ABSTRACT
First isolated and classified in the 1960s, Caulobacter crescentus has been instrumental in the study of bacterial cell biology and differentiation. C. crescentus is a Gram-negative alphaproteobacterium that exhibits a dimorphic life cycle composed of two distinct cell types: a motile swarmer cell and a nonmotile, division-competent stalked cell. Progression through the cell cycle is accentuated by tightly controlled biogenesis of appendages, morphological transitions, and distinct localization of developmental regulators. These features as well as the ability to synchronize populations of cells and follow their progression make C. crescentus an ideal model for answering questions relevant to how development and differentiation are achieved at the single-cell level. This review will explore the discovery and development of C. crescentus as a model organism before diving into several key features and discoveries that have made it such a powerful organism to study. Finally, we will summarize a few of the ongoing areas of research that are leveraging knowledge gained over the last century with C. crescentus to highlight its continuing role at the forefront of cell and developmental biology.
KEYWORDS: bacterial cell biology, Caulobacter, Caulobacter crescentus, model organism, cell cycle, differentiation, morphogenesis
INTRODUCTION
In a tap in Chicago, there lived a bacterium. An object of fascination for many bacteriologists in the early 20th century, Caulobacter spp. were initially described as curved rod-shaped bacteria that formed immobilized rosettes of cells (Fig. 1). These bacteria bore “flagella-like” appendages, later termed “stalks,” that extended into the center of these rosettes and divided transversely such that newly formed daughter cells were released. Caulobacter crescentus (here referred to as Caulobacter) is the primary model organism representing the family Caulobacteraceae in the order Caulobacterales (named from the Greek καυλος, meaning “stalk”). This order was first proposed in 1935 by Henrici and Johnson (1) in their observational work describing the existence of a novel stalked bacterial species (previously, and independently, observed by Mabel Jones [Fig. 1] [2] and Vasily Omeliansky [3]) that had adhered to a slide submerged in lake and tap water. Houwink and van Iterson (4) and Bowers et al. (5) then demonstrated the presence of a singular flagellum by electron microscopy and presented an initial protocol for maintaining laboratory cultures. Finally, Jeanne Poindexter isolated and optimized a protocol for cultivation of several Caulobacter species from fresh water (6). These isolates included a strain from a pond in California in 1960 that would become the ancestor to the currently used CB15 and CB15N/NA1000 lab strains (7). These efforts set the stage to develop Caulobacter as a model organism more than 60 years after its initial description.
FIG 1.
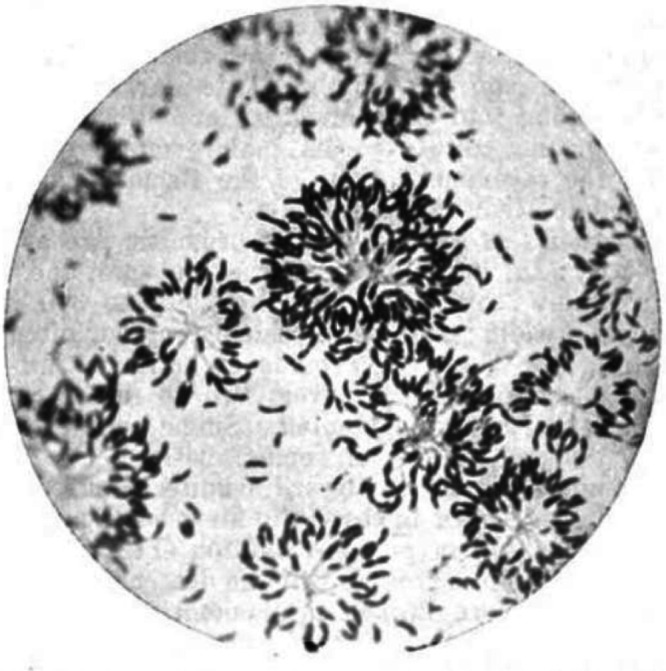
The first published image of Caulobacter-like cells. Film micrograph from 1905 depicting Caulobacter-like cells isolated by Mabel Jones from tap water in Chicago after 72 h in culture. Featured are rosettes of cells formed by descendants of single cells. Stalks are visible in the middle of rosettes, and predivisional and swarmer cells are both visible toward the periphery and in isolation. (Reproduced from reference 2).
A striking feature of Caulobacter is its asymmetry. As a Gram-negative vibrioid bacterium, Caulobacter exhibits both lateral and longitudinal asymmetry; it has an inner and outer curvature as well as distinct poles that can possess either a flagellum or a stalk. Cell division results in two distinct daughter cells, a flagellated “swarmer cell” and a “stalked cell” bearing the aforementioned stalk, which reenter the cell cycle at different points (Fig. 2). This asymmetric division was observed from the outset, and its implication for the survival strategy of Caulobacter is nicely summarized by Henrici and Johnson: “It is probable that the outer most cell is set free after cell division, and either swims or floats away until a new substrate is encountered, when it proceeds to secrete a stalk” (1). Indeed, studies since then have confirmed that the swarmer cell is motile, driven by its flagellum until pili-mediated initiation of surface adhesion, after which production of holdfast at the pole secures attachment (8). The flagellum is then shed, and the former swarmer cell differentiates into a division-competent stalked cell, although this differentiation does not strictly require surface attachment, as Caulobacter strains deficient in surface attachment are still culturable and progress through the cell cycle (7, 9). The well-defined, morphologically distinct life cycle of Caulobacter was immediately recognized as providing a useful model for studying cell cycle progression, and that is where our story really begins.
FIG 2.
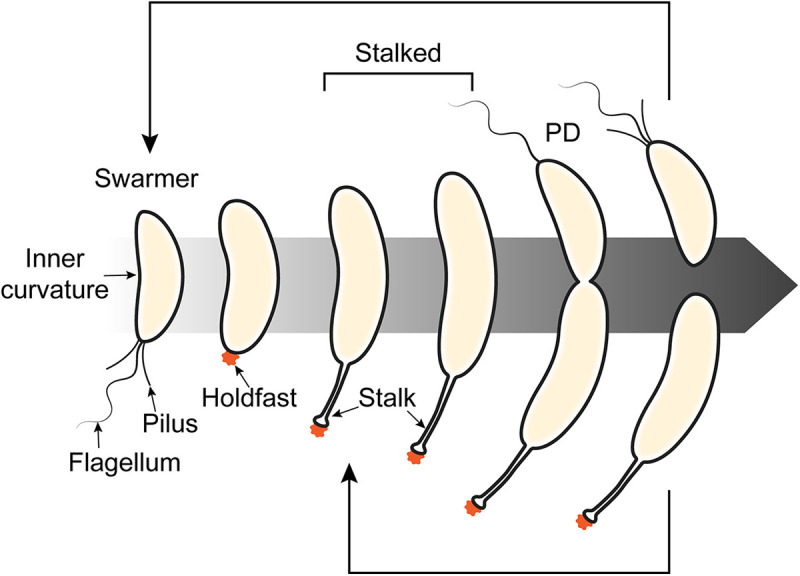
Caulobacter differentiates over the course of its cell cycle. A swarmer cell bearing a single flagellum and pili sheds or retracts these appendages and secretes holdfast (orange) coincident with differentiation to a stalked cell. The predivisional (PD) cell divides asymmetrically to yield swarmer and stalked daughter cells, both of which reenter the cell cycle at their respective stages. The new flagellum is formed concurrent with cell division, while pili are only detectable after completion of constriction.
THE DEVELOPMENT OF CAULOBACTER AS A MODEL ORGANISM
The study of any aspect of bacterial cell biology requires not only the ability to culture a bacterium of interest but also the ability to manipulate it to test hypotheses about its function. Recognizing the potential of Caulobacter to serve as a model for bacterial development, Lucy Shapiro outlined three necessary criteria for this kind of study: the organism must (i) have a simple and well-defined differentiation pattern, (ii) be culturable on defined media, and (iii) be genetically tractable. Shapiro proposed that Caulobacter exhibited all of these qualities, and its utility as a model was further potentiated by additional, as of yet unrecognized, technical advantages discussed below (10). Work with Caulobacter over the next 50 years, with significant initial contributions from the labs of Shapiro, Bert Ely, and Austin Newton, would lay the groundwork for the blossoming field of bacterial cell biology and establish Caulobacter as a powerhouse model organism for this type of study and beyond.
One of the exciting reasons for initially adapting Caulobacter as a model organism is the fact that it undergoes morphological differentiation throughout its cell cycle, unlike most bacterial models that had been studied up until that point. Along with the observations that genes are expressed at different levels at different times and in response to environmental stimuli, this suggested roles for regulated transcription, protein synthesis, and protein degradation as possible mechanisms for controlling progression through the cell cycle (10). To begin to answer questions about the details of this control, it was necessary to develop genetic tools to manipulate Caulobacter as well as methods for observing progression through the cell cycle and isolating cells at specific stages.
One of the first major innovations in the study of Caulobacter was the synchronization of cells with respect to cell cycle phase, arguably one of the most powerful tools for those who study the Caulobacter cell cycle. Synchrony relies on separation of cell types from one another, and protocols have evolved over time to increase both precision and yield. Cultures can be synchronized by one of (at least) two methods. Cultures can be incubated in glass petri plates to allow for adhesion of cells. Subsequent gentle agitation ensures that new swarmer cells are unable to adhere, allowing for their isolation from the culture medium (11). Alternatively, one can synchronize cells by subjecting a planktonic culture to centrifugation on a density gradient (12, 13), as stalked/predivisional cells synthesize a polysaccharide capsule that is absent in swarmer cells (14), allowing for separation by buoyancy (Fig. 3). After isolation, swarmer cells progress through the cell cycle synchronously, allowing for precise observation and measurement of cell cycle-dependent processes, which has made Caulobacter the premier organism for this type of analysis.
FIG 3.
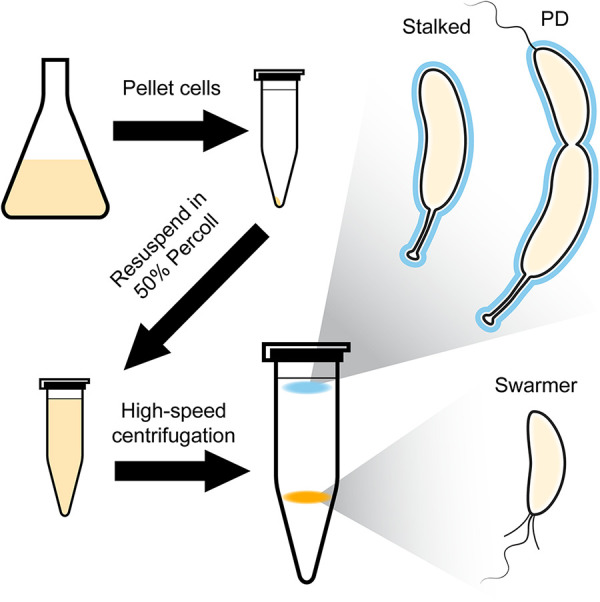
Schematic of swarmer cell isolation for synchrony of Caulobacter cultures. To isolate swarmer cells, a log-phase culture of Caulobacter is pelleted and resuspended in a solution of 50% Percoll in minimal medium, followed by high-speed centrifugation to segregate cells by buoyancy. Swarmer cells (orange band) are separated from stalked and predivisional (PD) cells (cyan band) due to a difference in buoyancy resulting from the production of capsule (cyan outline) in stalked and PD cells. The full protocol is available in reference 13.
