Abstract
Argonautes are small RNA-binding proteins, with some having small RNA-guided endonuclease (slicer) activity that cleaves target nucleic acids. One cardinal rule that is structurally defined is the inability of slicers to cleave target RNAs when nucleotide mismatches exist between the paired small RNA and the target at the cleavage site. Animal-specific PIWI clade Argonautes associate with PIWI-interacting RNAs (piRNAs) to silence transposable elements in the gonads, and this is essential for fertility. We previously demonstrated that purified endogenous mouse MIWI fails to cleave mismatched targets in vitro. Surprisingly, here we find using knock-in mouse models that target sites with cleavage-site mismatches at the 10th and 11th piRNA nucleotides are precisely sliced in vivo. This is identical to the slicing outcome in knock-in mice where targets are base-paired perfectly with the piRNA. Additionally, we find that pachytene piRNA-guided slicing in both these situations failed to initiate phased piRNA production from the specific target mRNA we studied. Instead, the two slicer cleavage fragments were retained in PIWI proteins as pre-piRNA and 17–19 nt by-product fragments. Our results indicate that PIWI slicing rules established in vitro are not respected in vivo, and that all targets of PIWI slicing are not substrates for piRNA biogenesis.
Keywords: MIWI, MILI, pachytene piRNA, slicer, PIWI, cleavage-site mismatch
INTRODUCTION
Argonautes are small RNA-binding proteins that function in most organisms and in all cells of multicellular organisms (Carmell et al. 2002). PIWI proteins belong to an animal-specific clade of Argonautes that are almost exclusively expressed in the gonads, and they associate with the small RNAs called PIWI-interacting RNAs (piRNAs) (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Vagin et al. 2006). One major target of piRNAs is the collection of transposable elements present within the genome, and they are repressed by posttranscriptional or cotranscriptional mechanisms mediated by the PIWI proteins. Consistently, animals (flies, fish, mice, and hamsters) that lack either PIWI proteins or piRNA pathway factors derepress transposable elements, leading to arrested gametogenesis and infertility (Ghildiyal and Zamore 2009; Luteijn and Ketting 2013).
Three mouse PIWI proteins are expressed in the male germline, with MIWI (PIWIL1) (Deng and Lin 2002; Reuter et al. 2011) and MILI (PIWIL2) (Kuramochi-Miyagawa et al. 2004; Aravin et al. 2006) being cytosolic, while MIWI2 (PIWIL4) is nuclear (Carmell et al. 2007; Aravin et al. 2008; Kojima-Kita et al. 2016). MIWI2 and MILI are expressed in the embryonic male germline where they associate with piRNAs that almost exclusively target transposable elements (Aravin et al. 2007, 2008; De Fazio et al. 2011; Xiol et al. 2012). In the postnatal meiotic pachytene spermatocytes and postmeiotic round spermatids, MILI and MIWI associate with a distinct set of small RNAs called the pachytene piRNAs (Aravin et al. 2006; Girard et al. 2006). They are composed of highly unique sequences that are largely depleted of transposable element sequences. Although pachytene piRNAs are very abundant, most lack any obvious perfectly complementary targets in the transcriptome. Searching for genuine targets of pachytene piRNAs with a view of explaining their functions has been a major preoccupation in the field.
Mouse MIWI is exclusively bound by pachytene piRNAs and its loss leads to male infertility (Deng and Lin 2002; Reuter et al. 2011). Pachytene piRNAs originate from ∼100 large genomic loci called piRNA clusters (Li et al. 2013). Pointing to the relevance of these small RNAs, loss of thousands of piRNAs from two such clusters leads to male infertility (Wu et al. 2020; Choi et al. 2021). However, not all clusters seem to be important for fertility when individually deleted (Homolka et al. 2015; Wu et al. 2020). Pachytene piRNAs are also shown to regulate germline gene expression by identifying mRNAs via partial complementarity pairing (via the 5′ seed sequence; piRNA nucleotides 2–8) to recruit the deadenylation machinery for promoting RNA decay (Gou et al. 2014), a mechanism similar to that used by microRNAs (Filipowicz et al. 2008).
One core function of some of the Argonaute proteins is their small RNA-guided endoribonuclease (slicer) activity (Liu et al. 2004; Meister et al. 2004; Meister 2013). This is true for MIWI (Reuter et al. 2011) and MILI (De Fazio et al. 2011), with loss of their slicer activities leading to male infertility. Pachytene piRNA-guided slicer activity is shown to cleave transposon transcripts (Reuter et al. 2011; Goh et al. 2015) and mRNAs (Goh et al. 2015; Wu et al. 2020). However, the number of such slicer targets—as defined by existing targeting rules—is very limited. PIWI slicing is also linked to piRNA biogenesis, where one of the target cleavage fragments matures as the so-called secondary piRNA (Brennecke et al. 2007; Gunawardane et al. 2007). Slicing can also initiate phased piRNA biogenesis, where additional piRNAs are generated downstream from the secondary piRNA by the action of the endonuclease Zucchini/MitoPLD (Izumi et al. 2020). Slicing-triggered phasing is demonstrated in the fly ovarian germline (Han et al. 2015; Homolka et al. 2015; Mohn et al. 2015; Wang et al. 2015), and by MILI in the mouse embryonic male germline (Yang et al. 2016; Wenda et al. 2017). A similar biogenesis role for pachytene piRNA-guided slicing of cluster precursor transcripts is proposed (Gainetdinov et al. 2018; Wu et al. 2020). Taken together, pachytene piRNA-guided PIWI slicing has a function in regulating target transcript levels and in initiating piRNA biogenesis.
