INTRODUCTION
Musculoskeletal (MSK) radiology is a subspecialty that focuses on evaluating bones, joints, and the surrounding tissues. MSK radiology assesses the normal anatomy, trauma, degenerative changes, cartilage abnormalities, meniscal tears, acute and chronic skeletal pain, infections and inflammation, neoplasms and metastases, osteoporosis and bone mineral measurement, arthritis, bone age assessment, and pediatric imaging [1,2]. Minimally invasive imaging-guided techniques including angiography, biopsy, ablation, and endoscopy are often used to address diagnosis and assessment of these diseases [3].
X-ray, computed tomography (CT), magnetic resonance imaging (MRI), and ultrasound are the mainly used imaging modalities in MSK radiology. Although CT is a preferred modality in trauma and emergent bone imaging, MRI is the method of choice when better soft-tissue contrast is required. MR image acquisition takes longer than CT acquisition but it provides excellent soft-tissue contrast, allowing in depth exploration of the soft-tissues (eg, muscle). MSK MRI can create thinner and higher quality images of complex body regions such as the knee or thigh [2].
The complicated interaction of biomechanics necessitates acquiring analytical abilities in MSK radiology [4]. Delivering definitive diagnosis in MSK imaging frequently requires knowledge and synthesis of the fundamental mechanics of the injuries with the imaging studies. The skeletal system’s bones, joints, and muscles act together, forming complex interdependent collective tissues whose activities depend on the skeletal system’s proximal and surrounding components [3]. As a result, when a system component is affected by a disease or abnormal condition, a domino of interrelated injuries is frequently caused by the subsequent malfunctioning of that particular component in the circuit. Radiologists already have the training required to apply these principles when interpreting the imaging, which increases their ability to make the correct diagnosis [3]. This motivates the development of computational tools such as artificial intelligence (AI) for MSK imaging and its applications. Similar strategies will be needed when computational tools are used in MSK imaging and its applications. Although the high-level reasonings are not completely available in AI tools yet, there is still a significant need for AI tools in MSK imaging and applications to automate measurements of tissues, organs, and pathologic conditions (or abnormalities).
Medical artificial intelligence
AI, specifically deep learning, becomes a key helper in radiology applications. Recent innovations in deep learning have generated software that enables automated and precise detection and quantification of disease in medical images [5]. Among many, the most commonly used deep learning implementations in MSK are detection of fractures, meniscal tears, cartilage lesions, osteoarthritis, degenerative and metastatic spinal lesions, body composition analysis, and identification of metabolic health problems [6]. In addition, AI can speed up the collecting of MRI data by converting low-quality data into high-quality images [7]. In this article, two of such applications are explored in detail: quantification of tissues (ie, thigh MRI tissue analysis in health and/or diseases) and quantification of abnormal conditions (ie, cartilage assessment from knee MRI).
Unique challenges for developing artificial intelligence in musculoskeletal
Contrary to other organs and systems, MSK diseases bring multiple obstacles that have affected the development of AI solutions [3]. For example, MSK images with complex relations between the joints, bones, and muscles pose a significant challenge for AI researchers to develop algorithms [3,8]. Hence, AI solutions for MSK imaging and its applications have unique challenges to be addressed [3]. First, MR images need to be preprocessed heavily before any AI operations due to high variations in the MRI field, noise, and acquisition differences in MRIs. Even for the same patients on the same day for 2 different scans with the same protocol, the same tissue may have different tissue intensities, putting a significant challenge in any computational modeling. Second, instead of a single object (tissue or abnormality/pathologic condition) analysis similar to most other medical AI applications, in MSK applications, there is a need to analyze multiple tissues and/or objects due to the complex relations between MSK system components (joints, bones, muscles, and so forth). Fig. 1 illustrates a general workflow of the AI integrated MSK imaging applications, where particular emphasis is given to preprocessing step. AI component is considered as a general framework in which, depending on the application type, one may conduct image-level or pixel level classifications (ie, diagnosis and quantification, respectively). In Section “Preprocessing–data cleaning/curation,” we explain how to “clean” MR images before AI tools. In Section “Artificial intelligence applications in musculoskeletal radiology,” we exemplify 2 important MSK AI applications and in Section “Discussion and concluding remarks,” we introduce the limitations of the current AI methods and discuss future trends.
FIG. 1.
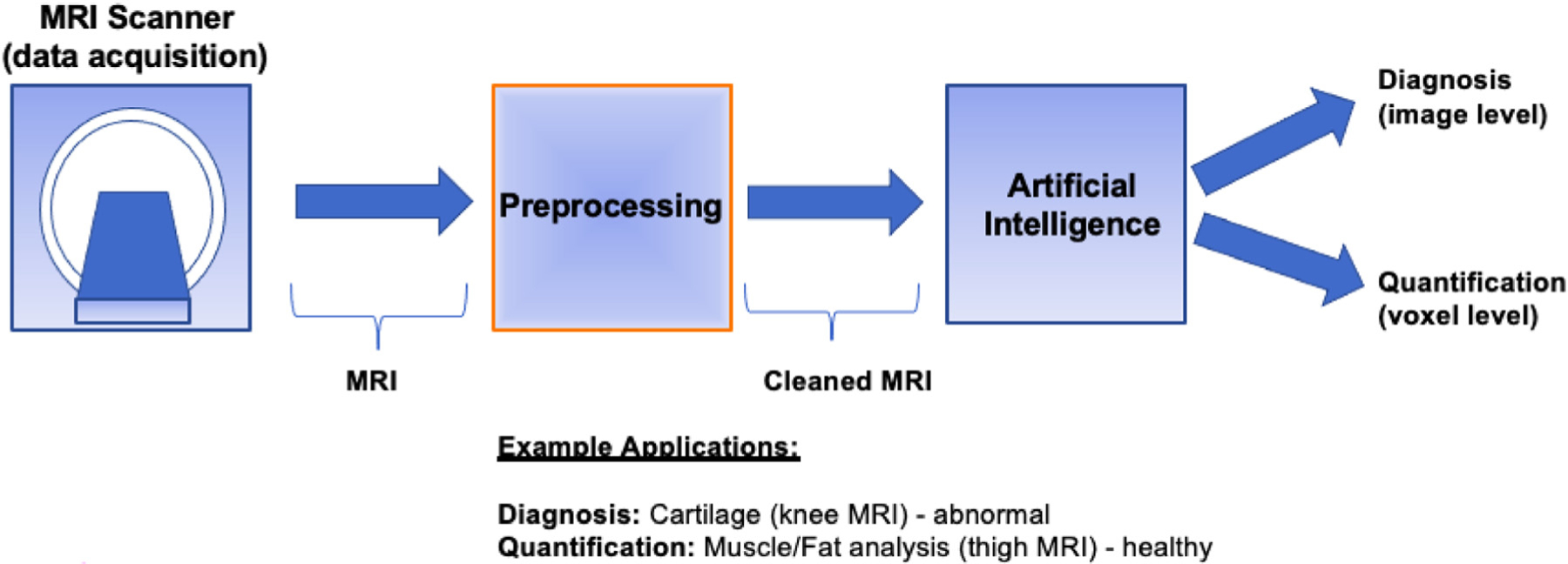
General workflow for MSK AI applications in a clinical setting.