Following the ability to synchronize populations, the first breakthrough in beginning to understand the connection between genetic regulation and development was the isolation of Caulobacter-specific bacteriophages. The ϕCb5 RNA (15) and ϕCbk DNA (16) bacteriophages were found to specifically infect Caulobacter and to only do so during certain parts of the cell cycle. ϕCb5 requires the presence of flagellum-adjacent pili for adsorption, which are only present on swarmer and late predivisional cells (17). ϕCbK, however, binds specifically to pili (18), and preliminary evidence suggests that it then binds to an uncharacterized lipopolysaccharide (LPS) receptor site that arises coincident with the transition to stalked cells (10). These discoveries allowed for the development of assays to follow progression through the cell cycle by infecting synchronized populations of Caulobacter with phage and observing the efficiency of infection over time, which resulted in the first approximation of the timing of each stage in the cell cycle (19). This innovation also allowed for the isolation of temperature-sensitive (TS) mutants that halted transition from one cell cycle state to another, opening the door for discovery of factors that are required for various stages of development (10). Suppressor screens using TS mutants allowed for the discovery of yet more crucial regulators of progression (20).
Further advancements in genetics (reviewed in reference 21) included the ability to generate and isolate spontaneous single mutants (22), the development of a conjugation system (23), a method for phage-based generalized transduction (24), the ability to transform a plasmid via electroporation (25), a method for chromosomal gene inactivation (26), and the use of the Tn5 transposon to integrate randomly in the genome without apparent bias (27). Finally, the Caulobacter genome map was constructed, first by a combination of conjugation and transduction (28) and later by restriction digest followed by pulsed-field electrophoresis (29). The entire genome sequence was published in 2001 (30). These advancements allowed for specific targeting of individual genes and regions, establishing a powerful and versatile genetic system that could be used to start answering questions about bacterial morphogenesis and cell cycle control.
After several years of work in multiple laboratories, a derivative of the original CB15 Caulobacter strain was isolated (called NA1000 or CB15N) that provided several technical advantages over CB15 (7). Genomic sequence analysis revealed several single nucleotide polymorphisms (SNPs), including a frameshift mutation in the holdfast synthesis gene hfsA and a mutation in the zwf promoter. Notably, NA1000 also carries a 26-kb mobile element harboring sequences with a lower GC content than the rest of the genome, indicating acquisition by horizontal transfer, and containing genes that are likely involved in capsule biosynthesis. As a result of these mutations, NA1000 exhibits faster growth, limited adhesion, and decreased lysis following infection with the ϕCr30 transducing phage compared to CB15. NA1000 also exhibits a lower propensity for stalked and predivisional cells to form a pellet after centrifugation, greatly enhancing the effectiveness of synchrony protocols. The combination of all these phenotypic differences, especially increased synchronization efficiency, resulted in the adoption of NA1000 as the primary Caulobacter strain used for research that does not require adhesion, and it remains the foremost studied strain today.
Beyond its utility as a model for differentiation, Caulobacter has also been a foundational model for the development and application of innovations in imaging. Fluorescence-based visualization of cells and subcellular components has been used since the invention and subsequent demonstration of immunofluorescence in 1941 and 1942, respectively (31, 32). A major improvement on this technique that allowed for visualization of proteins in live cells was the generation of fluorophore fusion constructs wherein a fluorescent protein is genetically conjugated to one of the termini of a protein of interest. One of the first demonstrations of this technique in bacteria was in Caulobacter, where Jacobs et al. demonstrated the localization of the histidine kinase CckA, which was tagged at its native locus with green fluorescent protein (GFP) (33). They found not only that it localizes to specific poles but also that its localization is dynamic over the course of the cell cycle. As another example of fluorescence-informed dynamic investigation, the first comprehensive study of bacterial chromosome organization was performed in synchronized populations of Caulobacter using a combination of fluorescence in situ hybridization and the fluorescence repressor operator system (34). This approach revealed that the positioning of chromosomal loci is stereotyped and that the segregation of the new chromosome following replication is tightly controlled with highly reproducible rates of movement and subcellular destinations for each locus.
The development of superresolution techniques has allowed for even further refinement of localization studies (35) and the investigation of the finer details of large cellular structures, such as the cytokinetic ring or Z-ring (36). Pushing the frontiers of imaging technologies, Caulobacter was one of the pioneer organisms in the development of electron cryotomography (cryo-ET) for probing fine structures and the three-dimensional organization of intact, unfixed bacterial cells with high resolution. Caulobacter was uniquely suited for this role early in the development of cryo-ET, as its cells are smaller than those of other well-studied bacterial models (e.g., Escherichia coli and Bacillus subtilis), allowing for visualization of cells in their entirety at high resolution (37). The first of these studies revealed that Caulobacter division concludes with constriction and fission events of the inner and outer membranes that are spatially and temporally distinct (38). Another demonstrated the presence of multiple classes of large filament bundles present in different orientations in Caulobacter cells (39). Ring-like and inner curvature-associated filaments were thought to be associated with the previously known cytoskeletal proteins FtsZ, MreB, and CreS, and these structures have since been studied in more detail to gain insight into their respective structures throughout the cell cycle. The structure of FtsZ filaments and their relationship to constriction were characterized in more detail using the unique morphology of a Caulobacter mutant overproducing the GTPase-deficient G105S FtsZ variant (40, 41).
Finally, as has become especially apparent in recent years with the exponential accumulation of data, the intimate study of a model organism is greatly aided by the existence of comprehensive databases. The first database-style resource for Caulobacter resulted from a high-throughput imaging screen to determine the distributions of nearly three-fourths of all Caulobacter proteins fused to fluorescent markers. This work yielded the Caulobacter crescentus localisome website, where users can find localization data tied to corresponding images for proteins of interest (42). Inspired by the recent acquisition of multiple “-omics”-level data sets, Lasker et al. endeavored to create CauloBrowser, a user-friendly, integrated online database that allows researchers to search for genes to gain information on their localization, expression profiles, interactions, etc. (43). Another important resource is the Fitness Browser, a multiorganism database that includes data on the relative fitness of various mutants under different conditions that allows users to gain insight on potential phenotypic effects and interactions among genes and proteins of interest (44). The development of these and other databases represents a significant step forward in the ability to easily integrate information to generate new hypotheses.
Caulobacter has a long and storied past, and its utility as a model for the study of dynamic, cell cycle-dependent processes has been demonstrated time and time again. For the remainder of this review, we will explore some of the notable features of and discoveries in Caulobacter that have made this such a powerful organism to investigate.
UNIQUE FEATURES AND NOTABLE DISCOVERIES
The right tool for the job.
Throughout its life cycle, Caulobacter uses distinct appendages to adapt and thrive under variable environmental conditions. Motile swarmer cells bear a single flagellum and pili at one pole. These cells can explore their environments then shed the flagellum and retract their pili, attach to a surface via the secretion of holdfast, and begin to differentiate into a stalked cell (Fig. 2). Stalked cells are competent for DNA replication, which occurs once per cell cycle (45), as well as division, which results in two morphologically and developmentally distinct daughter cells. The swarmer daughter can swim away and find a new environment in which to differentiate, and the stalked daughter immediately reenters the cell cycle at the point of DNA replication and cell division (Fig. 2).
The stalk has been noted from the earliest observations of Caulobacter. While early observers concluded that it might be a “flagella-like” appendage (1), transmission electron microscopy (TEM) experiments revealed that the stalk is an extension of the cell envelope, including both inner and outer membranes as well as peptidoglycan cell wall (4). Further observations suggested that the stalk is a biochemically distinct compartment, as evidenced by a lack of ribosomes and nucleoid and the presence of electron-dense crossbands along the stalk length (46). These crossbands create diffusion barriers that prevent exchange of proteins between the cell body and the stalk and between stalk compartments (47). Although the “swarmer-to-stalked” transition is a crucial step in the Caulobacter life cycle, the formation of the stalk is not strictly required for division to occur (48), suggesting that it functions in tandem with, but not as a regulator of, cell cycle progression.
The role of the stalk has been studied extensively in the decades since Caulobacter was isolated. Although holdfast, a strongly adhesive polysaccharide, is present at the tip of the stalk, holdfast synthesis precedes stalk biogenesis (49), and attachment is undertaken by swarmer cells, suggesting that the stalk does not play a role in surface attachment (46). Additionally, some stalked bacteria produce holdfast and stalks at separate locations (6), further indicating that surface attachment is not the stalk’s primary function. The stalk appears to have a role in sensing or responding to nutrient availability (50), as Caulobacter cells starved of phosphate grow elongated stalks (51, 52). This was initially hypothesized to be a mechanism for increasing surface area to facilitate nutrient uptake (53), but this model has been challenged by the apparent lack of protein diffusion along the stalk. A more recent hypothesis suggests that stalk elongation would allow for cells within a surface-attached biofilm to extend their cell body beyond the surface of the biofilm, increasing exposure to a more nutrient-rich medium (54). However, this model is difficult to apply to organisms like Asticcacaulis spp., where the site of adhesion is not located at the site of stalk extension (6, 46), suggesting that the role of stalks is more complicated than can explained by a single model (55). Finally, the stalk-localized protein StpX, which is required for proper stalk elongation, appears to protect cells from Zn2+ toxicity and plays a role in storage and utilization of Cu2+ (55). Additional studies in diverse organisms with stalks and stalk-like appendages are required to shed light on their function(s).
The same TEM experiments that granted insight into the structure of the Caulobacter stalk also allowed for initial observation of its flagellum (4). Caulobacter swarmer cells possess a single flagellum that is shed as the cell transitions to a stalked cell (Fig. 2). This flagellum is similar in structure to other canonical bacterial flagella and is composed of a stator complex, a hook, and a filament, all of which are visible by TEM (56). Flagellar gene expression is regulated in tandem with cell cycle progression, with genes falling into one of four different classes. Class I genes include global cell cycle regulators (e.g., dnaA and ctrA), which will be discussed in detail in a later section. These factors control expression of class II genes (57–59), which in turn regulate the expression of class III and IV genes, which encode structural components of flagella (reviewed in reference 60). Flagellins, the major component of the flagellar filament, are synthesized approximately 30 to 40 min before cell division in synchronized populations (61). Flagella are shed in swarmer cells before differentiation into stalked cells and can be observed in media or on surfaces previously occupied by cells (61).
At the flagellated pole, Caulobacter also synthesizes multiple type IV tight adherence (tad) pili, which are used for sensing and adhering to surfaces, are retracted following differentiation into stalked cells, and are synthesized once again on swarmer daughter cells following division (Fig. 2) (62). Tad pili, composed of a basal complex and a filamentous fiber of the pilin PilA, are dynamic, undergo cycles of extension and retraction, and are common across both bacteria and archaea. Surprisingly, extension and retraction of the pili are both facilitated in Caulobacter by a noncanonical, bifunctional ATPase motor CpaF (63).
Both flagella and pili mediate surface attachment and mechanosensation that trigger production of holdfast at the flagellated pole (64, 65). This signaling is primarily mediated by the second messenger 3′,5′-cyclic-di-GMP (c-di-GMP), which is present at low levels in swarmer cells and is synthesized in response to mechanical perturbation of the flagella and pili, activating pilus retraction and holdfast synthesis to facilitate adherence (66–68). Components of the flagellar machinery also mediate pilus biosynthesis, as loss of the former results in attenuated pilus formation (69). Additionally, transcription of the holdfast synthesis inhibitor gene hfiA is regulated both by flagellum assembly and nutrient availability (70, 71), providing a link between environmental cues and control of surface attachment.
