ABSTRACT
Gardnerella spp. are associated with bacterial vaginosis in which normally dominant lactobacilli are replaced with facultative and anaerobic bacteria, including Gardnerella spp. Co-occurrence of multiple species of Gardnerella is common in the vagina, and competition for nutrients such as glycogen likely contributes to the differential abundances of Gardnerella spp. Glycogen must be digested into smaller components for uptake, a process that depends on the combined action of glycogen-degrading enzymes. In this study, the ability of culture supernatants of 15 isolates of Gardnerella spp. to produce glucose, maltose, maltotriose, and maltotetraose from glycogen was demonstrated. Carbohydrate-active enzymes (CAZymes) were identified bioinformatically in Gardnerella proteomes using dbCAN2. Identified proteins included a single-domain α-amylase (EC 3.2.1.1) (encoded by all 15 isolates) and an α-amylase-pullulanase (EC 3.2.1.41) containing amylase, carbohydrate binding modules, and pullulanase domains (14/15 isolates). To verify the sequence-based functional predictions, the amylase and pullulanase domains of the α-amylase-pullulanase and the single-domain α-amylase were each produced in Escherichia coli. The α-amylase domain from the α-amylase-pullulanase released maltose, maltotriose, and maltotetraose from glycogen, and the pullulanase domain released maltotriose from pullulan and maltose from glycogen, demonstrating that the Gardnerella α-amylase-pullulanase is capable of hydrolyzing α-1,4 and α-1,6 glycosidic bonds. Similarly, the single-domain α-amylase protein also produced maltose, maltotriose, and maltotetraose from glycogen. Our findings show that Gardnerella spp. produce extracellular amylase enzymes as “public goods” that can digest glycogen into maltose, maltotriose, and maltotetraose that can be used by the vaginal microbiota.
IMPORTANCE Increased abundance of Gardnerella spp. is a diagnostic characteristic of bacterial vaginosis, an imbalance in the human vaginal microbiome associated with troubling symptoms, and negative reproductive health outcomes, including increased transmission of sexually transmitted infections and preterm birth. Competition for nutrients is likely an important factor in causing dramatic shifts in the vaginal microbial community, but little is known about the contribution of bacterial enzymes to the metabolism of glycogen, a major food source available to vaginal bacteria. The significance of our research is characterizing the activity of enzymes conserved in Gardnerella species that contribute to the ability of these bacteria to utilize glycogen.
KEYWORDS: Gardnerella, amylase, glycogen, human microbiome, pullulanase, vagina, vaginosis
INTRODUCTION
Gardnerella spp. are associated with bacterial vaginosis (BV), a condition characterized by the replacement of the usually dominant Lactobacillus community with a mixture of facultative and anaerobic bacteria, including Gardnerella spp. (1). BV is characterized by thin, white to grayish discharge, and unpleased “fishy” vaginal odor; however, a large majority of women diagnosed with BV do not have symptoms (2). Phenotypic diversity among Gardnerella spp. may explain why they are found in women presenting clinical signs of bacterial vaginosis and in asymptomatic or BV-negative women (3). The genus Gardnerella has been classified into four different species and at least 13 “genome species” (4). Co-occurrence of multiple Gardnerella species in the vaginal environment is common, and different species are dominant in different women (5). Given the potentially different roles of Gardnerella species in the vaginal microbiome, understanding the factors contributing to their population structure is important. Previous work has shown that interactions in cocultures of Gardnerella spp. are typical of a “scramble” competition (6). In scramble competition, one competitor outgrows others through its superior ability to use shared resources such as nutrients (7). Competition for nutrients could be an important factor contributing to the differential abundances of Gardnerella spp. in the vaginal microbiome.
Glycogen is an abundant carbon and energy source available to the vaginal microbiota, including Gardnerella. Glycogen is a glucose homopolysaccharide comprising linear α-1,4-linked glucose with α-1,6 branch points occurring approximately at every 8 to 13 residues to form a highly branched and complex structure. The sizes of glycogen molecules and branching patterns can vary based on the glycogen source (8). In the vagina, glycogen accumulates inside vaginal epithelial cells under the influence of estrogen (9) and is released into the vaginal lumen through the activity of bacterial cytolysins and/or lactic acid-mediated acidosis (10). The amount of free glycogen in vaginal fluid can range from undetectable to 32 μg/mL (11).
Glycogen must be digested into smaller products for uptake into bacterial cells. This complex and multistep process requires the combined action of enzymes collectively known as glycogen-degrading enzymes. The majority of glycogen-degrading bacterial amylases belong to the glycosyl hydrolase (GH) class within family 13 in the classification of carbohydrate-active enzymes (CAZymes). GH13 enzymes are further classified into more than 40 subfamilies based on their structure and substrate specificity (12). Previous studies have shown the presence and activity of human and/or bacterial amylases in vaginal fluid (13, 14), but the specific contribution of Gardnerella spp. to glycogen digestion is not clear.
This study aimed to assess the glycogen digestion ability of Gardnerella species isolated from the vaginal microbiome and to determine the distribution of extracellular glycogen-degrading enzymes among different species. In addition, we aimed to biochemically characterize the activities of the predicted functional domains of extracellular glycogen-degrading amylase enzymes identified. Our findings showed that Gardnerella spp. have α-amylase and α-amylase-pullulanase enzymes, which can digest glycogen into maltose and other oligosaccharides.
RESULTS
Amylase activity of Gardnerella culture supernatants.
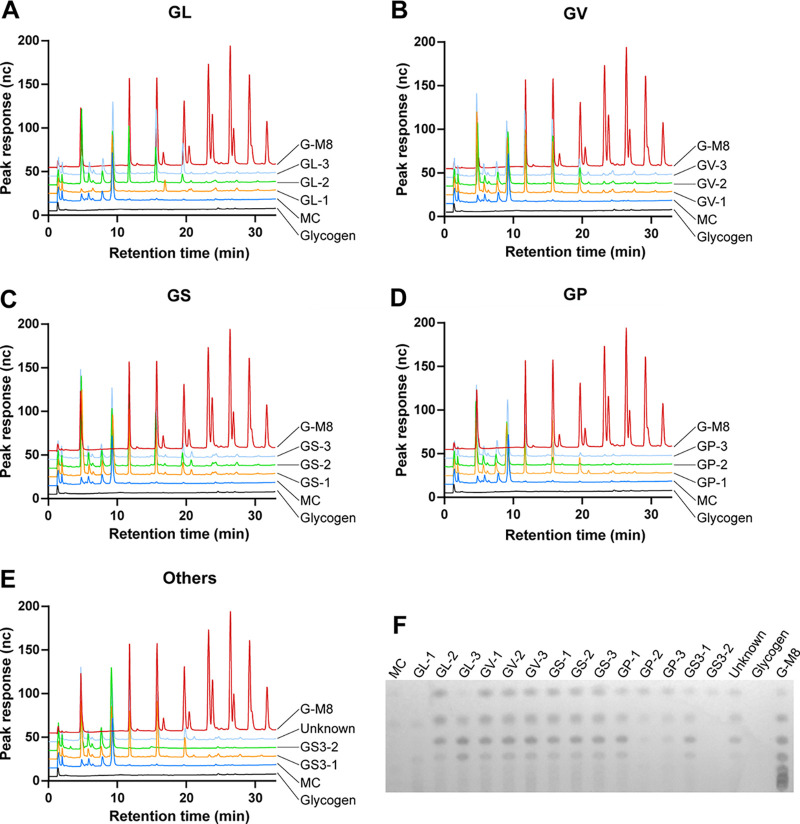
No growth was observed from filtered culture supernatants, confirming their sterility. Supernatants of all 15 isolates showed amylase activity in the agar plate assay (Fig. 1). Cell-free culture supernatants of 14 isolates produced glucose, maltose, maltotriose, and maltotetraose from glycogen, while glucose was the only product detected from N170 supernatant (Gardnerella genome sp. 3, GS3-2) (Fig. 2).
