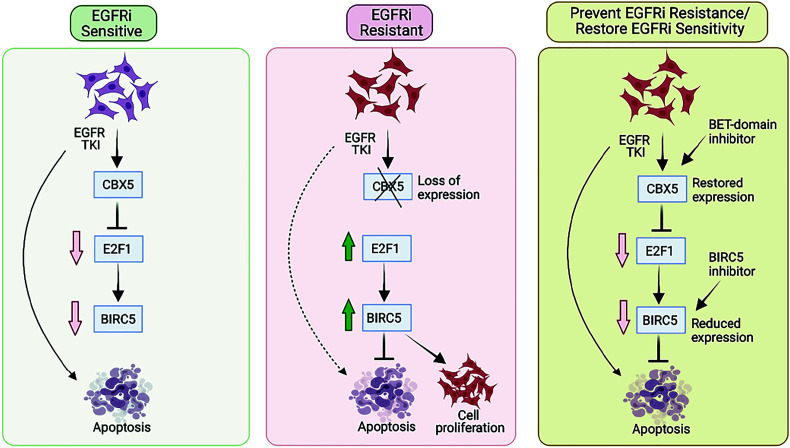
Fig. 7.
Proposed model for the mechanism by which EGFRi resistance in LUAD is regulated by the CBX5-E2F1-BIRC5 axis. Our findings are consistent with a model wherein loss of CBX5 promotes resistance to EGFRi via E2F1-mediated upregulation of BIRC5 expression, which leads to inhibition of EGFRi-induced apoptosis. CBX5 is down-regulated in EGFRi-resistant cells; however, expression can be restored by treatment with BETi. Consistent with this observation, combined treatment with EGFRi and BETi shows potential therapeutic benefits in cell culture and in a mouse model of EGFRi-resistant LUAD. In addition, similar therapeutic benefits can be achieved by treating EGFRi-resistant cells with a BIRC5 inhibitor.
BIRC5, a member of the IAP family that plays a role in cell cycle regulation and both apoptotic and nonapoptotic cell death, is a known target of E2F1 (33, 34, 52, 53). In particular, BIRC5 is known to be expressed in the G2/M phase of the cell cycle in a cell cycle-dependent manner (54). BIRC5 also associates with microtubules of the mitotic spindle, and disruption of BIRC5-microtubule interactions results in loss of the antiapoptotic function of BIRC5 (54). BIRC5 is overexpressed in some cancers (55, 56), and it is speculated that this overexpression thus potentially overcomes an antiapoptotic checkpoint and thereby facilitates progression of cancer cells through mitosis. Related to its role in mitosis, BIRC5 is also part of the so-called chromosome passenger complex that is essential for chromosome segregation during mitosis (57). BIRC5 has also been implicated in other processes relevant to its role in promoting tumor growth and progression, such as suppression of the host immune response (58). Similarly, nonapoptotic functions of BIRC5, such as enhancing aerobic glycolysis by altered regulation of mitochondrial fusion/fission machinery, have also been reported (59).
In addition, BIRC5 has been implicated in the progression and survival of several different cancer types (33, 60). Here, consistent with these findings and a previous study that reported altered regulation of BIRC5 in response to the disruption of RB/E2F family proteins (52), we found that BIRC5 is an E2F1 target in EGFR-mutant LUAD. BIRC5 was previously shown to be regulated by EGFR signaling and to modulate response to EGFRi (61). However, to our knowledge, its role as a mediator of CBX5-loss–driven EGFRi resistance has not been documented. In particular, our findings show that BIRC5 is a key downstream target of E2F1 that is up-regulated in CBX5-depleted cells and functions to promote EGFRi resistance by blocking EGFRi-mediated induction of apoptosis. Consistent with this model, we found that CBX5 is down-regulated, whereas both E2F1 and BIRC5 are up-regulated, in EGFRi-resistant cell lines. However, as indicated above, due to the role of BIRC5 in mitosis, it is possible that knockdown of BIRC5, or its pharmacological inhibition by YM155, induces apoptosis indirectly by regulating mitosis.