In this study, we sought to experimentally determine the in vivo consequence of pachytene piRNA-guided slicer activity on an endogenous mRNA. Using MIWI slicing rules established in vitro (Reuter et al. 2011), we designed two knock-in mouse lines (with a single binding site or ten binding sites) where the piRNA is perfectly complementary to the target site. This resulted in target slicing, as expected (Reuter et al. 2011). We also made two negative control knock-in mouse lines where the binding site for the piRNA on the target is mutated to result in a 2-nt mismatch at the cleavage site (nucleotides 10 and 11 of the piRNA) to prevent slicing, as previously shown in vitro (Reuter et al. 2011). Surprisingly, these negative control targets are also sliced in vivo, upending one of the sacrosanct rules of Argonaute/PIWI slicing. Our study should prompt a revision of the current piRNA-target engagement rules, which are largely based on in vitro slicing assays.
RESULTS AND DISCUSSION
Knock-in mice expressing mRNAs with pachytene piRNA binding sites
To study the in vivo consequence of pachytene piRNA-mediated slicing, we prepared knock-in mice where we placed binding sites for a single abundant pachytene piRNA in the 3′UTR of the Ythdc2 mRNA (Fig. 1A). The germline-specific RNA helicase Ythdc2 is expressed in meiotic germ cells, including pachytene spermatocytes (Wojtas et al. 2017; Jain et al. 2018). The piRNA we chose (piR-A) starts to be expressed at postnatal day 14 in mouse testes where pachytene spermatocytes are the dominant germ cell type (Girard et al. 2006; Reuter et al. 2011). It continues to increase in abundance in postmeiotic round spermatids in P20 mice and is detected in adult mouse testes. Analysis of PIWI-bound small RNAs from P14, P20 and adult mouse testes shows that piR-A is present in both MIWI and MILI complexes (Reuter et al. 2011).
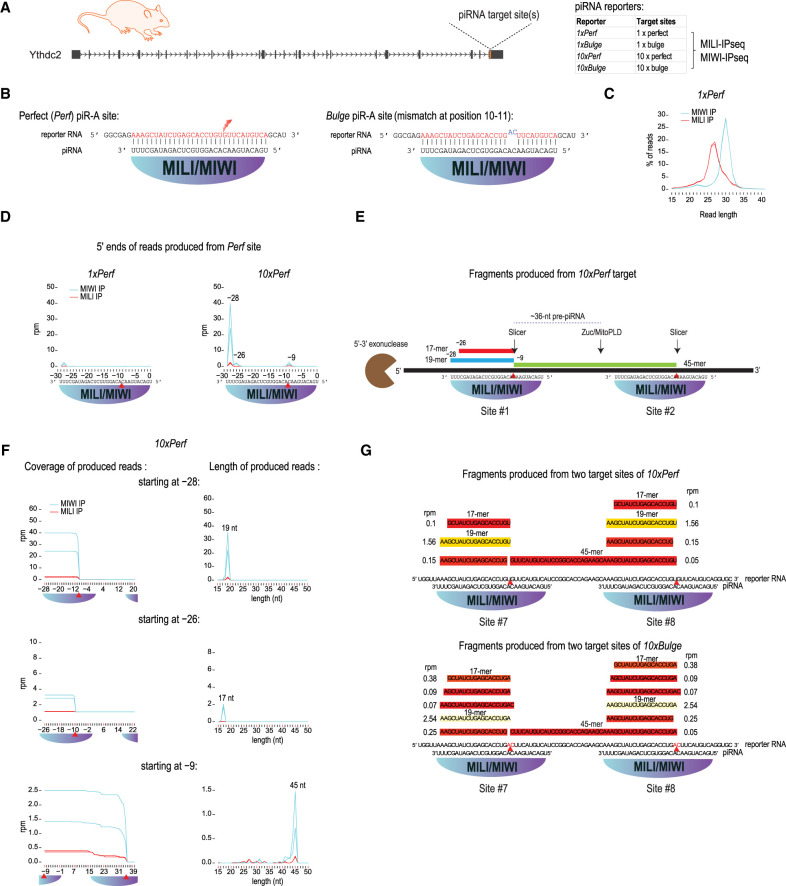
FIGURE 1.