FIG 1.
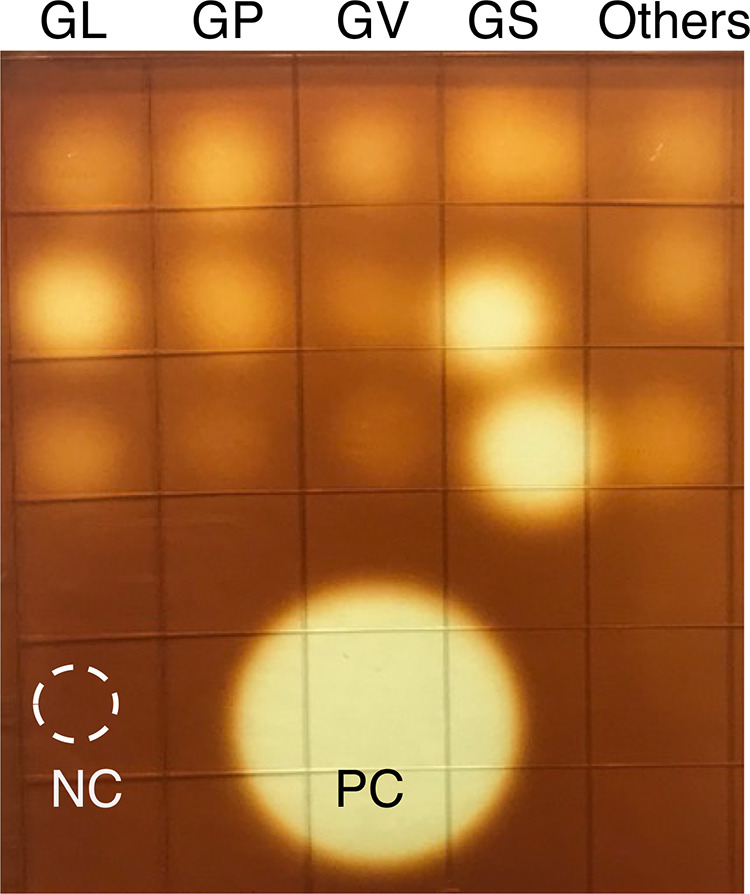
Amylase activity of cell-free culture supernatants on glycogen agar. Each spot represents 1 isolate, and a total of 15 different isolates, 3 each from G. leopoldii (GL), G. piotii (GP), G. vaginalis (GV), G. swidsinskii (GS), and others (2 isolates from Gardnerella genome sp. 3 [top and middle] and 1 isolate from unknown genome species [bottom] [corresponds to the subgroup D based on the cpn60 classification system]) are shown. NC, mNYC, PC, B. licheniformis amylase (0.05 mg/mL).
FIG 2.
Identification of glycogen breakdown products produced after 24 h by culture supernatants of 15 Gardnerella isolates by HPAEC-PAD (A to E) and TLC (F). GL, G. leopoldii; GV, G. vaginalis; GS, G. swidsinskii; GP, G. piotii; and others (2 isolates from Gardnerella genome sp. 3 [top and middle] and 1 isolate from unknown genome species [bottom] [corresponds to the subgroup D based on the cpn60 classification system]); MC, media control; G-M8, glucose-to-malto-octaose standards.
Identification of genes encoding carbohydrate-active enzymes in Gardnerella genomes.
Out of 20,017 predicted protein sequences from 15 isolates uploaded into the dbCAN2 webserver, 886 sequences had at least one carbohydrate-active domain. Results were filtered to include only glycosyl hydrolase (GH) domains associated with a signal peptide. Only 52/836 sequences identified as belonging to the GH class had an associated signal peptide. Results were further filtered to identify predicted extracellular glycogen-degrading enzymes (protein containing at least one GH13 domain associated with signal peptide), resulting in identification of 29 sequences.
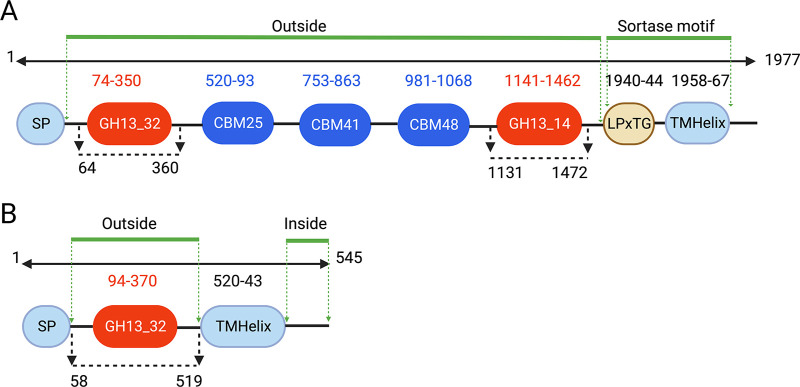
A multidomain enzyme containing an N-terminal α-amylase domain (GH13_32), followed by 2 to 4 carbohydrate binding modules (CBM; types CBM25, CBM41, or CBM48) and a C-terminal pullulanase domain (GH13_14), was identified in all isolates except N170. This enzyme also included an N-terminal signal peptide domain and a potential sortase sorting signal that included an LPxTG motif (Fig. 3A). This multidomain enzyme is traditionally referred to as type II amylopullulanase; however, recently, it has been suggested that these enzymes are more appropriately classified as type II α-amylase-pullulanase (15) and so, the term “α-amylase-pullulanase” is used throughout this paper. The domain organization of Gardnerella leopoldii NR017 α-amylase-pullulanase (GenPept accession number RFT33565) is shown in Fig. 3A. Similarly, a single α-amylase (GH13_32) domain containing an α-amylase enzyme with an N-terminal signal peptide and a C-terminal transmembrane domain was identified in all 15 isolates (GenPept accession number RFT33564 for NR017) (Fig. 3B).
FIG 3.
Predicted domains of G. leopoldii NR017 α-amylase-pullulanase (1,977 amino acids) (A) and α-amylase (545 amino acids) (B). Amylase and pullulanase domains belonging to GH13 subfamilies 32 and 14 are indicated in red, while three carbohydrate binding modules (CBM) are in blue. SP, signal peptide; sorting signal, LPxTG motif and a hydrophobic transmembrane helix domain (TMHelix) domain. Numbering indicates amino acid positions. The regions used for recombinant protein expression are indicated with black broken line with arrows. Predicted extracellular and intracellular regions are indicated with green lines.
Phylogenetic analysis of GH13_32 amylase domains and relationship with other amylases.
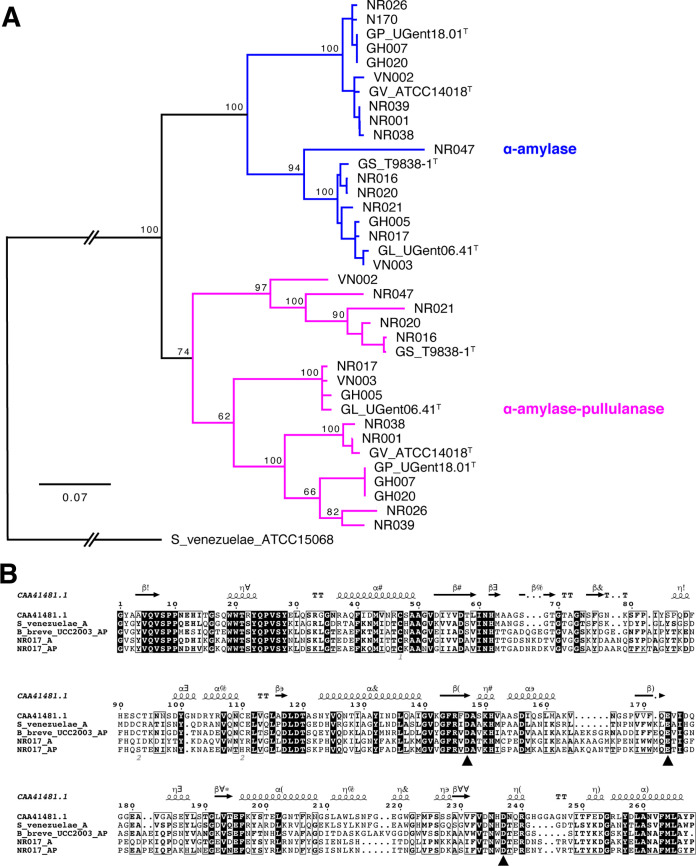
GH13_32 amylase domain sequences from the single-domain α-amylase and α-amylase-pullulanase from all study isolates formed two distinct clusters (Fig. 4A). Intracluster amino acid sequence identity of amylase domains within single-domain α-amylase and α-amylase-pullulanase enzymes was 75 to 100% and 66 to 100%, respectively, whereas intercluster identity was 64 to 74%.