In addition to the CBX5-E2F1-BIRC5 axis, there are likely other pathways downstream of CBX5 that play a role in mediating EGFRi resistance in LUAD. In this regard, we note that our RNA-seq analysis identified several other biological pathways that were enriched among genes up- or down-regulated in CBX5-knockdown cells, including those related to oxidative phosphorylation, mitochondrial dysfunction, sirtuin signaling, and GADD45 signaling (SI Appendix, Fig. S6A). Future work systematically evaluating the role of other CBX5 targets may shed light on additional pathways that mediate the downstream function of CBX5 in conferring EGFRi resistance in LUAD.
Based on our elucidation of the CBX5-E2F1-BIRC5 axis, we explored several therapeutic strategies for targeting CBX5-loss–driven EGFRi resistance in LUAD. Notably, we found that treatment with BETi restores CBX5 expression in EGFRi-resistant LUAD, and this resensitizes EGFRi-resistant cells to BETi/EGFRi-based combination therapy. Moreover, when used preemptively in conjunction with EGFRi, BETi also forestalls development of EGFRi resistance. These findings are reminiscent of results from a previous study that identified a role for regulators of histone acetylation and histone methylation in controlling the emergence of drug-tolerant cells in response to various targeted and chemotherapeutic agents (26). Lastly, consistent with the role of BIRC5 as an IAP downstream of CBX5, we found that treatment with the BIRC5 inhibitor YM155 suppresses growth of EGFRi-resistant LUAD both in cell culture and in mice.
Taken together, our findings uncover a CBX5-loss–driven antiapoptotic pathway that confers resistance to EGFRi. Moreover, we identify two unique therapeutic approaches for treating EGFRi-resistant LUAD, that is, by blocking BIRC5 expression with a BIRC5 inhibitor or by restoring CBX5 expression via BETi treatment. Therefore, collectively, these results provide a hope for LUAD patients who have failed EGFRi treatment due to the emergence of acquired resistance.
Materials and Methods
Cell Culture.
Human LUAD cell lines (HCC827, HCC2935, and PC9) and HEK293T cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). HEK293T cells were grown in Dulbecco’s Modified Eagle Medium with high glucose and L-glutamine, supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA) and 1× penicillin/streptomycin (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA). HCC827, HCC2935, and PC9 cells were grown in Roswell Park Memorial Institute (RPMI)-1640 media, supplemented with 10% FBS and penicillin/streptomycin. All cells were grown under 5% CO2 at 37 °C.
To establish erlotinib-resistant cell lines, HCC827, HCC2935, and PC9 cells were treated with increasing concentrations of erlotinib (1 to 5 μM) for 2 to 3 mo, changing the medium every 3 d and adding fresh drug. Resistant colonies were pooled, and resistance was confirmed using MTT and clonogenic assays with different concentrations of erlotinib and assessing growth compared to parental cell lines.
Large-Scale Epigenome-Wide shRNA Screen.
HCC827 cells were infected with eight lentiviral shRNA pools containing 1,862 shRNAs targeting 363 known or predicted chromatin modifier genes, or as a control a lentivirus containing an NS shRNA, at 0.2 MOI to prevent superinfection and ensure that each cell received no more than one shRNA. After infection, HCC827 cells were selected with puromycin (0.6 μg/mL) for 1 to 2 wk to enrich for cells expressing the shRNAs. After selection, cells were grown in erlotinib (2 μM) for 4 wk. Surviving colonies were collected, and genomic DNA was isolated. Integrated shRNAs were PCR-amplified using primers specific to the shRNA vector (pLKO.1), listed in SI Appendix, Table S3. Samples were sequenced using primer SP6 to identify candidate shRNAs. The epigenetic regulator shRNA library was obtained from the University of Massachusetts Chan Medical School RNAi Core Facility.
Drug Treatments.