Products of target RNA slicing remain bound to PIWI complexes in mouse testes. (A) Perfect or bulge piR-A target sites were inserted into the last exon of Ythdc2. Single or 10 tandem sites with spacers were inserted. MILI and MIWI loaded sequences were identified by sequencing of immunoprecipitated RNAs. (B) Perfect (Perf) piR-A sites were designed to be fully complementary to endogenous piR-A piRNA. The bulge piR-A sites were designed to contain noncomplementary bases to piR-A nucleotides at positions 10 and 11. (C) Length profiles of immunoprecipitated small RNAs from 1xPerf mouse. (D) The position (=distance from 5′ nt of targeting piR-A) and abundance of 5′ ends of reads produced from reporter target sites. In the case of 10xPerf, the metaplot summarizes the read counts from all ten sites. Red triangle refers to expected position of MILI/MIWI cleavage guided by piR-A. (E) Schematic overview of RNA fragments identified in MILI/MIWI immunoprecipitations from 10xPerf mice. (F) Coverage of reads is shown separately for RNAs with their 5′ end starting at a specific distance from the first piR-A target nucleotide (=0 distance). These RNAs are of specific lengths, with the 3′ ends always created by MILI/MIWI slicing at a subsequent site. Metaplots summarize the read counts from 10 sites. (G) The RNAs produced from sites 7 and 8 in 10xPerf and 10xBulge are shown together with their abundance. Reads from MILI and MIWI immunoprecipitations are combined. Only reads with abundance ≥0.05 rpm are shown.
We placed a single perfectly (1xPerf) complementary binding site for piR-A in the 3′UTR of Ythdc2 (Fig. 1B). To increase the chance of interaction between the target and the piRNA/PIWI complexes, we also prepared mice with ten perfectly (10xPerf) complementary binding sites that are separated with spacer sequences between them. In vitro slicer assays with immunopurified MIWI complexes isolated from mouse testes previously showed that a target RNA that is perfectly complementary to piR-A is cleaved (Reuter et al. 2011). Since MILI is also a slicer (De Fazio et al. 2011), the Ythdc2 mRNA with 1xPerf and 10xPerf target sites are expected to be sliced by MILI too. As a noncleaving control, we prepared knock-in mice with a single (1xBulge) or ten (10xBulge) mutated binding sites (Fig. 1B). The mutations result in mismatches with the 10th and 11th nucleotide of the base-paired piR-A, creating the “bulge” and preventing slicing as previously demonstrated in vitro (Reuter et al. 2011).
Knockout of Ythdc2 causes early spermatogenic arrest in P8 mouse testes, at a time when germ cells enter meiosis (Bailey et al. 2017; Wojtas et al. 2017; Jain et al. 2018), while conditional knockout in meiotic pachytene spermatocytes also results in arrested spermatogenesis (Liu et al. 2021). Slicing of an RNA target via small RNA-guided Argonaute leads to degradation of the RNA, reducing gene expression (Meister 2013). Slicer targets of pachytene piRNAs have lower transcript levels (Goh et al. 2015; Wu et al. 2020), as the RNA is either degraded or consumed during piRNA biogenesis (Han et al. 2015; Homolka et al. 2015; Mohn et al. 2015). However, we did not observe any impact on fertility for any of the four homozygous knock-in lines with piRNA binding sites in Ythdc2 mRNA. Although we occasionally noted reduced litter size with the 10xBulge line which also displayed reduced expression of postmeiotic genes (Supplemental Fig. 1C), all lines were maintained in the laboratory by crossing the homozygous lines. Also, there wasn't any dramatic change in target Ythdc2 mRNA levels (Supplemental Fig. 1B). We conclude that presence of multiple binding sites for an abundant pachytene piRNA in the 3′UTR of Ythdc2 did not result in gross fertility defects.
Sliced mRNA target fragments retained within PIWI complexes
To examine whether slicing of the 3′UTR of the Ythdc2 mRNA results in generation of new piRNAs, we prepared total testicular small RNA libraries (20–40 nt). We used P14 homozygous knock-in 1xPerf and 1xBulge mice but did not detect any target-derived small RNAs (data not shown). Since the level of piR-A is higher in later stages of spermatogenesis we next examined adult mouse testes (Girard et al. 2006; Reuter et al. 2011). To potentially enrich for any low levels of generated small RNAs, we immunoprecipitated MILI and MIWI complexes from knock-in mouse lines and wildtype controls. Deep sequencing of the associated RNAs revealed the expected peak read length of ∼26 nt for MILI piRNAs and ∼30 nt for MIWI piRNAs (Fig. 1C).
To detect target-derived RNAs, we mapped the reads to the Ythdc2 3′UTR target sequences expressed in the 1xPerf and the 10xPerf knock-in mice (Fig. 1D). MIWI/MILI slicing of the target sequence should result in a cleavage after the 10th nucleotide (at position −9) of the targeting piRNA (Fig. 1E; Reuter et al. 2011). This is expected to generate a pre-piRNA fragment with a 5′ monophosphate that matures into a new piRNA, and a by-product fragment with a 3′ hydroxyl group at the site of cleavage that is normally degraded. Mapping of the 5′ ends identify reads that start at position −9, consistent with their origin from slicer cleavage, and others with their 5′ ends at position −26 and −28 nt downstream from the 5′ nucleotide of the targeting piRNA (Fig. 1D). Since very low levels of target-derived sequences are detected in the 1xPerf mice, we focused our analyses on data obtained from the 10xPerf mice.