Finally, like many other bacteria, Caulobacter cells are enveloped by a periodic proteinaceous S-layer that is continuously present throughout all stages of the cell cycle (72, 73). The sole S-layer protein of Caulobacter, RsaA, is continuously produced throughout the cell cycle and is homologous to S-layer proteins from other organisms (74). RsaA forms a hexagonal lattice structure that encompasses the entire cell and binds to lipopolysaccharide, as observed by high-resolution cryo-ET that was aided by the thin diameter of Caulobacter stalks (75, 76). Proposed functions of the S-layer in various bacterial species include adhesion to surfaces and protection against chemical and biological threats (reviewed in reference 77). Additionally, the transducing phage ϕCr30 requires the S-layer for adsorption and subsequent infection, indicating the presence of more complicated roles in the environment and for Caulobacter evolution (78).
A living integrated circuit.
Most bacteria possess numerous mechanisms for exerting both spatial and temporal control, but this task is exquisitely complicated in a bacterium like Caulobacter that undergoes differentiation throughout its cell cycle. As we have explored so far, progression through the cell cycle is dependent on physical and chemical cues in the environment as well as internal timing. Moreover, in Caulobacter, cell cycle events are intimately coupled to polar development; but how are each of these steps regulated? Caulobacter possesses four global regulators (DnaA [79], GcrA [80], CtrA [81, 82], and CcrM [83]) that are produced sequentially and out of phase with one another (84) (Fig. 4) and together regulate over 200 genes required for progression to the next stages of the cell and developmental cycle, including each other (reviewed in references 85–87). Discovery of the extent of their regulatory networks began with development of the first Caulobacter microarray, made possible by the acquisition of the complete sequence of the genome (30). In this landmark study, temporal microarray measurements revealed that gene expression is coordinated with the function of respective gene products (e.g., expression of DNA replication machinery and divisome components is activated before their respective events in the cell cycle) (88). This study also revealed that the expression of genes whose products make up complexes are typically coregulated and that regulation of biogenesis of large structures is regulated by integrated signal cascades. More recent work has suggested that the global integrated circuit that controls cell cycle progression includes genetically separable subcircuits that control distinct aspects of the cell cycle, such as CtrA/MucR in swarmer cells (89), GcrA/CcrM and DnaA during DNA replication (90, 91), and CtrA/SciP during cell division (92) (reviewed in reference 93). Further studies implicated global regulation of translation (94), phosphorylation (33, 95–98), and protein degradation (96, 99) as well as localization and production of small-molecule second messengers (100) as additional mechanisms of cell cycle regulation. These findings made it clear that Caulobacter acts as a biological integrated circuit, complete with interacting positive and negative cues that give rise to both temporal and spatial regulation (95, 101).
FIG 4.
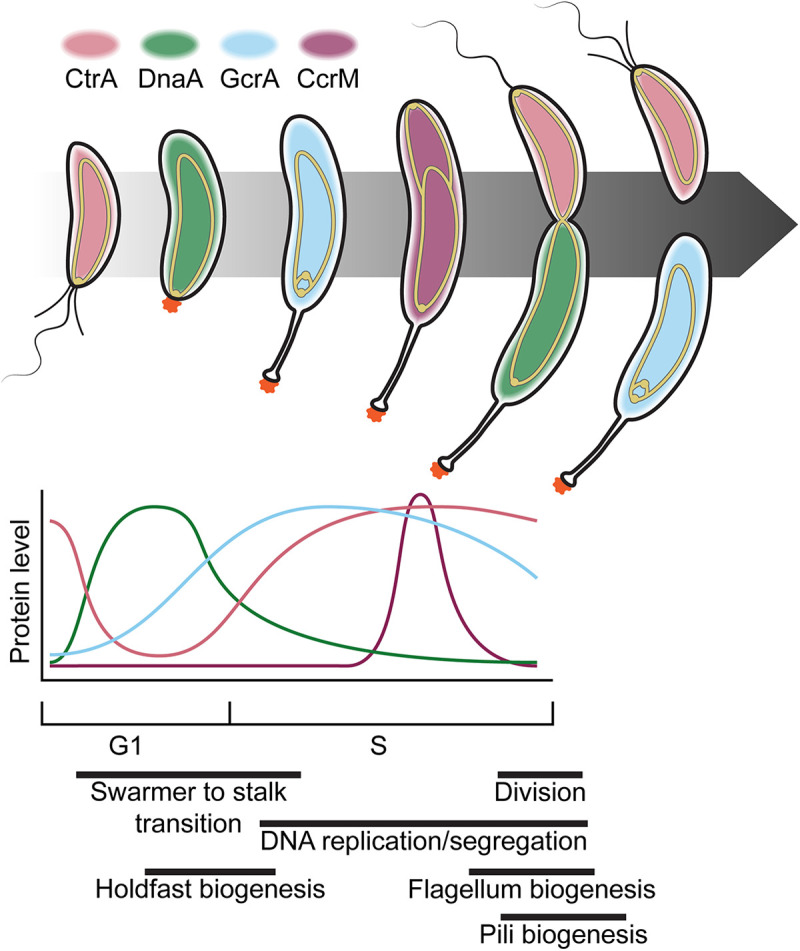
Caulobacter differentiation is mediated by global regulators. Progression through the cell cycle is shown as in Fig. 2, with the most abundant global regulator (CtrA, pink; DnaA, green; GcrA, cyan; CcrM, plum) present for each cell type indicated by the color of the cell. Relative levels of each global regulator throughout the cell cycle are shown in the graph below. The chromosome, shown as a gold-outlined ellipse (origins are indicated by circles at the ends), is anchored to the flagellar pole in swarmer cells (G1 phase) and begins replication after the transition to a stalked cell (S phase). The origin of the newly replicated DNA is translocated to the nascent swarmer pole, and the terminus is directed to midcell, where the two chromosomes are distributed between the daughter cells coincident with cell division. Relative timescales of various morphogenetic events are indicated below (94, 101, 192).
The CtrA protein is a response regulator that controls expression of over one-fourth of cell cycle-regulated genes, including those involved in flagellum and pilus biogenesis, DNA replication and methylation, signaling, and cell division (88). Phosphorylated CtrA (CtrA~P) inhibits replication initiation (102) and gcrA expression (84) in swarmer cells by binding to the origin of replication and the gcrA promoter, respectively. CtrA~P is later dephosphorylated and degraded at the stalked pole, allowing the replication initiator DnaA to bind to the origin, resulting in initiation of replication and production of GcrA (reviewed in reference 103). Following initiation, DnaA activity is inhibited by HdaA (104), which stimulates conversion of DnaA-ATP to DnaA-ADP, to prevent reinitiation, and DnaA-ATP is regenerated via degradation of DnaA-ADP and production of newly synthesized DnaA (90) (reviewed in reference 105). DnaA is primarily degraded by Lon protease in the event of nutrient limitation or proteotoxic stress (106, 107) and by ClpXP or ClpAP under starvation conditions (106) or during the stationary phase (108), respectively, to ensure cell cycle arrest under stress (also summarized in reference 105). GcrA, a cofactor of the σ70 transcription factor (80), activates the expression of ctrA in its hemimethylated state (i.e., only when the replication fork has passed and before the newly synthesized strand is methylated) (81, 109). Newly synthesized CtrA then regulates its own expression to rapidly increase CtrA levels in the cell. CtrA~P drives expression of ccrM, which encodes the methyltransferase CcrM that methylates the newly replicated genome, shutting down ctrA (81, 109) and ccrM (110) expression.
Cell identity is tightly controlled by an asymmetric logic system that integrates with the regulatory mechanisms described above. An overarching principle of this system is the dynamic interplay of interconnected signaling networks that work in tandem to control both pole identity and progression through the cell cycle (reviewed in references 82 and 111). The first component of this system that was discovered was pleC, the mutation of which resulted in loss of stalks and phage receptors as well as inactive flagella (46). The pleC-mutant phenotype provided a foundation for the discovery of other cell cycle regulators, including PleD (112), DivJ, DivK, and DivL (113) by isolation of bypass mutants that restored either motility or phage susceptibility to pleC-mutant cells. Localization and activity of the diguanylate cyclase PleD are regulated by its phosphorylation state. The phosphatase PleC inactivates PleD at the swarmer pole, and the kinase DivJ activates PleD at the stalked pole (114, 115) where it synthesizes c-di-GMP to signal for shedding of the flagellum and transition from swarmer to stalked cells (115–117).
The response regulator DivK provides a link between cell cycle progression and cell fate determination pathways. Like PleD, DivK is phosphorylated by DivJ at the stalked pole and dephosphorylated by PleC at the swarmer pole in predivisional cells. This results in differential localization of phosphorylated DivK (DivK~P) and DivK in the stalked and swarmer compartments, respectively, as division proceeds (118, 119). DivK~P inactivates DivL, a pseudokinase that activates the hybrid histidine kinase CckA, which phosphorylates and activates CtrA (120–122). Chromosome segregation is also required for this process, as the histidine phosphotransferase ChpT (93) is only recruited to the swarmer pole after successful segregation. ChpT localization completes the CckA-ChpT-CtrA phosphorelay to activate CtrA signaling and allow for progression through cell division (123). High levels of c-di-GMP produced by PleD in the stalked compartment (124) induce CckA to dephosphorylate CtrA (97, 125, 126) and lead to CtrA degradation by ClpXP (127). Swarmer pole identity is reestablished by the polar marker TipN (128, 129), which localizes to the nascent swarmer pole and recruits the phosphodiesterase TipF (flagellar assembly [128]) and PleC (48, 129), allowing for the relocalization of swarmer cell determinants and flagellar and pili assembly machinery (130).
A model for progress.
Caulobacter is a premier bacterial model for studying cell cycle progression. The presence of morphological markers and clearly defined asymmetry at every step in the cell cycle means that cells at any point in the cell cycle can be readily identified, even in unsynchronized populations. This, along with the ability to isolate swarmer cells and follow their progression, allows for powerful analysis of events that are coordinated with the cell cycle.
The Caulobacter cell cycle includes several distinct stages (Fig. 2). One of the key developmental stages in the Caulobacter life cycle is the differentiation from a swarmer cell to a stalked cell, which is coincident with entry into S phase. As one would predict based on the necessity of available nutrients for propagation, the swarmer-to-stalked (and G1 to S) transition is slowed under nutrient limitation. After starvation, Caulobacter initiates the stringent response, mediated by the small molecule alarmones (p)ppGpp, the synthesis of which delays this transition and regulates expression of numerous genes involved in metabolism, genome replication, growth, and division (131–133).
In the presence of nutrients, swarmer cells transition to a stalked fate and enter S phase, which is marked by replication and simultaneous segregation of the chromosome. This occurs exactly once per cell cycle in logarithmically growing cells, and its regulation is tightly coordinated with cell division. The Caulobacter origin of replication is anchored to the stalked pole via indirect interactions with the polar organizing protein PopZ (134, 135) and is acted upon primarily by the replication activator DnaA and the response regulator CtrA, as described in the previous section. PopZ is a scaffolding protein containing an intrinsically disordered region (IDR) and structured regions that together modulate interactivity (136, 137). PopZ forms a biomolecular condensate to create ribosome-free microdomains at the poles that selectively regulate entry of CckA, the phosphotransferase ChpT, and CtrA to regulate and enhance the phosphosignaling underlying cell differentiation (138, 139).