EGFRi (erlotinib, gefitinib, and osimertinib), BETi (JQ1 and PFI-1), and the BIRC5 inhibitor YM155 were purchased from Selleck Chemicals, LLC. All inhibitors were dissolved in hybridoma-grade dimethyl sulfoxide (DMSO; Sigma-Aldrich) at a stock concentration of 10 mM. Cells were treated with DMSO or drugs at the concentrations indicated in the figure legends. An epigenetic inhibitor library containing 33 small-molecule compounds that target 25 epigenetic genes was purchased from SGC Chemicals (Bang Sue, Krung Thep, Thailand), and cells were treated with indicated concentrations of inhibitors listed in SI Appendix, Table S2. For mouse studies, erlotinib and PFI-1 were dissolved in 2% DMSO in 0.5% methylcellulose, osimertinib was dissolved in 0.5% carboxy propyl cellulose and 0.1% Tween80, JQ1 was dissolved in 5% DMSO in 5% dextrose solution, and YM155 was dissolved in 0.9% saline.
shRNAs, Lentivirus Preparation, and Stable Cell Line Generation.
Gene-specific shRNAs were obtained from Open BioSystems (Thermo Fisher Scientific); catalog numbers for the shRNAs used in this study are listed in SI Appendix, Table S3. For lentivirus production, shRNAs and the packaging plasmids PDM2.G and psPAX2 were transfected into HEK293T cells using Effectene Transfection Reagent (QIAGEN). After 48 h, lentivirus-containing supernatants were harvested, filtered, and used for infections. Lentiviral shRNA-infected cells were selected using puromycin, at the following concentrations: 0.6 μg/ml for HCC827 cells and 0.70 μg/ml for HCC2935 and P9 cells. For E2F1/CBX5 and BIRC5/CBX5 double-knockdown experiments, PC9 or HCC827 cells were first infected with an E2F1 shRNA, BIRC5 shRNA, or TRC NS shRNA. Cells were selected with puromycin (0.70 μg/mL), and knockdowns were validated by immunoblot analysis. Cells expressing an E2F1, BIRC5, or TRC NS shRNA were then infected with PZIPZ NS shRNA or CBX5 shRNA and GFP-positive cells were sorted by fluorescence-activated cell sorting (FACS) using a BD FACSAria Flow Cytometer. E2F1/CBX5 and BIRC5/CBX5 double-knockdown cells were validated by immunoblot analysis.
clonogenic Assays.
Cells were seeded at 5 × 103 cells per well in 6-well plates and allowed to attach overnight. After 2 d, cells were treated with the indicated concentrations of EGFRi (erlotinib, gefitinib, or osimertinib), BIRC5 inhibitor (YM155), or BETi (JQ1 and PFI1), alone or in combination, as indicated in the text and figure legends. Cell culture media was changed every 3 d, adding fresh drug each time. After 2 wk of treatment, surviving colonies were fixed in a solution containing 50% methanol and 10% acetic acid and then stained with 0.05% Coomassie Brilliant Blue-R250 (Sigma-Aldrich).
Mouse Tumorigenesis Experiments.
Animal experiments were conducted under the ethical guidelines and protocols approved by the Institutional Animal Care and Usage Committee at the University of Alabama at Birmingham. NOD scid gamma (NSG) mice (Stock No. 005557) were obtained from The Jackson Laboratory (Bar Harbor, ME, USA). In all mouse experiments, tumor size was measured once a week using calipers. The average tumor volume in each experimental group was expressed in cubic millimeters and calculated using the following formula: π / 6 × (large diameter) × (small diameter)2. The relevant details for individual experiments are presented below.
Mouse Experiments with Erlotinib or Osimertinib.
A total of 5 × 106 PC9 cells expressing either a CBX5 or NSshRNA were suspended in 200 μL Matrigel/phosphate-buffered saline (PBS), combined at a 1:1 ratio, and subcutaneously injected into the flanks of 5- to 6-wk-old NSG mice. Once tumor volumes reached ~100 mm3, mice were randomly split into two groups (n = 5); one group received vehicle control and the other received an EGFRi. For erlotinib treatment, 25 mg/kg erlotinib was administered intraperitoneally (i.p.), three times per week until the end of the experiment; the vehicle control was 0.5% methylcellulose. For osimertinib treatment, 2.5 mg/kg osimertinib was administered orally three times per week until the end of the experiment; the vehicle control was 0.5% carboxy propyl cellulose and 0.1% Tween80.
CBX5/E2F1 and CBX5/BIRC5 Double-Knockdown Mouse Experiments.
