ABSTRACT
FlgM, an antagonist of FliA (also known as σ28), inhibits transcription of bacterial class 3 flagellar genes. It does so primarily through binding to free σ28 to prevent it from forming a complex with core RNA polymerase. We recently identified an FliA homolog (FliATd) in the oral spirochete Treponema denticola; however, its antagonist FlgM remained uncharacterized. Herein, we provide several lines of evidence that TDE0201 functions as an antagonist of FliATd. TDE0201 is structurally similar to FlgM proteins, although its sequence is not conserved. Heterologous expression of TDE0201 in Escherichia coli inhibits its flagellin gene expression and motility. Biochemical and mutational analyses demonstrate that TDE0201 binds to FliATd and prevents it from binding to the σ28-dependent promoter. Deletions of flgM genes typically enhance bacterial class 3 flagellar gene expression; however, deletion of TDE0201 has an opposite effect (e.g., the mutant has a reduced level of flagellins). Follow-up studies revealed that deletion of TDE0201 leads to FliATd turnover, which in turn impairs the expression of flagellin genes. Swimming plate, cell tracking, and cryo-electron tomography analyses further disclosed that deletion of TDE0201 impairs spirochete motility and alters flagellar number and polarity: i.e., instead of having bipolar flagella, the mutant has flagella only at one end of cells. Collectively, these results indicate that TDE0201 is a FlgM homolog but acts differently from its counterparts in other bacteria.
IMPORTANCE Spirochetes are a group of bacteria that cause several human diseases. A unique aspect of spirochetes is that they have bipolar periplasmic flagella (PFs), which bestow on the spirochetes a unique spiral shape and distinct swimming behaviors. While the structure and function of PFs have been extensively studied in spirochetes, the molecular mechanism that regulates the PFs’ morphogenesis and assembly is poorly understood. In this report, FlgM, an anti-σ28 factor, is identified and functionally characterized in the oral spirochete Treponema denticola. Our results show that FlgM regulates the number and polarity of PFs via a unique mechanism. Identification of FliA and FlgM in T. denticola sets a benchmark to investigate their roles in other spirochetes.
KEYWORDS: spirochetes, Treponema, flagella, sigma factors, anti-sigma factors
INTRODUCTION
Bacterial flagella are complex molecular machines that are important for motility, biofilm formation, and pathogenesis. More than 30 different proteins are coordinately assembled to constitute three distinct parts of the flagella: the basal body (motor), hook (joint), and filament (propeller) (1–3). The basal body is embedded within the cell envelope and works as a reversible rotary motor (4–6). The hook acts as a flexible joint that connects the basal body and filament, a long helical structure composed of tens of thousands of subunits of a single protein called flagellin (7). Flagellin proteins are synthesized in the cytoplasm and exported through the hook-basal body (HBB) (8). To cope with sophisticated flagellar structure, most bacteria have evolved a complex transcriptional hierarchy to control their flagellar gene expression both temporally and spatially (1, 2). Within this regulatory hierarchy, the operons that encode flagellar, motility, and chemotaxis proteins can be divided into three transcriptional classes. Class 1 genes encode FlhC and FlhD, two master regulators that initiate the expression of class 2 genes, which mainly encode HBB structural proteins and two transcriptional regulators, FliA (also called σ28) and FlgM (anti-σ28). As a flagellum-specific alternative σ factor, FliA initiates the expression of class 3 genes, such as those encoding flagellin and chemotaxis proteins. As an antagonist of FliA, FlgM negatively regulates the expression of class 3 genes through direct interaction with FliA, preventing it from forming a complex with core RNA polymerase (RNAP) (9, 10). FlgM can also attack and destabilize the FliA-RNAP holoenzyme and inhibit transcription initiation (11). FlgM exerts its inhibitory role on flagellar gene expression through a partner-switching mechanism in response to HBB assembly and completion (12, 13). Specifically, FlgM and FliA remain bound as a complex in the cytoplasm until HBB assembly is completed, whereupon FlgM is exported out of cells, releasing FliA to initiate transcription of class 3 genes. This regulatory paradigm was first established in the two model organisms Escherichia coli and Salmonella enterica serovar Typhimurium (1, 2, 13). Later, a similar regulatory scheme was found in numerous motile bacteria, such as Campylobacter jejuni (14), Helicobacter pylori (15, 16), Pseudomonas aeruginosa (17), Vibrio cholerae (18), and Bacillus subtilis (13, 19). Deletions of fliA led to mutants deficient in flagellar filaments and nonmotile, highlighting the gene’s essential role in bacterial flagellar synthesis and motility (16, 18, 20–22). In contrast, deletions of flgM typically increase class 3 flagellar gene expression: e.g., deletions of the flgM gene in H. pylori and S. Typhimurium enhance flagellin gene expression by several fold (16, 23).
Spirochetes are a group of bacteria that cause several human diseases: e.g., Lyme disease (Borrelia burgdorferi), syphilis (Treponema pallidum), leptospirosis (Leptospira interrogans), and periodontal disease (Treponema denticola) (24–27). A unique aspect of spirochetes is that they have bipolar flagella encased within the periplasmic space. Therefore, spirochetal flagella are named endoflagella or periplasmic flagella (PFs) (28, 29). Spirochetes are diverse in terms of flagellar number and length and whether or not PFs overlap at the middle of cells (29). For example, L. interrogans has a single short flagellum inserted at each pole that does not overlap the other (30). In contrast, B. burgdorferi has 7 to ~11 long PFs subterminally inserted at the cell poles. These PFs form two bundles of ribbon-like structures that overlap at the middle regions of the cells (31, 32). PFs confer upon spirochetes a distinctive flat-wave or spiral morphology and unique form of screw-like motility that empower them to translocate through viscous substrates and tissues to access otherwise inaccessible host niches (29, 33). Therefore, in pathogenic spirochetes, motility facilitates invasion and dissemination and is essential for infection (26, 34). Like external flagellates, PFs are also composed of a motor, hook, and filament; however, they are more complex in terms of motor structures and flagellar filament compositions (35). For example, PFs have a unique structure called the “collar,” a large multiprotein complex consisting of an inner core and an outer turbine-like structure. The collar has a remarkable structural plasticity essential not only for assembly of flagellar motors in the highly curved membrane of spirochetes but also for generation of the high torque necessary for spirochete motility (36–38). The flagellar hook protein FlgE in spirochetes self-catalyzes the formation of an unusual intersubunit lysinoalanine cross-link critical for cell motility (39, 40). Spirochetes are complex in terms of flagellar filament compositions. Most spirochetes have multiple flagellin proteins (28, 29). For instance, the flagellar filaments of T. denticola and T. pallidum comprise at least one sheath protein, FlaA, and three core FlaB proteins (41–45). The flagellar filament of Leptospira is even more complex as it is composed of two FlaA proteins, at least one FlaB protein, and two novel Leptospira-specific flagellar filament proteins, FcpA and FcpB (30, 46, 47). It is noteworthy that FlaB proteins are homologs of FliC. FlaA proteins are unique to spirochetes and do not share sequence similarity with bacterial flagellin proteins (28).
While the structure and function of PFs have been extensively studied in spirochetes (29, 30), the molecular mechanism that regulates PFs’ morphogenesis is poorly understood. Thus far, it remains unclear if spirochetes have evolved the aforementioned transcriptional hierarchy to regulate flagellar gene expression and assembly. In our recent studies, we have been seeking to fill this gap by using T. denticola as a model organism. Similar to other spirochetes, the flagellar filaments of T. denticola are composed of at least one FlaA protein and three FlaB proteins (FlaB1, FlaB2, and FlaB3). The genes encoding these four proteins are regulated by different promoters (41, 44). For instance, a conserved σ28-dependent promoter is mapped upstream of flaB2, and a σ70-dependent promoter is mapped upstream of flaA and flaB1 of T. denticola (41). Additionally, we recently identified and functionally characterized an FliA homolog (FliATd) in T. denticola and found that it functions as a flagellum-specific σ28 factor that positively regulates the flagellin genes of T. denticola (48). However, FlgM, an antagonist of FliA, has not been identified in T. denticola or other spirochetes. In this report, by using the approaches of bioinformatics, genetics, biochemistry, and cryo-electron tomography (cryo-ET), we provide several lines of evidence that TDE0201 is an FlgM homolog that regulates the flagellin gene expression and flagellar number and polarity of T. denticola.
RESULTS AND DISCUSSION
TDE0201 is an FlgM homolog.
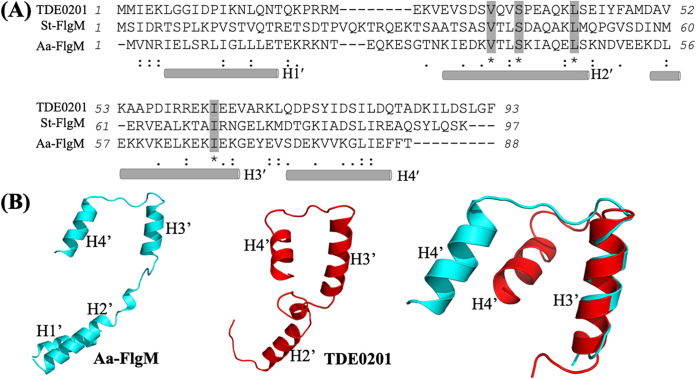
Our recent study shows that TDE2683 is a FliA (σ28) homolog that regulates the later class 3 flagellar genes of T. denticola (48), yet its antagonist FlgM has not been identified. FlgM proteins (PF04316 and PF05998) are not conserved, and their sizes are diverse, ranging from 65 to 131 amino acids (aa) (49). The function of FlgM has been well characterized in S. Typhimurium, and its three-dimensional (3D) structure was determined in Aquifex aeolicus as a complex with σ28 by X-ray crystallography (11, 50, 51). Using these two bacterial FlgM proteins as queries, we searched the genome of T. denticola for their homologs and found that TDE0201 has 15.73% shared sequence identity with S. Typhimurium FlgM (FlgMSt) and 20.48% shared identity with A. aeolicus FlgM (FlgMAa). We also performed large-scale sequence alignment analyses and found that TDE0201 shares a very limited sequence identity to its counterparts from other bacteria out of the Treponema genus, indicating that TDE0201 is not conserved. This is also true for other bacterial FlgM homologs: e.g., H. pylori FlgM (HP1122) is ~11% identical to the E. coli homolog (15). TDE0201 consists of 93 aa, and its predicted molecular weight (MW) is 10.54 kDa. Previous structural studies indicate that both FlgMAa and FlgMSt are composed of four α-helices (H1′, H2′, H3′, and H4′) (50, 51). Sequence alignment analysis revealed that, similar to FlgMAa, TDE0201 also comprises four α-helices, among which H2′ and H3′ align well and contain several conserved residues (e.g., Val33, Ser36, Leu42, and Ile63), although their overall conservation is relatively low (Fig. 1A). To further determine if TDE0201 is an FlgM homolog, we generated its structural model using chain D from the cocrystal structure of σ28/FlgMAa complex (PDB code 1rp3) (50) as a template and found that the two proteins share a similar structural architecture and their H3′ helices are well aligned (Fig. 1B). The alignment also uncovered significant differences in the positioning of the N-terminal helices, likely due to their disordered nature in the absence of binding to the σ28 factors (51). Collectively, sequence alignment and homology modeling analyses suggest that TDE0201 is an FlgM homolog; thus, here, it is denoted FlgMTd.