Replication continues until the genome is completely duplicated, during which a single origin remains at the stalked pole, while the second origin is translocated to the nascent swarmer pole by the parABS system (Fig. 5A). ParB is a DNA-binding CTPase that recognizes and binds to the origin-adjacent parS sequence, forming a ParB complex that spreads along the DNA (140). ParB binds to ParA, an ATPase that binds sequence nonspecifically to the chromosome, and regulates its nucleotide cycle to establish a ParA-ATP gradient (141–143). The combined biochemical activities and interactions of ParA and ParB result in processive diffusion of one copy of the ParA-ParB-parS complex along the length of the predivisional cell to the nascent swarmer pole, with regulatory input from TipN and PopZ (142, 144, 145). PopZ captures the newly segregated ParB-parS complex and anchors it at the swarmer pole (134, 135). The parABS system is broadly conserved for both genome and plasmid partitioning, having been identified in 69% of bacterial genomes surveyed (146), and the insights gained from the study of chromosome segregation in Caulobacter have been instrumental in establishing a mechanistic paradigm for its function.
FIG 5.
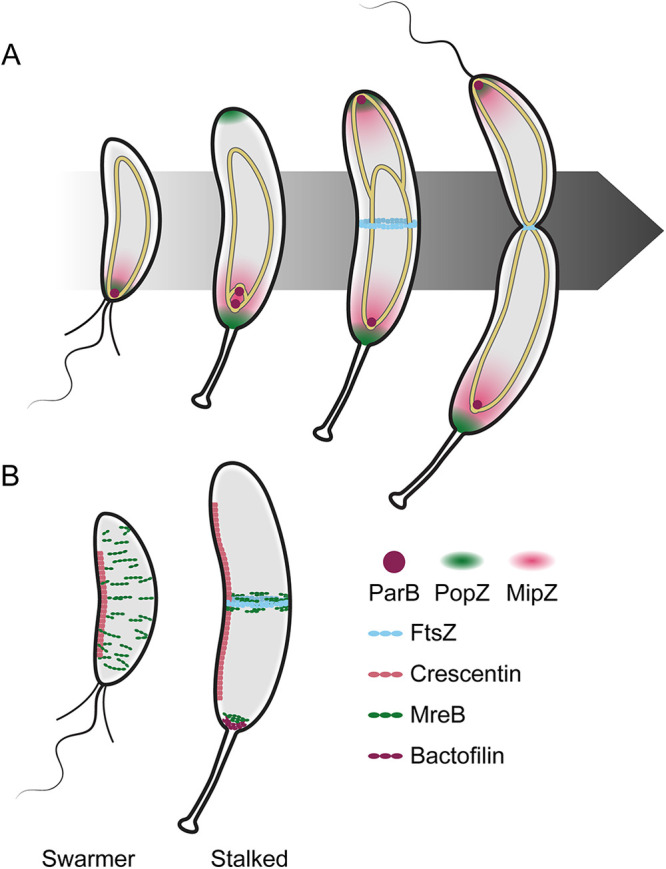
Spatial regulation of morphogenesis in Caulobacter. (A) FtsZ localization is driven by interactions with the chromosome (gold-outlined ellipse). ParB (plum circle) binds the parS sequence near the origin (circles at the ends of the chromosomes) and associates with PopZ (green cloud) at the pole. ParB translocates with the newly synthesized origin and anchors it to the nascent swarmer pole (top of cells). MipZ (pink cloud) binds to ParB and to the DNA to form a gradient, limiting FtsZ (cyan) polymerization to midcell in stalked and predivisional cells. (B) Localization of four major cytoskeletal proteins (crescentin, pink; MreB, green; FtsZ, cyan; bactofilin, plum) shown in swarmer (left) and stalked (right) cells. Crescentin directs curvature in both cell types. MreB directs elongasome localization and global cell wall insertion in swarmer cells and midcell insertion in stalked cells to facilitate elongation. FtsZ directs divisome assembly and cell wall remodeling at midcell in stalked cells to facilitate constriction. Bactofilins and MreB recruit and regulate stalk elongation machinery during the swarmer-to-stalk transition and remain localized at the base of the stalk in stalked cells.
Cell division, the final major step in the Caulobacter cell cycle, is unsurprisingly closely linked with DNA replication and segregation. This is important for two reasons: (i) to ensure that each daughter cell contains a copy of the genome and (ii) to ensure that the genome does not interfere with the process of constriction. Constriction activation begins with the tubulin homolog FtsZ, which localizes approximately to midcell to form a ring-like structure marking the division plane (Fig. 5, right) (147, 148) (reviewed in reference 149). FtsZ then recruits downstream components of the divisome, a multiprotein machinery responsible for performing cell division, including cell wall synthases that modify the cell wall to drive a change in shape that eventually results in constriction (150).
FtsZ localization to midcell is crucial for proper cell division to occur and is driven by interactions with MipZ, which binds to ParB and interferes with FtsZ polymerization (Fig. 5A) (151). MipZ forms a gradient emanating from the poles, resulting in an area of low concentration at midcell, allowing for FtsZ polymerization and the formation of the Z-ring there. Importantly, production of several divisome factors, including FtsZ, is tightly regulated throughout the cell cycle. The expression of ftsZ, inhibited by CtrA~P, increases coincident with differentiation into a stalked cell, and FtsZ levels peak just before the start of division (152). Following the initiation of constriction, FtsZ is degraded rapidly by ClpXP and ClpAP (153, 154), leading to clearance in the nascent swarmer cell and maintenance of low levels of FtsZ in the stalked cell until the next round of cell division (152). Like in the case of DNA replication, tight regulation of divisome components ensures that division only occurs at the correct time in the cell cycle (i.e., after DNA replication and cell differentiation have progressed sufficiently). Interference of envelope constriction by the nucleoid is also regulated by the protein FtsK. FtsK is composed of two domains, a C-terminal DNA translocation domain and an N-terminal transmembrane domain associated with activation of cell wall synthases. Both domains are essential for proper chromosome segregation and constriction, suggesting that it serves as another link between these processes to ensure timely completion of segregation before the completion of constriction (155, 156).
Following FtsZ localization to midcell, the rest of the divisome must assemble, and many of these recruitment events are dependent on the correct localization of upstream factors. This epistatic pathway has been observed in other bacteria, but the actual timing of each component’s arrival is difficult to ascertain without the ability to synchronize a population of cells. Leveraging Caulobacter synchrony to this end, Goley et al. observed the timing of midcell localization of roughly 20 factors involved in Caulobacter division (157) with a level of precision that was previously unavailable, giving critical insight into the timing of divisome assembly and its regulation.
Shape matters.
As one would expect in a vibrioid, asymmetric bacterium, Caulobacter encodes numerous factors that play a role in morphogenesis and shape determination (reviewed in reference 158). The divisome, described above, is the most broadly conserved morphogenetic system, primarily controlling cell length (reviewed in reference 150). The next system is known as the elongasome, organized by the actin-like protein MreB, which associates with the Rod complex to guide cell wall insertion and drive elongation, generating the cell’s rod-like shape (159). Unlike FtsZ, however, MreB levels are relatively constant throughout the cell cycle (88, 94), consistent with the notion that the elongasome functions constantly during cell growth. MreB filaments assemble along the inner membrane perpendicular to the long axis of the cell, resulting in dynamic, broadly distributed patches early in the cell cycle that are localized to midcell in an FtsZ-dependent manner before division (Fig. 5B) (159, 160). The divisome and elongasome are conserved in many other species, which has allowed for the undertaking of comparative studies to understand how these systems function in species-specific manners.
Both the divisome and the elongasome act on the peptidoglycan cell wall, with cytoskeletal proteins directing construction and modification of this rigid macromolecule to effect changes on overall cell shape (reviewed in reference 161). Similarly, the curvature of Caulobacter is mediated by crescentin (encoded by creS), the first intermediate filament-like protein described in bacteria. Crescentin assembles into curved filaments along the inner curvature of the cytoplasmic membrane and is required for cell curvature (Fig. 5B) (162). Crescentin is thought to exert its effects on cell shape by mechanically regulating cell wall insertion such that more cell wall synthesis occurs distal to crescentin localization than occurs proximal to crescentin. Ultimately this asymmetry in cell wall synthesis yields a curved cell shape. Curvature has been associated with increased motility in bacteria (163) (e.g., in Vibrio cholerae [164]). Findings that Caulobacter exhibits low flagellar motor torque (165) and helical body movement (166), both of which result in increased swimming efficiency, suggest that this is a key adaptation for a microbe that inhabits low-nutrient environments. Curvature has also been observed to play a role in Caulobacter adhesion to surfaces when dividing under flow (167). The discovery of crescentin inspired investigation into the shape determinants in other vibrioid cells, and, indeed, proteins such as CrvA in V. cholerae appear to perform a similar function with a similar proposed mechanism (168).
Like the systems described above, stalk morphogenesis is partly facilitated by cytoskeletal proteins. Bactofilins, which were first described in Caulobacter, are widely conserved, bacterium-specific cytoskeletal proteins that form membrane-associated polymers or sheets with a variety of functions across species (169). The Caulobacter bactofilins, BacA and BacB, accumulate at the nascent stalked pole before stalk formation and are continuously present at the base of the stalk (Fig. 5B, right). These bactofilins interact with the cell wall synthase PbpC, which is required for the recruitment of the stalk elongation modulator StpX (55, 170), and recruit it to the stalk. The absence of any of these factors results in a decrease in stalk length but do not change overall stalk structure, indicating roles specifically in stalk extension (171). MreB and RodA, components of the elongasome, also localize to the base of the stalk (Fig. 5B, right) and are necessary for stalk formation, as depletion of either protein results in a stalk elongation defect (172). Further studies implicated the autolytic enzymes DipM, SdpAB, and CrbA, which are also associated with divisome function, and LdpA, the gene of which lies in an operon with bacA, in stalk morphogenesis. MreB inhibition results in a failure of each of the autolytic enzymes, but not BacA, to localize to the stalked pole. Although the ultimate determinant of polar localization for these complexes remains unknown, these interactions suggest a mechanism involving the combined regulation of multiple scaffolding proteins and cell wall remodeling complexes for effecting stalk formation and elongation (173). Finally, the aforementioned crossbands that limit diffusion along the length of the stalk are composed of a complex of four proteins, StpABCD, with StpA directing the recruitment of the rest of the complex (47). Stalk morphology in related stalked organisms (e.g., Asticcacaulis and Brevundimonas) (46, 174) resembles that of Caulobacter. However, while BacA, the elongasome, and hydrolytic enzymes are generally well conserved in this group, several stalk-related genes, including bacB, pbpC, stpX, and stpABCD, are not well conserved outside Caulobacter, suggesting differences between the mechanisms of stalk morphogenesis among these species (175).
CURRENT/FUTURE QUESTIONS
While the bulk of this review has focused on the features and discoveries of Caulobacter that have led to its establishment as one of the premier model organisms for bacterial cell biology, we would be remiss if we failed to highlight some of the ongoing work and future directions for this exciting microorganism. One of these areas is the study of Caulobacter spp. in their native environments, taking the knowledge that we have acquired in the laboratory and applying it to understanding how the niche(s) that Caulobacter spp. occupy shape its biology (176, 177) and how Caulobacter spp. have evolved to interact with other organisms in those environments. Recent findings indicate that Caulobacter is far more abundant in soils than aquatic environments, in contrast to early predictions about the environmental range of Caulobacter, demonstrating its ability to survive in oligotrophic environments and indicating roles in decomposition and dispersal dynamics (178). Additional studies have showcased the discovery of multiple novel Caulobacter-targeting phages, including the first lysogenic phage found for Caulobacter to date (179), suggesting that there is a great deal more to discover about Caulobacter and its interaction with phages. Another phage was isolated from the rhizosphere, corroborating previous reports indicating that some Caulobacter species can localize to regions of plant growth and could fulfill roles as plant growth-promoting bacteria (180). There are numerous open questions about the role(s) that Caulobacter plays in these communities and how those roles might depend on features specific to a dimorphic, differentiating bacterium.