Reads with their 5′ ends at position −9 are not in the piRNA size-range (26–30 nt), indicating that these are not mature piRNAs. Instead, they have a fixed size of 45 nt (Fig. 1E,F) and their 5′ and 3′ ends precisely map to the slicer cleavage sites on two consecutive piRNA targeting sites (Fig. 1G). This shows that while PIWI slicer activity creates the 5′ end of the pre-piRNA fragment, the downstream cleavage at ∼36 nt that is normally expected to be generated by Zuccini/MitoPLD during phased piRNA biogenesis in mice (Yang et al. 2016; Wenda et al. 2017; Gainetdinov et al. 2018; Izumi et al. 2020), did not take place. A defining feature of abundant piRNAs is a uridine (U1) at the 5′ end, with non-U1 piRNAs being rare (Genzor et al. 2021). The pre-piRNA generated from the 10xPerf target RNA has a guanosine (G1) (Fig. 1B,G), which may be suboptimally loaded into a PIWI protein to enable efficient piRNA biogenesis (Kawaoka et al. 2011). Alternatively, we may conclude that not all MIWI/MILI slicer cleavages can lead to piRNA biogenesis.
The reads with their 5′ ends at positions −26 and −28 have defined lengths of 17 and 19 nt, respectively (Fig. 1F), with their 3′ end at the slicer cleavage site (Fig. 1E,G). This identifies these fragments as the by-product fragments whose 3′ end is generated by MIWI/MILI slicing, while the 5′ end is created by an unknown nuclease that shapes it according to the footprint of the targeting PIWI protein (Fig. 1E). Recombinant MIWI/piRNA complex remains bound to its cleavage products, thereby protecting the two cleaved ends generated by slicer activity (Arif et al. 2022). It is also shown that for similar target-small RNA pairing configurations, sponge and Silkworm PIWI remain bound to their targets much longer than AGO clade Argonautes (Anzelon et al. 2021). We speculate that 5′ → 3′ exonuclease activity like that of XRN1 may act on such a stalled postcatalytic complex, generating the 5′ monophosphate end of the by-product fragment. Taken together, pachytene piRNAs are perfectly complementary to a target mRNA guide slicing in vivo, but the cleavage fragments remain bound to the PIWI complex.
Unexpected MILI/MIWI slicer activity on targets with cleavage site mismatches
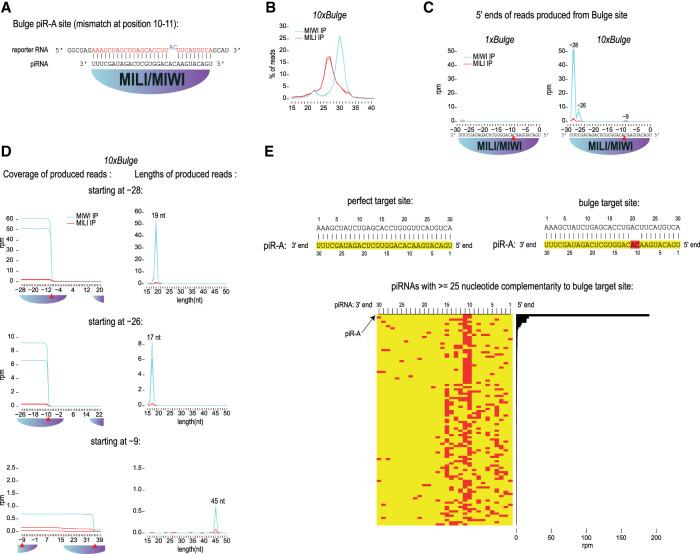
Immunopurified endogenous MIWI fails to slice a target RNA when a 2 nt mismatch (at positions 10 and 11 of the piRNA) exists between the guiding piRNA and the cleavage site (Reuter et al. 2011). Surprisingly, mapping of the sequencing reads from MIWI and MILI complexes isolated from the 10xBulge knock-in mice revealed the tell-tale signatures of slicer activity (Fig. 2A–C). First, the bulged target is cleaved after the 10th nucleotide of the targeting piRNA, as indicated by the presence of the pre-piRNA fragment with the 5′ nucleotide at a distance −9 from the first nucleotide of the piRNA (Fig. 2C). Second, the 17 and 19 nt by-product fragments are also detected with a common 3′ end at the site of slicer cleavage (Fig. 2D). We examined whether any of the other thousands of pachytene piRNAs can perfectly pair with the mutated binding site but found none (Fig. 2E). This suggests that mismatches at the cleavage site are not inhibitory for slicing of a target RNA by a paired piRNA/PIWI complex in mouse testes. This in vivo observation is in direct contradiction to the piRNA targeting rules previously established for MIWI by in vitro slicer assays (Reuter et al. 2011). This is also very different to the situation with AGO clade Argonautes which are unable to cleave a target with a central mismatch both in vitro and in vivo (Meister 2013).
FIGURE 2.