FIG 1.
TDE0201 is a homolog of FlgM. (A) Sequence alignment of FlgM homologs. The numbers show the positions of amino acids of T. denticola TDE0201 (WP_002666518), S. Typhimurium FlgM (St-FlgM) (NP_460143), and A. aeolicus FlgM (Aa-FlgM) (WP_010880181.1). The rods represent four helices of FlgM, including H1′, H2′, H3′, and H4′. The alignments were conducted using the program Clustal Omega. (B) Homology modeling of TDE0201. The structural model was generated using chain D from the cocrystal structure of A. aeolicus σ28/FlgM complex (PDB code 1rp3) (50) as a template with MODELLER version 10.0 (83) and the MPI Bioinformatics Toolkit (84). The H3′ helices of TDE0201 and FlgMAa were superimposed.
The flgMTd gene is regulated by a σ70-dependent promoter.
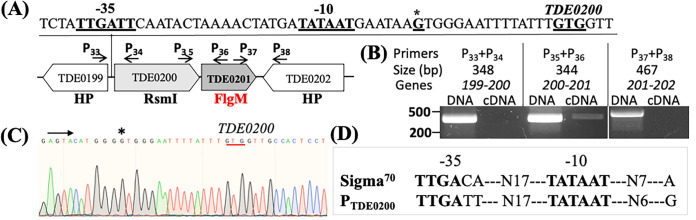
The flgMTd gene (TDE0201) and its upstream TDE0200 gene are in the same transcriptional orientation (Fig. 2A). TDE0200 encodes a rRNA small subunit methyltransferase I (RsmI) responsible for the 2′-O-methylation of cytidine 1402 in bacterial 16S rRNA (52). Co-reverse transcription-PCR (co-RT-PCR) analysis showed that only the pair of primers bridging TDE0200 and TDE0201 yielded a positive band, whereas the other two pairs of primers across the intergenic regions of TDE0199-TDE0200 and TDE0201-TDE0202 were both negative (Fig. 2B), suggesting that TDE0201 is cotranscribed with its upstream TDE0200 gene but not with its downstream TDE0199 gene, a gene in an opposite transcriptional direction. By using 5′ rapid amplification of cDNA ends (5′-RACE), we identified a transcriptional start site (TSS) at the upstream region of TDE0200, 21 bp from its start codon (Fig. 2C). The −10 and −35 regions from the TSS contain a consensus sequence of σ70-dependent promoter (TTGATT-N17-TATAAT) (Fig. 2D), suggesting that the expression of flgMTd is most likely controlled by σ70.
FIG 2.
TDE0201 resides in a gene cluster that is regulated by a σ70 promoter. (A) Diagram showing the genes adjacent to TDE0201. Arrows represent the relative positions and orientations of RT-PCR primers that span the intergenic regions between individual genes as labeled. (B) RT-PCR analysis. For each pair of primers, chromosomal DNA was used as a positive control. The numbers below the primers are predicted sizes of RT-PCR and PCR products. (C) 5′-RACE analysis. The arrow shows the sequencing direction, and an asterisk indicates the transcriptional start site (TSS). The red underlined sequence (GTG) is the start codon of TDE0200. (D) Sequence comparison between the canonical E. coli σ70 promoter sequence and that identified upstream of TDE0200.
FlgMTd inhibits E. coli motility and fliC expression.
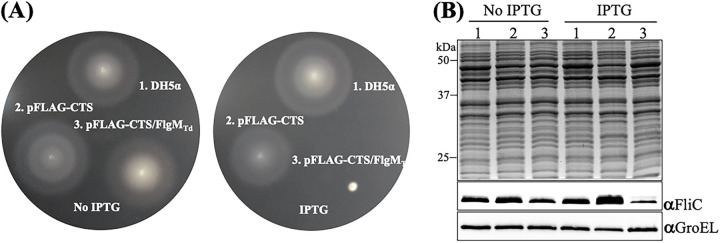
FlgMTd shares about 16% sequence identity and a similar structural topology with E. coli FlgM (FlgMEc). We speculated that if FlgMTd functions as an anti-σ28 factor, overexpressing this gene in E. coli may impair its late flagellar gene expression (e.g., the flagellin gene fliC) and motility. To determine if this is the case, we ectopically expressed flgMTd in an E. coli DH5α strain and then assessed its impact on motility using swimming plate assays and the level of FliC using immunoblotting analysis. Supporting our speculation, these two experiments showed that induction of flgMTd with IPTG (isopropyl-β-d-1-thiogalactopyranoside) substantially inhibited E. coli motility (Fig. 3A) and expression of FliC (Fig. 3B). FlgM proteins are exported when HBB assembly is completed. To determine if FlgMTd can be exported in E. coli, after addition of IPTG, we monitored the presence of FlgMTd in the growth medium of DH5α by immunoblotting with the FLAG antibody (anti-Flag). We detected FlgMTd in the whole-cell lysates of DH5α but not in the supernatants concentrated from the growth medium (data not shown), suggesting that no FlgMTd was secreted into the growth medium via the flagellar export apparatus of E. coli or that it was secreted but its level in the growth medium was too low to be detected. This result is understandable because the domain essential for FlgMEc export lies in its N-terminal region (53), which is not conserved in FlgMTd (Fig. 1A). Altogether, the heterologous expression of FlgMTd in E. coli indicates that it shares some common characteristics with FlgMEc (i.e., binding to FliA and repressing flagellin gene expression and motility).
FIG 3.
Heterologous expression of flgMTd inhibits E. coli motility and fliC expression. (A) Swimming plate assays with/without IPTG induction. This assay was carried out on agar plates containing 1% tryptone, 0.5% NaCl, and 0.3% Bacto agar as previously documented (86). The plates were incubated at 30°C overnight. Numbers on the plates represent the following conditions: 1, DH5a alone; 2, DH5a transformed with the empty vector pFLAG-CTS; and 3, DH5a transformed with the vector pFLAG-CTS containing flgMTd. (B) Detection of FliC by immunoblotting analysis. For this experiment, equivalent amounts (~10 μg) of whole-cell lysates of three E. coli strains were analyzed by SDS-PAGE and then probed with specific antibodies against E. coli GroEL (αGroEL) and FliC (αFliC). GroEL was used as a loading control.
FlgMTd interacts with FliATd.
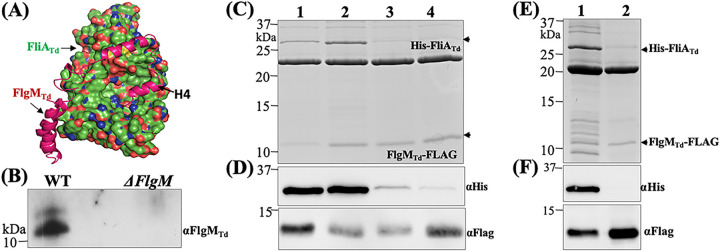
As an anti-σ28 factor, FlgM exerts its inhibitory role on class 3 flagellar gene expression via binding to FliA (11, 13). To determine if FlgMTd interacts with FliATd, we first conducted protein-protein docking analysis by comparing their homology models, which were generated from the cocrystal structure of the σ28/FlgM complex in A. aeolicus (PDB code 1rp3) (50). The docking analysis suggested that FlgMTd binds to FliATd primarily through its C-terminal H3′-H4′ α-helices, whereby H4′ is buried in a deep hydrophobic pocket of FliATd (Fig. 4A). To experimentally test the interaction between FliATd and FlgMTd in vivo, we carried out coimmunoprecipiation (co-IP) assays using T. denticola whole-cell lysates and a specific antibody against FliATd (anti-FliATd). The resulting samples were then subjected to immunoblotting probed against anti-FlgMTd to determine if FlgMTd was precipitated along with FliATd. FlgMTd was detected in the eluates from the wild type (WT) but not in those from the ΔflgM mutant (Fig. 4B), suggesting that FlgMTd and FliATd form a complex; otherwise, it would not have been pulled down by anti-FliATd. To corroborate this result, we coexpressed His-FliATd and FlgMTd-FLAG in E. coli BL21 cells as previously described (54). After induction with IPTG, two recombinant proteins were copurified under native conditions using FLAG beads. The resulting coeluates were then subjected to SDS-PAGE, followed by immunoblotting probed against either anti-His or anti-Flag antibodies. Our results show that His-FliATd was copurified along with FlgMTd-FLAG (Fig. 4C and D). Previous structural and mutagenesis studies indicate that FlgM interacts with FliA mainly via its C-terminal H3′-H4′ helices, which bind to the σ4 domain of FliA (11, 50, 53). To determine if the interaction between FlgMTd and FliATd is specific, we mutated E221 and V231, two conserved residues in the σ4 domain of FliATd (48), and truncated the H4′ helix (79 to 93 aa) of FlgMTd. We first repeated the copurification experiment using three mutated FliATd proteins (E221D, V231E, and E221D V231E) and found that the single mutation in V231 and double mutations in both E221 and V231 nearly abolished the interaction of two proteins, whereas the mutation in E221 had no impact (Fig. 4C and D). We then repeated this experiment using the truncated FlgMTd (ΔH4′) and found that the truncation completely abolished the interaction between FlgMTd and FliATd (Fig. 4E and F). Based on these results, we infer that FlgMTd binds to FliATd mainly through its C-terminal H4′ helix.