PREPROCESSING–DATA CLEANING/CURATION
Medical imaging investigations have revealed a wide range of artifacts that have negatively affected the diagnostic data collected from the scans. To improve image quality, each imaging modality uses a range of techniques called “preprocessing.” In MRI, these are collected under 3 headings: (1) noise, (2) artifacts such as bias field (inhomogeneity), and (3) nonstandardness in medical images (order matters as inhomogeneity correction algorithm enhances noises). Anatomy and pathologic condition variations are already imposing significant constraints in AI models. Having additional variations due to scanners and acquisition differences, noise, and other artifacts increase the complexity of AI models unnecessarily, resulting in suboptimal findings. Therefore, preparing “clean data” for AI algorithms is critical [9,10]. In this section, we show preprocessing results (before and after cleaning) on three-dimensional (3D) MR cartilage knee [11] and 3D MR thigh images [12]. We apply both denoising [13] and inhomogeneity correction techniques [14] as well as intensity standardization [15] to “clean” the MR images before delineation of the tissues.
Inhomogeneity correction
Inhomogeneity, or bias field, is a major artifact in MRI. Bias field changes true value (intensity) of pixels and the same tissue region starts to have different quantitative values, often slowly changing across the different parts of the image and hard to capture visually when the changes are small. Bias field affects the diagnosis and quantification by radiologists and degrades the AI algorithms’ performance significantly due to such changes in intensities, often causing over-segmentations or undersegmentations. We correct the field inhomogeneity induced by the RF coil with a widely recognized postprocessing method called “generalized-scale,” [16,17] which takes less than 10 seconds in our preliminary studies [18,19]. Although, nowadays, different scanner vendors have their preprocessing software for removing bias field during image reconstruction, there is still a need for a preprocessing step because the MR images still include inhomogeneities even after reconstruction-based corrections.
Denoising
Noise is ubiquitous in MRI. Considering that minor anomalies in MSK disorders or injuries can easily be exacerbated by noise, it is critical to remove or minimize them for autoanalysis. Moderate-to-high amounts of noise is typically expected in MRI scans due to varying scanner type, scanning time, patient size, and other factors including bias field correction methods. Based on our [20] previous experience in smoothing medical images while preserving critical details, we perform an edge-preserving denoising procedure to minimize the noise in scans.
Intensity standardization
To address acquisition-to-acquisition signal intensity variations, we apply the “intensity standardization” algorithm [21] that has been widely adopted by different vendors and the research community (performed in less than a second). The outcome is a “clean MRI” where variability across images (within and across centers) is minimized. Intensity standardization is simply based on representing the foreground and background of the images with the same intensity intervals.
Figs. 2 and 3 show the same subject’s MR images of the thigh and knee before and after cleaning operation (inhomogeneity correction and denoising), respectively. We first remove inherent bias with the generalized scale method [16,17] followed by the nonuniform nonpara-metric intensity normalization technique [14] to ensure there is no artifact left in the MR images. Then, for keeping tissue structures and image sharpness, we apply an edge-preserving diffusive filter [13].
FIG. 2.
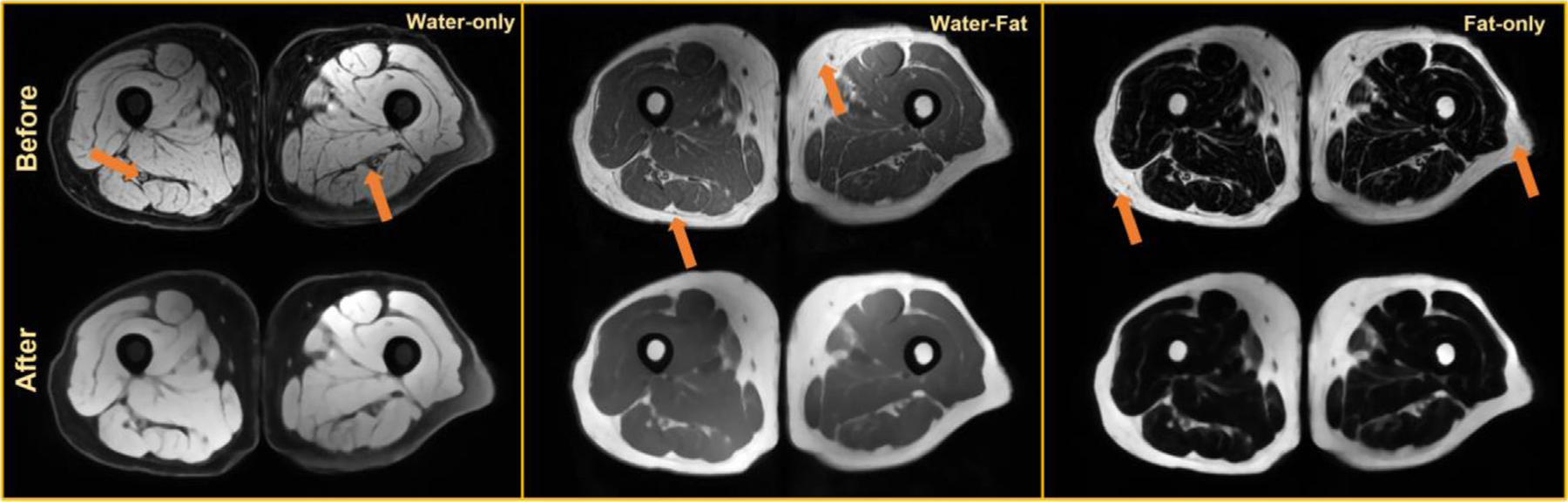
Preprocessing results for an axial thigh MRI [22] before and after preprocessing inherent noise and bias-field (marked with orange arrows), with water-only, water-fat, and fat-only contrasts.