RNA targets with cleavage-site mismatches are sliced by PIWI proteins in vivo. (A) The bulge piR-A sites were designed to contain noncomplementary bases to piR-A nucleotides at positions 10 and 11. (B) Length profiles of immunoprecipitated small RNAs from 10xBulge mouse. (C) The position (=distance from 5′ nt of targeting piR-A) and abundance of 5′ ends of reads produced from reporter target sites. In the case of 10xBulge, the metaplot summarizes the read counts from all ten sites. Red triangle refers to expected position of MILI/MIWI cleavage guided by piR-A. (D) Coverage of reads is shown separately for RNAs with their 5′ end starting at a specific distance from the first piR-A target nucleotide (=0 distance). These RNAs are of specific lengths, with the 3′ ends always created by MILI/MIWI slicing at a subsequent site. (E) Complementary piRNAs potentially targeting the bulge site. The heatmap depicts position of mismatches (in red) between the targeting piRNA and the target site sequence, while the barplot shows the abundance of such piRNAs.
As in 10xPerf, the pre-piRNA fragment originating from 10xBulge has a length of 45 nt consistent with its 5′ and 3′ ends being generated by slicer activity from two consecutive bulged piRNA-targeting sites (Figs. 1G, 2D). This observation again reinforces the fact that slicer activity by MIWI and MILI fails to initiate generation of new piRNAs from the targets we examined. The absence of U1 as the 5′ nucleotide (which is a cytidine here) (Figs. 1G, 2) could be the reason for failed entry of the pre-piRNA into the piRNA biogenesis pathway (Kawaoka et al. 2011; Genzor et al. 2021) or it just shows that not all RNA targets of PIWI slicing enter the piRNA biogenesis pathway.
Immunopurified MIWI from mouse testes (Reuter et al. 2011) or a recombinant MIWI/piRNA complex (Arif et al. 2022) is shown to slice RNA targets when the guiding piRNA has a minimum of 21 nt complementarity (positions 2–22 of the piRNA). In vivo studies with mice expressing human pachytene piRNAs showed that signatures of target slicing were observed even when some mismatches (at positions 12–21) existed between the guiding piRNA and the target, as long as guide-target pairing was maintained between positions 2–11 (Goh et al. 2015). Recombinant sponge PIWI/guide RNA complex is also shown to require extensive complementarity (positions 2–16 of the guide) between the 25 nt guide RNA and target for slicing (Anzelon et al. 2021). In fact, increasing the pairing toward the 3′ end (from positions 2–16 to 2–21 of the guide) enhances the slicing activity of sponge PIWI, with no more than 2 nt mismatches (within positions 11–18) tolerated for efficient cleavage (Anzelon et al. 2021). Recent studies indicate that PIWI proteins on their own are inefficient slicers, with the zinc-finger protein GTSF1 being able to enhance the rate of slicing in vitro (Arif et al. 2022). Interestingly, GTSF1 can also reduce the requirement for extensive complementarity by promoting MIWI slicing with just 2–16 nt of the piRNA being paired with the target (Arif et al. 2022). Finally, a recent preprint study reports that GTSF1 can further relax piRNA/MIWI targeting rules, with even targets having a central mismatch being cleaved (Gainetdinov et al. 2022). Taken together, using the four knock-in mouse models we have uncovered important aspects of in vivo PIWI slicer activity that were not previously appreciated, and demonstrated that PIWI slicing rules in vivo are different from those currently established in vitro. The ability of accessory factors to modulate PIWI slicer activity, potentially via affecting PIWI structure, likely explains this deviation in vivo.
MATERIALS AND METHODS
Animal work
The transgenic Mouse Facility of the University of Geneva was used to generate the four knock-in mouse lines utilized in this project. This study was approved by the Canton of Geneva (GE-162-19).
Generation of knock-in mouse lines
Knock-in mice were created by inserting one or ten complementary binding sites for a piRNA (piR-A) in the 3′ UTR of the Ythdc2 gene (MGI: 2448561; NCBI Gene: 240255). Binding sites are perfectly complementary (1xPerf or 10xPerf) or contain two mismatches (1xBulge or 10xBulge) on the 10th and 11th position relative to the guide RNA. To create these reporter mouse lines, the Ythdc2 locus of interest was targeted in single cell mouse embryos of the B6D2F1/J hybrid line (also called B6D2; The Jackson Laboratory, stock no. 100006).