FIG 4.
FliATd interacts with FlgMTd. (A) Protein-protein docking analysis of FliATd and FlgMTd. The homology models of FliATd and FlgMTd were first generated using the cocrystal structure of A. aeolicus s28/FlgM complex (PDB code 1rp3), and then protein-protein docking analysis was conducted on the ClusPro server, with FliATd shown as a molecular surface and FlgMTd shown as a backbone ribbon. (B) Coimmunoprecipitation (co-IP). This assay was performed using T. denticola whole-cell lysates and a polyclonal antibody against FliATd (αFliATd). The final co-IP eluates were probed using anti-FlgMTd. A deletion mutant of flgMTd (ΔflgM) was used as a negative control. (C to F) Copurification of N-terminal His-tagged FliATd (His-FliATd) and C-terminal FLAG-tagged FlgMTd (FlgMTd-FLAG). The two tagged proteins and their corresponding site-directed mutated or truncated proteins were coexpressed in the E. coli BL21 Star(DE3) strain and then purified using FLAG beads. The resulting samples were subjected to SDS-PAGE (C and E), followed by immunoblotting analysis (D and F) using either anti-FLAG (αFlag) or anti-His (αHis) antibodies. In panels C and D, in addition to the wild-type FliATd protein (lane 1), three site-directed mutated recombinant proteins were included: E221D (lane 2), V231E (lane 3), and a double mutation of E221D andV231E (lane 4). In panels E and F, lane 1 shows the wild-type FlgMTd protein and lane 2 shows a truncated FlgMTd protein without the H4′ helix (79 to 93 aa).
FlgMTd prevents FliATd from binding to the σ28-dependent promoter.
Our recent study shows that FliATd binds to σ28-dependent promoters, such as the flaB2 promoter (PflaB2) (48). We reasoned that if FlgMTd acts as an anti-σ28 factor, it should sequester FliATd and prevent it from binding to σ28-dependent promoters. To test this hypothesis, we carried out an electrophoretic mobility shift assay (EMSA) using biotin-labeled PflaB2 as a DNA probe. Our results showed that the recombinant FliATd (rFliATd) bound to PflaB2, which was titrated by the recombinant FlgMTd (rFlgMTd) in a dose-dependent manner (Fig. 5). Notably, rFlgMTd alone did not bind to PflaB2. Along with the above result, we conclude that FlgMTd functions as an anti-σ28 factor that interacts with FliATd and prevents it from binding to the σ28-dependent promoter.
FIG 5.
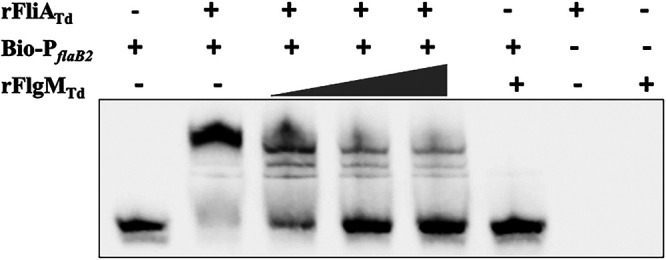
FlgMTd sequesters FliATd from binding to the flaB2 promoter. EMSA was performed using biotin-labeled flaB2 promoter (Bio-PflaB2) as a DNA probe. For this assay, Bio-PflaB2 was incubated with a fixed amount of recombinant FliATd protein (rFliATd) and then titrated with increasing concentrations of recombinant FlgMTd protein (rFlgMTd) as labeled.
FlgMTd controls T. denticola flagellar filament gene expression.
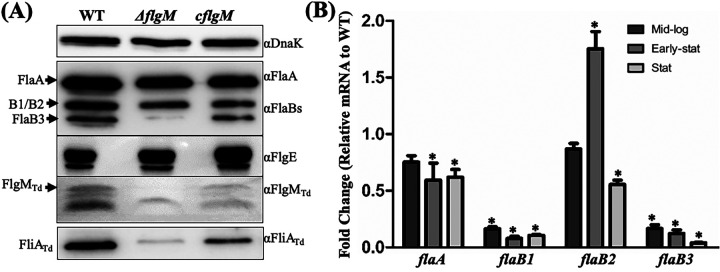
To investigate the role of FlgMTd in T. denticola, we constructed an flgMTd deletion mutant (ΔflgM) and its isogenic complemented strain (cflgM) using a strategy illustrated in Fig. S1 in the supplemental material. Immunoblotting assays with anti-FlgMTd showed that the production of FlgMTd was abolished in the ΔflgM mutant and then restored in the cflgM strain (Fig. 6A). Deletions of flgM genes typically increased class 3 flagellar gene expression (16, 23). Unexpectedly, our immunoblotting results uncovered that deletion of flgMTd significantly reduced the level of four flagellar filament proteins in T. denticola, including FlaA, FlaB1, FlaB2, and FlaB3, whereas the level of the hook protein FlgE remained unchanged (Fig. 6A). The result from FlgE is expected because it belongs to the class 2 flagellar genes, which are regulated by σ70 in the flagellar transcriptional cascade of E. coli and S. Typhimurium (2, 55). To elucidate the mechanism underpinning the reduction of four flagellar filament proteins in the ΔflgM mutant, we monitored their transcript (mRNA) levels during growth by using quantitative RT-PCR (qRT-PCR) and found that the expression of flaA, flaB1, and flaB3 genes was constantly impaired in the ΔflgM mutant at the mid-log, early stationary, and stationary phases (Fig. 6B). Compared to the wild type, expression of the flaB2 gene in the ΔflgM mutant was more diverse (Fig. 6B), ranging from a slight reduction at log phase, transient increase at early stationary phase, and ~50% reduction at stationary phase. We repeated this experiment three times with different biological samples and found a similar pattern. This result indicates that the reduction of four flagellar filament proteins in the ΔflgM mutant mainly occurs at the transcriptional level.
FIG 6.
Deletion of flgMTd impairs T. denticola flagellar filament gene expression. (A) Immunoblotting analysis. Equivalent amounts of whole-cell lysates of the wild-type (WT), ΔflgM, and cflgM strains were analyzed by SDS-PAGE and then probed with specific antibodies against the following proteins: DnaK (loading control), FlaA, FlaBs, FlgE, FlgMTd, and FliATd. (B) qRT-PCR analysis. For this experiment, the levels of four flagellar filament genes (flaA, flaB1, flaB2, and flaB3) were measured by qRT-PCR as previously described (41, 48). The dnaK gene transcript was used as an internal control to normalize the readouts of qRT-PCR. The results are expressed as the level of individual gene transcripts in the ΔflgM mutant relative to that of the wild type at three different growth phases, including the mid-logarithmic (Mid-log), early stationary (Early-stat), and stationary (Stat) phases. Asterisks indicate that the difference between the wild-type and ΔflgM strains was statistically significant at a P value of <0.01 (one-way ANOVA followed by Tukey’s multiple-comparison test).
Our recent reports show that the flaB2 gene is positively regulated by FliATd (41, 48). Thus, we expected that deletion of FlgMTd, an anti-σ28 factor, should increase its expression. Along with this expectation, we found that the level of flaB2 mRNA was transiently elevated at the early stationary phase of the ΔflgM mutant. However, its expression level was attenuated at the stationary phase, which is contradictory to the role of FlgMTd as an anti-σ28 factor. To explore its underlying mechanism, we first measured the level of FliATd by immunoblotting with anti-FliATd and found that its level significantly decreased in the mutant (Fig. 6A). FlgM interacts with FliA, forming a complex in the cytoplasm (50). We reasoned that deletion of FlgMTd may affect the stability of FliATd, which in turn impairs flagellin gene expression in the mutant. To test this possibility, we carried out protein turnover assays to monitor the stability of FliATd. Our results uncovered that while FliATd was stable in wild type during the course of 24 h, it was rapidly degraded in the ΔflgM mutant: e.g., 6 h after arresting protein synthesis with spectinomycin, FliATd was almost undetectable (Fig. 7A). This result can explain the reduction of flaB2 mRNA, a gene regulated by FliATd (σ28). It is possible that without FlgMTd, free FliATd in the cytoplasm is degraded by certain proteases that are activated when T. denticola enters the stationary phase, which in turn impairs the expression of the flaB2 gene. Supporting this possibility, a similar scenario is reported in E. coli, whereby FlgM protects FliA from proteolysis, which mainly depends on the Lon protease (56).
FIG 7.
Deletion of flgMTd leads to FliATd degradation and increases expression of the csrA gene. (A) Protein turnover assays of the wild type (WT) and the ΔflgM mutant. Protein translation was arrested by adding spectinomycin to the mid-log cultures. Samples were collected at the indicated time points and analyzed by immunoblotting. Equivalent amounts of whole-cell lysates were analyzed by SDS-PAGE and then probed with specific antibodies to DnaK (αDnaK) and FliATd (αFliATd), and DnaK was used as a loading control. (B) Detection of csrA (TDE2355) gene transcripts using qRT-PCR. Similar to the above qRT-PCR analysis, dnaK was used as an internal control to normalize the readouts of qRT-PCR, and the results are expressed as the level of individual gene transcripts in the ΔflgM mutant relative to those of the wild type at the mid-log, early stationary, and stationary phases. Asterisks indicate that the difference between the wild-type and ΔflgM strains was statistically significant at a P value of <0.01.