Many of the morphological features and subcellular complexes studied in Caulobacter are conserved across Alphaproteobacteria, making Caulobacter a useful model for engaging in comparative morphology and evolutionary studies to determine how these systems arise. Combining genomic sequence data with high-throughput culturing and imaging methods, it is possible to study the evolution of various morphological phenomena (e.g., polar growth, stalk formation, etc.) and identify the molecular determinants underlying their development (174). Having a well-studied, easily tractable organism like Caulobacter in hand allows for the generation of hypotheses about how various morphological systems evolved and are regulated across species. For example, our understanding of stalk formation and development in Caulobacter aided in the evolutionary and morphogenetic analysis of stalk placement and biogenesis in two Asticcacaulis species that form nonpolar stalks, resulting in the discovery of a novel function for the developmental regulator SpmX in these species (181).
The condensate-forming protein PopZ has promising future applications in the study of cell biology and biophysics. A PopZ-based recruitment assay (analogous to bacterial two-hybrid assays) was recently developed to enable the study of exogenous protein-protein interactions using E. coli as a host (182). Another study has recently indicated the potential of PopZ as a tunable model for generating synthetic condensates for the investigation of general properties in vitro and in mammalian cells (136).
Outside the field of cell biology, Caulobacter and its relatives are powerful organisms for the development of new technological advances. Caulobacter holdfast has been measured among the strongest known biological adhesives (183), and holdfast from the closely related Hirschia baltica exhibits an even greater degree of adhesion via an increased tolerance to ionic strength (184, 185), presenting opportunities for the development of versatile medically or commercially relevant adhesives (186). The Caulobacter S-layer is also being developed as a platform for synthesis and secretion of novel materials. The first study to demonstrate the feasibility of using Caulobacter’s S-layer secretion system to produce recoverable recombinant proteins was published in 1997 (187). This innovation, in combination with efforts to characterize S-layer assembly (188) and Caulobacter behavior and adaptability in a dense biofilm setting (189), has led to the development of a robust system for generating engineered living materials that have potential applications in human health, energy, and the environment (190, 191). From basic cell biology to the production of advanced industrial materials, Caulobacter remains at the forefront of bacterial research as we continue to learn more about this fascinating organism.
ACKNOWLEDGMENTS
We thank the members of the Goley lab and Lucy Shapiro, Bert Ely, and Peter Chien for helpful discussion and feedback on this article.
Work in the Goley laboratory on Caulobacter was supported by the National Institutes of Health (grant numbers R35GM136221 [to E.D.G.] and T32GM007445 [training grant support of J.M.B.]).
We declare no conflict of interest.
Contributor Information
Erin D. Goley, Email: egoley1@jhmi.edu.
George O’Toole, Geisel School of Medicine at Dartmouth.
REFERENCES
- 1.Henrici AT, Johnson DE. 1935. Studies of freshwater bacteria: II. Stalked bacteria, a new order of Schizomycetes. J Bacteriol 30:61–93. 10.1128/jb.30.1.61-93.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones M. 1905. A peculiar microorganism showing rosette formation. Zentr Bakteriol Parasitenk Abt II 14:459–463. [Google Scholar]
- 3.Omeliansky VL. 1914. A new bacillus: Bacillus flagellatus. Zh Mikrobiol Epidemiol Immunolobiol 1:24. [Google Scholar]
- 4.Houwink AL, van Iterson W. 1950. Electron microscopical observations on bacterial cytology; a study on flagellation. Biochim Biophys Acta 5:10–44. 10.1016/0006-3002(50)90144-2. [DOI] [PubMed] [Google Scholar]
- 5.Bowers LE, Weaver RH, Grula EA, Edwards OF. 1954. Studies on a strain of Caulobacter from water. I. Isolation and identification as Caulobacter vibrioides Henrici and Johnson with emended description. J Bacteriol 68:194–200. 10.1128/jb.68.2.194-200.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poindexter JS. 1964. Biological properties and classification of the Caulobacter group. Bacteriol Rev 28:231–295. 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marks ME, Castro-Rojas CM, Teiling C, Du L, Kapatral V, Walunas TL, Crosson S. 2010. The genetic basis of laboratory adaptation in Caulobacter crescentus. J Bacteriol 192:3678–3688. 10.1128/JB.00255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodenmiller D, Toh E, Brun YV. 2004. Development of surface adhesion in Caulobacter crescentus. J Bacteriol 186:1438–1447. 10.1128/JB.186.5.1438-1447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yun C, Ely B, Smit J. 1994. Identification of genes affecting production of the adhesive holdfast of a marine caulobacter. J Bacteriol 176:796–803. 10.1128/jb.176.3.796-803.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapiro L, Agabian-Keshishian N, Bendis I. 1971. Bacterial differentiation. Science 173:884–892. 10.1126/science.173.4000.884. [DOI] [PubMed] [Google Scholar]
- 11.Degnen ST, Newton A. 1972. Chromosome replication during development in Caulobacter crescentus. J Mol Biol 64:671–680. 10.1016/0022-2836(72)90090-3. [DOI] [PubMed] [Google Scholar]
- 12.Stove JL, Stanier RY. 1962. Cellular differentiation in stalked bacteria. Nature 196:1189–1192. 10.1038/1961189a0. [DOI] [Google Scholar]
- 13.Schrader JM, Shapiro L. 2015. Synchronization of Caulobacter crescentus for investigation of the bacterial cell cycle. J Vis Exp 2015:52633. 10.3791/52633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ardissone S, Fumeaux C, Bergé M, Beaussart A, Théraulaz L, Radhakrishnan SK, Dufrêne YF, Viollier PH. 2014. Cell cycle constraints on capsulation and bacteriophage susceptibility. eLife 3:e03587. 10.7554/eLife.03587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bendis I, Shapiro L. 1970. Properties of Caulobacter ribonucleic acid bacteriophage φCb5. J Virol 6:847–854. 10.1128/JVI.6.6.847-854.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agabian-Keshishian N, Shapiro L. 1970. Stalked bacteria: properties of deoxriybonucleic acid bacteriophage φCbK. J Virol 5:795–800. 10.1128/JVI.5.6.795-800.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt JM. 1966. Observations on the adsorption of Caulobacter bacteriophages containing ribonucleic acid. J Gen Microbiol 45:347–353. 10.1099/00221287-45-2-347. [DOI] [PubMed] [Google Scholar]
- 18.Skerker JM, Shapiro L. 2000. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J 19:3223–3234. 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shapiro L, Agabian-Keshishian N. 1970. Specific assay for differentiation in the stalked bacterium Caulobacter crescentus. Proc Natl Acad Sci USA 67:200–203. 10.1073/pnas.67.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohta N, Ninfa AJ, Allaire A, Kulick L, Newton A. 1997. Identification, characterization, and chromosomal organization of cell division cycle genes in Caulobacter crescentus. J Bacteriol 179:2169–2180. 10.1128/jb.179.7.2169-2180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ely B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol 204:372–384. 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 22.Johnson RC, Ely B. 1977. Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics 86:25–32. 10.1093/genetics/86.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ely B. 1979. Transfer of drug resistance factors to the dimorphic bacterium Caulobacter crescentus. Genetics 91:371–380. 10.1093/genetics/91.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ely B, Johnson RC. 1977. Generalized transduction in Caulobacter crescentus. Genetics 87:391–399. 10.1093/genetics/87.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilchrist A, Smit J. 1991. Transformation of freshwater and marine caulobacters by electroporation. J Bacteriol 173:921–925. 10.1128/jb.173.2.921-925.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minnich SA, Ohta N, Taylor N, Newton A. 1988. Role of the 25-, 27-, and 29-kilodalton flagellins in Caulobacter crescentus cell motility: method for construction of deletion and Tn5 insertion mutants by gene replacement. J Bacteriol 170:3953–3960. 10.1128/jb.170.9.3953-3960.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ely B, Croft RH. 1982. Transposon mutagenesis in Caulobacter crescentus. J Bacteriol 149:620–625. 10.1128/jb.149.2.620-625.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett JT, Rhodes CS, Ferber DM, Jenkins B, Kuhl SA, Ely B. 1982. Construction of a genetic map for Caulobacter crescentus. J Bacteriol 149:889–896. 10.1128/jb.149.3.889-896.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ely B, Ely TW. 1989. Use of pulsed field gel electrophoresis and transposon mutagenesis to estimate the minimal number of genes required for motility in Caulobacter crescentus. Genetics 123:649–654. 10.1093/genetics/123.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nierman WC, Feldblyum T.v, Laub MT, Paulsen IT, Nelson KE, Eisen J, Heidelberg JF, Alley MRK, Ohta N, Maddock JR, Potocka I, Nelson WC, Newton A, Stephens C, Phadke ND, Ely B, DeBoy RT, Dodson RJ, Durkin AS, Gwinn ML, Haft DH, Kolonay JF, Smit J, Craven MB, Khouri H, Shetty J, Berry K, Utterback T, Tran K, Wolf A, Vamathevan J, Ermolaeva M, White O, Salzberg SL, Venter JC, Shapiro L, Fraser CM. 2001. Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci USA 98:4136–4141. 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coons AH, Creech HJ, Jones RN. 1941. Immunological properties of an antibody containing a fluorescent group. Exp Biol Med 47:200–202. 10.3181/00379727-47-13084P. [DOI] [Google Scholar]
- 32.Coons AH, Creech J, Norman R, Berliner E. 1942. The demonstration of pneumococcal antigen in tissues by the use of fluorescent antibody. J Immunology 45:159–170. 10.4049/jimmunol.45.3.159. [DOI] [Google Scholar]
- 33.Jacobs C, Domian IJ, Maddock JR, Shapiro L. 1999. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell 97:111–120. 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- 34.Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. 2004. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci USA 101:9257–9262. 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biteen JS, Thompson MA, Tselentis NK, Bowman GR, Shapiro L, Moerner WE. 2008. Super-resolution imaging in live Caulobacter crescentus cells using photoswitchable EYFP. Nat Methods 5:947–949. 10.1038/nmeth.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holden SJ, Pengo T, Meibom KL, Fernandez C, Collier J, Manley S. 2014. High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc Natl Acad Sci USA 111:4566–4571. 10.1073/pnas.1313368111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen GJ, Briegel A. 2007. How electron cryotomography is opening a new window onto prokaryotic ultrastructure. Curr Opin Struct Biol 17:260–267. 10.1016/j.sbi.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Judd EM, Comolli LR, Chen JC, Downing KH, Moerner WE, McAdams HH. 2005. Distinct constrictive processes, separated in time and space, divide Caulobacter inner and outer membranes. J Bacteriol 187:6874–6882. 10.1128/JB.187.20.6874-6882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Briegel A, Dias DP, Li Z, Jensen RB, Frangakis AS, Jensen GJ. 2006. Multiple large filament bundles observed in Caulobacter crescentus by electron cryotomography. Mol Microbiol 62:5–14. 10.1111/j.1365-2958.2006.05355.x. [DOI] [PubMed] [Google Scholar]