FIG. 3.
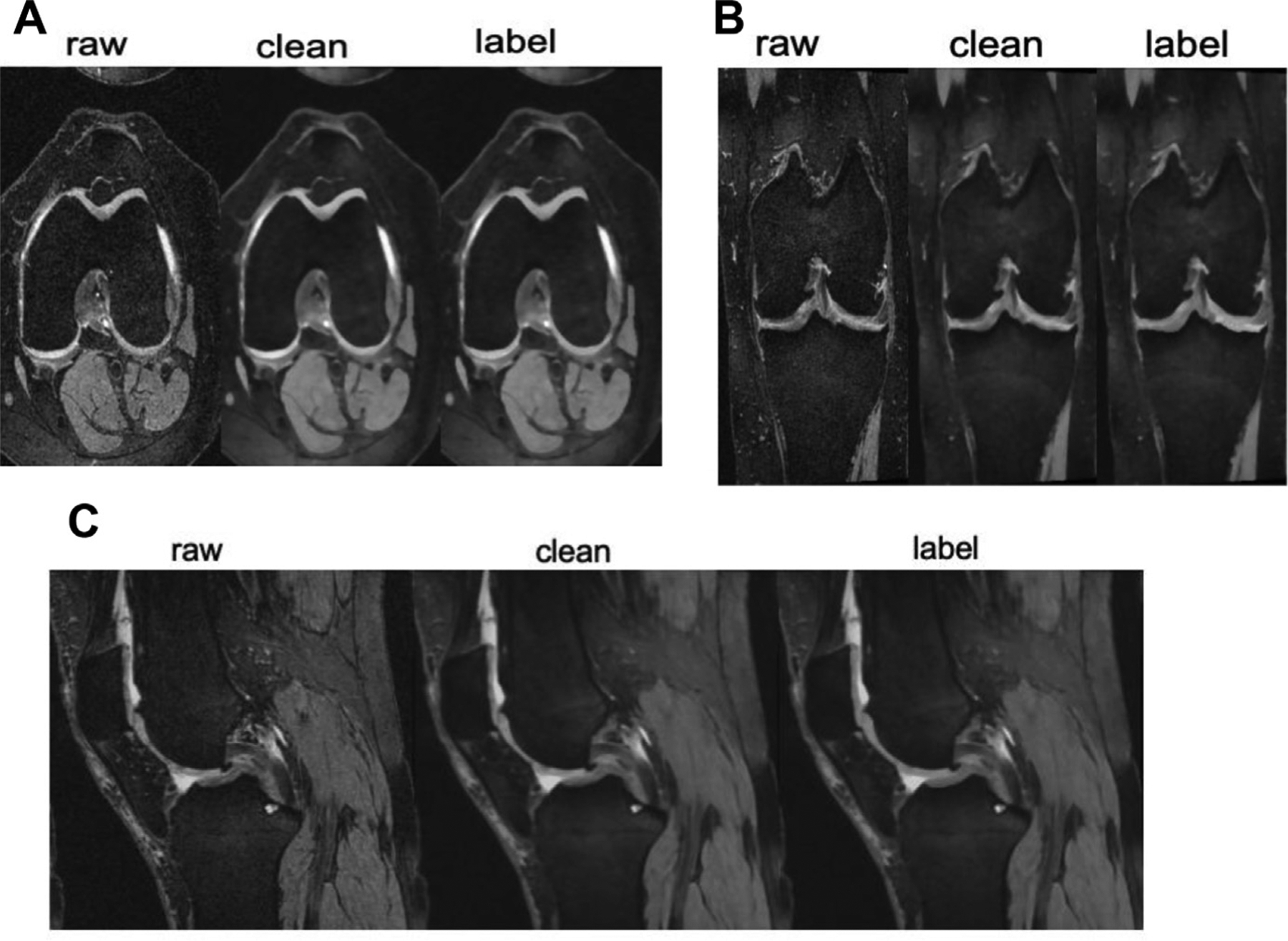
Axial (A), coronal (B), and sagittal (C) knee MRI. Raw demonstrates the images before preprocessing. Clean demonstrates the images after preprocessing. Tissue coloring for segmentation is referred to as the “labeled” image.
ARTIFICIAL INTELLIGENCE APPLICATIONS IN MUSCULOSKELETAL RADIOLOGY
AI, specifically deep learning, algorithms are well suited to the task of detecting disease or injury, which may be the most crucial part of an imaging examination. For this task, it is needed to use 1 of 2 approaches. Classification of images begins with training a deep learning algorithm to recognize features in a collection of images and then using that information to provide a single diagnosis. A disadvantage of this approach is the lack of a localizer to assist in human interpretation or verification. Computational methods such as “heatmaps” have been created to illustrate the portions of an image that are most important to the classifier while reaching its conclusion. In medical imaging, the effectiveness of these techniques for pinpointing the location of disease has been varied. An object-detection training model evaluates the entire image and coordinates the areas where the model predicts a disease or condition can be recognized. Radiologists can benefit from using these models because they can help them determine the location of an illness of interest. They also benefit from internal explainability (interpretability) in their decision to forecast the presence or absence of a disease or condition. A significant limitation of this model is the demand for images with annotations at the structure level, which necessitates the human insertion of frames on the particular disease, which is a long-lasting job [1,3,4].
AI models can assist basic activities that an MSK radiologist is responsible for interpreting imaging results. These tasks can be divided into 3 categories, which are as follows: (1) detection and characterization of disease (image level labeling) as described above, (2) segmentation (voxel level labeling), and 3) better quality of images (acquisition/reconstruction) [1,3]. In this article, 2 MSK applications are explored under the segmentation subfield: quantification of tissues (thigh MRI tissue analysis in health and diseases) and quantification of abnormal conditions (cartilage assessment from knee MRI).
Musculoskeletal application #1: tissue quantification via segmentation
Herein, we demonstrate how we address the challenging segmentation problem of tissues in thigh MRI. Separation of fat, muscle, and other tissues in MR images is challenging due to the overlap of intensity values in different tissues (similar appearances). Conventional methods for the tissue delineations and quantifications mostly depend on manual or semiautomated algorithms with limited accuracy, efficiency, and reproducibility. In this regard, the existing autosegmentation methods fall short especially when dealing with challenging tissues such as intermuscular adipose tissue (IMAT).