A total of 5 × 106 PC9 cells with shRNA-mediated double knockdown of E2F1/CBX5 or BIRC5/CBX5, or cells expressing an NS (TRC/PZIPZ) shRNA were suspended in 200 μL Matrigel/PBS (1:1) and subcutaneously injected into the flanks of 4 to 5-wk-old NSG mice. Mice were randomly split into two groups (n = 5); one received vehicle control (0.5% methylcellulose), and the other received 25 mg/kg erlotinib (i.p.), three times per week until the end of the experiment.
Mouse Experiments with BETi.
A total of 5 × 106 EGFRi-resistant PC9 cells (PC9-R2) were suspended in 200 μL Matrigel/PBS (1:1) and subcutaneously injected in the flanks of 5- to 6-wk-old NSG mice. Mice were randomized to receive vehicle control (0.5% methylcellulose), 25 mg/kg erlotinib alone (i.p.), 50 mg/kg JQ1 alone (i.p.), 5 mg/kg PFI-1 alone (i.p.), a combination of erlotinib + JQ1, or a combination of erlotinib + PFI-1, three times per week until the end of the experiment.
Mouse Experiments with the BIRC5 Inhibitor YM155.
A total of 1 × 107 EGFRi-resistant HCC827 cells (HCC827-R2) were suspended in 200 μL Matrigel/PBS (1:1) and subcutaneously injected in the flanks of 4 to 5-wk-old of NSG mice. Mice were randomly split into two groups; one group received vehicle control (0.9% saline), and the other received 3.5 mg/kg YM155 (i.p.), five times per week until the end of the experiment.
Statistical Analysis.
All experiments were conducted with at least three biological replicates. Results for individual experiments are expressed as the mean ± SEM. P-values for all other experiments were calculated using the two-tailed unpaired Student’s t test in GraphPad Prism v.9.0 for Macintosh (GraphPad Software). P-values < 0.05 were considered statistically significant, with significance indicated as follows: ns, not significant; *P < 0.05; **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (PDF)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Acknowledgments
We gratefully acknowledge grants from the NIH: R01CA257046 (N.W.), R01CA218008 (N.W. and M.R.G.), R03CA248913 (R.G.), and R01CA233481 (R.G.).
Author contributions
S.B., R.G., M.R.G., and N.W. designed research; S.B. performed research; R.G., M.R.G., and N.W. contributed new reagents/analytic tools; S.B., Y.J.K.E., R.G., M.R.G., and N.W. analyzed data; and S.B., R.G., M.R.G., and N.W. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
Reviewers: I.O., University of Southern California; B.R., University of Illinois at Chicago college of medicine; and S.S., University of South Alabama Mitchell Cancer Institute.
Contributor Information
Michael R. Green, Email: Michael.Green@umassmed.edu.
Narendra Wajapeyee, Email: nwajapey@uab.edu.
Data, Materials, and Software Availability
RNA-seq data have been deposited in [GEO] (GSE114563) (62). All additional data shown in the paper are available in the main manuscript and/or SI Appendix.
Supporting Information
References
- 1.Herbst R. S., Heymach J. V., Lippman S. M., Lung cancer. N. Engl. J. Med. 359, 1367–1380 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herbst R. S., Morgensztern D., Boshoff C., The biology and management of non-small cell lung cancer. Nature 553, 446–454 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Skoulidis F., Heymach J. V., Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat. Rev. Cancer 19, 495–509 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.N. Cancer Genome Atlas Research, Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.da Cunha Santos G., Shepherd F. A., Tsao M. S., EGFR mutations and lung cancer. Annu. Rev. Pathol. 6, 49–69 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Sharma S. V., Bell D. W., Settleman J., Haber D. A., Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer 7, 169–181 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Pao W., et al. , EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. U.S.A. 101, 13306–13311 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sequist L. V., Lynch T. J., EGFR tyrosine kinase inhibitors in lung cancer: An evolving story. Annu. Rev. Med. 59, 429–442 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Lynch T. J., et al. , Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 350, 2129–2139 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Shepherd F. A., et al. , Erlotinib in previously treated non-small-cell lung cancer. N. Engl. J. Med. 353, 123–132 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Ramalingam S. S., et al. , Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N. Engl. J. Med. 382, 41–50 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Soria J. C., et al. , Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N. Engl. J. Med. 378, 113–125 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Lim S. H., et al. , Comparison of clinical outcomes following gefitinib and erlotinib treatment in non-small-cell lung cancer patients harboring an epidermal growth factor receptor mutation in either exon 19 or 21. J. Thorac. Oncol. 