FIG 4.
(A) Phylogenetic tree of all α-amylase domains from 15 Gardnerella isolates and 4 reference strains (indicated with T after the strain name). The tree is rooted with Streptomyces venezuelae ATCC 15068. Trees are consensus trees of 100 bootstrap iterations and constructed using the neighbor-joining method using the Tamura-Nei distance model. The number at major branch points represents the percentage of bootstrap support. (B) Sequence alignment of GH13_32 catalytic domains of G. leopoldii NR017 α-amylase (NR017_A) and α-amylase-pullulanase (NR017_AP) with other functionally characterized GH13_32 members (α-amylases from Pseudoalteromonas haloplanktis A23 [GenPept accession number CAA41481] and S. venezuelae ATCC 15068 and amylopullulanase from Bifidobacterium breve UCC2003). Black triangles indicate conserved catalytic residues.
GH13 enzymes have a conserved catalytic triad of two aspartate residues (which act as a nucleophile and stabilize the transition state) and a glutamate residue (general acid-base) (16). In silico analysis of GH13_32 amylase domains of G. leopoldii NR017 with GH13_32 amylase domains of functionally characterized α-amylase and α-amylase-pullulanase revealed conserved catalytic residues (Fig. 4B). The catalytic triad was conserved across all GH13_32 amylase domains from 15 Gardnerella isolates (see Fig. S1 in the supplemental material).
Protein expression and purification.
The entire open reading frame of G. leopoldii NR017 α-amylase-pullulanase (GH13_32 and GH13_14) comprises 5,933 bp encoding a 1,977-amino-acid protein with a predicted mass of 215.3 kDa (Fig. 3A). The gene segments encoding the α-amylase (amino acids 64 to 360) and pullulanase domains (amino acids 1131 to 1472) were each PCR amplified and ligated into pQE-80L vector to express as N-terminal 6×His-tagged proteins in Escherichia coli (Fig. 5A, left). Expressed proteins were solubilized using 1% SDS and purified using nickel-nitriloacetic acid (Ni-NTA) columns (Fig. S2). Apparent molecular weights of the 6×His-tagged amylase and pullulanase domains were 35.7 and 40.5 kDa, respectively (Fig. 5B, left and middle).
FIG 5.
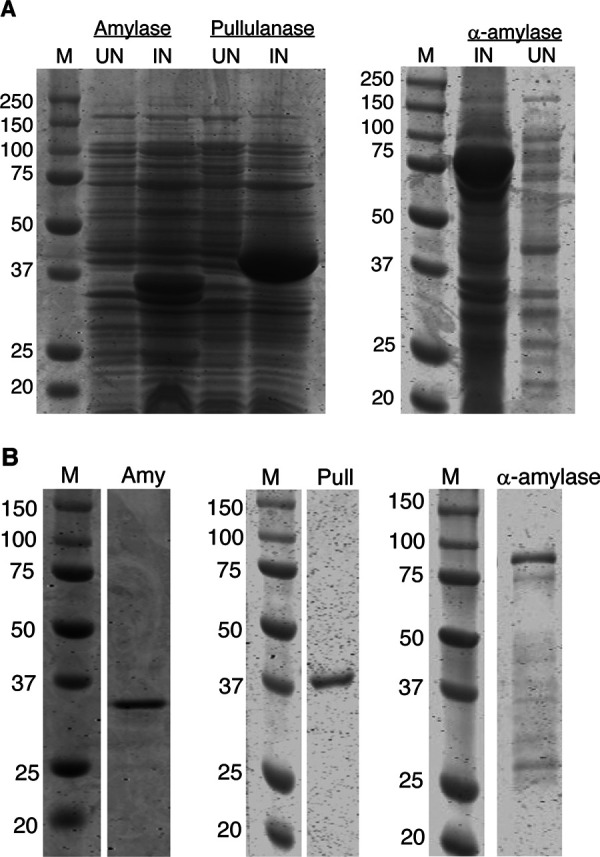
(A) Protein profiles of uninduced (UN) and induced (IN) E. coli cells containing pQE80L-AMY and pQE80L-PULL (top left) and pET41a-CO-AMY (top right) recombinant plasmids. (B) Purified 6×His-tagged amylase (Amy) (bottom left) and pullulanase (Pull) (bottom middle) and GST-6×His-tagged α-amylase (bottom right). M, molecular weight markers.
The G. leopoldii NR017 α-amylase enzyme (1,635 bp encoding a 545-amino-acid protein with a predicted mass of 60.5 kDa) has a predicted N-terminal signal peptide (amino acids 1 to 57) and C-terminal transmembrane domain (amino acids 520 to 545) (Fig. 3B). A codon-optimized version of the truncated α-amylase gene sequence encoding amino acids 58 to 519 was expressed as a glutathione S-transferase (GST) fusion protein with an apparent molecular weight of 79.9 kDa in E. coli BL21(DE3) cells (Fig. 5A, right). The recombinant protein was solubilized using 1% SDS. Attempts to purify this GST fusion protein using glutathione beads resulted in a very low recovery of target protein due to poor binding to the beads (Fig. S3). The pET41a-amylase construct includes an N-terminal hexa-histidine tag between the GST tag and the amylase sequence. Purification of the fusion protein using a Ni-NTA column resulted in partial protein purification (Fig. 5B, right). Additional lower-molecular-weight peptides observed in the column eluate were identified as degraded peptide fragments of the fusion protein by Western blotting with anti-His antibodies (Fig. S4).
Activity of the purified proteins.