We recently proposed a novel deep learning algorithm, called SegCaps [10], for general biomedical image segmentation problems. Specifically, we showed its efficacy in multiple different challenging problems in medical images and obtained state-of-the-art results in the literature. Herein, we summarize how we can apply this algorithm to the thigh muscle and adipose (fat) tissue segmentation from MRI scans. Our capsule-based algorithm works as follows. Compared with the commonly used convolutional neural network (CNN) algorithms, capsule uses communications of neurons with vectors instead of scalars. Briefly, a capsule consists of several neurons, and different from CNNs, spatial organization of the objects in the images are better captured with capsules thanks to their vector representations and lack of pooling operations. Technically, we showed that the capsules can be used effectively for object segmentation with high accuracy and heightened efficiency compared with the state-of-the-art segmentation method [10]. The proposed convolutional–deconvolutional capsule network, called SegCaps, showed strong results for the task of object segmentation with substantial decrease in parameter space. For segmentation of thigh tissues, we first established deconvolutional capsules and created a novel deep convolutional–deconvolutional capsule architecture, deeper than the original 3 layer capsule network (Fig. 4) [10]. Between different layers of the SegCaps, there is a routing algorithm called locally constrained dynamic routing, allowing a less number of parameters to be used and more robust training of neural networks while still providing better accuracy for the delineation (segmentation) process.
FIG. 4.
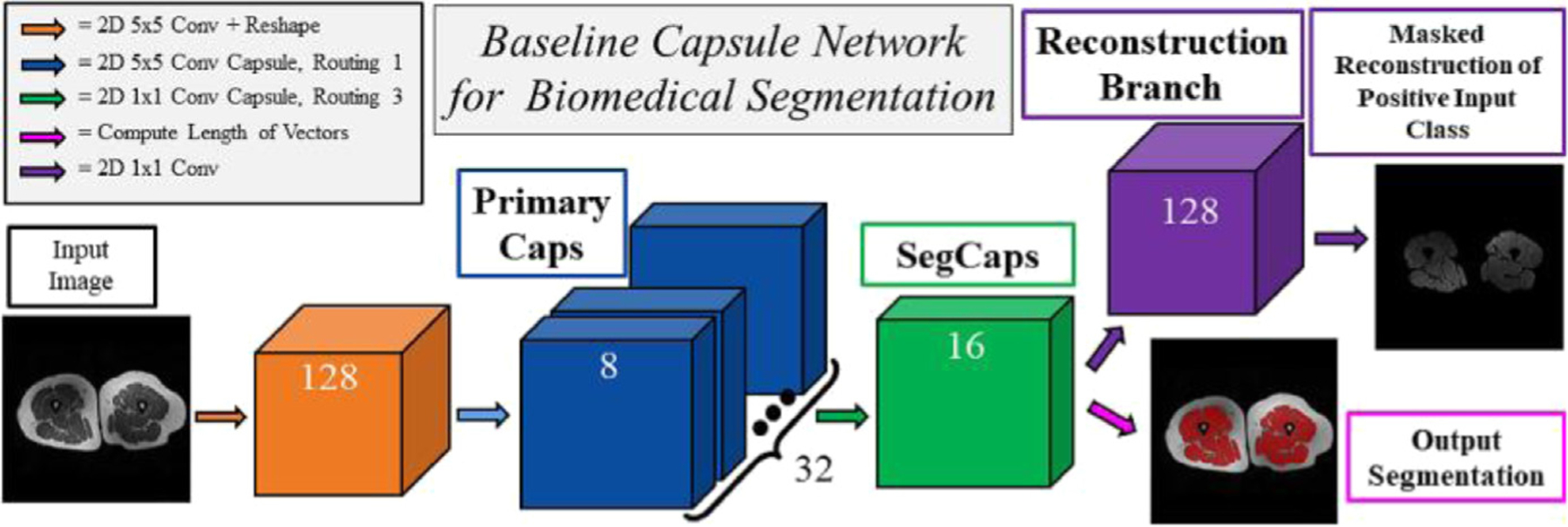
The segmentation capsule network in thigh segmentation.
Segmentation algorithms are often evaluated with (at least) 2 quantitative metrics: dice coefficient (ie, DSC) to measure how similar a segmented object is to its ground truths in terms of percentage of overlapped regions (ie, manually segmented object by radiologists) and a shape similarity metric showing how the boundary of a segmented object differs from its ground truth correspondence (Hausdorff Distance-HD is often used). A DSC value of 100% and HD value of 0 show perfect delineation (higher DSC and lower HD indicate a superior segmentation algorithm). Other metrics such as sensitivity and specificity can also be used for showing the success of a segmentation algorithm. We validated the efficacy of SegCaps on 150 MRI scans at 3 different contrasts (water-suppressed, fat-suppressed, water-fat) for thigh muscle and adipose (fat) tissue segmentation, and reported the best DSC and HD values in the literature compared with several other state-of-the-art deep segmentation algorithms [10].
It is also worth mentioning that before our SegCaps study, in another study, we suggested another technical innovation for segmenting muscle and fat tissue in the thigh area of whole-body MRI imaging using the fuzzy connectivity idea (pre-deep learning image processing method). This study represented the highly accurate delineation results in terms of Dice score applications for fat (DSC = 98.16%) and muscle (DSC = 96.78%) [23].
Additionally, we demonstrated that the semisupervised deep learning system (based on conventional U-Net variant system [9]) could be highly effective for training on both labeled and unlabeled data and outperformed existing state-of-the-art methods. Although supervised deep learning requires labeled data, our motivation was based on the lack of labeled data. Hence, we used a semisupervised approach [8]. In a total of 150 scans from 50 subjects, we obtained dice scores of 97.52%, 94.61%, 80.14%, 95.93%, and 96.83% for muscle, fat, IMAT, bone, and bone marrow segmentation, respectively [9]. Our approach was the first study to demonstrate that the thigh tissue segmentation problem can be solved using a combination of semisupervised deep learning and multicontrast MRI data [9], and comparable to the fully supervised deep segmentation methods even though the labels were not complete.
In summary, current techniques in MSK radiology are focused on deep learning algorithms and specifically U-Net and its variants. However, U-Net algorithms are known for challenges in their robustness and generalization. In other words, different image appearances (scanners, time, gender, patient group, age difference, or other factors) can easily cause these algorithms to fail. Newer algorithms are focused on addressing such challenges by developing more robust and generalized algorithms. Moreover, current algorithms often use fully supervised algorithms, where ground truth labeling of the tissues (and pathologic conditions) in MSK radiology scans are available for training deep learning algorithms. However, emerging deep learning algorithms are aiming at using imaging data with minimal supervision because it is quite laborious and expensive to get labeling.