- 40.Li Z, Trimble MJ, Brun YV, Jensen GJ. 2007. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J 26:4694–4708. 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Jones BD, Brun YV. 2001. A set of ftsZ mutants blocked at different stages of cell division in Caulobacter. Mol Microbiol 40:347–360. 10.1046/j.1365-2958.2001.02395.x. [DOI] [PubMed] [Google Scholar]
- 42.Werner JN, Chen EY, Guberman JM, Zippilli AR, Irgon JJ, Gitai Z. 2009. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc Natl Acad Sci USA 106:7858–7863. 10.1073/pnas.0901781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lasker K, Schrader JM, Men Y, Marshik T, Dill DL, McAdams HH, Shapiro L. 2016. CauloBrowser: a systems biology resource for Caulobacter crescentus. Nucleic Acids Res 44:D640–D645. 10.1093/nar/gkv1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price MN, Wetmore KM, Waters RJ, Callaghan M, Ray J, Liu H, Kuehl J.v, Melnyk RA, Lamson JS, Suh Y, Carlson HK, Esquivel Z, Sadeeshkumar H, Chakraborty R, Zane GM, Rubin BE, Wall JD, Visel A, Bristow J, Blow MJ, Arkin AP, Deutschbauer AM. 2018. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557:503–509. 10.1038/s41586-018-0124-0. [DOI] [PubMed] [Google Scholar]
- 45.Marczynski GT. 1999. Chromosome methylation and measurement of faithful, once and only once per cell cycle chromosome replication in Caulobacter crescentus. J Bacteriol 181:1984–1993. 10.1128/JB.181.7.1984-1993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stove Poindexter JL, Cohen-Bazire G. 1964. The fine structure of stalked bacteria belonging to the family Caulobacteraceae. J Cell Biol 23:587–607. 10.1083/jcb.23.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlimpert S, Klein EA, Briegel A, Hughes V, Kahnt J, Bolte K, Maier UG, Brun YV, Jensen GJ, Gitai Z, Thanbichler M. 2012. General protein diffusion barriers create compartments within bacterial cells. Cell 151:1270–1282. 10.1016/j.cell.2012.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fukuda A, Iba H, Okada Y. 1977. Stalkless mutants of Caulobacter crescentus. J Bacteriol 131:280–287. 10.1128/jb.131.1.280-287.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levi A, Jenal U. 2006. Holdfast formation in motile swarmer cells optimizes surface attachment during Caulobacter crescentus development. J Bacteriol 188:5315–5318. 10.1128/JB.01725-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner JK, Setayeshgar S, Sharon LA, Reilly JP, Brun YV. 2006. A nutrient uptake role for bacterial cell envelope extensions. Proc Natl Acad Sci USA 103:11772–11777. 10.1073/pnas.0602047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt JM, Stanier RY. 1966. The development of cellular stalks in bacteria. J Cell Biol 28:423–436. 10.1083/jcb.28.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gonin M, Quardokus EM, O’Donnol D, Maddock J, Brun YV. 2000. Regulation of stalk elongation by phosphate in Caulobacter crescentus. J Bacteriol 182:337–347. 10.1128/JB.182.2.337-347.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pate JL, Ordal EJ. 1965. The fine structure of two unusual stalked bacteria. J Cell Biol 27:133–150. 10.1083/jcb.27.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klein EA, Schlimpert S, Hughes V, Brun YV, Thanbichler M, Gitai Z. 2013. Physiological role of stalk lengthening in Caulobacter crescentus. Commun Integr Biol 6:e24561. 10.4161/cib.24561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hughes HV, Lisher JP, Hardy GG, Kysela DT, Arnold RJ, Giedroc DP, Brun YV. 2013. Co-ordinate synthesis and protein localization in a bacterial organelle by the action of a penicillin-binding-protein. Mol Microbiol 90:1162–1177. 10.1111/mmi.12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson RC, Walsh MP, Ely B, Shapiro L. 1979. Flagellar hook and basal complex of Caulobacter crescentus. J Bacteriol 138:984–989. 10.1128/jb.138.3.984-989.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anderson DK, Ohta N, Wu J, Newton A. 1995. Regulation of the Caulobacter crescentus rpoN gene and function of the purified σ54 in flagellar gene transcription. Mol Gen Genet 246:697–706. 10.1007/BF00290715. [DOI] [PubMed] [Google Scholar]
- 58.Reisenauer A, Quon K, Shapiro L. 1999. The CtrA response regulator mediates temporal control of gene expression during the Caulobacter cell cycle. J Bacteriol 181:2430–2439. 10.1128/JB.181.8.2430-2439.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gorbatyuk B, Marczynski GT. 2001. Physiological consequences of blocked Caulobacter crescentus dnaA expression, an essential DNA replication gene. Mol Microbiol 40:485–497. 10.1046/j.1365-2958.2001.02404.x. [DOI] [PubMed] [Google Scholar]
- 60.Brun YV, Marczynski G, Shapiro L. 1994. The expression of asymmetry during Caulobacter cell differentiation. Annu Rev Biochem 63:419–450. 10.1146/annurev.bi.63.070194.002223. [DOI] [PubMed] [Google Scholar]
- 61.Shapiro L, Maizel JV, Jr. 1973. Synthesis and structure of Caulobacter crescentus flagella. J Bacteriol 113:478–485. 10.1128/jb.113.1.478-485.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sommer JM, Newton A. 1988. Sequential regulation of developmental events during polar morphogenesis in Caulobacter crescentus: assembly of pili on swarmer cells requires cell separation. J Bacteriol 170:409–415. 10.1128/jb.170.1.409-415.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ellison CK, Kan J, Chlebek JL, Hummels KR, Panis G, Viollier PH, Biais N, Dalia AB, Brun YV. 2019. A bifunctional ATPase drives tad pilus extension and retraction. Sci Adv 5:eaay2591. 10.1126/sciadv.aay2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Snyder RA, Ellison CK, Severin GB, Whitfield GB, Waters CM, Brun YV. 2020. Surface sensing stimulates cellular differentiation in Caulobacter crescentus. Proc Natl Acad Sci USA 117:17984–17991. 10.1073/pnas.1920291117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hershey DM, Fiebig A, Crosson S. 2021. Flagellar perturbations activate adhesion through two distinct pathways in Caulobacter crescentus. mBio 12:e03266-20. 10.1128/mBio.03266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hug I, Deshpande S, Sprecher KS, Pfohl T, Jenal U. 2017. Second messenger-mediated tactile response by a bacterial rotary motor. Science 358:531–534. 10.1126/science.aan5353. [DOI] [PubMed] [Google Scholar]
- 67.Ellison CK, Kan J, Dillard RS, Kysela DT, Ducret A, Berne C, Hampton CM, Ke Z, Wright ER, Biais N, Dalia AB, Brun YV. 2017. Obstruction of pilus retraction stimulates bacterial surface sensing. Science 358:535–538. 10.1126/science.aan5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sangermani M, Hug I, Sauter N, Pfohl T, Jenal U. 2019. Tad pili play a dynamic role in Caulobacter crescentus surface colonization. mBio 10:e01237-19. 10.1128/mBio.01237-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ellison CK, Rusch DB, Brun YV. 2019. Flagellar mutants have reduced pilus synthesis in Caulobacter crescentus. J Bacteriol 201:e00031-19. 10.1128/JB.00031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berne C, Ellison CK, Agarwal R, Severin GB, Fiebig A, Morton RI, Waters CM, Brun YV. 2018. Feedback regulation of Caulobacter crescentus holdfast synthesis by flagellum assembly via the holdfast inhibitor HfiA. Mol Microbiol 110:219–238. 10.1111/mmi.14099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fiebig A, Herrou J, Fumeaux C, Radhakrishnan SK, Viollier PH, Crosson S. 2014. A cell cycle and nutritional checkpoint controlling bacterial surface adhesion. PLoS Genet 10:e1004101. 10.1371/journal.pgen.1004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smit J, Agabian N. 1982. Cell surface patterning and morphogenesis: biogenesis of a periodic surface array during Caulobacter development. J Cell Biol 95:41–49. 10.1083/jcb.95.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smit J, Grano DA, Glaeser RM, Agabian N. 1981. Periodic surface array in Caulobacter crescentus: fine structure and chemical analysis. J Bacteriol 146:1135–1150. 10.1128/jb.146.3.1135-1150.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fisher JA, Smit J, Agabian N. 1988. Transcriptional analysis of the major surface array gene of Caulobacter crescentus. J Bacteriol 170:4706–4713. 10.1128/jb.170.10.4706-4713.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bharat TAM, Kureisaite-Ciziene D, Hardy GG, Yu EW, Devant JM, Hagen WJH, Brun YV, Briggs JAG, Löwe J. 2017. Structure of the hexagonal surface layer on Caulobacter crescentus cells. Nat Microbiol 2:17059. 10.1038/nmicrobiol.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.von Kügelgen A, Tang H, Hardy GG, Kureisaite-Ciziene D, Brun YV, Stansfeld PJ, Robinson CV, Bharat TAM. 2020. In situ structure of an intact lipopolysaccharide-bound bacterial surface layer. Cell 180:348–358. 10.1016/j.cell.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gerbino E, Carasi P, Mobili P, Serradell MA, Gómez-Zavaglia A. 2015. Role of S-layer proteins in bacteria. World J Microbiol Biotechnol 31:1877–1887. 10.1007/s11274-015-1952-9. [DOI] [PubMed] [Google Scholar]
- 78.Edwards P, Smit J. 1991. A transducing bacteriophage for Caulobacter crescentus uses the paracrystalline surface layer protein as a receptor. J Bacteriol 173:5568–5572. 10.1128/jb.173.17.5568-5572.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marczynski GT, Shapiro L. 1992. Cell-cycle control of a cloned chromosomal origin of replication from Caulobacter crescentus. J Mol Biol 226:959–977. 10.1016/0022-2836(92)91045-q. [DOI] [PubMed] [Google Scholar]
- 80.Haakonsen DL, Yuan AH, Laub MT. 2015. The bacterial cell cycle regulator GcrA is a σ 70 cofactor that drives gene expression from a subset of methylated promoters. Genes Dev 29:2272–2286. 10.1101/gad.270660.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Quon KC, Marczynski GT, Shapiro L. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83–93. 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 82.van Teeseling MCF, Thanbichler M. 2020. Generating asymmetry in a changing environment: cell cycle regulation in dimorphic alphaproteobacteria. Biol Chem 401:1349–1363. 10.1515/hsz-2020-0235. [DOI] [PubMed] [Google Scholar]
- 83.Zweiger G, Marczynski G, Shapiro L. 1994. A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J Mol Biol 235:472–485. 10.1006/jmbi.1994.1007. [DOI] [PubMed] [Google Scholar]
- 84.Collier J, Murray SR, Shapiro L. 2006. DnaA couples DNA replication and the expression of two cell cycle master regulators. EMBO J 25:346–356. 10.1038/sj.emboj.7600927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brown PJB, Hardy GG, Trimble MJ, Brun YV. 2008. Complex regulatory pathways coordinate cell-cycle progression and development in Caulobacter crescentus. Adv Microb Physiol 54:1–101. 10.1016/S0065-2911(08)00001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kirkpatrick CL, Viollier PH. 2012. Decoding Caulobacter development. FEMS Microbiol Rev 36:193–205. 10.1111/j.1574-6976.2011.00309.x. [DOI] [PubMed] [Google Scholar]