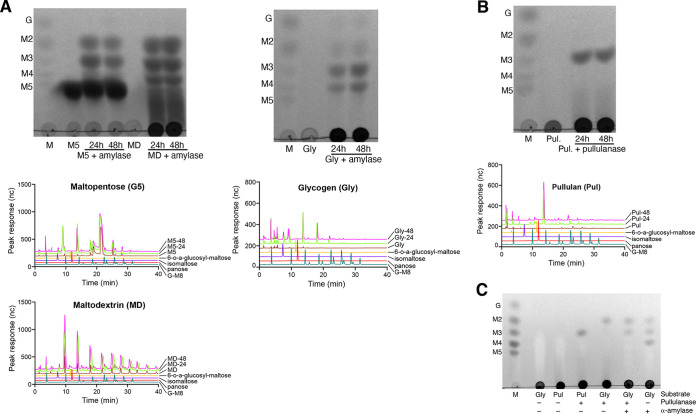
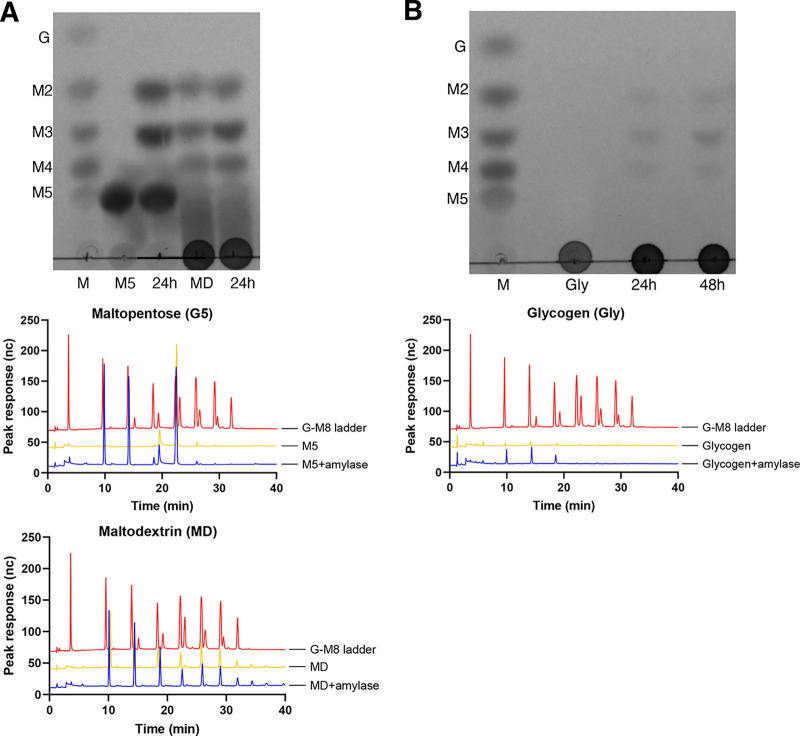
The α-amylase domain from the α-amylase-pullulanase released maltose and maltotriose from maltopentose, while maltose, maltotriose, and maltotetraose were identified as the major products from maltodextrin (dextrose equivalent 13 to 17) and glycogen (Fig. 6A). A small peak corresponding to isomaltose was observed in the high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) results from 24- and 48-h digests of glycogen with α-amylase domain (Fig. 6A, middle right). The pullulanase domain produced maltotriose as the only product from pullulan (Fig. 6B), while maltose was produced from glycogen (Fig. 6C). Combined activities of the α-amylase and pullulanase domains on glycogen released maltose and maltotriose (Fig. 6C) with no detectable maltotetraose. This suggests that the pullulanase domain can hydrolyze maltotetraose to release maltose but is less active (or inactive) on the smaller maltotriose substrate, perhaps because it is a poorer fit for the active site. To test this, the pullulanase domain was incubated with maltotriose, maltotetraose, and maltopentose. The results showed that pullulanase hydrolyzed maltopentose (into maltose and maltotriose) and, to a lesser extent, maltotetraose (into maltose) and maltotriose (into glucose and maltose) (Fig. S6). Overall, our results show that α-amylase and pullulanase domains from α-amylase-pullulanase can hydrolyze α-1,4, and α-1,4 and α-1,6 glycosidic bonds in glycogen, respectively.
FIG 6.
(A) Identification of products released by α-amylase domain (GH13_32) of α-amylase-pullulanase from maltopentose (M5) and maltodextrin (MD) and glycogen (Gly) after 24 and 48 h of incubation using TLC (top) and HPAEC-PAD (middle and bottom). (B) Products released from pullulan by the pullulanase domain (GH13_14) after 24 and 48 h of incubation were identified using TLC (top) and HPAEC-PAD (bottom). (C) Identification of products released after 24 h from pullulan or glycogen by pullulanase, α-amylase, or a mixture of α-amylase and pullulanase domains. Standards in lane M of each panel as follows: G, glucose; M2, maltose; M3, maltotriose; M4, maltotetraose; and M5, maltopentose.
Since the α-amylase domain from the single-domain α-amylase enzyme was expressed as a GST-6×His fusion, we initially confirmed that no amylase activity was detected from the GST-6×His tag (Fig. S5). Maltose and maltotriose were produced when maltopentose was incubated with single-domain α-amylase (Fig. 7A, top and middle). Similarly, maltose, maltotriose, and maltotetraose were identified as the major products in the reaction mixture of glycogen with single-domain α-amylase (Fig. 7B). As expected, the maltodextrin substrate included maltose, maltotriose, and maltotetraose; however, an increase in intensity of maltose, maltotriose, and maltotetraose spots was observed in thin-layer chromatography (TLC) results in the reaction mixture of single-domain α-amylase with maltodextrin compared to a substrate-only control (Fig. 7A, top). HPAEC-PAD analysis of products from maltodextrin also showed larger peaks of maltose, maltotriose, and maltotetraose relative to the control (Fig. 7A, bottom).
FIG 7.
(A) Identification of products released by the purified single-domain α-amylase enzyme from maltopentose (M5) and maltodextrin (MD) using TLC (top) and HPAEC-PAD (bottom) after 24 h of incubation. (B) Products released from glycogen by the purified single-domain α-amylase enzyme after 24 h of incubation were identified using TLC (top) and HPAEC-PAD (bottom). Standards in lane M of each panel as follows: G, glucose; M2, maltose; M3, maltotriose; M4, maltotetraose; and M5, maltopentose.
DISCUSSION
Dysbiosis in the vaginal microbiome has significant consequences that can affect the quality of life and reproductive health of women. There are several hypotheses about how dysbiosis is initiated and maintained (17, 18), but the ecological factors determining microbial population dynamics in the vaginal microbiome are not well understood. In addition to direct antagonistic (19) or synergistic (20) interactions among microbiota, changes in environmental conditions due to pregnancy status, hormone levels, infection, sexual behavior, and hygiene practices certainly play a role in determining the fitness of vaginal microbiota and thus microbial community composition. Gardnerella spp. have been implicated in the initial stages of transition to bacterial vaginosis (18), and so, factors affecting their fitness, including their interaction with vaginal glycogen, are of particular interest.
Glycogen breakdown is a complex and multistep process that requires the collective action of several glycogen-degrading enzymes. Vaginal secretions contain host-derived pancreatic α-amylase and α-glucosidases that can digest glycogen into smaller sugars (13). Until recently, the contribution of bacterial enzymes in this process was unclear. Recently, van der Veer et al. showed that vaginal isolates of Lactobacillus crispatus are capable of growing in glycogen-supplemented medium and encode type I pullulanase in their genomes (21). This study further reported that strains showing less efficient or no growth on glycogen have an N-terminal deletion mutation in their putative type I pullulanase gene, suggesting its role in extracellular glycogen digestion. Forney et al. identified eight putative amylases from vaginal metagenomes of Bifidobacterium vaginale, Bifidobacterium lacrimalis, Lactobacillus gasseri, Lactobacillus crispatus, Lactobacillus iners, and Lactobacillus jensenii and further reported that multiple microbial amylases are present in vaginal fluid (14). Similarly, Woolston et al. identified putative glycogen-degrading enzymes in genomes of vaginal bacteria, including Gardnerella spp., and biochemically characterized the glycogen degradation ability of six GH13 hydrolase enzymes (22) identified in those species. Together, these findings suggest that bacterial enzymes contribute to the breakdown of vaginal glycogen.
The genus Gardnerella was a monospecific genus until recently when it was classified into four species and additional genome species that have not yet been named due to insufficient evidence (4). The role of Gardnerella spp. in extracellular glycogen digestion and whether the species in this genus differ in digesting glycogen is not clear. Although earlier studies reported that Gardnerella isolates can utilize glycogen (23–25), it is not clear if all newly defined species are capable of digesting extracellular glycogen. In this study, we used cell-free culture supernatants from genetically characterized isolates to test the extracellular glycogen breakdown ability of different Gardnerella spp.