Fig. 5 demonstrates thigh segmentation qualitative examples with the proposed algorithm (SegCaps). Further, the first row shows uncleaned images where noise and inhomogeneity is observed, whereas the middle row indicates a smoother and cleaner version of the raw data.
FIG. 5.
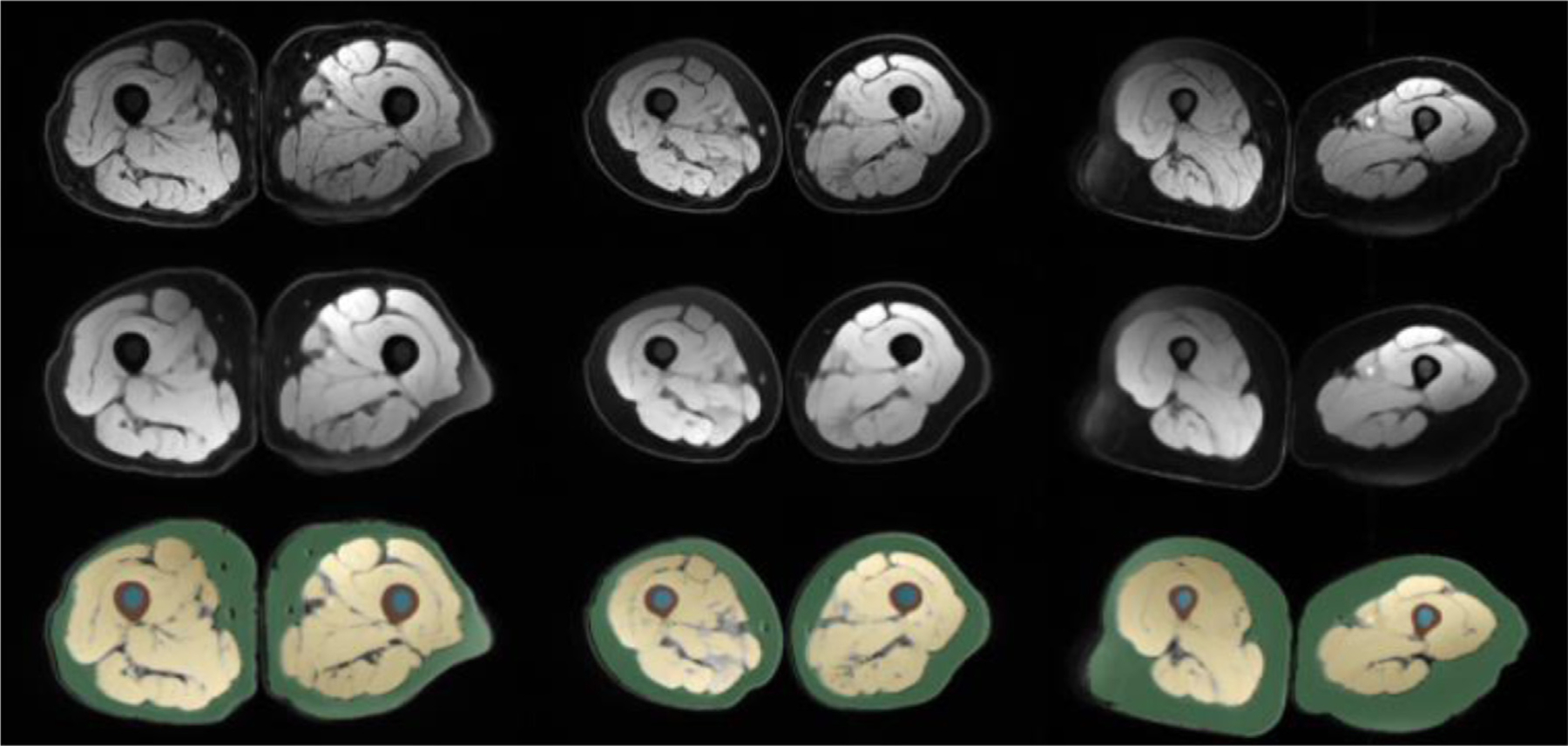
The figure demonstrates thigh MRI segmentation (last row), cleaned MRI (middle row), and raw images (first row).
Musculoskeletal application #2: cartilage quantification via segmentation
Many efforts have been put into segmenting knee cartilage because it is thicker than the cartilage in other joints and can be scanned using standard MR sequences [3]. The literature in knee cartilage quantification is denser than the thigh MRI tissue assessment. Liu and his colleagues have developed an automated, deep learning-based approach for detecting cartilage. An area under the curve (AUC) of 0.917% and a sensitivity of 88% was achieved using the same datasets as experienced radiologists, with 660 images for training and 1320 images for testing [24]. CNN-based deep neural network (DenseNet) designed by Pedoia and colleagues [25] used T2-voxel-based relaxometry data to detect cartilage abnormalities and diagnose osteoarthritis with an AUC of 0.83. There are two CNNs in this cartilage-detection CNN model: one for detection and one for recognition. First, a segmentation of the image was performed, and then a follow-up segmentation was performed to find any anomalies in the cartilage [6]. Liu and colleagues published a method for segmenting knee (bone and cartilage) based on Badrinarayanan and colleagues SegNet’s model [23,26]. Researchers have used SegNet to accomplish accurate knee cartilage segmentation to reduce computing costs to a minimum. Another 60 training and 40 testing examples from the SK10 Challenge’s 250-test case dataset were also used (sk10.org) [27]. For femoral and tibial cartilage segmentation, their SegNet model outperformed other state-of-the-art approaches and was the best performer in the femur segmentation categorization overall [6].
Other studies compared CNN-based automated segmentation applications for knee menisci (DSC = 83–86%) [26,28] adipose tissue (DSC = 96%), and the proximal femur (DSC = 94%) [29]. It is envisaged that the accuracy of lesion identification and classification would improve when the precision of tissue segmentation improves due to deep learning technologies [6]. Segmenting the proximal femur was done using images of volumetric structural MR scans taken from the bones of 86 subjects. The autosegmentation DSC was 0.94 [29].