Targeting of the locus was achieved by injecting two different guide RNAs (gRNAs from Integrated DNA Technologies) complexed with the Cas9 protein. The first gRNA (CAUAACGCUCUUCCAUAGCCGUUUUAGAGCUAUGCU) targeted the reverse strand at position chr18: 44887792. The second gRNA (GUGCAUUCUGUAGCUACAACGUUUUAGAGCUAUGCU) targeted the reverse strand at position chr18: 44887815. Combination of these two gRNAs and Cas9 endonuclease created a deletion of 23 bp in the last exon (exon 30) of Ythdc2. Two ssDNA oligonucleotide templates (Integrated DNA Technologies; 195 bp) containing the required single piRNA binding sites were coinjected. The two multiple binding sites reporter alleles were created by separately coinjecting two pUC57-Kan plasmids containing 10 piR-A binding sites (perfect or bulge) separated by spacers and flanked by homology arms to the Ythdc2 locus of interest (Supplemental Fig. 2). The gRNAs were composed of two parts: one tracrRNA and one crRNA (sequences as above). The tracrRNA is 67 bases long (Integrated DNA Technologies, Cat. No. 1072533) and is the common part of the guide RNA that binds the Cas9. A mixture of 200 pmol of tracrRNA and crRNA were annealed by incubating 5 min at 95°C in 1xTE buffer, pH 7.5 (Integrated DNA Technologies; Cat. No. 11-05-01-05) and cooling down at room temperature for 30 min. The final volume was 10 µL. Annealed gRNAs were stored at −20°C and then used for mouse single cell embryo injection.
The single cell embryo injection mix was prepared right before the injection. Cas9 protein at 30 ng/µL final concentration (Integrated DNA Technologies; Cat. No. 1081058) was mixed with the annealed gRNAs at 0.6 pmol/µL final concentration. Template DNA was also added to the mix at 20 ng/µL final concentration and the volume was adjusted to 100 µL with TE buffer (pH 7.5). Complex formation was achieved by incubating the mix 10 min at room temperature. Finally, the mix was centrifuged at 13,000 rpm for 5 min at 4°C, and 50 µL of supernatant were collected and placed on ice.
Genotyping
Primers (Forward: CTCAGGTTGGGGAACAGTTG, Reverse: ACTCTATGCCTCAAATCCACTC A) flanking the targeted insertion region in the mouse Ythdc2 gene were used to genotype the knock-in alleles. The expected PCR product sizes were 281 bp for WT, 320 bp for 1xPerf/1xBulge and 716 bp for 10xPerf/10xBulge. PCR reactions were run with Phire Green Hot Start II PCR Master Mix (Thermo) under the following conditions: 98°C for 30 sec, 35 cycles of (98°C for 5 sec, 63°C for 5 sec and 72°C for 10 sec), 72°C for 60 sec, and lastly at 4°C to hold the reaction.
PIWI immunoprecipitations
Immunoprecipitation from adult mouse testes was conducted as previously reported using anti-MILI and anti-MIWI rabbit polyclonal antibodies (Reuter et al. 2011). MILI- and MIWI-loaded piRNAs were extracted from the bead complexes by phenol–chloroform extraction, after proteinase K treatment.
RNA library preparation and sequencing
piRNA libraries were prepared with NEBNext Multiplex Small RNA Library Prep Set for Illumina (New England Biolabs, Ref. E7300L) and sequenced at EMBL Genomic Core Facilities in Heidelberg using the NextSeq 500 platform (Illumina).
Total RNA extracted from adult whole testes was used as starting material for the testicular transcriptome sequencing. The Stranded Total RNA Ribo-Zero Plus kit from Illumina was used for the library preparation with 500 ng of total RNA as input. Library molarity and quality were assessed with the Qubit and Tapestation (DNA High sensitivity chip). Libraries were sequenced on a NovaSeq 6000 Illumina sequencer for SR100 reads (iGE3 Genomics Platform, University of Geneva).
Quantification and statistical analysis of small RNA sequencing
Reads were sorted into individual libraries based on the barcodes, and the 3′ adapter sequences were clipped from the reads using cutadapt (Martin 2011) with only sequences of at least 15 nt left for further analysis (cutadapt parameters: -a AGATCGGAAGAGCACACGTCT -m 15 -e 0.2 -O 4 -q 10 –match-read-wildcards). The reads from small RNA-seq libraries were mapped to the mouse genome (GRCm38: Ensembl release 95) using STAR (Dobin et al. 2013) (with parameters: –runThreadN 10 -outFilterType BySJout –limitOutSJcollapsed 50000000 –limitIObufferSize 1500000000) and length distributions of the reads were plotted. Read counts were normalized to library sizes.
To see whether any piRNAs are produced from the reporters, the reads were mapped by bowtie (Langmead et al. 2009) (with parameters: -v 0 -a –best –strata) to the Ythdc2 transcript sequence containing individual reporters: 1xPerf, 10xPerf, 1xBulge and 10xBulge (Supplemental Fig. 2). We calculated 5′ end and body coverages of the reads produced from individual reporters as well as coverages targeting the reporters. Counts of reads were divided by number of mapped sites and normalized to library sizes (rpm—reads per million). To characterize the overall production of piRNAs from the ten sites of 10xPerf and 10xBulge reporters, we also created the metaplots where we summarized the counts of piRNAs produced from individual positions of all ten target sites. The positions refer to the distance from the 5′ end of targeting piR-A piRNA. Therefore, nucleotide at position 0 refers to the nucleotide which pairs with the first nucleotide of piR-A. The position −9 refers to the nucleotide pairing with 10th piR-A nucleotide and is the position of the 5′ end of secondary piRNAs created by MILI or MIWI cleavage.