Selection of an appropriate culture medium to make culture supernatants is a challenge. As our main interest was to identify the sugars released from glycogen, differentiating the background sugars that are already present in media from those obtained from glycogen hydrolysis is crucial. Gardnerella is typically cultured in brain heart infusion (BHI) broth or NYCIII (NYCIII medium contains [per L] 4 g HEPES, 15 g proteose peptone, 5 g NaCl, 3.75 g yeast extract, 5 g glucose, 100 mL heat inactivated horse serum), and both media contain glucose. In addition, the complement-inactivated horse serum (obtained by heat treatment at 56°C for 20 min) added to NYCIII can itself digest glycogen. It is reported that heat treatment at 70°C for 20 min inhibits the maltase and amylase activity of fetal bovine serum but not horse serum (26). For this reason, in our experiments, a modified NYCIII medium supplemented with heat-inactivated (70°C for 20 min) fetal bovine serum instead of horse serum and no added glucose was used to prepare culture supernatants. Cell-free culture supernatants of all isolates tested showed amylase activity in a glycogen agar plate assay, suggesting that glycogen degradation is a conserved property in the genus Gardnerella. Glucose, maltose, maltotriose, and maltotetraose were identified as the major products released from glycogen by culture supernatants of all Gardnerella isolates, except the N170 isolate, which produced only glucose. Consistent with this observation, α-amylase-pullulanase genes were detected in all isolates except N170. Overall, no species-specific patterns of glycogen breakdown products were found in Gardnerella spp.
Of the two extracellular glycosyl hydrolases identified by dbCAN2, the α-amylase-pullulanase was predicted to contain separate amylase and pullulanase catalytic domains, a conformation historically referred to as type II amylopullulanase (27). With the availability of more sequence data, these enzymes are now classified as type II α-amylase-pullulanase, and they differ from amylopullulanases, which have only one catalytic domain capable of hydrolyzing both α-1,4 and α-1,6 glycosidic bonds (15). The amylase and pullulanase domains of the identified Gardnerella α-amylase-pullulanase flanked 2 to 4 CBMs. CBMs appended to GH13 enzymes are important for binding to different α-glucans, such as amylose, amylopectin, glycogen, pullulan, and oligosaccharides, derived from these polysaccharides (28, 29). CBMs increase the concentration of enzyme on the substrate surface, and their removal can significantly reduce enzyme activity. It is reported that removal of CBMs reduced the activity of Eubacterium rectale α-amylase on starch by ~40-fold compared to the wild-type protein (30). Similarly, compared to a full-length enzyme, removal of CBM domains reduced the activity of B. adolescentis P2P3 type II pullulanase on glycogen (31). We did not investigate the roles of the CBMs in the α-amylase-pullulanase characterized in this study since our attempts to express full-length α-amylase-pullulanase were unsuccessful.
Recently, Woolston et al. (22) characterized a full-length amylopullulanase (2,013 amino acids) from a vaginal isolate of Gardnerella vaginalis and reported maltose and maltotriose as the major products from glycogen. In this report, the authors described the enzyme as containing “α-amylase domains,” rather than amylase and pullulanase catalytic domains as we describe here. By expressing the two catalytic domains separately, we have demonstrated that they have distinct activities, hydrolyzing α-1,4, and α-1,4 and α-1,6 bonds, respectively.
Pullulanase-degrading enzymes are classified into two major types, pullulanases (type I and type II) and pullulanase hydrolases (type I, II, and III) based on substrate specificities and reaction products. While type I pullulanases hydrolyze only α-1,6 glycosidic bonds, type II pullulanases are capable of hydrolyzing α-1,4 and α-1,6 glycosidic bonds. Amylopullulanases, a subtype of type II pullulanase, are mainly reported in thermophilic bacteria and have a single catalytic domain, whereas α-amylase-pullulanase has dual catalytic domains (α-amylase, for α-1,4 glycosidic bonds, and pullulanase, for α-1,6 glycosidic bonds) and are reported in mesophilic bacteria (15). α-Amylase-pullulanases have been characterized in Bifidobacterium breve UCC2003, Bifidobacterium adolescentis P2P3 (31, 32), and a few other species (15). Our results showed that the pullulanase domain of the G. leopoldii α-amylase-pullulanase hydrolyzes α-1,4 as well as α-1,6 glycosidic bonds, which, to our knowledge, has not been reported before. Kim et al. reported that the pullulanase domain of an α-amylase-pullulanase from B. adolescentis P2P3 is capable of hydrolyzing α-1,6 glycosidic bonds and releasing maltotriose from pullulan; however, no products were detected with amylose, indicating a lack of activity on α-1,4 linkages (31). The demonstrated activity of the G. leopoldii pullulanase domain on α-1,4, and α-1,6 glycosidic bonds would explain why maltose was the only detectable product released from glycogen (Fig. 6C) and suggests that this domain might better be described as an amylopullulanase. It is important to note, however, that the lack of CBMs associated with the domains we investigated in the current study could affect their activity and the range of products released from different substrates, and so, further study of the influence of CBMs is warranted.
Both α-amylase and α-amylase-pullulanase enzymes are active against oligosaccharides and glycogen. Although the overall sequence identity between the two amylase domains characterized in this study was only 64 to 74%, predicted catalytic residues were perfectly conserved (Fig. 4B). Either domain would thus be capable of acting on the malto-oligosaccharides produced by the debranching activity of the pullulanase domain to generate glucose, maltose, and other small oligosaccharides. This type of functional redundancy where two enzymes have similar biochemical activities is common in bacteria (33).
The Gardnerella α-amylase and α-amylase-pullulanase are predicted to be extracellular based on the presence of N-terminal signal peptides. The α-amylase contains a C-terminal transmembrane helix, suggesting that the protein is inserted in the cell membrane. In the α-amylase-pullulanase sequence, an LPxTG motif was identified immediately N-terminal to the predicted transmembrane helix (Fig. 3A), suggesting this protein is anchored to the cell wall through the sortase mechanism. The presence of these features suggests that both enzymes are tethered to the cell envelope. Cell envelope-anchored proteins can be released into the extracellular environment during bacterial growth as a by-product of cell wall turnover, cell death, or proteolytic cleavage (34), which is consistent with our detection of amylase activity in sterile culture supernatants. Some Gardnerella species also produce cell wall-anchored sialidases, NanH2 and NanH3, that are also released into the supernatant after cell death or by proteolytic cleavage (35).
Although glucose was identified as one of the major products in glycogen digests of Gardnerella culture supernatants, the purified α-amylase domain and single-domain α-amylase proteins did not produce glucose from glycogen. TLC and HPAEC-PAD analysis showed that glucose was neither produced by the activity of culture media itself nor present in glycogen substrate (Fig. 2F and G). Another possibility is that the activity of a full-length α-amylase-pullulanase protein on glycogen might generate different product profiles than the isolated domains. A full-length type II pullulanase enzyme (containing two separate α-amylase and pullulanase domains) from Bifidobacterium adolescentis P2P3 was reported to produce maltose only from glycogen, while the purified amylase domain protein from the same enzyme produced a mixture of glucose, maltose, and maltotriose (31). Glucose may also be produced by the activity of intracellular α-glucosidase on malto-oligosaccharides generated from glycogen hydrolysis that are released by dead/lysed bacteria in the culture. We have previously shown that Gardnerella spp. have conserved intracellular α-glucosidase that can release glucose from smaller oligosaccharides (36).
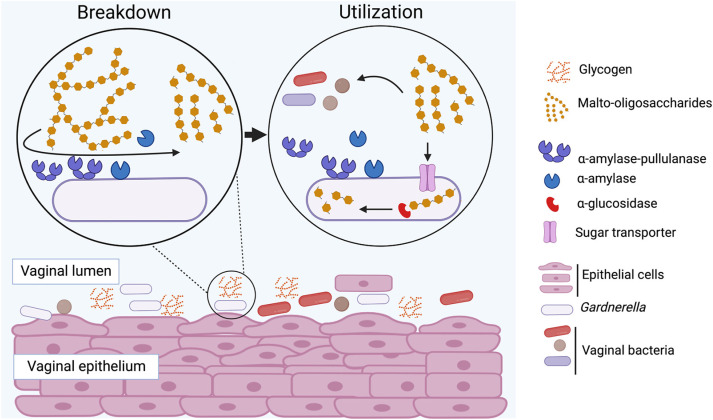
Our findings show that all Gardnerella species produce extracellular α-amylase and α-amylase-pullulanase enzymes that are “public goods” in the vaginal microbiome, breaking down glycogen to produce similar fingerprints of glycogen-derived products and contributing to the nutrient pool available to the vaginal microbiota (Fig. 8). Determining if these Gardnerella spp. differentially utilize and compete for these breakdown products will provide insights into whether competition for malto-oligosaccharides plays a role in determining Gardnerella community structure in the vaginal microbiome.