Meniscus, quadriceps, patellar tendon, and infrapatellar fat pad were all segmented using CNN (VGG16) and a mean DSC of 80% to 90% in their MRI study of the knee [30]. The challenge also tested the clinical efficacy of automatic segmentation methods on osteoarthritis initiative (OAI) Datasets. The study comprised 88 patients with Kellgren-Lawrence osteoarthritis grades 1 to 4 who had 176 3D MRIs twice a year. Six teams provided entries with no differences in any issue segmentation metrics (P = .99) [11].
By joining the segmentation challenge and using the same MRI data as summarized above, we first implemented a 2D multiview encoder-decoder that adaptively fused sagittal, axial, and coronal views to enforce high-level 3D semantic consistency on Knee MRI [31]. Fig. 6 shows our deep network architecture for segmenting knee cartilage. Details of the proposed U-Net variant segmentation algorithm are as follows: our network is U-Net type encoder/decoder architecture with corresponding contracting and expanding paths consisting of dense blocks. There are 10 dense blocks in the network. A growth rate of 12 is used for the dense blocks. There are 6, 8, 11, 15, 19, 19, 14, 10, 7, and 5 convolutions in respective dense blocks. A filter size of 3 × 3 is used for the dense block convolution operations including the final output convolution. First convolution operation uses a 7 × 7 convolution filter size. Dense block convolution operation is followed by ReLU activation and Batch Normalization. There are skip connections between corresponding dense blocks between contracting and expanding paths to enrich the feature richness for the expanding path. Our training learning rate is 0.0001 while we trained for 150 epochs. We used Adam Optimizer with beta1 = 0.9, beta2 = 0.999 and cross entropy loss has been used. We used 2 × 2 pooling for contracting paths with a stride size of 2 × 2 while bilinear interpolation is used for up sampling. For the implementation of the network, we used the TensorFlow library. Fig. 7 shows a sample knee cartilage segmentation result, and our dice scores of femoral, tibial, patellar cartilage, and meniscus, respectively are 87%, 85%, 81%, and 83% (See Fig. 7) [11].
FIG. 6.
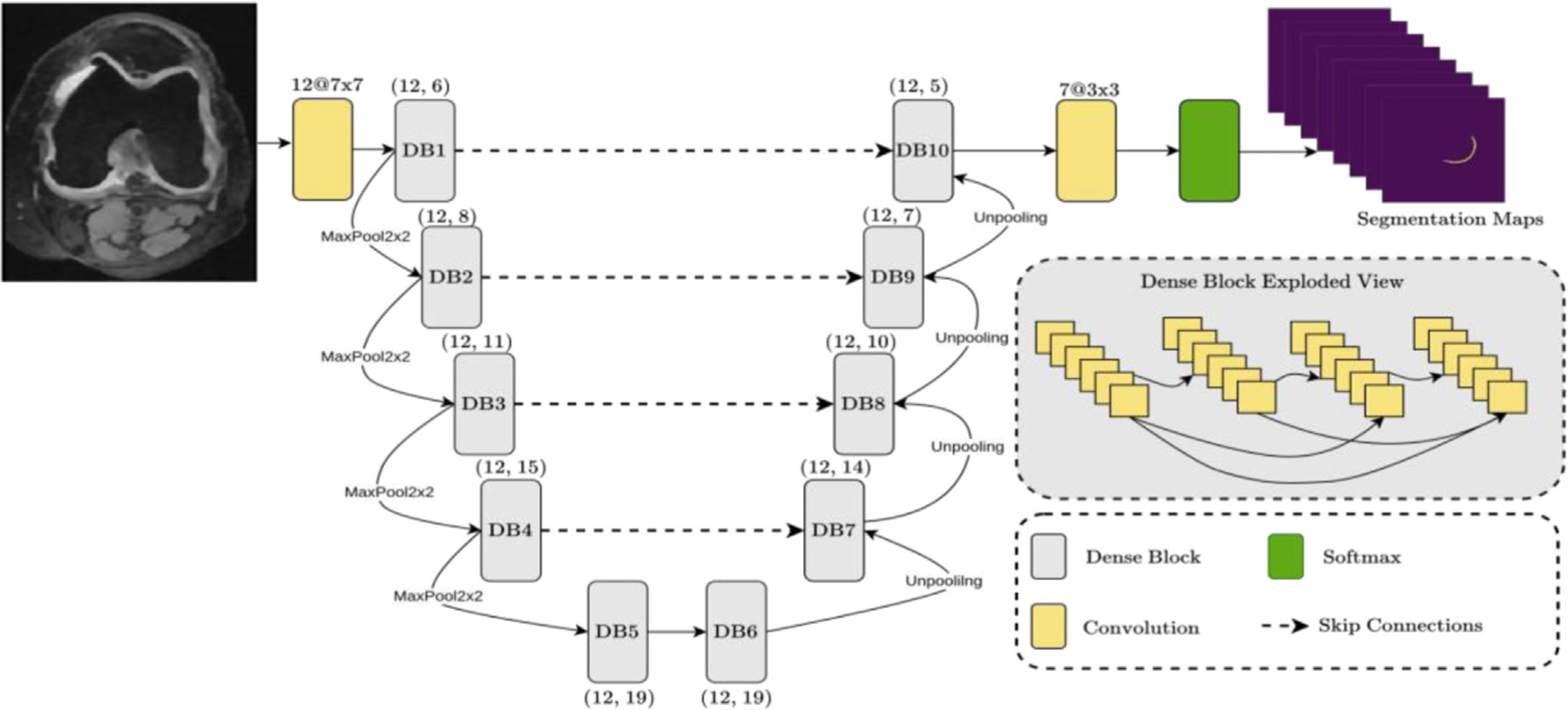
U-Net variant (with attention) architecture for knee cartilage segmentation.
FIG. 7.