- 87.Goley ED, Toro E, McAdams HH, Shapiro L. 2009. Dynamic chromosome organization and protein localization coordinate the regulatory circuitry that drives the bacterial cell cycle. Cold Spring Harbor Symp Quant Biol 74:55–64. 10.1101/sqb.2009.74.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 290:2144–2148. 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- 89.Fumeaux C, Radhakrishnan SK, Ardissone S, Théraulaz L, Frandi A, Martins D, Nesper J, Abel S, Jenal U, Viollier PH. 2014. Cell cycle transition from S-phase to G1 in Caulobacter is mediated by ancestral virulence regulators. Nat Commun 5:4081. 10.1038/ncomms5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jonas K, Chen YE, Laub MT. 2011. Modularity of the bacterial cell cycle enables independent spatial and temporal control of DNA replication. Curr Biol 21:1092–1101. 10.1016/j.cub.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Murray SM, Panis G, Fumeaux C, Viollier PH, Howard M. 2013. Computational and genetic reduction of a cell cycle to its simplest, primordial components. PLoS Biol 11:e1001749. 10.1371/journal.pbio.1001749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gora KG, Tsokos CG, Chen YE, Srinivasan BS, Perchuk BS, Laub MT. 2010. A cell-type-specific protein-protein interaction modulates transcriptional activity of a master regulator in Caulobacter crescentus. Mol Cell 39:455–467. 10.1016/j.molcel.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Panis G, Murray SR, Viollier PH. 2015. Versatility of global transcriptional regulators in alpha-proteobacteria: from essential cell cycle control to ancillary functions. FEMS Microbiol Rev 39:120–133. 10.1093/femsre/fuu002. [DOI] [PubMed] [Google Scholar]
- 94.Schrader JM, Li G-W, Childers WS, Perez AM, Weissman JS, Shapiro L, McAdams HH. 2016. Dynamic translation regulation in Caulobacter cell cycle control. Proc Natl Acad Sci USA 113:E6859–E6867. 10.1073/pnas.1614795113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. 2006. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 444:899–904. 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- 96.Domian IJ, Quon KC, Shapiro L. 1997. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90:415–424. 10.1016/S0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 97.Wu J, Ohta N, Zhao J-L, Newton A. 1999. A novel bacterial tyrosine kinase essential for cell division and differentiation. Proc Natl Acad Sci USA 96:13068–13073. 10.1073/pnas.96.23.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Coppine J, Kaczmarczyk A, Petit K, Brochier T, Jenal U, Hallez R. 2020. Regulation of bacterial cell cycle progression by redundant phosphatases. J Bacteriol 202:e00345-20. 10.1128/JB.00345-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gorbatyuk B, Marczynski GT. 2004. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol Microbiol 55:1233–1245. 10.1111/j.1365-2958.2004.04459.x. [DOI] [PubMed] [Google Scholar]
- 100.Xu C, Weston BR, Tyson JJ, Cao Y. 2020. Cell cycle control and environmental response by second messengers in Caulobacter crescentus. BMC Bioinformatics 21:408. 10.1186/s12859-020-03687-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Laub MT, Shapiro L, McAdams HH. 2007. Systems biology of Caulobacter. Annu Rev Genet 41:429–441. 10.1146/annurev.genet.41.110306.130346. [DOI] [PubMed] [Google Scholar]
- 102.Quon KC, Yang B, Domian IJ, Shapiro L, Marczynski GT. 1998. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci USA 95:120–125. 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ozaki S. 2019. Regulation of replication initiation: lessons from Caulobacter crescentus. Genes Genet Syst 94:183–196. 10.1266/ggs.19-00011. [DOI] [PubMed] [Google Scholar]
- 104.Collier J, Shapiro L. 2009. Feedback control of DnaA-mediated replication initiation by replisome-associated HdaA protein in Caulobacter. J Bacteriol 191:5706–5716. 10.1128/JB.00525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Felletti M, Omnus DJ, Jonas K. 2019. Regulation of the replication initiator DnaA in Caulobacter crescentus. Biochim Biophys Acta Gene Regul Mech 1862:697–705. 10.1016/j.bbagrm.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 106.Jonas K, Liu J, Chien P, Laub MT. 2013. Proteotoxic stress induces a cell-cycle arrest by stimulating Lon to degrade the replication initiator DnaA. Cell 154:623–636. 10.1016/j.cell.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Leslie DJ, Heinen C, Schramm FD, Thüring M, Aakre CD, Murray SM, Laub MT, Jonas K. 2015. Nutritional control of DNA replication initiation through the proteolysis and regulated translation of DnaA. PLoS Genet 11:e1005342. 10.1371/journal.pgen.1005342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu J, Francis LI, Jonas K, Laub MT, Chien P. 2016. ClpAP is an auxiliary protease for DnaA degradation in Caulobacter crescentus. Mol Microbiol 102:1075–1085. 10.1111/mmi.13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Reisenauer A, Shapiro L. 2002. DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO J 21:4969–4977. 10.1093/emboj/cdf490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stephens CM, Zweiger G, Shapiro L. 1995. Coordinate cell cycle control of a Caulobacter DNA methyltransferase and the flagellar genetic hierarchy. J Bacteriol 177:1662–1669. 10.1128/jb.177.7.1662-1669.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ausmees N, Jacobs-Wagner C. 2003. Spatial and temporal control of differentiation and cell cycle progression in Caulobacter crescentus. Annu Rev Microbiol 57:225–247. 10.1146/annurev.micro.57.030502.091006. [DOI] [PubMed] [Google Scholar]
- 112.Sommer JM, Newton A. 1989. Turning off flagellum rotation requires the pleiotropic gene pleD: pleA, pleC, and pleD define two morphogenic pathways in Caulobacter crescentus. J Bacteriol 171:392–401. 10.1128/jb.171.1.392-401.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sommer JM, Newton A. 1991. Pseudoreversion analysis indicates a direct role of cell division genes in polar morphogenesis and differentiation in Caulobacter crescentus. Genetics 129:623–630. 10.1093/genetics/129.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Aldridge P, Paul R, Goymer P, Rainey P, Jenal U. 2003. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol Microbiol 47:1695–1708. 10.1046/j.1365-2958.2003.03401.x. [DOI] [PubMed] [Google Scholar]
- 115.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev 18:715–727. 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hecht GB, Newton A. 1995. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J Bacteriol 177:6223–6229. 10.1128/jb.177.21.6223-6229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Aldridge P, Jenal U. 1999. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol Microbiol 32:379–391. 10.1046/j.1365-2958.1999.01358.x. [DOI] [PubMed] [Google Scholar]
- 118.Jacobs C, Hung D, Shapiro L. 2001. Dynamic localization of a cytoplasmic signal transduction response regulator controls morphogenesis during the Caulobacter cell cycle. Proc Natl Acad Sci USA 98:4095–4100. 10.1073/pnas.051609998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lam H, Matroule J-Y, Jacobs-Wagner C. 2003. The asymmetric spatial distribution of bacterial signal transduction proteins coordinates cell cycle events. Dev Cell 5:149–159. 10.1016/s1534-5807(03)00191-6. [DOI] [PubMed] [Google Scholar]
- 120.Iniesta AA, Hillson NJ, Shapiro L. 2010. Cell pole-specific activation of a critical bacterial cell cycle kinase. Proc Natl Acad Sci USA 107:7012–7017. 10.1073/pnas.1001767107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tsokos CG, Perchuk BS, Laub MT. 2011. A dynamic complex of signaling proteins uses polar localization to regulate cell-fate asymmetry in Caulobacter crescentus. Dev Cell 20:329–341. 10.1016/j.devcel.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Childers WS, Xu Q, Mann TH, Mathews II, Blair JA, Deacon AM, Shapiro L. 2014. Cell fate regulation governed by a repurposed bacterial histidine kinase. PLoS Biol 12:e1001979. 10.1371/journal.pbio.1001979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Guzzo M, Sanderlin AG, Castro LK, Laub MT. 2021. Activation of a signaling pathway by the physical translocation of a chromosome. Dev Cell 56:2145–2159. 10.1016/j.devcel.2021.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Abel S, Bucher T, Nicollier M, Hug I, Kaever V, Abel Zur Wiesch P, Jenal U. 2013. Bi-modal distribution of the second messenger c-di-GMP controls cell fate and asymmetry during the Caulobacter cell cycle. PLoS Genet 9:e1003744. 10.1371/journal.pgen.1003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lori C, Ozaki S, Steiner S, Böhm R, Abel S, Dubey BN, Schirmer T, Hiller S, Jenal U. 2015. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature 523:236–239. 10.1038/nature14473. [DOI] [PubMed] [Google Scholar]
- 126.Mann TH, Childers WS, Blair JA, Eckart MR, Shapiro L. 2016. A cell cycle kinase with tandem sensory PAS domains integrates cell fate cues. Nat Commun 7:11454. 10.1038/ncomms11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jenal U, Fuchs T. 1998. An essential protease involved in bacterial cell-cycle control. EMBO J 17:5658–5669. 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. 2006. Bacterial birth scar proteins mark future flagellum assembly site. Cell 124:1025–1037. 10.1016/j.cell.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 129.Lam H, Schofield WB, Jacobs-Wagner C. 2006. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell 124:1011–1023. 10.1016/j.cell.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 130.Viollier PH, Sternheim N, Shapiro L. 2002. A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. EMBO J 21:4420–4428. 10.1093/emboj/cdf454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Boutte CC, Crosson S. 2013. Bacterial lifestyle shapes stringent response activation. Trends Microbiol 21:174–180. 10.1016/j.tim.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gonzalez D, Collier J. 2014. Effects of (p)ppGpp on the progression of the cell cycle of Caulobacter crescentus. J Bacteriol 196:2514–2525. 10.1128/JB.01575-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Woldemeskel SA, Daitch AK, Alvarez L, Panis G, Zeinert R, Gonzalez D, Smith E, Collier J, Chien P, Cava F, Viollier PH, Goley ED. 2020. The conserved transcriptional regulator CdnL is required for metabolic homeostasis and morphogenesis in Caulobacter. PLoS Genet 16:e1008591. 10.1371/journal.pgen.1008591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L. 2008. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134:945–955. 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. 2008. A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell 134:956–968. 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lasker K, Boeynaems S, Lam V, Scholl D, Stainton E, Briner A, Jacquemyn M, Daelemans D, Deniz A, Villa E, Holehouse AS, Gitler AD, Shapiro L. 2022. The material properties of a bacterial-derived biomolecular condensate tune biological function in natural and synthetic systems. Nat Commun 13:5643. 10.1038/s41467-022-33221-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Holmes JA, Follett SE, Wang H, Meadows CP, Varga K, Bowman GR. 2016. Caulobacter PopZ forms an intrinsically disordered hub in organizing bacterial cell poles. Proc Natl Acad Sci USA 113:12490–12495. 10.1073/pnas.1602380113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bowman GR, Comolli LR, Gaietta GM, Fero M, Hong S-H, Jones Y, Lee JH, Downing KH, Ellisman MH, McAdams HH, Shapiro L. 2010. Caulobacter PopZ forms a polar subdomain dictating sequential changes in pole composition and function. Mol Microbiol 76:173–189. 10.1111/j.1365-2958.2010.07088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lasker K, von Diezmann L, Zhou X, Ahrens DG, Mann TH, Moerner WE, Shapiro L. 2020. Selective sequestration of signalling proteins in a membraneless organelle reinforces the spatial regulation of asymmetry in Caulobacter crescentus. Nat Microbiol 5:418–429. 10.1038/s41564-019-0647-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mohl DA, Gober JW. 1997. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell 88:675–684. 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- 141.Easter J, Gober JW. 2002. ParB-stimulated nucleotide exchange regulates a switch in functionally distinct ParA activities. Mol Cell 10:427–434. 10.1016/s1097-2765(02)00594-4. [DOI] [PubMed] [Google Scholar]
