Abstract
Z-RNA is a higher-energy, left-handed conformation of RNA, whose function has remained elusive. A growing body of work alludes to regulatory roles for Z-RNA in the immune response. Here, we review how Z-RNA features present in cellular RNAs—especially containing retroelements—could be recognized by a family of winged helix proteins, with an impact on host defense. We also discuss how mutations to specific Z-contacting amino acids disrupt their ability to stabilize Z-RNA, resulting in functional losses. We end by highlighting knowledge gaps in the field, which, if addressed, would significantly advance this active area of research.
Keywords: adenosine deaminase acting on RNA (ADAR1), E3L, innate immune response, retroelement, Zα, Z-D/RNA-binding protein 1 (ZBP1), Z-RNA
INTRODUCTION
The perceived relevance of left-handed double-stranded RNA (dsRNA) is currently undergoing a paradigm shift. Historical studies demonstrated that some regions within RNA molecules may adopt a left-handed conformation under certain high salt conditions (Hall et al. 1984), similarly to DNA (Jovin et al. 1987). Under more physiological conditions, this zig-zag double helix called “Z-RNA” can be achieved, for example, if the RNA is modified at certain positions (Uesugi et al. 1984; Nakamura et al. 1985; Rao and Kollman 1986; Teng et al. 1989). The overall unstable character of Z-RNA has raised concerns about its biological relevance that endure to this day.
Over time though, observations were made that have begun to lift the controversy over the existence of Z-RNA in cells. In particular, out of the many proteins recognizing nucleic acids, several recognize Z-conformations of DNA and RNA specifically using a similar winged helix Zα domain (Gajiwala and Burley 2000; Placido et al. 2007; Zhang et al. 2020). Notably, these Z-binding proteins exclusively participate in viral infections and the innate immune response (Athanasiadis 2012). RNA that binds to antibodies raised against Z-RNA was detected in the cytoplasm, and Z-binding proteins were reported in stress-related cytoplasmic membraneless condensates (Zarling et al. 1987; Ng et al. 2013). Most recently, altering the ability of these proteins to recognize Z-RNA in cancer cell lines and mouse models was shown to be associated with hyper-inflammation phenotypes (de Reuver et al. 2022; Hubbard et al. 2022; Zhang et al. 2022). What emerges from this collection of findings is evidence for a regulatory role of this transient conformation of RNA, particularly when cells have to defend against invaders.
In this mini-review, we assess how our knowledge of Z-RNA formation and stabilization in vitro could be applied to support the presence of Z-RNA in cellular and viral RNAs. We also examine how Z-RNA fits within innate immune response mechanisms and how certain viruses bypass these. We end by highlighting a few questions that, if addressed, could help propel this latecomer of a field even further.
WHAT DID WE LEARN FROM STUDYING Z-RNA IN VITRO?
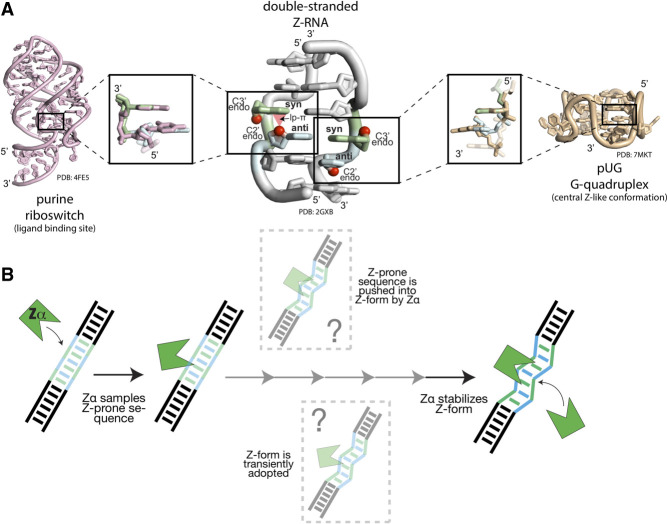
Double-stranded Z-RNA, like Z-DNA, is a succession of nucleotides with nucleobases alternating between syn/anti conformations, and sugars alternating between C3′/C2′-endo conformations (Fig. 1A; Wang et al. 1979, 1981; Hall et al. 1984; Ho and Mooers 1997). This arrangement leads to a lone pair–π contact only found within Z geometry (involving the O4′ of the C2′-endo sugar and the syn base, Kruse et al. 2020; Zirbel and Auffinger 2022). While most of the in vitro characterization of Z-RNA was carried out with GC-rich dsRNAs (Hall et al. 1984; Davis et al. 1990; Placido et al. 2007), Z-form geometry can also be supported by other sequences and other structural contexts. Z-RNA was, for example, proposed (Feng et al. 2011; Herbert 2019) and demonstrated (Nichols et al. 2021) to form within GU- and AU-rich dsRNAs belonging to inverted Alu repeat foldbacks and ribosomal RNA stem–loops. Furthermore, Z-RNA geometry was observed in non-GC single-stranded regions important for function within riboswitches, ribozymes and other structured RNAs with known functions (D'Ascenzo et al. 2016), as well as within atypical quadruplexes (Fig. 1A; Roschdi et al. 2022). These findings suggest that certain regions within cellular RNAs (i.e., alternating purine-pyrimidine sequences) could be prone to adopt Z conformations.
FIGURE 1.
Left-handed Z-conformations can be found within a variety of RNA functional sites. (A) Double-stranded Z-RNA (middle) is best described as being composed of dinucleotide building blocks, where the nucleobase alternates between the anti- and syn-conformations, along with alternating ribose sugar puckers (C2′-endo/C3′-endo conformations). The weak but characteristic lone pair–π contact is shown as a red triangle. The particular Z-geometry with sugar pointing in opposite directions is found in other structural contexts, such as, for example, the ligand binding site of the purine riboswitch (PDB: 4FE5, Batey et al. 2004), and the core of the pUG G-quadruplex (PDB: 7MKT, Roschdi et al. 2022). (B) Potential mechanisms for Z-RNA formation and stabilization in vivo. First, Zα binds nonspecifically to A-RNA near a Z-prone region. From here, Zα may allosterically push the Z-prone region into the Z-conformation or may have to stabilize transiently sampled Z-conformations. Each of these mechanisms may involve one or more intermediate steps.