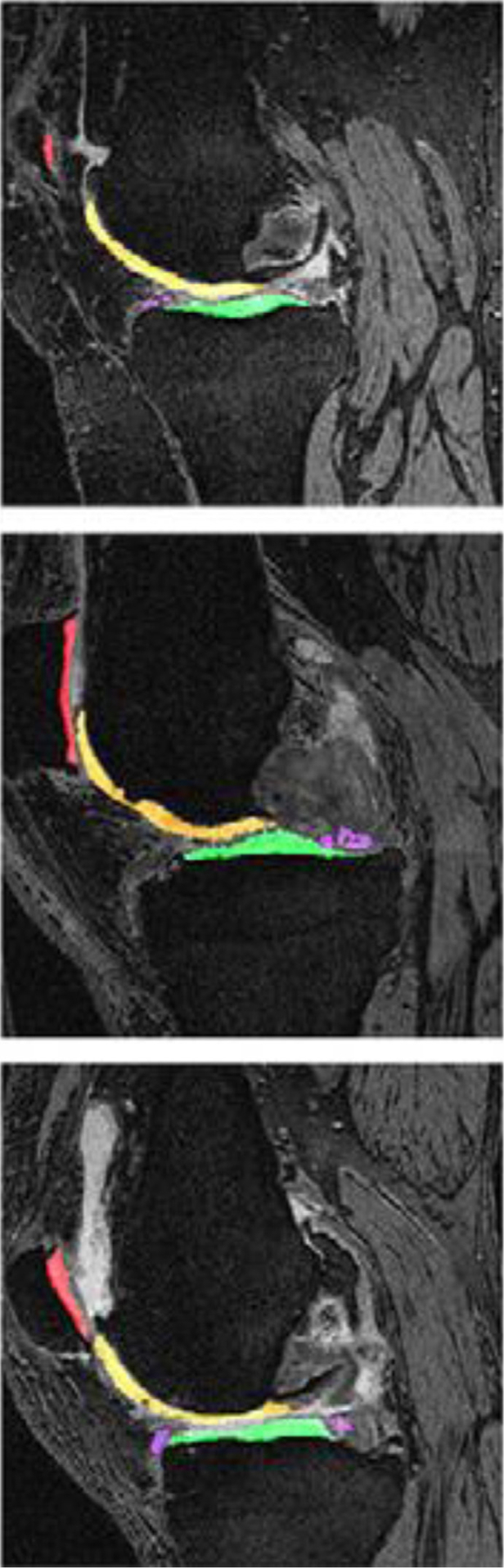
Sample segmentations of the knee in patients with Kellgren-Lawrence osteoarthritis grade 2 to 4 (first, second, and third image, respectively). The following tissues were segmented and colored: femoral cartilage, tibial cartilage (green), patellar cartilage (red), and meniscus (purple).
DISCUSSION AND CONCLUDING REMARKS
We acknowledge the current challenges in MRI segmentation in MSK radiology applications, particularly in the knee and thigh for quantification purposes. We introduce the preprocessing step as a key player in AI algorithm development in MSK applications, especially when MRI is involved. We demonstrate qualitative and quantitative results for tissue segmentation (fat, muscle, bone) from thigh MRI and cartilage segmentation from knee MRI with the U-Net variant as well as capsule-based (SegCaps) segmentation methods, the state-of-the-art algorithms for obtaining the best accuracy and efficiency. Based on the current promising results and trends of AI in MSK applications, we conclude that deep learning has an increased role in radiology AI applications, and specifically in MSK applications, and there is a need for developing better algorithms to mimic unique challenges of MSK imaging such as the complicated interaction of biomechanics of the soft tissues, joints, and bones.
In this article, we briefly demonstrate two important MSK applications but there are other MSK applications worth mentioning that would benefit from AI assistance: task assistance, measuring the length of the bone in the extremities, assessment of muscle volumes, accelerating MRI acquisition, enabling modality-to-modality conversion, improving the quality of images, and others. Deep learning segmentation models (CNNs) have been extensively used on MR imaging of the knee to directly estimate the severity of cartilage disease or abnormalities and the composition of the human body [2]. Using CNNs, Chaudhari and colleagues developed a superresolution method (Deep-Resolve) for thick slice MRI data that improved its usability. In DeepResolve, images of the knee are sliced into 0.7 mm-thick slices. This model was trained on 124 patients from the Osteoarthritis Initiative’s training dataset. DeepResolve was shown to produce high-quality knee MR images that met or exceeded the diagnostic standards of two radiologists and a quantitative assessment [32].
In a multi-view knee MRI dataset of 1370 MRIs, the classification performance of multiple CNN designs were evaluated, and one of the methods was developed by our group in this comparison. Using ResNet-18, AUC was 0.8787 and sensitivity was 0.7855. Using GoogleNET, AUC was 0.8579 and sensitivity was 0.8596. The study’s promising results demonstrate that multiview deep learning-based classification of MSK abnormalities may be used in routine clinical assessment [33].
One limitation of the current AI algorithms for MSK applications is that when using a trained algorithm, it is only possible to evaluate a single diagnosis at a time. Another drawback of the current AI system is that despite the sophistication of many deep learning applications, no deep learning model has yet been described capable of doing complete multisequence joint MRI interpretation (or even single view radiograph interpretation). There is a strong need for considering multimodal imaging and multiobject interpretations because this is a particular need for the daily practice of MSK radiology. Although deep learning can perform at the level of an expert in all 4 categories of activities, imaging investigations are still in the process of being fully understood [6]. Researchers, radiology professionals, and industry leaders are all interested in deep learning, and these developments are expected to impact daily MSK radiology practice soon. After deep learning has been integrated into the clinical workflow, it will be essential to achieve diagnostic accuracy [34]. As our reliance on deep learning expands, radiologists will be expected to have a thorough knowledge of the mechanisms and difficulties associated with AI in the scope of patient assessments [35].
KEY POINTS.
Deep learning has become one of the major approaches in artificial intelligence (AI).
AI implementations in musculoskeletal (MSK) radiology have many unique challenges due to the complication interaction of biomechanics.
AI introduces new stages in MSK radiology, facilitating detection, segmentation, quantification, and preprocessing of radiology scans.
Deep learning is promising in magnetic resonance imaging analysis in MSK applications, overcoming the obstacles of segmentation and quantification without human labor.
DISCLOSURE
Dr U. Bagci discloses Ther-AI LLC. This work is partially supported by the NIH NCI funding: R01-CA246704 and R01-CA240639.