It was unexpected that deletion of flgMTd led to the reduction of flaA, flaB1, and flaB3 transcripts (Fig. 6) because they are regulated by σ70 rather than σ28 (41, 57). CsrA (carbon storage regulator A) is a small RNA binding protein that represses flagellin gene expression in several bacteria both transcriptionally and translationally (32, 58, 59). CsrA is also a global regulator that orchestrates vast gene expression, including σ factors such as FliA (60). The genome of T. denticola encodes a CsrA homolog (TDE2355) (61), and its binding site is found at the upstream untranslated regions (UTRs) of the flaA, flaB1, and flaB3 genes (data not shown); thus, we speculated that deletion of flgMTd may increase expression of TDE2355 which, in turn, represses expression of the flaA, flaB1, and flaB3 genes. To determine if this possibility stands, we measured the TDE2355 mRNA using qRT-PCR and found that its level was significantly increased in the mutant at the log, early stationary, and stationary phases (Fig. 7B), which coincides with the reduction of flaA, flaB1, and flaB3 mRNA. A similar phenotype is also reported in Yersinia pseudotuberculosis (62). Taken together, these results indicate that while FlgMTd is an anti-σ28 factor, its role and mechanism of action are more complex than those of its counterparts in other bacteria. This unique regulatory mechanism may allow the spirochete to finely govern the level of multiple flagellin proteins to assemble flagellar filaments, which are structurally more complex than most external flagellates.
FlgMTd is essential for the motility of T. denticola.
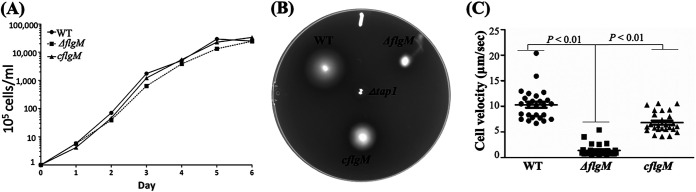
Deletion of flgMTd has no impact on T. denticola growth because the growth rate of the ΔflgM mutant was similar to those of the wild-type and cflgM strains (Fig. 8A). Under dark-field microscopy, we found that the ΔflgM mutant swam poorly and was unable to displace in 1% methylcellulose (Videos S1 to S3), suggesting that its motility was impaired. To confirm this observation, we carried out swimming plate assays (Fig. 8B) and found that the swimming rings of the ΔflgM mutant (5.25 ± 0.17 mm; n = 16 plates) were significantly smaller than those of the wild type (18.75 ± 0.24 mm; n = 16 plates) and the complemented strain (11.62 ± 0.38 mm; n = 16 plates) (Fig. 8C). This result was further confirmed by bacterial motion tracking analysis, wherein individual bacterial cells were tracked for at least 30 s in 1% methylcellulose, and their average velocities (μm/s, n = 26 cells) were measured as follows: wild type, 10.31 ± 0.6 μm/s; ΔflgM strain, 1.39 ± 0.23 μm/s; and cflgM strain, 6.82 ± 0.39 μm/s. These results indicate that FlgMTd is indispensable for T. denticola motility, in contrast to its counterparts in other bacteria, where deletions of flgM genes either have no significant impact on motility or lead to abnormal swimming behaviors (16, 23, 63, 64). We noticed that the motility of cflgM was not fully restored. This may be because the expressional level or timing of flgMTd in the complemented strain does not fully coincide with that of wild type. This strain was created by cis-complementation. We also tried to complement the ΔflgM mutant using pBFC, a shuttle vector of T. denticola (65, 66); however, this vector is poorly transformable in the parental wild-type ATCC 35405 strain (65).
FIG 8.
Deletion of flgMTd has no impact on T. denticola growth but inhibits its motility. (A) Growth curves of the wild-type (WT), ΔflgM, and cflgM strains. For this experiment, 105 T. denticola cells were inoculated into the TYGVS medium and enumerated every 24 h for up to 6 days using a Petroff-Hausser counting chamber. (B) Swimming plate assays. This assay was carried out on 0.35% agarose containing the TYGVS medium diluted 1:1 with PBS. The plates were incubated anaerobically at 37°C for 3 days. The Δtap1 strain, a nonmotile mutant, was used as a control to determine initial inoculum sizes. (C) Cell tracking analysis of the wild-type, ΔflgM, and cflgM strains. T. denticola cells were tracked in the presence of 1% methylcellulose as previously described (57). The results are expressed as average cell velocities (micrometers per second) of at least 20 cells. The data were analyzed by one-way ANOVA, followed by Tukey’s multiple-comparison test at P < 0.01.
Deletion of flgMTd alters the flagellar polarity of T. denticola.
To determine if deletion of flgMTd impairs T. denticola flagellar assembly, we applied cryo-ET to directly visualize PFs inside the cells. In total, we generated 20 whole-cell reconstructions, including wild-type (3 cells), ΔflgM (10 cells), and cflgM (7 cells) strains. We also enumerated the number of flagellar motors and measured the length of PFs (Table 1). As in our previous reports (41, 42, 48, 57), T. denticola wild-type cells uniformly have bipolar flagella (two PFs at both cell poles), which form two helical bundles wrapping around the cell cylinders (Fig. 9A). Interestingly, the two bundles of PFs are asymmetrical in terms of length (i.e., the PFs from one end of the cells are usually longer than those from the opposite end). Jutras et al. uncovered that T. denticola forms a single division zone and divides near the middle of the cells (67); thus, a nascent daughter cell might inherit one bundle of PFs from the old cell pole and newly assemble the other bundle of PFs at the new cell pole. Therefore, we referred to the pole with long PFs as the “old pole” and the one with short PFs as the “new pole.” We measured the lengths of PFs and found that those from “old pole” (5.0 ± 0.6 μm; n = 6 PFs) are nearly two times longer than those from “new pole” (2.9 ± 0.7 μm; n = 6 PFs). A similar phenotype was also observed in the complemented strain (Table 1). The phenotype of the ΔflgM mutant is quite striking. Instead of having bipolar PFs, as observed in both the wild-type and cflgM strains, the majority of mutant cells (8/10) had two PFs at only one end of cells (Fig. 9B and Table 1). Of the remaining two mutant cells, one had a single flagellum at each cell end, and the other had no PFs. We found that the number of flagellar motors (3.3 motors per cell) in the mutant is slightly lower than that of wild type (4 motors per cell) but not statistically significant (Table 1). We also measured the length of PFs in situ and found that the remaining two PFs in the mutant (5.2 ± 1.1 μm; n = 18 PFs) are longer (P < 0.05) than those in the wild type (3.9 ± 1.3 μm; n = 12 PFs). Occasionally, those PFs were too long and even protruded from the cells (Fig. 9B). These results indicate that deletion of flgMTd has limited impact on the number of flagellar motors but does alter the flagellar filament number, length, and polarity in T. denticola. To our knowledge, a similar phenotype has not been reported in other bacteria.
TABLE 1.
Cryo-ET analysis of the wild type, ΔflgM, and cflgM strains
| Strains | No. of motors | PF length(s) (μm) ina: |
|
|---|---|---|---|
| New pole | Old pole | ||
| Wild type | |||
| Cell 1 | 4 | 2.1 and 2.1 | 4.8 and 6.1 |
| Cell 2 | 4 | 3.5 and 2.5 | 5.0 and 5.5 |
| Cell 3 | 4 | 3.4 and 3.5 | 4.6 and 4.6 |
| Avg | 4 | 2.9 ± 0.7 (n = 6) | 5.1 ± 0.6 (n = 6) |
| ΔflgM mutant | |||
| Cell 1 | 3 | 2.5 | 5.0 |
| Cell 2 | 4 | ND | ND |
| Cell 3 | 3 | ND | 5.5 and 5.5 |
| Cell 4 | 4 | ND | 6.0 and 6.0 |
| Cell 5 | 3 | ND | 6.5 and 6.5 |
| Cell 6 | 3 | ND | 4.0 and 4.5 |
| Cell 7 | 3 | ND | 4.0 and 6.0 |
| Cell 8 | 3 | ND | 5.8 and 6.0 |
| Cell 9 | 3 | ND | 4.0 and 6.0 |
| Cell 10 | 4 | ND | 5.0 and 6.0 |
| Avg | 3.3 | ND | 5.6 ± 0.7 (n = 17) |
| cflgM complemented clone | |||
| Cell 1 | 4 | 4.0 and 4.0 | 5.0 and 5.0 |
| Cell 2 | 4 | 3.0 and 4.5 | 5.0 and 5.0 |
| Cell 3 | 4 | 4.5 and 5.0 | 5.0 and 5.0 |
| Cell 4 | 4 | 1.5 and 1.5 | 6.0 and 6.0 |
| Cell 5 | 4 | 0.5 and 1.5 | 5.5 and 5.5 |
| Cell 6 | 4 | 3.0 and 6.5 | 6.5 and 6.5 |
| Cell 7 | 4 | 6.5 and 5.0 | 6.5 and 6.0 |
| Avg | 4 | 3.6 ± 1.9 (n = 14) | 5.6 ± 0.7 (n = 14) |
ND, no PFs detected; n, number of PFs.
FIG 9.
Whole-cell cryo-ET analysis of the wild-type, ΔflgM, and cflgM cells. Shown are projection images of a wild-type cell (A), ΔflgM cell (B), and cflgM cell (C). PFs originating from one end of cells are colored in yellow and those from the opposite end in orange.
Summary.