How is double-stranded Z-RNA achieved and stabilized in solution? Z-RNA is a higher-energy, and thus, unstable conformation of RNA, which requires stabilization by high salt concentrations or protein binding, for example (Hall et al. 1984; Davis et al. 1986; Brown et al. 2000; Bae et al. 2013). Single-molecule FRET studies reported that Z-conformations are sampled transiently, supporting a conformational capture model (Bae et al. 2011). However, other evidence suggests that Z-forming RNAs (and DNAs) adopt multiple intermediate states between the initial binding of a winged helix Zα domain to A- (or B-) form followed by Z formation (Kang et al. 2009; Lee et al. 2011, 2016, 2019). Therefore, a hybrid model may be more suitable to explain Z-RNA formation in some cases, wherein an active binding event between Zα and a Z-conformation-forming sequence pushes the nucleic acid into an intermediate state that Zα then locks into the Z-conformation (Fig. 1B; Kim et al. 2018b). The relatively wide range of dissociation constants reported (from ∼1 nM to low µM; Schade et al. 1999a; Kang et al. 2014; Nichols et al. 2021) between nucleic acids (DNA has been more studied than RNA) and Z-binding proteins could be accounted for by differences in sequence and structural contexts.
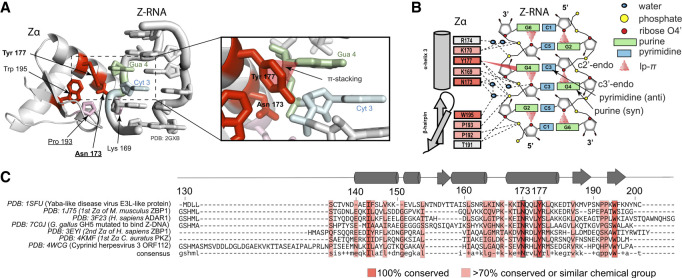
Z-RNA recognition requires Zα to make some key contacts with Z-RNA structural features (Brown et al. 2000; Placido et al. 2007). Notably, Tyr 177 (human ADAR1 numbering) is critical as it allows for the recognition of the nucleotide in the syn conformation (Fig. 2A,B; Schwartz et al. 1999; Placido et al. 2007). Asn 173 and Pro 193 contact the Z-RNA phosphate backbone, and they form interaction networks at the RNA–protein interface (Fig. 2A,B; Schwartz et al. 1999; Placido et al. 2007). These amino acids are all >95% conserved in Zα domains solved in complex with Z-DNA or Z-RNA (Fig. 2C), further hinting at their importance for Z-RNA recognition. In addition, in vitro site-directed mutagenesis of Asn 173 to Ala, or Tyr 177 to any other amino acid, causes a complete loss in the ability of Zα to stabilize the Z-conformation (Feng et al. 2011; Jeong et al. 2014; Kim et al. 2014; Nichols et al. 2021), while leaving the protein fold intact (Feng et al. 2011). Similar mutations in vivo alter the normal function of these proteins (see below).
FIGURE 2.
Z-RNA is a natural ligand of Zα domains. (A) Zα amino acids interacting with a r(CpG)3 duplex (PDB ID: 2GXB). Underlined residues are mutated in cases of Aicardi Goutières syndrome (Herbert 2019). (B) Cartoon schematic of the key residues of Zα and their interactions with the r(CpG)3 duplex in the Z-conformation. (C) Sequence alignment between Zα domains from different Zα-containing proteins and species. Only sequences from Zα domains with structures solved in complex with Z-DNA or Z-RNA were used for the alignment. The residue numbering is based off ADAR1 Zα. Consensus sequence: uppercase, 100% conserved; lowercase, one letter with high frequency; +, two letters with high-frequency; −, no consensus.
If Z-RNA could be adopted by some RNAs in the cell—even transiently—its recognition by Zα-containing proteins like ADAR1 (adenosine deaminase acting on RNA 1) and ZBP1 (Z/D-RNA binding protein 1) would explain how it could be stabilized in vivo. Because Zα domains stabilize Z-RNA by decreasing the energetic penalties of Z-form adoption through binding to the unique left-handed shape (Fig. 2A,B; Ha et al. 2008), any cellular condition favoring sampling of Z-conformations or Z-RNA stabilization would encourage Zα-containing proteins to accumulate on those RNAs, according to the relative energy costs of the targeted sequence. Here, it is tempting to speculate that the relatively immobile RNA “tangles” due to RNA–RNA interactions within biological condensates (Anderson and Kedersha 2006; Protter and Parker 2016; Van Treeck et al. 2018) may induce Z-RNA formation through torsional and/or mechanical stress (potentially through eIF4A recruitment and activity within stress granules, Tauber et al. 2020), as widely seen for Z-DNA in other contexts (Yi et al. 2022). In general, helicase activity as generated by dsRNA sensors or viral helicases could facilitate Z-RNA adoption (Herbert 2021). Z-RNA formation in condensates is consistent with the observation that functional Z-recognition amino acids are required and sufficient to localize Zα-containing proteins to condensates (Ng et al. 2013; Gabriel et al. 2021). Finally, other factors stabilizing Z-RNA in vitro comprise methylation or oxidation, which are chemical modifications that may be relevant to also account for Z-RNA stabilization in cells. These modifications can destabilize A-form RNA and provoke translational errors (Dai et al. 2018).
Z-RNA WITHIN PAIRED RETROELEMENTS
Recent studies have begun to sort out which RNA molecules may adopt Z conformations in cells, through pulldown experiments involving Z-RNA antibodies or Zα domains typically from ADAR1 and ZBP1 (Schwartz et al. 1999; Ha et al. 2008; Athanasiadis 2012). Pulldown experiments were performed using a Z-RNA antibody in mouse embryonic fibroblast cells, where ADAR1 and ZBP1 had been knocked out to prevent shielding of potential Z-RNA targets (Zhang et al. 2022). Anti-Z-DNA/RNA antibodies have not been tested for cross-reactivity to unique RNA structural folds but have been thoroughly shown to have no cross-reactivity with B-DNA or A-RNA (Zarling et al. 1990; Zhang et al. 2022). Zhang and colleagues uncovered mostly protein-coding mRNAs that contained short interspersed nuclear elements (SINE) foldbacks in their 3′ untranslated regions (3′UTRs) (Alu elements in primates [Chung et al. 2018; Sun et al. 2021]), and B1, B2, and B4 repeats in mice (de Reuver et al. 2022; Zhang et al. 2022). Another study carried out ZBP1 pulldowns, revealing endogenous dsRNAs that were speculated to be self-complementary B2 and Alu elements (Jiao et al. 2020). These experimental results gave further support to earlier reports that SINEs are heavily edited by ADAR1 (Athanasiadis et al. 2004; Levanon et al. 2004).