FIG 8.
Proposed mechanisms of extracellular glycogen utilization in Gardnerella spp. Glycogen is digested into malto-oligosaccharides by α-amylase-pullulanase and α-amylase, and these breakdown products are transported inside and can be further digested to glucose by other glycosyl hydrolases, including a previously characterized α-glucosidase enzyme (36). Extracellular glycogen breakdown products can also be utilized by other resident microbiota.
MATERIALS AND METHODS
Gardnerella isolates.
Isolates of Gardnerella used in this study were from a previously described culture collection (37) and included a total of 15 Gardnerella species isolates (3 representative strains each from G. leopoldii [GH005, VN003, and NR017], G. piotii [VN002, GH007, and GH020], G. vaginalis [NR038, NR001, and NR039], and G. swidsinskii [NR016, NR021, and NR020], 2 isolates from Gardnerella genome sp. 3 [NR026 and N170], and 1 from unknown genome species [NR047, subgroup D based on the cpn60 classification system]).
Amylase activity assay.
The amylase activity of Gardnerella isolates was assessed using a glycogen iodine test (38). Isolates were revived from −80°C storage on Columbia blood agar plates containing 5% sheep blood and incubated anaerobically using the GasPak system (BD, USA) at 37°C for 48 h. Isolated colonies were transferred into modified NYCIII broth (obtained by replacing horse serum with bovine serum and omitting glucose in NYCIII medium) and incubated anaerobically for 24 h at 37°C. Overnight broth cultures were centrifuged at 10,000 × g for 10 min, and the cell-free supernatant was passed through a 0.45-μm filter. Filtered supernatant (10 μL) was spotted onto a 1% (wt/vol) glycogen (bovine liver glycogen; Sigma) agar plate and incubated anaerobically at 37°C for 24 h. The filtered supernatant was also streaked on Columbia sheep blood agar to confirm sterility. Plates were flooded with 1% (wt/vol) Lugol’s iodine solution to detect evidence of amylase activity. α-Amylase from Bacillus licheniformis (0.05 mg/mL) (Sigma-Aldrich, Oakville, ON; catalog no. A455) and modified NYCIII broth were used as positive and negative controls, respectively.
The activity of cell culture supernatants on glycogen was assessed by incubating culture supernatants with glycogen and examining the products. Briefly, 20 μL of culture supernatant was added to 180 μL of 0.1% bovine liver glycogen in 50 mM sodium phosphate buffer, pH 7.0, and incubated at 37°C. Aliquots (50 μL) of the reaction mixture were collected at 24 h and frozen immediately until analysis by thin-layer chromatography (TLC) and high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD). TLC was performed on silica plates in a 1-butanol, acetic acid, and distilled water (2:1:1 [vol/vol/vol]) mobile phase and stained using 0.3% of N-naphthyl ethylenediamine dihydrochloride (Sigma) as described previously (39). HPAEC-PAD samples were first precipitated with 80% ethanol to remove large polysaccharides and diluted (to 1:20 for G5 and maltodextrin or 1:10 for glycogen) and resolved on a Dionex PA20 column in a mobile phase of 30 mM NaOH and gradient of 10 mM to 120 mM sodium acetate over 40 min at a flow rate of 0.5 mL min−1. Data were analyzed using Chromeleon v6.80 chromatography data system software.
Identification of the carbohydrate-active enzymes.
The Gardnerella proteomes, predicted from the RAST annotation, were uploaded into the dbCAN2 web server (40). All three available domain prediction tools (HMMER, eCAMI, and DIAMOND) were used to annotate the carbohydrate active domain(s) in protein sequences. The dbCAN2 uses SignalP to predict whether a signal peptide is present or absent in the protein in which each carbohydrate-active domain is located. To identify secreted glycogen-degrading glycosyl hydrolases, dbCAN2 output was filtered to include only GH13 domain(s) associated with signal peptide. TMHMM 2.0 (41) and LocateP (42) were used to predict transmembrane and cell wall-anchored domains, respectively.
Phylogenetic analysis of GH13_32 amylase domains.
Domains belonging to subfamily 32 within the family 13 of glycosyl hydrolases class are referred to as GH13_32 amylase domains. Amino acid sequences corresponding to the GH13_32 amylase domain were extracted from predicted extracellular glycosyl hydrolases of all study isolates and four Gardnerella reference strains (G. leopoldii [GenBank accession number CP029984], G. piotii [GenBank accession number QJUV00000000], G. vaginalis [GenBank accession number QJUZ00000000], and G. swidsinskii [GenBank accession number QJVB00000000]) (4). Multiple-sequence alignments were performed using ClustalW, and a neighbor-joining consensus tree was built in Geneious Prime version 2022.1.1 (https://www.geneious.com). The final tree was visualized using FigTree (v1.4.4).
To identify the conserved catalytic residues in GH13_32 domains, amino acid sequences of representative GH13_32 domains of G. leopoldii NR017 were combined with GH13_32 domain sequences from functionally characterized α-amylases (Pseudoalteromonas haloplanktis A23 [GenPept accession number CAA41481] and Streptomyces venezuelae ATCC 15068 [GenPept accession number AAB36561]) and amylopullulanase (Bifidobacterium breve UCC2003 [GenPept accession number AAY89038]) deposited in the CAZy database. Sequences were aligned using ClustalW, and alignments were visualized in ESPript (43).
Expression and purification of α-amylase and pullulanase domains from α-amylase-pullulanase.
To characterize activities of the amylase and pullulanase domains of the α-amylase-pullulanase, G. leopoldii (NR017) was chosen as a representative isolate. Genomic DNA was extracted from G. leopoldii NR017 using a modified salting-out procedure. Primers (see Table S1 in the supplemental material) were designed to amplify the nucleotide sequences encoding the predicted α-amylase (corresponding to amino acids 74 to 350 of GenPept accession number RFT33565) and pullulanase (corresponding to amino acids 1141 to 1462 of GenPept accession number RFT33565) domains of the NR017 α-amylase-pullulanase gene along with 30 additional flanking nucleotides each on the 5′ and 3′ ends. PCR was carried out in PCR buffer (0.2 M Tris-HCl [pH 8.4], 0.5 M KCl), a 200-mM concentration of each deoxynucleoside triphosphate (dNTP), 2.5 mM MgCl2, a 400-nM concentration of each primer (α-amylase, JH0810-F and JH0811-R, and pullulanase, JH0812-F and JH0813-R), 1 U/reaction of Platinum Taq DNA polymerase, high fidelity (5 U/mL in 50% glycerol, 20 mM Tris-HCl, 40 mM NaCl, 0.1 mM EDTA, and stabilizers) (Life Technologies), and 2 mL of template DNA, in a final volume of 50 mL. PCR was performed with the following parameters: initial denaturation at 94°C for 3 min, 40 cycles of denaturation at 94°C for 15 s, annealing at 60°C for 15 s, and extension at 72°C for 1 min (for α-amylase domain) or 1.5 min (for pullulanase domain), and final extension at 72°C for 3 min. Purified PCR products were digested with BamHI and KpnI and ligated into expression vector pQE-80L treated with the same restriction endonucleases. The pQE-80L-amylase domain plasmid (henceforth referred to as pQE80L-AMY) was used to transform chemically competent E. coli Rosetta DE3 cells (Novagen, Sigma-Aldrich), and the pQE-80L-pullulanase domain plasmid (henceforth referred to as pQE80L-PULL) was used to transform One Shot TOP10 chemically competent E. coli cells (Invitrogen, Carlsbad, CA). Insertion of amylase and pullulanase domain-encoding sequences in frame with the N-terminal His tag was confirmed by sequencing of the purified plasmid.