The above studies provide several lines of evidence that TDE0201 is a FlgM homolog: (i) it has a structural topology similar to that of its counterparts from other bacteria (Fig. 1), (ii) it represses E. coli motility and fliC expression (Fig. 2), and (iii) it interacts with the T. denticola σ28 (FliATd) and prevents it from binding to the σ28-dependent promoters (Fig. 4 and 5). Identification of FlgMTd and FliATd indicates that T. denticola has also evolved to regulate its later flagellar gene expression through the partner-switching mechanism between σ28 and its antagonist FlgM. We also searched the genomes of other spirochetes using FliATd and FlgMTd as queries and identified their homologs in T. pallidum, Brachyspira, and Leptospira species: e.g., FliATd and FlgMTd share 69% and 39% sequence identities with TP_0709 and TP_0974 of T. pallidum (68), respectively, suggesting that a similar regulatory mechanism also exists in other spirochetes. Therefore, the results presented in this report also shed light on the role of FliA and FlgM homologs in other spirochetes. Our results also demonstrate that FlgMTd acts differently from its counterparts in other bacteria in several ways: (i) FlgMTd protects FliATd from being degraded (e.g., deletion of flgMTd leads to FliATd rapid turnover) (Fig. 7), (ii) deletion of flgMTd represses flagellin gene expression in T. denticola (Fig. 6), rather than increasing flagellin gene expression as reported in other bacteria, (iii) FlgMTd interplays with other regulators, such as CsrA (Fig. 7), and (iv) deletion of flgMTd alters the flagellar polarity from bipolar to monopolar (Fig. 9). We found that the reduction of flagellin gene expression in the ΔflgM mutant is mainly due to FliATd turnover and CsrA upregulation. However, the mechanism by which FlgMTd regulates flagellar polarity remains unknown. Previous studies have shown that small GTPase FlhF and FlhG act cooperatively to harness flagellar number and placement in several polar flagellates, such as B. burgdorferi (31), C. jejuni (69), and Vibrio alginolyticus (70). Additionally, FapA (flagellar assembly protein A) controls the flagellar polarity of Vibrio vulnificus (71, 72). We recently found FlhF (TDE2686), FlhG (TDE2685), and FapA (TDE2682) homologs in a large motility gene operon of T. denticola (48). Therefore, it is possible that FlgMTd regulates flagellar polarity by directly or indirectly modulating these proteins. We are planning to perform chromatin immunoprecipitation sequencing (ChIP-seq) to map the promoters recognized by FliATd and transcriptome sequencing (RNA-seq) to determine genes coordinately regulated by FliATd and FlgMTd. Results from these studies will provide insights into the role that FlgMTd plays in control of T. denticola flagellar polarity.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and oligonucleotide primers.
Treponema denticola ATCC 35405 (wild type [WT]) was used in this study (61). Cells were grown in tryptone-yeast extract-gelatin-volatile fatty acids-serum (TYGVS) medium at 37°C in an anaerobic chamber in the presence of 85% nitrogen, 5% carbon dioxide, and 5% hydrogen (73). T. denticola isogenic mutants were grown with appropriate antibiotic for selective pressure as needed: erythromycin (50 μg/mL) and gentamicin (20 μg/mL). The Escherichia coli DH5α strain (New England Biolabs, Ipswich, MA) was used for DNA cloning. BL21 Star(DE3) (Invitrogen, Waltham, MA) was used for preparing recombinant proteins. The E. coli strains were cultivated in lysogeny broth (LB) supplemented with appropriate concentrations of antibiotics for selective pressure as needed: erythromycin, 400 μg/mL; gentamicin, 20 μg/mL; kanamycin 50 μg/mL; and ampicillin, 100 μg/mL. The oligonucleotide primers for PCR and RT-PCR used in this study are listed in Table S1 in the supplemental material. These primers were synthesized in Integrated DNA Technologies (IDT, Coralville, IA).
Electrophoresis and immunoblotting analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting analyses were carried out as previously described (48). For immunoblotting, T. denticola cells were harvested in the mid-logarithmic (mid-log) phase (~5 × 108 cells/mL). Equal amounts of whole-cell lysates (1 to 5 μg) were separated by SDS-PAGE and then transferred to polyvinylidene difluoride (PVDF) membranes. The immunoblots were probed with specific antibodies against T. pallidum FlaB and T. denticola FlaA, DnaK, FlgE, and FliA as described in previous publications (48, 74, 75). The antibody against T. denticola TDE0201 was generated in rats in this study (see below for details). His tag and FLAG tag antibodies were purchased from Thermo Fisher Scientific (Waltham, MA) and Sigma-Aldrich (St. Louis, MO). E. coli flagellin (FliC) and GroEL antibodies were purchased from Abcam (Cambridge, United Kingdom). Immunoblots were developed using a horseradish peroxidase-conjugated secondary antibody with an enhanced chemiluminescence (ECL) assay, and signals were quantified using the Molecular Imager ChemiDoc system with the Image Lab software (Bio-Rad Laboratories, Hercules, CA) as previously described (57).
Preparation of TDE0201 recombinant protein and its antiserum.
The gene encoding the full-length TDE201 protein was PCR amplified with primers P1/P2 using Pfx DNA polymerase (Life Technologies, Grand Island, NY) and then cloned into the pET200/d-TOPO expression vector (Life Technologies), which encodes a 6-histidine-tag (6×His) at the N terminus. The resulting plasmids were then transformed into BL21 Star(DE3) (Invitrogen). The expression of TDE0201 was induced using 1 mM isopropyl-β-d-1-thiogalactopyranodside (IPTG). For antiserum preparation, the recombinant TDE0201 protein was purified using Ni-nitrilotriacetic acid (NTA) agarose (Qiagen, Valencia, CA) under denaturing conditions according to the manufacturer’s protocol (Qiagen). For the electrophoretic mobility shift assay (EMSA), the recombinant proteins were purified using Ni-NTA agarose (Qiagen) under native conditions in the presence of 1% Sarkosyl according to the manufacturer’s protocol (Qiagen). The purified proteins were then dialyzed in 20 mM Tris-HCl buffer (pH 8.0) at 4°C overnight using 3.0-kDa molecular-weight-cutoff Spectra/Por dialysis bags (Spectrum Laboratories, Rancho Dominguez, CA). The concentrations of purified proteins were determined using a Bio-Rad Protein assay kit (Bio-Rad). The antibody against TDE0201 was produced by General Bioscience (Brisbane, CA) as previously described (48). In brief, 5 mg of purified recombinant TDE0201 was used to immunize two rats following a standard immunization procedure. The primers for preparation of the recombinant proteins are listed in Table S1.
Heterologous expression of TDE0201 in E. coli.
The gene encoding the full-length TDE201 protein was PCR amplified with primers P40/P41, and the resulting amplicon was first cloned into pGEM-T Easy vector (Promega, Madison, WI) and then subcloned into pFLAG-CTS expression vector (Sigma-Aldrich). The resulting plasmid (pFLAG-CTS-TDE0201) and pFLAG-CTS (negative control) were transformed into the E. coli DH5α strain, respectively. The resulting DH5α strains were subjected to immunoblotting analysis with FLAG antibody (anti-Flag) for detection of TDE0201 and anti-FliC for detection of E. coli FliC expression, as well as to swimming plate assays for assessing the impact of TDE0201 on E. coli motility. Swimming plate assays of E. coli were performed on soft agar plates containing 1% tryptone, 0.5% NaCl, and 0.3% agar as previously described (48). In brief, overnight E. coli cultures were diluted to 1:100. After 8 h of growth at 37°C, 1 μL of the culture was inoculated onto the soft agar plates with or without IPTG (1 mM), and then the plates were incubated at 30°C for 16 h. The average diameters of swimming rings were calculated from four independent plates; the results are represented as the mean of diameters ± standard error of the mean (SEM). The data were statistically analyzed by one-way analysis of variance (ANOVA), followed by Tukey’s multiple-comparison test at P < 0.01.
Construction of a TDE0201 deletion mutant and its complemented strain.
The TDE0201::erm (Fig. S1A) plasmid was constructed to replace the entire open reading frame of TDE0201 with a previously documented erythromycin resistance cassette (ERM-F/AM) (76). The vector TDE0201com (Fig. S1B) was constructed to cis-complement the TDE0201 deletion mutant. These two plasmids were constructed by two-step PCR and DNA cloning. To construct TDE0201::erm, the TDE0201 upstream region, ERM-F/AM, and the TDE0201 downstream region were PCR amplified with primers P3/P4, P5/P6, and P7/P8, respectively. The resulting PCR fragments were cloned into the pGEM-T Easy vector (Promega, Madison, WI). The upstream and downstream regions were ligated. The resulted fragment was then inserted by ERM-F/AM at an engineered XbaI cut site, generating TDE0201::erm, as illustrated in Fig. S1A. To delete TDE0201, TDE0201::erm was linearized and transformed into T. denticola wild-type competent cells via heat shock as previously described (77). The deletion was confirmed by PCR and immunoblotting analyses. The resulting mutant was designated the ΔflgM strain. To construct TDE0201com, the full-length TDE0201 gene along with its upstream region and the aacC1 cassette were PCR amplified with primers P3/P9 and P10/P11, respectively, and then fused together with primers P3/P11, generating TDE0201-aacC1. The downstream region of TDE0201 was PCR amplified with primers P12/P8 and then fused to TDE0201-aacC1 by PCR using primers P3/P8 (Fig. S1B). The obtained DNA fragment was cloned into the pGEM-T Easy vector, generating the vector TDE0201com. To restore the expression of TDE0201, TDE0201com was linearized and transformed into the ΔflgM mutant via heat shock (77). The resulting complemented clones were confirmed by PCR and immunoblotting analyses. One complemented clone (cflgM) was selected for further characterizations. The primers for constructing these two plasmids are listed in Table S1.
Coimmunoprecipitation.
Coimmunoprecipitation (co-IP) was carried out as previously described (57). Briefly, 500 mL of the mid-log-phase T. denticola wild-type and ΔflgM mutant (negative control) cultures (~5 × 108 cells/mL) was centrifuged at 5,000 × g for 20 min at 4°C. The obtained cell pellets were washed three times with phosphate-buffered saline (PBS [pH 7.4]) and then resuspended in 2 mL of TSEA buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 0.05% sodium azide [pH 7.5]). The resulting cell suspensions were sonicated on ice, followed by centrifugation (14,800 × g for 30 min at 4°C) to collect the cell supernatants. For co-IP, ~500 μL supernatant was incubated with 100 μL of FliATd antibody (anti-FliATd) for 16 h at 4°C in the presence of 1% bovine serum albumin (BSA). After the incubation, ~50 μL of protein A agarose (Sigma-Aldrich) was added, and the mixture was further incubated for 16 h at 4°C. The resulting samples were centrifuged (1,600 × g at 4°C) and then washed three times with 1 mL of PBS buffer containing 0.05% Tween 20. The final pellets were suspended in 50 μL of electrophoresis sample buffer, boiled for 5 min, and then briefly centrifuged as before to collect the supernatants, which were then subjected to SDS-PAGE and immunoblotting analysis.
Coexpression of recombinant TDE2683 (FliATd) and TDE0201 (FlgMTd) proteins.