Pulldown approaches have also helped identify possible Z-forming RNAs in other contexts, such as putative dumbbell structures from 3′-UTRs with presumed Zα-binding sites (Zhang et al. 2022), as well as all eight segments of the influenza A virus (IAV) genome within IAV-infected cells (Thapa et al. 2016; Zhang et al. 2020). Together, these findings suggest that Z-RNA formation within certain dsRNA regions—especially belonging to paired retroelements—would help target ADAR1, ZBP1, and other potentially Zα-containing proteins to these RNAs (Balachandran and Mocarski 2021). However, binding as inferred from pulldown experiments does not rule out the possibility of Z-RNA-independent binding, especially since positively charged Zα domains could bind A-form RNA nonspecifically with relatively low affinity (control pulldowns with mutant Zα domains that cannot stabilize Z-RNA would help distinguish true Z-RNA targets).
To further support Z-RNA adoption within retroelements as necessary for specific recognition by Zα domains, A-to-I editing profiles were compared between wild-type ADAR1 and ADAR1 point mutants in which Z-RNA binding was impaired. Consistently with prior findings that mapped editing sites to introns and untranslated regions (Athanasiadis et al. 2004; Kim et al. 2004; Levanon et al. 2004), differential editing sites localize to introns and 3′-UTRs, within SINEs and in close proximity to inverted SINEs, allowing for the formation of dsRNA foldbacks (Chung et al. 2018; de Reuver et al. 2021; Tang et al. 2021). These results are consistent with findings from the direct Z-RNA pulldown experiments. Surprisingly, Zα-dependent editing sites make up a minority (∼8%) of the total ADAR1p150 editing in the cell (Tang et al. 2021), but this small number of sites appears to be crucial for preventing MDA5 (melanoma differentiation-associated protein 5) (Nakahama et al. 2021) activation, the mechanism of which is poorly understood. Furthermore, mutating the Zα domain causes an increase in editing near the predicted ends of the dsRNA regions, but a decrease in editing closer to regions predicted to form Z-RNA, which could suggest that Z-RNA forming regions recruit Zα for specific editing of certain RNAs (Nakahama et al. 2021), as predicted earlier (Koeris et al. 2005). Further work is needed to determine whether Zα binding sites overlap with known Zα-dependent editing sites, or if Zα-binding protects certain regions from editing, and to generally localize and validate Z-RNA forming elements within 3′UTRs.
A bias for Z-RNA in SINEs and other repetitive elements could be advantageous for the cell, as SINEs represent a large source of endogenous dsRNA. Targeting proteins with Zα domains to these RNAs could ultimately help the cell better differentiate self- from non-self RNAs (Uggenti and Crow 2018). Additionally, repeat elements may contain sequence biases which make them more prone to adopt Z-RNA as compared to other RNAs, such as the putative Z-Box (a sequence with pyrimidine-purine repeats) within Alu elements (Herbert 2020). In any case, regions switching to Z-RNA within larger RNAs would generate A–Z junctions (Z-RNA within the context of a larger A-form helix) (Kim et al. 2009; Nichols et al. 2021), in a similar fashion to the B–Z junctions observed in DNA (Kim et al. 2018a).
A BIOLOGICAL FUNCTION FOR Z-RNA: RECRUITING WINGED HELIX PROTEINS TO MODULATE dsRNA SENSOR ACTIVATION
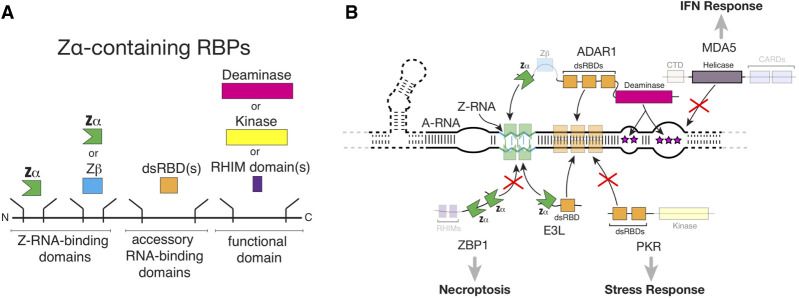
One of the central mechanisms of the innate immune system is the sensing of viral infection by recognizing foreign dsRNA in the cytoplasm (Hur 2019). In mammals, this is primarily carried out through dsRNA sensors (Fig. 3A,B). In particular, the retinoic acid-inducible gene-like receptors (RLR) family of dsRNA sensors, including RIG-I and MDA5, trigger the interferon (IFN) pathway after sensing dsRNA via their helicase domain (Hur 2019). In parallel, protein kinase R (PKR) senses dsRNA to initiate stress granule formation and translational shutdown (Anderson and Kedersha 2006; Protter and Parker 2016; Hur 2019). Another dsRNA sensor is ZBP1, which is unique in that it senses dsRNA via two Zα domains (Hur 2019; Jiao et al. 2020; Balachandran and Mocarski 2021). ZBP1 initiates apoptosis and necroptosis cell death pathways (Snyder and Oberst 2021) through its RHIM domains (Fig. 3A,B). When the innate immune system needs to be turned off, ADAR1 inhibits these responses through shielding of dsRNA regions, preventing ZBP1 and PKR activation (Fig. 3B; Okonski and Samuel 2013; Chung et al. 2018; de Reuver et al. 2021, 2022; Karki et al. 2021; Zhang et al. 2022). A general consensus is that ADAR1 also helps stop MDA5 activation by converting adenosines to inosines in dsRNA regions, which weakens their helical structure (Fig. 3B; Okonski and Samuel 2013; Mannion et al. 2014; Pestal et al. 2015; Yu et al. 2015; Ahmad et al. 2018; Chung et al. 2018; de Reuver et al. 2021; Nakahama et al. 2021).
FIGURE 3.
Competition for Z-RNA by dsRNA sensors modulates the innate immune response. (A) General domain architecture of Zα-containing RBPs, all of which have one or more Zα domains on their amino terminus, followed by one or more dsRBDs, and finally by a functional domain (such as a deaminase or kinase domain). (B) Summary of our current understanding of the interactions between Zα-containing proteins and Z-RNA within a hypothetical folded molecule, and how these interactions modulate pathways that depend on dsRNA sensor activation. Direct shielding of dsRNA and Z-RNA by viral E3L as well as host ADAR1 proteins prevent recognition by ZBP1 and PKR, preventing activation of necroptosis and the stress response. In addition, editing of dsRNA by ADAR1 prevents MDA5 activation. Abbreviations are explained within the text, except for CARDs (caspase recruitment domains), which mediate interactions with downstream signaling proteins, and RHIM (receptor-interaction protein [RIP] homotypic interaction motifs).