9, 506–511 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Camidge D. R., Pao W., Sequist L. V., Acquired resistance to TKIs in solid tumours: Learning from lung cancer. Nat. Rev. Clin. Oncol. 11, 473–481 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Lin J. J., Shaw A. T., Resisting resistance: Targeted therapies in lung cancer. Trends Cancer 2, 350–364 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chong C. R., Janne P. A., The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat. Med. 19, 1389–1400 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wheeler D. L., Dunn E. F., Harari P. M., Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat. Rev. Clin. Oncol. 7, 493–507 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z., et al. , Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin. Cancer Res. 24, 3097–3107 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Fan P. D., et al. , YES1 amplification is a mechanism of acquired resistance to EGFR inhibitors identified by transposon mutagenesis and clinical genomics. Proc. Natl. Acad. Sci. U.S.A. 115, E6030–E6038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wajapeyee N., Gupta R., Epigenetic alterations and mechanisms that drive resistance to targeted cancer therapies. Cancer Res. 81, 5589–5595 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bannister A. J., et al. , Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Hall I. M., et al. , Establishment and maintenance of a heterochromatin domain. Science 297, 2232–2237 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Smallwood A., Esteve P. O., Pradhan S., Carey M., Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes. Dev. 21, 1169–1178 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao S., et al. , A genetic interaction analysis identifies cancer drivers that modify EGFR dependency. Genes. Dev. 31, 184–196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forloni M., et al. , Oncogenic EGFR represses the TET1 DNA demethylase to induce silencing of tumor suppressors in cancer cells. Cell Rep. 16, 457–471 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma S. V., et al. , A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141, 69–80 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karachaliou N., Fernandez-Bruno M., Bracht J. W. P., Rosell R., EGFR first- and second-generation TKIs-there is still place for them in EGFR-mutant NSCLC patients. Transl. Cancer Res. 8, S23–S47 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mok T. S., et al. , Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N. Engl. J. Med. 376, 629–640 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.John T., et al. , EGFR mutation analysis for prospective patient selection in AURA3 phase III trial of osimertinib versus platinum-pemetrexed in patients with EGFR T790M-positive advanced non-small-cell lung cancer. Lung. Cancer 126, 133–138 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Sato H., et al. , Combined inhibition of MEK and PI3K pathways overcomes acquired resistance to EGFR-TKIs in non-small cell lung cancer. Cancer Sci. 109, 3183–3196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang W., et al. , PI3K-AKT-mTOR pathway alterations in advanced NSCLC patients after progression on EGFR-TKI and clinical response to EGFR-TKI plus everolimus combination therapy. Transl. Lung Cancer Res. 9, 1258–1267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ligresti G., et al. , CBX5/G9a/H3K9me-mediated gene repression is essential to fibroblast activation during lung fibrosis. JCI Insight 5, e127111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altieri D. C., survivin, cancer networks and pathway-directed drug discovery. Nat. Rev. Cancer 8, 61–70 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Wheatley S. P., Altieri D. C., survivin at a glance. J. Cell Sci. 132, jcs223826 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Filippakopoulos P., et al. , Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fish P. V., et al. , Identification of a chemical probe for bromo and extra C-terminal bromodomain inhibition through optimization of a fragment-derived hit. J. Med. Chem. 55, 9831–9837 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Picaud S., et al. , PFI-1, a highly selective protein interaction inhibitor, targeting BET Bromodomains. Cancer Res. 73, 3336–3346 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakahara T., et al. , YM155, a novel small-molecule survivin suppressant, induces regression of established human hormone-refractory prostate tumor Xenografts. Cancer Res. 67, 8014–8021 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Xu R., Shao H., Zhu J., Ju Q., Shi H., Combination strategies based on epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors for cancer patients: Pooled analysis and subgroup analysis of efficacy and safety. Medicine (Baltimore) 98, e14135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pal A. S., et al. , Loss of KMT5C promotes EGFR inhibitor resistance in NSCLC via LINC01510-mediated upregulation of MET. Cancer Res. 82, 1534–1547 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lachner M., O’Carroll D., Rea S., Mechtler K., Jenuwein T., Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001). [DOI] [PubMed] [Google Scholar]