To determine if FlgMTd binds to FliATd, these two proteins were coexpressed in E. coli with either a His tag (His-FliATd) or a FLAG tag (FlgMTd-FLAG) as previously described (58, 78). Briefly, the genes encoding the full-length FliATd and FlgMTd were amplified with primers P13/P14 and P15/P16, respectively, and then fused together with primers P13/P16. Of note, the sequence encoding FLAG was added to the 3′ end of TDE0201 and a stop codon was added between these two genes. The resulting amplicons were cloned into pGEM-T Easy vector (Promega) and then subcloned into pQE80L expression vector (Qiagen), which encodes the 6×His tag at the N terminus. The resulting plasmids were transformed into BL21 Star(DE3) (Invitrogen). Recombinant His-FliATd and FlgMTd-FLAG proteins were induced using 1 mM isopropyl β-d-1-thiogalactosidase (IPTG) and then purified using either Ni-NTA agarose (Qiagen) or anti-Flag M2 affinity gel (Sigma-Aldrich) under native conditions according to the manufacturers’ protocols.
Bacterial swimming plate assay and motion tracking analysis.
Swimming plate assays were performed as previously described (41). Briefly, 4 μL of T. denticola cultures (109 cells/mL) was inoculated onto 0.35% agarose containing TYGVS medium diluted 1:1 with PBS. The plates were incubated anaerobically at 37°C for 3 days to allow the cells to swim out. The diameters of swimming rings were measured in millimeters. As a negative control, a previously constructed nonmotile mutant of T. denticola (Δtap1) was included to determine the initial inoculum size (79). The average diameters of each strain were calculated from at least four independent plates; the results are represented as the mean of diameters ± standard error of the mean (SEM). The velocity of bacterial cells was measured using a computer-based bacterial tracking system as previously described (57). Briefly, 100 μL of mid-log-phase T. denticola cultures was first diluted (1:50) in oral bacterial growth medium (OBGM), and then 10 μL of diluted cultures was mixed with an equal volume of 2% methylcellulose with a viscosity of 4,000 cp (80). T. denticola cells were videotaped and tracked. For each bacterial strain, 26 cells were recorded for up to 30 s. The average cell swimming velocities (micrometers per second) of tracked cells were calculated.
Measuring T. denticola growth rates.
For this study, a total of 105 cells/mL of late-log-phase T. denticola cultures was inoculated into 5 mL TYGVS medium and then were enumerated every 24 h using a Petroff-Hausser counting chamber (Hausser Scientific, Horsham, PA). Each growth curve is representative of at least three independent cultures, and the results are represented as the mean of cell numbers ± standard error of mean (SEM).
Measuring the level of FlgMTd throughout the growth phase of T. denticola.
For this study, the wild-type strain and a previously constructed flgE-deficient mutant (ΔflgE) of T. denticola were used (61, 76). A total of 2 × 106 cells/mL of late-log-phase T. denticola cultures was inoculated into the TYGVS medium. T. denticola cells in the cultures were enumerated every 24 h using a Petroff-Hausser counting chamber (Hausser Scientific, Horsham, PA). Samples were harvested at different time points and subjected to immunoblotting analysis.
RNA preparations, qRT-PCR, and 5′-RACE.
RNA isolations were performed as previously described (42). Briefly, a total of 105 cells/mL of late-log-phase T. denticola cultures was inoculated into 100 mL TYGVS medium. T. denticola cells in the cultures were enumerated every 24 h using a Petroff-Hausser counting chamber (Hausser Scientific). Samples were harvested at different time points. Total RNA was extracted using TRI reagent (Sigma-Aldrich), following the manufacturer's instructions. The resulting samples were treated with Turbo DNase I (Thermo Fisher Scientific) at 37°C for 2 h to eliminate genomic DNA contamination. The resulting RNA samples were reextracted using acid phenol-chloroform (Ambion), precipitated in isopropanol, and washed once with 70% ethanol. The RNA pellets were resuspended in RNase-free water. cDNA was generated from the purified RNA (1 μg) using a SuperScript IV VILO cDNA synthesis kit (Thermo Fisher Scientific). qRT-PCR was performed using iQ SYBR green supermix and an MyiQ thermal cycler (Bio-Rad). The gene encoding DnaK (TDE0628) was used as an internal control to normalize the qRT-PCR data as previously described (42). The results were expressed as the normalized difference of threshold cycles (ΔΔCT) between the wild type and ΔflgM mutant. 5′-RACE analysis was performed using the SMART RACE 5′/3′ kit (TaKaRa Bio USA) according to the manufacturer’s protocol. The primers for RT-PCR, qRT-PCR, and 5′-RACE are listed in Table S1.
Protein turnover assay.
The protein turnover assay was carried out as previously described (42, 57). Briefly, T. denticola strains were first grown to the mid-log phase (~5 × 108 cells/mL). Subsequently, spectinomycin (100 μg/mL) was added into the cultures, followed by incubation at 37°C. Samples (10 mL) were harvested at the indicated time points and subjected to immunoblotting analysis.
EMSA.
The electrophoretic mobility shift assay (EMSA) was performed using recombinant proteins FliATd (rFliATd) and FlgMTd (rFlgMTd), as well as the flaB2 promoter (PflaB2) as a DNA probe, as previously described (48). The DNA probe was synthesized by PCR using primers P17/P18 (Table S1) and labeled with biotin using the Biotin DecaLabel DNA labeling kit (Thermo Fisher Scientific). EMSA was performed by adding increasing amounts of rFlgMTd (25.8 μM, 65 μM, and 130 μM) to biotin-labeled PflaB2 (2 mM) in binding buffer (10 mM Tris-HCl [pH 7.5]). The mixtures were incubated at room temperature for 30 min. After the incubation, 25.8 μM rFliATd was added to the reaction mixtures and the mixtures were further incubated for another 1 h. DNA loading dye was then added to the reaction mixtures immediately before loading. The reaction mixtures were separated on 4 to 20% Mini-Protean TGX gels using 0.5× Tris Borate EDTA (TBE) buffer and then transferred to a nylon membrane and cross-linked using UV light. The detection was performed using a Chemiluminescent nucleic acid detection module kit (Thermo Fisher Scientific).
Cryo-ET sample preparation, data collection, and image processing.
The freeze-hydrated specimens of T. denticola were prepared as previously described (42). Briefly, T. denticola cultures were mixed with 10-nm colloidal gold solutions and then deposited on a freshly glow-discharged, holey carbon grid for about 1 min. The grids were blotted with a small piece of filter paper for ~4 s and then rapidly plunged into liquid ethane using a gravity-driven plunger apparatus. The grids were then transferred to a 300-kV Krios electron microscope (Thermo Fisher Scientific) equipped with a field emission gun, Volta phase plate (VPP), and a direct detection detector (Gatan). To generate three-dimensional (3D) reconstructions of whole bacterial cells, SerialEM (81) was used to acquire multiple tilt series along the cell at a ×19,000 magnification. The pixel size at the specimen level is 5.4 Å. Tilt series were collected in the low-dose mode with the VPP at an ~0.5-μm defocus. A total dose of 60 e−/Å2 was distributed among 33 tilt images covering angles from −48° to +48° at tilt steps of 3°. The tilt series were aligned and reconstructed by IMOD (82). Multiple reconstructions from different segments of the same cell were integrated into one large reconstruction. In total, we generated over 90 tomograms to build 20 reconstructions of the entire cells from the wild-type (3 cells), ΔflgM (10 cells), and cflgM (7 cells) strains.
Bioinformatics and statistical analyses.
Sequence alignment analyses were performed using Clustal Omega. Homology modeling was generated using MODELLER version 10.0 (83), as incorporated in the MPI Bioinformatics Toolkit (84), and protein-protein docking analysis was conducted using the ClusPro server (85). Statistical significance was analyzed by one way ANOVA followed by Tukey’s multiple-comparison test at P < 0.01.
ACKNOWLEDGMENTS
This research was supported by funding from the National Institute of Dental and Craniofacial Research (DE023080 to C. Li) and National Institutes of Allergy and Infectious Diseases (AI078958 to C. Li, AI087946 to J. Liu, and AI148844 to Brian Crane and C. Li), National Institutes of Health (NIH). Cryo-ET data were collected at Yale Electron Cryo-Microscopy Resources, which is funded in part by NIH grant 1S10OD023603-01A1.
We thank Jennifer Aronson for critical reading of the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Chunhao Li, Email: cli5@vcu.edu.