Many viruses, including those from the influenza, poxvirus and herpesvirus families, have evolved strategies to avoid the innate immune response pathways so they can successfully replicate their genomes (Balachandran and Mocarski 2021). Of particular interest, these viruses use viral proteins with their own Zα domains, including E3L (or ORF112 in fish herpesviruses, Kuś et al. 2015), whose primary function is to outcompete ZBP1's Zα domains for binding to Z-RNA (Koehler et al. 2021), thereby inhibiting the necroptosis signaling pathway (Fig. 3B; Koehler et al. 2017; Balachandran and Mocarski 2021). In fact, both E3L and ORF112 have been reported to outcompete binding of a Z-RNA antibody to its epitope (Diallo et al. 2022). E3L and E3L-like proteins are particularly important for preventing ZBP1 activation during early viral infection when viral RNAs are being heavily transcribed (Koehler et al. 2021). Interestingly, the Zα and dsRNA binding domains (dsRBDs) of E3L and E3L-like proteins act cooperatively to inhibit dsRNA sensor activation. Mutating the Zα domain but not the dsRBD causes enhanced ZBP1 activation and increased signal from a Z-RNA antibody, suggesting Z-RNA accumulation (Koehler et al. 2021).
POINT MUTATIONS TO Zα DOMAINS CAUSE ABERRANT BIOLOGICAL FUNCTIONS
Mutations to Zα prevent cells from turning off the innate immune response, and such mutations have been observed in human diseases. For example, the N173S or P193A mutations within the Zα domain of ADAR1 result in the interferonopathies known as bilateral striatal necrosis/dystonia and Aicardi Goutières syndrome (Herbert 2019; Rice et al. 2012). These diseases are characterized by a deficiency in ADAR1 function, causing MDA5-dependent interferon signaling and spontaneous ZBP1 activation, ultimately causing cell death (Rice et al. 2012; Herbert 2019; Nakahama et al. 2021). While not observed in human diseases, mutations of Tyr 177 or Trp 195 (critical for stabilizing the Zα core [Fig. 2A,B; Schade et al. 1999b; Schwartz et al. 1999; Placido et al. 2007; Kim et al. 2014]) are also known to result in spontaneous MDA5- and ZBP1-dependent interferon production, embryonic death, and developmental defects in mice (Pestal et al. 2015; de Reuver et al. 2021, 2022; Karki et al. 2021; Nakahama et al. 2021; Tang et al. 2021; Zhang et al. 2022). Mutations of Asn 122 and Tyr 126 (analogous to Asn 173 and Tyr 177 in ADAR1) within Zα2 of ZBP1 similarly disrupt biological function. These mutations prevent ZBP1 from binding to retroelements, whose expression is increased upon viral infection and other stressors (Wang et al. 2020; Karki et al. 2021; de Reuver et al. 2022; Zhang et al. 2022), and abolish necroptosis signaling in the presence of viral infection (Thapa et al. 2016; Zhang et al. 2020; Balachandran and Mocarski 2021). Thus, in vivo ZBP1 mutants allow certain viruses to replicate unhindered.
Point mutations within Zα domains also disrupt Zα localization to biological condensates (Ng et al. 2013; Gabriel et al. 2021). For example, mutating Z-RNA-stabilizing amino acids within ORF112 leads to a disruption of the liquid–liquid phase separation necessary for the formation of condensates (Diallo et al. 2022). Mutating Lys 169 or Tyr 177 to Ala in the Zα domain of ADAR1 or the equivalent residues in E3L (Lys 40 and Tyr 48) leads to impaired localization of ADAR1 and E3L to condensates (Ng et al. 2013). Similarly, double mutants of the two Zα domains of human ZBP1 (Asn 46/Tyr 50 and Asn 141/Tyr 145 to Ala) affect ZBP1 localization, leading to a loss of binding to other nucleic acid-binding proteins through RNA–protein mediated contacts (Gabriel et al. 2021). Although these mutations have not been observed in diseases so far, these observations hint at further biological functions associated with RNA recognition by Zα in the process of condensate formation and regulation.
Finally, point mutants within viral Zα domains disrupt viral function, usually to the benefit of the infected host. In particular, mutation of the conserved Asn and Tyr amino acids within the E3L Zα domain from Vaccinia virus results in the accumulation of available targets for ZBP1, leading to activation of necroptosis (Koehler et al. 2017, 2021; Balachandran and Mocarski 2021). Although E3L contains a dsRBD (Fig. 3B), disrupting its ability to recognize Z-RNA renders it unable to properly shield its viral RNAs, leading to a robust innate immune response through ZBP1 (Koehler et al. 2021). Thus, Zα-containing proteins require functional Z-RNA-stabilizing residues to properly function. While not direct proof, these findings are highly suggestive that the RNA targets of these proteins contain Z-RNA fragments.
WHAT'S NEXT?
The existence of winged helix proteins that specifically recognize the left-handed Z-form geometry suggests that Z-RNA plays a role in biology. These proteins compete for binding to RNA, which directly impacts the innate immune response. In particular, an imbalance between ADAR1 and ZBP1 leads to inflammation and autoimmune diseases. However compelling the evidence for Z-RNA adoption in cells, it is important to point out that many of our current insights are indirect. Furthermore, it seems likely that stretches of dsRNAs would adopt Z-conformations in response to changes in the cellular environment. But figuring out exactly which sequence and structure would switch to Z-RNA, and under what conditions they would do so, remains an important future direction of research. We propose below a few directions for tackling these and additional aspects pertaining to a now alluring Z-RNA biology.
Do Zα domains actually bind to Z-RNA in cells?
Z-conformations have been investigated in cells through the binding of Z/D-RNA recognizing domains and antibodies, which offer an indirect read-out of Z-conformations. In addition, these proteins may be inducing a Z-conformation rather than capturing it (which would nonetheless support the hypothesis that Z-RNA can form in cells). Methodology which could directly identify Z-conformations in cells would push the field forward significantly. Currently, no direct method is available to conduct in-cell structural studies at the required resolution. Therefore, the best possible way to gain information about RNA structure in cells is to infer it from in vitro studies, as is routinely and successfully done in RNA structural biology (Cruz and Westhof 2009; Vicens and Kieft 2022; Xue et al. 2022).
What regions adopt a Z-RNA conformation?
The experiments attempting to identify Z-RNA in cells have mostly consisted of pulldowns using Z-conformation antibodies or Zα-containing proteins, which do not give the resolution needed to identify the specific sites being bound. Experiments such as HITS-CLIP-Seq or PAR-CLIP-Seq (Hafner et al. 2021) would help to identify the targeted Z-RNA regions. In addition to revealing what sequences adopt a Z-conformation in cells, such approaches would help correlate, for example, Zα binding sites to A-to-I editing events mediated by ADAR1.
What cellular conditions promote Z-RNA?
We do not fully understand the role of different cellular conditions and environments in Z-RNA adoption. Future work to explore if Z-conformations are enriched in certain areas under specific conditions (such as the stress response) would help answer this question. The creation of a Z-RNA reporter RNA that would give a particular signal when in the Z-conformation would be useful for such studies. This reporter could theoretically be localized to different cellular environments, including condensates, to monitor where Z-RNA is adopted.