Quantification and statistical analysis of long RNA sequencing
The reads from long RNA-seq libraries were mapped to the mouse genome (GRCm39 - Ensembl release 104) using salmon (Patro et al. 2017) (salmon quant with options -1 A –validateMappings–gcBias). Further analysis was performed using R (R Core Team 2017) and Bioconductor (Huber et al. 2015). The DESeq2 (Love et al. 2014) package was used to obtain normalized read counts for individual genes. The Volcano plots were plotted using the EnhancedVolcano function from the EnhancedVolcano 1.3.5 package (https://github.com/kevinblighe/EnhancedVolcano). To visualize expression of dysregulated genes across spermatogenic populations (NCBI: PRJNA317251), we calculated the z-scores of log2 expression (reads per million) for each of those genes and plotted the boxplots for individual samples. Similarly, the heatmap of z-scores was plotted using pheatmap 1.0.12.
DATA DEPOSITION
The sequencing data generated in the study are deposited with the Gene Expression Omnibus (GEO: GSE219200).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Pei-Hsuan Wu, University of Geneva, for discussions. We thank the EMBL Genomics Core Facility, Heidelberg, Germany, and the University of Geneva iGE3 Genomics Core Facility for sequencing. Mice were created at the Transgenic mouse facility of the University of Geneva. This work was supported by grants to R.S.P. from the Swiss National Science Foundation, Switzerland (ERC Transfer grant #CRETP3_166923, Project grant #310030_207468, and the NCCR RNA & Disease #51NF40_182880). Work in the Pillai laboratory is supported by the Republic and Canton of Geneva.
Author contributions: M.D. performed mouse experiments, PIWI immunoprecipitation and small RNA library preparation; computational analyses were done by D.H.; N.R. did testicular transcriptome sequencing; P.G. helped with mouse experiments; R.R.P. initiated the study by generating mouse lines; coordination and manuscript preparation were done by R.R.P. and R.S.P. with input from others.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079349.122.
Freely available online through the RNA Open Access option.
REFERENCES
- Anzelon TA, Chowdhury S, Hughes SM, Xiao Y, Lander GC, MacRae IJ. 2021. Structural basis for piRNA targeting. Nature 597: 285–289. 10.1038/s41586-021-03856-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207. 10.1038/nature04916 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747. 10.1126/science.1142612 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799. 10.1016/j.molcel.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif A, Bailey S, Izumi N, Anzelon TA, Ozata DM, Andersson C, Gainetdinov I, MacRae IJ, Tomari Y, Zamore PD. 2022. GTSF1 accelerates target RNA cleavage by PIWI-clade Argonaute proteins. Nature 608: 618–625. 10.1038/s41586-022-05009-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AS, Batista PJ, Gold RS, Chen YG, de Rooij DG, Chang HY, Fuller MT. 2017. The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. Elife 6: e26116. 10.7554/eLife.26116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. 10.1016/j.cell.2007.01.043 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. 2002. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev 16: 2733–2742. 10.1101/gad.1026102 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514. 10.1016/j.devcel.2007.03.001 [DOI] [PubMed] [Google Scholar]
- Choi H, Wang Z, Dean J. 2021. Sperm acrosome overgrowth and infertility in mice lacking chromosome 18 pachytene piRNA. PLoS Genet 17: e1009485. 10.1371/journal.pgen.1009485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O'Carroll D. 2011. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480: 259–263. 10.1038/nature10547 [DOI] [PubMed] [Google Scholar]
- Deng W, Lin H. 2002. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2: 819–830. 10.1016/s1534-5807(02)00165-x [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114. 10.1038/nrg2290 [DOI] [PubMed] [Google Scholar]
- Gainetdinov I, Colpan C, Arif A, Cecchini K, Zamore PD. 2018. A single mechanism of biogenesis, initiated and directed by PIWI proteins, explains piRNA production in most animals. Mol Cell 71: 775–790.e775. 10.1016/j.molcel.2018.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainetdinov I, Cecchini K, Vega-Badillo J, Bagci A, Colpan C, Arif A, Wu P-H, Zamore PD. 2022. Relaxed targeting rules allow PIWI-clade Argonaute proteins to silence ever-mutating transposons. bioRxiv 10.1101/2022.08.04.502788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genzor P, Konstantinidou P, Stoyko D, Manzourolajdad A, Marlin Andrews C, Elchert AR, Stathopoulos C, Haase AD. 2021. Cellular abundance shapes function in piRNA-guided genome defense. Genome Res 31: 2058–2068. 10.1101/gr.275478.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. 2009. Small silencing RNAs: an expanding universe. Nat Rev Genet 10: 94–108. 10.1038/nrg2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202. 10.1038/nature04917 [DOI] [PubMed] [Google Scholar]