Michael Y. Galperin, NCBI, NLM, National Institutes of Health
REFERENCES
- 1.Erhardt M, Namba K, Hughes KT. 2010. Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb Perspect Biol 2:a000299. 10.1101/cshperspect.a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465. 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armitage JP, Berry RM. 2020. Assembly and dynamics of the bacterial flagellum. Annu Rev Microbiol 74:181–200. 10.1146/annurev-micro-090816-093411. [DOI] [PubMed] [Google Scholar]
- 4.Meister M, Berg HC. 1987. The stall torque of the bacterial flagellar motor. Biophys J 52:413–419. 10.1016/S0006-3495(87)83230-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meister M, Lowe G, Berg HC. 1987. The proton flux through the bacterial flagellar motor. Cell 49:643–650. 10.1016/0092-8674(87)90540-x. [DOI] [PubMed] [Google Scholar]
- 6.Atsumi T, McCarter L, Imae Y. 1992. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature 355:182–184. 10.1038/355182a0. [DOI] [PubMed] [Google Scholar]
- 7.Samatey FA, Imada K, Nagashima S, Vonderviszt F, Kumasaka T, Yamamoto M, Namba K. 2001. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 410:331–337. 10.1038/35066504. [DOI] [PubMed] [Google Scholar]
- 8.Berg HC. 2000. Constraints on models for the flagellar rotary motor. Philos Trans R Soc Lond B Biol Sci 355:491–501. 10.1098/rstb.2000.0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feklistov A, Darst SA. 2011. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase sigma subunit. Cell 147:1257–1269. 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi W, Zhou W, Zhang B, Huang S, Jiang Y, Schammel A, Hu Y, Liu B. 2020. Structural basis of bacterial sigma(28)-mediated transcription reveals roles of the RNA polymerase zinc-binding domain. EMBO J 39:e104389. 10.15252/embj.2020104389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chadsey MS, Hughes KT. 2001. A multipartite interaction between Salmonella transcription factor sigma28 and its anti-sigma factor FlgM: implications for sigma28 holoenzyme destabilization through stepwise binding. J Mol Biol 306:915–929. 10.1006/jmbi.2001.4438. [DOI] [PubMed] [Google Scholar]
- 12.Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262:1277–1280. 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 13.Helmann JD. 2019. Where to begin? Sigma factors and the selectivity of transcription initiation in bacteria. Mol Microbiol 112:335–347. 10.1111/mmi.14309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wosten MM, van Dijk L, Veenendaal AK, de Zoete MR, Bleumink-Pluijm NM, van Putten JP. 2010. Temperature-dependent FlgM/FliA complex formation regulates Campylobacter jejuni flagella length. Mol Microbiol 75:1577–1591. 10.1111/j.1365-2958.2010.07079.x. [DOI] [PubMed] [Google Scholar]
- 15.Colland F, Rain JC, Gounon P, Labigne A, Legrain P, De Reuse H. 2001. Identification of the Helicobacter pylori anti-sigma28 factor. Mol Microbiol 41:477–487. 10.1046/j.1365-2958.2001.02537.x. [DOI] [PubMed] [Google Scholar]
- 16.Josenhans C, Niehus E, Amersbach S, Horster A, Betz C, Drescher B, Hughes KT, Suerbaum S. 2002. Functional characterization of the antagonistic flagellar late regulators FliA and FlgM of Helicobacter pylori and their effects on the H. pylori transcriptome. Mol Microbiol 43:307–322. 10.1046/j.1365-2958.2002.02765.x. [DOI] [PubMed] [Google Scholar]
- 17.Potvin E, Sanschagrin F, Levesque RC. 2008. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol Rev 32:38–55. 10.1111/j.1574-6976.2007.00092.x. [DOI] [PubMed] [Google Scholar]
- 18.Prouty MG, Correa NE, Klose KE. 2001. The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol Microbiol 39:1595–1609. 10.1046/j.1365-2958.2001.02348.x. [DOI] [PubMed] [Google Scholar]
- 19.Chen YF, Helmann JD. 1992. Restoration of motility to an Escherichia coli fliA flagellar mutant by a Bacillus subtilis sigma factor. Proc Natl Acad Sci USA 89:5123–5127. 10.1073/pnas.89.11.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eichelberg K, Galan JE. 2000. The flagellar sigma factor FliA (sigma(28)) regulates the expression of Salmonella genes associated with the centisome 63 type III secretion system. Infect Immun 68:2735–2743. 10.1128/IAI.68.5.2735-2743.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heuner K, Dietrich C, Skriwan C, Steinert M, Hacker J. 2002. Influence of the alternative sigma(28) factor on virulence and flagellum expression of Legionella pneumophila. Infect Immun 70:1604–1608. 10.1128/IAI.70.3.1604-1608.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lertsethtakarn P, Ottemann KM, Hendrixson DR. 2011. Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol 65:389–410. 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kutsukake K, Iino T. 1994. Role of the FliA-FlgM regulatory system on the transcriptional control of the flagellar regulon and flagellar formation in Salmonella typhimurium. J Bacteriol 176:3598–3605. 10.1128/jb.176.12.3598-3605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radolf JD, Deka RK, Anand A, Smajs D, Norgard MV, Yang XF. 2016. Treponema pallidum, the syphilis spirochete: making a living as a stealth pathogen. Nat Rev Microbiol 14:744–759. 10.1038/nrmicro.2016.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosa PA, Tilly K, Stewart PE. 2005. The burgeoning molecular genetics of the Lyme disease spirochaete. Nat Rev Microbiol 3:129–143. 10.1038/nrmicro1086. [DOI] [PubMed] [Google Scholar]
- 26.Picardeau M. 2017. Virulence of the zoonotic agent of leptospirosis: still terra incognita? Nat Rev Microbiol 15:297–307. 10.1038/nrmicro.2017.5. [DOI] [PubMed] [Google Scholar]
- 27.Holt SC, Ebersole JL. 2005. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex,” a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000 38:72–122. 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- 28.Li C, Motaleb A, Sal M, Goldstein SF, Charon NW. 2000. Spirochete periplasmic flagella and motility. J Mol Microbiol Biotechnol 2:345–354. [PubMed] [Google Scholar]
- 29.Charon NW, Cockburn A, Li C, Liu J, Miller KA, Miller MR, Motaleb MA, Wolgemuth CW. 2012. The unique paradigm of spirochete motility and chemotaxis. Annu Rev Microbiol 66:349–370. 10.1146/annurev-micro-092611-150145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.San Martin F, Fule L, Iraola G, Buschiazzo A, Picardeau M. 2022. Diving into the complexity of the spirochetal endoflagellum. Trends Microbiol 10.1016/j.tim.2022.09.010. [DOI] [PubMed] [Google Scholar]
- 31.Zhang K, He J, Cantalano C, Guo Y, Liu J, Li C. 2020. FlhF regulates the number and configuration of periplasmic flagella in Borrelia burgdorferi. Mol Microbiol 113:1122–1139. 10.1111/mmi.14482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sze CW, Morado DR, Liu J, Charon NW, Xu H, Li C. 2011. Carbon storage regulator A (CsrA(Bb)) is a repressor of Borrelia burgdorferi flagellin protein FlaB. Mol Microbiol 82:851–864. 10.1111/j.1365-2958.2011.07853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolgemuth CW. 2015. Flagellar motility of the pathogenic spirochetes. Semin Cell Dev Biol 46:104–112. 10.1016/j.semcdb.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Motaleb MA, Liu J, Wooten RM. 2015. Spirochetal motility and chemotaxis in the natural enzootic cycle and development of Lyme disease. Curr Opin Microbiol 28:106–113. 10.1016/j.mib.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao X, Zhang K, Boquoi T, Hu B, Motaleb MA, Miller KA, James ME, Charon NW, Manson MD, Norris SJ, Li C, Liu J. 2013. Cryoelectron tomography reveals the sequential assembly of bacterial flagella in Borrelia burgdorferi. Proc Natl Acad Sci USA 110:14390–14395. 10.1073/pnas.1308306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang Y, Xu H, Motaleb MA, Liu J. 2021. Characterization of the flagellar collar reveals structural plasticity essential for spirochete motility. mBio 12:e02494-21. 10.1128/mBio.02494-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moon KH, Zhao X, Xu H, Liu J, Motaleb MA. 2018. A tetratricopeptide repeat domain protein has profound effects on assembly of periplasmic flagella, morphology and motility of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 110:634–647. 10.1111/mmi.14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moon KH, Zhao X, Manne A, Wang J, Yu Z, Liu J, Motaleb MA. 2016. Spirochetes flagellar collar protein FlbB has astounding effects in orientation of periplasmic flagella, bacterial shape, motility, and assembly of motors in Borrelia burgdorferi. Mol Microbiol 102:336–348. 10.1111/mmi.13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lynch MJ, Miller M, James M, Zhang S, Zhang K, Li C, Charon NW, Crane BR. 2019. Structure and chemistry of lysinoalanine crosslinking in the spirochaete flagella hook. Nat Chem Biol 15:959–965. 10.1038/s41589-019-0341-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller MR, Miller KA, Bian J, James ME, Zhang S, Lynch MJ, Callery PS, Hettick JM, Cockburn A, Liu J, Li C, Crane BR, Charon NW. 2016. Spirochaete flagella hook proteins self-catalyse a lysinoalanine covalent crosslink for motility. Nat Microbiol 1:16134. 10.1038/nmicrobiol.2016.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurniyati K, Chang Y, Liu J, Li C. 2022. Transcriptional and functional characterizations of multiple flagellin genes in spirochetes. Mol Microbiol 118:175–190. 10.1111/mmi.14959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurniyati K, Kelly JF, Vinogradov E, Robotham A, Tu Y, Wang J, Liu J, Logan SM, Li C. 2017. A novel glycan modifies the flagellar filament proteins of the oral bacterium Treponema denticola. Mol Microbiol 103:67–85. 10.1111/mmi.13544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norris SJ, Charon NW, Cook RG, Fuentes MD, Limberger RJ. 1988. Antigenic relatedness and N-terminal sequence homology define two classes of periplasmic flagellar proteins of Treponema pallidum subsp. pallidum and Treponema phagedenis. J Bacteriol 170:4072–4082. 10.1128/jb.170.9.4072-4082.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li C, Sal M, Marko M, Charon NW. 2010. Differential regulation of the multiple flagellins in spirochetes. J Bacteriol 192:2596–2603. 10.1128/JB.01502-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li C, Wolgemuth CW, Marko M, Morgan DG, Charon NW. 2008. Genetic analysis of spirochete flagellin proteins and their involvement in motility, filament assembly, and flagellar morphology. J Bacteriol 190:5607–5615. 10.1128/JB.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wunder EA, Jr, Slamti L, Suwondo DN, Gibson KH, Shang Z, Sindelar CV, Trajtenberg F, Buschiazzo A, Ko AI, Picardeau M. 2018. FcpB is a surface filament protein of the endoflagellum required for the motility of the spirochete Leptospira. Front Cell Infect Microbiol 8:130. 10.3389/fcimb.2018.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gibson KH, Trajtenberg F, Wunder EA, Brady MR, San Martin F, Mechaly A, Shang Z, Liu J, Picardeau M, Ko A, Buschiazzo A, Sindelar CV. 2020. An asymmetric sheath controls flagellar supercoiling and motility in the Leptospira spirochete. eLife 9:e53672. 10.7554/eLife.53672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurniyati K, Chang Y, Liu J, Li C. 2022. Identification and characterization of the alternative sigma(28) factor in Treponema denticola. J Bacteriol 204:e00248-22. 10.1128/jb.00248-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pons T, Gonzalez B, Ceciliani F, Galizzi A. 2006. FlgM anti-sigma factors: identification of novel members of the family, evolutionary analysis, homology modeling, and analysis of sequence-structure-function relationships. J Mol Model 12:973–983. 10.1007/s00894-005-0096-5. [DOI] [PubMed] [Google Scholar]