What does Z-RNA look like within the context of full Zα-containing proteins?
Most of our current knowledge comes from biochemical and biophysical studies of Zα domains in isolation bound to GC-rich RNAs. However, Zα domains are not always found as the only RNA binding domain within RNA binding proteins, suggesting they may have different sequence specificities in various proteins. Additional biochemical and structural studies would be very informative to elucidate how Zα contributes to the binding of ZBP1/ADAR1 to larger RNA substrates (such as fragments of SINEs), in particular to reveal the structure of A–Z junctions.
How large is the repertoire of Z-RNA-recognizing proteins?
Additional protein families like PKZ in fish have been studied that contain Zα domains (Rothenburg et al. 2005; Kim et al. 2014; Xu et al. 2019). This finding of a PKR analog with Zα domains shows that recognition of Z-RNA is potentially an evolutionarily widespread mechanism for dealing with viral infections. Putative Zα domains have also been identified in mammals, fish and single-celled eukaryotes (Grice and Degnan 2015; Bartas et al. 2022). A rigorous biochemical and structural characterization of these proteins would be worthwhile, to expose further clues about Z-RNA recognition. A wider pool of Zα domains may offer additional model systems for exploring Z-RNA biology, as exemplified most recently with a study of ORF112 from a fish herpesvirus (Diallo et al. 2022).
When does Z-RNA play a role in the innate immune response?
Host Zα-containing proteins are all interferon-induced, while those from viruses are expressed heavily during infection. So, is it that Z-RNA accumulates during the innate immune response because of the surge of Zα domains, or that innate immune response proteins contain Zα domains because Z-RNA becomes prevalent during the interferon response due to other mechanisms? Further studies would also be needed to illuminate the interplay between the editing-related and editing-unrelated roles of Zα domains.
ACKNOWLEDGMENTS
We gratefully acknowledge Jillian Ramos, Conner Langeberg, and Jeffrey Kieft for careful reading of earlier versions of the manuscript; Jeffrey Kieft for support (to P.N. and Q.V.); the National Science Foundation for support (award #2153787 to Q.V. and B.V.); and the National Institutes of Health for support (award #1F31AI167396-01 to P.N.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079429.122.
Freely available online through the RNA Open Access option.
REFERENCES
- Ahmad S, Mu X, Yang F, Greenwald E, Park JW, Jacob E, Zhang CZ, Hur S. 2018. Breaching self-tolerance to Alu duplex RNA underlies MDA5-mediated inflammation. Cell 172: 797–810. 10.1016/j.cell.2017.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. 2006. RNA granules. J Cell Biol 172: 803–808. 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadis A. 2012. Zα-domains: at the intersection between RNA editing and innate immunity. Semin Cell Dev Biol 23: 275–280. 10.1016/j.semcdb.2011.11.001 [DOI] [PubMed] [Google Scholar]
- Athanasiadis A, Rich A, Maas S. 2004. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2: e391. 10.1371/journal.pbio.0020391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S, Kim D, Kim KK, Kim YG, Hohng S. 2011. Intrinsic Z-DNA is stabilized by the conformational selection mechanism of Z-DNA-binding proteins. J Am Chem Soc 133: 668–671. 10.1021/ja107498y [DOI] [PubMed] [Google Scholar]
- Bae S, Kim Y, Kim D, Kim KK, Kim YG, Hohng S. 2013. Energetics of Z-DNA binding protein-mediated helicity reversals in DNA, RNA, and DNA-RNA duplexes. J Phys Chem B 117: 13866–13871. 10.1021/jp409862j [DOI] [PubMed] [Google Scholar]
- Balachandran S, Mocarski ES. 2021. Viral Z-RNA triggers ZBP1-dependent cell death. Curr Opin Virol 51: 134–140. 10.1016/j.coviro.2021.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartas M, Slychko K, Brázda V, Červeň J, Beaudoin CA, Blundell TL, Pečinka P. 2022. Searching for new Z-DNA/Z-RNA binding proteins based on structural similarity to experimentally validated Zα domain. Int J Mol Sci 23: 768. 10.3390/ijms23020768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Gilbert SD, Montange RK. 2004. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432: 411–415. 10.1038/nature03037 [DOI] [PubMed] [Google Scholar]
- Brown BA II, Lowenhaupt K, Wilbert CM, Hanlon EB, Rich A. 2000. The Zα domain of the editing enzyme dsRNA adenosine deaminase binds left-handed Z-RNA as well as Z-DNA. Proc Natl Acad Sci 97: 13532–13536. 10.1073/pnas.240464097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Calis JJA, Wu X, Sun T, Yu Y, Sarbanes SL, Dao Thi VL, Shilvock AR, Hoffmann HH, Rosenberg BR, et al. 2018. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell 172: 811–824. 10.1016/j.cell.2017.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz JA, Westhof E. 2009. The dynamic landscapes of RNA architecture. Cell 136: 604–609. 10.1016/j.cell.2009.02.003 [DOI] [PubMed] [Google Scholar]
- D'Ascenzo L, Leonarski F, Vicens Q, Auffinger P. 2016. “Z-DNA like” fragments in RNA: a recurring structural motif with implications for folding, RNA/protein recognition and immune response. Nucleic Acids Res 44: 5944–5956. 10.1093/nar/gkw388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai DP, Gan W, Hayakawa H, Zhu JL, Zhang XQ, Hu GX, Xu T, Jiang ZL, Zhang LQ, Hu XD, et al. 2018. Transcriptional mutagenesis mediated by 8-oxoG induces translational errors in mammalian cells. Proc Natl Acad Sci 115: 4218–4222. 10.1073/pnas.1718363115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis PW, Hall K, Cruz P, Tinoco I, Neilson T. 1986. The tetraribonucleotide rCpGpCpG forms a left-handed Z-RNA double-helix. Nucleic Acids Res 14: 1279–1291. 10.1093/nar/14.3.1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis PW, Adamiak RW, Tinoco I Jr. 1990. Z-RNA: the solution NMR structure of r(CGCGCG). Biopolymers 29: 109–122. 10.1002/bip.360290116 [DOI] [PubMed] [Google Scholar]