- 42.De Koning L., et al. , Heterochromatin protein 1alpha: A hallmark of cell proliferation relevant to clinical oncology. EMBO Mol. Med. 1, 178–191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirschmann D. A., et al. , Down-regulation of HP1Hsalpha expression is associated with the metastatic phenotype in breast cancer. Cancer Res. 60, 3359–3363 (2000). [PubMed] [Google Scholar]
- 44.Yu Y. H., et al. , Network biology of tumor stem-like cells identified a regulatory role of CBX5 in lung cancer. Sci. Rep. 2, 584 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henley S. A., Dick F. A., The retinoblastoma family of proteins and their regulatory functions in the mammalian cell division cycle. Cell Div. 7, 10 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H. Z., Tsai S. Y., Leone G., Emerging roles of E2Fs in cancer: An exit from cell cycle control. Nat. Rev. Cancer 9, 785–797 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giacinti C., Giordano A., RB and cell cycle progression. Oncogene 25, 5220–5227 (2006). [DOI] [PubMed] [Google Scholar]
- 48.McNair C., et al. , Differential impact of RB status on E2F1 reprogramming in human cancer. J. Clin. Invest. 128, 341–358 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korotayev K., Ginsberg D., Many pathways to apoptosis: E2F1 regulates splicing of apoptotic genes. Cell Death Differ. 15, 1813–1814 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Iaquinta P. J., Lees J. A., Life and death decisions by the E2F transcription factors. Curr. Opin. Cell Biol. 19, 649–657 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dimova D. K., Dyson N. J., The E2F transcriptional network: Old acquaintances with new faces. Oncogene 24, 2810–2826 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Jiang Y., Saavedra H. I., Holloway M. P., Leone G., Altura R. A., Aberrant regulation of survivin by the RB/E2F family of proteins. J. Biol. Chem. 279, 40511–40520 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Huang C. L., et al. , E2F1 overexpression correlates with thymidylate synthase and survivin gene expressions and tumor proliferation in non small-cell lung cancer. Clin. Cancer Res. 13, 6938–6946 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Li F., et al. , Control of apoptosis and mitotic spindle checkpoint by survivin. Nature 396, 580–584 (1998). [DOI] [PubMed] [Google Scholar]
- 55.Small S., Keerthivasan G., Huang Z., Gurbuxani S., Crispino J. D., Overexpression of survivin initiates hematologic malignancies in vivo. Leukemia 24, 1920–1926 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Faldt Beding A., Larsson P., Helou K., Einbeigi Z., Parris T. Z., Pan-cancer analysis identifies BIRC5 as a prognostic biomarker. BMC Cancer 22, 322 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carmena M., Wheelock M., Funabiki H., Earnshaw W. C., The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 13, 789–803 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Asanuma K., Tsuji N., Endoh T., Yagihashi A., Watanabe N., survivin enhances Fas ligand expression via up-regulation of specificity protein 1-mediated gene transcription in colon cancer cells. J. Immunol. 172, 3922–3929 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Hagenbuchner J., Kuznetsov A. V., Obexer P., Ausserlechner M. J., BIRC5/survivin enhances aerobic glycolysis and drug resistance by altered regulation of the mitochondrial fusion/fission machinery. Oncogene 32, 4748–4757 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Chen X., Duan N., Zhang C., Zhang W., survivin and tumorigenesis: Molecular mechanisms and therapeutic strategies. J. Cancer 7, 314–323 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okamoto K., et al. , Role of survivin in EGFR inhibitor-induced apoptosis in non-small cell lung cancers positive for EGFR mutations. Cancer Res. 70, 10402–10410 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Bugide S., Wajapeyee N., CBX5 loss drives EGFR inhibitor resistance and results in therapeutically actionable vulnerabilities in lung cancer. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE114563. Deposited 16 May 2018. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (PDF)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Data Availability Statement
RNA-seq data have been deposited in [GEO] (GSE114563) (62). All additional data shown in the paper are available in the main manuscript and/or SI Appendix.