- 142.Schofield WB, Lim HC, Jacobs-Wagner C. 2010. Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. EMBO J 29:3068–3081. 10.1038/emboj.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Surovtsev IV, Lim HC, Jacobs-Wagner C. 2016. The slow mobility of the ParA partitioning protein underlies its steady-state patterning in Caulobacter. Biophys J 110:2790–2799. 10.1016/j.bpj.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Toro E, Hong S-H, McAdams HH, Shapiro L. 2008. Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc Natl Acad Sci USA 105:15435–15440. 10.1073/pnas.0807448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lim HC, Surovtsev IV, Beltran BG, Huang F, Bewersdorf J, Jacobs-Wagner C. 2014. Evidence for a DNA-relay mechanism in ParABS-mediated chromosome segregation. eLife 3:e02758. 10.7554/eLife.02758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Livny J, Yamaichi Y, Waldor MK. 2007. Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. J Bacteriol 189:8693–8703. 10.1128/JB.01239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bi E, Lutkenhaus J. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161–164. 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 148.Quardokus E, Din N, Brun YV. 1996. Cell cycle regulation and cell type-specific localization of the FtsZ division initiation protein in Caulobacter. Proc Natl Acad Sci USA 93:6314–6319. 10.1073/pnas.93.13.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Barrows JM, Goley ED. 2021. FtsZ dynamics in bacterial division: what, how, and why? Curr Opin Cell Biol 68:163–172. 10.1016/j.ceb.2020.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mahone CR, Goley ED. 2020. Bacterial cell division at a glance. J Cell Sci 133:jcs237057. 10.1242/jcs.237057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Thanbichler M, Shapiro L. 2006. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126:147–162. 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 152.Kelly AJ, Sackett MJ, Din N, Quardokus E, Brun YV. 1998. Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev 12:880–893. 10.1101/gad.12.6.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bhat NH, Vass RH, Stoddard PR, Shin DK, Chien P. 2013. Identification of ClpP substrates in Caulobacter crescentus reveals a role for regulated proteolysis in bacterial development. Mol Microbiol 88:1083–1092. 10.1111/mmi.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Williams B, Bhat N, Chien P, Shapiro L. 2014. ClpXP and ClpAP proteolytic activity on divisome substrates is differentially regulated following the Caulobacter asymmetric cell division. Mol Microbiol 93:853–866. 10.1111/mmi.12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang SCE, West L, Shapiro L. 2006. The bifunctional FtsK protein mediates chromosome partitioning and cell division in Caulobacter. J Bacteriol 188:1497–1508. 10.1128/JB.188.4.1497-1508.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mahone CR, Yang X, McCausland JW, Payne IP, Xiao J, Goley ED. 2022. Integration of cell wall synthesis activation and chromosome segregation during cell division in Caulobacter. bioRxiv. 10.1101/2022.11.05.515301. [DOI] [PMC free article] [PubMed]
- 157.Goley ED, Yeh Y-C, Hong S-H, Fero MJ, Abeliuk E, McAdams HH, Shapiro L. 2011. Assembly of the Caulobacter cell division machine. Mol Microbiol 80:1680–1698. 10.1111/j.1365-2958.2011.07677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Woldemeskel SA, Goley ED. 2017. Shapeshifting to survive: shape determination and regulation in Caulobacter crescentus. Trends Microbiol 25:673–687. 10.1016/j.tim.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Figge RM, Divakaruni AV, Gober JW. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol 51:1321–1332. 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- 160.Gitai Z, Dye N, Shapiro L. 2004. An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci USA 101:8643–8648. 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Daitch AK, Goley ED. 2020. Uncovering unappreciated activities and niche functions of bacterial cell wall enzymes. Curr Biol 30:R1170–R1175. 10.1016/j.cub.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ausmees N, Kuhn JR, Jacobs-Wagner C. 2003. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell 115:705–713. 10.1016/s0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- 163.Schuech R, Hoehfurtner T, Smith DJ, Humphries S. 2019. Motile curved bacteria are pareto-optimal. Proc Natl Acad Sci USA 116:14440–14447. 10.1073/pnas.1818997116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fernandez NL, Hsueh BY, Nhu NTQ, Franklin JL, Dufour YS, Waters CM. 2020. Vibrio cholerae adapts to sessile and motile lifestyles by cyclic di-GMP regulation of cell shape. Proc Natl Acad Sci USA 117:29046–29054. 10.1073/pnas.2010199117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Li G, Tang JX. 2006. Low flagellar motor torque and high swimming efficiency of Caulobacter crescentus swarmer cells. Biophys J 91:2726–2734. 10.1529/biophysj.106.080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liu B, Gulino M, Morse M, Tang JX, Powers TR, Breuer KS. 2014. Helical motion of the cell body enhances Caulobacter crescentus motility. Proc Natl Acad Sci USA 111:11252–11256. 10.1073/pnas.1407636111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Persat A, Stone HA, Gitai Z. 2014. The curved shape of Caulobacter crescentus enhances surface colonization in flow. Nat Commun 5:3824. 10.1038/ncomms4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bartlett TM, Bratton BP, Duvshani A, Miguel A, Sheng Y, Martin NR, Nguyen JP, Persat A, Desmarais SM, VanNieuwenhze MS, Huang KC, Zhu J, Shaevitz JW, Gitai Z. 2017. A periplasmic polymer curves Vibrio cholerae and promotes pathogenesis. Cell 168:172–185. 10.1016/j.cell.2016.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cho H. 2015. The role of cytoskeletal elements in shaping bacterial cells. J Microbiol Biotechnol 25:307–316. 10.4014/jmb.1409.09047. [DOI] [PubMed] [Google Scholar]
- 170.Hughes HV, Huitema E, Pritchard S, Keiler KC, Brun YV, Viollier PH. 2010. Protein localization and dynamics within a bacterial organelle. Proc Natl Acad Sci USA 107:5599–5604. 10.1073/pnas.0909119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kühn J, Briegel A, Mörschel E, Kahnt J, Leser K, Wick S, Jensen GJ, Thanbichler M. 2010. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO J 29:327–339. 10.1038/emboj.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wagner JK, Galvani CD, Brun YV. 2005. Caulobacter crescentus requires RodA and MreB for stalk synthesis and prevention of ectopic pole formation. J Bacteriol 187:544–553. 10.1128/JB.187.2.544-553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Billini M, Biboy J, Kühn J, Vollmer W, Thanbichler M. 2019. A specialized MreB-dependent cell wall biosynthetic complex mediates the formation of stalk-specific peptidoglycan in Caulobacter crescentus. PLoS Genet 15:e1007897. 10.1371/journal.pgen.1007897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kysela DT, Randich AM, Caccamo PD, Brun YV. 2016. Diversity takes shape: understanding the mechanistic and adaptive basis of bacterial morphology. PLoS Biol 14:e1002565. 10.1371/journal.pbio.1002565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Curtis PD. 2017. Stalk formation of Brevundimonas and how it compares to Caulobacter crescentus. PLoS One 12:e0184063. 10.1371/journal.pone.0184063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hentchel KL, Reyes Ruiz LM, Curtis PD, Fiebig A, Coleman ML, Crosson S. 2019. Genome-scale fitness profile of Caulobacter crescentus grown in natural freshwater. ISME J 13:523–536. 10.1038/s41396-018-0295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Heinrich K, Leslie DJ, Morlock M, Bertilsson S, Jonas K. 2019. Molecular basis and ecological relevance of Caulobacter cell filamentation in freshwater habitats. mBio 10:e01557-19. 10.1128/mBio.01557-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wilhelm RC. 2018. Following the terrestrial tracks of Caulobacter—redefining the ecology of a reputed aquatic oligotroph. ISME J 12:3025–3037. 10.1038/s41396-018-0257-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ely B, Berrios L, Thomas Q. 2022. S2B, a temperate bacteriophage that infects Caulobacter crescentus strain CB15. Curr Microbiol 79:98. 10.1007/s00284-022-02799-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Berrios L, Ely B. 2019. The isolation and characterization of Kronos, a novel Caulobacter rhizosphere phage that is similar to lambdoid phages. Curr Microbiol 76:558–565. 10.1007/s00284-019-01656-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Jiang C, Brown PJB, Ducret A, Brun YV. 2014. Sequential evolution of bacterial morphology by co-option of a developmental regulator. Nature 506:489–493. 10.1038/nature12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lim HC, Bernhardt TG. 2019. A PopZ-linked apical recruitment assay for studying protein-protein interactions in the bacterial cell envelope. Mol Microbiol 112:1757–1768. 10.1111/mmi.14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tsang PH, Li G, Brun YV, Freund LB, Tang JX. 2006. Adhesion of single bacterial cells in the micronewton range. Proc Natl Acad Sci USA 103:5764–5768. 10.1073/pnas.0601705103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chepkwony NK, Berne C, Brun YV. 2019. Comparative analysis of ionic strength tolerance between freshwater and marine Caulobacterales adhesins. J Bacteriol 201:e00061-19. 10.1128/JB.00061-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Chepkwony NK, Brun YV. 2021. A polysaccharide deacetylase enhances bacterial adhesion in high-ionic-strength environments. iScience 24:103071. 10.1016/j.isci.2021.103071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nyarko A, Barton H, Dhinojwala A. 2016. Scaling down for a broader understanding of underwater adhesives—a case for the Caulobacter crescentus holdfast. Soft Matter 12:9132–9141. 10.1039/c6sm02163h. [DOI] [PubMed] [Google Scholar]
- 187.Bingle WH, Nomellini JF, Smit J. 2000. Secretion of the Caulobacter crescentus S-layer protein: further localization of the C-terminal secretion signal and its use for secretion of recombinant proteins. J Bacteriol 182:3298–3301. 10.1128/JB.182.11.3298-3301.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Herdman M, von Kügelgen A, Kureisaite-Ciziene D, Duman R, el Omari K, Garman EF, Kjaer A, Kolokouris D, Löwe J, Wagner A, Stansfeld PJ, Bharat TAM. 2022. High-resolution mapping of metal ions reveals principles of surface layer assembly in Caulobacter crescentus cells. Structure 30:215–228. 10.1016/j.str.2021.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Smit J, Sherwood CS, Turner RF. 2000. Characterization of high density monolayers of the biofilm bacterium Caulobacter crescentus: evaluating prospects for developing immobilized cell bioreactors. Can J Microbiol 46:339–349. 10.1139/w99-145. [DOI] [PubMed] [Google Scholar]
- 190.Charrier M, Li D, Mann VR, Yun L, Jani S, Rad B, Cohen BE, Ashby PD, Ryan KR, Ajo-Franklin CM. 2019. Engineering the S-layer of Caulobacter crescentus as a foundation for stable, high-density, 2D living materials. ACS Synth Biol 8:181–190. 10.1021/acssynbio.8b00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Molinari S, Tesoriero RF, Li D, Sridhar S, Cai R, Soman J, Ryan KR, Ashby PD, Ajo-Franklin CM. 2022. A de novo matrix for macroscopic living materials from bacteria. Nat Commun 13:5544. 10.1038/s41467-022-33191-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Govers SK, Jacobs-Wagner C. 2020. Caulobacter crescentus: model system extraordinaire. Curr Biol 30:R1151–R1158. 10.1016/j.cub.2020.07.033. [DOI] [PubMed] [Google Scholar]