- de Reuver R, Dierick E, Wiernicki B, Staes K, Seys L, De Meester E, Muyldermans T, Botzki A, Lambrecht BN, Van Nieuwerburgh F, et al. 2021. ADAR1 interaction with Z-RNA promotes editing of endogenous double-stranded RNA and prevents MDA5-dependent immune activation. Cell Rep 36: 109500. 10.1016/j.celrep.2021.109500 [DOI] [PubMed] [Google Scholar]
- de Reuver R, Verdonck S, Dierick E, Nemegeer J, Hessmann E, Ahmad S, Jans M, Blancke G, Van Nieuwerburgh F, Botzki A, et al. 2022. ADAR1 prevents autoinflammation by suppressing spontaneous ZBP1 activation. Nature 607: 784–789. 10.1038/s41586-022-04974-w [DOI] [PubMed] [Google Scholar]
- Diallo MA, Pirotte S, Hu Y, Morvan L, Rakus K, Suarez NM, PoTsang L, Saneyoshi H, Xu Y, Davison A, et al. 2022. A fish herpesvirus highlights functional diversities among Zα domains related to phase separation induction and A-to-Z conversion. Nucleic Acids Res gkac761. 10.1093/nar/gkac761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Li H, Zhao J, Pervushin K, Lowenhaupt K, Schwartz TU, Dröge P. 2011. Alternate rRNA secondary structures as regulators of translation. Nat Struct Mol Biol 18: 169–176. 10.1038/nsmb.1962 [DOI] [PubMed] [Google Scholar]
- Gabriel L, Srinivasan B, Kus K, Mata JF, Amorim MJ, Jansen LET, Athanasiadis A. 2021. Enrichment of Zα domains at cytoplasmic stress granules is due to their innate ability to bind to nucleic acids. J Cell Sci 134: jcs258446. 10.1242/jcs.258446 [DOI] [PubMed] [Google Scholar]
- Gajiwala KS, Burley SK. 2000. Winged helix proteins. Curr Opin Struct Biol 10: 110–116. 10.1016/S0959-440X(99)00057-3 [DOI] [PubMed] [Google Scholar]
- Grice LF, Degnan BM. 2015. The origin of the ADAR gene family and animal RNA editing. BMC Evol Biol 15: 4. 10.1186/s12862-015-0279-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha SC, Kim D, Hwang HY, Rich A, Kim YG, Kim KK. 2008. The crystal structure of the second Z-DNA binding domain of human DAI (ZBP1) in complex with Z-DNA reveals an unusual binding mode to Z-DNA. Proc Natl Acad Sci 105: 20671–20676. 10.1073/pnas.0810463106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Katsantoni M, Köster T, Marks J, Mukherjee J, Staiger D, Ule J, Zavolan M. 2021. CLIP and complementary methods. Nat Rev Methods Prim 1: 20. 10.1038/s43586-021-00018-1 [DOI] [Google Scholar]
- Hall K, Cruz P, Tinoco I, Jovin TM, Van De Sande JH. 1984. “Z-RNA”: a left-handed RNA double helix. Nature 311: 584–586. 10.1038/311584a0 [DOI] [PubMed] [Google Scholar]
- Herbert A. 2019. Z-DNA and Z-RNA in human disease. Commun Biol 2: 7. 10.1038/s42003-018-0237-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert A. 2020. ALU non-B-DNA conformations, flipons, binary codes and evolution. R Soc Open Sci 7: 200222. 10.1098/rsos.200222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert A. 2021. To “Z” or not to “Z”: Z-RNA, self-recognition, and the MDA5 helicase. PLoS Genet 17: e1009513. 10.1371/journal.pgen.1009513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PS, Mooers BH. 1997. Z-DNA crystallography. Biopolymers 14: 65–90. [DOI] [PubMed] [Google Scholar]
- Hubbard N, Ames JM, Maurano M, Chu LH, Somfleth KY, Gokhale NS, Werner M, Snyder JM, Lichauco K, Savan R, et al. 2022. ADAR1 mutation causes ZBP1-dependent immunopathology. Nature 607: 769–775. 10.1038/s41586-022-04896-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur S. 2019. Double-stranded RNA sensors and modulators in innate immunity. Annu Rev Immunol 37: 349–375. 10.1146/annurev-immunol-042718-041356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong M, Lee AR, Kim HE, Choi YG, Choi BS, Lee JH. 2014. NMR study of the Z-DNA binding mode and B-Z transition activity of the Zα domain of human ADAR1 when perturbed by mutation on the α3 helix and β-hairpin. Arch Biochem Biophys 558: 95–103. 10.1016/j.abb.2014.06.026 [DOI] [PubMed] [Google Scholar]
- Jiao H, Wachsmuth L, Kumari S, Schwarzer R, Lin J, Eren RO, Fisher A, Lane R, Young GR, Kassiotis G, et al. 2020. Z-nucleic-acid sensing triggers ZBP1-dependent necroptosis and inflammation. Nature 580: 391–395. 10.1038/s41586-020-2129-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovin TM, Soumpasis DM, Mcintosh LP. 1987. The transition between B-DNA and Z-DNA. Annu Rev Phys Chem 38: 521–560. 10.1146/annurev.pc.38.100187.002513 [DOI] [Google Scholar]
- Kang YM, Bang J, Lee EH, Ahn HC, Seo YJ, Kyeong KK, Kim YG, Choi BS, Lee JH. 2009. NMR spectroscopic elucidation of the B-Z transition of a DNA double helix induced by the Zα domain of human ADAR1. J Am Chem Soc 131: 11485–11491. 10.1021/ja902654u [DOI] [PubMed] [Google Scholar]
- Kang HJ, Le TVT, Kim K, Hur J, Kim KK, Park HJ. 2014. Novel interaction of the Z-DNA binding domain of human ADAR1 with the oncogenic c-Myc promoter G-quadruplex. J Mol Biol 426: 2594–2604. 10.1016/j.jmb.2014.05.001 [DOI] [PubMed] [Google Scholar]
- Karki R, Sundaram B, Sharma BR, Lee SJ, Malireddi RKS, Nguyen LN, Christgen S, Zheng M, Wang Y, Samir P, et al. 2021. ADAR1 restricts ZBP1-mediated immune response and PANoptosis to promote tumorigenesis. Cell Rep 37: 109858. 10.1016/j.celrep.2021.109858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DDY, Kim TTY, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. 2004. Widespread RNA editing of embedded Alu elements in the human transcriptome. Genome Res 14: 1719–1725. 10.1101/gr.2855504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Reddy S, Kim DY, Rich A, Lee S, Kim KK, Kim YG. 2009. Base extrusion is found at helical junctions between right- and left-handed forms of DNA and RNA. Nucleic Acids Res 37: 4353–4359. 10.1093/nar/gkp364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Hur J, Park K, Bae S, Shin D, Ha SC, Hwang HY, Hohng S, Lee JH, Lee S, et al. 2014. Distinct Z-DNA binding mode of a PKR-like protein kinase containing a Z-DNA binding domain (PKZ). Nucleic Acids Res 42: 5937–5948. 10.1093/nar/gku189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Hur J, Han JH, Ha SC, Shin D, Lee S, Park S, Sugiyama H, Kim KK. 2018a. Sequence preference and structural heterogeneity of BZ junctions. Nucleic Acids Res 46: 10504–10513. 