- 50.Sorenson MK, Ray SS, Darst SA. 2004. Crystal structure of the flagellar sigma/anti-sigma complex sigma(28)/FlgM reveals an intact sigma factor in an inactive conformation. Mol Cell 14:127–138. 10.1016/S1097-2765(04)00150-9. [DOI] [PubMed] [Google Scholar]
- 51.Daughdrill GW, Hanely LJ, Dahlquist FW. 1998. The C-terminal half of the anti-sigma factor FlgM contains a dynamic equilibrium solution structure favoring helical conformations. Biochemistry 37:1076–1082. 10.1021/bi971952t. [DOI] [PubMed] [Google Scholar]
- 52.Zhao M, Zhang H, Liu G, Wang L, Wang J, Gao Z, Dong Y, Zhang L, Gong Y. 2016. Structural insights into the methylation of C1402 in 16S rRNA by methyltransferase RsmI. PLoS One 11:e0163816. 10.1371/journal.pone.0163816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iyoda S, Kutsukake K. 1995. Molecular dissection of the flagellum-specific anti-sigma factor, FlgM, of Salmonella typhimurium. Mol Gen Genet 249:417–424. 10.1007/BF00287103. [DOI] [PubMed] [Google Scholar]
- 54.Chang Y, Zhang K, Carroll BL, Zhao X, Charon NW, Norris SJ, Motaleb MA, Li C, Liu J. 2020. Molecular mechanism for rotational switching of the bacterial flagellar motor. Nat Struct Mol Biol 27:1041–1047. 10.1038/s41594-020-0497-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wozniak CE, Hughes KT. 2008. Genetic dissection of the consensus sequence for the class 2 and class 3 flagellar promoters. J Mol Biol 379:936–952. 10.1016/j.jmb.2008.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barembruch C, Hengge R. 2007. Cellular levels and activity of the flagellar sigma factor FliA of Escherichia coli are controlled by FlgM-modulated proteolysis. Mol Microbiol 65:76–89. 10.1111/j.1365-2958.2007.05770.x. [DOI] [PubMed] [Google Scholar]
- 57.Kurniyati K, Liu J, Zhang JR, Min Y, Li C. 2019. A pleiotropic role of FlaG in regulating the cell morphogenesis and flagellar homeostasis at the cell poles of Treponema denticola. Cell Microbiol 21:e12886. 10.1111/cmi.12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mukherjee S, Yakhnin H, Kysela D, Sokoloski J, Babitzke P, Kearns DB. 2011. CsrA-FliW interaction governs flagellin homeostasis and a checkpoint on flagellar morphogenesis in Bacillus subtilis. Mol Microbiol 82:447–461. 10.1111/j.1365-2958.2011.07822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dugar G, Svensson SL, Bischler T, Waldchen S, Reinhardt R, Sauer M, Sharma CM. 2016. The CsrA-FliW network controls polar localization of the dual-function flagellin mRNA in Campylobacter jejuni. Nat Commun 7:11667. 10.1038/ncomms11667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Potts AH, Vakulskas CA, Pannuri A, Yakhnin H, Babitzke P, Romeo T. 2017. Global role of the bacterial post-transcriptional regulator CsrA revealed by integrated transcriptomics. Nat Commun 8:1596. 10.1038/s41467-017-01613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seshadri R, Myers GS, Tettelin H, Eisen JA, Heidelberg JF, Dodson RJ, Davidsen TM, DeBoy RT, Fouts DE, Haft DH, Selengut J, Ren Q, Brinkac LM, Madupu R, Kolonay J, Durkin SA, Daugherty SC, Shetty J, Shvartsbeyn A, Gebregeorgis E, Geer K, Tsegaye G, Malek J, Ayodeji B, Shatsman S, McLeod MP, Smajs D, Howell JK, Pal S, Amin A, Vashisth P, McNeill TZ, Xiang Q, Sodergren E, Baca E, Weinstock GM, Norris SJ, Fraser CM, Paulsen IT. 2004. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc Natl Acad Sci USA 101:5646–5651. 10.1073/pnas.0307639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ding L, Wang Y, Hu Y, Atkinson S, Williams P, Chen S. 2009. Functional characterization of FlgM in the regulation of flagellar synthesis and motility in Yersinia pseudotuberculosis. Microbiology (Reading) 155:1890–1900. 10.1099/mic.0.026294-0. [DOI] [PubMed] [Google Scholar]
- 63.Caramori T, Barilla D, Nessi C, Sacchi L, Galizzi A. 1996. Role of FlgM in sigma D-dependent gene expression in Bacillus subtilis. J Bacteriol 178:3113–3118. 10.1128/jb.178.11.3113-3118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Correa NE, Barker JR, Klose KE. 2004. The Vibrio cholerae FlgM homologue is an anti-sigma28 factor that is secreted through the sheathed polar flagellum. J Bacteriol 186:4613–4619. 10.1128/JB.186.14.4613-4619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bian J, Li C. 2011. Disruption of a type II endonuclease (TDE0911) enables Treponema denticola ATCC 35405 to accept an unmethylated shuttle vector. Appl Environ Microbiol 77:4573–4578. 10.1128/AEM.00417-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chi B, Chauhan S, Kuramitsu H. 1999. Development of a system for expressing heterologous genes in the oral spirochete Treponema denticola and its use in expression of the Treponema pallidum flaA gene. Infect Immun 67:3653–3656. 10.1128/IAI.67.7.3653-3656.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jutras BL, Scott M, Parry B, Biboy J, Gray J, Vollmer W, Jacobs-Wagner C. 2016. Lyme disease and relapsing fever Borrelia elongate through zones of peptidoglycan synthesis that mark division sites of daughter cells. Proc Natl Acad Sci USA 113:9162–9170. 10.1073/pnas.1610805113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fraser CM, Norris SJ, Weinstock GM, White O, Sutton GG, Dodson R, Gwinn M, Hickey EK, Clayton R, Ketchum KA, Sodergren E, Hardham JM, McLeod MP, Salzberg S, Peterson J, Khalak H, Richardson D, Howell JK, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton MD, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Sandusky M, Weidman J, Smith HO, Venter JC. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375–388. 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 69.Balaban M, Joslin SN, Hendrixson DR. 2009. FlhF and its GTPase activity are required for distinct processes in flagellar gene regulation and biosynthesis in Campylobacter jejuni. J Bacteriol 191:6602–6611. 10.1128/JB.00884-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kusumoto A, Shinohara A, Terashima H, Kojima S, Yakushi T, Homma M. 2008. Collaboration of FlhF and FlhG to regulate polar-flagella number and localization in Vibrio alginolyticus. Microbiology (Reading) 154:1390–1399. 10.1099/mic.0.2007/012641-0. [DOI] [PubMed] [Google Scholar]
- 71.Park S, Yoon J, Lee CR, Lee JY, Kim YR, Jang KS, Lee KH, Seok YJ. 2019. Polar landmark protein HubP recruits flagella assembly protein FapA under glucose limitation in Vibrio vulnificus. Mol Microbiol 112:266–279. 10.1111/mmi.14268. [DOI] [PubMed] [Google Scholar]
- 72.Park S, Park YH, Lee CR, Kim YR, Seok YJ. 2016. Glucose induces delocalization of a flagellar biosynthesis protein from the flagellated pole. Mol Microbiol 101:795–808. 10.1111/mmi.13424. [DOI] [PubMed] [Google Scholar]
- 73.Ohta K, Makinen KK, Loesche WJ. 1986. Purification and characterization of an enzyme produced by Treponema denticola capable of hydrolyzing synthetic trypsin substrates. Infect Immun 53:213–220. 10.1128/iai.53.1.213-220.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ruby JD, Li H, Kuramitsu H, Norris SJ, Goldstein SF, Buttle KF, Charon NW. 1997. Relationship of Treponema denticola periplasmic flagella to irregular cell morphology. J Bacteriol 179:1628–1635. 10.1128/jb.179.5.1628-1635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kurniyati K, Li C. 2016. pyrF as a counterselectable marker for unmarked genetic manipulations in Treponema denticola. Appl Environ Microbiol 82:1346–1352. 10.1128/AEM.03704-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H, Ruby J, Charon N, Kuramitsu H. 1996. Gene inactivation in the oral spirochete Treponema denticola: construction of an flgE mutant. J Bacteriol 178:3664–3667. 10.1128/jb.178.12.3664-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kurniyati K, Li C. 2021. Genetic manipulations of oral spirochete Treponema denticola. Methods Mol Biol 2210:15–23. 10.1007/978-1-0716-0939-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oshiro RT, Rajendren S, Hundley HA, Kearns DB. 2019. Robust stoichiometry of FliW-CsrA governs flagellin homeostasis and cytoplasmic organization in Bacillus subtilis. mBio 10:e00533-19. 10.1128/mBio.00533-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Limberger RJ, Slivienski LL, Izard J, Samsonoff WA. 1999. Insertional inactivation of Treponema denticola tap1 results in a nonmotile mutant with elongated flagellar hooks. J Bacteriol 181:3743–3750. 10.1128/JB.181.12.3743-3750.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Orth R, O'Brien-Simpson N, Dashper S, Walsh K, Reynolds E. 2010. An efficient method for enumerating oral spirochetes using flow cytometry. J Microbiol Methods 80:123–128. 10.1016/j.mimet.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 81.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 82.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116:71–76. 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 83.Webb B, Sali A. 2016. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics 54:5.6.1–5.6.37. 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, Gabler F, Soding J, Lupas AN, Alva V. 2018. A completely reimplemented MPI Bioinformatics Toolkit with a new HHpred server at its core. J Mol Biol 430:2237–2243. 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 85.Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, Beglov D, Vajda S. 2017. The ClusPro web server for protein-protein docking. Nat Protoc 12:255–278. 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wolfe AJ, Berg HC. 1989. Migration of bacteria in semisolid agar. Proc Natl Acad Sci USA 86:6973–6977. 10.1073/pnas.86.18.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 and Fig. S1 and S2. Download jb.00463-22-s0001.pdf, PDF file, 0.5 MB (558.8KB, pdf)
Video S3 (The complemented strain). Download jb.00463-22-s0002.mpg, MPG file, 0.9 MB (900KB, mpg)
Video S2 (The flgM deletion mutant). Download jb.00463-22-s0003.mpg, MPG file, 0.9 MB (832KB, mpg)
Video S1 (The wild-type strain). Download jb.00463-22-s0004.mpg, MPG file, 0.9 MB (902KB, mpg)