E. coli Rosetta cells containing pQE80L-AMY were grown in 800 mL of LB broth containing ampicillin (100 mg/mL) and chloramphenicol (25 mg/mL), whereas E. coli TOP10 cells containing pQE80L-PULL were grown in the same volume of LB broth with ampicillin (100 mg/mL) only. Once the culture reached an optical density at 600 nm (OD600) of 0.4 to 0.6, protein expression was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside). After 4 h of incubation at 37°C with shaking at 200 rpm, cells were harvested by centrifugation at 8,500 × g for 20 min at 4°C, and pellets were stored at −20°C until further processing. Proteins were solubilized using anionic detergent as described previously (44) with a slight modification during sonication (6 min total run time, 2 min on, 1 min off at 4°C). Eluted proteins were dialyzed against 1× phosphate-buffered saline (PBS; pH 7.4) at 4°C using a semipermeable membrane with a cutoff size of 12 kDa for 24 to 48 h. The diluted proteins were concentrated using a 30-kDa molecular-weight-cutoff protein concentrator (Thermo Fisher Scientific). The final protein concentration was measured using a NanoDrop spectrophotometer before storing at −20°C.
Expression and purification of single amylase domain containing α-amylase.
A codon-optimized DNA construct corresponding to the full-length open reading frame of single-domain α-amylase from G. leopoldii NR017 (GenPept accession number RFT33564) was synthesized, and a region encoding amino acids 58 to 519 (excluding N-terminal signal peptide and C-terminal transmembrane domain) was amplified using JH0857 and JH0858 primers (Table S1). The 1,463-bp product was ligated into MfeI- and HindIII-digested pET41a(+) vector to express as an N-terminal GST-His tag fusion protein (79.9 kDa). The recombinant plasmid (pET41a-CO-AMY) was used to transform chemically competent E. coli DH5a cells. The presence of the insert in frame with the N-terminal GST-His tag was confirmed by Sanger sequencing of the purified plasmid.
The purified recombinant plasmid (pET41a-CO-AMY) was used to transform E. coli BL21(DE3) cells for protein production. E. coli BL21(DE3) cells containing recombinant plasmid were grown in 1.8 L of LB broth containing kanamycin (30 mg/mL). When culture reached an OD600 0.4 to 0.6, IPTG was added to a final concentration of 0.5 mM to induce protein expression at 37°C for 4 h with shaking at 200 rpm. Cells were harvested by centrifugation at 8,500 × g for 20 min. Expressed protein was solubilized using anionic detergent as described previously (44) with slight modification of the sonication protocol (80% duty cycle, 8 min total run time, 10 s on, 5 s off). The recombinant amylase protein was purified using a nickel column. Eluted proteins were dialyzed against 1× PBS (pH 7.4) at 4°C overnight using a semipermeable membrane of cutoff size 12 kDa, and dialyzed protein was concentrated using a 30-kDa molecular-weight-cutoff protein concentrator (Thermo Fisher Scientific). The final protein concentration was measured using a NanoDrop spectrophotometer before storing at −20°C.
The region encoding the GST and 6×His tags was expressed from empty pET41a(+) vector in E. coli BL21(DE3) cells and was purified using Ni-affinity chromatography for use as a negative control in activity experiments.
Enzyme activity on different substrates.
The activity of purified enzymes was tested by incubating purified enzymes with different substrates. Briefly, 250 μL of the purified amylase domain (22.3 μM) or α-amylase (13.0 mM) was incubated with 1% glycogen, maltodextrin (dextrose equivalent 13 to 17), or maltopentose, while purified pullulanase (25.3 μM) was incubated with 1% maltotriose, maltotetraose, maltopentose, pullulan, and glycogen. To test the combined activity of α-amylase and pullulanase domains, an equimolar (25.3 μM) mixture of both enzymes was incubated with glycogen (1%). All substrates were prepared in 50 mM sodium phosphate buffer at pH 6.0. Enzyme substrate reaction mixtures were incubated at 37°C for 24 h and stored at −20°C until further analysis by TLC or HPAEC-PAD as described above.
Data availability.
Whole-genome sequence data for all isolates were retrieved from NCBI BioProject accession number PRJNA394757.
ACKNOWLEDGMENTS
This research was supported by a Natural Sciences and Engineering Research Council of Canada Discovery Grant (J.E.H.) and Agriculture and Agri-Food Canada (project no. J-002262 to D.W.A.). P.B. is supported by a Devolved Scholarship from the University of Saskatchewan.
We thank Champika Fernando for technical support, Shubham Dutta for guidance with Western blotting, and Noreen Rapin and Tony Ruzzini for providing reagents.
Footnotes
Supplemental material is available online only.
Contributor Information
Janet E. Hill, Email: Janet.Hill@usask.ca.
Laurie E. Comstock, University of Chicago
REFERENCES
- 1.Hillier SL. 1993. Diagnostic microbiology of bacterial vaginosis. Am J Obstet Gynecol 169:455–459. 10.1016/0002-9378(93)90340-o. [DOI] [PubMed] [Google Scholar]
- 2.Muzny CA, Schwebke JR. 2020. Asymptomatic bacterial vaginosis: to treat or not to treat? Curr Infect Dis Rep 22:32. 10.1007/s11908-020-00740-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van de Wijgert JHHM, Borgdorff H, Verhelst R, Crucitti T, Francis S, Verstraelen H, Jespers V. 2014. The vaginal microbiota: what have we learned after a decade of molecular characterization? PLoS One 9:e105998. 10.1371/journal.pone.0105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaneechoutte M, Guschin A, Van Simaey L, Gansemans Y, Van Nieuwerburgh F, Cools P. 2019. Emended description of Gardnerella vaginalis and description of Gardnerella leopoldii sp. nov., Gardnerella piotii sp. nov. and Gardnerella swidsinskii sp. nov., with delineation of 13 genomic species within the genus Gardnerella. Int J Syst Evol Microbiol 69:679–687. 10.1099/ijsem.0.003200. [DOI] [PubMed] [Google Scholar]
- 5.Hill JE, Albert AYK. the VOGUE Research Group. 2019. Resolution and cooccurrence patterns of Gardnerella leopoldii, G. swidsinskii, G. piotii, and G. vaginalis within the vaginal microbiome. Infect Immun 87:e00532-19. 10.1128/IAI.00532-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan S, Voordouw MJ, Hill JE. 2019. Competition among Gardnerella subgroups from the human vaginal microbiome. Front Cell Infect Microbiol 9:374. 10.3389/fcimb.2019.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8:15–25. 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adeva-Andany MM, González-Lucán M, Donapetry-García C, Fernández-Fernández C, Ameneiros-Rodríguez E. 2016. Glycogen metabolism in humans. BBA Clin 5:85–100. 10.1016/j.bbacli.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruickshank R, Sharman A. 1934. The biology of the vagina in the human subject. BJOG:An Int J O&G 41:208–226. 10.1111/j.1471-0528.1934.tb08759.x. [DOI] [Google Scholar]
- 10.Nasioudis D, Beghini J, Bongiovanni AM, Giraldo PC, Linhares IM, Witkin SS. 2015. α-Amylase in vaginal fluid: association with conditions favorable to dominance of Lactobacillus. Reprod Sci 22:1393–1398. 10.1177/1933719115581000. [DOI] [PubMed] [Google Scholar]