- Goh WS, Falciatori I, Tam OH, Burgess R, Meikar O, Kotaja N, Hammell M, Hannon GJ. 2015. piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev 29: 1032–1044. 10.1101/gad.260455.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou LT, Dai P, Yang JH, Xue Y, Hu YP, Zhou Y, Kang JY, Wang X, Li H, Hua MM, et al. 2014. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res 24: 680–700. 10.1038/cr.2014.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. 2006. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 20: 1709–1714. 10.1101/gad.1434406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590. 10.1126/science.1140494 [DOI] [PubMed] [Google Scholar]
- Han BW, Wang W, Li C, Weng Z, Zamore PD. 2015. Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348: 817–821. 10.1126/science.aaa1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homolka D, Pandey RR, Goriaux C, Brasset E, Vaury C, Sachidanandam R, Fauvarque MO, Pillai RS. 2015. PIWI slicing and RNA elements in precursors instruct directional primary piRNA biogenesis. Cell Rep 12: 418–428. 10.1016/j.celrep.2015.06.030 [DOI] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. 2015. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12: 115–121. 10.1038/nmeth.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi N, Shoji K, Suzuki Y, Katsuma S, Tomari Y. 2020. Zucchini consensus motifs determine the mechanism of pre-piRNA production. Nature 578: 311–316. 10.1038/s41586-020-1966-9 [DOI] [PubMed] [Google Scholar]
- Jain D, Puno MR, Meydan C, Lailler N, Mason CE, Lima CD, Anderson KV, Keeney S. 2018. ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2. Elife 7: e30919. 10.7554/eLife.30919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka S, Izumi N, Katsuma S, Tomari Y. 2011. 3′ end formation of PIWI-interacting RNAs in vitro. Mol Cell 43: 1015–1022. 10.1016/j.molcel.2011.07.029 [DOI] [PubMed] [Google Scholar]
- Kojima-Kita K, Kuramochi-Miyagawa S, Nagamori I, Ogonuki N, Ogura A, Hasuwa H, Akazawa T, Inoue N, Nakano T. 2016. MIWI2 as an effector of DNA methylation and gene silencing in embryonic male germ cells. Cell Rep 16: 2819–2828. 10.1016/j.celrep.2016.08.027 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. 2004. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131: 839–849. 10.1242/dev.00973 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, Han BW, Xu J, Moore MJ, Schimenti JC, Weng Z, et al. 2013. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell 50: 67–81. 10.1016/j.molcel.2013.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441. 10.1126/science.1102513 [DOI] [PubMed] [Google Scholar]
- Liu R, Kasowitz SD, Homolka D, Leu NA, Shaked JT, Ruthel G, Jain D, Keeney S, Luo M, Pillai RS. 2021. YTHDC2 is essential for pachytene progression and prevents aberrant microtubule-driven telomere clustering in male meiosis. Cell Rep. 37: 110110. 10.1016/j.celrep.2021.110110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn MJ, Ketting RF. 2013. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet 14: 523–534. 10.1038/nrg3495 [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Meister G. 2013. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet 14: 447–459. 10.1038/nrg3462 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197. 10.1016/j.molcel.2004.07.007 [DOI] [PubMed] [Google Scholar]
- Mohn F, Handler D, Brennecke J. 2015. Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 348: 812–817. 10.1126/science.aaa1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS. 2011. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480: 264–267. 10.1038/nature10672 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324. 10.1126/science.1129333 [DOI] [PubMed] [Google Scholar]
- Wang W, Han BW, Tipping C, Ge DT, Zhang Z, Weng Z, Zamore PD. 2015. Slicing and binding by Ago3 or Aub trigger piwi-bound piRNA production by distinct mechanisms. Mol Cell 59: 819–830. 10.1016/j.molcel.2015.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenda JM, Homolka D, Yang Z, Spinelli P, Sachidanandam R, Pandey RR, Pillai RS. 2017. Distinct roles of RNA helicases MVH and TDRD9 in PIWI slicing-triggered mammalian piRNA biogenesis and function. Dev Cell 41: 623–637.e629. 10.1016/j.devcel.2017.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtas MN, Pandey RR, Mendel M, Homolka D, Sachidanandam R, Pillai RS. 2017. Regulation of m6A transcripts by the 3′→5′ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol Cell 68: 374–387.e312. 10.1016/j.molcel.2017.09.021 [DOI] [PubMed] [Google Scholar]
- Wu PH, Fu Y, Cecchini K, Ozata DM, Arif A, Yu T, Colpan C, Gainetdinov I, Weng Z, Zamore PD. 2020. The evolutionarily conserved piRNA-producing locus pi6 is required for male mouse fertility. Nat Genet 52: 728–739. 10.1038/s41588-020-0657-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiol J, Cora E, Koglgruber R, Chuma S, Subramanian S, Hosokawa M, Reuter M, Yang Z, Berninger P, Palencia A, et al. 2012. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol Cell 47: 970–979. 10.1016/j.molcel.2012.07.019 [DOI] [PubMed] [Google Scholar]
- Yang Z, Chen KM, Pandey RR, Homolka D, Reuter M, Janeiro BK, Sachidanandam R, Fauvarque MO, McCarthy AA, Pillai RS. 2016. PIWI slicing and EXD1 drive biogenesis of nuclear piRNAs from cytosolic targets of the mouse piRNA pathway. Mol Cell 61: 138–152. 10.1016/j.molcel.2015.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.