10.1093/nar/gky784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Lim SH, Lee AR, Kwon DH, Song HK, Lee JH, Cho M, Johner A, Lee NK, Hong SC. 2018b. Unveiling the pathway to Z-DNA in the protein-induced B-Z transition. Nucleic Acids Res 46: 4129–4137. 10.1093/nar/gky200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler H, Cotsmire S, Langland J, Kibler K V, Kalman D, Upton JW, Mocarski ES, Jacobs BL. 2017. Inhibition of DAI-dependent necroptosis by the Z-DNA binding domain of the vaccinia virus innate immune evasion protein, E3. Proc Natl Acad Sci 114: 11506–11511. 10.1073/pnas.1700999114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler H, Cotsmire S, Zhang T, Balachandran S, Upton JW, Langland J, Kalman D, Jacobs BL, Mocarski ES. 2021. Vaccinia virus E3 prevents sensing of Z-RNA to block ZBP1-dependent necroptosis. Cell Host Microbe 29: 1266–1276.e5. 10.1016/j.chom.2021.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeris M, Funke L, Shrestha J, Rich A, Maas S. 2005. Modulation of ADAR1 editing activity by Z-RNA in vitro. Nucleic Acids Res 33: 5362–5370. 10.1093/nar/gki849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse H, Mrazikova K, D'Ascenzo L, Sponer J, Auffinger P. 2020. Short but weak: the Z-DNA lone-pair⋅⋅⋅π conundrum challenges standard carbon van der Waals radii. Angew Chem Int Ed Engl 59: 16553–16560. 10.1002/anie.202004201 [DOI] [PubMed] [Google Scholar]
- Kuś K, Rakus K, Boutier M, Tsigkri T, Gabriel L, Vanderplasschen A, Athanasiadis A. 2015. The structure of the Cyprinid herpesvirus 3 ORF112-Zα·Z-DNA complex reveals a mechanism of nucleic acids recognition conserved with E3L, a poxvirus inhibitor of interferon response. J Biol Chem 290: 30713–30725. 10.1074/jbc.M115.679407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EH, Seo YJ, Kim HE, Lee YM, Kim CM, Lee JH. 2011. Population analysis of the intermediate complex states during B-Z transition of non-CG-repeat DNA duplexes induced by the Zα domain of human ADAR1. Bull Korean Chem Soc 32: 719–721. 10.5012/bkcs.2011.32.2.719 [DOI] [Google Scholar]
- Lee AR, Park CJ, Cheong HK, Ryu KS, Park JW, Kwon MY, Lee J, Kim KK, Choi BS, Lee JH. 2016. Solution structure of the Z-DNA binding domain of PKR-like protein kinase from Carassius auratus and quantitative analyses of the intermediate complex during B-Z transition. Nucleic Acids Res 44: 2936–2948. 10.1093/nar/gkw025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AR, Hwang J, Hur JH, Ryu KS, Kim KK, Choi BS, Kim NK, Lee JH. 2019. NMR dynamics study reveals the Zα domain of human ADAR1 associates with and dissociates from Z-RNA more slowly than Z-DNA. ACS Chem Biol 14: 245–255. 10.1021/acschembio.8b00914 [DOI] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. 2004. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol 22: 1001–1005. 10.1038/nbt996 [DOI] [PubMed] [Google Scholar]
- Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellåker C, Vesely C, Ponting CP, McLaughlin PJ, et al. 2014. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep 9: 1482–1494. 10.1016/j.celrep.2014.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahama T, Kato Y, Shibuya T, Inoue M, Kim JI, Vongpipatana T, Todo H, Xing Y, Kawahara Y. 2021. Mutations in the adenosine deaminase ADAR1 that prevent endogenous Z-RNA binding induce Aicardi-Goutieres-syndrome-like encephalopathy. Immunity 54: 1976–1988. 10.1016/j.immuni.2021.08.022 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Fujii S, Urata H, Uesugi S, Ikehara M, Tomita K. 1985. Crystal structure of a left-handed RNA tetramer, r(C-br8G)2. Nucleic Acids Symp Ser (no.16) 29–32. [PubMed] [Google Scholar]
- Ng SK, Weissbach R, Ronson GE, Scadden ADJ. 2013. Proteins that contain a functional Z-DNA-binding domain localize to cytoplasmic stress granules. Nucleic Acids Res 41: 9786–9799. 10.1093/nar/gkt750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols PJ, Bevers S, Henen M, Kieft JS, Vicens Q, Vögeli B. 2021. Recognition of non-CpG repeats in Alu and ribosomal RNAs by the Z-RNA binding domain of ADAR1 induces A-Z junctions. Nat Commun 12: 793. 10.1038/s41467-021-21039-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonski KM, Samuel CE. 2013. Stress granule formation induced by measles virus is protein kinase PKR dependent and impaired by RNA adenosine deaminase ADAR1. J Virol 87: 756–766. 10.1128/JVI.02270-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB. 2015. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity 43: 933–944. 10.1016/j.immuni.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placido D, Brown BA II, Lowenhaupt K, Rich A, Athanasiadis A. 2007. A left-handed RNA double helix bound by the Zα domain of the RNA-editing enzyme ADAR1. Structure 15: 395–404. 10.1016/j.str.2007.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DSW, Parker R. 2016. Principles and properties of stress granules. Trends Cell Biol 26: 668–679. 10.1016/j.tcb.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SN, Kollman PA. 1986. Conformations of the 8-methylated and unmethylated ribohexamer r(CGCGCG)2. J Am Chem Soc 108: 3048–3053. 10.1021/ja00271a039 [DOI] [Google Scholar]
- Rice GI, Kasher PR, Forte GMA, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, et al. 2012. Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat Genet 44: 1243–1248. 10.1038/ng.2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschdi S, Yan J, Nomura Y, Escobar CA, Petersen RJ, Bingman CA, Tonelli M, Vivek R, Montemayor EJ, Wickens M, et al. 2022. An atypical RNA quadruplex marks RNAs as vectors for gene silencing. Nat Struct Mol Biol 29: 1113–1121. 10.1038/s41594-022-00854-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenburg S, Deigendesch N, Dittmar K, Koch-Nolte F, Haag F, Lowenhaupt K, Rich A. 2005. A PKR-like eukaryotic initiation factor 2α kinase from zebrafish contains Z-DNA binding domains instead of dsRNA binding domains. Proc Natl Acad Sci 102: 1602–1607. 10.1073/pnas.0408714102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schade M, Behlke J, Lowenhaupt K, Herbert A, Rich A, Oschkinat H. 1999a. A 6 bp Z-DNA hairpin binds two Zα domains from the human RNA editing enzyme ADAR1. FEBS Lett 458: 27–31. 10.1016/S0014-5793(99)01119-9 [DOI] [PubMed] [Google Scholar]