- 11.Mirmonsef P, Hotton AL, Gilbert D, Gioia CJ, Maric D, Hope TJ, Landay AL, Spear GT. 2016. Glycogen levels in undiluted genital fluid and their relationship to vaginal pH, estrogen, and progesterone. PLoS One 11:e0153553. 10.1371/journal.pone.0153553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spear GT, French AL, Gilbert D, Zariffard MR, Mirmonsef P, Sullivan TH, Spear WW, Landay A, Micci S, Lee B-H, Hamaker BR. 2014. Human α-amylase present in lower-genital-tract mucosal fluid processes glycogen to support vaginal colonization by Lactobacillus. J Infect Dis 210:1019–1028. 10.1093/infdis/jiu231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nunn KL, Clair GC, Adkins JN, Engbrecht K, Fillmore T, Forney LJ. 2020. Amylases in the human vagina. Amylases in the Human Vagina mSphere 5:e00943-20. 10.1128/mSphere.00943-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kahar UM, Latif NA, Amran SI, Liew KJ, Goh KM. 2022. A bibliometric analysis and review of pullulan-degrading enzymes—past and current trends. Catalysts 12:143. 10.3390/catal12020143. [DOI] [Google Scholar]
- 16.Uitdehaag JCM, Mosi R, Kalk KH, van der Veen BA, Dijkhuizen L, Withers SG, Dijkstra BW. 1999. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the α-amylase family. Nat Struct Biol 6:432–436. 10.1038/8235. [DOI] [PubMed] [Google Scholar]
- 17.Muzny CA, Schwebke JR. 2016. Pathogenesis of bacterial vaginosis: discussion of current hypotheses. J Infect Dis 214:S1–S5. 10.1093/infdis/jiw121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muzny CA, Taylor CM, Swords WE, Tamhane A, Chattopadhyay D, Cerca N, Schwebke JR. 2019. An updated conceptual model on the pathogenesis of bacterial vaginosis. J Infect Dis 220:1399–1405. 10.1093/infdis/jiz342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atassi F, Brassart D, Grob P, Graf F, Servin AL. 2006. Lactobacillus strains isolated from the vaginal microbiota of healthy women inhibit Prevotella bivia and Gardnerella vaginalis in coculture and cell culture. FEMS Immunol Med Microbiol 48:424–432. 10.1111/j.1574-695X.2006.00162.x. [DOI] [PubMed] [Google Scholar]
- 20.Castro J, Rosca AS, Cools P, Vaneechoutte M, Cerca N. 2020. Gardnerella vaginalis enhances Atopobium vaginae viability in an in vitro model. Front Cell Infect Microbiol 10:83. 10.3389/fcimb.2020.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Veer C, Hertzberger RY, Bruisten SM, Tytgat HLP, Swanenburg J, de Kat Angelino-Bart A, Schuren F, Molenaar D, Reid G, de Vries H, Kort R. 2019. Comparative genomics of human Lactobacillus crispatus isolates reveals genes for glycosylation and glycogen degradation: implications for in vivo dominance of the vaginal microbiota. Microbiome 7:49. 10.1186/s40168-019-0667-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woolston BM, Jenkins DJ, Hood-Pishchany MI, Nahoum SR, Balskus EP. 2021. Characterization of vaginal microbial enzymes identifies amylopullulanases that support growth of Lactobacillus crispatus on glycogen. BioRxiv. 10.1101/2021.07.19.452977. [DOI]
- 23.Edmunds PN. 1962. The biochemical, serological and haemagglutinating reactions of “Haemophilus vaginalis.” J Pathol Bacteriol 83:411–422. 10.1002/path.1700830211. [DOI] [PubMed] [Google Scholar]
- 24.Piot P, Van Dyck E, Totten PA, Holmes KK. 1982. Identification of Gardnerella (Haemophilus) vaginalis. J Clin Microbiol 15:19–24. 10.1128/jcm.15.1.19-24.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunkelberg WE, McVeigh I. 1969. Growth requirements of Haemophilus vaginalis. Antonie Van Leeuwenhoek 35:129–145. 10.1007/BF02219124. [DOI] [PubMed] [Google Scholar]
- 26.Huffman RD, Nawrocki LD, Wilson WA, Brittingham A. 2015. Digestion of glycogen by α-glucosidase released by Trichomonas vaginalis. Exp Parasitol 159:151–159. 10.1016/j.exppara.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Hii SL, Tan JS, Ling TC, Ariff AB. 2012. Pullulanase: role in starch hydrolysis and potential industrial applications. Enzyme Res 2012:1–14. 10.1155/2012/921362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boraston AB, Healey M, Klassen J, Ficko-Blean E, van Bueren AL, Law V. 2006. A structural and functional analysis of α-glucan recognition by family 25 and 26 carbohydrate-binding modules reveals a conserved mode of starch recognition. J Biol Chem 281:587–598. 10.1074/jbc.M509958200. [DOI] [PubMed] [Google Scholar]
- 29.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238. 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cockburn DW, Suh C, Medina KP, Duvall RM, Wawrzak Z, Henrissat B, Koropatkin NM. 2018. Novel carbohydrate binding modules in the surface anchored α‐amylase of Eubacterium rectale provide a molecular rationale for the range of starches used by this organism in the human gut. Mol Microbiol 107:249–264. 10.1111/mmi.13881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim S-Y, Kim H, Kim Y-J, Jung D-H, Seo D-H, Jung J-H, Park C-S. 2021. Enzymatic analysis of truncation mutants of a type II pullulanase from Bifidobacterium adolescentis P2P3, a resistant starch-degrading gut bacterium. Int J Biol Macromol 193:1340–1349. 10.1016/j.ijbiomac.2021.10.193. [DOI] [PubMed] [Google Scholar]
- 32.O'Connell Motherway M, Fitzgerald GF, Neirynck S, Ryan S, Steidler L, van Sinderen D. 2008. Characterization of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl Environ Microbiol 74:6271–6279. 10.1128/AEM.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghosh S, O'Connor TJ. 2017. Beyond paralogs: the multiple layers of redundancy in bacterial pathogenesis. Front Cell Infect Microbiol 7:467. 10.3389/fcimb.2017.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siegel SD, Reardon ME, Ton-That H. 2016. Anchoring of LPXTG-like proteins to the gram-positive cell wall envelope, p 159–175. In Bagnoli F, Rappuoli R (ed), Protein and sugar export and assembly in Gram-positive bacteria. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 35.Agarwal K, Lewis AL. 2021. Vaginal sialoglycan foraging by Gardnerella vaginalis: mucus barriers as a meal for unwelcome guests? Glycobiology 31:667–680. 10.1093/glycob/cwab024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhandari P, Tingley JP, Palmer DRJ, Abbott DW, Hill JE. 2021. Characterization of an α-glucosidase enzyme conserved in Gardnerella spp. isolated from the human vaginal microbiome. J Bacteriol 203:e00213-21. 10.1128/JB.00213-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schellenberg JJ, Paramel Jayaprakash T, Withana Gamage N, Patterson MH, Vaneechoutte M, Hill JE. 2016. Gardnerella vaginalis subgroups defined by cpn60 sequencing and sialidase activity in isolates from Canada, Belgium and Kenya. PLoS One 11:e0146510. 10.1371/journal.pone.0146510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan SM, Fitzgerald GF, van Sinderen D. 2006. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl Environ Microbiol 72:5289–5296. 10.1128/AEM.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cockburn D, Koropatkin N. 2015. Product analysis of starch active enzymes by TLC. Bioprotocol J 5:1–8. 10.21769/BioProtoc.1621. [DOI] [Google Scholar]
- 40.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y. 2018. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46:W95–W101. 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Cohen J Mol Biol 305:567–580. 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 42.Zhou M, Boekhorst J, Francke C, Siezen RJ. 2008. LocateP: genome-scale subcellular-location predictor for bacterial proteins. BMC Bioinformatics 9:173. 10.1186/1471-2105-9-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlager B, Straessle A, Hafen E. 2012. Use of anionic denaturing detergents to purify insoluble proteins after overexpression. BMC Biotechnol 12:95. 10.1186/1472-6750-12-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S6. Download jb.00393-22-s0001.pdf, PDF file, 2.8 MB (2.9MB, pdf)
Data Availability Statement
Whole-genome sequence data for all isolates were retrieved from NCBI BioProject accession number PRJNA394757.