REFERENCES
- [1].Laur O, Wang B. Musculoskeletal trauma and artificial intelligence: current trends and projections. Skeletal Radiol 2022;51(2):257–69. [DOI] [PubMed] [Google Scholar]
- [2].Gorelik N, Gyftopoulos S. Applications of artificial intelligence in musculoskeletal imaging: from the request to the report. Can Assoc Radiol J 2021; 72(1):45–59. [DOI] [PubMed] [Google Scholar]
- [3].Mutasa S, Yi PH. Clinical artificial intelligence applications: musculoskeletal. Radiol Clin North Am 2021; 59(6):1013–26. [DOI] [PubMed] [Google Scholar]
- [4].Mutasa S, Yi PH. Deciphering musculoskeletal artificial intelligence for clinical applications: how do I get started? Skeletal Radiol 2022;51(2):271–8. [DOI] [PubMed] [Google Scholar]
- [5].Strohm L, Hehakaya C, Ranschaert ER, et al. Implementation of artificial intelligence (AI) applications in radiology: hindering and facilitating factors. Eur Radiol 2020;30(10):5525–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chea P, Mandell JC. Current applications and future directions of deep learning in musculoskeletal radiology. Skeletal Radiol 2020;49(2):183–97. [DOI] [PubMed] [Google Scholar]
- [7].Del Grande F, Guggenberger R, Fritz J. Rapid Musculo-skeletal MRI in 2021: value and Optimized Use of Widely Accessible Techniques. AJR Am J Roentgenol 2021;216(3):704–17. [DOI] [PubMed] [Google Scholar]
- [8].Hirschmann A, Cyriac J, Stieltjes B, et al. Artificial intelligence in musculoskeletal imaging: review of current literature, challenges, and trends. Semin Musculoskelet Radiol 2019;23(3):304–11. [DOI] [PubMed] [Google Scholar]
- [9].Anwar SM, Irmakci I, Torigian DA, et al. Semi-supervised deep learning for multi-tissue segmentation from multi-contrast MRI. J Signal Process Syst 2022; 94:497–510. [Google Scholar]
- [10].LaLonde R, Xu Z, Irmakci I, et al. Capsules for biomedical image segmentation. Med Image Anal 2021;68: 101889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Desai AD, Caliva F, Iriondo C, et al. The international workshop on osteoarthritis imaging knee MRI segmentation challenge: a multi-institute evaluation and analysis framework on a standardized dataset. Radiol Artif Intelligence 2021;3(3):e200078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ferrucci L The baltimore longitudinal study of aging (BLSA): a 50-year-long journey and plans for the future. J Gerontol Ser A Biol Sci Med Sci 2008; 63(12):1416–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Aurich V, Weule J. Non-linear Gaussian filters performing edge preserving diffusion. In: Mustererkennung 1995. Springer; 1995. p. 538–45. [Google Scholar]
- [14].Tustison NJ, Avants BB, Cook PA, et al. N4ITK: Improved N3 Bias correction. IEEE Trans Med Imaging 2010;29(6):1310–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nyúl LG, Udupa JK, Zhang X. New variants of a method of MRI scale standardization. IEEE Trans Med Imaging 2000;19(2):143–50. [DOI] [PubMed] [Google Scholar]
- [16].Souza A, Udupa JK, Madabhushi A. Image filtering via generalized scale. Med image Anal 2008;12(2):87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Madabhushi A, Udupa JK. New methods of MR image intensity standardization via generalized scale. Med Phys 2006;33(9):3426–34. [DOI] [PubMed] [Google Scholar]
- [18].Bağcı U, Udupa JK, Bai L. The role of intensity standardization in medical image registration. Pattern Recognition Lett 2010;31(4):315–23. [Google Scholar]
- [19].Bagci U, Chen X, Udupa JK. Hierarchical scale-based multiobject recognition of 3-D anatomical structures. IEEE Trans Med Imaging 2011;31(3):777–89. [DOI] [PubMed] [Google Scholar]
- [20].Xu Z, Gao M, Papadakis GZ, et al. Joint solution for PET image segmentation, denoising, and partial volume correction. Med Image Anal 2018;46:229–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ge Y, Udupa JK, Nyul LG, et al. Numerical tissue characterization in MS via standardization of the MR image intensity scale. J Magn Reson Imaging 2000;12(5):715–21. [DOI] [PubMed] [Google Scholar]
- [22].Ferrucci L The Baltimore Longitudinal Study of Aging (BLSA): a 50-year-long journey and plans for the future. J Gerontol A Biol Sci Med Sci 2008. Dec;63(12):1416–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Liu F, Zhou Z, Jang H, et al. Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med 2018;79(4):2379–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu F, Zhou Z, Samsonov A, et al. Deep learning approach for evaluating knee MR images: achieving high diagnostic performance for cartilage lesion detection. Radiology 2018;289(1):160–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pedoia V, Lee J, Norman B, et al. Diagnosing osteoarthritis from T2 maps using deep learning: an analysis of the entire Osteoarthritis Initiative baseline cohort. Osteoarthritis Cartilage 2019;27(7):1002–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Badrinarayanan V, Kendall A, Cipolla R. Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell 2017;39(12):2481–95. [DOI] [PubMed] [Google Scholar]
- [27].Heimann T, Morrison BJ, Styner MA, Niethammer M, Warfield S. Segmentation of knee images: a grand challenge. Paper presented at: Proc. MICCAI Workshop on Medical Image Analysis for the Clinic 2010. [Google Scholar]
- [28].Tack A, Mukhopadhyay A, Zachow S. Knee menisci segmentation using convolutional neural networks: data from the osteoarthritis initiative. Osteoarthritis Cartilage 2018;26(5):680–8. [DOI] [PubMed] [Google Scholar]
- [29].Deniz CM, Xiang S, Hallyburton RS, et al. Segmentation of the proximal femur from MR images using deep convolutional neural networks. Sci Rep 2018;8(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhou Z, Zhao G, Kijowski R, et al. Deep convolutional neural network for segmentation of knee joint anatomy. Magn Reson Med 2018;80(6):2759–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mortazi A, Karim R, Rhode K, Burt J, Bagci U. Cardiac-NET: Segmentation of left atrium and proximal pulmonary veins from MRI using multi-view CNN. Paper presented at: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, Cham, 2017. pp. 377–385. [Google Scholar]
- [32].Chaudhari AS, Fang Z, Kogan F, et al. Super-resolution musculoskeletal MRI using deep learning. Magn Reson Med 2018;80(5):2139–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Irmakci I, Anwar SM, Torigian DA, Bagci U. Deep learning for musculoskeletal image analysis. Paper presented at: 2019 53rd Asilomar Conference on Signals, Systems, and Computers2019. [Google Scholar]
- [34].Shin Y, Kim S, Lee YH. AI musculoskeletal clinical applications: how can AI increase my day-to-day efficiency? Skeletal Radiol 2022;51(2):293–304. [DOI] [PubMed] [Google Scholar]
- [35].Gorelik N, Chong J, Lin DJ. Pattern Recognition in Musculoskeletal Imaging Using Artificial Intelligence. Semin Musculoskelet Radiol 2020;24(1):38–49. [DOI] [PubMed] [Google Scholar]