- Schade M, Turner CJ, Lowenhaupt K, Rich A, Herbert A. 1999b. Structure-function analysis of the Z-DNA-binding domain Zα of dsRNA adenosine deaminase type I reveals similarity to the (α + β) family of helix-turn-helix proteins. EMBO J 18: 470–479. 10.1093/emboj/18.2.470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz T, Rould MA, Lowenhaupt K, Herbert A, Rich A. 1999. Crystal structure of the Zα domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science 284: 1841–1845. 10.1126/science.284.5421.1841 [DOI] [PubMed] [Google Scholar]
- Snyder AG, Oberst A. 2021. The antisocial network: cross talk between cell death programs in host defense. Annu Rev Immunol 39: 77–101. 10.1146/annurev-immunol-112019-072301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Yu Y, Wu X, Acevedo A, Luo JD, Wang J, Schneider WM, Hurwitz B, Rosenberg BR, Chung H, et al. 2021. Decoupling expression and editing preferences of ADAR1 p150 and p110 isoforms. Proc Natl Acad Sci 118: e2021757118. 10.1073/pnas.2021757118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Rigby RE, Young GR, Korning-Hvidt A, Davis T, Kit Tan T, Bridgeman A, Townsend AR, Kassiotis G, Rehwinkel J. 2021. Adenosine-to-inosine editing of endogenous Z-form RNA by the deaminase ADAR1 prevents spontaneous MAVS-dependent type I interferon responses. Immunity 54: 1961–1975. 10.1016/j.immuni.2021.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber D, Tauber G, Khong A, Van Treeck B, Pelletier J, Parker R. 2020. Modulation of RNA condensation by the DEAD-box protein eIF4A. Cell 180: 411–426. 10.1016/j.cell.2019.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng MK, Liaw YC, van der Marel GA, van Boom JH, Wang AH. 1989. Effects of the O2′ hydroxyl group on Z-DNA conformation: structure of Z-RNA and (araC)-[Z-DNA]. Biochemistry 28: 4923–4928. 10.1021/bi00438a001 [DOI] [PubMed] [Google Scholar]
- Thapa RJ, Ingram JP, Ragan KB, Nogusa S, Boyd DF, Benitez AA, Sridharan H, Kosoff R, Shubina M, Landsteiner VJ, et al. 2016. DAI senses influenza A virus genomic RNA and activates RIPK3-dependent cell death. Cell Host Microbe 20: 674–681. 10.1016/j.chom.2016.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesugi S, Ohkubo M, Urata H, Ikehara M, Kobayashi M, Kyogoku Y. 1984. Ribooligonucleotides, r(C-G-C-G) analogues containing 8-substituted guanosine residues, form left-handed duplexes with Z-form-like structure. J Am Chem Soc 106: 3675–3676. 10.1021/ja00324a047 [DOI] [Google Scholar]
- Uggenti C, Crow YJ. 2018. Sort your self out! Cell 172: 640–642. 10.1016/j.cell.2018.01.023 [DOI] [PubMed] [Google Scholar]
- Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, Parker R. 2018. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci 115: 2734–2739. 10.1073/pnas.1800038115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicens Q, Kieft JS. 2022. Thoughts on how to think (and talk) about RNA structure. Proc Natl Acad Sci 119: e2112677119. 10.1073/pnas.2112677119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AHJ, Quigley GJ, Kolpak FJ, Crawford JL, Van Boom JH, Van Der Marel G, Rich A. 1979. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 282: 680–686. 10.1038/282680a0 [DOI] [PubMed] [Google Scholar]
- Wang AJ, Quigley GJ, Kolpak FJ, van der Marel G, van Boom JH, Rich A. 1981. Left-handed double helical DNA: variations in the backbone conformation. Science 211: 171–176. 10.1126/science.7444458 [DOI] [PubMed] [Google Scholar]
- Wang R, Li H, Wu J, Cai ZY, Li B, Ni H, Qiu X, Chen H, Liu W, Yang ZH, et al. 2020. Gut stem cell necroptosis by genome instability triggers bowel inflammation. Nature 580: 386–390. 10.1038/s41586-020-2127-x [DOI] [PubMed] [Google Scholar]
- Xu X, Li M, Wu C, Li D, Jiang Z, Liu C, Cheng B, Mao H, Hu C. 2019. The fish-specific protein kinase (PKZ) initiates innate immune responses via IRF3- and ISGF3-like mediated pathways. Front Immunol 10: 582. 10.3389/fimmu.2019.00582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Lenz S, Zimmermann-Kogadeeva M, Tegunov D, Cramer P, Bork P, Rappsilber J, Mahamid J. 2022. Visualizing translation dynamics at atomic detail inside a bacterial cell. Nature 610: 205–211. 10.1038/s41586-022-05255-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, Yeou S, Lee NK. 2022. DNA bending force facilitates Z-DNA formation under physiological salt conditions. J Am Chem Soc 144: 13137–13145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Chen T, Cao X. 2015. RNA editing by ADAR1 marks dsRNA as “self.” Cell Res 25: 1283–1284. 10.1038/cr.2015.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarling DA, Calhoun CJ, Hardin CC, Zarling AH. 1987. Cytoplasmic Z-RNA. Proc Natl Acad Sci 84: 6117–6121. 10.1073/pnas.84.17.6117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarling DA, Calhoun CJ, Feuerstein BG, Sena EP. 1990. Cytoplasmic microinjection of immunoglobulin Gs recognizing RNA helices inhibits human cell growth. J Mol Biol 211: 147–160. 10.1016/0022-2836(90)90017-G [DOI] [PubMed] [Google Scholar]
- Zhang T, Yin C, Boyd DF, Quarato G, Ingram JP, Shubina M, Ragan KB, Ishizuka T, Crawford JC, Tummers B, et al. 2020. Influenza virus Z-RNAs induce ZBP1-mediated necroptosis. Cell 180: 1115–1129. 10.1016/j.cell.2020.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Yin C, Fedorov A, Qiao L, Bao H, Beknazarov N, Wang S, Gautam A, Williams RM, Crawford JC, et al. 2022. ADAR1 masks the cancer immunotherapeutic promise of ZBP1-driven necroptosis. Nature 606: 594–602. 10.1038/s41586-022-04753-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirbel CL, Auffinger P. 2022. Lone pair…π contacts and structure signatures of r(UNCG) tetraloops, Z-turns, and Z-steps: a WebFR3D survey. Molecules 27: 4365. 10.3390/molecules27144365 [DOI] [PMC free article] [PubMed] [Google Scholar]